
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 23 June 2022
Sec. Plant Systems and Synthetic Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.893095
This article is part of the Research Topic Insights in Plant Systems and Synthetic Biology: 2021 View all 5 articles
The evolution of Crassulacean acid metabolism (CAM) by plants has been one of the most successful strategies in response to aridity. On the onset of climate change, expanding the use of water efficient crops and engineering higher water use efficiency into C3 and C4 crops constitute a plausible solution for the problems of agriculture in hotter and drier environments. A firm understanding of CAM is thus crucial for the development of agricultural responses to climate change. Computational models on CAM can contribute significantly to this understanding. Two types of models have been used so far. Early CAM models based on ordinary differential equations (ODE) reproduced the typical diel CAM features with a minimal set of components and investigated endogenous day/night rhythmicity. This line of research brought to light the preponderant role of vacuolar malate accumulation in diel rhythms. A second wave of CAM models used flux balance analysis (FBA) to better understand the role of CO2 uptake in flux distribution. They showed that flux distributions resembling CAM metabolism emerge upon constraining CO2 uptake by the system. We discuss the evolutionary implications of this and also how CAM components from unrelated pathways could have integrated along evolution.
Crassulacean acid metabolism is a photosynthetic adaptation syndrome of carbon concentration that has evolved multiple times in plants (Crayn et al., 2004; Winter and Smith, 2012). It consists in the nocturnal fixation of CO2 into malate, which is stored in the vacuole and decarboxylated the next day providing CO2 for Rubisco (Dodd et al., 2002). It is commonly associated with conditions of water scarcity in which plants close their stomata during the day to avoid water loss. Keeping stomata closed during the day while concentrating CO2 at night permits a high water use efficiency, allowing plants to remain productive even under dry conditions. A great interest in CAM crops has recently developed, given the potential of extending their use in semi-arid and degraded lands (Liu et al., 2018). In natural ecosystems, CAM plants are less affected by increased nighttime temperatures and rainfall variability than C4 grasses (Huang et al., 2020) and are increasing their cover in many arid regions (Yu and D’Odorico, 2015). Global climate models predict drier regimes and decrease in soil moisture in large areas of the world (Dai, 2013). In many of today’s agricultural lands, CAM crops could be the only crops able to grow in a fairly close future. Understanding CAM evolution is especially relevant for CAM engineering into C3 crops. The multiple occurrences of CAM in numerous lineages across the plant kingdom and different levels of CAM expression suggest that the evolutionary path from C3 to CAM is relatively straightforward (Schiller and Bräutigam, 2021). Finding the key events that led to CAM emergence can potentially pinpoint the key transformations necessary for engineering CAM into C3 plants.
Mathematical models on CAM metabolism provide valuable information. Two types of models have been used to study CAM: ordinary differential equations (ODE) and flux balance analysis (FBA) models. ODE-based models can accurately describe metabolite concentrations and their change over time at the cost of requiring quantitative knowledge on the parameters and enzyme activities of the pathways being modeled. On the other hand, FBA-based models require no quantitative a priori knowledge. They can be used to study steady state flux distributions but provide no information on concentrations or transient dynamics.
ODE-based models have determined the components and regulatory interactions sufficient to bring about CAM behavior, while FBA models have shown that many CAM defining features —such as nocturnal malate accumulation—are the result of flux optimization upon limited CO2. Here, we review the findings of both groups of models. Although most of the works were not conceived with evolution in mind, several of their findings have direct evolutionary implications, in particular for the ongoing discussion on the C3-to-CAM evolutionary continuum (Cushman and Bohnert, 1999; Dodd et al., 2002; Silvera et al., 2010; Bräutigam et al., 2017).
Crassulacean acid metabolism is divided into four archetypical phases. Phase I occurs at night when the stomata are open and starch reserves are used to synthesize phosphoenolpyruvate (PEP). High amounts of PEP are needed to fix CO2 overnight, a reaction catalyzed by PEP carboxylase (PEPC). PEP carboxylase is reversibly phosphorylated during the night to reduce malate feedback inhibition (Carter et al., 1991). Oxaloacetate (OAA) is the product of PEPC and is quickly converted by malate dehydrogenase into malate (Cuevas and Podestá, 2000) which is transported into the vacuole. A vacuolar H+-ATPase moves large quantities of H+ into the vacuole generating a proton gradient that energizes malate transport (Cheffings et al., 1997). Phase II occurs during the first hours of dawn. In this phase, the transition from PEPC-mediated to Rubisco-mediated CO2 fixation occurs, and for a period of time CO2 is fixed by both carboxylases (Lüttge, 2015). Phase III takes place during the day while stomata are closed. During this phase, CO2 is freed from malate and re-fixed by Rubisco. For this to happen, malic acid is re-mobilized by passive efflux and decarboxylated. There are two alternative pathways for malate decarboxylation, depending on the species. One starts with the release of CO2 by the conversion of malate to pyruvate by NAD(P)-ME, followed by the regeneration of PEP by PPDK (Kondo et al., 2000). In a second pathway, NAD(P)-MDH converts malate to OAA, followed by PEPCK, regenerating PEP, and releasing CO2 (Cushman and Bohnert, 1999). Finally, phase IV takes place late in the afternoon once malate reserves have been depleted. At this time, stomata open again and allow for the uptake of CO2, which is processed directly by Rubisco (Dodd et al., 2002).
The occurrence of CAM four phases varies from species to species and depends on the developmental stage or the severity of environmental stress. Young tissues of all CAM plants perform C3 photosynthesis (Winter, 2019). Upon drought stress, phases II and IV can be lost or reduced in constitutive CAM plants (Dodd et al., 2002). The proportion of PEPC-fixed CO2 also increases with drought in species with a C3-CAM intermediate metabolism, while facultative CAM species can revert completely to C3 after rewatering (Winter and Holtum, 2014). In addition, several CAM modes are possible; including CAM cycling, consisting in respiratory CO2 re-fixation, but with stomatal opening during the day; archetypical CAM; and CAM idling, which happens upon severe drought, in which only respiratory CO2 is re-fixed and stomata remain closed day and night (Borland et al., 2011).
The functioning of CAM metabolism results in the diurnal oscillation of malate content and CO2 uptake. As the oscillations continue despite leaving the plants in continuous light, or darkness and CO2-free air (Wilkins, 1959, 1960; Nuernbergk, 1960), it became clear that understanding these dynamics would require a mathematical framework. Nungesser et al. (1984) translated a basic scheme of reactions known to be relevant for CAM metabolism into a set of ordinary differential equations (ODE). This so-called skeleton model would become the basis for a series of models that would continue for two more decades, dealing with the perturbation of the rhythmicity as well as with finding the dynamically essential components (Table 1).
