
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 11 March 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.859374
This article is part of the Research Topic Protein Posttranslational Modifications in Plant Responses to Abiotic Stress - Women in Plant Science Series View all 8 articles
Nitric oxide (NO), which is ubiquitously present in living organisms, regulates many developmental and stress-activated processes in plants. Regulatory effects exerted by NO lies mostly in its chemical reactivity as a free radical. Proteins are main targets of NO action as several amino acids can undergo NO-related post-translational modifications (PTMs) that include mainly S-nitrosylation of cysteine, and nitration of tyrosine and tryptophan. This review is focused on the role of protein tyrosine nitration on NO signaling, making emphasis on the production of NO and peroxynitrite, which is the main physiological nitrating agent; the main metabolic and signaling pathways targeted by protein nitration; and the past, present, and future of methodological and strategic approaches to study this PTM. Available information on identification of nitrated plant proteins, the corresponding nitration sites, and the functional effects on the modified proteins will be summarized. However, due to the low proportion of in vivo nitrated peptides and their inherent instability, the identification of nitration sites by proteomic analyses is a difficult task. Artificial nitration procedures are likely not the best strategy for nitration site identification due to the lack of specificity. An alternative to get artificial site-specific nitration comes from the application of genetic code expansion technologies based on the use of orthogonal aminoacyl-tRNA synthetase/tRNA pairs engineered for specific noncanonical amino acids. This strategy permits the programmable site-specific installation of genetically encoded 3-nitrotyrosine sites in proteins expressed in Escherichia coli, thus allowing the study of the effects of specific site nitration on protein structure and function.
Plant developmental programs as well as responses to stress involve a wide variety of processes affecting gene expression that include transcription, post-transcriptional processing, and alternative splicing, translation, and post-translational modifications (PTMs; recently reviewed in Zhang et al., 2021). After translation, proteins are often targets of a wide array of PTMs that may alter their 3D structure, cofactor- or ligand-binding, stability to proteolysis, or intracellular localization, and therefore, their function and fate. PTMs are thus potential modifiers of protein function that when occurring in response to environmental or endogenous cues and affect to signaling proteins have potential in regulating signal transduction pathways. Despite the variety of responses triggered by different stresses, a major common response involves the production of reactive oxygen species (ROS), nitric oxide (NO), and other reactive nitrogen species (RNS), and reactive sulfur species (RSS; Del Río, 2015; Choudhury et al., 2017; De Gara and Foyer, 2017; Hancock, 2019). The combined action of NO, ROS, and RNS regulate many developmental and stress-related processes in plants including pollen and seed germination, elongation of shoots and roots, root architecture, stomatal closure and related abiotic stresses, leaf greening, mitochondrial function, several developmental transitions including flowering or senescence, and cell death. Many of these regulatory functions are exerted directly on varied protein targets that undergo different PTMs such as carbonylation (Tola et al., 2021); sulfenylation and persulfidation of cysteines (Aroca et al., 2021; Wang et al., 2021); methionine sulfoxidation (Drazic and Winter, 2014); S-nitrosylation (Lindermayr et al., 2005); and tyrosine nitration (Bottari, 2015; Kolbert et al., 2017; Arasimowicz-Jelonek and Floryszak-Wieczorek, 2019), phosphorylation at Ser, Thr, or Tyr residues (Gow et al., 1996; Kong et al., 1996); sumoylation (Miura et al., 2009; Niu et al., 2019); and polyubiquitination (Walsh and Sadanandom, 2014). Many of these PTMs are reversible but some of them, such as carbonylation and tyrosine nitration are irreversible, and frequently associated to protein inactivation and degradation. Among PTMs, S-nitrosylation and nitration are linked to the production of NO and often involved in signaling processes downstream NO sensing (Astier and Lindermayr, 2012).
Nitric oxide is a gas molecule ubiquitously present in all living organisms, from prokaryotes to eukaryotes, that is involved in the regulation of a wide array of biological processes from development to stress-activated responses (Hancock, 2020). NO and their target processes have been extensively documented in humans, other mammals, and vertebrates, invertebrates, plants, low eukaryotic organisms such as protozoan and yeast, and bacteria (Torreilles, 2001). NO is endogenously synthesized through different pathways depending on the organism. Since early 90s, it is well-known that NO is produced through the oxidation of the reduced nitrogen molecule arginine, catalyzed by NO synthases (NOS) in animals (Marletta, 1993; Moncada, 1993; Palmer, 1993). NOS are homodimer enzymes that utilize L-arginine as the substrate, molecular oxygen, and reduced nicotinamide-adenine-dinucleotide phosphate (NADPH) as co-substrates, and flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and (6R-) 5,6,7,8-tetrahydro-L-biopterin (BH4) as cofactors (Förstermann and Sessa, 2012). Many prokaryotes also generate NO through arginine-dependent NOS-like systems (Adak et al., 2002; Sudhamsu and Crane, 2009). However, unlike mammalian NO synthases, bacterial NO synthases do not contain a reductase domain (Filippovich, 2010). Although NOS activity has been measured in yeast and plants, no NOS ortholog has been identified to date. NO synthesis and sensing in plants differ from animals, and many controversial issues remain unsolved (León and Costa-Broseta, 2020). To date, no such NOS enzymes have been identified in plants (Jeandroz et al., 2016), though a NOS protein has been identified in the marine green alga Ostreococcus tauri (Foresi et al., 2010, 2015). Therefore, although NOS activity has been reported in plants it remains unclear whether this oxidative pathway operates (Kolbert et al., 2019). Instead, plants synthesize NO under normoxic conditions mostly from the oxidized nitrogen molecule nitrate through two sequential reduction steps to nitrite and further to NO catalyzed by nitrate reductase (NR), thus as a branch of the nitrate assimilatory pathway (Lozano-Juste and León, 2010; Astier et al., 2018; León and Costa-Broseta, 2020). NO-related differences between organisms are not restricted to their biosynthetic pathways but also apply to the way is sensed and signaled. Animal and early prokaryotes share structural similarity in NO-sensing proteins with a heme-containing domain that enables NO sensing by the heme NO/oxygen (H-NOX) motif. In mammals, NO is sensed through soluble guanylate cyclase (GC) receptor that transduces the initial NO signal to the secondary messenger cyclic guanosine monophosphate (cGMP; Arnold et al., 1977; Zhao et al., 1999). Effectors, such as cGMP-dependent protein kinases, ion channels, and phosphodiesterases are downstream regulated by cGMP, then controlling varied physiological processes such as regulation of blood vessel dilatation, immune function, and neurotransmission in the brain and peripheral nervous system. Although soluble GCs have been identified both in plants and yeast (Kuo et al., 1998), none of them have the biochemical features enabling their functions as NO sensors. Unlike animals, plants do not mediate NO signaling through the classical NO/cGMP signaling module, suggesting that the evolution of nitric oxide signaling diverged between animal and green lineages (Astier et al., 2019). The way NO is sensed in plants differs from that reported for mammals as they lack a specific receptor. However, it has been recently reported that plant proteins not related to GCs but containing the H-NOX motif might be involved in NO sensing, thus proposing the existence of an evolutionary conserved heme-based NO sensor (Wong et al., 2021). Future work will address whether these H-NOX motif-containing proteins are true NO sensors and, if they are, whether they are organ-, tissue-, or process-specific or instead behave as general plant NO sensors. Nevertheless, a NO sensing mechanism not related to H-NOX motif has been demonstrated to act as a plant NO sensor in plants. Many NO-regulated physiological processes in plants are controlled through a proteolytic mechanism determining the fate of several regulatory proteins. This proteolytic pathway depends on the N-terminal sequence of the target proteins that constitutes a so-called N-degron. Proteins containing N-degrons are specifically recognized by E3 ubiquitin (UBQ) ligases called recognins that polyubiquitinate them. The Cys/Arg branch of N-degron pathway acts on proteins containing a Cys in position 2, which must be oxidized in a process requiring both O2 and NO and catalyzed by Plant Cysteine Oxidases (PCO). After the subsequent N-terminal arginylation, substrates are further polyubiquitinated. The group VII of ethylene response factors (ERFVIIs) are substrates of this pathway and are polyubiquitinated by the recognin Proteolysis6 (PRT6) that prepares them for the proteolytic degradation by the 26S proteasome. This process has been characterized as a NO sensor controlling multiple physiological processes including seed germination, seedling establishment, hypocotyl elongation, stomata aperture, apical hook opening, and leaf greening in Arabidopsis (Gibbs et al., 2014; Abbas et al., 2015). Moreover, such mechanism has been also characterized as a general sensor of abiotic stresses and pathogen responses (Vicente et al., 2017, 2019). However, it remains controversial whether the NO essential requirement for ERFVII degradation operates at Cys2 oxidation step or later during ERFVII arginylation or polyubiquitination. In yeast, the expression of Arabidopsis PCOs seems to be sufficient to trigger the N-terminal cysteine oxidation, thus pointing to the dispensability of nitric oxide at this step (Puerta et al., 2019). Nevertheless, it must be assessed whether this also happens in plants, in which some PCO encoding genes are induced by NO (Castillo et al., 2018). A comparative scheme showing the differences between NO biosynthesis and sensing in mammals and plants is shown in Figure 1. Nevertheless, the N-degron-mediated regulation of ERFVII levels is crucial to control many NO-related processes as it determines the available transcription factor able to transactivate a wide array of secondary regulatory proteins, including ABI5 and other hormone-related transcription factors that finally regulate multiple developmental and stress-related responses (Figure 1).
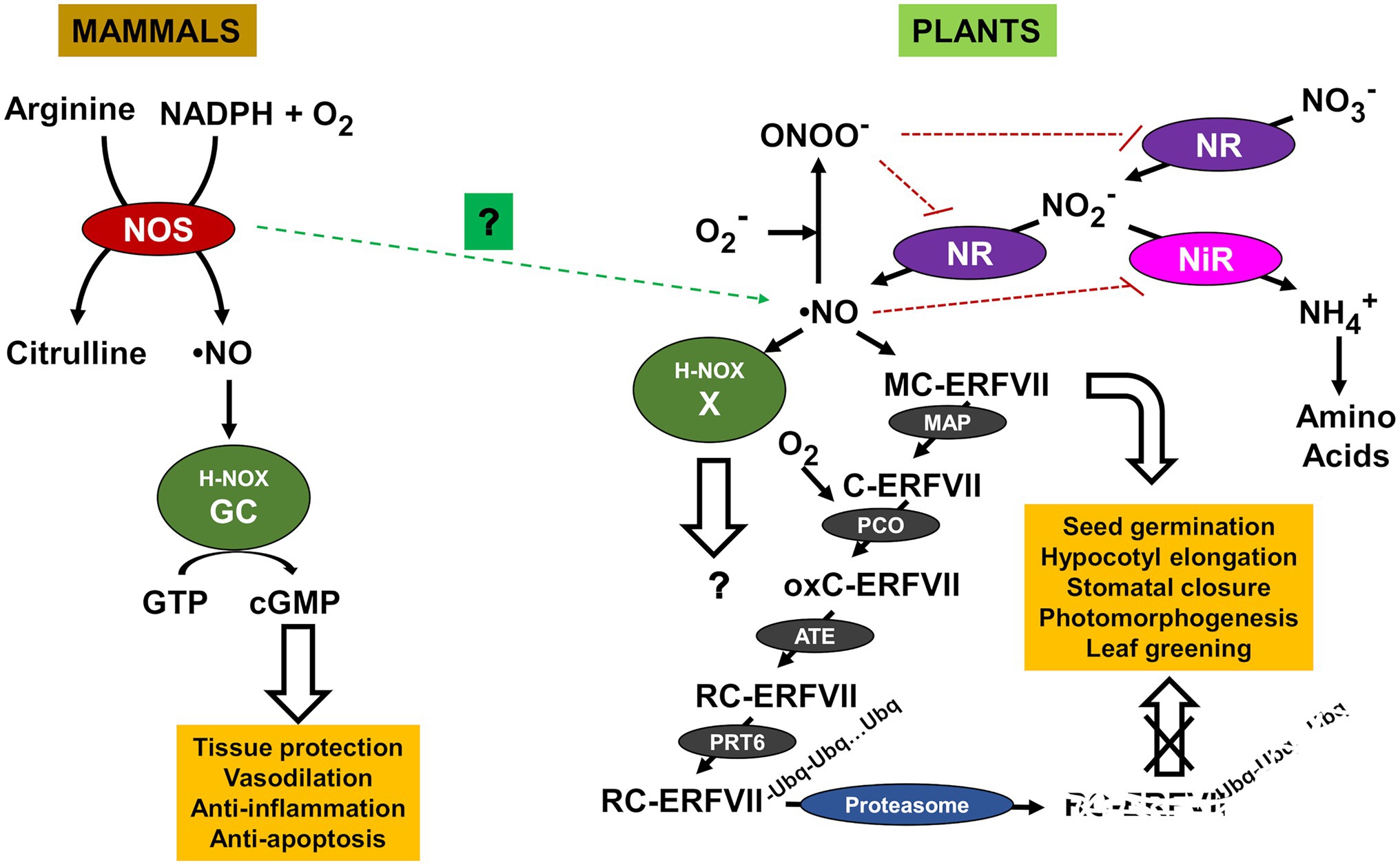
Figure 1. Nitric oxide (NO) production and sensing pathways in mammals and plants. Arginyl transferase (ATE), group VII Ethylene Response Factor (ERFVII), Cyclic guanosine monophosphate (cGMP), guanosine triphosphate (GTP), guanylate cyclase (GC), heme NO/oxygen motif (H-NOX), methionine aminopeptidase (MAP), nitrate reductase (NR), nitrite reductase (NiR), NO synthase (NOS), oxidized cysteine ERFVII (oxC-ERFVII), peroxynitrite (ONOO−), plant cysteine oxidase (PCO), E3 ubiquitin ligase Proteolysis6 (PRT6), arginylated cysteine ERFVII (RC-ERFVII), and unknown NO sensor (X). NO- and peroxinitrite-triggered inactivation are shown by blunt-ended red dotted lines. Still uncertain involvement of NOS in NO production is shown with green dotted arrow.
