
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 11 April 2022
Sec. Plant Biotechnology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.850924
This article is part of the Research Topic Targeted Genome Editing for Crop Improvement View all 20 articles
 Haiyan Zhang1
Haiyan Zhang1 Yuqin Shi1
Yuqin Shi1 Mengdan Sun1
Mengdan Sun1 Xuezhi Hu1
Xuezhi Hu1 Mengyu Hao1
Mengyu Hao1 Yu Shu1
Yu Shu1 Xue-Rong Zhou2
Xue-Rong Zhou2 Qiong Hu1
Qiong Hu1 Chao Li1*
Chao Li1* Desheng Mei1*
Desheng Mei1*
Tocopherols are essential nutrients for human health known as vitamin E. Vitamin E deficiency can have a profound effect on human health, including the central nervous system and cardiovascular and immune protection. Multiple enzymatic steps are involved in the conversion between different forms of tocopherols. Among them, γ-tocopherol methyltransferase encoded by gene VTE4 catalyzes the conversion of γ- to α-tocopherol or δ- to β-tocopherol isoforms. However, the gene copies and their functional contribution of VTE4 homologs in Brassica napus were not elucidated. To this end, different mutation combinations of four putative BnVTE4 homologous copies were generated by using CRISPR/Cas9 genome editing technology. Editing of those BnVTE4 homologs led to a significant change of the α-tocopherol content and the ratio between α- and γ-tocopherol compared with wide-type control. Analysis of the different combinations of BnVTE4-edited homologs revealed that the contribution of the BnVTE4 individual gene displayed obvious functional differentiation in α-tocopherol biosynthesis. Their contribution could be in order of VTE4.C02-2 (BnaC02G0331100ZS) > VTE4.A02-1 (BnaA02G0247300ZS) > VTE4.A02-2 (BnaA02G0154300ZS). Moreover, the VTE4.A02-1 and VTE4.A02-2 copies might have severe functional redundancies in α-tocopherol biosynthesis. Overall, this study systemically studied the different effects of BnVTE4 homologs, which provided a theoretical basis for breeding high α-tocopherol content oilseed rape.
Tocopherols and tocotrienols are also called vitamin E. In this study, we focus on the analysis of tocopherols only. Tocopherols are lipid-soluble strong antioxidants that are involved in the protection of oxidative damage of membrane lipids by scavenging singlet oxygen and other reactive oxygen species (ROS) (Trebst et al., 2002; Schneider, 2005; Fritsche et al., 2017). Thus, tocopherols are presumed to be key scavengers of senescence or stress-induced lipid radicals and ROS in plants. Their strong antioxidant capacity can reduce lipid peroxy radicals to corresponding hydrogen peroxide to avoid lipid peroxidation of polyunsaturated fatty acids. The high antioxidative property exerts a protective role in multiple plant stress responses such as cold and drought stress (Janeczko et al., 2018; Ma et al., 2020). Most importantly, this effect can also be extended to human health. Vitamin E is an essential nutrient in the human daily diet. Vitamin E deficiency primarily causes muscle atrophy and reproductive and neurologic dysfunctions (Martin et al., 2013; Kumar et al., 2018), whereas the adequate intake of vitamin E can prevent neurological diseases, cataracts, coronary heart disease, atherosclerosis, diabetes, Parkinson’s disease, Alzheimer’s disease, and vision diseases (Lloret et al., 2019; Rozanowska et al., 2019). Thus, biofortification of vitamin E in crop plants is not only beneficial for human health but also for plant stress response.
Based on the different numbers and positions of methyl substituents on the aromatic ring, tocopherols are defined as four isoforms, namely α-, β-, γ-, and δ-tocopherol (α-T, β-T, γ-T, and δ-T in short). Although those tocopherols have similar antioxidant activities in vitro, the vitamin E activity in vivo is significantly different. Among them, α-T possesses the highest vitamin E activity in vivo and is also a common type of tocopherol in the European diet (Galmés et al., 2018). It has been proven that α-T supplementation can improve cell-mediated immunity, and only α-T is selected to set the recommended dietary allowance (RDA) of vitamin E for Americans (Meydani et al., 1990; Meydani et al., 1997; Institute of Medicine (US) Panel on Dietary Antioxidants and Related Compounds, 2000; Ranard and Erdman, 2018). Tocopherols are only synthesized on the inner chloroplast membrane of photosynthetic organisms, including plants, green algae, and some cyanobacteria. Undoubtedly, a daily supplement of α-T derived from plant-based food is a safe and natural way to ensure human health.
Oilseed rape (Brassica napus L., AACC, 2n = 38) is one of the most important resources of edible vegetable oil in the world, accounting for about 16% of the total global vegetable oil production (Cheng et al., 2019). High-quality rapeseed oil, containing multiple beneficial nutrients including vitamin E, is the dominant edible vegetable oil in China, Europe, and Canada. A daily supplement of vitamin E via rapeseed oil is the safest and most effective way to keep the nutritional requirement for the human body. Therefore, genetic improvement of vitamin E content has been considered a key breeding objective (Cheng et al., 2019; Yan et al., 2021). However, oilseed rape is a typical allotetraploid crop, and most of the genes have multiple homologous copies with redundant or diverse functions (Chalhoub et al., 2014; Braatz et al., 2017; Zaman et al., 2019, 2021; Li et al., 2021). Clarification of the detailed roles of each homologous copy is the basis of the further breeding application.
The biosynthesis pathway of tocopherols has been extensively studied in the model plant Arabidopsis thaliana (Valentin et al., 2006; Vom Dorp et al., 2015). Tocopherol biosynthesis begins with the formation of homogentisic acid (HGA), which is catalyzed by p-hydroxyphenylpyruvic acid dioxygenase (HPPD/PDS1) and is derived from the deamination of tyrosine (Norris et al., 1995, 1998; Tsegaye et al., 2002). Phytyldiphosphate (PDP) is formed by phytol kinase, and phytyl—P kinase catalyzes the formation of phytol (Valentin et al., 2006; Vom Dorp et al., 2015). HGA and PDP are condensed to 2-methyl-6-phytyl-1,4-benzoquinol (MPBQ) catalyzed by the enzyme homogentisatephytyltransferase (HPT), which is encoded by VTE2 gene (Venkatesh et al., 2006). The 2-methyl-6-phyty-1,4-benzoquinol methyltransferase (MPBQ MT) methylates MPBQ to 2,3-dimethyl-6-phytyl-1,4-benzoquinone (DMPBQ), while MPBQ and DMPBQ are transformed into γ- and δ-tocopherol, respectively, by tocopherol cyclase (TC). TC is encoded by the VTE1 gene, and MPBQ MT is encoded by VTE3 gene (Porfirova et al., 2002; Semchuk et al., 2009). Then, the γ-Tmethyltransferase (γ-TMT) encoded by VTE4 gene catalyzes the conversion of δ- to β-T and γ- to α-T (Porfirova et al., 2002; Bergmüller et al., 2003; DellaPenna and Pogson, 2006; Hunter and Cahoon, 2007; Figure 1). Thus, VTE4 directly affects the content of α-T and γ-T. However, the putative functional differentiation of the VTE4 gene in B. napus is still unclear.
