- 1State Key Laboratory of North China Crop Improvement and Regulation, College of Plant Protection, Hebei Agricultural University, Baoding, China
- 2Hubei Hongshan Laboratory, College of Plant Science & Technology, Huazhong Agricultural University, Wuhan, China
- 3International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico
- 4Campo Experimental Valle de México the National Institute of Forestry, Agricultural and Livestock Research (INIFAP), Texcoco, Mexico
- 5Commonwealth Scientific and Industrial Research Organisation (CSIRO) Plant Industry, Canberra, ACT, Australia
Leaf rust and stripe rust are important wheat diseases worldwide causing significant losses where susceptible varieties are grown. Resistant cultivars offer long-term control and reduce the use of hazardous chemicals, which can be detrimental to both human health and the environment. Land races have been a valuable resource for mining new genes for various abiotic and biotic stresses including wheat rusts. Afghan wheat landrace “KU3067” displayed high seedling infection type (IT) for leaf rust and low IT for stripe rust; however, it displayed high levels of field resistance for both rusts when tested for multiple seasons against the Mexican rust isolates. This study focused on identifying loci-conferring seedling resistance to stripe rust, and also loci-conferring adult plant resistance (APR) against the Mexican races of leaf rust and stripe rust. A backcrossed inbred line (BIL) population advanced to the BC1F5 generation derived from the cross of KU3067 and Apav (triple rust susceptible line) was used for both, inheritance and QTL mapping studies. The population and parents were genotyped with Diversity Arrays Technology-genotyping-by-sequencing (DArT-Seq) and phenotyped for leaf rust and stripe rust response at both seedling and adult plant stages during multiple seasons in Mexico with relevant pathotypes. Mapping results identified an all-stage resistance gene for stripe rust, temporarily designated as YrKU, on chromosome 7BL. In total, six QTL-conferring APR to leaf rust on 1AS, 2AL, 4DL, 6BL, 7AL, and 7BL, and four QTL for stripe rust resistance on 1BS, 2AL, 4DL, and 7BL were detected in the analyses. Among these, pleiotropic gene Lr67/Yr46 on 4DL with a significantly large effect is the first report in an Afghan landrace-conferring resistance to both leaf and stripe rusts. QLr.cim-7BL/YrKU showed pleiotropic resistance to both rusts and explained 7.5–17.2 and 12.6–19.3% of the phenotypic variance for leaf and stripe rusts, respectively. QYr.cim-1BS and QYr.cim-2AL detected in all stripe environments with phenotypic variance explained (PVE) 12.9–20.5 and 5.4–12.5%, and QLr.cim-6BL are likely to be new. These QTL and their closely linked markers will be useful for fine mapping and marker-assisted selection (MAS) in breeding for durable resistance to multiple rust diseases.
Introduction
Wheat leaf rust and stripe rust caused by obligate biotrophic fungus Puccinia triticina (Pt) and P. striiformis f. sp. tritici (Pst), respectively, are the most important foliar diseases of wheat worldwide. Leaf rust is the most commonly occurring disease and can cause yield losses up to 40% under favorable conditions (Knott, 1989). Stripe rust occurs in cool temperate regions and can cause yield losses ranging from 10 to 70% and up to 100% in highly susceptible cultivars (Chen, 2005). Although fungicides can effectively control wheat rusts, growing resistant cultivars is a more efficient, economic, environment-friendly, and long-term strategy to minimize losses.
Resistance to wheat rusts can be broadly classified either as race-specific or as race non-specific resistances (Johnson, 1988). Race-specific resistance (or) seedling resistance (or) all-stage resistance is often characterized by a strong to moderate immune response usually associated with the hypersensitive response that fully curtails fungal infection and sporulation at all developmental stages if the pathogen possesses a corresponding avirulence gene (Flor, 1942). These resistance genes are usually effective against a single race or a few races of the pathogen. This kind of resistance may lose effectiveness when new virulent pathotypes arise through mechanisms of mutation or recombination. Race non-specific resistance on the other hand is under polygenic control and usually expressed susceptible response at the seedling stage and expressed quantitatively at post-seedling growth stages either as slow rusting or partial resistance, or adult plant resistance (APR). It is usually characterized by lower frequencies of infection, longer latency period, smaller uredinium, and less urediniospore production (Caldwell, 1968). The phenotypic effects of such genes are usually minor; however, several of such genes with additive effects can be combined leading to near levels of immunity (Singh et al., 2012). These genes usually are race non-specific and are effective against multiple races of the pathogen and compared with race-specific resistance, therefore, conditioning broader effectiveness and enhanced durability.
To date, 80 leaf rust resistance genes and 83 stripe rust resistance genes are officially cataloged in wheat (Li et al., 2020; Kumar et al., 2021) and most of the identified genes showed race-specific resistance. Only a few genes, for example, Lr34/Yr18/Pm38/Sr57 (Singh et al., 2012), Lr46/Yr29/Pm39/Sr58 (Singh et al., 2013), Lr67/Yr46/Pm46/Sr55 (Herrera-Foessel et al., 2011), and Lr68 (Herrera-Foessel et al., 2012) are also known to confer pleiotropic effect on the resistance. In addition to the formally named genes, 249 leaf rust and 327 stripe rust resistance QTL have been reported on every chromosome (Wang and Chen, 2017; Pinto da Silva et al., 2018). Even though multiple genes have been characterized and cataloged only a handful of genes, conferring adequate levels of resistance to prevalent rust races have made an impact in rust resistance breeding as the majority of them are effective to specific races (or) do not provide adequate levels of resistance (or) are associated with negative linkage drag with yield and other traits (Bhavani et al., 2019). The advent of molecular markers and the availability of reference maps and completely annotated wheat genome has greatly facilitated rapid gene discovery and characterization (Clavijo et al., 2017; Rosewarne et al., 2013).
Molecular markers have been widely used for mapping APR genes for rust resistance through QTL analysis. Several high-throughput genotyping technologies, such as single nucleotide polymorphism (SNP) and genotyping-by-sequencing (GBS) have greatly facilitated the identification and characterization of genomic regions of complex traits such as rust resistance (Bhavani et al., 2021). Diversity arrays technology sequencing (DArT-Seq) developed by the Diversity Arrays Technology Pty Ltd (Canberra, Australia) is a new approach based on traditional DArT complexity reduction methods and Next Generation Sequencing (NGS) techniques (http://www.diversityarrays.com/dart-application-dartseq).
Diversity arrays technology sequencing offers affordable genome profiling through the generation of high-density SNPs as well as PAV (presence/absence variation) markers. This technology can be better used in linkage map construction and accelerates high-resolution mapping and detailed genetic dissection of traits (Raman et al., 2014) and has been extensively used to study the diversity of wheat accessions in the CIMMYT gene bank (Sansaloni et al., 2020).
The wheat line “KU3067,” a landrace from Afghanistan, was stored in the National Bio-Resource Project of Japan in 1956 (Tanaka et al., 2008). It displays high levels of resistance to leaf rust and stripe rust in the Mexico's field conditions. This study aimed to determine the genetic basis of leaf rust, stripe rust resistance in KU3067, and identify molecular markers linked to these QTL that can be used in breeding.
Materials and Methods
Plant Materials and Pathotypes
The mapping population comprised of 148 BC1F5 lines (backcross-inbred lines, BILs) was derived from the cross of KU3067 and Apav (Apav was used as the recurrent susceptible parent). KU3067 showed a high level of APR to both rusts in field trials, whereas Apav derived from the “Avocet-YrA/Pavon 76” mapping population was completely susceptible to the three rusts. Predominant Mexican Pst pathotype MEX14.191 and two Pt pathotypes (MBJ/SP and MCJ/SP) were used to test the BILs in the greenhouse and field. These Pt and Pst pathotypes were available at the greenhouse at the International Maize and Wheat Improvement Center (CIMMYT).
Evaluation of Seedling Responses to Leaf and Stripe Rust in the Greenhouse
Seedling evaluations of KU3067, Apav and the BIL population were conducted in the greenhouse using Pst pathotype Mex14.191 (avirulence/virulence: Yr1, 4, 5a, 10, 15, 24, 26, 5b, Poll/Yr2, 3, 6, 7, 8, 9, 17, 27, 31, A) (Randhawa et al., 2018), and Pt pathotypes MBJ/SP [avirulence/virulence: Lr2a, 2b, 2c, 3ka, 9, 16, 19, 21, 24, 25, 28, 29, 30, 32, 33, 36/1, 3, 3bg, 10, 11, 12, 13, 14a, 14b, 15, 17a, 18, 20, 23, (26), 27+31] and MCJ/SP (additional virulence to Lr16) [avirulence/virulence: Lr2a, 2b, 2c, 3ka, 9, 19, 21, 24, 25, 28, 29, 30, 32, 33, 36/1, 3, 3bg, 10, 11, 12, 13, 14a, 14b, 15, 16,17a, 18, 20, 23, (26), 27+31] (Huerta-Espino et al., 2020). In total, thirty differential lines with known stripe rust resistance genes (mostly in the Avocet background) were also included in the seedling experiment, and a set of forty-eight lines with known LR genes, mostly in “Thatcher” background, were also included for leaf rust evaluations. Seedlings were inoculated at the two-leaf stage by spraying urediniospores suspended in the lightweight mineral oil Soltrol-170 (Chempoint.com) at a concentration of 2–3 mg/ml using an atomizer. Inoculated plants were placed in a dew chamber at 7°C for 18 h, and then transferred to the greenhouse maintained at 15–18°C. Infection types (ITs) were recorded 12–14 days after inoculation. Leaf rust was evaluated according to the Stakman 0 to 4 scale as modified by Roelfs et al. (1992); for stripe rust, ITs were recorded according to the 0 to 9 scale as modified by McNeal et al. (1971). BILs with ITs of 0–4, 5–6, and 7–9 to Pst are categorized as resistant, intermediate, and susceptible, respectively.
