
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 March 2022
Sec. Plant Breeding
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.860034
This article is part of the Research Topic Soybean Molecular Breeding and Genetics View all 25 articles
 He Wei1
He Wei1 Yun Lian1
Yun Lian1 Jinying Li1
Jinying Li1 Haichao Li1
Haichao Li1 Qijian Song2
Qijian Song2 Yongkang Wu1
Yongkang Wu1 Chenfang Lei1
Chenfang Lei1 Shiwei Wang1
Shiwei Wang1 Hui Zhang1
Hui Zhang1 Jinshe Wang1*
Jinshe Wang1* Weiguo Lu1*
Weiguo Lu1*
Soybean cyst nematode (SCN; Heterodera glycines Ichinohe) is a highly destructive pathogen for soybean production worldwide. The use of resistant varieties is the most effective way of preventing yield loss. Handou 10 is a commercial soybean variety with desirable agronomic traits and SCN resistance, however genes underlying the SCN resistance in the variety are unknown. An F2:8 recombinant inbred line (RIL) population derived from a cross between Zheng 9525 (susceptible) and Handou 10 was developed and its resistance to SCN HG type 2.5.7 (race 1) and 1.2.5.7 (race 2) was identified. We identified seven quantitative trait loci (QTLs) with additive effects. Among these, three QTLs on Chromosomes 7, 8, and 18 were resistant to both races. These QTLs could explain 1.91–7.73% of the phenotypic variation of SCN’s female index. The QTLs on chromosomes 8 and 18 have already been reported and were most likely overlapped with rhg1 and Rhg4 loci, respectively. However, the QTL on chromosome 7 was novel. Candidate genes for the three QTLs were predicted through genes functional analysis and transcriptome analysis of infected roots of Handou 10 vs. Zheng 9525. Transcriptome analysis performed also indicated that the plant–pathogen interaction played an important role in the SCN resistance for Handou 10. The information will facilitate SCN–resistant gene cloning, and the novel resistant gene will be a source for improving soybeans’ resistance to SCN.
Soybean cyst nematode (SCN, Heterodera glycines Ichinohe) is a devastating pest affecting soybean (Glycine max [L.] Merr.) production worldwide (Riggs, 1977; Koenning and Wrather, 2010; Tylka and Marett, 2014; Mitchum, 2016; Miraeiz et al., 2020; Peng et al., 2021). SCN has caused approximately 36% of yield losses in the total soybean production from 1996–2014 in the United States (Kim et al., 2016). In Chifeng area in China, the highest yield reduction rate reached 44.43% because of SCN during 2014–2019 (Li et al., 2020). Moreover, a highly virulent of SCN has been observed in China (Lian et al., 2019). SCN is a soil-borne pathogen and pest management is difficult. SCN management includes crop rotation, pesticide application, biological control, pest-resistant varieties, etc.; however, breeding for resistant varieties is the most effective method (Usovsky et al., 2021). Riggs and Schmitt (1988) reported 16 physiological races that could be differentiated, and the pathogenicity of each race was different. As in Missouri in the United States, the dominant race in the Huanghuai Valley in China is race 2, which was evolved from race 1 (Lu et al., 2006; Mitchum et al., 2007; Lian et al., 2016; Howland et al., 2018). Accordingly, race 2 was used to screen new varieties for SCN resistance in the Huanghuai Valley, unfortunately, most varieties were susceptible to SCN race 2.1
Most studies showed that SCN resistance is a quantitative trait and controlled by multiple genes (Concibido et al., 2004; Wang, 2019); more than 200 QTLs have been mapped on 20 chromosomes.2 Two major QTLs, rhg1 (Peking-type rhg1-a and PI88788-type rhg1-b) and Rhg4 (GmSHMT08), were cloned and functionally analyzed (Cook et al., 2012; Liu et al., 2012; Liu et al., 2017). Three genes (Glyma.18G022500, Glyma.18G022500, and Glyma.18G022700) around the rhg1-b locus and the gene GmSHMT (Glyma.08G108900) around the Rhg4 locus were identified as major-effect gene to SCN resistance (Cook et al., 2012; Liu et al., 2012; Guo et al., 2020). The PI88788-type requires at least 5.6 copies of rhg1-b (Cook et al., 2012; Patil et al., 2019), whereas the Peking-type requires rhg1-a and Rhg4 for SCN resistance (Patil et al., 2019). In the United States, most SCN resistant cultivars are from PI88788 and this has reduced the effectiveness of SCN prevention (Mitchum, 2016). For breeding resistant cultivars, it is vital to identify new quantitative trait loci (QTL) and genes underlying resistance (Liu et al., 2019) and to broaden the genetic basis for improving soybeans’ resistance to SCN. In recent years, Numerous researchers have identified genes (Guo et al., 2020) and reveled a complex regulatory network involved in SCN resistance (Guo et al., 2020; Kofsky et al., 2021; Shi et al., 2021).
Handou 10 was first identified as being resistant against SCN race 1 in routine cultivar testing in 2008 by our team and was registered by the Hebei Variety Approval Committee in 2011. The yield of the variety was significantly better than the control, having the desired agronomic traits in the Hebei uniform test and pre-releasing tests in 2007–2010. However, the origin and inheritance of the SCN resistance in Handou 10 was unknown. The objective of this study was to identify the QTL and candidate genes controlling the resistance and provide the basis for molecular marker development and marker-assisted breeding.
A recombinant inbred line (RIL) mapping population of 392 F2:8 lines was developed by single seed descent (SSD) from the cross between Zheng 9525 and Handou 10. Zheng 9525 was cultivated by Henan Academy of Agricultural Sciences, whereas Handou 10 was cultivated by Henan Jintun Seed Industry Co., Ltd., and Handan Academy of Agricultural Sciences. The seeds of nine differential cultivars, PI88788, Peking, PI437654, PI209332, PI548316, PI89772, PI90763, Pickett, and Lee, were obtained from Henan Academy of Agricultural Sciences.
Handou 10, Zheng 9525, and nine differential cultivars were evaluated for the resistance to SCN [HG types 2.5.7 (race 1), 1.2.5.7 (race 2), and 1.2.3.5.6.7 (race 4)] in a climate room in Henan Academy of Agricultural Sciences. Plastic cups (Ø 6 cm × h 12 cm) were filled with soil infected by SCN [HG types 2.5.7 (race 1), 1.2.5.7 (race 2), and 1.2.3.5.6.7 (race 4)], respectively. Lees were planted in SCN-infested soil, and after 30 days, we collected cysts from the roots using a 710–250 μm sieve tower. Cysts were collected from the 250 μm sieve and rinsed. The eggs were collected by breaking open cysts with a rubber stopper and collecting the eggs on a sieve stack consisting of 250 μm – 75 μm – 25 μm sieves. The mixture from the 25 μm sieve was backwashed into a 50 mL plastic conical tube. A 40% sucrose solution was added to the tubes, stirred, and centrifuged at 2,000 rpm for 5 min. Eggs in the middle layer or supernatant were then collected over a 25 μm sieve. Handou 10, Zheng 9525, and nine differential cultivars were transplanted with five replicates. Each replicate was one plant in a plastic cup (Ø 6 cm × h 12 cm). Five days after transplantation, seedlings were inoculated with about 4,000 eggs per cup. The plants grew at 70 to 80% relative humidity, 28–24 (L/D), and a photoperiod of 16 h: 8 h (L:D) and were watered daily.