Nungesser et al. (1984) skeleton model included six regulatory steps: inhibition of PEPCase by malate, activation of this last reaction by Glc6P, inhibition of starch consumption by PEP, activation of photosynthesis by light intensity, inhibition of CO2 uptake by increased internal concentration of CO2 (Ci), and activation of malate transport from the vacuole to the cytosol by light (Figure 1). The predicted relative content of Glc6P, PEP, malic acid, starch, Ci, and net CO2 exchange along the 24 h cycle were remarkably in agreement with experimental data. This shows that the enzymes and metabolite pools considered in the model are sufficient to explain the basic features of CAM.
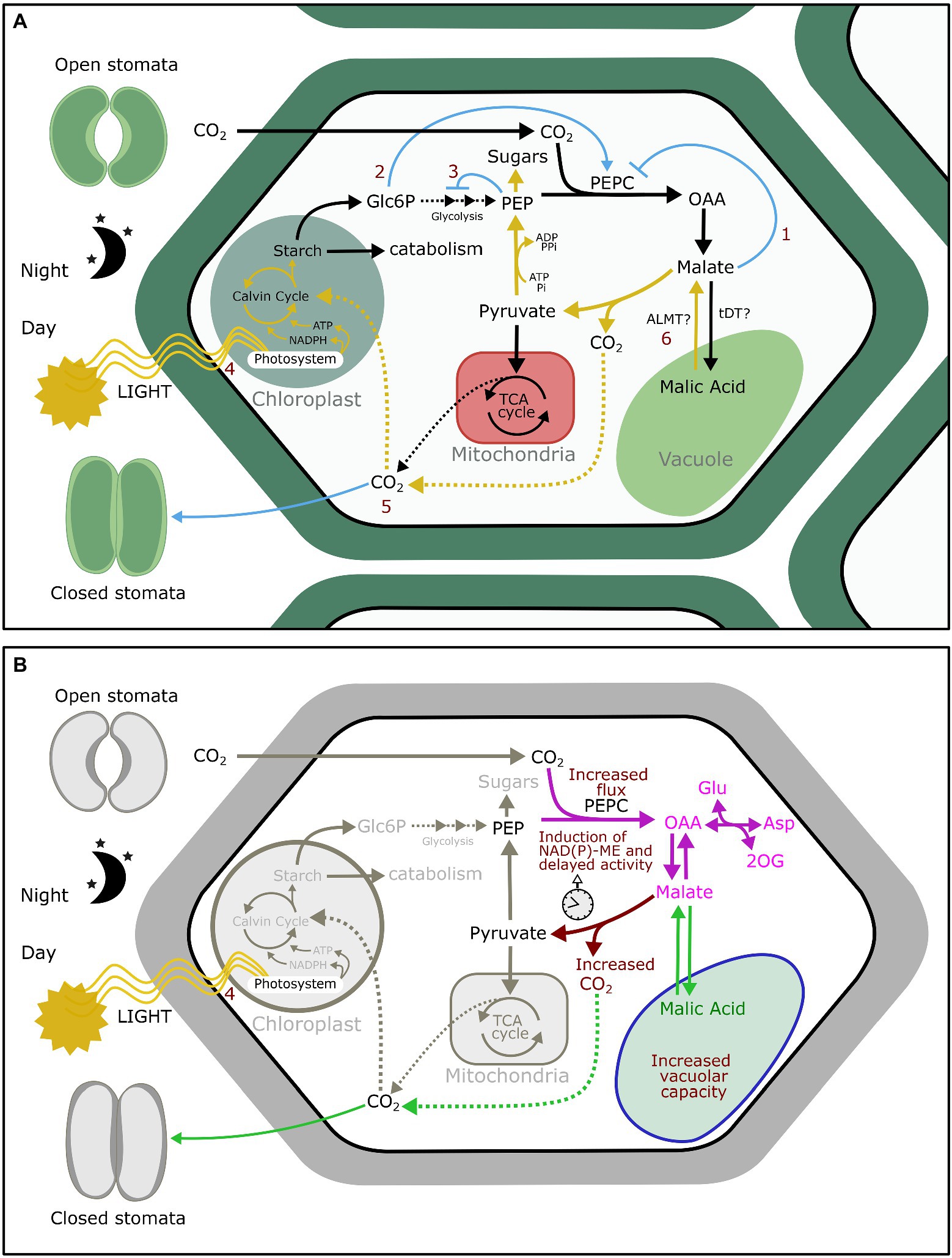
Figure 1. (A) Regulatory interactions sufficient to give rise to diel oscillations of malate accumulation and CO2 uptake adapted from Nungesser et al. (1984) skeleton model: malate inhibition of PEPC (1); Glc6P activation of PEPC (2); PEP inhibition of glycolysis (3); light activation of photosynthesis (4); induction of stomatal closure by high Ci (5); and the autonomous vacuolar oscillator (6). (B) Pathways that articulated during CAM evolution. The anapleurotic fixation of CO2 by PEPC in unicellular algae occurs to provide carbon skeletons for amino acid synthesis (magenta). The same pathway is found in C3 plants, which store in the vacuole malic acid resulting from CO2 fixation during the night to be used for amino acid synthesis the next day. C3 plants are also able to close stomata upon high Ci (green). Three events could have closed the cycle to give rise to CAM (brown): an increased flux through PEPC; an increase in decarboxylation activity, e.g., NAD(P)-ME, and its delay to not overlap with PEPC fixation; and finally, an increased vacuolar capacity that allows the concentration of CO2.
Nungesser et al. (1984) assumed that the transport of malate between the vacuole and the cytoplasm was stimulated by light. However, oscillations in malate content under continuous light challenged this idea. To solve this issue, they added an oscillating element—a hysteresis switch—controlling malate transport through the tonoplast that would be used in the future by other authors. Lüttge and Beck (1992) refined the model by adjusting several parameters to stabilize oscillations under prolonged runs, kept the hysteresis switch, and eliminated the direct influence of light on malate transport. The changes allowed the simulations to reproduce the endogenous malate oscillations under continuous light.
Three subsequent refinements improved model fitting to the observed effect of irradiance and temperature on oscillations (Blasius et al., 1997, 1998, 1999). The original six pools of metabolites were reduced to only three: cytoplasmic and vacuolar malate pools, and Ci. This model assumes an active influx filling the vacuole with malate during the night. At a critical concentration, the hysteresis switch kicks in and vacuolar malate is rapidly expelled. Ci is the carbon source for malate production, while malate consumption and CO2 assimilation connect the oscillatory behavior with the rest of metabolism. Blasius et al. (1999) improved the mechanistic details that generated the hysteresis switch by considering membrane properties and simulating tonoplast permeability as a function of membrane order.
The precise malate transporter responsible for the hysteresis switch remains to be identified. Meyer et al. (2011) found that malate channel ALMT6 shifts from inward to outward rectification in Arabidopsis guard cells at a low vacuolar pH. It was recently suggested that a similar mechanism could operate in CAM plants (Ceusters et al., 2021). Thus, it might be a drop in pH and not a critical vacuolar turgor that triggers the influx-to-efflux shift to begin malate decarboxylation. Transcriptomics data from the CAM species Agave americana suggest the malate/citrate antiporter tDT might also play a role in overnight malate accumulation (Yin et al., 2018).
