- 1College of Forestry, Basic Forestry and Proteomics Research Center, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
Light is one of the most important environmental factors affecting growth and geographic distribution of forestry plants. Moso bamboo is the largest temperate bamboo on earth and an important non-woody forestry species that serves not only important functions in the economy of rural areas but also carbon sequestration in the world. Due to its decades-long reproductive timing, the germplasm of moso bamboo cannot be readily improved by conventional breeding methods, arguing for a greater need to study the gene function and regulatory mechanisms of this species. We systematically studied the photoregulatory mechanisms of the moso bamboo (Phyllostachys edulis) cryptochrome 1, PheCRY1. Our results show that, similar to its Arabidopsis counterpart, the bamboo PheCRY1s are functionally restricted to the blue light inhibition of cell elongation without an apparent activity in promoting floral initiation. We demonstrate that PheCRY1s undergo light-dependent oligomerization that is inhibited by PheBIC1s, and light-dependent phosphorylation that is catalyzed by PhePPKs. We hypothesize that light-induced phosphorylation of PheCRY1s facilitate their degradation, which control availability of the PheCRY1 proteins and photosensitivity of bamboo plants. Our results demonstrate the evolutionary conservation of not only the function but also photoregulatory mechanism of PheCRY1 in this monocot forestry species.
Introduction
Bamboo is one of the most important non-timber forestry plants, covering 30 million hectares worldwide. Moso bamboo belongs to the grass family of Gramineae, but it is considered and often cultivated as an agroforestry and in recent years a carbon sequestering plant species. It has some remarkably unique characteristics, including the “fastest growing plant” record of any plant on the earth, among the tallest bamboo species, the long vegetative phase of more than 50 years before flowering, and the unique underground rhizome with a monopodial growth habit (Wilson and Loomis, 1967). There is a great need to improve bamboo agroforestry traits, but new bamboo cultivars cannot be readily generated through classical breeding methods, due to its long vegetative phase. A better understanding of the genes and gene functions of this species is critical to its future improvement.
Light is arguably the most critical environmental factor essential to both photosynthesis and plant development. Plants depend on blue light, red/far-red light, and UV-B photosensory receptor systems to control photosynthesis and development. Cryptochromes (CRY) are blue light receptors that control plant growth and development by mediating light-dependent regulation of gene expression (Wang and Lin, 2020). The prototype plant CRYs are Arabidopsis CRY1 and CRY2, which mediate primarily blue light inhibition of cell elongation and photoperiodic promotion of floral initiation (Ahmad and Cashmore, 1993; Mockler et al., 2003). It is previously reported that Arabidopsis CRY1 and CRY2 undergo these photoreactions: light-induced photo-oligomerization that is inhibited by the Blue-light Inhibitor of Cryptochromes (BICs), blue light-induced phosphorylation that is catalyzed by Photoregulatory Protein Kinases (PPKs), and polyubiquitination that is catalyzed by the E3 ubiquitin ligase Cul4COP1/SPAs and Cul3LRBs (Shalitin et al., 2002, 2003; Wang et al., 2016; Liu et al., 2017, 2020, 2022; Ma et al., 2020, 2021; Shao et al., 2020; Chen et al., 2021; Miao et al., 2021). Biological functions of CRYs have been reported in various plant species, including tomato (Ninu et al., 1999), soybean (Zhang et al., 2008; Lyu et al., 2021), Brassica (Chatterjee et al., 2006), pea (Platten et al., 2005), poplar (Mao et al., 2014), rice (Matsumoto et al., 2003), barley (Szucs et al., 2006), sorghum (Zhou et al., 2018), apple (Li et al., 2013a,b), and wheat (Xu et al., 2009). However, the photoregulatory mechanism of cryptochromes have not been studied in those species, and no photoreceptor has been previously reported in the monocot forestry species, such as bamboo. In the present study, we investigated different isoforms of moso bamboo (Phyllostachys edulis) PheCRY1. We show evolutionary conservation in not only function but also photoregulatory mechanisms of moso bamoo CRY1.
Materials and Methods
Accession Numbers
Moso bamboo gene sequences are obtained from the BambooGDB database1 under the accession numbers in Supplementary Table 1.
Plasmid Construction
In-Fusion Cloning methods were used to generate the plasmids used in this study.2 The sequences subcloned into plasmids were verified by Sanger sequencing. The human cell expression vectors used in this study were pQCMV-Flag-GFP, pQCMV-GFP, and pCMV-Myc, which were described previously (Liu et al., 2017; Chen et al., 2021). pQCMV-YFP and pQCMV-mTagRFP-T vectors were modified from pQCMV-Flag-GFP by replacing GFP with YFP or mTagRFP-T (Supplementary Figure 1). To generate plasmids expressing Flag-PheCRY1c, Flag-PheCRY1d, and Flag-PhePPKb, the coding sequences (CDS) of PheCRY1c or PheCRY1d were amplified from moso bamboo cDNA, and the purified PCR products were then subcloned into SpeI/KpnI-digested pQCMV-Flag-GFP vector through in-fusion. Myc-PheCRY1c and Myc-PheCRY1d plasmids were prepared by cloning the CDS of the genes into the BamHI site of pCMV-Myc vector. For plasmids expressing PheCRY1c-YFP and PheCRY1d-YFP, the CDS of PheCRY1c and PheCRY1d were PCR-amplified from plasmids made before, and introduced into SpeI digested pQCMV-YFP vector by in-fusion. The AtBIC1-mTagRFP and PheBIC1a-mTagRFP plasmids were prepared by cloning the CDS of the AtBIC1 or PheBIC1a genes into the SpeI-digested pQCMV-mTagRFP-T vector by in-fusion. The gene-specific primers for constructs used for expressing recombinant proteins in HEK293T cells are listed in Supplementary Table 2.
pFGFP binary vectors was used for creating overexpression transgenic lines (Chen et al., 2021). The pFGFP binary vector was modified from pCambia3301, which allows the interested genes to express under control of ACTIN 2 promoter and fuse with 2 × Flag and GFP tags. For generation of the plant binary plasmids of FGFP-PheCRYs and FGFP-PheBIC1a, the CDS of PheCRY1s and PheBIC1a were cloned into BamHI-digested pFGFP vector. The primers for constructs used for plant transformation are included in Supplementary Table 2.
Plant Materials and Growth Conditions
The wild-type Arabidopsis in this study is rdr6-11, which suppresses gene silencing (Peragine et al., 2004). The cry1cry2rdr6 was described previously (Chen et al., 2021). Arabidopsis transgenic lines were generated via Agrobacterium tumefaciens-mediated the floral dip method (Clough, 2005). Plasmids of FGFP-PheCRY1c and FGFP-PheCRY1d were introduced into cry1cry2rdr6 background to generate FGFP-PheCRY1c and FGFP-PheCRY1d overexpressing plants. FGFP-BIC1a overexpressing Arabidopsis was prepared in rdr6-11 background. The transgenic lines were screened on nutrient soil with 25 mg/L Glufosinate-ammonium and lines with a relative high protein expression level were used for phenotype analysis.
For hypocotyl phenotype analysis, seedlings were grown on Murashige and Skoog medium (MS) plates with 1% sucrose at 20∼22°C for 5 days under different light conditions. The same representative images and quantification results of control seedlings, cry1cry2, wild-type (WT), GFP-AtCRY1, GFP-AtCRY2, were used for Figures 1, 5. For endogenous CRY1 and CRY2 degradation analysis in WT, FGFP-PheBIC1a, seedlings were grown in darkness on MS plates with 1% sucrose for 7 days, then subjected to 30 μmol m–2 s–1 blue light for the indicated time. For FGFP-PheCRY1c and FGFP-PheCRY1d and degradation analysis, seedlings were grown in darkness on MS plates with 1% sucrose for 6 days, then subjected to 100 μmol m–2 s–1 blue light for the indicated time. For flowering time measurements, seeds were sown in compound soil, stratified at 4°C in darkness for 3 days and then were transferred to long day (LD; 16 h light/8 h darkness) walk-in chambers.
