- 1Key Laboratory of Genetics, Breeding and Multiple Utilization of Crops, Ministry of Education, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Fujian Key Laboratory of Crop Breeding by Design, Fujian Agriculture and Forestry University, Fuzhou, China
The evening complex (EC) plays a critical role in photoperiod flowering in Arabidopsis. Nevertheless, the underlying functions of individual components and coordinate regulation mechanism of EC genes in rice flowering remain to be elucidated. Here, we characterized the critical role of LUX ARRHYTHMO (LUX) in photoperiod perception and coordinating vegetative growth and flowering in rice. Non-functional alleles of OsLUX extremely extended vegetative phase, leading to photoperiod-insensitive late flowering and great increase of grain yield. OsLUX displayed an obvious diurnal rhythm expression with the peak at dusk and promoted rice flowering via coordinating the expression of genes associated with the circadian clock and the output integrators of photoperiodic flowering. OsLUX combined with OsELF4a and OsELF3a or OsELF3b to form two ECs, of which the OsLUX-OsELF3a-OsELF4a was likely the dominant promoter for photoperiodic flowering. In addition, OsELF4a was also essential for promoting rice flowering. Unlike OsLUX, loss OsELF4a displayed a marginal influence under short-day (SD) condition, but markedly delayed flowering time under long-day (LD) condition. These results suggest that rice EC genes share the function of promoting flowering. This is agreement with the orthologs of SD plant, but opposite to the counterparts of LD species. Taken together, rice EC genes display similar but not identical function in photoperiodic flowering, probably through regulating gene expression cooperative and independent. These findings facilitate our understanding of photoperiodic flowering in plants, especially the SD crops.
Introduction
Heading date is a main agronomic trait for cereal crops to adapt to specific cropping seasons and sophisticate regional environments. Photoperiod pathway is the main regulation pattern for flowering determined by synchronizing the exogenous environmental signals and the endogenous signaling cascades of photoreceptors, circadian clock systems, and floral integrator genes in rice. In Arabidopsis, a facultative long-day (LD) plant, the photoperiodic flowering is determined by the GI (GIGANTEA)-CO (CONSTANS)-FT (FLOWERING LOCUS T) pathway. FT is a major output of the photoperiodic flowering pathway, and CO is a crucial integrator of inducting the photoperiodic expression of FT, while GI acts both directly and indirectly to stimulate flowering, primarily by promoting CO and FT expression (Putterill et al., 1995; Park et al., 1999; Suárez-López et al., 2001; Turck et al., 2008; Sawa and Kay, 2011; Shim and Imaizumi, 2015). Although rice normally flowers under short-day (SD) condition, it has also developed the ability of flowering under LD condition. Extensive studies in rice have uncovered the multiple major genetic components and the mechanisms for regulating heading date, including the OsGI-Hd1 (Heading date 1)-Hd3a (Heading date 3a) pathway under SD condition (similar to Arabidopsis GI-CO-FT), and an unique Ghd7 (Grain number, plant height, and heading date 7)-Ehd1 (Early heading date 1)-Hd3a/RFT1 (RICE FLOWERING LOCUS T1) pathway under LD condition (Yano et al., 2000; Kojima et al., 2002; Hayama et al., 2003; Xue et al., 2008; Tsuji et al., 2011; Song et al., 2015; Wei et al., 2020). Unlike CO, which promotes flowering in Arabidopsis, Hd1 promotes flowering under SD condition, but represses flowering under LD condition through interaction with Ghd7 (Yano et al., 2000; Ishikawa et al., 2011; Nemoto et al., 2016).
The circadian clock acts as a hub of photoperiodic flowering in Arabidopsis, which directly links flowering to different developmental processes (Bendix et al., 2015). More than 20 clock or clock-associated components have been identified in Arabidopsis, which compose multiple interlocking transcription–translation feedback loops (Fogelmark and Troein, 2014; Shim and Imaizumi, 2015). The circadian network is likely conserved in higher plants, but the molecular circuitry that comprises the circadian clock in other plants than Arabidopsis has just begun to be understood. In rice, several clock-associated genes have been well characterized, but the circadian clock system still remains largely unclear. OsGI contributes to flowering time and the circadian clock and is important for proper regulation of genes within and outside the circadian clock in rice (Izawa, 2012; Shrestha et al., 2014). Knockout mutants of OsGI significantly delay flowering under SD condition, but the effect is weak under LD condition (Izawa et al., 2011; Lee and An, 2015), whereas overexpression of OsGI in rice causes late flowering regardless of day length, which is accompanied by increased expression of Hd1 and reduced expression of Hd3a (Hayama et al., 2003). Rice OsPRR37 (Pseudo-Response Regulator37) /Hd2/DTH7 downregulates Hd3a expression to suppress flowering under LD condition (Koo et al., 2013), and its mutation causes late flowering under SD condition but early flowering under LD condition (Lin et al., 2000). Nonetheless, it is still unclear whether other OsPRR genes are required for heading date control. The rice ELF3 (EARLY FLOWERING 3) orthologs, OsELF3-1/Hd17/Ef7 and OsELF3-2/OsEF3, are likely the component of evening complex (EC). Here, we rename OsELF3-1 and OsELF3-2 as OsELF3a and OsELF3b, respectively.
In Arabidopsis, EC is a trimeric protein consisting of LUX-ELF3-ELF4 (Nusinow et al., 2011; Silva et al., 2020). LUX is recognized as a transcription factor (Onai and Ishiura, 2005), while ELF3 and ELF4 are unique plant-specific proteins without conserved functional domains (Hicks, 2001; Doyle et al., 2002). EC components showed similar expression profiles, and their mutant alleles share multiple similar phenotypes, including an arrhythmic circadian oscillator, abnormal hypocotyl growth under diurnal cycles, and early flowering (Nusinow et al., 2011). EC is a direct regulator of genes. It represses the expression of circadian genes GI, PRR7 and PRR9 in the late evening by binding to the conserved LUX binding site in their promoters (Dixon et al., 2011; Helfer et al., 2011; Chow et al., 2012; Herrero et al., 2012; Mizuno et al., 2014). Meanwhile, EC also represses itself near dawn through inhibition of LUX, which allows the clock regulatory cycle to repeat the next day. This self-negative-feedback regulation of LUX forms an additional loop of EC (Helfer et al., 2011; Chow et al., 2012; Mizuno et al., 2014).
All of the EC components can be found in the genomes of land plant lineages. So far, most of our understanding of EC function comes from Arabidopsis, while the information about EC orthologs remains limited in other plants (Nusinow et al., 2011; Huang and Nusinow, 2016). However, it is likely that the ECs in other species than Arabidopsis will interact with different proteins/pathways or regulate new outputs. Therefore, molecular understanding of how EC gene orthologs function in other plants, especially in SD crops, such as rice, corn, and sorghum, will be necessary for elucidating their roles in circadian- and photo-regulation of physiology (Huang and Nusinow, 2016). Recently, two putative evening complexes have been identified and the accurate function of partial components has been characterized in soybean (Liew et al., 2017; Bu et al., 2021; Fang et al., 2021). In rice, two ELF3 orthologs have been characterized. Loss of OsELF3a function disrupts photoperiod-dependent control of key flowering time genes, and produces a photoperiod-insensitive flowering type (Matsubara et al., 2012; Saito et al., 2012; Zhao et al., 2012; Yang et al., 2013), whereas the contribution of OsELF3b to either flowering time or the circadian clock remains controversial (Fu et al., 2009; Zhao et al., 2012).
In this study, we identified OsLUX in rice through map-based cloning, characterized the role and interactions of OsLUX in the rice flowering pathway, and identified two rice ternary repressive protein complexes composed of OsLUX OsELF4a, and OsELF3a or OsELF3b. We found that the complex OsLUX-OsELF3a-OsELF4a is likely the dominant promoter for photoperiodic flowering. In addition, we characterized the role of OsELF4a in the control of rice heading date using CRISPR/Cas9. Thus, rice is another species apart from Arabidopsis and soybean with the components of EC identified and well characterized. The findings of this study provide new insights into the EC function and may facilitate our understanding of the mechanisms of photoperiodic flowering in other SD species.
Materials and Methods
Plant Materials
Rice cultivars Nipponbare (japonica), DZ60 (tropical japonica), long vegetative phase2-1 (lvp2-1) mutant and eight editing lines including lvp2-2 to lvp2-7, Oself4a-1, and Oself4a-2 were used in the study. The lvp2-1 mutant was obtained from the progeny of EMS mutagenesis of Nipponbare; lvp2-2 to lvp2-7, Oself4a-1, and Oself4a-2 were generated from Nipponbare using the CRISPR/Cas9 system.
Identification of LVP2/OsLUX Gene
To map LVP2 locus, an F2 population was developed from the cross between lvp2-1 and DZ60. To avoid the interfering effects of other flowering genes, a total of 389 progeny lines (F2:3) which display extremely late flowering and other phenotypic characteristics similar to lvp2-1 were selected for gene mapping. LVP2 was mapped to a rice chromosome by using the publicly available RM-series rice microsatellite markers (McCouch et al., 2002) and bulked segregant analysis (BSA) method. Furthermore, mapping of LVP2 gene was also performed by deep sequencing-based bulked segregant analysis (BSA-seq) based on an F3 population developed from the cross between lvp2-1 and Nipponbare. To make the wild-type DNA pool and the mutant DNA pool, equal amount of fresh leaves from 30 homozygous wild-type lines (5 plants/line) and 30 mutant lines randomly selected from the F3 population were mixed for genomic DNA extraction, respectively. The two DNA pools were sequenced using the Illumina PE Genome Analyzer. Based on the sequence information (read length = 300 bp, sequencing depth > 30×) of the two pools, the LVP2 locus was mapped.
Vector Construction and Plant Transformation
For genetic complementation test, a 3,420-bp genomic DNA sequence covering the promoter region, gene region, and downstream region of OsLUX was amplified from Nipponbare, and inserted into the binary vector pCAMBIA1300. The plasmid was introduced into lvp2-1 mutant embryonic calli. To confirm the function of OsLUX and to identify the function of OsELF4a, the CRISPR/Cas9 system was used, and the CRISPR targets for OsLUX and OsELF4a were selected as described by Miao et al. (2013). The vector construction was performed according to the manufacturer’s instruction of regent kit (VIEWSOLID Biotech, China). The vectors were then transferred into Nipponbare. The genomic regions surrounding the CRISPR target sites for OsLUX and OsELF4a were amplified by PCR using primer pairs 5′-tcgagtccccgatttggttc/5′-gcttcacgtagaggcgatact and 5′-atggaaggtgatagcttct/5′-gccgggccggacacgcttc, respectively, and were sequenced to identify mutants. For overexpression test, a 717-bp cDNA of OsLUX was amplified and fused into the downstream of maize Ubiquitin promoter of the binary vector pTCK303. The plasmid for overexpression was introduced into Nipponbare. To determine the expression pattern of OsLUX, a 2021-bp promoter upstream of the coding region of OsLUX was amplified from Nipponbare, and fused into the GUS reporter gene with the nopaline synthase terminator in pCAMBIA1391Z. The plasmid was introduced into Nipponbare. Histochemical assay for GUS activity in transgenic plants was performed as described (Jefferson et al., 1987).
All constructs were confirmed by sequencing, introduced into Agrobacterium tumefaciens strain EHA105, and transferred into the rice variety by Agrobacterium-mediated transformation. The primers used for vector construction are listed in Supplementary Table S1.
