
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 25 April 2022
Sec. Plant Proteomics and Protein Structural Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.865361
This article is part of the Research Topic Seed Proteomics View all 11 articles
Seeds are essential for the reproduction and dispersion of spermatophytes. The seed life cycle from seed development to seedling establishment proceeds through a series of defined stages regulated by distinctive physiological and biochemical mechanisms. The role of histone modification and chromatin remodeling in seed behavior has been intensively studied in recent years. In this review, we summarize progress in elucidating the regulatory network of these two kinds of epigenetic regulation during the seed life cycle, especially in two model plants, rice and Arabidopsis. Particular emphasis is placed on epigenetic effects on primary tissue formation (e.g., the organized development of embryo and endosperm), pivotal downstream gene expression (e.g., transcription of DOG1 in seed dormancy and repression of seed maturation genes in seed-to-seedling transition), and environmental responses (e.g., seed germination in response to different environmental cues). Future prospects for understanding of intricate interplay of epigenetic pathways and the epigenetic mechanisms in other commercial species are also proposed.
Well-developed seeds assure species dispersion of parent plants and serve as important sources of human food. Progression from seed development to seedling establishment is the crucial phase of ontogenesis in spermatophytes. It involves a series of sequential physio-morphological state changes and includes several biological stages. The process starts with seed development and followed by maturation, when seed desiccation and seed dormancy are achieved in some species. After that, seeds germinate in a suitable environment, which marks the initiation of seedling establishment.
Proper seed development is inseparable from the organized establishment of all tissues (such as embryo, endosperm and seed coat), which is coordinated by changes in hormone levels and gene expression (Zhou et al., 2013; Figueiredo and Köhler, 2018). Seed maturation proteins in the LAFL regulatory network—LEAFY COTYLEDON 1 (LEC1), ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3), and LEAFY COTYLEDON 2 (LEC2)—play predominant roles in triggering and maintaining embryonic cell fate by fine-tuning the expression of genes involved in the accumulation of storage protein and lipid reserves in the embryo (Giraudat et al., 1992; Keith et al., 1994; Lotan et al., 1998; Stone et al., 2001; Yamamoto et al., 2009; Lepiniec et al., 2018).
In some species, during the maturation stage of seed development, dormancy gradually increases, peaking in freshly matured seeds. Dormancy enables seeds to adapt to the environment and plants to maintain reproduction (Née et al., 2017). DELAY OF GERMINATION1 (DOG1) is the master regulator of primary dormancy in Arabidopsis (Arabidopsis thaliana); the encoded protein is a temperature detector that directs dormancy cycling in response to seasonal changes (Alonso-Blanco et al., 2003; Bentsink et al., 2006; Nakabayashi et al., 2012; Graeber et al., 2013; Footitt et al., 2015).
In appropriate condition, seeds can germinate once exposed to water. Germination behaviors, including germination rate and efficiency, differ between species and varieties in response to environment cues or abiotic stress. These differences are mediated mainly through the antagonistic roles of the plant hormones gibberellic acid (GA) and abscisic acid (ABA) (Jacobsen et al., 2002; Oh et al., 2007; Holdsworth et al., 2008; Seo et al., 2009; Vaistij et al., 2013; Shu et al., 2016).
Seed germination marks the initiation of the seed-to-seedling developmental transition. In this process, the sources of seedling nutrition and energy acquisition gradually transition from consumption of seed storage substances to photoautotrophy, in conjunction with significant alteration of biosynthetic and signaling pathways (Zanten et al., 2013; Jia et al., 2014). Correspondingly, suppression of seed maturation genes, the LAFL, and activation of those involved in vegetative growth is indispensable to avoid ectopic proliferation of embryonic tissues and thus maintain the normal vegetative morphology of seedlings (Parcy and Giraudat, 1997; Lotan et al., 1998; Stone et al., 2001; Gazzarrini et al., 2004; Braybrook et al., 2006; Yang et al., 2013).
The different stages of the seed life cycle are not isolated and are each under precise control. Accurate DNA processing and subsequent gene transcript levels are tightly linked to chromatin status, which is regulated by epigenetic modification. Epigenetic changes, including DNA methylation, histone modifications, chromatin remodeling, and the activities of small RNAs, affect plants in many ways (Goldberg et al., 2007). Research on the effects of epigenetic regulation in seed biology has recently increased. This review focuses mainly on the effects of two types of key regulatory factors, histone modifiers and chromatin remodelers, on the pivotal phase of the seed life cycle (Table 1).

Table 1. List of histone modifiers, chromatin remodelers and associated regulators involved in seed life cycle.
Histone modifications, which are usually added to the N-terminus of the histone protein tail, can either regulate chromatin state directly or act as hotspots for the recruitment of other effectors to chromatin. Different histone modifications, including acetylation, methylation, ubiquitylation, and phosphorylation, comprise a “histone code” that provides a flexible method of governing gene transcription in response to developmental or environmental cues (Strahl and Allis, 2000; Turner, 2000; Dutnall, 2003). Histone modifications are reversible, being added and removed by “writer” and “eraser” enzyme complexes, respectively, that execute distinct functions on chromatin, promoting either active transcription or gene silencing (Kumar et al., 2021). For example, trimethylation of histone H3 lysine 4 (H3K4me3) and 36 (H3K36me3) are usually associated with gene activation, whereas H3K27me3, which is directly regulated by the classical polycomb repressive complex 2 (PRC2), correlates with heterochromatinization and transcriptional silencing (Berger, 2007; Zhang et al., 2007a,b, 2009; Mozgova and Hennig, 2015; Liu et al., 2019).
ATP-dependent chromatin remodeling complexes, with members of DNA-dependent ATPases as core subunits, utilize energy from ATP hydrolysis to disrupt the contacts between histones and DNA, thereby regulating dynamic access to packaged DNA. They mediate DNA replication, damage repair, and gene expression by changing the positions and occupancy of nucleosomes, introducing histone variants, and cooperating with histone-modifying factors (Goldberg et al., 2007; Hargreaves and Crabtree, 2011; Li et al., 2016). In eukaryotes, different epigenetic regulations often work coordinately to achieve cooperative or antagonistic modes of regulation (Li et al., 2015).
In this review, we summarize the histone modification and chromatin remodeling that occur during the seed life cycle, from seed development to seedling establishment. We hope to thereby pave the way toward a fundamental understanding and integration of the complex networks of epigenetic regulation acting in seed biology.
The production of viable seed is important for plant dispersal, and is a major focus of crop breeders because of its direct association with grain yield. Seed development involves the sequential and orderly formation of various structures, including the embryo, endosperm, and seed coat. Among angiosperms, monocotyledonous and dicotyledonous plants show both similarities and differences in seed development (Zhou et al., 2013). In this section, we primarily discuss epigenetic effects on seed formation in Arabidopsis and rice (Oryza sativa), the model dicot and monocot species, respectively (Figure 1A).
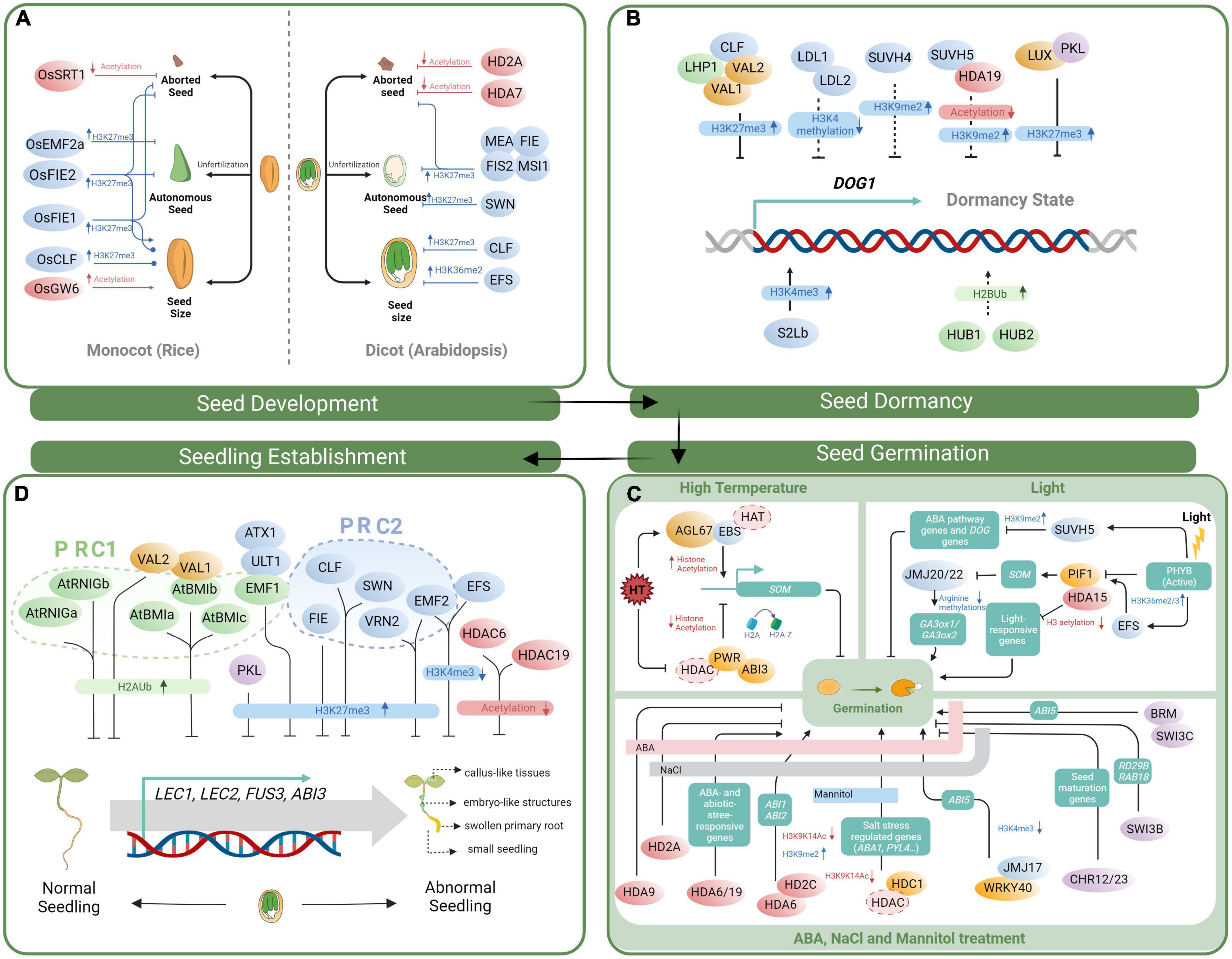
Figure 1. Model of histone modification and chromatin remodeling during various stages of the seed life cycle. (A) Epigenetic regulators act of pivotal seed development in Arabidopsis and rice. Line with a dot in tail end represent an unclear regulatory pathway. (B) Chromatin regulation of seed dormancy through mediating the expression of DOG1. Solid lines represent regulator that can modulate DOG1 expression directly. Dash lines represent those regulatory mechanisms that are still unclear. (C) The roles of histone modifiers and chromatin remodelers in regulating seed germination in response to light, temperature and abiotic stress. In the box below, the bold line with pink, gray and blue colors represent ABA, NaCl and mannitol treatment, respectively. Ellipse with dotted outline represents that the specific regulator is not clear. (D) Epigenetic factors act in seed-to-seedling transition. Illustrated histone modifiers and chromatin remodelers act in repression of ectopic expression of LAFL genes, thereby maintaining normal vegetative morphology of seedlings. The histone modifiers for histone acetylation, methylation, and ubiquitination are marked in red, blue and green, respectively. Chromatin remodelers and epigenetic associated regulators are marked in purple and orange, respectively. Downstream genes are shown in boxes with aquamarine blue.
