
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 09 June 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.899079
This article is part of the Research TopicAdvances in genetic engineering strategies for fruit crop breeding, Volume IIView all 5 articles
Elaeagnus L. is found in wild or grown as ornamental plants and is increasingly regarded as underutilized berry shrubs by breeders. This genus has cosmopolitan distribution with various species widely distributed in China, Europe, the United States, and Canada. Interspecific hybrids, which have been reported several times, have attracted intense interest from plant breeders attempting to develop a fruit crop of Elaeagnus. Orthogonal projections to latent structures discriminant analysis (OPLS-DA) is a powerful statistical modeling tool that provides insights into separations between experimental groups. In this study, the molecular phylogeny of Elaeagnus species was first discussed using the ITS and matK sequences for guiding the construction of a genetic basis pool. A morphological OPLS-DA clustering model based on the genetic divergence was also constructed for the first time, which effectively realized the morphological grouping of Chinese Elaeagnus species. The results showed that a total of 10 wild species widely distributed in China have the potential to develop fruit crops. Particularly, Elaeagnus conferta has the potential to provide a founder species with a large fruit size, while Elaeagnus Gonyanthes has the potential to provide important genetic resources with long pedicel. Elaeagnus lanceolata and Elaeagnus delavayi could be used to domesticate hybrids without spines, and the other five climbing shrubs could be used to develop high-yield crown-type commercial cultivars for automated field management. The top five contributing morphological traits affecting the current clustering model were V9 (flower color), V1 (flowering), V5 (evergreen or deciduous), V3 (leaf size), and V2 (fruiting). Furthermore, the grouping analysis indicated that the V9 was the most important factor affecting morphological clustering. Thereafter, the temporally calibrated phylogeny inferred from the matK sequence was used to reconstruct the origin and evolution of the genus Elaeagnus, and the results inferred an interesting geographic distribution pattern and potential cross-species interactions of Elaeagnus species at low latitudes in China. Our study also highlighted dispersal pattern investigation and genetic background analysis to improve future practices and policies related to species introduction of genetic basis pool.
Elaeagnus L., a genus of the Elaeagnaceae, with about 100 recognized wild species, is cultivated as an ornamental or a fruit crop for its dense shrub-like structure, fragrant flowers, and lycopene-rich ripe fruits (Sun and Lin, 2010; Alexandrov and Karlov, 2021). The genus Elaeagnus is native to temperate and subtropical regions of Asia, Australia, southern Europe, and North America (Ye et al., 2012). In order to better understand the worldwide use of this genus, a detailed table has been performed and gives a clear representation of medicinal and edible applications in China, South Korea, India, Australia, and other countries. As shown in Table 1, there are 15 species, including an endangered woody oil species, Elaeagnus mollis, in this genus that has been widely reported to have valuable edible or medicinal properties. As an endemic berry fruit, only seven juicy berries are considered underutilized fruits, including Elaeagnus conferta (Dandge et al., 2011; Jaafar et al., 2018), Elaeagnus kologa (Sasikumar et al., 2012; Patel, 2015), Elaeagnus latifolia (Dasila and Singh, 2021), Elaeagnus multiflora (Lee et al., 2011), Elaeagnus pyriformis (Banerjee et al., 2022), Elaeagnus trifloral (Ramadevi and Narayana, 1990), and Elaeagnus umbellata (Khattak, 2012). The red berries are known as oleaster, silverberry, autumn olive, thorn olive, Russian olive, Persian olive, and wild olive (Patel, 2015). Currently, these species are usually planted in gardens or around courtyards as ornamental shrubs in most countries and regions, and the sweet, rare fruits are nowadays only regarded as a kind of accessory product. Surprisingly, few varieties have been developed for pomology purposes although this genus has the potential to develop into a great fruit crop for the benefit of the world.
Nevertheless, it is interesting that the selection and breeding based on the excellent genetic material of E. conferta have been developed for nearly 20 years in mainland China (Gill and Gupta, 2018; Liu et al., 2019). E. conferta has an absolute advantage in fruit size, but distribution is limited in China’s low latitude subtropical regions, such as Yunnan and Guangxi Provinces. Except for the populations of E. conferta, nearly 55 species of this genus are also widely distributed in China, spread from the Hexi Corridor to the Yangtze River Basin to mountain areas of southern China (Sun and Lin, 2010). Self-incompatibility is a common feature of Elaeagnus plants, which offers the possibility of creating new cultivars through interspecific hybridization. In fact, the World Flora Online (WFO) and the Plant List (TPL) and recent reference (Abdalla, 2019) have recorded 11 hybrids with three accepted new hybrid cultivars of the genus Elaeagnus, namely E. × reflexa E. Morren & Decne., E. × submacrophylla Servett., and E. × maritima Koidz. A popular horticultural hybrid (Li Y. L. et al., 2015), gold-marginatus hybrid E. × ebbingei Boom., “Gilt Edge” (syn. E. × submacrophylla) could provide an inspiring example for creating new hybrid cultivars. It’s reported that the gold-marginatus hybrid E. × ebbingei (Chen and Wu, 2019) was first created by interspecific hybridization in Netherlands in 1929, and the two parents selected were Elaeagnus pungens and E. macrophylla, which are both widely distributed across multiple latitudes in the southeast coast of China (Alexandrov and Karlov, 2021). Therefore, the creation of new fruit crops of Elaeagnus is inseparable from the ingenious collection and selection of wild resources under a superb scientific design.
Hongwen et al. (2021) pointed out that conventional breeding methods, especially natural mutant selection, hybrid breeding, and systematic breeding are still the preferred ways for the initial domestication of new fruit crops. Among them, recurrent selection (Payne et al., 1986; Cobb et al., 2019) is an important breeding method to improve the population of crops, especially the population of cross-pollinated species. Since large-scale geographically adaptive distribution is an essential condition for the selection of hybrid fruit crops with desirable characteristics, an effective basis pool filled with more genetic resources needs to be established to select and breed the next generational fruit-type Elaeagnus crops (Cornille et al., 2015). Therefore, how to rapidly and efficiently construct primitive populations by using the local and cross-regional relatives in the same genus has become an urgent consideration for selection design. It is the general consensus now that the progeny of interspecific hybridization is difficult to be obtained in a wide cross because of their strong cross-incompatibility (Lexer et al., 2003; Maune et al., 2018). In order to achieve fertile recombinant progenies, the primary condition of parental selection is to overcome this strong barrier. Experimental crosses, as a tool for studying breeding in the wild, provide a way to overcome natural barriers, as the traits most important for adaptation and speciation may be fixed in wild populations (Lexer et al., 2003). However, the selection of wild resources of perennial shrubs requires a lot of time for resource collection and selective mating experiments. Which traits contribute most strongly to assortative mating specifically and to reproductive isolation more generally?
In most cases, molecular phylogenetic analysis and morphological clustering are reliable methods for providing genetic relationships and predicting reproduction compatibility among plant species (Baral and Bosland, 2004; Kubota et al., 2012). Currently, several phylogenetic studies have provided credible insights for understanding the evolutionary relationships of Elaeagnus, and the consensus trees supported the monophyletic origin of the genus Elaeagnus and Hippophae (Bartish et al., 2002; Kim et al., 2020; Zhao et al., 2020). Son et al. (2014) provided the molecular phylogenetic relationships of nine common Asian Elaeagnus species based on nrDNA ITS sequences. Their study reported that different clustering methods can be used for obtaining similar divergent results, except for the ambiguity in the divergence time of the outer taxa in the genera Shepherdia and Hippophae. Besides, the strict consensus trees based on cpDNA and morphological characters of the Elaeagnaceae showed that the genera Elaeagnus and Hippophaeare are similar in chloroplast DNA sequence (Zhao et al., 2020). Phylogenetic trees constructed by the maximum likelihood (ML) method based on complete chloroplast or plastid genomes also showed that the genus Elaeagnus has a close relationship with the genus Hippophae (Choi et al., 2015; Liu et al., 2019; Cheng et al., 2020; Kim et al., 2020). At present, more phylogenetic and molecular clock analysis of Chinese Elaeagnus using DNA marker regions in the chloroplast genome, such as matK, rbcL, trnH-psbA, and trnL-F, has not been reported yet. A more rigorous and phylogenetic analysis is very important for further resource evaluation. Recently, many studies (Yu et al., 2011; Cabelin and Alejandro, 2016) have reported that the matK (cpDNA) sequence has significant stability in phylogenetic analysis, and the constructed genetic relationship is closer to the real plant taxonomy. In this study, DNA markers in the nuclear genome, such as ITS, and that in the chloroplast genome, such as matK, should both be used to perform comparative analysis to obtain biparental and maternal characteristics of Elaeagnus species, and provide the theoretical basis for further crossbreeding. Moreover, because the phylogenetic signals provided by molecular phylogenetic results are prone to contradictory taxonomic results (Eernisse and Kluge, 1993), morphological clustering analysis is also important to infer the genetic relationship among the Elaeagnus species. Additionally, morphological clustering under a grouping condition of strict molecular phylogenetic clades may provide a new means to infer the interspecies cross-compatibility. Principal component analysis (PCA) and orthogonal projections to latent structures discriminant analysis (OPLS-DA) are powerful statistical modeling tools that provide insights into separations between experimental groups. OPLS-DA has spawned a general pattern in many fields. When PCA fails to identify significant separation between experimental groups, analysts may move to construct OPLS-DA models. OPLS-DA provides an avenue for predicting group membership based on a set of high-dimensional measurements and holds many advantages over PCA. So, we used the clades directly supported by the phylogenetic tree as the experimental grouping for morphological data grouping, which provided a potential training dataset for building OPLS-DA models.
In this study, phylogenetic analysis was conducted for the genetic grouping of wild species in the genus Elaeagnus by using the public sequencing data of DNA marker regions in the chloroplast genome. Then, an OPLS-DA model based on 12 morphological traits was established for all Chinese Elaeagnus species. In addition, the geographic dispersion of multilateral co-evolution was analyzed for the first time and was expected to be used to guide the selection of founder parents and the construction of basis pools for the selection and breeding of new Elaeagnus fruit crops.
