
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 April 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.869432
Monoterpenes are one of the most common groups belonging to the terpenoid family, with a C10 structure comprising of two isoprene units. Most of monoterpenes are volatile plant compounds, and they act as signaling molecules between plants and the environment, particularly as defensive compounds against herbivores and pathogens. In this study, 1,8-cineole synthase (SgCINS) gene was identified and cloned from the leaves of Salvia guaranitica plant. To examine the role of SgCINS in insect resistance, we transformed and expressed this gene into tobacco leaves. The metabolic analysis revealed that the production of various types and amount of terpenoid was increased and decreased in SgCINS overexpression and control lines, respectively, suggesting that overexpressing SgCINS in transgenic tobacco plants lead to an increase in the production of various types of terpenoids and other phytochemical compounds. These results indicated why transgenic tobacco was highly resistant against cotton worm than the highly susceptible control plants. Our results demonstrate that the SgCINS gene can play an important role in plants against cotton worm insect attack, and pave the way for using terpenoids genes for improving resistance to insect attack in higher plants.
Terpenes (i.e., terpenoids or isoprenoids) are considered the largest ecophysiologically active secondary metabolites with over 40,000 compounds known (Liu et al., 2014; Srividya et al., 2015; Ali et al., 2017, 2018, 2021). Terpenoids compounds have various structures and sizes and play numerous functional roles in primary and secondary metabolism (Rashad et al., 2022; Ali et al., 2021; Ali et al., 2017, 2018). In nature, terpenes play crucial roles in plant defense against a variety of organisms (Bakkali et al., 2008; Pavela and Benelli, 2016; Nobsathian et al., 2019; Rashad et al., 2022). In particular, their demonstrated toxicity against different insect pests makes terpenes suitable candidates for the development of eco-friendly pesticides (Isman, 2000; Miresmailli and Isman, 2014). Thus, various plant essential oils (EOs) and their terpenes have proven to have contact and fumigant toxic effects against mosquitoes (Culiseta longiareolata) (Maheswaran and Ignacimuthu, 2012; Pavela, 2015; Govindarajan et al., 2016), cotton leafworms (Spodoptera littoralis) (Liu et al., 2014), house flies (Musca domestica) (Rossi et al., 2012; Rossi and Palacios, 2013; Rossi and Palacios, 2015; Zhang et al., 2017; Benelli et al., 2017, 2018a,2018b), lepidoptera larvae (Lepidoptera genitalia) (Pavela, 2014), stored grain pests (e.g., Tribolium castaneum, Rhyzopertha dominica, Sitophilus oryzae, and Sitophilus zeamais) (Rajendran and Sriranjini, 2008; Herrera et al., 2015), and several other insect pests. The biological activity of plant EOs depends on the qualitative and quantitative analyses of terpene components (Bekele and Hassanali, 2001; Pavela, 2008).
Salvia is a genus from the Lamiaceae family, in which 1,000 species are distributed in Central and South America, East Asia, and West Asia, while the other species are distributed throughout the world (Alziar, 1988/1993; Ali et al., 2017, 2018; Sarrou et al., 2017). Many of salvia species are used in Chinese folk medicine, some of which are considered endemic, cultivated, or wild, including, Salvia przewalskii, S. japonica, S. fruticosa, S. tuxtlensis, S. epidermindis, S. miltiorrhiza, S. aureus, S. santolinifolia, S. hydrangea, S. nipponica, S. tomentosa, S. lavandulifolia, S. chloroleuca, S. glabrescens, S. isensis, S. allagospadonopsis, S. macrochlamys, and S. recognita. Salvia EO is composed of a mixture of terpenoids (i.e., mono-, sesqui-, di-, ses-, and triterpenoids), which are released from different parts of these species and play crucial roles in plant especially in plants’ defense against insect herbivores (Liu et al., 2014; Ali et al., 2017, 2018; Sarrou et al., 2017).
Plant genetic transformation with desired genes encoding factors that are related to resistance to phytophagous insects is a novel and attractive alternative from the synthetic chemical insecticides in the control of various aggressive plant pests (Meeusen and Warren, 1989; Estruch et al., 1997). Over the years, Agrobacterium tumefaciens-mediated transformation system has become a powerful tool to investigate the function of genes that are involved in primary and secondary metabolism (Wu et al., 2008; Zhang et al., 2013; Ali et al., 2017, 2018). Moreover, various studies have successfully used this system to uncover the role of different genes, such as GmAOS and GmNES, encoding allene oxide synthase and a novel monoterpene synthase, respectively, from soybean and determined the roles of these genes to protect plants against insect attack (Wu et al., 2008; Zhang et al., 2013). In this study, we performed the characterization and cloning of SgCINS gene involving 1,8-cineole synthase from S. guaranitica and determined its biological role in resistance against cotton worm through force-feeding-preference tests. These results suggest that overexpression of SgCINS gene in transgenic tobacco may be a better way of enhancing tolerance against the cotton worm insect attack.
Seedlings of S. guaranitica L. were collected from the Wuhan Botanical Garden, China, and grown at National Research Center, Cairo, Egypt. Different tissues were sampled from 2-year-old S. guaranitica plants. For gene cloning, four biological replicates from leaves were sampled and handled. Each replicate consisted of three young and three old leaves from the same plant. In addition, for quantitative real-time PCR (qRT-PCR), three biological replicates were collected from the following six parts (i.e., young leaves, old leaves, stems, bud flowers, flowers, and roots). All samples were directly frozen in liquid nitrogen and then stored at −20°C until RNA extraction.
The nucleotide sequence of SgCINS was selected from our previous RNA-Seq data (Ali et al., 2018). The potential transit peptide and physiochemical properties were analyzed for SgCINS gene using the PROTPARAM software1 and bioinformatics tool2. Furthermore, a phylogenetic tree of SgCINS gene was constructed using PhyML---Phylogeny.fr website3. Moreover, the multiple sequence alignment was carried out using the Clustal Omega software with default parameters4. Putative tissue expression profile from forty-nine Arabidopsis tissues (e.g., dry seed, imbibed seed, 24 h, 1st node, flower stage 12, stamens, Cauline leaf, cotyledon, root, entire rosette after, transition to flowering, flower stage 9, flower stage 10/11, flower stage 12, flower stage 15, flower stage 12-carpels, flower stage 12-petals, flower stage 12-sepals, flower stage 15-carpels, flower stage 15-petals, flower stage 15-sepals, flower stage 15-stamen, flowers stage 15-pedicels, leaf 1 + 2, leaf 7, petiole, leaf 7, distal half, leaf 7, proximal half hypocotyl, root, rosette leaf 2, rosette leaf 4, rosette leaf 6, rosette leaf 8, rosette leaf 10, rosette leaf 12, senescing leaf, shoot apex-inflorescence, shoot apex-transition, shoot apex-vegetative, stem, 2nd internode, mature pollen, seeds stage 3 w/siliques, seeds stage 4 w/siliques, seeds stage 5 w/siliques, seeds stage 6 w/o siliques, seeds stage 7 w/o siliques, seeds stage 8 w/o siliques, seeds stage 9 w/o siliques, seeds stage 10 w/o siliques, vegetative rosette, stem epidermis top of stem, and stem epidermis bottom of stem) were extracted from the public RNA-Seq Atlas of Arabidopsis5. Putative subcellular localization of SgCINS gene products from S. guaranitica was inferred from their sequence similarity to characterized Arabidopsis protein at the Arabidopsis Information Resource6. Subcellular localization profile images were built using Cell Electronic Fluorescent Pictograph Browsers (Cell eFP7).
Total RNA from four biological replicates of S. guaranitica were extracted from the leaf for gene cloning using the TRIzol reagent (Invitrogen, CA, United States) according to the manufacturer’s methods and instructions. Moreover, total RNAs from three biological replicates from each of the plant parts (e.g., young leaves, old leaves, stems, bud flowers, flowers, and roots) were extracted for qRT-PCR. Additionally, total RNAs from three biological replicates for each transgenic tobacco lines were extracted for semiquantitative RT-PCR. Total RNA samples were treated with DNase I (Takara, China). RNA quality was examined on 1.2% agarose gels, and the purity and concentration were analyzed using a Nano-Photometer spectrophotometer (IMPLEN, CA, United States). cDNA synthesis for gene cloning and qRT-PCR was performed with a 10 μg total RNA pool produced by mixing equal volumes of the three RNA replicates in a tube using a commercial reverse transcription kit (M-MLV, China) according to the manufacturer’s protocol (Hussain et al., 2017).
Quantitative RT-PCR was performed by an IQ5 Multicolor Real-Time PCR Detection System (Bio-Rad, United States) as described previously (Ali et al., 2017, 2018, 2021; Hussain et al., 2017) with SYBR Green Master (ROX) (Newbio Industry, China) following the manufacturer’s instructions at a total reaction volume of 20 μl. A gene-specific primer for SgACTIN forward 5′-TGGTTGTGACTTTTGGTCC CA-3′ and reverse 5′-ACAAACCCACGCTTGAGATCC-3′ was used as a reference gene with 155 bp, and SgCINS forward 5′-CTC ACAGCTCTTGGATTCAG-3′ and reverse 5′-GGAAAGATGC TTCGTAGAGTT-3′ with 152 bp length; all the primers were designed using the primer designing tools of IDTdna8, while semiquantitative real-time PCR was performed on an Eppendorf PCR (Mastercycler Nexus PCR Machine from Eppendorf, United Kingdom) system with a total reaction volume of 25 μl. A gene-specific primer for NtEF-1α forward 5′-TGGTTGTGACT TTTGGTCCCA-3′ and reverse 5′-ACAAACCCACGCTTGAGA TCC-3′ was used as a reference gene with 155 bp and SgCINS forward and reverse. The semiquantitative RT-PCR conditions were as follows: predenaturation step at 95°C for 4 min, 35 cycles of amplification (95°C for 30 s, 60°C for 30 s, and 72°C for 1 min), and a final extension step at 72°C for 10 min. The PCR products for semiquantitative RT-PCR were resolved on 1.6% agarose gel, and the expression levels of NtEF-1α and SgCINS genes were detected.
