
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 April 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.861193
This article is part of the Research Topic Flowering Time Control in Agricultural and Horticultural Crops View all 10 articles
Chrysanthemum is a well-known ornamental plant with numerous uses. WRKY is a large family of transcription factors known for a variety of functions ranging from stress resistance to plant growth and development. Due to the limited research on the WRKY family in chrysanthemums, we examined them for the first time in Chrysanthemum lavandulifolium. A total of 138 ClWRKY genes were identified, which were classified into three groups. Group III in C. lavandulifolium contains 53 members, which is larger than group III of Arabidopsis. The number of introns varied from one to nine in the ClWRKY gene family. The “WRKYGQK” motif is conserved in 118 members, while other members showed slight variations. AuR and GRE responsive cis-acting elements were located in the promoter region of WRKY members, which are important for plant development and flowering induction. In addition, the W box was present in most genes; the recognition site for the WRKY gene may play a role in autoregulation and cross-regulation. The expression of the most variable 19 genes in terms of different parameters was observed at different stages. Among them, 10 genes were selected due to the presence of CpG islands, while nine genes were selected based on their close association with important Arabidopsis genes related to floral traits. ClWRKY36 and ClWRKY45 exhibit differential expression at flowering stages in the capitulum, while methylation is detected in three genes, including ClWRKY31, ClWRKY100, and ClWRKY129. Our results provide a basis for further exploration of WRKY members to find their functions in plant growth and development, especially in flowering traits.
Chrysanthemum lavandulifolium is one of the original species of Chrysanthemum × morifolium and uses as a model plant. It is a major ornamental plant, originated in China, an important cut flower with diversified petal color and shape having a variety of flower structures, which makes it a highly valuable ornamental plant in the floral industry worldwide (Nguyen and Lim, 2019). Chrysanthemum is among the four most important ornamental plants in the world used for beautification and aesthetic value (Shinoyama et al., 2012). The flower of chrysanthemum is also known for its health benefits, and it contains important antioxidants and phenolic compounds (Lin and Harnly, 2010; Yue et al., 2018).
Due to a simple diploid (2n = 2x = 18) genetic background, C. lavandulifolium dominates over C. morifolium (Huang et al., 2012; Dong-ru et al., 2019). The wild ancestors having desired qualities against biotic and abiotic cues can be used for the genetic improvement of cultivated varieties. Identification and incorporation of such genes into commercial varieties reduces pesticide use, improves plant health, and induces photoperiod-controlling flowering mechanisms (Wu et al., 2010; Fu et al., 2013; Yang et al., 2017).
Genetic architectural study is one of the major research directions in any plant. By narrowing the path further, transcription activation and silencing is the major field of concern. Many transcription factors (TFs) have been explored to switch on and off against certain environmental conditions. WRKY TF is one of the major families first identified in plants in 1994 by the name of DNA-binding protein in sweet potato, which is labeled as SWEET POTATO FACTOR1 (SPF1) (Ishiguro and Nakamura, 1994). WRKY TFs play important roles in resistance against biotic (fungal or bacterial pathogen) and abiotic stresses, growth, and development. Several WRKY TFs have been proven to provide resistance against biotic stresses exerted by fungal or bacterial pathogens by influencing other associated genes (Turck et al., 2004; Duan et al., 2007; Zhou et al., 2008, 2011; Li et al., 2009, 2011; Chen et al., 2010; Scarpeci et al., 2013; Phukan et al., 2016; Yokotani et al., 2018; Warmerdam et al., 2020). The name WRKY has been given to this family for their distinguishing characteristic, the WRKY domain, which is composed of highly conserved 60 amino acids and a zinc finger motif. WRKY TFs are classified into three groups based on the number of (WRKYGQK) domain, and zinc-finger motif. Group I has two domains, and groups II and III have one domain. Groups I and II have C2H2 type zinc finger motifs while group III has a C2HC zinc finger motif. Members of group II were further classified into subgroups (Rushton et al., 2010).
WRKY TFs are extensively studied in Arabidopsis and most other plants. WRKY7 and OsWRKY11 expression resulted in blooming delay in Arabidopsis and rice (Cai et al., 2014; Chen et al., 2019), WRKY13 has an adverse effect on flowering time, and WRKY12, GsWRKY20 and CsWRKY50 promote flowering, possibly through the influence of gibberellic acid under short-day conditions in Arabidopsis (Luo et al., 2013; Kumar et al., 2016; Li et al., 2016), GhWRKY22 regulating pollen development (Wang et al., 2019; Ramos et al., 2021). While different WRKY family TFs play an important role in chrysanthemum, they showed resistance to salt stress (Liu et al., 2013, 2014; Li et al., 2015b; Jaffar et al., 2016; Liang et al., 2017; Wang K. et al., 2017; He et al., 2018; Zhang et al., 2020), and CmWRKY1 and CmWRKY15 transgenic lines in C. morifolium interact with SA and ABA and altered plant development (Fan et al., 2015, 2016; Bi et al., 2021). CmWRKY48 showed resistance against aphid infestation (Li et al., 2015a). They also play a regulatory role in the flowering stages. The complex network of VQ20, WRKY2, and WRKY34 interaction influences the plant gametogenesis and plays a crucial part in the development of pollen and pollen tube growth, modulating flowering (Lei et al., 2017; Ma et al., 2020).
Flowering time is a complex character regulated by a network of different external and internal signals. Different pathways have been identified, including photoperiod pathway, vernalization pathway, gibberellin-regulated pathways, and autonomous floral initiation pathway (Soares et al., 2020). Therefore, flowering induction studies are more crucial in ornamental plants, especially in chrysanthemum, where all the ornamental characteristics are associated with flowering, i.e., flower color, shape, and size. Chrysanthemum is a short-day plant and needs a specific photoperiod to induce flowering, which makes it relatively expensive as well as limits the annual production of chrysanthemum flowers. We explored the WRKY gene family in C. lavandulifolium for the first time, prior, the WRKY gene family has only been studied in the C. morifolium (Song A. et al., 2014). The size of the WRKY family in C. morifolium comprised 15 members, which were named from CmWRKY1 to CmWRKY15 (GenBank: KC615355–KC615369). As WRKY is a large family, influencing floral characteristics, we need to explore them deeply. Our present study was a part of such investigation in which we tried to explore the WRKY gene family in C. lavandulifolium comprised structural gene analysis, protein motifs, phylogenetic relatedness of C. lavandulifolium and Arabidopsis thaliana, cis-acting elements, recognition of CpG rich regions, the methylation status of the genes, and expression of 19 important genes in different tissues at different stages to check their role in flowering-related traits. This investigation will lead us to identify variation in transcription regions in the WRKY family for future chrysanthemum research.
The plant materials of C. lavandulifolium were obtained from Beijing Forestry University. The plants’ G1 lines were grown under controlled environmental conditions in the growth chamber. All the plants used in this research came from the G1 line. The plants were propagated by cuttings and grown in pots. The plants were kept under long-day conditions (14 h L/10 h D) light/dark at a 25°C temperature. The photoperiod conditions were changed to short-day conditions (12 h L/12 h D) when the plants attained maturity (more than 14 leaves). The temperature was kept constant. The plants were watered every 7 days. Stem, root, leaf, buds, and capitulum samples for quantitative real-time polymerase chain reaction (qRT-PCR) were taken at different growth stages, including young, mature, and flowering. The plants were considered at a young stage when they attained eight leaves. The plants were called into the adult stage when the number of leaves was 14, and they were transferred to short-day conditions. The flower develops in short conditions, and samples were taken at different stages of flower development (Supplementary Figure 1). The little buds (1–3 mm in diameter) appeared after 7 days in short-day conditions and were taken for RNA extraction. The median buds (5–8 mm in diameter) were taken after a few days before flower opening. The first flower samples were taken when the flowers had just opened.
The C. lavandulifolium genome has been sequenced (Wen et al., 2022). All the protein and genomic sequences of ClWRKY members were obtained from PRJNA681093, National Center for Biotechnology Information (NCBI). The genomic information of C. lavandulifolium was completed by Henan University and Beijing Forestry University, and related papers have been published (Wen et al., 2022). The redundant sequences, which lack the WRKY domain, were removed after the sequence analysis using manual inspection in the Mega X software. A total of 138 definite ClWRKY sequences were taken for further study. The ClWRKY members were named from ClWRKY1 to ClWRKY138, according to the source provider (Supplementary Table 3).
Multiple protein sequence alignments of C. lavandulifolium and A. thaliana WRKY were made using ClustalX and MEGA-X version 10.2.6 (Molecular Evolutionary Genetics Analysis) (Kumar et al., 2018). The phylogenetic tree based on amino acid (aa) sequences of ClWRKY and AtWRKY conserved WRKY domains was constructed following the neighbor-joining method with 1,000 bootstraps to determine the close association of both gene families.
The gene structure (intron-exon) of ClWRKY members, full-length gene, and coding (CDS) sequences were analyzed in the online database, Gene Structure Display Server (GSDS)1 as described (Hu et al., 2015). Analysis of the conserved protein motifs was performed using the NCBI CDD batch2 and the TBtools software (Chen et al., 2020).
The promoter sequences of all 138 ClWRKY members were submitted to the PlantCARE software to analyze their promoter region for cis-acting elements (Lescot et al., 2002). For this purpose, a 2,000 bp sequence upstream of the gene was taken to identify cis-acting elements. The MEME suite software was used to identify de novo motifs and known enriched motifs (Bailey et al., 2015). The de novo motif analysis was performed to identify significant patterns (Bailey and Elkan, 1994).3 The known enriched motifs were identified via simple enrichment analysis using the MEME software (Bailey and Grant, 2021).4 Tomtom motif comparison tool was used to quantify the similarity between motifs (Gupta et al., 2007).
For CpG analysis, promoter regions from the WRKY gene family were sequenced. For the exploration of CpG islands, the “Methyl Primer Express version 1.0” software was used (Methyl Primer Express Software v1.0 Quick Reference Card).5 All the sequences were submitted to the software one by one to identify CpG islands or rich regions. The minimum length of the island was kept at 200 bp, while the maximum length was 2,000 bp. The criterion of “C + G/Total bases” was 50%. The CpG observed/CpG expected ratio was 0.6.