Nitric oxide signaling downstream ERFVII-mediated sensing is highly dependent on the levels and function of these transcription factors. ERFVIIs regulate different target processes that includes responses to hypoxia (Gibbs et al., 2011; Gasch et al., 2016; Loreti et al., 2016) and ABA-regulated seedling establishment (Zhang et al., 2018). Specific ERFVIIs such as RAP2.12 regulates carbon metabolism (Paul et al., 2016), and together with RAP2.3 negatively regulates NO biosynthesis and responses through a rheostat-like mechanism in Arabidopsis (León et al., 2020). Despite the numerous processes that have been reported to be regulated by NO through ERFVII stability, the mechanism underlying such regulatory functions is often unknown. In addition to the ERFVII-mediated NO signaling, the reactive nature of NO makes it to react with other molecules as a primary action. Such NO reaction with proteins ends with PTMs that include the S-nitrosylation of Cys (Gupta et al., 2020) and, upon previous reaction with superoxide to form peroxynitrite (Liaudet et al., 2009; Vandelle and Delledonne, 2011), the nitration of Tyr, Trp, and to a lesser extent also Phe and His (Sabadashka et al., 2021). Besides peroxynitrite, the •NO2 radical formed from nitrite through peroxidase-mediated oxidation under certain conditions is also an efficient nitrating agent of proteins (Jones, 2012). The effect of nitrating agents on proteins are somehow regulated through the action of diverse antioxidant systems that includes glutathione peroxidases (Sies et al., 1997) as well as the most relevant plant antioxidant systems such as glutathione, ascorbic acid, tocopherols, and flavonoids (Arasimowicz-Jelonek and Floryszak-Wieczorek, 2011). By using mass spectrometry (MS) techniques, many S-nitrosylated and nitrated plant proteins have been identified (Lozano-Juste et al., 2011; Feng et al., 2019). Moreover, several algorithms have been developed to predict the probability of identifying S-nitrosylation and nitration sites in target proteins (Kolbert and Lindermayr, 2021; Nilamyani et al., 2021). However, the in vivo identification of S-nitrosylation and nitration sites represents a difficult task, and the characterization of the effects of nitration and S-nitrosylation on protein function are significantly less studied. For protein nitration, these studies are mainly limited by the low proportion of in vivo nitrated peptides compared to non-modified ones, the unstable nature of nitro-derivatives that tend to be converted to amino-derivatives in reductive environments, and by the lack of specificity in artificial nitration systems used to increase the amount of nitrated peptides for further MS-based analysis.
In contrast to Cys S-nitrosylated proteins, which can be readily reversed to unmodified protein by enzyme catalyzed processes (Benhar et al., 2009; Malik et al., 2011; Sengupta and Holmgren, 2013), Tyr-nitrated proteins were initially thought to be not reversible and ineluctably targeted for degradation (Souza et al., 2000; Zhang et al., 2019; Lee et al., 2020). However, it has been reported that animal cells have the capacity to denitrate Tyr-nitrated proteins, though the nature of the enzyme involved in denitration remains unclear (Gow et al., 1996; Kuo et al., 1999; Görg et al., 2007; Kang and Akbarali, 2008; Abello et al., 2009; Deeb et al., 2013; Ferrer-Sueta et al., 2018). A denitrating activity has not been reported yet in plants. Instead, a reversibility mechanism, which is slower than the enzyme-catalyzed denitration, could be based on the rapid protein degradation coupled to de novo protein synthesis. The protein nitration-induced proteolysis that has been reported in human cells (Souza et al., 2000; Zhang et al., 2019; Lee et al., 2020) has also been shown functionally relevant in the regulation of the ABA perception and nitrate-related NO synthesis in Arabidopsis (Castillo et al., 2015; Costa-Broseta et al., 2020, 2021). Nitration-induced proteasomal degradation of PYR/PYL/RCAR receptors have been reported as a potential rapid mechanism to control ABA signaling under certain conditions (Castillo et al., 2015). Moreover, NO also regulates ABA signaling through the S-nitrosylation and the subsequent KEEP ON GOING E3 ligase- and CULLIN4-mediated polyubiquitination of ABI5 transcription factor (Albertos et al., 2015). Besides, the function of ABI5 is modulated through sumoylation by the SUMO E3 ligase SIZ1 (Miura et al., 2009), which was previously identified as an in vivo nitration target (Lozano-Juste et al., 2011). It is worth mentioning that the NO biosynthetic enzyme NR has been reported to be relocated to nuclei by SIZ1 (Kim et al., 2018), thus potentially representing and additional link between NO and ABA signaling. SIZ1 plays also key regulation on salicylic acid (SA) signaling (Miura et al., 2010) and plant immunity (Gou et al., 2017; Niu et al., 2019), thus suggesting SIZ1 could be a relevant node in NO, ABA, and SA signaling through nitration, sumoylation, and polyubiquitination (Figure 2). Moreover, SIZ1 has also been recently characterized as an important regulator of many biological processes in plants including the repression of histone deacetylase and further regulation of flowering time (Gao et al., 2021), phosphate deficiency-related responses (Zheng and Liu, 2021), photomorphogenesis (Zhang et al., 2020), and cell wall formation (Liu et al., 2019). This could be a good example of the impact of Tyr nitration on a signaling node due to the regulatory relevance of SIZ1. However, more work is needed to explore the functional relevance of SIZ1 nitration, and specifically the identification of Tyr nitration sites will be essential for a better understanding on nitration-sumoylation functional interactions controlling multiple plant processes.
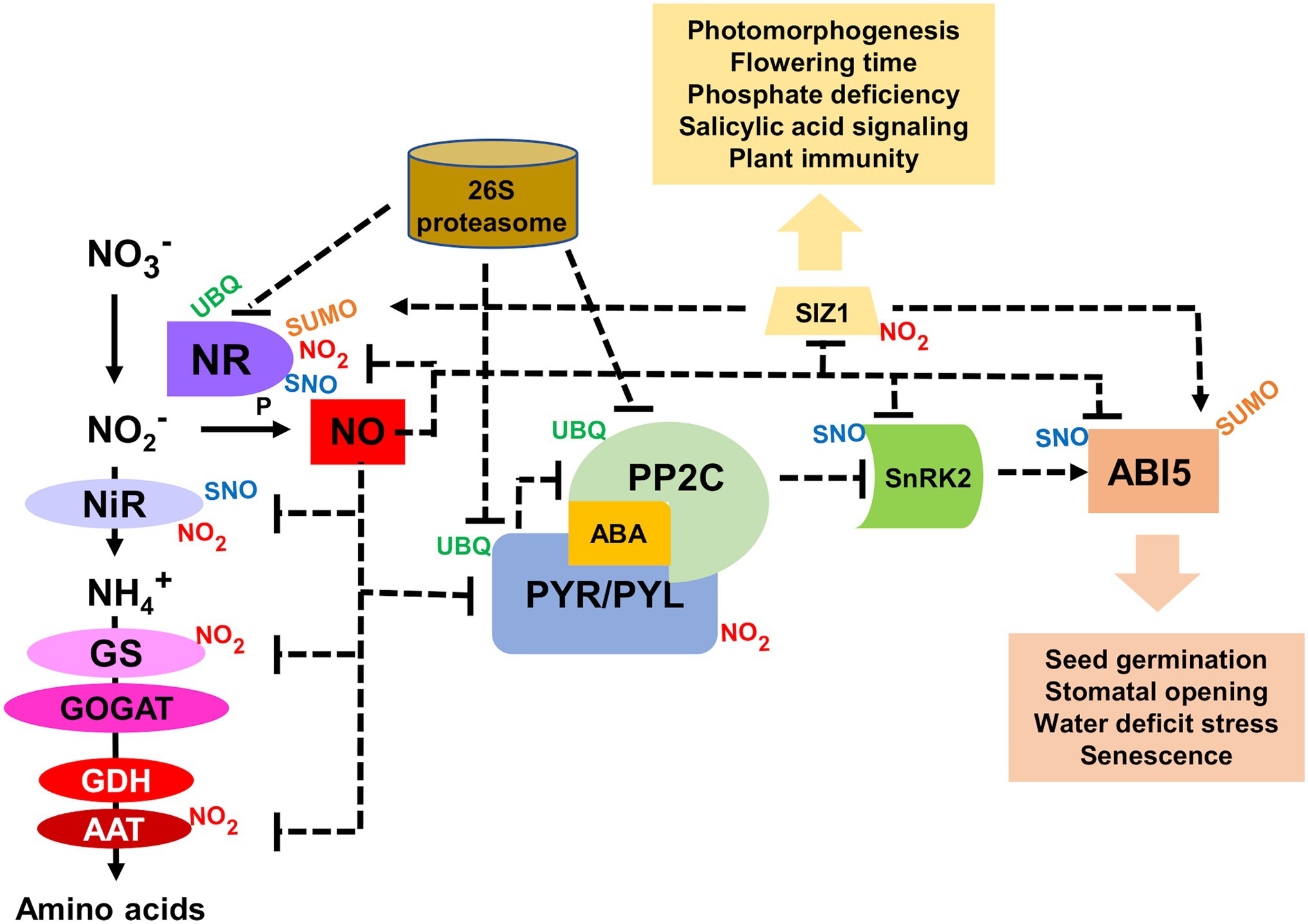
Figure 2. Functional interaction between NO and ABA signaling through NO-related post-translational modifications (PTMs). Aspartate aminotransferase (AAT), ABA-insensitive 5 (ABI5), glutamine synthetase (GS), glutamate dehydrogenase (GDH), glutamate synthase (GOGAT), nitrate reductase (NR), nitrite reductase (NiR), type 2C protein phosphatase (PP2C), Pyrabactin resistant ABA receptor (PYR), PYR-like (PYL), SAP and MIZ1 domain-containing ligase1 (SIZ1), sucrose non-fermenting1-related protein kinase 2 (SnRK2), small ubiquitin-like modifier (SUMO), nitrosothiol (SNO), and ubiquitin (UBQ). Dotted arrows and blun-ended lines represent activation and inactivation, respectively.
Nitric oxide also plays key roles in pathogen-triggered responses in plants (Jedelská et al., 2021), and under oxidative conditions, peroxynitrite is synthesized and functions as an effector of NO-mediated signaling (Alamillo and García-Olmedo, 2001; Saito et al., 2006; Vandelle and Delledonne, 2011). During the plant hypersensitive disease resistance response, protein tyrosine nitration has emerged as an important node between NO and ROS underlying a mechanism of co-operative interaction (Cecconi et al., 2009). Many plant-pathogen interactions are accompanied by the enhanced expression of Pathogenesis-Related (PR) proteins with diverse anti-pathogenic activities. Several PR proteins have been reported to be targets of Tyr nitration (Lozano-Juste et al., 2011; Tanou et al., 2012; Takahashi et al., 2016; Arasimowicz-Jelonek et al., 2016). Tyr nitration of proteins was also activated in tomato plants infected with leaf mold disease, though no specific targets have been identified (Chen et al., 2017). The effect of PR protein nitration on function/activity remains unknown and it should be further studied to know the functional relevance of this PTM on anti-pathogenic activity. Moreover, since PR proteins are frequently secreted to the apoplast, nitration may occur both at the cytoplasm and apoplast and when the modification takes place in the cytoplasm could also interfere with the final localization of these proteins. The involvement of protein nitration is not restricted to the interaction of plants with bacteria or fungi. Increased S-nitrosylation and nitration of proteins was detected during the infection of Arabidopsis roots with beet cyst nematodes (Labudda et al., 2020).
