- College of Horticulture, Nanjing Agricultural University, Nanjing, China
High temperature stress seriously affects the growth of cucumber seedlings, and even leads to a decline in yield and quality. miRNAs have been shown to be involved in regulating the response to stress in plants, but little is known about its effects on cucumber high temperature stress tolerance. Here, we found that high temperature stress induced the expression of miR9748 in cucumber. Overexpression of cucumber miR9748 in Arabidopsis improved high temperature tolerance. Transcriptome analysis revealed that miR9748 might mediate high temperature tolerance through plant hormone signal pathway. 5′ RNA ligase-mediated rapid amplification of cDNA ends (5′ RLM-RACE) and transient transformation technology demonstrated that CsNPF4.4 was the target gene of miR9748. CsNPF4.4 overexpression plants decreased high temperature tolerance accompanied by reducing the content of jasmonic acid (JA), but alleviated by foliar application of methyl jasmonate, indicating that CsNPF4.4 negatively regulated high temperature stress tolerance through inhibition JA signal pathway. Furthermore, high temperature stress also increased the expression level of CsbZIP2. Yeast one-hybrid and dual-luciferase assays showed that CsbZIP2 directly bound to the promoter of MIR9748 to induce its expression. Taken together, our results indicated that CsbZIP2 directly regulated miR9748 expression to cleave CsNPF4.4 to mediate high temperature tolerance through JA pathway.
Introduction
The protected cultivation area of cucumber (Cucumis sativus L.) is the second in China, which reached 105 million hectares in 2018 (Ji et al., 2020). With the global warming, high temperature has become a key limited effect for protected cucumber cultivation in China. Studies have shown that high temperature inhibits the growth and development of cucumber plants, as indicated by inhibited the bioaccumulation, decreased chlorophyll content, increased the lipid peroxidation level, delayed flowering, and appeared dropping flower and fruit (Zhou et al., 2016; Wang et al., 2018a; Chen et al., 2021b). Cucumber plants have evolved a series of physiological and molecular mechanisms in response to high temperature stress, such as increasing the content of proline, activating antioxidant defense system, inducing the expression of heat shock proteins, and regulating plant hormone signal pathways (Wei et al., 2019; He et al., 2020; Chen et al., 2021b). Furthermore, calcium signal, transcription factors, and post-transcription regulation also mediate cucumber high temperature stress tolerance (Zhou et al., 2013; Xu et al., 2015; Yu et al., 2018; Wang et al., 2018b, 2020a).
Plant miRNA is a kind of non-coding RNA and regulates the expression of target mRNA by cleavage or translation inhibition (Chen et al., 2018). miRNA is transcribed by RNA Polymerase II into primary miRNAs (pri-miRNA) with typical stem-loop structure. Subsequently, pri-miRNA is cleaved twice by DICER-LIKE1 (DCL1) to produce miRNA/miRNA* double strands. The methylated double strands are transported from the nucleus to the cytoplasm by HASTY. In the cytoplasm, miRNA is loaded into ARGONAUTE 1 protein to form an active RNA-induced silencing complex, and miRNA* is degraded (Wang et al., 2020b). miRNA plays critical roles in plant adaptation to high temperature stress (Wang et al., 2018b; Bhogireddy et al., 2021). miR156 is induced by high temperature stress to regulate the expression of SPL gene and promote the continuous expression of genes in response to high temperature stress (Cui et al., 2014; Stief et al., 2014). In contrast to miR156, the expression of miR172 is downregulated under high temperature stress, while the expression of target genes TARGET OF EAT1 (TOE1) and TOE2 is upregulated (May et al., 2013; Li et al., 2014). High temperature rapidly increases the expression level of miR398, which positively regulates heat stress tolerance through downregulation the expression of CSD1 (copper/zinc superoxide dismutase), CSD2, and CCS (a copper chaperone for CSD1 and CSD2; Guan et al., 2013). In addition, miRNA might mediate spermidine-induced high temperature stress tolerance in cucumber (Wang et al., 2018b). These results demonstrate that miRNA can help plants adapt to high temperature stress by regulating the expression of target genes.
miR9748 is a particularly conservative miRNA family, which mediates plant growth and stress response. It has been shown that EIN3 is targeted by miR9748 in radish to regulate anthocyanin accumulation by mediating sucrose signal pathway (Sun et al., 2017). miR9748 also regulates the formation of adventitious roots in lotus by affecting the expression of downstream genes and participating in the metabolic process of brassinosteroid and upregulating the expression of BRI1 (Cheng et al., 2019). In addition, miR9748 participates in regulation the expression of MYC2 and HSP90 in Astragalus chrysochlorus (Cakir et al., 2016), indicating that it might mediates plants response to stress. In our previous work, we constructed the competing endogenous RNA (ceRNA) networks of long non-coding RNAs (lncRNAs), circular RNAs (circRNAs), miRNAs, and mRNAs under high temperature stress, and found that miR9748 is the central molecule of cucumber heat stress response ceRNA network (He et al., 2020), but the response mechanism of miR9748 to high temperature stress has not been understood. Here, we found that transcription factor CsbZIP2 bound to the promoter of MIR9748 to induce its expression to further degrade CsNPF4.4. Overexpression of miR9748 in Arabidopsis enhanced high temperature stress tolerance, but CsNPF4.4 overexpression plants were hypersensitivity to high temperature stress, along with inhibiting the genes expression related to jasmonic acid (JA) synthesis and decreasing the JA content. However, foliar application of methyl jasmonate (MeJA) to CsNPF4.4 overexpression plants increased high temperature stress tolerance. Thus, CsbZIP2 directly regulated miR9748 expression to cleave CsNPF4.4 to mediate high temperature stress tolerance through JA pathway.
Materials and Methods
Plant Materials and Treatments
Cucumber (Cucumis sativus L, Jinchun No. 2) was used in this experiments, and the seeds were purchased from Tianjin Kernel Cucumber Research Institute (Tianjin, China). The germinated seeds were sown in plastic pots (10 cm × 7 cm × 8 cm) filled with peat and vermiculite (2,1, v:v). The growth conditions were maintained as follow: 25/18°C day/night, 60–70% relative air humidity, 300 μmol m−2 s−1 photosynthetic photon flux density (PPFD), and 14/10 h light/dark cycle. When the third leaves were fully expanded, the seedlings were treated with 42/32°C (day/night) as high temperature stress. The leaf samples were harvested at 0, 1, 2, 4, 6, 12, 24, and 48 h and frozen in liquid nitrogen and stored in −80°C.
Construction of MIR9748 and CsNPF4.4 Overexpression Plants
A 430 bp sequence containing the precursor of miR9748 was synthesized by General Biological Systems Co., LTD. (Chuzhou, China), and inserted into pFGC1008 vector. The full-length coding DNA sequence (CDS) of CsNPF4.4 was amplified using cucumber cDNA as template with the specific primers (Supplementary Table 1). The PCR fragment was ligated into the plant transformation vector pFGC1008 using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The constructed pFGC1008-MIR9748 and pFGC1008-CsNPF4.4 plasmids were transformed into Agrobacterium tumefaciens strain EHA105 and transformed Arabidopsis Col-0 wild-type (WT) plants using floral dip method (Clough and Bent, 1998). The transformed plants were selected and verified using qPCR and the homozygous lines of the T3 progeny were used for high temperature stress as the same method of cucumber.
Transcriptome Analysis of MIR9748 Transgenic Plants
Arabidopsis leaves were collected at 8 h of high temperature stress and the total RNA was extracted from the leaves of WT and miR9748 overexpression (OE3) Arabidopsis plants using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). The RNA quality was measured by a Nanodrop 2000 (Thermo Fisher Scientific, Rockford, IL, United States), and the high quality and integrity RNA samples were selected to construct RNA libraries. The quality and quantity of the library were verified using an Agilent 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) and ABI StepOnePlus real-time PCR System (Applied Biosystems, Foster, CA, United States), respectively. Then, the libraries were sequenced on a HiSeq 2000 platform (Illumina, San Diego, CA, United States) by the BGI, Shenzhen, China. After the original readings, adapter sequences and low-quality readings were removed, all of the clean reads were mapped to the Arabidopsis reference genome using HISAT2 (V2.0.4; Kim et al., 2015). The gene expression level was calculated using the fragments per kilobase of exon per million fragments (Trapnell et al., 2012). The differentially expressed genes (DEGs) were recognized according to the false discovery rate (FDR) value less than 0.01 and |log2(fold change)| ≥ 2. Gene ontology (GO) analysis was performed by WEGO (Ashburner et al., 2000) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using KOBAS 2.0 (Xie et al., 2011).
miR9748 Target Gene Prediction, and GO Pathway Enrichment Analysis
Target Finder and psRobot software were used to predict the target gene of miR9748 as previously described (Allen et al., 2005; Wu et al., 2012), and the co-predicted genes were selected as its target genes. The predicted target genes were employed for GO pathway analysis as above described.
5′ RNA Ligase-Mediated Rapid Amplification of cDNA Ends
To verify the cleavage relationship of miR9748 to CsaV3_5G039430, 5′ RLM-RACE was performed using the FirstChoice™ RLM-RACE Kit (AM1700, Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The correct PCR reaction products were cloned into pMD-19 T vector, and all positive clones were confirmed by PCR. The clones were sequenced by General Biological Systems Co., LTD. (Chuzhou, China).
GUS Histochemical Staining Analysis
The full-length CDS of CsaV3_5G039430 was amplified with the specific primers (Supplementary Table 1) and inserted into pBI121 using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China) to obtain 35S:: CsaV3_5G039430-GUS vector. For mutation 6 bases of miR9748 binding sites in CsaV3_5G039430, the CDS was amplified and inserted into pBI121 vector using the ClonExpress MultiS One Step Cloning Kit (Vazyme, Nanjing, China) to obtain 35S:: CsaV3_5G039430M-GUS vector. The recombinant plasmids were transformed into A. tumefaciens strain EHA105, and transiently transformed into the leaves of Nicotiana benthamiana as previously described method (Wang et al., 2020c). After 2 d transformation, the leaves were stained with GUS staining kit (Solarbio, Beijing, China) and photographed.
Subcellular Localization of CsNPF4.4
Subcellular localization of CsNPF4.4 was performed as previously described (Liu et al., 2021). The full-length CDS of CsNPF4.4 was amplified with the primers (Supplementary Table 1) and inserted into pFGC5941-GFP vector to generate a CsNPF4.4-GFP fusion expression vector. Subsequently, the pFGC5941-CsNPF4.4-GFP and pFGC5941-GFP empty vector were transformed into A. tumefaciens strain EHA105, and infiltrated into the leaves of N. benthamiana that expresses a H2B-RFP as a marker for nucleus (Mei et al., 2020). After inoculation for 48 h, the GFP and RFP fluorescence signals were observed under an LSM800 confocal microscope (Zeiss, Oberkochen, Germany).
Plant Hormone Treatment
For plant hormone treatment, 100 μmol MeJA was sprayed on 35 d Arabidopsis seedlings and distilled water was used as the control. After 12 h of pretreatment, high temperature treatment was performed as above described. After 2 d of treatment, the leaves of Arabidopsis were collected for determination of physiological indexes.
Yeast One-Hybrid Assay
The yeast one-hybrid assays were performed as the method previously described (Wang et al., 2019). The promoter sequence of MIR9748 was cloned using the specific primers (Supplementary Table 1) and inserted into the pAbAi vector. The recombinant plasmid was linearized by BstB I (Thermo Fisher Scientific, Rockford, IL, United States) and transformed into Y1HGold yeast strain. The full-length CDS of CsbZIP2, CsMYB44, CsMYCI, CsHBP-1b, CsTGA2, CsTGA2.2, and CsTGA10 was amplified with the specific primers (Supplementary Table 1) and ligated into the pGADT7 vector, respectively. The pGADT7 empty vector or pGADT7 harboring the transcription factors was transformed into the positive strains containing the bait vector, respectively, and cultured on SD/Leu solid medium containing 200 ng ml−1 aureobasidin A (AbA) for 3–5 d at 30°C to detect DNA–protein interactions.