Subcellular Localization
OsLUX-GFP fusion was made by in-frame fusion of the 717-bp full-length coding sequence of OsLUX into vector pRTVcGFP (He et al., 2018). OsLUX-GFP and GFP alone (as control) were transferred into rice protoplasts. NLS-RFP was co-transfected for nuclear localization control. The samples were observed with a confocal laser scanning microscope (LEICA-SP8). Primer pairs for amplifying OsLUX cDNA are listed in Supplementary Table S1.
Yeast Two-Hybrid (Y2H)
For Y2H assay, the coding sequences of OsLUX and OsELF4a were amplified from Nipponbare and inserted into pGBKT7. The coding region of OsELF3a and OsELF3b were amplified from Nipponbare and inserted into pGADT7. Different combinations of bait and prey were used to co-transform the yeast competent cell Y187. The transformed yeast cells were grown on non-selective SD media (−Trp-Leu/−LT) and selective SD media (−Trp-Leu-His-Ade/-LTHA) with X-α-Gal. A positive interaction was judged by the growth of yeast colonies on selective media. The primers used for vector construction are listed in Supplementary Table S1.
Bimolecular Fluorescence Complementation Assay
The CDSs of OsLUX, OsELF3a, OsELF3b, OsELF4a, OsELF4b, and OsELF4c (without termination codon) were amplified and inserted into the entry vector pDONR207 via a BP reaction, and then transferred into the N-terminal and the C-terminal of pSPYNE or pSPYCE (Choi et al., 2012) via a LR reaction, to form fusion protein NE-LUX, CE-LUX, NE-ELF3a, NE-ELF3b, CE-ELF4a, CE-ELF4b, and CE-ELF4c, respectively. Equal concentration mixture of NE-ELF3a and CE-LUX, NE-ELF3b and CE-LUX, NE-ELF3a and CE-ELF4a, NE-ELF3b and CE-ELF4a, NE-ELF3a and CE-ELF4b, NE-ELF3b and CE-ELF4b, NE-ELF3a and CE-ELF4c, NE-ELF3b and CE-ELF4c, and NE-LUX and CE-ELF4a were used to co-transform rice protoplasts. The vector combinations of CE + NE-LUX, NE + CE-LUX, CE + NE-ELF3a, CE + NE-ELF3b, NE + CE-ELF4a, NE + CE-ELF4b, and NE + CE-ELF4c were used as negative controls to verify the specificity of the interactions. Fluorescence in transformed protoplast was examined and imaged using a confocal laser scanning microscope (LEICA-SP8). The primers used for vector construction are listed in Supplementary Table S1.
Co-immunoprecipitation (Co-IP) Assays
To further validate the interaction between OsLUX and interacted partners in vivo, Co-IP assay was conducted. The ORFs of OsLUX and OsELF4a were amplified and inserted into pRTVcGFP (He et al., 2018) to generate expression vectors pRTVcGFP-OsLUX and pRTVcGFP-OsELF4a. Full-length cDNAs of ELF3a and ELF3b were amplified and cloned into the psuper1300-MYC. The Co-IP assay was performed as described (Kong et al., 2019). Briefly, the plasmids were used to transiently co-transform rice protoplasts from ten-day-old etiolated seedlings. After incubation for 16 h, total proteins were extracted from the protoplasts. Lysis buffer (50 mM Tris-MES pH = 0.8, 0.5Msucrose, 1 mM MgCl2, 10 mM EDTA, 1 mM PMSF, 0.5 mM DTT, 0.1% Trixon-100) was added to protease inhibitor cocktail (Sigma-Aldrich, P9599). The supernatant was collected after centrifugation 16,000 g for 30 min at 4°C and precleared with GFP-Nanoab-Agarose (Lablead, GNA-20-400) for 1 h. The GFP-Nanoab-Agarose was washed four times in 0.5 ml lysis buffer and then boiled in the 2XSDS-PAGE sample buffer. The immunoprecipitation products were detected by SDS-PAGE and western blot using anti-Myc (MBL, M047-3) or anti-GFP (MBL, 598) antibody. The primers used for vector construction are listed in Supplementary Table S1.
RNA Isolation and qRT-PCR Analysis
RNA isolation and qRT-PCR analysis were performed as described (Duan et al., 2019). Briefly, total RNA was isolated using TRIzol reagent kit (Invitrogen, Carlsbad, CA, United States). Reverse transcription of total RNA was performed using Primescript™ RT reagent kit (Takara, China). The cDNA samples were diluted to 8 ng/μl and 2 ng/μl. Triplicate quantitative assays were performed using the SYBR Premix Ex Taq II (Takara, China) with a Mastercycler ep realplex4 sequence detection system (Eppendorf). Amplification of UBQ was used as internal control to normalize all data. Primers used for qRT-PCR analysis are listed in Supplementary Table S2.
Accession Numbers
Sequence data from this article for the mRNA, cDNA, and genomic DNA can be found in the GenBank/EMBL/Gramene data libraries or Web site under accession numbers:
OsLUX, LOC_Os01g74020; OsGI, LOC_Os01g08700; OsLHY, LOC_Os08g06110; OsPRR37, LOC_Os07g49460; OsPRR95, LOC_Os09g36220; OsELF3a, LOC_Os06g05060; OsELF3b, LOC_Os01g38530; OsELF4a, LOC_Os11g40610; OsELF4b, LOC_Os03g29680; OsELF4c, LOC_Os08g27860; Hd1, LOC_Os06g16370; Hd3a, LOC_Os06g06320; Ghd7, LOC_Os07g15770; Ehd1, LOC_Os10g32600.
Results
lvp2-1 Exhibited Extremely Late Flowering Regardless of Day Length
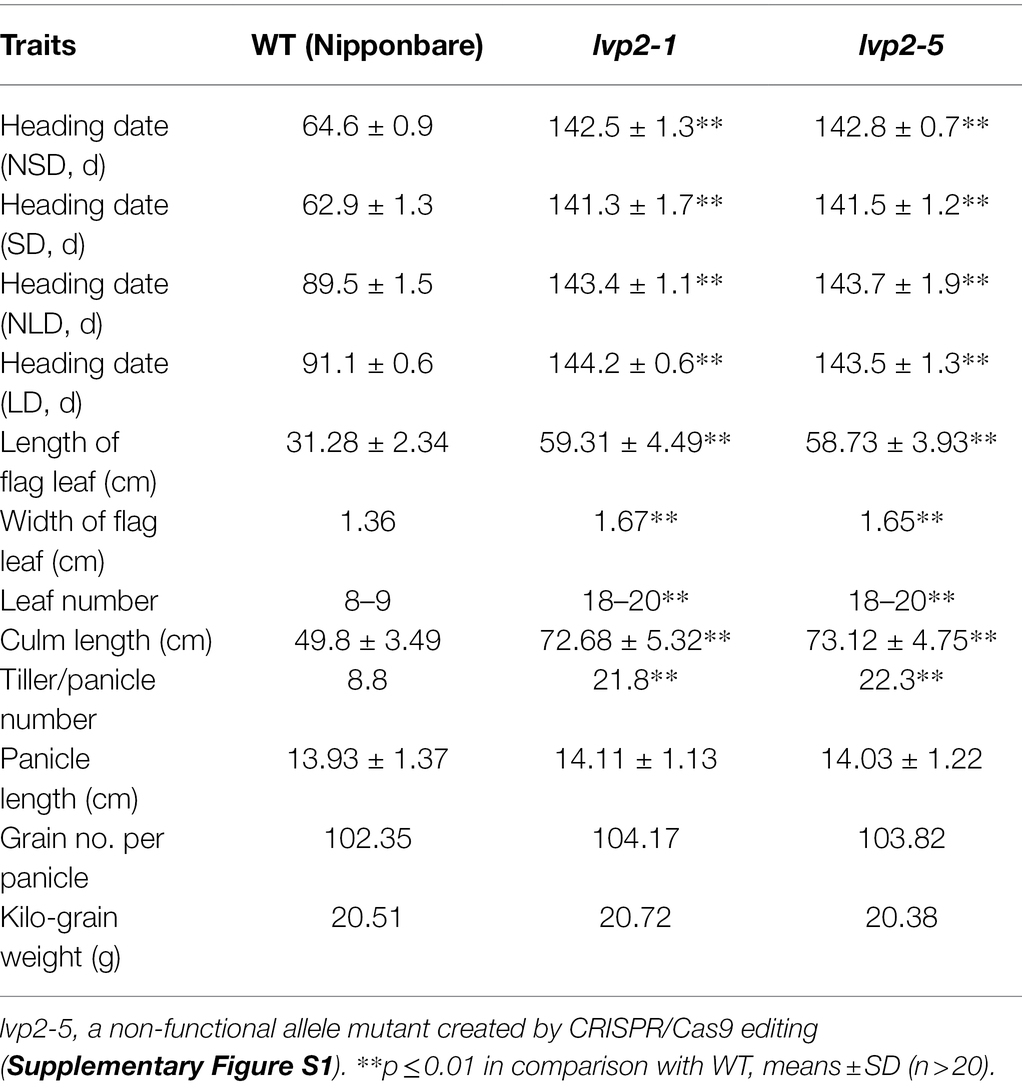
The lvp2-1 mutant, which displayed a long staying-green period and extremely late flowering similar to that of previously reported lvp2 mutant (Sun et al., 2012), was identified from the progeny of EMS mutagenesis of Nipponbare, a japonica rice variety (Figure 1A). To identify the photoperiod response of lvp2-1, we examined the heading date of lvp2-1 under the natural LD from July to September (NLD, ≥14 h light) condition in Fuzhou (26°08′) and the natural SD from January to March (NSD, ≤10 h light) condition in Sanya (18°10′). The wild type (WT) headed 24.9 days earlier in Sanya (64.6 days) than in Fuzhou (89.5 days; Table 1), whereas the mutant showed extremely late flowering in both Fuzhou (143.4 days) and Sanya (142.5 days), with only about 1 days delay under NLD (Table 1). To confirm this result, we grew lvp2-1 and WT under both SD (10/14 h light/dark) and LD (14/10 h light/dark) conditions in the growth chamber. The photoperiod response of lvp2-1 in the growth chamber was completely consistent with that under the natural field conditions (Table 1). These observations indicated that lvp2-1 was photoperiod-insensitive, suggesting that LVP2 plays an important role in increasing photoperiod sensitivity and acts as a flowering promoter under all photoperiod conditions in rice.

Figure 1. Phenotypes of lvp2-1 and WT. (A) WT and lvp2-1 under natural day-length condition. (B,C) Plants (B), roots and tillers (C) of WT and lvp2-1 at 45 days after sowing under NLD condition. (D) Sizes and shapes of WT and lvp2-1 seeds. (E) Seeds per plant of WT and lvp2-1. Scale bars = 10 cm in (A-C) and 1 cm in (D).
lvp2-1 Plant Grew Vigorously With Increased Biomass and Grain Yield
lvp2-1 exhibited vigorous growth throughout the entire vegetative phase under both SD and LD conditions. We compared several morphological traits of lvp2-1 with those of WT under the natural day length condition in Fuzhou, including size of leaf, number of leaves per plant, number of tillers per plant, culm length, and so on. This mutant showed similar phenotypes (Table 1). Compared with WT, lvp2-1 had significantly longer and wider leaves and taller culms (Figures 1A,B; Table 1), and produced much more leaves and tillers (Table 1), but its leaf emergence rate and tiller emergence rate remained almost the same as those of WT (Figures 1B,C). In addition, the mutant generated more lateral roots than WT (Figure 1C), which would possibly enable itself to obtain more nutrition for growth. These observations suggest that LVP2 controls biomass-driven growth. In the reproductive phase, no remarkable differences were found between lvp2-1 and WT in panicle length, number of grains per panicle, seed-setting rate, grain size, grain shape, and 1,000-grain weight (Table 1). However, lvp2-1 produced over twice as many grains as those produced by WT due to its dramatic increase in effective tillers/panicles (Figure 1E; Table 1), suggesting that lvp2-1 may have the potential to be used for increasing yield in breeding programs. Besides, LVP2 showed the function of repressing awn development. Most of the lvp2-1 grains had a long awn, while the WT grains were hardly awned (Figure 1D). In summary, LVP2 had pleiotropic effects on vegetative growth and reproductive development, and loss of LVP2 function would dramatically increase the biomass and grain yield of the plant.
