
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 May 2022
Sec. Plant Pathogen Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.870882
This article is part of the Research Topic Identification and Functional Dissection of Stress-responsive Genes in Cotton View all 26 articles
Teosinte branched1/Cincinnata/proliferating cell factor (TCP) transcription factors play important roles in plant growth and defense. However, the molecular mechanisms of TCPs participating in plant defense remain unclear. Here, we characterized a cotton TCP4-like fine-tuned by miR319b, which could interact with NON-EXPRESSER OF PATHOGEN-RELATED GENES 1 (NPR1) to directly activate isochorismate synthase 1 (ICS1) expression, facilitating plant resistance against Verticillium dahliae. mRNA degradome data and GUS-fused assay showed that GhTCP4-like mRNA was directedly cleaved by ghr-miR319b. Knockdown of ghr-miR319b increased plant resistance to V. dahliae, whereas silencing GhTCP4-like increased plant susceptibility by the virus-induced gene silencing (VIGS) method, suggesting that GhTCP4-like is a positive regulator of plant defense. According to the electrophoretic mobility shift assay and GUS reporter analysis, GhTCP4-like could transcriptionally activate GhICS1 expression, resulting in increased salicylic acid (SA) accumulation. Yeast two-hybrid and luciferase complementation image analyses demonstrated that GhTCP4-like interacts with GhNPR1, which can promote GhTCP4-like transcriptional activation in GhICS1 expression according to the GUS reporter assay. Together, these results revealed that GhTCP4-like interacts with GhNPR1 to promote GhICS1 expression through fine-tuning of ghr-miR319b, leading to SA accumulation, which is percepted by NPR1 to increase plant defense against V. dahliae. Therefore, GhTCP4-like participates in a positive feedback regulation loop of SA biosynthesis via NPR1, increasing plant defenses against fungal infection.
Teosinte branched1/Cincinnata/proliferating cell factor (TCP) proteins are plant-specific transcription factor families, whose names are derived from the first characterized members of the plant developmental pathway, TEOSINTE BRANCHED1 (TB1) in Zea mays, CYCLOIDEA (CYC) in Antirrhinum majus, and PROLIFERATING CELL FACTOR (PCF) in Oryza sativa (Cubas et al., 1999; Kosugi and Ohashi, 2002; Nag et al., 2009). TCP transcription factors (TFs) contain a TCP domain with a nonstandard basic helix-loop-helix (bHLH) motif of 59 amino acids, which is responsible for DNA binding and protein-protein interactions (Heim et al., 2003; Nicolas and Cubas, 2016). TCP proteins are classified into two groups: class I and class II. Class I includes the PCF subclade, and class II is further divided into the CIN and CYC/TB1 subclades (Martin-Trillo and Cubas, 2010). Class I and class II proteins preferentially bind to the consensus elements KHGGGVC and GTGGNCCC, respectively (Aggarwal et al., 2010; Daviere et al., 2014). In Arabidopsis, 5 out of 11 class II TCP genes belonging to CIN types, TCP2, TCP3, TCP4, TCP10, and TCP24, can be targeted by miR319 (Nath et al., 2003; Palatnik et al., 2003). TCP family members have been reported to play vital roles in many plant growth and development processes, including hormone signal transduction, gametophyte development, seed germination, leaf shape, internode length, and flower development (Aguilar-Martinez et al., 2007; Giraud et al., 2010; Liu et al., 2018).
Recent studies have shown that TCP proteins participate in plant immunity (Kim et al., 2014; Bao et al., 2019). For example, effectors from multiple pathogens can target TCP TFs to affect plant immune responses (Mukhtar et al., 2011; Wessling et al., 2014). TCP TFs interact with SUPPRESSOR OF rps4-RLD1 (SRFR1, a negative immune regulator), contributing to effector-triggered immunity (Kim et al., 2014). Arabidopsis TCP8, TCP9, and other TCP proteins co-ordinately regulate ISOCHORISMATE SYNTHASE 1 (ICS1) expression to determine salicylic acid (SA) biosynthesis (Wang et al., 2015). The type III effector HopBB1 targets Arabidopsis TCP14 to promote its degradation, leading to increased disease susceptibility (Yang et al., 2017). Arabidopsis TCP15 interacts with MODIFIER OF snc1-1 (MOS1) to participate in plant immune response during cell cycle progression (Zhang et al., 2018). However, the key TCP TFs, especially Class II proteins, that function in plant immunity remain to be identified.
SA plays a key role in plant immunity and induces systemic acquired resistance (SAR) (Fu and Dong, 2013). In plants, two SA biosynthesis pathways use chorismate as the initial substrate via a series of enzymatic reactions with phenylalanine ammonia-lyase (León et al., 1995; Mauch-Mani and Slusarenko, 1996; Coquoz et al., 1998; Ribnicky et al., 1998) and ICS (Verberne et al., 2000; Wildermuth et al., 2001). The ICS-dependent SA biosynthesis pathway is the major pathway (Catinot et al., 2008; Lee et al., 2010; An and Mou, 2011). ICS1 can be activated in plants infected with pathogens to promote SA accumulation, which acts a major defense regulator non-expressor of PR gene 1 (NPR1), leading to the growth-to-defense transition (Zhang and Li, 2019). Accumulating evidence suggests that ICS1 expression may be an early regulatory node of the network in plant immunity. For instance, SAR-deficient 1 (SARD1), calmodulin-binding protein 60 g (CBP60g), and WRKY28/46 are positive regulators of Arabidopsis ICS1 expression, whereas ethylene insensitive 3 (EIN3), EIN3-like1 (EIL1), MUR3, and NAC (for NAM, ATAF1, 2, and CUC2) 19/NAC55/NAC72 negatively regulate ICS1 expression (Verberne et al., 2000; Tedman-Jones et al., 2008; Chen et al., 2009; Zhang et al., 2010b; Van Verk et al., 2011; Zheng et al., 2012). NPR1 is a key regulator of SA signaling and is required for the regulation of ICS1 expression (Zhang et al., 2010a). Recently, Arabidopsis TCP8 was confirmed to be a direct positive regulator of ICS1 expression during an immune response (Wang et al., 2015). Therefore, a sophisticated network regulates ICS1 expression. Nevertheless, novel regulators of ICS1 expression remain to be explored in plant immunity.
In Arabidopsis, NPR1 is required for SA to induce PR1, PR2, and PR5 expression (Cao et al., 1994, 1997). NPR1 lacks a DNA-binding domain to interact with DNA, but it contains two protein-protein interaction domains that act as cofactors that interact with TFs to participate in SA-related downstream gene expression (Rochon et al., 2006; Boyle et al., 2009). For example, the interaction between NPR1 and TGAs regulates PR1 expression (Fan and Dong, 2002). Recently, NPR1 was shown to interact with three class I TCPs. TCP8, TCP14, and TCP15 enhance downstream gene expression, redundantly contributing to SAR establishment (Li et al., 2018). However, NPR1 interaction with other TCPs, especially class II TCPs, remains elusive.
Cotton is an important cash crop that provides natural fiber, plant oil, and protein feedstuff (Zhang et al., 2015). However, the Verticillium wilt caused by Verticillium dahliae resulted in huge losses to cotton production every year in China (Klimes et al., 2015). Identification and application of defense genes are important in cotton breeding. Here, we characterized cotton TCP4-like, fine-tuned by ghr-miR319b, that interacts with NPR1 to co-ordinately activate GhICS1 expression to promote SA accumulation, a feedback loop of SA biosynthesis that is percepted by NPR1 to enhance plant resistance to V. dahliae infection. When plants are infected by this fungus, GhTCP4-like can act as a key protein responsible for SA biosynthesis, possibly regulating the transition from plant growth to defense.
The V. dahliae-resistant G. hirsutum cv. Zhongzhimian 2, susceptible variety Jimian 11, and general resistant variety CCRI 35 were used to analyse the expression of GhTCP4-like and ghr-miR319b after V. dahliae infection. CCRI 35 was used in VIGS, STTM, and other experiments. Cotton plants were cultivated in a greenhouse with a photoperiod of 16-h-light/8-h-dark at 28/25°C (light/dark) and ~70% humidity.
Nicotiana benthamiana plants were grown in pots with 80% nutrient soil and 20% vermiculit with a photoperiod of 16-h-light/8-h-dark at 25°C with 70% humidity.
The coding sequence of GhTCP4-like and precursor sequence of ghr-miR319b was amplified from cotton cDNA and genomic DNA, respectively. The MEGA7 program was used for evolutionary tree construction using the neighbor-joining method.
To determine the expression levels of GhTCP4-like and ghr-miR319b in different cotton tissues, the roots, stems, and leaves of cotton were harvested at the three-leaf stage and stored at −80°C after quick freezing with liquid nitrogen. The cotton plants were inoculated with V. dahliae strain V991 by the root-dipped method at the three-leaf stage (Gao et al., 2010), and the control group was treated with sterile water. Roots from six individual seedlings of each treatment were collected at 0, 1, 4, 7, 10, and 13 dpi, and gene expression levels were measured after all samples were collected.
Total RNA was extracted using the RNeasy Plant Mini Kit (TSINGKE, Beijing, China), and miRNA was extracted using miRcute (TIANGEN, Beijing, China). RNA concentration was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Two micrograms of total RNA were used for reverse transcription using First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). The products were diluted 10-times, 2 μl of which was used for qPCR experiments with Green qPCR SuperMix UDG (TransGen Biotech, Beijing, China) using the Light Cycler 480 system (Bio-Rad, Foster City, USA). As previously described, stem-loop qPCR was performed for miRNA quantification (Varkonyi-Gasic et al., 2007). All experiments were repeated three times independently, with biological and technical repeats. GhUB7 was used as an internal control for coding genes, and GhU6 was used as an internal control for miRNA quantification. Relative gene expression levels of three biologically independent samples were evaluated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). Primers used in this study are listed in Supplementary Table 1.
