- 1Key Laboratory of Green Prevention and Control of Tropical Plant Disease and Pests, Ministry of Education and School of Plant Protection, Hainan University, Haikou, China
- 2Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
- 3State Key Laboratory of Crop Stress Biology for Arid Areas and College of Plant Protection, Northwest A&F University, Yangling, China
Fusarium wilt of banana (Musa spp.), a typical vascular wilt disease caused by the soil-borne fungus, Fusarium oxysporum f. sp. cubense race 4 (Foc4), seriously threatens banana production worldwide. Pathogens, including vascular wilt fungi, secrete small cysteine-rich proteins during colonization. Some of these proteins are required for pathogenicity. In this study, 106 small secretory proteins that contain a classic N-terminal signal peptide were identified using bioinformatic methods in Foc4. Among them, 11 proteins were selected to show transient expressions in tobacco. Interestingly, transient expression of FoSsp1 in tobacco, an uncharacterized protein (of 145 aa), induced necrotic cell death reactive oxygen burst, and callous deposition. Furthermore, the expression of FoSSP1 in Foc4 wild type (WT) was up-regulated during the stage of banana roots colonization. A split-marker approach was used to knock out FoSSP1 in the Foc4 WT strain. Compared with the WT, the deletion mutant Fossp1 was normal in growth rate but increased in conidiation and virulence. RT-qPCR analysis showed that the expression of four conidiation regulator genes in the Fossp1 deletion mutant was significantly decreased compared to the WT strain. In addition, the expression of four pathogenesis-related genes of bananas infected with Fossp1 deletion mutant was down-regulated in comparison with that of the WT. In summary, these results suggested that FoSSP1 is a putative elicitor that negatively regulates conidiation and pathogenicity in Foc4.
Introduction
Fusarium wilt of bananas is known as “Panama disease”, damaging the banana production around the world (Hwang and Ko, 2004). The causing agent Fusarium oxysporum f. sp. cubense (Foc) can be further divided into three races (Foc1, 2, and 4) based on pathogenicity differences to banana cultivars. Among them, Foc4 is the most virulent one which can infect all Cavendish banana cultivars in addition to all the cultivars that are susceptible to Foc1 and Foc2 (Ploetz et al., 2015). Foc4 can be further divided into tropical race 4 (TR4) and subtropical race 4 (STR4), of which TR4 is more virulent (Butler, 2013). TR4 was reclassified into the new species F. odoratissimum, based on the genetic diversity of Foc isolates in the Indonesian center of origin (Maryani et al., 2019). As a soil-borne fungus, Foc usually infects the root first. Conidia or hyphae move from soil to the surface of the root and rhizome, and then hyphae penetrate the root xylem and spread to the pseudostem xylem and even the outer leaf sheaths. At last, the infected plant shows symptoms (Dita et al., 2018). Pathogen spreading in the vascular bundle of bananas blocks nutrients and water transportation, resulting in leaves yellowing and plant death (Michielse and Rep, 2009).
In nature, there is a constant arm race between plants and pathogens. These pathogens use numerous virulence factors for pathogenicity or fitness in the plant (Chisholm et al., 2006). Therefore, plants need to mount effective defense mechanisms to protect themselves, including two innate immune systems (Boller and Felix, 2009; Dou and Zhou, 2012). When pathogens attack host cells, the pattern-recognition receptors (PRRs) of cell membranes are the first to detect the pathogen by recognizing pathogen-associated molecular patterns (PAMPs) of pathogens (Bigeard et al., 2015). This nonspecific immunity called Pattern-Triggered Immunity (PTI) can halt further colonization (Jones and Dangl, 2006). This is the first line of defense in plants against pathogens. In response, the pathogen secretes effectors which can interfere with PTI and result in effector-triggered susceptibility (ETS) (Bu et al., 2014). At the same time, the resistance protein of the plants detects the effectors to trigger the specific immunity of the plant, which is the second line of defense (known as ETI, Effector-Triggered Immunity) (Tsuda and Katagiri, 2010).
Currently, it has been reported that small secreted proteins play an important role in the interactions between plants and the pathogen. There are more and more studies on small secreted proteins, which share the same characteristics: generally, <300 amino acids, lack transmembrane domain, contain an N-terminal signal peptide (SP), and rich-in cysteine (Niu et al., 2021). Fungal small secreted proteins involved in the interaction between pathogens and plants have been referred to as effectors or elicitors. Recently, those effectors or elicitors have been identified in many fungal pathogens. PstSCR1 as an elicitor in wheat rust pathogen, Puccinia striiformis f. sp. tritici, can activate the surface-mediated immunity in Nicotiana benthamiana which contributes to the non-host resistance to oomycete pathogens (Dagvadorj et al., 2017). As an effector, CfEC92 inhibits plant immunity and participates in the early infection stage of Colletotrichum fructicola to glomeruli (Shang et al., 2020). VmE02 has been reported as a novel cross-kingdom PAMP widely spread in fungi and oomycetes, which exhibited cell-death-inducing activity in N. benthamiana. However, deletion of VmE02 in Valsa mali has no effect on pathogenicity but significantly reduces conidia production (Nie et al., 2019b).
Here, in this study, 106 small secretory proteins that contain a classic N-terminal signal peptide, were identified by bioinformatic methods in Foc4. Among them, 11 proteins were selected for transient expression in tobacco. Interestingly, transient expression of FoSsp1, an uncharacterized protein (of 145 aa), induced necrotic cell death reactive oxygen burst, and callous deposition in N. benthamiana. Furthermore, the expression of FoSSP1 was up-regulated during the stage of banana roots colonization. A split-marker approach was used to knock out FoSSP1 in the Foc4 wild type (WT) strain. Compared with the wild type, the deletion mutant Fossp1 was normal in growth rate but increased in conidiation and virulence. RT-qPCR analysis showed that the expression of four conidiation regulator genes in the Fossp1 deletion mutant was significantly decreased compared to the WT strain. In addition, the expression of four pathogenesis-related (PR) genes of bananas infected with Fossp1 deletion mutant was down-regulated in comparison with that of the wild type. In summary, these results suggested that FoSSP1 is a putative elicitor that negatively regulates conidiation and pathogenicity in Foc4.
Materials and Methods
Strains and Plant Growth Conditions
The WT strain of F. oxysporum f. sp. cubense used in the experiment was Foc4. The mycelium was maintained on potato dextrose Agar (PDA) medium and the conidial suspension was stored in 24% glycerol at −80 °C. The knockout and complement transformants were cultured on a medium containing 50 mg/mL hygromycin B or G418, respectively. E. coli DH5a, BL21 and A. tumefaciens GV3101, AGL1 were grown on a lysogeny broth (LB) medium containing corresponding antibiotics and were stored in 24% glycerol at −80°C. N. benthamiana was grown in a greenhouse with 8/16 (light/dark) per day at 24°C. Musa AAA, as warm-loving wet crops, was cultivated in a warm shed with a temperature of 30°C.
Plasmid Construction
To investigate whether FoSsp1 can induce hypersensitivity response in N. benthamiana, Phusion Plus DNA polymerase (Thermo Fisher Scientific) was used to amplify the coding region of FoSSP1 and the coding region without signal peptide FoSSP1ΔSP from the cDNA of Foc4 and cloned into the plant expression vector pBin-eGFP, respectively. The same sequence was amplified by PCR and cloned into a pSUC2 vector using EcoRI and XhoI restriction sites by ClonExpress® II One Step Cloning Kit (Vazyme Biotech) for verifying the secretion activity of signal peptide (SP) (Oh et al., 2009). The encoding region and promoter sequence of FoSSP1 were amplified from WT genomic DNA and cloned into the pFL2 vector to obtain the complementary vector pFL2-HB (Chen et al., 2014).
Total RNA Extraction and Quantitative Reverse Transcription PCR
To verify the expression of FoSSP1 during the process of the plant infection, roots of banana seedlings were inoculated with conidial suspension with a concentration of 108/mL. Total RNAs were extracted from the inoculated roots (12, 24, 36, 48, 60, and 72 hpi). The relative expression of FoSSP1 in conidia was considered as the 0 hpi control. The extraction method refers to Eastep® Super Total RNA Extraction Kit (Promega), following the manufacturer's instructions. The cDNA was synthesized with the PrimeScript 1st strand cDNA Synthesis kit (Takara), according to the manufacturer's instructions.