All DNA sequences including ITS and matK of Elaeagnus L. were downloaded from GenBank in NCBI. Sequence alignment was performed by using the BioEditor, and all sequences were aligned using Clustal W. Sequence deletions or errors exceeding 5% of the total bases were considered low-quality sequences and were discarded. Gaps were treated as missing data. The same species retained all haplotypes for further phylogenetic analysis. The head and tail of the aligned sequences were cut off. In order to ensure DNA sequence data were correctly linked taxonomically to the assigned species, we only retained haplotypes with more than 99% sequence similarity of the same species for further analysis. Thereafter, as shown in Supplementary Table 1 and Table 2, 56 ITS sequences of at least 541 bp and 53 matK sequences of at least 689 bp were selected for further processing, the full range of ITS and matK were shown in the Supplementary Table 2. Among them, the sequence of outgroup was given by a member of the Rhamnaceae (e.g., Ziziphus jujuba Mill.), 50 ITS sequences of Elaeagnus species were used for phylogenetic analysis together with five ITS sequences of Hippophanae species, and 53 matK sequences of Elaeagnus species were used for phylogenetic analysis together with eight matK sequences of Hippophanae species. Additionally, three hybrids and one environmental variety were the main focus in order to find pieces of evidence of cross-compatibility and interspecific hybridization. Finally, the software Geneious_11 was used for constructing the molecular matrix (Kearse et al., 2012).
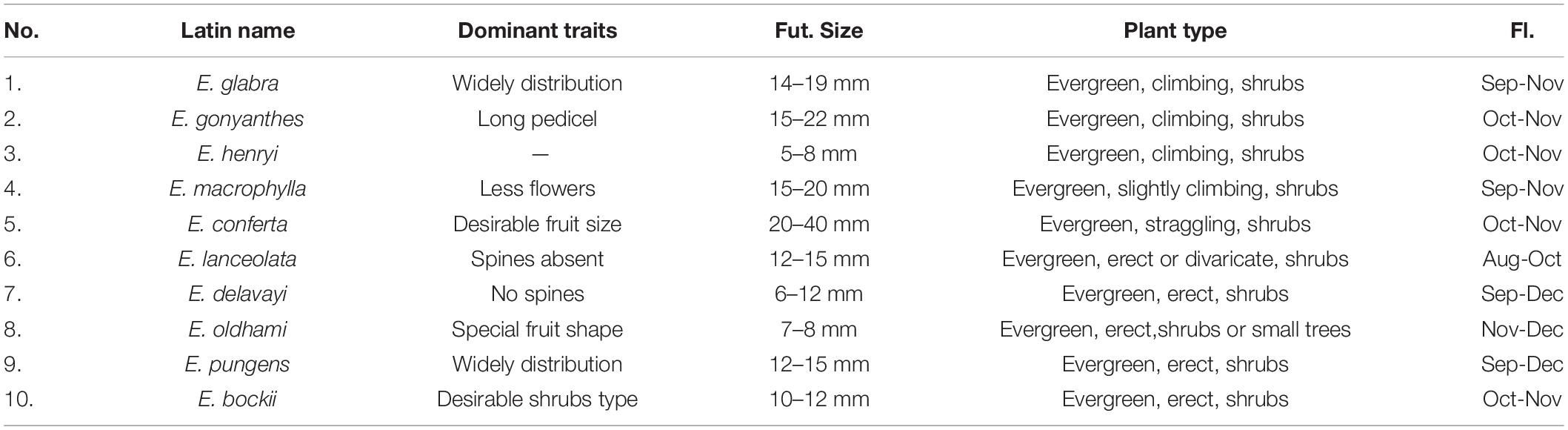
Table 2. Ten resources of Chinese wild Elaeagnus L. species with the potential to develop into fruit crops.
Referring to “Flora of China (FOC)” (Flora of China Editorial Committee, 2018), “Higher plants in China,” and “Image of higher plants in China,” all taxonomic descriptions were quantitatively summarized and analyzed. Operationally, we first extracted all morphological traits recorded in FOC, and after classification, quantification, and data standardization, we finally identified 12 traits related to life form, ecotype, pollination ecology, and reproductive ecology of Elaeagnus species. A total of 12 investigated traits were flowering, fruiting, the minimum leaf size, the maximum leaf size, evergreen or deciduous, plant height, spines or absent, climbing or not, flower color, the length of calyx tube, the style villous, and the flower number. The quantitative methods by chronological order or degree were used to obtain the quantitative data. In this section, 55 species were investigated by using high-resolution specimen pictures online. Specimens of the same species from at least six different regional sources were used to extract and quantify trait information. SPSS 22 (IBM, China) was used for data standardization (Leech et al., 2014).
All DNA sequences including ITS and matK were first aligned with MEGA-X (iGEM, France). Since many sequences have a small number of base deletions (often denoted as “NNNN…”). Then, the consensus sequences were assembled and visually checked for quality using SeqScape version 2.5 (Applied Biosystems, Foster, United States). Phylogenetic reconstruction was performed by using maximum parsimony (MP), ML, and Bayesian inference methods to resolve the interspecific and intergeneric phylogenetic relationships. For ITS and matK sequences, MEGA-X (iGEM, France) was used for conducting MP or ML analyses with 1,000 bootstrap replicates (Kumar et al., 2018), and the best model for constructing an ML tree by using MEGA-X (iGEM, France) is Hasegawa–Kishino–Yano + Gamma Distributed (G) model or Kimura 2-parametric (K2P) model, relying on the calculation results from Find Best DNA/Protein Models (Srivathsan and Meier, 2012). The time-calibrated phylogeny inferred from the matK sequences was using BEAST 2.2 (Bouckaert et al., 2019) with an uncorrelated rates model. BEAST 2.2 analysis was conducted under the guidance of the online tutorial. First, the Bayesian Evolutionary Analysis Utility (BEAUti) was used for setting the evolutionary model and options for the Markov chain Monte Carlo (MCMC) analysis. Because K2P models are not embedded within BEAST 2.2, the BEAUti interface was used in which a selected model (GTR + G) for the aligned dataset (nex or fasta file) was applied with a Yule speciation tree prior and an uncorrelated lognormal molecular clock model. Two runs of 100 million generations of MCMC chains were produced, sampling every 1,000 generations. Then, BEAST was running using the input file that contains the data, model, and settings. We used the following three constraints for fossil calibration (all with a normal prior distribution): Elaeagnus stem node 30 ± 6 Ma (Akgiin and Sözbilir, 2001), Hippophanaecrown node 0.39–0.1 Ma (Ruixue, 2013), and Elaeagnus crown node 11.2 ± 4 Ma (Su et al., 2014). Following convergence, the resulting trees were combined using TreeAnnotator (BEAST Developers). The final phylogenetic tree was visualized using the FigTree (Softpedia, Romania). The phylogenetic tree was visualized by EVOLVIEW (EvolGenus Info., Online) (Subramanian et al., 2019).
According to phylogenetic grouping, normalized data of 12 morphological traits were used to construct the OPLS-DA model. PCA without group setting was used to compare and support the advantages of the OPLS-DA method. The receiver operating characteristic (ROC) curve was typically used to evaluate the effectiveness of the grouping boundary, and the area under the ROC curve (AUC) is regarded as a measure of the overall performance of a diagnostic test and is interpreted as the average value of sensitivity for all possible values of specificity (Obuchowski, 2003; Zhou et al., 2009). AUC is often presented along with its 95% confidence interval (CI). Therefore, if the lower bound of the 95% CI of AUC for a test is greater than 0.5, then the test is statistically significantly better. The variable importance for the projection (VIP) summarized the importance of the variables and was sorted to display larger VIPs to the left. The top 5 contributing quantitative traits affecting the current clustering model were further analyzed by the F-test and the t-test.
Refer to FOC and the specimen records in “Chinese Virtual Herbarium”1 and as shown in Supplementary Table 3, specimen collection sites were counted, and the frequency of occurrence of specimens in the same geographic area was used to determine the distribution center of each species. The main distribution centers of different species in different groups separated by the OPLS-DA model were analyzed using ArcGIS 10.8 (Esri, United States). Neural network analysis based on Gephi 0.9.2 (Gephi.org, Atlanta, GA, United States) was used to better visualize the geographic dispersion of Elaeagnus species in China. The nodes include Latin names of species, group names, and five geographical regions of China, such as East China, Central China, South China, Southwest, and North China.
Excel as a basic tool was used to record and edit the data and convert file formats. Graphpad Prism 7 software (GraphPad Software Inc, San Diego, CAï United States) was used to analyze the data. All numerical values are presented as mean ± SD. R and open-source analysis packages were used for difference analysis, such as the t-test, f-test, mean, and the Kruskal–Wallis test. Statistically significant differences were determined by pair using a t-test, with p-values < 0.05 or 0.01.
The public sequences of Elaeagnus and Hippophae were collected for molecular phylogenetic analysis so as to facilitate the comparative analysis with traditional plant taxonomy. Two characteristic DNA marker sequences, including matK and ITS sequences, were obtained as the basic data to construct phylogenetic trees. In this study, the matK sequences of 21 species and ITS sequences of 25 species were used to construct the matrixes and perform molecular phylogenetic analysis. Among them, 109 parsimony characters were found in the ITS matrix, and 45 parsimony characters were found in the matK matrix. With Z. jujuba as the outgroup, the MP based on the ITS and the ML based on the ITS and matK were used for constructing the strict consensus trees (Supplementary Figure 1). Both trees revealed that Hippophae L. was divided into an independent clade with a bootstrap value of more than 95%. Therefore, both trees were invaluable for further analysis of genetic classification. Since the ITS is biparental and the matK is maternal, so the molecular phylogenetic results based on ITS sequences could better reflect the genetic relationship between hybrids and their parents. ITS tree analysis by MP or ML methods showed that three hybrids and one environmental variety were divided into one clade together with E. pungens, E. glabra, E. macrophylla, E. conferta, E. angustifolia, E. multiflora, and E. umbellata, which were mentioned in Table 1 and considered to be underutilized fruits or widely used cash crops. Therefore, the concerned seven species have the potential to become the founder species or precious wild resources for future hybrid breeding. In contrast, the matK tree did not provide any phylogenetic information for hybrids but revealed a more reliable phylogenetic relationship conforming to the characteristics of traditional taxonomy. For convenience, a circular phylogenetic tree combining the main plant taxonomic features was constructed. As shown in the following Figure 1 and Supplementary Figure 1, E. commutata formed a distinct clade alone with a 98% bootstrap; E. angustifolia and E. mollis formed a clade with only 60% bootstrap; E. umbellata, E. multiflora, and E. Montana also formed a distinct clade with a 90% bootstrap; E. bockii, E. henryi, E. pungens, E. conferta, and E. lanceolata formed a distinct clade with a 95% bootstrap; and the remaining species of E. glabra, E. macrophylla and another haplotype of E. pungens formed a clade. Referring to the basic taxonomic features of plants such as deciduous, evergreen, flowering, and fruiting, a phylogenetic grouping model was constructed to divide the Elaeagnus species into four groups. Along the molecular phylogeny, the results showed that the maternal inheritance of Elaeagnus was characterized by the evolution of plant life forms from deciduous shrubs to erect or climbing evergreen plants. Thus, the characteristic DNA sequences reflected the dispersion of major morphological features.
