- 1Department of Bioinformatics, College of Life Sciences, Zhejiang University, Hangzhou, China
- 2National Institute for Genomics and Advanced Biotechnology, National Agricultural Research Centre, Islamabad, Pakistan
Ribonucleic acid editing (RE) is a post-transcriptional process that altered the genetics of RNA which provide the extra level of gene expression through insertion, deletions, and substitutions. In animals, it converts nucleotide residues C-U. Similarly in plants, the role of RNA editing sites (RES) in rice under alkaline stress is not fully studied. Rice is a staple food for most of the world population. Alkaline stress cause reduction in yield. Here, we explored the effect of alkaline stress on RES in the whole mRNA from rice chloroplast and mitochondria. Ribonucleic acid editing sites in both genomes (3336 RESs) including chloroplast (345 RESs) and mitochondria (2991 RESs) with average RES efficiency ∼55% were predicted. Our findings showed that majority of editing events found in non-synonymous codon changes and change trend in amino acids was hydrophobic. Four types of RNA editing A-G (A-I), C-T (C-U), G-A, and T-C were identified in treated and untreated samples. Overall, RNA editing efficiency was increased in the treated samples. Analysis of Gene Ontology revealed that mapped genes were engaged in many biological functions and molecular processes. We also checked the expression of pentatricopeptide repeat (PPR), organelle zinc-finger (OZI), and multiple organellar RNA editing factors/RNA editing factor interacting proteins genes in control and treatment, results revealed upregulation of PPR and OZ1 genes in treated samples. This induction showed the role of these genes in RNA editing. The current findings report that RNA editing increased under alkaline stress which may contribute in adaptation for rice by changing amino acids in edited genes (88 genes). These findings will provide basis for identification of RES in other crops and also will be useful in alkaline tolerance development in rice.
Introduction
Post-transcriptional mechanisms such as RNA editing, cause the change from DNA to RNA by means of insertion, deletions, and substitutions. This modification was first reported in mitochondrial genome many decades before (Benne et al., 1986), later on many studies identified the RNA editing sites in mitochondria and chloroplast (plastids) of animals and plants. In the case of plants, alteration in hereditary material through nucleotide conversion, deletion, or insertion leads to change in cytosine to uracil (C to U) in the messenger RNA of functional genes is the central dogma of RNA editing (Keller et al., 1999). Previously, RES was thought to be a proofreading mechanism because it corrected the mitochondrial mRNA through deletion/addition of uridine (Golden and Hajduk, 2006). With the passage of time and inventions of new sequencing techniques such as RNA sequencing (RNA-seq) enables the researcher to its importance in plants developments under the abiotic stress conditions (Duruvasula et al., 2019). This type of RNA editing is dominant, occurred in nucleus, cytosol, plastids, and mitochondria. The RNA editing plays a key role in the multiple plant growth and developmental processes, plant adaption to environmental influences, and signal transduction (Fujii and Small, 2011; Hammani and Giegé, 2014). Hence, RNA editing helps plant to cope with different environmental stresses, i.e., drought, heat, salt, etc. (Rieder et al., 2015; Rodrigues et al., 2017; Riemondy et al., 2018). In some cases, impaired RNA editing leads to abnormal plant ideotypes such as stunted plant growth and poor adoptability, even lethal phenotypes as impaired embryo development (Yan et al., 2018). This suggests that the RE plays a crucial role in growth and plant development. The RNA editing process has been already demonstrated in Arabidopsis (Arabidopsis thaliana), goatgrass (Aegilops tauschii), rice (Oryza sativa), maize (Zea mays), tobacco (Nicotiana tabacum), and grape (Vitis vinifera) (Wang et al., 2017; Chen et al., 2018; Rodrigues et al., 2020).
The most common editing types in non-flowering land plants are adenosine (A) to inosine (I) in tRNA, cytidine (C) to uridine (U) in messenger tRNA and RNA, and uridine (U) to cytidine (C) in mRNA (Chateigner-Boutin and Small, 2010; Zhang et al., 2016; Rasool et al., 2022). Protein function at the RNA level is maintain by RE events, e.g., failure of editing of the plastid ATPase alpha-subunit mRNA causes the pigment deficiency in tobacco cybrids (Sun F. et al., 2015). To find the RNA editing sites (RES), it is very important to get the knowledge of such post-transcriptional changes at the whole genome/transcriptome level of an organism to detect the mutations or polymorphism, and experimental evidence including high-throughput DNA/RNA sequencing is required. Previously, it is reported that RNA-seq can be used in the identification of RES in different crops (Chateigner-Boutin and Small, 2010; Eisenberg et al., 2010; Ichinose and Sugita, 2017).
The RNA editing in plants is primarily intervened by altering edifices including different altering factors, including organelle RNA recognition motif-containing (ORRM) protein, plant polyphenol oxidase (PPO), PPR, OZ, and RIP/MORF (Takenaka et al., 2012; Wang et al., 2019). It is documented that an RNA editing involves the deamination of C-U with specified PPR and other genes both inside the nuclear genome (Yan et al., 2018). Studies showed the presence of motif of ∼35–40 amino acids in PPR proteins are the responsible for editing (Ichinose and Sugita, 2017). Many members of PPR family such as MEF9, CLB19, MEF32, and nad7-200 are required for the editing process (Takenaka et al., 2012). More and more members of PPR proteins are reported but the exact number of involved PPR is still missing (Schmitz-Linneweber and Small, 2008). Similarly, in kiwifruit, MORF2 and MORF9 are well documented; they interact with NADH4 and are very important for RNA editing of chloroplast genome (Xiong et al., 2022) while MORF8 interact with MORF1 and MORF2 (Zehrmann et al., 2015; Huang et al., 2019).
Rice is a very important cereal in the world, providing nutrition to the human. It faces different kinds of environmental stresses such as heat, drought, cold, salinity, insects, and pest attacks (Uzair et al., 2022). Being a high demand crop, it is necessary to increase the production of rice. Under salt stress, the processing of RNA due to RNA editing is still not well explored in rice. There are many types of RESs in chloroplast and mitochondrial transcriptomes but their effects in alkaline tolerant and susceptible genotypes is not reported. In this research, we analyzed the impact of alkaline stress on RNA processing by using RNA-seq data. We found more RNA editing events in the tolerant genotype, which might be due to stability of editing factors. Alkaline stress promotes the RNA editing at transcript level through the change in amino acids. These changes in amino acids in the genes will help the plants to cope with the environmental stresses.
Materials and Methods
Data Acquisition, Quality, and Mapping
Two rice genotypes Caidao (CD) which was alkaline-sensitive and WD20342 (WD) an alkaline-tolerant were assessed in control and alkaline environments (Li et al., 2018). The transcriptome-related data of these samples were publicly available and assessed from National Center Biotechnology Information (NCBI1) with the BioProject PRJNA414178.2 Reference genomes and annotation files of mitochondria (BA000029.3) and chloroplast (KT289404.1) were also accessed from NCBI in fasta format. Quality of sequence reads was assessed by using FastQC tool3 to identify low-quality reads and adapter sequences (Andrews, 2010). Hisat2 (v2.1.0) was used for index building and alignment of paired-end and clean reads with reference genome (Kim et al., 2019). Number of reads mapping to each gene were calculated with FeatureCounts (v2.0.0) (Liao et al., 2014). The fragments per kilobase of transcript per million mapped reads (FPKM) was calculated using the gene length and reads count mapped to that gene. Log fold change (LogFC) was applied on FPKM values and Heatmaps were generated by using “pheatmap” package of R.
Differential Genes Expression Analysis
Two genotypes under control and stress were used to highlight differentially expressed genes and this was achieved by using the DESeq R package (Love et al., 2014); DESeq utilizes negative binomial distribution statistical model to predict differential expression. The p-values were adjusted (Benjamini and Hochberg, 1995). This was done to minimize the false discovery rate. Genes discovered by DESeq with a p-adjusted less than 0.05 were labeled as differentially expressed.
Gene Ontology Analysis
Gene ontology (GO) analysis was performed to identify over-represented biological processes (BP), molecular functions (MF), and cellular components (CC) using the identified genes. The gene list was submitted to the ClueGO is a Cytoscape4 plug-in that combines Gene Ontology to produce a well-organized GO/pathway annotation (Shannon et al., 2003; Bindea et al., 2009). Gene ontology terms with p-values less than 0.05 were denoted significant enriched by DEGs.
Analysis and Detection of RNA Editing Sites
RNA editing sites were predicted as previously described by Lv et al. (2018). In details, clean reads of RNA-seq data of the two rice (alkaline tolerant and susceptible) genotypes were aligned to the reference genomes of rice chloroplast and mitochondria. This was achieved with the help of SPRINT (SnP-free RNA editing Identification Toolkit) tool5 (Zhang et al., 2017). For SNP identification between RNA-seq and reference genomes of chloroplast and mitochondria, Genome Analysis Tool Kit (GATK6) which utilize BAM files were used for SNP calling (Van der Auwera and O’Connor, 2020). The overlapped findings of these two methods were used for further analysis. After this, samples of control of each genotype were used as a background, the SNPs difference between control and stressed samples were counted as RNA editing sites. The RNA-seq data from the control samples of the same genotype were used to avoid genotype-specific genomic SNP polymorphisms. At the end, the screening of data was done on the basis of (1) the mapped reads with more than five edited sites, (2) the ratio of edited reads/total mapped reads was more than 50%, and (3) RES were predicted in all three replicates. The Ensemble Variant Effect Predictor (VEP) tool7 with default parameters (chromosome number, start and end position, allele, and strand) was used to annotate the RNA editing site (McLaren et al., 2016). For this purpose, O. sativa specie was used.
