
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 25 April 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.880919
 Ruosi Li1†
Ruosi Li1† Zhen Li1,2†
Zhen Li1,2† Jing Ye3†
Jing Ye3† Yingying Yang1
Yingying Yang1 Juahua Ye1
Juahua Ye1 Siliang Xu1
Siliang Xu1 Junrong Liu1,2
Junrong Liu1,2 Xiaoping Yuan1
Xiaoping Yuan1 Yiping Wang1
Yiping Wang1 Mengchen Zhang1
Mengchen Zhang1 Hanyong Yu1
Hanyong Yu1 Qun Xu1
Qun Xu1 Shan Wang1
Shan Wang1 Yaolong Yang1
Yaolong Yang1 Shu Wang2*
Shu Wang2* Xinghua Wei1*
Xinghua Wei1* Yue Feng1*
Yue Feng1*Grain size, grain number per panicle, and grain weight are key agronomic traits that determine grain yield in rice. However, the molecular mechanisms coordinately controlling these traits remain largely unknown. In this study, we identified a major QTL, SMG3, that is responsible for grain size, grain number per panicle, and grain weight in rice, which encodes a MYB-like protein. The SMG3 allele from M494 causes an increase in the number of grains per panicle but produces smaller grain size and thousand grain weight. The SMG3 is constitutively expressed in various organs in rice, and the SMG3 protein is located in the nucleus. Microscopy analysis shows that SMG3 mainly produces long grains by increasing in both cell length and cell number in the length direction, which thus enhances grain weight by promoting cell expansion and cell proliferation. Overexpression of SMG3 in rice produces a phenotype with more grains but reduces grain length and weight. Our results reveal that SMG3 plays an important role in the coordinated regulation of grain size, grain number per panicle, and grain weight, providing a new insight into synergistical modification on the grain appearance quality, grain number per panicle, and grain weight in rice.
Rice feeds nearly half of the world’s population. The yield per plant in rice is determined by four components: number of panicles, number of grains per panicle, grain weight, and ratio of filled grain (Li et al., 2013). The grain size of rice directly determines the grain weight, which in turn affects the yield of rice. In addition, grain size is an important appearance quality trait, which directly affects the milling quality of rice. Grain number per panicle is determined by the number of primary and secondary branches in rice (Xing and Zhang, 2010).
In recent years, using molecular markers to construct genetic linkage maps and QTL mapping for complex traits in rice has become a routine approach. Up to now, more than 500 QTLs related to rice grain size have been mapped,1 and several genes controlling grain size have been identified using natural variation populations. For example, GS3 was the first cloned QTL responsible for grain size, which encodes a transmembrane protein containing four putative domains and negatively regulates grain length and grain weight (Mao et al., 2010). GL3.1/qGL3 encodes a putative protein phosphatase (OsPPKL1) containing the Kelch repeat domains and regulates rice grain size and yield by influencing phosphorylation of Cyclin-T1;3 (Qi et al., 2012; Zhang et al., 2012; Gao et al., 2019). GL7 was a major QTL for grain size in rice, and the tandem duplication of a 17.1-kb at the GL7 locus leads to upregulation of GL7 expression, thus increasing grain length and improved appearance quality of rice (Wang et al., 2015a,b). GS9 encodes a protein without known conserved functional domain, which regulates grain shape by altering cell division (Zhao et al., 2018). TGW2 was a novel grain size QTL, encoding the CELL NUMBER REGULATOR 1 (OsCNR1), that interacts with KRP1, a regulator of cell cycles in plants, to negatively regulate grain width and weight (Ruan et al., 2020). Recently, a novel miR167a-OsARF6-OsAUX3 module which regulates grain length and weight was reported in rice (Qiao et al., 2021).
In rice, several genes associated with grain number have been cloned and characterized (Ashikari et al., 2005; Kurakawa et al., 2007; Huang et al., 2009; Oikawa and Kyozuka, 2009; Tabuchi et al., 2011; Wu et al., 2016; Huo et al., 2017; Guo et al., 2018; Ren et al., 2018). Rice has a determinate inflorescence, in which inflorescence meristems initiate primary branch meristems that are attached to the central rachis of the inflorescence and then produce several secondary branch meristems (Zhang and Yuan, 2014). During panicle development, the primary and secondary branch meristems differentiate into spikelets, which determine the number of grains per panicle (Tabuchi et al., 2011; Guo et al., 2018). The spikelet hull consists of a palea and a lemma, which determine the final grain size in rice (Li and Li, 2016). However, the underlying molecular mechanisms coordinating grain size and grain number in rice remain largely unclear.
In this study, we detected fourteen QTLs affecting grain size, grain weight, and grain number per panicle on six chromosomes. In addition, we fine mapped and cloned of a QTL, SMG3, coordinately regulating grain size, grain number per panicle, and grain weight near the centromere on chromosome 3. We demonstrated that SMG3 acted as a negative regulator of grain length, grain weight, and functioned as a positive regulator of grain number per panicle. These results laid a foundation for further genetic manipulation and trade-offs between grain size and grain number in rice.
The Indian small-grain rice M494 (japonica) and long-grain rice Zhong9 B (Z9B; indica) were both derived from the National Rice Mid-term Gene Bank of China. The F2 population consisting of 144 lines was developed from a cross between M494 and Z9B. Based on the QTL mapping results, we selected two long-grain lines in which GS3 was fixed with homozygous M494, but the SMG3 region was heterozygous in the F2:3 population. The two long-grain lines were backcrossed with M494 for three successive generations to obtain BC3F2 population. At first, 80 long-grain BC3F2 individual plants and 2,850 BC3F3 recessive single plants were genotyped with two markers Y3-53 and Y3-103 to fine map the grain size QTL-SMG3 (Supplementary Table 1). Seven recombinant plants were further selected and selfed to produce BC3F3 lines for phenotyping and substitution mapping. Two sets of NIL-SMG3 (NIL-SMG3M494 and NIL-SMG3Z9B) were constructed (Supplementary Figure 1).
Field planting was carried out according to the conventional management mode of rice planting, the F2 population was planted in Fuyang experimental fields of China National Rice Research Institute (CNRRI; 30°32′ N, 120°12′ E) in the summer of 2016, and the F2:3 population was planted in Lingshui station of CNRRI (18°48′ N, 110°02′ E) in the winter of 2016. The BC3F2 population and NILs were planted in Fuyang experimental fields of CNRRI in the summer of 2019 and 2020. The BC3F3 lines were planted in Lingshui station of CNRRI in the winter of 2019.
The F2 and BC3F2 populations were planted with 20 cm between plants and 25 cm between rows. The BC3F3 lines and NILs were grown in a randomized complete block design with two replications, five rows per plot, and eight plants per row.
At mature stage, the seeds were individually harvested for phenotypic investigation. The full grains were selected, and thousand grain weight (TGW) was weighed on the electronic balance. Each single plant was weighed three times, and the average value was taken. Grain length (GL), grain width (GW), and grain number per panicle (GNP) were measured by an automatic seed counting and analyzing instrument (Model SC-G, Wanshen Ltd., Hangzhou, China). The grain length-width ratio (LWR) is equal to GL divided by its GW. In addition, the percentage of chalky grains and degree of chalkiness of milled rice were measured using a grain appearance analyzer instrument (Model SC-E, Wanshen Ltd., Hangzhou, China).
