- Department of Microbiology and Cell Biology, Indian Institute of Science, Bengaluru, India
Stem cell homeostasis by the WUSCHEL–CLAVATA (WUS-CLV) feedback loop is generally conserved across species; however, its links with other meristem regulators can be species-specific, rice being an example. We characterized the role of rice OsbZIP47 in vegetative and reproductive development. The knockdown (KD) transgenics showed meristem size abnormality and defects in developmental progression. The size of the shoot apical meristem (SAM) in 25-day OsbZIP47KD plants was increased as compared to the wild-type (WT). Inflorescence of KD plants showed reduced rachis length, number of primary branches, and spikelets. Florets had defects in the second and third whorl organs and increased organ number. OsbZIP47KD SAM and panicles had abnormal expression for CLAVATA peptide-like signaling genes, such as FON2-LIKE CLE PROTEIN1 (FCP1), FLORAL ORGAN NUMBER 2 (FON2), and hormone pathway genes, such as cytokinin (CK) ISOPENTEYLTRANSFERASE1 (OsIPT1), ISOPENTEYLTRANSFERASE 8 (OsIPT8), auxin biosynthesis OsYUCCA6, OsYUCCA7 and gibberellic acid (GA) biosynthesis genes, such as GRAIN NUMBER PER PANICLE1 (GNP1/OsGA20OX1) and SHORTENED BASAL INTERNODE (SBI/OsGA2ox4). The effects on ABBERANT PANICLE ORGANIZATION1 (APO1), OsMADS16, and DROOPING LEAF (DL) relate to the second and third whorl floret phenotypes in OsbZIP47KD. Protein interaction assays showed OsbZIP47 partnerships with RICE HOMEOBOX1 (OSH1), RICE FLORICULA/LEAFY (RFL), and OsMADS1 transcription factors. The meta-analysis of KD panicle transcriptomes in OsbZIP47KD, OsMADS1KD, and RFLKD transgenics, combined with global OSH1 binding sites divulge potential targets coregulated by OsbZIP47, OsMADS1, OSH1, and RFL. Further, we demonstrate that OsbZIP47 redox status affects its DNA binding affinity to a cis element in FCP1, a target locus. Taken together, we provide insights on OsbZIP47 roles in SAM development, inflorescence branching, and floret development.
Introduction
The post-embryonic shoot development in flowering plants depends on the balance between stem cell renewal in the central zone of above ground meristems and the adoption of specific differentiation programs in cells from the peripheral zone. The genetic framework of the basic WUSCHEL–CLAVATA (WUS-CLV) pathway for meristem maintenance is largely conserved in monocots and dicots, yet some functional differences are reported among cereal grass models, such as maize and rice. In maize, functions for TASSEL DWARF1 (TD1, CLV1 ortholog) and FASCIATED EAR2 (FEA2, CLV2 ortholog) in the shoot apical meristem (SAM) are not obvious in the respective mutants, yet mutants in these genes have significant and somewhat differential effects on the female vs. male inflorescence meristems (IMs) (Bommert et al., 2005; Dodsworth, 2009; Pautler et al., 2013; Chongloi et al., 2019). In rice, FLORAL ORGAN NUMBER 1 (FON1) is the CLV1 ortholog, while FON2/FON4, FON2 SPARE1 (FOS1) and FON2-LIKE CLE PROTEIN1 (FCP1) encode CLV3 peptide paralogs. FON2 signaling through FON1 majorly regulates the homeostasis of IMs whereas FCP1 triggered signaling regulates SAM through effects on WUSCHEL RELATED HOMEOBOX (OsWOX4), functionally related to AtWUS (Nagasawa et al., 1996; Suzaki et al., 2004, 2006; Ohmori et al., 2013). These tissue-specific effects of maize TD1, FEA2, and of rice CLV3-like genes exemplify species-specific innovations in signaling components of this core meristem regulatory circuit. In the SAM of Arabidopsis, AtWUS activates SHOOT MERISTEMLESS (STM), and both directly regulate CLV3 expression to maintain constant stem cell number (Su et al., 2020). Integration of CLV-WUS pathway with the roles of class I KNOTTED-1-LIKE HOMEOBOX (KNOX) genes, Arabidopsis STM, rice HOMEOBOX1 (OSH1), and maize KNOTTED1 (KN1) in meristem maintenance is conserved across species (Vollbrecht et al., 2000; Brand et al., 2002; Tsuda et al., 2011). Similarly, the interlinking of WUS-CLV pathway with phytohormone-based meristem control, by cytokinin (CK), auxin (IAA/AUX), gibberellin (GA), brassinosteriod (BR) actions, is also conserved (Kurakawa et al., 2007; Gordon et al., 2009; Lee et al., 2009; Zhao et al., 2010; Yamaki et al., 2011; Somssich et al., 2016). OSH1 positively autoregulates itself directly by binding to evolutionarily conserved cis-elements within its locus (Tsuda et al., 2011). Further, OSH1 induces the expression of CK biosynthesis genes, such as ADENOSINE PHOSPHATE ISOPENTENYL TRANSFERASE 2 (OsIPT2) and OsIPT3 (Sakamoto et al., 2006). Interestingly, CK treatment of callus activates the transcription of rice KNOX genes (Tsuda et al., 2011). AtWUS and OsWOX4 also regulate CK signaling in Arabidopsis and rice, respectively (Leibfried et al., 2005; Ohmori et al., 2013). In the floral meristem center, the timing of the termination of stem cell activity is co-incident with carpel/ovule specification. This creates a determinate floral meristem for normal reproduction. Meristem termination is mediated by the concerted activity of floral organ identity genes (Class A, C, and E) whose regulatory effects on WUS-CLV and WUS-KNOX pathway genes operate in both monocots and eudicots [reviewed by Tanaka et al. (2013), Callens et al. (2018), and Chongloi et al. (2019)]. These genes are in turn spatially and temporally regulated. For example, Arabidopsis LEAFY (LFY) directly activates APETALA1 (AP1) and WUS (Lenhard et al., 2001; Lohmann et al., 2001) while repressing the shoot meristem identity gene, TERMINAL FLOWER1 (TFL1) (Moyroud et al., 2009, 2010). Furthermore, in young floral meristems, LFY together with UNUSUAL FLORAL ORGAN (UFO) and WUS activate APETALA3 (AP3) and AGAMOUS (AG) gene expression in the third and fourth whorls of the developing meristem (Parcy et al., 1998; Busch et al., 1999; Wagner et al., 1999; Lenhard et al., 2001; Lohmann et al., 2001). In the later stages of floral meristem development, AG directly activates KNUCKLES (KNU) which leads to the repression of WUS by the recruitment of Polycomb group (PcG) chromatin modifiers (Ming and Ma, 2009; Sun et al., 2009, 2014; Liu et al., 2011; Zhang, 2014). Aside from LFY, the AG expression is also influenced by PERIANTHIA (PAN) that encodes a bZIP class TF, whose orthologs are Oryza sativa basic LEUCINE ZIPPER 47 (OsbZIP47) and maize FASCIATED EAR4 (ZmFEA4). Floral meristem size and organ patterning defects in the Arabidopsis pan-3 lfy-31 double the mutant, and in transgenics with modified PAN fusion proteins (repressive vs. activated forms) show roles of AtPAN in floral determinacy, meristem size, and floral organ patterning (Running and Meyerowitz, 1996; Chuang et al., 1999; Das et al., 2009; Maier et al., 2009, 2011). Maize ZmFEA4 activates the expression of genes involved in AUX pathway and lateral organ differentiation and also regulates both SAM and IM size homeostasis (Pautler et al., 2015). Unlike Arabidopsis and maize, OsbZIP47 (LOC_Os06g15480) is not well-characterized, and its interacting partners are largely unknown. Further, how inflorescence BM identity and transition regulators intersect with the two meristem maintenance pathways (CLV-WUS and KNOX1) is not much explored in rice. Here, functional characterization of OsbZIP47 by RNA interference (dsRNAi)-based knockdown (KD) and identification of some meristem regulators, such as OsbZIP47 interacting partners sheds light on its role in meristem size and meristem developmental progression. Further, our transcriptome and meta-analysis uncovered downstream pathways that can be co-regulated by OsbZIP47 and OSH1, OsMADS1, or RFL.
Materials and Methods
Plasmid Constructs Generation and Rice Transformation
For siRNA (interference) mediated KD of endogenous OsbZIP47, a gene-specific 226bp 3′UTR DNA fragment was cloned in the sense and in the antisense orientation in pBluescript vector, and were separated by a 270-bp linker. Subsequently, the insert in recombinant pBluescript was re-cloned in the binary rice expression vector, pUN downstream to the maize ubiquitin promoter for the expression of OsbZIP47 hairpin RNAs (Supplementary Figure 1; Prasad et al., 2001). For the over-expression of OsbZIP47, the full length cDNA was cloned at BamHI (blunted)-KpnI sites in the pUN vector to create pUbi:OsbZIP47 (Supplementary Figure 4). These constructs for KD and over-expression of OsbZIP47 were transformed into the Agrobacterium tumefaciens strain, LBA4404 and then co-cultivated with embryogenic calli from TP309 WT (O. Sativa var japonica) seeds as described by Prasad et al. (2001). Transgenic plants, dsRNAi OsbZIP47 and Ox-OsbZIP47, were grown in IISc, Bangalore, Green house condition, approximately at 27°C during the months of January–May and July–October always with wild-type (WT) controls.
Phenotypic Characterization of Knockdown Transgenic
The transgenic plants, such as dsRNAi OsbZIP47 and Ox-OsbZIP47 were selected on half-strength MS medium containing 50 mg L–1 of hygromycin. Phenotypic analysis was done with T3 dsRNAi OsbZIP47 transgenics and T1 Ox-OsbZIP47 lines. Eosin-hematoxylin stained 25 DAG seedling tissue sections (7 μm, Lecia microtome; RM2045) were imaged by Apotome2 Zeiss. The cell size in the SAM was measured by ImageJ. The seedling height was measured at age eight DAG. Adult plant height, lamina joint angle, panicle length, branch characteristics, and spikelet numbers were measured after panicle booting. Pre-anthesis florets, in panicles prior to emergence from the flag leaf, were imaged using Leica Wild M3Z stereomicroscope.
RNA-Sequencing and RT-qPCR
Next Generation Sequencing (NGS) of RNA from OsbZIP47 KD panicles (0.1–0.5 cm) was done for two biological replicates with matched WT panicles as controls. The total RNA was extracted using Trizol Reagent (Sigma) according to manufacturer’s instructions. About 1 μg of total RNA was used for library preparation using rRNA depletion-based NEB Next UltraII RNA kit. The NGS was performed using Illumina Hi-Seq, pair-end 2 × 150 bp chemistry. After quality check (using FastQC and multiQC software), the reads were mapped against indexed O. sativa ssp. japonica cv. reference genome (RAP-DB)1 using STAR2 (v2.5.3a). Further, differential gene expression (DGE) of read counts between WT and transgenics were computed using edgeR (v3.28.0) package with the absolute log2 fold change ≥ 1 with p-value ≤ 0.05. For real-time qPCR experiments, oligo(dT)-primed cDNAs were synthesized using 2 ug of total RNA with MMLV (reverse transcriptase, NEB). The qRT-PCR reactions were set up with 50–70 ng of cDNA, 250 nM gene-specific primers, and FastStart Universal Sybr Green Master (Rox) mix (Roche) in CFX384 real-time system (Biorad) or Applied Biosystems ViiA 7 system. Fold change in the transcript levels of deregulated genes was calculated as a difference in cycle threshold value between transgenic and wild type. To obtain normalized threshold value (ΔΔCt), first ΔCt value was calculated by subtracting the Ct value for internal control; Ubiquitin5, from the Ct value for each gene of interest (Gene Ct-Ubi5 Ct). Then ΔΔCt was calculated by subtracting the WT ΔCt value from the ΔCt value obtained from the transgenic tissue. The fold-change was calculated as 2^–(ΔΔCt). Primers used and their sequences are listed in Supplementary Table 2. The RNA sequencing raw data files used in this study have been deposited to Gene Expression Omnibus (GEO) database under the accession number GSE196747.
RNA in situ Hybridization
To generate OsbZIP47 riboprobes, a gene-specific 226bp DNA fragment from 3′UTR (1329-1555 bp) was PCR-amplified and cloned in the pBluescript KS + vector. Sense and antisense Digoxigenin-labeled (DIG-UTP, Roche) riboprobes were prepared by in vitro transcription using T3 and T7 RNA polymerases (NEB), respectively. Tissue processing and probe hybridizations was done according to the study by Prasad et al. (2005). Signal was developed using anti-digoxygenin-alkaline phosphatase (AP) conjugated antibodies (Roche) and 5-Bromo-4-chloro-3-indolyl phosphate (BCIP)-nitro blue tetrazolium (NBT) chromogenic substrates (Roche). Images were captured by Apotome2 Zeiss microscope system.
Bacterial Expression of OsbZIP47 Full-Length Protein and Studies of Oligomeric Status
For OsbZIP47 protein expression and purification from bacteria, OsbZIP47 full-length (FL) CDS was cloned in the pET32a vector. Thioredoxin-His-tagged OsbZIP47FL was expressed from Rosetta (DE3) bacterial strain induced with 0.2 mM of isopropyl β- d-1-thiogalactopyranoside (IPTG) for 3 h at 37°C. Oligomeric states of OsbZIP47 protein was determined by analytical size-exclusion chromatography (SEC) performed at 4oC on a Superdex 200 increase column Cytiva (Formerly, GE Healthcare Life Sciences), Marlborough, United States pre-equilibrated with a buffer (25 mM of sodium phosphate (pH 7.4), 100 mM of NaCl, and 5% of glycerol). Approximately, 400 μg of protein, (∼2 mg/ml) was injected into AKTA purifiers Cytiva (Formerly, GE Healthcare Life Sciences), Marlborough, United States connected to the column. The flow rate was maintained at 0.3 ml/min and the protein elution profile was at 220 nm. The molecular weight was calculated using a standard plot. Molecular weight was calculated using the equation: Y = −0.602X + 4.6036, where Y = Ve/Vo (Ve = Elution volume; Vo = Void volume) and X = Log of molecular weight in Dalton.
Electrophoretic Mobility Shift Assays
Escherichia coli rosetta (DE3) bacterial lysates with the Trx-His-OsbZIP47 FL was prepared in a buffer: 10mM of HEPES-KOH, pH 7.8, 50mM of NaCl, 0.5% of Non-idet P-40, 0.5 mM of EDTA, 1 mM of MgCl2, 10% of glycerol, 0.5 mM of DTT, and 1x protease inhibitor cocktail (Sigma). About 1–4 μl of lysate was incubated with 5’end P32 labeled DNA oligonucleotide probes for 30 min at 4°C in 1× EMSA buffer (20 mM of HEPES-KOH pH 7.8, 100 mM of KCl, 2 mM of DTT, 1 mM of ETDA, 0.1% of BSA, 10 ng of Herring sperm DNA, 10% of glycerol, 1× protease inhibitor cocktail) in 15 μl reactions. After binding, the reaction constituents were resolved on an 8% of native-PAGE gel in 0.5× Tris-borate EDTA (TBE) buffer at room temperature. Gel autoradiography was done in a phoshorimager (GE; Typhoon FLA 9500). The DNA probe sequences are listed in Supplementary Table 2.
