- Laboratory of Horticultural Science, Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan
Grape (Vitis vinifera L.) is an important fruit crop in the world. It is used as a table grape and is also used for raisin and wine production. Grape berries accumulate secondary metabolites, such as anthocyanins, tannins, and resveratrol, which are known as functional compounds for human health. Multidrug and toxic compound extrusion transporter (MATEs) transport secondary metabolites. MATEs also transport other solutes, including organic acids, and toxic xenobiotics, depending on cation gradient and play various roles in plants. MATE comprises 300–500 amino acid residues and possesses a MATE domain and 8–12 transmembrane domains. In the present study, 59 MATE genes were identified in the grape genome, and phylogenetic analysis revealed the presence of four groups of grape MATEs (Group 1–4). Their information, such as gene structures, protein motifs, predicted subcellular localizations, and gene IDs of four genome annotations, that is, CRIBI v1, CRIBI v2, Genoscope, and Vcost v3, were annotated. The transport substrates and physiological functions of grape MATEs were estimated based on their homology with the analyzed MATEs in other plant species. Group 1 may transport toxic compounds and alkaloids, Group 2 may transport polyphenolic compounds, Group 3 may transport organic acids, and Group 4 may transport plant hormones related to signal transduction. In addition to the known anthocyanin transporters, VvMATE37 and VvMATE39, a novel anthocyanin transporter, VvMATE38 in Group 2, was suggested as a key transporter for anthocyanin accumulation in grape berry skin. VvMATE46, VvMATE47, and VvMATE49 in Group 3 may contribute to Al3+ detoxification and Fe2+/Fe3+ translocation via organic acid transport. This study provides helpful and fundamental information for grape MATE studies and resolves the confusion of gene IDs in different genome annotations.
Introduction
Grape (Vitis vinifera L.) is an important crop grown worldwide for table grape use and wine production. Grapes contain various secondary metabolites. For example, anthocyanins are responsible for the color of berries, catechins are responsible for the astringency of fruit taste, and resveratrol is responsible for disease resistance. These secondary metabolites also function as antioxidants and health-promoting compounds in humans. Plants produce these secondary metabolites and accumulate them strategically, defending against environmental stresses and pathogens.
The accumulation of secondary metabolites occurs via a series of synthetic enzymes and transport from the synthesis site to the accumulation site by the responsible transporters. Secondary metabolite transporters have been reported to belong to the following families (Nogia and Pati, 2021): ATP-binding cassette (ABC) transporter family (Rea, 2007), multidrug and toxic compound extrusion (MATE) transporter family (Omote et al., 2006), major facilitator superfamily (MFS; Nino-Gonzalez et al., 2019), NPF transporter family (Wen et al., 2020) and purine uptake permeases (PUPs; Dastmalchi et al., 2019). Some MATEs had been reported as antiporters using cation (H+/Na+) gradient and transport substrates from cytosol to apoplast or vacuole (Nogia and Pati, 2021). MATE comprises 300–500 amino acid residues and possesses a MATE domain and 8–12 transmembrane domains (Shoji, 2014). MATE is also called detoxification protein (DTX) in Arabidopsis (Li et al., 2002) and is involved in the detoxification mechanisms of toxic compounds and heavy metals (Diener et al., 2001; Li et al., 2002), tolerance to aluminum toxicity (Liu et al., 2009), iron homeostasis (Wang et al., 2017), and disease resistance (Ishihara et al., 2008).
MATEs are conserved in bacteria, fungi, animals, and plants (Hvorup et al., 2003). Genome-wide analyses, that is, identification and characterization of gene families using genome data, provide helpful and fundamental information on gene families, including evolutionary history and diversity of gene families and their members, characteristics of encoded proteins, and their physiological functions. Genome-wide analysis of MATEs and gene numbers in plants had been reported, including 56 in Arabidopsis thaliana (Li et al., 2002), 53 in Oryza sativa (Tiwari et al., 2014), 71 in Populus trichocarpa (Li et al., 2017), 117 in soybean (Liu et al., 2016), 48 in potato (Li et al., 2019), 67 in tomato (Dos Santos et al., 2017), and 73 in Linum usitassimum (Ali et al., 2021).
In grapes, only VvMATE1 and VvMATE2, which transport proanthocyanidins (Perez-Diaz et al., 2014), and VvAM1 and VvAM3, which transport cyanidin 3-glycosides (Gomez et al., 2009), have been reported. The numbers and functions of other grape MATEs are still unclear. Therefore, this study performed a genome-wide analysis of grape MATEs. As a result, 59 genes encoding MATEs were identified in the grape genome, obtaining useful information, including the gene IDs of different genome annotations, gene and protein structures, protein motifs, and gene expression profiles. In addition, the physiological functions of some MATEs in grapes were discussed with their gene expressions and transport substrates were estimated based on the homology with the analyzed MATEs in other plant species.
Materials and Methods
Identification of MATEs in Grape Genome
To obtain candidates of grape MATEs, BLASTP search (e ≤ 0.1) was performed against the CRIBI v2.1 genome annotation based on the V. vinifera 12X.0 genome assembly (Adam-Blondon, 2014) in Phytozome v131 (Goodstein et al., 2012) using the amino acid sequences of Arabidopsis MATEs (Li et al., 2002) as queries. Against obtained non-redundant candidates of grape MATEs, the presence of conserved MATE domain was confirmed by Pfam2 (El-Gebali et al., 2019) and by the Conserved Domain Database of NCBI (CDD,3 Shennan Lu et al., 2020). Among the candidates, the proteins with predicted MATE domain by both Pfam and CDD were identified as grape MATEs. Molecular weight and isoelectric point were estimated by ProtParam4 (Gasteiger et al., 2005). The number of transmembrane domains was predicted by the Simple Modular Architecture Research Tool (SMART,5 Letunic et al., 2021).
MATEs in other grape genome annotations, that is, the CRIBI v1, the Genoscope, and the Vcost.v3 were identified as follows. BLASTP was performed against the CRIBI v1 genome annotation based on the V. vinifera 12X.0 genome assembly in EnsemblPlants.6 Whole protein sequences of the Genoscope genome annotation based on the V. vinifera 12X.0 genome assembly were downloaded from URGI7 or those of the Vcost.v3 genome annotation based on the V. vinifera 12X.2 genome assembly (Canaguier et al., 2017) were downloaded from URGI8 and in house BLASTP was performed.
Multiple Sequence Alignment and Phylogenetic Analysis
Amino acid sequences of MATEs of grape and other plant species (Li et al., 2002; Takanashi et al., 2014; Biala-Leonhard et al., 2021) were aligned using the ClustalW program.9 The phylogenetic tree was created by MEGA X (Kumar et al., 2018) using the neighbor-joining model.
