- 1Key Laboratory of Soybean Molecular Design Breeding, Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, Changchun, China
- 2College of Advanced Agricultural Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3Zhejiang Lab, Hangzhou, China
The generation of oxygen and organic matter in plants mainly depends on photosynthesis, which directly affects plant growth and development. The chloroplast is the main organelle in which photosynthesis occurs. In this study, a Glycine max pale green leaf 3-1 (Gmpgl3-1) mutant was isolated from the soybean mutagenized population. The Gmpgl3-1 mutant presented with decreased chlorophyll contents, reduced chloroplast stroma thylakoids, reduced yields, and decreased numbers of pods per plant. Bulked segregant analysis (BSA) together with map-based cloning revealed a single-nucleotide non-synonymous mutation at the 341st nucleotide of the first exon of the chloroplast development-related GmTic110a gene. The phenotype of the knockout plants was the same as that of the mutant. The GmTic110a gene was highly expressed in the leaves at various developmental stages, and its protein was localized to the inner chloroplast membrane. Split luciferase complementation assays and coimmunoprecipitation (co-IP) experiments revealed that GmTic110a interacted with GmTic20, GmTic40a, and GmTic40b in tobacco leaves. These results indicated that the GmTic110a gene plays an important role in chloroplast development.
Introduction
Plant leaves are the most important tissues for photosynthesis. Chlorophyll is an important pigment involved in photosynthesis in chloroplasts (Waters and Langdale, 2009), and the development of plant chloroplasts positively correlates with the chlorophyll content in leaves (Davis and Fajer, 1979; Wang et al., 2003) and leaf photosynthesis rates (Peng et al., 2008). Mutations in chlorophyll synthesis-related genes can directly or indirectly affect chlorophyll biosynthesis or degradation pathways, leading to the loss of plant chlorophyll, thereby affecting the photosynthesis of plants and causing yellow leaves, albinism, striped leaf spots, purple–brown patches, or other characteristics of chlorophyll-deficient mutants (Awan et al., 1980). To date, researchers have studied chlorophyll-deficient mutants of Arabidopsis thaliana (Carol et al., 1999), tobacco (Okabe and Straub, 1977), corn (Lonosky et al., 2004), rice (Ki-Hong et al., 2003), soybean (Stockinger and Walling, 1994), pea (Highkin et al., 1969), wheat (Cao et al., 2006), barley (Preiss and Thornber, 1995) and other plant species. Chlorophyll-deficient mutants usually present with a decreased photosynthesis rate and reduced yields, and death can occur in severe cases.
Plant chloroplasts synthesize important amino acids through photosynthesis and are the main sources of energy for plant cells. Chloroplasts play an important role in plant growth and cellular metabolism (Tiller and Bock, 2014). The transport of substances in and out of chloroplasts depends on the translocon at the outer envelope membrane of chloroplasts (TOC) and the translocon at the inner envelope membrane of chloroplasts (TIC; Soll, 2004; Jocelyn and Paul, 2005; Jarvis, 2008). The TOC and TIC form a complex to facilitate this process. The chloroplast transport proteins on the outer chloroplast membrane identified to date include Toc159 (Waegemann and Soil, 1991; Schnell et al., 1994), Toc34 (Kessler et al., 1994; Schnell et al., 1994), Toc75 (Tranel et al., 1995; Sveshnikova et al., 2000) and Toc64 (Sohrt and Soll, 2000; Becker et al., 2004). Recent extensive studies have significantly updated our understanding of the components and mechanisms of the chloroplast translocon machinery (Kikuchi et al., 2013, 2018; Nakai, 2015, 2018, 2020). These studies have significantly revised the long-accepted “classical” model for chloroplast protein import: In the classical model, Tic110 (Ishida and Terakura, 1987; Inaba et al., 2005), Tic40 (Stahl et al., 1999; Chou et al., 2003), Tic20 (Kouranov and Schnell, 1997; Kasmati et al., 2011), and Tic21 (Vitale et al., 2015) are the main components, but they are not found in the translocon proposed by Nakai (2015, 2018, 2020). In the revised model, the 1-megadalton TIC complex consists of Tic214 (ycf1; de Vries et al., 2007; Bölter and Soll, 2017), Tic100 (Oshima et al., 1987; Ramundo et al., 2020), Tic56 (Köhler et al., 2015, 2016), Tic20 (Kikuchi et al., 2009), and Tic21 (Kouranov and Schnell, 1997; Teng et al., 2006), which functionally and physically cooperate with the ATP-driven import motor YCF2/FTSHI complex (Kikuchi et al., 2013, 2018; Thomson et al., 2020).
Tic110 is an important chloroplast inner membrane protein (Schnell et al., 1994; Inaba et al., 2005; Balsera et al., 2009). It has been reported that the Tic110 protein interacts with several molecular chaperones, such as Hsp93 and Hsp70, to form an inner membrane transport channel scaffold that ensures the successful import of various proteins into the chloroplast to perform cell biological functions (Inaba et al., 2003). Tic110 proteins interact with Tic32 proteins to perform redox functions and to regulate Ca2+ homeostasis in the chloroplast (Hormann et al., 2004). In addition, Tsai et al. (2013) reported that Tic110 is most likely a scaffolding component important for protein–protein interactions to recruit other translocon components and chaperones in the stroma (Tsai et al., 2013). In Arabidopsis and soybean, a lack of Tic110 blocks the transport of the inner and outer chloroplast membranes, affecting the development of chloroplasts and resulting in yellow leaves (Inaba et al., 2005; Sandhu et al., 2016). Tic110 assists in the formation of a scaffold for the assembly of the ATP-dependent import motor in the stroma (Richardson and Schnell, 2020). Soybean is an important source of grain and oil. This species is also the main source of high-quality protein for human diets and animal feed. As such, soybean occupies an important position in grain production worldwide. Obstruction of plant chloroplast development could lead to yellow leaves, which severely affects photosynthesis and plant yield; in severe cases, this results in dwarf-type plants or even plants that fail to produce harvestable yields. Therefore, it is of great scientific importance to study the regulatory molecular mechanisms of chloroplast membrane transport proteins.
In this study, we report the characterization of a Glycine max pale green leaf mutant (Gmpgl3-1). The chloroplast development-related gene GmTic110a encodes a chloroplast inner membrane protein. Gmpgl3-1, Gmpgl3-2, and Gmpgl3-3 are allelic mutants of the GmTic110a gene. The biological function of the GmTic110a gene was preliminarily analyzed in this research. We used split luciferase complementation and coimmunoprecipitation (co-IP) analyses, and the results indicated that GmTic110a proteins can interact with other GmTic proteins in tobacco. Our research lays a theoretical foundation for studies of the molecular mechanism underlying soybean chloroplast development.
Materials and Methods
Plant Material
All plants used in this study were grown at the Changchun Agricultural Station, Northeast Institute of Geography and Agroecology, Chinese Academy of Science (CAS), China. A Gmpgl3-1 mutant was isolated from an M2 population induced by ethylmethanesulfonate (EMS). The Gmpgl3-1 mutant was backcrossed to Williams 82 five times from 2014 to 2018 to purify the genetic background of the Gmpgl3-1 mutant. For protoplast isolation, Arabidopsis seeds of the Columbia ecotype (Col-0) were surface sterilized, vernalized, and then sown on 1/2-strength Murashige and Skoog (MS) media until the seedlings reached the four-leaf stage. Then, the seedlings were grown in pots containing peat moss and vermiculite (1/1, v/v) in a growth chamber under 150 μmol m−2 s−1 irradiance and a 14 h dark/10 h light photoperiod at 25°C, and the relative humidity was maintained at 60–75%. Leaves were collected from 3- to 4-week-old seedlings for transfection assays.
Mapping of GmTic110a via Bulked Segregant Analysis
Three F2 populations derived from a cross between the Gmpgl3-1 mutant and the Chinese soybean cultivar Hedou 12 were used to map the GmTic110a gene. DNA from 50 F2 individuals with the Gmpgl3-1 mutant phenotype and 50 F2 individuals with the wild-type phenotype were bulked into mutant and wild-type pools, respectively. Insertion–deletion (INDEL) markers for preliminary mapping were used according to a previously described method (Song et al., 2015). New molecular markers for fine mapping were generated; these markers are listed in Supplementary Table S4. The candidate genomic regions were identified via BSA of the F2 population at a depth of approximately 30× using an Illumina HiSeq 2000 device (Illumina Inc., San Diego, CA, United States). The Genome Analysis Toolkit (GATK, version 3.8) was used to detect single-nucleotide polymorphisms (SNPs; Mckenna et al., 2010). Genomic regions in which Δ(SNP index) was >0.5 were selected as candidate regions.
Database Searching and Phylogenetic Analysis
GmTic110a homologs were identified by querying the GmTic110a sequence in the NCBI1 database via the BLASTP program. Multiple sequence alignments were performed using ClustalX version 2.0 (Larkin et al., 2007) and were manually corrected. The obtained sequence was used as input to construct an unrooted phylogenetic tree with the neighbor-joining algorithm via the Molecular Evolutionary Genetics Analysis version 7.0 (MEGA 7.0) phylogenetic program (Sudhir et al., 2016). Bootstrap analysis was performed using 1,000 replicates. The protein motifs of GmTic110a-like genes were subsequently profiled by Multiple Expectation maximization for Motif Elicitation (MEME; Bailey et al., 2009).
Determination of Pigment Contents and Chlorophyll Fluorescence Analysis
To determine pigment contents, leaves of 21-day-old Gmpgl3-1 mutants and Williams 82 plants were collected and measured according to a previously reported procedure (Gregor and Marsálek, 2004). The pigment contents were calculated according to the following formulas: chlorophyll a = 13.95*A665-6.88*A649; chlorophyll b = 24.96*A649-7.32*A665; and carotenoids = (1,000*A470-2.05*Ca-114.8*Cb)/245. The photosynthesis rate (Pn), stomatal conductance (Gs), intercellular CO2 concentration (Mckenna et al.), and transpiration rate (Tr) of the leaves were measured using an LI-6400 photosynthesis system (LI-COR, Lincoln, NE, United States; Yamori et al., 2011), and the initial fluorescence (F0), maximal fluorescence (Fm), and variable fluorescence (Fv) values were measured using an OS-30p chlorophyll fluorometer (Opti-Sciences, Hudson, NY, United States). The maximum quantum yield of photosystem II (Fv/Fm) and the maximum photochemical yield of PSII (Fv/F0) were calculated as previously described (Genty et al., 1989). The plants were dark-adapted for 30 min before measurement. All the measurements involved the use of ten plants and were performed from 11:00 am to noon during the beginning of the flowering period. The operation of the machine and subsequent analysis were performed according to the manufacturer’s instructions.
