- College of Biology, Hunan University, Changsha, China
The CCHC-type zinc finger proteins (CCHC-ZFPs) play versatile roles in plant growth, development and adaptation to the environment. However, little is known about functions of CCHC-ZFP gene family memebers in Triticum aestivum. In the present study, we identified a total of 50 TaCCHC-ZFP genes from the 21 wheat chromosomes, which were phylogenetically classified into eight groups based on their specific motifs and gene structures. The 43 segmentally duplicated TaCCHC-ZFP genes were retrieved, which formed 36 segmental duplication gene pairs. The collinearity analyses among wheat and other eight mono/dicots revealed that no gene pairs were found between wheat and the three dicots. The promoter analyses of the TaCCHC-ZFP genes showed that 636 environmental stress-responsive and phytohormone-responsive cis-elements. The gene ontology enrichment analysis indicated that all the TaCCHC-ZFP genes were annotated under nucleic acid binding and metal ion binding. A total of 91 MicroRNA (miRNA) binding sites were identified in 34 TaCCHC-ZFP genes according to the miRNA target analysis. Based on the public transcriptome data, the 38 TaCCHC-ZFP genes were identified as differentially expressed gene. The expression profiles of 15 TaCCHC-ZFP genes were verified by the quantitative real-time PCR assays, and the results showed that these genes were responsive to drought or heat treatments. Our work systematically investigated the gene structures, evolutionary features, and potential functions of TaCCHC-ZFP genes. It lays a foundation for further research and application of TaCCHC-ZFP genes in genetic improvement of T. aestivum.
Introduction
The CCHC-type zinc finger proteins (CCHC-ZFPs) are one of the largest transcription factors in plants, which play versatile roles in a variety of physiological processes. The CCHC-ZFPs regulate the expression of their target genes by directly or indirectly recognizing and binding the promoters (Takatsuji, 1999; Kiełbowicz-Matuk, 2012). As one type of zinc finger proteins, the CCHC-ZFPs contain at least one CCHC motif, which is also called zinc knuckle, sharing the consensus sequence CX2CX4HX4C (X for any amino acid, numbers for the number of residues, C and H for cysteine and histidine, respectively; Summers, 1991). The CCHC motifs with high affinity for DNA and RNA usually consist of a short helix and two short β-strands joined through a zinc knuckle, which function during transcriptional activation, RNA packaging, DNA recognition, and regulation of apoptosis (Laity et al., 2001; Wang et al., 2021). The first CCHC-ZFP was identified in the murine leukemia virus and Rous avian sarcoma virus, and subsequently in antigen proteins of retroviral nucleocapsids and eukaryotic retrotransposons (Summers, 1991). Comprehensive studies of CCHC-ZFP gene family were carried out in humans (34), Arabidopsis (69), and yeast (7), while the CCHC-ZFPs in wheat have not been reported so far (Aceituno-Valenzuela et al., 2020).
The CCHC-ZFP genes were extensively founded in plant genomes, which played central roles in seed development, and plant growth mediated by phytohormones, such as indole-3-acetic acid (IAA), gibberellins (GA), abscisic acid (ABA), and methyl jasmonate (MeJA). In rice, OsZFP regulates lateral root development via IAA signaling pathways (Cui et al., 2017). AtCSP2 negatively modulates seed germination by adjusting GA and ABA contents (Sasaki et al., 2015). Additionally, a great number of CCHC-ZFP genes also participate in regulating plant tolerance to environmental stress. Overexpressing OsZFP6, a NaCl, H2O2, and NaHCO3 responsive gene, increases the tolerance to H2O2 and NaHCO3 in Arabidopsis (Guan et al., 2014). Similarly, BrCSDP3 is a positive regulator of seed germination and seedling growth during dehydration and salinity treatment (Choi et al., 2015). The transcription of OsRZ1, OsRZ2, and OsRZ3 is upregulated by low temperature treatment, but they show no response to high salinity and drought stress (Kim et al., 2010). In Pak-choi, BcCSP1 plays a key role in responses to cold and ABA treatments (Huang et al., 2016). Besides, some CCHC-ZFP genes also participate in the regulation of plant defense to biotic stress (Amorim et al., 2017). Ectopic expression of wheat TaRZ1 in Arabidopsis confers the transgenic plants enhanced resistance against bacterial (Xu et al., 2015). The up-regulation of AdRSZ21 under MeJA treatment and pathogen infection indicates that CCHC-ZFP genes might take part in plant defense (Kumar and Kirti, 2012).
Bread wheat (Triticum aestivum L., A, B, and D sub-genome) was obtained by natural hybridization between Triticum dicoccoides (A and B sub-genome) and Aegilops tauschii (D sub-genome), which was a valuable material for evolutionary research due to the specificity of heterohexaploid (Ozkan et al., 2001; Petersen et al., 2006). The growth and development of wheat are susceptible to complex and variable environments, leading to the reduction of yield (Fujita et al., 2006). Considering the roles of CCHC-ZFP genes in various biological processes, a comprehensive investigation of TaCCHC-ZFP gene family will contribute to wheat stress resistance breeding and gene function research. In this study, we identified the CCHC-ZFP genes by bioinformatic methods from wheat genome and analyzed the chromosomal location, subcellular localization, phylogenetic relationships, gene structures, proteins interaction network, and expression patterns of them. The promoter cis-elements and the MicroRNA (miRNA) potentially targeting TaCCHC-ZFP genes were predicted to study the transcriptional regulatory network. These works will provide the basis for further analyses and application of CCHC-ZFP genes in wheat.
Materials and Methods
Plant Materials and Abiotic Stress Treatments
Bread wheat cultivar Fielder was used throughout this study. Seeds were placed in Petri dishes with wet filter paper at 4°C for 5 days. Then, the germinated seedlings were cultured in an incubator at 22°C (8 h dark and 16 h light period, about 60–70 μmol⋅m–2⋅s–1, and 50% relative humidity) with half strength Murashige and Skoog liquid medium. Two-week seedlings of Fielder were treated by drought stress [20% (m/V) PEG-6000], heat stress (40°C), or combined drought and heat stress (20% PEG-6000 and 40°C) for 1 or 6 h, respectively, while the seedlings under normal growing conditions (22°C, watered) were used as a control. Leaves were collected at 1 and 6 h after treatments, frozen in liquid nitrogen immediately, and stocked at −80°C for further study. All the experiments were conducted in parallel, and three biological replications were performed for each timepoint.
Data Retrieval and Identification of CCHC Genes
The protein sequences and reference genomes for all the species in this study were available from the Ensemble Plants1. To identify the CCHC-ZFP family members, the Hidden Markov Model (HMM) profile of CCHC conserved motif (PF00098) from Pfam2 was applied to search against all of the protein sequences through the HMMER with the E-value < 1e–4 (Wheeler and Eddy, 2013; Mistry et al., 2021). After cleaning out the redundant sequences, candidate genes were subjected to Simple Modular Architecture Research Tool (SMART)3 to further verify CCHC-ZFP members (Letunic et al., 2021). The theoretical isoelectric point (pI), aliphatic index (AI), molecular weight (MW), instability index, and grand average of hydropathicity (GRAVY) were calculated using the ExPasy site4 (Gasteiger, 2003). The subcellular localization of each CCHC-ZFP was forecast using the Cell-PLoc 2.05 (Chou and Shen, 2010). The secondary structure of TaCCHC-ZFPs was predicted using SOPMA secondary structure prediction6 (Geourjon and Deléage, 1995).
Sequence Analysis and Structural Characterization of the CCHC Proteins in Wheat
Multiple protein sequence alignment of the characterized CCHC-ZFPs was carried out via ClustalX2 (Larkin et al., 2007). Then, depending on the full-length protein sequence alignment, the phylogenetic tree was constructed using MEGA 7.0 with the neighbor-joining method based on Poisson model, 1000 bootstrap replications and pairwise deletion (Kumar et al., 2016). The MEME online program7 was applied to identify the conserved motifs of CCHC-ZFPs in wheat (Bailey et al., 2009). Then, the conserved motif of wheat CCHC was extracted and visualized by WebLogo8 (Crooks et al., 2004). The wheat genome annotation file (GFF3 file) of wheat was retrieved from the Ensemble Plants (see text footnote 1) for analyzing the exon-intron structures of TaCCHC-ZFP genes. Finally, the prepared files were imported into TBtools for visualizing the protein motifs and gene structures (Chen et al., 2020a).
Chromosome Distribution, Collinearity Analysis, and Ka/Ks Analysis
According to the information of chromosome location obtained from the Ensemble Plants, the TaCCHC-ZFP genes were mapped into the wheat chromosome by MapGene2Chrom V29 (Chao et al., 2015). Subsequently, the gene duplication events and synteny of wheat CCHC-ZFP genes were analyzed using MCScanX and DIAMOND with the default parameters, and the figure was displayed by the Circos (Krzywinski et al., 2009; Wang et al., 2012; Buchfink et al., 2015). Additionally, the collinearity relationships and segmental duplication events of TaCCHC-ZFP gene pairs from other species were also performed similarly. The species evolution tree was drawn by using TimeTree online tool10 (Kumar et al., 2017). Then, the TBtools was adopted to calculated Ks (synonymous) and Ka (non-synonymous) of the duplicated gene pairs for further estimating duplication events (Chen et al., 2020a). The time (T) of duplication in millions of years (Mya) was estimated with the formula T = Ks/2λ × 10–6 Mya (λ = 6.5 × 10–9).
Cis-Acting Element Analysis and Gene Ontology Annotation of TaCCHC Family Genes
In order to investigate the cis-acting elements in the promoter of TaCCHC-ZFP genes, the 1.5-kb upstream genomic DNA sequences of the transcription start codon were submitted to the PlantCARE database11 (Lescot et al., 2002). Then, the Gene Structure Display Server (GSDS)12 was adopted to visualized the cis-element distribution (Hu et al., 2015). The gene ontology (GO) analysis of TaCCHC genes was predicted for functional annotation using Omicshare Tools13.
Prediction of Protein Interaction Network and MicroRNA Targets
The wheat CCHC-ZFPs were committed to the STRING database14 to analyze the protein-protein interaction network with high confidence (0.700; Szklarczyk et al., 2019). Then, The Cytoscape was adopted to visualize the interaction network with default parameters (Shannon et al., 2003). To predict the miRNAs targeting TaCCHC-ZFP genes, mature miRNA sequences and TaCCHC-ZFP gene sequences of wheat were submitted to the psRNATarget tool15, filtered at an expectation level ≤ 5.0 (Dai et al., 2018).
Expression Analyses of TaCCHC Genes
The gene expression patterns of TaCCHC under abiotic stresses were available from the expVIP16 (Liu et al., 2015; Ramírez-González et al., 2018). Subsequently, the edgeR package was performed to identified the differentially expressed genes (DEGs) with fold change ≥ 2 and q-value ≤ 0.5 (Robinson et al., 2009). The TBtools was applied to create the gene expression heatmap (Chen et al., 2020a). Finally, EVenn17 was adapted to construct Venn diagrams.
