
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 23 June 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.881342
This article is part of the Research Topic Discovery, Biosynthesis, Regulation, Transport, Release and Engineering of Plant Natural Products View all 13 articles
 Ting-Yan Qiang1
Ting-Yan Qiang1 Jiu-Shi Liu1,2
Jiu-Shi Liu1,2 Yu-Qing Dong1
Yu-Qing Dong1 Xin-Lu Mu1
Xin-Lu Mu1 Yu Chen1
Yu Chen1 Hong-Mei Luo1,2
Hong-Mei Luo1,2 Ben-Gang Zhang1,2
Ben-Gang Zhang1,2 Hai-Tao Liu1,2*
Hai-Tao Liu1,2*Schisandra chinensis owes its therapeutic efficacy to the dibenzocyclooctadiene lignans, which are limited to the Schisandraceae family and whose biosynthetic pathway has not been elucidated. Coniferyl alcohol is the synthetic precursor of various types of lignans and can be acetylated to form coniferyl acetate by coniferyl alcohol acyltransferase (CFAT), which belongs to the BAHD acyltransferase family. This catalytic reaction is important because it is the first committed step of the hypothetical biosynthetic pathway in which coniferyl alcohol gives rise to dibenzocyclooctadiene lignans. However, the gene encoding CFAT in S. chinensis has not been identified. In this study, firstly we identified 37 ScBAHD genes from the transcriptome datasets of S. chinensis. According to bioinformatics, phylogenetic, and expression profile analyses, 1 BAHD gene, named ScBAHD1, was cloned from S. chinensis. The heterologous expression in Escherichia coli and in vitro activity assays revealed that the recombinant enzyme of ScBAHD1 exhibits acetyltransferase activity with coniferyl alcohol and some other alcohol substrates by using acetyl-CoA as the acetyl donor, which indicates ScBAHD1 functions as ScCFAT. Subcellular localization analysis showed that ScCFAT is mainly located in the cytoplasm. In addition, we generated a three-dimensional (3D) structure of ScCFAT by homology modeling and explored the conformational interaction between protein and ligands by molecular docking simulations. Overall, this study identified the first enzyme with catalytic activity from the Schisandraceae family and laid foundations for future investigations to complete the biosynthetic pathway of dibenzocyclooctadiene lignans.
Schisandra chinensis (Turcz.) Baill. (Schisandraceae), a deciduous woody vine plant is mainly distributed in north-eastern China, Korea, Japan, and the eastern part of Russia (Hancke et al., 1999). The fruits of S. chinensis have been used as traditional Chinese medicine and functional food for 1,000 of years for the effects of astringent, tonifying qi, replenishing production of body fluid, and nourishing kidney to calm heart (Zhai et al., 2018). In addition, the fruits of S. chinensis have long been documented in the pharmacopeias of Russia (1990), America (1999), Korea (2002), Japan (2006), WHO (2007), and Europe (2008) (Szopa et al., 2017).
Extensive studies have indicated that the major bioactive components of S. chinensis are dibenzocyclooctadiene lignans (aka schisandra lignans; Szopa et al., 2017), which exhibit a wide variety of biological activities, such as anti-liver injury, anti-oxidant, anti-asthmatic, anti-inflammatory, sedation on the central nervous system activities, and so on (Opletal et al., 2004). The hepatoprotective effect is considered to be the principal one, and the structure-activity relationship suggests that the hepatoprotective activity depends on the configuration of biphenyls and the methylenedioxy substituent on the phenyl nucleus (Hikino et al., 1984). In China, two novel anti-hepatitis drugs (bifendate and bicyclol) were sequentially created through synthesizing the analogs of schizandrin C, which is the most effective component against liver injury of S. chinensis (Zhu et al., 2019). Apart from the significant pharmacological activities, the phylogenetic distribution of lignan-producing plant species displays that dibenzocyclooctadiene lignans are only found in the Schisandraceae family (Umezawa, 2003).
Accordingly, the identification of genes and elucidation of the biosynthetic pathway of dibenzocyclooctadiene lignans have received widespread interest due to the uniqueness of structure, the significance of effect, and the limitation of distribution. Since Erdtman first suggested that the lignan structure was formed by the coupling of two phenylpropanoid monomer units (Erdtman, 1933), the biosynthetic pathway of several typical lignans has been established, such as furofuran, furan, dibenzylbutane, dibenzylbutyrolactone, and aryltetralin lignans (Ono et al., 2006; Lau and Sattely, 2015). In addition, coniferyl alcohol has been proved to be a synthetic precursor of lignans (Suzuki and Umezawa, 2007; Kim et al., 2009). Dexter et al. discovered that coniferyl alcohol could be acetylated to coniferyl acetate by a novel petunia BAHD acyltransferase (PhCFAT; Dexter et al., 2007). And isoeugenol synthase 1 (PhIGS1) was shown to use coniferyl acetate and NADPH as substrates to form E-isoeugenol (Koeduka et al., 2006). Previously, Lopes et al. had revealed that E-isoeugenol was converted to a furan lignan verrucosin in Virola surinamensis through stable-isotope labeling and in vivo feeding experiment (Lopes et al., 2004). And there are indeed furan lignans (chicanine and d-epigalbacin) in S. chinensis (Liu et al., 1981; Hancke et al., 1999). Based on the existing research results, the comparison of chemical structures and the similarity of lignan biosynthesis, Suzuki et al. proposed the unique and authoritative biosynthetic pathway for dibenzocyclooctadiene lignans (Figure 1; Suzuki and Umezawa, 2007). What is more, Suzuki et al. also proposed that yatein was the precursor of deoxypodophyllotoxin in 2007 (Suzuki and Umezawa, 2007), which was proved to be right in 2015 (Lau and Sattely, 2015). In conclusion, we believe the biosynthetic pathway for dibenzocyclooctadiene lignans is also right and we are striving to clone functional genes according to this pathway.
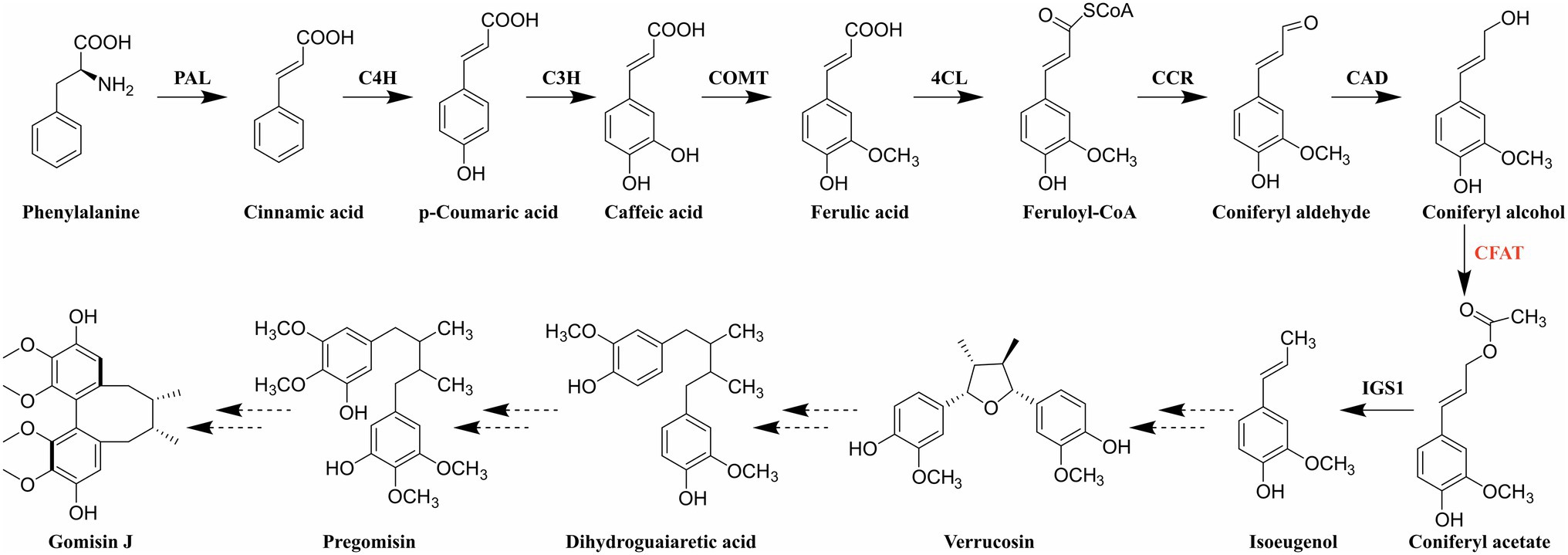
Figure 1. Biosynthetic pathway of dibenzocyclooctadiene lignans. PAL: phenylalanine ammonia-lyase; C4H: cinnamate-4-hydroxylase; C3H: coumarate-3-hydroxylase; COMT: caffeic acid O-methyltransferase; 4CL: 4-hydroxycinnamate CoA ligase; CCR: cinnamoyl-CoA reductase; CAD: cinnamyl alcohol dehydrogenase; CFAT: coniferyl alcohol acyltransferase; and IGS1: isoeugenol synthase 1.
