- Shanghai Key Laboratory of Plant Molecular Sciences, College of Life Sciences, Shanghai Normal University, Shanghai, China
AtRsmD was recently demonstrated to be a chloroplast 16S rRNA methyltransferase (MTase) for the m2G915 modification in Arabidopsis. Here, its function of AtRsmD for chloroplast development and photosynthesis was further analyzed. The AtRsmD gene is highly expressed in green photosynthetic tissues. AtRsmD is associated with the thylakoid in chloroplasts. The atrsmd-2 mutant exhibited impaired photosynthetic efficiency in emerging leaves under normal growth conditions. A few thylakoid lamellas could be observed in the chloroplast from the atrsmd-2 mutant, and these thylakoids were loosely organized. Knockout of the AtRsmD gene had minor effects on chloroplast ribosome biogenesis and RNA loading on chloroplast ribosomes, but it reduced the amounts of chloroplast-encoded photosynthesis-related proteins in the emerging leaves, for example, D1, D2, CP43, and CP47, which reduced the accumulation of the photosynthetic complex. Nevertheless, knockout of the AtRsmD gene did not cause a general reduction in chloroplast-encoded proteins in Arabidopsis grown under normal growth conditions. Additionally, the atrsmd-2 mutant exhibited more sensitivity to lincomycin, which specifically inhibits the elongation of nascent polypeptide chains. Cold stress exacerbated the effect on chloroplast ribosome biogenesis in the atrsmd-2 mutant. All these data suggest that the AtRsmD protein plays distinct regulatory roles in chloroplast translation, which is required for chloroplast development and chloroplast function.
Introduction
In all living organisms, ribosomal RNAs (rRNAs) constitute the functional center of the ribosome. These RNAs are extensively modified, and the modified nucleotides are usually located in the regions crucial to the function of the ribosome (Urlaub et al., 1997). In Escherichia coli, the majority of modified nucleotides in rRNAs belong to various types of base and ribose methylated residues (Sergiev et al., 2011), and there are 11 modified nucleotides in 16S rRNA (Andersen and Douthwaite, 2006). The three-dimensional locations of these modifications on ribosome crystal structures indicate that these nucleotides are distributed within several discrete regions related to essential ribosomal functions (Brimacombe et al., 1993; Ban et al., 2000; Wimberly et al., 2000; Harms et al., 2001; Yusupov et al., 2001; Schuwirth et al., 2005). rRNA modifications play important roles in efficient protein synthesis (Krzyzosiak et al., 1987; Green and Noller, 1999; Khaitovich et al., 1999). The corresponding enzymes for these sites have been determined and characterized in E. coli (Andersen and Douthwaite, 2006). For example, the yebU gene encodes a putative m5C RNA MTase for the methylation of nucleotide 1407 in 16S rRNA, and the yebU knockout strain displays slower growth and reduced fitness in competition with wild-type cells. The RsmH gene encodes a conserved MTase that is a specific AdoMet-dependent MTase responsible for the N(4)-methylation of C1402 in 16S rRNA in almost all species of bacteria (Wei et al., 2012). Two Escherichia coli guanine-N2 (m2G) MTases, rsmC and rsmD, modify nucleotides G1207 and G966 in 16S rRNA, respectively, and they possess a common MTase domain related to the YbiN family of hypothetical MTases in their C-terminus and a variable region in their N-terminus. Knockout of either rsmC or rsmD results in a mild reduction in cellular growth in E. coli (Bujnicki and Rychlewski, 2002; Kimura and Suzuki, 2010). Characterization of these MTase for 16S rRNA methylation has provided a comprehensive understanding of ribosome biogenesis in prokaryotes.
Chloroplasts in plant cells are derived from an endosymbiotic event in which a photosynthetic cyanobacterium was engulfed by an early eukaryotic cell (Keeling, 2013). During host-endosymbiont coevolution, the chloroplast retained a small-scale genome, while many genes were transferred to the nuclear genome (Abdallah et al., 2000). The chloroplast has both the transcriptional and translational machinery for the expression of chloroplast genes. The translational machinery is a bacterial-type 70S ribosome that is composed of a large 50S subunit and a small 30S subunit. The small 30S subunit comprises 25 ribosomal proteins and chloroplast 16S rRNA, while the 50S large subunit consists of 33 ribosomal proteins and three other types of chloroplast rRNAs (23S rRNA, 4.5S rRNA, and 5S rRNA) (Graf et al., 2017; Zoschke and Bock, 2018). Not only do chloroplast ribosomal proteins exhibit high similarities to their counterparts in E. coli, but rRNA constituents are also conserved between chloroplasts and E. coli (Zoschke and Bock, 2018). Analogous to methylation in E. coli rRNAs, this modification occurs in chloroplast rRNAs, and the modification sites and the corresponding enzymes are highly conserved (Zou et al., 2020; Manduzio and Kang, 2021; Ngoc et al., 2021a,b). In contrast, research on rRNA modification in chloroplasts lags behind that in E. coli. Two independent groups recently reported that the chloroplast-localized CMAL protein, an ortholog of the bacterial MraW/RsmH protein, is involved in the m4C methylation of C1352 in chloroplast 16S rRNA, which plays essential roles in chloroplast development and hormonal responses in higher plants (Zou et al., 2020; Ngoc et al., 2021a). Ngoc et al. (2021b) reported that chloroplast-localized AtRsmD, an ortholog of the MTase for the N2-methylguanosine (m2G) modification of 16S rRNA in E. coli, is involved in m2G methylation at position 915 in chloroplast 16S rRNA in Arabidopsis. The atrsmd mutant exhibited no observable differences in seed germination or seedling growth when grown under normal growth conditions; however, the atrsmd mutant was hypersensitive to both cold stress and translation inhibitors (Ngoc et al., 2021b). This finding indicates that AtRsmD is a chloroplast 16S rRNA MTase for the m2G915 modification and that it plays a role in the adaptation of Arabidopsis to cold stress (Ngoc et al., 2021b). Nevertheless, the role of the AtRsmD protein in chloroplast development remains to be further explored.
In this study, we further characterized the AtRsmD gene and the corresponding AtRsmD deletion mutant, atrsmd-2. Knockout of the AtRsmD gene impaired chloroplast development and reduced photosynthetic efficiency in emerging leaves in Arabidopsis under normal growth conditions. It did not cause in a general reduction in chloroplast-encoded proteins in Arabidopsis when grown under normal culture conditions but affected the amounts of photosynthetic proteins, such as D1, D2, CP43, and CP47. This reduction interrupted the accumulation of the photosynthetic complex in the atrsmd-2 mutant. These data suggested that the AtRsmD protein plays distinct regulatory roles in the rapid synthesis of photosynthesis-related proteins, which are required for chloroplast development and chloroplast function.
Materials and Methods
Plant Materials and Growth Conditions
The Arabidopsis thaliana Col-0 ecotype was used in this study. The T-DNA insertional line (CS832131) was obtained from the Arabidopsis Biological Resource Centre (ABRC; Ohio State University, United States). Plants were grown in a growth chamber with a 16-h light/8-h dark photoperiod at a constant temperature of 22°C. The light intensity was 120 μmol m–2 s–1. For growth on agar plates, the seeds were surface-sterilized with 75% alcohol and sown on Murashige-Skoog (MS) medium and 0.7% (w/v) phytoagar with or without the corresponding antibiotics.
For the genomic complementation experiment, the 2,594 bp wild-type genomic fragment of the AtRsmD (AT3G28460) gene was amplified with the gene-specific primers listed Supplementary Table 1 using high-fidelity KOD plus polymerases (TOYOBO)1 and then subcloned into the modified pCAMBIA1300 binary vector (CAMBIA)2 with a three-tandem FLAG tag. The construct was introduced into the atrsmd-2 mutant via Agrobacterium tumefaciens GV3101 (Clough and Bent, 1998). Transgenic lines were screened on MS medium with 80 mg/L hygromycin B (Roche).3
Chlorophyll Fluorescence and Chlorophyll Content Measurements
Chlorophyll fluorescence was determined using an Imaging PAM fluorometer (Walz, Germany). Measurement of chlorophyll fluorescence was performed according to Li et al.’s (2019) report. For chlorophyll content determination, equal fresh weights of Arabidopsis leaves were extracted with equal volumes of 80% (v/v) acetone, and the absorbance of the supernatant was measured at 663 and 646 nm. Both chlorophyll a and b contents were calculated using the equation of Porra (2002).
Transmission Electron Microscopy Observation
Young primary leaves grown under normal conditions or cold treatments were fixed and embedded as described in a previous report (Yu et al., 2013). Thin sections were prepared with an ultramicrotome and then stained with uranyl acetate and lead citrate. The ultrastructure was examined using a Tecnai Spirit G2 BioTWIN transmission electron microscope.
Subcellular Localization Analysis
The full-length coding sequence of the AtRsmD gene was amplified by high-fidelity KOD plus polymerases (TOYOBO)4 with the corresponding gene-specific primers and fused in frame with the enhanced green fluorescence protein (eGFP) of the modified pRIN101:eGFP vector (TOYOBO, see text footnote 4) to produce AtRsmD:eGFP. Both the construct and free pRIN101:eGFP vector were transformed into Arabidopsis protoplasts prepared as described in a previous report, respectively (Yoo et al., 2007). The fluorescence signals were imaged by a laser-scanning confocal microscope (Zeiss, Oberkochen, Germany) with excitation and emission wavelengths of 488 and 545 nm, respectively.
Chloroplast fractions were isolated as described in a previous report (Xiong et al., 2020). Three-week-old leaves from the AtRsmD:eGFP stable transgenic lines were homogenized in precooled buffer I (0.33 M sorbitol; 0.02 M Tricine/KOH, pH 8.4; 5 mM EGTA, pH 8.35; 5 mM EDTA, pH 8.0; 10 mM NaHCO3). After filtration with a double layer of Miracloth, the homogenate was further filtered through 100- and 40-μm sieves, in order, and the filtrate was centrifuged at 2,000 g for 5 min at 4°C to obtain intact chloroplasts. Following centrifugation, the intact chloroplasts were then resuspended in buffer II (0.33 M sorbitol; 5 mM MgCl2; 2.5 M EDTA, pH 8.0; 20 mM HEPES/KOH, pH 7.6) and in buffer III (5 mM MgCl2-6H2O, 25 M EDTA pH 8.0, 20 mM HEPES/KOH pH 7.6), successively. After centrifugation at 4°C, the supernatant contained stroma, while the sediment contained the thylakoid fractions. The corresponding antibodies were used to perform immunoblotting analysis.
Sequence Alignment and Phylogenetic Tree Construction
Protein sequences were aligned with ClustalW, and the alignment results were displayed with the BoxShade Server.5 The phylogenetic tree was constructed and tested using MEGA 3.16 based on the neighbor-joining method.
Nucleic Acid Isolation, cDNA Synthesis, and qPCR Analysis
For genomic DNA isolation, samples were homogenized in lysis buffer [200 mM Tris-HCl, pH 7.5; 25 mM NaCl; 25 mM EDTA; and 0.5% (w/v) SDS], and the homogenate was isolated with phenol/chloroform (1:1, v/v). After centrifugation, the supernatant was collected, and genomic DNA was precipitated by adding ice-cold isopropyl alcohol. After washing with 70% (v/v) ethanol, the genomic DNA was air-dried and dissolved in double-distilled water. For total RNA isolation, samples were ground in liquid nitrogen, and total RNA was isolated using an RNA isolation kit (Tiangen)7 according to the manufacturer’s instructions.
Total RNA (5 μg) was used to synthesize first-strand cDNA with Trans-Script® Fly First-Strand cDNA Synthesis Super-Mix. Quantitative PCR (qPCR) analysis was performed using the gene-specific primers listed in Supplementary Table 1 and SYBRGREEN I master mix reagent (Toyobo, see text footnote 1) on a real-time RT–PCR system (ABI7300, United States). Reactions were performed in triplicate for each sample, and expression levels were normalized against TUBLIN4.