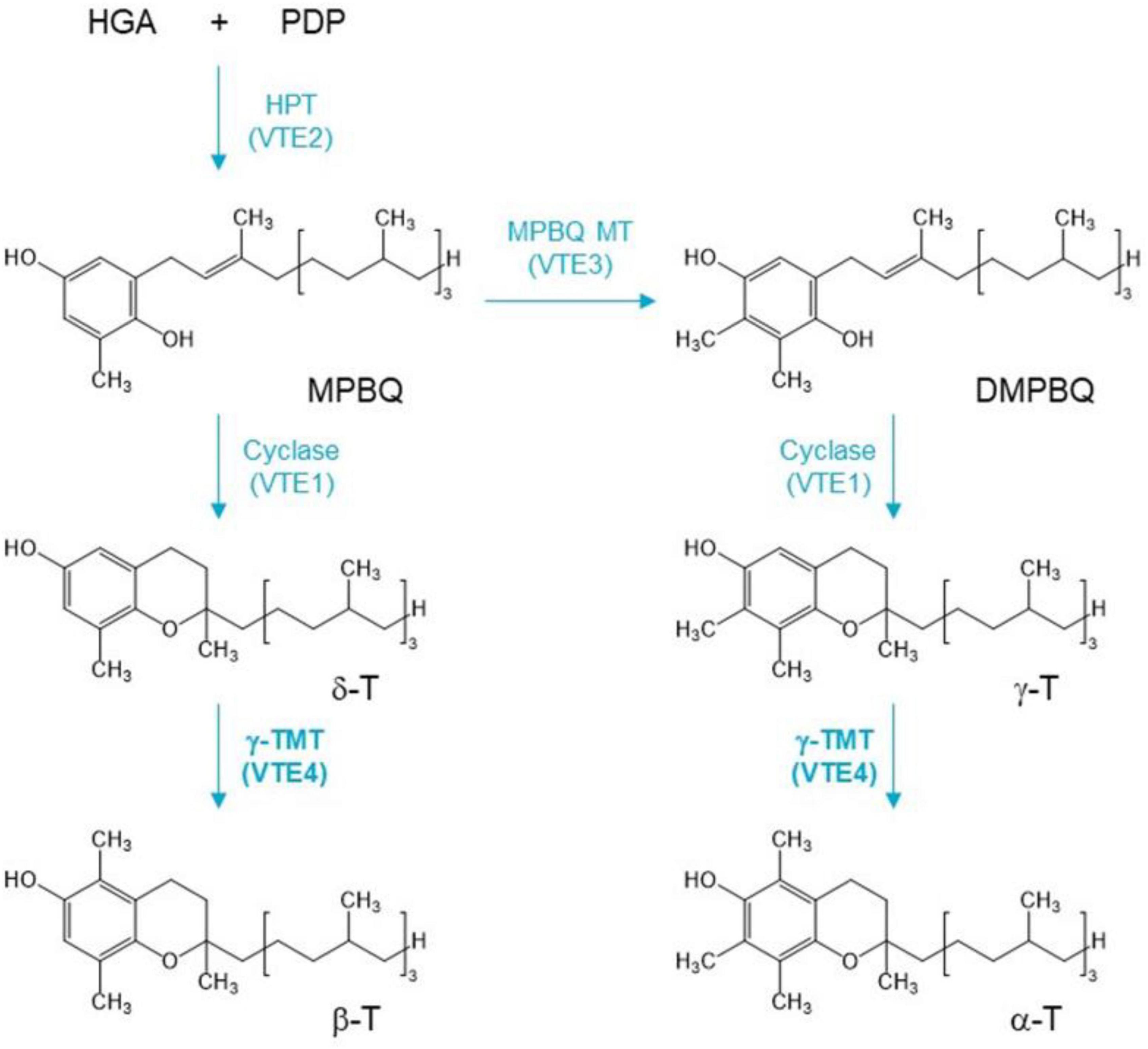
Figure 1. A simplified pathway of vitamin E isoform conversion. HGA, homogentisic acid; PDP, phytyldiphosphate; MPBQ, 2-methyl-6-phytyl-1,4-benzoquinol; DMPBQ, 2,3-dimethyl-6-phytyl-1,4-benzoquinone; α-T, β-T, γ-T, and δ-T are α, β, γ, and δ isoforms of tocopherol. VTE genes are described in the text. γ-Tocopherol methyltransferase (γ-TMT) gene VTE4 (bold) is the target gene to be edited in this study.
In this study, four putative copies of the BnVTE4 gene have been identified from the latest oilseed rape genome database. The functional contribution of BnVTE4 homologs in the α-T biosynthesis is studied by generating different mutation types using CRISPR/Cas9 genome editing technology. This study will shed new light on the breeding application of high α-T content in oilseed rape.
Four homologous copies of the BnVTE4 gene were retrieved in the B. napus genome database,1 namely BnaC02G0197500ZS (VTE4.C02-1), BnaC02G0331100ZS (VTE4.C02-2), BnaA02G0247300ZS (VTE4.A02-1), and BnaA02G015430ZS (VTE4.A02-2). Two sgRNAs with minimal off-target effects were designed using CRISPR-P 2.02 at the conserved sequence positions of the third and fourth exons, namely S1 (GGTGAGCATATGCCTGACA) and S2 (CCATGGGAGCAGAACCTCT). The sgRNA assembly and vector construction were performed as a previous report (Xing et al., 2014; Ma et al., 2015).
The qualified genome editing vector was transferred into Agrobacterium tumefaciens strain (GV3101) by the heat shock method. The vector containing two sgRNAs was introduced into B. napus L. variety “Zhongshuang 6” by the Agrobacterium-mediated transformation (Li et al., 2018). The selection marker was Kanamycin. The T0 generation mutants were planted in the artificial climate room and grown under a photoperiod of the 16 h light/8 h dark at a temperature of 22°C, and the T1 generation was planted in the field of the Hanchuan transgenic base, Hubei, China.
Plant genomic DNA was extracted from leaves by the CTAB (hexadecyltriethyl ammonium bromide) method. We used NPTII gene-specific primers NPTII-F (5′-GATGGATTGCACGCAGGT-3′) and NPTII-R (5′-TCGTCAAGAAGGCGATAGA-3′) for PCR reaction to identify positive transgenic plants.
