
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 April 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.865132
This article is part of the Research TopicAn Update on Brassinosteroids: Homeostasis, Crosstalk, and Adaptation to Environmental Stress, Volume IIView all 7 articles
 Min Song1,2†
Min Song1,2† Bin Linghu2†
Bin Linghu2† Shuhua Huang1
Shuhua Huang1 Fang Li1,2
Fang Li1,2 Ran An1
Ran An1 Changgen Xie2
Changgen Xie2 Yantao Zhu1
Yantao Zhu1 Shengwu Hu2
Shengwu Hu2 Jianxin Mu1*
Jianxin Mu1* Yanfeng Zhang1*
Yanfeng Zhang1*
The leucine-rich repeat receptor-like protein kinase (LRR-RLK) family represents the largest group of RLKs in plants and plays vital roles in plant growth, development and the responses to environmental stress. Although LRR-RLK families have been identified in many species, they have not yet been reported in B. napus. In this study, a total of 444 BnLRR-RLK genes were identified in the genome of Brassica napus cultivar “Zhongshuang 11” (ZS11), and classified into 22 subfamilies based on phylogenetic relationships and genome-wide analyses. Conserved motifs and gene structures were shared within but not between subfamilies. The 444 BnLRR-RLK genes were asymmetrically distributed on 19 chromosomes and exhibited specific expression profiles in different tissues and in response to stress. We identified six BnBRI1 homologs and obtained partial knockouts via CRISPR/Cas9 technology, generating semi-dwarf lines without decreased yield compared with controls. This study provides comprehensive insight of the LRR-RLK family in B. napus. Additionally, the semi-dwarf lines expand the “ideotype” germplasm resources and accelerate the breeding process for B. napus.
Receptor-like protein kinases (RLKs) serve as receptors for signaling transduction pathways that regulate many biological process (Clark et al., 1997; Li, 2010; Xun et al., 2020; Lu et al., 2021). Leucine-rich repeat receptor-like protein kinases (LRR-RLKs) comprise one of the largest groups of RLKs (Liu et al., 2017). There are 200–300 LRR-RLKs in Arabidopsis, tomato, rice, potato, and maize, respectively (Song et al., 2015; Wei et al., 2015; Bettembourg et al., 2017; Sun J. et al., 2017; Li et al., 2018). LRR-RLKs usually contain an extracellular, tandemly organized LRR domain (20–30 amino acid residues), a single-pass transmembrane domain, and functional protein kinase domain (Mishra et al., 2021).
LRR-RLKs are highly conserved, widely distributed and play vital roles in plant growth, development and stress responses. For instance, SERK3/BAK1 function in the plant immunity, growth, and cell death (Zhou et al., 2019). HSL3 function in regulating plant stomatal closure and the drought stress response through modulate hydrogen peroxide homeostasis (Liu et al., 2020). AtPXL1 functions in signal transduction pathways that respond to temperature fluctuations (Jung et al., 2015). XIP1/CEPR1 and CEPR2 are involved in the regulation of lateral root growth in Arabidopsis (Dimitrov and Tax, 2018). OsDOCS1 plays critical roles in plant root cap development (Bettembourg et al., 2017). OsSTLK regulates salt stress tolerance, potentially by regulating the ROS scavenging system, Na+/K+ ratio and MAPK signaling pathway (Lin et al., 2020).
The LRR-RLK BRI1 encodes a receptor serine/threonine kinase and has an extracellular domain that contains 25 leucine-rich repeats. BRI1 and BAK1 interact and regulate brassinosteroid signaling in Arabidopsis, and BRI1 can phosphorylate BAK1. Arabidopsis overexpressing BRI1 are highly sensitive to brassinosteroid and have higher brassinosteroid binding activity (Nam and Li, 2002). BRI1 is an important plasma-membrane receptor for plant steroids, as shown by immunoblotting and brassinolide (BL)-induced BRI1 phosphorylation (Wang et al., 2001). In wheat, knockdown of TaBRI1 reduces photosynthesis, the responses to light and temperature stresses, and yield (Fang et al., 2020). In potato, knockdown of BRI1 attenuates brassinosteroid signaling and decreases plant height (Huang et al., 2021). In Brachypodium distachyon, down-regulation of BdBRI1 expression results in reduced plant height, shortened internodes, as well as narrow and short leaves (Feng et al., 2015). In rice, loss-of-function of OsBRI1 reduced internode elongation and bending of the lamina joint (Yamamuro et al., 2000).
The rapeseed (Brassica napus L., AACC, 2n = 4x = 38) originated from the spontaneous hybridization of B. oleracea and B. rapa. It is the second largest oilseed crop in the world, and plays a crucial role in the production of edible oil (Zhu et al., 2016). Plant height and branch angle are key factors of rapeseed architecture, which is a major determinant of plant yield (Fan et al., 2021; Stanic et al., 2021). Taller rapeseed is not only unfavorable for mechanized harvesting, but also reduces the overall yield. We propose that an “ideotype” for rapeseed includes a semi-dwarf and compact architecture (Fu et al., 2016).
CRISPR/Cas9-mediated genome editing technology creates the opportunity to establish powerful germplasm resources (Zhang et al., 2021). However, there are few reports on germplasm resources of optimal plant-type, and no systematic study of LRR-RLK family in B. napus. In our study, we identified LRR-RLK genes in the rapeseed genome, and performed a systematic analysis of their phylogenetic relationship, chromosomal locations, gene structure, conserved protein domains, tandem and segmental duplication events, collinearity, and expression profiles. In addition, we used a CRISPR-Cas9 strategy to knockout BnBRI1 homologues to create ideotype mutants with semi-dwarf structure, good yield, and increased breeding potential.
A total of 225 AtLRR-RLKs protein sequences were download from the TAIR database1 (Sun J. et al., 2017). Protein sequences of the semi-winter rapeseed cultivar ‘‘Zhongshuang11’’ (ZS11) were download from the National Genomics Data Center (NGDC),2 and the accession number was PRJCA002883. The longest transcript of each gene was used for subsequent identification. Hidden Markov Model (HMM) and BLASTP search were used to classify the BnLRR-RLKs. The HMM profiles of LRR domain LRR1 (PF00560), LRR3 (PF07725), LRR4 (PF12799), LRR5 (PF13306), LRR6 (PF13516), LRR8 (PF13855), LRR9 (PF14580), LRR10 (PF18805), LRR11 (PF18831), LRR12 (PF18837), LRRNT-2 (PF08263), and Pkinase domain (PF00069) as well asPkinase_Tyr (PF07714) were obtained from the Pfam database.3
HUMMER3.1 software of Linux version was used to take the intersection of LRR and Pkinase domain, and the threshold was E-value < e–10. The 225 AtLRR-RLK protein sequences were used as queries to perform a BLASTP search in the local protein database of ZS11 with an E-value < e–10 and identity ≥ 50% (Camacho et al., 2009), and the putative BnLRR-RLKs were obtained by taking the intersection of the HUMMER and Blastap methods. Finally, these BnLRR-RLKs were submitted to the NCBI-CDD server4 and the SMART database5 to screen for proteins with the LRR and Pkinase domains. In addition, the theoretical isoelectric points (pI) and molecular weight (MW) of the LRR-RLKs were calculated by using the ExPASy database.6
Multiple sequence alignment of the full-length BnLRR-RLK proteins were performed with the FFT-NS-I method of the MAFFT software (Katoh et al., 2019). Fasttree software was used as the maximum likelihood method for constructing phylogenetic trees (Price et al., 2009). The EvolView software was used to visualize the phylogenetic tree (Zhang et al., 2012). Conserved motifs (the maximum is set to 10) were predicted using the MEME v5.2.0 with the maximum number of motifs and the optimum width of each motif falls between 10 and 200 residues (Bailey et al., 2009). The structures of the BnLRR-RLK genes were displayed based on GFF annotation files by TBtools software (Chen et al., 2020).