Expression Profiling of Selected Genes Through Real-Time PCR for Quantification
Seeds of rice genotype (IRRI6) were sterilized with 2.5% sodium hypochlorite for 35 min followed by three washes and placed in 37°C Owen for 1 day. Uniformly germinated seeds were grown under normal conditions (16-h light/8-h dark at 26 ± 2°C) for 2 weeks. For this purpose, 96-wells plates supported by a container filled with nutrients media (Yoshida) were used. After this, one batch was kept as a control and in other batch 0.5% Na2CO3 solution (pH = 11.3) was applied for 36 h to create alkaline stress. The leaf samples were collected in triplicates. The total RNA was extracted with the help of TRIzol method and complementary DNA was prepared by using reverse transcriptase-III, first strand cDNA Synthesis Kit (K1691, Thermo Scientific Revert Aid). Real-time PCR for quantification (qRT-PCR) of selected genes of PPR, OZ1, and MORF/RIP gene family members was performed by using StepOne RT-PCR (Applied Biosystems® 7900 HT Fast RT-PCR). Three biological replicates from control and treated samples were used. OsActin was used as an internal control and for expression calculations 2ΔΔ CT method was used (Uzair et al., 2021). List of gene-specific primers are provided in Supplementary Table 1.
Results
Alignment of RNA-Seq Data
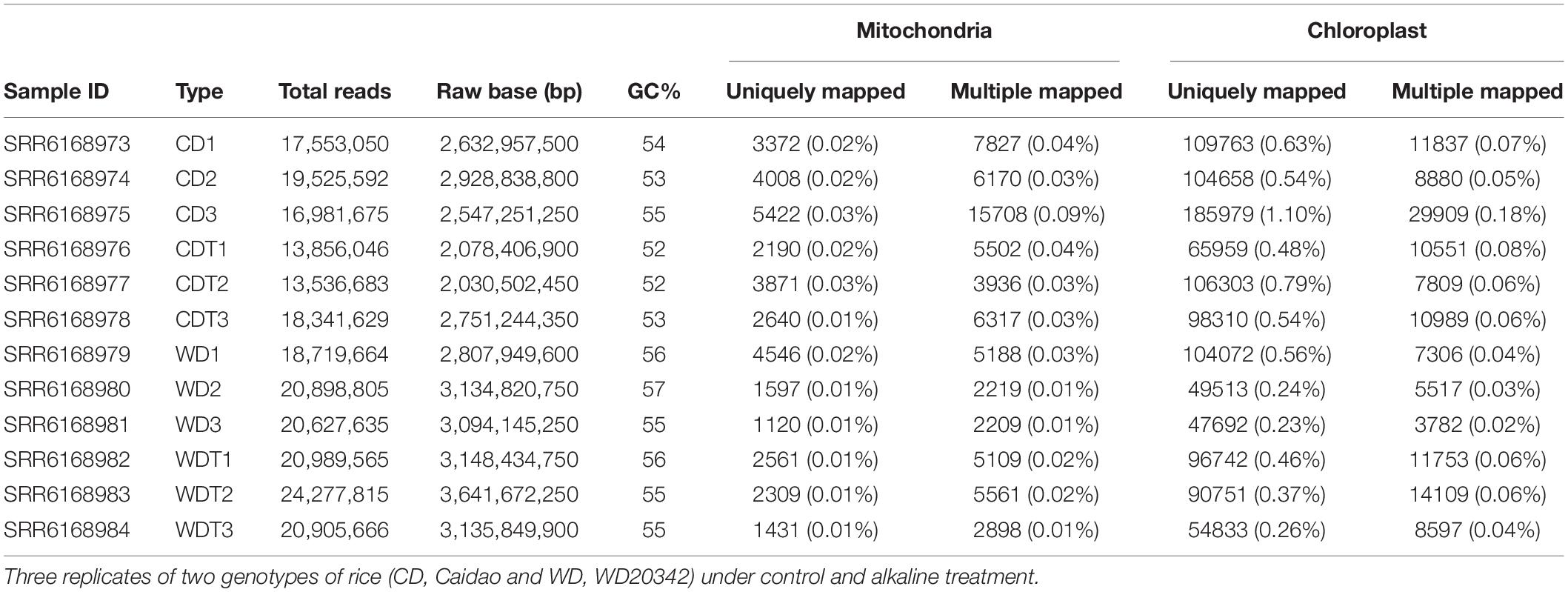
Plants of two rice genotypes Caidao (CD) and WD20342 (WD) were raised under normal and stress conditions. The genome size of rice mitochondrial and chloroplast genomes is 490,520 and 134,556 bp with number of genes 81 and 129, respectively (Notsu et al., 2002; Asaf et al., 2017; Feng et al., 2021). The genome of mitochondria has a smaller number of genes as compared to chloroplast genome. An average of ∼2,915 reads were aligned with the reference mitochondrial genomes with percentage of 0.01% (0.002 std). Similarly, for chloroplast, the average ∼92,881 reads aligned with the mapping on reference 0.51% (0.07 std). In general, chloroplast have more genes as compared with mitochondria. We expected that mapping reads should more in chloroplast and we found the expected results (Table 1 and Figure 1A).
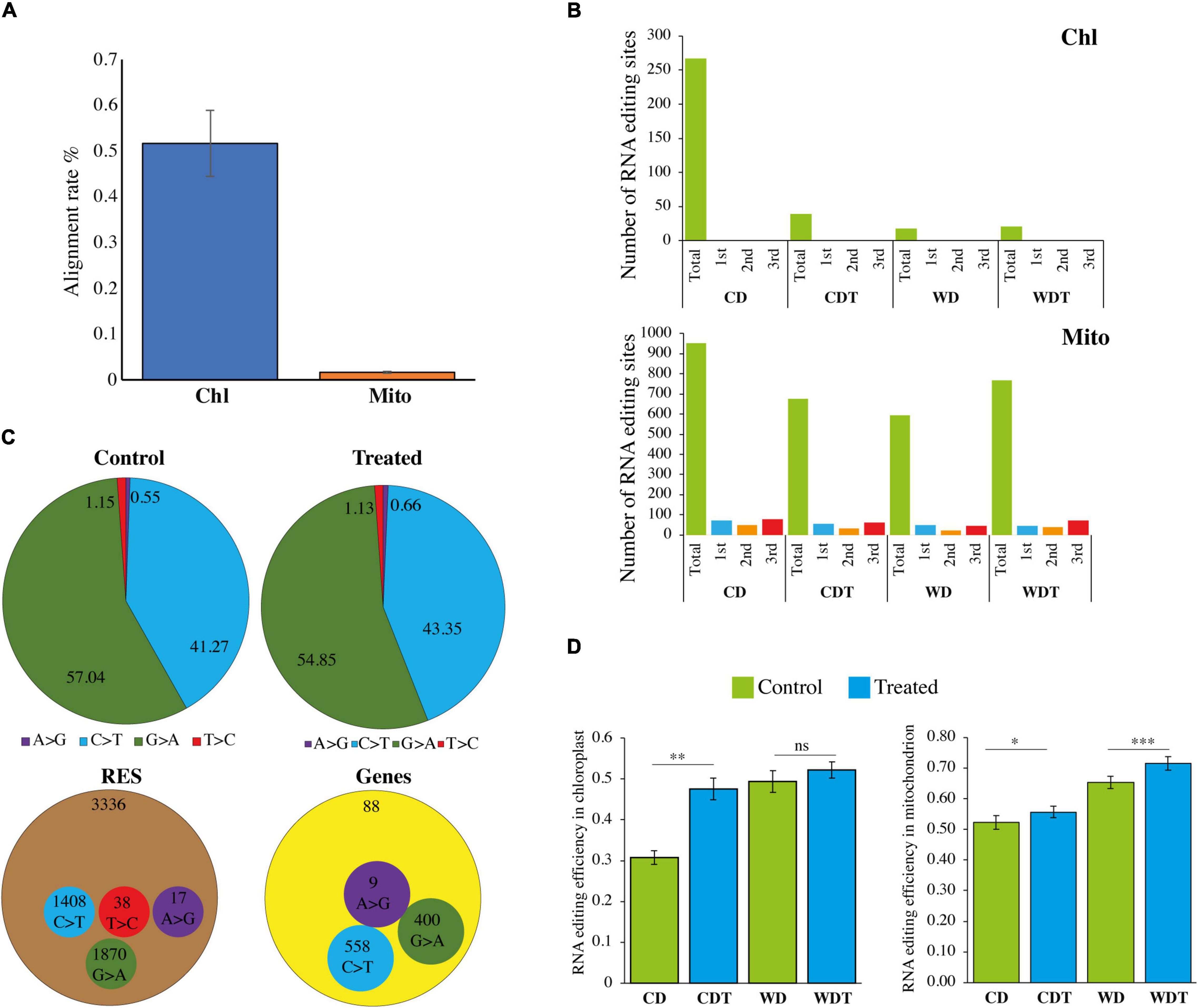
Figure 1. Summary of RNA-seq data. (A) Mapping rate of RNA-seq data of chloroplast (Chl) and mitochondria (Mito). (B) Codon position of RNA editing sites in two genotypes (CD, Caidao and WD, WD20342) of rice, chloroplast (Chl) genome, and mitochondrion (Mito) genomes. (C) Comparison of single nucleotide conversion in control and treated samples by RNA-seq approach. Values are shown by percentage. Number of total RES and edited genes. (D) Editing efficiency of all the RNA sites in rice chloroplast and mitochondrion genomes. For comparison, t-test was used. ***p < 0.001; **p < 0.01; *p < 0.05 was used for significant while ns, non-significant.
