
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 July 2022
Sec. Plant Systems and Synthetic Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.898541
This article is part of the Research Topic Insights in Plant Systems and Synthetic Biology: 2021 View all 5 articles
 Hang Zhang1,2†
Hang Zhang1,2† Enliang Liu3,4†
Enliang Liu3,4† Xinzheng Huang4†
Xinzheng Huang4† Junfeng Kou5
Junfeng Kou5 Dong Teng2
Dong Teng2 Beibei Lv2
Beibei Lv2 Xiaoqiang Han1*
Xiaoqiang Han1* Yongjun Zhang2*
Yongjun Zhang2*When attacked by insect herbivores, plants initiate sophisticated defenses mediated by complex signaling networks and usually release a blend of functional volatiles such as terpenes against infestation. The extra-long staple cotton Gossypium barbadense cultivated worldwide as natural textile fiber crop is frequently exposed to a variety of herbivores, such as cotton bollworm Helicoverpa armigera. However, little is known about insect-induced transcriptional changes and molecular mechanisms underlying subsequent defense responses in G. barbadense. In the current study, transcriptome changes in G. barbadense infested with chewing H. armigera larvae were investigated, and we identified 5,629 differentially expressed genes (DEGs) in the infested cotton leaves compared with non-infested controls. H. armigera feeding triggered complex signaling networks in which almost all (88 out of 90) DEGs associated with the jasmonic acid (JA) pathway were upregulated, highlighting a central role for JA in the defense responses of G. barbadense against target insects. All DEGs involved in growth-related photosynthesis were downregulated, whereas most DEGs associated with defense-related transcript factors and volatile secondary metabolism were upregulated. It was noteworthy that a terpene synthase gene in the transcriptome data, GbTPS1, was strongly expressed in H. armigera-infested G. barbadense leaves. The upregulation of GbTPS1 in qPCR analysis also suggested an important role for GbTPS1 in herbivore-induced cotton defense. In vitro assays showed that recombinant GbTPS1 catalyzed farnesyl pyrophosphate and neryl diphosphate to produce three sesquiterpenes (selinene, α-gurjunene, and β-elemene) and one monoterpene (limonene), respectively. Moreover, these catalytic products of GbTPS1 were significantly elevated in G. barbadense leaves after H. armigera infestation, and elemene and limonene had repellent effects on H. armigera larvae in a dual-choice bioassay and increased larval mortality in a no-choice bioassay. These findings provide a valuable insight into understanding the transcriptional changes reprogramming herbivore-induced sesquiterpene biosynthesis in G. barbadense infested by H. armigera, which help elucidate the molecular mechanisms underlying plant defense against insect pests.
Of the two cultivated cotton species, the extra-long staple cotton Gossypium barbadense produces superior-quality fibers and is widely planted across the world. However, G. barbadense is frequently subject to attack by a wide range of insect pests such as aphids, mirid bugs, and cotton bollworm Helicoverpa armigera (Ayubov and Abdurakhmonov, 2018; Llandres et al., 2018). Cotton bollworm has caused considerable costs and yield losses as well as overuse of chemical pesticides (Li et al., 2020). In China, Bacillus thuringiensis (Bt)-transgenic cotton cultivars have been widely planted for managing serious lepidopteran pests including H. armigera, and provided efficient and lasting control of target pests. However, sole reliance on Bt-transgenic crops can lead to pest resistance to specific Bt proteins (Lu et al., 2022). Consequently, it is urgently needed to develop new environmentally friendly pest management strategies utilizing the plant’s own defense system.
Plants have evolved sophisticated, diverse systems against a wide array of insect herbivores herbivory that can be triggered by insect feeding, herbivore egg deposition and even insect walking (Wu and Baldwin, 2010; Turlings and Erb, 2018). The defense system in plants is initiated by a series of early signaling components such as Ca2+, reactive oxygen species, and MAP kinases, then a dynamic and complex transcriptional reprogramming is mediated by signaling networks including phytohormones and transcription factors (Erb and Reymond, 2019), which finally leads to the synthesis and accumulation of toxic chemicals (direct defenses) and release of diverse herbivore−induced plant volatiles that attract natural enemies (indirect defenses).
In response to insect infestation, cotton plants emit complex blends of terpene volatiles as an important part of the plant defense (Rodriguez-Saona et al., 2002; Degenhardt et al., 2011). It was found that Gossypium hirsutum immediately emits constitutive terpene volatiles such as α−pinene and (E)−β−caryophyllene in response to herbivores including H. armigera, Spodoptera exigua, and Apolygus lucorum, whereas several other terpenes including limonene, (E)−β−ocimene and linalool are synthesized de novo (Huang et al., 2018, 2022; Arce et al., 2021). These terpenes play crucial ecological roles in direct and indirect defenses in cotton plants. For instance, the homoterpene compound (3E)-4,8-dimethyl-1,3,7-non-atriene (DMNT) released from herbivore-induced G. hirsutum suppresses olfactory−triggered orientation of the moth Spodoptera littoralis to host plants and sex partners (Hatano et al., 2015). Similarly, a behavioral bioassay showed that DMNT and (3R)−linalool in volatile blends of G. hirsutum had repellent effects on females and males of S. littoralis (Borrero-Echeverry et al., 2015). Apolygus lucorum-induced cotton volatile blends dominated by terpenes significantly attracted the parasitoid Peristenus spretus (Huang et al., 2022). In addition, enhanced (E)-β-caryophyllene emission in transgenic G. hirsutum has a deterrent effect on A. lucorum, Aphis gossypii, and H. armigera, and significantly attracts the parasitoids P. spretus and Aphidius gifuensis (Zhang et al., 2020).
Volatile terpenes dominated by mono−, sesqui−, and diterpenes are synthesized by terpene synthases/cyclases (TPSs) using a few acyclic prenyl diphosphate precursors such as neryl diphosphate (NPP), geranyl diphosphate (GPP), farnesyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP) as substrates (Zhou and Pichersky, 2020). Based on phylogenetic relationships, plant TPS family fall into eight subfamilies (TPS-a to TPS-h). In angiosperms, TPS−a subfamily contains primarily sesquiterpene synthases, TPS−b comprises cyclic mono- and hemiterpene synthases, members of TPS−f produce diterpenes and TPS−g generally contains acyclic monoterpenes (Booth et al., 2020). The published genomic sequence data of G. barbadense (Liu et al., 2015; Hu et al., 2019; Wang et al., 2019) revealed that there are 115 putative TPS genes including 44 monoterpene, 59 sesquiterpene, and eight diterpene synthases. However, little is known about the defense roles of TPSs in G. barbadense against insects until now.
In the current study, transcriptional changes in G. barbadense after a 36-h infestation by H. armigera were investigated to identify key pathways involved in the activation of cotton plant defense and candidate terpene synthases. Furthermore, the sesquiterpene synthase gene GbTPS1 was newly identified based on RNA-seq. And then the recombinantGbTPS1 was heterologously expressed in Escherichia coli. The catalytic products of the recombinant GbTPS1 were determined using an in vitro enzymatic assay and gas chromatography-mass spectrometry (GC-MS) analysis. What is more, the impacts of β-elemene and limonene on feeding behavior and growth of H. armigera larvae were also evaluated. These findings should contribute to developing markers for breeding new cotton cultivars with improved resistance to insect pests and further designing novel strategies for cotton pest control through the genetic modification of herbivore-induced terpene biosynthesis.
Cotton plants (G. barbadense cv. Hai7124) were sown in plastic pots (height, 14 cm; diameter, 16 cm). Seedlings were grown in a greenhouse compartment (29/25°C, 50 ± 10% RH, 16:8 L:D regime), and water was added every 2 days. All cotton plants were used in experiments at the 6–7 fully expanded true leaf stage (Huang et al., 2018). Laboratory colonies of cotton bollworm H. armigera were established from field−collected individuals. Larvae of H. armigera were reared on an artificial diet in a climate-controlled room (27 ± 2°C, 75 ± 10% RH, 14:10 L:D regime) (Liang et al., 2008). The 3rd instar larvae were used for all treatments.