Chromosome location information of the BnLRR-RLK genes was extracted from the GFF file, and plotted by the MapChart version 3.0 software (Voorrips, 2002). In order to accurately analyze BnLRR-RLK duplication events, the e-value of e–100 was used to align all of the protein sequences in ZS11 with the BLASTP program (Camacho et al., 2009). The duplication pattern of these genes were detected with the default parameters of the MCScanX software, and divided into tandem duplication and segmental duplication (Krzywinski et al., 2009). Similarly, the e-value of e–100 was used to align all of the protein sequences between ZS11 and Arabidopsis with the BLASTP program, and all syntenic blocks were mapped with JCVI software (Tang et al., 2008).
RNA-seq data from 12 tissues (root, stem, leaf, flower, silique, sepal, pistil, stamen, ovule, pericarp, wilting pistil, and blossomy pistil) of ZS11 were downloaded from the NCBI database (project ID: PRJNA394926) (Sun F. M. et al., 2017). RNA-seq data of ZS11 leaf under abiotic stress, including dehydration, salt, ABA and cold stress treatments, were downloaded from the NGDC database (project ID: CRA001775) (Zhang et al., 2019). These transcriptome data were mapped to the ZS11 reference genome using HISAT2 software (Kim et al., 2015). The TPM values (Transcripts Per Million) were calculated by FeatureCounts R package and the heatmaps were presented using TBtools software (Liao et al., 2014; Chen et al., 2018).
To construct the CRISPR/Cas9 recombinant plasmid, a common target sequence of six BnBri1 genes was designed (Supplementary Table 1), and this target dsDNA was generated via direct annealing of two oligonucleotides primers, and assembled into the pHSE401 vector (Xing et al., 2014). The resulting pHSE401-BRI1 constructs were transformed into the agrobacterium tumefaciens strain GV3101 by electroporation and subsequently transformed into the selfing line K407 of B. napus using the method described previously (Bhalla and Singh, 2008).
Genomic DNA extracted from T3 plants was used for PCR amplification with genotyping primers (Supplementary Table 1), the PCR products were conducted by sanger sequencing, and cloned into a pEASY-Blunt cloning vector (TransGen Biotech, Beijing, China), positive single clones were sequenced to identify mutations (Aoke Biotech, Beijing, China).
To analyze the hypocotyl variation of the gene-edited mutants, the seeds were sown in humus soil under 16 h light/8 h dark photoperiod at 22°C, and the hypocotyl length was measured after 6 days. To verify the response to Brassinozole (BRZ), the seeds of mutants were sterilized (75% alcohol for 30 s, 15% NaClO for 8 min, rinse 5 times with sterile water) and bunch planting at 1/2 strength Murashige and Skoog solid medium (1/2 MS), which contained 0 and 1,000 nM BRZ, respectively (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan). After 5 days of darkness cultivation at 22°C, the hypocotyl length was measured. Each treatment was repeated three times.
To detect the expression level of BnBRI1, total RNA was extracted from seedlings of the L18, L24 and WT by RNAiso Plus kit (TaKaRa, Dalian, China). One microgram of RNA was reverse-transcribed to cDNA using One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). qRT-PCR was performed using a SYBR Green Master Mix kit (TaKaRa, Dalian, China). The BnUBC21 gene was used as an internal reference, the primer pairs are listed in Supplementary Table 1. Each sample contains 3 biological replicates and 3 technical replicates.
All data were analyzed with One-way analysis of variance (ANOVA) of SPSS 26 software. GraphPad prism (version 8.0.2) was used for drawing graphs. Each experiment was repeated at least three times, and significant differences are indicated at the significance levels of P ≤ 0.05.
We detected a total of 444 BnLRR-RLK genes in the genome of B. napus cultivar ZS11, which is nearly double the number of LRR-RLK genes in Arabidopsis (Table 1). Specifically, we uncovered 215 LRR-RLK genes in the A subgenome and 225 LRR-RLK genes in the C subgenome. We renamed all of the BnLRR-RLK genes on the basis of chromosomal localization. Gene position and other details are presented in Supplementary Table 2. We found that BnLRR-RLK protein lengths were between 459 and 1,303 amino acids (aa). In addition, the protein molecular weights (MV) ranged from 51.31 to 143.19 kDa, and the isoelectric point (pI) from 4.77 to 10.27.
To evaluate the evolutionary relationships between LRR-RLK genes in B. napus and A. thaliana, we constructed a phylogenetic tree using the 444 BnLRR-RLK full-length proteins together with 225 AtLRR-RLK proteins. The 444 BnLRR-RLKs were subdivided into 22 groups (I–XV) (Figure 1). The largest group, LRR-RLK-III, contained 99 genes. In contrast with A. thaliana, no genes were identified in group I-2. There were at least twice as many LRR-RLK genes in each subgroup of B. napus vs. A. thaliana, except for groups I-1, VI-2, VIII-1, and XV, which had a similar number of LRR-RLK genes in B. napus and A. thaliana. These data suggest that allopolyploidization contributed to the expansion of LRR-RLKs in B. napus, but that not every subfamily has an increased the number of genes.

Figure 1. Phylogenetic tree analysis of BnLRR-RLK gene family in B. napus. Fasttree software was used as the maximum likelihood method for constructing phylogenetic trees with the protein sequences of B. napus and A. thaliana. The color clade illustrates the different subfamilies.
In addition, we analyzed conserved motifs and gene structures to define BnLRR-RLK family characteristics. Using MEME tools, we identified 10 putative motifs (Supplementary Table 3). We found that the type, number, and arrangement of the N-terminus determined the classification of different subfamilies, and expanded the variety and quantity of LRR-RLKs (Supplementary Figure 1). The N-termini of these proteins were enriched for many different tandem repeat motifs, such as motif 1, motif 3 and motif 4. Intriguingly, motif 2 and motif 5 were often found at the C-terminus.
The gene structure characteristics revealed that different subfamilies shared different intron-exon distribution, whereas members within the same subfamily had a similar gene structure, suggesting a similar evolution within subgroups but not between subgroups, based on GFF annotation files (Supplementary Figure 1). The large number of intron-exons in subgroups I-1, II, XIII, V, and VI-2, indicate a complex gene structure, whereas other subfamilies contained few intron-exons and showed simpler gene structures.
Knowledge of the chromosome distribution of gene family members is essential for studying family member duplication and collinearity. We found that the 444 BnLRR-RLK genes were widely distributed on 19 chromosomes, and the distribution was extremely uneven, with about 40 genes on chromosome 3C and 13 genes on chromosomes 4A and 8A, respectively (Supplementary Figure 2). We did not observe any preference for the distribution of LRR-RLK genes on each chromosome, with the exception that there were no LRR-RLK genes on the upper arm of chromosome 4A.
Gene duplication is a major mechanism underlying the expansion of gene family members. Based on BLAST and MCScanX software, we identified a total of 6 tandem duplication events and 395 segmental duplications in the ZS11 genome (Figure 2 and Supplementary Tables 4, 5). Only 9 events were detected within the same chromosome, whereas 386 segmental duplications occurred across chromosomes, which suggests that segmental duplication events across chromosomes plays a key role in LRR-RLKs expansion. Further analysis showed that 51 duplication events occurred on the AA subgenome, 55 events on the CC subgenome, whereas 289 events occurred across AA/CC subgenomes. Considering that B. napus (AACC genome) derived from a spontaneous hybridization between B. rapa (AA genome) and B. oleracea (CC genome), and contains the complete diploid chromosome sets of both parental genomes. Therefore, we infer that allopolyploidization plays a significant role in the expansion of the LRR-RLKs in B. napus.