The fact that skeleton models are sufficient to reproduce CAM dynamics suggests that CAM can originate from the emergence of such metabolic circuits during evolution. After a quick literature survey based on the model by Nungesser et al. (1984), we found evidence that not only enzymes but also regulatory interactions are already in place in C3 plants (Figure 1). These regulatory interactions play roles in different primary metabolism pathways and date back to different stages in the evolution of life. PEP inhibition of glycolytic enzymes is a feedback regulation mechanism that is shared with heterotrophic bacteria and animals (Plaxton, 1996; Ogawa et al., 2007; Grüning et al., 2014). Malate inhibition of PEPC has been documented in unicellular green algae and is crucial for coordinating carbon metabolism with nitrogen assimilation (Peak and Peak, 1981; Schuller et al., 1990). The activation of photosynthesis by light and the subsequent carbon fixation by Rubisco is common for all photosynthetic organisms (McKay et al., 1991; Portis et al., 2008; Tsai et al., 2015). Stomatal closure induction by high Ci is known to occur in early vascular plants and is thought to have been essential for the colonization of land by plants (Chater et al., 2011). Glc6P activation of PEPC is found in C3, C4, and CAM plants (Chollet et al., 1996; Blonde and Plaxton, 2003). In C3 plants, this regulation plays a role in gluconeogenesis by regulating the rate of starch mobilization with the production of dicarboxylic acids (Law and Plaxton, 1995). Finally, there is evidence that the autonomous vacuolar oscillator is also present in C3 plants. As malate can be toxic for the cell in large quantities and is essential for a number of biochemical pathways, its amount in the cytosol is tightly regulated by storing excess malate in the vacuole (Fernie and Martinoia, 2009). Vacuoles of C3 plants switch from malate influx to efflux depending on a critical concentration of cytosolic malate (Gout et al., 1993). As in CAM plants, malate in the vacuole of C3 plants fluctuates diurnally (Gerhardt et al., 1987).
The accumulated information on biochemical reactions together with the availability of genomic data made the first FBA genome-wide metabolic models possible (Schilling and Palsson, 1998; Schilling et al., 1999; Edwards and Palsson, 2000). In plants, the first FBA whole-genome (FBA-WG) metabolic reconstruction was done for Arabidopsis (Poolman et al., 2009), followed by maize (Saha et al., 2011). Later, Cheung et al. (2013) incorporated transport and maintenance costs into the Arabidopsis whole-genome FBA model, allowing for a more realistic prediction of flux distribution in central metabolism. Building on this improvement, Cheung et al. (2014) incorporated the interaction between diurnal and nocturnal metabolism (Table 2). Essentially, this was done by creating two alternative models corresponding to each phase. The models were coupled by adding “transport” reactions between them that would pass on accumulated metabolites. A specified amount of sucrose and amino acids was set to be exported to the phloem, thus requiring the cell to be productive. The flux distribution of both models was simultaneously optimized to decrease the overall amount of material flowing through metabolism. Since the study addressed diel metabolic changes, it was natural to use the model to compare C3 and CAM scenarios. This was done by setting CO2 exchange to zero during the night for CAM. Remarkably, the simulations are in agreement with knowledge on plant metabolism. The C3 simulation predicted citrate to accumulate during the night and to be used during the day for amino acid biosynthesis. For CAM, the nocturnal accumulation of malate was predicted with PEPC as the carboxylating enzyme.
Shameer et al. (2018) simplified Cheung’s FBA-WG model (Cheung et al., 2014) to a core stoichiometric model of central plant metabolism and considered explicitly organellar pH and the charged state of metabolites. The modifications allowed the improved model to match experimental measurements of starch and malate and revealed that CAM metabolism is more productive—higher phloem export rates—than C3, compensating the higher energetic costs of CAM metabolism. These results are highly valuable as a long-sought goal of biotechnologists has been the implementation of CAM into C3 crops by means of genetic engineering.
Shameer et al. (2018) model was the basis for two further works (Töpfer et al., 2020; Tay et al., 2021). Töpfer et al. (2020) extended the model by allowing for differences in vacuolar storage capacity, making gas exchange dependent on temperature and relative humidity, and considering the hourly progression of metabolism during a day. These last two additions allowed for CO2 uptake variations that resulted from a compromise between optimizing CO2, metabolic demand, and minimizing water loss. Interestingly, under a C3-like vacuole size restriction, flux optimization leads to reduced CO2 uptake during the hottest hours of the day—equivalent to midday depression of photosynthesis. A similar simulation under a CAM-like vacuole size restriction increased CO2 uptake at the end of the night together with carboxylic acid accumulation.
Tay et al. (2021) used the Shameer et al. (2018) model to study the C3-to-CAM continuum. They simulated CAM idling and CAM cycling—both considered as weak CAM—by impeding CO2 uptake during the night and during the whole cycle, respectively. The modified models predicted flux distribution intermediate between C3 and CAM metabolism in terms of malate and starch accumulation. In a second analysis, the authors performed a series of simulations decreasing CO2 uptake gradually. They observed a steady shift from C3 to CAM photosynthesis with a respective increase in flux through the starch/malate cycle.
Adaptive evolution reshapes metabolism to maximize fitness (Long and Antoniewicz, 2018). If we accept this, given adequately identified optimality criteria, mathematical models can be useful to understand and predict the course of evolution (Bordbar et al., 2014; de Visser and Krug, 2014). FBA models maximize fitness theoretically by translating a selective pressure into an objective function and have provided remarkably accurate predictions (Varma and Palsson, 1994; Harcombe et al., 2013). The set of FBA models studying the metabolic interactions between day and night suggests that simulating longer periods of stomatal closure are enough to trigger a flux distribution typical for CAM metabolism. Remarkably, CAM-like fluxes emerge upon constraining CO2 uptake on models derived from a C3 whole-genome metabolic reconstruction. Regardless of whether stomatal closure happens during the day or the night and regardless if the stomatal closure is full or partial, flux distributions resembling CAM metabolism are evident. Therefore, assuming that evolution selects for efficient responses to selective pressures (Harcombe et al., 2013), the models provide formal evidence for limited CO2 as the main driver of CAM evolution, a hypothesis long proposed and widely accepted (Lüttge, 2002).
Reduced CO2 uptake can result from limited water availability, as CAM-like fluxes appear when CO2 uptake is compromised by maximizing water saving (Töpfer et al., 2020). At low air humidity or when roots experience water deficit, different signals such as jasmonates and ABA induce stomatal closure (Bauer et al., 2013; De-Ollas et al., 2018). At hot dry hours, C3 and C4 plants close their stomata and undergo what is called midday depression of photosynthesis (Correia et al., 1990; Hirasawa and Hsiao, 1999). Töpfer et al. (2020) model was able to recapitulate this phenomenon. When water saving was maximized, a prolonged stomatal closure during the day was predicted. It is possible that as plants colonized dry areas or whole regions of the world became arid, the midday depression of photosynthesis widened, narrowing the time period at which CO2 exchange could occur. Extended hours behind closed stomata could have exerted a significant selective pressure to develop CAM so that plants could remain productive even under limited CO2 exchange.