The SCN resistance of the 392 RILs and the parents (Zheng 9525 and Handou 10) were evaluated for SCN resistance against HG type 1.2.5.7 (race 2) and HG type 2.5.7 (race 1) in a climate room. Five days after sowing, two plants of each line were transplanted into a plastic cup (Ø 6 cm × h 12 cm) with three replicates, each replicate had two plants. Five days after transplantation, seedlings were inoculated with about 4,000 eggs per cup. The growth conditions were the same as the above.
Thirty days after inoculation, nematode cysts were collected from the roots of each replicate and counted by an image analysis software (Wang et al., 2014). A female index (FI) was calculated as follows: FI (%) = (average number of cysts on each line/average number of cysts on Lee) × 100. FI was used as phenotype data for QTL analysis. The lines were rated as resistant (FI < 10), moderately resistant (10 ≤ FI < 30), moderately susceptible (30 ≤ FI < 60), or susceptible (FI ≥ 60) to classify the response to SCN.
Leaf samples of each progeny line and their parents were collected at the seedling stage. DNA was extracted by the plant genomic DNA kit [TIANGEN Biotech (Beijing) Co., Ltd.]. Zheng 9525 and Handou 10 were sequenced by Illumina HiSeq 4000, whereas 392 RIL2:8 lines from Zheng 9525 × Handou 10 were sequenced by HiSeq X Ten at Huada Gene Technology Co., Ltd. (Shenzhen, China). The sequences of the parents were aligned with the reference genome (Gmax_275_Wm82.a2) using SOAP2 (Li et al., 2009a) and single-nucleotide polymorphisms (SNPs) between the parents were detected by SOAPsnp (Li et al., 2009b). The SNPs between the parents were identified and filtered: (1) Mass value is greater than 20; (2) At least three reads are supported; (3) Heterozygous sites are removed. The pseudomolecules of parental genome sequences were obtained by SNPs between parents with the reference genome. The reads of the progeny population were compared with pseudomolecules of the parental genome sequences using SOAPaligner.
Instead of using the SNPs as such for linkage mapping, a sliding window-based approach was used to identify bin markers where consecutive SNPs were merged into one bin. We extract the bin area on each chromosome as bin marker, only the bin markers without segregation distortion are selected to construct a map. A genetic map with the bin markers was constructed with JoinMap 4.0 using the maximum likelihood mapping algorithm (Van Ooijen, 2006). Groups were created depending on LOD scores ≥ 3.0 and a maximum distance of 50 cM. The resulting linkage groups were assigned to specific chromosomes according to the reference genome (Gmax_275_Wm82.a2). Regression mapping was used as the mapping algorithm with Kosambi’s mapping function to convert recombination frequency into map distance.
Additive QTLs of SCN resistance with FI to HG types 2.5.7 (race 1) and 1.2.5.7 (race 2), were analyzed using the composite interval mapping (CIM) method in WinQTLCart 2.5 software (Wang et al., 2012). The walking speed for CIM was 1 cM and the LOD threshold at the 5% probability level was determined by a 1,000 permutation test.
The seedlings with consistent growth were selected to transplant into a plastic cup with sterile soil after Handou 10 and Zheng 9525 were sown in vermiculite, with one plant per cup. Five days later, well-developed plants of the same size were selected to inoculate 4,000 or 0 SCN HG type (1.2.5.7) (race 2) eggs per cup, with three replicates per genotype of each treatment, and five plants for each replicate. Ten days after inoculation, roots were collected, washed, frozen in liquid nitrogen and stored at -80°C until use. Total RNA was extracted by TRIzol (Invitrogen). Each biological replicate contained pooled roots from five individual plants. The RNA transcriptome sequencing and preliminary data analyzes were carried out by Shenzhen Huada Gene Technology Co., Ltd. (China). For each replicate, a mRNA library was constructed and sequenced using the DNBSEQ platform. Adaptor reads, reads with an unknown base N greater than 5%, and low-quality reads (bases with a quality value of less than 15 account for more than 20% of the total bases in the reads) were filtered out of the raw data to obtain high-quality (clean) reads using SOAPnuke (v1.4.0) (Chen et al., 2018). Clean reads were mapped to the soybean reference genome (G.max Wm82.a2.v1) by HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts, V2.1.0) (Kim et al., 2015) and aligned using Bowtie 2 (v2.2.5) (Langmead and Salzberg, 2012). The genes and transcripts were calculated using RSEM (v1.2.8) (Li and Dewey, 2011). Significant differentially expressed genes (DEGs) were obtained with false discovery rates (FDRs) of ≤ 0.05.
Significant DEGs were annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG).3 The statistical enrichment of DEGs in KEGG pathways was accomplished with the R (version 3.1.1) function phyper.4 Then FDR was performed on p-value, and q-values of ≤ 0.05 was considered to be significantly enriched.
The QTLs that could be detected for the resistance of Handou 10 to SCN HG type 2.5.7 (race 1) and 1.2.5.7 (race 2) were considered major QTLs. The physical positions of the major QTL intervals were identified according to the above genetic map and the reference genome (“Williams 82.a2.v1”) (Jiang et al., 2018). The DEGs between the infected roots of Handou 10 and Zheng 9525 in the major QTL intervals were the candidate genes controlling SCN resistance in Handou 10.
DNA was extracted by the plant genomic DNA kit [TIANGEN Biotech (Beijing) CO., Ltd]. DNA concentration and quality were measured with NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, United States). According to the SNPs between Zheng 9525 and Handou 10, the KASP (Kompetitive allele-specific PCR) markers were designed by LGC Science (Shanghai) Ltd. The primer sequences of KASP markers linked to rhg1 and Rhg4 were referred to the reports from Shi et al. (2015) and Kadam et al. (2016). The PCR reaction mixture was prepared according to the instructions of the KBioscience (Herts, United Kingdom). The program was set to hold at 94°C for 15 min, followed by 10 touch-down cycles of 20 s at 94°C and 1 min at 65–59°C (dropping 0.6°C per cycle), and then 23 cycles of 20 s at 94°C, 1 min at 57°C. The PCR amplification product was read with a PHERAstar SNP typing detector, and SNP alleles were determined based on the ratio of fluorescence signals.
Handou 10, Zheng 9525, and nine differential cultivars (with Lee as a susceptible control) were screened for SCN resistance. Handou 10 was resistant to SCN HG type 2.5.7 (race 1), moderately resistant to SCN HG type 1.2.5.7 (race 2) and moderately susceptible to HG type 1.2.3.5.6.7 (race 4) (Table 1). Among the 392 RILs, 3.83, 4.34, 9.18, and 82.65% were resistant, moderately resistant, moderately susceptible and susceptible to race 1, respectively, whereas 2.55, 5.61, and 91.84% of individuals were moderately resistant, moderately susceptible and susceptible to race 2, respectively. The FI ranged from 0.16 to 253.80%, and 13.87 to 245.05% for HG type 2.5.7 (race 1) and HG type 1.2.5.7 (race 2), respectively (Figure 1 and Table 2). None of the 392 RILs was resistant to SCN HG type 1.2.5.7 (race 2) (Figure 1).
