
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 31 May 2022
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.882228
This article is part of the Research Topic Effects of Plant-Microbiome Interactions on Phyto- and Bio-Remediation Capacity, Volume II View all 7 articles
The integration of phytoremediation and biostimulation can improve pollutant removal from the environment. Plant secondary metabolites (PSMs), which are structurally related to xenobiotics, can stimulate the presence of microbial community members, exhibiting specialized functions toward detoxifying, and thus mitigating soil toxicity. In this study, we evaluated the effects of enrichment of 4-chloro-2-methylphenoxyacetic acid (MCPA) contaminated soil (unplanted and zucchini-planted) with syringic acid (SA) on the bacterial community structure in soil, the rhizosphere, and zucchini endosphere. Additionally, we measured the concentration of MCPA in soil and fresh biomass of zucchini. The diversity of bacterial communities differed significantly between the studied compartments (i.e., unplanted soil, rhizospheric soil, and plant endosphere: roots or leaves) and between used treatments (MCPA or/and SA application). The highest diversity indices were observed for unplanted soil and rhizosphere. Although the lowest diversity was observed among leaf endophytes, this community was significantly affected by MCPA or SA: the compounds applied separately favored the growth of Actinobacteria (especially Pseudarthrobacter), while their simultaneous addition promoted the growth of Firmicutes (especially Psychrobacillus). The application of MCPA + SA together lead also to enhanced growth of Pseudomonas, Burkholderia, Sphingomonas, and Pandoraea in the rhizosphere, while SA increased the occurrence of Pseudomonas in leaves. In addition, SA appeared to have a positive influence on the degradative potential of the bacterial communities against MCPA: its addition, followed by zucchini planting, significantly increased the removal of the herbicide (50%) from the soil without affecting, neither positively nor negatively, the plant growth.
The presence of plant secondary metabolites (PSMs) such as flavonoids, coumarins, phenolic compounds, and terpenes, is known to modify the chemical and physical properties of soils. PSMs also serve as allelochemicals, protect plants against pathogens, act as substrates, and can induce pollutant catabolic pathways of soil microorganisms (Pilon-Smits, 2005; Singer, 2006; Zhou and Wu, 2012; Pino et al., 2016). They also shape the structure and function of plant-associated bacterial communities, i.e., rhizobacteria and endophytes, favoring the growth of suitable pollutant consumers (Uhlik et al., 2013; Qin et al., 2014; Eevers et al., 2016), which directly use their degradative capabilities to metabolize a given pollutant (Glick, 2010; Pawlik et al., 2015; Sauvêtre and Schroder, 2015; Khare et al., 2018; Wu et al., 2018). Thus, the role of PSMs in the degradation of pollutants can be substantial.
Due to their structural similarity, PSMs may have a considerable influence on the removal of structurally related pollutants (Uhlik et al., 2013; Hu et al., 2014). PSMs typically provide the energy for microorganisms to perform cometabolism, while the structurally similar pollutant is degraded as a secondary substrate (Musilova et al., 2016). Common examples of cometabolites are biphenyl and PCBs (Singer et al., 2003). However, PSMs can also be used as a primary source of carbon and energy by bacterial communities to support their growth and stimulate the expression of desirable genes involved in the catabolism of structurally similar pollutant. The degree of similarity between a PSM and a pollutant has been found to influence the rate of pollutant removal (Mierzejewska et al., 2019; Urbaniak et al., 2019a).
Earlier studies described syringic acid (SA) as a characteristic PSM for cucurbits (Blum et al., 2000; Campos et al., 2009; Kruczek et al., 2015; Shi et al., 2016), which are themselves known as effective phytoremediators of organic pollutants (Eevers et al., 2018; Urbaniak et al., 2019b,2020). The addition of SA to bacterial cultures not only enhanced MCPA removal but more importantly increased the number of detected functional genes responsible for the initiation of phenoxy herbicide biodegradation (Mierzejewska et al., 2019; Urbaniak et al., 2019a). Additionally, SA application and zucchini cultivation was found to decrease the toxicity of MCPA-contaminated soil (Mierzejewska et al., 2022).
4-chloro-2-methylphenoxyacetic acid (MCPA) is one of the most commonly used herbicides in Europe, and it is particularly persistent under the low winter temperatures, low soil organic carbon content, and acidic pH that are typical for Europe (Paszko et al., 2016). In areas with high contamination levels, heavy rainstorms facilitate the transport of MCPA residues from soil to water, resulting in the contamination of aquatic ecosystems (Matamoros et al., 2012; Rheinheimer dos Santos et al., 2020) to levels exceeding permissible threshold concentrations (Ignatowicz and Struk-Sokołowska, 2004; Rippy et al., 2017). These are believed to account for the adverse effects on living organisms reported in several worldwide studies (Pereira and Cerejeira, 2000; Nielsen and Dahllof, 2007; Palma et al., 2018; Morton et al., 2020).
Despite the large body of research performed on the effects of MCPA on soil- and water-inhabiting organisms, little is known about its influence on soil and plant-associated bacterial communities. Also, little attention has been devoted to the role of PSMs in the remediation of MCPA-contaminated soil. While our prior research has demonstrated that SA treatment enhanced MCPA transformation (Mierzejewska et al., 2019; Urbaniak et al., 2019a), no studies have examined the effects of such treatment on the structure of plant-associated bacterial communities (rhizobacteria and endophytes).
Consequently, this study aimed to determine whether adding SA, which is structurally like the herbicide, to unplanted and zucchini-planted soil contaminated with MCPA shapes the bacterial communities within the unplanted soil, rhizosphere, and plant endosphere (i.e., endophytic communities in roots and leaves). The obtained results could be applied to develop nature-based solutions for the removal of MCPA residues and to prevent their dispersal in the environment.
The potting soil was obtained from a certified supplier: Substral OSMOCOTE. Its composition was described as universal soil for plant growth, containing a mixture of peat, fertilizer, expanded clay, and silica and with the following properties: pH 4.3; 34.3% C, 2.2% N; 1.5 g P kg–1 soil, 2.5 g K kg–1 soil. This soil was selected based on previous experiments with cucurbits (Mierzejewska et al., 2022).
Based on previous investigations on the uptake of toxic organic pollutants by the cucurbits (Urbaniak et al., 2016; Wyrwicka et al., 2019; Mierzejewska et al., 2022), C. pepo L. “Atena Polka” (zucchini) was chosen for this study. The seeds were purchased from a certified supplier of garden seeds (W. Legutko) and were germinated under stable conditions in perlite for 5 days and grown in unamended potting soil for 7 days to select seedlings at the same growth stage.
MCPA, C9H9ClO3 (≥95.0% purity, molecular weight 200.61 g mol–1, 0.825 g L–1 water solubility at 20°C, pKa value 3.07) was obtained from Sigma-Aldrich. The PSM used in this study was syringic acid (SA), C9H10O5 (≥95.0% purity, molecular weight 198.17 g mol–1, 5.78 mg ml–1 water solubility at 25°C, pKa value 4.34), also obtained from Sigma-Aldrich. The concentrations of MCPA (0.05 mM) and SA (0.125 mM) were selected based on earlier studies (Urbaniak et al., 2019a; Mierzejewska et al., 2022). Stock solutions of MCPA and SA were prepared in methanol.
Each treatment was prepared in six replicates, with one seedling (zucchini) per pot, resulting in a total of six plants for each treatment variant. Unplanted soil variants were also included as an unplanted reference (Table 1). Soil humidity was adjusted to 60% v/w, and MCPA and SA were applied to the potting soil. The soil variants were kept for 1 h under the fume hood to allow the solvent to evaporate. Following this, the zucchini seedlings were planted into the potting soil.
All variants were cultivated in 500 cm3 soil pots, in a growth chamber at 23 ± 0.5°C based on a 16 h light/8 h dark cycle, with a photon flux density of 250 μmol m–2 s–1 during the light period, and 60% w/v soil humidity (Wyrwicka and Urbaniak, 2016). All variants were watered daily. The experiments were running for 20 days. Samples of unplanted soil and rhizosphere (rhizospheric soil and roots) soil were collected and stored at 4°C for further analyses. The aboveground biomass of the plants (stems and leaves) was determined subsequently. The leaves were collected and stored at 4°C for further analyses.
The MCPA concentrations were determined in all Soil variants (unplanted and planted) at the beginning of the experiment and after 20 days (Us + MCPA; Us + MCPA + SA; Zu + MCPA; Zu + MCPA + SA). MCPA was determined according to the sample preparation method published by the European Commission (EURL-SRM, 2015, 2020). Briefly, 5.00 g ± 0.05 g of soil was weighed in a 50 ml Teflon centrifuge tube, and 10 ml of deionized water was added. Then, a 10 ml sample extraction solvent (1% formic acid in acetonitrile) was added. After shaking this mixture for 15 min using the QuEChERS Hand Motion Shaker (Eberbach model EL 680.Q.25 QuEChERS) with 450 osc., 4 g of magnesium sulfate and 1 g of sodium chloride were added, followed by shaking for 1 min. The suspension was centrifuged for phase Separation at 8,100 rpm for 5 min. The extract was then diluted (200 μl of extract with 700 μl of water, 50 μl of acetonitrile, and 50 μl of internal standard MCPA D6). This mixture was vortexed and filtered through a 0.22 μm PTFE directly into an amber HPLC vial.
The MCPA concentrations in the extracts were determined by highly selective liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS—Agilent 1260 HPLC + 6460 Triple Quad LC/MS) (Agilent Technologies). The chromatography and mass spectrometry conditions are presented in Supplementary Tables 1, 2. The suitability of the method for analyzing MCPA residues in soil has been confirmed: the selectivity/specificity, limit of detection and quantification, linearity, precision, recovery, and expanded uncertainty of the method were validated. The obtained validation parameters met the criteria described in the document SANTE/12682/2019 (European Committee for Standardization, 2018).
The rhizospheric soil and roots were separated in 15 ml Falcon tubes containing 2.5 ml sterile PBS-S buffer amended with Tween 0.2 ml L–1 to separate rhizospheric soil tightly attached to roots. Subsequently, the rhizospheric soil was shaken off on the platform for 20 min at 180 rpm. The roots were then transferred to Falcon tubes containing sterile 2.5 ml of PBS-S buffer, rinsed, and subjected to surface sterilization. Rhizospheric soil was centrifuged for 20 min at 3,500 rpm, the supernatant was discarded and the resulting pellet was defined as a rhizosphere compartment (Bulgarelli et al., 2012).
To isolate the DNA of endophytic bacterial communities, the roots and leaves were subjected to surface sterilization. The tissues were first rinsed in sterile distilled water to wash off the bulk dust. Subsequently, tissues were incubated in 70% ethanol (1 min), NaOCl (1% roots, 2.5% leaves; 1 min), 70% ethanol (1 min) for surface sterilization. Afterward, the samples were rinsed three times in sterile dH2O. Finally, 100 μl of the third rinsing water was transferred to a Petri dish containing an undiluted 869 medium (Mergeay et al., 1985) to check for sterility.
Total DNA from bulk and rhizospheric soil was isolated according to the DNeasy PowerSoil Pro Kit (Qiagen, Venlo, Netherlands). The surface-sterilized plant tissues (roots and leaves) were lyophilized before DNA extraction and snap-frozen in liquid nitrogen. Endophytic DNA was isolated according to PowerPlant® Pro DNA Isolation Kit. DNA samples were quality checked using Nanodrop 2000 (Thermo Fisher Scientific, Wilmington, DE, United States) and stored at −20°C.