To confirm that ghr-miR319b directly cleaved the GhTCP4-like transcript at 980-nt, we performed a 5′-RACE assay. In brief, leaves of two-week-old cotton plants inoculated with V. dahliae for 4 d were harvested. Total RNA was extracted, and the 5′-RACE assay was performed with a HiScript-TS 5′/3′ RACE Kit (Vazyme, Nanjing, China) according to the manufacturer's instructions.
For the effector construct, the coding sequences of GhTCP4-like and the target mutant forms of GhTCP4-like (GhTCP4-likem) were cloned into plasmid pBI121 and fused with GUS under the control of the CaMV 35S promoter. GhMIR319b was amplified from cotton genomic DNA and inserted into the vector pCAMBIA1300 to generate the effector construct. All plasmids were electro-transformed into the Agrobacterium tumefaciens strain GV3101. Equal amounts of A. tumefaciens cultures carrying different effector or reporter vectors were mixed (OD600 = 0.8) and injected into the N. benthamiana leaf cells. GUS staining was performed as previously described using X-GLUC substrate (Sigma) (Jefferson et al., 1987). GUS enzyme activity in the different treatments was determined using the 4-methylumbelliferyl-β-D-glucuronide (4-MUG) substrate.
Cotton leaf crumple virus (CLCrV) and tobacco rattle virus (TRV)-induced silencing systems have been employed to knock down GhTCP4-like (Liu et al., 2002; Gu et al., 2014). A 300 bp fragment specific to GhTCP4-like was cloned into pCLCrVA and pTRV2 to generate pCLCrV-GhTCP4-like and pTRV-GhTCP4-like constructs, respectively. For the STTM assay, two imperfect target sequences for ghr-miR319b were separated using a 48-nt artificial designed linker and cloned into plasmids CLCrVA and pTRV2e, respectively. All plasmids were transformed into A. tumefaciens strain GV3101, One-week-old cotton seedlings were used for infection experiments, according to a previous report (Gao et al., 2011). At least 30 seedlings were injected with each construct. Silence efficiency was detected ~2 weeks after the injection. Successfully silenced plants were used in subsequent disease resistance analyses. Primers used in this study are listed in Supplementary Table 1.
The V. dahliae strain V991, a highly aggressive defoliating pathogen, was used, and activation and infection assays were performed as previously described (Hu et al., 2018). In brief, V991 was cultured on PDA for 4–5 d at 25°C; the mycelium was picked into Czapek's medium for 5–7 d, after gauze filtration, the spore concentration was adjusted to 106 spores per ml with ddH2O. Cotton plants were inoculated using the root-dipped method as previously described (Gao et al., 2010). Disease rate and index were calculated according to a previous report (Cheng et al., 2016). Fungal recovery culture experiments were performed as previously described (Tang et al., 2019). The relative fungal biomass was determined by qPCR assay as previously reported (Xiong et al., 2021).
The roots of TRV:00 and GhTCP4-like plants 1 d after V. dahliae inoculation were used to measure total SA content by high-performance liquid chromatography-tandem mass spectrometry (HPLC/MS/MS) system (AB SCIEX QTRAP 4500, Foster, CA, USA) according to a previously described protocol (Xiong et al., 2021). In brief, approximately 200 mg of root samples from corresponding plants were ground to powder with liquid nitrogen and extracted twice with pre-cold 90% methanol overnight at 4°C, centrifuged at 13,000 rpm for 15 min, and the supernatant was dried with nitrogen using a speedvac and then dissolved in 300 μL of 100% methanol. After filtering through a 0.22 μm size filter membrane, total SA was detected by HPLC/MS/MS, and pure SA (Sigma, St. Louis, MO, USA) was used as an external reference to establish the standard curve.
For Y2H assays, the coding sequences of GhTCP4-like and GhNPR1 were cloned into plasmids pGADT7 and pGBKT7, respectively, and co-transformed into the yeast strain AH109 using the polyethene glycol (PEG)/LiAc-based method with a yeast transformation kit (TaKaRa, Dalian, China). The monoclonal on SD-/-Trp/-Leu medium was validated by PCR. Positive clones were transferred to SD/-Leu/-Trp/-His/-Ade (with 20 mg/ml X-α-gal) medium to verify the interaction between GhTCP4-like and GhNPR1. The pGBKT7-p53 and pGADT7-largeT pairs were used as positive controls, and pGBKT7-laminC and pGADT7-largeT pairs were used as negative controls (Zhou et al., 2018).
The LCI assay was performed according to a previously descripted article (Chen et al., 2008), using an Agrobacterium-mediated transient expression system. In brief, the coding sequences of GhTCP4-like and GhNPR1 were cloned into the vectors pCAMBIA1300-CLuc and pCAMBIA1300-NLuc, respectively. The recombinant vectors were then transformed into A. tumefaciens strain GV3101. A. tumefaciens solution carrying the recombinant plasmid was mixed at a ratio of 1:1. After resting for 3 h at room temperature, the mixture was injected into N. benthamiana lower epidermal cells using a sterile, needle-free syringe. After 12 h in the dark, N. benthamiana was transferred to a light incubator for 36 h. Luciferase was detected by D-luciferin sodium salt (Solarbio, Beijing, China) using a low-light cooled charge-coupled device camera (Night owl LB985, Germany). A. tumefaciens carrying an empty vector mixed with another driven vector was used as a negative control.
The CDS sequence of GhTCP4-like was cloned into the pCAMBIA1302 vector between Nco I and Spe I upstream of the GFP tag to generate the recombinant plasmid GhTCP4-like-GFP, and the CDS of AtWRKY22 was fused with mCherry under the control of the CaMV 35S promoter to obtain a nuclear localization marker (Wang et al., 2019), which was electrically transferred to A. tumefaciens GV3101. After overnight culture, the precipitate was collected by centrifugation and resuspended in MMA (10 mM MES [pH 5.6], 10 mM MgCl2, 100 μM acetosyringone) buffer, diluted to an OD600 of 0.8, and the either A. tumefaciens carrying GFP empty vector or GhTCP4-like-GFP recombinant vector were mixed with A. tumefaciens carrying mcherry-tagged fusion vector in equal ratio. After 3 h at room temperature, the bacterial solution was injected into the epidermis of N. benthamiana leaves with a sterile syringe; after 12 h in the dark, N. benthamiana were transferred to a light incubator for 36 h. Fluorescence signals in the leaf epidermal cells were visualized using a confocal microscope (SP8, Leica, Germany).
The sequence 2 kb upstream of the start codon of GhICS1 was used for cis-element analysis by online database Plant CARE [PlantCARE, a database of plant promoters and their cis-acting regulatory elements (ugent.be)].
The EMSA was performed as previously described (Jiao et al., 2020). Briefly, biotin-labeled probes were synthesized by the TSINGKE Biotechnology Company. The full-length coding sequence of GhTCP4-like was cloned into the pGEX6p-1vector to generate a GST-tagged recombinant vector and transformed into E. coli BL21. After inducible expression with IPTG (0.5 mM) at 28°C, the fusion protein was purified using a GST-tag Protein Purification Kit (Beyotime, Nanjing, China). The Chemiluminescent EMSA Kit (Beyotime, Nanjing, China) was used to visualize the free probe and protein-probe complex, following the manufacturer's instructions.
Our previous research showed that a cotton-known miRNA, ghr-miR319b, showed a significant differential change in plants infected with V. dahliae (Hu et al., 2020). ghr-miR319b includes 21 nucleotides (5′-UUGGACUGAAGGGAGCUCCCU-3′), whose precursor is similar to Arabidopsis MIR319b (Supplementary Figure 1A). ghr-miR319b is located at the 3′ terminus of GhMIR319b and possesses a typical stem-loop structure (Supplementary Figure 1B). Previous mRNA degradome data have documented that the target gene of ghr-miR319b is Gh_A13G1272 (Supplementary Figure 1C) (Hu et al., 2020). According to amino acid sequence alignment and phylogenetic tree analysis, the Gh_A13G1272 protein showed high identity with Arabidopsis TCP4 (Supplementary Figure 1D). In the Gossypium hirsutum genome, the Gh_A13G1272 protein has high homology with Gh_D13G1576 with 99.02% identity (Supplementary Figure 2). Therefore, they can be regarded as potential genes for research. GhTCP4-like contains a nonstandard bHLH domain belonging to the CIN subgroup of Class II TCP family proteins (Figure 1A).