RNA Extraction and Quantitative Real-Time PCR Analyses
The total RNA from wheat leaves was extracted using TRIzol reagent (Vazyme Biotech Co., Ltd). For quantitative real-time PCR (qRT-PCR) analyses, RNA concentration was assessed by the NanoDrop 2000 spectrophotometer (ND-2000, Thermo Fisher Scientific, Inc.). Total RNAs were reverse transcribed with the HiScript II 1st Strand cDNA Synthesis Kit (+gDNA wiper; Vazyme Biotech Co., Ltd). The expression of 15 stress-responsive TaCCHC-ZFP genes were examined by qRT-PCR analyses, while TaRP15 was served as a reference gene. The reaction system was composed of 5 μL of 2 × ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd), 2 μL of template, 0.2 μL of each prime, and 2.6 μL of ddH2O. The reaction was performed as follows: pre-denaturation at 95°C for 30 s (step 1), denaturation at 95°C for 10 s (step 2), primer annealing/extension and collection of fluorescence signal at 60°C for 30 s (step 3). The next 40 cycles started at step 2. Each sample was performed in three biological replications and three technical replications. Subsequently, the data from qRT-PCR analyses was calculated with the 2–ΔΔCT method. Primer sequences used in this study were listed in detail in Supplementary Table 10.
Results
Identification and Characterization of the CCHC Gene Family
In this study, a total of 50 putative CCHC genes in wheat were retrieved based on the HMMER search. After SMART searched, 50 wheat proteins sharing the CCHC conserved motifs were obtained, which were consistent to the predictions. Meanwhile, several important dicotyledonous and monocotyledonous plants were selected for reference analyses. We identified 38 CCHC-ZFP genes in T. dicoccoides, 46 CCHC-ZFP genes in Ae. tauschii, 17 CCHC-ZFP genes in Hordeum vulgare, 26 CCHC-ZFP genes in Oryza sativa, 33 CCHC-ZFP genes in Zea mays, 22 CCHC-ZFP genes in Arabidopsis thaliana, 95 CCHC-ZFP genes in Glycine max, and 67 CCHC-ZFP genes in Solanum tuberosum in the same method (Supplementary Table 1).
Subsequently, physicochemical properties of TaCCHC-ZFPs were analyzed, including the length of proteins, MW, AI, pI, instability index, GRAVY, and the subcellular localization (Supplementary Table 2). Among the 50 TaCCHC-ZFPs, TaCCHC14 is identified to be the smallest protein with 162 residues of amino acids (aa), while TaCCHC31 with 1,149 residues of amino acids is the largest one. The pI ranges from 5.31 (TaCCHC40) to 11.63 (TaCCHC41), and AI fluctuates from 20.06 (TaCCHC25) to 74.88 (TaCCHC5), and instability index varies from 25.67 (TaCCHC22) to 117.51 (TaCCHC41). Besides, the GRAVY values of all TaCCHC-ZFPs are negative, implying that TaCCHC-ZFPs may be hydrophilic proteins. Additionally, the subcellular localization predictions showed that 28 TaCCHC-ZFPs were located both in the cell nucleus and chloroplast, whereas 20 and 2 TaCCHC-ZFPs were only located in the nucleus or chloroplast, respectively. Additionally, all TaCCHC-ZFPs are composed of four secondary structures, including alpha helix (0.62–40.61%), extended strand (5.99–26.53%), beta turn (3.05–24.62%), and random coil (43.32–83.6%), of which random coil accounts for the main part of protein secondary structure (Supplementary Table 3).
Subsequently, we obtained the amino acid sequences of the conserved motif CCHCs using the MEME tool. As shown in Figure 1, the CCHC conserved motif from wheat has the consensus sequence CX2CX4HX4C, which has high affinity to nucleic acids. Except for the completely conserved histidine (H) and cysteine (C) residues in the positions 6, 9, 14, 19, the conserved substituted glycine (G) residue occurs in the positions 10, 13, and hydrophobic or aromatic residues are found in the positions 7, 15 (Figure 1).
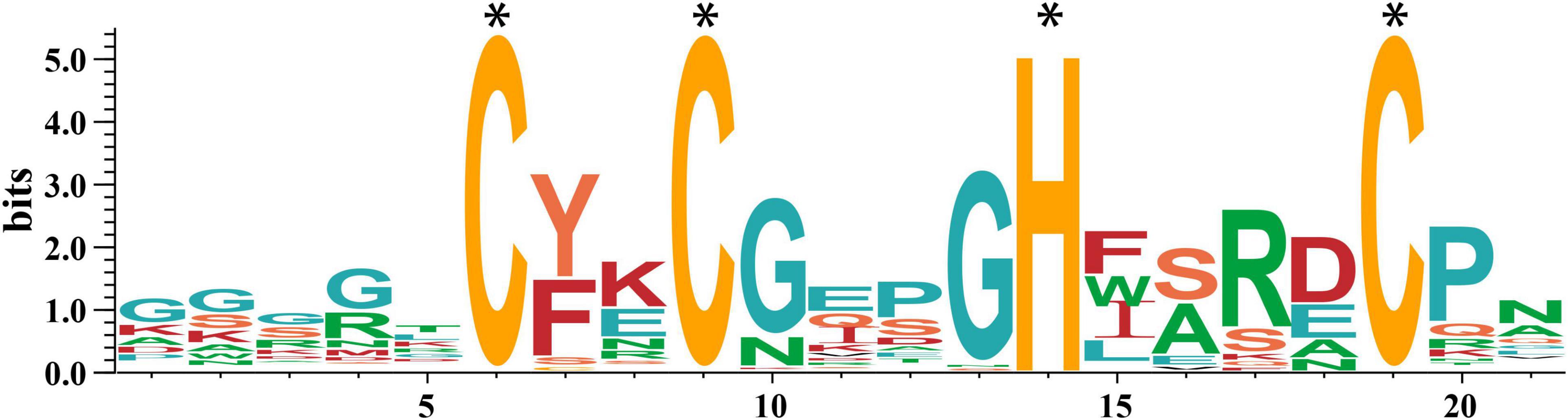
Figure 1. Sequence of the CCHC motifs in wheat. The height of the letter at each location (in bits) represents the conservation of the sequences, and the height of every single letter means the relative frequency of the corresponding amino acid of that position. The * represents the completely conserved residues.
Phylogenetic Tree and Sequence Structure Analysis
To study the evolutionary relationship of the CCHC-ZFP genes, a phylogenetic tree was constructed using the protein sequences of CCHC-ZFPs from both wheat and rice. These CCHC-ZFP genes are classified into nine groups, named as groups I to IX, which are distributed unevenly in each group (Figure 2). Except for the group IX, the others all possess CCHC-ZFP genes from both wheat and rice. The groups I and III both contain the most members of 13, and the group III is also the group with most members of 11 CCHC-ZFP genes in wheat. In addition, the group VIII possesses the fewest members, two from wheat and one from rice. Based on the phylogenetic analysis, TaCCHC-ZFP genes are classified into eight groups (groups I to VIII) for further analyses (Figure 3A).
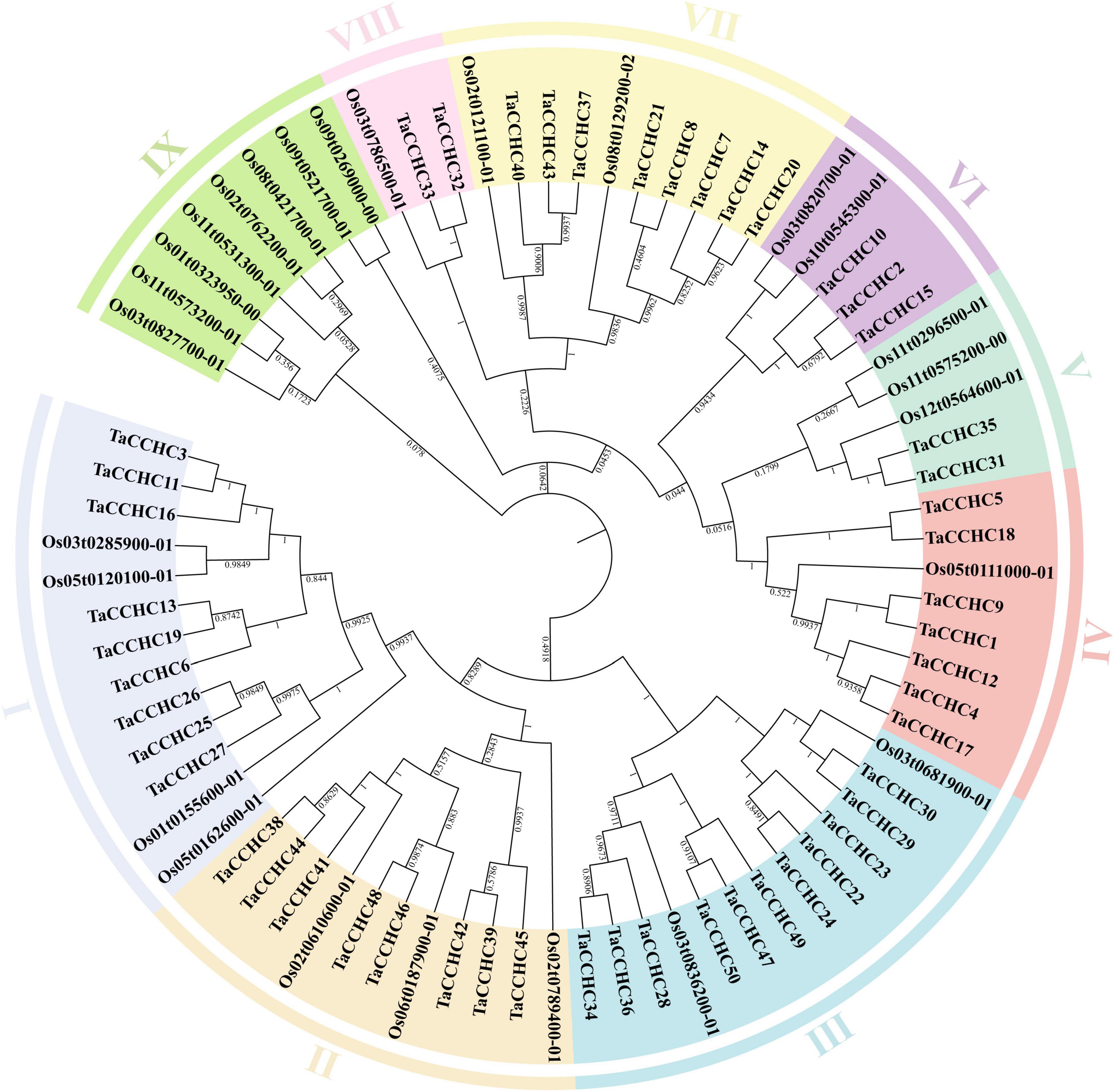
Figure 2. Phylogenetic tree of CCHC-ZFPs in wheat and rice. The protein sequences were aligned using the ClustaX2, and the Neighbor-joining (NJ) phylogenetic tree was built using the MEGA 7.0 based on Poisson model. Different groups are indicated by different colors, respectively.