The isolated DNA was used for the 16S rRNA gene amplification. DNA was isolated from unplanted variants, rhizospheric soil, surface-sterilized roots, and leaves.
All DNA samples were subjected to bacterial 16S rRNA gene amplicon PCR. In the first round of 16S rRNA gene PCR, an amplicon of 291 bp was generated, using primers 515F-GTGYCAGCMGCCGCGGTAA and 806R- GGACTACNVGGGTWTCTAAT (Walters et al., 2016), with an Illumina adapter overhang nucleotide sequence, resulting in the following sequences, 515F-adapter: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3′ and 806R-adapter: 5′- GTCTCGTGGGCTCGGAGATGTGTATAAG AGACAG-3′. For the first round of PCR, the Q5 High-Fidelity DNA Polymerase system (M0491, NEB), a reaction volume of 25 μl per sample was prepared containing 1 μl of extracted DNA (final DNA-concentration per reaction 1–10 ng), 1x Q5 Reaction Buffer with 2 mM MgCl2, 200 μM dNTP mix, 1x Q5 High GC Enhancer (for the roots endophytes samples), 0.25 μM forward or reverse primer, and 0.02 U/μl Q5 High-Fidelity DNA polymerase, and for the plant endophytic samples (roots and leaves), additionally 2.5 μl mitoPNA blocker (PNA Bio PCR Blockers, 2021) (5 μM final concentration added from a 50 μM stock), 2.5 μl plastidPNA blocker (5 μM final concentration from 50 μM stock) (Kusstatscher et al., 2021) were used.
The PCR program started with an initial denaturation stage (3 min at 98°C), followed by a second denaturation (10 s at 98°C). Following this, 30 cycles were performed of the following three steps: for root and leaf endophytes, PNA clamping (10 s at 75°C), followed by annealing for V3V4 (30 s at 56°C) and extension (30 s at 72°C). The reaction was ended by a final 7 min extension at 72°C. The amplified DNA was purified using AMPure XP beads (Beckman Coulter) and a MagMax magnetic particle processor (ThermoFisher, Leuven, Belgium).
Subsequently, 5 μl of the cleaned PCR product was used for a second PCR attaching the Nextera indices (Nextera XT Index Kit v2 Set A; FC-131-2001, and D; FC-131-2004, Illumina, Belgium). For these PCR reactions, 5 μl of the purified PCR product was used in a 25 μl reaction volume and prepared following the 16S Metagenomic Sequencing Library Preparation Guide. The PCR conditions were the same as described above, but the number of cycles was reduced to 20, and a 55°C annealing temperature was used. The PCR products were cleaned with the Agencourt AMPure XP kit, and then quantified using the Qubit dsDNA HS assay kit (Invitrogen) and the Qubit 2.0 Fluorometer (Invitrogen).
Once the molarity of the sample was determined, the samples were diluted down to 4 nM using 10 mM Tris pH 8.5 before sequencing on the Illumina Miseq. The samples were sequenced using the Miseq Reagent Kit v3 (600 cycle) (MS-102-3003) and 15% PhiX Control v3 (FC-110-3001). For quality control, a DNA extraction blank and PCR blank were included throughout the process, and the ZymoBIOMICS Microbial Mock Community Standard (D6300) was used to test the efficiency of DNA extraction (Zymo Research).
The sequences were demultiplexed using Illumina Miseq software; they were subsequently quality trimmed and the primers were removed using DADA2 1.10.1 (Callahan et al., 2016) in R version 3.5.1. The parameters for length trimming were set to keep the first 290 bases of the forward read and 200 bases of the reverse read, maxN = 0, MaxEE = (2,5) and PhiX removal. Error rates were inferred and the filtered reads were dereplicated and denoized using the DADA2 default parameters. After merging paired reads and removal of chimeras via the removeBimeraDenovo function, an amplicon sequence variant (ASV) table was built and taxonomy assigned using the SILVA v138 training set (Quast et al., 2012; Yilmaz et al., 2014). The resulting ASVs and taxonomy tables were combined with the metadata file into a Phyloseq object (Phyloseq, version 1.26.1) (McMurdie and Holmes, 2013). The pollutants were removed from the dataset using the Decontam package (version 1.2.1) applying the prevalence method with a 0.5 threshold value (Davis et al., 2018). A phylogenetic tree was constructed using a DECIPHER/Phangorn pipeline as described previously (Murali et al., 2018).
The ASV table was further processed by removing organelles (chloroplast, mitochondria), and prevalence was filtered using a 2% inclusion threshold (unsupervised filtering) as described by Callahan et al. (2016). The unfiltered data were subjected to alpha-diversity metrics, such as observed ASV count, Simpson’s and Shannon’s diversity index, using scripts from the MicrobiomeSeq package. Hypothesis testing was performed using analysis of variance (ANOVA) and Tukey’s HSD method using Statistica. For beta-diversity, the Bray–Curtis distances were calculated on unfiltered data using the vegan package (version 2.5.4), and the data were visualized using principal coordinate analysis (PCoA). Hypothesis testing was done by the Adonis function (vegan, version 2.5.5). To assess the homogeneity of variance, the Betadisper function (vegan, version 2.5.4) was used. Relative abundances were calculated and visualized in bar charts using Phyloseq. Differential abundance testing was done on unfiltered data using DESeq2 (version 1.22.2) (Bijnens et al., 2021). Hierarchical cluster analysis was performed to determine the differences between studied variants using PAST 4.0. All performed statistical tests were corrected for multiple testing, and p < 0.05 was considered statistically significant.
The MCPA content in the soils was determined at the beginning of the experiment (T0) and after 20 days (Tf) of incubation (Figure 1). The mean concentration of MCPA at T0 was 6.71 ± 0.270 mg g–1 in the soil amended with MCPA (Us + MCPA) and 6.93 ± 0.156 mg g–1 in soil amended with MCPA and SA (Us + MCPA + SA), respectively. After 20 days (Tf), the MCPA concentration in the Us + MCPA variant decreased to 5.63 ± 0.708 mg g–1 indicating about 18% reduction. In the soil amended with SA (Us + SA), the concentration of MCPA diminished to 4.73 ± 0.784 mg g–1, i.e., about 30% lower than at T0. The cultivation of zucchini in the MCPA-contaminated soil (Zu + MCPA) did not affect the MCPA concentration in the soil (7.77 ± 0.617 mg g–1). However, a substantial reduction of the MCPA concentration (∼50%) was observed in planted soil enriched with SA (Zu + MCPA + SA), where the final concentration of the herbicide was reduced to 3.53 mg g–1.
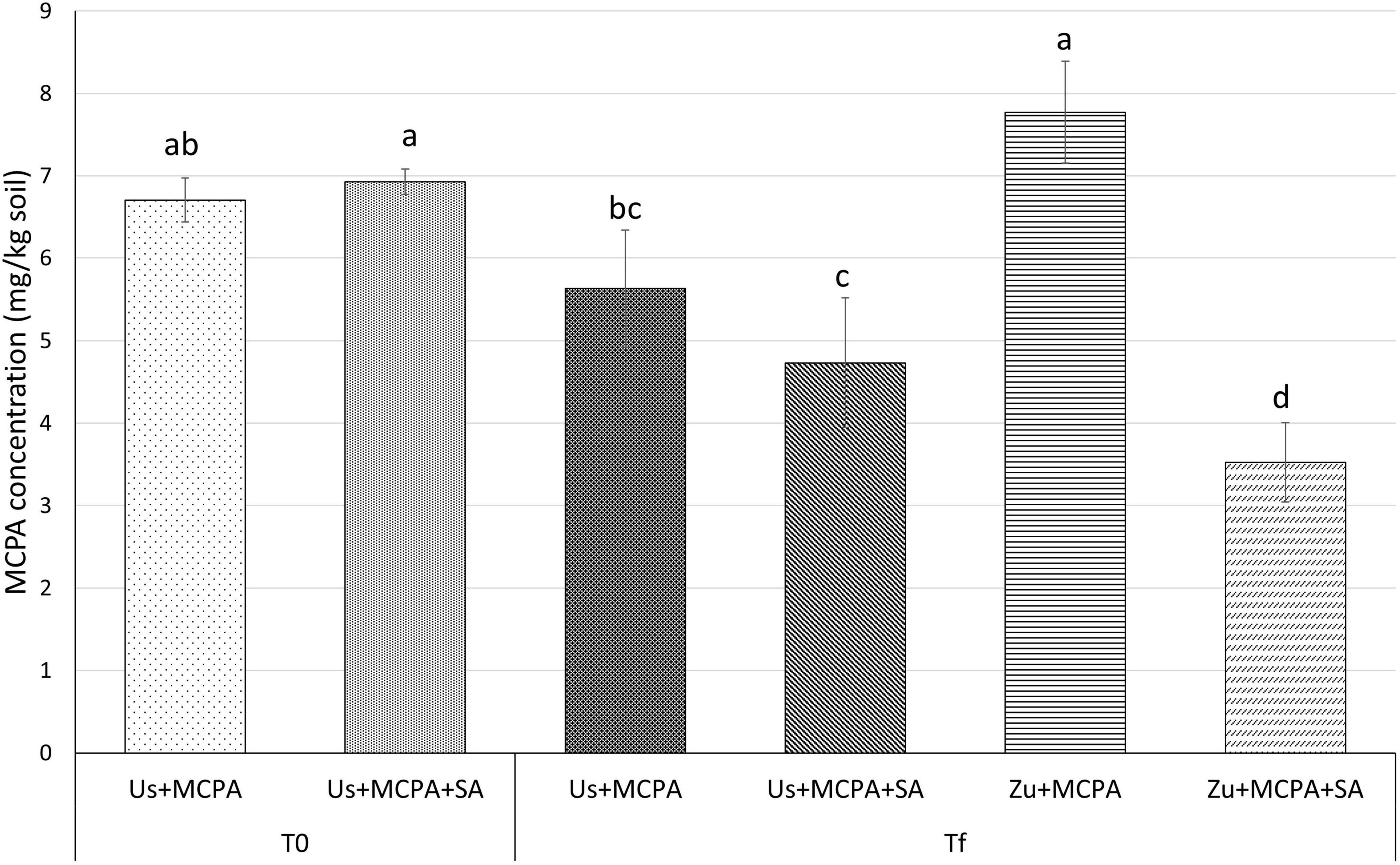
Figure 1. Concentrations of MCPA at the beginning of the experiment (T0) and after 20 days of incubation (Tf) in unplanted soil and in soil after zucchini cultivation. Variants: Us + MCPA, unplanted soil treated with MCPA; Us + MCPA + SA, unplanted soil treated with MCPA and SA; Zu + MCPA, soil after zucchini cultivation treated with MCPA; Zu + MCPA + SA, soil after zucchini cultivation treated with MCPA and SA. Different letters (a-d) indicate significant differences (Tukey’s HSD test, p < 0.05).
Plant fresh weight was significantly affected by the addition of MCPA (Table 2). Significantly higher aboveground biomasses were observed for the zucchini grown in soils without MCPA than those grown in soil amended with MCPA (Table 2). The same pattern was observed for fresh weights of leaves and stems measured separately, these values being significantly lower for variants amended with MCPA compared to unamended soil. The addition of SA alone had no significant effect on the plant’s fresh weight.