For glume cell observation, the spikelets of NIL-SMG3M494 and NIL-SMG3Z9B were collected at maturity stage. The samples were fixed in FAA solution (formalin: glacial acetic acid: ethanol in 1:1:18 ratio by volume) at 4°C for 24 h, then were dehydrated and dried as described by Feng et al. (2021). The outer and inner surfaces of the spikelet glumes were observed under the scanning electron microscope (Hitachi, S-3400).
The RNA was extracted from various plant tissues using a MiniBEST Plant RNA Extraction Kit (Takara Bio Inc., Japan). First-strand cDNAs were synthesized using Prime Script RT Master Mix (TAKARA Bio Inc.). Quantitative RT-PCR analysis was performed on the Applied Biosystems 7,500 Real-Time PCR system with a 2 × SYBR Green PCR Master Mix (Applied Biosystems). The rice Actin gene was used as an internal control. Each measurement was replicated at least three times with three biological samples. The qRT-PCR primers used in these assays are presented in Supplementary Table 1.
For complementation test of SMG3, using the high-purity DNA extracted from the NIL-SMG3M494 as the template, the gene-specific primers were used to amplify the DNA fragment, including the 2-kb region upstream of the transcription start site and 1.5-kb downstream of the termination site of SMG3/LOC_Os03g29614, and cloned into the pCAMBIA1300 vector to generate pSMG3::SMG3M494 plasmid. The plasmid was introduced into the callus of NIL-SMG3Z9B by Agrobacterium-mediated transformation.
To generate the overexpression construct, the full-length coding sequence (CDS) of LOC_Os03g29614 was amplified from NIL-SMG3M494 and cloned into the vector pCAMBIA1300S. The plasmid was introduced into the callus of Nipponbare by Agrobacterium-mediated transformation.
To analyze the expression pattern of SMG3, a 2072-bp promoter fragment was cloned from NIL-SMG3M494 and was introduced into the pCAMBIA1301 vector. The constructed plasmid SMG3::GUS was then transformed into the rice variety Nipponbare by the Agrobacterium-mediated method. GUS staining was performed on some parts (root, base, stem, node, flag leave, pulvinus, leaf sheath, young panicle, and flowering panicle) of positive plants (Jefferson, 1989). After decolorization, several parts were placed in the scanner for scanning and observation. The primers used are listed in Supplementary Table 1.
To determine the subcellular localization of SMG3, the Pro35S::SMG3-GFP plasmid was introduced into the Agrobacterium tumefaciens GV3101 and injected into rice protoplasts. The nuclear protein GHD7 fused with RFP was used as a nuclear marker (Xue et al., 2008; Fu et al., 2019). GFP signal was observed by confocal laser microscope. The primers are listed in Supplementary Table 1.
Genetic map was constructed using Mapmaker/EXP version 3.0 of SSR marker genotype data. The Kosambi Mapping Function was used to convert the recombination frequency to cM. The QTLs were named as described by McCouch et al. (1997).
All data were collected from at least three independent biological replicates and presented as means ± SD. Statistical analysis was performed using the SAS version 9.2. Statistical significance was determined using Student’s t-test for comparison of the two groups.
The M494 and Z9B differed significantly in grain size, grain weight, and grain number (Figures 1A–H). To dissect the genetic basis of these traits, we conducted a quantitative trait locus (QTL) analysis using an F2 population derived from a cross between M494 and Z9B. Fourteen QTLs affecting grain size, grain weight, and grain number per panicle (GNP), including one for grain length (GL), six for grain width (GW), three for grain length to width ratio (LWR), one for thousand grain weight (TGW), and three for GNP, were identified in this population (Supplementary Table 2).
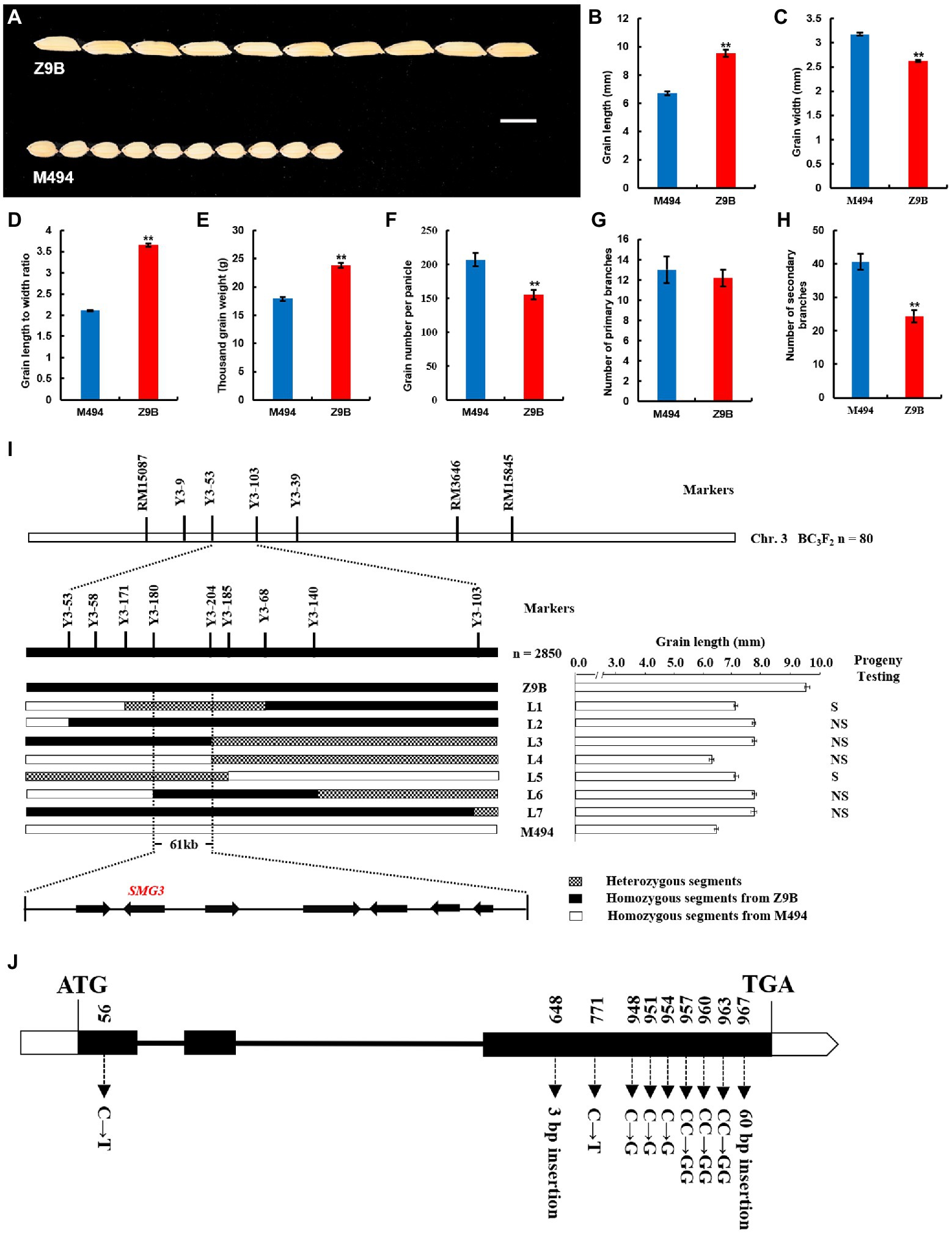
Figure 1. Map-based cloning of SMG3. (A) Z9B and M494 rice grains. Scale bar: 6 mm. (B–H) Comparison of Z9B and M494 grain length (n = 20; B), grain width (n = 20; C), grain length to width ratio (n = 20; D), thousand grain weight (n = 20; E), grain number per panicle (n = 20; F), number of primary branches (n = 20; G), and number of secondary branches (n = 20; H). Data are given as means ± SD. **indicates p < 0.01 by Student’s t-test. (I) Fine mapping of SMG3 and candidate gene analysis. S, segregation; NS, no segregation. (J) Gene structure and allelic variations of LOC_Os03g29614 (SMG3) between Z9B and M494. Filled boxes represent exons, black lines represent introns. The start codon (ATG) and the stop codon (TGA) are indicated.
