- 1Crop Improvement Division, ICAR-Indian Institute of Pulses Research (ICAR-IIPR), Kanpur, India
- 2All India Coordinated Research Project on Chickpea, ICAR-IIPR, Kanpur, India
- 3Indian Council of Agricultural Research, New Delhi, India
- 4Department of Field Crops, Faculty of Agricultural, Cukurova University, Adana, Turkey
Pea (Pisum sativum L.) is one of the most important and productive cool season pulse crops grown throughout the world. Biotic stresses are the crucial constraints in harnessing the potential productivity of pea and warrant dedicated research and developmental efforts to utilize omics resources and advanced breeding techniques to assist rapid and timely development of high-yielding multiple stress-tolerant–resistant varieties. Recently, the pea researcher’s community has made notable achievements in conventional and molecular breeding to accelerate its genetic gain. Several quantitative trait loci (QTLs) or markers associated with genes controlling resistance for fusarium wilt, fusarium root rot, powdery mildew, ascochyta blight, rust, common root rot, broomrape, pea enation, and pea seed borne mosaic virus are available for the marker-assisted breeding. The advanced genomic tools such as the availability of comprehensive genetic maps and linked reliable DNA markers hold great promise toward the introgression of resistance genes from different sources to speed up the genetic gain in pea. This review provides a brief account of the achievements made in the recent past regarding genetic and genomic resources’ development, inheritance of genes controlling various biotic stress responses and genes controlling pathogenesis in disease causing organisms, genes/QTLs mapping, and transcriptomic and proteomic advances. Moreover, the emerging new breeding approaches such as transgenics, genome editing, genomic selection, epigenetic breeding, and speed breeding hold great promise to transform pea breeding. Overall, the judicious amalgamation of conventional and modern omics-enabled breeding strategies will augment the genetic gain and could hasten the development of biotic stress-resistant cultivars to sustain pea production under changing climate. The present review encompasses at one platform the research accomplishment made so far in pea improvement with respect to major biotic stresses and the way forward to enhance pea productivity through advanced genomic tools and technologies.
Introduction
Pea (Pisum sativum L.), being cultivated throughout the world, either for food, fodder, and feed, is considered an important winter season food legume (Rubiales et al., 2019; Parihar et al., 2020). Cotyledons’ color of pea grains varies from yellow, green, and orange that are used in the human diet in different forms such as dal, stew, chhola, vegetables, snacks, soup, chat, and flour, while whole seeds are mainly used as animal feed (Mahajan et al., 2018; Singh et al., 2018). Nutritionally, pea seeds are considered to have about 21–33% protein and 56–74% carbohydrate, with an average iron, selenium, zinc, and molybdenum of about 97, 42, 41, and 12 ppm, respectively (Parihar et al., 2016, 2021). Therefore, it serves as an important ingredient in providing nutritional security for resources poor people in developing countries. Moreover, its consumption minimizes the risk of several chronic diseases such as diabetes (Marinangeli and Jones, 2011), subsides blood cholesterol levels (Ekvall et al., 2006), improves cardiovascular health (Singh et al., 2013), possesses cancer prevention attributes (Kalt, 2001; Steer, 2006), administers body weight, and improves gastrointestinal affairs (Fernando et al., 2010; Lunde et al., 2011).
It is being cultivated widely across many countries in the world (Parihar et al., 2021). Its worldwide cultivated area has increased from 6.58 to 8.09 mha and production from 10.44 to 16.21 mt since 2010. Canada, Russia, China, India, and the United States are the major pea-producing countries (Parihar et al., 2020); however, the United States shares the highest total production of pea (39.33%), followed by Europe (36.98%) and Asia (18.09%). At present, its average productivity is about 2.0 t/ha globally, which recorded an increase of about 36% in a decade (2007–2017), but the potential productivity of this crop is up to 5.0 t/ha in several countries including Netherland, Denmark, Belgium, Germany, and Finland harvests about 3.45–5.01 t/ha (Toker and Mutlu, 2011). However, countries such as India, China, Australia, and Myanmar are recording very low productivity of less than 2.00 t/ha (FAO, 2021). During the past few decades, the gain in yield of pea (15.3 kg/ha/year) is relatively low as compared to other crops, which could be majorly attributed to the least investment in the pea research program (Rubiales et al., 2019). Also, the susceptibility of a pea toward many abiotic/biotic stress is another reason for low productivity which becomes a serious threat to its sustainable productivity especially under changing climatic conditions (Parihar et al., 2020). The most devastating diseases that affect the productivity of pea are powdery mildew (PM), ascochyta blight (AB), rust (PR), wilt (FW), and root rots (Parihar et al., 2013; Mahajan et al., 2018), of which PM caused by Erysiphe pisi (DC.), E. baeumleri (Magnus) (U. Braun & S. Takam.), and E. trifolii (Grev.) has the potential of reducing seed yield by 25–80% (Warkentin et al., 1996; Ghafoor and McPhee, 2012). PR caused by Uromyces viciae-fabae (Pers.) J. Schröt. or U. pisi (Pers.) de Bary is reported to cause yield losses up to 30% (Barilli et al., 2010, 2018; Singh et al., 2015) while, AB, results due to a mixture of fungal species [Ascochyta pisi (Lib.), Peyronellaea pinodes (Berk. & A. Bloxam), Phoma medicaginis var. pinodella (L.K. Jones), P. Koolunga (Davidson), and P. glomerata (Corda) (Wollenw. & Hochapfel)], is one of the most complex and severe diseases worldwide (Bretag et al., 2006; Tran et al., 2014) with a potential of reducing grain yield by about 60% (Liu et al., 2016). Fusarium root rot (FRR) incited by Fusarium solani f. sp. pisi (W.C. Snyder & H.N. Hansen), which may occur in both dry and wet field conditions, reduces yield significantly (Porter, 2010). Similarly, fusarium wilt (FW) caused by F. oxysporum f. sp. pisi (W.C. Snyder & H.N. Hansen) has about 11 different races (Gupta and Gupta, 2019), of which races 1 and 2 are distributed widely affecting the productivity of pea significantly, whereas races 5 and 6 are sporadically distributed (Infantino et al., 2006; Bani et al., 2018). A disease caused by Aphanomyces euteiches (Drechsler) is common root rot (CRR) and is prevalent in the United States, Europe, and Canada causes wilting of the roots (Wicker et al., 2003; Pilet Nayel et al., 2005; Chatterton et al., 2015; Desgroux et al., 2016; Wu et al., 2018). Several insect pests such as pod borer complex [Helicoverpa armigera (Hübner), Etiella zinckenella (Treitschke), and Polyommatus boeticus L.], bruchid (Bruchus pisorum L.) pea leaf weevil (Sitona lineatus L.), leaf miners [Chromatomyia horticola (Goureau)], stem fly [Melanagromyza phaseoli (Vanschuytbroeck)], aphids [Acyrthospihon pisum (Harris)], and cut worms [Agrotis ipsilon (Hufnagel)] seriously reduce the yield of pea by affecting the crop growth (Sharma, 2000; Yadav and Patel, 2015; Yadav et al., 2019). Pod damage of about 40% has been observed in pea due to pod borer complex infestation (Dahiya and Naresh, 1993).
The development of resistant cultivars to the biotic and abiotic stresses is an outstanding tactic to enhance the productivity of any crop including pea. Therefore, knowledge of the genetics of disease and pest resistance is essentially required to breed the resistant/tolerant cultivars. In addition to this, genomic advances especially the accessibility of draft genome sequence of pea (Kreplak et al., 2019) have facilitated the identification of the genes responsible for disease and pest resistance/tolerance and also helped in uncovering the genetics of quantitatively inherited resistance of several major diseases and pests. Moreover, genomics has also facilitated modernizing the conventional breeding for rapid and precise development of resistant cultivars in crop plants including pea. Information on genetics, genomics, and breeding of biotic stress resistance in pea is scattered and only limited attempts were made to review the different aspects of biotic stress resistance (Fondevilla and Rubiales, 2012; Smýkal et al., 2012; Rubiales et al., 2015; Tayeh et al., 2015a). Recently, Mahajan et al. (2018) discussed the genetic improvement in pea in relation to biotic stresses; however, the information provided was largely related to legumes in general and in brief about pea. Thus, an effort is made through this review to make available the comprehensive information pertaining to genetic and genomic advancement at one platform as well as to share a futuristic road map using modern genomic and genetic tools in pea breeding that could aid the crop breeders in developing high-yielding multiple stress resilient pea cultivars.
Current Status of Genetic Resources
Genetic improvement in a target crop species requires availability and judicious exploitation of genetic resources. Globally, more than 98,000 pea accessions, comprised of advanced breeding lines (13%), landraces (38%), mutant stocks (5%), wild species (2.6%), and cultivars (34%), are available and conserved in diverse genebanks (Smýkal et al., 2015; Warkentin et al., 2015; Rubiales et al., 2019; Coyne et al., 2020). The National Institute for Agricultural Research (INRA), France, Australian Grains Genebank (AGG), N.I. Vavilov Research Institute of Plant Industry, Russia, US Department of Agriculture (USDA), United States, Leibniz Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany, and International Center for Agricultural Research in the Dry Areas (ICARDA), Lebanon are the six leading active pea germplasm repositories in the world with about 8,839, 7,432, 6,790, 6,827, 5,343, and 4,596 accessions, respectively (Figure 1). The National Germplasm Repositories of various countries also hold a good number of pea accessions such as 4,558 accessions in Italy, 3,837 in China, 4,484 in India, 3,298 in the United Kingdom, 2,896 in Poland, 2,849 in Sweden, 2,311 in Ukraine, and 2,110 in Aberystwyth University, United Kingdom. Besides, seven other countries hold > 1,000 accessions of Pisum in their national germplasm treasury (Figure 1). Interestingly, the National Genebank of Israel possesses a collection of crop wild relatives (CWRs) such as Pisum fulvum and P. sativum subsp. elatius var. pumilio, which contributes to about 2% of the entire preserved germplasm (Smýkal et al., 2013, 2015; Warkentin et al., 2015). This share of CWR has accessions to P. fulvum (706), P. s. subsp. elatius (624), P. s. subsp. sativum (syn. P. humile/syriacum; 1562), and P. abyssinicum (540) (Smýkal et al., 2013). Besides CWR and cultivated accessions, 575 and 122 accessions of pea mutant stocks are also available at the John Innes Collection, the United Kingdom and the Institute of Plant Genetics Resources Collection, Bulgaria, respectively (Smýkal et al., 2015). A Targeted Induced Local Lesions in Genomes (TILLING) population of 9,000 lines (Coyne et al., 2020) and fast neutron generated deletion mutant resources (around 3,000 lines) are also available, which are being exploited to identify various developmental genes (Smýkal et al., 2015). Internationally, several web-portals have been developed using the database of pea genetic resources such as the European Cooperative Program on Plant Genetic Resources, Cool Season Food Legume Database, Genetic Resources Information Network and System-wide Information Network for Genetic Resources, and KnowPulse for keeping records and disseminating the information related to pea genetic resources.
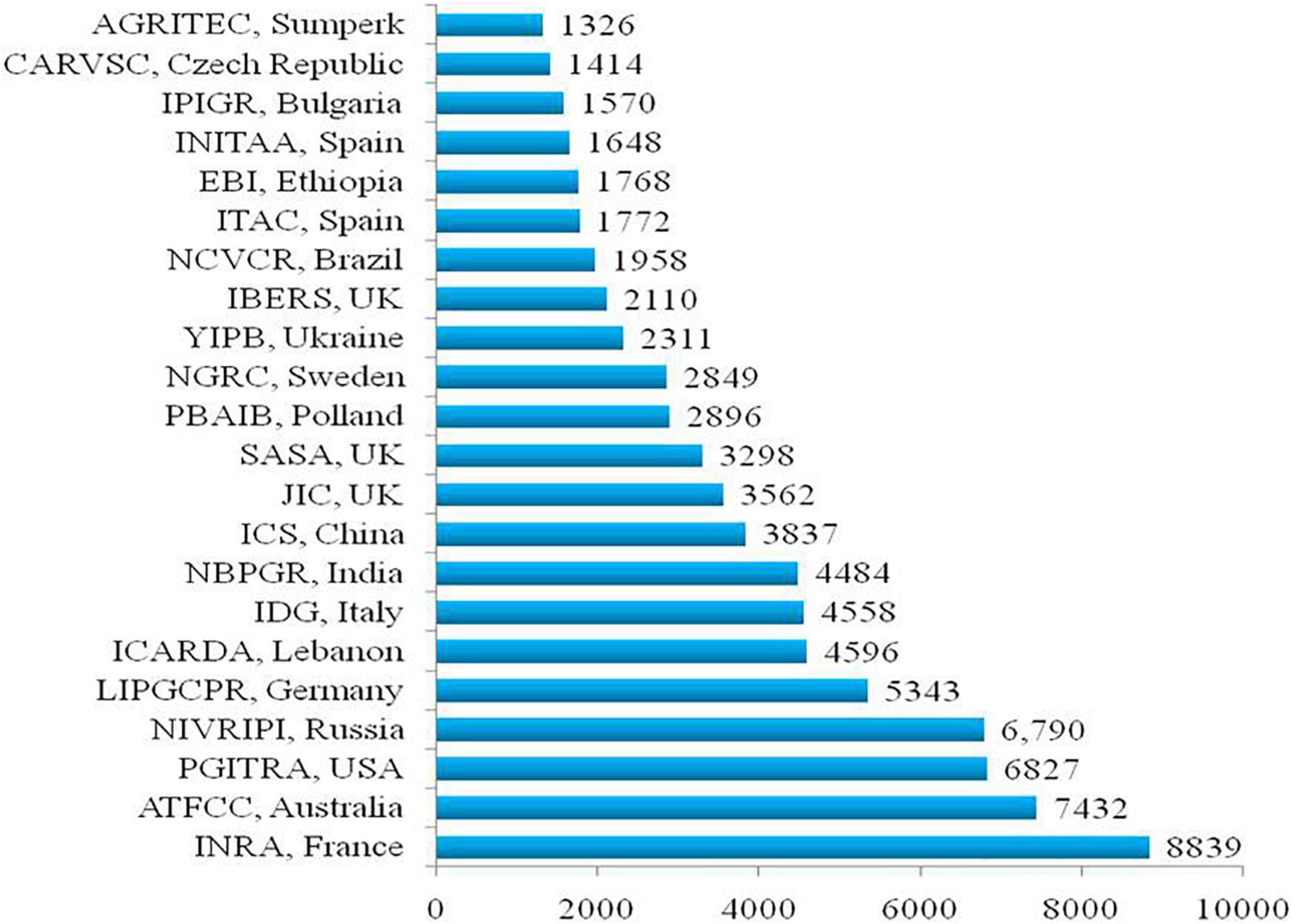
Figure 1. Major pea germplasm holding organizations worldwide (Warkentin et al., 2015; https://www.genesys-pgr.org/).
Crop wild relatives that include Pisum species and subspecies are in general a source of countless fascinating traits including various yield attributing parameters (Mikić et al., 2013). Besides, it is a source of resistance to several biotic stresses, e.g., pea seed weevil (Clement et al., 2002, 2009), PM (Fondevilla et al., 2007b; Esen et al., 2019), PR (Barilli et al., 2010), AB (Jha et al., 2012), and broomrape (Fondevilla et al., 2005). The significance of CWR has been demonstrated by successfully introducing a novel dominant gene (Er3), responsible for resistance to E. pisi from P. fulvum (Sharma and Yadav, 2003; Fondevilla et al., 2008a). Moreover, some P. fulvum accessions were reported to show resistance against bruchid, broomrape, and Mycosphaerella pinodes and are subsequently being utilized in hybridization programs (Fondevilla et al., 2005; Coyne et al., 2020). Similarly, resistance to PR (Barilli et al., 2010, 2018) and AB (Fondevilla et al., 2005; Jha et al., 2012) has been observed in P. fulvum. Diversity for the eIF4E gene and novel alleles for virus resistance has also been identified from CWR (Ashby et al., 2011; Konečná et al., 2014). In a recent report, the relationship between neoplasm and pea weevil (Bruchus pisorum L.) damage was not established in F1 and F2 derived from the inter-subspecific crosses of P. sativum subsp. sativum (with neoplasm) and P. sativum subsp. elatius (without neoplasm) in field conditions (Sari et al., 2020).
Interestingly, the germplasm with the least commercial acceptance in terms of colored seed coat and flowers was accredited as a wonderful resistance source for root rot diseases (Grunwald et al., 2003; Weeden and Porter, 2007) and Aphanomyces (Hamon et al., 2011). Most significantly, the resistance to different biotic stresses can also be transferred from Lathyrus species that are harbored in the tertiary pea gene pool (Patto et al., 2007, 2009), preferably through the utilization of contemporary biotechnological techniques. Most recently, super-early progeny derived from an interspecific cross between P. sativum and P. fulvum flowered in 13–17 days and set pod in 18–29 days after emergence. Such progeny could be used as a complementary to “speed breeding,” to generate more than six generations per year in an appropriate climate compartment (Sari et al., 2021). Significant contributions have been made toward the identification of resistant genetic resources for major biotic stresses in pea (Table 1), which might be utilized in breeding programs and further genetic analysis for the identification of new resistance genes.