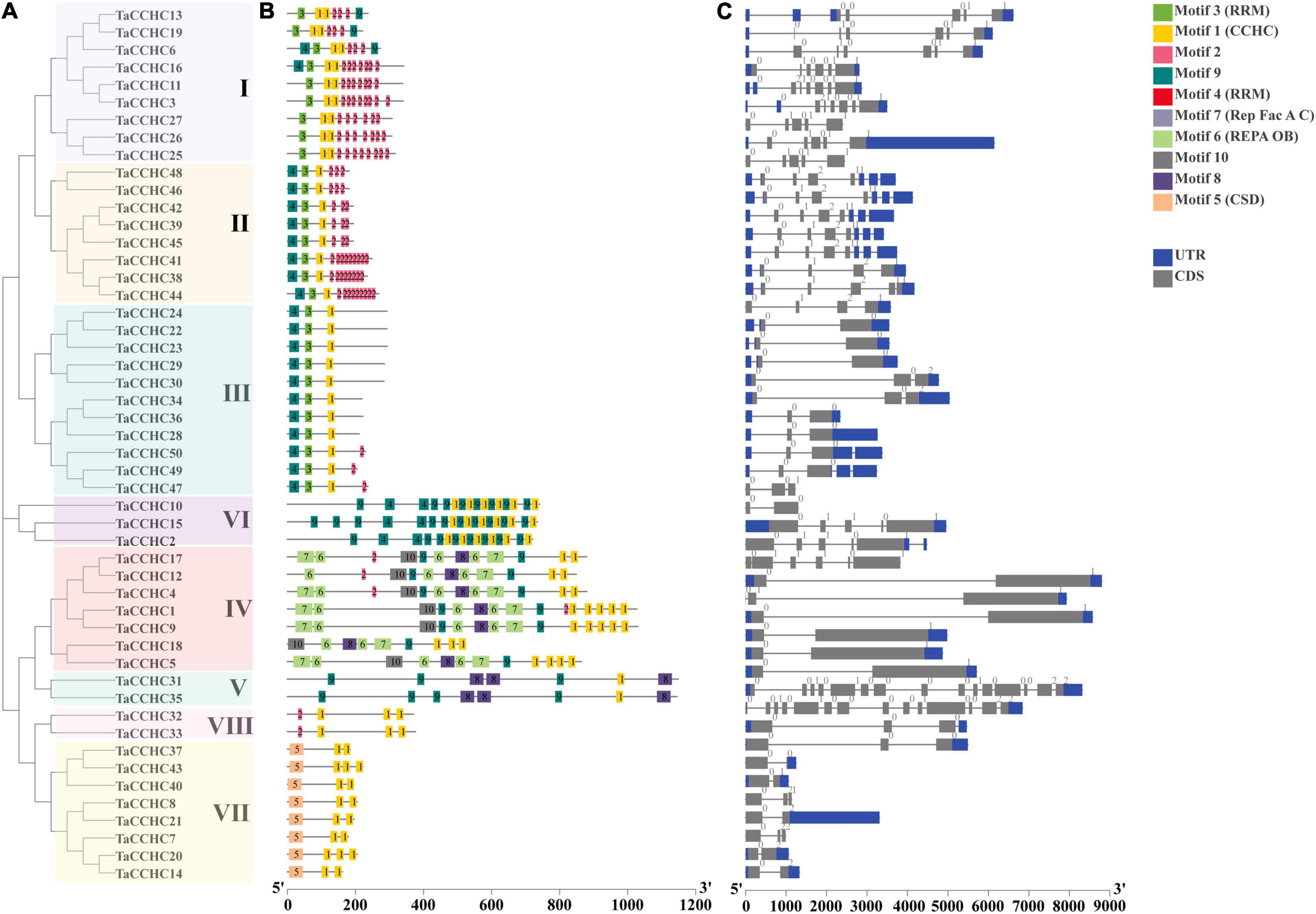
Figure 3. Comparative analyses of the phylogenetic relationships, protein conserved motifs, and gene structures of CCHC-ZFP family in wheat. (A) Phylogenetic tree of 50 TaCCHC-ZFPs was constructed by using MEGA 7.0. Each group was marked by a different color. (B) Motif composition of wheat CCHC-ZFPs. MEME was adopted to identify the conserved motifs of the TaCCHC-ZFPs. The motifs are displayed by different-colored boxes with the corresponding number in the center of the motifs. (C) Gene structures of TaCCHC-ZFP genes. The black lines represent the introns, while the blue and gray boxes represent the untranslated regions (UTRs) and exons, respectively. The numbers represent the phases of corresponding introns.
A schematic diagram displaying the motifs of TaCCHC-ZFPs was constructed with the MEME tool (Bailey et al., 2009). Through the annotations of the Pfam (PF00098) database, we found that the motif 1 was the CCHC domain, and the motif 3 and 4 both were RRM domains, and the motif 5, 6, and 7 were CSD domain, REPA OB domain, and Rep Fac-A C domain, respectively, (Supplementary Table 4; Mistry et al., 2021). As shown in Figure 3B, the motif 1 was extensively distributed in TaCCHC-ZFPs. Moreover, TaCCHC-ZFPs in one group generally tend to have a similar motif composition. For instance, the motif 5 only exists in the group VII, while the motifs 6, 7 and 10 are specific to the group IV. Similarly, the motifs 8 is unique to the group IV and V, and the motif 3 only occurs in the groups I, II, and III. As a result, the motif patterns of TaCCHC-ZFPs in a group are similar, suggesting that the protein structure is conserved within a specific group. The roles of the conserved motifs remain to be elucidated, which may be relevant to specific biological functions.
Additionally, the exon-intron structures of TaCCHC-ZFP genes were investigated to understand the evolution of the TaCCHC-ZFP family. The gene structures of TaCCHC-ZFPs in different groups are changeable in the number of exons (ranging from 2 to 15; Figure 3C). However, the TaCCHC-ZFP genes in the same group usually share similar numbers of exons as expected, suggesting that they are evolutionarily conserved. For instance, all members of the group VII contain two or three exons, while seven TaCCHC-ZFP genes of the group II possess four exons. In contrast, some of the more closely related members were also observed to share similar length of exons. In general, the diverse gene structures of TaCCHC-ZFP genes may be related to the involvement of TaCCHC-ZFP genes in many plant biological processes.
Chromosomal Location and Collinearity Analysis of the TaCCHC Genes
MapGene2Chrom V2 was adopted to create the chromosome map of the TaCCHC-ZFP genes based on the physical location information (Figure 4A; Chao et al., 2015). The TaCCHC-ZFP genes are unevenly spread across wheat chromosomes, with the number of the genes on each chromosome varying from one (2A, 2B, 2D, 3A, 3B, 3D, 4A, 4B, 4D, and 7D) to eight (1A; Figure 4B). Interestingly, we also found that the numbers of the TaCCHC-ZFP genes on each chromosome were not relevant to chromosome size. For instance, the smallest chromosome (6D, 473.6 Mb) encodes three TaCCHC-ZFP genes, while the largest chromosome (3B, 830.8 Mb) contains only one TaCCHC-ZFP gene. The TaCCHC-ZFP genes spread roughly equally in the three sub-genomes of wheat (sub-genome A, 18; sub-genome B, 16; and sub-genome D, 16), which may cause redundant functions with genes on sub-genome A, indicating some TaCCHC-ZFP genes may experience gene loss event during the evolution with low purifying selection. We also found that the 50 TaCCHC-ZFPs formed 19 homoeologous groups, of which 12 homoeologous groups contained three homoeologous genes with strict 1: 1: 1 correspondence, while the other 7 homoeologous groups were referred as dyads (1: 1: 0, 1: 0: 1, 0: 1: 1; Figures 3, 4A). Meanwhile, the homoeologous genes, TaCCHC28 (4A), TaCCHC34 (5B), TaCCHC36 (5D), were not located in the same homologous chromosomes, suggesting that the TaCCHC-ZFPs were involved in the structural rearrangements of the 4A-5A chromosomes during the evolution of wheat (Chen et al., 2020b).
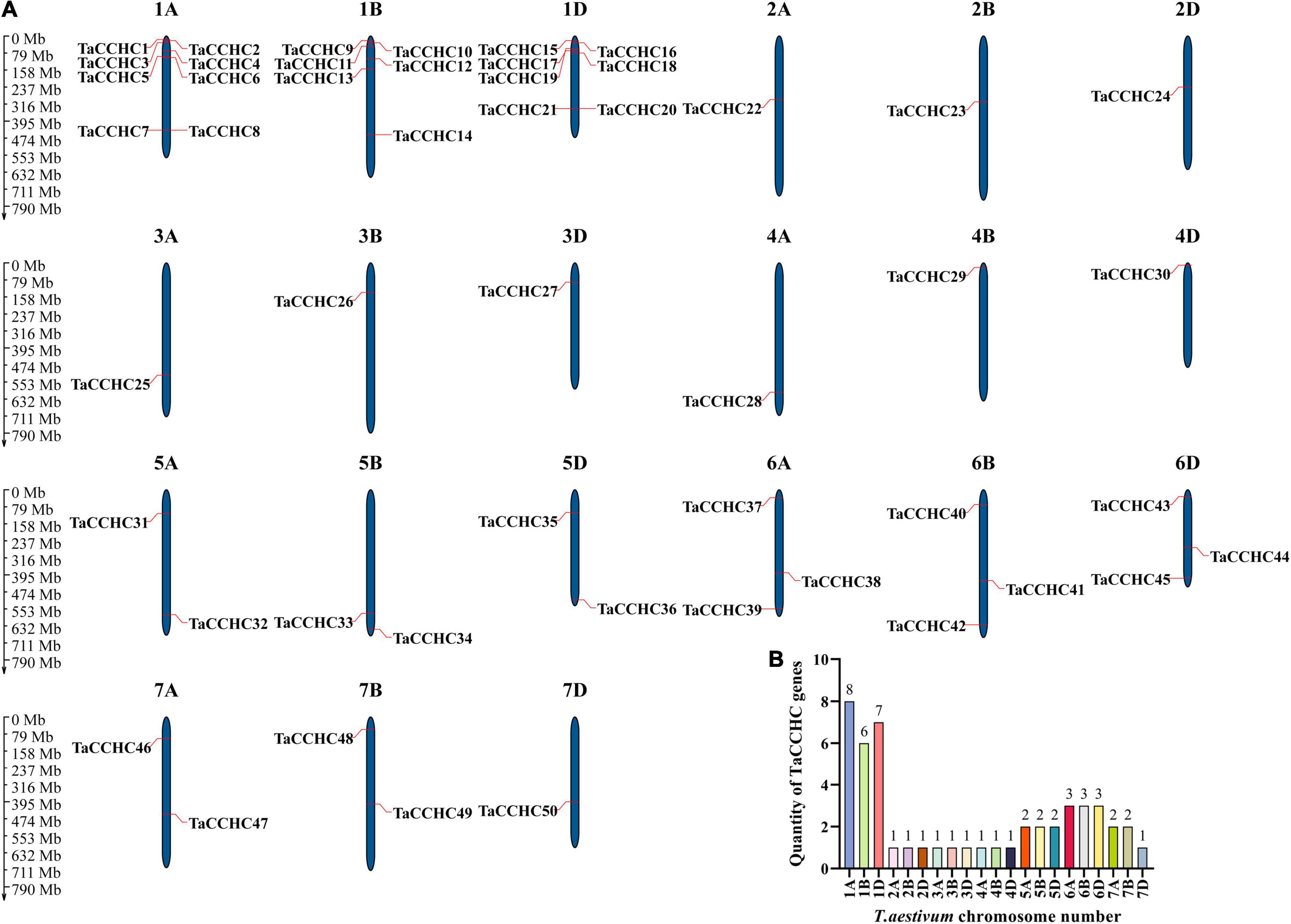
Figure 4. Chromosome distribution of the wheat CCHC-ZFP genes. (A) Chromosomal localization of the TaCCHC-ZFP genes. The dark blue columns indicate wheat chromosomes with the scale in megabases (Mb). The chromosome number is displayed at the top of each chromosome. (B) Numbers of TaCCHC-ZFP genes on each T. aestivum chromosome.