Histone deacetylases (HDACs) control seed-setting rate through affecting acetylation levels of target genes. AtHD2A, a member of the HD2 subfamily of HDAC proteins, is highly expressed in flowers and siliques. Silencing of AtHD2A expression aborts seed development (Wu et al., 2000). Similarly, the Athda7-2 mutant causes both degeneration of micropilar nuclei at the four-nucleate embryo sac stage and an overall delay of embryo development, ultimately decreasing seed fertility (Cigliano et al., 2013). Meanwhile, OsSRT1, a NAD + -dependent HDAC in rice, represses the expression of RICE STARCH REGULATOR1 (RSR1) and amylase genes, thus maintaining starch accumulation in developing seeds (Zhang et al., 2016). In maize, increases in acetylated histones H3 and H4 accompanied by decreases in H3K9me2 are observed in hda108 mutants, resulting in a wide range of plant damage, including impaired fertility of cobs (Forestan et al., 2018).
As a nutritional supply tissue, the endosperm is indispensable for seed development. Disordered timing of endosperm development leads to seed failure in interploid and interspecific hybrids, directly impeding crop breeding (Walia et al., 2009; Ishikawa et al., 2011; Kradolfer et al., 2013; Sekine et al., 2013). During cell proliferation, the PRC2 complex adds the repressive mark H3K27me3 on endosperm-related transcripts. In Arabidopsis, mutations of genes encoding members of the FERTILIZATION-INDEPENDENT SEED (FIS)–PRC2 complex [FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), MEDEA (MEA), FIS2, MULTICOPY SUPRESSOR OF IRA1 (MSI1)] decrease H3K27me3 accumulation and impair cellularization of endosperm, and the mutants are characterized by a gametophytic maternal effect. After fertilization, embryonic cell proliferation and morphogenesis are inhibited in these mutants, decreasing their seed-setting rate. Notably, endosperm proliferation can also initiate in mutants in the absence of fertilization, but with arrested embryo development, producing non-functional autonomous seeds (Chaudhury et al., 1997; Grossniklaus et al., 1998; Kiyosue et al., 1999; Ohad et al., 1999; Yadegari et al., 2000; Sørensen et al., 2001; Köhler et al., 2003a,b). The downstream genes regulated by the FIS–PRC2 complex include type I MADS-box transcription factor genes, which encode key regulators in endosperm formation (Kang et al., 2008; Hehenberger et al., 2012; Figueiredo et al., 2015; Zhang et al., 2018). Moreover, the seed-abortion phenotype of a mea mutant can be alleviated by reducing the expression of the type I MADS-box gene PHERES1 (PHE1) (Köhler et al., 2003b). Similarly, maternal loss of AGAMOUS-LIKE62 (AGL62) can also rescue delayed cellularization of endosperm cells, normalizing seed development in a fis2 mutant (Hehenberger et al., 2012). Aside from the FIS–PRC2 complex, SWINGER (SWN), a subunit of the EMBRYONIC FLOWER 1 (EMF)–PRC2 complex also participates in the initiation of endosperm development. Although a swn mutant shows no identifiable developmental defect, a swn mea double mutant has an enhanced autonomous seed formation phenotype compared to the mea single mutant, indicating that SWN and MEA work redundantly (Wang et al., 2006).
In cereal, the PRC2 complex is evolutionarily conserved, but shows differences in combination of subunits and in specific function compared with that in Arabidopsis, as exemplified by the absence of some homologs of Arabidopsis FIS genes (MEA and FIS2) in cereal genomes (Rossi et al., 2001; Springer et al., 2002; Danilevskaya et al., 2003; Spillane et al., 2007; Luo et al., 2009; Rodrigues et al., 2010). Rice has two FIE homologs, OsFIE1 and OsFIE2, with different expression patterns: OsFIE1 is specifically expressed in endosperm, whereas OsFIE2 is expressed in all tissues tested. In earlier research, no autonomous endosperm development was observed in OsFIE1 and OsFIE2 loss-of-function plants with emasculated florets (Luo et al., 2009; Nallamilli et al., 2013). However, other studies revealed that the autonomous endosperm phenotype could be occasionally detected in unfertilized lines with OsFIE2 defect, along with impaired cellularization, suggesting that OsFIE2 may retain functional similarity to its Arabidopsis homolog (Li et al., 2014; Cheng et al., 2020).
Moreover, rice also has two homologs of the Arabidopsis PcG gene EMBRYONIC FLOWER2 (EMF2): EMF2a and EMF2b. Arabidopsis EMF2 is required to maintain vegetative development in Arabidopsis (Moon et al., 2003). Rice EMF2a, a maternally expressed gene in the endosperm, is indispensable for early seed development; delayed cellularization of endosperm and subsequent autonomous endosperm is observed in emasculated spikelets of an osemf2a mutant. Not surprisingly, the loss of OsEMF2a function reduces H3K27me3 modifications at various type I MADS-box genes, several of which (e.g., OsMADS77 and OsMADS89) may control the timing of cellularization in endosperm in a similar manner to AGL62 and PHE1 in Arabidopsis (Luo et al., 2009; Cheng et al., 2021; Tonosaki et al., 2021).
Seed size and weight are crucial agronomical traits, tightly linked with grain yield in crop breeding (Sweeney and McCouch, 2007). In Arabidopsis, the inner space of mature seed is mostly occupied by the embryo, and nutrients are mainly stored in the cotyledons (Zhou et al., 2013). Mutation of EARLY FLOWERING IN SHORT DAYS (EFS), also called SDG8, encoding the major contributor to H3K36 methylation, leads to the formation of larger embryos, resulting in enlarged seeds in Arabidopsis (Cheng et al., 2018). Similarly, larger and heavier seeds are also observed in clf-28 lines (mutants of CURLY LEAF [CLF], which encodes the core unit of the PRC2 complex), along with a large-scale, dynamic change in H3K27me3 level during embryonic development (Liu J. et al., 2016). Unlike in dicotyledons, endosperm in monocotyledon crops is not gradually consumed during seed maturation, but filled with large amounts of starch and nutrients for storage (Olsen, 2004; Agarwal et al., 2011; Zhou et al., 2013). In rice, unlike in the Arabidopsis clf-28 mutant, which has enlarged seeds, OsSDG711 (OsCLF) downregulation lines have smaller seeds, accompanied by altered expression of starch-related genes (Liu et al., 2021). A similar phenotype of smaller seed size and reduced contents of multiple storage proteins is also observed in reduction lines of OsFIE2 or OsFIE1 (Nallamilli et al., 2013; Huang et al., 2016; Liu X. et al., 2016). However, overexpression of SDG711 or OsFIE1 also decrease seed size, but the regulatory mechanisms are not fully elucidated (Folsom et al., 2014; Liu et al., 2021). Another tissue with important effects on grain size and weight is the spikelet hull. In rice, the grain is physically restricted by the size of the hull. The quantitative trait locus (QTL) GRAIN WEIGHT ON CHROMOSOME 6 (GW6a), also called OsglHAT1, encodes a histone acetyltransferase that regulates grain weight, hull size, yield, and plant biomass. Elevated GW6a expression enhances grain yield by enlarging spikelet hulls via increasing cell number and accelerating grain filling (Song et al., 2015).