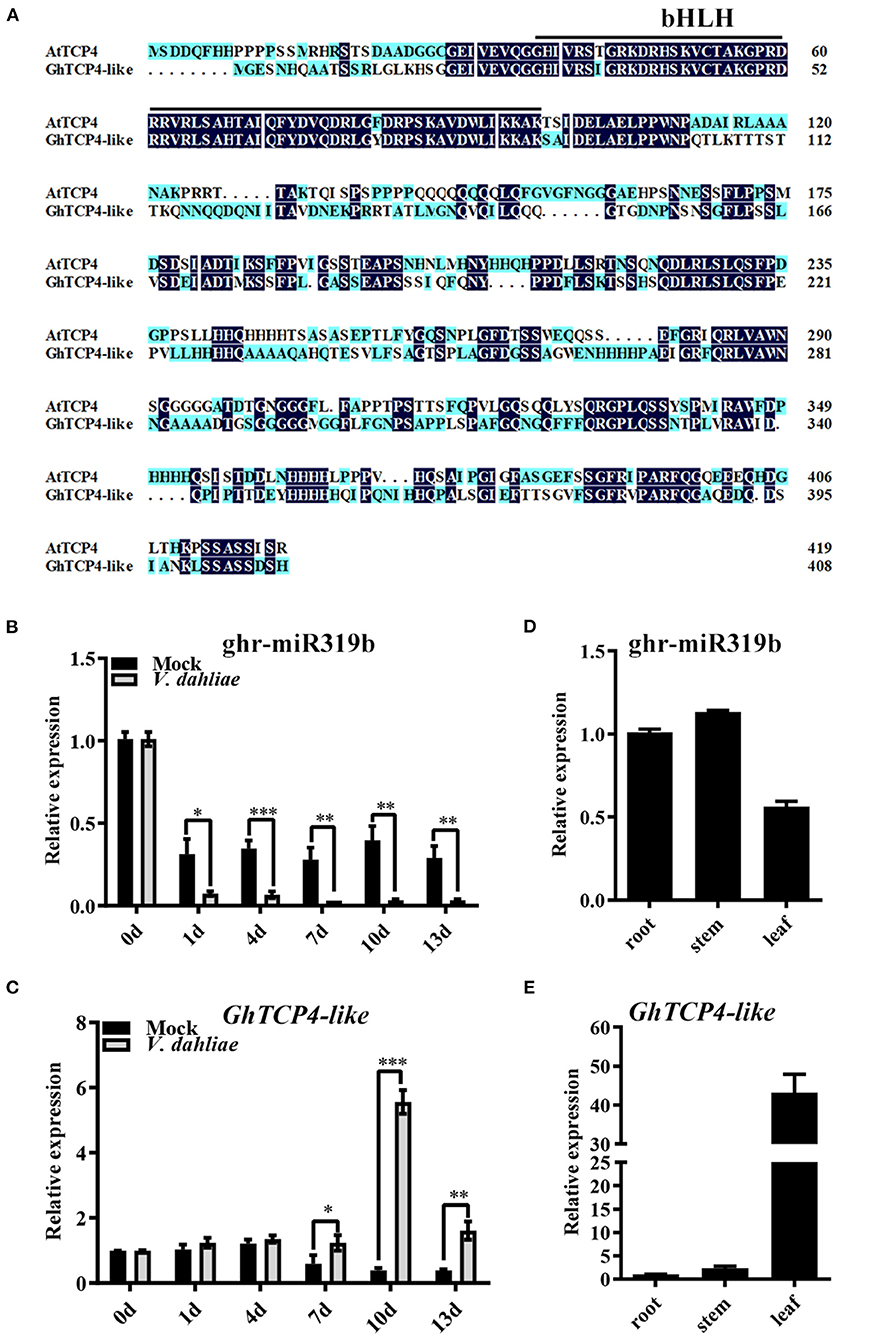
Figure 1. GhTCP4-like characterization and expression patterns of ghr-miR319b and GhTCP4-like. (A) Amino acid sequence alignment of GhTCP4-like and AtTCP4. The solid black underline indicates the TCP domain (a non-standard bHLH domain). (B,C) qPCR analysis of ghr-miR319b and GhTCP4-like expression levels at 0, 1, 4, 7, 10, and 13 d after V. dahliae infection. Three-leaf stage cotton seedlings cultured in Gamborg's B5 (B5) liquid medium were infected with V. dahliae (106 spores/ml) using the root-dipped method. Error bars represent the standard deviation (SD) of three biological replicates. Student's t-test was performed, *p < 0.05, **p < 0.01, ***p < 0.001. (D,E) qPCR analysis of ghr-miR319b and GhTCP4-like expression levels in different tissues. Error bars represent the SD of three biological replicates.
Given the prolonged response of ghr-miR319b to V. dahliae infection reported by Hu et al. (2020), we performed qPCR analysis to confirm the response from the beginning of inoculation to 13 days post-inoculation (dpi). In G. hirsutum elite cultivar “CCRI 35,” the expression profile of ghr-miR319b was significantly lower when infected by V. dahliae than the mock treatment over time (Figure 1B). Notably, the target gene GhTCP4-like showed higher expression levels in plants infected with this fungus after 7 dpi compared to mock treatment, seemingly exhibiting a contrary trend with ghr-miR319b accumulation (Figure 1C). The same experiment was performed for the other two cotton cultivars, “Zhongzhimian (ZZM) 2” with high resistance and “Jimian (JM) 11,” which are highly susceptible to V. dahliae. As shown in Supplementary Figure 3, ghr-miR319b accumulation and GhTCP4-like expression levels in “ZZM 2” and “JM 11” were similar, consistent with “CCRI35” response to V. dahliae infection. ghr-miR319b and GhTCP4-like genes were constitutively expressed in roots, stems, and leaves (Figures 1D,E). These results suggest that ghr-miR319b regulates GhTCP4-like expression, which participates in the plant response to V. dahliae infection.
Our previous study showed that GhTCP4-like mRNA was first recognized by ghr-miR319b through base complementarity and was directedly cut at 980-nucleotide (nt) according to mRNA degradome sequencing data (Supplementary Figure 1C). A 5′-RACE analysis was performed to confirm GhTCP4-like transcript cleavage using the ghr-miR319b guide. Sequencing analysis revealed that 13 out of 15 clones of GhTCP4-like mRNA were cleaved at the 980-nt through sequencing analysis (Figure 2A). GUS-fused reporter analysis was conducted to verify the cleavage of the GhTCP4-like transcript directed by ghr-miR319b. The GhTCP4-like coding sequence (CDS) and the corresponding mutant in the target sequence were fused with the GUS gene under the control of the CaMV 35S promoter and were constructed as reporter vectors, 35S:GhTCP4-like-GUS and 35S:GhTCP4-likem-GUS. GhMIR319b was inserted into the plant expression vector under control of the CaMV 35S promoter as an effector vector, 35S:GhMIR319b (Figure 2B). Co-expression analysis in Nicotiana benthamiana leaf cells was performed using the agroinfiltration technique. As shown in Figure 2C, the leaves agroinfiltrated with 35S:GhTCP4-like-GUS or 35S:GhTCP4-likem-GUS showed normal blue staining by GUS. The tobacco leaves equally infiltrated with 35S:GhMIR319b and 35S:GhTCP4-like-GUS showed a weak blue color, whereas the leaves treated with 35S:GhMIR319b and 35S:GhTCP4-likem-GUS exhibited normal blue color. The GUS staining analysis revealed that the expression levels of GhTCP4-like in various leaf spots treated with combinations of vectors (Figure 2C) showed that GhTCP4-like can be directedly cleaved by ghr-miR319b in tobacco cells (Figure 2D). These results demonstrated that GhTCP4-like mRNA can be directedly cleaved by ghr-miR319b through a post-transcriptional process.
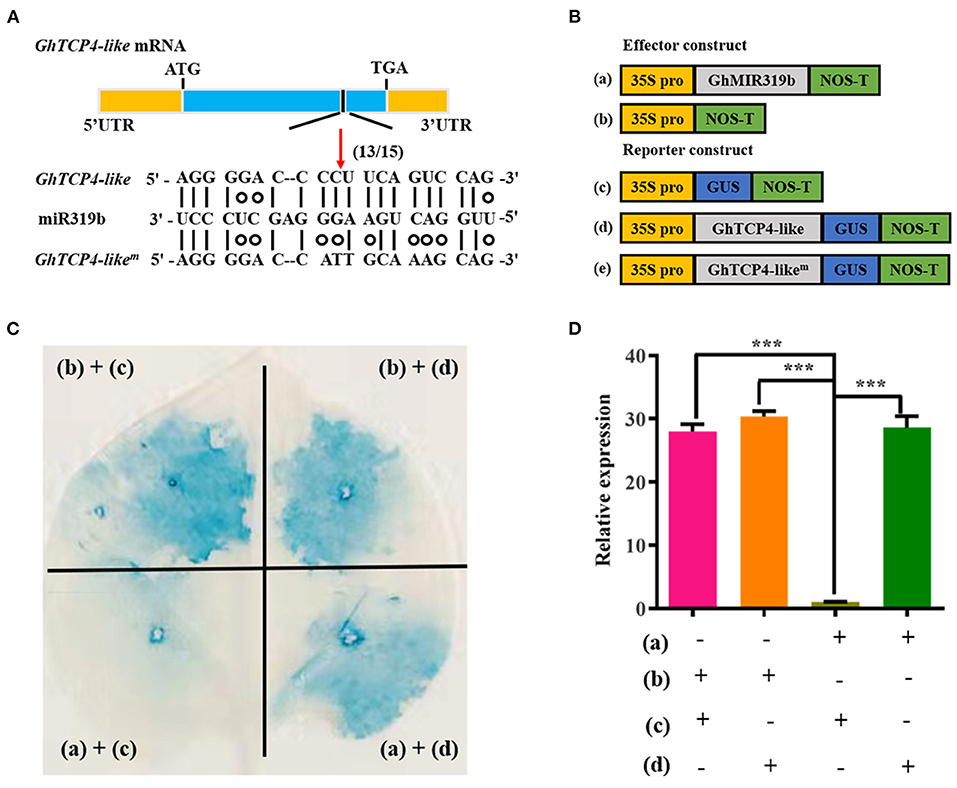
Figure 2. GhTCP4-like directedly cleaved by ghr319b. (A) Diagram of the structure of GhTCP4-like transcript and 5′-RACE analysis. GhTCP4−likem is an equivocal mutation sequence of GhTCP4-like mRNA on the ghr-miR319b target sequence. The black box points to the predicted cleavage site at 980-nt of GhTCP4-like mRNA. (13/15) represents that 13 out of 15 clones are cleaved in target sequence by sequencing. (B) Schematic diagram of the constructs for transient expression assay. (a), (b), (c) and (d) represent 35S:GhMIR319b, pCAMBIA1300, 35S:GhTCP4-GUS and 35S:GhTCP4-likem-GUS, respectively. (C) GUS staining assay. The combinations of A. tumefaciens containing vectors labeled in the edge indicated in (B) were co-injected into the tobacco leaf cells. (D) GhTCP4-like relative expression in treated leaf spots as indicated in (C). Error bar means SD of three independent biological replications. Student's t-test was performed, ***p < 0.001. Leaves were harvested 48 h after agroinfiltration. All experiments were replicated at least three times.
