
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 April 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.890052
GRAS transcription factors play crucial roles in plant growth and development and have been widely explored in many plant species. Garlic (Allium sativum L.) is an important crop owing to its edible and medicinal properties. However, no GRAS transcription factors have been identified in this crop. In this study, 46 garlic GRAS genes were identified and assigned to 16 subfamilies using the GRAS members of Arabidopsis thaliana, Oryza sativa, and Amborella trichopoda as reference queries. Expression analysis revealed that garlic GRAS genes showed distinct differences in various garlic tissues, as well as during different growth stages of the bulbs. Five of these 46 genes were identified as DELLA-like protein-encoding genes and three of which, Asa2G00237.1/Asa2G00240.1 and Asa4G02090.1, responded to exogenous GA3 treatment, and showed a significant association between their transcription abundance and bulb traits in 102 garlic accessions, thereby indicating their role in regulating the growth of garlic bulbs. These results will lay a useful foundation for further investigation of the biological functions of GRAS genes and guiding the genetic breeding of garlic in the future.
The expression of most eukaryotic genes relies on the specific transcription factors (TFs) that bind to or modulate the DNA structure in the regulatory region of genes by activating RNA polymerase to initiate transcription. One of the most important families of transcription factors in plants is the GRAS family, which is named after its first three identified members: gibberellic acid intensive ( GAI), repressor of GAI-3 mutant ( RGA ), and Scarecrow ( SCR). GRAS proteins are typically composed of 400–700 amino acid residues with a variable N-terminal structure. They contain five critical and conserved domains: leucine-heptad repeat I (LHR I), Val-His-Ile-Ile-Asp (VHIID), leucine-heptad repeat II (LHR II), Pro-Phe-Tyr-Arg-Glu (PFYRE), and Ser-Ala-Trp (SAW; Pysh et al., 1999). The representatives of dicotyledons and monocotyledons, Arabidopsis thaliana and rice contain 33 and 57 GRAS members, respectively; of these, eight GRAS subfamilies, namely, LISCL, PAT1, SCL3, DELLA, SCR, SHR, LS, and HAM, are common in both plants (Tian et al., 2004). Recently, more GRAS members consisting of 17 subfamilies have been identified in eight species (Cenci and Rouard, 2017). A total of 9,304 GRAS genes have been added to the Plant transcription factor database (PlantTFDB) so far (Jin et al., 2017), indicating that this family is one of the largest gene families.
GRAS TFs are multifunctional proteins that play various roles in plant growth and development, including gibberellin and (Sun and Gubler, 2004) phytochrome A signal transduction (Bolle et al., 2000), axillary meristem initiation (Tanaka et al., 2015), shoot meristem maintenance (Jha et al., 2020), and root radial patterning (Sabatini et al., 2003). Members of the LS subfamily mainly function as regulators of bud outgrowth; for example, rice MONOCULM 1 (MOC1) is essential for forming axillary meristems and controlling tiller number (Schumacher et al., 1999; Greb et al., 2003). DELLA-like proteins are another type of GRAS TFs with conserved domains of DELLA and TVHYNP and act as repressors of GA signaling, thereby regulating plant growth, including stem elongation (Sun and Gubler, 2004), secondary cell wall biosynthesis (Huang et al., 2015), and stress response (Maggio et al., 2010). DELLA proteins also regulate the MOC1 degradation in rice to control tiller number (Liao et al., 2019). In addition, members of the HAM subfamily are involved in the WUS-CLV3 interaction module, which coordinates with auxin and cytokinin to control the maintenance of stem cells and the differentiation of shoot apical meristems (SAM; Zhou et al., 2015, 2018). In Arabidopsis, SCR and SHR regulate the radial patterning of roots (Di Laurenzio et al., 1996; Helariutta et al., 2000), whereas PAT1 responds to light signaling (Bolle et al., 2000).
Being cultivated for over 5,000 years, Garlic (A. sativum) is not only widely consumed as green nutritional vegetable but also used effectively in medicinal and nutraceutical industries (Martin and Ernst, 2003; Kamenetsky et al., 2015). Bulbs are the main consumed organ of garlic and consist of several cloves, which are abnormal buds that undergo enlarged growth. GRAS members are known to play crucial roles in regulating axillary meristem initiation and bud outgrowth (Sun et al., 2012). Furthermore, a recent study indicated the potential association between genes involved in SAM development and garlic bulb growth (Sun et al., 2020). Therefore, GRAS members probably play a role in the bulb growth of garlic and identification and characterization of GRAS members will be helpful for further research on bulb growth in this Allium crop. However, none of the GRAS members have been identified and reported in garlic. In the present study, we conducted a systematic investigation of GRAS members in garlic and the results provide a basis for analyzing their function in the future.
The GRAS protein sequences in Arabidopsis were downloaded from the TAIR database1 (Tian et al., 2004) and those in rice and Amborella trichopoda were collected and published by Alberto et al. (Cenci and Rouard, 2017). The GRAS proteins of the three plant species were used as reference queries to identify GRAS proteins in garlic using BLASTP (Chen et al., 2020). Subsequently, the preliminarily identified proteins were subjected to conserved domain region analysis using the CDD program in NCBI2 (Lu et al., 2020) and MEME3 (Bailey et al., 2009). After manual verification, only proteins with lengths greater than 200 amino acids and specific GRAS domains were selected for subsequent analysis. The basic features of GRAS proteins, including molecular weight (MW), coding sequence length (CDS), and isoelectric point (pI), were predicted using the ExPASy software4 (Gasteiger et al., 2005), and subcellular localizations were predicted by Localizer 1.0.4 (Sperschneider et al., 2017).
The annotation of garlic genome (Sun et al., 2020), locastion of GRAS genes on the chromosomes/scaffolds was visualized using TBtools (v1.098696; Chen et al., 2020). Multiple sequence alignments of all GRAS proteins were conducted using the JalView (version 2.10.3) software to investigate the protein domains (Waterhouse et al., 2009). The conserved motifs were analyzed using two online tools: MEME (Bailey et al., 2009) and batch CD-Search (Lu et al., 2020). Protein structures and motifs were visualized using TBTools (Chen et al., 2020).
Tissue-specific expression pattern of AsGRAS genes was analyzed by extracting the expression data from the reported transcriptome analysis of garlic (Sun et al., 2020) and an expression heatmap was obtained using TBtools (Chen et al., 2020). Zhu et al. (2019) completed the mRNA sequencing of enlarged bulbs from 102 accessions and introduced an associated transcriptomic method to identify genes related to transcript abundance and bulb traits (Zhu et al., 2019). Based on the expression data of these 102 accessions and the associated transcriptomic method, we performed a correlation analysis of bulb traits and transcript abundance of garlic DELLA genes. Orthologous gene pairs between AsGRASs and AtGRASs were identified and the interaction network of orthologous GRAS genes was predicted using the STRING 11.5 database (von Mering et al., 2005).
