- College of Horticulture, Shenyang Agricultural University, Shenyang, Liaoning, China
Clubroot, caused by Plasmodiophora brassicae, is a major disease of crucifers. Effector proteins are important virulence factors in host recognition of pathogens and the interactions between pathogens and hosts. Secretory proteins, as effector candidates, have been studied in the interaction between Plasmodiophora brassicae and its hosts. In this study, 518 secretary proteins were screened from the Plasmodiophora brassicae genome. A total of 63 candidate effectors that induce or suppress cell death were identified using agroinfiltration-mediated transient expression in Nicothiana benthamiana. The candidate effectors, Pb4_102097 and Pb4_108104 showed high expressing level in the stage of rest spore maturity, could induce cell death and were associated with H2O2 accumulation in N. benthamiana leaves. In addition, 55 candidate effectors that could suppress BAX (Bcl-2-associated X protein) induced cell death, and 21 out of which could suppress the immunity caused by bacterial pathogen Pseudomonas syringae pv. tomato strain DC3000 expressing avrRps4 in Arabidopsis. Based on the expression pattern in different stages, 28 candidate effectors showed high expression levels during the primary and secondary infection stage. Five candidate effectors containing the RXLR motif functioned in the cytoplasm and cell membrane.
Introduction
Plants rely on innate immunity and the transmission of systemic signals from pathogen infection sites to resist pathogen invasion (Dangl and Jones, 2001; Chisholm et al., 2006; Jones and Dangl, 2006). Pathogens have conserved pathogen-associated molecular patterns (PAMPs), such as flagellin, that lead to PAMP-triggered immunity (PTI). Pathogens can secrete effector proteins targeted to plant apoplasts or delivered inside the host cytoplasm to suppress PTI and contribute to successful infection. Plants have an internal immune receptor-disease resistance gene (R-gene) encoding a nucleotide-binding leucine-rich repeat (NB-LRR) protein to recognize effectors and induce defense responses. These responses include programmed cell death (PCD) and reactive oxygen species (ROS) bursts and are known as effector-triggered immunity (ETI). Plasmodiophora brassicae is a well-adapted pathogen of Brassica but PTI and ETI are not well-characterized in the host-P. brassicae pathosystem (Pérez-López et al., 2018).
P. brassicae is an intracellular obligate biotrophic plant pathogen responsible for clubroot disease in the Brassicaceae (Kageyama and Asano, 2009). The life cycle of P. brassicae has three stages. In the soil survival stage, resting spores germinate to primary zoospores under optimal conditions. In the primary infection stage, primary zoospores colonize root hairs and form primary plasmodia and cleave into zoosporangia. Secondary zoospores released from zoosporangia penetrate the cortical tissues. The secondary infection develops into secondary plasmodia, which proliferate and produce characteristic swollen gall or club-shaped plant roots (Kageyama and Asano, 2009; Feng et al., 2012). The infection stages are critical periods for the host to recognize the pathogen and establish an immune defense system (McDonald et al., 2014). However, the recognition mechanism and interaction between P. brassicae and hosts harboring clubroot resistance genes is unknown.
To facilitate host plant colonization many pathogens secrete proteins and other molecules, collectively known as effectors, in the hosts to disable the plant defenses (Hogenhout et al., 2009). Many effectors have been characterized in bacteria, fungi, oomycetes and nematodes using agroinfiltration of Nicotiana benthamiana (Pitino et al., 2016). The development of bioinformatics and ultra-deep sequencing of plant pathogenic microbes has enabled genome-wide putative effectors genes to be cataloged based on special motifs (Huang et al., 2005; Liu et al., 2014). The basic criteria used to identify candidate secreted effector proteins include proteins with a signal peptide (within the initial 60 amino acids at the N-terminus), no trans-membrane domains and a size of 300 to 450 amino acids (Pérez-López et al., 2020). Many predicted effectors in each pathogen can be identified using bioinformatics analysis but assessing the functions of the candidate effectors is challenging. In the combined secretome of Plasmopara viticola, 51 predicted effectors with the RXLR motif were identified and 23 fully suppressed programmed cell death that was elicited by cell death-inducing proteins in N. benthamiana (Ling et al., 2015; Xiang et al., 2016). In fungi, 18 of 30 randomly selected putative effectors of Ustilaginoidea virens suppressed the Burkholderia glumae-triggered hypersensitive reaction (HR) at different levels in N. benthamiana (Zhang et al., 2014). In the cereal cyst nematode, Heterodera avenae, 95 candidate effector genes were evaluated and 78 effectors suppressed BT-PCD in N. benthamiana leaves (Lin et al., 2016).
A limited number of P. brassicae effectors have been functionally characterized based on the genome of sequences (Rolfe et al., 2016). A total of 553 secreted proteins in P. brassicae isolate e3 were predicted by bioinformatics analysis. A secretory methyltransferase can methylate salicylic acid and alter host susceptibility to P. brassicae (Schwelm et al., 2015). During primary infection, 23 of 33 secretory proteins suppressed mouse Bcl-2-associated X protein (BAX)-induced cell death and two effectors could induce cell death in N. benthamiana (Chen et al., 2019). And endomembrane-targeting P. brassicae effectors were reported to modulate PAMP and triggered immune responses in plants (Hossain et al., 2021). However, the other putative effectors at different infection stages and isolates have not been identified and characterized.
In this study, secreted protein genes were initially screened and predicted by bioinformatics software. A total of 63 candidate effectors of P. brassicae were selected and cloned. The capacities of these secretory proteins to induce or suppress cell death were assessed. The results of these experiments helped to reveal the pathogenic mechanism of P. brassicae and its interactions with host plants.
Materials and methods
Plant materials, Plasmodiophora brassicae isolate, and culture conditions
Nicotiana benthamiana and susceptible Chinese cabbage ‘91–12’ (Brassica rapa) were grown at 24°C and a 16:8 h (L:D) photoperiod. N. benthamiana was used for transient agro-Infiltration assays. The susceptible ‘91–12’ was inoculated with P. brassicae and the infected root was collected for P. brassicae RNA extraction. The P. brassicae isolate used in this study was collected from a B. rapa field located in Xinmin, Liaoning Province, China. This pathotype was Pb4 based on the result of the Williams system. The club roots were stored at −80°C until used.