Microscale Thermophoresis Assay
Escherichia coli rosetta (DE3)-expressed proteins (Trx-His-OsbZIP47 FL and Trx-His; Supplementary Figure 9) were added to the buffer: 10mM of HEPES-KOH pH 8.0, 50 mM of NaCl, 0.5% of TWEEN-20, 0.5 mM of EDTA, 1 mM of MgCl2, 10% of glycerol, 2 mM of beta mercaptoethanol, and 1 mM of PMSF. About 10 mM of each protein was labeled with 5 mM of Red-NHS 2nd Generation primary amine labeling dye (NanoTemper GmbH, Cat# MO-L011), and then eluted in the buffer: 10 mM of HEPES-KOH pH 8.0, 50 mM of NaCl, 0.5% of TWEEN-20, 0.5 mM of EDTA, 1 mM of MgCl2, 10% of glycerol, 1X PIC (ROSCHE), and a reducing/oxidizing agent (20 mM of DTT/1 mM of oxidized glutathione). The labeled protein was incubated with dsOsFCP1 oligonucleotides which was serially diluted from 100 μM to 3.05 nM in 16 steps and fluorescence was measured using Monolith NT.115Pico (NanoTemper GmbH). The excitation power was varied between 1 and 50% to obtain measurable fluorescence signal. The MST power was varied between medium to high, to achieve high signal to noise ratio. For OsbZIP47 FL, the initial fluorescence was measured which is indicative of rapid binding. For Trx-His tag, the response evaluation was done at default on time. MO Control v1.6.1 (NanoTemper GmbH) and MO Affinity Analysis v2.6 (NanoTemper GmbH) were used for the analysis.
Yeast Two-Hybrid Assays
The full length CDS of OsbZIP47 was amplified from KOME clone, AK109719 using gene-specific primers, cloned into pBSKS vector and validated by restriction digestions and Sanger sequencing. The CDS was subsequently cloned into yeast two hybrid vectors, pGBDUC1 and pGADC1. Similarly, all the CDS fragments that would encode prey proteins, such as OsMADS1, OsETTIN1/2, RFL, OSH1, OsMADS15, OsMADS2, OSH15, and OsMADS15 were PCR-amplified from either KOME cDNA clones or from cDNA made from panicle tissue RNA, and subcloned into pGBDUC1 and pGADC-1 vectors. The bait clone, pGBDUC1 OsbZIP47 and indicated prey recombinants in pGADC1 were co-transformed into the yeast, pJ69-4A yeast two-hybrid (Y2H) strain (James et al., 1996). Transformants were selected on synthetic drop out media lacking leucine and uracil. Protein interactions were assessed in at least five purified transformants by serial dilution spotting of broth cultures onto SD/-Leu-Ura-His plates supplemented with 10 mM of 3AT and by the ONPG assay (Supplementary Materials and Methods).
Bimolecular Fluorescence Complementation Assays
The OsbZIP47 cDNA with a truncated C domain (amino acid 199–385) was cloned into pSPYCE (M) (C-terminal fusion) and pSPYNE (R) 173 (N-terminal fusion) bimolecular fluorescence complementation (BiFC) vectors (Waadt et al., 2008). Similarly, the full-length CDS encoding prey proteins, such as OsMADS1, OsETTIN 2, RFL, OSH1, and OsMADS15 were subcloned into pSPYNE (R) 173 vector. Six combinations of cEYFP and nEYFP fusions, including positive and negative controls, were transiently co-expressed in onion (Allium cepa) epidermal cells by Agrobacterium. tumefaciens (C58C1) infiltration as described by Xu et al. (2014). Co-transformed tissues were incubated at 25°C in dark for 48 h before being assayed for YFP activity. Fluorescence images were screened using a confocal laser microscope (Zeiss LSM880, Airyscan) with 2AU 480 nm excitation and 520 nm emission for the detection of YFP signal.
Meta-Analysis
The published transcriptome datasets in dsRNAiOsMADS1 and dsRNAiRFL panicles were adopted in this study to compare them with that of OsbZIP47KD transcriptome dataset. The differentially expressed genes (DEGs) from each dataset was taken up for pair-wise comparison to identify unique, or commonly (upregulated, or downregulated) downstream genes. The deregulated genes were also corelated with the published data on OSH1 genome-wide binding (Supplementary Materials and Methods). To align genes from the diverse datasets, i.e., transcriptomes downstream to OsMADS1, RFL, and genes bound by OSH1 for meta-analysis and for GO enrichment analyses, the gene IDs as per RAP-dB (see text footnote 1) were converted to their corresponding gene ID in MSU-TIGR v7.2 After this curation, among the 2,800 RAP-dB ID genes, only 2,210 genes were also annotated in MSU-TIGR v7. The list of RAP-dB gene IDs and their corresponding MSU v7 LOC_IDs are presented in Supplementary Dataset 1.
Results
OsbZIP47 Knockdown Plants Have Enlarged Shoot Apical Meristem Size
To investigate the developmental roles of OsbZIP47, we generated thirteen independent OsbZIP47 KD transgenics (OsbZIP47KD) by RNA interference (dsRNAi, Figure 1A) specific to a unique region of OsbZIP47 3′UTR. Based on the degree of KD and seed viability in primary T0 transgenic lines, we chose two lines; OsbZIP47KD line #10 and OsbZIP47KD line #14 for detailed phenotypic analysis in T3 generation. As a representative, phenotypic data from the OsbZIP47KD line #14 is further discussed here. In pooled panicle tissues (0.1–0.5 cm) from this line, qRT-PCR showed approximately 24-fold downregulation of the endogenous OsbZIP47 transcripts (Supplementary Figure 1). The earliest phenotype noted was the seedling height at 8 days after germination (DAG), which was significantly reduced in OsbZIP47KD as compared to the WT (Figures 1B,C and Supplementary Table 1). This observation led us to examine SAM in the histological sections of seedlings aged 25 days after germination (DAG) from both WT and OsbZIP47KD plants. First, we examined SAM size and found that SAM area was increased as compared to WT (Figures 1D,F). Consistent with SAM enlargement in OsbZIP47KD plants, SAM width and height showed significant and marginal increase, respectively (Supplementary Table 1). To understand the cellular differences that underlie meristem size abnormalities, the cell size of L1 layer in the upper central and the peripheral zone of the meristem and the internal cells underlying the L1 layer were measured. Intriguingly, the size of these cells was increased as compared to WT (Figure 1E). Further, the spatial distribution of dividing cells in 25DAG SAMs was assessed by RNA in situ hybridization for the cell cycle S-phase marker, HISTONE4 (H4) (Supplementary Figure 2). Compared to WT, the overall H4 transcript signal in the median longitudinal SAM sections of OsbZIP47KD seedlings was higher. Altogether, these results suggest that the increased SAM area in OsbZIP47KD seedlings is attributed to an increase in cell size and number. To understand some molecular corelates for SAM phenotypes, transcript levels for few known regulators of SAM size homeostasis were tested (Figure 1G) using SAM tissues from 25 DAG seedlings. The downregulation of FCP1 and FON2 (rice homologs of CLV3), APO1 (UFO1 homolog), CYP734A4, and YUCCA6 was observed. Also, CUC1, the lateral meristem boundary marker (Aida et al., 1997; Takeda et al., 2011), showed a marginal reduction in expression. The downregulation of rice CLV3 homologs in OsbZIP47KD SAM may contribute to the overall enlarged SAM size. Further, the increased cell size in L1 layer and its underlying cells could be attributed to a reduction in CYP734A4 expression in SAM of OsbZIP47KD plants. Of note is the report that SAM cells in CYP734A RNAi plants are more vacuolated as compared to the WT which was suggested to indicate premature cell differentiation (Tsuda et al., 2014). Altogether, these gene expression effects of OsbZIP47 can be related to abnormal SAM size homeostasis on its KD with novel effects on the components in the CLV-WUS pathway, on other meristem regulators, AUX, and BR phytohormone pathways.
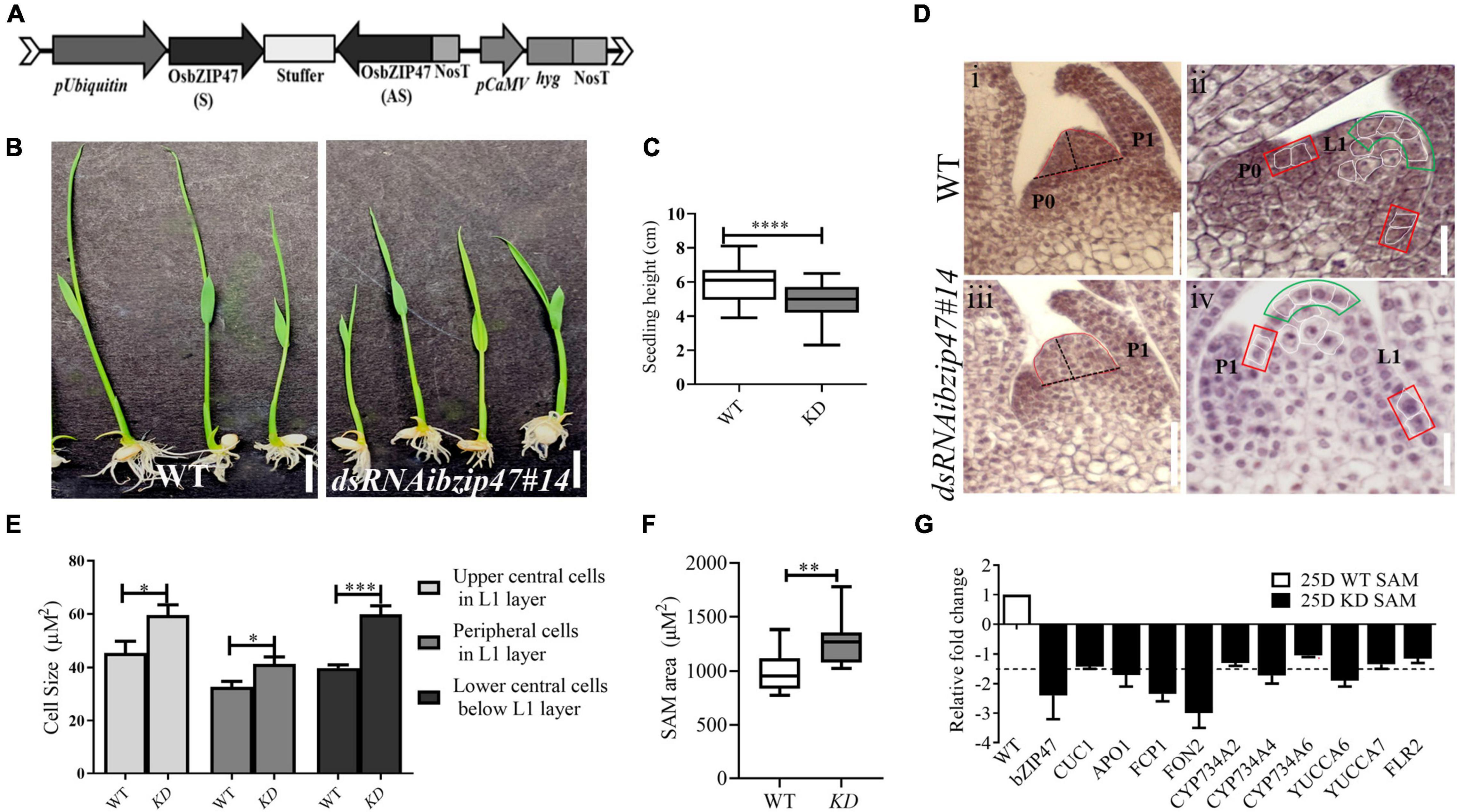
Figure 1. OsbZIP47 knockdown (KD) vegetative phenotypes. (A) Schematic representation of dsRNAiOsbZIP47 transgene T-DNA segment. (B) Seedling height of 8-day OsbZIP47KD (dsRNAibZIP47#14) plants is shorter than that of wild-type (WT) plants. Scale bar = 1 cm. (C) Seedling height data shown as mean ± s.d., Student’s t-tests, ****P < 0.0001, n = 30. (D) Representative histological sections of SAMs from WT and dsRNAibZIP47#14. SAM area in 25 days after germination (DAG) seedlings marked by red outline in panels (i,iii). SAM height and width are marked by dashed black lines. Measured cells are marked with white outlines in panels ii and iv. Scale bar in panes (i,iii) are of 50 μM, and panels (ii,iv) are of 20 μM. (E) Comparison of L1 cell size in the central, and peripheral cells, and in the lower central cells underlying the central L1 layer. The statistical analysis of cell size, for cells in different regions of the meristem, was derived from 10 to 12 different sections. Data is shown as mean ± s.d. Student’s t-tests. *P < 0.05, ***P < 0.001. (F) Comparison of SAM area in OsbZIP47KD (dsRNAibZIP47#14) vs. WT in 25 DAG seedlings. Data is shown as mean ± s.d. Student’s t tests, **P < 0.01, n = 10. (G) RT-qPCR analyses of CUC1, APO1, FCP1, FON2, CYP734A2, CYP734A4, CYP734A6, YUCCA6, DRUS2/FLR2, and FON2 transcripts in SAM from 25 DAG seedlings. Fold-change values were determined by comparing the normalized expression levels in OsbZIP47KD plants to WT plants.
Late Heading Date and Altered Panicle Architecture of OsbZIP47 Knockdown Plants
OsbZIP47KD plants are delayed by 20 days for SAM to IM transition. At this stage, OsbZIP47KD plant height was reduced compared to WT (Figures 2A–D) suggesting that in the WT, OsbZIP47 promotes developmental transition from the vegetative to reproductive phase. The shorter plant height was due to poor stem internode elongation in the KD transgenics without change in internode number (Figure 2F). The panicle of KD plants showed developmental abnormalities, i.e., reduced inflorescence axis (panicle rachis) length (Figure 2E), reduced number of primary branches, and spikelets (Figures 3A,L,M and Supplementary Table 1). Together, these phenotypes indicate early progression of primary branch meristems to spikelets in OsbZIP47KD plants and point to a possible role of OsbZIP47 in the temporal control of branch meristem indeterminacy. Moreover, OsbZIP47KD plants had greater flag leaf lamina joint angle as compared to the WT (Figures 3B,C,K). KD plants of CYP734A4 (Tsuda et al., 2014), a gene with reduced expression in OsbZIP47KD plants (Figure 1G), also share this phenotype and have abnormal meristems. Plant architecture, flowering time, leaf angle, and inflorescence architectures all impact yield, grain shape, and size (Harder and Prusinkiewicz, 2013; Sakuma and Schnurbusch, 2020). Interestingly, seeds from OsbZIP47KD plants were altered for the length/width (L/W) ratio as compared to WT (Figures 3I,J), suggesting OsbZIP47 impedes cell proliferation in the grain width direction, which is supported by a recent finding of Hao et al. (2021). In later sections of our study on dsRNAiOsbZIP47 lines, we identify some molecular links underlying cell proliferation in the developing grain. We propose that the role of OsbZIP47 in restriction of cell proliferation is likely attributed to the positive regulation of EL2, encoding a plant cyclin-dependent kinase inhibitor, and negative regulation of some key cell-cycle regulators i.e., CYCLIN-D7-1 (CYCD7;1) and MITOGEN-ACTIVATED PROTEIN KINASE KINASE 10 (MKK10-1) in WT panicles (Supplementary Dataset 1). Rice EL2 cell cycle inhibitory functions are proposed to link cell cycle progression with biotic and abiotic stress responses (Peres et al., 2007). In Arabidopsis, AtCYCD7 expression is transcriptionally regulated by cell type-specific transcription factors that confine its expression to appropriate developmental window as ectopic expression triggered division (Weimer et al., 2018). The OsMKK10-1 paralog, OsMKK10-2 phosphorylates OsMPK6 in vivo (Ma et al., 2017). This is interesting since mutations of OsMPK6 impair differentiation of L1 layer cells during early embryogenesis (Yi et al., 2016).