To identify whether the BnVTE4 gene of the positive transgenic plants had been edited, gene-specific primers (Supplementary Table 1) were used to amplify the DNA sequence containing the target site by PCR, and then Sanger sequencing was used to identify the mutants. The heterozygous mutants were determined by the Hi-TOM platform (Liu et al., 2019). Hi-TOM sequencing consists of two rounds of PCR. In the first round of PCR, gene-specific primers (Supplementary Table 1) were used to amplify the genomic sequence of about 500–2,000 bp around the 4 copies of the target site. In the second round of PCR, gene-specific primers containing Hi-TOM adaptor primers (Supplementary Table 1) were used to amplify the 80–300 bp genomic region around the target site. The products of the second round were sequenced by the company.3
Tocopherol extraction was performed according to the reported method with slight modification (Yu et al., 2016; Xu et al., 2019). A total of 200 mg seeds were placed in a 2 ml centrifuge tube with 1 steel bead of a 5-mm diameter and grounded for 5 min at 60 Hz using a rapid grinder. An accurate 60 mg aliquot was weighed from the ground seeds. Three biological replicates were set up. Tocopherols were extracted by adding 1.5-ml hexane. The mixture was sealed and shaken for 4 h in the dark and then extracted at 4°C for 12 h. The mixture was centrifuged at 10,000 rpm for 10 min, and the supernatant was filtered through a.22-μm organic membrane.
Determination of tocopherols was carried out on high-performance liquid chromatography (HPLC, Waters). Agilent liquid chromatography column ZORBAX RX-SIL (4.6 mm × 250 mm) was used, and the temperature was set at 30°C. The mobile phase was n-Hexane:isopropanol (99:1, v/v) at a flow rate of 1 ml/min. The sample composition was determined qualitatively and quantitatively by UV light at 292 nm. Standards (95%, pure HPLC) for α- and γ-T were purchased from Merck, and all the standards and samples were in 5 μl injection volumes.
Statistical software SPSS v22.0 was used to analyze the data, and one-way ANOVA was employed to comparatively analyze the differences between α- and γ-T of different copy mutation combination materials and wild-type rape seeds (p < 0.05).
Genomic sequence analysis showed that the BnVTE4 gene possesses four homologous copies in B. napus, each of which is composed of six exons and five introns. Two sgRNAs (named S1 and S2) were designed in their conserved sequence regions located in the third and fourth exons, respectively (Figures 2A,B).
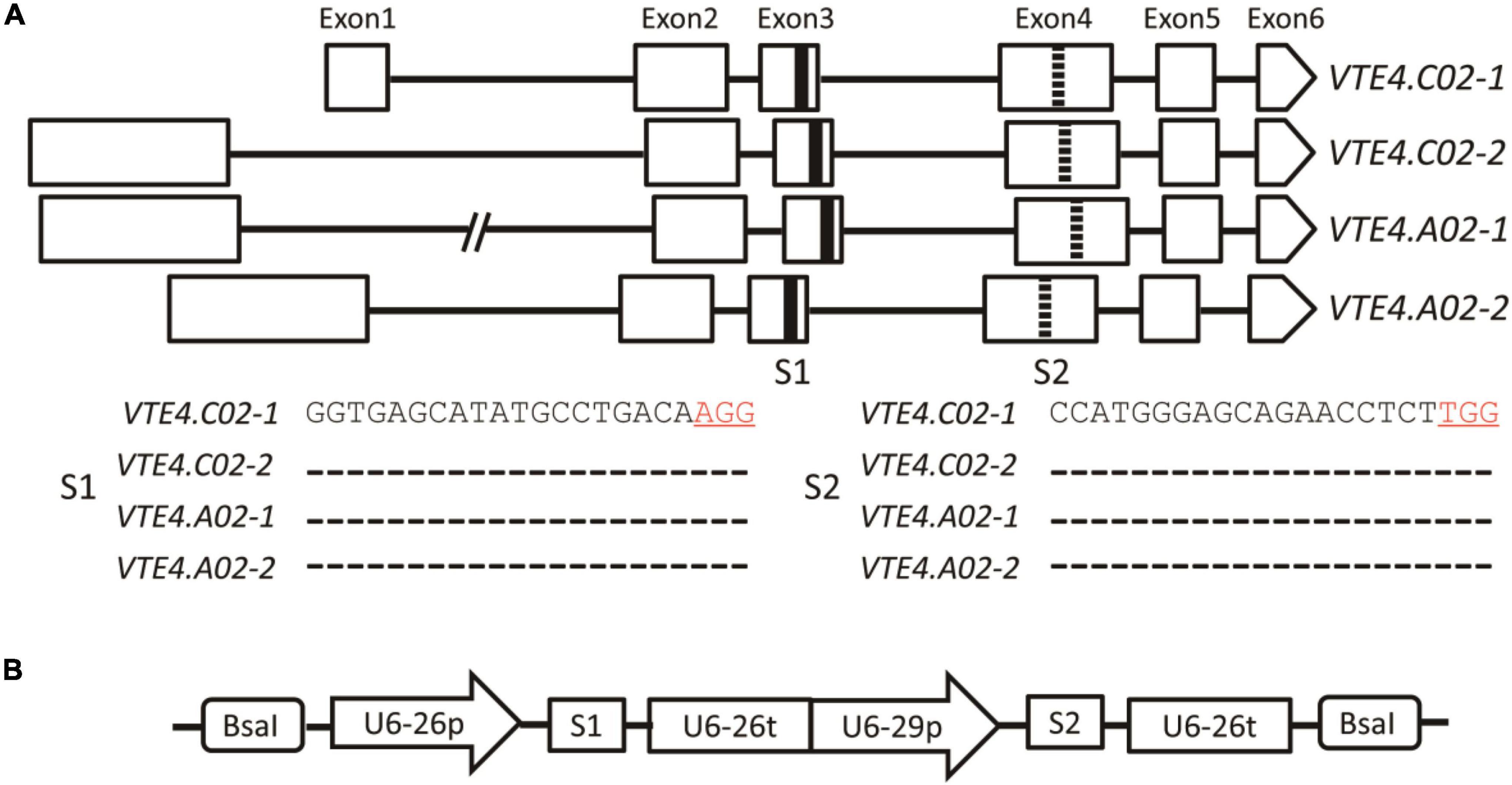
Figure 2. A schematic diagram of BnVTE4 gene editing vector construction. (A) Rectangular boxes indicate exons of VTE4.C02-1, VTE4.C02-2, VTE4.A02-1, and VTE4.A02-2 copies; black horizontal lines represent introns; thickened black vertical lines show sgRNA1 (S1) targeting Exon3; thickened dashed lines indicate sgRNA2 (S2) targeting Exon4. (B) A schematic diagram of the BnVTE4 gene editing vector. sgRNA1 is initiated and terminated by U6-26p and U6-26t, respectively, sgRNA2 is initiated and terminated by U6-29p and U6-26t, respectively.