Field Experiments
Leaf rust field evaluations were carried out in CIMMYT (International Maize and Wheat Improvement Center) research stations in Mexico at El Batan (19.5277° N, 98.8569° W, and 2,249 masl) during the 2016 and 2017 cropping seasons where leaf rust screening can be carried out successfully (hereafter referred to as LR16B and LR17B, respectively), and at CENEB-Campo Experimental Norman E. Borlaug (27.3710° N, 109.9305° W, and 39 masl) located in Ciudad Obregon during the 2016–2017 and 2017–2018 growing seasons which is also the favorable environment for leaf rust (LR17O and LR18O). Stripe rust trials were conducted at Toluca which is a disease phenotyping platform for yellow rust and Septoria (19.5562° N, 99.2675° W, and 2,640 masl) Mexico, during the 2017 and 2018 crop seasons (YR17, YR18 experiments). Field plots consisted of 0.7-m paired rows with approximately 60 plants on each line. Avocet near-isolines (NILs) carrying Yr24 and Yr26 were used as spreaders in leaf rust studies, whereas a mixture of Morocco, Avocet NIL carrying Yr31, and six lines possessing the Yr27 gene, derived from the cross Avocet/Attila, were used as stripe rust spreaders. The spreader cultivars were planted as hills in the middle of a 0.3-m pathway on one side of each plot and planted perpendicular and adjacent to the test rows. Artificial inoculations on spreader rows and hills were carried out twice at weekly intervals by spraying aqueous suspensions of urediniospores of equal mixtures of Pt pathotypes MBJ/SP and MCJ/SP for leaf rust and Mex14.191 for stripe rust which were suspended in Soltrol 170 at a concentration of 2–3 mg/ml and dispensed onto the spreader rows at the tillering stage (Feekes growth stage 5; Large, 1954). The avirulence/virulence formulas of MBJ/SP and MCJ/SP were described in Herrera-Foessel et al. (2012) and of race Mex14.191 in Zhang et al. (2019). The host response to infection in adult plants was determined according to Roelfs et al. (1992). Disease severities were scored following the 0–100% visual ratings two or three times at weekly intervals according to the Modified Cobb Scale (Peterson et al., 1948). The area under the disease progress curve (AUDPC) was calculated using the method suggested by Bjarko and Line (1988).
where Xi is the disease severity on assessment date i, Ti is the number of days after inoculation on assessment date i, and n is the number of disease assessments. Maximum disease severity (MDS, %) data and AUDPC were used for QTL analysis.
Genetic and Statistical Analyses
Correlation analysis between MDS in different environments was conducted using bivariate two-tailed Pearson's correlation coefficients by IBM SPSS Statistics 21.0 (IBM Corp., Armonk, NY). The number of genes were estimated using chi-squared tests using the expected and observed segregation ratios. Based on disease severity and infection response, BILs were broadly classified into three phenotypic categories–homozygous parental type resistant (HPTR), homozygous parental type susceptible (HPTS), and intermediate types (others)–following Singh and Rajaram (1992). Chi-squared tests were carried out to test the best fit for different gene segregation ratios.
Linkage Map Construction and QTL Detection
Genomic DNA of the parents and BILs were isolated from non-infected tissues by the CTAB method (Sharp et al., 1988). DNA concentration was measured using Thermo Scientific NanoDrop 8000. The 148 BILs and parents were genotyped with DarT-Seq in CIMMYT's Biotech Laboratory. The linkage map was constructed using the MAP function in IciMapping 4.1 (http://www.isbreeding.net/software/?type=detail&id=18, Li et al., 2007). QTL analysis was conducted with the ICIM–ADD function in BIP using the software QTL IciMapping 4.1 through 1,000 permutations at P = 0.01 (Li et al., 2007). Stepwise regression analysis was used to detect the percentages of phenotypic variance explained (PVE, R2) by individual QTL and additive effects at the LOD peaks. The linkage map was drawn using MapChart 2.3 (http://www.earthatlas.mapchart.com/, Voorrips, 2002). The sequences of all the markers were subjected to the BLAST against the Chinese Spring reference sequence (version 2.0 https://urgi.versailles.inra.fr/blast_iwgsc/blast.php, IWGSC, 2018) in order to determine physical positions.
Phenotypic distributions of stripe rust and leaf rust MDS were also compared between two groups of BILs that were classified based on the presence or absence of a QTL. Finally, all the BILs were grouped into different QTL genotypic classes by the flanking markers to check the additive effect, and disease severity was calculated by averaging rust scores within a QTL group across environments.
Results
Seedling Response to Leaf Rust and Stripe Rust
In the seedling test, KU3067 and Apav displayed susceptible IT (3+ and 4) against both Pt races MBJ/SP and MCJ/SP based on a 0–4 scale. For the stripe rust, KU3067 expressed resistant IT 3 when tested with Pst Mex14.191, whereas Apav produced susceptible IT 7/8 based on the 0–9 scale. When tested on 148 BIL lines, 42 lines were found resistant, 101 lines were susceptible, and 6 lines showed segregation. Chi-squared analysis conformed to the expected frequency for a single stripe rust resistance gene in the seedling test against Pst Mex14.191 (Table 1). The resistance gene was temporarily designated as YrKU.
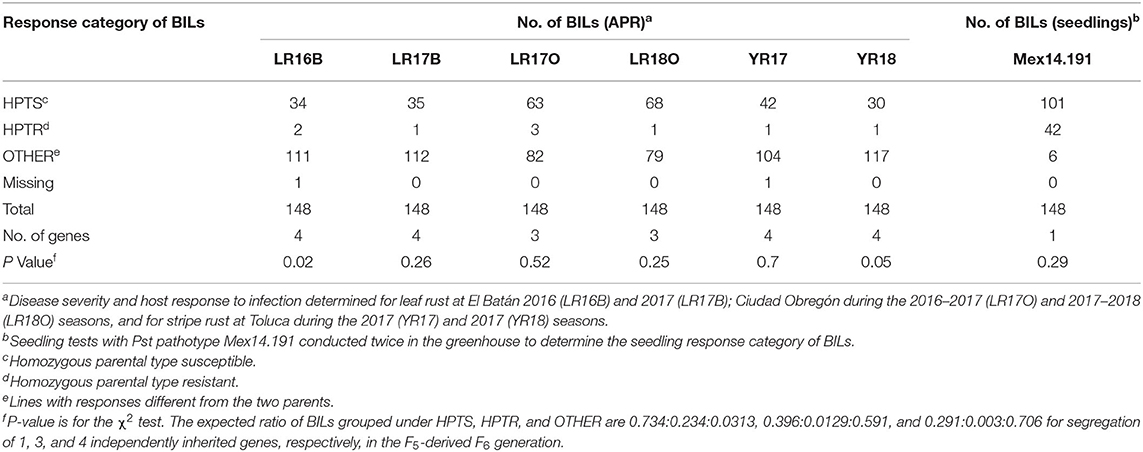
Table 1. Estimated number of resistance genes that confer seedling resistance to stripe rust and adult plant resistance to leaf rust and stripe rust in 148 KU3067 × Apav BILs based on Mendelian segregation analysis.
Characterization of Leaf Rust and Stripe Rust Resistance in the Field
In the field trials, the MDS and IT reaction to leaf rust was 1MSS for KU3067 and 100S for Apav across all the seasons. Mean leaf rust severities on BILs ranged from 50.1 to 79.0% during all the leaf rust trials (Table 2). The frequency distribution of BILs for leaf rust severity was continuous over the four environments (Figure 1A), indicating the polygenic inheritance of APR to leaf rust in the population. Genetic analyses by Mendelian segregation analysis (Table 1) indicated the presence of 3–4 APR genes that confer resistance to the leaf rust in the population.
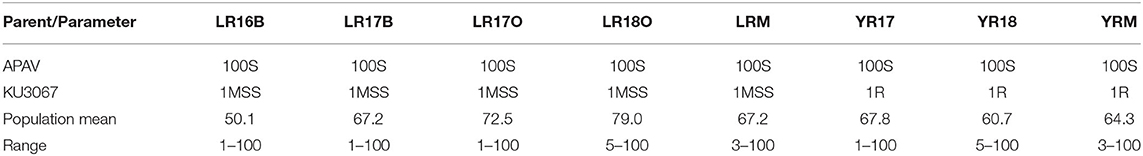
Table 2. Summary of MDS in the KU3067 × Apav BIL population phenotyped for leaf rust and stripe rust.