Full-length cDNAs of SgCINS (GenBank: KX893964.1) was obtained by PCR amplification with short and long gene-specific primers designed based on our transcriptome sequencing data (Ali et al., 2018). Leaf cDNA was used as a template for the first PCR, which was performed with short primers, such as SgCINS forward 5′-ATGTGTACCATTAGCATGCATGTATC-3′ and reverse 5′-TTACATTTCTTAAAACCGTGCTGG-3′ with the KOD-Plus DNA polymerase (Toyobo, Japan) with the following cycling conditions: an initial step of 4 min at 95°C followed by 34 cycles of denaturation for 10 s at 98°C; 30 s at 60.4°C and an extension for 2 min at 68°C, and a final extension step for 11 min at 68°C. The first PCR products were used as templates for PCR cloning using long primers, such as SgCINS forward 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTC ATGTGTACCATTAGCATG-3′ and reverse 5′-GGGGACCA CTTTGTACAAGAAAGCTGGGTTTACATTTCTTAAAACCG -3′ with the and KOD-Plus DNA polymerase. The amplified PCR products were purified using the QIAEX II Gel Extraction Kit, China and cloned into the Gateway entry vector pDONR221 using BP Clonase (Gateway BP Clonase II Enzyme mix, Invitrogen). The resulting pDONR221 construct harboring target gene was sequenced, and the LR Clonase (Gateway LR Clonase Enzyme mix, Invitrogen) was used for recombination into the destination vector pB2GW7 for Nicotiana tabacum transformation. Sanger sequencing confirmed that all final constructs contained SgCINS cDNAs. The construct was introduced into A. tumefaciens strain “GV3101” by direct electroporation.
The pB2GW7-SgCINS vector was introduced into A. tumefaciens strain GV3101 using the direct electroporation method. The transformation procedure was performed using the leaf disk method as described previously (Horsch et al., 1984; Lloyd et al., 1986; Burow et al., 1990; Ali et al., 2017) with a few modifications. The transgenic tobacco plants were regenerated from transformed leaf disk following selection on 55 mg/L hygromycin. More than 16 individual transgenic tobacco lines were generated and examined with RT-PCR for positive transgenic lines. The positive plants with good and healthy roots were transferred to the greenhouse for acclimatization. Finally, the transgenic tobacco plants were analyzed for the insect resistance assay and terpenoid profiling.
Experiments were performed on transgenic tobacco and control plants as described in De las Mercedes Dana et al. (2006), Singla-Pareek et al. (2006), Wu et al. (2008), and Liu et al. (2014) with a few modifications. For the force-feeding tests, four mature full green leaves from transgenic tobacco and control plants with nearly similar size were placed on moist filter paper in Petri dishes, and ten third-instar larvae of cotton leafworms (Prodenia litura, Fabricius) were placed on the leaf surface and allowed to feed on transgenic tobacco and control leaves. The relative growth rate (RGR) = (the weight of cotton worm after fed by leaf—the weight before fed)/(the weight of cotton worm before fed), which was monitored by three independent bioassays at different times (e.g., 0, 3, 6, 12, and 24 h) after the start of feeding.
All terpenoid compounds from transgenic tobacco leaves containing SgCINS gene and non-transgenic tobacco leaves (control) at different times (e.g., 0, 3, 6, 12, and 24 h) after the start of insect feeding were extracted and isolated. For this, leaves of transgenic tobacco and control were collected at different times after the start of feeding and were homogenized to a powder using liquid nitrogen with a pestle and mortar, and then the powder was soaked in amber storage bottles [60 ml screw-top vials with silicone/polytetrafluoroethylene (PTFE) septum lids]9, containing n-hexane as a solvent. Later, amber storage bottles were incubated at 37°C along with shaking at 210 rpm for 70 h. Then, the supernatant solvent was collected by centrifugation at 5,000 rpm and 4°C for 10 min, then pipetted into glass vials, and mixed with 1.5 ml of concentrated oils under a stream of nitrogen gas using an evaporator (Organomation; Toption-China-WD-12). The concentrated oils were transferred to a fresh 1.5-ml crimp vial amber glass and were placed on the autosampler of the gas chromatography mass spectrometry (GC-MS) system for the analysis as described previously by Ali et al. (2017, 2018, 2021).
Data for control and transgenic tobacco for the leaf feeding assay experiment were collected. The RGR was monitored by three independent bioassays at different times (e.g., 0, 3, 6, 12, and 24 h) after the start of feeding (Liu et al., 2014 and Wu et al., 2008). These data were analyzed using the SAS 9.2 software (SAS Institute, 2008).
The complete ORF of SgCINS gene from S. guaranitica with 2,263 bp encoded a 731 amino acid protein with 86.636 kDa of molecular mass and 9.34 of predicted theoretical isoelectric point (pI). An analysis using the “iPSORT” program revealed that the SgCINS has a 30-amino-acid sequence (MCTISMHVSILSKPLNSLHRSERRSSNSWP) at the N-terminal, which means our target protein is localized in the chloroplast (i.e., plastid) and mitochondria, where terpene synthases (TPSs) are originated and their biosynthesis takes place. The phylogenetic tree was built using our target gene and TPS from Lamiaceae family and other plant species using PhyML-Phylogeny.fr website. The phylogenetic analysis result classified the SgCINS into the TPS-b subfamily of angiosperm monoterpene synthases (Figure 1). The putative function of SgCINS gene was initially predicted based on the sequence alignment with well-known other TPS sequences and conserved motifs from Lamiaceae family and other plants. Based on these predictions, SgCINS protein has various motifs such as RR (X8) W (residues 55–65), LYEAS (residues 194–199), RXR (residues 311–313), aspartate-rich-DDxxD (residues 348–352), and GTLxEL (residues 355–360) region, which are dominant at similar TPS genes (Zhao et al., 2021; Abbas et al., 2019; Su-Fang et al., 2014; Degenhardt et al., 2009; Ali et al., 2017, 2018; Supplementary Figure 1). Compared to other TPS genes, SgCINS has six domains that were specified by the InterPro protein sequence analysis and classification (InterPro10) database. So, SgCINS protein has six TPS family domains at different amino acid positions such as terpene cyclases, class 1, plant (IPR044814: from 55 to 369); TPS, N-terminal domain (IPR001906: from 65 to 242); TPS, metal-binding domain (IPR005630: from 273 to 386); terpenoid cyclases/protein prenyltransferase alpha-alpha toroid (IPR008930: from 64 to 267); isoprenoid synthase domain superfamily (IPT08949: from 269 to 409) and TPS, and N-terminal domain superfamily (IPR036965: from 65 to 282) (Figure 2). Finally, each protein sequence has one or two or all of these conserved domains belonging to the TPS family (Zhao et al., 2021; Abbas et al., 2019; Su-Fang et al., 2014; Degenhardt et al., 2009; Ali et al., 2017, 2018).
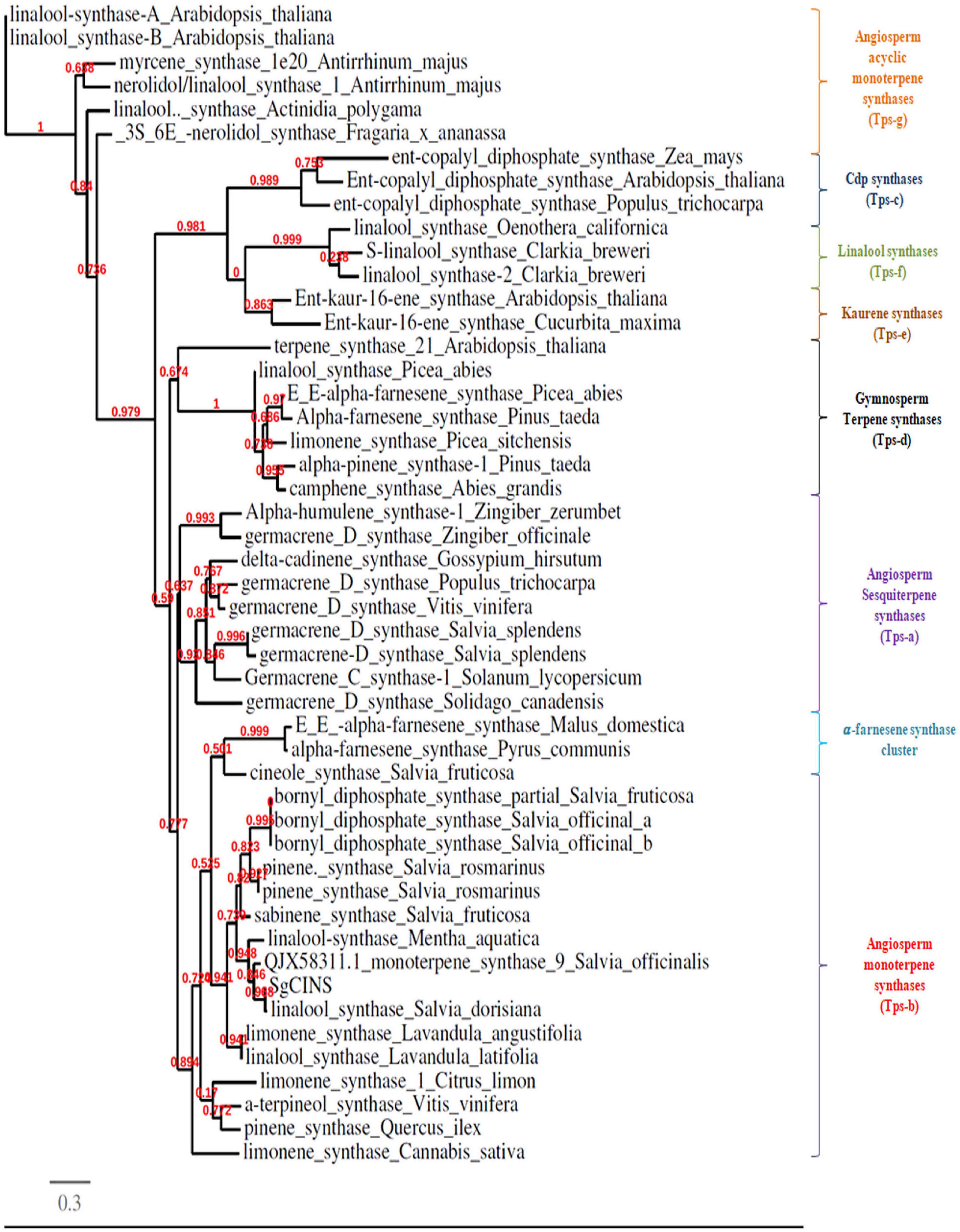
Figure 1. Phylogenetic tree of 1,8-cineole synthase (SgCINS) gene from S. guaranitica with selected terpene synthases genes from other plant species. Bootstrap values based on maximum likelihood are reported at the nodes.