Dual-Luciferase Assay
The dual-luciferase assay was performed as previously described (Yang et al., 2021). The promoter sequence and CDS of MIR9748 and CsbZIP2 was amplified with specific primers (Supplementary Table 1), and inserted into the pGreenII 0800-LUC and pFGC5941-GFP vector, respectively. A. tumefaciens strain GV3101 (pSoup-p19) containing the indicated recombinant plasmids injected into the leaves of N. benthamiana. After injection for 48 h, luciferase luminescence was detected using a Tanon 5200Multi Image System (Tanon, Shanghai, China).
RNA Extraction and Gene Expression Analysis
miRNAs were extracted from leaves of all treated samples using the miRcute miRNA Extraction Kit (Tiangen, Beijing, China). Reverse transcription of miRNA into cDNA was performed using a Mir-X miRNA first-strand Synthesis Kit (Takara, Dalian, China). The obtained cDNA was used for qPCR analysis with the TB Green Advantage qPCR Premix (Takara, Dalian, China). The U6 gene was selected as an internal reference for standardized data, and the primers are listed in Supplementary Table 2.
Total RNA was extracted from leaves of treated samples using RNA Simple Total RNA Kit (Tiangen, Beijing, China). Total RNA was reverse transcribed into cDNA using the HiScript II Q RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme, Nanjing, China). qPCR was performed with the ChamQ SYBR qPCR Master Mix (Vazyme, Nanjing, China) on the StepOnePlus™ Real-Time PCR System (Applied Biosystems, United States) and the specified primers (Supplementary Table 2) were designed according to gene CDS sequence. Actin gene was selected as an internal control and the relative gene expression was calculated as previously described (Livak and Schmittgen, 2001).
Chlorophyll, Proline, H2O2, Malondialdehyde Content, and Electrolyte Leakage Measurement
After 2 d of high temperature treatment, the contents of chlorophyll, proline, and H2O2 and the value of electrolyte leakage were determined. The content of chlorophyll was determined by 80% acetone extracts method (Arnon, 1949). The content of proline was determined by the method previously described (Bates et al., 1973). H2O2 content and the value of electrolyte leakage were measured as previously described (Zhang et al., 2021). The content of malondialdehyde (MDA) in leaves was determined by thiobarbituric acid method (Hodges et al., 1999).
Determination of Abscisic Acid, JA, and Ethylene Content
0.2 g leaves of Arabidopsis seedlings were weighed and determined using ELISA kit (Shanghai Renjie Biotechnology Co., LTD.) according to the manufacturer’s instructions.
Statistical Analysis
At least 3 independent replicates were used for each determination. Analysis of variance (ANOVA) was used to test for significance. Different letters above the bars indicate significant differences with Tukey’s test at p < 0.05.
Results
High Temperature Stress Induces the Expression of miR9748 in Cucumber Leaves
To investigate the role of miR9748 in cucumber, we first analyzed its expression patterns in different tissues. Tissue expression analysis revealed that miR9748 was expressed in different tissues, with the lowest expression in flowers, the higher expression in fruits, roots, and stems, and the highest expression in leaves, suggesting that miR9748 was mainly expressed in cucumber leaves (Figure 1A). Therefore, we further analyzed the response of miR9748 in cucumber leaves under high temperature stress. The expression level of miR9748 was upregulated after high temperature treatment and reached the peak at 2 h, which was approximately 4.5 times that of 0 h (Figure 1B), indicating that the expression level of miR9748 in cucumber leaves was induced by high temperature.
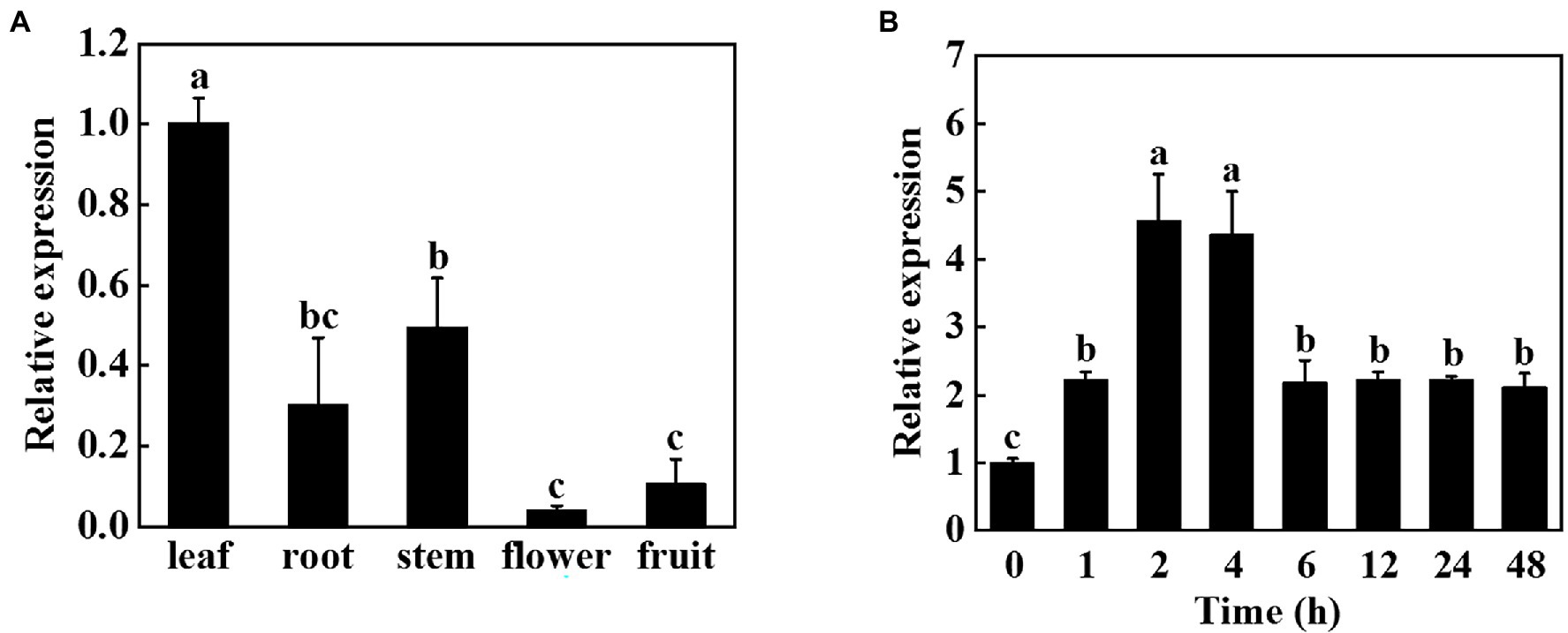
Figure 1. Expression patterns of cucumber miR9748 in different tissues and in response to high temperature stress. (A) qPCR analysis the expression of miR9748 in cucumber leaves, roots, stems, flowers, and fruits. The expression level of miR9748 in leaves was set to 1.0. (B) qPCR analysis the expression level of miR9748 under high temperature stress. The leaf samples were harvested at the indicated time points and analyzed by qPCR. The results represent the mean ± SD (n = 3). Means with the same letter did not significantly differ at p < 0.05 according to Tukey’s test.
Ectopic Overexpression of miR9748 in Arabidopsis Improves High Temperature Stress Tolerance
In order to verify whether miR9748 plays a vital role in cucumber response to high temperature stress, we predicted the precursor sequence of miR9748, and then constructed the miR9748 overexpression plants in Arabidopsis thaliana (OE1, OE2, and OE3). The expression level of miR9748 in overexpression plants was 4.8- to10.9-fold of WT plants (Supplementary Figure 1A). The miR9748 overexpression plant was smaller than WT plant under normal growth condition, as indicated by lower fresh and dry weight (Supplementary Figures 1B,C). After 2 d of high temperature treatment, the wilting degree of leaves in WT plants was more obvious than that of miR9748 overexpression plants (Figure 2A). High temperature treatment resulted in 38.8, 22.1, 22.3, and 20.0% decrease in the fresh weight of WT, OE1, OE2, and OE3 plants, respectively (Supplementary Figure 1B), and the dry matter accumulation decreased by 36.8, 24.4, 18.7, and 19.7%, respectively, compared with their own control plants (Supplementary Figure 1C).
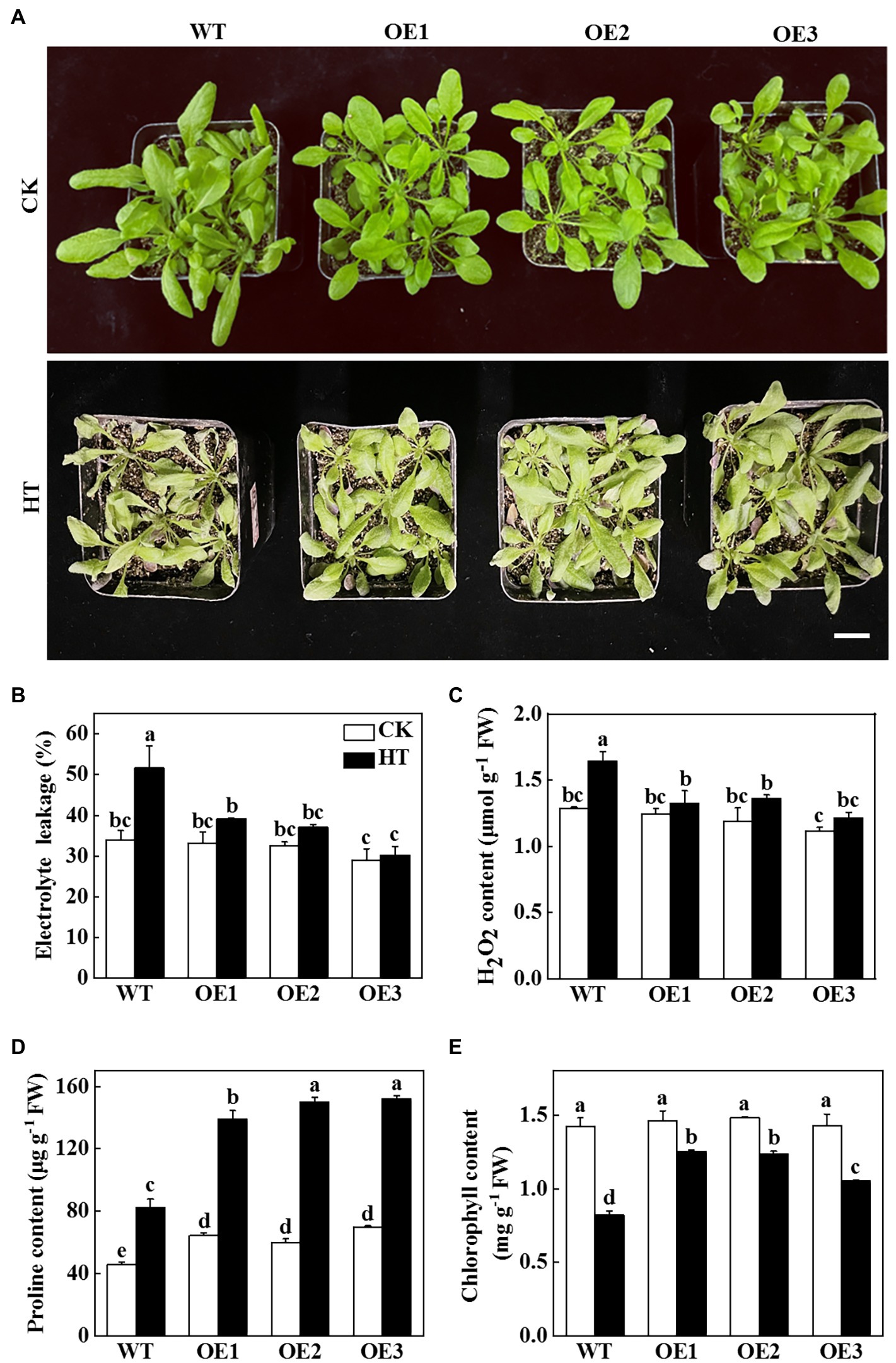
Figure 2. Functional analysis of miR9748 in response to high temperature stress. (A) Overexpression of cucumber miR9748 in Arabidopsis improved high temperature stress tolerance. Bar: 1 cm. (B) Electrolyte leakage. (C) H2O2 content in leaves. (D) Proline content in leaves. (E) Chlorophyll content. 35-d-old Arabidopsis seedlings were subjected to high temperature stress for 2 d, and the phenotype, electrolyte leakage, H2O2, proline, and chlorophyll content were measured. The results represent the mean ± SD of 3 replicates. Means with the same letter did not significantly differ at p < 0.05 according to Tukey’s test. CK, control; HT, high temperature; FW, fresh weight.