Nitrate together with ammonium are the most abundant nitrogen (N) source on Earth (Crawford and Glass, 1998). Both N sources present complex interactions that determine their transport/uptake, their allocation and assimilation (Hachiya and Sakakibara, 2017). Nitrate is photosynthetically assimilated by plants through a reductive linear pathway that converts nitrate into nitrite, and then to ammonium, requiring the sequential catalysis by NR and nitrite reductases (NiR; Campbell and Kinghorn, 1990). After reduction, ammonium is then incorporated to 2-oxoglutarate through the glutamine synthetase (GS)—glutamine oxoglutarate aminotransferase (GOGAT)/glutamate synthase metabolic cycle (Krapp, 2015) as well as the activities of aspartate aminotransferases (AATs), glutamate dehydrogenase (GDH), and asparagine synthetases (Kishorekumar et al., 2020). These enzymes enable the initial N incorporation to the amino acids Gln, Glu, Asn, and Asp, and then by transamination to the other amino acids. NRs, together with their main function catalyzing the conversion of nitrate to nitrite, can also reduce nitrite to NO (Maia and Moura, 2015; Chamizo-Ampudia et al., 2017), thus making NO a side product of nitrate assimilation. Most of the enzymes involved in nitrate assimilation have been identified as nitrated in Tyr residues. Arabidopsis NRs NIA1 and NIA2 as well as the only NiR1 enzyme has been recently reported to be Tyr-nitrated (Costa-Broseta et al., 2020, 2021). In Arabidopsis NRs, nitration occurs at two Tyr residues that coordinates flavin co-factor thus potentially hampering its efficient binding, and therefore the proper electron transfer flow throughout the enzyme (Costa-Broseta et al., 2021). For NiR1 nitration might not be the most relevant NO-triggered PTM, as it has been reported the S-nitrosylation of two Cys residues involved in coordinating the 4Fe-4S cluster and siroheme, which are key co-factors for the NiR redox activity of NiR1 (Costa-Broseta et al., 2020). Moreover, nitration not only affects to the first two enzymes of nitrate assimilation, but also GS has been identified as targets of nitration (Lozano-Juste et al., 2011). GS enzymes from mammals and fungi seem to be nitrated and inactivated, thus affecting the metabolism of Glu and ammonia in liver (Frieg et al., 2021) and the secondary metabolism in fungi (Zhu et al., 2021), respectively. Plant GDH seem to be involved mainly in Glu oxidation to supply cells with carbon skeletons under carbon limiting conditions (Robinson et al., 1991). Similarly, in mammals, GDH catalyzes the reversible oxidative deamination of Glu into 2-oxoglutarate and ammonium, which is a N source for the urea cycle. The enzyme from renal cortex cell mitochondria has been reported to be nitrated (Ishii et al., 2013). In turn, no reports on plant GDH nitration are available. Regarding other N metabolism-related enzymes, AATs catalyze the conversion of aspartate and 2-oxoglutarate to oxaloacetate and glutamate, thus playing mostly a transaminase function. The chloroplastic isoform from Arabidopsis was found nitrated in vivo (Lozano-Juste et al., 2011), and both the cytosolic and mitochondrial mammalian enzymes were inactivated upon nitration (Scandurra et al., 1975; DiCola et al., 1976), thus representing a potential regulatory target of NO in regulating the distribution of reduced N between amino acids.
The degradation of nitrated proteins involved in nitrate assimilation and NO production requires an additional PTM as targets should be previously polyubiquitinated in Lys residues before being degraded by the proteasome, therefore connecting nitration with polyubiquitination. NRs are polyubiquitinated (Costa-Broseta et al., 2021) and are translocated to the nucleus by the SUMO E3 ligase SIZ1 (Kim et al., 2018). Remarkably, a recent report showed that proteasomal-mediated cleavage at C-terminal of nitroTyr residues in proteins was reduced when compared to the unmodified residues, making nitration a possible factor that decreases the turnover of oxidized proteins (Ott et al., 2021), and casting doubts on the relationship between protein nitration and proteasome-mediated degradation. However, polyubiquitination is not the only PTM functionally connected to Tyr nitration. Another way Tyr-nitration may have an impact on cell signaling is through interference with a well-known signaling pathway mediated by Tyr-phosphorylation (Gow et al., 1996; Kong et al., 1996). Although Tyr-nitration and Tyr-phosphorylation might be mutually exclusive, some proteins can be simultaneously nitrated and phosphorylated in Tyr residues thus suggesting both PTMs are more competitive than exclusive (Monteiro, 2002; Joshi et al., 2015). Moreover, the nitrating agent peroxynitrite can also act as an inducer of Tyr phosphorylation-mediated signaling events (Takakura et al., 1999; Monteiro, 2002), thus suggesting both PTMs are connected in a complex way. Phosphorylation is very relevant for the regulation of NR and GS activities (Riedel et al., 2001; Lillo et al., 2004), but it occurs in key Ser but not Tyr residues, thus pointing to no phosphorylation-nitration competition in these enzymes. Therefore, protein that are targets of NO-related PTMs can be simultaneously or sequentially modified by other PTMs, which can modify their functions by triggering conformational changes, hampering ligand or cofactor binding, translocating them to different subcellular localization, promoting their preteasome-mediated degradation, or even altering the functional interaction with other signaling pathways (Figure 3).
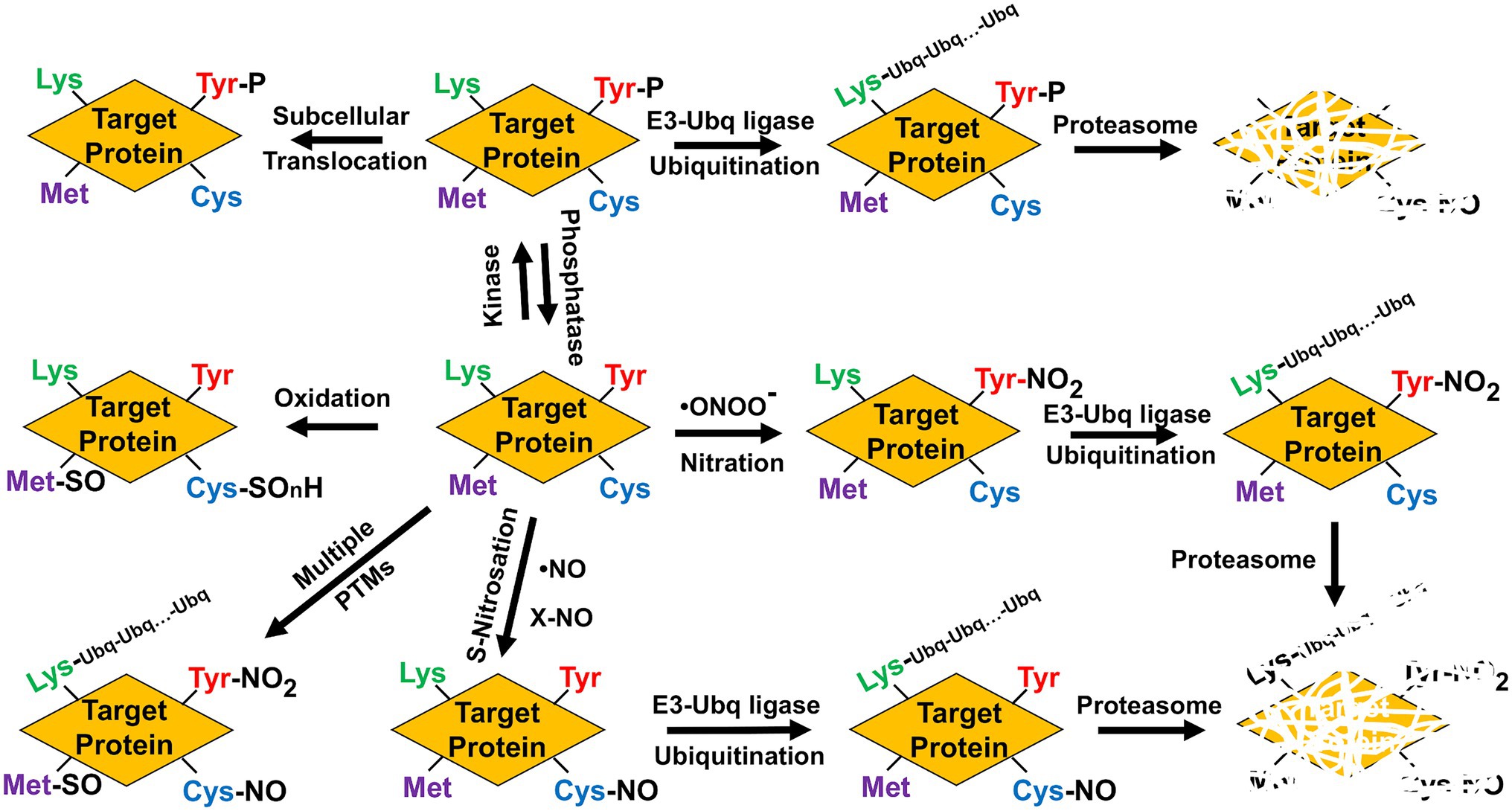
Figure 3. Multiple post-translational modifications potentially alter function/activity, localization, and stability of target proteins. Phosphorylation (P), methionine sulfoxide (Met-SO), Cys oxidation to sulphonic, sulphenic, or sulphinic group (SOnH), nitrosoCys (Cys-NO), nitroTyr (Tyr-NO2), Ubiquitin (Ubq), and polyubiquitinated lysines (Lys-Ubq-Ubq…-Ubq).
Stress conditions that rise NO and ABA levels are usually accompanied by ROS such as superoxide anion, hydroxyl radicals, and hydrogen peroxide, which together are involved in multiple signaling pathways (Neill et al., 2002). The levels of ROS are controlled by the activity of antioxidant enzymes, mainly superoxide dismutase (SOD) and catalase (CAT) that metabolize superoxide and hydrogen peroxide, respectively. These enzymes are regulated by NO-related PTMs (Begara-Morales et al., 2016). Nitration and S-nitrosylation of catalases and SODs have been reported (Lozano-Juste et al., 2011; Holzmeister et al., 2015; Begara-Morales et al., 2016; Palma et al., 2020). For Arabidopsis SODs, the nitration-mediated inhibition of their activities was specifically exerted in some members of the different families (Holzmeister et al., 2015). Regarding CAT, the enzymes of pepper fruits were inhibited by Tyr nitration during ripening (Chaki et al., 2015), though specific target residues have not yet been identified.
Hydrogen peroxide levels are also regulated through the so-called ascorbate-glutathione antioxidant cycle involving the activity of ascorbate peroxidases (APX), monodehydroascrobate reductases (MDAR) and dehydroascrobate reductases (DHAR), and glutathione reductases (GR; Noctor and Foyer, 1998). As for SODs and CATs, also enzymes of ascorbate-glutathione cycle have been identified as nitration targets (Lozano-Juste et al., 2011; Begara-Morales et al., 2016). The Tyr nitration sites for pea APX and MDAR have been identified (Begara-Morales et al., 2014, 2015). DHAR and GR have been also identified as Tyr nitration targets in citrus and sunflower, respectively (Chaki et al., 2009; Tanou et al., 2012), but nitration sites remain to be determined. It is worth mentioning that nitration of GR did not affect to their activity (Begara-Morales et al., 2015), thus representing one of the few cases where nitration did not lead to enzyme inactivation. The rest of antioxidant enzymes are inhibited by Tyr nitration thus representing a very relevant node of regulation of ROS metabolism by NO (Figure 4).