To elucidate the function of ghr-miR319b and GhTCP4-like in the plant response to V. dahliae infection, ghr-miR319b- and GhTCP4-like-silenced plants were generated using the VIGS method, which is a robust tool for identifying the function of genes associated with plant defense and development, especially at the juvenile stage. Cotton leaf crumple virus (CLCrV)-derived vectors and/or short tandem target mimic (STTM) technology were used to develop ghr-miR319b-silenced plants (CLCrV:STTM319b) and GhTCP4-like-silenced plants (CLCrV:GhTCP4-like). Phytoene desaturase (PDS)-silenced plants showed a photobleaching phenotype as a VIGS marker about 14 d post-agroinfiltration (Supplementary Figure 4) upon examining ghr-miR319b accumulation and the level of GhTCP4-like expression. The results showed that ghr-miR319b accumulation in silenced plants was significantly reduced by 50% compared to plants agroinfiltrated with empty vectors (CLCrV:00, hereafter referred to as the control), in which GhTCP4-like expression levels were significantly higher (Figure 3A). GhTCP4-like expression in GhTCP4-like-silenced plants was significantly decreased by 74% compared to control (Figure 3B).
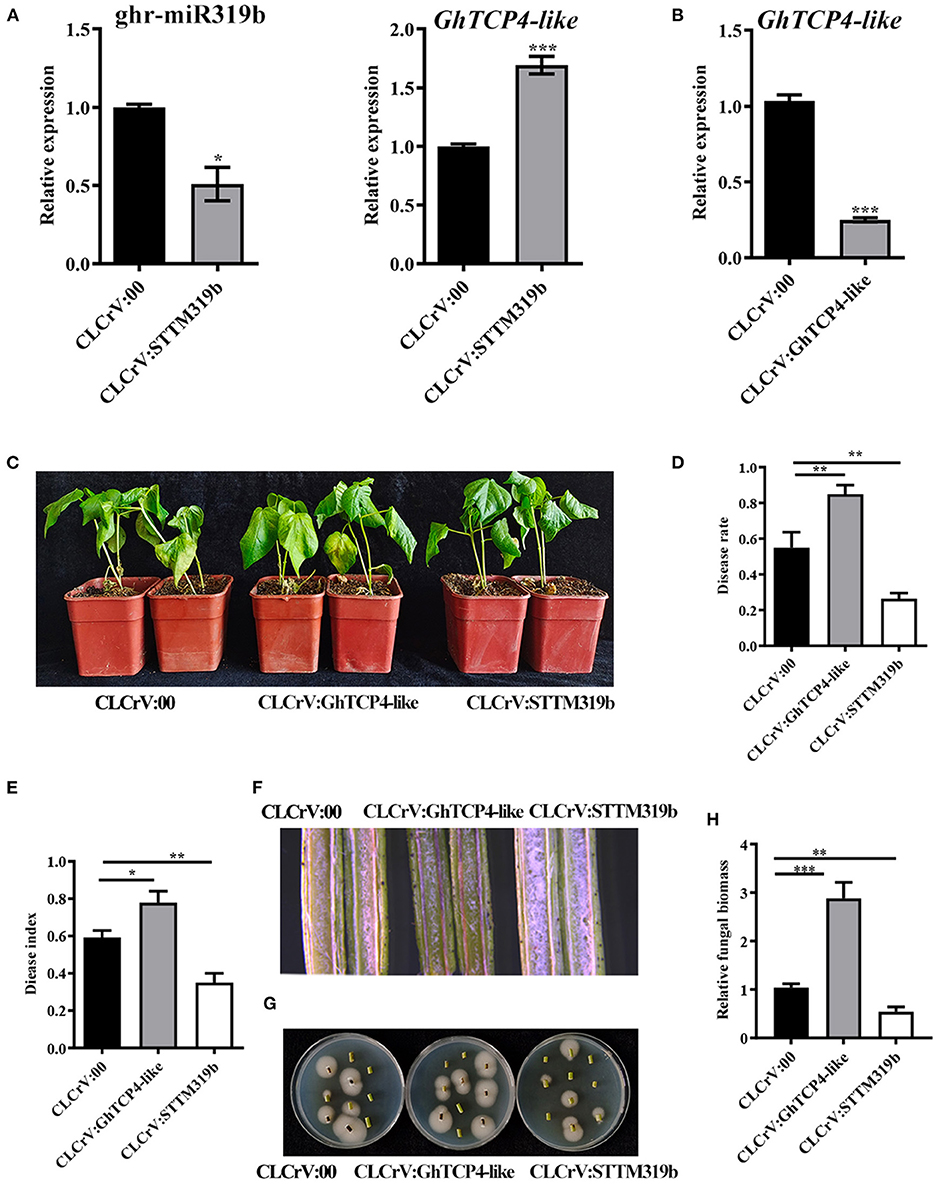
Figure 3. Resistance analysis of ghr-miR319b and GhTCP4-like against V. dahlia. (A) Transcript level of ghr-miR319b and GhTCP4-like in the control and STTM plants determined by qPCR. Total RNA was extracted from the second true leaf 14 d after agroinfiltration. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05, ***p < 0.001. (B) The transcript level of GhTCP4-like in control and GhTCP4-like-silenced plants was determined by qPCR. Error bar means SD of three independent biological replications. Student's t-test was performed, ***p < 0.001. (C) Disease symptom of CLCrV:00, CLCrV:GhTCP4-like and CLCrV:STTM319b plants were photographed at 14 dpi. (D,E) Disease rate and disease index were calculated at 14 dpi. More than 30 plants per treatment were sampled. Error bar means SD. Student's t-test was performed, *p < 0.05, **p < 0.01. (F) Color intensity of longitudinal sections of the stem of CLCrV:00, CLCrV:GhTCP4-like and CLCrV:STTM319b plants 14 dpi. Photos were taken by the body microscope. (G) Fungal discovery culture assay. V. dahliae growth condition on PDA medium from stems of CLCrV:00, CLCrV:GhTCP4-like, CLCrV:STTM319b plants 14 dpi. Photographs were taken after 5 days of culture at 25°C. (H) The relative fungal biomass assay. Stems from CLCrV:00, CLCrV:GhTCP4-like, CLCrV:STTM319b plants were used to extract DNA at 14 dpi, which were used as templates for relative fungal biomass analysis by qPCR. Error bar means SD of three independent biological replications. Student's t-test was performed, **p < 0.01, ***p < 0.001.
To evaluate ghr-miR319b-GhTCP4-like function in plant defense, the VIGS plants were inoculated with V. dahliae. As shown in Figure 3C, ghr-miR319b-silenced plants showed higher resistance to this fungus than the control with less yellow and wilt leaves, whereas GhTCP4-like-silenced plants exhibited higher susceptibility to V. dahliae infection, possessing more yellow and wilt leaves. Statistical analysis showed that the disease rate and disease index in CLCrV:STTM319b plants were lower than those in control, whereas, they were higher in CLCrV: GhTCP4-like plants (Figures 3D,E). Stem longitudinal sections in ghr-miR319b-silenced plants were lighter than those in control, indicating less damage. However, longitudinal sections of the stems in GhTCP4-like-silenced plants were heavier than those in the control (Figure 3F). Fewer fungal colonies were recovered from stem sections of ghr-miR319b-silenced plants in the media compared to the control, while more fungal colonies appeared from stem sections of GhTCP4-like-silenced plants, which was supported with the results of relative fungal biomass assay (Figures 3G,H). To further verify ghr-miR319b-GhTCP4-like the function in plant resistance to V. dahliae infection, a parallel experiment was performed using another set of tobacco rattle virus (TRV)-induced gene silencing (TRVIGS) plants. As with the results of CLCrV-derived VIGS (CLCrVIGS) plants, TRVIGS plants with ghr-miR319b knockdown showed higher resistance to this fungus compared to control (TRV:00), while those with GhTCP4-like knockdown showed significantly increased susceptibility (Supplementary Figures 5, 6). The above results demonstrate that GhTCP4-like fine-tuned by ghr-miR319b is a positive regulator of plant resistance to V. dahliae infection.
To elucidate the molecular mechanism of ghr-miR319b-GhTCP4-like module function in plant's response to V. dahliae, we tested whether this module affects SA- and JA-related gene expression levels. The results showed that three SA-related synthesis genes, GhICS1, GhEDS1, and GhPAD4, were significantly upregulated in ghr-miR319b-silenced plants compared to the control under V. dahliae infection, whereas they were significantly downregulated in GhTCP4-like-silenced plants (Figure 4A). Three other SA response genes, GhPR1, GhPR2, and GhPR5, showed higher expression levels in ghr-miR319b-silenced plants than in control and exhibited lower expression levels in GhTCP4-like-silenced plants (Figure 4A). The results suggested that GhTCP4-like knockdown possibly reduced SA biosynthesis by repressing expression of GhICS1, GhEDS1 and GhPAD4, leading to down-regulation of GhPR1, GhPR2 and GhPR5 under the pathogen infection. However, the JA-related genes GhAOS, GhLOX2, GhOPR3, GhPDF1.2, and GhVSP showed slight changes in expression levels among ghr-miR319b-silenced plants, GhTCP4-like-silenced plants, and the control inoculated with V. dahliae (Figure 4B), suggesting that the ghr-miR319-GhTCP4-like module participates in the plant response to this fungal infection and is not involved in the JA signaling pathway. These results suggest that the ghr-miR319-GhTCP4-like module is associated with SA synthesis and signaling pathways that regulate plant resistance to V. dahliae infection.
