- 1Guangdong Key Laboratory for New Technology Research of Vegetables, Vegetable Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Guangdong Laboratory for Lingnan Modern Agriculture, Guangzhou, China
Cucurbitacins are extremely bitter compounds mainly present in Cucurbitaceae, where Luffa belongs. However, there is no comprehensive analysis of cucurbitacin biosynthesis in Luffa fruit. Therefore, this study analyzed bitter (WM709) and non-bitter (S1174) genotypes of Luffa to reveal the underlying mechanism of cucurbitacin biosynthesis by integrating metabolome and transcriptome analyses. A total of 422 metabolites were detected, including vitamins, essential amino acids, antioxidants, and antitumor substances. Of these, 131 metabolites showed significant differences between bitter (WM709) and non-bitter (S1174) Luffa fruits. The levels of isocucurbitacin B, cucurbitacin D, 23,24-dihydro cucurbitacin E, cucurbitacin F were significantly higher in bitter than in non-bitter Luffa. Transcriptome analysis showed that Bi, cytochromes P450s (CYP450s), and acyltransferase (ACT) of the cucurbitacin biosynthesis pathway, were significantly up-regulated. Moreover, drought stress and abscisic acid (ABA) activated genes of the cucurbitacin biosynthesis pathway. Furthermore, dual-luciferase reporter and yeast one-hybrid assays demonstrated that ABA-response element binding factor 1 (AREB1) binds to the Bi promoter to activate Bi expression. Comparative analysis of the Luffa and cucumber genomes showed that Bi, CYP450s, and ACT are located in the conserved syntenic loci, and formed a cucurbitacin biosynthesis cluster. This study provides important insights into major genes and metabolites of the cucurbitacin biosynthetic pathway, deepening the understanding of regulatory mechanisms of cucurbitacin biosynthesis in Luffa.
Introduction
Luffa [Luffa acutangula (L.) Roxb.] is a member of the Cucurbitaceae family, with a long history of cultivation in tropical and subtropical regions (Kalloo, 1993; Wu et al., 2016). The Luffa genus has nine species, but only L. acutangula Roxb. and L. cylindrica Roem. are domesticated (Walters, 1989; Gautam, 2017; Wu et al., 2020). The crop is highly nutritious, rich in vitamins (A, B, and C), minerals (P, K, Ca, Na, S, Fe, Zn, and Mn), and amino acids (aspartic acid, glutamic acid, alanine, cysteine, and tyrosine; Swetha and Muthukumar, 2016; Shendge and Belemkar, 2018; Zhang et al., 2020). Moreover, Luffa is an important medicinal plant. Luffa has anti-HIV-1, anticancer, hepatoprotective, antidiabetic, antiulcer, antioxidant, fungistatic, analgesic, larvicidal, and immunomodulatory activities (Ng et al., 2011; Kuppast and Mankani, 2012; Shendge and Belemkar, 2018). Recent research also shows that compounds isolated from Luffa spp. function as anti-SARS-CoV-2 main protease (Cao et al., 2021).
Cucurbitacins are oxygenated tetracyclic triterpenoids divided into 12 categories of extremely bitter compounds, mainly in the Cucurbitaceae family (Vieira et al., 2019; Ma et al., 2022). Many cucurbitacins have antitumor activities, but the activities are different because of their varied structural characteristics (Iwanski et al., 2010; Jevtić et al., 2016; Yang et al., 2020). Various Cucurbitaceae crops have different cucurbitacins, namely Cucurbitacins B (CuB) in melon, CuC in cucumber, CuE and CuI in bottle gourd, and CuE in watermelon (Shang et al., 2014; Mashilo et al., 2018; Gong et al., 2021; Xu et al., 2022).
Typically, cucurbitacin synthesis starts with cyclization of 2,3-oxidosqualene to cucurbitadienol in Cucurbitaceae plants, catalyzed by oxidosqualene cyclases (OSCs or Bi; Shang et al., 2014; Zhou et al., 2016). However, a basic helix–loop–helix (bHLH) transcription factor in bitter fruit (Bt) regulates Bi (Shang et al., 2014), while CYP450s oxidize cucurbitadienol, which is further acetylated by ACT to produce various cucurbitacin (Shibuya et al., 2004; Shang et al., 2014; Zhou et al., 2016). Comparative analyses of cucumber, melon, and watermelon genomes showed that Bi, CYP450s, and ACT are located in the conserved syntenic loci, and formed a cucurbitacin cluster with biosynthesis-related functionality (Shang et al., 2014; Zhou et al., 2016; Xu et al., 2022). In cucumber, the UDP-glucosyltransferase UGT73AM3 regulates glucosylation of bitter triterpenoid (Zhong et al., 2017; Yang et al., 2019).
Biotic and abiotic stress (i.e., diseases, insect pests, salt, heat, and drought stress) induce cucurbitacin biosynthesis in plants (Shang et al., 2014; Yoshida et al., 2014; Pan et al., 2022). The plant hormone ABA, produced in abiotic-stressed plants, is critical for mediating plant responses to stressful environmental conditions (Yoshida et al., 2014; Li et al., 2021b). Moreover, ABA-response element binding factors (AREBs) are activated in an ABA-dependent pathway and bind to the ABA-responsive elements (ABREs) in the promoters of downstream genes, thus, activating downstream genes (Yoshida et al., 2014; Li et al., 2021a). AREB1 regulates the ABRE-dependent ABA signaling that enhances abiotic tolerance in Arabidopsis (Uno et al., 2000; Furihata et al., 2006). Moreover, AREB1 has similar functions in many plants, including rice, maize, tomato, and Populus trichocarpa (Uno et al., 2000; Schütze et al., 2008; Orellana et al., 2010; Yoshida et al., 2010; Li et al., 2018). In cucumber, cucurbitacin synthesis-related genes Bt, Bi, CYP450s, and ACT, were up-regulated under drought, cold, or ABA treatments, promoting cucurbitacin C biosynthesis (Shang et al., 2014).
Nevertheless, there is no comprehensive report on cucurbitacin biosynthesis combining the metabolome and transcriptome of Luffa fruits. Besides, the relationship between cucurbitacin biosynthesis and the ABA signaling pathway remains unknown. Therefore, this study combined transcriptome and metabolome analyses to comprehensively analyze the characteristics of metabolic components and molecular mechanisms underlying cucurbitacin biosynthesis in bitter Luffa. Furthermore, the study established a direct relationship between cucurbitacin synthesis and abiotic stress.
Materials and Methods
Plant Materials
S1174, a Luffa inbred line with non-bitter fruit, and WM709, bitter fruit germplasm that has been selfed for multiple generations were used in this study. Both varieties were cultivated in an open field in Baiyun experimental base under normal field conditions, including irrigation, fertilization, pest, and disease control. In brief, organic matter (3,000 kg/ha) and NPK (500 kg/ha) fertilizers were added to the soil before planting, plus two to three additions of NPK compound fertilizer (50 kg/ha) during the growing season. The plants were irrigated at 4–5-day intervals. Fruit flies were controlled by spraying plants with 1.8% avermectin EC. Downy mildew was controlled with 75% chlorothalonil WP 600 solution, while powdery mildew was controlled with 20% triadimefon 2000 solution. Finally, the fruits were collected 10 days after pollination, cut into three sections, wrapped with tin foil, and immediately frozen in liquid nitrogen. This study had three replicates with five fruits per replicate.
Metabolomic Analysis
The Wuhan Metware Biotechnology Co., Ltd.1 extracted, detected, and quantified the metabolites following standard procedures (Chen et al., 2013; Wang et al., 2019). Briefly, the metabolites in freeze-dried fruits were extracted overnight in 70% methanol at 4°C. Afterward, the samples were centrifuged at 10,000 g for 10 min, and the extracts were absorbed and filtered for further analysis on an LC-ESI-MS/MS system (UPLC, Shim-pack UFLC SHIMADZU CBM30A system, Kyoto, Japan; MS, Applied Biosystems 4500 Q TRAP, Applied Biosystems, MA, United States).
All detected metabolites were annotated on the MetWare database and quantified using the multiple reaction monitoring (MRM) method. Metabolite data analysis was conducted using Analyst 1.6.1 software. An orthogonal projection to latent structures discriminant analysis (OPLS-DA) model was established using multiple supervision methods. The relative importance of each metabolite to the OPLS-DA model was checked using the variable importance in projection (VIP) value. Metabolites with significantly different contents were determined using the VIP ≥ 1 and |log2(fold change) | ≥ 1 thresholds. After that, the differential metabolites were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2007) to identify significantly enriched pathways.