Figure 2. Phenotypic effects of OsbZIP47KD on plant growth and floral transition. (A) Flowering timing (SAM to inflorescence meristem (IM)/panicle meristem transition) in OsbZIP47KD plants is delayed by approximately 22 days as compared to the WT. White arrow indicates the booted panicle in WT and red arrow points to the absence of the booted panicle in a OsbZIP47KD plant of the same age. (B) Height of mature flowering plants shows that as compared to the WT, OsbZIP47KD plants are shorter. Quantitation of heading dates (C), plant height (D), and panicle rachis length (E) in WT and OsbZIP47KD plants. Data are shown as the mean ± s.d. (Student’s t-tests, **P < 0.01, ****P < 0.0001, n = 10). (F) Internodes in mature flowering WT and OsbZIP47KD plants are displayed and are numbered from I to V from the apical end. P is the panicle bearing node and internode. Bar, 2.5 cm. Shorter internodes from I to V in OsbZIP47KD contributes to the reduced height.
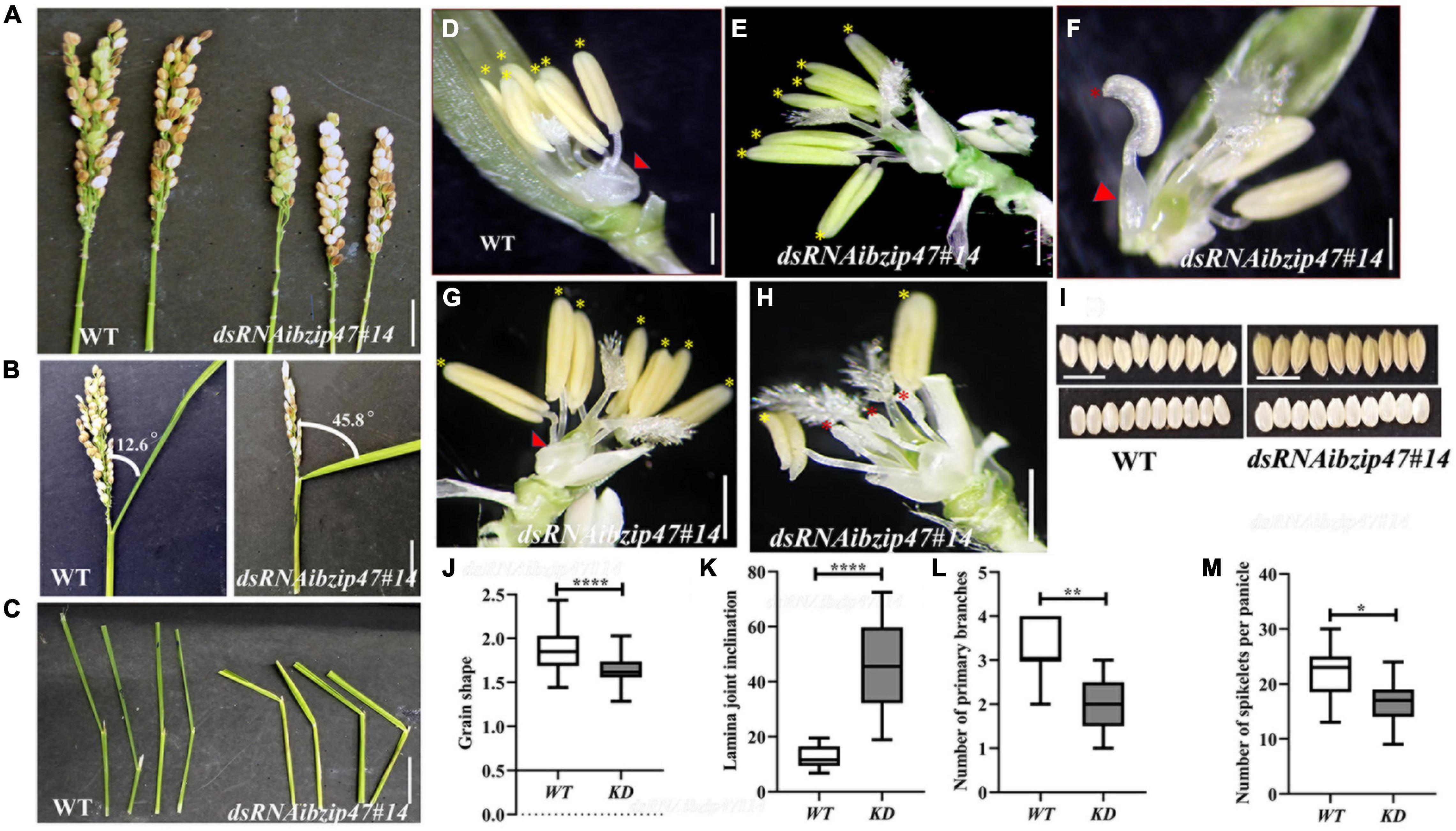
Figure 3. Floret organ numbers and organ development in OsbZIP47KD plants. (A) Shorter panicle OsbZIP47KD with reduced number of primary branches and spikelet number per panicle as compared to the WT plant. (B,C) Flag leaf angle in fully headed panicles shows increased lamina joint angle in OsbZIP47KD plant as compared to the WT. (D) WT floret organs shown after removal of lemma and the spikelet sterile lemmas. Red arrow points to the pair of lodicules and yellow asterisks to the six normal stamens. (E–H) Organ phenotypes in OsbZIP47KD florets. (E) Floret with seven stamens (yellow asterisks) with normal filaments and anthers. (F) Floret with mildly deformed lodicule (red arrowhead), two of its six normal stamens (others were removed) and a chimeric second whorl organ with lodicule and stamenoid features (red asterisk). (G) Floret with deformed lodicule and seven normal stamens (yellow asterisk). (H) Floret with slightly deformed lodicule, short stamens, and shrunken anthers (red asterisk), two near normal stamens (yellow asterisk). (I,J) Grain morphology in OsbZIP47KD and WT plants. The length/width (L/W) ratio of OsbZIP47KD seeds was lower than that of WT seeds, suggesting increased grain width. Scale bar = 1 cm in panels (A–I). (J–M) Statistical analysis (mean ± s. d.) of grain shape, lamina joint angle, primary branch number, and number of spikelets per panicles. Data in panels (J) [n = 40 in panel (J)], (K) (n = 10), (L) (n = 11), (M) (n = 9), Student’s t-tests, *P < 0.05, **P < 0.01, ***P < 0.001.
OsbZIP47 Contributes to Second and Third Whorls, Lodicule, and Stamen Organ Development
OsbZIP47KD floret phenotypes were largely restricted to lodicules and stamens (Figures 3D–H). The organ defects were grouped into four classes. Class I, representing 40% of OsbZIP4KD florets, had mild deformed lodicule (distal elongation) with normal stamen number (Supplementary Figure 3). In class II (∼28%), mild lodicule elongation occurred with abnormal short stamens and poorly developed anthers (Figure 3H). Florets of class III (∼20%) had partially deformed lodicules with an increase in stamen number to 7 (Figures 3E,G). In class IV (∼12%) florets had mildly elongated lodicules and chimeric organs with lodicule and stamen characteristics (Figure 3F). Also, in most florets of all classes, the lodicules were abnormally fused with lemma making dissection of the lemma from the floret difficult. Altogether, these data suggest that OsbZIP47 contributes to floral organ development in the second and third whorls. In a complementary analysis, we examined consequences of ubiquitous overexpression of full-length OsbZIP47cDNA in transgenic rice. Surprisingly, none of the OsbZIP47OX lines had any notable phenotypic changes from the WT despite ∼10-fold overexpression in OsbZIP47OX panicle tissues (Supplementary Figure 4). A speculation is that OsbZIP47 functions may depend on partners or that some post-translational modifications (PTMs) may modulate its functions, as was concluded from overexpression studies of Arabidopsis AtPAN (Chuang et al., 1999).
Tissue Expression Profile of OsbZIP47 Through Development
RNA in situ hybridization was performed to examine spatial distribution of OsbZIP47 mRNA in various above ground meristems. These experiments confirmed transcripts in meristems that is consistent with the phenotypes of OsbZIP47KD plants. In SAM of wild type young seedlings (5 DAG and 25 DAG), transcripts were evenly distributed (Figures 4A,B). This pattern is somewhat different from maize FEA4 where the signals are excluded from SAM stem cell niche and from incipient P0 leaf primordium (Pautler et al., 2015). During reproductive development, high levels of OsbZIP47 transcripts are shown at the apical end of growing IM/rachis and at the ends of branch meristem (PBM and SBM, Figures 4C,D) which may relate to the poorly branched inflorescence of knockdown plants. In elongating primary and secondary branches (Figure 4D), transcript signal is mild and spatially uniform. In spikelet meristem (SM, Sp2, Figure 4E), and in floral meristems (Sp4-Sp6, Figure 4F), the signal is high and spatially uniform. However, in mature florets, OsbZIP47 RNA was confined to the lodicule, stamen and carpel organ primordia, and differentiating organs (Figure 4G). Additionally, hybridization signal in carpel wall (c) and ovule (o) was observed (Figure 4H). Arabidopsis pan mutant flowers occasionally have multiple carpels up to three with deviated gynoecium (Running and Meyerowitz, 1996). We speculate OsbZIP47 may have a minor role in carpel development or could be functionally redundant with floral C-class function genes. Thus, OsbZIP47 is expressed in various above-ground meristems reflecting its diverse roles in different meristems.
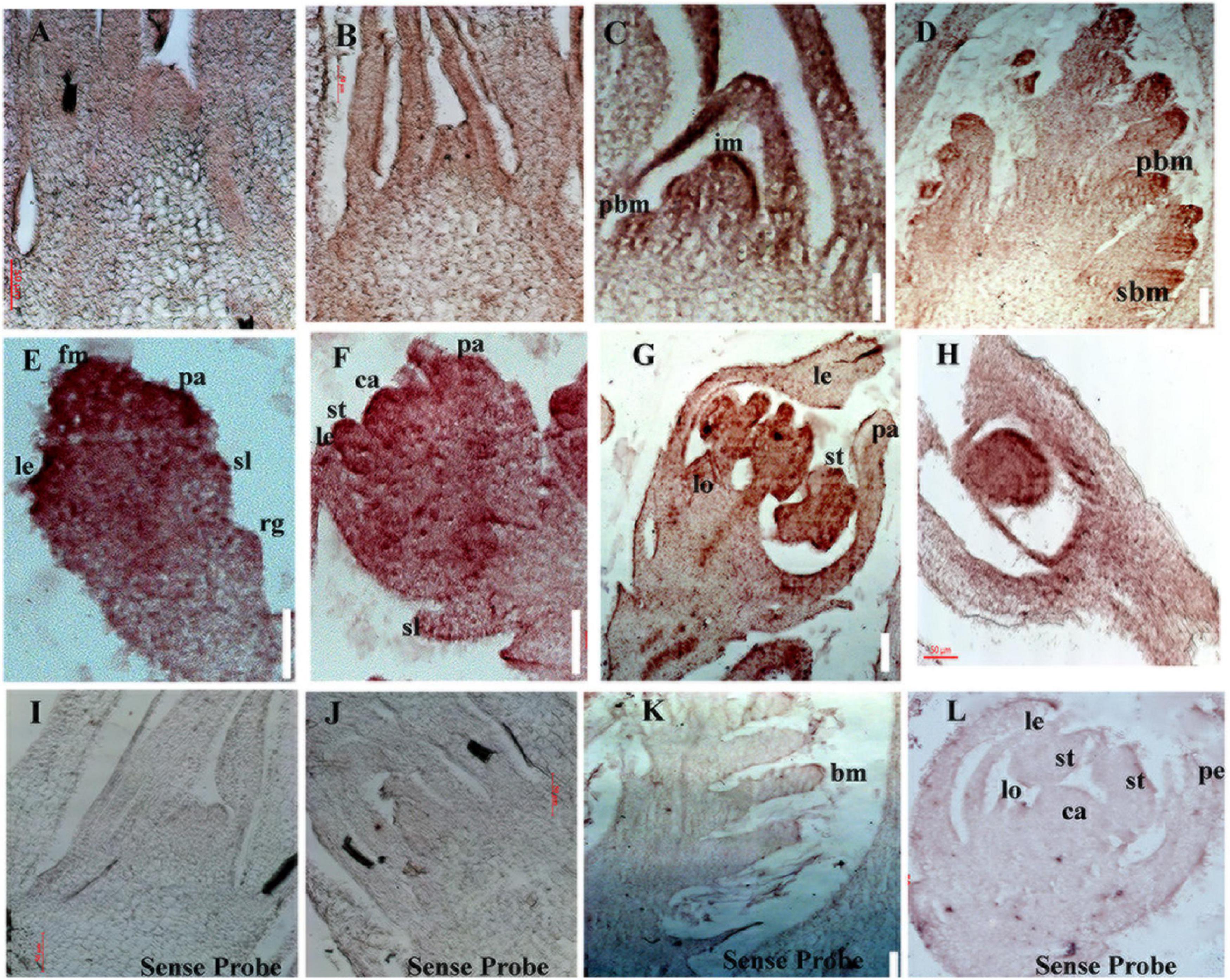
Figure 4. Spatial distribution of OsbZIP47 transcripts in meristems and florets. (A,B) OsbZIP47 in situ RNA hybridization signal in SAM tissues from 5 DAG and 25 seedlings. Expression in SAM and in emerging leaf primordia. (C,D) Inflorescence meristem (IM) with emerging primary branch meristems show expression of OsbZIP47 in the elongation meristem and primary branch meristems (prb), secondary branch meristems (srb), and in young leaves. (E) OsbZIP47 transcripts in very early floret meristem (FM) with uniform spatial distribution of signal. (F) Floret uniform signal in the well-formed stamens (st), lemma/palea (le and pa) organ primordia, and in the central early carpel primordia. (G) High level of OsbZIP47 expression in the lodicule and in stamens, and lower signal in the near mature lemma and palea. (H) OsbZIP47 transcripts in the ovary wall and ovule. (I,J) SAM in 5 DAG and 25 DAG plants probe with sense probe as a negative control. (K,L) IM and near mature floret, respectively probed with sense RNA.