A reduction in atmospheric CO2 has also been proposed to trigger CAM evolution, as CAM radiation events coincide with declining CO2 in the geological records (Keeley and Rundel, 2003; Silvera et al., 2009). Tay et al. (2021) tested this hypothesis by gradually decreasing the simulated concentration of CO2 and observed CAM flux distributions as well. A sustained condition of limited CO2 could have selected for more efficient metabolic fluxes over time that resulted in what we know as CAM. Tay et al. (2021) model provided several predictions for the metabolic transitions during CAM evolution worth testing experimentally by further works.
Although conceived with a different purpose, ODE models addressing CAM make also a significant contribution to the understanding of CAM evolution. Our quick literature survey found that not only the enzymes, but the regulatory interactions in CAM skeleton models are already in place in C3 plants. These regulatory interactions date back to different times in evolution and belong to different pathways in primary metabolism. Borrowing existing reactions is commonplace in the evolution of new pathways since it is faster and energetically less expensive than generating de novo reactions. The Krebs cycle is believed to have arisen from primitive pathways for amino acid synthesis (Meléndez-Hevia et al., 1996). Interestingly, CAM has also been proposed to be derived from an amino acid synthesis pathway (Bräutigam et al., 2017). In C3 plants, the accumulation of organic acids during the night fuels amino acid synthesis the following day (Gauthier et al., 2010). In this pathway, PEPC carboxylase is active during the night and catalyzes the synthesis of PEP resulting from carbohydrate degradation (Nimmo, 2003). In agreement with this, interactions (1), (2), and (3) of CAM skeleton model (Figure 1) can coordinate carbon flux toward amino acid synthesis in C3 plants: Glc6P is a feedforward activator of PEPC, while malate and PEP are feedback inhibitors of PEPC and glycolysis, respectively. Excluding vacuolar storage, this pathway is also found in unicellular green algae, in which PEPC activity is regulated to coordinate carbon metabolism with nitrogen assimilation (Peak and Peak, 1981).
The crucial step that diverts the pathway from amino acid synthesis to a new type of photosynthetic metabolism is malate decarboxylation. Malate decarboxylation connects the pathway regulated by interactions (1), (2), and (3) with carbon re-fixation by Rubisco during the day (4) and the accumulation of CO2 that induces stomatal closure (5; Figure 1). Either NAD(P)-ME or PEPCK are responsible for the release of CO2, depending on the pathway. Neither enzyme was originally part of photosynthetic metabolism (Sanwal and Smando, 1969; Boles et al., 1998; Drincovich et al., 2001), but we can presume that mutations that led to the induction of NAD(P)-ME and PEPCK activity during the day could have been evolutionarily advantageous by alleviating CO2 starvation during extended periods of stomatal closure. Both enzymes have been recruited multiple times during evolution when CO2 is limiting (Reiskind and Bowes, 1991; Magnin et al., 1997; Casati et al., 2000; Hibberd and Quick, 2002), besides their well-known role in C4 and CAM metabolism (Bovdilova et al., 2019; Lim et al., 2019). For the CAM subtypes using NAD(P)-ME, NAD(P)-ME induction could have been a direct consequence of an initial increase in PEPC activity. In heterotrophic tissues relying primarily on oxidation, PEPC and NAD(P)-ME work in concert to provide reducing power in the form of NAD(P)H (O'Leary et al., 2011). Transgenic C3 plants overexpressing PEPC exhibit an important increase in NAD(P)-ME expression and NAD(P)-ME enzymatic activity, probably triggered by an excess of cytosolic malic acid (Häusler et al., 2001). It has been previously proposed that during CAM evolution NAD(P)-ME and PEPC activities became coordinated to avoid futile CO2 cycling between both enzymes (Dodd et al., 2002). If high cytosolic malic acid induces NAD(P)-ME, the storage of malate during the night and the switch from malate influx to efflux in the vacuole during the day could theoretically sort out the issue of the overlap of both enzymatic activities, although this possibility has not been tested yet.
In order for CAM skeleton models to run, enough amounts of malate need to accumulate in the vacuole so that decarboxylation builds up a high Ci that induces stomatal closure, and thus, the reverse stomatal patterns appear. Vacuolar size is critical for CAM function (Griffiths et al., 2008). Considering this parameter explicitly, Töpfer et al. (2020) showed that the typical malate storage capacity of C3 plants is not sufficient to allow the emergence of CAM flux distributions. Thus, changes that lead to an increased malate storage are necessary in order to develop a proper CAM pathway, these could include H+-ATPase (Ratajczak et al., 1994), malate channels (Pantoja and Smith, 2002), and malate/citrate antiporters (Frei et al., 2018).
So far, the proposed ODE models consider only the control of Ci on stomatal aperture. Although there is substantial evidence for this regulation (Cockburn et al., 1979; Males and Griffiths, 2017), we know now that it is not the only one. In order to undergo the transition from diurnal to nocturnal stomatal aperture, C3 plants need to rewire the circadian control on stomata (Hassidim et al., 2017). Comparative genomic studies have shown that genes involved in stomatal regulation have shifted their peak expression to the evening together with clock or clock-entraining genes (Yang et al., 2017; Yin et al., 2018). Thus, it is also possible that the inverse pattern of stomatal aperture was achieved before attaining a high Ci during the night.
Another theoretical possibility is that the circadian control on stomatal aperture was lost during CAM evolution and stomatal aperture relied mostly on the autonomous vacuolar oscillator (6). This way autonomous oscillations of malate content could drive stomatal closure during the day on their own. More research is needed to understand how autonomous oscillations in malate content and the circadian clock are integrated to control CAM rhythmicity. Currently, it is known that both, impeding the circadian input on CAM and silencing CAM, alter oscillations in CO2 uptake and the circadian clock itself (Boxall et al., 2017, 2020). Further ODE models integrating the clock with CAM metabolism could not only shed light on CO2 uptake regulation, but on the overall interplay between carbon metabolism and the clock.
An ongoing discussion is the C3-to-CAM continuum itself (Edwards, 2019; Yang et al., 2019; Schiller and Bräutigam, 2021; Winter and Smith, 2022). On one hand, the remarkable phenotypic plasticity observed in CAM plants generates a wide gradient in δ13C—an indicator of the amount of carbon fixed by CAM—in several plant lineages (Silvera et al., 2010). On the other hand, there is a clear bimodal distribution showing there are only a few intermediate phenotypes in comparison with C3 and strong CAM phenotypes, which might indicate mechanistic constraints for intermediate metabolism (Borland et al., 2011). Yang et al. (2019) argue that the continuum hypothesis underestimates the number of changes needed to achieve CAM, since available genomics data indicate at least 60 genes are involved in CAM evolution. Winter and Smith (2022) highlight that the change to nocturnal malate accumulation would involve a qualitative leap in CAM evolution since for many C3 species malate accumulates during the day.
