
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 02 June 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.893896
This article is part of the Research TopicAbiotic Stress: Molecular Genetics and Genomics, Volume IIView all 32 articles
The vital roles of R2R3-MYB transcription factors (TFs) in regulating stress response and phytohormone signaling have been thoroughly studied in numerous plant species, but the functions of these TFs in rubber tree are poorly understood. Rubber tree is the most important source of natural rubber but often suffers from various abiotic and biotic stresses that cause severe yield losses each year. In this study, we reported a novel MYB44 gene in rubber tree (named HbMYB44) and revealed its biological function. HbMYB44 was highly similar to AtMYB44 and clustered into subgroup 22. Transient expression indicated that HbMYB44 is a nuclear localized protein and displays transactivation activity at the C-terminus. HbMYB44 was ubiquitously expressed in rubber tree, and its expression was strongly induced by multiple phytohormones, drought stress, wounding, and H2O2 treatments. Furthermore, overexpression of HbMYB44 in Arabidopsis (OE) demonstrated that OE plants significantly enhanced stress tolerance, i.e., salt stress, osmotic stress, and drought stress. Additionally, HbMYB44 promoted recovery from root growth inhibition of OE plants caused by exogenous phytohormones (including abscisic acid, methyl jasmonic acid, gibberellic acid 3, and salicylic acid), but the opposite effect was present in response to ethephon. Interestingly, HbMYB44 increased the expression of its homologous genes and interacting protein-encoding genes in OE plants. Overall, HbMYB44 plays versatile functions in modulating multiple phytohormone signaling pathways and stress tolerance.
The first MYB transcription factor (TF) was identified as a protein encoded by a viral oncogene (v-MYB) in birds (Wang X. et al., 2018). In plants, MYB TFs compose one of the most important families of TFs and regulate various plant development processes and multiple stress responses (Ampomah-Dwamena et al., 2019). MYB TFs can be classified into four groups according to their repeats (R) with 50–53 amino acid residues: 1R-, R2R3-, 3R-, and 4R-MYBs (Zhang T. et al., 2018). R2R3-MYB TFs play important roles in regulating plant development, abiotic and biotic stress responses, etc. For example, the kiwifruit MYB7 modulates chlorophyll and carotenoid accumulation by activating the promoter of lycopene beta-cyclase (AdLCY-β) (Ampomah-Dwamena et al., 2019). Overexpression of wild soybean GsMYB1 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis (Shen et al., 2018). Recently, advances in genome and transcriptome sequencing techniques have facilitated the identification of the MYB family in non-model plant species, such as peach (Zhang C. et al., 2018) and Pleurotus ostreatus (Wang L. et al., 2018).
The Arabidopsis R2R3-MYB family can divide into 25 subgroups based on the conserved motifs present in the C-terminus to the MYB domain. AtMYB44, together with AtMYB70, AtMYB73, and AtMYB77, belongs to subgroup 22, which contains two conserved motifs, namely, 22.1 (TGLYMSPxSP) and 22.2 (GxFMxVVQEMIxxEVRSYM) (Stracke et al., 2001). The MYBs of subgroup 22 play essential roles in regulating plant stress response and phytohormone signaling. For instance, AtMYB73 expression was found to be upregulated under salt stress conditions in Arabidopsis, and it negatively regulated SOS1 and SOS3 transcripts (Kim et al., 2013). AtMYB44 confers abiotic stress tolerance and non-specific phytohormonal induction in Arabidopsis (Jung et al., 2008, 2010). Moreover, by activating EIN2 expression, AtMYB44 functions in various biotic stress responses, such as resistance to Myzus persicae Sulzer and Plutella xylostella L. (Lu et al., 2013). On the one hand, AtMYB44 regulates salicylic acid (SA)- and jasmonic acid (JA)-mediated defense responses by directly regulating WRKY70 expression; thus, this TF acts as an integrator of cross-talk between SA and JA in plant defense responses (Shim and Choi, 2013; Shim et al., 2013). On the other hand, AtMYB44 interacts with the abscisic acid (ABA) receptors PYL9 and PYL8 and regulates ABA signaling (Shim et al., 2013; Li et al., 2014; Zhao et al., 2014). Additionally, AtMYB44 is regulated by MPK3-mediated phosphorylation (Persak and Pitzschke, 2013). Recently, chromatin immunoprecipitation showed that several features present in the promoter region of AtMYB44 facilitate the binding of different TFs involving various signals, including a low density of nucleosomes, no DNA methylation, and multiple TF-binding elements (Nguyen et al., 2012; Nguyen and Cheong, 2018a,b). Although the function of MYB44 has been reported in the model plant species Arabidopsis, its function in non-model plant species is poorly understood. To improve our knowledge of its transcriptional regulation mechanism, the functions of MYB44 in different species need to be characterized.
Rubber tree (Hevea brasiliensis Müll. Arg) is the most important plant species for the production of natural rubber (NR) (Puskas et al., 2006). Latex within NR is produced in the cytoplasm of laticifer cells and harvested by regularly tapping the bark of rubber trees. Ethephon (ET) is widely used in NR production to increase the latex yield. Rubber trees often suffer from abiotic stress, wounding (caused by frequent tapping), drought, low temperature, and biotic stress, especially the destructive disease South American leaf blight (SALB), which is still a serious threat to plantations. From a biological point of view, NR is important for disease and insect defense and high-temperature stress responses in rubber trees (Sharkey and Yeh, 2001). Although important roles of MYBs have been revealed in plants, few MYB TFs in rubber tree have been characterized. HbMyb1 can suppress cell death induced by stress conditions in tobacco (Peng et al., 2011). The expression of HbSM1 (a 1R-MYB) was shown to be induced by phytohormones and wounding (Qin et al., 2014). Recently, 44 MYBs (HblMYB1-HblMYB44) were identified from the laticifer transcriptome database and the genome database of rubber tree. Among them, HblMYB44 (belonging to a new group G3) and HblMYB19 (belonging to a new group G2) were shown to bind to the promoters of HRT1, FDPS1, and SRPP in yeast and increase their expression in transient expression assays of tobacco (Wang et al., 2017). Until now, the function of MYBs of subgroup 22 has remained unknown in rubber tree. Due to the important functions of MYB44 in the plant stress response and phytohormone signaling, we assumed that MYB44 homologs may play a vital role in the stress response in rubber tree. In this study, HbMYB44 was cloned, and its function was characterized in rubber tree and transgenic Arabidopsis. These results will lay a good foundation for elucidating the transcriptional regulatory mechanism underlying the stress response in rubber tree.
The rubber tree clone CATAS7-33-97 was planted at the experimental farm of Danzhou, Hainan Province, China. At the flowering stage, samples of leaves, bark, latex, and flowers were collected from the same plants simultaneously. Tissue culture-generated seedlings were used for different stress and phytohormone treatments according to our reported method (Qin et al., 2015). Briefly, plants were subjected to drought stress treatment by continuously withholding water for 10 days, and the control plants were well-watered, with samples collected at the same time points. Wounds were made on the leaves by pinching with forceps, and untreated plants were used as controls. For chemical treatments, whole plants were sprayed with 2% (v/v) H2O2, 1.0% (v/v) ET, 200 mM methyl jasmonate (MeJA), 3 mM gibberellic acid 3 (GA3), 200 μM ABA, and 5 mM SA, respectively. All the chemicals were diluted in distilled water that included 0.05% (v/v) ethanol, and the control plants were sprayed with distilled water that included 0.05% (v/v) ethanol. Six plants were included in each treatment, and leaves from the six plants were harvested at each time point. Three different biological replicates were analyzed. The harvested samples were immediately frozen in liquid nitrogen and stored at −80°C until analysis.
The AtMYB44 (AT5G67300) sequence was subjected to tBLASTn searches of the transcriptome shotgun assembly (TSA) and genome database of H. brasiliensis. According to the obtained sequence, primers (MYB44-F1 and MYB44-R1, Table 1) were designed to amplify the full-length open reading frame (ORF) of HbMYB44 from the rubber tree.
The National Center for Biotechnology Information (NCBI) conserved domain searches1 and SMART2 were used to analyze the structure of HbMYB44. Sequences of HblMYBs identified from the laticifer transcriptome database and the genome database of H. brasiliensis (Wang et al., 2017), and Arabidopsis MYBs obtained from the phytozome,3 were used for the phylogenetic analysis. Information on H. brasiliensis and Arabidopsis MYBs is listed in Supplementary Table 1. Sequence alignments were conducted using the DNAMAN version 8.0 and ClustalX (Thompson et al., 2002). The phylogenetic analysis was performed using the MEGA version X based on the neighbor-joining method with a Poisson correction model and a bootstrap test of 1,000 replicates (Tamura et al., 2013).