Transmission Electron Microscopy Analysis
Williams 82 and Gmpgl3-1 mutant plants grown for 21 days were selected. The leaves were cut into rectangular pieces that were approximately 2 mm*1 mm, and the plant materials were vacuum fixed in 2.5% glutaraldehyde solution with 0.2 mol of phosphate buffer. The samples were postfixed for 3 h in 1% osmium tetroxide at 4°C. The samples were then treated according to previously described methods (Kowalewska et al., 2016). Ultrathin sections were obtained using an MT-X (RMC, Tucson, AZ, United States) ultramicrotome and stained with uranyl acetate for 20 min followed by lead citrate for 10 min. Observations of the samples and recording of images were performed using a Hitachi H-7650 electron microscope (Tokyo, Japan).
CRISPR/Cas9 Vector Construction and Soybean Transformation
To obtain GmTic110a-knockout plants, the CRISPR/Cas9 gene editing system for targeted genome modification of plants was used (Shan et al., 2013). Several 20-nt single-guide RNAs (sgRNAs) highly specific for Cas9 target sites were identified using the web-based tool CRISPR-P version 2.02 (Lu et al., 2017). A pair of 24-bp long oligonucleotides (5′-GATTGCGGCGGCTGGATACGGCCT-3′ and 5′-AAACAGGCCGTATCCAGCCGCCGC-3′) specific to the GmTic110a sequence were annealed and cloned into a modified VK005-04-soU6-2-GmUbi3 knockout expression vector (Du et al., 2016). The resulting recombinant plasmid (VK005-GmTic110a) was introduced into Agrobacterium tumefaciens EHA105, which was then used to transform Williams 82 cotyledonary explants (Zhao et al., 2016). Three independent GmTic110a-knockout transgenic plants were obtained for further phenotypic analysis.
Analysis of the Expression Profile of the GmTic110a Gene
New leaves at the VE (emergence) stage; stem tips, stems, and roots at the V1 (first unrolled trifoliate leaf) stage; leaves and flowers at the R1 (beginning bloom) stage; leaves and flowers at the R2 (full bloom) stage; and leaves at the R3 (beginning of pod development) stage were collected. Total RNA was subsequently extracted using TRIzol reagent (Tiangen, Lot 118,721; China) according to the manufacturer’s instructions. The integrity of the RNA was determined through agarose gel electrophoresis, and complementary DNA (cDNA) was synthesized using 5 μg of RNA with oligo(dT)18 primers and Moloney murine leukemia virus (M-MLV) reverse transcriptase (TransGen Lot N31204; China) according to the manufacturer’s protocol. Relative transcript levels of GmTic110a were analyzed through real-time quantitative PCR (qRT–PCR) on an Mx3005P instrument (Stratagene, La Jolla, CA, United States) in conjunction with SYBR Green Master Mix (Genstar Lot 9 BC01; China). The PCR parameters were 95°C for 30 s (1 cycle), 95°C for 5 s, and 60°C for 20 s (40 cycles), which was followed by a melting curve analysis at 95°C for 60 s, 55°C for 30 s, and 95°C for 30 s. The internal control gene GmActin11 (Glyma.18G290800) was used for normalization of the transcript levels of GmTic110a in the samples (Hu et al., 2009). The relative fold differences were calculated via the 2-ΔΔCt method. Three independent biological replicates were used to confirm the expression profiles. The specific primer pairs used are listed in Supplementary Table S4.
Subcellular Localization Analysis
We next sought to determine the subcellular localization of GmTic110a, GmTic110aG114A, Gmtic110aT805S, GmTic110aCR1, GmTic110aCR2, and GmTic110aCR3 from the knockout transgenic strains. For this analysis, the full-length cDNA sequence and the mutated and knockout sequences of GmTic110a were cloned into pUC19-GFP (Zheng et al., 2017), and the resulting recombinant plasmid was transiently introduced into Arabidopsis (Col-0) protoplasts using 20% polyethylene glycol (Yu et al., 2015). The fluorescence signals were visualized using a Nikon C2 confocal laser scanning microscope (Japan) under a 488 nm excitation wavelength and 495–540 nm emission wavelengths to determine the subcellular localization of GmTic110a, GmTic110aG114A, Gmtic110aT805S, GmTic110aCR1, GmTic110aCR2, and GmTic110aCR3. Chloroplast autofluorescence was detected at wavelengths of 488 nm (excitation) and 680–700 nm (emission). Image processing was performed with ImageJ.3 The specific primer pairs used are listed in Supplementary Table S4.
Luciferase Complementation Assays
Luciferase complementation assays were performed as described previously (Chen et al., 2008), with minor modifications. The coding DNA sequences (CDSs) of GmTic110a, GmTic20, GmTic40a, and GmTic40b were cloned into either a pCAMBIA1300-NLuc or a pCAMBIA1300-CLuc vector. pCAMBIA1300-GmTic110a-NLuc, pCAMBIA1300-GmTic20-CLuc, pCAMBIA1300-GmTic40a-CLuc, and pCAMBIA1300-GmTic40b-CLuc in various combinations were transferred into Nicotiana benthamiana leaves by A. tumefaciens-mediated transformation. Agrobacterium cells with N-Luc and C-Luc vectors were resuspended in infiltration buffer (pH 5.6; 10 mm 2-(N-morpholino) ethanesulfonic acid, 10 mm MgCl2, and 150 mm acetosyringone) to reach an optimal optical density at 600 nm in the range of 0.9 to 1. After 3 h of incubation at room temperature, the suspensions were infiltrated into the leaves of 4-week-old N. benthamiana plants, which were then cultivated for 2 days at 23°C. To inject tobacco leaves with 1 mmol of luciferin (115144–35-9, GoldBio) for measurements of luciferase activity, the leaves were maintained in the dark for 5 min. Images were captured using a chemiluminescence image analysis system (4600SF, Tanon). The sequences of the primers used are listed in Supplementary Table S4.
Co-IP Assays
Co-IP assays were performed as described previously (Zhou et al., 2015; Liu et al., 2020; Wang et al., 2020), with minor modifications. The CDSs of GmTic110a, GmTic20, GmTic40a, and GmTic40b were cloned into either a pCAMBIA1300-FLAG or pCAMBIA1300-HA vector, resulting in pCAMBIA1300-GmTic110a-FLAG, pCAMBIA1300-GmTic20-HA, pCAMBIA1300-GmTic40a-HA, and pCAMBIA1300-GmTic40b-HA vectors. To measure protein–protein interactions, A. tumefaciens strain EHA105 containing pairs of these constructs together with pCAMBIA1300-GmTic110a-FLAG, pCAMBIA1300-GmTic20-HA, pCAMBIA1300-GmTic40a-HA, and pCAMBIA1300-GmTic40b-HA were coinfiltrated into the leaves of 4-week-old N. benthamiana plants. Samples (1 g each) were then collected at 3 days after infiltration, ground in liquid nitrogen and homogenized in 1.5 ml of extraction buffer (50 mm Tris–HCl [pH 7.5], 150 mm NaCl, 1 mm EDTA [pH 8.0], 0.2% [v/v] Triton X-100, 20% [v/v] glycerol, and 1× protease inhibitor cocktail [pH 7.5]). The lysates were incubated at 4°C for 30 min and subsequently centrifuged at 15,000 × g for 30 min at 4°C. After instantaneous centrifugation, the supernatants were added to 500-μl suspensions of anti-DDDDK-tag-FLAG magnetic beads (No. M185-11R, Medical and Biological Laboratories), incubated at 4°C for 4 h, and then washed 4 times with extraction buffer. The proteins were eluted from the beads with 30 μl of 1 × Protein Loading Buffer, boiled for 5 min, and then centrifuged at 8,000 × g for 1 min at room temperature. The supernatants were electrophoretically separated via 10% SDS–PAGE and transferred to a nitrocellulose membrane (No. q0600003, GE Healthcare Life Sciences). Immunoblots were performed using an anti-FLAG antibody (1:5000; No. M180-5, Medical and Biological Laboratories) for probing pCAMBIA1300-GmTic110a-FLAG and an anti-HA antibody (1/5000, No. M180-3, Medical and Biological Laboratories) for probing pCAMBIA1300-GmTic20-HA, pCAMBIA1300-GmTic40a-HA, or pCAMBIA1300-GmTic40b-HA. The sequences of the primers used are listed in Supplementary Table S4.
Results
Phenotypic Characterization of the Chloroplast Development-Related Mutant Gmpgl3
Compared with the wild-type Williams 82, the Gmpgl3-1 mutant showed a pale green leaf phenotype from the seedling stage to the mature stage (Figure 1A). Moreover, on the basis of their phenotypes and genotypes, we identified two allelic mutants named Gmpgl3-2 and Gmpgl3-3 from within the mutant library. The pale green leaf phenotype was observed for the Gmpgl3-2 mutant (Figure 1A), while the pale green leaf phenotype was not observed for the Gmpgl3-3 mutant (Supplementary Figure S1). We analyzed mature Williams 82, Gmpgl3-1, and Gmpgl3-2 plants separately. The results showed that the overall heights of the Gmpgl3-1 and Gmpgl3-2 mutants were reduced by 7 and 2.5%, respectively, compared with Williams 82, while the number of nodes was reduced by 17.3%. Moreover, compared with Williams 82, the number of pods per plant for the Gmpgl3-1 and Gmpgl3-2 mutants was reduced by 30.8 and 39.5%, respectively; the number of grains per plant for the Gmpgl3-1 and Gmpgl3-2 mutants was reduced by 39.5 and 39.2%, respectively; the grain weight per plant for the Gmpgl3-1 and Gmpgl3-2 mutants was reduced by 50.8 and 48.2%, respectively; and the 100-seed weight for the Gmpgl3-1 and Gmpgl3-2 mutants was reduced by 18.1 and 14.8%, respectively (Supplementary Table S1).