Bioinformatic identification of Plasmodiophora brassicae secreted effector candidates
The putative effector genes were mined from the genome sequence of P. brassicae (PRJNA851541). The candidate secreted effector proteins were screened according to the basic criteria: proteins with a signal peptide, no trans-membrane domains and size less than 450 amino acids. The N-terminal signal peptides were predicted using SignalP 4.11 with Default D-cutoff values (Petersen et al., 2011). The proteins with a transmembrane domain or mitochondrial signal peptides were removed after analyzing with TMHMM and TargetP (Krogh et al., 2001; Emanuelsson et al., 2007), respectively. Proteins less than 450 amino acids, with signal peptides but lacking any transmembrane domains were regarded as candidate secreted effectors. The PFAM2 (Finn et al., 2014) online software was employed to characterize the candidate effector domains of P. brassicae.
RNA extraction, cDNA synthesis, and RT-PCR expression analysis
Tissue samples from both P. brassicae inoculated or non-inoculated B. rapa ‘91–12’ roots at 5, 14, 21 and 28 DPI (days post inoculation) were collected in liquid nitrogen. Total RNA was extracted using Monzol (Monad, SuZhou, China). cDNA synthesis was carried out using MonScript™ RTIII All-in-One Mix with dsDNase kit (Monad, SuZhou, China) following manufacturer instructions. RT-PCR was performed using SuperReal PreMix Plus (SYBR Green) in a 20 μl final volume. Information on primers is provided in Supplementary Table S1. GraphPad Prism (Swift, 1997) software was employed to construct the heat map based on the expression pattern in different stages.
RNA extraction and plasmid construction
RNA were extracted from P. brassicae-infected roots using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s recommended protocol. First-strand cDNA was synthesized using an RT-PCR system (Promega, Madison, WI, United States) following the manufacturer’s instructions. Primers were designed according to the instructions on the In-Fusion® HD Cloning kit based on the coding sequence in Supplementary Table S2. The PCR amplifications were purified with TIANGEN DNA purification kit (DP214), and cloned to the vector pGR106. BAX and GFP genes were also cloned to vector pGR106 according to the method described above.
Subcellular localization
The PCR-generated open reading frame of the candidate effector genes without stop codons was cloned in-frame upstream of the GFP gene in the binary potato virus X (PVX) vector pGR106-GFP. The constructed vectors were transformed in Agrobacterium GV3101 and cultivated with LB medium supplemented with 50 μg ml−1 of rifampicin and 50 μg ml−1 kanamycin and then used to transform N. benthamiana for transient expression studies. GFP signal was detected at room temperature after 24 h of expression using confocal fluorescence microscopy (Zeiss, LSM510 Meta, Carl Zeiss Germany).
Agrobacterium co-infiltration and cell death assay
Agroinfiltration-mediated transient expression in N. benthamiana was performed as described by Bourras et al. (2015). The constructed vectors connected with the putative effector ORF sequence (with signal peptide) were transformed into Agrobacterium tumefaciens strain GV3101. To identify the putative effectors that induced cell death or suppressed cell death triggered by BAX, the OD600 value of the A. tumefaciens suspension was adjusted to 0.4, and A. tumefaciens carrying pGR106-BAX was injected 12 h after the injection of A. tumefaciens carrying putative effector genes. The phenotypic character of injection sites was observed after 4–5 days of agro-infiltration. Agrobacterium cultures carrying pGR106-GFP or pGR106-BAX were infiltrated in parallel as controls.
Detection of H2O2 accumulation in the injection sites
To clarify the accumulation of reactive oxygen species (ROS) induced by the candidate effectors in the process of Agrobacterium inducing N. benthamiana leaves (Thordal-Christensen et al., 1997). The accumulation of H2O2 was detected by 3,3′-diaminobenzidine (DAB). The N. benthamiana leaves were harvested at 3 DPI with A. tumefaciens, and soaked in 1 mg/ml DAB solution at room temperature for 3 days. Then, the leaves were discolored by boiling in 99.9% ethanol for 25 min, and transparency with saturated trichloroacetic aldehyde solution and photographed.
Plasmodiophora Syringae inoculation infection
The P. syringae strain Pst DC3000 (AvrRps4) was cultivated in King’s B medium containing rifampicin (50 μg/ml). After overnight culture with shaking, the bacteria were harvested by centrifugation at 4,000 rpm/min, washed, resuspended in 10 mM MgCl2, and diluted to the required density. Pst DC3000 (AvrRps4) was hand-infiltrated into the abaxial air space of the 4-week-old leaves 12 h after the injection of A. tumefaciens carrying putative effector genes or GFP as control. And the seedlings were cultured for 3 days for evaluate the bacterial populations. Six leaves were selected for evaluate for bacterial disease assay. And the methods followed the procedures described by Yuan and Xin (2021).
Results
Sixty-three putative Plasmodiophora brassicae effectors were selected
The genomic DNA of rest spore DNA from P. brassicae strain used in this study was extracted and re-sequenced with PacBio technic. The 25.25 MB genome was de novo assembled into 28 scaffolds with an N50 size of 1.23 MB. Total 13,036 genes were predicted with Augustus software (PRJNA851541). In order to mine the putative effectors, all the annotated genes were screened with SignalP 4.1, TMHMM and TargetP software. A total of 518 secreted proteins with a signal peptide with no trans-membrane domains and a size of less than 450 amino acids were identified and assigned as candidate effectors (Supplementary Table S2). And 449 proteins showed high similarity with the annotated proteins of P. brassicae. Another 17 proteins were homologous with other organism, such as Pseudomonas, Chlorobium, and Acanthamoeba castellanii. The left proteins were probably P. brassicae-specific, which have no assigned function or information in NCBI database (Supplementary Table S2).
Among these possible effectors, 225 had annotated domains, including Chitin-binding domain, Leucine Rich repeats Pkinase domain after blast against the pfam database (Supplementary Table S3). The number of candidate effectors containing the RXLR, LXAR, and LXLFLAK motifs were identified as effectors in some oomycetes and fungi was 28, 11, and 12, respectively (Supplementary Table S3). Considering the special character of P. brassicae, total 63 candidate effectors with different domains, containing an RXLR, LXAR, LXLFLAK motif or even only signal peptide were broad selected for further experimental verification (Table 1).