On the chromosome 3, we found there were two LOD peaks for grain size and weight at approximately 79 cM and 92 cM in the F2 population (Supplementary Figure 2), which suggested that there existed two QTLs for grain size and weight in the 79–92 cM region of chromosome 3. One peak indicated that is the previously cloned grain length gene GS3, and the other might be a novel QTL for grain size and weight closely linked to GS3. Therefore, first, we use the functional marker SF28 of GS3 gene to screen out the individual plant with genotype A of GS3 and a long-grain phenotype relative to M494 in the F2:3 population (Fan et al., 2009). Then, we constructed a BC3F2 population with M494 as a recurrent parent in the target region flanked by RM15087 and RM3646. The QTL named SMG3 for grain length was mapped to the region between the STS marker Y3-53 and the Indel marker Y3-103 using 80 long-grain BC3F2 individual plants and their progenies. To fine map the position of the SMG3 locus, we selected 2,850 BC3F3 individuals heterozygous between marker Y3-53 and Y3-103. Seven recombinants were identified, which were genotyped with seven newly designed indel markers, progenies exhibiting long-grain phenotype (Supplementary Table 1). A precise phenotyping of grain length was performed by progeny testing, which also enabled the determination of segregation patterns of SMG3 in each recombinant line. The L4 progeny line showed short grain length, indicating that the L4 recombinant line was M494 homozygous for SMG3. The L2, L3, L6, and L7 progeny lines at SMG3 locus were Z9B homozygous due to their progenies exhibited long-grain phenotype. The L1 and L5 progeny lines displayed segregating grain length, indicating that the L1 and L5 recombinant lines were heterozygous at SMG3 locus (Figure 1I). These results allowed us to delimite SMG3 to a region of approximately 61 kb between the markers Y3-180 and Y3-204.
In the 61 kb target region, seven genes are annotated. Among these genes, LOC_Os03g29600 encodes a putative transposon protein, LOC_Os03g29614 encodes a protein containing a putative myb-like DNA-binding domain, LOC_Os03g29630 encodes an ulp1 protease family protein, LOC_Os03g29650 and LOC_Os03g29660 encode putative retrotransposon proteins, LOC_Os03g29680 encodes a putative early flowering protein, and LOC_Os03g29690 encodes a putative expressed protein. Interestingly, the LOC_Os03g29614 encodes the myb-related protein OsMYB3 and was identified as a negative regulator of grain length (Suzuki et al., 1997; Li et al., 2020), which suggested that the LOC_Os03g29614 is the most likely candidate gene for SMG3.
The LOC_Os03g29614 gene contains 3 exons and 2 introns (Figure 1J). We sequenced the LOC_Os03g_29614 gene in the M494 and Z9B varieties. Comparative analysis of the coding regions of the LOC_Os03g_29614 gene revealed that the M494 contains eight polymorphisms compared with Z9B, including one nucleotide substitution (c.56 C to T) in the exon 1, one 3 bp insertion mutation (c.648 - to GCA), one synonymous mutation (c.771 C to T), six missense polymorphisms (c.948 C to G, c.951 C to G, c.954 C to G, c.957 CC to GG, c.960 CC to GG, and c.963 CC to GG) and a 60 bp insertion in the exon 3 (Figure 1J). The novel 60 bp insertion in the exon 3 of M494 resulted in a frame-shift mutation and may influence the grain size, grain weight and grain number per panicle.
To confirm whether the LOC_Os03g_29614 gene was the candidate gene for SMG3, we carried out a genetic complementation test. A NIL-SMG3M494 DNA fragment containing the 2-kb promoter region, the entire LOC_Os03g_29614 gene, and 1.5-kb downstream of the gene was cloned into the binary vector pCAMBIA1300 to generate the pSMG3 construct. The pSMG3 construct was transferred into the NIL-SMG3Z9B background. The positive transgenic plants produced shorter grains and reduced TGW compared with the NIL-SMG3Z9B (Figures 2A,B). In addition, we overexpressed NIL-SMG3M494 in the Nipponbare background and found that the SMG3-overexpressing plants exhibited a decrease in GL and TGW but increase in GNP (Figures 2C,D; Supplementary Figure 3). Our results indicated that the SMG3 gene is a new allele of OsMYB3, which functioned as a negative regulator for grain size and grain weight, but also as a positive regulator of GNP. This finding showed that the Indian variety M494 possessed a desirable allele of OsMYB3, which provided a valuable resource for rice genetics research and breeding.
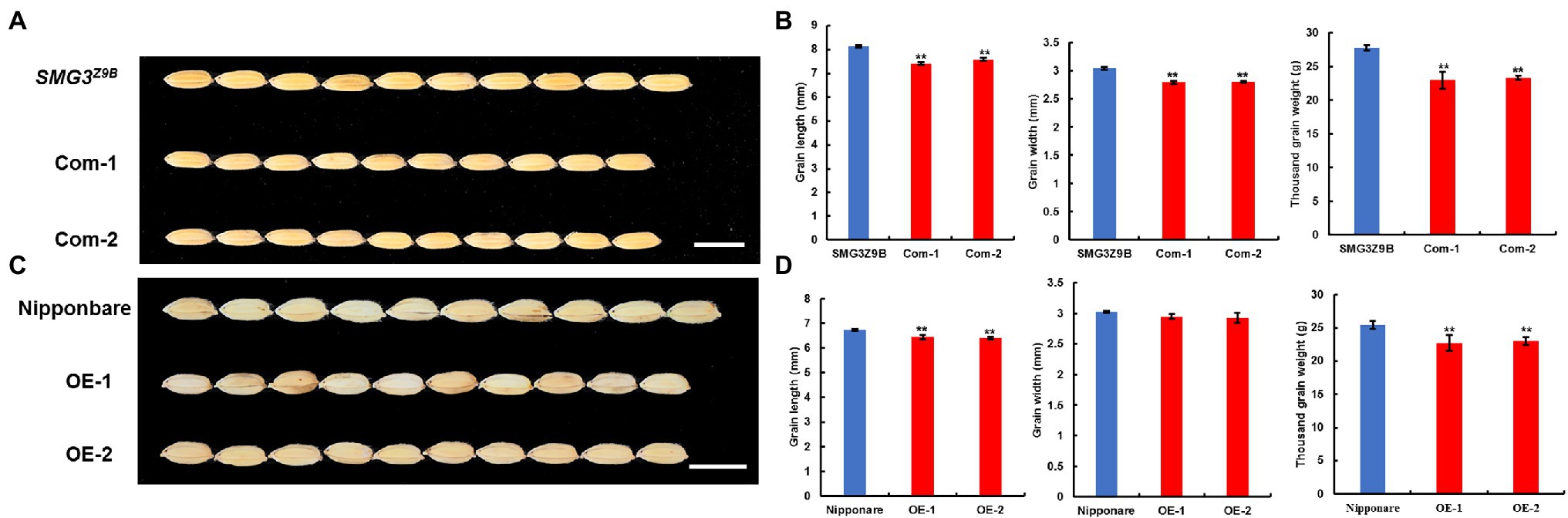
Figure 2. Complementary test and overexpression analysis. (A) Grains of NIL-SMG3Z9B and complementary (Com) transgenic plants. Scale bar: 8 mm. (B) Grain length, grain width, and thousand grain weight of NIL-SMG3Z9B and the complementary transgenic lines. (C) Grains of Nipponbare and overexpression (OE) transgenic plants. Scale bar: 8 mm. (D) Grain length, grain width, and thousand grain weight of Nipponbare and the SMG3 overexpression transgenic lines. Data in (B) and (D) are the means ± SD; ** indicates p < 0.01 by Student’s t-test.