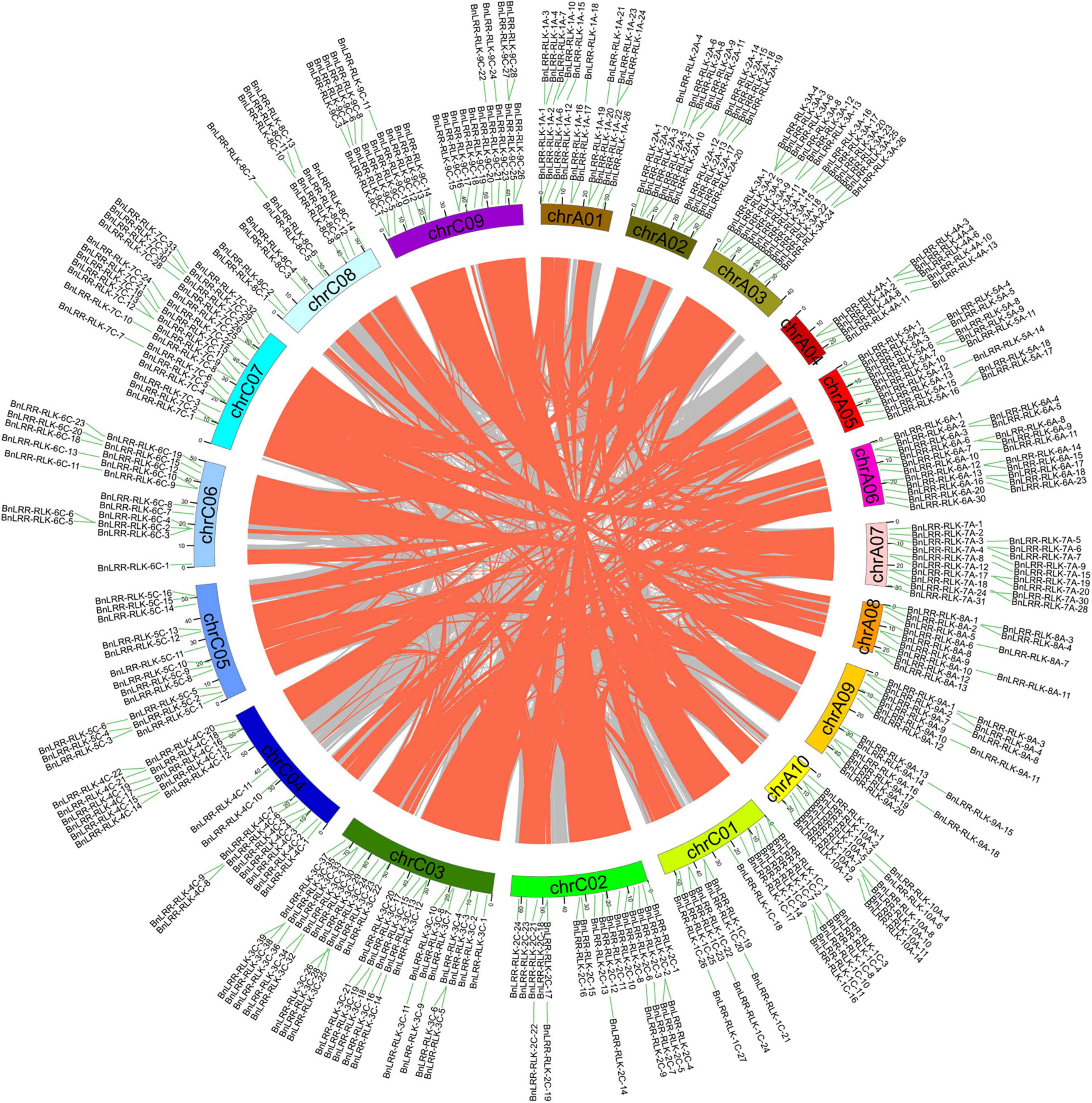
Figure 2. The chromosome locations associated with BnLRR-RLKs segmental duplication in B. napus. The gray and orange lines represent all synteny blocks in B. napus Cultivar ZS11 genome and segmental duplicate gene pairs, respectively. Different chromosomes are indicated by different colors.
To investigate the evolution of the LRR-RLKs, we analyzed their synteny between B. napus and A. Thaliana at the whole genome level. A total of 374 gene pairs were detected between the two genomes (Supplementary Figure 3 and Supplementary Table 6). Most AtLRR-RLK genes have multiple orthologous genes in B. napus. For instance, AtBRI1 (AT4G39400) corresponds to 6 BnBRI1 genes, indicating that BnBRI1 genes expanded in rapeseed. However, the 29 AtLRR-RLKs have only one collinear BnLRR-RLK gene.
The specific expression of genes in different tissues sheds light on gene function. In order to explore the function of the BnLRR-RLKs, we investigated their expression profiles in 12 different tissues, using public available RNA-seq data. We detected the expression of 426 BnLRR-RLKs in at least one tissue, whereas 18 genes were not expressed in any of the tissues analyzed (Figure 3 and Supplementary Table 7). We divided the 426 genes into two groups: group 1 displayed high expression levels in different tissues, whereas group 2 displayed low expression in tissues. For instance, BnLRR-RLK-6A-15 was highly expressed in ovule, BnLRR-RLK-8A-12 and BnLRR-RLK-8C-6 were highly expressed in root. These results reflected that the expression of LRR-RLKs vary in different tissues, suggesting that BnLRR-RLKs play functionally diverse roles in tissue development.
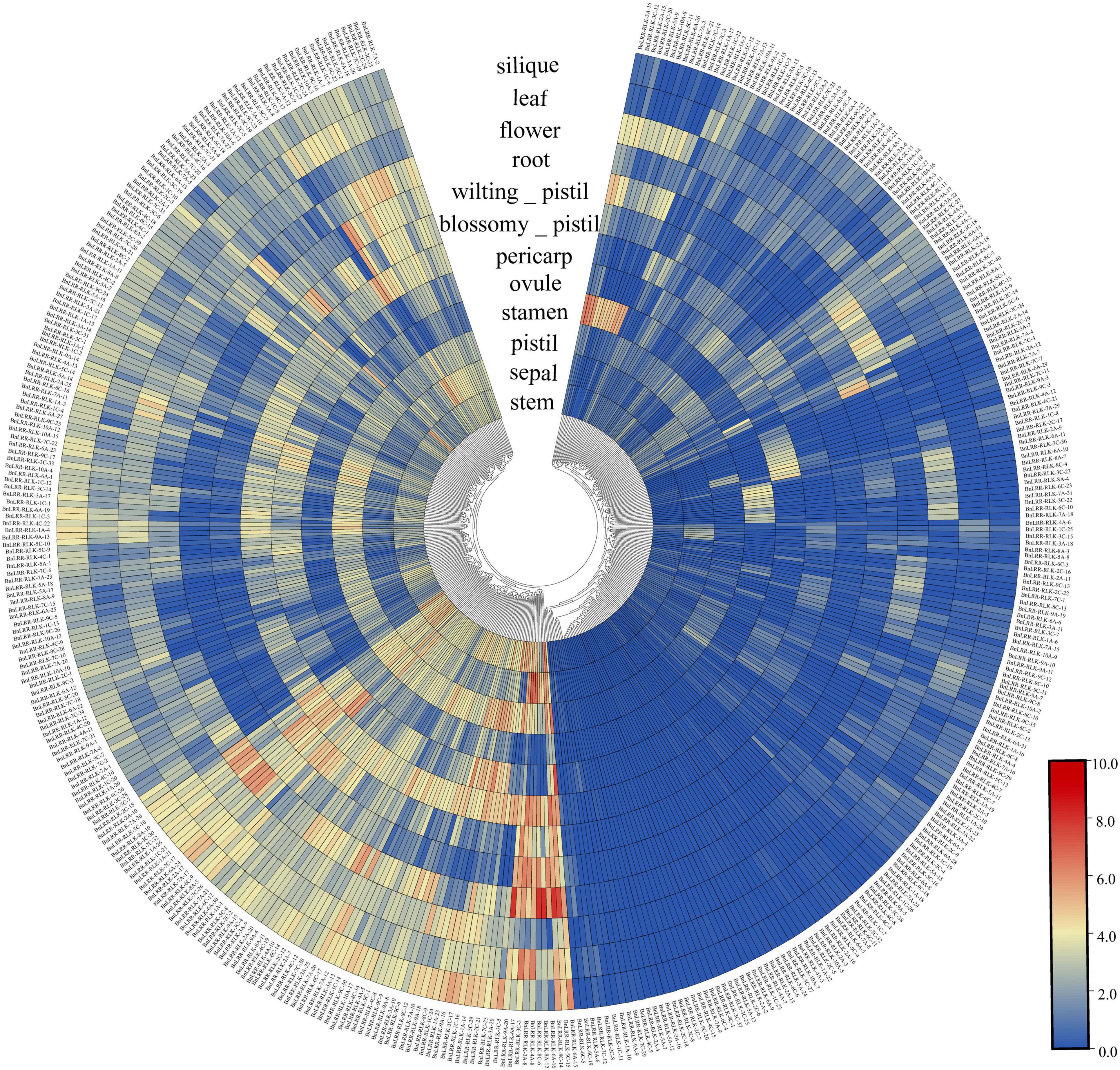
Figure 3. Expression profiles of BnLRR-RLKs in different tissues. Publicly available RNA-seq data of root, stem, leaf, flower, silique, sepal, pistil, stamen, ovule, pericarp, blossomy pistil, and wilting pistil from ZS11 were download for expression profile analysis. The expression level is equal to the mean values and transforms log2 values for normalization. Colors from blue to red represent relative expression levels from low to high.