The total DNA was extracted from different tissues of C. lavandulifolium. The tissues included root, stem, leaf, and capitulum at three different growth stages, i.e., young stage, adult stage, and flowering stage. Total genomic DNA was extracted using the “Plant Genomic DNA Kit” (TOLOBIO), according to the manufacturer’s instructions. DNA was quantified using Thermo Scientific NanoDrop™ 2000/2000c Spectrophotometers, and the quality of DNA was checked by 1.5% agarose gel electrophoresis.
Total genomic DNA from each sample was treated with sodium bisulfites using the “DNA Bisulfite Conversion Kit” (TIANGEN) according to the manufacturer’s instructions. The conversion efficiency and quality were measured using Thermo Scientific NanoDrop™ 2000/2000c Spectrophotometers.
Ten genes were selected on the basis of CpG analysis. Primers for all ten genes were designed using the “Methyl primer express version 1.0” software. For this purpose, two types of primers (methylated and unmethylated) were designed to check the possible methylated sites. The primer length ranged from 18 to 22 bp. The PCR amplicon length was between 100 and 175 bp. Methylation-Specific Polymerase Chain Reaction (MSP) was conducted to analyze the CpG island among the ten selected genes after treatment with bisulfite for methylation status. The MSP conditions were as follows: pre-denaturation for 3 min at 95°C, followed by 34 cycles of the 30s at 95°C and 30 s at 55°C, 1 min at 72°C, and a final extension for 5 min at 72°C. PCR products were resolved onto 1.5% agarose gel and observed under UV transilluminator.
Nineteen (Zhou et al., 2011) variable genes on the basis of CpG island and phylogenetic analysis were selected for gene expression patterns. Primers for all the 19 ClWRKY genes were designed using the “Primer Premier 5” software. RNA was extracted from different plant tissues, i.e., root, stem, and leaves at young, adult, and flowering stages, while the capitulum samples, which comprise little buds, median buds, and first flower (right at the time of flower opening), were taken at flowering stages. The RNA was extracted using the RNAprep Pure Plant Plus Kit (spin column) according to the manufacturer’s instructions. The cDNA was synthesized using the “StarScript II First-strand cDNA Synthesis Mix with gDNA Remover.” The expression of all the selected genes was observed in qRT-PCR. The qRT-PCR was carried out on light cycle “Quantageneq225 Fluorescent Quantitative PCR,” using “2 × RealStar Green Fast Mixture.” The conditions of PCR were as follows: denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 20 s, and extension at 65°C for 5 s. The actin gene of the chrysanthemum was used as an internal control. The data from qRT-PCR for relative expression were analyzed using the 2–Δ ΔCt method proposed by Livak and Schmittgen (2001).
WRKY genes possess a conserved “WRKYGQK” domain. The “WRKYGQK” heptapeptide is the most defining characteristic of WRKY TFs. The C. lavandulifolium WRKY sequences were obtained from PRJNA681093, NCBI. A total of 153 WRKY gene members have been selected; among them, 138 were exhibited for sequence analysis, and 15 were considered defective. The WRKY members are named from ClWRKY1 to ClWRKY138 according to their location on the chromosome. Protein motif analysis showed homology in most of the conserved regions. Among 138 ClWRKY members, 85.51% of members have homology in the “WRKYGQK” core domain, and 19 members have two conserved WRKY domains. However, 14.5% of WRKY members showed variation in the conserved heptapeptide domain and had substitutions in the core domain. Out of 14.5% variants, 13 members (ClWRKY37, 38, 39, 49, 50, 51, 52, 53, 54, 74, 76, 77, and 98) have “WRKYGKK,” substituting glutamine as a lysine aa. ClWRKY12 replaced histidine instead of lysine, two genes, ClWRKY20 and ClWRKY21, shared sixth and seventh as glutamine and asparagine substitutions. High diversity has been found in three genes, i.e., (ClWRKY27) “WKKYGEQK,” (ClWRKY97) “WKKYGEKK,” and (ClWRKY131) “WRKNGQN” WRKY domains, respectively (Figure 1).
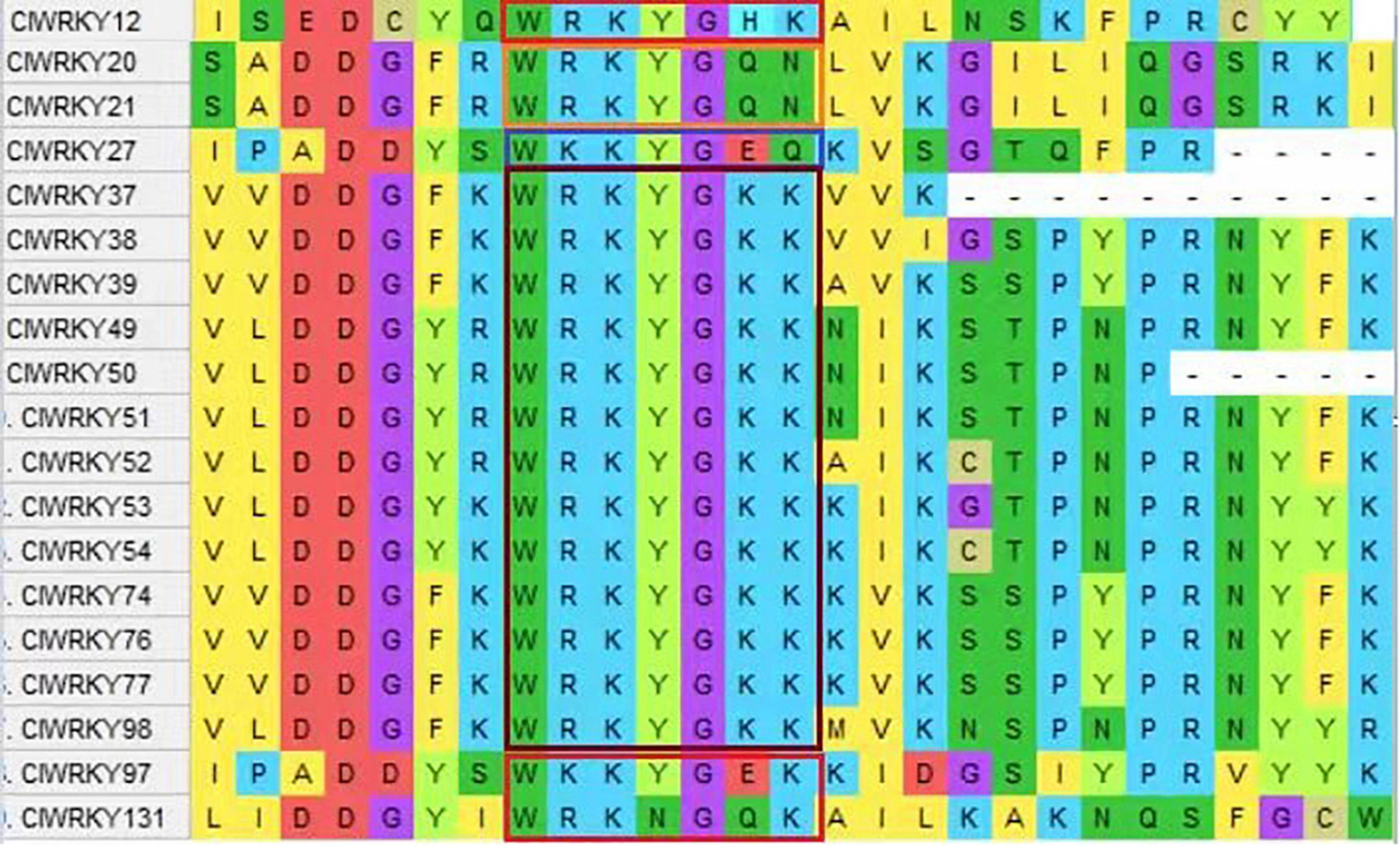
Figure 1. A screenshot of the MEGA X version 10.2.6, showing variation in the conserved domain (WRKYGQK) of ClWRKY proteins, and the variation of the conserved domain is encircled in different colors.
For the determination of important properties such as aa length, molecular weight, and isoelectric point, we thoroughly analyzed the WRKY gene. The average length of WRKY proteins was 313 aa, while 74 aa was the minimum and 697 aa was the maximum length. The WRKY members with the least and highest lengths were ClWRKY58 and ClWRKY36, respectively. The molecular weight ranged from 0.02 kDa (ClWRKY55) to 76.26 kDa (ClWRKY36). The pIs (isoelectric point) ranged from 4.73 (ClWRKY75) to 10.63 (ClWRKY97). Among 138 ClWRKY members, 82 were acidic (having less than 7 pI), while 56 were basic (having more than 7 pI). More details of ClWRKY features are available in Supplementary Table 1.
A phylogenetic tree was constructed between C. lavandulifolium and A. thaliana to investigate the relatedness of different WRKY genes and to understand the classification of different groups among WRKY members. Arabidopsis is a model plant for genetic study and has been explored for the function of WRKY genes. The MEGA-X software was used for agglomerative construction following the neighbor-joining method to find the correlation between WRKY members of C. lavandulifolium and A. thaliana. The WRKY members are divided into three groups or seven subgroups. Group I has 27 members, and group II has 58 members with 9 members in II a, 10 members in II b, 10 members in II c, 10 members in II d, and 19 members in II e. Group III has 53 members and was found to be the largest group in this classification (Figure 2). The majority of the ClWRKY members clustered in the corresponding groups. Some of the WRKY members (ClWRKY20, 21, 55, 56, and 58) had one WRKY domain, but still, clustered in group I; by the rules, group I members should have two domains. These WRKY members may evolve from two WRKY domain genes and lose one domain in the process of evolution. Previously, such exceptional cases of WRKY members with one WRKY domain clustering in group I have been reported (Bi et al., 2016).