To clarify the function of SMG3, we developed two near-isogenic lines (NILs) differing only at the SMG3 region. The NILs carrying the M494 and Z9B alleles at the SMG3 locus were designated NIL-SMG3M494 and NIL-SMG3Z9B, respectively. Compared with NIL-SMG3Z9B, NIL-SMG3M494 showed significantly decreased GL, GW, and TGW, increased grain number per plant, GNP, and the number of secondary branches (SB; Figures 3A–G,I). However, no significant differences were observed between the NILs in number of primary branches and grain yield per plant (GYP; Figures 3H,J). These results suggest that SMG3 plays an important role in coordinated regulation between grain size and grain number.
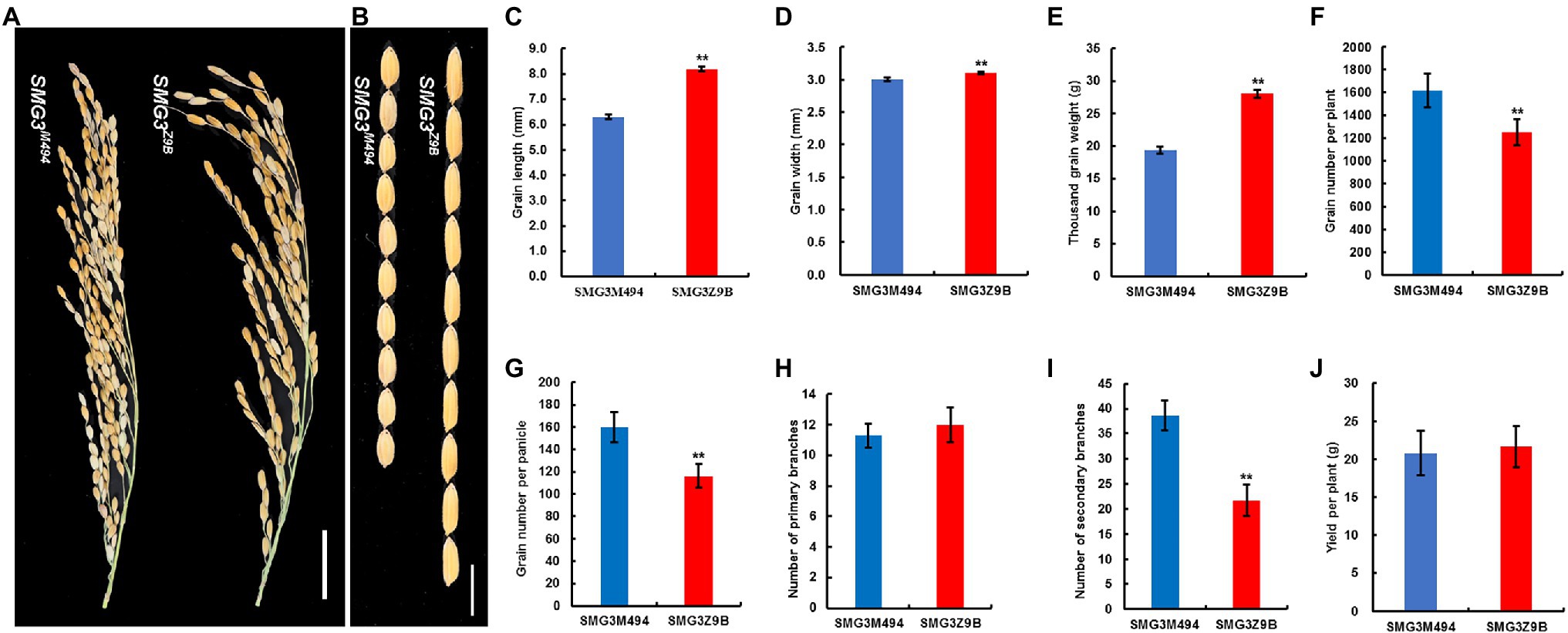
Figure 3. Comparison of the agronomic traits between NIL-SMG3Z9B and NIL-SMG3M494. (A) Panicle phenotype of NIL-SMG3Z9B and NIL-SMG3M494. Scale bar: 3 cm. (B) NIL-SMG3Z9B and NIL-SMG3M494 rice grains. Scale bar: 8 mm. (C–J) Grain length (C), grain width (D), thousand grain weight (E), grain number per plant (F), grain number per panicle (G), number of primary branches (H), number of secondary branches (I), and yield per plant (J) of NIL-SMG3Z9B and NIL-SMG3M494. Data are given as means ± SD. ** Indicates p < 0.01 by Student’s t-test.
In rice, grain size is restricted by the size of the spikelet hull (Song et al., 2015), which is controlled by the coordination of cell proliferation and expansion (Duan et al., 2014). Therefore, we examined the cell size and cell number in the spikelet hulls of the NIL-SMG3M494 and NIL-SMG3Z9B using the scanning electron microscope. Compared with NIL-SMG3Z9B, the average cell length of the outer and inner glumes was decreased in NIL-SMG3M494, but the average cell width was increased (Figures 4A–F). In addition, we measured the cell number of outer spikelet hulls along the length and width between NIL-SMG3Z9B and NIL-SMG3M494. Compared with NIL-SMG3Z9B, the total cell number along the length was significantly reduced in the NIL-SMG3M494, but the cell number along the width was significantly increased in the NIL-SMG3M494 (Figures 4G–I). These results suggested that SMG3 affects grain size by promoting cell expansion and cell proliferation in spikelet hulls.
