
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 18 May 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.898286
This article is part of the Research Topic Women in Plant Symbiotic Interactions: 2022 View all 8 articles
Qa-SNARE gene SYP132 (isoform α) was previously reported to affect arbuscular mycorrhizal (AM) symbiosis in the legume species Medicago truncatula. In non-legumes especially monocots, it remains unknown whether certain SNARE genes are also involved in AM symbiosis. In this work, we studied a rice orthologous gene OsSYP132, which showed induced expression in mycorrhizal roots and two paralogous genes OsSYP131a and OsSYP131b, which were not induced by the AM fungus Rhizophagus irregularis. After employing CRISPR/Cas9 technique to generate their mutants, the Ossyp131a homozygous mutant T0 plants exhibited a dwarf phenotype and produced no fertile seeds, indicating a required role of this gene in seed fertility. Unlike the case in legume, the Ossyp132 mutants exhibited normal mycorrhizal phenotype, so did the Ossyp131b mutants. In the Ossyp131b Ossyp132 double mutants, however, the colonization rate and arbuscule abundance level decreased markedly, indicating an impaired fungal proliferation ability in rice roots. Such a defect was further confirmed by the reduced expression levels of AM marker genes. Our results in rice therefore demonstrated that while SYP13II members showed evolutionary and induction patterns specific to symbiosis, AM symbiosis is in fact controlled by the combined action of both SYP13I and SYP13II clades, revealing a functional redundancy among SYNTAXIN genes in mutualism.
Arbuscular mycorrhiza (AM) symbiosis is a mutualistic endosymbiosis formed between most terrestrial plants and Glomeromycotina fungi (Wang and Qiu, 2006; Smith and Read, 2008; Spatafora et al., 2016). During the symbiosis, the fungal hyphae penetrate root inner cortical cells and develop highly branched, bush-like structures called arbuscules (Genre et al., 2008). Arbuscules are surrounded by the peri-arbuscular membrane (PAM), which is continuous to the cell plasma membrane. Through the interface of PAM and arbuscules, nutrient exchanges occur between symbiotic partners, where plants provide carbohydrates, mainly in the form of fatty acids, to fungi and fungi supply mineral nutrients, especially phosphorus and nitrogen, to plants (Smith and Smith, 2011; Bravo et al., 2017; Jiang et al., 2017; Keymer et al., 2017; Luginbuehl et al., 2017; Wang et al., 2017, 2020; Wipf et al., 2019).
In eukaryotic cells, exocytosis via membrane trafficking is a fundamental process involving the fusion of secretory vesicles with target membranes (Kanazawa and Ueda, 2017). It is mediated by a group of SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins that are divided into two classes, Q (glutamine)-SNAREs and R (arginine)-SNAREs (Fasshauer et al., 1998). When secretory vesicles move close to the target membrane, one R-SNARE located on the vesicle membrane (also called v-SNARE) and three types of Q-SNAREs (Qa, Qb, and Qc) on the target membrane (also called t-SNAREs) can form a stable SNARE complex to bridge and drive membrane fusion (Bock et al., 2001; Jahn and Scheller, 2006; Lipka et al., 2007).
During AM symbiosis, the formation of PAM requires the delivery of an enormous amount of membrane material to the proximity of branched fungal hyphae (Dickson and Kolesik, 1999; Harrison and Ivanov, 2017). Several specific SNAREs have been reported to be involved in this process. Two VAMP72 (vesicle-associated membrane protein 72) subfamily R-SNAREs, VAMP721d and VAMP721e in Medicago truncatula, were first revealed to be involved in the formation of the plant-fungus interface (Ivanov et al., 2012). Simultaneously silencing MtVAMP721d and MtVAMP721e by RNA interference (RNAi) led to a marked decrease of mature arbuscules and the accumulation of many abnormal arbuscules lacking fine-branches (Ivanov et al., 2012). Also, Lotus japonicus VTI12, encoding a Qb-SNARE, showed enhanced expression in mycorrhizal roots, and down-regulation of LjVTI12 resulted in small and distorted arbuscules (Lota et al., 2013). Moreover, a SYNTAXIN OF PLANTS 13 subfamily clade II (SYP13II) member, SYP132 encoding a Qa-SNARE in M. truncatula, is able to produce two isoforms, SYP132α and SYP132β via alternative cleavage and polyadenylation (APA). Relative to SYP132β, SYP132α exhibited higher expression in the arbuscule-containing cells. Silencing SYP132α caused many underdeveloped arbuscules, while silencing SYP132β seemed to inhibit root growth, thus indicating functional divergences of the two isoforms (Huisman et al., 2016; Pan et al., 2016).
Phylogenetic analyses showed that both MtVAMP721d/e and MtSYP132 belong to the AM host-specific ‘symbiotic’ clades that lack representative genes from many non-mycorrhizal species, such as Arabidopsis thaliana, Striga hermonthica and Beta vulgaris (Ivanov et al., 2012; Huisman et al., 2016; Pan et al., 2016). Since MtVAMP721d/e and MtSYP132α RNAi roots showed similar phenotypes of abnormal arbuscules, it was hypothesized that MtSYP132α can interact with MtVAMP721d/e to define a symbiosis-specific membrane trafficking pathway, which is used to deliver specific cargo to the plant-fungus interface (Huisman et al., 2016; Pan et al., 2016). However, investigating more Qa-SNARE genes of SYP11/12/13 subfamilies and R-SNARE genes of VAMP72 subfamily in M. truncatula revealed a rather unspecific pattern. When driven by the promoter of Medicago PT4 (Phosphate transporter 4), subcellular localization to the PAM were observed not only for symbiotic SNAREs, but also for non-symbiotic SNAREs such as SYP121, SYP131, SYP132β, VAMP721a and VAMP724 (Huisman et al., 2020). Also, non-symbiotic SNAREs can restore the arbuscule phenotypes in MtSYP132α or MtVAMP712d/e RNAi roots, if they were under the control of MtSYP132 or MtVAMP721e promoters (Huisman et al., 2020). It remains to be explored whether non-symbiotic SNAREs are involved in symbiosis if their own promoters were maintained.
To gain more understanding on the functional divergences of different SNAREs, in this study, we investigated three SYP13 genes in rice, including two genes, OsSYP131a and OsSYP131b in the non-symbiotic SYP13I clade and one gene OsSYP132 in the symbiotic SYP13II clade. By generating mutants for each of these genes via CRISPR/Cas9 technique and obtaining the OsSYP131b OsSYP132 double mutants, the potential roles of these Qa-SNARE-encoding genes in AM symbiosis and also in rice seed fertility were investigated. We discovered that OsSYP131b and OsSYP132 single mutants displayed apparently normal mycorrhization patterns, but AM symbiosis is markedly disrupted by removing OsSYP131b and OsSYP132 simultaneously. Our results suggest that both the constitutively expressed SYP13I clade and the symbiotically induced SYP13II clade of SYNTAXINs are involved in AM symbiosis.
