- 1Hainan Key Laboratory of Tropical Oil Crops Biology/Coconut Research Institute, Chinese Academy of Tropical Agricultural Sciences, Wenchang, China
- 2Institute of Tropical Agriculture and Forestry, Hainan University, Haikou, China
- 3Tropical Crops Genetic Resources Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
Oil palm (Elaeis guineensis Jacq.) is a tropical woody oil crop of the palm family and is known as “the oil king of the world,” but its palm oil contains about 50% palmitic acid, which is considered unhealthy for humans. Intron polymorphisms (IP) are highly efficient and easily examined molecular markers located adjacent to exon regions of functional genes, thus may be associated with targeted trait variation. In order to speed up the breeding of oil palm fatty acid composition, the current study identified a total of 310 introns located within 52 candidate genes involved in fatty acid biosynthesis in the oil palm genome. Based on the intron sequences, 205 primer pairs were designed, 64 of which showed polymorphism among 70 oil palm individuals. Phenotypic variation of fatty acid content in the 70 oil palm individuals was also investigated. Association analysis revealed that 13 IP markers were significantly associated with fatty acid content variation, and these IP markers were located on chromosomes 2, 5, 6, 8, 9, and 10 of oil palm. The development of such IP markers may be useful for the genetic improvement of fatty acid composition in oil palm.
Introduction
Oil palm (Elaeis guineensis, 2n = 32) is an important tropical oil crop and is often referred to as “the oil king of the world” because it has the highest oil yield per unit area among all oil crops (Singh et al., 2013; Huang, 2017). Global production of palm oil in 2017 was approximately 75.70 million tons (Mozzon et al., 2020). The two oil storage tissues of oil palm are the mesocarp and kernel, each of which produces oil with a different fatty acid composition. Palmitic acid (16:0) is the major fatty acid (about 50%) in oil from the oil palm mesocarp, while lauric acid (12:0) is the major fatty acid (about 50%) in kernel oil. As for many important oil crops, improving fatty acid content is a major breeding objective for this tropical oil crop, especially aiming to decrease the palmitic acid content and increase the oleic acid content. However, the breeding scheme is slow due to the long life cycle of oil palms. Developing molecular markers associated with fatty acid compositions could facilitate the breeding and selection of the tropical oil-seed crop.
Molecular markers such as amplified fragment length polymorphisms (AFLPs), random amplified polymorphic DNA (RAPD), and restriction fragment length polymorphisms (RFLPs) have been widely used for analyzing genetic diversity and population structure, identification of trait-associated markers, and genotype characterization in oil palm (Moretzsohn et al., 2000; Rance et al., 2001; Billotte et al., 2005; Cochard et al., 2009; Singh et al., 2009; Ting et al., 2013). In eukaryotic genomes, there are generally some introns across each gene (Hawkins, 1988; Deutsch and Long, 1999; Haas et al., 2005). Diversity of intron sequences between different individuals is abundant due to low selection pressure (Stoltzfus et al., 1997; Panjabi et al., 2008; Sharma et al., 2020). Therefore, numerous intron polymorphism (IP) markers are available in the eukaryotic genome (Badoni et al., 2016; Kita et al., 2016). Studies have identified different types of IP, including ILP (Intron Length Polymorphism; Wang et al., 2005; Sharma et al., 2020) and ISNP (Intron Single Nucleotide Polymorphism; Ferreira et al., 2009; Liu et al., 2018). IP markers are generally co-dominant and highly polymorphic, and are widely used for constructing genetic maps, diversity analysis, and quantitative trait locus mapping (Williams et al., 1990; Wei et al., 2005; Yang et al., 2007; Xia et al., 2017). In 2013, the genome sequence of oil palm was released, providing an opportunity to develop larger numbers of IP markers (Singh et al., 2013). Polymorphism markers located in the intron region can be efficient function markers in genic regions. Therefore, IP in targeted genes may be associated with targeted traits in E. guineensis.
In this study, candidate genes and their introns involved in the biosynthesis and metabolism of fatty acids were identified in the genome sequence of E. guineensis. Subsequently, IP markers were developed based on intron sequences among 70 oil palm individuals. Fatty acid composition was also investigated among the 70 oil palm individuals. Finally, associations between IP markers and fatty acid variation were analyzed. This study will provide a exhaustive understanding of fatty acid content in oil palm, and the IP makers and candidate genes detected will facilitate breeding for fatty acid content in oil palm. Provide a theoretical basis for increasing the content of oleic acid and reducing the content of palmitic acid in future oil palm breeding.
Materials and Methods
Plant Materials and DNA Extraction
A total of 70 oil palm individuals were selected from the oil palm germplasm resources of the Coconut Research Institute of Chinese Academy of Tropical Agricultural Sciences, Wenchang town, Hainan province, China. Detailed information for these oil palm individuals is listed in Supplementary Table 1. DNA samples were prepared from spear leaves using the mini-CTAB method (Murray and Thompson, 1980).
Extraction and Measurement of Fatty Acid Contents in Oil Palm Mesocarp Tissues
Three fruits per oil palm individual (three biological replicates) were harvested, and fatty acid extraction and analyses for each mesocarp tissue were performed in triplicate (three different
extractions as technical replicates). Approximately 60 mg mesocarp was used for extracting fatty acids according to methods described by Li-Beisson et al. (2013). Fatty acid composition was subsequently examined and measured using 7890A gas chromatograph equipped with a HP-5MS column (30 m by 250 μm, 0.25 μm). The heating procedure was as follows: initial temperature 180°C, followed by a temperature increase to 220°C at a rate of 10°C per min. The contents of decanoic acid (C10:0), lauric acid (12:0), myristic acid (C14:0), tripalmitelaidin acid (16:1), palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), and linoleic acid (18:2) were determined with reference to an internal standard using the following calculation: (1) total fatty acid = (total area of all measured fatty acid peaks × quantity of heptadecanoic acid-methyl ester)/(peak area of heptadecanoic acid-methyl ester × quantity of the sample); (2) relative fatty acid percentage = peak area of a specific fatty acid /total area of all measured fatty acid peaks. The peak area was calculated using Agilent software.
Identification of Candidate Genes Involved in Fatty Acid Biosynthesis
The whole-genome sequence of E. guineensis was downloaded from the National Center for Biotechnology Information (NCBI). Protein sequences involved in fatty acid biosynthesis were downloaded from the Arabidopsis Information Resource (TAIR), available at http://www.arabidopsis.org. The protein sequences from Arabidopsis were used as queries for BLASTx searches against the coding sequence (CDS) database from E. guineensis to identify candidate genes involved in fatty acid biosynthesis. Conserved motifs were also predicted by alignment with Conserved Domains Database (CDD)1 and PFAM databases.2
Identification of Intron Sequence and Primer Design
CDSs of candidate genes involving in fatty acid biosynthesis were used as queries for BLASTn searches against the genome sequence of E. guineensis to identify the boundary between exon and intron. The MEME program3 was used to identify the gene structures of candidate genes. Primers were designed based on the intron sequences using Primer3 software (Rozen and Skaletsky, 2000). The list of PCR primers used in this study is provided in Supplementary Table 2.
PCR Amplification and Electrophoresis
PCR amplification was performed in 10 μl reaction mixtures containing 100 ng genomic DNA, 1× PCR buffer, 2 mM MgCl2, 1 U Taq DNA polymerase (TaKaRa, China), 0.5 μM of each primer, and 0.2 mM dNTP mix. The PCR program comprised a denaturation step for 4 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 54.7°C, and 30 s at 72°C. PCR products were electrophoretically separated on 8% polyacrylamide denaturing gels and visualized by silver staining. Product sizes were determined by comparison with a 100 bp DNA ladder.