Each pot containing one cotton plant was placed in the greenhouse compartment. One H. armigera larva of was placed on a leaf of each cotton plant in a nylon mesh cage at 18:00 PM. After 36 h infestation, larvae were removed, and the damaged leaves were collected for RNA extraction. Non-infested plants in cages maintained under the same conditions were used as controls. For each treatment, three biological replicates were employed.
Total RNA was extracted using the easy-spin Total RNA Extraction Kit (Aidlab Biotechnologies Co., Ltd., Beijing, China). RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States). About 1 μg RNA per sample was used to construct a cDNA library using the NEBNext® Ultra™ RNA Library Prep Kit, and then was sequenced on an Illumina Hiseq 2500 platform (Gene Denovo Biotechnology Co., Ltd., Guangzhou, China) using a PE150 strategy. Clean reads were mapped to G. barbadense genome (Hu et al., 2019) using Hisat2 software.
Gene expression levels were calculated based on fragments per kilobase of transcript per million fragments mapped (FPKM). Genes with log2(fold change) > 1 and false discovery rate < 0.05 were defined as differentially expressed genes (DEGs). The DEGs were functionally annotated using the Gene Ontology (GO) database and Blast2GO.1 DEGs were assigned to KEGG pathways using KOBAS 2.0 software (Xie et al., 2011).
In the transcriptome, GbTPS1 (GB_D01G0996) was significantly expressed after H. armigera infestation. Subsequently, complete open reading frame (ORF) of GbTPS1 was fully sequenced and inserted into the expression vector pET-30a (+). The plasmid was then inserted into E. coli strain BL21 (DE3) for heterologous expression. Freshly transformed E. coli cells were cultured in 500 mL LB with kanamycin (50 mg mL–1) at 37°C to an OD600 of 0.6. Then, 1 mM isopropyl 1-thio-β-D-galactopyranoside (IPTG) was added, and the cultures were incubated at 18°C for 20 h. Bacterial cells were pelleted by centrifugation at 8,000 × g for 10 min at 4°C and resuspended in 4 mL of extraction buffer (50 mM MOPS [pH 7.0], 5 mM MgCl2, 5 mM sodium ascorbate, 5 mM dithiothreitol, 0.5 mM phenylmethanesulfonyl fluoride and 10% [v/v] glycerol). Finally, the cells were disrupted by sonication, and the suspension was centrifuged at 14,000 × g.
Catalytic activities of the recombinant GbTPS1 were assayed as reported previously (Huang et al., 2018). In screw-capped 20-mL glass vials, 500 μL of E. coli crude extract, 10 μM of geranyl pyrophosphate (GPP), NPP, farnesyl pyrophosphate (FPP) or GGPP as the substrate, 10 mM MgCl2, 0.2 mM Na2WO4, 0.05 mM MnCl2 and 0.1 mM NaF, 10 mM MOPS (pH 7.0), 1 mM dithiothreitol and 10% (v/v glycerol). Samples were incubated for 1 h at 30°C. Solid phase microextraction (SPME, SAAB-57330U, Sigma-Aldrich, Shanghai, China) handle with a 65 μm polydimethylsiloxane (PDMS) fiber was inserted into the vial at a distance of 1 cm from the liquid surface and absorbed at 30°C for 30 min. Then the handle containing volatiles samples was inserted into a QP2010 SE gas GC-MS system (Shimadzu, Kyoto, Japan) equipped with an Rxi-5Sil column (30 m × 0.25 mm × 0.25 μm). The oven temperature was set at 40°C for 1 min, then raised 4°C/min to 130°C and held for 5 min, then increased at 10°C/min to 250°C and held for 5 min. Identity of (R)-(+)-limonene (CAS:5989-27-5) and β-elemene (CAS: 515-13-9) was confirmed by comparing retention times and mass spectra with those of the authentic standard (Sigma-Aldrich) under the same conditions. However, selinene and gurjunene are not commercially available, so we performed a comparison of mass spectra with those reported in the NIST17 library. The raw extracts from E. coli expressing empty vector pET-30a (+) were set as controls.
cDNA was synthesized from the total RNA using the FastQuant RT Kit as described above. The quantitative real time PCR (qPCR) analysis was carried out using SuperReal PreMix Plus (SYBR Green) reagent kit (TianGen, Beijing, China) (Huang et al., 2018). GhACT4 (accession number: AY305726) was used as an endogenous control (Supplementary Table 1). Three biological replicates were performed.
The volatile terpenes emitted from H. armigera-infested or control G. barbadense were collected by headspace-SPME as described previously (Avellaneda et al., 2021). Briefly, cotton leaves were gently enclosed in 3 L polyethylene terephthalate containers. The headspace volatiles emitted from G. barbadense were collected for 4 h by SPME as described above. Ethyl nonanoate (17.2 mg, CAS: 123-29-5) was added as an internal standard. Then, the samples were analyzed using GC-MS as described above for enzymatic activity assay. Three biological replicates for each treatment were analyzed.
The choice behavior of H. armigera larvae to β-elemene or limonene at three different concentrations (0.1, 1, and 10 μg/μL) were assessed in plastic petri dishes (9 cm diameter) (Guo et al., 2021). A piece of 1 × 1 cm artificial diet containing 20 μL chemical samples was placed at one end of the petri dish, the control artificial diet containing n-hexane was placed at the other end. Fifty H. armigera larvae of third instar were tested in each compound and each concentration, and three biological replicates were analyzed. After 2 h, the choice of insects was recorded. Survival rate of H. armigera larvae on β-elemene or limonene at four different concentrations (0.05, 0.1, 1, and 10 μg/μL) on growth of H. armigera larvae was also investigated. Aliquots (20 μL) of β-elemene or limonene dissolved in hexane were added in artificial diet. Artificial diet containing n-hexane was used as control. One newly hatched larva per well (1.5 cm in diameter × 2 cm in height) were reared on 500 mg artificial diet in a 24-well plate and allowed to feed for 7 days. A total of 24 insects were tested in each compound and concentration, and five biological replicates were analyzed.
Data analyses were performed using SPSS Statistics software (version 17.0) (SPSS Inc., Chicago, IL, United States). Data are given as the mean ± standard error of the mean (SEM) and if needed, were transformed before analyses. Differences among treatment groups for target gene expression data and survival rate data were analyzed using a one-way analysis of variance (ANOVA) followed by Duncan’s multiple range test. In the behavioral assay, a chi-square test (50:50 distribution) was used to test for significant differences in larval choice between controls and treatments.
Sequencing of the transcriptome from G. barbadense leaves generated 40,502,916–46,404,650 raw reads per sample, and the quality control steps generated 40,457,602–46,358,304 high quality reads. A PCA plot based on the transcriptomic data was shown in Supplementary Figure 1. Among these genes, 5,629 were associated with a cutoff of q ≤ 0.05 and log2(fold change) ≥ 1, and thus identified as DEGs. Among these DEGs, 2,460 were significantly upregulated after infestation, while 3,169 were downregulated (Supplementary Table 2).
In the GO analysis, these genes were classified into biological process, cellular component and molecular function (Figure 1). In the biological process category, the top three terms were metabolic process (GO 0008152; 1013 up- and 1409 downregulated genes), cellular process (GO 0009987; 817 up- and 1314 downregulated genes), and single-organism process (GO 0044699; 775 up- and 1038 downregulated genes). In the category of cellular component, the most abundant were cell (GO 0005623; 383 up- and 839 downregulated genes) and cell part (GO 0044464; 383 up- and 839 downregulated genes), followed by organelle (GO 0043226; 303 up- and 690 downregulated genes), membrane (GO 0016020; 265 up- and 532 downregulated genes) and membrane part (GO 0044425; 230 up- and 434 downregulated genes). In the molecular function ontology, most DEGs were mapped to catalytic activity (GO 0003824; 977 up- and 1158 downregulated) and binding (GO 0005488; 685 up- and 1028 downregulated). To identify the key biological pathways activated by H. armigera infestation, the KEGG pathway analysis was conducted. Among the top 20 pathways (Figure 2), the top three pathways were metabolic pathways (641 genes), biosynthesis of secondary metabolites (408) and plant hormone signal transduction (143).