To further demonstrate the role of miR9748 under high temperature stress, physiological indices of high temperature tolerance in WT and miR9748 overexpression plants were analyzed. There was no significant difference in the values of electrolyte leakage in all of the plant under optimal growth temperature, but high temperature stress induced the increase of the level of electrolyte leakage, especially in WT plants, which was 24.1 to 41.4% higher than that in miR9748 overexpression plants (Figure 2B). The content of H2O2 in WT plants was 19.5 to 26.1% higher than that in miR9748 overexpression plants under high temperature stress (Figure 2C). However, the proline and chlorophyll content of WT plants were significantly lower than that of miR9748 overexpression plants under high temperature stress (Figures 2D,E). These results suggested that overexpression of miR9748 could improve the high temperature tolerance.
miR9748 Regulates the Expression of Genes Related to ABA, ETH, and JA Signaling Pathways
To further explore the molecular regulatory pathways of miR9748 under high temperature stress, RNA sequencing (RNA-seq) analysis was performed on WT and OE3 plants. Illumina sequencing was performed on 12 leaf cDNA libraries (WT and OE3 plants were treated at optimal temperature and high temperature, 3 replicates per treatment). After removing the low quality, adapter contamination, and unknown high N reads from the results, 23,131 clean reads were obtained, with an average net read of 90.81% for WT-CK, 90.98% for WT-HT, and 90.87% for OE3-CK. The average net read of OE-HT library was 92.38% (Supplementary Table 3). After the clean reads were obtained, HISAT2 was used to alignment clean reads to Arabidopsis reference genome database, and the results showed that over 92.0% of the reads were uniquely mapped to the genome (Supplementary Table 4). There were 348 DEGs of WT-CK vs. OE3-CK, of which 268 DEGs were upregulated and 80 DEGs were downregulated (Figures 3A,B). There were 13,726 DEGs of WT-CK vs. WT-HT, including 6,974 upregulated and 6,752 downregulated genes (Figures 3A,B). There were 13,504 genes with significant differential expression of OE3-CK vs. OE3-HT. Among these, 6,873 genes were upregulated and 6,631 genes were downregulated (Figures 3A,B). A total of 859 DEGs of WT-HT vs. OE3-HT were obtained, of which 354 DEGs were upregulated and 505 DEGs were downregulated (Figures 3A,B). GO enrichment pathways analysis showed that these DEGs were mainly related to abscisic acid (ABA), ethylene (ETH), and JA signal pathways (Figures 3C–E), indicating that these plant hormones signaling might mediate high temperature tolerance. KEGG pathway analysis showed that DEGs were mainly enriched in ribosomes, spliceosomes, ribosome biogenesis in eukaryotes, metabolic pathways, photosynthesis, and starch and sucrose metabolism (Figure 3F). Transcriptomic data analysis showed that overexpression of miR9748 might induce the expression of genes involved in ABA, ETH, and JA to participate in the response to high temperature stress.
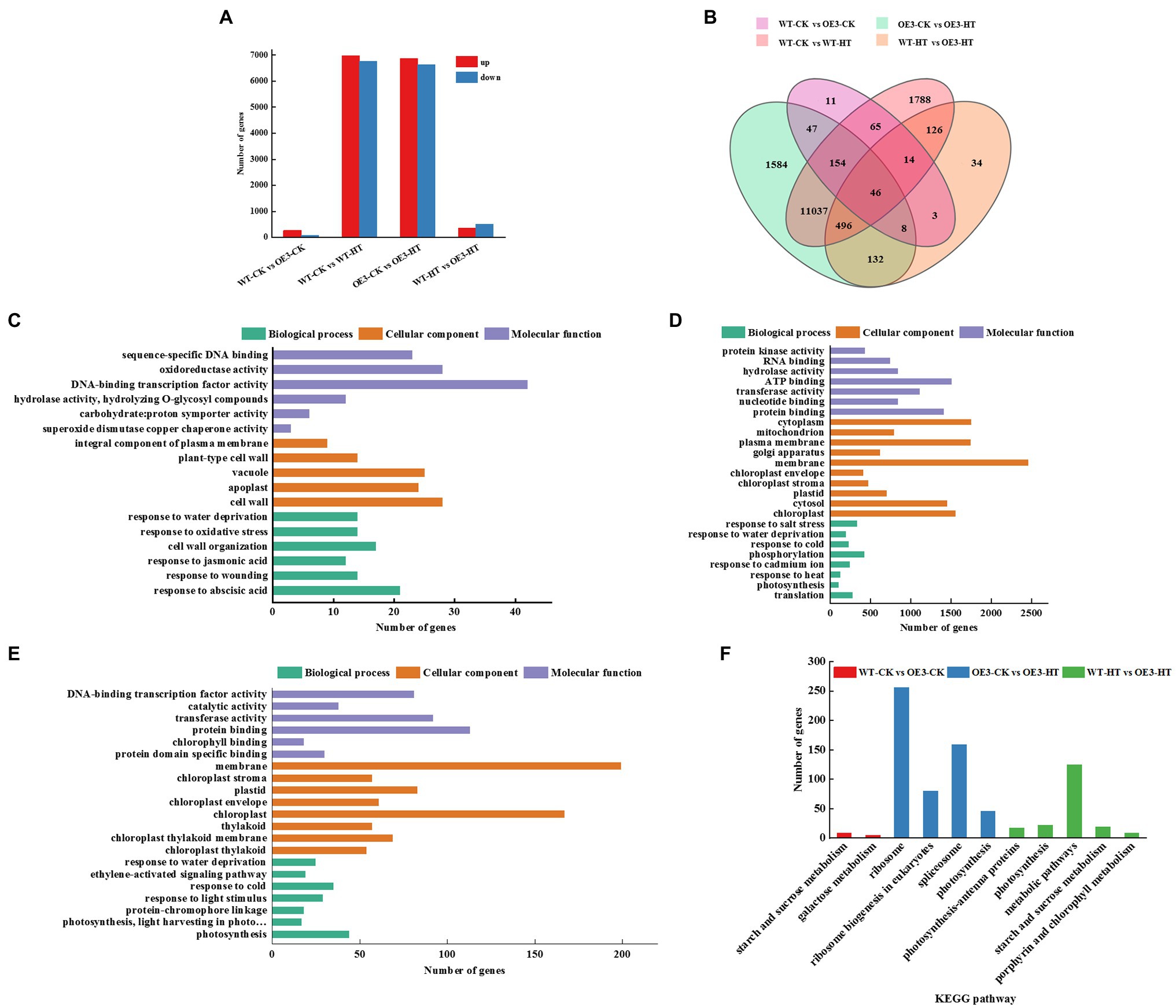
Figure 3. GO and KEGG pathway analysis of differentially expressed genes (DEGs) between wild-type (WT) and miR9748 overexpression (OE3) plants under high temperature stress. (A) The total number of upregulated and downregulated DEGs after high temperature stress. (B) Venn diagrams analysis of DEGs between WT and OE3 plants under high temperature stress. (C) GO enrichment analysis of DEGs of WT-CK vs. OE3-CK. (D) GO enrichment analysis of DEGs of OE3-CK vs. OE3-HT. (E) GO enrichment analysis of DEGs of WT-HT vs. OE3-HT. (F) KEGG pathway enrichment analysis of WT-CK vs. OE3-CK, OE3-CK vs. OE3-HT, and WT-HT vs. OE3-HT. CK, control; HT, high temperature.
miR9748 Target Gene Identification
To further investigate the function mechanism of miR9748 under high temperature stress, we predicted the target gene of miR9748 using psRobot and Target Finder software. PsRobot predicted 467 target genes and Target Finder predicted 649 target genes, containing a total of 233 commonly regulated target genes. GO enrichment analysis was performed on 233 possible target genes. It was found that these target genes were successfully assigned to the corresponding 17 GO items. In molecular function category, 146 target genes were mainly enriched, among which 77 and 83 target genes were significantly enriched in catalytic activity (GO:0003824) and binding (GO:0005488), respectively. In cellular component group classification, 133 target genes were mainly enriched, and 97 and 76 target genes were significantly enriched in cell (GO:0005623) and organelle (GO:0043226), respectively. There were 126 target genes in biological processes, among which cellular processes (GO:0065007) and metabolic processes (GO:0008152) were significantly enriched, with 97 and 94 target genes, respectively (Supplementary Figure 2). Furthermore, four predicted target genes of cucumber miR9748 (CsaV3_6G007840, CsaV3_1G045520, CsaV3_7G029600, and CsaV3_5G039430) were related to plant hormone signal transduction, which was closely related to high temperature. In order to understand whether they are involved in high temperature stress, we detected their expression patterns under high temperature, and found that CsaV3_6G007840 was upregulated and then downregulated under high temperature treatment, and reached the peak at 2 h, with 5 times of that in 0 h (Supplementary Figure 3). The expression of CsaV3_1G045520 was downregulated at 1 h and then upregulated again, and reached the peak at 2 h after treatment, which was 1.8 times of that in 0 h (Supplementary Figure 3). The expression of CsaV3_7G029600 was similar with that of CsaV3_1G045520 (Supplementary Figure 3). However, the expression level of CsaV3_5G039430 was always downregulated (Supplementary Figure 3), which was opposite to the expression pattern of miR9748, suggesting that it might be the target gene of miR9748 under high temperature stress.
It is well known that plant miRNAs negatively regulate the expression of their target genes through either mRNA cleavage or translational inhibition (Zhao et al., 2016; Chen et al., 2018). To investigate whether CsaV3_5G039430 was the target gene of miR9748, we used 5′ RLM-RACE to locate miR9748-directed cleavage sites in CsaV3_5G039430. The results showed that there was a cleavage site between the 10th and 11th base pairs at the miR9748 target site (Figure 4A). To further verify whether CsaV3_5G039430 was the true target of miR9748, the interaction between miR9748 and CsaV3_5G039430 was verified using Agrobacterium-mediated transient expression in tobacco leaves. The results showed that when the transient transformation of 35S::MIR9748 overexpression vector, tobacco leaves were white after GUS staining and alcohol decolorization (Figure 4B). When 35S::GUS or 35S::CsaV3_5G039430-GUS or 35S::MIR9748 and 35S::GUS were injected into tobacco leaves, GUS staining was found in tobacco leaves with blue color and large area, indicating that GUS was strongly expressed (Figure 4B). In contrast, when 35S::MIR9748 was co-transformed with 35S::CsaV3_5G039430-GUS overexpression vector, the blue color became lighter and the area became smaller (Figure 4B). However, when 35S::MIR9748 was co-transformed with the mutated sites overexpression vector (35S::CsaV3_5G039430M-GUS), tobacco had large blue area and dark color (Figure 4B). These results indicated that CsaV3_5G039430 was the true target gene of miR9748, and miR9748 negatively regulated the expression of CsaV3_5G039430 through cleavage.