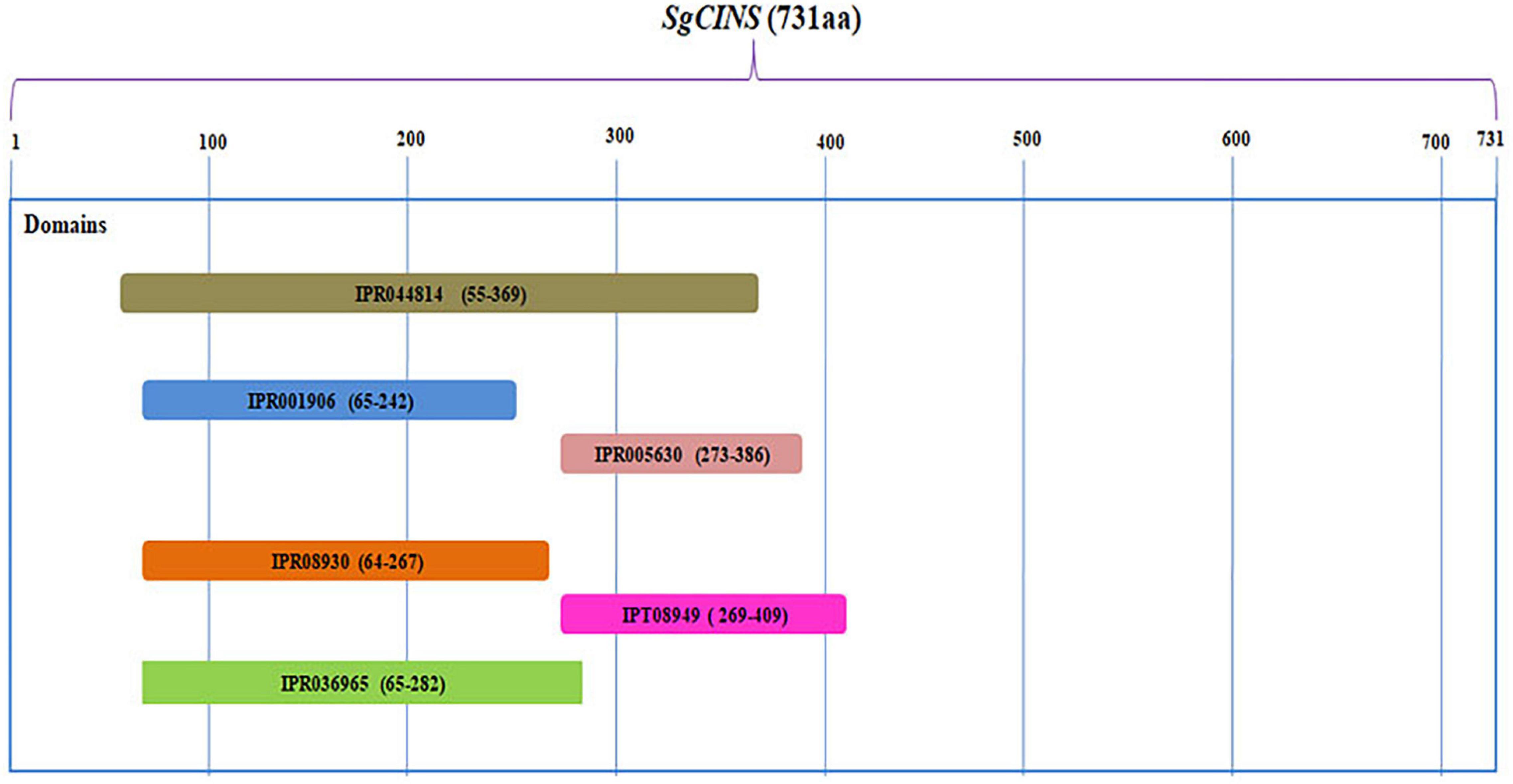
Figure 2. The putative domains structure for SgCINS gene. Six putative domains were detected in SgCINS at different amino acids positions such as, terpene cyclases, class 1, plant (IPR044814); terpene synthase, N-terminal domain (IPR001906); terpene synthase, metal-binding domain (IPR005630); terpenoid cyclases/protein prenyltransferase alpha-alpha toroid (IPR008930); isoprenoid synthase domain superfamily (IPT08949); and terpene synthase, N-terminal domain superfamily (IPR036965).
The putative expression patterns of SgCINS gene of S. guaranitica were uncovered based on their higher similarity with AT3G25830/TPS-CIN gene from A. thaliana, and by transcript analysis across forty-nine Arabidopsis tissues (e.g., dry seed, imbibed seed, 24 h, 1st node, flower stage 12, stamens, Cauline leaf, cotyledon, root, entire rosette after, transition to flowering, flower stage 9, flower stage 10/11, flower stage 12, flower stage 15, flower stage 12-carpels, flower stage 12-petals, flower stage 12-sepals, flower stage 15-carpels, flower stage 15-petals, flower stage 15-sepals, flower stage 15-stamen, flowers stage 15-pedicels, leaf 1 + 2, leaf 7, petiole, leaf 7, distal Half, leaf 7, proximal half hypocotyl, root, rosette leaf 2, rosette leaf 4, rosette leaf 6, rosette leaf 8, rosette leaf 10, rosette leaf 12, senescing leaf, shoot apex-inflorescence, shoot apex-transition, shoot apex-vegetative, stem, 2nd internode, mature pollen, seeds stage 3 w/siliques, seeds stage 4 w/siliques, seeds stage 5 w/siliques, seeds stage 6 w/o siliques, seeds stage 7 w/o siliques, seeds stage 8 w/o siliques, seeds stage 9 w/o siliques, seeds stage 10 w/o siliques, vegetative rosette, stem epidermis top of stem, and stem epidermis bottom of stem) using the Arabidopsis Electronic Fluorescent Pictograph Browsers [eFP browsers (see text footnote 5)]. Interestingly, we observed the highest expression levels of this gene in seeds stage 10 w/o siliques, seeds stage 8 w/o siliques, seeds stage 9 w/o siliques, imbibed seed, 24 h, hypocotyl, stem epidermis bottom of stem, and stem epidermis top of stem (Supplementary Figures 2A,B). Moreover, we further explored the potential subcellular localization of the SgCINS gene products from S. guaranitica based on Arabidopsis protein localization to recognize possible synthesis sites using the Cell eFP browsers (see text footnote 7). From this analysis, the SgCINS gene is localized mainly in the plastid, followed by the mitochondrion and nucleus (Supplementary Figure 2C).
To determine the spatial expression patterns of the SgCINS, we quantified the expression levels of SgCINS transcripts in S. guaranitica young leaves, old leaves, stems, bud flowers, flowers, and roots tissues using the qPCR-PCR (Figure 3). From our results, we found that the SgCINS gene is expressed in all tissues with distinct expression patterns. In roots, SgCINS transcripts gene showed the highest expression levels, followed by young leaves, old leaves, bud flowers, flowers, and stems (Figure 3).
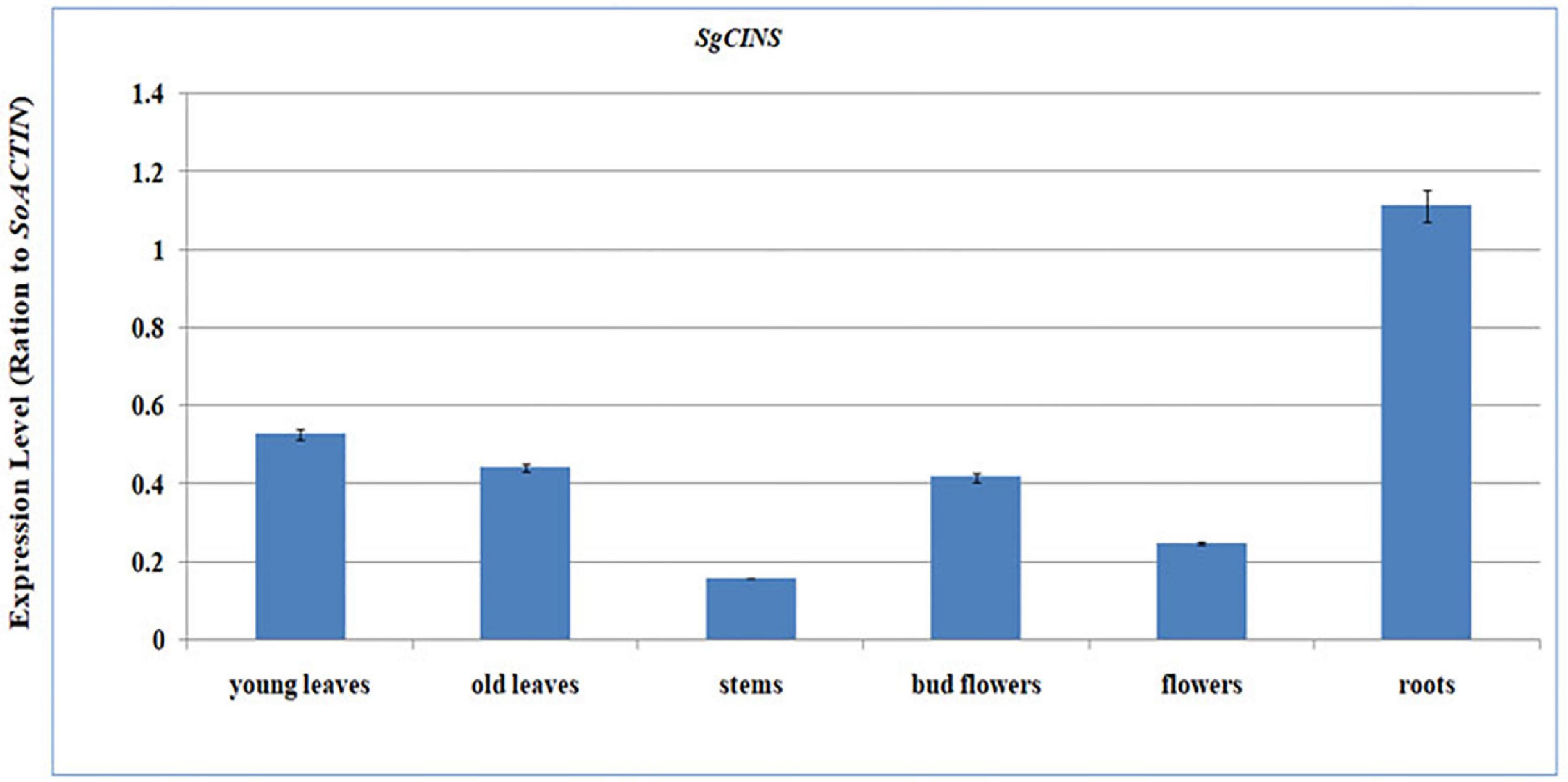
Figure 3. Quantitative RT-PCR validation of expression of SgCINS gene in S. guaranitica. Total RNAs were extracted from young leaves, old leaves, stem, bud flower, flower, and root samples, and the expression of gCINS gene was analyzed using quantitative RT-PCR. SgACTIN was used as the internal reference. The values are means ± SE of three biological replicates.