OsLUX Mutation Was Responsible for the lvp2-1 Phenotype
Genetic analysis showed that the phenotype of lvp2-1 was caused by a single gene mutation (Supplementary Table S3). To identify the LVP2 locus, we crossed lvp2-1 with variety DZ60 to develop an F2 population, and a total of 389 progeny lines (F2:3) which display extremely late flowering and other phenotypic characteristics similar to lvp2-1 were selected for gene mapping. Using the RM-series rice microsatellite markers, we mapped LVP2 on the terminal region of the long arm of chromosome 1, linked with RM414. To identify the candidate gene, we performed whole-genome resequencing of the DNA pools of WT and mutant, each consisting of 30 lines. Based on the sequence information obtained from resequencing, the LVP2 locus was also mapped on the terminal region of the long arm of chromosome 1, agreeing with the results of linkage-based mapping (Figure 2A). By comparing the nucleotide sequences of the genomic fragments in this region, we found that there was a single base transition from C (CGG) to T (TGG) in gene LOC_Os01g74020 (OsLUX), which resulted in a change from Arginine to Tryptophan at the 120th amino acid (aa) of OsLUX in lvp2-1 (Figures 2B,C). OsLUX is a MYB-like transcription factor consisting of 238 aa homologous to Arabidopsis LUX (Supplementary Figure S1). The mutation in lvp2-1 was located within the crucial GARP domain (117th–175th aa) in OsLUX (Supplementary Figure S2). The ORF sequences of all the other genes in this region remained unchanged between WT and lvp2-1. Expression analysis showed that the rhythm pattern of OsLUX expression in lvp2-1 was almost the same as that in WT, but the expression level in lvp2-1 was significantly downregulated (Supplementary Figure S3). These results strongly suggested that the lvp2-1 phenotype was caused by the mutation in OsLUX.
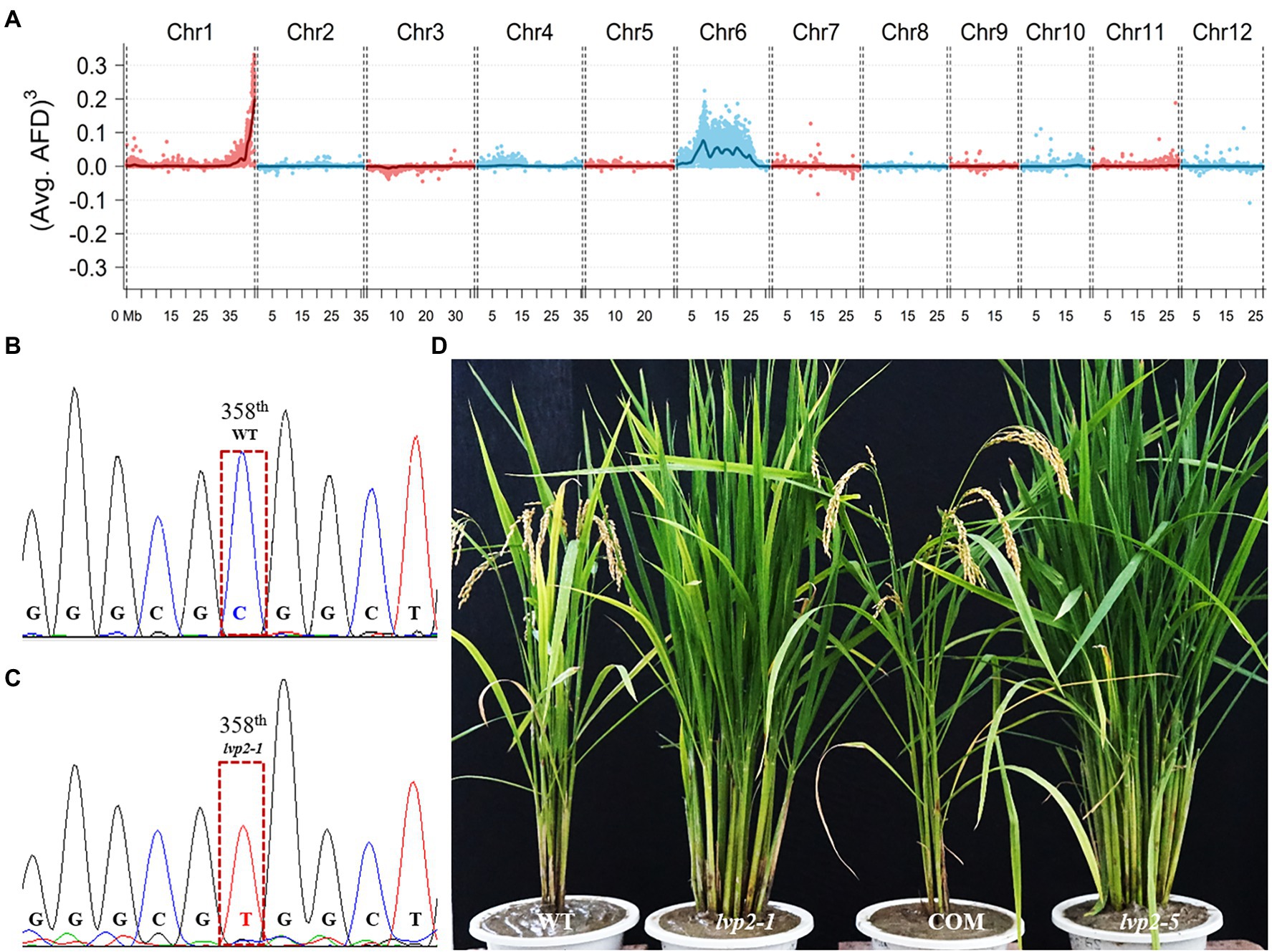
Figure 2. Identification and characterization of OsLUX function. (A) Mapping and screening the candidate gene of LVP2 by resequencing the DNA pools of WT and mutant. LVP2 was located on the terminal region of the long arm of chromosome1. (B,C) A single base transversion (C to T) was found in lvp2-1, which resulted in the change of the 120th aa from Arginine to Tryptophan. (D) Functional validation of OsLUX via genetic complementation and gene editing. The WT phenotype was recovered by genetic complementation in the transgenic lvp2-1 plant, whereas the loss-of-function mutant plant showed lvp2-1 phenotype. COM, a genetic complementation plant; lvp2-5, a non-functional allele generated by CRISPR/Cas9 editing, which encoded a truncated peptide of OsLUX consisting of only 18 amino acids.
To validate the candidate gene, we constructed a complementary plasmid pCAMBIA1300-OsLUX carrying a 3,420-bp wild-type genomic DNA, which covered the entire OsLUX gene (717-bp) and its upstream promoter region. By introducing the plasmid into lvp2-1, the mutant phenotype was rescued and restored to that of WT in all of 25 transgenic lines (Figure 2D; Supplementary Table S4), confirming that OsLUX is LVP2. To further validate and investigate OsLUX, we created the loss-of-function mutants of OsLUX from WT Nipponbare using CRISPR/Cas9 technology. Eleven effectively edited lines were obtained, which could be classified into six types, including one-base insertion between the 55th and the 56th bases (Types 1–3, 6 lines), deletion of the 55th base (Type 4, 3 lines), 16-base deletion of 42nd-57th base (Type 5, 1 line), and 274-base deletion of 30th-303rd base (Type 6, 1 line). We named these lines as lvp2-2 to lvp2-7. All of the edited alleles were predicted to encode a short peptide lacking the crucial GARP domain (Supplementary Table S5). Hence, all of them should be non-functional. As expected, all of the edited lines exhibited the same phenotype as that of lvp2-1 in photoperiod sensitivity, heading date, and other characteristics under various photoperiod conditions (Table 1; Figure 2D), indicating that lvp2-1 should be a non-functional allele as well. We also developed OsLUX overexpression lines, but the gene expression was not noticeably increased and no significant phenotype alteration was observed in the transgenic lines (Supplementary Figure S4), suggesting that OsLUX represses the expression of itself, similar to LUX in Arabidopsis.
OsLUX Showed a Constitutive Rhythmic Expression Pattern
To investigate whether the expression of OsLUX is rhythmic, we assayed its expression every 4 h in a 48-h time course under both LD (14 h light) and SD (10 h light) conditions. The transcript level of OsLUX showed an obvious diurnal circadian rhythm of the daily 24 h cycle, in which it started to accumulate at ZT8, reached the peak approximately at the dusk, and slowly decreased during the night under both LD and SD, but the peak under LD was almost twice as high as that under SD (Figure 3A). To examine whether OsLUX expression is organ-specific, quantitative real-time PCR (qRT-PCR) was performed for different organs. OsLUX expression was detected in all the organs examined, with the highest accumulation occurring in 5-mm-long young panicle and developing floral organs, followed by shoot and leaf, and weak in shoot, root, and stem (Figure 3B). To confirm the results of qRT-PCR analysis and intuitively display the spatio-temporal expression characteristic of OsLUX, we fused the promoter of OsLUX with GUS (β-glucuronidase) reporter gene and transferred the fused vector into rice with the mediation of Agrobaterium tumefaciens. Totally, 23 transgenic lines were obtained (Supplementary Figure S5). The results of GUS activity examination indicated that OsLUX was expressed in all organs including root, stem, leaf, young panicle, spikelet, and developing seed (Figures 3C–O). This was agreement with the results of qRT-PCR. However, GUS analysis indicated that OsLUX expression was not uniform within organs. In root, OsLUX expression was basically limited to the actively developing young roots and young root hairs, but little in the mature zones (Figure 3C). Developing leaf sheath displayed stronger expression than developing leaf blade (Figures 3D,E). Elongating stem showed higher expression than mature stem (Figure 3F). OsLUX expression was strong in developing young panicle and the first internode from top (Figure 3G), but decreased later (Figure 3H). In developing spikelet, OsLUX expression was observed in all floral organs, with the strongest in the lemma/palea and receptacle (Figures 3I–L). In addition, OsLUX expression was also detected in developing seed (Figures 3M,N) and the germ and radicle of germinating seed (Figure 3O).