Heterodimerization of OsbZIP47 With Other Floral Meristem Regulators
Heterodimerization of transcription factors can modulate genome wide gene expression by modifying specificity and affinity to target DNA binding sites, and by integrating independent pathways controlled by two or more factors. Interacting partners of OsbZIP47 or of its Arabidopsis and Maize homologs are largely unknown. To understand molecular mechanism of OsbZIP47, we investigated its interaction with different transcription factors. Among the co-occurring motifs in genome-wide loci bound by OsMADS1, the motif for bZIP factor binding is enriched (Khanday et al., 2016). This was the basis for our hypothesis that OsMADS1 and members of OsbZIP family could function in complexes in early floral meristems. Further, the temporal co-expression profiles of OsMADS1 and OsbZIP47 overlap in developing panicles (Arora et al., 2007; Nijhawan et al., 2008); hence we tested interaction among these proteins using the heterologous yeast two hybrid assay (Figure 5). Additionally, to investigate possibility of OsbZIP47 heterodimerization with other meristem regulators, we relied on reports from genetic studies in Arabidopsis, maize and rice to curate and choose candidates for interaction assays (Das et al., 2009; Khanday et al., 2013; Deshpande et al., 2015; Pautler et al., 2015). OSH1, OsH15, ETTIN1, ETTIN2, and RFL emerged as candidates. We re-visited reports on the panicle and floret expression patterns of these meristem regulators to deduce if spatial co-expression of OsbZIP47, OSH1 and OSH15 could occur. RNA in situ patterns of OSH1 in rice panicles and florets (Komatsu et al., 2001; Chu et al., 2006; Hu et al., 2015) and OsbZIP47 transcript spatial profile (Figure 4) point to an overlap of OsbZIP47 and OSH1 transcripts in primary and secondary branch primordia and in a broad range of developing spikelet/floret meristems (Sp2–Sp8) (Supplementary Figure 5). Further, mutant osh1 (Tsuda et al., 2011, 2014) and OsbZIP47KD plants share common phenotypes such as increased leaf lamina joint angle, short panicle with reduced number of spikelets and deformed stamens. Moreover, Pautler et al. (2015) reported that several gene loci are cobound by ZmKN1 (the ortholog of rice OSH1) and ZmFEA4 (ortholog of OsbZIP47). These findings together indicate likelihood of OsbZIP47 and OSH1 interactions for co-regulation of target genes. Also, OsH15, a closely related paralog of OSH1 (Tsuda et al., 2011), co-expresses with OsbZIP47 in spikelet meristem/floret meristem stage 6 (Yoon et al., 2017; Supplementary Figure 5). As OsbZIP47 KD caused abnormal floral phenotypes, it was intriguing to determine if OsbZIP47 could heterodimerize with OsH15. In Arabidopsis, pan ettin phenotypes suggest AtPAN and AtETTIN/AUXIN-RESPONSIVE FACTOR3 (ARF3) redundantly regulate floral organ numbers and patterns (Sessions et al., 1997). Additionally, rice ETTIN1 and ETTIN2 RNAi lines have aberrant plant height, compromised panicle branching, and defects in stamen and carpel development (Khanday et al., 2013). Some of these phenotypes resembled those observed in OsbZIP47KD plants. Similarly, for mutants in RFL, the rice AtLEAFY ortholog, the alleles apo2 and ssc, or the RNAi RFLKD (Kyozuka et al., 1998; Rao et al., 2008; Wang et al., 2017) plants share some phenotypes with OsbZIP47KD plants. The common phenotypes include shorter plant height, delayed flowering, reduced panicle rachis length, and branch complexity. Thus, we hypothesized that OsETTIN1, OsETTIN2, and RFL may interact with OsbZIP47 to modulate aspects of organ development. Based on these meta-analyses, protein partnership between OsbZIP47 and OSH1, OsH15, ETTIN1, ETTIN2, and RFL was tested by the Y2H assay. Moreover, we also tested the interactions of OsbZIP47 with OsMADS15 (an A Class APETALA1/FRUITFULL AP1/FUL-clade transcription factor) and OsMADS2 (a B class PISTILLATA/GLOBOSA-like protein). OsMADS15 regulates vegetative to reproductive floral transition and functions in specifying meristem identity (Kater et al., 2006; Kobayashi et al., 2012). OsMADS2 functions in partnership with OsMADS16/SUPERWOMAN1 (SPW) as a B-class complex (Lombardo et al., 2017; Kong et al., 2019). The delayed flowering phenotypes of OsbZIP47KD transgenics and the defects in the second and third whorl floral organs, justified our choice of OsMADS15 and OsMADS2, respectively. Since full length OsbZIP47 exhibited transcriptional transactivation activity in yeast (Supplementary Figure 6A), a C-terminal truncated version (OsZIP47ΔC) lacking 186-385 amino acids including the transcription activation domain was taken as bait protein in fusion with Gal4 BD. Prey proteins (OsMADS1, OSH1, OsH15, ETTIN1, ETTIN2, RFL, OsMADS15, or OsMADS2) were fused to GAL4 AD. The GAL4AD-OsMADS15 interaction with GAL4DB-OsMADS1 was taken as the positive control (Lim et al., 2000). In addition, homodimerization capability of OsbZIP47ΔC was tested. Growth pattern of transformed yeast cells on reporter media SD/-Leu-Ura-His + 10 mM3AT and the X-gal quantitative assays (Supplementary Figure 6B) suggested that OsbZIP47 can heterodimerize with OsMADS1, OSH1, and RFL. We also found a strong homodimerization of OsbZIP47ΔC protein (Figures 5A,B). Both OsMADS15 and OsH15 showed weak interactions with OsbZIP47 (Figure 5A), while ETTIN1, ETTIN2, and OsMADS2 showed no interaction. Further, we performed in-planta BiFC assays to substantiate the protein–protein interactions screened in Y2H assay. OsbZIP47ΔC was cloned upstream to the coding sequence of C-terminal region of split YFP to express OsbZIP47ΔC-cYFP fusion protein. The coding sequences of OsMADS1, OsbZIP47ΔC, RFL, OsETTIN2, and OSH1 were cloned in frame downstream to the coding sequence of the N-terminal split YFP (nYFP) to express nYFP fusion proteins. These six different combinations of nYFP and cYFP fusion proteins were transiently co-expressed in onion epidermal cells. Nuclear YFP fluorescence signals confirmed protein interaction of OsbZIP47 with OsMADS1, OSH1, and RFL (Figure 5B). Thus, we suggest that OsbZIP47 partnership with OsMADS1, OSH1, and RFL could contribute to meristem functions, inflorescence complexity, and floret development.
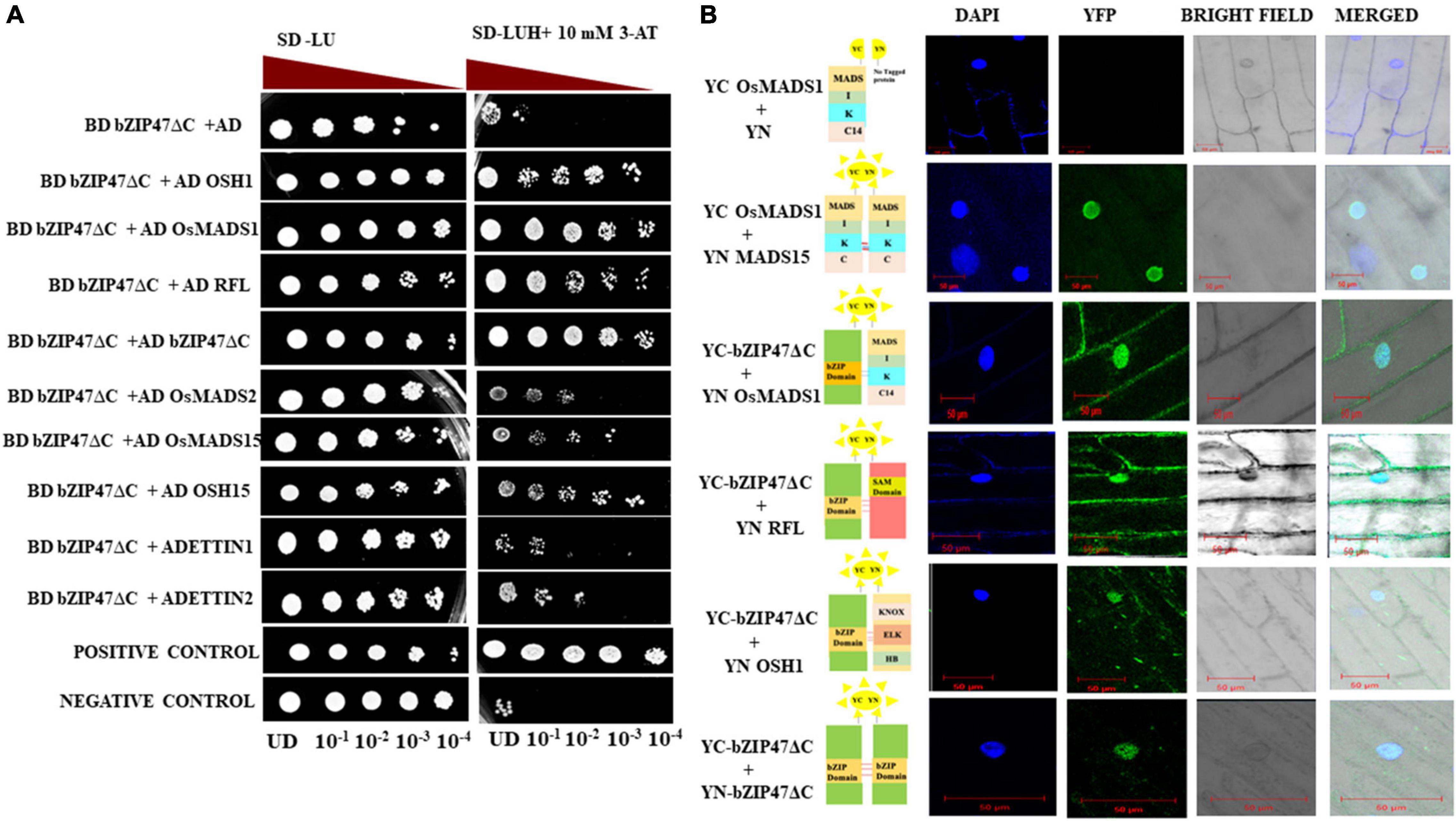
Figure 5. Interaction of OsbZIP47 with other meristem factors. (A) Yeast two hybrid (Y2H) assays with OsbZIP47ΔC protein (lacking 186–380 amino acids in C terminal) and predicted protein partners, such as OsMADS1 (MIKC14 domain), RFL, OsETTIN1, OsETTIN2, OSH1, OsMADS2, OsMADS15, and OSH15. Serial dilutions of yeast cells (PJ694A) spotted on media lacking histidine and supplemented with 10 mM 3-AT. Growth after five days at 30°C is shown. Yeast transformants with OsMADS15 in pGBDUC1 vector and OsMADS1 MIKC14 in pGADC1vector served as positive control for protein interaction. A combination of pGADC1 and pGBDUC1 empty vectors (without protein fusion) was the negative control. (B) Validation of Y2H protein interactions by Bimolecular Fluorescence Complementation (BiFC) assays in onion epidermal cells. YFP fluorescence was detected when OsbZIP47ΔC-cYFP fusion protein was co-expressed with OsMADS1-nYFP, RFL-nYFP, and OSH1 nYFP. Absence of YFP fluorescence in the negative control i.e., OsbZIP47ΔCYC and YN domain alone was the negative control. Bars = 50 μm.