LRR-RLK genes play a major role in abiotic stress responses. To explore the expression of LRR-RLK genes in response to abiotic stress, we examined their expression patterns under four abiotic stresses (dehydration, salt, abscisic acid (ABA), cold) using the published transcriptome data of B. napus. A total of 132 genes were extracted from the expression matrix based on STDEV value > 2 among different treatments (Figure 4 and Supplementary Table 8). These genes clustered into two groups.
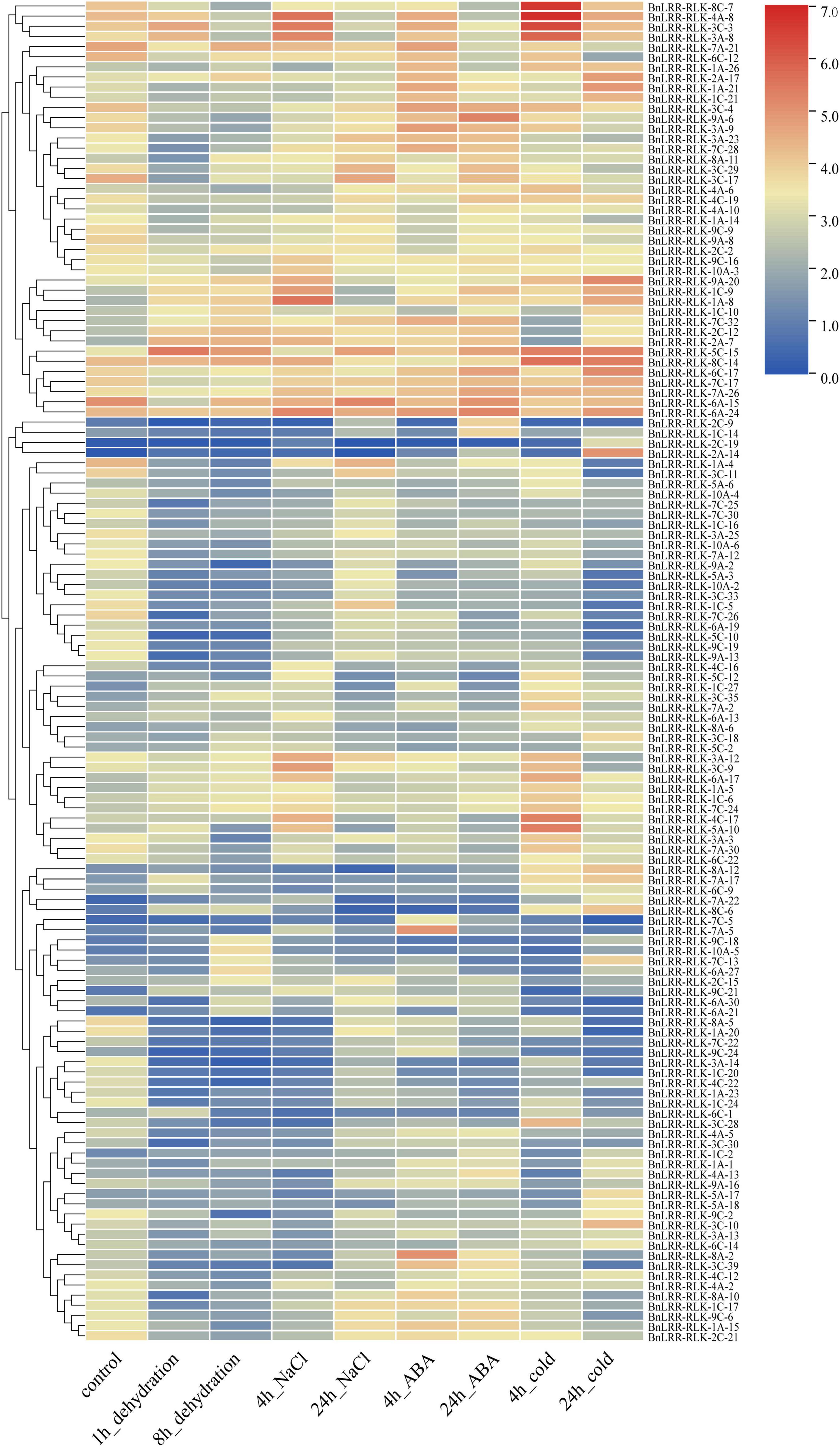
Figure 4. Expression profiles of BnLRR-RLKs under different stresses. Leaf RNA-seq data for ZS11 under abiotic stress, namely dehydration, salt, ABA and cold stress treatments, was downloaded from the NGDC database. Expression level is equal to the mean values and transforms log2 values for normalization. Colors from blue to red represent relative expression levels from low to high.
Group 1 contained 41 genes with significantly different expression patterns under different stress treatments. For example, BnLRR-RLK-4A-8, BnLRR-RLK-3C-3, and BnLRR-RLK-3A-8 were all up-regulated after 4 h-salt or 4 h-cold treatment, suggesting they may be involved in the related stress response. Interestingly, some genes displayed opposing expression profiles under different treatments. For instance, BnLRR-RLK-9A-6 was sharply down-regulated after 1 h-dehydration, but up-regulated after 4 h-ABA treatment. These data suggest that the responses to dehydration and ABA stress have opposing molecular mechanisms.
Group 2 contained 91 genes with low expression levels under each stress treatment. Unusually, part of the genes exhibited sharply changes in expression levels after a specific stress treatment, for instance, BnLRR-RLK-4C-17 and BnLRR-RLK-5A-10 were significantly up-regulated after 4 h-cold treatment, BnLRR-RLK-7A-5 and BnLRR-RLK-8A-2 were up-regulated after 4 h-ABA treatment, whereas BnLRR-RLK-1A-4 and BnLRR-RLK-3C-11 were strikingly down-regulated under dehydration treatment. The results provided useful information for studying the function of LRR-RLKs in response to abiotic stress.
Based on previous research in Arabidopsis and rice, we identified six BRI1 orthologs (renamed as BnBRI1.a∼BnBRI1.f), and six BRI1-Like (BRL) genes in B. napus (Figure 5A and Supplementary Table 9). The BnBRI1 and BnBRL genes both belonged to the Xb-1 subfamily, while the BnBRI1.b and the BnBRI1.f had higher homology with AtBRI1. Clustering analysis based on expression between 12 tissues showed that the six BnBRI1 genes clustered into one subclass and six BnBRL genes into another subclass, and the expression levels of BnBRI1 genes were generally higher than the BnBRL genes (Figure 5B). Furthermore, BnBRI1.c, BnBRI1.e, and BnBRI1.f had higher expression in various tissues compared to other BnBRI1 genes. It is noteworthy BnBRI1.f gene was highly expressed in all tissues.
