- 1Division of Plant Biosciences, School of Applied Biosciences, College of Agriculture and Life Science, Kyungpook National University, Daegu, South Korea
- 2Crop Breeding Division, National Institute of Crop Science, Rural Development Administration, Wanju, South Korea
- 3Department of Agronomy, Faculty of Agriculture, Jember University, Jember, Indonesia
Recent unpredictable climate change is the main reason for the decline in rice yield. In particular, drought stress is a major constraint in reducing yield and quality for rice at rainfed agriculture areas, such as Asia and South America. CRISPR/Cas9 provides an effective solution for gene function study and molecular breeding due to specific editing of targeted genome sequences. In addition, CRISPR/Cas9 application can significantly reduce the time required to develop new cultivars with improved traits compared to conventional complex and time-consuming breeding. Here, drought-induced gene Oryza sativa Senescence-associated protein (OsSAP) was edited by CRISPR/Cas9. To investigate the possible role of OsSAP in drought stress, genome-editing plants were subjected to drought stress until the soil moisture content reached 20%, and the reactive oxygen species (ROS) scavenging efficiency of genome-editing plants were decreased. When the genome-editing plants were subjected to drought stress, survival rate, shoot length, root length, content of chlorophyll number of tiller, and 1,000-grain weight decreased, and more H2O2 and O2− were detected in leaves. In addition, expression levels of several critical stress-related transcription factors were decreased in the OsSAP genome-editing plant. These results suggest that OsSAP function as a positive regulator during drought stress response in rice. We analyzed the expression of OsSAP and Cas9 in T0 and T1 plants as well as T2 seeds. As the course of generation advancement progressed, Cas9 expression remained stable or weakened but the OsSAP expression was continuously removed from the T0 plant. The coefficient of variation (CV) in both T1 plants and T2 seeds was lower than 5%. Overall, our results suggest that CRISPR/Cas9 could be a novel and important tool for efficiently generating specific and inheritable targeted genome editing in rice, with short breeding cycles.
Introduction
To respond to rapid climate change, it is necessary to improve cultivars of crops and develop rice with resistance to biotic and abiotic stress; this is a primary goal in plant breeding worldwide (Zhang et al., 2018). Recent rapid increases in the global temperature have raised awareness of the fragility of the world’s food supply, and solutions are needed (Siebert et al., 2017). It has recently been established that drought stress is the main cause of the reduction of rice yield, and in fact, drought can have a direct impact on future food shortages (Passioura, 2007; Prabnakorn et al., 2018). In addition, the area of regions subjected to drought stress is expanding, and as drought frequency and intensity increase, drought stress is a problem that must be resolved. In particular, in Asia, which accounts for most of the world’s rice production, annual production of rice suffers a loss of up to 40% due to drought stress (Guo et al., 2021). One specific problem in relation to drought is how to maintain stable yield, while minimizing the drought-related effects of climate change on various food crops. Plants undergo many structural and physiological changes during drought; thus, the breeding of drought stress-resistant crops has become a means by which to strengthen the plants (Yang et al., 2019).
CRISPR/Cas9 became the most powerful and common tool for gene functional analysis and agronomic studies in plant species, especially in rice (Zhou et al., 2017; Hu et al., 2018). This technology is important in studying the agronomic traits and has been widely used for physiological and molecular studies of plants (Tsai and Joung, 2016). In addition, CRISPR/Cas9 is a more precise and higher efficient technique to achieve the goal of improving agricultural traits (Fiaz et al., 2019). Application of CRISPR/Cas9 improved abiotic/biotic stress tolerance and agricultural traits. By genome editing of the flowering suppressor gene SP5G (Rehman et al., 2022), rice with promoted flowering time was developed, and rice with improved resistance to biotic stress due to genome-engineering was developed (Zhou et al., 2015). The genome-editing line created by CRISPR/Cas9 proved that YK17S is a superior yield potential gene in indica rice (Barman et al., 2019). In addition, CRISPR/Cas9 provides researchers with the opportunity to further research by increasing the understanding of various genes (Liu et al., 2020).