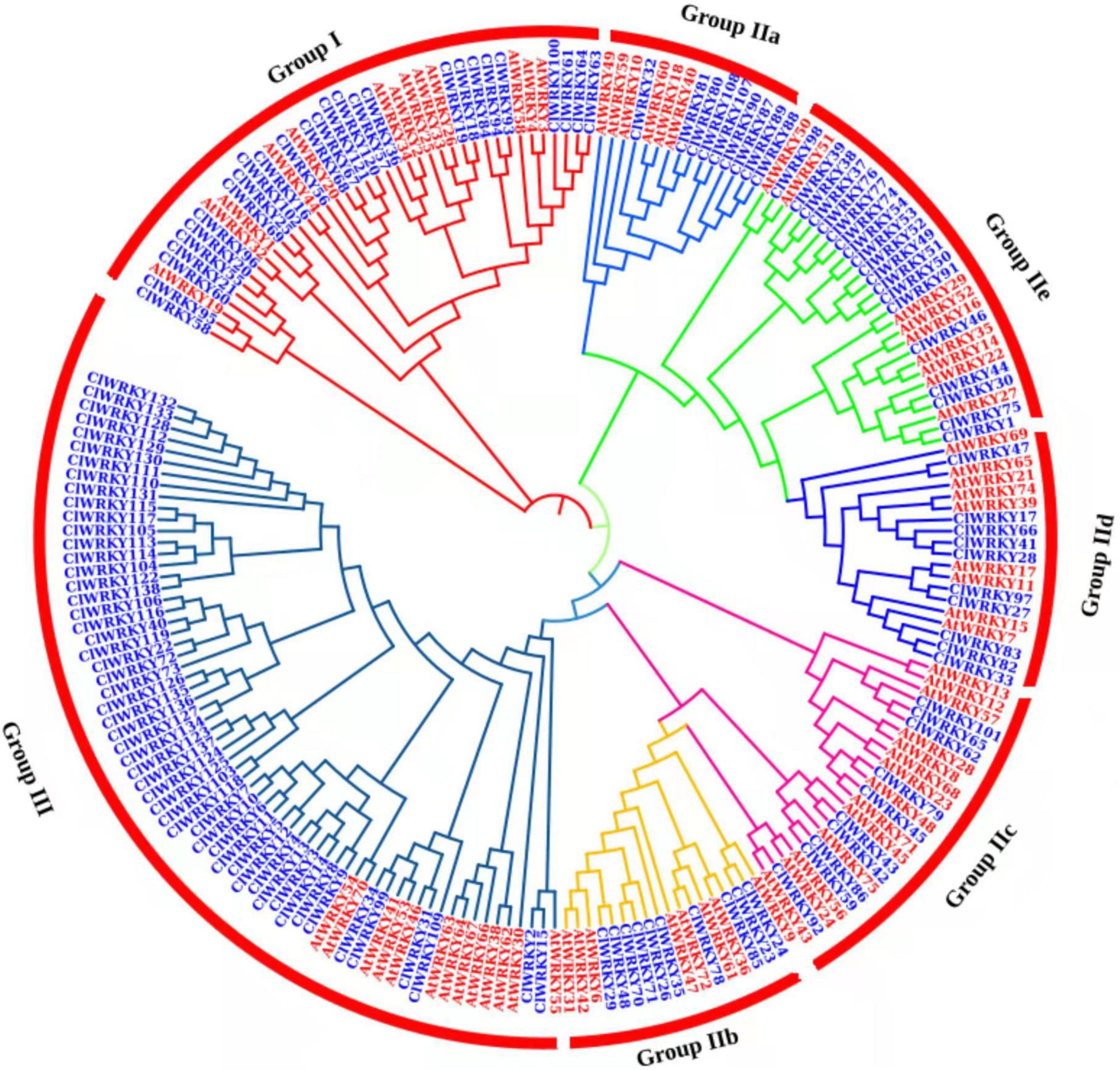
Figure 2. A phylogenetic tree of 138 ClWRKY proteins of Chrysanthemum lavandulifolium (blue) and 74 AtWRKY proteins of Arabidopsis thaliana (red) on the basis of amino acid sequences. The tree was constructed in MEGA X. The domains were clustered into three groups, namely, group I, group II, and group III. The group II was divided into five subgroups (a–e).
Some of the ClWRKY members clustered with important Arabidopsis WRKY genes that helped us identify the genes that are associated with flowering. ClWRKY36 is clustered with Arabidopsis AtWRKY2 and AtWRKY34 (Figure 2), both of which play an important role in pollen development through interaction with the VQ20 protein (Lei et al., 2017). Similarly, ClWRKY45 clustered with AtWRKY71 (Figure 2), which play a key role in the regulation of flowering, directly and indirectly, by influencing other flowering-associated genes (Yu et al., 2016, 2018).
For the determination of intron-exon structure similarity, Gene Structure Display Server (GSDS)6 database has been used. Variation among ClWRKY members was observed in terms of a number of introns. The minimal number of introns in a gene was one, which occurred in seven members including ClWRKY27, ClWRKY58, ClWRKY59, ClWRKY76, ClWRKY86, ClWRKY92, and ClWRKY125 while ClWRKY90 had 9 introns, which is the maximal in the whole family. The overall average number of introns in the ClWRKY family was three (Figure 3). The number of introns in group I ranged from 1 to 5, group II a varied from 3 to 9, group II b from 2 to 6, group II c from 1 to 2, group II d from 1 to 4, and group II e from 1 to 4. Group III varied from 1 to 6 (Supplementary Table 2). The minimal number of exons was two, which appeared in ClWRKY27, ClWRKY58, ClWRKY59, ClWRKY76, ClWRKY86, ClWRKY92, and ClWRKY125, while the maximal number of exons was 10, which occurred in ClWRKY87 and ClWRKY90 (Supplementary Table 2). The overall average number of exons in the ClWRKY family was four (Figure 3). The exon numbers varied within the groups; group I ranged from 2 to 6, group IIa from 4 to 10, group IIb from 3 to 7, group IIc from 2 to 3, group IId from 2 to 5, group IIe from 2 to 4, and group III from 2 to 7, respectively. The average number of exons within the groups is as follows: group I has 4, group IIa has 6, group IIb has 5, group IIc has 3, group IId has 3, group IIe has 3, and group III has 3, respectively (Supplementary Table 2). Protein motif analyses were performed on the ClWRKY gene family. The first two motifs represented by green and yellow colors are WRKY protein motifs (Figure 4). At least one or two WRKY protein motifs have been identified among all the gene sequences. This suggests that WRKY is a conserved domain.
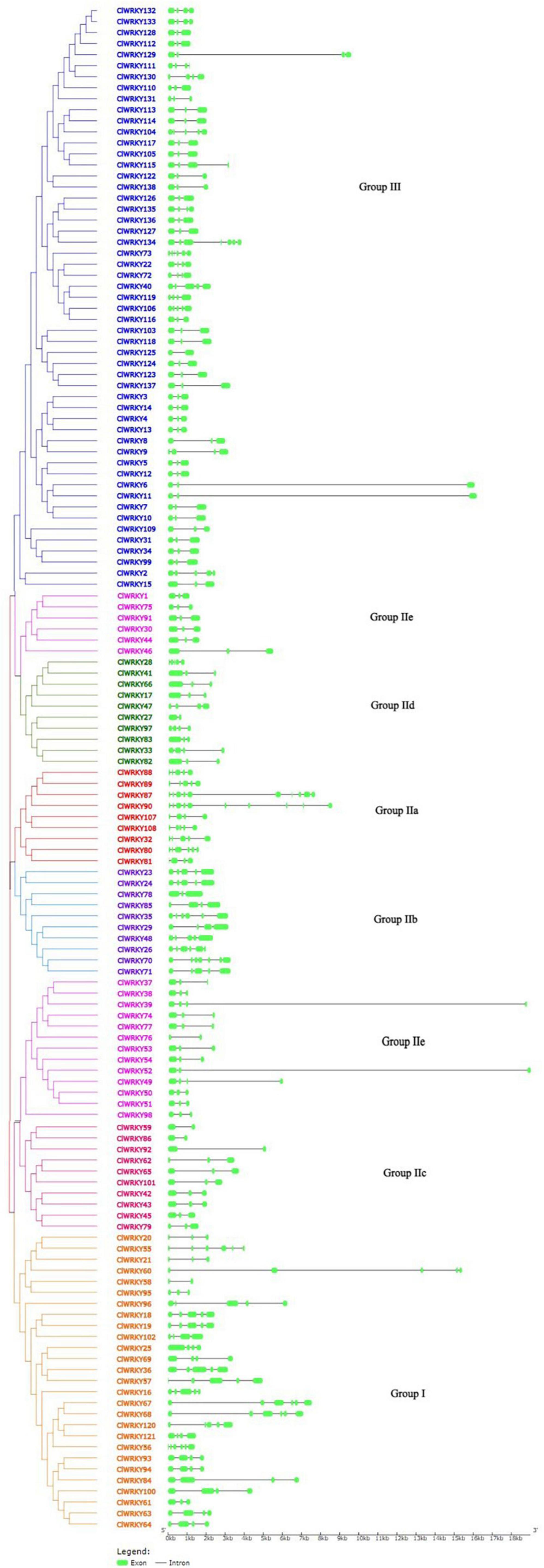
Figure 3. The intron-exon structure of 138 ClWRKY members according to the phylogenetic relationship. A phylogenetic tree was constructed with the ClWRKY protein sequences using MEGA X. The Gene Structure Display Server (GSDS) software was used to determine the intron-exon structure. Green bars represent exons in the gene. Different colors of WRKY members represent different groups in the ClWRKY family.
