
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 14 June 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.886268
This article is part of the Research TopicMolecular Interactions between Crops and Phytopathogens, Volume III: Vegetables and Other CropsView all 11 articles
Many Gram-negative bacteria use small signal molecules, such as N-acyl-homoserine lactones (AHLs), to communicate with each other and coordinate their collective behaviors. Recently, increasing evidence has demonstrated that long-chained quorum-sensing signals play roles in priming defense responses in plants. Our previous work indicated that a short-chained signal, N-3-oxo-octanoyl homoserine lactone (3OC8-HSL), enhanced Arabidopsis resistance to the hemi-biotrophic bacteria Pseudomonas syringae pv. tomato DC3000 through priming the salicylic acid (SA) pathway. Here, we found that 3OC8-HSL could also prime resistance to the necrotrophic bacterium Pectobacterium carotovorum ssp. carotovorum (Pcc) through the jasmonic acid (JA) pathway, and is dependent on auxin responses, in both Chinese cabbage and Arabidopsis. The subsequent Pcc invasion triggered JA accumulation and increased the down-stream genes’ expressions of JA synthesis genes (LOX, AOS, and AOC) and JA response genes (PDF1.2 and VSP2). The primed state was not observed in the Arabidopsis coi1-1 and jar1-1 mutants, which indicated that the primed resistance to Pcc was dependent on the JA pathway. The 3OC8-HSL was not transmitted from roots to leaves and it induced indoleacetic acid (IAA) accumulation and the DR5 and SAUR auxin-responsive genes’ expressions in seedlings. When Arabidopsis and Chinese cabbage roots were pretreated with exogenous IAA (10 μM), the plants had activated the JA pathway and enhanced resistance to Pcc, which implied that the JA pathway was involved in AHL priming by coordinating with the auxin pathway. Our findings provide a new strategy for the prevention and control of soft rot in Chinese cabbage and provide theoretical support for the use of the quorum-sensing AHL signal molecule as a new elicitor.
Many bacteria use small signal molecules to communicate with each other and modulate their collective behavior, a process called quorum sensing (Taga et al., 2003; Reading and Vanessa, 2010). The most common quorum-sensing signal molecules in Gram-negative bacteria are N-acyl-homoserine lactones (AHLs; Sharma et al., 2020). AHLs molecules have varied acyl chain lengths (from 4 to 18 carbons) and substitutions of hydroxyl (OH) or oxo (O) groups at the chain’s γ position (Sharma et al., 2020). To date, over 30 types of AHLs have been identified from more than 70 species of Gram-negative bacteria.
Accumulating evidence indicates that bacterial AHLs are perceived by plant cells and modulate plant growth and development, as well as the responses to abiotic and biotic stresses, particularly those involved in plant immunity (Palmer et al., 2014). Mathesius et al. (2003) reported that the treatment of Medicago truncatula with two AHLs, N-3-oxo-dodecanoyl-homoserine lactone (3OC12-HSL) and N-3-oxo-hexadecanoyl-homoserine lactone (3OC16-HSL), resulted in the differential expression of proteins involved in the processes of flavonoid synthesis, hormone metabolism, and oxidative stress by two-dimensional gel electrophoresis (2D-PAGE). Our previous proteomic analysis showed that differentially expressed proteins were involved in carbon metabolism, protein biosynthesis, and plant resistance after plants were pretreated with N-3-oxo-octanoyl homoserine lactone (3OC8-HSL; Miao et al., 2012). The exposure of Arabidopsis roots to N-hexanoyl-homoserine lactone (C6-HSL), N-3-oxo-hexanoyl-homoserine lactone (3OC6-HSL), and 3OC8-HSL promotes primary root growth, whereas treatment with N-decanoyl-homoserine lactone (C10-HSL) inhibits primary root growth, but promotes lateral root and root hair formation in Arabidopsis (Ortíz-Castro et al., 2008; Rad et al., 2008; Liu et al., 2012; Schenk et al., 2012; Zhao et al., 2016; Shrestha et al., 2020). Inoculation with AHL-producing Burkholderia graminis M14 enhances the ability of tomatoes to tolerate salt stress. Similarly, 3OC6-HSL enhances salt tolerance in Arabidopsis and wheat (Zhao et al., 2020). Exposure to AHLs can elicit plant immunity. Several long-chain AHLs have been shown to induce AHL-priming for enhancing resistance against biotrophic and hemi-biotrophic pathogens in Arabidopsis thaliana, Medicago truncatula, and Hordem vulgare (Mathesius et al., 2003; Schikora et al., 2011; Schenk et al., 2012, 2014; Zarkani et al., 2013; Shrestha et al., 2020). The N-3-oxo-tetradecanoyl-homoserine lactone (3OC14-HSL)-mediated resistance priming in plants involves mitogen-activated protein kinase 6 (MPK6) activation, phenolic compound accumulation, lignin, and callose deposition (Schikora et al., 2011; Schenk et al., 2012, 2014). Some short-chain AHLs such as C6-HSL and N-3-hydroxybutyl-homoserine (C4-HSL) increase the expressions of salicylic acid (SA)- and ethylene-responsive defense genes and the SA accumulation in tomato plants (Schuhegger et al., 2006). Root inoculation with C4-HSL- and C6-HSL-producing Serration plymuthica protects plants from infection by Botrytis cinerea (Pang et al., 2009).
Priming is regulated by a complex network, which allows plants to activate defense responses in a faster and stronger manner as a consequence of triggering stimuli (Mauch-Mani et al., 2017). Many chemicals can induce priming, such as SA, benzothiadiazole (BTH), β-aminobutyric acid, pipecolic acid, jasmonic acid (JA), and volatile organic compounds (VOCs; Conrath et al., 2002; Martinez-Medina et al., 2016). The SA was the first synthetic compound shown to prime defense responses (Kauss et al., 1992) and effectively induce resistance against major fungal and bacterial pathogens in various crops (Kessmann et al., 1994). BTH acts as a priming agent in plant defense leading to a reduction in the penetration and development of the root-knot nematode Meloidogyne incognita in susceptible tomato roots (Veronico et al., 2018). β-Aminobutyric acid is a non-protein amino acid that primes the plants defense system to protect plants from various microbial pathogens (Thevenet et al., 2016). In Arabidopsis, the SA-dependent signaling pathway is considered to be effective mainly against biotrophic pathogens, such as the oomycete Hyloperonospora, the fungus Erysiphe orontii, and the hemi-biotrophic bacterium Pseudomonas syringae, and the JA-dependent defense response is considered to be effective mainly against necrotrophic microbial pathogens, such as the fungus B. cinerea and the bacterium Pectobacterium carotovorum ssp. carotovorum (Pcc; Norman-Setterblad et al., 2000; Zimmerli et al., 2000; Friml et al., 2003).
Using AHL-producing and AHL-negative strains, researchers have demonstrated the important role of C4-HSL and C6-HSL in the induction of resistance against necrotrophic pathogens in plants (Schuhegger et al., 2006; Pang et al., 2009). 3OC14-HSL enhances plant systemic resistance to biotrophic and hemi-biotrophic pathogens, such as Golovinomyces orontii, Blumeria graminis f. sp.hordei, and P. syringae, but not to necrobiotrophic pathogens, including B. cinerea and Plectosphaerella cucumber (Schikora et al., 2011). In contrast, Hu et al. (2018) found that C10-HSL treatment induced systemic immunity and protected tomatoes from infection by the necrotrophic fungus B. cinerea. His results showed that C10-HSL-induced resistance against B. cinerea was mainly dependent on the JA-signaling pathway. These contradictory results may indicate the complexity of the interaction outcome between plants and bacteria modulated by AHLs. We recently demonstrated the function of 3OC8-HSL in priming against hemi-biotrophic bacterial pathogen (Liu et al., 2020). However, whether 3OC8-HSL primes plant resistance to necrotrophic bacteria and the mechanism by which 3OC8-HSL induces resistance remain unknown.
The Gram-negative bacterium Pcc is a species of necrotrophic pathogen that causes soft-rot disease in a wide variety of plants (Perombelon and Kelman, 1980). In the present study, we found that the expression levels of genes involved in JA and auxin pathways were induced by 3OC8-HSL treatment. Pretreatment in which 10 μM 3OC8-HSL was added to the roots for 48 h decreased the disease symptoms and the Pcc growth on leaves of both Arabidopsis and Chinese cabbage. We investigated the roles of auxin in the interactions between Arabidopsis thaliana plants and their necrotrophic pathogen Pcc after pretreatment with AHL. Our results suggested that AHL contributed to the enhanced resistance in systemic leaves and provided evidence supporting the hypothesis that the JA pathway is involved in AHL priming by coordinating with the auxin pathway.
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used throughout this study. The Arabidopsis mutants and transgenic lines used in the study are in the Col-0 background. Seeds of the T-DNA insertion null mutants of coronatine insensitive 1-1, coi1-1 (CS4144), and jar1-1 (CS8072) were obtained from The Arabidopsis Information Resource (TAIR)1. The jar1-1 mutant is compromised in the synthesis of jasmonic acid–isoleucine (JA-Ile), the active compound in JA signaling, whereas coi1-1 is defective in JA perception (Chini et al., 2007). Some of the transgenic plant materials have been described previously, as follows: DR5::GFP, DR5::GUS (Sun et al., 2010), PIN1::PIN1-GFP (Benková et al., 2003) and PIN3::PIN3-GFP (Blilou et al., 2005). The Chinese cabbage used is the homozygous inbred line ‘A03’ that has light green leaves. The cabbage is normally grown and cultivated in the greenhouse. The experiment was carried out on potted cabbage approximately 20 days after germination. For pathogenicity, transcriptional, and biochemical analyses, the plants were cultivated using a hydroponic system. Arabidopsis seeds were surface sterilized with 75% (v/v) ethanol for 1 min and 30% (v/v) NaClO for 5 min. After washing five times with distilled water, seeds were germinated and grown on agar plates containing Murashig and Skoog medium (MS) at pH 5.8. Plants were placed in a growth chamber having a 16-h light:8-h dark photoperiod and 4,000-Lux light intensity at 22 ± 2°C. When the seedlings were grown to the two-leaf stage and roots reached 2 cm in length, the plants were transplanted into a plastic basin (a modified Eppendorf holder covered with parafilm:18 cm × 11 cm) containing 400 ml of Hoagland medium, which was exchanged every 2 days. AHLs were added directly into the medium.
The four shorter acyl chain AHLs (C6-HSL, 3OC6-HSL, 3OC8-HSL, and N-octanoyl-homoserine lactone C8-HSL) were dissolved independently in distilled water and the two longer acyl-chain AHLs C10-HSL and 3OC14-HSL, detected in this study, were dissolved in acetone. They were all purchased from Sigma-Aldrich (Taufkirchen, Germany) and stored in dry condition. They were diluted independently into 10 mM stock solutions in distilled water or acetone and adjusted to pH 5.0 just before use. All the compound solutions were sterilized by passing them through a 0.22 μm filter. AHLs were added directly into the hydroponic system. Plants were pretreated for 2 days. All the experiments were performed using the unpretreated plants.