Population Structure and Genetic Diversity Analysis
Bayesian clustering was employed to analyze the population structure of 70 oil palm individuals using the software STRUCTURE (Pritchard et al., 2000). Ten independent calculations were performed for K value (K set from 2 to 7). Burn-in time and replication number were both set to 100,000 in each run. The maximum-likelihood method was applied to assign every individual to a cluster, and the cut-off probability was set to 0.6. The most probable number of populations (K) was identified by plotting ΔK values of K from 1 to 10 in replicate runs for each K and corresponded to the peak of the ΔK graph. Association analysis was conducted using the software Tassel4; statistical significance (value of p) was determined by 100,000 permutations (Bradbury et al., 2007).
Results
Identification of Candidate Genes Involved in de novo Synthesis of Fatty Acids
The protein sequences involved in de novo synthesis of fatty acids from Arabidopsis thaliana were used as queries to align against the protein database of E. guineensis. A total of 52 candidate genes that might participate in the de novo synthesis of fatty acids, carbon termination or dehydration were identified. These candidate genes were distributed across 16 different chromosomes. Among them, the maximum number of candidate genes were detected in chromosome 7, followed by chromosome 5, and then chromosome 9. Chromosomes 4 and 11 each contained only one candidate gene involved in fatty acid biosynthesis (Figure 1).
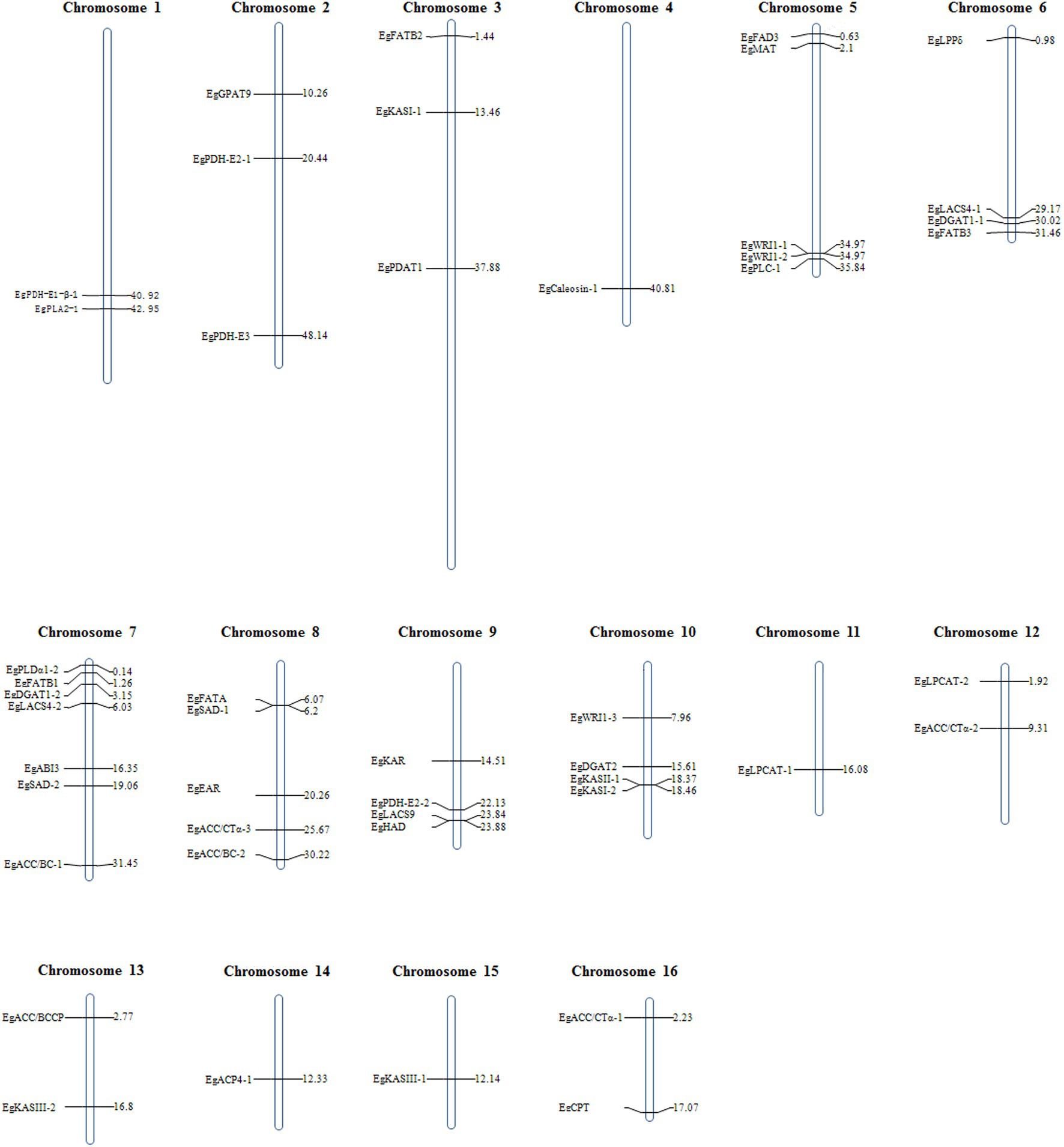
Figure 1. Chromosomal positon of candidate genes involved in the biosynthesis of fatty acid identified in the genome of E. guineensis.
CDSs of 52 candidate genes involved in fatty acid biosynthesis were used as queries to align with the E. guineensis genome to identify the sequence boundary between intron and exon. Almost all candidate genes contained at least two intron sequences. Among all the candidate genes, the EgPLA2-2 gene contained the maximum number of introns (22). A large proportion of candidate genes contained approximately 6–7 introns (Figure 2).
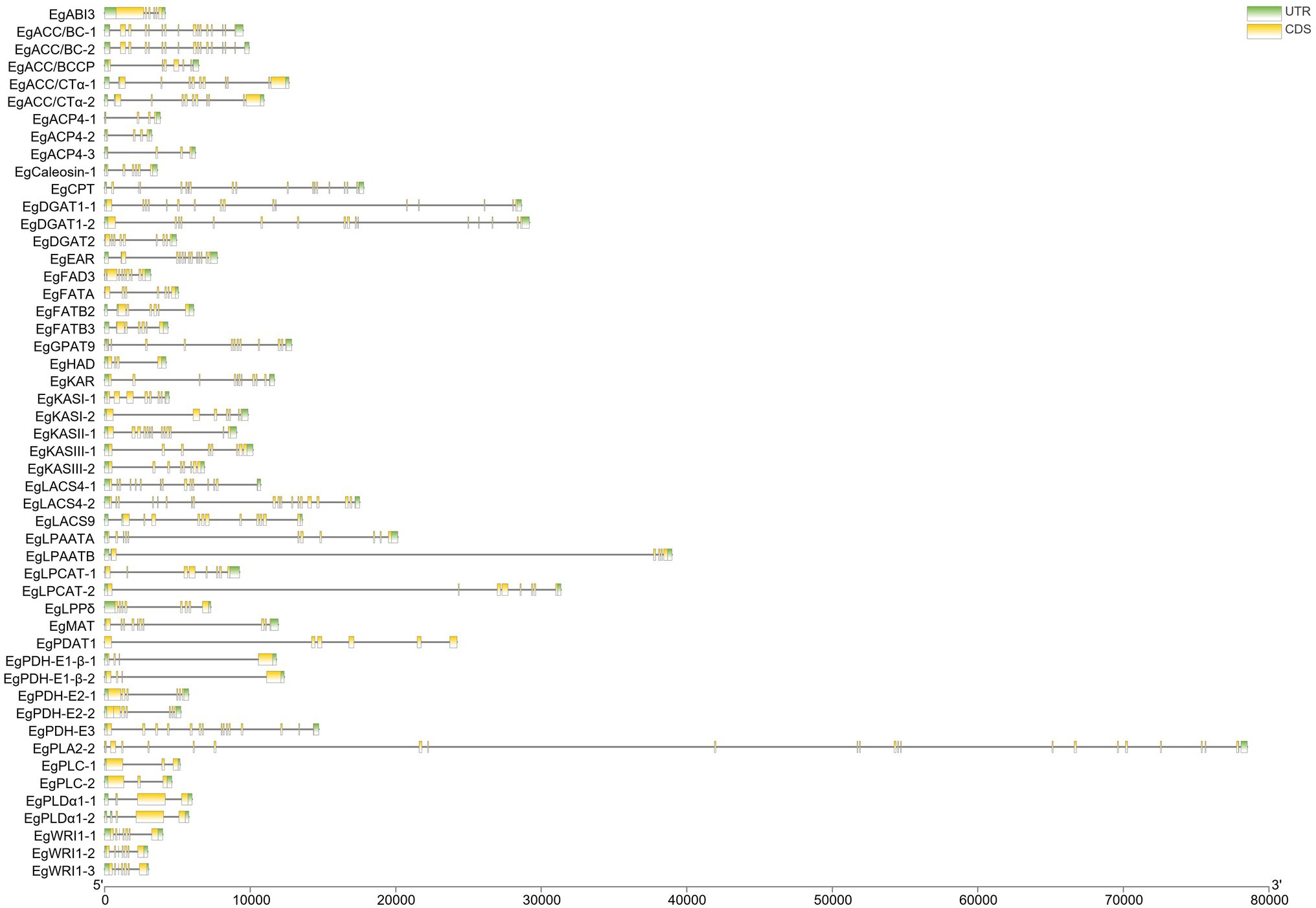
Figure 2. The gene structure of enzyme genes related to the biosynthesis of fatty acid in Elaeis guineensis.