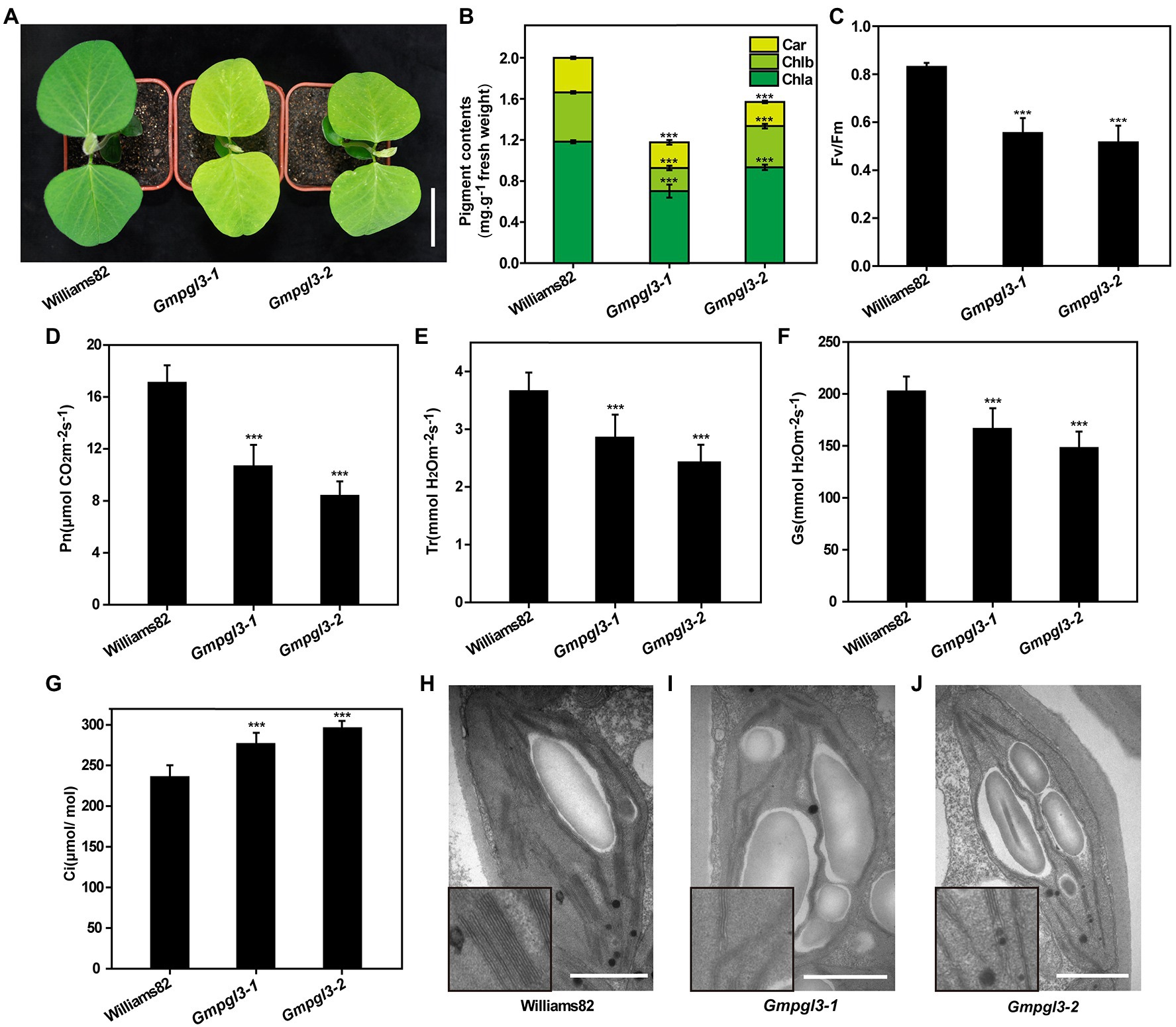
Figure 1. (A) Phenotypes of the Williams 82, Gmpgl3-1, and Gmpgl3-2 mutants. Scale bars, 5 cm. (B–G) Pigment contents and photosynthesis parameters of the Williams 82, Gmpgl3-1, and Gmpgl3-2 mutants. (B) Chlorophyll a (Chl a), chlorophyll b (Chl b) and carotenoid (Car). (C) Chlorophyll fluorescence parameter (Fv/FM). (D) Photosynthetic rate (Pn). (E) Transpiration rate (Tr). (F) Stomatal conductance (Gs). (G) Intercellular CO2 concentration (Ci). (H) Chloroplast structure in Williams 82. (I,J) Chloroplast structure in the Gmpgl3-1 and Gmpgl3-2 mutants. Scale bars, 1 μm. *** represents significant differences compared with the control (Williams 82) at p < 0.001, and the error bars represent standard deviations.
Because the Gmpgl3-1 and Gmpgl3-2 mutants exhibited a pale green leaf phenotype throughout the growth period, the chlorophyll and carotenoid contents of Williams 82, Gmpgl3-1, and Gmpgl3-2 leaves were measured spectrophotometrically, and the results showed that both chlorophyll a and chlorophyll b contents in the Gmpgl3-1 and Gmpgl3-2 mutants were significantly lower than those in the wild type (Figure 1B). This finding indicates that the GmTic110a gene mutation may affect the stability of Chl a/b. The chlorophyll content of the Gmtic110a mutant was reduced by 44.2%. The chloroplast fluorescence Fv/Fm values of Gmpgl3-1 and Gmpgl3-2 were significantly lower than those of Williams 82, which showed that the photosynthetic efficiency of Gmpgl3-1 and Gmpgl3-2 was lower than that of Williams 82 (Figure 1C). The photosynthetic rate (Pn) of the Gmpgl3-1 and Gmpgl3-2 mutants was reduced by 37.7 and 50.9%, respectively, compared with that of Williams 82. In addition, compared with those of the wild type, the stomatal conductance (Gs) of the Gmpgl3-1 and Gmpgl3-2 mutants decreased by 22 and 33.7%, respectively, the transpiration rate (Tr) of the Gmpgl3-1 and Gmpgl3-2 mutants decreased by 17.9 and 26.9%, respectively, and the intercellular CO2 concentration of the Gmpgl3-1 and Gmpgl3-2 mutants increased by 17.3 and 25.6%, respectively (Figures 1D–G). Taken together, these results showed that the decrease in the chlorophyll content in the Gmpgl3-1 and Gmpgl3-2 mutants significantly affected the photosynthetic ability of the leaves of those plants.
Compared with that in the wild-type (Williams 82) chloroplasts, the number of starch grains in the chloroplasts of the Gmpgl3-1 and Gmpgl3-2 mutants increased, and the basal thylakoids became thinner (Figures 1H–J). Because there are photosynthetic pigment components on the thylakoid membrane and because photosynthesis mainly occurs within thylakoids, both the chlorophyll a and chlorophyll b contents in the mutants were significantly reduced, which led to thinning of the basal thylakoid membrane and a reduction in the photosynthesis rate.
Genetic Mapping of the Gmpgl3-1 Mutation Locus From the F2 Population
The Gmpgl3-1 mutant was crossed with Hedou 12 to generate a segregating population for mapping the Gmtic110a gene. The F2 segregating population comprised 537 plants: 423 wild-type plants and 114 mutant plants. A 3:1 segregation ratio was observed for the three F2 segregating populations (χ2 = 1.89, df = 1, p = 0.17; Supplementary Table S2), indicating that the Gmpgl3-1 mutant is the result of a single recessive gene. The F2 population was used to identify the Gmpgl3-1 locus. A total of 60 INDEL markers covering all 20 chromosomes were used for mapping, and the mapping results showed that Gmpgl3-1 was restricted to a 2-Mb region (41–43 Mb) on chromosome 02 (Figures 2A,B). To finely map the Gmpgl3-1 locus, we developed 7 INDEL markers, that is, MOL3067, MOL4032, MOL3069, MOL3071, MOL3073, MOL2733, and MOL0699; the Gmpgl3-1 locus was further narrowed down to a 0.44-Mb region between 41.79 Mb and 42.23 Mb on chromosome 02, which harbors 18 annotated genes (Figure 2C). To identify the causal mutation, the DNA from 40 F2 individuals carrying the Gmpgl3-1 mutation under homozygous conditions and the DNA of 50 F2 individuals exhibiting the wild-type phenotype were pooled into a Gmpgl3-1 bulk and a Williams 82 bulk for further BSA. The Gmpgl3-1 mutant was resequenced to a depth of approximately 30× using an Illumina HiSeq 2000 device. We identified a G−341 to A−341 transition in the first exon of Glyma.02G233700 (Figure 2D), which caused a non-synonymous substitution of Gly−114 to Asp−114 in the predicted protein. No other mutations were discovered among the 18 genes in the candidate Gmpgl3-1 genomic region (Figure 2C; Supplementary Table S3). However, a single-base mutation (A–2413 to T–2413) was identified in the 14th exon of the GmTic110a gene of the Gmpgl3-2 mutant (Figure 2D). The expression level of Glyma.02G233700 decreased in the Gmpgl3-1 and Gmpgl3-2 mutants (Supplementary Figure S2), suggesting that Glyma.02G233700 is the GmTic110a gene.