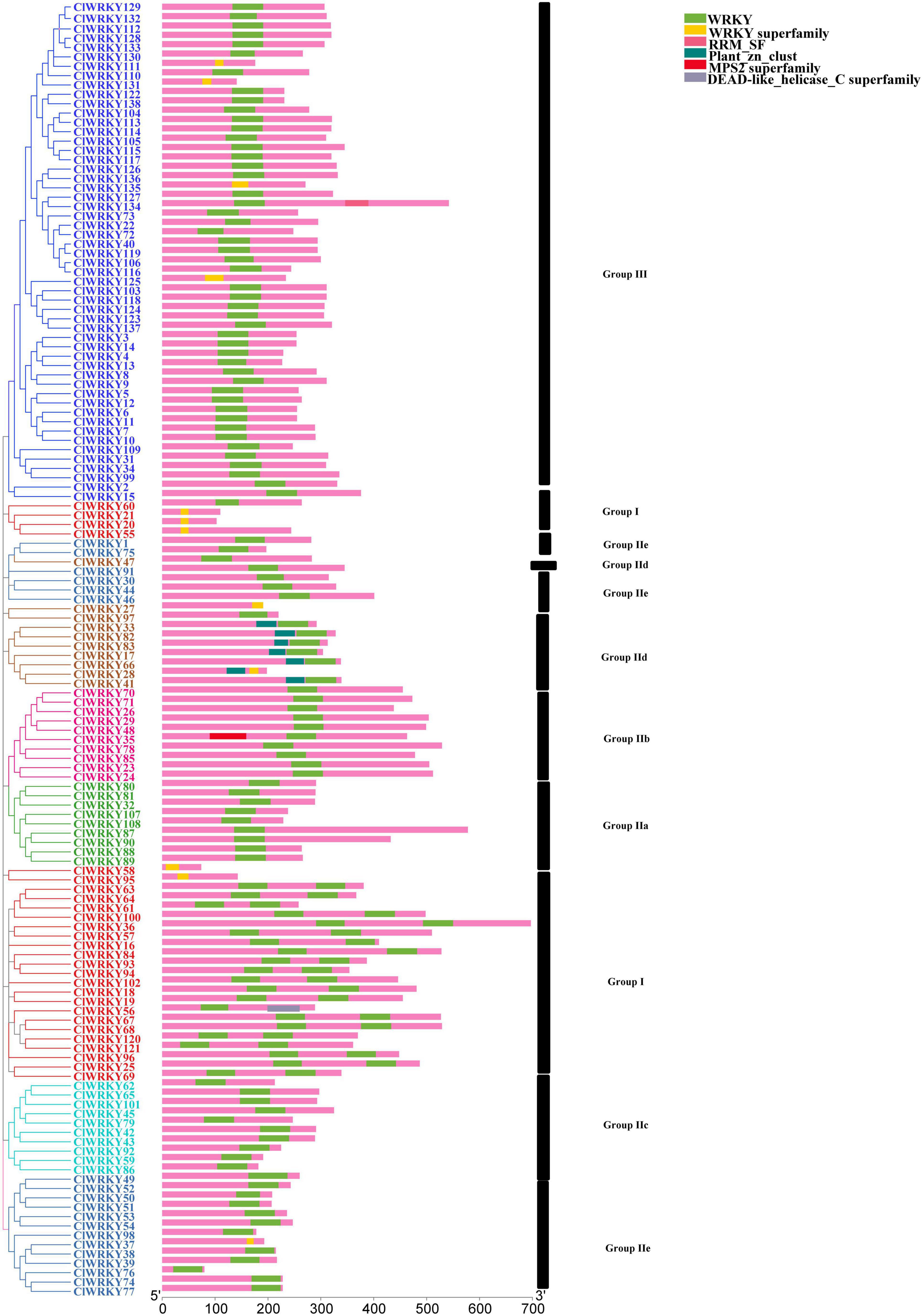
Figure 4. Schematic diagram of conserved motifs in ClWRKY proteins according to the phylogenetic relationship. Different color ClWRKY members indicate different groups. The conserved motifs were predicted using the TBtools software.
CpG island is the region of DNA that is rich in CpG sites and is often located near the promoters of genes. Ten members were found to have CpG islands; among them, seven members contained CpG in their promoter region, two in the cross-promoter and gene region, and one member was found to have it in the gene body. All the ten members had one CpG island. ClWRKY115 had the shortest island with 438 bp, while ClWRKY83 had the longest island with a length of 1,065 bp (Table 1). The genes with the CG-rich content are the obvious means of DNA methylation and a prominent source of epigenetic improvement.
Total genomic DNA of C. lavandulifolium was extracted from the root, stem, leaf, little buds, median buds, and first flowers (flowers at just opening time). These tissues were taken at different growth stages of the plant, which include the young stage, adult stage, and flowering stage. The methylation and unmethylation primers were designed for the ten different genes using the “Methyl Primer Express version 1.0” software. These genes were selected due to the presence of CpG islands. The MSP was carried out for the following ten genes: ClWRKY9, ClWRKY31, ClWRKY33, ClWRKY41, ClWRKY83, ClWRKY97, ClWRKY100, ClWRKY115, ClWRKY129, and ClWRKY130. The methylation status detected by MSP was analyzed through gel electrophoresis. The MSP results showed the presence of methylation in ClWRKY31, ClWRKY100, and ClWRKY129 at different stages in different tissues. These genes showed significant methylation status in all tissue samples at all growth stages of the plant (Figures 5A–C). However, the methylation status detection through bisulfite sequencing can give us more clear results compared with the MSP method.
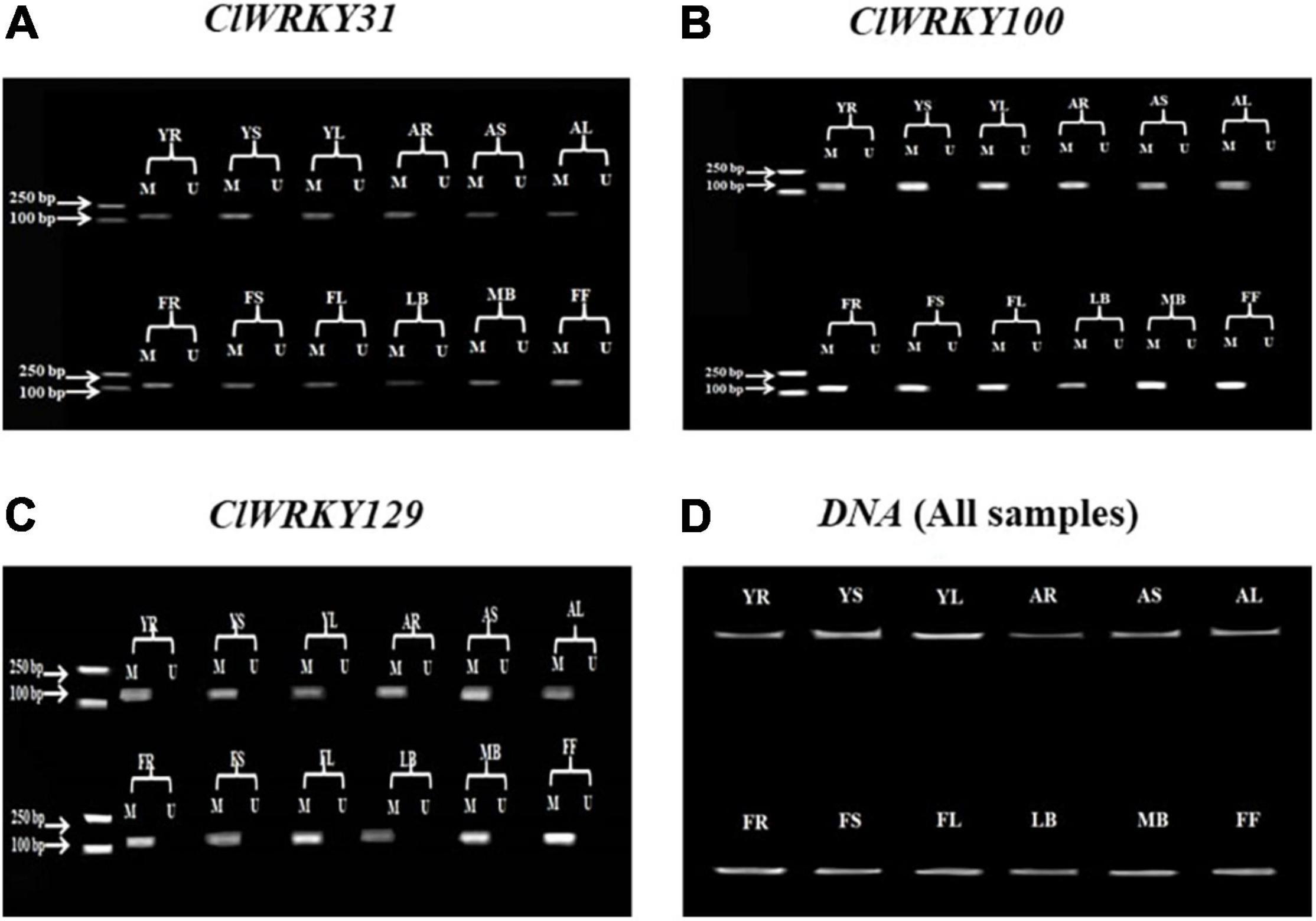
Figure 5. ClWRKY31, ClWRKY100, and ClWRKY129 Methylation-Specific Polymerase Chain Reaction (MSP) results (A–C). An MSP was conducted to determine the methylation status of selected genes. The PCR product length of the genes was as follows: ClWRKY31 = 156 bp, ClWRKY100 = 135 bp, and ClWRKY129 = 131 bp. “M” represents a methylated PCR product, and “U” represents an unmethylated PCR product. (D) The genomic DNA results for all samples. These results were verified through gel (1.5%) electrophoresis. The abbreviations of different growth stages and tissues are as follows: YR, young stage root; YS, young stage stem; YL, young stage leaf; AR, adult stage root; AS, adult stage stem; AL, adult stage leaf; FR, flowering stage root; FS, flowering stage stem; FL, flowering stage leaf; LB, little buds; MB, median buds; FF, first flower at just opening time.
The cis-acting elements in the promoter region are really important for gene expression (Rombauts et al., 1999). The 2,000 bp sequence upstream of the genes was submitted to the PlantCARE software and searched for cis-acting elements. A range of cis-acting elements was identified in the promoter region of the ClWRKY family, which were highly diverse.
A total of 110 cis-acting elements were found in the promoter region of ClWRKY members. Most of these elements have a diverse role in the plant life cycle. According to our objectives, we screened out the elements that can play a role in plant growth and development and particularly in flowering induction. Therefore, we selected 15 cis-acting elements on the basis of their possible role in flowering-related traits and the overall development of the plant. The detailed features of the 15 cis-acting elements are provided in Supplementary Table 4.
Most of the genes have important cis-acting elements, such as auxin responsiveness (AuR) and gibberellin responsive elements (GRE), associated with flowering-related traits (Figure 6). W-box was most widely available among the ClWRKY family. ClWRKY45 and ClWRKY36, the genes that resulted in high expression at the capitulum stage, had some important elements involved in endosperm expression, elements involved in auxin responsiveness, and ethylene-responsive elements (AuR, EE, and ERE), which are supposed to play a role in flower development. Methyl jasmonate (MeJAR), which plays important role in flowering induction, was present in most of the ClWRKY members, especially in group III members, which indicates that WRKY members may play a role in the overall development of a plant.