Seventeen-day-old seedlings were cultured in Hoagland medium with or without 10 μM 3OC8-HSL. Plants were harvested at 24 h after the 3OC8-HSL pretreatment. Total RNA was extracted from pretreated and unpretreated plants using the RNAiso Plus reagent (TaKaRa, dalian, China) and purified using a NucleoSpin RNA clean-up kit (Macherey Nagel) in accordance with the manufacturers’ instructions. The probes were prepared using a CapitalBio cRNA-amplified labeling kit (Capitalbio Corp.) and fluorescently labeled with Cy5-dCTP and Cy3-dCTP (GE Healthcare). The 29k Arabidopsis Genome Arrays (Capitalbio Corp.) were prepared in accordance with the A. thaliana Genome Oligo Set (version 3.0; Operon). After hybridization, the arrays were scanned using a LuxScan 10KA two-channel laser scanner (CapitalBio Corp.) and analyzed using the LuxScan 3.0 software (CapitalBio Corp.). Each data point represents the average of three independent experiments. A two-fold increase (ratio > 2.0) or a two-fold decrease (ratio < 0.5) in the expression of pretreated plants compared with untreated plants was considered as a differential expression, corresponding to the upregulation or downregulation, respectively, in response to the AHL. The gene annotation and functional classification were performed using the Molecule Annotation System v3.0 and the Gene Ontology tool at TAIR. The microarray data discussed in the present study have been deposited in NCBI GEO and were released as GEO Series accession number GSE197485. The 3OC6-HSL Microarray data were published and are accessible through the GEO Series accession number GSE78079. Gene expression profiling and functional analyses were plotted using http://www.bioinformatics.com.cn, a free online platform for data analysis and visualization.
Arabidopsis plants were inoculated with the bacterial pathogen. Pcc was cultured overnight in Luria-Bertani medium (LB) until colony forming units (CFUs) reached 109 CFU/ml. The cells were collected by centrifugation, washed in 10 mM MgCl2, and re-suspended in 10 mM MgCl2. The inoculation solution was adjusted to 107 CFU/ml. Plants grown in the hydroponic system were spray-inoculated with a bacterial solution containing 0.02% Silwet L-77 uniformly. After 1, 6, 12, 24, 36, and 48 h, 100 mg leaf tissue was harvested and homogenized in 1 mM MgCl2. Samples were diluted and plated for CFU counting. Each of the six independent biological experiments was conducted with three technical replications. Chinese cabbage was inoculated with the bacterial pathogen. Briefly, petioles of the third leaves (from inside to outside) of 7- to 8-leaf plants were lightly scored (through the epidermis) with a sterile scalpel and inoculated with 5–10 μL of a uniform bacterial suspension made from cultures, which were labeled “in vivo”. Similarly, the third leaves were cut into 5.5-cm-diameter disks and placed in closed 9-cm-diamater petri dishes containing two layers of moist filter paper to maintain high humidity and then inoculated with 5–10 μL of bacterium suspension. They were then placed in an incubator (at 28°C with 90% relative humidity). These cultures were designated as “in vitro”. The inoculation concentration was 108CFU/mL and the disease phenotype was investigated at 48 h after inoculation (Liu et al., 2019).
The 3OC8-HSL-pretreated or unpretreated Arabidopsis seedlings were collected at 0, 6, 12, and 24 h post-inoculation with Pcc. The total RNA of homogenized plant tissues was extracted using the RNA plus reagent purchased from TaKaRa. Briefly, the cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa) in accordance with the manufacturer’s instructions. For the relative quantification of gene expression, the comparative CT method (Livak and Schmittgen, 2001) with a 7,500 Real Time PCR System (Applied Biosystems, Foster City, CA, United States) was used. PCR amplification was performed in a total volume of 20 μL containing 5 μL diluted cDNA, 0.4 μL of each primer (10 μM), and 10 μL SYBR Premix Ex Taq™ (TaKaRa). The following qRT-PCR thermal cycling program was employed: 10 s at 95°C, 40 cycles of 5 s at 95°C, and 34 s at 60°C. The amount of target gene was normalized to the endogenous reference gene Actin2/8. Each data point represented the average of three independent experiments. For technical controls, each qRT-PCR experiment was repeated four times on the same 96-well plate. qRT-PCR was performed using the primers listed in Supplementary Table 1.
Extraction and quantification of free JA were performed using 3OC8-HSL-pretreated Arabidopsis seedlings grown in the hydroponic system at 24 h after inoculation with Pcc. Plant tissues were frozen and ground in liquid N2. As an internal standard, 5 μL of 10 μg/mL dihydro-JA was added to the frozen tissue (0.5 g). In addition, 5 mL of 80% cold methanol was added, the samples were vortexed for 1 min for fully dissolving the powder, and extracted at 4°C overnight. The samples were centrifuged at 4,000 g for 15 min at 4°C. The supernatant was removed with a 1.5 mL syringe and passed through a 0.22 μM organic filter. The filtrate was prepared for HPLC-MS analysis. The control was 1 μg/mL JA standard. Chromatography was performed on a Waters1525 HPLC system (Waters Technologies). Chromatographic separation was achieved on an Inertsil ODS C18 column (50 mm × 4.6 mm, 5 μm, GL Sciences, Tokyo, Japan).
Propidium iodide was used to stain the plant cell wall. Examination of Green Fluorescent Protein (GFP) fluorescence intensity was performed using a laser scanning confocal microscope (excitation, 488 nm; emission, 500-550 nm; Leica SP8).
For all the experiments, the overall data were statistically analyzed using the DPS v7.05 program. ANOVA test was used to determine plant defense responses to 3OC8-HSL in different genotypes, including wild-type (Col-0), coi1-1, and jar1-1. All the data were represented as mean ± SD of three or six independent experiments.
N-acyl-homoserine lactones confer resistance against biotrophic and hemi-biotrophic pathogens in host plants. However, the effects of AHLs in Arabidopsis against necrotrophic bacteria Pcc are still unknown. To evaluate the spectra of AHL-related actions, we pretreated Arabidopsis roots, which were grown in a hydroponic system, for 2 days with six types of AHLs having different chain lengths and modifications at the C3 position. The plant leaves were spray-inoculated with a Pcc cell suspension. The bacterial CFUs on the leaf tissues were counted at 24 h post-inoculation. The 3OC8-HSL pretreatment showed the strongest inhibitory effects on pathogen proliferation compared with the unpretreated plants. No significant differences in pathogen propagation were observed in plants pretreated with MgCl2, acetone, C6-HSL, C8-HSL, C10-HSL, or 3OC14-HSL (Figure 1A). Then 3OC8-HSL was selected for further analysis. In addition, detached leaves from soil-grown Arabidopsis were pretreated with 10 μM 3OC8-HSL for 2 days prior to spray-inoculation with 107 CFU/mL Pcc. The disease symptoms were recorded at 24 h after inoculation. The symptoms of 3OC8-HSL unpretreated leaves having yellow or water-soaked lesions were more serious than those of the 3OC8-HSL pretreated leaves (Figure 1B). To monitor the disease progression on the leaves of 3OC8-HSL-pretreated plants, we determined the CFUs at 48 h after pathogen infection. Pathogen proliferation was significantly inhibited in the 3OC8-HSL-pretreated plants from 24 to 48 h compared with unpretreated plants (Figure 1C). To analyze the effects of AHLs on Pcc bacteria, the different concentrations of 3OC8-HSL, ranging from 10 nM to 100 μM, were used as pretreatment and the numbers of Pcc CFU were determined. Pathogen proliferation was significantly inhibited in the 10 μM 3OC8-HSL-pretreated plants (Figure 1D).
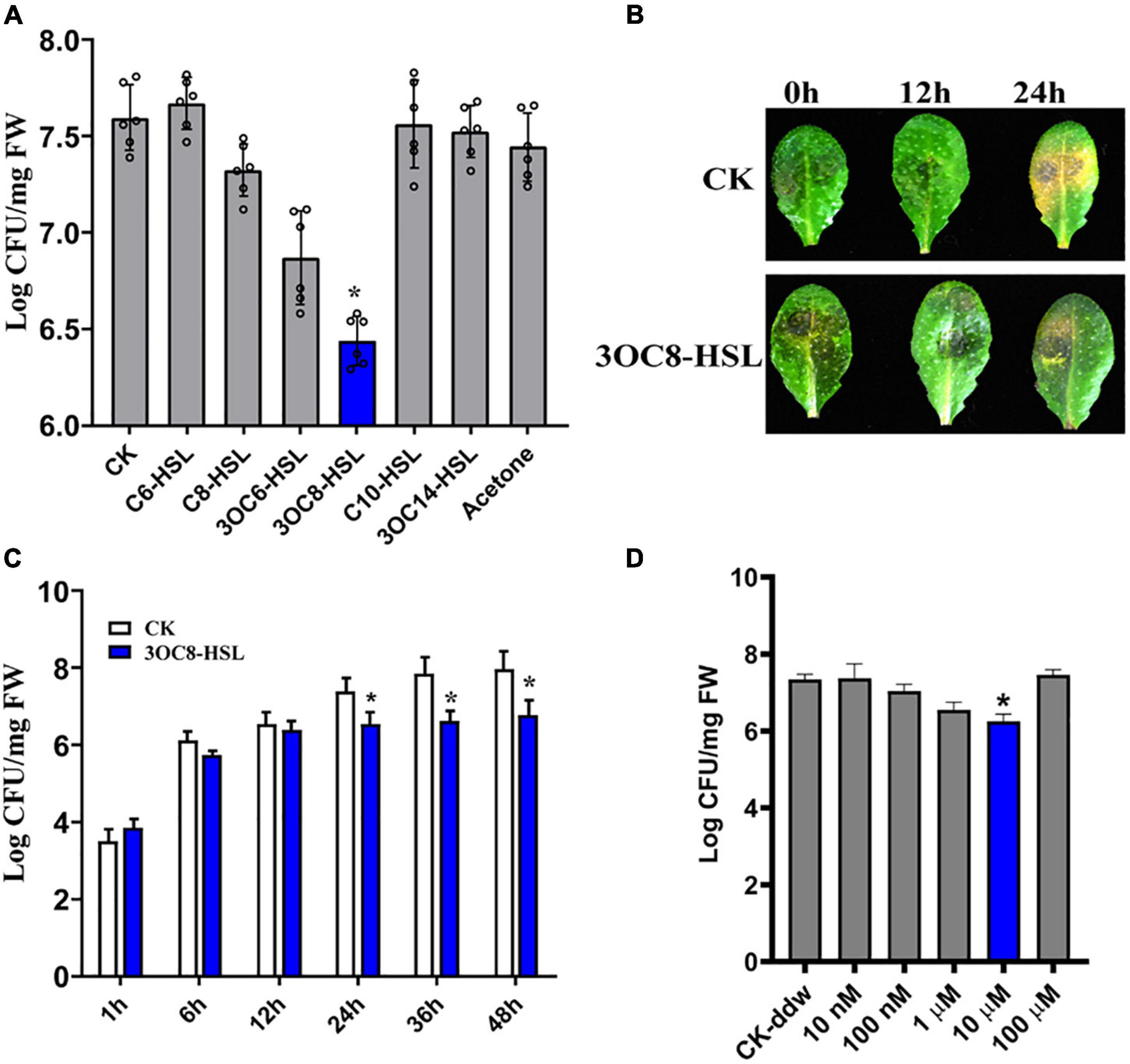
Figure 1. Enhanced resistance of 3OC8-HSL-treated Arabidopsis against Pcc. (A) Proliferation of Pcc in the leaves of Arabidopsis plants (grown in a hydroponic system) in which the roots were pretreated with 10 μM of different AHL compounds for 48 h, and the leaves were subsequently spray-inoculated with 107 CFU/mL Pcc. CFUs were counted at 24 h post-inoculation. “CK” is for wild type Arabidopsis Col-0 inoculated with MgCl2. The input of each sample and control were similar, around 103 CFU/g FW. (B) Symptoms of Pcc infection on wild-type Arabidopsis pretreated with 10 μM 3OC8-HSL. The disease symptoms were recorded at 24 h after inoculation. (C) Inhibitory effect of 3OC8-HSL on Pcc growth in Arabidopsis. The leaves (grown in hydroponic system) were inoculated with 107 CFU/mL Pcc at 48 h after pretreatment with 10 μM 3OC8-HSL at the roots. CFUs were counted at different hour intervals post-inoculation. Data represent the means of three independent biological replicates ± standard deviation (SD). (D) Different priming effects of different concentrations of 3OC8-HSL. The experiments were performed with six leaves per treatment, and similar results were obtained in three independent experiments. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of six independent experiments.