To further investigate the function of SgCINS gene in plant insect resistance, SgCINS gene from S. guaranitica was cloned and overexpressed in tobacco as a transgenic expression system. The stable constitutive expression of the SgCINS gene in tobacco was carried out by the infection of N. tabacum leaves using A. tumefaciens strain GV3101 carrying pB2GW7-SgCINS under the control of 35S promoter (Figure 4A). Transgenic tobacco plants were successfully generated, which have many big green leaves with elliptical leaf shape, while the non-transgenic tobacco (i.e., control) showed a few numbers of green leaves with a lanceolate leaf shape (Figure 4A). The expression of the SgCINS gene in positive transgenic tobacco was confirmed using semiquantitative RT-PCR (Figure 4B). To investigate the role of SgCINS gene expression in transgenic tobacco in response to insect resistance, force-feeding-preference tests were conducted to evaluate the insect resistance of transgenic tobacco plants with SgCINS gene. Full green and healthy leaves with nearly similar sizes from transgenic and non-transgenic tobacco (i.e., control) plants were placed on a moist filter paper in Petri dishes, and ten third-instar larvae of cotton leafworms (Prodenia litura, Fabricius) were placed on the leaf surface and allowed to feed on transgenic tobacco or control leaves (Figure 5A). The RGR was documented after 0, 3, 6, 12, and 24 h of feeding. Most of cotton leafworms preferred to feeding on the non-transgenic tobacco leaves rather than the SgCINS-expressing tobacco leaves (Figure 5B), suggesting that some substances and compounds in transgenic tobacco had a negative effect on cotton leafworms.
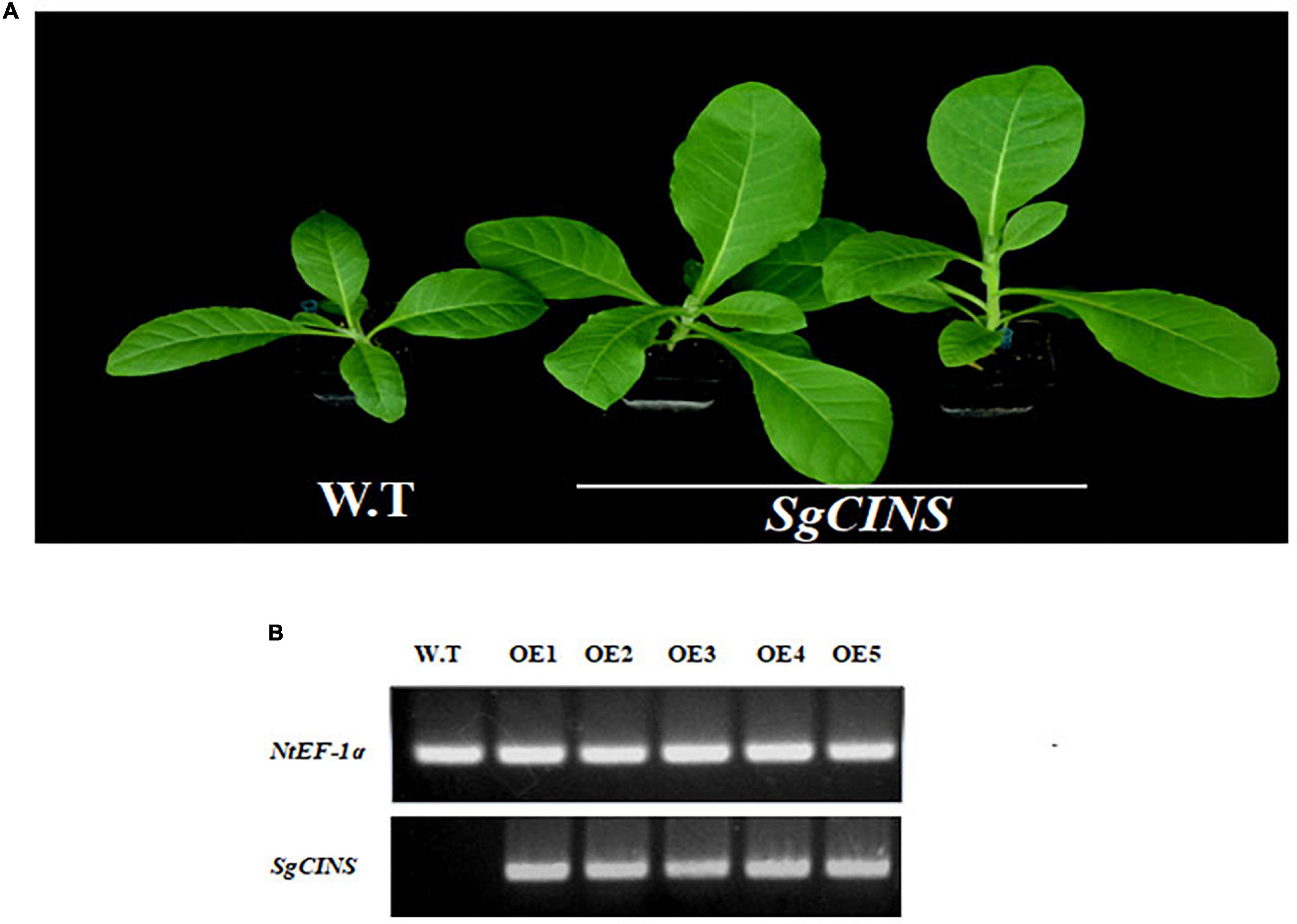
Figure 4. Overexpression of S. guaranitica SgCINS gene in transgenic tobacco. (A) Comparison of the phenotypes of the transgenic N. tabacum and wild-type N. tabacum. (B) Semiquantitative RT-PCR to confirm the expression of SgCINS gene.
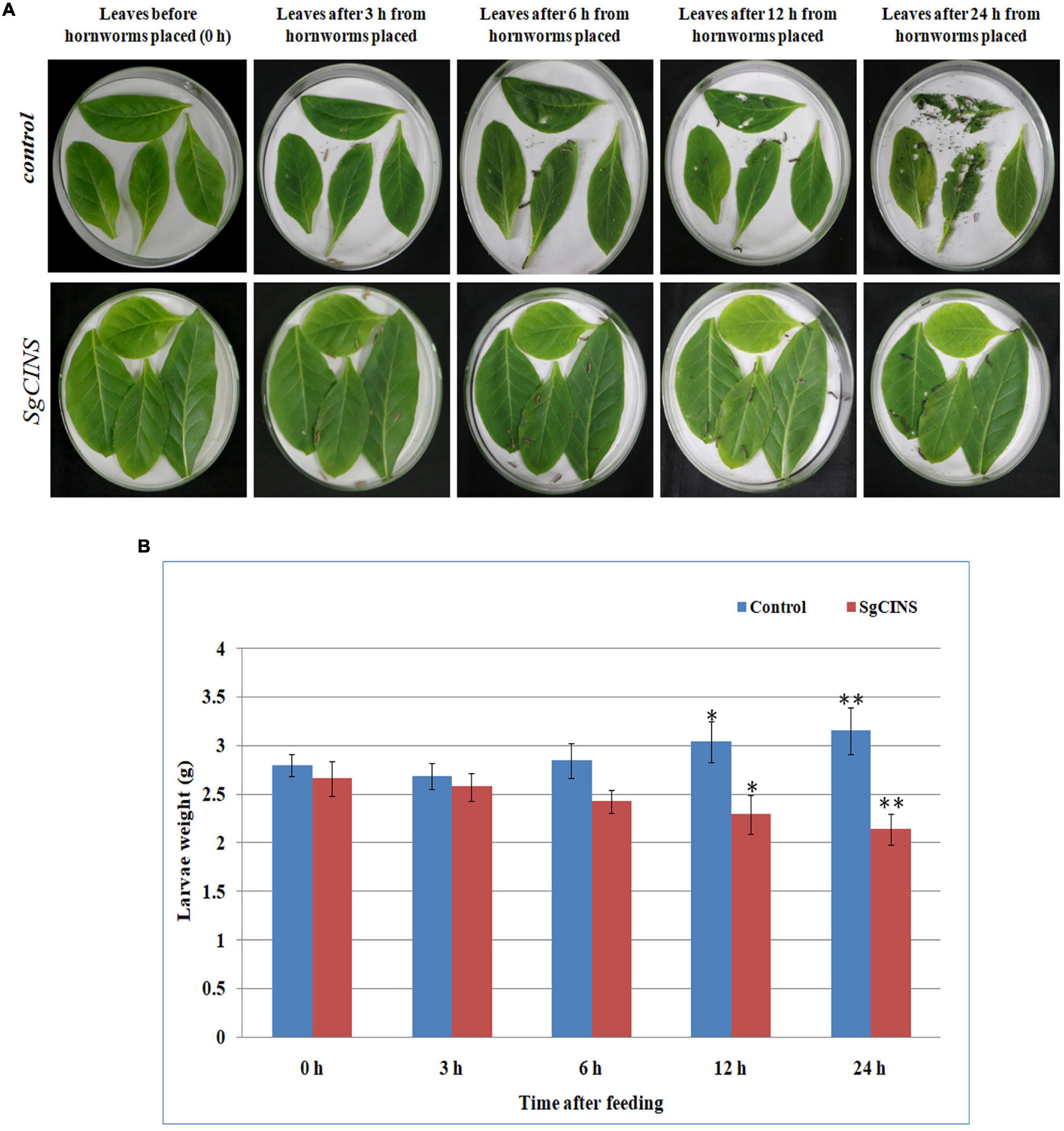
Figure 5. Force-feeding assays for insect resistance on control and transgenic plants after at different times (i.e., 0, 3, 6, 12, and 24 h). (A) Insect force-feeding assays with third-instar larvae of cotton leafworms (Prodenia litura) on N. tabacum. (B) The relative growth rate (RGR) for cotton leaf worms after feeding on control and T1 transgenic tobacco plants at different times (i.e., 0, 3, 6, 12, and 24 h). Each bar represents the mean and SD of the weight and significances are based on the Student’s t-test (*p < 0.05; **p < 0.01).