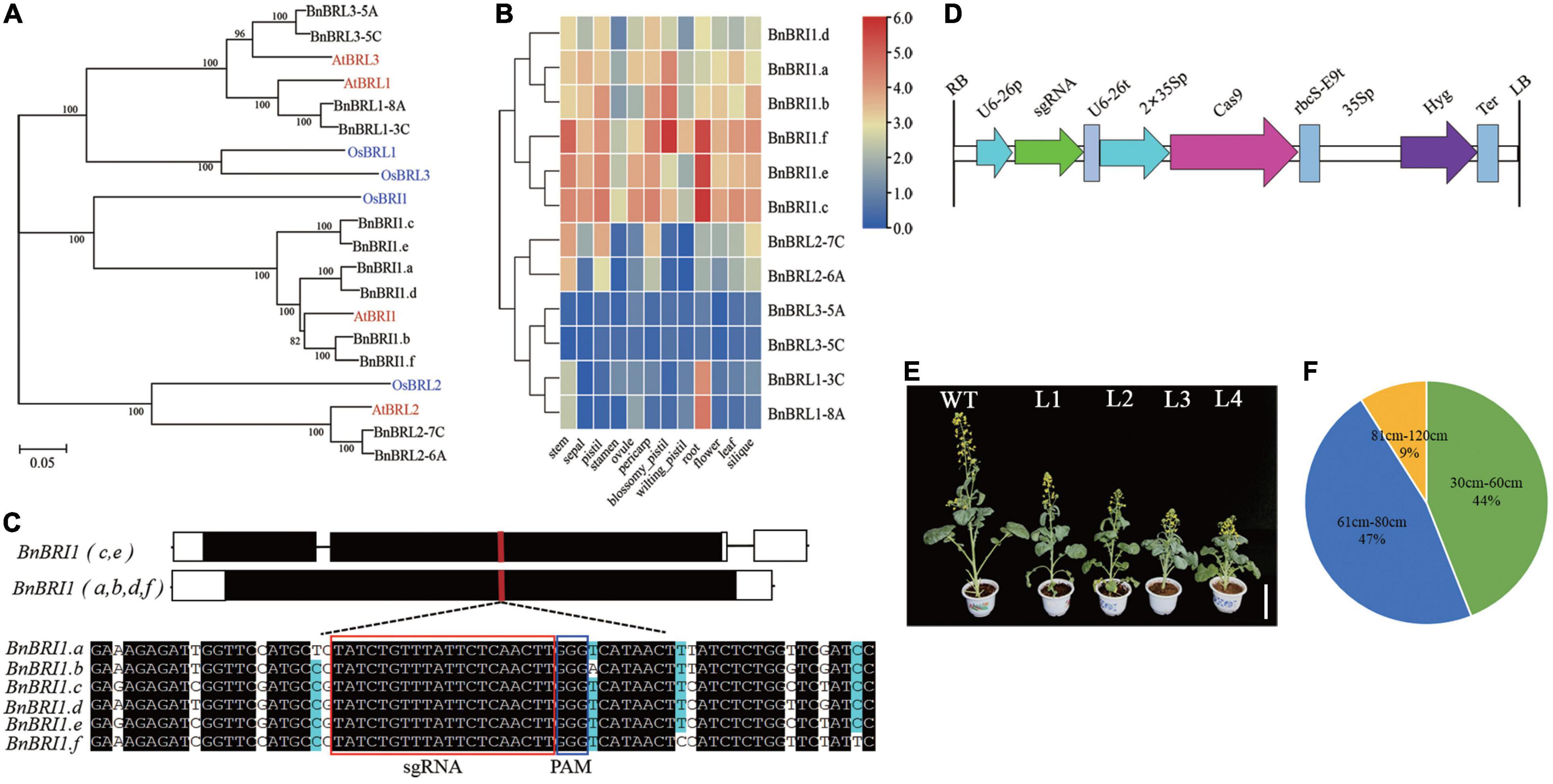
Figure 5. Evolutionary analysis of BRI1 and BRI1-like receptor family and CRISPR/cas9 targeted BnBRI1 gene mutation. (A) Phylogenetic tree analysis of BRI1-like receptor family in B. napus and A. thaliana. (B) Expression profiles of BnBRI1 and BnBRL genes in different tissues. (C) CRISPR/Cas9 sgRNA targets the exon of BnBRI1 genes. The red box highlights the target sequences in six BnBRI1 homologous sequences. The protospacer adjacent motif (PAM) is indicated in the blue box. (D) Physical map of CRISPR/cas9 vector pHSE401 between the RB and LB. (E) Phenotypes of CRISPR/Cas9-edited BnBRI1 mutants during flowering of rapeseed. From left to right are WT (K407), L1∼L4 strains, scale bars = 12 cm. (F) Pie chart of plant height from the gene edited Bnbri1 mutants in T0 generation. The green, blue, and yellow indicates the percentage of extreme dwarf, dwarf, and semi-dwarf phenotypes, respectively.
To generate new germplasm resources with a dwarf and optimized plant structure of B. napus, we designed single guide RNAs targeted to a common conserved region of BnBRI1 genes (Figure 5C), inserted these sgRNA into the pHSE401 expression vector and transformed into hypocotyls of rapeseed cultivar K407 (Figure 5D). We obtained a total of 130 transgenic positive plants (Figure 5E). Interestingly, more than 34 T0 plants displayed retarded growth with dark green rolling leaves at the stage of seedling growth (Supplementary Figure 4), consistent with phenotypes of previously reported classical bri1 mutants (Huang et al., 2021). About 42 plants (44%) exhibited an extreme dwarf phenotype (plant height 30–60 cm), and 45 plants (47%) showed dwarf phenotypes (61–80 cm), and 9 plants (9%) showed semi-dwarf phenotypes (81–120 cm) (Figure 5F).
To determine whether the transgenic lines had gene editing events, we selected the T3 generation plants of the semi-dwarf lines L18 and L24 for subsequent experiments (Figure 6A). The sequencing results of the single clones and PCR products near the sgRNA-targeted sites indicated that all target sites were edited in L24, except the BnBRI1.b and BnBRI1.f genes, similarly, all target sites were edited in L18, except the BnBRI1.f gene. The variations between the two lines were single base insertions of A, T or C (Figures 6B,C and Supplementary Figures 5, 6).