As shown in Figure 1, coniferyl alcohol is synthesized from phenylalanine through the general phenylpropanoid pathway. It should be noted that coniferyl alcohol is not only a synthetic precursor of lignans but also a kind of monolignol involved in the biosynthesis of lignin (Vanholme et al., 2010). Therefore, the biochemical steps before coniferyl alcohol are shared between dibenzocyclooctadiene lignan and lignin biosynthesis (Muhlemann et al., 2014). Consequently, coniferyl alcohol acyltransferase (CFAT), the first enzyme after the branch point, must play a vital role in the synthesis of dibenzocyclooctadiene lignans. CFAT is a member of the BAHD acyltransferase family, which has two conserved motifs, HXXXD and DFGWG (D’Auria, 2006). The BAHD acyltransferases depending on acyl-CoA are responsible for the acylation of compounds, which can improve the stability and solubility of compounds, protect substances from enzymatic degradation, help plants resist diseases, and so on (Yu et al., 2009). Since St-Pierre et al. first coined the name BAHD based on the first four enzymes (BEAT, AHCT, HCBT, and DAT; St-Pierre and Luca, 2000), the BAHD superfamily is fast-growing and found to play important roles in the biosynthesis of a variety of secondary metabolites, such as Catharanthus alkaloid vindoline (St-Pierre et al., 1998), the Papaver alkaloid morphine (Grothe et al., 2001), the diterpenoid alkaloid Taxol (Walker and Croteau, 2000a,b; Walker et al., 2000), anthocyanins (Suzuki et al., 2001, 2002), and floral volatiles (Dudareva et al., 1998), as well as some phytoalexins (Chakravarthy et al., 2010). However, to date, only two CFAT genes, PhCFAT and PmCFAT1, have been characterized from Petunia × hybrida and Prunus mume (Dexter et al., 2007; Zhang et al., 2019). The gene encoding CFAT has not been identified from S. chinensis.
Although some progress has been made in the biosynthesis of several types of lignans, it has been stymied to elucidate the biosynthetic pathway of dibenzocyclooctadiene lignans because the biosynthetic genes, the genetic transformation system, and the whole genome of S. chinensis are unavailable. In this study, to determine whether there is a gene encoding CFAT in S. chinensis, we confirmed the presence of isoeugenol in S. chinensis by GC–MS for the first time. Then, a total of 37 ScBAHD genes were obtained in a transcriptome-wide identification. Phylogenetic, conversed motifs, and expression pattern analyses were performed to screen the putative gene encoding CFAT in S. chinensis. Finally, ScBAHD1 was demonstrated to be ScCFAT through heterologous expression and in vitro enzymatic reaction. Furthermore, to clarify the molecular mechanism of ScCFAT, especially to improve its catalytic efficiency, bioinformatics (molecular modeling and docking simulations) were used in this work. In summary, we identified the first enzyme with catalytic activity from the Schisandraceae family, which is of guiding significance for fully revealing the biosynthetic pathway of dibenzocyclooctadiene lignans.
The fruits of S. chinensis were collected in the Beijing medicinal plant garden at IMPLAD (Institute of Medicinal Plant Development; 40°N and 116°E), Beijing, China. The air-dried samples were pulverized into powder, passed through a 50-mesh sieve, and stored at room temperature until analysis. In addition, the green and fresh fruits of S. chinensis were immediately frozen in liquid nitrogen and stored at −80°C for RNA extraction. Ethyl acetate (UPLC grade) was purchased from Merck (Merck KGaA, Darmstadt, Germany). Reference compounds, isoeugenol was purchased from Chengdu DeSiTe Biological Technology (Chengdu, China), and cinnamyl acetate was purchased from Shanghai Macklin Biochemical (Shanghai, China).
The powdered S. chinensis fruits (1.0000 g) were suspended in 50 ml of UPLC grade ethyl acetate in a 100 ml conical flask with a stopper and then was extracted in an ultrasonic bath for 60 min at room temperature. The extract was collected by filtering and the residue was washed with 5 ml of ethyl acetate three times. Then, the extract was vacuum evaporated to recover the solvent. Later the residue was dissolved in 10 ml of ethyl acetate and the sample solution was filtered through a 0.22 μm membrane filter into the vials for GC–MS analysis. The standard stock solution of isoeugenol was prepared by dissolving it in UPLC grade ethyl acetate and then diluted to the appropriate concentration for GC–MS analysis.
GC–MS analysis for identification of isoeugenol in the fruits of S. chinensis was performed using an Agilent Technologies 7890B gas chromatography instrument, combined with an Agilent 5977A MSD equipped with electron ionization (EI) and quadrupole analyzer, and an Agilent Chem Station data system. GC separation was performed on a 30 m HP-5MS Ultra Inert capillary column with an internal diameter of 0.25 mm and a film thickness of 0.25 μm (Agilent 19091S-433UI, Agilent Technologies). The carrier gas was helium (99.99%) with a flow rate of 1 ml/min. The temperature of the injector and detector was set at 280°C and 250°C, respectively. Spectra were obtained over a scan range of 50 to 550 amu at 2.9 scans/s. The GC program was set as follows: the initial temperature was 60°C and held for 2 min, then increased by 5°C/min to 135°C, then raised by 3°C/min to 180°C, and finally raised by 10°C/min to 200°C and held at 200°C for 10 min. The sample and isoeugenol solution (1 μl) were injected manually while maintaining a solvent delay of 4 min. Interpretation of the mass spectrum was made by comparing the peak distribution against the database of National Institute Standard and Technology (NIST MS 2.0, Gaithersburg, MD, USA).
The transcriptome datasets (PRJNA533953) of S. chinensis (Chen et al., 2020b) were downloaded from NCBI1 and used for the identification of ScBAHD genes. The Hidden Markov Model (HMM) profile of the transferase domain (PF02458) downloaded from the Pfam database (Mistry et al., 2021) was exploited for the identification of ScBAHD genes by using the simple HMM search program of TBtools (Eddy, 2004; Chen et al., 2020a). The NCBI Batch Web CD-Search Tool with default parameters2 was exploited to test for the presence of the transferase domain. The sequence integrity of ScBAHDs was analyzed by performing multiple sequence alignment analyses of all ScBAHDs by ClustalW (Chenna et al., 2003). Some ScBAHDs, whose number of amino acids is shorter than 400 or longer than 500 were removed (D’Auria, 2006). ExPASy3 was used to calculate the length of protein sequences, molecular weight (MW), theoretical isoelectric point (pI), and the grand average of hydropathicity (GRAVY; Artimo et al., 2012). Wolf PSORT4 was employed to predict the subcellular localization of ScBAHD proteins (Horton et al., 2007).
The peptide sequences of putative ScBAHD proteins were submitted to MEME 5.45 for the analysis of conserved motif composition (Bailey et al., 2015). The optimized parameters were set as follows: the maximum number of motifs was 10, the width of each motif comprised between 6 to 50 residues, and the remaining parameters were default. Putative ScBAHD protein sequences were aligned along with 75 biochemically characterized BAHD members (Supplementary Table S1) using MAFFT 7.4 (Katoh and Standley, 2013). The maximum likelihood phylogenetic tree was constructed by IQ-TREE 2 with 1,000 bootstrap replicates (Minh et al., 2020). And the iTOL online tool6 was used to visual embellish (Letunic and Bork, 2021).
The transcriptome datasets of S. chinensis fruits at three stages during fruit ripening (green, light red, and dark red) obtained from NCBI (PRJNA533953; Chen et al., 2020b) were used for expression pattern analysis. The expression level of each gene at three stages was calculated by using Bowtie2 (Langmead and Salzberg, 2012) and RSEM (Li and Dewey, 2011), and converted into transcripts per million reads (TPM). The data were visualized using an R package heatmap.
Total RNA was extracted from the fresh fruits of S. chinensis by using the EASYspin Plus Complex Plant RNA Kit (Aidlab Biotechnology, Beijing, China) following the manufacturer’s instructions. The quality of total RNA was verified by absorbance at 260 nm and 280 nm optical densities by using the NanoDrop 2000 Spectrophotometer (Thermo Scientific, San Jose, CA, USA) and by 1% agarose gel electrophoresis. The first-strand cDNA was synthesized by using the GoScript™ Reverse Transcription Kit (Promega, Beijing, China) according to the instructions. The cDNA was stored at −80°C for further gene clone and qPCR.
The results of RNAseq were validated by RT-qPCR to see if there is a good correlation between the expression of ScBAHD1 obtained by both techniques. After entering the fruiting period, we regularly picked the S. chinensis fruits with three biological replicates at four different developmental stages every 30 days. The samples were numbered as 1-1, 1-2, 1-3 (three replicates of the first period), 2-1, 2-2, 2-3 (three replicates of the second period), 3-1, 3-2, 3-3 (three replicates of the third period), and 4-1, 4-2, 4-3 (three replicates of the fourth period). Primers (ScBAHD1-qF and ScBAHD1-qR) designed by CLC Genomics Workbench are listed in Supplementary Table S2. The qPCR experiments were performed in triplicate using TB Green Premix Ex Taq (Takara Biotechnology, Dalian, China) in a Bio-Rad CFX96 Real-Time system. ScGAPDH was the reference gene, and the relative expression levels were calculated by the 2–ΔΔCt method from Ct values.