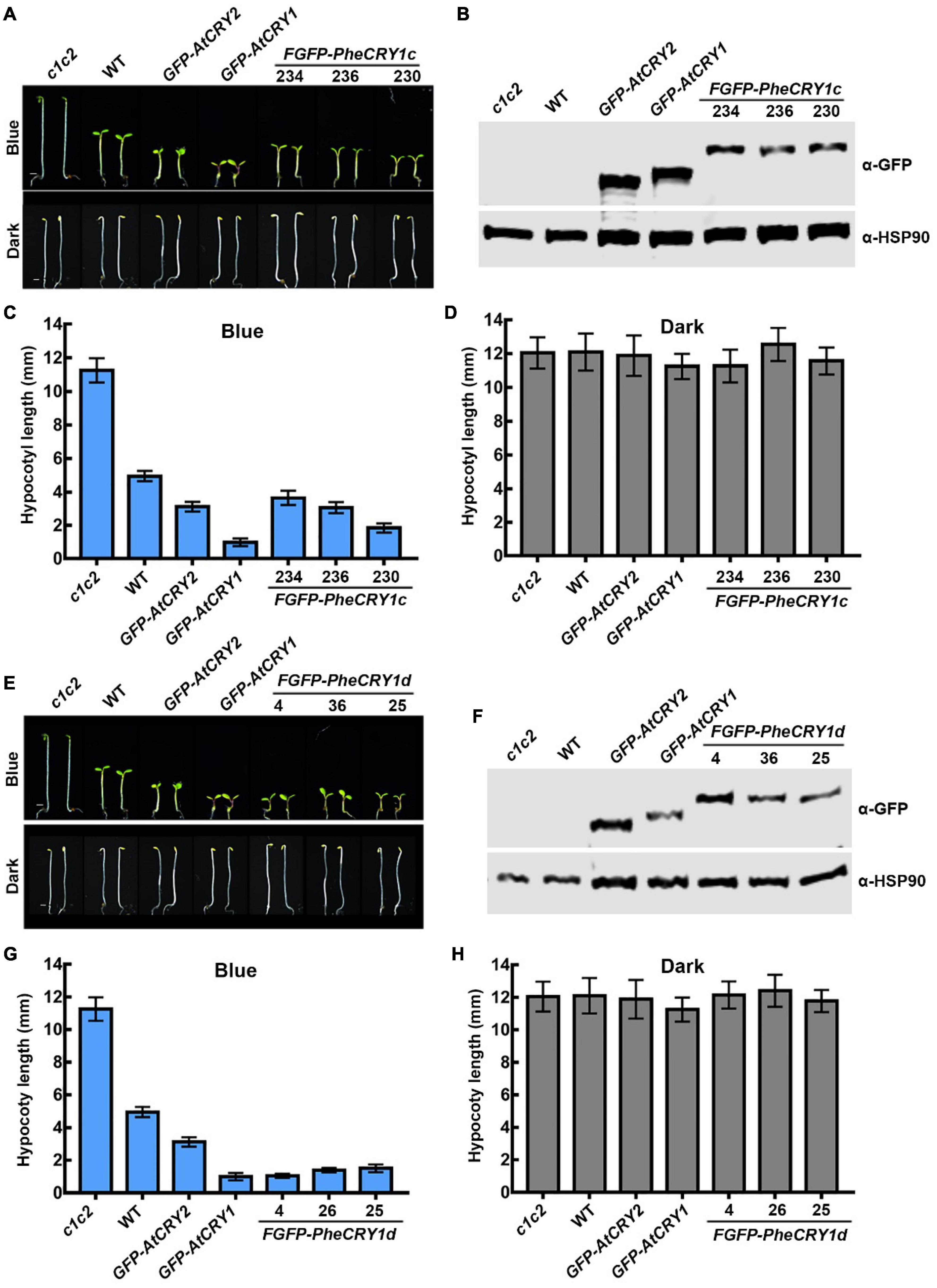
Figure 1. Bamboo PheCRY1s mediate blue light inhibition of hypocotyl elongation. (A,E) Images of 5-day-old Arabidopsis seedings grown under continuous blue light (10 μmolm–2s–1) or darkness. Scale bar, 1 mm. (B,F) Immunoblots showing the expression of FGFP-PheCRY1c (B) or FGFP-PheCRY1d (F) of seedlings shown in (A) or (E). PheCRY1 and HSP90 were detected with anti-GFP antibody or anti-HSP90 antibody, respectively. HSP90 is used as a loading control. (C,D) Measurements of hypocotyl length of seedlings shown in (A), (mean ± SD, n ≥ 20). (G,H) Measurements of hypocotyl length of seedlings shown in (E), (mean ± SD, n ≥ 20).
Protein Expression and Co-immunoprecipitation in HEK293T Cells
These methods were described previously (Chen et al., 2021).
Photobody Formation Analysis
For photobody observation in HEK293T cells, cells were grown in 20 mm glass bottom confocal dishes (Cat. # 801001, NEST), and fixed in 4% formaldehyde for 15 min before analysis under confocal microscope. Hoechst 33342 (Cat. # 14533, Sigma-Aldrich) solution, at the concentration of 10 μg/ml, was used for nuclei staining. Leica TCS SP8 confocal microscope with a HC PL APO CS2 63x/1.40 OIL objective was used to captured the microscopic images. YFP signals were detected at 500–550 nm with 488 nm as excitation, mTagRFP was excited with 554 nm laser and detected at 566–629 nm. Hoechst 33342 was excited with 405 nm laser and detected at 420–480 nm.
Blue Light-Induced Cryptochromes Degradation Assays
This method was described previously (Chen et al., 2021).
Antibodies Used in This Study
The primary antibodies used in this study are as following: anti-AtCRY1 (1:5,000, homemade) (Lin et al., 1996), anti-AtCRY2 (1:3,000, homemade) (Lin et al., 1998), anti-HSP90 (1:20,000, cat # AbM51099-31-PU, Beijing Protein Innovation), anti-GFP (1:2,000, cat # 598, MBL), anti-Myc (1:2,000, cat # 05-724, Millipore), anti-Flag (1:1,500, cat # F3165, Sigma-Aldrich).
The secondary antibodies used in this study are as following: anti-Mouse-HRP (1:15,000, cat # 31430, Thermo Fisher Scientific), anti-Rabbit-HRP (1:15,000, cat # 31460, Thermo Fisher Scientific), goat anti-mouse Alexa Fluor 790 (1:15,000, cat # A11357), goat anti-rabbit IgG-Alexa Fluor 790 (1:15,000, cat # A11369), goat anti-mouse IgG-Alexa Fluor 680 (1:15,000, cat # A21058), goat anti-rabbit IgG-Alexa Fluor 680 (1:15,000, cat # A21109).
Results
Function of Moso Bamboo Cryptochromes
To investigate possible roles that cryptochromes play in moso bamboo, we searched moso bamboo (Phyllostachys edulis) genome sequence database, and identified five moso bamboo cryptochrome-like genes (PheCRY), PheCRY1a, PheCRY1b, PheCRY1c, PheCRY1d, and PheCRY2 (Supplementary Figure 2A). PheCRY1a, PheCRY1b, PheCRY1c, and PheCRY1d show higher similarity to Arabidopsis cryptochrome 1 (AtCRY1, ∼ 72–84% identity) than that to Arabidopsis cryptochrome 2 (AtCRY2, ∼ 66–74% identity) (Supplementary Figure 2B). Similar to that found in cryptochromes of other plants, PheCRYs share more extensive sequence similarity in the N-terminal photolyase homologous chromophore-binding domains (PHR domain) than in the C-terminal domains (CCE domain) (Supplementary Figure 2C). We successfully cloned two representative PheCRY genes (PheCRY1c and PheCRY1d) for further biochemical and functional analysis.
Because the transformation technique has not been established for moso bamboo, we generated FGFP-PheCRY1c (with Flag and GFP tags fused to the N-terminus of PheCRY1c) and FGFP-PheCRY1d overexpression transgenic Arabidopsis in cry1cry2 mutant background to test whether the bamboo CRY1s may complement the Arabidopsis cry1cry2 mutant. Similar to previous studies of cryptochromes in other plants (Ninu et al., 1999; Matsumoto et al., 2003; Platten et al., 2005; Chatterjee et al., 2006; Szucs et al., 2006; Zhang et al., 2008; Xu et al., 2009; Li et al., 2013a,b; Mao et al., 2014; Zhou et al., 2018; Lyu et al., 2021), our study demonstrates that bamboo CRY1 serves similar function as the Arabidopsis CRYs in regulating cell elongation. Transgenic expression of FGFP-PheCRY1c and FGFP-PheCRY1d rescued the blue light-specific long hypocotyl phenotype of the Arabidopsis cry1cry2 mutant, resulted in hypersensitivity to blue light as the GFP-AtCRY1 and GFP-AtCRY2 transgenic plants, which showed hypocotyls obviously shorter than that of the wild-type seedlings (Figure 1). Given that light inhibition of cell elongation is likely an ancient cellular response, it is not surprising that this activity of cryptochromes seems universally conserved in different plant species.