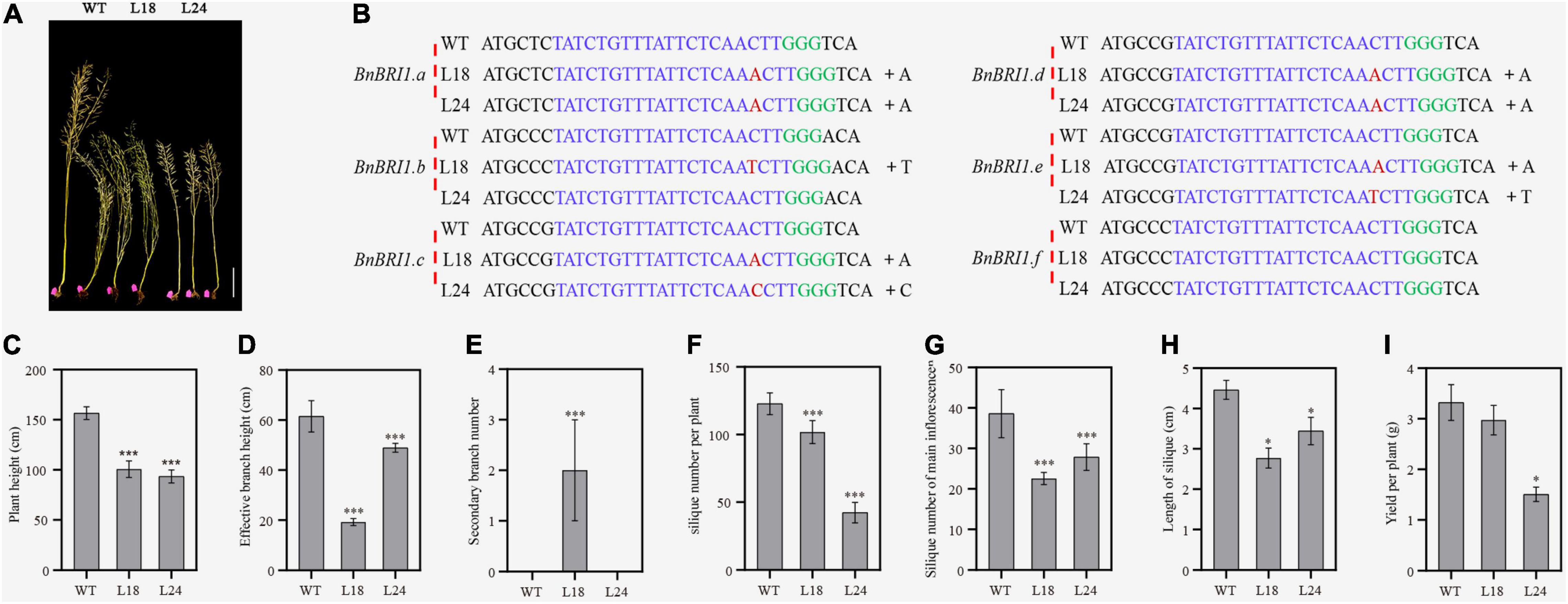
Figure 6. Sequencing results of BnBri1 mutants and their agronomic traits. (A) Semi-dwarf material obtained by editing BnBRI1 from left to right were WT (K407), L18 and L24 strains, scale bars, 20 cm. (B) Sequencing results of L18 and L24 strains. The sgRNA targeting sequences are highlighted in blue and the PAM sequences are green. (C–I) Statistical analysis of plant height, effective branch height, secondary branch number, silique number per plant, silique number of main inflorescence, yield per plant and length of silique in WT (K407), L18 and L24 homozygous plants from the greenhouse. Error bars standard deviation (n = 10). One-way analysis of variance was used for statistical analysis (“*,” corresponding to P < 0.05, and “***,” corresponding to P < 0.001).
Furthermore, qRT-PCR was conducted (Supplementary Figure 7), and showed that the BnBRI1 expression of the L18 and L24 were both reduced significantly. As a result, the hypocotyl length of both mutants shortened obviously (Supplementary Figures 8A,C). The results of the BRZ treatment showed that the hypocotyl length of the two mutants were significantly decreased under 1,000 nM, so the L18 and L24 were BRZ sensitive mutants (Supplementary Figures 8B,D). These verified that the both gene edited lines are indeed related to brassinostreoid signaling transduction pathway.
Agronomic traits were closely related to agricultural production. We planted the L18 and L24 lines in a greenhouse, and investigated the major agronomic traits, plant height, effective branch height, secondary branch number, silique number per plant, silique number of main inflorescence, length of silique and yield per plant (Figures 6C–I). We found that almost all traits exhibited significant differences between the control WT and the two gene edited lines. Specifically, compared with the control, L24 had fewer secondary branches, fewer silique number of main inflorescences, fewer siliques in the whole plant, and shorter siliques, resulting in a significantly lower yield than the control. In contrast, when the plant height and the branch length of L18 were significantly diminished compared to the control, the yield per plant was not reduced. Among yield-related traits, although L18 had shorter silique and a lower silique number of main inflorescence, the increased number of secondary branches increased the total number of silique and prevented a decline in yield.
LRR-RLKs sense extracellular signals and stimuli. In this study, we identified 444 LRR-RLK genes in the genome of ZS11, approximately twice as many as in A. thaliana. These 444 genes were further divided into 22 subfamilies, with LRR-RLK-III subfamily as the largest, whereas there were no genes in the I-2 subfamily. In general, a typical LRR-RLK consists of an extracellular tandem LRR domain, a transmembrane domain, and an intracellular protein kinase domain (Mishra et al., 2021), however, the I-1 subfamily also contains a malectin-like domain at the N-terminus. This domain is found in a different receptor-like kinase, known as the Catharanthus roseus receptor-like kinase 1-like proteins (CrRLK1Ls) (Franck et al., 2018). These findings highlight the domain similarity and functional complexity of LRR-RLKs family.
Gene duplication including tandem, tetraploid, segmental, and transpositional duplication, represents a major mechanism of new gene formation and gene family expansion (Freeling, 2009). Compared with the diploid Arabidopsis, many genes were significantly expanded in the allopolyploidization Brassica napus, likely due to genome polyploidy. Indeed, our research showed that 289 segmental duplications occurred across AA/CC subgenomes. Moreover, 51 and 55 duplication events occurred in AA and CC subgenomes, respectively. These repeated events within each subgenome may relate to their progenitors (Brassica rapa and Brassica oleracea). In addition, 6 tandem duplication events took place on chromosomes A04, A07, C04, C06, C07, and C09, respectively. Thus, at least three types of gene duplications appeared in the BnLRR-RLKs family: tetraploid, segmental and tandem duplication.
Gene expression patterns are closely related to their functions. We examined public RNA sequencing data to determine the expression patterns of BnLRR-RLK genes from 12 tissues in ZS11. We found that most BnLRR-RLKs have tissue-specific expression, for instance, BnLRR-RLK-3C-12 and BnLRR-RLK-3A-15 were highly expressed in stamens. Phylogenetic tree clustering suggested that both genes were clustered with a pollen tube related gene, AtPRK4 (At3G20190). Based on these findings we speculate that the two highly expressed genes in stamens are related to pollen tube elongation and fertilization (Chang et al., 2013; Duckney et al., 2017). Similarly, we also found that BnLRR-RLK-8A-12 and BnLRR-RLK-8C-6 are highly expressed in the roots; its homologous gene RLK7 (AT1G09970) in Arabidopsis is involved in the development of lateral roots, reflecting that the two genes are related to root morphogenesis (Toyokura et al., 2019).
Abiotic stress seriously compromises plant growth and development. We uncovered 132 BnLRR-RLKs from the expression matrix of various abiotic stresses, such as dehydration, NaCl, ABA, cold. We found that BnLRR-RLK-8C-7 was highly expressed after 4 h cold treatment, whereas its homologous SIF3 (AT1G51805), belongs to the STRESS INDUCED FACTOR (SIF) gene family, involved in plant basal defenses and pathogen resistance of Arabidopsis (Yuan et al., 2018). We propose that BnLRR-RLK-8C-7 probably relate to the early cold stress response. BnLRR-RLK-7A-5 was highly expressed only after a 4-h ABA treatment, and its homologous AtBARK1 is involved in the BR signaling pathway, suggesting that this gene may play a role in ABA signaling pathway (Kim et al., 2013). Given the importance of LRR-RLK family members for plant growth, development, and stress tolerance, our study of tissue-specific and abiotic stress expression patterns facilitates the discovery of LRR-RLK genes with important biological functions and their underlying regulatory mechanisms.
BR signaling requires the BRI1 and other LRR-RLKs. The six BnBRI1 genes have similar motif elements and intron-exon structure, supporting their homology and evolutionary conservation. Nevertheless, the six BnBRI1 have tissue-specific expression patterns: BnBRI1.c, BnBRI1.e, BnBRI1.f were highly expressed in various tissues; in particular, BnBRI1.f was highly expressed in all tested tissues. We propose that the six copies of BnBRI1 have different functions despite their similarities.
Gene editing technology can be used to improve agronomic trait in crops and accelerate the breeding process. This technology was used in rapeseed to knockout the BnaA03.BP gene, which generated semi-dwarf plants without affecting other traits (Fan et al., 2021). Knockout of the two BnaMAX1 homologous improved plant architecture and increased yield (Zheng et al., 2020). Editing the BnD14 gene resulted in a compact architecture and is expected to achieve high-density plants in production (Stanic et al., 2021).