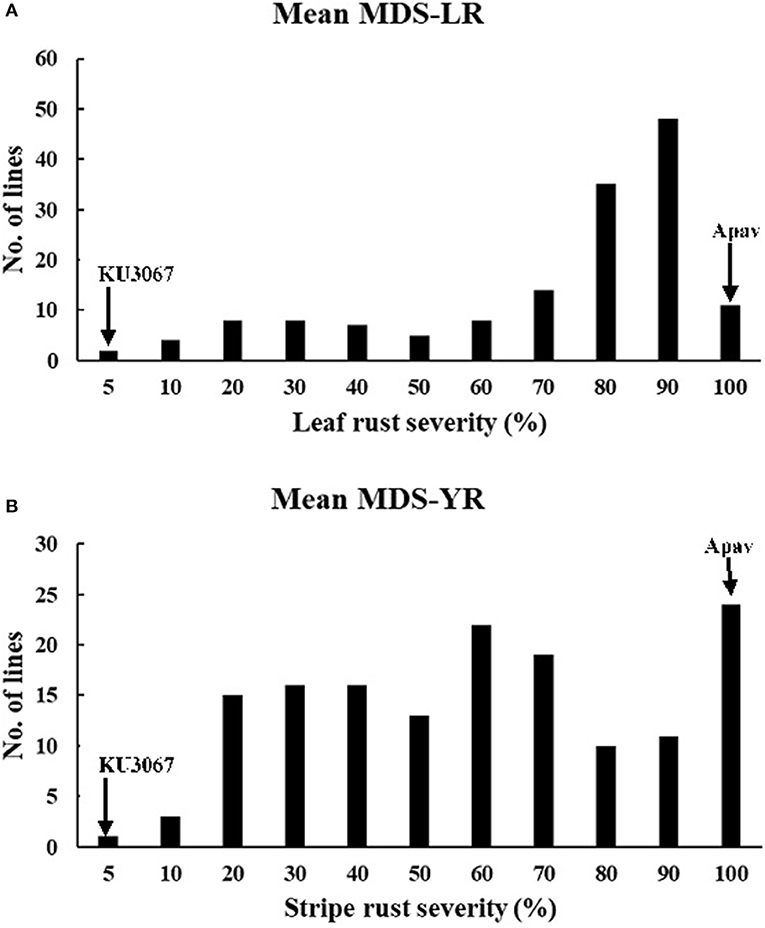
Figure 1. Frequency distributions of KU3067/Apav BILs lines for mean LR (A) and YR (B) MDS in the field conditions. Mean values for the parents, KU3067 and Apav, are indicated by arrows.
KU3067 and Apav displayed MDS and IT for stripe rust of 5R and 100S under field conditions. The mean MDS for all the BILs was 67.8% and 67.0% in YR2017 and YR2018, respectively. The stripe rust MDS scores for the 148 BILs across all environments also showed continuous distributions (Figure 1B). Four genes were estimated to provide resistance to stripe rust using the Mendelian segregation analysis (Table 1).
MDS scores for leaf rust across all the crop seasons were significantly correlated with correlation coefficients ranging from 0.73 to 0.88 (P < 0.001; Table 3). For stripe rust, the correlation coefficient for YR2017 and YR2018 was 0.84 (P < 0.001). The coefficients of the correlation between stripe rust and leaf rust disease scores ranged from 0.61 to 0.76 across experiments, indicating the presence of pleiotropic genes conferring resistance to both the rusts.
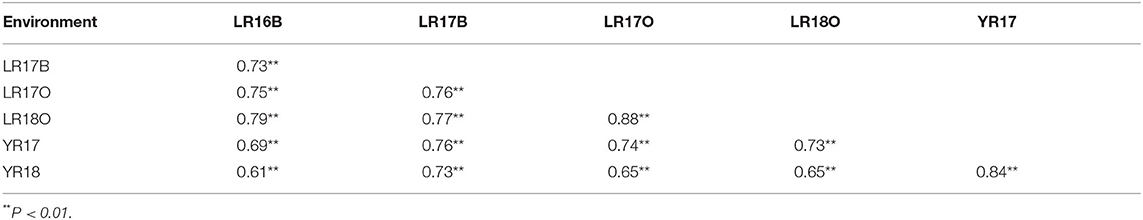
Table 3. Pearson correlation coefficients (r) for two-way comparisons of leaf rust and stripe rust severity data from different environments.
Linkage Map Construction and Mapping the Seedling Stripe Rust Resistance Gene YrKU
A total of 4,053 markers were used to construct the linkage map. The resulting linkage map comprised 40 linkage groups and a total map distance of 2,863.9 cM with A 1,190.5 cM, B 930.7 cM and D 742.7 cM, respectively (Supplementary Table 1). YrKU was mapped on 7BL and flanked by SNP markers 1070196|F|0–47:A>G and 5324909|F|0–54:T>C with genetic resistance 0.02 and 0.33 cM, respectively (Figure 2).
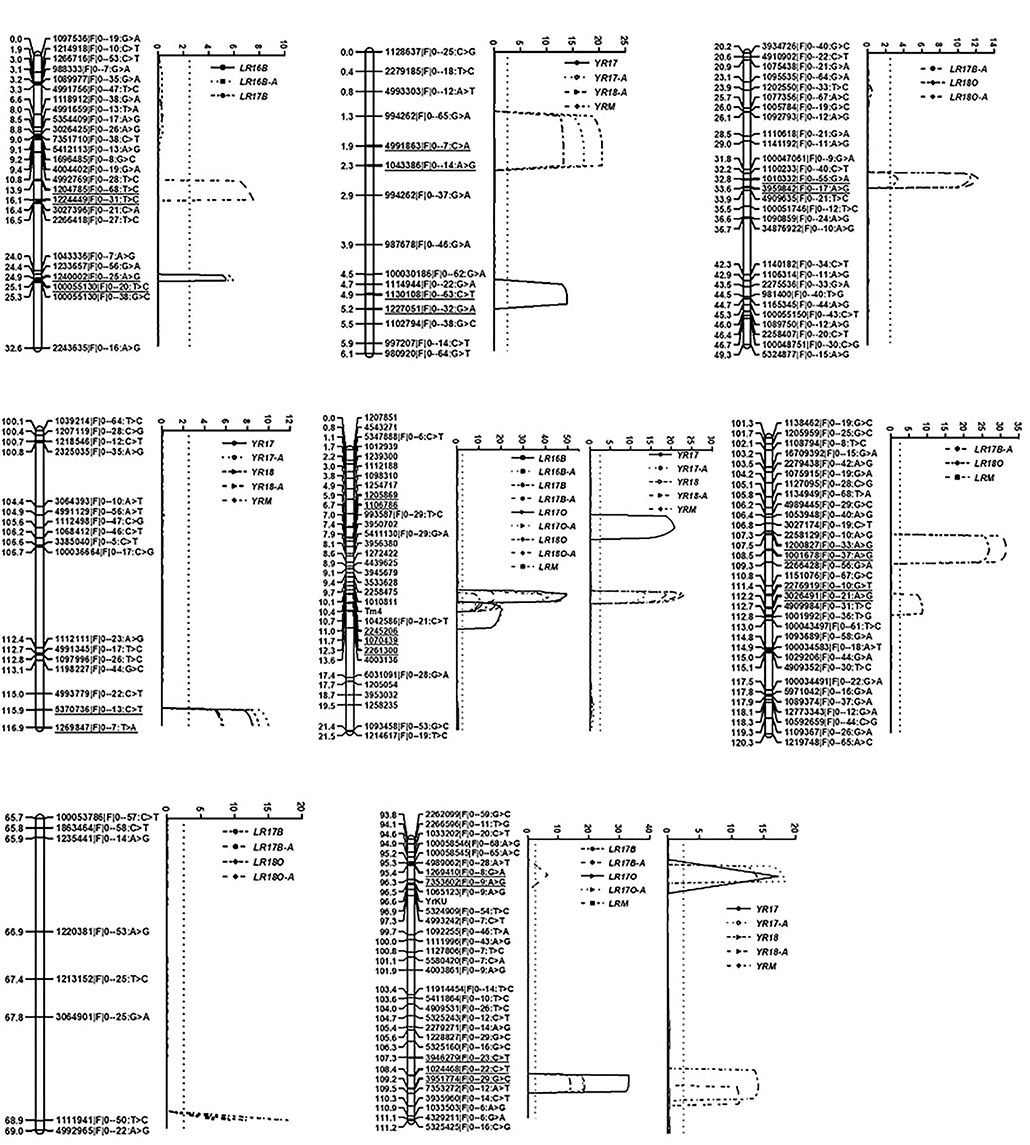
Figure 2. Likelihood plots of QTL for APR to leaf rust and stripe rust identified by IciMapping V4.1 in the KU3067 × Apav BILs population. The significant LOD threshold was detected based on 1,000 permutations. Positions (in cM) of cumulated genetic distances of linkage group along chromosomes are shown on the left axes and for molecular markers on the right. The marker interval of the QTL was underlined.
QTL Mapping for Leaf Rust and Stripe Rust Resistance
QTL Mapping of APR to Leaf Rust
Six leaf rust APR QTL were identified on chromosomes 1AS, 2AL, 4DL, 6BL, 7AL, and 7BL, and designated as QLr.cim-1AS, QLr.cim-2AL, QLr.cim-4DL, QLr.cim-6BL, QLr.cim-7AL, and QLr.cim-7BL, respectively (Table 4; Figure 2). All the resistance alleles were from KU3067. QLr.cim-1AS was detected in LR16B, LR16B-A (AUDPC for leaf rust at El Batán in 2016) and LR17B and explained 9.4–12.0% of the variation. QLr.cim-2AL with relatively smaller effects on disease response was identified in LR17B-A, LR18O, and LR18O-A with PVE ranging from 3.0 to 11.4%. QLr.cim-4DL with large effects was stably detected in all the environments and explained 10.9–56.4% of the phenotypic variances for leaf rust. QLr.cim-6BL was detected in LR17B-A, LR18O, and LRM (mean MDS across all the leaf rust environments) and explained 8.7–17.9% of the phenotypic variance. QLr.cim-7AL in the marker interval 4992965|F|0–22:A>G and 1111941|F|0–50:T>C was identified in LR17B, LR17B-A, LR18O, and LR18O-A with PVE ranging from 6.7–21.7%. QLr.cim-7BL was detected in LR17B, LR17B-A, LR17O, LR17O-A, and LRM and explained 7.5–17.2% of the phenotypic variance.