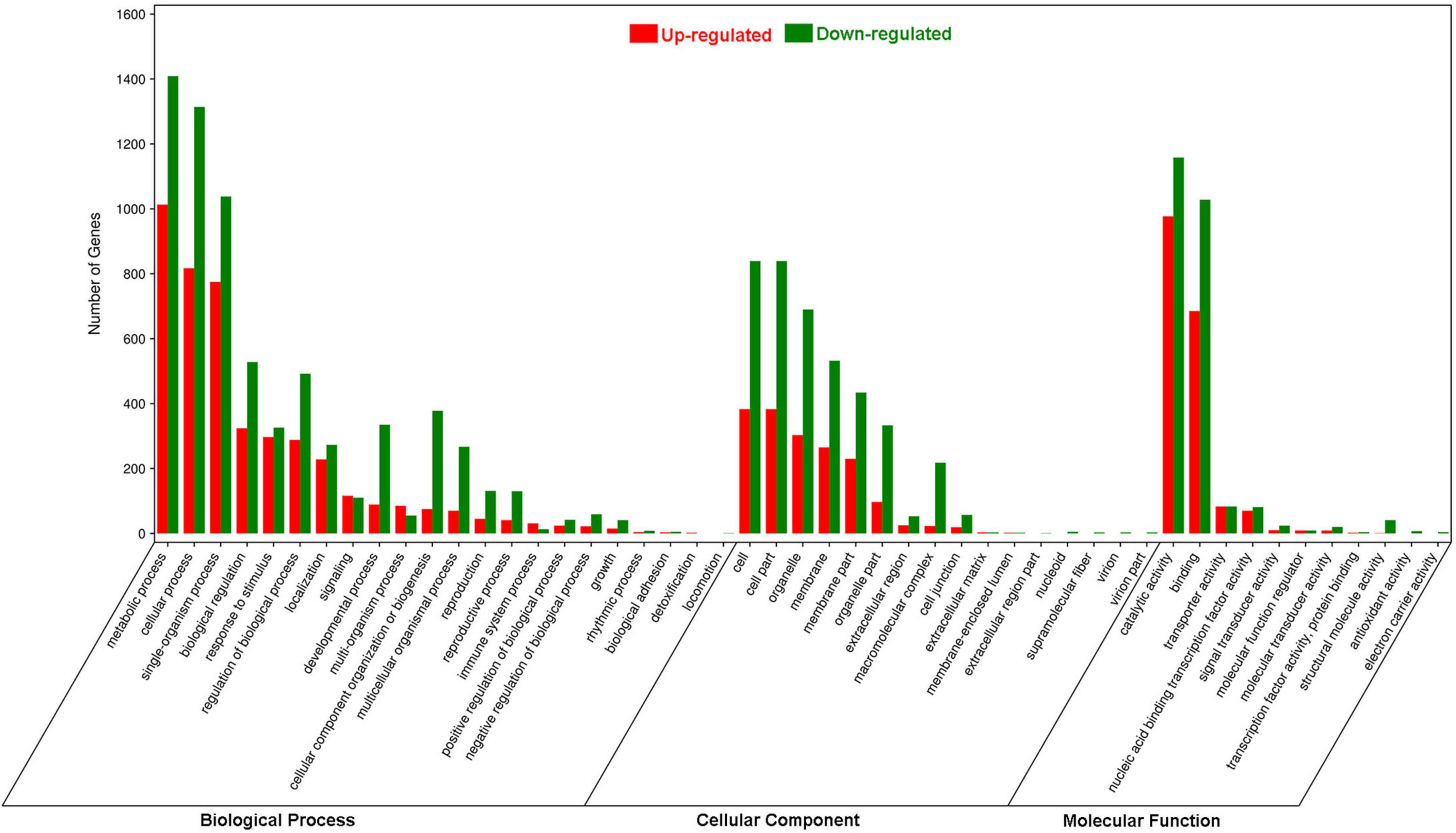
Figure 1. Gene Ontology enrichment analysis of differentially expressed genes (DEGs) in Gossypium barbadense leaves after Helicoverpa armigera infestation. The DEGs were judged as differentially expressed when log2| Ratio| ≥ 1 with FDR ≤ 0.05.
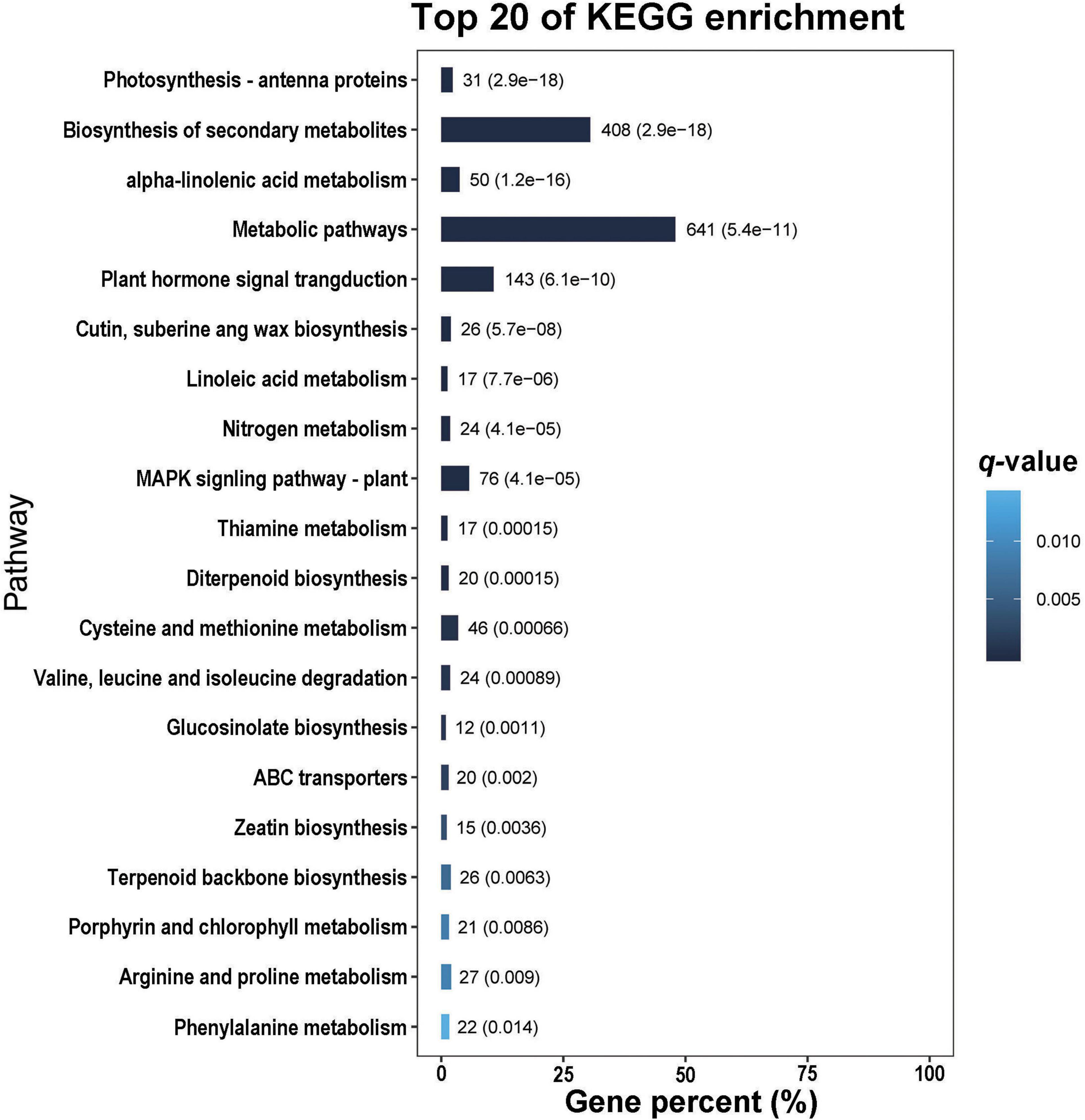
Figure 2. The top 20 pathways of differentially expressed genes (DEGs) identified in Gossypium barbadense leaves after Helicoverpa armigera infestation.