The specific primers (ScCFAT-F and ScCFAT-R) in Supplementary Table S2 were designed by using CLC Genomics Workbench. The full-length ScCFAT cDNA was amplified in a 25 μl system including 1 μl cDNA, 0.5 μl forward and reverse primer, 5 μl 5× TransStart FastPfu Buffer, 2 μl 2.5 mM dNTPs, 15.5 μl ddH2O, and 0.5 μl TransStart FastPfu DNA Polymerase (TransGen Biotech, Beijing, China). The amplification procedure was shown as follows: 95°C for 1 min, 40 cycles of 95°C for 20 s, 55°C for 20 s, 72°C for 45 s, and a final extension step for 5 min at 72°C. Subsequently, the amplification products of proper length were purified by using Gel DNA Mini Purification Kit (Aidlab Biotechnology, Beijing, China) and ligated into the pET-28a vector digested by QuickCut™ EcoRI through seamless cloning by using ClonExpress Ultra One Step Cloning Kit (Vazyme, Nanjing, China). The recombinant plasmid was transformed into Escherichia coli DH5α competent cell and sequenced in Tsingke Biotechnology (Beijing, China). Then, the right plasmid was isolated and transformed into E. coli BL21 (DE3) competent cell.
Escherichia coli BL21 (DE3), which was transformed into the recombinant plasmid, was grown in LB medium with 50 μg/ml kanamycin at 37°C until the OD600 of 0.6–0.8 was reached. The expression was induced by adding 0.1–0.2 mM Isopropylb-D-1-thiogalactopyranoside (IPTG) at 18°C for 18 h. After induction, the cells were collected, resuspended in 1× PBS buffer, and broken by sonication. The lysate was centrifuged at 4°C and the supernatant was collected for purification by using Hiper Ni-Agarose His tag soluble protein purification kit (Mei5 Biotechnology, Beijing, China). After denaturing with SDS loading dye at 100°C for 5 min, the eluted fractions were examined through 12% SDS-PAGE gel electrophoresis followed by staining of the gel with Coomassie brilliant blue.
The 200 μl reaction mixture contained 56 μl purified protein, 140 μM acetyl-CoA, and 120 μM alcohol substrate in assay buffer (50 mM citric acid, pH 6.0, 1 mM DTT or 50 mM Tris–HCL, pH 7.5, and 1 mM DTT). While in the control group, instead of purified protein, 56 μl protein of empty vector was added to the reaction mixture. After incubation in 25°C water bath for 15 min, added 200 μl of chromatographic grade methanol into the mixture and centrifugated at 13,000 rpm for 25 min. Then, 200 μl upper reaction mixture was absorbed into vials and the products were verified by using UPLC.
The pCAMBIA1302 vector driven by the cauliflower mosaic virus (CaMV) 35S promoter was used to identify the subcellular localization of ScCFAT by utilizing the transient expression system of Nicotiana benthamiana leaves. The entire coding sequence (CDS) of ScCFAT without its stop codon was amplified with primers (ScCFAT-LF and ScCFAT-LR) listed in Supplementary Table S2. The pCAMBIA1302 vector was digested by SpeI (TaKaRa Biotechnology, Dalian, China). Then, the CDS of ScCFAT was inserted into the digested vector to generate ScCFAT-GFP fusion protein. The resulting construct harboring ScCFAT-GFP gene was transferred into Agrobacterium tumefaciens strains GV3101 through the conventional freezing–thawing method, and the GV3101 strains harboring pCAMBIA1302-ScCFAT-GFP or empty pCAMBIA1302-GFP were transiently infiltrated into 5-week-old N. benthamiana leaves. After infection for 48 h, the green fluorescence protein (GFP) expression was observed under the Nikon C2-ER laser scanning confocal microscope.
The BLASTP search for the amino acid sequence of ScCFAT was performed to obtain the most suitable template in the Brookhaven Protein Data Bank (PDB).7 The homology model of ScCFAT structure was generated by using Modeller 10.2 software. Five models were generated and selected according to the DOPE (Discrete Optimized Protein Energy) score. The stereochemical property of the model was evaluated by PROCHECK (Laskowski et al., 1996) and VERIFY-3D (Lüthy et al., 1992). The docking of ScCFAT with its natural substrate acetyl CoA and coniferyl alcohol was simulated by using AutoDock, and the visualized results were drawn by PyMOL.
As shown in Figure 2, the total ion chromatogram (TIC) of the isoeugenol standard is presented in the red line and the TIC of the fruits of S. chinensis is presented in the black line. The mass profiles are presented in Supplementary Figure 1. The result indicates that S. chinensis contains both E-isoeugenol and Z-isoeugenol, and the former is dominant, which provides evidence that there must be a gene encoding CFAT in S. chinensis.
The previous transcriptome datasets (PRJNA533953; Chen et al., 2020b) were used to identify all BAHD genes in S. chinensis. After removing short/long sequences and redundant sequences, we spotted 37 BAHD genes and named them ScBAHD1 to ScBAHD37. As shown in Table 1, the length of 37 ScBAHD proteins varies from 425 to 489 amino acids, corresponding to Mw range of 43.40–52.66 kDa, the theoretical pI ranges from 4.68 to 8.38, and GRAVY varies from −0.310 to 0.076. Subcellular localization prediction results indicate that the majority of ScBAHD proteins (33 out of 37, 89%) localize in cytoplasm and chloroplast.
Phylogenetic relationships among 75 biochemically characterized BAHD acyltransferases from other species (Supplementary Table S1) and 37 putative ScBAHD acyltransferases were constructed using a maximum likelihood algorithm. As shown in Figure 3, the phylogenetic tree is divided into five clades. There are seven ScBAHDs (ScBAHD4, ScBAHD8, ScBAHD10, ScBAHD14, ScBAHD19, ScBAHD20, and ScBAHD37) clustered into clade I. ScBAHD9, ScBAHD11, ScBAHD18, ScBAHD22, and ScBAHD30 are clustered into clade II along with ZmGlossy2 and AtCER2. There are 12 ScBAHDs clustered into clade III, which contains the PhCFAT. And ScBAHD1, ScBAHD3, and ScBAHD7 are close to PhCFAT, which indicates that the three ScBAHD proteins may be CFAT in S. chinensis. In addition, there is not a ScBAHD clustered into clade IV with HvACT. The remaining ScBAHD sequences are clustered into clade V. The result of conserved motifs analysis indicates that the BAHD proteins in the same clade have similar motifs. Motif 1 contains the HXXXD domain and motif 3 contains the DFGWG domain. All 37 ScBAHD proteins have the motif HXXXD, and aside from nine ScBAHDs, the others have the conserved motif DFGWG. Motifs 1–7 and motif 9 are widely distributed, but they do not exist in certain clades. For example, motif 3 exists in all other clades except clade II. Motif 8 is merely present in clade II, IV, and V. And motif 10 is only present in clade III and V.

Figure 3. Maximum likelihood phylogenetic tree and conversed motif of 75 biochemically characterized BAHD acyltransferases and 37 putative ScBAHD acyltransferases. The five clades are shown in different colors.
To further investigate the expression profiles of 37 ScBAHD genes, we analyzed the expression levels of each ScBAHD gene during the ripening stages of the S. chinensis fruits, including the green stage, light red stage, and dark red stage. As shown in Figure 4, the clustering divides into three main clades. There are six ScBAHD genes (ScBAHD3, ScBAHD5, ScBAHD23, ScBAHD24, ScBAHD34, and ScBAHD36) primarily detected at the green stage. A total of 19 ScBAHD genes display higher expression at the green and light red stages than at the dark red stage. And 12 ScBAHD genes show high expression at the light red and dark red stages, but they are not detected at the green stage. ScBAHD1 shows higher expression at the light red and dark red stages than at the green stage. However, ScBAHD3 and ScBAHD7 are not detected at the light red and dark red stages. The expression of ScBAHD1 was further analyzed by RT-qPCR (Supplementary Figure 2), and the results were consistent with the RNAseq data.