In order to elucidate the possible functional differentiation of different homologous copies of the BnVTE4 gene during α-T synthesis, we screened different mutation types of BnVTE4 editing in the T1 generation. Five editing types with different mutation combinations were obtained, named bnvte4-1, bnvte4-2, bnvte4-3, bnvte4-4, and bnvte4-5, respectively. Sequencing results indicated that VTE4.C02-1 and VTE4.C02-2 copies were homozygous mutations or wild type (WT) (Figure 3A). All five editing types had a homozygous mutation in the VTE4.C02-1 copy with a single base insertion leading to a frame shift. The VTE4.C02-2 copy also had homozygous mutations with single-base insertions leading to a frame shift in bnvte4-1, bnvte4-3, bnvte4-4, and bnvte4-5, except for bnvte4-2 that was not mutated (Figure 3). VTE4.A02-1 and VTE4.A02-2 copies had heterozygous mutations in some editing types, and Hi-TOM high-throughput sequencing was employed to verify the editing frequency and amino acid changes at the targeted sites (Figures 3B–F and Supplementary Figures 1, 2). VTE4.A02-1 in bnvte4-1 and bnvte4-4 was unmutated (WT). One nucleotide deletion in the VTE4.A02-1 copy of bnvte4-2 caused 64% of the frame shifts (Figures 3A,C). Both bnvte4-3 and bnvte4-5 had one or several nucleotide deletions and one nucleotide insertion in the VTE4.A02-1 copy, 78 and 83%, respectively (Figures 3A,D,F), resulting in frame shifts or amino acid deletions. The VTE4.A02-2 copies of bnvte4-1 had a deletion of one nucleotide, resulting in frame shifts (Figures 3A,B). The VTE4.A02-2 copies of bnvte4-2 had a deletion of one nucleotide, resulting in frame shifts that accounted for only 6%, and the one nucleotide substitution without amino acid change accounted for only 1% (Figures 3A,C). Both bnvte4-3 and bnvte4-5 had one or more nucleotide deletion and one nucleotide insertion in the VTE4.A02-2 copy, accounting for 88 and 90%, respectively (Figures 3A,D,F), resulting in frame shifts or amino acid deletions. The VTE4.A02-2 copy of bnvte4-4 had one or more nucleotide deletions, resulting in frame shifts of just 12% (Figures 3A,E).
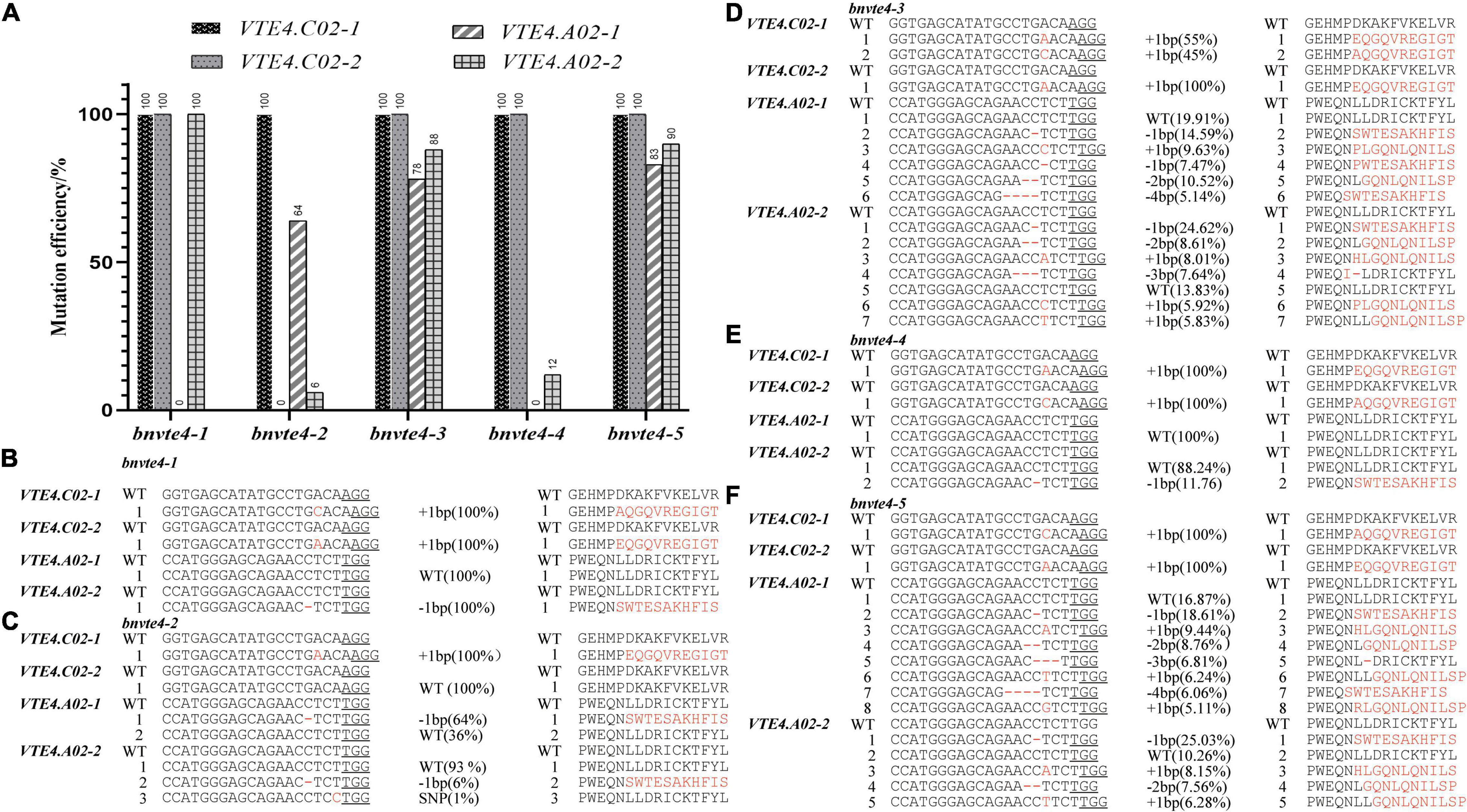
Figure 3. Mutation type, mutation efficiency, and amino acid alterations in homologous copies of the BnVTE4 gene in the T1 generation. (A) Mutation efficiency of each copy of the BnVTE4 mutant. (B) Mutation types and amino acid alterations in homologous copies in the BnVTE4 gene of bnvte4-1. (C) Mutation types and amino acid alterations in homologous copies in the BnVTE4 gene of bnvte4-2. (D) Mutation types and amino acid alterations in homologous copies in the BnVTE4 gene of bnvte4-3. (E) Mutation types and amino acid alterations in homologous copies in the BnVTE4 gene of bnvte4-4. (F) Mutation types and amino acid alterations in homologous copies in the BnVTE4 gene of bnvte4-5.
The five BnVTE4 T1-mutated lines and WT control were grown and harvested under the same condition with good growth and no significant difference from the control (Supplementary Figure 3), and mature seeds were used to determine the content and composition of tocopherols. HPLC results showed that the contents of α-T and γ-T were successfully detected in both WT control and T1 seeds (Figure 4). Consistent with a previous report (Zhang et al., 2007), the β- and δ-tocopherol contents were extremely low, which were neglected in the subsequence analysis.