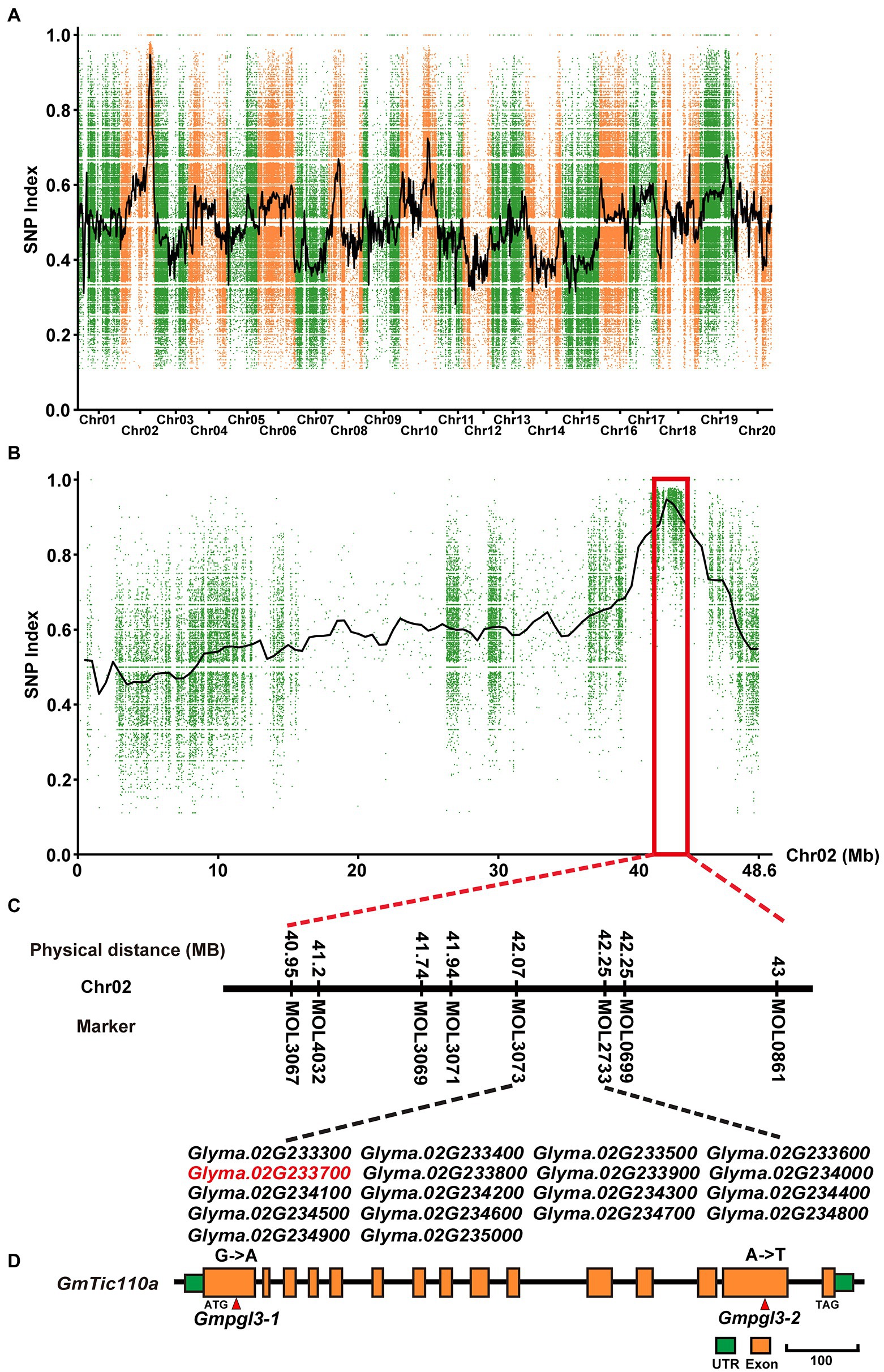
Figure 2. Map-based cloning of the GmTic110a locus. (A) SNP index plot of all chromosomes of the F2 plants. (B) SNP index plots of chromosome 02 of the Gmpgl3a mutant from the F2 population. (C) Physical position of the GmTic110a candidate gene. (D) Schematic diagram showing the structure of GmTic110a. The red lines indicate mutation sites within the GmTic110a gene in the two mutant lines.
GmTic110a Encodes an Inner Chloroplast Membrane Protein
BLAST searches revealed that GmTic110a, which encodes a chloroplast inner membrane protein, is highly homologous to the soybean GmTic110b protein (Glyma.14G201500; 98.4% amino acid similarity) and the Arabidopsis Tic110 protein (At1G06950; 84.3% amino acid similarity). In Medicago, the gene with the highest homology to GmTic110a is Medtr5g074690, whose sequence is 90.5% identical to that of GmTic110a and 86.4% identical to that of Medtr3g466170. Both of their proteins are 994 and 985 amino acids in length. Amino acid sequence analysis resulted in the generation of a phylogenetic tree composed of the GmTic110a homologous gene and its homologs from dicotyledonous plant species (G. max, A. thaliana, and Medicago sativa), monocotyledonous plant species (Oryza sativa, Zea mays, and Sorghum bicolor), Selaginella tamariscina, and Physcomitrella patens (Figure 3C). The results suggest that two GmTic110 homologs are evolutionarily conserved among plant species and share a common genomic structure in the observed plant species. The results of MEME analysis showed that GmTic110a contains 12 conserved motifs (Figure 3D). The mutation site of the Gmpgl3-1 mutant is located within the conserved TM2 domain, the mutation site of the Gmpgl3-2 mutant is located within the conserved chaperone-binding (co) domain, and the Gmpgl3-3 mutant site is located within the terminal TM1 domain (Figures 3A,B).
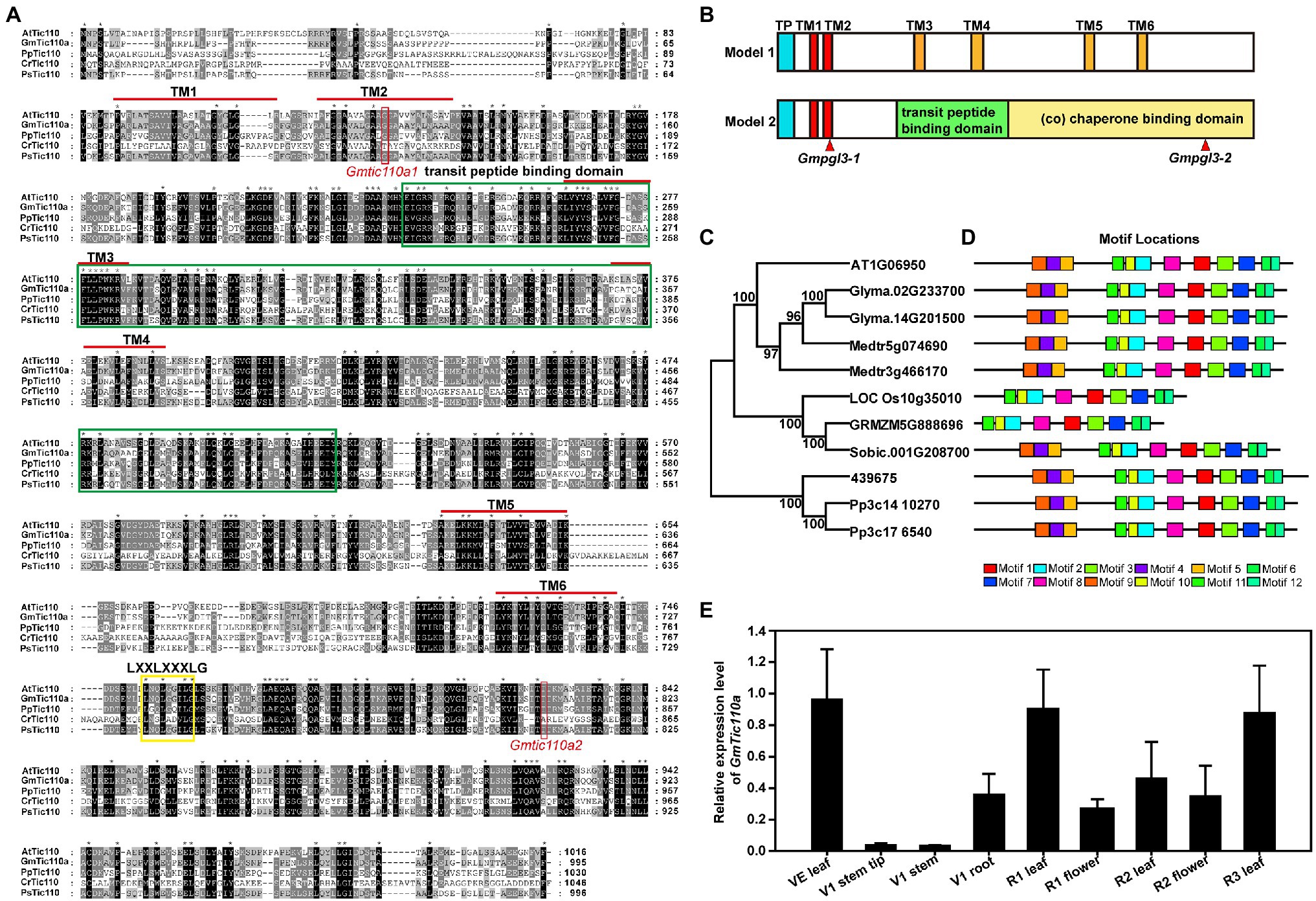
Figure 3. (A) Multiple sequence alignments of the Tic110 protein in Arabidopsis thaliana (At), Glycine max (Gm), Physcomitrella patens (Pp), Chlamydomonas reinhardtii (Cr), and Pisum sativum (Ps). The dark region represents identical amino acids, and the grey region represents similar amino acids. (B) Schematic diagram representations of the two structural models of the Tic110 protein. In terms of the locations of the proposed TM domains, the red boxes represent TM1 and TM2, the orange boxes represent TM3 to TM6, the blue boxes represent transit peptides (TPs), the green box represents TP binding, and the yellow box represents a co-domain. (C) Phylogenetic trees based on the multiple sequence alignments of the Tic110 proteins. Bootstrap values from 1,000 replicates are indicated at each node. (D) Conserved motifs of Tic110 proteins in G. max, A. thaliana, Medicago sativa, Oryza sativa, Zea mays, Sorghum bicolor, Selaginella tamariscina, and P. patens were identified using the MEME search tool. Different motifs (1–12) are represented by boxes with different colors. (E) Tissue-specific expression profiles were determined via qRT–PCR.
Using qRT–PCR, we examined the GmTic110a expression patterns in new leaves at the VE stage; tips, stems, and roots at the V1 stage; leaves and flowers at the R1 stage; leaves and flowers at the R2 stage; and leaves at the R3 stage. GmTic110a was slightly expressed in the tips and stems and had low expression in the roots and flowers. The highest expression levels were detected in the leaves at various stages (new leaves at the VE stage, leaves at the R1 stage, leaves at the R2 stage, and leaves at the R3 stage; Figure 3E), indicating that GmTic110a may play an important role in leaf development and regulatory processes. These results also explained why the GmTic110a mutation severely affected leaf growth at various stages.
CRISPR/Cas9-Mediated GmTic110a Gene Editing of Transgenic Plants
To confirm whether Gmpgl3-1 was the GmTic110a gene, loss-of-function transgenic lines were generated by inducing mutations in the GmTic110a gene using the CRISPR/Cas9 system. The resulting CRISPR/Cas9-induced mutations in three separate mutants led to the development of a GmTic110-specific mutant phenotype (Figure 4A). The results showed that the phenotypes of GmTic110aCR1, GmTic110aCR2, and GmTic110aCR3 were the same as those of Gmpgl3-1 and Gmpgl3-2. The GmTic110aCR1 mutant contains a 7-bp substitution corresponding to the CDS of the GmTic110a gene from the 261st bp to the 273rd bp. The GmTic110aCR2 mutant had a 2-bp deletion from the 271st to the 272nd bp, and the GmTic110aCR3 mutant had a 2-bp deletion at 269th bp of the CDS of GmTic110a and a 1-bp substitution (Gly to Ala) at the 272nd bp (Figure 4B). These data suggested that the GmTic110aCR1, GmTic110aCR2 and GmTic110aCR3 mutants have strong alleles and that complete loss of Gmpgl3 function strongly influences soybean development (Supplementary Table S1). Moreover, the expression levels of Glyma.02G233700 decreased in GmTic110aCR1, GmTic110aCR2, and GmTic110aCR3 (Figure 4C). These results further suggested that Glyma.02G233700 is the GmTic110a gene.