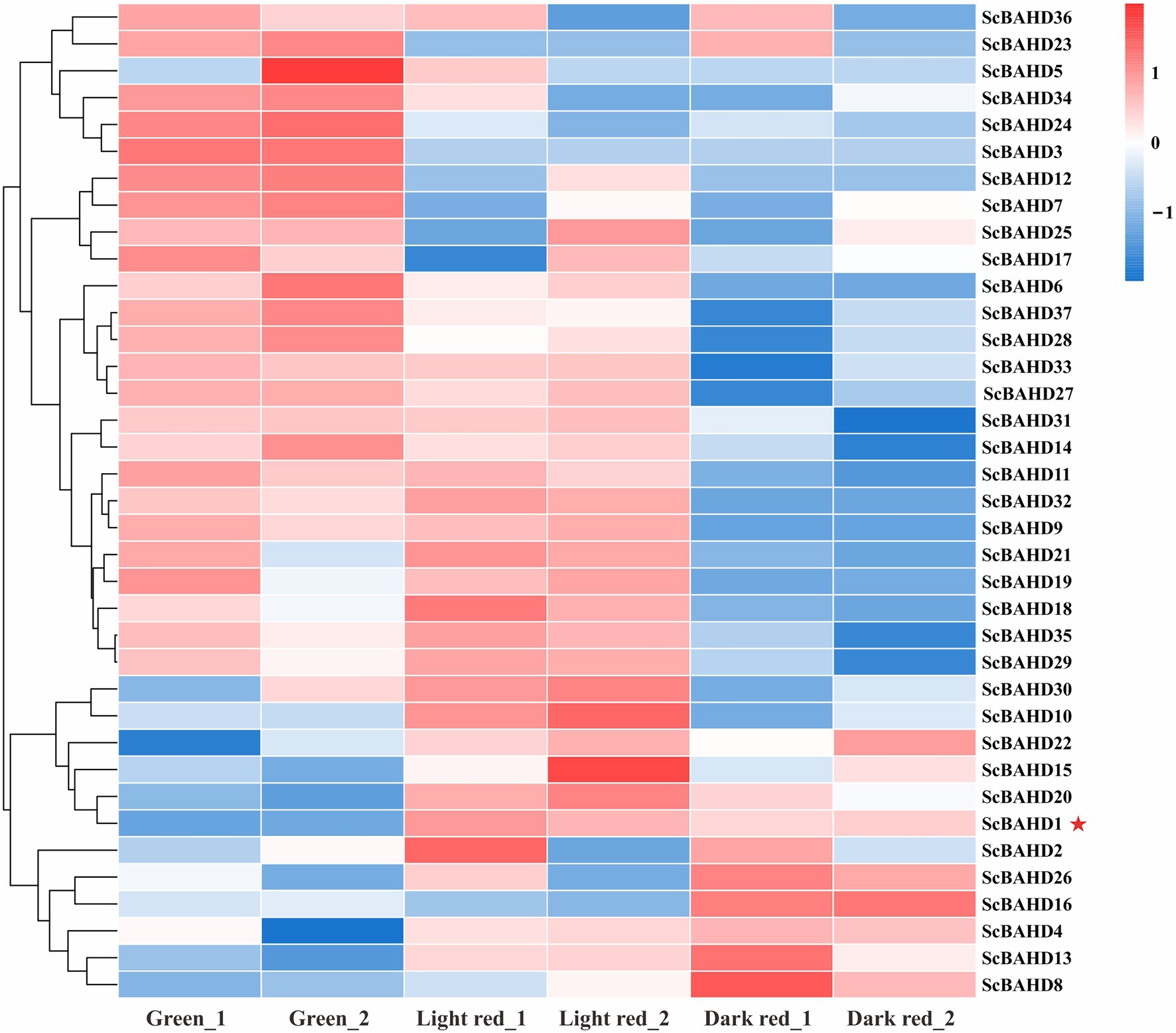
Figure 4. Heat map exhibits expression patterns of 37 ScBAHD genes during the ripening stages of S. chinensis fruits, including the green stage (Green_1 and Geen_2), light red stage (Light red_1 and Light red_2), and dark red stage (Dark red_1 and Dark red_2). Red indicates high relative gene expression, whereas blue indicates low relative gene expression.
ScBAHD1, named ScCFAT (1,368 bp, GenBank Accession Number, OM804184), encodes a protein of 455 amino acid residues, and the predicted molecular weight of ScCFAT is 50.34 kDa and the pI value is 5.10. ScCFAT was subcloned into the expression vector pET-28a using a restriction site to generate N-terminal hexa-histidine tagged recombinant proteins and the purified protein was assessed by SDS-PAGE (Figure 5A). The purified recombinant protein was evaluated for its ability to acetylate coniferyl alcohol as well as a variety of other alcohols by using acetyl-CoA as a source for the acetyl moiety. Assay products were analyzed by using UPLC. Because it has not been able to detect coniferyl acetate due to the instability (Koeduka et al., 2006), the product of ScCFAT catalyzing coniferyl alcohol was verified by using PhCFAT. As shown in Figure 5B, ScCFAT could catalyze coniferyl alcohol to the same product as PhCFAT catalyzed. In addition, ScCFAT could also catalyze cinnamyl alcohol into cinnamyl acetate (Figure 5B), which further proved the acetylation activity of ScCFAT. Apart from coniferyl alcohol and cinnamyl alcohol, ScCFAT exhibited detected activities with sinapyl alcohol, p-coumaryl alcohol, (E)-3-phenyl-2-methyl-2-propenol, benzyl alcohol, 4-hydroxybenzyl alcohol, and (+)-secoisolariciresinol as shown in Table 2 and Supplementary Figure 3.
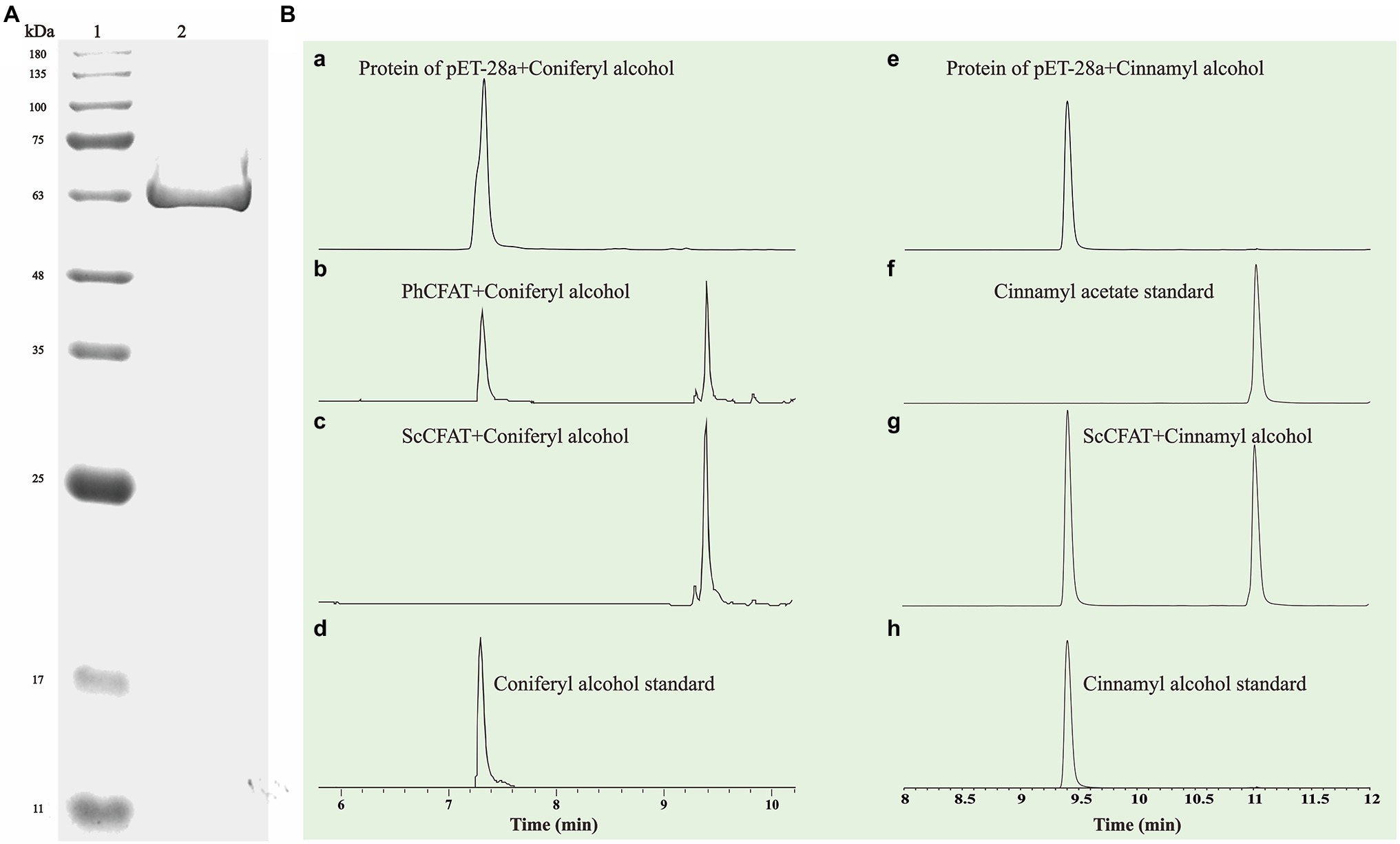
Figure 5. (A) SDS-PAGE analysis of recombinant ScCFAT. ScCFAT was separated using 12% SDS-PAGE gel electrophoresis and protein bands were visualized after staining with Coomassie brilliant blue. Lane 1: protein marker; Lane 2: purified ScCFAT. (B) In vitro analyses of ScCFAT and PhCFAT activities. (a) UPLC analysis of compounds found in a reaction mixture containing protein of pET-28a, coniferyl alcohol, and acetyl-CoA after 15 min of incubation. (b) UPLC analysis of compounds found in a reaction mixture containing purified PhCFAT, coniferyl alcohol, and acetyl-CoA after 15 min of incubation. (c) UPLC analysis of compounds found in a reaction mixture containing purified ScCFAT, coniferyl alcohol, and acetyl-CoA after 15 min of incubation. (d) UPLC analysis of coniferyl alcohol standard. (e) UPLC analysis of compounds found in a reaction mixture containing protein of pET-28a, cinnamyl alcohol, and acetyl-CoA after 15 min of incubation. (f) UPLC analysis of cinnamyl acetate standard. (g) UPLC analysis of compounds found in a reaction mixture containing purified ScCFAT, cinnamyl alcohol, and acetyl-CoA after 15 min of incubation. (h) UPLC analysis of cinnamyl alcohol standard.