Gene Structure and Motif Analyses
Gene structure (exon–intron structure) was schematized using coding sequences (CDS) and genomic sequence of the grape MATEs by the Gene Structure Display Server (GSDS,10 Hu et al., 2015). The Multiple Expectation Maximization for Motif Elicitation (MEME,11 Bailey et al., 2009) was used to identify the conserved motifs in the amino acid sequences of grape MATEs. The parameters were set as follows: site distribution on zero or one occurrence per sequences, the maximum number of motifs was 12, and motif width is between 6 and 50.
In silico Gene Expression Analysis
The gene expression data in various organs, tissues, and developmental stages of grape by microarray analysis were downloaded from Grape eFP Browser (Fasoli et al., 2012).12 The gene expression data were converted to base 10 logarithm and then heatmap was created by using HemI 1.0 (Deng et al., 2014).13
Results
Identification of MATEs in the Grape Genome
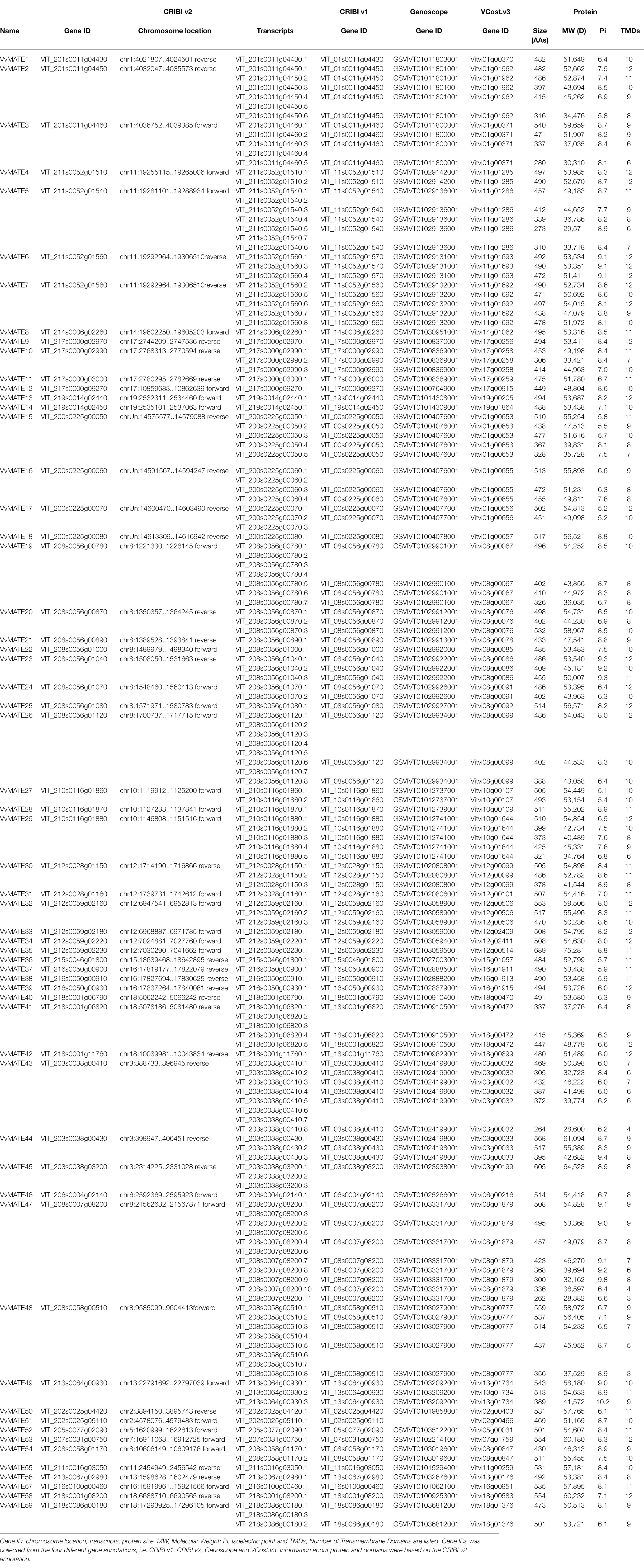
To identify MATEs in the grape genome, BLAST searches were performed against the grape genome assembly 12X.0 (Adam-Blondon, 2014) and genome annotation CRIBI v2.1 (Vitulo et al., 2014) using Arabidopsis MATEs as queries. Consequently, 78 genes were hit in the grape genome. The presence of MATE domains in the candidates was confirmed using domain search tools Pfam and CDD. As a result, two of the 78 hits did not have a motif predicted by both Pfam and CDD and were therefore excluded from further analyses. Fifty-eight candidates predicted to possess the MATE domain by both Pfam and CDD were identified as grape MATEs and analyzed in further study. The 18 candidates predicted by either Pfam or CDD to possess the MATE domain are listed in Supplementary Table S2.
The whole grape genome was sequenced and annotated in 2007 (Jaillon et al., 2007; Velasco et al., 2007). Since then, the sequence information was improved from 8X to 12X in 2009 (Adam-Blondon, 2014), and annotation was revised to CRIBI v1 (Jérôme et al., 2012), Genoscope, CRIBI v2 (Vitulo et al., 2014) and Vcost v3 (Canaguier et al., 2017). Different annotations were assigned different gene IDs, and they were not unified. This has led to confusion in grape research. Therefore, the corresponding gene IDs in the different annotations were linked in this study.
A BLAST search of the 58 MATE genes identified in CRIBI v2 to the genes in the three annotations (CRIBI v1, Genoscope, and Vcost v3) was performed. Most genes showed 1:1 correspondence; however, some genes corresponded to 2. These genes are discussed below.
Two genes (VIT_217s0000g02990 and VIT_217s0000g03000) in CRIBI v2 corresponded to one gene (GSVIVT01008369001) in Genoscope (Table 1), although these genes had a 1:1 correspondence in CRIBI v1 and Vcost v3. These suggest that the annotation of Genoscope may be miss annotation.
VIT_211s0052g01560 in CRIBI v2 corresponded to two genes (VIT_11s0052g01570 and VIT_11s0052g01560) in CRIBI v1, two genes (GSVIVT01029131001 and GSVIVT01029132001) in Genoscope, and two genes (Vitvi11g01693 and Vitvi11g01692) in Vcost v3. In the CRIBI v2 annotation, for VIT_211s0052g01560, eight splicing variants (VIT_211s0052g01560.1–8) were suggested, and VIT_211s0052g01560.1, 3, 4, and VIT_211s0052g01560.2, 5, 6, 7, and 8 were derived from different genome positions (Supplementary Figure S1). VIT_211s0052g01560.1, 3, and 4 correspond to VIT_11s0052g0157 in CRIBI v1, GSVIVT01029131001 in Genoscope, and Vitvi11g01693 in Vcost v3. In contrast, VIT_211s0052g01560.2, 5, 6, 7, and 8 corresponded to VIT_11s0052g0156 in CRIBI v1, GSVIVT01029132001 in Genoscope, and Vitvi11g01692 in Vcost v3, respectively (Supplementary Table S1). From these results, we concluded that the CRIBI v2 annotation VIT_211s0052g01560 contains two MATE genes, and 59 genes encoding MATEs are present in the grape genome.