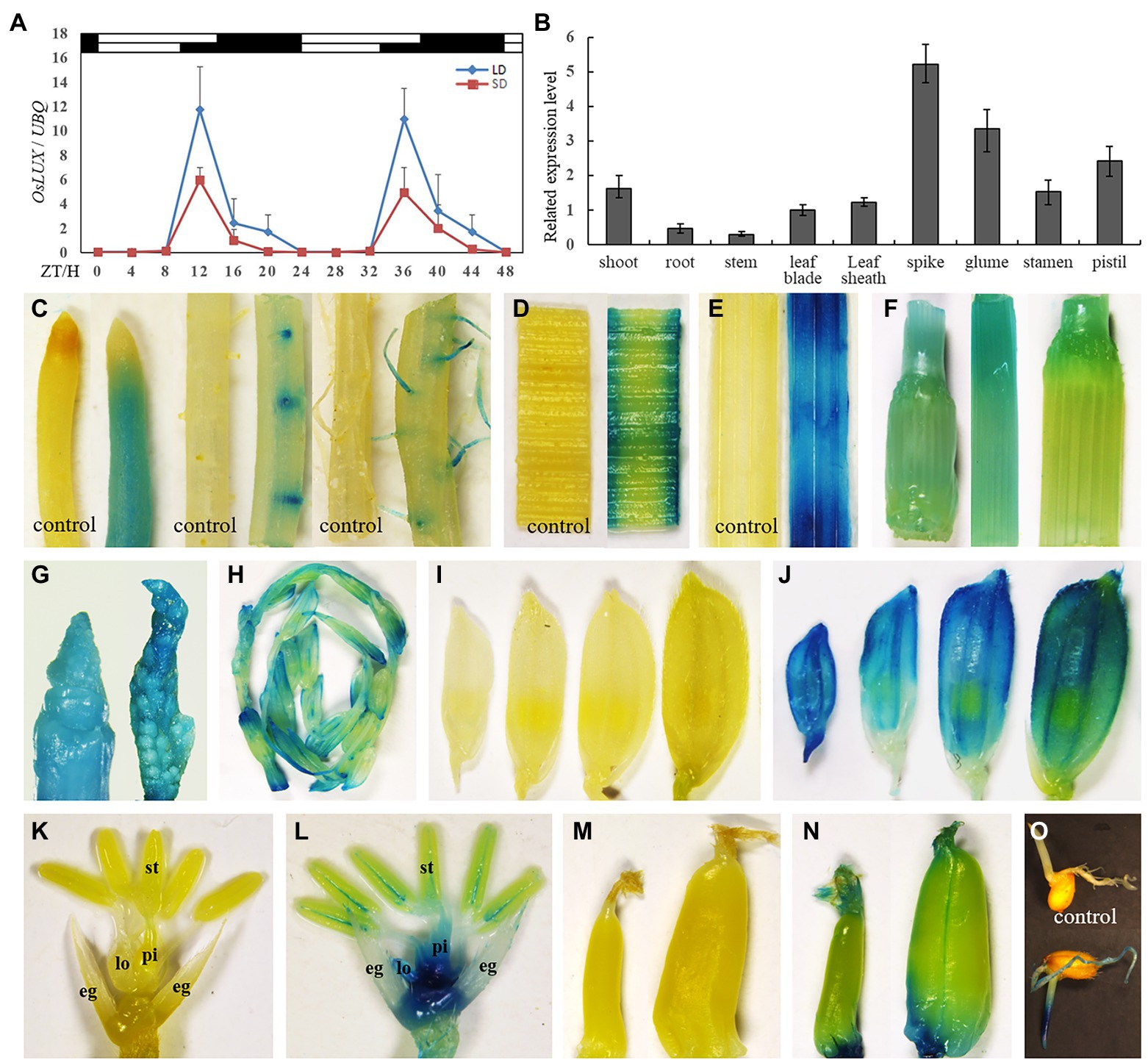
Figure 3. qRT-PCR and GUS analyses of OsLUX expression pattern. (A) The rhythmic expression of OsLUX under both LD and SD conditions in Nipponbare. Rice leaves were collected at the indicated time points from 35- and 70-day-old plants grown under SD and LD, respectively. The expression levels are relative to the UBQ mRNA. Values and error bars are the mean and SD of three replicates. The white bars and black bars at the top represent the light and dark periods, respectively. ZT, Zeitgeber time. (B) OsLUX transcript accumulation in different tissues and organs. Values and error bars are the mean and SD of three replicates. (C–O) GUS detection of OsLUX transcript accumulation in roots of different parts or developmental stages (C), young leaf blade (D) and leaf sheath (E), stems of different parts or developmental stages (F), young panicle (G,H), developing spikelet (J), floral organs with lemma and palea removed (L), developing seed (N), and the germ and radicle of germinating seed (O). (I), (K) and (M) were controls. eg, extra glums; lo, lodicule; st, stamen; pi, pistil.
OsLUX Regulated Genes Associated With the Circadian Clock
Arabidopsis LUX affects the expression of circadian clock genes. To investigate the effect of OsLUX mutation on the circadian clock, we compared the diurnal expression of several known or likely-to-be circadian clock genes in WT and that in lvp2-1 under both SD and LD, including five with known function (OsGI, OsPRR37, OsLHY, OsELF3a, and OsELF3b), one unidentified function (OsPRR95), and three speculative OsELF4 genes (OsELF4a, OsELF4b, and OsELF4c; Supplementary Figure S6). The rhythmic expression patterns of these genes in lvp2-1 were similar to those in WT under both SD and LD conditions, but the expression levels of the most genes changed (Figure 4). Among them, OsGI, OsPRR37, and OsPRR95 showed higher expression peaks in lvp2-1 than in WT (Figures 4A–F), suggesting that OsLUX negatively regulates the expression of these genes under both day-length conditions, whereas the expression of OsLHY, OsELF3a, OsELF3b, and OsELF4a were dramatically reduced in lvp2-1 mutant (Figures 4G–N), suggesting that OsLUX positively regulates the expression of these genes under both day-length conditions. However, OsELF4b and OsELF4c did not show significant change under SD and LD conditions (Figures 4O–R). These results suggest that OsLUX is essential to the circadian function through positive or negative regulation of the expression of genes associated with the circadian clock in rice.
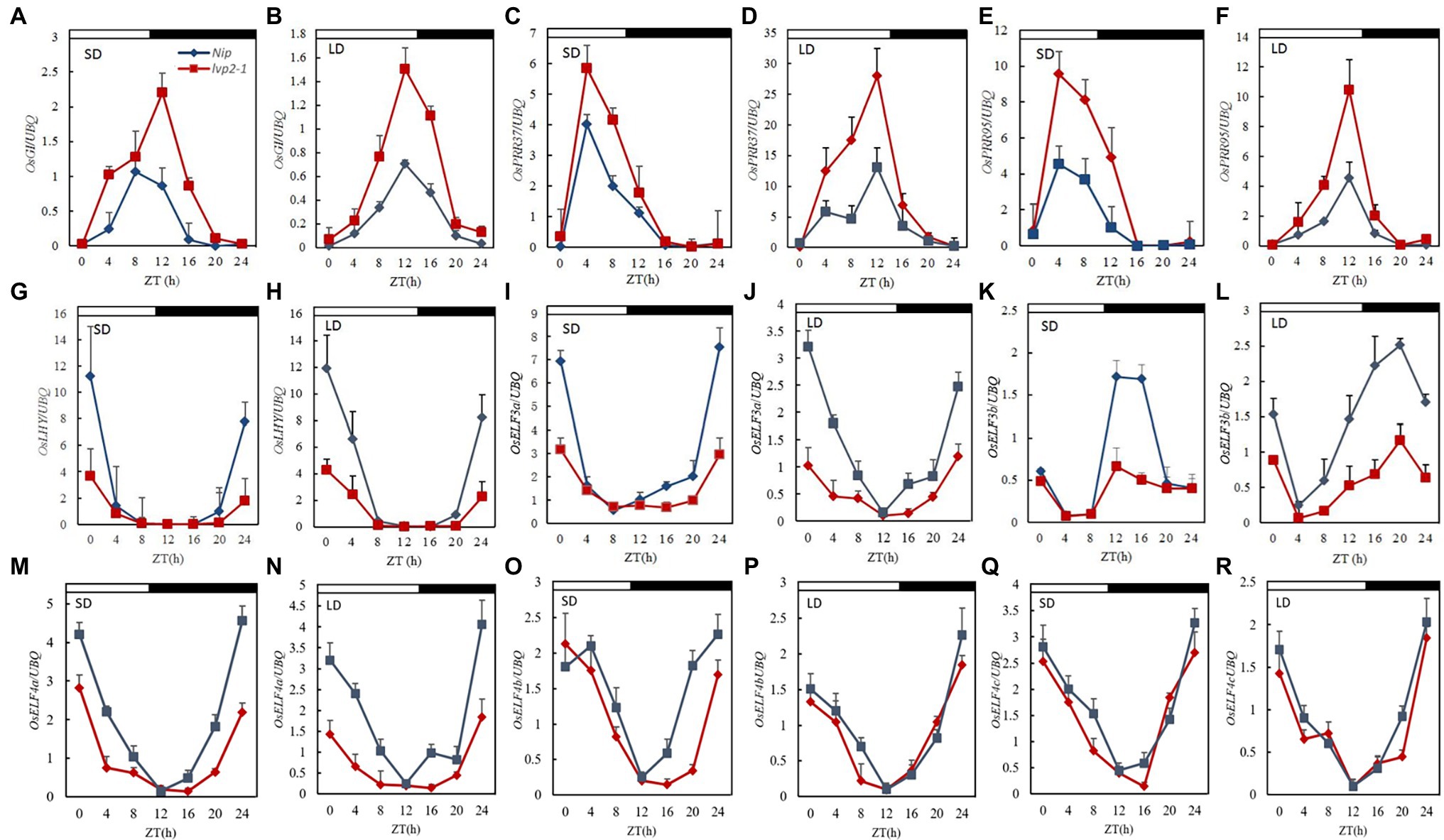
Figure 4. Diurnal expression of clock-associated genes in WT and lvp2-1 under both SD and LD conditions. Diurnal expression patterns of OsGI (A,B), OsPRR37 (C,D), OsPRR95 (E,F), OsLHY (G,H), OsELF3a (I,J) OsELF3b (K,L), OsELF4a (M,N), OsELF4b (O,P), and OsELF4c (Q,R) in Nipponbare (blue diamonds) and lvp2-1 mutant (red squares) plants in SD and LD detected by qRT-PCR analysis. Rice leaves were collected at the indicated time points from 35- and 70-day-old plants grown under LD (14 h light) and SD (10 h light) conditions, respectively. The expression levels are relative to the UBQ mRNA. Values and error bars are the mean and SD of three replicates. The white bars and black bars at the top represent the light and dark periods, respectively. Nip, Nipponbare; ZT, Zeitgeber time.
OsLUX Affected the Expression of Floral Integrator Genes
In rice, Ehd1 and Hd1 determine the degree of expression of florigenic genes Hd3a and RFT1 through distinct pathways under a given photoperiod. To identify potential downstream flowering time genes regulated by OsLUX, we examined the diurnal expression of five photoperiod flowering-related integrator genes, Hd1, Ghd7, Ehd1, Hd3a, and RFT1, under both SD and LD. Under SD, the peak of Hd1 expression was lower in lvp2-1 than in WT (Figure 5A), and almost no Hd3a expression could be detected in lvp2-1 (Figure 5B). This was likely to be directly responsible for the later flowering of lvp2-1 under SD. Meanwhile, the peak of Ghd7 expression was higher in lvp2-1 than in WT (Figure 5C), suggesting that OsLUX represses Ghd7 expression; while the peak of Ehd1 expression was lower in lvp2-1 than in WT (Figure 5D), due to the repression of Ghd7. As expected, the expression of these genes except RFT1 under LD (Figures 5E,J) showed similar alternation patterns to those under SD in lvp2-1 (Figures 5F–I), consistent with the mutant phenotype of photoperiod-insensitive late flowering (Figure 1). Under SD, very low RFT1 expression was detected in both WT and lvp2-1 (Figure 5E), whereas under LD, the peak of RFT1 expression was markedly lower in lvp2-1 than in WT (Figure 5J), which led to late flowering in lvp2-1. Taken together, OsLUX is essential for the photoperiodic control of rice flowering. OsLUX functions as a floral promoter by promoting Hd1 and Ehd1 expression and repressing Ghd7 expression under both SD and LD conditions, so as to regulate the expression of downstream florigenic genes Hd3a and RFT1.