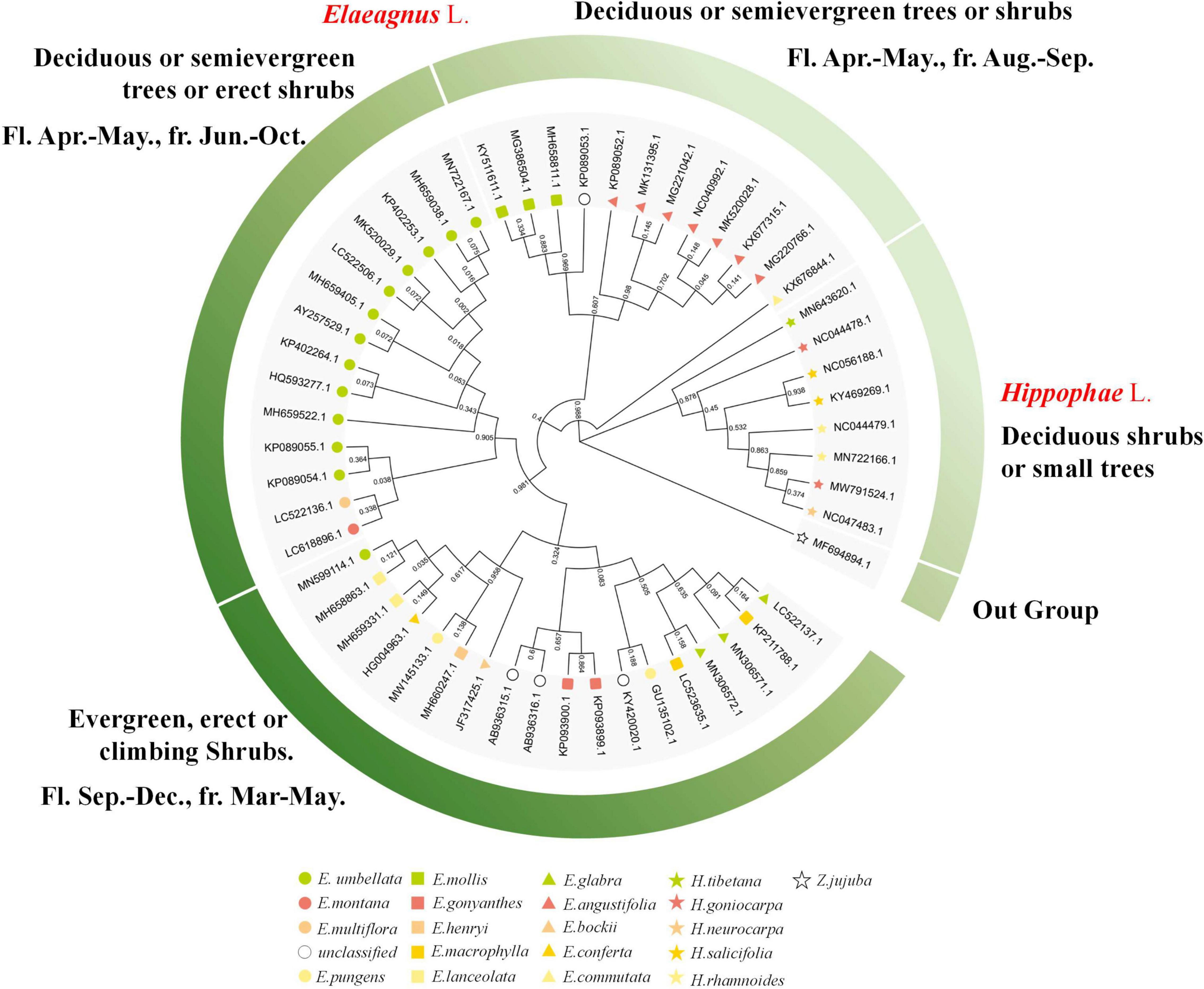
Figure 1. Phylogenetic tree combined with the main plant taxonomic features constructed with the matK sequences for Elaeagnus plants (Bootstrap values are expressed as decimals).
When the phylogenetic tree was constructed, we also checked in time that some species were displayed to not be monophyletic. It was not surprising but instead regarded as a reasonable result, because the public DNA sequences come from different countries and laboratories, and there would inevitably be analytical interferences within acceptable limits due to sequencing, species identification, or other issues. According to the formulated principle at the beginning of sequence analysis, we retained all haplotypes of the same species for the next phylogenetic analysis. Thus, a small number of potentially outlier sequences (errors may arise from species identification errors or sequencing errors) must be presented on the phylogenetic tree. Therefore, critical thinking required us to first decide whether the phylogenetic tree is credible and usable and whether the results of genetic analysis can positively promote the next step of morphological grouping work. Practically, morphological grouping models were constructed mainly based on the clades supported by the branches of the phylogenetic tree with high confidence. A bootstrap value is the most reliable parameter to evaluate the phylogenetic tree and reliability of the branches. In general practice, branches with a bootstrap value greater than 95% will be explicitly discussed in the results, while branches with a bootstrap value less than 70% are considered low confidence and not discussed. We checked again if the species seem to not be monophyletic. For the matK sequences, we only found that only one haplotype (E. umbellata, MN599114) formed a non-monophyletic with a bootstrap value above 95%; For ITS, all non-monophyletic branch bootstrap values that can be found in the revised high-resolution Supplementary Figure 1 are less than 70%. In conclusion, the clades supported by the phylogenetic tree are invaluable for further analysis of genetic classification.
The molecular phylogeny of the 14 Elaeagnus species presented a clear evolutionary relationship consistent with the plant taxonomy, and the divided clades could provide us with a basic grouping principle and an optimizing and training dataset for constructing a grouping model based solely on plant morphological data. The realization of this grouping model could undoubtedly provide us with an extremely practical breeding guide to deal with the dilemma that it is difficult to obtain more DNA sequences of larger-scale species in a short term. It is also believed that this dilemma cannot be easily solved with only sufficient financial support, because the field collection and species identification of more than 50 wild species cannot be completed in a short period of time. The preciousness of the breeder’s time is especially recognized in the perennial fruit tree breeding community. Therefore, we are looking forward to an invention and application of a morphological rapid grouping model based on molecular phylogenetic theory. In this study, a total of 12 morphological traits of 52 species, associated with sexual plant reproduction, were extracted and quantified according to the records of the FOC. Specimen information was used for a few species distributed abroad, such as E. commutata distributed in the United States and E. montana distributed in Japan. The 12 morphological traits were, respectively, related to the type of tree, leaf, the number, and color of the flowers, calyx shape, style character, and ecological type, among others. As shown in Figure 2A, the PCA method was first used, and the result was frustrating that the 56 species could not be clustered well for morphological grouping. However, the OPLS-DA model based on phylogenetic clades presented a clear clustering result. The result showed that 56 species were clustered well into three groups, which were marked with G1, G2, and G3 shown in Figure 2B. The results also pointed out the top 5 contributing morphological traits affecting the current clustering model, as shown in Figure 2C, which are V9 (flower color), V1 (flowering), V5 (evergreen or deciduous), V3 (leaf size), and V2 (fruiting), respectively. Among them, the flower color and flowering period played a decisive role in the morphological clustering of Chinese Elaeagnus. Furthermore, as shown in Figure 2D, the ROC curve revealed that the area under the ROC curve (AUC values) for both G1 and G2 both exceed 95%, indicating that the OPLS-DA model has a very well analysis effect for morphological grouping. Thus, the phylogenetic-based OPLS-DA model was proved to be an effective method for clustering the wild Elaeagnus plants as well as mining the key morphological features related to plant taxonomy.