However, bamboo CRY1 could not fully rescue the late-flowering phenotype of Arabidopsis cry1cry2 double mutant (Figure 2). At the time of flowering, FGFP-PheCRY1 transgenic Arabidopsis had less rosette leaves than that of cry1cry2 mutant plants, but had more rosette leaves than that of wild-type and GFP-AtCRY1 or GFP-AtCRY2 overexpressing plants (Figure 2). Consistently, FGFP-PheCRY1 transgenic Arabidopsis flowered at about the same time as the cry1cry2 mutant (Figure 2). One transgenic line of FGFP-PheCRY1d (line #4) seemed to rescue the late-flowering phenotype of Arabidopsis cry1cry2 mutant (Figure 2B), which is inconsistent with the flowering phenotype of other FGFP-PheCRY1d overexpressing lines and FGFP-PheCRY1c overexpressing lines. We reasoned that this inconsistence may cause by T-DNA insertion in the genomic region of a gene involving in flowering regulation, because all the transgenic lines of FGFP-PheCRY1c and FGFP-PheCRY1d showed similar physiological activity in hypocotyl inhibition. Since flowering time is a more recent evolutionary “invention” of angiosperm, the moso bamboo cryptochromes and Arabidopsis cryptochromes may have a distinct mode of action in the regulation of flowering time. It’s also possible that the flowering regulation mechanisms in moso bamboo are distinct from that of Arabidopsis, especially that bamboo has the extremely long vegetative phase and bamboo plants often take decades to flower (Numata, 1970; Lin et al., 2010). In Arabidopsis, CRY1 and CRY2 primarily mediate blue light inhibition of hypocotyl elongation and photoperiodic control of floral initiation, respectively (Ahmad and Cashmore, 1993; Mockler et al., 2003). It would also be interesting to investigate whether bamboo CRY2 may have the function in promoting floral initiation.
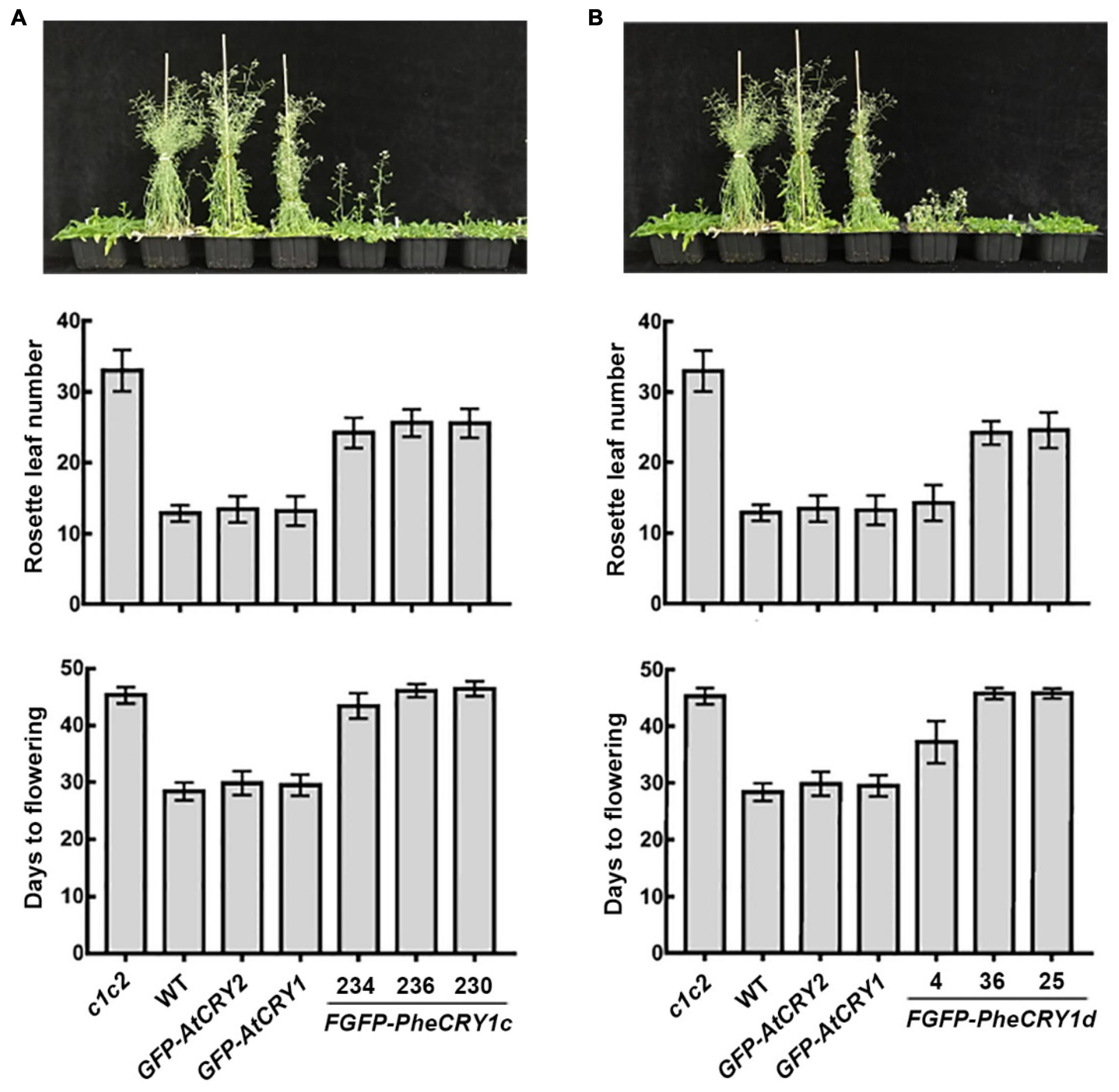
Figure 2. Bamboo PheCRY1 doesn’t function in the flowering time regulation in Arabidopsis. (A,B) Images of the flowering phenotypes of indicated plants grown in LD (16 h light/8 h dark) were shown. Rosette leaf numbers and days to flowering were recorded when flowering. Data were shown as mean ± SD, n ≥ 20.
Photoactivation of Moso Bamboo Cryptochromes
Having demonstrated the blue light-dependent activity of PheCRY1c and PheCRY1d in transgenic Arabidopsis plants, we next asked whether the PheCRY1 may undergo similar photochemical reactions as that of Arabidopsis cryptochromes. Photo-oligomerization is the early photoreaction necessary for photoactivation of both CRY1 and CRY2 (Sang et al., 2005; Rosenfeldt et al., 2008; Wang et al., 2016; Liu et al., 2020). To investigate if the moso bamboo PheCRYs undergo homo-oligomerization in response to blue light, we examined the PheCRY1 oligomerization by co-expressing Flag-PheCRY1 and Myc-PheCRY1 in human embryonic kidney 293T (HEK293T) cells and tested the interaction of the two differentially tagged PheCRY1 by co-immunoprecipitation (co-IP) assay. In cells with similar amounts of Flag-PheCRY1 and Myc-PheCRY1 expression, Flag-beads coprecipitated more Myc-PheCRY1 in blue light than that in the dark (Figures 3A,B), demonstrating the blue light-enhanced PheCRY1 homo-oligomerization in the absence of other bamboo proteins. These results also demonstrated that, similar to Arabidopsis CRY1 and CRY2, bamboo PheCRY1 undergo oligomerization to become active. Photoexcited AtCRY1 and AtCRY2 are known to oligomerize into photobodies (Yu et al., 2009; Bugaj et al., 2013; Ozkan-Dagliyan et al., 2013; Wang et al., 2021; Liu et al., 2022), which is another early photoreaction of cryptochromes. We further investigated the PheCRY1 photobody formation in HEK293T cells. Figures 3C,D shows that PheCRY1-YFP (fused to yellow fluorescent protein) formed photobodies in the nucleus with blue light treatment, whereas no photobodies were detected in the cells in the dark, demonstrating that photoexcited PheCRY1 is capable of homo-oligomerization into photobodies in the absence of other proteins. These observations are also consistent with a hypothesis that photo-oligomerization might be an evolutionarily conserved photoactivation mechanism of cryptochrome photoreceptors. Taken together, our results demonstrate that moso bamboo PheCRY1 undergoes blue light-dependent homo-oligomerization to become photoactivated.