The discussion truly appears as another instance of the old saltationism versus gradualism debate (Gould and Eldredge, 1977). However, it has opened the conversation on the pace of CAM evolution and the conditions that would facilitate either a smooth or sharp transition. Edwards (2019) analyzes the subject in terms of evolutionary accessibility and proposes there is smooth transition between C3 and weak CAM phenotypes, but for strong CAM to occur, the development of extreme succulence is necessary. Future modeling efforts on CAM metabolism could provide more evidence to discuss the subject. It would be highly informative to combine genomic information, evolutionary modeling and genome-wide models to investigate possible points of evolutionary contingency. In addition, mathematically controlled comparisons can be used to understand the evolutionary limits of the various metabolic types (Alves and Savageau, 2000; Schwacke and Voit, 2004). Another area of opportunity for CAM models is to aid in the rational implementation of CAM metabolism in C3 crops. Recently, a set of 13 CAM enzymes were overexpressed independently in Arabidopsis (Lim et al., 2019). Remarkably, the lines expressing the carboxylation module resulted in increased stomatal conductance and increased malate accumulation, and most lines from both carboxylation and decarboxylation modules had increased biomass. Feeding FBA models with experimental measurements of these plants can indicate how fluxes may be optimized after the overexpression and provide clues for novel bioengineering strategies.
Computational analysis and modeling of CAM metabolism have not only contributed to the understanding of how CAM basic components integrate for the emergence of an autonomous rhythm, and of how fluxes through central metabolism are reshaped, but may also contribute significantly to identify evolutionary selection processes. The convergence of different FBA models to CAM flux distributions provides formal mathematical evidence for low Ci as the main driver for the evolution of CAM. We believe that the use of these models together with evolutionary models and genomic data will shed more light on CAM evolution and assist rational engineering of CAM into C3 crops.
AB conceived the idea. AB, EM, EV, IM-C, and RA wrote the manuscript. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AB acknowledges Mexico’s National Council for Science and Technology (CONACYT) for its support under Cátedras-CONACYT project 939.
Alves, R., and Savageau, M. A. (2000). Extending the method of mathematically controlled comparison to include numerical comparisons. Bioinformatics 16, 786–798. doi: 10.1093/bioinformatics/16.9.786
Bauer, H., Ache, P., Lautner, S., Fromm, J., Hartung, W., Al-Rasheid, K. A., et al. (2013). The stomatal response to reduced relative humidity requires guard cell-autonomous ABA synthesis. Curr. Biol. 23, 53–57. doi: 10.1016/j.cub.2012.11.022
Blasius, B., Beck, F., and Lüttge, U. (1997). A model for photosynthetic oscillations in crassulacean acid metabolism (CAM). J. Theor. Biol. 184, 345–351. doi: 10.1006/jtbi.1996.0287
Blasius, B., Beck, F., and Lüttge, U. (1998). Oscillatory model of crassulacean acid metabolism: structural analysis and stability boundaries with a discrete hysteresis switch. Plant Cell Environ. 21, 775–784. doi: 10.1046/j.1365-3040.1998.00312.x
Blasius, B., Neif, R., Beck, F., and Lüttge, U. (1999). Oscillatory model of crassulacean acid metabolism with a dynamic hysteresis switch. Proc. R. Soc. Lond. Ser. B Biol. Sci. 266, 93–101. doi: 10.1098/rspb.1999.0608
Blonde, J. D., and Plaxton, W. C. (2003). Structural and kinetic properties of high and low molecular mass phosphoenolpyruvate carboxylase isoforms from the endosperm of developing castor oilseeds. J. Biol. Chem. 278, 11867–11873. doi: 10.1074/jbc.M211269200
Boles, E., de Jong-Gubbels, P., and Pronk, J. T. (1998). Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180, 2875–2882. doi: 10.1128/JB.180.11.2875-2882.1998
Bordbar, A., Monk, J. M., King, Z. A., and Palsson, B. O. (2014). Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120. doi: 10.1038/nrg3643
Borland, A. M., Barrera Zambrano, V. A., Ceusters, J., and Shorrock, K. (2011). The photosynthetic plasticity of crassulacean acid metabolism: an evolutionary innovation for sustainable productivity in a changing world. New Phytol. 191, 619–633. doi: 10.1111/j.1469-8137.2011.03781.x
Bovdilova, A., Alexandre, B. M., Höppner, A., Luís, I. M., Alvarez, C. E., Bickel, D., et al. (2019). Posttranslational modification of the NADP-malic enzyme involved in C4 photosynthesis modulates the enzymatic activity during the day. Plant Cell 31, 2525–2539. doi: 10.1105/tpc.19.00406
Boxall, S. F., Dever, L. V., Kneřová, J., Gould, P. D., and Hartwell, J. (2017). Phosphorylation of phospho enol pyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi. Plant Cell 29, 2519–2536. doi: 10.1105/tpc.17.00301
Boxall, S. F., Kadu, N., Dever, L. V., Kneřová, J., Waller, J. L., Gould, P. J., et al. (2020). Kalanchoë PPC1 is essential for crassulacean acid metabolism and the regulation of core circadian clock and guard cell signaling genes. Plant Cell 32, 1136–1160. doi: 10.1105/tpc.19.00481
Bräutigam, A., Schlüter, U., Eisenhut, M., and Gowik, U. (2017). On the evolutionary origin of CAM photosynthesis. Plant Physiol. 174, 473–477. doi: 10.1104/pp.17.00195
Carter, P. J., Nimmo, H. G., Fewson, C. A., and Wilkins, M. B. (1991). Circadian rhythms in the activity of a plant protein kinase. EMBO J. 10, 2063–2068. doi: 10.1002/j.1460-2075.1991.tb07737.x