To gain insight into why transgenic tobacco plants were less damaged by the cotton worms, repelled the cotton leafworms, and inhibited their growth. So, we analyzed the terpenoids compounds from non-transgenic and transgenic tobacco plants overexpressing SgCINS at different times after the start of feeding using the n-hexane extraction method and GC-MS. The GC-MS analysis showed a significant difference between non-transgenic and transgenic tobacco plants in producing different amounts and types of terpenoids at different times after the start of feeding (Table 1, Figure 6, and Supplementary Table 1). As expected, the leaves of transgenic tobacco plants emitted a high level of various terpenoids compared with non-transgenic tobacco leaves. In this study, the improved production of different amounts and types of terpenoids at different times after the start of feeding was observed in transgenic tobacco plants that may be the reason for enhanced tolerance to cotton leafworms attack in transgenic tobacco. For GC-MS analysis, 283 compounds were identified using n-hexane extracts from non-transgenic and transgenic tobacco plants overexpressing SgCINS at different times (e.g., 0, 3, 6, 12, and 24 h) after the start of feeding. The numbers of compounds obtained from non-transgenic and transgenic tobacco leaves at different times (e.g., 0, 3, 6, 12, and 24 h) were 48 and 67 (96.36 and 100%), 67 and 58 (100 and 96.71%), 46 and 57 (92.13 and 100%), 47 and 64 (99.9 and 96.53%), and 65 and 62 (96.5 and 100%), respectively. The quantitative and qualitative analyses of all compounds from the n-hexane extraction are reported in Table 1 and Supplementary Table 1.
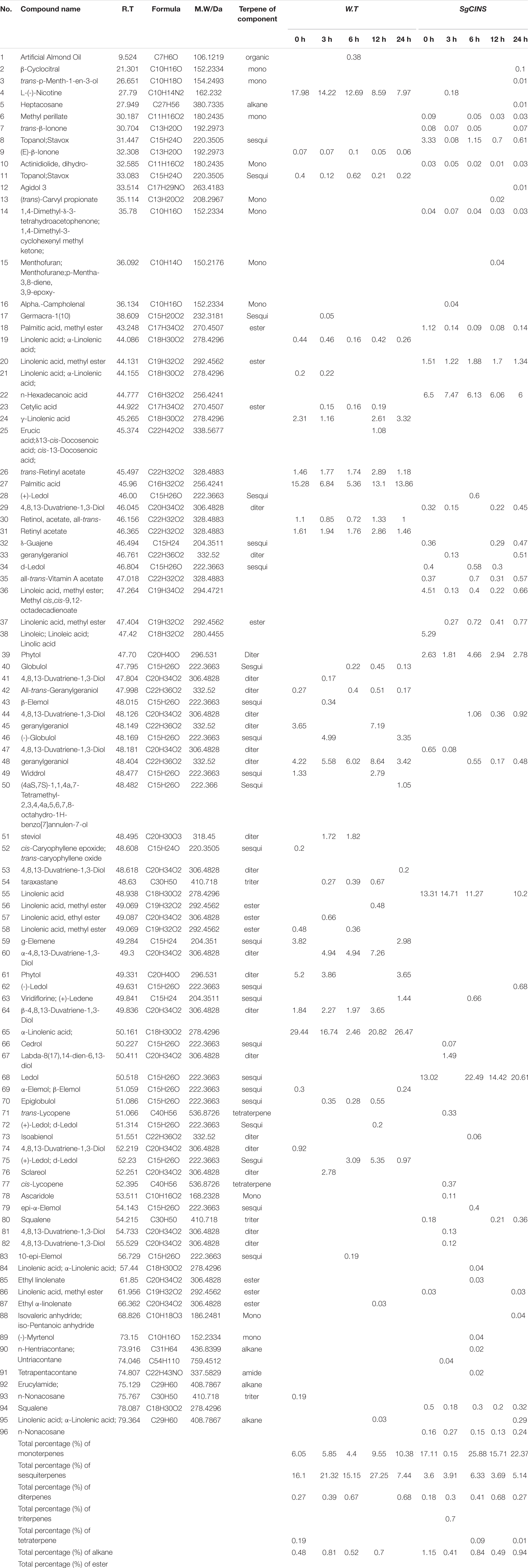
Table 1. The major chemical composition and terpenes from transgenic Nicotiana tabacum leaves at different times from insect force-feeding assays.
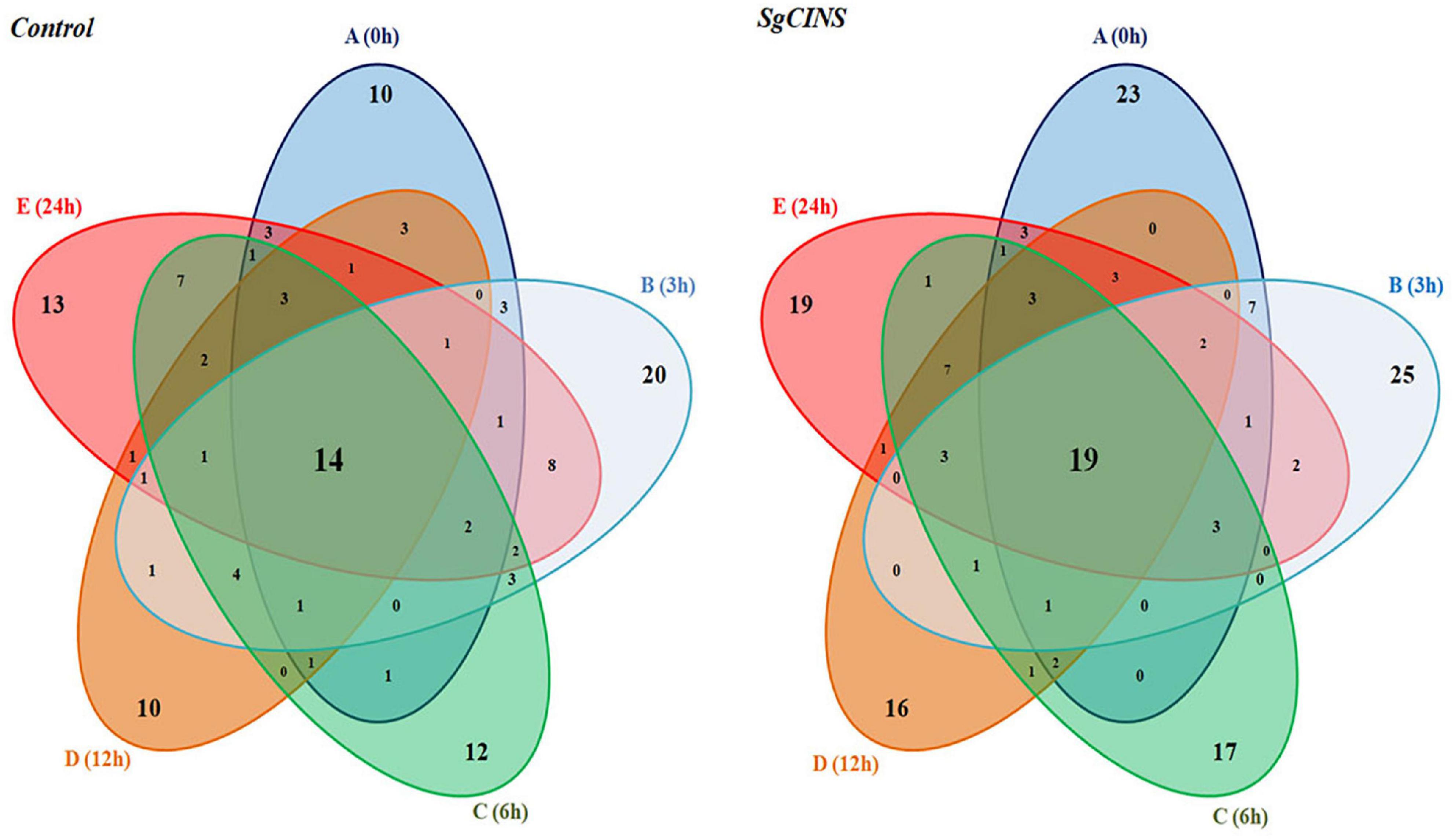
Figure 6. Five-way Venn diagram to show the number of unique and common compounds in the essential oil extracts from control and transgenic tobacco plants at different times from force-feeding assays for insect resistance [e.g., 0 h (A), 3 h (B), 6 h (C), 12 h (D), and 24 h (E)].