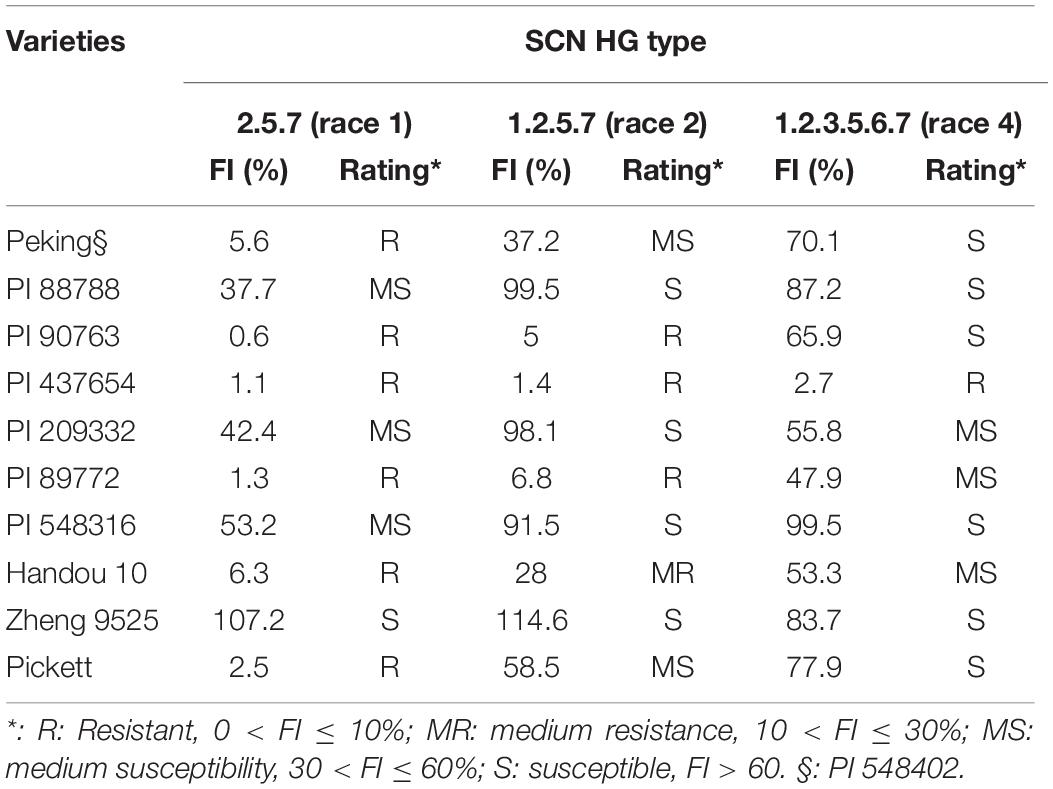
Table 1. Evaluation of Handou 10, Zheng 9525, and eight differential cultivars for resistance to three HG types of soybean cyst nematode (SCN).
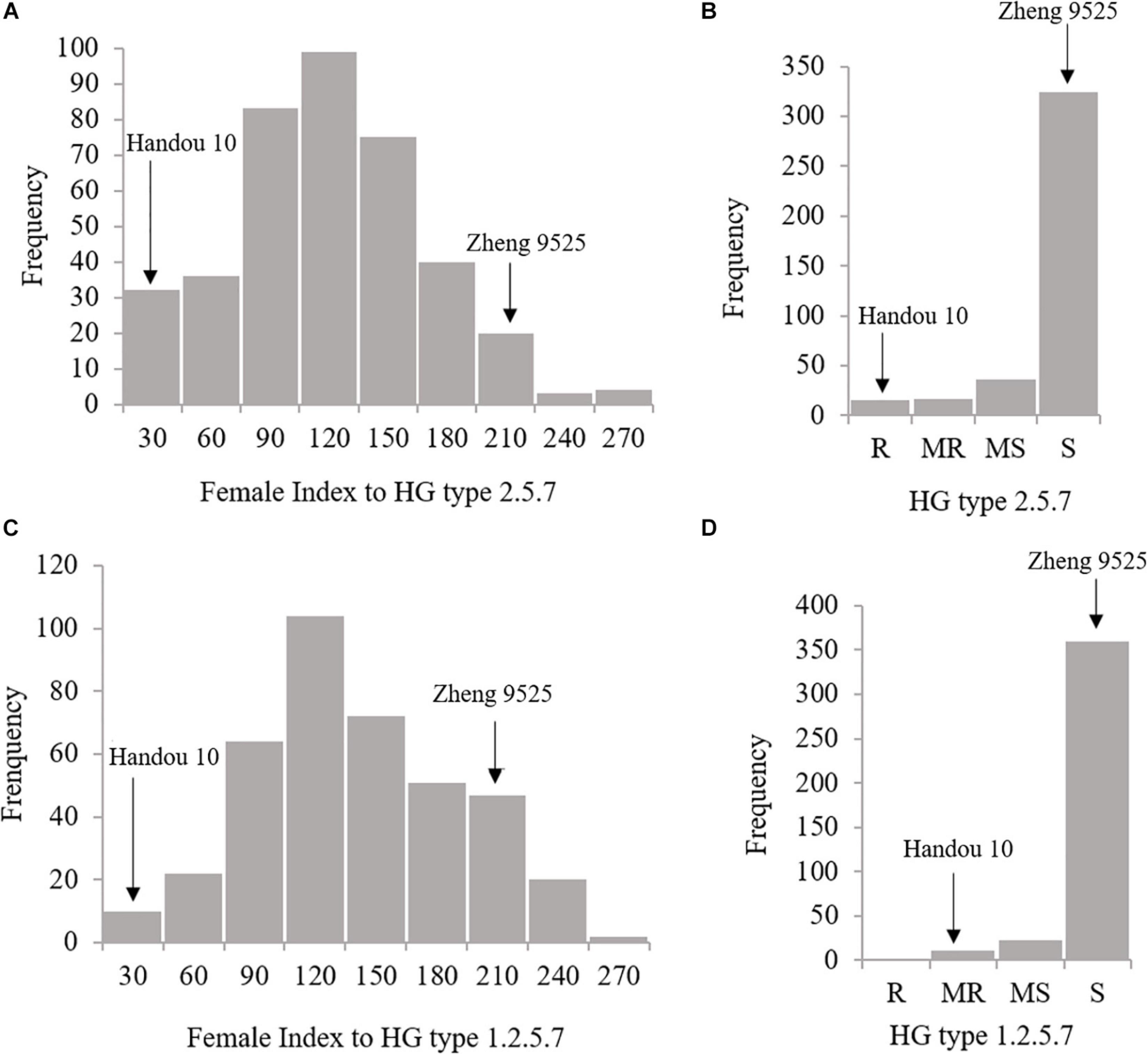
Figure 1. Female index (FI) and rating distributions of SCN resistance to HG type 2.5.7 and HG type 1.2.5.7 for the RILs derived from Zheng 9525×Handou 10, respectively. (A) The FI distribution of SCN resistance to HG types 2.5.7; (B) the rating of SCN resistance to HG types 2.5.7; (C) the FI distribution of SCN resistance to HG types 1.2.5.7; (D) the rating of SCN resistance to HG types 1.2.5.7.
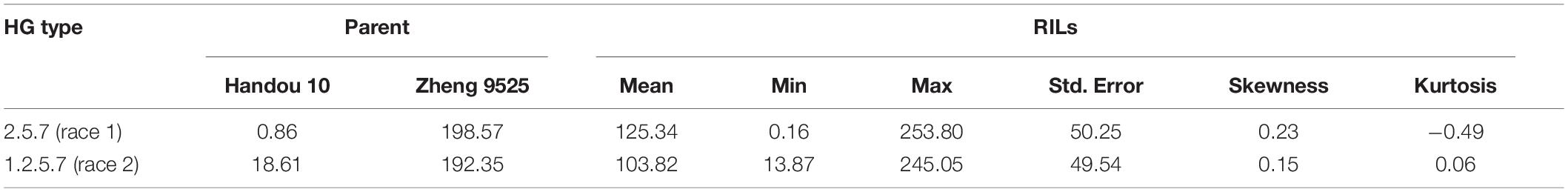
Table 2. Descriptive statistics of the female index (FI) of the parents and the 392 F2:8 RILs from Zheng 9525 × Handou10 after inoculation of SCN HG types 2.5.7 (race 1) and 1.2.5.7 (race 2), respectively.