In general, the epigenetic regulation of major seed traits in two model plants, Arabidopsis and rice, partially overlaps but also shows some divergence. The comparison of epigenetic machinery in seed development is also extended to other crop plants, like soybean and maize (Lu et al., 2015; Lin et al., 2017). Therefore, the similarity and divergence highlight the need for more knowledge of the complex network of epigenetic regulation influencing seed development programs in different species.
Seed dormancy is an innate state in which the seed is unable to germinate, even under favorable conditions. Entry into dormancy is determined primarily by genetic factors and is also influenced by the environment surrounding the mother plant (Finch-Savage and Leubner-Metzger, 2006). Seeds in the soil seed bank (SSB) can sense seasonal signals and continually adjust their dormancy levels in order to complete germination at a suitable time of the year (Baskin and Baskin, 2006; Walck et al., 2011; Finch-Savage and Footitt, 2017).
Several histone-modification genes regulating histone ubiquitination, methylation, and acetylation exhibit dynamic expression patterns in response to seasonal change that are correlated with dormancy cycling in the SSB. Moreover, the accumulation of two antagonistic histone marks, H3K4me3 and H3K27me3, on key dormancy genes changes dynamically accompanied by changes in dormancy level, suggesting that chromatin regulators play pivotal roles in this process (Footitt et al., 2015). DOG1 is a master regulator of primary dormancy that acts in concert with ABA and HEME to delay germination (Née et al., 2017; Carrillo-Barral et al., 2020). During release from dormancy, the active H3K4me3 mark on DOG1 chromatin is removed when the seeds are exposed to light; meanwhile, the repressive H3K27me3 mark accumulates on DOG1 in seeds of the SSB, and light exposure amplifies this accumulation (Müller et al., 2012; Footitt et al., 2015).
The components of polycomb-group proteins, including CLF and LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), are recruited by B3-domain-containing transcriptional repressors, HSI2/VAL1 (HIGH-LEVEL EXPRESSION OF SUGAR INDUCIBLE2/VIVIPAROUS-1/ABA3-LIKE1) and HSL1 (HSI2-LIKE1)/VAL2, to RY elements in the DOG1 promoter. Hence, they accelerate the deposition of H3K27me3 marks and subsequent repression of DOG1 expression (Chen N. et al., 2020). PICKLE (PKL) is an ATP-dependent chromatin-remodeling factor that promotes the deposition of H3K27me3 (Zhang et al., 2008, 2012). LUX ARRHYTHMO (LUX), a member of the evening complex (EC) of the circadian clock, physically interacts with PKL and recruits it to the chromatin region of DOG1. Correspondingly, levels of the repressive mark H3K27me3 at specific DOG1 chromatin loci are greatly reduced in the lux and pkl mutants, increasing dormancy compared with the wild type. Moreover, these phenotypes are abolished when the mother plants are grown under continuous light. Thus, there may exist a regulatory mechanism in which EC proteins coordinate with PKL to transmit circadian signals, thereby directly regulating DOG1 expression and seed dormancy during seed maturation (Zha et al., 2020). On the other hand, the binding site of LUX is close to the transcriptional start site of the non-coding antisense transcript asDOG1, which suppresses the expression of the DOG1 sense transcript, and asDOG1 transcription is decreased in the pkl-1 mutant (Fedak et al., 2016; Zha et al., 2020). It suggests that the effect of PKL-LUX repression of DOG1 transcription maybe much more elaborate than might be expected. In contrast to the H3K27me3 deposition functions of the LUX-PKL regulatory complex, RELATIVE OF EARLY FLOWERING6 (REF6), a key H3K27me3 demethylase that binds directly to the ABA catabolism genes CYP707A1 and CYP707A3, is responsible for reducing their H3K27me3 levels. Correspondingly, the seeds of ref6 mutants display enhanced dormancy, associated with increased endogenous ABA content (Chen H. et al., 2020).
Beyond H3K27me3, H3K9 and H3K4 methylation are also involved in DOG1 and seed dormancy control. The global accumulation of H3K9 dimethylation is catalyzed by KYP/SUVH4, a Su(var)-type methyltransferase. Mutations in the KYP increase DOG1 and ABI3 expression, promoting seed dormancy. On the other hand, the sensitively of seed germination to ABA and paclobutrazol (PAC) is also increased in kyp-2 mutant (Zheng et al., 2012). SUVH5, a homolog of SUVH4, interacts with the histone deacetylase HDA19 in vivo and in vitro. Mutants of both SUVH5 and HDA19 increase histone H3 acetylation (H3ac) but decrease H3K9me2, therefore enhancing DOG1 expression and seed dormancy (Zhou et al., 2020). LYSINE-SPECIFIC DEMETHYLASE 1-LIKE1 (LDL1) and LDL2, two Arabidopsis histone demethylases, reduce the level of the histone H3-Lys 4 methylation in chromatin. They act redundantly to repress genes related to seed dormancy, including DOG1, ABA2, and ABI3, and LDL1 or LDL2 overexpression lines cause reduced seed dormancy (Zhao et al., 2015).
In yeast, H3K4me3 deposition is regulated by the SET1 histone methyltransferase (HMT) embedded in a so-called COMPlex of Proteins Associated with Set1 (COMPASS), in a process dependent on H2B monoubiquitination (H2Bub) (Miller et al., 2001; Sun and Allis, 2002). Similarly, Arabidopsis possesses homologs of all known COMPASS subunits, which potentially form several COMPASS-like complexes (Jiang et al., 2009, 2011; Fletcher, 2017). Genetic knockout of SWD2-LIKE b (S2Lb), the Arabidopsis homolog of Swd2 axillary subunit of yeast COMPASS, triggers pleiotropic developmental phenotypes, including reduced fertility and seed dormancy, accompanied by decreased H3K4me3 deposition and barely detectable DOG1 expression (Fiorucci et al., 2019). However, even though H2B monoubiquitination regulated by HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2 also increases DOG1 expression and seed dormancy, the classical H2Bub–H3K4me3 trans-histone crosstalk seems to be lacking in Arabidopsis, since global H3K4me3 enrichment and the occupancy of an S2Lb-GFP (green fluorescent protein) fusion on target genes do not show obvious differences in the hub1-3 background as compared with that in wild-type (Liu et al., 2007; Fiorucci et al., 2019).
Unlike direct regulators of histone modification, histone readers are involved in recognizing these marks and transferring the information to subsequent regulator units. The reader EARLY BOLTING IN SHORT DAYS (EBS) specifically recognizes the H3K4me2/3 mark and interacts with HDAC proteins such as HDA6 to modulate gene expression. Loss of function mutation of EBS reduces seed dormancy, and mutation of EBS homolog SHORT LIFE (SHL) deepens the seed dormancy alteration. However, EBS acts independently of two other types of dormancy regulators, HUB proteins and ARABIDOPSIS TRITHORAX-RELATED7 (ATXR7), and it does not affect the expression of DOG1 (Liu et al., 2007, 2011; Narro-Diego et al., 2017).
Overall, studies of the epigenetic regulation of seed dormancy have mainly focused on DOG1 expression (Figure 1B), and other regulatory pathways need further investigation to further complete the picture.
Seed germination is an important physiological event that marks a transition from the quiet status of seeds to the active statues of seedlings, during which many processes are reprogrammed. The condensed chromatin state may diminish during seed germination (van Zanten et al., 2011; Zanten et al., 2013), providing a suitable environment for activating gene expression and physiological metabolisms that facilitate the process. Seeds can adjust their germination strategies in response to external environmental cues. In this section, we highlight current knowledge of the roles of histone modifiers and chromatin remodelers in regulating seed germination in response to light, temperature, and abiotic stresses (Figure 1C).
Light and temperature are two main exogenous factors determining plant growth, development, and productivity, including seed germination. In this phase, one of the classical light signal transport chains is the PHYTOCHROME B (PHYB)–PHYTOCHROME INTERACTING FACTOR1 (PIF1)/PIL5 pathway. In Arabidopsis, PHYB destabilizes PIF1 to regulate light-responses seed germination through affecting genes expression in ABA and GA pathways. SOMNUS (SOM) is an important PIF1 downstream target that negatively regulates seed germination (Oh et al., 2006; Kim et al., 2008; Luo et al., 2014; Vaistij et al., 2018). Chromatin remodeling and histone modification that participate in light-regulated seed germination mainly function by affecting this PHYB-dependent pathway.
SUVH5, an H3K9 methyltransferase, acts as a positive regulator of PHYB-dependent seed germination in Arabidopsis. It functions by repressing the transmission of ABA signal and ABA biosynthesis, as well as suppressing the expression a family of DOG1 genes via H3K9me2 in imbibed seeds (Gu et al., 2019). Mutation of EFS/SDG8, another HMT gene, decreases H3K36me2 and H3K36me3 levels at the PIF1 locus, resulting in reduced PIF1 expression in imbibed seeds (Lee et al., 2014). Meanwhile, HDA15 can be recruited by PIF1 and to form a repression module that regulates light-dependent seed germination by decreasing histone H3 acetylation levels and the corresponding transcription of light-responsive genes (Gu et al., 2017). JMJ20 and JMJ22, two histone demethylation enzymes, act redundantly as positive regulators of seed germination. When PHYB is inactive, JMJ20 and JMJ22 are directly suppressed by the zinc-finger protein SOM, and the repression will be released upon PHYB activation by light. Derepressed JMJ20/JMJ22 increase seed germination rate through the removal of repressive histone arginine methylations at GIBBERELLIN 3-OXIDASE1 (GA3ox1) and GA3ox2 (Cho et al., 2012). Therefore, light treatment promotes seed germination in a process that may be partially regulated by the PHYB–PIF1–SOM–JMJ20/JMJ22 pathway.