Transcriptome of Developing Inflorescences of OsbZIP47 Knockdown Lines
To capture gene expression landscape in OsbZIP47KD panicles, high throughput RNA-sequencing was carried out in two biological replicates of OsbZIP47KD and WT panicles (In2-In4, 1 mm to 5 mm panicles), and the DEGs (greater than two-fold change, p-value < 0.05) were extracted (Supplementary Dataset 1, Supplementary Materials and Methods). Further, DEGs were examined for gene ontology pathway enrichment. Among the DEGs, 1,945 genes were upregulated, and 855 genes were downregulated in OsbZIP47KD panicles (Supplementary Figure 7, Supplementary Dataset 1). Gene Ontology (GO) analysis of positively regulated gene set (Figures 6A,B and Supplementary Materials and Methods, Supplementary Dataset 2) revealed enrichment of RNA (regulation of transcription), lipid, CHO metabolism, signaling, development, and hormone metabolism pathways (Figure 6A). Whereas in the negatively regulated set, genes related to secondary metabolism, transport, cell wall, signaling, stress, hormone metabolism, and miscellaneous factors were overrepresented (Figure 6B). Not surprisingly, genes involved in hormone signaling and metabolism were controlled by OsbZIP47, both positively and negatively (Figures 6C,D). Specifically, jasmonate (JA) and abscisic acid (ABA) pathway genes are overrepresented in the positively regulated gene set. CK degradation and ethylene signal transduction genes are notable in the negatively regulated gene set. Interestingly, genes of GA pathway were enriched in the positively regulated gene set, whereas genes for GA-synthesis-degradation were enriched in the negatively regulated gene set (Figure 6D). Examples of OsbZIP47 downstream genes that could interlink hormone pathways for panicle and floret development include APETALA-2-LIKE TRANSCRIPTION FACTOR39 (OsAP2-39), 9- cis-epoxycarotenoid dioxygenase3 (OsNCED3), and ELONGATED UPPER MOST INTERNODE1 (OsEUI1). AP2-39 balances the antagonistic relation between ABA and GA by modulating the expression levels of OsNCED3 and OsEUI1 to regulate plant height, yield, and seed germination (Yaish et al., 2010; Shu et al., 2016). We observed positive regulation of OsAP2-39 and OsNCED3, and negative regulation of EUI1 by OsbZIP47. Thus, we suggest that OsbZIP47 may enhance ABA biosynthesis and modulate GA biosynthesis possibly to regulate plant height and panicle rachis elongation (Figures 2D,E). Examples of other OsbZIP47 downstream genes linked to different phenotypes in OsbZIP47KD plants are further discussed. Interestingly, in OsbZIP47KD inflorescences, whole transcriptome analyses showed higher transcript levels for the CLV3 paralog genes, FON2/4 and FCP1. Moreover, genes from the KNOX-WUS pathway, and isopentenyl-transferases, IPT6, and IPT8 that encode CK-biosynthesis rate limiting enzymes were also upregulated. These results together, suggest the roles of OsbZIP47 in the regulation of panicle primary branch meristems and secondary branch meristems. Reduced transcript expression was observed for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 7 (SPL7), OsMADS16, and YABBY domain factor-DROOPING LEAF (DL) in OsbZIP47KD. The SPL7 regulates inflorescence meristem and spikelet transition (Dai et al., 2018). OsMADS16 and DL regulate lodicule, stamen, and carpel development (Nagasawa et al., 2003; Yamaguchi et al., 2004). Moreover, the F-box gene, APO1 with roles in spikelet and floret development was also deregulated (Supplementary Dataset 1 and Figure 6E; Ikeda et al., 2005, 2007). These findings corelate with the OsbZIP47KD inflorescence branching and floret organ defects (Figures 2, 3). The positive regulation of CUC1 by OsbZIP47 supports plausible mechanism for its influence on organ whorl boundaries (Takeda et al., 2011; Figure 6E and Supplementary Dataset 1) and could explain the development of chimeric floral organs in OsbZIP47KD transgenics. Transcription factors control the dynamics of hormone signaling pathways by modulating gene expression levels. Transcription factors genes that are deregulated in OsbZIP47KD panicles include bHLH gene members (22 genes), Co-like Zn finger (5 genes), TCP class 1 (2 genes), and TCP class 2 (1 gene), trihelix (4 genes), C2H2 zinc (20 genes) (Figure 6C and Supplementary Dataset 2). The genes for transcription factors that are positively regulated by OsbZIP47 are from NAC class, WRKY class, and MYB-related class genes. Among the latter class is OsLHY (LATE ELONGATED HYPOCOTYL)/CCA1 (CIRCADIAN CLOCK ASSOCIATED1) which functions in photoperiodic flowering, plant tillering, and grain yield (Wang et al., 2020; Sun et al., 2021). We speculate that the delayed flowering phenotype of OsbZIP47KD plants can be associated with the positive regulation of OsLHY by OsbZIP47. We also speculate that positive regulation of DWARF AND RUNTISH SPIKELET2/FERONIA like Receptor 2 (DRUS2/FLR2) may contribute to architecture, fertility, and seed yield (Li et al., 2016; Figure 6E and Supplementary Dataset 1). To obtain a predicted list of candidate direct genes and targets of OsbZIP47, we queried the dataset of deregulated genes in OsbZIP47KD panicles for the occurrence of cis motif typical to the TGA-subclade within the large family of bZIP factors in the rice, Arabidopsis, and maize genomes. Arabidopsis TGA sub-family includes AtPAN, the homolog of OsbZIP47, and AtPAN binds to the core cis regulatory element TGACG (Gutsche and Zachgo, 2016). Among the 2,210 differentially expressed annotated (MSU-TIGR v7) genes, a large number displayed the core motif TGACG in their TSS-promoter proximal regions (-500 bp to + 100 bp from TSS; Supplementary Dataset 3). Interestingly, the TGACG core motif occurred three times in this region of the FCP1 locus hinting that FCP1 deregulation is likely a direct effect of OsbZIP47. Other predicted direct targets that relate to developmental functions of OsbZIP47 are APO1, GNP1 (GA20Ox1), CYCD7, OsSPL7, OsIAA20, and GRX6, to name a few. Other gene targets could be regulated by degenerate cis elements related to “core motif” or by “core motif” in other distal regions of these loci. This in silico prediction of downstream targets of OsbZIP47 provides an extra level of confidence to the transcriptome-based deregulated gene set and can facilitate DNA–protein interaction studies. Overall, these results give a snapshot of OsbZIP47 molecular functions in inflorescence tissues and give leads to its unique vs. evolutionarily conserved roles for panicle meristem transitions and floral organ development.
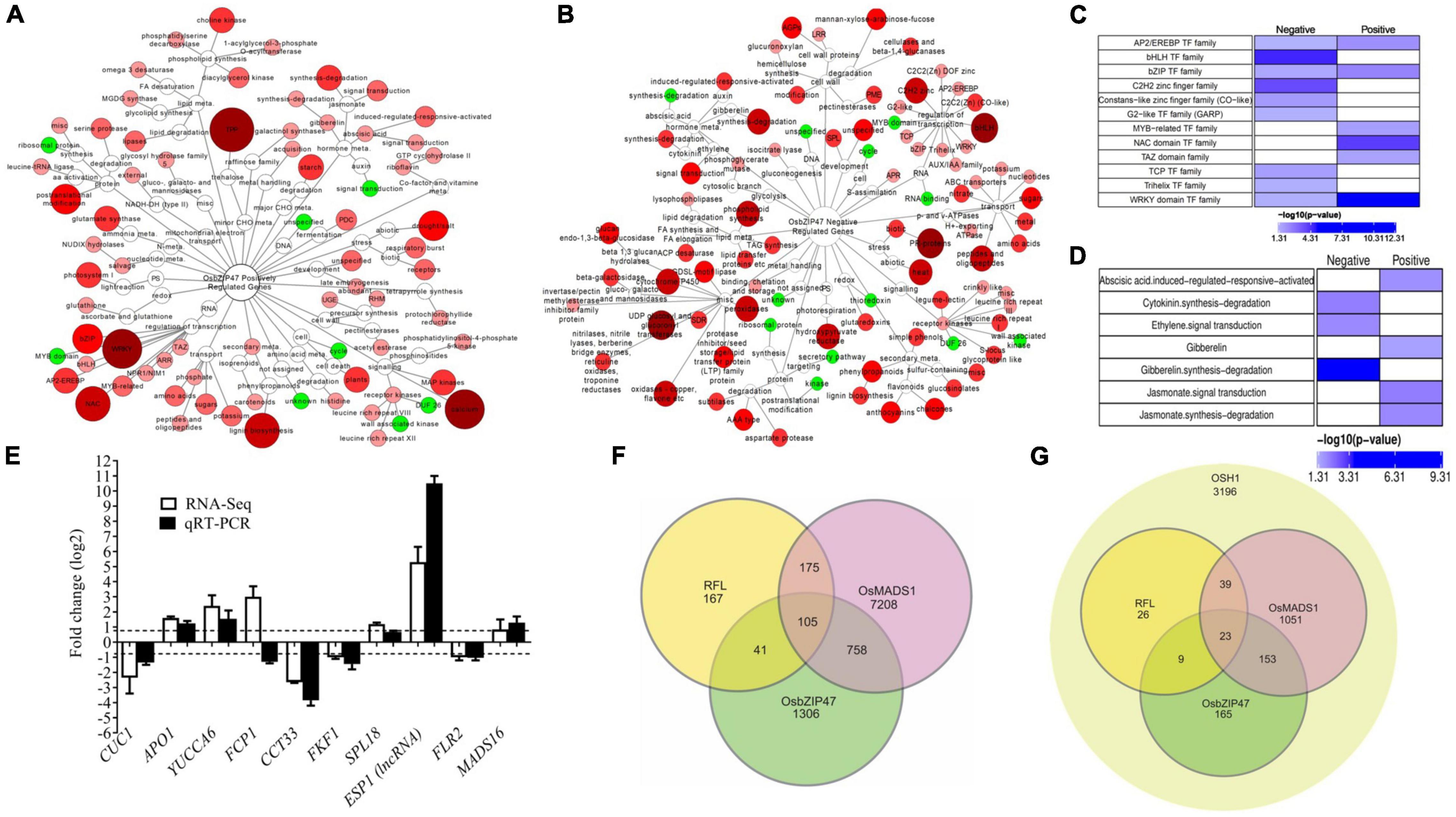
Figure 6. Differentially expressed genes (DEGs) and GO enrichment analysis of pathways downstream to OsbZIP47. Enrichment networks depicting pathways regulated by OsbZIP47. (A) positively regulated, and (B) negatively regulated pathways. The red and green color nodes represent enriched and depleted functional categories, respectively (P < 0.05). The size and color intensity of the node correlates with the over-representation of genes within the given class. (C) Enrichment map of transcription factor (TF) genes shows those encoding members from bHLH, G2-like, C2H2 zinc finger, TCP, Trihelix are underrepresented, whereas TFs, such as AP2/EREBP, MYB-related, NAC domain, TAZ, WRKY, and bZIP were overrepresented. (D) The 2,210 DEGs annotated in MSU-TIGR v7 were taken for GO-enrichment analysis for phytohormone metabolism and signaling. Cytokinin (CK) degradation, ethylene signal transduction genes, and gibberellin (GA)-synthesis-degradation were enriched in negatively regulated dataset, whereas, abscisic acid (ABA), GA pathway, and jasmonate (JA) pathways predominate in the positively regulated set. (E) RT-qPCR analysis of the relative fold-change for transcripts from several candidate target genes of OsZIP47 in 0.1 to 0.5 cm panicles from OsbZIP47KD as compared to WT tissues. The normalization of transcript level of each gene was done using UBQ5 transcript levels. Error bars represent the standard deviation. (F) Venn diagram showing unique and overlapping sets of differentially expressed genes (DEGs) derived from OsMADS1, OsbZIP47, and RFL transcriptome datasets (Rao et al., 2008; Khanday et al., 2013; this study). (G) Genes implicated to be coregulated by OsMADS1, OsbZIP47, and RFL and bound by OSH1 (Rao et al., 2008; Khanday et al., 2013; Tsuda et al., 2014; this study).
Comprehensive Datamining of Transcriptome Datasets of OsMADS1, OsbZIP47, RFL, and Genome Binding Dataset for OSH1
Extending our findings of OsbZIP47 interaction with OsMADS1, RFL, and OSH1, we carried out meta-analyses of published transcriptome datasets affected in mutants of these partner proteins. To identify candidate genes for co-regulation by these factors, the differential transcriptome in dsRNAiOsbZIP47KD, dsRNAiOsMADS1, and dsRNAiRFL panicles were examined (Rao et al., 2008; Khanday et al., 2013). First, we aligned genes from three different transcriptomic datasets for this meta-study (Supplementary Materials and Methods). The 2,210 (as annotated by MSU-TIGR v7) deregulated genes in dsRNAiOsbZIP47KD panicles were examined for overlap with 8,246 affected genes in dsRNAiOsMADS1KD transcriptome (Figure 6F and Supplementary Dataset 4; Khanday et al., 2013). Among 758 candidate genes co-regulated by both OsbZIP47 and OsMADS1 (Figure 6F and Supplementary Dataset 4), 204 genes were downregulated in both OsbZIP47KD and OsMADS1KD lines. These genes included Gibberellin-regulated protein precursor expressed (GASR3), auxin-responsive SMALL AUXIN-UP RNA 11 (OsSAUR11), DRUS2/FLR2, and JASMONATE ZIM-DOMAIN12 (TIFY11D/OsJAZ12) (ZIM domain transcription factor). Another group of genes (386 out of 758 genes) were upregulated in both OsbZIP47KD and OsMADS1KD panicle datasets. This sub-set includes OsMADS16, GNP1 (GRAIN NUMBER PER PANICLE1) and genes that regulate hormone signaling, such as OsIAA20, YUCCA7, PROBABLE AUXIN EFFLUX CARRIER COMPONENT 5B (PIN5B), and ETHYLENE INSENSITIVE LIKE 4 (EIL). Among the 758 candidate genes co-regulated by OsbZIP47 and OsMADS1, a subgroup of 153 genes (Figure 6G) are also bound by OSH1 (Tsuda et al., 2014). Striking among this sub-set are: IAA20, YUCCA7, SPLIT-HULL (SPH), and DRUS2/FLR2. Similarly, 41 genes (Figure 6F) are common to the DEGs in OsbZIP47KD panicles (RNA-Seq) and a low-density microarray study of panicles from dsRNAiRFL KD plants (Rao et al., 2008). Interestingly, 32 out of 41 genes were downregulated in both these datasets including ethylene signaling gene, ACO1 which regulates internode elongation (Iwamoto et al., 2011), JA signaling gene, TIFY11D (Kim et al., 2009), and the EMBRYOSAC1 (OsEMSA1) involved in embryo sac development (Zhu Q. et al., 2017). Finally, we identified 105 DEGs common to the differential transcriptome in RFLKD, OsMADS1KD, and OsbZIP47KD panicles (Figure 6F). The notable genes include ETHYLENE INSENSITIVE-LIKE GENE 2 (EIL2), ALLENE OXIDE SYNTHASE (AOS1), and GIBBERELLIN 2-OXIDASE 9 (GA2OX9). A subset of 23 genes are potentially regulated by OSH1 (Figure 6G; Tsuda et al., 2014). We infer that these transcription regulators possibly multimerize in one or more forms of complexes to regulate meristem development in rice. Overall, these findings hint that protein complexes, plausibly heterogeneous, with the combinations of OsbZIP47 and its varied partner factors may spatially and temporally co-ordinate downstream gene expression during panicle development, spikelet, and floret development.
Redox-Dependent DNA Binding of OsbZIP47
DNA binding by Arabidopsis AtPAN is redox-sensitive due to the five Cysteine residues in the extended N-terminal domain (Gutsche and Zachgo, 2016; Supplementary Figure 8). Unlike AtPAN homologs from diverse species, rice OsbZIP47 lacks this domain. All proteins share a conserved Cys in the C terminal transcription transactivation domain (AtPAN Cys340/OsbZIP47 Cys269). Cys17 in OsbZIP47 is conserved in monocot species, while Cys196 is unique to OsbZIP47. Thus, though the rice and Arabidopsis proteins are homologs, they differ in size and in the overall number of cysteine residues. To examine OsbZIP47 oligomerization and the effects of its redox status on binding to target gene cis DNA elements, the full length (FL) OsbZIP47 protein was expressed in bacteria. The DNA binding activity of AtPAN is regulated by S-glutathionylation of the conserved Cys340 by AtROXY1, a glutaredoxin redox enzyme (Li et al., 2009; Gutsche and Zachgo, 2016). The corresponding conserved Cys 269 in OsbZIP47 may also render the rice protein to be redox-sensitive for biochemical activity. To determine if OsbZIP47 FL protein forms higher order self-oligomers, SEC with the purified Trx-His OsbZIP47 (62 Kda) protein was done and the elution of the protein in the column void volume (Figure 7A) suggested either aggregation or the formation of high order oligomers in the given condition. To examine the latter possibility, the purified OsbZIP47-His-Trx protein was treated with 2 mM of diamide, an oxidizing agent. In parallel, another aliquot of the protein was treated with 20 mM DTT, a reducing agent, and both treated protein fractions were analyzed on a non-reducing SDS-PAGE gel. The oxidized OsbZIP47-His-Trx sample had slower mobility whereas the reduced OsbZIP47-His-Trx sample migrated with the expected mobility for a ∼68 Kda protein. Importantly, we found that the effects of the oxidizing agent (diamide) can be reversed by DTT treatment. These data show that OsbZIP47 oligomerization is affected by its redox status (Figure 7B). Recently, OsbZIP47 maize ortholog, FEA4 was shown to switch its oligomerization status following redox change (Yang et al., 2021). Next, we tested the DNA binding affinity of OsbZIP47 to the TGACGT cis motif (predicted for OsbZIP47 DNA binding) present at around -371 bp upstream of the start codon in the OsFCP1 locus (Figure 7C). The latter is a downstream gene target whose expression levels are affected in the SAM and in the panicles of OsbZIP47KD plants. The OsbZIP47 FL protein status was altered by incubation with the reducing agent DTT (20 mM) or with the oxidant diamide (2 mM) for 30 min prior to the incubation with the TGACGT motif containing DNA substrate. OsbZIP47 FL protein bound to the TGACGT motif under reducing conditions. Interestingly, incubation with diamide decreased OsbZIP47 FL-DNA interaction. Thus, electrophoretic mobility shift assays showed redox-sensitive DNA-binding of the OsbZIP47 FL protein (Figure 7D). Further, we quantified the binding affinity of OsbZIP47 FL with cis element from the OsFCP1 locus using microscale thermophoresis (MST). To this end, OsbZIP47 FL protein was labeled with the RED-NHS 2nd Generation Dye (MO-L011, Nanotemper GmbH) and was mixed with increasing concentrations of OsFCP1 oligos until saturation. The fluorescent signals obtained with increasing ligand concentrations followed a clear sigmoidal binding curve. As expected, OsbZIP47 FL protein displayed a stronger binding affinity to the OsFCP1 locus in its reduced state, with a dissociation constant Kd of 815 nM, as compared to Kd of 2.16 uM in the oxidized state (Figure 7E). The differential Kd values confirm a redox sensitive DNA–protein interaction. The Trx-His tag protein was taken as a negative control in these assays and no interaction between protein and ligand was detected (Supplementary Figure 9). Overall, the results of our qualitative and quantitative data suggest that affinity of OsbZIP47 FL binding to DNA was redox-dependent despite the absence of the extended Cys rich N-terminal domain commonly found in homologs from other species. Recently, OsbZIP47 close ortholog, ZmFEA4 was shown to interact with three GRX proteins to modulate its redox status and DNA accessibility (Yang et al., 2021).
