
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 May 2022
Sec. Aquatic Photosynthetic Organisms
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.896138
This article is part of the Research Topic Insights in Marine and Freshwater Plants: 2021 View all 6 articles
The Raphidophyceae is an ecologically important eukaryotic lineage of primary producers and predators that inhabit marine and freshwater environments worldwide. These organisms are of great evolutionary interest because their plastids are the product of eukaryote-eukaryote endosymbiosis. To obtain deeper insight into the evolutionary history of raphidophycean plastids, we sequenced and analyzed the plastid genomes of three freshwater and three marine species. Our comparison of these genomes, together with the previously reported plastid genome of Heterosigma akashiwo, revealed unexpected variability in genome structure. Unlike the genomes of other analyzed species, the plastid genome of Gonyostomum semen was found to contain only a single rRNA operon, presumably due to the loss of genes from the inverted repeat (IR) region found in most plastid genomes. In contrast, the marine species Fibrocapsa japonica contains the largest IR region and overall plastid genome for any raphidophyte examined thus far, mainly due to the presence of four large gene-poor regions and foreign DNA. Two plastid genes, tyrC in F. japonica and He. akashiwo and serC in F. japonica, appear to have arisen via lateral gene transfer (LGT) from diatoms, and several raphidophyte open reading frames are demonstrably homologous to sequences in diatom plasmids and plastid genomes. A group II intron in the F. japonica psbB gene also appears to be derived by LGT. Our results provide important insights into the evolutionary history of raphidophyte plastid genomes via LGT from the plastids and plasmid DNAs of diatoms.
The Raphidophyceae, a class of photosynthetic stramenopile algae, is an evolutionarily diverse and ecologically important lineage whose members thrive in both marine and freshwater environments. The common marine genera Chattonella and Heterosigma are known to be toxic and capable of forming red tides (Hara and Chihara, 1982). Raphidophyte cells are flagellated with two unequal emergent flagella, numerous discoid chloroplasts, and mucocysts (Heywood, 1982, 1990). Three freshwater genera (Gonyostomum, Merotricha, and Vacuolaria) and seven marine genera (Chattonella, Chlorinimonas, Fibrocapsa, Haramonas, Heterosigma, Psammamonas, and Viridilobus) have been formally described (Fott, 1968; Heywood, 1990; Horiguchi, 1996). Most marine members possess fucoxanthin as their main accessory photosynthetic pigment and have yellowish brown-colored plastids. All freshwater species described thus far are green in color due to the absence of fucoxanthin (Fiksdahl et al., 1984; Bjørnland and Liaaen-Jensen, 1989). Little is known about the extent to which differences in habitat and pigment composition have contributed to the evolutionary diversification of raphidophycean algae.
The plastid of photosynthetic stramenopiles stems from rhodophytes via secondary or possibly tertiary endosymbiosis and is very different from those of the primary plastid-bearing green algae, land plants, glaucophytes, and rhodophytes. Recent phylogenomic studies of photosynthetic stramenopiles have included plastid genome data from Bacillariophyceae (diatoms), Bolidophyceae, Chrysophyceae, Dictyochophyceae, Eustigmatophyceae, Olisthodiscophyceae, Phaeophyceae (brown algae), Pelagophyceae, Xanthophyceae, and Raphidophyceae, as well as plastid-bearing alveolates (Cattolico et al., 2008; Ong et al., 2010; Ruck et al., 2014; Ševčíková et al., 2015; Han et al., 2019; Kim et al., 2019, 2020; Sassenhagen and Rengefors, 2019; Barcytė et al., 2021). The genus Olisthodiscus has recently been transferred from Raphidophyceae to the newly erected class Olisthodiscophyceae based on phylogenomics of plastid sequences (Barcytė et al., 2021).
To better understand the relationships among raphidophycean algae as well as the broader evolution of plastids in stramenopiles, we sequenced six raphidophyte plastid genomes, two of which are from the freshwater species Gonyostomum semen and Vacuolaria virescens, and carried out detailed comparative genomic and phylogenomic analyses of these data, as well as the published plastid genome of the raphidophycean Heterosigma akashiwo [pre- existing but incomplete plastid genomic data for G. semen and V. virescens were not included (Sassenhagen and Rengefors, 2019)]. We discovered instances of gene loss/gain, duplication/reduction of the inverted repeat (IR) region, and gene rearrangements in raphidophycean plastid genomes. We also show that their genomes have been impacted by lateral gene transfer (LGT) from diatoms.
Strains of Gonyostomum semen, Vacuolaria virescens, Chattonella marina, and Fibrocapsa japonica were collected from natural habitats: G. semen from freshwater, Geumsan, Chungcheongnam-do, Korea (36° 10′ 53.9” N 127° 27′ 38.4″ E), V. virescens from freshwater, Changnyeong, Gyeongsangnam-do, Korea (35° 31′ 15.7” N 128° 23′ 08″ E), C. marina from marine, Kunsan, Jeollanam-do, Korea (35° 56′ 11.1’’ N, 126° 31′ 39.4″ E), and F. japonica from marine, Masanman, Gyeongsangnam-do, Korea (35° 12′ 02.7” N, 128° 34′ 40.3″ E). The G. semen and V. virescens strains are available from the culture collection at the Chungnam National University Korea. The C. marina and F. japonica strains are available from the culture collections at the Kunsan National University or Chonnam National University Korea. Cultured strains of Merotricha bacillata NIES-1809 and Haramonas pauciplastida NIES-1870 were obtained from the Microbial Culture Collection (MCC), National Institute for Environmental Studies (NIES), Japan. All cultures studied herein were grown in AF-6 or f/2 medium (Andersen et al., 2005) with distilled water for the freshwater strains or distilled seawater for the marine strains and were maintained at 20°C under a 14:10 light:dark cycle with 30 μmol photons·m−2·s−1 from cool white fluorescent tubes. All strains were derived from single-cell isolation for unialgal cultivation. Total genomic DNAs were extracted using the QIAGEN DNEasy Blood Mini Kit (QIAGEN, Valencia, CA, United States) following the manufacturer’s instructions. Next-generation sequencing was carried out using a MiSeq (Illumina, San Diego, CA, United States). Amplified DNAs were fragmented and tagged using the NexteraXT protocol (Illumina), indexed, size selected, and pooled for sequencing using the small amplicon targeted resequencing run, which performs paired end 2 × 300 bp sequencing reads using the MiSeq Reagent Kit v3 (Illumina), according to the manufacturer’s recommendations.
Sequence data were trimmed (base = 80 bp, error threshold = 0.05, n ambiguities = 2) prior to de novo assembly with the default option (automatic bubble size, minimum contig length = 1,000 bp). The raw reads were assembled using the SPAdes 3.7 assembler1 and mapped to the assembled contigs (similarity = 95%, length fraction = 75%), excluding contigs <1,000 bp (assembly statistics are summarized in Supplementary Table S1). Contigs were deemed to be of plastid genome origin as follows: (1) BLAST searches against the entire assembly using commonly known plastid genes as queries resulted in hits to these contigs using Genome Search Plotter (Ejigu et al., 2021) and (2) the predicted genome sizes were similar to the previously published 160 Kbp plastid genome of He. akashiwo NIES-293 (NC_010882).
To aid in gene annotation, we created a database of protein-coding genes, rRNA, and tRNA genes using data from previously sequenced raphidophycean plastid genomes. Preliminary annotation of protein-coding genes was performed using AGORA (Jung et al., 2018) and GeneMarkS.2 The final annotation file was checked in Geneious Prime3 using the ORF Finder program with genetic code 11 (Bacterial, Archaeal, and Plant Plastid Code). The predicted ORFs were checked manually and the corresponding ORFs (and predicted functional domains) in the genome sequences were annotated accordingly.
The tRNA genes were identified from the tRNAscan-SE version 1.21 server4 with the default settings using the “Mito/Chloroplast” model. To help identify rRNA gene sequences, a set of known plastid rRNA sequences from the public database was used as query sequences to search new genomic data using BLASTn. We used RNAweasel5 to search for, and classify introns. Physical maps were visualized with the OrganellarGenomeDRAW program.6 For structural and synteny comparisons, the genomes were aligned using Mauve Genome Alignment version 2.2.0 (Darling et al., 2004) and geneCo (Jung et al., 2019) with default settings. Genome sequences were deposited in the NCBI GenBank database under the accession numbers shown in Table 1.