Approximately 100 garlic cloves (cv. Ershuizao) were planted in a pot and grown in a greenhouse with a 12 h light period. After 1 month, half of the garlic seedlings were treated by spraying exogenous GA3 and the other half served as the control group. Briefly, GA3 (200 mg/l) and sterile distilled water were sprayed on the leaves of treatment and control groups, respectively. Leaf samples were collected in triplicates from both groups after 0, 4, and 16 h of treatment and were pre-ground to a powder in liquid nitrogen. Total RNA extraction was performed using SteadyPure plant RNA extraction kit AG21019 [Accurate Biotechnology (Hunan) Co., Ltd.] and resultant RNA population was qualified by gel electrophoresis followed by first-strand cDNA biosynthesis [Evo M-MLV RT premix for qPCR AG11706, Accurate Biotechnology (Hunan) Co., Ltd.]. qRT-PCR analysis was performed using CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, United States) with SYBR green Premix Pro Taq HS qPCR Kit AG11701 [Accurate Biotechnology (Hunan) Co., Ltd.]. Primer sequences are listed in Supplementary Table S1.
A total of 61 GRAS candidate genes were obtained from the garlic genome according to the BLAST results. Of these, 15 encoded a protein with <200 amino acids residues and/or without GRAS domains and were filtered. Finally, 46 GRAS members were identified which typically comprised of five conserved domains: LHRI, VHIID, LHRII, PFYRE, and SAW (Supplementary Figure S1; Supplementary Table S2). The 46 identified GRAS genes were distributed on seven of eight garlic chromosomes (except for chromosome 1) and their distribution varied among chromosomes, ranging from 2 to 15 genes per chromosome (Figure 1).
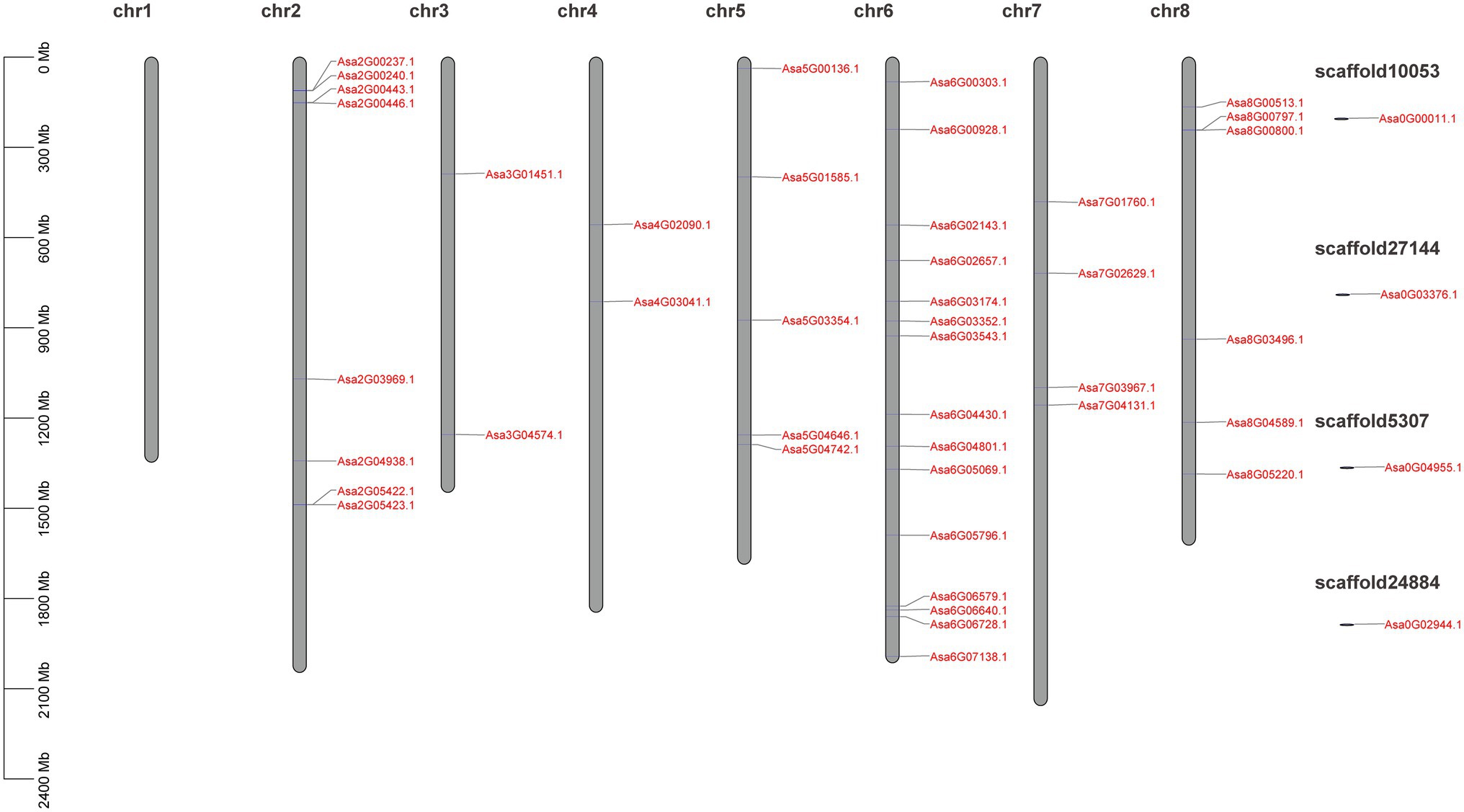
Figure 1. Schematic representation of the chromosomal distribution of the AsGRAS genes. Vertical bars represent garlic chromosomes. The chromosome number is mentioned above each chromosome. The scale on the left represents chromosome length (Mb).
The length of the putative GRAS proteins ranged from 202 to 705 amino acids and the predicted molecular weights ranged from 22 to 80 kDa (Table 1). The theoretical pI varied from 4.87 to 9.76. Except for Asa4G03041.1, all the proteins had a hydropathicity value of <0, indicating the hydrophilic nature of these GRAS proteins. Additionally, only seven proteins showed an instability index value of <40.0 (i.e., threshold value; Table 1), suggesting that most of the garlic GRAS proteins probably had an unstable structure. Subcellular localization prediction identified between 16 and 2 GRAS proteins located in the nucleus and chloroplast, respectively (Table 1). Interestingly, the Asa3G01451.1 protein was predicted with position signals in both the nucleus and chloroplast.
Phylogenetic analysis was performed for 46 AsGRAS members, together with 123 known GRAS proteins from Arabidopsis (33 members), O. sativa (56 members), and A. trichopoda (34 members). The results indicated that 46 AsGRAS members were assigned to 16 of the 17 known GRAS subfamilies (Figure 2). The protein structure investigation indicated that 46 AsGRAS proteins had conserved domains of the GRAS or GRAS superfamily, comprising of a conserved motif structure with LHRI-VHIID-LHRII-PRYRE-SAW domains (Table 1; Figure 3A). However, protein motifs showed larger differences between members from different subfamilies than from the same subfamily, suggesting a diversification of motif sequences among the garlic GRAS subfamily (Figure 3B; Supplementary Figure S2). The details of structural organization revealed that 32 of 46 AsGRAS genes had no introns; in particular, all genes of the LISCL, DELLA, SCL3, SHR, and HAM subfamilies lacked any introns, whereas all genes of the PAT1, RAM1, and SCR subfamilies had more than two exons (Figure 3C).