A pair of specific (SSP1-RT-F/R) primers of the gene was used for qRT-PCR analysis with 100 ng cDNA as a template (Supplementary Table S1). The qRT-PCR was performed using the SuperReal PreMix Plus kit (Tiangen Biotech), following the manufacturer's instructions. The actin gene was used as an endogenous reference to normalize the expression of FoSSP1 in different periods and the relative expression levels of genes were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001). Select one of the sets of data and arbitrarily set its relative expression to 1. The qRT-PCR conditions were as follows: an initial 95°C denaturation step for 3 min, followed by denaturation for 15 s at 95°C, annealing for 15 s at 57°C, and extension for 20 s at 72°C for 35 cycles (Wu et al., 2010). For the relative expression of PR genes in infected bananas, the actin gene (Degradi et al., 2021) of bananas was used as an endogenous reference to normalize the expression. One-way ANOVA was used to compare the significance of the difference between the two groups at the same time point.
Signal Peptide Secretion Activity Verification
The recombinant vectors pSUC2-FoSSP1 and pSUC2-FoSSP1ΔSP were constructed and transferred into the YTK12 strain (Gu et al., 2011). The CMD-W medium (0.08% tryptophan dropout supplement, 0.65% yeast nitrogen base without amino acids, 2% sucrose, 0.1% glucose, and 2% agar) was used to screen the positive colonies from all the transformants. The successful transformant was cultured on a YPRAA medium to verify its secretion activity. To further test the secretion activity of the signal peptide, the reduction of 2, 3, 5-triphenyltetrazolium chloride (TTC) to insoluble red-colored 1, 3, 5-triphenylformazan (TPF) was monitored.
Culture Conditions and Fungal Transformation
Gene replacement was carried out according to homologous recombination (Yang et al., 2018b). The upstream and downstream flanking sequences of FoSSP1 were cloned with primer pairs of UF/UR and DF/DR (containing the Hyg tag) [(Supplementary Table S1) Polynucleotide chain reaction primers used in this study], using the DNA of Foc4 as a template. The upstream and downstream of hygromycin resistant gene fragment was amplified with PCR primer pair of HYG-F/R and pEX2 vector as DNA template. Fusion fragment for transformation was generated by overlap PCR as described (Goswami, 2012). Purified fragments for protoplast transformation using Gel Extraction Kit (Omega) according to the manufacturer's instructions.
The hyphae were digested in enzymatic solution (25 g/L Driselase, 5 g/L Lysing Enzymes in 1.2 M KCl) and protoplasts were released until the concentration is 107 protoplasts/ml (Yun et al., 2014). The DNA fragments or plasmids for transformation were mixed with protoplasts and incubated on ice for 20 min. Then, 1 ml PTC solution (0.6 M KCl, 50 mM Tris-HCl pH 8.0, 50 mM CaCl2, and 40% PEG 4000) was added to the mixture and incubated at room temperature for 30 min. The mixture was transferred to a TB3 medium for further culture for 20 h. The transformed mixture was inoculated on a 15ml PDA medium containing 50 μg/ml hygromycin B or G418, then covered with PDA agar modified with 75 μg/ml hygromycin B or G418. Transformants harboring hygromycin B or G418 resistance were identified by PCR.
Transient Expression Assay in N. benthamiana
The recombinant vectors pBin-FoSSP1 and pBin-FoSSP1ΔSP were transferred into Agrobacterium tumefaciens GV3101 by electroporation and cultured in LB containing 20 mg/ml rifampicin and 50 mg/ml kanamycin for 48 h. Bacteria were harvested and resuspended with injection buffer (1% MgCl2·6H2O, 1% MES, 150 μM acetosyringone) to the OD600 of 1.0. The suspension was incubated at room temperature for 2–3 h to release the plasmids in the injection buffer. A plant of N. benthamiana at the stage of four leaves was selected and infiltrated the buffer from the back of the leaves (Wei et al., 2020). The empty pBin-eGFP vector was used as a negative control, and the vector containing BAX was used as a positive control. Leaves cell death symptoms were photographed both under natural light and under ultraviolet (wavelength 365 nm) at 6 days post-inoculation. Each assay was repeated three times.
Protein Extraction and Western Blot
Leaves were thoroughly ground in liquid nitrogen. The process of protein separation refers to the previous (Yang et al., 2012). The 20 μl total protein was separated with the 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (Shenggong Biotech) and transferred to the polyvinylidene difluoride (PVDF) membrane. Rabbit anti-GFP pAb (ABclonal) and rabbit anti-HA pAb (Shenggong Biotech) were used as primary antibodies and HRP Goat anti-rabbit IgG (ABclonal) as secondary antibodies.
Phenotypic Characterization of the Foc Mutants
To explore the growth rate, Fossp1, WT, and Fossp1-C were inoculated on the PDA plates, respectively, and cultured at 28°C for 6 days. The size of fungus mycelium was measured by crossing every day to calculate the mycelium growth rate. WT, Fossp1, and Fossp1-C were cultured in a PDB medium at 28°C. Up to the fourth day, the conidia numbers were counted daily using a hemocytometer.
WT, Fossp1, and Fossp1-C were cultured in a PDB medium for 4 days, and the spores were collected at least 108 conidia/ml. The roots of banana seedlings were cleaned and root wounds were made with an art knife, then immersed in the prepared spore solution for 30 min. The seedlings were transplanted into flowerpots. And 50 mL conidial suspension was poured into each pot.
To evaluate the disease index, the corm of banana seedlings at 20 dpi was cut to measure the lesion area. The lesion area was used for disease grade, disease grade for each plant ranging from 0 to 4 (0, no lesion in corm; 1, 1–10% lesion area in corm; 2, 11–30% lesion area in corm; 3, 31–50% lesion area in corm; 4, more than 50% lesion area in corm). The disease index of infected banana seedlings was calculated as follows formula: Disease index (%) = [∑ (grade × number of plants in that grade)/ (4 × total number of assessed plants)] × 100% (Zhang et al., 2019).
Subcellular Localization in Tobacco Leaves
Agrobacterium containing recombinant vectors and pBin-FoSSP1ΔSP was re-suspended in buffer solution at OD600 of 0.001. The syringe was used to infiltrate Agrobacterium solutions into the tobacco leaves. The leaves were photographed and observed under a laser confocal microscope 2 days post infiltration. DAPI staining was conducted as previously described (Tarnowski et al., 1991). The empty vector was set as the negative control.
ROS Staining and Callose Deposition Detection
The accumulation of H2O2 was detected by staining tobacco leaves with 3, 3'-diamino benzidine (DAB). The leaves were immersed in A. tumefaciens for 3 days and stained in DAB for 12 h in the dark. Then the leaves were destained in 95% ethanol until translucent and photographed (Dong and Chen, 2013).
For callose staining, tobacco leaves were boiled and decolorized in 95% ethanol, and then immersed in 1% aniline blue solution in the dark for at least 2 h. The deposition of callose was detected under the microscope.
Result
FoSsp1 Is Conserved in Filamentous Fungi
A small secreted protein of plant pathogens is characterized by an N-terminal signal peptide, no transmembrane domain, rich-in cysteine, and no more than 300-amino acids (Zhang et al., 2020). Here, SignalP (Nielsen, 2017) (https://services.healthtech.dtu.dk/service.php?SignalP-5.0), TMHMM (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) and Big-PI Predictor (https://mendel.imp.ac.at/gpi/fungi_server.html) were used to predict the small secreted proteins of F. oxysporum f. sp. Cubense Race 4 (Foc4). More than 1% of cysteine is considered rich in cysteine. In total, 106 putative secreted proteins were obtained that met the above characteristics. Among them, 11 proteins were selected randomly for transient expression in tobacco (Supplementary Table S2). Small secreted proteins screened in this study). The encoding genes were constructed into a recombinant pGR107 vector and transiently expressed in N. benthamiana leaves by A. tumefaciens-mediated transformation. The results showed that FOIG_06406 (Gene ID: 42031581) which encodes a 145-amino acids protein can induce necrotic cell death in N. benthamiana (Figure 3A). We named it FoSSP1 (Small Secreted Protein) in this study. A BLASTp search against the NCBI Nr database was conducted using the FoSsp1 protein sequence (NCBI Reference Sequence: XP_031064189.1) as a query, results indicated that FoSsp1 homologs are conserved in the Fusarium genus and many other higher Ascomycetes. Interestingly, FoSsp1 homologs were also found in Pterula gracilis, which belongs to Basidiomycetes. These results indicate that FoSsp1 was conserved in fungi (Figure 1).
