
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 May 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.873621
This article is part of the Research Topic Horticultural Crops: Interaction with Fungal Community View all 6 articles
 Yun Jiang1,2*
Yun Jiang1,2* Yu Song1
Yu Song1 Chengyang Jiang1
Chengyang Jiang1 Xiang Li1
Xiang Li1 Tingting Liu1
Tingting Liu1 Jiarui Wang1
Jiarui Wang1 Changqing Chen2,3*
Changqing Chen2,3* Jie Gao2,3*
Jie Gao2,3*A bacterial strain JI39 that had plant growth-promoting traits was isolated from the rhizosphere soil of Panax ginseng. It had the ability to produce high indole-3-acetic acid (13.1 μg/ml), phosphate solubilization (164.2 μg/ml), potassium solubilization (16.1 μg/ml), and nitrogen fixation. The strain JI39 was identified to be Arthrobacter nicotinovorans based on morphological, physiological, and biochemical traits and through 16S rDNA sequence analysis. The optimal culture environment for strain growth was 1.0% NaCl, 30°C, pH 6.0, and without UV irradiation. The strain can produce cellulase and protease. The strain JI39 can significantly promote the growth of ginseng. After ginseng seeds were treated with 3 × 108 CFU/ml of JI39 bacterial suspension, the shoot's length was significantly increased by 64.61% after 15 days. Meanwhile, the fresh weight of 2-year-old ginseng roots was significantly increased by 24.70% with a treatment by the 108 CFU/ml bacterial suspension after 150 days in the field. The gene expression of phenylalanine ammonia-lyase (PAL), β-1.3 glucanase (β-1,3-GA), chitinase (CHI), superoxide dismutase (SOD), and peroxidase (POD) of ginseng was upregulated, and it also can improve the soil urease, phosphatase, invertase, and catalase activity. In conclusion, the bacterial strain JI39 could efficiently promote the growth of ginseng and has the potential to be a good microbial fertilizer for ginseng.
For thousands of years, Chinese ginseng (Panax ginseng C.A. Meyer) has been utilized for its medicinal value in China, Korea, and Japan. Ginseng is used as a health tonic and helps in maintaining natural energy, enhancing mental and physical abilities, mood improvement, and promoting general health and wellbeing (Wang, 1965). With the need for P. ginseng rising, the scale of artificial ginseng planting has also rapidly expanded in recent years.
The wide application of chemical fertilizers in agricultural production can increase crop yields and contribute to ensure food security. However, its long-term and unreasonable application has not only affected the quality of crops but also destroyed the soil environment. With the over-reliance on chemical fertilizers in agricultural production, the amount of chemical fertilizer used has greatly increased which further impacted its utilization rate (Bruna et al., 2018). In the past years, to reduce to use the chemical fertilizer, many countries have considered the microbial fertilizer to be an efficient way as its' advantages, such as good fertilizer efficiency, better crop yields and crop quality, and resilience of soil. In microbial fertilizer, suitable bacteria could improve the plant growth by enhancing the resistance of the plant to pathogens, improving the soil environment, providing nutrient elements, etc. In the research and development of microbial fertilizer, it has become a hot topic to study and screen the high-efficient plant growth-promoting bacteria (Supratim et al., 2017). Plant growth-promoting rhizobacteria (PGPRs) have many characteristics that could significantly promote plant growth, such as auxin (IAA) secretion, nitrogen fixation, phosphate solubilizing, and potassium solubilizing (Xu et al., 2001; Furnkranz et al., 2009; Shahab et al., 2009; Michele et al., 2013; Faisal et al., 2016; Ahmad et al., 2019). Recently, PGPRs have been frequently studied to find the high-efficient plant growth-promoting bacteria. As an important Chinese medicine, ginseng has high economic value and market potential. However, its production is almost limited in the Changbai Mountain region in China as its high requirements for soil and environmental conditions. It is key to protect the ecological environment in the planting area to improve the output and quality of ginseng and ensure the sustainable development of the ginseng industry. Therefore, it would be of great significance to research and apply the environmentally friendly microbial bacterial fertilizer to benefit the ginseng industry.
Ginseng rhizosphere bacteria are an important type of microbial resource, and previous studies have reported its potential in the prevention and treatment of ginseng diseases (Jiang et al., 2013). However, the ginseng growth-promoting bacteria have been rarely reported. The objective of this study was to identify the taxonomy position and evaluate the ginseng growth promotion ability of the bacterial strain JI39, which can be expected to promote the development and application of microbial fertilizer in the ginseng production.
The bacterial strain JI39 was isolated and screened from P. ginseng rhizosphere soil in Ji'an city, at Jilin province, China. This strain was inoculated in the NA medium (Beef extract 3.0 g/L, peptone 5.0 g/L, glucose 2.5 g/L, agar 18 g/L, and pH 7.0 ± 0.1), incubated at 28°C for 24 h, and then stored at 4°C.
The strain JI39 was inoculated in the Ashby's nitrogen-free agar medium and incubated at 30°C for 24 h. Then the nitrogen fixation potency was determined using the method described by Xu et al. (2001). Phosphate-solubilizing activity testing was done using the plate-screening method as described by Pikovskaya (1948). The strain was cultured on Aleksandrow's agar medium, and the potassium solubilizing ability was determined using a flame atomic absorption spectrometer (Tian et al., 2014; Bhagya et al., 2017). Salkowski's colorimetric method was used to screen the strain JI39 with high IAA production (Deepmala et al., 2019; Mohamad et al., 2020). The siderophores production was detected by the Chrome Azurol S (CAS) method (Xu et al., 2001; Paola et al., 2020). The 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase activity was detected by the Honma method (Sarita et al., 2016; Silva et al., 2019).
Bacterial strain JI39 was inoculated in NA plates and incubated at 28°C for 18 h. Then, the morphological traits of the colony were observed. Cellular morphology was observed by the Gram's stain and transmission electron microscope (Imane et al., 2019). Briefly, the strain JI39 was cultured in Luria-Bertani (LB) liquid medium for 24 h, and after centrifugation, the precipitate was washed repeatedly with sterile water to prepare the samples for negative staining. The copper mesh was dipped in the strain JI39 bacterial suspension, and after standing for 2 min, the remaining liquid was drained from the edge by filter paper. Then, the copper mesh was rinsed with purified water, and the extra water would be drained again with filter paper. Thus, the negative staining was carried out by 2% phosphotungstic acid for 5 min, and then we observed the phenotype under a JEM-1200EXII transmission electron microscope after drying.
The biochemical characteristics of the strain JI39 were determined according to the recommended partial media with the method of Systematic Identification of Common Bacteria (Dong and Cai, 2001) and Berger's Manual for Systematic Identification (Garrity et al., 2004).