Identification and Characterization of RNA Editing Sites
To build the sequencing profundity and unwavering quality of editing sites, the subsequent bam documents of three replicates of each genotype under each condition were converged for resulting distinguishing proof of RESs. After the SNPs calling, we found total 3,336 RNA editing sites in both genomes including 345 in chloroplast and 2,991 in mitochondria, respectively (Table 2). We notice that some of the genes expressed in specific conditions such as control and alkaline stress. The chloroplast genome showed only seven edited genes. The data revealed that the higher editing sites were G-to-A (150 out of 345); 137 out of 345 editing sites were C-to-T, the second most common type in chloroplast. Similarly, T-C were 38 and A-G were 20 out of 345 (Supplementary Table 2). In chloroplast genome, in control samples (CD = 3 genes and WD = 1 gene) showed a larger number of edited genes while in stress conditions (CDT = 2 genes and WDT = 1 gene), less genes were edited (Table 3). In mitochondrial genome, two types of editing sites C-to-T and G-to-A were found; most edited sites 1,720 out of 2,991 editing sites were G-to-A and the second most common type of editing, 1,271 out of 2,991, was C-to-T (Supplementary Table 3). Mitochondrial genome showed that total 81 edited genes. In control samples (CD = 19 genes and WD = 19 genes) showed a smaller number of edited genes, while in stress conditions (CDT = 23 genes and WDT = 20 genes) more genes were edited in mitochondrial genome (Table 3). These results showed that more genes were edited in mitochondrial genome as compared to chloroplast and similarly under stress condition as compared to control samples. This increase is might be due to biasness of the sequencing; mapping rate increase in stress samples as compared to control one which might lead to new editing sites. Second, upregulation of genes (PPR and OZ1) involved in RNA editing.
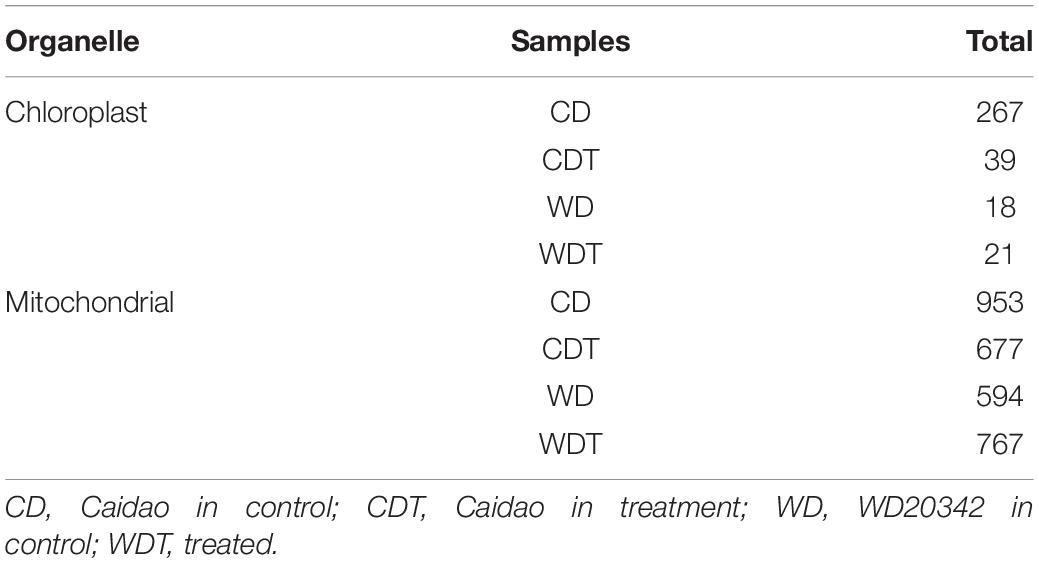
Table 2. Summary of identified RNA editing sites in chloroplast and mitochondrion genome of rice under alkaline stress.
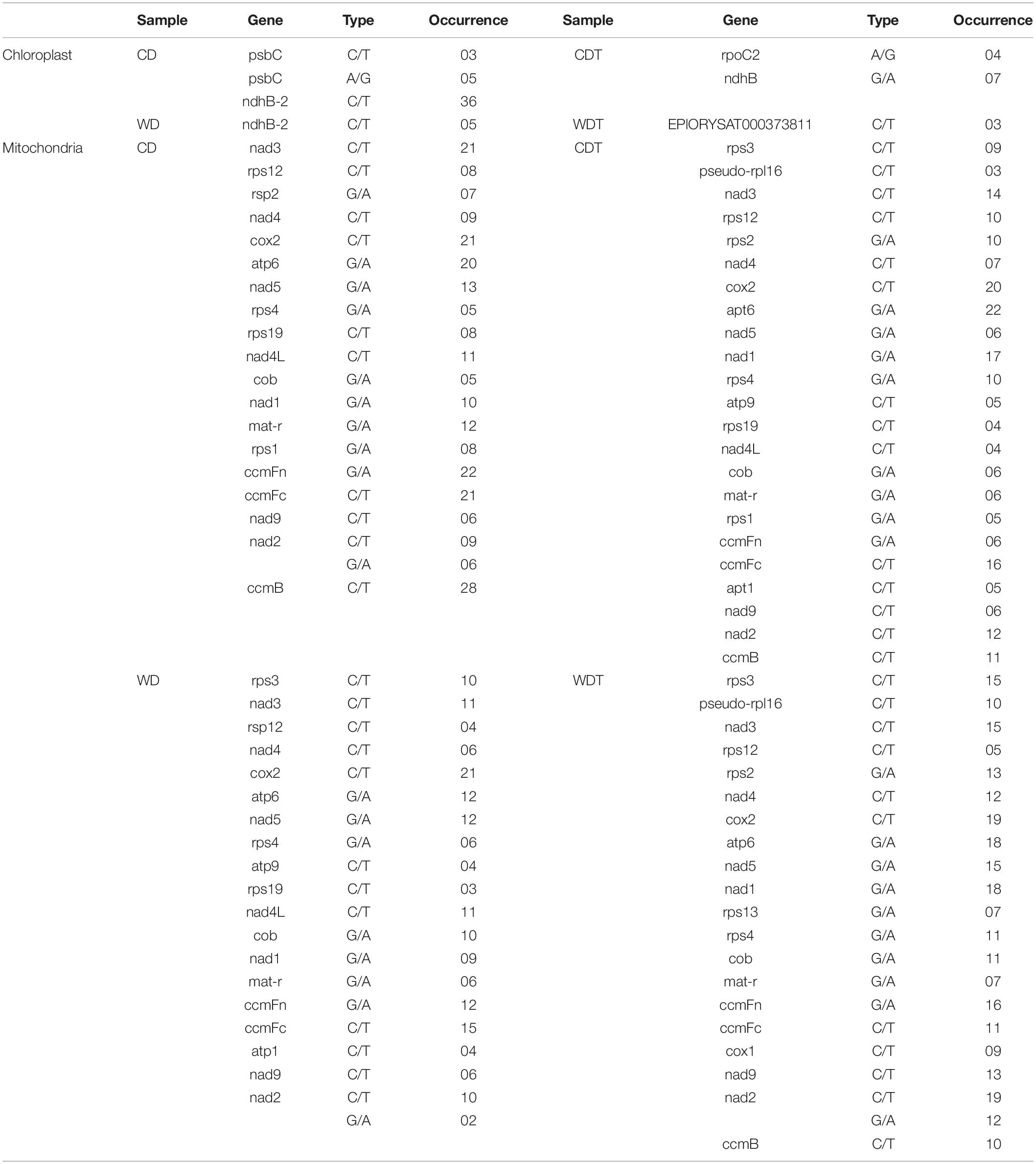
Table 3. Mitochondrial RNA editing sites in genes, type of editing, and their occurrence in four samples of rice.