BRI1 mutations caused developmental defects, including extreme dwarfism and male sterility (Clouse et al., 1996; Yamamuro et al., 2000; Feng et al., 2015; Huang et al., 2021). We used CRISPR/cas9 technology to edit the six BnBRI1 genes in rapeseed, and obtained 130 gene-edited rapeseed plants, some of which exhibited a dwarf or semi-dwarf phenotype. Target identification in the two semi-dwarf strains revealed that all target genes were edited except BnBRI1.b and BnBRI1.f in L24, and BnBRI1.f in L18. Although the BnBRI1.f gene was not edited, we found one nucleotide variation in this gene compared to other five BnBRI1 genes that causes a F689S amino acid change, which probably relates to the phenotypes of L18 and L24. Among yield-related traits, although L18 had shorter silique and decreased silique number of main inflorescence, the increased number of secondary branches led to more silique and ultimately prevented a decline in yield. As a semi-dwarf line with excellent agronomic characteristics, L18 could be used to construct an ideotype plant structure of rapeseed.
This study systematically analyzed the LRR-RLKs family in B. napus. A total of 444 genes were detected in the genome of cultivar ZS11, which were divided into 22 groups based on phylogenetic relationships. These genes were widely and asymmetrically distributed across 19 chromosomes, and exhibited tissue- and stress- specific expression profiles. Moreover, we identified and knocked out BnBRI1 homologs to generate semi-dwarf lines with increased yield. This study lays the foundation to investigate LRR-RLK family gene function, expands the “ideotype” germplasm resources for field breeding, and accelerates the process of rapeseed breeding.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YFZ and JM designed the study. MS and BL performed the analysis and wrote the manuscript. RA, YTZ, FL, and SHH contributed to genetic transformation of Brassica napus. SWH and CX revised the manuscript. All authors have read and approved the final manuscript version.
This research was funded by the West Light Foundation of the Chinese Academy of Sciences (XAB2018AW16), the Shaanxi Creative Talents Promotion Plan-Technological Innovation Team (2020TD-051), the Scientific Research and Sharing Platform Construction project of Shaanxi Province (2020PT-036), and the National Key Research and Development Program of China (2016YFD0101900).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Chen Qijun (China Agricultural University) for providing the CRISPR/Cas9 system.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.865132/full#supplementary-material
Supplementary Figure 1 | Phylogenetic relationship, conserved motifs and gene structures of BnLRR-RLK proteins. (A) Phylogenetic relationship of BnLRR-RLKs. (B) Conserved motifs of BnLRR-RLKs, boxes of different colors represent different motifs. (C) Gene structure of BnLRR-RLKs. The green and yellow boxes represent the coding sequences (CDSs) and untranslated regions (UTRs), respectively, and black lines represent introns.
Supplementary Figure 2 | Chromosomal localization of BnLRR-RLK genes across B. napus chromosomes. Words marked in red indicate tandem duplications. The location of each gene was determined based on its physical location on the chromosome.
Supplementary Figure 3 | The synteny of BnLRR-RLK genes in genomes of Arabidopsis and Brassica napus. The light-gray lines represent all collinear blocks between B. napus and A. thaliana, and light-purple lines represent orthologous relationships between BnLRR-RLKs and AtLRR-RLKs.
Supplementary Figure 4 | Phenotypes of CRISPR/Cas9 edited BnBRI1 strong mutantats at seedling stage. The bri1 strong mutants showed significant growth retardation with dark green and edge valgus thick leaves, and were eventually lost due to failure to harvest seeds, scale bars, 1.2 cm.
Supplementary Figure 5 | Sequencing chromatograms of positive single clones in T3 generation. Each single clone was evaluated by Sanger sequencing. The inserted nucleotides are highlighted with yellow colors, pointing by black arrows.
Supplementary Figure 6 | Sequencing chromatograms of PCR products in T3 generation. Each PCR product was purified and sequenced. The inserted nucleotides are highlighted by black box.
Supplementary Figure 7 | Expression of BnBRI1genes in WT, L18 and L24. The BnUBC21 gene was used as an internal control. Error bars represent SE (standard errors) of three independent replicates. Different lowercase letters indicate values significant differences compared to WT at the level P < 0.05.
Supplementary Figure 8 | Hypocotyl length and response to BRZ in gene edited lines. (A) Phenotypes of the gene edited lines at seedling stage. (B) The gene edited lines under BRZ treatment. (C) Hypocotyl length of seedlings shown in (A). Data are the mean ± SD (n = 15), scale bars, 1 cm. (D) Hypocotyl length of seedlings at different BRZ concentrations shown in (B), scale bars, 1.5 cm. Different letters indicate significant differences based on one-way ANOVA (P < 0.05).
Bailey, T. L., Mikael, B., Buske, F. A., Martin, F., Grant, C. E., Luca, C., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bettembourg, M., Dal-Soglio, M., Bureau, C., Vernet, A., Dardoux, A., Portefaix, M., et al. (2017). Root cone angle is enlarged in docs1 LRR-RLK mutants in rice. Rice 10, 1–8. doi: 10.1186/s12284-017-0190-1
Bhalla, P. L., and Singh, M. B. (2008). Agrobacterium-mediated transformation of Brassica napus and Brassica oleracea. Nat. Protoc. 3, 181–189. doi: 10.1038/nprot.2007.527
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Chang, F., Gu, Y., Ma, H., and Yang, Z. (2013). AtPRK2 promotes ROP1 activation via RopGEFs in the control of polarized pollen tube growth. Mol. Plant. 6, 1187–1201. doi: 10.1093/mp/sss103
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant. 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Rui, X., Hao, C., and He, Y. (2018). TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. BioRxiv [Preprint] BioRxiv. 289660, doi: 10.1101/289660
Clark, S. E., Williams, R. W., and Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/s0092-8674(00)80239-1
Clouse, S. D., Langford, M., and McMorris, T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. doi: 10.1104/pp.111.3.671
Dimitrov, I., and Tax, F. E. (2018). Lateral root growth in Arabidopsis is controlled by short and long distance signaling through the LRR RLKs XIP1/CEPR1 and CEPR2. Plant Signal. Behav. 13:e1489667. doi: 10.1080/15592324.2018.1489667
Duckney, P., Deeks, M. J., Dixon, M. R., Kroon, J., Hawkins, T. J., and Hussey, P. J. (2017). Actin-membrane interactions mediated by NETWORKED2 in Arabidopsis pollen tubes through associations with pollen receptor-like kinase 4 and 5. New Phytol. 216, 1170–1180. doi: 10.1111/nph.14745
Fan, S., Zhang, L., Tang, M., Cai, Y., Liu, J., Liu, H., et al. (2021). CRISPR/Cas9-targeted mutagenesis of the BnaA03.BP gene confers semi-dwarf and compact architecture to rapeseed (Brassica napus L.). Plant Biotechnol. J. 19, 2383–2385. doi: 10.1111/pbi.13703
Fang, J., Zhu, W., and Tong, Y. (2020). Knock-down the expression of brassinosteroid receptor TaBRI1 reduces photosynthesis, tolerance to high light and high temperature stresses and grain yield in Wheat. Plants (Basel) 9:840. doi: 10.3390/plants9070840
Feng, Y., Yin, Y., and Fei, S. (2015). Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 234, 163–173. doi: 10.1016/j.plantsci.2015.02.015