Next, the synteny analyses were performed to evaluate the gene duplication events in T. aestivum. Interestingly, we didn’t identify any tandem duplication events in these TaCCHC-ZFP genes. Nevertheless, a total of 36 segmental duplication events with 43 TaCCHC-ZFP genes were identified, indicating that segmental duplication events were the major driver for the evolution of TaCCHC-ZFP genes (Figure 5). All duplicated genes in a pair belong to the same TaCCHC-ZFP genes group. Furthermore, the Ka/Ks values of the TaCCHC-ZFP gene pairs were computed to explore the evolutionary constraints. The Ka/Ks values of the 36 gene pairs in wheat are generally less than 1, implying that the replicated TaCCHC-ZFP genes could experience strong purification selection pressure (Supplementary Table 5). The Ks values were adopted to assess the divergence time (T) based on the formula T = Ks/2λ × 10–6 Mya (λ = 6.5 × 10–9). The divergence time of these genes diverged between 0.994 and 19.055 Mya (average 6.735, 34 values in 36 earlier than 2.26), mostly before the early Gramineae whole-genome duplication event.
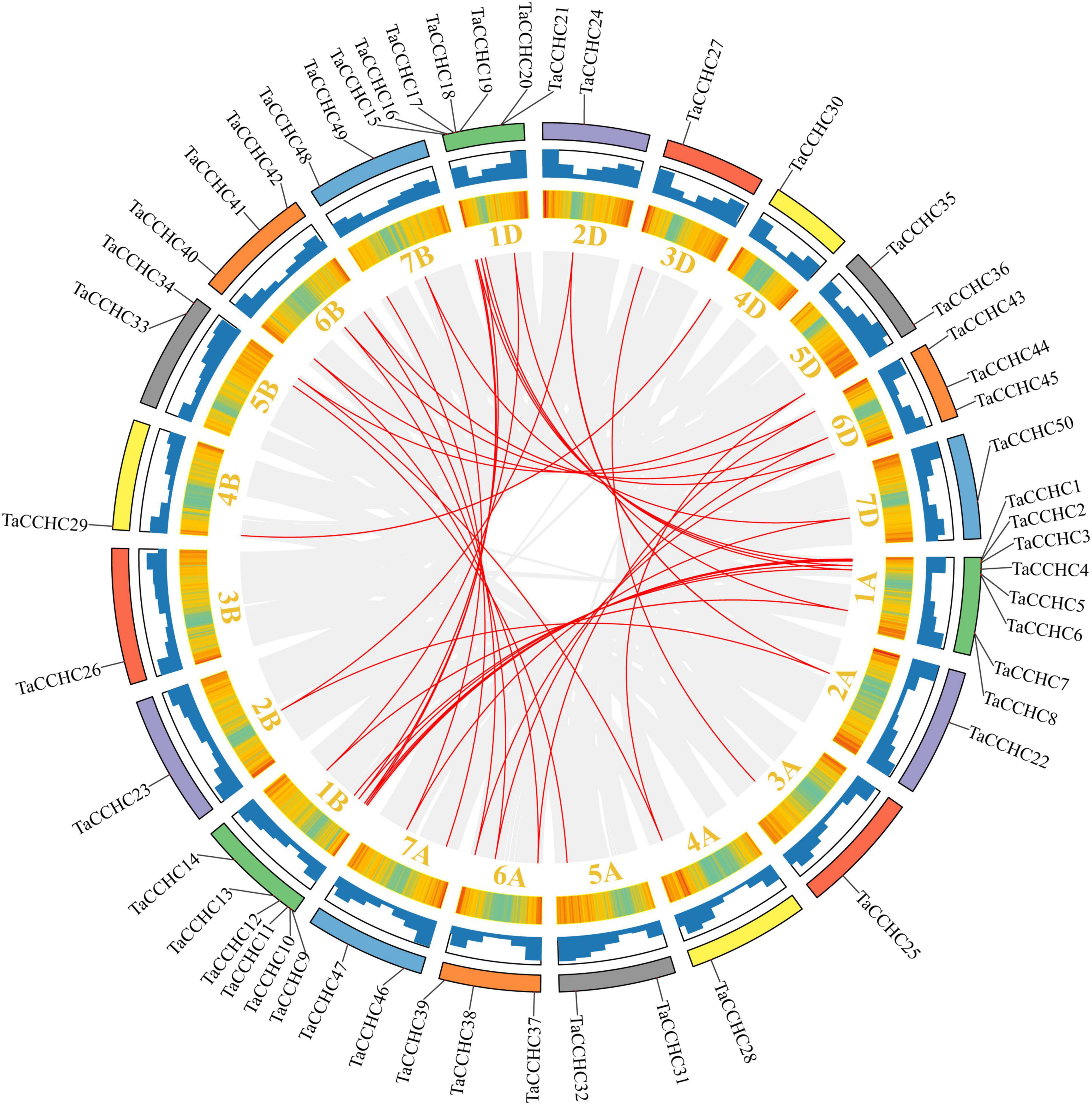
Figure 5. Synteny analysis of the CCHC-ZFP family in wheat. The gray lines indicate all synteny blocks within the wheat genome, while the red lines represent duplicated CCHC-ZFP gene pairs. Wheat chromosomes are displayed by rectangles with different colors, and the heatmaps and histograms along the rectangles show the gene density of each chromosome.
Synteny Analyses of CCHC Members From Wheat and Eight Other Plant Species
To further investigate the evolutionary mechanisms and homologous genes of TaCCHC-ZFPs, comparative syntenic maps were constructed by comparing eight representative species with wheat, including five monocots (T. dicoccoides, Ae. tauschii, Z. mays, O. sativa, and H. vulgare) and three dicots (A. thaliana, S. tuberosum, and G. max; Figure 6A). A total of 46 TaCCHC-ZFP genes show collinearity relationships with 15 CCHC-ZFP genes in Ae. tauschii, 29 in T. dicoccoides, 12 in O. sativa, 8 in H. vulgare, and 8 in Z. mays, respectively, while no this relationship among wheat and the three dicots analyzed was found, suggesting the closer phylogenetic relationships with the monocots than the dicots (Figure 6B). Therefore, 31, 76, 28, 16, 17 orthologous gene pairs among wheat and Ae. tauschii, T. dicoccoides, O. sativa, Z. mays, and H. vulgare were identified, respectively, (Supplementary Table 6). Hexaploid wheat (A, B, and D sub-genome) was obtained by natural hybridization between T. dicoccoides (A and B sub-genome) and Ae. tauschii (D sub-genome). Compared to T. dicoccoides and Ae. tauschii, more wheat CCHC-ZFP genes were derived from T. dicoccoides based on the number of orthologous CCHC-ZFP gene pairs. Among the three sub-genomes of wheat, 36 gene pairs (14 between the A and B sub-genomes, 11 between the A and D sub-genomes, 11 between the B and D sub-genomes) were identified, which were less than that between wheat and the sub-genome donors (Figures 5, 6A). This might be related to either the gene lost or chromosomal recombination during the polyploidization and evolution. Additionally, three TaCCHC-ZFP genes (TaCCHC37, TaCCHC46, and TaCCHC48) were observed in all of five syntenic maps, indicating that these TaCCHC genes were relatively conserved in the evolution. However, some wheat TaCCHC-ZFP genes identified were collinear with genes from only one species. For instance, TaCCHC35 was identified to have a collinearity relationship with Os12t0564600-01, while there was no collinearity with the CCHC-ZFP genes from the other four species, implying that TaCCHC35 might have been lost in the rest four plants and remained in wheat and rice.
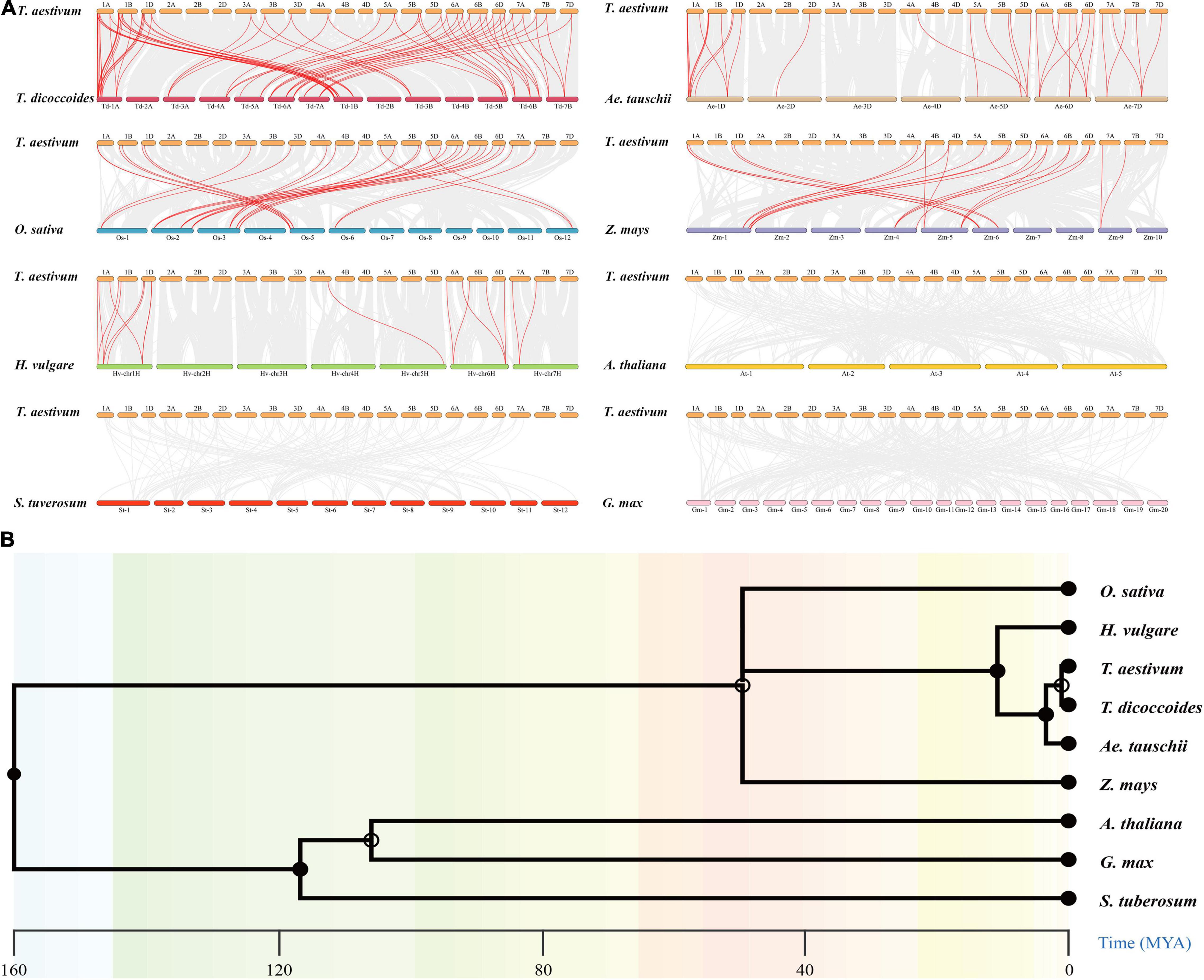
Figure 6. Synteny analyses of the CCHC-ZFP family between wheat and other species. (A) Collinearity analysis of the TaCCHC-ZFP family with other eight representative species. The gray lines in the background represent the collinear blocks in the genome of wheat and other species, while the red lines indicate the syntenic CCHC-ZFP gene pairs. (B) Species evolution tree of wheat and other eight species.