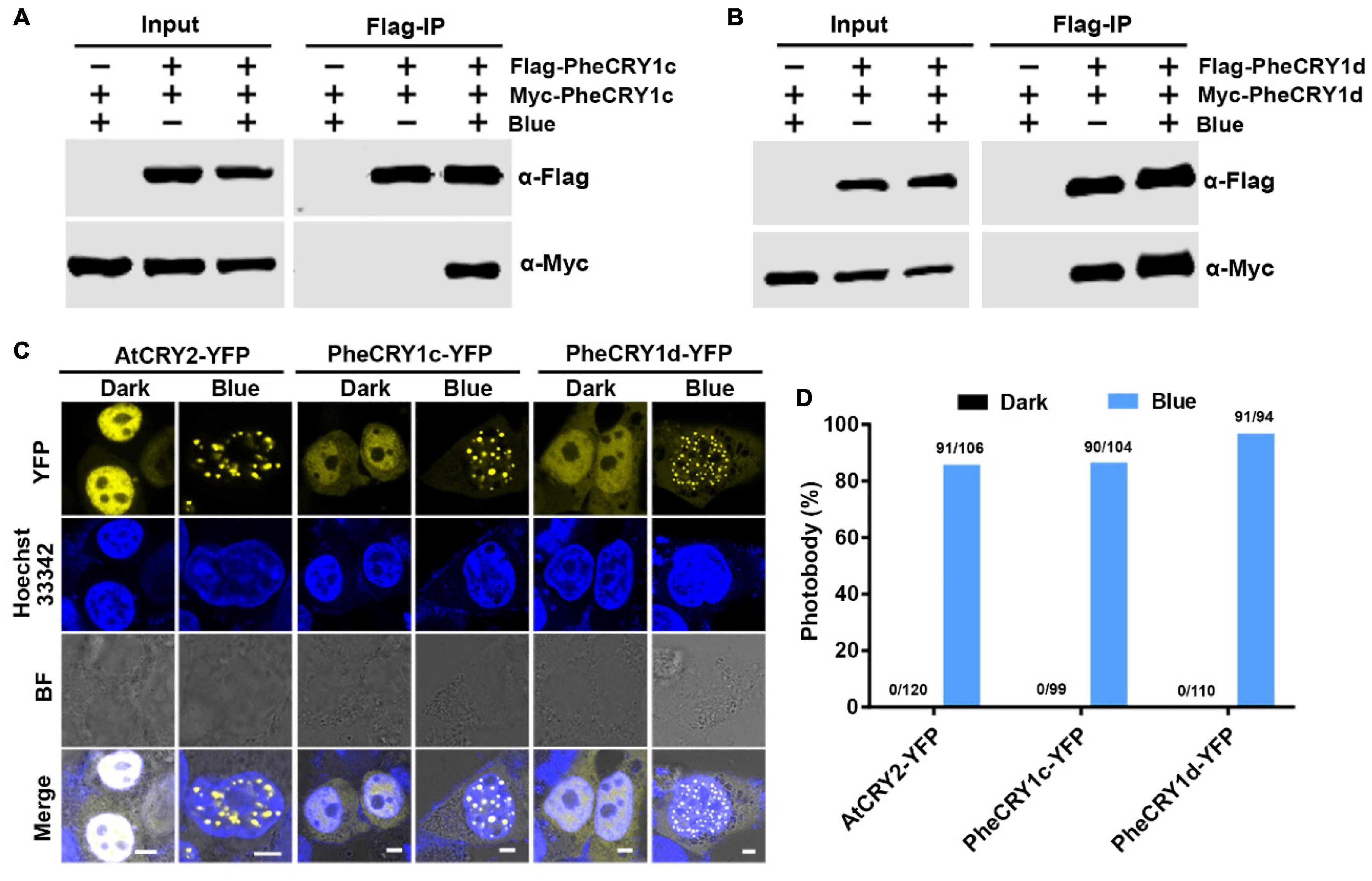
Figure 3. Bamboo PheCRY1s undergo blue light-enhanced homo-oligomerization. (A,B) Co-immunoprecipitation (co-IP) assays showing the blue light-enhanced homo-oligomerization of PheCRY1 in HEK293T cells. Cells co-expressing Flag-PheCRY1 and Myc-PheCRY1 were kept in the dark (Blue –) or exposed to 100 μmol–2s–1 of blue light for 2 h (Blue +). Immunoprecipitations (IP) were performed with Flag-conjugated beads. The IP and co-IP products were detected with anti-Flag and anti-Myc antibodies, respectively. (C) Confocal images showing the photobody formation of PheCRY1 in response to blue light in HEK293T cells. Cells were kept in the dark or exposed to blue light (100 μmol m–2 s–1) for 1 h. Before imaging, cells were fixed in 4% formaldehyde. Hoechst staining shows the nuclei. BF, bright field; scale bar, 5 μm. (D) Quantitative analysis of PheCRY1-YFP photobody formation in (C). Photobody (%) is the percent of nuclei with photobodies among counted nuclei. The number of nuclei with photobodies and total counted nuclei was indicated above the columns.
Inactivation of Moso Bamboo Cryptochromes
Photoreceptor inactivation is an important photoregulatory mechanism for the maintenance of a sustained photosensitivity of the cells. Arabidopsis cryptochromes are inactivated by at least three mechanisms: BIC inhibition of oligomerization, dark-dependent monomerization, and blue light-dependent proteolysis (Yu et al., 2007; Wang et al., 2016; Liu et al., 2020, 2022; Chen et al., 2021). In Arabidopsis, the blue light-dependent cryptochrome oligomerization is suppressed by two closely related cryptochrome inhibitory proteins, BIC1 and BIC2. We examined whether there are BIC proteins in moso bamboo and whether the putative bamboo BIC proteins might suppress photo-oligomerization of the bamboo cryptochromes. Two highly homologous PheBIC1 genes, PheBIC1a and PheBIC1b, were identified from the moso bamboo genome sequence database (Supplementary Figure 3). PheBIC1a and PheBIC1b share over 80% sequence identity and share more extensive sequence similarity in the CID domain (CRY-interacting domain) to Arabidopsis BIC1 and BIC2 (Supplementary Figure 3C), indicating that PheBICs may also function as cryptochrome inhibitors in moso bamboo. To test this hypothesis, we first examined the interaction of PheBIC1 and PheCRY1 in HEK293T cells co-expressing epitope-tagged PheBIC1 and PheCRY1 using co-IP assays. Our results show that blue light enhances PheBIC1 and PheCRY1 interaction. Both PheCRY1c and PheCRY1d coimmunoprecipitated PheBIC1a in HEK293T cells exposed to blue light, but little PheBIC1-PheCRY1 complex was coprecipitated in the dark (Figures 4A,B). This result suggests that PheBIC1 might interact with photoactivated PheCRY1 to inhibit their activities. We next tested this possibility by examining the effects of PheBIC1 on blue light-dependent PheCRY1 photobody formation by co-expressing PheBIC1-mTagRFP (fused to red fluorescent protein) and PheCRY1-YFP in HEK293T cells. AtCRY2-YFP and AtBIC1-mTagRFP co-expression cells were included as controls. Our results shows that no PheCRY1 photobodies were detected in the cells co-expressing PheBIC1 in blue light (Figures 4C,D), confirming our hypothesis that PheBIC1 act as cryptochrome-specific inhibitors in moso bamboo.
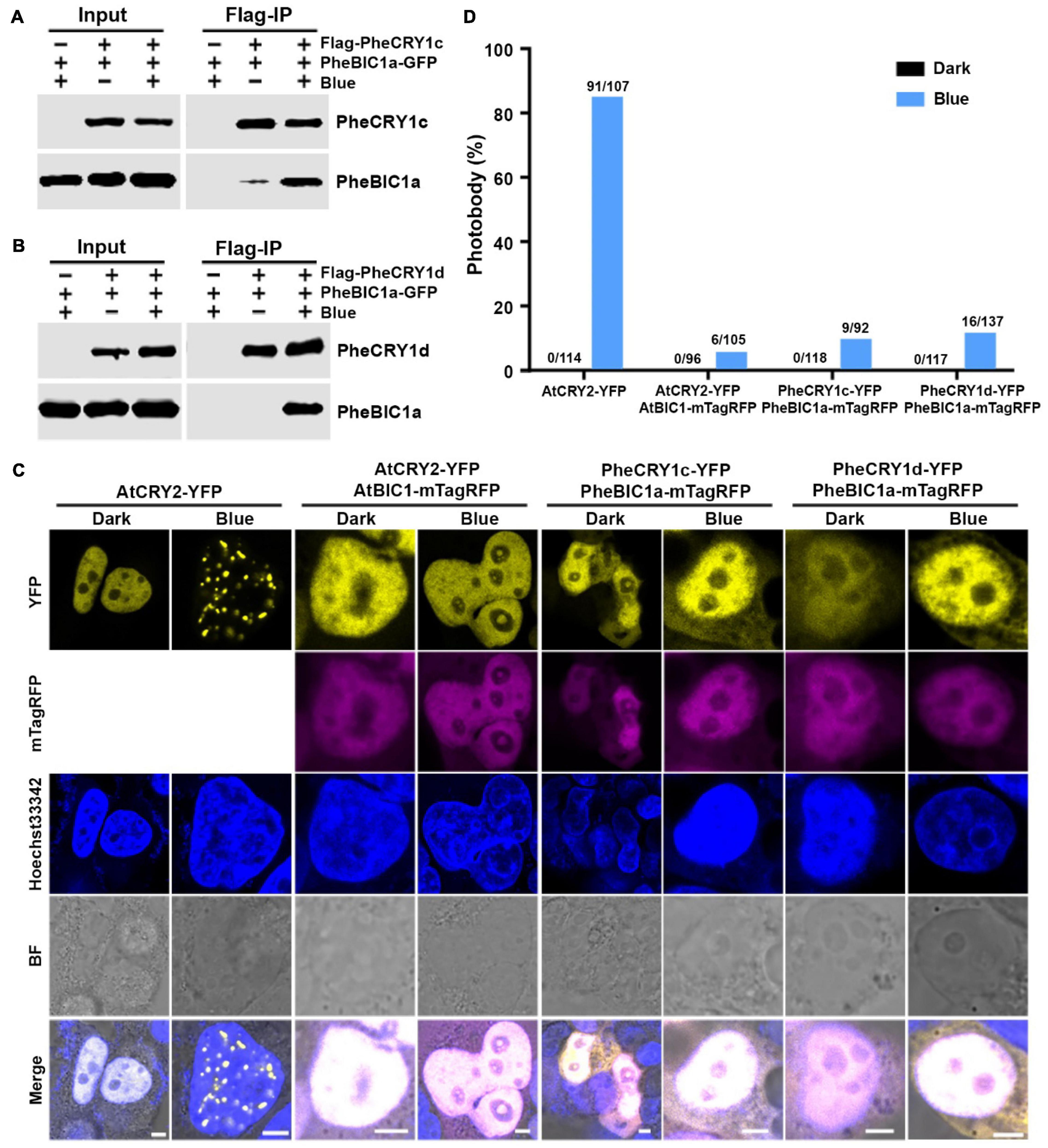
Figure 4. Bamboo PheBIC1 interacts with PheCRY1s to inhibit the photo-oligomerization of PheCRY1. (A,B) Co-immunoprecipitation (co-IP) assays showing the blue light-dependent interaction of PheCRY1 and PheBIC1 in HEK293T cells. Cells co-expressing Flag-PheCRY1 and PheBIC1-GFP were kept in the dark (Blue –) or exposed to 100 μmol–2s–1 of blue light for 2 h (Blue +). IP were performed with Flag-conjugated beads. The IP and co-IP products were detected with anti-Flag and anti-GFP antibodies, respectively. (C) Confocal images showing the inhibition of PheBIC1 on PheCRY1 photobody formation in HEK293T cells. Cells expressing indicated plasmid pairs were kept in the dark or exposed to blue light (100 μmol m–2 s–1) for 1 h. Cells were fixed in 4% formaldehyde before imaging. Hoechst staining shows the nuclei. BF, bright field; scale bar, 5 μm. (D) Quantitative analysis of the photobody formation in (C). Photobody (%) is the percent of nuclei with photobodies among counted nuclei. The number of nuclei with photobodies and total counted nuclei was indicated above the columns.