Current Knowledge on Genetics for Disease Resistance
Knowledge of genes controlling disease resistance is important to accelerate the success of any breeding program (Shashikumar et al., 2010). Understanding gene action/effects operating in a particular breeding population helps to select a suitable parent for hybridization and breeding procedure for making genetic improvements of resistance against that disease (Sharma et al., 2013). Notably, the pea is acknowledged as the original model organism and was utilized in the finding of Mendel’s laws of inheritance, which laid the foundation for modern plant genetics. In the recent years, inheritance has been studied for resistance attributes of disease in pea by several researchers (Lamprecht, 1948; Yarnell, 1962; Blixt, 1974; Gritton, 1980; Kalloo and Bergh, 1993; Kumar et al., 2006; Amin et al., 2010), and genes were identified and mapped using conventional gene mapping approaches. Varieties with inbuilt resistance are the most appropriate, competent, and economic strategies for tackling biotic stresses. Therefore, comprehensive efforts have been made to understand the inheritance of biotic stresses. Inheritance study for PM revealed that it is being operated by two recessive genes (er1 and er2) and one dominant gene (Er3) (Fondevilla et al., 2007a). A recent report illustrated that PM resistance is operated via er1 owing to the non-functioning of gene PsMLO1 (Humphry et al., 2011). The gene er2 is reported to provide complete resistance to PM but is efficient only in location-specific breeding (Tiwari et al., 1997; Fondevilla et al., 2006), while gene Er3 confers resistance in P. fulvum (Fondevilla et al., 2007a,2010).
With regard to PR resistance, it was reported to be operated by a single dominant gene (Ruf) (Tyagi and Srivastava, 1999); however, the polygenic nature of gene action (Singh and Ram, 2001) and partial dominance of a single gene in conjunction with minor and additive genes (2–3) (Singh et al., 2012) have also been found recently. A single dominant gene governs resistance toward races 1 and 2 of F. oxysporum f. pisi, pea enation mosaic virus, F. solani f. sp. pisi, brown root rot, bacterial blight, downy mildew, and other root rot diseases of pea, whereas a recessive gene regulates resistance to pea seed borne mosaic virus (sbm), yellow bean mosaic virus (mo), pea mosaic virus (pmv), and bean virus (Amin et al., 2010; Mohan et al., 2013). However, Davidson et al. (2004) reported downy mildew to be controlled by a single dominant gene and two complementary recessive genes. The nature of inheritance of AB and FRR resistance has been reported to be regulated by many genes (Kraft, 1992; Fondevilla et al., 2007b; Carrillo et al., 2014b; Jha et al., 2017). The pod resistance for pea weevil is quantitatively controlled whereas the seed resistance is operated by three (pwr1, pwr2, and pwr3) major recessive alleles (Byrne et al., 2008). The neoplasm appearance on pods is controlled by a single dominant gene and its expressivity is influenced by one or a combination of environmental factors (Sari et al., 2020).
Exploitation of Genetic Knowledge Through Traditional Breeding Approaches for Biotic Stress Resistance
Numerous biotic stresses including FW, AB, PM, PR, FRR, and CRR are serious threats to pea production (Bohra et al., 2014). These diseases are reported to occur in a severe form in almost all the pea growing countries. Therefore, efforts have been made to exploit the available genetic knowledge of resistance through conventional breeding for these key biotic stresses for developing resistant cultivars (Fondevilla and Rubiales, 2012; Ghafoor and McPhee, 2012). To develop high yielding pea cultivars possessing PM resistance, three genes, namely, er1, er2, and Er3 have been exploited successfully using conventional breeding approaches (Heringa et al., 1969; Fondevilla et al., 2007c). The er1 gene has the highest existence in resistant pea accessions followed by the er2 gene, which is harbored in restricted accessions (Tiwari et al., 1997). Therefore, the er1 gene that provides resistance through the pre-penetration resistance mechanism has been largely exploited in most pea improvement programs worldwide (Fondevilla et al., 2006). PR is another serious disease, scattered across the countries where the pea is being cultivated. Resistance to PR has been reported to be polygenic (Singh et al., 2012) and oligogenic (Vijayalakshmi et al., 2005). AB or black spot disease is one of the most devastating diseases of peas causing yield setbacks of up to 60% (Xue et al., 1996; Liu et al., 2016). Being seed borne, the rate of transmission from seed to sapling for A. pisi and P. pinodes is 40–100% (Maude, 1966; Xue, 2000), with an ability to remain viable on seeds for 5–7 years (Wallen, 1955). To date, the absolute resistant source for AB has not been identified; however, a prominent scale of resistance was found in accession (P651) of P. fulvum, which is being actively utilized in pea improvement (Wroth, 1998; Sindhu et al., 2014). The polygenic inheritance pattern of AB makes the development of resistant cultivars through conventional breeding very difficult. The FRR is considered a serious bottleneck in harnessing the full potential of a cultivar (Bisby, 1918; Jones, 1923). The condensed soil with a temperature of 18–24°C is the ideal thermal regime for the proliferation of FRR (Kraft and Boge, 2001). Unfortunately, complete resistance to this disease is yet to be explored; however, genetic sources carrying partial tolerance to this disease are available in pea (Gretenkort and Helsper, 1993; Porter, 2010). Noteworthy, the majority of the colored flower accessions portrayed a good level of resistance to FRR as compared to white colored flower accessions (Grunwald et al., 2003). Also, the polygenic inheritance of this disease has made the development of resistant varieties more complicated (Muehlbauer and Kraft, 1973; Kraft, 1992). FW is another severe production menace scattered around the world caused by Fusarium oxysporum. f. sp. pisi and causes absolute yield loss under appropriate environmental circumstances (Aslam et al., 2019). The most favorable soil temperature for FW disease development is 23–27°C. In total, 11 different races of fusarium have been discovered considering its virulence (Gupta and Gupta, 2019); of them, races 1 and 2 have become cosmopolitan; on the contrary, races 5 and 6 are prevailing in some areas (Bani et al., 2018). Among these races, race 1 is considered the most devastating and dominating (Kraft and Pfleger, 2001). Being a soil-borne pathogen, it may outlast for a prolonged period below the ground without pea crop (Gupta and Gupta, 2019). McPhee et al. (1999) recognized resistance sources against races 1 and 2 and used them to breed resistant cultivars. Interestingly, one CWR accession (PI 344012) having resistance to races 1 and 2 has been identified. Knowledge of inheritance is vital for incorporating any attribute of interest in the targeted genotype. Therefore, the inheritance pattern of resistance to Fop races 1, 5, and 6 have been studied and confirmed that it is monogenic with dominance in nature, while resistance to race 2 is regulated quantitatively (McPhee et al., 1999, 2012; Rispail and Rubiales, 2014; Bani et al., 2018). The monogenic dominant resistance is successfully introgressed in many pea cultivars (McPhee, 2003). The integration of quantitatively operated resistance in a targeted background is a cumbersome task wherein molecular markers can support significantly to accelerate the introgression process. For such traits, visual selection always remains long-lasting and labor exhaustive. Thus, modern genomic tools and techniques have paved a way for questing, utilizing, and choosing the naturally available sources of resistance against FW in pea (McClendon et al., 2002; Smýkal et al., 2012).
In pea under congruent circumstances particularly under excess moisture in the soil, CRR reduces grain yield significantly by severe damage to the root framework and subsequent wilting of the infected plant (Wu et al., 2018). Unfortunately, the existing old school disease management approaches such as crop rotation and seed treatments are incapable of controlling this disease completely, owing to the prolonged persistence of the pathogen in the form of oospores, which can contaminate crops at any phase. Consequently, resistant cultivar development has been advocated as an ultimate aim in the pea breeding scheme. Few accessions of pea having moderate resistance to CRR have been identified and subsequently used in breeding programs for developing cultivars (Pilet Nayel et al., 2002, 2005; Roux-Duparque et al., 2004; Moussart et al., 2007; Pilet Nayel et al., 2007; Hamon et al., 2011; McGee et al., 2012; Conner et al., 2013; Hamon et al., 2013; Lavaud et al., 2015). However, polygenic inheritance of this disease and its linkage with some objectionable attributes such as lengthy internodes, anthocyanin content, and delayed-flowering made it difficult to breed CRR-tolerant cultivars (Marx et al., 1972; Pilet Nayel et al., 2002).
Toward Genomic-Based Disease and Insect-Pest Resistance Breeding
Mapping Gene/Quantitative Trait Loci Using Molecular Markers
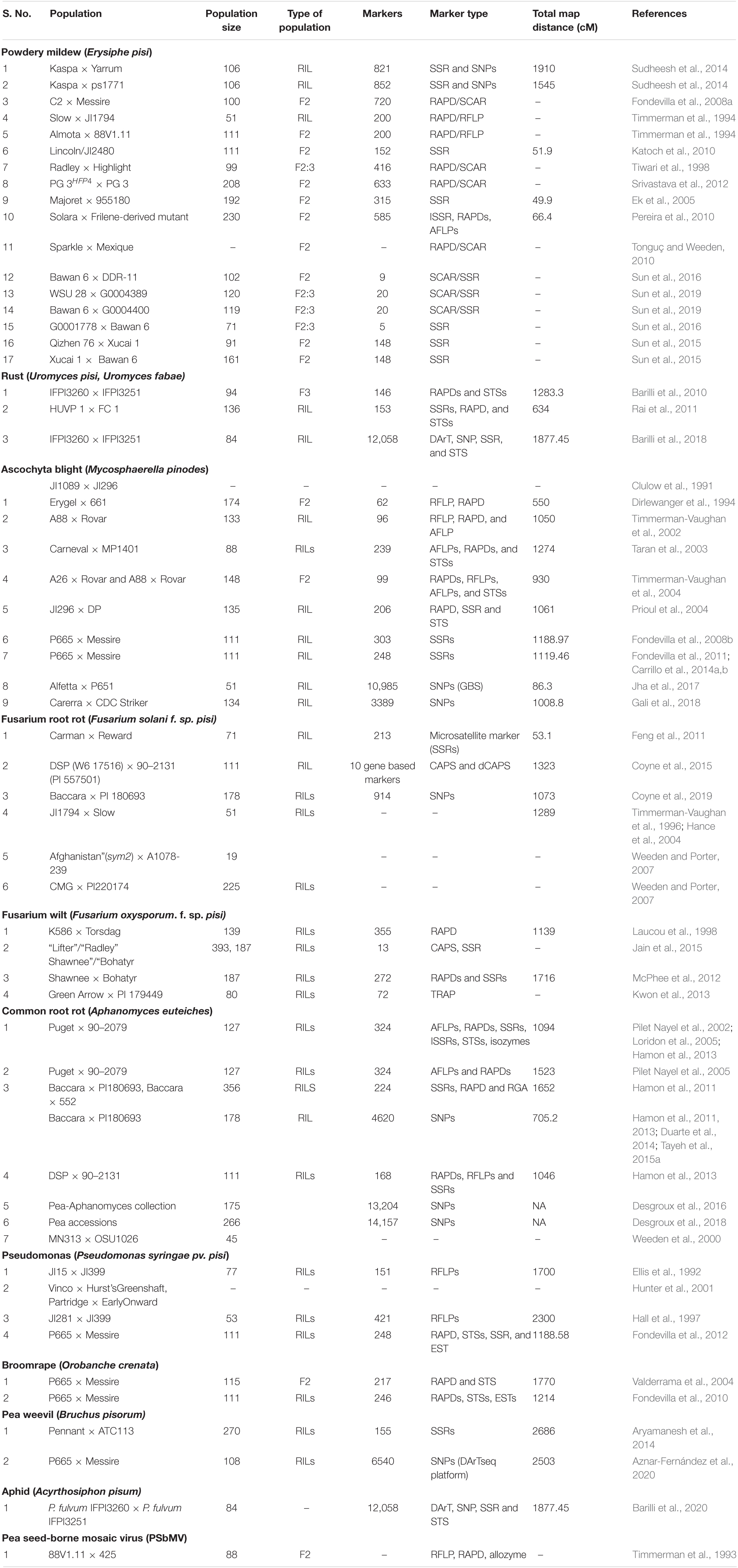
Traditional gene mapping could not be used widely to map the genes/quantitative trait loci (QTLs) regulating disease resistance because of narrow variability and their polygenic inheritance pattern. Moreover, quantitatively inherited traits are highly influenced by environmental conditions; therefore, the DNA-based markers are widely exploited to map genes/QTLs regulating quantitatively inherited traits in pea. In this crop, DNA-based markers that include STMS (Haghnazari et al., 2005); ISSR (Lázaro and Aguinagalde, 2006), SRAP (Esposito et al., 2007), SNP (Duarte et al., 2014), IRAP (Smýkal et al., 2008a), RBIP (Smýkal et al., 2008b), EST-SSR (Teshome et al., 2015), and SSR (Handerson et al., 2014; Negisho et al., 2017; Mohamed et al., 2019) have been developed and successfully utilized to compute genetic variations. However, similar to other crop species, only SSR makers have become popular owing to their low cost, rapidness, polymorphism, and reliable (Snowdon and Friedt, 2004). More recently, next-generation sequencing has authorized the quick discovery of SNPs and the development of an array for genotyping in pea (Leonforte et al., 2013; Duarte et al., 2014; Sindhu et al., 2014). The initial linkage maps were developed in pea utilizing various molecular markers, which were further used in mapping genes/QTLs controlling biotic stress tolerance. The genes such as er 1, er2, and Er3 and their alleles conferring resistance to PM have been mapped using different types of markers (Table 2). In pea, sequencing of cDNA belonging to PsMLO1 has identified a new allele er1-6 of gene er1 that has been validated by a closely linked specific SSR marker (Sun et al., 2016). In addition to this, alleles, namely, er1-8 and er1-9 have been mapped using co-dominant functional markers and validated in pea (Sun et al., 2019). The single dominant gene controlling FW resistance has also been mapped using dominant and co-dominant markers (Jiang, 2013), which were not appropriate for marker-assisted selection (MAS) due to their poor linkage with gene and dominant nature. Thus, Jain et al. (2015) recently designed a co-dominant CAPS marker with 94% accuracy and found that it was helpful in the selection of resistance toward F. oxysporum race 1. QTL mapping has been followed for genes regulating partial or intricate inherited resistance and recognized major or minor QTLs for biotic stress tolerance in pea. For example, molecular mapping has identified one major gene (Ruf)/QTL (Up1, Qruf) and one minor QTL (Qruf1) for PR resistance (Vijayalakshmi et al., 2005; Barilli et al., 2010; Rai et al., 2011). However, markers associated with these genes/QTLs were not close enough (>5.0-cm distance) for utilization in MAS. Further validation of markers linked with QTL Qruf and Qruf1 did not show complete discrimination between PR susceptible and resistant genotypes limiting their application for marker-assisted breeding (MAB) (Singh et al., 2015). However, high-density molecular maps based on SNP makers and the use of isogenic lines (NILs) and heterogeneous inbred family (HIF) populations have provided opportunities for fine mapping of the genes/QTLs and identified more closely linked makers for precise MAS (Mohan et al., 1997; Tuinstra et al., 1997). The SNP marker-mediated linkage mapping has identified three QTLs (UpDSII, UpDSIV, and UpDSIV.2) for PR resistance (Barilli et al., 2018). For AB resistance, various QTL mapping studies have recognized various genomic regions concerned with the regulation of resistance (Table 3; Timmerman-Vaughan et al., 2002; Taran et al., 2003; Fondevilla et al., 2008b). Recently, Jha et al. (2015) have identified SNPs within the linked genes, namely, RGA-G3A (RGA-G3Ap103) and PsDof1 (PsDof1p308), which displayed a noteworthy relationship with AB resistance. Correspondingly in another report association of nine QTLs with resistance to AB has been reported in an interspecific population derived by crossing P. sativum (Alfetta) and P. fulvum (P651), of which, only QTLs abIII-1 and abI-IV-2 were found to be stable over the locations/years (Jha et al., 2016), which were further fine mapped in HIF populations (Jha et al., 2017). Furthermore, selective genotyping was done utilizing genotyping-by-sequencing (GBS) in RILs recognizing eight novel SNP markers within the abI-IV-2 QTL with no extra SNPs in the QTL abIII-1. Similarly, several QTLs explaining phenotypic variation up to 53.4% for polygenic inherited FRR resistance have been recognized using SSR and SNP markers (Coyne et al., 2019). The genome-wide association study (GWAS) refined or validated the previously reported QTLs and identified new loci for resistance to A. euteiches (Desgroux et al., 2016), which identified 52 QTLs including six previously identified QTLs for its resistance. However, Desgroux et al. (2018) employed a comparative GWAS approach for resistance to A. euteiches in a large set of contrasting pea genotypes (266) using 14,157 SNP markers and identified 11 genomic intervals having significant association with resistance to A. euteiches and also confirmed numerous QTLs reported previously. One SNP marker, mapped to the major QTL Ae-Ps7.6, was linked with disease resistance and root system architecture, which can be employed in regular pea breeding programs to reduce root rot incidence in pea.