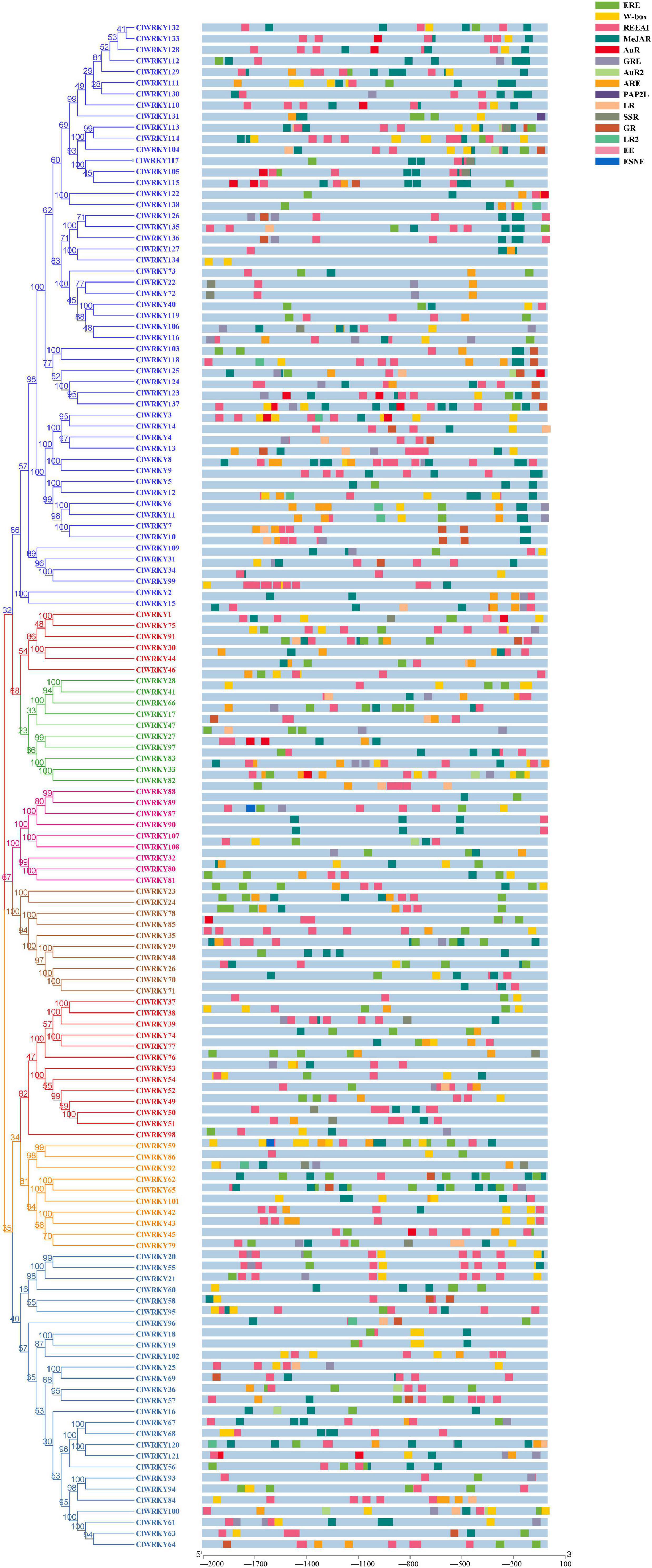
Figure 6. Cis-acting element analysis in the promoter region of the ClWRKY family. The PlantCARE software was used to search the cis-acting elements. Abbreviation of important cis-acting elements are as follows: EE, endosperm expression; ARE, auxin-responsive elements; GR, gibberellin responsiveness; LR, light responsiveness; REEAI, regulatory element essential for anaerobic induction; AuR, elements involved in auxin responsiveness; SSR, seed-specific regulation; MeJAR, methyl jasmonate responsiveness; ERE, ethylene-responsive elements; GRE, gibberellin responsive elements; ESNE, endosperm specific negative expression; LR2, elements involved in light response; PAP2L, plant-AP2 such as AuR2, part of auxin responsiveness; and W-box, recognition site of WRKY TFs.
The de novo motif analysis in MEME identified 15 motifs (Supplementary Figure 2). Tomtom tool comparison showed homology with some important motifs, such as motifs similar to MEME12 and MEME14, which are involved in seed germination, pollen tube growth, and WRKY binding sites (Supplementary Table 5). The enriched motifs identified in MEME are mostly different from the motifs found in the PlantCare software. The simple enrichment analysis (SEA) in MEME identified 95 known enriched motifs. Among them, 49 motifs encoded ethylene, which is essential for plant growth and development. Other important motifs included are WRKY binding sites, TEOSINTE BRANCHED, cycloidea and PCF (TCP) binding sites, and so on (Supplementary Table 6).
Different CIWRKY genes having special characters were selected on the basis of previous analysis. Total RNA was extracted from different tissues (root, stem, leaves, and flowers) of C. lavandulifolium at different stages (young, adult, and flowering). The expression pattern of 19 WRKY genes was observed in all tissues at different stages of the plant’s life span (Figure 7). Among them, 10 genes were selected due to the presence of CG-rich regions, and nine genes were selected as they showed close association with important Arabidopsis WRKY genes that are associated with floral traits. Most of the genes exhibited higher expression at the flowering stage in the capitulum tissues. The highest expression in the capitulum tissues at the flowering stage was exhibited by ClWRKY36, ClWRKY45, ClWRKY57, ClWRKY59, and ClWRKY82 (Figure 7). The expression of these genes was higher in the flowering stage, especially in the first flowering (FF) stage and median bud stage, which suggests their role in flowering regulation. These genes were selected for qRT-PCR as they clustered with flowering-related AtWRKY genes (Figure 2). On the basis of previous analysis, ClWRKY36 clustered with CIWRKY71, so it might have a basic role in the control of flowering time and plant development, as WRKY71 homolog was found to promote flowering in woodland strawberries (Lei et al., 2020). ClWRKY31, ClWRKY33, ClWRKY41, ClWRKY97, ClWRKY100, ClWRKY129, and ClWRKY130, which were selected on the basis of having CpG rich regions (Supplementary Table 1), showed higher expression in capitulum tissues (Figure 7). In addition, ClWRKY31, ClWRKY100, and ClWRKY129 were detected with DNA methylation (Figure 5), which is linked with flowering regulation and is one of the basic sources of epigenetics. Interestingly, ClWRKY83 remains stable across all the tissues and stages as it is constantly expressed in all samples of C. lavandulifolium. It might play a role in adaptability. Moreover, ClWRKY83 showed close association with important AtWRKY7 of Arabidopsis. The other WRKY member’s expressions showed less or non-significant changes at different stages in all tissues.
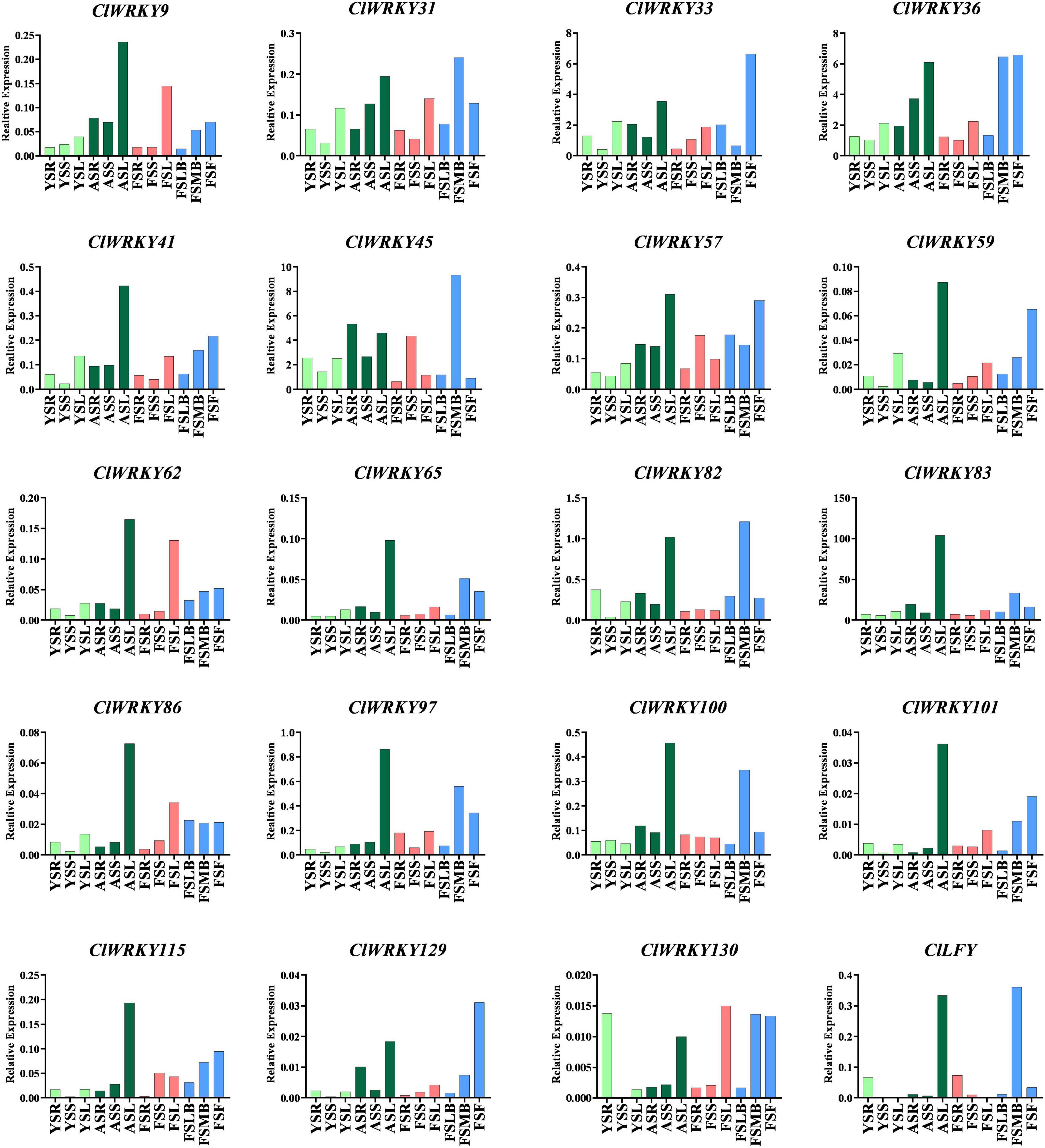
Figure 7. Expression analysis of the selected ClWRKY genes in different tissues (capitulum, leaf, stem, and root) at different stages (young, adult, and flowering). Expression of 19 genes: 10 genes were selected due to the presence of CpG island, nine genes were selected based on their close association with important Arabidopsis genes found in the phylogenetic tree built between C. lavandulifolium and A. thaliana. Abbreviations of different growth stages and tissues are as follows: YSR, young stage root; YSS, young stage stem; YSL, young stage leaf; ASR, adult stage root; ASS, adult stage stem; ASL, adult stage leaf; FSR, flowering stage root; FSS, flowering stage stem; FSL, flowering stage leaf; FSLB, Flowering stage little buds; FSMB, Flowering stage buds; FSF, Flowering stage first flower.