When plants are exposed to drought, they are subjected to many simultaneous negative effects, such as apoptosis, cell aging, reactive oxygen species (ROS) accumulation, and the deterioration of growth development (Chen et al., 2018; Liu et al., 2019; Singh et al., 2019). Recent research has shown that ROS play an important role in plants as signaling factors that transmit in response to major plant growth stimuli, such as stress, pathogen intrusion, and programmed cell death (Qi et al., 2017; Fichman and Mittler, 2020). According to evolutionary theory, photosynthetic life has played an important role in the development of almost all life on Earth due to oxygen release, which is also unfortunately the source of ROS (Cezary et al., 2018). Plants can convert oxygen into ROS in organelles, such as mitochondria and chlorophyll through various common metabolic reactions (Munro and Treberg, 2017). Cellular senescence due to plant ROS accumulation begins with the onset of senescence during the development of all tissues; it decreases photosynthesis, chlorophyll degradation, and protein synthesis ability, leading to the loss of cellular homeostasis and ultimately apoptosis (Huang et al., 2019). In addition to developmental aging, the overall aging process of plants is affected by various biotic and abiotic stresses, including nutrient deficiency, temperature, pathogen attacks, and humidity. Senescence-associated genes respond to drought and cause substantial yield loss (Uji et al., 2017; Jiang et al., 2018). Plants are typically settled in one place; thus, they cannot escape environmental stresses that are unfavorable to their growth. Due to their immobility and exposure to stress, plants have developed numerous defense mechanisms that minimize or eliminate the damage they receive (Ilyas et al., 2021). In particular, plants overcome stress by synthesizing substances, such as proline, raffinose, and glycine betaine under different stress situations including drought; these substances help stabilize cell structure and remove excessively generated ROS including hydrogen peroxide (H2O2), singlet oxygen, and superoxide anions (Nahar et al., 2018; Verma et al., 2019).
Traditional breeding and transformants have been effectively used to breed stress-tolerant cultivars; indeed, such techniques have facilitated plant growth during drought and solved several drought-related problems (Bai et al., 2018). However, developing cultivars that can improve various stresses resistant and maintain a stable yield requires a deep understanding of plant genetics. In addition, traditional breeding is time-consuming, labor-intensive, and costly. However, novel plant breeding techniques applying CRISPR/Cas9 have been proposed as a new breeding method that can accurately and rapidly improve crop traits (Fiaz et al., 2021a). In previous studies, several quantitative trait loci and genes that induce apoptosis during drought were discovered (Shamloo-Dashtpagerdi et al., 2020; Ganie and Ahammed, 2021); however, validating these results during drought conditions is a complex process. Biotechnology holds the key to solving these problems.
CRISPR/Cas9 is an important genome-editing technology used to create plant varieties with a wide range of traits that solve various problems (Li et al., 2017, 2020). In addition, the application of CRISPR/Cas9 made it possible to understand the functions of various previously unresolved genes (Fiaz et al., 2021b). In the present study, to characterize the functional role of Oryza sativa SAP (OsSAP) in rice during drought, genome-edited rice plant was developed using CRISPR/Cas9 technology. The morphological and molecular changes in this gene editing plant during drought were compared with the changes in the rice cultivar, O. sativa spp. japonica cv. Ilmi (Ilmi). Consequently, we found that OsSAP induced cellular senescence and apoptosis during drought. Specifically, our results suggest that OsSAP accelerates cellular senescence during exposure to drought stress and acts as a negative regulator of cells. Additionally, OsSAP was found to function in processes related to apoptosis and cellular senescence during various environmental stressors. Our findings will facilitate further research to elucidate the underlying mechanism of these effects.
Materials and Methods
Plant Materials and Development of Genome-Editing Rice
GeD lines were generated using Ilmi as a background. The generated GeD, and Ilmi were cultivated at 28/26°C during a 16/8-h light/dark photoperiod. Three sgRNAs (sgRNA 1–3) were designed using the CRISPR RGEN Tools1 program to target the first exon of OsSAP (Oryza sativa Senescence-associated protein, Accession no. AB734097) and edit the genome (Park et al., 2015); these sgRNAs were integrated into the pRGEB32 vector, which is capable of expressing Cas9 in plants (Mikami et al., 2015). The obtained binary vector was transformed through calli derived from Ilmi using Agrobacterium tumefaciens EHA105 and the rice plant was regenerated as previously described (Yukoh and Toshihiko, 2008). We also used OsSAP-T6 lines for compared GeD lines related to drought tolerance were acquired from Prof. Ubaidillah (Ubaidillah et al., 2013).