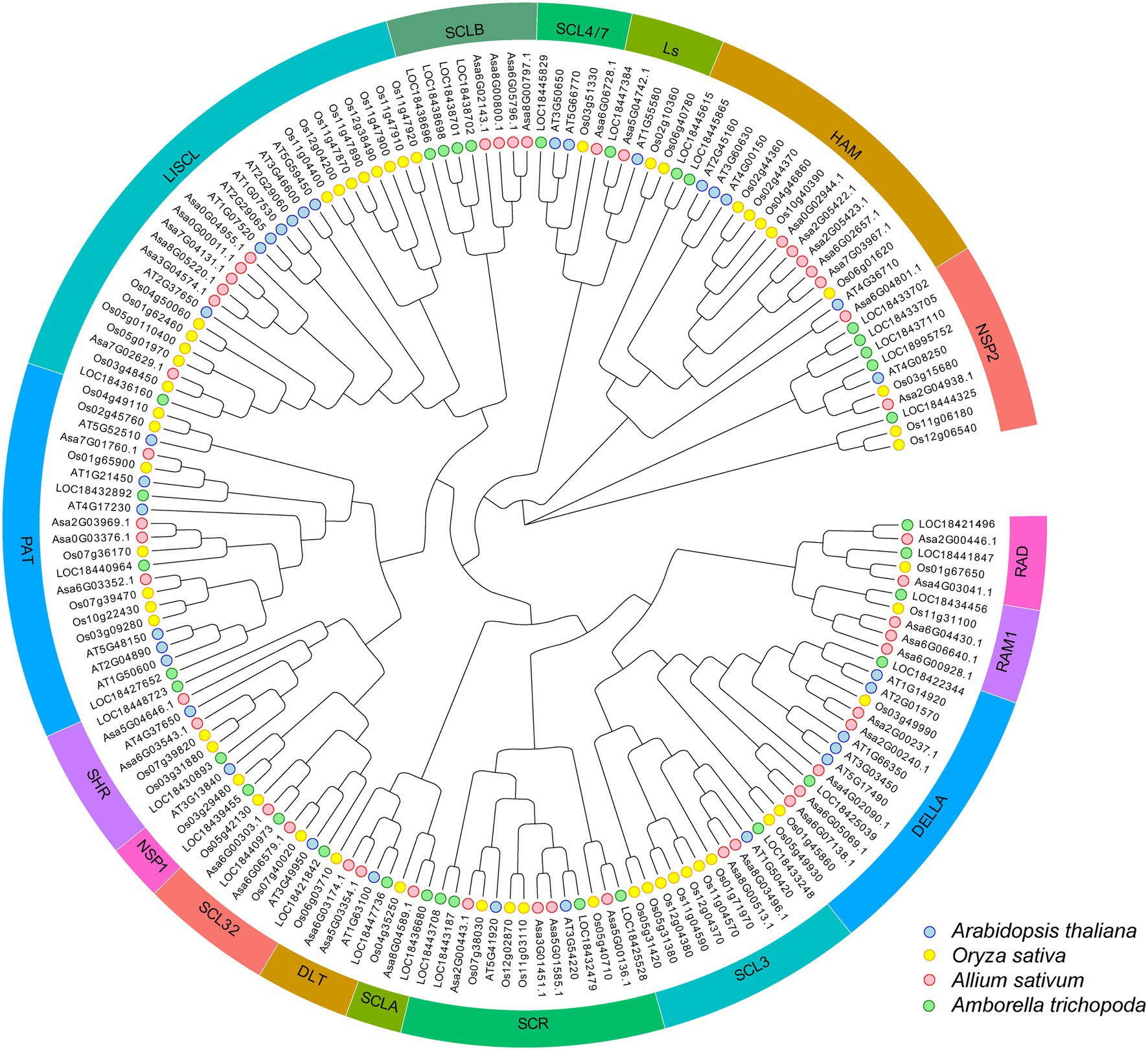
Figure 2. Unrooted phylogenetic tree representing relationships among GRAS families of four plant species. The phylogenetic tree was constructed using the NJ method and shows 17 subfamilies. GRAS proteins from Allium sativum L., Arabidopsis, Oryza sativa, and Amborella trichopoda are marked with red, blue, yellow, and green, respectively.
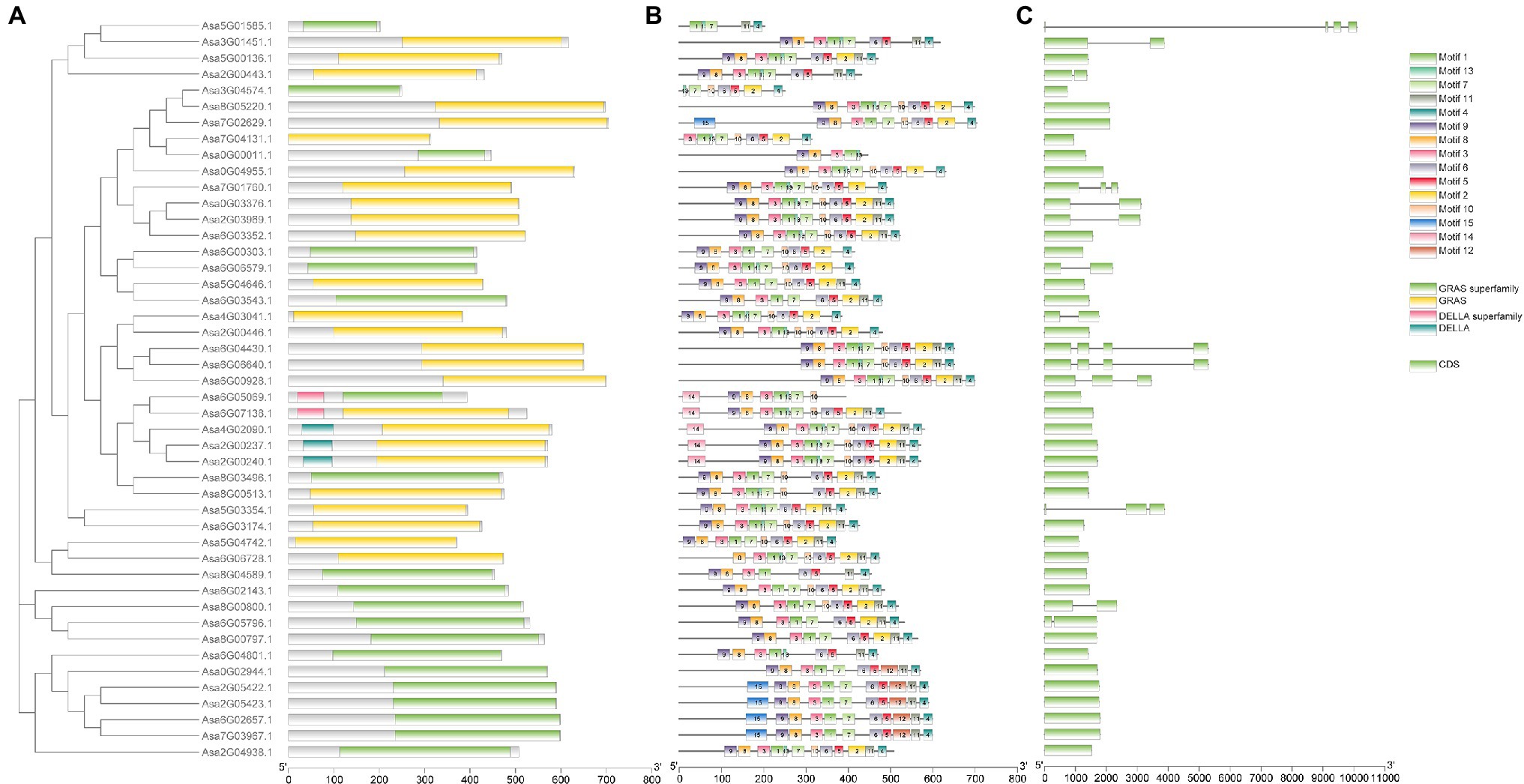
Figure 3. Phylogenetic relationship, gene structure analysis, and motif distribution of garlic GRAS genes. (A) Conserved domain structures of 46 AsGRASs. (B) Amino acid motifs in the AsGRAS proteins are represented by colored boxes. The black lines indicate relative protein lengths. (C) Exons and introns are indicated by rectangles and gray lines, respectively.