We also investigated CILFY (leafy), as it not only promotes flowering time but also influences other genes that regulate flowering-related traits (Yang et al., 2016). The expression of this gene was high in the median bud and adult leaf stages, but low in stem and root (Figure 7). Besides, ClLFY may promote flowering time in C. lavandulifolium by making association with ClWRKY36 and ClWRKY45.
WRKY TFs are one of the largest transcription families, playing an important role in plant development and resistance against different biotic and abiotic stresses. WRKY TFs are spread broadly across higher plants (Li et al., 2014). WRKY family investigation is limited to C. morifolium with 15 genes (Song A. et al., 2014). Being a model plant for chrysanthemums (Dong-ru et al., 2019), the characterization of the WRKY gene family in C. lavandulifolium is of great importance. For the first time, we identified 138 WRKY members in C. lavandulifolium. Out of 138 members, 118 were properly conserved in the “WRKYGQK” domain. Substitution of glutamine occurs instead of lysine in thirteen WRKY heptapeptide domains. This WRKYGKK sequence is reported to be a major variant in many studies (Song H. et al., 2014; Song et al., 2016a,b). ClWRKY12 (WRKYGHK) “Q” replaced by “H” ClWYKY20 and 21 (WRKYGQN) “K” replaced by “N,” ClWRKY27 (WKKYGEQK) “K” replaced by “Q,” ClWRKY97 (WKKYGEK) “Q” replaced by “E,” and ClWRKY131 (WRKNGQK) “Y” replaced by “N” as their respective domains were also reported in carrot (Nan and Gao, 2019). The variation in the heptapeptide domain of WRKY genes has been largely demonstrated (Yamasaki et al., 2005; Brand et al., 2013; Rinerson et al., 2015; Yang et al., 2020); however, variations in aa sequence were different from our findings. The variation in the WRKYGQK conserved domain mainly occurs from Q to K aa. The same kind of substitution was observed in the WRKY gene family of Camelina sativa (Song et al., 2020).
The ClWRKY gene family is composed of 138 genes, which is bigger than the AtWRKY gene family (Rushton et al., 2010). The ClWRKY family was classified into three groups. The classification of ClWRKY members was according to the classification of the WRKY family in Arabidopsis. Most of the ClWRKY members clustered in the corresponding groups. However, some of the ClWRKY members with one WRKY domain clustered in group I, which included, ClWRKY58, 95, 60, 56, 20, 21, and ClWRKY55. Among them, ClWRKY20, 21, and 55 possessed a unique pattern “WNSIVV” which was conserved in these three members. Clustering of one domain WRKY of factors in group I is consistent with the study of the WRKY TF family in Linum usitatissimum (Yuan et al., 2021). The same results are reported in Cicer arietinum (Kumar et al., 2016). Some WRKY members with one conserved domain and clustered into group I have been reported in peanut (Arachis hypogaea L.) (Zhao et al., 2020).
The intron-exon structure seems to be diverse, starting from 1 to 9 introns. Diversification in intron-exon structure may play an important role in the evolution of genes (Xu et al., 2012). In this study, we have identified different intron constitutions within gene groups. Group I has four introns on average, group IIa has 5, group IIb has 4, and all other groups have an average of three introns. This suggests that the latter groups originated from group I and earlier subgroups of group II. The same trend of intron presence in different groups of WRKY members in eggplant was reported (Yang et al., 2020). A total of six motifs were detected; among them, two motifs were considered to be WRKY domains. The conserved domain “WRKYGQK” was almost included in all of the members; however, some of the WRKY members showed a slight variation as “WRKYGKK” domain as suggested by Liu et al. (2021) and Qu et al. (2021).
The cis-acting elements are involved in plant development and flowering induction. There are several elements, such as auxin, gibberellins, and jasmonic acid, which affect flowering-related traits. Jasmonates, cytokinins, and auxin control second bud growth in chrysanthemum (Sun et al., 2021). Auxin controls plant growth and development (Singla et al., 2006). The petal is the most important factor influencing the chrysanthemum’s ornamental value. Auxin signaling is associated with petal growth in chrysanthemums (Wang J. et al., 2017). AuR is found in most ClWRKY members and may play a role in plant development and flowering induction. GRE was abundant in ClWRKY, an important hormone associated with flowering induction in chrysanthemum (Dong et al., 2017). CmBBX24 regulates flowering in chrysanthemum in conjunction with gibberellin synthesis (Zhu et al., 2020). In addition, the key recognition site for the WRKY gene, the W-box containing the TGAC sequence, was observed in most ClWRKY members. The W-box contributes under abiotic stress conditions (Dhatterwal et al., 2019) by influencing the early senescence of rice flag leaves (Liu et al., 2016). The presence of flower-associated cis-acting elements in most genes indicates the potential of ClWRKY to play a role in flowering control in C. lavandulifolium.
DNA methylation has an impact on growth and development, with CG content, or CpG, being the most abundant source of DNA methylation (Dong-ru et al., 2019). Various methods such as methylation-sensitive amplification polymorphism and MSP have been applied to methylated genes (Herman et al., 1996; Coronel et al., 2018). In this study, we have identified ten genes with strong CpG content in the promoter region (Table 1), which may cause epigenetic changes in C. lavandulifolium (Chip and Dresselhaus, 2019). Three genes having methylated sites, i.e., ClWRKY31, ClWRKY100, and ClWRKY129, were identified on the basis of MSP results. These genes can regulate gene expression and may play a role in flowering-related traits, as they are expressed significantly in capitulum tissues at the flowering stage. CmMET1 can affect methylation and regulate candidate genes associated with flowering in C. morifolium (Kang et al., 2021). DNA methylation changes in the promoter and gene body regions can influence gene expression and function of the protein (Miguel and Marum, 2011), the morphology of floral characters, photoperiod-related sensitivity, and fruit ripening (Sun et al., 2014; Xiao et al., 2020). We studied only four stages to detect methylation, and it will be better to explore the methylation status in more stages in the future.
This study focuses on the expression of ClWRKY members to find their role in plant development, especially their influence over flower induction. The expression analysis revealed variations of different genes in different organs. ClWRKY45 showed significant expression at the flowering stage in the capitulum tissue, which may be related to some morphological characters and floral organ development. In the same fashion, ClWRKY45 clustered with Arabidopsis AtWRKY71, a prominent gene that controls flowering characteristics (Yu et al., 2016, 2018), providing strong evidence for our results. Moreover, ClWRKY36 and ClWRKY57 are significantly expressed in capitulum at the flowering stages and have close homology with Arabidopsis AtWRKY2 and AtWRKY34, indicating the role of CIWRKY36 and ClWRKY57 in flowering, as Arabidopsis homologs are the key regulators of pollen development (Lei et al., 2017). ClWRKY59 is expressed significantly in the first flower tissue and can positively regulate flowering, as ClWRKY59 homolog, AtWRKY75, promoted flowering in Arabidopsis (Zhang et al., 2018). ClWRKY82 exhibited higher expression in the median bud stage, which indicates its potential to play a role in flowering regulation. AtWRKY7, a ClWRKY82 homolog, regulates plant growth (Chen et al., 2019). The qRT-PCR results indicate that ClWRKY83 is a stable gene as it expresses across all the tissues at different stages of plant life and may play a role in adaptability. The higher expression of three genes in capitulum tissues, including ClWRKY31, ClWRKY100, and ClWRKY129, which are methylated as well, can play an important role in flowering-related traits. These results suggest that ClWRKY33, ClWRKY36, and ClWRKY45, along with ClWRKY31, ClWRKY100, and ClWRKY129, can play a crucial role in the genetic and epigenetic improvement of the chrysanthemum. ClWRKY45, ClWRKY36, and ClWRK129, in particular, have the potential to alter flowering-related traits. The overall investigation of the CIWRKY domain is providing theoretical insight into C. lavandulifolium transcriptional activation and inactivation mechanisms for the improvement of plant development.
This study aimed to identify the WRKY gene family in C. lavandulifolium and explore the potential of WRKY genes for genetic improvement. A total of 138 WRKY genes were analyzed in C. lavandulifolium. All the data results are unique to explore the underlying mechanism of the WRKY TF in C. lavandulifolium plant improvement. The gene structure analysis suggested the vast variation among WRKY members. The WRKY domain remains conserved across all the members of this family. Ten WRKY genes possessed CG-rich content in the promoter region where three genes exhibit methylation status. This research can provide a basis to study the role of WRKY genes in the improvement of chrysanthemum plants, especially in flowering traits.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WZ designed the research project, coordinated, and supervised the study. MA performed all the experiments and wrote the manuscript. MA and KD did most of the experimental work and analyzed the data. WuY, AP, and WaY performed specific experiments during the research. All authors read and approved the final manuscript.
This research was supported by the National Key Research and Development Plan (2018YFD1000403).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank all the colleagues who helped them a lot during this research. We specially thank WZ and KD for their support and timely response.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.861193/full#supplementary-material
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Bailey, T. L., and Grant, C. E. (2021). SEA: Simple Enrichment Analysis of motifs. bioRxiv [Preprint]. doi: 10.1101/2021.09.02.458722
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME Suite. Nucleic Acids Res. 43, W39–W49.