Casati, P., Lara, M. V., and Andreo, C. S. (2000). Induction of a C4-Like mechanism of CO2 fixation in Egeria densa, a submersed aquatic species. Plant Physiol. 123, 1611–1622. doi: 10.1104/pp.123.4.1611
Ceusters, N., Borland, A. M., and Ceusters, J. (2021). How to resolve the enigma of diurnal malate remobilisation from the vacuole in plants with crassulacean acid metabolism? New Phytol. 229, 3116–3124. doi: 10.1111/nph.17070
Chater, C., Kamisugi, Y., Movahedi, M., Fleming, A., Cuming, A. C., Gray, J. E., et al. (2011). Regulatory mechanism controlling stomatal behavior conserved across 400 million years of land plant evolution. Curr. Biol. 21, 1025–1029. doi: 10.1016/j.cub.2011.04.032
Cheffings, C. M., Pantoja, O., Ashcroft, F. M., and Smith, J. A. C. (1997). Malate transport and vacuolar ion channels in CAM plants. J. Exp. Bot. 48, 623–631. doi: 10.1093/jxb/48.Special_Issue.623
Cheung, C. M., Poolman, M. G., Fell, D. A., Ratcliffe, R. G., and Sweetlove, L. J. (2014). A diel flux balance model captures interactions between light and dark metabolism during day-night cycles in C3 and crassulacean acid metabolism leaves. Plant Physiol. 165, 917–929. doi: 10.1104/pp.113.234468
Cheung, C. M., Williams, T. C., Poolman, M. G., Fell, D. A., Ratcliffe, R. G., and Sweetlove, L. J. (2013). A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J. 75, 1050–1061. doi: 10.1111/tpj.12252
Chollet, R., Vidal, J., and O'Leary, M. H. (1996). Phospho enol pyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu. Rev. Plant Biol. 47, 273–298. doi: 10.1146/annurev.arplant.47.1.273
Cockburn, W., Ting, I. P., and Sternberg, L. O. (1979). Relationships between stomatal behavior and internal carbon dioxide concentration in crassulacean acid metabolism plants. Plant Physiol. 63, 1029–1032. doi: 10.1104/pp.63.6.1029
Correia, M. J., Chaves, M. M. C., and Pereira, J. S. (1990). Afternoon depression in photosynthesis in grapevine leaves—evidence for a high light stress effect. J. Exp. Bot. 41, 417–426. doi: 10.1093/jxb/41.4.417.hi
Crayn, D. M., Winter, K., and Smith, J. A. C. (2004). Multiple origins of crassulacean acid metabolism and the epiphytic habit in the Neotropical family Bromeliaceae. Proc. Natl. Acad. Sci. 101, 3703–3708. doi: 10.1073/pnas.0400366101
Cuevas, I. C., and Podestá, F. E. (2000). Purification and physical and kinetic characterization of an NAD+-dependent malate dehydrogenase from leaves of pineapple (Ananas comosus). Physiol. Plant. 108, 240–248. doi: 10.1034/j.1399-3054.2000.108003240.x
Cushman, J. C., and Bohnert, H. J. (1999). Crassulacean acid metabolism: molecular genetics. Annu. Rev. Plant Biol. 50, 305–332. doi: 10.1146/annurev.arplant.50.1.305
Dai, A. (2013). Increasing drought under global warming in observations and models. Nat. Clim. Chang. 3, 52–58. doi: 10.1038/NCLIMATE1633
de Visser, J. A. G. M., and Krug, J. (2014). Empirical fitness landscapes and the predictability of evolution. Nat. Rev. Genet. 15, 480–490. doi: 10.1038/nrg3744
De-Ollas, C., Arbona, V., Gómez-Cadenas, A., and Dodd, I. C. (2018). Attenuated accumulation of jasmonates modifies stomatal responses to water deficit. J. Exp. Bot. 69, 2103–2116. doi: 10.1093/jxb/ery045.corre
Dodd, A. N., Borland, A. M., Haslam, R. P., Griffiths, H., and Maxwell, K. (2002). Crassulacean acid metabolism: plastic, fantastic. J. Exp. Bot. 53, 569–580. doi: 10.1093/jexbot/53.369.569
Drincovich, M. F., Casati, P., and Andreo, C. S. (2001). NADP-malic enzyme from plants: a ubiquitous enzyme involved in different metabolic pathways. FEBS Lett. 490, 1–6. doi: 10.1016/S0014-5793(00)02331-0
Edwards, E. J. (2019). Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytol. 223, 1742–1755. doi: 10.1111/nph.15851
Edwards, J. S., and Palsson, B. O. (2000). The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc. Natl. Acad. Sci. 97, 5528–5533. doi: 10.1073/pnas.97.10.5528
Fernie, A. R., and Martinoia, E. (2009). Malate. Jack of all trades or master of a few? Phytochemistry 70, 828–832. doi: 10.1016/j.phytochem.2009.04.023
Frei, B., Eisenach, C., Martinoia, E., Hussein, S., Chen, X. Z., Arrivault, S., et al. (2018). Purification and functional characterization of the vacuolar malate transporter tDT from Arabidopsis. J. Biol. Chem. 293, 4180–4190. doi: 10.1074/jbc.RA117.000851
Gauthier, P. P., Bligny, R., Gout, E., Mahé, A., Nogués, S., Hodges, M., et al. (2010). In folio isotopic tracing demonstrates that nitrogen assimilation into glutamate is mostly independent from current CO2 assimilation in illuminated leaves of Brassica napus. New Phytol. 185, 988–999. doi: 10.1111/j.1469-8137.2009.03130.x
Gerhardt, R., Stitt, M., and Heldt, H. W. (1987). Subcellular metabolite levels in spinach leaves: regulation of sucrose synthesis during diurnal alterations in photosynthetic partitioning. Plant Physiol. 83, 399–407. doi: 10.1104/pp.83.2.399
Gould, S. J., and Eldredge, N. (1977). Punctuated equilibria: the tempo and mode of evolution reconsidered. Paleobiology 3, 115–151. doi: 10.1017/S0094837300005224
Gout, E., Bligny, R., Pascal, N., and Douce, R. (1993). 13C nuclear magnetic resonance studies of malate and citrate synthesis and compartmentation in higher plant cells. J. Biol. Chem. 268, 3986–3992. doi: 10.1016/S0021-9258(18)53568-7
Grams, T. E., Beck, F., and Lüttge, U. (1996). Generation of rhythmic and arrhythmic behaviour of crassulacean acid metabolism in Kalanchoë daigremontiana under continuous light by varying the irradiance or temperature: measurements in vivo and model simulations. Planta 198, 110–117.
Grams, T. E. E., Borland, A. M., Roberts, A., Griffiths, H., Beck, F., and Luttge, U. (1997). On the mechanism of reinitiation of endogenous crassulacean acid metabolism rhythm by temperature changes. Plant physiology 113, 1309–1317.