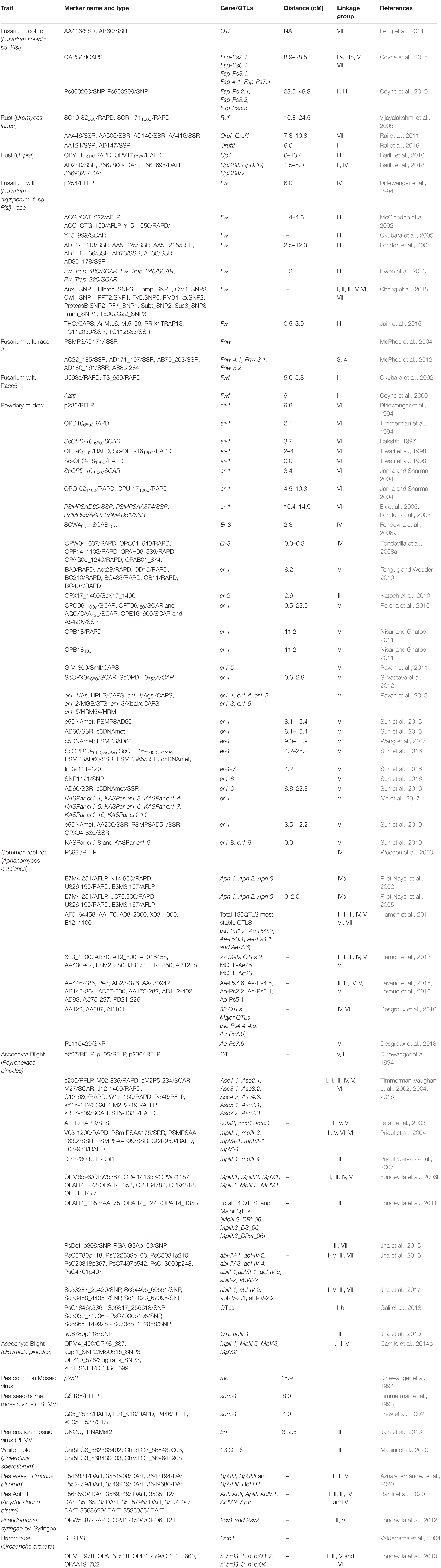
Table 3. Genomic region or markers associated with resistance to different biotic stresses in field pea (Pisum sativum L.).
Marker-Assisted Selection
A close association of markers with a trait of interest is the prerequisite of MAS, which identifies the target traits without assessing their phenotype in the early generation (Tayeh et al., 2015a). Both biparental and association mapping approaches have been utilized in the identification of closely associated markers with genes controlling disease resistance in pea. Such gene-linked markers control resistance to PM (Lakshmana Reddy et al., 2015), pea enation or seed borne mosaic virus (Swisher Grimm and Porter, 2020), FW (Jiang, 2013; Kwon et al., 2013), PR (Singh et al., 2015; Barilli et al., 2018), AB (Carrillo et al., 2014b; Jha et al., 2015, 2017), FRR (Coyne et al., 2019), and CRR (Lavaud et al., 2015; Desgroux et al., 2016) and are available for MAB. The marker-assisted backcrossing (MABC) has been successfully used for the introgression of QTLs for Aphanomyces root rot (ARR) resistance into several recipient genotypes (Hamon et al., 2013; Lavaud et al., 2015). During the recent years, efforts were made to identify markers closely linked with disease resistance genes. However, such markers are not being widely used in the MAB program for developing resistant cultivars due to their poor linkage with target traits. These efforts have proved the utility of MABC and MAS in pea improvement. Accessibility of the reference genome will pave the way toward finding the genes of interest and understanding the genetic background of individuals at the genome level by deploying molecular markers responsive to high-throughput genotyping.
Genomics for Understanding the Complex Genetics of Biotic Stress Response and Identification of Candidate Genes
Resistance in the host plant can occur at different stages during compatibility interaction between pathogen and host. Therefore, many mechanisms, metabolic pathways, and proteins are involved in the host plant and pathogen compatibilities. Thus, many genes have to be expressed to control these metabolic pathways or proteins for completing the infectivity of the pathogen with the host plant. Functional knowledge of these genes can help to understand the genetics involved in host plant resistance, which can further be utilized to develop resistant cultivars against a disease. During the recent years, genomic advances have made it possible to know the candidate genes involved in plant resistance by analyzing transcripts of genes expressed during host–pathogen interaction.
Transcriptomics
Transcriptome analysis has been used to know functional genes responsible for resistance in host plants in many food legumes including pea. In pea, different approaches have been used to recognize the genes responsible for disease and pest resistance (Fondevilla et al., 2011). In the case of white mold [Sclerotinia sclerotiorum (Lib.) de Bary], 2,840 host expressed sequence tags (ESTs) (pea) and 996 pathogen ESTs (S. sclerotiorum) were identified manifesting exclusively amid the host–pathogen interface, of which about 10% of pea ESTs demonstrated their alliance with genes concerned to its defense against various biotic or abiotic stress, whereas about 9% of S. sclerotiorum ESTs exhibited their association with genes reguating pathogenicity or virulence (Zhuang et al., 2012). In another study, microarray analysis investigated gene expression alteration associated with contagion with D. pinodes in pea where 346 genes were found to be regulated differentially between resistant and susceptible response, which was responsible mainly for cell wall build-up, phytoalexin and phenylpropanoid metabolism, genes encoding pathogenesis-associated (PR) proteins, and detoxification processes (Fondevilla et al., 2011). The use of deepSuperSAGE identified 17,561 different UniTags, of which about 70% were known sequences from pea or other plants. Among these, 509 UniTags were differentially articulated (Fondevilla et al., 2014). A similar approach was adopted to identify the candidate genes controlling resistance to bacterial blight infection and found a set of about 651 UniTags that expressed differentially between the resistant and susceptible genotypes (Martín-Sanz et al., 2016). In another study, a transcriptome analysis was used to identify the genes and understand the resistance mechanism against P. pisi and A. euteiches and identified nearly 574 and 817 genes, respectively that were differentially articulated in response to A. euteiches contamination at 6 h post-inoculation (hpi) and 20 hpi, respectively, whereas 544 and 611 genes were expressed differentially against P. pisi at 6 and 20 hpi, respectively (Hosseini et al., 2015). These genes were associated with phenylpropanoid metabolism, strengthening of the cell wall, and hormonal (jasmonic acid, auxin, and ethylene) signaling (Hosseini et al., 2015). In a comparative transcriptome analysis, contrast responding genotypes to E. pisi infection have identified 2,755 transcripts suggesting altered gene expression between the susceptible and resistant genotypes. This study further identified glycolysis as the major pathway of ATP production during pathogen growth and identified genes responsible for putative receptor and regulatory sequences involved in the defense system of resistant genotypes (Bhosle and Makandar, 2021). This information of disease resistant candidate genes can further be utilized for the development of functional markers for MAB.
Proteomics
Disease and pest infestation trigger changes in the protein profile of the host plant. Knowledge of such protein profiles responsible for compatible interaction between host and pathogen can help in better understanding the host plant resistance mechanism at the molecular level. In addition to this, the abundance of specific proteins can be used as the markers for differentiating resistant and susceptible genotypes, which can be utilized in resistance breeding. Therefore, during the recent years, efforts have been made on proteomic analysis for diseases and pests in pea. Resistance to AB is a complex trait, and infection of this disease alters proteins and their abundance. First protein markers linked to AB resistance have been depicted utilizing resistant and susceptible genotypes. Subsequently, quantitative estimation of these proteins was done in a mapping population for the detection of putative protein markers linked with AB resistance and explored its possible use in breeding (Castillejo et al., 2020). This study eventually developed a group of potential protein markers for resistance to AB and advocated a molecular mechanism against AB resistance in pea. Previously, the proteomic approach identified changes in host proteins during infection of downy mildew in a susceptible cultivar of pea (Amey et al., 2008), of which the levels of eight proteins [PI176 (protein accession number P13239), ABR17 (protein accession number Q06931), glycine-rich RNA-binding protein (protein accession number P49311), cytosolic GAPDH (protein accession number P34922), chloroplastic GAPDH (protein accession number P12858), photosystem I reaction center subunit II (protein accession number Q9S7H1), ATP synthase epsilon chain (protein accession number P05039), and photosystem I iron sulfur center (protein accession number P10793)] increased significantly in the infected leaves of the susceptible plant. Identification of these proteins provided the base for the advancement to reveal molecular defense mechanisms to P. viciae infection (Amey et al., 2008). In another study, proteomic analysis of PM susceptible and resistant genotypes resulted in the identification of proteins concerned with photosynthetic activity and carbon metabolism, signal transduction functions, protein synthesis, and protein degradation, which aids in understanding the mechanisms of E. pisi resistance in pea (Curto et al., 2006). Similarly, in a recent study, proteomic analysis was done for PM isolates infecting susceptible pea cultivar and identified proteins involved in virulence and pathogenesis through signal transduction, secondary metabolite formation, and stress functions (Bheri et al., 2019). For understanding the resistance mechanism to Acyrthosiphon pisum (pea aphid), a serious pest of pea, proteomic analysis between contrasting genotypes identified the proteins mostly corresponding to amino acid metabolism, carbohydrate metabolism, folding or degradation, stress response, photosynthesis, signal transduction, and transcription or translation suggesting the role of different metabolic pathways in controlling resistance to this pest (Carrillo et al., 2014a). Thus, proteomic analysis has provided better insight into the molecular mechanism underlying disease and pest resistance in pea, and hence, it is further required to enhance the understanding of the molecular mechanism of quantitatively inherited diseases and pests resistance in pea.
Future Breeding Strategies for Developing Cultivars Resistant to Biotic Stresses
Development of Functional Markers
Poor association of molecular markers with genes/QTLs controlling disease resistance has led to their limited use for MAS in pea breeding programs. Therefore, the development of the functional markers within targeted genes/QTLs controlling the disease resistance is important for this purpose. Earlier, few efforts have been made to develop functional markers for the er1 gene controlling PM in pea (Sun et al., 2016, 2019). A functional co-dominant CAPS marker with 94% accuracy was found useful for the selection of resistance genes responsible for F. oxysporum race 1 (Jain et al., 2015). Furthermore, next-generation sequencing also assisted in developing functional SNP markers from genes/QTLs governing resistance to different diseases in pea. For example, SNP markers within two candidate genes (PsDof1 and RGA-G3A) were identified for AB resistance (Jha et al., 2015). Association mapping with a large number of SNP markers developed through next-generation sequencing identified SNP marker, associated with a major QTL Ae-Ps7.6 responsible for reducing ARR severity and root system architecture (RSA). Therefore, the identified genes for RSA could be utilized in improving ARR incidence in pea. Furthermore, the availability of a reference genome sequence of pea along with a high-throughput next-generation genotyping platform provides the opportunity to identify the candidate genes for targeted traits and development of functional markers linked with disease resistance genes for marker-assisted breeding in pea.
Toward Genomic Selection in Pea
For obtaining maximum genetic gain with more accuracy, genomic selection (GS) using molecular markers is a promising approach. This can help to improve biotic stress resistance, which is a primary breeding objective of the pea genetic improvement program. This approach is more useful for improving quantitatively inherited disease resistance in pea. It uses genome-wide molecular markers associated with resistance genes for predicting and selecting high breeding value lines. In a recent review, different models used in GS were discussed in detail; particularly, the use of multivariate GS models (MTGS) over single trait GS (STGS) was presented (Budhlakoti et al., 2019). Multi-trait GS (MTGS) methods may provide more accurate genomic-estimated breeding values (GEBVs). Several MTGS methods were used for GS, e.g., the multivariate mixed model approach (Jia and Jannink, 2012; Klápšě et al., 2020), Bayesian multi-trait model (Jia and Jannink, 2012; Cheng et al., 2018), multivariate regression with covariance estimation (MRCE) (Rothman et al., 2010), and conditional Gaussian graphical model (cGGM) (Chiquet et al., 2017). Jia and Jannink (2012) presented three multivariate linear models (i.e., GBLUP, Bayes A, and Bayes Cπ) and compared them with univariate models. Most of the successful events of the utilization of GS in biotic stress resistance were in cereal crops. In wheat, GS was used for three types of rust, Fusarium head blight, septoria tritici blotch, PMD, tan spot, and Stagonospora nodorum blotch (Budhlakoti et al., 2022). The genomic prediction accuracies for these diseases ranged from 0.14 to 0.85 (Daetwyler et al., 2010; Rutkoski et al., 2012; Mirdita et al., 2015; Juliana et al., 2019; Sarinelli et al., 2019). Similarly, in the case of rice, GS has been used in blast disease tolerance (Huang et al., 2019). In maize, GS has been used against Stenocarpella maydis causing ear rot (Dos Santos et al., 2016) and heavy infestation of Striga (Badu-Apraku et al., 2019). In the case of barley, for Fusarium head blight, the prediction accuracy was 0.72 (Lorenz et al., 2012; Sallam and Smith, 2016). Though limited reports of the use of genomic selection to improve biotic stresses in pea are available, efforts have been made to know the impact of the marker density, statistical method, and/or the training population size for evaluating genomic prediction accuracy using the number of seeds per plant, thousand seed weight, and flowering time. Such information provides opportunities for developing GS strategies (Tayeh et al., 2015b), which is important for biotic stress tolerance in pea.
Mining Allelic Variants for Resistance Genes
Breeding for improving a trait requires ample availability of diversity in germplasm for the targeted traits. In pea, a large collection of genetic resources is available, which are a reservoir of undiscovered allelic variants for many traits (Tanksley and McCouch, 1997; Smýkal et al., 2012). This large collection may have new resistant allele(s) of the gene(s) controlling disease incidence in pea. For mining such alleles from germplasm, there is a need to test the entire germplasm for their response following a specific screening protocol, which is not only time-consuming but also expensive. However, current genomic tools have provided an opportunity to uncover the allelic variation, especially for those monogenic traits for which candidate genes are already known (Robaglia and Caranta, 2006; Hofinger et al., 2011; Reeves et al., 2012). The use of such genomic tools increases the identification of allelic variants for resistance genes by screening the wild and cultivated germplasm in several crops (Bhullar et al., 2009). In pea, eukaryotic translation initiation factor 4E provides resistance against many potyviruses. Therefore, gene eIF4E encoding this factor has been used for the identification of allelic diversity among 2,803 pea accessions, which resulted in the identification of four eIF4EA-B-C-S variants, whose distribution was geographically linked, suggesting its independent evolution (Konečná et al., 2014). This study has opened an avenue of research for the identification of new allelic variants for complex diseases of a pea.
Toward Epigenetic Breeding
Transgenerational epigenetic variation, which transfers steadily to the next generation, becomes one of the important strategies for breeding climate-resilient cultivars in crop plants. These variations cause alteration in gene expression through DNA methylation or histone modification (Kumar et al., 2019). Identification or genome-wide mapping of epigenetic markers can help the breeder to manipulate epigenomic variability toward the development of climate resilient crop varieties. This epigenetic variation was detected in host plant resistance against a broad array of plant pathogens such as fungi, bacteria, viruses, nematodes, oomycetes, and herbivorous insects (Espinas et al., 2016; Ramirez-Prado et al., 2018; Alonso et al., 2019). For example, in soybean, methylome has been identified for compatible interaction of roots with cyst nematodes (Rambani et al., 2015). In pea, differences have been detected for methylations among plants, which were propagated through in vitro culture for a long time (Smýkal et al., 2007). Artificially induced and naturally occurring epigenetic variations controlling plant disease resistance were identified, and similar efforts are required to identify epigenetic variation responsible for polygenetically inherited disease resistance in pea. In pea, no potential genetic sources for resistance are available so far for many serious diseases, and hence, new epigenetic alleles can be generated using promising approaches such as induced gene-specific DNA methylation and epigenome editing (Zhi and Chang, 2021). Thus, epigenetic breeding has a great potential for improving disease resistance in pea.
Genome Editing
In pea, insect pests and diseases are the major yield-limiting factors and hence pose a substantial threat to food security globally. In recent years, genome editing or modification has revolutionized the functional analyses of genes and the introduction of new alleles for the trait of interest into commercial crop plants (Mushtaq et al., 2019). Different approaches of genome editing have been developed for this purpose; however, clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (CRISPR-Cas9), meganucleases, transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases (ZNFs) are being used extensively for genetic improvement (Mushtaq et al., 2019). In crop plants, susceptibility (S) or resistance (R) genes have been considered eventual targets intended for escalating crop protection (Singh et al., 2016; Ren et al., 2017). These genes were identified as the best candidate for gene editing for conferring disease or pest resistance in a crop (Das et al., 2019b). In addition to this, editing of most conserved regions of multiple viral genomes using multiplex CRISPR/Cas9 system also helped in conferring disease resistance in various crops by interfering with their duplication and progress (Iqbal et al., 2016). In pea, the transcriptomic analysis provides elucidation of the genes and pathways concerned with disease or pest resistance. Moreover, the study of expression alteration, modification, and interaction of protein during the plant-pathogen interface provided knowledge of key proteins involved in pathogenesis. This information is a useful repository for editing or modification of the genome of a crop or realtered pathogen toward the development of resistant cultivars (Barakate and Stephens, 2016). In addition to this, genome editing can be used to alter epi-alleles or to generate new epi-alleles involved in disease resistance (Latutrie et al., 2019).