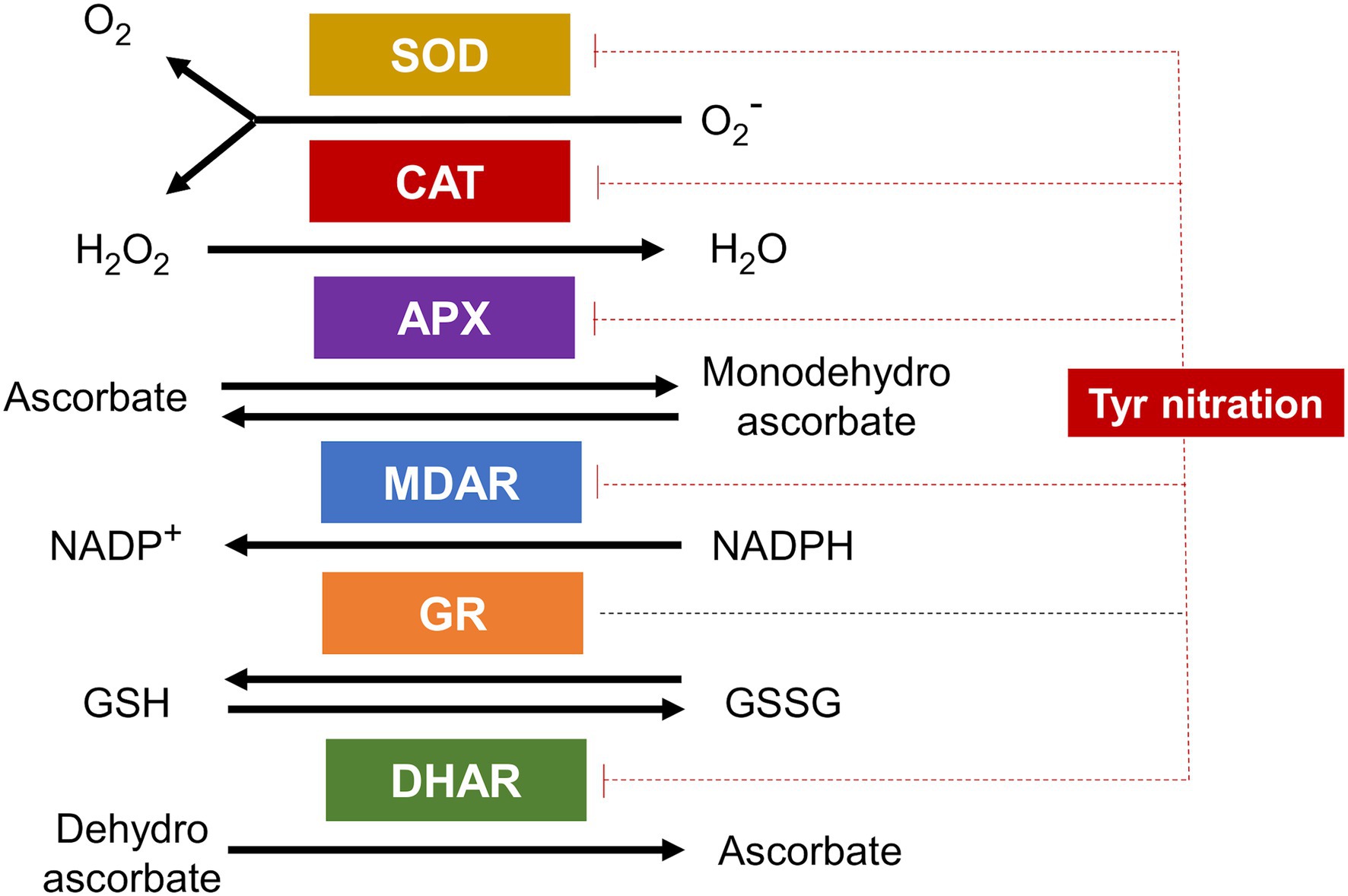
Figure 4. Antioxidant enzymes are targets of Tyr nitration. Superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), monodehydroascorbate reductase (MDAR), glutathione reductase (GR), and dehydroascorbate reductase (DHAR) are all targets of Tyr nitration and all but GR inactivated by this PTM (blunt ended red dotted lines).
A large overrepresentation of protein with organelle localization was identified in the in vivo nitrated proteome of Arabidopsis that identified 127 nitrated proteins (Lozano-Juste et al., 2011). Around 20, 5, and 3% of the identified nitrated proteins were chloroplastic, mitochondrial, and peroxisomal, respectively (Lozano-Juste et al., 2011). It has been reported that the Arabidopsis thylakoid proteome contains more than 100 nitrated proteins (Galetskiy et al., 2011a). Chloroplast metabolism is regulated by Tyr nitration among other PTMs in plastid proteins (Lehtimäki et al., 2015). Photosynthesis-related nitrated proteins involved in the Calvin-Benson cycle have been also identified in Citrus aurantium (Tanou et al., 2012). In sunflower and Arabidopsis, two key enzymes of photosynthetic carbon metabolism such as glyceraldehyde phosphate dehydrogenase and carbonic anhydrase were inactivated by nitration (Chaki et al., 2009; Lozano-Juste et al., 2011). In addition to central metabolism enzymes, several proteins of the photosystems have been also identified as targets of Tyr nitration (Galetskiy et al., 2011b), which are likely altered in the electron transfer chain and redox signaling. Mitochondria typically contain larger amounts of nitrated proteins with respect to other cellular compartments in different organisms. This is likely due to the high levels of NO and ROS production, thus acting as a continuous source of peroxynitrite (Radi et al., 2002). Peroxisomal enzymes such as serine hydroxymethyl transferase, catalase, alanine:glyoxylate aminotransferase, glycolate oxidase, and malate dehydrogenase have been also identified as nitrated proteins (Lozano-Juste et al., 2011; Sandalio et al., 2019). Some of the identified nitrated plant peroxisome enzymes are involved in reactive oxygen species metabolism thus altering, like above mentioned for chloroplastic proteins, redox signaling (Corpas et al., 2021).
Liquid chromatography followed by tandem mass spectrometry (LC–MS/MS) is likely the most used methodology for free and protein-bound 3-nitro-L-tyrosine analyses in biological samples (Larsen et al., 2006; Tsikas and Duncan, 2014). The overwhelming evidence suggesting that the ratio of nitrated vs. non-nitrated protein is usually extremely low likely makes protein nitration identification a very difficult task. It has been calculated that in human plasma the nitroTyr to unmodified Tyr ratio could be around one nitrated molecule per 1 million of non-nitrated molecules (Tsikas and Duncan, 2014). Therefore, the use of enrichment techniques is crucial for the success of subsequent MS-based analysis. For decades, the most widespread methodology for the study of protein nitration has been based on the immunoprecipitation (IP) of proteins with anti-3-nitroTyr antibodies followed by the subsequent identification by LC–MS/MS. These methods have been extensively used in mammals (Zhan and Desiderio, 2009) and plants (Chaki et al., 2009; Lozano-Juste et al., 2011). However, the low proportion of nitrated protein isoforms in vivotogether with the naturally unstable nature of this PTM made the identification of nitrated protein a challenging task. Because 3-nitro-Tyr are easily reduced to 3-amino-Tyr, MS-based methodologies has required working under strict non-reducing conditions. Unfortunately, it is difficult to ensure that non-reducing conditions are kept during sample manipulation and processing. An alternative methodology is based on the previous full reduction of 3-nitroTyr to 3-aminoTyr. High-Performance Liquid Chromatography (HPLC) technique coupled to electrochemical detection of the N-acetylated, dithionite-reduced derivative of 3-nitroTyr has been proposed for the analysis of nitrated proteins (Shigenaga, 1999). Based on the same principle also the specific enrichment of a targeted nitrotyrosine-containing peptides before MS has been proposed. This method starts by blocking all primary amines by acetylation, reduction of nitroTyr to aminoTyr with dithiothreitol and hemin, and affinity chromatography with an N-hydroxysuccinimide-ester-functionalized stationary phase (Yang, 2017). Another method, using IP with anti-3-nitroTyr followed by reduction of the immunoprecipitated proteins, ligation of biotin tags, streptavidin-based enrichment and final MS detection has been also used (Nikov et al., 2003). The direct MS-based analysis searching for mass shifts of +45 Da in nitrated peptides has been also used to identify nitrated proteins in vitro (Lozano-Juste et al., 2011; Medeiros et al., 2021). However, there is abundant evidence pointing to the need of having extreme caution when making assignments based on MS/MS spectra alone, especially when MS/MS resolution or mass accuracy are not good enough. It has been reported an automated method for the validation of endogenous Tyr nitration called rPTMDetermine that enhances the verification through similarity scoring of tandem MS/MS comparisons between modified peptides and their unmodified analogs (Dong et al., 2020). On the other hand, the low levels of nitrated proteins detected in vivo has been tried to overcome by exogenous treatment with nitrating agents that largely increase the number and quantity of detected nitrated proteins (Table 1). However, this approach does not allow the identification of proteins that are only nitrated under physiological conditions. Alternatively, genetic strategies based on the use of mutant plants with increased peroxynitrite content should also lead to enhanced levels of protein nitration (Corpas et al., 2009). However, the lack of specificity of nitrating agents such as peroxynitrite leads to the common identification of nitration sites that are not physiologically relevant. This drawback must be solved by using methodologies allowing the specific targeting of nitration events.
Likely the most used specific methodology to study the effects of PTMs on protein function has been the site-specific mutagenesis. In most of the cases, the strategies were based on substitution of amino acids for others that mimic a specific PTM. Ser and Thr change to Glu or Asp has been used to mimic phosphorylation. However, this approach just allows imitating the changes in charge and steric effects caused by PTMs, but it is often far from the real PTM. A potential alternative would be the chemical synthesis of PTM-related functional groups into the protein backbone, a procedure that allows site-specific modifications of a desired protein and afford the product in large quantities for biochemical and structural analyses (Siman and Brik, 2012). This technique has been used for glycosylation, phosphorylation, ubiquitination, acetylation, and lipidation (Siman and Brik, 2012), but to our knowledge nor for nitration of S-nitrosylation. In any case, this strategy relies on the stability of the modified protein and on the chemistry used for synthesis that commonly involves the thiol group of Cys residues, which is the target residue for S-nitrosylation. An alternative method is based on the capacity of bacteria to perform genetically encoded synthesis of proteins containing non-proteinogenic amino acids including 3-nitroTyr (Beyer et al., 2020; Jang et al., 2020; Chen and Tsai, 2021), therefore representing a valuable platform for studying specific protein nitration effects. These genetic code expansion methodologies allow the co-translational incorporation of non-canonical amino acids into proteins (Figure 5). The method requires de expression of an orthogonal aminoacyl-tRNA synthetase/t-RNA pair for the modified amino acid of interest. The resulting aminoacylated orthogonal tRNA is used by ribosomes to decode and amber stop codon at the position, where the non-proteinogenic amino acid should be incorporated (Nödling et al., 2019). This genetic code expansion technologies are useful for incorporating nitroTyr residues to proteins by using 3-nitroTyr tRNA synthetases (Cooley et al., 2014; Beyer et al., 2020). Most of the uses of orthogonal aminoacyl-tRNA synthetase/t-RNA pairs have been reported for mammal cells. By using this genetically encoding 3-nitro-tyrosine into the tyrosine nitration sites of human indoleamine 2,3-dioxygenase 1 and α-synuclein, the native nitrated proteins have been obtained and used for assessing the effects of nitration of different Tyr sites on the structure, function, and enzyme activity (Gerding et al., 2019; Zheng et al., 2020). Site-specific nitration of calmodulin at its two Tyr residues using genetic code expansion technology allowed assessing the effects of these alterations on calcium binding by calmodulin, and on the subsequent binding and activation of human endothelial NOS (eNOS; Porter et al., 2020). By substituting a key Tyr residue by nitroTyr using genetic code expansion technologies, the primary electron transfer in the reaction centers of the photosynthetic bacteria Rhodobacter sphaeroides has been studied and further engineered (Weaver et al., 2021). This genetically encoded synthesis of proteins containing 3-nitroTyr residues opens a potentially valuable strategy to assess the functional relevance of site-specific nitration on diverse target proteins, but to date no such application has been reported for any plant protein.