Franck, C. M., Westermann, J., and Boisson-Dernier, A. (2018). Plant malectin-like receptor kinases: from cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 69, 301–328. doi: 10.1146/annurev-arplant-042817-040557
Freeling, M. (2009). Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60, 433–453. doi: 10.1146/annurev.arplant.043008.092122
Fu, D. H., Jiang, L. Y., Masons, A. S., Xiao, M. L., Zhu, L. R., Li, L. Z., et al. (2016). Research progress and strategies for multifunctional rapeseed: a case study of China. J. Integr. Agric. 15, 1673–1684. doi: 10.1016/S2095-3119(16)61384-9
Huang, S. H., Zheng, C. Y., Zhao, Y., Li, Q., Liu, J. W., Deng, R., et al. (2021). RNA interference knockdown of the brassinosteroid receptor BRI1 in potato (Solanum tuberosum L.) reveals novel functions for brassinosteroid signaling in controlling tuberization. Sci. Hortic. Amsterdam 290:110516. doi: 10.1016/j.scienta.2021.110516
Jung, C. G., Hwang, S. G., Park, Y. C., Park, H. M., Kim, D. S., Park, D. H., et al. (2015). Molecular characterization of the cold- and heat-induced Arabidopsis PXL1 gene and its potential role in transduction pathways under temperature fluctuations. J. Plant Physiol. 176, 138–146. doi: 10.1016/j.jplph.2015.01.001
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, M. H., Kim, Y., Kim, J. W., Lee, H. S., Lee, W. S., Kim, S. K., et al. (2013). Identification of Arabidopsis BAK1-associating receptor-like kinase 1 (BARK1) and characterization of its gene expression and brassinosteroid-regulated root phenotypes. Plant Cell Physiol. 54, 1620–1634. doi: 10.1093/pcp/pct106
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Li, J. (2010). Multi-tasking of somatic embryogenesis receptor-like protein kinases. Curr. Opin. Plant Biol. 13, 509–514. doi: 10.1016/j.pbi.2010.09.004
Li, X. X., Ahmad, S., Guo, C., Yu, J., Cao, S. X., Gao, X. M., et al. (2018). Identification and characterization of LRR-RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells 7:120. doi: 10.3390/cells7090120
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lin, F., Li, S., Wang, K., Tian, H., Gao, J., Zhao, Q., et al. (2020). A leucine-rich repeat receptor-like kinase, OsSTLK, modulates salt tolerance in rice. Plant Sci. 296:110465. doi: 10.1016/j.plantsci.2020.110465
Liu, P. L., Du, L., Huang, Y., Gao, S. M., and Yu, M. (2017). Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol. Biol. 17:47. doi: 10.1186/s12862-017-0891-5
Liu, X. S., Liang, C. C., Hou, S. G., Wang, X., Chen, D. H., Shen, J. L., et al. (2020). The LRR-RLK protein HSL3 regulates stomatal closure and the drought stress response by modulating hydrogen peroxide homeostasis. Front. Plant Sci. 11:1789. doi: 10.3389/fpls.2020.548034
Lu, K., Chen, T., Yao, S., Liang, W. H., Wei, X. D., Zhang, Y. D., et al. (2021). Functional analysis on four receptor-like protein kinases under salt stress in Rice. Chin. J. Rice Sci. 35, 103–111. doi: 10.16819/j.1001-7216.2021.0905
Mishra, D., Suri, G. S., Kaur, G., and Tiwari, M. (2021). Comprehensive analysis of structural, functional, and evolutionary dynamics of leucine rich repeats-RLKs in Thinopyrum elongatum. Int. J. Biol. Macromol. 183, 513–527. doi: 10.1016/j.ijbiomac.2021.04.137
Nam, K. H., and Li, J. (2002). BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. doi: 10.1016/s0092-8674(02)00814-0
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Song, W., Wang, B., Li, X., Wei, J., Chen, L., Zhang, D., et al. (2015). Identification of immune related LRR-containing genes in Maize (Zea mays L.) by genome-wide sequence analysis. Int. J. Genomics 2015:231358. doi: 10.1155/2015/231358
Stanic, M., Hickerson, N. M. N., Arunraj, R., and Samuel, M. A. (2021). Gene-editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola). Plant Biotechnol. J. 19, 639–641. doi: 10.1111/pbi.13513
Sun, F. M., Fan, G. Y., Hu, Q., Zhou, Y. M., Guan, M., Tong, C. B., et al. (2017). The high-quality genome of Brassica napus cultivar ‘ZS11’ reveals the introgression history in semi-winter morphotype. Plant J. 92, 452–468. doi: 10.1111/tpj.13669
Sun, J., Li, L., Wang, P., Zhang, S., and Wu, J. (2017). Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Rosaceae genomes. BMC Genomics 18:763. doi: 10.1186/s12864-017-4155-y
Tang, H., Bowers, J. E., Wang, X., Ming, R., Alam, M., and Paterson, A. H. (2008). Synteny and collinearity in plant genomes. Science 320, 486–488. doi: 10.1126/science.1153917
Toyokura, K., Goh, T., Shinohara, H., Shinoda, A., Kondo, Y., Okamoto, Y., et al. (2019). Lateral inhibition by a peptide hormone-receptor cascade during Arabidopsis lateral root founder cell formation. Dev. Cell 48, 64–75. doi: 10.1016/j.devcel.2018.11.031
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, Z. Y., Seto, H., Fujioka, S., Yoshida, S., and Chory, J. (2001). BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383. doi: 10.1038/35066597
Wei, Z., Wang, J., Yang, S., and Song, Y. (2015). Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome 58, 121–134. doi: 10.1139/gen-2015-0035
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xun, Q., Wu, Y., Li, H., Chang, J., Ou, Y., He, K., et al. (2020). Two receptor-like protein kinases, MUSTACHES and MUSTACHES-LIKE, regulate lateral root development in Arabidopsis thaliana. New Phytol. 227, 1157–1173. doi: 10.1111/nph.16599
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Takatsuto, S., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. doi: 10.1105/tpc.12.9.1591
Yuan, N., Yuan, S., Li, Z., Zhou, M., Wu, P., Hu, Q., et al. (2018). STRESS INDUCED FACTOR 2, a leucine-rich repeat kinase regulates basal plant pathogen defense. Plant Physiol. 176, 3062–3080. doi: 10.1104/pp.17.01266
Zhang, H., Gao, S., Lercher, M. J., Hu, S., and Chen, W. H. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. doi: 10.1093/nar/gks576
Zhang, S., Zhang, R., Gao, J., Song, G., Li, J., Li, W., et al. (2021). CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol. J. 19, 1684–1686. doi: 10.1111/pbi.13647
Zhang, Y. T., Ali, U., Zhang, G. F., Yu, L. Q., Fang, S., Iqbal, S., et al. (2019). Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 39, 1–19. doi: 10.1007/s11032-019-1052-x
Zheng, M., Zhang, L., Tang, M., Liu, J., Liu, H., Yang, H., et al. (2020). Knockout of two BnaMAX1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18, 644–654. doi: 10.1111/pbi.13228
Zhou, J., Wang, P., Claus, L. A. N., Savatin, D. V., Xu, G., Wu, S., et al. (2019). Proteolytic processing of SERK3/BAK1 regulates plant immunity, development, and cell death. Plant Physiol. 180, 543–558. doi: 10.1104/pp.18.01503
Keywords: Brassica napus, LRR-RLK, CRISPR/Cas9, BnBRI1, semi-dwarf
Citation: Song M, Linghu B, Huang S, Li F, An R, Xie C, Zhu Y, Hu S, Mu J and Zhang Y (2022) Genome-Wide Survey of Leucine-Rich Repeat Receptor-Like Protein Kinase Genes and CRISPR/Cas9-Targeted Mutagenesis BnBRI1 in Brassica napus. Front. Plant Sci. 13:865132. doi: 10.3389/fpls.2022.865132
Received: 29 January 2022; Accepted: 14 March 2022;
Published: 12 April 2022.
Edited by:
Qian-Feng Li, Yangzhou University, ChinaReviewed by:
Jian Wu, Yangzhou University, ChinaCopyright © 2022 Song, Linghu, Huang, Li, An, Xie, Zhu, Hu, Mu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxin Mu, anhtc3h5Y0AxNjMuY29t; Yanfeng Zhang, emhhbmd5ZmNsQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.