The identified compounds are listed based on the name of compounds, retention time, compounds mass, and percentage of peak area (Table 1 and Supplementary Table 1). First, in non-transgenic tobacco leaves, the sesquiterpene compounds were shown as the main group after 24 h (10.38%), followed by 12 h (9.55%), 0 h (6.05%), 3 h (5.85%), and 6 h (4.4%). Furthermore, the diterpene compounds were reported as the main group after 12 h (27.25%), followed by 3 h (21.32%), 0 h (16.1%), 6 h (15.15%), and 24 h (7.44%). In contrast, the triterpene compounds were shown as the main group after 24 h (0.68%), followed by 6 h (0.67%), 3 h (0.39%), and 0 h (0.27%). In addition, one alkane compound was detected after 0 h (0.19%). Moreover, the ester compounds were shown as the main group after 3 h (0.81%), followed by 6 h (0.52%), 3 h (0.48%), and 0 h (0.7%). Finally, the leaves of non-transgenic tobacco after all the times did not produce any monoterpene or tetraterpene compounds (Table 1 and Supplementary Table 1).
Second, in transgenic tobacco leaves overexpressing SgCINS, the monoterpene compounds were shown as the main group after 3 h (0.27%), followed by 24 h (0.24%), 0 h (0.16%), 6 h (0.15%), and 12 h (0.13%). Furthermore, after 6 h, the sesquiterpene compounds were observed to be the main group (25.88%), followed by 24 h (22.37%), 0 h (17.11%), 12 h (15.71%), and 3 h (0.15%). Moreover, the diterpene compounds were reported as the main group after 6 h (6.33%), followed by 24 h (5.14%), 3 h (3.91%), 12 h (3.69%), and 0 h (3.6%), while the triterpenes compounds were shown as the main group after 12 h (0.68%), followed by 6 h (0.41%), 3 h (0.3%), 24 h (0.27%), and 0 h (0.18%). Also, the tetraterpene compound group was detected only in transgenic tobacco leaves after 3 h (0.7%). After 12 h, the alkane compounds were reported as the main group (0.1%), followed by 6 h (0.09%) and 24 h (0.01%). Finally, the ester compounds were shown as the main group after 0 h (1.15%), followed by 24 h (0.94%), 6 h (0.84%), 12 h (0.49%), and 3 h (0.41%) (Table 1 and Supplementary Table 1). In contrast, the twelve hexane extracts from the non-transgenic and transgenic tobacco leaves at different times (e.g., 0, 3, 6, 12, and 24 h) after the start of feeding have unique, common, and major compounds (Figure 6). For example, the unique compounds in non-transgenic and transgenic tobacco leaves at different times were 10 and 23 (0 h), 20 and 25 (3 h), 12 and 17 (6 h), 10 and 16 (12 h), and 13 and 19 (24 h), respectively (Figure 6). Additionally, we found some other common compounds shared among all the times in non-transgenic and transgenic tobacco leaves (14 and 16), respectively.
Salvia is considered one of the largest genera of plants in the Lamiaceae family. Salvia spp. terpenoids were analyzed several decades ago and were found to contain various types and amounts of mono-, sesqui-, di-, ses-, and triterpenes (Aziz et al., 2008; Loizzo et al., 2010; Fateme et al., 2013; Ali et al., 2017, 2018). Despite this, there are only a few number of recent reports that describe the role and the function of terpene genes in insect resistance (Zhang et al., 2013; Liu et al., 2014). In this study, we selected and identified the SgCINS gene from our RNA-Seq sequence data of S. guaranitica (Ali et al., 2018). The phylogenetic tree analysis showed that SgCINS from S. guaranitica forms a clade homology with linalool synthase from S. dorisiana and monoterpene synthase 9 from S. officinalis, and this clade belongs to angiosperm-specific TPS-b, which encodes to monoterpene synthase (Chen et al., 2011; Liu et al., 2014; Yu et al., 2020; Mehmood et al., 2021; Chen et al., 2021; Figure 1). To know the physiological role of SgCINS, we analyzed the putative expression patterns of our gene in forty-nine Arabidopsis tissues based on their higher similarity with AT3G25830/TPS-CIN gene from A. thaliana. This SgCINS gene was detected in all the tissues, and predominantly expressed in seeds stage 10 w/o siliques, seeds stage 8 w/o siliques, seeds stage 9 w/o siliques, imbibed seed, 24 h, hypocotyl, stem epidermis bottom of stem, and stem epidermis top of stem (Supplementary Figures 2A,B). GmTPS21, SoHUMS, SoLINS2, SoNEOD, SgTPSV, SgFARD, and SgGERIS genes from G. max, S. officinalis, and S. guaranitica were reported with higher expression levels in roots and seeds by Liu et al. (2014), Ali et al. (2017, 2018), respectively. Moreover, SgCINS was reported to be localized in the plastid (Supplementary Figure 2C). These results are in line with studies by Taniguchi et al. (2014), Chen et al. (2018), Ali et al. (2021), Wang et al. (2022) who reported that most of TPSs genes were targeted to the plastid or other cell organelles such as mitochondrion and nucleus. Additionally, an organ-specific expression pattern of SgCINS was determined using mRNA samples of 2-year-old S. guaranitica plants using the qRT-PCR. As shown in Figure 3, SgCINS expressions were observed higher in roots (1.13-fold), young leaves (0.5-fold), old leaves (0.42-fold), and bud flowers (0.41-fold) rather than in the flowers (0.22-fold) and stems (0.18-fold). Similar results were obtained by Ali et al. (2017), of which the highest SoCINS expression was found in young leaves, followed by old leaves. We further cloned the full-length cDNAs of SgCINS from S. guaranitica and overexpressed this gene in transgenic tobacco leaves to evaluate the function of SgCINS gene in resistance to cotton leafworms.
Terpenoids are considered from the major active secondary metabolites that may prevent insect and pathogen invasion by forming various chemical barriers (Dudareva et al., 2006; Chen et al., 2018). A number of terpenoid components from many plant species (e.g., Commiphora erythraea; Valeriana jatamansi Jones, Valeriana officinalis L., Nardostachys chinensis Bat, Valeriana officinalis L. var. latifolia Miq., Oryza sativa, and G. max) have been well studied, and most of them have been reported to be insect resistance such as, 1,8-cineole, nerol, (S)-limonene, geraniol, citronellal, citronellic acid, citronellol, linalool, (R)-limonene, (R)-α-pinene, (S)-β-pinene, (R)-pulegone, α-terpinene, γ-terpinene, thymol, camphene, and bornyl acetate (Zhang et al., 2013; Scalerandi et al., 2018; Chen et al., 2018; Feng et al., 2020; Muturi et al., 2020). SgCINS, identified as a 1,8-cineole synthase, was characterized to play an important role in the defense of transgenic tobacco plants against the third-instar larvae of cotton leafworms. In the leaf force-feeding tests, most cotton leafworms preferred the non-transgenic tobacco leaves over the SgCINS-expressing tobacco leaves. The significant differences in RGR of cotton worms (Figure 5B) indicate that the SgCINS overexpressors in transgenic tobacco had stronger resistance against cotton leafworms than non-transgenic tobacco, which had a weaker resistance against cotton leafworms, implying that they may be substances in these transgenic tobacco plants which interfered with the normal growth and development of cotton leafworms. It is sensible to attribute the various levels of resistance of transgenic and non-transgenic tobacco plants against cotton leafworms insect to the different types and amounts of terpenoids they produced (Figure 6 and Table 1).
A number of TPSs genes from a range of plant species have been well characterized, and most of them have the ability to convert the substrate into a single or diverse products during various reaction cycles, which are considered one of the unique traits of this type of gene (Aharoni et al., 2003; Yu and Utsumi, 2009; Ali et al., 2017, 2018; Chen et al., 2018). In this study, we found that the overexpression of SgCINS gene increased the accumulation of different types and amounts of terpenoids. Furthermore, the level of ester, not alkane, increased in overexpression tobacco plants. To understand the mechanisms underlying the SgCINS-related accumulation of terpenoids, we predicted the conserved motifs and domains using the InterPro protein sequence analysis and classification11 database (Ali et al., 2017, 2018). The amino acid sequence analysis indicated that SgCINS has various motifs [e.g., RR (X8) W, LYEAS, RXR, spartate-rich-DDxxD, and GTLxEL] and six domains at different amino acids positions (e.g., terpene cyclases, class 1, plant, TPS, N-terminal domain, TPS, metal-binding domain, terpenoid cyclases/protein prenyltransferase alpha-alpha toroid, isoprenoid synthase domain superfamily, and TPS, N-terminal domain superfamily). In previous studies, each protein sequence has one or two or some of these conserved motifs and domains belonging to the TPS family (Zhao et al., 2021; Chen et al., 2020; Abbas et al., 2019; Su-Fang et al., 2014; Degenhardt et al., 2009; Ali et al., 2017, 2018). As described above, this property is found in most of the characterized TPS synthases genes. For example, overexpression of A. grandis γ-humulene synthase, a sesquiterpene synthase gene with two DDxxD motifs that located on the opposite sides, can generate 52 compounds from sesquiterpenes (Steele et al., 1998; Ali et al., 2018). In addition, ectopic expression of rice OsTPS19 can catalyze the formation of 16 compounds from monoterpenes such as, α-thujene, α-pinene, cis-ocimene, sabinene, myrcene, α-phellandrene, (S)-limonene, trans-ocimene, α-terpinene, γ-terpinene, cis-sabinene hydrate, trans-sabinene hydrate, α-terpinolene, terpinen-4-ol, neo alloocimene, and α-terpineol hydrate (Chen et al., 2018). In another example, (+)-sabinene synthase from S. officinalis catalyzes the formation of (+)-sabinene, terpinene, terpinolene, limonene, and myrcene in in vitro assays (Wise et al., 1998; Ali et al., 2018). Also, overexpression of lemon α-zingiberene synthase catalyzed the formation of α-zingiberene and other sesquiterpenes and monoterpenes in tomatoes (Davidovich-Rikanati et al., 2008). In our previous studies, we found that the overexpression of terpenoids and TPS genes, such as SoLINS, SoNEOD, SoTPS6,SoSABS, SoCINS, SgGPS, SgFPPS, and SgLINS from S. officinalis and S. guaranitica increased the accumulation of sesquiterpenes and monoterpenes in N. tabacum and A. thaliana plants (Ali et al., 2017, 2018). Finally, this property is found in our target gene, which catalyzes the formation of different types and amounts of terpenoids.