The expression patterns of 46 AsGRAS genes in seven tissues and eight bulb-developmental stages were investigated and revealed significant differences (Figure 4). Notably, DELLA-like Asa4G02090.1 showed the highest expression level in all the tissues except roots, whereas nine AsGRAS genes were not expressed at all in the 15 samples including, Asa6G00928.1, Asa4G03041.1, Asa6G04430.1, Asa6G06640.1, Asa2G00446.1, Asa8G00797.1, Asa6G02143.1, Asa6G00303.1, and Asa7G03967.1.
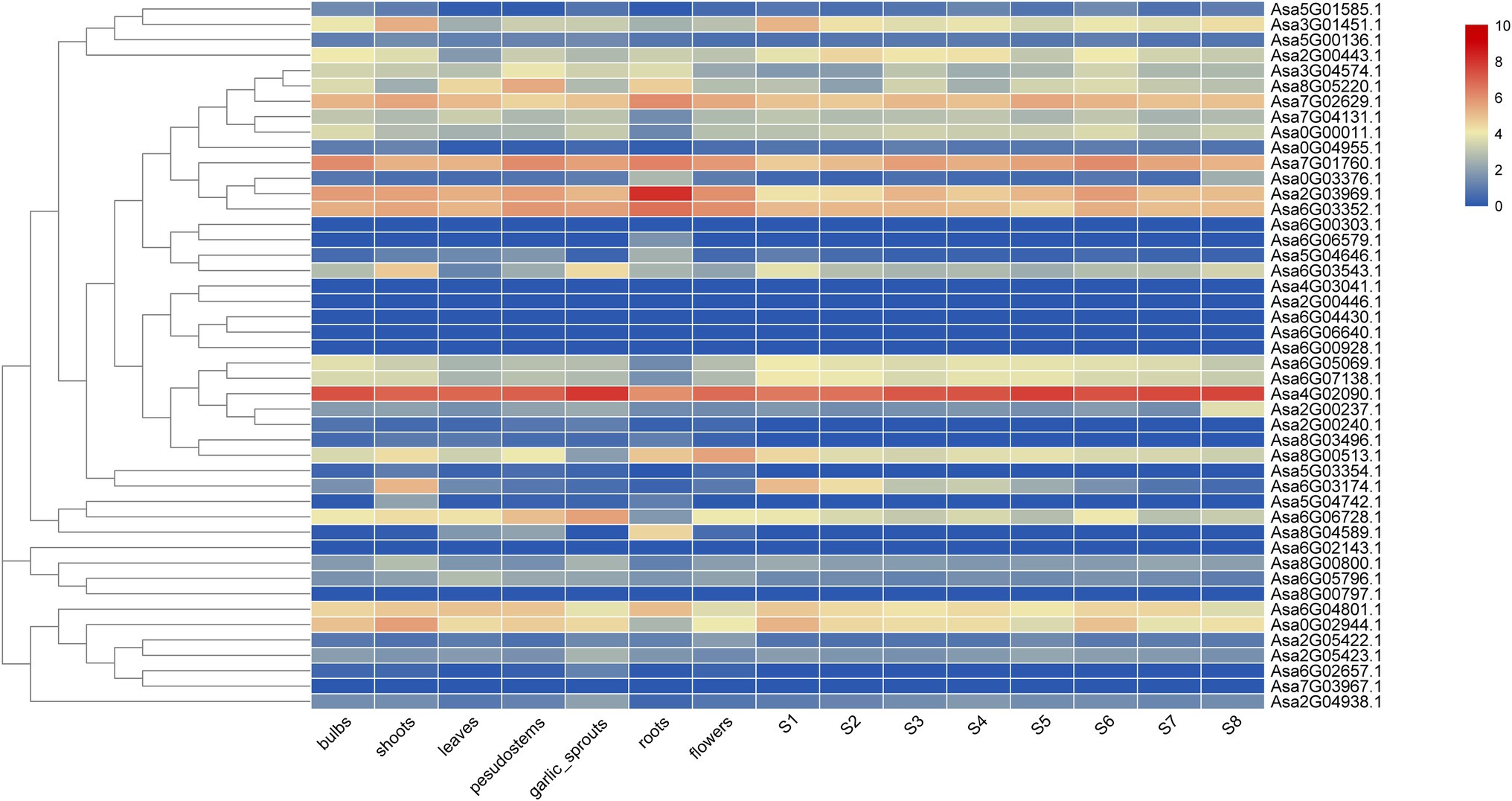
Figure 4. Expression heatmap of garlic GRAS genes in seven tissues and eight bulb growth stages. The colored scale on the right represents the degree of expression, which increases from blue to red.
To understand the biological function of AsGRAS TFs further, their protein–protein interactions were predicted using the ortholog-based method. The results showed that 46 AsGRAS members were orthologs of 20 Arabidopsis GRAS proteins that constituted an interacting network (Figure 5). According to the prediction, AsGRAS proteins interacted with PIF, BZR1, BIN2, and GID1 proteins, which are involved in GA and brassinolide signal transduction mechanism, and lighting-responsive PHYA and PHYB proteins. Furthermore, some members of SCL subfamilies showed a putative interaction with cell cycle regulatory DPA and E2F proteins, indicating a potential role of these garlic GRAS proteins in the regulation of cell cycle.