The genomic DNA of strain JI39 was extracted by the conventional cetyl trimethylammonium bromide (CTAB) method (Jiang et al., 2012b). The 16S rDNA was amplified using the universal primers 27F (5′-TACGGYTACCTTGTTACG ACTT-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) in Eppendorf Mastercycler 5333 PCR amplifier by the following PCR conditions: 94°C for 5 min, followed by 32 cycles of 94°C denaturations for 50 s, 54°C annealings for 50 s, 72°C extensions for 90 s, and then, 72°C incubation for 10 min (Lane, 1991). The PCR products were sequenced by Sangon Biotech (Shanghai, China). The DNA sequence was deposited into the NCBI database. In addition, similar bacterial sequences were searched and downloaded using the blastn tool. The retrieved sequences and the internal transcribed spacer (ITS) sequence of JI39 were used for phylogenetic analysis using MEGA 7.0 software by the neighbor-joining method with bootstrapping 1,000 replicates (Sudhir et al., 2016).
The cellulase production capacity of strain JI39 was determined by cellulase screening medium, and its protease production capacity was determined by skim milk screening medium. In addition, the amylase screening medium was used to test the amylase production capacity, and β-1,3-glucanase production capacity was determined by the poria cocos screening medium (Chen et al., 2004; Shahab et al., 2009; Rania et al., 2016; Silva et al., 2019).
The effects of temperature, pH, sodium chloride concentration, and UV irradiation on strain JI39 growth were investigated (Bhagya et al., 2017; Motahhareh et al., 2019; Silva et al., 2019).
An agar plug of strain JI39 was put into LB medium and incubated in the shaker with 160 rpm at 28°C for 12 h. Then, the medium was centrifuged at 12,000 rpm for 5 min and the sedimentary bacterial cells were resuspended in sterile distilled water. The ginseng seeds were dipped into the bacterial suspension at the concentration of 3 × 108 CFU/ml for 3 h (Tian et al., 2014). Then these seeds were dried naturally and placed on the Petri dishes, which contained 40 g of sand and 10 ml of sterile distilled water. Each petri dish has 15 seeds, and the seeds processed only with sterile distilled water were set as control. All seeds were cultured in the dark at 20°C for 15 days, and each treatment had three replicates.
The effect of growth promotion of strain JI39 on ginseng root was evaluated in the field at Liuhe County, Tonghua City, Jilin province, China from 2019 to 2020. The pH value of black soil was 5.5, and the high content of organic matter was with an average of 28.94 g/kg. In late April, the 2-year ginseng plants with uniform size were planted at 5 cm depth in the soil and fertilized consistently. After the full expansion of the leaf, the bacterial suspension was applied to the ginseng root according to the root irrigation method in early June. The bacterial suspension was prepared at the concentration of 1 × 108 CFU/ml (Tian et al., 2014; Wang et al., 2019). In each treatment, 60 ginseng plants were planted in four rows and each row was irrigated by 500 ml bacterial suspension. The 100 × 107 CFU/ml of YIWEI solution, which was a commercial biological agent produced by Shandong Jingqing Agricultural Technology Co., Ltd., and water were used as the controls, respectively. The ginseng plants were irrigated every 15 days with a total of three times. In late October, waiting for the stems and leaves of ginseng to fell off, the ginseng roots of each treatment were harvested to weigh the fresh weight (Yacine et al., 2014; Avishek et al., 2019; Sangeeta and Shikha, 2020).
The rhizosphere soil samples were collected at 0, 3, 7, 15, 30, 45, and 60 d after strain JI39 treatment at the concentration of 1 × 108 CFU/ml in a pot. Soil urease was determined using the method described by Kandeler and Gerber (1988). The catalase activity was determined according to Lúcia et al. (2016). The alkaline phosphatase activity was determined according to the protocol described by Rahul and Anita (2016). The invertase activity was measured using the method described by Sun et al. (2021).
Three years old healthy ginseng roots were inoculated by bacterial suspension of JI39 strain at the concentration of 1 × 108 CFU/ml in a pot and the water inoculation was used as the control. The ginseng roots were sampled at 0, 3, 6, 9, 12, 18, and 30 d respectively after bacteria treatment and were soaked in liquid nitrogen.
The total RNA of ginseng roots was extracted by using the RNAiso Plus method (reference required). Reverse transcription was conducted by using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase [Takara Biotechnology (Dalian) Co., Ltd. China]. The genes encoding phenylalanine ammonia-lyase (PAL), β-1.3 glucanase (β-1,3-GA), chitinase (CHI), superoxide dismutase (SOD), and peroxidase (POD) were selected as target genes, and the β-actin gene was employed as the internal reference. Primers were designed by Primer Primier 5.0 software (Table 1) and synthesized by Sangon Biotech (Shanghai) Co., Ltd. in China. Quantitative RT-PCR was conducted on ABI 7500 system (ABI) using the SYBR Premix Ex Taq mixture. The 10 μl PCR reaction system is cDNA 1 μl, 10 μmol/L upstream and downstream primers are 0.2 μl, 2 × Fast SYBR Mixture 5 μl, 50 × Low ROX 0.2 μl, ddH2O 3.4 μl, and the final volume is 10 μl. The qRT-PCR reaction conditions are 95°C pre-denaturation for 10 min, 95°C denaturation for 10 s, 60°C annealing, and extension for 30 s, a total of 40 cycles. The qPCR of each gene was conducted with three biological and technical replicates, respectively. The relative gene expression was analyzed using the 2−ΔΔCt (Jiang et al., 2018; Kim et al., 2019).
All the tests were arranged with three replications in each treatment. For each trial, results obtained were subjected to analysis of variance (ANOVA) and treatments means were compared by Fisher's protected least significant difference (LSD) test at 5% probability. Before the ANOVA was performed, the normal distribution of residuals and homogeneity of variance were verified. An independent sample t-test was used to test the expression of defense enzyme genes and soil enzyme activity. The statistical analyses were carried out by SPSS 26.0 (Sun et al., 2019; Ning et al., 2022).
The four growth-promoting abilities of strain JI39 are shown in Figure 1. Bacterial strain JI39 can grow normally on the Ashby's agar medium, which suggests a potential ability for nitrogen fixation. On the Pikovskaya (PVK) agar medium, JI39 can grow with a transparent zone, suggesting an ability to solubilize phosphate. In addition, the strain JI39 could grow on Aleksandrow's agar medium with a transparent zone with the potassium solubilizing activity of 16.1 μg/ml and the high IAA production activity with the concentration of 13.1 μg/ml in the LB medium. However, no activity of siderophores production and the ACC deaminase was observed.