Transcriptome Analysis
Total RNA was extracted from six samples (three replicates each of S1174 and WM709) using the TRIZOL reagent (TaKaRa, Kusatsu-Shiga, Japan) following the manufacturer’s instructions. The RNA quality was confirmed using the Agilent 2100 Bioanalyzer (Agilent, CA, United States). The cDNA libraries were sequenced on the Illumina Hiseq2500 platform following the 150 bp paired-end protocol, and reads were processed using Trimmomatic (Bolger et al., 2014). Clean reads were assembled and aligned to the S1174 reference genome (unpublished) using Hisat2 (Kim et al., 2019). DESeq2 (Love et al., 2014) was used to identify differentially expressed genes (DEGs) using the |log2(fold change)| ≥ 1 and padj <0.05 threshold. The functions of DEGs were determined using the Gene Ontology (GO; The Gene Ontology Consortium, 2018) and KEGG databases (Kanehisa et al., 2007).
Synteny Analysis
For synteny analysis, the cucumber genomic FASTA and feature files (.gff files) were downloaded from the CuGenDB2 and used as input files in the one-step MCScanX (Wang et al., 2012) algorithm of TBTools (Chen et al., 2020) toolkit.
Drought Stress and ABA Treatment
For drought stress, Luffa was grown at 26 ± 2°C with a 16/8 h light/dark cycle in a climate-controlled room. The 12-day-old Luffa seedlings were not watered, while control plants were watered at 2–3-day intervals to keep the soil moist. The leaves began to wilt on the fourth day and were sampled the same day. For the ABA treatment, 12-day-old seedlings were sprayed with a 100 μM ABA solution, while control plants were sprayed with mock solution (water). Leaves were sampled 6 h after treatment (ABA or mock). All the samples were frozen in liquid nitrogen for qRT-PCR analyses. One biological replicate consisted of five plant samples, and three biological replicates were analyzed.
Quantitative Real-Time PCR
Total RNA was isolated from the test samples and used to generate cDNA. The qRT-PCR analysis was performed in the Bio-Rad CFX96 machine (Bio-Rad Laboratories, CA, United States) using the SYBR Premix Ex Taq Kit (TaKaRa, Kusatsu-Shiga, Japan). The 2−△△CT method was used to calculate the fold change of gene expression, and the data were log2 transformed before analysis (Liu et al., 2017). The gene-specific primers used are listed in Supplementary Table S1, and 18s rRNA was the internal reference gene.
Dual-Luciferase Reporter Assays
Here, full-length open reading frames (ORFs) of AtAREB1 and LaAREB1 were amplified from the Arabidopsis thaliana and S1174 cDNA and cloned into pGreenII 62-SK (Hellens et al., 2005). The -2000--1201 region of LaBi promoter fragment was ligated into pGreen II 0800-LUC (Hellens et al., 2005). Arabidopsis protoplasts were prepared and transformed with the described vectors, following a previous protocol (Yoo et al., 2007). The firefly luciferase (LUC) and renilla luciferase (REN) activity assays were performed via the dual-luciferase reporter assay system (Promega, WI, United States). The promoter activity was shown as the ratio of LUC to REN.
Yeast One-Hybrid Assays
Full-length ORFs of AtAREB1 and LaAREB1 were amplified from the A. thaliana, S1174 cDNA, and cloned into pGADT7. The -2000--1201 region of the LaBi promoter fragment was ligated into pHis2. Yeast one-hybrid (Y1H) analysis (Clontech Laboratories Inc., CA, United States) was performed to examine AtAREB1 and LaAREB1 binding to the LaBi promoter.
Results
Identification of Metabolites in the Fruits of Two Luffa Varieties
Metabolome analysis was performed to unravel the metabolite differences between S1174 and WM709 varieties of Luffa. The total ions current (TIC) and multiple reactions monitoring (MRM) profiles detected 422 metabolites, including 73 lipids, 67 phenolic acids, 62 amino acids and derivatives, 58 other compounds, 42 nucleotides and derivatives, 41 organic acids, 34 flavonoids, 24 alkaloids, 14 terpenoids and seven lignans and coumarins in the two varieties (Figure 1A; Supplementary Table S2). A principal component analysis (PCA) of the detected metabolites showed that the first two principal components explain 49.68% (PC1) and 21.89% (PC2) of the samples’ variance, respectively (Figure 1B). Besides, a heatmap analysis clearly distinguished the metabolites in the two varieties (Figure 1C). Thus, the six samples were separated into two groups according to their OPLS-DA (Supplementary Figure S1A). R2X, R2Y, and Q2 were 0.728, 0.999, and 0.895, respectively (Supplementary Figure S1B). The R2 and Q2 values were close to 1.0, suggesting a satisfactory model with a reliable predictive ability.
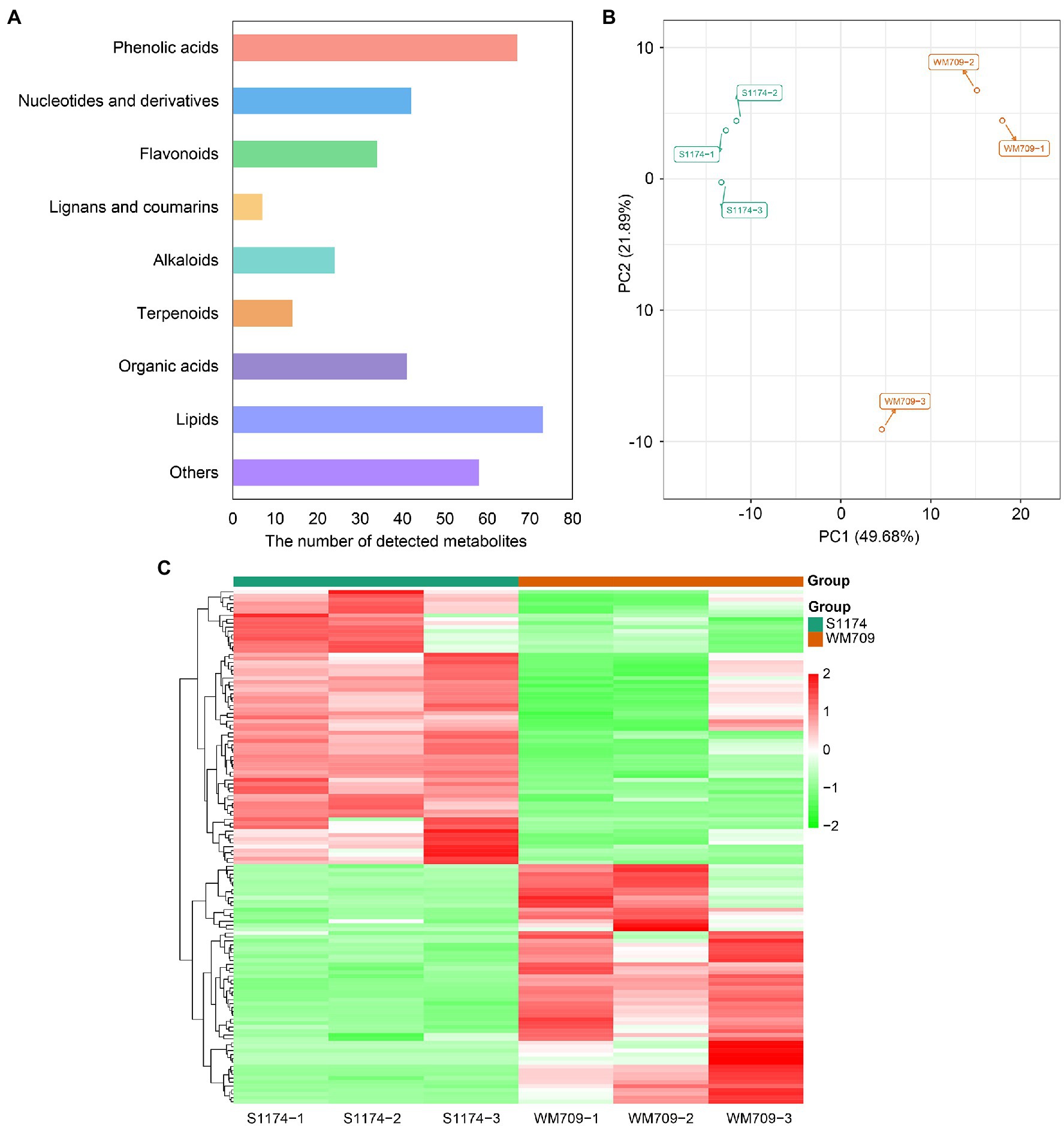
Figure 1. Preliminary analysis of metabolomics data. (A) Classification of detected metabolites. (B) Principal component analysis (PCA) of detected metabolites. (C) Clustered heatmap based on the metabolites between S1174 and WM709.