Polysome Association Analysis
Polysome association assays were performed as described previously (Zhang et al., 2018). Total extracts from the young leaves of 2-week-old plants were fractionated in 15–55% sucrose gradients through centrifugation at 45,000 rpm (246,000 g) at 4°C for 65 min. After centrifugation, 10 fractions (0.5 mL each fraction) were collected successively from the sucrose gradients. Then, the total RNA in each fraction was isolated, and an equal proportion of RNA was separated using RNA gel blot analysis.
RNA Gel-Blot Hybridization
RNA hybridization was performed according to the Roche manual as described previously (Yu et al., 2013). Total RNA (5 μg) from the wild type and atrsmd-2 mutant was separated using formamide denaturing agarose gel electrophoresis, transferred onto nylon membranes (MILLIPORE), and subjected to RNA gel blotting with digoxigenin (DIG)-labeled nucleic acid probes (DIG Easy Hyb system; Roche). The probes were prepared using a PCR DIG synthesis kit (Roche, see text footnote 3) with the specific primers listed in Supplementary Table 1. Total RNA was detected by ethidium bromide staining, and the signals on the RNA gel blot were visualized with a LuminoGraph WSE-6100 (ATTO).
Chloroplast Blue Native PAGE and Two-Dimensional Analysis
Chloroplast BN-PAGE and 2-D analysis were performed according to a previous report (Zhang et al., 2018). Briefly, 1 g of 2-week-old seedlings was homogenized in precooled HMSN buffer (0.4 M sucrose, 10 mM NaCl, 5 mM MgCl2-6H2O, and 10 mM HEPES) and then filtered through a multilayer microcloth. The isolated thylakoid pellets were resuspended in buffer [25 mM Bis-Tris-HCl, pH 7.0, 1% n-dodecyl b-D-maltoside (w/v), and 20% glycerol (w/v)] at 1.0 mg chlorophyll/mL and incubated at 4°C for 5 min followed by centrifugation at 12,000 g for 10 min. After one-tenth volume of loading buffer [100 mM Bis-Tris-HCl, pH 7.0, 0.5 M 6-amino-n-caproic acid, 5% Serva blue G (w/v), and 30% glycerol (w/v)] was added to the supernatant, and the samples were separated on 0.75-mm 4–12% acrylamide gradient gels in a Tannon vertical electrophoresis device at 4°C. For two-dimensional analysis, excised BN-PAGE lanes were soaked in SDS sample buffer for 30 min and layered onto 1-mm 10% SDS polyacrylamide gels containing 6 M urea. After electrophoresis, the proteins were stained with Coomassie bright blue.
Total Protein Extraction and Immunoblotting Analysis
Total protein was isolated from the young leaves of 2-week-old Arabidopsis seedlings according to a previous report (Zou et al., 2020). Briefly, samples were ground in extraction buffer [25 mM Tris-HCl (pH 7.9), 50 mM NaCl, 1 mM EDTA, 5% glycerol (v/v), and 2% SDS] and subsequently centrifuged at 12,000 g for 10 min. The total protein in the supernatant was quantified using the DC Protein Assay Kit according to the manufacturer’s instructions (Bio–Rad Laboratories). Approximately 10 μg of total protein was separated on 10% SDS polyacrylamide gels and transferred onto PVDF membranes. Immunoblotting analysis was performed with specific primary antibodies, and the signals from the secondary conjugated antibodies were detected with enhanced chemiluminescence and a TANNON imaging system.
Results
Knockout of the AtRsmD Gene Impaired Photosynthetic Efficiency in Arabidopsis
The chlorophyll fluorescence parameter Fv/Fm reflects the maximum quantum efficiency of photosystem II (PSII) photochemistry and has been widely used for early stress detection in plants. To obtain novel insights into the regulation of photosynthetic efficiency in Arabidopsis, we screened Arabidopsis T-DNA insertion lines on a large scale to detect their Fv/Fm values. We found that the Fv/Fm value of the homozygous T-DNA insertion line CS832131 was only 0.643 ± 0.02, which was lower than the 0.788 ± 0.02 value found for the control (Figures 1A,B). The homozygous mutants exhibited no obviously visible phenotype (Figure 1B). Measurement of the total chlorophyll contents indicated that the chlorophyll contents in the homozygous mutant were slightly lower than those in the wild type (Figure 1C). PCR sequencing analysis confirmed that the T-DNA is inserted in the fourth intron of the AT3G28460 gene. RT-PCR analysis showed that the full-length transcript of the AT3G28460 gene was not detected in the homozygous mutant but was detected in the wild type (Figure 1D), which demonstrated that the line is another mutant for the locus. Therefore, we named the line as the atrsmd-2 mutant, while a previously reported line (Ngoc et al., 2021b) was named as the atrsmd-1 mutant.
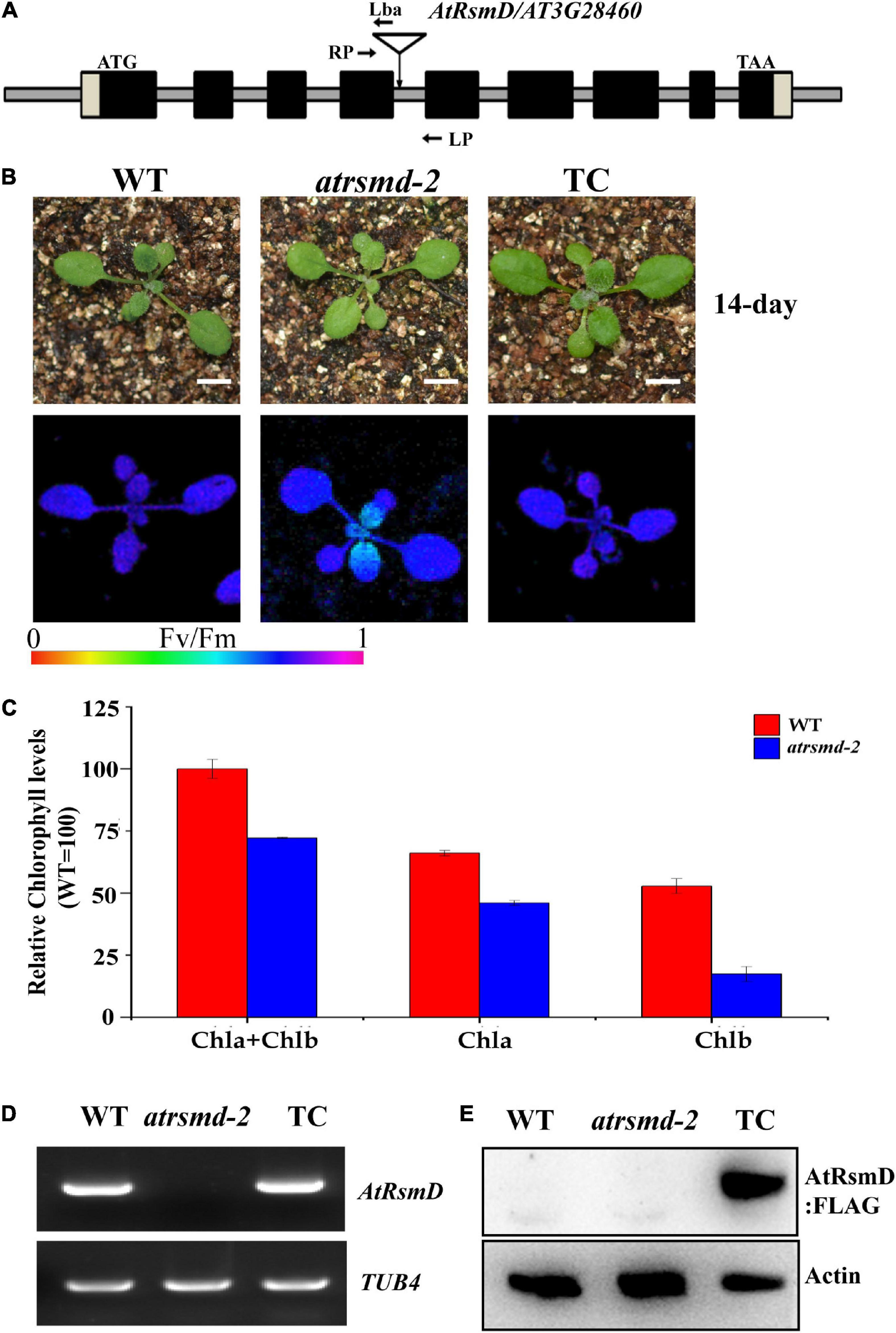
Figure 1. Knockout of the AtRsmD gene impaired photosynthetic efficiency in Arabidopsis. (A) Schematic illustrating the genomic structures of AtRsmD and the location of the T-DNA insertion. Black boxes and striped boxes indicate exons and introns, respectively. The T-DNA insertion site is indicated by an inverted triangle. Lba represents the left border primer of the T-DNA insertion. LP and RP represent the left and right genomic primers around the T-DNA insertion site, respectively. (B) Phenotype and Fv/Fm values of 14-day-old seedlings the wild type (WT), atrsmd-2 mutant and complemented lines (TC). Bar represents 1 cm. (C) Chlorophyll content in the emerging leaves from the 14-day-old wild type and atrsmd-2 mutant seedlings. (D) RT–PCR analysis of the AtRsmD gene in the wild type, atrsmd-2 mutant, and complemented line (TC). Tubulin 4 was used as a control. (E) Immunoblotting analysis of AtRsmD:FLAG in the wild type, atrsmd-2 mutant, and complemented line (TC) with the anti-FLAG antibody. Actin was used as a control.
To verify that the AT3G28460 locus is responsible for the impaired photosynthetic efficiency in the atrsmd-2 mutant, we produced a construct carrying the genomic sequence of the AT3G28460 gene fused with the coding sequence of a three-tandem FLAG tag before its stop codon that was driven by its native promoter, and we transformed it into the atrsmd-2 mutant via Agrobacterium tumefaciens (Clough and Bent, 1998). Three independent transgenic lines carrying the AtRsmD genomic fragment were identified to have the atrsmd-2/- genotype. RT-PCR results indicated that AtRsmD transcripts could be detected in the complemented line (Figure 1D). Immunoblotting analysis detected a protein with the predicted size of 35 kDa in the complemented line, but this protein was absent in both the wild type and atrsmd-2 mutant (Figure 1E). The values of Fv/Fm in the complemented line were restored to the levels of the wild type (Figure 1B). These data showed that knockout of the AtRsmD gene impaired the photosynthesis efficiency of photosystem II under normal conditions. Taken together, our data demonstrated that the AtRsmD:FLAG fusion protein is able to complement the atrsmd-2 mutant, and the fusion protein is functional.
Expressional Pattern of the AtRsmD Gene and Subcellular Localization of Its Product in Arabidopsis
To examine the expression pattern of the AtRsmD gene in different tissues, we performed reverse-transcription (RT)-PCR analysis of various tissues from wild-type plants. Our results showed that the AtRsmD transcripts were present in all tissues (Figures 2A,B). The AtRsmD transcripts were much more abundant in seedlings and leaves than in the other tissues. These data indicated that the AtRsmD gene was highly expressed in these green tissues.