Annotation of Editing Sites
We employed VEP tool for the identification of characteristics summary of RNA editing sites. The results revealed that there was no RNA editing site detected in the genes of chloroplast while in mitochondrial genome RNA editing in third codon position was mainly occurred (Figure 1B) whereas editing in first codon position was second the most common type. In synonymous variant, we did not find any codon changes at the second position of the amino acid. We compared the control samples with treated ones and found that both have four (A > G, C > T, G > A, and T > C) types of editing (Figure 1C), respectively. In control samples, we found 57.04% G > A, and 41.27% C > T types of editing. Similarly, in the treated samples, G > A was 54.85% and C > T was 43.35%. We found a total of 3,336 RNA editing sites in both genomes and has four types of RNA editing types (Figure 1C). The G > A was the most dominant type 1,870 out of 3,336 RESs. Similarly, C > T was the second most type 1 408 out of 3 336 RES, followed by T > C (38 RES), and A > G (17 RES). The results showed that there were 88 total edited genes and these genes have three types C > T (558 RES), G > A (400 RES), and A > G (9 RES) of editing types (Figure 1C). In addition, larger editing events resulted in as degenerative codon. We found that most of the neutral amino acid changes to hydrophobics followed by neutral to hydrophilic (Supplementary Table 4). Previously, Yan et al. (2018) and Zhang et al. (2020) also described that RNA editing increased the hydrophobicity of the newly synthesized proteins.
Based on the genetic properties, we divided RNA editing sites in the mitochondrial genome into six different types (Supplementary Table 5). These types include upstream variants, downstream variants, missense variants, synonymous variants, start lost, and stop gained. The results revealed that CD owned the 44% downstream gene variants followed by upstream gene variant 27%, missense 17%, synonymous variants 11%, start lost 1%, and stop gained 2%. In CDT, 50% downstream gene variants followed by upstream gene variant 25%, missense 14%, synonymous variants 10%, and 4% stop gained. Similarly, in tolerant genotype WD have 51% downstream gene variants, 26% upstream gene variant, 14% missense variants, 9% synonymous variants, start lost 1%, and 2% stop gained under control condition. Similarly, under stress condition, WDT have 46% downstream gene variants, 36% upstream gene variant, 10% missense variants, 9% synonymous variants, and 2% stop gained. Interestingly, we did not find start lost in CDT and WDT.
Ribonucleic Acid Editing Efficiency With and Without Stress
We checked the statistics of RNA editing efficiency. In total, the average editing efficiency of RESs was about 0.54. In chloroplast, the genome efficiency was 0.31, 0.48, 0.49, and 0.52 (Figure 1D). While in mitochondria, RE efficiency was 0.52, 0.56, 0.65, and 0.72 (Figure 1D). In chloroplast, there was a significant (p < 0.01) difference in CD untreated and treated samples while no difference was found in WD treated and untreated samples. In mitochondria, both genotypes CD and WD under treatment showed induction in editing efficiency. Overall, the tolerant genotype showed increased efficiency.
Cluster Analysis of RNA Editing Efficiency
We performed cluster profiling of the chloroplast (Figure 2A) and mitochondria (Figure 2B) genomes in samples of CD, CDT, WD, and WDT. The results revealed that RE efficiency was increased in the CDT and WDT under treatment. This increased in RE efficiency is related to tolerance of the genotypes and also showed that RE is a process which helps the plants to survived in the stress environment. These results are the further confirmation of the previous results (Figure 1D). All the information related to RE efficiency was presented in Supplementary Table 6.
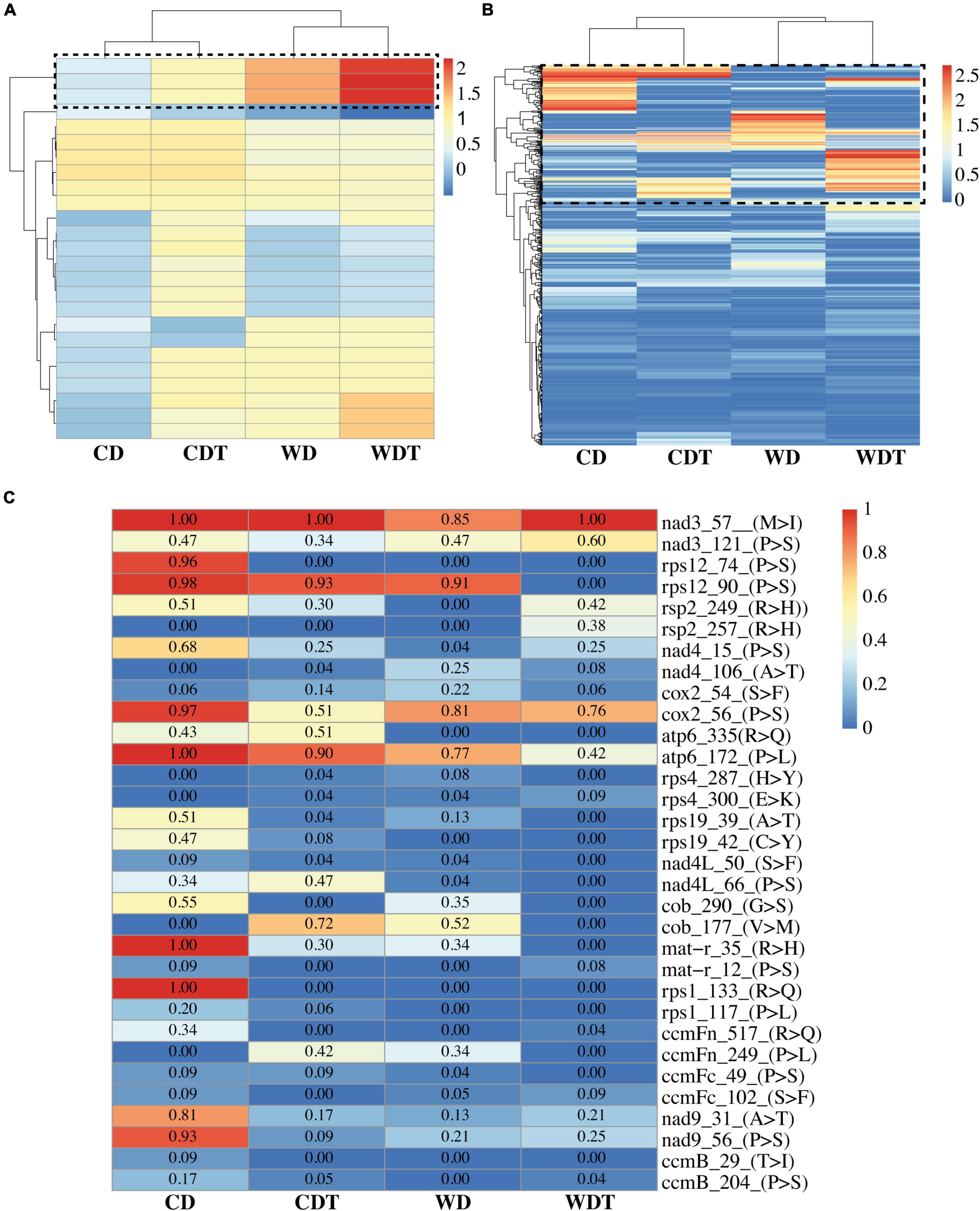
Figure 2. Heatmap of RNA editing efficiency of all the RNA sites in two genotypes of rice (CD, Caidao and WD, WD20342), (A) Chloroplast genome; (B) mitochondrion genome. The x-axis shows the genotypes under control (CD and WD) and treated (CDT and WDT) while the y-axis shows the editing sites. Black dotted box representing the RNA editing sites increased efficiency. (C) Reduced RNA editing efficiency in genes of mitochondria. Values insides the boxes were showing RNA editing efficiency. The x-axis shows the genotypes under control (CD and WD) and treatment (CDT and WDT). Gene names with amino acid position and type of changes were shown on the y-axis.
Effects of RNA Editing Efficiency on Genes
Findings of annotations showed that 81 genes out of total 88 genes were annotated. All the annotated genes were belonging to mitochondrial genome (Figure 2C and Supplementary Tables 5, 7). The genes in chloroplast were not showed any annotation. In mitochondrial genome, many genes were showing more editing sites, especially ccmB showed 49 editing sites in all the samples of two genotypes. All the editing types were C-T (C-U) showing decreasing trend from CD to WDT; amino acid changes were A (Ala)–T (Thr), P (Pro)–L (Leu), P (Pro)–S (Ser), and T (Thr)–I (Ile). Overall trend for these amino acids was neutral to hydrophobic. Similarly, the ndh2 showed two types of RNA editing: (1) C-T (C-U = 50) and (2) G-A (20) but not annotated. Different genes such as NADH dehydrogenase gene family (nad3/4/4L/9), ribosomal genes family (rps1/2/3/4/12/19), ATPase family (atp1/6), heme trafficking system membrane genes family (ccmB/ccmFn/ccmFc), mitochondrial cytochrome c oxidase gene family (cox1 and 2), and maturase R (matR) were detecting in this study and heatmap of randomly selected genes were generated (Figure 2C).
It was previously reported that the ribosomal genes family played a role in protein synthesis, growth development in plants, and proper functioning of chloroplasts (Zhou et al., 2015; Uzair et al., 2021). Members of the mitochondrial cytochrome c oxidase (COX) helps in aerobic energy development (Timón-Gómez et al., 2018). The NADH dehydrogenase genes involved in plant growth in response against the environmental stress (Hashida et al., 2009). Similarly, in chloroplast genome, ndhB-2 gene was edited which help in fixation of C-to-U back mutations through the DNA proofreading mechanism (Martín and Sabater, 2010). In tobacco ndhB genes improves the photosynthetic ability, stress resistance, and energy transformation (Horváth et al., 2000). The molecular function of maturase R (matR) is not fully clear but it converts the edited amino acid to its original form. In angiosperm, one matR was reported in the fourth intron of nad1 gene (Sultan et al., 2016). Subsequently, alteration in amino acids sites due to evolution or RES might adjusts through maturase implementation.