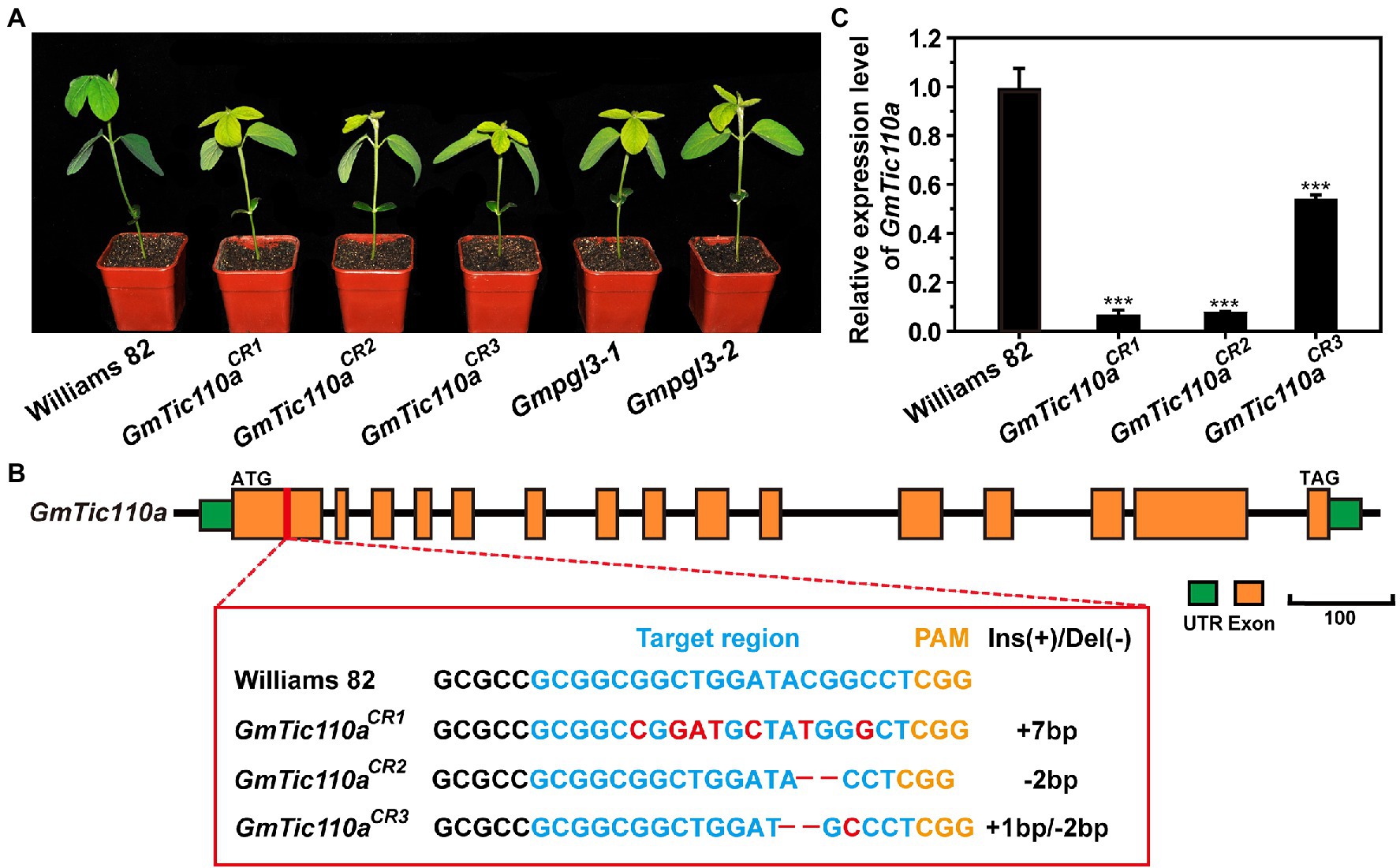
Figure 4. (A) Phenotypes of Williams 82, the Gmpgl3 mutant, and CRISPR/Cas9-edited plants (GmTic110aCR1, GmTic110aCR,2 and GmTic110aCR3). Scale bar = 1 cm. (B) sgRNA target sequences of Williams 82, GmTic110aCR1, GmTic110aCR2, and GmTic110aCR3. The sgRNA target sequence is shown in blue letters, and the protospacer-adjacent motif (PAM) site is shown in yellow letters. The red letters indicate a single-base substitution. – indicates a deletion of the corresponding nucleotide. (C) Relative expression of the GmTic110a gene in unifoliate leaves of Williams 82, the Gmpgl3 mutant, and CRISPR/Cas9-edited plants. The asterisks indicate statistically significant differences, as determined by Student’s t-test (***, p < 0.001), and the error bars represent the standard deviations.
The GmTic110a Protein Localizes to the Chloroplast Inner Membrane
To confirm the subcellular localization of GmTic110a, the colocalization of green fluorescent proteins (GmTic110a-GFP) and the AtPIC1-mCherry marker protein (localization to the inner envelope of chloroplasts; Duy et al., 2007) was analyzed. As shown in Supplementary Figure S3, the GmTic110a protein targeted the inner chloroplast membrane (Figure 5A). At the same time, a single transformation of an empty vector that contained GFP was used as a positive control, and the results revealed that the fluorescent signal of the empty vector was detected throughout the entire cell protoplast (Figure 5G). The results were consistent with the bioinformatics predictions of the subcellular localization of GmTic110a (Supplementary Table S5). To determine the effects of Gmpgl3 mutations on protein localization, we transiently transformed PUC19-GFP-GmTic110aG114A, PUC19-GFP-Gmtic110aT805S, PUC19-GFP-GmTic110aCR1, PUC19-GFP-GmTic110aCR2, and PUC19-GFP-GmTic110aCR3 into Arabidopsis protoplast cells. The results showed that proteins resulting from a mutated or knocked out GmTic110a gene were located in the inner chloroplast membrane, but the GFP fluorescence signal remained diffuse (Figures 5B–F), indicating that GmTic110a mutation or knockout altered the structure of the GmTic110a protein, thereby affecting the subcellular localization of the protein.
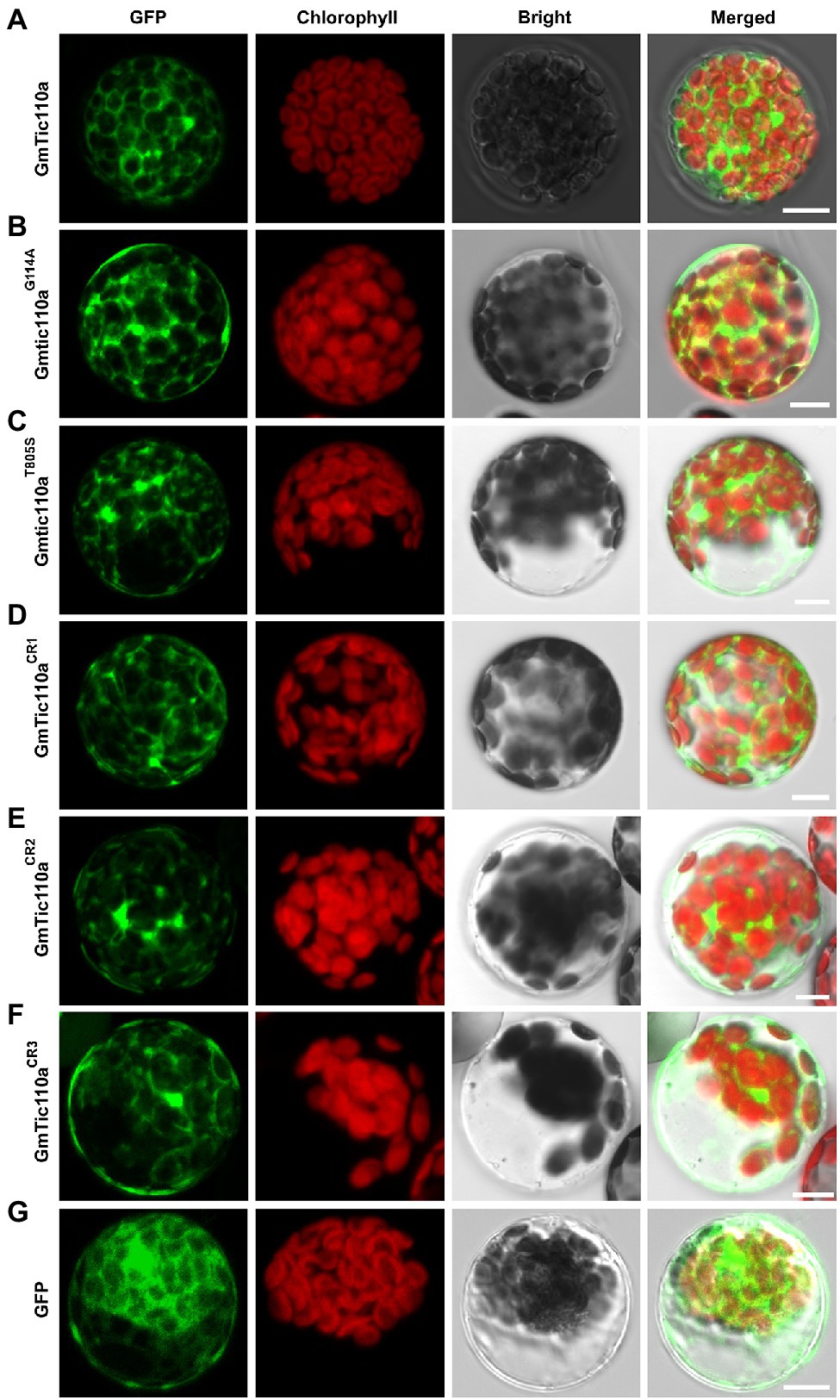
Figure 5. Subcellular localization of GmTic110a, GmTic110aG114A, Gmtic110aT805S, GmTic110aCR1, GmTic110aCR2, and GmTic110aCR3. (A-G) Transient expression of GFP-GmTic110a, GFP-GmTic110aG114A, GFP-Gmtic110aT805S, GFP-GmTic110aCR1, GFP-GmTic110aCR2, GFP-GmTic110aCR3 and GFP in Arabidopsis protoplasts. GFP, GFP fluorescence; Chlorophyll, chlorophyll autofluorescence; Bright, bright field. Merged, merged image of GFP fluorescence, chlorophyll autofluorescence and bright field images. Scale bars = 10 μm.