To further investigate the evolutionary constraints of the TaCCHC-ZFP genes, the Ka/Ks ratios of the CCHC-ZFP gene pairs were computed. The Ka/Ks ratios of nearly all orthologous CCHC-ZFP gene pairs were less than 1, indicating that the TaCCHC-ZFP genes might undergone purifying selection during the evolution to eliminate harmful mutations at the protein level (Supplementary Table 6). The divergence time of these duplicated orthologous TaCCHC-ZFP gene pairs were approximately 4.872 Mya (T. dicoccoides), 5.143 Mya (Ae. tauschii), 11.074 Mya (H. vulgare), 44.765 Mya (O. sativa), and 60.761 Mya (Z. mays), respectively, which were close to the result of the species evolution tree (Figure 6B).
Cis-Acting Elements and Gene Ontology Enrichment Analyses of TaCCHC-ZFP Genes
Transcription factors bind the cis-acting elements of the promoter regions to regulate transcription. Thus, the 1.5-kb upstream promoter regions of all TaCCHC-ZFP genes were submitted to the PlantCARE to study the potential biological functions of TaCCHC-ZFP genes. A total of 636 cis-elements associated with environmental stress signal and phytohormone responsiveness were found in the promoter regions of TaCCHC-ZFP genes (Figure 7A and Supplementary Table 7). Among them, 152 MeJA-responsive elements (TGAGG-motif and CGTCA-motif) and 128 ABA-responsive elements (ABRE) were found, respectively, which were the two most cis-acting elements of TaCCHC-ZFP genes. The result suggested that MeJA and ABA might take part in the transcriptional regulation of TaCCHC-ZFP genes. Moreover, 40 auxin-responsive cis-acting elements (AuxRR-core, TGA-element), 40 ethylene-responsive elements (ERE), 27 gibberellin-responsive elements (P-box, GARE-motif, and TATC-box), 16 salicylic acid-responsive elements (SARE and TCA-element) were identified in 40, 27, 16, and 14 TaCCHC-ZFP genes, respectively. Meanwhile, four types of cis-elements associated with biotic or abiotic stress responsiveness were identified, such as 45 drought responsive elements (MBS), 60 low-temperature responsive elements (LTR), 8 wound responsive elements (WUN-motif), 9 defense and stress responsive elements (TC-rich repeats). Additionally, except for TaCCHC31, TaCCHC44, TaCCHC49, and TaCCHC50, the anaerobic induction (ARE) or anoxic specific inducibility element (GC-motif) were found in the rest 46 TaCCHC-ZFP genes. In brief, the cis-acting elements identified in the promoter regions indicate that TaCCHC-ZFP genes may participate in the transcriptional regulation of phytohormone signaling and biotic/abiotic stress responses.
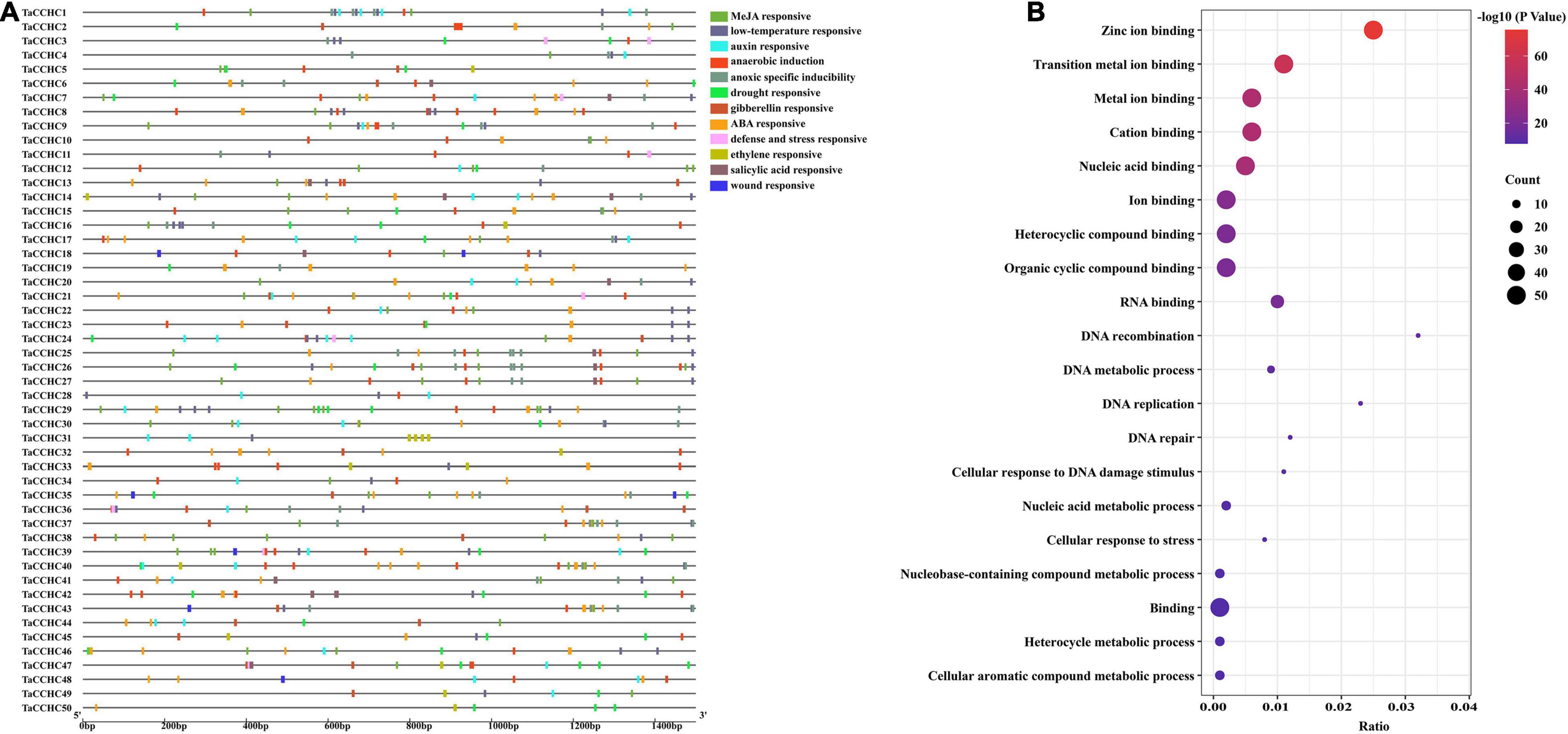
Figure 7. Analyses of cis-acting elements and functional annotation of TaCCHC-ZFP genes. (A) Distribution of predicted cis-acting elements in the promoter regions of TaCCHC-ZFP genes. The color blockers indicate different cis-acting elements and their locations in these TaCCHC-ZFP genes. (B) GO enrichment analysis of the TaCCHC-ZFP genes.
Additionally, the GO enrichment of all TaCCHC-ZFP genes was constructed to further explore the gene functions. The GO terms consist of three categories: cellular component, biological process (BP), and molecular function (MF). The enrichment results of the MF category revealed that all 50 TaCCHC-ZFP genes were annotated under nine GO terms, including zinc ion binding, metal ion binding, transition metal ion binding, cation binding, nucleic acid binding, ion binding, heterocyclic compound binding, organic cyclic compound binding, and binding, all of which belonged to the MF category, suggesting that they might act as zinc finger proteins to regulate gene expression through DNA or RNA binding (Figure 7B). The enrichment results of the BP category revealed that 12 TaCCHC genes participated in four kinds of metabolic processes, such as nucleic acid metabolic process and cellular aromatic compound metabolic process. Moreover, seven TaCCHC-ZFP genes shared five GO terms, which were DNA recombination, DNA replication, cellular response to DNA damage stimulus, DNA repair, and cellular response to stress, implying the potential roles of them during stress responses.
Protein Interaction Network and MicroRNA Targets Analysis
Proteins that perform similar functions or participate in the same pathway are more likely to exhibit interaction networks, forming gene modules or clusters in proteins interaction networks. To further understand the interaction relationships and biological functions among TaCCHC-ZFPs, the STRING database was adopted to map the protein-protein networks within TaCCHC-ZFP family. As shown in Figure 8, 24 TaCCHC-ZFPs were found to be involved in the protein interaction networks with 202 branches, suggesting that they might perform similar function.
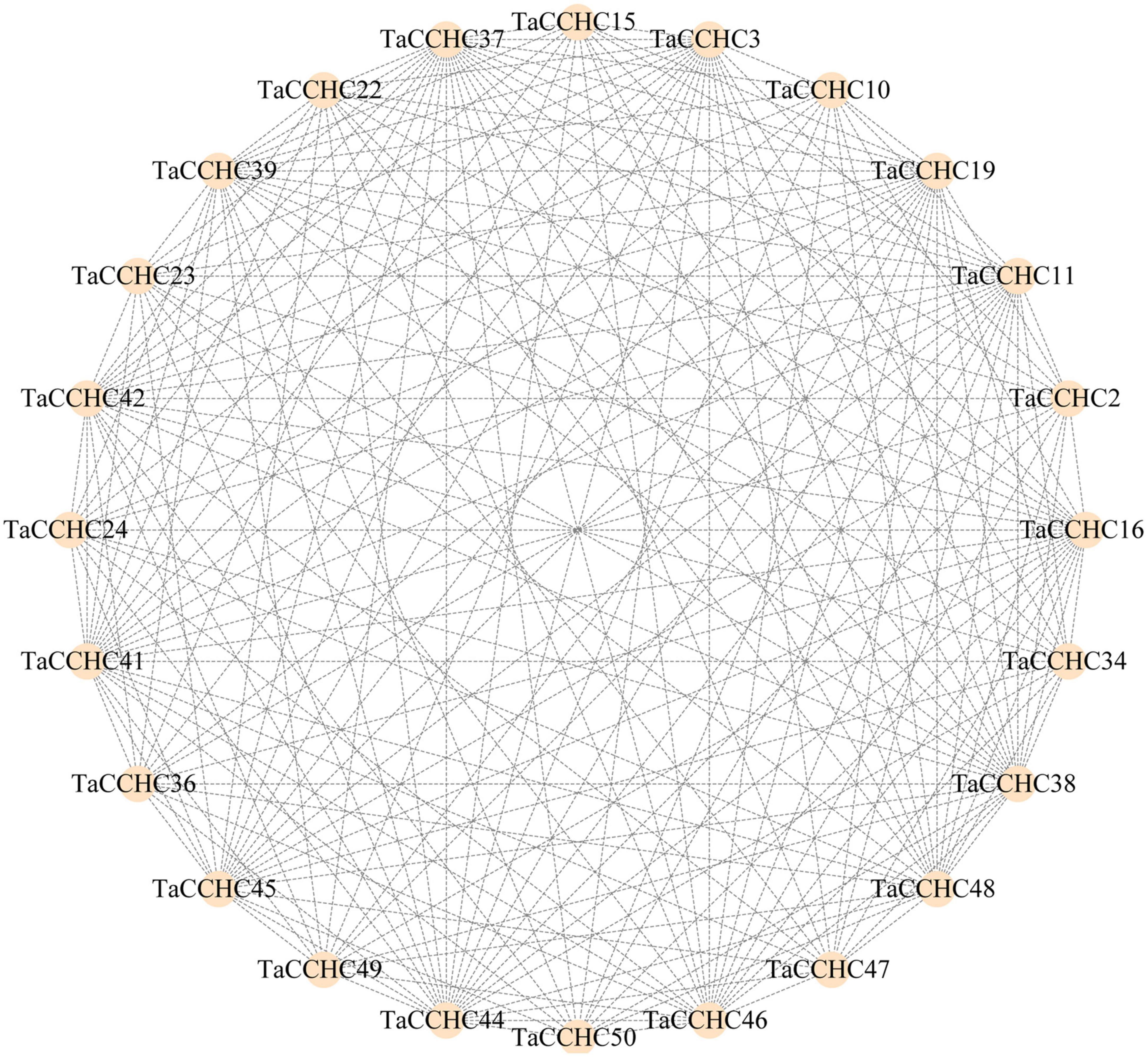
Figure 8. Interaction network of TaCCHC-ZFPs. A total of 202 interactions are displayed among 24 TaCCHC-ZFPs. The protein-protein interaction networks of wheat CCHC-ZFPs were predicted using the STRING tools with high confidence (0.700; Szklarczyk et al., 2019), and was used to visualized by the Cytoscape with default parameters (Shannon et al., 2003).