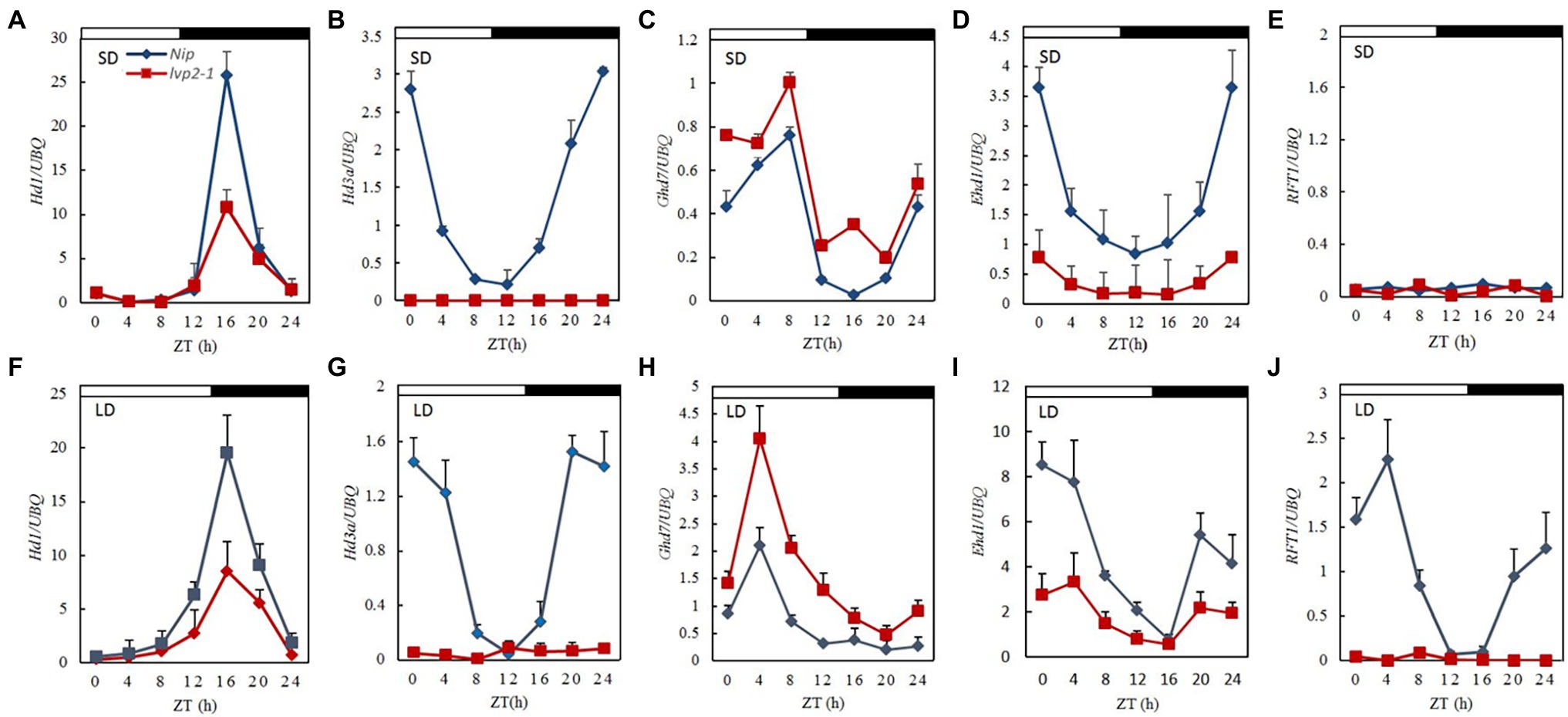
Figure 5. Diurnal expression of photoperiodic flowering pathway genes in lvp2-1 mutant under both SD and LD conditions. Diurnal expression patterns of Hd1 (A,F), Hd3a (B,G), Ghd7 (C,H), Ehd1 (D,I) and RFT1 (E,J) in Nipponbare (blue diamonds) and lvp2-1 (red squares) plants under LD (14 h light) and SD (10 h light) detected by qRT-PCR analysis. Rice leaves were collected at the indicated time points from 35- and 70-day-old plants grown under SD and LD conditions, respectively. The expression levels are relative to the UBQ mRNA. Values and error bars are the mean and SD of three replicates. The white bars and black bars at the top represent the light and dark periods, respectively. ZT means Zeitgeber time.
OsLUX Located in Nucleus and Formed ECs With OsELF4a and OsELF3a or OsELF3b
To investigate the intracellular location of OsLUX, we constructed an expressional vector of the fusion gene of GFP (green fluorescent protein) and OsLUX to transform rice protoplasts. The green fluorescent signal was observed specifically in nucleus (Supplementary Figure S7), suggesting that OsLUX was anchored in nucleus.
In rice, two ELF3 orthologs, OsELF3-1/OsELF3a and OsELF3-2/OsELF3b, have been identified, but no apparent ELF4 ortholog has been found (Murakami et al., 2007). To identify the putative rice ELF4, we used the full-length sequence of ELF4 to perform BLAST analyses. Four ELF4 homologues were found from the rice genome sequence, namely, LOC_Os11g40610, LOC_Os03g29680, LOC_Os08g27860, and LOC_Os08g27870, which showed considerable similarities to ELF4 in amino acid sequence. Protein sequence alignment and phylogenetic analysis showed that these ELF4 homologues contained a function-unknown evolutionarily conserved domain DUF1313 (Supplementary Figure S6; Kolmos et al., 2010; Lin et al., 2019). Among these genes, LOC_Os08g27870 transcript was undetectable in rice leaf in this study. Hence, we named LOC_Os11g40610, LOC_Os03g29680 and LOC_Os08g27860 as OsELF4a, OsELF4b and OsELF4c, respectively.
To investigate the components of EC in rice, we performed bimolecular fluorescence complementation (BiFC) assay to test the interactions of OsLUX with two OsELF3s and three OsELF4s. The coding sequences of OsLUX, OsELF3a, OsELF3b, OsELF4a, OsELF4b, and OsELF4c were fused into the N- and/or C-terminal of vectors pSPYNE and pSPYCE, respectively. No fluorescence was observed in the mixed carrier of all the combinations of NE-LUX+CE, NE + CE-LUX, NE-ELF3a + CE, NE-ELF3b + CE, NE + CE-ELF4a, NE + CE-ELF4b, and NE + CE-ELF4c (data not shown). Green fluorescence was observed from the nuclei of rice protoplasts co-transformed with NE-OsELF3a + CE-OsLUX (Figure 6A), NE-OsELF3b + CE-OsLUX (Figure 6B), NE-OsELF3a + CE-OsELF4a (Figure 6C), and NE-OsELF3b + CE-OsELF4a (Figure 6D). However, no green fluorescence was observed using the mixed carrier of NE-OsLUX+CE-OsELF4a (Figure 6E), NE-OsELF3a + CE-OsELF4b (Figure 6F), NE-OsELF3b + CE-OsELF4b (Figure 6G), NE-OsELF3a + CE-OsELF4c (Figure 6H), and NE-OsELF3b + CE-OsELF4c (Figure 6I). The interactions between OsLUX and OsELF3a, OsLUX and OsELF3b, OsELF4a and OsELF3a, and OsELF4a and OsELF3b were further confirmed by yeast two-hybrid assay (Figure 6K) and in vivo Co-IP assay (Figures 6L–O). These results indicated that OsLUX forms two ECs with OsELF4a and OsELF3a or OsELF3b, respectively; but OsELF4b and OsELF4c cannot form EC with OsLUX and OsELF3a or OsELF3b. We named the two ECs as LUX-ELF3a-ELF4a and LUX-ELF3b-ELF4a in rice.
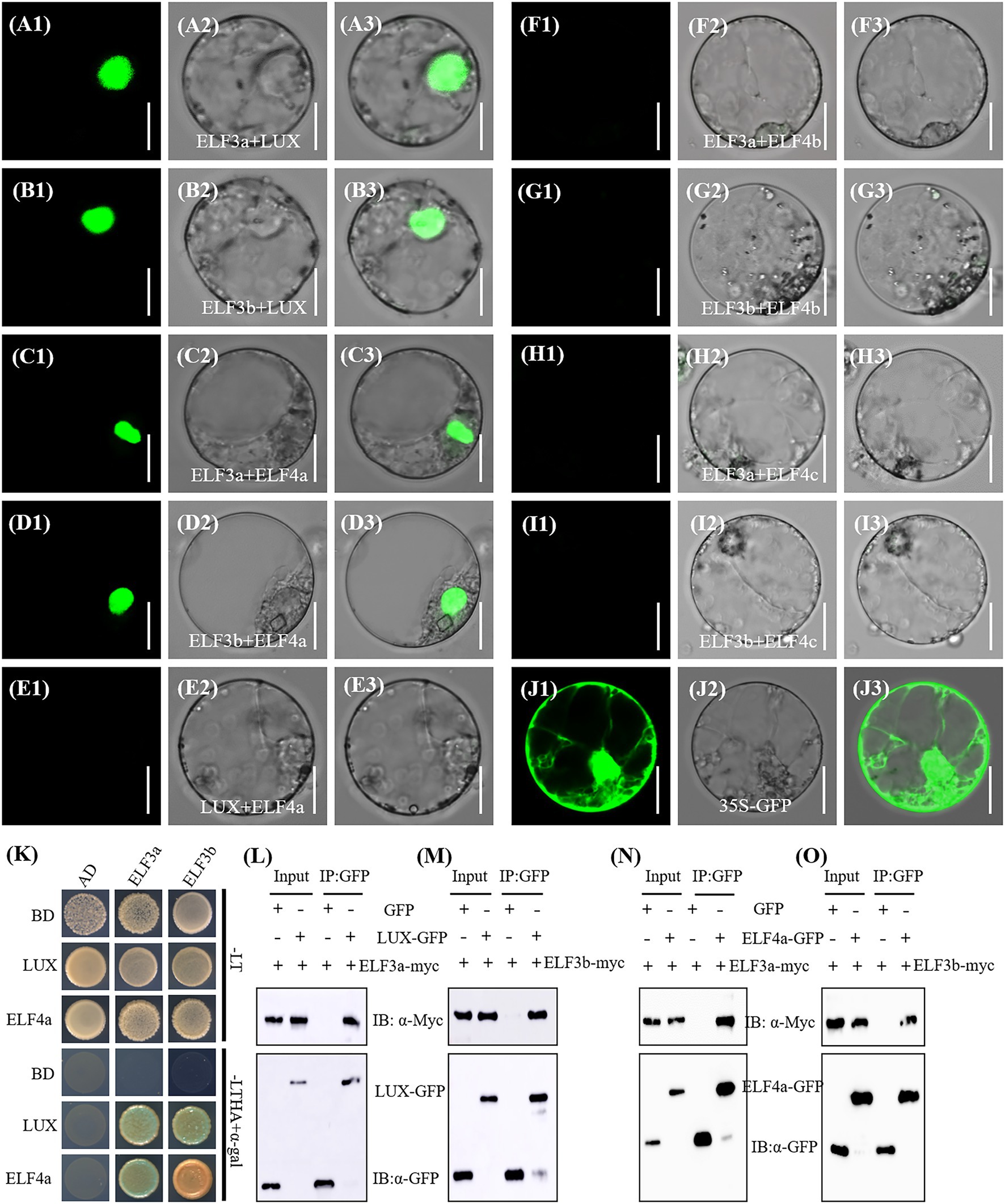
Figure 6. Detection of interactions among OsLUX, OsELF3s and OsELF4s. (A-I) Bimolecular fluorescence complementation (BiFC) assay of the interaction between OsELF3a and OsLUX (A), OsELF3b and OsLUX (B), OsELF3a and OsELF4a (C), OsELF3b and OsELF4a (D), OsLUX and OsELF4a (E), OsELF3a and OsELF4b (F), OsELF3b and OsELF4b (G), OsELF3a and OsELF4c (H), and OsELF3b and OsELF4c (I). (J) Ubi-GFP as the control. (A1-J1) GFP filter; (A2-J2) bright field; (A3-J3) merged images. Scale bars = 10 μm. (K) yeast two-hybrid assay the interactions between LUX or ELF4a with ELF3a or ELF3b, respectively. Transformed yeast cells were grown on non-selective SD media (−Trp-Leu/−LT) and selective SD media (−Trp-Leu-His-Ade/-LTHA) with X-α-Gal. (L-O) Co-IP analysis confirms that OsLUX interacts with OsELF3a (L) and OsELF3b (M), and OsELF4a interacts with OsELF3a (N) and OsELF3b (O), respectively.