For the 59 identified grape MATEs, information was collected based on the 12X.0 genome assembly and CRIBI v2.1 annotation. The number of amino acid residues, molecular weight, isoelectric point, and the number of transmembrane domains of MATE proteins and splicing variants are summarized in Table 1. The predicted subcellular localizations are listed in Supplementary Table S1. The number of amino acids was 262–689, molecular weights were 28,382–75,281, isoelectric points were 5–10, and the number of transmembrane domains was 3–12.
Phylogenetic Analysis and Nomenclature
A phylogenetic tree of the amino acid sequences of 59 grape MATEs, 56 Arabidopsis MATEs, and the analyzed MATEs of various plants was generated (Figure 1). The tree revealed four groups of grape MATEs: 14 grape MATEs in Group 1, 28 in Group 2, 7 in Group 3, and 10 in Group 4. The 59 grape MATEs were named according to the gene ID in each group: VvMATE1-14 in Group 1, VvMATE15-42 in Group 2, VvMATE43-49 in Group 3, and VvMATE50-59 in Group 4 (Supplementary Table S1).
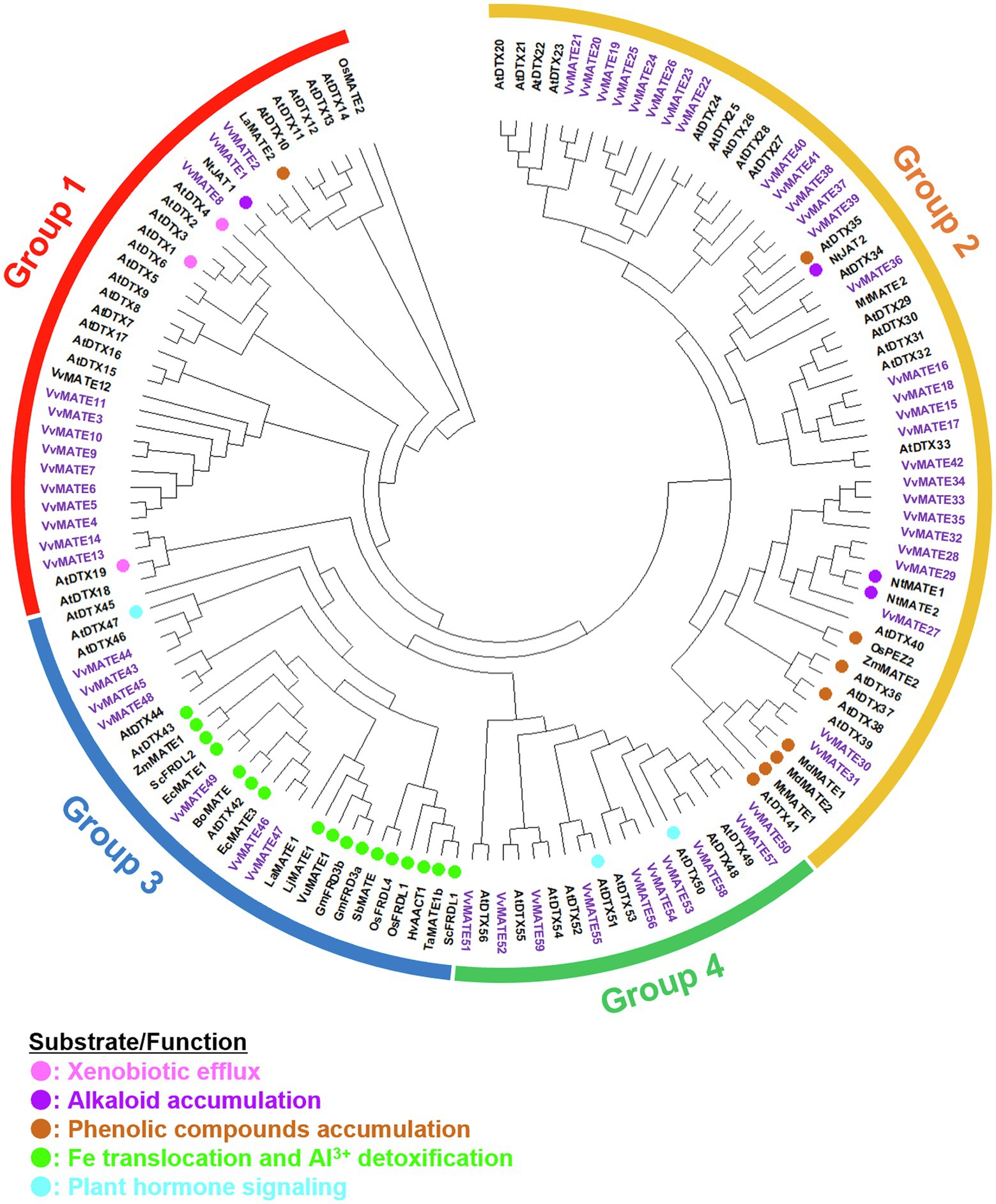
Figure 1. Phylogenetic tree of the MATEs of grape and those of other plant species reported previously. Amino acid sequences of the MATEs of grape and those of other plant species (Li et al., 2002; Takanashi et al., 2014; Biala-Leonhard et al., 2021) were aligned using the ClustalW program. The phylogenetic tree was created by MEGAX using the neighbor-joining model. The grape MATEs are indicated with purple and the MATEs of other plant species are indicated with circle (different colors show different physiological functions, Takanashi et al., 2014).
Gene Structure and Protein Motif
Exon-intron structures and protein motifs were shown with the phylogenetic tree generated by amino acid sequence homology (Figure 2A). The gene structures of grape MATEs were schematized in Figure 2C. The exon–intron structures were conserved in each group. The MATEs in group 1 had 5–8 exons and tended to have a long intron between the first and second exons. The MATEs in Group 2 have 7–9 exons, with long introns between the first and second exons and between the second and third exons. The MATEs in Group 3 had a larger number of exons than the other groups, that is, 10–13 exons. In contrast to Group 3, the MATEs in Group 4 have smaller exon numbers than the other groups, that is, 1–3 exons.

Figure 2. Conserved protein motifs and gene structures of the grape MATEs. (A) The neighbor-joining tree of Grape MATEs. (B) Protein motifs of the grape MATEs. Conserved motifs were identified by the MEME web server. Different motifs are represented by different colored boxes. (C) Exon–intron structures of the grape MATEs. Exons are shown as green boxes, introns are shown as black lines and untranslated regions (UTRs) are shown as blue boxes.