Transgenic Technology
In pea, limited resistance sources are available among cross-compatible germplasm for several devastating diseases and insect pests such as FRR, CRR, PR, alfalfa mosaic virus, and bruchids. Therefore, transferring resistance genes from other non-cross-compatible species is one of the ways to develop resistant cultivars, possibly by developing transgenic plants. However, genetic transformation in pea is not easy when compared to other legume crops due to difficulties in transformation and plant regeneration (Svabova et al., 2005; Warkentin et al., 2015). Although, during the recent years, advances in biotechnology have made possible the development of transgenics in pea for diseases and insect pests. For example, transgenic lines with two chimeric genes encoding the coat protein (CP) of alfalfa mosaic virus (AMV) strain NZ1 have been developed and tested under green house and field conditions for improved AMV resistance in pea. However, results showed partial virus resistance of transgenic lines having genetically modified AMV CP sequences (Timmerman-Vaughan et al., 2001). In another study, two antifungal genes (chitinase and glucanase) for resistance to fungal diseases have been transferred using genetic transformation, and transgenic pea has been developed by stacking these genes (Amian et al., 2011). Weevils are the most devastating insect of food legumes including pea. Genetic resistance to this insect is not available currently in cross-compatible germplasm. However, a gene for alpha-amylase inhibitor-1 (αAI) has been identified in the common bean that completely protects from weevil destruction. This has been transferred through a genetic transformation in pea, and developed transgenic lines showed resistance to this pest. Moreover, αAI transgenic peas are found to be less allergenic than beans or non-transgenic peas in mice (Reiner et al., 2013).
In a more recent study, four antifungal genes, 1-3 β glucanase (G), endochitinase (C) (belonging to the PR proteins family), polygalacturonase inhibiting proteins (PGIPs) (P), and stilbene synthase (V), have been transformed for disease tolerance in European pea cultivars. This resulted in the development of transgenic lines having an individual antifungal gene or all four genes that were stacked through hybridization. However, the resistance of these transgenic lines against FRR was not consistent over the years in confined field trials probably due to lower relative gene expression in the roots (Kahlon et al., 2018). Although, these studies showed the possibility of developing transgenic pea against major diseases and insect pests. Thus, transgenic technologies have great promise but the economic benefits of genetically modified (GM) pea will need to surpass the regulatory costs, time, and labor involved in bringing a GM crop to market. In addition to this, more research experiments are required on issues associated with genetically modified crops, such as discrete changes in the molecular architecture, cellular function, and antigenicity of the expressed protein translated from the transferred gene in the transgenic plants. In pea, transgenic expression of a plant protein (alpha-amylase inhibitor-1) from the common bean, which is a non-native host of pea, led to the synthesis of a structurally modified form of this inhibitor. The effect of this modified protein has been studied in mice and found that non-native proteins in transgenic plants may lead to structural modification with altered immunogenicity (Prescott et al., 2005).
Speed Breeding
Environmental conditions play an instrumental role in making crop plants susceptible to biotic stresses. The changing environmental condition due to global warming provides opportunities for evolving new races and pathogens, which has significantly raised concern for meeting global food security. Therefore, there is an urgent need of developing resistant cultivars within a short period of time. However, present breeding approaches take several years to develop the resistant cultivars, and hence, the current improvement rate is inadequate to meet the future food demands. Elongated generation advancement time of crops is one of the key reasons for delay in the development of improved resistant cultivars against biotic stresses. Therefore, in recent years, speed breeding has emerged as a powerful tool for accelerating crop research and breeding as several workers have developed speed breeding protocols in pea for shortening the breeding time (Ghosh et al., 2018; Watson et al., 2018; Cazzola et al., 2020). These speed breeding techniques along with new biotechnological tools available in pea can accelerate the development of resistant cultivars against new emerging pathogens or races due to climate changes in the following way:
• Taking 4–5 breeding generations in a year could substantially reduce the time span to release a variety.
• Development of RIL mapping populations within a short period of time using speed breeding can help in the rapid identification of QTLs for disease resistance and their use in the breeding program for developing improved resistant cultivars.
• The MABC for introgression of QTLs/genes controlling disease resistance can be faster through speed breeding leading to the rapid development of improved and resistant cultivars.
• The amalgamation of speed breeding with other modern breeding and biotechnological techniques such as genome editing, genomic selection, and high-throughput genotyping has great potential for accelerating the genetic gain toward the development of biotic stress-tolerant cultivars.
Conclusion and Perspectives
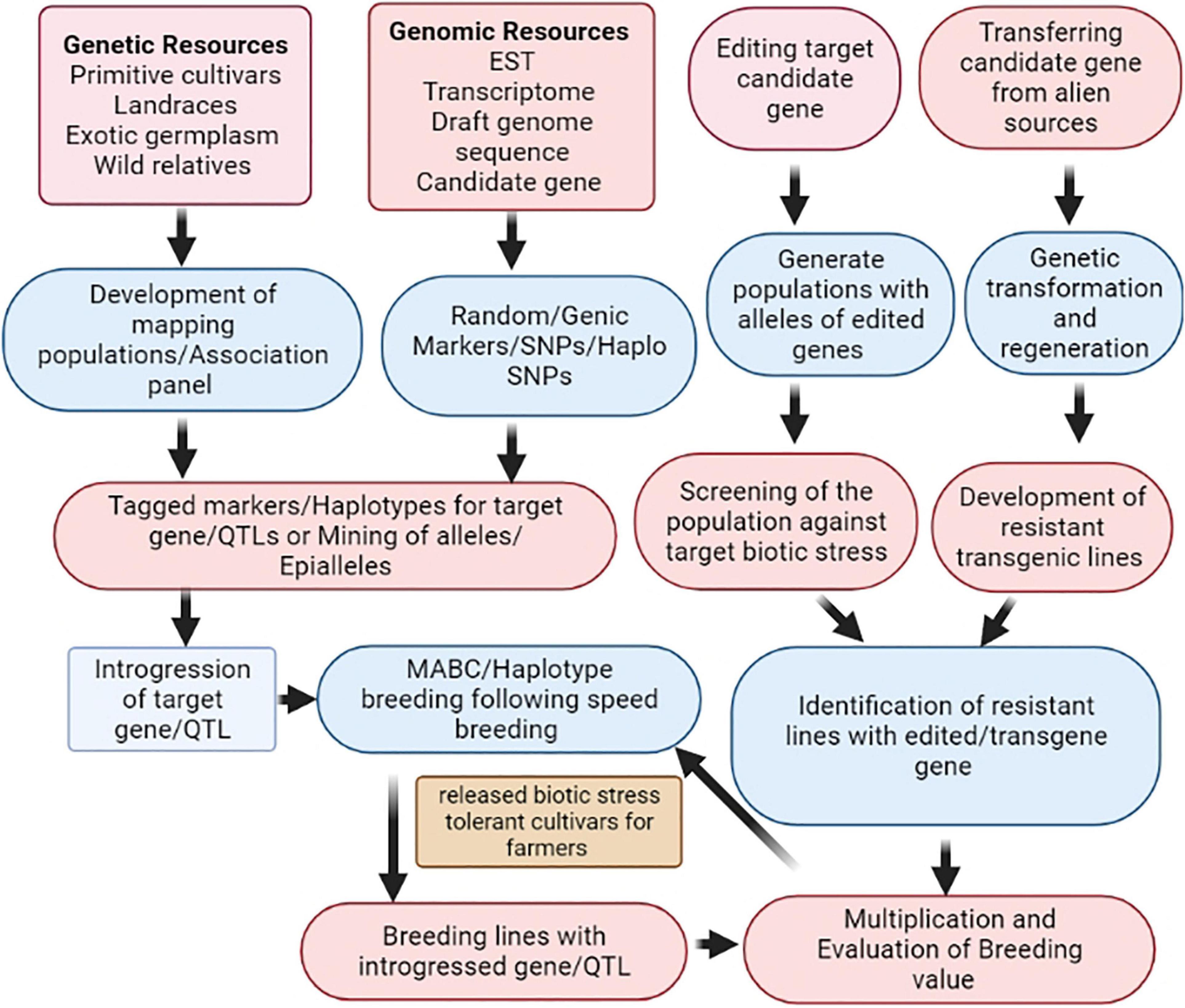
Pea is an important and exceptionally high-yielding cool season pulse crop in the world. Numerous biotic stresses are the key constraints in harnessing the full production potential of a pea, of which fungal diseases such as PM, FW, FRR, AB, CRR, and PR causing infection during different growth stages are devastating to the crop. Nevertheless, sincere efforts have been made to elevate the productivity and production of pea, but many more milestones are yet to be achieved for making it a resilient crop to upcoming challenges. Several major and minor genes/QTLs governing important biotic stresses in pea have been dissected and mapped using existing genomic tools, nevertheless, not utilized to a large extent in regular pea breeding programs. The reliable DNA markers flanking the genes/QTLs of interest could accelerate the introgression of resistance from the resistance sources using the genomic-assisted protocol to speed up the pea breeding program accomplishments more efficiently and precisely. Updated research efforts are warranted for the amalgamation of next-generation genomics and phenomics in pea improvement programs. The schematic diagram explains how different genomic approaches can be combined to accelerate the success of a pea breeding program (Figure 2). This figure also explains the combined use of genetic resources, genomic resources, and advanced biotechnological tools in the pea improvement program for the development of biotic stress-resistant cultivars. Underlying resistance mechanisms for AB, PM, and pea aphids have been elucidated using different pathogenic resistance proteins pertinent to the genes and pathways involved in pathogen resistance. However, more concentrated efforts are needed in the future on proteomic and transcriptomic analyses to untangle the disease and pest resistance mechanism in pea at the molecular level and to validate the sequencing results at the functional level for the identification of candidate genes controlling biotic stress resistance. This information will be certainly useful for editing or modification of crop genomes or realtered pathogens to develop resistant cultivars. Genome-wide association and genomic selection, which elucidate specific genetic variations at the genome scale, should be judiciously used for the identification of several gene(s)/QTLs exerting smaller effects on the biotic stress resistance. The transgenic technology should be exploited to let researchers utilize the variability existing outside the crop’s primary/secondary gene pool and also offer an opportunity to conquer crossability constraints. In addition, induced gene-specific DNA methylation and epigenome editing can be exploited to generate new epigenetic alleles for different biotic stresses. Most recently, speed breeding or rapid generation advancement protocols developed for shortening breeding times (4–5 cycles/year) have emerged as a potent technology for accelerating genetic gain in pea. Though, several tools and technologies are in hand judicious use to reap the best of them is challenging, certainly, there is a huge scope to achieve new heights in productivity enhancement by breeding biotic stress-resistant pea cultivars.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Funding
This publication has received funding support for open access fee payment from Cukurova University, Agricultural Faculty, Field Crops Department, Adana, Turkey.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alonso, C., Medrano, M., Perez, R., Canto, A., ParraTabla, V., and Herrera, C. M. (2019). Interspecific variation across angiosperms in global DNA methylation: phylogeny, ecology and plant features in tropical and Mediterranean communities. New Phytol. 224, 949–960. doi: 10.1111/nph.16046
Amey, R. C., Schleicher, T., Slinn, J., Lewis, M., Macdonald, H., Neill, S. J., et al. (2008). “Proteomic analysis of a compatible interaction between Pisum sativum (pea) and the downy mildew pathogen Peronospora viciae,” in The Downy Mildews-Genetics, Molecular Biology and Control, eds A. Lebeda, P. T. N. Spencer-Phillips, and B. M. Cooke (Dordrecht: Springer), 41–55. doi: 10.1007/978-1-4020-8973-2_5
Amian, A. A., Papenbrock, J., Jacobsen, H. J., and Hassan, F. (2011). Enhancing transgenic pea (Pisum sativum L.) resistance against fungal diseases through stacking of two antifungal genes (chitinase and glucanase). GM Crops 2, 104–109. doi: 10.4161/gmcr.2.2.16125
Amin, A., Mushtaq, F., Singh, P. K., Wani, K. P., Spaldon, S., and Nazir, N. (2010). Genetics and breeding of pea-a review. Int. J. Curr. Res. 10, 28–34. doi: 10.3923/ijpbg.2015.28.31
Aryamanesh, N., Zeng, Y., Byrne, O., Hardie, D. C., Al-Subhi, A. M., Khan, T., et al. (2014). Identification of genome regions controlling cotyledon, pod wall/seed coat and pod wall resistance to pea weevil through QTL mapping. Theor. Appl. Genet. 127, 489–497. doi: 10.1007/s00122-013-2234-2
Ashby, J. A., Stevenson, C. E. M., Jarvis, G. E., Lawson, D. M., and Maule, A. J. (2011). Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea. PLoS One 6:e15873. doi: 10.1371/journal.pone.0015873
Aslam, S., Ghazanfar, M. U., Munir, N., and Hamid, M. I. (2019). Managing fusarium wilt of pea by utilizing different application methods of fungicides. Pak. J. Phytopathol. 31, 81–88. doi: 10.33866/phytopathol.031.01.0482
Assen, K. Y. (2020). Diversity analysis and identification of promising powdery mildew resistance genotypes in field pea (Pisum sativum L.). Amer J. Biol. Environ. Stat. 6, 7–16. doi: 10.11648/j.ajbes.20200601.12
Azmat, M. A., Khan, A. A., Saeed, A., Ashraf, M., and Niaz, S. (2012). Screening pea germplasm against Erysiphe polygoni for disease severity and latent period. Int. J. Veg. Sci. 18, 153–160. doi: 10.1080/19315260.2011.592571
Aznar-Fernández, T., Barilli, E., Cobos, M. J., Kilian, A., Carling, J., and Rubiales, D. (2020). Identification of quantitative trait loci (QTL) controlling resistance to pea weevil (Bruchus pisorum) in a high-density integrated DArTseq SNP-based genetic map of pea. Sci. Rep. 10:33. doi: 10.1038/s41598-019-56987-7
Badu-Apraku, B., Talabi, A. O., Fakorede, M. A. B., Fasanmade, Y., Gedil, M., Magorokosho, C., et al. (2019). Yield gains and associated changes in an early yellow bi-parental maize population following genomic selection for Striga resistance and drought tolerance. BMC Plant Biol. 19:129. doi: 10.1186/s12870-019-1740-z
Bani, M., Pérez De Luque, A., Rubiales, D., and Rispail, N. (2018). Physical and chemical barriers in root tissues contribute to quantitative resistance to Fusarium oxysporum f. sp. pisi in pea. Front. Plant Sci. 9:199. doi: 10.3389/fpls.2018.00199
Bani, M., Rubiales, D., and Rispail, N. (2012). A detailed evaluation method to identify sources of quantitative resistance to Fusarium oxysporum f. sp. pisi race 2 within a Pisum spp. germplasm collection. Plant Pathol. 61, 532–542. doi: 10.1111/j.1365-3059.2011.02537.x
Barakate, A., and Stephens, J. (2016). An overview of CRISPR-based tools and their improvements: new opportunities in understanding plant pathogen interactions for better crop protection. Front. Plant Sci. 7:765. doi: 10.3389/fpls.2016.00765
Barilli, E., CarrilloPerdomo, E., Cobos, M. J., Kilian, A., Carling, J., and Rubiales, D. (2020). Identification of potential candidate genes controlling pea aphid tolerance in a Pisum fulvum high density integrated DArTseq SNP based genetic map. Pest Manag. Sci. 76, 1731–1742. doi: 10.1002/ps.5696
Barilli, E., Cobos, M. J., Carrillo, E., Kilian, A., Carlin, J., and Rubiales, D. (2018). A high-density integrated DArTseq SNP-based genetic map of Pisum fulvum and identification of QTLs controlling rust resistance. Front. Plant Sci. 9:167. doi: 10.3389/fpls.2018.00167
Barilli, E., Satovic, Z., Rubiales, D., and Torres, A. M. (2010). Mapping of quantitative trait loci controlling partial resistance against rust incited by Uromyces pisi (Pers.) Wint. in a Pisum fulvum L. intraspecific cross. Euphytica 175, 151–159. doi: 10.1007/s10681-010-0141-z
Bheri, M., Bhosle, S. M., and Makandar, R. (2019). Shotgun proteomics provides an insight into pathogenesis-related proteins using anamorphic stage of the biotroph, Erysiphe pisi pathogen of garden pea. Microbiol. Res. 222, 25–34. doi: 10.1016/j.micres.2019.02.006
Bhosle, S. M., and Makandar, R. (2021). Comparative transcriptome of compatible and incompatible interaction of Erysiphe pisi and garden pea reveals putative defense and pathogenicity factors. FEMS Microbiol. Ecol. 97:fiab006. doi: 10.1093/femsec/fiab006
Bhullar, N. K., Street, K., Mackay, M., Yahiaoui, N., and Keller, B. (2009). Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc. Natl. Acad. Sci. U.S.A. 106, 9519–9524. doi: 10.1073/pnas.0904152106
Blixt, S. (1974). “The pea,” in Handbook of Genetics, Vol. 2, ed. R. C. King (New York, NY: Plenum Press), 181–221.