Detection of Genome-Edited Rice and Assay of Transgene-Free Plant Lines
Genomic DNA was extracted from rice leaves using a DNeasy Plant Mini Kit (QIAGEN, Cat. 69104, Hilden, Germany) according to the manufacturer’s instructions. To amplify the target genome region of CRISPR/Cas9, PCR analysis was performed with primers using these region sequences. The amplified PCR product was purified and mutants for which the target gene was genome-edited were detected through Ilmi sequences and alignment via sequencing chromatogram analysis. For all regeneration GeD plants, each line of the T0 generation was analyzed for the target region of OsSAP.
RNA Isolation and Gene Expression Analysis
Roots, stems, and leaves were sampled at the seedling stage of rice, whereas the roots, stems, leaves, and panicles at the heading stage were sampled. Total RNA was extracted from the sampled organs using the RNeasy Micro Kit (QIAGEN, Cat. 74004, Hilden, Germany) according to the manufacturer’s instructions. Subsequently, cDNA was synthesized using 1 μg of total RNA from each sample with the qPCRBIO cDNA Synthesis Kit (PCRBIOSYSTEMS, United States), used according to the manufacturer’s instructions. The synthesized cDNA was diluted 1:10 (v/v) using deionized water and then used for qPCR analysis together with the housekeeping gene OsActin. qPCR analysis was performed using an Eco™ Real-Time PCR System (Illumina, Cat. EC-900-1001, CA, United States) with 2X Multi-Star OneStep qRT-PCR Master Mix (BIOFACT, Cat. RQ351-50h, Seoul, Korea); relative expression levels were analyzed according to the manufacturer’s instructions. Cycle threshold values for each sample were normalized using OsActin as a standard (Ariani et al., 2004). Samples were each analyzed in five independent biological replicates. The list of primer sets used for the analysis is listed in Supplementary Table S1.
Drought Stress Treatment and Phenotype Evaluation After Stress
GeD lines, Ilmi, and OsSAP-T6 lines were grown by filling a Wagner pot with soil. Each pot used in the experiment was filled with the same weight of soil, and water was supplied equally to each pot at a height of 5 cm from the soil. Drought conditions were created by not watering plants when they reached the fourth leaf stage; the water supply was stopped until the soil moisture content reached 20%. When the soil moisture content reached 20%, the plants were watered for 7 days and the survival rate was analyzed (Li et al., 2013). To analyze growth, shoot length, root length, chlorophyll content, number of tillers, and 1,000-grain weight, data were collected when the soil moisture content was 20%.
Detection and Measurement of H2O2
To detect H2O2 after drought treatment, the 3,3′-diaminobenzidine (DAB) staining method of Liu et al. (2014) was modified and used. After drought treatment, leaves were cut to a length of 5 cm and then submerged in 1-mg/ml DAB (pH 3.8) to be dyed in darkness at 25°C for 16 h. To prevent the penetration of DAB solution from being disturbed by foreign substances on the leaves, they were cut and their surface was washed with deionized water. After decolorization using 100% ethanol, the reaction of H2O2 on each leaf was analyzed through newly created brown spots. DAB staining was analyzed on leaves of five different plants for each line and representative images were collected. In addition, to analyze the content of H2O2 contained in leaves, H2O2 was extracted using the method proposed by Rao et al. (2000). The content of H2O2 contained in plant leaves was determined via the Quantitative Peroxide Assay Kit (Thermo Fisher, Cat. 23280, Seoul, Korea), used according to the manufacturer’s instructions. Leaf extracts were measured at OD560 on a spectrometer. The absorbance values measured for each leaf were calibrated with the standard curve generated with from a known H2O2 concentration.
NBT Staining
To evaluate the generation of O2− in the leaves of plants after drought stress, 0.1% nitro blue tetrazolium (NBT) solution was added to 10-mM potassium phosphate buffer (pH 7.8), and then leaf samples were cut to a length of 5 cm and precipitated (Fitzgerald et al., 2004). They were incubated in darkness at 25°C for 16 h. To analyze the blue spots generated by the reaction of NBT and O2−, the chlorophyll contained in the leaves was removed. To remove chlorophyll, the leaves were fixed in alcohol lactophenol (2:1:1, 95% ethanol: lactic acid: phenol) at 65°C for 30 min before they were rinsed with 50% ethanol. Afterward, NBT was found to react with superoxide on the surface of the leaf to form blue spots.