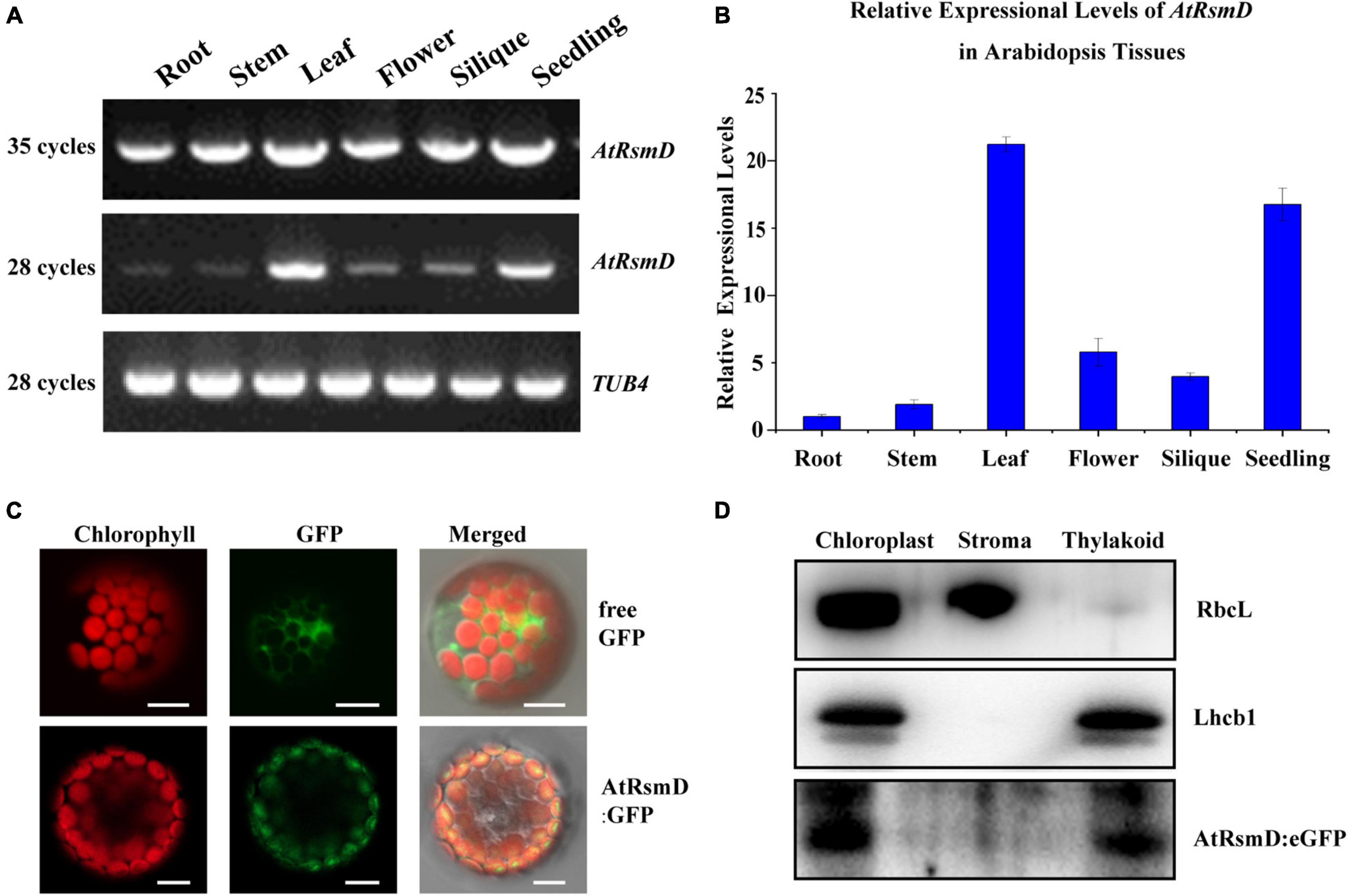
Figure 2. Expression pattern of the AtRsmD gene and subcellular localization of its product in Arabidopsis. (A) Semi-RT–PCR and (B) RT–qPCR analysis of AtRsmD mRNA in different Arabidopsis tissues and organs, including roots, stems, leaves, flowers, siliques and seedlings. Tubulin 4 was used as a control. (C) Localization of the AtRsmD:eGFP fusion protein in Arabidopsis protoplasts. Free eGFP in Arabidopsis protoplasts was used as a control. Green fluorescence shows eGFP fluorescence, red fluorescence indicates chloroplast autofluorescence, and orange/yellow fluorescence shows overlapping images of the two types of fluorescence. Scale bars: 5 μm. (D) The AtRsmD:eGFP protein is associated with thylakoid membranes in chloroplasts. The fractionation of Arabidopsis seedling chloroplasts was performed with anti-GFP, anti-Lhcb1, and anti-RbcL antibodies.
TargetP software predicted that the AtRsmD protein contains a potential chloroplast transpeptide at the N-terminus,8 and a very recent study demonstrated that the AtRsmD protein is localized to the chloroplast (Ngoc et al., 2021b). We made a construct in which the full-length encoding sequence of the AtRsmD protein was fused into the coding sequence of eGFP and introduced it into Arabidopsis protoplasts. AtRsmD:eGFP fluorescence colocalized with chlorophyll autofluorescence (Figure 2C), demonstrating that the AtRsmD protein is localized to the chloroplast. We also checked whether AtRsmD is associated with thylakoids. Chloroplast stroma and thylakoids were isolated from the stable AtRsmD:eGFP transgenic lines, and then immunoblotting analysis was carried out with the corresponding antibodies. Our immunoblotting analysis showed that AtRsmD:eGFP was detected in both chloroplasts and thylakoid fragments (Figure 2D). All these data showed that the AtRsmD protein is associated with thylakoids in chloroplasts.
Absence of the AtRsmD Protein Impaired the Accumulation of Chloroplast-Encoded Photosynthetic Proteins
The atrsmd-2 mutant exhibited impaired photosynthetic efficiency (Figure 1B), which could be due to reduced protein levels of photosynthetic complex elements. We investigated whether the absence of AtRsmD affects the accumulation of photosynthetic complex elements. Thylakoid membranes were solubilized with 1% dodecyl-β-D-maltopyranoside (DM), and then membrane protein complexes were separated by blue native PAGE (BN-PAGE). Our results showed that the amounts of supercomplex elements, such as the PSII supercomplex, PSI-PSII dimer, PSII monomer, and LhcII trimers, were clearly reduced in the atrsmd-2 mutant compared with those of the wild type (Figure 3A). The complexes resolved by BN-PAGE were then separated into their subunits in the second dimension by electrophoresis on SDS–PAGE gels and stained with Coomassie brilliant blue R 250. We found that the accumulation of the core subunits of PSII, CP43, CP47, D1, and D2 in the complex was clearly reduced in the atrsmd-2 mutant (Figure 3B) compared with their counterparts in the wild type. These results showed that assembly of the photosynthetic complex was seriously reduced in the atrsmd-2 mutant.
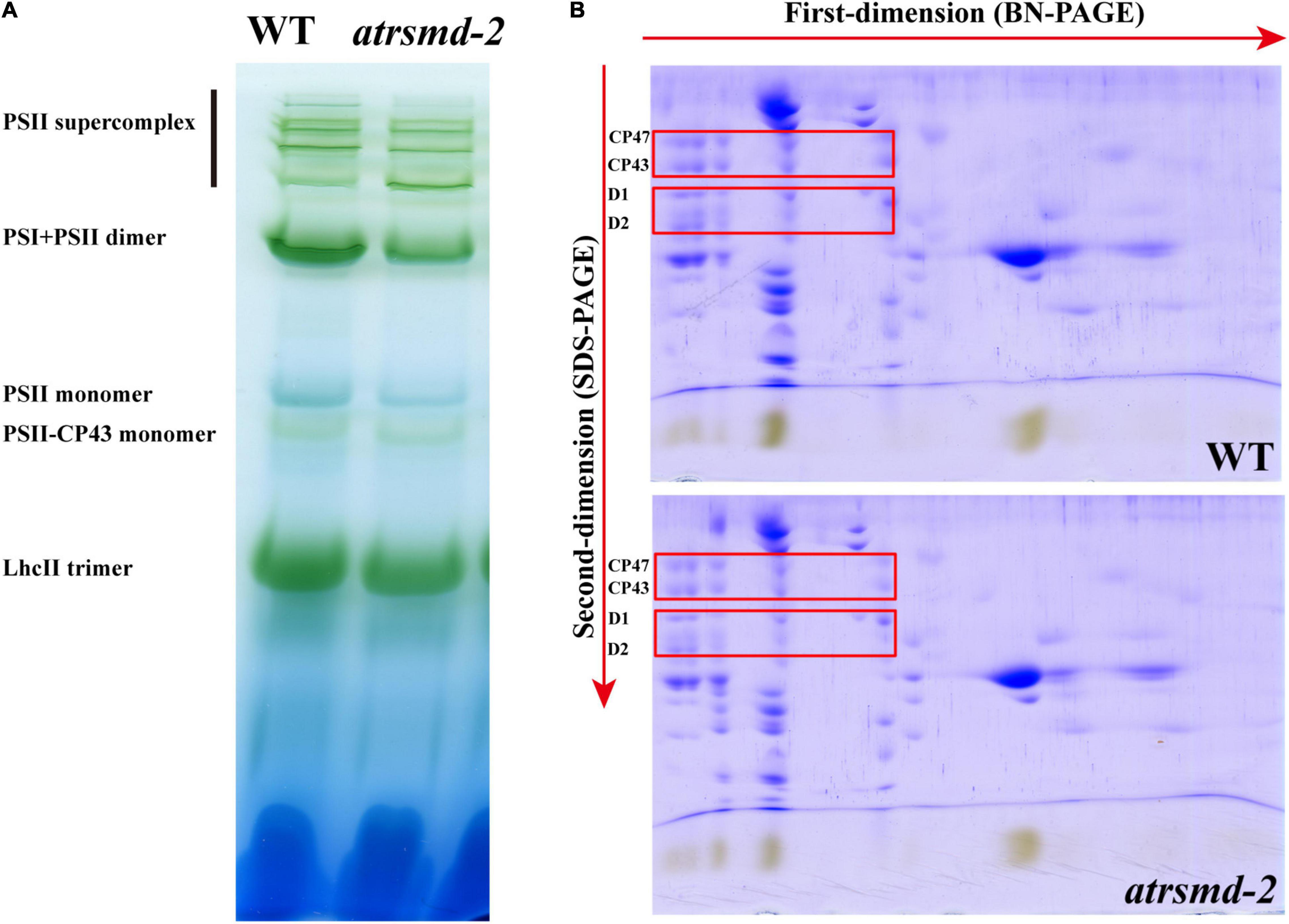
Figure 3. Analysis of photosystem complexes from the wild-type and atrsmd-2 mutant grown under normal growth conditions. (A) BN-gel analysis of the thylakoid membrane protein complexes in the wild type (WT) and the atrsmd-2 mutant. Representative unstained BN-PAGE gel electrophoresis is shown. Thylakoid membranes from the wild type (WT) and atrsmd-2 mutant were solubilized with 1% DM and separated by native PAGE. A sample with an equal amount of chlorophyll (16 μg) was loaded in each lane. The bands for supercomplexes are marked on the left. (B) 2-D, BN/SDS-urea-PAGE analysis of the thylakoid membrane complexes. Thylakoid complexes were separated by BN-PAGE and further subjected to 2D SDS–PAGE. The gels were stained with Coomassie Brilliant Blue. The locations of the plastid-encoded core proteins of PSII (CP43, CP47, D1, and D2) are indicated by red boxes on the gels, following to a previous report (Li et al., 2019).
To examine the steady-state levels of the thylakoid membrane proteins, immunoblotting analysis was performed with antibodies against specific subunits of the photosynthetic thylakoid membrane complex. Our results showed that the amounts of plastid-encoded photosynthesis-related proteins, including D1, D2, CP43, CP47, and AtpF, were 25–50% those of the wild type (Figure 4). In contrast, other photosynthesis-related proteins, such as AtpA, AtpB, RbcL, PetA, PetD, OEC33, and Lhcb1, accumulated to similar levels as the wild type (Figure 4), while the abundance of the PsaA protein increased in the mutant. These results indicated that knockout of the AtRsmD gene interrupted the accumulation of photosynthesis-related proteins, including chloroplast-encoded photosystem II proteins, and affected the assembly of the photosynthetic complex, which probably impaired photosynthetic efficiency in the atrsmd-2 mutant.