Griffiths, H., Robe, W. E., Girnus, J., and Maxwell, K. (2008). Leaf succulence determines the interplay between carboxylase systems and light use during Crassulacean acid metabolism in Kalanchoë species. J. Exp. Bot. 59, 1851–1861. doi: 10.1093/jxb/ern085
Grüning, N. M., Du, D., Keller, M. A., Luisi, B. F., and Ralser, M. (2014). Inhibition of triosephosphate isomerase by phosphoenolpyruvate in the feedback-regulation of glycolysis. Open Biol. 4:130232. doi: 10.1098/rsob.130232
Harcombe, W. R., Delaney, N. F., Leiby, N., Klitgord, N., and Marx, C. J. (2013). The ability of flux balance analysis to predict evolution of central metabolism scales with the initial distance to the optimum. PLoS Comput. Biol. 9:e1003091. doi: 10.1371/journal.pcbi.1003091
Hassidim, M., Dakhiya, Y., Turjeman, A., Hussien, D., Shor, E., Anidjar, A., et al. (2017). CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and the circadian control of stomatal aperture. Plant Physiol. 175, 1864–1877. doi: 10.1104/pp.17.01214
Häusler, R. E., Rademacher, T., Li, J., Lipka, V., Fischer, K. L., Schubert, S., et al. (2001). Single and double overexpression of C4-cycle genes had differential effects on the pattern of endogenous enzymes, attenuation of photorespiration and on contents of UV protectants in transgenic potato and tobacco plants. J. Exp. Bot. 52, 1785–1803. doi: 10.1093/jexbot/52.362.1785
Hibberd, J. M., and Quick, W. P. (2002). Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415, 451–454. doi: 10.1038/415451ª
Hirasawa, T., and Hsiao, T. C. (1999). Some characteristics of reduced leaf photosynthesis at midday in maize growing in the field. Field Crop Res. 62, 53–62. doi: 10.1016/S0378-4290(99)00005-2
Huang, H., Yu, K., and D’Odorico, P. (2020). CAM plant expansion favored indirectly by asymmetric climate warming and increased rainfall variability. Oecologia 193, 1–13. doi: 10.1007/s00442-020-04624-w
Keeley, J. E., and Rundel, P. W. (2003). Evolution of CAM and C4 carbon-concentrating mechanisms. Int. J. Plant Sci. 164, S55–S77. doi: 10.1086/374192
Kondo, A., Nose, A., Yuasa, H., and Ueno, O. (2000). Species variation in the intracellular localization of pyruvate, Pi dikinase in leaves of crassulacean-acid-metabolism plants: an immunogold electron-microscope study. Planta 210, 611–621. doi: 10.1007/s004250050051
Law, R. D., and Plaxton, W. C. (1995). Purification and characterization of a novel phosphoenolpyruvate carboxylase from banana fruit. Biochem. J. 307, 807–816. doi: 10.1042/bj3070807
Lim, S. D., Lee, S., Choi, W. G., Yim, W. C., and Cushman, J. C. (2019). Laying the foundation for crassulacean acid metabolism (CAM) biodesign: expression of the C4 metabolism cycle genes of CAM in Arabidopsis. Front. Plant Sci. 10:101. doi: 10.3389/fpls.2019.00101.4
Liu, D., Palla, K. Y., Hu, R., Moseley, R. C., Mendoza, C., Chen, M., et al. (2018). Perspectives on the basic and applied aspects of crassulacean acid metabolism (CAM) research. Plant Science 274, 394–401.
Long, C. P., and Antoniewicz, M. R. (2018). How adaptive evolution reshapes metabolism to improve fitness: recent advances and future outlook. Curr. Opin. Chem. Eng. 22, 209–215. doi: 10.1016/j.coche.2018.11.001
Lüttge, U. (2002). CO2-concentrating: consequences in crassulacean acid metabolism. J. Exp. Bot. 53, 2131–2142. doi: 10.1093/jxb/erf081
Lüttge, U., and Beck, F. (1992). Endogenous rhythms and chaos in crassulacean acid metabolism. Planta 188, 28–38. doi: 10.1007/BF00198936
Magnin, N. C., Cooley, B. A., Reiskind, J. B., and Bowes, G. (1997). Regulation and localization of key enzymes during the induction of Kranz-less, C4-type photosynthesis in Hydrilla verticillata. Plant Physiol. 115, 1681–1689. doi: 10.1104/pp.115.4.1681
Males, J., and Griffiths, H. (2017). Stomatal biology of CAM plants. Plant Physiol. 174, 550–560. doi: 10.1104/pp.17.00114
McKay, R. M. L., Gibbs, S. P., and Vaughn, K. C. (1991). RuBisCo activase is present in the pyrenoid of green algae. Protoplasma 162, 38–45. doi: 10.1007/BF01403899
Meléndez-Hevia, E., Waddell, T. G., and Cascante, M. (1996). The puzzle of the Krebs citric acid cycle: assembling the pieces of chemically feasible reactions, and opportunism in the design of metabolic pathways during evolution. J. Mol. Evol. 43, 293–303. doi: 10.1007/BF02338838
Meyer, S., Scholz-Starke, J., De Angeli, A., Kovermann, P., Burla, B., Gambale, F., et al. (2011). Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 67, 247–257. doi: 10.1111/j.1365-313X.2011.04587.x
Nimmo, H. G. (2003). Control of the phosphorylation of phosphoenolpyruvate carboxylase in higher plants. Arch. Biochem. Biophys. 414, 189–196. doi: 10.1016/S0003-9861(03)00115-2
Nuernbergk, E. L. (1960). Endogener Rhythmus und CO2-stoffwechsel bei Pflanzen mit diurnalem Säurerhythmus. Planta 56, 28–70. doi: 10.1007/BF01894844
Nungesser, D., Kluge, M., Tolle, H., and Oppelt, W. (1984). A dynamic computer model of the metabolic and regulatory processes in Crassulacean acid metabolism. Planta 162, 204–214. doi: 10.1007/BF00397441
Ogawa, T., Mori, H., Tomita, M., and Yoshino, M. (2007). Inhibitory effect of phosphoenolpyruvate on glycolytic enzymes in Escherichia coli. Res. Microbiol. 158, 159–163. doi: 10.1016/j.resmic.2006.11.003
O'Leary, B., Park, J., and Plaxton, W. C. (2011). The remarkable diversity of plant PEPC (phosphoenolpyruvate carboxylase): recent insights into the physiological functions and post-translational controls of non-photosynthetic PEPCs. Biochem. J. 436, 15–34. doi: 10.1042/BJ20110078
Owen, N. A., and Griffiths, H. (2013). A system dynamics model integrating physiology and biochemical regulation predicts extent of crassulacean acid metabolism (CAM) phases. New Phytologist 200, 1116–1131.