Bohra, A., Sahrawat, K. L., Kumar, S., Joshi, R., Parihar, A. K., Singh, U., et al. (2014). Genetics and genomics-based interventions for nutritional enhancement of grain legume crops: status and outlook. J. Appl. Genet. 56, 151–161. doi: 10.1007/s13353-014-0268-z
Bretag, T. W., Keane, P. J., and Price, T. V. (2006). The epidemiology and control of ascochyta blight in field peas: a review. Aust. J. Agric. Res. 57, 883–902. doi: 10.1071/ar05222
Budhlakoti, N., Kushwaha, A. K., Rai, A., Chaturvedi, K. K., Kumar, A., Pradhan, A. K., et al. (2022). Genomic selection: a tool for accelerating the efficiency of molecular breeding for development of climate resilient crops. Front. Genet. 13:832153. doi: 10.3389/fgene.2022.832153
Budhlakoti, N., Mishra, D. C., Rai, A., Lal, S. B., Chaturvedi, K. K., and Kumar, R. R. (2019). A comparative study of single-trait and multi-trait genomic selection. J. Comput. Biol. 26, 1100–1112. doi: 10.1089/cmb.2019.0032
Byrne, O. M., Hardie, D. C., Khan, T. N., Speijers, J., and Yan, G. (2008). Genetic analysis of pod and seed resistance to pea weevil in a Pisum sativum × P. fulvum interspecific cross. Aust. J. Agric. Res. 59, 854–862. doi: 10.1071/ar07353
Carrillo, E., Rubiales, D., and Castillejo, M. A. (2014b). Proteomic analysis of pea (Pisum sativum L.) response during compatible and incompatible interactions with the pea aphid (Acyrthosiphon pisum H.). Plant Mol. Bio. Rep. 32, 697–718. doi: 10.1007/s11105-013-0677-x
Carrillo, E., Satovic, Z., Aubert, G., Boucherot, K., Rubiales, D., and Fondevilla, S. (2014a). Identification of quantitative trait loci and candidate genes for specific cellular resistance responses against Didymella pinodes in pea. Plant Cell Rep. 33, 1133–1145. doi: 10.1007/s00299-014-1603-x
Castillejo, M. Á, Fondevilla-Aparicio, S., Fuentes-Almagro, C., and Rubiales, D. (2020). Quantitative analysis of target peptides related to resistance against Ascochyta blight (Peyronellaea pinodes) in pea. J. Proteome Res. 19, 1000–1012. doi: 10.1021/acs.jproteome.9b00365
Cazzola, F., Bermejo, C. J., Guindon, M. F., and Cointry, E. (2020). Speed breeding in pea (Pisum sativum L.), an efficient and simple system to accelerate breeding programs. Euphytica 216, 1–11.
Chatterton, S., Bowness, R., and Harding, M. W. (2015). First report of root rot of field pea caused by Aphanomyces euteiches in Alberta. Can. Plant Dis. 99, 288. doi: 10.1094/PDIS-09-14-0905-PDN
Chaudhary, R. G., and Naimuddin. (2000). “Pea diseases in Indian perspective and their economic management,” in Advances in Plant Disease Management, eds U. Narain, K. Kumar, and M. Srivastava (New Delhi: Advance Publishing Concept), 47–60.
Cheng, H., Kizilkaya, K., Zeng, J., Garrick, D., and Fernando, R. (2018). Genomic prediction from multiple-trait bayesian regression methods using mixture priors. Genetics 209, 89–103. doi: 10.1534/GENETICS.118.300650/-/DC1
Cheng, P., Holdsworth, W., Ma, Y., Coyne, C. J., Mazourek, M., Grusak, M. A., et al. (2015). Association mapping of agronomic and quality traits in USDA pea single-plant collection. Mol. Breed. 35, 1–13.
Chiquet, J., Mary-Huard, T., Robin, S., and Robin, S. (2017). Structured regularization for conditional gaussian graphical models. Stat. Comput. 27, 789–804. doi: 10.1007/s11222-016-9654-1
Clement, S. L., Hardie, D. C., and Elberson, L. R. (2002). Variation among accessions of Pisum fulvum for resistance to pea weevil. Crop Sci. 42, 2167–2173. doi: 10.2135/cropsci2002.2167
Clement, S. L., McPhee, K. E., Elberson, L. R., and Evans, M. A. (2009). Pea weevil, Bruchus pisorum L. (Coleoptera: Bruchidae), resistance in Pisum sativum x Pisum fulvum interspecific crosses. Plant Breed. 128, 478–485. doi: 10.1111/j.1439-0523.2008.01603.x
Clulow, S. A., Lewis, B. G., and Matthews, P. (1991). A pathotype classification for Ascochyta pinodes. J. Phytopathol. 131, 322–332. doi: 10.1111/j.1439-0434.1991.tb01203.x
Conner, R. L., Chang, K. F., Hwang, S. F., Warkentin, T. D., and McRae, K. B. (2013). Assessment of tolerance for reducing yield losses in field pea caused by Aphanomyces root rot. Can. J. Plant Sci. 93, 473–482. doi: 10.4141/cjps2012-183
Coyne, C. J., Inglis, D. A., Whitehead, S. J., McClendon, M. T., and Muehlbauer, F. J. (2000). Chromosomal location of Fwf, the Fusarium wilt race 5 resistance gene in Pisum sativum. Pisum Genet. 32, 20–22.
Coyne, C. J., Kumar, S., von Wettberg, E. J., Marques, E., Berger, J. D., Redden, R. J., et al. (2020). Potential and limits of exploitation of crop wild relatives for pea, lentil, and chickpea improvement. Legum. Sci. 2:e36.
Coyne, C. J., Pilet-Nayel, M. L., McGee, R. J., Porter, L. D., Smýkal, P., and Grünwald, N. J. (2015). Identification of QTL controlling high levels of partial resistance to Fusarium solani f. sp. pisi in pea. Plant Breed. 134, 446–453. doi: 10.1111/pbr.12287
Coyne, C. J., Porter, L. D., Boutet, G., Ma, Y., McGee, R. J., Lesné, A., et al. (2019). Confirmation of Fusarium root rot resistance QTL Fsp-Ps 2.1 of pea under controlled conditions. BMC Plant Biol. 19:98. doi: 10.1186/s12870-019-1699
Curto, M., Camafeita, E., Lopez, J. A., Maldonado, A. M., Rubiales, D., and Jorrín, J. V. (2006). A proteomic approach to study pea (Pisum sativum) responses to powdery mildew (Erysiphe pisi). Proteomics 6, S163–S174. doi: 10.1002/pmic.200500396
Daetwyler, H. D., Hickey, J. M., Henshall, J. M., Dominik, S., Gredler, B., Van Der Werf, J. H. J., et al. (2010). Accuracy of estimated genomic breeding values for wool and meat traits in a multi-breed sheep population. Anim. Prod. Sci. 50, 1004–1010. doi: 10.1071/an10096
Dahiya, B., and Naresh, J. S. (1993). “Bio-efficacy of some insecticides against pea pod borer in field pea,” in Proceedings of the National Conference on Ecofriendly Approches in the Management of Pests, Diseases and Industrial Effluents, (Kanpur: University of Agriculture and Technology), 20–22.
Das, A., Parihar, A. K., Saxena, D., Singh, D., Singha, K. D., Kushwaha, K. P. S., et al. (2019a). Deciphering genotype-by-environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum L.). Front Plant Sci. 10:825. doi: 10.3389/fpls.2019.00825
Das, A., Sharma, N., and Prasad, M. (2019b). CRISPR/Cas9: a novel weapon in the arsenal to combat plant diseases. Front Plant Sci. 9:2008. doi: 10.3389/fpls.2018.02008
Davidson, J. A., Krysinska-Kaczmarek, M., Kimber, R. B. E., and Ramsey, M. D. (2004). Screening field pea germplasm for resistance to downy mildew (Peronospora viciae) and powdery mildew (Erysiphe pisi). Aus. Plant Pathol. 33, 413–417. doi: 10.1071/ap04040
Desgroux, A., Baudais, V. N., Aubert, V., Le Roy, G., de Larambergue, H., Miteul, H., et al. (2018). Comparative genome-wide-association mapping identifies common loci controlling root system architecture and resistance to Aphanomyces euteiches in pea. Front Plant Sci. 8:2195. doi: 10.3389/fpls.2017.02195
Desgroux, A., L’anthoëne, V., Roux-Duparque, M., Rivière, J. P., Aubert, G., Tayeh, N., et al. (2016). Genome-wide association mapping of partial resistance to Aphanomyces euteiches in pea. BMC Genomics 17:124. doi: 10.1186/s12864-016-2429-4
Dhall, R. K. (2015). Breeding for biotic stresses resistance in vegetable crops: a review. J. Crop Sci. Tech. 4, 13–27.
Dirlewanger, E., Isaac, P., Ranade, S., Belajouza, M., Cousin, R., and Devienne, D. (1994). Restriction fragment length polymorphism analysis of loci associated with disease resistance genes and developmental traits in Pisum sativum (L.). Theor. Appl. Genet. 88, 17–27. doi: 10.1007/BF00222388
Dixit, G. P., and Gautam, N. K. (2015). “Fieldpea (Pisum sativum L.),” in Breeding Indian Field Crops, ed. D. N. Bharadwaj (Jodhpur: Agrobios), 195–216.
Dos Santos, J. P. R., Pires, L. P. M., de Castro Vasconcellos, R. C., Pereira, G. S., Von Pinho, R. G., and Balestre, M. (2016). Genomic selection to resistance to Stenocarpella maydis in maize lines using DArTseq markers. BMC Genet. 17:86. doi: 10.1186/s12863-016-0392-3
Duarte, J., Rivière, N., Baranger, A., Aubert, G., Burstin, J., Cornet, L., et al. (2014). Transcriptome sequencing for high throughput SNP development and genetic mapping in Pea. BMC Genomics. 15:126. doi: 10.1186/1471-2164-15-126
Ek, M., Eklund, M., Von Post, R., Dayteg, C., Henriksson, T., Weibull, P., et al. (2005). Microsatellite markers for powdery mildew resistance in pea (Pisum sativum L.). Hereditas 142, 86–91. doi: 10.1111/j.1601-5223.2005.01906.x
Ekvall, J., Stegmark, R., and Nyman, M. (2006). Content of low molecular weight carbohydrates in vining peas (Pisum sativum) related to harvest time, size and brine grade. Food Chem. 94, 513–519. doi: 10.1016/j.foodchem.2004.11.044
Ellis, T. H., Turner, L., Hellens, R. P., Lee, D., Harker, C. L., Enard, C., et al. (1992). Linkage maps in pea. Genetics 130, 649–663. doi: 10.1093/genetics/130.3.649
Esen, A., Sari, H., Erler, F., Adak, A., Sari, D., Eker, T., et al. (2019). Screening and selection of accessions in the genus Pisum L. for resistance to pulse beetle (Callosobruchuschinensis L.). Euphytica 215, 1–9. doi: 10.1007/978-3-319-23534-9_1
Espinas, N. A., Saze, H., and Saijo, Y. (2016). Epigenetic control of defense signaling and priming in plants. Front Plant Sci. 7:1201. doi: 10.3389/fpls.2016.01201
Esposito, M. A., Martin, E. A., Cravero, V. P., and Cointry, E. (2007). Characterization of pea accessions by SRAP’s markers. Sci. Hortic. 113, 329–335. doi: 10.1016/j.scienta.2007.04.006
FAO (2021). Food and Agriculture Organization Statistics. Available online at: https://www.fao.org/faostat/en/#data/QCL (accessed October, 2021).
Feng, J., Hwang, R., Chang, K. F., Conner, R. L., Hwang, S. F., Strelkov, S. E., et al. (2011). Identification of microsatellite markers linked to quantitative trait loci controlling resistance to Fusarium root rot in field pea. Can. J. Plant Sci. 91, 199–204. doi: 10.4141/cjps09176
Fernando, W. M. U., Hill, J. E., Zello, G. A., Tyler, R. T., Dahl, W. J., and Van Kessel, A. G. (2010). Diets supplemented with chickpea or its main oligosaccharide component raffinose modify fecal microbial composition in healthy adults. Benef. Microbes 1, 197–207. doi: 10.3920/bm2009.0027
Fondevilla, S., Almeida, N. F., Satovic, Z., Rubiales, D., Patto, M. C. V., Cubero, J. I., et al. (2011). Identification of common genomic regions controlling resistance to Mycosphaerella pinodes, earliness and architectural traits in different pea genetic backgrounds. Euphytica 182, 43–52. doi: 10.1007/s10681-011-0460-8
Fondevilla, S., and Rubiales, D. (2012). Powdery mildew control in pea: a review. Agron Sustain. Dev. 32, 401–409. doi: 10.1007/s13593-011-0033-1
Fondevilla, S., Avila, C. M., Cubero, J. I., and Rubiales, D. (2005). Response to Ascochyta pinodes in a germplasm collection of Pisum spp. Plant Breed. 124, 313–315. doi: 10.1111/j.1439-0523.2005.01104.x
Fondevilla, S., Carver, T. L. W., Moreno, M. T., and Rubiales, D. (2006). Macroscopical and histological characterization of genes er1 and er2 for powdery mildew resistance in pea.Eur. J. Plant Pathol. 115, 309–321. doi: 10.1007/s10658-006-9015-6
Fondevilla, S., Torres, A. M., Moreno, M. T., and Rubiales, D. (2007b). Identification of a new gene for resistance to powdery mildew in Pisum fulvum, a wild relative of pea. Breed. Sci. 57, 181–184. doi: 10.1270/jsbbs.57.181
Fondevilla, S., Carver, T. L. W., Moreno, M. T., and Rubiales, D. (2007a). Identification and characterization of sources of resistance to Erysiphe pisi Syd. in Pisum spp. Plant Breed. 126, 113–119. doi: 10.1111/j.1439-0523.2006.01312.x
Fondevilla, S., Cubero, J. I., and Rubiales, D. (2007c). Inheritance of resistance to Mycosphaerella pinodes in two wild accessions of Pisum. Eur. J. Plant Pathol. 119, 53–58. doi: 10.1007/978-1-4020-6065-6_6
Fondevilla, S., Fernández-Aparicio, M., Satovic, Z., Emeran, A. A., Torres, A. M., Moreno, M. T., et al. (2010). Identification of quantitative trait loci for specific mechanisms of resistance to Orobanche crenata Forsk. in pea (Pisum sativum L.). Mol. Breed. 25, 259–272. doi: 10.1007/s11032-009-9330-7
Fondevilla, S., Krajinski, F., Kuster, H., Cubero, J. I., and Rubiales, D. (2011). Identification of genes differentially expressed in a resistance reaction to Mycospherella pinodes in pea using micro-array technology. BMC Genomics 12:28. doi: 10.1186/1471-2164-12-28
Fondevilla, S., Martín-Sanz, A., Satovic, Z., Fernández-Romero, M. D., Rubiales, D., and Caminero, C. (2012). Identification of quantitative trait loci involved in resistance to Pseudomonas syringae pv. syringae in pea (Pisum sativum L.). Euphytica 186, 805–812. doi: 10.1007/s10681-011-0592-x
Fondevilla, S., Rotter, B., Krezdorn, N., Jüngling, R., Winter, P., and Rubiales, D. (2014). Identification of genes involved in resistance to Didymella pinodes in pea by deepSuperSAGE transcriptome profiling. Plant Mol. Biol. Rep. 32, 258–269. doi: 10.1007/s11105-013-0644-6
Fondevilla, S., Rubiales, D., Moreno, M. T., and Torres, A. M. (2008a). Identification and validation of RAPD and SCAR markers linked to the gene Er3 conferring resistance to Erysiphe pisi DC in pea. Mol. Breed. 22, 193–200. doi: 10.1007/s11032-008-9166-6
Fondevilla, S., Rubiales, D., Zatovic, S., and Torres, A. M. (2008b). Mapping of quantitative trait loci for resistance to Mycosphaerella pinodes in Pisum sativum subsp. syriacum. Mol. Breed. 21, 439–454. doi: 10.1007/s11032-007-9144-4
Frew, T. J., Russell, A. C., and Timmerman-Vaughan, G. M. (2002). Sequence tagged site markers linked to the sbm1 gene for resistance to pea seed borne mosaic virus in pea. Plant Breed. 121, 512–516.
Gali, K. K., Liu, Y., Sindhu, A., Diapari, M., Shunmugam, A. S., Arganosa, G., et al. (2018). Construction of high-density linkage maps for mapping quantitative trait loci for multiple traits in field pea (Pisum sativum L.). BMC Plant Biol. 18:172. doi: 10.1186/s12870-018-1368-4
Ghafoor, A., and McPhee, K. (2012). Marker assisted selection (MAS) for developing powdery mildew resistant pea cultivars. Euphytica 186, 593–607.