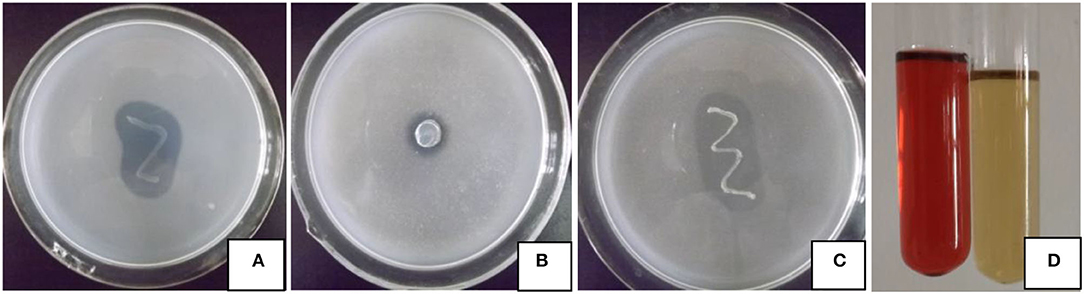
Figure 1. Plant growth-promoting traits of strain JI39. (A) Nitrogen fixation; (B) phosphate solubilizing; (C) potassium solubilizing; (D) IAA production, treatment at left, control at right.
On the NA medium, the colony of strain JI39 was a circle, pale yellow, opaque, and with neat edges (Figure 2A). Under the transmission electron microscope, the cells of strain JI39 were found to be rod shaped with several flagellums (Figure 2B).
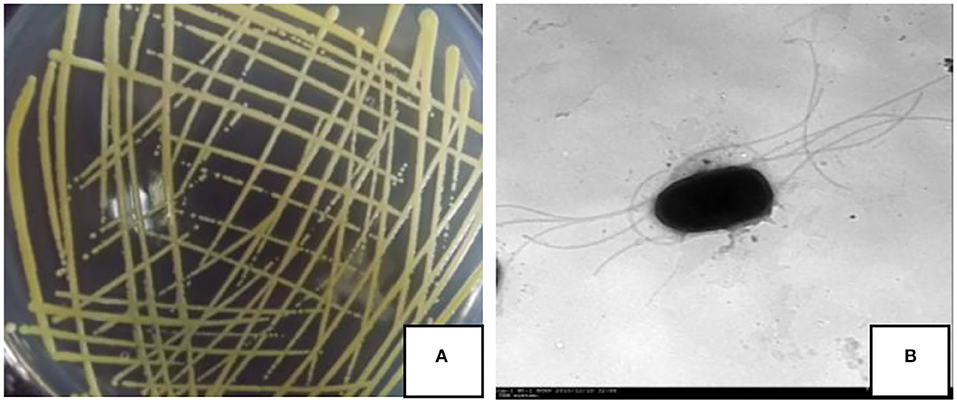
Figure 2. Colony morphology (A) and transmission electron microscopy observation (B) of the strain JI39.
Major physiological and biochemical characteristics of strain JI39 are listed in Table 2. This strain is Gram-stain-negative. It is positive under the Oxidase test, Catalase test, and V-P test, and it can use citrate, malonic acid to reduce nitrate. Besides, this strain is negative for amylohydrolysis and lipid hydrolysis, but positive for Gelatin liquefaction. The strain JI39 could produce acid in litmus milk and grow in the highest NaCl concentration at 7%. In addition, JI39 can utilize glucose, mannitol, D-galactose, and D-fructose as their sole carbon source.
The 16S rDNA of strain JI39 was sequenced and deposited into the GenBank database (KU587789), which shows 99.68 and 99.57% similarity to Arthrobacter nicotinovorans (A. nicotinovorans) strain T258 (KC764990) and A. nicotinovorans strain KNUC601 (HM047516), respectively. The phylogenetic tree showed that strain JI39 was closely related to A. nicotinovorans and fell into the same clade (Figure 3). In conclusion, the strain JI39 was identified to be A. nicotinovorans that is based on the culture characteristics, physiological and biochemical characteristics, and molecular biological identification.
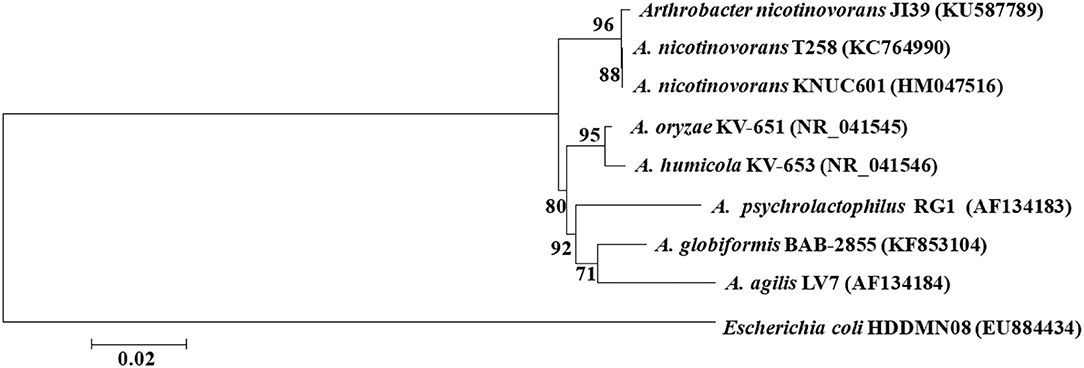
Figure 3. Unrooted neighbor-joining tree based on 16S rRNA gene sequences of strain JI39 (Bootstrap value).
There were halos around the colony of strain JI39 in the cellulase screening medium and the skim milk screening medium (Figure 4). Hence, these results indicated that strain JI39 could produce cellulase and protease.
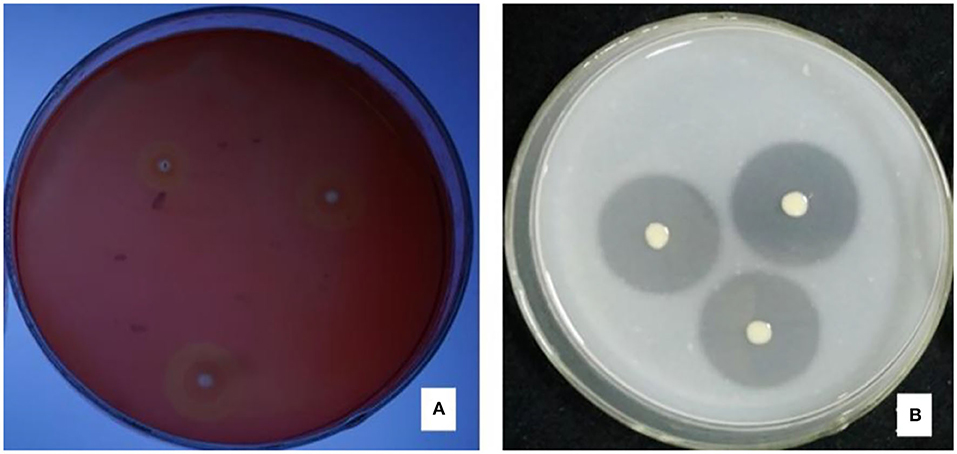
Figure 4. The enzyme production activity of strain JI39. (A) Cellulase screening medium; (B) Skim milk screening medium.