We cloned a SgCINS gene that could encode a 1,8-cineole synthase and catalyze the biosynthesis of monoterpene from leaves of S. guaranitica plant. To examine the role of SgCINS in insect resistance, we transformed and expressed this gene into tobacco leaves. The availability of transgenic tobacco plants with overexpression of SgCINS made it possible to determine the biological function of SgCINS. In this study, we were particularly interested in the potential role of SgCINS in defense against cotton leafworms. At different times after the start of feeding (e.g., 0, 3, 6, 12, and 24 h), the RGR and the metabolic analysis were quantified. The RGR for cotton leafworms feeding on SgCINS overexpression tobacco was smaller than those on non-transgenic tobacco. In contrast, the levels of terpenoids accumulation from transgenic tobacco were larger than those from non-transgenic tobacco plants. Moreover, detecting high levels of terpenoids in transgenic tobacco might provide evidence that the various types of terpenoids are the principal compound that inhibits the growth and development of cotton leafworms. The results in this study contribute to our understanding the role of the SgCINS gene in tobacco defense against cotton leafworms. Most importantly, our study paves the way for engineering transgenic plants with enhanced insect resistance and reduces the application of pesticides.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, KX893964.1.
MA conceived and designed the study. MA, DD, AA, NE, and DA performed the experiments. MA wrote the draft for the article. DD, AA, NE, and DA reviewed the final draft of the manuscript. All authors discussed the results and commented on the manuscript and participated in the analysis of the data, reading and, approving the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This study was under the activity of the Egyptian Desert Gene Bank (EDGB), Genetic Resources Department, and Desert Research Center (DRC), Egypt. Our special thanks to Ahmed Ali at the Department of Plant Agricultural, Faculty of Agriculture Science, Al-Azhar University, Assiut, Egypt, for valuable support. We also owe thank to Mohamed Hamdy Amar and Wael Moussa at Desert Research Center (DRC) for constructive comment and help. We also acknowledge Samah Elsayed Ali for her constructive comment and help.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.869432/full#supplementary-material
Abbas, F., Yanguo, K., Rangcai, Y., and Yanping, F. (2019). Functional characterization and expression analysis of two terpene synthases involved in floral scent formation in Lilium ‘Siberia’. Planta 249, 71–93. doi: 10.1007/s00425-018-3006-7
Aharoni, A., Giri, A. P., Deuerlein, S., Griepink, F., de Kogel, W. J., Verstappen, F. W., et al. (2003). Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell. 15, 2866–2884. doi: 10.1105/tpc.016253
Ali, M., Li, P., She, G., Chen, D., Wan, X., and Zhao, J. (2017). Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis). Sci. Rep. 7:16074. doi: 10.1038/s41598-017-15478-3
Ali, M., Hussain, R. M., Rehman, N. U., She, G., Li, P., Wan, X., et al. (2018). De novo transcriptome sequencing and metabolite profiling analyses reveal the complex metabolic genes involved in the terpenoid biosynthesis in blue anise sage (Salvia guaranitica L.). DNA Res. 25, 597–617. doi: 10.1093/dnares/dsy028
Ali, M., Miao, L., Hou, Q., Darwish, D. B., Alrdahe, S. S., Ali, A., et al. (2021). Overexpression of terpenoid biosynthesis genes from garden sage (Salvia officinalis) modulates rhizobia interaction and nodulation in soybean. Front. Plant Sci. 12:783269. doi: 10.3389/fpls.2021.783269
Alziar, G. (1988/1993). Catalogue synonymique des Salvia L. dumonde (Lamiaceae). I.–VI. Biocosme Mesoge’en 5, 87–136.
Aziz, R. A., Hamed, F. K., and Abdulah, N. A. (2008). Determination of the main components of the essential oil extracted from Salvia fruticosa by sing GC and GC-MS DAMASCUS. J. AGR. SCI. 24, 223–236.
Bakkali, F., Averbeck, S., Averbeck, D., and Idaomar, M. (2008). Biological effects of essential oils–a review. Food Chem. Toxicol. 46, 446–475. doi: 10.1016/j.fct.2007.09.106
Bekele, J., and Hassanali, A. (2001). Blend effects in the toxicity of the essential oil constituents of Ocimum kilimandscharicum and Ocimum kenyense (Labiateae) on two post-harvest insect pests. Phytochemistry 57, 385–391. doi: 10.1016/S0031-9422(01)00067-X
Benelli, G., Pavela, R., Giordani, C., Casettari, L., Curzi, G., Cappellacci, L., et al. (2018a). Acute and sub-lethal toxicity of eight essential oils of commercial interest against the filariasis mosquito Culex quinquefasciatus and the housefly Musca domestica. Ind. Crops Prod. 112, 668–680. doi: 10.1016/j.indcrop.2017.12.062
Benelli, G., Pavela, R., Petrelli, R., Cappellacci, L., Santini, G., Fiorini, D., et al. (2018b). The essential oil from industrial hemp (Cannabis sativa L.) by-products as an effective tool for insect pest management in organic crops. Ind. Crops Prod. 122, 308–315. doi: 10.1016/j.indcrop.2018.05.032
Benelli, G., Pavela, R., Ricciutelli, M., Lupidi, G., and Maggi, F. (2017). Efficacy of the volatile Oil from water celery (Helosciadium nodiflorum, Apiaceae) against the filariasis vector Culex quinquefasciatus, the housefly Musca domestica, and the African Cotton Leafworm Spodoptera littoralis. Chem. Biodivers. 14:e1700376. doi: 10.1002/cbdv.201700376
Burow, M. D., Chlan, C. A., Sen, P., Lisca, A., and Murai, N. (1990). High-frequency generation of transgenic tobacco plants after modified leaf disk cocultivation with Agrobacterium tumefaciens. Plant Mol. Biol. Rep. 8, 124–139. doi: 10.1007/bf02669766
Chen, F., Tholl, D., Bohlmann, J., and Pichersky, E. (2011). The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229. doi: 10.1111/j.1365-313X.2011.04520.x
Chen, Q., Li, J., Liu, Z., Mitsuhashi, T., Zhang, Y., Liu, H., et al. (2020). Molecular basis for sesterterpene diversity produced by plant terpene synthases. Plant Commun. 1:100051. doi: 10.1016/j.xplc.2020.100051
Chen, X., Chen, H., Yuan, J. S., Köllner, T. G., Chen, Y., Guo, Y., et al. (2018). The rice terpene synthase gene OsTPS19 functions as an (S)-limonene synthase in planta, and its overexpression leads to enhanced resistance to the blast fungus Magnaporthe oryzae. Plant Biotechnol. J. 16, 1778–1787. doi: 10.1111/pbi.12914
Chen, Z., Qi, X., Yu, X., Zheng, Y., Liu, Z., Fang, H., et al. (2021). Genome-Wide analysis of terpene synthase gene family in Mentha longifolia and catalytic activity analysis of a single terpene synthase. Genes. 12:518. doi: 10.3390/genes12040518
Davidovich-Rikanati, R., Lewinsohn, E., Bar, E., Iijima, Y., Pichersky, E., and Sitrit, Y. (2008). Overexpression of the lemon basil alpha-zingiberene synthase gene increases both mono- and sesquiterpene contents in tomato fruit. Plant J. 56, 228–238. doi: 10.1111/j.1365-313X.2008.03599.x
De las Mercedes Dana, M., Pintor-Toro, J. A., and Cubero, B. (2006). Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant physiol. 142, 722–730. doi: 10.1104/pp.106.086140
Degenhardt, J., Köllner, T. G., and Gershenzon, J. (2009). Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70, 1621–1637. doi: 10.1016/j.phytochem.2009.07.030
Dudareva, N., Negre, F., Nagegowda, D. A., and Orlova, I. (2006). Plant volatiles: recent advances and future perspectives. Crit. Rev. Plant Sci. 25, 417–440. doi: 10.1080/07352680600899973
Estruch, J. J., Carozzi, N. B., Desai, N., Duck, N. B., Warren, G. W., and Koziel, M. G. (1997). Transgenic plants: an emerging approach to pest control. Nat. Biotechnol. 15, 137–141. doi: 10.1038/nbt0297-137
Fateme, A. M., Mohammad, H. F., Abdolhossein, R., Ali, Z., and Maryam, S. (2013). Volatile constituents of Salvia compressa and logochilus macranthus, two labiatae herbs growing wild in iran. Res. J. Recent Sci. 2, 66–68.