Analysis of MDA, CAT, POD, SOD, and Proline Content
Proline content was determined using the method from Bates et al. (1973). After drought treatment on GeD, and OsSAP-T6 lines, 0.5 g of leaf was ground, and homogenized with 3% sulfosalicylic acid (w/v) at 100°C for 10 min. This was then centrifuged at 4,500 RPM for 30 min after 10 min. Subsequently, 2 ml acidninhydrin and 2-ml glacial acetic acid were added to 2 ml of the supernatant, boiled at 100°C for 40 min, and then cooled using ice. Proline levels in the drought-treated samples were determined at OD520. MDA content was analyzed via the method outlined by Heath and Packer (1968). A 1-g leaf sample was placed in 10 ml of 10% trichloroacetic (v/v) reagent and was homogenized, after which it was centrifuged at 4,500 RPM for 10 min. Next, 2 ml of the supernatant was reacted with 2 ml of thiobarbituric acid at 100°C for 15 min and then cooled with ice before it was analyzed at OD450, OD532, and OD600 to determine MDA concentration. Finally, the MDA content was determined by applying the formula (μmol L−1) = 6.45 (OD532 − OD600) − 0.56 OD450, which was proposed by Shi et al. (2012). Catalase (CAT) activity was analyzed by modifying the method of Beers and Sizer (1952). The 3.0 ml reaction mixture contained 2 ml of sodium phosphate buffer (50 mM at pH 7.0), 0.5 ml of H2O2 (40 mM), and 0.5 ml of enzyme extract. The decomposition of H2O2 at OD240 was analyzed. The activity of superoxide dismutase (SOD), an antioxidant enzyme, under drought stress was analyzed according to the protocol of Duan et al. (2012). The unit of SOD activity was determined as the minimum amount of enzyme required to inhibit the photochemical reduction of NBT chloride by 50% at OD560. Peroxidase (POD) activity was analyzed by the guaiacol oxidation method proposed by Weydert and Cullen (2010). The POD reaction mixture (3 ml) contained 50 mM sodium acetate buffer (pH 5.0), 20-mM guaiacol, 40-mM H2O2, and 0.1-ml enzyme extract. The prepared POD reaction mixture was analyzed at OD240 at 1 min intervals; data were recorded before 3 min had passed after the start of the reaction.
Strategy Establishment for Short Breeding Cycle of CRISPR/Cas9 GE Rice
Oryza sativa Senescence-associated protein genome-edited plants with CRISPR/Cas9 were analyzed through genomic sequencing. Regenerated plants from callus were named T0 plant, and seeds harvested from T0 plant were named T1 seed. Also, when T1 seeds are sowing and transplanted into the field, they are named T1 plants, and the seeds harvested from T1 plants become T2 seeds. Among genome-edited plants, homozygous types were selected from T0. An accession number was assigned to each tiller of the T0 plants, and T1 seeds were harvested, and each was assigned an accession number. T1 seeds were transplanted in one row for each line in the field, and 25 plants were transplanted for each row. Twenty-five plants were planted in each line, and the plants that died after growth were replaced with purple rice and were not used for subsequent analysis. In T1 plants, T2 seeds were harvested in bulk. In each generation, the expression level of Cas9 and the target gene, OsSAP, was analyzed using OsActin as a control.
Statistical Analysis
All data were statistically analyzed using one-way ANOVA and Duncan’s multiple range tests via the SPSS program (IBM SPSS Statistics, version 22, NC, United States). Value of p < 0.05 was considered statistically significant.