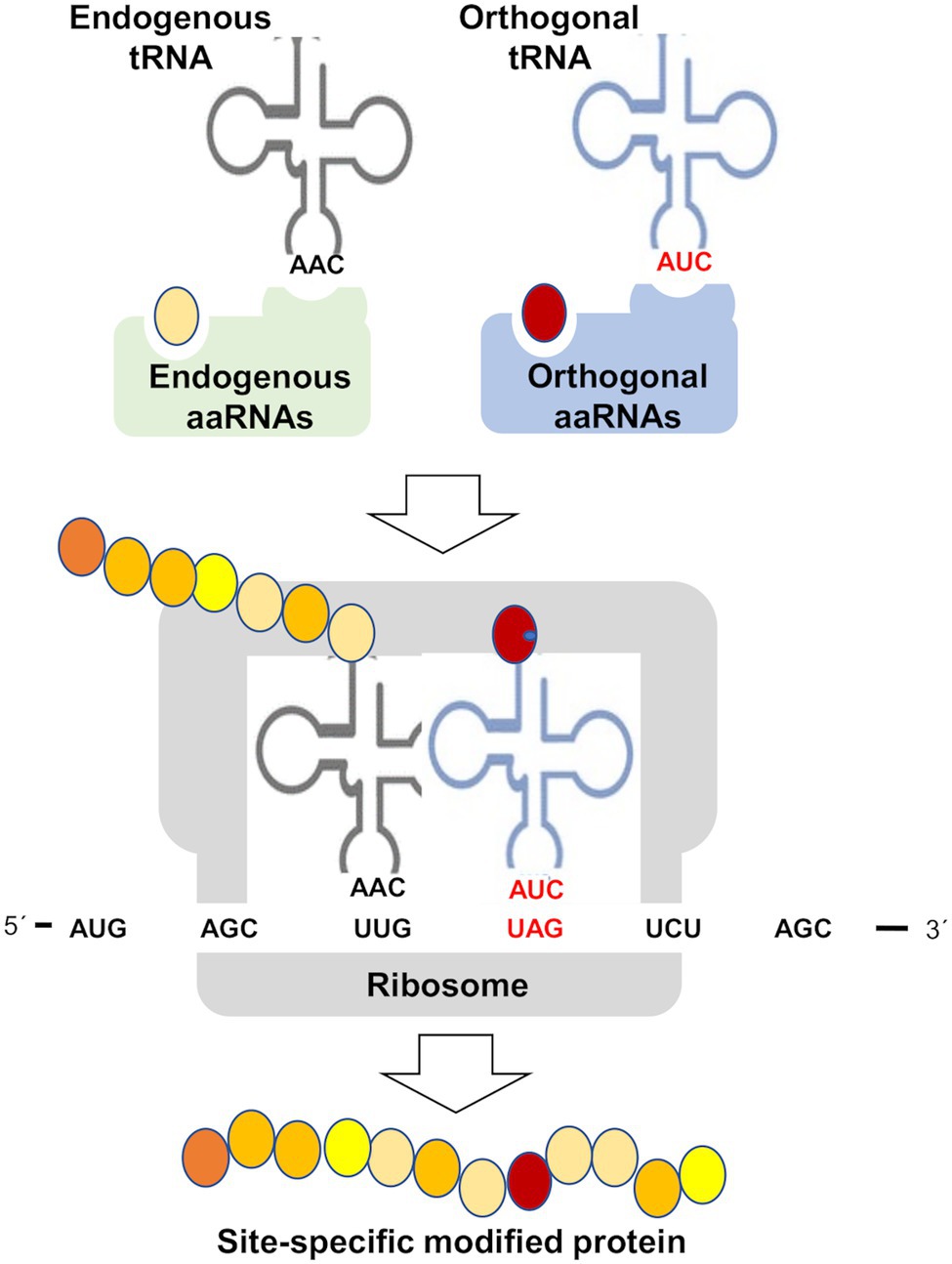
Figure 5. Genetic code expansion methodologies for site-specific incorporation of non-canonical amino acids. Aminoacyl-tRNA synthetase (aaRNAs), transfer RNA (tRNA), and amber stop codon (UAG). This methodology is based on the specificity of aaRNAs/tRNA pairs in such a way that non-canonical amino acid (red circle) and orthogonal tRNA are not substrates of endogenous aaRNAs and vice versa orthogonal aaRNAs do not use canonical amino acids (yellow and orange circles) and endogenous tRNAs. This method generates a site-specific modified protein by incorporation of a non-canonical amino acid, which may be 3-nitroTyr, thus becoming a potentially useful tool to study Tyr nitration or any other PTM specific effects on target proteins.
Besides MS-based methods, a wide array of chromatographic, immunochemical, and bio-sensing techniques has been reported to be useful for the identification and quantification of protein Tyr-nitration (recently reviewed in Bandookwala et al., 2020). After HPLC, nitrated proteins have been quantified also by UV detection (Yang et al., 2010); by electrochemical detection with sensitivity and selectivity compared to immunodetection methods (Vujacic-Mirski et al., 2020); by fluorescence detection after derivatization (Pourfarzam et al., 2013); and also by photodiode array detection (Teixeira et al., 2017). Size exclusion chromatography coupled to reverse phase-HPLC with diode array detection has been also developed to evaluate protein nitration (Liu et al., 2017). Nitrated protein identification has benefited also from other spectroscopic techniques. It has been recently reported the use of Surface enhanced Raman spectroscopy (SERS) to identify some 3-nitroTyr-containing neurodegenerative disease-related proteins that are converted to azobenzene containing peptides using a protocol based on silver nanoparticles stabilized by citrate (Niederhafner et al., 2021). Protein Tyr nitration has been also detected by electron paramagnetic resonance (EPR) spin trapping (Romero et al., 2003; Chen et al., 2004).
The identification of nitrated proteins and the involvement of this PTM on the regulation of diverse plant physiological processes has gained increasing interest in the last decades. However, much more work will be needed to fill the multiple gaps remain in our knowledge of the regulatory functions exerted by Tyr protein nitration in plants. The low proportion of nitrated peptides in the in vivo proteomic analyses together with its inherent instability make the identification of nitration sites a difficult task. The use of artificial nitration procedures is likely not the best strategy for nitration site identification due to the lack of specificity of nitrating systems. Therefore, more work must be performed on the application of genetic code expansion technologies based on the use of orthogonal aminoacyl-tRNA synthetase/tRNA pairs engineered for specific non-canonical amino acids. These methodologies permit the programmable site-specific installation of genetically encoded 3-NT sites in proteins expressed in Escherichia coli thus assessing the effects of specific nitration. These recombinant proteins containing PTMs will represent a very valuable tool for providing insights into how specific modifications regulate protein structure and function. Genetic code expansion methodologies will open not only the possibility to work with bacteria-expressed site-specific modified recombinant proteins in vitro, but also will allow to engineer plants by expressing the orthogonal aRNAs/tRNA pairs and supplying the non-canonical amino acid of interest, thus allowing the expression of specifically modified protein versions with agro-biotechnological advantages. However, this strategy will require significant advances on our knowledge of these processes and also to explore the feasibility of application for plants. In any case, a deeper knowledge on the function of Tyr nitration of plant proteins will certainly help to understand the functional relevance of this PTM not only in stress-related conditions, when this modification is more prevalent, but also under non-stress conditions, when nitrated targets may play or stop playing decisive regulatory roles on growth and development. Therefore, better known Tyr nitration targets and mechanisms should allow also a deeper knowledge on the signaling nodes involved in controlling plant physiology.
JL compiled and organized the reported information and wrote the article.
The work on hypoxia in the JL laboratory is supported by grants BIO2017-82945-P and PID2020-112618GBI00 funded by MCIN/AEI/10.13039/501100011033 and by the European Union.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The author acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
Abbas, M., Berckhan, S., Rooney, D. J., Gibbs, D. J., Conde, J. V., Correia, C. S., et al. (2015). Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25, 1483–1488. doi: 10.1016/j.cub.2015.03.060
Abello, N., Kerstjens, H. A., Postma, D. S., and Bischoff, R. (2009). Protein tyrosine nitration: selectivity, physicochemical and biological consequences, denitration, and proteomics methods for the identification of tyrosine-nitrated proteins. J. Proteome Res. 8, 3222–3238. doi: 10.1021/pr900039c
Adak, S., Aulak, K. S., and Stuehr, D. J. (2002). Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J. Biol. Chem. 277, 16167–16171. doi: 10.1074/jbc.M201136200
Alamillo, J. M., and García-Olmedo, F. (2001). Effects of urate, a natural inhibitor of peroxynitrite-mediated toxicity, in the response of Arabidopsis thaliana to the bacterial pathogen Pseudomonas syringae. Plant J. 25, 529–540. doi: 10.1046/j.1365-313x.2001.00984.x
Albertos, P., Romero-Puertas, M. C., Tatematsu, K., Mateos, I., Sánchez-Vicente, I., Nambara, E., et al. (2015). S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6:8669. doi: 10.1038/ncomms9669
Alvarez, C., Lozano-Juste, J., Romero, L. C., García, I., Gotor, C., and León, J. (2011). Inhibition of Arabidopsis O-acetylserine(thiol)lyase A1 by tyrosine nitration. J. Biol. Chem. 286, 578–586. doi: 10.1074/jbc.M110.147678
Anderson, G. P., Draheim, J. E., and Gross, E. L. (1985). Plastocyanin conformation: the effect of the oxidation state on the pKa of nitrotyrosine-83. Biochim. Biophys. Acta 810, 123–131. doi: 10.1016/0005-2728(85)90127-6
Arasimowicz-Jelonek, M., and Floryszak-Wieczorek, J. (2011). Understanding the fate of peroxynitrite in plant cells–from physiology to pathophysiology. Phytochemistry 72, 681–688. doi: 10.1016/j.phytochem.2011.02.025
Arasimowicz-Jelonek, M., and Floryszak-Wieczorek, J. (2019). A physiological perspective on targets of nitration in NO-based signaling networks in plants. J. Exp. Bot. 70, 4379–4389. doi: 10.1093/jxb/erz300
Arasimowicz-Jelonek, M., Floryszak-Wieczorek, J., Izbianska, K., Gzyl, J., and Jelonek, T. (2016). Implication of peroxynitrite in defense responses of potato to Phytophthora infestans. Plant Pathol. 65, 754–766. doi: 10.1111/ppa.12471
Arnold, W. P., Mittal, C. K., Katsuki, S., and Murad, F. (1977). Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. U. S. A. 74, 3203–3207. doi: 10.1073/pnas.74.8.3203
Aroca, A., Zhang, J., Xie, Y., Romero, L. C., and Gotor, C. (2021). Hydrogen sulfide signaling in plant adaptations to adverse conditions: molecular mechanisms. J. Exp. Bot. 72, 5893–5904. doi: 10.1093/jxb/erab239
Astier, J., Gross, I., and Durner, J. (2018). Nitric oxide production in plants: an update. J. Exp. Bot. 69, 3401–3411. doi: 10.1093/jxb/erx420
Astier, J., and Lindermayr, C. (2012). Nitric oxide-dependent posttranslational modification in plants: an update. Int. J. Mol. Sci. 13, 15193–15208. doi: 10.3390/ijms131115193
Astier, J., Mounier, A., Santolini, J., Jeandroz, S., and Wendehenne, D. (2019). The evolution of nitric oxide signalling diverges between animal and green lineages. J. Exp. Bot. 70, 4355–4364. doi: 10.1093/jxb/erz088
Bandookwala, M., Thakkar, D., and Sengupta, P. (2020). Advancements in the analytical quantification of nitroxidative stress biomarker 3-nitrotyrosine in biological matrices. Crit. Rev. Anal. Chem. 50, 265–289. doi: 10.1080/10408347.2019.1623010
Begara-Morales, J. C., Chaki, M., Sánchez-Calvo, B., Mata-Pérez, C., Leterrier, M., Palma, J. M., et al. (2013). Protein tyrosine nitration in pea roots during development and senescence. J. Exp. Bot. 64, 1121–1134. doi: 10.1093/jxb/ert006
Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., Mata-Pérez, C., Valderrama, R., Padilla, M. N., et al. (2015). Differential molecular response of monodehydroascorbate reductase and glutathione reductase by nitration and S-nitrosylation. J. Exp. Bot. 66, 5983–5996. doi: 10.1093/jxb/erv306
Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., Valderrama, R., Mata-Pérez, C., López-Jaramillo, J., et al. (2014). Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 65, 527–538. doi: 10.1093/jxb/ert396
Begara-Morales, J. C., Sánchez-Calvo, B., Chaki, M., Valderrama, R., Mata-Pérez, C., Padilla, M. N., et al. (2016). Antioxidant systems are regulated by nitric oxide-mediated post-translational modifications (NO-PTMs). Front. Plant Sci. 7:152. doi: 10.3389/fpls.2016.00152