Loss of OsELF4a Function Delayed Flowering Time Under LD Condition
In Arabidopsis, disruption of ELF4 results in early flowering and attenuation of rhythmicity, while overexpression of ELF4 delays flowering (Doyle et al., 2002; Kikis et al., 2005). In addition, two ELF4-Like genes, EFL1 and EFL3, are involved in flowering time regulation. Overexpression of EFL1 in elf4 shows normal rhythmicity and delays flowering; overexpression of EFL3 in elf4 partially rescues the early flowering phenotype of elf4 (Lin et al., 2019). To characterize the function of OsELF4a, we created the loss-of-function mutants of OsELF4a from Nipponbare (WT) using the CRISPR/Cas9 method. In total, five transgenic lines with effective editing in the ORF of OsELF4a were obtained (Supplementary Table S6), which could be classified into two types. Four lines harbored a 44-base deletion from 107th to 150th, and one line harbored a 43-base deletion from 107th to 149th. The two types of deletions were both predicted to encode a truncated short polypeptide consisting of 35 aa plus a missense sequence, leading to complete loss of the conserved DUF1313 domain (Supplementary Figure S6, Supplementary Table S6). Thus, both of them should be null alleles of OsELF4a. There was no significant phenotypic difference between these two types of edited lines. We named them Oself4a-1 and Oself4a-2, respectively.
To investigate whether OsELF4a is involved in flowering time regulation, we planted Oself4a mutants and examined their heading dates under the NLD of summer in Fuzhou and the NSD of winter in Sanya. The Oself4a mutants showed a marginal (1 or 2 days of) delay in flowering under NSD (Figure 7A), but much greater delay (9 ± 0.45 days, n > 30) under NLD compared with WT (Figure 7B). Other main agronomic traits all remained unchanged between WT and the Oself4a mutants under both NSD and NLD (Figures 7A,B). To confirm this result, we grew the Oself4 mutants and WT under SD (10/14 h light/dark) and LD (14/10 h light/dark) conditions in the growth chamber. The results agreed with those under the natural field conditions (Figures 7C,D). These observations reveal that OsELF4a functions as a floral promoter under LD condition.

Figure 7. Oself4a and WT plants grown under different day-length conditions. (A,B) in the field; (C,D) in the growth chamber. Scale bar = 10 cm.
Discussions
In this study, we identified rice EC composition and characterized its two components OsLUX and OsELF4a. OsLUX is the sole rice LUX ortholog and is critical for regulating heading date. Disruption OsLUX extremely extended the vegetative phase and caused photoperiod-insensitive late flowering and significantly higher grain yield (Figure 1). OsLUX interacted with OsELF4a and OsELF3a or OsELF3b to form two ternary repressive complexes (Figure 6). Unlike OsLUX and ELF3a, OsELF4a functioned as a floral promoter only under LD condition (Figure 7). Our results indicate that the EC genes in rice share the function of promoting flowering. This is agreement with the orthologs of short-day plants, but opposite to the counterparts of long-day species. Taken together, it is likely that rice EC genes display similar but not identical function in controlling flowering time by cooperatively and independently regulating the expression of genes associated with the circadian clock and the output integrators of photoperiodic flowering genes (Figure 8). These findings facilitate our understanding of photoperiodic flowering in plants, especially the short-day crops. We have noticed that when we are preparing this manuscript, a paper of an independent study on the rice EC role in salt tolerance and heading regulation has recently been accepted for publication, in which the findings about the EC composition and the OELF4a role in heading date are almost identical to ours, but those about the function of OsLUX in heading date are significantly different from ours, which may be due to the weak allele used in their study, in which a T-DNA was inserted upstream of its transcription initiation site and reduced the expression of OsLUX, resulting in a slightly delay in heading date under SD condition, but not under LD condition (Wang et al., 2021; in press).
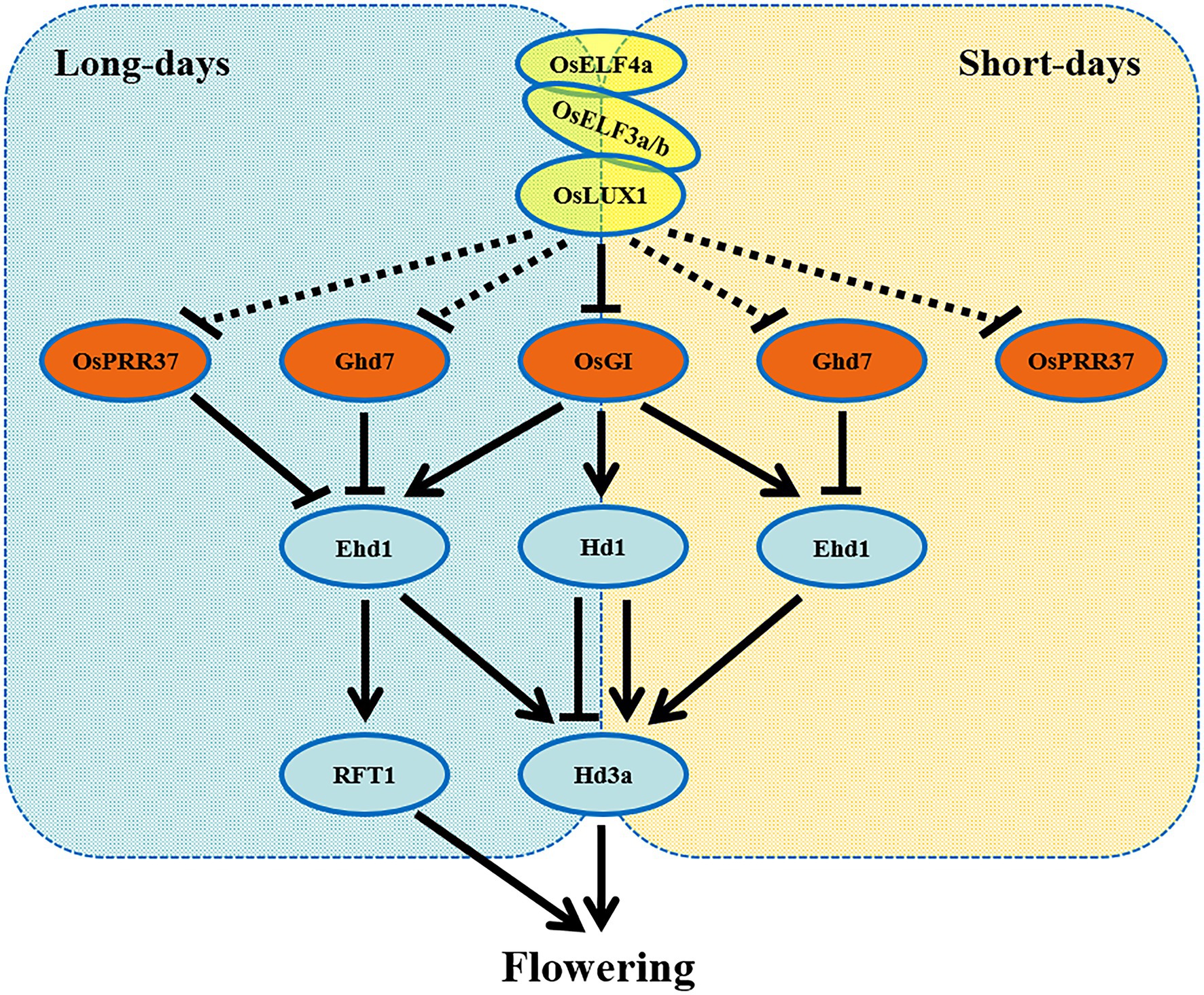
Figure 8. A model for EC-mediated photoperiodic control of flowering in rice. The EC, composed of OsLUX, OsELF3a or OsELF3b, and OsELF4a, is an integrant of repressing flowering in rice. EC controls photoperiodic flowering via repressing the expression of PRR37, OsGI, and Ghd7 under both LD and SD conditions, so as to regulate the expression of the critical output integrators Hd1, Ehd1, Hd3a, and RFT1 in rice.
EC Genes Show Conserved and Divergent Roles Across Species
The EC components LUX, ELF3, and ELF4 can be found in the genomes of land plant lineages, and increasing evidence has shown that EC genes function in flowering time. In Arabidopsis, EC members share a similar expression pattern and almost conserved molecular function. Disruption of any of its components leads to common phenotypic defects, displaying arrhythmic circadian period, long hypocotyl, and photoperiod-insensitive early flowering (Nusinow et al., 2011). Meanwhile, overexpression of either ELF3 or LUX complements the elf4 mutant phenotype (Herrero et al., 2012). These cross-gene complementation in Arabidopsis reveals an identical functions of EC genes. Interestingly, cross-species complementation for Arabidopsis mutants have also been observed in EC gene orthologs of other plants. For example, both the ELF3 orthologs from monocot Brachypodium and Setaria (Huang et al., 2017) and those from dicot LD plant pea (Weller et al., 2012) and SD plant soybean (Lu et al., 2017) can rescue Arabidopsis elf3 phenotype. Similar to ELF3, soybean LUX2 can complement Arabidopsis lux mutants as well (Liew et al., 2017; Fang et al., 2021). These cross-species complementation between monocots and dicots, and LD and SD plants reveal a widely conserved function in EC genes across plant species.
In addition, the potential divergent roles of EC genes have been observed across species as well. In rice, four EC gene orthologs have been characterized. We have seen that OsLUX, OsELF3a/b, and OsELF4a are essential for promoting photoperiodic flowering, but their mutant phenotypes in flowering time and photoperiodic response are significantly different (Figures 1, 7; Saito et al., 2012; Zhao et al., 2012; Yang et al., 2013; Wang et al., 2021). Apart from affecting photoperiodic flowering, OsLUX also showed profound effects on the global regulation of growth and development in this study. The non-functional alleles altered some pivotal agronomic traits, displaying extremely long juvenile phase and vigorous biomass-driven growth, which led to producing more leaves, tillers, and lateral roots (Figures 1, 2E; Table 1) and directly contributed to the high biomass and grain yield (Figure 1E). OsELF3a has pleiotropic effects as well, but its mutation mainly influences the traits of panicle number and panicle length (Saito et al., 2012), which are markedly different from those of lvp2 (Figures 1, 2; Table 1). Disruption of OsELF4 did not cause significant changes in other traits in all day length. These observations showed that, unlike the case in Arabidopsis, the fundamental functions of the four rice EC genes, OsLUX, OsELF4, OsELF3a/3b, are not completely consistent with each other in most agronomic characters including flowering time.