Development of IP Markers Based on the Intron Region of Candidate Genes
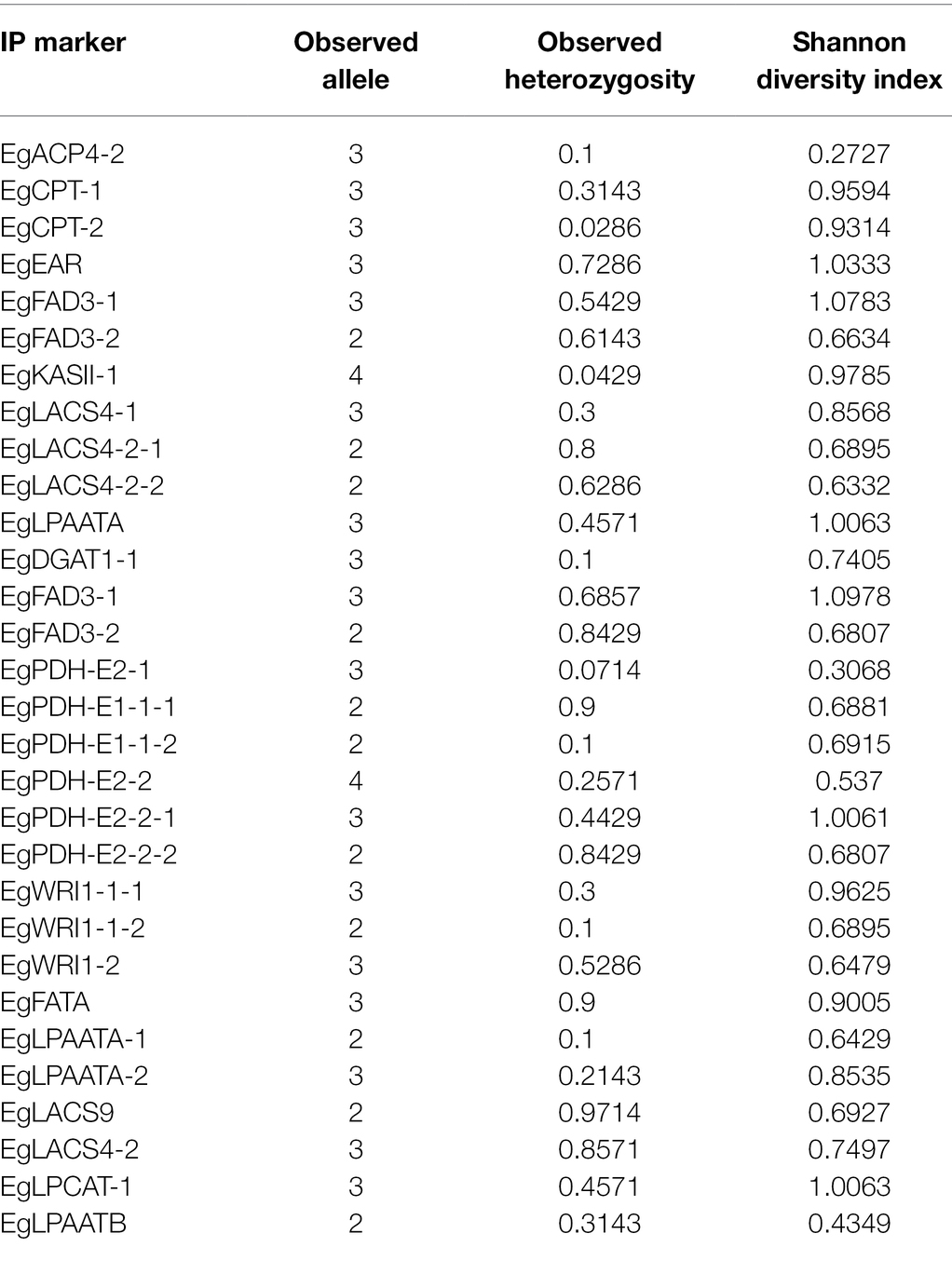
A total of 205 primer pairs were designed based on intron region of candidate genes and were used to amplify the DNA templates of different 70 oil palm individuals (Figure 3). Amplification products from 64 primer pairs showed polymorphisms between oil palm individuals; the observed heterozygosity varied from 0.0286 to 0.9714, with an average of 0.4514. Meanwhile, a total of 81 alleles were identified, with an average of 2.7 alleles per locus. Sizes of the amplification products varied from 120 bp to 1,300 bp, with an average of 424 bp per PCR product. The 81 alleles were located on the intron region of 18 candidate genes involved in fatty acid biosynthesis, were ACP4, CPT, EAR, FAD3, KASII-1, LACS4-1, LACS4-2, LPAATA, DGAT1-1, PDH-E2, PDH-E1, WRI1, WRI2, FatA, LACS9, LACS4-2, LPCAT-1, and LPAATB (Table 1).