Results
Generation of OsSAP Genome-Editing Lines Using CRISPR/Cas9 System
To evaluate the role of OsSAP in drought conditions, we developed GeD0 (Genome-editing Drought gene OsSAP 0 generation) lines through genome editing of OsSAP using CRISPR/Cas9. In the CRISPR/Cas9 vector, which can be expressed in rice, the OsSAP guide RNA is regulated by the U3 promoter (Figure 1A). The guide RNA is inserted into the BsaI site of the pRGEB32 vector (Figure 1B). Three guide RNAs were designed, and the GC contents were set at 50–70% (Figure 1C). Afterward, the CRISPR/Cas9 vector, which the sgRNA was inserted into, was transformed into the callus by Agrobacterium medium (Figures 2A–F). For callus inoculation, callus cultured for 21 days were selected (Figure 2A). After co-culture with Agrobacterium for 3 days, green spots were investigated after 7 days in the regeneration medium (Figure 2B). After 45 days on the regeneration plate, shoot growth was almost completed (Figure 2C), and culture was performed for rooting on a new regeneration plate for 30 days (Figure 2D). After 3 days of acclimatization (Figure 2E), T0 plants were transplanted into pots for T1 seed harvesting (Figure 2F). A total of 237 regenerated plants were developed, but only 104 regenerated plants survived when transplanted into soil after acclimatization. Among 104 regenerated plants, there were 45 regenerated plants with genome editing via OsSAP-sgRNA1, and T-DNA insertions were analyzed in 23 of them (Figure 2G). There were 38 regenerated OsSAP-sgRNA2 plants, and T-DNA insertion was analyzed in 15 of them. When OsSAP-sgRNA3 was used, only 15 out of 21 regenerated plants had T-DNA insertion. Around 1,000 Ilmi seeds were used for callus culture, and 237.0 ± 12.8 regenerated plants were developed. Among them, T-DNA was successfully inserted in 53 regenerated plants (Figure 