To confirm the reliability and repeatability of the RNA-seq data, seven DEGs involved in JA, salicylic acid (SA), ethylene pathway, photosynthesis and terpene volatiles biosynthesis were evaluated using qPCR measurement. It was found that these DEGs in qPCR results were highly consistent with the data obtained from RNA-seq (Figure 3), indicating the reliability of RNA-seq data.
In the present study, 51 genes involved in the JA biosynthesis pathway and 39 genes involved in the JA signaling pathway were differentially expressed in leaves of G. barbadense after H. armigera infestation, and most (88 of 90) of these DEGs were upregulated. In addition, most of the DEGs involved in ethylene (22 of 23), gibberellic acid (11 of 16), SA (10 of 15), and abscisic acid (35 of 54) were upregulated, whereas genes associated with brassinosteroids (19 of 25), cytokinin (16 of 30), and auxin (42 of 71) were mostly downregulated (Supplementary Table 3).
Of 344 genes for TFs that were differentially expressed, 200 were upregulated and 144 were downregulated after H. armigera infestation. Among these DEGs, most belonged to the gene family bHLH (33 up- and 43 downregulated), followed by ERF (56 up- and 12 downregulated genes), MYB (35 up- and 17 downregulated genes), and WRKY (24 up- and 6 downregulated) (Supplementary Table 4).
After H. armigera infestation, many genes involved in amino acid metabolism (113 up- and 48 downregulated), carbohydrate metabolism (123 up- and 106 downregulated), and nucleotide metabolism (28 up- and 26 downregulated) were differentially expressed. Notably, all 54 DEGs related to photosynthesis were downregulated (Supplementary Table 5).
Numerous genes contributing to plant defenses such as cytochrome P450 (CYP) (38 up- and 33 downregulated), glutathione S-transferases (GSTs) (5 up- and 6 downregulated), and UDP-dependent glycosyltransferases (UGTs) (20 up- and 6 downregulated) were differentially expressed (Supplementary Table 6). Additionally, several genes involved in volatile secondary metabolite pathways were also differentially expressed after the larvae infestation. Six of 8 DEGs involved in green leaf volatiles production and 34 of 49 DEGs for phenylpropanoid or benzenoid biosynthesis were upregulated (Supplementary Table 6). Most DEGs related to terpenoid volatile biosynthesis (39 of 41 DEGs) were upregulated. It is also important to note that all 13 TPSs, the genes for key enzymes involved in terpene biosynthesis, were upregulated (Supplementary Table 6).
The GbTPS1 (GB_D01G0996) in transcriptome was strongly expressed after H. armigera infestation. This gene caught our attention and was selected for further functional characterization. GbTPS1 has an ORF of 1653 bp, which encodes 551 amino acids with a predicted molecular mass of 64.18 kDa. Phylogenetic tree analysis showed that GbTPS1 was clustered within the TPS−a subfamily (Figure 4). The amino acid sequence of GbTPS1 has the typical DDxxD motif (NSE/DTE motif), the modified RR(x)8W motif in the N-terminal region, and the RxR motif located 35 amino acids upstream of the DDxxD motif (Huang et al., 2018).
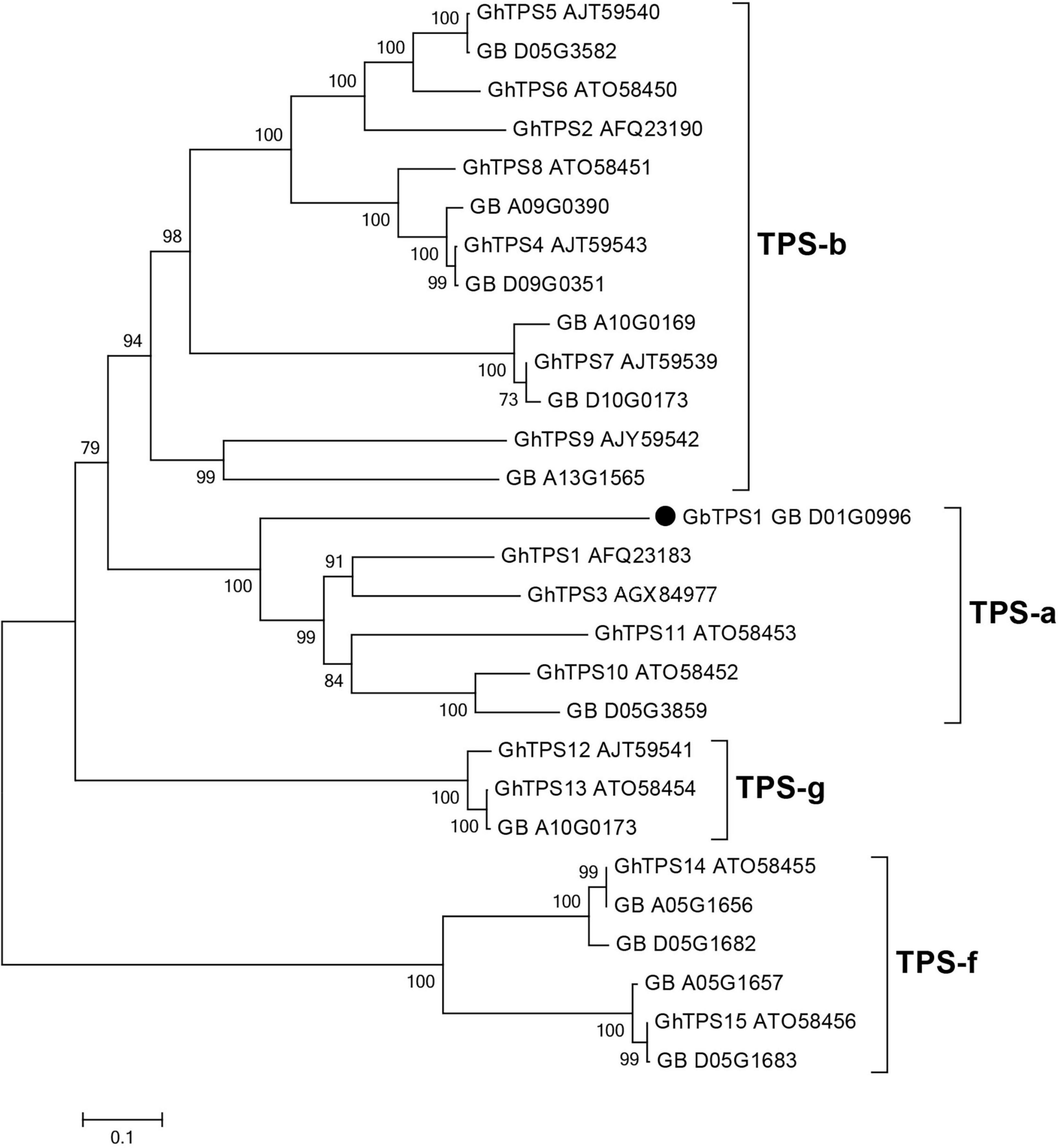
Figure 4. Phylogenetic tree of amino acid sequences of 13 TPSs identified in the transcriptome from G. barbadense leaves and previously characterized TPSs (GhTPS1-GhTPS15) from Gossypium hirsutum. The tree was constructed with the neighbor-joining method and 1,000 replications for bootstrapping.