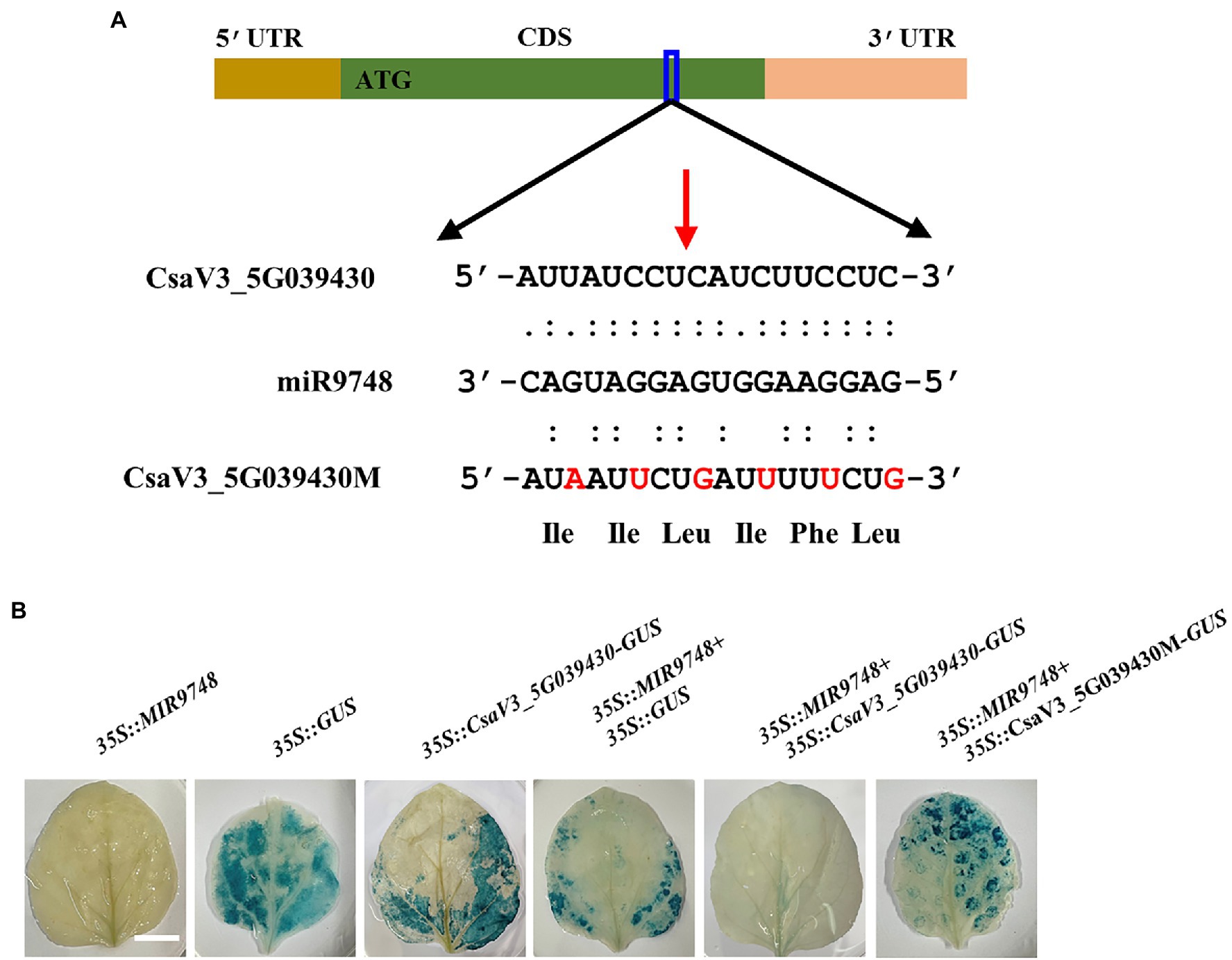
Figure 4. CsaV3_5G039430 is the target gene of miR9748. (A) 5′ RLM-RACE analysis of miR9748 cleavage site in CsaV3_5G039430, and the cleavage site is indicated by red arrow. (B) Transient GUS expression verification of miR9748 cleavage of CsaV3_5G039430. Agrobacterium tumefaciens containing the indicated plasmids were infiltrated into tobacco leaves, and the leaves were stained after infiltration for 2 d. Bar: 1 cm.
Functional Analysis of CsaV3_5G039430 Under High Temperature Stress
To further investigate the role of CsaV3_5G039430 under high temperature stress, we searched this gene in cucurbitaceae genome database and found that it was a NRT1/PTR family 4.4 protein (Supplementary Table 5), and an orthologous of AtNRT1.13/NPF4.4, which is lack of the conserved proline residue between the 10th and 11th transmembrane domains, resulting in only binding nitrate and without transport activity (Chen et al., 2021a). Therefore, the protein encoded by CsaV3_5G039430 was named as CsNPF4.4, which shared 63.30% sequence identity and had the conserved domains and similar molecular weight and theoretical isoelectric point (pI) with AtNPF4.4 (Supplementary Figure 4; Supplementary Table 5). Tissue expression analysis showed that CsNPF4.4 had lower expression in flowers, fruits, and leaves and higher expression in roots and stems (Supplementary Figure 5A). The expression level of CsNPF4.4 in roots was approximately 5-fold that in leaves and 2.5-fold that in stems (Supplementary Figure 5A), indicating that CsNPF4.4 was predominantly expressed in cucumber roots. Subcellular localization results showed that tobacco cells transformed with 35S::CsNPF4.4-GFP detected green fluorescence both in cell membrane, cytosol, and nucleus (Supplementary Figure 5B).
To further test the role of CsNPF4.4 under high temperature stress, the 35S:: CsNPF4.4 overexpression vector was constructed and transformed into Arabidopsis. The expression level of CsNPF4.4 in overexpression lines (OE) was 18-fold higher than that in WT plants (Supplementary Figure 6A). Before high temperature treatment, OE plants had more and larger leaves than WT plants (Figure 5A; Supplementary Figure 6B). After 2 d of high temperature treatment, OE plants were more sensitive to high temperature than WT plants (Figure 5A). The fresh weight of WT and OE plants decreased by 19.34 and 27.51%, and the dry weight decreased by 15.87 and 23.87%, respectively, compared with their own control plants (Supplementary Figures 6B,C). High temperature stress increased the value of electrolyte leakage of WT and OE plants, as indicated increased by 28.2% in WT plants and 34.7% in OE plants (Figure 5B). Furthermore, high temperature promoted the accumulation of proline, but the proline content in OE plants was still lower than that in WT plants (Figure 5C). Although high temperature stress reduced chlorophyll content both in WT and OE plants, the chlorophyll content in WT plants was significantly higher than that in OE plants (Figure 5D). These results indicated that CsNPF4.4 overexpression plants suffered more damage under high temperature treatment, and had the opposite effect of miR9748.
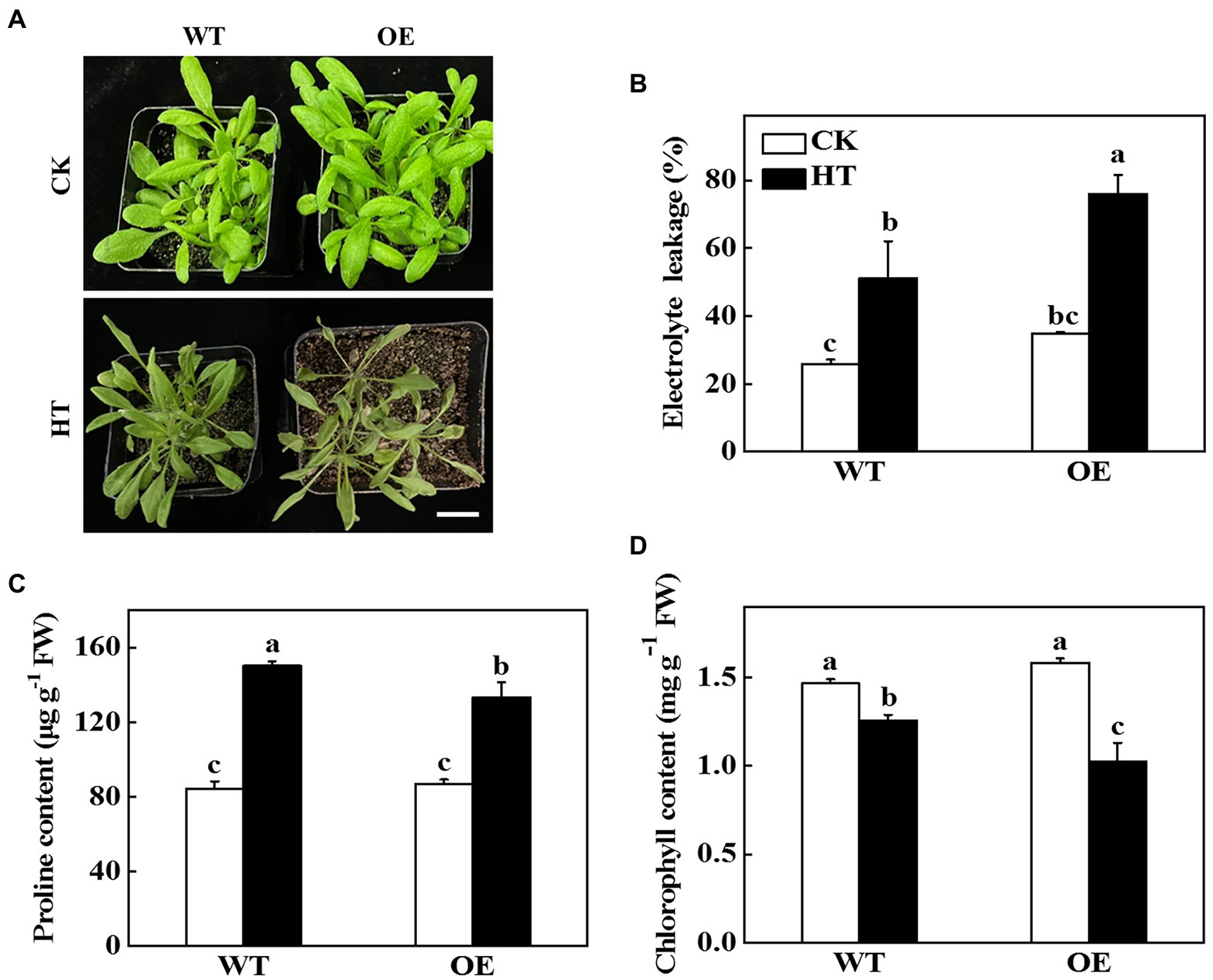
Figure 5. Functional analysis of CsNPF4.4 in response to high temperature stress. (A) Overexpression of cucumber CsNPF4.4 in Arabidopsis reduced high temperature stress tolerance. Bar: 1 cm. (B) Electrolyte leakage. (C) Proline content in leaves. (D) Chlorophyll content. 35-d-old Arabidopsis seedlings were subjected to high temperature stress for 2 d, and the phenotype, electrolyte leakage, proline, and chlorophyll content were measured. The results represent the mean ± SD (n = 3). Means with the same letter did not significantly differ at p < 0.05 according to Tukey’s test. CK, control; HT, high temperature; FW, fresh weight.