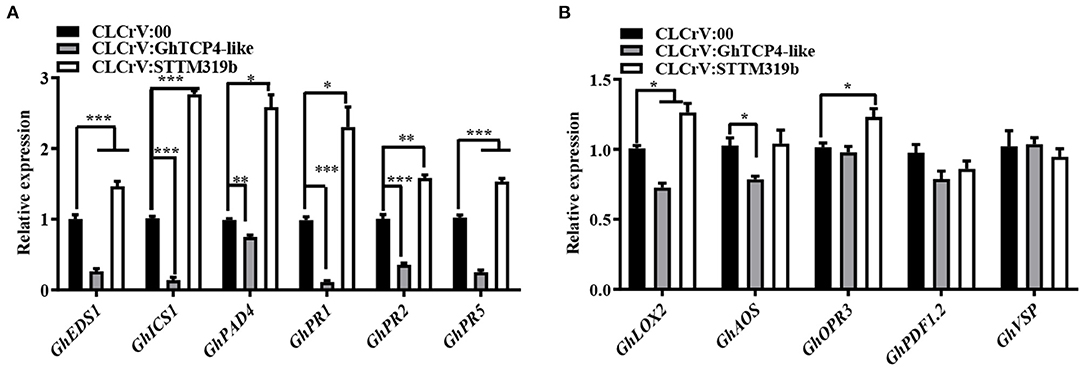
Figure 4. Expression levels of SA- and JA-related genes in CLCrV:00, CLCrV:GhTCP4-like and CLCrV:STTM319b plants under V. dahliae infection. (A,B) The expression analysis of SA biosynthesis-related genes (GhEDS1, GhICS1 and GhPAD4) and SA responsive genes (GhPR1, GhPR2, and GhPR5) in various plants 1-day post-inoculation (dpi). (B) The expression analysis of JA biosynthesis-related genes (GhLOX2, GhAOS, and GhOPR3) and JA responsive genes (GhPDF1.2 and GhVSP) in various plants 1 dpi. Root samples were collected from various treatments. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05, **p < 0.01, ***p < 0.001.
Given that GhTCP4-like knockdown significantly downregulated the expression of SA biosynthesis-related genes, we hypothesized that GhTCP4-like would affect SA biosynthesis. A recent study also reported that Arabidopsis TCP8/9 proteins could co-ordinately regulate ICS1 expression to participate in plant immunity (Wang et al., 2015), which is similar to our results of GhTCP4-like knockdown regulating GhICS1 expression during V. dahliae infection (Figure 4A). A subcellular location was investigated using a GFP reporter system in N. benthamiana leaf cells to characterize the GhTCP4-like transcription factor. As shown in Figure 5A, the green fluorescence of the GhTCP4-like-GFP protein located in the nucleus of tobacco leaf cells merged with a red color from the nucleus protein mCherry. These results suggested that GhTCP4-like proteins are distributed in the nucleus.
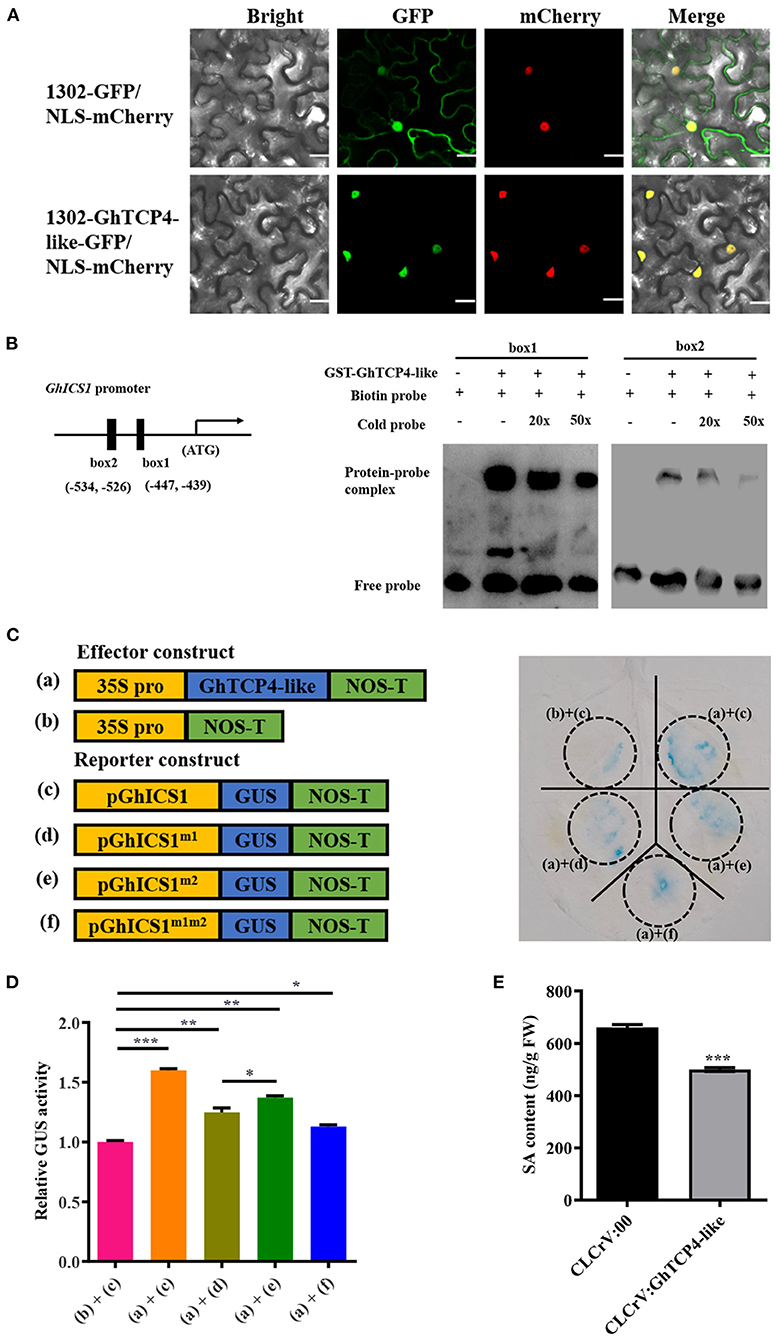
Figure 5. GhTCP4-like directly transactivates GhICS1 expression. (A) Subcellular localization of GhTCP4-like in tobacco leaf cells. NLS-mCherry was a nuclear location marker. Scale bar = 20 μm. (B) Electrophoretic mobility shift assay (EMSA). Structure of GhICS1 promoter in the left panel. Solid black boxes indicate predicted two GhTCP4-like binding sites (GTGGCACC) in the GhICS1 promoter. GhTCP4-like bound to GTGGCACC motif from GhICS1 promoter in the right panel. The lower bands represent unbound probes; lagging bands indicate the complexity of protein and probe. (C) GUS reporter analysis for GhTCP4-like transactivating GhICS1 expression. Schematic diagram of the constructs for GUS reporter assays in tobacco leaf cells in the left panel. (a), (b), (c), (d), (e), and (f) represent p35S:GhTCP4-like, pCAMBIA1300, pGhICS1:GUS, pGhICS1m1:GUS, pGhICS1m2:GUS and pGhICS1m1m2:GUS, respectively. Tobacco leaves were harvested at 48 h after agroinfiltration. (D) GUS enzyme activity assayed by 4-MU measurement of treated leaf spots indicated in (C). “+” indicates that the corresponding treatment has been added, “–“ indicates that the corresponding treatment has not been added. Tobacco leaves were harvested at 48 h after agroinfiltration. (E) Determination of total SA content. SA of GhTCP4-like-silenced and control plants 1 d after V. dahliae infection was measured by HPLC-MS/MS. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05, **p < 0.01, ***p < 0.001. All experiments were repeated more than three times with similar results.
The cis-element bond of GhTCP4-like is generally the GTGGNCCC box previously reported (Martin-Trillo and Cubas, 2010), and we found that there are two putative GhTCP4-like binding sequences (GTGGCACC) in the GhICS1 promoter, namely box1 and box2, located−447 to−439 and−534 to−526 upstream of the translation initiation site (ATG), respectively (Figure 5B). The two fragments containing box1 or box2 and their flanking sequences were chemically synthesized and labeled with biotin at the 5′ terminal end as probes to perform an electrophoretic mobility shift assay (EMSA). The box1/box2 probe incubated with a combination of GhTCP4-like showed one upshifted bond in the gel, which weakened with increasing cold probe levels (Figure 5B). Therefore, box1 and box2 were verified to be specifically bound to GhTCP4-like.
To further elucidate the transcriptional activity of GhTCP4-like to GhICS1, a GUS reporter assay in N. benthamiana leaf cells was conducted. A 1,200 bp of GhICS1 promoter was isolated to drive GUS gene expression as a reporter vector (pGhICS1:GUS), and the GhICS1 promoter mutated in box1 and/or box2 was used to control GUS gene expression as promoter mutant reporter vectors (pGhICS1m1:GUS, pGhICS1m2:GUS, and pGhICS1m1m2:GUS). GhTCP4-like was expressed under the control of the CaMV 35S promoter as an effector vector, p35S:GhTCP4-like (Figure 5C). As shown in Figure 5C, tobacco leaves agroinfiltrated with pGhICS1:GUS showed normal blue staining by GUS, indicating that the GhICS1 promoter can drive GUS gene expression. When p35S:GhTCP4-like and pGhICS1:GUS were co-infiltrated with tobacco leaves, they were bluer than the leaves agroinfiltrated with pGhICS1:GUS. Tobacco leaves co-injected with p35S:GhTCP4-like and pGhICS1m1:GUS or pGhICS1m2:GUS were lighter blue than those co-injected with p35S:GhTCP4-like and pGhICS1:GUS. However, there was less blue color in tobacco leaves agroinfiltrated with p35S:GhTCP4-like and pGhICSm1m2:GUS, suggesting that box1 and box2 in the GhICS1 promoter have additive effects on GhTCP4-like activation of GhICS1 transcription. In line with these results, the GUS activities tested by the 4-MU assay showed that the leaves co-infected with p35S:GhTCP4-like and pGhICS1:GUS had the highest enzyme activity compared to those co-injected with p35S:GhTCP4-like and pGhICS1m1:GUS, p35S:GhTCP4-like and pGhICS1m2:GUS, and p35S:GhTCP4-like and pGhICS1m1m2:GUS (Figure 5D). Notably, the enzyme activity of sample treated with p35S:GhTCP4-like and pGhICS1m1:GUS significantly lower than that with p35S:GhTCP4-like and pGhICS1m2:GUS, indicated that box1 is more essential than box2 to promote GhICS1 expression by GhTCP4-like. These results demonstrate that GhTCP4-like can activate GhICS1 expression through the additive effects of the box1 and box2 cis-elements.