The prediction of protein subcellular localization with the WoLF PSORT Server showed that ScCFAT was located in the cytoplasm and nucleus with a probability of 26.32%. To investigate the intracellular localization of this protein, the nucleotide sequence without stop codon and with homologous sequences of restriction enzyme SpeI was obtained and ligated into the digested pCAMBIA1302 vector by homologous recombination. Transient transformation of N. benthamiana leaves was used to examine protein localization with the empty vector as a control. At 48 h after injection, the tobacco leaves were collected to observe the green fluorescence. As shown in Figure 6, the fluorescent signals of ScCFAT-GFP fusion protein are distributed predominantly within the cytoplasm of the cells. Therefore, ScCFAT appears to be primarily a cytoplasmic protein.
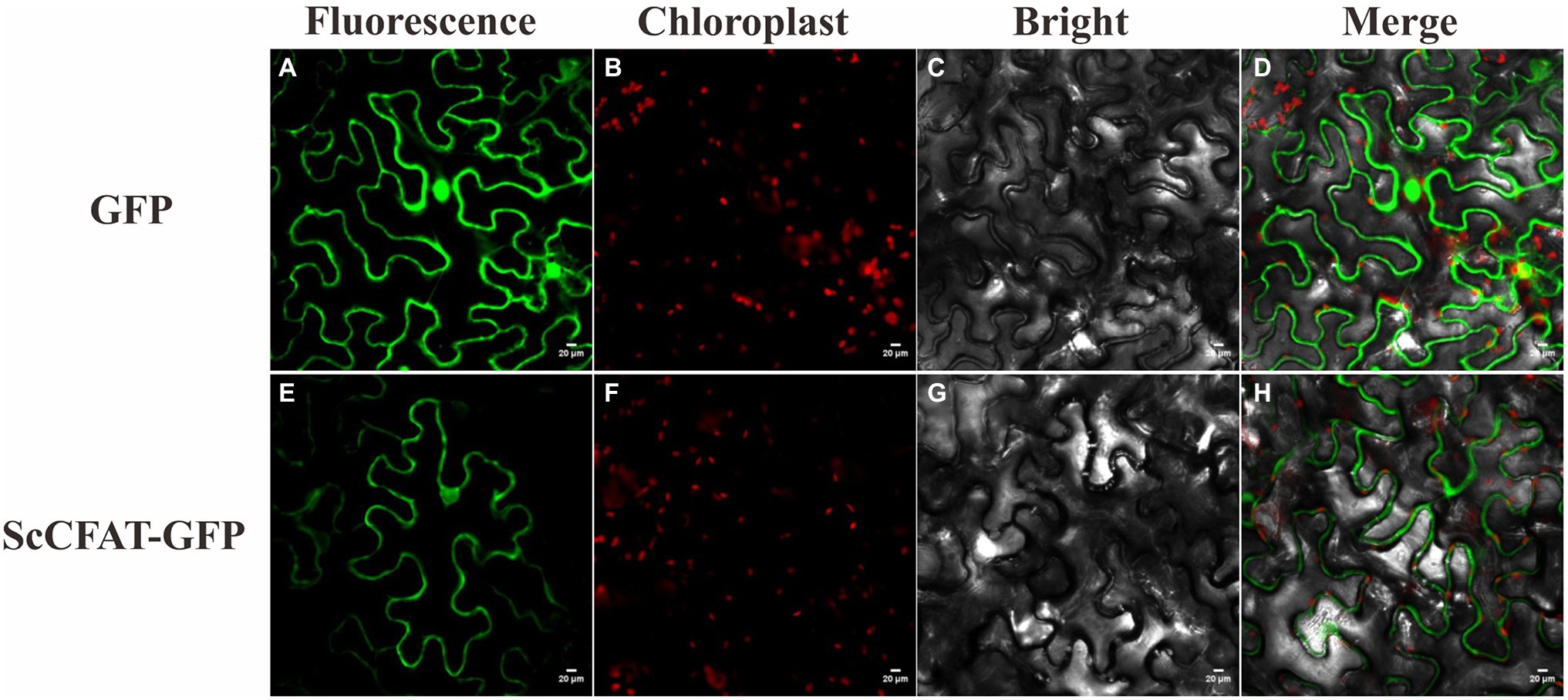
Figure 6. Subcellular localization of ScCFAT. The fusion proteins were observed under a confocal laser scanning microscope. a, e show the green fluorescence channel; b, f show the chloroplast autofluorescence channel; c, g show the bright field channel; and d, h were created from the images shown in the first three panels. Scale bar = 20 μm.
Due to the divergent evolution of the BAHD family from one ancestral gene, this family displays low overall sequence identity (25–34%; St-Pierre and Luca, 2000), but these proteins possess highly similar three-dimensional (3D) structures (Walker et al., 2013). The best template for modeling the ScCFAT structure is the X-ray crystal structure of rosmarinic acid synthase from Coleus blumei (PDB accession number: 6MK2) with a sequence identity of 28.71%. The final unperturbed conformations of the model were acquired by progressively relaxing parts of the initial model. The stereochemical quality of the ScCFAT model was analyzed with PROCHECK. According to the Ramachandran plot, the most favored regions correspond to 81.7% of the structure, and only 1.8% of residues are placed in disallowed regions (Supplementary Figure 4). An analysis using Verify-3D shows that 81.32% of the residues have averaged 3D-1D score ≥ 0.2 (Supplementary Figure 5). The final structure of ScCFAT protein was accepted for subsequent analyses.
The predicted ScCFAT structure consists of two nearly equal domains connected through a loop as shown in Figure 7A. A solvent channel runs through the ScCFAT molecule, allowing the substrate and co-substrate to bind independently. Results of docking simulation showed that the natural substrate acetyl-CoA was located in the solvent channel and docked with His158 as shown in Supplementary Figure 6. To identify the putative residues participating in the binding and/or catalysis of acetyl-CoA or coniferyl alcohol, molecular docking was conducted (Figures 7B,C). And the residues within 4 Å distance from the substrate position were selected as “hot spots” for the coming functional analysis.
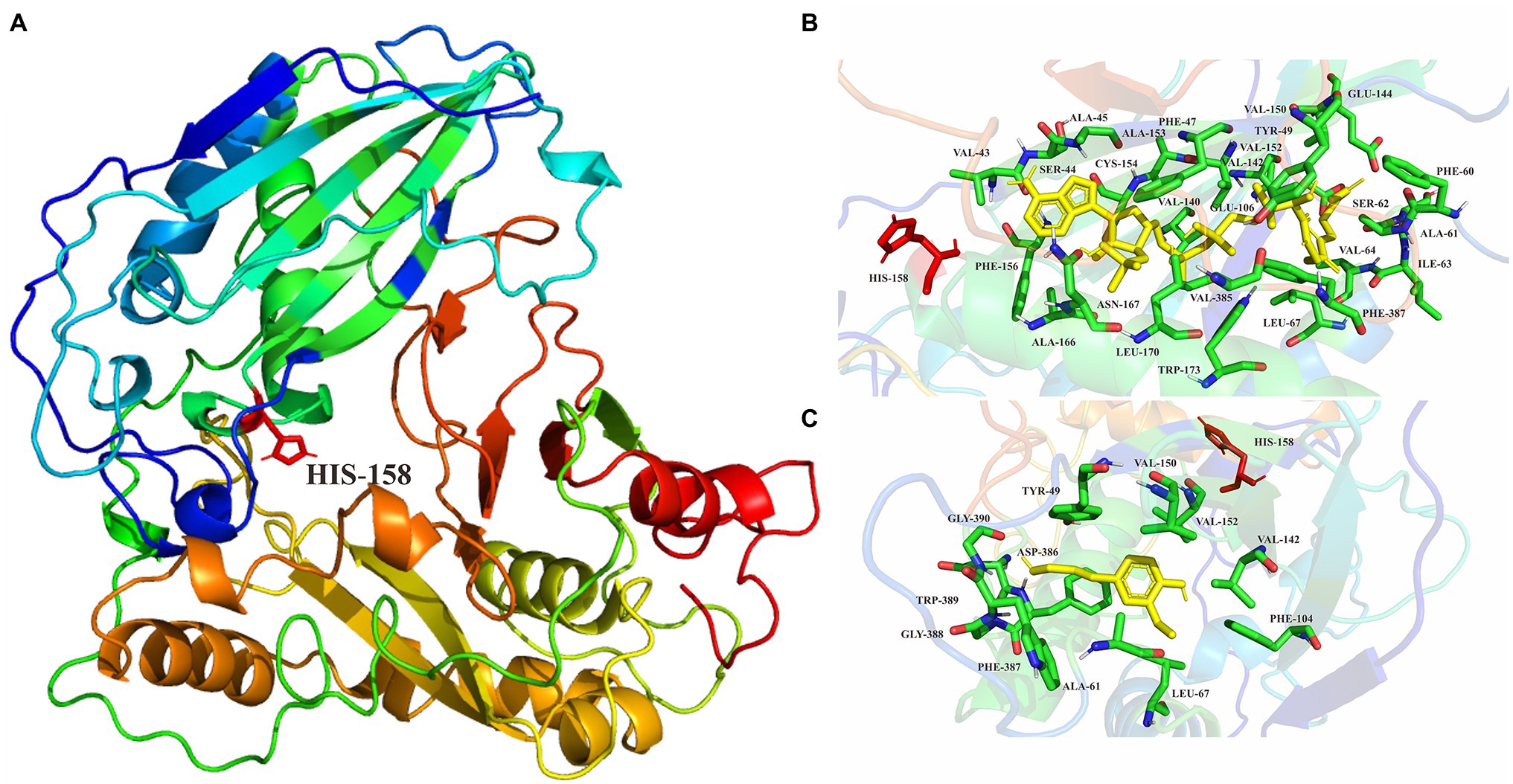
Figure 7. (A) Homology modeling of ScCFAT structure constructed with Modeller. His158 is shown in stick representation. (B) Enlarged views of molecular docking of ScCFAT with acetyl-CoA. (C) Enlarged views of molecular docking of ScCFAT with coniferyl alcohol.