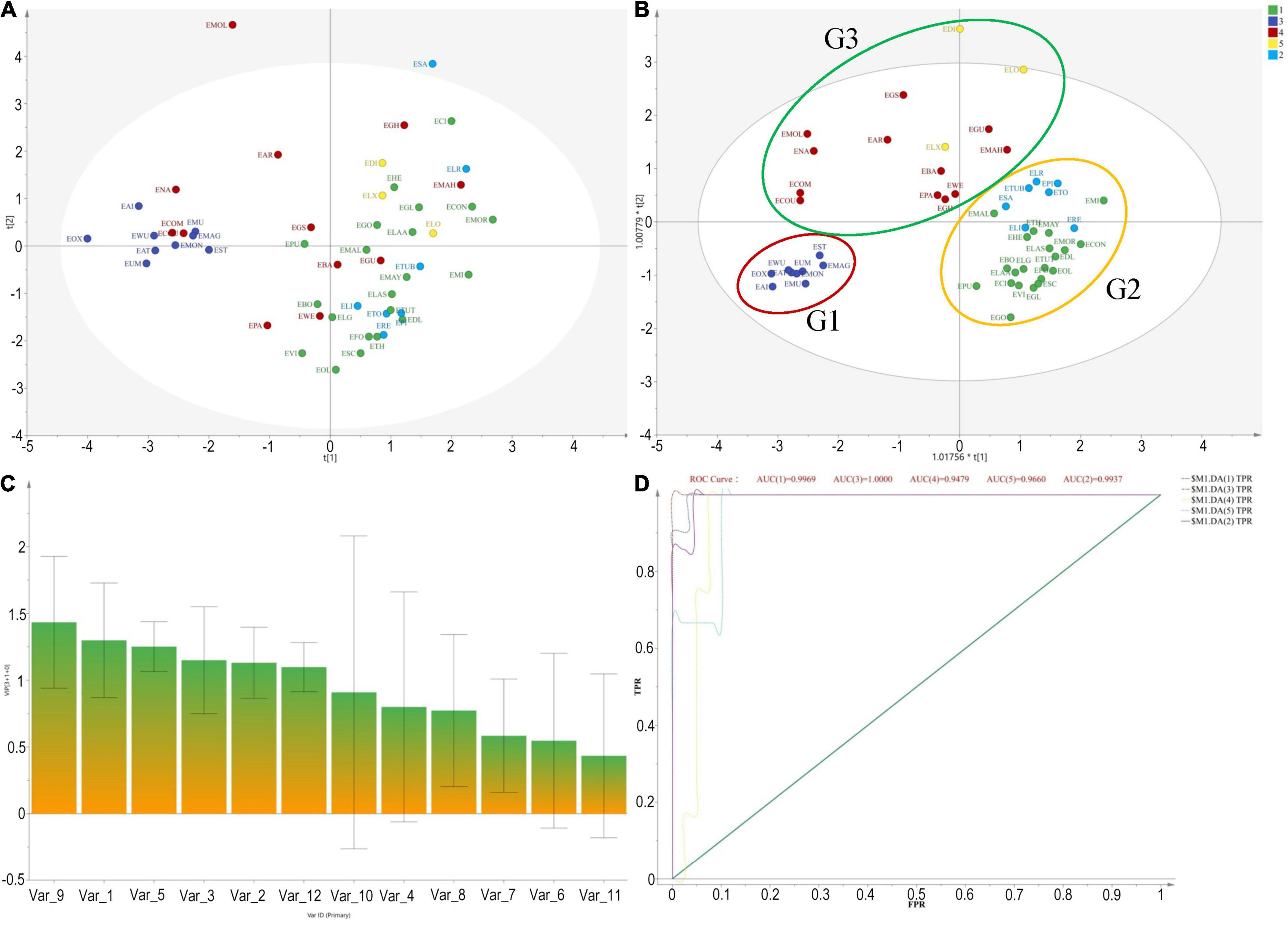
Figure 2. Cluster analysis based on morphological characteristics and molecular phylogenetic tree for grouping 56 Elaeagnus species in China. (A) Pure principal component analysis performed by using morphological traits, associated with sexual plant reproduction, were extracted and quantified according to the records of the Flora of China; (B) With the orthogonal partial least squares discriminant analysis (OPLS-DA) based on the phylogenetic tree of matK sequences, 56 species were well clustered into 3 groups, which were marked with G1, G2, and G3; (C) The variable importance for the projection (VIP) summarizing the importance of the variables, and this plot is sorted to display larger VIPs to the left. The top 5 contributing quantitative traits affecting the current clustering model are V9 (flower color), V1 (flowering), V5 (evergreen or deciduous), V3 (leaf size), and V2 (fruiting), respectively, (VIP > 1); and (D) The receiver operating characteristic (ROC) curve was used to evaluate the effectiveness of the grouping boundary.
The data of morphological traits were normalized, and the results were consistent with the above OPLS-DA results. As shown in Figure 3 and Supplementary Figure 3, among the 12 morphological traits, five traits were significantly different among the three groups. First of all, the flower colors of the three groups were significantly different (ANOVA, p = 0.0013) and gradually changed from yellow to white from G1 to G3; the species divided into G2 had a relatively independent flowering and fruiting, and the flowering is concentrated in the fourth quarter (t-test, p < 0.0001), and the fruiting is in the first quarter of the next year (t-test, p < 0.05). Flowering uniformity was indeed a prerequisite for natural interspecific gene recombination, which directly leads to the rich species diversity of the G2 group. The results also implicated negatively that the types of calyx tube and style villous did not significantly contribute to group formation (ANOVA, p = 0.9729).
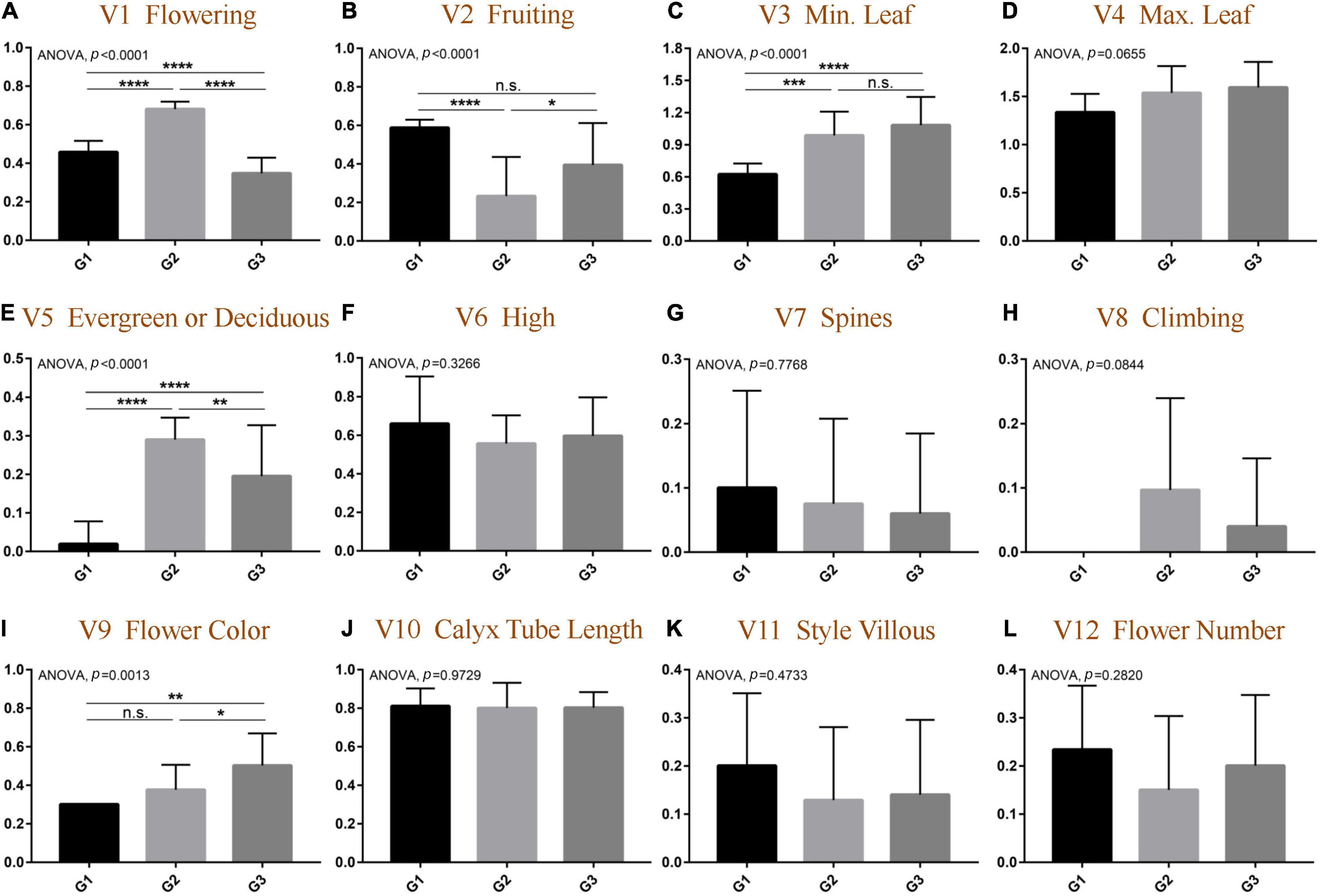
Figure 3. Comparative analysis of morphological differentiation at quantitative traits among different groups of genus Elaeagnus. (A) Flowering (V1); (B) Fruiting (V2); (C) Minimum leaf size (V3); (D) Maximum leaf size (V4); (E) Evergreen or deciduous (V5); (F) Plant height (V6); (G) Spines or absent (V7); (H) Climbing or not (V8); (I) Flower color (V9); (J) The length of calyx tube (V10); (K) Style villous (V11); and (L) the flower number (V12). (N > 9; t-test, ****p < 0.0001; ***p < 0.001; **p < 0.01; and *p < 0.05).
In summary, the most significant trait dispersion among different groups is flowering, while the G2 group of plants has better wild conditions for natural hybridization in China, mainly including the consistency of pollinating cycles and the cross-distribution of multiple species. Nonetheless, the geographic dispersion of different groups of Elaeagnus plants in China remains unclear. Further analysis of geographical dispersion is believed to be helpful in assessing cross-compatibility and uncovering potential geographic barriers.
This study illustrated that there is no obvious geographic isolation among the three groups, except for a few species that were distributed on the plateau or crossed the Taiwan Strait. As shown in Supplementary Table 3 and Figure 4, the three geographic nodes include South China, Central China, and East China, which divided all species into upper and lower parts. Group G2 contained 28 species in the upper left corner of Figure 4B, and the groups G1 and G3 contained 24 species in the bottom right of Figure 4B. Therefore, the result is also consistent with some reported articles that the Elaeagnus species is native to Eurasia, northeastern Australia, and North America and distributes into the two river basins and the central plains of China along the Hexi corridor, and then to the southeast coast, southwest and north China (Qin et al., 2007; Du et al., 2020). The tree species dominated by deciduous shrubs formed the G3 group with narrower leaves, which were mainly distributed in high altitude or high latitude regions, including Gansu, Yunnan, and Guizhou provinces. The G1 group was also formed by some deciduous shrubs, but it was mainly distributed at low altitude regions (Figure 4A), and its leaf shape was significantly larger than that of the G3 group (Figure 3). The G2 group had reached the summit of molecular phylogeny and was distributed widely in the south of the Yellow River. Evergreen or semievergreen plants formed the G2 group. It was gratifying that the G2 group seems to have gathered all the Chinese underutilized fruits in Elaeagnus L., such as E. pungens, E. glabra, E. macrophylla, Elaeagnus oldhami, and E. conferta. In addition, the G2 group also figured out that a total of 10 wild species widely distributed in China have the potential to develop fruit crops. As shown in Table 2, there are three evergreen climbing shrubs, including E. glabra, E. gonyanthes, and E. henryi; two evergreen slightly climbing shrubs, including E. macrophylla and E. conferta; and five erect shrubs, including E. lanceolata, E. delavayi, E. oldhami, E. pungens, and E. bockii. Here, we have also sorted out the excellent dominant traits to prepare for the next collection of these wild resources. First, E. conferta could provide a founder species with a large fruit size, while E. gonyanthes could provide an important genetic resource with a long pedicel. E. lanceolata and E. delavayi could be used to domesticate hybrids without spines, and the five climbing shrubs could be used to develop high-yield crown type commercial cultivars that facilitate automated field management.