Protein motifs of grape MATEs were analyzed and schematized, as shown in Figure 2B. Twelve motifs were identified in grape MATEs. Generally, the motif patterns in Groups 1, 2, and 4 were similar. The motif patterns of Group 3 were different from those of the other groups, suggesting that the evolution and function of Group 3 were different from those of the other groups.
Transport Substrate and Physiological Function
As for plant species, such as rice (Wang et al., 2016a) and flax (Ali et al., 2021), the grape MATEs were classified into four groups. Previous reports (Wang et al., 2016a) suggested that group members share characteristics and transport similar substrates.
Group 1 included AtDTX19, AtDTX1, and AtDTX6 in Arabidopsis, NtJAT1 in tobacco, and LaMATE2 in white lupin (Figure 1). AtDTX19 transports tetramethylammonium and protects the roots from toxic compounds in the soil (Diener et al., 2001). AtDTX1 transports alkaloids, antibiotics, and toxic compounds (Li et al., 2002). AtDTX6 transports paraquat and relates to paraquat tolerance (Xia et al., 2021). NtJAT1 transports alkaloids, such as nicotine (Morita et al., 2009). LaMATE2 is responsible for exporting isoflavonoids, such as genistein, from the roots to recruit Rhizobia (Biala-Leonhard et al., 2021). These results suggested that grape MATEs in Group 1 are involved in the transport of toxic compounds, alkaloids, and flavonoids. VvMATE1 and VvMATE2 have high homology with LaMATE2, suggesting that VvMATE1 and VvMATE2 have the potential to transport flavonoids.
Group 2 included AtDTX41, AtDTX35, AtDTX33, and AtDTX25 of Arabidopsis and NtJAT2, NtMATE1, and NtMATE2 of tobacco (Figure 1). AtDTX41 is responsible for accumulating proanthocyanidin precursors in vacuoles and for anthocyanin transport (Marinova et al., 2007). AtDTX35 is involved in flavonoid transport (Thompson et al., 2010), and also reported to be a chloride channel (Zhang et al., 2017). AtDTX33 also reported to be a chloride channel (Zhang et al., 2017). AtDTX25 transports ascorbate to vacuole and relates to Fe transport to seeds (Hoang et al., 2021). NtJAT2, NtMATE1, and NtMATE2 are involved in the transport of alkaloids, such as nicotine (Morita et al., 2009; Shoji et al., 2009; Shitan et al., 2014). Based on these reports, the grape MATEs in Group 2 are considered to transport flavonoids and alkaloids. The possibility that the grape MATEs in Group 2 were involved in the transport of polyphenolic compounds will be discussed later.
Group 3 included AtDTX43 and AtDTX43 in Arabidopsis and ZmMATE1 in maize (Figure 1). AtDTX43 contributes to iron and zinc homeostasis by transporting citric acid (Pineau et al., 2012). ZmMATE1 and AtDTX42 are responsible for the aluminum detoxification by transporting citrate (Maron et al., 2010; Wang et al., 2016b). These results suggested that the grape MATEs in Group 3 were responsible for organic acid transport.
Group 4 included AtDTX50 and AtDTX51 in Arabidopsis (Figure 1). AtDTX50 is responsible for the efflux of abscisic acid (ABA; Zhang et al., 2014). AtDTX51 has been reported to contribute to auxin and salicylic acid (SA) signaling (Sun et al., 2011; Li et al., 2014). These results suggested that some grape MATEs in Group 4 function in the transport or signaling of phytohormones, including ABA, auxin, and SA, and play an important role in the grapes’ growth and defense response.
In silico Gene Expression Analysis
To understand the functions of MATEs in the growth and berry development of grapes, the gene expression in various organs, tissues, and growth stages of berries was profiled. Microarray data were obtained from the Grape eFP Browser, and a heat map was created (Figure 3). Although the genes of most grape MATEs were uniformly expressed in various organs, tissues, and growth stages of berries, some genes were expressed in specific organs, tissues, or growth stages of berries.
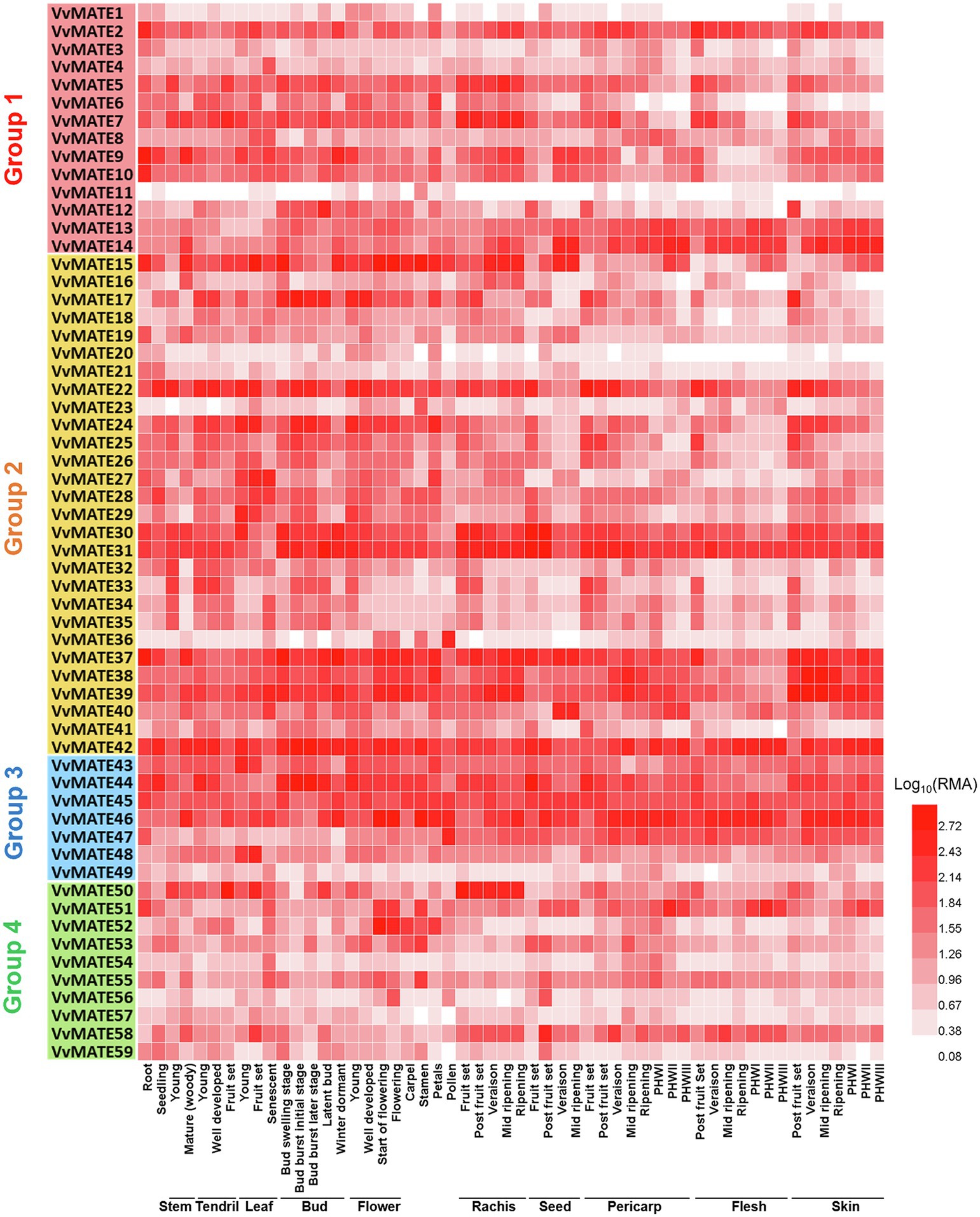
Figure 3. Gene expression profiles of the grape MATEs in various organs, tissues, and developmental stages. The gene expression data of Vitis vinfera “Corvina” (GEO Accession: GSE36128) were normalized using logarithm with the base of 10. The heat maps were created by HemI (Deng et al., 2014). PHWI: post-harvest withering I, PHWII: post-harvest withering II, PHWIII: berry flesh post-harvest withering III.