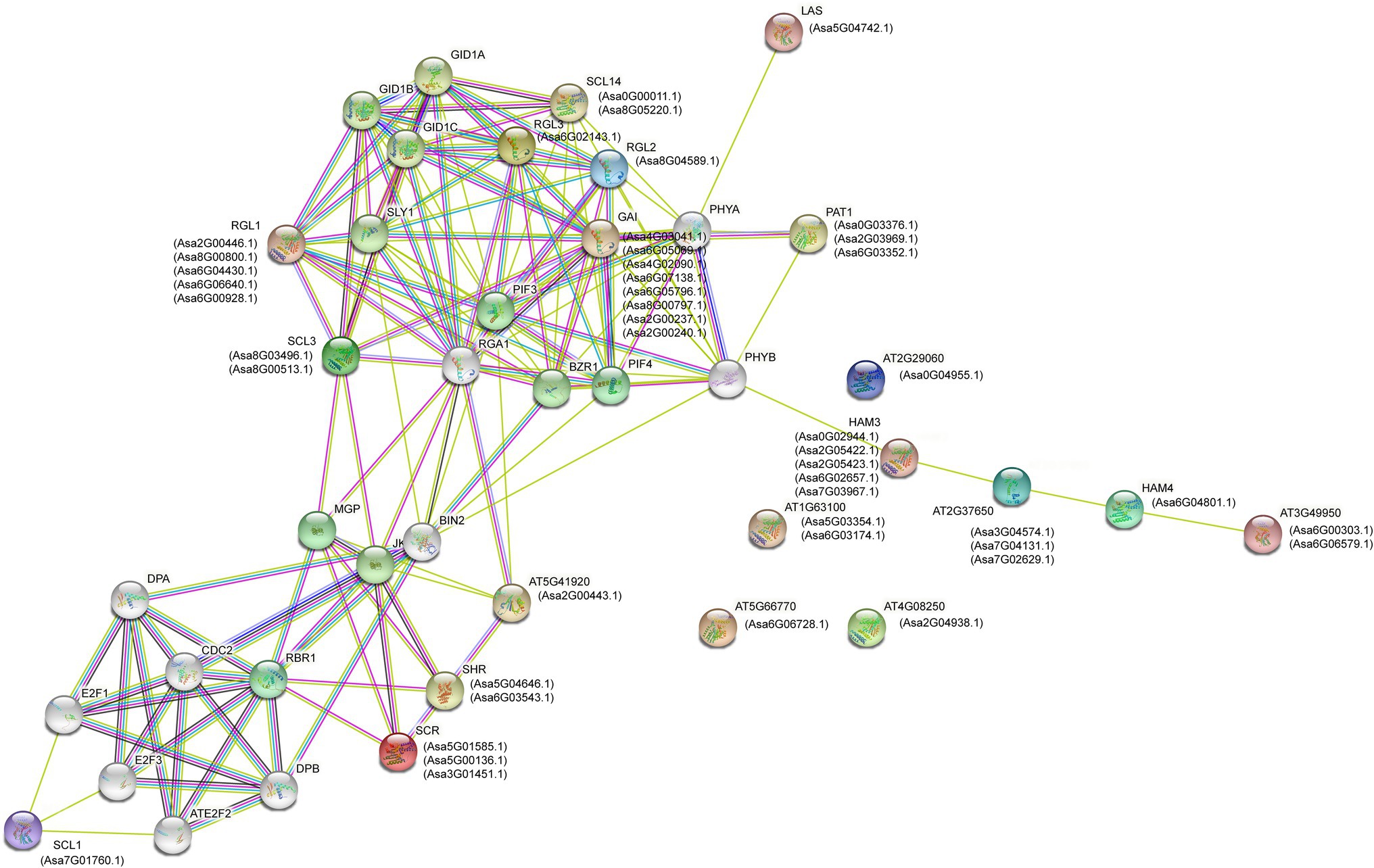
Figure 5. Protein–protein interaction network of the GRAS proteins in garlic with the orthologous ones in Arabidopsis.
As a subfamily of the AsGRAS family, the DELLA subfamily has been frequently studied as these proteins are the central repressors of gibberellin (GA) response (Park et al., 2013). Bioinformatics prediction identified five garlic DELLA members as Asa2G00237.1, Asa2G00240.1, Asa4G02090.1, Asa6G05069.1, and Asa6G07138.1. Of these DELLA-like genes, Asa2G00237.1 and Asa2G00240.1 showed a tandem repeat distribution in the genomic regions, with a complete identical coding sequence; whereas Asa6G05069.1 and Asa6G07138.1 displayed almost identical sequences in their coding regions.
The bulb is the main consumed organ of garlic and consists of several cloves. GA3 treatment can increase the number of cloves in garlic (Liu et al., 2019). In this study, we investigated the expression response of DELLA-like genes to GA3 treatment. Because a complete identical coding sequence makes a challenge to distinguish their transcripts, we performed the expression analysis for these two genes as a whole. Consequently, we found that Asa4G02090.1 and Asa2G00237.1/Asa2G00240.1 showed distinct expression response to GA3 treatment (Figure 6; Supplementary Table S3). Interestingly, the expression of these three genes in response to GA3 treatment was observed during the transition from dark to light, but not in constant light or dark conditions. Furthermore, the associated transcriptomic analysis revealed a significant correlation between the transcript abundance of DELLA-like genes, Asa4G02090.1, and bulb weight and diameter (p = 0.003784 and 0.001473, respectively). Also, the expression of Asa2G00237.1/Asa2G00240.1 displayed a significant association with the clove number trait in 102 garlic accessions (p = 0.03147; Figure 7), indicating a role of these three DELLA-like genes in bulb growth. Collectively, our results indicate that GA3 could result in varied expression of DELLA-like Asa4G02090.1 and Asa2G00237.1/Asa2G00240.1, thereby influencing bulb growth.