To test whether the bamboo BIC1 and their physical interaction with the bamboo CRY1 may be physiologically relevant, we prepared FGFP-PheBIC1a overexpression transgenic Arabidopsis in the wild-type background. The PheBIC1a overexpressing lines are insensitive to blue light, showing a long hypocotyl phenotype in blue light, resembling that of the Arabidopsis cry1cry2 double mutant (Figures 5A–D). This result suggests that PheBIC1 may inhibit the function of Arabidopsis cryptochromes. We then investigated this hypothesis by comparing the phosphorylation and degradation of Arabidopsis endogenous CRY1 and CRY2 in the wild-type and PheBIC1a overexpressing plants. As expected, the blue light-dependent phosphorylation of both AtCRY1 and AtCRY2 were not detectable in PheBIC1a overexpressing Arabidopsis (Figures 5E,F), and the blue light-dependent degradation of AtCRY2 was also impaired in PheBIC1a overexpressing plants (Figure 5F). In the wild-type seedlings, obvious AtCRY1 phosphorylation was detected within 15 min of blue light exposure, while no AtCRY1 phosphorylation was detected even after 60 min of blue light exposure in the PheBIC1a overexpressing Arabidopsis (Figure 5E). The phosphorylation of AtCRY2 was detected within 10 min of blue light exposure and AtCRY2 protein was no detectable after 60 min of blue light exposure in the wild-type seedings (Figure 5F). However, neither phosphorylation nor degradation of AtCRY2 were detected in the PheBIC1a overexpressing Arabidopsis in response to blue light (Figure 5F). These results are consistent with our previously published results that overexpression of Arabidopsis BICs suppressed all known photobiochemical and photophysiological activities of AtCRY1 and AtCRY2, resulted in unrestrictive hypocotyl growth in blue light (Wang et al., 2016). Results of these experiments also indicate that PheBICs may be the PheCRYs-specific inhibitors to regulate the activity of PheCRYs in moso bamboo.
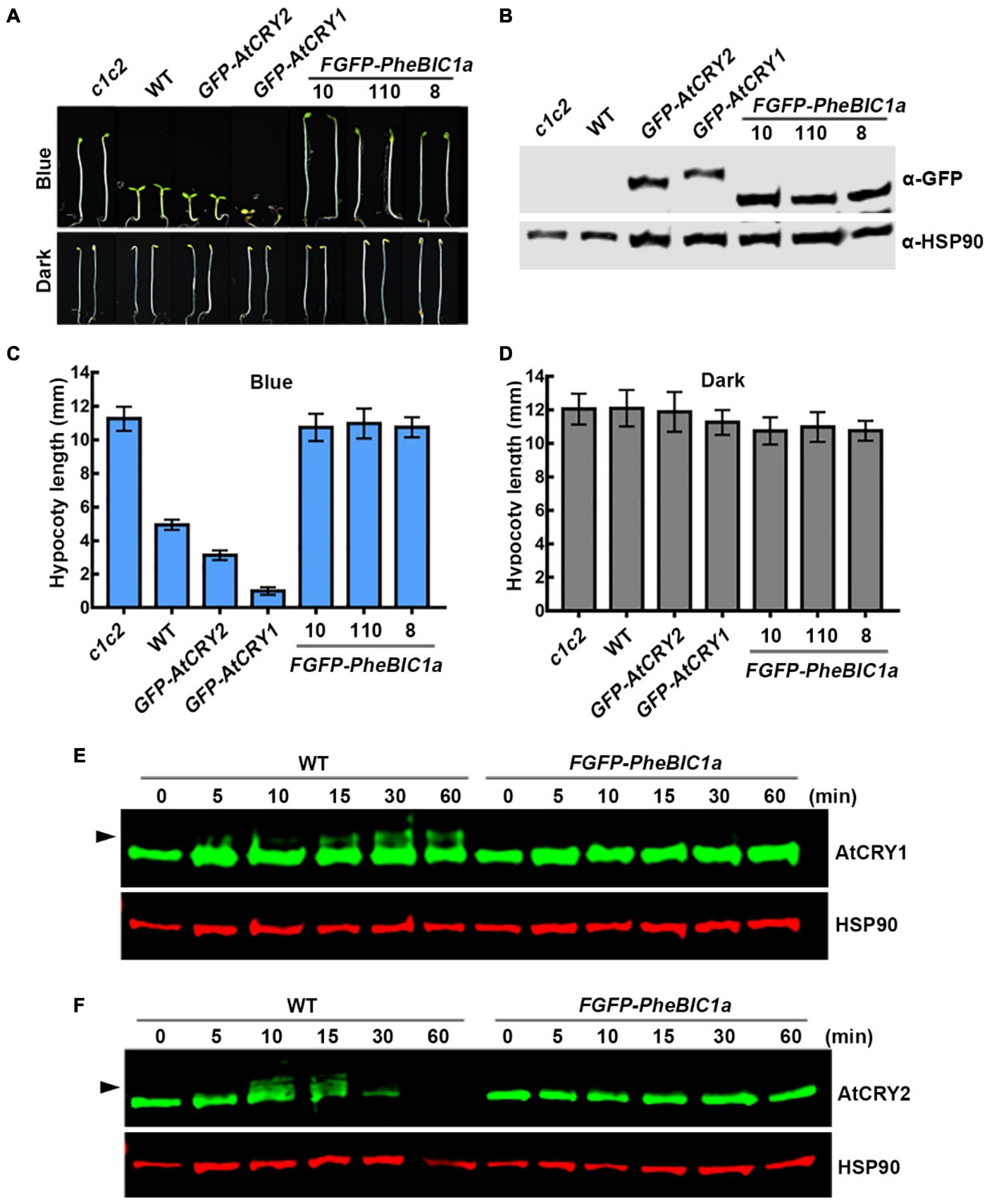
Figure 5. Bamboo PheBIC1 promotes hypocotyl elongation in blue light. (A) Images of 5-day-old Arabidopsis seedings grown under continuous blue light (10 μmolm–2s–1) or darkness. Scale bar, 1 mm. (B) Immunoblots showing the expression of FGFP-PheBIC1a of seedlings shown in (A). PheBIC1 and HSP90 were detected with anti-GFP antibody or anti-HSP90 antibody, respectively. HSP90 is used as a loading control. (C,D) Measurements of hypocotyl length of seedlings shown in (A), (mean ± SD, n ≥ 20). (E,F) Immunoblots showing the phosphorylation and degradation of endogenous Arabidopsis CRY1 and CRY2 in the wild-type or FGFP-PheBIC1a transgenic Arabidopsis. 7-day-old etiolated seedlings were irradiated with 30 μmolm–2 s–1 of blue light for the indicated time. Anti-AtCRY1, anti-AtCRY2 and anti-HSP90 antibodies were used to probe AtCRY1, AtCRY2 or HSP90. HSP90 is used as a loading control.