Zheng 9525 and Handou 10 were re-sequenced using an Illumina HiSeq4000 platform with sequencing average depths of 11.94× and 16.94×, respectively (Supplementary Table 1). Approximately 79.25 and 111.92 M reads, and 11.84 and 16.74 G bases of raw data were obtained from the two parents, respectively; a total of 78.25 reads from Zheng 9525 and 110.09 M reads from Handou 10 were mapped, and the mapped bases of each genotype were 11.74 and 16.64 G, respectively. The genome coverage of these two cultivars was 91.77 and 93.43%, respectively (Supplementary Table 1). A total of 1,062,100 SNPs between Zheng 9525 and Williams 82, and a total of 946,194 SNPs between Handou 10 and Williams 82 were identified using SOAPsnp, and a total of 732,008 SNPs could be identified between the parents.
The average sequencing depth of 392 RILs was 1.94× and the coverage rate was 62.38% (Supplementary Table 2). The sequencing data showed that the average mapped reads was 12.74 M and the average bases were 1.91 G. A total of 607,635 SNPs were identified in the RIL population and 8,593 breaking points were detected using the sliding window approach (Supplementary Table 3).
A high-density genetic map was constructed by joining 5,233 bin markers (missing < 20%, Supplementary Table 4). The map comprised 4763.23 cM with an average distance of 0.91 cm between adjacent markers (Table 3). Chromosome 18 had the highest number of bin markers (405 markers). The linkage map length was the largest for chromosome 5 (325.35 cM), and the smallest was for chromosome 4 (89.47 cM) (Table 3).
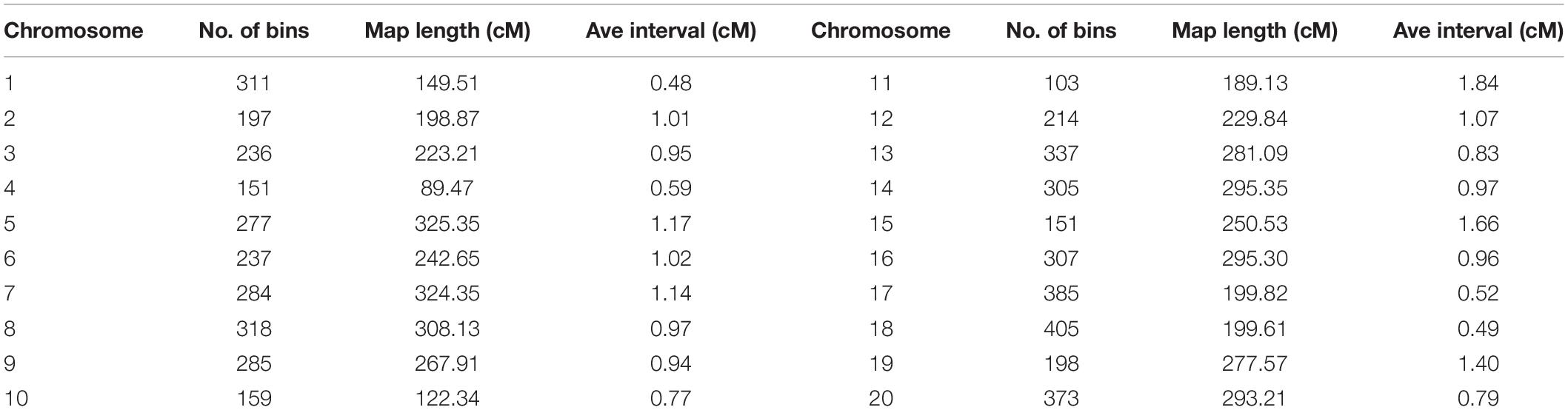
Table 3. Distribution of polymorphic bins for each chromosome of the 392 RILs mapping population of the cross Zheng 9525 × Handou 10.
Seven QTLs for SCN resistance with FI were detected on six chromosomes by CIM, which were designated as SCN_7_1, SCN_8_2, SCN_12_3, SCN_15_4, SCN_18_5, SCN_18_6, and SCN_20_7, respectively (Table 4). The resistance alleles of five QTLs (SCN_7_1, SCN_8_2, SCN_18_5, SCN_18_6, and SCN_20_7) were derived from the resistant parent Handou 10, and the resistance alleles of the remaining two QTLs (SCN_12_3 and SCN_15_4) were derived from the susceptible parent. Three QTLs (SCN_7_1, SCN_8_2, and SCN_18_6) were detected in race 1 and race 2. The percentage of the explained variance of the identified QTLs varied from 1.91 to 7.73%. According to their physical position in the genome, the QTL intervals of SCN_8_2 (6.75–10.35 Mb) and SCN_18_6 (0.05–3.16 Mb) overlapped with the rhg1 and Rhg4 loci (see Footnote 2), respectively.
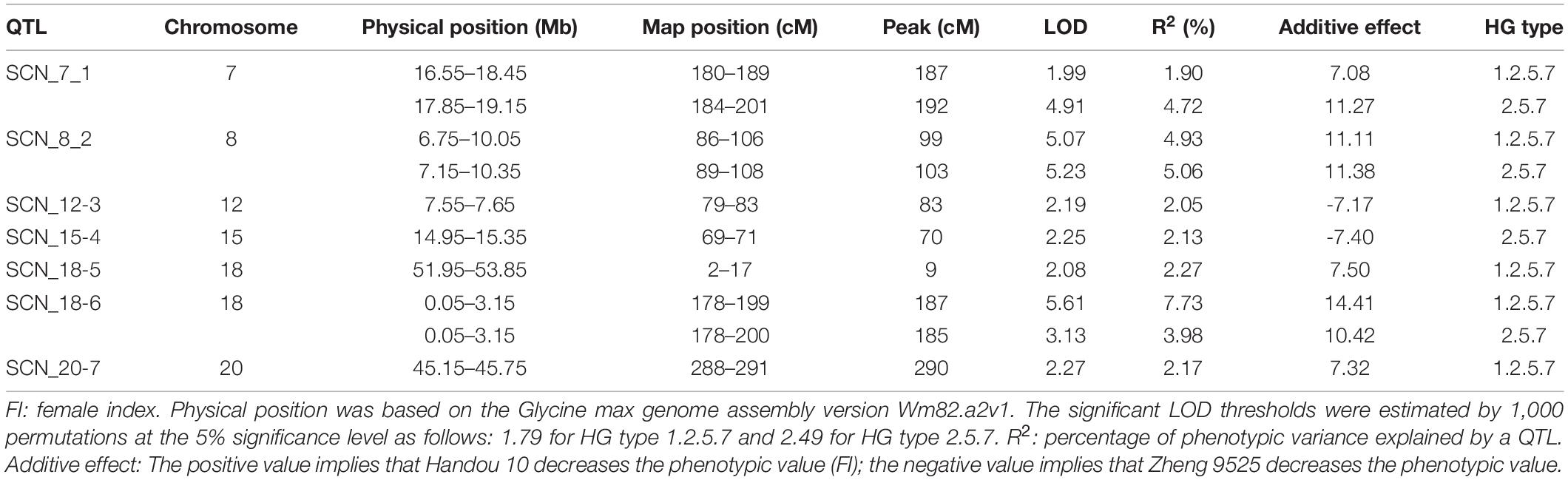
Table 4. Summary of additive QTLs for SCN resistance detected in the mapping population of 392 RILs derived from the cross between susceptible parent Zheng 9525 and resistant parent Handou 10 using composite interval mapping (CIM) with the female index of HG types 2.5.7 (race 1) and 1.2.5.7 (race 2).