Given that GhTCP4-like can directly regulate GhICS1 expression, we hypothesized that SA levels would be altered when GhTCP4-like expression is disrupted. HPLC-MS/MS analysis showed that GhTCP4-like-silenced plants had significantly lower SA content than the control (Figure 5E). These data confirm that GhTCP4-like transcriptionally activates GhICS1 expression, increasing SA content.
To explore whether the regulation of GhTCP4-like to GhICS1 transcription is associated with other regulators, we need to search for its partners in cells. Several reports have shown that the NPR1 is a partner of TCPs (Li et al., 2018). Therefore, we performed a yeast two-hybrid assay to verify whether GhTCP4-like interactes with GhNPR1. The GhTCP4-like coding sequence (CDS) was isolated and inserted into the AD vector, and the GhNPR1 CDS was constructed into a BD vector. As shown in Figure 6A, yeast co-transformed with GhTCP4-like and GhNPR1 grew on SD/-Trp/-Leu/-His/-Ade medium at different dilution concentrations and was stained blue as well as the positive control, while yeasts treated with BD and AD-TCP4-like, AD and BD-NPR1, or the negative control, did not grow. These results indicated that GhTCP4-like directly interacts with GhNPR1. To confirm the GhTCP4-like interacts with GhNRP1, we conducted a luciferase complementation image (LCI) analysis in tobacco cells. The CDSs of GhTCP4-like and GhNPR1 were fused to the C-terminus and N-terminus of LUC, respectively, generating cLUC-TCP4-like and nLUC-NPR1 vectors. As shown in Figure 6B, leaf spots agroinfiltrated with cLUC-TCP4-like and nLUC-NPR1 showed LUC fluorescence, while other leaf spots injected with various vectors, as indicated in the left panel of Figure 6B did not. The results of the LCI analysis suggested that GhTCP4-like interacts with GhNPR1, similar to the interaction of Arabidopsis TCP8/14/15 with NPR1 reported by Li et al. (2018).
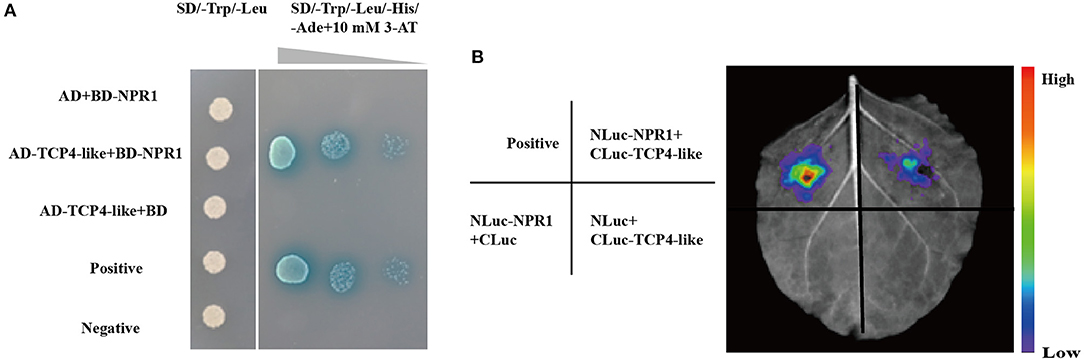
Figure 6. GhTCP4-like interact with GhNPR1. (A) Yeast two-hybrid (Y2H) assay shows GhTCP4-like interaction with GhNPR1. All recombination yeast cells were grown on SD/-Trp/-Leu medium. After gradient dilution and sprayed 10 mM 3-AT and 20 mg/ml X-α-Gal on SD/-Trp/-Leu /-His/-Ade medium. Yeast transformed with paired plasmid AD-largeT and BD-p53 were used as a positive control, AD-largeT and BD-laminC were used as a negative control. Photos were taken after 3 d at 28°C. (B) LCI assays to detect the association between GhTCP4-like and GhNPR1. Agrobacterium strains containing the indicated vectors were co-expressed in leaf cells of N. benthamiana. The luminescence signal was detected by applying 1 mM luciferin after 48 h of agroinfiltration. The positive control was constructed according to Chen et al. (2008). The colored scale indicates the intensity of luciferin activity. All experiments were repeated more than three times with similar results.
Because GhNPR1 can interact with GhTCP4-like, we investigated whether GhNPR1 affects GhTCP4-like transcriptional activity in GhICS1 expression. The GhNPR1 CDS was cloned into a plant expression vector under the control of the CaMV 35S promoter to carry out GUS reporter analysis. As shown in Figure 7A, the leaf spots co-injected with p35S:GhTCP4-like and pGhICS1:GUS showed a normal blue color under GUS staining treatment, whereas the leaf spots co-injected with p35S:GhNPR1, p35S:GhTCP4-like, and pGhICS1:GUS exhibited a heavier blue color than the former leaf spots. The GUS content tested with 4-MU supported the hypothesis that GhNPR1 promotes GhTCP4-like activation of GhICS1 transcription (Figure 7B). GhNPR1-silenced plants were generated using the VIGS method to elucidate GhNPR1 response to V. dahliae infection (Figure 7C). As expected, GhNPR1 knockdown significantly reduced GhICS1 expression levels compared to control after V. dahliae inoculation (Figure 7D). Moreover, GhNPR1-silenced plants showed higher susceptibility to V. dahliae infection than the control, with more yellow and wilting leaves, higher disease rate, disease index, and fungal biomass in infected stems (Figures 7E–J). Therefore, GhNPR1 co-ordinately participates in plant resistance to pathogen infection via two pathways. First, GhNPR1, one of the receptors, can generally activate SA signaling to increase plant defense against pathogens. Second, GhNPR1 interacts with GhTCP4-like co-ordinately promotes GhICS1 transcription to increase SA biosynthesis in the feedback regulation of SA biosynthesis.
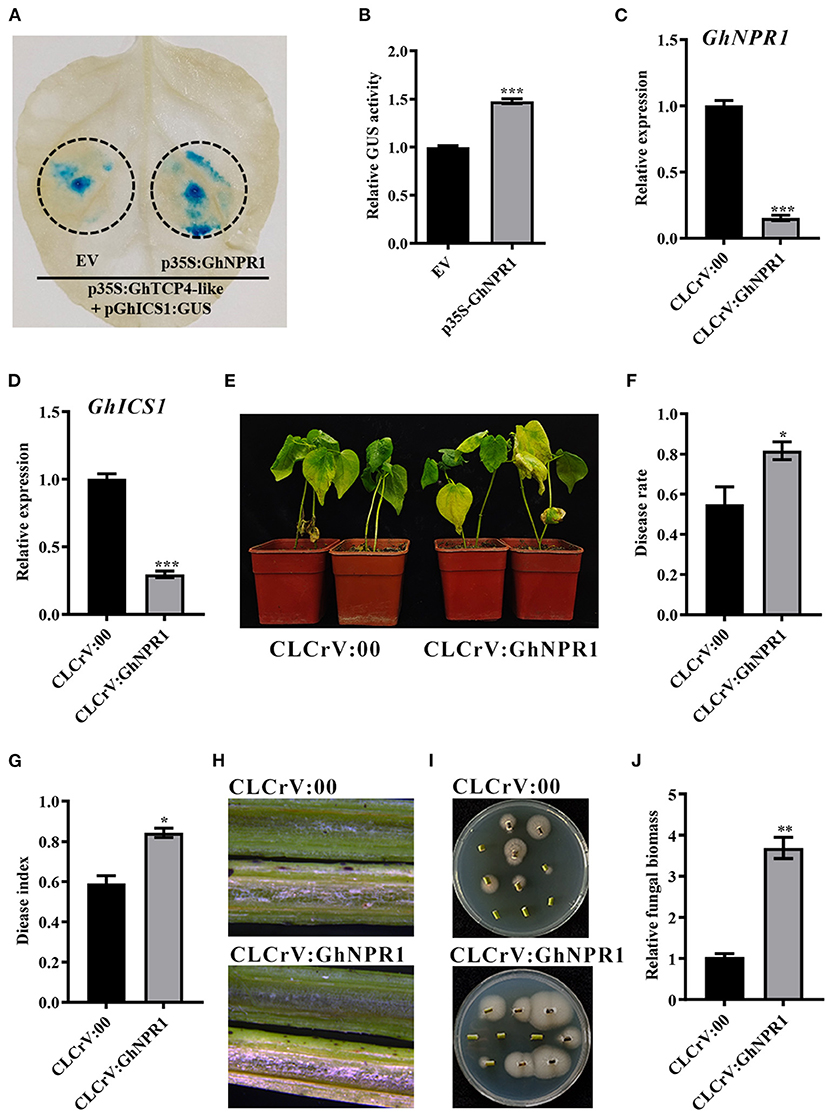
Figure 7. GhNPR1 promotes transcriptional activating activity of GhTCP4-like for GhICS1 expression. (A) GUS staining assay. EV represents pCAMBIA1300. Tobacco leaves were harvested at 48 h after corresponding treatments. (B) GUS enzyme activity measurement. Error bar means SD of three independent biological replications. Student's t-test was performed, ***p < 0.001. (C) GhNPR1 expression level in CLCrV:00 and CLCrV:GhNPR1 plants. Total RNA was extracted 14 d after agroinfiltration. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05. (D) Expression of GhICS1 in CLCrV:00 and CLCrV:GhNPR1 plants 1 dpi. Error bar means SD of three independent biological replications. Student's t-test was performed, ***p < 0.001. (E) Disease symptoms of CLCrV:00 and CLCrV:GhNPR1 plants after V. dahliae infection. Photos were taken at 21 dpi. (F,G) Disease rate and disease index of CLCrV:00 and CLCrV:GhNPR1 plants 21 dpi. Error bar means SD of three independent biological replications. Student's t-test was performed, *p < 0.05. (H) Color intensity of longitudinal sections of the stem of CLCrV:00 and CLCrV:GhNPR1 plants 21 dpi. Photos were taken by the body microscope. (I) Fungus discovery culture assay. Stems from CLCrV:00 and CLCrV:GhNPR1 plants 21 dpi were sprayed on PDA medium. Photos were taken after 5 d of culture at 25°C. (J) Relative fungal biomass in CLCrV:00 and CLCrV:GhNPR1 plants were determined by qPCR at 21 dpi. Error bar means SD of three independent biological replications. Student's t-test was performed, **p < 0.01.