A phylogenetic analysis of diverse eukaryotic algae was carried out on a concatenated set of 91 proteins (18,216 amino acid sites in total) encoded in 135 plastid genomes (Supplementary Figure S1). The sequences of six Viridiplantae and one glaucophyte species were used as outgroup taxa for rooting purposes. The concatenated proteins were aligned using MacGDE2.6 with manual refinement (Smith et al., 1994). Alignments for protein-specific phylogenetic analyses were constructed for tsg1, serC1/serC2, tyrC, the reverse transcriptase domain of group II intron-encoded proteins, and various conserved hypothetical ORFs. For each protein, homologs were retrieved from the NCBI non-redundant database using BLASTp (e-value cutoff = 1e−5) with raphidophyte proteins as queries. ML phylogenetic analyses of individual protein alignments and concatenated alignments were conducted using IQ-TREE Ver. 1.5.2 (Nguyen et al., 2015) with 1,000 bootstrap replicates. The best evolutionary model for each tree was automatically selected using the –m LG + I + G option incorporated in IQ-TREE. Trees were visualized using FigTree v.1.4.2.7
Six plastid genomes (ptDNA) of representative raphidophycean strains from freshwater and marine habitats were sequenced (Table 1). The structure and coding capacity of these ptDNAs were compared to the previously published genome of the raphidophycean alga He. akashiwo NIES-293 (Cattolico et al., 2008). Plastid genome sizes ranged from 122 Kbp (G. semen) to 249 Kbp (F. japonica) with GC content ranging from 26.5% (V. virescens) to 34.8% (F. japonica). The plastid genomes contained 141 (G. semen) to 199 (F. japonica) protein-coding genes (including hypothetical proteins), 3 rRNAs, and 27 ~ 35 tRNAs. Minor variation was found in tRNA gene content. For instance, trnEUUC is present in G. semen and V. virescens, and trnRACG is present in G. semen as a pseudogene. The raphidophyte plastid genomes generally showed canonical structure in possessing a large single-copy region (LSC), a small single-copy region (SSC), and two inverted repeats (IRs) with protein-coding genes and ribosomal RNA operons. The only exception is G. semen, which is highly unusual in possessing a single ribosomal RNA operon (Figures 1, 2; Supplementary Figure S2). As described in the sections that follow, the increase in plastid genome size in some raphidophyte species is mainly caused by the expansion of gene-poor regions and acquisition of foreign sequences from the plastid or plasmid DNAs of diatoms (red-dashed boxes in Figure 2). Remarkably, the foreign sequences in the F. japonica plastid genomes make it almost twice the size of the G. semen genome.
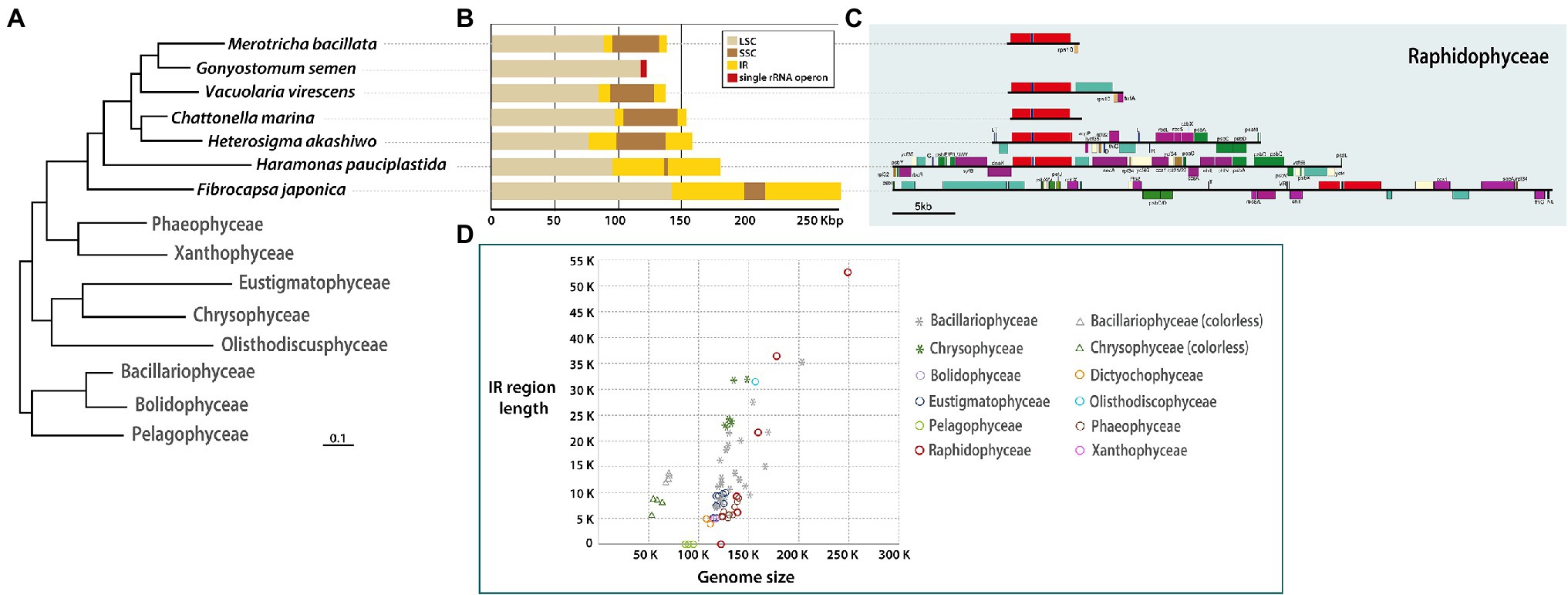
Figure 1. (A) Phylogenetic tree based on plastid genes (see Supplementary Figure S1). (B) Sequence length of SSC, LSC, IR regions. (C) Detailed gene contents in IR region of each raphidophycean species. (D) Distribution graph for length of IR region vs. total plastid genome among stramenopiles.
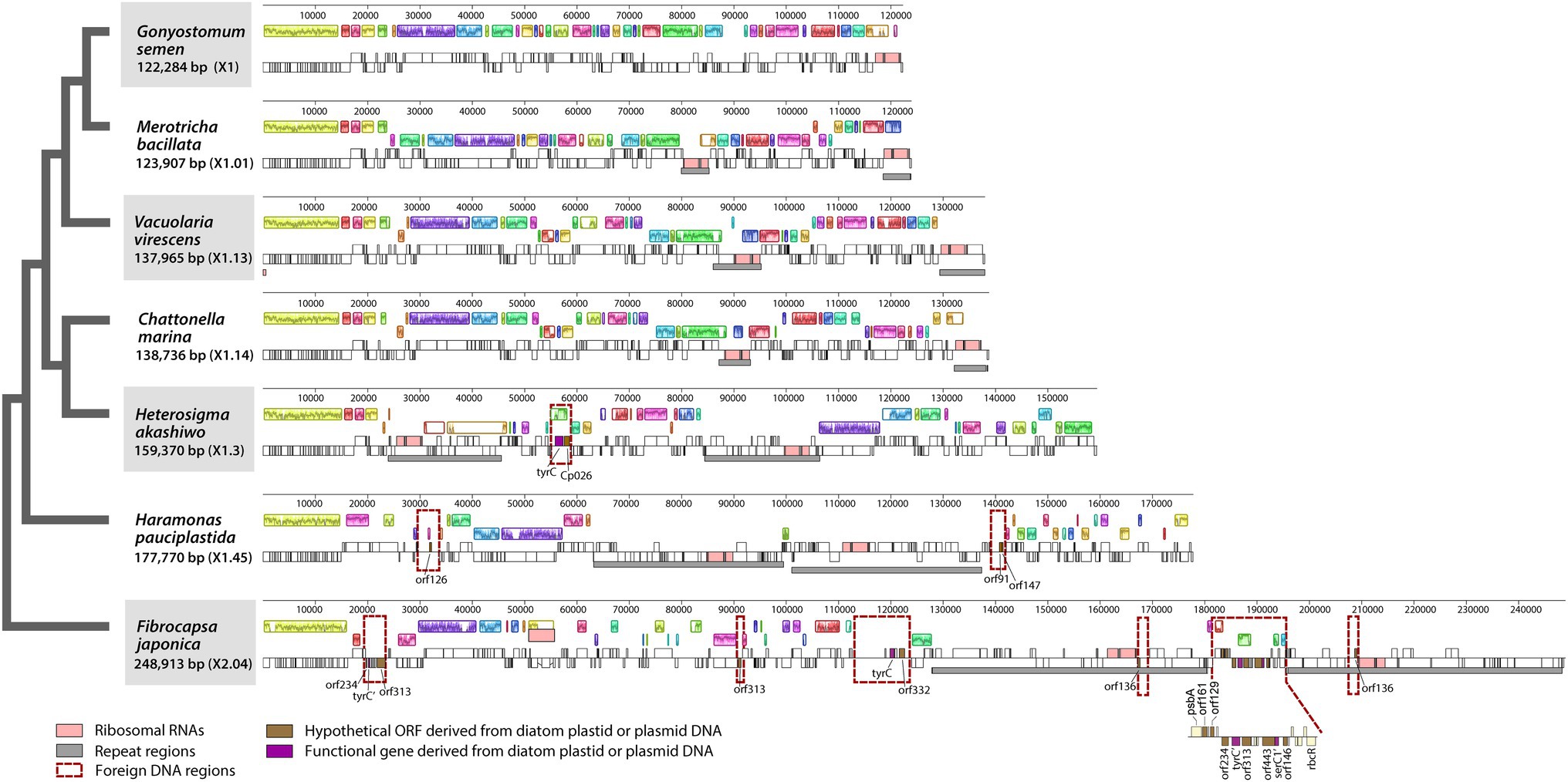
Figure 2. Linearized maps of the plastid genomes of Raphidophyceae. Foreign DNA-containing regions are highlighted with red-dashed lines.
The newly sequenced raphidophycean plastid genomes show evidence of species-specific gene order and content expansion, contraction, and loss in their IR regions (Figures 1B,C). The IR sequence length was found to range from 0 Kbp (i.e., no repeat region) to 52.43 Kbp with functional protein-coding genes, hypothetical ORFs, three rRNAs, and tRNAs. The gene content in the plastid genome of G. semen was distinct from all other raphidophycean algal plastid genomes. The small single-copy (SSC) region varies in length, ranging from 1,418 bp (Ha. pauciplastida) to 38,834 bp (He. akashiwo; Figures 1B,C).