MicroRNAs are small non-coding RNAs that function in RNA silencing and post-transcriptional regulation of gene expression. Thus, the potential miRNA targets of TaCCHC-ZFP genes were predicted to provide support information about the regulatory mechanism of the TaCCHC-ZFP genes. The results revealed that a total of 91 miRNA target sites were identified in 34 TaCCHC-ZFP genes, with each gene corresponding to one to seven miRNAs (Supplementary Table 8). Among the 34 wheat CCHC-ZFP genes, TaCCHC36 and TaCCHC50 had the most targets with seven miRNA target sites, followed by TaCCHC21 and TaCCHC26 with six miRNA target sites, implying that the expression of TaCCHC-ZFP genes might be regulated by multiple miRNAs. At the same time, tae-miR9652-5p had the most target sites (nine TaCCHC-ZFP genes) among the 47 wheat miRNAs, followed by tae-miR9782 targeting six genes.
Expression Patterns of TaCCHC Genes Under Different Stresses
To further dissect the function of TaCCHC genes under abiotic stresses, the expression profiles of 50 TaCCHC-ZFP genes during different treatments (drought, heat, drought, and heat) were analyzed in this study (Figure 9A). Among the 50 genes, 38 DEGs were screened out via the edgeR package from five kinds of treatments [drought stress for 1 (6) h: DS-1 (6) h, heat stress for 1 (6) h: HS-1 (6) h, and combined drought and heat stress for 1 (6) h: DHS-1 (6) h], while no DEGs were identified under the DS-1h treatment (Supplementary Table 9). As shown in Figure 9B, 32 TaCCHC-ZFP genes responded to at least two treatments, while six TaCCHC-ZFP genes only responded to one treatment. For instance, TaCCHC34, TaCCHC42, and TaCCHC47 showed decreased expression under the HS-1h and DHS-1h treatments, while TaCCHC11 exhibited increased expression under the HS-1h and DHS-1h treatments, implying that these genes were sensitive to heat and drought. As expected, some genes with close evolutionary relationships showed similar expression patterns. The expression of TaCCHC22, TaCCHC23, and TaCCHC24 were downregulated under the HS-1h and DHS-1h treatments, while the expression of TaCCHC32 and TaCCHC33 were upregulated under the HS-6h and DHS-6h treatments. Additionally, the expression patterns of TaCCHC-ZFP genes under cold and phosphorous starvation treatments were also analyzed (Supplementary Figure 1). Only eight TaCCHC-ZFP genes responded to cold treatment, while no DEGs were identified under the phosphorous starvation treatment (Supplementary Table 9).
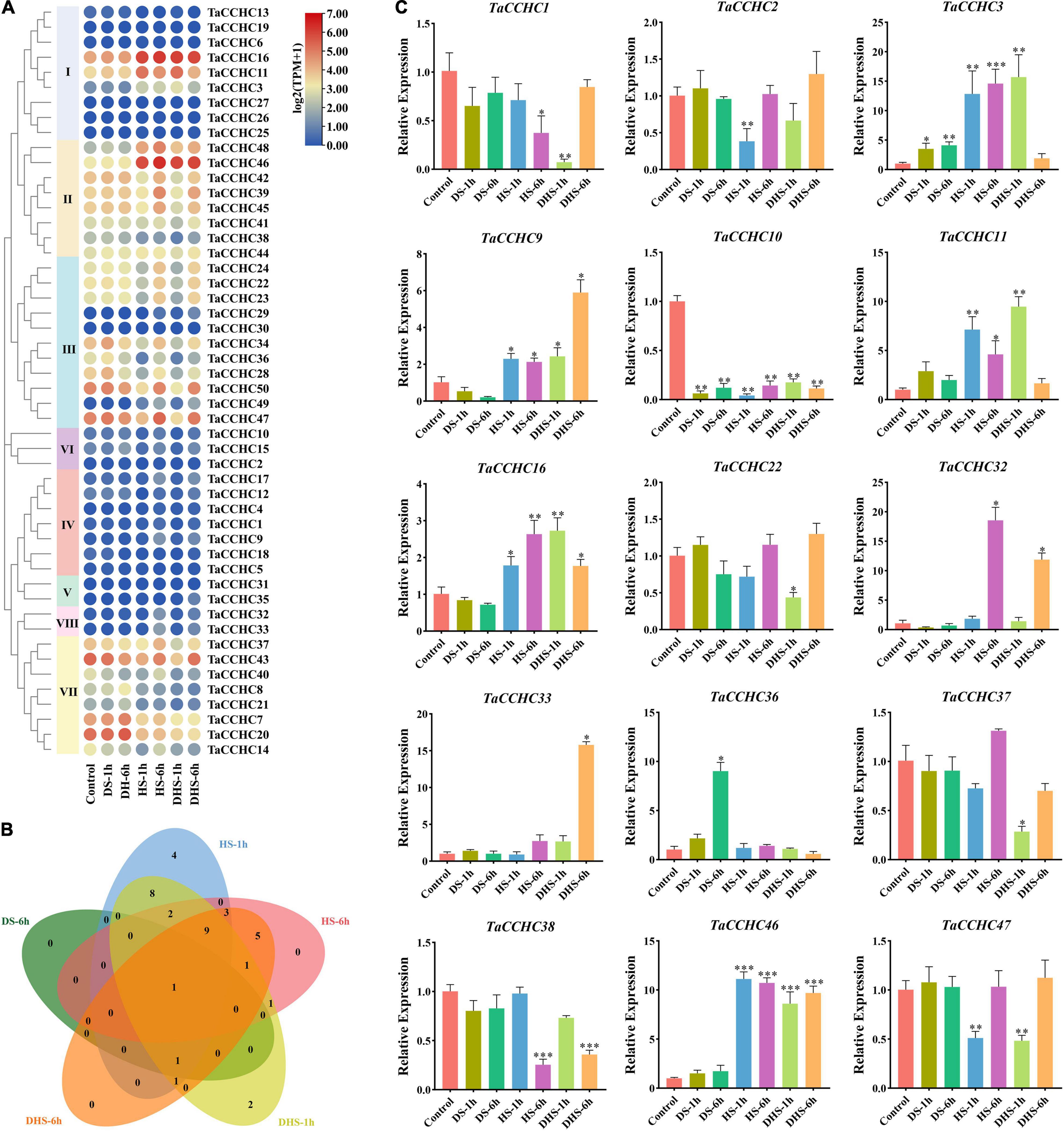
Figure 9. Expression profiles of wheat CCHC-ZFP genes under different conditions. (A) Expression profiles of 50 TaCCHC-ZFP genes under different stress treatments. HS-1 (6) h: heat stress for 1 (6) h; DS-1 (6) h: drought stress for 1 (6) h; DHS-1 (6) h: combined drought and heat stress for 1 (6) h. The color in the heat map reflects TaCCHC-ZFP genes expression level. (B) Venn diagrams of DEGs under different treatments. (C) Expression analyses of 15 TaCCHC-ZFP genes in response to different treatments by qRT-PCR. Data were normalized to actin-TaRP15 and error bars represent standard deviation among three independent replicates (*P < 0.05, **P < 0.01, ***P < 0.001, and Student’s t-test).
To confirm the transcription profiles of the TaCCHC-ZFP genes derived from the transcriptome data, 15 TaCCHC-ZFP genes from the eight groups were selected to analyze their expression level under different treatments by qRT-PCR. As shown in Figure 9C, the expression profiles of most TaCCHC-ZFP genes are congruent with the previously published data according to the results of qRT-PCR. Overall, the expression of TaCCHC-ZFP genes could be influenced by multiple treatments.
Discussion
As one of the most important food crops, wheat is subject to environmental stresses, resulting in the reduction of yield. Zinc finger protein transcription factors play vital roles in plant growth and development, and biotic and abiotic stress responses (Li et al., 2013). Some previous studies revealed that TaCCHC-ZFP genes regulated plant growth and stress responses, such as AtRZ-1a, Mt-Zn-CCHC, and NTT (Kim et al., 2005; Crawford et al., 2007; Lee and Kang, 2016; Radkova et al., 2019). Thus, the comprehensive bioinformatic analyses of TaCCHC-ZFP gene family were conducted to better study the gene functions of TaCCHC-ZFP genes due to the limited work on TaCCHC-ZFP gene family.
In this study, we identified 50 TaCCHC-ZFPs from wheat and extracted CCHC motif sequences of these members, in which the conserved sites were consistent with the previous study (Figure 1; Armas and Calcaterra, 2013). Studies revealed that CCHC motif was a kind of nucleic acid binding domain, which contributed to RNA binding, DNA regulation, or protein-protein interactions (Matthews et al., 2000; Espinosa, 2003; Ganie, 2020). Meanwhile, the results of GO enrichment showed that all 50 TaCCHC-ZFP genes were annotated under nucleic acid binding term and zinc ion binding term (Figure 7B), implying that TaCCHC-ZFPs might function by binding DNA or RNA. Previous study showed that WCSP1 (TaCCHC7 in this study) was capable of binding dsDNA, ssDNA, and RNA homopolymers, whereas its ability to bind dsDNA was almost eliminated in the absence of C-terminal CCHC motif (Karlson et al., 2002). In Arabidopsis, CSDP1, homologous to TaCCHC14, which possesses seven tandem repeated CCHC motifs in the C-terminal half, acts as an RNA chaperone in the response to cold stress, helping to export mRNA from the nucleus to the cytoplasm (Park et al., 2009).