CsNPF4.4 Negatively Regulates High Temperature Stress Tolerance Through Inhibition JA
The transcriptome sequencing results of Arabidopsis suggested that overexpression of miR9748 regulated the expression of genes related to ABA, ETH, and JA (Figure 3). CsNPF4.4 acted as the target gene of miR9748, it might regulate plant hormones to mediate high temperature stress tolerance. Therefore, we measured the content of ABA, ETH, and JA in WT and OE plants. The ABA and ETH contents in OE plants were significantly higher than those of WT plants under high temperature stress (Figures 6A,B). High temperature stress induced the accumulation of JA in WT plants, but decreased in OE plants, as indicated by the content of JA significantly lower than that of WT plants (Figure 6C). Furthermore, high temperature induced the expression of JA synthesis genes, such as LOX2, AOC4, AOS, and JAR1, in WT plants (Figure 6D). However, the expression level of LOX2, AOC4, and JAR1 significantly decreased in OE plants under high temperature stress compared with the control plants (Figure 6D). Thus, CsNPF4.4 might negatively regulate high temperature tolerance by downregulating the expression of genes involved in JA synthesis and reducing JA content.
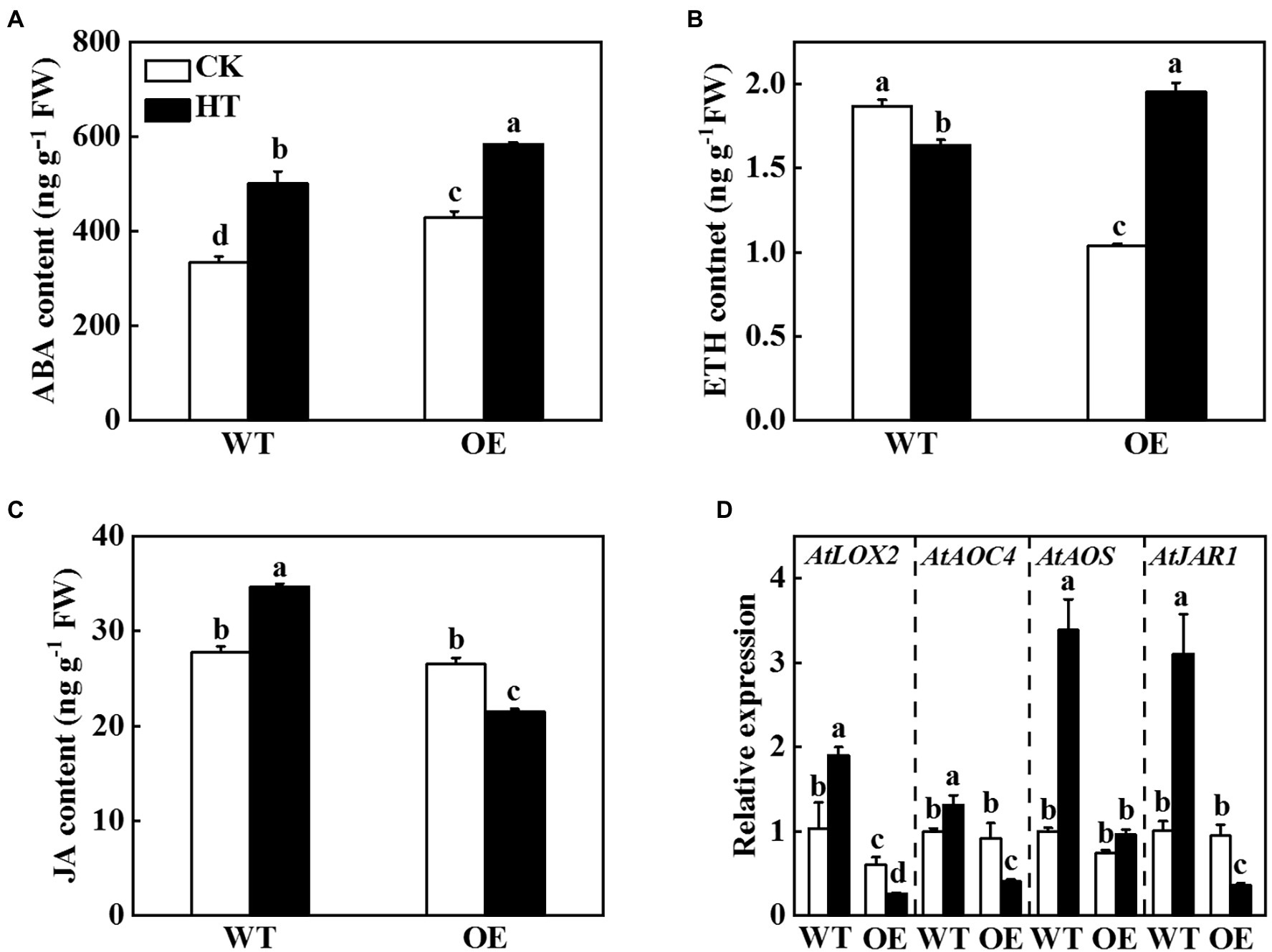
Figure 6. Effects of CsNPF4.4 on plant hormone content and the expression of genes related to jasmonic acid (JA) synthesis under high temperature stress. (A) Abscisic acid content. (B) Ethylene content. (C) Jasmonic acid content. 35-d-old Arabidopsis seedlings were subjected to high temperature stress for 2 d, and the plant hormone contents were measured. (D) Effects of CsNPF4.4 on the expression of genes related to JA synthesis under high temperature. The leaf samples were harvested after high temperature stress for 24 h and analyzed by qPCR. The results represent the mean ± SD (n = 3). Means with the same letter did not significantly differ at p < 0.05 according to Tukey’s test. CK, control; HT, high temperature; FW, fresh weight.
To investigate whether JA was involved in CsNPF4.4-mediated response to high temperature stress, we sprayed WT and OE plants with 100 μmol MeJA and then analyzed their high temperature stress tolerance. After 2 d of high temperature stress, WT plants-treated with distilled water showed wilting and coking of leaf margins, but the OE plants were more serious (Figure 7A). However, the WT and OE plants were alleviated after spraying MeJA (Figure 7A). Furthermore, sprayed with MeJA inhibited high temperature-induced the increase of electrolyte leakage, MDA and H2O2 content, and also increased proline content both in WT and OE plants compared with the distilled water-treated plants (Figures 7B–E), indicating that exogenous spraying of MeJA could ameliorate the high temperature sensitivity of CsNPF4.4 overexpression plants.
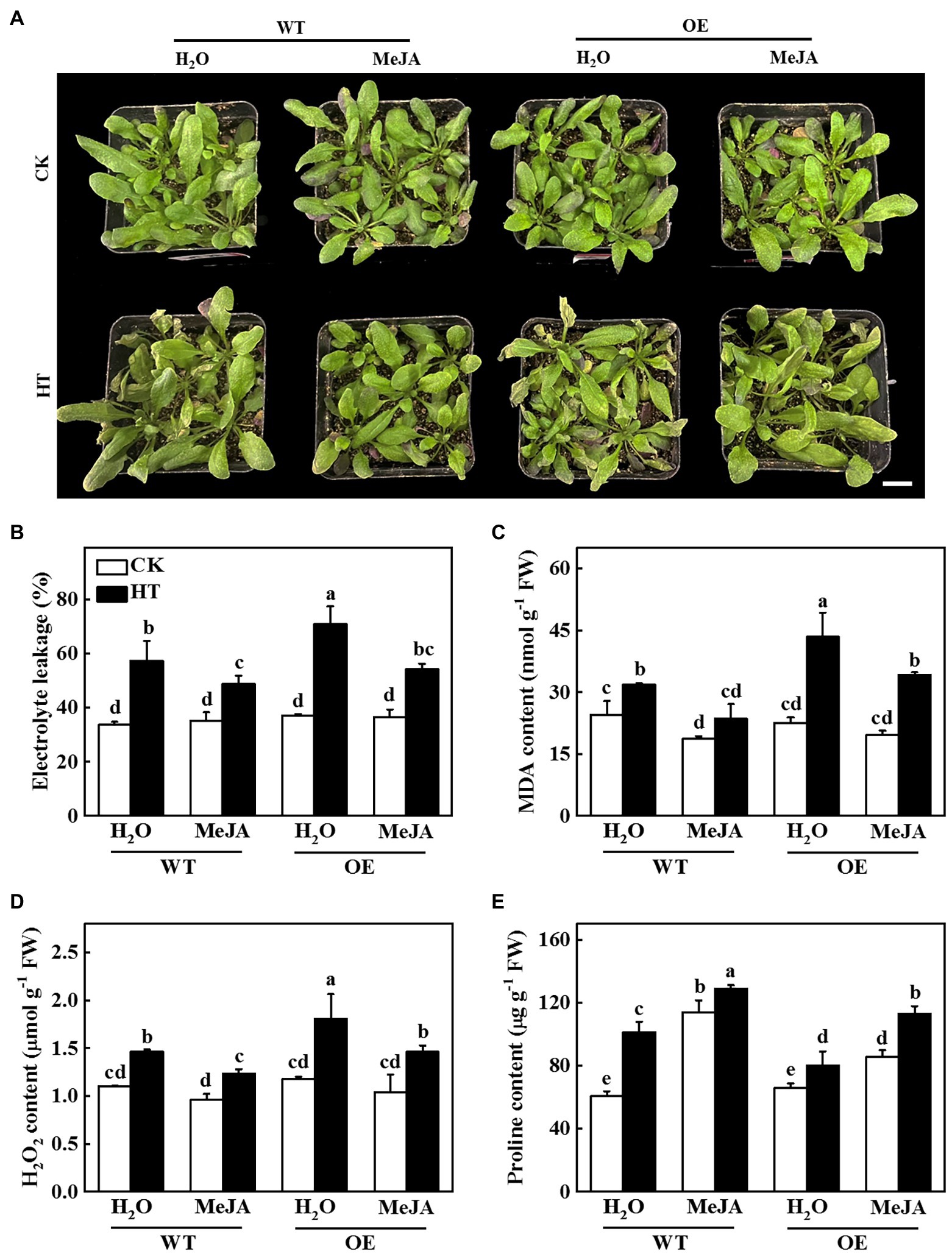
Figure 7. Functional analysis of methyl jasmonate (MeJA) on CsNPF4.4 overexpression plants in response to high temperature stress. (A) Exogenous spraying of MeJA alleviated the damage of high temperature to CsNPF4.4 overexpression plants. Bar: 1 cm. (B) Electrolyte leakage. (C) Malondialdehyde (MDA) content. (D) H2O2 content. (E) Proline content. 35-d-old Arabidopsis seedlings were subjected to high temperature stress for 2 d, and the phenotype, electrolyte leakage, MDA, and proline content were measured. The results represent the mean ± SD of 3 replicates. Means with the same letter did not significantly differ at p < 0.05 according to Tukey’s test. CK, control; HT, high temperature; FW, fresh weight.
CsbZIP2 Binds to the Promoter of MIR9748 to Promote Its Transcription
In order to identify the transcription factors that regulate the expression of MIR9748 gene, we first used the online software PlantCARE to analyze the MIR9748 promoter sequence, and found that there was multiple ABA response element (ABRE), JA response elements (TGACG-motif, TGACG-motif, CGTCA-motif) and a variety of light-responsive elements (G-box, GATA-motif), and MYB, MYC, ERE, and other functional elements (Figure 8A).