To determine the function of 3OC8-HSL in plant-immunity, we used the 29k Arabidopsis Genome Array to profile the gene expressions of Arabidopsis seedlings planted in greenhouse. Seventeen-day-old seedlings were cultured in Hoagland medium with or without 10-μM 3OC8-HSL. Plants were harvested at 24 h after the 3OC8-HSL pretreatment. The transcriptional chip analysis identified a total of 2,589 of the differentially expressed genes (DEGs), including 1,013 upregulated and 1,576 downregulated [National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) accession number GSE197485]. The data suggested that approximately 9% of the genes were 3OC8-HSL responsive. The DEGs were grouped into 15 functional categories of TAIR. The AHLs were mainly involved in carbohydrate transport and metabolism, signal transduction mechanisms, plant hormone signal transduction, biosynthesis, transport and catabolism of secondary metabolites, defense mechanisms, and large enzyme family mechanisms (Supplementary Figures 1A,B). Notably, ∼90 transcription factors including AP2/ERF-ERF, NAC, WRKY, MYB, C2H2, and bHLH participated in 3OC8-HSL response, and these were similar to those involved in 3OC6-HSL response (Supplementary Table 1). Here, we identified 28 JA- and 13 auxin-related genes responsive to both 3OC8-HSL and 3OC6-HSL.
In molecular function analysis of the microarray data, the expression level-changed transcription factors were listed according to the fold-change value. The expression of RVE1 (A MYB-like transcription factor that regulates hypocotyl growth by regulating free auxin levels in a time-of-day specific manner) was increased by 26.7 times, the expression of MYB44 was increased by 3.56 times, and the expression of WRKY70 was downregulated by 0.44-fold. Several studies have indicated that AtMYB44 is induced by a number of phytohormones, including abscisic acid, gibberellin acid, JA, ethylene, and auxin. Our previous research suggests that AtMYB44 may play a role in 3OC6-HSL-mediated primary root elongation by regulating the expressions of auxin- and cytokinin-related genes (Zhao et al., 2016). Our qRT-PCR data showed that MYB44 responded to 3OC8-HSL, but did not change at the early stage of Pcc infection (Supplementary Figure 1C). We also detected the expressions of the genes related to SA synthesis, ICS1 (encodes isochorismate synthase1), CBP60g (encodes calmodulin binding protein 60-like), and SARD1 (encodes SAR deficient 1), which were involved in 3OC8-HSL response. These results suggested that the SA synthesis seemed to have nothing to do with 3OC8-HSL-induced resistance to Pcc (Supplementary Figures 1D-F).
WRKY70 is negatively regulated in the Arabidopsis response to Pcc (Li et al., 2017). A WRKY70 deficiency enhances resistance to necrotrophic pathogens by enhancing PDF1.2 expression through the activation of the JA pathway (Li et al., 2004). Our data suggested that WRKY70 was significantly reduced after 3OC8-HSL pretreatment and subsequent Pcc inoculation (Supplementary Figure 1G). These results suggested that 3OC8-HSL mediated the JA response through WRKY70 and induced disease resistance. RVE1 is a MYB-like transcription factor that regulates hypocotyl growth by regulating free auxin levels, and RVE1 was elevated after 3OC8-HSL pretreatment (Supplementary Figure 1H). Thus, both JA and auxin pathways may be involved in 3OC8-HSL priming.
To further investigate the effects of 3OC8-HSL on the JA pathway, we monitored the expressions of LOX (encodes a lipoxygenase that catalyze the oxygenation of fatty acids), AOS (encodes an enzyme that catalyzes the dehydration of the hydroperoxide to an unstable allene oxide in JA biosynthesis), and AOC (encodes an enzyme that catalyzes an essential step in JA biosynthesis), which encode key enzymes in the JA biosynthetic pathway (Kazan and Manners, 2013). The pretreatment of roots with 3OC8-HSL resulted in increased expression levels of LOX, AOS, and AOC at 6 h after inoculation with Pcc (Figure 2A). Additionally, the expression levels of the JA-signaling marker genes PDF1.2 (encodes an ethylene- and JA-responsive plant defensin), VSP2 (has acid phosphatase activity dependent on the presence of divalent cations), and Thi2.1 (encodes a thionin, which is a cysteine-rich protein having antimicrobial properties) increased dramatically in the leaves of plants primed with 3OC8-HSL when compared with unpretreated plants. The induction of gene expressions by 3OC8-HSL reached a maximum at 6 h after Pcc infection. MYC2 (encodes a MYC-related transcriptional activator with a typical DNA binding domain of a basic helix-loop-helix leucine zipper motif), the transcription factor of JA-response genes, was crucial to the JA-signaling pathway. JAZs (encodes jasmonate-zim-domain protein) bind to MYC2 and inhibit their dissociation. Transcriptional levels of MYC2, JAZ1, and JAZ6 were also examined. After AHL-pretreatments, the expression of MYC2 increased 5.8 times at 6 h after inoculation compared with the unpretreated plants. Additionally, the expression of JAZ1 increased rapidly to a 30-fold increase at 6 h after inoculation. JAZ6 expression also increased, but the changes were not greater than those of JAZ1 (Figures 2B,C). This implied that JA signaling is involved in 3OC8-HSL priming in Arabidopsis.
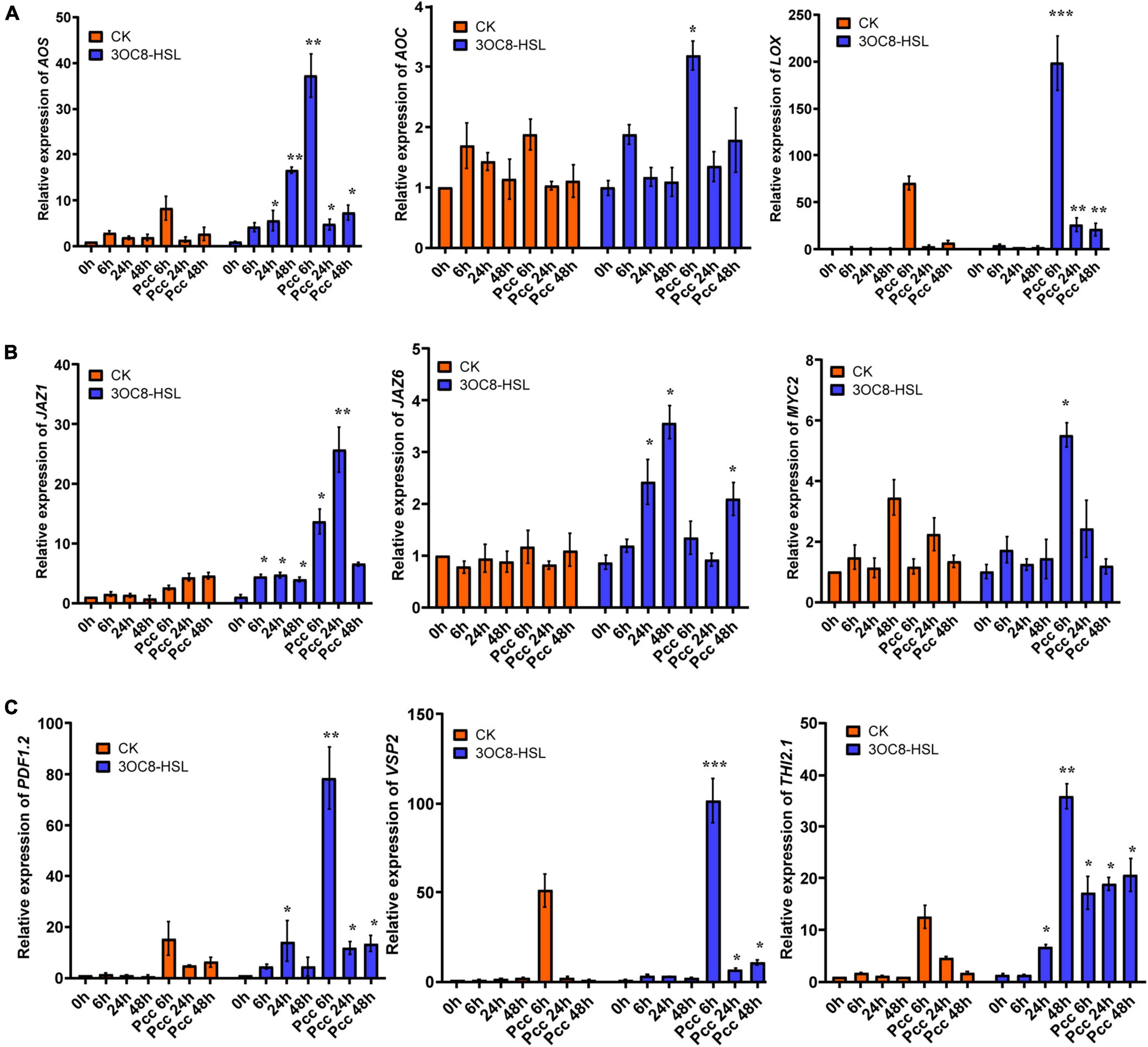
Figure 2. Expressions of the JA-related genes in Arabidopsis plants pretreated with 3OC8-HSL and challenged with Pcc. (A) Expressions of JA synthesis genes AOS, AOC, and LOX. (B) Expressions of JA pathway transcriptional regulators genes JAZ1, JAZ6, and MYC2. (C) Expressions of JA downstream cascade genes PDF1.2, VSP2, and Thi2.1. At least five independent experiments were performed, each of which with three technical repeats. Total RNA was extracted from Arabidopsis Col-0 seedlings pretreated with 10 μM 3OC8-HSL at the roots for 48 h followed by Pcc inoculation. Samples were collected at the indicated time points (hpi). Real-time PCR was performed using gene-specific primers, and the relative expression levels of the induced resistance marker genes are shown. Values are means ± SD of five independent experiments. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05; **P < 0.01, ***P < 0.001).