Blue light-dependent proteolysis serve as another inactivation mechanism of crytochromes by removing the activated cryptochromes in cells. Depending on the plant species, CRY1, CRY2, and other types of cryptochromes have all been shown to undergo light-dependent proteolysis (Lin et al., 1998; Imaizumi et al., 2000; Reisdorph and Small, 2004; Chatterjee et al., 2006; Hirose et al., 2006). In Arabidopsis, both AtCRY1 and AtCRY2 undergo blue light-dependent degradation in the nucleus (Yu et al., 2007; Liu et al., 2022). We next examined the protein degradation of PheCRY1 in transgenic Arabidopsis. Figure 6 showed that in etiolated seedlings exposed to blue light, the PheCRY1c and PheCRY1d protein exhibits a pronounced decrease after 12 h of blue light treatment. The rate of blue light-induced degradation of PheCRY1c and PheCRY1d were slower than that of the AtCRY2, which showed apparent degradation after 2 h of blue light treatment. We reasoned that PheCRY1c and PheCRY1d might be more closely related to Arabidopsis CRY1, which requires higher intensities of blue light and longer blue light treatment time to induce its degradation. Our results also suggest that the light-dependent proteolysis may be another evolutionary conserved mechanism for inactivation of PheCRYs in moso bamboo, and the E3 ubiquitin ligases, CUL3LRBs and CUL4COP1/SPAs, responsible for Arabidopsis cryptochromes degradation may also be conserved between Arabidopsis and moso bamboo. However, this hypothesis remains to be tested.
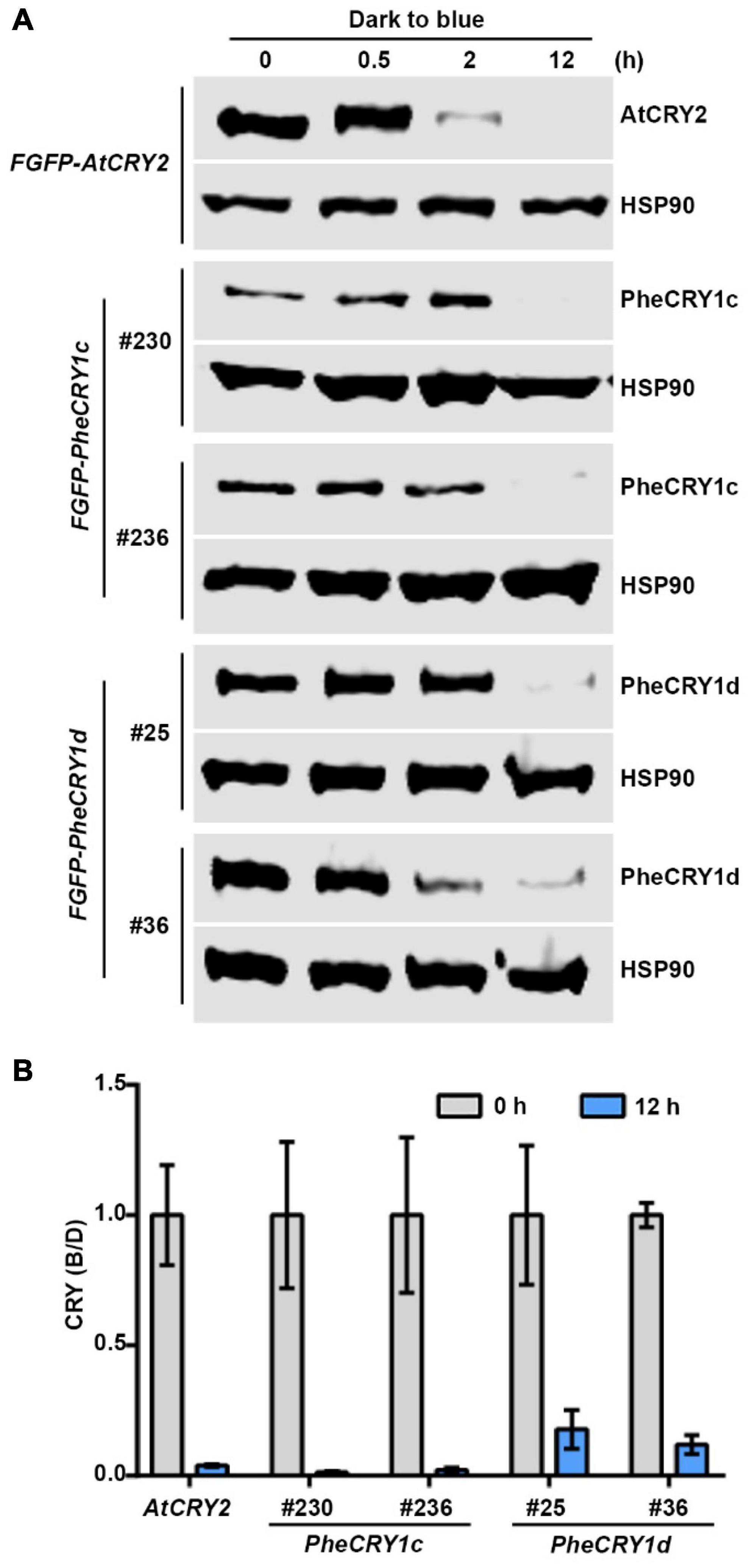
Figure 6. Bamboo PheCRY1s undergo blue light-dependent proteolysis. (A) Immunoblots showing the degradation of PheCRY1 in transgenic Arabidopsis. 6-day-old etiolated seedlings were irradiated with 100 μmolm–2 s–1 of blue light for the indicated time. Two independent transgenic lines were used for analysis. Anti-Flag antibody was used to detect AtCRY2 and PheCRY1. Anti-HSP90 antibody was used to detect HSP90. HSP90 is used as a loading control. (B) Quantitative analysis of CRY protein degradation at time 0 and 12 h of blue light treatment from immunoblots shown in (A). CRY (B/D) = [CRY/HSP90]blue/[CRY/HSP90]dark. Data are presented as mean ± SD (n = 3 individual immunoblots).
Phosphorylation of Moso Bamboo Cryptochromes
Photoexcited Arabidopsis CRY1 and CRY2 further undergo blue light-dependent phosphorylation to both enhance their activities and facilitate their ubiquitination and degradation in the nucleus (Shalitin et al., 2002, 2003; Yu et al., 2007; Weidler et al., 2012; Liu et al., 2016, 2022; Miao et al., 2021). Four closely related plant-specific PPKs (PPK1, PPK2, PPK3, and PPK4) have been shown to catalyze the phosphorylation of Arabidopsis CRY1 and CRY2 in blue light (Liu et al., 2017; Ni et al., 2017). To test whether moso bamboo PheCRYs undergo blue light-induced phosphorylation and whether PhePPKs are the protein kinases catalyzing light-induced phosphorylation of PheCRYs, we searched moso bamboo genome sequence database, and found 10 closely related PhePPKs (PhePPKa–PhePPKj) genes in moso bamboo (Supplementary Figure 4). Similar to Arabidopsis PPKs, PhePPKs have two domains, a N-terminal Ser/Thr kinase domain which is homolog to casein kinase 1 (CK1) (Liu et al., 2017), and a C-terminal domain that are highly conserved within the PPK gene families (Supplementary Figure 4C). We cloned PhePPKb and examine its catalytic characteristics in HEK293T cells. We co-expressed Arabidopsis CRY2 or moso bamboo PheCRY1 with AtPPK1 or PhePPKb in HEK293T cells to investigate the catalytic activity of PPK on CRY by electrophoretic mobility upshift immunoblot assays (Shalitin et al., 2002, 2003; Wang et al., 2016). AtCRY2, PheCRY1c, and PheCRY1d exhibit retarded migration in cells co-expressing PhePPKb or AtPPK1 treated with blue light, whereas no migration upshift was detected in cells treated with green light, red light or far-red light (Figures 7A–C and Supplementary Figures 5A–C). The blue light-specific upshift migration of CRYs disappeared after protein phosphatase treatments (Figures 7D–F and Supplementary Figures 5D–F), confirming the slow migration bands were phosphorylated PheCRY1. We also found that AtCRY2 can be phosphorylated by moso bamboo PhePPK1, and AtPPK1 can phosphorylate bamboo PheCRY1 (Figures 7A,D and Supplementary Figures 5B,C,E,F), indicating that PPK-CRY (enzyme-substrate) pairing is more likely determined by structure-based mechanisms regardless of plant species. In summary, our results demonstrate that moso bamboo CRY1s undergo blue-light dependent phosphorylation in vitro, and PhePPKs are the protein kinases that catalyze the blue-light dependent phosphorylation of moso bamboo cryptochromes, indicating that the molecular basis for light-dependent phosphorylation of cryptochrome is also evolutionary conserved between Arabidopsis and moso bamboo.
