
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 April 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.882946
This article is part of the Research Topic Flowering Time Control in Agricultural and Horticultural Crops View all 10 articles
Gossypium hirsutum is most broadly cultivated in the world due to its broader adaptation to the environment and successful breeding of early maturity varieties. However, how cotton responds to environmental cues to adjust flowering time to achieve reproductive success is largely unknown. SOC1 functions as an essential integrator for the endogenous and exogenous signals to maximize reproduction. Thus we identified six SOC1-like genes in Gossypium that clustered into two groups. GhSOC1-1 contained a large intron and clustered with monocot SOC1s, while GhSOC1-2/3 were close to dicot SOC1s. GhSOC1s expression gradually increased during seedling development suggesting their conserved function in promoting flowering, which was supported by the early flowering phenotype of 35S:GhSOC1-1 Arabidopsis lines and the delayed flowering of cotton silencing lines. Furthermore, GhSOC1-1 responded to short-day and high temperature conditions, while GhSOC1-2 responded to long-day conditions. GhSOC1-3 might function to promote flowering in response to low temperature and cold. Taken together, our results demonstrate that GhSOC1s respond differently to light and temperature and act cooperatively to activate GhLFY expression to promote floral transition and enlighten us in cotton adaptation to environment that is helpful in improvement of cotton maturity.
Cotton is an important cash crop that produce the natural textile fiber, which supports the life of an estimated 150 million people (Amrouk et al., 2021). The upland cotton (Gossypium hirsutum L. AADD, 2n = 52) is broadestly cultivated to provide the world major cotton fibers. Cotton origins from subtropics. Consistent to the unnecessary adaption to winter, the main regulator in vernalization, FLC (FLOWERING LOCUS C), was absent in the Gossypium genomes (Nardeli et al., 2018). It evolved with a broader adaptation to the environment and the sensitiveness to short-day condition is lost during domestication (Hao et al., 2008; Ren et al., 2017), which largely extent planting area of cotton. Epigenome and GWAS analysis have identified genetic and epigenetic changes contributing to loss sensitivity of upland cotton to short-day photoperiod (Song et al., 2017; Li et al., 2021b). However, the knowledge for cotton flowering time control is deficient. The question how cotton response to environment to initiate floral transition at the appropriate time remains unknown.
Floral transition divides plant growth into the vegetative and reproductive stages. Plant only grows leaves, stems and roots in the vegetative growth until floral transition when the reproductive organs initiate. In Arabidopsis, CONSTANS (CO) acts as the central regulator of photoperiod pathway. Overexpression of CO causes early flowering phenotype, which could be rescued by mutation in a gene named SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1). SOC1 integrates the signals from photoperiod, age, temperature and GA to initiate the differentiation of flower bud, which is essential for plants to flowering at the appropriate time to achieve the maximal reproductive success (Lee and Lee, 2010). SOC1 transcription can be activated by effectors of photoperiod and GA signaling (Yoo et al., 2005), and is suppressed by SHORT VEGETATIVE PHASE (SVP) that responses to the autonomous and GA pathway to avoid low propagation resulting from early flowering (Turck et al., 2008). The SOC1-SPL model governs the signals from photoperiod and GA to control flowering (Jung et al., 2012). During vernalization, FLC is fine tuned to directly inhibit SOC1 expression, thus the plants can overcome the cold winter (Li et al., 2008). In the shoot apical meristem (SAM), SOC1 and AGL24 abundance is maintained by positivist feedback loop to ensure the entrance of SOC1 into nucleus with the help of AGL24, where the SOC1/AGL24 complex directly activates LFY transcription (Lee et al., 2008). Furthermore, SOC1 binds loci of a large number of flowering time regulators including the majority of its own repressors to establish the double-negative feedback loop (Immink et al., 2012). It is more likely that SOC1 situates in the center of the flowering regulatory network containing multiple regulatory and feedback loops that serves as a molecular switch of floral transition.
In Arabidopsis, soc1 mutant flowers late, while overexpression of SOC1 accelerates flowering (Borner et al., 2008; Lee and Lee, 2010). SOC1-like genes in other species also function as flowering time promoters, such as orchard DoSOC1 (Ding et al., 2013) and peony PsSOC1 (Wang et al., 2015). Ectopic expression of the K domain of blue berry VcSOC1 gene also promotes flowering of tobacco, suggesting that the K-box is the functional domain of VcSOC1 (Song et al., 2013). Although the function of SOC1 homologs is conserved, there are evidence supporting functional divergence of SOC1-like genes. FaSOC1 from cultivated strawberry promotes flowering, while FvSOC1 from wild strawberry suppresses flowering to maintain the vegetative growth (Lei et al., 2013; Mouhu et al., 2013). Ectopic expression of the SOC1 homolog from Gerbra hybrida cannot affect flowering time, but cause defects of floral organs (Ruokolainen et al., 2011). Functional and transcriptional analysis of SOC1-like genes in Actinidia chinensis suggest that they are involved in seed dormancy and may evolved to loss the function of flowering time control (Voogd et al., 2015). SOC1 also responds to photoperiod, draft, cold and high temperature to regulate growth, stomatal opening and chloroplast biogenesis (Ryu et al., 2009; Richter et al., 2013; Kimura et al., 2015; Wang et al., 2019).
Considering the comprehensive response of SOC1 to endogenous and exogenous signals, it is imperative to identify the function of SOC1 homologs in Gossypium. In previous study, GhSOC1 was cloned from CCRI36, which could promote flowering of Arabidopsis when overexpressed. But overexpression of GhSOC1 in cotton only affected floral organ development rather than flowering time (Zhang et al., 2016). Therefore, the clarification of functions of different SOC1-like genes is vital for understanding the regulatory mechanism of cotton flowering time control. In this study, SOC1-like genes were identified in Gossypium genomes and their evolution were analyzed. Then the expression characters of different GhSOC1s were investigation and GhSOC1-1 function was further investigated by overexpression and silencing. The results showed that cotton SOC1-like genes evolved divergently to respond differently to light and temperature and promote flowering in a cooperative way, which indicates genomic changes among GhSOC1 locus relating to adaptation. Our findings draw a dynamic regulatory model participated by homologies in tetraploid cotton, which help us to understand the mechanism of cotton flowering time control in response to the environment.
The protein sequences of different versions in diploid and tetraploid cotton were downloaded from CottonGen.1 SOC1 homologs were obtained through tBLSATp searches using AtSOC1 (AT2G45660) as query against the diploid and tetraploid genomes, respectively. The conserved MADS and K-box domains were further confirmed by NCBI CD-Search.2 The identified cotton SOC1 homologs were listed in Supplementary Table 1.
To construct the phylogenetic tree, we aligned full-length protein sequences of all cotton SOC1-like genes together with the other eighteen species that were listed in Supplementary Table 2 and used MEGA 7.0 to construct the phylogenetic tree (Neighbor join, Bootstrap 1,000 and 50% cutoff values) (Kumar et al., 2016). DNAMAN was applied for alignment.
The gene structure was analyzed on GSDS.3 WebLogo3 was applied online4 to build weblogo diagram. The three-dimensional structure of proteins was predicted on the website SWISS-MODEL.5 The cis-elements were predicted on Plant CARE.6 The CarG-box elements were searched by tbtools software and visualized according to the position. Transcriptome data of floral organs was downloaded from the NCBI SRA (Sequence Read Archive) database (Genome sequencing project accession: PRJNA248163). The FPKM of each GhSOC1 gene was normalized to log2 (1+FPKM), and expression heat maps were drawn using TBtools software.
G. hirsutum L. TM-1 were grown in the green house (28°C, 16 h light/8 h dark). The roots, stems and true leaves were collected, respectively, when the first true leaf flattened, and the flowers and fibers were sampled at 0 and 10 days post anthesis, respectively. These samples were used for tissue specific expression analysis. The aerial parts of true leaves were collected when the third, fourth and fifth true leaf flattened, respectively, for temporal expression analysis. When the cotton seedlings grew to the third true leaf stage under long-day (LD), they were shifted to short-day (SD, 28°C), high temperature (35°C) and low temperature (18°C) conditions and the whole seedlings were collected after 48 h for quick response analysis. Upland cotton TM-1 was continuously cultivated under different day length (28°C) and temperatures (18, 22, 28, 35°C under LD) and the whole seedling were sampled at the third and fifth true-leaf stages for expression comparison.
Arabidopsis plants were grown under 23°C, 16 h light/8 h dark. The rosette leaves of the T1 transgenic lines were collected for DNA extraction followed by genotyping with gene specific primers. The seedlings of T3 homozygous lines were collected 9 days after germination for expression analysis.
All the samples were frozen in liquid nitrogen and save under −80°C before RNA extraction.
The coding region of GhSOC1-1 without stop codon was cloned from G. hirsutum TM-1 using gene-specific primers listed in Supplementary Table 3 and fused with GFP driven by 35S promoter. The resulting vector was transformed into Agrobacterium by electroporation. The Agrobacterium harboring vector 35S:GhSOC1-1-GFP were infiltrated into tobacco leaves. And the GFP signal was observed under confocal microscope Olympus FV1000.
The full length CDS were cloned and constructed to vector pGreen0229 with 35S promoter. The resulting construct 35S:GhSOC1-1 was subsequently transformed into wild type Arabidopsis plants via Agrobacterium mediated transformation using floral dip method. The transgenic plants were selected with basta for positive lines which were verified by PCR examination on genome DNA with primers 35Spro and pgp2 (Supplementary Table 3). The expression of transgene was examined in the leaves of positive lines. The flowering phenotype was observed among at least 25 individuals of each line.
For the VIGS assay, specific primers were designed to clone the 404 bp fragment of GhSOC1-1 including 340 bp C-termina and 64 bp 3′ UTR into the PTRV2 vector. Then, Agrobacterium carrying the plasmids of PTRV2:GhSOC1-1 and PTRV1, respectively, were co-infiltrated into cotyledons of NDM8 after sowing for 7 days. TRV:CLA1 and TRV:00 were utilized as positive and negative control, respectively. The leaves were collected from each GhSOC1-1 silencing lines to detect silence efficiency when albinism was obvious on the positive control. The flowering time of silencing lines were record when the first square appeared.