The recombinant GbTPS1 were able to convert FPP into three sesquiterpenes selinene (48.20%), α-gurjunene (32.65%), and elemene (19.15%). When NPP was the substrate the catalytic product of recombinant GbTPS1 is a single limonene (Figure 5). No product was detected when GbTPS1 was incubated with GPP or GGPP as substrate. Meanwhile, cells expressing empty vector PET-30 did not produce target chemicals (Figure 5).
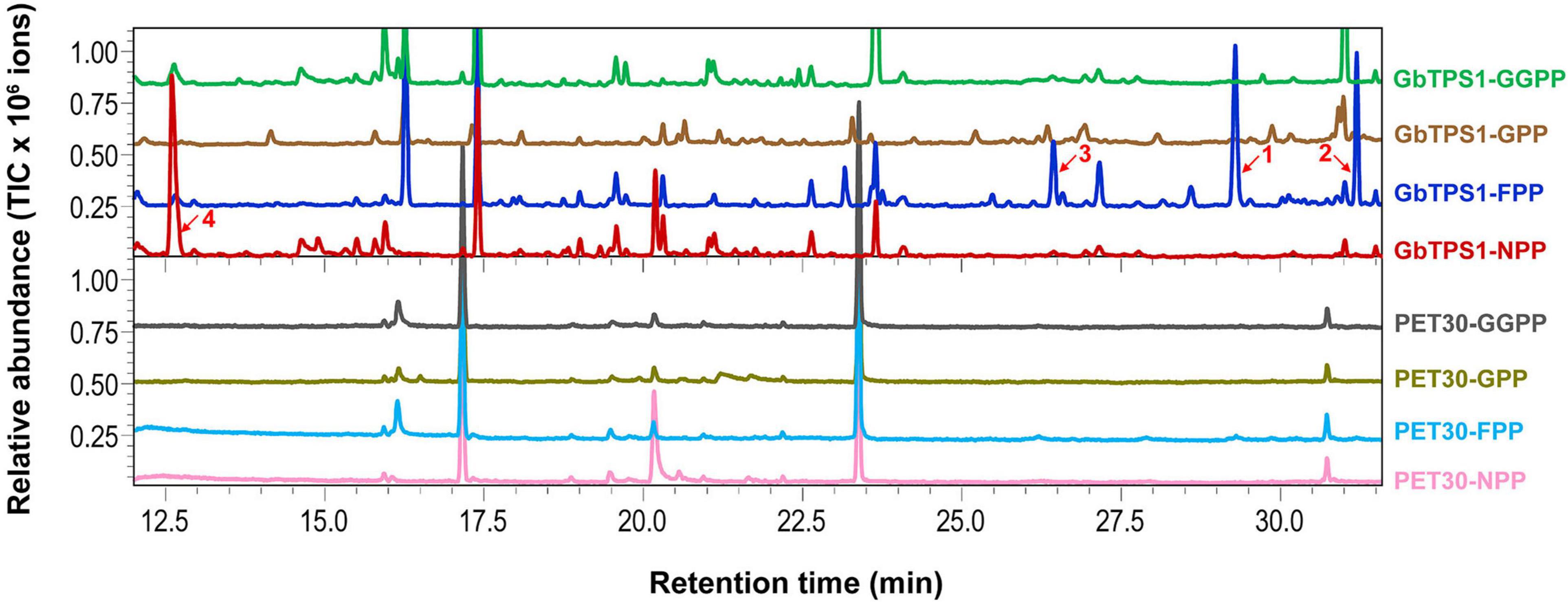
Figure 5. Gas chromatography-mass spectrometry analysis of the products catalyzed by recombinant GbTPS1. 1, selinene; 2, α-gurjunene; 3, β-elemene; 4, limonene. Other peaks are contamination.
Helicoverpa armigera-induced G. barbadense produced relatively high levels of volatile terpenes including six monoterpenes, 12 sesquiterpenes and two homoterpenes, whereas undamaged plants emitted only a trace amount of β−caryophyllene. Interestingly, in vitro products of GbTPS1–selinene, α-gurjunene, β-elemene, and limonene–significantly increased in the headspace volatiles from H. armigera-induced G. barbadense compared with those from undamaged plants (Figure 6; Table 1).
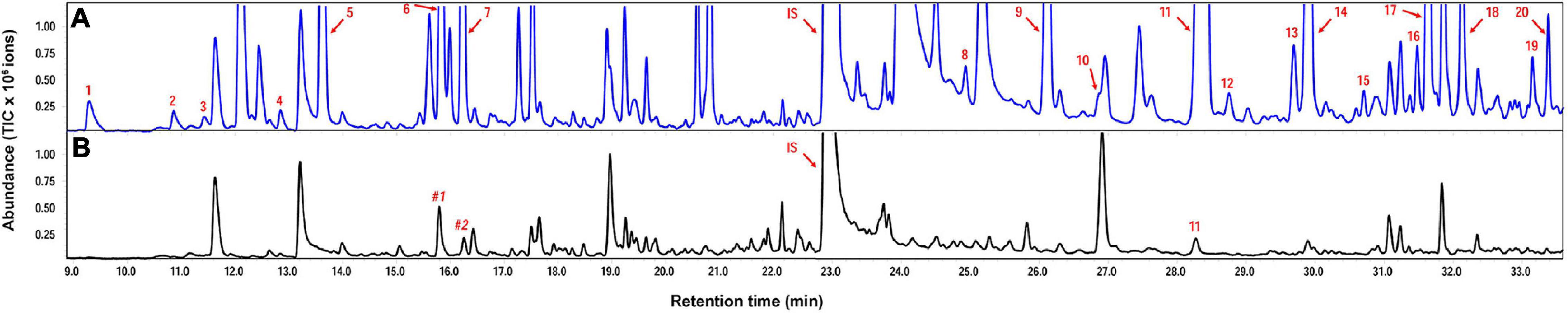
Figure 6. Representative chromatograms of volatile terpenes from Gossypium barbadense infested by Helicoverpa armigera (A) and from undamaged control plants (B). Numbered compounds correspond to those in Table 1. IS, internal standard (ethyl nonanoate); #1, Non-anal; #2, 1,2,4,5-tetramethylbenzene.