For the growth of strain JI39, the optimal temperature was 30°C and the optimal pH value was 7.0. The optimal sodium chloride concentration was 1% for the growth of strain JI39. We found that the UV irradiation could inhibit the JI39 growth with the longer UV exposure time, the inhibition effect would be stronger (Figure 5).
The average length of seed buds treated with strain JI39 bacterial suspension was 20.51 ± 0.51 mm, which was significantly longer than that of the sterile distilled water control group (12.46 ± 0.39 mm; t = 12.54, p < 0.01; Figure 6). Since the shoot length of the treatment group increased by 64.61% under the JI39 treatment, it may have the effect of promoting the ginseng seed germination.
The strain JI39 had a significant effect on the promotion growth of ginseng root. The 2-year average fresh weight of 120 ginseng roots treated by 10 × 107 CFU/ml of JI39 fermentation broth (2256.67 ± 55.22 g) was significantly higher than that of other treatments, i.e., 100 × 107 CFU/ml of YIWEI treatment (1978.33 ± 19.01) and the control group (1819.17 ± 20.71 g; Table 3).
The soil urease, phosphatase, and invertase enzyme activities were changed after the inoculation of JI39 bacterial suspension (Figures 7A–C). Urease and phosphatase accumulated significantly from 3 to 30 dpi, with urease of the treatment reaching a maximum at 30 dpi and phosphatase of the treatment at 15 dpi, respectively (Figures 7A,B). Invertase activity occurred at a significantly higher level from 7 to 30 dpi and with the highest at 7 dpi (Figure 7C). For catalase, the enzyme activity has remained stable, and there is no significant difference between the treatment group and the control group (Figure 7D).
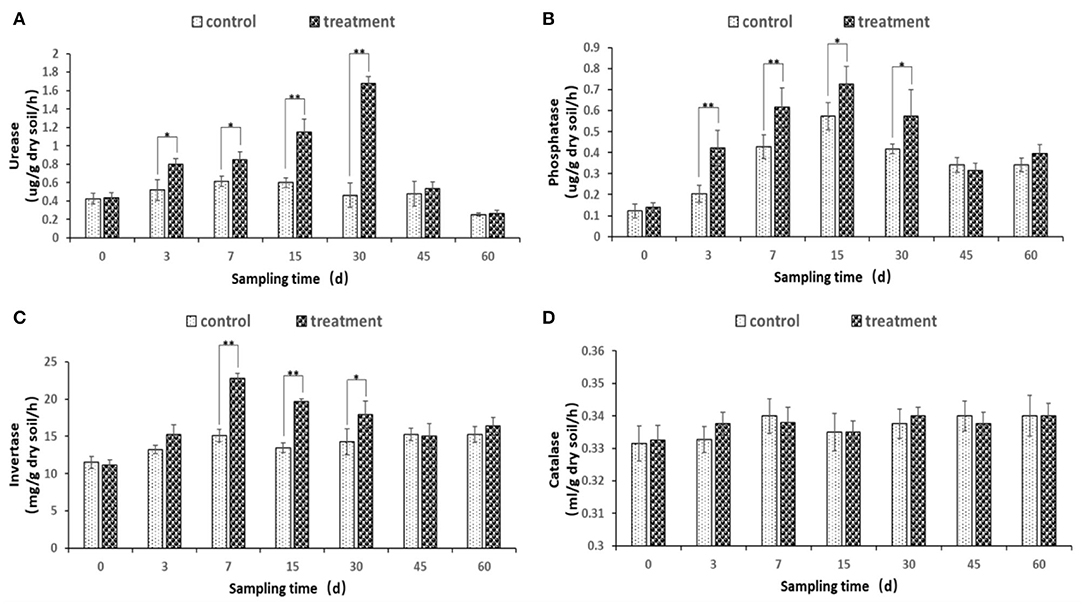
Figure 7. Changes of soil enzyme activity. (A) urease; (B) phosphatase; (C) invertase; (D) catalase. Statistical analysis was conducted using one-way analysis of independent sample t-test. Bars represent the mean and standard error (SE) with three biological replicates with two experiments (n = 6). Values of p were indicated by * symbol: **p < 0.01; *p < 0.05. Blank, not significant.
The expression of all the five defense genes of β-1.3-GA, CHI, PAL, SOD, and POD from ginseng root inoculated by bacterial suspension of strain JI39 showed upregulation trends as compared to the control (Figure 8). The transcripts of three β-1.3-GA, CHI, and PAL genes were significantly accumulated to a higher degree in the ginseng roots with A. nicotinovorans JI39 strain from 3 to 12 dpi and reached their peaks at 6 and 9 dpi, respectively (Figures 8A–C). For the two other genes of SOD and POD, both were induced significantly from 3 to 30 and with the peaks at 9 and 12 dpi, respectively (Figures 8D,E).
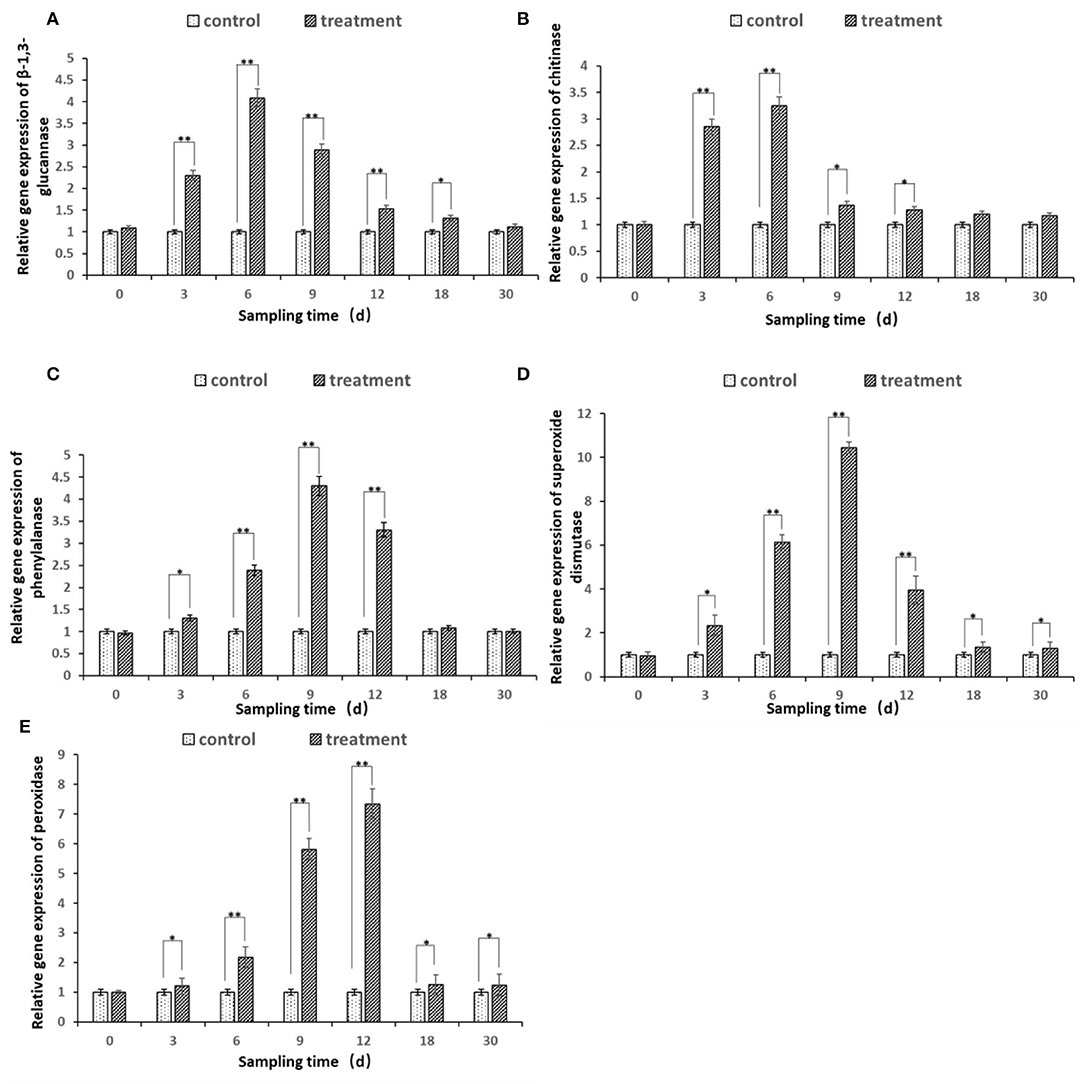
Figure 8. Differential expression of five defense enzyme genes in ginseng root treated by A. nicotinovorans JI39 strain. (A) β-1.3 glucanase; (B) chitinase; (C) phenylalanine ammonia-lyase; (D) superoxide dismutase; (E) peroxidase. Statistical analysis was conducted using one-way analysis of independent sample t-test. Bars represent the mean and standard error (SE) with three biological replicates with two experiments (n = 6). Values of p were indicated by * symbol: **p < 0.01; *p < 0.05. Blank, not significant.
In recent 20 years, the study and application of microbial fertilizer had developed greatly with the development of biotechnology and the efficiency of plant growth-promoting rhizobacteria (PGPRs) had become a hot topic. Since Kloepper proposed the concept of PGPR in 1978, a lot of strains had been screened from the rhizosphere and endophyte of different plants (Kloepper and Schroth, 1978).
Due to the low availability of nitrogen, phosphorus, and potassium in the natural soil, it restricts plant growth and affects the yield and quality of crops. The development and application of PGPR strain, which has the ability to fix nitrogen, dissolve phosphorus and potassium, have attracted the interest of researchers. Yang et al. (2019) found that the single application of potassium-releasing bacteria could significantly reduce the salt content of the arched continuous cropping soil, increase the soil nutrient content, and promote the growth and production of peppers. Wang et al. (2021) used Pseudomonas bacteria with phosphate solubilizing to improve the reclaimed soil, which greatly increased the content