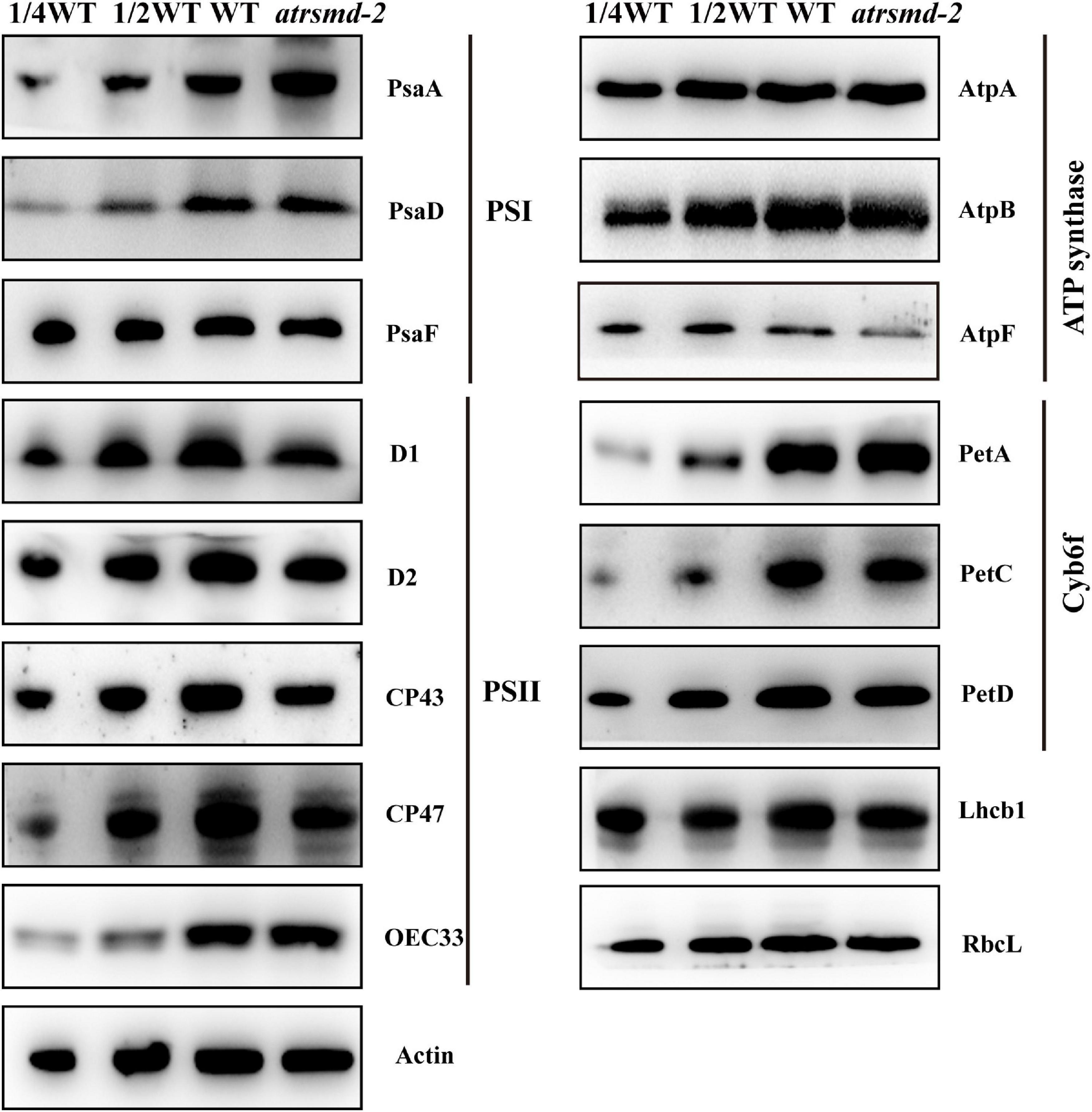
Figure 4. Immunoblot analysis of the photosynthetic proteins from the wild type (WT) and atrsmd-2 mutant. Total protein samples were prepared from 14-day-old emerging leaves of the wild type and atrsmd-2 mutant and then separated by SDS–PAGE. Immunoblot analysis of the photosynthetic proteins PsaA, PsaD, PsaF, D1, D2, CP43, CP47, OEC33, AtpA, AtpB, AtpF, PetA, PetC, PetD, Lhcb1, and RbcL using the corresponding antibodies was performed. Total proteins from the WT samples were loaded at three different concentrations (2.5, 5, and 10 μg), and total proteins from the mutant samples were loaded at 10 μg.
Absence of AtRsmD Affects Chloroplast Transcripts
Since the amounts of several chloroplast-encoded photosynthetic proteins, including D1, D2, CP43, CP47, and AtpF, were clearly reduced in the atrsmd mutant, we investigated whether the absence of the AtRsmD gene affected the accumulation of their corresponding chloroplast transcripts through RT–qPCR analysis. Our results showed that the chloroplast transcripts psbA, psbB, psbC, psbD, and atpF were 1-6-fold higher in the atrsmd-2 mutant than those in the wild type (Figure 5A), although the corresponding photosynthetic proteins were clearly reduced in the mutant. Additionally, we observed that the abundances of the chloroplast transcripts atpA, atpB, petA, petD, and rbcL were increased. We further investigated the accumulation of the PEP complex in the atrsmd-2 mutant. The amounts of the core subunit, RpoB, and the essential component, PAP8/pTAC6, were checked in this mutant, and our results showed that the abundance of these core subunits in the atrsmd-2 mutant was slightly increased compared with those in the wild type (Figure 5B). All these results indicated that knockout of the AtRsmD gene increased the accumulation of the PEP complex and altered the expression of these chloroplast transcripts in Arabidopsis, which is probably the result of feedback regulation.
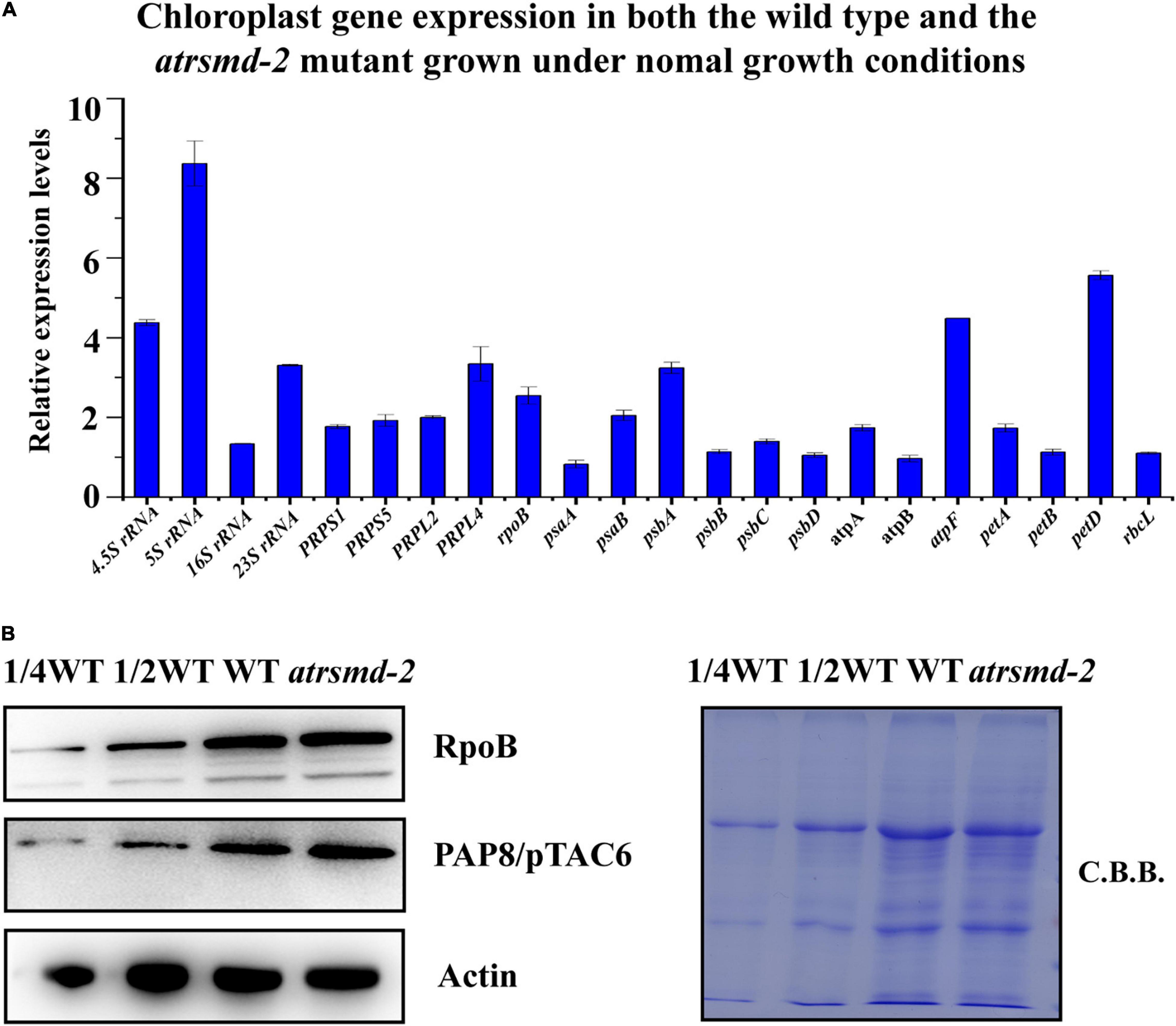
Figure 5. Absence of AtRsmD affects chloroplast transcripts in the wild type and atrsmd-2 mutant. (A) Relative expression levels of chloroplast-related genes in the wild type and atrsmd-2 mutant. (B) Immunoblot analysis of the RpoB and PAP8/pTAC6 proteins of the PEP complex from the wild type (WT) and atrsmd-2 mutant was performed. Total proteins from the WT samples were loaded at three different concentrations (2.5, 5, and 10 μg), and total proteins from the mutant samples were loaded at 10 μg.
Absence of AtRsmD Affects the Accumulation of Chloroplast Ribosomes
A previous report indicated that the AtRsmD protein is responsible for the methylation of 16S rRNA m2G915, and this methylation pattern was missing in the knockout line of the AtRsmD gene (Ngoc et al., 2021b). We further investigated the possible impacts of the absence of the AtRsmD protein on chloroplast ribosomal biogenesis. The levels of two components of the ribosomal 30S small subunit, PRPS1 (SMALL RIBOSOMAL SUBUNIT 1, bS1c) and PRPS5 (SMALL RIBOSOMAL SUBUNIT 5, uS5c), and two components of the ribosomal 50S large subunit, PRPL4 (LARGE RIBOSOMAL SUBUNIT 4, uL4c) and PRPL2 (LARGE RIBOSOMAL SUBUNIT 2, uL2c), were slightly reduced in the atrsmd-2 mutant grown under normal growth conditions, compared with those in the wild type (Figure 6A). Then, the levels of four chloroplast rRNA transcripts (16S rRNA, 23S rRNA, 5S rRNA, and 4.5S rRNA) were also investigated in the atrsmd-2 mutant using RNA blot analysis (Figures 6B,C). Our results indicated that the amount of the mature form of chloroplast 16S rRNA (1.5-knt molecule) in the atrsmd-2 mutant was reduced, whereas the amount of the precursor rRNA (1.7-kb) molecule was increased (Figure 6C). Levels of the 3. 2-, 2. 8-, 2.4- and 1.7-knt 23S rRNA species were increased in the atrsmd-2 mutant. Additionally, the level of one shorter form of the 1.3-knt species was decreased (Figure 6C), while the 1.1-knt form accumulated to a level similar as that in the wild type. In contrast, the levels of 4.5S and 5S rRNA transcripts were increased in the atrsmd-2 mutant. These data suggest that knockout of the AtRsmD gene affected the accumulation of chloroplast rRNAs and chloroplast ribosomal proteins. Nevertheless, knockout of the AtRsmD gene has a minor effect on chloroplast ribosome biogenesis.