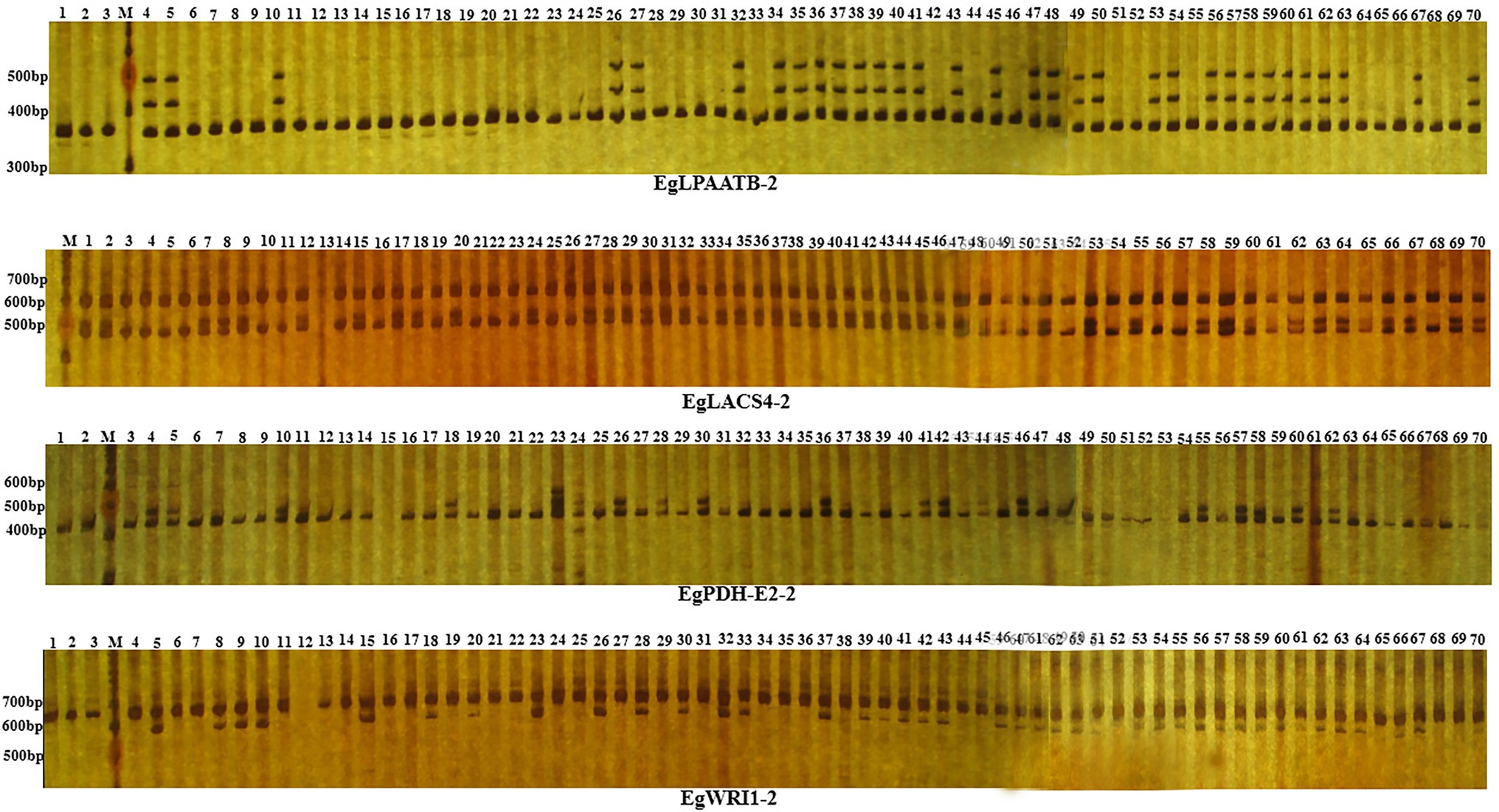
Figure 3. PCR products of IP marker EgLPAATB-2, EgLACS4-2, EgPDH-E2-2 and EgWRI1-2 in 70 oil palm samlpes separated by electrophoresis on 6% non-denaturing PAGE. Lanes 1–70 represent the 70 oil palm samples. M represents 100 bp ladder.
Analysis on the Variation of Fatty Acid Content in 70 Oil Palm Individuals
Fatty acid components of the 70 oil palm individuals were analyzed. Myristic acid content varied from 0.54 to 1.99%, with an average of 1.03%; palmitic acid content varied from 32.7 to 44.32%, with an average of 39.21%; oleic acid content varied from 35.86 to 53.42%, with an average of 46.13%; linoleic acid content varied from 7.17 to 18.07%, with an average of 11.61%; stearic acid content varied from 0.24 to 1.26%, with an average of 0.65%, and lauric acid content varied from 0.03 to 1.97%, with an average of 0.44% (Table 2; Figure 4).
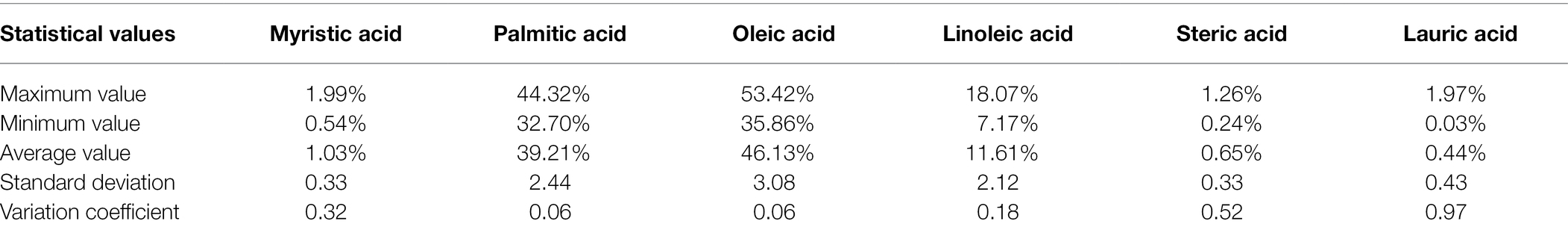
Table 2. Statistical values of different fatty acid components in the population of Elaeis guineensis.
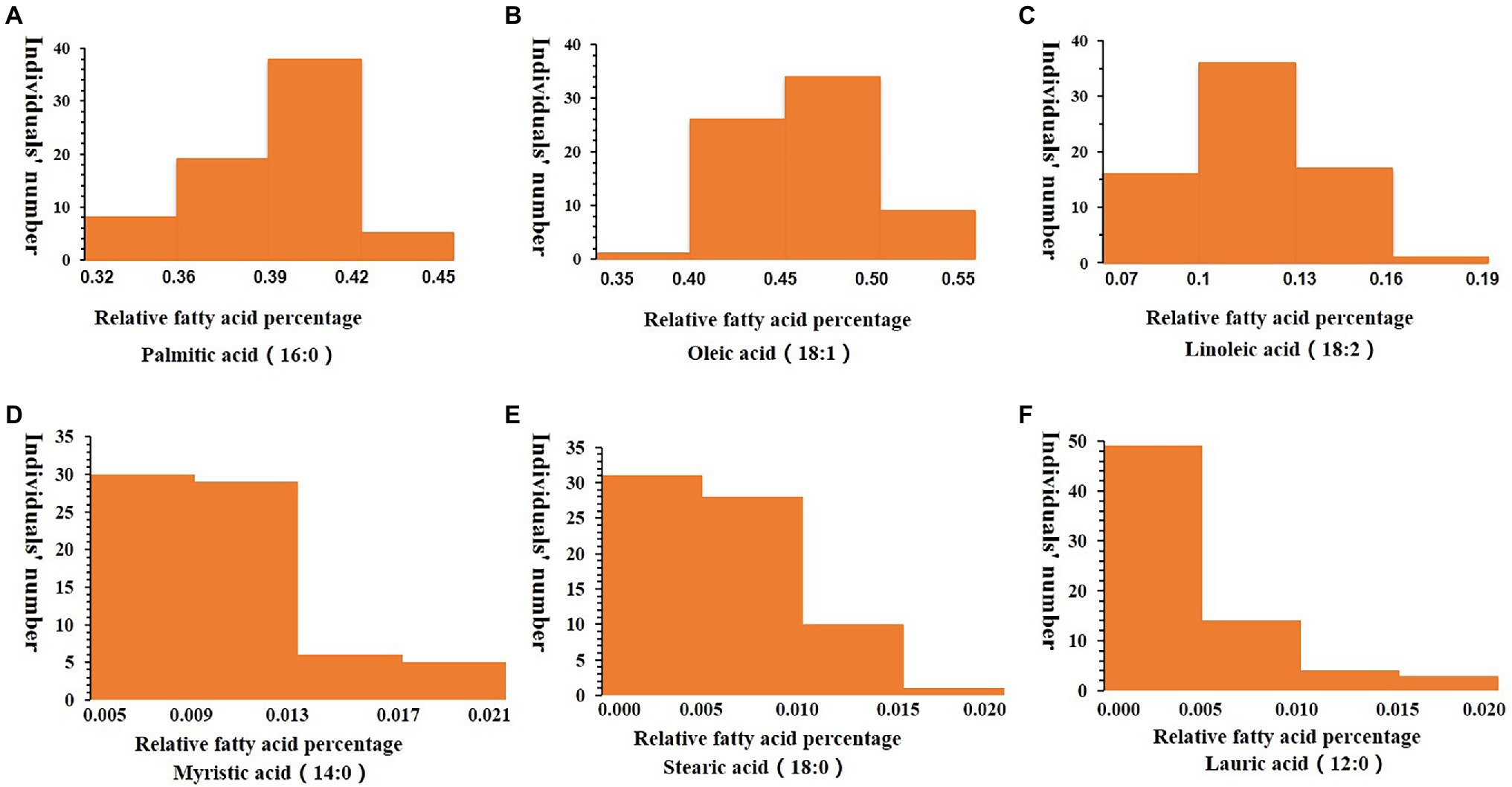
Figure 4. Frequency distribution of relative fatty acid percentages in the mesocarp of 70 oil palm individuals, including palmitic acid (16:0; A), oleic acid (18:1; B), linoleic acid (18:2; C), Myristic acid (14:0; D), Stearic acid (18:0; E) and Lauric acid (12:0; F). The x-axis represents the trait value, and the y-axis represents the number of oil palm individuals.