TCPs are transcription factors that regulate plant growth and development, including gametophyte development, seed germination, leaf shape, internode length, and flower development (Aguilar-Martinez et al., 2007; Giraud et al., 2010; Liu et al., 2018). Recently, accumulating evidence has shown that TCPs play a role in plant immunity against many pathogenic infections (Mukhtar et al., 2011; Wang et al., 2015; Yang et al., 2017; Li et al., 2018). However, it is necessary to identify novel TCPs and reveal their potential mechanisms in plant responses to pathogen infections. In this study, we characterized the cotton TCP4-like functions in plant immunity. GhTCP4-like can be directedly cleaved by ghr-miR319b through a post-transcriptional process and activates GhICS1 transcription and promotes SA biosynthesis, resulting in increased plant resistance to V. dahliae infection via GhNPR1 coordination.
GhTCP4-like, a class II protein, was fine-tuned by ghr-miR319 through a post-transcriptional process, which participates in plant response to V. dahliae infection. Knockdown of GhTCP4-like significantly reduced plant resistance to this fungal infection compared with the control, whereas ghr-miR319b-silenced plants showed a higher resistance. In Arabidopsis, TCP8 and TCP9 participate in plant responses to pathogens (Wang et al., 2015). AtTCP15 regulates plant immune response during cell cycle progression (Zhang et al., 2018). Arabidopsis TCP8, TCP9, and TCP15 belong to class I proteins, while GhTCP4-like is a class II protein and fine-tuned by ghr-miR319b. Therefore, GhTCP4-like, targeted by ghr-miR319, is a novel module that participates in plant immunity.
In this study, GhTCP4-like activated GhICS1 transcription, which promoted SA accumulation and increased the expression levels of SA-related genes, resulting in plant resistance to V. dahliae infection. Several transcription factors and regulators have transcriptionally regulated ICS1 in plant immunity. Four positive regulators, SARD1, CBP60g, and WRKY28/46, have been shown to activate ICS1 expression in Arabidopsis to promote SA biosynthesis. In contrast, EIN3, EIL1, MUR3, and NAC19/NAC55/NAC72 negatively regulates ICS1 expression during plant immunity (Verberne et al., 2000; Tedman-Jones et al., 2008; Chen et al., 2009; Zhang et al., 2010b; Van Verk et al., 2011; Zheng et al., 2012). A similar report showed that, in Arabidopsis, TCP8 could directly activate ICS1 expression during the immune response (Wang et al., 2015). Therefore, our results further explain that ICS1 transcription is a knot of the network that is regulated by several factors in plant resistance to pathogen infection.
GhTCP4-like interacts with GhNPR1, which co-ordinately regulates GhICS1 transcription to promote SA biosynthesis. Knockdown of GhNPR1 significantly reduced GhICS1 expression, resulting in increased susceptibility to V. dahliae infection compared with the control. Similar reports have shown that the NPR1 interacts with TCP proteins to participate in plant immunity. For example, NPR1 interacts with TCP15 to promote its transcriptional activation activity of PR1 in Arabidopsis but does not participate in SA biosynthesis like GhTCP4-like (Li et al., 2018). Therefore, in plants, NPR1 can interact with TCP proteins to regulate ICS1 transcription to promote SA accumulation or act downstream gene expression, facilitating plant response to pathogen infection partially due to NPR1 and TGA protein interaction to promote defense-related expression.
In the present study, we addressed GhTCP4-like targets of ghr-miR319b in response to V. dahliae infection. The Ghr-miR319b-GhTCP4-like module regulates plant resistance to pathogen challenge by regulating SA biosynthesis. GhTCP4-like interactions with GhNPR1 co-ordinately activate GhICS1 transcription to promote SA accumulation, resulting in increased plant resistance to V. dahliae infection, possibly through GhNPR1 interaction with GhTGAs. Our results deepen the understanding of NPR1 interaction with TCPs in plant immunity through feedback regulation of SA biosynthesis (Figure 8).
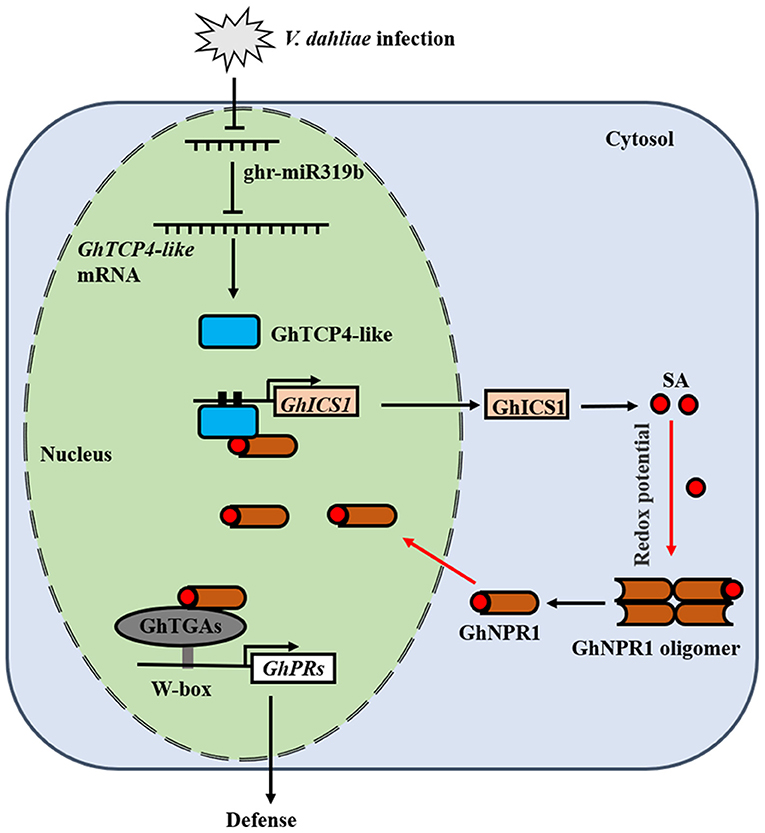
Figure 8. Working model diagram of ghr-miR319b-GhTCP4-like module in defense against V. dahliae infection. V. dahliae invasion releases the effectors or PAMPs to inhibit ghr-miR319, increasing GhTCP4-like expression, which transactivates GhICS1 expression to promote SA biosynthesis. GhNPR1 interacting with GhTCP4-like enhances activation activity of GhTCP4-like to GhICS1 transcription in feedback regulation loop of SA biosynthesis, facilitating in plant high resistance to V. dahliae infection. NPR1 interacting with TGAs promotes defense-related gene expression to increase plant resistance to pathogens. The black arrows represent regulation positively, the T-shaped arrows represent regulation negatively, the red arrows represent diffusion.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
JW and QP conceived and designed the experiments. PJ, YT, and GH performed the experiments. PJ, YQ, and AC constructed the vectors and data analysis. PJ, NZ, and JW wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (31971905), and sponsored by State Key Laboratory of Cotton Biology Open Fund (CB2021B02).