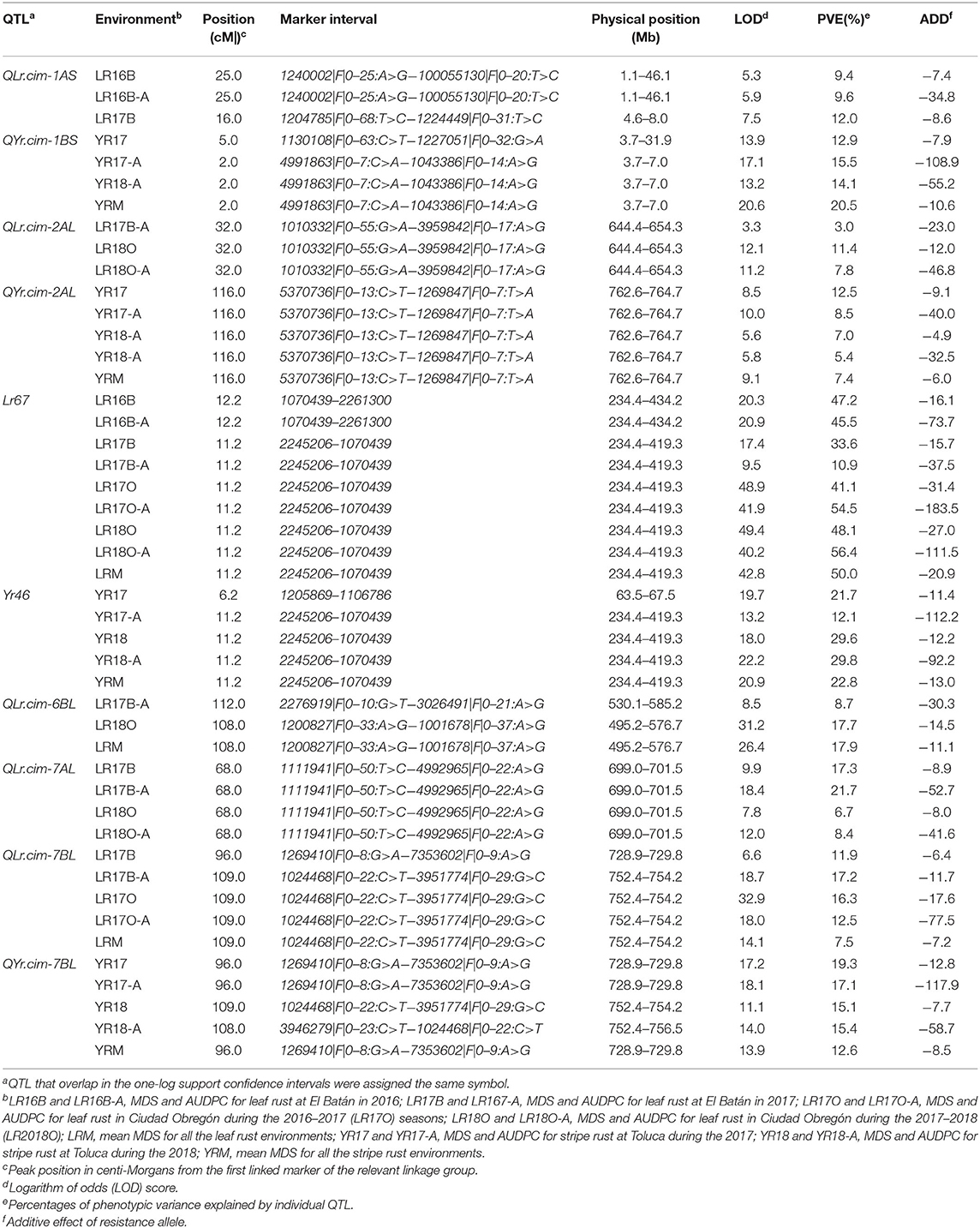
Table 4. Quantitative trait locus/loci for MDS to leaf rust and stripe rust by ICIM in the BIL population from Ku3067/Apav.
QTL Mapping for APR to Stripe Rust
Four QTL, QYr.cim-1BS, QYr.cim-2AL, QYr.cim-4DL, and QYr.cim-7BL, with the resistant allele from KU3067 were identified. QYr.cim-1BS was detected in YR17, YR17A, YR18-A, and YR-M and explained 12.9–20.5% of the phenotypic variance. QYr.cim-2AL was detected in all the stripe rust environments with PVE of 5.4–12.5%. QYr.cim-4DL was stably detected in all the stripe rust environments and explained 12.1–29.8% of the phenotypic variances. QYr.cim-7BL was identified in all the stripe rust environments with PVE 12.6–19.3% (Table 4).
Possible Pleiotropic Rust Resistance QTL
Among the QTL detected, two showed possible pleiotropic resistance for both rusts. The first one, QLr.cim-4DL/QYr.cim-4DL, proved to be Lr67/Yr46 based on the closely linked marker Tm4 (Moore et al., 2015) the marker was validated on the parent Ku3067, thus, confirming the presence of the gene Lr67. Another QTL, QLr.cim-7BL/QYr.cim-7BL located on 7BL also showed pleiotropic resistance. QYr.cim-7BL overlapped the Yr seedling resistance gene YrKU and showed all-stage resistance to stripe rust.
Overall, a total of six QTL for leaf rust and four QTL for stripe rust were identified in the population (Table 4; Figure 2). The total phenotypic variance explained by detected QTLs ranged from 55.1 to 83.9% across the environments for leaf rust and 44.4 to 71.6% for stripe rust, confirming their significant effect in reducing rust severity.
Average Effects of Two Potentially Pleiotropic Rusts QTL and Additive Effects of Leaf Rust and Stripe Rust QTL
Average effects of two potentially pleiotropic QTL were estimated in RIL carrying QTL and RIL lacking QTL were compared based on the closely linked flanking markers (Figure 3; Table 5). Lr67/Yr46 showed significantly large effect in reducing leaf rust and stripe rust by 37.9–66.9 and 35.1–48.6%, respectively, in four leaf rust trials and two stripe rust trials (Table 5). Leaf rust severities in the presence of Lr67 ranged from 4.3–60.1% compared with 21.2–100% in its absence. For QLr.cim-7BL/QYr.cim-7BL, there was a significant reduction in mean leaf rust and stripe rust severity by 20.0–35.0 and 29.8–40.9%, respectively. The mean leaf rust and stripe rust severities of RIL carrying the 7BL QTL, ranged from 3.4–90.0 and 5.3–65.0% compared with 7.5–100 and 17.5–100% in its absence, respectively (Figure 3).
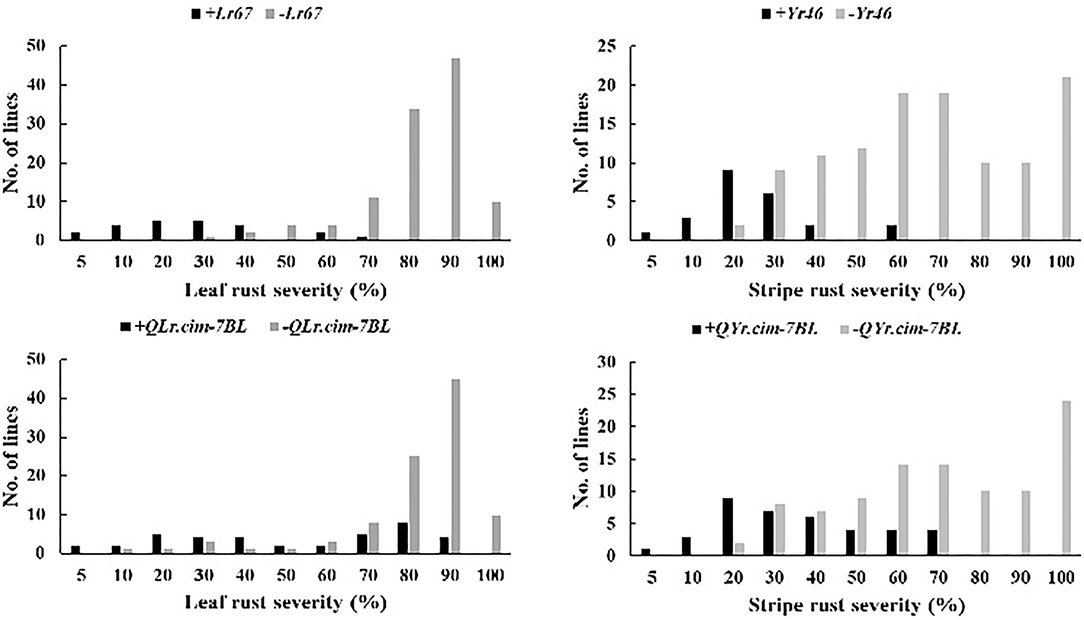
Figure 3. Comparison of KU3067/Apav BILs for mean MDS of LR and YR in the presence or absence of resistance allele for Lr67/Yr46 and QLr.cim-7BL/QYr.cim-7BL in field trials.