Moreover, another JmjC-domain demethylase JMJ17 participates in ABA response in seed germination through co-regulation with WRKY DNA-BINDING PROTEIN 40 (WRKY40), HYPOCOTYL5 (HY5), and ABI5. An elevated level of H3K4me3 at ABI5 has been detected in jmj17 and wrky40 mutants. In the presence of ABA, WRKY40 and JMJ17 are released from ABI5 chromatin, which allows HY5 to induce ABI5 expression. Because HY5 is another crucial factor that helps promote photomorphogenesis, the transcriptional switch composed of JMJ17–WRKY40 and HY5–ABI5 modules may play an essential role in the integration of light and ABA signaling (Wang et al., 2021).
Temperature is another critical environmental cue affecting seed germination (Toh et al., 2008). SOM participates in thermoinhibition of seed germination by altering ABA and GA metabolism (Park et al., 2011; Lim et al., 2013; Chang et al., 2018). EBS, the histone mark reader, can be recruited by AGL67 to the SOM locus, thus recognizing H3K4me3 at the SOM promoter. Under high temperature (HT), the AGL67–EBS complex is highly enriched around the SOM promoter, leading to deposition of the activation mark H4K5 acetylation on SOM and ultimately inhibiting seed germination (Li et al., 2020). POWERDRESS (PWR), a protein with a SANT-domain, interacts with ABI3 and HDAC proteins to modify histone acetylation status and the level of nucleosome histone H2A.Z incorporation in the target loci. The complex inhibits SOM expression by reducing H4 acetylation deposition and increasing nucleosome H2A.Z content at the SOM locus, thus promoting the thermotolerance of seed germination. Under HT, the PWR transcript decreased, resulting in releasing of SOM from repression state (Yang et al., 2019).
ABA, as a barrier to germination, plays a pivotal role in plant response to abiotic stresses, such as drought and salt (Zhu, 2016). Members in the SWITCH2(SWI2)/SNF2 chromatin-remodeling complexes affect seed germination under ABA treatment. BRAHMA (BRM), the core SWI2/SNF2 ATPases within the complex, directly repress the expression of ABI5, the brm-3 mutant presents ABA hypersensitivity in seed germination (Han et al., 2012). SWITCH SUBUNIT3 (SWI3) proteins (called SWI3A–D) in Arabidopsis are also important subunits of SWI2/SNF2-dependent chromatin-remodeling complexes (Sarnowski et al., 2005). BRM and SWI3C show strong direct physical interaction and null swi3c-2 mutants show an ABA-hypersensitive phenotype similar to brm. These observations suggest that SWI2C may be a dedicated BRM complex component (Hurtado et al., 2006; Han et al., 2012). In contrast, mutants of SWI3B show reduced sensitivity to ABA-mediated inhibition of seed germination, with reduced expression of the ABA-responsive genes RAB18 and RD29B (Saez et al., 2008). Furthermore, in overexpression lines of AtCHR12 or AtCHR23, another two SWI2/SNF2 ATPase genes, the phenotype of reduced germination is pronounced under ABA and NaCl treatment, coinciding with increased transcription of seed maturation genes (Leeggangers et al., 2015).
In addition to chromatin remodelers, HDAC proteins are also involved in abiotic-stress-responsive seed germination. In Arabidopsis, germination of the hd2c mutant is restrained under ABA and salinity stress, while the hd2a mutant is insensitive to ABA (Colville et al., 2011). HD2C interacts with HDA6 and binds to histone H3. The expressions of ABI1 and ABI2 are decreased along with increased H3K9K14Ac and decreased H3K9me2 modification in hda6, hd2c, and hda6 hd2c-1 (Luo et al., 2012). Moreover, HDA6 and HDA19 may play redundant roles in modulating seed germination response to abiotic stress by increasing the expression of ABA- and abiotic-stress-responsive genes (Chen and Wu, 2010; Chen L. T. et al., 2010). Arabidopsis HISTONE DEACETYLATION COMPLEX1 (HDC1) is the rate-limiting component of the histone deacetylation complex that physically interacts with HDAC proteins to desensitize plant germination to salt, mannitol, ABA, and PAC treatments (Perrella et al., 2013). By contrast, hda9-1 and hda9-2 mutants show increased germination in response to ABA treatment and HDA9 forms a complex with ABI4 to regulate the expression of the ABA catabolic genes CYP707A1 and CYP707A2 (Baek et al., 2020). In rice, plants overexpressing HDA705 or HDT701 show not only delayed germination under ABA, NaCl, or polyethylene glycol (PEG) treatment, but also stronger resistance to drought stress as seedlings (Zhao et al., 2014, 2016).
After seeds germination, plants undergo an irreversible transition from embryo to seedling development, accompanied by repression of embryonic traits and emergence of vegetative tissue. Expression change of the seed-maturation genes collectively known as LAFL is important for the switch of the developmental program. In loss-of-function mutants of these genes, embryos skip late-embryonic development and enter the vegetative program prematurely (Keith et al., 1994; West et al., 1994; Nambara et al., 2000). However, when some of these genes are misexpressed in vegetative tissues, abnormally developed seedlings emerge that show induced ectopic deposition of seed storage proteins and even somatic embryo or callus formation (Parcy and Giraudat, 1997; Lotan et al., 1998; Stone et al., 2001; Gazzarrini et al., 2004; Braybrook et al., 2006; Yang et al., 2013). The factors involved in chromatin remodeling and histone modification protect normal seedling morphology mainly by repressing the transcription of LAFL genes (Figure 1D).
In the PRC1 complex, two types of ring-finger proteins, AtRING1s and AtBMI1s, are major subunits that directly catalyze H2A monoubiquitination (H2Aub). The mutants Atring1a Atring1b and Atbmi1a Atbmi1b show ectopic expression of seed-maturation genes and indeterminate embryonic traits at the seedling stage (Bratzel et al., 2010; Chen D. et al., 2010; Yang et al., 2013). Moreover, mutations of EMF1 or EMF2, two PcG proteins, can strengthen the phenotype of Atbmi1a Atbmi1b and expression of the seed maturation genes increased obviously in the emf1 mutant, suggesting these regulators may collaborate in repression of the maturation program after germination (Moon et al., 2003; Kim et al., 2012; Yang et al., 2013). VAL proteins are B3-type transcription factors that interact with AtBMI1 proteins. val1/2 seedlings display phenotypic defects similar to those of Atbmi1a/b/c mutants, accompanied by strongly reduced H2Aub levels at seed-maturation genes and concomitant derepressed gene transcription (Suzuki et al., 2007; Yang et al., 2013). On the other hand, the levels of H3K27me3 at LEC1, FUS3, and ABI3 are also strongly decreased in val1/2 and Atbmi1a/b/c mutants (Yang et al., 2013). VAL proteins can recruit the PRC2 subunit CLF and promote the placement of H3K27me3 on target loci (Chen N. et al., 2020; Yuan et al., 2021), indicative of genetic and physical interaction between the PRC1 and PRC2 complexes.
The Arabidopsis PRC2 complex, which catalyzes H3K27me3 addition, represses seed maturation genes, as evidenced by somatic embryo emergence in vegetative tissues of double mutants deficient in redundant PRC2 subunits, i.e., CLF and SWN or EMF2 and VERNALIZATION2 (VRN2) (Chanvivattana et al., 2004; Schubert et al., 2005; Makarevich et al., 2006; Yang et al., 2013). A single mutant of Arabidopsis FIE also gives rise to the degeneration of vegetative cells into neoplastic, callus-like structures in seedlings with abolished H3K27me3 deposition (Kinoshita et al., 2001; Bouyer et al., 2011). It should be noted that EMF2 or FIE are mainly worked in repressing flower formation upon germination, though discussion of this function is outside the scope of this manuscript (Kinoshita et al., 2001; Moon et al., 2003). Furthermore, the chromatin remodeler PKL, which has a potential role in the retention of H3K27me3, acts throughout the seedling, repressing embryonic traits. Loss of PKL function reduces levels of H3K27me3 and ectopic expression of the embryo-specific genes LEC1, LEC2, and FUS3, resulting in seedlings with swollen primary roots, referred to as pickle roots (Ogas et al., 1997, 1999; Dean Rider et al., 2003; Henderson et al., 2004; Li et al., 2005; Carter et al., 2018).
Therefore, the PRC1 and PRC2 repressive system are tightly integrated in the transition from seed to seedling. Moreover, many other chromatin regulators also participate in this process, and some show crosstalk with the PcG working program.
In Arabidopsis, trithorax group (trxG) proteins catalyze H3K4 methylation, which play roles opposite to that of H3K27me3. Correspondingly, the trxG members (ATX1) and ULTRAPETALA1 (ULT1) counteract the effect of CLF in floral repression (Carles and Fletcher, 2009; Alvarez-Venegas, 2010). However, removal of either or both ATX1 and ULT1 fails to rescue the defects exhibited by an emf1 mutant, but promote H3K27me3, causing a swollen, pickle-like root phenotype in seedlings of emf1 atx1 ult1 triple mutants. Yeast two-hybrid assays reveal that ULT1 physically interacts with ATX1 and EMF1, and both ATX1 and ULT1 are able to bind the chromatin of seed genes, including LEC2 and ABI3 (Xu et al., 2018). This suggests a new, more complex framework whereby trxG acts in concert with PcG to maintain chromatin integrity and prevent seed maturation gene expression after germination. A mutation of SDG8/EFS, encoding an HMT that mainly mediates H3K36 methylation, acts synergistically with emf2 to induce the deposition of the active mark H3K4me3 on seed-maturation loci, leading to the emergence of embryonic traits (Tang et al., 2012). However, the mechanism and the specific pathway whereby the activating H3K4me3 marks are deposited in the sdg8 emf2 double mutant is still unclear.