The changes in alpha diversity are presented in Table 3. Calculated indices (InvSimpson; Obs.ASVs; Shannon) were slightly lower in Us + SA than in Bs. The amendment of soil with SA increased the InvSimpson/Obs ASVs in the rhizosphere (ZuRh + SA) and roots endosphere (ZuRo + SA), and slightly increased the Shannon index in the roots endosphere (ZuRo + SA). Higher InvSimspon scores were observed in unamended ZuLe than in the SA-amended variant ZuLe + SA.
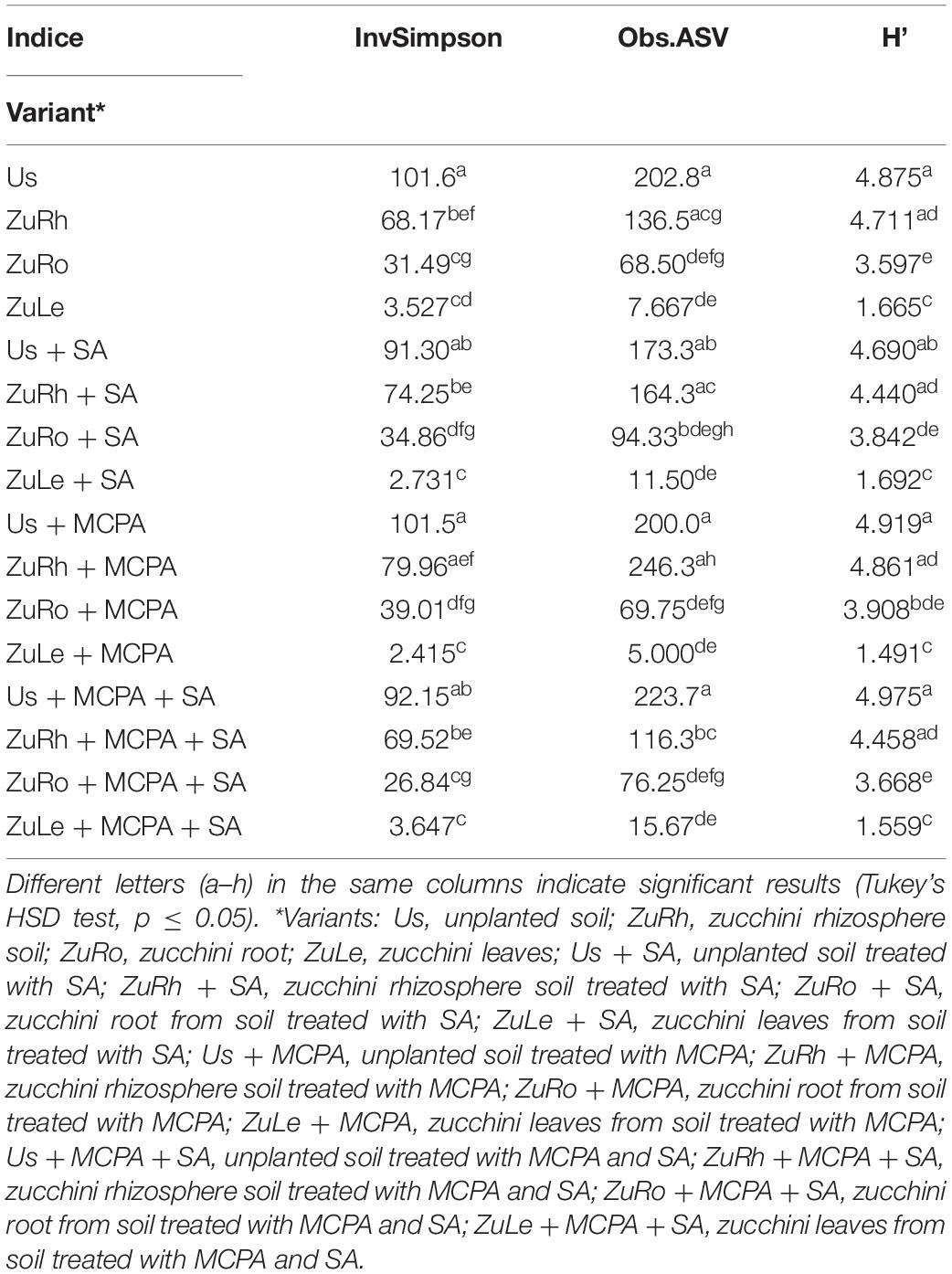
Table 3. Alpha-diversity analyses: three alpha-diversity indices [viz. InvSimpson: inverse of Simpson’s diversity index; obs.ASV: the observed amount of operational taxonomic units (ASV); H’–Shannon’s diversity index].
The addition of SA alone, without MCPA, influenced the composition of bacterial communities, both in soils and plant tissues (Figures 2A,B). A high abundance of Proteobacteria was observed in Us + SA (66.88%). However, the phyla Acidobacteriota, Gemmatimonadota, Planctomycetoota, Firmicutes, and Chloroflexi were not detected after the amendment of unplanted soil with SA (Us + SA). Firmicutes were also not detected in ZuRh + SA. After SA addition, the abundance of Proteobacteria in ZuLe + SA was three times lower (25.58%) than in ZuLe (77.81%), while the abundance of Actinobacteria was almost five times higher in ZuLe + SA (63.27%) than in ZuLe (14.67%).
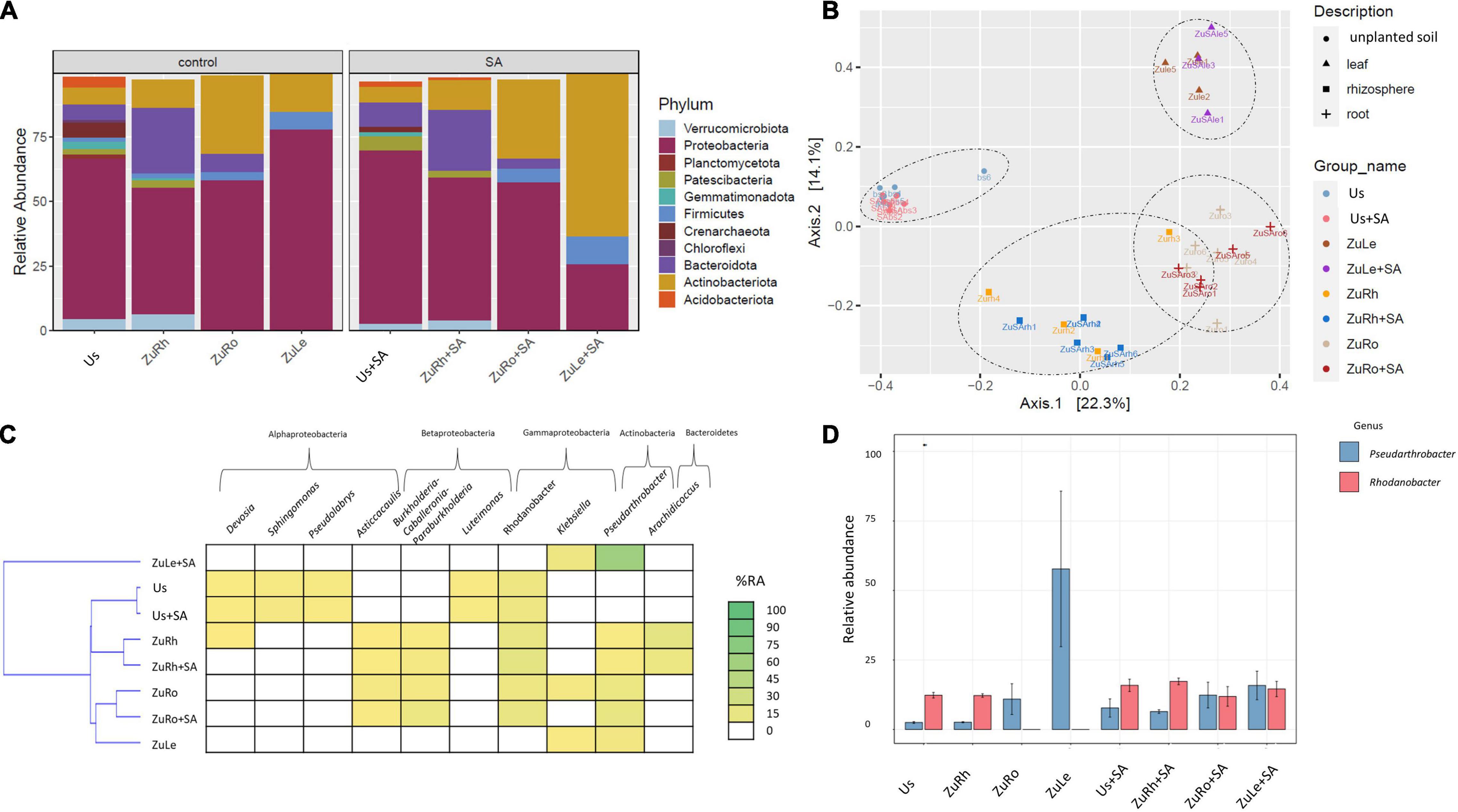
Figure 2. Bacterial communities structure based on 16S rRNA gene sequence for SA-amended variants in comparison to untreated variants: (A) relative abundances (%) of the bacterial community at the phylum level; (B) PcoA ordination plots of the results based on the Bray–Curtis distances of the classified 16S rRNA gene sequences; (C) Heat map of top 10 genera in each treatment; (D) Relative abundance (%) of Pseudarthrobacter and Rhodanobacter in the different treatments. Variants: Us, unplanted soil; ZuRh, zucchini rhizosphere soil; ZuRo, zucchini root; ZuLe, zucchini leaves; Us + SA, unplanted soil treated with SA; ZuRh + SA, zucchini rhizosphere soil treated with SA; ZuRo + SA, zucchini root from soil treated with SA; ZuLe + SA, zucchini leaves from soil treated with SA.
The relative abundance of certain genera was also influenced by SA (Table 4 and Figures 2C,D). In contrast to the unamended variant (Bs), Candidatus udaeobacter was not detected in the Us + SA. Devosia and Candidatus xiphinematobacter genera were not detected in the ZuRh + SA. In turn, the relative abundance of Mucilaginibacter was six times higher in ZuRh + SA (6.588%) than in ZuRh (1.000%). SA had significant effects on the presence of several genera, especially in the roots endosphere: the relative abundance of Dyella was three times higher in ZuRo + SA (3.966%) than in ZuRo (1.000%). Although the presence of the genera Ralstonia, Rhizobium, Massilia, Luteimonas, Klebsiella, Streptomyces, and Mucilaginibacter was confirmed in ZuRo, they were not detected in ZuRo + SA. SA exerted a particularly strong effect on the dominant genus in leaves endosphere (Figure 2D), where the abundance of Pseudarthrobacter was significantly higher (61.50%) than in other variants. SA lowered the prevalence of Paenibacillus in ZuLe + SA five times (1.000%) in comparison to ZuLe (5.786%). Also, the relative abundance of Paeniglutamicibacter was four times lower in ZuLe + SA (1%) than in ZuLe (4.284%). In turn, higher abundances of Pseudomonas (13.72%), Klebsiella (11.77%), Exiguobacterium (5.138%), and Paenisporosarcina (4.417%) were observed in leaves endosphere after SA treatment (ZuLe + SA).