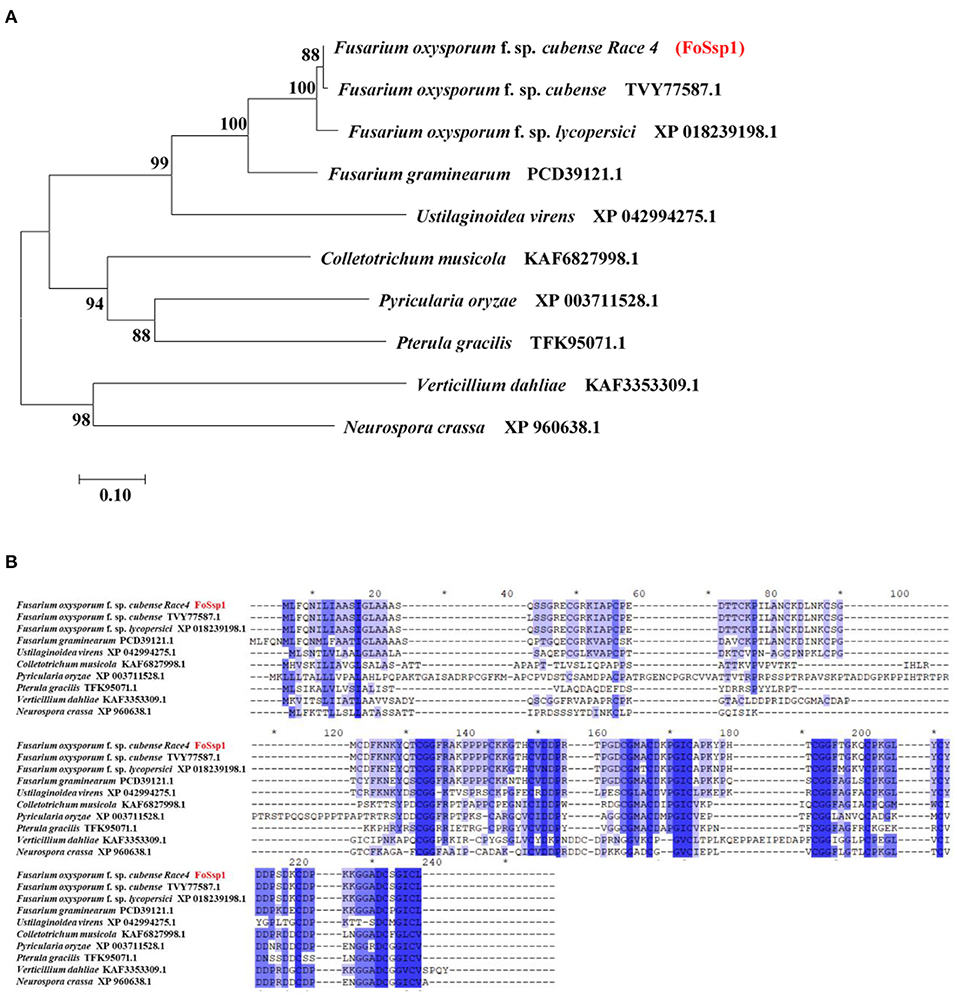
Figure 1. Homology analysis of Ssp1 protein among filamentous fungi. (A) The unrooted tree of full-length protein was generated with the MEGA7 program by the neighbor-joining method. Numbers on major branches indicate percentage boot strap confident scores (1,000 replications). The genetic distance corresponding to the scale is 0.10. (B) Comparative analysis of protein sequences of FoSsp1 and its homologous proteins. The protein sequences were compared and edited with GeneDoc, and the different shades of blue reflected the level of amino acid identity at each location. Identical and conserved catalytic residues are marked with an asterisk (*).
The Expression of FoSSP1 Was Up-Regulated in Foc4 Infected Banana Roots
Fungal secreted proteins usually participate in pathogen infection. To explore the relative expression of FoSSP1, qRT-PCR was conducted in banana roots infected with Foc4. Compared with the expression of FoSSP1 in conidia (0 hpi, hour post-inoculation), the expression of FoSSP1 was significantly up-regulated at 60 and 72 hpi (hours post-inoculation). There was no significant difference between the expression levels in other periods and the gene expression levels in spores (Figure 2). This result indicates that FoSSP1 is induced in plant infection, implying its function in the plant-pathogen interaction.
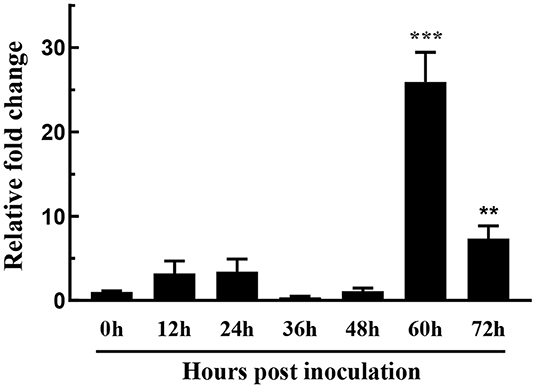
Figure 2. The expression level of FoSSP1 in Foc4 WT during different times after being inoculated in banana roots (0, 12, 24, 36, 48, 60, and 72 hpi). The relative expression of FoSSP1 in conidia (0 h control) was arbitrarily set to 1. The results are representative of three independent experiments. Error bars represent standard error. Asterisks indicate significant differences in one-way analysis of variance (p < 0.01).
FoSsp1 Can Induce Cell Death of N. benthamiana
To dissect the effect of FoSsp1 on plant defense, transient expression of FoSsp1 in N. benthamiana was conducted. The cell death triggering protein Bax (Gene ID: 581) (Lacomme and Santa Cruz, 1999), was set as a positive control. Given that signal peptide is often thought to be cleaved to form mature proteins (Owji et al., 2018), recombinant vectors of pBin-eGFP-FoSSP1 and pBin-eGFP-FoSSP1ΔSP were constructed respectively. Results indicate that Bax, pBin-eGFP-FoSSP1, and pBin-eGFP-FoSSP1ΔSP induced cell death in leaves of N. benthamiana after 5 days post agroinfiltration, whereas the EV (empty vector) did not (Figure 3A). Proper protein expression in N. benthamiana leaves was further confirmed by Western Blot analysis (Figure 3B). The experiment was repeated three times with the same results. The above results showed that FoSsp1 could induce cell death in N. benthamiana.
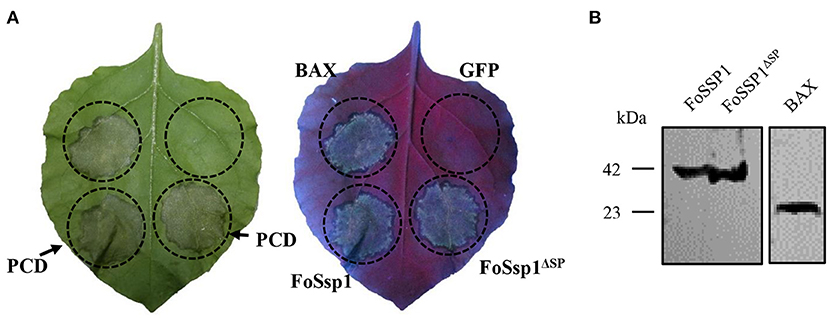
Figure 3. FoSsp1 was transiently expressed in the N. benthamiana by A. tumefaciens-mediated transformation. (A) FoSsp1 induced cell death in tobacco leaves. The recombinant vectors of pBin-FoSSP1 and pBin-FoSSP1ΔSP were infiltrated into leaves by A. tumefaciens-mediated transformation. Empty vector pBin-eGFP (EV) was used as negative control and Bax as positive control. After 5 days of infiltration, the hypersensitive response was indicated by ultraviolet (UV) light. The experiment was repeated at least four times. (B) Western blot analysis was used to detect the protein expression.