We analyzed the transcriptomes of the resistant and susceptible parents infected with race 2. Each sample produced 6.45 G data on average (Supplementary Table 5). The average comparison rate between each sample and reference genome was 80.70%. A total of 912 and 981 significant DEGs were detected between the infected and uninfected roots in Handou 10 and Zheng 9525, respectively (Figure 2). In Handou 10, 312 and 600 up- and down- regulated genes were detected, respectively. In Zheng 9525, 395 and 586 up- and down- regulated genes were detected, respectively. A total of 4424 DEGs were detected between the infected roots of Handou 10 and Zheng 9525.
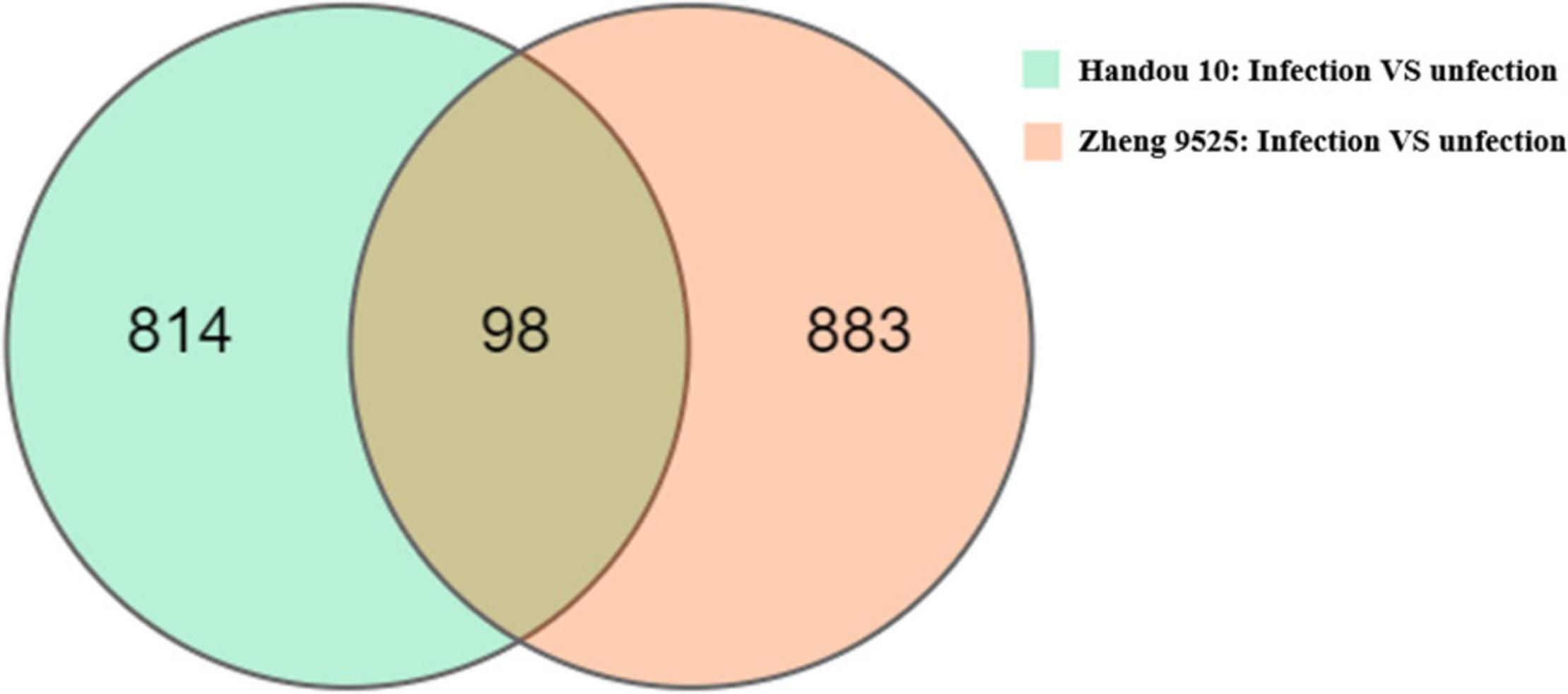
Figure 2. Different expressed genes (DEGs) in Handou 10 and Zheng 9525. S: Zheng 9525 with SCN infected; R: Handou 10 with SCN infected; CK_R: mock-inoculated control from Handou 10; CK_S: mock-inoculated control from Zheng 9525.
The KEGG analysis showed that 2100 DEGs were assigned to 132 pathways, and the plant–pathogen interaction pathway (ko04626) was the top enriched pathway (Figure 3) with 213 DEGs between the SCN infected roots of Handou 10 and Zheng 9525. The results suggested that the plant–pathogen interaction played the most important role in the SCN resistance for Handou 10. MPK signaling-plant (ko04016), phenylpropanoid biosynthesis (ko00940), plant hormone signal transduction (ko04075) and other pathways also played important roles in the resistance to SCN (Figure 3).
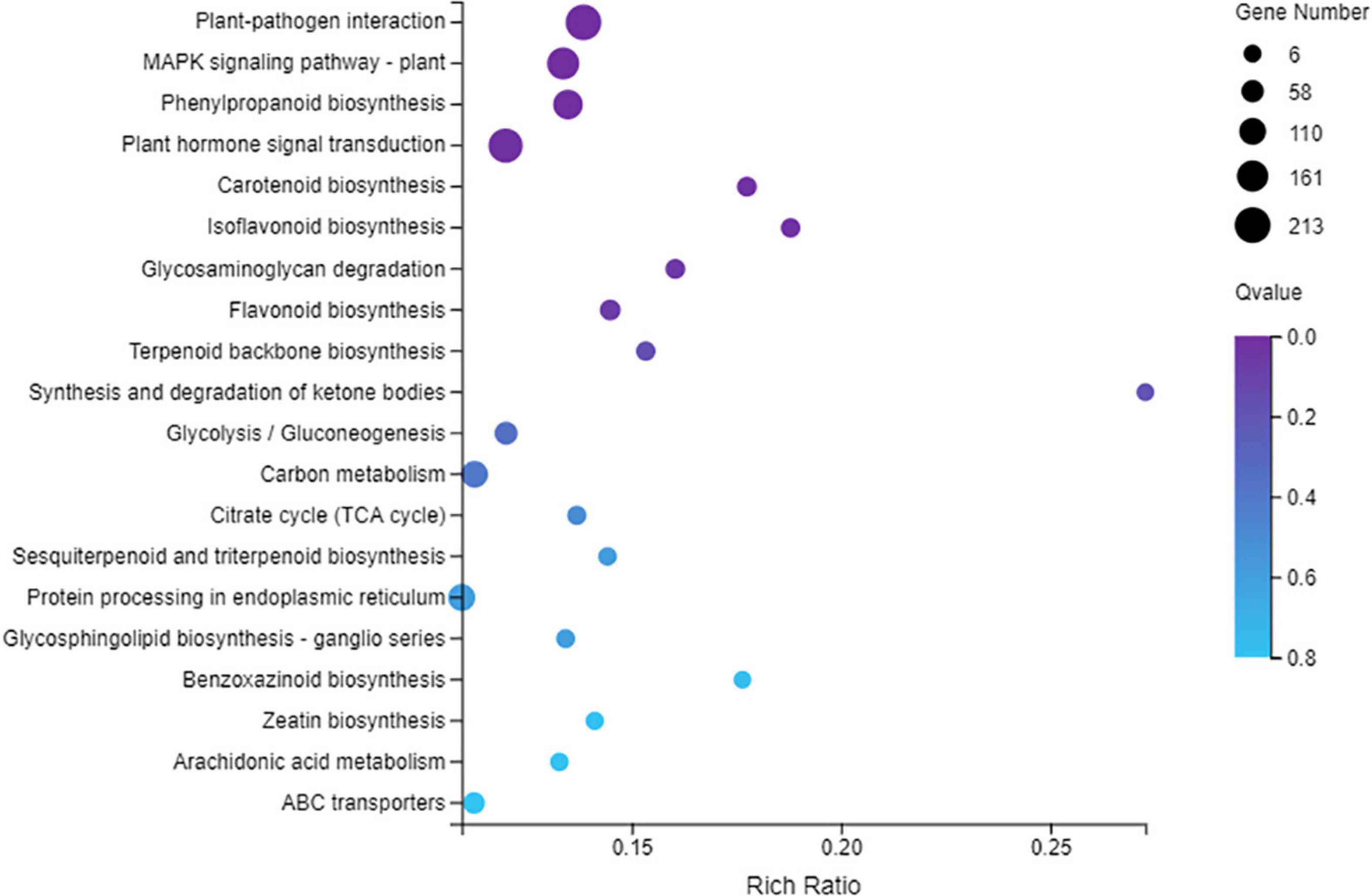
Figure 3. Statistics of KEGG Pathway Enchrichment scatter diagram of DEGs between infected roots of Handou 10 and Zheng 9525. The 20 most enriched pathways are displayed.