Ghosh, S., Watson, A., Gonzalez-Navarro, O. E., Ramirez-Gonzalez, R. H., Yanes, L., Mendoza-Suárez, M., et al. (2018). Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 13, 2944–2963. doi: 10.1038/s41596-018-0072-z
Gretenkort, M. A., and Helsper, J. P. F. G. (1993). Disease assessment of pea lines with resistance to foot rot pathogens: protocols for in vitro selection. Plant Pathol. 42, 676–685. doi: 10.1111/j.1365-3059.1993.tb01552.x
Gritton, E. T. (1980). “Field pea,” in Hybridization of Crop Plants, eds W. R. Fehr and H. H. Hadley (Madison, WI: American Society of Agronomy : Crop Science Society of America), 347–356. doi: 10.2135/1980.hybridizationofcrops.c23
Grunwald, N. J., Coffman, V. A., and Kraft, J. M. (2003). Sources of partial resistance to Fusarium root rot in the Pisum core collection. Plant Dis. 87, 1197–1200. doi: 10.1094/PDIS.2003.87.10.1197
Gupta, S. K., and Gupta, M. (2019). Fusarium wilt of pea-A mini reviews. Plant Dis. Res. 34, 1–9. doi: 10.5958/2249-8788.2019.00001.5
Hagedorn, D. J., and Gritton, E. T. (1973). Inheritance of resistance to the pea seed-borne mosaic virus. Phytopathology 63, 1130–1133. doi: 10.1094/phyto-63-1130
Haghnazari, A., Samimifard, R., Najafi, J., and Mardi, M. (2005). Genetic diversity in pea (Pisum sativum L.) accessions detected by sequence tagged microsatellite markers. J. Gen. Breed. 59:145.
Hall, K. J., Parker, J. S., and Ellis, T. H. (1997). The relationship between genetic and cytogenetic maps of pea. I. Standard and translocation karyotypes. Genome 40, 744–754. doi: 10.1139/g97-797
Hamon, C., Baranger, A., Coyne, C. J., McGee, R. J., LeGoff, I., L’Anthoëne, V., et al. (2011). New consistent QTL in pea associated with partial resistance to Aphanomyces euteiches in multiple French and American environments. Theor. Appl. Genet. 123, 261–281. doi: 10.1007/s00122-011-1582-z
Hamon, C., Coyne, C. J., McGee, R. J., Lesné, A., Esnault, R., Mangin, P., et al. (2013). QTL meta-analysis provides a comprehensive view of loci controlling partial resistance to Aphanomyces euteiches in four sources of resistance in pea. BMC Plant Biol. 13:45. doi: 10.1186/1471-2229-13-45
Hance, S. T., Grey, W., and Weeden, N. F. (2004). Identification of tolerance to Fusarium solani in Pisum sativum ssp. elatius. Pisum Genet. 36, 9–13.
Handerson, C., Noren, S. K., Wricha, T., Meetei, N. T., Khanna, V. K., Pattanayak, A., et al. (2014). Assessment of genetic diversity in pea (Pisum sativum L.) using morphological and molecular markers. Indian J. Genet. Plant Breed. 74:205. doi: 10.5958/0975-6906.2014.00157.6
Hardie, D. C., Baker, G. J., and Marshall, D. R. (1995). Field screening of Pisum accessions to evaluate their susceptibility to the pea weevil (Coleoptera: Bruchidae). Euphytica 84, 155–161. doi: 10.1007/bf01677954
Heringa, R. J., Van Norel, A., and Tazelaar, M. F. (1969). Resistance to powdery mildew (Erysiphe polygoni D.C.) in peas (Pisum sativum L.). Euphytica 18, 163–169. doi: 10.1007/bf00035687
Hofinger, B. J., Russell, J. R., Bass, C. G., Baldwin, T., Dos Reis, M., Hedley, P. E., et al. (2011). An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol. Ecol. 20, 3653–3668. doi: 10.1111/j.1365-294X.2011.05201.x
Hosseini, S., Malin, E., Fredrik, H., Dan Funck, J., and Magnus, K. (2015). Deciphering common and specific transcriptional immune responses in pea towards the oomycete pathogens Aphanomyces euteiches and Phytophthora pisi. BMC Genom. 16:627. doi: 10.1186/s12864-015-1829-1
Huang, M., Balimponya, E. G., Mgonja, E. M., McHale, L. K., Luzi-Kihupi, A., Wang, G. L., et al. (2019). Use of genomic selection in breeding rice (Oryza sativa L.) for resistance to rice blast (Magnaporthe oryzae). Mol. Breed. 39, 1–16.
Humphry, M., Reinstädler, A., Ivanov, S., Bisseling, T., and Panstruga, R. (2011). Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant Pathol. 12, 866–867. doi: 10.1111/j.1364-3703.2011.00718.x
Hunter, P. J., Ellis, N., and Taylor, J. D. (2001). Association of dominant loci for resistance to Pseudomonas syringae pv. pisi with linkage groups II, VI and VII of Pisum sativum. Theor. Appl. Genet. 103, 129–135. doi: 10.1007/s001220100566
Infantino, A., Kharrat, M., Riccioni, L., Coyne, C. J., McPhee, K. E., and Grunwald, N. J. (2006). Screening techniques and sources of resistance to root diseases in legumes. Euphytica 147, 201–221. doi: 10.1007/s10681-006-6963-z
Iqbal, Z., Sattar, M. N., and Shafiq, M. (2016). CRISPR/Cas9: a tool to circumscribe cotton leaf curl disease. Front. Plant Sci. 7:475. doi: 10.3389/fpls.2016.00475
Jain, S., Weeden, N. F., Kumar, A., Chittem, K., and McPhee, K. (2015). Functional codominant marker for selecting the Fw gene conferring resistance to Fusarium wilt race 1 in pea. Crop Sci. 55, 2639–2646. doi: 10.2135/cropsci2015.02.0102
Jain, S., Weeden, N. F., Porter, L. D., Eigenbrode, S. D., and McPhee, K. (2013). Finding linked markers for efficient selection of pea enation mosaic virus resistance in pea. Crop Sci. 53, 2392–2399.
Janila, P., and Sharma, B. (2004). RAPD and SCAR markers for powdery mildew resistance gene er in pea. Plant Breed. 123, 271–274. doi: 10.1111/j.1439-0523.2004.00963.x
Jha, A. B., Gali, K. K., Banniza, S., and Warkentin, T. D. (2019). Validation of SNP markers associated with ascochyta blight resistance in pea. Can. J. Plant Sci. 99, 243–249. doi: 10.1139/cjps-2018-0211
Jha, A. B., Gali, K. K., Taran, B., and Warkentin, T. D. (2017). Fine mapping of QTLs for ascochyta blight resistance in pea using heterogeneous inbred families. Front. Plant Sci. 8, 765. doi: 10.3389/fpls.2017.00765
Jha, A. B., Taran, B., Diapari, M., Sindhu, A., Shunmugam, A., Bett, K., et al. (2015). Allele diversity analysis to identify SNPs associated with ascochyta blight resistance in pea. Euphytica 202, 189–197. doi: 10.1007/s10681-014-1254-6
Jha, A. B., Taran, B., Stonehouse, R., and Warkentin, T. D. (2016). Identification of QTLs associated with improved resistance to ascochyta blight in an interspecific pea recombinant inbred line population. Crop Sci. 56, 2926–2939. doi: 10.2135/cropsci2016.01.0001
Jha, A. B., Warkentin, T. D., Gurusamy, V., Taran, B., and Banniza, S. (2012). Identification of mycosphaerella blight resistance in wild Pisum species for use in pea breeding. Crop Sci. 52, 2462–2468. doi: 10.2135/cropsci2012.04.0242
Jia, Y., and Jannink, J. L. (2012). Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics 192, 1513–1522. doi: 10.1534/genetics.112.144246
Jiang, G. L. (2013). “Molecular markers and marker-assisted breeding in plants,” in Plant Breeding from Laboratories to Fields, ed. S. B. Andersen (Rijeka: InTech), 45–83.
Jones, F. R. (1923). Stem and root rot of peas in the United States caused by species of Fusarium. Agric. Res. 26, 459–476.
Juliana, P., Poland, J., Huerta-Espino, J., Shrestha, S., Crossa, J., Crespo-Herrera, L., et al. (2019). Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nat. Gen. 51, 1530–1539. doi: 10.1038/s41588-019-0496-6
Kahlon, J. G., Jacobsen, H. J., Chatterton, S., Hassan, F., Bowness, R., and Hall, L. M. (2018). Lack of efficacy of transgenic pea (Pisum sativum L.) stably expressing antifungal genes against Fusarium spp. in three years of confined field trials. GM Crops Food 9, 90–108. doi: 10.1080/21645698.2018.1445471
Kalloo, G., and Bergh, B. O. (1993). “Pea (Pisum sativum L.),” in Genetic Improvement of Vegetable Crops, eds G. Kallo and B. O. Bergh (Oxford: Pergamon Press Ltd), 409–425.
Kalt, W. (2001). Interspecific variation in anthocyanins, phenolics and antioxidant capacity among genotypes of high bush and low bush blueberries (Vaccinium section cyanococcus spp.). J. Agric. Food Chem. 49, 4761–4767. doi: 10.1021/jf010653e
Katoch, V., Sharma, S., Pathania, S., Banayal, K., Sharma, S. K., and Rathour, R. (2010). Molecular mapping of pea powdery mildew resistance gene er2 to pea linkage group III. Mol. Breed. 25, 229–237. doi: 10.1007/s11032-009-9322-7
Klápště, J., Dungey, H. S., Telfer, E. J., Suontama, M., Graham, N. J., Li, Y., et al. (2020). Marker selection in multivariate genomic prediction improves accuracy of low heritability traits. Front. Genet. 11:499094. doi: 10.3389/fgene.2020.499094
Konečná, E., Šafářová, D., Navratil, M., Hanáček, P., Coyne, C., Flavell, A., et al. (2014). Geographical gradient of the eIF4E alleles conferring resistance to potyviruses in pea (Pisum) germplasm. PLoS One 9:e90394. doi: 10.1371/journal.pone.0090394
Kraft, J. M. (1992). Registration of 90-2079, 90-2131 and 90-2322 pea germplasms. Crop Sci. 32, 1076. doi: 10.2135/cropsci1992.0011183x003200040063x
Kraft, J. M., and Boge, W. (2001). Root characteristics in pea in relation to compaction and Fusarium rootrot. Plant Dis. 85, 936–940. doi: 10.1094/PDIS.2001.85.9.936
Kraft, J. M., and Pfleger, F. L. (2001). Compendium of Pea Diseases and Pests, 2nd Edn. St. Paul, MN: The American Phytopathological Society.
Kreplak, J., Madoui, M. A., Cápal, P., Novák, P., Labadie, K., Aubert, G., et al. (2019). A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 51, 1411–1422. doi: 10.1038/s41588-019-0480-1
Kumar, J., Choudhary, A. K., Gupta, D. S., and Kumar, S. (2019). Towards exploitation of adaptive traits for climate-resilient smart pulses. Inter. J. Mol. Sci. 20:2971. doi: 10.3390/ijms20122971
Kumar, S., Srivastava, R. L., and Singh, R. (2006). Combining ability for yield and its component traits in field pea. Ind. J. Pulses Res. 19, 173–175.
Kwon, S. J., Smýkal, P., Hu, J., Wang, M., Kim, S. J., McGee, R. J., et al. (2013). User-friendly markers linked to Fusarium wilt race 1 resistance Fw gene for marker-assisted selection in pea. Plant Breed. 132, 642–648. doi: 10.1111/pbr.12085
Lakshmana Reddy, D. C., Preethi, B., Wani, M. A., Aghora, T. S., Aswath, C., and Mohan, N. (2015). Screening for powdery mildew (Erysiphe pisi DC) resistance gene-linked SCAR and SSR markers in five breeding lines of Pisum sativum L. J. Hortic. Sci. Biotech. 90, 78–82. doi: 10.1080/14620316.2015.11758545
Lamprecht, H. (1948). The variation of linkage and the course of crossing over. Agric. Hortic. Genet. 6:10. doi: 10.1371/journal.pcbi.1002462
Latutrie, M., Gourcilleau, D., and Pujol, B. (2019). Epigenetic variation for agronomic improvement: an opportunity for vegetatively propagated crops. Am. J. Bot. 106:1281. doi: 10.1002/ajb2.1357
Laucou, V., Haurogné, K., Ellis, N., and Rameau, C. (1998). Genetic mapping in pea. 1. RAPD-based genetic linkage map of Pisum sativum. Theor. Appl. Genet. 97, 905–915. doi: 10.1007/s001220050971
Lavaud, C., Baviere, M., Le Roy, G., Hervé, M. R., Moussart, A., Delourme, R., et al. (2016). Single and multiple resistance QTL delay symptom appearance and slow down root colonization by Aphanomyces euteiches in pea near isogenic lines. BMC Plant Biol. 16:166. doi: 10.1186/s12870-016-0822-4
Lavaud, C., Lesné, A., Piriou, C., Le Roy, G., Boutet, G., Moussart, A., et al. (2015). Validation of QTL for resistance to Aphanomyces euteiches in different pea genetic backgrounds using near isogenic lines. Theor. Appl. Genet. 128, 2273–2288. doi: 10.1007/s00122-015-2583-0
Lázaro, A., and Aguinagalde, I. (2006). Genetic variation among Spanish pea landraces revealed by Inter Simple Sequence Repeat (ISSR) markers: its application to establish a core collection. J. Agric. Sci. 144, 53–61. doi: 10.1017/s0021859605005848
Leonforte, A., Sudheesh, S., Cogan, N. O. I., Salisbury, P. A., Nicolas, M. E., Materne, M., et al. (2013). SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant Biol. 13:161. doi: 10.1186/1471-2229-13-161
Liu, N., Xu, S., Yao, X., Zhang, G., Mao, W., Hu, Q., et al. (2016). Studies on the control of Ascochyta blight in field peas (Pisum sativum L.) caused by Ascochyta pinodes in Zhejiang province, China. Front. Microbiol. 7:481. doi: 10.3389/fmicb.2016.00481
Liu, S. M., O’Brien, L., and Moore, S. G. (2003). A single recessive gene confers effective resistance to powdery mildew of field pea grown in northern New South Wales. Aust. J. Exp. Agric. 43, 373–378. doi: 10.1071/ea01142
Lorenz, A. J., Smith, K. P., and Jannink, J. L. (2012). Potential and optimization of genomic selection for Fusarium head blight resistance in six-row barley. Crop Sci. 52, 1609–1621. doi: 10.2135/cropsci2011.09.0503
Loridon, K., McPhee, K. E., Morin, J., Dubreuil, P., Pilet-Nayel, M. L., Aubert, G., et al. (2005). Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor. Appl. Genet. 111, 1022–1031. doi: 10.1007/s00122-005-0014-3
Lunde, M. S., Hjellset, V. T., Holmboe-Ottesen, G., and Høstmark, A. T. (2011). Variations in postprandial blood glucose responses and satiety after intake of three types of bread. J. Nutr. Metab. 2011:437587. doi: 10.1155/2011/437587
Ma, Y., Coyne, C. J., Main, D., Pavan, S., Sun, S., Zhu, Z., et al. (2017). Development and validation of breeder-friendly KASPar) markers for er1, a powdery mildew resistance gene in pea (Pisum sativum L.). Mol. Breed. 37:151. doi: 10.1007/s11032-017-0740-7
Mahajan, R., Dar, A. A., Mukthar, S., Zargar, S. M., and Sharma, S. (2018). “Pisum improvement against biotic stress: current status and future prospects,” in Pulse Improvement, eds S. H. Wani and M. Jain (Cham: Springer), 109–136. doi: 10.1007/978-3-030-01743-9_6
Mahini, R. A., Kumar, A., Elias, E. M., Fiedler, J. D., Porter, L. D., and McPhee, K. E. (2020). Analysis and identification of QTL for resistance to Sclerotinia sclerotiorum in pea (Pisum sativum L.). Front. Genet. 11:1436. doi: 10.3389/fgene.2020.587968
Marinangeli, C. P., and Jones, P. J. (2011). Whole and fractionated yellow pea flours reduce fasting insulin and insulin resistance in hypercholesterolaemic and overweight human subjects. Br. J. Nutr. 105, 110–117. doi: 10.1017/S0007114510003156
Martín-Sanz, A., Aparicio, T., Santana, J. C., García, P., Winter, P., Caminero, C., et al. (2016). Mapping genes for resistance to bacterial blight (Pseudomonas syringae pv. pisi) in pea and identification of genes involved in resistance by DeepsuperSAGE transcriptome profiling. Euphytica 210, 375–392. doi: 10.1007/s10681-016-1700-8
Martín-Sanz, A., de la Vega, M. P., Murillo, J., and Caminero, C. (2012). Genetic, biochemical and pathogenic diversity of Pseudomonas syringae pv. pisi strains. Plant Pathol. 61, 1063–1072. doi: 10.1111/j.1365-3059.2012.02604.x
Marx, G. A., Schroeder, W. T., Provvidenti, R., and Mishanec, W. (1972). A genetic study of tolerance in pea (Pisum sativum L.) to Aphanomyces root rot. J. Am. Soc. Hortic. Sci. 97, 619–621.