FoSSP1 Deletion Mutant Was Normal in Growth Rate but Increased in Conidiation
Split-marker approach was used to generate the deletion mutant of FoSSP1 in Foc4. The nucleotide sequence of a segment containing the FoSSP1 gene (NW_022158695.1: 156,986-157,890 nt) was replaced by a hygromycin-resistance gene in Foc4. The resulting transformants harboring the hygromycin B resistance were further confirmed by PCR. In total, five strains of Fossp1 deletion mutant were generated and FT1 was chosen for further experiments [(Supplementary Figure S1) PCR Test of Fossp1 deletion mutants]. To generate the complement strain of the Fossp1 mutant, the promoter and the encoding region of FoSSP1 (NW_022158695.1: 157,201-157,734 nt) were constructed into the pFl2 plasmid and transformed into the Fossp1 deletion mutant by PEG-mediated protoplast transformation. The resulting transformants harboring neomycin (G418) resistance were screened by PCR. In total, six complement strains were generated.
To examine the growth rate, wild-type strain of Foc4, Fossp1 deletion mutant, and complement strain of Fossp1-C were cultured on PDA agar plates for 6 days. The results showed that no significant difference in growth rate among WT (0.59 ± 0.01 cm/d), Fossp1-C (0.61 ± 0.01 cm/d) and Fossp1 deletion mutant (0.59 ± 0.01 cm/d) (Figures 4A,B).
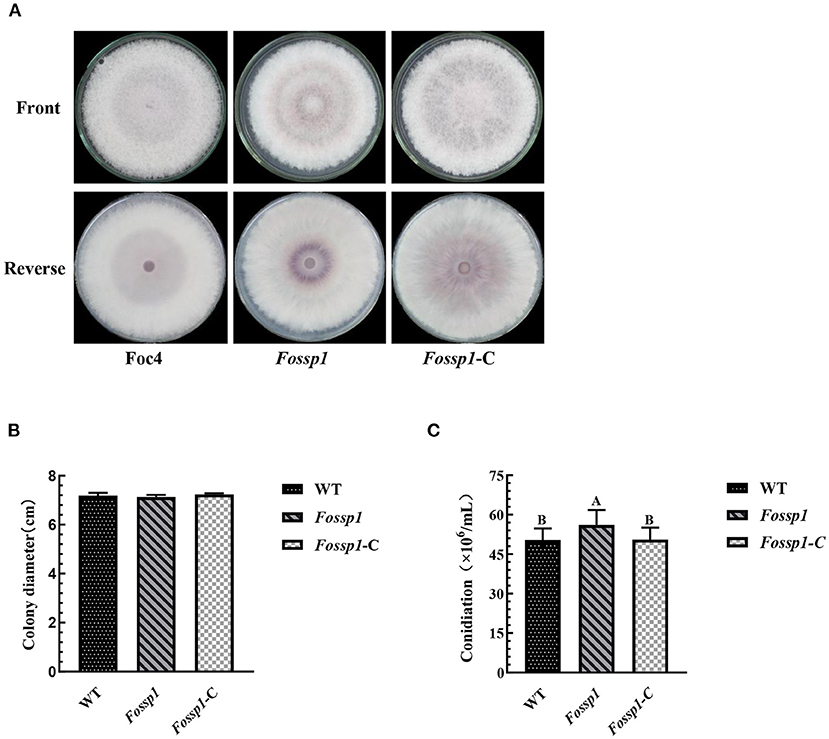
Figure 4. Effects of Fossp1 deletion on colony morphology, growth rate, and spore yield of Foc4. (A,B) Colony morphology and growth rate of WT, Fossp1, and Fossp1-C cultures on PDA medium for 6 days. (C) Conidiation of WT, Fossp1, and Fossp1-C cultures in PDB medium for 2 days. The results are representative of three independent experiments. The error bars represent the standard error. Identical and conserved catalytic residues are marked with letters.
The conidia production of Foc4, Fossp1-C, and Fossp1 deletion mutant were calculated in PDB medium 2 days after inoculation. Interestingly, the conidial yield of Fossp1 mutant (5.61 ± 0.35 × 105 conidia/ml) was significantly higher than that of WT strain (5.04 ± 0.24 × 105 conidia/ml) and complement strain Fossp1-C (5.05 ± 0.21 × 105 conidia/ml) (Figure 4C). These results suggest that the FoSSP1 gene regulates the sporulation of Foc4.
Deletion Mutant of Fossp1 Increased Virulence to Banana
To analyze the pathogenicity of FoSSP1, it was knocked out and complementary successfully (Supplementary Figure S1). The knockout mutant Fossp1, complement strain Fossp1-C, and the WT Foc4 on banana seedlings (Cavendish banana var. Brazil (Musa AAA) was tested. The conidia suspension (1 × 108 conidia/ml) of Fossp1 mutant, complement strain Fossp1-C, and WT Foc4 were inoculated to the banana plantlets. At 10 dpi (days post inoculation), no symptoms presented on leaves and external bulbs of the Fossp1, Fossp1-C, and Foc4 (Figure 5A upper and middle panel). However typical discoloration internal of bulbs appeared when observed in the vertical section of the bulbs of Fossp1 deletion mutant, but not of WT and complement strain (Figure 5A, lower panel). At 20 dpi, typical yellowing leaves of seedlings infected by Foc4 were observed. In comparing, two or more yellowing leaves of each seedling inoculated by the Fossp1 deletion mutant, while of the wild-type and the complement strains yellowing leaves up to one of each seedling (Figure 5B). To quantify the disease severity of the infected banana seedlings with different Foc4 strains, the disease index of infected banana seedlings at 20 dpi was calculated. The disease index of banana plants infected with Fossp1 deletion mutant was significantly increased than that of the WT strain Foc4 and complement Fossp1-C at 20 dpi (Figure 5D). When observed banana seedlings at 30 dpi, all plants infected by Foc4 displayed leaf yellowing, even the death of Fossp1 (Figure 5C, upper panel). In addition, the pseudostem and root displayed typical discoloration of infected seedlings (Figure 5C, lower panel). Discoloration length of pseudostem was measured at 30 dpi, in comparison to WT Foc4 (4.15 ± 0.17 cm) and the complement strain (7.55 ± 0.31 cm), discoloration length of pseudostem of Fossp1 (4.20 ± 0.30 cm) increased by about 50%. These results indicated that deletion of FoSSP1 enhances the virulence of Foc4 in banana seedlings.
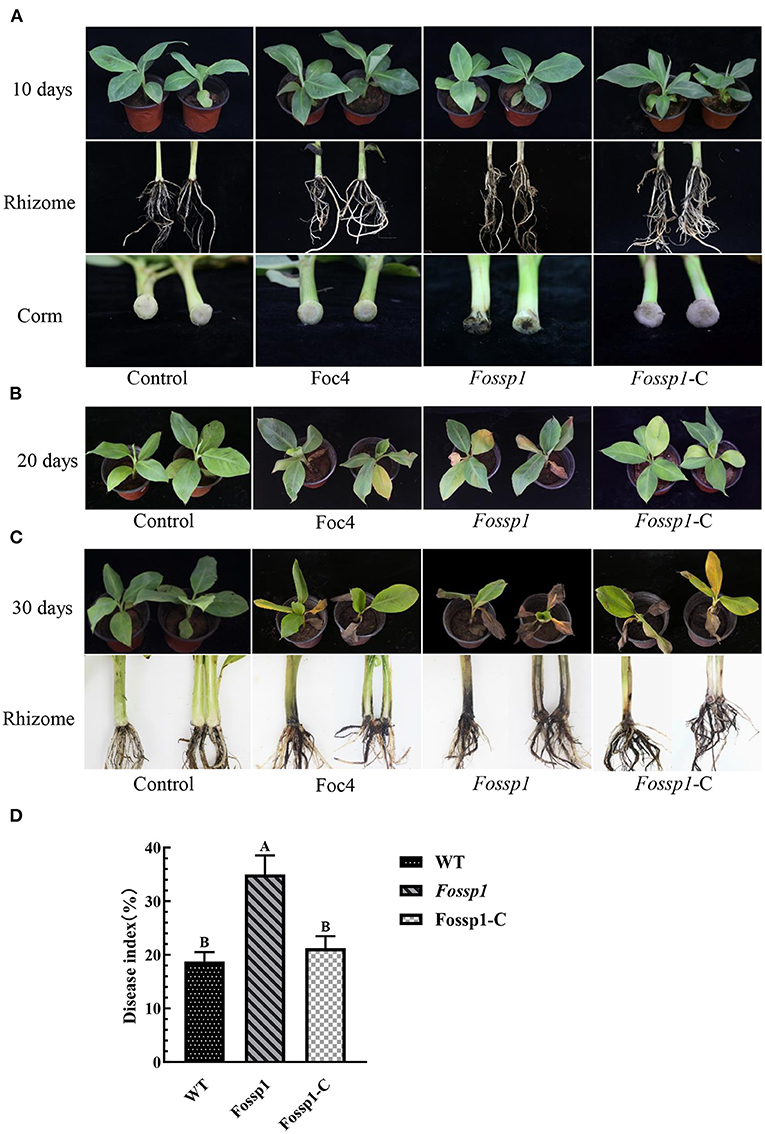
Figure 5. Deletion of FoSSP1 increased the virulence of Foc4 on roots and rhizomes of bananas. Compared with the WT, the corms of banana seedlings inoculated with Fossp1 showed obvious brown spots after 10 days (A), and leaf yellowing appeared after 20 days (B). The pseudostem and root displayed typical discoloration of infected seedlings after 30 days (C). The experiment was repeated 3 times with 10 replicates for each of the three strains. (D) The disease index of bananas was scored 20 days post inoculation.