Functional Analysis of Metabolites
The differentially accumulated metabolites (DAMs) between the two Luffa varieties were determined based on the variable importance in the projection (VIP) ≥ 1 and a | Log2Fold Change | ≥ 1. Essentially, 131 metabolites were significantly different between the varieties, with 61 up-regulated and 70 down-regulated (Supplementary Table S3). The DAMs included 26 lipids, 25 phenolic acids, 24 nucleotides and derivatives, 11 terpenoids, 11 flavonoids, nine organic acids, nine other compounds, six amino acids and derivatives, five alkaloids, and five lignans and coumarins.
KEGG pathway analysis for the DAMs showed that they were enriched in metabolism, ABC transporters, and aminoacyl−tRNA biosynthesis pathways (Figure 2A; Supplementary Table S4). The top five significantly enriched pathways were glycerophospholipid (ko00564), alpha−Linolenic acid (ko00592), purine (ko00230), pyrimidine (ko00240), and linoleic acid metabolism (ko00591). In addition, the pathway of ubiquinone and terpenoid-quinone biosynthesis metabolite (ko00130) was significantly enriched. In bitter Luffa S1174, the high level of metabolites enriched purine metabolism (ko00230), pyrimidine metabolism (ko00240), and ABC transporters (ko02010; Supplementary Figure S2).
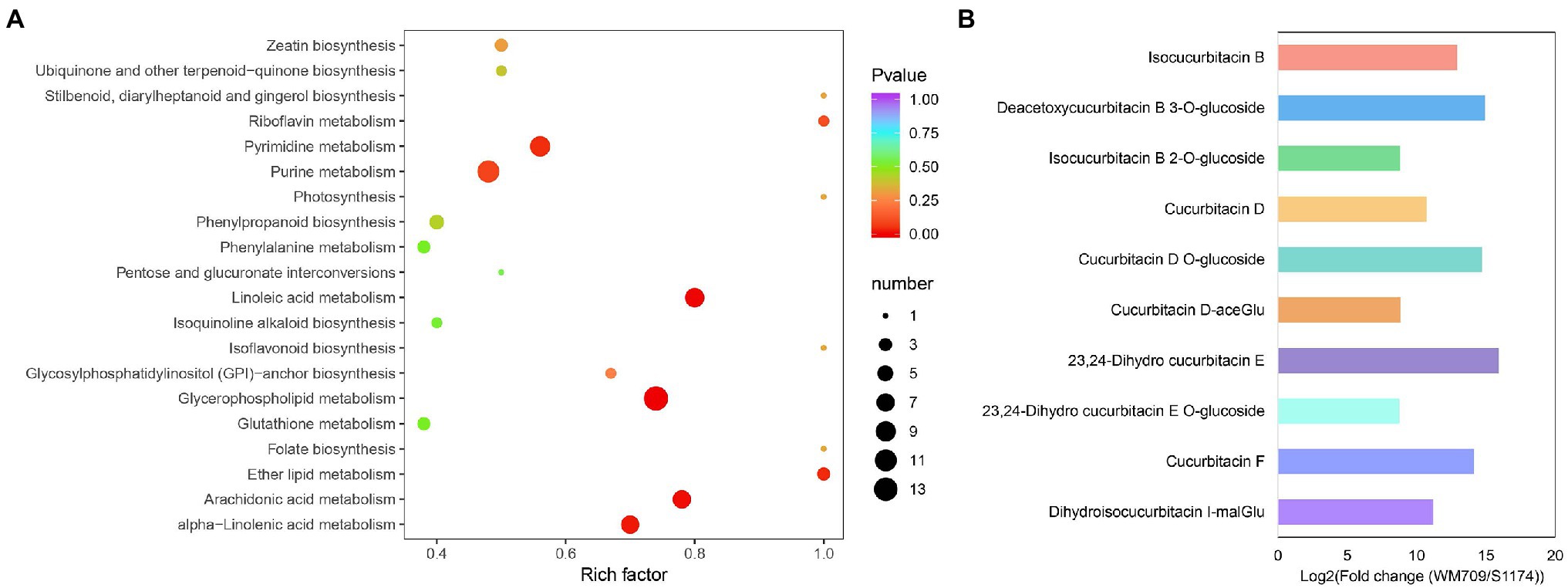
Figure 2. Analyses of differentially accumulated metabolites (DAMs) between WM709 and S1174. (A) The top 20 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. (B) Detected cucurbitacins in WM709.
Moreover, the contents of terpenoids, including cucurbitacin D, cucurbitacin F, 23,24-dihydro cucurbitacin E, isocucurbitacin B, deacetoxycucurbitacin B 3-O-glucoside, cucurbitacin D O-glucoside, isocucurbitacin B 2-O-glucoside, cucurbitacin D-aceGlu, 23,24-dihydro cucurbitacin E O-glucoside, and dihydroisocucurbitacin I-malGlu, were significantly higher in bitter Luffa than in non-bitter Luffa (Figure 2B; Supplementary Table S4).
Differentially Expressed Genes in the Fruits of Two Luffa Varieties
The transcriptome sequence of S1174 and WM709 generated 54.20 Gb of clean data, with >94.19% of the bases scoring Q30 (Supplementary Table S5). Over 95.42% of clean reads were mapped to the reference genome, generating >92.76% uniquely mapped sequences (Supplementary Table S6). The correlation coefficients (Supplementary Figure S3A) and PCA (Supplementary Figure S3B) indicated expression pattern similarities between replicate samples. Finally, 2,634 DEGs were identified, including 1,455 up-regulated and 1,179 down-regulated genes (Supplementary Table S7). The transcriptome analysis identified 222 DEGs (98 up-regulated and 124 down-regulated) belonging to 37 families of transcription factors (TFs) between WM709 and S1174 (Supplementary Figure S4). The MYB, AP2/ERF, bHLH, C2C2, and NAC families were the top five TF among the DEGs (Supplementary Figure S4).
Functional Analysis of DEGs
The GO enrichment analysis identified 135 significantly enriched GO terms (Figure 3A; Supplementary Table S8). The significantly enriched GO terms included 82 biological processes, such as secondary metabolic process (GO:0019748), flavonoid biosynthetic process (GO:0009813), and photosynthesis (GO:0015979). The 38 significantly enriched molecular functions included galactosidase activity (GO:0015925), glucosidase activity (GO:0015926), chlorophyll binding (GO:0016168), and UDP-glycosyltransferase activity (GO:0008194). Fifteen GO terms were significantly enriched in the cellular component, including thylakoids (GO:0009579) and photosystem (GO:0009521).