Maude, R. B. (1966). Pea seed infection by Mycosphaerella pinodes and Ascochyta pisi and its control by seed soaks in thiram and captan suspensions. Ann. Appl. Biol. 57, 193–200. doi: 10.1111/j.1744-7348.1966.tb03814.x
McClendon, M. T., Inglis, D. A., McPhee, K. E., and Coyne, C. J. (2002). DNA markers for fusarium wilt race 1 resistance gene in pea. J. Am. Soc. Hortic. Sci. 127, 602–607. doi: 10.21273/jashs.127.4.602
McGee, R. J., Coyne, C. J., Pilet Nayel, M. L., Moussart, A., Tivoli, B., Baranger, A., et al. (2012). Registration of pea germplasm lines partially resistant to aphanomyces root rot for breeding fresh or freezer pea and field pea types.J. Plant Regist. 6, 203–207.1.
McPhee, K. (2003). Field pea production and breeding – a mini-review. Food Agric. Environ. 1, 64–69.
McPhee, K. E., Inglis, D. A., and Coyne, C. J. (2004). “Linkage map location of Fusarium wilt race 2 (Fnw) in pea,” in Proceedings of the 5th European Conference on Grain Legumes, June 7–11, 2004, Dijon, 342.
McPhee, K. E., Inglis, D. A., Gundersen, B., and Coyne, C. J. (2012). Mapping QTL for fusarium wilt race 2 partial resistances in pea (Pisum sativum). Plant Breed. 131, 300–306. doi: 10.1111/j.1439-0523.2011
McPhee, K. E., Tullu, A., Kraft, J. M., and Muehlbauer, F. J. (1999). Resistance to Fusarium wilt race 2 in the Pisum core collection. J. Am. Soc. Hortic. Sci. 124, 28–31. doi: 10.21273/jashs.124.1.28
Mikić, A., Smýkal, P., Kenicer, G., Vishnyakova, M., Sarukhanyan, N., Akopian, J., et al. (2013). The bicentenary of the research on ‘beautiful’vavilovia (Vavilovia formosa), a legume crop wild relative with taxonomic and agronomic potential. Bot. J. Linn. Soc. 172, 524–531. doi: 10.1111/boj.12060
Mirdita, V., He, S., Zhao, Y., Korzun, V., Bothe, R., Ebmeyer, E., et al. (2015). Potential and limits of whole genome prediction of resistance to Fusarium head blight and Septoria tritici blotch in a vast Central European elite winter wheat population. Theor. Appl. Genet. 128, 2471–2481. doi: 10.1007/s00122-015-2602-1
Mohamed, A., Garcia-Martinez, S., Carbonell, M. L. P., Jose ìRuiz, J., and Boubaker, M. (2019). Assessment of genetic diversity among local pea (Pisum sativum L.) accessions cultivated in the arid regions of Southern Tunisia using agro-morphological and SSR molecular markers. Genet. Resour. Crop Evol. 66, 1189–1203. doi: 10.1007/s10722-019-00784-8
Mohan, M., Nair, S., Bhagwat, A., Krishna, T. G., Yano, M., Bhatia, C. R., et al. (1997). Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol. Breed. 3, 87–103. doi: 10.1023/A:1009651919792
Mohan, N., Aghora, T. S., Wani, M. A., and Divya, B. (2013). Garden pea improvement in India. J.Hortic. Sci. 8, 125–164.
Moussart, A., Devaux, C., Muel, F., Pilet-Nayel, M., Baranger, A., Tivoli, B., et al. (2007). “Improving partial resistance to Aphanomyces root rot in GSP breeding program,” in Proceedings of the 3rd International Aphanomyces Workshop on Legumes, Rennes.
Muehlbauer, F. J., and Kraft, K. M. (1973). Evidence of heritable resistance to Fusarium solani f. sp. pisi and Phythium ultimum in peas. Crop Sci. 13, 34–36. doi: 10.2135/cropsci1973.0011183x001300010011x
Mushtaq, M., Sakina, A., Wani, S. H., Shikari, A. B., Tripathi, P., Zaid, A., et al. (2019). Harnessing genome editing techniques to engineer disease resistance in plants. Front. Plant Sci. 10:550. doi: 10.3389/fpls.2019.00550
Negisho, K., Teshome, A., and Keneni, G. (2017). Genetic diversity in Ethiopian field pea (Pisum sativum L.) germplasm collections as revealed by SSR markers. Ethiop. J. Agric. Sci. 27, 33–47.
Nisar, M., and Ghafoor, A. (2011). Linkage of a RAPD marker with powdery mildew resistance er-1 gene in Pisum sativum L. Rus. J. Genet. 47, 300–304. doi: 10.1134/s102279541102013x
Nisar, M., Ghafoor, A., Khan, M. R., and Qureshi, A. S. (2006). Screening of Pisum sativum L. germplasm against Erysiphe pisi Syd. Acta. Biol. Cracoviensia Ser. Bot. 48, 33–37.
Okubara, P. A., Inglis, D. A., Muehlbauer, F. J., and Coyne, C. J. (2002). A novel RAPD marker linked to the Fusarium wilt race 5 resistance gene (Fwf) in Pisum sativu. Pisum Genet. 34, 115–120.
Okubara, P. A., Keller, K. E., McClendon, M. T., Inglis, D. A., McPhee, K. E., and Coyne, C. J. (2005). Y15_999Fw, a dominant SCAR marker linked to the Fusarium wilt race 1 (Fw) resistance gene in pea. Pisum Genet. 37, 30–33.
Parihar, A. K., Bohra, A., and Dixit, G. P. (2016). “Nutritional benefits of winter pulses with special emphasis on peas and rajmash,” in Biofortification of Food Crops, eds U. Singh, C. S. Praharaj, S. S. Singh, and N. P. Singh (New Delhi: Springer), 61–71. doi: 10.1007/978-81-322-2716-8_6
Parihar, A. K., Dixit, G. P., and Chaturvedi, S. K. (2013). Diseases resistance breeding in fieldpea — a review. Progress. Res. 8, 1–13.
Parihar, A. K., Dixit, G. P., Bohra, A., Gupta, D. S., Singh, A. K., Kumar, N., et al. (2020). “Genetic advancement in field pea (Pisum sativum L.): retrospect and prospect,” in Accelerated Plant Breeding, eds S. S. Gosal and S. H. Wani (Cham: Springer), 283–341. doi: 10.1007/978-3-030-47306-8_10
Parihar, A. K., Dixit, G. P., Singh, U., Singh, A. K., Kumar, N., and Gupta, S. (2021). “Potential of field pea as a nutritionally rich food legume crop,” in Breeding for Enhanced Nutrition and Bio-Active Compounds in Food Legumes, eds D. S. Gupta, S. Gupta, and J. Kumar (Cham: Springer), 47–82. doi: 10.1007/978-3-030-59215-8_3
Patto, M. V., Fernández-Aparicio, M., Moral, A., and Rubiales, D. (2007). Resistance reaction to powdery mildew (Erysiphe pisi) in a germplasm collection of Lathyrus cicera from Iberian origin. Gen. Resour.Crop.Evo. 54, 1517–1521. doi: 10.1007/s10722-006-9142-0
Patto, M. V., Fernández-Aparicio, M., Moral, A., and Rubiales, D. (2009). Pre and posthaustorial resistance to rusts in Lathyrus cicera L. Euphytica 165, 27–34. doi: 10.1007/s10681-008-9737-y
Pavan, S., Schiavulli, A., Appiano, M., Marcotrigiano, A. R., Cillo, F., Visser, R. G., et al. (2011). Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Gen. 123, 1425–1431. doi: 10.1007/s00122-011-1677-6
Pavan, S., Schiavulli, A., Appiano, M., Miacola, C., Visser, R. G. F., Bai, Y. L., et al. (2013). Identification of a complete set of functional markers for the selection of er1 powdery mildew resistance in Pisum sativum L. Mol. Breed. 31, 247–253. doi: 10.1007/s11032-012-9781-0
Pereira, G., Marques, C., Ribeiro, R., Formiga, S., Dâmaso, M., Sousa, M. T., et al. (2010). Identification of DNA markers linked to an induced mutated gene conferring resistance to powdery mildew in pea (Pisum sativum L.). Euphytica 171, 327–335.
Pilet Nayel, M. L., Coyne, C., Hamon, C., Lesne’, A., Le Goff, I., Esnault, R., et al. (2007). “Understanding genetics of partial resistance to Aphanomyces root rot in pea for new breeding prospects,” in Proceedings of the 3rd international Aphanomyces workshop on legumes, Rennes, 36.
Pilet Nayel, M. L., Muehlbauer, F. J., McGee, R. J., Kraft, J. M., Baranger, A., and Coyne, C. J. (2005). Consistent quantitative trait loci in pea for partial resistance to Aphanomyces euteiches isolates from the United States and France. Phytopathology 95, 1287–1293. doi: 10.1094/PHYTO-95-1287
Pilet Nayel, M. L., Muehlbauer, F. J., McGee, R. J., Kraft, J. M., Baranger, A., and Coyne, C. J. (2002). Quantitative trait loci for partial resistance to Aphanomyces root rot in pea. Theor. Appl. Genet. 106, 28–39. doi: 10.1007/s00122-002-0985-2
Porter, L. D. (2010). Identification of tolerance to Fusarium root rot in wild pea germplasm with high levels of partial resistance. Pisum Genet. 42, 1–6.
Prescott, V. E., Campbell, P. M., Moore, A., Mattes, J., Rothenberg, M. E., Foster, P. S., et al. (2005). Transgenic expression of bean alpha-amylase inhibitor in peas results in altered structure and immunogenicity. J. Agric. Food Chem. 53, 9023–9030. doi: 10.1021/jf050594v
Prioul, S., Frankewitz, A., Deniot, G., Morin, G., and Baranger, A. (2004). Mapping of quantitative trait loci for partial resistance to Mycosphaerellapinodes in pea (Pisum sativum L.) at the seedling and adult plant stages. Theor. Appl. Genet. 108, 1322–1334. doi: 10.1007/s00122-003-1543-2
Prioul-Gervais, S., Deniot, G., Receveur, E. M., Frankewitz, A., Fourmann, M., Rameau, C., et al. (2007). Candidate genes for quantitative resistance to Mycosphaerella pinodes in pea (Pisum sativum L.). Theor. Appl. Gen. 114, 971–984. doi: 10.1007/s00122-006-0492-y
Rai, R., Singh, A. K., Chand, R., Srivastava, C. P., Joshi, A. K., and Singh, B. D. (2016). Genomic regions controlling components of resistance for pea rust caused by Uromyces fabae (Pers.) de-Bary. J. Plant Biochem. Biotech. 25, 133–141. doi: 10.1007/s13562-015-0318-6
Rai, R., Singh, A. K., Singh, B. D., Joshi, A. K., Chand, R., and Srivastava, C. P. (2011). Molecular mapping for resistance to pea rust caused by Uromyces fabae (Pers.) de-Bary. Theor Appl Genet 123, 803–813. doi: 10.1007/s00122-011-1628-2
Rakshit, S. (1997). Biochemical and molecular analyses of powdery mildew resistance in pea(Pisum sativum L.). Ph. D. Dissertation. New Delhi: Indian Agricultural Research Institute.
Rambani, A., Rice, J. H., Liu, J., Lane, T., Ranjan, P., Mazarei, M., et al. (2015). The methylome of soybean roots during the compatible interaction with the soybean cyst nematode. Plant Physiol. 168, 1364–1377. doi: 10.1104/pp.15.00826
Ramirez-Prado, J. S., Abulfaraj, A. A., Rayapuram, N., Benhamed, M., and Hirt, H. (2018). Plant immunity: from signaling to epigenetic control of defense. Trends Plant. Sci. 23, 833–844. doi: 10.1016/j.tplants.2018.06.004
Rana, J. C., Banyal, D., Sharma, K. D., Sharma, M. K., Gupta, S. K., and Yadav, S. K. (2013). Screening of pea germplasm for resistance to powdery mildew. Euphytica 189, 271–282. doi: 10.1007/s10681-012-0798-6
Reeves, P. A., Panella, L. W., and Richards, C. M. (2012). Retention of agronomically important variation in germplasm core collections: implications for allele mining. Theor.Appl. Gen. 124, 1155–1171. doi: 10.1007/s00122-011-1776-4
Reiner, D., Lee, R., Higgins, T. J., and Epstein, M. M. (2013). Genetically modified α-amylase inhibitor peas are not specifically allergenic in mice. Clin. Transl. Allergy 3:1. doi: 10.1371/journal.pone.0052972
Ren, Y., Singh, R. P., Basnet, B. R., Lan, C. X., Huerta-Espino, J., Lagudah, E. S., et al. (2017). Identification and mapping of adult plant resistance loci to leaf rust and stripe rust in common wheat cultivar Kundan. Plant Dis. 101, 456–463. doi: 10.1094/PDIS-06-16-0890-re
Rispail, N., and Rubiales, D. (2014). Identification of sources of quantitative resistance to Fusarium oxysporum f. sp. medicaginis in Medicago truncatula. Plant Dis. 98, 667–673. doi: 10.1094/pdis-03-13-0217-re
Robaglia, C., and Caranta, C. (2006). Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. doi: 10.1016/j.tplants.2005.11.004
Rothman, A. J., Levina, E., and Zhu, J. (2010). Sparse multivariate regression with covariance estimation. J. Comput. Graph. Stat. 19, 947–962. doi: 10.1198/jcgs.2010.09188
Roux-Duparque, M., Boitel, C., Decaux, B., Moussart, A., Alamie, J., Pilet-Nayel, M. L., et al. (2004). “Breeding peas for resistance to Aphanomyces root rot: current main outputs of three breeding programmes,” in Proceedings of the 5th European Conference on Grain Legumes, Dijon, 133.
Rubiales, D., Fondevilla, S., Chen, W., Gentzbittel, L., Higgins, T. J., Castillejo, M. A., et al. (2015). Achievements and challenges in legume breeding for pest and disease resistance. Crit. Rev. Plant Sci. 34, 195–236. doi: 10.1080/07352689.2014.898445
Rubiales, D., González Bernal, M. J., Warkentin, T., Bueckert, T., VazPatto, M. C., McPhee, K., et al. (2019). “Advances in pea breeding,” in The Achieving Sustainable Cultivation of Vegetables, ed. G. Hochmuth (Cambridge: BurleighDodds Science Publishing).
Rutkoski, J., Benson, J., Jia, Y., Brown-Guedira, G., Jannink, J. L., and Sorrells, M. (2012). Evaluation of genomic prediction methods for Fusarium head blight resistance in wheat. Plant Genome 5, 51–61. doi: 10.3835/plantgenome2012.02.0001
Sallam, A. H., and Smith, K. P. (2016). Genomic selection performs similarly to phenotypic selection in barley. Crop Sci. 56, 2871–2881. doi: 10.2135/CROPSCI2015.09.0557
Sari, H., Sari, D., Eker, T., and Toker, C. (2021). De novo super-early progeny in interspecific crosses Pisum sativum L.× P. fulvum Sibth. et Sm. Sci. Rep. 11:19706. doi: 10.1038/s41598-021-99284-y
Sari, H., Sari, D., Eker, T., Aydinoglu, B., Canci, H., Ikten, C., et al. (2020). Inheritance and expressivity of neoplasm trait in crosses between the domestic pea (Pisum sativum subsp. sativum) and tall wild pea (Pisum sativum subsp. elatius). Agronomy 10:1869. doi: 10.3390/agronomy10121869
Sarinelli, J. M., Murphy, J. P., Tyagi, P., Holland, J. B., Johnson, J. W., Mergoum, M., et al. (2019). Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor. Appl. Genet. 132, 1247–1261. doi: 10.1007/S00122-019-03276-6
Sharma, B., and Yadav, Y. (2003). Pisum fulvum carries a recessive gene for powdery mildew resistance. Pisum Genet. 35:31.
Sharma, S. D. (2000). Insect pests of pea (Pisum sativum L.) in Himachal Pradesh. Insect Environ. 6:113.