VvMATE31, VvMATE42, and VvMATE46 showed extremely high expression in all organs, tissues, and growth stages of berries, suggesting that they function as the major MATEs in grapes. VvMATE22 was highly expressed in seed and berry tissues at early developmental stages, and its expression decreased with development, suggesting that VvMATE22 function is important in young grape berries. VvMATE36 was highly expressed in pollen, and VvMATE50 expression was higher in the rachis than in other organs, although their expression was not specific.
Discussion
Grapes are important fruits not only for eating but also for wine production. Polyphenolic compounds, including anthocyanins, catechins, and resveratrol, are important for the berries’ skin color and astringency and are functional compounds for human health. Therefore, grape berry content has a significant effect on grape quality.
As mentioned above, grape MATEs belonging to Group 2 may potentially transport polyphenolic compounds. Indeed, VvMATE1 and VvMATE2 (VvMATE31 and VvMATE30 in this study) have been reported to transport proanthocyanidins (Perez-Diaz et al., 2014), and VvAM1 and VvAM3 (VvMATE39 and VvMATE37 in this study) have been reported to transport anthocyanins, that is, cyanidin 3-glycosides (Gomez et al., 2009). Uncharacterized VvMATE38 in the same clade as VvMATE37 and VvMATE39 (Figure 4) had the potential to transport anthocyanins. The gene expression of VvMATE38 increased dramatically at the veraison stage, whereas those of VvMATE37 and VvMATE39 did not (Supplementary Figure S2A). These results suggested that VvMATE38 functions as an anthocyanin transporter for grape berry skin coloration.
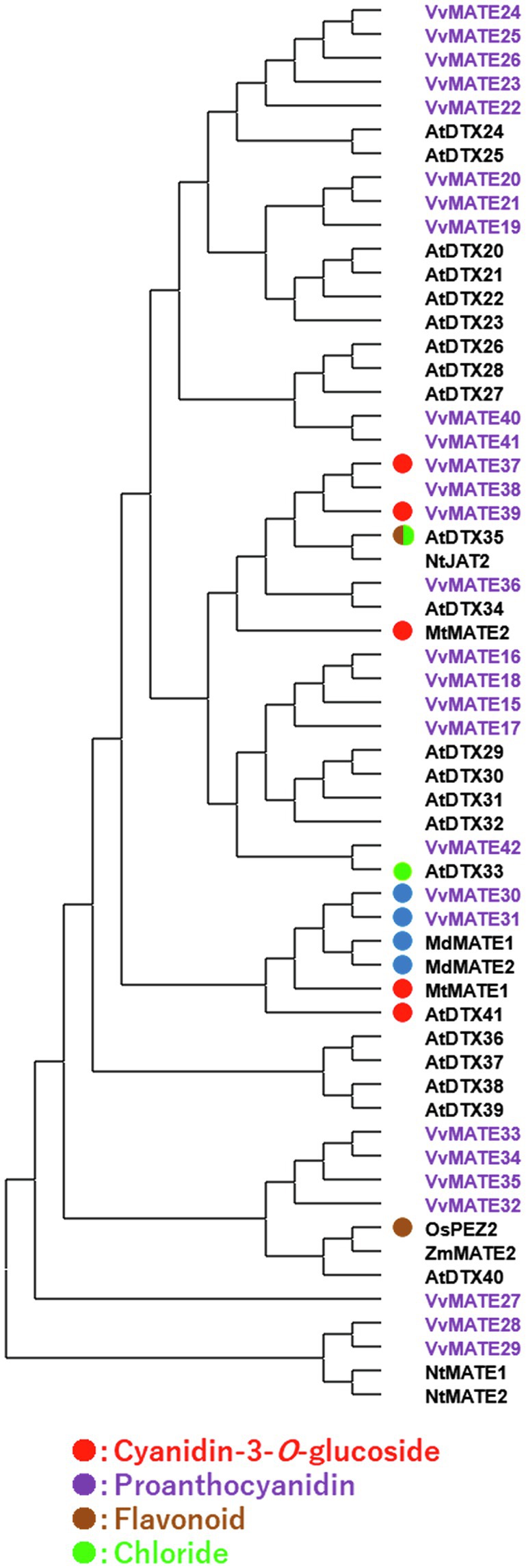
Figure 4. Phylogenetic tree of the group 2 MATEs of grape, Arabidopsis, and those of other plant species reported previously. Amino acid sequences of the MATEs of grape and those of other plant species (Li et al., 2002; Takanashi et al., 2014) were aligned using the ClustalW program. The phylogenetic tree was created by MEGAX using the neighbor-joining model. The group2 of grape MATEs are indicated with purple. Substrates of the MATEs with circles have been reported previously (different colors show different substrate, Takanashi et al., 2014; Zhang et al., 2017).
VvMATE3 and VvMATE1 (VvMATE30 and VvMATE 31 in this study) have been reported to transport proanthocyanidins. The gene expression levels of VvMATE3 and VvMATE1 in seeds, where proanthocyanidin accumulation occurs, were high before veraison and decreased rapidly at the veraison stage, whereas those in berry skin did not change before and after veraison (Perez-Diaz et al., 2014). These trends were consistent with the gene expression of VvMATE30 and VvMATE 31 in the in silico gene expression analysis shown in Supplementary Figure S2B. Based on these results, VvMATE30 and VvMATE31 were considered to play important roles in the accumulation of proanthocyanidins in grape seeds.