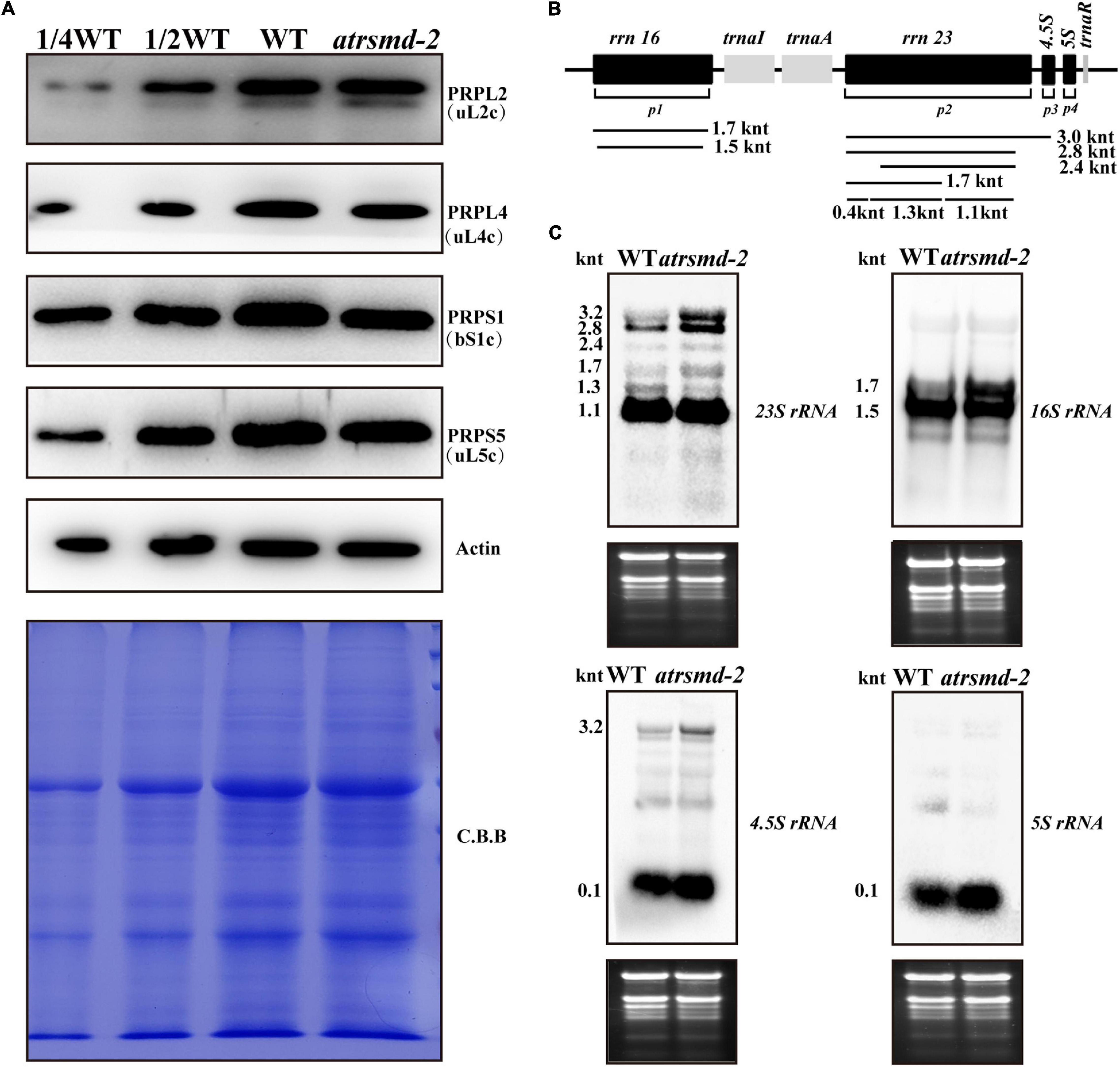
Figure 6. Accumulation of chloroplast ribosomal proteins and rRNAs in the wild type and atrsmd-2 mutant grown under normal conditions. (A) Immunoblot analysis of the chloroplast ribosomal proteins from the wild type (WT) and atrsmd-2 mutant was performed. PRPS1/bS1c,PRPS5/uS5c, PRPL4/uL4c, and PRPL2/uL2cwere checked in the wild type and atrsmd-2 mutant. Total proteins from the WT samples were loaded at three different concentrations (2.5, 5, and 10 μg), and total proteins from the mutant samples were loaded at 10 μg. (B) Schematic diagram of Arabidopsis chloroplast rRNA operon and the probe positions used RNA gel blot analyses. Sizes of different transcript forms of chloroplast rRNA are indicated below the diagram. p1, p2, p3, and p4 indicate positions of the probes for 16S rRNA, 23S rRNA, 4.5S rRNA, and 5S rRNA, respectively. (C) RNA gel analysis of chloroplast rRNAs in the wild type and atrsmd-2 mutant. Five micrograms of total RNA was loaded in each lane with the corresponding probes as shown above. Total RNA was stained with ethidium bromide (EtBr) and used as a loading control. The sizes of different transcript forms of chloroplast rRNAs are shown.
We subsequently investigated the effect of AtRsmD gene knockout on chloroplast translation initiation and termination. We examined the association of several plastid-encoded RNAs with chloroplast ribosomes. Polysome-enriched samples were isolated from the emerging young leaves of both the wild-type and atrsmd-2 mutant under polysome-preserving conditions and further fractionated through sucrose density-gradient centrifugation (Figure 7). Total RNA was isolated from 10 fractions and subjected to denaturing gel electrophoresis and RNA gel blot analyses using selected plastid transcript probes. As shown in Figure 7, we found that the amount of 16S rRNA in the polysomal fragments was slightly greater than that in the monosomal fragments in the atrsmd-2 mutant compared with that in the wild type. Similar trends were also observed for other transcripts, including 23S rRNA, psbA, and rbcL. These results suggested that the knockout of AtRsmD had minor effects on the RNA loading of chloroplast ribosomes.
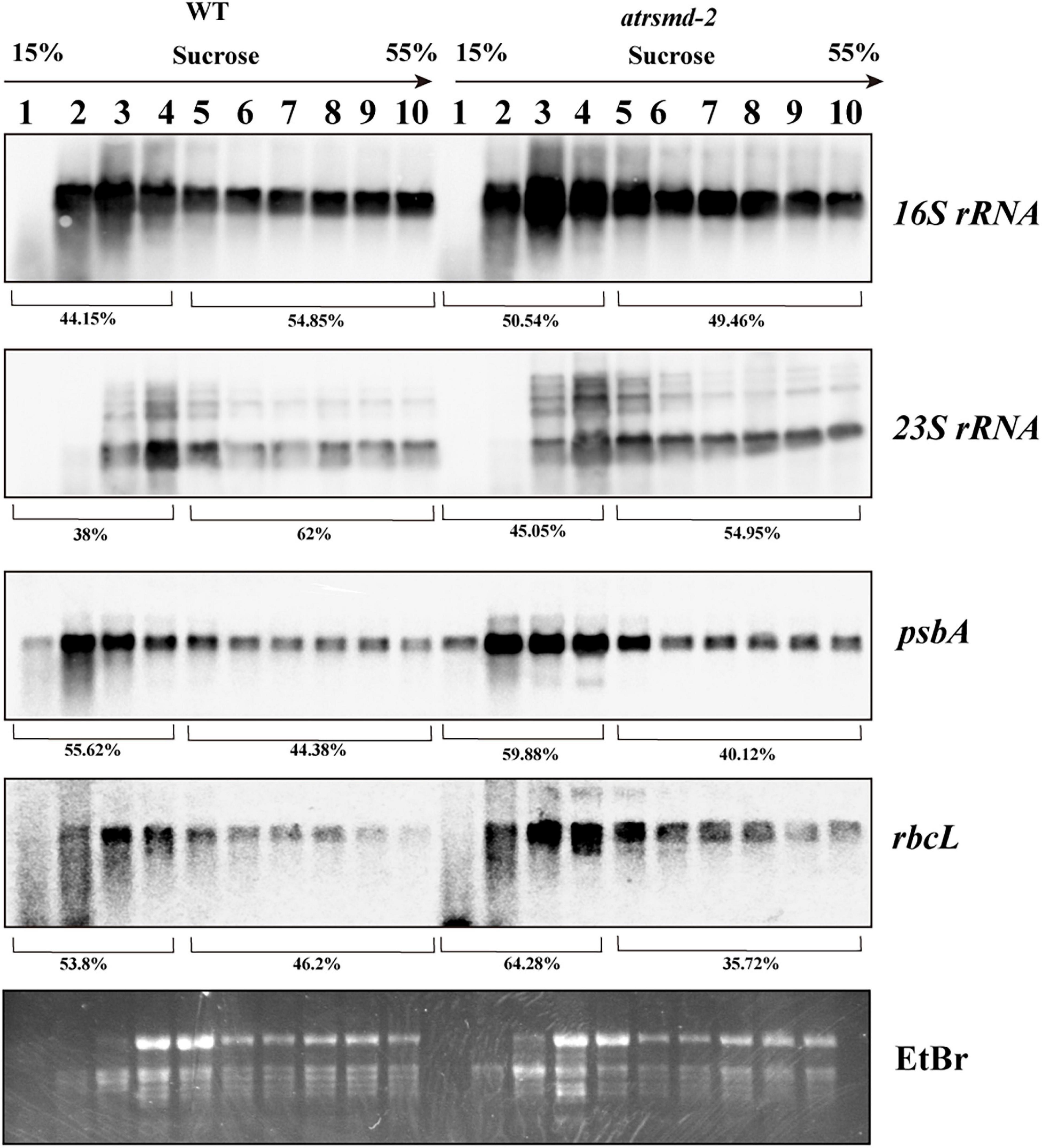
Figure 7. Chloroplast polysome analysis in the wild type and atrsmd-2 mutant. RNA gel-blot hybridization of fractions obtained following sucrose density-gradient centrifugation under polysome-preserving conditions. Free ribosomes (monosomes) are found in fractions 1–4, whereas fractions 5–10 contain RNA-polysome complexes. Membranes were hybridized with probes specific for 16S rRNA, 23S rRNA, psbA, and rbcL. Ethidium bromide staining was used as a loading control. Signals of the polysomal fractions (fractions 1–4) and monosomal/free RNA fraction (fractions 5–10) were quantified with the ImageJ software, and their relative quantities were indicated as percentage of the total signal over the 10 fractions.
Absence of the AtRsmD Protein Enhances the Sensitivity to Cold Stress and Chloroplast Translation Inhibitors
A recent study revealed that loss-of-function mutation of the AtRsmD gene enhanced its sensitivity to cold stress and prokaryotic translation inhibitors (Ngoc et al., 2021b). Lincomycin (LIN) and chloramphenicol (CAP) specifically inhibit the elongation of nascent polypeptide chains or block mRNA binding to the ribosome in prokaryotes. Here, we also investigated the effect of these two prokaryotic translation inhibitors on the atrsmd-2 mutant. The atrsmd-2 mutant was grown on MS medium plates in the presence of different LIN or CAP concentrations under sterile conditions, while the wild-type line was used as a control. Five days after germination, the atrsmd-2 mutant clearly exhibited hypersensitivity to lincomycin, as indicated by the enhanced loss of leaf coloration relative to the wild-type control. In contrast, the atrsmd-2 mutant is less sensitive to chloramphenicol than it is to lincomycin. Quantification of chlorophyll levels showed that LIN-treated atrsmd-2 seedlings accumulated significantly less of these pigments than wild-type seedlings (Supplementary Figure 1). These results demonstrated that knockout of AtRsmD enhanced the sensitivity to chloroplast translation inhibitors, especially lincomycin, indicating that the AtRsmD knockout line has compromised chloroplast ribosome activity.