The collected samples were grinded for total RNA extracted with RNA prep Pure Plant Kit (TIANGEN), followed by synthesis of cDNA using PrimeScript™ 1st strand cDNA Synthesis kit (Solarbio). Gene transcription was detected with AugeGreen qPCR Master Mix (US Everbright) on an ABI Q5 machine. GhHIS and AtTUB2 were set as the internal control. Gene specific primers for qPCR were listed in Supplementary Table 3. The relative expression level was calculated using the 2–△CT formular (Livak and Schmittgen, 2001). There biological repeats were applied on each sample and three technical repeats were performed on each reaction.
The protein sequence of AtSOC1 (AT2G45660) was obtained from TAIR7 and blast against the cotton genomes on Cotton-FGD.8 After CD-search for the conserved domains, six SOC1-like genes were confirmed in G. hirsutum considering the most recent genome of NDM8 and other five genomes of TM-1, which were named according to their chromosome location (Supplementary Table 1). The sequence similarity of homologs located in the A and D subgenomes was up to 95% (Supplementary Table 5), suggesting their redundant function between A and D subgenome. The length of GhSOC1 proteins varied little with only 23 amino acid difference, resulting in similar molecular weight (23.200–25.713 kDa). The isoelectric point of GhSOC1-3D was lower than the other GhSOC1s that showed similar pI within 9.01–9.33, which was related to the sequence variation (Supplementary Table 4). GhSOC1-2A and GhSOC1-2D were highly conserved with the published GhSOC1 gene (Zhang et al., 2016).
Gene structure analysis of SOC1 homologs showed that they were composed of seven exons and six introns. The length of Exon 1, 4, 5 and 6 were highly conserved and varied within Exon 7. The variations of intron length contributed to the gene length (Figure 1A). Noticeably, GhSOC1-1 contained a large Intron 1, which distinguished from AtSOC1 and other GhSOC1s. Then the MADS and K-box domains were compared in detail. GhSOC1s contained highly conserved MADS-box domain and less conserved K-box domain, and diverse C terminal including a typical SOC1-motif (Figure 1B).
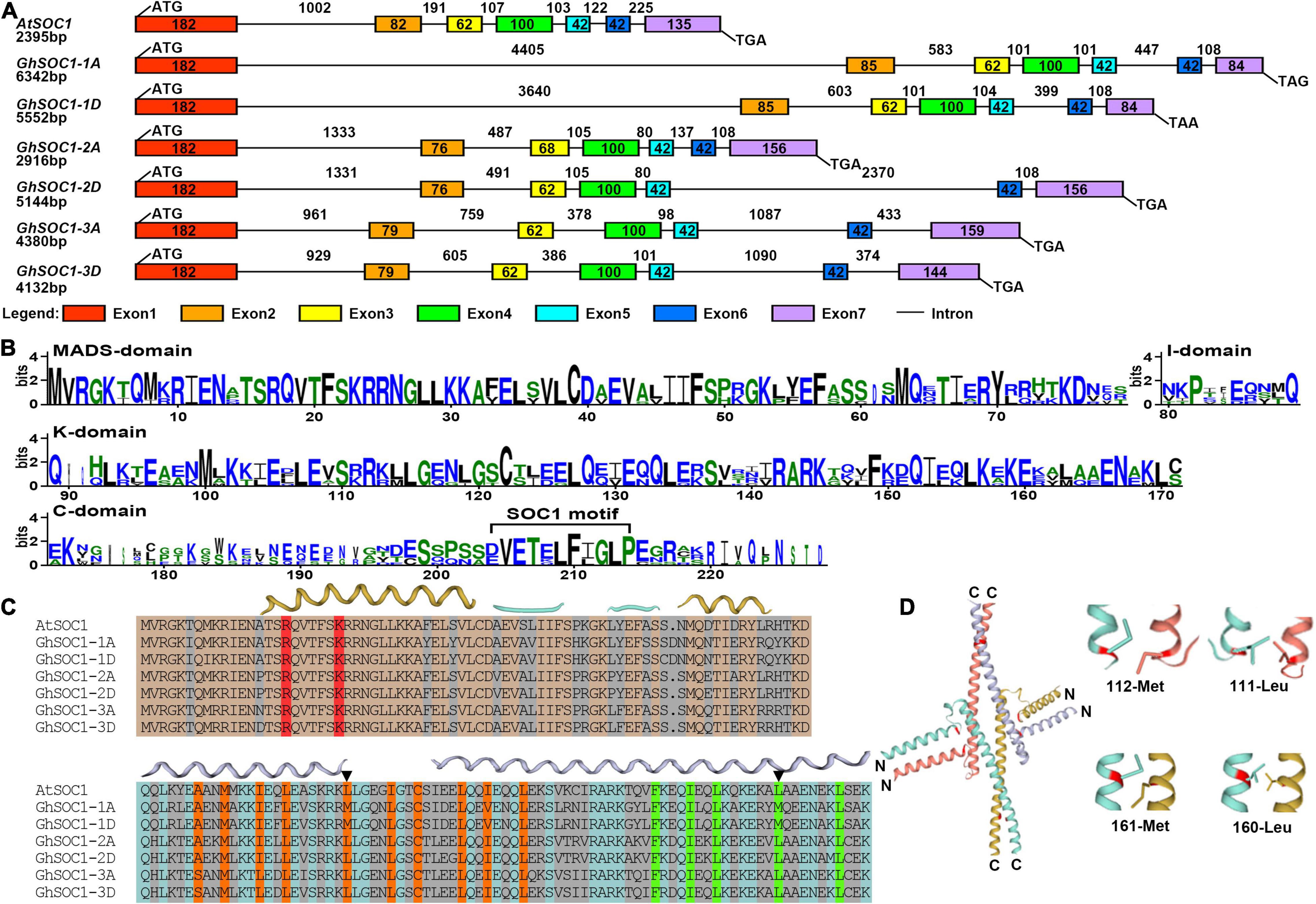
Figure 1. Sequence analysis of GhSOC1s. (A) Gene structure of GhSOC1s. (B) Sequence conservation of the M, I, K, C domain of GhSOC1s. (C) Structure prediction of GhSOC1s. The interface of the MADS domain with DNA groove are marked in red. The orange color highlights the important residues for dimerization and the green color highlights the important residues for tetramerization. Triangles mark the varied Leucine residues. (D) 3-D structure of the K domain. The varied Leucine residues of GhSOC1-1 are highlighted in red. Close-up display the varied interacting residues depicted as sticks.
Oligomerization is common for MADS transcription factors to recognize different targets. The K domain is critical for dimerization and tetramerization capabilities. The K domain contains two helices that helix 1 stabilized the tetramer facilitated by helix 2 (Puranik et al., 2014). We then predicted the structure of GhSOC1s and found that the MADS domain interface with DNA groove were residue Arginine and Lysine that were conserved among GhSOC1s (Figure 1C). It has been reported that the Leucine residues in the K domain are essential for co-operative DNA binding of MADS proteins (Rumpler et al., 2018). Two variations of the Leucine were identified in GhSOC1-1 (Figure 1C). The variation 112-met was at the end of Helix 1 that might affect dimerization. The variation 161-met comprised the tetramerization interface (Figure 1D). The variations of Leucine indicated the different binding sites or binding affinity of GhSOC1-1.
In 2020, it was reported that G. hirsutum might be originated from the genome polyploidization of G. raimondi and A0 (Huang et al., 2020). To study the evolution of SOC1-like genes, we further identified SOC1 homologies in G. raimondii, G. arboreum and the other tetraploid cotton G. barbadense. Then other SOC1-like genes were obtained from dicots (Arabidopsis thaliana, Actinidia chinensis, Lactuca sativa, Vitis vinifera, Fragaria vesca, Fragaria × ananassa, Medicago truncatula, Glycine max, Orchidaceae, Theobroma cacao, Brassica rapa, Nelumbo nucifera) and monocots (Oryza sativa, Sorghum bicolor, Zea mays, Musa nana Lour., Triticum aestivum, Saccharum spontaneum) (Supplementary Table 2). The sequences of the homologies were analyzed in detail using neighbor-joint method to build an unrooted phylogenetic tree (Figure 2).