FoSSP1 Regulates the Expression of Foc4 Conidiation Regulator Genes and Fossp1 Deletion Mutant Suppresses the Expression of Host Pathogenesis-Related Genes
Deletion of FoSSP1 in Foc4 leads to the enhancement of sporulation capability. We hypothesized that FoSSP1 might regulate Foc4 sporulation-related genes. Firstly, RNA of mycelia grown in a PDB medium for 2 days was extracted. Then, the transcription levels of the sporulation-related genes in WT and Fossp1 deletion mutant were tested by quantitative reverse transcription PCR (qRT-PCR). Compared with the WT, the key genes in the sporulation pathway, BrlA, Fgb, wet, and AbaA were significantly down-regulated (Figure 6A).
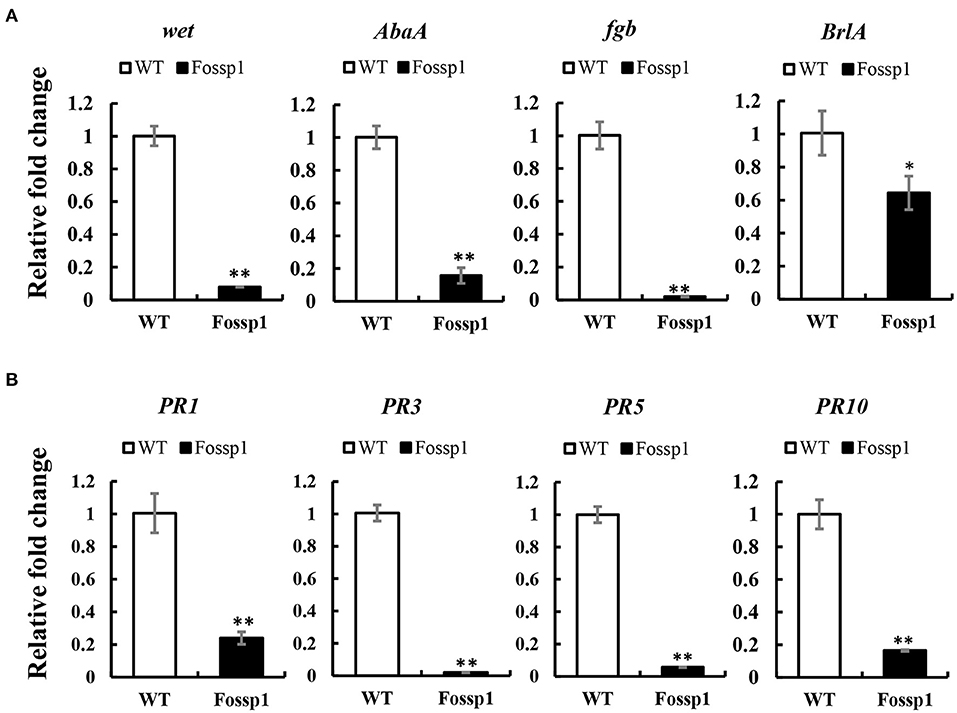
Figure 6. FoSSP1 participates in the sporulation process and plant defense process. (A) The total RNAs were extracted from the spore suspension of WT and Fossp1 cultured for 2 days, and the relative expression of four sporulation related genes was analyzed by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR), actin gene in Foc4 was used as reference gene. (B) The total RNAs were extracted from banana roots inoculated with WT and Fossp1 spore suspension for 3 days, and the expression of 4 defense-related genes in banana was analyzed by qRT-PCR. Actin gene in banana was used as an internal reference gene in qRT-PCR. The experiment was repeated at least three times. The error bars represent the standard error. Identical and conserved catalytic residues are marked with asterisks (**).
Plant pathogenesis-related genes play a crucial role in plant defense against the pathogen (Joshi et al., 2021). Deletion of Fossp1 to analyze differential expression of pathogenesis-related genes, the roots of banana seedlings were inoculated with a spore suspension of Foc4 and Fossp1, respectively, and total RNA was extracted 3 days post-inoculation to detect transcriptional levels of pathogenesis-related genes. RT-qPCR showed that the transcription level of PRs gene, PR1b, PR2b, PR5, and PR10 in root infected by Fossp1 was significantly lower than that did the WT (Figure 6B). The results of two qRT-PCR showed that FoSSP1 induced the expression of host defense genes during the pathogen infection of roots. Furthermore, in the process of conidiation, FoSSP1 participates in the central regulatory pathway controlling conidiation.
The Predicted Secretory Peptide of FoSsp1 Is Functional
Secreted proteins can be divided into typical secreted proteins and atypical secreted proteins according to the presence or absence of signal peptides. A total of 23 amino acids (aa 1-23) at the N-terminal of FoSsp1 were predicted to be a signal peptide by SignalP5.0. The secretory function of the FoSsp1 signal peptide was verified using the yeast secretion system (Gu et al., 2011). The YTK12 strain was transformed with the reconstructed pSUC2 vector to express the signal peptide of pSUC2-FoSsp1 and pSUC2- FoSsp1ΔSP. The signal peptide of a known secreted protein, Avr1b, was used as a positive control (Tian et al., 2021). Untransformed YTK12 and empty pSUC2 vectors were used as negative controls. Similar to the Avr1b signal peptide, the FoSsp1 but not the FoSsp1ΔSP or the empty vector, allowed YTK12 to grow on the YPRAA medium (Figure 7A). The strain transformed with the FoSsp1 with its signal peptide displayed secreted invertase enzyme activity that catalyzed the reduction of TTC to a red-colored compound (Figure 7A). These results suggest that FoSsp1 is a secreted protein, and its signal peptide is required for its secretion.