Identification of A-I and C-U RNA Editing Sites
We checked the RESs in chloroplast and mitochondria of four samples (Supplementary Tables 2, 3). Both genotypes (CD and WDT) were used under control and treatment which making four samples (CD, CDT, WD, and WDT). In the chloroplast genomes, the genotypes CD showed total 10 A-I RES all were A-G associated with psbC gene and 122 RES were C-T (C-U). In CDT samples, total identified A-I RES were 10, from this 10 RES were A-G associated with the gene rpoC2 and 11 RES were C-T (C-U). A total of 9 out of 20 RES were A-I in both of these genes. We did not find any RES in WD samples under control and under treatment. We found only five C-T (C-U) RES in WD and 16 C-T (C-U) RES in WDT. We also checked the mitochondrial genome also, but unluckily we cannot find any A-I RES in any sample. However, we found 386 C-T (C-U) RES in CD, 311 RES in CDT, 243 RES in WT, and 331 RES in WDT.
Gene Ontology Analysis of Mapped Genes
To check the functions of the genes in which RESs were detected, all REGs in all sample were annotated to the terms in the GO database. Results revealed that 12 and 48 significantly enriched functions identified in chloroplast and mitochondria, respectively. In chloroplast we found only 12 cellular components (Figure 3A). Most enriched components in each sample including organelle ribosome and mitochondrial small ribosomal subunit were associated with genes of ribosomes such as rpl16, rpl2, rpl20, rps11, rps12, rps14, rps16, rps19, rps2, rps3, and rps7.
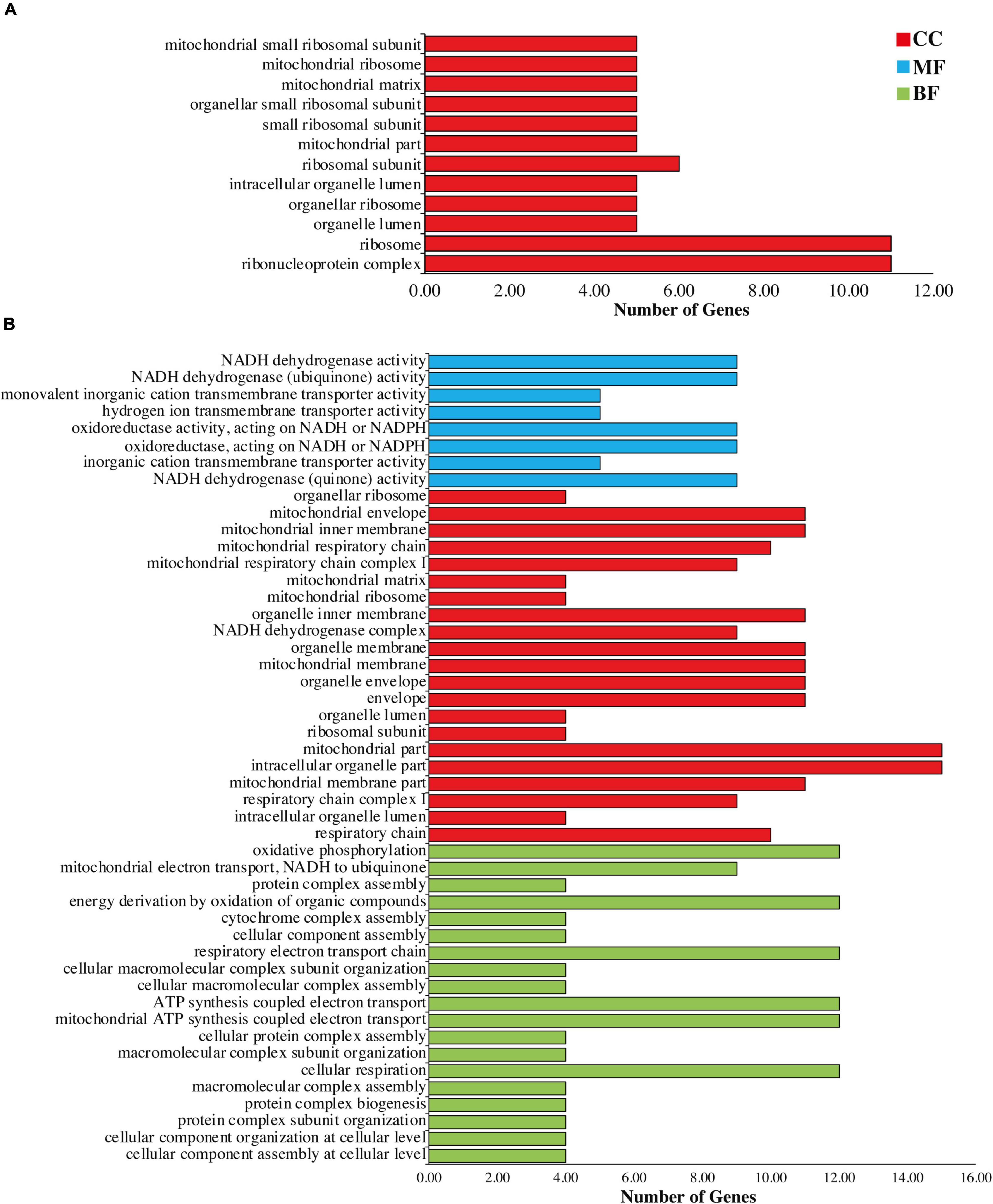
Figure 3. Gene Ontology annotation of genes of (A) chloroplast and (B) mitochondria. Red color showing cellular components (CC), blue color representing molecular functions (MF), and green color showing biological functions (BF).
In mitochondria genome, most enriched processes were mitochondrial ribosome, NADH dehydrogenase activity, cytochrome complex assembly, protein complex assembly, oxidative phosphorylation, cellular component organization at cellular level, ribosomal subunit, mitochondrial membrane, ATP synthesis coupled electron transport, and mitochondrial electron transport NADH to ubiquinone (Figure 3B and Supplementary Table 8).
Expression Profiling of PPR, OZ1, MORF/RIP, and RNA Edited Genes
For the comparison among the edited genes and genes involved in editing was done through transcriptome analysis. We checked the expression of RNA edited genes, but there were no significant differences observed under treated and untreated samples (Supplementary Figure 1). This indicates that alkaline stress only influences the RE events and has no role in the expression of these genes. To find the reason of reduction of RE efficiency in edited genes we analyzed the expression of genes involved in RE-like pentatricopeptide repeat proteins (PPR), Organelle Zinc finger 1 (OZ1), and multiple organellar RNA editing factors (MORF/RIP) genes. After the blast results, we found a total of 482 genes of PPR family, 12 genes of OZ1 family, and 7 MORF/RIP genes in rice. Previously, it was reported that rice have 477 PPR genes but one more study reported 491 PPR genes in rice (O’Toole et al., 2008; Chen et al., 2018). Similarly, seven MORF genes are reported in rice (Zhang et al., 2019). The results showed that there was a reduction of expression with the treatment (Figure 4A). Some of the PPR genes were also upregulated (Supplementary Table 9). The expression of OZ1 genes and MORF/RIP genes were also reduced for most of the genes (Figures 4B,C). To confirmed these findings, we selected already reported PPR genes LOC_Os03g53170, LOC_Os05g30240, and LOC_Os01g58080 (Chen et al., 2018), and MORF genes LOC_Os09g04670, LOC_Os06g02600, and LOC_Os08g04450 (Zhang et al., 2019). While OZ1 genes LOC_Os07g30821, LOC_Os02g07070, and LOC_Os01g59980 were selected randomly for expression analysis through qPCR. The results showed that there was induction in PPR and OZ1 genes under alkaline stress, while the genes of MORF were downregulated (Figure 4D). These results were with the agreement of previous studies. Overall, results showed that there was a positive association among the RNA editing and PPR genes.