The protein sequences of AtSYP131 (At3G03800), AtSYP132 (At5G08080) and MtSYP132 (Medtr2g088700) were used as query sequences to run BLASTP searches against the proteomes of 29 representative seed plants, including 4 gymnosperms and 25 angiosperms (for species list and genome information, see Supplementary Table 1). For each surveyed species, the obtained sequences showing an E-value < 10–4 and > 50% length coverage were kept and further examined for their resemblance with AtSYP131 rather than AtSYP111 (At1G08560) and AtSYP121 (At3G11820). A total of 58 sequences closely-related to the SYP13 subfamily were obtained from the surveyed species and aligned together with AtSYP111 and AtSYP121 (as outgroup) by ClustalW and Muscle programs, as implemented in MEGA v7.0 (Kumar et al., 2016). Among them, the rice gene OsSYP132 (LOC4343440 in NCBI database)1 was not annotated in the surveyed O. sativa ssp. japonica cv. Nipponbare genome (v7.0), but an identical gene (OsKitaake07g168700) was annotated in the O. sativa ssp. japonica cv. Kitaake genome at Phytozome v132. After manual examination for site homology, the obtained alignment was used to reconstruct a maximum-likelihood tree of SYP13 subfamily under the GTR + F + I + G4 model via IQ-TREE, and the robustness of internal branches were examined by conducting the Shimodaira-Hasegawa approximate likelihood ratio test (SH-aLRT) and by performing 1,000 ultrafast bootstrap replicates (Guindon et al., 2010; Trifinopoulos et al., 2016).
The rice (O. sativa ssp. japonica) wild-type and transgenic plants used in this study were in the cv. Nipponbare background. Rice seeds were surface-sterilized with 2.5% sodium hypochlorite and 0.1% Tween20 for 15 min, 70% alcohol for 2 min, rinsed 3 times with sterile water, and germinated on a 1/2 modified Murashige-Skoog (MS) medium in a growth chamber with 12/12 h day/night cycle at 28°C/22°C and 70% relative humidity for two weeks. For AM colonization, seedlings were transplanted in pots (height 9 cm, diameter 12 cm) containing an autoclaved sand/perlite (3:1 [v/v]) mixture with three seedlings per pot. Rhizophagus irregularis fungal spores were freshly extracted from transgenic hairy carrot root cultures and were resuspended in water. Approximately 120 spores were added to the rhizosphere of each seedling. Plants were grown under a 16/8 h day/night cycle at 28°C/22°C and were fertilized twice a week with 1/2 Hoagland solution containing 20 μM phosphates. Plant roots were harvested at 3-week post inoculation (wpi) and 6-wpi, where half of the samples were used for staining and mycorrhizal phenotype examination and the other half were frozen in liquid nitrogen for RNA extraction.
For hydroponic culture, surface-sterilized rice seeds were germinated in sterile water for 2 days at 37°C, then transferred into hydroponic boxes (length 12.6 cm, width 8.5 cm, height 11 cm, 10 seedlings/box). The plants were grown in sterile water for one week, then in 1/4 Hoagland solution for another two weeks. To compare the root morphology between wild-type rice and the mutants, the aboveground height, primary root length (PRs), crown root (CRs) and lateral root numbers (LRs) were recorded each week.
The total RNA of rice roots was extracted by the Trizol method (Invitrogen, Carlsbad, CA, United States). The RNA purity and integrity were examined by RNA electrophoresis in 2.5% (wt/vol) agarose gel. RNA (1 μg) was treated with DNase I prior to cDNA synthesis using a Revert-Aid First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative RT-PCR (qRT-PCR) was performed by measuring the intensity of SYBR Green Fluorescent dye conjugated to double stranded DNA molecules using a C1000 Thermal Cycler Real-Time PCR detection system (Bio-Rad). Gene expression values were normalized to the rice housekeeping gene, ubiquitin 1 (OsUBI1) (Chen et al., 2008), and analyzed by the 2–Δ CT method (Livak and Schmittgen, 2001). Statistical differences were tested via the Student’s t-test (p < 0.05). All the qRT-PCR primers used in this study are provided in Supplementary Table 2.
To generate single mutants for three OsSYP13 genes, the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 technique was employed. Single-guide RNAs (sgRNAs) targeting OsSYP131a, OsSYP131b and OsSYP132, respectively, were designed using the CRISPR-PLANT3 and CRISPRdirect4 online tools (Xie et al., 2014; Naito et al., 2015). For each sgRNA, a pair of complementary primers with sticky ends were synthesized, annealed, and ligated with a linear CRISPR vector, pRGEB31, which was pre-digested by the restriction enzyme, BsaI. After transforming rice calli via agrobacteria containing the pRGEB31-sgRNA vectors (Liu et al., 2022), the editing alleles of OsSYP13 were examined through PCR and sequencing with specific primers, OsSYP131a edit-F/R, OsSYP131b edit-F/R and OsSYP132 edit-F/R, respectively, which all covered the editing regions (Supplementary Figure 2 and Supplementary Table 2). In subsequent generations, plants carrying desired homozygous mutations without the pRGEB31 construct, which can be detected by pRGEB31-Cas9-F/R, were selected for reproduction (Supplementary Table 2). Finally, the T2 generations of the mutant lines were used for further studies.
Ossyp131b Ossyp132 double mutant lines were obtained by hybridization between homozygous Ossyp132-2 and Ossyp131b-1 or Ossyp131b-2 single mutants. The doubly heterozygous F1 progeny were grown to maturity and allowed to self-pollinate, and seeds containing the segregating F2 embryos were obtained. Because the OsSYP131b and OsSYP132 genes are located on separate chromosomes and are hence genetically unlinked, in such a cross, we would expect to find a 15: 1 ratio of F2 progeny with other genotypes to that of the Ossyp131b Ossyp132 doubly homozygous mutant. The doubly homozygous F2 progeny were selected for reproduction and the obtained F3 generation mutants were used for further studies.
For wheat germ agglutinin (WGA) staining (Schaffer and Peterson, 1993), fresh rice roots were fixed in 50% ethanol for > 4 h and cleared in 20% KOH (w/v) for 2–3 d at room temperature. Next, the roots were acidified in 0.1 M HCl for 20 min and rinsed 5 times with phosphate-buffered saline (PBS) solution (pH 7.4). After incubating overnight in PBS solution containing 1 μg/mL WGA-Alexa Fluor 488 (Invitrogen) in the dark and washing with PBS, the stained roots were examined under a confocal Olympus FV10-ASM microscope (Olympus, Tokyo, Japan) and representative images were captured. For ink-vinegar staining (Vierheilig et al., 1998), rice roots were cleared with 20% KOH for 40 to 120 min at 65°C, acidified by 5% acetic acid for 5 min at room temperature, incubated in ink-vinegar solution for 30 min at 65°C, and decolored in tap water for 14 h. The stained roots were examined by a Nikon E100 light microscope (Nikon, Tokyo, Japan). To quantify the fungal colonization rate, a modified gridline intersect method (McGonigle et al., 1990) was used to observe and record fungal structures, including hyphopodia, intraradical hyphae, arbuscules, and vesicles. For arbuscule size measurement, after ink-vinegar staining, the images of colonized root fragments were taken by JieDa microscopic imaging software and the longitudinal lengths of arbuscules were measured by using a ruler implemented in the software. For either wild-type rice or the mutants, at least 15 root fragments from three individuals were measured at 3- and 6-wpi. The sizes of arbuscules were determined by measuring the length of all visible arbuscules in independent infection units (from hyphopodia to the running edge of hyphae toward the root tip) and classified into eight clusters from 0 to 10 μm to > 70 μm. The numbers in different clusters were divided by the number of all measured arbuscules from each genotype to get the percentage of arbuscule size classes.
After identifying a total of 58 SYP13 subfamily members from 29 surveyed seed plants (Supplementary Table 1), a maximum-likelihood (ML) tree was reconstructed (see Materials and Methods). One SYP13 gene was found in each of the four surveyed gymnosperms and these sequences formed a monophyletic group sister to all the angiosperm SYP13 sequences, suggesting that there was one ancestral SYP13 gene in the common ancestor of seed plants (Figure 1). In angiosperms, however, the ancestral gene had further duplicated into two lineages, namely SYP13I and SYP13II clades (Figure 1).