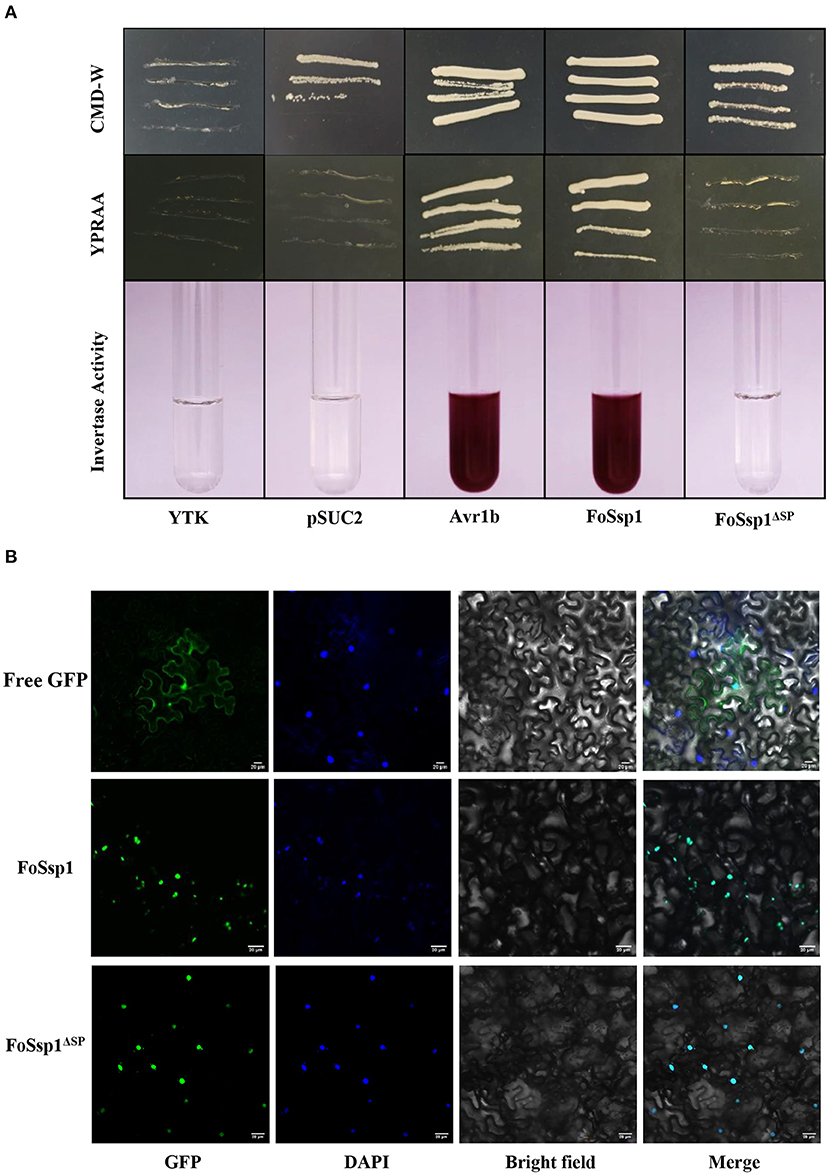
Figure 7. The functional validation of signaling peptide of FoSsp1 and the subcellular localization of FoSsp1 in tobacco epidermal cells. (A) Empty vector pSUC2 was used as negative control, SP sequence of Avr1b from Phytophthora soja was the positive control. The growth of yeast strains in the YPRAA and CMD-W medium were recorded. The change in color put down to the reduction of 2, 3, 5-triphenyltetrazolium chloride (TTC) to insoluble red-colored 1, 3, 5-triphenylformazan (TPF) was monitored. (B) DAPI used plant nuclear staining as nuclear control. The recombinant vector containing green fluorescent protein permeated tobacco leaves mediated by A. tumefaciens. Images were taken 2 days post infiltration. Bars, 20 μm. All experiments were repeated at least three times.
Subcellular Localization of FoSsp1 in Tobacco Leaf Cell
To explore the subcellular localization of FoSsp1 in plant cells, the recombinant vector pBin-eGFP-FoSSP1 and pBin-eGFP-FoSSP1ΔSP were transiently expressed in tobacco leaves, and pBin-eGFP was used as a negative control. The plant nucleus was labeled with DAPI (2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride) staining. When observed under the microscope, bright green fluorescence in the leaf cell nucleus of tobacco was overlapped with the blue fluorescence of the DAPI staining (Figure 7B). Interestingly, a weak fluorescence signal of pBin-eGFP-FoSSP1 was also distributed in extracellular and intracellular tobacco cells (Supplementary Figure S2). The result showed that FoSsp1 localized in the nucleus of a tobacco leaf cell.
FoSsp1 Activates Plant Immune Response
Due to the increased virulence phenotype of the Fossp1 deletion mutant, we speculate that FoSsp1 is an elicitor that can stimulate the plant immune response. Therefore, the accumulation of H2O2 and the callose deposition in tobacco were detected after A. tumefaciens infiltration. FoSsp1 accumulates more H2O2 and callose than GFP control (Figure 8A). The results showed that FoSsp1 can activate the defense response of host plants, promoting oxygen burst and callose deposition.
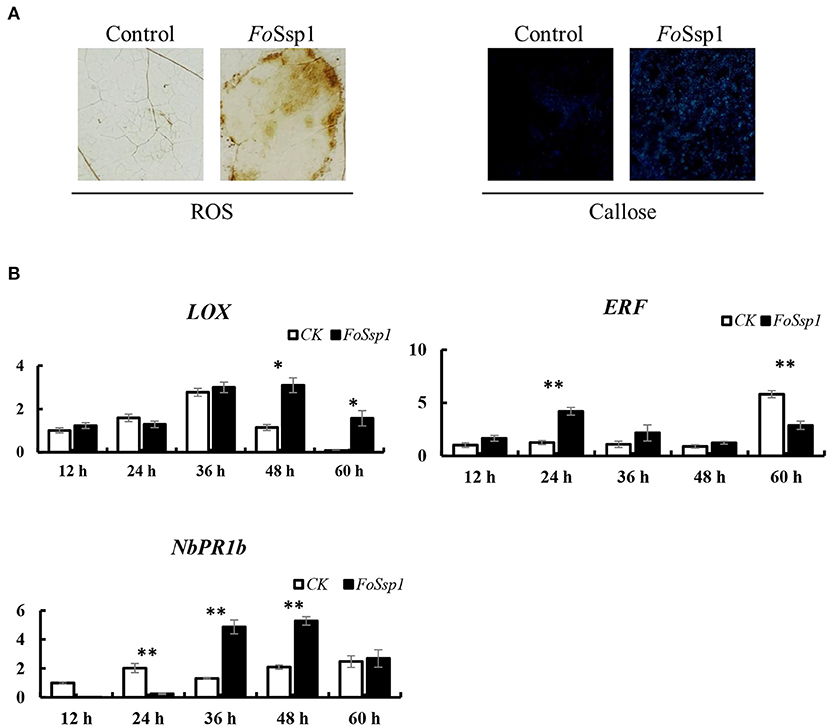
Figure 8. FoSsp1 can induce basic immune response in plants. (A) Tobacco leaves were infiltrated by A. tumefaciens-mediated transformation and stained with DAB and aniline blue 2 days post infiltration. Oxygen accumulation and callose depositions were visualized in tobacco leaves. The representative images were captured at 48 hpi. (B) The expression levels of 3 genes related to immune response were analyzed by qRT-PCR. The pBin-eGFP was the control group, and actin in tobacco was the internal reference gene. The expression of CK and FoSsp1 at the same time point were compared using one-way ANOVA. Asterisks represent significant differences in one-way ANOVA (p < 0.01). The results are representative of three independent experiments.
To further explore the defense response of N. benthamiana activated by FoSsp1, the expression of defense-related genes was detected using qRT-PCR. The genes pathogenesis-related protein 1B (PR1b), pathogenesis-related protein 2B (PR2b), ethylene-responsive transcription factor 1 (ERF1), and linoleate 9S-lipoxygenase 5 (LOX) related to salicylic acid, ethylene, and jasmonic acid signal pathways were detected (Wang et al., 2000; Lorenzo et al., 2003; Wu et al., 2018). There were statistically significant differences in the expression of these genes. From the results, it is clear that PR1b and LOX showed higher expression levels at 48 hpi, while ERF1 expression significantly increased at 24 hpi compared with control (Figure 8B).
Discussion
Fusarium wilt of bananas is a devastating soil-borne disease that has put the worldwide banana industry under threat (Dita et al., 2018). F. oxysporum f. sp. cubense race4 (Foc4) infects the root of banana, colonizes the vascular and blocks water and nutrient transportation, resulting in banana wilt (Chisholm et al., 2006). So far, however, genetic regulation of fungal virulence mechanisms remains elusive, with only a few virulence factors being reported. For example, loss of FGA1 and FGA3 resulted in decreased intracellular cyclic AMP levels and increased heat resistance, suggesting that FGA1 and FGA3 may regulate growth and development, pathogenicity, and heat resistance through the cAMP-dependent protein kinase A pathway (Guo et al., 2016). In this study, we identified a small secreted protein FoSsp1 that elicits plant defense reactions and negatively regulates plant infection. The results provide insights into fungal virulence mechanisms, and more importantly, offer implications for strategies aiming at sustainable control of fusarium wilt of bananas.