2H). The editing type of OsSAP was analyzed with each sgRNA (Figure 3). When OsSAP-sgRNA1 was used, deletion occurred in 12 of 23 regenerated plants. There were also two insertions, and nine did not respond to genome editing (Figure 3A). Among the regenerated plants, the editing was homozygous in three plants (Figure 3B). In OsSAP-sgRNA2 plants, there were eight deletions, two insertions, and six did not respond to editing. In one of these plants, the homozygous edited type was analyzed. In OsSAP-sgRNA3 plants, there were four deletions, one insertion, and 10 were not edited. Among editing plants, two plants were edited homozygous. For continuous research, three plants (designated as GeD0 1–1, GeD0 3–1, and GeD0 3–2) were selected for the generation of GeD lines.
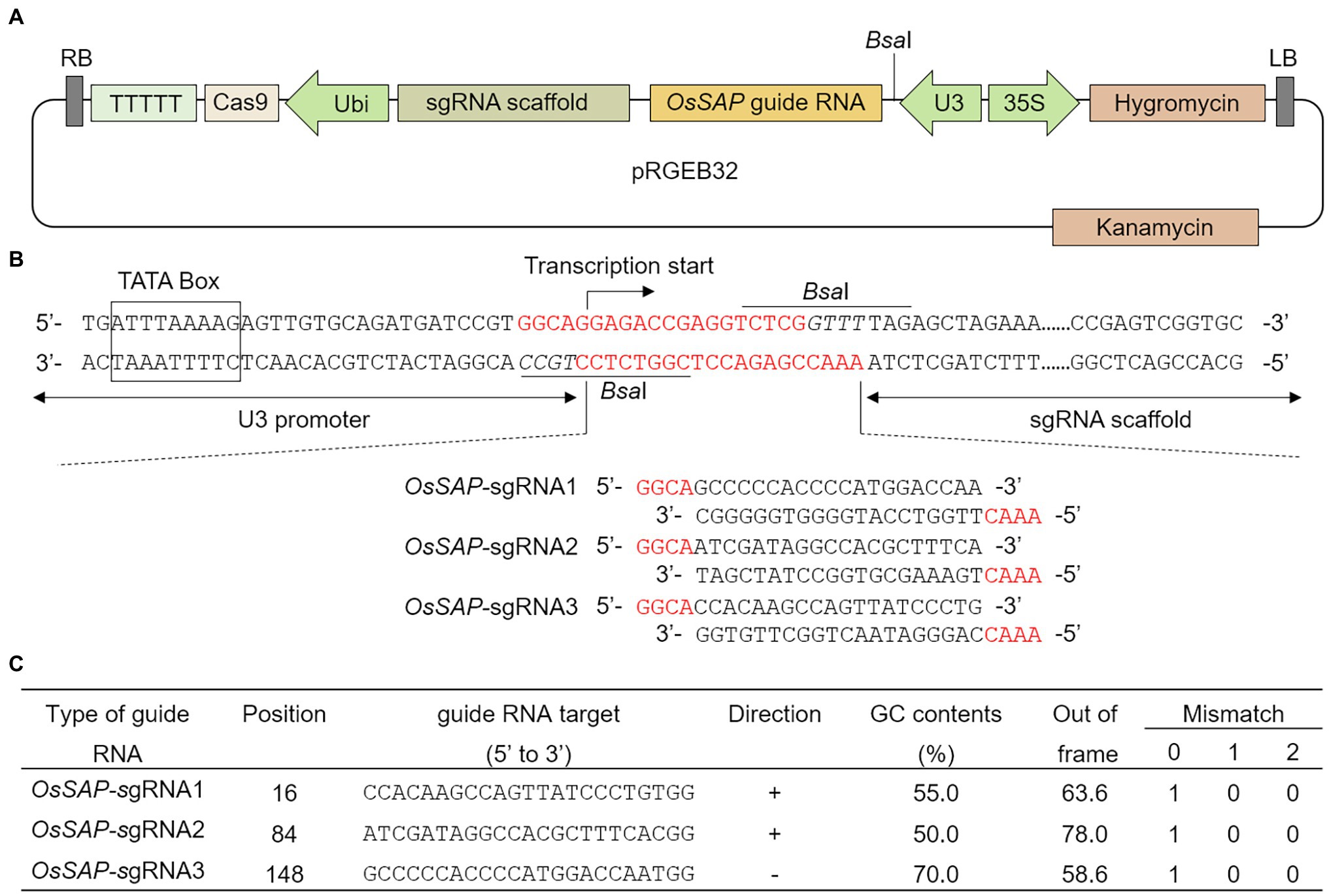
Figure 1. CRISPR/Cas9 vector construct and guide RNA design for Oryza sativa Senescence-associated protein (OsSAP) genome editing. (A) Schematic diagram of the pRGEB32 vector used for genome editing of OsSAP through CRISPR/Cas9 expression in rice. The guide RNA and sgRNA scaffold are expressed by the U3 promoter. Cas9 is expressed by the ubiquitin promoter (Ubi). Hygromycin (HPT) is expressed by the CaMV35S promoters (35S). LB and RB: left border and right border, respectively. (B) In the CRISPR/Cas9 vector, OsSAP guide RNA inserted at the site of the BsaI restriction enzyme site. (C) Three sgRNAs were designed for genome editing of OsSAP. The GC content of guide RNAs was set to 50–70%. Out-of-frame and mismatch values indicate the possibility that the guide RNA will act off-target. The out-of-frame value was set to 60 or more, and a mismatch of 1-0-0 meant that the designed guide RNA does not have the same sequence as OsSAP.
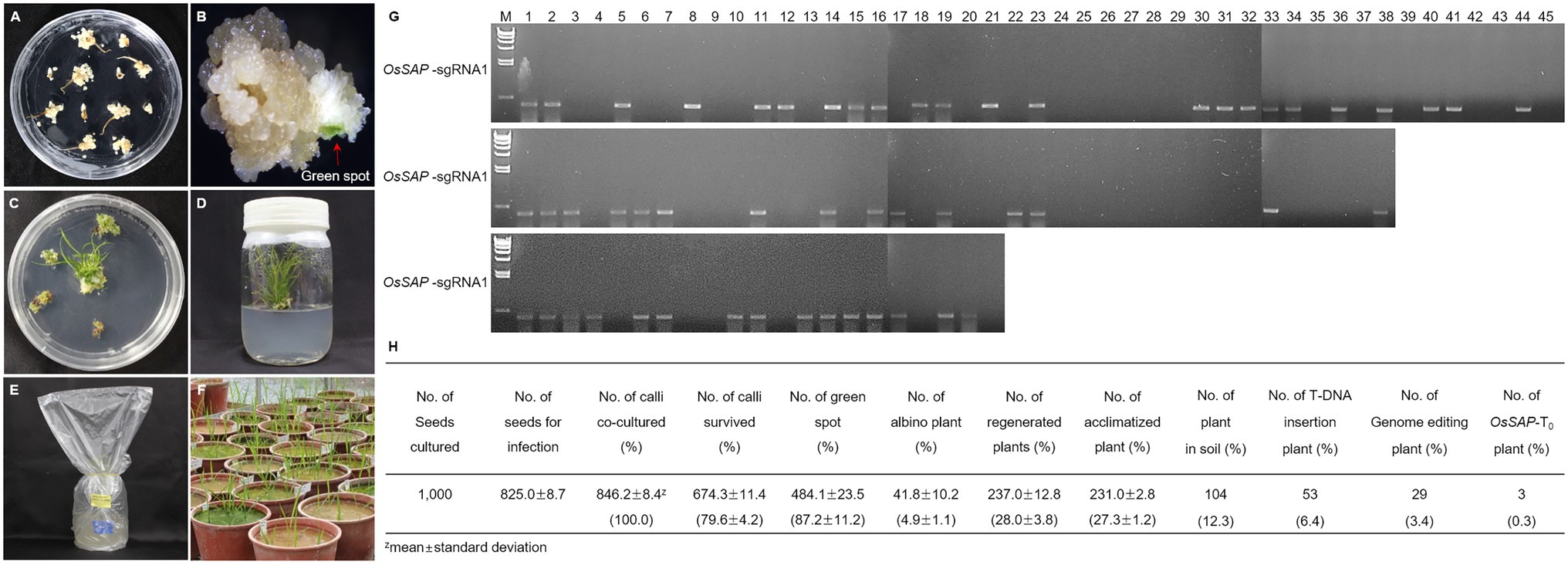