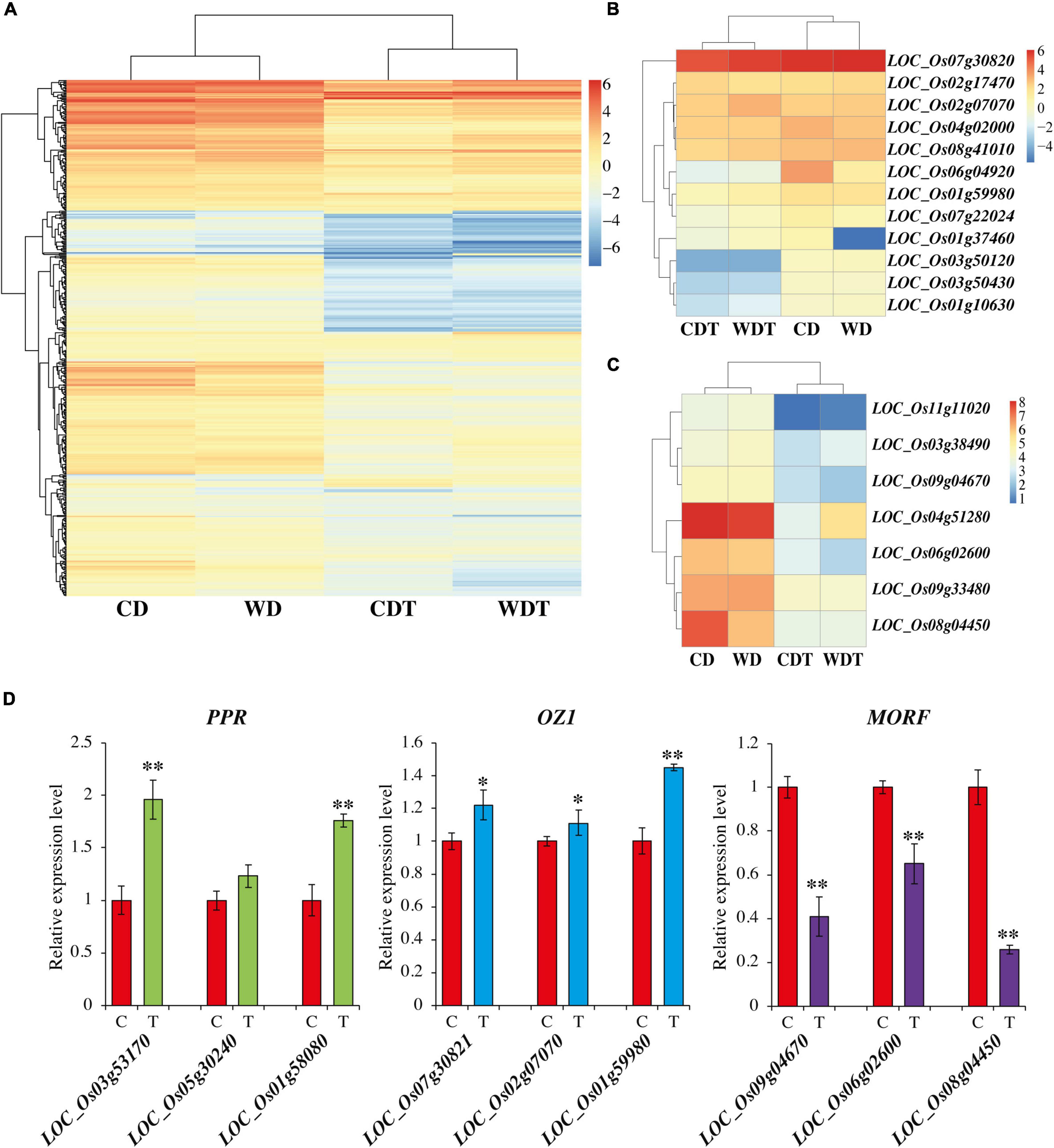
Figure 4. Expression of PPR genes (A), OZ1 genes (B), and MORF/RIP genes (C) in two genotypes of rice (CD, Caidao and WD, WD20342) under control and treatment. (D) Expression analysis of PPR, OZ1, and MORF/RIP genes through qRT-PCR in control (C) and treatment (T). Mean of three replicates with ±SE were used. For comparison t-test was used (**p < 0.01 and *p < 0.05).
Discussion
The RNA editing (RE) is a post-transcriptional mechanism in which transcripts modified by different mutations types such as insertion, deletion, and substitutions. The RE was firstly reported in kinetoplastid protozoa and then identified in higher plants and animals (Maslov et al., 1994). It can change the gene structure which cause the RNA splicing (Farré et al., 2012). For proper functioning of the RNA, its structure is very important. The thermodynamics of secondary structure are well studied under salt stress (Serra and Turner, 1995; Zuker, 2003) and folding of RNA is very sensitive to salt concentrations (Tan and Chen, 2011). Editing events played role in RNA degradation and also helps in microRNA regulation. The tertiary structure of the RNA helps in the arrangement of the secondary structure (Thirumalai, 1998). Our study suggests that the RNA editing due to secondary and tertiary structures are affected by alkaline stress. For confirmation of molecular mechanism of this RNA editing needs future experimentations. The RNA editing improves the plant growth and adoptability (Liu et al., 2013; Sun T. et al., 2015). Previously, it is reported that changes in mitochondrial genome at specific sites cause harmful effects such as growth, seed production, development, and fertility (Toda et al., 2012; Yap et al., 2015). Some studies also reported that abiotic and biotic stresses also affect RE and it have role in evolution of the species (Nakajima and Mulligan, 2001; Fujii and Small, 2011; Hammani and Giegé, 2014). In grapes it was proved that RNA editing is sensitive to temperature (Zhang et al., 2020). Similarly, in wheat RNA editing improve the drought tolerance (Rasool et al., 2022). Furthermore, in maize RNA editing is necessary for seed germination and development (Liu et al., 2013).
With the innovation of new sequencing approaches such as RNA sequencing, RESs were distinguished mainly in living species, particularly in plants. Current study, reported many RESs, and the statics of altering types demonstrated that RE normally happens as G-A, T-C, C-U, and A-I changes in deciphered areas of cellular organs mRNAs. We found that changes in coding regions of the proteins were found at third and first positions which changed the physiochemical properties of the amino acids (Khosravi and Jantsch, 2021; Din et al., 2022). Most of the amino acids were changed to hydrophobic which are agreement to the previous research (Zhang et al., 2020). This indicates that for the production of functional proteins RNA editing acts as a proofreading mechanism in plants (Ichinose and Sugita, 2017). In this study, induction of RE in alkaline tolerant genotypes proved the relationship of abiotic stress and RNA editing. Previously many studies showed the relationship of abiotic stress and RNA editing (Ruwe et al., 2013; Hajrah et al., 2017). It is reasonable that RNA editing helps the plants to survive in the stressed environment by regulating gene functions. This increase is necessary for keeping the homeostasis among the functions of the genes and RNA editing is proved to be very important for crop vigor (He et al., 2018). Similar findings also reported in soybean, in which RNA editing efficiency increased due to salt stress (Rodrigues et al., 2017). In other study of NADH dehydrogenase subunit 7 (nad7) in barley, salinity enhanced the RNA editing efficiency (Ramadan et al., 2021).
In plants, the RNA editing plays a crucial role for transcript maturation by conversion and insertion/deletion. Different genes families such as pentatricopeptide repeat proteins (PPR), organelle zinc finger 1 (OZ1), organelle RNA recognition motif-containing protein (ORRM), protoporphyrinogen IX oxidase (PPO), and multiple organellar RNA editing factors/RNA editing factor interacting protein (MORF/RIP) genes are reported in the RNA editing (Zhang et al., 2014; Haag et al., 2017; Tian et al., 2019; Wang et al., 2019). Out of these gene families, PPR, OZ1, and MORF/RIP are the important part of editosome and they are directly or indirectly involved in RNA editing (Okudaira et al., 2021). Pentatricopeptide repeat proteins played important role in editing and more than 450 members are reported in the different crops such as Arabidopsis and rice (Lurin et al., 2004; Chen et al., 2018). Pentatricopeptide repeat proteins are reported that they played role in growth, development, and found in plastids and mitochondria. The mutants of PPR showed different abnormal defects related to growth (Saha et al., 2007). The target sites of the RNA are predefined and they have specific sites for the attachment of PPR. So, the RNA editing is the best tool to understand this mechanism (Manna, 2015). In our study, the expression of PPR genes was increased under stress condition which indicates that RES are responsive to alkaline stress. It was previously reported that MORF genes family have role in abiotic stress and involved in RNA editing (Wang et al., 2019). Pentatricopeptide repeat proteins contains a C-terminal domain and interact with MORF genes (Takenaka et al., 2012; Brehme et al., 2014). Latest research reported that MORF involved in plant growth, development, stress response such as survival of seedling rice, drought resistance in poplar, and pathogen resistance in tobacco (Wang et al., 2019; Zhang et al., 2019; Yang et al., 2020). Our results showed the downregulation of MORF genes under alkaline stress and agreement with the findings of Zhang et al. (2019) and Xiong et al. (2022). Similarly, NbMORF8 which is present in the mitochondria and negatively enhance the immunity to Phytophthora pathogens in tobacco (Yang et al., 2020). More research needs to be involved to clarify the role of such gene family’s involvement or not in RNA editing.
Conclusion
The post-transcriptional changes such as C-U are conserved among animals as well as in plants. We used alkaline treated and control samples for comparison. We found four types of RNA editing A-G (A-I), C-T (C-U), G-A, and T-C, and most editing events were identified in non-synonymous codons which increased the hydrophobicity. Overall, RNA editing efficiency was increased due to stability of editing factors in the alkaline stressed samples. Furthermore, RNA editing changes the amino acids in the edited genes which help the plants to cope abiotic situations. These outcomes will contribute to improve understanding the molecular mechanism of dynamics of gene regulation at post-transcriptional level in rice. These findings will be useful in designing alkaline tolerance breeding programs in rice.
Data Availability Statement
The RNA-seq data was publically available at the followimg link https://www.ncbi.nlm.nih.gov//bioproject/PRJNA414178.