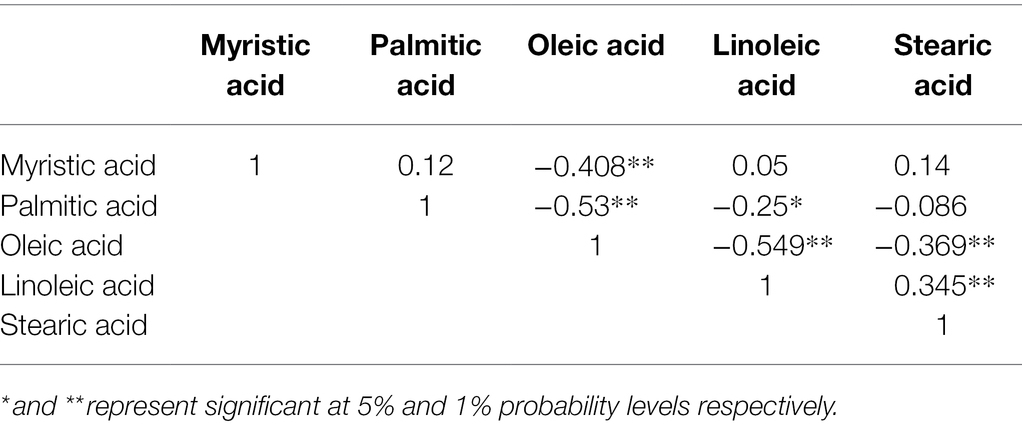
The relationships between different fatty acid components were analyzed using SPSS software. Oleic acid content was negatively correlated with myristic acid content (r = −0.408** and p = 0.00). Meanwhile, palmitic acid content showed significant negative correlation with oleic acid content (r = −0.53** and p = 0.00), indicating that decreasing palmitic acid content in oil palm may enhance oleic acid content. Palmitic acid content and oleic acid content both showed significant negative correlation with linoleic acid content (r = −0.25* and p = 0.04, and r = −0.549** and p = 0.00 for palmitic acid and oleic acid, respectively). Oleic acid content showed significant negative correlation with stearic acid content (r = −0.369** and p = 0.002), while linoleic acid content showed significant positive correlation with stearic acid content (r = 0.345** and p = 0.003; Table 3).
Association Between IP Markers and Fatty Acid Components
The developed IP markers were used to evaluate the population structure of 70 individuals of E. guineensis. When the method of Evanno et al. (2005) was applied to identify the most likely number of ‘true population’, K = 3 genetic groups were found. The STRUCTURE assignment procedure revealed that the largest genetic group comprised 34 oil palm individuals, the second group comprised 19 oil palm individuals, and the remaining group only included 17 oil palm individuals (Figure 5).
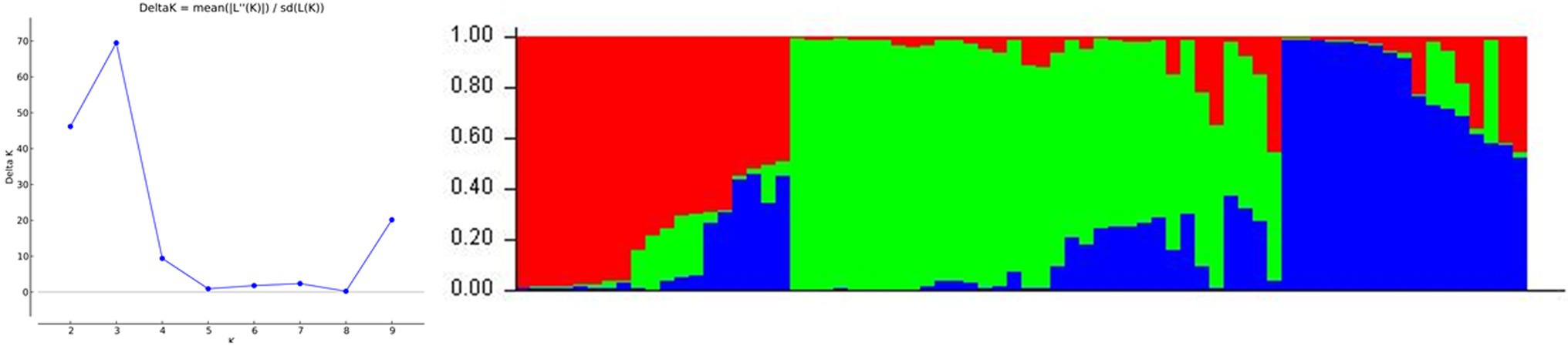
Figure 5. Inferred population structure of 70 oil palm germplasm. Each oil palm individual is represented by a single vertical line. Each color represents one culster. The length of the colored segment indicates the proportion of an individual assigned into one genetic group. The left diagram indicates the true number of genetic clusters.