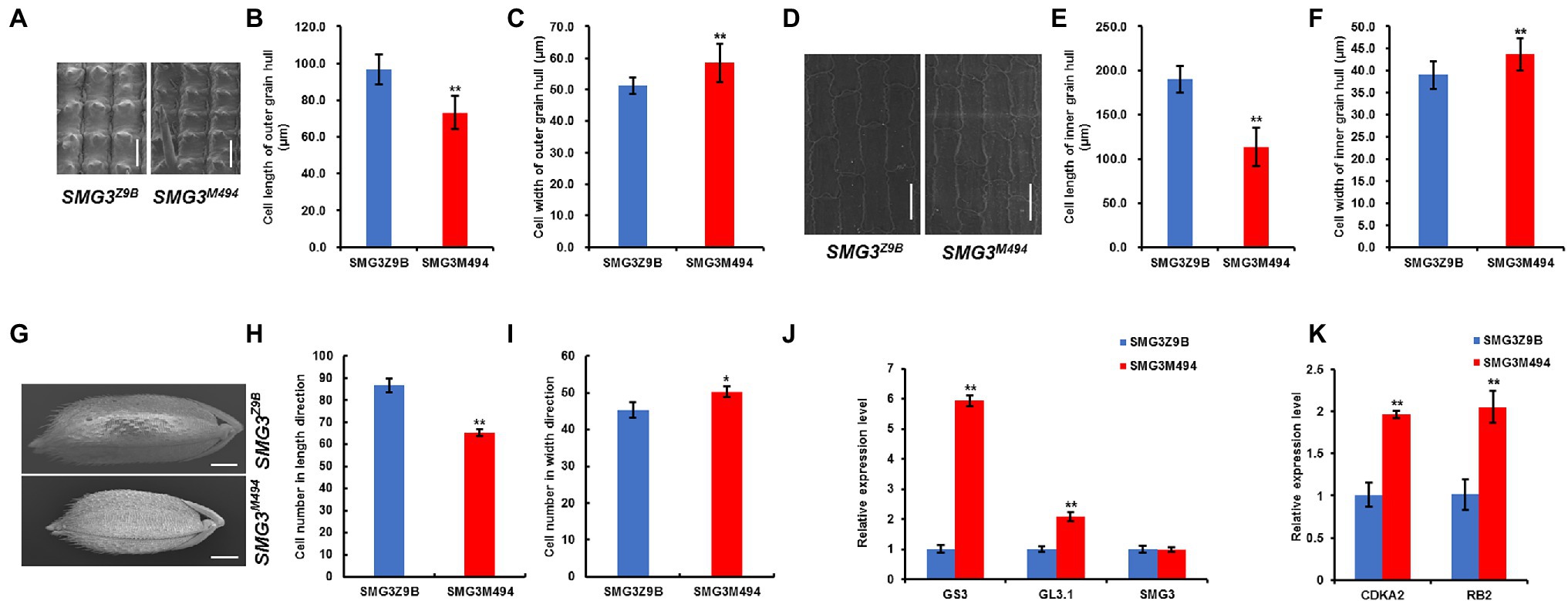
Figure 4. Histological and qPCR analysis between NIL-SMG3Z9B and NIL-SMG3M494. (A) Scanning electron microscope analysis of the outer surfaces of the grain hulls in SMG3Z9B and SMG3M494. Scale bar: 50 μm. (B,C) The average length and width of outer epidermal cells. (D) Scanning electron microscope analysis of the inner surfaces of the grain hulls in NIL-SMG3Z9B and NIL-SMG3M494. Scale bar: 50 μm. (E,F) The average length and width of inner epidermal cells. (G) Scanning electron micrographs of the outer surfaces of glumes in NIL-SMG3Z9B and NIL-SMG3M494. Scale bar: 1 mm. (H,I) the cell number in the outer glumes along the length and width direction between NIL-SMG3Z9B and NIL-SMG3M494. (J) qPCR analysis of GS3, GL3.1, and SMG3 genes in the NIL-SMG3Z9B and NIL-SMG3M494. (K) qPCR analysis of CDKA2 and RB2 genes between NIL-SMG3Z9B and NIL-SMG3M494. Data are given as means ± SD. * and ** indicate p < 0.05 and p < 0.01 by Student’s t-test.
Several genes/QTLs, such as GL7/GW7 and GS2, are known to control grain size by regulating cell expansion, and GS3, GL3.1/qGL3, and GS5 are involved in regulation of cell proliferation. To reveal how SMG3 regulates cell expansion and cell proliferation in spikelet hulls, we detected their expression levels in the NIL-SMG3M494 and NIL-SMG3Z9B panicles. Compared with NIL-SMG3Z9B, only the expression levels of GS3 and GL3.1/qGL3 were significantly raised in the NIL-SMG3M494 (Figure 4J), and the expression levels of GL7/GW7, GS2, and GS5 were similar between NIL-SMG3M494 and NIL-SMG3Z9B (data not shown). A similar difference was observed between Nipponbare and the SMG3 overexpression plants (Supplementary Figure 4).
Furthermore, we also detected the expression of genes involved in the cell cycle, such as CYCA2.1, CDKA2, CYCD4.2, CDKC1, CYCU3.1, and RB2, and found that besides CYCA2.1, CYCD4.2, CDKC1, and CYCU3.1 (data not shown), CDKA2 and RB2 were downregulated in NIL-SMG3Z9B, suggesting that the increase in cell length in NIL-SMG3Z9B might result from lower expression of these two genes promoting cell proliferation (Figure 4K). In addition, we found that the CDKA2 and RB2 expression levels were upregulated in the SMG3 overexpression plants compared with Nipponbare (Supplementary Figure 5). Taken together, these results supported that SMG3 regulated grain size by promoting cell expansion and cell proliferation in spikelet hulls.
Although the grain size and grain weight of NIL-SMG3M494 were smaller than NIL-SMG3Z9B, the final grain yield per plant was identical (Figure 3J). As expected, milled rice from NIL-SMG3Z9B was more slender than that of NIL-SMG3M494 (Figure 5A). More importantly, the chalky grain percentage and chalkiness degree of milled rice of NIL-SMG3M494 were significantly decreased than NIL-SMG3Z9B (Figures 5B,C). These findings suggested that the potential for application of the SMG3 allele of M494 to improve grain appearance quality in high yield but low quality rice varieties.