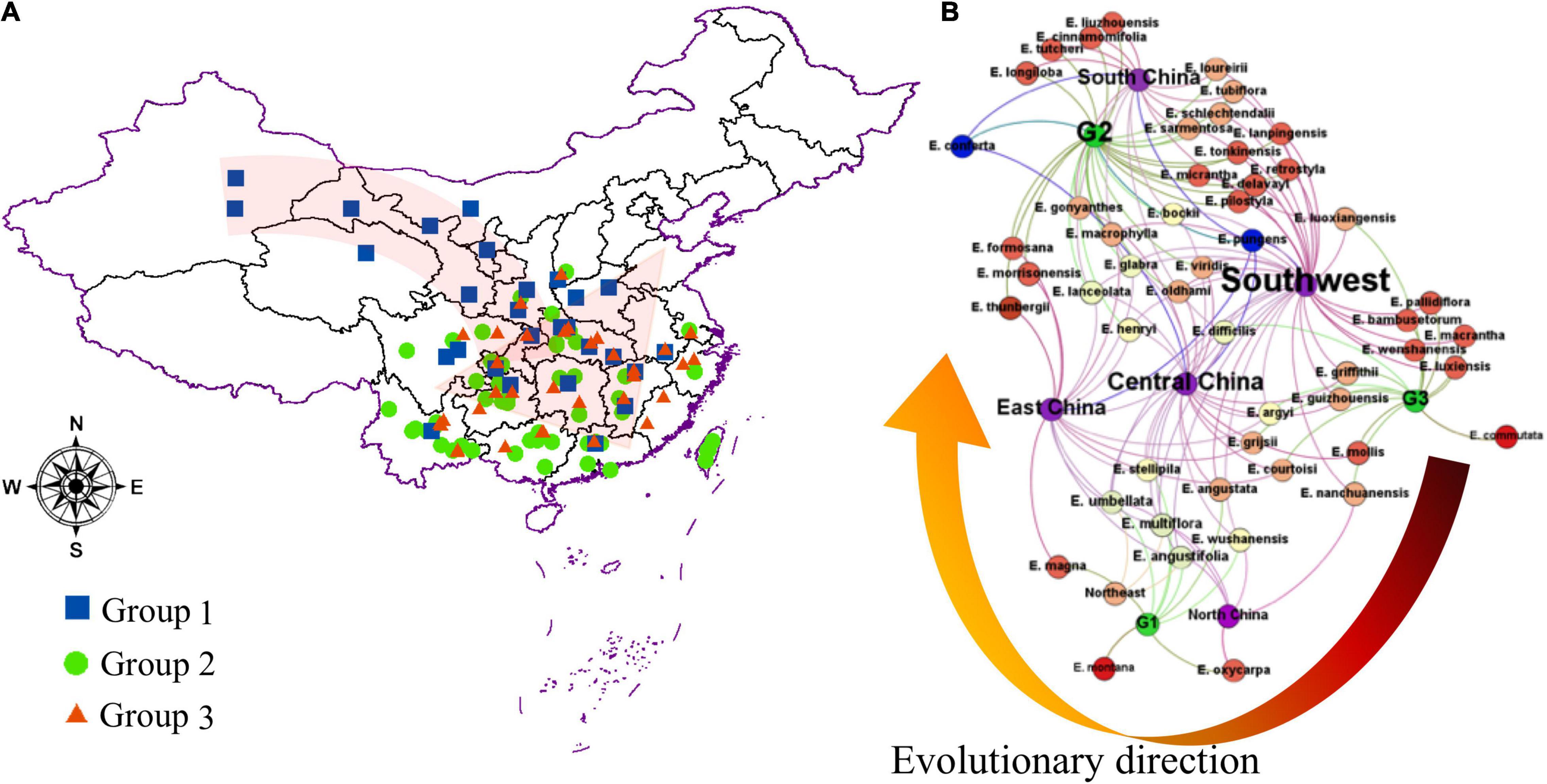
Figure 4. The geographical dispersion of the genus Elaeagnus in China. (A) Distribution of the main Chinese genus Elaeagnus, and three groups were divided by using the OPLS-DA model; (B) Neural Network analysis for distribution of different groups of genus Elaeagnus in China.
Genetic diversity estimates were calculated by using the matK sequence. Then, we used the uncorrelated log-normal relaxed-clock model and the GTR + G model of DNA sequence evolution for each partition to accommodate mean branch rate variation among gene trees. As shown in Figure 5, a time-calibrated phylogeny was reconstructed by BEAST. E. commutata (KX676844) is native to Minnesota, North America and is an independent species as it formed a separate group with a bootstrap value of 98%, without combining with other groups. E. angustifolia and E. mollis formed a separate group and represented the deciduous clade together with E. commutata (Clade I). E. umbellata formed a separate deciduous group (Clade II). Elaeagnus montana and E. multiflora formed the last deciduous clade (Clade III). The remaining species formed a richly divided evergreen group. Among them, E. lanceolata, E. conferta, E. pungens, E. henryi, and E. bockii formed an evergreen clade (Clade IV-Part 1). E. gonyanthes, E. macrophylla, and E. glabra formed another evergreen clade (Clade IV-Part 2). The phylogeny of extant and fossil species showed support for our divergence time evaluation (Bell et al., 2010). The molecular clock inferred Elaeagnus originated between the early Oligocene and the middle Miocene at 11.22 Ma (95% HPD: 17.1–7.3 Ma).
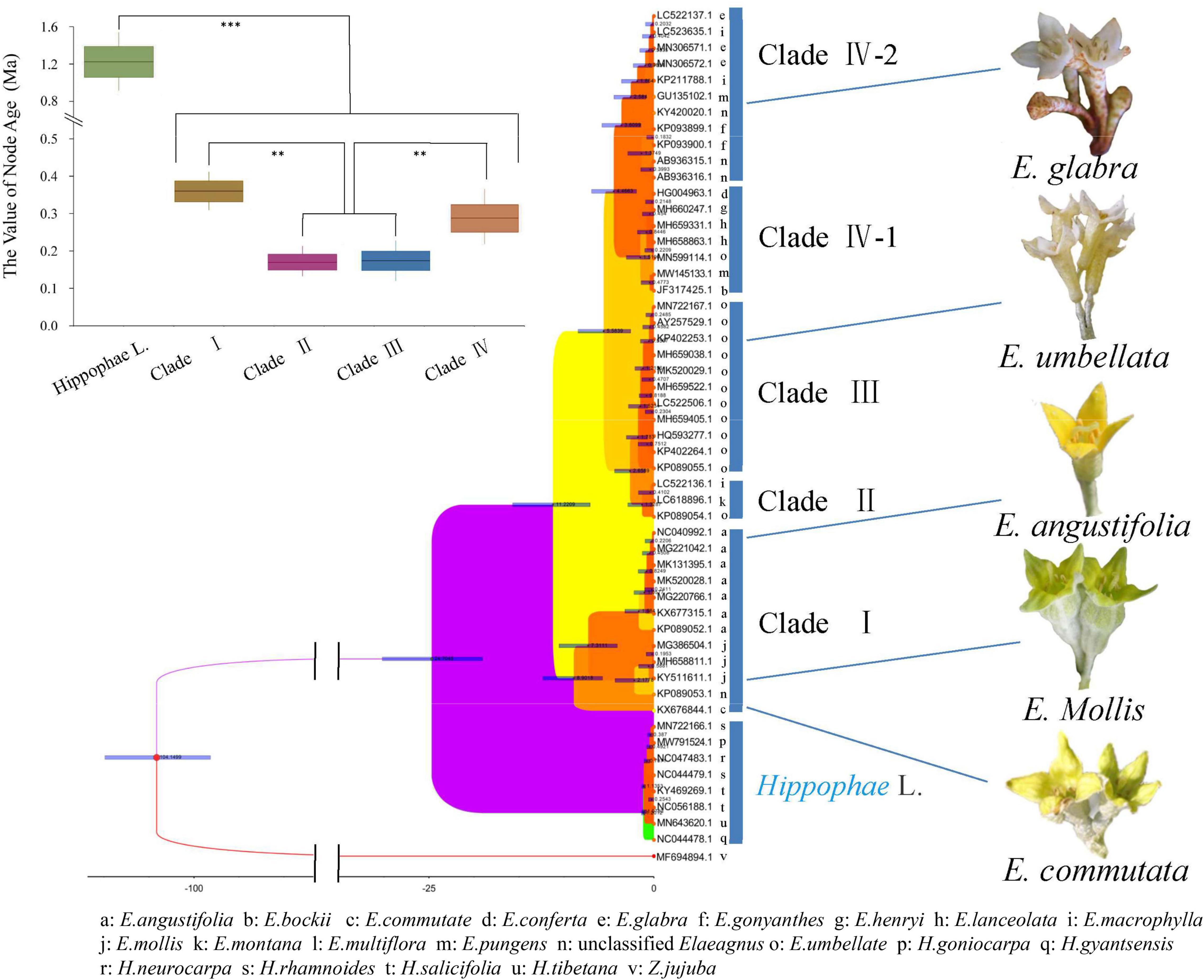
Figure 5. Time-calibrated Bayesian tree inferred from the matK sequences of the family Elaeagaceae and one outgroup species of Acer using BEAST. t-test, ***p < 0.001 and **p < 0.01.