Figure 2. Tissue culture system for development of OsSAP genome-editing rice. (A) When selecting seeds that induced callus, the seeds were clean and uncontaminated. After 21 days, callus was used for inoculation. (B) Agrobacterium was used to inoculate the callus and green spots formed in the regeneration media after 7 days. (C) Shoots from the green spot and a regenerated plant developed after 45 days in regeneration medium. (D) Shooting callus is transferred to new regeneration media for rooting for 30 days. (E) To acclimatize the regenerated plants, the lid of the medium was opened and covered with a plastic bag for 3 days. (F) After acclimatization, regeneration plants were transplanted into soil. After 90 days the T1 seed can be harvested. (G) To analyze T-DNA insertion in regenerated plants, PCR amplification was performed using a hygromycin primer set. The hygromycin sequence was amplified in regeneration plants. The amplified products were loaded on 0.8% agarose gel. M: λ/Hind III size marker, Numeric: number of regenerated plants. (H) Plant regeneration of the OsSAP genome-editing plant.
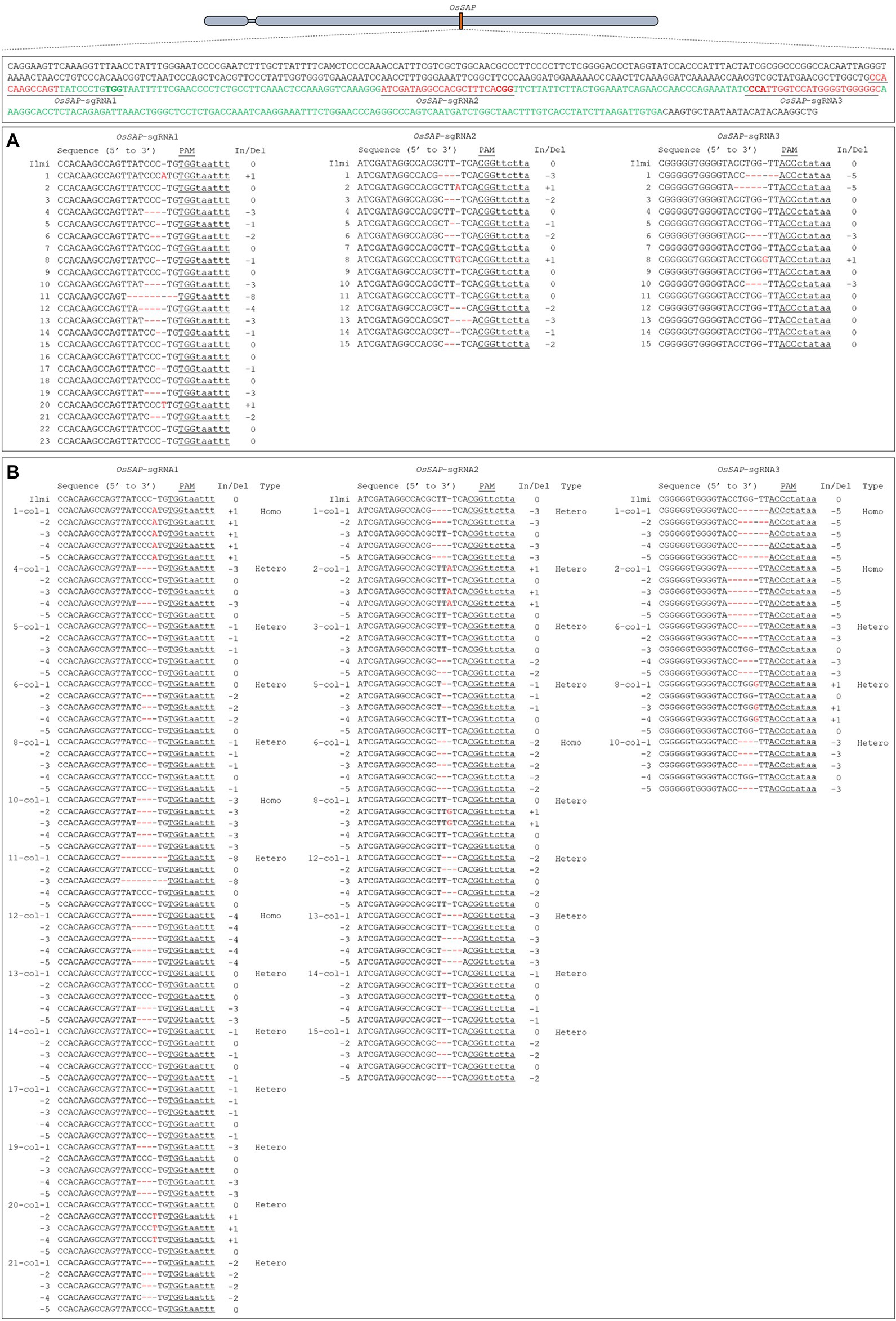
Figure 3. Guide RNA design and genome-editing analysis of target sequence for OsSAP genome editing. (A) The insertion or deletion sequences of target regions are ranked as hyphens (−) with deletion, or plus (+) with insertion. When OsSAP-sgRNA1 was used, 14 genome editing regenerated lines were edited; with OsSAP-sgRNA2, 10 were edited, and with OsSAP-sgRNA3, five regenerated lines were analyzed. (B) Analysis of CRISPR/Cas9-mediated genome-editing type. When guide OsSAP-sgRNA1 was used, editing occurred in 14 genome-editing lines, of which three regenerated lines were homozygous and 11 were heterozygote. In OsSAP-sgRNA2, out of 10 genome-editing lines, only one was homozygous and the rest were heterozygote. For OsSAP-sgRNA3, five genome-editing lines were edited, with two of those lines being homozygous, and the others were heterozygote. Red color: OsSAP-sgRNAs sequence. Green color: Motif sequence of OsSAP.