According to the physical positions of the major QTL intervals, there were 6, 23, and 12 candidate genes from the DEGs between the infected roots of Handou 10 and Zheng 9525 for SCN_7_1, SCN_8_2, and SCN_18_6, respectively (Table 5). The candidate gene Glyma.08G108900 at SCN_8_2 and candidate genes Glyma.18G022400, Glyma.18G022500, and Glyma.18G022700 at SCN_18_6 have been reported for SCN resistance (Guo et al., 2019; Cook et al., 2012; Liu et al., 2012). Glyma.08G108900 was located in the Rhg4 locus. Glyma.18G022400, Glyma.18G022500, and Glyma.18G022700 were located in the rhg1 locus. These three genes (Glyma18g02580, Glyma18g02590, and Glyma18g02610 in Wm82.a1) in rhg1 were all expressed in the resistance of Handou 10 according to the transcriptome data (Table 5). The genotypes of KASP markers linked to rhg1 (Rhg1-2, Rhg1-5, DD381, and DD383) and Rhg4 (Rhg4-3, Rhg4-5, and DD191) for Handou 10 were the same as those in Peking, whereas the genotypes of those (DD7_1, DD7_5, DD7_8, DD7_9, DD7_11, and DD7_16) located at SCN_7_1 for Handou 10 were consistent with those in PI437654 (Table 6 and Supplementary Table 6). There is no relevant report about the candidate genes at SCN_7_1 and these are likely novel.
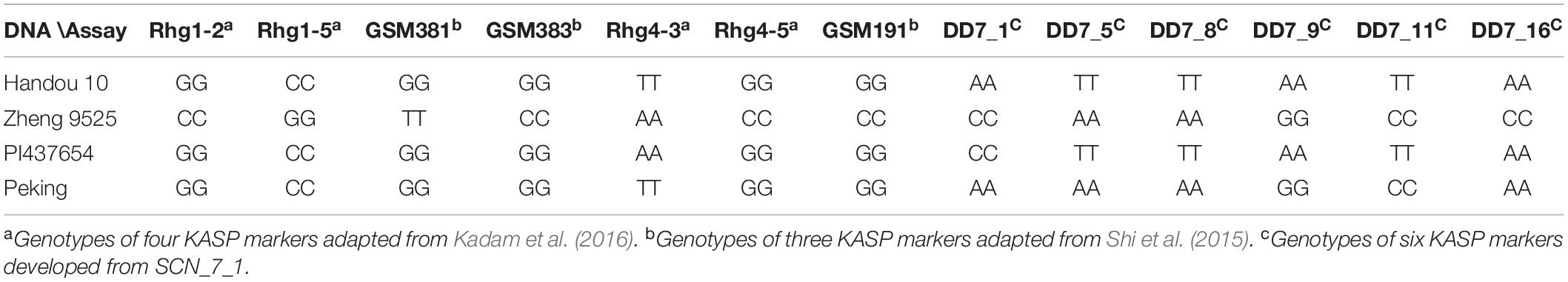
Table 6. Detection and analysis of soybean germplasm by KASP markers located at major QTL intervals (SCN_8_2, SCN_18_6, and SCN_7_1).
Handou 10 is a variety with a broad–spectrum resistant to SCN. Our recent experiment result showed that Handou 10 was resistant to SCN HG type 7 (race 3). In this study, Handou 10 was identified as being resistant against HG type 2.5.7 (race 1), and moderately resistant against HG type 1.2.5.7 (race 2); additionally, its resistance was identified as being Peking-type based on the analysis of the QTL (Table 4), KASP markers (Table 6) and the CNV at the rhg1/Rhg4 (Supplementary Figure 1). SCN_8_2 and SCN_18_6 overlapped with the resistance loci Rhg4 and rhg1-a, respectively. This two additive QTLs to SCN resistance with FI were also detected by IciMapping 4.2 software (Supplementary Table 7). SCN_8_2 and SCN_18_6 were interactive for SCN resistance to HG types 2.5.7 (race 2) (Supplementary Table 8). According to the CNV analyzes (Suvakov et al., 2021) for the whole-genome re-sequencing, the CNV at the rhg1 and Rhg4 for Handou 10 is 2.95 and 2.05, respectively (Supplementary Figure 1). The resistance to SCN and CNV at the rhg1/Rhg4 in Handou 10 is similar to PI404166, PI 437679, and PI089772 (Patil et al., 2019). The rhg1-a, sometimes in combination with the Rhg4, provides strong resistance to SCN (Guo et al., 2019). Three genes (Glyma.18G022400, Glyma.18G022500, and Glyma.18G022700) are responsible for the resistance provided by rhg1-b (Cook et al., 2012). In our study, these three candidate genes in rhg1-a might be regulate the SCN resistance of Handou 10. However, the mechanisms of rhg1-a and rhg1-b to SCN resistance are not same (Cook et al., 2012; Patil et al., 2019). For rhg1-b, overexpression of the individual genes in roots was ineffective and SCN resistance is conferred by copy number variation. The Peking-type requires rhg1-a and Rhg4 for SCN resistance (Patil et al., 2019). In this study, the expression of Glyma.08G108900 located in the Rhg4 locus was verified by qPCR (Supplementary Figure 2) and this was basically consistent with transcriptome analysis. Glyma.08G108900 was responsible for the resistance to SCN and the function has been verified. The resistant mechanism of Handou 10 is complicate and we are breeding new superior lines with it now.
SCN_7_1 is an important QTL to SCN resistance for Handou 10. Some QTLs (Webb et al., 1995; Ferreira et al., 2011; Abdelmajid et al., 2014; Chang et al., 2016; Vuong et al., 2015; Li et al., 2016) related to SCN resistance on chromosome 7 were reported at www.soybase.org. The physical location of SCN_7_1 was basically the same as the marker php02301a mapped in PI437654 (Webb et al., 1995), but far from the QTLs detected by other researchers (see Footnote 2). There are six DEGs in the SCN_7_1 region and these candidate genes are not previously described as SCN resistance genes. Among the DEGs at SCN_7_1, Glyma.07G139700 and Glyma.07G139800 both code Glutathione S–transferases (GSTs), which were up–regulated between infected and uninfected SCN in Handou 10 and down–regulated in Zheng 9525 according to transcriptome data. Recently, the expression of Glyma.07G139800 was verified by qPCR (Supplementary Figure 2) and this was basically consistent with transcriptome analysis in our study. GSTs are multifunctional enzymes which play a crucial role in cellular detoxification and oxidative stress tolerance (Rezaei et al., 2013). GST was elevated in the SCN–infected roots relative to uninoculated roots (Alkharouf et al., 2004). The over-expression of a GST gene from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco (Ji et al., 2010). However, the relationship between glutathione metabolism and the disease resistance of Handou 10 still needs to be further studied.