IR-related plastid genome dynamics is a well-studied phenomenon (Goulding et al., 1996; Wang et al., 2008). Contractions, expansions, and small-scale changes in IR and SSC regions have been documented in the plastid genomes of diatoms, chrysophytes, and green algae, sometimes giving rise to changes in gene content (Jansen and Ruhlman, 2012; Sabir et al., 2014; Turmel et al., 2015; Kim et al., 2019, 2020). Expansions and contractions of the IR region have occurred during the evolutionary history of Raphidophyceae as well, leading to changes in gene content and length (Figures 1, 2; Supplementary Figure S2). The size of the IR region correlates strongly with total plastid genome size among stramenopile lineages. The Bacillariophyceae and Chrysophyceae genomes separate into two clusters because of genome reduction in non-photosynthetic groups (stars and triangles in Figure 1D). The pelagophycean algae studied thus far have only single-copy genes in their plastid genomes (Ong et al., 2010). Interestingly, the raphidophycean algae have both IR-lacking and IR-containing plastid genome types and show the largest plastid genome size variation among stramenopile lineages studied thus far (Figure 1D, red circles).
Previous work has shown that in red alga-derived complex plastids, most of the lineage-specific genes show complex distribution patterns suggestive of independent losses across a broad range of phylogenetic depths (Kim et al., 2017, 2019). Although the plastid genomes of Raphidophyceae studied herein are generally conserved in structure and gene content, a handful of genes were identified as being lineage-specific (Figures 3, 4A). To better understand the evolutionary distribution and phylogenetic relationships of these patchily distributed genes among eukaryotes, we performed comparative genomic and phylogenetic analyses of plastid homologs from all the major photosynthetic eukaryotic groups relative to their homologs in non-plastid genomes (Figures 4–6). As we shall see, the results support widespread differential loss in primary- and secondary/tertiary plastid-bearing organisms, but also that LGT has resulted in lineage-specific gene gain.
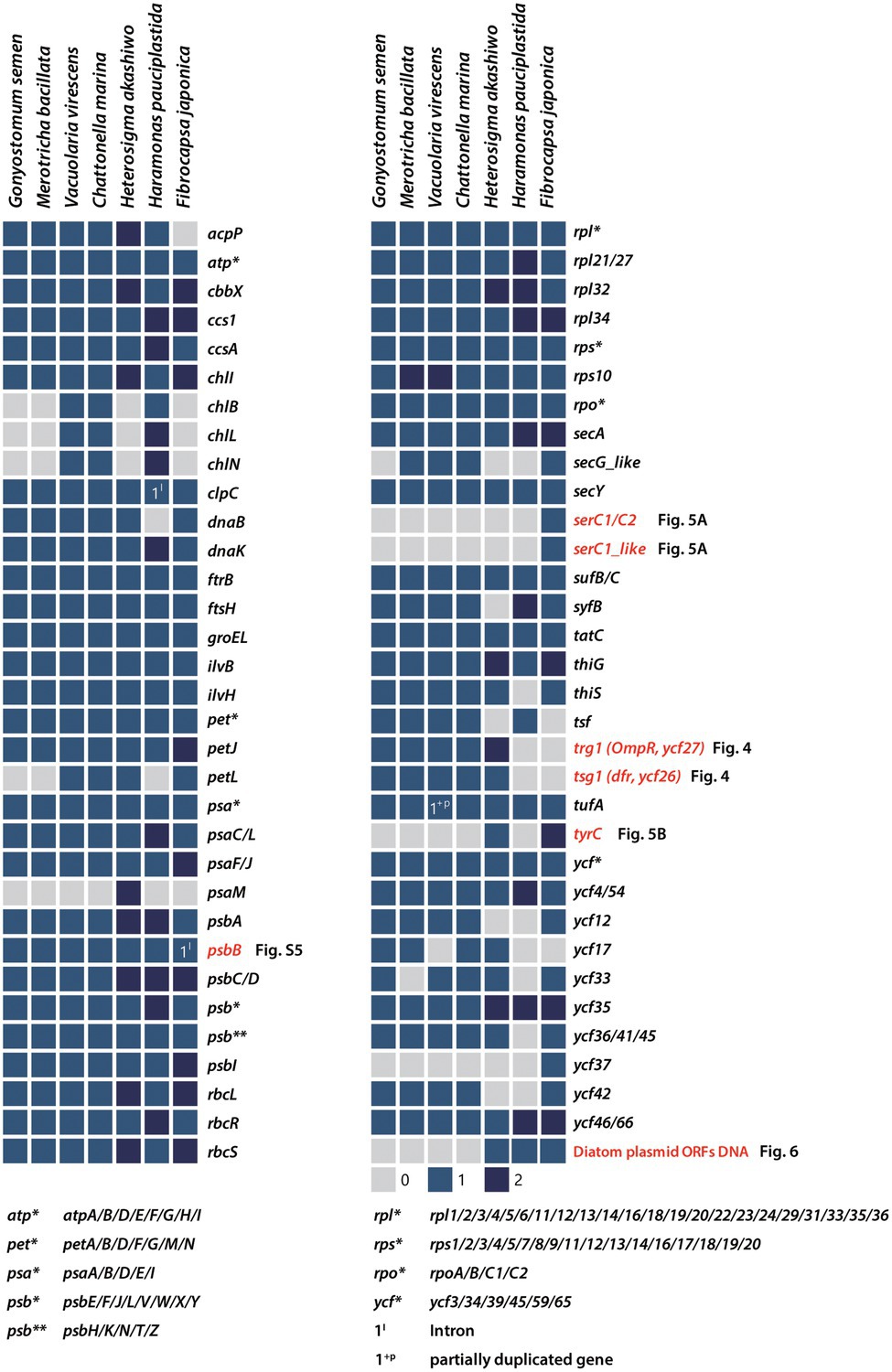
Figure 3. Presence/absence of plastid-encoded genes in Raphidophyceae. Filled boxes indicate the status of each gene (grey = absent, blue = present, dark blue = 2 copies). Patchily distributed genes (i.e., SerC1, serC1-like, trg1, tsg1, tyrC, and various hypothetical ORFs) were detected in the plastid genomes and phylogenies were constructed to infer their evolutionary history.
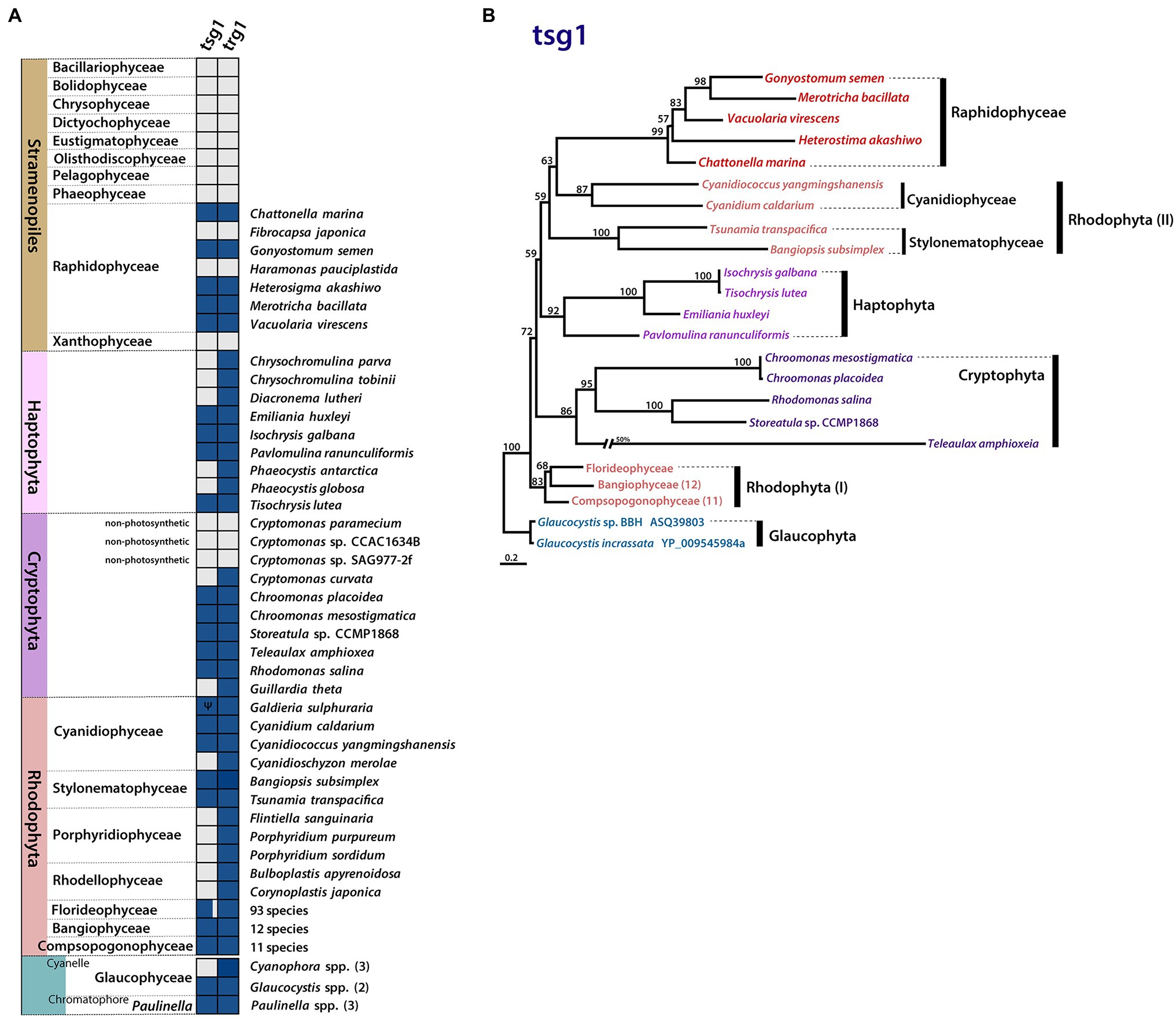
Figure 4. Plastid gene gain and loss in Raphidophyceae. (A) Presence/absence of plastid-encoded tsg1 and trg1 genes. Filled boxes indicate the status of each gene (grey = absent, blue = present, Ψ = pseudogene). (B) Phylogenetic tree of tsg1 protein sequences in Rhodophyta, red alga-derived lineages, and Glaucophyta. Support values on branches are from IQ-Tree UFBoot. The scale bar shows the inferred number of amino acid substitutions per site.