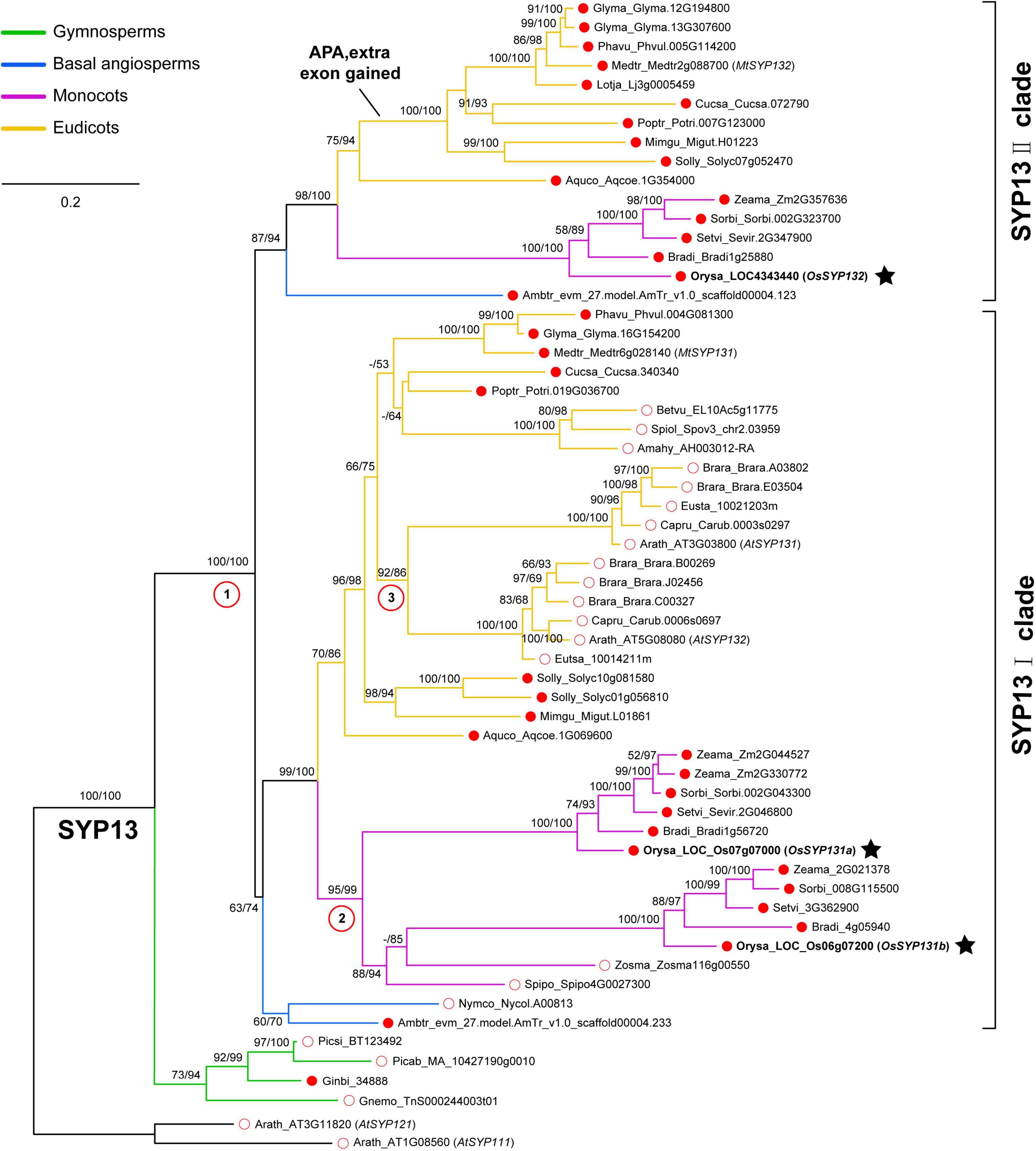
Figure 1. Phylogenetic analyses of SYNTAXIN OF PLANTS 13 (SYP13) subfamily including 29 seed plants. The phylogeny was reconstructed using a maximum-likelihood method under the GTR + F + I + G4 model via IQ-TREE. Branch support values were examined by conducting the Shimodaira-Hasegawa approximate likelihood ratio test (SH-aLRT) and by performing 1,000 ultrafast bootstrap replicates. Two Arabidopsis thaliana SYP1 genes (AtSYP111 and AtSYP121) were used as the outgroup. There were three duplication events in angiosperms marked by red circles with numbers. The ancestral SYP13 gene was first duplicated into two clades, SYP13I and SYP13II; then two independent duplications occurred in the common ancestor of Brassicaceae or that of monocots, respectively. Three rice SYP13 genes identified in this work were indicated by black stars. Red dot and red circle indicate AMS host plants and AMS non-host plants, respectively. The sequences from gymnosperms, basal angiosperm, monocots and eudicots were showed in different colors.
Within the SYP13I clade, representative genes were found in both mycorrhizal and non-mycorrhizal species. The two basal angiosperm species Amborella trichopoda and Nymphaea colorata and many dicot species have all maintained one member in this clade (Figure 1). However, two independent duplication events likely occurred in the common ancestor of Brassicaceae species and in the common ancestor of monocots, respectively. As a result, A. thaliana possesses two SYP13I genes, which were previously named as AtSYP131 and AtSYP132, and their encoded proteins share ∼71% identities. Similarly, two SYP13I genes were present in the rice (Oryza sativa) genome. In this study, we referred to gene Os07g07000 as OsSYP131a and gene Os06g07200 as OsSYP131b (Figure 1). Their protein sequences share ∼61% identities.
Within the SYP13II clade, representative genes were not detected in the 10 surveyed non-mycorrhizal angiosperms, including the aquatic basal angiosperm N. colorata, two aquatic monocot species Spirodela polyrhiza and Zostera marina, three Amaranthaceae species and four Brassicaceae species (Supplementary Table 1). The remaining 15 mycorrhizal angiosperms all possess one SYP13II gene, except soybean (Glycine max), with two copies. An extra exon 14 of SYP13II could be found in all the mycorrhizal dicot species except A. coerulea, suggesting that these dicot species can produce two isoforms of SYP13II gene via APA as in M. truncatula. Differently, the basal angiosperm A. trichopoda, the five mycorrhizal monocot species and the basal eudicot species A. coerulea all possess a SYP13II gene without the extra exon. In rice, one gene OsSYP132 (LOC4343440) belongs to this clade and it encodes a protein sharing ∼58% sequence identities to OsSYP131a and ∼55% identities to OsSYP131b.
A previous research provided a detailed transcriptome data for various rice organs (roots, leaves, flowers and seeds) at different developmental stages (Wang et al., 2015). According to this study, OsSYP132 was barely expressed in leaves, flowers, seeds and was expressed in roots at a low level before flowering, while OsSYP131a was expressed in most tissues including flowers, and OsSYP131b was expressed at a rather high level in roots and leaves (Supplementary Figure 1).
To explore the expression patterns of OsSYP13 genes during AM symbiosis, the root samples of rice individuals inoculated by the AM fungus (AMF) Rhizophagus irregularis or by mock-solution were respectively collected at two different times points, 3- and 6-weeks post-inoculation (wpi). Quantitative RT-PCR (qRT-PCR) experiments were further conducted. At 3-wpi, the relative expression level of OsSYP132 in mycorrhizal roots was only slightly higher than in control roots (Figure 2A, p > 0.05), while at 6-wpi, it became ∼25-fold higher than control (Figure 2A, p < 0.02). On the contrary, for OsSYP131a or OsSYP131b, similar relative expression levels were observed between mycorrhizal and non-mycorrhizal roots at both 3- and 6-wpi (Figures 2B,C).