YQ and AC were employed by Join Hope Seeds Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.870882/full#supplementary-material
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., and Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22, 1174–1189. doi: 10.1105/tpc.109.066647
Aguilar-Martinez, J. A., Poza-Carrion, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
An, C., and Mou, Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x
Bao, S., Zhang, Z., Lian, Q., Sun, Q., and Zhang, R. (2019). Evolution and expression of genes encoding TCP transcription factors in Solanum tuberosum reveal the involvement of StTCP23 in plant defence. BMC Genet. 20, 91–106. doi: 10.1186/s12863-019-0793-1
Boyle, P., Le Su, E., Rochon, A., Shearer, H. L., Murmu, J., Chu, J. Y., et al. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21, 3700–3713. doi: 10.1105/tpc.109.069971
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.2307/3869945
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Catinot, J., Buchala, A., Abou-Mansour, E., and Metraux, J. P. (2008). Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 582, 473–478. doi: 10.1016/j.febslet.2007.12.039
Chen, H., Xue, L., Chintamanani, S., Germain, H., Lin, H., Cui, H., et al. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21, 2527–2540. doi: 10.1105/tpc.108.065193
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Cheng, H. Q., Han, L. B., Yang, C. L., Wu, X. M., Zhong, N. Q., Wu, J. H., et al. (2016). The cotton MYB108 forms a positive feedback regulation loop with CML11 and participates in the defense response against Verticillium dahliae infection. J. Exp. Bot. 67, 1935–1950. doi: 10.1093/jxb/erw016
Coquoz, J. L., Buchala A Fau-Metraux, J. P., and Metraux, J. P. (1998). The biosynthesis of salicylic acid in potato plants. Plant Physiol. 117, 1095–1101. doi: 10.1104/pp.117.3.1095
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Daviere, J. M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gao, F., Zhou, B. J., Li, G. Y., Jia, P. S., Li, H., Zhao, Y. L., et al. (2010). A glutamic acid-rich protein identified in Verticillium dahliae from an insertional mutagenesis affects microsclerotial formation and pathogenicity. PLoS ONE 5:e15319. doi: 10.1371/journal.pone.0015319
Gao, X., Britt, R. C. Jr., Shan, L., and He, P. (2011). Agrobacterium-mediated virus-induced gene silencing assay in cotton. J. Vis. Exp. 54:e2938. doi: 10.3791/2938
Giraud, E., Ng, S., Carrie, C., Duncan, O., Low, J., Lee, C. P., et al. (2010). TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22, 3921–3934. doi: 10.1105/tpc.110.074518
Gu, Z., Huang, C., Li, F., and Zhou, X. (2014). A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 12, 638–649. doi: 10.1111/pbi.12169
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., and Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Hu, G., Hao, M., Wang, L., Liu, J., Zhang, Z., Tang, Y., et al. (2020). The Cotton miR477-CBP60A module participates in plant defense against Verticillium dahlia. Mol. Plant Microbe Interact. 33, 624–636. doi: 10.1094/MPMI-10-19-0302-R
Hu, Q., Zhu, L., Zhang, X., Guan, Q., Xiao, S., Min, L., et al. (2018). GhCPK33 negatively regulates defense against Verticillium dahliae by phosphorylating GhOPR3. Plant Physiol. 178, 876–889. doi: 10.1104/pp.18.00737
Jefferson, R. A., Kavanagh Ta Fau - Bevan, M. W., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jiao, X., Wang, H., Yan, J., Kong, X., Liu, Y., Chu, J., et al. (2020). Promotion of BR biosynthesis by miR444 is required for ammonium-triggered inhibition of root growth. Plant Physiol. 182, 1454–1466. doi: 10.1104/pp.19.00190
Kim, S. H., Son, G. H., Bhattacharjee, S., Kim, H. J., Nam, J. C., Nguyen, P. D., et al. (2014). The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 78, 978–989. doi: 10.1111/tpj.12527
Klimes, A., Dobinson, K. F., Thomma, B. P., and Klosterman, S. J. (2015). Genomics spurs rapid advances in our understanding of the biology of vascular wilt pathogens in the genus Verticillium. Annu. Rev. Phytopathol. 53, 181–198. doi: 10.1146/annurev-phyto-080614-120224
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313X.2002.01294.x
Lee, S., Kim, S. G., and Park, C. M. (2010). Salicylic acid promotes seed germination under high salinity by modulating antioxidant activity in Arabidopsis. New Phytol. 188, 626–637. doi: 10.1111/j.1469-8137.2010.03378.x
León, J., Shulaev, V., Yalpani, N., Lawton, M. A., and Raskin, I. (1995). Benzoic acid 2-hydroxylase, a soluble oxygenase from tobacco, catalyzes salicylic acid biosynthesis. Proc. Nat. Acad. Sci.USA 92, 10413–10417. doi: 10.1073/pnas.92.22.10413
Li, M., Chen, H., Chen, J., Chang, M., Palmer, I. A., Gassmann, W., et al. (2018). TCP transcription factors interact with NPR1 and contribute redundantly to systemic acquired resistance. Front. Plant Sci. 9:1153. doi: 10.3389/fpls.2018.01153
Liu, Y., Guan, X., Liu, S., Yang, M., Ren, J., Guo, M., et al. (2018). Genome-wide identification and analysis of TCP transcription factors involved in the formation of leafy head in Chinese cabbage. Int. J. Mol. Sci. 19, 847–862. doi: 10.3390/ijms19030847
Liu, Y., Schiff, M., and Dinesh-Kumar, S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31, 777–786. doi: 10.1046/j.1365-313X.2002.01394.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Mauch-Mani, B., and Slusarenko, A. J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of Arabidopsis to Peronospora parasitica. Plant Cell 8, 203–212. doi: 10.2307/3870265
Mukhtar, M. S., Carvunis, A. R., Dreze, M., Epple, P., Steinbrenner, J., Moore, J., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601. doi: 10.1126/science.1203659
Nag, A., King, S., and Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 106, 22534–22539. doi: 10.1073/pnas.0908718106
Nath, U., Crawford, B. C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354
Nicolas, M., and Cubas, P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. doi: 10.1016/j.pbi.2016.05.006
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. doi: 10.1038/nature01958
Ribnicky, D. M., Shulaev, V., and Raskin, I. (1998). Intermediates of Salicylic Acid Biosynthesis in Tobacco1. Plant Physiol. 118, 565–572. doi: 10.1104/pp.118.2.565
Rochon, A., Boyle, P., Wignes, T., Fobert, P. R., and Despres, C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18, 3670–3685. doi: 10.1105/tpc.106.046953
Tang, Y., Zhang, Z., Lei, Y., Hu, G., Liu, J., Hao, M., et al. (2019). Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front Plant Sci. 10:526. doi: 10.3389/fpls.2019.00526
Tedman-Jones, J. D., Lei, R., Jay, F., Fabro, G., Li, X., Reiter, W. D., et al. (2008). Characterization of Arabidopsis mur3 mutations that result in constitutive activation of defence in petioles, but not leaves. Plant J. 56, 691–703. doi: 10.1111/j.1365-313X.2008.03636.x
Van Verk, M. C., Bol Jf Fau - Linthorst, H. J. M., and Linthorst, H. J. (2011). WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol. 11, 89–101. doi: 10.1186/1471-2229-11-89
Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., and Hellens, R. P. (2007). Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3:12. doi: 10.1186/1746-4811-3-12
Verberne, M. C., Verpoorte R Fau - Bol, J. F., Bol Jf Fau - Mercado-Blanco, J., Mercado-Blanco J Fau - Linthorst, H. J., and Linthorst, H. J. (2000). Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18, 779–783. doi: 10.1038/77347
Wang, X., Gao, J., Zhu, Z., Dong, X., Wang, X., Ren, G., et al. (2015). TCP transcription factors are critical for the coordinated regulation of isochorismate synthase 1 expression in Arabidopsis thaliana. Plant J. 82, 151–162. doi: 10.1111/tpj.12803
Wang, Y., Li, Y., He, S. P., Gao, Y., Wang, N. N., Lu, R., et al. (2019). A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol. Biochem. 141, 231–239. doi: 10.1016/j.plaphy.2019.06.005
Wessling, R., Epple, P., Altmann, S., He, Y., Yang, L., Henz, S. R., et al. (2014). Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16, 364–375. doi: 10.1016/j.chom.2014.08.004
Wildermuth, M. C., Dewdney J Fau - Wu, G., Wu G Fau - Ausubel, F. M., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Xiong, X. P., Sun, S. C., Zhu, Q. H., Zhang, X. Y., Liu, F., Li, Y. J., et al. (2021). Transcriptome analysis and RNA interference reveal GhGDH2 regulating cotton resistance to Verticillium Wilt by JA and SA Signaling Pathways. Front. Plant Sci. 12:654676. doi: 10.3389/fpls.2021.654676
Yang, L., Teixeira, P. J. P. L., Biswas, S., Finkel, O. M., He, Y., Salas-Gonzalez, I., et al. (2017). Pseudomonas syringae Type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21, 156–168. doi: 10.1016/j.chom.2017.01.003
Zhang, N., Wang, Z., Bao, Z., Yang, L., Wu, D., Shu, X., et al. (2018). MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. Plant J. 93, 66–78. doi: 10.1111/tpj.13757
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, X., Chen, S., and Mou, Z. (2010a). Nuclear localization of NPR1 is required for regulation of salicylate tolerance, isochorismate synthase 1 expression and salicylate accumulation in Arabidopsis. J. Plant Physiol. 167, 144–148. doi: 10.1016/j.jplph.2009.08.002
Zhang, Y., and Li, X. (2019). Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. doi: 10.1016/j.pbi.2019.02.004
Zhang, Y., Xu, S., Ding, P., Wang, D., Cheng, Y. T., He, J., et al. (2010b). Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA. 107, 18220–18225. doi: 10.1073/pnas.1005225107
Zheng, X. Y., Spivey, N. W., Zeng, W., Liu, P. P., Fu, Z. Q., Klessig, D. F., et al. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11, 587–596. doi: 10.1016/j.chom.2012.04.014
Keywords: Gossypium hirsutum, Verticillium dahlia, GhTCP4-like, GhICS1, ghr-miR319b
Citation: Jia P, Tang Y, Hu G, Quan Y, Chen A, Zhong N, Peng Q and Wu J (2022) Cotton miR319b-Targeted TCP4-Like Enhances Plant Defense Against Verticillium dahliae by Activating GhICS1 Transcription Expression. Front. Plant Sci. 13:870882. doi: 10.3389/fpls.2022.870882
Received: 07 February 2022; Accepted: 15 April 2022;
Published: 20 May 2022.
Edited by:
Yangrong Cao, Huazhong Agricultural University, ChinaReviewed by:
Muhammad Sajjad, Chinese Academy of Agricultural Sciences (CAAS), ChinaCopyright © 2022 Jia, Tang, Hu, Quan, Chen, Zhong, Peng and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiahe Wu, d3VqaWFoZUBpbS5hYy5jbg==; Qingzhong Peng, cXpwZW5nanN1QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.