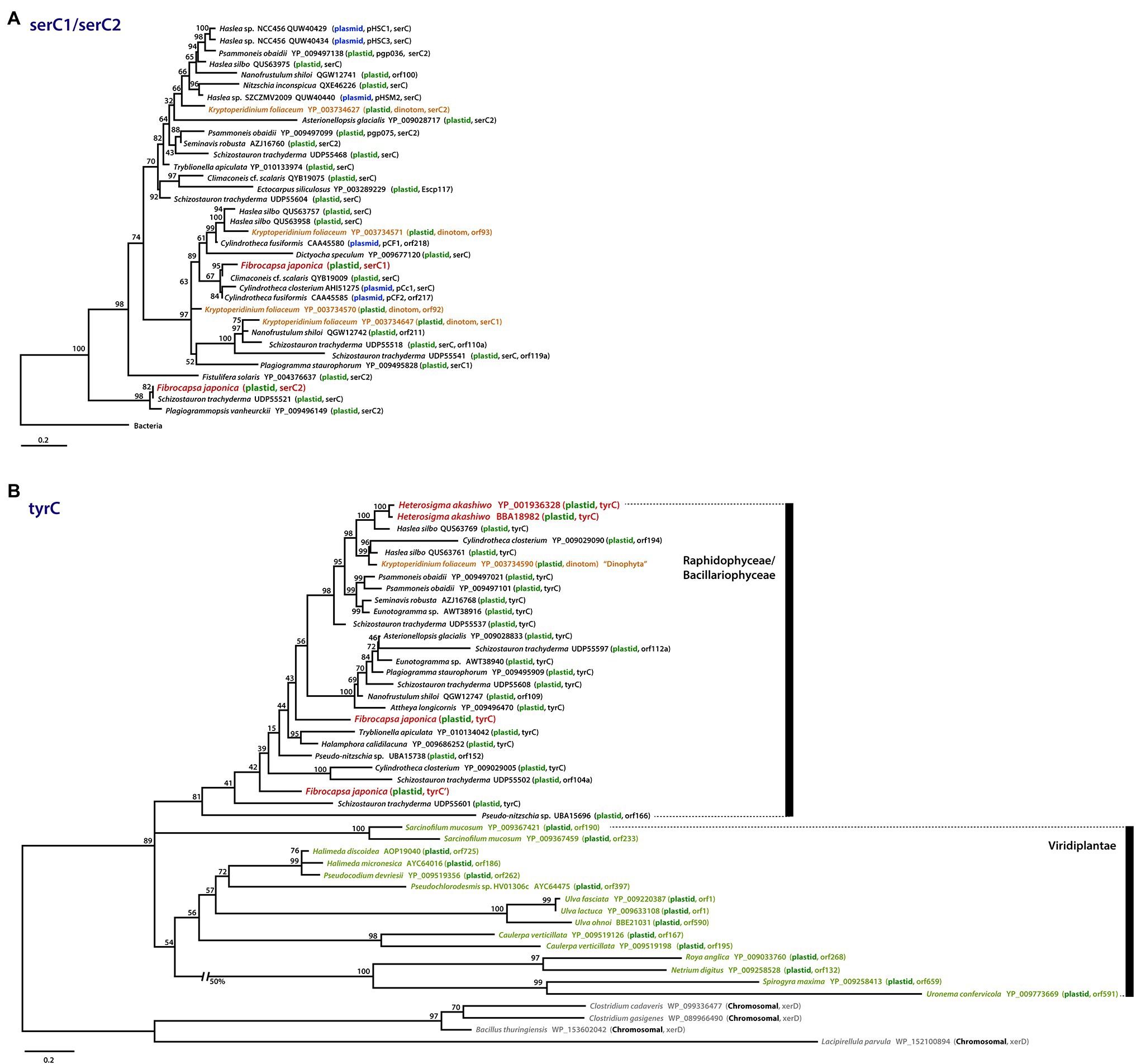
Figure 5. Phylogenies of patchily distributed genes in raphidophyte plastid genomes and their homologs in other organisms. (A) Phylogenetic tree of serC1 and serC2 proteins. (B) Phylogenetic tree of tyrC proteins. Sequences are color-coded according to their genomic location (plastid, plasmid, bacterial chromosome) and the organismal lineage in which they reside. Scale bars indicate the inferred number of amino acid sequences per site.
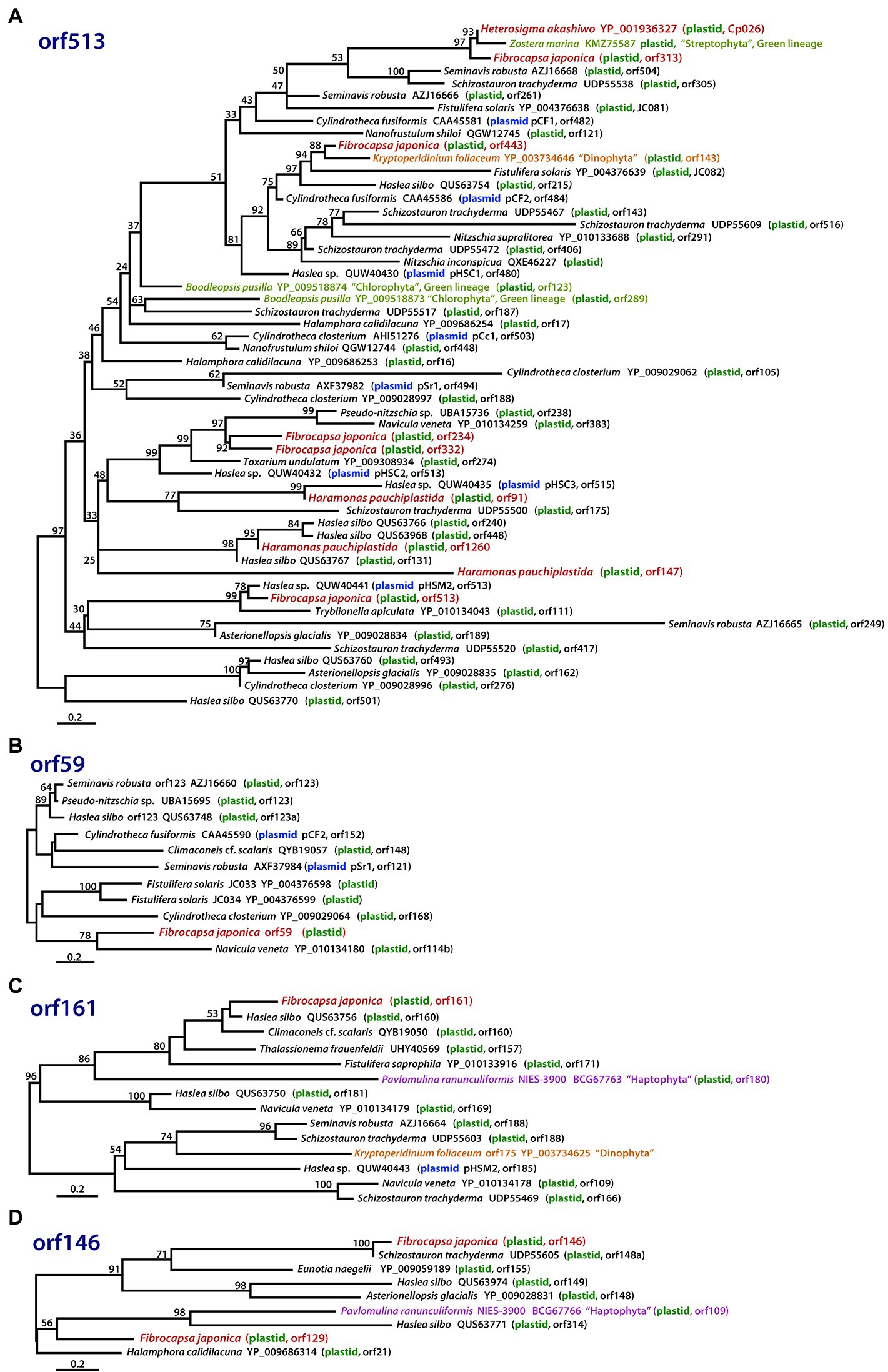
Figure 6. Phylogenies of hypothetical ORFs derived via LGT in raphidophytes, diatoms, and other organisms. The phylogenies shown are of proteins with homology to (A) ORF513, (B) ORF59, (C) ORF161, and (D) ORF146 in the Fibrocapsa japonica plastid genome. Numbers on branches are IQ-Tree UFBoot support values. The scale bar shows the inferred number of amino acid substitutions per site.