CFAT, which must play an essential part in the biosynthetic pathway of dibenzocyclooctadiene lignans, is unknown in S. chinensis. Before studying ScCFAT, we first needed to confirm whether isoeugenol is produced in S. chinensis since the biosynthetic pathway of dibenzocyclooctadiene lignans is hypothetical and coniferyl acetate is unstable and undetected. However, no literature has confirmed the presence of isoeugenol in S. chinensis. Here, we report the extraction method, instrument parameters, and the first detection of isoeugenol in the fruits of S. chinensis by using GC–MS for the first time. The presence of isoeugenol not only proves that there must be a gene encoding CFAT in S. chinensis, but also indicates that the biosynthetic pathway of dibenzocyclooctadiene lignans shown in Figure 1 may be authentic.
PhCFAT, the first identified coniferyl alcohol acyltransferase from Petunia × hybrida, is critical to the production of isoeugenol and can catalyze the formation of coniferyl acetate from coniferyl alcohol and acetyl-CoA (Dexter et al., 2007). To pick out the ScCFAT gene, transcriptome-wide identification of BAHD genes in S. chinensis was performed and a total of 37 ScBAHD genes were obtained. Bioinformatics analyses were conducted and ScBAHDs were diverse based on their sequence and physiochemical properties. Then, we constructed a phylogenetic tree of the 37 putative ScBAHD proteins and 75 identified BAHD acyltransferases from other species. The phylogenetic tree is consistent with that of D’Auria (2006), who sorted 46 functionally characterized BAHD acyltransferases into five clades. The BAHD acyltransferases in clade I are mainly involved in the modification of anthocyanins (anthocyanin acyltransferases, AATs). Apart from HXXXD and DFGWG, most AATs share the sequence Tyr-Phe-Gly-Asn-Cys (YFGNC), which has been used as defining characteristic to clone new AATs by homology-based strategy (Nakayama et al., 2003). Although there are seven ScBAHDs clustered into clade I, only ScBAHD8, ScBAHD10, and ScBAHD14 have the motif YFGNC. Therefore, the three ScBAHDs are most likely to participate in the acylation of anthocyanins in S. chinensis. The BAHD acyltransferases in Clade III can acetylate a diverse range of alcohol substrates, and PhCFAT is clustered into this clade. ScBAHD1, ScBAHD3, and ScBAHD7 are close to PhCFAT, which shows that these three proteins might function as CFAT in S. chinensis. In addition, the BAHD proteins clustered into clade V can be subdivided into two well-supported groups corresponding to Tuominen’s clade Va and Vb (Tuominen et al., 2011).
Gene expression patterns can provide significant clues for predicting the biological function of genes. The content of dibenzocyclooctadiene lignans in the fruits of S. chinensis gradually increases as the fruit matures and reaches the maximum until the half-mature period (Zhao and Wang, 2011). To explore the expression profiles of 37 ScBAHD genes, the transcriptome datasets of S. chinensis fruits at the green, light red, and dark red stages were employed. The results indicate that the expression profile of ScBAHD1 showed higher expression at the light red and dark red stages than at the green stage, which was consistent with the accumulation of dibenzocyclooctadiene lignans. Therefore, based on the phylogenetic analysis and expression profile, it is suggested that ScBAHD1 is most likely to function as ScCFAT involved in the biosynthesis of dibenzocyclooctadiene lignans in S. chinensis.
The recombinant protein of ScBAHD1 was incubated with acetyl-CoA and coniferyl alcohol to assay its activity. Because coniferyl acetate is unstable, the product was detected by UPLC and verified with PhCFAT as a control. Functional analysis in vitro confirmed that ScBAHD1 catalyzed coniferyl alcohol to the same product as PhCFAT catalyzed, which indicated the ScBAHD1 we cloned from the S. chinensis fruits is ScCFAT. Subsequently, we screened the candidate genes coding IGS1 in S. chinensis through bioinformatics analyses and will conduct functional studies. PhCFAT is most efficient with coniferyl alcohol among the alcohol substrates tested and involved in the isoeugenol biosynthesis through RNAi-induced gene silencing (Dexter et al., 2007). From the size of the substrate peak area observed by UPLC (Figure 5B), we preliminarily found ScCFAT may have a higher activity with coniferyl alcohol than PhCFAT.
To further test the acetylation activity of ScCFAT, cinnamyl alcohol was used as an acceptor substrate with acetyl-CoA as a thioester donor. The product was verified to be cinnamyl acetate by using the authentic standard based on the same retention time. The ScCFAT enzyme was then further tested with various alcohol acceptors in combination with acetyl-CoA. Besides coniferyl alcohol and cinnamyl alcohol, ScCFAT exhibited detectable activity on sinapyl alcohol, p-coumaryl alcohol, (E)-3-phenyl-2-methyl-2-propenol, benzyl alcohol, 4-hydroxybenzyl alcohol, and (+)-secoisolariciresinol, but no activity was monitored for other tested substances. These results indicate that ScCFAT functionally prefers alcohol hydroxyl acetylation rather than phenol hydroxyl. Anyway, to determine whether coniferyl alcohol acts as the substate of ScCFAT in vivo, it is necessary to observe if there will be a decrease in the synthesis of isoeugenol after the suppression of the expression of ScCFAT via RNAi-induced gene silencing. However, at present, we lack an effective genetic transformation system of S. chinensis, which is time-consuming to establish. But we are working actively to carry out this part of work. In addition, the result of subcellular localization shows that ScCFAT is predominantly located in the cytoplasm, which is consistent with the prediction of BAHD acyltransferases as cytosolic soluble enzymes due to the lack of transit peptides or other sequences leading to localization in organelles or secretion (D’Auria, 2006).
Ma et al. crystallized and solved the X-ray crystal structure of vinorine synthase, which is the first representative of the BAHD superfamily (Ma et al., 2005). The structural analyses combined with biochemical and mutagenesis studies demonstrate that the HXXXD motif is involved in catalysis at the active site and is indispensable for acyltransferase activity, and the DFGWG motif is unique for BAHD enzymes and plays an important structural role (Suzuki et al., 2003; Bayer et al., 2004; Ma et al., 2005). Here, we generated a 3D structure of ScCFAT with the structure of C. blumei RAS as the template, thereby obtaining a reasonable model with 81.7% of the structure in the most favored regions. After molecular docking, the points within 4 Å distance from acetyl-CoA and coniferyl alcohol are selected as “hot spots” for individual mutations of the residues to L-alanine to identify putative residues participating in the binding and/or catalysis. Furthermore, site-directed mutagenesis and saturation mutagenesis of ScCFAT can be used to improve its catalytic efficiency against coniferyl alcohol, which potentially improves the yield of dibenzocyclooctadiene lignans in S. chinensis.
In addition, to determine the apparent optimal conditions for the enzymatic activity of ScCFAT, reactions need to be conducted at different temperatures, pH, and reaction times. Then, under the determined apparent optimal conditions, besides acetyl-CoA, the thioesters of malonyl-, benzoyl-, p-hydroxybenzoyl-, p-coumaroyl-, and feruloyl-CoAs are also needed to test as the potential substrates. We learned by consulting pieces of literature that the enzyme activity assay of CFAT should be measured by determining how much of the 14C-labeled acetyl group of acetyl-CoA is transferred to the side chain of acceptor substrate and the radioactive product is quantified through liquid scintillation counting (Dexter et al., 2007; Carlos et al., 2016; Zhao et al., 2021). We will further study the relative activities and the kinetics parameters of ScCFAT, crystallize and solve its X-ray crystal structure, and conduct a comprehensive study on its characteristics in combination with biochemical and mutagenic studies.
S. chinensis is indispensable as the source of dibenzocyclooctadiene lignans for medicines, food supplements, and cosmetics, but the Schisandra plants grow slowly. Chemical synthesis of dibenzocyclooctadiene lignans is feasible in theory, but it is multistage and in low overall yields (Chang et al., 2005). The core and unique features of dibenzocyclooctadiene lignans are biphenyl moiety and cyclooctadiene ring, which have long been regarded as interesting and challenging synthetic targets by organic chemists. The plant biotechnology method of in vitro culture of S. chinensis has potential usefulness. However, the amounts of dibenzocyclooctadiene lignans in vitro cultures were lower than that in the fruits (Szopa et al., 2016). According to biogenesis theory, biosynthesis is the most effective process in terms of energy consumption and yield. In recent years, the types and throughput of plant natural products synthesized by microorganisms are increasing, such as artemisinin (Paddon et al., 2013), tanshinones (Zhou et al., 2012; Guo et al., 2013), aglycons of ginsenosides (Dai et al., 2014), gastrodin (Bai et al., 2016), and so on.