Soybean resistance to SCN is regulated by multiple genes. In our study the plant-pathogen interaction pathway was the most enriched KEGG pathway between infected Handou 10 and Zheng 9525, and between infected Zheng 9525 and uninfected Zheng 9525, whereas the MAPK signaling pathway was the most enriched KEGG pathway between infected and uninfected Handou 10. Plants and pathogens should be studied together as an interacting system (Peyraud et al., 2017). Soybean root cells undergo dramatic morphological and biochemical changes after being infected with SCN (Khan et al., 2004). The development of cyst nematodes in the infected SCN roots in Handou 10 was slower than in Zheng 9525 after 10 days of inoculation (Supplementary Figure 3). Many DEGs may be related to the development of soybean cyst nematode. These DEGs genes (Glyma.07G139700, Glyma.07G139800, Glyma.08G108900, Glyma.08G118900, Glyma.08G119200, Glyma.18G022400, Glyma.18G022500, and Glyma.18G022700) might be the important candidate gene to SCN resistance according to KEGG analysis and genes function.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NGDC CAR005982.
WL, JW, and HW conceived the project and designed the experiments. HW, YL, JL, HL, YW, CL, SW, and HZ performed the experiments. HW, WL, JW, and QS participated in the data analysis and manuscript revised. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (NSFC Grant Nos. 31901501 and 31371652).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Kurt Koch Georg (Henan Academy of Crops Molecular Breeding) for editing and suggestions on the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.860034/full#supplementary-material
Abdelmajid, K., Ramos, L., Hyten, D., Bond, J., Bendahmane, A., Arelli, P. R., et al. (2014). Quantitative trait loci (QTL) that underlie SCN resistance in soybean [Glycine max (L.) Merr.] PI438489B by ‘Hamilton’ recombinant inbred line (RIL) population. Atlas J. Plant Biol. 1, 29–38. doi: 10.5147/ajpb.2014.0140
Alkharouf, N., Khan, R., and Matthews, B. (2004). Analysis of expressed sequence tags from roots of resistant soybean infected by the soybean cyst nematode. Genome 47, 380–388. doi: 10.1139/g03-114
Chang, H. X., Lipka, A. E., Domier, L. L., and Hartman, G. L. (2016). Characterization of disease resistance loci in the USDA soybean germplasm collection using genome-wide association studies. Phytopathology 106, 1139–1151. doi: 10.1094/PHYTO-01-16-0042-FI
Chen, Y. X., Chen, Y. S., Shi, C. M., Huang, Z. B., Zhang, Y., Li, S. K., et al. (2018). SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 7, 1–6. doi: 10.1093/gigascience/gix120
Concibido, V. C., Diers, B. W., and Arelli, P. R. (2004). A decade of QTL mapping for cyst nematode resistance in soybean. Crop Sci. 44, 1121–1131. doi: 10.2135/cropsci2004.1121
Cook, D. E., Lee, T. G., Guo, X. L., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
Ferreira, M. F. D. S., Cervigni, G. D. L., Ferreira, A., Schuster, I., Santana, F. A., Pereira, W. D., et al. (2011). QTL for resistance to soybean cyst nematode races 3, 9, and 14 in cultivar Hartwig. Pesq. Agropec. Bras. 46, 420–428. doi: 10.1590/S0100-204X2011000400012
Guo, W., Chen, J. S., Zhang, F., Li, Z. Y., Chen, H. F., Zhang, C. J., et al. (2020). Characterization of Pingliang xiaoheidou (ZDD 11047), a soybean variety with resistance to soybean cyst nematode Heterodera glycines. Plant Mol. Biol. 103, 253–267. doi: 10.1007/s11103-020-00990-4
Guo, W., Zhang, F., Bao, A., You, Q., Li, Z., Chen, J., et al. (2019). The soybean Rhg1 amino acid transporter gene alters glutamate homeostasis and jasmonic acid-induced resistance to soybean cyst nematode. Mol. Plant Pathol. 20, 270–286. doi: 10.1111/mpp.12753
Howland, A., Monnig, N., Mathesius, J., Nathan, M., and Mitchum, M. G. (2018). Survey of Heterodera glycines population densities and virulence phenotypes during 2015-2016 in Missouri. Plant Dis. 102, 2407–2410. doi: 10.1094/PDIS-04-18-0650-SR
Ji, W., Zhu, Y., Li, Y., Yang, L., Zhao, X., Cai, H., et al. (2010). Over-expression of a glutathione S-transferase gene, GsGST, from wild soybean (Glycine soja) enhances drought and salt tolerance in transgenic tobacco. Biotechnol. Lett. 32, 1173–1179. doi: 10.1007/s10529-010-0269-x
Jiang, H., Li, Y., Qin, H., Li, Y., Qi, H., Li, C., et al. (2018). Identification of major QTLs associated with first pod height and candidate gene mining in soybean. Front. Plant Sci. 9:1280. doi: 10.3389/fpls.2018.01280
Kadam, S., Vuong, T. D., Qiu, D., Clinton, G., Meinhardt, C. G., Song, L., et al. (2016). Genomic-assisted phylogenetic analysis and marker development for next generation soybean cyst nematode resistance breeding. Plant Sci. 242, 342–350. doi: 10.1016/j.plantsci.2015.08.015
Khan, R., Alkharouf, N., Beard, H., Macdonald, M., Chouikha, I., Meyer, S., et al. (2004). Microarray analysis of gene expression in soybean roots susceptible to the soybean cyst nematode two days post invasion. J. Nematol. 36, 241–248.
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, K. K., Vuong, T. D., Qiu, D., Robbins, R. T., Shannon, G. J., Li, Z. L., et al. (2016). Advancements in breeding, genetics, and genomics for resistance to three nematode species in soybean. Theor. Appl. Genet. 129, 2295–2311. doi: 10.1007/s00122-016-2816-x
Koenning, S. R., and Wrather, J. A. (2010). Suppression of soybean yield potential in the continental United States from plant diseases estimated from 2006 to 2009. Plant Health Prog. 11:5. doi: 10.1094/PHP-2010-1122-01-RS
Kofsky, J., Zhang, H., and Song, B. (2021). Novel resistance strategies to soybean cyst nematode (SCN) in wild soybean. Sci. Rep. 11:7967. doi: 10.1038/s41598-021-86793-z
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, F., Wang, X. L., Hu, H. B., Zhou, X. C., Na, R. N., Xiu, M., et al. (2020). Analysis on occurrence regularity of soybean cyst nematode in Chifeng. Biol. Disaster Sci. 43, 373–377.