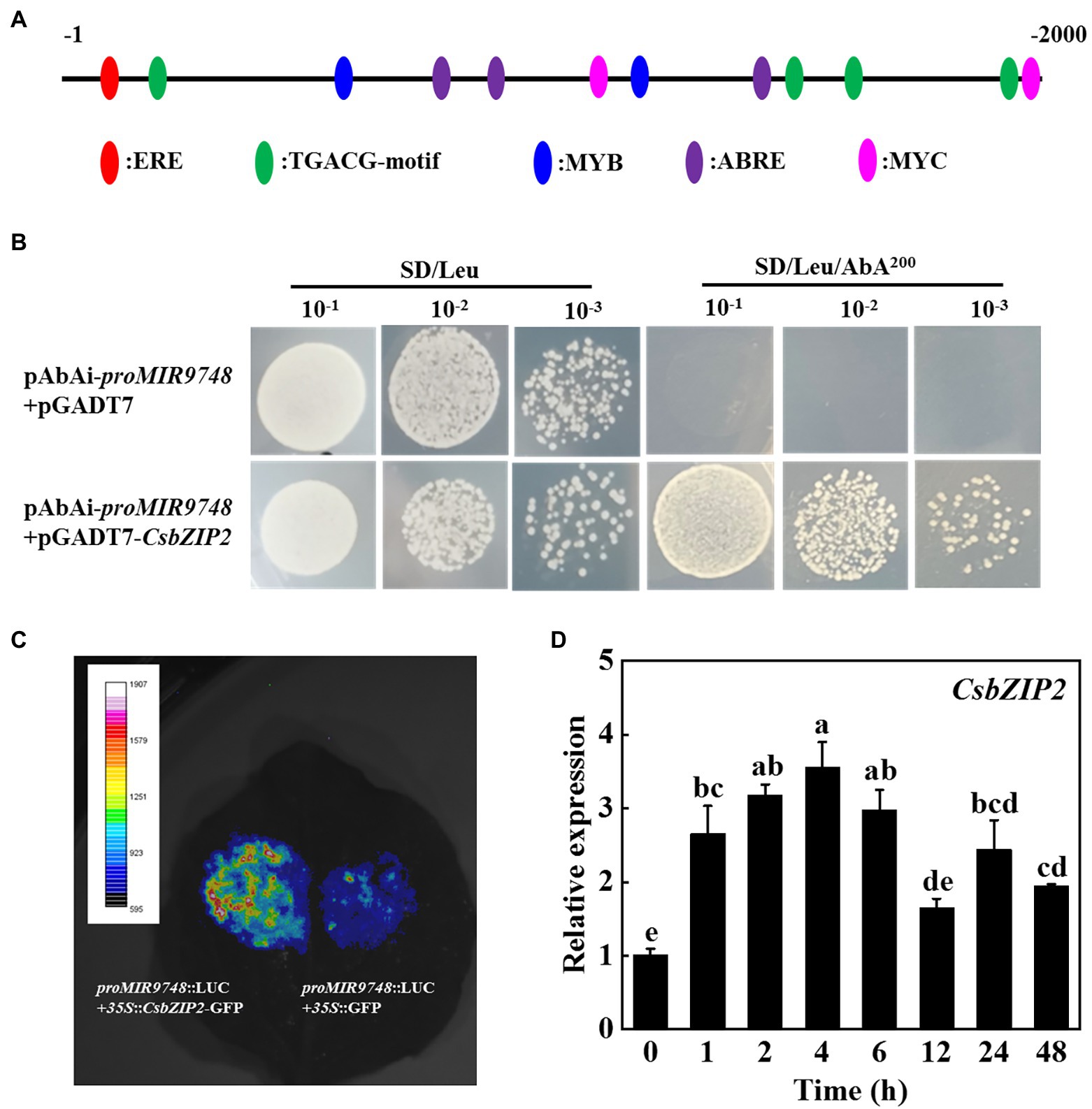
Figure 8. CsbZIP2 binds to the promoter of MIR9748 and high temperature stress induces the expression of CsbZIP2. (A) Analysis of cis-acting elements in MIR9748 promoter. Numbering is from predicted transcriptional start sites. (B) Yeast one-hybrid assay showing the binding of CsbZIP2 to MIR9748 promoter. Yeast cells with DNA–protein interactions were grown on SD/Leu plates with 200 ng ml−1 aureobasidin A. (C) Luciferase assay showing transient overexpression of CsbZIP2 enhancement the expression of MIR9748 in Nicotiana benthamiana leaves. The empty vector was used as a control. (D) qPCR analysis the expression level of CsbZIP2 under high temperature stress. The leaf samples were harvested at the indicated time points and analyzed by qPCR. The results represent the mean ± SD (n = 3). Means with the same letter did not significantly differ at p < 0.05 according to Tukey’s test.
We used yeast one-hybrid experiments to screen the transcription factors that bound to the promoter of MIR9748. The results showed that yeast cells containing the bait vector harboring MIR9748 promoter sequence grew on the SD/Leu medium containing 200 ng ml−1 AbA when transformed with pGADT7-CsbZIP2, but those transformed with pGADT7 vector or other transcription factors could not grow on the same selection medium (Figure 8B; Supplementary Figure 7), indicating that CsbZIP2 bound to the promoter of MIR9748 in vitro. To further validate this results, we performed dual-luciferase assay using the tobacco transient transformation system. As shown in Figure 8C, the fluorescence signal was stronger when co-injected with proMIR9748-LUC and 35S::CsbZIP2 than that co-injected with proMIR9748-LUC and 35S::GFP. Furthermore, the transcription level of CsbZIP2 was significantly increased under high temperature treatment, and reached the maximum value at 4 h of high temperature stress, which increased by 2.5-fold compared with that of 0 h (Figure 8D), indicating that CsbZIP2 responded to high temperature stress. Thus, the transcription factor CsbZIP2 was upregulated under high temperature stress to promote the expression of MIR9748 gene in cucumber leaves to negatively regulate the expression of CsNPF4.4, resulting in improving high temperature tolerance.
Discussion
It has been demonstrated that miR9748 is involved in the regulation of plant growth and development and abiotic stress response (Cakir et al., 2016; Sun et al., 2017; Cheng et al., 2019). Tomato miR9748 might regulate the expression of ERD (early response to dehydration-like), DREB (dehydration response element binding), and DI19 (dehydration/drought-induced 19 protein) in response to drought stress (Candar-Cakir et al., 2016). Cowpea (Vigna unguiculata L.) miR9748 targets heat shock proteins expression in response to heat stress (Gul et al., 2017). Selenium treatment induced the expression of miR9748 in A. chrysochlorus to further regulate the expression of MYC2 and HSP90 (Cakir et al., 2016). Here, we found that cucumber miR9748 was significantly upregulated under high temperature treatment (Figure 1B), indicating that miR9748 might play a critical role in high temperature. Indeed, overexpression of cucumber miR9748 in Arabidopsis enhanced the resistance to high temperature stress compared with WT plants, as indicated by miR9748 overexpression plants maintaining higher level of proline and chlorophyll content, and lower H2O2 content and electrolyte leakage (Figure 2). Our previous study revealed that miR9748 may interact with lncRNAs and circRNAs to mediate high temperature stress through plant hormone signal transduction pathways (He et al., 2020). In the present study, transcriptome data showed that the DEGs were mainly related to the response to ABA, ETH, and JA (Figure 3), revealing that miR9748 might increase high temperature stress tolerance via regulating these plant hormones signal pathways.
Plant miRNAs mediate growth and development, and response to various stresses through negative regulation target genes expression or translation (Chen et al., 2018; Bhogireddy et al., 2021). miR1432 negatively regulates the expression of target gene OsACOT (Acyl-CoA thioesterase) and increases grain filling amount to regulate rice yield (Zhao et al., 2019). Defense against rice streak virus invasion leads to a reduction of miR528 in rice, alleviating miR528-mediated degradation of L-ascorbate oxidase mRNA, and enhancing rice antiviral activity (Yao et al., 2019). In addition, numerous of miRNAs are identified in the regulation of high temperature response in plants (Sailaja et al., 2014; Mangrauthia et al., 2017; Ravichandran et al., 2019). ABA inhibits the expression of miR159b, which downregulates the expression of its target genes, such as GAMYB1, MYB29-like, and HSP70, to enhance high temperature tolerance in grafted cucumber plants (Li et al., 2016). Heat stress induces the expression of miR4200 to degrade the mRNA of HSFB4a, a negative regulator in heat stress, thereby exhibiting higher heat stress tolerance in tomato (Rao et al., 2022). Similarly, HSFA1b and HSFA7b induce miR398 to increase thermotolerance of Arabidopsis through downregulation the expression of CSD1, CSD2, and CCS (Guan et al., 2013). Furthermore, miR156-SPL module regulates the response to recurring heat stress in Arabidopsis (Cui et al., 2014; Sailaja et al., 2014). In this study, it was demonstrated that the expression of miR9748 and CsNPF4.4 was upregulated and downregulated, respectively, under high temperature stress (Figure 1B; Supplementary Figure 3). 5′ RLM-RACE technology and tobacco transient co-transformation experiments found that CsNPF4.4 was a target gene of miR9748 in cucumber (Figure 4). Our results suggested that miR9748 was involved in the response to high temperature by precisely cleaving CsNPF4.4. CsNPF4.4 is a possible peptide/nitrate transporter involved in peptide/nitrate transport. Studies have shown that NRT1.1 in Arabidopsis regulates NO3− distribution to roots by coordinating the accumulation of Cd2+ in root vacuoles, thereby promoting Cd2+ detoxification (Jian et al., 2019). Furthermore, NRT1.1 suppresses lateral root development through inhibiting the expression of auxin synthesis and auxin influx carrier genes and promoting basipetal auxin transport out of the lateral root primordia at low-nitrate availability in Arabidopsis (Krouk et al., 2010; Zhang et al., 2019; Maghiaoui et al., 2020). In Medicago truncatula, NPF6.8 mediates high nitrate-induced repression of primary root growth via ABA (Pellizzaro et al., 2014). Moreover, nrt2 mutants increase the tolerance to Pseudomonas syringae pv tomato DC3000 through inducing the accumulation of salicylic acid (Camanes et al., 2012). These results suggest that NRT not only regulates the uptake and transport of nitrate, but also mediates the homeostasis of plant hormones to response to environmental stresses. AtNPF4.4, an orthologous of CsNPF4.4, lacks of the conserved proline between the 10th and 11th transmembrane domain and cannot transport nitrate but can bind it (Chen et al., 2021a). Deficient of AtNPF4.4 exhibits delayed flowering, enhanced node number, inhibited branch outgrowth, and lateral nitrate allocation to nodes under low-nitrate conditions (Chen et al., 2021a). Amino acid sequence alignment showed that CsNPF4.4 also did not contain this conserved proline (Supplementary Figure 4), indicating that CsNPF4.4 might have the similar function in nitrate transport. Interestingly, overexpression of CsNPF4.4 promoted plants growth under the normal growth conditions, as indicated by higher fresh and dry weight compared with WT plants (Supplementary Figures 6B,C). However, their roles in high temperature response are largely unknown. Here, we found that CsNPF4.4 overexpression plants were more sensitive to high temperature stress accompanied by inhibition JA accumulation (Figures 5, 6C). It has been shown that NRT displays transport plant hormones, including auxin, ABA, gibberellin, and jasmonoyl-isoleucine (Chiba et al., 2015). Interestingly, low nitrogen stress enhances lateral root number and JA content in wheat (Triticum aestivum L.), while high nitrate inhibits root growth in maize (Zea mays L.) and decreases JA content (Saiz-Fernández et al., 2020; Lv et al., 2021). Here, we found that JA content in CsNPF4.4 overexpression plants was no significant difference with that in WT plants under normal growth conditions (Figure 6C). However, the expression of JA synthesis genes and JA content in CsNPF4.4 overexpression plants dramatically decreased under high temperature stress (Figures 6C,D). These results indicate that NRT is involved in JA synthesis under adverse conditions, but the specific functional mechanism remains further investigation.
It has been demonstrated that JA is involved in the regulation of plant response to high temperature stress (Xia et al., 2015; Balfagon et al., 2019). Exogenous spraying of MeJA improves high temperature tolerance of plants through regulation osmotic adjustment, antioxidant defense, maintenance the stability of photosynthesis proteins, and inducing the expression of JA-responsive genes (Fatma et al., 2021; Su et al., 2021). Silencing of WRKY6 increases the susceptibility to heat stress in pepper by downregulation of JA-induced gene expression (Cai et al., 2015). HsfA1b mediates heat resistance via OPR3 and JA signal pathway in wheat and Arabidopsis (Tian et al., 2020). High temperature stress significantly induces the expression of JA pathway genes and the production of JA, thereby improving the adaptability to heat stress (Su et al., 2021). Nevertheless, CsNPF4.4 overexpression plants compromised the expression of JA synthesis genes and the increase of JA under high temperature stress (Figures 6C,D), resulting in reduced the tolerance to high temperature. Exogenous application of MeJA significantly alleviated the damage of high temperature to CsNPF4.4 overexpression plants (Figure 7). Therefore, miR9748 directly cleaved the target gene CsNPF4.4 to increase the content of JA, resulting in enhancing the tolerance to high temperature stress.