Total RNA was extracted from different tissues using a Total RNA Isolation kit (OMEGA Bio-tek, Norcross, GA, United States). cDNA was synthesized with ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Shanghai, China) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on a CFX96™ Real-Time System (Bio-Rad Laboratories, Hercules, CA, United States) using the AceQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) according to the manufacturers’ instructions. Specific primers MYB44-F2 and MYB44-R2 were used for amplifying HbMYB44 (Table 1). HbActin (GenBank accession number: HO004792) and AtActin2 (GenBank accession number: NM_001338358.1) were used as housekeeping genes in the rubber tree and Arabidopsis, respectively. The relative expression levels of target genes were calculated using the 2–ΔΔCT method (Nolan et al., 2006; Schmittgen and Livak, 2008). All the data were analyzed by one-way ANOVA, and multiple comparisons were performed by the Tukey’s test at the P < 0.01 level. Figures were constructed by OriginPro 2018 (OriginLab Corporation, Northampton, MA, United States).
To determine the subcellular localization of HbMYB44, a fluorescent GFP tag was fused to the C-terminus of HbMYB44. The ORF of HbMYB44 was amplified with primers (MYB44-EcoR I-F1 and MYB44-BamH I-R1, Table 1) and subsequently inserted in the EcoR I/BamH I restriction sites of the vector 35S: XXGFP (provided by Dr. Xingliang Hou). The recombinant plasmid 35S:HbMYB44:mGFP was then transformed into Agrobacterium tumefaciens strain GV3101, and then, the GV3101 strain harboring the recombinant plasmid was transiently expressed in tobacco (Nicotiana benthamiana) leaf epidermal cells according to the methods of our previous study (Qin et al., 2019). The fluorescent protein expression was examined by fluorescence imaging together with confocal microscopy (Leica, Wetzlar, Germany) after 1 day of dark/light cultivation post infiltration. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) as previously reported (Kapuscinski, 1995) and visualized by confocal laser scanning microscopy.
The complete ORF of HbMYB44 (amplified by MYBY44-EcoR I-F1 and MYBY44-BamH I-R2) and fragments of HbMYB44 encoding the N-terminus [amino acids (aa) 1-200, amplified by MYBY44-EcoR I-F1 and MYBY44-▲N-BamH I-R3] or the C-terminus (aa 201-345, amplified by MYBY44-▲C-EcoR I-F2 and MYBY44-BamH I-R2), were amplified and cloned into the EcoR I/BamH II restriction sites of the vector pGBKT7,4 respectively. The target fragment was fused to the 3′ end of the GAL4 DNA-binding domain (DNA-BD) of the vector pGBKT7 between the EcoR I and BamH I restriction sites of MCS, such that a GAL4 DNA-BD-HbMYB44 fusion protein would be produced when the yeast cells were transformed. The pGBKT7-HbMYB44, pGBKT7-▲N-HbMYB44, and pGBKT7-▲ C-HbMYB44 constructs; the negative control pGBKT7; and the positive control pGBKT7-HbNAC24 were used (HbNAC24 is an NAC TF of the rubber tree and has been shown to have transcriptional activation activity) (Kang et al., 2014). These constructs were individually transformed into yeast strain Y2H Gold using a Yeastmaker™ Yeast Transformation System 2 (Clontech, Mountain View, CA, United States) according to the manufacturer’s instructions. If the fusion protein had transactivation activity, the colonies containing the pGBKT7+fusion protein construct in Y2H Gold would grow on SD/-Trp/-His/-Ade (TDO) media and become blue on TDO supplemented with X-α-Gal (TDO/X) media. All the yeast transformants were grown at 30°C for 2–3 days on SD media lacking tryptophan (SD/-Trp). Then, the positive clones were transferred to TDO media or TDO/X media and cultured at 30°C for 2–3 days (Fujita et al., 2004; Zhang et al., 2014). The experiment was repeated three times.
GV3101 cells harboring the recombinant plasmid 35S:HbMYB44:mGFP were used in Arabidopsis transformation for the overexpression of HbMYB44 (OE). Arabidopsis thaliana Col-0 wild type (WT) was transformed via the floral-dip method as previously described (Guan et al., 2014). All the transgenic plants were selected on Murashige and Skoog (MS) media that included 10 mg L–1 Basta. Then, the resistant plants of the T2 generation were tested with the primers MYB44-EcoR I-F1 and MYB44-BamH I-R1 of HbMYB44 and used for the following studies.
The expression of HbMYB44, its homologous genes (including AtMYB44, AtMYB73, AtMYB70, and AtMYB77), and genes (including AtWRKY70, AtPYL8, and AtPYL9) encoding the interacting proteins of AtMYB44 in OE and WT plants were analyzed using qRT-PCR according to the method mentioned above. The primers used are listed in Table 1.
The seeds of the T3 generation and WT seeds were sterilized with 10% bleach and 0.001% Triton X-100 for 10 min and washed five times with sterile water. The sterilized seeds were sown on MS media, stratified at 4°C in darkness for 2 days, and then transferred to a plant growth chamber (MMM/Climacell 707, Germany) set at 22°C with a 16 h light/8 h dark photoperiod (light intensity of 13,000 lux) for germination for 1–2 days. Then, the germinated seeds were transferred to MS media supplemented with 100 mM NaCl, 250 mM mannitol, 15% (w/v) PEG4000, 0.3 μM ET, 10 μM ABA, 20 μM MeJA, 50 μM GA3, and 10 μM SA. The plates were placed vertically in the illumination incubator under the same conditions used for germination, and the root length of the seedlings was measured after 10–13 days of treatments. For each treatment, at least six plants were tested in triplicate, and three different biological replicates were performed. The final value was the average of three batches of measurement data.
The rubber tree MYB44 homolog HbMYB44 (GenBank accession number: AZZ09361.1) was cloned using RT-PCR. The ORF of HbMYB44 is 1,038 bp in length and encodes a protein of 345 aa. No introns were found in the genomic sequence of HbMYB44. The deduced HbMYB44 protein contains 2 SANT domains (one is from the 11th to 60th aa, and the other is from the 63rd to 111th aa; Figures 1A,B), which indicated that HbMYB44 is an R2R3-MYB TF. A BLASTp search of the NCBI non-redundant (NR) protein database revealed that HbMYB44 was highly similar to MYB44 homologs from Ricinus communis, Populus euphratica, Glycine max, and A. thaliana with 72, 71, 61, and 55% identities, respectively (Figure 1B).
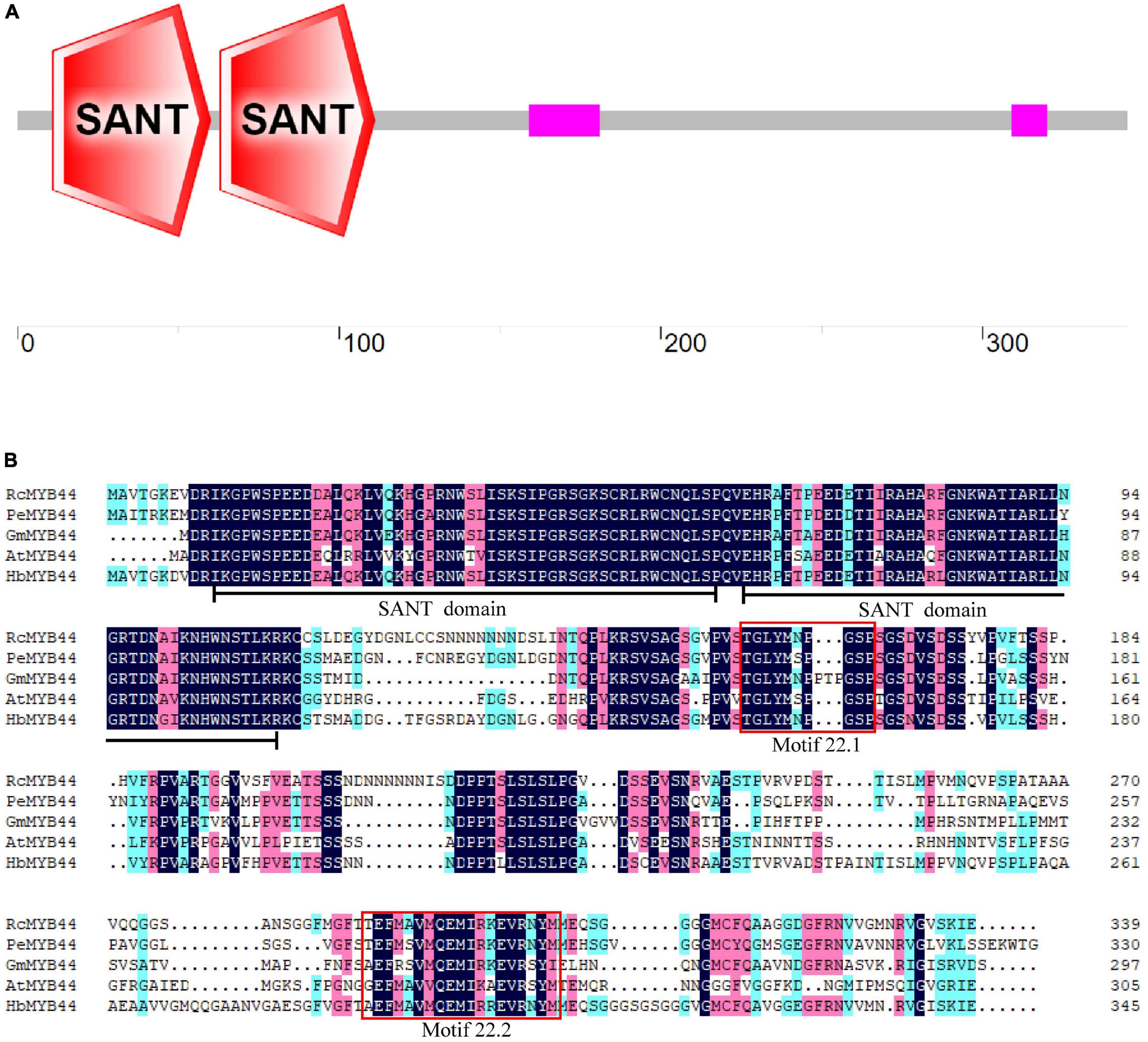
Figure 1. Analysis of HbMYB44 protein structure and multiple sequence alignment. (A) Protein architecture of HbMYB44. (B) Multiple sequence alignment of HbMYB44 and its homologous proteins. The proteins and their accession numbers are as follows, HbMYB44 (Hevea brasiliensis, JN038190), RcMYB44 (Ricinus communis, XP_002524926.1), PeMYB44 (Populus euphratica, XP_011006987.1), GmMYB44 (Glycine max, NP_001238087.1), and AtMYB44 (Arabidopsis thaliana, AT5G67300). Identical amino acids are marked with a dark background and well-conserved residues are marked with a pink background. Two SANT domains are underlined and two conserved motifs 22.1 and 22.2 are shown in red boxes.