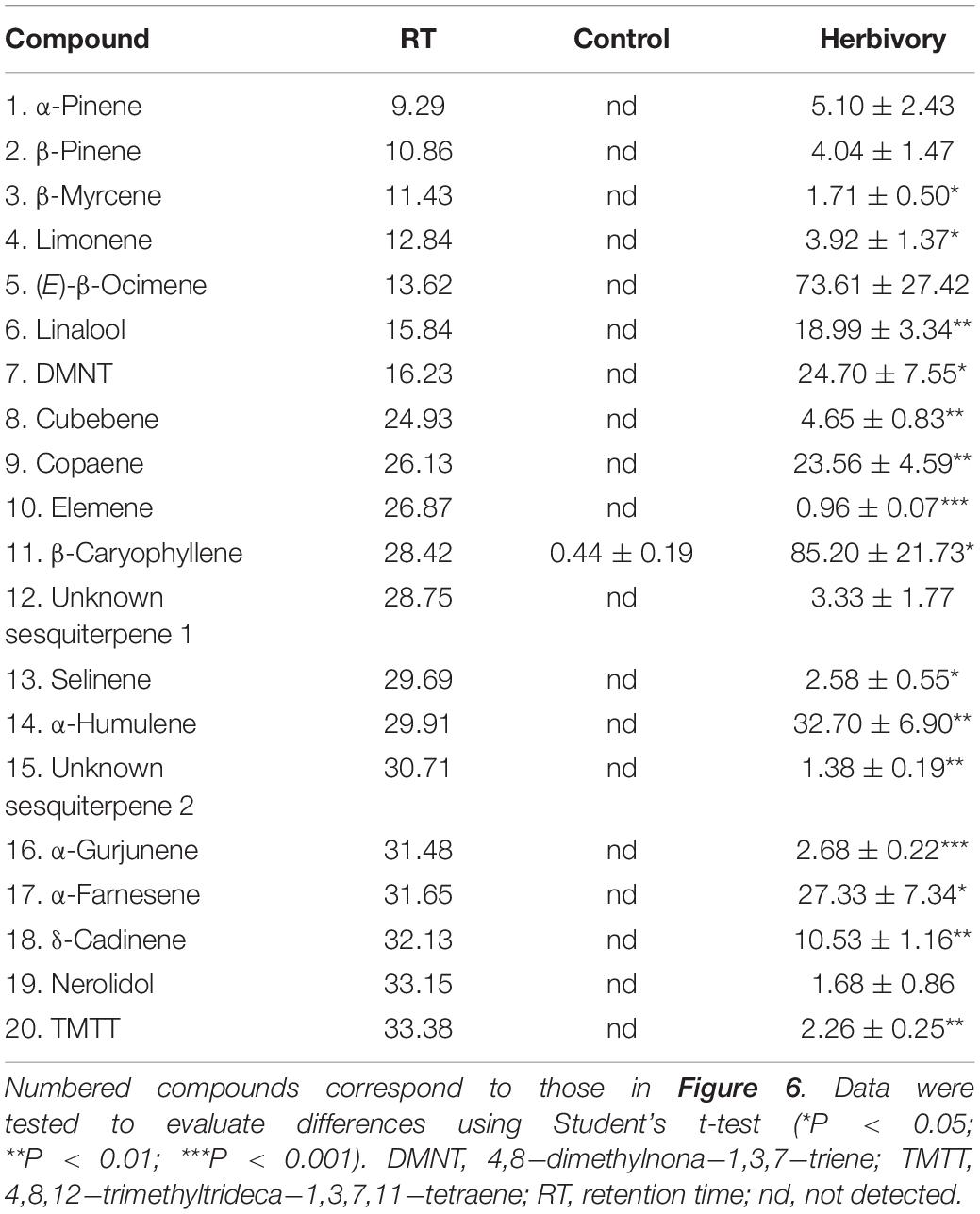
Table 1. Proportions (% of internal standard ethyl nonanoate) of volatile terpenes from Gossypium barbadense infested by Helicoverpa armigera and from undamaged control plants.
Helicoverpa armigera larvae showed significantly preference for control diet over those containing β-elemene or limonene at the three concentrations (Figure 7; elemene: 0.1 μg/μL, χ2 = 16.658, P < 0.001; 1 μg/μL, χ2 = 26.036, P < 0.001; 10 μg/μL, χ2 = 61.364, P < 0.001; limonene: 0.1 μg/μL: χ2 = 7.567, P = 0.006; 1 μg/μL: χ2 = 11.836, P = 0.001; 10 μg/μL: χ2 = 6.818, P = 0.009). In the no-choice bioassay, the H. armigera larvae feeding on β-elemene and limonene at the four tested concentrations (0.05, 0.1, 1, and 10 μg/μL) showed significantly higher mortalities compared with those feeding on control diet (Figure 8).
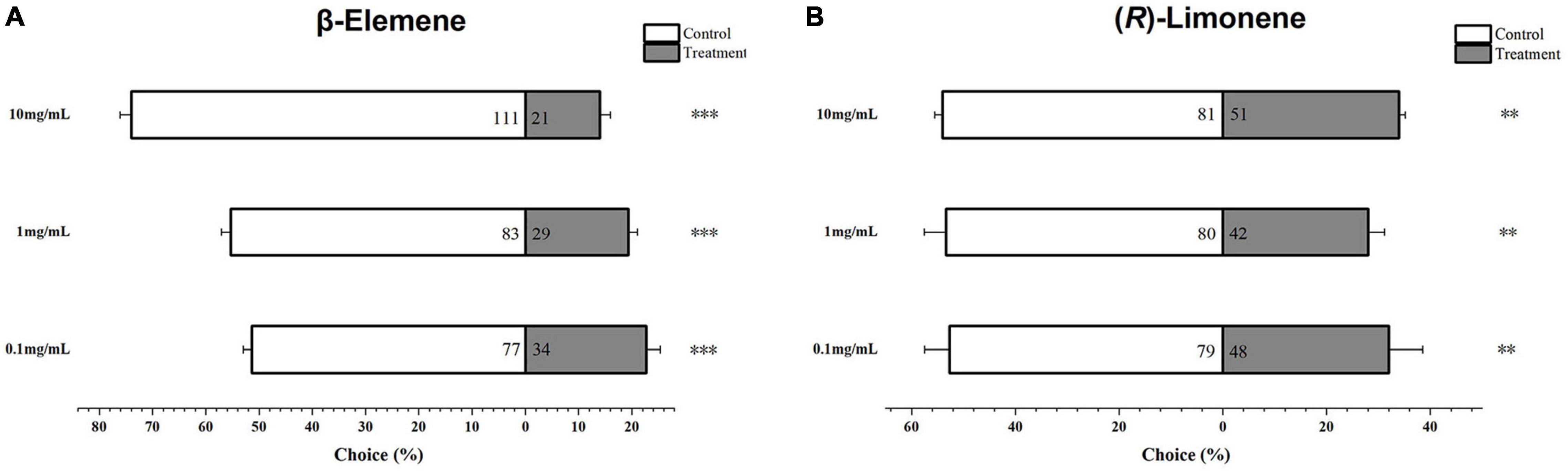
Figure 7. Behavioral preference of Helicoverpa armigera larvae to β-elemene (A) and limonene (B). Data were tested for significant differences using χ2 analysis (**P < 0.01; ***P < 0.001).
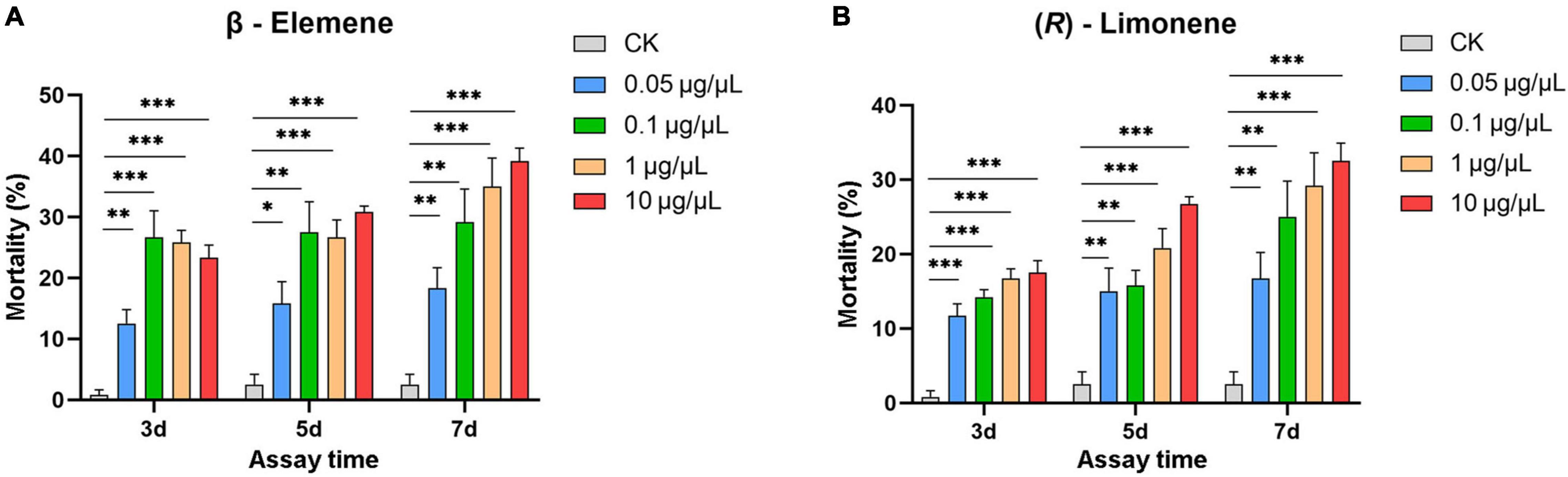
Figure 8. Larval mortality of Helicoverpa armigera caused by β-elemene (A) and limonene (B). Data were tested to evaluate differences using Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.001).