Bi, C., Xu, Y., Ye, Q., Yin, T., and Ye, N. (2016). Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. PeerJ 4:e2437. doi: 10.7717/peerj.2437
Bi, M., Li, X., Yan, X., Liu, D., Gao, G., Zhu, P., et al. (2021). Chrysanthemum WRKY15-1 promotes resistance to Puccinia horiana Henn. via the salicylic acid signaling pathway. Hortic Res. 8:6. doi: 10.1038/s41438-020-00436-4
Brand, L. H., Fischer, N. M., Harter, K., Kohlbacher, O., and Wanke, D. (2013). Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 41, 9764–9778. doi: 10.1093/nar/gkt732
Cai, Y., Chen, X., Xie, K., Xing, Q., Wu, Y., Li, J., et al. (2014). Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS One 9:e102529. doi: 10.1371/journal.pone.0102529
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., and Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10:281. doi: 10.1186/1471-2229-10-281
Chen, W., Hao, W. J., Xu, Y. X., Zheng, C., Ni, D. J., Yao, M. Z., et al. (2019). Isolation and characterization of CsWRKY7, a subgroup IId WRKY transcription factor from Camellia sinensis, linked to development in arabidopsis. Int. J. Mol. Sci. 20:2815. doi: 10.3390/ijms20112815
Chip, I., and Dresselhaus, T. (2019). Chapter 12 and Reproductive Development in Cereals Using Chromatin. Cereal Genomics2072, 141–156. doi: 10.1007/978-1-4939-9865-4_12
Coronel, C. J., González, A. I., Ruiz, M. L., and Polanco, C. (2018). Analysis of somaclonal variation in transgenic and regenerated plants of Arabidopsis thaliana using methylation related metAFLP and TMD markers. Plant Cell Rep. 37, 137–152. doi: 10.1007/s00299-017-2217-x
Dhatterwal, P., Basu, S., Mehrotra, S., and Mehrotra, R. (2019). Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci Rep. 9:1681. doi: 10.1038/s41598-019-38757-7
Dong, B., Deng, Y., Wang, H., Gao, R., Stephen, G. K., Chen, S., et al. (2017). Gibberellic acid signaling is required to induce flowering of chrysanthemums grown under both short and long days. Int. J. Mol. Sci. 18:1259. doi: 10.3390/ijms18061259
Dong-ru, K., Si-lan, D., Kang, G., Fan, Z., and Hong, L. (2019). Morphological variation of Chrysanthemum lavandulifolium induced by 5-azaC treatment. Sci. Hortic. 257:108645. doi: 10.1016/j.scienta.2019.108645
Duan, M. R., Nan, J., Liang, Y. H., Mao, P., Lu, L., Li, L., et al. (2007). DNA binding mechanism revealed by high resolution crystal structure of Arabidopsis thaliana WRKY1 protein. Nucleic Acids Res. 35, 1145–1154. doi: 10.1093/nar/gkm001
Fan, Q., Song, A., Jiang, J., Zhang, T., Sun, H., Wang, Y., et al. (2016). CmWRKY1 enhances the dehydration tolerance of chrysanthemum through the regulation of ABA-aociated genes. PLoS One 11:e0150572. doi: 10.1371/journal.pone.0150572
Fan, Q., Song, A., Xin, J., Chen, S., Jiang, J., Wang, Y., et al. (2015). CmWRKY15 facilitates Alternaria tenuissima infection of chrysanthemum. PLoS One 10:e0143349. doi: 10.1371/journal.pone.0143349
Fu, J., Wang, Y., Huang, H., Zhang, C., and Dai, S. (2013). Reference gene selection for RT-qPCR analysis of Chrysanthemum lavandulifolium during its flowering stages. Mol. Breed. 31, 205–215. doi: 10.1007/s11032-012-9784-x
Gupta, S., Stamatoyannopoulos, J. A., Bailey, T. L., and Noble, W. S. (2007). Quantifying similarity between motifs. Genome Biol. 8:R24, doi: 10.1186/gb-2007-8-2-r24
He, L., Wu, Y. H., Zhao, Q., Wang, B., Liu, Q. L., and Zhang, L. (2018). Chrysanthemum DgWRKY2 gene enhances tolerance to salt stress in transgenic chrysanthemum. Int. J. Mol. Sci. 19:2062. doi: 10.3390/ijms19072062
Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D., and Baylin, S. B. (1996). Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands (DNA methylation/tumor suppressor genes/pl6/p15). Med. Sci. 93, 9821–9826. doi: 10.1073/pnas.93.18.9821
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, H., Cao, H., Niu, Y., and Dai, S. (2012). Expression Analysis of Nudix Hydrolase Genes in Chrysanthemum lavandulifolium. Plant Mol. Biol. Rep. 30, 973–982. doi: 10.1007/s11105-011-0401-7
Ishiguro, S., and Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5’ upstream regions of genes coding for sporamin and β-amylase from sweet potato. MGG Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jaffar, M. A., Song, A., Faheem, M., Chen, S., Jiang, J., Liu, C., et al. (2016). Involvement of CmWRKY10 in drought tolerance of chrysanthemum through the ABA-signaling pathway. Int. J. Mol. Sci. 17:693. doi: 10.3390/ijms17050693
Kang, D. R., Zhu, Y., Li, S. L., Ai, P. H., Khan, M. A., Ding, H. X., et al. (2021). Transcriptome analysis of differentially expressed genes in chrysanthemum MET1 RNA interference lines. Physiol. Mol. Biol. Plants 27, 1455–1468. doi: 10.1007/s12298-021-01022-1
Kumar, K., Srivastava, V., Purayannur, S., Kaladhar, V. C., Cheruvu, P. J., and Verma, P. K. (2016). WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): Comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene(s). DNA Res. 23, 225–239. doi: 10.1093/dnares/dsw010
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lei, R., Li, X., Ma, Z., Lv, Y., Hu, Y., and Yu, D. (2017). Arabidopsis WRKY2 and WRKY34 transcription factors interact with VQ20 protein to modulate pollen development and function. Plant J. 91, 962–976. doi: 10.1111/tpj.13619
Lei, Y., Sun, Y., Wang, B., Yu, S., Dai, H., Li, H., et al. (2020). Woodland strawberry WRKY71 acts as a promoter of flowering via a transcriptional regulatory cascade. Hortic Res. 7:137. doi: 10.1038/s41438-020-00355-4
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, H. L., Guo, D., Yang, Z. P., Tang, X., and Peng, S. Q. (2014). Genome-wide identification and characterization of WRKY gene family in Hevea brasiliensis. Genomics 104, 14–23. doi: 10.1016/j.ygeno.2014.04.004
Li, P., Song, A., Gao, C., Jiang, J., Chen, S., Fang, W., et al. (2015a). The over-expression of a chrysanthemum WRKY transcription factor enhances aphid resistance. Plant Physiol. Biochem. 95, 26–34. doi: 10.1016/j.plaphy.2015.07.002
Li, P., Song, A., Gao, C., Wang, L., Wang, Y., Sun, J., et al. (2015b). Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep. 34, 1365–1378. doi: 10.1007/s00299-015-1793-x
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Li, S., Fu, Q., Huang, W., and Yu, D. (2009). Functional analysis of an Arabidopsis transcription factor WRKY25 in heat stress. Plant Cell Rep. 28, 683–693. doi: 10.1007/s00299-008-0666-y
Li, W., Wang, H., and Yu, D. (2016). Arabidopsis WRKY Transcription Factors WRKY12 and WRKY13 Oppositely Regulate Flowering under Short-Day Conditions. Mol. Plant 9, 1492–1503. doi: 10.1016/j.molp.2016.08.003
Liang, Q. Y., Wu, Y. H., Wang, K., Bai, Z. Y., Liu, Q. L., Pan, Y. Z., et al. (2017). Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 7:4799. doi: 10.1038/s41598-017-05170-x
Lin, L. Z., and Harnly, J. M. (2010). Identification of the phenolic components of chrysanthemum flower (Chrysanthemum morifolium Ramat). Food Chem. 120, 319–326. doi: 10.1016/j.foodchem.2009.09.083
Liu, J., Wang, X., Chen, Y., Liu, Y., Wu, Y., Ren, S., et al. (2021). Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides. Genomics 113, 3294–3309. doi: 10.1016/j.ygeno.2021.05.011
Liu, L., Xu, W., Hu, X., Liu, H., and Lin, Y. (2016). W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci. Rep. 6:20881. doi: 10.1038/srep20881
Liu, Q. L., Xu, K. D., Pan, Y. Z., Jiang, B. B., Liu, G. L., Jia, Y., et al. (2014). Functional Analysis of a Novel Chrysanthemum WRKY Transcription Factor Gene Involved in Salt Tolerance. Plant Mol. Biol. Rep. 32, 282–289. doi: 10.1007/s11105-013-0639-3
Liu, Q. L., Zhong, M., Li, S., Pan, Y. Z., Jiang, B. B., Jia, Y., et al. (2013). Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, intobacco enhances tolerance to salt stress. Plant Physiol. Biochem. 69, 27–33. doi: 10.1016/j.plaphy.2013.04.016
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, X., Sun, X., Liu, B., Zhu, D., Bai, X., Cai, H., et al. (2013). Ectopic Expression of a WRKY Homolog from Glycine soja Alters Flowering Time in Arabidopsis. PLoS One 8:e73295. doi: 10.1371/journal.pone.0073295
Ma, Z., Li, W., Wang, H., and Yu, D. (2020). WRKY transcription factors WRKY12 and WRKY13 interact with SPL10 to modulate age-mediated flowering. J. Integr. Plant Biol. 62, 1659–1673. doi: 10.1111/jipb.12946
Miguel, C., and Marum, L. (2011). An epigenetic view of plant cells cultured in vitro: Somaclonal variation and beyond. J. Exp. Bot. 62, 3713–3725. doi: 10.1093/jxb/err155
Nan, H., and Gao, L. Z. (2019). Genome-wide analysis of WRKY genes and their response to hormone and mechanic stresses in carrot. Front. Genet. 10:363. doi: 10.3389/fgene.2019.00363
Nguyen, T. K., and Lim, J. H. (2019). Tools for Chrysanthemum genetic research and breeding: Is genotyping-by-sequencing (GBS) the best approach? Hortic. Environ. Biotechnol. 60, 625–635. doi: 10.1007/s13580-019-00160-6