Begara-Morales, J. C., Sánchez-Calvo, B., Gómez-Rodríguez, M. V., Chaki, M., Valderrama, R., Mata-Pérez, C., et al. (2019). Short-term low temperature induces nitro-oxidative stress that deregulates the NADP-malic enzyme function by tyrosine nitration in Arabidopsis thaliana. Antioxidants 8:448. doi: 10.3390/antiox8100448
Benhar, M., Forrester, M. T., and Stamler, J. S. (2009). Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732. doi: 10.1038/nrm2764
Beyer, J. N., Hosseinzadeh, P., Gottfried-Lee, I., Van Fossen, E. M., Zhu, P., Bednar, R. M., et al. (2020). Overcoming near-cognate suppression in a release factor 1-deficient host with an improved nitro-tyrosine tRNA synthetase. J. Mol. Biol. 432, 4690–4704. doi: 10.1016/j.jmb.2020.06.014
Bottari, S. P. (2015). Protein tyrosine nitration: a signaling mechanism conserved from yeast to man. Proteomics 15, 185–187. doi: 10.1002/pmic.201400592
Campbell, W. H., and Kinghorn, K. R. (1990). Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem. Sci. 15, 315–319. doi: 10.1016/0968-0004(90)90021-3
Castillo, M. C., Coego, A., Costa-Broseta, Á., and León, J. (2018). Nitric oxide responses in Arabidopsis hypocotyls are mediated by diverse phytohormone pathways. J. Exp. Bot. 69, 5265–5278. doi: 10.1093/jxb/ery286
Castillo, M. C., Lozano-Juste, J., González-Guzmán, M., Rodriguez, L., Rodriguez, P. L., and León, J. (2015). Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci. Signal. 8:ra89. doi: 10.1126/scisignal.aaa7981
Cecconi, D., Orzetti, S., Vandelle, E., Rinalducci, S., Zolla, L., and Delledonne, M. (2009). Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 30, 2460–2468. doi: 10.1002/elps.200800826
Chaki, M., de Morales, P. Á., Ruiz, C., Begara-Morales, J. C., Barroso, J. B., Corpas, F. J., et al. (2015). Ripening of pepper (Capsicum annuum) fruit is characterized by an enhancement of protein tyrosine nitration. Ann. Bot. 116, 637–647. doi: 10.1093/aob/mcv016
Chaki, M., Valderrama, R., Fernández-Ocaña, A. M., Carreras, A., López-Jaramillo, J., Luque, F., et al. (2009). Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J. Exp. Bot. 60, 4221–4234. doi: 10.1093/jxb/erp263
Chamizo-Ampudia, A., Sanz-Luque, E., Llamas, A., Galvan, A., and Fernandez, E. (2017). Nitrate reductase regulates plant nitric oxide homeostasis. Trends Plant Sci. 22, 163–174. doi: 10.1016/j.tplants.2016.12.001
Chen, Y. R., Chen, C. L., Chen, W., Zweier, J. L., Augusto, O., Radi, R., et al. (2004). Formation of protein tyrosine ortho-semiquinone radical and nitrotyrosine from cytochrome c-derived tyrosyl radical. J. Biol. Chem. 279, 18054–18062. doi: 10.1074/jbc.M307706200
Chen, J., and Tsai, Y. H. (2021). Applications of genetic code expansion in studying protein post-translational modification. J. Mol. Biol. 28:167424. doi: 10.1016/j.jmb.2021.167424
Chen, S., Zhao, H., Wang, M., Li, J., Wang, Z., Wang, F., et al. (2017). Overexpression of E3 ubiquitin ligase gene AdBiL contributes to resistance against chilling stress and leaf mold disease in tomato. Front. Plant Sci. 8:1109. doi: 10.3389/fpls.2017.01109
Choudhury, F. K., Rivero, R. M., Blumwald, E., and Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Cooley, R. B., Feldman, J. L., Driggers, C. M., Bundy, T. A., Stokes, A. L., Karplus, P. A., et al. (2014). Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry 53, 1916–1924. doi: 10.1021/bi5001239
Corpas, F. J., González-Gordo, S., and Palma, J. M. (2021). Nitric oxide (NO) scaffolds the peroxisomal protein-protein interaction network in higher plants. Int. J. Mol. Sci. 22:2444. doi: 10.3390/ijms22052444
Corpas, F. J., Hayashi, M., Mano, S., Nishimura, M., and Barroso, J. B. (2009). Peroxisomes are required for in vivo nitric oxide accumulation in the cytosol following salinity stress of Arabidopsis plants. Plant Physiol. 151, 2083–2094. doi: 10.1104/pp.109.146100
Corpas, F. J., Leterrier, M., Begara-Morales, J. C., Valderrama, R., Chaki, M., López-Jaramillo, J., et al. (2013). Inhibition of peroxisomal hydroxypyruvate reductase (HPR1) by tyrosine nitration. Biochim. Biophys. Acta 1830, 4981–4989. doi: 10.1016/j.bbagen.2013.07.002
Costa-Broseta, Á., Castillo, M., and León, J. (2020). Nitrite reductase 1 is a target of nitric oxide-mediated post-translational modifications and controls nitrogen flux and growth in Arabidopsis. Int. J. Mol. Sci. 21:7270. doi: 10.3390/ijms21197270
Costa-Broseta, Á., Castillo, M., and León, J. (2021). Post-translational modifications of nitrate reductases autoregulates nitric oxide biosynthesis in Arabidopsis. Int. J. Mol. Sci. 22:549. doi: 10.3390/ijms22020549
Crawford, N. M., and Glass, A. D. (1998). Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. doi: 10.1016/S1360-1385(98)01311-9
De Gara, L., and Foyer, C. H. (2017). Ying and Yang interplay between reactive oxygen and reactive nitrogen species controls cell functions. Plant Cell Environ. 40, 459–461. doi: 10.1111/pce.12936
Deeb, R. S., Nuriel, T., Cheung, C., Summers, B., Lamon, B. D., Gross, S. S., et al. (2013). Characterization of a cellular denitrase activity that reverses nitration of cyclooxygenase. Am. J. Physiol. Heart Circ. Physiol. 305, H687–H698. doi: 10.1152/ajpheart.00876.2012
Del Río, L. A. (2015). ROS and RNS in plant physiology: an overview. J. Exp. Bot. 66, 2827–2837. doi: 10.1093/jxb/erv099
DiCola, D., Polidoro, G., Di Ilio, G., Del Boccia, C., Politi, L., and Scandurra, R. (1976). Role of tyrosine residues in cytoplasmic aspartate aminotransferase from beef kidney. Mol. Cell. Biochem. 11, 97–101. doi: 10.1007/BF01792790
Dong, N., Spencer, D. M., Quan, Q., Le Blanc, J. C. Y., Feng, J., Li, M., et al. (2020). rPTMDetermine: a fully automated methodology for endogenous tyrosine nitration validation, site-localization, and beyond. Anal. Chem. 92, 10768–10776. doi: 10.1021/acs.analchem.0c02148
Drazic, A., and Winter, J. (2014). The physiological role of reversible methionine oxidation. Biochim. Biophys. Acta 1844, 1367–1382. doi: 10.1016/j.bbapap.2014.01.001
Feng, J., Chen, L., and Zuo, J. (2019). Protein S-nitrosylation in plants: current progresses and challenges. J. Integr. Plant Biol. 61, 1206–1223. doi: 10.1111/jipb.12780
Ferrer-Sueta, G., Campolo, N., Trujillo, M., Bartesaghi, S., Carballal, S., Romero, N., et al. (2018). Biochemistry of peroxynitrite and protein tyrosine nitration. Chem. Rev. 118, 1338–1408. doi: 10.1021/acs.chemrev.7b00568
Filippovich, S. Y. (2010). Bacterial NO synthases. Biochemistry 75, 1217–1224. doi: 10.1134/s0006297910100032
Foresi, N., Correa-Aragunde, N., Parisi, G., Caló, G., Salerno, G., and Lamattina, L. (2010). Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22, 3816–3830. doi: 10.1105/tpc.109.073510
Foresi, N., Mayta, M. L., Lodeyro, A. F., Scuffi, D., Correa-Aragunde, N., García-Mata, C., et al. (2015). Expression of the tetrahydrofolate-dependent nitric oxide synthase from the green alga Ostreococcus tauri increases tolerance to abiotic stresses and influences stomatal development in Arabidopsis. Plant J. 82, 806–821. doi: 10.1111/tpj.12852
Förstermann, U., and Sessa, W. C. (2012). Nitric oxide synthases: regulation and function. Eur. Heart J. 33, 829–837. doi: 10.1093/eurheartj/ehr304
Frieg, B., Görg, B., Gohlke, H., and Häussinger, D. (2021). Glutamine synthetase as a central element in hepatic glutamine and ammonia metabolism: novel aspects. Biol. Chem. 402, 1063–1072. doi: 10.1515/hsz-2021-0166
Galetskiy, D., Lohscheider, J. N., Kononikhin, A. S., Popov, I. A., Nikolaev, E. N., and Adamska, I. (2011a). Mass spectrometric characterization of photooxidative protein modifications in Arabidopsis thaliana thylakoid membranes. Rapid Commun. Mass Spectrom. 25, 184–190. doi: 10.1002/rcm.4855
Galetskiy, D., Lohscheider, J. N., Kononikhin, A. S., Popov, I. A., Nikolaev, E. N., and Adamska, I. (2011b). Phosphorylation and nitration levels of photosynthetic proteins are conversely regulated by light stress. Plant Mol. Biol. 77, 461–473. doi: 10.1007/s11103-011-9824-7
Gao, S., Zeng, X., Wang, J., Xu, Y., Yu, C., Huang, Y., et al. (2021). Arabidopsis SUMO E3 ligase SIZ1 interacts with HDA6 and negatively regulates HDA6 function during flowering. Cell 10:3001. doi: 10.3390/cells10113001
Gasch, P., Fundinger, M., Müller, J. T., Lee, T., Bailey-Serres, J., and Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180. doi: 10.1105/tpc.15.00866
Gerding, H. R., Karreman, C., Daiber, A., Delp, J., Hammler, D., Mex, M., et al. (2019). Reductive modification of genetically encoded 3-nitrotyrosine sites in alpha synuclein expressed in E. coli. Redox Biol. 26:101251. doi: 10.1016/j.redox.2019.101251
Gibbs, D. J., Isa, N., Movahedi, M., Lozano-Juste, J., Mendiondo, G. M., Berckhan, S., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53, 369–379. doi: 10.1016/j.molcel.2013.12.020
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Görg, B., Qvartskhava, N., Voss, P., Grune, T., Häussinger, D., and Schliess, F. (2007). Reversible inhibition of mammalian glutamine synthetase by tyrosine nitration. FEBS Lett. 581, 84–90. doi: 10.1016/j.febslet.2006.11.081
Gou, M., Huang, Q., Qian, W., Zhang, Z., Jia, Z., and Hua, J. (2017). Sumoylation E3 ligase SIZ1 modulates plant immunity partly through the immune receptor gene SNC1 in Arabidopsis. Mol. Plant-Microbe Interact. 30, 334–342. doi: 10.1094/MPMI-02-17-0041-R