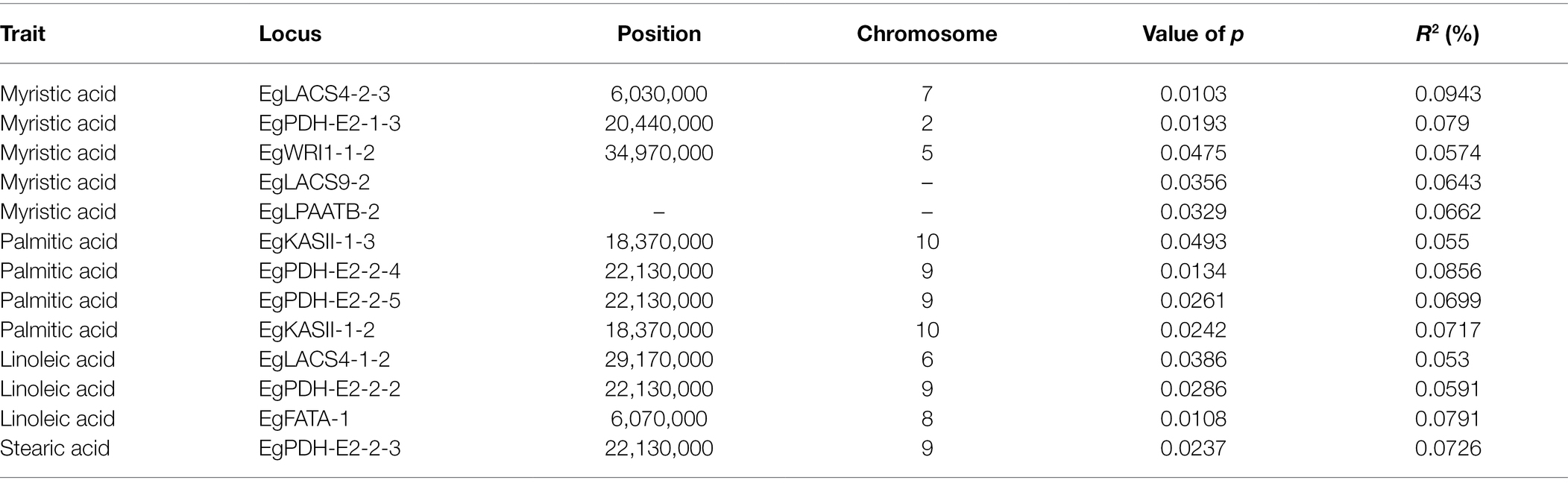
A simple linear model (SLM) was used to identify the association between IP markers and fatty acid components. Five IP markers were significantly associated with the variation in myristic acid content, including EgLACS9-2 (p = 0.0356 and r2 = 0.0643), EgPDH-E2-1-3 (p = 0.0193 and r2 = 0.079), EgLACS4-2-3 (p = 0.0103 and r2 = 0.0943), EgWRI1-1-2 (p = 0.0475 and r2 = 0.0574) and EgLPAATB-2 (p = 0.0329 and r2 = 0.0662). Three IP markers showed significant association with linoleic acid, including EgLACS4-1-2 (p = 0.0386 and r2 = 0.053), EgPDH-E2-2-2 (p = 0.0286 and r2 = 0.0591), and EgFATA-1(p = 0.0108 and r2 = 0.0791). One IP marker, EgPDH-E2-2, showed significant association with stearic acid content (p = 0.0237 and r2 = 0.0726). Furthermore, four IP markers were significantly associated with palmitic acid content, including EgPDH-E2-2 -4 (p = 0.0134 and r2 = 0.0856), EgPDH-E2-2 -5 (p = 0.0261 and r2 = 0.0699), EgKASII-1-2 (p = 0.0242 and r2 = 0.0717), and EgKASII-1-3 (p = 0.0493 and r2 = 0.055; Table 4).
Expression Analysis of Candidate Genes
To analyze the expression patterns of the candidate genes in different tissues, transcriptomic raw reads data were downloaded from the SRA (Short Read Archive) database of the NCBI website, including SSR851069 (mesocarp 10 weeks after anthesis), SRR190698 (mesocarp 15 weeks after anthesis), SRR190699 (mesocarp 17 weeks after anthesis), SRR190700 (mesocarp 19 weeks after anthesis), SRR190701 (mesocarp 21 weeks after anthesis), SRR190702 (mesocarp 23 weeks after anthesis), SSR851068 (kernel 10 weeks after anthesis), SSR851068 (kernel 15 weeks after anthesis), SSR851099 (pollen), SRR851103 (shoot), and SRR851110 (root). Results of the analysis revealed that almost all candidate genes had a high level of expression in mesocarp and kernel tissues compared with other tissues, except for the gene EgLACS4-2. Among them, EgLPAATB had higher expression in oil palm kernel compared with that in other tissues. However, seven other candidate genes had a higher expression level in mesocarp and kernel compared with the expression in three other tissues; these genes were EgKASII-1, EgPDH-E2-2, EgLACS9, EgFATA, Eglacs4-1, EgWRI1-1, and EgPDH-E2-1 (Figure 6).
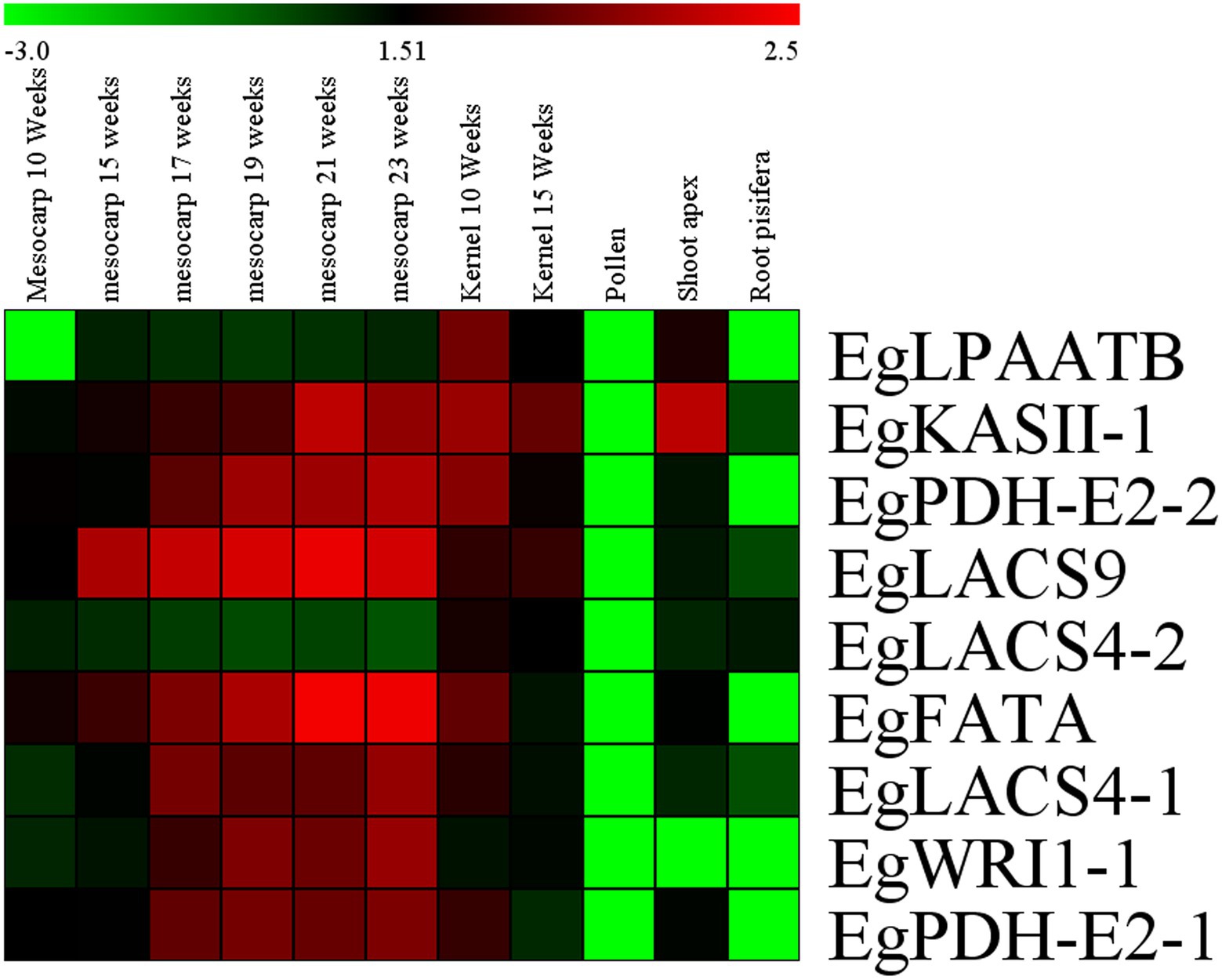
Figure 6. Heat map of candidate genes expression in different tissues of Elaeis guineensis. Log10RPKM value were sued to construct the heat map with clustering.