Plant bZIP transcription factors play an indispensable role in the regulation of plant high temperature stress response (Droege-Laser et al., 2018). Overexpression of OsbZIP46 in rice and overexpression of wheat TabZIP60 in Arabidopsis improve the high temperature tolerance of transgenic plants (Chang et al., 2017; Geng et al., 2018). Knockdown the expression of bZIP60 in maize increases the hypersensitivity to high temperature stress and compromised high temperature-induced the expression of genes related to HSFs, chlorophyll metabolism and chloroplast protein turnover (Li et al., 2020). Furthermore, maize bZIP4 is induced by high temperature stress (Ma et al., 2018). Similarly, CsbZIP2 was induced by high temperature, and the transcription level of CsbZIP2 was significantly increased under high temperature (Figure 8D). Interestingly, yeast one-hybrid and dual-luciferase reporter assays found that the transcription factor CsbZIP2 could bind to the promoter of MIR9748 to promote its transcription (Figure 8).
In conclusion, high temperature stress induced the expression of CsbZIP2, which bound to the promoter of MIR9748 to promote its transcription to form mature miR9748. miR9748 negatively regulated target gene CsNPF4.4 through direct cleavage. Overexpression of CsNPF4.4 decreased high temperature tolerance and subdued high temperature-induced the increase of JA, while foliar application of MeJA mitigated the sensitivity of CsNPF4.4 overexpression plants to high temperature stress. Thus, high temperature stress induced CsbZIP2 to trigger the expression of miR9748, which negatively regulated target gene CsNPF4.4 through direct cleavage, to promote the accumulation of JA, resulting in enhancing high temperature tolerance (Figure 9). Our results provide a new perspective for elucidating the response mechanism of cucumber to high temperature stress.
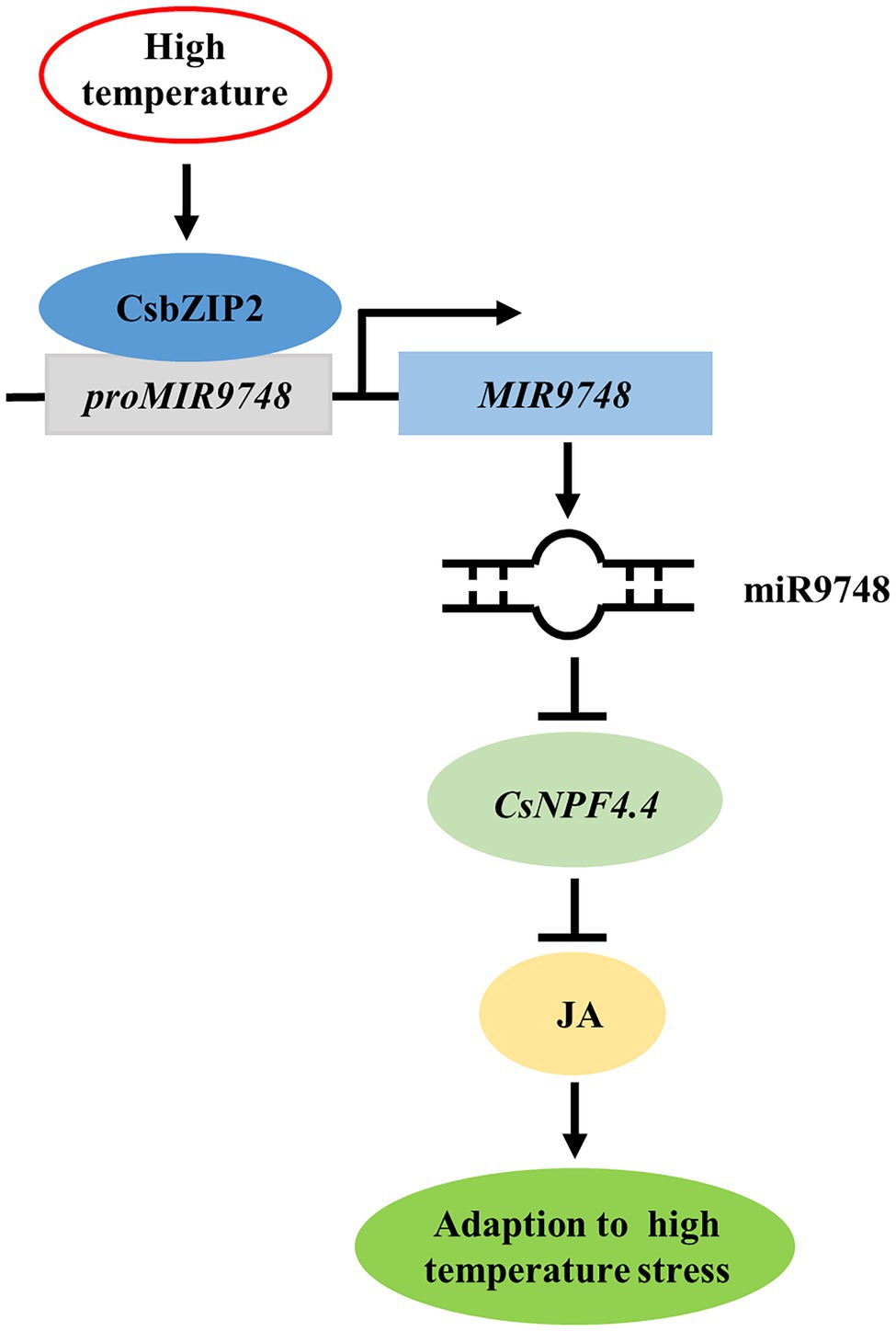
Figure 9. Proposed model of CsbZIP2-miR9748-CsNPF4.4 module in response to high temperature stress. High temperature induced the expression of CsbZIP2, which bound to the promoter of MIR9748 to induce its transcription to form mature miR9748. miR9748 negatively regulated target gene CsNPF4.4 through direct cleavage, to promote the accumulation of JA, resulting in enhancing high temperature tolerance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JS and YW designed the experiment. LL, GC, and MY performed the experiments. LL and SG analyzed the data. LL and YW wrote the manuscript. JS revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFD1001900), the China Agriculture Research System (CARS-23), and the Postdoctoral Research Funding Scheme of Jiangsu Province (2019K071).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Yuehua Ma (Central Laboratory of College of Horticulture, Nanjing Agricultural University) for assistance in using LSM 800 confocal microscope.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.883876/full#supplementary-material
References
Allen, E., Xie, Z. X., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Balfagon, D., Sengupta, S., Gomez-Cadenas, A., Fritschi, F. B., Azad, R. K., Mittler, R., et al. (2019). Jasmonic acid is required for plant acclimation to a combination of high light and heat stress. Plant Physiol. 181, 1668–1682. doi: 10.1104/pp.19.00956
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bhogireddy, S., Mangrauthia, S. K., Kumar, R., Pandey, A. K., Singh, S., Jain, A., et al. (2021). Regulatory non-coding RNAs: a new frontier in regulation of plant biology. Funct. Integr. Genomics 21, 313–330. doi: 10.1007/s10142-021-00787-8
Cai, H., Yang, S., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Cakir, O., Candar-Cakir, B., and Zhang, B. (2016). Small RNA and degradome sequencing reveals important microRNA function in Astragalus chrysochlorus response to selenium stimuli. Plant Biotechnol. J. 14, 543–556. doi: 10.1111/pbi.12397
Camanes, G., Pastor, V., Cerezo, M., Garcia-Andrade, J., Vicedo, B., Garcia-Agustin, P., et al. (2012). A deletion in NRT2.1 attenuates pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 158, 1054–1066. doi: 10.1104/pp.111.184424
Candar-Cakir, B., Arican, E., and Zhang, B. (2016). Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 14, 1727–1746. doi: 10.1111/pbi.12533
Chang, Y., Ba Hoanh, N., Xie, Y., Xiao, B., Tang, N., Zhu, W., et al. (2017). Co-overexpression of the constitutively active form of OsbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in rice. Front. Plant Sci. 8:1102. doi: 10.3389/fpls.2017.01102
Chen, H. Y., Lin, S. H., Cheng, L. H., Wu, J. J., Lin, Y. C., and Tsay, Y. F. (2021a). Potential transceptor AtNRT1.13 modulates shoot architecture and flowering time in a nitrate-dependent manner. Plant Cell 33, 1492–1505. doi: 10.1093/plcell/koab051
Chen, L., Yun, M., Cao, Z., Liang, Z., Liu, W., Wang, M., et al. (2021b). Phenotypic characteristics and transcriptome of cucumber male flower development under heat stress. Front. Plant Sci. 12:758976. doi: 10.3389/fpls.2021.758976
Chen, C., Zeng, Z., Liu, Z., and Xia, R. (2018). Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 5:63. doi: 10.1038/s41438-018-0072-8
Cheng, L., Liu, H., Han, Y., and Li, S. (2019). Transcriptome analysis of miRNAs expression reveals novel insights into adventitious root formation in lotus (Nelumbo nucifera Gaertn.). Mol. Biol. Rep. 46, 2893–2905. doi: 10.1007/s11033-019-04749-z
Chiba, Y., Shimizu, T., Miyakawa, S., Kanno, Y., Koshiba, T., Kamiya, Y., et al. (2015). Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 128, 679–686. doi: 10.1007/s10265-015-0710-2
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., and Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Droege-Laser, W., Snoek, B. L., Snel, B., and Weiste, C. (2018). The Arabidopsis bZIP transcription factor family-an update. Curr. Opin. Plant Biol. 45, 36–49. doi: 10.1016/j.pbi.2018.05.001
Fatma, M., Iqbal, N., Sehar, Z., Alyemeni, M. N., Kaushik, P., Khan, N. A., et al. (2021). Methyl jasmonate protects the PS II system by maintaining the stability of chloroplast D1 protein and accelerating enzymatic antioxidants in heat-stressed wheat plants. Antioxidants 10:1216. doi: 10.3390/antiox10081216
Geng, X., Zang, X., Li, H., Liu, Z., Zhao, A., Liu, J., et al. (2018). Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 274, 252–260. doi: 10.1016/j.plantsci.2018.05.029
Guan, Q., Lu, X., Zeng, H., Zhang, Y., and Zhu, J. (2013). Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 74, 840–851. doi: 10.1111/tpj.12169
Gul, Z., Barozai, M. Y. K., and Din, M. (2017). In-silico based identification and functional analyses of miRNAs and their targets in cowpea (Vigna unguiculata L.). AIMS Genet. 4, 138–165. doi: 10.3934/genet.2017.2.138
He, X., Guo, S., Wang, Y., Wang, L., Shu, S., and Sun, J. (2020). Systematic identification and analysis of heat-stress-responsive lncRNAs, circRNAs and miRNAs with associated co-expression and ceRNA networks in cucumber (Cucumis sativus L.). Physiol. Plantarum 168, 736–754. doi: 10.1111/ppl.12997
Hodges, D. M., DeLong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1007/s004250050524
Ji, F., Wei, S., Liu, N., Xu, L., and Yang, P. (2020). Growth of cucumber seedlings in different varieties as affected by light environment. Int. J. Agric. Biol. Eng. 13, 73–78. doi: 10.25165/j.ijabe.20201305.5566
Jian, S., Luo, J., Liao, Q., Liu, Q., Guan, C., and Zhang, Z. (2019). NRT1.1 regulates nitrate allocation and cadmium tolerance in Arabidopsis. Front. Plant Sci. 10:384. doi: 10.3389/fpls.2019.00384
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18, 927–937. doi: 10.1016/j.devcel.2010.05.008
Li, S., Liu, J., Liu, Z., Li, X., Wu, F., and He, Y. (2014). Heat-induced tas1 target1 mediates thermotolerance via heat stress transcription factor A1a-directed pathways in Arabidopsis. Plant Cell 26, 1764–1780. doi: 10.1105/tpc.114.124883
Li, Z., Tang, J., Srivastava, R., Bassham, D. C., and Howell, S. H. (2020). The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell 32, 3559–3575. doi: 10.1105/tpc.20.00260
Li, H., Wang, Y., Wang, Z., Guo, X., Wang, F., Xia, X. J., et al. (2016). Microarray and genetic analysis reveals that csa-miR159b plays a critical role in abscisic acid-mediated heat tolerance in grafted cucumber plants. Plant Cell Environ. 39, 1790–1804. doi: 10.1111/pce.12745
Liu, W., Chen, G., Chen, J., Jahan, M. S., Guo, S., Wang, Y., et al. (2021). Overexpression of 7-hydroxymethyl chlorophyll a reductase from cucumber in tobacco accelerates dark-induced chlorophyll degradation. Plan. Theory 10:1820. doi: 10.3390/plants10091820