We also investigated whether the atrsmd-2 mutant is sensitive to cold stress. Two-week-old atrsmd-2 mutants grown at 22°C were transferred to 4°C and grown for another 2 weeks, while the wild-type line was used as a control. For the atrsmd-2 mutant, yellowing became visible after 1 week of cold treatment and only occurred in the emerging leaves (Supplementary Figure 2A). In contrast, the wild type did not show obvious yellowing of the leaves (Supplementary Figure 2A). Transmission electron microscopy observations showed that a few grana and thylakoids could be observed in the chloroplasts of the atrsmd-2 mutant, and these membrane systems did not connect well. In contrast, the well-organized stroma and thylakoid systems could be observed in the wild-type chloroplasts (Supplementary Figure 2B). Chloroplasts from pale green leaves have more scattered grana structures in the atrsmd-2 mutant, and the thylakoid membranes were loosely organized. In contrast, well-organized thylakoids could be observed in the wild type (Supplementary Figure 2B). We then checked the abundance of photosynthesis-related proteins, and our results revealed that the amounts of the photosynthesis-related proteins PetD, D1, CP43, OEC33, and AtpF were reduced to 10–50% of those in the wild-type (Supplementary Figure 2C). These results showed that the accumulation of photosynthesis-related proteins was reduced in the atrsmd-2 mutant under cold stress conditions. We further examined the amount of chloroplast ribosome proteins, including PRPS1 (bS1c), PRPS5 (uS5c), PRPL2 (uL2c), and PRPL4 (uL4c), in the wild type and atrsmd-2 mutant. Our immunoblotting data revealed that the amounts of these chloroplast ribosomal proteins in the atrsmd-2 mutant were approximately 10–20% of those in the wild type under cold stress (Figure 8A). The levels of chloroplast rRNA transcripts were also investigated using RNA blot analysis in the atrsmd-2 mutant. Our results indicated that the amount of the mature form of chloroplast 16S rRNA (1.5-knt molecule) in the atrsmd-2 mutant was half of that in the wild type, whereas the amount of the precursor rRNA (1.7-knt) molecule was increased (Figure 8B). The levels of the three 3.2-knt 23S rRNA species were increased in atrsmd-2, while the level of a shorter form of the 1.3-knt transcript was decreased (Figure 8B). The levels of both the 4.5S and 5S rRNA transcripts were clearly reduced in the atrsmd-2 mutant. These data suggest that the effects of AtRsmD gene knockout on chloroplast ribosome assembly and chloroplast protein accumulation were exacerbated, which enhances sensitivity to cold stress.
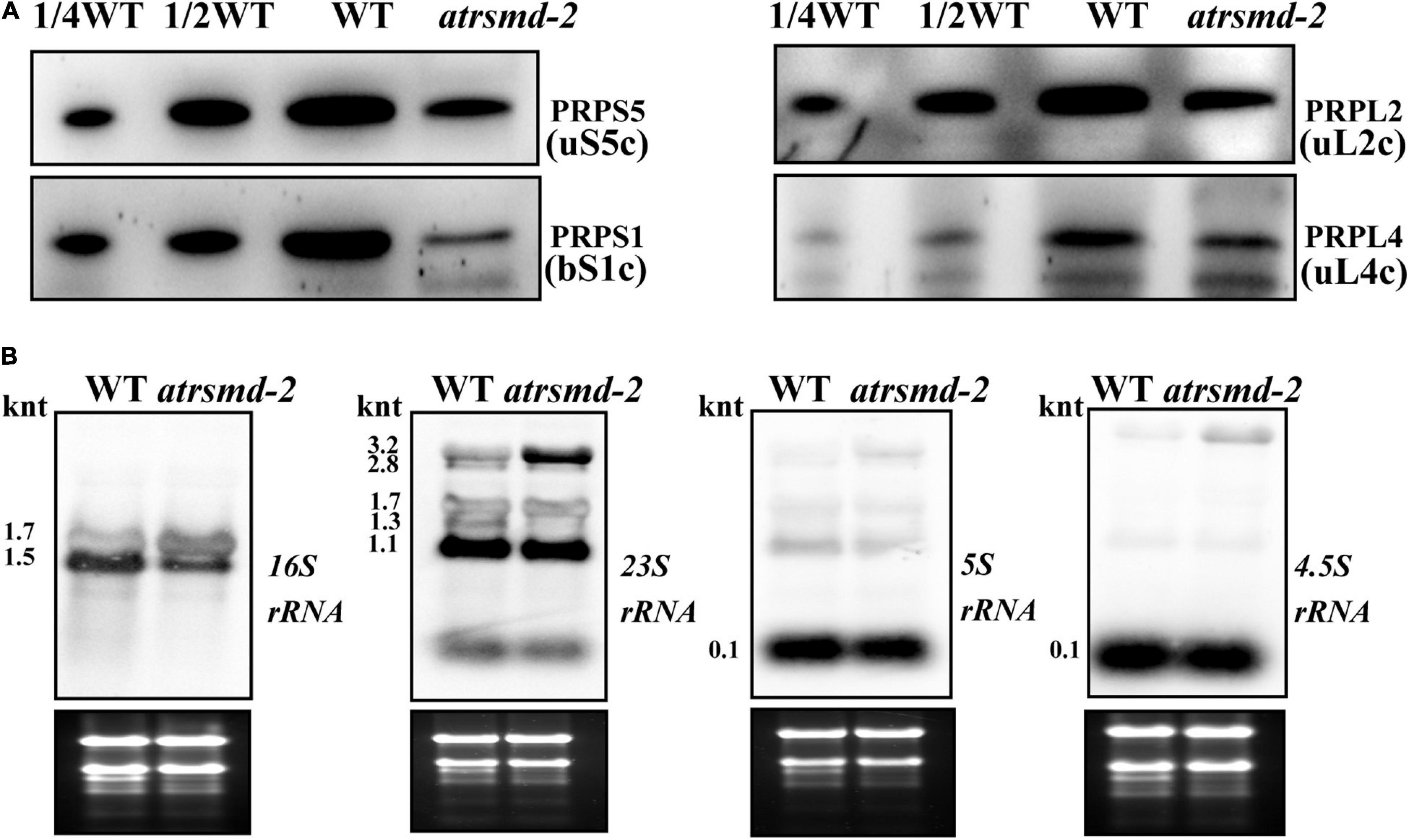
Figure 8. Accumulation of chloroplast ribosomal proteins and chloroplast rRNAs in the wild type and atrsmd-2 mutant treated with cold stress. (A) Immunoblot analysis of chloroplast ribosomal proteins, including PRPS1/bS1c, PRPS5/uS5c, PRPL4/uL4c and PRPL2/uL2c, from the wild type (WT) and atrsmd-2 mutant treated with cold stress. Total proteins from the WT samples were loaded at three different concentrations (2.5, 5, and 10 μg), and total proteins from the mutant samples were loaded at 10 μg. (B) RNA gel analysis of chloroplast rRNA species in the wild type and atrsmd-2 mutant treated with cold stress. Total RNA (5 μg) was loaded in each lane with the corresponding probes as shown above.
Discussion
AtRsmD encodes a chloroplast-localized MTase for N2-methylguanosine (m2G) modification of 16S rRNA at position 915 in Arabidopsis chloroplasts, which plays a role in the adaptation of Arabidopsis to cold stress. There were no obvious differences in seedling growth in the knockout mutant when grown under normal growth conditions (Ngoc et al., 2021b). This work extends the previous study and further characterizes the atrsmd-2 mutant. Our current study sheds light on the important roles of the AtRsmD protein in chloroplast development and chloroplast function. Fewer thylakoid membranes were observed in the atrsmd-2 mutant (Supplementary Figure 2B). Notably, the loss-of-function atrsmd-2 mutant exhibited impaired photosynthetic efficiency under normal growth conditions, although no visible phenotype could be observed in the atrsmd-2 mutant (Figure 1). Impaired photosynthesis is a possible consequence of the reduced levels of chloroplast-encoded photosynthesis-related proteins that hindered the assembly of the photosynthetic super complex (Figures 4, 5). Deletion of the AtRsmD gene had minor effects on chloroplast ribosome biogenesis and the RNA loading of chloroplast ribosomes (Figures 6, 7), but the effect on chloroplast ribosome biogenesis and chloroplast development was aggravated when the mutated line was treated with cold stress (Figure 8 and Supplementary Figure 2). Our data and those of a recent report (Ngoc et al., 2021b) together suggested that the AtRsmD protein for the m2G915 modification of 16S rRNA plays important regulatory roles in chloroplast development and chloroplast function.
16S rRNA is a constituent of the 30S small subunit in the prokaryotic-type 70S ribosome that is widely present in prokaryotes and eukaryotic organelles (mitochondria and chloroplasts). The nucleotide residue of 16S rRNA tends to be guanosine in bacteria and chloroplasts (Lesnyak et al., 2007), while it is usually uridine in archaea and adenosine in mitochondria (Lesnyak et al., 2007). Accordingly, homologous proteins of E. coli RsmD are present in higher plant chloroplasts (Supplementary Figures 3, 4) but are completely absent in both Archaea and Eukarya. The AtRsmD protein shares highly conserved motifs, such as Dx(F/G/Y)xGxG and (D/N/T)PP(F/Y), with those in E. coli; these motifs are required for S-adenosyl-L-methionine (SAM) binding (Supplementary Figure 3; Rana et al., 2013; Ngoc et al., 2021b). Importantly, a recent report demonstrated that AtRsmD is a MTase responsible for chloroplast 16S rRNA m2G 915 methylation, which is absent in the atrsmd-1 mutant (Ngoc et al., 2021b). Although we did not investigate modifications to the atrsmd-2 mutant in this study, genetic analysis demonstrated that the phenotype of atrsmd-2 is caused by the deletion of AtRsmD. Thus, it is possible that chloroplast 16S rRNA m2G 915 methylation is lacking in the atrsmd-2 mutant. Chloroplast ribosomes are attached to the thylakoid membrane (Yamamoto et al., 1981). A chloroplast ribosome proteome study revealed that the homologous protein of RsmD in Chlamydomonas reinhardtii comigrated with the 30S ribosome (Westrich et al., 2021). In E. coli, the RsmD enzyme methylates the nucleotide of 16S rRNA and is involved the assembly 30S subunit of ribosome (Lesnyak et al., 2007). AtRsmD also methylates 16S rRNA in chloroplast ribosomes (Ngoc et al., 2021b). An early plastid nucleoid proteomics study has shown that the homolog protein of RsmD in maize is a plastid nucleoid protein (Majeran et al., 2012). Our result also indicates that AtRsmD is associated with the thylakoids (Figure 2D). The thylakoid-associated localization of AtRsmD protein and the high levels of the AtRsmD transcript in green tissues are consistent with its putative function.