The analyses of phylogenetic relationships, protein motifs, and gene structures showed that the homologous TaCCHC-ZFP genes in sub-genomes A, B, D shared similar gene structures and conserved motifs, indicating the functions of TaCCHC-ZFP genes were conservative during the evolution (Figures 3A–C). The motif 1 (CCHC motif) is conserved in all TaCCHC-ZFPs. It is noteworthy that some motifs are distributed in specific groups, such as RRM, CSD, REPA OB, and Rep Fac-A C, which may participate in various biological processes based on the different functions of TaCCHC-ZFP genes (Supplementary Table 4; Bochkarev et al., 1997; Daubner et al., 2013; Budkina et al., 2020). For instance, AtGRP2 containing two CCHC zinc fingers and one CSD motifs may be involved in cold-response and flower development (Fusaro et al., 2007). Besides, RRM exists in groups I, II, and III, which can bind single-strand RNA and participate in the regulation of flowering and adaptation to heat stress (Pi et al., 2018). AtSF1, a protein containing RRM, takes part in regulating heat stress response by affecting the alternative splicing of the pre-mRNA of the heat shock transcription factor HsfA2 (Lee et al., 2017). Meanwhile, to better understand the homoeologous relationships, we analyzed homoeologous groups of wheat TaCCHC-ZFPs in detail (Figures 3, 4A). Approximately 36% of wheat genes are presented in homoeologous groups of three (1: 1: 1), while the remaining 64% have a more complex homologous relationship (e.g., 1: 1: 0 or 1: 1: N; Glover et al., 2016; Juery et al., 2021). By contrast, about 63% of wheat TaCCHC-ZFP genes identified are presented as triads, which is considerably above the average homoeologous retention rate in wheat (36%). On the other hand, the loss of one homoeolog is less in TaCCHC-ZFP genes (37 vs 64%), suggesting that the high homoelog retention rate could partly explain the abundance of TaCCHC-ZFP genes.
Previous researches revealed that gene families normally experienced tandem duplication events or segmental duplication events to expand gene family members in the process of evolution (Cannon et al., 2004). Subsequently, syntenic analyses were carried out in this study (Figures 5, 6A). Wheat has undergone two major polyploid evolutionary events, accompanied by segmental duplication, tandem duplication, and transposition events (Peng et al., 2011). However, the number of TaCCHC-ZFP genes in a specific sub-genome was severely reduced during the transition from tetraploid to hexaploidy through the identification of CCHC-ZFP genes in wheat and its sub-genomes donors, T. dicoccoides and Ae. tauschii (for A sub-genome, from 20 to 18 genes; B sub-genome, from 18 to 16 genes; D sub-genome, from 46 to 16 genes), proving that gene loss during hexaploidy wheat formation occurred extensively (Berkman et al., 2013). Generally, the Ka/Ks ratios for all the homologous CCHC gene pairs are less than 1, indicating that TaCCHC-ZFP genes may have undergone purifying selection pressure and the functions of these gene pairs do not diverge much after the two polyploidization events (Supplementary Tables 5, 6).
Cis-acting elements and miRNAs are involved in the regulation of gene expression at the transcriptional and post-transcriptional levels, respectively, (Hausser and Zavolan, 2014; Hernandez-Garcia and Finer, 2014). Therefore, we predicted the cis-acting elements in the promoter regions of wheat CCHC-ZFP genes and miRNAs targeting TaCCHC genes. Plenty of studies showed that cis-elements were essential factors of modulating gene expression under biotic and abiotic stress. For instance, PbrMYB21 could interact with the MYB-recognizing cis-element in the promoter region of PbrADC to modulate polyamine synthesis via regulating ADC expression, improving drought tolerance (Li et al., 2017). In this study, a lot of cis-acting elements associated with environmental stress and phytohormone responsiveness were identified, indicating that TaCCHC-ZFP genes might take part in multiple signaling pathways (Figure 7A and Supplementary Table 7; Liu et al., 2014). Additionally, plant miRNAs are associated with cell biology processes and response to stress, which can regulate gene expression at the post-transcriptional level through splicing mRNA or inhibiting translation. In this study, we found 47 wheat miRNAs target with 34 TaCCHC-ZFP genes, including tae-miR9652-5p, tae-miR9782, tae-miR156, tae-miR159a/b, tae-miR164, and tae-miR167, etc. (Supplementary Table 8). Previous studies reported that some plant miRNAs, such as miR156, miR159a/b, miR164, miR319, and miR399, played an important role in modulating plant developmental time, the differentiation of tissues, and response to environmental stresses (Willmann and Poethig, 2007). The miR156-overexpression alfalfa showed significant improvement in drought tolerance with reduced water loss and higher survival compared with the wild-type control (Arshad et al., 2017). Moreover, ABA induced the accumulation of miR159 to mediate the cleavage of MYB33 and MYB101 transcripts in geminating Arabidopsis seeds (Reyes and Chua, 2007). In brief, cis-acting elements and miRNAs may be regulators of TaCCHC-ZFP gene expression.
Previous studies showed that CCHC-ZFP genes responded to multiple stresses. For example, the cold resistance of atRZ-1a-overexpressing transgenic Arabidopsis plants was enhanced compared to wild-type plants, with earlier germination and better seedling growth under cold treatment as well (Kim et al., 2005). Drought and heat are the main environmental stresses affecting wheat growth and development, often resulting in the decline of wheat yield. In this study, we investigated the potential functions of TaCCHC-ZFP genes under drought and heat treatments, and 38 DEGs were screened out (Figures 9A–C and Supplementary Table 9). Previous research revealed that AtCSP3-overexpressing transgenic Arabidopsis plants exhibited higher survival rates under the drought and salt treatment, whereas the atcsp3 mutant displayed lower survival rate (Kim et al., 2013). TaCCHC20, homologous to Arabidopsis At4g36020.1 (AtCSP3), was downregulated under the DHS-6h treatment, indicating that they might have similar functions under the drought and heat stresses. TaRZ2 (TaCCHC49 in this study) can negatively regulate seed germination and seedling growth under the salt or dehydration treatments but contribute to enhancing cold tolerance of transgenic Arabidopsis (Xu et al., 2014). Meanwhile, TaCCHC20 and TaCCHC49 were found to share similar cis-elements, such as TGA-element, LTR, and so on. Overall, these results suggested that TaCCHC-ZFP genes might be involved in the plant responses to drought and heat stresses.
Conclusion
CCHC-ZFPs are involved in multiple physiological processes, such as seed development, plant growth, and responses to biotic and abiotic stresses. In this study, a total of 50 TaCCHC-ZFP genes were identified from wheat by bioinformatics tools. Subsequently, these TaCCHC-ZFP genes were categorized into eight groups with specific motifs and gene structures. Interestingly, only segmental duplication events were identified in TaCCHC-ZFP genes, suggesting that the segmental duplication events were the major driver for TaCCHC-ZFP genes evolution. In addition, collinearity relationships among wheat and eight other representative plants were analyzed and no gene pairs were identified between wheat and the three dicots. Plenty of cis-acting elements related to environmental stress were found in the promoters of TaCCHC-ZFP genes. GO enrichment results showed that all TaCCHC-ZFP genes were annotated under nucleic acid binding and metal ion binding. The analyses of miRNA targets suggested that the TaCCHC-ZFP genes could be regulated by the miRNAs. Furthermore, the expression patterns of TaCCHC-ZFP genes and qRT-PCR verification showed that some TaCCHC-ZFP genes participated in the responses to drought and heat stresses.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
XG and WX designed the experiments. AS and XG wrote the main manuscript text. YLL and AS conducted the experiments. AS, YLL, YH, XZ, FC, RJ, CY, KY, and YL collected and analyzed phenotype data. AS, YL, and WX prepared Figures 1–9. All authors read and approved the manuscript.
Funding
This work was supported by grants from National Natural Science Foundation of China (31872866), Changsha Natural Science Foundation (kq2202149), and China Postdoctoral Science Foundation (2021M701160).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are grateful Shuai Hu from School of life sciences, Tsinghua University for critical review of this manuscript. This manuscript has been preprinted (DOI: 10.21203/rs.3.rs-900125/v1 and 10.21203/rs.3.rs-900125/v2).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.892105/full#supplementary-material
Abbreviations
CCHC-ZFP, CCHC-type zinc finger protein; GA, gibberellins; IAA, indole-3-acetic acid; ABA, abscisic acid; MeJA, methyl jasmonate; MW, molecular weight; AI, aliphatic index; pI, isoelectric point; GRAVY, grand average of hydropathicity; aa, amino acids; REPA OB, Replication protein A OB; Rep Fac-A C, Replication factor-A C terminal domain; Ka, Non-synonymous; Ks, Synonymous; Mya, Mya millions of years; SA, salicylic acid; BP, biological process; MF, molecular function; CC, cellular component; miRNA, MicroRNA; DS-1 (6) h, drought stress for 1 (6) h; HS-1 (6) h, heat stress for 1 (6) h; DHS-1 (6) h, combined drought and heat stress for 1 (6) h; DEG, differentially expressed gene; qRT-PCR, quantitative real-time PCR; HMM, Hidden Markov Model; NJ, neighbor-joining; SMART, Simple Modular Architecture Research Tool; GSDS, Gene Structure Display Server; GO, gene ontology; UTR, untranslated regions.
Footnotes
- ^ http://plants.ensembl.org/index.html
- ^ http://pfam.xfam.org
- ^ http://smart.emblheidelberg.de/
- ^ http://web.expasy.org/protparam/
- ^ http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/
- ^ https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html
- ^ https://meme-suite.org/meme/tools/meme
- ^ http://weblogo.threeplusone.com/
- ^ http://mg2c.iask.in/mg2c_v2.0
- ^ http://www.timetree.org
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html
- ^ http://gsds.gao-lab.org
- ^ https://www.omicshare.com
- ^ https://string-db.org
- ^ https://www.zhaolab.org/psRNATarget/
- ^ http://www.wheat-expression.com
- ^ http://www.ehbio.com/test/venn/#/
References
Aceituno-Valenzuela, U., Micol-Ponce, R., and Ponce, M. R. (2020). Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans. Cell Mol. Life Sci. 77, 3991–4014. doi: 10.1007/s00018-020-03518-7
Amorim, L., Santos, R., Neto, J., Guida-Santos, M., Crovella, S., and Benko-Iseppon, A. M. (2017). Transcription factors involved in plant resistance to pathogens. Curr. Prot. Pept. Sci. 18, 335–351. doi: 10.2174/1389203717666160619185308
Armas, P., and Calcaterra, N. B. (2013). Retroviral zinc knuckles in eukaryotic cellular proteins. Nova Sci. Publish. 1, 51–80.