GmTic110a Interacts With GmTic20, GmTic40a, and GmTic40b
Previous studies have shown that AtTic110 mediates transport across the inner membrane via interactions with AtTic40 and other proteins (Kovacheva et al., 2005; Chiu and Li, 2008; Yuan et al., 2021). Because the GmTic40 protein contains a highly conserved transmembrane (TM) motif, we investigated whether GmTic40 functions in conjunction with AtTic40. We used split luciferase complementation assays to confirm whether GmTic110a proteins could interact with GmTic20, GmTic40a, and GmTic40b (Figures 6A,C,E). The interactions of GmTic110a with GmTic20, GmTic40a, and GmTic40b were verified by co-IP analyses. As shown in Figure 6, we transiently coexpressed GmTic110a with GmTic20, GmTic40a, and GmTic40b in N. benthamiana leaves. Total proteins were isolated, after which they and anti-FLAG magnetic beads were incubated together to immunoprecipitate anti-FLAG. The results showed that GmTic110a, GmTic20, GmTic40a, and GmTic40b were present in the immunoprecipitate (Figures 6B–F), indicating that GmTic110a could interact with GmTic20, GmTic40a, and GmTic40b in vivo.
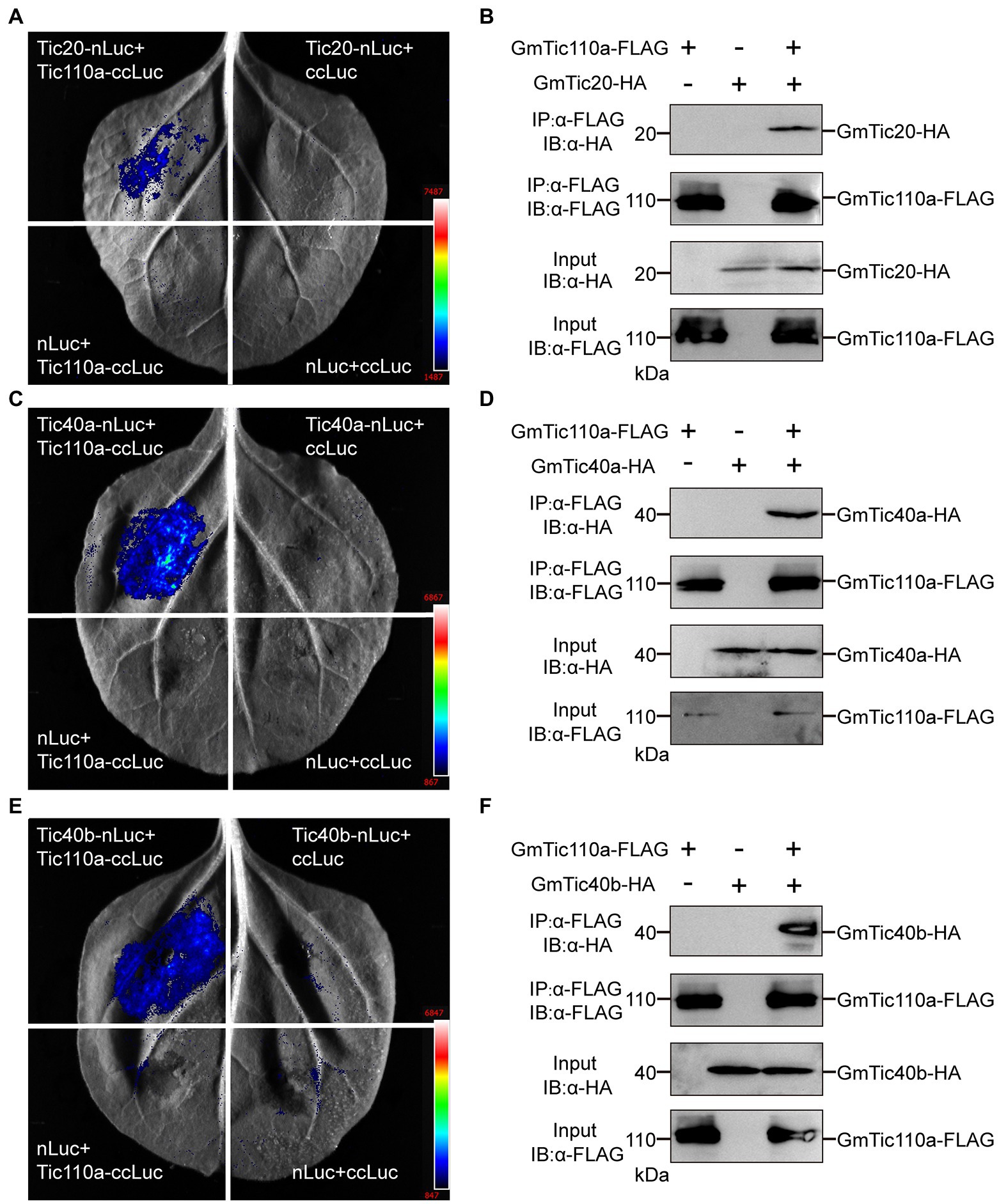
Figure 6. GmTic110a interacts with GmTic20, GmTic40a, and GmTic40b. (A,C,E) Luciferase complementation assay showing that GmTic110a interacts with GmTic20, GmTic40a, and GmTic40b in Nicotiana benthamiana. Luciferase activity was detected 3 days after injection. (B,D,F) Interactions with GmTic20, GmTic40a, and GmTic40b in N. benthamiana according to a Co-IP assay. Immunoblots of the total protein extracts (20% input) and the immunoprecipitation product were performed using an anti-HA antibody (a-HA) or an anti-FLAG antibody (a-FLAG), respectively.
Discussion
Currently, there are two hypotheses concerning the Arabidopsis Tic110 protein structure. Hypothesis 1 proposes that the Tic110 protein is composed of 6 TM domains (TM1, TM2, TM3, TM4, TM5, and TM6), while hypothesis 2 proposes that the Tic110 protein consists of 2 TM domains, one transit peptide-binding domain and 1 co-domain. We compared the amino acid sequences of the Tic110 proteins of A. thaliana, G. max, P. patens, C. reinhardtii, and P. sativum and identified six TM domains, transit peptide-binding domain domains, and co-domains (Figure 3A). By analyzing the structural site of the Gmpgl3 mutant, we confirmed that the mutation site of the Gmpgl3-1 mutant lies in the TM domain of TM2 (position 113 aa), the mutation site of the Gmpgl3-2 mutant lies in the co-domain (amino acid position 805 aa), and the mutation site of the Gmpgl3-3 mutant lies in the TM domain of TM1 (amino acid position 94 aa; Figure 3A). Phenotypic observations revealed that both the Gmpgl3-1 and Gmpgl3-2 mutants showed a pale green leaf phenotype (Figure 1A); Gmpgl3-3 did not display a pale green leaf phenotype (Supplementary Figure S1). We speculated that the site variation of the Gmpgl3-1 mutant affected the TM domain of TM2, indicating that it is the key site of protein function. In contrast, the mutation site of the Gmpgl3-3 mutant is an amino acid at the end of the TM domain of TM1, and the mutation did not cause phenotypic abnormalities, indicating that this site is not a key site of the TM domain. The phenotype of the Gmpgl3-2 mutant is similar to that of Gmpgl3-1, as both show a pale green leaf phenotype. The gene mutation site of Gmpgl3-2 is in the co-domain. Mutations in this site may affect the GmTic110a gene and its function, which may impact the development of chloroplasts. The GmTic110a protein was knocked out by CRISPR/Cas9 technology, and the resulting three plants with point mutations or deletions in the TM1 domain all showed similar phenotypes, which further confirmed that the phenotype was caused by a mutation in this gene. The knockout experiment using CRISPR/Cas9 technology also showed that the protein structural change due to the mutation is the main cause of the pale green leaf phenotype.
A protein must be in a suitable subcellular location to perform its function. Therefore, studying protein subcellular location is highly important for understanding protein function. This study used the online tool WoLF PSORT (Horton et al., 2006) and Target-P 1.1 Server (Emanuelsson et al., 1999) to predict the subcellular location of GmTic110a, which showed that the N-terminal domain contains chloroplast transit peptides, indicating that TIC110a is located on chloroplasts (Supplementary Table S5). To confirm whether the GmTic110a protein is located on the chloroplast, we extracted protoplasts from A. thaliana and transiently expressed GmTic110a-GFP constructs. The results showed that GmTic110a is located mainly on the chloroplast membrane (Figure 5A), which is consistent with the results predicted by the online tool. These results also indicated that the soybean GmTic110a protein is on the inner membrane of the chloroplast and is responsible for the TM transport of proteins into the chloroplast to perform normal functions. It was reported that the Arabidopsis tic110 mutant exhibits pale green leaves and an albino phenotype. Trypsin digestion was used to prove that the Arabidopsis TIC110 fusion protein is located on the chloroplast membrane (Duy et al., 2007; Yuan et al., 2021). In this study, we confirmed that Tic110 proteins of soybean and Arabidopsis have the same subcellular localization. To confirm that the function of the GmTic110a protein is related to its specific membrane localization, we compared the GmTic110a protein localization patterns between mutant, deletion, and wild-type GmTic110a proteins. Point mutations or deletions of the GmTic110a protein alter the normal localization of the protein, and the protein is diffusely distributed throughout the chloroplast membrane (Figures 5B–F). Mutation or deletion of the TM1 domain of GmTic110a affected protein localization and normal function in the cells. The mutation or deletion position occurs after the leader peptide (amino acids 33–56 aa) in TM1, in the TM2 domain or in the co-domain, which does not affect the positioning of the GmTic110a protein in the chloroplast membrane. Point mutations or deletions in these conserved domains may affect the TM positioning or interactions with the protein, thereby affecting the chloroplast membrane localization and function of the GmTic110a protein.