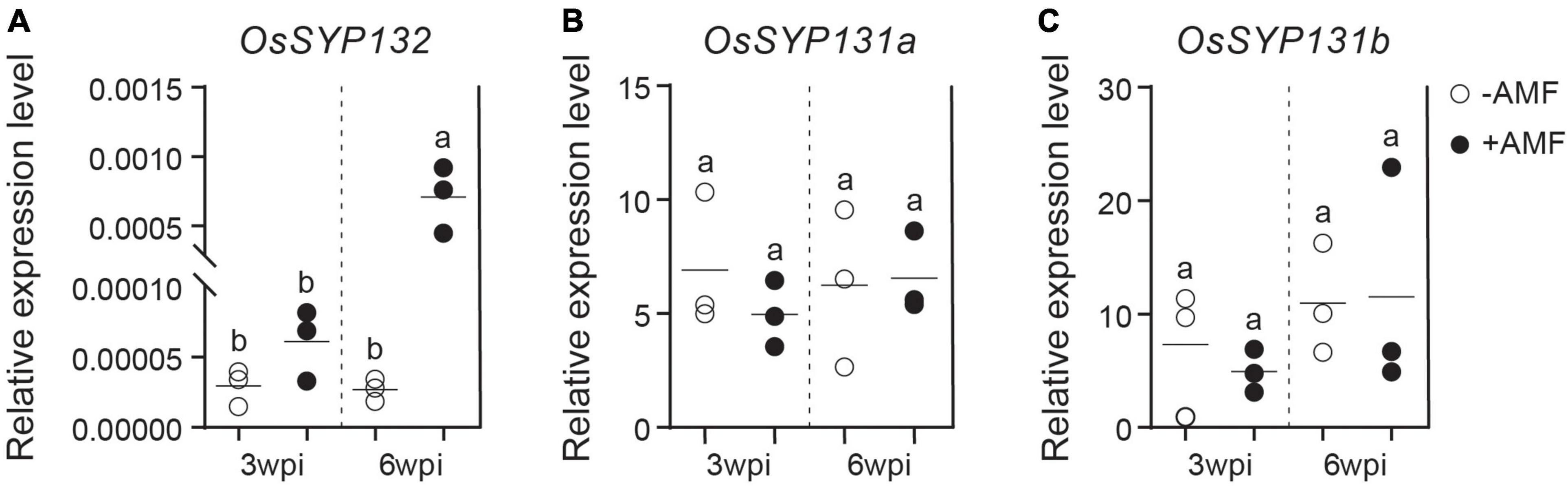
Figure 2. The expression patterns of three OsSYP13 genes in rice roots during arbuscular mycorrhizal (AM) symbiosis (A–C). The relative expression levels of OsSYP132, OsSYP131a and OsSYP131b were measured by Quantitative RT-PCR (qRT-PCR). Rice wild-type (WT) roots were either mock inoculated (–AMF) or colonized by Rhizophagus irregularis (+ AMF) were harvested at 3- and 6-weeks post-inoculation (wpi). The gene expression values were normalized to the rice housekeeping gene, ubiquitin 1 (OsUBI1). Data are individual values from three biological replicates and the horizontal line shows the mean value. Different letters indicate significant statistical differences between groups evaluated by the Tukey’s honest significance test (between the pairs, P < 0.05 or P > 0.05). This is a representative experiment that was repeated three times.
To explore the potential roles of these three OsSYP13 genes in AM symbiosis and/or in plant growth and development, we tried to obtain their null mutants using CRISPR/Cas9 technique (see Materials and Methods). For each gene, four to six mutated alleles were detected in the T0 generation (Supplementary Figure 2). Two alleles per gene were then chosen to obtain their homozygotes in subsequent generations, meanwhile eliminating the pRGEB31 vector. By introducing a 1-nucleotide deletion in exon 6, Ossyp132-1 allele showed a premature stop codon, thus encoding a truncated protein only 123 aa in length (Supplementary Figure 2A). Similarly, the Ossyp132-2 allele had a 1-nucleotide deletion in exon 8 and also produced a premature stop codon, encoding a truncated protein of 169 aa (Supplementary Figure 2A). Due to different 1-nucleotide insertions in exon 8, both Ossyp131a-1 and Ossyp131a-2 alleles had altered reading frame, consequently encoding truncated proteins only 155 aa in length (Supplementary Figure 2B). As for the OsSYP131b gene, Ossyp131b-1 had a 1-nucleotide deletion and Ossyp131b-2 showed a 1-nucleotide insertion in exon 6. Premature stop codons were produced, consequently encoding truncated proteins of 136 aa and 121 aa, respectively (Supplementary Figure 2C). Since all these mutated alleles resulted in truncated proteins that lack both SNARE domain and the transmembrane domain, it can be expected that these mutated alleles would lose the functions of OsSYP13 genes.
To preclude off-target effects in CRISPR/Cas9-mediated genome editing, we further examined the relative expression levels of three OsSYP13 genes in the Ossyp131b or Ossyp132 mutants by qRT-PCR at 6 wpi by R. irregularis. The results showed that although the mutated gene was expressed at markedly reduced levels, the relative expression levels of the other two homologous genes were not affected (Supplementary Figure 3).
In the T0 generation, compared to the control plants transformed with an empty vector (EV), the Ossyp131a-1 homozygous mutant exhibited a dwarf phenotype (Figure 3A). Moreover, unlike the healthy grains seen on control plants, only shriveled infertile grains were produced on the Ossyp131a-1 T0 homozygous mutant, indicating a required role of OsSYP13a in rice seed fertility (Figures 3B,C). A heterozygous T0 plant (WT/Ossyp131a-2), instead, showed normal vegetative growth phenotype, and produced both healthy and infertile grains (Figures 3D-G). Germinating the produced healthy seeds and further examining their genotypes via sequencing then revealed a 1:2 ratio of WT to WT/Ossyp131a-2 heterozygotes at the T1 generation (n = 56, 19: 37, χ2 = 0.009, P > 0.05), indicating that the Ossyp131a-2 homozygous grains were also infertile.

Figure 3. The Ossyp131a T0 mutants showed defects in growth and seed fertility (A–C). The growth phenotype (A) and mature panicles (B) of a control T0 rice plant (with an empty vector, EV) and a Ossyp131a-1 T0 homozygous mutant; (C) An amplified circle region in (B); (D,E), The growth phenotype (D) and mature panicles (E) of a control T0 rice plant (EV) and a Ossyp131a-2 T0 heterozygous mutant; (F–G) The rice grain morphology of control T0 rice plant (EV), the Ossyp131a-1 T0 homozygous mutant and the Ossyp131a-2 T0 heterozygous mutant, either with husks (F) or without husks (G). Scale bars = 10 cm (A,D), 1 cm (B,E–G) or 0.5 cm (C).
After obtaining the homozygous Ossyp131b and Ossyp132 mutants in the T2 generation, we first performed hydroponic cultivation experiments to examine their growth phenotypes under asymbiotic condition (see Materials and Methods). After 3-weeks cultivation, the Ossyp131b and Ossyp132 mutants all showed similar values to wild-type rice in terms of above-ground height, maximum length of primary root, number of crown roots and number of lateral roots (Supplementary Figure 4), indicating a normal growth phenotype of these mutants under our experimental condition.