Two candidate effectors can induce cell death and H2O2 accumulation in Nicothiana benthamiana
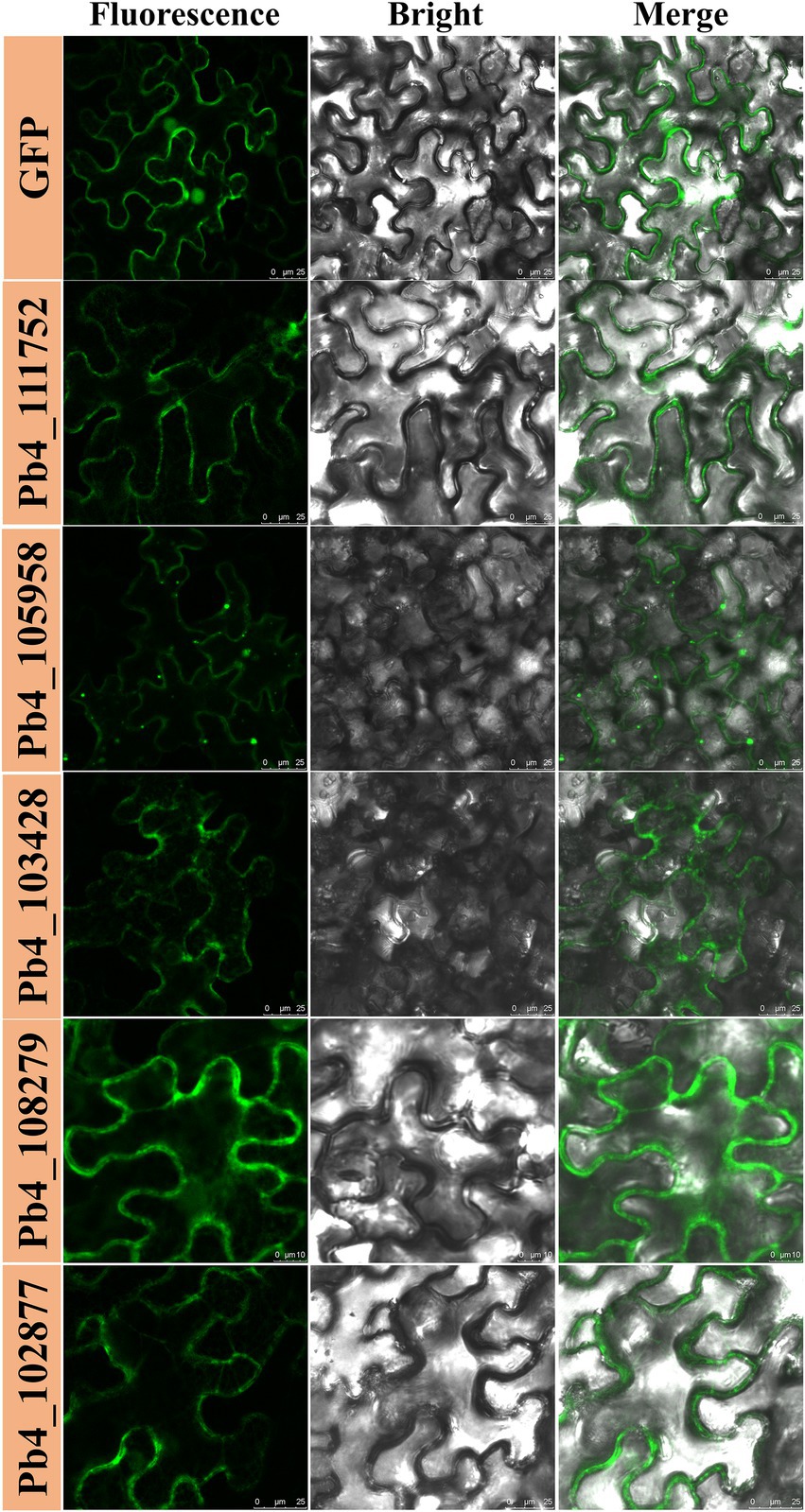
To determine if any of the 63 proteins could induce cell death, all the candidate effector genes were transiently expressed in the leaves of N. benthamiana with the Agrobacterium-mediated plant virus transient expression method. Leaf necrosis was observed at 7 DPI. Necrosis was only found on the N. benthamiana leaves expressing the candidate effectors encoded by the Pb4_102097 and Pb4_108104 genes but not in other putative effectors, et Pb4_110503 (Figure 1; Supplementary Figure S1). These data indicated that the two candidate effectors could strongly activate the immune system of N. benthamiana. However, the degree of cell death was lower than the positive control (leaves expressing BAX), which indicated that the proteins encoded by these two genes induced cell death in N. benthamiana less effectively than BAX Although a slight yellowing phenotype was found in negative control (leaves expressing eGFP), no obvious necrosis was found.
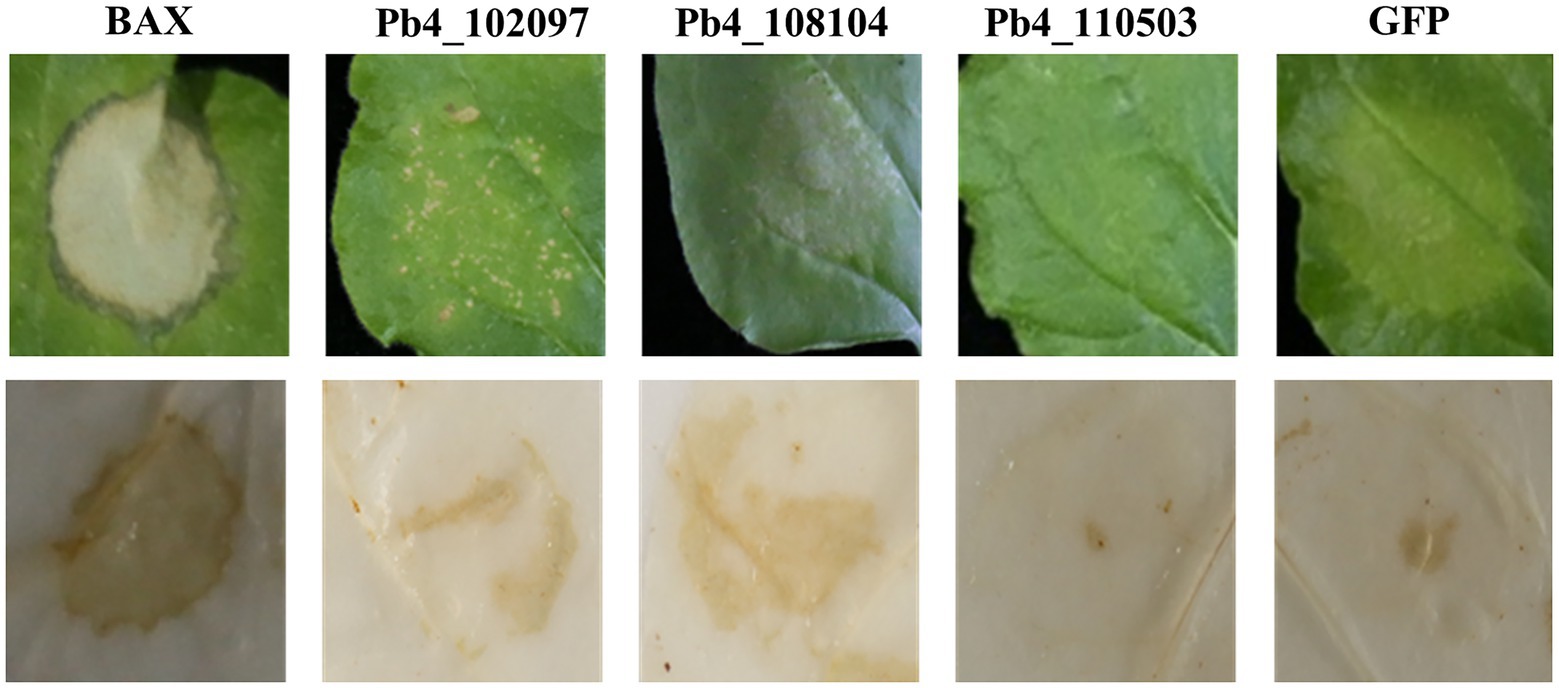
Figure 1. Putative effectors of Plasmodiophora brassicae induced programmed cell death and H2O2 accumulation in Nicothiana benthamiana. Necrosis and H2O2 accumulation in N. benthamiana leaves expressing BAX, Pb4_102097, Pb4_108104, Pb4_110503, and GFP (from left to right). HR cell death were observed four to five days after the infiltration.