Fruits of Hippophae L. (Sea buckthorn) in the family Elaeagnaceae have been domesticated and cultivated in orchards, especially in China, Europe, Canada, and the United States with a long development history (Zhao, 1997; Rongsen et al., 2013). Fruits of Elaeagnus L. have similar characteristics to that of sea buckthorn (Li and Schroeder, 1996). Why did we spend so much energy on the cultivation and domestication of the future fruit crops of the genus Elaeagnus L.? First, sea buckthorn occurs as a native plant distributed only throughout the arid area of the northwestern part of the northern temperate zone, which directly restricted the large-scale cultivation as a fruit crop (Li et al., 2005); second, sea buckthorn is considered to be drought resistant (Zhang et al., 2022) is, therefore, considered an ideal plant that has been used for fighting soil erosion and also used in land reclamation (Ruan et al., 2013; Li G. et al., 2015), so the fruit used is just to supplement a by-product in many countries. Additionally, one of the major factors restricting the development of sea buckthorn is the berries type with difficult to transport and the limited shelf life of the fresh fruit. Whereas, the fruits of Elaeagnus L. have the potential to be popularized in a wide range of regions around the world due to their advantages in geographical dispersion and cross-latitude adaptability. The natural distribution of Elaeagnus L. species can extend from the northern temperate zone to subtropical and even tropical zones. This feature has practical implications for new approaches to fruit breeding in mid-low latitudes.
In general, traditional breeding methods, such as genetic selection or systematic breeding, are suitable for the initial domestication of wild plant resources (Dempewolf et al., 2017). Accurate characterization of the available genetic basis pool is therefore important in breeding programs and essential for the protection of future property right over new cultivars (Adebayo et al., 2009; Viruel et al., 2021). When the founder species has been determined, how to assess the taxonomic and phylogenetic position and finding the closely related species is very important to examine the cross-compatibility between them. In this study, Chinese Elaeagnus species were investigated first by using DNA marker regions of ITS and matK sequences. The traditional plant taxonomy was used to correct the results of molecular phylogenetic inferences. Phylogenetic relationships reconstructed with the matK sequences of the chloroplast genome suggested a clear characteristic of maternal inheritance. The present results may be interpreted as limited pieces of evidence, which supported that the phylogenetic analysis and the clustering of morphological traits may help to construct an effective population pool and revealed the genetic basis of a set of elite parents for breeding practice. The construction of genetic pools mainly relies on the breeder’s clear ideas and the grasp of breeding objectives, as well as the genetic collection of wild relatives with excellent morphological traits (Mars, 2000). However, in the absence of sufficient DNA sequences, how do we analyze the dispersive pattern only by using the morphological traits? In this study, the novel clustering model innovative provided excellent insights for understanding the evolution and biogeography of Elaeagnus L. in China. First, the phylogenetic positions of successful hybrids provided reference examples to gain the genetic characteristics and distribution patterns of hybrid compatible species and second, statistics and OPLS-DA model clustering 12 quantitative traits indicated natural dispersion of different species clusters. In addition, the current inference directly excavated 10 potential wild species in line with the breeding aims for further improvement and domestication.
Previous phylogenetic studies using several gene regions revealed three well-supported clades in the genus Elaeagnus L. (Liu et al., 2019; Alexandrov and Karlov, 2021). Similarly, our results indicated that both MP and ML trees showed three or four well-supported clades in the genus Elaeagnus L. For the first time, we used the matK sequence of the DNA marker region of the chloroplast genome, and the obtained molecular phylogeny was concordant with the morphological taxonomy. As shown in Figure 5, the molecular clock inferred Elaeagnus originated between the early Oligocene and the middle Miocene at 11.22 Ma. In a macrofossil record, a single living and fossil species of Elaeagnus, namely E. commutata, is native to western and boreal North America, suggesting that the lineage had Asian ancestry and entered North America via the Bering land bridge (Su et al., 2014). As the first confirmed leaf fossil record in Elaeagnus, Elaeagnus tibetensis demonstrated that this genus was already distributed in the Qinghai-Tibet Plateau by the late Miocene (about 10∼5 Ma; Wang et al., 2006; Su et al., 2014). The diversity of Elaeagnus in this region may be closely associated with the uplift of the Qinghai-Tibet Plateau. This is consistent with our estimate that the evergreen group began to separate from deciduous shrubs at 5.58 Ma years ago. The Chinese current evergreen Elaeagnus species originated at 0.3 Ma. The uplift of the Qinghai-Tibet Plateau gradually strengthened the monsoonal climate in eastern Asia during the Neogene (Zhisheng et al., 2001; Liu and Yin, 2002). Because of the strongly seasonal precipitation, Elaeagnus tend to have a high density of scales on both sides of young leaves, but the adaxial scales tend to detach when the leaves are fully expanded. The hydrophobic of leaf properties may be an important condition for the migration of Elaeagnus from high latitudes to low latitudes, but how the interspecies evolution after living in low latitudes has not been reported now. Interestingly, intraspecific diversity was observed in this study. E. pungens, E. macrophylla, and E. glabra have abundant haplotypes and are clustered within different phylogenetic branches (Supplementary Figure 1) in Clade IV. Additionally, as shown in Figure 5, the perianth color of Elaeagnus evolved from dark yellow or green to white, and the eight species we investigated in Clade IV all represent flowers of the same or similar white color (Supplementary Figure 2). Thus, the clade IV is a large evergreen type woody shrub, and the white flowers are hermaphrodite and are pollinated by honey bees. Hypothetically, pollinator diversity (Ollerton, 2017) at low latitudes may facilitate the exchange of genetic information across Elaeagnus species. In conclusion, the phylogenetic analysis reconstructed the preliminary pieces of evidence for the geographic dispersion and cross-species interactions at low-latitude of Elaeagnus species in China.
The widely distributed wild germplasm resources of Elaeagnus L. in China are the core conditions for developing new berry crops of fruit trees. Aiming at how to efficiently construct competent genetic basis pools, in this study, we constructed a phylogenetic tree using the DNA marker regions in the chloroplast genome and a novel OPLS-DA model for morphological clustering according to the phylogenetic tribes. In this study, the Chinese Elaeagnus species were divided into three groups, and we have pioneered the phylogenetic position of the hybrid species, which can have far-reaching significance for the construction of the basic population pools. The results summarized that a total of 10 wild species widely distributed in China have the potential to develop fruit crops. Furthermore, the results here are the first time to visualize the natural co-evolution of Elaeagnus species in low-latitude of China from a new perspective through phylogenetic analysis and geographical dispersion. We also analyzed the phylogenetic status of the parental materials of the reported interspecific hybridization and its distribution in China. In conclusion, we have provided a list of species that are solemnly recommended for systematic selection or cross-breeding to create new fruit-type Elaeagnus crops.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
CC, ZW, and HH: conceptualization. CC, CW, LY, and SF: methodology. CC, CW, and SF: software. LY and SF: validation. SF: formal analysis and visualization. CC: investigation, data curation, writing—original draft preparation, and project administration. SF and CC: resources. CC and HH: writing—review and editing. HH: supervision and funding acquisition. All authors have read and agreed to the published version of the manuscript.
This research was funded by the Plant Germplasm Innovation Program, the Chinese Academy of Sciences (Grant No. KFJ-BRP-007-001), and the Science and Technology Development Fund of Lushan Botanical Garden, Chinese Academy of Sciences (2021ZWZX08).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Lisong Wang and Dr. Jie Liu (Lushan Botanical Garden) for their help with genetics related comments, taxonomic advices and specimen analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.899079/full#supplementary-material
Abdalla, T. E. (2019). “Some wild elaeagnus species: overview, description, biochemistry, and utilization,” in Wild Fruits: Composition, Nutritional Value and Products, ed. A. Mariod (Cham: Springer), 507–521. doi: 10.1007/978-3-030-31885-7_38
Adebayo, O., Bola, O., Opeyemi, W., Gloria, M., and Temitope, O. (2009). Phylogenetic and genomic relationships in the genus Malus based on RAPDs. Afr. J. Biotechnol. 8, 3387–3391.
Ahmadiani, A., Hosseiny, J., Semnanian, S., Javan, M., Saeedi, F., Kamalinejad, M., et al. (2000). Antinociceptive and anti-inflammatory effects of Elaeagnus angustifolia fruit extract. J. Ethnopharmacol. 72, 287–292. doi: 10.1016/s0378-8741(00)00222-1
Akgiin, F., and Sözbilir, H. (2001). A palynostratigraphic approach to the SW Anatolian molasse basin: Kale-Tavas molasse and Denizli molasse. Geodin. Acta. 14, 71–93. doi: 10.1080/09853111.2001.11432436
Alexandrov, O. S., and Karlov, G. I. (2021). The Development of new species-specific molecular markers based on 5S rDNA in Elaeagnus L. Species. Plants. 10, 2713. doi: 10.3390/plants10122713
Banerjee, S., Kar, P., Sarkar, I., Chhetri, A., Mishra, D. K., Dutta, A., et al. (2022). Structural elucidation and chemical characterization of underutilized fruit silverberry (Elaeagnus pyriformis) silver nanoparticles playing a dual role as anti-cancer agent by promoting apoptosis and inhibiting ABC transporters. S. Afr. J. Bot. 145, 243–257. doi: 10.1016/j.sajb.2021.06.029
Baral, J. B., and Bosland, P. W. (2004). Unraveling the species dilemma in Capsicum frutescens and C. chinense (Solanaceae): a multiple evidence approach using morphology, molecular analysis, and sexual compatibility. J. Am. Soc. Horticult. Sci. 129, 826–832. doi: 10.21273/JASHS.129.6.0826
Bartish, I. V., Jeppsson, N., Nybom, H., and Swenson, U. (2002). Phylogeny of Hippophae (Elaeagnaceae) inferred from parsimony analysis of chloroplast DNA and morphology. Syst. Bot. 27, 41–54.