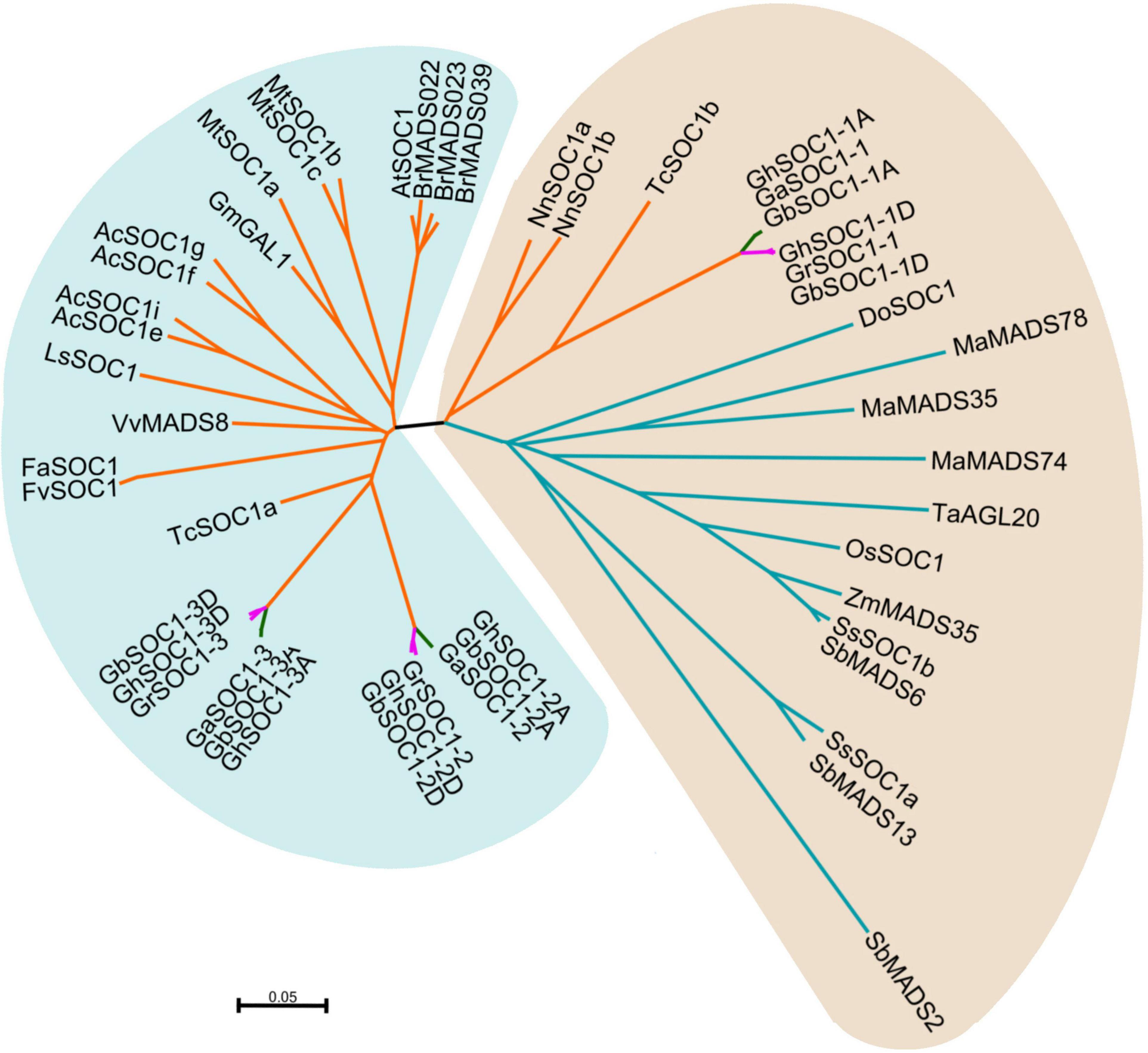
Figure 2. Phylogenetic analysis of SOC1-like genes in cotton and other plants. SsSOC1a and SsSOC1b in Saccharum spontaneum was download from the article (Fatima et al., 2020). Other SOC1 sequences were downloaded from NCBI and their accession numbers are given in Supplementary Table 2.
Phylogenetic analysis showed that cotton SOC1-like genes were more conserved within the A genomes, but evolved differently between D genomes. The phylogenetic tree was mainly clustered into two groups. Cotton SOC1-2 and SOC1-3 genes were clustered with other SOC1-like genes in dicots. However, cotton SOC1-1 genes together with monocot SOC1 genes formed a monophyletic group (Figure 2). Lotus is an ancient dicot, whose SOC1-like genes were also clustered with the monocot SOC1 genes. Cacao is considered as an evolutionary close specie with Gossypium, whose genome contained two SOC1-like genes that one was conserved with monocot SOC1 genes and the other evolved with dicot SOC1 genes. These findings suggested that Gossypium genomes retained the conserved ancient SOC1 gene and evolved other SOC1-like genes with dicots.
Cis-elements are the binding sites for transcription regulators. We predicted the cis-elements on the 2 kb upstream sequences of GhSOC1s. The promoters of GhSOC1s contained plenty of light responsive elements, including 3-AF1 binding site, ABRE, AE-box, AT1-motif, ATCT-motif, Box 4, chs-CMA1a, GATA-motif, G-box, GT1-motif, I-box, MRE and CT-motif. The GhSOC1-3D promoter harbored unique elements relating to photoperiod (circadian). A low temperature responsive element LTR was found on promoters of GhSOC1s. Salicylic acid and auxin responsive elements were predicted on GhSOC1-1/2/3 promoter region. GhSOC1-1 and GhSOC1-2 were predicted to response to abscisic acid. GhSOC1-2 and GhSOC1-3 might be regulated by gibberellin. And jasmonate responsive elements only existed on GhSOC1-1 promoters. SOC1-like genes are important regulators in plant growth and development which could be disturbed and interrupted by biotic and abiotic stresses. Although little is known for SOC1-like genes in response to stress, TC-rich repeats that are involved in defense and stress response were identified on GhSOC1-1 and GhSOC1-2 promoter.
Since the GhSOC1-1 genome comprised a large Intron 1, we further compared the cis-elements located in this region. The results showed that light responsive elements were all predicted in Intron 1 of GhSOC1s, but GhSOC1-1 possessed more abundant regulatory elements in Intron 1 (Figure 3). This region of GhSOC1-2 included JA and drought regulatory elements, whereas ABA, SA, GA and low-temperature responsive elements were also found in Intron 1 of GhSOC1-1. The large number of cis-elements indicated regulatory importance of Intron 1 in GhSOC1-1 genome.
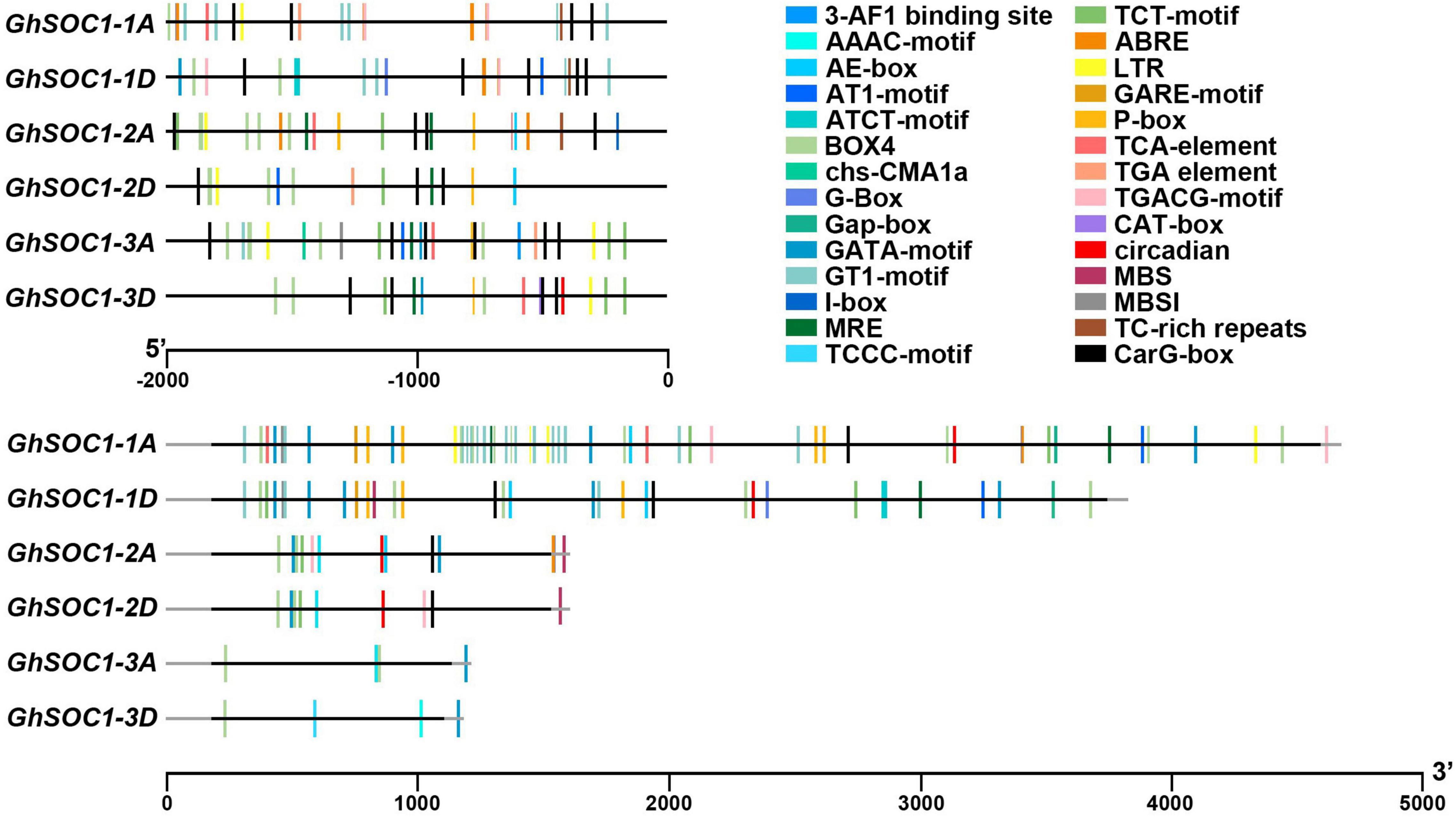
Figure 3. Prediction of cis-acting elements in GhSOC1s genome. The coding region and non-coding region are presented in gray and black lines.
The diversity of cis-elements on genome region of GhSOC1s suggested differences in transcription and regulatory mechanism. The existence of abundant light related elements indicated a regulation of GhSOC1s by light effectors, which might contribute to the loss of light sensitivity of upland cotton during domestication.
Expression of GhSOC1s were examined in roots, stems, leaves, flower and fiber. GhSOC1s expressed ubiquitously in different organs (Figure 4A). GhSOC1-1 transcripts were most abundant in all the examined tissues (Figures 4A,B). GhSOC1-3 was transcribed less abundantly with relatively high expression in the vegetative organs. The expression of GhSOC1-1 and GhSOC1-2 was higher in flowers. Therefore, GhSOC1s transcriptions were compared between different parts of flowers according to the transcriptome data (Figure 4B). Both GhSOC1-1 and GhSOC1-2 were expressed higher in the calyx, and GhSOC1-3 demonstrated an expression preference in torus. GhSOC1-1A also expressed in the petals, stamens and pistils. Then the expression patterns were detected in developing seedlings. The GhSOC1s expression increased gradually during seedling development (Figure 4C). Especially, their expression was upregulated sharply at the fourth true leaf stage, indicating the occurrence of floral transition.