Gossypium barbadense has excellent traits such as high resistance to biotic stresses. However, little is known about transcriptional changes in G. barbadense infested by herbivores. It is well known that herbivores often trigger complicated transcriptional responses and cross-talk between signal-transduction pathways such as JA, SA, ethylene, and abscisic acid in plants. The complex signaling networks link early signaling networks and downstream transcriptional reprogramming in plant defense responses against insect herbivore attack. Generally, JA pathway play a central role with synergistic or antagonistic effects of other phytohormone pathways, especially stress-related SA, ethylene, and abscisic acid pathways (Erb and Reymond, 2019). Consistently, our investigation showed that H. armigera larvae primarily activated JA signaling pathway. After H. armigera infestation, a total of 90 DEGs including 51 involved in the JA biosynthesis and 39 involved in the JA signaling were differentially expressed, of which almost all (88 DEGs) were upregulated (Supplementary Table 3). Notably, three DEGs (GB_A06G0079, GB_D06G0081, and GB_A05G1944) annotated as JA carboxyl methyltransferase (JMT) were upregulated in H. armigera-infested cotton plants. JMT catalyzes the formation of the volatile ester methyl jasmonate from JA and has been reported to be involved in herbivore-induced defense responses (Qi et al., 2016). In addition to JA pathway, a number of DEGs associated with other phytohormone pathways including ethylene, gibberellic acid, SA, abscisic acid, brassinosteroids, cytokinin, and auxin were also differentially expressed (Supplementary Table 3). These findings suggested that H. armigera infestation activated a complex signaling network in G. barbadense. Similar, in G. hirsutum, our group found that H. armigera infestation induced the expression of genes associated with JA and ET, and a complex regulatory pattern for genes involved in phytohormone pathways such as auxin, cytokinin, and brassinosteroids (Huang et al., 2015). Thus, a crosstalk between these signaling pathways might contribute to induced cotton defenses against H. armigera, and thereby lead to the synthesis and accumulation of secondary defense compounds.
Transcription factors that regulate expression levels of the target genes are known to be very important in activating downstream plant defense responses against insect herbivore attack. In this study, 344 TFs consisting mainly of bHLH (76 genes), ERF (68 genes), MYB (52 genes), and WRKY (30 genes) were differentially expressed in response to H. armigera infestation (Supplementary Table 4), which might contribute to the activation of different pathways of cotton plants to initiate sophisticated defense responses against H. armigera. TFs are involved in the biosynthesis of insect-induced volatile terpenes such as (E)-β-caryophyllene in Arabidopsis thaliana (Hong et al., 2012), (E)-geraniol and valencene in Citrus sinensis (Shen et al., 2016; Li et al., 2017), linalool in Freesia hybrida (Yang et al., 2020), α-farnesene in apple (Wang et al., 2020), (E)-β-farnesene and (E)-α-bergamotene in Zea mays (Li et al., 2015). In cotton, GaWRKY1 is involved in the accumulation of δ-cadinene, the essential precursor in the gossypol biosynthetic pathway (Xu et al., 2004). H. armigera-induced TFs, in particular those with an expression pattern correlated with the upregulation of 13 TPSs in transcriptome, likely play roles in the transcriptional regulation of the H. armigera-induced TPS genes.
Many genes associated with proteins or enzymes are involved in biosynthetic pathways of natural antibiotics. For example, CYP (71 genes), GSTs (11 genes), and UGTs (26 genes) were differentially expressed (Supplementary Table 6), suggesting possible roles in cotton defense against H. armigera. It is noteworthy that many genes associated with the biosynthetic pathways of volatile terpenoids (39 genes) and HPL-derived green leaf volatiles (6 genes) and shikimic acid−derived volatiles (34 genes) were upregulated after H. armigera infestation, whereas all 54 DEGs related to photosynthesis were downregulated (Supplementary Table 5), as resources were redirected toward the production of defense metabolites, as found for G. hirsutum after H. armigera infestation (Huang et al., 2015). Thus, the suppression of photosynthesis might be a general plant response to herbivore attack and contribute to reallocation of limited resources to hamper herbivore performance (Zhou et al., 2015; Huang et al., 2022).
In the transcriptomic data, all 13 TPSs were upregulated (Supplementary Table 6). Consistently, H. armigera-infested G. barbadense produced a high diversity of volatile terpenes (Figure 6). The upregulation of GaTPS1 after H. armigera infestation was consistent with the elevated release of in vitro products of GaTPS1 including selinene, α-gurjunene, β-elemene, and limonene, which suggested that sesquiterpene synthase GbTPS1 is responsible for herbivore−induced terpene volatiles and contribute to anti-herbivore defenses in G. barbadense. The role of GbTPS1 in terpene formation in planta is additionally strengthened by the fact that the ratios of selinene, α-gurjunene, and β-elemene (∼2.7:2.8:1) in the headspace volatiles from H. armigera-induced G. barbadense (Figure 6) appear similar to the product distributions of GbTPS1 (∼1.8:2.5:1) when FPP was the substrate (Figure 5). Moreover, insect bioassay data showed elemene and limonene had repellent effects against H. armigera larvae (Figure 7) and increased mortalities of H. armigera (Figure 8). These findings suggested that GaTPS1 play a role in cotton plant direct defenses. Our findings not only enrich the understanding of terpenoid diversity in cotton plants, but also facilitate the discovery of new TPSs through transcriptome mining and homology-based searches.
In summary, the leaves of G. barbadense underwent extensive transcriptomic reprogramming that was regulated by complex phytohormonal signaling networks responding to H. armigera infestation. Almost all (88 out of 91) DEGs in the JA pathway were upregulated, highlighting that the H. armigera infestation mainly activated JA-mediated defense responses, thereby defense-related genes and pathways such as fatty acid-derived, shikimic acid−derived and terpenoid-related secondary metabolism were activated. The upregulation of GbTPS1 highlights its potential role in herbivore-induced cotton defense. Recombinant GbTPS1 catalyzed FPP and NPP as substrates to produce selinene as the major product and the single product limonene, respectively. What is more, β-elemene and limonene showed impacts on choice behavior and growth of H. armigera larvae. These findings provide valuable insights into understanding induced plants defense of G. barbadense against chewing caterpillars and help design novel pest management strategy.
The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number: PRJNA802699.
XQH and YZ conceived and designed the experiments. HZ, EL, XZH, JK, DT, and BL performed the experiments and analyzed the data. HZ, EL, XZH, XQH, and YZ refined the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31701800, 31972338, and 31772176), the National Key Research and Development Program of China (2019YFD0300100), and the Open Fund Project of State Key Laboratory for Biology of Plant Diseases and Insect Pests (SKLOF201901).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.898541/full#supplementary-material
Arce, C. M., Besomi, G., Glauser, G., and Turlings, T. C. J. (2021). Caterpillar-induced volatile emissions in cotton: the relative importance of damage and insect-derived factors. Front. Plant Sci. 12:709858. doi: 10.3389/fpls.2021.709858
Avellaneda, J., Díaz, M., Coy-Barrera, E., Rodríguez, D., and Osorio, C. (2021). Rose volatile compounds allow the design of new control strategies for the western flower thrips (Frankliniella occidentalis). J. Pest Sci. 94, 129–142. doi: 10.1007/s10340-019-01131-7
Ayubov, M. S., and Abdurakhmonov, I. Y. (2018). “The cotton-insect interactive transcriptome–molecular elements involved in plant-insect interactions,” in The Biology of Plant-Insect Interactions–A Compendium for the Plant Biotechnologist, ed. C. Emani (Boca Raton, FL: CRC Press), 62–73.