Aluminum ions (Al3+) in acidic soils, which account for approximately 50% of the world’s cultivated land, are toxic to most plants. In contrast, iron ions are essential for plants, and their homeostasis is important for various enzymatic reactions, including respiration and electron transfer systems in photosynthesis (Briat et al., 2015). Al3+ and iron ions are chelated by organic acids, such as citric acid and malic acid, produced by plants. Chelation is important for Al3+detoxification and Fe ion facilitation (Kochian et al., 2015). As mentioned above, the grape MATEs in Group 3 have a potential to detoxify Al3+ and promote the translocation of iron ions via organic acid transport. Protein motif analysis showed that the grape MATEs in Group 3 had a unique structural feature (Figure 2B). This structural feature has also been observed in soybeans (Liu et al., 2016), poplars (Li et al., 2017) and flax (Ali et al., 2021). This suggested that the structural features of the MATEs in Group 3 are important for the transport of organic acids.
In the same clade, VvMATE 46, VvMATE 47, and VvMATE 49 were present with the MATEs of other plants, which are related to Al3+ detoxification or iron ion translocation via citrate transport (Figure 5). VvMATE46, VvMATE47, and VvMATE49 were expressed in the whole plant body, including roots (Figure 3). These results suggested that VvMATE46, VvMATE47, and VvMATE49 are involved in Al3+ detoxification in roots and iron ion translocation in the plant body. The gene expression of VvMATE46, VvMATE47, and VvMATE49 were detected in berries (Figure 3), suggesting that they have a potential to transport organic acids and play a role in organic acid accumulation in grape berries.
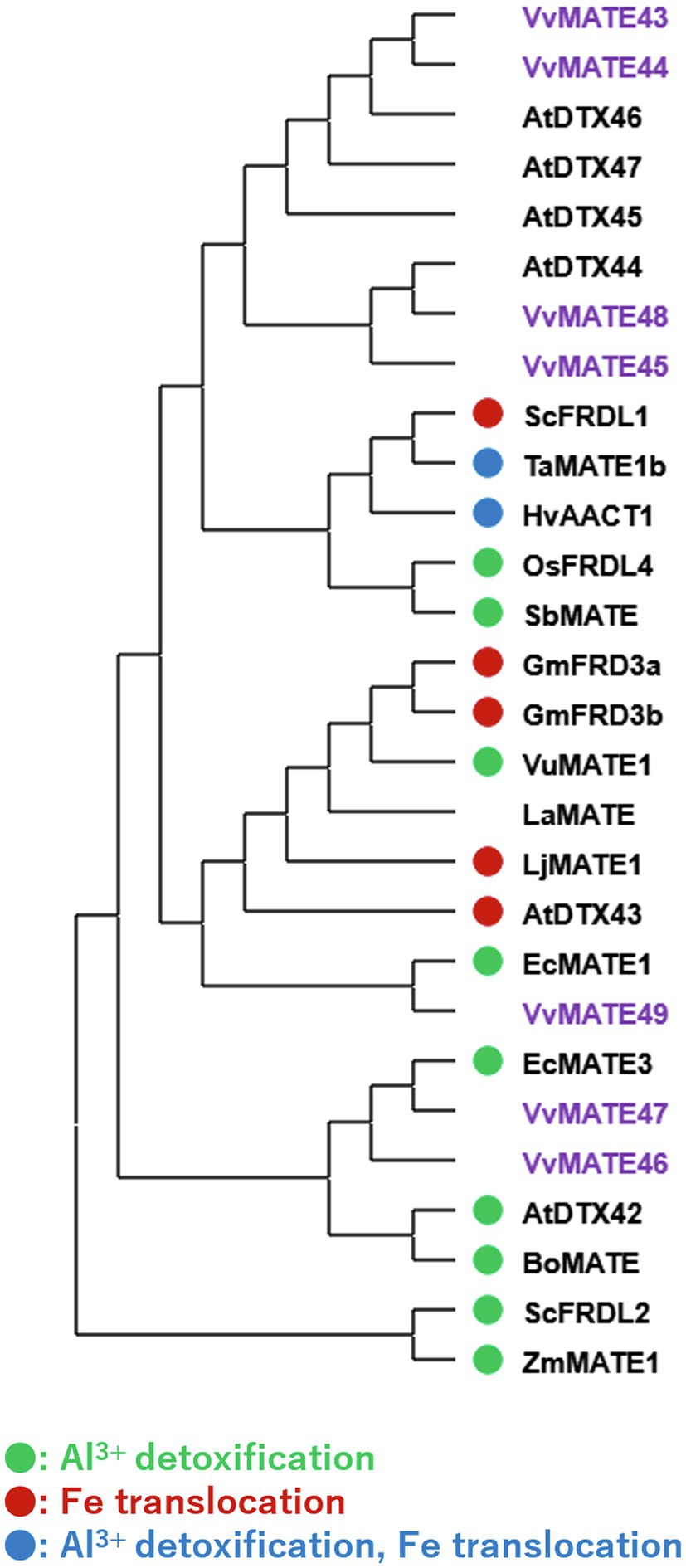
Figure 5. Phylogenetic tree of the group 3 MATEs of grape, Arabidopsis, and those of other plant species reported previously. Amino acid sequences of the MATEs of grape and those of other plant species (Li et al., 2002; Takanashi et al., 2014) were aligned using the ClustalW program. The phylogenetic tree was created by MEGAX using the neighbor-joining model. The group3 of grape MATEs are indicated with purple. MATEs with circle are reported their substrate previously (different colors show different substrates, Takanashi et al., 2014).