Author Contributions
OR, MU, and HC performed the analysis. OR and MU wrote the original draft. MK provided research facilities and guided during the whole study. MC supervised the whole study, reviewed, and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (32070677); the 151 Talent Project of Zhejiang Province (first level); Jiangsu Collaborative Innovation Center for Modern Crop Production; and Collaborative Innovation Center for Modern Crop Production cosponsored by Province and Ministry.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We appreciate the research facilities provided by MC’s Lab, China and NIGAB, Pakistan. We acknowledge the China Scholarship Council for providing fully funded Ph.D. to OR.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.892729/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov
- ^ https://www.ncbi.nlm.nih.gov//bioproject/PRJNA414178
- ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ https://cytoscape.org/
- ^ http://sprint.tianlab.cn
- ^ https://gatk.broadinstitute.org/hc/en-us
- ^ https://asia.ensembl.org/info/docs/tools/vep/index.html
References
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Bioinformatics, Babraham Institute.
Asaf, S., Waqas, M., Khan, A. L., Khan, M. A., Kang, S.-M., Imran, Q. M., et al. (2017). The complete chloroplast genome of wild rice (Oryza minuta) and its comparison to related species. Front. Plant Sci. 8:304. doi: 10.3389/fpls.2017.00304
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300.
Benne, R., Van Den Burg, J., Brakenhoff, J. P., Sloof, P., Van Boom, J. H., and Tromp, M. C. (1986). Major transcript of the frameshifted coxll gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46, 819–826. doi: 10.1016/0092-8674(86)90063-2
Bindea, G., Mlecnik, B., Hackl, H., Charoentong, P., Tosolini, M., Kirilovsky, A., et al. (2009). ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093. doi: 10.1093/bioinformatics/btp101
Brehme, N., Zehrmann, A., Verbitskiy, D., Härtel, B., and Takenaka, M. (2014). Mitochondrial RNA editing PPR proteins can tolerate protein tags at E as well as at DYW domain termini. Front. Plant Sci. 5:127. doi: 10.3389/fpls.2014.00127
Chen, G., Zou, Y., Hu, J., and Ding, Y. (2018). Genome-wide analysis of the rice PPR gene family and their expression profiles under different stress treatments. BMC Genomics 19:1–14. doi: 10.1186/s12864-018-5088-9
Din, M. S. U., Wang, X., Alamery, S., Fiaz, S., Rasheed, H., Khan, M. A., et al. (2022). Identification of CT novel polymorphism in 3rd exon of OsSPL14 gene governing seed sequence in rice. PLoS One 17:e0264478. doi: 10.1371/journal.pone.0264478
Duruvasula, S., Mulpuri, S., and Kandasamy, U. (2019). Mapping of plastid RNA editing sites of Helianthus and identification of differential editing in fungal infected plants. Curr. Plant Biol. 18:100109.
Eisenberg, E., Li, J. B., and Levanon, E. Y. (2010). Sequence based identification of RNA editing sites. RNA Biol. 7, 248–252. doi: 10.4161/rna.7.2.11565
Farré, J.-C., Aknin, C., Araya, A., and Castandet, B. (2012). RNA editing in mitochondrial trans-introns is required for splicing. PLoS One 7:e52644. doi: 10.1371/journal.pone.0052644
Feng, Q., Song, W.-C., Zhang, Y.-J., and Shi, C. (2021). The complete chloroplast genome sequence of Oryza sativa Temperate japonica. Mitochondrial DNA B Resour. 6, 927–928. doi: 10.1080/23802359.2021.1888331
Fujii, S., and Small, I. (2011). The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191, 37–47. doi: 10.1111/j.1469-8137.2011.03746.x
Golden, D. E., and Hajduk, S. L. (2006). The importance of RNA structure in RNA editing and a potential proofreading mechanism for correct guide RNA: pre-mRNA binary complex formation. J. Mol. Biol. 359, 585–596. doi: 10.1016/j.jmb.2006.03.041
Haag, S., Schindler, M., Berndt, L., Brennicke, A., Takenaka, M., and Weber, G. (2017). Crystal structures of the Arabidopsis thaliana organellar RNA editing factors MORF1 and MORF9. Nucleic Acids Res. 45, 4915–4928. doi: 10.1093/nar/gkx099
Hajrah, N. H., Obaid, A. Y., Atef, A., Ramadan, A. M., Arasappan, D., Nelson, C. A., et al. (2017). Transcriptomic analysis of salt stress responsive genes in Rhazya stricta. PLoS One 12:e0177589. doi: 10.1371/journal.pone.0177589
Hammani, K., and Giegé, P. (2014). RNA metabolism in plant mitochondria. Trends Plant Sci. 19, 380–389. doi: 10.1016/j.tplants.2013.12.008
Hashida, S.-N., Takahashi, H., and Uchimiya, H. (2009). The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 103, 819–824. doi: 10.1093/aob/mcp019
He, P., Xiao, G., Liu, H., Zhang, L., Zhao, L., Tang, M., et al. (2018). Two pivotal RNA editing sites in the mitochondrial atp1 mRNA are required for ATP synthase to produce sufficient ATP for cotton fiber cell elongation. New Phytol. 218, 167–182. doi: 10.1111/nph.14999
Horváth, E. M., Peter, S. O., Joët, T., Rumeau, D., Cournac, L., Horváth, G. V., et al. (2000). Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 123, 1337–1350. doi: 10.1104/pp.123.4.1337
Huang, C., Li, Z.-R., Yu, Q.-B., Ye, L.-S., Cui, Y.-L., Molloy, D. P., et al. (2019). MORF2 tightly associates with MORF9 to regulate chloroplast RNA editing in Arabidopsis. Plant Sci. 278, 64–69. doi: 10.1016/j.plantsci.2018.10.020
Ichinose, M., and Sugita, M. (2017). RNA editing and its molecular mechanism in plant organelles. Genes 8:5. doi: 10.3390/genes8010005
Keller, W., Wolf, J., and Gerber, A. (1999). Editing of messenger RNA precursors and of tRNAs by adenosine to inosine conversion. FEBS Lett. 452, 71–76. doi: 10.1016/s0014-5793(99)00590-6
Khosravi, H. M., and Jantsch, M. F. (2021). Site-directed RNA editing: recent advances and open challenges. RNA biology 18, 41–50. doi: 10.1080/15476286.2021.1983288
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Li, N., Liu, H., Sun, J., Zheng, H., Wang, J., Yang, L., et al. (2018). Transcriptome analysis of two contrasting rice cultivars during alkaline stress. Sci. Rep. 8, 1–16. doi: 10.1038/s41598-018-27940-x
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liu, Y.-J., Xiu, Z.-H., Meeley, R., and Tan, B.-C. (2013). Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25, 868–883. doi: 10.1105/tpc.112.106781
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Lurin, C., Andreés, C., Aubourg, S., Bellaoui, M., Bitton, F., BruyeÌRe, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103. doi: 10.1105/tpc.104.022236
Lv, S., Feng, K., Peng, S., Wang, J., Zhang, Y., Bian, J., et al. (2018). Comparative analysis of the transcriptional response of tolerant and sensitive wheat genotypes to drought stress in field conditions. Agronomy 8:247.
Manna, S. (2015). An overview of pentatricopeptide repeat proteins and their applications. Biochimie 113, 93–99. doi: 10.1016/j.biochi.2015.04.004
Martín, M., and Sabater, B. (2010). Plastid ndh genes in plant evolution. Plant Physiol. Biochem. 48, 636–645. doi: 10.1016/j.plaphy.2010.04.009
Maslov, D. A., Avila, H. A., Lake, J. A., and Simpson, L. (1994). Evolution of RNA editing in kinetoplastid protozoa. Nature 368, 345–348.
McLaren, W., Gil, L., Hunt, S. E., Riat, H. S., Ritchie, G. R., Thormann, A., et al. (2016). The ensembl variant effect predictor. Genome Biol. 17, 1–14.
Nakajima, Y., and Mulligan, M. R. (2001). Heat stress results in incomplete C-to-U editing of maize chloroplast mRNAs and correlates with changes in chloroplast transcription rate. Curr. Genet. 40, 209–213. doi: 10.1007/s002940100249
Notsu, Y., Masood, S., Nishikawa, T., Kubo, N., Akiduki, G., Nakazono, M., et al. (2002). The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Genet. Genom. 268, 434–445. doi: 10.1007/s00438-002-0767-1
O’Toole, N., Hattori, M., Andres, C., Iida, K., Lurin, C., Schmitz-Linneweber, C., et al. (2008). On the expansion of the pentatricopeptide repeat gene family in plants. Mol. Biol. Evol. 25, 1120–1128. doi: 10.1093/molbev/msn057
Okudaira, C., Sakari, M., and Tsukahara, T. (2021). Genome-wide identification of U-To-C RNA editing events for nuclear genes in Arabidopsis thaliana. Cells 10:635. doi: 10.3390/cells10030635
Ramadan, A. M., Said, O. A., and Abushady, A. M. (2021). Salinity stress reveals three types of RNA editing sites in mitochondrial Nad7 gene of wild barley both in silico and in qRT-PCR experiments. Theor. Exp. Plant Physiol. 34, 13–22.