Multiple sequence alignments between rubber tree and A. thaliana MYB family members indicated that HbMYB44 shared the highest similarity with rubber tree HblMYB7 (with 82% identity) and Arabidopsis AtMYB44 and AtMYB73 (with 55% identity). The phylogenetic analysis demonstrated that HbMYB44 clustered into subgroup 22 together with AtMYB44, AtMYB70, AtMYB73, AtMYB77, rubber tree HblMYB7, HblMYB9, HblMYB10, and HblMYB35 (Figure 2). Moreover, two specific motifs of subgroup 22, namely, motif 22.1 (TGLYMSPxSP) and motif 22.2 (GxFMxVVQEMIxxEVRSYM) were found to be present in HbMYB44 (Figure 1B, indicated by red boxes).
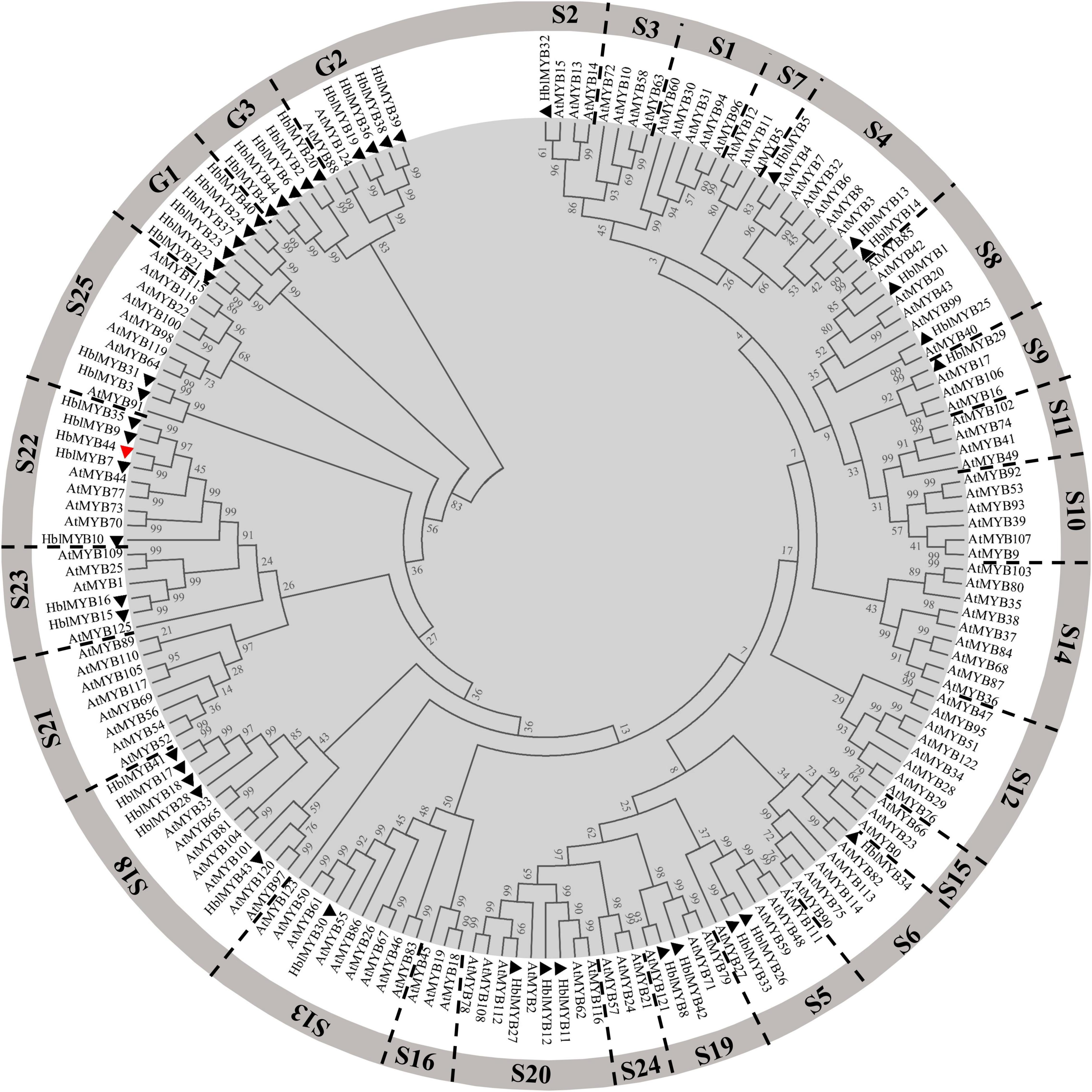
Figure 2. Phylogenetic tree analysis of HbMYB44 and the MYB family members of Arabidopsis thaliana and Hevea brasiliensis. HblMYBs identified from the laticifer transcriptome database and the genome database of Hevea brasiliensis (Wang et al., 2017) are indicated by triangles. Information on Hevea brasiliensis and Arabidopsis MYBs are listed in Supplementary Table 1.
To determine the subcellular localization of HbMYB44, a fusion protein HbMYB44-GFP driven by the 35S promoter was generated and transformed into tobacco epidermal cells. The nucleus dyed with DAPI was blue, and HbMYB44-GFP fusion protein was green, both of which were cyan after being merged. The results demonstrated that HbMYB44-GFP was exclusively localized in the nucleus of tobacco epidermal cells, while the control empty vector free-GFP was distributed throughout the cells (Figure 3A). These results demonstrated that HbMYB44 is a nuclear protein.