Arrested growth and the formation of embryo-like structures on vegetative tissues can also be observed in a hda6 hda19 double mutant. Moreover, the disturbed cell fate seen in the hda6 mutant upon treatment with the HDAC inhibitor trichostatin A (TSA) is rescued by mutations of lec1, fus3, and abi3, indicating that acetylation also has effects at the seedling development stage (Tanaka et al., 2008). In animals, CHD3 chromatin remodelers are components of RPD3-containing HDAC complexes (Tong et al., 1998; Zhang et al., 1998; Fukaki et al., 2006). Therefore, whether there are connections between the HDACs and the CHD3 protein PKL that influence the repression of embryonic properties is a question in need of further study.
As plants progress from seed development through seedling establishment, gene expression is dynamically affected by histone modifications and chromatin states. These epigenetic regulations include a combination of synergistic and antagonistic crosstalk between histone-modifying enzymes through specific connecting factors. By screening and analyzing regulators in the nodes of the epigenetic network, it is possible to uncover the comprehensive changes in the epigenetic modifications of pivotal genes during development.
Additionally, loss of function of epigenetic regulatory genes often gives rise to pleiotropic effects, with changes in chromatin state at the whole-genome level. To some extent, these extensive effects are mediated by various specific co-regulators that participate in diverse biological processes. Further studies to identify the working partners of epigenetic regulatory proteins will thus provide further information about the processes governing specific pathways at the epigenetic level.
Furthermore, gene transcription regulation mediated by histone modifiers or chromatin remodelers might be the later step in the cascades of environmental signal transition. Specific knowledge about how the epigenetic regulators receive these signals needs to be uncovered.
Finally, investigations to date of the effects of histone modification and chromatin remodeling on seed developmental programs have mainly focused on the model plant Arabidopsis, with few studies performed in commercial species, such as grain, fruit, and vegetable crop species. Therefore, research on classical epigenetic regulatory pathways and associated components in model plants need to be extended to other, economically important species to accelerate its application to molecular breeding for agricultural production. Moreover, better understanding of the conserved and diverse regulatory mechanisms acting in different plant species will enhance knowledge of the complex epigenetic regulatory mechanisms controlling this process.
WJ: writing-original draft. YX, XD, and XJ: review and editing. YX: supervision. All authors contributed to the article and approved the submitted version.
This research was supported by the Key-Area Research and Development Program of Guangdong Province (Grant No. 2021B0707010006), China Postdoctoral Science Foundation (Grant No. 2019M650914), The Science, Technology and Innovation Commission of Shenzhen Municipality (Grant Nos. KCXFZ20201221173203009 and JCYJ20200109150713553), and Dapeng District Industry Development Special Funds (Grant Nos. KJYF202101-09 and RCTD20180102).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We apologize to those whose work was not cited because of space constrains.
Agarwal, P., Kapoor, S., and Tyagi, A. K. (2011). Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 33, 189–202. doi: 10.1002/bies.201000107
Alonso-Blanco, C., Bentsink, L., Hanhart, C. J., Blankestijn-de Vries, H., and Koornneef, M. (2003). Analysis of natural allelic variation at seed dormancy loci of Arabidopsis thaliana. Genetics 164, 711–729. doi: 10.1093/genetics/164.2.711
Alvarez-Venegas, R. (2010). Regulation by polycomb and trithorax group proteins in Arabidopsis. Arabidopsis Book 8:e0128. doi: 10.1199/tab.0128
Baek, D., Shin, G., Kim, M. C., Shen, M., Lee, S. Y., and Yun, D. J. (2020). Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant. Sci. 11:143. doi: 10.3389/fpls.2020.00143
Baskin, C. C., and Baskin, J. M. (2006). The natural history of soil seed banks of arable land. Weed Sci. 54, 549–557. doi: 10.1614/WS-05-034R.1
Bentsink, L., Jowett, J., Hanhart, C. J., and Koornneef, M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 17042–17047. doi: 10.1073/pnas.0607877103
Berger, S. L. (2007). The complex language of chromatin regulation during transcription. Nature 447, 407–412. doi: 10.1038/nature05915
Bouyer, D., Roudier, F., Heese, M., Andersen, E. D., Gey, D., Nowack, M. K., et al. (2011). Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 7:e1002014. doi: 10.1371/journal.pgen.1002014
Bratzel, F., López-Torrejón, G., Koch, M., Del Pozo, J. C., and Calonje, M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20, 1853–1859. doi: 10.1016/j.cub.2010.09.046
Braybrook, S. A., Stone, S. L., Park, S., Bui, A. Q., Le, B. H., Fischer, R. L., et al. (2006). Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 3468–3473. doi: 10.1073/pnas.0511331103
Carles, C. C., and Fletcher, J. C. (2009). The SAND domain protein ULTRAPETALA1 acts as a trithorax group factor to regulate cell fate in plants. Genes Dev. 23, 2723–2728. doi: 10.1101/gad.1812609
Carrillo-Barral, N., Rodríguez-Gacio, M. D. C., and Matilla, A. J. (2020). Delay of germination-1 (DOG1): a key to understanding seed dormancy. Plants (Basel) 9:480. doi: 10.3390/plants9040480
Carter, B., Bishop, B., Ho, K. K., Huang, R., Jia, W., Zhang, H., et al. (2018). The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell 30, 1337–1352. doi: 10.1105/tpc.17.00867
Chang, G., Wang, C., Kong, X., Chen, Q., Yang, Y., and Hu, X. (2018). AFP2 as the novel regulator breaks high-temperature-induced seeds secondary dormancy through ABI5 and SOM in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 501, 232–238. doi: 10.1016/j.bbrc.2018.04.222
Chanvivattana, Y., Bishopp, A., Schubert, D., Stock, C., Moon, Y. H., Sung, Z. R., et al. (2004). Interaction of polycomb-group proteins controlling flowering in Arabidopsis. Development 131, 5263–5276. doi: 10.1242/dev.01400
Chaudhury, A. M., Ming, L., Miller, C., Craig, S., Dennis, E. S., and Peacock, W. J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94, 4223–4228. doi: 10.1073/pnas.94.8.4223
Chen, D., Molitor, A., Liu, C., and Shen, W. H. (2010). The Arabidopsis PRC1-like ring-finger proteins are necessary for repression of embryonic traits during vegetative growth. Cell Res. 20, 1332–1344. doi: 10.1038/cr.2010.151
Chen, H., Tong, J., Fu, W., Liang, Z., Ruan, J., Yu, Y., et al. (2020). The H3K27me3 demethylase RELATIVE OF EARLY FLOWERING6 suppresses seed dormancy by inducing abscisic acid catabolism. Plant Physiol. 184, 1969–1978. doi: 10.1104/pp.20.01255
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, L. T., and Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Chen, N., Wang, H., Abdelmageed, H., Veerappan, V., Tadege, M., and Allen, R. D. (2020). HSI2/VAL1 and HSL1/VAL2 function redundantly to repress DOG1 expression in Arabidopsis seeds and seedlings. New Phytol. 227, 840–856. doi: 10.1111/nph.16559
Cheng, L., Shafiq, S., Xu, W., and Sun, Q. (2018). EARLY FLOWERING IN SHORT DAYS (EFS) regulates the seed size in Arabidopsis. Sci. China Life Sci. 61, 214–224. doi: 10.1007/s11427-017-9236-x
Cheng, X., Pan, M., E, Z., Zhou, Y., Niu, B., and Chen, C. (2020). Functional divergence of two duplicated fertilization independent endosperm genes in rice with respect to seed development. Plant J. 104, 124–137. doi: 10.1111/tpj.14911
Cheng, X., Pan, M., E, Z., Zhou, Y., Niu, B., and Chen, C. (2021). The maternally expressed polycomb group gene OsEMF2a is essential for endosperm cellularization and imprinting in rice. Plant Commun. 2:100092. doi: 10.1016/j.xplc.2020.100092
Cho, J. N., Ryu, J. Y., Jeong, Y. M., Park, J., Song, J. J., Amasino, R. M., et al. (2012). Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 22, 736–748. doi: 10.1016/j.devcel.2012.01.024
Cigliano, R. A., Cremona, G., Paparo, R., Termolino, P., Perrella, G., Gutzat, R., et al. (2013). Histone deacetylase AtHDA7 is required for female gametophyte and embryo development in Arabidopsis. Plant Physiol. 163, 431–440. doi: 10.1104/pp.113.221713
Colville, A., Alhattab, R., Hu, M., Labbé, H., Xing, T., and Miki, B. (2011). Role of HD2 genes in seed germination and early seedling growth in Arabidopsis. Plant Cell Rep. 30, 1969–1979. doi: 10.1007/s00299-011-1105-z