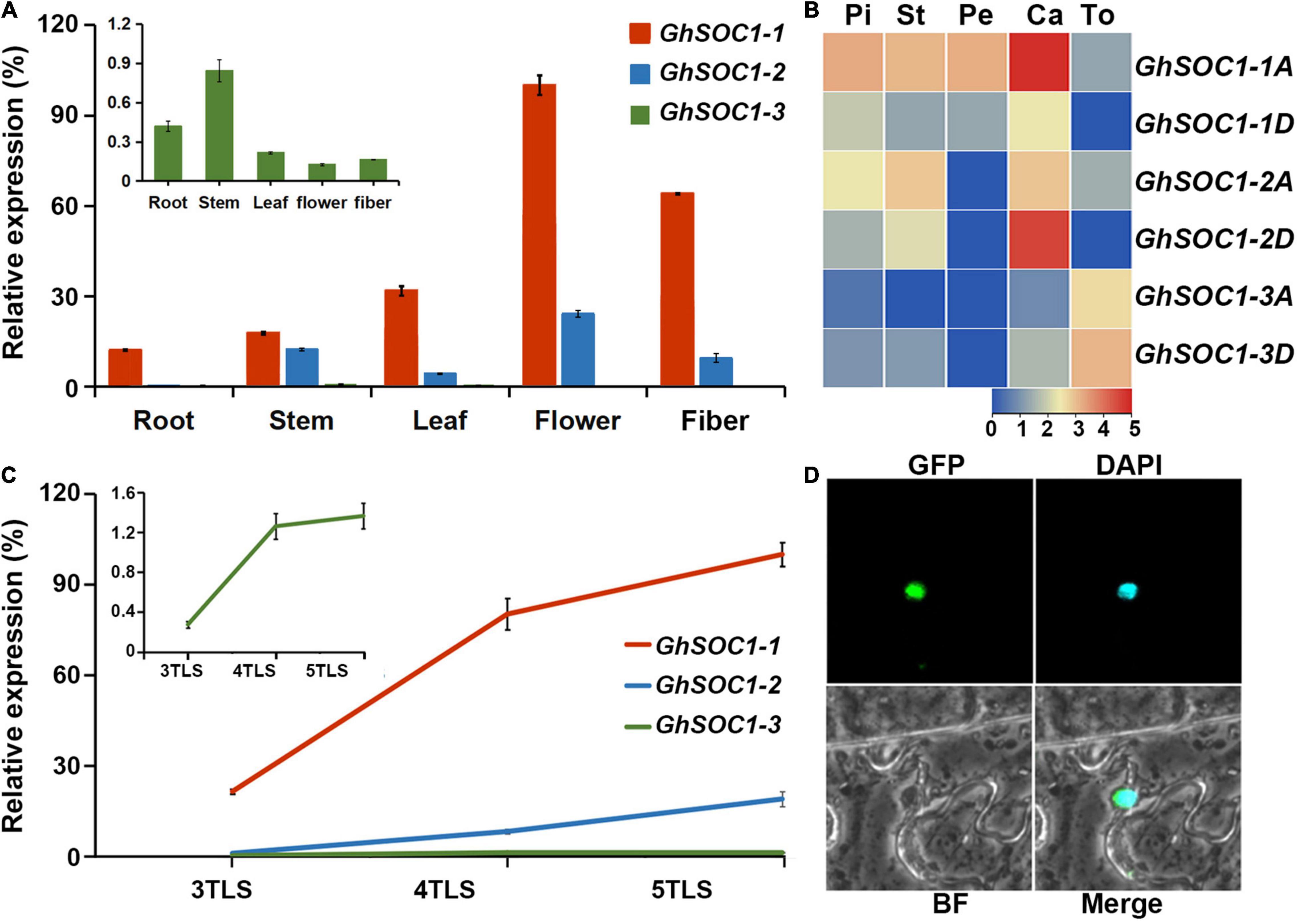
Figure 4. Expression analysis of GhSOC1s. (A) Tissue specific expression of GhSOC1s in G. hirsutum. (B) Expression pattern of GhSOC1s in a flower. Pi, pistil; St, stamen; Pe, petal; Ca; calyx; To, torus. (C) Expression changes of GhSOC1s during seedling development. 3TLS, 4TLS, and 5TLS demonstrates the third, fourth and fifth true leaf stages, respectively. The expression is normalized to GhHIS and the maximum expression is set as 100%. Error bars represent the standard deviations of three biological replicates. (D) Subcellular localization of GhSOC1-1 in tobacco leaves. GFP, GFP fluorescence; DAPI, fluorescence of 40,6-diamino-2-phenylindole; BF, Bright field; Merge, merge of GFP, DAPI, and BF images.
Further, the coding region of GhSOC1-1 was cloned from TM-1 because it was clustered in different branch from other GhSOC1s in the phylogenetic tree and its transcript was most abundant. The amplification displayed the same sequence as GhSOC1-1 in genome of NDM8 (Ma et al., 2021). The coding region was subsequently ligated with GFP sequence to construct GhSOC1-1-GFP fusion protein. The fluorescence was observed in the nucleus of the tobacco epidermal cells (Figure 4D), which distinguished from AtSOC1 that only localized in the nuclear with existence of AtAGL24 (Lee et al., 2008). The nuclear localization of GhSOC1-1 supported its role as a transcription factor.
In order to study the function of GhSOC1-1, 35S:GhSOC1-1 vector was constructed, and transformed into Arabidopsis wild-type plants. The positive lines were confirmed by amplification of the exogenous gene from genome DNA (Figure 5A). Finally, twenty-nine individual transgenic lines were obtained that displayed consistent early flowering under long day condition (Figure 5B). Four transgenic lines were randomly selected for GhSOC1-1 expression detection. The expression levels related with the number of rosette leaves (Figure 5C), suggesting that GhSOC1-1 functioned in a dosage-dependent way to promote flowering. The function of GhSOC1-1 in flowering time control was consistent with its temporal expression pattern.
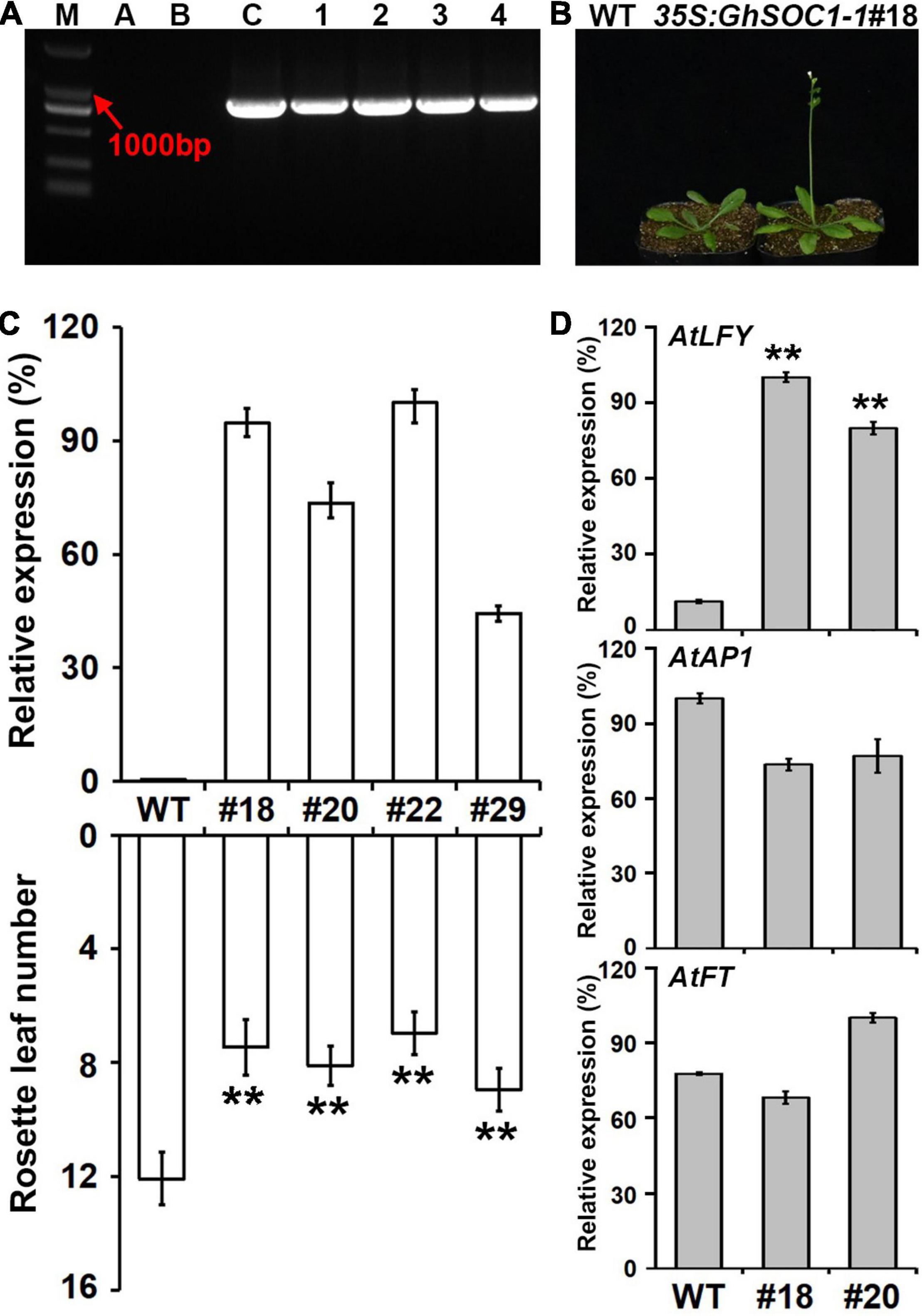
Figure 5. Overexpression of GhSOC1-1 promotes flowering of Arabidopsis. (A) Genotype verification of positive plants in T1 generation. M, DNA molecular weight marker DL 2000; A-C, water control, WT control and positive control; 1-4, PCR detection of positive plants. (B) Phenotype of T3 generation line. (C) Relative expression of GhSOC1-1 and statistics of flowering time in transgenic lines and wild-type Arabidopsis. Asterisks indicate statistically significant differences according to Student’s t-test (**P < 0.01). (D) Expression of AtLFY, AtAP1, and AtFT in transgenic and wild-type Arabidopsis. The expression is normalized to AtTUB2 and the maximum expression is set as 100%. Error bars represent the standard deviations of three biological replicates.