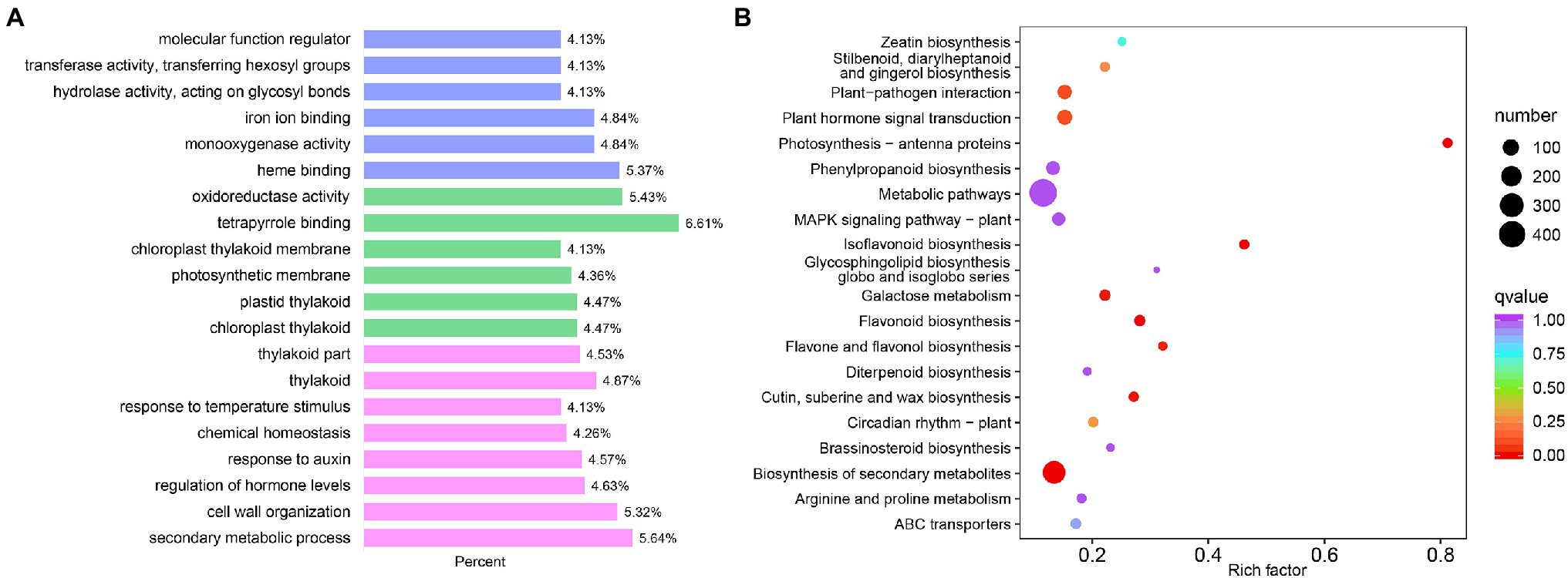
Figure 3. Differentially expressed genes between WM709 and S1174. (A) Top 20 of Gene Ontology (GO) annotations of differentially expressed genes (DEGs). Blue bar, biological process; Green bar, molecular function; and Pink bar, cellular components. (B) Top 20 enriched GO annotations using the DEGs.
Besides, the KEGG enrichment analysis showed that the DEGs were mainly involved in metabolic pathways (ko01100), biosynthesis of secondary metabolites (ko01110), photosynthesis-antenna proteins (ko00196), flavonoid biosynthesis (ko00941), flavone and flavanol biosynthesis (ko00944; Figure 3B; Supplementary Table S9). Additionally, terpenoid synthesis, including terpenoid backbone biosynthesis (ko00900), monoterpenoid biosynthesis (ko00902), diterpenoid biosynthesis (ko00904), sesquiterpenoid and triterpenoid biosynthesis (ko00909), ubiquinone, and another terpenoid-quinone biosynthesis (ko00130) were significantly enriched.
Expression of Cucurbitacin Biosynthetic Pathway Genes
Considering the cucurbitacin biosynthesis and regulatory pathway in cucumber, we used the amino acid sequences of cucumber genes as the query. Thus, a BLASTP search was performed for Bt, Bi, ACT, CYP450s, and UDP-Glucosyltransferases (UGTs) homologous genes in Luffa (Supplementary Table S10). The results showed that both S1174 and WM709 had undetectable levels of LaBt (Lac02g016030; Supplementary Table S7). The LaBi (Lac05g013070) gene, homologous to cucumber Bi, encodes an oxidosqualene cyclase (OSC) responsible for the first committed step in the generation of cucurbitane skeleton, catalyzing 2,3-oxidosqualene to cucurbitadienol, was up-regulated in WM709 than S1174 (Figure 4A; Supplementary Table S7). Besides, five CYP450s (La160, La180, La710, La890, and La490) and ACT gene were significantly up-regulated (Figure 4B; Supplementary Table S7). The expression of La170 gene (located in chromosome 5) was not detected in S1174 and WM709 Luffa varieties (Supplementary Tables S7 and S10). Notably, Cs530, Cs540, Cs550, and Cs560 were homologous to La530 (Figure 4A). The expression levels of La530 and La510 were undetectable in S1174 and WM709 (Figure 4A; Supplementary Table S7). A Bi cluster containing LaBi, LaACT, and three CYP450s (Figure 4A) was in the synteny block between chromosome 6 of cucumber and chromosome 5 of Luffa (Figure 4B). Besides, the metabolome levels of five cucurbitacin glucosides increased in bitter Luffa WM709 compared to non-bitter Luffa S1174 (Figure 2B; Supplementary Table S4). Sixty-six UGT genes were identified in the Luffa genome, including 16 significantly regulated genes (14 up-regulated and two down-regulated; Figure 4C; Supplementary Table S7).
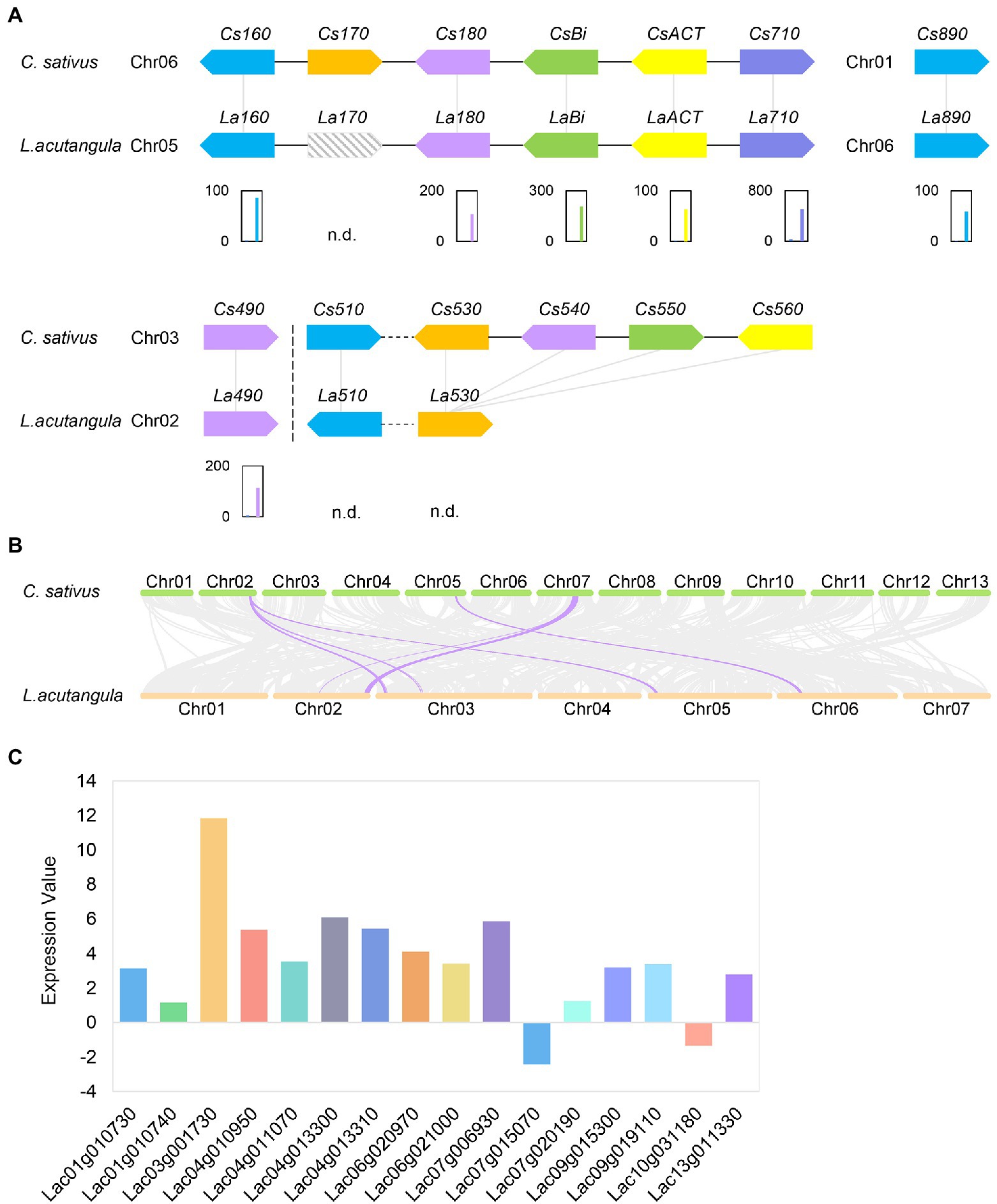
Figure 4. Cucurbitacin biosynthesis genes. (A) The distribution and expression of cucurbitacin biosynthesis genes are shown by the fragments per kilobase of transcript per million (FPKM) values. Left bar, S1174 and Right bar, WM709. n.d., not detected. (B) Synteny blocks between cucumber and Luffa. (C) Expression analysis of UGTs between WM709 and S1174 Luffa varieties. The height of the columns represents the log2 [fold change (WM709/S1174)].