Bell, C. D., Soltis, D. E., and Soltis, P. S. (2010). The age and diversification of the angiosperms re-revisited. Am. J. Bot.. 97, 1296–1303. doi: 10.3732/ajb.0900346
Bouckaert, R., Vaughan, T. G., Barido-Sottani, J., Duchêne, S., Fourment, M., Gavryushkina, A., et al. (2019). BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15:e1006650. doi: 10.1371/journal.pcbi.1006650
Cabelin, V. L. D., and Alejandro, G. J. D. (2016). Efficiency of matK, rbcL, trnH-psbA, and trnL-F (cpDNA) to molecularly authenticate Philippine ethnomedicinal Apocynaceae through DNA barcoding. Pharmacogn. Mag. 12:S384. doi: 10.4103/0973-1296.185780
Chen, C.-Y., Chen, C., Yeh, H., Li, H., and Huang, G. (2019). Secondary metabolites from the leaves of Elaeagnus glabra. Chem. Nat. Compd. 55, 724–725. doi: 10.1007/s10600-019-02790-9
Chen, H., and Wu, C. (2019). Preliminarystudyonleaf color change of a new leaf color mutant of Elaeagnus×ebbingei ‘Gilt Edge’. J. Jiangsu For. Sci. Technol. 46, 49–53. (In Chinese),
Cheng, Y., Yang, Y., Fu, X., Liu, L., Jiang, Z., and Cai, J. (2020). Plastid genomes of Elaeagnus mollis: comparative and phylogenetic analyses. J. Genet.. 99, 1–10.
Choi, K. S., Son, O., and Park, S. (2015). The chloroplast genome of Elaeagnus macrophylla and trnH duplication event in Elaeagnaceae. PLoS One 10:e0138727. doi: 10.1371/journal.pone.0138727
Chwil, M., and Weryszko-Chmielewska, E. (2011). Micromorphology of the floral elements, the structure of the nectary, and the apicultural value of Elaeagnus commutata Bernh. ex Rydb. Acta Agrobot. 64, 27–34. doi: 10.5586/aa.2011.004
Cobb, J. N., Juma, R. U., Biswas, P. S., Arbelaez, J. D., Rutkoski, J., Atlin, G., et al. (2019). Enhancing the rate of genetic gain in public-sector plant breeding programs: lessons from the breeder’s equation. Theor. Appl. Genet. 132, 627–645. doi: 10.1007/s00122-019-03317-0
Cornille, A., Feurtey, A., Gélin, U., Ropars, J., Misvanderbrugge, K., Gladieux, P., et al. (2015). Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evol. Appl. 8, 373–384. doi: 10.1111/eva.12250
Corns, W. G., and Schraa, R. J. (1962). Dormancy and germination of seeds of silverberry (Elaeagnus commutata Bernh.). Can. J. Bot. 40, 1051–1055. doi: 10.1139/b62-095
Dandge, P., Kasabe, P., and Patil, R. (2011). Evaluation of medicinal and nutritional components from the Eleagnus conferta fruit. Sci. Res. Rep. 1, 56–60.
Dasila, K., and Singh, M. (2022). Bioactive compounds and biological activities of Elaegnus latifolia L.: an underutilized fruit of North-East Himalaya, India. S. Afr. J. Bot. 145, 177–185. doi: 10.1016/j.sajb.2021.07.020
Dempewolf, H., Baute, G., Anderson, J., Kilian, B., Smith, C., and Guarino, L. (2017). Past and future use of wild relatives in crop breeding. Crop Sci. 57, 1070–1082. doi: 10.2135/cropsci2016.10.0885
Du, S., Ye, Z., Hu, X., Liu, S., Duan, A., Yu, W., et al. (2020). Phylogeographic investigation of Elaeagnus mollis revealed potential glacial refugia and allopatric divergence in central China. Plant Syst. Evol. 306, 1–9. doi: 10.1007/s00606-020-01696-2
Eernisse, D. J., and Kluge, A. G. (1993). Taxonomic congruence versus total evidence, and amniote phylogeny inferred from fossils, molecules, and morphology. Mol. Biol. Evol. 10, 1170–1195. doi: 10.1093/oxfordjournals.molbev.a040071
Fordham, I. M., Clevidence, B. A., Wiley, E. R., and Zimmerman, R. H. J. H. (2001). Fruit of autumn olive: a rich source of lycopene. HortScience 36, 1136–1137. doi: 10.21273/HORTSCI.36.6.1136
Gao, R., Hou, J., Zhao, R., Yang, X., Hou, X., Huo, L., et al. (2021). Seed dormancy and germination of a critically endangered plant, Elaeagnus mollis, on the Loess Plateau of China. Eur. J. For. Res. 140, 451–461. doi: 10.1007/s10342-020-01342-z
Gill, N. S., and Gupta, M. J. (2018). Elaeagnus conferta: a comprehensive review. Res. J. Pharm. Technol. 11, 2667–2671. doi: 10.5958/0974-360X.2018.00494.8
Hongwen, H., Shuaiyu, Z., and Chunsong, C. (2021). Domestication and breeding strategy of wild fruit trees on track of plant introduction and domestication history. J. Plant Genet. Resour. 22, 1463–1473.
Hu, H., Cheng, Z., Li, L., Chen, J., and Yang, L. (2016). Determination of carotenoids in fruit of five Elaeagnus spp. genotypes by high performance liquid chromatography. Chine. Bull. Bot. 51:306.
Ismail, M., Hussain, M., Mahar, S., and Iqbal, S. (2015). Investigation on total phenolic contents of Elaeagnus multiflora. Asian J. Chem. 27:4587. doi: 10.14233/ajchem.2015.19244
Jaafar, H., Ain, M. F., and Ahmad, Z. A. (2018). Performance of E. conferta and G. atroviridis fruit extracts as sensitizers in dye-sensitized solar cells (DSSCs). Ionics 24, 891–899. doi: 10.1007/s11581-017-2244-1
Kar, P., Dey, P., Misra, A. K., Chaudhuri, T. K., and Sen, A. (2016). Phytometabolomic fingerprinting of selected actinorhizal fruits popularly consumed in North-East India. Symbiosis 70, 159–168. doi: 10.1007/s13199-016-0415-x
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Khattak, K. F. (2012). Free radical scavenging activity, phytochemical composition and nutrient analysis of Elaeagnus umbellata berry. J. Med. Plants Res. 6, 5196–5203. doi: 10.5897/JMPR11.1128
Khomdram, S., Barthakur, S., and Devi, G. S. (2014). Biochemical and molecular analysis of wild endemic fruits of the Manipur region of India. Int. J. Fruit Sci.. 14, 253–266. doi: 10.1080/15538362.2013.818483
Kim, Y., Shin, J., Kim, D. W., Lee, H. S., and Choi, C. (2020). Complete chloroplast genomes of E. umbellata Thunb., E. multiflora Thunb., E. macrophylla Thunb., and E. glabra Thunb.(Elaeagnaceae). Mitochond. DNA B Resour. 5, 2490–2492. doi: 10.1080/23802359.2020.1779142
Klich, M. G. (2000). Leaf variations in Elaeagnus angustifolia related to environmental heterogeneity. Environ. Exp. Bot. 44, 171–183. doi: 10.1016/s0098-8472(00)00056-3
Kubota, S., Konno, I., Kanno, A., and Genetics, A. (2012). Molecular phylogeny of the genus Asparagus (Asparagaceae) explains interspecific crossability between the garden asparagus (A. officinalis) and other Asparagus species. Theor. Appl. Genet. 124, 345–354. doi: 10.1007/s00122-011-1709-2
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547. doi: 10.1093/molbev/msy096
Lee, J. H., Seo, W. T., and Cho, K. M. (2011). Determination of phytochemical contents and biological activities from the fruits of Elaeagnus multiflora. Prev. Nutr. Food Sci.. 16, 29–36.
Leech, N. L., Barrett, K. C., and Morgan, G. A. (2014). IBM SPSS for Intermediate Statistics: Use and Interpretation. Milton Park: Routledge.
Lexer, C., Randell, R. A., and Rieseberg, L. H. J. E. (2003). Experimental hybridization as a tool for studying selection in the wild. Ecology 84, 1688–1699.
Li, C., Yang, Y., Junttila, O., and Palva, E. T. J. P. S. (2005). Sexual differences in cold acclimation and freezing tolerance development in sea buckthorn (Hippophae rhamnoides L.) ecotypes. Plant Sci. 168, 1365–1370.
Li, G., Du, S., and Guo, K. (2015). Evaluation of limiting climatic factors and simulation of a climatically suitable habitat for Chinese sea buckthorn. PLoS One. 10:e0131659. doi: 10.1371/journal.pone.0131659
Li, L. H., Baek, I. K., Kim, J. H., Kang, K. H., Koh, Y. S., Jung, Y. D., et al. (2009). Methanol extract of Elaeagnus glabra, a Korean medicinal plant, inhibits HT1080 tumor cell invasion. Oncol. Rep. 21, 559–563.
Li, T. S., and Schroeder, W. R. (1996). Sea buckthorn (Hippophae rhamnoides L.): a multipurpose plant. HortTechnology 6, 370–380. doi: 10.21273/HORTTECH.6.4.370
Li, Y. L., Lu, X. Q., Wang, C. Y., Cai, X. L., and Chen, H. J. (2015). Study on introduction, propagation and cultivation techniques of gold-marginatus hybrid Elaeagnus pungens (In Chinese). Chin. Hortic. Abstr. 31, 144–147.
Liang, S., Yang, R., Dong, C., and Yang, Q. (2015). Physicochemical properties and fatty acid profiles of Elaeagnus mollis Diels nut oils. J. Oleo Sci. 64, 1267–1272. doi: 10.5650/jos.ess15158
Liao, C. R., Ho, Y. L., Huang, G. J., Yang, C. S., Chao, C. Y., Chang, Y. S., et al. (2013). One lignanoid compound and four triterpenoid compounds with anti-inflammatory activity from the leaves of Elaeagnus oldhamii maxim. Molecules 18, 13218–13227. doi: 10.3390/molecules181113218
Liao, C.-R., Kuo, Y.-H., Ho, Y.-L., Wang, C.-Y., Yang, C.-S., Lin, C.-W., et al. (2014). Studies on cytotoxic constituents from the leaves of Elaeagnus oldhamii Maxim. in non-small cell lung cancer A549 cells. Molecules 19, 9515–9534. doi: 10.3390/molecules19079515
Liu, J., Gong, L. D., Qi, L., Liu, Z. Y., Niu, Y. F., and Shi, C. J. (2019). The complete chloroplast genome of Elaeagnus conferta Roxb (Elaeagnaceae). Mitochond. DNA B Resour. 4, 2035–2036. doi: 10.1080/23802359.2019.1617074
Liu, X., and Yin, Z. Y. (2002). Sensitivity of East Asian monsoon climate to the uplift of the Tibetan Plateau. Palaeogeogr. Palaeoclimatol. Palaeoecol. 183, 223–245. doi: 10.1016/S0031-0182(01)00488-6
Lizardo, R. C. M., Cho, H. D., Lee, J. H., Won, Y. S., and Seo, K. I. (2020). Extracts of Elaeagnus multiflora Thunb. fruit fermented by lactic acid bacteria inhibit SW480 human colon adenocarcinoma via induction of cell cycle arrest and suppression of metastatic potential. J. Food Sci. 85, 2565–2577. doi: 10.1111/1750-3841.15300
Mars, M. (2000). Pomegranate plant material: genetic resources and breeding, a review. Options Mediterr. Ser. A. 42, 55–62.