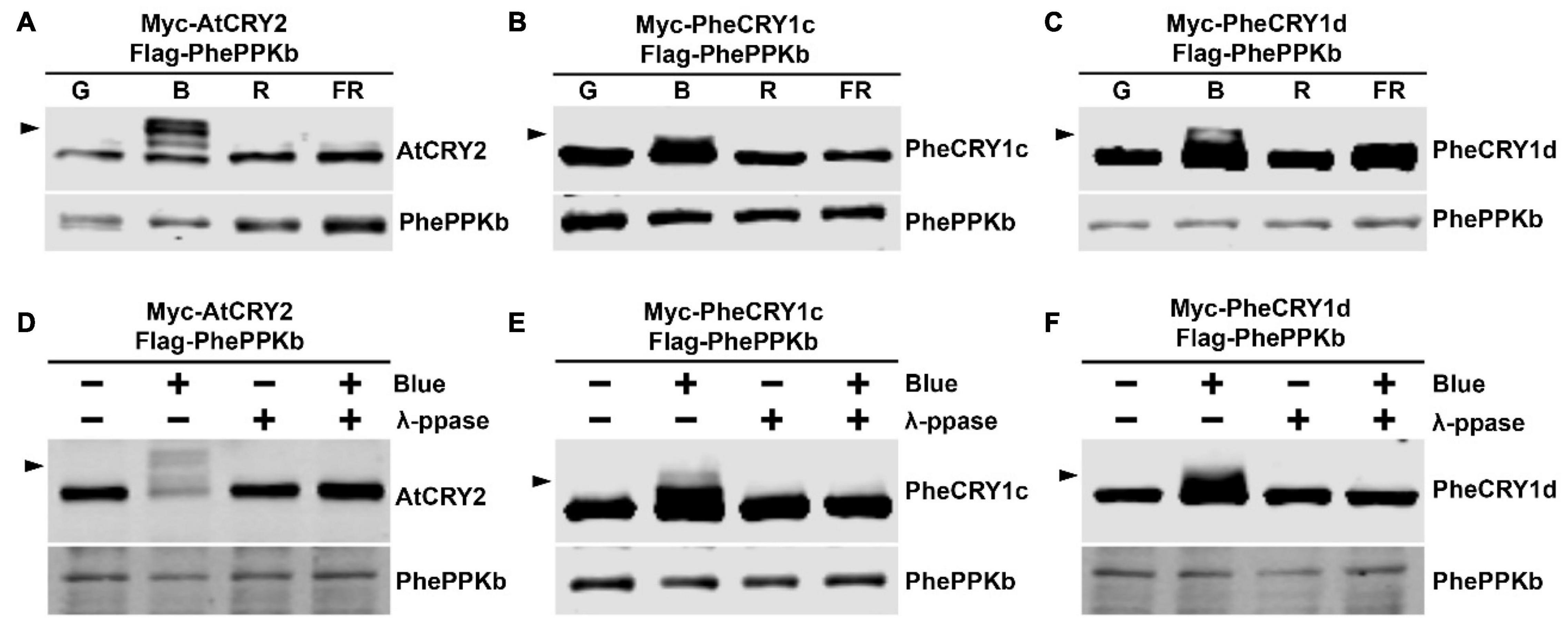
Figure 7. Bamboo PhePPKb catalyzes the blue light-dependent phosphorylation of bamboo PheCRY1 in HEK293T cells. (A–C) Cells co-expressing indicated plasmid pairs were exposed to green light (G, 50 μmolm–2 s–1), blue light (B, 50 μmolm–2 s–1), red light (R, 50 μmolm–2 s–1), or far-red light (FR, 5 μmolm–2 s–1) for 60 min. Immunoblots were probed with the anti-Myc or anti-Flag antibodies. (D–F) Cells co-expressing indicated plasmids were kept in the dark (Blue –) or treated with 100 μmolm–2 s–1 of blue light for 2 h (Blue +). Lysates were treated without (- λ-PPase) or with λ-PPase (+ λ-PPase), and analyzed by immunoblots probed with the anti-Flag or anti-Myc antibodies. Arrowheads indicate the phosphorylated CRY.
Discussion
Although the function of CRYs have been reported in many plant species, the photoregulatory mechanisms of CRYs have been study for primarily the Arabidopsis CRYs. A major goal of our present study is to address the question whether the photoregulatory mechanism of plant CRYs are evolutionarily conserved. Our results demonstrate that at least three photoregulatory mechanisms of bamboo CRY1 are evolutionarily conserved. First, bamboo CRY1 undergo blue light-dependent homo-oligomerization as that of Arabidopsis CRY2, and this photoreaction is inhibited by the negative regulator of bamboo BICs. Second, bamboo CRY1 also undergoes blue light-induced phosphorylation that can be catalyzed by the bamboo PPK kinases. Third, bamboo CRY1 proteins undergo light-induced proteolysis as that of the Arabidopsis CRYs. Our results that bamboo CRY1 are phosphorylated and degraded in Arabidopsis and that bamboo PPKb catalyze light-induced phosphorylation of Arabidopsis CRY1 demonstrate the evolutionary conservation of this photoregulatory mechanism. Based on these results, we hypothesize that the photoregulatory mechanism of plant CRYs are predominantly conserved during evolution.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
ZC performed most of the experiments and wrote part of the manuscript. ML generated genetic materials and collected the phenotypic data. SL helped with photobody formation analysis. XC and WZ repeated some results. QZ helped with bamboo transformation. MK provided financial supports. QW conceived the project, designed the experiments, and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the Natural Science Foundation of Fujian Province (2019J01421 to MK) and Natural Science Foundation of Fujian Province (2019J06014 to QW).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Chentao Lin for his contribution to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.866057/full#supplementary-material
Footnotes
References
Ahmad, M., and Cashmore, A. R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. doi: 10.1038/366162a0
Bugaj, L. J., Choksi, A. T., Mesuda, C. K., Kane, R. S., and Schaffer, D. V. (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Meth. 10, 249–252. doi: 10.1038/nmeth.2360
Chatterjee, M., Sharma, P., and Khurana, J. P. (2006). Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol. 141, 61–74. doi: 10.1104/pp.105.076323
Chen, Y., Hu, X., Liu, S., Su, T., Huang, H., Ren, H., et al. (2021). Regulation of Arabidopsis photoreceptor CRY2 by two distinct E3 ubiquitin ligases. Nat. Commun. 12:2155. doi: 10.1038/s41467-021-22410-x
Clough, S. J. (2005). Floral dip: agrobacterium-mediated germ line transformation. Methods Mol. Biol. 286, 91–102. doi: 10.1385/1-59259-827-7:091
Hirose, F., Shinomura, T., Tanabata, T., Shimada, H., and Takano, M. (2006). Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol. 47, 915–925. doi: 10.1093/pcp/pcj064
Imaizumi, T., Kanegae, T., and Wada, M. (2000). Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern adiantum capillus-veneris. Plant Cell 12, 81–96. doi: 10.1105/tpc.12.1.81
Li, Y. Y., Mao, K., Zhao, C., Zhang, R. F., Zhao, X. Y., Zhang, H. L., et al. (2013a). Molecular cloning of cryptochrome 1 from apple and its functional characterization in Arabidopsis. Plant Physiol. Biochem. 67, 169–177. doi: 10.1016/j.plaphy.2013.02.031
Li, Y. Y., Mao, K., Zhao, C., Zhao, X. Y., Zhang, R. F., Zhang, H. L., et al. (2013b). Molecular cloning and functional analysis of a blue light receptor gene MdCRY2 from apple (Malus domestica). Plant Cell Rep. 32, 555–566. doi: 10.1007/s00299-013-1387-4
Lin, C., Ahmad, M., and Cashmore, A. R. (1996). Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J. 10, 893–902. doi: 10.1046/j.1365-313x.1996.10050893.x
Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A. R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95, 2686–2690. doi: 10.1073/pnas.95.5.2686
Lin, X. C., Chow, T. Y., Chen, H. H., Liu, C. C., Chou, S. J., Huang, B. L., et al. (2010). Understanding bamboo flowering based on large-scale analysis of expressed sequence tags. Genet. Mol. Res. 9, 1085–1093. doi: 10.4238/vol9-2gmr804
Liu, Q., Su, T., He, W., Ren, H., Liu, S., Chen, Y., et al. (2020). Photooligomerization determines photosensitivity and photoreactivity of plant cryptochromes. Mol. Plant 13, 398–413. doi: 10.1016/j.molp.2020.01.002
Liu, Q., Wang, Q., Deng, W., Wang, X., Piao, M., Cai, D., et al. (2017). Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat. Commun. 8:15234. doi: 10.1038/ncomms15234
Liu, Q., Wang, Q., Liu, B., Wang, W., Wang, X., Park, J., et al. (2016). The blue light-dependent polyubiquitination and degradation of Arabidopsis cryptochrome2 requires multiple E3 Ubiquitin Ligases. Plant Cell Physiol. 57, 2175–2186. doi: 10.1093/pcp/pcw134
Liu, S., Zhang, L., Gao, L., Chen, Z., Bie, Y., Zhao, Q., et al. (2022). Differential photoregulation of the nuclear and cytoplasmic CRY1 in Arabidopsis. New Phytol. 2022:7. doi: 10.1111/nph.18007
Lyu, X., Cheng, Q., Qin, C., Li, Y., Xu, X., Ji, R., et al. (2021). GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 14, 298–314. doi: 10.1016/j.molp.2020.11.016