We then examined the mycorrhizal phenotypes of the Ossyp131b and Ossyp132 mutants. At 3-wpi by R. irregularis, an average of ∼18% wild-type rice lateral roots was colonized, including ∼13% root length forming arbuscules (Figure 4A). Rather surprisingly, comparable total colonization levels (10-19%) and arbuscule abundance (7-12%) were observed in the examined Ossyp131b and Ossyp132 mutants, with arbuscular morphology unaffected (Figure 4A and Supplementary Figure 5). At 6-wpi, the total colonization levels still showed no statistically significant differences between wild-type and the mutants (∼72% vs. 67-68%). Neither did the arbuscule abundance levels, which was ∼57% in WT and 53-58% in the mutants (Figure 4A).
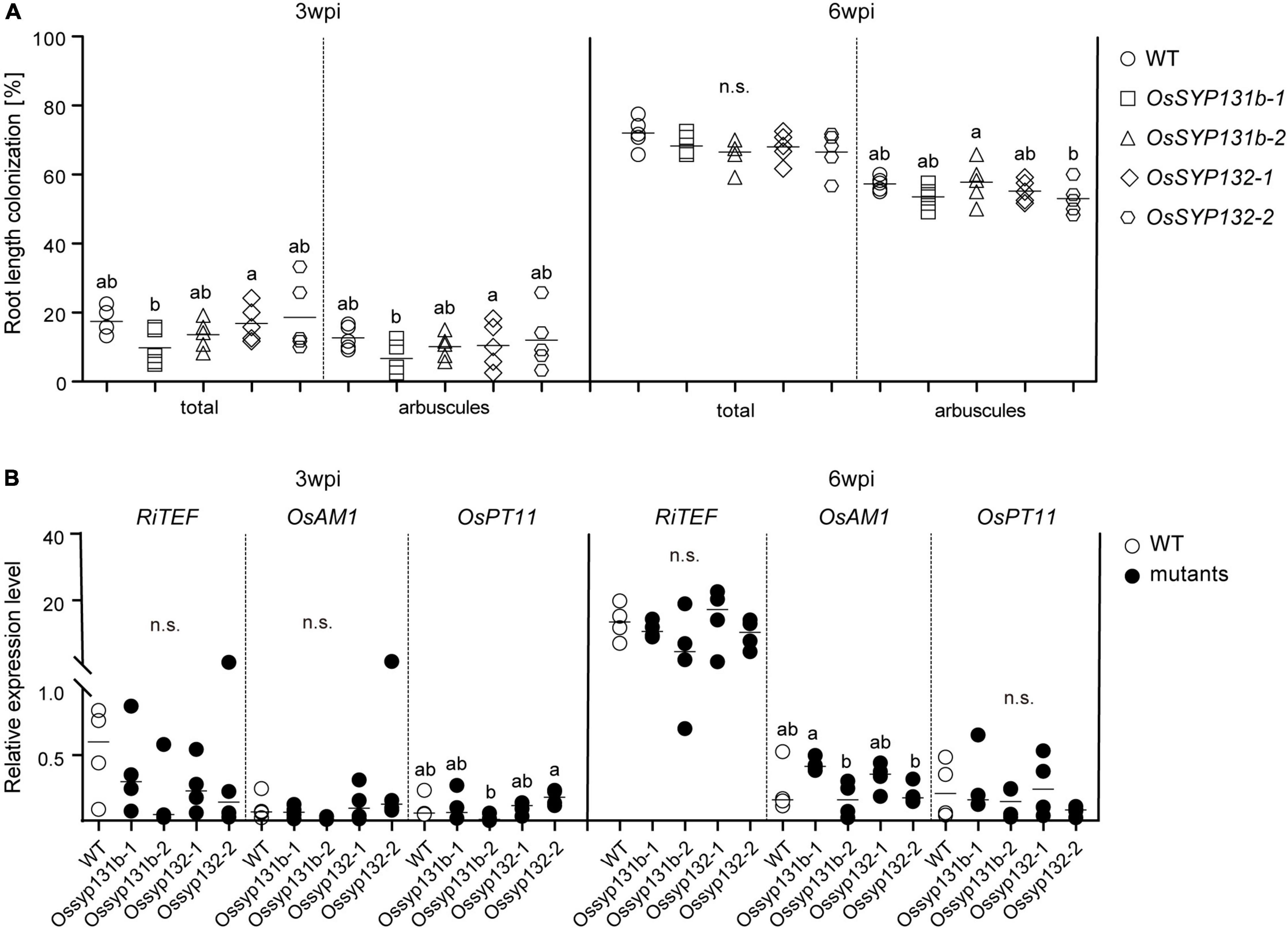
Figure 4. AM symbiosis was normal in the Ossyp131b and Ossyp132 single mutants. (A) The total root length colonization rates and arbuscule abundance (%) of WT rice and Ossyp131b, Ossyp132 mutants at 3- and 6-wpi by R. irregularis were determined by the gridline intersect method. Data were individual values from five biological replicates and the horizontal line showed the mean value. Different letters indicate statistical differences between genotypes evaluated by the Tukey’s honest significance test (all pairs between WT and mutants, P > 0.05, n.s., no significance). (B) Transcript accumulation of OsRiTEF (Translation Elongation Factor), OsAM1 and OsPT11 (Phosphate transporter 11) were measured by qRT-PCR in WT roots and mutants in the presence of R. irregularis at 3- and 6-wpi. The housekeeping gene, OsUBI1, was used for normalization. Data were individual values from four biological replicates and the horizontal line showed the mean value. Different letters indicate statistical differences evaluated by the Tukey’s test, too (all pairs between WT and mutants, P > 0.05, n.s., no significance). This is a representative experiment that was repeated three times.
The unaffected AMF colonization phenotype in the Ossyp131b and Ossyp132 mutants was then confirmed by measuring the relative expression levels of three AM marker genes, RiTEF (Translation Elongation Factor), OsAM1 and OsPT11 (Phosphate transporter 11). The fungal housekeeping gene RiTEF represents a molecular marker correlated with the abundance of intraradical fungal structures, while locating to the PAM. OsAM1 is early expressed in rice cells colonized by small arbuscules and in cortex cells flanking intercellularly growing hyphae. OsPT11 is mainly responsible for transferring phosphate from fungi to plants. For all the three marker genes, comparable relative expression levels were detected between wild-type rice and the examined mutants, thus supporting that AM symbiosis was successfully established in the Ossyp131b and Ossyp132 mutants as in wild-type (Figure 4B).
To further test whether OsSYP131b and OsSYP132 play redundant roles in AM symbiosis, a homozygous Ossyp132-2 mutant was crossed with the Ossyp131b-1 or Ossyp131b-2 mutants, respectively. The double mutants were identified from F2 population and further generated homozygous Ossyp131b-1 Ossyp132-2 and Ossyp131b-2 Ossyp132-2 double mutants at F3 generation. Hydroponic cultivation experiments were also performed for the double mutants, which showed normal growth phenotypes compared to wild-type (Supplementary Figure 4).
We then assessed the mycorrhizal phenotypes. This time, the total colonization rates by AMF in the Ossyp131b Ossyp132 double mutants were only 4∼5% at 3-wpi, which was much lower than the average rate observed in wild-type (∼25%, p < 0.05). The arbuscule abundance level also greatly decreased from ∼14% in wild-type to only 1-4% in the double mutants (p < 0.05, Figures 5A,B), indicating that both fungal colonization and arbuscule formation were greatly delayed in the double mutants at early inoculation process. At 6-wpi, the AMF colonization rate in the double mutants increased to 43-44%, which was still much lower than in wild-type (∼61%, p < 0.05). Similarly, fewer arbuscules were observed in the double mutants comparing to wild-type (31-35% vs. 51%, p < 0.05). However, the mature arbuscules observed in the double mutants can occupy the colonized cells completely (Figure 5A). We then quantified and compared the arbuscular size distribution pattern between the double mutant and wild-type. As a result, no statistically significant differences on arbuscular size distribution were observed (Supplementary Figure 6), indicating that loss functions of both OsSYP131b and OsSYP132 genes would not impair a specific stage of arbuscular development. Therefore, the rather fewer arbuscules and lower colonization rates observed in the double mutants were more likely due to a less efficient but still working membrane trafficking pathway, which affected fungal proliferation in rice roots.