SOC1, FT, and LFY are floral integrator in the network of flowering time control. Therefore, the expression of FT and LFY were detected in overexpression lines to investigate their regulatory relationship. The results showed that overexpression of GhSOC1-1 did not affect FT and AP1 expression, while LFY expression was greatly upregulated (Figure 5D).
To further confirm the function of GhSOC1-1 in cotton, the Virus-induced gene silencing (VIGS) was employed using TRV containing a fragment of the C terminal of GhSOC1-1 and silencing of CLA1 provided a visible reporter of silencing effects. The silence efficiency was detected when the TRV:CLA1 plants demonstrated obvious leaf photobleaching (Figure 6A). GhSOC1-1 expression was only a quarter of that in the control plants infected with empty TRV vectors, meanwhile the expression of other GhSOC1 genes also decreased by nearly half (Figure 6C), resulting from the sequence similarity. The first square appeared on the eighth branch of the control plants, while the square grew out on the tenth branch of the GhSOC1-silenced plants (Figures 6B,D,E). The flowering time was delayed significantly by downregulation of GhSOC1s, suggesting their consistent roles in promoting floral transition. Furthermore, the expression changes of GhFT, GhAP1, and GhLFY were consistent with the results of Arabidopsis that only GhLFY displayed significant decrease (Figure 6F).
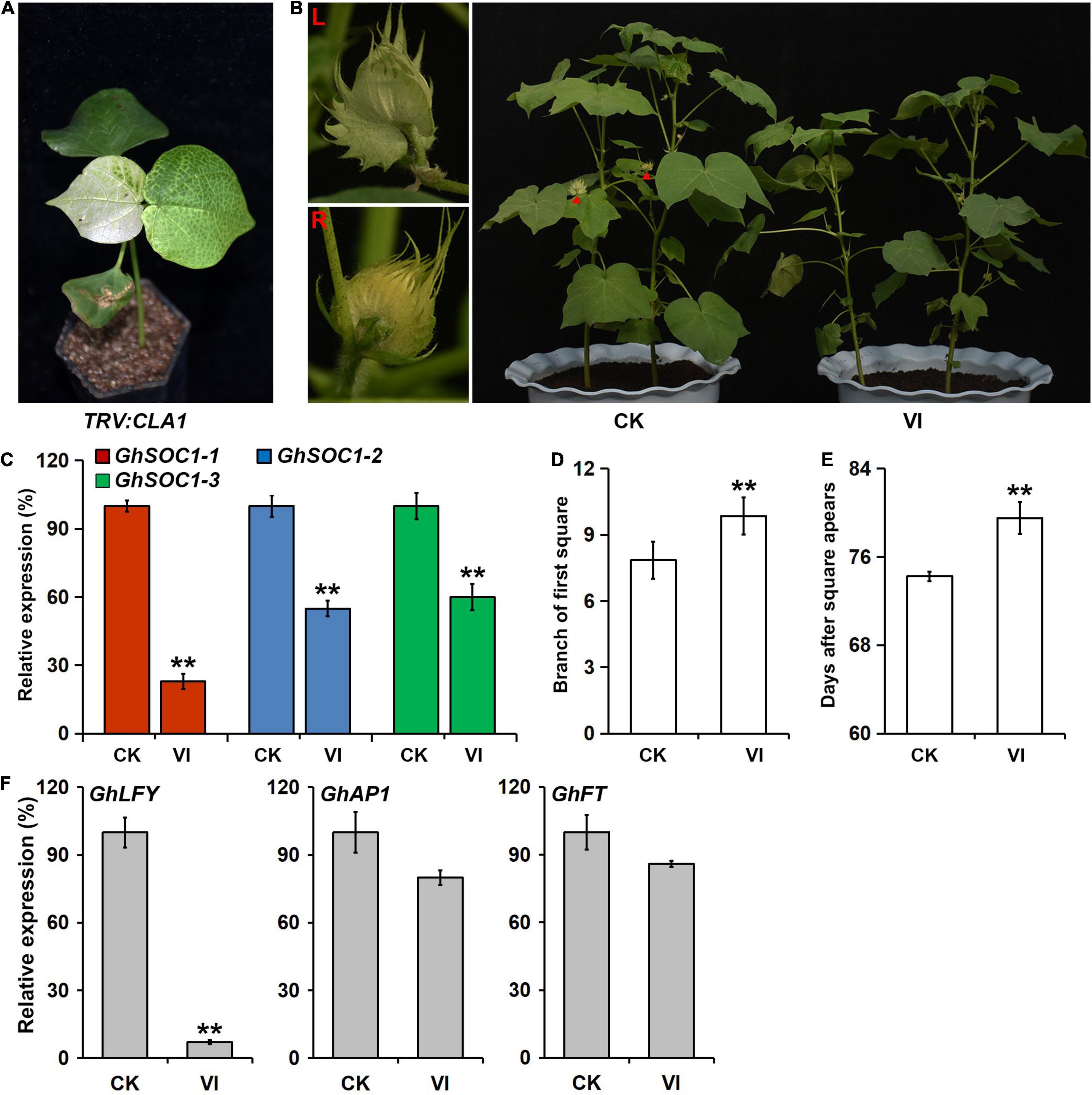
Figure 6. VIGS of GhSOC1s in G. hirsutum delays flowering time. (A) Albino phenotype of TRV:CLA1 as a control. (B) Delayed flowering phenotype of VIGS plants. Red triangles point the flower buds displayed in the left. CK, empty vector control; VI, silence plants. (C) Detection of the efficiency of gene silencing. The expression was normalized to GhHIS and the maximum expression is set as 100%. Error bars represent the standard deviations of three biological replicates. (D,E) Maturity traits of VIGS plants in branch of first square (D) and days from sowing until square appears (E). Asterisks indicate statistically significant differences among 15 individuals according to Student’s t-test (**P < 0.01). (F) Expression of GhLFY, GhAP1, and GhFT in Silencing and control plants. The expression is normalized to GhHIS and the maximum expression is set as 100%. Error bars represent the standard deviations of three biological replicates.
Considering the existence of light and temperature responsive cis-elements in the genome region of GhSOC1s, we examined their transcriptional level under different environmental conditions. Firstly, GhSOC1-3 expression was lowest under long-day condition and GhSOC1-1 transcription was higher than GhSOC1-2 (Figure 4A). When the plants were shifted to short-day for 48 h, their expression remained unchanged (Figure 7A). Differently, GhSOC1-1 and GhSOC1-2 responded to temperature change quickly (48 h), and their response to high temperature were violent (Figure 7B). But GhSOC1-3 didn’t response to temperature change in a short time. These results suggested that GhSOC1-1 and GhSOC1-2 might involve in the ambient temperature pathway to regulate flowering time.
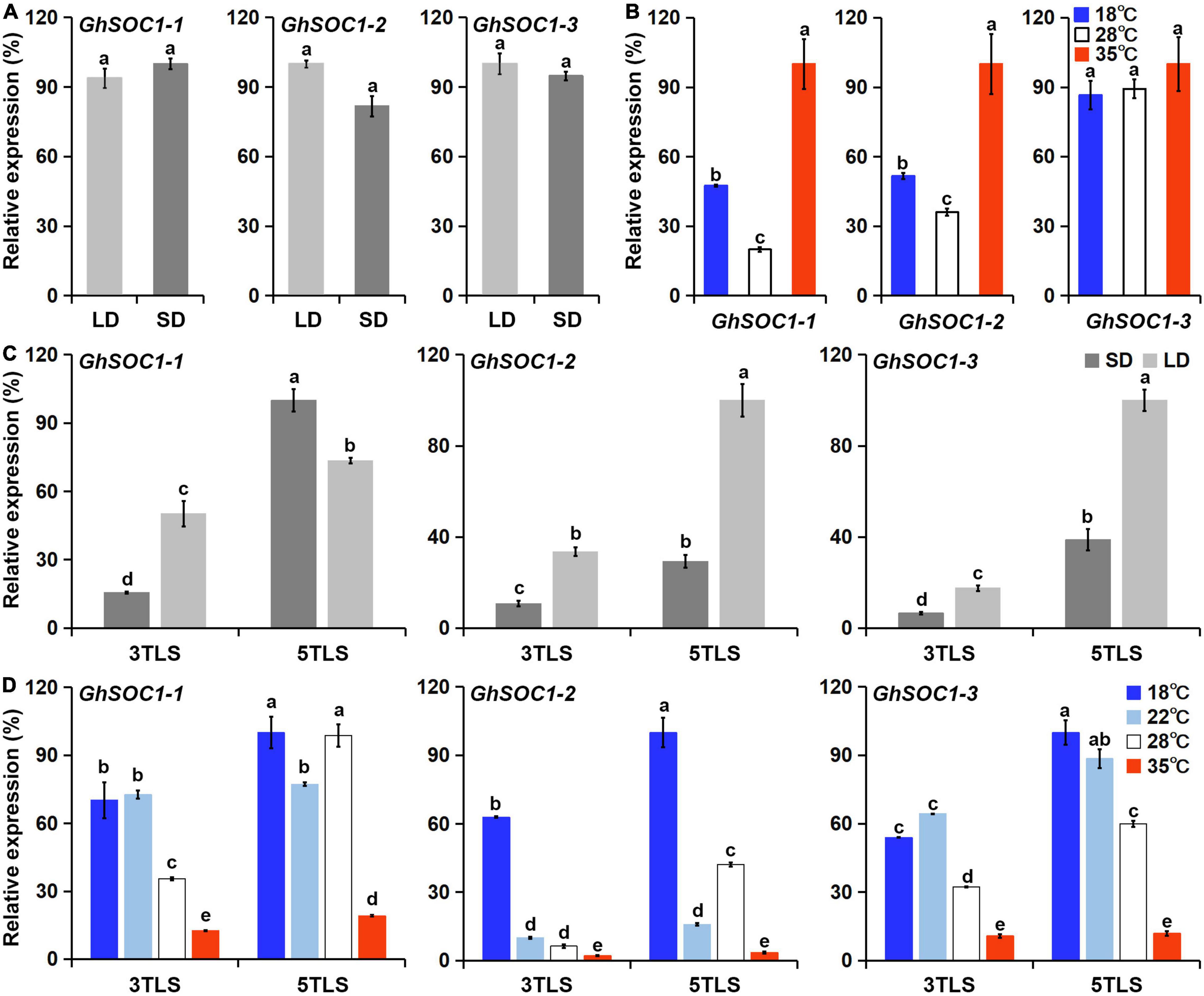
Figure 7. Responses of GhSOC1s to light and temperature. (A) Quick response of GhSOC1s to day length shift from long-day to short-day for 48 h under 28°C. (B) Quick response of GhSOC1s to temperature shift for 48 h examined in seedlings grown under 28°C and long-day condition. (C) GhSOC1s expression is examined under continues long-day and short-day conditions under 28°C at 3TLS and 5TLS. (D) GhSOC1s expression is examined under different temperature conditions under long-day at 3TLS and 5TLS. The expression is normalized to GhHIS. Error bars represent the standard deviations of three biological replicates. Different letters suggest significant differences calculated by Student’s t-test (P < 0.01).