Tic20, Tic110, and Tic40 are considered components of the TIC import machinery in the chloroplast; however, Kikuchi et al. (2013) reported that only Tic56, Tic100, and Tic214 were isolated from the 1-megadalton complex when using a tagged form of Tic20. Ramundo et al. (2020) also demonstrated that the TIC complex contains Tic20, Tic56, Tic100, and Tic214 by combining transcriptomic, biochemical, and genetic tools in the green alga Chlamydomonas, indicating that the complex is widely conserved among photosynthetic organisms (Ramundo et al., 2020). This result conflicts with our finding of the GmTic110a interaction with GmTic20 in the above studies, even though a similar interaction was also reported previously (Kouranov et al., 1998; Chen et al., 2002; Inaba et al., 2003). We suspected that these results were more likely caused by method limitations, and further physical interaction experiments will clear this confusion in the future. It is still unclear how Tic110 and Tic40 interact with the 1-megadalton complex, as they might be recruited to coordinate chaperone functions during later stages and/or are only required for the import of some preproteins (Lee and Hwang, 2018; Nakai, 2018; Thomson et al., 2020). Lee et al. reported that there were no differences in the import of preprotein via the wild-type transit peptide between tic40 and wild-type protoplasts of Arabidopsis thaliana, while the import of N-terminal mutants of the RbcS protein (RbcS-nt) was dependent on Tic40; however, HA (hemagglutinin)-tagged Tic40 showed an intermediate form present in the stroma of tic40 protoplasts (Lee and Hwang, 2019). In this study, we determined that GmTic110a interacted with GmTic20, GmTic40a, and GmTic40b in tobacco leaves (Figure 6); however, GmTic110a may also interact with other unknown partners. Thus, in the future, additional biochemical experiments will be performed to evaluate whether there is a direct interaction between them.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
XF, SY, and HY designed the research. HY, QW, ZZ, TW, and XY performed the experiments. XZ, YY, and JL analyzed the data. HY, QW, and XF wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 31700213 and U21A20215), Science and Technology Development Plan Project of Jilin Province of China (grant nos. 20210302005NC), and Zhejiang Lab (grant nos. 2021PE0AC04).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.892077/full#supplementary-material
Abbreviations
TOC, Translocon at the Outer Envelope Membrane of Chloroplasts; TIC, Translocon at the Inner Envelope Membrane of Chloroplasts; EMS, Ethyl Methanesulfonate; BSA, Bulked Segregation Analysis; SNP, Single-Nucleotide Polymorphism; NCBI, National Center for Biotechnology Information; MEGA 7.0, Molecular Evolutionary Genetics Analysis Software, Version 7.0; MEME, Multiple Expectation Maximization for Motif Elicitation; Pn, Photosynthetic Rate; Gs, Stomatal Conductance; Ci, Intercellular CO2 Concentration; Tr, Transpiration Rate; F0, Initial Fluorescence; Fm, Maximal Fluorescence; Fv, Variable Fluorescence; cDNA, Complementary DNA; qRT–PCR, Real-Time Quantitative PCR.
Footnotes
References
Awan, M. A., Konzak, C. F., Rutger, J. N., and Nilan, R. A. (1980). Mutagenic effects of sodium azide in rice. Crop Sci. 20, 663–668. doi: 10.2135/cropsci1980.0011183X002000050030x
Bailey, T. L., Mikael, B., Buske, F. A., Martin, F., Grant, C. E., Luca, C., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Balsera, M., Goetze, T. A., Kovács-Bogdán, E., Schürmann, P., Wagner, R., Buchanan, B. B., et al. (2009). Characterization of Tic110, a channel-forming protein at the inner envelope membrane of chloroplasts, unveils a response to Ca2+ and a stromal regulatory disulfide bridge. J. Biol. Chem. 284, 2603–2616. doi: 10.1074/jbc.M807134200
Becker, T., Hritz, J., Vogel, M., Caliebe, A., Bukau, B., Soll, J., et al. (2004). Toc12, a novel subunit of the intermembrane space preprotein translocon of chloroplasts. Mol. Biol. Cell 15, 5130–5144. doi: 10.1091/mbc.E04-05-0405
Bölter, B., and Soll, J. (2017). Ycf1/Tic214 is not essential for the accumulation of plastid proteins. Mol. Plant 10, 219–221. doi: 10.1016/j.molp.2016.10.012
Cao, L., Wang, H., Sun, D., and Feng, Y. (2006). Photosynthesis and chlorophyll fluorescence characters of xantha wheat mutants. Acta Botan. Boreali-Occiden. Sin. 26, 2083–2087. doi: 10.3321/j.issn:1000-4025.2006.10.020
Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Mache, R., et al. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. doi: 10.1105/tpc.11.1.57
Chen, X., Smith, M. D., Fitzpatrick, L., and Schnell, D. J. (2002). In vivo analysis of the role of at Tic20 in protein import into chloroplasts. Plant Cell 14, 641–654. doi: 10.1105/tpc.010336
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.2307/40065847
Chiu, C. C., and Li, H. M. (2008). Tic40 is important for reinsertion of proteins from the chloroplast stroma into the inner membrane. Plant J. 56, 793–801. doi: 10.1111/j.1365-313X.2008.03638.x
Chou, M. L., Fitzpatrick, L. M., Tu, S. L., Budziszewski, G., Potter-Lewis, S., Akita, M., et al. (2003). Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J. 22, 2970–2980. doi: 10.1093/emboj/cdg281
Davis, M. S., and Fajer, A. F. (1979). Ligated chlorophyll cation radicals: their function in photosystem II of plant photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 76, 4170–4174. doi: 10.1073/pnas.76.9.4170
de Vries, J., Sousa, F. L., Bölter, B., Soll, J., and Gould, K. (2007). YCF1: a green TIC?. Plant Cell 27, 1827–1833. doi: 10.1105/tpc.114.135541
Du, H. Y., Zeng, X. R., Zhao, M., Cui, X. P., Wang, Q., Yang, H., et al. (2016). Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 217, 90–97. doi: 10.1016/j.jbiotec.2015.11.005
Duy, D., Wanner, G., Meda, A. R., Von Wiren, N., Soll, J., and Philippar, K. (2007). PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19, 986–1006. doi: 10.1105/tpc.106.047407
Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Genty, B., Briantais, J. M., and Baker, N. R. (1989). The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta Gen. Subj. 990, 87–92. doi: 10.1016/S0304-4165(89)80016-9
Gregor, J., and Marsálek, B. (2004). Freshwater phytoplankton quantification by chlorophyll a: a comparative study of in vitro, in vivo and in situ methods. Water Res. 38, 517–522. doi: 10.1016/j.watres.2003.10.033
Highkin, H. R., Boardman, N. K., and Goodchild, D. J. (1969). Photosynthetic studies on a pea-mutant deficient in chlorophyll. Plant Physiol. 44, 1310–1320. doi: 10.1104/pp.44.9.1310
Hormann, F., Kuchler, M., Sveshnikov, D., Oppermann, U., Li, Y., and Soll, J. (2004). Tic32, an essential component in chloroplast biogenesis. J. Biol. Chem. 279, 34756–34762. doi: 10.1074/jbc.M402817200
Horton, P., Park, K. J., Obayashi, T., and Nakai, K. (2006). Protein subcellular localisation prediction with WoLF PSORT. In Proceedings of the 4th Asia-Pacific Bioinformatics Conference 2006.
Hu, R., Fan, C., Li, H., Zhang, Q., and Fu, Y. F. (2009). Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol. Biol. 10:93. doi: 10.1186/1471-2199-10-93
Inaba, T., Alvarez-Huerta, M., Li, M., Bauer, J., Ewers, C., Kessler, F., et al. (2005). Arabidopsis Tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17, 1482–1496. doi: 10.1105/tpc.105.030700
Inaba, T., Li, M., Alvarez-Huerta, M., Kessler, F., and Schnell, D. J. (2003). AtTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J. Biol. Chem. 278, 38617–38627. doi: 10.1074/jbc.M306367200
Ishida, H., and Terakura, K. (1987). Lattice dynamics of TiC(110) and TiC(111) surfaces. Phys. Rev. B Condens. Matter 36, 4403–4406. doi: 10.1103/physrevb.36.4403
Jarvis, P. (2008). Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 179, 257–285. doi: 10.2307/25150450
Jocelyn, B., and Paul, J. (2005). Recognition and envelope translocation of chloroplast preproteins. J. Exp. Bot. 56, 2287–2320. doi: 10.1093/jxb/eri243
Kasmati, A. R., Töpel, M., Patel, R., Murtaza, G., and Jarvis, P. (2011). Molecular and genetic analyses of Tic20 homologues in Arabidopsis thaliana chloroplasts. Plant J. 66, 877–889. doi: 10.1111/j.1365-313X.2011.04551.x
Kessler, F., Blobel, G., Patel, H. A., and Schnell, D. J. (1994). Identification of two GTP-binding proteins in the chloroplast protein import machinery. Science 266, 1035–1039. doi: 10.1126/science.7973656
Ki-Hong, J., Junghe, H., Choong-Hwan, R., Youngju, C., Yong-Yoon, C., Akio, M., et al. (2003). Characterization of a rice chlorophyll-deficient mutant using the T-DNA gene-trap system. Plant Cell Physiol. 44, 463–472. doi: 10.1093/pcp/pcg064
Kikuchi, S., Asakura, Y., Imai, M., Nakahira, Y., Kotani, Y., Hashiguchi, Y., et al. (2018). A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell 30, 2677–2703. doi: 10.1105/tpc.18.00357
Kikuchi, S., Be Dard, J., Hirano, M., Hirabayashi, Y., Oishi, M., Imai, M., et al. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339, 571–574. doi: 10.1126/science.1229262
Kikuchi, S., Oishi, M., Hirabayashi, Y., Dong, W. L., Hwang, I., and Nakai, M. (2009). A 1-megadalton translocation complex containing Tic20 and Tic21 mediates chloroplast protein import at the inner envelope membrane. Plant Cell 21, 1781–1797. doi: 10.1105/tpc.108.063552
Köhler, D., Helm, S., Agne, B., and Baginsky, S. (2016). Importance of translocon subunit Tic56 for rRNA processing and chloroplast ribosome assembly. Plant Physiol. 172, 2429–2444. doi: 10.1104/pp.16.01393
Köhler, D., Montandon, C., Hause, G., Majovsky, P., Kessler, F., Baginsky, S., et al. (2015). Characterization of chloroplast protein import without Tic56, a component of the 1-megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol. 167, 972–990. doi: 10.1104/pp.114.255562
Kouranov, A., Chen, X., Fuks, B., and Schnell, D. J. (1998). Tic20 and Tic22 are new components of the protein import apparatus at the chloroplast inner envelope membrane. J. Cell Biol. 143, 991–1002. doi: 10.1083/jcb.143.4.991
Kouranov, A., and Schnell, D. J. (1997). Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J. Cell Biol. 139, 1677–1685. doi: 10.1083/jcb.139.7.1677
Kovacheva, S., Bédard, J., Patel, R., Dudley, P., Twell, D., Ríos, G., et al. (2005). In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J. 41, 412–428. doi: 10.1111/j.1365-313X.2004.02307.x
Kowalewska, M., Mazur, R., Suski, S., Garstka, M., and Mostowska, A. (2016). Three-dimensional visualization of the tubular-lamellar transformation of the internal plastid membrane network during runner bean chloroplast biogenesis. Plant Cell 28, 875–891. doi: 10.1105/tpc.15.01053
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Lee, D. W., and Hwang, I. (2018). Evolution and design principles of the diverse chloroplast transit peptides. Mol. Cells 41, 161–167.