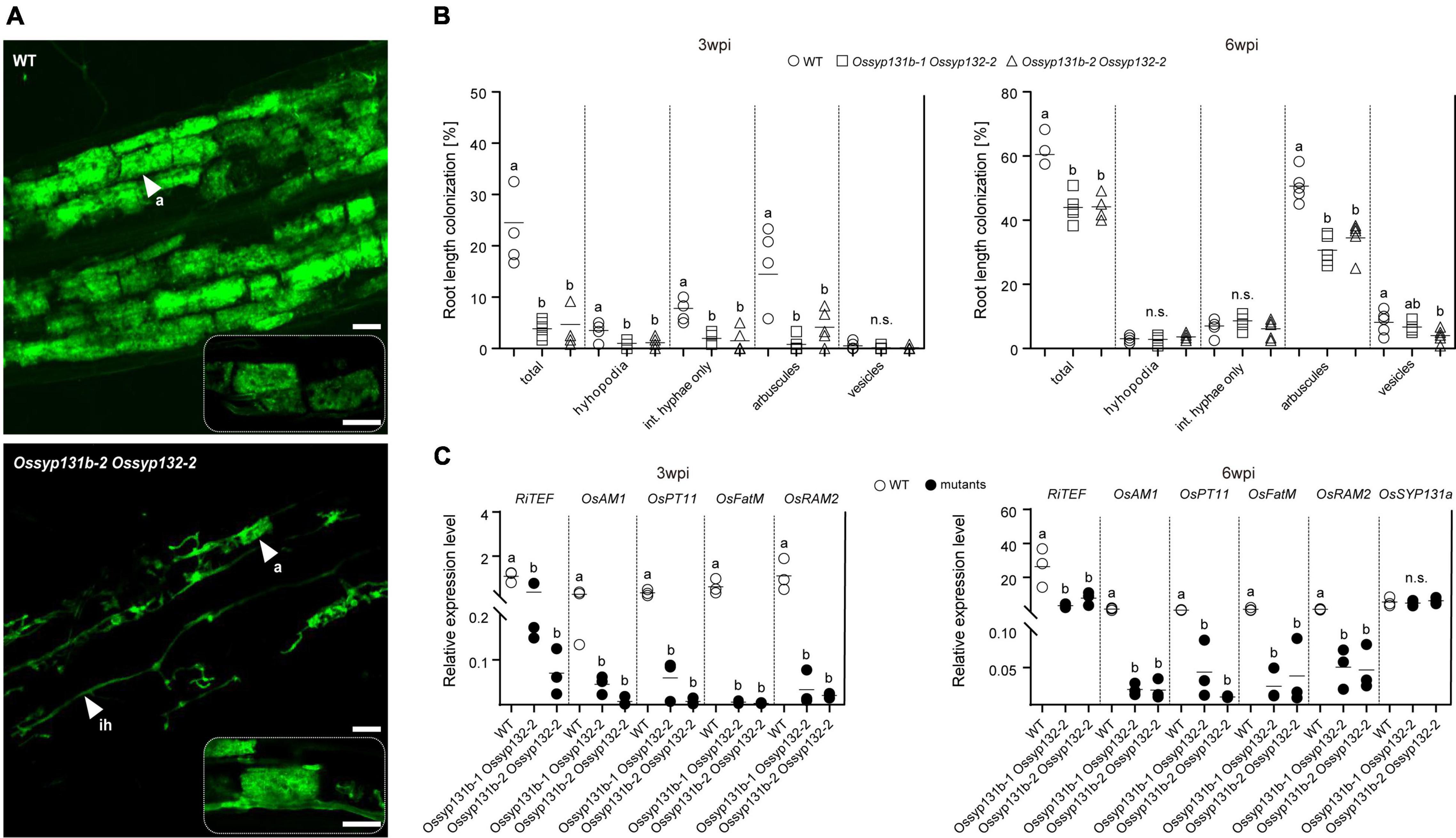
Figure 5. AM fungal colonization was impaired in the Ossyp131b Ossyp132 double mutants. (A) Laser scanning confocal images showing fungal structures stained by WGA-Alexa Fluor 488 in WT rice roots and the Ossyp131b Ossyp132 double mutants colonized by R. irregularis at 6-wpi. White arrows indicate mycorrhizal structures: a, arbuscule; ih, intraradical hyphae. Scale bars = 20 μm (above images), 50 μm (below images), or 25 μm (rectangle images with dotted lines). (B) The root length colonization rates (%) containing total, hyphopodia, intraradical hyphae, arbuscules, and vesicles of WT rice and the Ossyp131b Ossyp132 double mutants by R. irregularis at 3- and 6-wpi were determined by the gridline intersect method. Data were individual values from five biological replicates and the horizontal line showed the mean value. Different letters indicate significant statistical differences between genotypes evaluated by the Tukey’s honest significance test (between the pairs, P < 0.05 or P > 0.05, n.s., no significance). Int. hyphae, intraradical hyphae. (C) The transcript accumulation of AM marker genes RiTEF, OsAM1, OsPT11, OsFatM (fat required for AM symbiosis) and OsRAM2 (required for arbuscular mycorrhization2) as well as OsSYP131a were measured by qRT-PCR in the WT roots and Ossyp131b Ossyp132 double mutants by R. irregularis. The housekeeping gene, OsUBI1, was used for normalization. Data are individual values from three biological replicates and the horizontal line shows the mean value. Different letters indicate levels of statistical differences evaluated by the Tukey’s test, too (all pairs except the OsSYP131a gene, P < 0.05). This is a representative experiment that was repeated three times with similar results.
Consistent with the impaired mycorrhizal phenotype, qRT-PCR results also demonstrated that the five marker genes RiTEF, OsPT11, OsAM1, OsFatM (fat required for AM symbiosis) and OsRAM2 (required for arbuscular mycorrhization2) (Wang et al., 2012; Bravo et al., 2017; Jiang et al., 2017; Dai et al., 2022; Liu et al., 2022) were expressed at markedly reduced levels in the Ossyp131b Ossyp132 double mutants than in WT plants at both examined time points (Figure 5C). The relative expression level of OsSYP131a gene was not affected in the double mutants, though (Figure 5C).
An interesting feature of AM symbiosis is that the fungal hyphae do not actually enter into the host cytoplasm, but are closely surrounded by host cell plasma membrane. As fungal hyphae continuously branch to form arbuscules in the inner cortical cells, the PAM also expands quickly (up to 10-fold) in order to completely surround the hyphal branches (Alexander et al., 1989). Multiple SNARE proteins likely involved in PAM formation have been identified in legume species, including MtVAMP721d and MtVAMP721e from a ‘symbiotic’ VAMP72II clade and MtSYP132 from a ‘symbiotic’ SYP13II clade (Ivanov et al., 2012; Huisman et al., 2016, 2020; Pan et al., 2016). It was wondered whether functional divergences could have occurred for SNAREs of the ‘symbiotic’ clades, consequently defining a symbiosis-specific membrane trafficking pathway.