Booth, J. K., Yuen, M. M., Jancsik, S., Madilao, L. L., Page, J. E., and Bohlmann, J. (2020). Terpene synthases and terpene variation in cannabis sativa. Plant Physiol. 184, 130–147. doi: 10.1104/pp.20.00593
Borrero-Echeverry, F., Becher, P. G., Birgersson, G., Bengtsson, M., Witzgall, P., and Saveer, A. M. (2015). Flight attraction of Spodoptera littoralis (Lepidoptera, Noctuidae) to cotton headspace and synthetic volatile blends. Front. Ecol. Evol. 3:56. doi: 10.3389/fevo.2015.00056
Degenhardt, D., Greene, J., Khalilian, A., and Reeves, R. B. (2011). Volatile emissions from developing cotton bolls in response to hemipteran feeding damage. J. Entomol. Sci. 46, 177–190. doi: 10.18474/0749-8004-46.3.177
Erb, M., and Reymond, P. (2019). Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 70, 527–557. doi: 10.1146/annurev-arplant-050718-095910
Guo, M., Du, L., Chen, Q., Feng, Y., Zhang, J., Zhang, X., et al. (2021). Odorant receptors for detecting flowering plant cues are functionally conserved across moths and butterflies. Mol. Biol. Evol. 38, 1413–1427. doi: 10.1093/molbev/msaa300
Hatano, E., Saveer, A. M., Borrero-Echeverry, F., Strauch, M., Zakir, A., Bengtsson, M., et al. (2015). A herbivore-induced plant volatile interferes with host plant and mate location in moths through suppression of olfactory signalling pathways. BMC Biol. 13:75. doi: 10.1186/s12915-015-0188-3
Hong, G. J., Xue, X. Y., Mao, Y. B., Wang, L. J., and Chen, X. Y. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 24, 2635–2648. doi: 10.1105/tpc.112.098749
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Huang, X., Kou, J., Jing, W., Han, X., Liu, D., Ghasemzadeh, S., et al. (2022). Transcriptomic and metabolomic reprogramming in cotton after Apolygus lucorum feeding implicated in enhancing recruitment of the parasitoid Peristenus spretus. J. Pest. Sci. 95, 249–262. doi: 10.1007/s10340-021-01369-0
Huang, X. Z., Chen, J. Y., Xiao, H. J., Xiao, Y. T., Wu, J., Wu, J. X., et al. (2015). Dynamic transcriptome analysis and volatile profiling of Gossypium hirsutum in response to the cotton bollworm Helicoverpa armigera. Sci. Rep. 5:11867. doi: 10.1038/srep11867
Huang, X. Z., Xiao, Y. T., Köllner, T. G., Jing, W. X., Kou, J. F., Chen, J. Y., et al. (2018). The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 41, 261–274. doi: 10.1111/pce.13088
Li, S., Wang, H., Li, F., Chen, Z., Li, X., Zhu, L., et al. (2015). The maize transcription factor EREB58 mediates the jasmonate-induced production of sesquiterpene volatiles. Plant J. 84, 296–308. doi: 10.1111/tpj.12994
Li, X., Xu, Y., Shen, S., Yin, X., Klee, H., Zhang, B., et al. (2017). Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 68, 4929–4938. doi: 10.1093/jxb/erx316
Li, Y., Hallerman, E. M., Wu, K., and Peng, Y. (2020). Insect-resistant genetically engineered crops in China: development, application, and prospects for use. Annu. Rev. Entomol. 65, 273–292. doi: 10.1146/annurev-ento-011019-025039
Liang, G., Wu, K., Yu, H., Li, K., Feng, X., and Guo, Y. (2008). Changes of inheritance mode and fitness in Helicoverpa armigera (Hübner) (Lepidoptera: Noctuidae) along with its resistance evolution to Cry1Ac toxin. J. Invertebr. Pathol. 97, 142–149. doi: 10.1016/j.jip.2007.09.007
Liu, X., Zhao, B., Zheng, H.-J., Hu, Y., Lu, G., Yang, C.-Q., et al. (2015). Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5:14139. doi: 10.1038/srep14139
Llandres, A. L., Almohamad, R., Brévault, T., Renou, A., Téréta, I., Jean, J., et al. (2018). Plant training for induced defense against insect pests: a promising tool for integrated pest management in cotton. Pest Manag. Sci. 74, 2004–2012. doi: 10.1002/ps.5039
Lu, Y., Wyckhuys, K. A., Yang, L., Liu, B., Zeng, J., Jiang, Y., et al. (2022). Bt cotton area contraction drives regional pest resurgence, crop loss, and pesticide use. Plant Biotechnol. J. 20, 390–398. doi: 10.1111/pbi.13721
Qi, J., Li, J., Han, X., Li, R., Wu, J., Yu, H., et al. (2016). Jasmonic acid carboxyl methyltransferase regulates development and herbivory-induced defense response in rice. J. Integr. Plant Biol. 58, 564–576. doi: 10.1111/jipb.12436
Rodriguez-Saona, C., Crafts-Brandner, S. J., Williams, L., and Paré, P. W. (2002). Lygus hesperus feeding and salivary gland extracts induce volatile emissions in plants. J. Chem. Ecol. 28, 1733–1747. doi: 10.1023/A:1020552932566
Shen, S. L., Yin, X. R., Zhang, B., Xie, X. L., Jiang, Q., Grierson, D., et al. (2016). CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 67, 4105–4115. doi: 10.1093/jxb/erw189
Turlings, T. C. J., and Erb, M. (2018). Tritrophic interactions mediated by herbivore-induced plant volatiles: mechanisms, ecological relevance, and application potential. Annu. Rev. Entomol. 63, 433–452. doi: 10.1146/annurev-ento-020117-043507
Wang, M., Tu, L., Yuan, D., Zhu, D., Shen, C., Li, J., et al. (2019). Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat. Genet. 51, 224–229. doi: 10.1038/s41588-018-0282-x
Wang, Q., Liu, H., Zhang, M., Liu, S., Hao, Y., and Zhang, Y. (2020). MdMYC2 and MdERF3 positively co-regulate α-Farnesene biosynthesis in apple. Front. Plant Sci. 11:512844. doi: 10.3389/fpls.2020.512844
Wu, J., and Baldwin, I. T. (2010). New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 44, 1–24. doi: 10.1146/annurev-genet-102209-163500
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, Y., Wang, J., Wang, S., Wang, J., and Chen, X. (2004). Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 135, 507–515. doi: 10.1104/pp.104.038612
Yang, Z., Li, Y., Gao, F., Jin, W., Li, S., Kimani, S., et al. (2020). MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 71, 4140–4158. doi: 10.1093/jxb/eraa184
Zhang, L., Lu, G., Huang, X., Guo, H., Su, X., Han, L., et al. (2020). Overexpression of the caryophyllene synthase gene GhTPS1 in cotton negatively affects multiple pests while attracting parasitoids. Pest Manag. Sci. 76, 1722–1730. doi: 10.1002/ps.5695
Zhou, F., and Pichersky, E. (2020). The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 226, 1341–1360. doi: 10.1111/nph.16431
Keywords: extra-long staple cotton, transcriptome sequencing, sesquiterpene biosynthesis, GbTPS1, β-elemene, Helicoverpa armigera, induced defense
Citation: Zhang H, Liu E, Huang X, Kou J, Teng D, Lv B, Han X and Zhang Y (2022) Characterization of a Novel Insect-Induced Sesquiterpene Synthase GbTPS1 Based on the Transcriptome of Gossypium barbadense Feeding by Cotton Bollworm. Front. Plant Sci. 13:898541. doi: 10.3389/fpls.2022.898541
Received: 17 March 2022; Accepted: 23 June 2022;
Published: 13 July 2022.
Edited by:
Reuben J. Peters, Iowa State University, United StatesReviewed by:
Soheil Mahmoud, The University of British Columbia, CanadaCopyright © 2022 Zhang, Liu, Huang, Kou, Teng, Lv, Han and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqiang Han, aGFuc2h6QHNoenUuZWR1LmNu; Yongjun Zhang, eWp6aGFuZ0BpcHBjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.