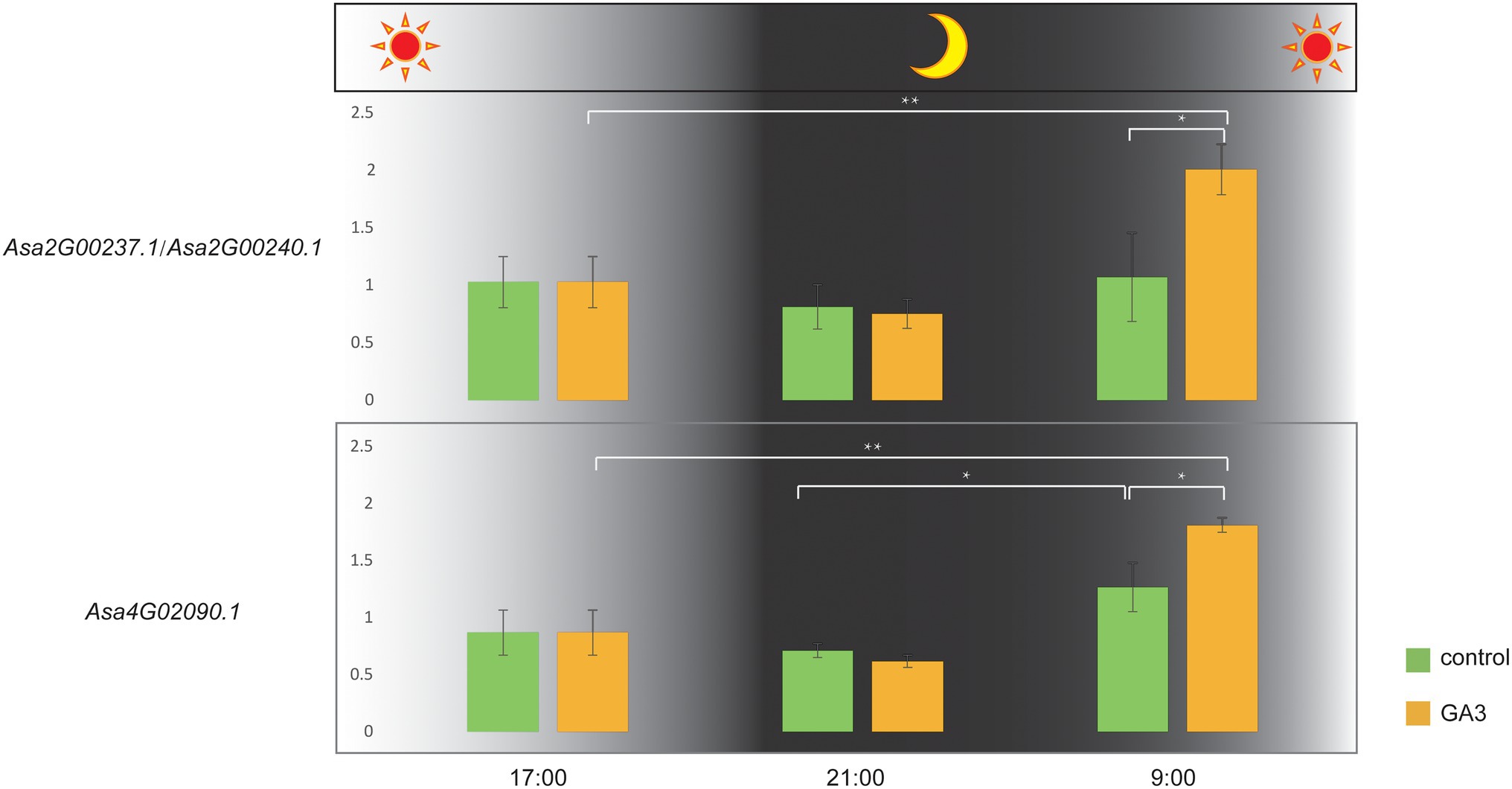
Figure 6. Expression differences of DELLA subfamily genes in one-month-old garlic seedlings sprayed with GA3 solution (sampling times: 0, 4, and 16 h after treatment). * and ** indicate significant differences at the 0.05 and 0.01 levels, respectively. Control: the same amount of distilled water treatment, GA3: 200 mg/l GA3 treatment.
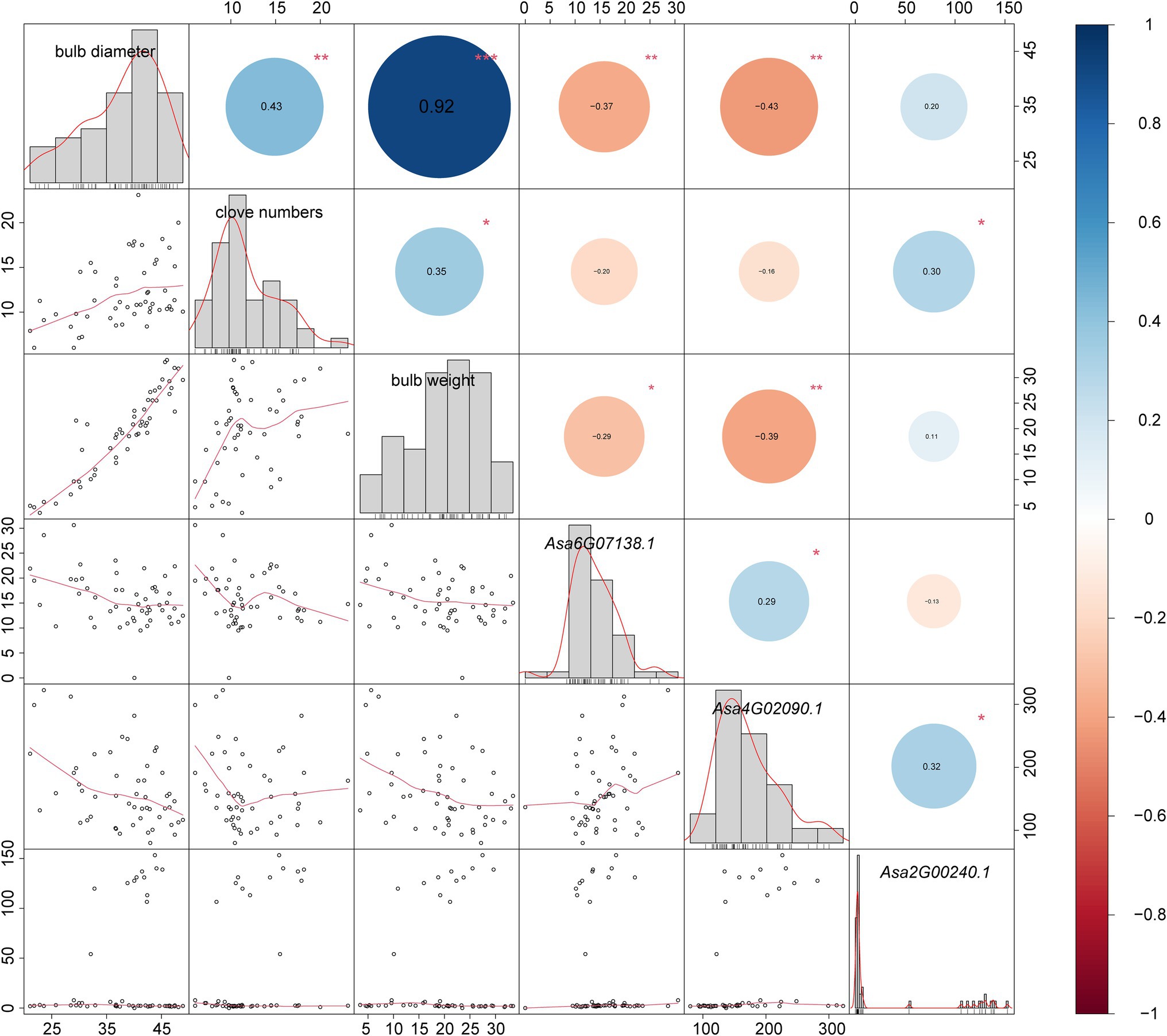
Figure 7. Correlation diagrams between garlic bulb traits (bulb diameter, bulb weight, and clove numbers) and DELLA genes. The correlation diagrams are represented by bar graphs, red curves, and scatter plots simultaneously. Numbers in colored dots are correlation coefficients, dot size indicates correlation degree, red indicates negative correlation, and blue indicates positive correlation. *, **, and *** indicate significant correlations at the 0.05, 0.01, and 0.001 levels, respectively. As the CDS sequences are identical, a pair of tandem repeat genes are represented by one gene ID.