The light-independent protochlorophyllide oxidoreductase (LIPOR) genes involved in the light-independent synthesis of chlorophyll (Shi and Shi, 2006) are present in red-algal plastid genomes. LIPOR arose in anoxygenic photosynthetic bacteria, likely from an ancestral nitrogenase enzyme (Fujita and Bauer, 2003; Muraki et al., 2010). Unlike the gene for POR light-dependent protochlorophyllide oxidoreductase, which has been transferred to the nucleus (or acquired by LGT), LIPOR genes, when present, remain in the plastid (Hunsperger et al., 2015). In lineages with red alga-derived plastids, the three LIPOR genes (chlB, chlL, and chlN) are patchily distributed. They are absent from the plastid genome of the raphidophyte He. akashiwo but present in Chattonella subsalsa (Hunsperger et al., 2015) and in three of the raphidophycean species examined herein (V. virescens, C. marina, and Ha. pauciplastida; Figure 3), and present as pseudogenes in some but not all sequenced cryptophyte plastid genomes (Fong and Archibald, 2008; Hunsperger et al., 2015; Kim et al., 2017). These results underscore the dynamic evolution of the LIPOR subunit genes in algae with red alga-derived plastids, the functional significance of which is presently unclear (Hunsperger et al., 2015).
The sensor kinase/response regulator protein subunits tsg1 and trg1 are generally thought to function together in two-component His-to-Asp signal transduction (Duplessis et al., 2007). The presence of this gene pair is linked to a variety of adaptive responses to environmental cues. Although one or both of these genes are found in most rhodophyte plastid genomes, the genes show a variable presence–absence pattern in lineages with red alga-derived secondary/tertiary plastids (Figure 4A). Most plastid genomes of cryptophytes and haptophytes retain both tsg1 and trg1, while most stramenopile lineages possess neither gene; the raphidophytes are an interesting exception.
Five of the seven raphidophycean plastid genomes analyzed contain the His-to-Asp sensor kinase gene tsg1 (transcriptional sensor gene 1); it is absent in the Ha. pauciplastida and F. japonica plastid genomes. The presence of a plastid-encoded tsg1 gene is not universal in red alga-derived plastid-containing lineages (Figure 4A). A plastid tsg1 homolog is found in the haptophytes Emiliania huxleyi, Isochrysis galbana, Tisochrysis lutea, and Pavlomulina ranunculiformis (annotated as dfr) but missing in Chrysochromulina, Diacronema, and Phaeocystis. In cryptophytes, the tsg1 gene is present in five species but missing in the plastid genomes of Cryptomonas spp. and Guillardia theta. In stramenopiles, tsg1 is only found in raphidophyceans; its absence is notable in the bulk of sequenced plastid-bearing members (bacillariophytes, Bolidophyceae, Chrysophyceae, Dictyochophyceae, Eustigmatophyceae, Olisthodiscophyceae, Pelagophyceae, Phaeophyceae, Xanthophyceae). In rhodophytes, the tsg1 gene (annotated as ycf26 or dfr) is encoded in the plastid genomes of Bangiophyceae, Compsopogonophyceae, Stylonematophyceae, most florideophycean species, and some Cyanidiophyceae (Cyanidium caldarium, Cyanidiococcus yangmingshanensis, as a pseudogene in Galdieria sulphuraria, missing in Cyanidioschyzon merolae), but missing in other rhodophycean algal groups (e.g., Porphyridiophyceae and Rhodellophyceae). In glaucophytes, the gene is present in Glaucocystis spp. but missing in studied Cyanophora species. The tsg1 gene has also been identified in the “chromatophore” genome of the rhizarian testate amoeba Paulinella (the chromatophore is a photosynthetic organelle that evolved independent of canonical plastids; Macorano and Nowack, 2021).
Together with tsg1, trg1 (transcriptional response regulator gene 1, annotated as trg1, ycf27, orf27, or ompR) was found in five raphidophycean species, but missing in Ha. pauciplastida and F. japonica, which also lack tsg1 (Figure 4A). Beyond the raphidophytes, the distribution of trg1 is varied among disparate taxa, but generally present more often than is the tsg1 gene in red alga-derived secondary/tertiary plastids. As with tsg1, the trg1 gene is missing in other stramenopiles. The raphidophycean tsg1 gene/protein appears more closely related to homologs in Cyanidiophyceae and Stylonematophyceae (Rhodophyta) than to Cryptophyta and Haptophyta, although the phylogenetic tree topology is not robust in this regard (Figure 4B; Supplementary Figure S3).
The presence–absence patterns of tsg1 and trg1 in sequenced plastid genomes is complex. The presumptive loss of the sensor kinase in some plastid genomes suggests that under such circumstances, the regulatory protein may be governed by one or more nuclear-encoded sensor kinases or by as-yet undescribed accessory proteins that are either of nuclear or plastid origin. Given that most of the raphidophyte plastid genomes sequenced herein encode a single response regulator and its cognate sensor kinase, but other stramenopile plastid genomes lack both genes, the extent to which nuclear genes participate in this signal transduction system in stramenopiles is unclear.
The serC gene encodes a phosphoserine aminotransferase, which in algae has thus far only been found in red alga-derived secondary plastid genomes, specifically those of diatoms (Figure 5A; Brembu et al., 2014; Ruck et al., 2014; Hamsher et al., 2019; Li et al., 2019; Gastineau et al., 2021). Interestingly, the diatom plastid serC has strong similarity to plasmid DNAs of several diatoms (e.g., Cylindrotheca fusiformis and Haslea sp.; Figure 5A, blue) and the diatom-derived plastid genome of the “dinotom” Kryptoperidinium foliaceum (Figure 5A, orange-brown). Unexpectedly, we found a serC homolog in the plastid genome of the marine raphidophyte F. japonica that in a protein phylogeny branches specifically with the plasmid-borne sequences of C. fusiformis and C. closterium and a plastid homolog in Climaconeis cf. scalaris. Fibrocapsa japonica also has a “serC2-like” gene with very high similarity to the plastid serC2 of the diatom Plagiogrammopsis vanheurckii and one of four serC homologs in Schizostauron trachyderma (Figure 5A). One explanation for the presence of these genes in F. japonica is that they were acquired recently by LGT from a diatom plastid and/or plasmid DNA, perhaps by mixotrophic feeding. This scenario may also explain the existence of the serC homolog in K. foliaceum. From where exactly the plasmid and plastid serC homologs of diatoms originated is unclear; these sequences are embedded within the plastid and plasmid clade, very distinct from the bacterial homologs (e-value cutoff = 1e−5, word size = 6) used to root the serC1 / serC2 tree (Figure 5A; Supplementary Figure S4).
The tyrC gene encodes a putative site-specific tyrosine recombinase in the He. akashiwo and F. japonica plastid genomes (Figures 3, 5B). The tyrC gene has also been found in other diatoms, “dinotoms,” green algae, and select bacteria; in phylogenetic analyses, the raphidophyte homologs are more similar to sequences in pennate diatoms than to green algal homologs (Figure 5B). As is the case for serC, the presence of tyrC in the He. akashiwo and F. japonica genomes (as well as a tyrC pseudogene in F. japonica) is consistent with LGT from a diatom, though it is worth noting that the He. akashiwo and F. japonica sequences are not monophyletic. In our phylogenies, two He. akashiwo tyrC homologs are nested deeply within a strongly supported clade of diatom sequences (and the “dinotom” of K. foliaceum), far away from two F. japonica sequences, which are themselves not monophyletic; some or all of these sequences may have been acquired independent of one another. Ulva fasciata (Chlorophyta), Netrium digitus (Streptophyta), and various other green algae possess tyrC genes in their plastid genomes (Figure 5B; Brouard et al., 2008; Civáň et al., 2014). In bacteria, xerC/D family tyrosine recombinases with a lower degree of sequence similarity (e-value cutoff = 1e−5, word size = 6) to tyrC are found primarily in Firmicutes. The specific evolutionary connections between the raphidophyte, diatom, and green algal tyrC homologs to one another and to bacterial xerC/D are unclear.