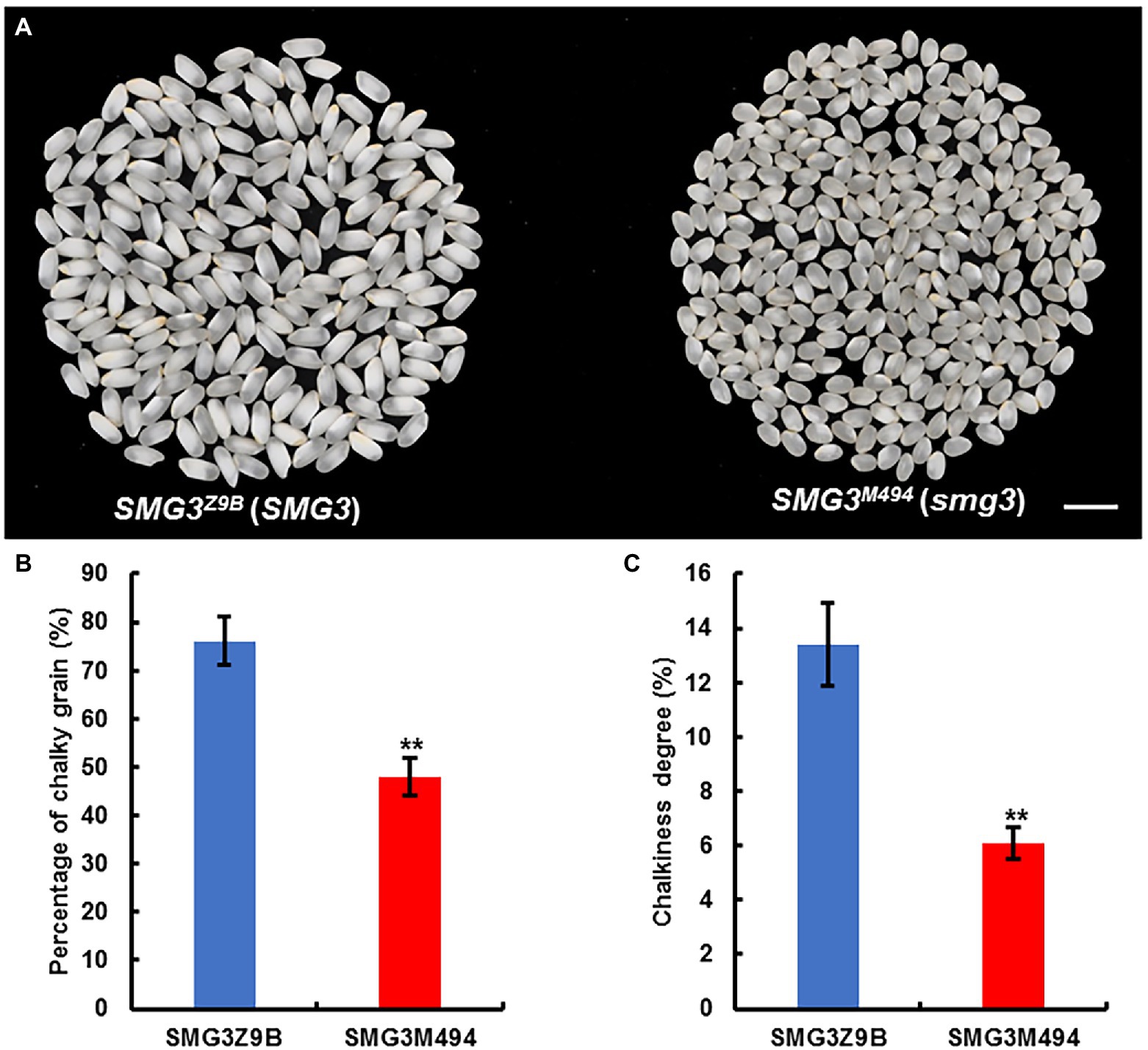
Figure 5. Improved appearance quality of milled rice by introgression of SMG3 allele from M494. (A) Comparison of the appearance of milled rice between NIL-SMG3Z9B and NIL-SMG3M494, carrying the homozygous for Z9B and M494 allele, respectively. Scale bar: 8 mm. (B,C) The percentage of milled rice with chalkiness (B) and the chalkiness degree of milled rice (C) from NIL-SMG3Z9B and NIL-SMG3M494. Data are given as means ± SD. ** Indicates p < 0.01 by Student’s t-test.
To examine the temporal and spatial expression pattern of SMG3, total RNA from nine organs of the SMG3::GUS transgenic plants, including root, base, stem, node, flag leave, pulvinus, leaf sheath, young panicle, and flowering panicle was extracted. qRT-PCR analysis showed that SMG3 was constitutively expressed in various rice organs, with high expression in the leaf sheath, base, and flowering panicle (Figure 6A). In the SMG3::GUS transgenic plants, GUS activity of old panicle was slightly stronger than in young panicle (Figures 6B,C). The expression pattern of SMG3 is consistent with its role in spikelet hull development.
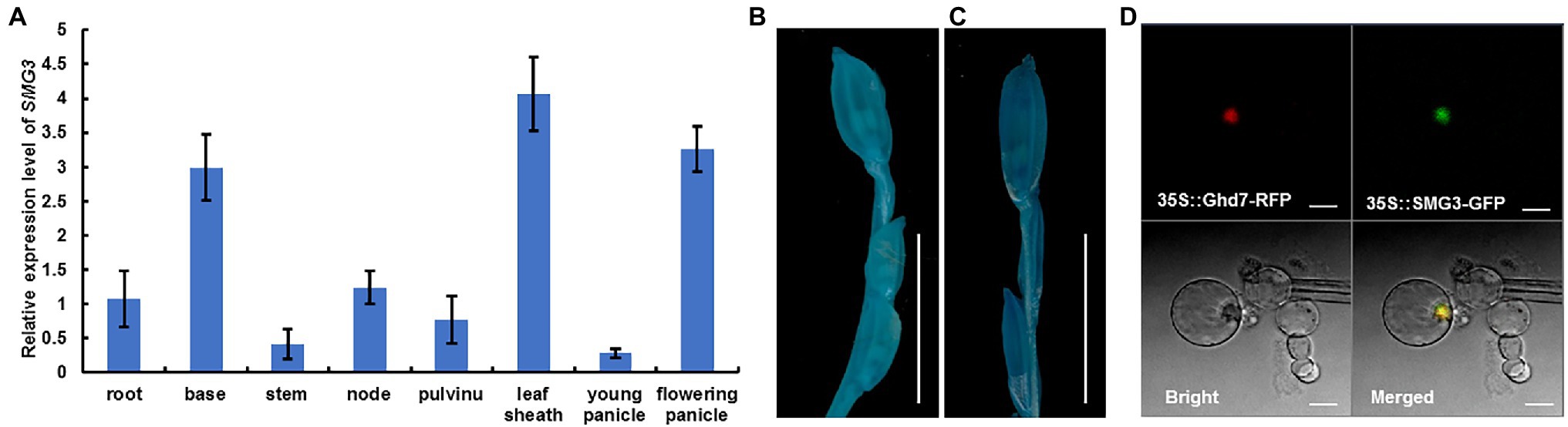
Figure 6. Expression pattern of SMG3 and subcellular localization of SMG3 protein. (A) Relative expression levels of SMG3 in different tissues analyzed by qRT-PCR. Values are means ± SD of three independent experiments. (B,C) Histochemical analysis of GUS activity in caryopsis of 4–5 cm (B) and 13–14 cm (C) long panicle. Scale bars: 1 cm. (D) Subcellular localization of the SMG3 protein by GFP assays. The subcellular localization was performed with red fluorescent protein (RFP) as a nuclear marker. The 35S::SMG3-GFP and 35S::Ghd7-RFP were co-expressed in rice protoplasts. Scale bar: 10 μm.
To determine the subcellular localization of SMG3, we constructed SMG3::GFP and Ghd7-RFP fusions driven by the 35S promoter. The two fusions were transferred together into rice protoplasts. We observed that the SMG3::GFP fusion protein was localized in the nucleus of rice protoplasts (Figure 6D). This result indicated that SMG3 encodes a nuclear protein, consistent with its proposed role as a transcription factor (Li et al., 2020).