The garlic genome is one of the most complex genomes with a large size (16.9 Gb), high heterozygosity (1.69%), and a high ratio of repetitive sequences (91.3%). Recently, the assembly of garlic genome was completed using a repertoire of five advanced sequencing methodologies (Sun et al., 2020), which made it feasible to identify the genes important for garlic growth and development in Allium crops. Consequently, several gene families, such as GH19, PR, and KNOX, have been systemically characterized (Anisimova et al., 2021; Filyushin et al., 2021; Zhang et al., 2021). The GRAS family is one of the most important transcription factor families that participate widely in the plant growth regulation and development. However, GRAS genes were not identified in garlic so far. In the present study, we identified 46 GRAS members in the garlic genome. The GRAS members were slight fewer than in rice (57; Tian et al., 2004) and maize (86; Guo et al., 2017), indicating a slight contraction of this gene family in garlic genome. According to the difference of grouping criterion, the number of subfamily varied across previous studies, such as eight groups identified in Arabidopsis, rice, and maize (Tian et al., 2004; Guo et al., 2017), and 17 in the report of Cenci and Rouard (2017). To obtain a fine classification, this study analyzed the distribution of garlic GRAS members in 17 reported subfamilies and revealed that 16 of these 17 groups had garlic members. Furthermore, we systematically characterized these GRAS regulators by studying their sequence alignment, gene structure, expression, chromosomal distribution, protein domains encoded by them and their respective interactions, and subcellular localizations. The identification and characterization of AsGRAS members provide an important basis for further investigation of their function in the future.
The garlic bulb is composed of several cloves that are essentially buds in morphology. Previous studies have shown that GAs play a key role in bud formation (Zhang et al., 2020) and the DELLA protein is an essential repressor of gibberellin signal transduction (Ikeda et al., 2001; Sun and Gubler, 2004; Park et al., 2013). For example, SLR1 is the DELLA protein of rice which promotes bud outgrowth by inhibiting the degradation of MOC1 protein, leading to an increase in tiller number (Liao et al., 2019). Similarly, StGA20 induces tuber formation by reducing gibberellin activity in potatoes (Carrera et al., 2000). In addition, exogenous application of GA3 could increase tillers in Welsh onion (Yamazaki et al., 2015) and clove numbers in garlic (Liu et al., 2019). Therefore, gibberellin has a potential role in the development of garlic bulbs. However, the mechanism that gibberellin regulates bulb formation remains unclear. In the present study, we identified three DELLA genes, Asa4G02090.1 and Asa2G00237.1/Asa2G00240.1, whose expression was observed in response to exogenous GA3 treatment and was associated with bulb traits in 102 garlic accessions. These results indicate the possible role of these three DELLA-like genes in the bulb growth. This observation provides an important evidence to explain a previous finding that spraying exogenous GAs could increase the clove number in garlic (Liu et al., 2019).
Light and gibberellins (GAs) are two essential signals that trigger plant developmental processes and their signaling pathways show great overlap (Feng et al., 2008). Light signals can promote the accumulation of DELLA proteins by reducing the GA levels (Achard et al., 2007). In Arabidopsis and potato, the expression of GA 20-oxidase responds to photoperiod to regulate the GA biosynthesis in vivo (Carrera et al., 2000; Porri et al., 2014). PIL5 is a light-labile bHLH TF that interacts with phytochrome, directly binds to the promoter of DELLA, and increases the expression of the DELLA gene in the dark (Oh et al., 2007). In garlic, long daylight can promote bulb enlargement (Wu et al., 2016). GA3 treatment dramatically stimulates lateral bud formation but inhibits the growth of garlic plants and bulbs (Liu et al., 2019). Probably, enlarged size and growth of garlic bulbs are regulated by gibberellin signaling and photoperiod. However, the combined underlying mechanism of gibberellin and daylight in garlic bulb development remains poorly understood. In this study, although none of garlic DELLA-like genes showed an expression response to light treatment, the GA3 response of three DELLA-like genes were promoted in the dark environment, indicating that a potential cross-talk between the signals of GA3 and light. These findings provide insights into the response and adaptation of garlic crops to environmental changes.
A total of 46 garlic GRAS genes were identified and phylogenetically divided into 16 subfamilies. There were five members in the DELLA family, three of which showed a response to exogenous GA3 treatment, with a significant association between their transcription abundances and bulb traits in 102 garlic accessions. Therefore, these three DELLA members have been proposed to be associated with bulb growth. These findings provide a valuable foundation for further studies on the functions of GRAS members in growth and development, especially bulb growth.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
TL coordinated the project, conceived and designed experiments, and corrected the manuscript. XZ performed experiments and wrote the manuscript. XY revised the manuscript. QH and YW contributed to data analysis and managed reagents. XY and GL contributed with valuable discussions. All authors contributed to the article and approved the submitted version.
This research was financially supported by the National Agricultural Science and Technology Innovation Program of China (CAAS-ASTIP-IBFC) and Central Public-interest Scientific Institution Basal Research Fund (1610242021002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Chengjie Chen for kindly providing adaptive modification in analysis tools (TBtools) about family analysis of huge genome.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.890052/full#supplementary-material
1. ^www.arabidopsis.org/index.jsp
2. ^https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
Achard, P., Liao, L., Jiang, C., Desnos, T., Bartlett, J., Fu, X., et al. (2007). DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172. doi: 10.1104/pp.106.092254
Anisimova, O. K., Shchennikova, A. V., Kochieva, E. Z., and Filyushin, M. A. (2021). Pathogenesis-related genes of pr1, pr2, pr4 and pr5 families are involved in the response to fusarium infection in garlic (Allium sativum L.). Int. J. Mol. Sci. 22, 1–20. doi: 10.3390/ijms22136688
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bolle, C., Koncz, C., and Chua, N. H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278. doi: 10.1101/gad.14.10.1269
Carrera, E., Bou, J., Garcia-Martínez, J. L., and Prat, S. (2000). Changes in GA 20-oxidase gene expression strongly affect stem length, tuber induction and tuber yield of potato plants. Plant J. 22, 247–256. doi: 10.1046/j.1365-313X.2000.00736.x
Cenci, A., and Rouard, M. (2017). Evolutionary analyses of GRAS transcription factors in angiosperms. Front. Plant Sci. 8, 1–15. doi: 10.3389/fpls.2017.00273
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Di Laurenzio, L., Wysocka-Diller, J., Malamy, J. E., Pysh, L., Helariutta, Y., Freshour, G., et al. (1996). The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86, 423–433. doi: 10.1016/S0092-8674(00)80115-4
Feng, S., Martinez, C., Gusmaroli, G., Wang, Y., Zhou, J., Wang, F., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479. doi: 10.1038/nature06448
Filyushin, M. A., Anisimova, O. K., Kochieva, E. Z., and Shchennikova, A. V. (2021). Genome-wide identification and expression of chitinase class i genes in garlic (Allium sativum L.) cultivars resistant and susceptible to fusarium proliferatum. Plan. Theory 10:720. doi: 10.3390/plants10040720
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., et al. (2005). “The Proteomics Protocols Handbook,” in Protein Identification and Analysis Tools on the ExPASy Server. ed. J. M. Walker (New York, NY: Humana Press), 571–607.