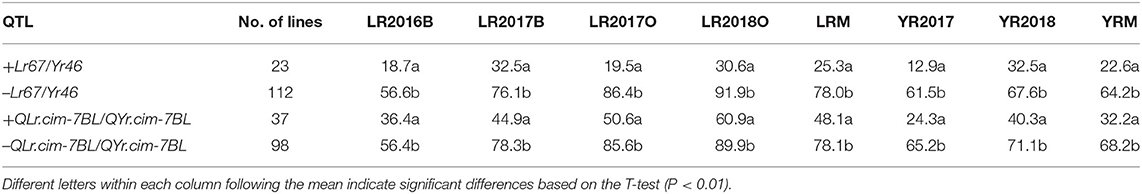
Table 5. T-tests for the comparison of MDS for leaf rust and stripe rust in KU3067 × Apav BILs with and without the resistance alleles.
The BILs were divided into 24 and 15 genotypes based on the presence of six-leaf rust and four stripe rust resistance QTL, and the additive nature of identified QTLs is shown in Figure 4. More QTLs imparted higher rust resistance in the BILs. For leaf rust, one line contained five QTL (4D + 7B + 1A + 2A + 6B) with mean MDS 6.3%. The mean MDS of the -QTL group reached 83.3%. For stripe rust, four lines with all four YR QTL had a mean MDS of 7.3%. In total, 46 RIL's without any of the reported stripe rust QTL had an MDS of 86.6%.
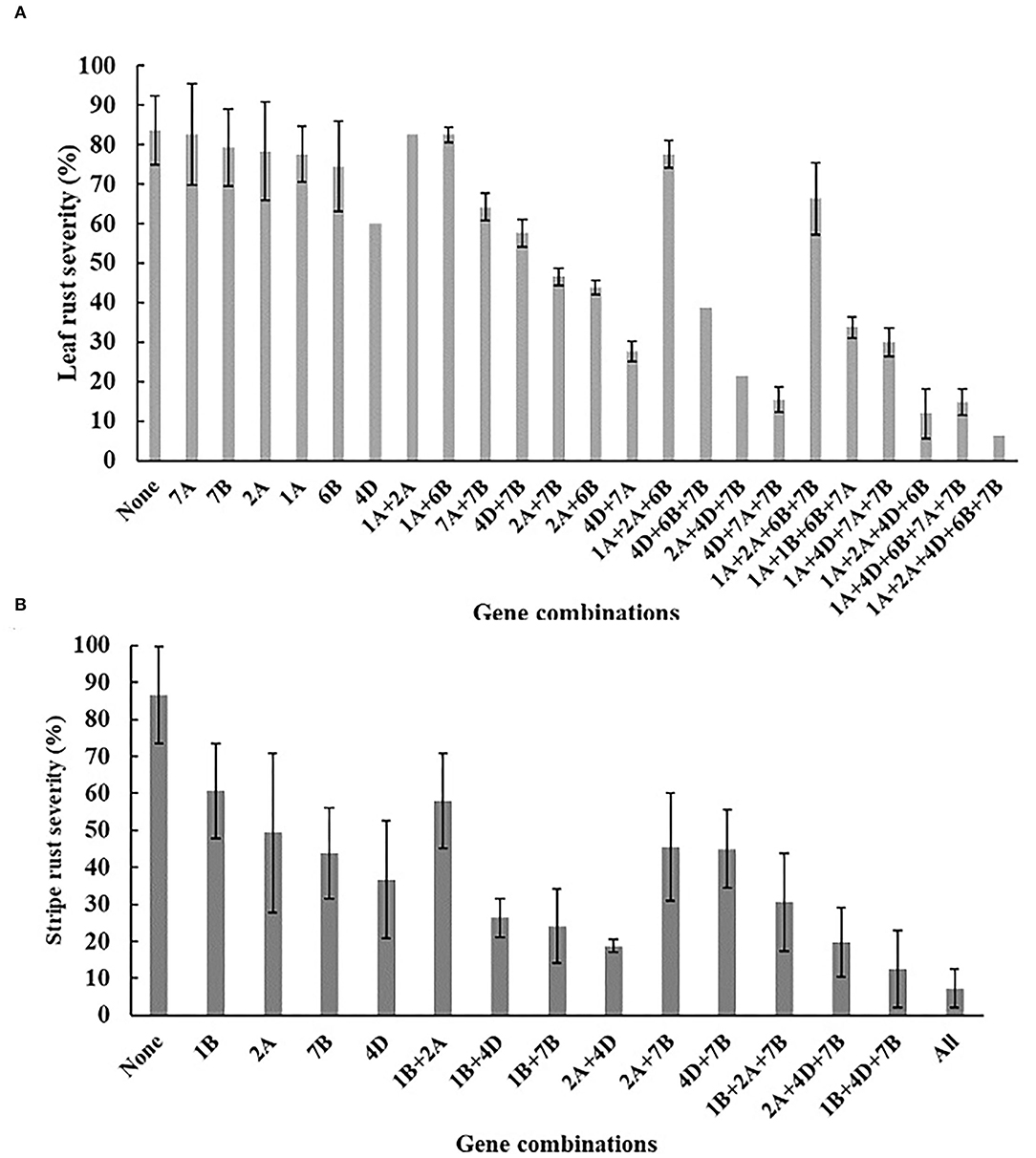
Figure 4. Mean MDS of lines carrying different combinations of QTLs. Lines containing the different QTL combinations were grouped together and the corresponding rust severities were averaged over environments. (A) Corresponds leaf rust combinations, (B) Corresponds to stripe rust evaluations. Means and standard errors of the means are shown.
Discussion
Genetic analyses indicated that four genes/loci controlled the APR resistance to leaf and stripe rusts in KU3067. In this mapping study, six QTL for leaf rust resistance and four for stripe rust resistance were identified using ICIM. The result of stripe rust was consistent with the estimated gene number, while there is a slight discrepancy with the genetic analyses for leaf rust. The Mendelian genetic approach for estimating gene numbers is based on the theoretical assumptions of polygenetic quantitative inheritance, where each gene is considered to contribute equally to the phenotype. However, it is common that variable effects of different QTL on phenotypes occur in the wheat genotypes, which results in segregating distortion of various phenotypic classes. Therefore, it is often observed that the estimated number of genes varies from the number of QTL identified. Yang et al. (2013) and Lan et al. (2015) also reported the discrepancy on the population Sujata/Avocet-YrA and Chapio/Avocet RIL populations, respectively. The estimated gene number usually represents the minimum number of polygenic loci segregating in a population, as is also evident in this study.
Lr67/Yr46 on 4DL
In this study, the most consistent QTL across all the environments was Lr67/Yr46 on 4DL. Lr67/Yr46 with a large effect explained 10.9–56.4 and 12.1–29.8% of the phenotypic variation for leaf and stripe rusts, respectively. The gene also confers resistance to stem rust and powdery mildew (Herrera-Foessel et al., 2014). Lr67/Yr46 was originally transferred from PI250413, a Pakistani wheat accession, to Thatcher-derived line RL6077 (Dyck and Samborski, 1979). Earlier studies also confirm a larger phenotypic effect of Lr67/Yr46 in Indian wheat lines Sujata (Lan et al., 2015) and New Pusa 876 (Ponce-Molina et al., 2018). This study is the first to report the presence of Lr67/Yr46 in an Afghan landrace, which suggests a broad range of deployment of this gene in diverse wheat germplasm. This gene has been effective for over 60 years and remains a key gene in the development of lines with durable rust resistance in many breeding programs, including CIMMYT's program.
QLr.cim-7BL/QYr.cim-7BL
QLr.cim-7BL/QYr.cim-7BL overlapped the Yr seedling resistance gene YrKU, indicating that YrKU showed all-stage resistance in the study. To date, six known Yr resistance genes Yr39 (Lin and Chen, 2007), Yr52 (Ren et al., 2012), Yr59 (Zhou et al., 2014), Yr67 (Li et al., 2009), YrZH84 (Li et al., 2006), and YrSuj (Lan et al., 2015) as well as three leaf rust resistance genes Lr14a, Lr14b (McIntosh et al., 1995), and Lr68 (Herrera-Foessel et al., 2012) have been reported on 7BL. The physical position of the QLr.cim-7BL/QYr.cim-7BL gene and closely linked markers is shown in Supplementary Table 2. YrKU (719.8–741.5 Mb) mapped at a similar position where Yr52 (732.4 Mb), Yr59 (723.9 Mb), Yr67 (699.9–718.4 Mb), YrSuj (718.3–734.8 Mb), and YrZH84 (718.3–723.9 Mb) are reported. As Yr52 and Yr59 are high-temperature adult-plant (HTAP) genes with susceptible infection type at the seedling stage, suggesting they are different from YrKU. YrZH84, a stripe rust seedling resistance gene, was identified in Chinese wheat cultivar “Zhou 8425B” however, does not show pleiotropic resistance to the leaf rust. YrSuj mapped in the Indian bread wheat cultivar Sujata (Lan et al., 2015) with the pleiotropic effect on APR to leaf rust is similar to our findings. Lan et al. (2015) also reported that YrSuj and Yr67 from Indian variety C591 might be same (Lan et al., 2015). The relationship among YrKU, Yr67, and YrSuj will be confirmed through the allelism test in the next step to confirm if it is a known gene or a new gene at that locus.