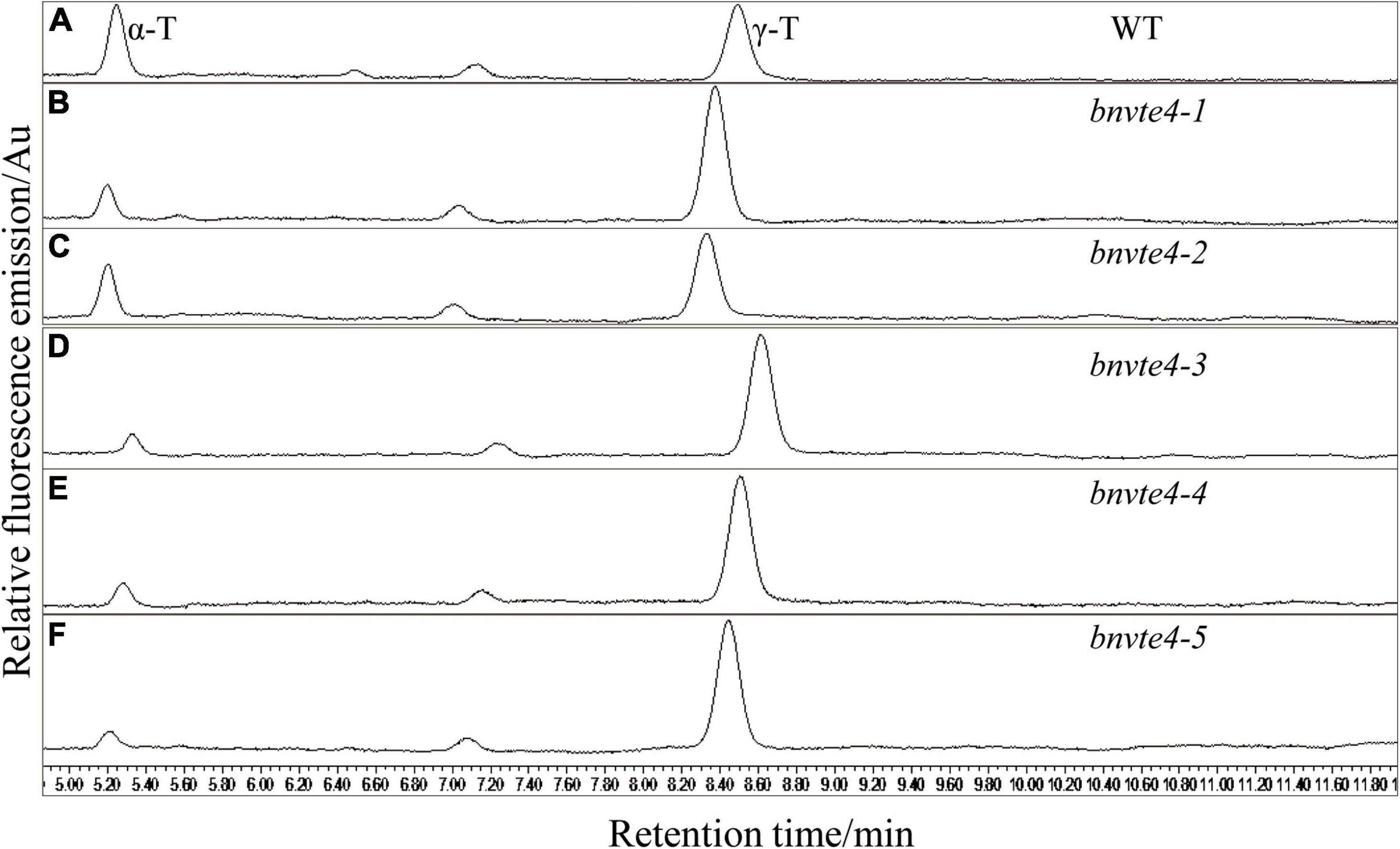
Figure 4. Determination of the components and content of tocopherol by HPLC. The peak at 5.2 min indicated α-T, the peak at 8.5 min indicated γ-T and the peak area indicated the content of tocopherol. (A) Two types of tocopherols in the wild type control (WT) were determined by HPLC. (B) Two types of tocopherols in bnvte4-1 were determined. (C) Two types of tocopherols in bnvte4-2 were determined. (D) Two types of tocopherols in bnvte4-3 were determined. (E) Two types of tocopherols in bnvte4-4 were determined. (F) Two types of tocopherols in bnvte4-5 were determined.
As shown in Figure 4, α-T content in BnVTE4 mutant lines was substantially decreased and significantly lower than that in WT (p < 0.05, Figure 5A). The reduction of α-T content in the bnvte4-2 mutant type was the lowest one among these five mutant types. The reduced α-T content was accompanied by a significant increase in γ-T content compared to the WT (p < 0.05, Figure 5B), except for the bnvte4-2 mutant type, which had a similar level of γ-T compared to WT. Nevertheless, the ratios of α- to γ-T (α-/γ-T) in all five BnVTE4 mutant types were only 0.1– 0.5, which were significantly lower than 0.67 in WT oilseed rape (p < 0.05, Figure 5C). This result confirmed that the mutations in the BnVTE4 gene can significantly affect the conversion of γ-T to α-T.
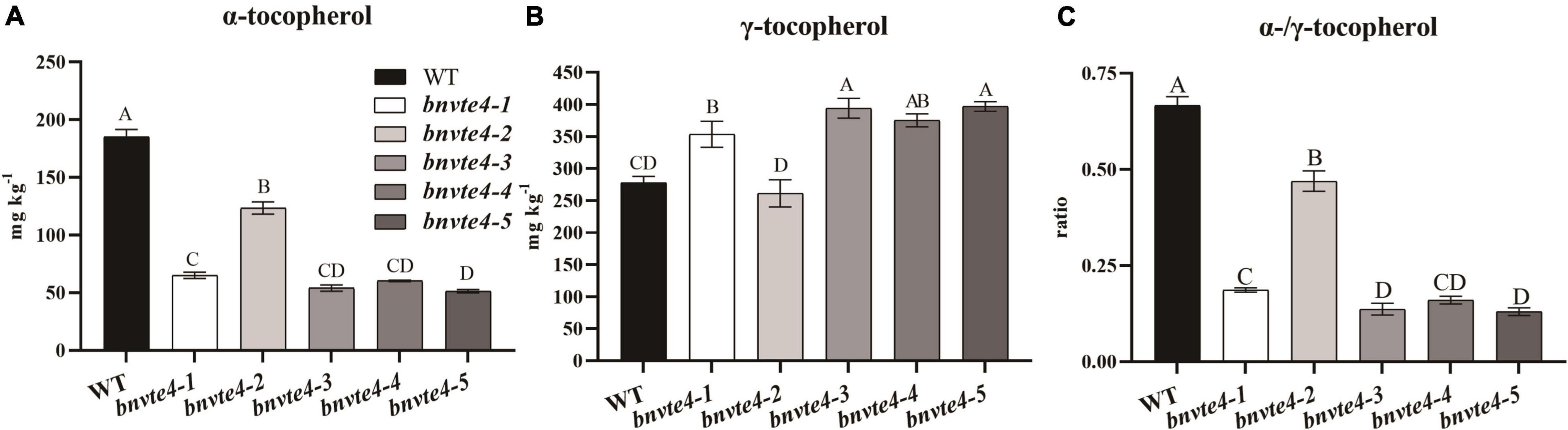
Figure 5. The α-and γ-T content and the ratio in T1 transgenic mutants. (A) The α-T content of the mutants. (B) The γ-T content of the mutants. (C) The α-/γ-T ratios of mutants.