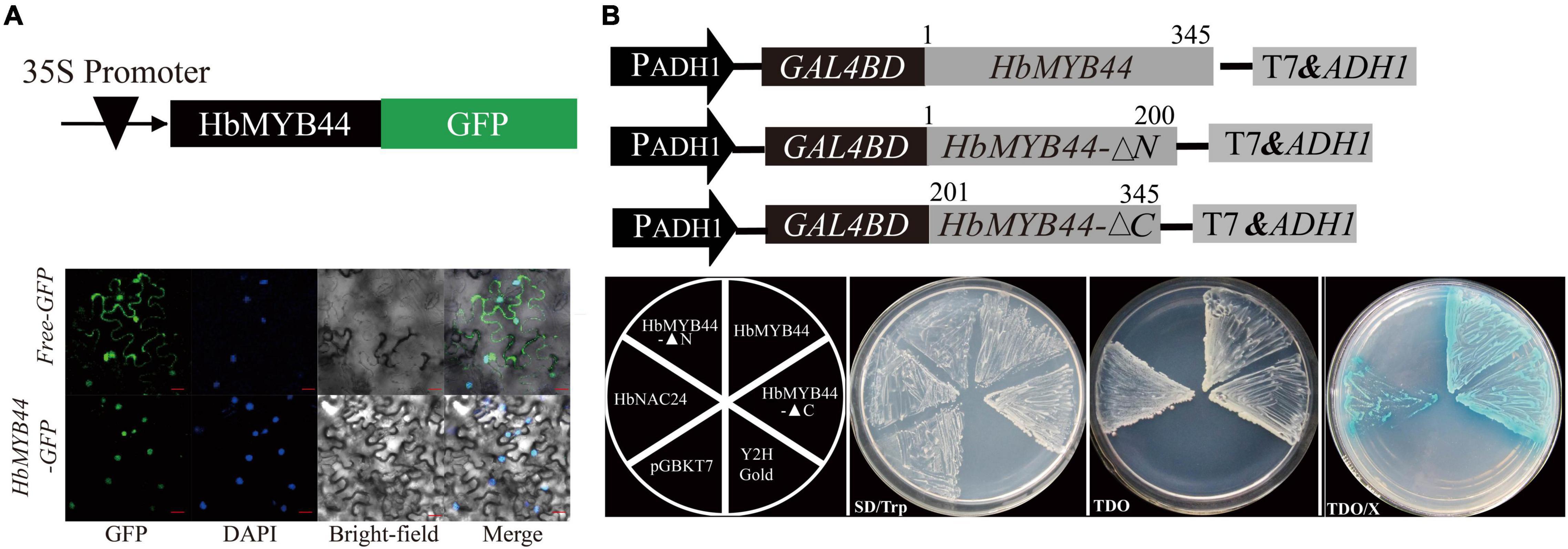
Figure 3. Subcellular localization and transactivation activity of HbMYB44. (A) Subcellular localization of HbMYB44 in the leaf epidermal cells of tobacco. Bars = 30 μm. (B) Analysis of the transactivation activity of HbMYB44 in yeast. The full length ORF of HbMYB44, and fragments encoding the N-terminus (1st–200th aa, HbMYB44-▲N) or the C-terminus (201st–345th aa, HbMYB44-▲C) of HbMYB44 were fused to the GAL4 DNA-binding domain of the pGBKT7 vector for the assay. TDO, SD/-Trp/-His/-Ade media; TDO/X, TDO media supplemented with X-α-Gal.
Since MYB proteins function as TFs (Li et al., 2006), we tested the transactivation activity of HbMYB44. GAL4 DNA-BD-HbMYB44 fusion proteins were expressed in yeast, and their ability to activate transcription from the GAL4 upstream activation sequence and to promote yeast growth in defective media was analyzed. The full-length ORF of HbMYB44 (HbMYB44), the N-terminal (encoding the 1st–200th aa, HbMYB44-▲N) and the C-terminal (encoding the 201st–345th aa, HbMYB44-▲C) fragments of HbMYB44 were individually fused to the GAL4 DNA-BD within the pGBKT7 vector (Figure 3B) and transformed into yeast strain Y2H Gold for transactivation activity analysis as previously reported (Zhao T. et al., 2016). As shown in Figure 3B, all constructs (including pGBKT7-HbMYB44, pGBKT7-HbMYB44-▲N, pGBKT7-HbMYB44-▲C, negative control pGBKT7, and positive control pGBKT7-HbNAC24) grew on the SD/-Trp media, but only pGBKT7-HbNAC24, pGBKT7-HbMYB44, and pGBKT7-HbMYB44-▲C grew on the TDO media and appeared blue on TDO/X media. The full-length ORF of HbMYB44 fused with the DNA-BD of GAL4 can self-activate the reporter genes in yeast cells. There was no effect on its transactivation activity when 200 aa within the N-terminus was deleted. However, truncation of 145 aa within the C-terminus resulted in the loss of transactivation activity. These results showed that HbMYB44 is a nuclear protein whose last 145 aa of the C-terminus have transactivation activity.
The expression pattern of HbMYB44 in rubber tree was measured using qRT-PCR. HbMYB44 was expressed in all the tested tissues, but its expression in flowers was higher than that in other tissues; the expression in the flowers was nearly 6.5-fold higher than that in the bark (Figure 4A). Since MYBs of subgroup 22 play important roles in regulating the stress response and phytohormone signaling in plants, HbMYB44 expression following different phytohormones and stress treatments was analyzed systematically. As shown in Figure 4B, HbMYB44 expression was rapidly induced by wounding, and it was upregulated by 45.9-fold at 2 h after treatment. Under drought stress, HbMYB44 expression was dramatically increased and peaked at 10 days after treatment, the level of which was 34.1-fold greater than that at 0 day (before treatment) (Figure 4C). After H2O2 treatment, HbMYB44 expression was increased by 12.7-fold at 6 h (Figure 4D). Furthermore, HbMYB44 expression was strongly induced in response to ET, MeJA, and ABA and peaked at 6 h, the levels of which were increased to 31.2-fold, 9.9-fold, and 28.4-fold, respectively (Figure 4E). In response to SA treatment, the HbMYB44 expression level was upregulated by 2.8-fold at 2 h (Figure 4E). Interestingly, HbMYB44 expression was rapidly triggered by GA3 application. HbMYB44 expression peaked at 0.5 h, which was an increase of approximately 83-fold, but then quickly fell back at 2 h (Figure 4E). These results indicated that the HbMYB44 expression was induced by multiple stress conditions and exogenous phytohormones.
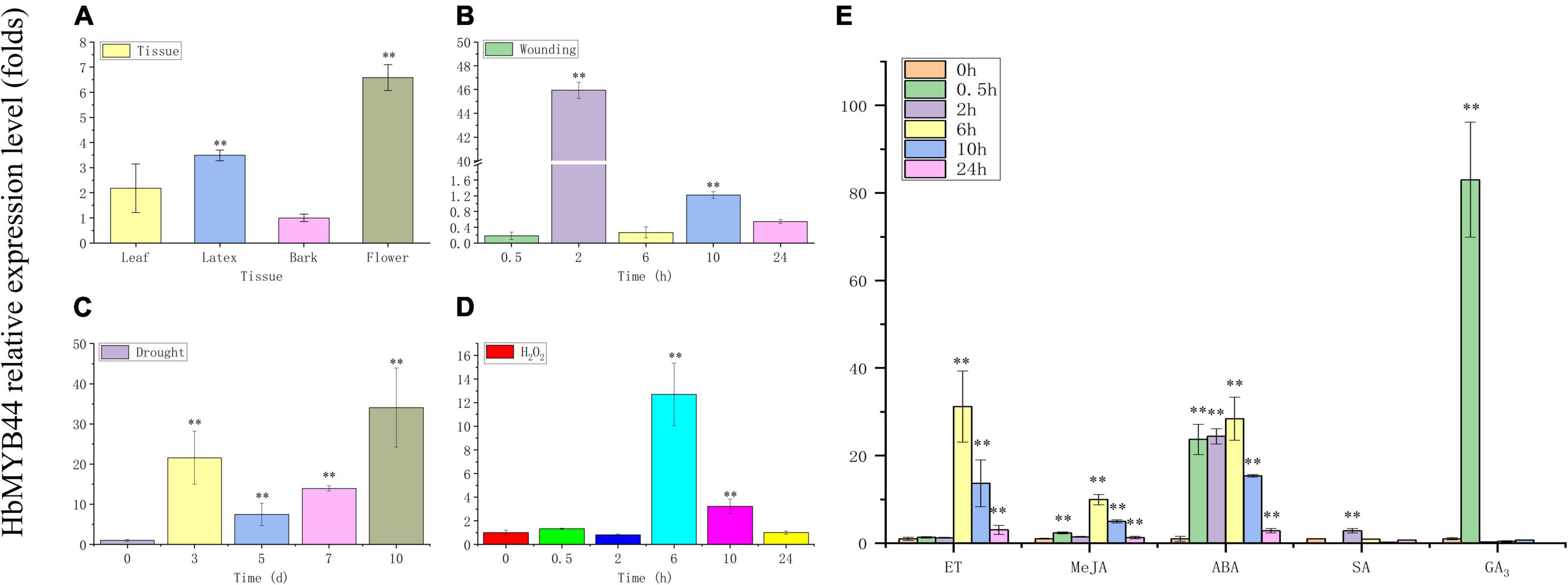
Figure 4. Expression of HbMYB44 in different tissues and in response to stress and phytohormone treatments in rubber tree. The expression of HbMYB44 was determined in different tissues of tapping trees (A) and in the leaves of tissue culture-generated seedlings subjected to wounding (B), drought stress (C), H2O2 (D), and phytohormones (E). The relative expression levels were compared with those at 0 h or 0 day. **Indicates a significant differences at the P < 0.01 level.
To verify the function of HbMYB44 in response to stress and phytohormones, transgenic Arabidopsis lines overexpressing HbMYB44 (OE) were generated. The OE plants of the T2 generation were tested using PCR with the specific primers of HbMYB44, the results of which indicated that all 10 OE lines contained the target gene (Figure 5A). Then, the expression level of HbMYB44 in OE lines was confirmed using qRT-PCR. There were differences in the expression abundance of HbMYB44 among different transgenic lines, and OE2 with the highest expression was used for further analysis (Figure 5B).