For leaf rust, Lr14a and Lr14b, located on 7BL, are seedling resistance genes and seedling tests of the RIL population displayed infection type “4” or “3+,” indicating the two genes are ineffective against the Pt races that were tested, and hence, they are different from QLr.cim-7BL. Lr68 is a slow-rusting resistance gene flanked by markers cs7BLNLRR and Xgwm146 was mapped in CIMMYT line Arula (Herrera-Foessel et al., 2012). QLr.cim-7BL (729.4–753.8 Mb) mapped at a similar position with Lr68 (752.2–752.8 Mb); but the absence of Lr68 in KU3067 was confirmed by the linked molecular markers csGS and CAPS marker cs7BLNLRR (Herrera-Foessel et al., 2012). Hence QLr.cim-7BL may not be Lr68.
QYr.cim-1BS
In total, six designated seedling Yr resistance genes, Yr9 (Mettin et al., 1978), Yr10 (Metzger and Silbaugh, 1970), Yr15 (Gerechter-Amitai et al., 1989), Yr24 (McIntosh and Lagudah, 2000), Yr64 (Cheng et al., 2014), and Yr65 (Cheng et al., 2014) were mapped on 1BS. Yr10, Yr15, Yr24/Yr26, Yr64, and Yr65 were mapped at 5.5, 46.8–78.8, 218.9, 78.8, and 221.9–228.6 Mb, respectively, compared with QYr.cim-1BS at 3.7–7.0 Mb (Supplementary Table 2). In addition, the seedling tests showed that plants carrying Yr10, Yr15, and Yr24/Yr26 were immune (IT 0) to Pst isolate Mex14.191, and plants with Yr9 were susceptible (IT 8) while KU3067 showed IT 3, 4. Molecular markers of Yr9, Yr10, Yr15, and Yr24/Yr26 also confirmed the absence of these genes in KU3067 (data not shown). Based to the chromosome position, seedling reaction and molecular markers detection, it can be concluded that QYr.cim-1BS is different from these genes and might be new.
QYr.cim-2AL
Two known seedling resistance Yr genes, Yr1 (Zadoks 1961), and Yr32 (Eriksen et al., 2004), were mapped on 2AL. Yr1 (Zadoks, 1961; Lupton and Macer, 1962) from winter wheat genotype Chinese 166 confers seedling resistance typically with IT 1 to pathotype Mex14.191, which is obviously different from IT 34 of KU3067. Yr32 was originally characterized in cultivar Carstens V, and the seedling reaction of Yr32 was unknown to Pst Mex14.191. Yr32 was closely linked to wmc198 at 711.5 Mb with genetic distance 2 cM compared with QYr.cim-2AL at 764.7–762.6 Mb. It can be concluded that QYr.cim-2AL is different from Yr1 and Yr32. In addition, five QTL on 2AL for stripe rust resistance were reported in wheat varieties Récital (Dedryver et al., 2009), Camp Remy (Boukhatem et al., 2002) Triticum monococcum acc. pau14087 (Chhuneja et al., 2008), Solist (Christiansen et al., 2006), and Wasmo (Christiansen et al., 2006). QYr.inra-2AL and QYR2 from Récital and Camp Remy, respectively, were closely linked to Xgwm382 at 774.3 Mb (Supplementary Table 2). QTL identified in Solist, Wasmo, and pau14087 were linked to Xwmc170 at 715.3 Mb (Supplementary Table 2). In this study, QYr.cim-2AL at 764.7–762.6 Mb was 12 Mb away from QYr.inra-2AL and QYR2 and might be potentially new.
QLr.cim-1AS
The known seedling resistance gene Lr10 (Feuillet et al., 2003) and two QTL, QLr.cim-1AS from the Indian bread wheat cultivar Sujata (Lan et al., 2015) and QLr.cau-1AS from the American wheat cultivar Luke (Du et al., 2015), were mapped on 1AS. Lr10 expressed IT 3 and 4 to Pt pathotypes MBJ/SP and MCJ/SP in the seedling test indicating it is ineffective against the tested Pt. QLr.cim-1AS (Lan et al., 2015) and QLr.cau-1AS (Du et al., 2015) were mapped at the distal end of 1AS with physical positions of 1.7–8.5 and 9.0 Mb (Supplementary Table 2). The physical position of KU3067 QTL QLr.cim-1AS (1.1–46.1 Mb) overlapped the latter two QTL (Supplementary Table 2). QLr.cim-1AS from Sujata showed pleiotropic resistance to stripe rust; however, we did not detect any effect of QLr.cim-1AS on stripe rust in this study. The relationship of these three QTL needs further analysis in the next step.
QLr.cim-2AL
Leaf rust resistance gene Lr38 introgressed from Thinopyrym intermedium to common wheat was mapped on 2AL (Friebe et al., 1992). There is no previous report suggesting that land race KU3067 might possibly carry Lr38. In addition, six QTL for leaf rust resistance, QLr.cimmyt-2AL in Avocet (Rosewarne et al., 2013), QLr.sfr-2AL in Forno (Schnurbusch et al., 2004), QLr.ubo-2A in Lloyd (Maccaferri et al., 2008), QLr.hebau-2AL in Chinese Spring (Zhang et al., 2017), QLr.ifa-2AL in Capo (Buerstmayr et al., 2014), and QTL-2AL in Opata 85 (Nelson et al., 1997), were mapped on 2AL. These QTL mapped in the different physical positions with the KU3067 QTL based on the position of closely linked markers (Supplementary Table 2), except for QLr.cimmyt-2AL in Avocet in an unknown position. It is currently not known if QLr.cimmyt-2AL is in the same region as the KU3067 QTL.
QLr.cim-6BL
Four known QTL viz. QLr.fcu-6BL (Chu et al., 2009), QLr.cimmyt-6BL.1 (Rosewarne et al., 2013), QLr.cim-6BL (Lan et al., 2017), and QLr.cimmyt-6BL.2 (William et al., 2006) with resistance allele from synthetic hexaploid wheat TA4152-60, and CIMMYT bread wheats Pastor, Bairds, and Pavon 76, respectively, were mapped on 6BL. The Pastor QTL QLr.cimmyt-6BL.1 (Rosewarne et al., 2013) and Pavon 76 QTL QLr.cimmyt-6BL.2 (William et al., 2006) were effective against both leaf rust and stripe rust. However, no pleiotropic resistance was detected for the QTL identified in KU3067. QLr.cim-6BL in this study mapped at 495.2–576.7 Mb and is obviously different from 17.1 Mb for QLr.fcu-6BL, 706.7 Mb for QLr.cim-6BL, and 426.4 Mb for QLr.cimmyt-6BL.2 (Supplementary Table 2). It can be concluded that QLr.cim-6BL from KU3067 might be potentially new.
QLr.cim-7AL
Till date, only one known Lr gene, Lr20 (Browder, 1972), was mapped at about 668.0 Mb on 7AL. Lr20 was a seedling resistance gene and was susceptible to the two Pt pathotypes tested indicating it is ineffective. Two minor QTL, viz. QLr.hwwg-7AL (Lu et al., 2017) and QLr.mma-7AL (Tsilo et al., 2014), were mapped at 701.9 and 722.9 Mb on 7AL, respectively (Supplementary Table 2). QLr.cim-7AL was located at 699.0–701.5 Mb near to QLr.hwwg-7AL.
Potential Application of QTL for Leaf Rust and Stripe Rust in Wheat Breeding
In this study, six QTL for leaf rust and four QTL for stripe rust were identified in the KU3067 RIL population. The leaf rust and stripe rust resistance of KU3067 had remained effective for more than 60 years. Despite their inferior agronomic traits (e.g., low grain yield, lodging), KU3067 and lines with multiple QTLs and combinations in the population can serve as a valuable source of resistance for common wheat improvement. The tightly linked SNP markers identified in this study can be converted to KASP assays (Semagn et al., 2014) for marker-assisted selection (MAS) and pyramiding of APR genes to improve leaf rust and stripe rust resistance in wheat-breeding programs.
Data Availability Statement
The data used to present the findings reported in this study is available upon request through the corresponding author.