MCPA had little effect on the alpha diversity in unplanted soil, rhizosphere, endosphere of roots, and leaves (Table 3). Additionally, in roots endosphere, MCPA enhanced the InvSimpson and Shannon indices. However, in the leaf, MCPA treatment lowered the InvSimpson value compared to the unamended variant.
MCPA caused similar changes in the structure of the bacterial communities as SA (Figures 3A,B). The most abundant phylum in Us + MCPA was Proteobacteria (69.26%). In Us + MCPA, the relative abundance of Acidobacteriota (1.695%), Crenarchaeota (2.366%), and Verrumicrobiota (2.851%) was approximately two times lower than in Us (being: 4.124, 5.563, and 2.851%, respectively). In contrast, the abundance of Patescibacteria was two times higher in Us + SA (4.644%) than in Us (2.256%). Proteobacteria also predominated the bacterial community in ZuRh + MCPA (61.20%). In contrast, the relative abundance of Actinobacteria in ZuRh + MCPA increased by a factor of two (2.135%) in comparison to ZuRh (1.000%). A three times lower abundance of Verrumicrobiota was observed for ZuRh + MCPA (1.966%) than in ZuRh (6.469%). The relative abundance of Actinobacteria and Firmicutes in ZuLe + MCPA increased after MCPA treatment (78.90 and 18.41%, respectively), in comparison to ZuLe (14.67 and 7.10%, respectively). In contrast, the abundance of Proteobacteria from ZuLe + MCPA (77.81%) was approximately thirty times lower than in ZuLe (2.443%).
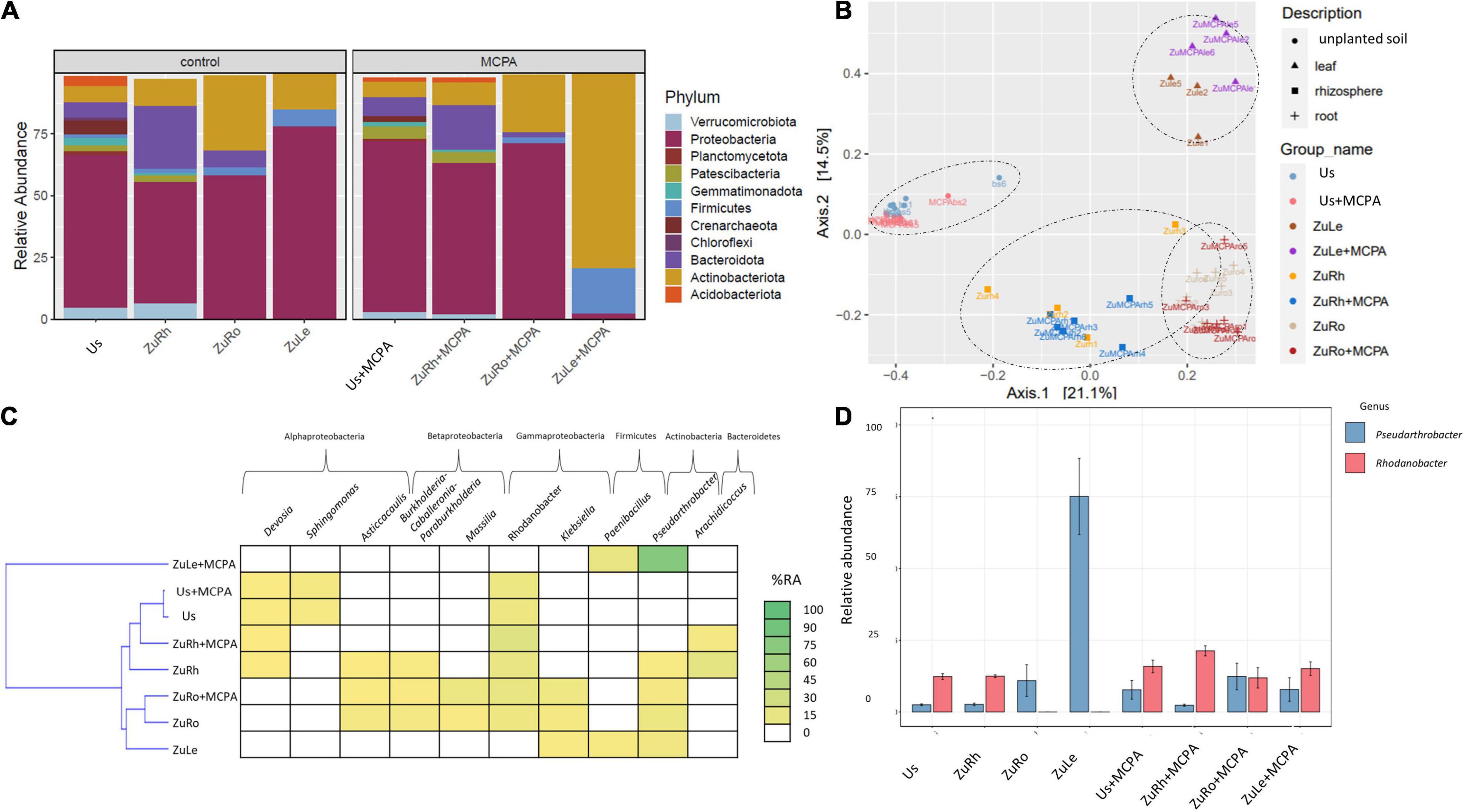
Figure 3. Bacterial community structure based on 16S rRNA gene sequence for MCPA-amended variants in comparison to untreated variants: (A) Relative abundances (%) of the bacterial community at the phylum level; (B) PcoA ordination plots of the results based on the Bray–Curtis distances of the classified 16S rRNA gene sequences; (C) Heat map of top 10 genera in each treatment; (D) Relative abundance (%) of Pseudarthrobacter and Rhodanobacter in the different treatments. Variants: Us, unplanted soil; ZuRh, zucchini rhizosphere soil; ZuRo, zucchini root; ZuLe, zucchini leaves; Us + MCPA, unplanted soil treated with MCPA; ZuRh + MCPA, zucchini rhizosphere soil treated with MCPA; ZuRo + MCPA, zucchini root from soil treated with MCPA; ZuLe + MCPA, zucchini leaves from soil treated with MCPA.
MCPA also changed the prevalence of individual genera (Table 4 and Figures 3C,D). The addition of MCPA to unplanted soil decreased the abundance of Candidatus udaeobacter by five times from 5.852% in Us + MCPA + SA to 1.000% in Us + MCPA. The genera Asticcacaulis, Burkholderia, and Candidatus xiphinematobacter were not detected in ZuRh + MCPA. In turn, the abundance of Methylophilus was six and half times higher in ZuRh + MCPA (6.510%) than in ZuRh (1.000%). Furthermore, higher values of the abundance of Rhodanobacter were observed for ZuRh + MCPA (20.52%) than in ZuRh (14.73%). In roots endosphere (ZuRo + MCPA), the abundances of Dyella and Salmonella (5.410 and 3.419%, respectively) were higher than in ZuRo (1.000 and 1.000%, respectively). In contrast, after the amendment of soil with MCPA Mucilaginibacter, Ralstonia and Rhizobium were not detected in roots endosphere (ZuRo + MCPA). MCPA had a particularly strong effect on the dominant genera in leaves endosphere (Figures 3C,D): the abundance of Pseudarthrobacter was almost eight times higher in ZuLe + MCPA (78.09%) than in ZuLe (10.12%). Furthermore, the genus Paeniglutamicibacter was not detected in ZuLe + MCPA.
Statistically significant differences in alpha diversity were found between the studied compartments, particularly regarding the InvSimpson index (Table 3). For unplanted soil, the InvSimpson was slightly lower in Us + MCPA + SA than in Bs, and for roots endosphere, the index for ZuRo + MCPA + SA was lower than in ZuRo. However, in rhizosphere and leaves, the values were higher in the MCPA-treated variants than in untreated ones. In addition, a lower value of Obs. ASV was observed in ZuRh + MCPA + SA than in ZuRh.
The simultaneous addition of MCPA and SA to the soil affected both the soil and endophytic bacterial communities (Figures 4A–D). The predominant phylum in Us + MCPA + SA was Proteobacteria (69.26%). In turn, in US + MCPA + SA Firmicutes, Planctomycetota, and Crenarchaeota were not detected. Proteobacteria was also the predominant phylum in the rhizosphere ZuRh + MCPA (61.20%). The prevalence of Bacteroidota in ZuRh + MCPA + SA (12.32%) was two times higher in ZuRh (25.23%) than in ZuRh + MCPA + SA. Also, the relative abundance of Proteobacteria in roots endosphere (ZuRo + MCPA + SA) was higher (71.14%) than the abundance of other phyla. The prevalence of Actinobacteria was two times higher in ZuRo (30.23%) than in ZuRo + MCPA + SA (15.79%). The phylum Firmicutes was not detected in ZuRo + MCPA + SA, whereas its presence was confirmed in ZuRo (3.41%). In turn, Firmicutes dominated in ZuLe + MCPA + SA. The relative abundance of Firmicutes in ZuLe + MCPA + SA (65.39%) was almost seven times higher than in ZuLe (7.097%). The abundance of Actinobacteria was two times higher in ZuLe + MCPA + SA (29.68%) than in ZuLe (14.67%).