To explore this question, we reconstructed a phylogeny of plant SYP13 subfamily (Figure 1). It illustrated that there was an ancestral SYP13 gene in the common ancestor of seed plants, which further duplicated into two lineages in angiosperms, namely the ‘non-symbiotic’ SYP13I clade (including both mycorrhizal and non-mycorrhizal host species) and the ‘symbiotic’ SYP13II clade that includes only mycorrhizal species (Figure 1). Within the SYP13I clade, two independent duplication events had occurred in the common ancestor of monocots and in the common ancestor of Brassicaceae, respectively (Figure 1). Moreover, an unequal crossover event likely occurred between homologous members of the two SYP13 clades in the dicot lineage after the split of A. coerulea (Huisman et al., 2016), resulting in chimeric SYP13II genes that can produce two isoforms in most dicot species. These evolutionary events had complicated gene relationships among different plant species (especially in dicots), making it difficult to explore functional similarities or divergences among genes of the two SYP13 clades.
Having lost the representative gene from the SYP13II clade, A. thaliana has two representative genes, AtSYP131 and AtSYP132 in the SYP13I clade. Their encoded proteins were both revealed to be located on the plasma membrane, like other Qa-SNAREs in SYP11 and SYP12 subfamilies (Uemura et al., 2004). AtSYP131 was specifically expressed in mature pollen grains, while AtSYP132 exhibit a ubiquitous expression pattern in all tissues at all developmental stages (Enami et al., 2009). A T-DNA insertion in the promoter region of AtSYP132 resulted in bushy plants with almost no seeds, and the seedlings also formed adventitious roots instead of a single primary root, demonstrating that AtSYP132 likely plays essential roles in plant growth and development (Park et al., 2018). Downregulating AtSYP132 via artificial microRNA also revealed that this gene plays an important role in cytokinesis (Park et al., 2018).
In the legume M. truncatula, gene MtSYP131 belongs to the SYP13I clade, while gene MtSYP132 of the SYP13II clade can produce two isoforms MtSYP132α and MtSYP132β. The function of MtSYP131 is not known yet, although this gene was found expressed in arbusculated cells together with other SYP11 and SYP12 members (Huisman et al., 2020). Silencing MtSYP132α caused underdeveloped arbuscule phenotype, suggesting a specific role of this isoform in symbiosis; meanwhile, silencing MtSYP132β largely inhibited root growth, which is analogous to the lethal phenotype of AtSYP132 (Huisman et al., 2016; Pan et al., 2016). It should be noticed, however, silencing MtSYP132α seemed not only decreased the transcript abundance levels of MtSYP132α, but also reduced MtSYP132β expression at least in one of the previous studies (Pan et al., 2016), making it unclear whether both isoforms are involved in AM symbiosis. Moreover, after deleting the alternatively spliced exon of Mtsyp132α, the constitutively spliced MtSYP132β (with elevated expression levels than in wild-type) can functionally replace MtSYP132α to restore normal arbuscule morphology (Huisman et al., 2020). It therefore seemed to suggest that both isoforms are involved in regulating AM symbiosis. However, direct genetic evidence is lacking since silencing MtSYP132β was lethal and inhibited root growth.
Without forming chimeric genes, the single copy of SYP13II gene in monocot species, such as rice OsSYP132 serves as an ideal candidate to investigate whether it has evolved specific role in AM symbiosis. The results of qRT-PCR also showed that OsSYP132 was barely expressed in control roots, but became strongly upregulated in mycorrhizal roots (Figure 2A). On the contrary, both OsSYP131a and OsSYP131b of the SYP13I clade were stably expressed in both the mock-inoculated and mycorrhizal rice roots (Figures 2B,C). Therefore, both phylogenetic and expression data suggested that OsSYP132 likely have a specific role in AM symbiosis. However, our OsSYP132 knockout revealed no apparent AM developmental and morphological defects (Figure 4A and Supplementary Figure 5), and it was further confirmed by the normal expressions of three AM marker genes (Figure 4B).
The strict evolutionary conservation of a symbiosis-specific form of SYP132 is in sharp contrast to the lack of a significant AM symbiosis defect when this gene is removed. It therefore pointed to another possibility, that is, both SYP13I and SYP13II genes are involved in regulating AM symbiosis; meanwhile SYP13I genes are also involved in plant growth and development, so they are retained in non-mycorrhizal hosts. To verify this possibility, we further obtained the Ossyp131b mutants and the Ossyp131b Ossyp132 double mutants. Although the OsSYP131b mutants showed no defective mycorrhizal phenotype (Figure 4 and Supplementary Figure 5), the loss of functions of both OsSYP131b and OsSYP132 strongly inhibited AMF colonization rate and arbuscule abundance levels (Figure 5). This finding reveals that successful AM symbiosis employed SNARE proteins from not only SYP13II but also SYP13I clades. Possibly the active vesicle trafficking in arbuscular cells in higher plants need the combined coding capacity of multiple SYP13 genes, whose protein products can all partner with symbiotically induced VAMP72 proteins. It remains to be seen whether a similar requirement exists in other species, especially dicots that employ the APA mechanism to produce a ‘symbiotic’ SYP132 transcript.
As for OsSYP131a, our results showed that its T0 homozygous mutants exhibited a dwarf phenotype and were not able to develop fertile seeds (Figure 3), indicating a function of this gene in plant growth and in seed fertility, which is to some extents, analogous to the AtSYP132 phenotype (Park et al., 2018). However, since no fertile mutant seeds could be obtained for this gene and further used to test the mycorrhizal phenotype, its potential involvement in AM symbiosis cannot be determined. It should be realized that the arbuscule morphology was normal and the AM colonization was not completely blocked in the Ossyp131b Ossyp132 double mutants (Figure 5A and Supplementary Figure 6), implying that additional SNAREs encoded by either OsSYP131a or other syntaxin genes can still make membrane trafficking work in the colonized cells, though in a less efficient way.