Pantoja, O., and Smith, J. A. C. (2002). Sensitivity of the plant vacuolar malate channel to pH, Ca2+ and anion-channel blockers. J. Membr. Biol. 186, 31–42. doi: 10.1007/s00232-001-0132-z
Peak, J. G., and Peak, M. J. (1981). Heterotrophic carbon dioxide fixation by Euglena: function of phosphoenolpyruvate carboxylase. Biochimica et Biophysica Acta (BBA)-General Subjects 677, 390–396. doi: 10.1016/0304-4165(81)90251-8
Plaxton, W. C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Biol. 47, 185–214. doi: 10.1146/annurev.arplant.47.1.185
Poolman, M. G., Miguet, L., Sweetlove, L. J., and Fell, D. A. (2009). A genome-scale metabolic model of Arabidopsis and some of its properties. Plant Physiol. 151, 1570–1581. doi: 10.1104/pp.109.141267
Portis, A. R. Jr., Li, C., Wang, D., and Salvucci, M. E. (2008). Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59, 1597–1604. doi: 10.1093/jxb/erm240
Ratajczak, R., Richter, J., and Lüttge, U. (1994). Adaptation of the tonoplast V-type H+-ATPase of Mesembryanthemum crystallinum to salt stress, C3–CAM transition and plant age. Plant Cell Environ. 17, 1101–1112. doi: 10.1111/j.1365-3040.1994.tb02008.x
Reiskind, J. B., and Bowes, G. (1991). The role of phosphoenolpyruvate carboxykinase in a marine macroalga with C4-like photosynthetic characteristics. Proc. Natl. Acad. Sci. 88, 2883–2887. doi: 10.1073/pnas.88.7.2883
Saha, R., Suthers, P. F., and Maranas, C. D. (2011). Zea mays i RS1563: a comprehensive genome-scale metabolic reconstruction of maize metabolism. PLoS One 6:e21784. doi: 10.1371/journal.pone.0021784
Sanwal, B. D., and Smando, R. (1969). Malic enzyme of Escherichia coli: diversity of the effectors controlling enzyme activity. J. Biol. Chem. 244, 1817–1823. doi: 10.1016/S0021-9258(18)91755-2
Schiller, K., and Bräutigam, A. (2021). Engineering of crassulacean acid metabolism. Annu. Rev. Plant Biol. 72, 77–103. doi: 10.1146/annurev-arplant-071720-104814
Schilling, C. H., Edwards, J. S., and Palsson, B. O. (1999). Toward metabolic phenomics: analysis of genomic data using flux balances. Biotechnol. Prog. 15, 288–295. doi: 10.1021/bp9900357
Schilling, C. H., and Palsson, B. O. (1998). The underlying pathway structure of biochemical reaction networks. Proc. Natl. Acad. Sci. 95, 4193–4198. doi: 10.1073/pnas.95.8.4193
Schuller, K. A., Plaxton, W. C., and Turpin, D. H. (1990). Regulation of Phosphoenol pyruvate carboxylase from the Green alga Selenastrum minutum: properties associated with replenishment of Tricarboxylic acid cycle intermediates during ammonium assimilation. Plant Physiol. 93, 1303–1311. doi: 10.1104/pp.93.4.1303
Schwacke, J. H., and Voit, E. O. (2004). Improved methods for the mathematically controlled comparison of biochemical systems. Theor. Biol. Med. Model. 1, 1–18. doi: 10.1186/1742-4682-1-1
Shameer, S., Baghalian, K., Cheung, C. M., Ratcliffe, R. G., and Sweetlove, L. J. (2018). Computational analysis of the productivity potential of CAM. Nat. Plants 4, 165–171. doi: 10.1038/s41477-018-0112-2
Silvera, K., Neubig, K. M., Whitten, W. M., Williams, N. H., Winter, K., and Cushman, J. C. (2010). Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 37, 995–1010. doi: 10.1071/FP10084
Silvera, K., Santiago, L. S., Cushman, J. C., and Winter, K. (2009). Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 149, 1838–1847. doi: 10.1104/pp.108.132555
Tay, I. Y., Odang, K. B., and Cheung, C. Y. (2021). Metabolic modeling of the C3-CAM continuum revealed the establishment of a starch/sugar-malate cycle in CAM evolution. Front. Plant Sci. 11:197. doi: 10.3389/fpls.2020.573197
Töpfer, N., Braam, T., Shameer, S., Ratcliffe, R. G., and Sweetlove, L. J. (2020). Alternative crassulacean acid metabolism modes provide environment-specific water-saving benefits in a leaf metabolic model. Plant Cell 32, 3689–3705. doi: 10.1105/tpc.20.00132
Tsai, Y. C. C., Lapina, M. C., Bhushan, S., and Mueller-Cajar, O. (2015). Identification and characterization of multiple rubisco activases in chemoautotrophic bacteria. Nat. Commun. 6, 8883–8810. doi: 10.1038/ncomms9883
Varma, A., and Palsson, B. O. (1994). Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl. Environ. Microbiol. 60, 3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994
Wilkins, M. B. (1959). An endogenous rhythm in the rate of carbon dioxide output of Bryophyllum: I. Some preliminary experiments. J. Exp. Bot. 10, 377–390. doi: 10.1093/jxb/10.3.377
Wilkins, M. B. (1960). An endogenous rhythm in THE rate of CO2 output of Bryophyllum: II. THE EFFECTS OF LIGHT AND DARKNESS ON THE PHASE AND PERIOD OF THE RHYTHM. J. Exp. Bot. 11, 269–288. doi: 10.1093/jxb/11.2.269
Winter, K. (2019). Ecophysiology of constitutive and facultative CAM photosynthesis. J. Exp. Bot. 70, 6495–6508. doi: 10.1093/jxb/erz002
Winter, K., and Holtum, J. A. (2014). Facultative crassulacean acid metabolism (CAM) plants: powerful tools for unravelling the functional elements of CAM photosynthesis. J. Exp. Bot. 65, 3425–3441. doi: 10.1093/jxb/eru063
Winter, K., and Smith, J. A. C. (Eds.). (2012). Crassulacean acid Metabolism: Biochemistry, Ecophysiology and Evolution. Germany: Springer Science & Business Media.
Winter, K., and Smith, J. A. C. (2022). CAM photosynthesis: the acid test. New Phytol. 233, 599–609. doi: 10.1111/nph.17790
Wyka, T. P., Bohn, A., Duarte, H. M., Kaiser, F., and Lüttge, U. E. (2004). Perturbations of malate accumulation and the endogenous rhythms of gas exchange in the Crassulacean acid metabolism plant Kalanchoë daigremontiana: testing the tonoplast-as-oscillator model. Planta 219, 705–713.
Yang, X., Hu, R., Yin, H., Jenkins, J., Shu, S., Tang, H., et al. (2017). The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 8, 1899–1815. doi: 10.1038/s41467-017-01491-7
Yang, X., Liu, D., Tschaplinski, T. J., and Tuskan, G. A. (2019). Comparative genomics can provide new insights into the evolutionary mechanisms and gene function in CAM plants. J. Exp. Bot. 70, 6539–6547. doi: 10.1093/jxb/erz408
Yin, H., Guo, H. B., Weston, D. J., Borland, A. M., Ranjan, P., Abraham, P. E., et al. (2018). Diel rewiring and positive selection of ancient plant proteins enabled evolution of CAM photosynthesis in Agave. BMC Genomics 19:588. doi: 10.1186/s12864-018-4964-7
Keywords: CAM, CAM evolution, ODE models, FBA models, carbon concentration mechanism
Citation: Burgos A, Miranda E, Vilaprinyo E, Meza-Canales ID and Alves R (2022) CAM Models: Lessons and Implications for CAM Evolution. Front. Plant Sci. 13:893095. doi: 10.3389/fpls.2022.893095
Received: 10 March 2022; Accepted: 23 May 2022;
Published: 23 June 2022.
Edited by:
Zoran Nikoloski, Max Planck Institute of Molecular Plant Physiology, GermanyReviewed by:
Kailiang Yu, University of Virginia, United StatesCopyright © 2022 Burgos, Miranda, Vilaprinyo, Meza-Canales and Alves. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asdrubal Burgos, YXNkcnViYWwuYnVyZ29zQGFjYWRlbWljb3MudWRnLm14
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.