Drought Stress and ABA Treatment Activate Cucurbitacin Biosynthesis Genes
Luffa seedlings were subjected to drought stress and ABA treatment to verify whether stress activates cucurbitacin genes. The results showed that drought stress and ABA treatment significantly increased the expression of Bi, ACT, and CYP450s (Figures 5A,B). Indeed, the Bi promoters in Luffa contained ABREs (Supplementary Table S11). AREB1 might directly regulate LaBi expression; therefore, subsequent dual-luciferase reporter assays were performed to determine whether LaBi is a direct target gene of the LaAREB1 protein. The results showed that co-expressing 35S::LaAREB1 and LaBipro::LUC drastically increased the luminescence intensity, indicating that LaAREB1 binds to the LaBi promoter. These results were further confirmed by co-expressing 35S::AtAREB1 and LaBipro::LUC (Figure 5C), and performing yeast one-hybrid assays. As indicated in Figure 5D, co-transformed yeast strains harboring pGADT7-LaAREB1 and pHIS2-LaBi exhibited normal growth in a selective medium, confirmed by pGADT7-AtAREB1. These results show that AREB1 positively regulates LaBi expression by directly binding to its promoter.
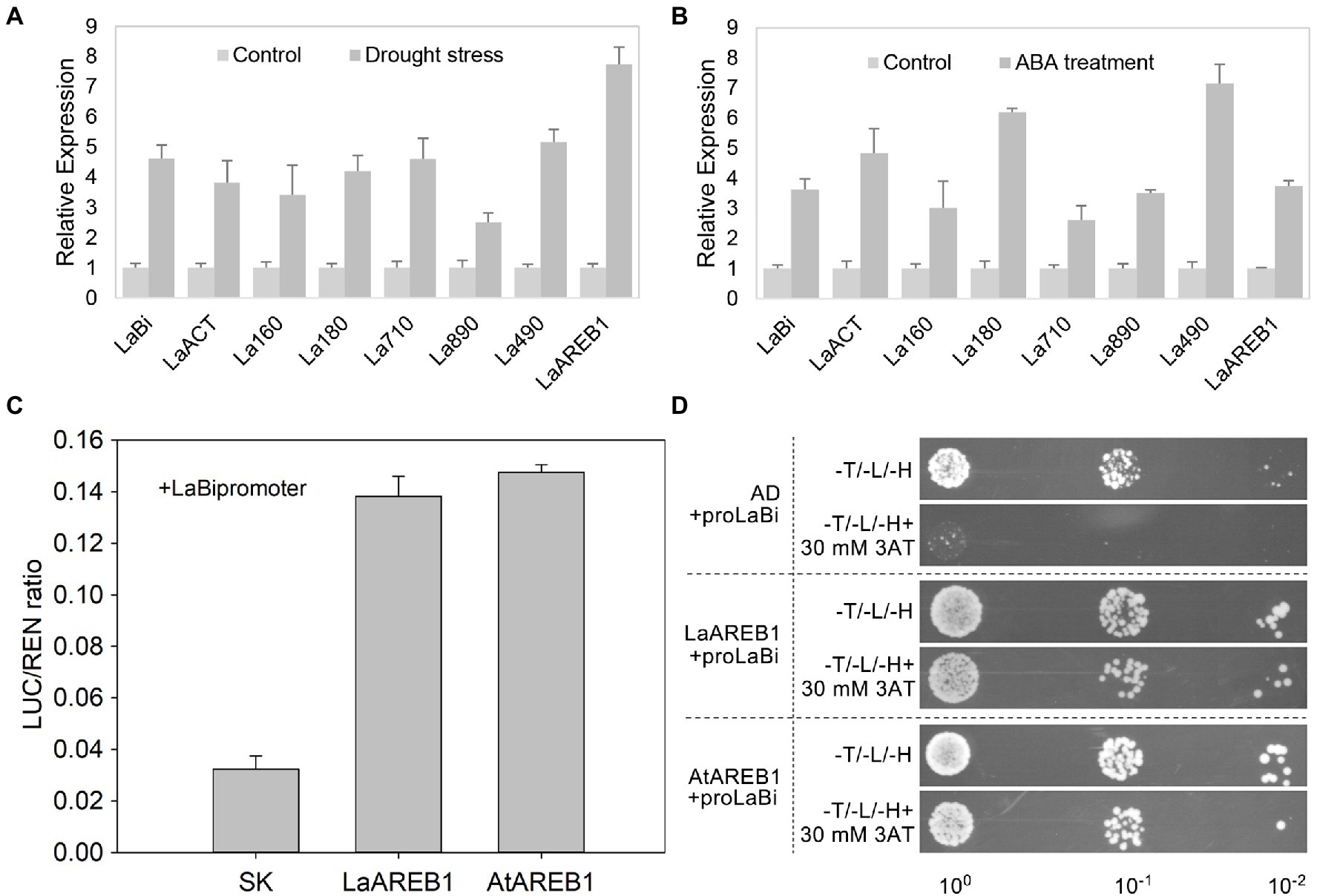
Figure 5. Stress-activated cucurbitacin biosynthesis genes. (A) Expression of Bi, ACT, and CYP450s after drought stress. (B) Expression of Bi, ACT, and CYP450s after ABA treatment. (C) Yeast one-hybrid analysis of the interactions between the LaAREB1 and AtAREB1 proteins and the LaBi promoters. (D) Dual-Luciferase reporter assays show the interactions between LaAREB1 and AtAREB1 proteins and the LaBi promoters.
Discussion
Luffa Is an Important Dietary and Medicinal Plant
Luffa, a common vegetable in the daily diet, contains substantial amounts of phytochemicals (Zhang et al., 2020). The fruit is rich in dietary fiber, essential amino acids, vitamins, minerals, polyphenols, flavonoids, saponin, triterpenoids, oleanolic acid, carotenoids, and other nutrients (Zhang et al., 2021). In this study, we conducted a comprehensive targeted metabolomic analysis to determine the metabolite compositions of two Luffa varieties. Notably, human essential amino acids and their derivatives, including L- (+)-Lysine, L- (−)-Tyrosine, L-Phenylalanine, L-Tryptophan, L-Valine, and L-Leucine. Nicotinic acid (Vitamin B3), pyridoxine (Vitamin B7), riboflavin (Vitamin B2), and D-pantothenic acid (Vitamin B5) were detected in the fruits of the two Luffa varieties. Besides, the Luffa fruits had γ-Vitamins such as linolenic acid and their derivatives, punic acid, and α-essential unsaturated fatty acids, including linolenic, eicosenoic, and octadecatetraenoic acid. The fruits are also rich in saccharides, including sucrose, glucose, mannose, and galactose. As Luffa is rich in nutrients, it has become an important dietary vegetable.
Several studies have demonstrated that consuming Luffa considerably supplies antioxidant, anti-inflammation, anticancer, and anti-bacterial constituents to the human body (Du et al., 2006). In this study, abundant methyl ferrate, syringic acid, 1-O-[(E)-caffeoyl], β-D-glucopyranose, punic acid, diosmetin, cymaroside, cucurbitacin B, D, E, I, and their glucosides were detected in the Luffa fruits. These compounds have important roles in antimicrobial, antioxidant, and anti-inflammatory activities and can inhibit cancer proliferation (Du et al., 2006; Patel et al., 2013; Winkler-Moser et al., 2015; Aruna et al., 2016; Srinivasulu et al., 2018; Sikander et al., 2019). Therefore, Luffa fruits can be considered an essential dietary food and medicinal plant (Partap et al., 2012).