Gow, A. J., Duran, D., Malcolm, S., and Ischiropoulos, H. (1996). Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 385, 63–66. doi: 10.1016/0014-5793(96)00347-X
Gupta, K. J., Kolbert, Z., Durner, J., Lindermayr, C., Corpas, F. J., Brouquisse, R., et al. (2020). Regulating the regulator: nitric oxide control of post-translational modifications. New Phytol. 227, 1319–1325. doi: 10.1111/nph.16622
Gusenkov, S., and Stutz, H. (2018). Top-down and bottom-up characterization of nitrated birch pollen allergen bet v 1a with CZE hyphenated to an Orbitrap mass spectrometer. Electrophoresis 39, 1190–1200. doi: 10.1002/elps.201700413
Hachiya, T., and Sakakibara, H. (2017). Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 68, 2501–2512. doi: 10.1093/jxb/erw449
Hancock, J. T. (2019). Hydrogen sulfide and environmental stresses. Environ. Exp. Bot. 161, 50–56. doi: 10.1016/j.envexpbot.2018.08.034
Hancock, J. T. (2020). Nitric oxide signaling in plants. Plan. Theory 9:1550. doi: 10.3390/plants9111550
Holzmeister, C., Gaupels, F., Geerlof, A., Sarioglu, H., Sattler, M., Durner, J., et al. (2015). Differential inhibition of Arabidopsis superoxide dismutases by peroxynitrite-mediated tyrosine nitration. J. Exp. Bot. 66, 989–999. doi: 10.1093/jxb/eru458
Ishii, N., Carmines, P. K., Yokoba, M., Imaizumi, H., Ichikawa, T., Ikenagasa, H., et al. (2013). Angiotensin-converting enzyme inhibition curbs tyrosine nitration of mitochondrial proteins in the renal cortex during the early stage of diabetes mellitus in rats. Clin. Sci. 124, 543–552. doi: 10.1042/CS20120251
Jang, H. S., Gu, X., Cooley, R. B., Porter, J. J., Henson, R. L., Willi, T., et al. (2020). Efficient site-specific prokaryotic and eukaryotic incorporation of halotyrosine amino acids into proteins. ACS Chem. Biol. 15, 562–574. doi: 10.1021/acschembio.9b01026
Jeandroz, S., Wipf, D., Stuehr, D. J., Lamattina, L., Melkonian, M., Tian, Z., et al. (2016). Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal. 9:re2. doi: 10.1126/scisignal.aad4403
Jedelská, T., Luhová, L., and Petřivalský, M. (2021). Nitric oxide signalling in plant interactions with pathogenic fungi and oomycetes. J. Exp. Bot. 72, 848–863. doi: 10.1093/jxb/eraa596
Jones, L. H. (2012). Chemistry and biology of biomolecule nitration. Chem. Biol. 19, 1086–1092. doi: 10.1016/j.chembiol.2012.07.019
Joshi, M. S., Mihm, M. J., Cook, A. C., Schanbacher, B. L., and Bauer, J. A. (2015). Alterations in connexin 43 during diabetic cardiomyopathy: competition of tyrosine nitration versus phosphorylation. J. Diabetes 7, 250–259. doi: 10.1111/1753-0407.12164
Kang, M., and Akbarali, H. I. (2008). Denitration of L-type calcium channel. FEBS Lett. 582, 3033–3036. doi: 10.1016/j.febslet.2008.07.042
Kim, J. Y., Park, B. S., Park, S. W., Lee, H. Y., Song, J. T., and Seo, H. S. (2018). Nitrate reductases are relocalized to the nucleus by AtSIZ1 and their levels are negatively regulated by COP1 and ammonium. Int. J. Mol. Sci. 19:1202. doi: 10.3390/ijms19041202
Kishorekumar, R., Bulle, M., Wany, A., and Gupta, K. J. (2020). An overview of important enzymes involved in nitrogen assimilation of plants. Methods Mol. Biol. 2057, 1–13. doi: 10.1007/978-1-4939-9790-9_1
Kolbert, Z., Barroso, J. B., Brouquisse, R., Corpas, F. J., Gupta, K. J., Lindermayr, C., et al. (2019). A forty year journey: the generation and roles of NO in plants. Nitric Oxide 93, 53–70. doi: 10.1016/j.niox.2019.09.006
Kolbert, Z., Feigl, G., Bordé, Á., Molnár, Á., and Erdei, L. (2017). Protein tyrosine nitration in plants: present knowledge, computational prediction and future perspectives. Plant Physiol. Biochem. 113, 56–63. doi: 10.1016/j.plaphy.2017.01.028
Kolbert, Z., and Lindermayr, C. (2021). Computational prediction of NO-dependent posttranslational modifications in plants: current status and perspectives. Plant Physiol. Biochem. 167, 851–861. doi: 10.1016/j.plaphy.2021.09.011
Kong, S. K., Yim, M. B., Stadtman, E. R., and Chock, P. B. (1996). Peroxynitrite disables the tyrosine phosphorylation regulatory mechanism: lymphocyte-specific tyrosine kinase fails to phosphorylate nitrated cdc2(6-20)NH2 peptide. Proc. Natl. Acad. Sci. U. S. A. 93, 3377–3382. doi: 10.1073/pnas.93.8.3377
Krapp, A. (2015). Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr. Opin. Plant Biol. 25, 115–122. doi: 10.1016/j.pbi.2015.05.010
Kuo, W. N., Kanadia, R. N., and Mcnabb, M. (1998). Soluble guanylate cyclase in Saccharomyces cerevisiae. Biochem. Mol. Biol. Int. 45, 125–131. doi: 10.1080/15216549800202492
Kuo, W. N., Kanadia, R. N., Shanbhag, V. P., and Toro, R. (1999). Denitration of peroxynitrite-treated proteins by ‘protein nitratases’ from rat brain and heart. Mol. Cell. Biochem. 201, 11–16. doi: 10.1023/A:1007024126947
Labudda, M., Różańska, E., Gietler, M., Fidler, J., Muszyńska, E., Prabucka, B., et al. (2020). Cyst nematode infection elicits alteration in the level of reactive nitrogen species, protein S-nitrosylation and nitration, and nitrosoglutathione reductase in Arabidopsis thaliana roots. Antioxidants 9:795. doi: 10.3390/antiox9090795
Larsen, M. R., Trelle, M. B., Thingholm, T. E., and Jensen, O. N. (2006). Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques 40, 790–798. doi: 10.2144/000112201
Lee, C. M., Wilderman, P. R., Park, J. W., Murphy, T. J., and Morgan, E. T. (2020). Tyrosine nitration contributes to nitric oxide-stimulated degradation of CYP2B6. Mol. Pharmacol. 98, 267–279. doi: 10.1124/molpharm.120.000020
Lehtimäki, N., Koskela, M. M., and Mulo, P. (2015). Posttranslational modifications of chloroplast proteins: an emerging field. Plant Physiol. 168, 768–775. doi: 10.1104/pp.15.00117
León, J., and Costa-Broseta, Á. (2020). Present knowledge and controversies, deficiencies, and misconceptions on nitric oxide synthesis, sensing, and signaling in plants. Plant Cell Environ. 43, 1–15. doi: 10.1111/pce.13617
León, J., Costa-Broseta, Á., and Castillo, M. C. (2020). RAP2.3 negatively regulates nitric oxide biosynthesis and related responses through a rheostat-like mechanism in Arabidopsis. J. Exp. Bot. 71, 3157–3171. doi: 10.1093/jxb/eraa069
Liaudet, L., Vassalli, G., and Pacher, P. (2009). Role of peroxynitrite in the redox regulation of cell signal transduction pathways. Front. Biosci. 14, 4809–4814. doi: 10.2741/3569
Lillo, C., Meyer, C., Lea, U. S., Provan, F., and Oltedal, S. (2004). Mechanism and importance of post-translational regulation of nitrate reductase. J. Exp. Bot. 55, 1275–1282. doi: 10.1093/jxb/erh132
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Liu, F., Reinmuth-Selzle, K., Lai, S., Weller, M. G., Pöschl, U., and Kampf, C. J. (2017). Simultaneous determination of nitrated and oligomerized proteins by size exclusion high-performance liquid chromatography coupled to photodiode array detection. J. Chromatogr. A 1495, 76–82. doi: 10.1016/j.chroma.2017.03.015
Liu, C., Yu, H., and Li, L. (2019). SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet. 15:e1007928. doi: 10.1371/journal.pgen.1007928
Loreti, E., van Veen, H., and Perata, P. (2016). Plant responses to flooding stress. Curr. Opin. Plant Biol. 33, 64–71. doi: 10.1016/j.pbi.2016.06.005
Lozano-Juste, J., Colom-Moreno, R., and León, J. (2011). In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 62, 3501–3517. doi: 10.1093/jxb/err042
Lozano-Juste, J., and León, J. (2010). Enhanced abscisic acid-mediated responses in nia1nia2noa1-2 triple mutant impaired in NIA/NR- and AtNOA1-dependent nitric oxide biosynthesis in Arabidopsis. Plant Physiol. 152, 891–903. doi: 10.1104/pp.109.148023
Maia, L. B., and Moura, J. J. (2015). Nitrite reduction by molybdoenzymes: a new class of nitric oxide-forming nitrite reductases. J. Biol. Inorg. Chem. 20, 403–433. doi: 10.1007/s00775-014-1234-2
Malik, S. I., Hussain, A., Yun, B. W., Spoel, S. H., and Loake, G. J. (2011). GSNOR-mediated de-nitrosylation in the plant defence response. Plant Sci. 181, 540–544. doi: 10.1016/j.plantsci.2011.04.004
Marletta, M. A. (1993). Nitric oxide synthase: function and mechanism. Adv. Exp. Med. Biol. 338, 281–284.
Medeiros, R., Sousa, B., Rossi, S., Afonso, C., Bonino, L., Pitt, A., et al. (2021). Identification and relative quantification of 3-nitrotyrosine residues in fibrinogen nitrated in vitro and fibrinogen from ischemic stroke patient plasma using LC-MS/MS. Free Radic. Biol. Med. 165, 334–347. doi: 10.1016/j.freeradbiomed.2021.01.049
Melo, P. M., Silva, L. S., Ribeiro, I., Seabra, A. R., and Carvalho, H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. doi: 10.1104/pp.111.186056
Méndez, A. A. E., Mangialavori, I. C., Cabrera, A. V., Benavides, M. P., Vázquez-Ramos, J. M., and Gallego, S. M. (2020). Tyr-nitration in maize CDKA;1 results in lower affinity for ATP binding. Biochim. Biophys. Acta, Proteins Proteomics 1868:140479. doi: 10.1016/j.bbapap.2020.140479
Miura, K., Lee, J., Jin, J. B., Yoo, C. Y., Miura, T., and Hasegawa, P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U. S. A. 106, 5418–5423. doi: 10.1073/pnas.0811088106
Miura, K., Lee, J., Miura, T., and Hasegawa, P. M. (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51, 103–113. doi: 10.1093/pcp/pcp171
Moncada, S. (1993). The L-arginine: nitric oxide pathway, cellular transduction and immunological roles. Adv. Second Messenger Phosphoprotein Res. 28, 97–99.