Author Contributions
PZ, SB, and RPS contributed to the manuscript preparation, phenotyping, and QTL analysis. CL and JH-E contributed to population development. ZL contributed to experimental design and analysis. EL contributed to manuscript preparation and critical review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Australian Grains Research and Development Corporation (GRDC) with funding to the Australian Cereal Rust Control Program (ACRCP) and Accelerating Genetic Gains in Maize and Wheat (AGG) project funded by Bill and Melinda Gates Foundation (BMGF), the UK's Foreign, Commonwealth and Development Office (FCDO), The United States Agency for International Development (USAID), The Foundation for Food and Agricultural Research (FFAR), and National Natural Science Foundation of China (32001538 and 32161143007) for supporting this research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.894528/full#supplementary-material
References
Bhavani, S., Hodson, D. P., Huerta-Espino, J., Randhawa, M. S., and Singh, R. P. (2019). Progress in breeding for resistance to Ug99 and other races of the stem rust fungus in CIMMYT wheat germplasm. Front. Agric. Sci. Eng. 6, 210–224. doi: 10.15302/J-FASE-2019268
Bhavani, S., Singh, P. K., Qureshi, N., He, X., Biswal, A. K., Juliana, P., et al. (2021). Globally important wheat diseases: status, challenges, breeding and genomic tools to enhance resistance durability. Genomic Des. Biot. Stress Resist. Cereal Crop 59–128. doi: 10.1007/978-3-030-75879-0_2
Bjarko, M. E., and Line, R. F. (1988). Heritability and number of genes controlling leaf rust resistance in four cultivars of wheat. Phytopathology 78, 457–461. doi: 10.1094/Phyto-78-457
Boukhatem, N., Baret, P. V., Mingeot, D., and Jacquemin, J. M. (2002). Quantitative trait loci for resistance against yellow rust in two wheat-derived recombinant inbred line populations. Theor. Appl. Genet. 104, 111–118. doi: 10.1007/s001220200013
Browder, L. E. (1972). Designation of two genes for resistance to Puccinia recondita in Triticum aestivum. Crop Sci. 12, 705–706. doi: 10.2135/cropsci1972.0011183X001200050049x
Buerstmayr, M., Matiasch, L., Mascher, F., Vida, G., Ittu, M., Robert, O., et al. (2014). Mapping of quantitative adult plant field resistance to leaf rust and stripe rust in two European winter wheat populations reveals colocation of three QTL conferring resistance to both rust pathogens. Theor. Appl. Genet. 127, 2011–2028. doi: 10.1007/s00122-014-2357-0
Caldwell, R. M. (1968). “Breeding for general and/or specific plant disease resistance,” in Proceedings of the 3rd International Wheat Genetics Symposium, eds K. W. Findlay, and K. W. Shepherd (Canberra, ACT: Australian Academy of Science), 263–272.
Chen, X. M. (2005). Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Cheng, P., Xu, L. S., Wang, M. N., See, D. R., and Chen, X. M. (2014). Molecular mapping of genes Yr64 and Yr65 for stripe rust resistance in hexaploid derivatives of durum wheat accessions PI 331260 and PI 480016. Theor. Appl. Genet. 127, 2267–2277. doi: 10.1007/s00122-014-2378-8
Chhuneja, P., Kaur, S., Garg, T., Ghai, M., Kaur, S., Prashar, M., et al. (2008). Mapping of adult plant stripe rust resistance genes in diploid A genome wheat species and their transfer to bread wheat. Theor. Appl. Genet. 116, 313–324. doi: 10.1007/s00122-007-0668-0
Christiansen, M. J., Feenstra, B., Skovgaard, I. M., and Andersen, S. B. (2006). Genetic analysis of resistance to yellow rust in hexaploid wheat using a mixture model for multiple crosses. Theor. Appl. Genet. 112, 581–591. doi: 10.1007/s00122-005-0128-7
Chu, C. G., Friesen, T. L., Xu, S. S., Faris, J. D., and Kolmer, J. A. (2009). Identification of novel QTLs for seedling and adult plant leaf rust resistance in a wheat doubled haploid population. Theor. Appl. Genet. 119, 263–269. doi: 10.1007/s00122-009-1035-0
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27, 885–896. doi: 10.1101/GR.217117.116/-/DC1
Dedryver, F., Paillard, S., Mallard, S., Robert, O., Trottet, M., Nègre, S., et al. (2009). Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat ’Renan'. Phytopatholgy 99, 968–973. doi: 10.1094/PHYTO-99-8-0968
Du, Z., Che, M., Li, G., Chen, J., Quan, W., Guo, Y., et al. (2015). A QTL with major effect on reducing leaf rust severity on the short arm of chromosome 1A of wheat detected across different genetic backgrounds and diverse environments. Theor. Appl. Genet. 128, 1579–1594. doi: 10.1007/s00122-015-2533-x
Dyck, P. L., and Samborski, D. J. (1979). Adult plant resistance in PI250413, an introduction of common wheat. Can. J. Plant Sci. 59, 329–332. doi: 10.4141/cjps79-053
Eriksen, L., Afshari, F., Christiansen, M. J., McIntosh, R. A., Jahoor, A., and Wellings, C. R. (2004). Yr32 for resistance to stripe (yellow) rust present in the wheat cultivar carstens V. Theor. Appl. Genet. 108, 567–575. doi: 10.1007/s00122-003-1456-0
Feuillet, C., Travella, S., Stein, N., Albar, L., Nublat, A., and Keller, B. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U.S.A. 100, 15253–15258. doi: 10.1073/pnas.2435133100
Friebe, B., Zeller, F. J., Mukai, Y., Forster, B. P., Bartos, P., and McIntosh, R. A. (1992). Characterization of rust-resistant wheat-Agropyron intermedium derivatives carrying resistance against leaf, stripe and stem rust by C-banding, in situ hybridization and isozyme analysis. Theor. Appl. Genet. 83, 775–782. doi: 10.1007/BF00226697
Gerechter-Amitai, Z. K., van Silfhout, C. H., Grama, A., and Kleitman, F. (1989). Yr15-a new gene for resistance to Puccinia striiformis in Triticum dicoccoides sel. G-25. Euphytica 43, 187–190. doi: 10.1007/BF00037912
Herrera-Foessel, S. A., Lagudah, E. S., Huerta-Espino, J., Hayden, M. J., Bariana, H. S., Singh, D., et al. (2011). New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor. Appl. Genet. 122, 239–249. doi: 10.1007/s00122-010-1439-x
Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Rosewarne, G. M., Periyannan, S. K., Viccars, L., et al. (2012). Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor. Appl. Genet. 124, 1475–1486. doi: 10.1007/s00122-012-1802-1
Herrera-Foessel, S. A., Singh, R. P., Lillemo, M., Huerta-Espino, J., Bhavani, S., Singh, S., et al. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127, 781–789. doi: 10.1007/s00122-013-2256-9
Huerta-Espino, J., Singh, R., Crespo-Herrera, L. A., Villaseñor-Mir, H. E., Rodriguez-Garcia, M. F., Dreisigacker, S., et al (2020). Adult plant slow rusting genes confer high levels of resistance to rusts in bread wheat cultivars from Mexico. Front. Plant Sci. 11, 824. doi: 10.3389/fpls.2020.00824
International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Johnson, R. (1988). “Durable resistance to yellow (stripe) rust in wheat and its implications in plant breeding,” in Breeding Strategies for Resistance to the Rusts of Wheat, eds N. W. Simmonds, and S. Rajaram (Mexico: CIMMYT).
Knott, D. R. (1989). The Wheat Rusts—Breeding for Resistance. Berlin: Springer-Verlag. doi: 10.1007/978-3-642-83641-1
Kumar, S., Bhardwaj, S. C., Gangwar, O. P., Sharma, A., Qureshi, N., Kumaran, V. V., et al. (2021). Lr80: A new and widely effective source of leaf rust resistance of wheat for enhancing diversity of resistance among modern cultivars. Theor. Appl. Genet. 134, 849–858. doi: 10.1007/s00122-020-03735-5
Lan, C., Basnet, B. R., Singh, R. P.;, Huerta-Espino, J., Herrera-Foessel, S. A.;, Ren, Y., et al. (2017). Genetic analysis and mapping of adult plant resistance loci to leaf rust in durum wheat cultivar bairds. Theor. Appl. Genet. 130, 609–619. doi: 10.1007/s00122-016-2839-3
Lan, C., Zhang, Y., Herrera-Foessel, S. A., Basnet, B. R., Huerta-Espino, J., Lagudah, E. S., et al. (2015). Identification and characterization of pleiotropic and co-located resistance loci to leaf rust and stripe rust in bread wheat cultivar sujata. Theor. Appl. Genet. 128, 549–561. doi: 10.1007/s00122-015-2454-8
Large, E. C. (1954). Growth stages in cereals illustration of the feekes scale. Plant Pathol. 3, 128–129. doi: 10.1111/j.1365-3059.1954.tb00716.x
Li, H. H., Ye, G. Y., and Wang, J. K. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374. doi: 10.1534/genetics.106.066811
Li, J., Dundas, I., Dong, C., Li, G., Trethowan, R., Yang, Z., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, Y., Niu, Y. C., and Chen, X. M. (2009). Mapping a stripe rust resistance gene YrC591 in wheat variety C591 with SSR and AFLP markers. Theor. Appl. Genet. 118, 339–346. doi: 10.1007/s00122-008-0903-3
Li, Z. F., Zheng, T. C., He, Z. H., Li, G. Q., Xu, S. C., Li, X. P., et al. (2006). Molecular tagging of stripe rust resistance gene YrZH84 in Chinese wheat line Zhou 8425B. Theor. Appl. Genet. 112, 1098–1103. doi: 10.1007/s00122-006-0211-8
Lin, F., and Chen, X. M. (2007). Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar alpowa. Theor. Appl. Genet. 114, 1277–1287. doi: 10.1007/s00122-007-0518-0
Lu, Y., Bowden, R. L., Zhang, G., Xu, X., Fritz, A. K., and Bai, G. (2017). Quantitative trait loci for slow-rusting resistance to leaf rust in doubled-haploid wheat population CI13227 x Lakin. Phytopathology 107, 1372–1380. doi: 10.1094/PHYTO-09-16-0347-R
Lupton, F. G. H., and Macer, R. C. F. (1962). Inheritance of resistance to yellow rust (Puccinia glumarum) in seven varieties of wheat. Br. Mycol. Soc. 45, 21–45. doi: 10.1016/S0007-1536(62)80032-1
Maccaferri, M., Mantovani, P., Tuberosa, R., DeAmbrogio, E., Giuliani, S., Demontis, A., et al. (2008). A major QTL for durable leaf rust resistance widely exploited in durum wheat breeding programs maps on the distal region of chromosome arm 7BL. Theor. Appl. Genet. 117, 1225–1240. doi: 10.1007/s00122-008-0857-5
McIntosh, R. A., and Lagudah, E. S. (2000). Cytogenetic studies in wheat XVIII. Gene Yr24 for resistance to stripe rust. Plant Breed. 119, 81–83. doi: 10.1046/j.1439-0523.2000.00449.x
McIntosh, R. A., Wellings, C. R., and Park, R. F. (1995). Wheat Rusts: An Atlas of Resistance Genes. Melbourne, VIC: CSIRO Publishing. doi: 10.1071/9780643101463
McNeal, F. H., Konzak, C. F., Smith, E. P., Tate, W. S., and Russell, T. S. (1971). A uniform system for recording and processing cereal research data. US Dept. Agric. Res. Serv. ARS 34, 34–121.