Cucurbitacin Biosynthesis Pathway in Luffa
The Bi cyclization of 2,3-oxidosqualene to cucurbitadienol initiates cucurbitacin biosynthesis. Bi is considered the first key pathway-specific enzyme for cucurbitacin biosynthesis in cucurbits (Shibuya et al., 2004; Shang et al., 2014). CYP450 genes are often part of gene clusters in specialized metabolism, especially for mono-, di-, and tri-terpenoid synthesis in cucurbits and tomato (Dar et al., 2021). CYP450s tailor the cucurbitacin core skeleton and key structural variations among cucurbitacins B, C, and E in Cucurbitaceae (Zhou et al., 2016). Moreover, ACT acetylates cucurbitadienol to form various cucurbitacin (Shang et al., 2014; Zhou et al., 2016). Bt activates Bi and regulates CuC biosynthesis in cucumber fruit (Shang et al., 2014). In cucumber, melon, and watermelon fruits, Bt, Bi, CYP450s, and ACT form gene clusters to regulate cucurbitacin synthesis. Indeed, these genes were up-regulated in bitter cucumber fruit (Shang et al., 2014; Zhou et al., 2016). In this study, the expression of LaBt (Lac02g016030) was undetected in S1174 and WM709 Luffa varieties (Supplementary Table S7). However, LaBi, LaACT, La160, La180, La710, La890, and La490 were up-regulated in the bitter variety (WM709; Figure 4A; Supplementary Table S7), suggesting that they potentially participate in cucurbitacin synthesis in Luffa. Notably, these genes formed a Bi cluster in the synteny block between cucumber and Luffa (Figure 4B). However, La170, La540, La550, and La560 were lacking in Luffa (Figure 4A), and the expression of La510 and La530 was undetectable in WM709 and S1174 varieties (Figure 4A), suggesting that they may not participate in cucurbitacin synthesis in Luffa, unlike cucumber, watermelon, and melon (Zhou et al., 2016).
Cycloartenol synthase (CAS) converts 2,3-oxidosqualene to cycloartenol through the protosteryl cation intermediate, and was the basal plant OSC from which others derived (Phillips et al., 2006). Some studies have shown that abiotic stress can up-regulate the CAS genes (Babiychuk et al., 2008; Shirazi et al., 2019). However, some studies reported no significant change in CAS expression under drought and salt stress (Basyuni et al., 2009; Nasrollahi et al., 2014). In this study, BLASTp algorithm identified CSA gene (Lac05g022090) in Luffa genome using Arabidopsis homologs for CSA (AT2G07050) as query sequences. Notably, there was no difference in CSA expression between S1174 and WM709 Luffa varieties (Supplementary Figure S5A). However, a slight up-regulation of the CAS gene was observed after ABA and drought treatments (Supplementary Figure S5B). Further studies are needed to determine whether CAS mediates abiotic stress response in Luffa.
UGTs Regulate Cucurbitacin Glucosylation
In cucumber, UGTs regulate cucurbitacin glucosylation (Zhong et al., 2017). In this study, cucurbitacins and their glucosides were detectable and exhibited significant accumulations, with 14 up-regulated UGTs in WM709 (Figure 4C). Perhaps these UGTs convert cucurbitacin to cucurbitacin glucoside in Luffa. These results suggest that the cucurbitacin biosynthesis mechanism in Luffa may be similar to that in other Cucurbitaceae plants.
Abiotic Stress Activates the Expression of Cucurbitacin Biosynthesis Genes
Cucurbitacins are biosynthesized in response to abiotic stress in Cucurbitaceae plants (Kim et al., 2020). In cucumber, exogenous ABA, drought, and cold stress up-regulated Bi, ACT, and CYP450s (Shang et al., 2014). However, the relationship between these genes and stress conditions has not been studied in Luffa. In this study, drought stress and ABA treatment up-regulated LaBi, LaACT, and LaCYP450s in Luffa (Figures 5A,B). Further analysis showed that LaBi promoters contain ABRE elements. Furthermore, luciferase trans-activation and yeast one-hybrid assays demonstrated that Luffa and A. thaliana AREB1 activate LaBi expression by directly binding to its promoter (Figures 5C,D). These results indicate that drought stress and exogenous ABA activates Bi expression, which promotes cucurbitacin biosynthesis, resulting in bitter fruit production in Luffa.
Conclusion
Integrated metabolome and transcriptome analysis has revealed that Luffa is an important medicinal plant with high nutritional value. In this study, 11 terpenoids, including cucurbitacin D, cucurbitacin F, and 23,24-Dihydro cucurbitacin E, were up-regulated in bitter Luffa. The cucurbitacin biosynthesis genes, Bi, CYP450s, and ACT, were significantly up-regulated and formed gene clusters, thereby maintaining effective biosynthesis of cucurbitacins. AREB1 binds to the Bi promoter to activate the expression of Bi in stress-exposed Luffa (Figure 6). These findings uncover the direct link between cucurbitacin biosynthesis and abiotic stress response and deepen the understanding of regulatory mechanisms underlying cucurbitacin biosynthesis in Luffa.
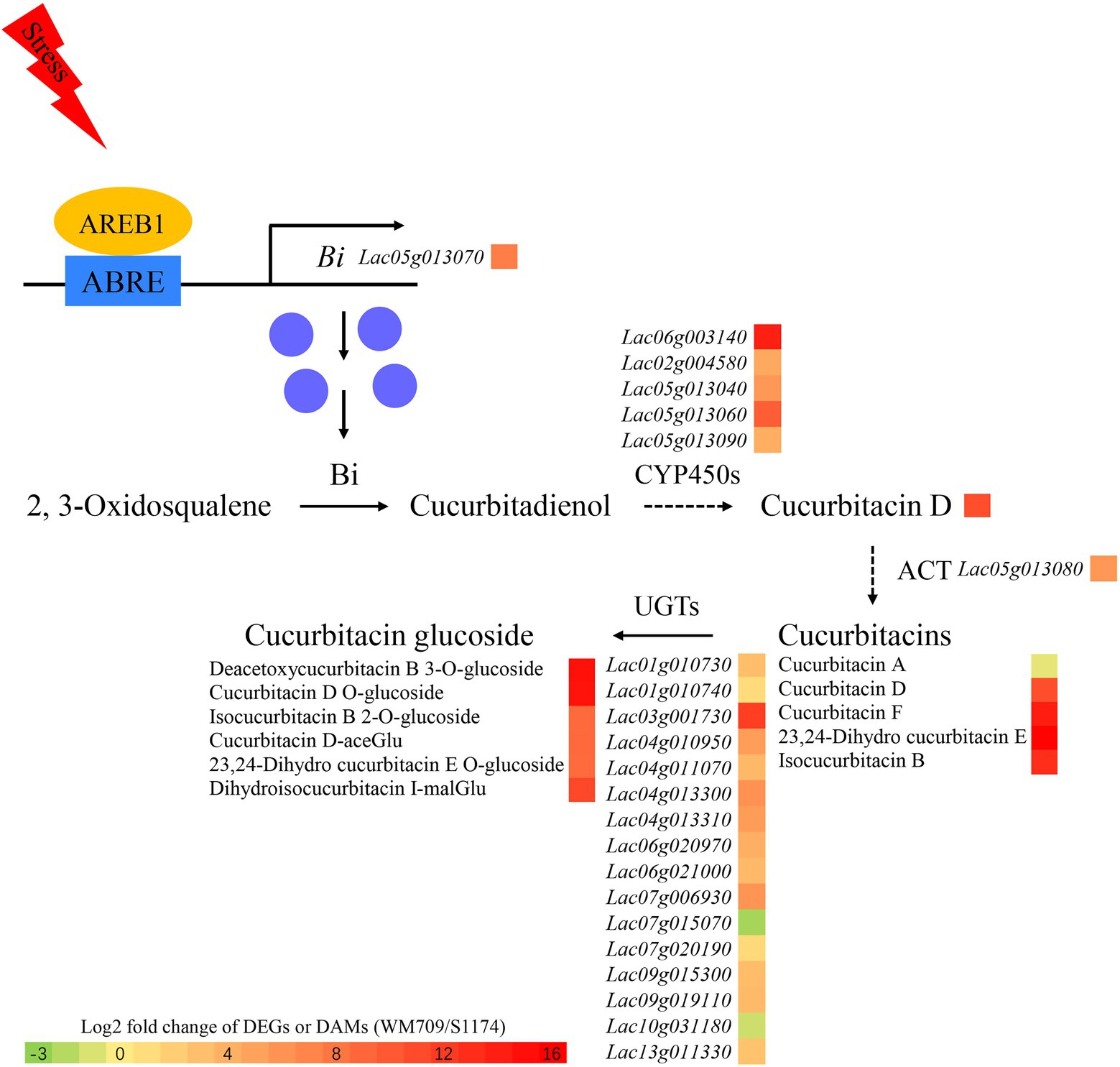
Figure 6. Regulatory network of predicted cucurbitacin biosynthesis. CYP450s, cytochromes P450s; ACT, acyltransferase; and UGTs, UDP-Glucosyltransferases. Green boxes indicate down-regulated transcripts and metabolites, red boxes indicate up-regulated transcripts and metabolites.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA810936.