Cell death induced by effectors is often associated with H2O2 accumulation. N. benthamiana leaves expressing Pb4_102097, Pb4_108104, Pb4_110503, BAX, and eGFP were harvested for DAB staining. Brown substances showed that H2O2 accumulation was present in leaves expressing Pb4_102097, Pb4_108104, and BAX but not in leaves expressing eGFP and Pb4_110503 (Figure 1). Leaves expressing Pb4_102097, Pb4_108104 were stained a light color compared with leaves expressing BAX. These results demonstrated that both Pb4_102097 and Pb4_108104 could induce cell death and produce H2O2 accumulation.
Twenty-one candidate effectors suppressed the immunity induced by BAX in Nicothiana benthamiana and avrRps4 in Arabidopsis
To identify all the remaining 61 effectors’ possible roles in suppressing plant immunity, A. tumefaciens carrying pGR106-BAX was injected 12 h after the injection of A. tumefaciens carrying putative effector genes. Then, the presence of necrosis on N. benthamiana leaves was observed and the accumulation of H2O2 was determined. Severe necrosis was observed on the positive control leaves, which were injected with Agrobacterium carrying pGR-GFP and pGR-BAX, respectively. Necrosis was observed only in N. benthamiana leaves expressing Pb4_102877, Pb4_107399, Pb4_108186, Pb4_103507, and Pb4_108519 (Figure 2A). However, the degree of cell death and accumulation of H2O2 in the leaves expressing Pb4_103507, Pb4_108519 were lower than the positive control (Figure 2A). In contrast, the level of cell death of Pb4_102877, Pb4_107399, and Pb4_108186 were greater than the positive control (Figure 2A). The accumulation of H2O2 was also greater than the positive control. This result indicated that these putative effectors above could not inhibit the plant immunity induced by BAX (Figure 2A). However, 55 putative effectors could strongly suppress the cell death and accumulation of H2O2 induced by BAX, such as Pb4_108437 and Pb4_108237 (Figure 2A; Supplementary Figure S2).
Figure 2. Analysis the putative effectors function in suppressing the immunoreaction in Nicothiana benthamiana and Arabidopsis. (A) Symptoms of transient expression of putative effectors in N. benthamiana leaves. The red font were the effectors which could not suppress BAX-induced cell death. The black font were the effectors significantly suppress BAX-induced cell death. HR cell death were observed four to five days after the infiltration. (B) Bacterial growth quantification of Pst DC3000 (AvrRps4) in the Arabidopsis leaves expressing the putative effector. 4-week-old plants were infiltrated with OD600 = 0.0001 after the injection of Agrobacterium tumefaciens carrying putative effector genes or GFP as control and the samples were collected at 0 (yellow bars) and 3 dpi (green bars) for assay. Error bars represent SD of the mean of six samples. Significance difference between treatment and control groups (t-test): *p < 0.05; **p < 0.01.
In order to confirm the responsibility of these 55 putative effectors to plant immunity, bacteria growth assay was explored with Pst DC3000 (AvrRps4) in Arabidopsis leaves. The amount of bacteria growth in the leaves expression the 28 putative effectors were higher than that of control (p < 0.05), 21 of 28 showed significant difference (p < 0.01, Figure 2B; Supplementary Figure S3). However, there were no significant in the number of bacteria present in the leaves expression the left 7 effectors compared with control. The which suppressed the cell death induced by BAX in N. benthamiana leaves were significant higher than that of control leaves (p < 0.01). These results demonstrated that the majority of candidate effectors play a role in suppressing host immunity.
Expression characteristics of candidate effectors regulating plant defenses
Transcription levels of the 63 putative effectors were evaluated at 5, 14, 24, and 28 days post inoculation of B. rapa. The putative effectors were divided into three categories based on expression pattern (Figure 3A). During primary infection, 13 effector genes showed high expression levels. Of these, 11 continued to decrease up to 28 DPI, except for Pb4_110950 and Pb4_109005. During the second infection, the expression levels of 16 effectors, including Pb_106352, Pb_109599 and Pb_110503, increased significantly at 7 DPI, then gradually decreased after 14 DPI. The majority of effector expression levels, including Pb_102097 and Pb_108104, increased during resting spore formation (after 14 DPI; Figure 3B).