Comparing the mutations in individual copies of the BnVTE4 gene (Figure 3A) with the change in tocopherol composition (Figure 5), we found that the homologous copies of the BnVTE4 gene had different contributions in α-T biosynthesis. bnvte4-3 and bnvte4-5 lines had both homozygous mutations in VTE4.C02-1 and VTE4.C02-2 copies, and the editing efficiency in VTE4.A02-1 and VTE4.A02-2 copies were both more than 50%. There was no significant difference in α-T and γ-T content, as well as the α-/γ-T ratio between these two lines. The bnvte4-1 and bnvte4-5 differed in the mutation of the VTE4.A02-1 copy. The bnvte4-1 had no mutation in VTE4.A02-1, while bnvte4-5 had 83% editing efficiency in this gene. The bnvte4-1 showed significant differences in α- and γ-T contents and the α-/γ-T ratio compared with bnvte4-5, implying the contribution of the VTE4.A02-1 copy to α-T synthesis.
The bnvte4-1 and bnvte4-4 were only different at the mutation in the VTE4.A02-2 copy. The VTE4.A02-2 copy in bnvte4-1 was completely mutated, while the editing efficiency of bnvte4-4 was only 12%, but the α-T and γ-T contents and the α-/γ-T ratio were not significantly different. This suggested that the contribution of the VTE4.A02-2 copy might be low. Alternatively, the wild-type copy of VTE4 in bnvte4-1 and bnvte4-4 might dominate the contribution, leading to the effect of the VTE4.A02-2 mutant being insignificant.
VTE4.C02-1 and VTE4.A02-2 copies in bnvte4-2 and bnvte4-4 had similar mutation patterns, while the other two VTE4 genes had different mutation profiles. The VTE4.C02-2 copy in bnvte4-2 was WT, and its VTE4.A02-1 copy had an editing efficiency of 64%, while VTE4.C02-2 in bnvte4-4 was completely mutated, and its VTE4.A02-1 copy was not mutated. The α-T and γ-T contents and the α-/γ-T ratio between bnvte4-2 and bnvte4-4 were significantly different. The α-/γ-T ratio of bnvte4-2 was higher than that of bnvte4-4, suggesting that the contribution of VTE4.C02-2 might be greater than that of VTE4.A02-1.
The α-T and γ-T contents, and the α-/γ-T ratio of bnvte4-3, bnvte4-4, and bnvte4-5 were not significantly different. In terms of mutation type, bnvte4-3 and bnvte4-5 were similar. VTE4.C02-1 and VTE4.C02-2 copies in bnvte4-4 were fully mutated, same as in bnvte4-3 and bnvte4-5. However, VTE4.A02-1 and VTE4.A02-2 copies did not exhibit mutation type, which again demonstrated that VTE4.A02-1 and VTE4.A02-2 copies might have a minor contribution.
It is particularly worth mentioning the obviously different tocopherol profile of bnvte4-2 to other mutant types. bnvte4-2 had similar mutations to other lines except for the VTE4.C02-2 copy that was WT other than homozygous mutation in other lines. Considering the lowest reduction of α-T in bnvte4-2 (from 185 mg/kg in WT to 123 mg/kg), while other lines had reduced α-T to ∼60 mg/kg, the key difference was the WT copy of VTE4.C02-2 compared to a homozygous mutation in other lines. This again suggested the great contribution of VTE4.C02-2 in α-T synthesis.
Tocopherol, especially α-tocopherol, is not only an important scavenger of stress-induced oxidative damage but an essential nutrient for human health. Genetic improvement of high-quality rapeseed oil with multiple vitamins such as α-tocopherol is an effective way to ensure the daily nutritional requirement of the human body. However, the complex genomic structure of oilseed rape that most of the genes have multiple homologous gene copies with putative redundant or diverse gene functions is one of the open questions for breeding application.
CRISPR/Cas9 technology has several inherent advantages in the precisely studying gene functions and subsequent application in crop plants (Lee et al., 2018; Hirohata et al., 2019; Zhai et al., 2019), especially polyploidy crops such as oilseed rape. In our previous studies, the highly efficient genome editing platform has been established to dissect the functional diversity of different homologs in oilseed rape (Li et al., 2018; Zaman et al., 2019; Cheng et al., 2021). Many independent case studies suggested that the homologous gene copies, although possess similar genomic information, often exert different effects in a particular trait (Okuzaki et al., 2018; Zhai et al., 2019, 2020; Ahmar et al., 2021; Chen et al., 2021).
To elucidate the contribution of the BnVTE4 gene in α-T biosynthesis, its four homologous gene copies were studied in detail by using CRISPR/Cas9 genome technology. To generate different mutation combinations of BnVTE4 homologs, two sgRNAs were designed to target their conserved regions in exon 3 and exon 4 of the coding sequences. Sequencing data demonstrated that the majority of BnVTE4-mutated lines showed homozygous mutation types in VTE4.C02-1 and VTE4.C02-2 homologous copies (Figure 3A), which suggested that our designed sgRNA1 had high mutation capacity on the genomic region of BnVTE4 homologs. However, it was very difficult to obtain homozygous mutation types in VTE4.A02-1 and VTE4.A02-2 homologs, even dozens of positive transgenic T0 lines and plenty of T2 plants had been performed by mutation screening. One of our hypotheses is that those four homologous copies probably have functional diversification in α-tocopherol biosynthesis or other key developmental processes such as seed vigor. Similar to this result, simultaneous mutation of five BnJAG homologs drastically affected the seed development, and its seeds are hard to survive, whereas the single mutation of BnJAG.A08-NUB homologous copy displays a pod-shattering resistance phenotype (Zaman et al., 2019).
To further evaluate the contribution of BnVTE4 homologs in α-tocopherol biosynthesis, the mutation frequency of VTE4.A02-1 and VTE4.A02-2 homologs was quantified by the Hi-TOM high-throughput sequencing method. Hi-TOM data suggested that the mutation frequency of VTE4.A02-1 and VTE4.A02-2 homologous copies exhibited a significant difference in bnvte4-1 and bnvte4-5 mutation lines. In addition, sequencing data showed that the most common mutation types were –1/+1 bp indels, and the amino acid sequence was completely changed due to a frame shift (Figures 3B–F). Thus, the designed sgRNAs can effectively generate targeted mutagenesis in all BnVTE4 homologous copies. This result was further verified by subsequence analysis of tocopherols content using HPLC, which demonstrated that the α- and γ-tocopherol contents and α-/γ-tocopherol ratios of these mutated lines showed significant alteration compared to WT control.