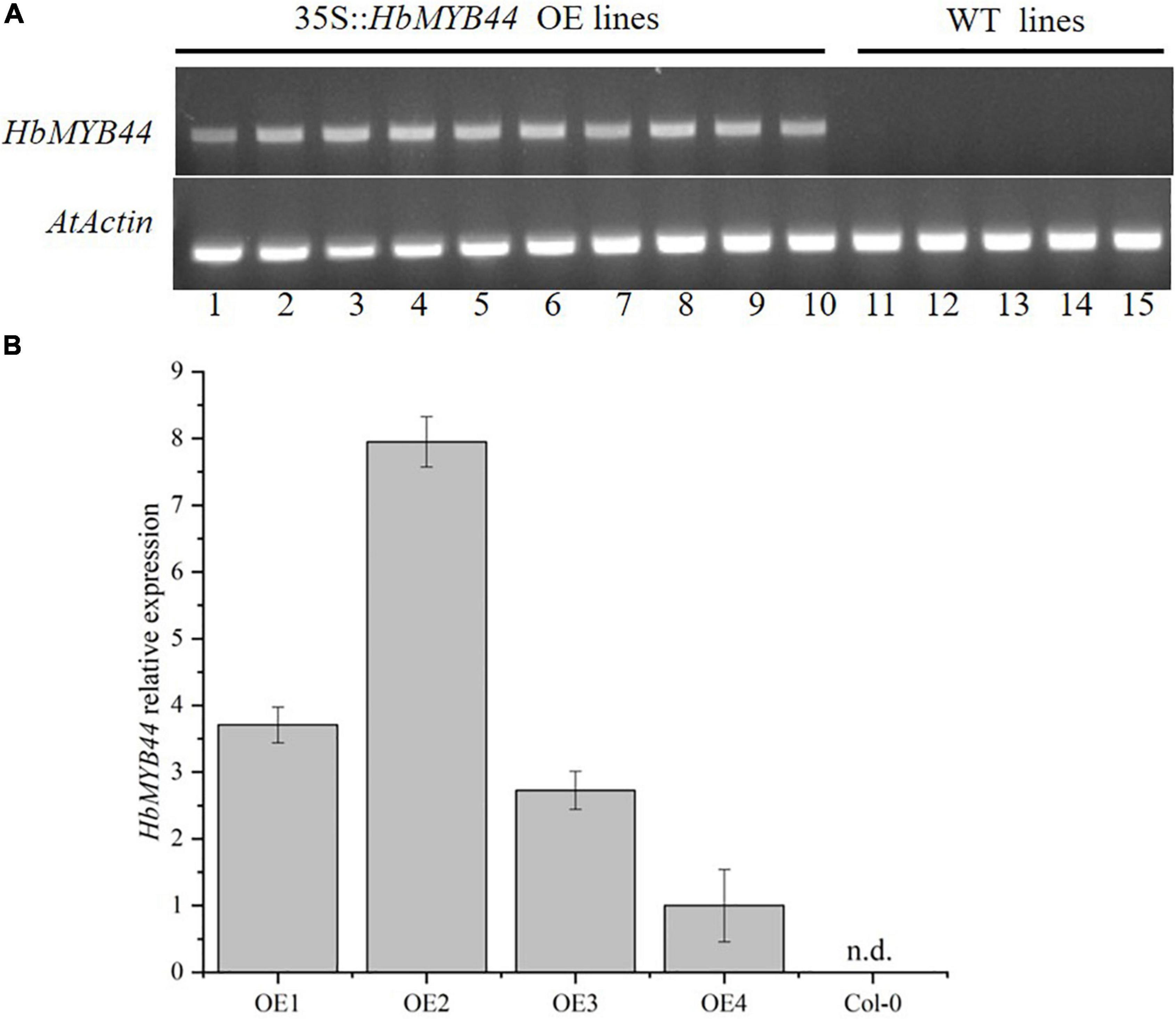
Figure 5. Verification of HbMYB44 expression in transgenic Arabidopsis. (A) Validation of HbMYB44 integration into the genome of transgenic plants. Arabidopsis AtActin2 was used as a control. (B) Analysis of HbMYB44 expression levels in transgenic plants by qRT-PCR. AtActin2 was used as a housekeeping gene. Each value is the average of six plants tested in triplicate. n.d. means no detection.
Since HbMYB44 functions as a TF with transactivation activity (Figure 3B), and its sequence is highly homologous to those of Arabidopsis MYBs of subgroup 22 (Figure 2), we analyzed whether HbMYB44 altered the expression of its homologous genes and their interacting proteins in transgenic Arabidopsis plants by qRT-PCR. As shown in Figure 6, except for that of AtMYB77, the expression of genes of subgroup S22, including AtMYB44, AtMYB70, and AtMYB73, was obviously increased in the HbMYB44-overexpressing plants compared with the WT plants. Moreover, the expression of AtWRKY70 and AtPYL8 but not AtPYL9 was also obviously increased in OE plants. These results indicated that HbMYB44 enhanced the expression of its homologous genes and interacting protein-encoding genes in transgenic Arabidopsis.
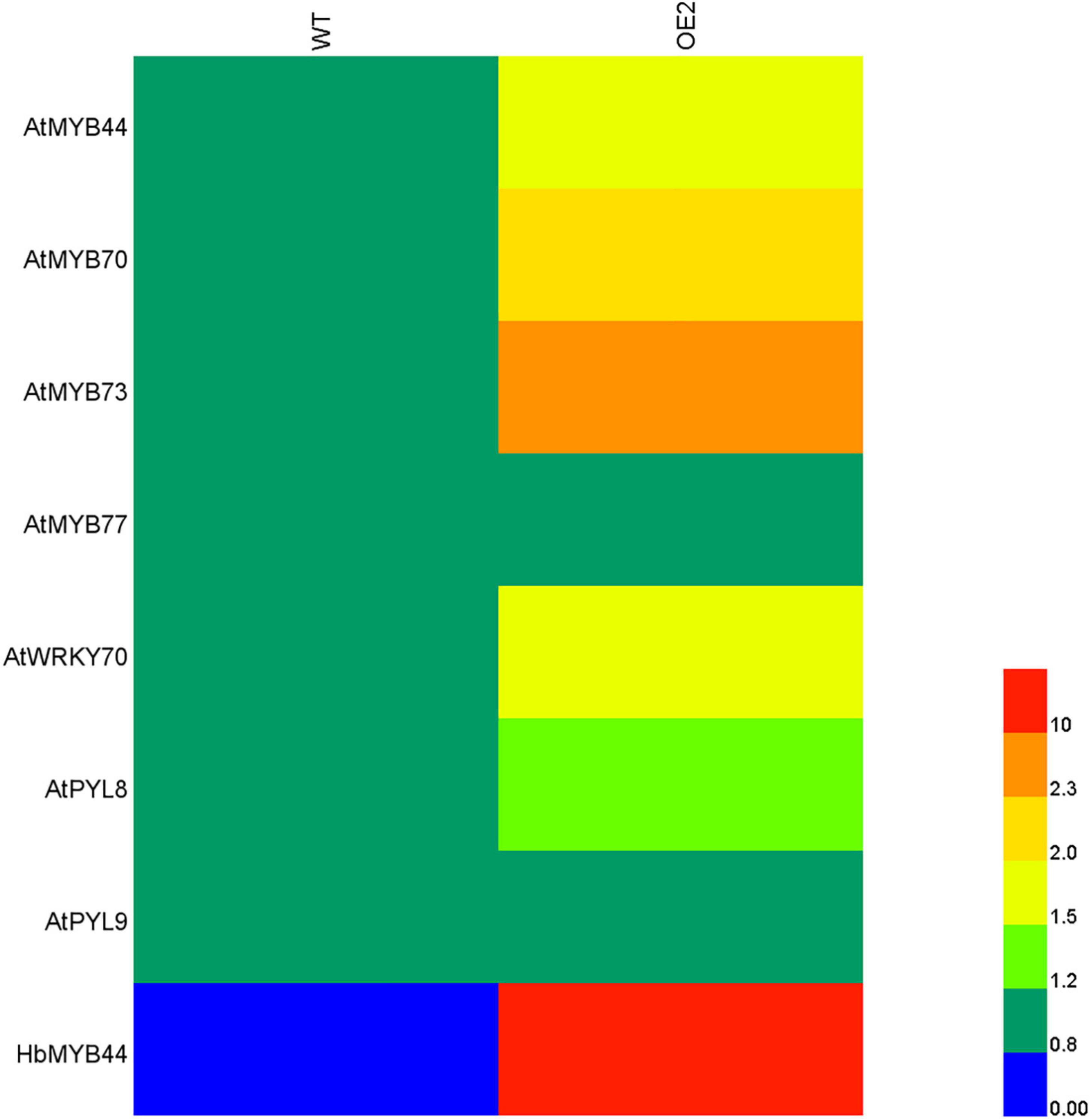
Figure 6. Expression of the subgroup S22 MYB and related genes in the HbMYB44 overexpression line and WT. AtActin2 was used as a housekeeping gene. Each value was the average of six plants tested in triplicate.