To further explore the role of JA in 3OC8-HSL-treated plants, the content of JA was detected by HPLC using the internal standard method. Plant roots were pretreated with 3OC8-HSL for 48 h. Afterward, the JA content was slightly induced, whereas further inoculations of Pcc dramatically promoted JA and JA-Ile accumulations in leaves (Figure 3A and Supplementary Figure 2). The results indicated that 3OC8-HSL was primarily involved in plants responding against Pcc by quickly elevating the accumulation of JA, which was in agreement with the enhanced accumulation of JA at this time point after the subsequent inoculation of Pcc (Figure 3A).
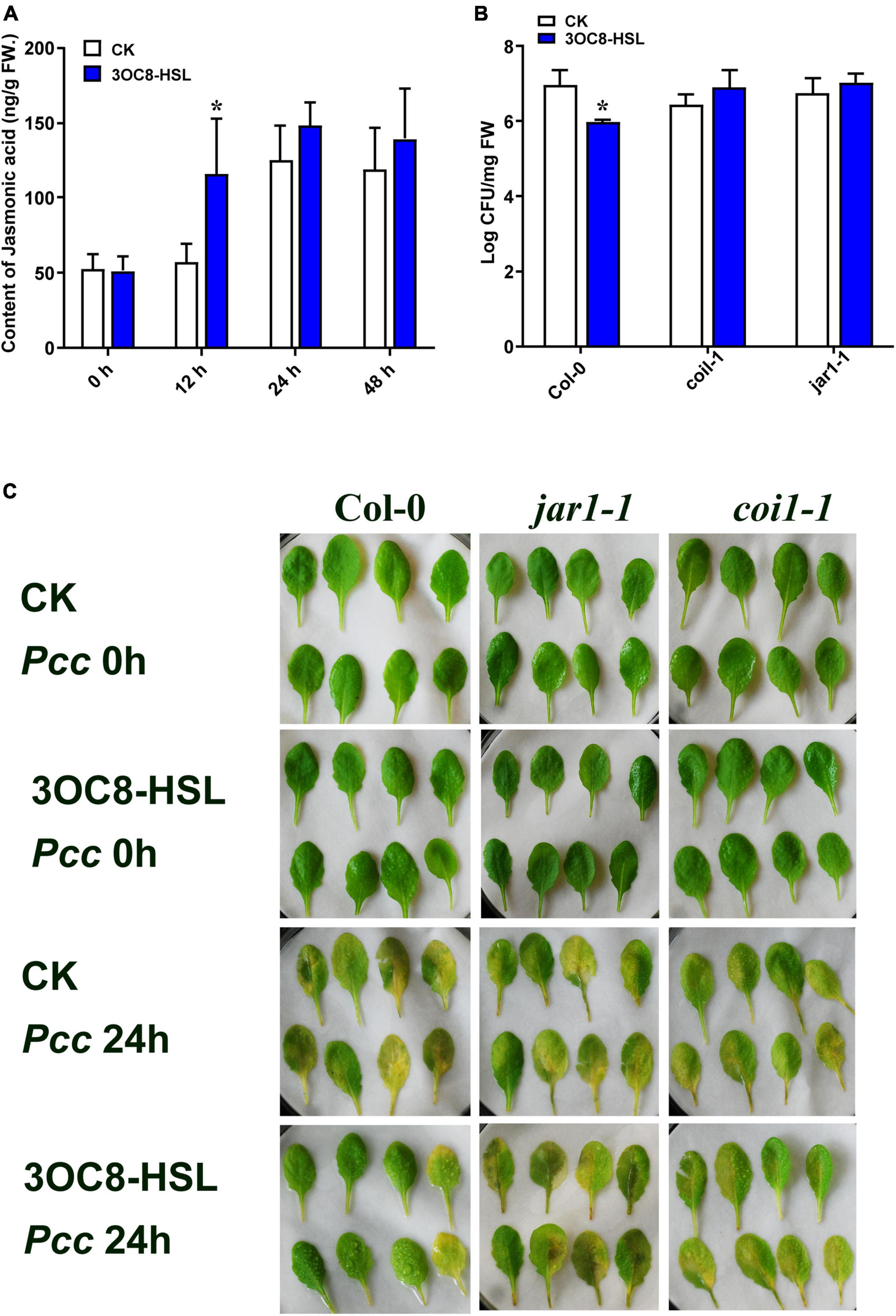
Figure 3. Effects of 3OC8-HSL application on the accumulation of JA and effects of 3OC8-HSL on Pcc growth in wild-type Arabidopsis Col-0, coi1-1, and jar1-1. (A) Accumulation of free JA measured by HPLC in Arabidopsis plants in which the roots were pretreated with 10 μM 3OC8-HSL for 48 h and the leaves were subsequently spray-inoculated with 107 CFU/mL Pcc. (B) Proliferation of Pcc in the leaves of Col-0, coi1-1, and jar1-1. (C) Symptoms of Pcc infection Col-0, coi1-1, and jar1-1. At least five independent experiments were performed, each of which with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of six independent experiments.
To further investigate the role of the JA-defense signaling cascade in 3OC8-HSL-induced plants, we compared the resistance of wild-type Arabidopsis plants (Col-0) and mutants impaired in the JA-signaling pathway. The jar1-1 mutant is defective in the synthesis of JA-Ile, the active compound in JA signaling (Chini et al., 2007), whereas coi1-1 is defective in JA perception (Yang et al., 2019). Unlike wild-type plants, which showed significantly reduced bacterial proliferation levels after pretreatment with 3OC8-HSL, the jar1-1 and coi1-1 mutants exhibited no difference in Pcc proliferation levels, independent of the 3OC8-HSL pretreatment, which suggested that the 3OC8-HSL-induced resistance required JAR1 and COI1 (Figures 3B,C). Thus, our findings demonstrated that the perception or the synthesis of the active JA-Ile was required for the 3OC8-HSL-primed resistance to Pcc in Arabidopsis. We also examined the expression levels of the JA transcriptional regulators JAZ1 and MYC2, as well as the response genes PDF1.2 and VSP2 in the coi1-1 mutant. In COI1-deficient mutant, the JAZ1 expression level was the same as in Col-0. In addition, the expression of MYC2, which increased four-fold in Col-0, showed no changes in coi1-1 mutant (Figures 4A,B). Downstream marker genes PDF1.2 and VSP2 also increased significantly, suggesting that the 3OC8-HSL priming of Pcc resistance was dependent on JA-Ile perception (Figures 4C,D).
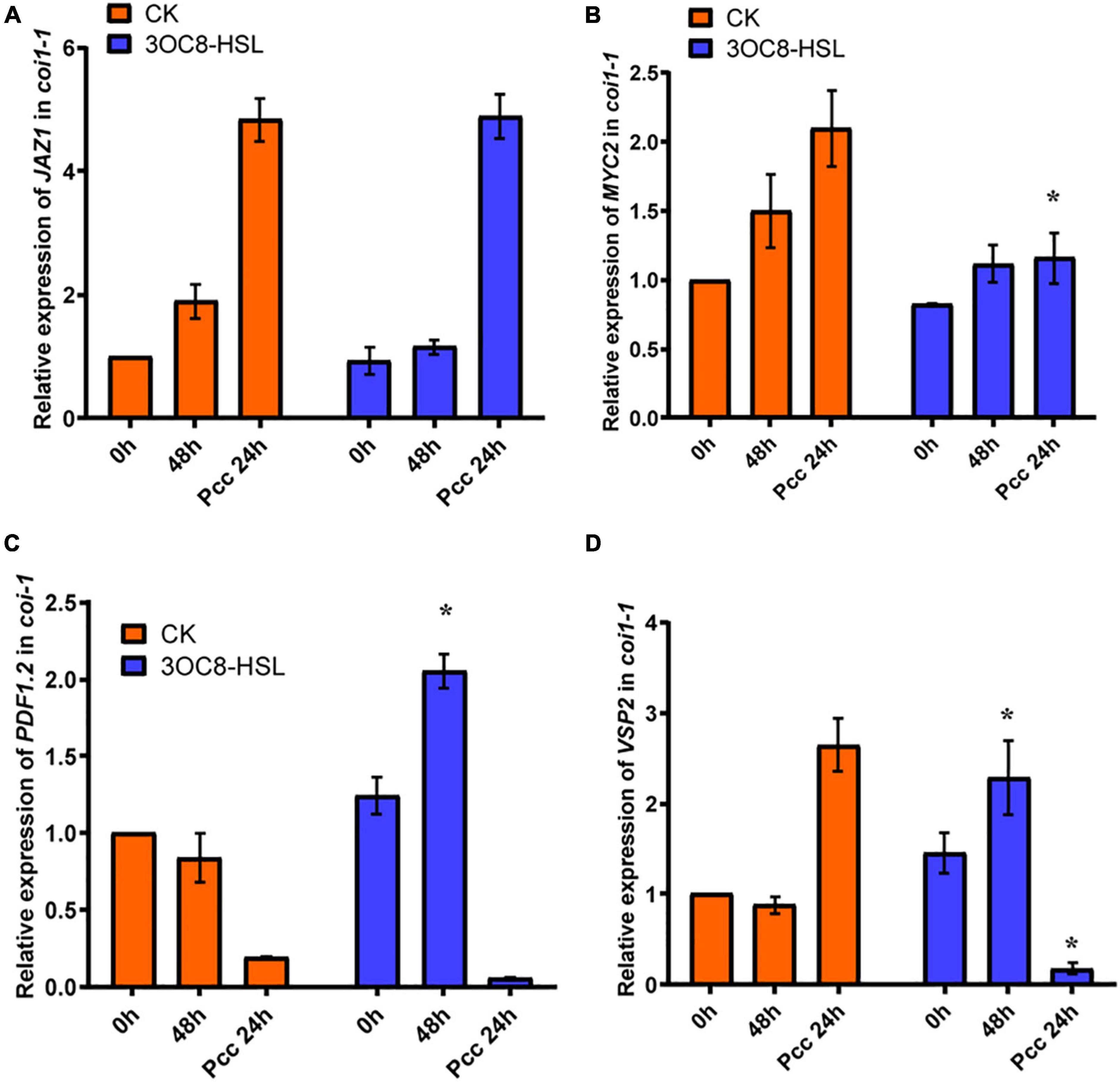
Figure 4. Expressions of JA regulatory genes and response genes in JA pathway mutant coi1-1. (A) Expression of JAZ1 in coi1-1 after 3OC8-HSL pretreated or not. (B) Expression of MYC2 in coi1-1 after 3OC8-HSL pretreated or not. (C) Expression of PDF1.2 in coi1-1 after 3OC8-HSL pretreated or not. (D) VSP2 in coi1-1 after 3OC8-HSL pretreated or not. At least five independent experiments were performed, each of which with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of five independent experiments.