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Qu, R., Cao, Y., Tang, X., Sun, L., Wei, L., and Wang, K. (2021). Identification and expression analysis of the WRKY gene family in Isatis indigotica. Gene 783:145561. doi: 10.1016/j.gene.2021.145561
Ramos, R. N., Martin, G. B., Pombo, M. A., and Rosli, H. G. (2021). WRKY22 and WRKY25 transcription factors are positive regulators of defense responses in Nicotiana benthamiana. Plant Mol Biol. 105, 65–82. doi: 10.1007/s11103-020-01069-w
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., and Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15:66. doi: 10.1186/s12870-015-0456-y
Rombauts, S., Déhais, P., Van Montagu, M., and Rouzé, P. (1999). PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 27, 295–296. doi: 10.1093/nar/27.1.295
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., and Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol Biol. 83, 265–277. doi: 10.1007/s11103-013-0090-8
Shinoyama, H., Aida, R., Ichikawa, H., Nomura, Y., and Mochizuki, A. (2012). Genetic engineering of chrysanthemum (Chrysanthemum morifolium): Current progress and perspectives. Plant Biotechnol. 29, 323–337. doi: 10.5511/plantbiotechnology.12.0521a
Singla, B., Chugh, A., Khurana, J. P., and Khurana, P. (2006). An early auxin-responsive Aux/IAA gene from wheat (Triticum aestivum) is induced by epibrassinolide and differentially regulated by light and calcium. J. Exp. Bot. 57, 4059–4070. doi: 10.1093/jxb/erl182
Soares, J. M., Weber, K. C., Qiu, W., Stanton, D., Mahmoud, L. M., Wu, H., et al. (2020). The vascular targeted citrus FLOWERING LOCUS T3 gene promotes non-inductive early flowering in transgenic Carrizo rootstocks and grafted juvenile scions. Sci. Rep. 10:21404. doi: 10.1038/s41598-020-78417-9
Song, A., Li, P., Jiang, J., Chen, S., Li, H., Zeng, J., et al. (2014). Phylogenetic and transcription analysis of chrysanthemum WRKY transcription factors. Int. J. Mol. Sci. 15, 14442–14455. doi: 10.3390/ijms150814442
Song, H., Wang, P., Hou, L., Zhao, S., Zhao, C., Xia, H., et al. (2016a). Global analysis of WRKY genes and their response to dehydration and salt stress in soybean. Front. Plant Sci. 7:9. doi: 10.3389/fpls.2016.00009
Song, H., Wang, P., Lin, J. Y., Zhao, C., Bi, Y., and Wang, X. (2016b). Genome-wide identification and characterization of WRKY gene family in peanut. Front. Plant Sci. 7:534. doi: 10.3389/fpls.2016.00534
Song, H., Wang, P., Nan, Z., and Wang, X. (2014). The WRKY transcription factor genes in lotus japonicus. Int. J. Genom. 2014:420128. doi: 10.1155/2014/420128
Song, Y., Cui, H., Shi, Y., Xue, J., Ji, C., Zhang, C., et al. (2020). Genome-wide identification and functional characterization of the Camelina sativa WRKY gene family in response to abiotic stress. BMC Genom. 21:786. doi: 10.1186/s12864-020-07189-3
Sun, D., Zhang, L., Yu, Q., Zhang, J., Li, P., Zhang, Y., et al. (2021). Integrated Signals of Jasmonates, Sugars, Cytokinins and Auxin Influence the Initial Growth of the Second Buds of Chrysanthemum after Decapitation. Biology 10:440 doi: 10.3390/biology10050440
Sun, H., Guo, Z., Gao, L., Zhao, G., Zhang, W., Zhou, R., et al. (2014). DNA methylation pattern of Photoperiod-B1 is associated with photoperiod insensitivity in wheat (Triticum aestivum). New Phytol. 204, 682–692. doi: 10.1111/nph.12948
Turck, F., Zhou, A., and Somssich, I. E. (2004). Stimulus-dependent, promoter-specific binding of transcription factor WRKY1 to its native promoter and the defense-related gene PcPR1-1 in parsley. Plant Cell 16, 2573–2585. doi: 10.1105/tpc.104.024810
Wang, J., Wang, H., Ding, L., Song, A., Shen, F., Jiang, J., et al. (2017). Transcriptomic and hormone analyses reveal mechanisms underlying petal elongation in Chrysanthemum morifolium ‘Jinba.’ Plant Mol. Biol. 93, 593–606. doi: 10.1007/s11103-017-0584-x
Wang, K., Wu, Y. H., Tian, X. Q., Bai, Z. Y., Liang, Q. Y., Liu, Q. L., et al. (2017). Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front. Plant Sci. 8:1592. doi: 10.3389/fpls.2017.01592
Wang, Y., Li, Y., He, S. P., Gao, Y., Wang, N. N., Lu, R., et al. (2019). A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol. Biochem. 141, 231–239. doi: 10.1016/j.plaphy.2019.06.005
Warmerdam, S., Sterken, M. G., Sukarta, O. C. A., Van Schaik, C. C., Oortwijn, M. E. P., Lozano-Torres, J. L., et al. (2020). The TIR-NB-LRR pair DSC1 and WRKY19 contributes to basal immunity of Arabidopsis to the root-knot nematode Meloidogyne incognita. BMC Plant Biol. 20:73. doi: 10.1186/s12870-020-2285-x
Wen, X., Li, J., Wang, L., Lu, C., Gao, Q., Xu, P., et al. (2022). The chrysanthemum lavandulifolium genome and the molecular mechanism underlying diverse capitulum types. Hortic Res. 9:uhab022. doi: 10.1093/hr/uhab022
Wu, J., Zhang, Y., Zhang, H., Huang, H., Folta, K. M., and Lu, J. (2010). Whole genome wide expression profiles of Vitis amurensis grape responding to downy mildew by using Solexa sequencing technology. BMC Plant Biol. 10:234. doi: 10.1186/1471-2229-10-234
Xiao, K., Chen, J., He, Q., Wang, Y., Shen, H., and Sun, L. (2020). DNA methylation is involved in the regulation of pepper fruit ripening and interacts with phytohormones. J. Exp. Bot. 71, 1928–1942. doi: 10.1093/jxb/eraa003
Xu, G., Guo, C., Shan, H., and Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. U.S.A. 109, 1187–1192. doi: 10.1073/pnas.1109047109
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., et al. (2005). Solution structure of an Arabidopsis WRKY DNA binding domain. Plant Cell 17, 944–956. doi: 10.1105/tpc.104.026435
Yang, C., Ye, Y., Song, C., Chen, D., Jiang, B., and Wang, Y. (2016). Cloning and functional identification of the AcLFY gene in Allium cepa. Biochem. Biophys. Res. Commun. 473, 1100–1105. doi: 10.1016/j.bbrc.2016.04.022
Yang, L., Fu, J., Qi, S., Hong, Y., Huang, H., and Dai, S. (2017). Molecular cloning and function analysis of ClCRY1a and ClCRY1b, two genes in Chrysanthemum lavandulifolium that play vital roles in promoting floral transition. Gene 617, 32–43. doi: 10.1016/j.gene.2017.02.020
Yang, Y., Liu, J., Zhou, X., Liu, S., and Zhuang, Y. (2020). Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 8:e8777. doi: 10.7717/peerj.8777
Yokotani, N., Shikata, M., Ichikawa, H., Mitsuda, N., Ohme-Takagi, M., Minami, E., et al. (2018). OsWRKY24, a blast-disease responsive transcription factor, positively regulates rice disease resistance. J. Gen. Plant Pathol. 84, 85–91. doi: 10.1007/s10327-018-0768-5
Yu, Y., Liu, Z., Wang, L., Kim, S. G., Seo, P. J., Qiao, M., et al. (2016). WRKY71 accelerates flowering via the direct activation of FLOWERING LOCUS T and LEAFY in Arabidopsis thaliana. Plant J. 85, 96–106. doi: 10.1111/tpj.13092
Yu, Y., Wang, L., Chen, J., Liu, Z., Park, C. M., and Xiang, F. (2018). WRKY71 Acts Antagonistically Against Salt-Delayed Flowering in Arabidopsis thaliana. Plant Cell Physiol. 59, 414–422. doi: 10.1093/pcp/pcx201
Yuan, H., Guo, W., Zhao, L., Yu, Y., Chen, S., Tao, L., et al. (2021). Genome-wide identification and expression analysis of the WRKY transcription factor family in flax (Linum usitatissimum L.). BMC Genom. 22:375. doi: 10.1186/s12864-021-07697-w
Yue, J., Zhu, C., Zhou, Y., Niu, X., Miao, M., Tang, X., et al. (2018). Transcriptome analysis of differentially expressed unigenes involved in flavonoid biosynthesis during flower development of Chrysanthemum morifolium ‘Chuju.’ Sci. Rep. 8:13414. doi: 10.1038/s41598-018-31831-6
Zhang, L., Chen, L., and Yu, D. (2018). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhang, W., Gao, T., Li, P., Tian, C., Song, A., Jiang, J., et al. (2020). Chrysanthemum CmWRKY53 negatively regulates the resistance of chrysanthemum to the aphid Macrosiphoniella sanborni. Hortic Res. 7:109, doi: 10.1038/s41438-020-0334-0
Zhao, N., He, M., Li, L., Cui, S., Hou, M., Wang, L., et al. (2020). Identification and expression analysis of WRKY gene family under drought stress in peanut (Arachis hypogaea L.). PLoS One 15:e0231396. doi: 10.1371/journal.pone.0231396
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Zhou, X., Jiang, Y., and Yu, D. (2011). WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 31, 303–313. doi: 10.1007/s10059-011-0047-1
Keywords: WRKY transcription factors, ornamental, motifs, cis-acting elements, CpG islands, methylation, flowering traits
Citation: Ayoub Khan M, Dongru K, Yifei W, Ying W, Penghui A and Zicheng W (2022) Characterization of WRKY Gene Family in Whole-Genome and Exploration of Flowering Improvement Genes in Chrysanthemum lavandulifolium. Front. Plant Sci. 13:861193. doi: 10.3389/fpls.2022.861193
Received: 24 January 2022; Accepted: 02 March 2022;
Published: 26 April 2022.
Edited by:
Yang Zhu, Zhejiang University, ChinaReviewed by:
Yifeng Xu, Nanjing Agricultural University, ChinaCopyright © 2022 Ayoub Khan, Dongru, Yifei, Ying, Penghui and Zicheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wang Zicheng, WkNXYW5nQGhlbnUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.