Secondly, the expression analysis suggested the occurrence of floral transition of TM-1 at 4TLS (Figure 4C), and thus GhSOC1s expression changes before and after floral transition in response to photoperiod and ambient temperature were compared at 3TLS and 5TLS. The results showed that GhSOC1-1 expression elevated dramatically under short-day during floral transition, while expression elevation of GhSOC1-2 and GhSOC1-3 were more violent under long-day compared with short-day, suggesting that GhSOC1-2 and GhSOC1-3 were more influential to floral transition in response to long-day condition (Figure 7C). The differences of expression elevation during floral transition revealed distinct participation of GhSOC1s in response to photoperiod.
Thirdly, the responses of GhSOC1s to ambient temperature were different from the quick response to temperature change. GhSOC1s expression under high temperature was the lowest compared to normal temperature (28°C) and only GhSOC1-1 expression significantly increased during floral transition under 35°C (Figure 7D), suggesting that GhSOC1-1 rather than GhSOC1-2/3 acts as an effector of high ambient temperature to accelerate flowering. Under 18 and 22°C, GhSOC1s expression was promoted at the vegetative growth stage and GhSOC1-3 increased significantly during floral transition. Transcription of GhSOC1-2 was dramatically induced under 18°C, suggesting a unique role of GhSOC1-2 in response to cold. It was likely that GhSOC1s played cooperatively under low ambient temperature to promote flowering and ensure the success of reproductive growth.
Gossypium origins from the tropical and subtropical area that is adapted to short-day photoperiod and the megathermal climate. The cultivated species has changed after domestication. The widely planted upland cotton losses the seed dormancy and sensitivity to short-day photoperiod and produces more fibers with increased length and whiteness (Chen et al., 2020). The application of GWAS has revealed the molecular mechanism of domestication on some traits and identified some locus contributing to maturity (Ma et al., 2018; Li et al., 2021a). However, our knowledge on cotton flowering time control is limited, restricting the co-improvement of cotton on maturity, yield, quality and resistance.
Previous studies have identified several SOC1 homologs in tetraploid upland cotton (Nardeli et al., 2018), one of which was cloned as GhSOC1 (GhSOC1-2). In this study, six SOC1 homologous were identified in upland cotton genome with the conserved domains (MADS-box and K-box) and SOC1-motif (Figure 1B). But they formed two clades in the phylogenetic tree that GhSOC1-1 sequences were more similar to monocot SOC1 sequences and clustered in the different group from the dicot SOC1 subclade including GhSOC1-2 and GhSOC1-3 (Figure 2). Consistent with the phylogenetic tree, the gene structure of GhSOC1-1 characterized by a large first intron is similar as OsSOC1 (Tadege et al., 2003).
Whole-genome duplication (WGDs) is significant for transformative evolution. The gamma (γ) WGD occurred after the derivation of monocots from the core dicots (Vekemans et al., 2012). The Vitis species only experienced this ancient whole-genome triplication, during which SOC1 genes triplicated (Vekemans et al., 2012; Zhang et al., 2020). Cocoa, the lineage close specie of Gossypium, also experienced the γ WGD. But, the early diverging dicot lineage of lotus (Nelumbo nucifera) did not experience this whole-genome triplication (Zhang et al., 2020). Therefore, cotton SOC1-1 genes that were close to NnSOC1 and monocot SOC1 genes should be the ancient loci retained in the Gossypium genomes, and triplicated during the γ WGD after which cotton SOC1-2 and SOC1-3 genes evolved as the dicot SOC1 genes. The conserved gene structure further supported the above inference.
The early flowering phenotype caused by overexpression of GhSOC1-1 (Figures 5B,C) or GhSOC1-2 (Zhang et al., 2016) in Arabidopsis suggested functional conservation in flowering time control. And the consistent sharp increase of GhSOC1s during the occurrence of floral transition (Figure 4C) further suggested their conserved function in promoting flowering. Corporate silencing of GhSOC1-1/2/3 delayed cotton flowering, confirming the functional overlap of GhSOC1s as flowering promoters. Another commonality was observed in their ubiquitous expression in vegetative organs (Figure 4A), which is consistent with other SOC1 homologs as flowering regulators (Lee et al., 2000; Wei et al., 2016; Liu et al., 2020). Moreover, SOC1 act as a floral integrator to function in the downstream of the flowering time regulation network to mainly activate LFY expression (Lee and Lee, 2010). The expression changes of LFY in both GhSOC1-1 overexpression and silencing plants revealed that GhSOC1-1 promote flowering via GhLFY.
However, the diversity of cis-elements in the promoter of different GhSOC1s suggest discrepancy of their transcription regulation (Figure 3), which was supported by their expression differences (Figure 4A). First, their transcription abundance varied greatly. The actively transcribed GhSOC1-1 may play a dominant role, while minimal expression of GhSOC1-3 was detected prevalently in roots and stems indicating a functional divergence. Although the coding region determines gene function, the introns are proved to contain cis-acting elements. For example, the 3.5 kb first intron of AtFLC are critical for the epigenetic repression (Sheldon et al., 2002). The large intron might contribute to the abundant transcription of GhSOC1-1 (Figure 1A). The failure in promoting flowering of constitutive expression of GhSOC1-2 in cotton is evidence for functional divergency in flowering time control (Zhang et al., 2016). Moreover, ABF3 and ABF4 bind to SOC1 promotor to mediate drought-accelerated flowering (Hwang et al., 2019). The ABRE elements were predicted in GhSOC1-1 and GhSOC1-2 regulatory region, but drought responsive elements only existed in GhSOC1-1 genome (Figure 3). As functions of ABFs diverge and overlap (Finkelstein et al., 2005), GhSOC1-1 with more ABRE elements was more likely to respond to drought to promote flowering.
GhSOC1s expressed differently in floral organs. GhSOC1-3 showed higher expression in vegetative organs, but GhSOC1-1 and GhSOC1-2 were most abundantly expressed in flower. Detailed expression of GhSOC1s in flowers demonstrated that they were accordantly expressed highest in calycale, and GhSOC1-1 also accumulated in the inner whorls, indicating their participation in flower development. It was supported that 35S:GhSOC1-2 causes floral defects in cotton (Zhang et al., 2016). In contrary, the cotton plants produced normal flowers when GhSOC1s were silenced (data not shown), which is the same as soc1-2 mutant of Arabidopsis. But triple mutants of soc1 svp agl24 display sever floral defects (Liu et al., 2009). The Gerbera hybrida SOC1-like gene is only expressed in flowers and overexpression caused partial defects of petal on color and shape without changes in flowering time (Ruokolainen et al., 2011). Therefore, GhSOC1s might be involved in the development of different floral organs with other MADS cooperators. Further, the function in floral development needs to be induces. As FBP21/22 and AtSOC1 affects petal color and development only under high temperature (Wang et al., 2019).
Additionally, quick response of GhSOC1-1 and GhSOC1-2 to high and low temperature indicated involvement in stress resistance (Figure 7B), which is supported by the existence of ABA responsive and defense related cis-elements in their genome rather than GhSOC1-3 (Figure 3).
In the model plant, SOC1 integrates the environmental and endogenous signals to initiate floral transition (Lee and Lee, 2010). And SOC1 homologs diverged in role of flowering time control among species. TgSOC1-like1 and TgSOC1-like2 functions opposite in flowering time control (Leeggangers et al., 2017). Four SOC1-like genes (AcSOC1e, AcSOC1f, AcSOC1i, AcSOC1g) from Actinidia sinensis promote flowering of Arabidopsis in varying degrees, but respond differently to winter chilling to regulate woody perennials (Voogd et al., 2015). The Medicago truncatula genome possesses three SOC1 genes (MtSOC1a—MtSOC1c) that are expressed differently in response to day-length and vernalization to promote flowering (Fudge et al., 2018). Similarly, GhSOC1s respond differently to photoperiod although their genomes all possess a large number of light responsive elements (Figures 3, 7C). GhSOC1-1 was greatly induced by short-day condition, while GhSOC1-2 and GhSOC1-3 respond to long-day. These suggested that GhSOC1-1 evolved conserved as discussed above, while GhSOC1-2 and GhSOC1-3 contribute to the insensitivity of day-length during domestication.