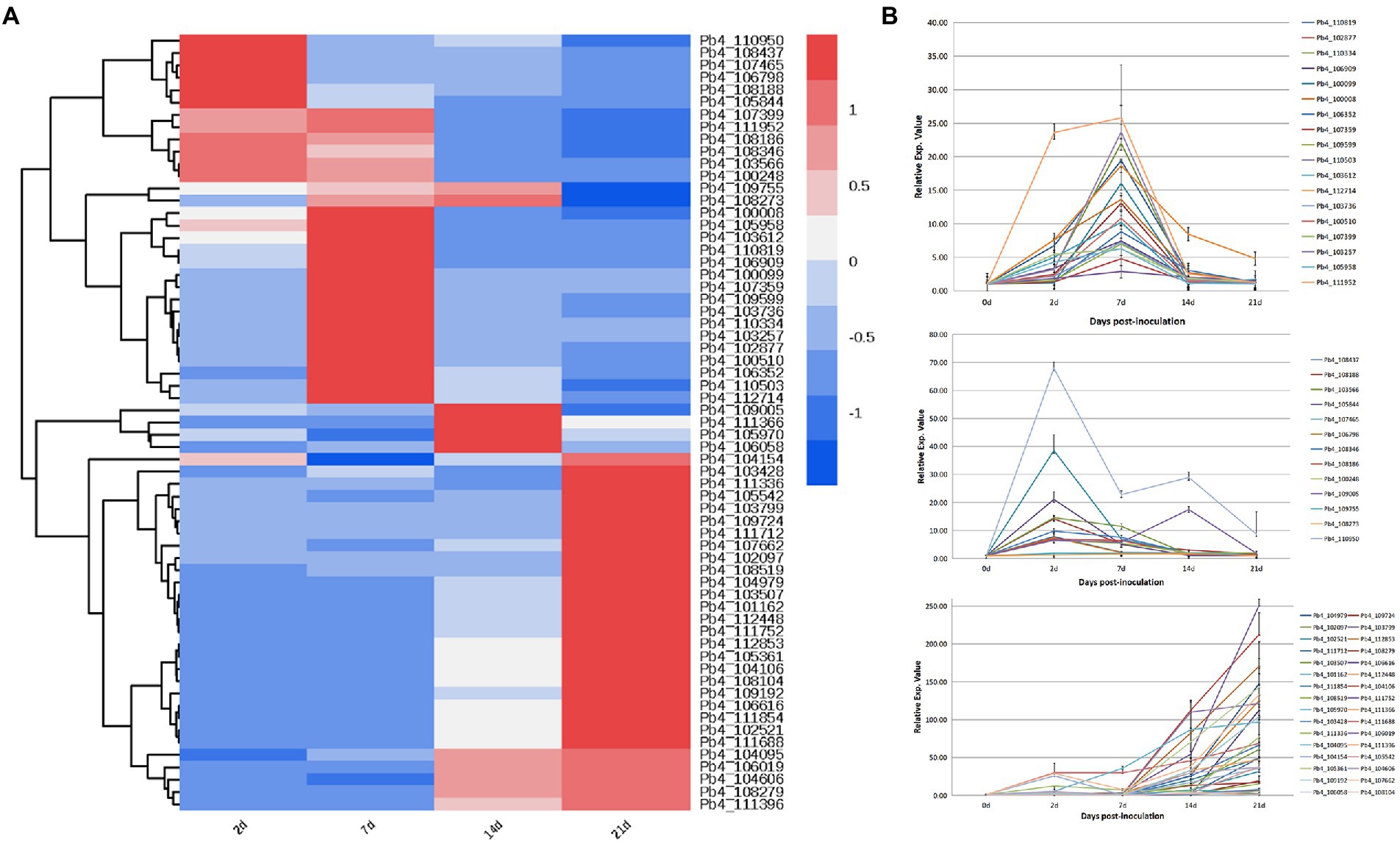
Figure 3. Expression profiles of putative effector genes in the root samples of Brassica rapa infected with P. brassicae. (A) Cluster analysis heat map of all P. brassicae effector genes. (B) The expression level of these effector genes in different categories.
Subcellular localization of nine RXLR candidate effectors in Nicothiana benthamiana
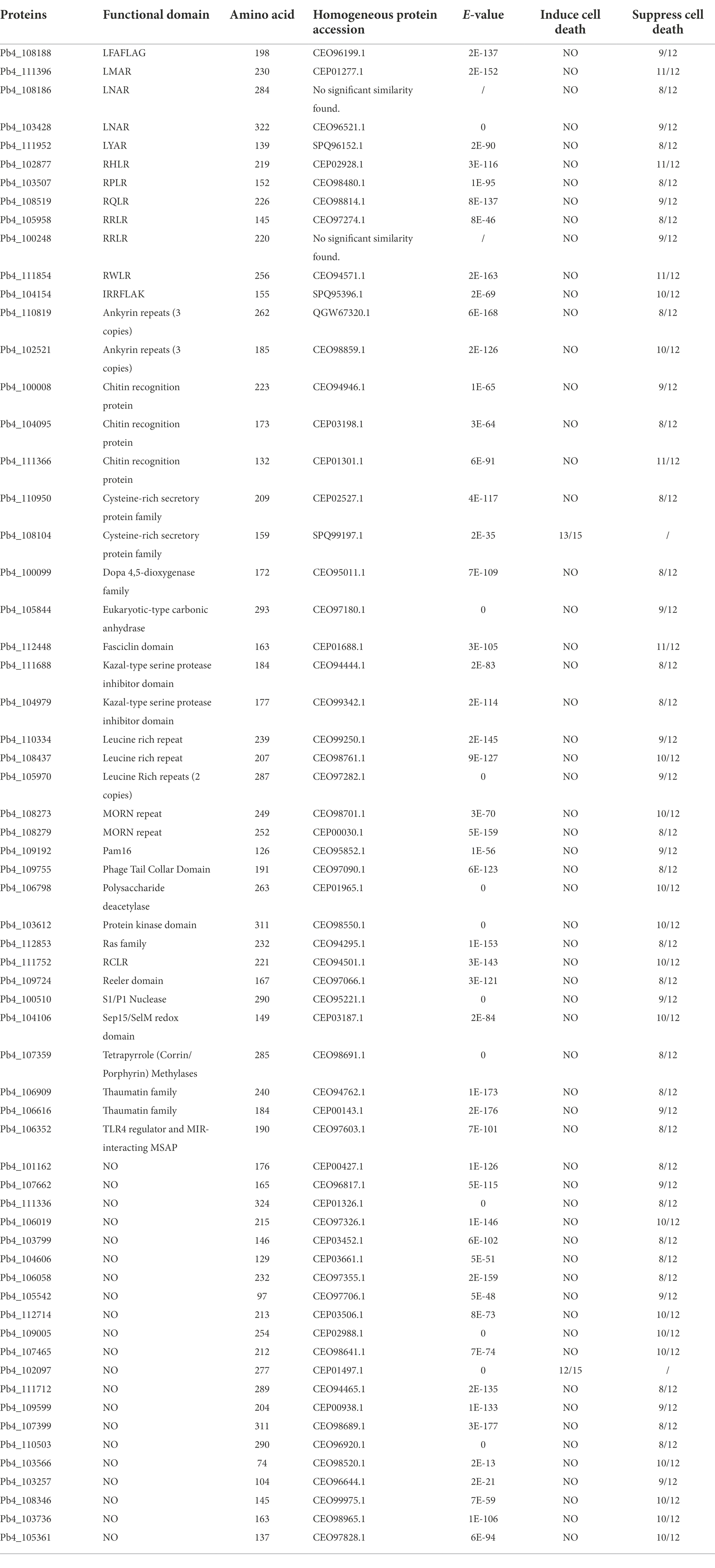
The localization of pathogen effectors after entering a plant cell could indicate their mode of action (Lamond and Spector, 2003; Spector and Lamond, 2011). To characterize the subcellular localization of five candidate effectors containing RXLR, C-terminal eGFP-tagged constructs were generated and expressed in N. benthamiana via agroinfiltration. All these effectors were expressed in the cytoplasm and cytomembrane. The weak green fluorescence signal of Pb4_108279-GFP was also detected in the nucleus. We observed predominant green fluorescence spots of Pb4_105958 dispersed in the cytoplasm, which was probably caused by polymerization of Pb4_105958-GFP proteins (Figure 4). The subcellular localization of these RXLR effectors implied different functions in response to plant immunity.