Grain size and weight are important determinants of yield production and appearance quality in rice. Several genes regulating grain size and weight such as GS3, GS5, GSE5, GW2, GS2, GL7/GW7, and TGW2 (Song et al., 2007; Mao et al., 2010; Li et al., 2011; Hu et al., 2015; Wang et al., 2015a,b; Duan et al., 2017; Ruan et al., 2020) have been identified, but the molecular mechanisms underlying grain size and weight control in rice remain elusive. In the present study, we reported QTL mapping results for grain size, grain weight, and grain number per panicle with an F2 population from a cross between a japonica small-grain variety (M494) and a long-grain indica variety (Z9B). A total of fourteen putative QTLs for grain size, grain weight, and grain number per panicle were detected in the F2 population (Supplementary Table 2). Especially, the main QTL cluster on chromosome 3 was responsible for GL, GW, LWR, and TGW in the F2 population, which covers two QTL peaks, one is the previously cloned grain length gene GS3, and the other is the QTL for grain length and weight, SMG3 (Supplementary Figure 2). To fine map the hidden QTL-SMG3 nearby GS3, we generated a large BC3F3 population and selected recombinant individuals in the region around SMG3. Using a chromosome fragment substitution strategy, the SMG3 gene was narrowed down to a 61 kb region. Genetic complementation test demonstrated that SMG3 is a novel allele of OsMYB3. Interestingly, The NIL-SMG3M494 showed a significant increase in GNP compared to NIL-SMG3Z9B, and similar differences were observed between SMG3 overexpression positive and negative plants (Figure 3G; Supplementary Figure 3). Our results demonstrated SMG3 was a negative regulator of GL and grain weight but a positive regulator of GNP. This finding was consistent with the previous study that showed that an increase in GNP usually causes a decrease in grain size and weight (Huang et al., 2009).
Grain size is determined by cell proliferation and cell expansion (Duan et al., 2014). Our results revealed that the length of epidermal cells of the outer and inner glumes was increased in NIL-SMG3Z9B compared with those in NIL-SMG3M494, and the total cell number along the length direction was also significantly increased in the NIL-SMG3Z9B (Figures 4B,E,H). In addition, we found that two cell cycle genes, namely, CDKA2 and RB2, were upregulated in NIL-SMG3M494 compared with NIL-SMG3Z9B (Figure 4K). The previous studies showed that RB2 could bind to E2 promoter binding factor (E2F) and inhibits the activity of E2F transcription factor, preventing cell cycle progression (Dyson, 1998; Desvoyes et al., 2006; Shi et al., 2019), and the high level expression of RB2 which was in the NIL-SMG3M494 may inhibit cell elongation and proliferation in the length direction in spikelet hulls and result in small grains in the present study. As is known, Cylin-Dependent Kinase (CDK)-related genes play functional roles in cell cycle regulation of seed size and development in plants (Jiang et al., 2012; Sabelli et al., 2013; Dante et al., 2014; Chen et al., 2015; Li et al., 2019; Liu et al., 2021). In this study, downregulation of CDKA2 may contribute to increase in cell expansion and proliferation in the length direction in spikelet hulls and lead to longer grains in NIL-SMG3Z9B. Collectively, our results suggested that SMG3 regulated grain size by promoting both cell expansion and cell proliferation.
Cereal crops had been selected and domesticated to satisfy human needs (Sang, 2009). However, the process usually led to a decrease in offspring genetic diversity, and lost some useful genetic loci during domestication. We analyzed the 120 rice re-sequenced varieties by Li et al. (2020) and found that the 60 bp insertion at nucleotide 967 in exon 3 was specific in the M494 accession, which indicated that it was a novel mutation of OsMYB3 and might not have been selected by breeders. Compared with SG3 (Li et al., 2020), SMG3 is a new allele of OsMYB3, and not only negatively affected grain length and grain weight, but positively regulated grain number per panicle. In addition, our results showed that the NIL-SMG3M494 carrying the M494 allele could reduce the chalky grain percentage and chalkiness degree of milled rice compared to NIL-SMG3Z9B, and the final grain yield per plant was equivalent. These results revealed that the M494 allele of SMG3 has the potential for grain quality improvement and could be used to combine with other genes for grain size to regulate rice grain appearance in breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
RL, ZL, SW, XW, and YF conceived and designed the experiments. RL, ZL, JY, YiY, JL, YW, and MZ performed the experiments. HY, QX, SW, and YaY analyzed the data. RL and YF wrote the manuscript and other authors revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the Key Technology Research and Development Program of Zhejiang Province (2021C02056), the Key Research and Development Project of Hainan Province (ZDYF2021XDNY170), the Fundamental Research Funds for Central Public Welfare Research Institutes of Chinese Rice Research Institute (CPSIBRF-CNRRI-202101), and the Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2013-CNRRI).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at https://www.frontiersin.org/articles/10.3389/fpls.2022.880919/full#supplementary-material
QTL, Quantitative Trait Locus; Z9B, Zhong 9B; MYB, Myeloblastosis; NIL, Near-Isogenic Line; GL, Grain Length; GW, Grain Width; LWR, Grain Length-Width Ratio; TGW, Thousand Grain Weight; GNP, Grain Number per Panicle; cM, Centimorgan; RT-PCR, Reverse Transcription Polymerase Chain Reaction; DNA, Deoxyribo Nucleic Acid; RNA, Ribo Nucleic Acid; GFP, Green Fluorescent Protein; GUS, β-Glucuronidase; CDS, Coding Sequence; SB, Secondary Branches; CDK, Cylin-Dependent Kinase.