Because the plant grows slowly, methods for constructing mutants and transgenic experiments are laborious, the discovery of biosynthetic genes involved in the biosynthesis of dibenzocyclooctadiene lignans is pioneering and challenging. Although Suzuki et al. proposed the biosynthetic pathway for dibenzocyclooctadiene lignans back in 2007, there has been no progress on functionally characterized enzymes of S. chinensis apart from ScDIR (GenBank accession number: ADR30610; Kim et al., 2012). The current research focus of our group is to decipher the pathway of dibenzocyclooctadiene lignans and we speculated on the enzymes involved in the four major uncharacterized steps shown in Figure 1. First, we consider that isoeugenol is converted to verrucosin under the action of an auxiliary oxidase and dirigent protein (DIR; Halls et al., 2004). And we identified DIR gene family in S. chinensis and screened candidate genes (Dong et al., 2021). Verrucosin is converted to dihydroguaiaretic acid under the action of pinoresinol/lariciresinol reductase (PLR; Katayama et al., 1992a,b). Then, dihydroguaiaretic acid is likely to be catalyzed by CYP450 and O-methyltransferases (OMT) to form pregomisin (Lau and Sattely, 2015; Nett et al., 2020). It is CYP450 catalyzing pregomisin to gomisin J (Ikezawa et al., 2008; Gesell et al., 2009).
Because ScCFAT is the first known and committed enzyme in the biosynthetic pathway of dibenzocyclooctadiene lignans, the subject of this study is to identify and obtain the sequence information of ScCFAT, which can be used to screen candidate genes coding DIR, PLR, CYP450, and OMT via co-expression analysis combined with gene family analyses. Therefore, ScCFAT is critical to the elucidation of the biosynthetic pathway of dibenzocyclooctadiene lignans. Subsequently, we will imitate the approach of podophyllotoxin biosynthetic pathway elucidation and use Agrobacterium-mediated transient expression in N. benthamiana to test the candidate genes (Lau and Sattely, 2015).
The sequence of ScCFAT reported in this article has been deposited in NCBI GenBank with accession number OM804184.
H-TL, B-GZ, and J-SL conceived and designed the study. T-YQ, X-LM, and YC carried out the experiments. T-YQ wrote the manuscript. Y-QD performed the bioinformatic analyses. H-ML and J-SL revised the manuscript. T-YQ, Y-QD, and J-SL analyzed the data. All authors contributed to the article and approved the submitted version.
This research was funded by the Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources (No. 2060302-2002-05), the National Natural Science Foundation of China (No. 81872965), and the CAMS Initiative for Innovative Medicine (No. 2021-1-I2M-031).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.881342/full#supplementary-material
Supplementary Figure 1 | The mass profiles of the S. chinensis fruits and the isoeugenol standard.
Supplementary Figure 2 | Gene expression analysis of ScBAHD1 by RT-qPCR at different fruit developmental stages.
Supplementary Figure 3 | UPLC analysis of compounds found in a reaction mixture containing purified PhCFAT, alcohol acceptors, and acetyl-CoA after 15 min of incubation.
Supplementary Figure 4 | Evaluation of the predicted structure of ScCFAT by Ramachandran plot.
Supplementary Figure 5 | Verify-3D graph drawn for the ScCFAT model.
Supplementary Figure 6 | Molecular docking of substrate acetyl-CoA with modeled ScCFAT structure in the active site pocket.
1. ^https://www.ncbi.nlm.nih.gov/
2. ^https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi
3. ^https://web.expasy.org/protparam/
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., Castro, E. D., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Bai, Y., Yin, H., Bi, H., Zhuang, Y., Liu, T., and Ma, Y. (2016). De novo biosynthesis of gastrodin in Escherichia coli. Metab. Eng. 35, 138–147. doi: 10.1016/j.ymben.2016.01.002
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bayer, A., Ma, X., and Stockigt, J. (2004). Acetyltransfer in natural product biosynthesis-functional cloning and molecular analysis of vinorine synthase. Bioorg. Med. Chem. 12, 2787–2795. doi: 10.1016/j.bmc.2004.02.029
Carlos, N. R., Carlos, G. E., Raúl, H., Julio, C., and Jans, H. A. M. (2016). Structural and affinity determinants in the interaction between alcohol acyltransferase from F. x ananassa and several alcohol substates: a computational study. PLoS One 11:e0153057. doi: 10.1371/journal.pone.0153057
Chakravarthy, S., Velásquez, A. C., Ekengren, S. K., Collmer, A., and Martin, G. B. (2010). Identification of Nicotiana benthamiana genes involved in pathogen-associated molecular pattern-triggered immunity. Mol. Plant-Microbe Interact. 23, 715–726. doi: 10.1094/MPMI-23-6-0715
Chang, J., Reiner, J., and Xie, J. (2005). Progress on the chemistry of dibenzocyclooctadiene lignans. Chem. Rev. 105, 4581–4609. doi: 10.1021/cr050531b
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020a). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Liu, S., Yan, Y., Yin, L., Di, P., Liu, H., et al. (2020b). Candidate genes involved in the biosynthesis of lignan in Schisandra chinensis fruit based on transcriptome and metabolomes analysis. Chin. J. Nat. Med. 1, 1–12.
Chenna, R., Sugawara, H., Koike, T., Lopez, R., Gibson, T. J., Higgins, D. J., et al. (2003). Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500. doi: 10.1093/nar/gkg500
Dai, Z., Wang, B., Liu, L., Shi, M., Wang, D., Zhang, X., et al. (2014). Producing aglycons of ginsenosides in bakers’ yeast. Sci. Rep. 4:3698. doi: 10.1038/srep03698
D’Auria, J. C. (2006). Acyltransferases in plants: a good time to be BAHD. Curr. Opin. Plant Biol. 9, 331–340. doi: 10.1016/j.pbi.2006.03.016
Dexter, R., Qualley, A., Kish, C. M., Ma, C. J., Koeduka, T., Nagegowda, D. A., et al. (2007). Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 49, 265–275. doi: 10.1111/j.1365-313X.2006.02954.x
Dong, Y., Qiang, T., Liu, J., Li, B., Wei, X., Qi, Y., et al. (2021). Identification and characterization of DIR gene family in Schisandra chinensis. Chin J Chin Mater Med. 46, 5270–5277. doi: 10.19540/j.cnki.cjcmm.20210723.101
Dudareva, N., D’Auria, J. C., Nam, K. H., Raguso, R. A., and Pichersky, E. (1998). Acetyl-CoA: benzylalcohol acetyltransferase-an enzyme involved in floral scent production in Clarkia breweri. Plant J. 14, 297–304. doi: 10.1046/j.1365-313X.1998.00121.x
Eddy, S. R. (2004). What is a hidden Markov model? Nat. Biotechnol. 22, 1315–1316. doi: 10.1038/nbt1004-1315
Erdtman, H. (1933). Dehydrierungen in der coniferylreihe. (I). dehydrodi-eugenol and dehydrodiisoeugenol. Biochem. Z. 258, 172–180.