Discussion
Identification of effector proteins is key to understanding the interaction between P. brassicae and its susceptible host plants. A bioinformatics pipeline approach enables screening of putative effectors based on the genome sequence of P. brassicae. However, it is difficult to clarify all functions of the putative effectors identified by bioinformatics. Transient expression assays in the N. benthamiana model system employing agro-infiltration have been used to identify effectors in bacteria, oomycetes, fungi and nematodes (Wang et al., 2011; Ali et al., 2015; Fang et al., 2016; Choi et al., 2017). In this study, 518 secretory proteins were screened from the genome of P. brassicae strain used in this study. The number of secretory proteins was slightly less than that (558) predicted by Arne (Schwelm et al., 2015). Although, the secretory proteins with special motif, such as RXLR, LXAR and CRN probably function as effectors, some proteins without special candidate motif were also identified in different pathogens (Kemen et al., 2005; Dodds et al., 2009; Malcova et al., 2021). Considering the special relationship between pathogen and host that P. brassicae covered by the membrane structure and parasitized in the root cells, there must be active exchange between them, including the secretary proteins. Therefore, the relative broad selection from the putative secretary proteins for function verification was necessary. In this study, 63 putative effectors were cloned based on the bioinformatics analysis of the P. brassicae genome. We found that 55 putative effectors could suppress plant immunity triggered by BAX in N. benthamiana. And 21 out of 55 could significant suppress the proliferation of Pst DC3000 (AvrRps4) in Arabidopsis. P. brassicae, an intracellular biotrophic parasite, needs to absorb nutrients from living host cells. Therefore, it secretes a large number of proteins to suppress plant immunity. This phenomenon has also been found in other biotrophic pathogens. Of the 83 putative RXLR proteins of P. viticola, 62 could suppress cell death triggered by elicitin in N. benthamiana (Xiang et al., 2016). Additionally, 78 of the 95 putative effectors of H. avenae could suppress programmed cell death triggered by BAX in N. benthamiana (Chen et al., 2013). The universal rule in the biotrophic pathogens probably benefit its propagation and colonization.
Surprisingly, two putative effectors, Pb4_102097 and Pb4_108104, which strongly expressed in the stage of rest spore maturity and release could successfully induce cell death accompanied by H2O2 accumulation in N. benthamiana. These secretary proteins were necessary for the life cycle of P. brassicae. The obligate pathogens try their best to hide from host’s immune-response system to hijack the nutrition and populate inside a host (Dimitrios et al., 2015). However, they are waiting an opportunity to escape from host, when their life cycle is coming to end. Pb4_102097 and Pb4_108104 showed high expressing level in the stage of rest spore maturity, which probably play an important in facility the rest spore release to the soil.
Many pathogen effectors were screened from secretary proteins identified with agroinfiltration of Nicotiana benthamiana method. The best example of plant-specific recognition is the recognition of avirulence effectors either directly or indirectly by nucleotide-binding-leucine-rich-repeat receptors (NLRs), which results in the activation of plant programmed cell death, and cessation of further pathogen growth (Whitham et al., 1994; Tang et al., 1996; Dodds et al., 2004; Rehmany et al., 2005). In B. rapa, several clubroot resistant (CR) loci have been mapped, and two resistant genes CRa, Crr1a are cloned. Both are typical resistance proteins with TIR-NBS-LRR domains (Ueno et al., 2012). Considering the interaction models between other pathogens and hosts, there must be effectors recognized by these CR genes. However, little research on the interaction mechanism between P. brassicae effectors and CR genes has been reported. Therefore, it is challenging to determine the interaction mechanism between effectors of P. brassicae and clubroot resistant genes.
Effectors delivered into host cells deregulate host immunity in a variety of subcellular compartments (Rovenich et al., 2014). The AvrPiz-t effector of Magnaporthe oryzae targets proteasome activity through interaction with the RING E3 ubiquitin ligase APIP6, leading to their mutual degradation and suppression of host immunity (Park et al., 2012). Hyaloperonospora arabidopsis directly delivers its effector to the nucleus of the host to interact with the mediator complex that controls interactions between transcription regulators and RNA polymerase (Caillaud et al., 2012). Five of the putative effectors containing the RXLR motif in this study functioned in the cytoplasm of N. benthamiana. One effector also functioned in the nucleus. Some RXLR effectors require nuclear localization to trigger or suppress cell death (Du et al., 2015; Xiang et al., 2017; Liu et al., 2018; Qi et al., 2019). Transcriptome analysis revealed a variety of genes responding to P. brassicae infection in B. rapa. These included the SWEET family responsible for glucose transporter. A large amount of nutrients are required for the propagation of P. brassicae, which is probably directly controlled by the effectors delivered into the host nucleus.