Further evidence for the role of LGT in the evolution of raphidophyte plastid genomes comes from our discovery of a group II intron in the psbB gene of F. japonica, a feature not described in raphidophytes thus far (Figure 3). Interestingly, the intron contains an intron-encoded protein (IEP) with a reverse transcriptase (RT) domain that is clearly related to IEPs in other red alga-derived plastid genomes, such as those in the group II introns of the dnaK, psaA and trnM genes of rhodophytes, chlB, psaA, and psbB in cryptophytes, in the psaJ gene of the green alga-derived plastid genome of euglenoids, as well as various plastid and/or mitochondrial genes in other stramenopiles, fungi, and rhizarians (Supplementary Figure S5). This is consistent with earlier studies documenting the patchy distribution of group II introns in cyanobacteria and diverse eukaryotes (e.g., Odom et al., 2004; Sheveleva and Hallick, 2004; Khan and Archibald, 2008; Kim et al., 2022; Suzuki et al., 2022). While our phylogenetic analyses failed to identify an obvious donor of the F. japonica psbB intron, it is noteworthy that previous investigation of the mitochondrial genome of the raphidophyte Chattonella marina identified a putative LGT of a group II intron from diatoms to raphidophytes (Kamikawa et al., 2009). More specifically, phylogenetic analysis of the RT domain of the mitochondrial cox1 IEP in C. marina showed that it clustered with cox1 IEPs of the diatom Thalassiosira pseudonana and the pennate diatom obligate endosymbiont in the dinoflagellate Kryptoperidinium foliaceum. We also discovered a 2,472 nt intron found only in the clpC gene of the marine raphidophycean species Ha. pauciplastida, but its origin is unknown as it does not contain an obvious and analyzable IEP (the intron nucleotide sequence shows no obvious significant similarity to known introns). Detailed speculation on the frequency and directionality of group II intron transfers in eukaryotes is beyond the scope of our study. But we should emphasize that mitochondrial IEPs, such as those analyzed by Kamikawa et al. (2009) and Kim et al. (2022), were only rarely retrieved in our BLAST sequence searches (e-value cutoff = 1e−5, word size = 6) and are thus poorly represented in the RT / IEP phylogenetic analyses presented herein (the F. japonica plastid psbB intron and the C. marina mitochondrial cox1 IEP are too distant from one another to allow for meaningful comparison). All these caveats aside, our results are consistent in showing intriguing connections between mobile genetic elements in diatoms and marine raphidophytes.
Examples of LGT-derived plastid genes and introns in algae are increasingly well known, such as in rhodophytes (Janouškovec et al., 2010), cryptophytes (Khan and Archibald, 2008; Kim et al., 2017; Suzuki et al., 2022), haptophytes (Rice and Palmer, 2006), chrysophytes (Kim et al., 2019), diatoms (Ruck et al., 2014), eustigmatophytes (Yurchenko et al., 2016), and the raphidophyte He. akashiwo (Cattolico et al., 2008). Curiously, some of these foreign sequences appear to have been acquired from plasmid DNAs, portions of which are found inserted into the plastid genomes of several diatoms and encode putatively functional site-specific recombinase genes (Brembu et al., 2014; Ruck et al., 2014; Gastineau et al., 2021). They are also found in “dinotoms” (Imanian et al., 2010), raphidophytes (Cattolico et al., 2008), and red algae (Lee et al., 2016). Further investigation is required to determine if the plastid-encoded DNA recombinases in disparate algal lineages are actually involved in site-specific recombination and integration of foreign sequences.
The six gene-poor regions in the F. japonica plastid genome sequenced in our study were found to contain several hypothetical ORFs derived from various pennate diatom plastids or plasmid DNAs (Figure 2). In fact, hypothetical ORFs with similarity (e-value cutoff = 1e−5, word size = 6) to ORFs in the plasmid DNAs of the diatoms Cylindrotheca closterium, Cylindrotheca fusiformis, Seminavis robusta, and Haslea species are found not just in the plastid genome of F. japonica, but He. akashiwo and Ha. pauciplastida as well (Figures 6A–C). These sequences are also homologous to predicted genes in the plastids of some chlorophyte green algae (Figure 6A), the “dinotom” K. foliaceum (Figures 6A,C), and haptophytes (Figures 6C,D). Two hypothetical ORFs of F. japonica share homology with pennate diatom plastid sequences: orf136 is homologous to Seminavis robusta orf140 (e-value = 8.96e−25) and Toxarium undulatum orf127 (e-value = 6.92e−26), and orf153 is homologous to Asterionellopsis glacialis orf126 (e-value = 1.95e−02). How the plastid genomes of these disparate algae have come to contain these sequences is unclear, but it seems likely that plasmid-mediated gene exchange was somehow involved.
Raphidophyceae are mixotrophs that photosynthesize as well as feed on diverse bacteria and organic nutrients in freshwater and marine environments. The marine raphidophytes Chattonella spp., He. akashiwo, and Fibrocapsa spp. feed on heterotrophic and autotrophic bacteria by capturing prey cells/microspheres in mucus excreted by mucocysts (Jeong, 2011). We have shown that the freshwater raphidophycean taxa have small plastid genomes relative to those of marine species, and the comparative genomic and phylogenetic analyses carried out here and elsewhere (Cattolico et al., 2008) suggest a link between mixotrophy, plastid genome expansion, and LGT. How and how often LGT has occurred is still unclear. Nevertheless, based on our results we propose a model of raphidophycean plastid evolution (Figure 7). The red alga-type plastid of Raphidophyceae stems from a secondary or possibly tertiary endosymbiotic event in an ancestor shared with other plastid-bearing stramenopiles (Figure 7A; Supplementary Figure S1; Kim et al., 2017, 2019, 2020; Sibbald and Archibald, 2020). The presence and distribution of tsg1 and trg1 genes is consistent with tertiary endosymbiotic linkages between raphidophytes, haptophytes, and cryptophytes, although the most likely donor(s) and recipients are not clear from these data alone (Figure 4A). After the diversification of Raphidophyceae, some mixotrophic marine species appear to have taken up mobile elements in plastid and/or plasmid DNAs by feeding on eukaryotes, such as pennate diatoms, with genes being transferred to their plastid genomes (Figure 7B). Plastid genome expansion and gene rearrangement thus occurred as a result of—and/or was facilitated by—acquisition of foreign genetic material by LGT. This pattern has been detected in the “dinotom” Kryptoperidinium and Durinskia species (Imanian et al., 2010). As noted above, the evolution of the raphidophyte LGT-derived tyrC genes in He. akashiwo and F. japonica is unclear. Some of these genes could conceivably could have been acquired directly by feeding on tiny green algal cells, or indirectly from other sources (e.g., diatoms; Figure 7C). Together with LGT-associated plastid genome expansion in marine mixotrophic species, contraction of the IR region in the plastid genome of the freshwater species G. semen represents an example of genome reduction “in action,” which is also consistent with our model.
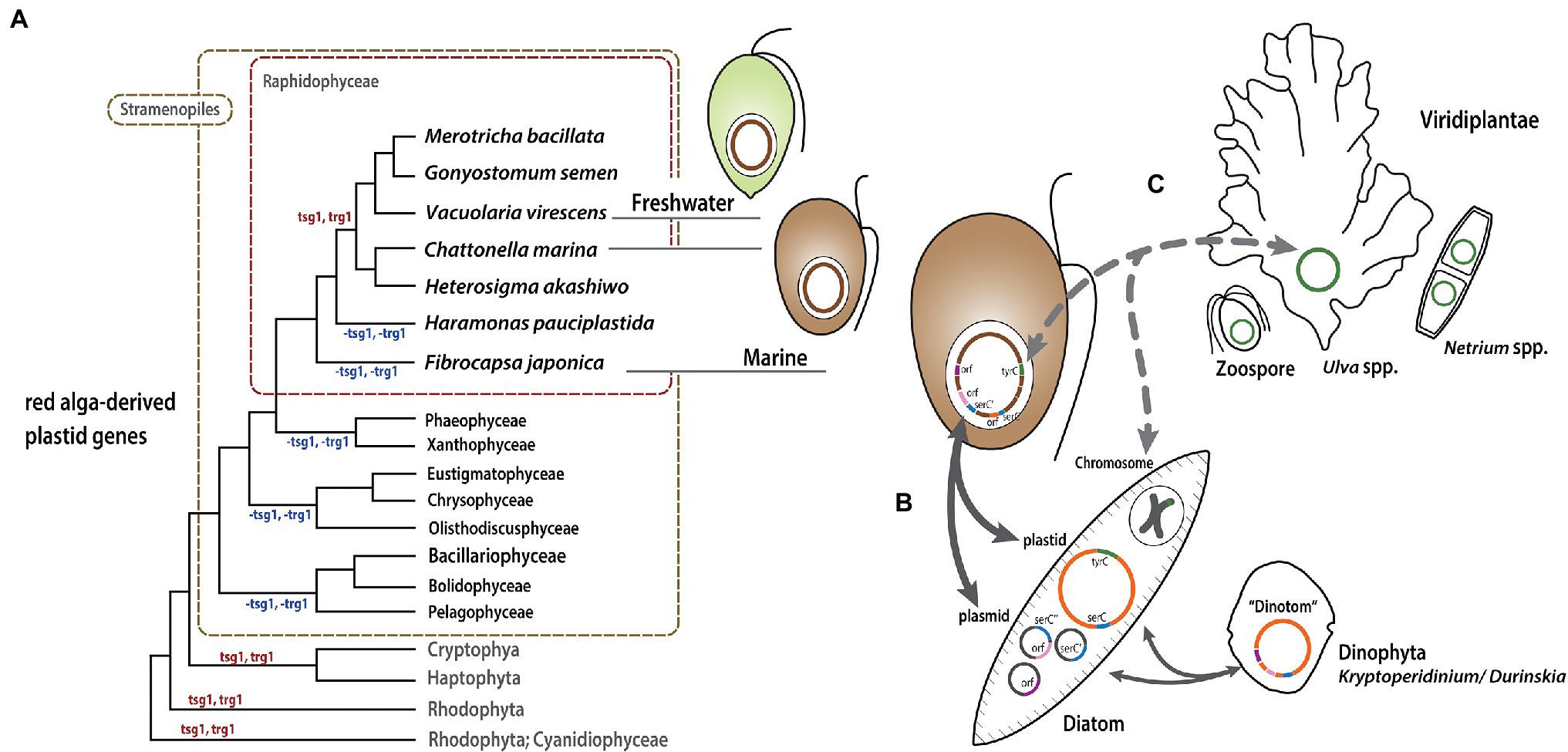
Figure 7. Model of Raphidophyceae plastid evolution. Putative gene gain/loss and LGT events are mapped onto a schematic tree (A) modified from the phylogeny inferred from plastid proteins shown in Supplementary Figure S1. (B) and (C) show possible routes of gene exchange between the organisms described in the text.