Discussion
This study generated a total of 64 polymorphic IP markers, based on intron sequences of candidate genes involved in fatty acid biosynthesis, for use in molecular breeding of oil palm. These markers were screened across 70 oil palm individuals and are predicted to be suitable for widespread use in oil palm breeding, particularly association analysis. This premise was validated by identifying 13 IP markers linked to different fatty acid compositions; these IP markers will be used immediately for marker-assisted selection.
Molecular markers such as simple sequence repeat (SSR), RAPDs, and AFLPs have been widely used for analyzing genetic diversity and population structure, identification of trait-associated markers, and genotype characterization in E. guineensis (Moretzsohn et al., 2000; Rance et al., 2001; Billotte et al., 2005; Cochard et al., 2009; Seng et al., 2011; Premkrishnan and Arunachalam, 2012; Myint et al., 2021). The release of the whole genome sequence of E. guineensis has provided an opportunity to develop IP markers with regards to targeted candidate genes. Among the various molecular markers, identifying RFLP markers in a genetically diverse population can be cumbersome as it is mainly based on molecular hybridization (Maizura et al., 2006). Moreover, AFLP markers are based on enzyme digestion and PCR amplification with adapter primers and random primers, which is generally unstable (Barcelos et al., 2002). In recent years, a large number of SNP markers have been identified by using next-generation sequencing technology. Xia et al. (2019) revealed 62 SNP markers that were signifcantly associated with fatty acid content, including palmitic acid content (32 SNPs), oleic acid content (4 SNPs), linoleic acid content (1 SNP), and total oil content (25 SNPs) in oil palm. However, SNP remains costly and consequently limits the broad applications of SNP markers for oil palm improvement using these strategies (Ithnin et al., 2021). In contrast, the polymorphisms obtained using IP markers were easily discernable on simple 1.5–2.0% agarose gels and this would enable rapid screening of a large diverse population. In this study, 64 IP markers were developed based on the intron sequences of candidate genes involved in fatty acid biosynthesis, 28.44% of which were polymorphic among different individuals in accordance with previous results (Xia et al., 2019). Moreover, among these IP markers, 20.31% were significantly associated with different fatty acid compositions, including EgLACS4-2-3, EgPDH-E2-1-3, EgWRI1-1-2, EgLACS9-2, EgLPAATB-2, EgFATA-1. The markers EgPDH-E2-2-4, EgPDH-E2-2-5, EgKASII-1-2, and EgKASII-1-3 were significantly associated with variation in palmitic acid content and located in the intron regions of different EgKASII genes that have an important role in carbon extension. Furthermore, EgLACS4-1-2, EgPDH-E2-2-2, and EgFATA-1 markers significantly associated with linoleic acid and located on the FatA gene, which is involved in carbon termination of unsaturated fatty acids.
In past several decades, several studies had been performed to identify and validate genes involved in fatty acid biosynthesis. For example, seed-specific RNAi-mediated down-regulation of KASII led to in dramatic increase of palmitic acid (Liu et al., 2017). Our research also showed the IP markers derived from EgKASII-1-2 and EgKASII-1-3 had significant association with palmitic acid composition (p = 0.0242 and 0.0493). Meanwhile, our study also indicated that the IP markers located on EgLACS4-1-2 were significantly associated with linoleic acid (p = 0.0386 and r2 = 0.053). LACS has been validated to play a role in linoleic acid biosynthesis in a previous study (Jang et al., 2015). Our results also demonstrated that IP markers obtained from EgWRI1-4 and EgLPAAT were significantly associated with myristic acid. In previous studies, LPAAT catalyzed 14:0-acyl-carrier protein specifically and resulted in high myristic acid content in Cyanothece. However, no documents showed the relationship between EgWRI1-4 and myristic acid.
Therefore, it is possible that some IP markers are closely linked with targeted genes that govern fatty acid content and subsequently show significant associations with targeted traits. In future, these candidate markers could be ideal targets for further study and may have potential application in marker-assisted selection for fatty acid composition.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YX and WX participated in the design of the study. JL and YY performed the statistical analysis. JL and PS conducted the major experimental work including the extraction and measurement of oil content and relative fatty acid contents. YX and XL wrote the first draft of the manuscript. YWa, YWu, RL, LZ, and XS wrote sections of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (no. 31870670).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.885418/full#supplementary-material
Footnotes
1. ^http://www.ncbi.nlm.nih.gov/cdd
4. ^http://www.maizegenetic.net/bioinforFormatics/tasselindex.htm
References
Badoni, S., Das, S., Sayal, Y. K., Gopalakrishnan, S., Singh, A. K., Rao, A. R., et al. (2016). Genome-wide generation and use of informative intron-spanning and intron-length polymorphism markers for high-throughput genetic analysis in rice. Sci. Rep. 6:23765. doi: 10.1038/srep23765
Barcelos, E., Amblard, P., Berthaud, J., and Seguin, M. (2002). Genetic diversity and relationship in american and african oil palm as revealed by RFLP and AFLP molecular markers. Pesquisa Agropecuária Brasileira 37, 1105–1114. doi: 10.1590/S0100-204X2002000800008
Billotte, N., Marseillac, N., Risterucci, A. M., Adon, B., Brottier, P., Baurens, F. C., et al. (2005). Microsatellite-based high density linkage map in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 110, 754–765. doi: 10.1007/s00122-004-1901-8
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cochard, B., Adon, B., Rekima, S., Billotte, N., de Desmier, C. R., and Koutou, A. (2009). Geographic and genetic structure of African oil palm diversity suggests new approaches to breeding. Tree Genet. Genomes 5, 493–504. doi: 10.1007/s11295-009-0203-3
Deutsch, M., and Long, M. (1999). Intron-exon structure of eukaryotic model organisms. Nucleic Acids Res. 27, 3219–3228. doi: 10.1093/nar/27.15.3219
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Ferreira, A. O., Cardoso, H. G., Macedo, E. S., Breviario, D., and Arnholdt-Schmitt, B. (2009). Intron polymorphism pattern in AOX1b of wild St John’s wort (Hypericum perforatum) allows discrimination between individual plants. Physiol. Plant. 137, 520–531. doi: 10.1111/j.1399-3054
Haas, B. J., Wortman, J. R., Ronning, C. M., Hannick, L. I., Smith, R. K., Maiti, R., et al. (2005). Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol. 3:7. doi: 10.1186/1741-7007-3-7
Hawkins, J. D. A. (1988). A survey on intron and exon lengths. Nucleic Acids Res. 16, 9893–9908. doi: 10.1093/nar/16.21.9893