Sharma, S., Upadhyaya, H. D., Varshney, R. K., and Gowda, C. L. L. (2013). Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legumes. Front. Plant Sci. 4:309. doi: 10.3389/fpls.2013.00309
Shashikumar, K. T., Pitchaimuthu, M., and Rawal, R. D. (2010). Generation mean analysis of resistance to downey mildew in adult muskmelon plants. Euphytica 173, 121–127. doi: 10.1007/s10681-010-0132-0
Sindhu, A., Ramsay, L., Sanderson, L. A., Stonehouse, R., Li, R., Condie, J., et al. (2014). Gene-based SNP discovery and genetic mapping in pea. Theor. Appl. Genet. 127, 2225–2241. doi: 10.1007/s00122-014-2375-y
Singh, A. K., Rai, R., Singh, B. D., Chand, R., and Srivastava, C. P. (2015). Validation of SSR markers associated with rust (Uromyces fabae) resistance in pea (Pisum sativum L.). Physiol. Mol. Biol. Plants 21, 243–247. doi: 10.1007/s12298-015-0280-8
Singh, A. K., Rai, R., Srivastava, C. P., Singh, B. D., Kushwaha, C., and Chand, R. (2012). A quantitative analysis of rust (Uromyces fabae) resistance in pea (Pisum sativum) using RILs. Ind. J. Agric. Sci. 82, 190–192.
Singh, J., Nadarajan, N., Basu, P. S., Srivastava, R. P., and Kumar, L. (2013). Pulses for Human Health and Nutrition. Kanpur: Indian Institute of Pulses Research, 1–35.
Singh, R., and Ram, H. (2001). Inheritance of days to flowering and rust resistance in peas. Res. Crops 2, 414–418.
Singh, R., Babu, S., Avasthe, R. K., Singh, A., Yadav, G. S., Pashte, V., et al. (2018). Screening of field pea varieties for rice-fallow areas under organic management conditions in NE Himalayas. Ann. Agric. Res. 39, 246–250.
Singh, V. K., Khan, A. W., Saxena, R. K., Kumar, V., Kale, S. M., Sinha, P., et al. (2016). Next-generation sequencing for identification of candidate genes for Fusarium wilt and sterility mosaic disease in pigeonpea (Cajanu scajan). Plant Biotechnol. J. 14, 1183–1194. doi: 10.1111/pbi.12470
Smýkal, P., Aubert, G., Burstin, J., Coyne, C. J., Ellis, N. T., Flavell, A. J., et al. (2012). Pea (Pisum sativum L.) in the genomic era. Agronomy 2, 74–115.
Smýkal, P., Clarice, C., Robert, R., and Nigel, M. (2013). “Peas,” in The Genetic and Genomic Resources of Grain Legume Improvement, eds M. Singh, H. D. Upadhyaya, and I. S. Bisht (Amsterdam: Elsevier), 41–80.
Smýkal, P., Coyne, C. J., Ambrose, M. J., Maxted, N., Schaefer, H., Blair, M. W., et al. (2015). Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 34, 43–104.
Smýkal, P., Horáček, J., Dostálová, R., and Hýbl, M. (2008a). Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J. Appl. Genet. 49, 155–166. doi: 10.1007/BF03195609
Smýkal, P., Hýbl, M., Corander, J., Jarkovský, J., Flavell, A. J., and Griga, M. (2008b). Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retro- transposon, microsatellite and morphological marker analysis. Theor.Appl. Genet. 117, 413–424. doi: 10.1007/s00122-008-0785-4
Smýkal, P., Valledor, L., Rodriguez, R., and Griga, M. (2007). Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.). Plant Cell Rep. 26, 1985–1998. doi: 10.1007/s00299-007-0413-9
Snowdon, R. J., and Friedt, W. (2004). Molecular markers in brassica oilseed breeding: current status and future possibilities. Plant Breed. 123, 1–8. doi: 10.1111/j.1439-0523.2003.00968.x
Srivastava, R. K., Mishra, S. K., Singh, A. K., and Mohapatra, T. (2012). Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene ‘er1’in pea (Pisum sativum L.). Euphytica 186, 855–866. doi: 10.1007/s10681-012-0650-z
Steer, T. E. (2006). Phytochemicals – a future in functional foods? Food Sci. Technol. Bull. 3, 23–29. doi: 10.1616/1476-2137.14379
Sudheesh, S., Lombardi, M., Leonforte, A., Cogan, N. O., Materne, M., Forster, J. W., et al. (2014). Consensus genetic map construction for field pea (Pisum sativum L.), trait dissection of biotic and abiotic stress tolerance and development of a diagnostic marker for the er1 powdery mildew resistance gene. Plant Mol. Biol. Rep. 33, 1391–1403. doi: 10.1007/s11105-014-0837-7
Sun, S., Deng, D., Duan, C., Zong, X., Xu, D., He, Y., et al. (2019). Two novel er1 alleles conferring powdery mildew (Erysiphe pisi) resistance identified in a worldwide collection of pea (Pisum sativum L.) germplasms. Int. J. Mol. Sci. 20:5071. doi: 10.3390/ijms20205071
Sun, S., Fu, H., Wang, Z., Duan, C., Zong, X., and Zhu, Z. (2016). Discovery of a novel er1 allele conferring powdery mildew resistance in Chinese pea (Pisum sativum L.) landraces. PLoS One 11:e0147624. doi: 10.1371/journal.pone.0147624
Sun, S., Wang, Z., Fu, H., Duan, C., Wang, X., and Zhu, Z. (2015). Resistance to powdery mildew in the pea cultivar Xucai 1 is conferred by the gene er1. Crop J. 3, 489–499. doi: 10.1016/j.cj.2015.07.006
Svabova, L., Smykal, P., Griga, M., and Ondrej, V. (2005). Agrobacterium-mediated transformation of Pisum sativum in vitro and in vivo. Biol. Plant. 49, 361–370. doi: 10.1007/s10535-005-0009-6
Swisher Grimm, K. D., and Porter, L. D. (2020). Development and validation of KASP markers for the identification of pea seedborne mosaic virus Pathotype P1 resistance in Pisum sativum. Plant Dis. 104, 1824–1830. doi: 10.1094/PDIS-09-19-1920-RE
Tanksley, S., and McCouch, S. (1997). Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
Taran, B., Warkentin, T., Somers, D., Miranda, D., Vandenberg, A., Blade, S., et al. (2003). Identification of quantitative trait loci for plant height, lodging resistance and reaction to mycosphaerella blight in pea (Pisum sativum L.) using an AFLP-based linkage map. Theor. Appl. Genet. 107, 1482–1491. doi: 10.1007/s00122-003-1379-9
Tayeh, N., Aluome, C., Falque, M., Jacquin, F., Klein, A., Chauveau, A., et al. (2015a). Development of two major resources for pea genomics: the GenoPea 13.2K SNP Array and a high density, high resolution consensus genetic map. Plant J. 84, 1257–1273. doi: 10.1111/tpj.13070
Tayeh, N., Aubert, G., Pilet-Nayel, M. L., Lejeune-Hénaut, I., Warkentin, T. D., and Burstin, J. (2015b). Genomic tools in pea breeding programs: status and perspectives. Front. Plant Sci. 6:1037. doi: 10.3389/fpls.2015.01037
Teshome, A., Bryngelsson, T., Dagne, K., and Geleta, M. (2015). Assessment of genetic diversity in Ethiopian field pea (Pisum sativum L.) accessions with newly developed EST-SSR markers. BMC Genetics 16:102. doi: 10.1186/s12863-015-0261-5
Timmerman-Vaughan, G. M., Moya, L., Frew, T. J., Murray, S. R., and Crowhurst, R. (2016). Ascochyta blight disease of pea (Pisum sativum L.): defence-related candidate genes associated with QTL regions and identification of epistatic QTL. Theor. Appl. Genet. 129, 879–896. doi: 10.1007/s00122-016-2669-3
Timmerman, G. M., Frew, T. J., Miller, A. L., Weeden, N. F., and Jermyn, W. A. (1993). Linkage mapping of sbm-1, a gene conferring resistance to pea seed-borne mosaic virus, using molecular markers in Pisum sativum. Theor. Appl. Genet. 85, 609–615. doi: 10.1007/BF00220920
Timmerman, G. M., Frew, T. J., Weeden, N. F., Miller, A. L., and Goulden, D. S. (1994). Linkage analysis of er-1, a recessive Pisum sativum gene for resistance to powdery mildew fungus (Erysiphe pisi DC). Theor. Appl. Genet. 88, 1050–1055. doi: 10.1007/BF00220815
Timmerman-Vaughan, G. M., Frew, T. J., Butler, R., Murray, S., Gilpin, M., Falloon, K., et al. (2004). Validation of quantitative trait loci for ascochyta blight resistance in pea (Pisum sativum L.), using populations from two crosses. Theor. Appl. Genet. 109, 1620–1631. doi: 10.1007/s00122-004-1779-5
Timmerman-Vaughan, G. M., Frew, T. J., Russell, A. C., Khan, T., Butler, R., Gilpin, M., et al. (2002). QTL mapping of partial resistance to field epidemics of ascochyta blight of pea. Crop Sci. 42, 2100–2111. doi: 10.2135/cropsci2002.2100
Timmerman-Vaughan, G. M., Mc Callum, J. A., Frew, T. J., Weeden, N. F., and Russell, A. C. (1996). Linkage mapping of quantitative trait loci controlling seed weight in pea (Pisum sativum L.). Theor.Appl. Genet. 93, 431–439. doi: 10.1007/BF00223187
Timmerman-Vaughan, G. M., Pither-Joyce, M. D., Cooper, P. A., Russell, A. C., Goulden, D. S., Butler, R., et al. (2001). Partial resistance of transgenic peas to alfalfa mosaic virus under greenhouse and field conditions. Crop Sci. 41, 846–853. doi: 10.2135/cropsci2001.413846x
Tiwari, K. R., Penner, G. A., and Warkentin, T. D. (1997). Inheritance of powdery mildew resistance in pea. Can. J. Plant Sci. 77, 307–310. doi: 10.4141/p96-157
Tiwari, K. R., Penner, G. A., and Warkentin, T. D. (1998). Identification of coupling and repulsion phase RAPD markers for powdery mildew resistance gene er-1 in pea. Genome 41, 440–444. doi: 10.1139/g98-014
Toker, C., and Mutlu, N. (2011). “Breeding for abiotic stresses,” in Biology and Breeding of Food Legumes, eds A. Pratap and J. Kumar (Wallingford: CABI), 241–261. doi: 10.1098/rstb.2007.2179
Tonguç, M., and Weeden, N. F. (2010). Identification and mapping of molecular markers linked to er1 gene in pea. J. Plant Mol. Biol. Biotechnol. 1, 1–5.
Tran, H. S., You, M. P., Lanoiselet, V., Khan, T. N., and Barbetti, M. J. (2014). First report of Phoma glomerata associated with the ascochyta blight complex on field pea (Pisum sativum) in Australia. Plant Dis. 98, 427. doi: 10.1094/PDIS-08-13-0809-PDN
Tuinstra, M. R., Ejeta, G., and Goldsbrough, P. B. (1997). Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor. Appl. Genet. 95, 1005–1011. doi: 10.1007/s001220050654
Tyagi, M. K., and Srivastava, C. P. (1999). Inheritance of powdery mildew and rust resistance in pea. Ann. Biol. 15, 13–16.
Valderrama, M. R., Román, B., Satovic, Z., Rubiales, D., Cubero, J. I., and Torres, A. M. (2004). Locating quantitative trait loci associated with Orobanche crenata resistance in pea. Weed Res. 44, 323–328. doi: 10.1111/j.1365-3180.2004.00406.x
Vijayalakshmi, S., Yadav, K., Kushwaha, C., Sarode, S. B., Srivastava, C. P., Chand, R., et al. (2005). Identification of RAPD markers linked to the rust (Uromyces fabae) resistance gene in pea (Pisum sativum). Euphytica 144, 265–274. doi: 10.1007/s10681-005-6823-2
Vishal, M., and Ram, U. (2005). Field screening of pea, Pisum sativum L. germplasm for resistance against major insect pests. J. Plant Prot. Environ. 2, 50–58.
Wallen, V. R. (1955). The effect of storage for several years on the viability of Ascochyta pisi in pea seed and on the germination of the seed and emergence. Plant Dis. Rep. 39, 674–677. doi: 10.15258/sst.2007.35.3.14
Wang, Z., Fu, H., Sun, S., Duan, C., Wu, X., Yang, X., et al. (2015). Identification of powdery mildew resistance gene in pea line X9002. Acta Agron. Sin. 41, 515–523. doi: 10.3724/sp.j.1006.2015.00515
Warkentin, T. D., Rashid, K. Y., and Xue, A. G. (1996). Fungicidal control of powdery mildew in field pea. Can. J. Plant Sci. 76, 933–935. doi: 10.4141/cjps96-156
Warkentin, T. D., Smykal, P., Coyne, C. J., Weeden, N., Domoney, C., Bing, D., et al. (2015). “Pea (Pisum sativum L.),” in The Grain Legumes. Series Handbook of Plant Breeding, ed. A. M. De Ron (New York, NY: Springer Science Business Media), 37–83.
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi: 10.1038/s41477-017-0083-8
Weeden, N. F., and Porter, L. (2007). The genetic basis of Fusarium root rot tolerance in the ‘Afghanistan’ pea. Pisum Genet. 39, 35–36.
Weeden, N. F., McGee, R., Grau, C. R., and Muehlbauer, F. J. (2000). A gene influencing tolerance to common root rot is located on linkage group IV. Pisum Genet. 32, 53–55.
Wicker, E., Moussart, A., Duparque, M., and Rouxel, F. (2003). Further contributions to the development of a differential set of pea cultivars (Pisum sativum) to investigate the virulence of isolates of Aphanomyces euteiches. Eur. J. Plant Pathol. 109, 47–60.
Wroth, J. M. (1998). Possible role for wild genotypes of Pisum spp. to enhance ascochyta blight resistance in pea. Aust. J. Exp. Agric. 38, 469–479. doi: 10.1071/ea98024
Wu, L., Chang, K. F., Conner, R. L., Strelkov, S., FreduaAgyeman, R., Hwang, S. F., et al. (2018). Aphanomyces euteiches: a threat to Canadian field pea production. Engineering 4, 542–551. doi: 10.1016/j.eng.2018.07.006
Xue, A. G. (2000). Effect of seed-borne Mycosphaerella pinodes and seed treatments on emergence, foot rot severity, and yield of field pea. Can. J. Plant Pathol. 22, 248–253. doi: 10.1080/07060660009500471
Xue, A. G., Warkentin, T. D., and Kenaschuk, E. O. (1996). “Mycosphaerella blight of field pea-potential damage and fungicide control,” in Proceedings of Manitoba Agri-Forum, Winnipeg, MB, 5–6.
Yadav, A., Singh, V., Yadav, A., and Singh, H. (2019). Studies on succession of insect pest complex associated with pea at Bikaner. J. Entomol. Zool. Stud. 7, 1606–1608.
Yadav, S. K., and Patel, S. (2015). Insect-pest complex on Pisum sativum L. and their natural enemies at Pantnagar. J. Plant Dev. Sci. 7, 839–841.
Yarnell, S. H. (1962). Cytogenetics of the vegetable crops. III. Legumes. A. Garden peas, Pisum sativum L. Bot. Rev. 28, 465–537. doi: 10.1007/bf02868757
Zhi, P., and Chang, C. (2021). Exploiting epigenetic variations for crop disease resistance improvement. Front. Plant Sci. 12:953. doi: 10.3389/fpls.2021.692328
Zhuang, L. L., Ambrose, M., Rameau, C., Weng, L., Yang, J., Hu, X. H., et al. (2012). LATHYROIDES, encoding a WUSCHEL-related Homeobox1 transcription factor, controls organ lateral growth, and regulates tendril and dorsal petal identities in garden pea (Pisum sativum L.). Mol. Plant 5, 1333–1345. doi: 10.1093/mp/sss067
Keywords: biotic stresses, genomics, proteomics, marker assisted breeding, speed breeding
Citation: Parihar AK, Kumar J, Gupta DS, Lamichaney A, Naik SJ S, Singh AK, Dixit GP, Gupta S and Toklu F (2022) Genomics Enabled Breeding Strategies for Major Biotic Stresses in Pea (Pisum sativum L.). Front. Plant Sci. 13:861191. doi: 10.3389/fpls.2022.861191
Received: 24 January 2022; Accepted: 28 March 2022;
Published: 18 May 2022.
Edited by:
Sukhjiwan Kaur, Agriculture Victoria, AustraliaReviewed by:
Cengiz Toker, Akdeniz University, TurkeySurinder Banga, Punjab Agricultural University, India
Babu Ram Pandey, Agriculture Victoria, Grains Innovation Park, Australia
Copyright © 2022 Parihar, Kumar, Gupta, Lamichaney, Naik SJ, Singh, Dixit, Gupta and Toklu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashok Kumar Parihar, YXNob2thLnBhcmloYXJAZ21haWwuY29t; Jitendra Kumar, aml0ZW5kcmE3M0BnbWFpbC5jb20=; Faruk Toklu, ZmFwZXRAY3UuZWR1LnRy
†These authors have contributed equally to this work
 Ashok Kumar Parihar
Ashok Kumar Parihar Jitendra Kumar
Jitendra Kumar Debjyoti Sen Gupta
Debjyoti Sen Gupta Amrit Lamichaney
Amrit Lamichaney Satheesh Naik SJ1
Satheesh Naik SJ1 Girish P. Dixit
Girish P. Dixit Sanjeev Gupta
Sanjeev Gupta Faruk Toklu
Faruk Toklu