In accordance with the gene chip data, we verified the upregulation of auxin-related transcription factors by 3OC8-HSL using qRT-PCR. We pretreated Col-0 plants with 3OC8-HSL for 48 h and detected the genes’ expression levels at 6, 24, and 48 h after inoculation. 3OC8-HSL-pretreated seedlings showed substantially increased transcript levels of the auxin biosynthetic genes ASB1 (encodes an anthranilate synthase beta subunit 1), CYP79B2 (encodes a cytochrome P450, family 79, subfamily B, polypeptide 2), and CYP79B3 (encodes a cytochrome P450, family 79, subfamily B, polypeptide 3) after inoculation (Figures 5A-C). We also analyzed the expression pattern of several genes involved in auxin perception and transport, as well as transcription factors after exposure to 3OC8-HSL. As shown in Figures 5D-F, after Col-0 plants were treated with 3OC8-HSL, the expression levels of TIR1 (encodes an auxin receptor that mediates auxin-regulated transcription), GH3 (encodes an IAA-amido synthase that conjugates Asp and other amino acids to auxin in vitro), and SAUR (encodes a small auxin-up RNA) were basically stable. However, after inoculation with Pcc, the expressions of TIR1, GH3, and SAUR increased within 48 h in all the AHL-pretreated seedlings (Figures 5D-F).
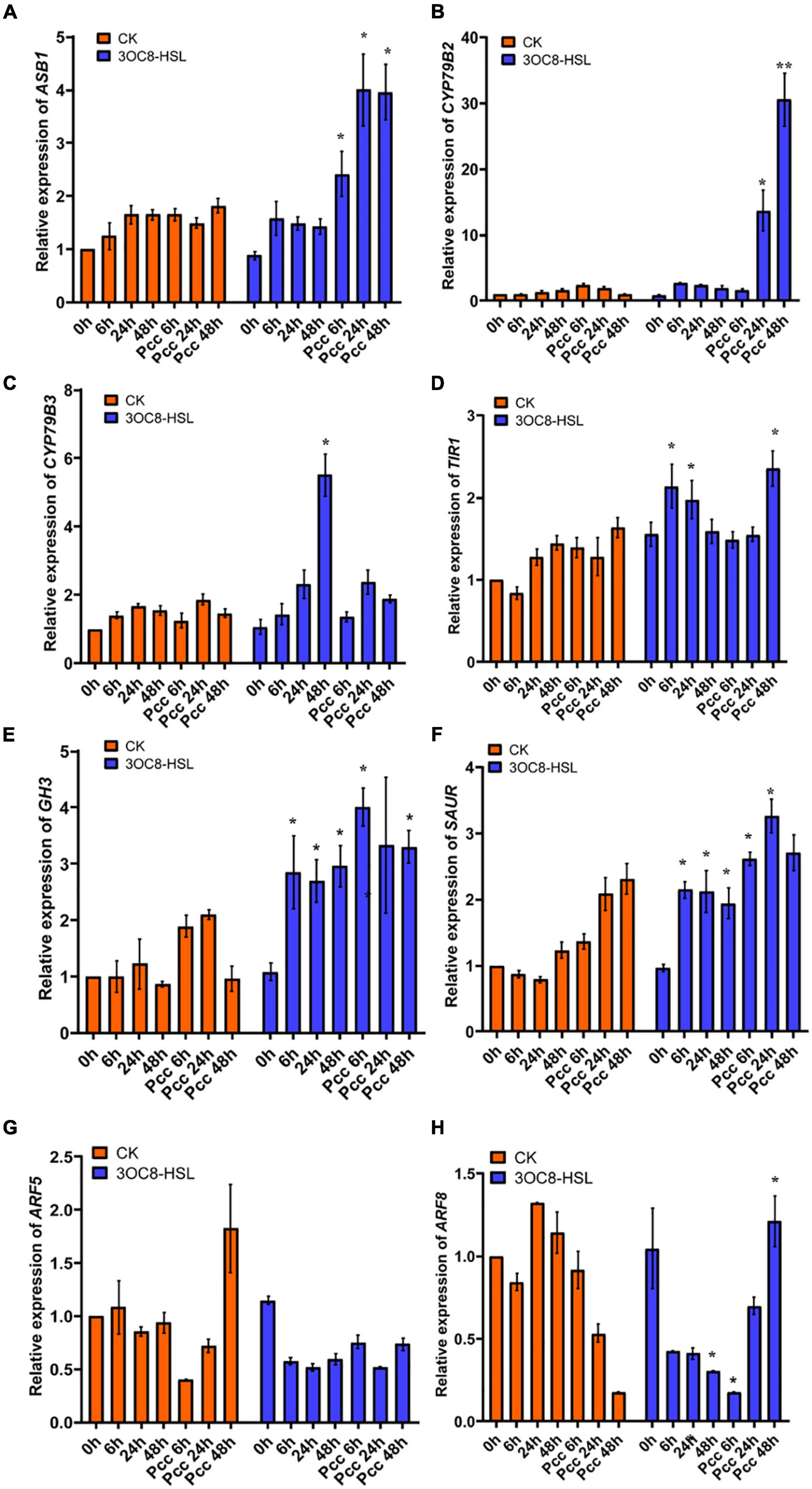
Figure 5. Expressions of the auxin-related genes in Arabidopsis plants pretreated with 3OC8-HSL and challenged with Pcc. (A) Expression of Auxin pathway transcriptional regulators genes ASB1. (B) Expression of auxin pathway transcriptional regulators gene CYP79B2. (C) Expression of auxin perception gene CYP79B3. (D) Expression of auxin synthesis gene TIR1. (E) Expression of auxin synthesis gene GH3. (F) Expression of auxin synthesis gene SAUR. (G) Expression of auxin downstream cascade gene ARF5. (H) Expression of auxin downstream cascade gene ARF7. At least five independent experiments were performed, each of which with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of five independent experiments.
Auxin response factors (ARFs) are transcriptional factors that bind to the specific DNA sequence 5’-TGTCTC-3’ found in auxin-responsive promoter elements. ARF5 mediates embryo axis formation and vascular tissues’ differentiation (Spaepen and Vanderleyden, 2011), and it did not dramatically change in AHL-pretreated plants compared with the unpretreated plants (Figure 5G). ARF8 has been reported to be required for JA biosynthesis (Werghi et al., 2021), and it had a lower expression level after unpretreated plants were inoculated with Pcc. The downregulation of ARF8 leads to a decrease in auxin responses and JA synthesis (Yang et al., 2019). However, AHL pretreatment prior to inoculation with Pcc rescued the repression of ARF8 in plants, resulting in increased resistance to Pcc (Figure 5H).
To investigate the influence of auxin on priming effect initiated by 3OC8-HSL, we measured the expression of the GFP reporter gene in DR5::GFP (the auxin-response marker) transgenic Arabidopsis. The results showed that the intensity of GFP fluorescence in primary root cells increased at 24 h after 10-μM 3OC8-HSL pretreatment (Figure 6A). These results indicate that 3OC8-HSL induced the accumulation of auxin in the primary root tips of Arabidopsis. The GFP expression level was significantly induced in seedlings by AHL pretreatment and increased more sharply after inoculation with Pcc bacteria, reaching five times the level in the unpretreated plants (Figure 6B). Similarly, the 10 μM 3OC8-HSL treatment induced a rapid increase in the GUS activity in DR5::GUS transgenic plants (Supplementary Figure 3).
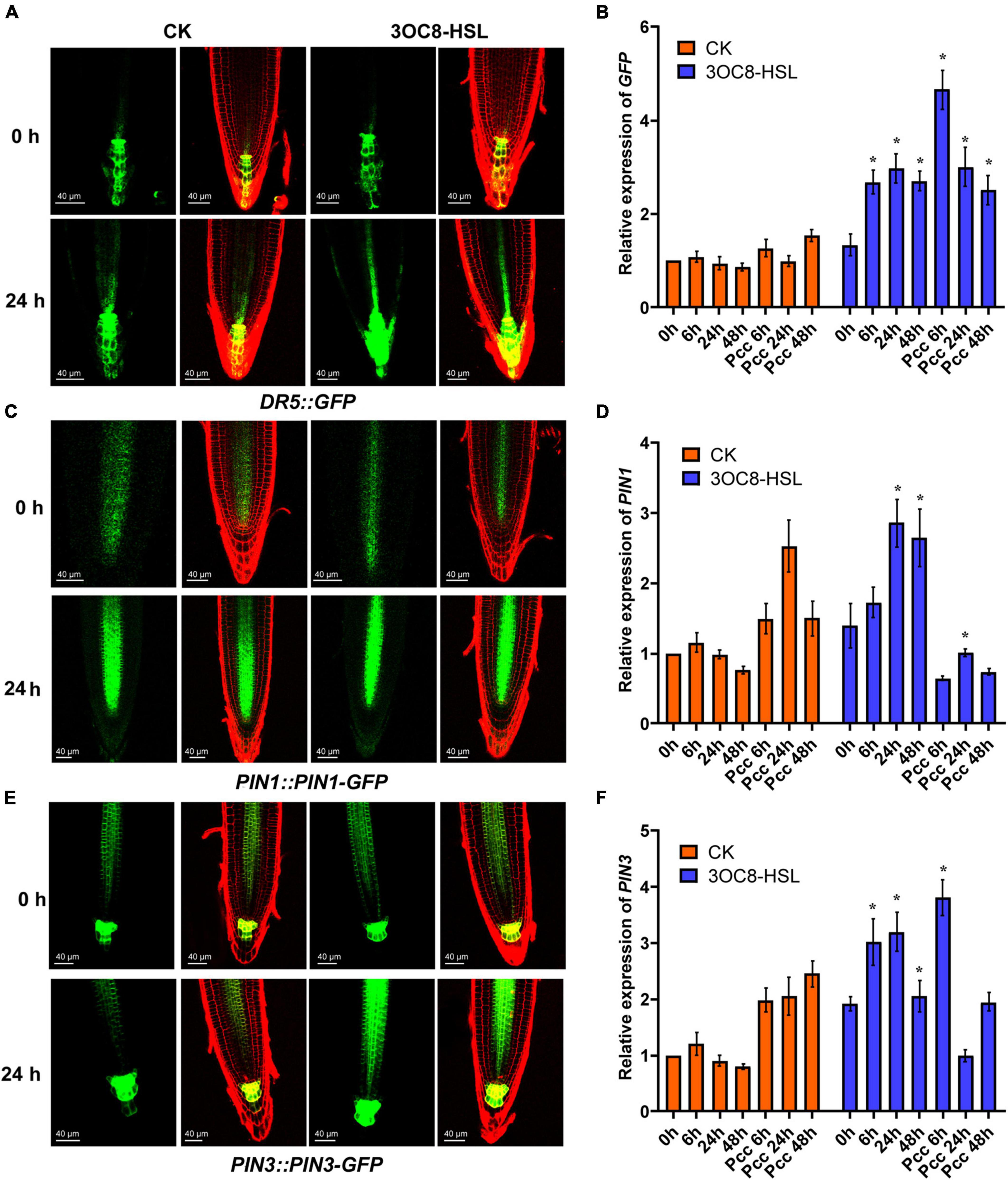
Figure 6. AHL-induced expressions of auxin response gene DR5, auxin transport gene PIN1 and PIN3. (A,C,E) GFP fluorescence of primary roots (PR), in 5-d-old DR5::GFP, PIN1::PIN1-GFP, PIN3::PIN3-GFP seedlings exposed to 10 μM 3OC8-HSL for 24h. DR5::GFP auxin response is increased in QC. Cell walls were stained with PI. We performed the experiment 5 times and 30 roots each treatment. The left panels show the GFP fluorescence, on the right is the merged image of PI stained cell wall with GFP fluorescence. (B,D,F) Expressions of GFP, PIN1, PIN3. At least five independent experiments were performed, each of which with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of five independent experiments.