Methylation of rRNAs has been shown to be a novel cellular regulatory mechanism that is intimately associated with chloroplast translation (Manduzio and Kang, 2021). Multiple nucleotide sites of 16S rRNA are methylated in E. coli, and these methylation sites play important regulatory roles in the function of ribosomes (Carrión et al., 1999; Lesnyak et al., 2007). In E. coli, C1352 methylation is essential for viability, and the gene mraW encoding the corresponding MTase is essential (Carrión et al., 1999). Additionally, the lack of C1352 methylation in 16S rRNA (Zou et al., 2020; Ngoc et al., 2021a) clearly reduced chloroplast translation (Zou et al., 2020), which seriously affected plant growth and development. Thus, methylation of the C1352 nucleotide is essential for maintaining chloroplast function. The G966 nucleotide of E. coli 16S rRNA is in direct contact with P-site-bound tRNA and forms a direct contact with the tip of the anticodon loop (Lesnyak et al., 2007). The nucleotide sequence of 16S rRNA is conserved between E. coli and chloroplasts (Supplementary Figure 5). Knockout of the rsmD gene is viable, and no visible phenotype can be observed in E. coli (Jemiolo et al., 1991; Lesnyak et al., 2007; Burakovsky et al., 2012). It seems that no vital function of the ribosome is affected when there is an absence of methylated G966 in E. coli (Lesnyak et al., 2007). Similarly, neither germination nor seedling growth of the Arabidopsis rsmD mutant were affected under normal growth conditions (Ngoc et al., 2021b). However, our data presented here clearly indicated that the photosynthesis efficiency was reduced in the atrsmd-2 mutant grown under normal conditions (Figure 1B), although no visible phenotype was observed (Figure 1B). Further observation showed that chloroplast development was impaired in the atrsmd-2 mutant (Supplementary Figure 2). In accordance with this finding, the chloroplast-encoded photosynthesis-related proteins D1, D2, CP43, CP47, and AtpF were obviously reduced in the atrsmd-2 mutant, and assembly of the photosynthesis complex was also decreased (Figures 3, 4). Thus, the AtRsmD protein is required for chloroplast development and chloroplast function. In contrast to the effect of the deletion of the CAML protein on chloroplast ribosome biogenesis (Zou et al., 2020), our data indicated that the knockout of the AtRsmD protein had minor effects on chloroplast ribosome biogenesis (Figure 6) and chloroplast RNA loading in Arabidopsis under normal growth conditions (Figure 7). Functional activity of the chloroplast ribosome is retained in the absence of the 16S rRNA m2G915 modification. Lack of the 16S rRNA m2G915 site did not have a major effect on the translation of photosynthetic proteins because the amounts of the chloroplast-encoded photosynthetic proteins D1 and D2 were reduced in the atrsmd-2 mutant; however, PsaA and PetA were not reduced (Figure 4). This result is different from the observation of the cmal mutant (Zou et al., 2020; Ngoc et al., 2021a), in which the absence of C1352 methylation seriously generally affected chloroplast translation events. Noticeably, the nucleotide modification of the G966 nucleotide influences Watson-Crick positions within the modified base in E. coli, since the nucleotide is located in the anticodon region (Moazed and Noller, 1990; Lesnyak et al., 2007). Lack of modification interrupts the hydrophobic interaction with the anticodon of the P-site-bound tRNA, which probably affects the fidelity of chloroplast translation. Nevertheless, it is difficult to check the fidelity of chloroplast translation in the atrsmd-2 mutant at present. Lack of the m2G915 modification in the 16S rRNA might affect the elongation of polypeptide chains because the atrsmd-2 mutant exhibited more sensitivity to lincomycin, which specifically inhibits the elongation of nascent polypeptide chains (Supplementary Figure 4). It is likely that the m2G915 modification of the 16S rRNA represents a novel regulatory mechanism for translation in chloroplast-encoded photosynthesis proteins rather than a general regulation of chloroplast translation. Mutants in which chloroplast translation is compromised exhibit sensitivity to cold stress (Tokuhisa et al., 1998; Rogalski et al., 2008; Liu et al., 2010; Fleischmann et al., 2011; Zhang et al., 2016; Pulido et al., 2018). Similar to the atrsmd-1 mutant described in Ngoc et al. (2021b), the atrsmd-2 mutant was found to be more sensitive to translational inhibitors (Supplementary Figure 1) and cold stress (Supplementary Figure 2) than the wild-type. Further characterization of the atrsmd-2 mutant treated with cold stress revealed that defects in chloroplast ribosomal biogenesis were aggravated under cold stress (Figure 8). Our study, together with the report by Ngoc et al. (2021b), showed that knockout of the AtRsmD protein for the methylation of 16S rRNA m2G915 in chloroplasts reduced the accumulation of several important chloroplast-encoded photosynthetic proteins, which could not meet the needs of rapid chloroplast development and thylakoid membrane formation in emerging leaves, and cold stress exacerbated the effect on chloroplast development (Figure 9). Overall, this study combined with a previous report (Ngoc et al., 2021b) showed that the AtRsmD protein involved in the methylation of 16S rRNA m2G915 in chloroplasts is required for chloroplast development and chloroplast function. This work extends our understanding of the significance of the methylation of chloroplast rRNAs in chloroplast development and chloroplast function.
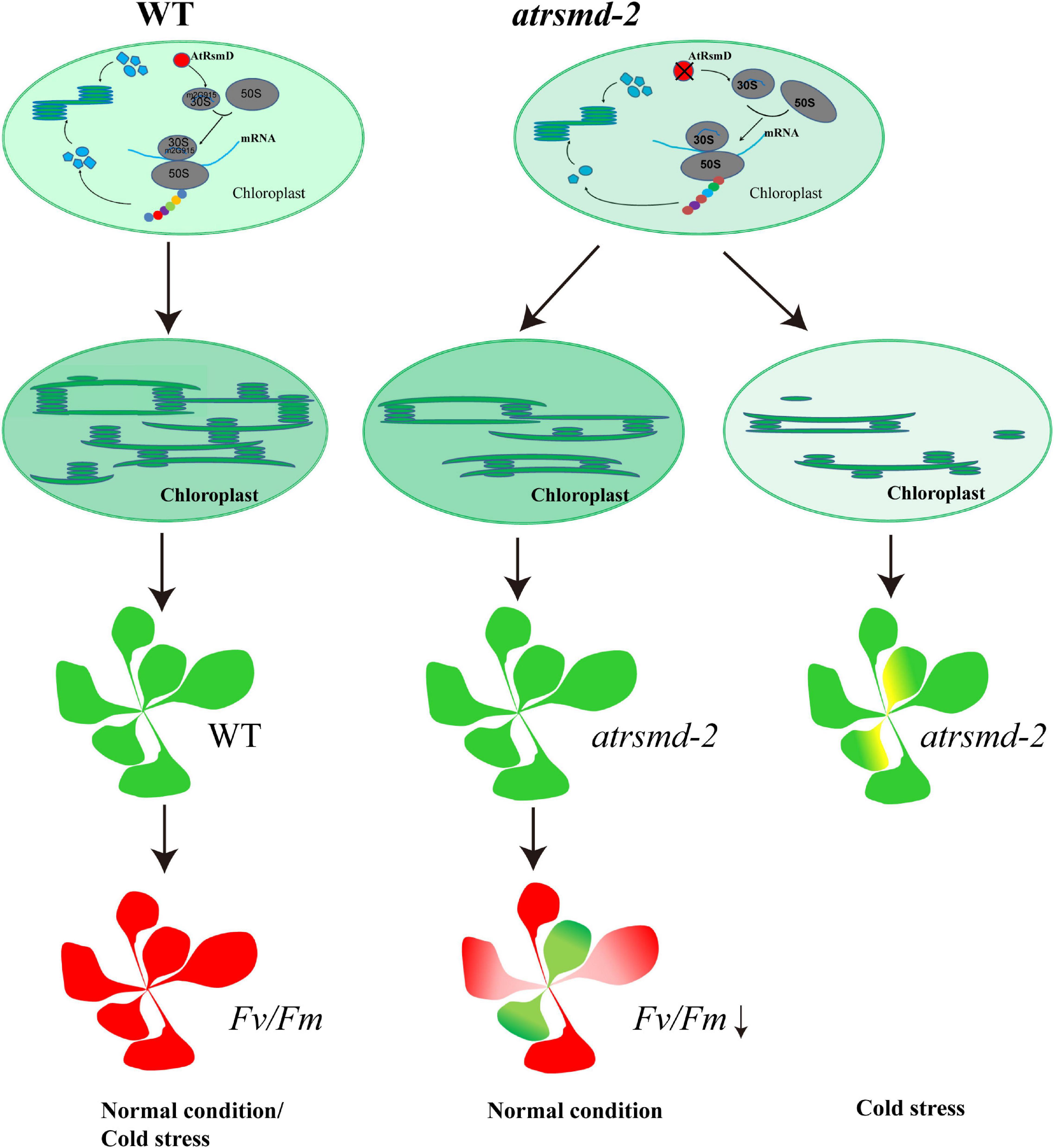
Figure 9. A putative working model for AtRsmD in chloroplasts. AtRsmD is essential for the methylation of the 16S rRNA m2G915 site (Ngoc et al., 2021b), which ensures the rapid synthesis of chloroplast-encoded photosynthetic proteins, such as D1, D2, CP43, and CP47. In the atrsmd-2 mutant, absence of AtRsmD might reduce the synthesis of these photosynthetic proteins and affect the formation of the thylakoids, which may cause decreased photosynthetic efficiency. Cold stress aggravated the effect of AtRsmD knockout on chloroplast translation, which blocked chloroplast development in the atrsmd-2 mutant.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
Q-BY designed the research and wrote the manuscript. Z-YW, W-TQ, TM, NZ, and N-YY performed the experiments. Z-NY, H-BX, X-FX, and Q-BY analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Scientific Foundation of China (grant no. 31570232) and funds from the Shanghai Key Laboratory of Plant Molecular Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the T-DNA insertion line (CS832131). We thank Prof. Lianwei Peng for help with BN-PAGE and polysome analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.860945/full#supplementary-material
Supplementary Figure 1 | Absence of AtRsmD enhances the effects of the translation inhibitors LIN and CAP. (A) Representative images of 5-day-old wild-type (WT) and atrsmd-2 mutant plants germinated on Murashige and Skoog (MS) medium containing the indicated concentrations of LIN. The quantification of the total chlorophyll levels (see section “Materials and Methods”) demonstrates the differences between wild-type and mutant plants grown in the presence of LIN. Relative data are shown (wild-type plants grown in the absence of LIN = 100%), and means ± se values (n = 3) are provided. (B) Representative images of 5-day-old wild-type (WT) and atrsmd-2 mutant plants germinated on Murashige and Skoog (MS) medium containing the indicated concentrations of CAP. The quantification of the total chlorophyll levels (see section “Materials and Methods”) demonstrates the differences between wild-type and mutant plants grown in the presence of CAP. Relative data are shown (wild-type plants grown in the absence of CAP = 100%), and means ± se values (n = 3) are provided.
Supplementary Figure 2 | Chloroplast development in the wild type and atrsmd-2 mutant treated with cold stress. (A) Phenotype of the wild type and atrsmd-2 mutant treated with cold stress. Bar: 1 cm. (B) Chloroplast ultrastructure observation in the wild type and atrsmd-2 mutant. Bar: 1 μm. (C) Immunoblot analysis of the photosynthetic proteins from the wild type (WT) and atrsmd-2 mutant that were treated with cold stress. Immunoblot analysis of the photosynthetic proteins PsaA, PsaF, D1, CP43, OEC33, AtpA, AtpC, AtpF, PetC, PetD, and RbcL was performed using the corresponding antibodies. Total proteins from the WT samples were loaded at three different concentrations (2.5, 5, and 10 μg), and total proteins from the mutant samples were loaded at 10 μg.
Supplementary Figure 3 | Alignment of E. coli RsmD homologous proteins in land plants. Alignment of the amino acid sequence of RsmD homologs in land plants. Sequence identifiers for RsmD homologs are as follows: Arabidopsis thaliana (NP_189487.2), Oryza sativa japonica group (EEE59885.1), and Escherichia coli (EGS2103359.1).
Supplementary Figure 4 | Phylogenetic analysis of the AtRsmD protein and its orthologs from different species. The orthologous proteins are listed as follows: Camelina sativa (XP_010514550.1), Brassica napus (XP_013711825.1), Capsella rubella (XP_023639147.1), Arabidopsis lyrata subsp. Lyrata (XP_002877109.1), Arabidopsis thaliana (NP_189487.2), Ricinus communis (XP_015573725.1), Vitis vinifera (XP_002281501.1), Oryza sativa Japonica Group (EEE59885.1), Zea mays (ACG38583.1), Physcomitrella patens (XP_024381071.1), Selaginella moellendorffii (EFJ21750.1), Ostreococcus lucimarinus CCE9901(XP_001417522.1), Micromonas pusilla CCMP1545(XP_003060663.1), Chloropicon primus (QDZ19162.1), Gonium pectoral (KXZ51239.1), Chlamydomonas reinhardtii (XP_042917255.1), Cyanidioschyzon merolae strain 10D (XP_005537355.1), Galdieria sulphuraria (XP_005703506.1), Gracilariopsis chorda (PXF40802.1), Fischerella thermalis (WP_102206177.1), Escherichia coli (EGS2103359.1), Oscillatoriales cyanobacterium MTP1(TAD77760.1), Synechococcus sp. PCC 73109 (WP_062431539.1), Cyanobacteria (WP_044151411.1), Synechocystis sp. PCC 7509 (WP_009631812.1), Nostoc sp. 106C (WP_086756145.1), and Phormidium ambiguum (WP_073593883.1).
Supplementary Figure 5 | Alignment of 16S rRNAs from different organisms. Sequence identifiers for RsmD homologs are as follows: Oryza sativa (MK348618.1), Brachypodium distachyon (LR537486.1), Hordeum vulgare, (MN171392.1), Sorghum bicolor (MK348612.1), Zea mays (MK348606.1), Arabidopsis thaliana (MK353213.1), Cyanobacterium (LN833508.1), Populus trichocarpa (AC208048.1), Glycine max (DQ317523.1), Chlamydomonas reinhardtii (MF083689.2), and Escherichia coli (CP056921.1).