Conclusion
This study performed a genome-wide analysis of grape MATEs and identified 59 genes in the grape MATE family. We collected and listed useful information, such as gene IDs, gene structures, protein sizes, motifs, predicted subcellular localization, and gene expression profiles. Phylogenetic analysis revealed the presence of four groups in the grape MATE family, and the transport substrates were estimated for each group. Physiological functions of grape MATEs were discussed together with the estimated transport substrates and gene expression profiles. From this discussion, the uncharacterized MATE, VvMATE38, was suggested as an anthocyanin transporter to color grape berry skin. Different genome annotations assigned different gene IDs for the grape genome, which led to confusion in grape research. In this study, the gene IDs by the different genome annotations were corresponded and the confusion was eliminated. The information provided in this study is essential for research on grape MATEs and will be useful in future studies.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
KS conceived and managed the research project. MW performed analyses. SO and SM provided scientific suggestions. MW and KS wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (24380019, 18H05361, 18H03950, 21H02184, and 21K19111) and by the research program on development of innovative technology grants (28001A and 28001AB) from the Project of the Bio-oriented Technology Research Advancement Institution (BRAIN).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.892638/full#supplementary-material
Footnotes
1. ^https://phytozome-next.jgi.doe.gov/
3. ^http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi/
4. ^http://www.expasy.org/tools/protparam.html
5. ^http://smart.embl-heidelberg.de/smart/batch.pl
6. ^https://plants.ensembl.org/Vitis_vinifera/Tools/Blast
7. ^https://urgi.versailles.inra.fr/Species/Vitis/Annotations
8. ^https://urgi.versailles.inra.fr/Species/Vitis/Annotations
9. ^https://www.genome.jp/tools-bin/clustalw
11. ^https://meme-suite.org/meme/tools/meme
References
Adam-Blondon, A. F. (2014). Grapevine genome update and beyond. Int. Con. Grapevine Breed. Gene. 1046, 311–318. doi: 10.17660/ActaHortic.2014.1046.42
Ali, E., Saand, M. A., Khan, A. R., Shah, J. M., Feng, S. M., Ming, C., et al. (2021). Genome-wide identification and expression analysis of detoxification efflux carriers (DTX) genes family under abiotic stresses in flax. Physiol. Plant. 171, 483–501. doi: 10.1111/ppl.13105
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Biala-Leonhard, W., Zanin, L., Gottardi, S., Francisco, R. D., Venuti, S., Valentinuzzi, F., et al. (2021). Identification of an isoflavonoid transporter required for the nodule establishment of the Rhizobium-Fabaceae symbiotic interaction. Front. Plant Sci. 12:758213. doi: 10.3389/fpls.2021.758213
Briat, J. F., Dubos, C., and Gaymard, F. (2015). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20, 33–40. doi: 10.1016/j.tplants.2014.07.005
Canaguier, A., Grimplet, J., Di Gaspero, G., Scalabrin, S., Duchene, E., Choisne, N., et al. (2017). A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genomics Data 14, 56–62. doi: 10.1016/j.gdata.2017.09.002
Dastmalchi, M., Chang, L. M., Chen, R. J., Yu, L. S., Chen, X., Hagel, J. M., et al. (2019). Purine permease-type benzylisoquinoline alkaloid transporters in opium poppy. Plant Physiol. 181, 916–933. doi: 10.1104/pp.19.00565
Deng, W. K., Wang, Y. B., Liu, Z. X., Cheng, H., and Xue, Y. (2014). HemI: A toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
Diener, A. C., Gaxiola, R. A., and Fink, G. R. (2001). Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13, 1625–1638. doi: 10.1105/tpc.010035
Dos Santos, A. L., Chaves-Silva, S., Yang, L. N., Maia, L. G. S., Chalfun, A., Sinharoy, S., et al. (2017). Global analysis of the MATE gene family of metabolite transporters in tomato. BMC Plant Biol. 17:185. doi: 10.1186/s12870-017-1115-2
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. doi: 10.1093/nar/gky995
Fasoli, M., Dal Santo, S., Zenoni, S., Tornielli, G. B., Farina, L., Zamboni, A., et al. (2012). The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 24, 3489–3505. doi: 10.1105/tpc.112.100230
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Marc, R. W., Appel, R. D., et al. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook. ed. J. M. Walker (United States: Humana Press), 571–607.
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournier-Level, A., Verries, C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.1104/pp.109.135624
Goodstein, D. M., Shu, S. Q., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Hoang, M. T. T., Almeida, D., Chay, S., Alcon, C., Corratge-Faillie, C., Curie, C., et al. (2021). AtDTX25, a member of the multidrug and toxic compound extrusion family, is a vacuolar ascorbate transporter that controls intracellular iron cycling in Arabidopsis. New Phytol. 231, 1956–1967. doi: 10.1111/nph.17526
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hvorup, R. N., Winnen, B., Chang, A. B., Jiang, Y., Zhou, X. F., and Saier, M. H. (2003). The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) exporter superfamily. Eur. J. Biochem. 270, 799–813. doi: 10.1046/j.1432-1033.2003.03418.x
Ishihara, T., Sekine, K. T., Hase, S., Kanayama, Y., Seo, S., Ohashi, Y., et al. (2008). Overexpression of the Arabidopsis thaliana EDS5 gene enhances resistance to viruses. Plant Biol. 10, 451–461. doi: 10.1111/j.1438-8677.2008.00050.x
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jérôme, G., Van Hemert, J., Carbonell-Bejerano, P., Díaz-Riquelme, J., Dickerson, J., Fennell, A., et al. (2012). Comparative analysis of grapevine whole-genome gene predictions, functional annotation, categorization and integration of the predicted gene sequences. BMC. Res. Notes 5:213. doi: 10.1186/1756-0500-5-213
Kochian, L. V., Pineros, M. A., Liu, J. P., and Magalhaes, J. V. (2015). Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598. doi: 10.1146/annurev-arplant-043014-114822
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, Y. Q., He, H., and He, L. F. (2019). Genome-wide analysis of the MATE gene family in potato. Mol. Biol. Rep. 46, 403–414. doi: 10.1007/s11033-018-4487-y
Li, L. G., He, Z. Y., Pandey, G. K., Tsuchiya, T., and Luan, S. (2002). Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368. doi: 10.1074/jbc.M108777200
Li, R. X., Li, J. R., Li, S. B., Qin, G. J., Novak, O., Pencik, A., et al. (2014). ADP1 affects plant architecture by regulating local auxin biosynthesis. PLoS Genet. 10:e1003954. doi: 10.1371/journal.pgen.1003954
Li, N. N., Meng, H. J., Xing, H. T., Liang, L., Zhao, X., and Luo, K. M. (2017). Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus. J. Exp. Bot. 68, 5669–5683. doi: 10.1093/jxb/erx370