The PIN proteins are important regulators involved in the establishment of the auxin gradient and the maximum auxin level in the root apex. We used the PIN1::PIN1-GFP and PIN3::PIN3-GFP transgenic lines to monitor their expression levels. As shown in Figure 6, the 3OC8-HSL pretreatment significantly promoted the expression level of PIN3, but not PIN1, compared with in the unpretreated plants (Figures 7B-D). These data indicated that PIN3 may involve in the 3OC8-HSL-induced changes in auxin accumulation and distribution in plant tissues.
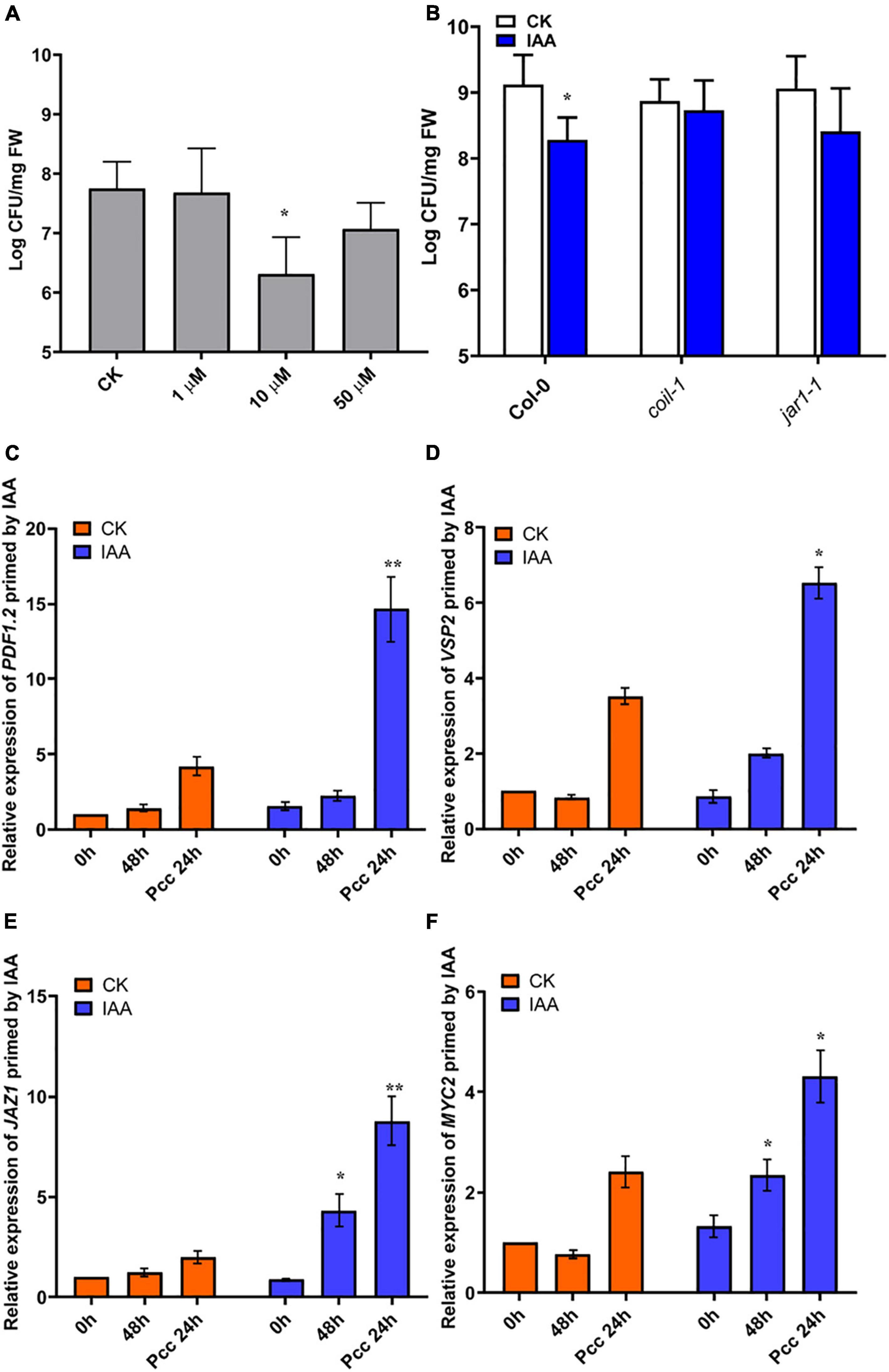
Figure 7. IAA induces resistance to Pcc in a JA-dependent manner. (A) Effects of different concentrations of IAA on the proliferation of Pcc in the leaves of Arabidopsis plants (grown in a hydroponic system). (B) Proliferation of Pcc in the leaves of Col-0, coi1-1, jar1-1(grown in a hydroponic system) in which the roots were pretreated with 10 μM IAA for 48 h and the leaves were subsequently spray-inoculated with 107 CFU/mL Pcc. (C–F) Expressions of JAZ1, MYC2, PDF1.2, and VSP2 induced by 10 μM IAA. At least five independent experiments were performed, each of which with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants ANOVA test, (*P < 0.05, **P < 0.01). Values are means ± SD of five independent experiments.
Our microarray data suggested that significant changes in auxin signaling pathway-related genes occurred after 3OC8-HSL pretreatment. Consequently, we measured plant resistance to Pcc after roots pretreatment with 10 μM IAA. We found that IAA significantly reduced Pcc count numbers in pretreated plants (Figure 7A). We also found that the IAA pretreatment could induce elevation in the expression levels of JA pathway genes, such as JAZ1, MYC2, PDF1.2, and VSP2. Thus, the JA pathway was activated by the IAA pretreatment. We detected bacterial cell numbers in coi1-1 and jar1-1 mutants after the IAA pretreatment, and the Pcc CFU was similar in the IAA unpretreated mutants (Figure 7B). This indicated that IAA-induced resistance to Pcc was dependent on the JA pathway. Because 3OC8-HSL can induce the JA and auxin pathways, the priming effect may result from the integration of the JA and auxin pathways.
To explore the application effects of 3OC8-HSL on Chinese cabbage resistance, detached leaves and potted plants were used. The lesion areas of the leaves treated with 3OC8-HSL were significantly smaller than those on leaves of plants not receiving AHL treatment. For the potted plants, the main veins of leaves from unpretreated plants were broken and slowly withered owing to the decay at the inoculation site (Figure 8A). The 3OC8-HSL pretreatment significantly reduced Pcc colonization in Chinese cabbage leaves (Figure 8B). Like 3OC8-HSL, IAA had a priming effect on Pcc, resulting in the reduced bacterial colonization of Chinese cabbage. In addition, the JA synthesis gene BraAOS (Bra035320), auxin-related gene BraTIR1, and BraJAZ1 in Chinese cabbage were detected. The expression levels of BraAOS, BraJAZ1, and BraTIR1 were significantly increased after AHL pretreatment (Figures 8D-F).