Danilevskaya, O. N., Hermon, P., Hantke, S., Muszynski, M. G., Kollipara, K., and Ananiev, E. V. (2003). Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15, 425–438. doi: 10.1105/tpc.006759
Dean Rider, S. Jr., Henderson, J. T., Jerome, R. E., Edenberg, H. J., Romero-Severson, J., and Ogas, J. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35, 33–43. doi: 10.1046/j.1365-313x.2003.01783.x
Dutnall, R. N. (2003). Cracking the histone code: one, two, three methyls, you’re out! Mol. Cell 12, 3–4. doi: 10.1016/s1097-2765(03)00282-x
Fedak, H., Palusinska, M., Krzyczmonik, K., Brzezniak, L., Yatusevich, R., Pietras, Z., et al. (2016). Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc. Natl. Acad. Sci U.S.A. 113, 7846–7855. doi: 10.1073/pnas.1608827113
Figueiredo, D. D., Batista, R. A., Roszak, P. J., and Köhler, C. (2015). Auxin production couples endosperm development to fertilization. Nat. Plants 1:15184. doi: 10.1038/nplants.2015.184
Figueiredo, D. D., and Köhler, C. (2018). Auxin: a molecular trigger of seed development. Genes. Dev. 32, 479–490. doi: 10.1101/gad.312546.118
Finch-Savage, W. E., and Footitt, S. (2017). Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 68, 843–856. doi: 10.1093/jxb/erw477
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Fiorucci, A. S., Bourbousse, C., Concia, L., Rougée, M., Deton-Cabanillas, A. F., Zabulon, G., et al. (2019). Arabidopsis S2Lb links AtCOMPASS-like and SDG2 activity in H3K4me3 independently from histone H2B monoubiquitination. Genome Biol. 20:100. doi: 10.1186/s13059-019-1705-4
Fletcher, J. C. (2017). State of the art: trxG factor regulation of post-embryonic plant development. Front. Plant. Sci. 8:1925. doi: 10.3389/fpls.2017.01925
Folsom, J. J., Begcy, K., Hao, X., Wang, D., and Walia, H. (2014). Rice fertilization-independent endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165, 238–248. doi: 10.1104/pp.113.232413
Footitt, S., Muller, K., Kermode, A. R., and Finch-Savage, W. E. (2015). Seed dormancy cycling in Arabidopsis: chromatin remodelling and regulation of DOG1 in response to seasonal environmental signals. Plant J. 81, 413–425. doi: 10.1111/tpj.12735
Forestan, C., Farinati, S., Rouster, J., Lassagne, H., Lauria, M., Dal Ferro, N., et al. (2018). Control of maize vegetative and reproductive development, fertility, and rRNAs silencing by HISTONE DEACETYLASE 108. Genetics 208, 1443–1466. doi: 10.1534/genetics.117.300625
Fukaki, H., Taniguchi, N., and Tasaka, M. (2006). PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J. 48, 380–389. doi: 10.1111/j.1365-313X.2006.02882.x
Gazzarrini, S., Tsuchiya, Y., Lumba, S., Okamoto, M., and McCourt, P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385. doi: 10.1016/j.devcel.2004.06.017
Giraudat, J., Hauge, B. M., Valon, C., Smalle, J., Parcy, F., and Goodman, H. M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. doi: 10.1105/tpc.4.10.1251
Goldberg, A. D., Allis, C. D., and Bernstein, E. (2007). Epigenetics: a landscape takes shape. Cell 128, 635–638. doi: 10.1016/j.cell.2007.02.006
Graeber, K., Voegele, A., Büttner-Mainik, A., Sperber, K., Mummenhoff, K., and Leubner-Metzger, G. (2013). Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium delay of germination1 genes. Plant Physiol. 161, 1903–1917. doi: 10.1104/pp.112.213298
Grossniklaus, U., Vielle-Calzada, J. P., Hoeppner, M. A., and Gagliano, W. B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. doi: 10.1126/science.280.5362.446
Gu, D., Chen, C. Y., Zhao, M., Zhao, L., Duan, X., Duan, J., et al. (2017). Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic. Acids Res. 45, 7137–7150. doi: 10.1093/nar/gkx283
Gu, D., Ji, R., He, C., Peng, T., Zhang, M., Duan, J., et al. (2019). Arabidopsis histone methyltransferase SUVH5 is a positive regulator of light-mediated seed germination. Front. Plant. Sci. 10:841. doi: 10.3389/fpls.2019.00841
Han, S. K., Sang, Y., Rodrigues, A., Wu, M. F., Rodriguez, P. L., and Wagner, D. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24, 4892–4906. doi: 10.1105/tpc.112.105114
Hargreaves, D. C., and Crabtree, G. R. (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420. doi: 10.1038/cr.2011.32
Hehenberger, E., Kradolfer, D., and Köhler, C. (2012). Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039. doi: 10.1242/dev.077057
Henderson, J. T., Li, H. C., Rider, S. D., Mordhorst, A. P., Romero-Severson, J., Cheng, J. C., et al. (2004). PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol. 134, 995–1005. doi: 10.1104/pp.103.030148
Leeggangers, H. A., Folta, A., Muras, A., Nap, J. P., and Mlynarova, L. (2015). Reduced seed germination in Arabidopsis over-expressing SWI/SNF2 ATPase genes. Physiol. Plant 153, 318–326. doi: 10.1111/ppl.12231
Holdsworth, M. J., Bentsink, L., and Soppe, W. J. J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179, 33–54. doi: 10.1111/j.1469-8137.2008.02437.x
Huang, X., Lu, Z., Wang, X., Ouyang, Y., Chen, W., Xie, K., et al. (2016). Imprinted gene OsFIE1 modulates rice seed development by influencing nutrient metabolism and modifying genome H3K27me3. Plant J. 87, 305–317. doi: 10.1111/tpj.13202
Hurtado, L., Farrona, S., and Reyes, J. C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62, 291–304. doi: 10.1007/s11103-006-9021-2
Ishikawa, R., Ohnishi, T., Kinoshita, Y., Eiguchi, M., Kurata, N., and Kinoshita, T. (2011). Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J. 65, 798–806. doi: 10.1111/j.1365-313X.2010.04466.x
Jacobsen, J. V., Pearce, D. W., Poole, A. T., Pharis, R. P., and Mander, L. N. (2002). Abscisic acid, phaseic acid and gibberellin contents associated with dormancy and germination in barley. Physiol. Plant 115, 428–441. doi: 10.1034/j.1399-3054.2002.1150313.x
Jia, H., Suzuki, M., and McCarty, D. R. (2014). Regulation of the seed to seedling developmental phase transition by the LAFL and VAL transcription factor networks. Wiley Interdiscip. Rev. Dev. Biol. 3, 135–145. doi: 10.1002/wdev.126
Jiang, D., Gu, X., and He, Y. (2009). Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21, 1733–1746. doi: 10.1105/tpc.109.067967
Jiang, D., Kong, N. C., Gu, X., Li, Z., and He, Y. (2011). Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 7:e1001330. doi: 10.1371/journal.pgen.1001330
Kang, I. H., Steffen, J. G., Portereiko, M. F., Lloyd, A., and Drews, G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20, 635–647. doi: 10.1105/tpc.107.055137
Keith, K., Kraml, M., Dengler, N. G., and McCourt, P. (1994). fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6, 589–600. doi: 10.1105/tpc.6.5.589
Kim, D. H., Yamaguchi, S., Lim, S., Oh, E., Park, J., Hanada, A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20, 1260–1277. doi: 10.1105/tpc.108.058859
Kim, S. Y., Lee, J., Eshed-Williams, L., Zilberman, D., and Sung, Z. R. (2012). EMF1 and PRC2 cooperate to repress key regulators of Arabidopsis development. PLoS Genet. 8:e1002512. doi: 10.1371/journal.pgen.1002512
Kinoshita, T., Harada, J. J., Goldberg, R. B., and Fischer, R. L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. U.S.A. 98, 14156–14161. doi: 10.1073/pnas.241507798
Kiyosue, T., Ohad, N., Yadegari, R., Hannon, M., Dinneny, J., Wells, D., et al. (1999). Control of fertilization-independent endosperm development by the MEDEA polycomb gene in Arabidopsis. Proc. Natl. Acad. Sci U.S.A. 96, 4186–4191. doi: 10.1073/pnas.96.7.4186
Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003b). The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. doi: 10.1101/gad.257403
Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003a). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO. J. 22, 4804–4814. doi: 10.1093/emboj/cdg444
Kradolfer, D., Wolff, P., Jiang, H., Siretskiy, A., and Köhler, C. (2013). An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev. Cell 26, 525–535. doi: 10.1016/j.devcel.2013.08.006
Kumar, V., Thakur, J. K., and Prasad, M. (2021). Histone acetylation dynamics regulating plant development and stress responses. Cell Mol. Life Sci. 78, 4467–4486. doi: 10.1007/s00018-021-03794-x
Lee, N., Kang, H., Lee, D., and Choi, G. (2014). A histone methyltransferase inhibits seed germination by increasing PIF1 mRNA expression in imbibed seeds. Plant J. 78, 282–293. doi: 10.1111/tpj.12467
Lepiniec, L., Devic, M., Roscoe, T. J., Bouyer, D., Zhou, D. X., Boulard, C., et al. (2018). Molecular and epigenetic regulations and functions of the LAFL transcriptional regulators that control seed development. Plant Reprod. 31, 291–307. doi: 10.1007/s00497-018-0337-2
Li, C., Gu, L., Gao, L., Chen, C., Wei, C. Q., Qiu, Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Li, H. C., Chuang, K., Henderson, J. T., Rider, S. D. Jr., Bai, Y., Zhang, H., et al. (2005). PICKLE acts during germination to repress expression of embryonic traits. Plant J. 44, 1010–1022. doi: 10.1111/j.1365-313X.2005.02602.x