Ashikari, M., Sakakibara, H., Lin, S. Y., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373
Chen, J., Gao, H., Zheng, X. M., Jin, M., Weng, J. F., Ma, J., et al. (2015). An evolutionarily conserved gene, FUWA, plays a role in determining panicle architecture, grain shape and grain weight in rice. Plant J. 83, 427–438. doi: 10.1111/tpj.12895
Dante, R. A., Larkins, B. A., and Sabelli, P. A. (2014). Cell cycle control and seed development. Front. Plant Sci. 5:493. doi: 10.3389/fpls.2014.00493
Desvoyes, B., Ramirez-Parra, E., Xie, Q., Chua, N. H., and Gutierrez, C. (2006). Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140, 67–80. doi: 10.1104/pp.105.071027
Duan, P. G., Rao, Y. C., Zeng, D. L., Yang, Y. L., Li, Y., Xu, R., et al. (2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4 (MKK4), influences grain size in rice. Plant J. 77, 547–557. doi: 10.1111/tpj.12405
Duan, P. G., Xu, J. S., Zeng, D. L., Zhang, B. L., Geng, M. F., Zhang, G. Z., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
Dyson, N. (1998). The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262. doi: 10.1101/gad.12.15.2245
Fan, C. C., Yu, S. B., Wang, C. R., and Xing, Y. Z. (2009). A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor. Appl. Genet. 118, 465–472. doi: 10.1007/s00122-008-0913-1
Feng, Y., Yuan, X. P., Wang, Y. P., Yang, Y. L., Zhang, M. C., Yu, H., et al. (2021). Validation of a QTL for grain size and weight using an introgression line from a cross between Oryza sativa and Oryza minuta. Rice 14:43. doi: 10.1186/s12284-021-00472-1
Fu, S., Fu, L. B., Zhang, X., Huang, J. J., Yang, G. Z., Wang, Z. G., et al. (2019). OsC2DP, a novel C2 domain-containing protein is required for salt tolerance in rice. Plant Cell Physiol. 60, 2220–2230. doi: 10.1093/pcp/pcz115
Gao, X. Y., Zhang, J. Q., Zhang, X. J., Zhou, J., Jiang, Z. S., Huang, P., et al. (2019). Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 31, 1077–1093. doi: 10.1105/tpc.18.00836
Guo, T., Chen, K., Dong, N. Q., Shi, C. L., Ye, W. W., Gao, J. P., et al. (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain NUMBER per panicle and grain size in rice. Mol. Plant 30, 871–888. doi: 10.1105/tpc.17.00959
Hu, J., Wang, Y. X., Fang, Y. X., Zeng, L. J., Xu, J., Yu, H. P., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Huang, X. Z., Qian, Q., Liu, Z. B., Sun, H. Y., He, S. Y., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Huo, X., Wu, S., Zhu, Z. F., Liu, F. X., Fu, Y. C., Cai, H. W., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8:1497. doi: 10.1038/s41467-017-01501-8
Jiang, Y. H., Bao, L., Jeong, S. Y., Kim, S. K., Xu, C. G., Li, X., et al. (2012). XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 70, 398–408. doi: 10.1111/j.1365-313X.2011.04877.x
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Li, Y. B., Fan, C. C., Xing, Y. Z., Jiang, Y. H., Luo, L. J., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Li, N., and Li, Y. H. (2016). Signaling pathways of seed size control in plants. Curr. Opin. Plant Biol. 33, 23–32. doi: 10.1016/j.pbi.2016.05.008
Li, S. C., Li, W. B., Huang, B., Cao, X. M., Zhou, X. Y., Ye, S. M., et al. (2013). Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat. Commun. 4:2793. doi: 10.1038/ncomms3793
Li, Q., Lu, L., Liu, H., Bai, X., Zhou, X., Wu, B., et al. (2020). A minor QTL, SG3, encoding an R2R3-MYB protein, negatively controls grain length in rice. Theor. Appl. Genet. 133, 2387–2399. doi: 10.1007/s00122-020-03606-z
Li, N., Xu, R., and Li, Y. H. (2019). Molecular networks of seed size control in plants. Annu. Rev. Plant Biol. 70, 70435–70463. doi: 10.1146/annurev-arplant-050718-095851
Liu, D. P., Zhao, H., Xiao, Y. H., Zhang, G. X., Cao, S. Y., Yin, W. C., et al. (2021). A cryptic inhibitor of cytokinin phosphorelay controls rice grain size. Mol. Plant 15, 293–307. doi: 10.1016/j.molp.2021.09.010
Mao, H. L., Sun, S. Y., Yao, J. L., Wang, C. R., Yu, S. B., Xu, C. G., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 19579–19584. doi: 10.1073/pnas.1014419107
McCouch, S. R., Cho, Y. G., Yano, M., Paul, E., Blinstrub, M., Morishima, H., et al. (1997). Report on QTL nomenclature. Rice Genet. Newsl. 13, 351–365. doi: 10.1007/s10142-013-0328-1
Oikawa, T., and Kyozuka, J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21, 1095–1108. doi: 10.1105/tpc.108.065425
Qi, P., Lin, Y. S., Song, X. J., Shen, J. B., Huang, W., Shan, J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Qiao, J. Y., Jiang, H. Z., Lin, Y. Q., Shang, L. G., Wang, M., Li, D. M., et al. (2021). A novel miR167a-OsARF6-OsAUX3 module regulates grain length and weight in rice. Mol. Plant 14, 1683–1698. doi: 10.1016/j.molp.2021.06.023
Ren, D. Y., Hu, J., Xu, Q. K., Cui, Y. J., Zhang, Y., Zhou, T. T., et al. (2018). FZP determines grain size and sterile lemma fate in rice. J. Exp. Bot. 69, 4853–4866. doi: 10.1093/jxb/ery264
Ruan, B. P., Shang, L. G., Zhang, B., Hu, J., Wang, Y. X., Lin, H., et al. (2020). Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 227, 629–640. doi: 10.1111/nph.16540
Sabelli, P. A., Liu, Y., Dante, R. A., Lizarraga, L. E., Nguyen, H. N., Brown, S. W., et al. (2013). Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. U. S. A. 110, E1827–E1836. doi: 10.1073/pnas.1304903110
Sang, T. (2009). Genes and mutations underlying domestication transitions in grasses. Plant Physiol. 149, 63–70. doi: 10.1104/pp.108.128827
Shi, C. L., Ren, Y. L., Liu, L. L., Wang, F., Zhang, H., Tian, P., et al. (2019). Ubiquitin specific protease 15 has an important role in regulating grain width and size in rice. Plant Physiol. 180, 381–391. doi: 10.1104/pp.19.00065
Song, X. J., Huang, W., Shi, M., Zhu, M. Z., and Lin, H. X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Song, X. J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U. S. A. 112, 76–81. doi: 10.1073/pnas.1421127112
Suzuki, A., Suzuki, T., Tanabe, F., Toki, S., Washida, H., Wu, C., et al. (1997). Cloning and expression of five myb-related genes from rice seed. Gene 198, 393–398. doi: 10.1016/S0378-1119(97)00344-2
Tabuchi, H., Zhang, Y., Hattori, S., Omae, M., Shimizu-Sato, S., Oikawa, T., et al. (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. doi: 10.1105/tpc.111.088765
Wang, S. K., Li, S., Liu, Q., Wu, K., Zhang, J. Q., Wang, S. S., et al. (2015a). The OSSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, Y. X., Xiong, G. S., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015b). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Wu, Y., Wang, Y., Mi, X. F., Shan, J. X., Li, X. M., Xu, J. L., et al. (2016). The QTL GNP1 encodes GA20ox1, which increases grain number and yield by increasing cytokinin activity in rice panicle meristems. PLoS Genet. 12:e1006386. doi: 10.1371/journal.pgen.1006386
Xing, Y., and Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209
Xue, W. Y., Xing, Y. Z., Weng, X. Y., Zhao, Y., Tang, W. J., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Zhang, X. J., Wang, J. F., Huang, J., Lan, H. X., Wang, C. L., Yin, C. F., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U. S. A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhang, D. B., and Yuan, Z. (2014). Molecular control of grass inflorescence development. Annu. Rev. Plant Biol. 65, 553–578. doi: 10.1146/annurev-arplant-050213-040104
Keywords: grain size, grain number per panicle, grain weight, QTL, rice
Citation: Li R, Li Z, Ye J, Yang Y, Ye J, Xu S, Liu J, Yuan X, Wang Y, Zhang M, Yu H, Xu Q, Wang S, Yang Y, Wang S, Wei X and Feng Y (2022) Identification of SMG3, a QTL Coordinately Controls Grain Size, Grain Number per Panicle, and Grain Weight in Rice. Front. Plant Sci. 13:880919. doi: 10.3389/fpls.2022.880919
Received: 22 February 2022; Accepted: 29 March 2022;
Published: 25 April 2022.
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Kiyosumi Hori, National Agriculture and Food Research Organization (NARO), JapanCopyright © 2022 Li, Li, Ye, Yang, Ye, Xu, Liu, Yuan, Wang, Zhang, Yu, Xu, Wang, Yang, Wang, Wei and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu Wang, c3dhbmcxMjNAc3lhdS5lZHUuY24=; Xinghua Wei, d2VpeGluZ2h1YUBjYWFzLmNu; Yue Feng, ZnlfNTU1NTAwQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.