Liu, J. G., Li, Y., Wang, W., and Gai, J. Y. (2016). Genome-wide analysis of MATE transporters and expression patterns of a subgroup of MATE genes in response to aluminum toxicity in soybean. BMC Genomics 17:223. doi: 10.1186/s12864-016-2559-8
Liu, J. P., Magalhaes, J. V., Shaff, J., and Kochian, L. V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57, 389–399. doi: 10.1111/j.1365-313X.2008.03696.x
Lu, S. N., Wang, J. Y., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Marinova, K., Pourcel, L., Weder, B., Schwarz, M., Barron, D., Routaboul, J. M., et al. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19, 2023–2038. doi: 10.1093/nar/gkz991
Maron, L. G., Pineros, M. A., Guimaraes, C. T., Magalhaes, J. V., Pleiman, J. K., Mao, C. Z., et al. (2010). Two functionally distinct members of the MATE (multi-drug and toxic compound extrusion) family of transporters potentially underlie two major aluminum tolerance QTLs in maize. Plant J. 61, 728–740. doi: 10.1111/j.1365-313X.2009.04103.x
Morita, M., Shitan, N., Sawada, K., Van Montagu, M. C. E., Inze, D., Rischer, H., et al. (2009). Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proc. Natl. Acad. Sci. U. S. A. 106, 2447–2452. doi: 10.1073/pnas.0812512106
Nino-Gonzalez, M., Novo-Uzal, E., Richardson, D. N., Barros, P. M., and Duque, P. (2019). More transporters, more substrates: The Arabidopsis major facilitatorsuperfamily revisited. Mol. Plant 12, 1182–1202. doi: 10.1016/j.molp.2019.07.003
Nogia, P., and Pati, P. K. (2021). Plant secondary metabolite transporters: diversity, functionality, and their modulation. Front. Plant Sci. 12:758202. doi: 10.3389/fpls.2021.758202
Omote, H., Hiasa, M., Matsumoto, T., Otsuka, M., and Moriyama, Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 27, 587–593. doi: 10.1016/j.tips.2006.09.001
Perez-Diaz, R., Ryngajllo, M., Perez-Diaz, J., Pena-Cortes, H., Casaretto, J. A., Gonzalez-Villanueva, E., et al. (2014). VvMATE1 and VvMATE2 encode putative proanthocyanidin transporters expressed during berry development in Vitis vinifera L. Plant Cell Rep. 33, 1147–1159. doi: 10.1007/s00299-014-1604-9
Pineau, C., Loubet, S., Lefoulon, C., Chalies, C., Fizames, C., Lacombe, B., et al. (2012). Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana. PLoS Genet. 8:e1003120. doi: 10.1371/journal.pgen.1003120
Rea, P. A. (2007). Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 58, 347–375. doi: 10.1146/annurev.arplant.57.032905.105406
Shitan, N., Minami, S., Morita, M., Hayashida, M., Ito, S., Takanashi, K., et al. (2014). Involvement of the leaf-specific multidrug and toxic compound extrusion (MATE) transporter Nt-JAT2 in vacuolar sequestration of nicotine in Nicotiana tabacum. PLoS One 9:e108789. doi: 10.1371/journal.pone.0108789
Shoji, T. (2014). ATP-binding cassette and multidrug and toxic compound extrusion transporters in plants: A common theme among diverse detoxification mechanisms. Int. Rev. Cell Mol. Biol. 309, 303–346. doi: 10.1016/B978-0-12-800255-1.00006-5
Shoji, T., Inai, K., Yazaki, Y., Sato, Y., Takase, H., Shitan, N., et al. (2009). Multidrug and toxic compound extrusion-type transporters implicated in vacuolar sequestration of nicotine in tobacco roots. Plant Physiol. 149, 708–718. doi: 10.1104/pp.108.132811
Sun, X. L., Gilroy, E. M., Chini, A., Nurmberg, P. L., Hein, I., Lacomme, C., et al. (2011). ADS1 encodes a MATE-transporter that negatively regulates plant disease resistance. New Phytol. 192, 471–482. doi: 10.1111/j.1469-8137.2011.03820.x
Takanashi, K., Shitan, N., and Yazaki, K. (2014). The multidrug and toxic compound extrusion (MATE) family in plants. Plant Biotechnol. 31, 417–430. doi: 10.5511/plantbiotechnology.14.0904a
Thompson, E. P., Wilkins, C., Demidchik, V., Davies, J. M., and Glover, B. J. (2010). An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J. Exp. Bot. 61, 439–451. doi: 10.1093/jxb/erp312
Tiwari, M., Sharma, D., Singh, M., Tripathi, R. D., and Trivedi, P. K. (2014). Expression of OsMATE1 and OsMATE2 alters development, stress responses and pathogen susceptibility in Arabidopsis. Sci. Rep. 4:3964. doi: 10.1038/srep03964
Velasco, R., Zharkikh, A., Troggio, M., Cartwright, D. A., Cestaro, A., Pruss, D., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2:e1326. doi: 10.1371/journal.pone.0001326
Vitulo, N., Forcato, C., Carpinelli, E. C., Telatin, A., Campagna, D., D'Angelo, M., et al. (2014). A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 14:99. doi: 10.1186/1471-2229-14-99
Wang, L. H., Bei, X. J., Gao, J. S., Li, Y. X., Yan, Y. M., and Hu, Y. K. (2016a). The similar and different evolutionary trends of MATE family occurred between rice and Arabidopsis thaliana. BMC Plant Biol. 16:207. doi: 10.1186/s12870-016-0895-0
Wang, J. J., Hou, Q. Q., Li, P. H., Yang, L., Sun, X. C., Benedito, V. A., et al. (2017). Diverse functions of multidrug and toxin extrusion (MATE) transporters in citric acid efflux and metal homeostasis in Medicago truncatula. Plant J. 90, 79–95. doi: 10.1111/tpj.13471
Wang, Z. Y., Qian, C. Z., Guo, X. C., Liu, E. L., Mao, K. L., Mu, C. J., et al. (2016b). ELS1, a novel MATE transporter related to leaf senescence and iron homeostasis in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 476, 319–325. doi: 10.1016/j.bbrc.2016.05.121
Wen, J., Li, P. F., Ran, F., Guo, P. C., Zhu, J. T., Yang, J., et al. (2020). Genome-wide characterization, expression analyses, and functional prediction of the NPF family in Brassica napus. BMC Genomics 21:871. doi: 10.1186/s12864-020-07274-7
Xia, J. Q., Nazish, T., Javaid, A., Ali, M., Liu, Q. Q., Wang, L., et al. (2021). A gain-of-function mutation of the MATE family transporter DTX6 confers paraquat resistance in Arabidopsis. Mol. Plant 14, 2126–2133. doi: 10.1016/j.molp.2021.09.004
Zhang, H. W., Zhao, F. G., Tang, R. J., Yu, Y. X., Song, J. L., Wang, Y., et al. (2017). Two tonoplast MATE proteins function as turgor-regulating chloride channels in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, E2036–E2045. doi: 10.1073/pnas.1616203114
Keywords: genome-wide analysis, grape, multidrug and toxic compound extraction transporter, MATE, secondary metabolite transporter, Vitis vinifera
Citation: Watanabe M, Otagaki S, Matsumoto S and Shiratake K (2022) Genome-Wide Analysis of Multidrug and Toxic Compound Extruction Transporters in Grape. Front. Plant Sci. 13:892638. doi: 10.3389/fpls.2022.892638
Edited by:
Markus Geisler, Université de Fribourg, SwitzerlandReviewed by:
Linfeng Sun, University of Science and Technology of China, ChinaHussam Hassan Nour-Eldin, University of Copenhagen, Denmark
Copyright © 2022 Watanabe, Otagaki, Matsumoto and Shiratake. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katsuhiro Shiratake, c2hpcmFAYWdyLm5hZ295YS11LmFjLmpw
 Megumi Watanabe
Megumi Watanabe Shungo Otagaki
Shungo Otagaki Shogo Matsumoto
Shogo Matsumoto Katsuhiro Shiratake
Katsuhiro Shiratake