Monteiro, H. P. (2002). Signal transduction by protein tyrosine nitration: competition or cooperation with tyrosine phosphorylation-dependent signaling events? Free Radic. Biol. Med. 33, 765–773. doi: 10.1016/s0891-5849(02)00893-6
Neill, S. J., Desikan, R., Clarke, A., Hurst, R. D., and Hancock, J. T. (2002). Hydrogen peroxide and nitric oxide as signalling molecules in plants. J. Exp. Bot. 53, 1237–1247. doi: 10.1093/jexbot/53.372.1237
Niederhafner, P., Šafařík, M., Neburková, J., Keiderling, T. A., Bouř, P., and Šebestík, J. (2021). Monitoring peptide tyrosine nitration by spectroscopic methods. Amino Acids 53, 517–532. doi: 10.1007/s00726-020-02911-7
Nikov, G., Bhat, V., Wishnok, J. S., and Tannenbaum, S. R. (2003). Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal. Biochem. 320, 214–222. doi: 10.1016/S0003-2697(03)00359-2
Nilamyani, A. N., Auliah, F. N., Moni, M. A., Shoombuatong, W., Hasan, M. M., and Kurata, H. (2021). PredNTS: improved and robust prediction of nitrotyrosine sites by integrating multiple sequence features. Int. J. Mol. Sci. 22:2704. doi: 10.3390/ijms22052704
Niu, D., Lin, X. L., Kong, X., Qu, G. P., Cai, B., Lee, J., et al. (2019). SIZ1-mediated SUMOylation of TPR1 suppresses plant immunity in Arabidopsis. Mol. Plant 12, 215–228. doi: 10.1016/j.molp.2018.12.002
Noctor, G., and Foyer, C. H. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Nödling, A. R., Spear, L. A., Williams, T. L., Luk, L. Y. P., and Tsai, Y. H. (2019). Using genetically incorporated unnatural amino acids to control protein functions in mammalian cells. Essays Biochem. 63, 237–266. doi: 10.1042/EBC20180042
Ott, C., Tomasina, F., Campolo, N., Bartesaghi, S., Mastrogiovanni, M., Leyva, A., et al. (2021). Decreased proteasomal cleavage at nitrotyrosine sites in proteins and peptides. Redox Biol. 46:102106. doi: 10.1016/j.redox.2021.102106
Palma, J. M., Mateos, R. M., López-Jaramillo, J., Rodríguez-Ruiz, M., González-Gordo, S., Lechuga-Sancho, A. M., et al. (2020). Plant catalases as NO and H(2)S targets. Redox Biol. 34:101525. doi: 10.1016/j.redox.2020.101525
Palmer, R. M. (1993). The L-arginine: nitric oxide pathway. Curr. Opin. Nephrol. Hypertens. 2, 122–128. doi: 10.1097/00041552-199301000-00018
Paul, M. V., Iyer, S., Amerhauser, C., Lehmann, M., van Dongen, J. T., and Geigenberger, P. (2016). Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 172, 141–153. doi: 10.1104/pp.16.00460
Porter, J. J., Jang, H. S., Haque, M. M., Stuehr, D. J., and Mehl, R. A. (2020). Tyrosine nitration on calmodulin enhances calcium-dependent association and activation of nitric-oxide synthase. J. Biol. Chem. 295, 2203–2211. doi: 10.1074/jbc.RA119.010999
Pourfarzam, M., Movahedian, A., Sarrafzadegan, N., Basati, G., and Samsamshariat, S. Z. (2013). Association between plasma myeloperoxidase and free 3-nitrotyrosine levels in patients with coronary artery disease. Int. J. Clin. Med. 4, 158–164. doi: 10.4236/ijcm.2013.43028
Puerta, M. L., Shukla, V., Carbonare, L. D., Weits, D. A., Perata, P., Licausi, F., et al. (2019). A ratiometric sensor based on plant n-terminal degrons able to report oxygen dynamics in Saccharomyces cerevisiae. J. Mol. Biol. 431, 2810–2820. doi: 10.1016/j.jmb.2019.05.023
Radi, R., Cassina, A., Hodara, R., Quijano, C., and Castro, L. (2002). Peroxynitrite reactions and formation in mitochondria. Free Radic. Biol. Med. 33, 1451–1464. doi: 10.1016/S0891-5849(02)01111-5
Reinmuth-Selzle, K., Ackaert, C., Kampf, C. J., Samonig, M., Shiraiwa, M., Kofler, S., et al. (2014). Nitration of the birch pollen allergen bet v 1.0101: efficiency and site-selectivity of liquid and gaseous nitrating agents. J. Proteome Res. 13, 1570–1577. doi: 10.1021/pr401078h
Riedel, J., Tischner, R., and Mäck, G. (2001). The chloroplastic glutamine synthetase (GS-2) of tobacco is phosphorylated and associated with 14-3-3 proteins inside the chloroplast. Planta 213, 396–401. doi: 10.1007/s004250000509
Robinson, S. A., Slade, A. P., Fox, G. G., Phillips, R., Ratcliffe, R. G., and Stewart, G. R. (1991). The role of glutamate dehydrogenase in plant nitrogen metabolism. Plant Physiol. 95, 509–516. doi: 10.1104/pp.95.2.509
Romero, N., Radi, R., Linares, E., Augusto, O., Detweiler, C. D., Mason, R. P., et al. (2003). Reaction of human hemoglobin with peroxynitrite. Isomerization to nitrate and secondary formation of protein radicals. J. Biol. Chem. 278, 44049–44057. doi: 10.1074/jbc.M305895200
Sabadashka, M., Nagalievska, M., and Sybirna, N. (2021). Tyrosine nitration as a key event of signal transduction that regulates functional state of the cell. Cell Biol. Int. 45, 481–497. doi: 10.1002/cbin.11301
Sainz, M., Calvo-Begueria, L., Pérez-Rontomé, C., Wienkoop, S., Abián, J., Staudinger, C., et al. (2015). Leghemoglobin is nitrated in functional legume nodules in a tyrosine residue within the heme cavity by a nitrite/peroxide-dependent mechanism. Plant J. 81, 723–735. doi: 10.1111/tpj.12762
Saito, S., Yamamoto-Katou, A., Yoshioka, H., Doke, N., and Kawakita, K. (2006). Peroxynitrite generation and tyrosine nitration in defense responses in tobacco BY-2 cells. Plant Cell Physiol. 47, 689–697. doi: 10.1093/pcp/pcj038
Sandalio, L. M., Gotor, C., Romero, L. C., and Romero-Puertas, M. C. (2019). Multilevel regulation of peroxisomal proteome by post-translational modifications. Int. J. Mol. Sci. 20:4881. doi: 10.3390/ijms20194881
Scandurra, R., Polidoro, G., Di Cola, D., Politi, L., and Riordan, J. F. (1975). Role of tyrosine residues in mitochondrial aspartate aminotransferase from beef kidney. Biochemistry 14, 3701–3706. doi: 10.1021/bi00687a029
Sengupta, R., and Holmgren, A. (2013). Thioredoxin and thioredoxin reductase in relation to reversible S-nitrosylation. Antioxid. Redox Signal. 18, 259–269. doi: 10.1089/ars.2012.4716
Shigenaga, M. K. (1999). Quantitation of protein-bound 3-nitrotyrosine by high-performance liquid chromatography with electrochemical detection. Methods Enzymol. 301, 27–40. doi: 10.1016/s0076-6879(99)01066-6
Sies, H., Sharov, V. S., Klotz, L. O., and Briviba, K. (1997). Glutathione peroxidase protects against peroxynitrite-mediated oxidations. A new function for selenoproteins as peroxynitrite reductase. J. Biol. Chem. 272, 27812–27817. doi: 10.1074/jbc.272.44.27812
Siman, P., and Brik, A. (2012). Chemical and semisynthesis of posttranslationally modified proteins. Org. Biomol. Chem. 10, 5684–5697. doi: 10.1039/c2ob25149c
Souza, J. M., Choi, I., Chen, Q., Weisse, M., Daikhin, E., Yudkoff, M., et al. (2000). Proteolytic degradation of tyrosine nitrated proteins. Arch. Biochem. Biophys. 380, 360–366. doi: 10.1006/abbi.2000.1940
Sudhamsu, J., and Crane, B. R. (2009). Bacterial nitric oxide synthases: what are they good for? Trends Microbiol. 17, 212–218. doi: 10.1016/j.tim.2009.02.003
Takahashi, M., Shigeto, J., Izumi, S., Yoshizato, K., and Morikawa, H. (2016). Nitration is exclusive to defense-related PR-1, PR-3 and PR-5 proteins in tobacco leaves. Plant Signal. Behav. 11:e1197464. doi: 10.1080/15592324.2016.1197464
Takahashi, M., Shigeto, J., Sakamoto, A., Izumi, S., Asada, K., and Morikawa, H. (2015). Dual selective nitration in Arabidopsis: almost exclusive nitration of PsbO and PsbP, and highly susceptible nitration of four non-PSII proteins, including peroxiredoxin II E. Electrophoresis 36, 2569–2578. doi: 10.1002/elps.201500145
Takakura, K., Beckman, J. S., MacMillan-Crow, L. A., and Crow, J. P. (1999). Rapid and irreversible inactivation of protein tyrosine phosphatases PTP1B, CD45, and LAR by peroxynitrite. Arch. Biochem. Biophys. 369, 197–207. doi: 10.1006/abbi.1999.1374
Tanou, G., Filippou, P., Belghazi, M., Job, D., Diamantidis, G., Fotopoulos, V., et al. (2012). Oxidative and nitrosative-based signaling and associated post-translational modifications orchestrate the acclimation of citrus plants to salinity stress. Plant J. 72, 585–599. doi: 10.1111/j.1365-313X.2012.05100.x
Teixeira, D., Prudencio, C., and Vieira, M. (2017). Development of a new HPLC-based method for 3-nitrotyrosine quantification in different biological matrices. J. Chromatogr. B 1046, 48–57. doi: 10.1016/j.jchromb.2017.01.035
Tola, A. J., Jaballi, A., and Missihoun, T. D. (2021). Protein carbonylation: emerging roles in plant redox biology and future prospects. Plan. Theory 10:1451. doi: 10.3390/plants10071451
Torreilles, J. (2001). Nitric oxide: one of the more conserved and widespread signaling molecules. Front. Biosci. 6, D1161–D1172. doi: 10.2741/torreill
Tsikas, D., and Duncan, M. W. (2014). Mass spectrometry and 3-nitrotyrosine: strategies, controversies, and our current perspective. Mass Spectrom. Rev. 33, 237–276. doi: 10.1002/mas.21396
Vandelle, E., and Delledonne, M. (2011). Peroxynitrite formation and function in plants. Plant Sci. 181, 534–539. doi: 10.1016/j.plantsci.2011.05.002
Vicente, J., Mendiondo, G. M., Movahedi, M., Peirats-Llobet, M., Juan, Y. T., Shen, Y. Y., et al. (2017). The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27, 3183–3190. doi: 10.1016/j.cub.2017.09.006
Vicente, J., Mendiondo, G. M., Pauwels, J., Pastor, V., Izquierdo, Y., Naumann, C., et al. (2019). Distinct branches of the N-end rule pathway modulate the plant immune response. New Phytol. 221, 988–1000. doi: 10.1111/nph.15387
Vujacic-Mirski, K., Bruns, K., Kalinovic, S., Oelze, M., Kröller-Schön, S., Steven, S., et al. (2020). Development of an analytical assay for electrochemical detection and quantification of protein-bound 3-nitrotyrosine in biological samples and comparison with classical, antibody-based methods. Antioxidants 9:388. doi: 10.3390/antiox9050388
Walsh, C. K., and Sadanandom, A. (2014). Ubiquitin chain topology in plant cell signaling: a new facet to an evergreen story. Front. Plant Sci. 5:122. doi: 10.3389/fpls.2014.00122
Wang, Y., Li, G., Chen, T., and Tian, S. (2021). Protein sulfenylation contributes to oxidative burst-triggered responses during the interaction between Botrytis cinerea and Nicotiana benthamiana. J. Proteome 251:104423. doi: 10.1016/j.jprot.2021.104423
Weaver, J. B., Lin, C. Y., Faries, K. M., Mathews, I. I., Russi, S., Holten, D., et al. (2021). Photosynthetic reaction center variants made via genetic code expansion show Tyr at M210 tunes the initial electron transfer mechanism. Proc. Natl. Acad. Sci. U. S. A. 118:e2116439118. doi: 10.1073/pnas.2116439118
Wong, A., Tian, X., Yang, Y., and Gehring, C. (2021). Identification of potential nitric oxide-sensing proteins using the H-NOX motif. Mol. Plant 14, 195–197. doi: 10.1016/j.molp.2020.11.015
Yang, Y. (2017). Specific enrichment of a targeted nitrotyrosine-containing peptide from complex matrices and relative quantification for liquid chromatography-mass spectrometry analysis. J. Chromatogr. A 1485, 90–100. doi: 10.1016/j.chroma.2017.01.036
Yang, H., Zhang, Y., and Pöschl, U. (2010). Quantification of nitrotyrosine in nitrated proteins. Anal. Bioanal. Chem. 397, 879–886. doi: 10.1007/s00216-010-3557-3
Zhan, X., and Desiderio, D. M. (2009). Mass spectrometric identification of in vivo nitrotyrosine sites in the human pituitary tumor proteome. Methods Mol. Biol. 566, 137–163. doi: 10.1007/978-1-59745-562-6_10
Zhang, H., Gannon, L., Jones, P. D., Rundle, C. A., Hassall, K. L., Gibbs, D. J., et al. (2018). Genetic interactions between ABA signalling and the Arg/N-end rule pathway during Arabidopsis seedling establishment. Sci. Rep. 8:15192. doi: 10.1038/s41598-018-33630-5
Zhang, X., Huai, J., Liu, S., Jin, J. B., and Lin, R. (2020). SIZ1-mediated SUMO modification of SEUSS regulates photomorphogenesis in Arabidopsis. Plant Commun. 1:100080. doi: 10.1016/j.xplc.2020.100080
Zhang, Y., Huang, X., Wang, J., Wang, X., Liu, X., Chen, Y., et al. (2019). Nitration-induced ubiquitination and degradation control quality of ERK1. Biochem. J. 476, 1911–1926. doi: 10.1042/BCJ20190240
Zhang, H., Zhu, J., Gong, Z., and Zhu, J. K. (2021). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, Y., Brandish, P. E., Ballou, D. P., and Marletta, M. A. (1999). A molecular basis for nitric oxide sensing by soluble guanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 96, 14753–14758. doi: 10.1073/pnas.96.26.14753
Zheng, Z., Guo, X., Yu, M., Wang, X., Lu, H., Li, F., et al. (2020). Identification of human IDO1 enzyme activity by using genetically encoded nitrotyrosine. Chembiochem 21, 1593–1596. doi: 10.1002/cbic.201900735
Zheng, Z., and Liu, D. (2021). SIZ1 regulates phosphate deficiency-induced inhibition of primary root growth of Arabidopsis by modulating Fe accumulation and ROS production in its roots. Plant Signal. Behav. 16:1946921. doi: 10.1080/15592324.2021.1946921
Zhu, J., Song, S., Sun, Z., Lian, L., Shi, L., Ren, A., et al. (2021). Regulation of glutamine synthetase activity by transcriptional and posttranslational modifications negatively influences ganoderic acid biosynthesis in Ganoderma lucidum. Environ. Microbiol. 23, 1286–1297. doi: 10.1111/1462-2920.15400
Keywords: nitration, nitric oxide, 3-nitro-tyrosine, post-translational modification, sensing, signaling
Citation: León J (2022) Protein Tyrosine Nitration in Plant Nitric Oxide Signaling. Front. Plant Sci. 13:859374. doi: 10.3389/fpls.2022.859374
Received: 21 January 2022; Accepted: 21 February 2022;
Published: 11 March 2022.
Edited by:
Cecilia Gotor, Institute of Plant Biochemistry and Photosynthesis (CSIC), SpainReviewed by:
Inmaculada Sánchez-Vicente, University of Salamanca, SpainCopyright © 2022 León. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: José León, amxlb25AaWJtY3AudXB2LmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.