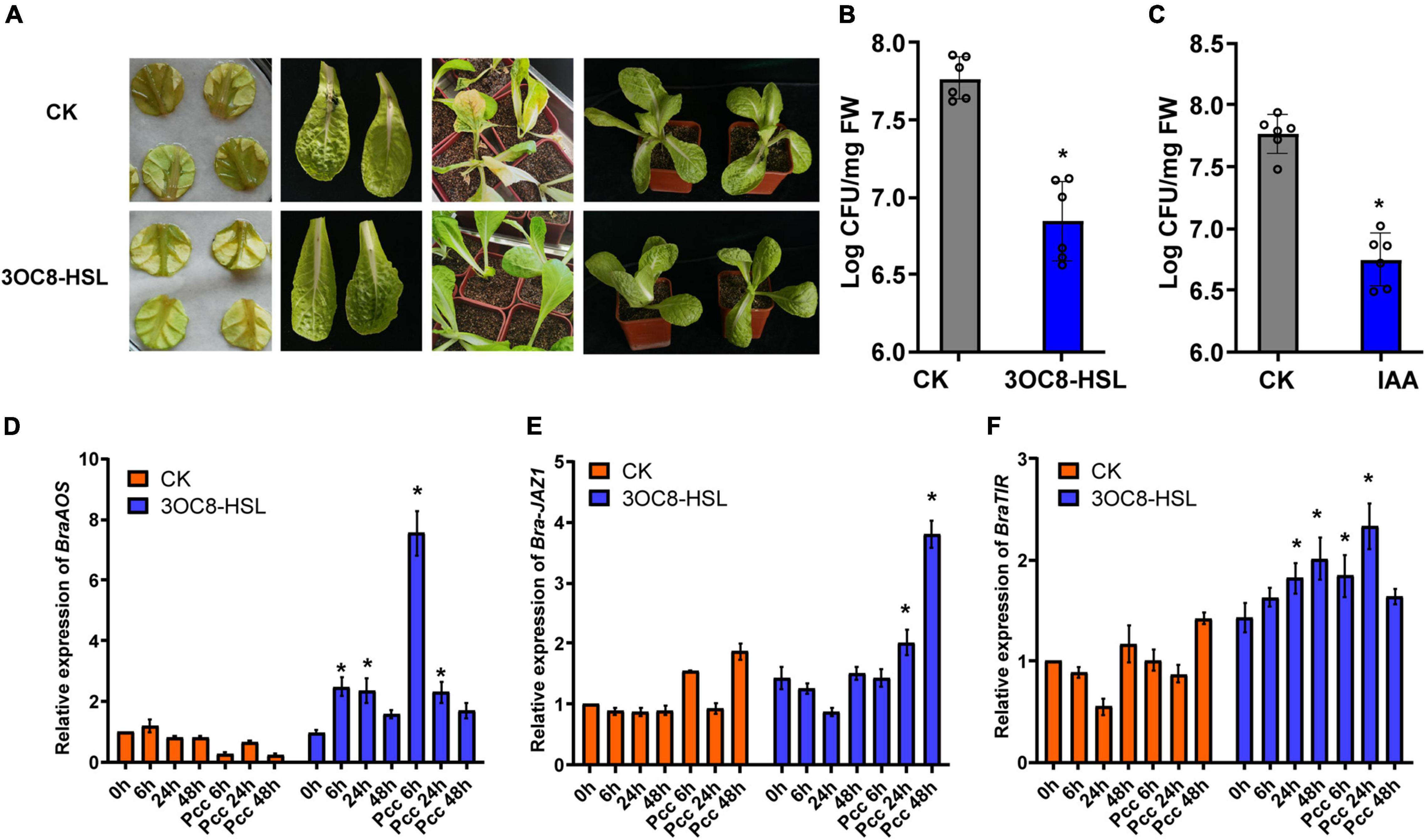
Figure 8. (A) Symptoms of Pcc infection in Chinese cabbage. The top line is untreated Chinese cabbage leaves inoculated with Pcc, and the bottom line is 3OC8 pretreated leaves inoculated with Pcc. (B) AHL priming effect in Chinese cabbage. The proliferation of Pcc in the leaves of Chinese cabbage (grown in pot) in which the roots were pretreated with 10 μM 3OC8-HSL for 48 h and the leaves were subsequently spray-inoculated with 107 CFU/mL Pcc. (C) IAA priming effect in Chinese cabbage. The proliferation of Pcc in the leaves of Chinese cabbage (grown in pot) in which the roots were pretreated with 10 μM IAA compound for 48 h and the leaves were subsequently spray-inoculated with 107 CFU/mL Pcc. (D–F) Expressions of BraAOS, BraJAZ1, and BraTIR1 induced by 10 μM IAA. At least five independent experiments were performed, each of which with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of five independent experiments.
Priming is defined as an induced state whereby a plant reacts more rapidly and more efficiently to stress. This is an adaptive strategy that improves the defensive capacity of plants (Tugizimana et al., 2018). The primed plants may respond to very low levels of stimuli in a faster and stronger manner than unprimed plants (Mauch-Mani et al., 2017). In this study, we demonstrated that 3OC8-HSL pretreated plants acted as an inducer of resistance against the necrotrophic pathogen Pcc in Arabidopsis (Figure 1). We found that 3OC8-HSL pretreatment did not affect the JA content and JA-related genes’ expression levels, whereas those of the auxin-response gene DR5 were slightly induced (Figures 2, 3, 6). After Pcc inoculation, there were dramatically induced expression levels of both auxin and JA pathways (Figures 2–5). These phenomena indicated that the 3OC8-HSL pretreatment only primed the plants to maintain a ready-to-go state that had no or minimal negative impacts on the host plants’ energy status. When encountering stresses, such as Pcc invasion, the primed plants defended against the bacterial infection more rapidly and dramatically than unprimed plants (Figure 8). Thus, we reported that the primed plants are more sensitive to stresses involved in induced systemic resistance and systemic acquired resistance activation (Katagiri, 2018). Consistent with these findings, the root pretreatment of the plants with 3OC8-HSL led to the upper leaves (distal tissues) being more resistant to the bacterial pathogen (Figure 7), but 3OC8-HSL was not detected in these leaves, which indicated that 3OC8-HSL triggers plant immunity by activating induced systemic resistance or systemic acquired resistance.
Numerous AHLs have been identified from 70 species of Gram-negative bacteria. It was reported that AHLs with short side chains (four to six carbons) regulate root growth and development, whereas long-chain AHLs such as C12- and C14-HSL induce plant resistance (Schenk and Schikora, 2014). In our study, we showed that 3OC8-HSL was a short-chain AHL that enhanced resistance against the necrotrophic pathogen Pcc in Arabidopsis and cabbage. The phenomena were similar to those found in 3OC14-HSL-pretreated plants, in which triggered defense does not respond. The 3OC14-HSL-pretreated plants showed faster and stronger activation of defense responses than unpretreated plants after inoculation with biotrophic and hemi-biotrophic pathogenic bacteria or a pathogenic elicitor flagellin peptide (flg22, Schenk et al., 2014). 3OC6-HSL also induces the priming of plants, which indicated that AHLs carbonylated at the C3-position of fatty acid side chains might play an important role in regulating plant disease resistance.
Our previous data indicate that 3OC8-HSL primes the Arabidopsis defense response to hemi-biotrophic bacterial infection and that 3OC8-HSL-primed resistance is dependent on the SA-signaling pathway (Liu et al., 2020). Cross-talk between the defense signaling hormones SA and JA, as well as growth regulators play significant roles in mediating the trade-off between growth and defense in plants. Recent studies have provided new insights into the role of auxin in plant defense. Although SA does not affect auxin biosynthesis, it represses the expressions of auxin receptor genes TIR1/AFBs to reduce the auxin response (Navarro et al., 2006; Wang et al., 2007). In contrast to the well-recognized antagonistic crosstalk between SA and auxin during plant resistance to biotrophic pathogens, Qi et al. (2012) demonstrated that Alternaria brassicicola infection led to an enhanced auxin response in host plants. This also provided molecular evidence that supported the hypothesis in which JA and auxin promoted plant resistance to necrotrophic pathogen coordinately. When plants are pretreated by exogenous auxin, the auxin–TIR–AUX/IAA–ARF signaling is activated, then JA synthesis increases, which leads to further up-regulation of auxin synthase genes and the auxin accumulation.
The plant hormone JA plays essential roles in many biological processes including plant defense against necrotrophic pathogens (Kazan and Manners, 2013). We found that the JA pathway was not affected by AHLs treatment, and those significant changes occurred after the plants were inoculated with the necrotrophic pathogen Pcc. The JA perception impaired mutant coi1-1 and JA synthesis impaired mutant jar1-1, did not show the priming effects. These results indicated that the JA pathway was essential in mediating resistance to necrotrophic pathogens. The rapidly increasing expression levels of JA-responsive genes, such as PDF1.2 and VSP2, in 3OC8-HSL-pretreated plants may contribute to the elevated response of primed plants. The 3OC8-HSL pretreatment also induced the expressions of auxin pathway genes, and consequently, IAA priming effects played the same roles in the resistance against Pcc, but the effects were absent in the coi1-1 or jar1-1 mutant, suggesting that IAA-induced resistance was dependent on the JA pathway. Consistently, Qi et al. (2012) demonstrated that JA and auxin interact by positively regulating plant resistance to the necrotrophic pathogen A. brassicicola, and that auxin signaling is activated through JA, which may contribute to plant resistance to necrotrophic pathogens. We deduced that the synergistic induction of JA and auxin, as well as the underlying molecular mechanism need to be further explored. These findings will expand our understanding of the mechanisms that plants use to respond to 3OC8-HSL and will provide insights into novel applications of these biological molecules in regulating crop growth and development.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, GSE197485.
FL and QZ performed the plant pathology analysis and molecular, genetic assay, respectively. ZJ performed the network analysis. SZ and JW helped with the acquisition, analysis, and interpretation of data for the work. YJ and SS were responsible for the design of the work. All authors gave approval to the final version.
This work was financially supported by a project from the National Basic Research Program of China (No. 2015CB150604), the National Natural Science Foundation of China (Nos. 31972233 and 31601144), and the Natural Science Foundation of Hebei (C2015302020).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Wenqiang Tang (Hebei Normal University in China) for providing the transgenic lines DR5::GFP, DR5::GUS, PIN1::PIN1-GFP, and PIN3::PIN3-GFP seeds, Shuxing Shen (Hebei Agricultural University in China) for providing Chinese cabbage Wild type A03 seeds and Pcc pathogen BC1. We would also like to thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the language of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.886268/full#supplementary-material
Supplementary Figure 1 | Enrichment analysis of differential genes expressions in microarray and expressions of key transcription factors. (A) GO classification plots and enrichment scatter plots of differentially expressed genes. The larger the enrichment factor indicates the more significant the enrichment level of differentially expressed genes in this pathway, the size of the circle indicates the number of genes enriched in the pathway, and the larger the circle indicates the more genes. (B) Pathway classification and enrichment of differentially expressed genes. (A) and (B) were plotted by http://www.bioinformatics.com.cn, a free online platform for data analysis and visualization. (C) Expression of the transcription factor gene MYB44. (D) Expression of the SA synthesis related gene ICS1. (E) Expression of the SA synthesis related gene SARD1. (F) Expression of the SA synthesis related gene CBP60g. (G) Expression of the transcription factor gene WRKY70. (H) Expression of the transcription factor gene RVE1. For genes’ expressions, Arabidopsis plants were pretreated with 3OC8-HSL and challenged with Pcc. The experiments were performed at least five independent experiments with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of five independent experiments.
Supplementary Figure 2 | Effects of 3OC8-HSL application on the accumulation of JA-Ile. Accumulation of JA-Ile was measured by ELISA in Arabidopsis plants in which the roots were pretreated with 10 μM 3OC8-HSL for 48 h and the leaves were subsequently spray-inoculated with 107 CFU/mL Pcc. The experiments were performed at least three independent experiments with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of three independent experiments.
Supplementary Figure 3 | AHL induced expression GUS in DR5::GUS plant. The experiments were performed at least five independent experiments with three technical repeats. Asterisks indicate a statistically significant difference between the AHL-pretreated and the water-treated plants (ANOVA test, *P < 0.05). Values are means ± SD of five independent experiments.