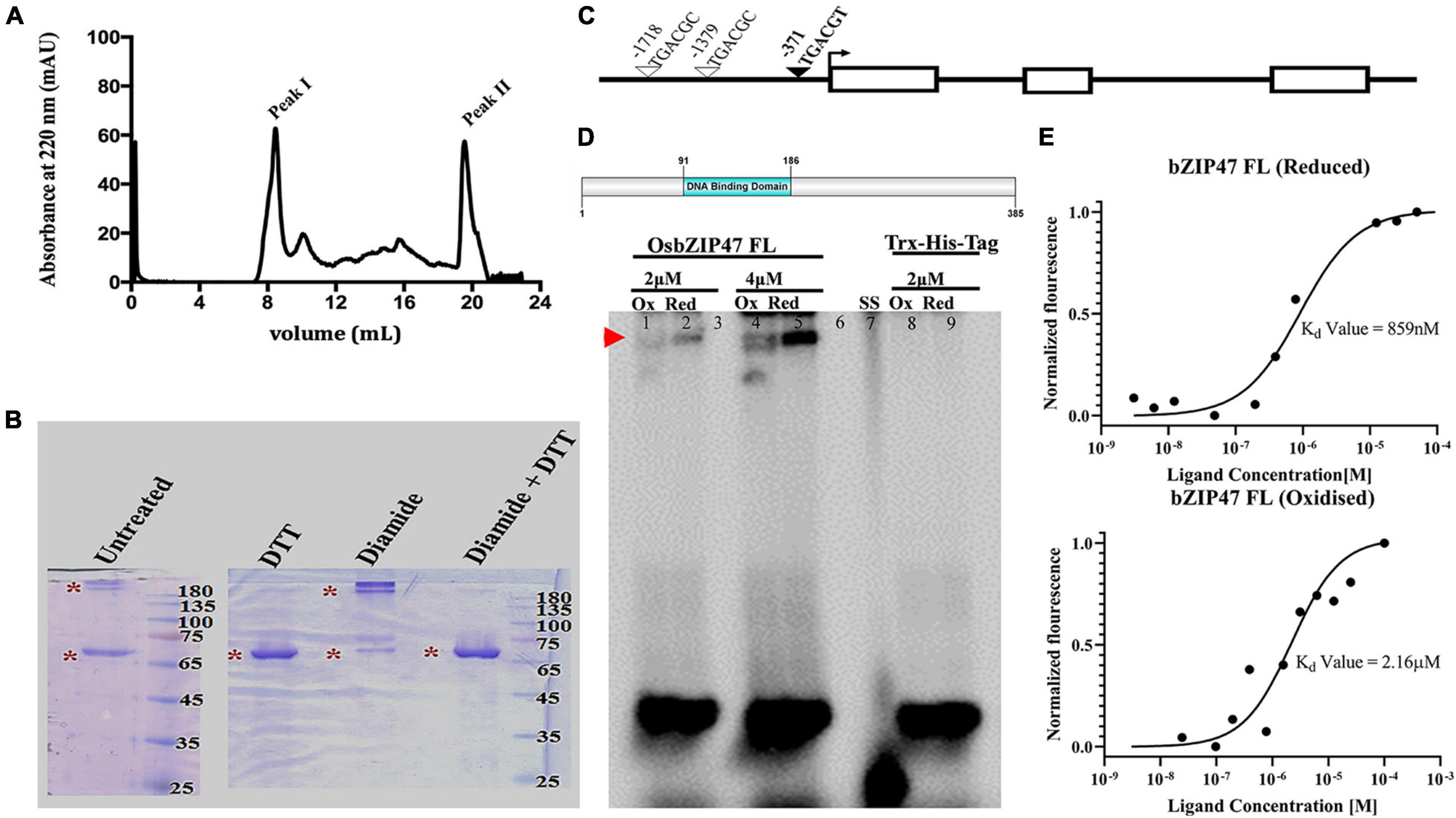
Figure 7. OsbZIP47 oligomerization and the effects of its redox status on DNA binding. (A) Size-exclusion chromatography (SEC) profile for OsbZIP47 protein/oligomer on a Superdex 200 increase column. (B) Mobility of purified OsbZIP47-Trx protein on non-reducing 10% of SDS-PAGE after treatment with 2 mM of diamide (an oxidizing agent), or with 20 mM DTT (a reducing agent), or with 2 mM diamide followed by 20 mM DTT. (C) Schematic representation of the FCP1 gene locus. Exons are open white boxes, introns are shown as black lines, and the predicted cis motifs for bZIP DNA binding domain (DBD) are represented as inverted triangles. The DNA binding properties of OsbZIP47FL protein was tested for the TGACGT cis motif located at -371bp of FCP1 (marked with a filled inverted triangle). (D) Schematic representation of OsbZIP47 FL protein (amino acid 1 to 385) used for EMSA and microscale thermophoresis assays. EMSA assays with full-length OsbZIP47 (62 Kda protein) shows retarded mobility of DNA-protein complex for the OsFCP1 locus (as indicated by red arrow). Lanes 1 and 2 correspond to the oxidized and reduced form of bZIP47 FL protein incubated with 2 μM of oligo. Lanes 4 and 5 represent the oxidized and reduced form of the same protein, incubated with 4 μM of oligo. Lanes 8 and 9 correspond to Trx-Tag protein incubated with 4 μM of probe. Lanes 3 and 6 are blank. Unbound dsDNA oligonucleotide is at the bottom. (E) Microscale thermophoresis-based assay to determine the dissociation constant for OsbZIP47 full-length and OsFCP1 dsDNA oligo interaction. 80 nM of fluorescently labeled protein was taken for binding with DNA sequences from OsFCP1 locus taken at a wide concentration range (100 μM to 3.05 nM). The OsbZIP47 full length protein was reduced with 20 mM DTT or oxidized with 1mM of oxidized glutathione. OsbZIP47 full length protein shows increased binding in the reduced state as compared to the oxidized state.
Discussion
Arabidopsis PAN, maize FEA4, and rice bZIP47 proteins share a significant degree of similarity throughout their lengths with the most conserved region being the DNA binding domain (Supplementary Figure 8; Chuang et al., 1999; Nijhawan et al., 2008). Species-specific developmental roles for these factors may therefore arise from interacting co-regulators, from protein modification of these transcriptional regulators, and through variations in their downstream genes and pathways. Arabidopsis AtPAN has pleiotropic vegetative and reproductive growth effects. Flowers in the pan mutant are characterized by an increase in the floral organ number without a corresponding increase in FM size (Running and Meyerowitz, 1996; Chuang et al., 1999). Another study reported early flowering of pan mutant plants in long-day and short-day grown plants having an enlarged SAM and inflorescence meristems (Maier et al., 2011). Maize mutant zmfea4 have enlarged vegetative SAMs, severely fasciated inflorescences, and florets with reduced stamen numbers (Pautler et al., 2015). We show that OsbZIP47KD plants have abnormalities in the shoot meristem size homeostasis similar to the enlarged SAM of maize fea4 mutant. Yet, other phenotypes are unique to rice OsbZIP47KD lines, for instance, delayed flowering, increased stamen numbers, chimeric floral organs, and subtle changes to grain size and shape. Hence, despite redundancy of bZIP transcription factors, the phenotypes of single mutants, such as atpan1, zmfea4, and OsbZIP47KD indicate OsbZIP47 has evolutionarily conserved as well as unique roles in the vegetative and reproductive development. The partnership of OsbZIP47, with meristem regulators, OsMADS1, RFL, and OSH1 (KNOX1/STM), its oligomeric and redox status could relate to its functions in different meristems. This partnership and the findings that emerge from the differential transcriptome in OsbZIP47KD panicles allowed us to map OsbZIP47-regulated downstream genes, and those potentially dependent on its co-regulators.
Meristem Development in Vegetative and Reproductive Phase
The enlarged SAM in OsbZIP47KD seedlings is superficially similar to that of maize fea4, yet there are underlying subtle differences in rice KD plants. A detailed phenotyping of SAMs from OsbZIP47KD seedlings showed increased cell size of L1 layer and its underlying cells suggesting precocious cell differentiation. Further, SAMs of OsbZIP47KD showed increased transcript signal of cell proliferation marker, H4 as indicated by in situ hybridization. The downregulation of FON2, FCP1, YUCCA6, CUC1, APO1, and CYP734A4 in SAM, suggests several complex pathways by which OsbZIP47 contributes to SAM size and plant growth by modulating cell proliferation and differentiation. Notable here is the regulation of CYP734A, which as a direct target of OSH1 is suggested to repress premature cell differentiation in meristems (Tsuda et al., 2014). This observation together with our data on protein interactions between OsbZIP47 and OSH1 supports a plausible mechanism by which OsbZIP47-OSH1 partnership could regulate CYP734A expression with ensuing effects on meristem and lateral primordia development. Further, phytohormones, CK, AUX, and GA are also essential for cell division and organ differentiation (Leibfried et al., 2005; Zhao et al., 2010; Su et al., 2011). Transcriptome profiling of OsbZIP47KD panicles shows deregulated expression levels for different phytohormone related genes including KNOTTED1-LIKE11, IPT1, IPT8, OsGA3OX2 OsGA2OX3, OsGA2OX4, OsGA20OX1, YUCCA6, and YUCCA7. Plant meristems are under redox control by reactive oxygen species (ROS), the by-products of aerobic metabolism. ROS levels control the expression of WUS and the activity of TCP class1 transcription factors to balance cell proliferation and differentiation in Arabidopsis SAM (Viola et al., 2013; Zeng et al., 2017). Like TCP class1, proteins containing cysteines with low pKa values are sensitive to cellular redox status (Martins et al., 2018). Glutaredoxins (GRX) reduce cellular ROS level and interact with different proteins including AtPAN1, ZmFEA4, and OsbZIP47 to modulate their activity by modifying their redox state (Li et al., 2009; Laporte et al., 2012; Schippers et al., 2016; Hao et al., 2021; Yang et al., 2021). Arabidopsis pan and maize fea4 mutants exhibited increased meristem size (Maier et al., 2011; Pautler et al., 2015). Contrastingly, single, double, and triple mutations of different GRX paralogs in maize showed progressive reduction of meristem size, proposing a model where GRXs balance redox status and activity of ZmFEA4 to control meristem size (Yang et al., 2021). Interestingly, in transcriptome profiling of OsbZIP47KD panicles, OsGRX6 was downregulated whereas OsGRX16 and OsGRX20 were upregulated (Supplementary Dataset 1), suggesting a mechanism by which OsbZIP47 could be regulating meristem development. We propose OsbZIP47 functions as an integrator of the WUS-CLV and KNOX pathways for meristem development as it regulates the expression levels of key signaling factors in both pathways. Also, genes predicted to have roles in floral organ primordia differentiation such as OsBLH1 (BEL1-like homeodomain), OsKANADI1, and CUC1 are deregulated in OsbZIP47KD panicle tissues. Interestingly, reproductive panicle branching phenotypes of OsbZIP47KD panicles resemble apo1 and apo2/rfl mutants (Ikeda et al., 2005, 2007; Rao et al., 2008; Ikeda-Kawakatsu et al., 2009; Deshpande et al., 2015). Thus, the interactions between RFL and OsbZIP47 could positively regulate the panicle meristem branch identity and its developmental transitions. One example of a target gene for co-regulation by OsbZIP47 and RFL is CUC1. Further, the elevated transcript levels of APO1 in OsbZIP47KD panicle tissues hints that OsbZIP47 in the WT panicle suppresses the expression of APO1 which we speculate may affect the partnership with APO2/RFL. From this, we anticipate that OsbZIP47 could have evolved to regulate some unique molecular pathways for vegetative and reproductive phase meristem growth and development.
Transition of Shoot Apical Meristem to Inflorescence Meristem
Knockdown of rice OsbZIP47 showed delayed flowering (Figure 2A). This trait is common in mutants or KD transgenics in OsMADS1 and APO2/RFL that encode OsbZIP47 protein partners (Jeon et al., 2000; Rao et al., 2008; Ikeda-Kawakatsu et al., 2012; Kannan et al., 2021). These observations support our hypothesis that these factors function in “one or more” complexes. Panicle transcript analysis in KD transgenics indicates that OsbZIP47 can promote flowering by fine-tuning the expression of several flowering time regulators that are upstream to florigens, HEADING DATE 3a (Hd3a) and RICE FLOWERING LOCUS (RFT), and by controlling the expression of circadian clock-associated genes. Examples of genes from these two categories are O. sativa LATE FLOWERING (OsLF), LATERAL ORGAN BOUNDARY DOMAIN (OsLBD38), INDETERMINATE DOMAIN 6 (OsIDD6), FLAVIN-BINDING, KELCH REPEAT F-BOX1 (OsFKF1), PHYTOCLOCK 1(OsPCL1), and OsLHY/CCA1. Several rice flowering time quantitative trait loci (QTLs) also influence grain traits (Chen et al., 2014; Zhu Y.J. et al., 2017; Ma et al., 2019). The effect of OsbZIP47KD on rice grain shape is not reported for maize fea kernels suggesting unique effects of OsbZIP47 on grain size and shape in rice as also reported by Hao et al. (2021). Our transcriptomic analysis identified a set of grain shape genes regulated by OsbZIP47. Examples of this category are LONG GRAIN 3 (OsLG3), GRAIN SHAPE GENE ON CHROMOSOME 9 (GS9), GRAIN WIDTH QTL on chromosome 7 (GW) and FLOURY ENDOSPERM 2 (FLO2). GS9 positively controls the grain size by altering the cell division along with BR signaling (Zhao et al., 2018). Interestingly, we noted increased expression of CYCD7;1 in OsbZIP47KD panicles (Supplementary Dataset 1). This is remarkable as in Arabidopsis, the tissue and stage-specific control of this G1-S phase cell cycle gene controls the cell division in different contexts, with ectopic expression driving increased cell division and expansion in the embryo and the endosperm (Collins et al., 2012; Weimer et al., 2018). With this, we postulate that OsbZIP47 links flowering time, cell cycle, and BR signaling to regulate grain shape.