However, the effect of different mutation combinations on α-T content was a significant difference among five mutated lines. Similar mutation types (bnvte4-3 and bnvte4-5) showed no significant difference in α-T content. There was no significant difference in the content of α-T when the mutation types of the other copies except VTE4.A02-2 were similar (bnvte4-1 and bnvte4-4), indicating that the VTE4.A02-2 copy did not play a major role in α-tocopherol biosynthesis. This conclusion was supported by comparing bnvte4-3 and bnvte4-5. The content of α-T was significantly different when the mutation types were similar for other copies except for VTE4.A02-1 (bnvte4-1 and bnvte4-5), indicating the VTE4.A02-1 copy was important. VTE4.C02-2 and VTE4.A02-2 mutation types of bnvte4-2 and bnvte4-3 were different, and the content of α-T was significantly varied. The functionality of the VTE4.C02-2 copy was revealed in BnVTE4-2 by comparing it to other lines. The significant difference in the content of α-T when the mutation types of other copies except VTE4.C02-2 are similar (bnvte4-2 and bnvte4-4) indicates that VTE4.C02-2 played an important role. Taken together, those data demonstrated that the four BnVTE4 gene homologs might have functional differentiation in α-T biosynthesis, and their contribution was likely VTE4.C02-2 (BnaC02G0331100ZS) > VTE4.A02-1 (BnaA02G0247300ZS) > VTE4.A02-2 (BnaA02G0154300ZS). This knowledge will shed new light on the cultivation of high α-T-content oilseed rape.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
QH, X-RZ, CL, and DM conceived and revised the manuscript. CL designed the experiments. HZ, YQS, MS, XH, MH, and YS performed the experiments. HZ and CL analyzed the data. HZ drafted the manuscript. All authors reviewed and approved the manuscript.
This work was supported by the National Natural Science Foundation of China (No. 31700316), the Central Public-Interest Scientific Institution Basal Research Fund (No. 1610172020001), and the Natural Science Foundation of Hubei Province (No. 2018CFB543).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Jin-Long Qiu and Kangquan Yin at the Institute of Microbiology CAS for valuable discussion.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.850924/full#supplementary-material
Ahmar, S., Zhai, Y., Huang, H., Yu, K., Khan, M. H. U., Shahid, M., et al. (2021). Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus L. Crop J. 10, 67–74. doi: 10.1016/j.cj.2021.03.023
Bergmüller, E., Porfirova, S., and DöRmann, P. (2003). Characterization of an Arabidopsis mutant deficient in γ-tocopherol methyltransferase. Plant Mol. Biol. 52, 1181–1190. doi: 10.1023/b:plan.0000004307.62398.91
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., and Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. doi: 10.1104/pp.17.00426
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, Y., Fu, M., Li, H., Wang, L., Liu, R., Liu, Z., et al. (2021). High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 19, 424–426. doi: 10.1111/pbi.13507
Cheng, H., Hao, M., Ding, B., Mei, D., Wang, W., Wang, H., et al. (2021). Base editing with high efficiency in allotetraploid oilseed rape by A3A-PBE system. Plant Biotechnol. J. 19, 87–97. doi: 10.1111/pbi.13444
Cheng, L., Zhong-Chao, F., Tang-Hua, X., Xiao-Min, M., Guang-Sheng, Z., FengHong, H., et al. (2019). Development, potential and adaptation of Chinese rapeseed industry. Chin. J. of Oil Crop Sci. 41, 485–489.
DellaPenna, D., and Pogson, B. J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57, 711–738. doi: 10.1146/annurev.arplant.56.032604.144301
Fritsche, S., Wang, X., and Jung, C. (2017). Recent advances in our understanding of tocopherol biosynthesis in plants: an overview of key genes, functions, and breeding of vitamin E improved crops. Antioxidants 6:99. doi: 10.3390/antiox6040099
Galmés, S., Serra, F., and Palou, A. (2018). Vitamin E metabolic effects and genetic variants: a challenge for precision nutrition in obesity and associated disturbances. Nutrients 10:1919. doi: 10.3390/nu10121919
Hirohata, A., Sato, I., Kaino, K., Iwata, Y., Koizumi, N., and Mishiba, K. I. (2019). CRISPR/Cas9-mediated homologous recombination in tobacco. Plant Cell Rep. 38, 463–473. doi: 10.1007/s00299-018-2320-7
Hunter, S. C., and Cahoon, E. B. (2007). Enhancing vitamin E in oilseeds: unraveling tocopherol and tocotrienol biosynthesis. Lipids 42, 97–108. doi: 10.1007/s11745-007-3028-6
Institute of Medicine (US) Panel on Dietary Antioxidants and Related Compounds (2000). Dietary Reference Intakes for Vitamin C, Vitamin E, Selenium, and Carotenoids. Washington, DC: National Academies Press (US).
Janeczko, A., Dziurka, M., and Pociecha, E. (2018). Increased leaf tocopherol and β-carotene content is associated with the tolerance of winter wheat cultivars to frost. J. Agron. Crop Sci. 204, 594–602. doi: 10.1111/jac.12287
Kumar, D. A., Kumar, J. S., Vignesh, M., Uttamrao, Z. R., Singh, C. H., Gulab, C., et al. (2018). Molecular diversity and genetic variability of kernel tocopherols among maize inbreds possessing favourable haplotypes of γ-tocopherol methyl transferase (ZmVTE4). J. Plant Biochem. Biotechnol. 28, 253–262. doi: 10.1007/s13562-018-0470-x
Lee, Z. H., Yamaguchi, N., and Ito, T. (2018). Using CRISPR/cas9 system to introduce targeted mutation in Arabidopsis. Methods Mol Biol. 1830, 93–108. doi: 10.1007/978-1-4939-8657-6_6
Li, C., Hao, M., Wang, W., Wang, H., Chen, F., Chu, W., et al. (2018). An Efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Front. Plant Sci. 9:442. doi: 10.3389/fpls.2018.00442
Li, C., Sang, S., Sun, M., Yang, J., Shi, Y., Hu, X., et al. (2021). Direct modification of multiple gene homoeologs in Brassica oleracea and Brassica napus using doubled haploid inducer-mediated genome-editing system. Plant Biotechnol. J. 19, 1889–1891. doi: 10.1111/pbi.13632
Liu, Q., Wang, C., Jiao, X., Zhang, H., Song, L., Li, Y., et al. (2019). Hi-TOM: a platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. doi: 10.1007/s11427-018-9402-9
Lloret, A., Esteve, D., Monllor, P., Cervera-Ferri, A., and Lloret, A. (2019). The effectiveness of vitamin E treatment in Alzheimer’s disease. Int J. Mol. Sci. 20:879. doi: 10.3390/ijms20040879