Maune, J. F., Camadro, E. L., and Erazzú, L. E. (2018). Cross-incompatibility and self-incompatibility: unrelated phenomena in wild and cultivated potatoes? Botany 96, 33–45. doi: 10.1139/cjb-2017-0070
Naumann, J. C., Bissett, S. N., Young, D. R., Edwards, J., and Anderson, J. E. (2010). Diurnal patterns of photosynthesis, chlorophyll fluorescence, and PRI to evaluate water stress in the invasive species, Elaeagnus umbellata Thunb. Trees 24, 237–245. doi: 10.1007/s00468-009-0394-0
Obuchowski, N. A. (2003). Receiver operating characteristic curves and their use in radiology. Radiology 229, 3–8. doi: 10.1148/radiol.2291010898
Olas, B. (2018). The beneficial health aspects of sea buckthorn (Elaeagnus rhamnoides (L.) A. Nelson) oil. J. Ethnopharmacol. 213, 183–190. doi: 10.1016/j.jep.2017.11.022
Ollerton, J. (2017). Pollinator diversity: distribution, ecological function, and conservation. Ann. Rev. Ecol. Evol. Syst. 48, 353–376. doi: 10.1146/annurev-ecolsys-110316-022919
Panja, S., Chaudhuri, D., Ghate, N. B., Le Minh, H., and Mandal, N. (2014). In vitro assessment of phytochemicals, antioxidant and DNA protective potential of wild edible fruit of Elaeagnus latifolia Linn. Fruits 69, 303–314. doi: 10.1051/fruits/2014019
Patel, R., Singh, A., and Deka, B. (2008). Soh-Shang (Elaeagnus latifolia): an underutilized fruit of North East region needs domestication. ENVIS Bull. Himalayan Ecol. 16, 1–2.
Patel, S. (2015). Plant genus Elaeagnus: underutilized lycopene and linoleic acid reserve with permaculture potential. Fruits 70, 191–199. doi: 10.1051/fruits/2015014
Paulsamy, S., Kumar, P. S., Kumar, A., and Kumar, P. S. (2010). Elaeagnus kologa schlecht.–an under utilized edible and endemic fruit plant in Nilgiris, the western ghats. Indian J. Nat. Prod. Resour. 1, 258–260.
Payne, T., Stuthman, D., Mcgraw, R., and Bregitzer, P. J. C. S. (1986). Physiological changes associated with three cycles of recurrent selection for grain yield improvement in oats 1. Crop Sci. 26, 734–736. doi: 10.2135/cropsci1986.0011183X002600040021x
Qi, X. L., He, J., Li, D. W., and Huang, L. (2021). First report of leaf spot on Elaeagnus pungens caused by Epicoccum latusicollum in China. Forest Pathol. 51:e12716.
Qin, H., Michael, G. J. B., Press, M. S., and Garden, M. B. (2007). Flora of China (English version, Vol. 13. Beijing: Science Press.
Ramadevi, D., and Narayana, L. J. P. P. S. (1990). Morphology of the flower and fruit ofHydrocera triflora Wight and Arn. emend Venkat. and Dutt—an elucidation. Proc. Plant Sci. 100, 43–49. doi: 10.1007/BF03053467
Rongsen, L., Ahani, H., Shaban, M., Esfahani, M., Alizade, G., Rostampour, M., et al. (2013). The genetic resources of Hippophae genus and its utilization. advance in agriculture and biology. Int. J. Scholary Res. Gate 1, 15–21.
Ruan, C.-J., Rumpunen, K., and Nybom, H. (2013). Advances in improvement of quality and resistance in a multipurpose crop: sea buckthorn. Crit. Rev. Biotechnol. 33, 126–144. doi: 10.3109/07388551.2012.676024
Ruixue, W. (2013). Phylogeography of Hippophae rhamnoides ssp. Sinensis (In Chinese). Lanzhou: Northwest Normal University.
Sasikumar, J., Patharaj, J., Adithya, E., Christabel, P. H., and Shamna, R. J. (2012). Antioxidant capacity and phenolic content of Elaeagnus kologa schlecht. an underexploited fruit from India. Free Radic. Antioxid. 2, 28–35. doi: 10.5530/ax.2012.3.4
Selvamuthukumaran, M., and Khanum, F. (2014). Development of spiced seabuckthorn [Elaeagnus rhamnoides (L.) a. Nelson syn. Hippophae rhamnoides L.] mixed fruit squash. Indian J. Tradit. Knowl. 13, 132–141.
Sohn, E., Lim, H. S., Kim, Y. J., Kim, B. Y., Kim, J. H., and Jeong, S. J. (2019). Elaeagnus glabra f. oxyphylla attenuates scopolamine-induced learning and memory impairments in mice by improving cholinergic transmission via activation of CREB/NGF signaling. Nutrients 11:1205. doi: 10.3390/nu11061205
Son, O., Yoon, C. Y., and Park, S. J. (2014). Phylogenetic relationships in Korean Elaeagnus L. based on nrDNA ITS sequences. Korean J. Plant Resour. 27, 671–679. doi: 10.7732/kjpr.2014.27.6.671
Srivathsan, A., and Meier, R. (2012). On the inappropriate use of Kimura-2-parameter (K2P) divergences in the DNA-barcoding literature. Cladistics 28, 190–194. doi: 10.1111/j.1096-0031.2011.00370.x
Su, T., Wilf, P., Xu, H., and Zhou, Z. K. (2014). Miocene leaves of Elaeagnus (Elaeagnaceae) from the qinghai-tibet Plateau, its modern center of diversity and endemism. Am. J. Bot. 101, 1350–1361. doi: 10.3732/ajb.1400229
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic acids Res. 47, 270–275. doi: 10.1093/nar/gkz357
Sun, M., and Lin, Q. (2010). A revision of Elaeagnus L.(Elaeagnaceae) in mainland China. J. Syst. Evol.. 48, 356–390. doi: 10.1111/j.1759-6831.2010.00085.x
Viruel, J., Kantar, M. B., Gargiulo, R., Hesketh-Prichard, P., Leong, N., Cockel, C., et al. (2021). Crop wild phylorelatives (CWPs): phylogenetic distance, cytogenetic compatibility and breeding system data enable estimation of crop wild relative gene pool classification. Bot. J. Linn. Soc. 195, 1–33. doi: 10.1093/botlinnean/boaa064
Wang, L., Lü, H., Wu, N., Li, J., Pei, Y., Tong, G., et al. (2006). Palynological evidence for Late miocene–pliocene vegetation evolution recorded in the red clay sequence of the central Chinese loess plateau and implication for palaeoenvironmental change. Palaeogeogr. Palaeoclimatol. Palaeoecol. 241, 118–128. doi: 10.1016/j.palaeo.2006.06.012
Wang, Y., Ma, Y., Jia, B., Wu, Q., Zang, D., and Yu, X. (2020). Analysis of the genetic diversity of the coastal and island endangered plant species Elaeagnus macrophylla via conserved DNA-derived polymorphism marker. PeerJ. 8:e8498. doi: 10.7717/peerj.8498
Ye, L., Song, Y., Yamada, K., Nakao, Y., and Nii, N. (2012). Anatomical and histological changes in developing silverberry (Elaeagnus multiflora var. gigantea L.) fruit. J. Horticult. Sci. Biotechnol. 87, 64–70. doi: 10.1080/14620316.2012.11512832
Yu, J., Xue, J. H., and Zhou, S. L. (2011). New universal matK primers for DNA barcoding angiosperms. J. Syst. Evol. 49, 176–181. doi: 10.3732/apps.1500137
Zhang, D., Yang, N., Dong, J., Wang, C., Li, Q., Wang, R., et al. (2022). Prediction of the sea buckthorn AQP gene structure and its spatiotemporal expression pattern under drought stress. J. Plant Biochem. Biotechnol. 31, 239–249. doi: 10.1007/s13562-021-00740-7
Zhao, H. J. W. F. R. (1997). A brief introduction to breeding Hippophae in Russia and its genetic improvement in China. World For. Res.. 10, 65–71.
Zhao, K. K., Wang, J. H., Zhu, Z. X., Shi, G. Z., Luo, S. X., and Wang, H. F. (2020). Complete plastome sequence of Elaeagnus glabra (Elaeagnaceae): an Asian endemic plant species. Mitochond. DNA B Resour. 5, 288–289. doi: 10.1080/23802359.2019.1702483
Zhisheng, A., Kutzbach, J. E., Prell, W. L., and Porter, S. C. (2001). Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature 411, 62–66. doi: 10.1038/35075035
Keywords: underutilized fruit, fruit crop, morphological clustering, molecular phylogeny, phylogenetic tree
Citation: Cheng C, Fan S, Wang C, Ye L, Wang Z and Huang H (2022) Phylogenetic Analysis of Elaeagnus L. in China: A Basis for Genetic Improvement of a Berry Crop. Front. Plant Sci. 13:899079. doi: 10.3389/fpls.2022.899079
Received: 18 March 2022; Accepted: 19 April 2022;
Published: 09 June 2022.
Edited by:
Kevin M. Folta, University of Florida, United StatesReviewed by:
Miao Sun, Huazhong Agricultural University, ChinaCopyright © 2022 Cheng, Fan, Wang, Ye, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunsong Cheng, Y2hlbmdjc0Bsc2JnLmNu; Hongwen Huang, aHVhbmdod0BzY2JnLmFjLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.