In this study, the functions of P. brassicae proteins expressed during the different infection stages in the induction and suppression of plant immunity were investigated. Only two putative effectors induced plant immunity and most of the secretory proteins suppressed plant immunity. All the effectors containing the RXLR motif functioned in the cytoplasm and the cytomembrane. These findings improved our understanding of the functions of P. brassicae effector interactions between P. brassicae and hosts. Whether the effects of these effectors in plant cell death are beneficial for P. brassicae infection and the mechanisms by which these effectors manipulate plant immunity are unclear and require further investigation.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
ZZ: analyzed the data, performed the experiments, and drafted the manuscript. SL, YY, and SL: contributed to perform the experiments. XL and ZP: conceived the study, participated in its coordination, and helped to draft the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by grants from the China Agriculture Research System (CARS-12).
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.881992/full#supplementary-material
Footnotes
References
Ali, S., Magne, M., Chen, S., Côté, O., Stare, B. G., Obradovic, N., et al. (2015). Analysis of putative apoplastic effectors from the nematode, Globodera rostochiensis, and identification of an expansin-like protein that can induce and suppress host defenses. PLoS One 10:e0115042. doi: 10.1371/journal.pone.0115042
Bourras, S., Rouxel, T., and Meyer, M. (2015). Agrobacterium tumefaciens gene transfer: how a plant pathogen hacks the nuclei of plant and nonplant organisms. Phytopathology 105, 1288–1301. doi: 10.1094/PHYTO-12-14-0380-RVW
Caillaud, M. C., Piquerez, S. J., Fabro, G., Steinbrenner, J., Ishaque, N., Beynon, J., et al. (2012). Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69, 252–265. doi: 10.1111/j
Chen, W., Li, Y., Yan, R., Xu, L., Ren, L., Liu, F., et al. (2019). Identification and characterization of Plasmodiophora brassicae primary infection effector candidates that suppress or induce cell death in host and nonhost plants. Phytopathology 109, 1689–1697. doi: 10.1094/PHYTO-02-19-0
Chen, S., Songkumarn, P., Venu, R. C., Gowda, M., Bellizzi, M., Hu, J., et al. (2013). Identification and characterization of in planta-expressed secreted effector proteins from Magnaporthe oryzae that induce cell death in rice. Mol. Plant-Microbe Interact. 26, 191–202. doi: 10.1094/MPMI-05-12-
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 24;124:803–814. doi:doi: 10.1016/j.cell.2006.02.008
Choi, S., Jayaraman, J., Segonzac, C., Park, H. J., Park, H., Han, S. W., et al. (2017). Pseudomonas syringae pv. Actinidiae type III effectors localized at multiple cellular compartments activate or suppress innate immune responses in Nicotiana benthamiana. Front Plant Sci. 8:2157. doi: 10.3389/fpls.2017.02157
Dangl, J. L., and Jones, J. D. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Dimitrios, K., Limsoon, W., Archer, J. A. C., et al. (2015). Hi-jack: a novel computational framework for pathway-based inference of host–pathogen interactions. Bioinformatics 31, 2332–2339. doi: 10.1093/bioinformatics/btv138
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Ayliffe, M. A., and Ellis, J. G. (2004). The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell 16, 755–768. doi: 10.1105/tpc.020040
Dodds, P. N., Rafiqi, M., Gan, P. H., Hardham, A. R., Jones, D. A., and Ellis, J. G. (2009). Effectors of biotrophic fungi and oomycetes: pathogenicity factors and triggers of host resistance. New Phytol. 183, 993–1000. doi: 10.1111/j.1469-8137.2009.02922.x
Du, Y., Berg, J., Govers, F., and Bouwmeester, K. (2015). Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 207, 735–747. doi: 10.1111/nph.13355
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Fang, A., Han, Y., Zhang, N., Zhang, M., Liu, L., Li, S., et al. (2016). Identification and characterization of plant cell death-inducing secreted proteins from Ustilaginoidea virens. Mol. Plant-Microbe Interact. 29, 405–416. doi: 10.1094/MPMI-09-15-0200-R
Feng, J., and Xiao, Q., Hwang, SF., Hwang, S.F., Strelkov, S. E., and Gossen, B. D. (2012). Infection of canola by secondary zoospores of Plasmodiophora brassicaeproduced on a nonhost. Eur. J. Plant Pathol. 132, 309–315. doi:doi: 10.1007/s10658-011-9875-2
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Hogenhout, S. A., Vander Hoorn, R. A., Terauchi, R., and Kamoun, S. (2009). Emerging concepts in effector biology of plant-associated organisms. Mol. Plant-Microbe Interact. 22, 115–122. doi: 10.1094/MPMI-22-2-0115
Hossain, M. M., Pérez-López, E., Todd, C. D., Wei, Y., and Bonham-Smith, P. C. (2021). Endomembrane-targeting plasmodiophora brassicae effectors modulate PAMP triggered immune responses in plants. Front. Microbiol. 12:1687. doi: 10.3389/fmicb.2021.651279
Huang, S., van der Vossen, E. A., Kuang, H., Vleeshouwers, V. G., Zhang, N., Borm, T. J., et al. (2005). Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251–261. doi: 10.1111/j.1365-313X.2005.02365.x
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 16;444:323–329. doi:doi: 10.1038/nature05286
Kageyama, K., and Asano, T. (2009). Life cycle of Plasmodiophora brassicae. J. Plant Growth Regul. 28, 203–211. doi: 10.1007/s00344-009-9101-z
Kemen, E., Kemen, A. C., Rafiqi, M., Hempel, U., Mendgen, K., Hahn, M., et al. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant-Microbe Interact. 18, 1130–1139. doi: 10.1094/MPMI-18-1130
Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 19;305:567–580. doi:doi: 10.1006/jmbi.2000.4315
Lamond, A. I., and Spector, D. L. (2003). Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612. doi: 10.1038/nrm1172