Under normal conditions, there was no marked difference in the morphology of WT and OE plants cultivated in MS media, but different phenotypes, especially in terms of root growth, were found under stress and treatments involving exogenous phytohormones (Figure 7A). As shown in Figure 7, the growth of leaves and roots of both in WT and OE2 plants was severely suppressed under stress and exogenous phytohormone conditions. In the media supplemented with exogenous ABA, MeJA, GA3, and SA, the root length of OE plants significantly increased compared with that of the WT plants under the same conditions. However, the root length of OE plants was reduced compared with that of the WT plants in the media supplemented with ET (Figures 7A,B). These results demonstrated that the root growth of OE plants is less sensitive to ABA, MeJA, GA3, and SA, but more sensitive to ET. Under NaCl-salt stress, mannitol-mediated osmotic stress, and PEG-simulated drought stress conditions, the root lengths were markedly increased in the OE plants compared with the WT; the roots especially increased more than 5-fold under NaCl conditions (Figures 7A,B). These results indicated that HbMYB44 enhanced tolerance to abiotic stress and promoted recovery from root growth inhibition in response to exogenous ABA, MeJA, GA3, and SA, suggesting that HbMYB44 is involved in a complex phytohormone signaling network.
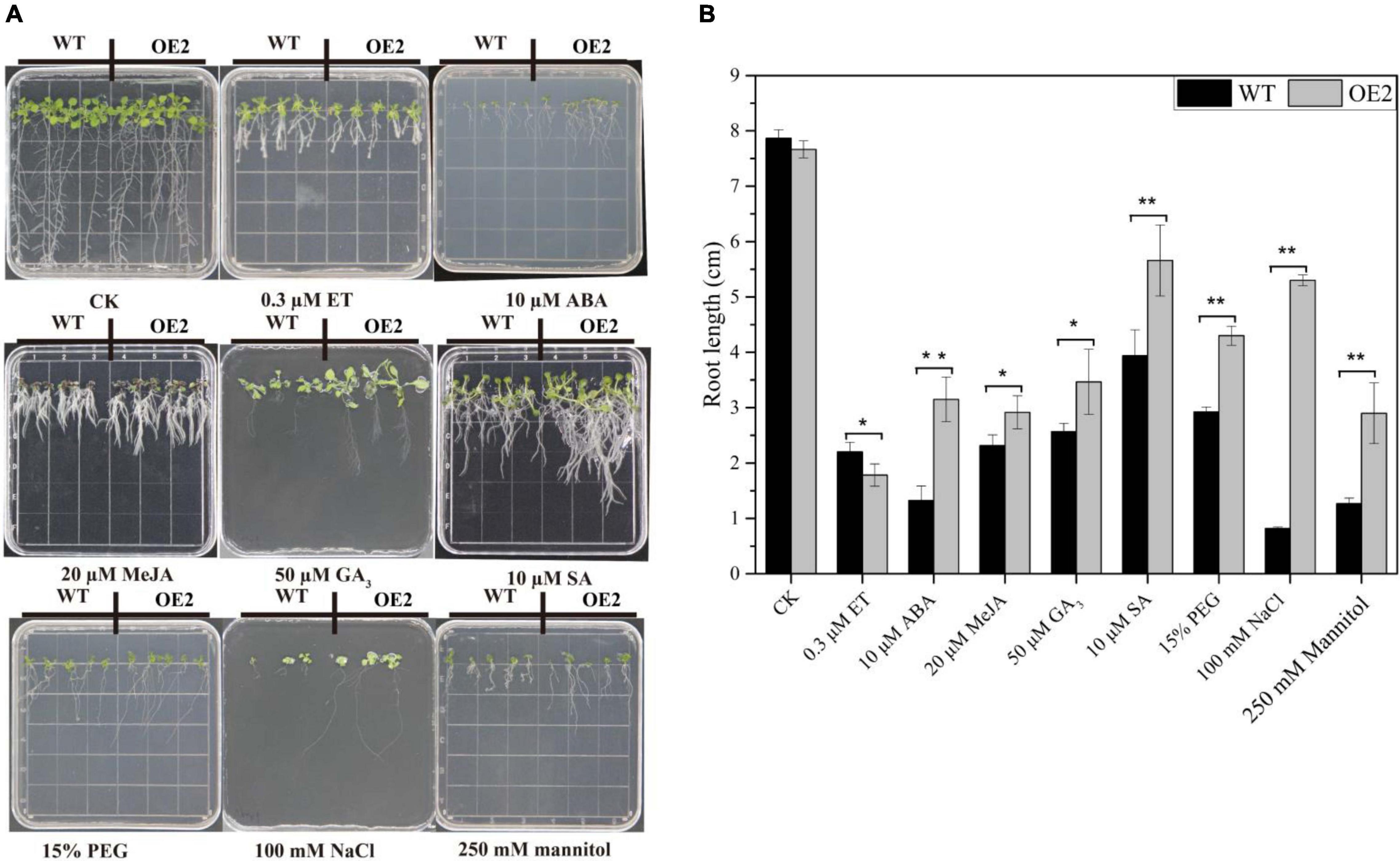
Figure 7. Phytohormone response and stress tolerance assays in the HbMYB44 overexpression line and wild type (WT). (A) Phenotypes of the HbMYB44 overexpression line and WT upon application of different phytohormones and stress conditions. (B) The root length of the seedlings under different phytohormone and stress treatments was measured after 10–13 days of treatments. Each value is the average of three batches of measurement data. The error bars represent ± SDs of three biological replicates, and the asterisks indicate significant differences between OE2 and WT plants (* and **Indicate P < 0.05 and P < 0.01, respectively).
The DNA-BD of MYB TFs is essential for DNA recognition and transactivation (Ogata et al., 1996). However, significant differences in plant and animal MYB TFs are present in the domains of these TFs (Williams and Grotewold, 1997). Most plant MYB functions are related to their DNA-BD (Millard et al., 2019). The phylogenetic analysis showed that 125 R2R3-MYBs identified from the genome of A. thaliana can be divided into 25 subgroups (Stracke et al., 2001). There are two typical SANT domains present in subgroup 22 MYB TFs, enabling DNA-binding specificities and DNA interactions (Millard et al., 2019). Additionally, the subgroup 22 MYB TFs contain two conserved motifs, namely, motif 22.1 and motif 22.2, which are the binding domains of interacting proteins (Stracke et al., 2001; Li et al., 2014). This study showed that HbMYB44 is a member of subgroup 22, as supported by the phylogenetic analysis in which HbMYB44 is categorized into subgroup 22 together with AtMYB44, AtMYB70, AtMYB73, AtMYB77 and with rubber tree HblMYB7, HblMYB9, HblMYB10, and HblMYB35 (Figure 2). The two conserved motifs (22.1 and 22.2) are also present in HbMYB44 (Figure 1B). Moreover, HbMYB44 is located to the nucleus and has transactivation activity at the C-terminus. The region encompassing aa 201-345 is indispensable for transcriptional activation (Figure 3). These results indicated that HbMYB44 is a TF with transcriptional activation activity. Based on the conserved function of the genes in subgroup 22, it is speculated that HbMYB44, HblMYB7, HblMYB9, HblMYB10, and HblMYB35 in this subgroup may play an important role in phytohormone signaling and stress responses. Thus, further study should be carried out to verify the functions of these genes, which would help elucidate phytohormone signaling networks involved in plant defense and NR biosynthesis in rubber tree.
In plants, MYB44 is a key factor functioning as a bHLH-MYB-WRKY70 transcriptional module in plant non-specific phytohormone induction (Jung et al., 2010), defense responses (Shim and Choi, 2013; Shim et al., 2013), root elongation (Zhao Q. et al., 2016), stress resistance (Park et al., 2012), etc. Genes encoding the subgroup 22 MYB TFs have similar expression patterns and are associated with resistance to disease and stress. AtMYB44 is expressed in different tissues throughout plants, with the highest abundance in the vasculature and stomata (Kirik et al., 1998). Moreover, AtMYB44 expression was shown to be inducted in response to drought, cold, high salinity, CdCl2, sugars, wounding, SA, MeJA, ET, GA3, ABA, brassinosteroid, and cytokinin (Chen et al., 2006; Jung et al., 2008, 2010). The expression patterns of AtMYB44, AtMYB73, and AtMYB77 were also shown to increase after wounding (Cheong et al., 2002), white-light (Ma et al., 2005) and cold stress treatments (Fowler and Thomashow, 2002). In this study, HbMYB44 was expressed in all tissues, and it was strongly induced by multiple stresses (including wounding, drought stress, and H2O2 treatments) in rubber tree (Figure 4). Moreover, its transcript rapidly accumulated in response to phytohormones, and a significant increase in HbMYB44 transcription was observed within 0.5 h after treatment with MeJA, ABA, and GA3 (Figure 4E). Therefore, HbMYB44 transcripts accumulated non-specifically under diverse stress conditions and in response to various phytohormones, the results of which are similar to those of AtMYB44 in A. thaliana, suggesting HbMYB44 has functions similar to those of AtMYB44.