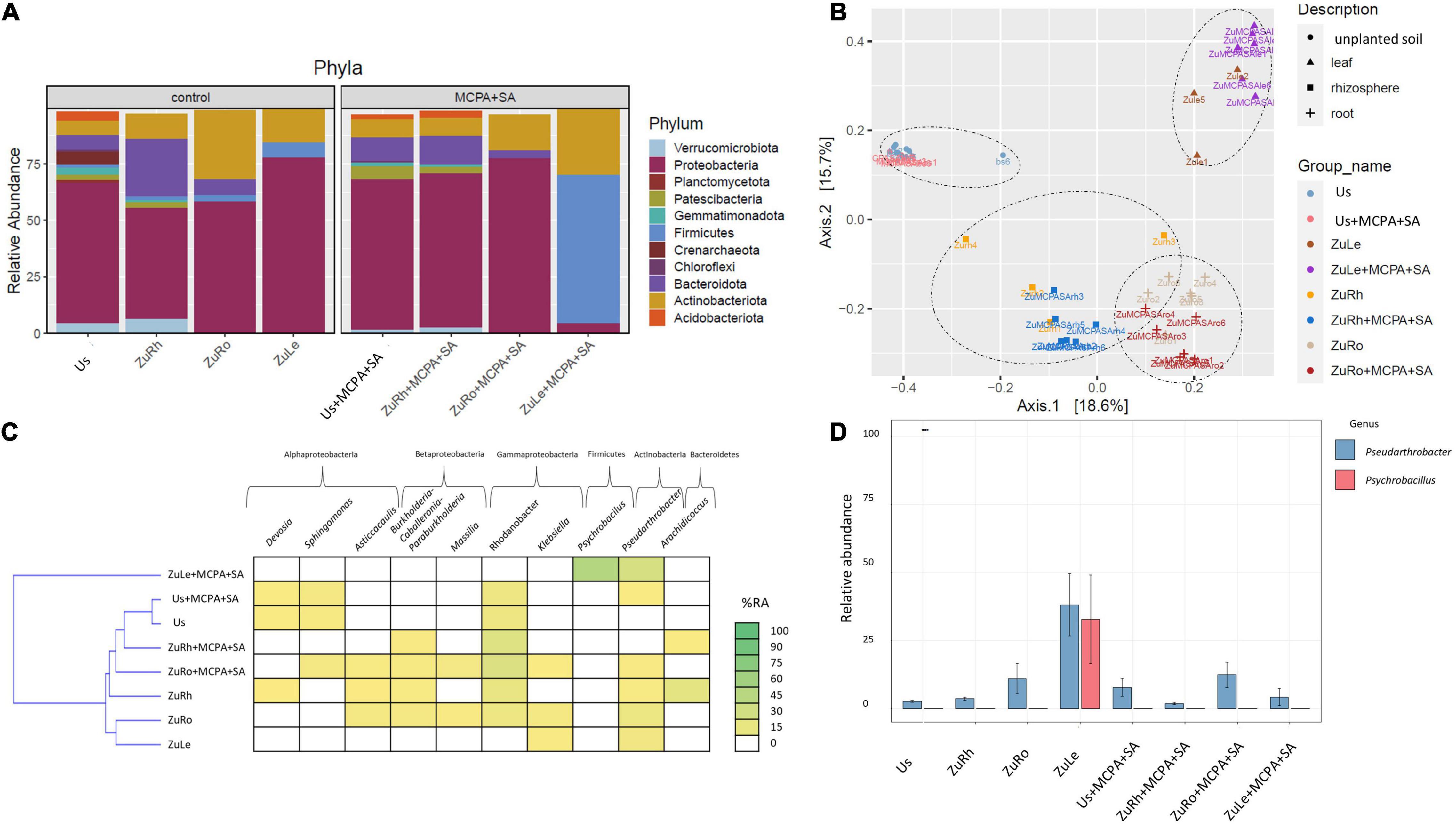
Figure 4. Bacterial community structure based on 16S rRNA gene sequence for MCPA + SA-amended variants in comparison to untreated variants: (A) Relative abundances (%) of the bacterial community at the phylum level; (B) PcoA ordination plots of the results based on the Bray–Curtis distances of the classified 16S rRNA gene sequences; (C) Heat map of top 10 genera in each treatment; (D) Relative abundance (%) of Pseudarthrobacter and Psychrobacillus in the different treatments. Variants: Us, unplanted soil; ZuRh, zucchini rhizosphere soil; ZuRo, zucchini root; ZuLe, zucchini leaves; Us + MCPA + SA, unplanted soil treated with MCPA and SA; ZuRh + MCPA + SA, zucchini rhizosphere soil treated with MCPA and SA; ZuRo + MCPA + SA, zucchini root from soil treated with MCPA and SA; ZuLe + MCPA + SA, zucchini leaves from soil treated with MCPA and SA.
The addition of MCPA + SA to the soil had substantial effects on the presence of certain genera (Table 4 and Figures 4C,D). The genera Pseudolabrys and Candidatus udaeobacter were not detected in Us + MCPA + SA. In ZuRh + MCPA + SA, the highest relative abundance was observed for Rhodanobacter (21.23%) (Figure 4C). The abundance of Methylophillus was eight times higher in ZuRh + MCPA + SA (7.953%) than in ZuRh (1.000%). In turn, Devosia, Asticcacaulis, Arachidococcus, and Candidatus xiphinematobacter genera were not detected in ZuRh + MCPA + SA. Further, the highest observed relative abundance in ZuRo + MCPA + SA was noted for Rhodanobacter (23.56%). The amendment of soil with MCPA and SA enhanced the occurrence of Sphingomonas (7.485%) and Pandoraea (3.100%) in roots endosphere (ZuRo + MCPA + SA), whereas these genera were not detected in ZuRo. Neither Ralstonia, Rhizobium, Streptomyces nor Mucilaginibacter genera were detected in roots after MCPA and SA amendment (ZuRo + MCPA + SA). In ZuLe + MCPA + SA, the genus Psychrobacillus (48.28%) predominated the bacterial community, followed by Pseudarthrobacter (21.56%) (Figure 4D). In ZuLe + MCPA + SA, the genera Pseudomonas (3.380%) and Solibacillus (3.611%) were also detected, while Klebsiella and Paeniglutamicibacter were not observed.
Considerable variations were observed in the alpha diversities of unplanted soil, rhizosphere soil, roots, and leaves endosphere (Table 3). The MANOVA analysis (Table 5) revealed that the diversity indices varied significantly in the function of the compartment (unplanted soil, rhizospheric soil, roots, and leaves endosphere). The highest diversity indices (Table 3) were observed in unplanted soil, ranging from 91.30 for Us + SA to 101.6 for Us (Inv. Simpson); from 173.3 for Us + SA up to 223.7 for Us + MCPA + SA (observed ASV); and from 4.690 for Us + SA up to 4.975 for Us + MCPA + SA (Shannon index).
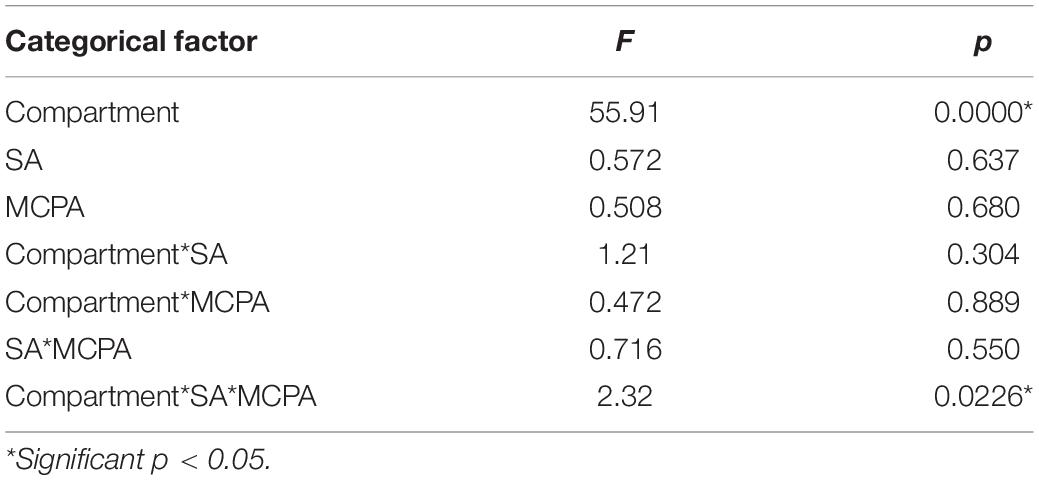
Table 5. Main effects of categorical factors (compartment vs. SA vs. MCPA treatment) on diversity indices, estimated using Multivariate Tests of Significance (MANOVA).
Slightly lower Inv. Simpson indices were observed in the rhizosphere, i.e., the highest value was found for ZuRh + MCPA (79.96) and the lowest for ZuRh (68.17). Lower diversity indices were observed in roots and leaves endosphere than in bulk and rhizospheric soil, ranging from 2.415 for ZuLe + MCPA to 39.01 for ZuRo + MCPA (Inv. Simpson), from 5.000 for ZuLe + MCPA to 94.33 for ZuRo + SA (observed ASV), and from 1.559 for ZuLe + MCPA + SA to 3.908 for ZuRo + MCPA (Shannon index).
Furthermore, the beta-diversity analysis indicated clustering (Figures 2B, 3B, 4B) of samples from different compartments. The MANOVA analysis demonstrated significant compartment specificity of the bacterial and fungal community structure (p < 0.001).
A compartment specificity was observed for the occurrence of specific genera irrespective of the treatment (Table 4). For example, the occurrence of Devosia, Sphingomonas, and Luteimonas was observed only in unplanted variants. Similarly, Asticaccaulis and Burkholderia were present in the roots endosphere of all variants. In turn, Rhodanobacter was observed in all variants of unplanted soil, rhizosphere, and roots endosphere, but not in the leaves endosphere.
Recent research has provided a comprehensive insight into the interactions between bacteria and plants in contaminated environments (Tardif et al., 2016; Thijs et al., 2018). It appears that the structure of bacterial communities in soils and plants is determined by the presence of pollutants. However, our knowledge about the effects of plant root exudates (including PSMs, such as SA) on the communities of bacteria in contaminated environments remains limited. Some studies have shown that certain phenolic compounds, such as benzoic acid, can enhance the biodegradative activity of bacteria in soil (Mandal et al., 2010; Zwetsloot et al., 2020). However, phenolic compounds can exert contrasting effects on microbial communities depending on the prevailing conditions (Zwetsloot et al., 2020).
While most studies have examined the biodegradation of phenoxy herbicides, especially 2,4-D (2,4-dichlorophenoxy acid) in soil, the most widely-used pesticide in European agriculture is MCPA. MCPA is usually sprayed as a commercial formulation in the form of amines, sodium salts, or esters to control perennial and broadleaf annual weeds (Paszko et al., 2016). Higher phenoxy herbicide (i.e., MCPA) concentrations are mostly detected in regions with ongoing intensive agriculture (Bianchi et al., 2017), although residuals of MCPA can be transported with intensive surface run-off and become a threat to non-target organisms. Monitoring studies showed that MCPA was present in 33–60% of the studied sites along the river tributaries in Poland (Zagibajło et al., 2017; Jarosiewicz et al., 2018). Thus, the development of methods for the prevention of MCPA residuals leaching is of the highest importance. Still, little attention has been paid to the potential of selected plants as phytoremediators of soils contaminated with phenoxy herbicides. The members of the Cucurbitaceae family are particularly promising candidates due to their ability to extract, translocate, and accumulate highly toxic persistent organic pollutants from soils (Hülster et al., 1994; White et al., 2003; Inui et al., 2008; Zhang et al., 2009; Wyrwicka et al., 2014; Urbaniak et al., 2016) and several cucurbit species, such as zucchini (C. pepo cv “Atena Polka”), have demonstrated resistance to MCPA (Mierzejewska et al., 2022).
Therefore, this study investigates the effects of a specific PSM (SA) and zucchini cultivation on the removal of MCPA from the soil. In unplanted MCPA-contaminated soil, amendment with SA led to a higher decrease of MCPA (30%) than in untreated soil (18%) (Figure 1). However, higher MCPA removal (50%) was observed in the planted condition amended with SA. Another study showed that the removal of 2,4-D from water, using the plant species Plectranthus neochilus, was 49% after 30 days (Ramborger et al., 2017). Furthermore, Germaine et al. (2006) demonstrated that endophyte-enhanced phytoremediation significantly improves the capacity for removal of 2,4-D. Our study showed that SA improves the removal of MCPA from unplanted soil. However, when SA application and zucchini cultivation are combined, the decrease of the herbicide concentration in the soil is significantly greater.
Although SA enhanced the removal of MCPA from soil (Figure 1), MCPA had a detrimental effect on fresh biomass (Table 2). Indeed, MCPA application was associated with 90% lower aboveground biomass in the planted variants. Phenoxy herbicides are transported to meristems, causing uncontrolled growth and consequently damaging the development of plant tissues (Grossmann, 2003). SA has been found to alleviate the toxic effects of MCPA (Mierzejewska et al., 2022), however, these studies investigated higher initial concentrations of both compounds. Kováčik et al. (2010) showed that certain phenolic acids, e.g., salicylic acid, can alleviate stress symptoms in plants. On the other hand, Zhou et al. (2014) suggested that SA can exhibit phytotoxic effects. Blum (1996) reported the concentration, composition, and synergism of individual phenolic compounds to be crucial for plant growth inhibition in the environment. Hence, we claim that despite improving MCPA-removal efficiency, the application of SA throughout the incubation time did not enhance the plant growth-promoting properties of the system.