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lv, X., Zhang, Y., Hu, L., Zhang, Y., Zhang, B., Xia, H., et al. (2021). Low-nitrogen stress stimulates lateral root initiation and nitrogen assimilation in wheat: roles of phytohormone signaling. J. Plant Growth Regul. 40, 436–450. doi: 10.1007/s00344-020-10112-5
Ma, H., Liu, C., Li, Z., Ran, Q., Xie, G., Wang, B., et al. (2018). ZmbZIP4 contributes to stress resistance in maize by regulating ABA synthesis and root development. Plant Physiol. 178, 753–770. doi: 10.1104/pp.18.00436
Maghiaoui, A., Bouguyon, E., Cuesta, C., Perrine-Walker, F., Alcon, C., Krouk, G., et al. (2020). The Arabidopsis NRT1.1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 71, 4480–4494. doi: 10.1093/jxb/eraa242
Mangrauthia, S. K., Bhogireddy, S., Agarwal, S., Prasanth, V. V., Voleti, S. R., Neelamraju, S., et al. (2017). Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 68, 2399–2412. doi: 10.1093/jxb/erx111
May, P., Liao, W., Wu, Y., Shuai, B., McCombie, W. R., Zhang, M. Q., et al. (2013). The effects of carbon dioxide and temperature on microRNA expression in Arabidopsis development. Nat. Commun. 4:2145. doi: 10.1038/ncomms3145
Mei, Y., Wang, Y., Li, F., and Zhou, X. (2020). The C4 protein encoded by stomato leaf curl Yunnan virus reverses transcriptional gene silencing by interacting with NbDRM2 and impairing its DNA-binding ability. PLoS Pathog. 16:e1008829. doi: 10.1371/journal.ppat.1008829
Pellizzaro, A., Clochard, T., Cukier, C., Bourdin, C., Juchaux, M., Montrichard, F., et al. (2014). The nitrate transporter MtNPF6.8 (MtNRT1.3) transports abscisic acid and mediates nitrate regulation of primary root growth in Medicago truncatula. Plant Physiol. 166, 2152–2165. doi: 10.1104/pp.114.250811
Rao, S., Das, J. R., Balyan, S., Verma, R., and Mathur, S. (2022). Cultivar-biased regulation of HSFA7 and HSFB4a govern high-temperature tolerance in tomato. Planta 255:31. doi: 10.1007/s00425-021-03813-y
Ravichandran, S., Ragupathy, R., Edwards, T., Domaratzki, M., and Cloutier, S. (2019). MicroRNA-guided regulation of heat stress response in wheat. BMC Genomics 20:488. doi: 10.1186/s12864-019-5799-6
Sailaja, B., Anjum, N., Prasanth, V. V., Sarla, N., Subrahmanyam, D., Voleti, S. R., et al. (2014). Comparative study of susceptible and tolerant genotype reveals efficient recovery and root system contributes to heat stress tolerance in rice. Plant Mol. Biol. Rep. 32, 1228–1240. doi: 10.1007/s11105-014-0728-y
Saiz-Fernández, I., Lacuesta, M., Pérez-López, U., Sampedro, M. C., Barrio, R. J., and De Diego, N. (2020). Interplay between 1-aminocyclopropane-1-carboxylic acid, γ-aminobutyrate and D-glucose in the regulation of high nitrate-induced root growth inhibition in maize. Plant Sci. 293:110418. doi: 10.1016/j.plantsci.2020.110418
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Baeurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Su, Y., Huang, Y., Dong, X., Wang, R., Tang, M., Cai, J., et al. (2021). Exogenous methyl jasmonate improves heat tolerance of perennial ryegrass through alteration of osmotic adjustment, antioxidant defense, and expression of jasmonic acid-responsive genes. Front. Plant Sci. 12:664519. doi: 10.3389/fpls.2021.664519
Sun, Y., Qiu, Y., Duan, M., Wang, J., Zhang, X., Wang, H., et al. (2017). Identification of anthocyanin biosynthesis related microRNAs in a distinctive Chinese radish (Raphanus sativus L.) by high-throughput sequencing. Mol. Gen. Genomics 292, 215–229. doi: 10.1007/s00438-016-1268-y
Tian, X., Wang, F., Zhao, Y., Lan, T., Yu, K., Zhang, L., et al. (2020). Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnol. J. 18, 1109–1111. doi: 10.1111/pbi.13268
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Wang, W. Q., Allan, A. C., and Yin, X. R. (2020b). Small RNAs with a big impact on horticultural traits. Crit. Rev. Plant Sci. 39, 30–43. doi: 10.1080/07352689.2020.1741923
Wang, Y., Cao, J. J., Wang, K. X., Xia, X. J., Shi, K., Zhou, Y. H., et al. (2019). BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 179, 671–685. doi: 10.1104/pp.18.01028
Wang, Y., Gong, X., Liu, W., Kong, L., Si, X., Guo, S., et al. (2020c). Gibberellin mediates spermidine-induced salt tolerance and the expression of GT-3b in cucumber. Plant Physiol. Biochem. 152, 147–156. doi: 10.1016/j.plaphy.2020.04.041
Wang, Y., Guo, S., Wang, L., Wang, L., He, X., Shu, S., et al. (2018b). Identification of microRNAs associated with the exogenous spermidine-mediated improvement of high-temperature tolerance in cucumber seedlings (Cucumis sativus L.). BMC Genomics 19:285. doi: 10.1186/s12864-018-4678-x
Wang, M., He, X., Peng, Q., Liang, Z., Peng, Q., Liu, W., et al. (2020a). Understanding the heat resistance of cucumber through leaf transcriptomics. Funct. Plant Biol. 47, 704–715. doi: 10.1071/FP19209
Wang, L., Zhou, H., Guo, S., An, Y., Shu, S., Lu, N., et al. (2018a). Exogenous spermidine maintains the chloroplast structure of cucumber seedlings and inhibits the degradation of photosynthetic protein complexes under high-temperature stress. Acta Physiol. Plant. 40:47. doi: 10.1007/s11738-018-2624-9
Wei, Y., Wang, Y., Wu, X., Shu, S., Sun, J., and Guo, S. (2019). Redox and thylakoid membrane proteomic analysis reveals the Momordica (Momordica charantia L.) rootstock-induced photoprotection of cucumber leaves under short-term heat stress. Plant Physiol. Biochem. 136, 98–108. doi: 10.1016/j.plaphy.2019.01.010
Wu, H. J., Ma, Y. K., Chen, T., Wang, M., and Wang, X. J. (2012). PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res. 40, W22–W28. doi: 10.1093/nar/gks554
Xia, X. J., Zhou, Y. H., Shi, K., Zhou, J., Foyer, C. H., and Yu, J. Q. (2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 66, 2839–2856. doi: 10.1093/jxb/erv089
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, X., Liu, M., Lu, L., He, M., Qu, W., Xu, Q., et al. (2015). Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Gen. Genomics 290, 1403–1414. doi: 10.1007/s00438-015-1002-1
Yang, K., An, J. P., Li, C. Y., Shen, X. N., Liu, Y. J., Wang, D. R., et al. (2021). The apple C2H2-type zinc finger transcription factor MdZAT10 positively regulates JA-induced leaf senescence by interacting with MdBT2. Hortic. Res. 8:159. doi: 10.1038/s41438-021-00593-0
Yao, S., Yang, Z., Yang, R., Huang, Y., Guo, G., Kong, X., et al. (2019). Transcriptional regulation of miR528 by OsSPL9 orchestrates antiviral response in rice. Mol. Plant 12, 1114–1122. doi: 10.1016/j.molp.2019.04.010
Yu, B., Yan, S., Zhou, H., Dong, R., Lei, J., Chen, C., et al. (2018). Overexpression of CsCaM3 improves high temperature tolerance in cucumber. Front. Plant Sci. 9:797. doi: 10.3389/fpls.2018.00797
Zhang, X., Cui, Y., Yu, M., Su, B., Gong, W., Baluska, F., et al. (2019). Phosphorylation-mediated dynamics of nitrate transceptor NRT1.1 regulate auxin flux and nitrate signaling in lateral root growth. Plant Physiol. 181, 480–498. doi: 10.1104/pp.19.00346
Zhang, Y., Wang, Y., Wen, W., Shi, Z., Gu, Q., Ahammed, G. J., et al. (2021). Hydrogen peroxide mediates spermidine-induced autophagy to alleviate salt stress in cucumber. Autophagy 17, 2876–2890. doi: 10.1080/15548627.2020.1847797
Zhao, J., He, Q., Chen, G., Wang, L., and Jin, B. (2016). Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 7:1213. doi: 10.3389/fpls.2016.01213
Zhao, Y. F., Peng, T., Sun, H. Z., Teotia, S., Wen, H. L., Du, Y. X., et al. (2019). miR1432-OsACOT (acyl-CoA thioesterase) module determines grain yield via enhancing grain filling rate in rice. Plant Biotechnol. J. 17, 712–723. doi: 10.1111/pbi.13009
Zhou, H., Guo, S., An, Y., Shan, X., Wang, Y., Shu, S., et al. (2016). Exogenous spermidine delays chlorophyll metabolism in cucumber leaves (Cucumis sativus L.) under high temperature stress. Acta Physiol. Plant. 38:224. doi: 10.1007/s11738-016-2243-2
Keywords: cucumber, high temperature stress, miR9748, jasmonic acid, CsbZIP2, CsNPF4.4
Citation: Li L, Chen G, Yuan M, Guo S, Wang Y and Sun J (2022) CsbZIP2-miR9748-CsNPF4.4 Module Mediates High Temperature Tolerance of Cucumber Through Jasmonic Acid Pathway. Front. Plant Sci. 13:883876. doi: 10.3389/fpls.2022.883876
Edited by:
Monika Chodasiewicz, King Abdullah University of Science and Technology, Saudi ArabiaReviewed by:
Chaoxing He, Institute of Vegetables and Flowers (CAAS), ChinaXueyi Xue, University of Illinois at Urbana–Champaign, United States
Copyright © 2022 Li, Chen, Yuan, Guo, Wang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu Wang, eXdhbmdAbmphdS5lZHUuY24=; Jin Sun, amluc3VuQG5qYXUuZWR1LmNu
 Lan Li
Lan Li Guangling Chen
Guangling Chen Mingzhu Yuan
Mingzhu Yuan Shirong Guo
Shirong Guo Yu Wang
Yu Wang Jin Sun
Jin Sun