The stress response and growth regulation of MYB TFs are related to the phytohormone signaling pathway. For instance, AtMYB44 regulates EIN2 expression, and EIN2 is the key gene in ethylene signaling and insect resistance. Moreover, AtMYB44 regulates plant insect resistance by activating EIN2 and its related defense responses in Arabidopsis (Lu et al., 2013). The ABA receptor RCAR3/PYL8 promotes lateral root growth by directly interacting with MYB77, MYB44, and MYB73, then enhancing MYB77-dependent transcription of auxin-responsive genes, to augment auxin signaling (Zhao et al., 2014). In the ABA signaling pathway, the ABA receptor RCAR1/PYL9 interacts with all the members of the subgroup 22 MYBs, i.e., AtMYB44, AtMYB70, AtMYB73, and AtMYB77 (Li et al., 2014). In Solanum melongena, the AtMYB44 homolog SmMYB44 enhances resistance to Ralstonia solanacearum by directly binding to the promoter of spermidine synthase (SPDS) gene SmSPDS and activating its expression (Qiu et al., 2019). These studies suggested that the members of subgroup 22 MYBs share similar functions and are functionally redundant. Through hormone crosstalk, AtMYB44 directly binds to the promoter of AtWRKY70, regulates AtWRKY70 expression, and modulates antagonistic interactions between SA and JA signaling (Shim et al., 2013). Phytohormones play important roles in the NR production of rubber trees. The application of ET to the bark of rubber tree increases NR yield per tapping (Kush, 1994), and JA treatment of the bark can promote secondary laticifer differentiation and subsequent NR biosynthesis (Haoji-Lin, 2000; Deng et al., 2018). Rubber tree plantations often suffer from various biotic stresses and abiotic stresses, especially wounding (caused by tapping in the bark for NR harvesting). Thus, the identification and functional characterization of stress response and phytohormone signaling genes are of great significance for improving plant stress resistance and NR yield in rubber trees. In this study, overexpression of HbMYB44 in Arabidopsis demonstrated that OE plants enhanced resistance to mannitol-mediated osmotic stress, NaCl-salt stress, and PEG-simulated drought stress (Figure 7). Moreover, HbMYB44 promoted recovery from root growth inhibition caused by ABA, MeJA, GA3, and SA (Figure 7). In addition, the leaf biomass of OE plants was higher than those of the WT plants in the media supplemented with GA3, and the root biomass of OE plants was higher than those of WT plants in the media supplemented with SA (Figure 7A). These results demonstrated that HbMYB44 is involved in a complex phytohormone signaling network. Interestingly, expression of the genes of subgroup S22 (including AtMYB44, AtMYB70, and AtMYB73) and interacting protein-encoding genes (AtWRKY70 and AtPYL8) were significantly increased in OE plants (Figure 6). These results suggested that HbMYB44 enhances stress tolerance and modulates phytohormone induction by increasing the expression of its homologous genes and interacting protein-encoding genes in Arabidopsis. Therefore, overexpression of HbMYB44 in the rubber tree will be further conducted to determine its function in NR biosynthesis, stress resistance, and phytohormone signaling.
The rubber tree HbMYB44 is a member of the subgroup 22 MYB TFs, which functions in the nucleus and has transactivation activity via its C-terminus. HbMYB44 expression was strongly induced in response to multiple phytohormones and stresses. The overexpression of HbMYB44 in Arabidopsis enhanced tolerance to abiotic stress and promoted recovery from root growth inhibition by exogenous phytohormones. This study suggested that HbMYB44 plays an important role in modulating multiple phytohormone signaling pathways and stress resistance of rubber tree.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
BQ was the experimental designer and the executor of the experimental research, completed the data analysis, and wrote the first draft of the manuscript. BQ, S-LF, H-YY, and Y-XL participated in experimental design, performed experimental research, and experimental result analysis. BQ and L-FW directed the design of the experiment, data analysis, and the modification of the manuscript. All authors read and approved the final manuscript.
This study was funded by the Major Science and Technology Plan of Hainan Province (ZDKJ2021004) and National Natural Science Foundation of China (Grant Nos. 31570591, 31270643, and 31970364).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Xing-Liang Hou (South China Botanical Garden, Chinese Academy of Sciences), providing the vector 35S: XXGFP, and Gui-Juan Kang (Rubber Research Institute, Chinese Academy of Tropical Agricultural Sciences) for the assistance in the transcriptional activation assay.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.893896/full#supplementary-material
aa, amino acids; ABA, abscisic acid; DAPI, 4’,6-diamidino-2-phenylindole; DNA-BD, DNA-binding domain; ET, ethephon; GA3, gibberellic acid 3; GFP, green fluorescent protein; MeJA, methyl jasmonic acid; NR, natural rubber; PEG, polyethylene glycol; SA, salicylic acid; TDO, SD/-Trp/-His/-Ade; TF, transcription factor; TSA, transcriptome shotgun assembly; X- α-Gal, 5-bromo-4-chloro-3-indolyl- α-d -galactopyranoside.
Ampomah-Dwamena, C., Thrimawithana, A. H., Dejnoprat, S., Lewis, D., Espley, R. V., and Allan, A. C. (2019). A kiwifruit (actinidia deliciosa) R2R3-MYB transcription factor modulates chlorophyll and carotenoid accumulation. New Phytol. 221, 309–325. doi: 10.1111/nph.15362
Chen, Y. H., Yang, X. Y., He, K., Liu, M. H., Li, J. G., Gao, Z. F., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Cheong, Y. H., Chang, H.-S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. doi: 10.1104/pp.002857
Deng, X., Guo, D., Yang, S., Shi, M., Chao, J., Li, H., et al. (2018). Jasmonate signalling in the regulation of rubber biosynthesis in laticifer cells of rubber tree, Hevea brasiliensis. J. Exp. Bot. 69, 3559–3571. doi: 10.1093/jxb/ery169
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Guan, Q. J., Wang, L. F., Bu, Q. Y., and Wang, Z. Y. (2014). The rice gene OsZFP6 functions in multiple stress tolerance responses in yeast and Arabidopsis. Plant Physiol. Biochem. 82, 1–8. doi: 10.1016/j.plaphy.2014.04.021
Haoji-Lin, B. Z. (2000). Laticifer differentiation in Hevea brasiliensis: induction by exogenous jasmonic acid and linolenic acid. Ann. Bot. 85, 37–43. doi: 10.1006/anbo.1999.0995
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant. Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Jung, C., Shim, J. S., Seo, J. S., Lee, H. Y., Kim, C. H., Choi, Y. D., et al. (2010). Non-specific phytohormonal induction of AtMYB44 and suppression of jasmonate-responsive gene activation in Arabidopsis thaliana. Mol. Cells 29, 71–76. doi: 10.1007/s10059-010-0009-z
Kang, G., Yu, L. I., and Zeng, R. (2014). Clone and expression of HbNAC24 gene from Hevea brasiliensis. Acta Bot. Boreali Occidentalia Sin. 34, 2374–2381.