The above-mentioned observations are as per earlier studies indicating that SA significantly enhanced the removal of MCPA from liquid cultures enriched with microorganisms derived from agricultural soil (Mierzejewska et al., 2019; Urbaniak et al., 2019a). Hence, this study also examines the effects of SA and MCPA, used individually or in combination, on the composition of bacterial communities within the soil, rhizosphere, and the zucchini plant itself.
The effects on the diversity of bacteria were found to be compartment-specific, with an interaction between SA and MCPA (Table 5). The highest diversity indices values were found for unplanted bulk and rhizospheric soil, and the lowest in leaves. Approximately, 10% lower values of diversity indices were observed after SA application (Table 3). Indeed, it has previously been reported that SA has antimicrobial activity against some bacterial strains (Shi et al., 2016). Our findings also indicate that the amendment of soil with MCPA did not affect diversity indices significantly (Table 3). Also, Ławniczak et al. (2016) found that the application of oligomeric herbicidal ionic liquids with MCPA did not cause any changes in the diversity indices. In variants amended with SA and MCPA diversity indices (i.e., Obs.ASV) were higher than in untreated variants. Also, Lipthay et al. (2004) reported higher species richness and Shannon index in herbicide-acclimated sediments. This effect can be explained by the direct and indirect effects of the applied compounds (MCPA and SA).
Formed metabolites and root exudates produced in response to stress conditions can also influence the structural diversity (Uhlik et al., 2013; Musilova et al., 2016). It is important to emphasize that interactions between the studied compartments and the application of both compounds, MCPA and SA, determined the diversity of bacteria.
After the addition of phenolic compounds (SA and MCPA), the bacterial communities in the unplanted soil, rhizospheric soil, and root endosphere were found to be dominated by Proteobacteria and differed only slightly from those of the untreated variant (Figures 2A, 3A, 4A). Also, the addition of DDE (0.0001 mg/L) to vermiculite led to the dominance of Proteobacteria in the roots endosphere of zucchini (Eevers et al., 2016). Similarly, Tejada et al. (2010) report that the use of MCPA (1.5 L/ha) did not appear to affect microbial structural diversity in soil. In contrast, it has been found that microbial communities isolated from the rhizosphere of soil contaminated with mecocrop (MCPP; 0.53 or 1.06 g/L) differed substantially from those isolated from unplanted soil (Lappin et al., 1985). The above-mentioned results show that in unplanted soil, rhizospheric soil, and roots endosphere, the amendment of soil with SA and MCPA not significantly affect the predominant taxa.
However, the application of MCPA and SA, either separately or together, considerably affected the composition of the endophytic bacterial community in leaves, although they demonstrated the lowest diversity indices (Figures 2–4). MCPA is absorbed through both leaves and roots and is translocated throughout the plant (via xylem and phloem) to the meristematic regions (Polit et al., 2014). The induced changes result in a series of biochemical and physiological processes which influence and disturb the morphology of plant roots, stems, and leaves (Grossmann, 2009). The observed changes in the composition of the plant endosphere microbiome can be due to the application of herbicides. Endophytes in plants can directly contribute to the detoxification of pollutants in plants or promote plant growth in stress conditions (Tétard-Jones and Edwards, 2016). However, the latter was not observed, since fresh weight was not higher after the simultaneous amendment of SA and MCPA. To the best of our knowledge, this is the first study showing that after the application of phenolic compounds (i.e., SA and MCPA) to soil, the most profound changes in structural diversity across the plant microbiome are observed in the leaves.
Further investigation of structural diversity showed that amendment of the soil with SA or MCPA led to the prevalence of the Actinobacteria (genus Pseudarthrobacter) in the leaves endosphere (Figures 2D, 3D). Similarly, Zhou et al. (2018) found that amendment of soil with the phenolic compound p-coumaric acid enhanced the relative abundances of Actinobacteria (i.e., Pseudarthrobacter) in the rhizosphere. Actinobacteria are described as common endophytic microorganisms, which can have multiple beneficial functions in plants, such as the production of antimicrobial agents (Musa et al., 2020) and biodegradation of petroleum and plastic compounds (Singh and Dubey, 2018). Combined MCPA and SA treatment, however, resulted in Firmicutes (genus Psychrobacillus) becoming the predominant taxon in leaves endosphere (Figure 4D). Regar et al. (2019) suggest that an increase in the abundance of Firmicutes can be a response to environmental stress such as herbicide contamination. Rangjaroen et al. (2019) mentioned that endophytic strains of Bacillus isolated from an herbicide-treated environment can enhance the resistance of plants to fungal pests. Our observations confirmed that the amendment of the soil with SA and MCPA favored the presence of Actinobacteria (genus Pseudarthrobacter) and Firmicutes (genus Psychrobacillus) in leaves.
The shifts in bacterial communities that occur after SA treatment could influence the degradative capacity against MCPA. Various genes belonging to the tfd cluster (responsible for initial steps of phenoxy herbicides biodegradation) have been identified in several taxa, e.g., Burkolderia, Pseudomonas, Achromobacter, Delftia, Bradyrhizobium, and Cupriavidus (Suwa et al., 1996; Kamagata et al., 1997; Baelum et al., 2010). Previous studies also demonstrated that SA doubled the presence of tfdA genes (Mierzejewska et al., 2019; Urbaniak et al., 2019a). In our study, the enrichment of soil with SA induced the occurrence of certain Proteobacteria genera such as Pseudomonas in leaves endosphere (Table 4), while the addition of MCPA + SA positively influenced the presence of Pseudomonas and Burkholderia in the rhizosphere (Table 4). Germaine et al. (2006) demonstrated that the endophytic strain Pseudomonas putida VM1450 enhanced the removal of 2,4-D from soil and demonstrated high biodegradative potential. In addition, the presence of Burkholderia in soils has been confirmed in our previous studies (Mierzejewska et al., 2019; Urbaniak et al., 2019a). Our findings indicate that combined MCPA + SA treatment induced the presence of Sphingomonas and Pandoraea in the roots endosphere (Table 4). Several Sphingomonas strains have been reported as being MCPA-degraders (Gözdereliler et al., 2013; Liu et al., 2013; Nielsen et al., 2013), and Pandoraea was found to substantially contribute to the degradation of lindane (HCH) in water and soil slurries (Okeke et al., 2002); however, to our knowledge, no studies have examined whether this genus was able to degrade phenoxy herbicides or related phenolic compounds. The above findings suggest that the amendment of the soil promoted the growth of specialized taxa; however, they were not the predominant taxa in the investigated compartments. Still, the functions of the identified strains need to be further confirmed by culturing techniques and molecular as well as metabolic analysis.
The above findings show that the addition of SA can enhance the removal of MCPA from unplanted soil. This removal is further enhanced by combining SA amendment with the cultivation of zucchini; although SA itself did not have a positive effect on the zucchini growth. The application of both phenolic compounds (i.e., MCPA and SA) led to changes in bacterial diversity: the highest bacterial diversity indices were observed for unplanted soil and rhizospheric soil, and the lowest in leaves. Despite the lowest diversity, the leaf endophytes were strongly affected by the addition of the studied phenolic compounds favoring the growth of the phyla Actinobacteria (especially Pseudarthrobacter spp.) and Firmicutes (especially Psychrobacillus). SA also promoted the growth of specific genera in the rhizosphere (Burkholderia, Pseudomonas, Sphingomonas) and in leaves endosphere (Pseudomonas), which were previously reported to harbor functional genes for MCPA biodegradation. Hence, it appears that SA not only influenced the structure of the bacterial communities in either MCPA-contaminated soil and zucchini, but also enhanced the bacterial degradation of the herbicide in the soil–plant system.
This is the first study to collectively examine different aspects of MCPA removal from soil: biostimulation (use of the PSM as an enhancer of MCPA removal) and phytoremediation (use of zucchini as a potential tool for removal of the herbicide) and to study the composition of soil and endophytic bacterial communities as an effect of addition of structurally related phenolic compounds (SA and MCPA). However, to disentangle the mechanisms behind the shifts in bacterial communities and their functions, additional research is needed.
The datasets presented in this study can be found in online repositories. The latest version of link to repository of our sequencing data is https://www.ncbi.nlm.nih.gov/bioproject/PRJNA809520.
EM, ST, MU, and JV designed the experimental setup and analysis and wrote the manuscript. EM and MU performed the experiments and prepared the samples. EM and ST prepared the samples for DNA extraction and sequencing and analyzed the results using bioinformatics tools. EM determined the fresh biomass of plants and analyzed the fresh biomass and MCPA removal data. KZ prepared the samples and measured the MCPA concentration in the soil. All authors agreed on the final version of the manuscript.
The Council of the National Science Centre in Poland ETIUDA 7–funded the project “Cucurbits and their plant secondary metabolites as stimulators of biological soil remediation contaminated with phenoxy herbicides” [No. 2019/32/T/NZ9/00403]. This work was also supported by the UHasselt Methusalem project 08M03VGRJ.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.882228/full#supplementary-material
Baelum, J., Jacobsen, C. S., and Holben, W. E. (2010). Comparison of 16S rRNA gene phylogeny and functional tfdA gene distribution in thirty-one different 2, 4-dichlorophenoxyacetic acid and 4-chloro-2-methylphenoxyacetic acid degraders. Syst. Appl. Microbiol. 33, 67–70. doi: 10.1016/j.syapm.2010.01.001
Bianchi, E., Lessing, G., Brina, K. R., Angeli, L., Andriguetti, N. B., Soares Peruzzo, J. R., et al. (2017). Monitoring the genotoxic and cytotoxic potential and the presence of pesticides and hydrocarbons in water of the Sinos River Basin, Southern Brazil. Arch. Environ. Contam. Toxicol. 72, 321–334. doi: 10.1007/s00244-016-0334-0
Bijnens, K., Thijs, S., Leynen, N., Stevens, V., McAmmond, B., Van Hamme, J., et al. (2021). Differential effect of silver nanoparticles on the microbiome of adult and developing planaria. Aquat. Toxicol. 230:105672. doi: 10.1016/j.aquatox.2020.105672
Blum, U., Staman, K. L., Flint, L. J., and Shafer, S. R. (2000). Induction and/or selection of phenolic acid-utilizing bulk-soil and rhizosphere bacteria and their influence on phenolic acid phytotoxicity. J. Chem. Ecol. 26, 2059–2078. doi: 10.1023/A:1005560214222
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Campos, F. M., Couto, J. A., Figueiredo, A. R., Tóth, I. V., Rangel, A. O. S. S., and Hogg, T. A. (2009). Cell membrane damage induced by phenolic acids on wine lactic acid bacteria. Int. J. Food Microbiol. 135, 144–151. doi: 10.1016/j.ijfoodmicro.2009.07.031
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6:226. doi: 10.1186/s40168-018-0605-2
Eevers, N., Hawthorne, J. R., White, J. C., Vangronsveld, J., and Weyens, N. (2018). Endophyte-enhanced phytoremediation of DDE-contaminated using Cucurbita pepo: a field trial. Int. J. Phytoremediation 20, 301–310. doi: 10.1080/15226514.2017.1377150
Eevers, N., Hawthorne, J. R. R., White, J. C. C., Vangronsveld, J., and Weyens, N. (2016). Exposure of Cucurbita pepo to DDE-contamination alters the endophytic community: a cultivation dependent vs a cultivation independent approach. Environ. Pollut. 209, 147–154. doi: 10.1016/j.envpol.2015.11.038
EURL-SRM (2015). Analysis of Acidic Pesticides Using QuEChERS (EN15662) and Acidified QuEChERS Method. Stuttgart: EURL-SRM.