Rasool, F., Ishtiaq, I., Uzair, M., Naz, A. A., Leon, J., and Khan, M. R. (2022). Genome-wide investigation and functional analysis of RNA editing sites in wheat. PLoS One 17:e0265270. doi: 10.1371/journal.pone.0265270
Rieder, L. E., Savva, Y. A., Reyna, M. A., Chang, Y.-J., Dorsky, J. S., Rezaei, A., et al. (2015). Dynamic response of RNA editing to temperature in Drosophila. BMC Biol. 13:1. doi: 10.1186/s12915-014-0111-3
Riemondy, K. A., Gillen, A. E., White, E. A., Bogren, L. K., Hesselberth, J. R., and Martin, S. L. (2018). Dynamic temperature-sensitive A-to-I RNA editing in the brain of a heterothermic mammal during hibernation. RNA 24, 1481–1495.
Rodrigues, N. F., Fonseca, G. C. D., Kulcheski, F. R., and Margis, R. (2017). Salt stress affects mRNA editing in soybean chloroplasts. Genet. Mol. Biol. 40, 200–208. doi: 10.1590/1678-4685-GMB-2016-0055
Rodrigues, N. F., Nogueira, F., Domont, G. B., and Margis, R. (2020). Identification of soybean trans-factors associated with plastid RNA editing sites. Genet. Mol. Biol. 43:e20190067. doi: 10.1590/1678-4685-GMB-2019-0067
Ruwe, H., Castandet, B., Schmitz-Linneweber, C., and Stern, D. B. (2013). Arabidopsis chloroplast quantitative editotype. FEBS Lett. 587, 1429–1433. doi: 10.1016/j.febslet.2013.03.022
Saha, D., Prasad, A., and Srinivasan, R. (2007). Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45, 521–534. doi: 10.1016/j.plaphy.2007.03.026
Schmitz-Linneweber, C., and Small, I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends in plant science 13, 663–670. doi: 10.1016/j.tplants.2008.10.001
Serra, M. J., and Turner, D. H. (1995). [11] Predicting thermodynamic properties of RNA. Methods Enzymol. 259, 242–261.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Sultan, L. D., Mileshina, D., Grewe, F., Rolle, K., Abudraham, S., Głodowicz, P., et al. (2016). The reverse transcriptase/RNA maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. Plant Cell 28, 2805–2829. doi: 10.1105/tpc.16.00398
Sun, F., Wang, X., Bonnard, G., Shen, Y., Xiu, Z., Li, X., et al. (2015). Empty pericarp7 encodes a mitochondrial E–subgroup pentatricopeptide repeat protein that is required for ccm FN editing, mitochondrial function and seed development in maize. Plant J. 84, 283–295. doi: 10.1111/tpj.12993
Sun, T., Shi, X., Friso, G., Van Wijk, K., Bentolila, S., and Hanson, M. R. (2015). A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genet. 11:e1005028. doi: 10.1371/journal.pgen.1005028
Takenaka, M., Zehrmann, A., Verbitskiy, D., Kugelmann, M., Härtel, B., and Brennicke, A. (2012). Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5104–5109. doi: 10.1073/pnas.1202452109
Tan, Z.-J., and Chen, S.-J. (2011). Salt contribution to RNA tertiary structure folding stability. Biophys. J. 101, 176–187. doi: 10.1016/j.bpj.2011.05.050
Thirumalai, D. (1998). Native secondary structure formation in RNA may be a slave to tertiary folding. Proc. Natl. Acad. Sci. U.S.A. 95, 11506–11508. doi: 10.1073/pnas.95.20.11506
Tian, F., Yu, J., Zhang, Y., Xie, Y., Wu, B., and Miao, Y. (2019). MORF9 functions in plastid RNA editing with tissue specificity. Int. J. Mol. Sci. 20:4635. doi: 10.3390/ijms20184635
Timón-Gómez, A., Nývltová, E., Abriata, L. A., Vila, A. J., Hosler, J., and Barrientos, A. (2018). Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 76, 163–178. doi: 10.1016/j.semcdb.2017.08.055
Toda, T., Fujii, S., Noguchi, K., Kazama, T., and Toriyama, K. (2012). Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J. 72, 450–460. doi: 10.1111/j.1365-313X.2012.05091.x
Uzair, M., Long, H., Zafar, S. A., Patil, S. B., Chun, Y., Li, L., et al. (2021). Narrow Leaf21, encoding ribosomal protein RPS3A, controls leaf development in rice. Plant Physiol. 186, 497–518. doi: 10.1093/plphys/kiab075
Uzair, M., Patil, S. B., Zhang, H., Kumar, A., Mkumbwa, H., Zafar, S. A., et al. (2022). Screening direct seeding-related traits by using an improved mesocotyl elongation assay and association between seedling and maturity traits in rice. Agronomy 12:975.
Van der Auwera, G. A., and O’Connor, B. D. (2020). Genomics in the Cloud: Using Docker, GATK, and WDL in Terra. Newton, MA: O’Reilly Media.
Wang, D., Meng, S., Su, W., Bao, Y., Lu, Y., Yin, W., et al. (2019). Genome-wide analysis of multiple organellar RNA editing factor family in poplar reveals evolution and roles in drought stress. Int. J. Mol. Sci. 20:1425. doi: 10.3390/ijms20061425
Wang, M., Liu, H., Ge, L., Xing, G., Wang, M., Weining, S., et al. (2017). Identification and analysis of RNA editing sites in the chloroplast transcripts of Aegilops tauschii L. Genes 8:13. doi: 10.3390/genes8010013
Xiong, Y., Fang, J., Jiang, X., Wang, T., Liu, K., Peng, H., et al. (2022). Genome-wide analysis of multiple organellar RNA editing factor (MORF) family in kiwifruit (Actinidia chinensis) reveals its roles in chloroplast RNA editing and pathogens stress. Plants 11:146. doi: 10.3390/plants11020146
Yan, J., Zhang, Q., and Yin, P. (2018). RNA editing machinery in plant organelles. Sci. China Life Sci. 61, 162–169.
Yang, Y., Fan, G., Zhao, Y., Wen, Q., Wu, P., Meng, Y., et al. (2020). Cytidine-to-uridine RNA editing factor NbMORF8 negatively regulates plant immunity to Phytophthora pathogens. Plant Physiol. 184, 2182–2198. doi: 10.1104/pp.20.00458
Yap, A., Kindgren, P., Colas Des Francs-Small, C., Kazama, T., Tanz, S. K., Toriyama, K., et al. (2015). AEF 1/MPR 25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 81, 661–669. doi: 10.1111/tpj.12756
Zehrmann, A., Härtel, B., Glass, F., Bayer-Császár, E., Obata, T., Meyer, E., et al. (2015). Selective homo-and heteromer interactions between the multiple organellar RNA editing factor (MORF) proteins in Arabidopsis thaliana. J. Biol. Chem. 290, 6445–6456. doi: 10.1074/jbc.M114.602086
Zhang, A., Jiang, X., Zhang, F., Wang, T., and Zhang, X. (2020). Dynamic response of RNA editing to temperature in grape by RNA deep sequencing. Funct. Integr. Genom. 20, 421–432. doi: 10.1007/s10142-019-00727-7
Zhang, F., Lu, Y., Yan, S., Xing, Q., and Tian, W. (2017). SPRINT: an SNP-free toolkit for identifying RNA editing sites. Bioinformatics 33, 3538–3548. doi: 10.1093/bioinformatics/btx473
Zhang, F., Tang, W., Hedtke, B., Zhong, L., Liu, L., Peng, L., et al. (2014). Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proc. Natl. Acad. Sci. U.S.A. 111, 2023–2028. doi: 10.1073/pnas.1316183111
Zhang, L., Yang, C.-S., Varelas, X., and Monti, S. (2016). Altered RNA editing in 3’ UTR perturbs microRNA-mediated regulation of oncogenes and tumor-suppressors. Sci. Rep. 6, 1–13. doi: 10.1038/srep23226
Zhang, Q., Shen, L., Ren, D., Hu, J., Chen, G., Zhu, L., et al. (2019). Characterization, expression, and interaction analyses of OsMORF gene family in rice. Genes 10:694. doi: 10.3390/genes10090694
Zhou, X., Liao, W.-J., Liao, J.-M., Liao, P., and Lu, H. (2015). Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7, 92–104. doi: 10.1093/jmcb/mjv014
Keywords: RNA editing, alkaline stress, rice, RNA-seq, PPR, OZ1, MORF
Citation: Rehman O, Uzair M, Chao H, Khan MR and Chen M (2022) Decoding RNA Editing Sites Through Transcriptome Analysis in Rice Under Alkaline Stress. Front. Plant Sci. 13:892729. doi: 10.3389/fpls.2022.892729
Received: 09 March 2022; Accepted: 18 May 2022;
Published: 23 June 2022.
Edited by:
Nasim Ahmad Yasin, University of the Punjab, PakistanReviewed by:
Waheed Akram, BECS Center for Research and Innovation, PakistanTalha Javed, Fujian Agriculture and Forestry University, China
Rubab Shabbir, University of Agriculture, Faisalabad, Pakistan
Copyright © 2022 Rehman, Uzair, Chao, Khan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Chen, bWNoZW5Aemp1LmVkdS5jbg==
 Obaid Rehman
Obaid Rehman Muhammad Uzair
Muhammad Uzair Haoyu Chao1
Haoyu Chao1 Muhammad Ramzan Khan
Muhammad Ramzan Khan Ming Chen
Ming Chen