Ambient temperature is another important environmental cue that affects flowering time. Cotton can grow normally under the temperature above 22°C. Low ambient temperature induced expression of GhSOC1s in the vegetative stage, and GhSOC1-3 expression further elevated during floral transition (Figure 7D). But only GhSOC1-1 respond to the high ambient temperature to activate floral transition (Figure 7D). The response of GhSOC1-1 to high ambient temperature together with the induction by short-day is reasonable for the dominant role in flowering time control as cotton origins in the subtropical area. When the temperature is not suitable for cotton growth, GhSOC1-2 was dramatically induced. And overexpression of GhSOC1-2 causes significant increase of FT and LFY expression (Zhang et al., 2016). We speculated that GhSOC1s act cooperatively to promote flowering via GhFT and GhLFY (Figure 8), which would be essential for seed generation under different growth conditions to ensure reproductive success.
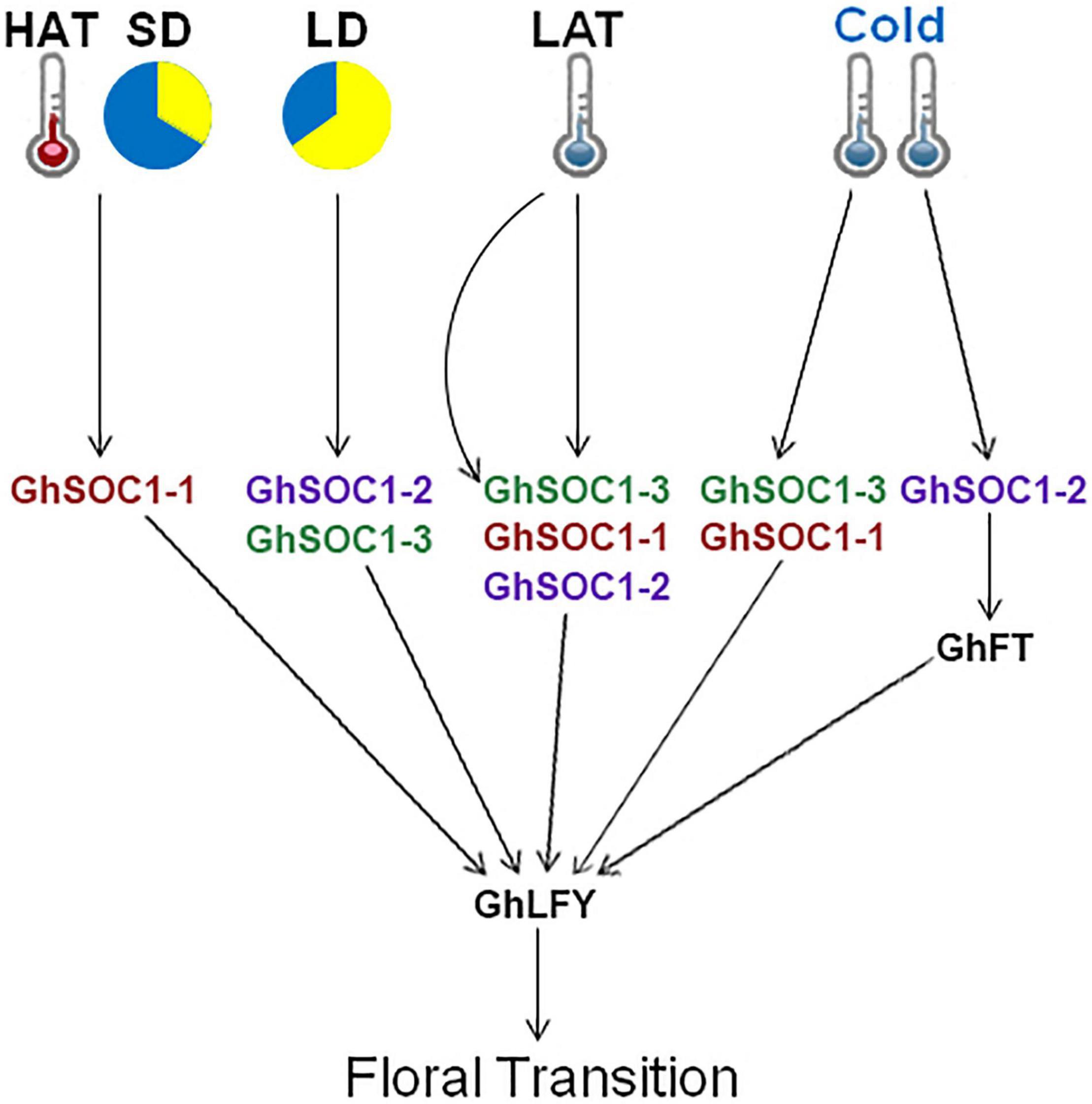
Figure 8. Proposed regulatory model of GhSOC1s. HAT, high ambient temperature; LAT, low ambient temperature; LD, long-day condition; SD, short-day condition.
Multimerization is common for MADS proteins contributing to a large protein-protein interaction network that fine tune the reproductive growth (Theissen et al., 2016). The formation of SOC1-AGL24 heterodimer is compulsory for entrance of SOC1 to the nucleus where SOC1-AGl24 binds to the target gene (Lee et al., 2008). Similarly, expression of FaSOC1 (Lei et al., 2013) or GmGAL1 (Zhong et al., 2012) (SOC1 homolog gene) alone, the resulting proteins are localized in the cytoplasm. Differently, GhSOC1-1 proteins were localized in the nucleus without any assistance to play its function (Figure 4D), which is consistent with LsSOC1 (Chen et al., 2018). Structure prediction found two variations of the essential Leucine residues in the K domain of GhSOC1-1 (Figure 1D). Single mutation at L115 and L164 is able to destroy the coiled-coil structure of the K domain, and one substitution mutation L164P or L115P reduces the ability to form tetramer resulting in altered DNA-binding, which is likely to be overcame by high protein concentration (Rumpler et al., 2018). Thus the interactions of GhSOC1-1 probably distinct from GhSOC1-2 and GhSOC1-3, and proper function of GhSOC1-1 depends on the concentration that is consistent with its abundant transcripts. The structure effects on DNA-binding provide an explanation for differences of the downstream gene. It has reported that GhSOC1-2 directly binds to the promotor of GhLFY to activate its transcription (Li et al., 2013). Overexpression of 35S:GhSOC1-1 or 35S:GhSOC1-2 upregulated LFY expression (Figure 5D; Zhang et al., 2016). Meanwhile, FT expression is greatly enhanced by overexpression of GhSOC1-2 (Zhang et al., 2016). However, FT transcription was not affected by GhSOC1-1 (Figure 5D), suggesting that GhSOC1-1 is the downstream of FT as in Arabidopsis. Thus, our results revealed that GhSOC1-1 acts downstream of FT to promote floral transition via LFY. Although GhSOC1-2 promotes FT expression in Arabidopsis, it failed to bind to FT genome (Zhang et al., 2016), suggesting another regulation pathway for GhSOC1-2 (Figure 8).
The original data presented in the study are included in the Supplementary Material, further inquiries can be directed to the corresponding author/s.
YY conceived and designed the experiment. LM performed the experiments. YY and LM analyzed the data and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This work was supported by the National Natural Science Foundation of China (grant no. 31801410), the Provincial Natural Science Foundation of Hebei (grant no. C2020204079), the Supporting Project of Hebei Agricultural University (grant nos. ZD201601 and PT2018004), and the Top Talent Project of Hebei Province to ZM (031601801). We are grateful to Prof. Ma Zhiying for his advise and helps on this work.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.882946/full#supplementary-material
Amrouk, E. M., Mermigkas, G., and Townsend, T. (2021). Recent Trends and Prospects in the World Cotton Market and Policy Developments. Rome: Food and Agriculture Organization of the United Nations, 72.
Borner, R., Kampmann, G., Chandler, J., Gleiûner, R., Wisman, E., Apel, K., et al. (2008). A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 24, 591–599. doi: 10.1046/j.1365-313x.2000.00906.x
Chen, Z., Zhao, W., Ge, D., Han, Y., Ning, K., Luo, C., et al. (2018). LCM-seq reveals the crucial role of LsSOC1 in heat-promoted bolting of lettuce (Lactuca sativa L.). Plant J. 95, 516–528. doi: 10.1111/tpj.13968
Chen, Z. J., Sreedasyam, A., Ando, A., Song, Q., De Santiago, L. M., Hulse-Kemp, A. M., et al. (2020). Genomic diversifications of five Gossypium allopolyploid species and their impact on cotton improvement. Nat. Genet. 52, 525–533. doi: 10.1038/s41588-020-0614-5
Ding, L., Wang, Y., and Yu, H. (2013). Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant Cell Physiol. 54, 595–608. doi: 10.1093/pcp/pct026
Fatima, M., Zhang, X., Lin, J., Zhou, P., Zhou, D., and Ming, R. (2020). Expression profiling of MADS-box gene family revealed its role in vegetative development and stem ripening in S. spontaneum. Sci. Rep. 10:20536. doi: 10.1038/s41598-020-77375-6
Finkelstein, R., Gampala, S. S., Lynch, T. J., Thomas, T. L., and Rock, C. D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 59, 253–267.