Analysis of six newly sequenced plastid genomes from Raphidophyceae has provided insight into organellar genome dynamics. The raphidophycean lineages evolved from a common ancestor shared with diatoms and other red alga plastid-bearing stramenopiles, with LGT having increased the coding capacity of mixotrophic species on multiple occasions during raphidophycean algal evolution. While the extent to which LGT has contributed to the plastid genomes of Raphidophyceae and other algae remains to be seen, our results indicate that plasmids may play an important role. Our understanding of the diversity and biology of such mobile elements in eukaryotes is still very limited, and the discovery and characterization of plasmids in diverse algae will hopefully provide the data with which to test this hypothesis.
The datasets presented in this study can be found in GenBank under the following accession numbers: ON228255–ON228260.
JIK and WS conceived and designed the experiments. JIK, BYJ, MGP, and YDY isolated cells and provided cultures. JIK performed the experiments and analyzed the data. JIK, WS, and JMA interpreted the data and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by funds from the National Research Foundation (NRF) of Korea awarded to JIK (NRF-2018R1D1A1B07050727 and 2021R1C1C2012996), BYJ (NRF-2014R1A1A3052827), MGP (NRF-2016R1A6A1A03012647), YDY (NRF-2021M 3I6A1091272), and WS (NRF-2019R1I1A2A01063159), and a Natural Sciences and Engineering Research Council of Canada grant (RGPIN-2019-05058) awarded to JMA.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank reviewers for their helpful comments on an earlier version of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.896138/full#supplementary-material
Supplementary Figure S1 | Phylogenetic tree of raphidophyte plastids. This tree was constructed using a dataset of 91 plastid genes (18,216 amino acids) from 135 taxa. The numbers on each node represent ultrafast bootstrap approximation (UFBoot) values calculated using IQ-Tree. The scale bar indicates the number of amino acid substitutions per site.
Supplementary Figure S2 | Circular map of newly sequenced plastid genomes of Raphidophyceae. The lineage-specific gene gains are labeled inside or outside of the circles.
Supplementary Figure S3 | (A) Expanded phylogenetic tree for Figure 4B based on tsg1 amino acid sequences. The tree was constructed using a dataset assembled by BLASTp (e-value cutoff = 1e−5) against the NCBI non-redundant database using the raphidophyte homologs as queries. (B) Protein alignment of homologs used for phylogenetic reconstruction.
Supplementary Figure S4 | (A) Expanded phylogenetic tree for Figure 5B based on tyrC amino acid sequences from F. japonica and detectable homologs in other organisms. The dataset was assembled using sequences retrieved by BLASTp (e-value cutoff = 1e−5) against the NCBI non-redundant database using raphidophyte homologs as queries. (B,C) Aligned amino acid sequences of serC and tyrC, respectively.
Supplementary Figure S5 | Phylogenetic tree based on the reverse transcriptase homologs of the intron-encoded protein in the psbB group II intron of F. japonica. The location of the intron is shown after the gene names [e.g., rnl-i1 = intron 1 in rnl or psbB (with single intron IEP)]. The tree was constructed using a dataset of homologs retreived by BLASTp (e-value cutoff = 1e−5) from the NCBI non-redundant database using the F. japonica psbB group II intron IEP as a query. Numbers (Bootstrap values > 50%) on branches are IQ-Tree UFBoot. The scale bar shows the inferred number of amino acid substitutions per site.
1. ^http://bioinf.spbau.ru/spades
2. ^http://exon.gatech.edu/genemark/genemarks.cgi
4. ^http://lowelab.ucsc.edu/tRNAscan-SE/
5. ^http://megasun.bch.umontreal.ca/cgi-bin/RNAweasel/RNAweaselInterface.pl
Andersen, R. A., Berges, J. A., Harrison, P. J., and Watanabe, M. M. (2005). “Appendix A. recipes for freshwater and seawater media,” in Algal Culturing Techniques. ed. R. A. Andersen (United States: Elsevier Academic Press), 429–538.
Barcytė, D., Eikrem, W., Engesmo, A., Seoane, S., Wohlmann, J., Horák, A., et al. (2021). Olisthodiscus represents a new class of Ochrophyta. J. Phycol. 57, 1094–1118. doi: 10.1111/jpy.13155
Bjørnland, T., and Liaaen-Jensen, S. (1989). “Distribution patterns of carotenoids in relation to chromophyte phylogeny and systematics,” in The Chromophyte Algae: Problems and Perspectives. eds. J. C. Green, B. S. C. Leadbeater, and W. L. Diver (Oxford: Clarendon Press), 37–60.
Brembu, T., Winge, P., Tooming-Klunderud, A., Nederbragt, A. J., Jakobsen, K. S., and Bones, A. M. (2014). The chloroplast genome of the diatom Seminavis robusta: new features introduced through multiple mechanisms of horizontal gene transfer. Mar. Genomics 16, 17–27. doi: 10.1016/j.margen.2013.12.002
Brouard, J.-S., Otis, C., Lemieux, C., and Turmel, M. (2008). Chloroplast DNA sequence of the green alga Oedogonium cardiacum (Chlorophyceae): unique genome architecture, derived characters shared with the Chaetophorales and novel genes acquired through horizontal transfer. BMC Genomics 9:290. doi: 10.1186/1471-2164-9-290
Cattolico, R. A., Jacobs, M. A., Zhou, Y., Chang, J., Duplessis, M., Lybrand, T., et al. (2008). Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics 9:211. doi: 10.1186/1471-2164-9-211
Civáň, P., Foster, P. G., Embley, M. T., Séneca, A., and Cox, C. J. (2014). Analyses of charophyte chloroplast genomes help characterize the ancestral chloroplast genome of land plants. Genome Biol. Evol. 6, 897–911. doi: 10.1093/gbe/evu061
Darling, A. C., Mau, B., Blattner, F. R., and Perna, N. T. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Duplessis, M. R., Karol, K. G., Adman, E. T., Choi, L. Y., Jacobs, M. A., and Cattolico, R. A. (2007). Chloroplast his-to-asp signal transduction: a potential mechanism for plastid gene regulation in Heterosigma akashiwo (Raphidophyceae). BMC Evol. Biol. 7:70. doi: 10.1186/1471-2148-7-70
Ejigu, G. F., Yi, G., Kim, J. I., and Jung, J. (2021). ReGSP: a visualized application for homology-based gene searching and plotting using multiple reference sequences. PeerJ 9:e12707. doi: 10.7717/peerj.12707
Fiksdahl, A., Withers, N., Guillard, R. R. L., and Liaaen-Jensen, S. (1984). Carotenoids of the Raphidophyceae–a chemosystematic contribution. Comp. Biochem. Physiol.Biochem. Mol. Biol. 78, 265–271. doi: 10.1016/0305-0491(84)90181-0
Fong, A., and Archibald, J. M. (2008). Evolutionary dynamics of light-independent protochlorophyllide oxidoreductase genes in the secondary plastids of cryptophyte algae. Eukaryot. Cell 7, 550–553. doi: 10.1128/EC.00396-07
Fott, B. (1968). “Chloromonadophyceae,” in Das Phytoplankton des Süßwassers. ed. G. Huber-Pestalozzi (Stuttgart: E. Schweizerbart’sche Verlagsbuchandlung), 79–93.
Fujita, Y., and Bauer, C. E. (2003). “The light-dependent protochlorophyllide reductase: a nitrogenase-like enzyme catalyzing a key reaction for greening in the dark,” in The Porphyrin Handbook. Vol. 13. eds. K. Kadish, K. Smith, and R. Guilard (San Diego: Elsevier Science), 109–156.
Gastineau, R., Hansen, G., Poulin, M., Lemieux, C., Turmel, M., Bardeau, J. F., et al. (2021). Haslea silbo, A novel cosmopolitan species of blue diatoms. Biology 10:328. doi: 10.3390/biology10040328
Goulding, S. E., Olmstead, R. G., Morden, C. W., and Wolfe, K. H. (1996). Ebb and flow of the chloroplast inverted repeat. Mol. Gen. Genet. 252, 195–206. doi: 10.1007/BF02173220
Hamsher, S. E., Keepers, K. G., Pogoda, C. S., Stepanek, J. G., Kane, N. C., and Kociolek, J. P. (2019). Extensive chloroplast genome rearrangement amongst three closely related Halamphora spp. (Bacillariophyceae), and evidence for rapid evolution as compared to land plants. PLoS One 14:e0217824. doi: 10.1371/journal.pone.0217824
Han, K. Y., Maciszewski, K., Graf, L., Yang, J. H., Andersen, R. A., Karnkowska, A., et al. (2019). Dictyochophyceae plastid genomes reveal unusual variability in their organization. J. Phycol. 55, 1166–1180. doi: 10.1111/jpy.12904
Hara, Y., and Chihara, M. (1982). Ultrastructure and taxonomy of Chattonella (class Raphidophyceae) in Japan. Jap. J. Phycol. 30, 47–56.