EURL-SRM (2020). SANTE/12682/2019: Analytical Quality Control and Method Validation Procedures for Pesticide Residues Analysis in Food and Feed. Stuttgart: EURL-SRM.
European Committee for Standardization (2018). CSN EN 15662: Foods of Plant Origin – Multimethod for the Determination of Pesticide Residues using GC- and LC-Based Analysis Following Acetonitrile Extraction/Partitioning and Clean-up by Dispersive SPE – Modular QuEChERS-Method. Brussel: CEN (European Committee for Standardization).
Germaine, K. J., Liu, X., Cabellos, G. G., Hogan, J. P., Ryan, D., and Dowling, D. N. (2006). Bacterial endophyte-enhanced phytoremediation of the organochlorine herbicide 2,4-dichlorophenoxyacetic acid. FEMS Microbiol. Ecol. 57, 302–310. doi: 10.1111/j.1574-6941.2006.00121.x
Glick, B. R. (2010). Using soil bacteria to facilitate phytoremediation. Biotechnol. Adv. 28, 367–374. doi: 10.1016/j.biotechadv.2010.02.001
Gözdereliler, E., Boon, N., Aamand, J., De Roy, K., Granitsiotis, M. S., Albrechtsen, H.-J., et al. (2013). Comparing metabolic functionalities, community structures, and dynamics of herbicide-degrading communities cultivated with different substrate concentrations. Appl. Environ. Microbiol. 79, 367–375. doi: 10.1128/AEM.02536-12
Grossmann, K. (2003). Mediation of herbicide effects by hormone interactions. J. Plant Growth Regul. 22, 109–122. doi: 10.1007/s00344-003-0020-0
Grossmann, K. (2009). Auxin herbicides: current status of mechanism and mode of action. Pest Manage. Sci. 66, 113–120. doi: 10.1002/ps.1860
Hu, C., Zhang, Y., Tang, X., and Luo, W. (2014). PCB biodegradation and bphA1 gene expression induced by salicylic acid and biphenyl with Pseudomonas fluorescence P2W and Ralstonia eutropha H850. Polish J. Environ. Stud. 23, 1591–1598.
Hülster, A., Müller, J. F., and Marschner, H. (1994). Soil–plant transfer of polychlorinated dibenzo-p-dioxins and dibenzofurans to vegetables of the cucumber family (Cucurbitaceae). Environ. Sci. Technol. 28, 1110–1115. doi: 10.1021/es00055a021
Ignatowicz, K., and Struk-Sokołowska, J. (2004). Sezonowe wahania zanieczyszczeń agrotechnicznych w rzece Narwi ze szczególnym uwzględnieniem herbicydów fenoksyoctowych. Środkowo-Pomorskie Tow. Nauk. Ochr. Środowiska 4, 189–205.
Inui, H., Wakai, T., Gion, K., Kim, Y., and Eun, H. (2008). Chemosphere differential uptake for dioxin-like compounds by zucchini subspecies. Chemosphere 73, 1602–1607. doi: 10.1016/j.chemosphere.2008.08.013
Jarosiewicz, P., Zagibajło, K., Rudziłski, K., Miszczak, A., and Zalewski, M. (2018). “The occurence of plant protection agents in pilica basin according to crop characteristics in river catchment (in Polish),” in Proceedings of the XXIV Congress of Polish Hydrobiologists “Hydrobiology in the Face of Climate Change”, Wrocław.
Kamagata, Y., Fulthorpe, R. R., Tamura, K., Takami, H., Forney, L. J., and Tiedje, J. M. (1997). Pristine environments harbor a new group of oligotrophic 2,4- dichlorophenoxyacetic acid-degrading bacteria. Appl. Environ. Microbiol. 63, 2266–2272. doi: 10.1128/aem.63.6.2266-2272.1997
Kováčik, J., Klejdus, B., Hedbavny, J., and Bačkor, M. (2010). Effect of copper and salicylic acid on phenolic metabolites and free amino acids in Scenedesmus quadricauda (Chlorophyceae). Plant Sci. 178, 307–311. doi: 10.1016/j.plantsci.2010.01.009
Kruczek, M., Ledwożyn-Smoleń, I., Kałużny, K., Kopeć, P., Nowicka-Połeć, A., and Kaszycki, P. (2015). Pumpkin (Cucurbita sp.) as a source of health-beneficial compounds with antioxidant properties dynia (Cucurbita sp.) jako źródło prozdrowotnych związków o charakterze antyoksydacyjnym. Przem. Chem. 1, 86–90. doi: 10.15199/62.2015.6.10
Khare, E., Mishra, J., and Arora, N. K. (2018). Multifaceted interactions between endophytes and plant: developments and prospects. Front. Microbiol. 9:2732. doi: 10.3389/fmicb.2018.02732
Kusstatscher, P., Adam, E., Wicaksono, W. A., Bernhart, M., Olimi, E., Müller, H., et al. (2021). Microbiome-assisted breeding to understand cultivar-dependent assembly in Cucurbita pepo. Front. Plant Sci. 12:642027. doi: 10.3389/fpls.2021.642027
Lappin, H. M., Greaves, M. P., and Slater, J. H. (1985). Degradation of the herbicide mecoprop [2-(2-methyl-4-chlorophenoxy)propionic acid] by a synergistic microbial community. Appl. Environ. Microbiol. 49, 429–433. doi: 10.1128/aem.49.2.429-433.1985
Ławniczak, Ł, Syguda, A., Borkowski, A., Cyplik, P., Marcinkowska, K., Wolko, Ł, et al. (2016). Influence of oligomeric herbicidal ionic liquids with MCPA and Dicamba anions on the community structure of autochthonic bacteria present in agricultural soil. Sci. Total Environ. 563–564, 247–255. doi: 10.1016/j.scitotenv.2016.04.109
Lipthay, J. R., Johnsen, K., Albrechtsen, H.-J., Rosenberg, P., and Aamand, J. (2004). Bacterial diversity and community structure of a sub-surface aquifer exposed to realistic low herbicide concentrations. FEMS Microbiol. Ecol. 49, 59–69. doi: 10.1016/j.femsec.2004.02.007
Liu, Y.-J., Liu, S., Drake, H. L., and Horn, M. A. (2013). Consumers of 4-chloro-2-methylphenoxyacetic acid from agricultural soil and drilosphere harbor cadA, r/sdpA, and tfdA -like gene encoding oxygenases. FEMS Microbiol. Ecol. 86, 114–129. doi: 10.1111/1574-6941.12144
Mandal, S. M., Chakraborty, D., and Dey, S. (2010). Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 5, 359–368. doi: 10.4161/psb.5.4.10871
Matamoros, V., Arias, C. A., Nguyen, L. X., Salvadó, V., and Brix, H. (2012). Occurrence and behavior of emerging contaminants in surface water and a restored wetland. Chemosphere 88, 1083–1089. doi: 10.1016/j.chemosphere.2012.04.048
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mergeay, M., Nies, D., Schlegel, H. G., Gerits, J., Charles, P., and Van Gijsegem, F. (1985). Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 162, 328–334. doi: 10.1128/jb.162.1.328-334.1985
Mierzejewska, E., Baran, A., Tankiewicz, M., and Urbaniak, M. (2019). Removal and ecotoxicity of 2,4-D and MCPA in microbial cultures enriched with structurally-similar plant secondary metabolites. Water 11:1451. doi: 10.3390/w11071451
Mierzejewska, E., Tołoczko, W., and Urbaniak, M. (2022). The effects of syringic acid on the properties of MCPA-contaminated soil and the growth of two cucurbit species. Int. J. Phytoremediation 24, 205–214. doi: 10.1080/15226514.2021.1932727
Morton, P. A., Fennell, C., Cassidy, R., Doody, D., Fenton, O., Mellander, P., et al. (2020). A review of the pesticide MCPA in the land-water environment and emerging research needs. Wires Water 7:e1402. doi: 10.1002/wat2.1402
Murali, A., Bhargava, A., and Wright, E. S. (2018). IDTAXA: a novel approach for accurate taxonomic classification of microbiome sequences. Microbiome 6:140. doi: 10.1186/s40168-018-0521-5
Musa, Z., Ma, J., Egamberdieva, D., Abdelshafy Mohamad, O. A., Abaydulla, G., Liu, Y., et al. (2020). Diversity and antimicrobial potential of cultivable endophytic actinobacteria associated with the medicinal plant Thymus roseus. Front. Microbiol. 11:191. doi: 10.3389/fmicb.2020.00191
Musilova, L., Ridl, J., Polivkova, M., Macek, T., and Uhlik, O. (2016). Effects of secondary plant metabolites on microbial populations: changes in community structure and metabolic activity in contaminated environments. Int. J. Mol. Sci. 17:1205. doi: 10.3390/ijms17081205
Nielsen, L., and Dahllof, I. (2007). Direct and indirect effects of the herbicides glyphosate, bentazone and MCPA on eelgrass (Zostera marina). Aquat. Toxicol. 82, 47–54. doi: 10.1016/j.aquatox.2007.01.004
Nielsen, T. K., Xu, Z., Gözdereliler, E., Aamand, J., Hansen, L. H., and Sørensen, S. R. (2013). Novel insight into the genetic context of the cadAB genes from a 4-chloro-2-methylphenoxyacetic acid-degrading Sphingomonas. PLoS One 8:e83346. doi: 10.1371/journal.pone.0083346
Okeke, B. C., Siddique, T., Arbestain, M. C., and Frankenberger, W. T. (2002). Biodegradation of γ-hexachlorocyclohexane (Lindane) and α-hexachlorocyclohexane in water and a soil slurry by a Pandoraea species. J. Agric. Food Chem. 50, 2548–2555. doi: 10.1021/jf011422a
Palma, P., Matos, C., Alvarenga, P., Köck-Schulmeyer, M., Simões, I., Barceló, D., et al. (2018). Ecological and ecotoxicological responses in the assessment of the ecological status of freshwater systems: a case-study of the temporary stream Brejo of Cagarrão (South of Portugal). Sci. Total Environ. 634, 394–406. doi: 10.1016/j.scitotenv.2018.03.281
Paszko, T., Muszyński, P., Materska, M., Bojanowska, M., Kostecka, M., and Jackowska, I. (2016). Adsorption and degradation of phenoxyalkanoic acid herbicides in soils: a review. Environ. Toxicol. Chem. 35, 271–286. doi: 10.1002/etc.3212
Pawlik, M., Płociniczak, T., and Piotrowska-Seget, Z. (2015). Endophytic bacteria and their significance in environmental protection, medicine and industry (in polish). Postep. Mikrobiol. 54, 115–122.
Pereira, T., and Cerejeira, M. J. (2000). Use of microbiotests to compare the toxicity of water samples fortified with active ingredients and formulated pesticides. Environ. Toxicol. 15, 401–405.