of available phosphorus to promote the growth of corn. In addition, IAA production is one of the most important factors that can promote plant growth. Once under the high IAA production, the bacteria can significantly promote plant growth through interaction with their host plants. Through treating varieties of plant seedlings with Azotobacter paspal that can produce IAA, the plant height and leaf area had increased significantly (Barea and Brown, 1974). In addition, the longer root lengths and the larger leaf areas were found by spraying maize and tomato seedlings with the fermentation filtrate of Enterobacter cloacae B8, which with high IAA production activity (Xu et al., 2001). The effect of PGPR on increasing the crop yield has been fully confirmed. For example, when applying 1 kg of phosphate solubilizing bacterial fertilizer per 667 m2, the maximum increase of eggplant root vigor was 35.9% (Jiang et al., 2012a). Bakhshandeh et al. (2017) inoculated three potassium-dissolving bacteria into the rhizosphere of rice under pot and field test conditions and found that the plant height, stem thickness, root length, leaf area, and the dry grain quality could be improved. Moreover, the JI39 strain has multiple growth-promoting characteristics, such as nitrogen fixation, phosphorus and potassium solubilization, IAA production, induction of upregulation of ginseng defense enzyme gene expression, and enhancement of soil enzyme activity.
There were few reports on the growth-promoting bacteria in ginseng plants. In our previous work, Jiang et al. (2018) obtained endophyte growth-promoting bacteria, Pantoea agglomerans, from ginseng, which had the ability to fix nitrogen, release potassium, produce IAA, siderophores, and increase yield on ginseng. In addition, Tian et al. (2014) determined an endophyte bacterium of Pseudomonas fluorescens from ginseng, which had the ability to produce 1-aminocyclopropane 1-carboxylic acid (ACC) deaminase-producing and siderophores, fix nitrogen, dissolve phosphorus, and promote the growth of ginseng in the field. In this study, both the seed germination experiment and a 2-year field trial proved that the A. nicotinovorans JI39 strain with three different concentrations could promote the growth of ginseng and increase significantly the roots yield of 2 years old ginseng with an average of 8.45–24.05% when compared with the control. This result is similar to published PGPR strains of the siderophore-producing Mesorhizobium panacihumi (Huo et al., 2021) and Rhizobium panacihumi (Kang et al., 2021), which could increase 2-year-old ginseng seedlings biomass.
Although the data on the effect of PGPR on the medicinal plants are relatively scarce as compared to that of an increase in some crops (Dos Santos et al., 2020; Wei et al., 2020), there were still studies of different PGPRs on medicinal plants except for Panax ginseng, especially in recent years, such as, Bacillus flexus on Limonium sinense (Xiong et al., 2020) and Mentha arvensis (Singh et al., 2019), Pseudomonas putida on Mentha piperita (Del Rosario Cappellari et al., 2019) and Papaver somniferum (Barnawal et al., 2017), Azotobacter chroococcum and Azospirillum brasilense on M. pulegium (Asghari et al., 2020), B. amyloliquefaciens on M. piperita (Chiappero et al., 2019) and Codonopsis pilosula (Zhao et al., 2016), Pantoea agglomerans and P. putida on M. piperita (Seif Sahandi et al., 2019), and Enterobacter tabaci and Paraburkholderia sp. on Aloe vera (Silva et al., 2019).
The bacterial strain JI39 in this study was determined for its enhancement of IAA production, fixing nitrogen, dissolving phosphorus, and dissolving potassium, which was screened among the 178 bacterial strains from rhizosphere soil of Panax ginseng. The bacterial strain JI39 was identified to be A. nicotinovorans based on the analysis of culture characteristics, physiological and biochemical characteristics, and 16S rDNA gene sequence. The previous studies about the soil source A. nicotinovorans mainly focused on the nicotine degradation, in vivo nicotinic dehydrogenase, and degradation of other chemicals in the soil (Möhler et al., 1972; Kazuya and Kentaro, 2007; Talaat et al., 2011; Roderich and Marius, 2020). This study firstly reported the plant growth-promoting activity of A. nicotinovorans and indicated that it could significantly promote seed germination and the root growth of ginseng.