Lee, D. W., and Hwang, I. (2019). Protein import into chloroplasts via the Tic40-dependent and independent pathways depends on the amino acid composition of the transit peptide. Biochem. Biophys. Res. Commun. 518, 66–71. doi: 10.1016/j.bbrc.2019.08.009
Liu, Z., Li, N., Zhang, Y., and Li, Y. (2020). Transcriptional repression of GIF1 by the KIX-PPD-MYC repressor complex controls seed size in Arabidopsis. Nat. Commun. 11:1846. doi: 10.1038/s41467-020-15603-3
Lonosky, P. M., Zhang, X., Honavar, V. G., Dobbs, D. L., Fu, A., and Rodermel, S. R. (2004). A proteomic analysis of maize chloroplast biogenesis. Plant Physiol. 134, 560–574. doi: 10.1104/pp.103.032003
Lu, X., Lai, Y. C., Du, W. G., Man, W. Q., Chen, S. Y., Zhang, J. S., et al. (2017). A PP2C-1 allele underlying a quantitative trait locus enhances soybean 100-seed weight. Mol. Plant 10, 670–684. doi: 10.1016/j.molp.2017.03.006
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Nakai, M. (2015). The TIC complex uncovered: The alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim. Biophys. Acta 1847, 957–967. doi: 10.1016/j.bbabio.2015.02.011
Nakai, M. (2018). New perspectives on chloroplast protein import. Plant Cell Physiol. 59, 1111–1119. doi: 10.1093/pcp/pcy083
Nakai, M. (2020). Reply: The revised model for chloroplast protein import. Plant Cell 32, 543–546. doi: 10.1105/tpc.19.00821
Okabe, K., and Straub, S. J. (1977). Genetic characterization and high efficiency photosynthesis of an Aurea mutant of tobacco. Plant Physiol. 60, 150–156. doi: 10.2307/4264941
Oshima, C., Aizawa, T., Wuttig, M., Souda, R., Otani, S., Ishizawa, Y., et al. (1987). Surface-phonon dispersion curves of TiC(100). Phys. Rev. B Condens. Matter 36, 7510–7517. doi: 10.1103/physrevb.36.7510
Peng, S., Khush, G. S., Virk, P., Tang, Q., and Zou, Y. (2008). Progress in ideotype breeding toincrease rice yield potential. Field Crops Res. 108, 32–38. doi: 10.1016/j.fcr.2008.04.001
Preiss, S., and Thornber, J. P. (1995). Stability of the apoproteins of light-harvesting complex I and II during biogenesis of thylakoids in the chlorophyll b-less barley mutant chlorina f2. Plant Physiol. 107, 709–717. doi: 10.1104/pp.107.3.709
Ramundo, S., Asakura, Y., Salomé, P. A., Strenkert, D., Boone, M., Mackinder, L. C. M., et al. (2020). Coexpressed subunits of dual genetic origin define a conserved supercomplex mediating essential protein import into chloroplasts. Proc. Natl. Acad. Sci. U. S. A. 117, 32739–32749. doi: 10.1073/pnas.2014294117
Richardson, L. G. L., and Schnell, D. J. (2020). Origins, function, and regulation of the TOC-TIC general protein import machinery of plastids. J. Exp. Bot. 71, 1226–1238. doi: 10.1093/jxb/erz517
Sandhu, D., Atkinson, T., Noll, A., Johnson, C., Espinosa, K., Boelter, J., et al. (2016). Soybean proteins GmTic110 and GmPsbP are crucial for chloroplast development and function. Plant Sci. 252, 76–87. doi: 10.1016/j.plantsci.2016.07.006
Schnell, D. J., Kessler, F., and Blobel, G. (1994). Isolation of components of the chloroplast protein import machinery. Science 266, 1007–1012. doi: 10.1126/science.7973649
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Sohrt, K., and Soll, J. (2000). Toc64, a new component of the protein translocon of chloroplasts. J. Cell Biol. 148, 1213–1222. doi: 10.1083/jcb.148.6.1213
Soll, J. (2004). Protein import into chloroplasts. Mol. Cell. Biol. 5, 529–535. doi: 10.1016/S1369-5266(02)00296-0
Song, X., Wei, H., Cheng, W., Yang, S., Zhao, Y., Li, X., et al. (2015). Development of INDEL markers for genetic mapping based on whole genome Resequencing in soybean. G three 5, 2793–2799. doi: 10.1534/g3.115.022780
Stahl, T., Glockmann, C., Soll, J., and Heins, L. (1999). Tic40, a new "old" subunit of the chloroplast protein import translocon. J. Biol. Chem. 274, 37467–37472. doi: 10.1074/jbc.274.52.37467
Stockinger, E. J., and Walling, L. L. (1994). A chlorophyll a/b-binding protein gene from soybean (Glycine max [L.] Merr.). Plant Physiol. 104, 1475–1476. doi: 10.1104/pp.104.4.1475
Sudhir, K., Glen, S., and Koichiro, T. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Sveshnikova, N., Grimm, R., Soll, J., and Schleiff, E. (2000). Topology studies of the chloroplast protein import channel Toc75. Biol. Chem. 381, 687–693. doi: 10.1515/BC.2000.089
Teng, Y. S., Su, Y. S., Chen, L. J., Lee, Y. J., and Li, H. H. M. (2006). Tic21 is an essential translocon component for protein translocation across the chloroplast inner envelope membrane. Plant Cell 18, 2247–2257. doi: 10.1105/tpc.106.044305
Thomson, S. M., Pulido, P., and Jarvis, R. P. (2020). Protein import into chloroplasts and its regulation by the ubiquitin-proteasome system. Biochem. Soc. Trans. 48, 71–82. doi: 10.1042/BST20190274
Tiller, N., and Bock, R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120. doi: 10.1093/mp/ssu022
Tranel, P. J., Froehlich, J., Goyal, A., and Keegstra, K. (1995). A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO J. 14, 2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x
Tsai, J. Y., Chu, C. C., Yeh, Y. H., Chen, L. J., Li, H. M., and Hsiao, C. D. (2013). Structural characterizations of the chloroplast translocon protein Tic110. Plant J. 75, 847–857. doi: 10.1111/tpj.12249
Vitale, A., Schnell, D. J., Raikhel, N. V., and Chrispeels, M. J. (2015). “Protein sorting and vesicle traffic,” in Biochemistry & Molecular Biology of Plants. 2nd Edn. eds. B. B. Buchanan, W. Gruissem, and R. L. Jones. (Chichester: John Wiley & Sons, Ltd.), 151–190.
Waegemann, K., and Soil, J. (1991). Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1, 149–158. doi: 10.1111/j.1365-313X.1991.00149.x
Wang, B., Lan, T., Wu, W. R., and Li, W. M. (2003). Mapping of QTLs controlling chlorophyll content in rice. Acta Genet. Sin. 30, 1127–1132. doi: 10.1016/S0891-0618(02)00103-5
Wang, D., Liang, X., Bao, Y., Yang, S., Zhang, X., Yu, H., et al. (2020). A malectin-like receptor kinase regulates cell death and pattern-triggered immunity in soybean. EMBO Rep. 21:e50442. doi: 10.15252/embr.202050442
Waters, M. T., and Langdale, J. A. (2009). The making of a chloroplast. EMBO J. 28, 2861–2873. doi: 10.1038/emboj.2009.264.
Yamori, W., Takahashi, S., Makino, A., Price, G. D., Badger, M. R., and Von Caemmerer, S. (2011). The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiol. 155, 956–962. doi: 10.1104/pp.110.168435
Yu, H., Jiang, W., Liu, Q., Zhang, H., Piao, M., Chen, Z., et al. (2015). Expression pattern and subcellular localization of the ovate protein family in rice. PLoS One 10:e0118966. doi: 10.1371/journal.pone.0118966
Yuan, H., Pawlowski, E. G., Yang, Y., Sun, T., Thannhauser, T. W., Mazourek, M., et al. (2021). Arabidopsis ORANGE protein regulates plastid pre-protein import through interacting with tic proteins. J. Exp. Bot. 72, 1059–1072. doi: 10.1093/jxb/eraa528
Zhao, B., Dai, A., Wei, H., Yang, S., Wang, B., Jiang, N., et al. (2016). Arabidopsis KLU homologue GmCYP78A72 regulates seed size in soybean. Plant Mol. Biol. 90, 33–47. doi: 10.1007/s11103-015-0392-0
Zheng, K., Wang, Y., Na, Z., Jia, Q., and Wang, S. (2017). Involvement of paclobutrazol resistance6/KIDARI, an atypical bHLH transcription factor, in auxin responses in arabidopsis. Front. Plant Sci. 8:1813. doi: 10.3389/fpls.2017.01813
Keywords: soybean, Gmpgl3 mutant, gene mapping, GmTic110a, GmTic20, GmTic40a/b
Citation: Yu H, Wang Q, Zhang Z, Wu T, Yang X, Zhu X, Ye Y, Leng J, Yang S and Feng X (2022) Genetic Mapping of the Gmpgl3 Mutant Reveals the Function of GmTic110a in Soybean Chloroplast Development. Front. Plant Sci. 13:892077. doi: 10.3389/fpls.2022.892077
Edited by:
Petr Smýkal, Palacký University Olomouc, CzechiaReviewed by:
Jean-David Rochaix, Université de Genève, SwitzerlandShinji Masuda, Tokyo Institute of Technology, Japan
Copyright © 2022 Yu, Wang, Zhang, Wu, Yang, Zhu, Ye, Leng, Yang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianzhong Feng, ZmVuZ3hpYW56aG9uZ0BpZ2EuYWMuY24=
 Hui Yu
Hui Yu Qiushi Wang1
Qiushi Wang1 Tao Wu
Tao Wu Jiantian Leng
Jiantian Leng Suxin Yang
Suxin Yang Xianzhong Feng
Xianzhong Feng