Mettin, D., Bluthner, W. D., and Weinrich, M. (1978). Studies on the nature and the possible origin of the spontaneously translocated 1B-1R chromosome in wheat. Wheat Inform. Serv. 47–48, 12–16.
Metzger, R. J., and Silbaugh, B. A. (1970). Inheritance of resistance to stripe rust and its association with brown glume colour in Triticum aestivum L. PI 178383. Crop Sci. 10, 567–568. doi: 10.2135/cropsci1970.0011183X001000050035x
Moore, J. W., Herrera-Foessel, S., Lan, C., Schnippenkoetter, W., Ayliffe, M., Huerta-Espino, J., et al. (2015). A recently evolved hexose transporter variant confers resistance to multiple pathogens in wheat. Nat. Genet. 47, 1494–1498. doi: 10.1038/ng.3439
Nelson, J. C., Singh, R. P., Autrique, J. E., and Sorrells, M. E. (1997). Mapping genes conferring and suppressing leaf rust resistance in wheat. Crop Sci. 37, 1928–1935. doi: 10.2135/cropsci1997.0011183X003700060043x
Peterson, R. F., Campbell, A. B., and Hannah, A. E. (1948). A dia-grammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Pinto da Silva, G. B., Zanella, C. M., Martinelli, J. A., Chaves, M. S., Hiebert, C. W., McCallum, B. D., et al. (2018). Quantitative trait loci conferring leaf rust resistance in exaploidy wheat. Phytopathology 108, 1344–1354. doi: 10.1094/PHYTO-06-18-0208-RVW
Ponce-Molina, L. J., Huerta-Espino, J., Singh, R. P., Basnet, B. R., Lagudah, E., Aguilar-Rincón, V. H., et al. (2018). Characterization of adult plant resistance to leaf rust and stripe rust in Indian wheat cultivar ’new pusa 876'. Crop Sci. 58, 630–638. doi: 10.2135/cropsci2017.06.0396
Raman, H., Raman, R., Kilian, A., Detering, F., Carling, J., and Coombes, N. (2014). Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PloS ONE 9, e101673. doi: 10.1371/journal.pone.0101673
Randhawa, M. S., Singh, R. P., Dreisigacker, S., Bhavani, S., Huerta-Espino, J., Rouse, M. N., et al. (2018). Identification and validation of a common stem rust resistance locus in two bi-parental populations. Front. Plant Sci. 871, 1788. doi: 10.3389/fpls.2018.01788
Ren, R. S., Wang, M. N., Chen, X. M., and Zhang, Z. J. (2012). Characterization and molecular mapping of Yr52 for high-temperature adult-plant resistance to stripe rust in spring wheat germplasm PI 183527. Theor. Appl. Genet. 125, 847–857. doi: 10.1007/s00122-012-1877-8
Roelfs, A. P., Singh, R. P., and Saari, E. E. (1992). Rust Diseases of Wheat: Concepts and Methods of Disease Management. Mexico: CIMMYT.
Rosewarne, G. M., Herrera-Foessel, S. A., Singh, R. P., Huerta-Espino, J., Lan, C. X., and He, Z. H. (2013). Quantitative trait loci of stripe rust resistance in wheat. Theor. Appl. Genet. 126, 2427–2449. doi: 10.1007/s00122-013-2159-9
Sansaloni, C., Franco, J., Santos, B., Percival-Alwyn, L., Singh, S., Petroli, C., et al. (2020). Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 2020 111 11, 4572. doi: 10.1038/s41467-020-18404-w
Schnurbusch, T., Paillard, S., Schori, A., Messmer, M., Schachermayr, G., Winzeler, M., et al. (2004). Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor. Appl. Genet. 108, 477–484. doi: 10.1007/s00122-003-1444-4
Semagn, K., Babu, R., Hearne, S., and Olsen, M. (2014). Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement. Mol. Breeding 33, 1–14. doi: 10.1007/s11032-013-9917-x
Sharp, P. J., Kreis, M., Shewry, P. R., and Gale, M. D. (1988). Location of b-amylase sequence in wheat and its relatives. Theor. Appl. Genet. 75, 286–290.
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Bariana, H., Bansal, U., McCallum, B., et al. (2012). “Lr34/Yr18/Sr57/Pm38/Bdv1/Ltn1 confers slow rusting, adult plant resistance to Puccinia graminis tritici,” in Proceedings of the 13th International Cereal Rusts and Powdery Mildews Conference, ed W. Q. Chen (Beijing).
Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C. X., Basnet, B. R., Bhavani, S., et al. (2013). “Pleiotropic gene Lr46/Yr29/Pm39/Ltn2 confers slow rusting, adult plant resistance to wheat stem rust fungus,” in Proceedings Borlaug Global Rust Initiative, 2013 Technical Workshop (New Delhi), 17.
Singh, R. P., and Rajaram, S. (1992). Genetics of adult-plant resistance to leaf rust in ’frontana' and three CIMMYT wheats. Genome 35, 24–31.
Tanaka, H., Morris, C., Haruna, M., and Tsujimoto, H. (2008). Prevalence of puroindoline alleles in wheat varieties from eastern Asia including the discovery of a new SNP in puroindoline b. Plant Genet. Resour. 6, 142–152. doi: 10.1017/S1479262108993151
Tsilo, T. J., Kolmer, J. A., and Anderson, J. A. (2014). Molecular mapping and improvement of leaf rust resistance in wheat breeding lines. Phytopathology 104, 865–870. doi: 10.1094/PHYTO-10-13-0276-R
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, M., and Chen, X. (2017). “Stripe rust resistance,” in Stripe Rust, eds X. M. Chen, and Z. S. Kang (Berlin: Springer), 353–558.
William, H. M., Singh, R. P., Huerta-Espino, J., Palacios, G., and Suenaga, K. (2006). Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49, 977–990. doi: 10.1139/g06-052
Yang, E. N., Rosewarne, G. M., Herrera-Foessel, S. A., Huerta-Espino, J., Tang, Z. X., Sun, C. F., et al. (2013). QTL analysis of the spring wheat “Chapio” identifies stable stripe rust resistance despite inter-continental genotype × environment interactions. Theor. Appl. Genet. 126, 1721–1732. doi: 10.1007/s00122-013-2087-8
Zadoks, J. C. (1961). Yellow rust on wheat. Studies on epidemiology and physiological specialization. J. Pl. Ziekten 67, 69–256.
Zhang, P., Yin, G., Zhou, Y., Qi, A., Gao, F., Xia, X., et al. (2017). QTL mapping of adult-plant resistance to leaf rust in the wheat cross Zhou 8425B/Chinese spring using high-density SNP markers. Front. Plant Sci. 8, 793. doi: 10.3389/fpls.2017.00793
Zhang, R., Singh, R. P., Lillemo, M., He, X., Randhawa, M. S., Huerta-Espino, J., et al. (2019). Two main stripe rust resistance genes identified in synthetic-derived wheat line Soru# 1. Phytopathology 109, 120–126. doi: 10.1094/PHYTO-04-18-0141-R
Keywords: genetic analysis, molecular mapping, wheat rusts, APR genes, landraces
Citation: Zhang P, Lan C, Singh RP, Huerta-Espino J, Li Z, Lagudah E and Bhavani S (2022) Identification and Characterization of Resistance Loci to Wheat Leaf Rust and Stripe Rust in Afghan Landrace “KU3067”. Front. Plant Sci. 13:894528. doi: 10.3389/fpls.2022.894528
Received: 11 March 2022; Accepted: 25 April 2022;
Published: 28 June 2022.
Edited by:
Feng Chen, Henan Agricultural University, ChinaReviewed by:
Francis Chuks Ogbonnaya, Grains Research and Development Corporation, AustraliaAwais Rasheed, Quaid-i-Azam University, Pakistan
Copyright © 2022 Zhang, Lan, Singh, Huerta-Espino, Li, Lagudah and Bhavani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sridhar Bhavani, Uy5CaGF2YW5pQGNnaWFyLm9yZw==
†These authors have contributed equally to this work
 Peipei Zhang
Peipei Zhang Caixia Lan
Caixia Lan Ravi P. Singh
Ravi P. Singh Julio Huerta-Espino
Julio Huerta-Espino Zaifeng Li1
Zaifeng Li1 Sridhar Bhavani
Sridhar Bhavani