Author Contributions
GZ, HW, and JLu conceived and designed the research. GZ, MW, CL, JLi, and XL performed the experiments. GZ, HG, and XZ analyzed the data. GZ wrote the article. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (31902011 and 31872093), the Laboratory of Lingnan Modern Agriculture Project (NZ2021008), the Science and Technology Program of Guangdong Province (2018B020202007, 2019A050520002, 201904020012, 2020A0505020006, 2021KJ110, and 2021A1515012500), the Science and Technology Program of Guangzhou of China (X20210201020), the Agricultural competitive industry discipline team building project of Guangdong Academy of Agricultural Sciences (202103TD and 202114TD), and Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science (R2020PY-JG004, R2018QD-035, and R2018QD-038).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.886870/full#supplementary-material
Supplementary Figure S1 | OPLS-DA analysis of detected metabolites. (A) Scores OPLA-DA plot. (B) R2X, R2Y, and Q2 plot.
Supplementary Figure S2 | KEGG enriched pathways from the up-regulated metabolites in S1174.
Supplementary Figure S3 | The correlation coefficients (A) and PCA (B) of DEGs.
Supplementary Figure S4 | The number of DEGs belonging to different transcription factor families detected in S1174 and WM709.
Supplementary Figure S5 | Expression profiles of LacCAS. (A) LacCAS read counts. (B) Relative expression of LacCAS after drought stress and ABA treatments.
Footnotes
References
Aruna, P., Venkataramanamma, D., Singh, A. K., and Singh, R. P. (2016). Health benefits of punicic acid: a review. Compr. Rev. Food Sci. Food Saf. 15, 16–27. doi: 10.1111/1541-4337.12171
Babiychuk, E., Bouvier-Navé, P., Compagnon, V., Suzuki, M., Muranaka, T., Van Montagu, M., et al. (2008). Allelic mutant series reveal distinct functions for Arabidopsis cycloartenol synthase 1 in cell viability and plastid biogenesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3163–3168. doi: 10.1073/pnas.0712190105
Basyuni, M., Baba, S., Inafuku, M., Iwasaki, H., Kinjo, K., and Oku, H. (2009). Expression of terpenoid synthase mRNA and terpenoid content in salt stressed mangrove. J. Plant Physiol. 166, 1786–1800. doi: 10.1016/j.jplph.2009.05.008
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cao, T. Q., Kim, J. A., Woo, M. H., and Min, B. S. (2021). SARS-CoV-2 main protease inhibition by compounds isolated from Luffa cylindrica using molecular docking. Bioorg. Med. Chem. Lett. 40:127972. doi: 10.1016/j.bmcl.2021.127972
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Gong, L., Guo, Z., Wang, W., Zhang, H., Liu, X., et al. (2013). A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780. doi: 10.1093/mp/sst080
Dar, M. S., Dholakia, B. B., Kulkarni, A. P., Oak, P. S., Shanmugam, D., Gupta, V. S., et al. (2021). Influence of domestication on specialized metabolic pathways in fruit crops. Planta 253:61. doi: 10.1007/s00425-020-03554-4
Du, Q., Xu, Y., Li, L., Zhao, Y., Jerz, G., and Winterhalter, P. (2006). Antioxidant constituents in the fruits of Luffa cylindrica (L.) Roem. J. Agric. Food Chem. 54, 4186–4190. doi: 10.1021/jf0604790
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
Gautam, A. (2017). Genetic divergence and character association studies in indegenous ridge gourd (Luffa Acutangula (L.) Roxb.) genotypes. J. Pharmacogn. Phytochem. 6, 1769–1774.
Gong, C., Zhu, H., Lu, X., Yang, D., Zhao, S., Umer, M. J., et al. (2021). An integrated transcriptome and metabolome approach reveals the accumulation of taste-related metabolites and gene regulatory networks during watermelon fruit development. Planta 254:35. doi: 10.1007/s00425-021-03680-7
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Iwanski, G. B., Lee, D. H., En-Gal, S., Doan, N. B., Castor, B., Vogt, M., et al. (2010). Cucurbitacin B, a novel in vivo potentiator of gemcitabine with low toxicity in the treatment of pancreatic cancer. Brit. J. Pharmacol. 160, 998–1007. doi: 10.1111/j.1476-5381.2010.00741.x
Jevtić, B., Djedović, N., Stanisavljević, S., Despotović, J., Miljković, D., and Timotijević, G. (2016). Cucurbitacin E potently modulates the activity of Encephalitogenic cells. J. Agric. Food Chem. 64, 4900–4907. doi: 10.1021/acs.jafc.6b00951
Kalloo, G. (1993). “Loofah: Luffa spp.,” in Genetic Improvement of Vegetable Crops. eds. G. Kalloo and B. O. Bergh (Pergamon Press), 265–266.
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kim, Y.-C., Choi, D., Cha, A., Lee, Y.-G., Baek, N.-I., Rimal, S., et al. (2020). Critical enzymes for biosynthesis of cucurbitacin derivatives in watermelon and their biological significance. Commun. Biol. 3:444. doi: 10.1038/s42003-020-01170-2
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kuppast, I. J., and Mankani, K. L. (2012). A review on Luffa acutangula. Nat. Prod. Ind. J. 8, 198–193.
Li, S., Lin, Y.-C. J., Wang, P., Zhang, B., Li, M., Chen, S., et al. (2018). The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 31, 663–686. doi: 10.1105/tpc.18.00437
Li, Y., Wu, X., Xu, W., Sun, Y., Wang, Y., Li, G., et al. (2021a). High-throughput physiology-based stress response phenotyping: advantages, applications and prospective in horticultural plants. Hortic. Plant J. 7, 181–187. doi: 10.1016/j.hpj.2020.09.004
Li, Y., Zhang, X., Jiang, J., Zhao, T., Xu, X., Yang, H., et al. (2021b). Virus-induced gene silencing of SlPYL4 decreases the drought tolerance of tomato. Hortic. Plant J. 8, 361–368. doi: 10.1016/j.hpj.2021.06.005
Liu, M., Yu, H., Zhao, G., Huang, Q., Lu, Y., and Ouyang, B. (2017). Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics 18:481. doi: 10.1186/s12864-017-3869-1
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, L., Wang, Q., Zheng, Y., Guo, J., Yuan, S., Fu, A., et al. (2022). Cucurbitaceae genome evolution, gene function and molecular breeding. Hortic. Res. 9:uhab057. doi: 10.1093/hr/uhab057
Mashilo, J., Odindo, A. O., Shimelis, H. A., Musenge, P., Tesfay, S. Z., and Magwaza, L. S. (2018). Photosynthetic response of bottle gourd [Lagenaria siceraria (Molina) Standl.] to drought stress: relationship between cucurbitacins accumulation and drought tolerance. Sci. Hortic. 231, 133–143. doi: 10.1016/j.scienta.2017.12.027
Nasrollahi, V., Mirzaie-asl, A., Piri, K., Nazeri, S., and Mehrabi, R. (2014). The effect of drought stress on the expression of key genes involved in the biosynthesis of triterpenoid saponins in liquorice (Glycyrrhiza glabra). Phytochemistry 103, 32–37. doi: 10.1016/j.phytochem.2014.03.004
Ng, Y. M., Yang, Y., Sze, K. H., Zhang, X., Zheng, Y. T., and Shaw, P. C. (2011). Structural characterization and anti-HIV-1 activities of arginine/glutamate-rich polypeptide Luffin P1 from the seeds of sponge gourd (Luffa cylindrica). J. Struct. Biol. 174, 164–172. doi: 10.1016/j.jsb.2010.12.007
Orellana, S., Yañez, M., Espinoza, A., Verdugo, I., González, E., Ruiz-Lara, S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33, 2191–2208. doi: 10.1111/j.1365-3040.2010.02220.x
Pan, L., Zhou, C., Jing, J., Zhuang, M., Zhang, J., Wang, K., et al. (2022). Metabolomics analysis of cucumber fruit in response to foliar fertilizer and pesticides using UHPLC-Q-Orbitrap-HRMS. Food Chem. 369:130960. doi: 10.1016/j.foodchem.2021.130960
Partap, S., Kumar, A., Sharma, N. K., and Jha, K. (2012). Luffa Cylindrica: an important medicinal plant. J. Nat. Prod. Plant Resour. 2, 127–134.