Arshad, M., Feyissa, B. A., Amyot, L., Aung, B., and Hannoufa, A. (2017). MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 258, 122–136. doi: 10.1016/j.plantsci.2017.01.018
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Berkman, P. J., Visendi, P., Lee, H. C., Stiller, J., Manoli, S., Lorenc, M. T., et al. (2013). Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol. J. 11, 564–571. doi: 10.1111/pbi.12044
Bochkarev, A., Pfuetzner, R. A., Edwards, A. M., and Frappier, L. (1997). Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385, 176–181. doi: 10.1038/385176a0
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Budkina, K. S., Zlobin, N. E., Kononova, S. V., Ovchinnikov, L. P., and Babakov, A. V. (2020). Cold shock domain proteins: structure and interaction with nucleic acids. Biochemistry 85, S1–S19. doi: 10.1134/S0006297920140011
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chao, J., Kong, Y., Wang, Q., Sun, Y., Gong, D., Lv, J., et al. (2015). MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan. 37, 91–97.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Song, W., Xie, X., Wang, Z., Guan, P., Peng, H., et al. (2020b). A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant. 13, 1694–1708. doi: 10.1016/j.molp.2020.09.019
Choi, M. J., Park, Y. R., Park, S. J., and Kang, H. (2015). Stress-responsive expression patterns and functional characterization of cold shock domain proteins in cabbage (Brassica rapa) under abiotic stress conditions. Plant Physiol. Biochem. 96, 132–140. doi: 10.1016/j.plaphy.2015.07.027
Chou, K., and Shen, H. (2010). Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 02, 1090–1103. doi: 10.4236/ns.2010.210136
Crawford, B. C., Ditta, G., and Yanofsky, M. F. (2007). The NTT gene is required for transmitting-tract development in carpels of Arabidopsis thaliana. Curr. Biol. 17, 1101–1108. doi: 10.1016/j.cub.2007.05.079
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cui, P., Liu, H., Ruan, S., Ali, B., Gill, R. A., Ma, H., et al. (2017). A zinc finger protein, interacted with cyclophilin, affects root development via IAA pathway in rice. J. Integr. Plant Biol. 59, 496–505. doi: 10.1111/jipb.12531
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
Daubner, G. M., Clery, A., and Allain, F. H. (2013). RRM-RNA recognition: NMR or crystallography.and new findings. Curr. Opin. Struct. Biol. 23, 100–108. doi: 10.1016/j.sbi.2012.11.006
Espinosa, J. (2003). Trypanosoma cruzi poly-zinc finger protein: a novel DNA/RNA-binding CCHC-zinc finger protein. Mol. Biochem. Parasitol. 131, 35–44. doi: 10.1016/S0166-6851(03)00187-7
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Fusaro, A. F., Bocca, S. N., Ramos, R. L., Barrôco, R. M., Magioli, C., Jorge, V. C., et al. (2007). AtGRP2, a cold-induced nucleo-cytoplasmic RNA-binding protein, has a role in flower and seed development. Planta 225, 1339–1351. doi: 10.1007/s00425-006-0444-4
Ganie, S. A. (2020). RNA chaperones: potential candidates for engineering salt tolerance in rice. Crop Sci. 60, 530–540. doi: 10.1002/csc2.20134
Gasteiger, E. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Geourjon, C., and Deléage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Glover, N. M., Redestig, H., and Dessimoz, C. (2016). Homoeologs: what are they and how do we infer them? Trends Plant Sci. 21, 609–621. doi: 10.1016/j.tplants.2016.02.005
Guan, Q., Wang, L., Bu, Q., and Wang, Z. (2014). The rice gene OsZFP6 functions in multiple stress tolerance responses in yeast and Arabidopsis. Plant Physiol Biochem. 82, 1–8. doi: 10.1016/j.plaphy.2014.04.021
Hausser, J., and Zavolan, M. (2014). Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat. Rev. Genet. 15, 599–612. doi: 10.1038/nrg3765
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 21, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, F., Tang, J., and Hou, X. (2016). Molecular cloning and characterization of BcCSP1, a Pak-choi (Brassica rapa ssp. chinensis) cold shock protein gene highly co-expressed under ABA and cold stimulation. Acta Physiol Plant. 38, 1–8. doi: 10.1007/s11738-015-2058-6
Juery, C., Concia, L., De Oliveira, R., Papon, N., Ramírez-González, R., and Benhamed, M. (2021). New insights into homoeologous copy number variations in the hexaploid wheat genome. Plant Genome 14:e20069. doi: 10.1002/tpg2.20069
Karlson, D., Nakaminami, K., Toyomasu, T., and Imai, R. (2002). A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J. Biol. Chem. 277, 35248–35256. doi: 10.1074/jbc.M205774200
Kiełbowicz-Matuk, A. (2012). Involvement of plant C2H2-type zinc finger transcription factors in stress responses. Plant Sci. 18, 78–85. doi: 10.1016/j.plantsci.2011.11.015
Kim, J. Y., Kim, W. Y., Kwak, K. J., Oh, S. H., Han, Y. S., Kang, H., et al. (2010). Zinc finger-containing glycine-rich RNA-binding protein in Oryza sativa has an RNA chaperone activity under cold stress conditions. Plant Cell Environ. 33, 759–768. doi: 10.1111/j.1365-3040.2009.02101.x
Kim, M., Sato, S., Sasaki, K., Saburi, W., Matsui, H., and Imai, R. (2013). COLD SHOCK DOMAIN PROTEIN 3 is involved in salt and drought stress tolerance in Arabidopsis. FEBS Open Bio 3, 438–442. doi: 10.1016/j.fob.2013.10.003
Kim, Y., Kim, J. S., and Kang, H. (2005). Cold-inducible zinc finger-containing glycine-rich RNA-binding protein contributes to the enhancement of freezing tolerance in Arabidopsis thaliana. Plant J. 42, 890–900. doi: 10.1111/j.1365-313X.2005.02420.x
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, K. R., and Kirti, P. B. (2012). Novel role for a serine/arginine-rich splicing factor, AdRSZ21 in plant defense and HR-like cell death. Plant Mol Biol. 80, 461–476. doi: 10.1007/s11103-012-9960-8
Kumar, S., Stecher, G., Suleski, M., and Hedges, S. B. (2017). TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812–1819. doi: 10.1093/molbev/msx116
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laity, J. H., Lee, B. M., and Wright, P. E. (2001). Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46. doi: 10.1016/s0959-440x(00)00167-6
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, K., and Kang, H. (2016). Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol. Cells 39, 179–185. doi: 10.14348/molcells.2016.2359
Lee, K. C., Jang, Y. H., Kim, S., Park, H., Thu, M. P., Lee, J. H., et al. (2017). RRM domain of Arabidopsis splicing factor SF1 is important for pre-mRNA splicing of a specific set of genes. Plant Cell Rep. 36, 1083–1095. doi: 10.1007/s00299-017-2140-1
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, K., Xing, C., Yao, Z., and Huang, X. (2017). PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 15, 1186–1203. doi: 10.1111/pbi.12708
Li, W., He, M., Wang, J., and Wang, Y. (2013). Zinc finger protein (ZFP) in plants-A review. Plant Omics 6, 474–480.
Liu, J., Peng, T., and Dai, W. (2014). Critical cis-acting elements and interacting transcription factors: key players associated with abiotic stress responses in plants. Plant Mol. Biol. Rep. 32, 303–317. doi: 10.1007/s11105-013-0667-z
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 15:8. doi: 10.1186/s12870-015-0511-8
Matthews, J. M., Kowalski, K., Liew, C. K., Sharpe, B. K., Fox, A. H., Crossley, M., et al. (2000). A class of zinc fingers involved in protein-protein interactions. Eur. J. Biochem. 267, 1030–1038. doi: 10.1046/j.1432-1327.2000.01095.x
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Ozkan, H., Levy, A. A., and Feldman, M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13, 1735–1747. doi: 10.1105/tpc.010082
Park, S. J., Kwak, K. J., Oh, T. R., Kim, Y. O., and Kang, H. (2009). Cold shock domain proteins affect seed germination and growth of Arabidopsis thaliana under abiotic stress conditions. Plant Cell Physiol. 50, 869–878. doi: 10.1093/pcp/pcp037
Peng, J. H., Sun, D., and Nevo, E. (2011). Domestication evolution, genetics and genomics in wheat. Mol. Breed. 28, 281–301. doi: 10.1007/s11032-011-9608-4
Petersen, G., Seberg, O., Yde, M., and Berthelsen, K. (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39, 70–82. doi: 10.1016/j.ympev.2006.01.023
Pi, B., He, X., Ruan, Y., Jang, J., and Huang, Y. (2018). Genome-wide analysis and stress-responsive expression of CCCH zinc finger family genes in Brassica rapa. BMC Plant Biol. 18:7. doi: 10.1186/s12870-018-1608-7
Radkova, M., Revalska, M., Kertikova, D., and Iantcheva, A. (2019). Zinc finger CCHC-type protein related with seed size in model legume species Medicago truncatula. Biotechnol. Biotechnol. Equip. 33, 278–285. doi: 10.1080/13102818.2019.1568914
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361:r6089. doi: 10.1126/science.aar6089
Reyes, J. L., and Chua, N. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2009). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Sasaki, K., Kim, M., Kanno, Y., Seo, M., Kamiya, Y., and Imai, R. (2015). Arabidopsis cold shock domain protein 2 influences ABA accumulation in seed and negatively regulates germination. Biochem. Biophys. Res. Commun. 456, 380–384. doi: 10.1016/j.bbrc.2014.11.092
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Summers, M. F. (1991). Zinc finger motif for single-stranded nucleic acids? Investigations by nuclear magnetic resonance. J. Cell Biochem. 45, 41–48. doi: 10.1002/jcb.240450110
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2019). STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Takatsuji, H. (1999). Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 39, 1073–1078. doi: 10.1023/a:1006184519697
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Y., Yu, Y., Pang, Y., Yu, H., Zhang, W., Zhao, X., et al. (2021). The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol. 18, 2107–2126. doi: 10.1080/15476286.2021.1909320
Wheeler, T. J., and Eddy, S. R. (2013). nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489. doi: 10.1093/bioinformatics/btt403
Willmann, M. R., and Poethig, R. S. (2007). Conservation and evolution of miRNA regulatory programs in plant development. Curr. Opin. Plant Biol. 10, 503–511. doi: 10.1016/j.pbi.2007.07.004
Xu, T., Gu, L., Choi, M. J., Kim, R. J., Suh, M. C., Kang, H., et al. (2014). Comparative functional analysis of wheat (Triticum aestivum) zinc finger-containing glycine-rich RNA-binding proteins in response to abiotic stresses. PLoS One 9:e96877. doi: 10.1371/journal.pone.0096877
Keywords: wheat, CCHC-ZFP genes, evolution, abiotic stress, expression analyses
Citation: Sun A, Li Y, He Y, Zou X, Chen F, Ji R, You C, Yu K, Li Y, Xiao W and Guo X (2022) Comprehensive Genome-Wide Identification, Characterization, and Expression Analysis of CCHC-Type Zinc Finger Gene Family in Wheat (Triticum aestivum L.). Front. Plant Sci. 13:892105. doi: 10.3389/fpls.2022.892105
Received: 08 March 2022; Accepted: 11 April 2022;
Published: 29 April 2022.
Edited by:
Fan Chen, Institute of Genetics and Developmental Biology (CAS), ChinaReviewed by:
Ji Huang, Nanjing Agricultural University, ChinaYingyin Yao, China Agricultural University, China
Copyright © 2022 Sun, Li, He, Zou, Chen, Ji, You, Yu, Li, Xiao and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhong Guo, Z3hoQGhudS5lZHUuY24=; Wenjun Xiao, eGlhb3dqOTBAaG51LmVkdS5jbg==
†These authors have contributed equally to this work
 Aolong Sun†
Aolong Sun† Wenjun Xiao
Wenjun Xiao Xinhong Guo
Xinhong Guo