This study had identified and characterized the bacterial strain JI39 as a kind of rhizospheric A. nicotinovorans, and it could promote the growth of Panax ginseng. This study contributed to the further study of the interaction between JI39 and its host ginseng. While ginseng is an important Chinese traditional medicine, it should be further researched on the effect of JI39 on stress resistance and the medicinal component of ginseng in the future.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
YJ and YS: data curation, formal analysis, methodology, and writing—original draft preparation. CJ and XL: data curation and resources. TL and JW: methodology and data curation. YJ and CC: writing—reviewing and editing. CC and JG: conceptualization, funding acquisition, project administration, and writing—reviewing. All authors agreed to be accountable for all aspects of work ensuring integrity and accuracy.
This work was funded by Jilin Science and Technology Development Project (20210202075NC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors thanks to Fusong Shenwang Plant Protection Co., Ltd., China for its support in field trials and ginseng samples.
Ahmad, M., Rio, A., Oguz, C. T., Ahmet, E. Y., Takeshi, T., and Ryota, K. (2019). High salt tolerant plant growth promoting rhizobacteria from the common ice-plant Mesembryanthemum crystallinum L. Rhizosphere. 9, 10–17. doi: 10.1016/j.rhisph.2018.10.004
Asghari, B., Khademian, R., and Sedaghati, B. (2020). Plant Growth Promoting Rhizobacteria (PGPR) confer drought resistance and stimulate biosynthesis of secondary metabolites in pennyroyal (Mentha pulegium L.) under water shortage condition. Sci. Hortic. 263, 0304–4238. doi: 10.1016/j.scienta.2019.109132
Avishek, B., Gautam, K., Dash., Padmini, S., Upendra, K., Subhra, K. M., et al. (2019). Application of rice (Oryza sativa L.) root endophytic diazotrophic Azotobacter sp. strain Avi2 (MCC 3432) can increase rice yield under green house and field condition. Microbiol. Res. 219, 944–5013. doi: 10.1016/j.micres.2018.11.004
Bakhshandeh, E., Pirdashti, H., and Lendeh, K. S. (2017). Phosphate and potassium-solubilizing bacteria effect on the growth of rice. Ecol. Eng. 103, 164–169. doi: 10.1016/j.ecoleng.2017.03.008
Barea, J. M., and Brown, M. E. (1974). Effects on plant growth by Azotobacter paspali related to synthesis of plant growth regulating substances. J. Appl. Microbiol. 37, 583–593. doi: 10.1111/j.1365-2672.1974.tb00483.x
Barnawal, D., Pandey, S. S., Bharti, N., Pandey, A., Ray, T., Singh, S., et al. (2017). ACC deaminase-containing plant growth-promoting rhizobacteria protect Papaver somniferum from Downy Mildew. J. Appl. Microbiol. 122, 1286–1298. doi: 10.1111/jam.13417
Bhagya, I., Mahendrapal, S. R., and Shalini, R. (2017). Effect of succinate on phosphate solubilization in nitrogen fixing bacteria harbouring chick pea and their effect on plant growth. Microbiol. Res. 202, 43–50. doi: 10.1016/j.micres.2017.05.005
Bruna, D. B., Paulo, T. L., Andrea, F., Natalia, S. T., Maria, L. B., Sarina, T., et al. (2018). Screening of tropically derived, multi-trait plant growth-promoting rhizobacteria and evaluation of corn and soybean colonization ability. Microbiol. Res. 206, 33–42. doi: 10.1016/j.micres.2017.09.007
Chen, F. S., Wang, R., and Chen, J. W. (2004). Study on high β-1,3-glucanase strain screening from soil and enzyme producing conditions. J. Food Sci. 5, 31–35. doi: 10.3321/j.issn:1002-6630.2004.05.002
Chiappero, J., del Rosario Cappellari, L., Sosa Alderete, L. G., Palermo, T. B., and Banchio, E. (2019). Plant growth promoting rhizobacteria improve the antioxidant status in Mentha piperita grown under drought stress leading to an enhancement of plant growth and total phenolic content. Ind. Crops Prod. 139, 926–6690. doi: 10.1016/j.indcrop.2019.111553
Deepmala, P., Rafiq, L., Salim, K., and Koul, K. K. (2019). Isolation, screening and molecular characterization of free-living bacteria of potato (Solanum tuberosum L.) and their interplay impact on growth and production of potato plant under Mycorrhizal association. Sci. Hortic. 252, 388–397. doi: 10.1016/j.scienta.2019.02.072
Del Rosario Cappellari, L., Santoro, M. V., Schmidt, A., Gershenzon, J., and Banchio, E. (2019). Induction of essential oil production in Menthax piperita by plant growth promoting bacteria was correlated with an increase in jasmonate and salicylate levels and a higher density of glandular trichomes. Plant Physiol. Biochem. 141, 142–153. doi: 10.1016/j.plaphy.2019.05.030
Dong, X. Z., and Cai, M. Y. (2001). Handbook of Systematic Identification of Common Bacteria. Beijing: Science Press.
Dos Santos, R. M., Diaz, P. A. E., Lobo, L. L. B., and Rigobelo, E. C. (2020). Use of plant growth-promoting rhizobacteria in maize and sugarcane: characteristics and applications. Front. Sustain. Food Syst. 4:136. doi: 10.3389/fsufs.2020.00136
Faisal, I., Tahira, Y., Muhammad, S. A., Muhammad, R., Sher, M. S., Qaiser, I., et al. (2016). Combined ability of chromium (Cr) tolerant plant growth promoting bacteria (PGPB) and salicylic acid (SA) in attenuation of chromium stress in maize plants. Plant Physiol. Biochem. 108, 456–467. doi: 10.1016/j.plaphy.2016.08.014
Furnkranz, M., Muller, H., and Berg, G. (2009). Characterization of plant growth promoting bacteria from crops in Bolivia. J. Plant Dis.protection. 116, 149–155. doi: 10.1007/BF03356303
Garrity, G. M., Bell, J. A., and Lilburn, T. G. (2004). Taxonomic Outline of the Prokaryotes. Bergey's Manual of Systema-Tic Bacteriology, 2nd edn, Release 5.0. New York, NY: Springer.