In this study, we investigated three rice SYP13 subfamily Qa-SNARE genes, OsSYP131a, OsSYP131b and OsSYP132. Among them, OsSYP131a is essential for rice seed embryogenesis; while OsSYP131b and OsSYP132 are functionally redundant in regulating AM symbiosis, probably by mediating membrane trafficking to form the plant-fungus interface in the colonized cortical cells.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
BW and Y-NL designed the project. Y-NL and C-CL performed most of the experiments. RG, LT, J-FC, and Y-NW assisted in experiments. DW participated in the discussion and helped in writing. Y-NL and BW analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
This research was financed by National Natural Science Foundation of China (31870203).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.898286/full#supplementary-material
Alexander, T., Toth, R., Meier, R., and Weber, H. C. (1989). Dynamics of arbuscule development and degeneration in onion, bean, and tomato with reference to vesicular arbuscular mycorrhizae in grasses. Can. J. Bot. 67, 2505–2513. doi: 10.1139/b89-320
Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839–841. doi: 10.1038/35057024
Bravo, A., Brands, M., Wewer, V., Dormann, P., and Harrison, M. J. (2017). Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol. 214, 1631–1645. doi: 10.1111/nph.14533
Chen, C. Y., Ane, J. M., and Zhu, H. Y. (2008). OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol. 180, 311–315. doi: 10.1111/j.1469-8137.2008.02612.x
Dai, H. L., Zhang, X. W., Zhao, B. Y., Shi, J. C., Zhang, C., Wang, G., et al. (2022). Colonization of mutualistic mycorrhizal and parasitic blast fungi requires OsRAM2-regulated fatty acid biosynthesis in rice. Mol. Plant Microbe Interact. 35, 178–186. doi: 10.1094/MPMI-11-21-0270-R
Dickson, S., and Kolesik, P. (1999). Visualisation of mycorrhizal fungal structures and quantification of their surface area and volume using laser scanning confocal microscopy. Mycorrhiza 9, 205–213. doi: 10.1007/s005720050268
Enami, K., Ichikawa, M., Uemura, T., Kutsuna, N., Hasezawa, S., Nakagawa, T., et al. (2009). Differential expression control and polarized distribution of plasma membrane-resident SYP1 SNAREs in Arabidopsis thaliana. Plant Cell Physiol. 50, 280–289. doi: 10.1093/pcp/pcn197
Fasshauer, D., Sutton, R. B., Brunger, A. T., and Jahn, R. (1998). Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 95, 15781–15786. doi: 10.1073/pnas.95.26.15781
Genre, A., Chabaud, M., Faccio, A., Barker, D. G., and Bonfante, P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20, 1407–1420. doi: 10.1105/tpc.108.059014
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Harrison, M. J., and Ivanov, S. (2017). Exocytosis for endosymbiosis: membrane trafficking pathways for development of symbiotic membrane compartments. Curr. Opin. Plant Biol. 38, 101–108. doi: 10.1016/j.pbi.2017.04.019
Huisman, R., Hontelez, J., Bisseling, T., and Limpens, E. (2020). SNARE complexity in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 11:354. doi: 10.3389/fpls.2020.00354
Huisman, R., Hontelez, J., Mysore, K. S., Wen, J. Q., Bisseling, T., and Limpens, E. (2016). A symbiosis-dedicated SYNTAXIN OF PLANTS 13II isoform controls the formation of a stable host-microbe interface in symbiosis. New Phytol. 211, 1338–1351. doi: 10.1111/nph.13973
Ivanov, S., Fedorova, E. E., Limpens, E., De Mita, S., Genre, A., Bonfante, P., et al. (2012). Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. Proc. Natl. Acad. Sci. U.S.A. 109, 8316–8321. doi: 10.1073/pnas.1200407109
Jahn, R., and Scheller, R. H. (2006). SNAREs: engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643. doi: 10.1038/nrm2002
Jiang, Y. N., Wang, W. X., Xie, Q. J., Liu, N., Liu, L. X., Wang, D. P., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Kanazawa, T., and Ueda, T. (2017). Exocytic trafficking pathways in plants: why and how they are redirected. New Phytol. 215, 952–957. doi: 10.1111/nph.14613
Keymer, A., Pimprikar, P., Wewer, V., Huber, C., Brands, M., Bucerius, S. L., et al. (2017). Lipid transfer from plants to arbuscular mycorrhiza fungi. Elife 6:e29107.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lipka, V., Kwon, C., and Panstruga, R. (2007). SNARE-ware: the role of SNARE-domain proteins in plant biology. Annu. Rev. Cell Dev. Biol. 23, 147–174. doi: 10.1146/annurev.cellbio.23.090506.123529
Liu, Y. N., Liu, C. C., Zhu, A. Q., Niu, K. X., Guo, R., Tian, L., et al. (2022). OsRAM2 function in lipid biosynthesis is required for arbuscular mycorrhizal symbiosis in rice. Mol. Plant Microbe Interact. 35, 187–199. doi: 10.1094/MPMI-04-21-0097-R
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lota, F., Wegmuller, S., Buer, B., Sato, S., Brautigam, A., Hanf, B., et al. (2013). The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12, a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicus. Plant J. 74, 280–293. doi: 10.1111/tpj.12120
Luginbuehl, L. H., Menard, G. N., Kurup, S., Van Erp, H., Radhakrishnan, G. V., Breakspear, A., et al. (2017). Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178. doi: 10.1126/science.aan0081
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular–arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Naito, Y., Hino, K., Bono, H., and Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Pan, H. R., Oztas, O., Zhang, X. W., Wu, X. Y., Stonoha, C., Wang, E. T., et al. (2016). A symbiotic SNARE protein generated by alternative termination of transcription. Nat. Plants 2:15197. doi: 10.1038/nplants.2015.197
Park, M., Krause, C., Karnahl, M., Reichardt, I., El Kasmi, F., Mayer, U., et al. (2018). Concerted action of evolutionarily ancient and novel SNARE complexes in flowering-plant cytokinesis. Dev. Cell 44, 500–511.e4. doi: 10.1016/j.devcel.2017.12.027
Schaffer, G. F., and Peterson, R. L. (1993). Modifications to clearing methods used in combination with vital staining of roots colonized with vesicular-arbuscular mycorrhizal fungi. Mycorrhiza 4, 29–35. doi: 10.1007/bf00203248
Smith, S. E., and Smith, F. A. (2011). Roles of arbuscular mycorrhizas in plant nutrition and growth: new paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 62, 227–250. doi: 10.1146/annurev-arplant-042110-103846
Spatafora, J. W., Chang, Y., Benny, G. L., Lazarus, K., Smith, M. E., Berbee, M. L., et al. (2016). A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046. doi: 10.3852/16-042
Trifinopoulos, J., Nguyen, L. T., von Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Uemura, T., Ueda, T., Ohniwa, R. L., Nakano, A., Takeyasu, K., and Sato, M. H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65. doi: 10.1247/csf.29.49
Vierheilig, H., Coughlan, A. P., Wyss, U., and Piche, Y. (1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64, 5004–5007. doi: 10.1128/AEM.64.12.5004-5007.1998
Wang, B., and Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363. doi: 10.1007/s00572-005-0033-6
Wang, E. T., Schornack, S., Marsh, J. F., Gobbato, E., Schwessinger, B., Eastmond, P., et al. (2012). A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Curr. Biol. 22, 2242–2246. doi: 10.1016/j.cub.2012.09.043
Wang, H., Niu, Q. W., Wu, H. W., Liu, J., Ye, J., Yu, N., et al. (2015). Analysis of non-coding transcriptome in rice and maize uncovers roles of conserved lncRNAs associated with agriculture traits. Plant J. 84, 404–416. doi: 10.1111/tpj.13018
Wang, S. S., Chen, A. Q., Xie, K., Yang, X. F., Luo, Z. Z., Chen, J. D., et al. (2020). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. U.S.A. 117, 16649–16659. doi: 10.1073/pnas.2000926117
Wang, W. X., Shi, J. C., Xie, Q. J., Jiang, Y. N., Yu, N., and Wang, E. T. (2017). Nutrient exchange and regulation in arbuscular mycorrhizal symbiosis. Mol. Plant 10, 1147–1158. doi: 10.1016/j.molp.2017.07.012
Wipf, D., Krajinski, F., van Tuinen, D., Recorbet, G., and Courty, P. E. (2019). Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. New Phytol. 223, 1127–1142. doi: 10.1111/nph.15775
Keywords: rice (Oryza sativa), arbuscular mycorrhiza (AM), SNARE, OsSYP13, functional redundancy
Citation: Liu Y-N, Liu C-C, Guo R, Tian L, Cheng J-F, Wu Y-N, Wang D and Wang B (2022) The Rice Qa-SNAREs in SYP13 Subfamily Are Involved in Regulating Arbuscular Mycorrhizal Symbiosis and Seed Fertility. Front. Plant Sci. 13:898286. doi: 10.3389/fpls.2022.898286
Received: 17 March 2022; Accepted: 28 April 2022;
Published: 18 May 2022.
Edited by:
Ertao Wang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Senjuti Sinharoy, National Institute of Plant Genome Research (NIPGR), IndiaCopyright © 2022 Liu, Liu, Guo, Tian, Cheng, Wu, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Wang, Ymlud2FuZ0BuanUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.