Li, R., Chang, Y., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009a). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Li, R., Li, Y., Fang, X., Yang, H., Wang, J., Kristiansen, K., et al. (2009b). SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132. doi: 10.1101/gr.088013.108
Li, Y. H., Shi, X. H., Li, H. H., Reif, J. C., Wang, J. J., Liu, Z. X., et al. (2016). Dissecting the genetic basis of resistance to soybean cyst nematode combining linkage and association mapping. Plant Genome 9, 1–11. doi: 10.3835/plantgenome2015.04.0020
Lian, Y., Wang, J. S., Li, H. C., Wei, H., Li, J. Y., Wu, Y. K., et al. (2016). Race Distribution of soybean cyst nematode in the main soybean producing area of Huang-Huai rivers valley. Acta Agron. Sin. 42, 1479–1486. doi: 10.3724/SP.J.1006.2016.01479
Lian, Y., Wei, H., Wang, J. S., Lei, C. F., Li, H. C., Li, J. Y., et al. (2019). Chromosome-level reference genome of X12, a highly virulent race of the soybean cyst nematode Heterodera glycines. Mol. Ecol. Resour. 19, 1637–1646. doi: 10.1111/1755-0998.13068
Liu, S. M., Ge, F. Y., Huang, W. K., Lightfoot, D. A., and Peng, D. L. (2019). Effective identification of soybean candidate genes involved in resistance to soybean cyst nematode via direct whole genome resequencing of two segregating mutants. Theor. Appl. Genet. 132, 2677–2687. doi: 10.1007/s00122-019-03381-6
Liu, S. M., Kandoth, P. K., Warren, S. D., Yeckel, G., Heinz, R., Alden, J., et al. (2012). Soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492, 256–260. doi: 10.1038/nature11651
Liu, S., Kandoth, P. K., Lakhssassi, N., Kang, J., Colantonio, V., Heinz, R., et al. (2017). The soybean GmSNAP18 gene underlies two types of resistance to soybean cyst nematode. Nat. Commun. 8:14822. doi: 10.1038/ncomms14822
Lu, W. G., Gai, J. Y., and Li, W. D. (2006). Sampling survey and identification of races of soybean cyst nematode (Heterodem glycines Ichinohe) in Huang-HuaiValleys. Sci. Agric. Sin. 39, 306–312. In Chinese,Google Scholar
Miraeiz, E., Chaiprom, U., Afsharifar, A., Karegar, A., Drnevich, J. M., and Hudson, M. E. (2020). Early transcriptional responses to soybean cyst nematode HG Type 0 show genetic differences among resistant and susceptible soybeans. Theor. Appl. Genet. 133, 87–102. doi: 10.1007/s00122-019-03442-w
Mitchum, M. G. (2016). Soybean resistance to the soybean cyst nematode Heterodera glycines: an update. Phytopathology 106, 1444–1450. doi: 10.1094/PHYTO-06-16-0227-RVW
Mitchum, M. G., Wrather, J. A., Heinz, R. D., Shannon, J. G., and Danekas, G. (2007). Variability in distribution and virulence phenotypes of Heterodera glycines in Missouri during 2005. Plant Dis. 91, 1473–1476. doi: 10.1094/PDIS-91-11-1473
Patil, G. B., Lakhssassi, N., Wan, J., Song, L., Zhou, Z., Klepadlo, M., et al. (2019). Whole-genome re-sequencing reveals the impact of the interaction of copy number variants of the rhg1 and Rhg4 genes on broad-based resistance to soybean cyst nematode. Plant Biotechnol. J. 17, 1595–1611. doi: 10.1111/pbi.13086
Peng, D., Jiang, R., Peng, H., and Liu, S. (2021). Soybean cyst nematodes: a destructive threat to soybean production in China. Phytopath. Res. 3:19. doi: 10.1186/s42483-021-00095-w
Peyraud, R., Dubiella, U., Barbacci, A., Genin, S., Raffaele, S., and Roby, D. (2017). Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant J. 90, 720–737. doi: 10.1111/tpj.13429
Rezaei, M. K., Shobbar, Z., Shahbazi, M., Abedini, R., and Zare, S. (2013). Glutathione S-transferase (GST) family in barley: identification of members, enzyme activity, and gene expression pattern. J. Plant Physiol. 170, 1277–1284. doi: 10.1016/j.jplph.2013.04.005
Riggs, R. D. (1977). Worldwide distribution of soybean-cyst nematode and its economic importance. J. Nematol. 9, 34–39. doi: 10.2307/1380029
Riggs, R. D., and Schmitt, D. P. (1988). Complete characterization of the race scheme for Heterodera glycines. J. Nematol. 20, 392–395.
Shi, X., Chen, Q., Liu, S., Wang, J., Peng, D., and Kong, L. (2021). Combining targeted metabolite analyses and transcriptomics to reveal the specific chemical composition and associated genes in the incompatible soybean variety PI437654 infected with soybean cyst nematode HG1.2.3.5.7. BMC Plant Biol. 21:217. doi: 10.1186/s12870-021-02998-4
Shi, Z., Liu, S., Noe, J., Arelli, P., Meksem, K., and Li, Z. (2015). SNP identification and marker assay development for high-throughput selection of soybean cyst nematode resistance. BMC Genomics 16:314. doi: 10.1186/s12864-015-1531-3
Suvakov, M., Panda, A., Diesh, C., Ian Holmes, I., and Abyzov, A. (2021). CNVpytor: a tool for copy number variation detection and analysis from read depth and allele imbalance in whole-genome sequencing. Gigascience 10:giab074. doi: 10.1093/gigascience/giab074
Tylka, G. L., and Marett, C. C. (2014). Distribution of the soybean cyst nematode, Heterodera glycines, in the United States and Canada: 1954 to 2014. Plant Health Prog. 15, 85–87. doi: 10.1094/PHP-BR-14-0006e
Usovsky, M., Ye, H., Vuong, T. D., Patil, G. B., Wan, J. R., Zhou, L. J., et al. (2021). Fine-mapping and characterization of qSCN18, a novel QTL controlling soybean cyst nematode resistance in PI 567516C. Theor. Appl. Genet. 134, 621–631. doi: 10.1007/s00122-020-03718-6
Van Ooijen, J. W. (2006). JoinMap® 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen: Kyazma, BV.
Vuong, T. D., Sonah, H., Meinhardt, C. G., Deshmukh, R., Kadam, S., Nelson, R. L., et al. (2015). Genetic architecture of cyst nematode resistance revealed by genome-wide association study in soybean. BMC Genomics 16:593. doi: 10.1186/s12864-015-1811-y
Wang, C. (2019). Research advances in genetic markers for resistance to soybean cyst nematodes (SCN). Plant Dis. Pests 10, 10–14. doi: 10.19579/j.cnki.plant-d.p.2019.01.003
Wang, J. S., Lu, W. G., Li, J. Y., Lian, Y., Wei, H., Li, H. C., et al. (2014). The Data Acquisition System on the Phenotype of Plant Diseases and Insect Pests. China. 2014SR060158 Beijing: Administration of the People’s Republic of China.
Wang, S., Basten, C. J., and Zeng, Z. B. (2012). Windows QTL Cartographer 2.5. Raleigh, NC: Department of Statistics, North Carolina State University.
Keywords: soybean, soybean cyst nematode resistance, candidate genes, QTL, whole genome re-sequencing, transcriptome
Citation: Wei H, Lian Y, Li J, Li H, Song Q, Wu Y, Lei C, Wang S, Zhang H, Wang J and Lu W (2022) Identification of Candidate Genes Controlling Soybean Cyst Nematode Resistance in “Handou 10” Based on Genome and Transcriptome Analyzes. Front. Plant Sci. 13:860034. doi: 10.3389/fpls.2022.860034
Received: 22 January 2022; Accepted: 21 February 2022;
Published: 15 March 2022.
Edited by:
Yuan-Ming Zhang, Huazhong Agricultural University, ChinaReviewed by:
Ainong Shi, University of Arkansas, United StatesCopyright © 2022 Wei, Lian, Li, Li, Song, Wu, Lei, Wang, Zhang, Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinshe Wang, d2pzMzMzMTRAMTI2LmNvbQ==; Weiguo Lu, MTIzYmVhbkAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.