Benková, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertová, D., and Jürgens, G. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., and Friml, J. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Chini, A., Fonseca, S., Fernandez, G., Adie, B., Chico, J. M., and Lorenzo, O. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Conrath, U., Pieterse, C., and Mauch-Mani, B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7, 210–216. doi: 10.1016/S1360-1385(02)02244-6
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., and Hamann, T. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. doi: 10.1038/nature02085
Hu, Z., Shao, S., Zheng, C., Sun, Z. H., Sh, J. Y., and Yu, J. Q. (2018). Induction of systemic resistance in tomato against Botrytis cinerea by N-decanoyl-homoserine lactone via jasmonic acid signaling. Planta 247, 1217–1227. doi: 10.1007/s00425-018-2860-7
Katagiri, F. (2018). Review: Plant immune signaling from a network perspective. Plant Sci. 276, 14–21. doi: 10.1016/j.plantsci.2018.07.013
Kauss, H., Theisinger-Hinkel, E., Mindermann, R., and Conrath, U. (1992). Dichloroisonicotinic and salicylic acid, inducers of systemic acquired resistance, enhance fungal elicitor responses in parsley cells. Plant J. 2, 655–660. doi: 10.1111/j.1365-313X.1992.tb00134.x
Kazan, K., and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Kessmann, H., Staub, T., Hofmann, C., Maetzke, T., and Herzog, J. (1994). Induction of systemic acquired disease resistance in plants by chemicals. Annu. Rev. Phytopathol. 32, 439–459. doi: 10.1146/annurev.py.32.090194.002255
Livak, K. J., and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–8. doi: 10.1006/meth.2001.1262
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, J., Zhong, R., and Palva, E. T. (2017). WRKY70 and its homolog WRKY54 negatively modulate the cell wall-associated defenses to necrotrophic pathogens in Arabidopsis. PLoS One 12:e0183731. doi: 10.1371/journal.pone.0183731
Liu, F., Bian, Z., Jia, Z., Zhao, Q., and Song, S. (2012). The GCR1 and GPA1 participate in promotion of Arabidopsis primary root elongation induced by N-acyl-homoserine lactones, the bacterial quorum-sensing signals. Mol. Plant Microbe Interact. 25, 677–683. doi: 10.1094/mpmi-10-11-0274
Liu, F., Zhao, Q., Jia, Z., Song, C., Huang, Y., and Ma, H. (2020). N-3-oxo-octanoyl-homoserine lactone-mediated priming of resistance to Pseudomonas syringae requires the salicylic acid signaling pathway in Arabidopsis thaliana. BMC Plant Biol. 20:38. doi: 10.1186/s12870-019-2228-6
Liu, M., Wu, F., Wang, S., Lu, Y., Chen, X., and Wang, Y. (2019). Comparative transcriptome analysis reveals defense responses against soft rot in Chinese cabbage. Hortic. Res. 1:68. doi: 10.1038/s41438-019-0149-z
Martinez-Medina, A., Flors, V., Heil, M., Mauch-Mani, B., Pieterse, C., and Pozo, M. J. (2016). Recognizing plant defense priming. Trends Plant Sci. 818–822. Epub ahead of print doi: 10.1016/j.tplants.2016.07.009
Mathesius, U., Mulders, S., Gao, M., Teplitski, M., Caetano-Anolles, G., and Rolfe, B. (2003). Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci.U. S. A. 100, 1444–1449. doi: 10.1073/pnas.262672599
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense Priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512. doi: 10.1146/annurev-arplant-042916-041132
Miao, C., Liu, F., Zhao, Q., Jia, Z., and Song, S. (2012). A proteomic analysis of Arabidopsis thaliana seedling responses to 3-oxo-octanoyl-homoserine lactone, a bacterial quorum-sensing signal. Biochem. Biophys. Res. Commun. 427, 293–298. doi: 10.1016/j.bbrc.2012.09.044
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., and Estelle, M. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Norman-Setterblad, C., Vidal, S., and Palva, E. T. (2000). Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora. Mol. Plant Microbe Interact 13, 430–438. doi: 10.1094/MPMI.2000.13.4.430
Ortíz-Castro, R., Martínez-Trujillo, M., and López-Bucio, J. (2008). N-acyl-L-homoserine lactones: a class of bacterial quorum-sensing signals alter post-embryonic root development in Arabidopsis thaliana. Plant Cell Environ. 31, 1497–1509. doi: 10.1111/j.1365-3040.2008.01863.x
Palmer, A. G., Senechal, A. C., Mukherjee, A., Ane, J. M., and Blackwell, H. E. (2014). Plant responses to bacterial N-acyl L-homoserine lactones are dependent on enzymatic degradation to L-homoserine. ACS Chem. Biol. 9, 1834–1845. doi: 10.1021/cb500191a
Pang, Y., Liu, X., Ma, Y., Chernin, L., Berg, G., and Gao, K. (2009). Induction of systemic resistance, root colonisation and biocontrol activities of the rhizospheric strain of Serratia plymuthica are dependent on N-acyl homoserine lactones. Eur. J. Plant Pathol. 124, 261–268. doi: 10.1007/s10658-008-9411-1
Perombelon, M. C. M., and Kelman, A. (1980). Ecology of the soft rot Erwinias. Annu. Rev. Phytopathol. 18, 361–387. doi: 10.1146/annurev.py.18.090180.002045
Qi, L. L., Yan, J., Li, Y. N., Jiang, H. L., Sun, J. Q., and Chen, Q. (2012). Arabidopsis thaliana plants differentially modulate auxin biosynthesis and transport during defense responses to the necrotrophic pathogen Alternaria brassicicola. New Phytol. 195, 872–882. doi: 10.1111/j.1469-8137.2012.04208.x
Rad, U. V., Klein, I., Dobrev, P. I., Kottova, J., Zazimalova, E., and Fekete, A. (2008). Response of Arabidopsis thaliana to N-hexanoyl-dl -homoserine-lactone, a bacterial quorum sensing molecule produced in the rhizosphere. Planta 229, 73–85. doi: 10.1007/s00425-008-0811-4
Reading, N. C., and Vanessa, S. (2010). Quorum sensing: the many languages of bacteria. FEMS Microbiol. Lett. 254, 1–11. doi: 10.1111/j.1574-6968.2005.00001.x
Schenk, S., Hernández-Reyes, C., Samans, B., Stein, E., Neumann, C., and Schikora, M. (2014). N-acyl-homoserine lactone primes plants for cell wall reinforcement and induces resistance to bacterial pathogens via the salicylic acid/oxylipin pathway. Plant Cell 26, 2708–2723. doi: 10.1105/tpc.114.126763
Schenk, S., Stein, E., Kogel, K., and Schikora, A. (2012). Arabidopsis growth and defense are modulated by bacterial quorum sensing molecules. Plant Signal. Behav. 7, 178–181. doi: 10.4161/psb.18789
Schenk, S., and Schikora, A. (2014). AHL-priming functions via oxylipin and salicylic acid. Front. Plant Sci. 5:784. doi: 10.3389/fpls.2014.00784
Schikora, A., Schenk, S. T., Stein, E., Molitor, A., and Kogel, Z. (2011). N-acyl-homoserine lactone confers resistance toward biotrophic and hemibiotrophic pathogens via altered activation of AtMPK6. Plant Physiol. 157, 1407–1418. doi: 10.1104/pp.111.180604
Schuhegger, R., Ihring, A., Gantner, S., Bahnweg, G., Knappe, C., and Vogg, G. (2006). Induction of systemic resistance in tomato by N-acyl-L-homoserine lactone-producing rhizosphere bacteria. Plant Cell Environ. 29, 909–918. doi: 10.1111/j.1365-3040.2005.01471.x
Sharma, A., Singh, P., Sarmah, B. K., and Nandi, S. P. (2020). Quorum sensing: its role in microbial social networking. Res. Microbiol. 171, 159–164. doi: 10.1016/j.resmic.2020.06.003
Shrestha, A., Grimm, M., Ojiro, I., Krumwiede, J., and Schikora, A. (2020). Impact of quorum sensing molecules on plant growth and immune system. Front. Microbiol. 11:e1545. doi: 10.3389/fmicb.2020.01545
Spaepen, S., and Vanderleyden, J. (2011). Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 3:a001438. doi: 10.1101/cshperspect.a001438
Sun, P., Tian, Q. Y., Chen, J., and Zhang, W. H. (2010). Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 61, 347–356. doi: 10.1093/jxb/erp306
Taga, M. E., Miller, S. T., and Bassler, B. L. (2003). Lsr-mediated transport and processing of AI-2 in salmonella typhimurium. Mol. Microbiol. 50, 1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x
Thevenet, D., Pastor, V., Baccelli, I., Balmer, A., Vallat, A., and Neier, R. (2016). The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. New Phytol. 213, 552–559. doi: 10.1111/nph.14298
Tugizimana, F., Mhlongo, M. I., Piater, L. A., and Dubery, I. A. (2018). Metabolomics in plant priming research: the way forward? Int. J. Mol. Sci. 19:1759. doi: 10.3390/ijms19061759
Veronico, P., Paciolla, C., Pomar, F., De Leonardis, S., García-Ulloa, A., and Melillo, M. T. (2018). Changes in lignin biosynthesis and monomer composition in response to benzothiadiazole and root-knot nematode Meloidogyne incognita infection in tomato. Plant Physiol. 230, 40–50. doi: 10.1016/j.jplph.2018.07.013
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H., and Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790. doi: 10.1016/j.cub.2007.09.025
Werghi, S., Herrero, F. A., Fakhfakh, H., and Gorsane, F. (2021). Auxin drives tomato spotted wilt virus (TSWV) resistance through epigenetic regulation of auxin response factor ARF8 expression in tomato. Gene 804:145905. doi: 10.1016/j.gene.2021.145905
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10:1349. doi: 10.3389/fpls.2019.01349
Zarkani, A., Stein, E., Röhrich, C., Schikora, M., Evguenieva-Hackenberg, E., and Degenkolb, T. (2013). Homoserine lactones influence the reaction of plants to rhizobia. Int. J. Mol. Sci. 14, 17122–17146. doi: 10.3390/ijms140817122
Zhao, Q., Li, M., Jia, Z., Liu, F., Ma, H., Huang, Y., et al. (2016). AtMYB44 positively regulates the enhanced elongation of primary roots induced by N-3-oxo-hexanoyl-homoserine lactone in Arabidopsis thaliana. Mol. Plant Microbe. Interact. 29:774. doi: 10.1094/MPMI-03-16-0063-R
Zhao, Q., Yang, X., Li, Y., Liu, F., Cao, X., and Jia, Z. (2020). N-3-oxo-hexanoyl-homoserine lactone, a bacterial quorum sensing signal, enhances salt tolerance in Arabidopsis and wheat. Bot. Stud. 61:8. doi: 10.1186/s40529-020-00283-5
Keywords: jasmonic acid, auxin, AHL, Pectobacterium carotovorum, priming
Citation: Liu F, Zhao Q, Jia Z, Zhang S, Wang J, Song S and Jia Y (2022) N-3-Oxo-Octanoyl Homoserine Lactone Primes Plant Resistance Against Necrotrophic Pathogen Pectobacterium carotovorum by Coordinating Jasmonic Acid and Auxin-Signaling Pathways. Front. Plant Sci. 13:886268. doi: 10.3389/fpls.2022.886268
Received: 28 February 2022; Accepted: 25 April 2022;
Published: 14 June 2022.
Edited by:
Xiaodong Wang, Agricultural University of Hebei, ChinaReviewed by:
Yinyue Deng, Sun Yat-sen University, ChinaCopyright © 2022 Liu, Zhao, Jia, Zhang, Wang, Song and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuishan Song, c2h1aXNoYW5zNjIwQDE2My5jb20=; Yantao Jia, amlheXRAaW0uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.