Gesell, A., Rolf, M., Ziegler, J., Chávea, M. L. D., Huang, F. C., and Kutchan, T. M. (2009). CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem. 284, 24432–24442. doi: 10.1074/jbc.M109.033373
Grothe, T., Lenz, R., and Kutchan, T. M. (2001). Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy Papaver somniferum. J. Biol. Chem. 276, 30717–30723. doi: 10.1074/jbc.M102688200
Guo, J., Zhou, Y., Hillwig, M. L., Shen, Y., Yang, L., Wang, Y., et al. (2013). CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc. Natl. Acad. Sci. U. S. A. 110, 12108–12113. doi: 10.1073/pnas.1218061110
Halls, S. C., Davin, L. B., Kramer, D. M., and Lewis, N. G. (2004). Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemist 43, 2587–2595. doi: 10.1021/bi035959o
Hancke, J. L., Burgos, R. A., and Ahumade, F. (1999). Schisandra chinensis (Turcz.) Baill. Fitoterapia 70, 451–471. doi: 10.1016/S0367-326X(99)00102-1
Hikino, H., Kiso, Y., Taguchi, H., and Ikeya, Y. (1984). Antihepatotoxic actions of lignoids from Schisandra chinensis fruits. Planta Med. 50, 213–218. doi: 10.1055/s-2007-969681
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Ikezawa, N., Iwasa, K., and Sato, F. (2008). Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C-C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 283, 8810–8821. doi: 10.1074/jbc.M705082200
Katayama, T., Davin, L. B., Chu, A., and Lewis, N. G. (1992b). Novel benzylic ether reductions in lignan biogenesis in Forsythia intermedia. Phytochemistry 33, 581–591. doi: 10.1016/0031-9422(93)85452-W
Katayama, T., Davin, L. B., and Lewis, N. G. (1992a). An extraordinary accumulation of (−)-pinoresinol in cell-free extracts of Forsythia intermedia: evidence for enantiospecific reduction of (+)-pinoresinol. Phytochemistry 31, 3875–3881. doi: 10.1016/S0031-9422(00)97545-9
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, K. W., Moinuddin, S. G. A., Atwell, K. M., Costa, M. A., Davin, L. B., and Lewis, N. G. (2012). Opposite stereoselectivities of dirigent proteins in Arabidopsis and Schizandra species. J. Biol. Chem. 287, 33957–33972. doi: 10.1074/jbc.M112.387423
Kim, H. J., Ono, E., Morimoto, K., Yamagaki, T., Okazawa, A., Kobayashi, A., et al. (2009). Metabolic engineering of lignan biosynthesis in Forsythia cell culture. Plant Cell Physiol. 50, 2200–2209. doi: 10.1093/pcp/pcp156
Koeduka, T., Fridman, E., Gang, D. R., Vassão, D. G., Jackson, B. L., Kish, C. M., et al. (2006). Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. U. S. A. 103, 10128–10133. doi: 10.1073/pnas.0603732103
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. doi: 10.1007/BF00228148
Lau, W., and Sattely, E. S. (2015). Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science 349, 1224–1228. doi: 10.1126/science.aac7202
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, B., and Dewey, C. N. (2011). RSEM accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 1–16. doi: 10.1186/1471-2105-12-323
Liu, J., Huang, M., and Gao, Y. (1981). The structure of chicanine, a new lignan from Schisandra sp. Can. J. Chem. 59, 1680–1684. doi: 10.1139/v81-250
Lopes, N. P., Yoshida, M., and Kato, M. J. (2004). Biosynthesis of tetrahydrofuran lignans in Virola surinamensis. Braz. J. Pharm. Sci. 40, 53–57. doi: 10.1590/S1516-93322004000100009
Lüthy, R., Bowie, J. U., and Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature 356, 83–85. doi: 10.1016/S0076-6879(97)77022-8
Ma, X., Koepke, J., Panjikar, S., Fritzsch, G., and Stockigt, J. (2005). Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem. 280, 13576–13583. doi: 10.1074/jbc.M414508200
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Haeseler, A. V., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L. L., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Muhlemann, J. K., Woodworth, B. D., Morgan, J. A., and Dudareva, N. (2014). The monolignol pathway contributes to the biosynthesis of volatile phenylpropenes in flowers. New Phytol. 204, 661–670. doi: 10.1111/nph.12913
Nakayama, T., Suzuki, H., and Nishino, T. (2003). Anthocyanin acyltransferases: specificities, mechanism, phylogenetics, and applications. J. Mol. Catal. B Enzym. 23, 117–132. doi: 10.1016/S1381-1177(03)00078-X
Nett, R. S., Lau, W., and Sattely, E. S. (2020). Discovery and engineering of colchicine alkaloid biosynthesis. Nature 584, 148–153. doi: 10.1038/s41586-020-2546-8
Ono, E., Nakai, M., Fukui, Y., Tomimori, N., Fukuchi-Mizutani, M., Saito, M., et al. (2006). Formation of two methylenedioxy bridges by a Sesamum CYP81Q protein yielding a furofuran lignan, (+)-sesamin. Proc. Natl. Acad. Sci. U. S. A. 103, 10116–10121. doi: 10.1073/pnas.0603865103
Opletal, L., Sovová, H., and Bártlová, M. (2004). Dibenzo[a,c]cyclooctadiene lignans of the genus Schisandra: importance, isolation and determination. J. Chromatogr. B 812, 357–371. doi: 10.1016/j.jchromb.2004.07.040
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi: 10.1038/nature12051
St-Pierre, B., Laflamme, P., Alarco, A. M., and Luca, V. D. (1998). The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 14, 703–713. doi: 10.1046/j.1365-313x.1998.00174.x
St-Pierre, B., and Luca, V. D. (2000). Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. Recent Adv. Phytochem. 34, 285–315. doi: 10.1016/S0079-9920(00)80010-6
Suzuki, H., Nakayama, T., and Nishino, T. (2003). Proposed mechanism and functional amino acid residues of malonyl-CoA: anthocyanin 5-O-glucoside-6′′′-O-malonyltransferase from flowers of Salvia splendens, a member of the versatile plant acyltransferase family. Biochemistry 42, 1764–1771. doi: 10.1021/bi020618g
Suzuki, H., Nakayama, T., Sakakibara, K. Y., Fukui, Y., Nakamura, N., Nakao, M., et al. (2001). Malonyl-CoA: anthocyanin 5-O-glucoside-6‴-O-malonyltransferase from scarlet sage (Salvia splendens) flowers. J. Biol. Chem. 276, 49013–49019. doi: 10.1074/jbc.M108444200
Suzuki, H., Nakayama, T., Sakakibara, K. Y., Fukui, Y., Nakamura, N., Yamaguchi, M., et al. (2002). cDNA cloning, heterologous expressions, and functional characterization of malonyl-coenzyme A: anthocyanidin 3-O-glucoside-6″-O-malonyltransferase from dahlia flowers. Plant Physiol. 130, 2142–2151. doi: 10.1104/pp.010447
Suzuki, S., and Umezawa, T. (2007). Biosynthesis of lignans and norlignans. J. Wood Sci. 53, 273–284. doi: 10.1007/s10086-007-0892-x
Szopa, A., Ekiert, R., and Ekiert, H. (2017). Current knowledge of Schisandra chinensis (Turcz.) Baill. (Chinese magnolia vine) as a medicinal plant species: a review on the bioactive components, pharmacological properties, analytical and biotechnological studies. Phytochem. Rev. 16, 195–218. doi: 10.1007/s11101-016-9470-4
Szopa, A., Kokotkiewicz, A., Marzec-Wróblewska, U., Bucinski, A., Luczkiewicz, M., and Ekiert, H. (2016). Accumulation of dibenzocyclooctadiene lignans in agar cultures and in stationary and agitated liquid cultures of Schisandra chinensis (Turcz.) Baill. Appl. Microbiol. Biotechnol. 100, 3965–3977. doi: 10.1007/s00253-015-7230-9
Tuominen, L. K., Johnson, V. E., and Tsai, C. J. (2011). Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12, 1–17. doi: 10.1186/1471-2164-12-236
Umezawa, T. (2003). Diversity in lignan biosynthesis. Phytochem. Rev. 2, 371–390. doi: 10.1023/B:PHYT.0000045487.02836.32
Vanholme, R., Demedts, B., Morreel, K., Ralph, J., and Boerjan, W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153, 895–905. doi: 10.1104/pp.110.155119
Walker, K., and Croteau, R. (2000a). Taxol biosynthesis: molecular cloning of a benzoyl CoA: taxane 2α-O-benzoyltransferase cDNA from Taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 97, 13591–13596. doi: 10.1073/pnas.250491997
Walker, K., and Croteau, R. (2000b). Molecular cloning of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97, 583–587. doi: 10.1073/pnas.97.2.583
Walker, A. M., Hayes, R. P., Youn, B., Vermerris, W., Sattler, S. E., and Kang, C. (2013). Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme a-dependent transferases and synthases. Plant Physiol. 162, 640–651. doi: 10.1104/pp.113.217836
Walker, K., Schoendorf, A., and Croteau, R. (2000). Molecular cloning of a Taxa-4(20),11(12)-dien-5α-ol-O-acetyl Transferase cDNA from Taxus and functional expression in Escherichia coli. Arch. Biochem. Biophys. 374, 371–380. doi: 10.1006/abbi.1999.1609
Yu, X., Guo, J., and Liu, C. (2009). BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression. Plant Mol. Biol. 70, 421–442. doi: 10.1007/s11103-009-9482-1
Zhai, J., Zhang, F., Gao, S., Chen, L., Feng, G., Yin, J., et al. (2018). Schisandra chinensis extract decreases chloroacetaldehyde production in rats and attenuates cyclophosphamide toxicity in liver, kidney and brain. J. Ethnopharmacol. 210, 223–231. doi: 10.1016/j.jep.2017.08.020
Zhang, T., Huo, T., Ding, A., Hao, R., Wang, J., Cheng, T., et al. (2019). Genome-wide identification, characterization, expression and enzyme activity analysis of coniferyl alcohol acetyltransferase genes involved in eugenol biosynthesis in Prunus mume. PLoS One 14:e0223974. doi: 10.1371/journal.pone.0223974
Zhao, Y., and Wang, B. (2011). Determination of five kinds of lignans in Schisandra chinensis at different growing period by HPLC. West China J. Pharm. Sci. 26, 172–174. doi: 10.13375/j.cnki.wcjps.2011.02.022
Zhao, Y., Yu, X., Lam, P. Y., Zhang, K., Tobimatsu, Y., and Liu, C. (2021). Monolignol acyltransferase for lignin p-hydroxybenzoylation in Populus. Nat Plants. 7, 1288–1300. doi: 10.1038/s41477-021-00975-1
Zhou, Y., Gao, W., Rong, Q., Jin, G., Chu, H., Liu, W., et al. (2012). Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Am. Chem. Soc. 134, 3234–3241. doi: 10.1021/ja2114486
Keywords: Schisandra chinensis, coniferyl alcohol, coniferyl alcohol acyltransferase, BAHD acyltransferase family, dibenzocyclooctadiene lignans
Citation: Qiang T-Y, Liu J-S, Dong Y-Q, Mu X-L, Chen Y, Luo H-M, Zhang B-G and Liu H-T (2022) Identification, Molecular Cloning, and Functional Characterization of a Coniferyl Alcohol Acyltransferase Involved in the Biosynthesis of Dibenzocyclooctadiene Lignans in Schisandra chinensis. Front. Plant Sci. 13:881342. doi: 10.3389/fpls.2022.881342
Received: 22 February 2022; Accepted: 24 May 2022;
Published: 23 June 2022.
Edited by:
Wei Zhou, Zhejiang Chinese Medical University, ChinaCopyright © 2022 Qiang, Liu, Dong, Mu, Chen, Luo, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai-Tao Liu, aHRsaXUwNzE4QDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.