Regulation of Inner Floral Organ Identity and Specification
Consistent with OsbZIP47 expression in the second and third whorl organs of near mature florets (Sp6–Sp8), we observed lodicule and stamen differentiation defects in dsRNAiOsbZIP47 florets. Increased stamen numbers, with degenerated anthers on short filaments and lodicule-stamen chimeric organs support roles for OsbZIP47 in organ differentiation. Interestingly, in OsbZIP47KD florets, the higher transcript abundance of OsMADS16 (homolog of AtAP3) and DL, a contributor to Class C function in rice florets (Supplementary Dataset 2), are indicative of some distinct effects in rice florets. Overexpression of OsMADS16 can increase stamen numbers and form stamenoid carpels without any effects on lodicules (Lee et al., 2003). More recently, rice transgenics with a modified repressive OsMADS16 (OsMADS16-SDX repressor domain fusion) exhibited indehiscent anthers (Sato et al., 2012). These phenotypes are akin to third whorl organ differentiation defects seen in OsbZIP47KD florets. Microsporogenesis in anthers of Arabidopsis flowers requires SPOROCYTELESS/NOZZLE (SPL/NZZ), a target of Class B and C organ identity factors (Ito et al., 2004). In line with this, we noted upregulation of OsSPL, possibly an effect of increased OsMADS16 in OsbZIP47KD florets. Since we did not detect OsbZIP47 interaction with OsMADS2 in the Y2H assay, we speculate that OsbZIP47 regulates stamen differentiation by modulating the expression of OsMADS16. Upstream regulators of OsMADS16 are OsDL and FON2 (Ikeda et al., 2007; Xu et al., 2017). Both are upregulated in OsbZIP47KD inflorescence. The upregulated transcript of FON2 in OsbZIP47KD inflorescence suggests that OsbZIP47KD stamen phenotypes are OsMADS16-mediated.
Biochemical Properties of OsbZIP47 Can Underlie Its Unique Functions and Downstream Effects
Multiple sequence alignment shows that OsbZIP47 shares nearly 50% of amino acid identity with homologs across diverse species. A common feature among many bZIP47 proteins, except OsbZIP47 and Bamboo PH01000727G0540, is a variably extended N terminal domain (Supplementary Figure 8). DNA binding activity of Arabidopsis PAN is redox-sensitive due to the presence of five Cysteine (Cys) amino acids in the extended N-ter domain and the conserved C-ter Cys340 in the transcription transactivation domain (Gutsche and Zachgo, 2016). OsbZIP47 protein has only three Cys (Cys17, Cys196, and Cys269). Cys17 is represented in all the monocot species. Cyse269 (Cys 340 of AtPAN) is conserved in all homologs compared here (Supplementary Figure 8). Cys196 is unique to OsbZIP47. Interestingly, among proteins compared here, wheat TAE56722G002 has the maximum number of 11 Cys. These observations hint that the number of Cys residues in this clade of bZIP proteins may have evolved for species-specific roles, plausibly for the adoption of unique structures with effects on tissue-specific target gene expression. Despite being a shorter protein with fewer Cys residues, OsbZIP47 showed redox-dependent DNA binding to OsFCP1, a downstream gene whose expression was upregulated in OsbZIP47KD panicles. Yang et al. (2021) demonstrated that OsbZIP47 maize ortholog, FEA4 interacts with three GRX proteins to modulate its redox status and DNA accessibility proposing a model by which redox status of FEA4 mediate meristem size. In rice, OsGRX19 or MICROSPORELESS1 (OsMIL1) is a potential glutaredoxin redox enzyme for OsbZIP47 as it is a homolog of the glutaredoxin redox enzyme, AtROXY1 and ZmMSCA1 from Arabidopsis and maize, respectively (Timofejeva et al., 2013; Yang et al., 2015). Interaction between OsMIL1 and TGA1 in yeast and the reduction of glutathionylation of OsbZIP47 by ROXY homolog, WG1 are both established (Hong et al., 2012; Hao et al., 2021). Indehiscent anthers phenotype is common to OsbZIP47KD and mil1 mutant (Hong et al., 2012) leading us to propose S-glutathionylation of OsbZIP47 could be important for the development of anther. Overall, we uncover conserved as well as unique functions and mechanisms of OsbZIP47 that support meristem growth and determinacy during vegetative and reproductive development leading to grain formation. Together, these functions make OsbZIP47 a potential locus for allele mining and crop improvement.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196747.
Author Contributions
UV, SP, RR, MZ, and OA designed the research. SP, RR, MZ, and OA performed research and experiments. RP performed bioinformatic analyses. Data analyses and manuscript preparation was done by SP, RR, MZ, RP, and UV. All authors read and approved the final manuscript.
Funding
This work was funded by the Department of Biotechnology, Ministry of Science and Technology, Government of India project entitled, Functional Analysis of Gene Regulatory Networks During Flower and Seed Development in Rice, Project number: BT/AB/FG-1 (PH-II)/2009 to UV. University Grants Commission, Delhi was acknowledged for DS Kothari postdoctoral research fellowship to RR; Project ID: No. F.42/2006 (BSR)/BL/1718/0151. Research fellowship to RP and MZ was from the Indian Institute of Science. Fellowship to OA was from the Council for Industrial and Scientific Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Inputs from Grace Chongloi and other members of the UVR laboratory during the course of the study are acknowledged. Divya is thanked for assistance with confocal imaging and Murthy and Jagadeesh are thanked for assistance in plant growth and care. We acknowledge the DBT-IISc Partnership Divisional Bio-imaging and Greenhouse Facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.865928/full#supplementary-material
Footnotes
References
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Arora, R., Agarwal, P., Ray, S., Singh, A. K., Singh, V. P., Tyagi, A. K., et al. (2007). MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8:242. doi: 10.1186/1471-2164-8-242
Bommert, P., Satoh-Nagasawa, N., Jackson, D., and Hirano, H. Y. (2005). Genetics and evolution of inflorescence and flower development in grasses. Plant Cell Physiol. 46, 69–78. doi: 10.1093/pcp/pci504
Brand, U., Grunewald, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129, 565–575. doi: 10.1104/pp.001867
Busch, M. A., Bomblies, K., and Weigel, D. (1999). Activation of a floral homeotic gene in Arabidopsis. Science 285, 585–587. doi: 10.1126/science.285.5427.585
Callens, C., Tucker, M. R., Zhang, D., and Wilson, Z. A. (2018). Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69, 2435–2459. doi: 10.1093/jxb/ery086
Chen, J. Y., Guo, L., Ma, H., Chen, Y. Y., Zhang, H. W., Ying, J. Z., et al. (2014). Fine mapping of qHd1, a minor heading date QTL with pleiotropism for yield traits in rice (Oryza sativa L.). Theor. Appl. Genet. 127, 2515–2524. doi: 10.1007/s00122-014-2395-7
Chongloi, G. L., Prakash, S., and Vijayraghavan, U. (2019). Regulation of meristem maintenance and organ identity during rice reproductive development. J. Exp. Bot. 70, 1719–1736. doi: 10.1093/jxb/erz046
Chu, H., Qian, Q., Liang, W., Yin, C., Tan, H., Yao, X., et al. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. doi: 10.1104/pp.106.086736
Chuang, C. F., Running, M. P., Williams, R. W., and Meyerowitz, E. M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13, 334–344. doi: 10.1101/gad.13.3.334
Collins, C., Dewitte, W., and Murray, J. A. (2012). D-type cyclins control cell division and developmental rate during Arabidopsis seed development. J. Exp. Bot. 63, 3571–3586. doi: 10.1093/jxb/ers015
Dai, Z., Wang, J., Yang, X., Lu, H., Miao, X., and Shi, Z. (2018). Modulation of plant architecture by the miR156f-OsSPL7-OsGH3.8 pathway in rice. J. Exp. Bot. 69, 5117–5130. doi: 10.1093/jxb/ery273
Das, P., Ito, T., Wellmer, F., Vernoux, T., Dedieu, A., Traas, J., et al. (2009). Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 136, 1605–1611. doi: 10.1242/dev.035436
Deshpande, G. M., Ramakrishna, K., Chongloi, G. L., and Vijayraghavan, U. (2015). Functions for rice RFL in vegetative axillary meristem specification and outgrowth. J. Exp. Bot. 66, 2773–2784. doi: 10.1093/jxb/erv092
Dodsworth, S. (2009). A diverse and intricate signalling network regulates stem cell fate in the shoot apical meristem. Dev. Biol. 336, 1–9. doi: 10.1016/j.ydbio.2009.09.031
Gordon, S. P., Chickarmane, V. S., Ohno, C., and Meyerowitz, E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Nat. Acad. Sci. U.S.A. 106, 16529–16534. doi: 10.1073/pnas.0908122106
Gutsche, N., and Zachgo, S. (2016). The N-Terminus of the floral Arabidopsis TGA transcription factor PERIANTHIA mediates redox-sensitive DNA-binding. PLoS One 11:e0153810. doi: 10.1371/journal.pone.0153810
Hao, J., Wang, D., Wu, Y., Huang, K., Duan, P., Li, N., et al. (2021). The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol. Plant 14, 1266–1280. doi: 10.1016/j.molp.2021.04.011
Harder, L. D., and Prusinkiewicz, P. (2013). The interplay between inflorescence development and function as the crucible of architectural diversity. Ann. Bot. 112, 1477–1493. doi: 10.1093/aob/mcs252
Hong, L., Tang, D., Zhu, K., Wang, K., Li, M., and Cheng, Z. (2012). Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24, 577–588. doi: 10.1105/tpc.111.093740
Hu, Y., Liang, W., Yin, C., Yang, X., Ping, B., Li, A., et al. (2015). Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol. Plant 8, 1366–1384. doi: 10.1016/j.molp.2015.04.009
Ikeda, K., Ito, M., Nagasawa, N., Kyozuka, J., and Nagato, Y. (2007). Rice ABERRANT PANICLE ORGANIZATION 1, encoding an F-box protein, regulates meristem fate. Plant J. 51, 1030–1040. doi: 10.1111/j.1365-313X.2007.03200.x
Ikeda, K., Nagasawa, N., and Nagato, Y. (2005). ABERRANT PANICLE ORGANIZATION 1 temporally regulates meristem identity in rice. Dev. Biol. 282, 349–360. doi: 10.1016/j.ydbio.2005.03.016
Ikeda-Kawakatsu, K., Maekawa, M., Izawa, T., Itoh, J., and Nagato, Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. doi: 10.1111/j.1365-313X.2011.04781.x
Ikeda-Kawakatsu, K., Yasuno, N., Oikawa, T., Iida, S., Nagato, Y., Maekawa, M., et al. (2009). Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 150, 736–747. doi: 10.1104/pp.109.136739
Ito, T., Wellmer, F., Yu, H., Das, P., Ito, N., Alves-Ferreira, M., et al. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430, 356–360. doi: 10.1038/nature02733
Iwamoto, M., Kiyota, S., Hanada, A., Yamaguchi, S., and Takano, M. (2011). The multiple contributions of phytochromes to the control of internode elongation in rice. Plant Physiol. 157, 1187–1195. doi: 10.1104/pp.111.184861
James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. doi: 10.1093/genetics/144.4.1425
Jeon, J. S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S. H., et al. (2000). leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. doi: 10.1105/tpc.12.6.871
Kannan, P., Chongloi, G. L., Majhi, B. B., Basu, D., Veluthambi, K., and Vijayraghavan, U. (2021). Characterization of a new rice OsMADS1 null mutant generated by homologous recombination-mediated gene targeting. Planta 253:39. doi: 10.1007/s00425-020-03547-3
Kater, M. M., Dreni, L., and Colombo, L. (2006). Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J. Exp. Bot. 57, 3433–3444. doi: 10.1093/jxb/erl097
Khanday, I., Das, S., Chongloi, G. L., Bansal, M., Grossniklaus, U., and Vijayraghavan, U. (2016). Genome-wide targets regulated by the OsMADS1 transcription factor reveals its DNA recognition properties. Plant Physiol. 172, 372–388. doi: 10.1104/pp.16.00789
Khanday, I., Yadav, S. R., and Vijayraghavan, U. (2013). Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol. 161, 1970–1983. doi: 10.1104/pp.112.212423
Kim, E. H., Kim, Y. S., Park, S. H., Koo, Y. J., Choi, Y. D., Chung, Y. Y., et al. (2009). Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 149, 1751–1760. doi: 10.1104/pp.108.134684
Kobayashi, K., Yasuno, N., Sato, Y., Yoda, M., Yamazaki, R., Kimizu, M., et al. (2012). Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell 24, 1848–1859. doi: 10.1105/tpc.112.097105
Komatsu, M., Maekawa, M., Shimamoto, K., and Kyozuka, J. (2001). The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev. Biol. 231, 364–373. doi: 10.1006/dbio.2000.9988
Kong, L., Duan, Y., Ye, Y., Cai, Z., Wang, F., Qu, X., et al. (2019). Screening and analysis of proteins interacting with OsMADS16 in rice (Oryza sativa L.). PLoS One 14:e0221473. doi: 10.1371/journal.pone.0221473
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Kyozuka, J., Konishi, S., Nemoto, K., Izawa, T., and Shimamoto, K. (1998). Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc. Nat. Acad. Sci. U.S.A. 95, 1979–1982. doi: 10.1073/pnas.95.5.1979
Laporte, D., Olate, E., Salinas, P., Salazar, M., Jordana, X., and Holuigue, L. (2012). Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J. Exp. Bot. 63, 503–515. doi: 10.1093/jxb/err301
Lee, B. H., Johnston, R., Yang, Y., Gallavotti, A., Kojima, M., Travencolo, B. A., et al. (2009). Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol. 150, 205–216. doi: 10.1104/pp.109.137034
Lee, S., Jeon, J.-S., An, K., Moon, Y.-H., Lee, S., Chung, Y.-Y., et al. (2003). Alteration of floral organ identity in rice through ectopic expression of OsMADS16. Planta 217, 904–911. doi: 10.1007/s00425-003-1066-8
Leibfried, A., To, J. P., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270
Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. doi: 10.1016/s0092-8674(01)00390-7