Patel, K., Gadewar, M., Tahilyani, V., and Patel, D. K. (2013). A review on pharmacological and analytical aspects of diosmetin: a concise report. Chin. J. Integr. Med. 19, 792–800. doi: 10.1007/s11655-013-1595-3
Phillips, D. R., Rasbery, J. M., Bartel, B., and Matsuda, S. P. T. (2006). Biosynthetic diversity in plant triterpene cyclization. Curr. Opin. Plant Biol. 9, 305–314. doi: 10.1016/j.pbi.2006.03.004
Schütze, K., Harter, K., and Chaban, C. (2008). Post-translational regulation of plant bZIP factors. Trends Plant Sci. 13, 247–255. doi: 10.1016/j.tplants.2008.03.002
Shang, Y., Ma, Y., Zhou, Y., Zhang, H., Duan, L., Chen, H., et al. (2014). Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346, 1084–1088. doi: 10.1126/science.1259215
Shendge, P. N., and Belemkar, S. (2018). Therapeutic potential of Luffa acutangula: a review onits traditional uses, phytochemistry, pharmacology and toxicological aspects. Front. Pharmacol. 9:1177. doi: 10.3389/fphar.2018.01177
Shibuya, M., Adachi, S., and Ebizuka, Y. (2004). Cucurbitadienol synthase, the first committed enzyme for cucurbitacin biosynthesis, is a distinct enzyme from cycloartenol synthase for phytosterol biosynthesis. Tetrahedron 60, 6995–7003. doi: 10.1016/j.tet.2004.04.088
Shirazi, Z., Aalami, A., Tohidfar, M., and Sohani, M. M. (2019). Triterpenoid gene expression and phytochemical content in Iranian licorice under salinity stress. Protoplasma 256, 827–837. doi: 10.1007/s00709-018-01340-4
Sikander, M., Malik, S., Chauhan, N., Khan, P., Kumari, S., Kashyap, V. K., et al. (2019). Cucurbitacin D reprograms glucose metabolic network in prostate cancer. Cancer 11:364. doi: 10.3390/cancers11030364
Srinivasulu, C., Ramgopal, M., Ramanjaneyulu, G., Anuradha, C. M., and Suresh Kumar, C. (2018). Syringic acid (SA): a review of its occurrence, biosynthesis, pharmacological and industrial importance. Biomed. Pharmacother. 108, 547–557. doi: 10.1016/j.biopha.2018.09.069
Swetha, M. P., and Muthukumar, S. P. (2016). Characterization of nutrients, amino acids, polyphenols and antioxidant activity of ridge gourd (Luffa acutangula) peel. J. Food Sci. Technol. 53, 3122–3128. doi: 10.1007/s13197-016-2285-x
The Gene Ontology Consortium (2018). The gene ontology resource: 20 years and still going strong. Nucleic Acids Res. 47, D330–D338. doi: 10.1093/nar/gky1055
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Vieira, E. F., Pinho, O., Ferreira, I. M. P. L. V. O., and Delerue-Matos, C. (2019). Chayote (Sechium edule): a review of nutritional composition, bioactivities and potential applications. Food Chem. 275, 557–568. doi: 10.1016/j.foodchem.2018.09.146
Walters, T. W. (1989). Historical overview on domesticated plants in China with special emphasis on the Cucurbitaceae. Econ. Bot. 43, 297–313. doi: 10.1007/BF02858729
Wang, S., Alseekh, S., Fernie, A. R., and Luo, J. (2019). The structure and function of major plant metabolite modifications. Mol. Plant 12, 899–919. doi: 10.1016/j.molp.2019.06.001
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Winkler-Moser, J. K., Hwang, H.-S., Bakota, E. L., and Palmquist, D. A. (2015). Synthesis of steryl ferulates with various sterol structures and comparison of their antioxidant activity. Food Chem. 169, 92–101. doi: 10.1016/j.foodchem.2014.07.119
Wu, H. B., He, X. L., Gong, H., Luo, S. B., Li, M. Z., Chen, J. Q., et al. (2016). Genetic linkage map construction and QTL analysis of two interspecific reproductive isolation traits in sponge gourd. Front. Plant Sci. 7:980. doi: 10.3389/fpls.2016.00980
Wu, H., Zhao, G., Gong, H., Li, J., Luo, C., He, X., et al. (2020). A high-quality sponge gourd (Luffa cylindrica) genome. Hortic. Res. 7:128. doi: 10.1038/s41438-020-00350-9
Xu, Y., Zhang, H., Zhong, Y., Jiang, N., Zhong, X., Zhang, Q., et al. (2022). Comparative genomics analysis of bHLH genes in cucurbits identifies a novel gene regulating cucurbitacin biosynthesis. Hortic. Res. 9:uhac038. doi: 10.1093/hr/uhac038
Yang, Z., Li, G., Tieman, D., and Zhu, G. (2019). Genomics approaches to domestication studies of horticultural crops. Hortic. Plant J. 5, 240–246. doi: 10.1016/j.hpj.2019.11.001
Yang, P., Liu, W., Fu, R., Ding, G.-B., Amin, S., and Li, Z. (2020). Cucurbitacin E Chemosensitizes colorectal cancer cells via mitigating TFAP4/Wnt/β-catenin signaling. J. Agric. Food Chem. 68, 14148–14160. doi: 10.1021/acs.jafc.0c05551
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Zhang, T., Wang, Y., Munir, S., Wang, T., Ye, Z., Zhang, J., et al. (2021). Cyclin gene SlCycB1 alters plant architecture in association with histone H3.2 in tomato. Hortic. Plant J. 8, 341–350. doi: 10.1016/j.hpj.2021.12.004
Zhang, L., Yue, Y., Shi, M., Tian, M., Ji, J., Liao, X., et al. (2020). Dietary Luffa cylindrica (L.) Roem promotes branched-chain amino acid catabolism in the circulation system via gut microbiota in diet-induced obese mice. Food Chem. 320:126648. doi: 10.1016/j.foodchem.2020.126648
Zhong, Y., Xue, X., Liu, Z., Ma, Y., Zeng, K., Han, L., et al. (2017). Developmentally regulated glucosylation of bitter triterpenoid in cucumber by the UDP-Glucosyltransferase UGT73AM3. Mol. Plant 10, 1000–1003. doi: 10.1016/j.molp.2017.02.005
Keywords: Luffa, Cucurbitaceae, metabolome, cucurbitacin, abiotic stress
Citation: Zhao G, Wang M, Luo C, Li J, Gong H, Zheng X, Liu X, Luo J and Wu H (2022) Metabolome and Transcriptome Analyses of Cucurbitacin Biosynthesis in Luffa (Luffa acutangula). Front. Plant Sci. 13:886870. doi: 10.3389/fpls.2022.886870
Edited by:
Mehar Hasan Asif, National Botanical Research Institute (CSIR), IndiaReviewed by:
Jacob Mashilo, Limpopo Department of Agriculture and Rural Development, South AfricaTessa Moses, University of Edinburgh, United Kingdom
Copyright © 2022 Zhao, Wang, Luo, Li, Gong, Zheng, Liu, Luo and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haibin Wu, d3VoYmhvcGVAeWVhaC5uZXQ=; Jianning Luo, bHVvam5AZ2RhYXMuY24=
 Gangjun Zhao
Gangjun Zhao Meng Wang1
Meng Wang1 Junxing Li
Junxing Li Xiaoxi Liu
Xiaoxi Liu Jianning Luo
Jianning Luo