Heywood, P. (1982). “Raphidophyceae (Chloromonadophyceae): introduction and bibliography,” in Selected Papers in Phycology. eds. J. R. Rosowski and B. C. Parker (Kansas: Phycological Society of America (Book Division), Lawrence), 719–722.
Heywood, P. (1990). “Raphidophyceae,” in Handbook of Protoctists. eds. L. Margulis, J. O. Corliss, M. Melkonian, and D. J. Chapman (Boston, MA: Jones and Bartlett Publishers), 318–325.
Horiguchi, T. (1996). Haramonas dimorpha gen. Et sp. nov. (Raphidophyceae), a new marine raphidophyte from Australian mangrove. Phycol. Res. 44, 143–150. doi: 10.1111/j.1440-1835.1996.tb00043.x
Hunsperger, H. M., Randhawa, T., and Cattolico, R. A. (2015). Extensive horizontal gene transfer, duplication, and loss of chlorophyll synthesis genes in the algae. BMC Evol. Biol. 15:16. doi: 10.1186/s12862-015-0286-4
Imanian, B., Pombert, J.-F., and Keeling, P. J. (2010). The complete plastid genomes of the two ‘Dinotoms’ Durinskia baltica and Kryptoperidinium foliaceum. PLoS One 5:e10711. doi: 10.1371/journal.pone.0010711
Janouškovec, J., Horák, A., Oborník, M., Lukeš, J., and Keeling, P. J. (2010). A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. U. S. A. 107, 10949–10954. doi: 10.1073/pnas.1003335107
Jansen, R. K., and Ruhlman, T. A. (2012). “Plastid genomes of seed plants,” in Genomics of Chloroplasts and Mitochondria. eds. R. Bock and V. Knoop (Germany: Springer), 103–126.
Jeong, H. J. (2011). Mixotrophy in red tide algae raphidophytes. J. Eukaryot. Microbiol. 58, 215–222. doi: 10.1111/j.1550-7408.2011.00550.x
Jung, J., Kim, J. I., Jeong, Y.-S., and Yi, G. (2018). AGORA: organellar genome annotation from the amino acid and nucleotide references. Bioinformatics 34, 2661–2663. doi: 10.1093/bioinformatics/bty196
Jung, J., Kim, J. I., and Yi, G. (2019). geneCo: a visualized comparative genomic method to analyze multiple genome structures. Bioinformatics 35, 5303–5305. doi: 10.1093/bioinformatics/btz596
Kamikawa, R., Masuda, I., Demura, M., Oyama, K., Yoshimatsu, S., Kawachi, M., et al. (2009). Mitochondrial group II introns in the raphidophycean flagellate Chattonella spp. suggest a diatom-to-Chattonella lateral group II intron transfer. Protist 160, 364–375. doi: 10.1016/j.protis.2009.02.003
Khan, H., and Archibald, J. M. (2008). Lateral transfer of introns in the cryptophyte plastid genome. Nucleic Acids Res. 36, 3043–3053. doi: 10.1093/nar/gkn095
Kim, J. I., Jeong, M., Archibald, J. M., and Shin, W. (2020). Comparative plastid genomics of non-photosynthetic Chrysophytes: genome reduction and compaction. Front. Plant Sci. 11:572703. doi: 10.3389/fpls.2020.572703
Kim, D., Lee, J., Cho, C. H., Kim, E. J., Bhattacharya, D., and Yoon, H. S. (2022). Group II intron and repeat-rich red algal mitochondrial genomes demonstrate the dynamic recent history of autocatalytic RNAs. BMC Biol. 20:2. doi: 10.1186/s12915-021-01200-3
Kim, J. I., Moore, C. E., Archibald, J. M., Bhattacharya, D., Yi, G., Yoon, H. S., et al. (2017). Evolutionary dynamics of cryptophyte plastid genomes. Genome Biol. Evol. 9, 1859–1872. doi: 10.1093/gbe/evx123
Kim, J. I., Shin, H., Škaloud, P., Jung, J., Yoon, H. S., Archibald, J. M., et al. (2019). Comparative plastid genomics of Synurophyceae: inverted repeat dynamics and gene content variation. BMC Evol. Biol. 19:20. doi: 10.1186/s12862-018-1316-9
Lee, J., Kim, K. M., Yang, E. C., Miller, K. A., Boo, S. M., and Bhattacharya, D. (2016). Reconstructing the complex evolutionary history of mobile plasmids in red algal genomes. Sci. Rep. 6:23744. doi: 10.1038/srep23744
Li, C., Gastineau, R., Turmel, M., Witkowski, A., Otis, C., Car, A., et al. (2019). Complete chloroplast genome of the tiny marine diatom Nanofrustulum shiloi (Bacillariophyta) from the Adriatic Sea. Mitochondrial DNA Part B. 4, 3374–3376. doi: 10.1080/23802359.2019.1673245
Macorano, L., and Nowack, E. C. M. (2021). Paulinella chromatophora. Curr. Biol. 31, R1024–R1026. doi: 10.1016/j.cub.2021.07.028
Muraki, N., Nomata, J., Ebata, K., Mizoguchi, T., Shiba, T., Tamiaki, H., et al. (2010). X-ray crystal structure of the light-independent protochlorophyllide reductase. Nature 465, 110–114. doi: 10.1038/nature08950
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Odom, O. W., Shenkenberg, D. L., Garcia, J. A., and Herrin, D. L. (2004). A horizontally acquired group II intron in the chloroplast psbA gene of a psychrophilic Chlamydomonas: in vitro self-splicing and genetic evidence for maturase activity. RNA 10, 1097–1107. doi: 10.1261/rna.7140604
Ong, H. C., Wilhelm, S. W., Gobler, C. J., Bullerjahn, G., Jacobs, M. A., McKay, J., et al. (2010). Analyses of the complete chloroplast genome sequences of two members of the pelagophyceae: Aureococcus anophagefferens CCMP1984 and Aureoumbra lagunensis CCMP1507. J. Phycol. 46, 602–615. doi: 10.1111/j.1529-8817.2010.00841.x
Rice, D. W., and Palmer, J. D. (2006). An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are siters. BMC Biol. 4:31. doi: 10.1186/1741-7007-4-31
Ruck, E. C., Nakov, T., Jansen, R. K., Theriot, E. C., and Alverson, A. J. (2014). Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol. Evol. 6, 644–654. doi: 10.1093/gbe/evu039
Sabir, J. S. M., Yu, M., Ashworth, M. P., Baeshen, N. A., Baeshen, M. N., Bahieldin, A., et al. (2014). Conserved gene order and expanded inverted repeats characterize plastid genomes of Thalassiosirales. PLoS One 9:e107854. doi: 10.1371/journal.pone.0107854
Sassenhagen, I., and Rengefors, K. (2019). Sequencing and phylogenetic analysis of chloroplast genes in freshwater raphidophytes. Genes 10:245. doi: 10.3390/genes10030245
Ševčíková, T., Horák, A., Klimeš, V., Zbránková, V., Demir-Hilton, E., Sudek, S., et al. (2015). Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 5:10134. doi: 10.1038/srep10134
Sheveleva, E. V., and Hallick, R. B. (2004). Recent horizontal intron transfer to a chloroplast genome. Nucleic Acids Res. 32, 803–810. doi: 10.1093/nar/gkh225
Shi, C., and Shi, X. (2006). Characterization of three genes encoding the subunits of light-independent protochlorophyllide reductase in Chlorella protothecoides CS-41. Biotechnol. Prog. 22, 1050–1055. doi: 10.1021/bp060087d
Sibbald, S., and Archibald, J. M. (2020). Genomic insights into plastid evolution. Genome Biol. Evol. 12, 978–990. doi: 10.1093/gbe/evaa096
Smith, S. W., Overbeek, R., Woese, C. R., Gilbert, W., and Gillevet, P. M. (1994). The genetic data environment an expandable GUI for multiple sequence analysis. Comput. Appl. Biosci. 10, 671–675. doi: 10.1093/bioinformatics/10.6.671
Suzuki, S., Matsuzaki, R., Yamaguchi, H., and Kawachi, M. (2022). What happened before losses of photosynthesis in cryptophyte algae? Mol. Biol. Evol. 39:msac001. doi: 10.1093/molbev/msac001
Turmel, M., Otis, C., and Lemieux, C. (2015). Dynamic evolution of the chloroplast genome in the green algal classes Pedinophyceae and Trebouxiophyceae. Genome Biol. Evol. 7, 2062–2082. doi: 10.1093/gbe/evv130
Wang, R. J., Cheng, C. L., Chang, C. C., Wu, C. L., Su, T. M., and Chaw, S. M. (2008). Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 8:36. doi: 10.1186/1471-2148-8-36
Keywords: genome reduction, lateral gene transfer, mosaicism, plastid genome, Raphidophyceae
Citation: Kim JI, Jo BY, Park MG, Yoo YD, Shin W and Archibald JM (2022) Evolutionary Dynamics and Lateral Gene Transfer in Raphidophyceae Plastid Genomes. Front. Plant Sci. 13:896138. doi: 10.3389/fpls.2022.896138
Received: 14 March 2022; Accepted: 09 May 2022;
Published: 26 May 2022.
Edited by:
Miroslav Obornik, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Richard Dorrell, École Normale Supérieure, FranceCopyright © 2022 Kim, Jo, Park, Yoo, Shin and Archibald. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woongghi Shin, c2hpbndAY251LmFjLmty; John M. Archibald, Sm9obi5BcmNoaWJhbGRAZGFsLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.