Li, P., Zhang, Q., He, D., Zhou, Y., Ni, H., Tian, D., et al. (2020). AGAMOUS-LIKE67 cooperates with the histone mark reader EBS to modulate seed germination under high temperature. Plant Physiol. 184, 529–545. doi: 10.1104/pp.20.00056
Li, S., Zhou, B., Peng, X., Kuang, Q., Huang, X., Yao, J., et al. (2014). OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 201, 66–79. doi: 10.1111/nph.12472
Li, X., Jiang, Y., Ji, Z., Liu, Y., and Zhang, Q. (2015). BRHIS1 suppresses rice innate immunity through binding to monoubiquitinated H2A and H2B variants. EMBO Rep. 16, 1192–1202. doi: 10.15252/embr.201440000
Lim, S., Park, J., Lee, N., Jeong, J., Toh, S., Watanabe, A., et al. (2013). ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. doi: 10.1105/tpc.113.118604
Lin, J. Y., Le, B. H., Chen, M., Henry, K. F., Hur, J., Hsieh, T. F., et al. (2017). Similarity between soybean and Arabidopsis seed methylomes and loss of non-CG methylation does not affect seed development. Proc. Natl. Acad. Sci U.S.A. 114, E9730–E9739. doi: 10.1073/pnas.1716758114
Liu, B., Liu, Y., Wang, B., Luo, Q., Shi, J., Gan, J., et al. (2019). The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment. Nat. Commun. 10:2999. doi: 10.1038/s41467-019-10850-5
Liu, J., Deng, S., Wang, H., Ye, J., Wu, H. W., Sun, H. X., et al. (2016). CURLY LEAF regulates gene sets coordinating seed size and lipid biosynthesis. Plant Physiol. 171, 424–436. doi: 10.1104/pp.15.01335
Liu, X., Luo, J., Li, T., Yang, H., Wang, P., Su, L., et al. (2021). SDG711 is involved in rice seed development through regulation of starch metabolism gene expression in coordination with other histone modifications. Rice (N Y) 14:25. doi: 10.1186/s12284-021-00467-y
Liu, X., Wei, X., Sheng, Z., Jiao, G., Tang, S., Luo, J., et al. (2016). Polycomb protein OsFIE2 affects plant height and grain yield in rice. PLoS One 11:e0164748. doi: 10.1371/journal.pone.0164748
Liu, Y., Geyer, R., van Zanten, M., Carles, A., Li, Y., Hörold, A., et al. (2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One 6:e22241. doi: 10.1371/journal.pone.0022241
Liu, Y., Koornneef, M., and Soppe, W. J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19, 433–444. doi: 10.1105/tpc.106.049221
Lotan, T., Ohto, M., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/s0092-8674(00)81463-4
Lu, X., Wang, W., Ren, W., Chai, Z., Guo, W., Chen, R., et al. (2015). Genome-wide epigenetic regulation of gene transcription in maize seeds. PLoS One 10:e0139582. doi: 10.1371/journal.pone.0139582
Luo, M., Platten, D., Chaudhury, A., Peacock, W. J., and Dennis, E. S. (2009). Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol. Plant 2, 711–723. doi: 10.1093/mp/ssp036
Luo, M., Wang, Y. Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Luo, Q., Lian, H. L., He, S. B., Li, L., Jia, K. P., and Yang, H. Q. (2014). COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell 26, 2441–2456. doi: 10.1105/tpc.113.121657
Makarevich, G., Leroy, O., Akinci, U., Schubert, D., Clarenz, O., Goodrich, J., et al. (2006). Different polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7, 947–952. doi: 10.1038/sj.embor.7400760
Miller, T., Krogan, N. J., Dover, J., Erdjument-Bromage, H., Tempst, P., Johnston, M., et al. (2001). COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci U.S.A. 98, 12902–12907. doi: 10.1073/pnas.231473398
Moon, Y. H., Chen, L., Pan, R. L., Chang, H. S., Zhu, T., Maffeo, D. M., et al. (2003). EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15, 681–693. doi: 10.1105/tpc.007831
Mozgova, I., and Hennig, L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Müller, K., Bouyer, D., Schnittger, A., and Kermode, A. R. (2012). Evolutionarily conserved histone methylation dynamics during seed life-cycle transitions. PLoS One 7:e51532. doi: 10.1371/journal.pone.0051532
Nakabayashi, K., Bartsch, M., Xiang, Y., Miatton, E., Pellengahr, S., Yano, R., et al. (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24, 2826–2838. doi: 10.1105/tpc.112.100214
Nallamilli, B. R., Zhang, J., Mujahid, H., Malone, B. M., Bridges, S. M., and Peng, Z. (2013). Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet. 9:e1003322. doi: 10.1371/journal.pgen.1003322
Nambara, E., Hayama, R., Tsuchiya, Y., Nishimura, M., Kawaide, H., Kamiya, Y., et al. (2000). The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 220, 412–423. doi: 10.1006/dbio.2000.9632
Narro-Diego, L., López-González, L., Jarillo, J. A., and Piñeiro, M. (2017). The PHD-containing protein EARLY BOLTING IN SHORT DAYS regulates seed dormancy in Arabidopsis. Plant Cell Environ. 40, 2393–2405. doi: 10.1111/pce.13046
Née, G., Xiang, Y., and Soppe, W. J. (2017). The release of dormancy, a wake-up call for seeds to germinate. Curr. Opin. Plant Biol. 35, 8–14. doi: 10.1016/j.pbi.2016.09.002
Ogas, J., Cheng, J. C., Sung, Z. R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94. doi: 10.1126/science.277.5322.91
Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci U.S.A. 96, 13839–13844. doi: 10.1073/pnas.96.24.13839
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. doi: 10.1105/tpc.107.050153
Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W. I., and Choi, G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139. doi: 10.1111/j.1365-313X.2006.02773.x
Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J. J., et al. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416. doi: 10.1105/tpc.11.3.407
Olsen, O. A. (2004). Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16(Suppl), S214–S227. doi: 10.1105/tpc.017111
Parcy, F., and Giraudat, J. (1997). Interactions between the ABI1 and the ectopically expressed ABI3 genes in controlling abscisic acid responses in Arabidopsis vegetative tissues. Plant J. 11, 693–702. doi: 10.1046/j.1365-313x.1997.11040693.x
Park, J., Lee, N., Kim, W., Lim, S., and Choi, G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23, 1404–1415. doi: 10.1105/tpc.110.080721
Perrella, G., Lopez-Vernaza, M. A., Carr, C., Sani, E., Gosselé, V., Verduyn, C., et al. (2013). Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell 25, 3491–3505. doi: 10.1105/tpc.113.114835
Rodrigues, J. C., Luo, M., Berger, F., and Koltunow, A. M. (2010). Polycomb group gene function in sexual and asexual seed development in angiosperms. Sex Plant Reprod. 23, 123–133. doi: 10.1007/s00497-009-0131-2
Rossi, V., Varotto, S., Locatelli, S., Lanzanova, C., Lauria, M., Zanotti, E., et al. (2001). The maize WD-repeat gene ZmRbAp1 encodes a member of the MSI/RbAp sub-family and is differentially expressed during endosperm development. Mol. Genet. Genomics 265, 576–584. doi: 10.1007/s004380100461
Saez, A., Rodrigues, A., Santiago, J., Rubio, S., and Rodriguez, P. L. (2008). HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20, 2972–2988. doi: 10.1105/tpc.107.056705
Sarnowski, T. J., Ríos, G., Jásik, J., Swiezewski, S., Kaczanowski, S., Li, Y., et al. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17, 2454–2472. doi: 10.1105/tpc.105.031203
Schubert, D., Clarenz, O., and Goodrich, J. (2005). Epigenetic control of plant development by polycomb-group proteins. Curr. Opin. Plant Biol. 8, 553–561. doi: 10.1016/j.pbi.2005.07.005
Sekine, D., Ohnishi, T., Furuumi, H., Ono, A., Yamada, T., Kurata, N., et al. (2013). Dissection of two major components of the post-zygotic hybridization barrier in rice endosperm. Plant J. 76, 792–799. doi: 10.1111/tpj.12333
Seo, M., Nambara, E., Choi, G., and Yamaguchi, S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69, 463–472. doi: 10.1007/s11103-008-9429-y
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Song, X. J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 76–81. doi: 10.1073/pnas.1421127112
Sørensen, M. B., Chaudhury, A. M., Robert, H., Bancharel, E., and Berger, F. (2001). Polycomb group genes control pattern formation in plant seed. Curr. Biol. 11, 277–281. doi: 10.1016/s0960-9822(01)00072-0
Spillane, C., Schmid, K. J., Laoueillé-Duprat, S., Pien, S., Escobar-Restrepo, J. M., Baroux, C., et al. (2007). Positive darwinian selection at the imprinted MEDEA locus in plants. Nature 448, 349–352. doi: 10.1038/nature05984
Springer, N. M., Danilevskaya, O. N., Hermon, P., Helentjaris, T. G., Phillips, R. L., Kaeppler, H. F., et al. (2002). Sequence relationships, conserved domains, and expression patterns for maize homologs of the polycomb group genes E(z), esc, and E(Pc). Plant Physiol. 128, 1332–1345. doi: 10.1104/pp.010742