Kapuscinski, J. (1995). DAPI: a DNA-specific fluorescent probe. Biotech Histochem. 70, 220–233. doi: 10.3109/10520299509108199
Kim, J. H., Nguyen, N. H., Jeong, C. Y., Nguyen, N. T., Hong, S. W., and Lee, H. (2013). Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant. Physiol. 170, 1461–1465. doi: 10.1016/j.jplph.2013.05.011
Kirik, V., Kolle, K., Misera, S., and Baumlein, H. (1998). Two novel MYB homologues with changed expression in late embryogenesis-defective Arabidopsis mutants. Plant Mol. Biol. 37, 819–827. doi: 10.1023/a:1006011002499
Li, D., Li, Y., Zhang, L., Wang, X., Zhao, Z., Tao, Z., et al. (2014). Arabidopsis ABA receptor RCAR1/PYL9 interacts with an R2R3-type MYB transcription factor, AtMYB44. Int. J. Mol. Sci. 15, 8473–8490. doi: 10.3390/ijms15058473
Li, J., Yang, X., Yan, W., Li, X., Gao, Z., Meng, P., et al. (2006). Two groups of MYB transcription factors share a motif which enhances trans-activation activity. Biochem. Biophys. Res. Commun. 341, 1155–1163. doi: 10.1016/j.bbrc.2006.01.077
Lu, B. B., Li, X. J., Sun, W. W., Li, L., Gao, R., Zhu, Q., et al. (2013). AtMYB44 regulates resistance to the green peach aphid and diamondback moth by activating EIN2-affected defences in Arabidopsis. Plant. Biol. (Stuttg) 15, 841–850. doi: 10.1111/j.1438-8677.2012.00675.x
Ma, L., Sun, N., Liu, X., Jiao, Y., Zhao, H., and Deng, X. W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138, 80–91. doi: 10.1104/pp.104.054783
Millard, P. S., Kragelund, B. B., and Burow, M. (2019). R2R3 MYB transcription factors - functions outside the DNA-binding domain. Trends Plant Sci. 24, 934–946. doi: 10.1016/j.tplants.2019.07.003
Nguyen, N. H., and Cheong, J. J. (2018a). H2A.Z-containing nucleosomes are evicted to activate AtMYB44 transcription in response to salt stress. Biochem. Biophys. Res. Commun. 499, 1039–1043. doi: 10.1016/j.bbrc.2018.04.048
Nguyen, N. H., and Cheong, J. J. (2018b). The AtMYB44 promoter is accessible to signals that induce different chromatin modifications for gene transcription. Plant Physiol. Biochem. 130, 14–19. doi: 10.1016/j.plaphy.2018.06.030
Nguyen, X. C., Hoang, M. H., Kim, H. S., Lee, K., Liu, X. M., Kim, S. H., et al. (2012). Phosphorylation of the transcriptional regulator MYB44 by mitogen activated protein kinase regulates Arabidopsis seed germination. Biochem. Biophys. Res. Commun. 423, 703–708. doi: 10.1016/j.bbrc.2012.06.019
Nolan, T., Hands, R. E., and Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582. doi: 10.1038/nprot.2006.236
Ogata, K., Kanei-Ishii, C., Sasaki, M., Hatanaka, H., Nagadoi, A., Enari, M., et al. (1996). The cavity in the hydrophobic core of Myb DNA binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Mol. Biol. 3, 178–187. doi: 10.1038/nsb0296-178
Park, J.-B., Sendon, P. M., Kwon, S. H., Seo, H. S., Park, S.-K., Kim, J. H., et al. (2012). Overexpression of stress-related genes, BrERF4 and AtMYB44, in Arabidopsis thaliana alters cell expansion but not cell proliferation during leaf growth. J. Plant Biol. 55, 406–412. doi: 10.1007/s12374-012-0114-y
Peng, S. Q., Wu, K. X., Huang, G. X., and Chen, S. C. (2011). HbMyb1, a Myb transcription factor from Hevea brasiliensis, suppresses stress induced cell death in transgenic tobacco. Plant Physiol. Biochem. 49, 1429–1435. doi: 10.1016/j.plaphy.2011.09.007
Persak, H., and Pitzschke, A. (2013). Tight interconnection and multi-level control of Arabidopsis MYB44 in MAPK cascade signalling. PLoS One 8:e57547. doi: 10.1371/journal.pone.0057547
Puskas, J. E., Gautriaud, E., Deffieux, A., and Kennedy, J. P. (2006). Natural rubber biosynthesis—a living carbocationic polymerization? Progr. Polymer Sci. 31, 533–548.
Qin, B., Wang, M., He, H. X., Xiao, H. X., Zhang, Y., and Wang, L. F. (2019). Identification and characterization of a potential candidate Mlo gene conferring susceptibility to powdery mildew in rubber tree. Phytopathology 109, 1236–1245. doi: 10.1094/PHYTO-05-18-0171-R
Qin, B., Zhang, Y., and Wang, M. (2014). Molecular cloning and expression of a novel MYB transcription factor gene in rubber tree. Mol. Biol. Rep. 41, 8169–8176. doi: 10.1007/s11033-014-3717-1
Qin, B., Zheng, F., and Zhang, Y. (2015). Molecular cloning and characterization of a Mlo gene in rubber tree (Hevea brasiliensis). J. Plant Physiol. 175, 78–85. doi: 10.1016/j.jplph.2014.10.019
Qiu, Z., Yan, S., Xia, B., Jiang, J., Yu, B., Lei, J., et al. (2019). The eggplant transcription factor MYB44 enhances resistance to bacterial wilt by activating the expression of spermidine synthase. J. Exp. Bot. 70, 5343–5354. doi: 10.1093/jxb/erz259
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Sharkey, T. D., and Yeh, S. (2001). Isoprene emission from plants. Ann. Rev. Plant Physiol. Plant Mol. Biol. 52, 407–436. doi: 10.1146/annurev.arplant.52.1.407
Shen, X. J., Wang, Y. Y., Zhang, Y. X., Guo, W., Jiao, Y. Q., and Zhou, X. A. (2018). Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int. J. Mol. Sci. 19:3958. doi: 10.3390/ijms19123958
Shim, J. S., and Choi, Y. D. (2013). Direct regulation of WRKY70 by AtMYB44 in plant defense responses. Plant Signal Behav. 8:e20783. doi: 10.4161/psb.24509
Shim, J. S., Jung, C., Lee, S., Min, K., Lee, Y. W., Choi, Y., et al. (2013). AtMYB44 regulates WRKY70 expression and modulates antagonistic interaction between salicylic acid and jasmonic acid signaling. Plant J. 73, 483–495. doi: 10.1111/tpj.12051
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant. Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2002). Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. Chapter 2:Unit23. doi: 10.1002/0471250953.bi0203s00
Wang, L., Gao, W., Wu, X., Zhao, M., Qu, J., Huang, C., et al. (2018). Genome-wide characterization and expression analyses of Pleurotus ostreatus MYB transcription factors during developmental stages and under heat stress based on de novo sequenced genome. Int. J. Mol. Sci. 19:2052. doi: 10.3390/ijms19072052
Wang, X., Angelis, N., and Thein, S. L. (2018). MYB - a regulatory factor in hematopoiesis. Gene 665, 6–17. doi: 10.1016/j.gene.2018.04.065
Wang, Y., Zhan, D. F., Li, H. L., Guo, D., Zhu, J. H., and Peng, S. Q. (2017). Transcriptome-wide identification and characterization of MYB transcription factor genes in the laticifer cells of Hevea brasiliensis. Front. Plant Sci. 8:1974. doi: 10.3389/fpls.2017.01974
Williams, C. E., and Grotewold, E. (1997). Differences between plant and animal Myb domains are fundamental for DNA binding activity, and chimeric Myb domains have novel DNA binding specificities. J. Biol. Chem. 272, 563–571. doi: 10.1074/jbc.272.1.563
Zhang, C., Ma, R., Xu, J., Yan, J., Guo, L., Song, J., et al. (2018). Genome-wide identification and classification of MYB superfamily genes in peach. PLoS One 13:e0199192. doi: 10.1371/journal.pone.0199192
Zhang, J., Chen, J., Yi, Q., Hu, Y., Liu, H., Liu, Y., et al. (2014). Novel role of ZmaNAC36 in co-expression of starch synthetic genes in maize endosperm. Plant Mol. Biol. 84, 359–369. doi: 10.1007/s11103-013-0153-x
Zhang, T., Zhao, Y., Wang, Y., Liu, Z., and Gao, C. (2018). Comprehensive analysis of MYB gene family and their expressions under abiotic stresses and hormone treatments in Tamarix hispida. Front. Plant. Sci. 9:1303. doi: 10.3389/fpls.2018.01303
Zhao, Q., Li, M., Jia, Z., Liu, F., Ma, H., Huang, Y., et al. (2016). AtMYB44 positively regulates the enhanced elongation of primary roots induced by N-3-oxo-hexanoyl-homoserine lactone in Arabidopsis thaliana. Mol. Plant Microbe Int. 29, 774–785. doi: 10.1094/MPMI-03-16-0063-R
Zhao, T., Huang, X., and Xia, Y. (2016). Human heart cell proteins interacting with a C-terminally truncated 2A protein of coxsackie B3 virus: identification by the yeast two-hybrid system. Virus Genes 52, 172–178. doi: 10.1007/s11262-015-1270-1
Keywords: rubber tree, MYB transcription factor, HbMYB44, phytohormone signaling, stress tolerance
Citation: Qin B, Fan S-L, Yu H-Y, Lu Y-X and Wang L-F (2022) HbMYB44, a Rubber Tree MYB Transcription Factor With Versatile Functions in Modulating Multiple Phytohormone Signaling and Abiotic Stress Responses. Front. Plant Sci. 13:893896. doi: 10.3389/fpls.2022.893896
Received: 11 March 2022; Accepted: 09 May 2022;
Published: 02 June 2022.
Edited by:
Prasanta Kumar Subudhi, Louisiana State University, United StatesReviewed by:
Deepanker Yadav, Guru Ghasidas Vishwavidyalaya, IndiaCopyright © 2022 Qin, Fan, Yu, Lu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Feng Wang, bGZ3YW5nQGNhdGFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.