Besides, we have also noticed that EC genes have inhibiting effect on flowering in LD species, such as pea, lentil, wheat, and barley. All mutant alleles of EC gene orthologs identified in these species show early flowering regardless of day length, resembling the mutant alleles of LUX, ELF3, and ELF4 in Arabidopsis (Liew et al., 2009; Faure et al., 2012; Mizuno et al., 2012; Weller et al., 2012; Zakhrabekova et al., 2012; Campoli et al., 2013; Gawroński et al., 2014; Boden et al., 2014; Liew et al., 2014; Mizuno et al., 2016). However, all mutant alleles of LUX, ELF3, and ELF4 orthologs identified so far in SD crops, such as rice and soybean, have later flowering time than wild-type lines (Lu et al., 2017; Bu et al., 2021; Fang et al., 2021; Wang et al., 2021; Figure 7), implying that EC genes promote flowering in SD plants, opposite to the counterparts in LD plants. The functional difference of EC components in SD plants and LD plants probably resides in the regulation or activity of downstream integrator components, rather than in the EC gene itself. This speculation, however, needs further experimental verification.
The Possible Pathway for EC’s Promoting Flowering in Rice
Photoperiodic flowering in plant is closely tied to EC, a core component of circadian clock. In Arabidopsis, EC plays a vital role in circadian rhythms and flowering. As an integrant of repressing flowering, EC represses the expression of circadian genes PRR7, PRR9, and GI directly and of output genes, such as CO, FLC, and FT indirectly, so as to control flowering time (Nusinow et al., 2011; Chow et al., 2012; Mizuno et al., 2014; Song et al., 2015; Silva et al., 2020). Unlike the case in Arabidopsis, where only one FT presents, there are two florigens in rice, Hd3a and RFT1, and thus two pathways, including the conserved OsGI-Hd1-Hd3a pathway (similar to the Arabidopsis GI-CO-FT) and a unique Ghd7-Ehd1-Hd3a/RFT1 pathway, have evolved to adapt to the photoperiodic flowering under SD or LD condition. Each pathway is regulated by the coincidence of the internal circadian clock and the external photoperiodic information (Song et al., 2015).
In rice, OsLUX interacts with OsELF4a and OsELF3a or OsELF3b to form two ECs (Figure 6). Nevertheless, the interaction between OsELF3a and OsLUX is stronger than that between OsELF3b and OsLUX (Wang et al., 2021). Two ELF3 orthologs are likely simultaneously required for the control of flowering time, but OsELF3a plays a more predominant role (Saito et al., 2012; Zhao et al., 2012; Yang et al., 2013), while the contribution of OsELF3b to flowering time remains controversy (Fu et al., 2009; Zhao et al., 2012). These findings suggest that the OsLUX-OsELF3a-OsELF4a is likely the dominant promoter for photoperiodic flowering.
We found that OsLUX participated the regulation of rice photoperiodic flowering via coordinating the expression of genes associated with the circadian clock and the output integrators under both LD and SD conditions. As a transcriptional repressor, OsLUX suppressed Ghd7 expression, while its non-functional mutation elevated the expression of Ghd7 under both LD and SD, leading to reduced expression of Ehd1, Hd3a, and RFT1 (Figures 4, 5). Besides, OsLUX also negatively regulated the expression of OsGI (Figures 4A,B) and OsPRR37 (Figures 4C,D), of which the former is responsible for Hd1 and Ehd1 expression (Hayama et al., 2003; Itoh et al., 2010), and the latter preferentially suppresses the expression of Ehd1 but not Hd1, making the expression of Hd3a and RFT1 reduced under LD, but not under SD (Gao et al., 2014; Liu et al., 2018). Interestingly, similar to that of OsLUX, OsELF3a also acts as a flowering activator under both LD and SD conditions, and disruption of OsELF3a function causes similar expression alteration of these genes associated with photoperiodic flowering under various conditions (Saito et al., 2012; Zhao et al., 2012). Nevertheless, OsELF4a displays a regulatory mechanism similar to those of OsLUX and OsELF3a under SD condition, but a more complicated regulatory mechanism under LD condition (Wang et al., 2021). In addition, Arabidopsis ELF4-like proteins do not appear in EC, but influence flowering time (Lin et al., 2019). Therefore, it will be of interest to further investigate the roles of ELF4-like genes in rice heading.
Taken together, as an integrant, OsLUX, OsELF3a, and OsELF4a promote rice flowering through the same pathway. It is likely that they regulate the expression of genes associated with photoperiodic flowering cooperatively and independently, similar to soybean GmLUX2 and GmELF3a (Fang et al., 2021). Based on previous findings (Wei et al., 2020; Wang et al., 2021) and our results, we draw a simple outline model to explain how EC regulates photoperiodic flowering in rice (Figure 8). As a complex of transcriptional repressor, EC acts at the upstream of PRR37 and Ghd7 (genetically) and OsGI (physically), repressing the expression of these genes under both LD and SD (Figures 4, 5; Wang et al., 2021), so as to regulate the expression of Ehd1 and Hd1. Ehd1 and Hd1 determine the expression level of florigenic genes Hd3a and RFT1 through distinct pathways under a given photoperiod. Hd1 is crucial for SD photoperiodic induction of Hd3a (Izawa, 2007). Ehd1 promotes flowering independent of Hd1 under SD, but also promotes flowering under LD when Hd1 represses Hd3a expression, whereas Ghd7 negatively regulates Ehd1 expression (Woods et al., 2014).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YD supervised the project. YD and WW conceived and designed the research plans. YD, ZC, YZ, WT, XC, CL, YL, and YY performed the experiments and collected and analyzed the data. YD and WW analyzed the data wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 31871600), Sci-Tech Innovation Special Fund of Fujian Agriculture and Forestry University (No. CXZX2018112 and CXZX2016160), the Natural Science Foundation of Fujian Province (No. 2021J01074), and Chinese Scholarship Council (201908350032).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Mathew M. S. Evans (Department of Plant Biology, Carnegie Institution for Science, United States) and the reviewers for offering valuable suggestions to improve the manuscript. We thank Yongquan Wang, Chuanlei Wang, and Qingan Peng (Fujian Agriculture & Forestry University, CHINA) for their helps in the experiment. No conflict of interest declared.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.853042/full#supplementary-material
References
Bendix, C., Marshall, C. M., and Harmon, F. G. (2015). Circadian clock genes universally control key agricultural traits. Mol. Plant 8, 1135–1152. doi: 10.1016/j.molp.2015.03.003
Boden, S. A., Weiss, D., Ross, J. J., Davies, N. W., Trevaskis, B., Chandler, P. M., et al. (2014). EARLY FLOWERING3 regulates flowering in spring barley by mediating gibberellin production and FLOWERING LOCUS T expression. Plant Cell. 26, 1557–1569. doi: 10.1105/tpc.114.123794
Bu, T., Lu, S., Wang, K., Dong, L., Li, S., Xie, Q., et al. (2021). A critical role of the soybean evening complex in the control of photoperiod sensitivity and adaptation. Proc. Natl. Acad. Sci. 118:e2010241118. doi: 10.1073/pnas.2010241118
Campoli, C., Pankin, A., Drosse, B., Casao, C. M., Davis, S. J., and Korff, M. (2013). HvLUX1 is a candidate gene underlying theearly maturity 10 locus in barley: phylogeny, diversity, and interactions with the circadian clock and photoperiodic pathways. New Phytol. 199, 1045–1059. doi: 10.1111/nph.12346
Choi, D. S., Hwang, I. S., and Hwang, B. K. (2012). Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 24, 1675–1690. doi: 10.1105/tpc.112.095869
Chow, B. Y., Helfer, A., Nusinow, D. A., and Kay, S. A. (2012). ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signal. Behav. 7, 170–173. doi: 10.4161/psb.18766
Dixon, L. E., Knox, K., Kozma-Bognar, L., Southern, M. M., Pokhilko, A., and Millar, A. J. (2011). Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in arabidopsis. Curr. Biol. 21, 120–125. doi: 10.1016/j.cub.2010.12.013
Doyle, M. R., Davis, S. J., Bastow, R. M., McWatters, H. G., Kozma-Bognár, L., Nagy, F., et al. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77. doi: 10.1038/nature00954
Duan, Y., Chen, Y., Li, W., Pan, M., Qu, X., Shi, X., et al. (2019). RETINOBLASTOMA-RELATED genes specifically control inner floral organ morphogenesis and pollen development in rice. Plant Physiol. 181, 1600–1614. doi: 10.1104/pp.19.00478
Fang, X., Han, Y., Liu, M., Jiang, J., Li, X., Lian, Q., et al. (2021). Modulation of evening complex activity enables north-to-south adaptation of soybean. Sci. China Life Sci. 64, 179–195. doi: 10.1007/s11427-020-1832-2
Faure, S., Turner, A. S., Gruszka, D., Christodoulou, V., Davis, S. J., von Korff, M., et al. (2012). Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. 109, 8328–8333. doi: 10.1073/pnas.1120496109
Fogelmark, K., and Troein, C. (2014). Rethinking transcriptional activation in the arabidopsis circadian clock. PLoS Comput. Biol. 10:e1003705. doi: 10.1371/journal.pcbi.1003705
Fu, C., Yang, X. O., Chen, X., Chen, W., Ma, Y., Hu, J., et al. (2009). OsEF3, a homologous gene of Arabidopsis ELF3, has pleiotropic effects in rice. Plant Biol. 11, 751–757. doi: 10.1111/j.1438-8677.2008.00156.x
Gao, H., Jin, M., Zheng, X., Chen, J., Yuan, D., Xin, Y., et al. (2014). Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. 111, 16337–16342. doi: 10.1073/pnas.1418204111
Gawroński, P., Ariyadasa, R., Himmelbach, A., Poursarebani, N., Kilian, B., Stein, N., et al. (2014). A distorted circadian clock causes early flowering and temperature-dependent variation in spike development in the Eps-3Am mutant of einkorn wheat. Genetics 196, 1253–1261. doi: 10.1534/genetics.113.158444
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
He, F., Zhang, F., Sun, W., Ning, Y., and Wang, G. (2018). Correction to: a versatile vector toolkit for functional analysis of rice genes. Rice 11:49. doi: 10.1186/s12284-018-0238-x
Helfer, A., Nusinow, D. A., Chow, B. Y., Gehrke, A. R., Bulyk, M. L., and Kay, S. A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the arabidopsis core clock. Curr. Biol. 21, 126–133. doi: 10.1016/j.cub.2010.12.021
Herrero, E., Kolmos, E., Bujdoso, N., Yuan, Y., Wang, M., Berns, M. C., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428–443. doi: 10.1105/tpc.111.093807