Huo, Y., Kang, J. P., Ahn, J. C., Kim, Y. J., Piao, C. H., Yang, D. U., et al. (2021). Siderophore-producing rhizobacteria reduce heavy metal-induced oxidative stress in Panax ginseng meyer. J. Ginseng Res. 45, 1226–8453. doi: 10.1016/j.jgr.2019.12.008
Imane, A., Kaoutar, T., Berraho, B., Mhamed, K., Imane, T. A., and Jamal, A. (2019). Screening of stress tolerant bacterial strains possessing interesting multiplant growth promoting traits isolated from root nodules of Phaseolus vulgaris L. Biocatal. Agric. Biotechnol. 20:101225. doi: 10.1007/bergeysoutline200405
Jiang, C. H., Liao, M. J., Wang, H. K., Zheng, M. Z., Xu, J. J., and Guo, J. H. (2018). Bacillus velezensis, a potential and efficient biocontrol agent in control of pepper gray mold caused by Botrytis cinerea. Biol. Control. 126, 147–157. doi: 10.1016/j.biocontrol.2018.07.017
Jiang, X. M., Xia, X. H., Yu, X. H., Ni, S. J., and Zhang, S. C. (2012a). Effect of microbial phosphorus-solubilizing bacteria fertilizer on the growth of eggplant in greenhouse and the utilization of soil available phosphorus. J. Zhejiang Univ. 39, 685–688. doi: 10.3785/j.issn.1008-9497.2012.06.016
Jiang, Y., Yin, W., Chen, C. Q., and Ni, R. Q. (2012b). 16S rDNA-RFLP Analysis of Culturable Endophytic Bacteria from Panax ginseng. J. Northeast For. Univ. 40, 34–39. doi: 10.13759/j.cnki.dlxb.2012.08.026
Jiang, Y., Yin, W., Chen, C. Q., Tian, L., and Xu, P. (2013). Identification and optimized fermentation condition of an Endophyte Antagonistic Bacteria NJ13. Agrochemicals. 52, 97–101. doi: 10.16820/j.cnki.1006-0413.2013.02.007
Kandeler, E., and Gerber, H. (1988). Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils. 6, 68–72. doi: 10.1007/BF00257924
Kang, J. P., Huo, Y., Yang, D. U., and Yang, D. C. (2021). Influence of the plant growth promoting rhizobium panacihumi on aluminum resistant in Panax ginseng. J. Ginseng Res. 45, 442–449. doi: 10.1016/j.jgr.2020.01.001
Kazuya, M., and Kentaro, M. (2007). Microbial resolution of dl-homoserine for the production of d-homoserine using a novel isolate, Arthrobacter nicotinovorans strain 2-3. Enzyme Microb. Technol. 41, 318–321. doi: 10.1016/j.enzmictec.2007.02.013
Kim, H., Rim, S. O., and Bae, H. (2019). Antimicrobial potential of metabolites extracted from ginseng bacterial endophyte Burkholderia stabilis against ginseng pathogens. Biol. Control. 128, 24–30. doi: 10.1016/j.biocontrol.2018.08.020
Kloepper, J. W., and Schroth, M. N. (1978). “In plant growth-promoting rhizobacteria on radishes,” in Proceedings of the 4th International Conference on Plant Pathogenic Bacteria (Angers), 879–882.
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt, M. Goodfellow (Chichester: Wiley), 115−175.
Lúcia, R. R. B., Newton, P. S., Lilia, G. W., Delson, L., Marcos, A. B. L., Samuel, M. M. M., et al. (2016). Cowpea resistance induced against Fusarium oxysporum f. sp. tracheiphilum by crustaceous chitosan and by biomass and chitosan obtained from Cunninghamella elegans. Biol. Control. 92, 45–54. doi: 10.1016/j.biocontrol.2015.09.006
Michele, T. H., Malkanthi, K. G., Kithsiri, W., Leslie, G., and Elizabeth, A. (2013). Endohyphal bacterium enhances production of indole-3-acetic acid by a foliar fungal endophyte. PLoS ONE 8, 1–8. doi: 10.1371/journal.pone.0073132
Mohamad, O., Ma, J. B., Liu, Y. H., Zhang, D., and Li, L. (2020). Beneficial endophytic bacterial populations associated with medicinal plant thymus vulgaris alleviate salt stress and confer resistance to Fusarium oxysporum. Front. Plant Sci. 11:47. doi: 10.3389/fpls.2020.00047
Möhler, H., Brühmüller, M., and Decker, K. (1972). Covalently bound flavin in D-6-hydroxynicotine oxidase from Arthrobacter oxidans identification of the 8,-(N-3-histidyl)-riboflavin-linkage between FAD and apoenzyme. Eur. J. Biochem. 37, 256–258. doi: 10.1111/j.1432-1033.1972.tb01969.x
Motahhareh, A., Hassan, E., and Hossein, A. A. (2019). Characterization of rhizosphere and endophytic bacteria from roots of maize (Zea mays L.) plant irrigated with wastewater with biotechnological potential in agriculture. Biotechnol. Rep. 20, 1–12. doi: 10.1016/j.btre.2019.e00305
Ning, Z. Y., Li, Y. L., Zhao, X. Y., Han, D., and Zhan, J. (2022). Comparison of leaf and fine root traits between annuals and perennials, implicating the mechanism of species changes in desertified grasslands. Front. Plant Sci. 12:1664-462X. doi: 10.3389/fpls.2021.778547
Paola, D. Q., Nadia, C. L., Viviana, A. R., and Raúl, O. P. (2020). The plant growth promoting bacteria Gluconacetobacter diazotrophicus and Azospirillum brasilense contribute to the iron nutrition of strawberry plants through siderophores production. Plant Growth Regul. 91, 185–199. doi: 10.1007/s10725-020-00598-0
Pikovskaya, R. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya. 17, 362–370.