Greb, T., Clarenz, O., Schafer, E., Herrero, R., Schmitz, G., and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis. Genes Dev. 17, 1175–1187. doi: 10.1101/gad.260703.differentially
Guo, Y., Wu, H., Li, X., Li, Q., Zhao, X., Duan, X., et al. (2017). Identification and expression of GRAS family genes in maize (Zea mays L.). PLoS One 12:e0185418. doi: 10.1371/journal.pone.0185418
Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., et al. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101, 555–567. doi: 10.1016/S0092-8674(00)80865-X
Huang, D., Wang, S., Zhang, B., Shang-Guan, K., Shi, Y., Zhang, D., et al. (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27, 1681–1696. doi: 10.1105/tpc.15.00015
Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., et al. (2001). Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. doi: 10.1105/tpc.13.5.999
Jha, P., Ochatt, S. J., and Kumar, V. (2020). WUSCHEL: a master regulator in plant growth signaling. Plant Cell Rep. 39, 431–444. doi: 10.1007/s00299-020-02511-5
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Kamenetsky, R., Faigenboim, A., Shemesh Mayer, E., Ben Michael, T., Gershberg, C., Kimhi, S., et al. (2015). Integrated transcriptome catalogue and organ-specific profiling of gene expression in fertile garlic (Allium sativum L.). BMC Genomics 16:12. doi: 10.1186/s12864-015-1212-2
Liao, Z., Yu, H., Duan, J., Yuan, K., Yu, C., Meng, X., et al. (2019). SLR1 inhibits MOC1 degradation to coordinate tiller number and plant height in rice. Nat. Commun. 10, 2738–2739. doi: 10.1038/s41467-019-10667-2
Liu, H., Deng, R., Huang, C., Cheng, Z., and Meng, H. (2019). Exogenous gibberellins of garlic (Allium sativum L.) bulb. Sci. Hortic. 246, 298–306. doi: 10.1016/j.scienta.2018.11.003
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Maggio, A., Barbieri, G., Raimondi, G., and de Pascale, S. (2010). Contrasting effects of GA3 treatments on tomato plants exposed to increasing salinity. J. Plant Growth Regul. 29, 63–72. doi: 10.1007/s00344-009-9114-7
Martin, K. W., and Ernst, E. (2003). Herbal medicines for treatment of bacterial infections: a review of controlled clinical trials. J. Antimicrob. Chemother. 51, 241–246. doi: 10.1093/jac/dkg087
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. doi: 10.1105/tpc.107.050153
Park, J., Nguyen, K. T., Park, E., Jeon, J. S., and Choi, G. (2013). Della proteins and their interacting ring finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25, 927–943. doi: 10.1105/tpc.112.108951
Porri, A., Torti, S., Mateos, J., Romera-Branchat, M., García-Martínez, J. L., Fornara, F., et al. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition Fernando Andrés1. Proc. Natl. Acad. Sci. U. S. A. 111, E2760–E2769. doi: 10.1073/pnas.1409567111
Pysh, L. D., Wysocka-Diller, J. W., Camilleri, C., Bouchez, D., and Benfey, P. N. (1999). The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. doi: 10.1046/j.1365-313X.1999.00431.x
Sabatini, S., Heidstra, R., Wildwater, M., and Scheres, B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17, 354–358. doi: 10.1101/gad.252503
Schumacher, K., Schmitt, T., Rossberg, M., Schmitz, G., and Theres, K. (1999). The lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. U. S. A. 96, 290–295. doi: 10.1073/pnas.96.1.290
Sperschneider, J., Catanzariti, A.-M., DeBoer, K., Petre, B., Gardiner, D. M., Singh, K. B., et al. (2017). LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 7:44598. doi: 10.1038/srep44598
Sun, T. P., and Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55, 197–223. doi: 10.1146/annurev.arplant.55.031903.141753
Sun, X., Jones, W. T., and Rikkerink, E. H. A. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem. J. 442, 1–12. doi: 10.1042/BJ20111766
Sun, X., Zhu, S., Li, N., Cheng, Y., Zhao, J., Qiao, X., et al. (2020). A chromosome-level genome assembly of garlic (Allium sativum L.) provides insights into genome evolution and allicin biosynthesis. Mol. Plant 13, 1328–1339. doi: 10.1016/j.molp.2020.07.019
Tanaka, W., Ohmori, Y., Ushijima, T., Matsusaka, H., Matsushita, T., Kumamaru, T., et al. (2015). Axillary meristem formation in rice requires the WUSCHEL ortholog TILLERS ABSENT1open. Plant Cell 27, 1173–1184. doi: 10.1105/tpc.15.00074
Tian, C., Wan, P., Sun, S., Li, J., and Chen, M. (2004). Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 54, 519–532. doi: 10.1023/B:PLAN.0000038256.89809.57
von Mering, C., Jensen, L. J., Snel, B., Hooper, S. D., Krupp, M., Foglierini, M., et al. (2005). STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 33, D433–D437. doi: 10.1093/nar/gki005
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wu, C., Wang, M., Cheng, Z., and Meng, H. (2016). Response of garlic (Allium sativum L.) bolting and bulbing to temperature and photoperiod treatments. Biol. Open 5, 507–518. doi: 10.1242/bio.016444
Yamazaki, H., Shiraiwa, N., Itai, A., and Honda, I. (2015). Involvement of gibberellins in the regulation of tillering in welsh onion (Allium fistulosum L.). Hortic. J. 84, 334–341. doi: 10.2503/hortj.MI-050
Zhang, S., Pan, Y., Zhi, C., Zheng, Y., Wang, X., Li, X., et al. (2021). Genome-wide identification and characterization of KNOTTED-like homeobox (KNOX) homologs in garlic (allium sativum L.) and their expression profilings responding to exogenous cytokinin and gibberellin. Int. J. Mol. Sci. 22:9237. doi: 10.3390/ijms22179237
Zhang, Q. Q., Wang, J. G., Wang, L. Y., Wang, J. F., Wang, Q., Yu, P., et al. (2020). Gibberellin repression of axillary bud formation in Arabidopsis by modulation of DELLA-SPL9 complex activity. J. Integr. Plant Biol. 62, 421–432. doi: 10.1111/jipb.12818
Zhou, Y., Liu, X., Engstrom, E. M., Nimchuk, Z. L., Pruneda, T., Paz, J. L., et al. (2015). Control of plant stem cell function by conserved interacting transcriptional regulators. Nature 517, 377–380. doi: 10.1038/nature13853
Zhou, Y., Yan, A., Han, H., Li, T., Geng, Y., Liu, X., et al. (2018). HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science 361, 502–506. doi: 10.1126/science.aar8638
Keywords: Allium sativum, GRAS gene family, GA3 treatment, lighting response, DELLA
Citation: Zhang X, Yang X, He Q, Wang Y, Liang G and Liu T (2022) Genome-wide Identification and Characterization of the GRAS Transcription Factors in Garlic (Allium sativum L.). Front. Plant Sci. 13:890052. doi: 10.3389/fpls.2022.890052
Received: 05 March 2022; Accepted: 22 March 2022;
Published: 12 April 2022.
Edited by:
Yongzhong Xing, Huazhong Agricultural University, ChinaReviewed by:
Ningyang Li, Shandong Agricultural University, ChinaCopyright © 2022 Zhang, Yang, He, Wang, Liang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Touming Liu, bGl1dG91bWluZ0BjYWFzLmNu; Guolu Liang, bGlhbmdnbEBzd3UuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.