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci U.S.A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Strahl, B. D., and Allis, C. D. (2000). The language of covalent histone modifications. Nature 403, 41–45. doi: 10.1038/47412
Sun, Z. W., and Allis, C. D. (2002). Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108. doi: 10.1038/nature00883
Suzuki, M., Wang, H. H., and McCarty, D. R. (2007). Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143, 902–911. doi: 10.1104/pp.106.092320
Sweeney, M., and McCouch, S. (2007). The complex history of the domestication of rice. Ann. Bot. 100, 951–957. doi: 10.1093/aob/mcm128
Tanaka, M., Kikuchi, A., and Kamada, H. (2008). The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 146, 149–161. doi: 10.1104/pp.107.111674
Tang, X., Lim, M. H., Pelletier, J., Tang, M., Nguyen, V., Keller, W. A., et al. (2012). Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J. Exp. Bot. 63, 1391–1404. doi: 10.1093/jxb/err383
Toh, S., Imamura, A., Watanabe, A., Nakabayashi, K., Okamoto, M., Jikumaru, Y., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146, 1368–1385. doi: 10.1104/pp.107.113738
Tong, J. K., Hassig, C. A., Schnitzler, G. R., Kingston, R. E., and Schreiber, S. L. (1998). Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature 395, 917–921. doi: 10.1038/27699
Tonosaki, K., Ono, A., Kunisada, M., Nishino, M., Nagata, H., Sakamoto, S., et al. (2021). Mutation of the imprinted gene OsEMF2a induces autonomous endosperm development and delayed cellularization in rice. Plant Cell 33, 85–103. doi: 10.1093/plcell/koaa006
Turner, B. M. (2000). Histone acetylation and an epigenetic code. Bioessays 22, 836–845. doi: 10.1002/1521-1878(200009)22:9<836::Aid-bies9<3.0.Co;2-x
Vaistij, F. E., Barros-Galvão, T., Cole, A. F., Gilday, A. D., He, Z., Li, Y., et al. (2018). MOTHER-OF-FT-AND-TFL1 represses seed germination under far-red light by modulating phytohormone responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 115, 8442–8447. doi: 10.1073/pnas.1806460115
Vaistij, F. E., Gan, Y., Penfield, S., Gilday, A. D., Dave, A., He, Z., et al. (2013). Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc. Natl. Acad. Sci U.S.A. 110, 10866–10871. doi: 10.1073/pnas.1301647110
van Zanten, M., Koini, M. A., Geyer, R., Liu, Y., Brambilla, V., Bartels, D., et al. (2011). Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation. Proc. Natl. Acad. Sci U.S.A. 108, 20219–20224. doi: 10.1073/pnas.1117726108
Walck, J. L., Hidayati, S. N., Dixon, K. W., Thompson, K. A., and Poschlod, P. (2011). Climate change and plant regeneration from seed. Glob. Change Biol. 17, 2145–2161. doi: 10.1111/j.1365-2486.2010.02368.x
Walia, H., Josefsson, C., Dilkes, B., Kirkbride, R., Harada, J., and Comai, L. (2009). Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr. Biol. 19, 1128–1132. doi: 10.1016/j.cub.2009.05.068
Wang, D., Tyson, M. D., Jackson, S. S., and Yadegari, R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci U.S.A. 103, 13244–13249. doi: 10.1073/pnas.0605551103
Wang, T. J., Huang, S., Zhang, A., Guo, P., Liu, Y., Xu, C., et al. (2021). JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol. 230, 567–584. doi: 10.1111/nph.17177
West, M., Yee, K. M., Danao, J., Zimmerman, J. L., Fischer, R. L., Goldberg, R. B., et al. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6, 1731–1745. doi: 10.1105/tpc.6.12.1731
Wu, K., Tian, L., Malik, K., Brown, D., and Miki, B. (2000). Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 22, 19–27. doi: 10.1046/j.1365-313x.2000.00711.x
Xu, F., Kuo, T., Rosli, Y., Liu, M. S., Wu, L., Chen, L. O., et al. (2018). Trithorax group proteins act together with a polycomb group protein to maintain chromatin integrity for epigenetic silencing during seed germination in Arabidopsis. Mol. Plant 11, 659–677. doi: 10.1016/j.molp.2018.01.010
Yadegari, R., Kinoshita, T., Lotan, O., Cohen, G., Katz, A., Choi, Y., et al. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12, 2367–2382. doi: 10.1105/tpc.12.12.2367
Yamamoto, A., Kagaya, Y., Toyoshima, R., Kagaya, M., Takeda, S., and Hattori, T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58, 843–856. doi: 10.1111/j.1365-313X.2009.03817.x
Yang, C., Bratzel, F., Hohmann, N., Koch, M., Turck, F., and Calonje, M. (2013). VAL- and AtBMI1-mediated H2Aub initiate the switch from embryonic to postgerminative growth in Arabidopsis. Curr. Biol. 23, 1324–1329. doi: 10.1016/j.cub.2013.05.050
Yang, W., Chen, Z., Huang, Y., Chang, G., Li, P., Wei, J., et al. (2019). Powerdress as the novel regulator enhances Arabidopsis seeds germination tolerance to high temperature stress by histone modification of SOM locus. Plant Sci. 284, 91–98. doi: 10.1016/j.plantsci.2019.04.001
Yuan, L., Song, X., Zhang, L., Yu, Y., Liang, Z., Lei, Y., et al. (2021). The transcriptional repressors VAL1 and VAL2 recruit PRC2 for genome-wide Polycomb silencing in Arabidopsis. Nucleic Acids Res. 49, 98–113. doi: 10.1093/nar/gkaa1129
Zanten, M.v, Liu, Y.-x, and Soppe, W. J. J. (2013). “Epigenetic signalling during the life of seeds,” in Epigenetic Memory and Control in Plants, eds G. Grafi and N. Ohad (Berlin: Springer), 127–153. doi: 10.1007/978-3-642-35227-0_7
Zha, P., Liu, S., Li, Y., Ma, T., Yang, L., Jing, Y., et al. (2020). The evening complex and the chromatin-remodeling factor PICKLE coordinately control seed dormancy by directly repressing DOG1 in Arabidopsis. Plant Commun. 1:100011. doi: 10.1016/j.xplc.2019.100011
Zhang, H., Bishop, B., Ringenberg, W., Muir, W. M., and Ogas, J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159, 418–432. doi: 10.1104/pp.112.194878
Zhang, H., Lu, Y., Zhao, Y., and Zhou, D. X. (2016). OsSRT1 is involved in rice seed development through regulation of starch metabolism gene expression. Plant Sci. 248, 28–36. doi: 10.1016/j.plantsci.2016.04.004
Zhang, H., Rider, S. D. Jr., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V., et al. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 28, 22637–22648. doi: 10.1074/jbc.M802129200
Zhang, S., Wang, D., Zhang, H., Skaggs, M. I., Lloyd, A., Ran, D., et al. (2018). FERTILIZATION-INDEPENDENT SEED-Polycomb repressive complex 2 plays a dual role in regulating type I MADS-Box genes in early endosperm development. Plant Physiol. 177, 285–299. doi: 10.1104/pp.17.00534
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10:R62. doi: 10.1186/gb-2009-10-6-r62
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007a). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., and Jacobsen, S. E. (2007b). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/nsmb1283
Zhang, Y., LeRoy, G., Seelig, H. P., Lane, W. S., and Reinberg, D. (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95, 279–289. doi: 10.1016/s0092-8674(00)81758-4
Zhao, J., Li, M., Gu, D., Liu, X., Zhang, J., Wu, K., et al. (2016). Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 470, 439–444. doi: 10.1016/j.bbrc.2016.01.016
Zhao, J., Zhang, J., Zhang, W., Wu, K., Zheng, F., Tian, L., et al. (2014). Expression and functional analysis of the plant-specific histone deacetylase HDT701 in rice. Front. Plant. Sci. 5:764. doi: 10.3389/fpls.2014.00764
Zhao, M., Yang, S., Liu, X., and Wu, K. (2015). Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating DELAY OF GERMINATION 1 and ABA signaling-related genes. Front. Plant. Sci. 6:159. doi: 10.3389/fpls.2015.00159
Zheng, J., Chen, F., Wang, Z., Cao, H., Li, X., Deng, X., et al. (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 193, 605–616. doi: 10.1111/j.1469-8137.2011.03969.x
Zhou, S. R., Yin, L. L., and Xue, H. W. (2013). Functional genomics based understanding of rice endosperm development. Curr. Opin. Plant Biol. 16, 236–246. doi: 10.1016/j.pbi.2013.03.001
Zhou, Y., Yang, P., Zhang, F., Luo, X., and Xie, J. (2020). Histone deacetylase HDA19 interacts with histone methyltransferase SUVH5 to regulate seed dormancy in Arabidopsis. Plant Biol. (Stuttg.) 22, 1062–1071. doi: 10.1111/plb.13158
Keywords: histone modification, chromatin remodeling, seed development, seed dormancy, seed germination, seedling establishment
Citation: Ding X, Jia X, Xiang Y and Jiang W (2022) Histone Modification and Chromatin Remodeling During the Seed Life Cycle. Front. Plant Sci. 13:865361. doi: 10.3389/fpls.2022.865361
Received: 29 January 2022; Accepted: 21 March 2022;
Published: 25 April 2022.
Edited by:
Bing Bai, University of Copenhagen, DenmarkReviewed by:
Henk Hilhorst, Wageningen University and Research, NetherlandsCopyright © 2022 Ding, Jia, Xiang and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenhui Jiang, amlhbmd3ZW5odWlAY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.