Footnotes
- ^ http://www.toyobo-global.com/
- ^ http://www.cambia.org
- ^ http://www.roche.com
- ^ http://www.toyobo.co.jp
- ^ http://arete.ibb.waw.pl/PL/html/boxshade.html
- ^ http://www.megasoftware.net
- ^ https://www.tiangen.com/
- ^ www.cbs.dtu.dk/services/TargetP-1.1/index.php
References
Abdallah, F., Salamini, F., and Leister, D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5, 141–142. doi: 10.1016/s1360-1385(00)01574-0
Andersen, N. M., and Douthwaite, S. (2006). YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 359, 777–786. doi: 10.1016/j.jmb.2006.04.007
Ban, N., Nissen, P., Hansen, J., Moore, P. B., and Steitz, T. A. (2000). The complete atomic structure of the large ribosomal subunit at 2.4 A° resolution. Science 289, 905–920. doi: 10.1126/science.289.5481.905
Brimacombe, R., Mitchell, P., Osswald, M., Stade, K., and Bochkariov, D. (1993). Clustering of modified nucleotides at the functional center of bacterial ribosomal RNA. FASEB J. 7, 161–167. doi: 10.1096/fasebj.7.1.8422963
Bujnicki, J. M., and Rychlewski, L. (2002). RNA:(guanine-N2) methyltransferases RsmC/RsmD and their homologs revisited–bioinformatic analysis and prediction of the active site based on the uncharacterized Mj0882 protein structure. BMC Bioinformatics 3:10. doi: 10.1186/1471-2105-3-10
Burakovsky, D. E., Prokhorova, I. V., Sergiev, P. V., Milón, P., Sergeeva, O. V., Bogdanov, A. A., et al. (2012). Impact of methylations of m2G966/m5C967 in 16S rRNA on bacterial fitness and translation initiation. Nucleic Acids Res. 40, 7885–7895. doi: 10.1093/nar/gks508
Carrión, M., Gómez, M. J., Merchante-Schubert, R., Dongarrá, S. R., and Ayala, J. A. (1999). mraW, an essential gene at the dcw cluster of Escherichia coli codes for a cytoplasmic protein with methyltransferase activity. Biochimie 81, 879–888. doi: 10.1016/s0300-9084(99)00208-4
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Fleischmann, T. T., Scharff, L. B., Alkatib, S., Hasdorf, S., Schöttler, M. A., and Bock, R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23, 3137–3155. doi: 10.1105/tpc.111.088906
Graf, M., Arenz, S., Huter, P., Dönhöfer, A., Novácek, J., and Wilson, D. N. (2017). Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 45, 2887–2896. doi: 10.1093/nar/gkw1272
Green, R., and Noller, H. F. (1999). Reconstitution of functional 50S ribosomes from in vitro transcripts of Bacillus stearothermophilus 23S rRNA. Biochemistry 38, 1772–1779. doi: 10.1021/bi982246a
Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., et al. (2001). High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688. doi: 10.1016/s0092-8674(01)00546-3
Jemiolo, D. K., Taurence, J. S., and Giese, S. (1991). Mutations in 16S rRNA in Escherichia coli at methyl-modified sites: G966, G967, and G1207. Nucleic Acids Res. 19, 4259–4265. doi: 10.1093/nar/19.15.4259
Keeling, P. J. (2013). The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu. Rev. Plant Biol. 64, 583–607. doi: 10.1146/annurev-arplant-050312-120144
Khaitovich, P., Tenson, T., Kloss, P., and Mankin, A. S. (1999). Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro transcribed rRNA. Biochemistry 38, 1780–1788. doi: 10.1021/bi9822473
Kimura, S., and Suzuki, T. (2010). Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 38, 1341–1352. doi: 10.1093/nar/gkp1073
Krzyzosiak, W., Denman, R., Nurse, K., Hellmann, W., Boubik, M., Gehrke, C. W., et al. (1987). In vitro synthesis of 16S ribosomal RNA containing single base changes and assembly into functional 30S ribosome. Biochemistry 26, 2353–2364. doi: 10.1021/bi00382a042
Lesnyak, D. V., Osipiuk, J., Skarina, T., Sergiev, P., Bogdanov, A. A., Edwards, A., et al. (2007). Methyltransferase that modifies guanine 966 of the 16S rRNA: functional identification and tertiary structure. J. Biol. Chem. 282, 5880–5887. doi: 10.1074/jbc.M608214200
Li, Y., Liu, B., Zhang, J., Kong, F., Zhang, L., Meng, H., et al. (2019). OHP1, OHP2, and HCF244 form a transient functional complex with the photosystem II reaction center. Plant Physiol. 179, 195–208. doi: 10.1104/pp.18.01231
Liu, X., Rodermel, S. R., and Yu, F. (2010). A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol. 10:287. doi: 10.1186/1471-2229-10-287
Majeran, W., Friso, G., Asakura, Y., Qu, X., Huang, M., Ponnala, L., et al. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158, 156–189. doi: 10.1104/pp.111.188474
Manduzio, S., and Kang, H. (2021). RNA methylation in chloroplasts or mitochondria in plants. RNA Biol. 18, 2127–2135. doi: 10.1080/15476286.2021.1909321
Moazed, D., and Noller, H. F. (1990). Binding of tRNA to the ribosomal A and P sites protects two distinct sets of nucleotides in 16S rRNA. J. Mol. Biol. 211, 135–145. doi: 10.1016/0022-2836(90)90016-F
Ngoc, L. N. T., Park, S. J., Huong, T. T., Lee, K. H., and Kang, H. (2021a). N4- methylcytidine rRNA methylation in chloroplasts is crucial for chloroplast function, development, and abscisic acid response in Arabidopsis. J. Integr. Plant Biol. 63, 570–582. doi: 10.1111/jipb.13009
Ngoc, L. N. T., Park, S. J., Cai, J., Huong, T. T., Lee, K., and Kang, H. (2021b). RsmD, a Chloroplast rRNA m2G Methyltransferase, Plays a Role in Cold Stress Tolerance by Possibly Affecting Chloroplast Translation in Arabidopsis. Plant Cell Physiol. 62, 948–958. doi: 10.1093/pcp/pcab060
Porra, R. J. (2002). The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73, 149–156. doi: 10.1023/A:1020470224740
Pulido, P., Zagari, N., Manavski, N., Gawronski, P., Matthes, A., Scharff, L. B., et al. (2018). CHLOROPLAST RIBOSOME ASSOCIATED supports translation under stress and interacts with the ribosomal 30S Subunit. Plant Physiol. 177, 1539–1554. doi: 10.1104/pp.18.00602
Rana, K. A., Chandr, S., Siddiqi, M. I., and Misra-Bhattacharya, S. (2013). Molecular Characterization of an rsmD-Like rRNA Methyltransferase from the Wolbachia Endosymbiont of Brugia malayi and Antifilarial Activity of Specific Inhibitors of the Enzyme. Antimicrob. Agents Chemother. 57, 3843–3856. doi: 10.1128/AAC.02264-12
Rogalski, M., Schöttler, M. A., Thiele, W., Schulze, W. X., and Bock, R. (2008). Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20, 2221–2237. doi: 10.1105/tpc.108.060392
Schuwirth, B. S., Borovinskaya, M. A., Hau, C. W., Zhang, W., Vila-Sanjurjo, A., Holton, J. M., et al. (2005). Structures of the bacterial ribosome at 3.5 A° resolution. Science 310, 827–834.
Sergiev, P., Golovina, A., Prokhorova, I., Sergeeva, O., Osterman, I., Nesterchuk, M., et al. (2011). “Modifications of ribosomal RNA: from enzymes to function,” in Ribosomes: Structure, Function, and Dynamics, eds M. V. Rodnina, W. Wintermeyer, and R. Green (Wien, NY: Springer), 97–110. doi: 10.1007/978-3-7091-0215-2_9
Tokuhisa, J. G., Vijayan, P., Feldmann, K. A., and Browse, J. A. (1998). Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA demethylase. Plant Cell 10, 699–711. doi: 10.1105/tpc.10.5.699
Urlaub, H., Thiede, B., Müller, E. C., Brimacombe, R., and Wittmann-Liebold, B. (1997). Identification and sequence analysis of contact sites between ribosomal proteins and rRNA in Escherichia coli 30 S subunits by a new approach using matrix-assisted laser desorption/ionization-mass spectrometry combined with N-terminal microsequencing. J. Biol. Chem. 272, 14547–14555. doi: 10.1074/jbc.272.23.14547
Wei, Y., Zhang, H., Gao, Z. Q., Wang, W. J., Shtykovac, E. V., Xu, J. H., et al. (2012). Crystal and solution structures of methyltransferase RsmH provide basis for methylation of C1402 in 16S rRNA. J. Struct. Biol. 179, 29–40. doi: 10.1016/j.jsb.2012.04.011
Westrich, L. D., Gotsmann, V. L., Herkt, C., Ries, F., Kazek, T., Trosch, R., et al. (2021). The versatile interactome of chloroplast ribosomes revealed by affinity purification mass spectrometry. Nucleic Acids Res. 49, 400–415. doi: 10.1093/nar/gkaa1192
Wimberly, B. T., Brodersen, D. E., Clemons, W. M. J., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., et al. (2000). Structure of the 30 S ribosomal subunit. Nature 407, 327–339.
Xiong, H. B., Wang, J., Huang, C., Rochaix, J. D., Lin, F. M., Zhang, J. X., et al. (2020). mTERF8, a member of the mitochondrial transcription termination factor family, is involved in the transcription termination of chloroplast Gene psbJ. Plant Physiol. 182, 408–423. doi: 10.1104/pp.19.00906
Yamamoto, T., Burke, J., Autz, G., and Jagendorf, A. T. (1981). Bound Ribosomes of Pea chloroplast thylakoid membranes: location and release in vitro by high salt, Puromycin, and RNase. Plant Physiol. 67, 940–949. doi: 10.1104/pp.67.5.940
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yu, Q. B., Lu, Y., Ma, Q., Zhao, T. T., Huang, C., Zhao, H. F., et al. (2013). TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol. Plant. 148, 408–421. doi: 10.1111/j.1399-3054.2012.01718.x
Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H., et al. (2001). Crystal structure of the ribosome at 5.5 A° resolution. Science 292, 883–896.
Zhang, J., Yuan, H., Yang, Y., Fish, T., Lyi, S. M., Thannhauser, T. W., et al. (2016). Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J. Exp. Bot. 67, 2731–2744. doi: 10.1093/jxb/erw106
Zhang, L., Pu, H., Duan, Z., Li, Y., Liu, B., Zhang, Q., et al. (2018). Nucleus-Encoded Protein BFA1 Promotes Efficient Assembly of the Chloroplast ATP Synthase Coupling Factor. Plant Cell 30, 1770–1788. doi: 10.1105/tpc.18.00075
Zoschke, R., and Bock, R. (2018). Chloroplast translation: structural and functional organization, operational control, and regulation. Plant Cell 30, 745–770. doi: 10.1105/tpc.18.00016
Keywords: AtRsmD, chloroplast, rRNA methylation, ribosome, photosynthesis
Citation: Wang Z-Y, Qu W-T, Mei T, Zhang N, Yang N-Y, Xu X-F, Xiong H-B, Yang Z-N and Yu Q-B (2022) AtRsmD Is Required for Chloroplast Development and Chloroplast Function in Arabidopsis thaliana. Front. Plant Sci. 13:860945. doi: 10.3389/fpls.2022.860945
Received: 24 January 2022; Accepted: 16 March 2022;
Published: 25 April 2022.
Edited by:
Hongbo Gao, Beijing Forestry University, ChinaCopyright © 2022 Wang, Qu, Mei, Zhang, Yang, Xu, Xiong, Yang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhong-Nan Yang, em55YW5nQHNobnUuZWR1LmNu; Qing-Bo Yu, eXVxaW5nOTg2MEBzaG51LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Zi-Yuan Wang
Zi-Yuan Wang Wan-Tong Qu
Wan-Tong Qu Tong Mei
Tong Mei Xiao-Feng Xu
Xiao-Feng Xu Zhong-Nan Yang
Zhong-Nan Yang Qing-Bo Yu
Qing-Bo Yu