Ma, J., Qiu, D., Gao, H., Wen, H., Wu, Y., Pang, Y., et al. (2020). Over-expression of a γ-tocopherol methyltransferase gene in vitamin E pathway confers PEG-simulated drought tolerance in alfalfa. BMC Plant Biol. 20:226. doi: 10.1186/s12870-020-02424-1
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A Robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Martin, C., Zhang, Y., Tonelli, C., and Petroni, K. (2013). Plants, diet, and health. Annu. Rev. Plant Biol. 64, 19–46. doi: 10.1146/annurev-arplant-050312-120142
Meydani, S. N., Barklund, M. P., Liu, S., Meydani, M., Miller, R. A., Cannon, J. G., et al. (1990). Vitamin E supplementation enhances cell-mediated immunity in healthy elderly subjects. Am. J. Clin. Nutr. 52, 557–563. doi: 10.1093/ajcn/52.3.557
Meydani, S. N., Meydani, M., Blumberg, J. B., Leka, L. S., Siber, G., Loszewski, R., et al. (1997). Vitamin E supplementation and in vivo immune response in healthy elderly subjects. A randomized controlled trial. JAMA 277, 1380–1386. doi: 10.1001/jama.1997.03540410058031
Norris, S. R., Barrette, T. R., and Dellapenna, D. (1995). Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7, 2139–2149. doi: 10.1105/tpc.7.12.2139
Norris, S. R., Shen, X., and Dellapenna, D. (1998). Complementation of the Arabidopsis pds1 mutation with the gene encoding p-hydroxyphenylpyruvate dioxygenase. Plant Physiol 117, 1317–1323. doi: 10.1104/pp.117.4.1317
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Porfirova, S., Bergmuller, E., Tropf, S., Lemke, R., and Dormann, P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 12495–12500. doi: 10.1073/pnas.182330899
Ranard, K. M., and Erdman, J. W. Jr. (2018). Effects of dietary RRR α-tocopherol vs all-racemic α-tocopherol on health outcomes. Nutr. Rev. 76, 141–153. doi: 10.1093/nutrit/nux067
Rozanowska, M., Edge, R., Land, E. J., Navaratnam, S., Sarna, T., and Truscott, T. G. (2019). Scavenging of retinoid cation radicals by urate, trolox, and α-, β-, γ-, and δ-tocopherols. Int. J. Mol. Sci. 20:2799. doi: 10.3390/ijms20112799
Schneider, C. (2005). Chemistry and biology of vitamin E. Mol. Nutr. Food Res. 49, 7–30. doi: 10.1002/mnfr.200400049
Semchuk, N. M., Lushchak, O. V., Falk, J., Krupinska, K., and Lushchak, V. I. (2009). Inactivation of genes, encoding tocopherol biosynthetic pathway enzymes, results in oxidative stress in outdoor grown Arabidopsis thaliana. Plant Physiol. Biochem. 47, 384–390. doi: 10.1016/j.plaphy.2009.01.009
Trebst, A., Depka, B., and Holländer-Czytko, H. (2002). A specific role for tocopherol and of chemical singlet oxygen quenchers in the maintenance of photosystem II structure and function in Chlamydomonas reinhardtii. FEBS Lett. 516, 156–160. doi: 10.1016/s0014-5793(02)02526-7
Tsegaye, Y., Shintani, D. K., and Dellapenna, D. (2002). Overexpression of the enzyme p-hydroxyphenolpyruvate dioxygenase in Arabidopsis and its relation to tocopherol biosynthesis. Plant Physiol. Biochem. 40, 913–920. doi: 10.1016/s0981-9428(02)01461-4
Valentin, H. E., Lincoln, K., Moshiri, F., Jensen, P. K., Qi, Q., Venkatesh, T. V., et al. (2006). The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18, 212–224. doi: 10.1105/tpc.105.037077
Venkatesh, T. V., Karunanandaa, B., Free, D. L., Rottnek, J. M., Baszis, S. R., and Valentin, H. E. (2006). Identification and characterization of an Arabidopsis homogentisate phytyltransferase paralog. Planta 223, 1134–1144. doi: 10.1007/s00425-005-0180-1
Vom Dorp, K., Hölzl, G., Plohmann, C., Eisenhut, M., Abraham, M., Weber, A. P., et al. (2015). Remobilization of phytol from chlorophyll degradation is essential for tocopherol synthesis and growth of Arabidopsis. Plant Cell 27, 2846–2859. doi: 10.1105/tpc.15.00395
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xu, J., Nwafor, C. C., Shah, N., Zhou, Y., and Zhang, C. (2019). Identification of genetic variation in Brassica napus seeds for tocopherol content and composition using near-infrared spectroscopy technique. Plant Breed. 138, 624–634. doi: 10.1111/pbr.12708
Yan, Y., Liang, Y., Xuekun, Z., Jingli, G., and Jijun, W. (2021). Status and countermeasure of the high-quality development of rapeseed industy in China. J. Agr. Sci. Tech Iran. 23, 1–7.
Yu, L., Li, G., Li, M., Xu, F., Beta, T., and Bao, J. (2016). Genotypic variation in phenolic acids, vitamin E and fatty acids in whole grain rice. Food Chem. 197, 776–782. doi: 10.1016/j.foodchem.2015.11.027
Zaman, Q. U., Chu, W., Hao, M., Shi, Y., Sun, M., Sang, S. F., et al. (2019). CRISPR/Cas9-mediated multiplex genome editing of JAGGED gene in Brassica napus L. Biomolecules 9:725. doi: 10.3390/biom9110725
Zaman, Q. U., Wen, C., Yuqin, S., Mengyu, H., Desheng, M., Jacqueline, B., et al. (2021). Characterization of SHATTERPROOF homoeologs and CRISPR-Cas9-mediated genome editing enhances pod-shattering resistance in Brassica napus L. CRISPR J. 4, 360–370. doi: 10.1089/crispr.2020.0129
Zhai, Y., Cai, S., Hu, L., Yang, Y., Amoo, O., Fan, C., et al. (2019). CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132, 2111–2123. doi: 10.1007/s00122-019-03341-0
Zhai, Y., Yu, K., Cai, S., Hu, L., Amoo, O., Xu, L., et al. (2020). Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. doi: 10.1111/pbi.13281
Keywords: α-tocopherol, BnVTE4, homologous copies, functional diversification, CRISPR/Cas9
Citation: Zhang H, Shi Y, Sun M, Hu X, Hao M, Shu Y, Zhou X-R, Hu Q, Li C and Mei D (2022) Functional Differentiation of BnVTE4 Gene Homologous Copies in α-Tocopherol Biosynthesis Revealed by CRISPR/Cas9 Editing. Front. Plant Sci. 13:850924. doi: 10.3389/fpls.2022.850924
Received: 08 January 2022; Accepted: 04 March 2022;
Published: 11 April 2022.
Edited by:
Maria Raffaella Ercolano, University of Naples Federico II, ItalyReviewed by:
Qing Liu, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2022 Zhang, Shi, Sun, Hu, Hao, Shu, Zhou, Hu, Li and Mei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chao Li, bGljaGFvMDFAY2Fhcy5jbg==; Desheng Mei, bWVpZGVzaGVuZ0BjYWFzLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.