Hicks, K. A. (2001). EARLY FLOWERING3 encodes a novel protein that regulates circadian clock function and flowering in Arabidopsis. Plant Cell Online 13, 1281–1292. doi: 10.1105/tpc.13.6.1281
Huang, H., Gehan, M. A., Huss, S. E., Alvarez, S., Lizarraga, C., Gruebbling, E. L., et al. (2017). Cross-species complementation reveals conserved functions for EARLY FLOWERING 3 between monocots and dicots. Plant Direct 1:e00018. doi: 10.1002/pld3.18
Huang, H., and Nusinow, D. A. (2016). Into the evening: complex interactions in the arabidopsis circadian clock. Trends Genet. 32, 674–686. doi: 10.1016/j.tig.2016.08.002
Ishikawa, R., Aoki, M., Kurotani, K., Yokoi, S., Shinomura, T., Takano, M., et al. (2011). Phytochrome B regulates heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol. Gen. Genomics. 285, 461–470. doi: 10.1007/s00438-011-0621-4
Itoh, H., Nonoue, Y., Yano, M., and Izawa, T. (2010). A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 42, 635–638. doi: 10.1038/ng.606
Izawa, T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58, 3091–3097. doi: 10.1093/jxb/erm159
Izawa, T. (2012). Physiological significance of the plant circadian clock in natural field conditions. Plant Cell Environ. 35, 1729–1741. doi: 10.1111/j.1365-3040.2012.02555.x
Izawa, T., Mihara, M., Suzuki, Y., Gupta, M., Itoh, H., Nagano, A. J., et al. (2011). Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 23, 1741–1755. doi: 10.1105/tpc.111.083238
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Kikis, E. A., Khanna, R., and Quail, P. H. (2005). ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 44, 300–313. doi: 10.1111/j.1365-313X.2005.02531.x
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., et al. (2002). Hd3a, a rice ortholog of the arabidopsis FT gene, promotes transition to flowering downstream of hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Kolmos, E., Nowak, M., Werner, M., Fischer, K., Schwarz, G., Mathews, S., et al. (2010). Integrating ELF4 into the circadian system through combined structural and functional studies. HFSP J. 3, 350–366. doi: 10.2976/1.3218766
Kong, L., Duan, Y., Ye, Y., Cai, Z., Wang, F., Qu, X., et al. (2019). Screening and analysis of proteins interacting with OsMADS16 in rice (Oryza sativa L.). PLoS One 14:e221473, e0221473. doi: 10.1371/journal.pone.0221473
Koo, B., Yoo, S., Park, J., Kwon, C., Lee, B., An, G., et al. (2013). Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol. Plant 6, 1877–1888. doi: 10.1093/mp/sst088
Lee, Y., and An, G. (2015). OsGI controls flowering time by modulating rhythmic flowering time regulators preferentially under short day in rice. J. Plant Biol. 58, 137–145. doi: 10.1007/s12374-015-0007-y
Liew, L. C., Hecht, V., Laurie, R. E., Knowles, C. L., Vander Schoor, J. K., Macknight, R. C., et al. (2009). DIE NEUTRALIS and LATE BLOOMER1 contribute to regulation of the pea circadian clock. Plant Cell 21, 3198–3211. doi: 10.1105/tpc.109.067223
Liew, L. C., Hecht, V., Sussmilch, F. C., and Weller, J. L. (2014). The pea photoperiod response GeneSTERILE NODES is an ortholog of LUX ARRHYTHMO. Plant Physiol. 165, 648–657. doi: 10.1104/pp.114.237008
Liew, L. C., Singh, M. B., and Bhalla, P. L. (2017). A novel role of the soybean clock gene LUX ARRHYTHMO in male reproductive development. Sci. Rep. 7:10605. doi: 10.1038/s41598-017-10823-y
Lin, H. X., Yamamoto, T., Sasaki, T., and Yano, M. (2000). Characterization and detection of epistatic interactions of 3 QTLs, Hd1, Hd2, and Hd3, controlling heading date in rice using nearly isogenic lines. Theor. Appl. Genet. 101, 1021–1028. doi: 10.1007/s001220051576
Lin, K., Zhao, H., Gan, S., and Li, G. (2019). Arabidopsis ELF4-like proteins EFL1 and EFL3 influence flowering time. Gene 700, 131–138. doi: 10.1016/j.gene.2019.03.047
Liu, C., Qu, X., Zhou, Y., Song, G., Abiri, N., Xiao, Y., et al. (2018). OsPRR37 confers an expanded regulation of the diurnal rhythms of the transcriptome and photoperiodic flowering pathways in rice. Plant Cell Environ. 41, 630–645. doi: 10.1111/pce.13135
Lu, S., Zhao, X., Hu, Y., Liu, S., Nan, H., Li, X., et al. (2017). Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat. Genet. 49, 773–779. doi: 10.1038/ng.3819
Matsubara, K., Ogiso-Tanaka, E., Hori, K., Ebana, K., Ando, T., and Yano, M. (2012). Natural variation in hd17, a homolog of arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 53, 709–716. doi: 10.1093/pcp/pcs028
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Mizuno, N., Kinoshita, M., Kinoshita, S., Nishida, H., Fujita, M., Kato, K., et al. (2016). Loss-of-function mutations in three homoeologous PHYTOCLOCK 1 genes in common wheat are associated with the extra-early flowering phenotype. PLoS One 11:e165618. doi: 10.1371/journal.pone.0165618
Mizuno, N., Nitta, M., Sato, K., and Nasuda, S. (2012). A wheat homologue of PHYTOCLOCK 1 is a candidate gene conferring the early heading phenotype to einkorn wheat. Genes Genet. Syst. 87, 357–367. doi: 10.1266/ggs.87.357
Mizuno, T., Nomoto, Y., Oka, H., Kitayama, M., Takeuchi, A., Tsubouchi, M., et al. (2014). Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol. 55, 958–976. doi: 10.1093/pcp/pcu030
Murakami, M., Tago, Y., Yamashino, T., and Mizuno, T. (2007). Comparative overviews of clock-associated genes of Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 110–121. doi: 10.1093/pcp/pcl043
Nemoto, Y., Nonoue, Y., Yano, M., and Izawa, T. (2016). Hd1, a CONSTANS ortholog in rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 86, 221–233. doi: 10.1111/tpj.13168
Nusinow, D. A., Helfer, A., Hamilton, E. E., King, J. J., Imaizumi, T., Schultz, T. F., et al. (2011). The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. doi: 10.1038/nature10182
Onai, K., and Ishiura, M. (2005). PHYTOCLOCK 1 encoding a novel GARP protein essential for the Arabidopsis circadian clock. Genes Cells 10, 963–972. doi: 10.1111/j.1365-2443.2005.00892.x
Park, D. H., Somers, D. E., Kim, Y. S., Choy, Y. H., Lim, H. K., Soh, M. S., et al. (1999). Control of circadian rhythms and photoperiodic flowering by the arabidopsis GIGANTEA gene. Science 285, 1579–1582. doi: 10.1126/science.285.5433.1579
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Saito, H., Ogiso-Tanaka, E., Okumoto, Y., Yoshitake, Y., Izumi, H., Yokoo, T., et al. (2012). Ef7 encodes an ELF3-like protein and promotes rice flowering by negatively regulating the floral repressor gene ghd7 under both short- and long-day conditions. Plant Cell Physiol. 53, 717–728. doi: 10.1093/pcp/pcs029
Sawa, M., and Kay, S. A. (2011). GIGANTEA directly activates flowering locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 108, 11698–11703. doi: 10.1073/pnas.1106771108
Shim, J. S., and Imaizumi, T. (2015). Circadian clock and photoperiodic response in Arabidopsis: from seasonal flowering to redox homeostasis. Biochemistry 54, 157–170. doi: 10.1021/bi500922q
Shrestha, R., Gómez-Ariza, J., Brambilla, V., and Fornara, F. (2014). Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 114, 1445–1458. doi: 10.1093/aob/mcu032
Silva, C. S., Nayak, A., Lai, X., Hutin, S., Hugouvieux, V., Jung, J. H., et al. (2020). Molecular mechanisms of evening complex activity in Arabidopsis. Proc. Natl. Acad. Sci. 117, 6901–6909. doi: 10.1073/pnas.1920972117
Song, Y. H., Shim, J. S., Kinmonth-Schultz, H. A., and Imaizumi, T. (2015). Photoperiodic flowering: time measurement mechanisms in leaves. Annu. Rev. Plant Biol. 66, 441–464. doi: 10.1146/annurev-arplant-043014-115555
Suárez-López, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Sun, C., Fang, J., Zhao, T., Xu, B., and Zhang, F. (2012). The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24, 3235–3247. doi: 10.1105/tpc.112.101436
Tsuji, H., Taoka, K., and Shimamoto, K. (2011). Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 14, 45–52. doi: 10.1016/j.pbi.2010.08.016
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS t moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Wang, X., He, Y., Wei, H., and Wang, L. (2021). A clock regulatory module is required for salt tolerance and control of heading date in rice. Plant Cell Environ. 44, 3283–3301. doi: 10.1111/pce.14167
Wei, H., Wang, X., Xu, H., and Wang, L. (2020). Molecular basis of heading date control in rice. Abiotech 1, 219–232. doi: 10.1007/s42994-020-00019-w
Weller, J. L., Liew, L. C., Hecht, V. F. G., Rajandran, V., Laurie, R. E., Ridge, S., et al. (2012). A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc. Natl. Acad. Sci. 109, 21158–21163. doi: 10.1073/pnas.1207943110
Woods, D. P., Ream, T. S., Minevich, G., Hobert, O., and Amasino, R. M. (2014). PHYTOCHROME c is an essential light receptor for photoperiodic flowering in the temperate grass, brachypodium distachyon. Genetics 198, 397–408. doi: 10.1534/genetics.114.166785
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yang, Y., Peng, Q., Chen, G., Li, X., and Wu, C. (2013). OsELF3 is involved in circadian clock regulation for promoting flowering under long-day conditions in rice. Mol. Plant 6, 202–215. doi: 10.1093/mp/sss062
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time GeneCONSTANS. Plant Cell 12, 2473–2483. doi: 10.1105/tpc.12.12.2473
Zakhrabekova, S., Gough, S. P., Braumann, I., Muller, A. H., Lundqvist, J., Ahmann, K., et al. (2012). Induced mutations in circadian clock regulator mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl. Acad. Sci. 109, 4326–4331. doi: 10.1073/pnas.1113009109
Keywords: rice, vegetative growth, photoperiodic flowering, LUX ARRHYTHMO, evening complex
Citation: Cai Z, Zhang Y, Tang W, Chen X, Lin C, Liu Y, Ye Y, Wu W and Duan Y (2022) LUX ARRHYTHMO Interacts With ELF3a and ELF4a to Coordinate Vegetative Growth and Photoperiodic Flowering in Rice. Front. Plant Sci. 13:853042. doi: 10.3389/fpls.2022.853042
Edited by:
Kun Lu, Southwest University, ChinaReviewed by:
Kiyosumi Hori, National Agriculture and Food Research Organization (NARO), JapanJun Fang, Key Laboratory of Soybean Molecular Design and Breeding, Northeast Institute of Geography and Agroecology (CAS), China
Ting Zhang, Southwest University, China
Zhen-Hua Zhang, China National Rice Research Institute (CAAS), China
Copyright © 2022 Cai, Zhang, Tang, Chen, Lin, Liu, Ye, Wu and Duan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiren Wu, d3V3ckBmYWZ1LmVkdS5jbg==; Yuanlin Duan, eWxkdWFuODYzQGZhZnUuZWR1LmNu
†These authors have contributed equally to this work
 Zhengzheng Cai
Zhengzheng Cai Yudan Zhang1,2†
Yudan Zhang1,2† Xuequn Chen
Xuequn Chen Weiren Wu
Weiren Wu Yuanlin Duan
Yuanlin Duan