Ma, L., Li, X., Zhao, Z., Hao, Y., Shang, R., Zeng, D., et al. (2021). Light-Response Bric-A-Brack/Tramtrack/Broad proteins mediate cryptochrome 2 degradation in response to low ambient temperature. Plant Cell 33, 3610–3620. doi: 10.1093/plcell/koab219
Ma, L., Wang, X., Guan, Z., Wang, L., Wang, Y., Zheng, L., et al. (2020). Structural insights into BIC mediated inactivation of Arabidopsis cryptochrome 2. Nat. Struct. Mol. Biol. 6, 1432–1438. doi: 10.1038/s41477-020-00800-1
Mao, K., Jiang, L., Bo, W., Xu, F., and Wu, R. (2014). Cloning of the cryptochrome-encoding PeCRY1 gene from Populus euphratica and functional analysis in Arabidopsis. PLoS One 9:e115201. doi: 10.1371/journal.pone.0115201
Matsumoto, N., Hirano, T., Iwasaki, T., and Yamamoto, N. (2003). Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol. 133, 1494–1503. doi: 10.1104/pp.103.025759
Miao, L., Zhao, J., Yang, G., Xu, P., Cao, X., Du, S., et al. (2021). Arabidopsis cryptochrome 1 undergoes COP1 and LRBs-dependent degradation in response to high blue light. New Phytol. 2021:695. doi: 10.1111/nph.17695
Mockler, T., Yang, H., Yu, X., Parikh, D., Cheng, Y. C., Dolan, S., et al. (2003). Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 100, 2140–2145. doi: 10.1073/pnas.0437826100
Ni, W., Xu, S. L., Gonzalez-Grandio, E., Chalkley, R. J., Huhmer, A. F. R., Burlingame, A. L., et al. (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8:15236. doi: 10.1038/ncomms15236
Ninu, L., Ahmad, M., Miarelli, C., Cashmore, A. R., and Giuliano, G. (1999). Cryptochrome 1 controls tomato development in response to blue light. Plant J. 18, 551–556. doi: 10.1046/j.1365-313x.1999.00466.x
Numata, M. (1970). Conservation implications of bamboo flowering and death in Japan. Biol. Conserv. 2, 227–229. doi: 10.1016/0006-3207(70)90120-5
Ozkan-Dagliyan, I., Chiou, Y.-Y., Ye, R., Hassan, B. H., Ozturk, N., and Sancar, A. (2013). Formation of Arabidopsis cryptochrome 2 photobodies in mammalian nuclei: application as an optogenetic dna damage checkpoint switch. J. Biol. Chem. 288, 23244–23251. doi: 10.1074/jbc.M113.493361
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379. doi: 10.1101/gad.1231804
Platten, J. D., Foo, E., Elliott, R. C., Hecht, V., Reid, J. B., and Weller, J. L. (2005). Cryptochrome 1 contributes to blue-light sensing in pea. Plant Physiol. 139, 1472–1482. doi: 10.1104/pp.105.067462
Reisdorph, N. A., and Small, G. D. (2004). The CPH1 gene of Chlamydomonas reinhardtii encodes two forms of cryptochrome whose levels are controlled by light-induced proteolysis. Plant Physiol. 134, 1546–1554. doi: 10.1104/pp.103.031930
Rosenfeldt, G., Viana, R. M., Mootz, H. D., von Arnim, A. G., and Batschauer, A. (2008). Chemically induced and light-independent cryptochrome photoreceptor activation. Mol. Plant 1, 4–12. doi: 10.1093/mp/ssm002
Sang, Y., Li, Q. H., Rubio, V., Zhang, Y. C., Mao, J., Deng, X. W., et al. (2005). N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17, 1569–1584.
Shalitin, D., Yang, H., Mockler, T. C., Maymon, M., Guo, H., Whitelam, G. C., et al. (2002). Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417, 763–767. doi: 10.1038/nature00815
Shalitin, D., Yu, X., Maymon, M., Mockler, T., and Lin, C. (2003). Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell 15, 2421–2429. doi: 10.1105/tpc.013011
Shao, K., Zhang, X., Li, X., Hao, Y., Huang, X., Ma, M., et al. (2020). The oligomeric structures of plant cryptochromes. Nat. Struct. Mol. Biol. 27, 480–488. doi: 10.1038/s41594-020-0420-x
Szucs, P., Karsai, I., von Zitzewitz, J., Meszaros, K., Cooper, L. L., Gu, Y. Q., et al. (2006). Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theor. Appl. Genet. 112, 1277–1285. doi: 10.1007/s00122-006-0229-y
Wang, Q., and Lin, C. (2020). Mechanisms of cryptochrome-mediated photoresponses in plants. Annu. Rev. Plant Biol. 71, 103–129. doi: 10.1146/annurev-arplant-050718-100300
Wang, Q., Zuo, Z., Wang, X., Gu, L., Yoshizumi, T., Yang, Z., et al. (2016). Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354, 343–347. doi: 10.1126/science.aaf9030
Wang, X., Jiang, B., Gu, L., Chen, Y., Mora, M., Zhu, M., et al. (2021). A photoregulatory mechanism of the circadian clock in Arabidopsis. Nat. Plants 7, 1397–1408. doi: 10.1038/s41477-021-01002-z
Weidler, G., ZurOvenKrockhaus, S., Heunemann, M., Orth, C., Schleifenbaum, F., Harter, K., et al. (2012). Degradation of Arabidopsis CRY2 is regulated by SPA proteins and phytochrome A. Plant Cell 24, 2610–2623. doi: 10.1105/tpc.112.098210
Xu, P., Xiang, Y., Zhu, H., Xu, H., Zhang, Z., Zhang, C., et al. (2009). Wheat cryptochromes: subcellular localization and involvement in photomorphogenesis and osmotic stress responses. Plant Physiol. 149, 760–774. doi: 10.1104/pp.108.132217
Yu, X., Klejnot, J., Zhao, X., Shalitin, D., Maymon, M., Yang, H., et al. (2007). Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19, 3146–3156. doi: 10.1105/tpc.107.053017
Yu, X., Sayegh, R., Maymon, M., Warpeha, K., Klejnot, J., Yang, H., et al. (2009). Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell 21, 118–130. doi: 10.1105/tpc.108.061663
Zhang, Q., Li, H., Li, R., Hu, R., Fan, C., Chen, F., et al. (2008). Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl. Acad. Sci. USA 105, 21028–21033. doi: 10.1073/pnas.0810585105
Keywords: Phyllostachys edulis, light signaling, cryptochrome, PhePPK, PheBIC
Citation: Chen Z, Li M, Liu S, Chen X, Zhang W, Zhu Q, Kohnen MV and Wang Q (2022) The Function and Photoregulatory Mechanisms of Cryptochromes From Moso Bamboo (Phyllostachys edulis). Front. Plant Sci. 13:866057. doi: 10.3389/fpls.2022.866057
Received: 30 January 2022; Accepted: 07 March 2022;
Published: 30 March 2022.
Edited by:
Jigang Li, China Agricultural University, ChinaReviewed by:
Ruohe Yin, Shanghai Jiao Tong University, ChinaZhou Mingbing, Zhejiang Agriculture and Forestry University, China
Copyright © 2022 Chen, Li, Liu, Chen, Zhang, Zhu, Kohnen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Markus V. Kohnen, S29obmVuLk1hcmt1c0B3ZWIuZGU=; Qin Wang, cWlud2FuZ0NSWUAxNjMuY29t
 Ziyin Chen
Ziyin Chen Min Li1
Min Li1 Siyuan Liu
Siyuan Liu Qiang Zhu
Qiang Zhu Qin Wang
Qin Wang