Small secretory proteins (SSPs) are important factors involved in plant host interaction (Xie et al., 2021). These proteins may facilitate infection by suppressing plant defenses, manipulating plant metabolism or signaling, or directly killing plant cells (Ramachandran et al., 2017). The identification and functional characterization of SIX8 support that SSPs could contribute toward the infection of vascular fungal pathogens as well (An et al., 2019). In this study, by combining bioinformatics analysis and functional analysis, we identified FoSsp1 to be a novel virulence-related SSP. Upon being transiently expressed in tobacco, FoSsp1 could induce cell death similar to BAX. Furthermore, FoSsp1 expression triggered obvious oxygen burst, callose accumulation, and the expressional up-regulation of plant immunity genes PR1b, PR2b, NbLOX, and NbERF1. Corresponding with such defense-eliciting activities on tobacco, deletion of the FoSSP1 gene increased fungal virulence toward bananas and reduced the expressional up-regulation of plant defense genes PR1b, PR2b, PR5, and PR10. Together, these results support that FoSsp1 is a novel defense-eliciting SSP that might enhance banana's resistance against fusarium wilt.
Elicitors can activate immune responses in plants and they can be engineered as plant immune inducers to preactivate the plant immune system to resist the invasion of pathogens, so as to promote plant disease resistance (Qiu et al., 2017; An et al., 2019). For example, BcGs1 enhanced bcGS1-induced resistance to Botrytis cinerea by activating basic plant defense responses and inducing lignin accumulation (Yang et al., 2018a). MoHrip2 stimulates the early defense response of rice seedlings through pressure-related pathways, induces up-regulated expression of the PR gene, activates SA and JA signaling pathways, and ultimately improves disease resistance of rice (Nie et al., 2019a). Identified elicitors are diverse in biochemical properties, and may be classified into oligosaccharides, lipids, aromatic compounds, or proteins (Shimizu et al., 2010; Ma et al., 2015; Clinckemaillie et al., 2017). A variety of elicitors are conserved molecular patterns that are indicative of pathogen presence and are perceived by plant cells via membrane-localized kinase receptors. For instance, PcCBP3 was labeled by mCherry and expressed in tobacco cells, and red fluorescence mainly was observed at the cell edge (Yin et al., 2021). Thus, elicitor perception represents an important plant immunity layer against pathogen infection. Functionally, FoSsp1 elicits plant defense reactions. However, our experimental evidence indicates that FoSsp1 might not function as a typical pathogen-associated molecular pattern (PAMP). First, deleting the predicted secretory peptide of FoSsp1 does not interfere with its death and defense triggering activity toward tobacco cells, indicating that the elicitor activity of FoSsp1 relies on perturbing intracellular plant cell activity. During infection, FoSsp1 might translocate inside a plant cell to exert elicitor functions. In accordance with this, our GFP-based subcellular localization experiment demonstrates FoSsp1 localizes within the nucleus upon transiently expressed on tobacco. Besides its subcellular localization, FoSsp1 also differs from a typical PAMP in that its deletion does not negatively affect pathogen growth, development, and virulence, indicating that FoSSP1 might be dispensable for the fungal pathogen. As said, it is worth noting that FoSSP1 homologs are widespread in ascomycetes, indicating a conserved biological function. Indeed, maintaining a defense-eliciting elicitor would be evolutionarily disadvantageous for a plant pathogen unless the elicitor provides additional benefits for the pathogen. In this study, deletion of FoSSP1 caused elevated conidia production, indicating that FoSSP1 may regulate proper conidial differentiation. In the future, characterizing the functions of FoSSP1 homologs in other fungi would be important to unravel the potentially multifaceted biological functions of the SSP family.
The identification of an elicitor protein from the banana-Fusarium pathosystem offers important insights for the sustainable control of fusarium wilt disease. Due to the cloning propagation nature of bananas, the natural banana population is the shortage of genetic diversity and the breeding utilization of potential natural resistance is technically challenging (Nyine et al., 2017), which has added considerable difficulties to the sustainable control of banana fusarium wilt disease. In the future, it might be feasible to take advantage of the elicitor activity of FoSsp1 for developing transgene plants or developing bioproducts with activated elicitor.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DC and LZ designed the experiments. YW, XZ, and TW performed the experiments. YW, SZ, XL, CX, and ZK analyzed the data. XL, DC, and LZ joined the discussion and gave the original ideas. YW, DC, and LZ wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by Hainan Provincial Natural Science Foundation of China (no. 320QN188), the Finance Science and Technology Project of Hainan Province (no. YSPTZX202018), Hainan Major Research Fund of Science and Technology (no. ZDKJ201817), Funding for the Construction of World First Class Discipline of Hainan University (no. RZZX201911), and the Scientific Research Foundation for Advanced Talents, Hainan University (no. KYQD(ZR)1873).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.873451/full#supplementary-material
References
An, B., Hou, X., Guo, Y., Zhao, S., Luo, H., He, C., et al. (2019). The effector SIX8 is required for virulence of Fusarium oxysporum f.sp. cubense tropical race 4 to Cavendish banana. Fungal Biol. 123, 423–430. doi: 10.1016/j.funbio.2019.03.001
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant. 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bu, B., Qiu, D., Zeng, H., Guo, L., Yuan, J., and Yang, X. (2014). A fungal protein elicitor PevD1 induces Verticillium wilt resistance in cotton. Plant Cell Rep. 33, 461–470. doi: 10.1007/s00299-013-1546-7
Chen, D., Wang, Y., Zhou, X., Wang, Y., and Xu, J. (2014). The Sch9 kinase regulates conidium size, stress responses, and pathogenesis in Fusarium graminearum. Plos ONE. 9, e105811. doi: 10.1371/journal.pone.0105811
Chisholm, S. T., Coaker, G., Day, B., and Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Clinckemaillie, A., Decroës, A., van Aubel, G., Carrola dos Santos, S., Renard, M. E., Van Cutsem, P., et al. (2017). The novel elicitor COS-OGA enhances potato resistance to late blight. Plant Pathol. 66, 818–825. doi: 10.1111/ppa.12641
Dagvadorj, B., Ozketen, A. C., Andac, A., Duggan, C., Bozkurt, T. O., and Akkaya, M. S. (2017). A Puccinia striiformis f. sp. tritici secreted protein activates plant immunity at the cell surface. Sci Rep. 7, 1141. doi: 10.1038/s41598-017-01100-z
Degradi, L., Tava, V., Kunova, A., Cortesi, P., Saracchi, M., and Pasquali, M. (2021). Telomere to Telomere Genome Assembly of Fusarium musae F31, Causal Agent of Crown Rot Disease of Banana. Mol. Plant Microbe. Interact. 34, 1455–1457. doi: 10.1094/MPMI-05-21-0127-A
Dita, M., Barquero, M., Heck, D., Mizubuti, E. S. G., and Staver, C. P. (2018). Fusarium wilt of banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 9, 1468. doi: 10.3389/fpls.2018.01468
Dong, J., and Chen, W. (2013). The role of autophagy in chloroplast degradation and chlorophagy in immune defenses during Pst DC3000 (AvrRps4) infection. Plos ONE. 8, e73091. doi: 10.1371/journal.pone.0073091
Dou, D., and Zhou, J. M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe. 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Goswami, R. S.. (2012). Targeted gene replacement in fungi using a split-marker approach. Methods Mol. Biol. 835, 255–269. doi: 10.1007/978-1-61779-501-5_16