Pilon-Smits, E. (2005). Phytoremediation. Annu. Rev. Plant Biol. 56, 15–39. doi: 10.1146/annurev.arplant.56.032604.144214
Pino, N. J., Muñera, L. M., and Peñuela, G. A. (2016). Root exudates and plant secondary metabolites of different plants enhance polychlorinated biphenyl degradation by rhizobacteria. Bioremediat. J. 20, 108–116. doi: 10.1080/10889868.2015.1124065
PNA Bio PCR Blockers (2021). mPNA &pPNA. Available online at: https://www.pnabio.com/products/PCR_blocker.htm (accessed date July, 22 2021).
Polit, J. T., Praczyk, T., Pernak, J., Sobiech, Ł, Jakubiak, E., and Skrzypczak, G. (2014). Inhibition of germination and early growth of rape seed (Brassica napus L.) by MCPA in anionic and ester form. Acta Physiol. Plant. 36, 699–711. doi: 10.1007/s11738-013-1448-x
Qin, H., Brookes, P. C., Xu, J., and Feng, Y. (2014). Bacterial degradation of Aroclor 1242 in the mycorrhizosphere soils of zucchini (Cucurbita pepo L.) inoculated with arbuscular mycorrhizal fungi. Environ. Sci. Pollut. Res. 21, 12790–12799. doi: 10.1007/s11356-014-3231-y
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramborger, B. P., Ortis Gularte, C. A., Rodrigues, D. T., Gayer, M. C., Sigal Carriço, M. R., Bianchini, M. C., et al. (2017). The phytoremediation potential of Plectranthus neochilus on 2,4-dichlorophenoxyacetic acid and the role of antioxidant capacity in herbicide tolerance. Chemosphere 188, 231–240. doi: 10.1016/j.chemosphere.2017.08.164
Rangjaroen, C., Lumyong, S., Sloan, W. T., and Sungthong, R. (2019). Herbicide-tolerant endophytic bacteria of rice plants as the biopriming agents for fertility recovery and disease suppression of unhealthy rice seeds. BMC Plant Biol. 19:580. doi: 10.1186/s12870-019-2206-z
Regar, R. K., Gaur, V. K., Bajaj, A., Tambat, S., and Manickam, N. (2019). Comparative microbiome analysis of two different long-term pesticide contaminated soils revealed the anthropogenic influence on functional potential of microbial communities. Sci. Total Environ. 681, 413–423. doi: 10.1016/j.scitotenv.2019.05.090
Rheinheimer dos Santos, D., Monteiro de Castro Lima, J. A., Paranhos Rosa de Vargas, J., Camotti Bastos, M., Santanna dos Santos, M. A., Mondamert, L., et al. (2020). Pesticide bioaccumulation in epilithic biofilms as a biomarker of agricultural activities in a representative watershed. Environ. Monit. Assess. 192:381. doi: 10.1007/s10661-020-08264-8
Rippy, M. A., Deletic, A., Black, J., Aryal, R., Lampard, J.-L., Tang, J. Y.-M., et al. (2017). Pesticide occurrence and spatio-temporal variability in urban run-off across Australia. Water Res. 115, 245–255. doi: 10.1016/j.watres.2017.03.010
Sauvêtre, A., and Schroder, P. (2015). Uptake of carbamazepine by rhizomes and endophytic bacteria of Phragmites australis. Front. Plant Sci. 6:83. doi: 10.3389/fpls.2015.00083
Shi, C., Sun, Y., Zheng, Z., Zhang, X., Song, K., Jia, Z., et al. (2016). Antimicrobial activity of syringic acid against Cronobacter sakazakii and its effect on cell membrane. Food Chem. 197, 100–106. doi: 10.1016/j.foodchem.2015.10.100
Singer, A. C. (2006). “The chemical ecology of pollutant biodegradation: bioremediation and phytoremediation from mechanistic and ecological perspectives,” in Phytoremediation Rhizoremediation, eds M. Mackova, D. Dowling, and T. Macek (Dordrecht: Springer). doi: 10.1007/978-1-4020-4999-4
Singer, A. C., Crowley, D. E., and Thompson, I. P. (2003). Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 21, 123–130. doi: 10.1016/S0167-7799(02)00041-0
Singh, R., and Dubey, A. K. (2018). Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 9:1767. doi: 10.3389/fmicb.2018.01767
Suwa, Y., Wright, A. D., Fukumori, F., Nummy, K. A., Hausinger, R. P., Holben, W. E., et al. (1996). Characterization of a chromosomally encoded 2,4-dichlorophenoxyacetic acid (2,4-D)/alpha-ketoglutarate dioxygenase from Burkholderia sp. RASC. Appl. Environ. Microbiol. 62, 2464–2469. doi: 10.1128/aem.62.7.2464-2469.1996
Tardif, S., Yergeau, É, Tremblay, J., Legendre, P., Whyte, L. G., and Greer, C. W. (2016). The willow microbiome is influenced by soil petroleum-hydrocarbon concentration with plant compartment-specific effects. Front. Microbiol. 7:1363. doi: 10.3389/fmicb.2016.01363
Tejada, M., García-Martínez, A. M., Gómez, I., and Parrado, J. (2010). Application of MCPA herbicide on soils amended with biostimulants: short-time effects on soil biological properties. Chemosphere 80, 1088–1094. doi: 10.1016/j.chemosphere.2010.04.074
Tétard-Jones, C., and Edwards, R. (2016). Potential roles for microbial endophytes in herbicide tolerance in plants. Pest Manage. Sci. 72, 203–209. doi: 10.1002/ps.4147
Thijs, S., Sillen, W., Truyens, S., Beckers, B., van Hamme, J., van Dillewijn, P., et al. (2018). The sycamore maple bacterial culture collection from a TNT polluted site shows novel plant-growth promoting and explosives degrading bacteria. Front. Plant Sci. 9:1134. doi: 10.3389/fpls.2018.01134
Uhlik, O., Musilova, L., Ridl, J., Hroudova, M., Vlcek, C., Koubek, J., et al. (2013). Plant secondary metabolite-induced shifts in bacterial community structure and degradative ability in contaminated soil. Appl. Microbiol. Biotechnol. 97, 9245–9256. doi: 10.1007/s00253-012-4627-6
Urbaniak, M., Baran, A., Lee, S., and Kannan, K. (2020). Utilization of PCB-contaminated Hudson River sediment by thermal processing and phytoremediation. Sci. Total Environ. 738:139841. doi: 10.1016/j.scitotenv.2020.139841
Urbaniak, M., Mierzejewska, E., and Tankiewicz, M. (2019a). The stimulating role of syringic acid, a plant secondary metabolite, in the microbial degradation of structurally-related herbicide, MCPA. PeerJ 7:e6745. doi: 10.7717/peerj.6745
Urbaniak, M., Wyrwicka, A., Siebielec, G., Siebielec, S., Kidd, P., and Zieliński, M. (2019b). The application of different biological remediation strategies to PCDDs/PCDFs contaminated urban sediments. Water 11:1962. doi: 10.3390/w11101962
Urbaniak, M., Wyrwicka, A., Zieliński, M., and Mankiewicz-Boczek, J. (2016). Potential for phytoremediation of PCDD/PCDF-contaminated sludge and sediments using Cucurbitaceae plants: a pilot study. Bull. Environ. Contam. Toxicol. 97, 401–406. doi: 10.1007/s00128-016-1868-6
Walters, W., Hyde, E. R., Berg-Lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2016). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009-15. doi: 10.1128/mSystems.00009-15
White, J. C., Wang, X., Gent, M. P. N., Iannucci-Berger, W., Eitzer, B. D., Schultes, N. P., et al. (2003). Subspecies-level variation in the phytoextraction of weathered p,p’ -DDE by Cucurbita pepo. Environ. Sci. Technol. 37, 4368–4373. doi: 10.1021/es034357p
Wu, T., Xu, J., Xie, W., Yao, Z., Yang, H., Sun, C., et al. (2018). Pseudomonas aeruginosa L10: a hydrocarbon-degrading, biosurfactant-producing, and plant-growth-promoting endophytic bacterium isolated from a reed (Phragmites australis). Front. Microbiol. 9:1087. doi: 10.3389/fmicb.2018.01087
Wyrwicka, A., Steffani, S., and Urbaniak, M. (2014). The effect of PCB-contaminated sewage sludge and sediment on metabolism of cucumber plants (Cucumis sativus L.). Ecohydrol. Hydrobiol. 14, 75–82. doi: 10.1016/j.ecohyd.2014.01.003
Wyrwicka, A., and Urbaniak, M. (2016). The different physiological and antioxidative responses of zucchini and cucumber to sewage sludge application. PLoS One 11:e0157782. doi: 10.1371/journal.pone.0157782
Wyrwicka, A., Urbaniak, M., and Przybylski, M. (2019). The response of cucumber plants (Cucumis sativus L.) to the application of PCB-contaminated sewage sludge and urban sediment. PeerJ 7:e6743. doi: 10.7717/peerj.6743
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zagibajło, K., Sikorski, P., and Miszczak, A. (2017). Control of Plants Protection Agents Residuals in Surface Waters (in Polish). Available online at: http://www.inhort.pl/files/aktualnosci/2017/nauka_praktyce/materialy/2.5_Poster_Kontrola_woda.pdf (accessed July 21, 2020).
Zhang, H., Chen, J., Ni, Y., Zhang, Q., and Zhao, L. (2009). Uptake by roots and translocation to shoots of polychlorinated dibenzo-p-dioxins and dibenzofurans in typical crop plants. Chemosphere 76, 740–746. doi: 10.1016/j.chemosphere.2009.05.030
Zhou, X. G., Wu, F. Z., and Xiang, W. S. (2014). Syringic acid inhibited cucumber seedling growth and changed rhizosphere microbial communities. Plant Soil Environ. 60, 158–164.
Zhou, X., and Wu, F. (2012). Effects of amendments of ferulic acid on soil microbial communities in the rhizosphere of cucumber (Cucumis sativus L.). Eur. J. Soil Biol. 50, 191–197. doi: 10.1016/j.ejsobi.2012.03.001
Zhou, X., Zhang, J., Pan, D., Ge, X., Jin, X., Chen, S., et al. (2018). p-Coumaric can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol. Fertil. Soils 54, 363–372. doi: 10.1007/s00374-018-1265-x
Keywords: 16S rRNA gene amplicons, 4-chloro-2-methylphenoxyacetic acid, zucchini, microbiome, syringic acid
Citation: Mierzejewska E, Urbaniak M, Zagibajło K, Vangronsveld J and Thijs S (2022) The Effect of Syringic Acid and Phenoxy Herbicide 4-chloro-2-methylphenoxyacetic acid (MCPA) on Soil, Rhizosphere, and Plant Endosphere Microbiome. Front. Plant Sci. 13:882228. doi: 10.3389/fpls.2022.882228
Received: 23 February 2022; Accepted: 02 May 2022;
Published: 31 May 2022.
Edited by:
Stefano Castiglione, University of Salerno, ItalyReviewed by:
Pankaj Bhatt, Purdue University, United StatesCopyright © 2022 Mierzejewska, Urbaniak, Zagibajło, Vangronsveld and Thijs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elżbieta Mierzejewska, ZWx6YmlldGEubWllcnplamV3c2thQGVkdS51bmkubG9kei5wbA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.