Huang, H. (2017). The industrial survey of Elaeis guineensis in Malaysia. World Trop. Agric. Inf. 7:7. doi: 10.3969/j.issn.1009-1726.2017.07.020
Ithnin, M., Vu, W. T., Shin, M. G., Suryawanshi, V., Sherbina, K., Zolkafli, S. H., et al. (2021). Genomic diversity and genome-wide association analysis related to yield and fatty acid composition of wild American oil palm. Plant Sci. 304:110731. doi: 10.1016/j.plantsci.2020.110731
Jang, Y. E., Kim, M. Y., Shim, S., Lee, J., and Lee, S. H. (2015). Gene expression profiling for seed protein and oil synthesis during early seed development in soybean. Genes Genomics 37, 409–418. doi: 10.1007/s13258-015-0269-2
Kita, T., Komatsu, K., Zhu, S., Iida, O., Sugimura, K., Kawahara, N., et al. (2016). Development of intron length polymorphism markers in genes encoding diketide-CoA synthase and curcumin synthase for discriminating Curcuma species. Food Chem. 194, 1329–1336. doi: 10.1016/j.foodchem.2015.08.034
Li-Beisson, Y., Shorrosh, B., Beisson, F., Andersson, M. X., Arondel, V., Bates, P. D., et al. (2013). Acyl-lipid metabolism, Acyl-Lipid Metabolism. Arabidopsis Book 11:e0161. doi: 10.1199/tab.0161
Liu, Q., Wu, M., Zhang, B., Shrestha, P., Petrie, J., Green, A. G., et al. (2017). Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding β-ketoacyl-ACP synthase II (KASII). Plant Biotechnol. J. 15, 132–143. doi: 10.1111/pbi.12598
Liu, Y., Xu, A., Liang, F., Yao, X., Wang, Y., Liu, X., et al. (2018). Screening of clubroot-resistant varieties and transfer of clubroot resistance genes to Brassica napus using distant hybridization. Breed. Sci. 68, 258–267. doi: 10.1270/jsbbs.17125
Maizura, I., Rajanaidu, N., Zakri, A. H., and Cheah, S. C. (2006). Assessment of genetic diversity in oil palm (Elaeis guineensis Jacq.) using restriction fragment length polymorphism (RFLP). Genet. Resour. Crop. Evol. 53, 187–195. doi: 10.1007/s10722-004-4004-0
Moretzsohn, M. C., Nunes, C. D. M., Ferreira, M. E., and Grattapaglia, D. (2000). RAPD linkage mapping of the shell thickness locus in oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 100, 63–70. doi: 10.1007/s001220050009
Mozzon, M., Foligni, R., and Mannozzi, C. (2020). Current knowledge on interspecific hybrid palm oils as food and food ingredient. Foods 9, 631–646. doi: 10.3390/foods9050631
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Myint, K. A., Yaakub, Z., Rafii, M. Y., Oladosu, Y., Samad, M., Ramlee, S. I., et al. (2021). Genetic diversity assessment of MPOB-Senegal oil palm germplasm using microsatellite markers. Biomed. Res. Int. 2021:6620645. doi: 10.1155/2021/6620645
Panjabi, P., Jagannath, A., Bisht, N. C., Padmaja, K. L., Sharma, S., Gupta, V., et al. (2008). Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9:113. doi: 10.1186/1471-2164-9-113
Premkrishnan, B. V., and Arunachalam, V. (2012). In silico RAPD priming sites in expressed sequences and iSCAR markers for oil palm. Comp. Funct. Genomics 2012:913709. doi: 10.1155/2012/913709
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
Rance, K. A., Mayes, S., Price, Z., Jack, P. L., and Corley, R. H. V. (2001). Quantitative trait loci for yield components oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 103, 1302–1310. doi: 10.1007/s122-001-8204-z
Rozen, S., and Skaletsky, H. (2000). Primer 3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132, 365–386. doi: 10.1385/1-59259-192-2:365
Seng, T.-Y., Mohamed Saad, S. H., Chin, C.-W., Ting, N.-C., Harminder Singh, R. S., Qamaruz Zaman, F., et al. (2011). Genetic linkage map of a high yielding FELDA deli×Yangambi oil palm cross. PLoS One 6:e26593. doi: 10.1371/journal.pone.0026593
Sharma, H., Bhandawat, A., Rahim, M. S., Kumar, P., Choudhoury, M. P., and Roy, J. (2020). Novel intron length polymorphic (ILP) markers from starch biosynthesis genes reveal genetic relationships in Indian wheat varieties and related species. Mol. Biol. Rep. 47, 3485–3500. doi: 10.1007/s11033-020-05434-2
Singh, R., Ong-Abdullah, M., Low, E. T., Manaf, M. A., Rosli, R., Nookiah, R., et al. (2013). Oil palm genome sequence reveals divergence of interfertile species in old and new worlds. Nature 500, 335–339. doi: 10.1038/nature12309
Singh, R., Tan, S. G., Panandam, J. M., Rahman, R. A., Ooi, L. C., Low, E. T., et al. (2009). Mapping quantitative trait loci (QTLs) for fatty acid composition in an interspecific cross of oil palm. BMC Plant Biol. 9:114. doi: 10.1186/1471-2229-9-114
Stoltzfus, A., Logsdon, J. M., Palmer, J. D., and Doolittle, W. F. (1997). Intron “sliding” and the diversity of intron position. Proc. Natl. Acad. Sci. U. S. A. 94, 10739–10744. doi: 10.1073/pnas.94.20.10739
Ting, N. C., Jansen, J., Nagappan, J., Ishak, Z., Chin, C. W., Tan, S. G., et al. (2013). Identification of QTLs associated with callogenesis and embryogenesis in oil palm using genetic linkage maps improved with SSR markers. PLoS One 8:e53076. doi: 10.1371/journal.pone.0053076
Wang, X., Zhao, X., Zhu, J., and Wu, W. (2005). Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa L.). DNA Res. 12, 417–427. doi: 10.1093/dnares/dsi019
Wei, H., Fu, Y., and Arora, R. (2005). Intron-flanking EST-PCR markers: from genetic marker development to gene structure analysis in Rhododendron. Theor. Appl. Genet. 111, 1347–1356. doi: 10.1007/s00122-005-0064-6
Williams, J. G., Kubelik, A. R., Livak, K. J., Rafalski, J. A., and Tingey, S. V. (1990). DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18, 6531–6535. doi: 10.1093/nar/18.22.6531
Xia, X., Luan, L. L., Qin, G., Yu, L. F., Wang, Z. W., Dong, W. C., et al. (2017). Genome-wide analysis of SSR and ILP markers in trees: diversity profiling, alternate distribution, and applications in duplication. Sci. Rep. 7:17902. doi: 10.1038/s41598-017-17203-6
Xia, W., Luo, T. T., Dou, Y. J., Zhang, W., Mason, A. S., Huang, D. Y., et al. (2019). Identification and validation of candidate genes involved in fatty acid content in oil palm by genome-wide association analysis. Front. Plant Sci. 10:1263. doi: 10.3389/fpls.2019.01263
Keywords: Elaeis guineensis, fatty acid, IP markers, candidate gene, association analysis
Citation: Li J, Yang Y, Sun X, Liu R, Xia W, Shi P, Zhou L, Wang Y, Wu Y, Lei X and Xiao Y (2022) Development of Intron Polymorphism Markers and Their Association With Fatty Acid Component Variation in Oil Palm. Front. Plant Sci. 13:885418. doi: 10.3389/fpls.2022.885418
Edited by:
Peter Poczai, University of Helsinki, FinlandReviewed by:
Wei Zhang, Zhejiang Normal University, ChinaRobert Jarret, United States Department of Agriculture, United States
Copyright © 2022 Li, Yang, Sun, Liu, Xia, Shi, Zhou, Wang, Wu, Lei and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xintao Lei, eHRsZWlAMjYzLm5ldA==; Yong Xiao, eGlhb3lvbmdfY29jb251dEAxNjMuY29t
 Jing Li
Jing Li Yaodong Yang
Yaodong Yang Xiwei Sun
Xiwei Sun Rui Liu
Rui Liu Wei Xia
Wei Xia Peng Shi
Peng Shi Lixia Zhou
Lixia Zhou Yong Wang
Yong Wang Yi Wu
Yi Wu Xintao Lei3*
Xintao Lei3* Yong Xiao
Yong Xiao