Gu, B., Kale, S. D., Wang, Q., Wang, D., Pan, Q., Cao, H., et al. (2011). Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains a RXLR-like motif sufficient for translocation into plant cells. Plos ONE. 6, e27217. doi: 10.1371/journal.pone.0027217
Guo, L., Yang, Y., Yang, L., Wang, F., Wang, G., and Huang, J. (2016). Functional analysis of the G-protein α subunits FGA1 and FGA3 in the banana pathogen Fusarium oxysporum f. sp. cubense. Physiol. Mol. Plant P. 94, 75–82. doi: 10.1016/j.pmpp.2016.04.003
Hwang, S. C., and Ko, W. H. (2004). Cavendish banana cultivars resistant to fusarium wilt acquired through somaclonal variation in Taiwan. Plant Dis. 88, 580–588. doi: 10.1094/PDIS.2004.88.6.580
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature. 444, 323–329. doi: 10.1038/nature05286
Joshi, V., Joshi, N., Vyas, A., and Jadhav, S. K. (2021). Pathogenesis-related proteins: Role in plant defense. Biocontrol Agents and Secondary Metabolites. 573–590. doi: 10.1016/B978-0-12-822919-4.00025-9
Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. P. Natl. Acad. Sci.U.S.A. 96, 7956–7961. doi: 10.1073/pnas.96.14.7956
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., and Solano, R. (2003). Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 15, 165–178. doi: 10.1105/tpc.007468
Ma, Z., Song, T., Zhu, L., Ye, W., Wang, Y., Shao, Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
Maryani, N., Lombard, L., Poerba, Y. S., Subandiyah, S., Crous, P. W., and Kema, G. H. J. (2019). Phylogeny and genetic diversity of the banana Fusarium wilt pathogen Fusarium oxysporum f. sp. cubense in the indonesian centre of origin. Stud. Mycol. 92, 155–194. doi: 10.1016/j.simyco.2018.06.003
Michielse, C. B., and Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10, 311–324. doi: 10.1111/j.1364-3703.2009.00538.x
Nie, H., Zhang, L., Zhuang, H., Yang, X., Qiu, D., and Zeng, H. (2019a). Secreted protein MoHrip2 is required for full virulence of Magnaporthe oryzae and modulation of rice immunity. Appl. Microbiol. Biot. 103, 6153–6167. doi: 10.1007/s00253-019-09937-2
Nie, J., Yin, Z., Li, Z., Wu, Y., and Huang, L. (2019b). A small cysteine-rich protein from two kingdoms of microbes is recognized as a novel pathogen-associated molecular pattern. New Phytol. 222, 995–1011. doi: 10.1111/nph.15631
Nielsen, H.. (2017). Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73. doi: 10.1007/978-1-4939-7015-5_6
Niu, X., Yang, G., Lin, H., Liu, Y., Li, P., and Zheng, A. (2021). A novel small cysteine-rich effector, RsSCR10 in Rhizoctonia solani is sufficient to trigger plant cell death. Front. Microbiol. 12, 684923. doi: 10.3389/fmicb.2021.684923
Nyine, M., Uwimana, B., Swennen, R., Batte, M., Brown, A., Christelov,á, P., et al. (2017). Trait variation and genetic diversity in a banana genomic selection training population. Plos ONE. 12, e0178734. doi: 10.1371/journal.pone.0178734
Oh, S. K., Young, C., Lee, M., Oliva, R., Bozkurt, T. O., Cano, L. M., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 21, 2928–2947. doi: 10.1105/tpc.109.068247
Owji, H., Nezafat, N., Negahdaripour, M., Hajiebrahimi, A., and Ghasemi, Y. (2018). A comprehensive review of signal peptides: structure, roles, and applications. Eur. J. Cell Biol. 97, 422–441. doi: 10.1016/j.ejcb.2018.06.003
Ploetz, R. C., Kema, G. H., and Ma, L. J. (2015). Impact of diseases on export and smallholder production of banana. Annu. Rev. Phytopathol. 53, 269–288. doi: 10.1146/annurev-phyto-080614-120305
Qiu, D., Dong, Y., Zhang, Y., Li, S., and Shi, F. (2017). Plant immunity inducer development and application. Mol. Plant Microbe. Interact. 30, 355–360. doi: 10.1094/MPMI-11-16-0231-CR
Ramachandran, S. R., Yin, C., Kud, J., Tanaka, K., Mahoney, A. K., Xiao, F., et al. (2017). Effectors from wheat rust fungi suppress multiple plant defense responses. Phytopathology 107, 75–83. doi: 10.1094/PHYTO-02-16-0083-R
Shang, S., Wang, B., Zhang, S., Liu, G., Liang, X., Zhang, R., et al. (2020). A novel effector CfEC92 of Colletotrichum fructicola contributes to glomerella leaf spot virulence by suppressing plant defences at the early infection phase. Mol. Plant Pathol. 21, 936–950. doi: 10.1111/mpp.12940
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two lysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Tarnowski, B. I., Spinale, F. G., and Nicholson, J. H. (1991). DAPI as a useful stain for nuclear quantitation. Biotech. Histochem. 66, 297–302. doi: 10.3109/10520299109109990
Tian, L., Huang, C., Zhang, D., Li, R., Chen, J., Sun, W., et al. (2021). Extracellular superoxide dismutase VdSOD5 is required for virulence in Verticillium dahliae. J. Integ. Agr. 20, 1858–1870. doi: 10.1016/S2095-3119(20)63353-6
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Wang, C., Zien, C. A., Afitlhile, M., Welti, R., Hildebrand, D. F., and Wang, X. (2000). Involvement of phospholipase D in wound-induced accumulation of jasmonic acid in Arabidopsis. Plant Cell 12, 2237–2246. doi: 10.1105/tpc.12.11.2237
Wei, Y., Liu, W., Hu, W., Yan, Y., and Shi, H. (2020). The chaperone MeHSP90 recruits MeWRKY20 and MeCatalase1 to regulate drought stress resistance in cassava. New Phytol. 226, 476–491. doi: 10.1111/nph.16346
Wu, C. H., Derevnina, L., and Kamoun, S. (2018). Receptor networks underpin plant immunity. Science. 360, 1300–1301. doi: 10.1126/science.aat2623
Wu, J., Zhang, Y., Zhang, H., Huang, H., Folta, K. M., and Lu, J. (2010). Whole genome wide expression profiles of Vitis amurensisgrape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 10, 234. doi: 10.1186/1471-2229-10-234
Xie, C., Shang, Q., and Mo, C. (2021). Early secretory pathway-associated proteins SsEmp24 and SsErv25 are involved in morphogenesis and pathogenicity in a filamentous phytopathogenic fungus. mBio. 12. doi: 10.1128/mBio.03173-21
Yang, C., Liang, Y., Qiu, D., Zeng, H., Yuan, J., and Yang, X. (2018a). Lignin metabolism involves Botrytis cinerea BcGs1- induced defense response in tomato. BMC Plant Biol. 18, 103. doi: 10.1186/s12870-018-1319-0
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018b). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Yang, Q., Yan, L., Gu, Q., and Ma, Z. (2012). The mitogen-activated protein kinase kinase kinase BcOs4 is required for vegetative differentiation and pathogenicity in Botrytis cinerea. Appl. Microbiol. Biot. 96, 481–492. doi: 10.1007/s00253-012-4029-9
Yin, Z., Wang, N., Duan, W., Pi, L., Shen, D., and Dou, D. (2021). Phytophthora capsici CBM1-containing protein CBP3 is an apoplastic effector with plant immunity-inducing activity. Mol. Plant Pathol. 22, 1358–1369. doi: 10.1111/mpp.13116
Yun, Y., Liu, Z., Zhang, J., Shim, W. B., Chen, Y., and Ma, Z. (2014). The MAPKK FgMkk1 of Fusarium graminearum regulates vegetative differentiation, multiple stress response, and virulence via the cell wall integrity and high-osmolarity glycerol signaling pathways. Environ. Microbiol. 16, 2023–2037. doi: 10.1111/1462-2920.12334
Zhang, L., Cenci, A., Rouard, M., Zhang, D., and Zheng, S. (2019). Transcriptomic analysis of resistant and susceptible banana corms in response to infection by Fusarium oxysporum f. sp. cubense tropical race 4. Sci. Rep. 9. doi: 10.1038/s41598-019-44637-x
Keywords: Fusarium oxysporum, secreted protein, conidiation, pathogenesis, elicitor
Citation: Wang Y, Zhang X, Wang T, Zhou S, Liang X, Xie C, Kang Z, Chen D and Zheng L (2022) The Small Secreted Protein FoSsp1 Elicits Plant Defenses and Negatively Regulates Pathogenesis in Fusarium oxysporum f. sp. cubense (Foc4). Front. Plant Sci. 13:873451. doi: 10.3389/fpls.2022.873451
Received: 10 February 2022; Accepted: 30 March 2022;
Published: 10 May 2022.
Edited by:
Guixia Hao, USDA/ARS/NCAUR, United StatesReviewed by:
Xiufang Xin, Institute of Plant Physiology and Ecology (CAS), ChinaLina Castano-Duque, Southern Regional Research Center, United States
Copyright © 2022 Wang, Zhang, Wang, Zhou, Liang, Xie, Kang, Chen and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daipeng Chen, Y2hlbi5kYWlwZW5nQDE2My5jb20=; Li Zheng, emhlbmdsaWhhcHB5MDYxN0AxMjYuY29t
 Yuhua Wang1
Yuhua Wang1 Daipeng Chen
Daipeng Chen Li Zheng
Li Zheng