Li, C., Wang, L., Cui, Y., He, L., Qi, Y., Zhang, J., et al. (2016). Two FERONIA-like receptor (FLR) genes are required to maintain architecture, fertility, and seed yield in rice. Mol. Breeding 36:151. doi: 10.1007/s11032-016-0580-x
Li, S., Lauri, A., Ziemann, M., Busch, A., Bhave, M., and Zachgo, S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21, 429–441. doi: 10.1105/tpc.108.064477
Lim, J., Moon, Y. H., An, G., and Jang, S. K. (2000). Two rice MADS domain proteins interact with OsMADS1. Plant Mol. Biol. 44, 513–527. doi: 10.1023/a:1026517111843
Liu, X., Kim, Y. J., Muller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23, 3654–3670. doi: 10.1105/tpc.111.091538
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/s0092-8674(01)00384-1
Lombardo, F., Kuroki, M., Yao, S. G., Shimizu, H., Ikegaya, T., Kimizu, M., et al. (2017). The superwoman1-cleistogamy2 mutant is a novel resource for gene containment in rice. Plant Biotechnol. J. 15, 97–106. doi: 10.1111/pbi.12594
Ma, H., Chen, J., Zhang, Z., Ma, L., Yang, Z., Zhang, Q., et al. (2017). MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 92, 557–570. doi: 10.1111/tpj.13674
Ma, X., Feng, F., Zhang, Y., Elesawi, I. E., Xu, K., Li, T., et al. (2019). A novel rice grain size gene OsSNB was identified by genome-wide association study in natural population. PLoS Genet. 15:e1008191. doi: 10.1371/journal.pgen.1008191
Maier, A. T., Stehling-Sun, S., Offenburger, S. L., and Lohmann, J. U. (2011). The bZIP transcription factor PERIANTHIA: a multifunctional hub for meristem control. Front. Plant Sci. 2:79. doi: 10.3389/fpls.2011.00079
Maier, A. T., Stehling-Sun, S., Wollmann, H., Demar, M., Hong, R. L., Haubeiss, S., et al. (2009). Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136, 1613–1620. doi: 10.1242/dev.033647
Martins, L., Trujillo-Hernandez, J. A., and Reichheld, J. P. (2018). Thiol based redox signaling in plant nucleus. Front. Plant Sci. 9:705. doi: 10.3389/fpls.2018.00705
Ming, F., and Ma, H. (2009). A terminator of floral stem cells. Genes Dev. 23, 1705–1708. doi: 10.1101/gad.1834409
Moyroud, E., Kusters, E., Monniaux, M., Koes, R., and Parcy, F. (2010). LEAFY blossoms. Trends Plant Sci. 15, 346–352. doi: 10.1016/j.tplants.2010.03.007
Moyroud, E., Tichtinsky, G., and Parcy, F. (2009). The LEAFY floral regulators in angiosperms: conserved proteins with diverse roles. J. Plant Biol. 52, 177–185. doi: 10.1007/s12374-009-9028-8
Nagasawa, N., Miyoshi, M., Kitano, H., Satoh, H., and Nagato, Y. (1996). Mutations associated with floral organ number in rice. Planta 198, 627–633. doi: 10.1007/BF00262651
Nagasawa, N., Miyoshi, M., Sano, Y., Satoh, H., Hirano, H., Sakai, H., et al. (2003). SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130, 705–718. doi: 10.1242/dev.00294
Nijhawan, A., Jain, M., Tyagi, A. K., and Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333–350. doi: 10.1104/pp.107.112821
Ohmori, Y., Tanaka, W., Kojima, M., Sakakibara, H., and Hirano, H. Y. (2013). WUSCHEL-RELATED HOMEOBOX4 is involved in meristem maintenance and is negatively regulated by the CLE gene FCP1 in rice. Plant Cell 25, 229–241. doi: 10.1105/tpc.112.103432
Parcy, F., Nilsson, O., Busch, M. A., Lee, I., and Weigel, D. (1998). A genetic framework for floral patterning. Nature 395, 561–566. doi: 10.1038/26903
Pautler, M., Eveland, A. L., LaRue, T., Yang, F., Weeks, R., Lunde, C., et al. (2015). FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 27, 104–120. doi: 10.1105/tpc.114.132506
Pautler, M., Tanaka, W., Hirano, H. Y., and Jackson, D. (2013). Grass meristems I: shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol. 54, 302–312. doi: 10.1093/pcp/pct025
Peres, A., Churchman, M. L., Hariharan, S., Himanen, K., Verkest, A., Vandepoele, K., et al. (2007). Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J. Biol. Chem. 282, 25588–25596. doi: 10.1074/jbc.M703326200
Prasad, K., Parameswaran, S., and Vijayraghavan, U. (2005). OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J. 43, 915–928. doi: 10.1111/j.1365-313X.2005.02504.x
Prasad, K., Sriram, P., Kumar, C. S., Kushalappa, K., and Vijayraghavan, U. (2001). Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev. Genes Evol. 211, 281–290. doi: 10.1007/s004270100153
Rao, N. N., Prasad, K., Kumar, P. R., and Vijayraghavan, U. (2008). Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc. Natl. Acad. Sci. U.S.A. 105, 3646–3651. doi: 10.1073/pnas.0709059105
Running, M. P., and Meyerowitz, E. M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261–1269. doi: 10.1242/dev.122.4.1261
Sakamoto, T., Sakakibara, H., Kojima, M., Yamamoto, Y., Nagasaki, H., Inukai, Y., et al. (2006). Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 142, 54–62. doi: 10.1104/pp.106.085811
Sakuma, S., and Schnurbusch, T. (2020). Of floral fortune: tinkering with the grain yield potential of cereal crops. New Phytol. 225, 1873–1882. doi: 10.1111/nph.16189
Sato, H., Yoshida, K., Mitsuda, N., Ohme-Takagi, M., and Takamizo, T. (2012). Male-sterile and cleistogamous phenotypes in tall fescue induced by chimeric repressors of SUPERWOMAN1 and OsMADS58. Plant Sci. 183, 183–189. doi: 10.1016/j.plantsci.2011.08.010
Schippers, J. H., Foyer, C. H., and van Dongen, J. T. (2016). Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol. 29, 121–128. doi: 10.1016/j.pbi.2015.11.009
Sessions, A., Nemhauser, J. L., McColl, A., Roe, J. L., Feldmann, K. A., and Zambryski, P. C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491. doi: 10.1242/dev.124.22.4481
Shu, K., Chen, Q., Wu, Y., Liu, R., Zhang, H., Wang, P., et al. (2016). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. Plant J. Cell Mol. Biol. 85, 348–361. doi: 10.1111/tpj.13109
Somssich, M., Je, B. I., Simon, R., and Jackson, D. (2016). CLAVATA-WUSCHEL signaling in the shoot meristem. Development 143, 3238–3248. doi: 10.1242/dev.133645
Su, Y. H., Liu, Y. B., and Zhang, X. S. (2011). Auxin-cytokinin interaction regulates meristem development. Mol. Plant 4, 616–625. doi: 10.1093/mp/ssr007
Su, Y. H., Zhou, C., Li, Y. J., Yu, Y., Tang, L. P., Zhang, W. J., et al. (2020). Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Nat. Acad. Sci. U.S.A. 117, 22561–22571. doi: 10.1073/pnas.2015248117
Sun, B., Looi, L. S., Guo, S., He, Z., Gan, E. S., Huang, J., et al. (2014). Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343:1248559. doi: 10.1126/science.1248559
Sun, B., Xu, Y., Ng, K. H., and Ito, T. (2009). A timing mechanism for stem cell maintenance and differentiation in the Arabidopsis floral meristem. Genes Dev. 23, 1791–1804. doi: 10.1101/gad.1800409
Sun, C., Zhang, K., Zhou, Y., Xiang, L., He, C., Zhong, C., et al. (2021). Dual function of clock component OsLHY sets critical day length for photoperiodic flowering in rice. Plant Biotechnol. J. 19, 1644–1657. doi: 10.1111/pbi.13580
Suzaki, T., Sato, M., Ashikari, M., Miyoshi, M., Nagato, Y., and Hirano, H. Y. (2004). The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development 131, 5649–5657. doi: 10.1242/dev.01441
Suzaki, T., Toriba, T., Fujimoto, M., Tsutsumi, N., Kitano, H., and Hirano, H. Y. (2006). Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 47, 1591–1602. doi: 10.1093/pcp/pcl025
Takeda, S., Hanano, K., Kariya, A., Shimizu, S., Zhao, L., Matsui, M., et al. (2011). CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 66, 1066–1077. doi: 10.1111/j.1365-313X.2011.04571.x
Tanaka, W., Pautler, M., Jackson, D., and Hirano, H. Y. (2013). Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54, 313–324. doi: 10.1093/pcp/pct016
Timofejeva, L., Skibbe, D. S., Lee, S., Golubovskaya, I., Wang, R., Harper, L., et al. (2013). Cytological characterization and allelism testing of anther developmental mutants identified in a screen of maize male sterile lines. G3 (Bethesda) 3, 231–249. doi: 10.1534/g3.112.004465
Tsuda, K., Ito, Y., Sato, Y., and Kurata, N. (2011). Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice. Plant Cell 23, 4368–4381. doi: 10.1105/tpc.111.090050
Tsuda, K., Kurata, N., Ohyanagi, H., and Hake, S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26, 3488–3500. doi: 10.1105/tpc.114.129122
Viola, I. L., Guttlein, L. N., and Gonzalez, D. H. (2013). Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol. 162, 1434–1447. doi: 10.1104/pp.113.216416
Vollbrecht, E., Reiser, L., and Hake, S. (2000). Shoot meristem size is dependent on inbred background and presence of the maize homeobox gene, knotted1. Development 127, 3161–3172. doi: 10.1242/dev.127.14.3161
Waadt, R., Schmidt, L. K., Lohse, M., Hashimoto, K., Bock, R., and Kudla, J. (2008). Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 56, 505–516. doi: 10.1111/j.1365-313X.2008.03612.x
Wagner, D., Sablowski, R. W., and Meyerowitz, E. M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285, 582–584. doi: 10.1126/science.285.5427.582
Wang, F., Han, T., Song, Q., Ye, W., Song, X., Chu, J., et al. (2020). The rice circadian clock regulates tiller growth and panicle development through strigolactone signaling and sugar sensing. Plant Cell 32, 3124–3138. doi: 10.1105/tpc.20.00289
Wang, J., Wang, R., Wang, Y., Zhang, L., Zhang, L., Xu, Y., et al. (2017). Short and Solid Culm/RFL/APO2 for culm development in rice. Plant J. 91, 85–96. doi: 10.1111/tpj.13548
Weimer, A. K., Matos, J. L., Sharma, N., Patell, F., Murray, J. A. H., Dewitte, W., et al. (2018). Lineage- and stage-specific expressed CYCD7;1 coordinates the single symmetric division that creates stomatal guard cells. Development 145:dev160671. doi: 10.1242/dev.160671
Xu, K., Huang, X., Wu, M., Wang, Y., Chang, Y., Liu, K., et al. (2014). A rapid, highly efficient and economical method of Agrobacterium-mediated in planta transient transformation in living onion epidermis. PLoS One 9:e83556. doi: 10.1371/journal.pone.0083556
Xu, W., Tao, J., Chen, M., Dreni, L., Luo, Z., Hu, Y., et al. (2017). Interactions between FLORAL ORGAN NUMBER4 and floral homeotic genes in regulating rice flower development. J. Exp. Bot. 68, 483–498. doi: 10.1093/jxb/erw459
Yaish, M. W., El-Kereamy, A., Zhu, T., Beatty, P. H., Good, A. G., Bi, Y. M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. doi: 10.1371/journal.pgen.1001098
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H. Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16, 500–509. doi: 10.1105/tpc.018044
Yamaki, S., Nagato, Y., Kurata, N., and Nonomura, K. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 351, 208–216. doi: 10.1016/j.ydbio.2010.12.006
Yang, F., Bui, H. T., Pautler, M., Llaca, V., Johnston, R., Lee, B. H., et al. (2015). A maize glutaredoxin gene, abphyl2, regulates shoot meristem size and phyllotaxy. Plant Cell 27, 121–131. doi: 10.1105/tpc.114.130393
Yang, R. S., Xu, F., Wang, Y. M., Zhong, W. S., Dong, L., Shi, Y. N., et al. (2021). Glutaredoxins regulate maize inflorescence meristem development via redox control of TGA transcriptional activity. Nat. Plants 7, 1589–1601. doi: 10.1038/s41477-021-01029-2
Yi, J., Lee, Y. S., Lee, D. Y., Cho, M. H., Jeon, J. S., and An, G. (2016). OsMPK6 plays a critical role in cell differentiation during early embryogenesis in Oryza sativa. J. Exp. Bot. 67, 2425–2437. doi: 10.1093/jxb/erw052
Yoon, J., Cho, L. H., Antt, H. W., Koh, H. J., and An, G. (2017). KNOX protein OSH15 induces grain shattering by repressing lignin biosynthesis genes. Plant Physiol. 174, 312–325. doi: 10.1104/pp.17.00298
Zeng, J., Dong, Z., Wu, H., Tian, Z., and Zhao, Z. (2017). Redox regulation of plant stem cell fate. EMBO J. 36, 2844–2855. doi: 10.15252/embj.201695955
Zhang, X. (2014). Delayed gratification—waiting to terminate stem cell identity. Science 343, 498–499. doi: 10.1126/science.1249343
Zhao, D.-S., Li, Q.-F., Zhang, C.-Q., Zhang, C., Yang, Q.-Q., Pan, L.-X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhao, Z., Andersen, S. U., Ljung, K., Dolezal, K., Miotk, A., Schultheiss, S. J., et al. (2010). Hormonal control of the shoot stem-cell niche. Nature 465, 1089–1092.
Zhu, Q., Zhang, X. L., Nadir, S., DongChen, W. H., Guo, X. Q., Zhang, H. X., et al. (2017). A LysM domain-containing gene OsEMSA1 involved in embryo sac development in rice (Oryza sativa L.). Front. Plant Sci. 8:1596. doi: 10.3389/fpls.2017.01596
Keywords: Oryza sativa, shoot apical meristem, panicle, floret, PERIANTHIA, FASCIATED EAR4, OsMADS1
Citation: Prakash S, Rai R, Zamzam M, Ahmad O, Peesapati R and Vijayraghavan U (2022) OsbZIP47 Is an Integrator for Meristem Regulators During Rice Plant Growth and Development. Front. Plant Sci. 13:865928. doi: 10.3389/fpls.2022.865928
Received: 30 January 2022; Accepted: 09 March 2022;
Published: 13 April 2022.
Edited by:
José Manuel Pérez-Pérez, Miguel Hernández University of Elche, SpainReviewed by:
Ram Kishor Yadav, Indian Institute of Science Education and Research Mohali, IndiaWeibing Yang, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences (CAS), China
Copyright © 2022 Prakash, Rai, Zamzam, Ahmad, Peesapati and Vijayraghavan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Usha Vijayraghavan, dXZyQGlpc2MuYWMuaW4=; dXZyMTIzQGdtYWlsLmNvbQ==
†These authors share first authorship
‡These authors share second authorship
 Sandhan Prakash
Sandhan Prakash Rashmi Rai
Rashmi Rai Mohamed Zamzam
Mohamed Zamzam Owais Ahmad
Owais Ahmad Raghavaram Peesapati
Raghavaram Peesapati Usha Vijayraghavan
Usha Vijayraghavan