Lin, B., Zhuo, K., Chen, S., Hu, L., Sun, L., Wang, X., et al. (2016). A novel nematode effector suppresses plant immunity by activating host reactive oxygen species-scavenging system. New Phytol. 209, 1159–1173. doi: 10.1111/nph.13701. Epub 2015 Oct 20
Ling, Y., Xinlong, L., Jiang, X., et al. (2015). Characterization of the secretome of plasmopara viticola by de novo transcriptome analysis. Physiol. Mol. Plant Pathol. 91, 1–10. doi: 10.1016/j.pmpp.2015.05.002
Liu, Y., Lan, X., Song, S., Yin, L., Dry, I. B., Qu, J., et al. (2018). In Planta functional analysis and subcellular localization of the Oomycete pathogen Plasmopara viticola candidate RXLR effector repertoire. Front. Plant Sci. 9:286. doi: 10.3389/fpls.2018.00286
Liu, X. Y., Min, Y., Wang, K. M., Wan, Z. Y., Zhang, Z. G., Cao, C. X., et al. (2014). Draft genome sequence of bacillus amyloliquefaciens HB-26. Stand. Genomic Sci. 9, 775–782. doi: 10.4056/sigs.4978673
Malcova, I., Bumba, L., Uljanic, F., Kuzmenko, D., Nedomova, J., and Kamanova, J. (2021). Lipid binding by the N-terminal motif mediates plasma membrane localization of Bordetella effector protein BteA. J. Biol. Chem. 296:100607. doi: 10.1016/j.jbc.2021.100607
McDonald, M. R., Sharma, K., Gossen, B. D., Deora, A., Feng, J., and Hwang, S. F. (2014). The role of primary and secondary infection in host response to Plasmodiophora brassicae. Phytopathology 104, 1078–1087. doi: 10.1094/PHYTO-07-13-0189-R
Park, C. H., Chen, S., Shirsekar, G., Zhou, B., Khang, C. H., Songkumarn, P., et al. (2012). The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell 24, 4748–4762. doi: 10.1105/tpc.112.105429
Pérez-López, E., Hossain, M. M., Tu, J., Waldner, M., Todd, C. D., Kusalik, A. J., et al. (2020). Transcriptome analysis identifies Plasmodiophora brassicae secondary infection effector candidates. J. Eukaryot. Microbiol. 67, 337–351. doi: 10.1111/jeu.12784
Pérez-López, E., Waldner, M., Hossain, M., Kusalik, A. J., Wei, Y., Bonham-Smith, P. C., et al. (2018). Identification of Plasmodiophora brassicae effectors: A challenging goal. Virulence 9, 1344–1353. doi: 10.1080/21505594.2018.1504560
Petersen, T. N., Brunak, S., von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 29;8:785–786. doi:doi: 10.1038/nmeth.1701
Pitino, M., Armstrong, C. M., Cano, L. M., and Duan, Y. (2016). Transient expression of Candidatus Liberibacter Asiaticus effector induces cell death in Nicotiana benthamiana. Front. Plant Sci. 7:982. doi: 10.3389/fpls.2016.00982
Qi, T., Guo, J., Liu, P., He, F., Wan, C., Islam, M. A., et al. (2019). Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol. Plant 2;12:1624–1638. doi:doi: 10.1016/j.molp.2019.09.010
Rehmany, A. P., Gordon, A., Rose, L. E., Allen, R. L., Armstrong, M. R., Whisson, S. C., et al. (2005). Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. Plant Cell 17, 1839–1850. doi: 10.1105/tpc.105.031807
Rolfe, S. A., Strelkov, S. E., Links, M. G., Clarke, W. E., Robinson, S. J., Djavaheri, M., et al. (2016). The compact genome of the plant pathogen plasmodiophora brassicae is adapted to intracellular interactions with host brassica spp. BMC Genomics 17:272. doi: 10.1186/s12864-016-2597-2
Rovenich, H., Boshoven, J. C., and Thomma, B. P. (2014). Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr. Opin. Plant Biol. 20, 96–103. doi: 10.1016/j.pbi.2014.05.001
Schwelm, A., Fogelqvist, J., Knaust, A., Jülke, S., Lilja, T., Bonilla-Rosso, G., et al. (2015). The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci. Rep. 5:11153. doi: 10.1038/srep11153
Spector, D. L., and Lamond, A.I. (2011). Nuclear speckles. Cold Spring Harb. Perspect. Biol. 1;3:a000646. doi:doi: 10.1101/cshperspect.a000646
Swift, M. L. (1997). GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Model. 37, 411–412.
Tang, X., Frederick, R. D., Zhou, J., Halterman, D. A., Jia, Y., and Martin, G. B. (1996). Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science 20;274:2060–2063. doi:doi: 10.1126/science.274.5295.2060
Thordal-Christensen, H., Zhang, Z., Wei, Y., and Collinge, D. B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Ueno, H., Matsumoto, E., Aruga, D., Kitagawa, S., Matsumura, H., and Hayashida, N. (2012). Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Mol. Biol. 80, 621–629. doi: 10.1007/s11103-012-9971-5
Wang, Q., Han, C., Ferreira, A. O., Yu, X., Ye, W., Tripathy, S., et al. (2011). Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. Plant Cell 23, 2064–2086. doi: 10.1105/tpc.111.086082. Epub 2011 Jun 7
Whitham, S., Dinesh-Kumar, S. P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cells 23;78:1101–1115. doi:doi: 10.1016/0092-8674(94)90283-6
Xiang, J., Li, X., Wu, J., Yin, L., Zhang, Y., and Lu, J. (2016). Studying the mechanism of Plasmopara viticola RxLR effectors on suppressing plant immunity. Front. Microbiol. 7:709. doi: 10.3389/fmicb.2016.00709
Xiang, J., Li, X., Yin, L., Liu, Y., Zhang, Y., Qu, J., et al. (2017). A candidate RxLR effector from Plasmopara viticola can elicit immune responses in Nicotiana benthamiana. BMC Plant Biol. 14;17:75. doi:doi: 10.1186/s12870-017-1016-4
Yuan, M., and Xin, X. F. (2021). Bacterial infection and hypersensitive response assays in Arabidopsis-pseudomonas syringae Pathosystem. Bio-protocol. 11:e4268. doi: 10.21769/BioProtoc.4268
Keywords: clubroot disease, Plasmodiophora brassicae, secretory proteins, plant immunity, effectors
Citation: Zhan Z, Liu H, Yang Y, Liu S, Li X and Piao Z (2022) Identification and characterization of putative effectors from Plasmodiophora brassicae that suppress or induce cell death in Nicotiana benthamiana. Front. Plant Sci. 13:881992. doi: 10.3389/fpls.2022.881992
Edited by:
Jacqueline Batley, University of Western Australia, AustraliaCopyright © 2022 Zhan, Liu, Yang, Liu, Li and Piao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongyun Piao, enlwaWFvQHN5YXUuZWR1LmNu; Xiaonan Li, Z3JhY2VzbGVleG44M0BzeWF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Zongxiang Zhan
Zongxiang Zhan Huishan Liu†
Huishan Liu† Shuang Liu
Shuang Liu Xiaonan Li
Xiaonan Li