Fudge, J. B., Lee, R. H., Laurie, R. E., Mysore, K. S., Wen, J., Weller, J. L., et al. (2018). Medicago truncatula SOC1 genes are up-regulated by environmental cues that promote flowering. Front. Plant Sci. 9:496. doi: 10.3389/fpls.2018.00496
Hao, J. J., Yu, S. X., Ma, Q. X., Fan, S. L., and Song, M. Z. (2008). Inheritance of time of flowering in upland cotton under natural conditions. Plant Breed. 127, 383–390. doi: 10.1111/j.1439-0523.2007.01474.x
Huang, G., Wu, Z., Percy, R. G., Bai, M., Li, Y., Frelichowski, J. E., et al. (2020). Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat. Genet. 52, 516–524. doi: 10.1038/s41588-020-0607-4
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., and Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505. doi: 10.1016/j.molp.2019.01.002
Immink, R. G., Pose, D., Ferrario, S., Ott, F., Kaufmann, K., Valentim, F. L., et al. (2012). Characterization of SOC1’s central role in flowering by the identification of its upstream and downstream regulators. Plant Physiol. 160, 433–449. doi: 10.1104/pp.112.202614
Jung, J. H., Ju, Y., Seo, P. J., Lee, J. H., and Park, C. M. (2012). The SOC1-SPL module integrates photoperiod and gibberellic acid signals to control flowering time in Arabidopsis. Plant J. 69, 577–588. doi: 10.1111/j.1365-313X.2011.04813.x
Kimura, Y., Aoki, S., Ando, E., Kitatsuji, A., Watanabe, A., Ohnishi, M., et al. (2015). A flowering integrator, SOC1, affects stomatal opening in Arabidopsis thaliana. Plant Cell Physiol. 56, 640–649. doi: 10.1093/pcp/pcu214
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, H., Suh, S. S., Park, E., Cho, E., Ahn, J. H., Kim, S. G., et al. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14, 2366–2376. doi: 10.1101/gad.813600
Lee, J., and Lee, I. (2010). Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 61, 2247–2254. doi: 10.1093/jxb/erq098
Lee, J., Oh, M., Park, H., and Lee, I. (2008). SOC1 translocated to the nucleus by interaction with AGL24 directly regulates leafy. Plant J. 55, 832–843. doi: 10.1111/j.1365-313X.2008.03552.x
Leeggangers, H. A., Nijveen, H., Bigas, J. N., Hilhorst, H. W., and Immink, R. G. (2017). Molecular regulation of temperature-dependent floral induction in Tulipa gesneriana. Plant Physiol. 173, 1904–1919. doi: 10.1104/pp.16.01758
Lei, H. J., Yuan, H. Z., Liu, Y., Guo, X. W., Liao, X., Liu, L. L., et al. (2013). Identification and characterization of FaSOC1, a homolog of SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from strawberry. Gene 531, 158–167. doi: 10.1016/j.gene.2013.09.036
Li, D., Liu, C., Shen, L., Wu, Y., Chen, H., Robertson, M., et al. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15, 110–120. doi: 10.1016/j.devcel.2008.05.002
Li, J., Fan, S. L., Song, M. Z., Pang, C. Y., Wei, H. L., Li, W., et al. (2013). Cloning and characterization of a FLO/LFY ortholog in Gossypium hirsutum L. Plant Cell Rep. 32, 1675–1686. doi: 10.1007/s00299-013-1479-1
Li, J., Yuan, D., Wang, P., Wang, Q., Sun, M., Liu, Z., et al. (2021a). Cotton pan-genome retrieves the lost sequences and genes during domestication and selection. Genome Biol. 22:119. doi: 10.1186/s13059-021-02351-w
Li, L., Zhang, C., Huang, J., Liu, Q., Wei, H., Wang, H., et al. (2021b). Genomic analyses reveal the genetic basis of early maturity and identification of loci and candidate genes in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 19, 109–123. doi: 10.1111/pbi.13446
Liu, C., Xi, W., Shen, L., Tan, C., and Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Liu, Z., Wu, X., Cheng, M., Xie, Z., Xiong, C., Zhang, S., et al. (2020). Identification and functional characterization of SOC1-like genes in Pyrus bretschneideri. Genomics 112, 1622–1632. doi: 10.1016/j.ygeno.2019.09.011
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, Z., He, S., Wang, X., Sun, J., Zhang, Y., Zhang, G., et al. (2018). Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat. Genet. 50, 803–813. doi: 10.1038/s41588-018-0119-7
Ma, Z., Zhang, Y., Wu, L., Zhang, G., Sun, Z., Li, Z., et al. (2021). High-quality genome assembly and resequencing of modern cotton cultivars provide resources for crop improvement. Nat. Genet. 53, 1385–1391. doi: 10.1038/s41588-021-00910-2
Mouhu, K., Kurokura, T., Koskela, E. A., Albert, V. A., Elomaa, P., and Hytonen, T. (2013). The Fragaria vesca homolog of suppressor of overexpression of constans1 represses flowering and promotes vegetative growth. Plant Cell 25, 3296–3310. doi: 10.1105/tpc.113.115055
Nardeli, S. M., Artico, S., Aoyagi, G. M., De Moura, S. M., Da Franca Silva, T., Grossi-De-Sa, M. F., et al. (2018). Genome-wide analysis of the MADS-box gene family in polyploid cotton (Gossypium hirsutum) and in its diploid parental species (Gossypium arboreum and Gossypium raimondii). Plant Physiol. Biochem. 127, 169–184. doi: 10.1016/j.plaphy.2018.03.019
Puranik, S., Acajjaoui, S., Conn, S., Costa, L., Conn, V., Vial, A., et al. (2014). Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell 26, 3603–3615. doi: 10.1105/tpc.114.127910
Ren, Z., Yu, D., Yang, Z., Li, C., Qanmber, G., Li, Y., et al. (2017). Genome-wide identification of the MIKC-Type MADS-Box gene family in Gossypium hirsutum L. unravels their roles in flowering. Front. Plant Sci. 8:384. doi: 10.3389/fpls.2017.00384
Richter, R., Bastakis, E., and Schwechheimer, C. (2013). Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 162, 1992–2004. doi: 10.1104/pp.113.219238
Rumpler, F., Theissen, G., and Melzer, R. (2018). A conserved leucine zipper-like motif accounts for strong tetramerization capabilities of SEPALLATA-like MADS-domain transcription factors. J. Exp. Bot. 69, 1943–1954. doi: 10.1093/jxb/ery063
Ruokolainen, S., Ng, Y. P., Albert, V. A., Elomaa, P., and Teeri, T. H. (2011). Over-expression of the Gerbera hybrida At-SOC1-like1 gene Gh-SOC1 leads to floral organ identity deterioration. Ann. Bot. 107, 1491–1499. doi: 10.1093/aob/mcr112
Ryu, C. H., Lee, S., Cho, L. H., Kim, S. L., Lee, Y. S., Choi, S. C., et al. (2009). OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 32, 1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x
Sheldon, C. C., Conn, A. B., Dennis, E. S., and Peacock, W. J. (2002). Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14, 2527–2537. doi: 10.1105/tpc.004564
Song, G. Q., Walworth, A., Zhao, D., Hildebrandt, B., and Leasia, M. (2013). Constitutive expression of the K-domain of a Vaccinium corymbosum SOC1-like (VcSOC1-K) MADS-box gene is sufficient to promote flowering in tobacco. Plant Cell Rep. 32, 1819–1826. doi: 10.1007/s00299-013-1495-1
Song, Q., Zhang, T., Stelly, D. M., and Chen, Z. J. (2017). Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol. 18:99. doi: 10.1186/s13059-017-1229-8
Tadege, M., Sheldon, C. C., Helliwell, C. A., Upadhyaya, N. M., Dennis, E. S., and Peacock, W. J. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1, 361–369. doi: 10.1046/j.1467-7652.2003.00034.x
Theissen, G., Melzer, R., and Rumpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Vekemans, D., Proost, S., Vanneste, K., Coenen, H., Viaene, T., Ruelens, P., et al. (2012). Gamma paleohexaploidy in the stem lineage of core eudicots: significance for MADS-box gene and species diversification. Mol. Biol. Evol. 29, 3793–3806. doi: 10.1093/molbev/mss183
Voogd, C., Wang, T., and Varkonyi-Gasic, E. (2015). Functional and expression analyses of kiwifruit SOC1-like genes suggest that they may not have a role in the transition to flowering but may affect the duration of dormancy. J. Exp. Bot. 66, 4699–4710. doi: 10.1093/jxb/erv234
Wang, S., Beruto, M., Xue, J., Zhu, F., Liu, C., Yan, Y., et al. (2015). Molecular cloning and potential function prediction of homologous SOC1 genes in tree peony. Plant Cell Rep. 34, 1459–1471. doi: 10.1007/s00299-015-1800-2
Wang, Z., Shen, Y., Yang, X., Pan, Q., Ma, G., Bao, M., et al. (2019). Overexpression of particular MADS-box transcription factors in heat-stressed plants induces chloroplast biogenesis in petals. Plant Cell Environ. 42, 1545–1560. doi: 10.1111/pce.13472
Wei, J., Liu, D., Liu, G., Tang, J., and Chen, Y. (2016). Molecular cloning, characterization, and expression of misoc1: a homolog of the flowering gene SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 from mango (Mangifera indica L). Front. Plant Sci. 7:1758. doi: 10.3389/fpls.2016.01758
Yoo, S. K., Chung, K. S., Kim, J., Lee, J. H., Hong, S. M., Yoo, S. J., et al. (2005). CONSTANS activates suppressor of overexpression of constans 1 through flowering locus T to promote flowering in Arabidopsis. Plant Physiol. 139, 770–778. doi: 10.1104/pp.105.066928
Zhang, L., Wu, S., Chang, X., Wang, X., Zhao, Y., Xia, Y., et al. (2020). The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant Cell Environ. 43, 2847–2856. doi: 10.1111/pce.13898
Zhang, X., Wei, J., Fan, S., Song, M., Pang, C., Wei, H., et al. (2016). Functional characterization of GhSOC1 and GhMADS42 homologs from upland cotton (Gossypium hirsutum L.). Plant Sci. 242, 178–186. doi: 10.1016/j.plantsci.2015.05.001
Keywords: Gossypium hirsutum L., SOC1-like gene, flowering time control, environmental response, evolution
Citation: Ma L and Yan Y (2022) GhSOC1s Evolve to Respond Differently to the Environmental Cues and Promote Flowering in Partially Independent Ways. Front. Plant Sci. 13:882946. doi: 10.3389/fpls.2022.882946
Received: 24 February 2022; Accepted: 23 March 2022;
Published: 20 April 2022.
Edited by:
Yang Zhu, Zhejiang University, ChinaReviewed by:
Geng-Qing Huang, Central China Normal University, ChinaCopyright © 2022 Ma and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuanyuan Yan, c2VsaW5hMzAwMTYzMDAxNkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.