Feng, Y. X., Wang, Y., Geng, Z. F., Zhang, D., Almaz, B., and Du, S. S. (2020). Contact toxicity and repellent efficacy of valerianaceae spp. to three stored-product insects and synergistic interactions between two major compounds camphene and bornyl acetate. Ecotoxicol. Environ. Saf. 190:110106. doi: 10.1016/j.ecoenv.2019.110106
Govindarajan, M., Rajeswary, M., Hoti, S. L., Bhattacharyya, A., and Benelli, G. (2016). Eugenol, α-pinene and β-caryophyllene from Plectranthus barbatus essential oil as eco-friendly larvicides against malaria, dengue and Japanese encephalitis mosquito vectors. Parasitol. Res. 115, 807–815. doi: 10.1007/s00436-015-4809-0
Herrera, J. M., Zunino, M. P., Dambolena, J. S., Pizzolitto, R. P., Gañan, N. A., Lucini, E. I., et al. (2015). Terpene ketones as natural insecticides against Sitophilus zeamais. Ind. Crops Prod. 70, 435–442. doi: 10.1016/j.indcrop.2015.03.074
Horsch, R. B., Fraley, R. T., Rogers, S. G., Sanders, P. R., Lloyd, A., and Hoffmann, N. (1984). Inheritance of functional foreign genes in plants. Science. 223, 496–498. doi: 10.1126/science.223.4635.496
Hussain, R. M., Ali, M., Feng, X., and Li, X. (2017). The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 17:55. doi: 10.1186/s12870-017-1001-y
Isman, M. B. (2000). Plant essential oils for pest and disease management. Crop Protect. 19, 603–608. doi: 10.1016/S0261-2194(00)00079-X
Liu, J., Huang, F., Wang, X., Zhang, M., Zheng, R., Wang, J., et al. (2014). Genome-wide analysis of terpene synthases in soybean: functional characterization of GmTPS3. Gene. 544, 83–92. doi: 10.1016/j.gene.2014.04.046
Lloyd, A. M., Barnason, A. R., Rogers, S. G., Byrne, M. C., Fraley, R. T., and Horsch, R. B. (1986). Transformation of Arabidopsis thaliana with Agrobacterium tumefaciens. Science. 234, 464–466. doi: 10.1126/science.234.4775.464
Maheswaran, R., and Ignacimuthu, S. (2012). A novel herbal formulation against dengue vector mosquitoes Aedes aegypti and Aedes albopictus. Parasitol. Res. 110, 1801–1813. doi: 10.1007/s00436-011-2702-z
Meeusen, R. L., and Warren, G. (1989). Insect control with genetically engineered crops. Ann. Rev. Entomol. 34, 373–381. doi: 10.1146/annurev.en.34.010189.002105
Mehmood, N., Yuan, Y., Ali, M., Ali, M., Iftikhar, J., Cheng, C., et al. (2021). Early transcriptional response of terpenoid metabolism to Colletotrichum gloeosporioides in a resistant wild strawberry Fragaria nilgerrensis. Phytochemistry 181, 112590. doi: 10.1016/j.phytochem.2020.112590
Miresmailli, S., and Isman, M. B. (2014). Botanical insecticides inspired by plant–herbivore chemical interactions. Trend. Plant Sci. 19, 29–35. doi: 10.1016/j.tplants.2013.10.002
Muturi, E. J., Selling, G. W., Doll, K. M., Hay, W. T., and Ramirez, J. L. (2020). Leptospermum scoparium essential oil is a promising source of mosquito larvicide and its toxicity is enhanced by a biobased emulsifier. PLoS One 15:e0229076. doi: 10.1371/journal.pone.0229076
Loizzo, M. R., Menichini, F., Tundis, R., Bonesi, M., Nadjafi, F., Saab, A. M., et al. (2010). Comparative chemical composition and antiproliferative activity of aerial parts of Salvia leriifolia Benth. and Salvia acetabulosa L. essential oils against human tumor cell in vitro models. J. Med. Food 13, 62–69. doi: 10.1089/jmf.2009.0060
Nobsathian, S., Ruttanaphan, T., and Bullangpoti, V. (2019). Insecticidal Effects of triterpene glycosides extracted from Holothuria atra (Echinodermata: Holothuroidea) against Spodoptera litura (Lepidoptera: Noctuidae). J. Econ. Entomol. 112, 1683–1687. doi: 10.1093/jee/toz075
Pavela, R. (2008). Acute and synergistic effects of some monoterpenoid essential oil compounds on the house fly (Musca domestica L.). J. Essent. Oil Bear. Plants 11, 451–459. doi: 10.1080/0972060X.2008.10643653
Pavela, R. (2014). Acute, synergistic and antagonistic effects of some aromatic compounds on the Spodoptera littoralis boisd. (Lep., Noctuidae) larvae. Ind. Crops Prod. 60, 247–258. doi: 10.1016/j.indcrop.2014.06.030
Pavela, R. (2015). Acute toxicity and synergistic and antagonistic effects of the aromatic compounds of some essential oils against Culex quinquefasciatus say larvae. Parasitol. Res. 114, 3835–3853. doi: 10.1007/s00436-015-4614-9
Pavela, R., and Benelli, G. (2016). Essential oils as ecofriendly biopesticides? challenges and constraints. Trend. Plant Sci. 21, 1000–1007. doi: 10.1016/j.tplants.2016.10.005
Rajendran, S., and Sriranjini, V. (2008). Plant products as fumigants for stored-product insect control. J. Stored Prod. Res. 44, 126–135. doi: 10.1016/j.jspr.2007.08.003
Rashad, Y. M., Abdel Razik, E. S., and Darwish, D. B. (2022). Essential oil from Lavandula angustifolia elicits expression of three SbWRKY transcription factors and defense-related genes against sorghum damping-off. Sci. Rep. 12:857. doi: 10.1038/s41598-022-04903-x
Rossi, Y. E., and Palacios, S. M. (2013). Fumigant toxicity of Citrus sinensis essential oil on Musca domestica L. adults in the absence and presence of a P450 inhibitor. Acta. Trop. 127, 33–37. doi: 10.1016/j.actatropica.2013.03.009
Rossi, Y. E., and Palacios, S. M. (2015). Insecticidal toxicity of Eucalyptus cinerea essential oil and 1,8-cineole against Musca domestica and possible uses according to the metabolic response of flies. Ind. Crops Prod. 63, 133–137. doi: 10.1016/j.indcrop.2014.10.019
Rossi, Y. E., Canavoso, L., and Palacios, S. M. (2012). Molecular response of Musca domestica L. to Mintostachys verticillata essential oil, (4R) (+)-pulegone and menthone. Fitoterapia. 83, 336–342. doi: 10.1016/j.fitote.2011.11.019
Sarrou, E., Ioannis, G., Aliki, X., Domenico, M., Stefan, M., Panagiotis, M., et al. (2017). Genetic diversity and metabolic profile of Salvia officinalis populations: implications for advanced breeding strategies. Planta 246, 201–215. doi: 10.1007/s00425-017-2666-z
Scalerandi, E., Flores, G. A., Palacio, M., Defagó, M. T., Carpinella, M. C., Valladares, G., et al. (2018). Understanding synergistic toxicity of terpenes as insecticides: contribution of metabolic detoxification in Musca domestica. Front. Plant Sci. 9:1579. doi: 10.3389/fpls.2018.01579
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., and Sopory, S. K. (2006). Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 140, 613–623. doi: 10.1104/pp.105.073734
Srividya, N., Davis, E. M., Croteau, R. B., and Lange, B. M. (2015). Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proc. Natl. Acad. Sci. U.S.A. 112, 3332–3337. doi: 10.1073/pnas.1501203112
Steele, C. L., Crock, J., Bohlmann, J., and Croteau, R. (1998). Sesquiterpene synthases from grand fir (Abies grandis) – comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J. Biol. Chem. 273, 2078–2089. doi: 10.1074/jbc.273.4.2078
Su-Fang, E., Zeti-Azura, M., Roohaida, O., Noor, A. S., Ismanizan, I., and Zamri, Z. (2014). Functional Characterization of Sesquiterpene Synthase from Polygonum minus. ScientificWorldJournal 2014:840592. doi: 10.1155/2014/840592
Taniguchi, S., Miyoshi, S., Tamaoki, D., Yamada, S., Tanaka, K., Uji, Y., et al. (2014). Isolation of jasmonate-induced sesquiterpene synthase of rice: product of which has an antifungal activity against Magnaporthe oryzae. J. Plant Physiol. 171, 625–632. doi: 10.1016/j.jplph.2014.01.007
Wang, Y., Yang, Q., Zhu, Y., Zhao, L., Ju, P., Wang, G., et al. (2022). MrTPS3 and MrTPS20 are Responsible for β-Caryophyllene and α-Pinene Production, Respectively, in Red Bayberry (Morella rubra). Front. Plant Sci 12:798086. doi: 10.3389/fpls.2021.798086
Wise, M. L., Savage, T. J., Katahira, E., and Croteau, R. (1998). Monoterpene synthases from common sage (Salvia officinalis) – cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1, 8-cineole synthase, and (+)-bornyl diphosphate synthase. J. Biol. Chem. 273, 14891–14899. doi: 10.1074/jbc.273.24.14891
Wu, J., W, Q., Wu, Q., Gai, J., and Yu, D. (2008). Constitutive overexpression of AOS-like gene from soybean enhanced tolerance to insect attack in transgenic tobacco. Biotechnol. Lett. 30, 1693–1698. doi: 10.1007/s10529-008-9742-1
Yu, F., and Utsumi, R. (2009). Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell. Mol. Life Sci. 66, 3043–3052. doi: 10.1007/s00018-009-0066-7
Yu, Z., Zhao, C., Zhang, G., Teixeira da Silva, J. A., and Duan, J. (2020). Genome-wide identification and expression profile of TPS gene family in Dendrobium officinale and the role of DoTPS10 in linalool biosynthesis. Int. J. Mol. Sci. 21:5419. doi: 10.3390/ijms21155419
Zhang, M., Liu, J., Li, K., and Yu, D. (2013). Identification and characterization of a novel monoterpene synthase from soybean restricted to neryl diphosphate precursor. PLoS One. 8:e75972. doi: 10.1371/journal.pone.0075972
Zhang, Z., Xie, Y., Wang, Y., Lin, Z., Wang, L., and Li, G. (2017). Toxicities of monoterpenes against housefly, Musca domestica L. (Diptera: Muscidae). Environ. Sci. Pollut. Res. Int. 24, 24708–24713. doi: 10.1007/s11356-017-0219-4
Keywords: Salvia guaranitica, terpene synthase (TPS), transgenic Nicotiana tabacum, Spodoptera littoralis (cotton leafworm), 1-8-cineole synthase (SgCINS)
Citation: Ali M, Alshehri D, Alkhaibari AM, Elhalem NA and Darwish DBE (2022) Cloning and Characterization of 1,8-Cineole Synthase (SgCINS) Gene From the Leaves of Salvia guaranitica Plant. Front. Plant Sci. 13:869432. doi: 10.3389/fpls.2022.869432
Received: 04 February 2022; Accepted: 28 February 2022;
Published: 15 April 2022.
Edited by:
Ahmad M. Alqudah, Aarhus University, DenmarkReviewed by:
Ihteram Ullah, Gomal University, PakistanCopyright © 2022 Ali, Alshehri, Alkhaibari, Elhalem and Darwish. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammed Ali, bW9oYW1tZWRhbGlkcmNAZ21haWwuY29t, Mjg0ODQ2NjE2MUBxcS5jb20=, orcid.org/0000-0001-9232-1781
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.