Rahul, J., and Anita, P. (2016). Soil enzymes and microbial endophytes as indicators of climate variation along an altitudinal gradient with respect to wheat rhizosphere under mountain ecosystem. Rhizosphere. 2, 75–84. doi: 10.1016/j.rhisph.2016.07.007
Rania, A. B. A., Sonia, M. T., Ahlem, N., Hayfa, J. K., and Mejda, D. R. (2016). Biocontrol of Fusarium wilt and growth promotion of tomato plants using endophytic bacteria isolated from Nicotiana glauca organs. Biol. Control. 97, 80–88. doi: 10.1016/j.biocontrol.2016.03.005
Roderich, B., and Marius, M. (2020). A soil bacterial catabolic pathway on the move: Transfer of nicotine catabolic genes between Arthrobacter genus megaplasmids and invasion by mobile elements. J. Biosci. 58:45. doi: 10.1007/s12038-020-00030-9
Sangeeta, P., and Shikha, G. (2020). Diversity analysis of ACC deaminase producing bacteria associated with rhizosphere of coconut tree (Cocos nucifera L.) grown in Lakshadweep islands of India and their ability to promote plant growth under saline conditions. J. Biotechnol. 234, 0168–1656. doi: 10.1016/j.jbiotec.2020.10.024
Sarita, T., Bijaya, K. S., and Sanjog, T. (2016). Identification of arsenic resistant endophytic bacteria from Pteris vittata roots and characterization for arsenic remediation application. J. Environ. Manage. 180, 359–365. doi: 10.1016/j.jenvman.2016.05.029
Seif Sahandi, M., Mehrafarin, A., Naghdi Badi, H., Khalighi-Sigaroodi, F., and Sharifi, M. (2019). Improving growth, phytochemical, and antioxidant characteristics of peppermint by phosphate-solubilizing bacteria along with reducing phosphorus fertilizer use. Ind. Crops Prod. 141, 0926–6690. doi: 10.1016/j.indcrop.2019.111777
Shahab, S., Ahmed, N., and Khan, N. S. (2009). Indole acetic acid production and enhanced plant growth promotion by indigenous PSBs. Afr. J. Agric. Res. 4, 1312–1316. doi: 10.1021/jf902355m
Silva, C. F., Vitorinob, M. A. C., Mendoncab, W. L., Araujoc, M. N., Albuquerqued, L. C., Soares, E. L., et al. (2019). Screening of plant growth-promoting endophytic bacteria from the roots of the medicinal plant Aloe vera. S. Afr. J. Bot. 19, 1–14. doi: 10.1016/j.sajb.2019.09.019
Singh, S., Tripathi, A., Maji, D., Awasthi, A., Vajpayee, P., and Kalra, A. (2019). Evaluating the potential of combined inoculation of Trichoderma harzianum and Brevibacterium halotolerans for increased growth and oil yield in mentha arvensis under greenhouse and field conditions. Ind. Crops Prod. 131, 173–181. doi: 10.1016/j.indcrop.2019.01.039
Sudhir, K., Glen, S., and Koichiro, T. (2016). MEGA 7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Sun, X. D., Ye, Y. Q., Ma, Q. X., Guan, Q. W., and Jones, D. L. (2021). Variation in enzyme activities involved in carbon and nitrogen cycling in rhizosphere and bulk soil after organic mulching. Rhizosphere 19:100376. doi: 10.1016/j.rhisph.2021.100376
Sun, Z., Yang, L. M., Han, M., Han, Z. M., Yang, L., Cheng, L., et al. (2019). Biological control ginseng grey mold and plant colonization by antagonistic bacteria isolated from rhizospheric soil of Panax ginseng Meyer. Biol. Control. 138, 1049–9644. doi: 10.1016/j.biocontrol.2019.104048
Supratim, B., Roel, R., and Sangeeta, N. (2017). Towards a better greener future - an alternative strategy using biofertilizers. I: plant growth promoting bacteria. Plant Gene 12, 43–49. doi: 10.1016/j.plgene.2017.07.004
Talaat, E. S., Marion, D. L., Frédérique, C., Nadine, R., and Fabrice, M. L. (2011). Evidence of atrazine mineralization in a soil from the Nile Delta: isolation of Arthrobacter sp. TES6, an atrazine-degrading strain. Int. Biodeterior. Biodegrad. 65, 1249–1255. doi: 10.1016/j.ibiod.2011.05.011
Tian, L., Jiang, Y., Chen, C. Q., Zhang, G. J., Li, T., Tong, B., et al. (2014). Screening and identification of a ginseng endogenous 1-aminocyclopropane-1-1-carboxylic acid (ACC) deaminase activity bacteria Effects on host growth. Acta Microbiol. 54, 760–769. doi: 10.13343/j.cnki.wsxb.2014.07.006
Wang, X., Zhang, D. N., Wang, C. W., Chen, X., Yang, L. N., Lu, B. H., et al. (2019). Colonization characteristics of Bacillus amyloliquefaciens FS6 in ginseng and its disease resistance to ginseng. J. Northwest A&F Univ. 47, 125–138. doi: 10.13207/j.cnki.jnwafu.2019.07.017
Wang, X. Y., Gao, J. H., Meng, H. S., Zhang, J., Wu, X., and Hong, J. P. (2021). GFP marking of two phosphorus resolving bacteria and their colonization in reclaimed soil. Shanxi Agri. Sci. 49, 976–982. doi: 10.3969/j.issn.1002-2481.2021.08.15
Wei, M., Zhang, M., Huang, G., Yuan, Y., Fu, C., and Yu, L. (2020). Coculture with two bacillus velezensis strains enhances the growth of anoectochilus plants via promoting nutrient assimilation and regulating rhizosphere microbial community. Ind. Crops Prod. 154, 0926–6690. doi: 10.1016/j.indcrop.2020.112697
Xiong, Y. W., Li, X. W., Wang, T. T., Gong, Y., Zhang, C. M., Xing, K., et al. (2020). Root exudates-driven rhizosphere recruitment of the plant growth-promoting rhizobacterium Bacillus flexus KLBMP 4941 and its growth-promoting effect on the coastal halophyte limonium sinense under salt stress. Ecotoxicol. Environ. Saf. 194, 0147–6513. doi: 10.1016/j.ecoenv.2020.110374
Xu, Y. P., Zang, R. C., Chen, W. L., and Lou, Q. C. (2001). Promoting plant growth and IAA analysis of Enterobacter cloacae B8 fermentation liquid. J. Zhejiang Univ. 27, 282–284.
Yacine, G., Omrane, T., Amine, Y., Nasserdine, S., Florence, M., and Abdelghani, Z. (2014). Biocontrol of Rhizoctonia solani damping-off and promotion of tomato plant growth by endophytic actinomycetes isolated from native plants of Algerian Sahara. Microbiol. Res. 169, 0944–5013. doi: 10.1016/j.micres.2013.06.014
Yang, D. Y., Gao, J. X., Sang, T., Wang, X. M., and Xie, H. (2019). Effects of phosphorus and potassium solubilizing bacteria on the growth and soil nutrient content of continuous cropping pepper in arched shed. Northern Hortic. 6, 1–6. doi: 10.11937/bfyy.20182358
Keywords: Panax ginseng, rhizosphere bacteria, plant growth promotion, Arthrobacter nicotinovorans, soil enzyme
Citation: Jiang Y, Song Y, Jiang C, Li X, Liu T, Wang J, Chen C and Gao J (2022) Identification and Characterization of Arthrobacter nicotinovorans JI39, a Novel Plant Growth-Promoting Rhizobacteria Strain From Panax ginseng. Front. Plant Sci. 13:873621. doi: 10.3389/fpls.2022.873621
Received: 11 February 2022; Accepted: 28 March 2022;
Published: 09 May 2022.
Edited by:
Zhongwei Zou, University of Manitoba, CanadaReviewed by:
Linlin Dong, China Academy of Chinese Medical Sciences, ChinaCopyright © 2022 Jiang, Song, Jiang, Li, Liu, Wang, Chen and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Jiang, eXVuakBqbGF1LmVkdS5jbg==; Jie Gao, amllZ2FvMTE1QDE2My5jb20=; Changqing Chen, Y2hlbmNoYW5ncWluZ0BqbGF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.