CRISPR/Cas9-Mediated OsSAP Genome-Editing Lines Exhibited Reduced Drought Tolerance
To evaluate the response of OsSAP to drought conditions, the soil moisture content of the GeD, and OsSAP-T6 lines was reduced to 20%. At this level, the survival rate, shoot length, root length, chlorophyll content, number of tillers, and 1,000-grain weight of each line were investigated (Supplementary Figure S1). These variables showed no significant differences in all lines of GeD, and OsSAP-T6. The survival rates of GeD lines (GeD0 1–1, GeD0 3–1, and GeD0 3–2) under drought stress were lower than that of OsSAP-T6 (43.5% vs. 17.4, 16.8, and 15.6%, respectively). The overall shoot length showed a trend under drought stress: GeD lines (GeD0 1–1, GeD0 3–1, and GeD0 3–2) had shoot lengths of 6.4, 5.8, and 6.1 cm, respectively, which were lower than that of OsSAP-T6 (15.1 cm). Under drought stress, the root lengths (2.8 cm, 3.5 cm, 4.9 cm, respectively) of GeD lines (GeD0 1–1, GeD0 3–1, and GeD0 3–2) were shorter than or equal to that of OsSAP-T6 lines (9.1 cm). The chlorophyll content of OsSAP-T6 plants under drought conditions was 42.1 SPAD; that of the GeD lines (GeD0 1–1, GeD0 3–1, and GeD0 3–2) was lower than that of the OsSAP-T6 Lines. Drought stress treatment reduced the number of tillers; the tiller number of OsSAP-T6, 12.3, was larger than that of the GeD lines at (GeD0 1–1, GeD0 3–1, and GeD0 3–2), 2.3, 2.3, and 2.0, respectively. Finally, when 1,000-grain weights were measured in each line after drought stress, the weight of OsSAP-T6 lines was 20.3 g, whereas the weights of the GeD lines (GeD0 1–1, GeD0 3–1, and GeD0 3–2) were lower at 9.5, 7.8, and 7.1 g, respectively.
Increasing the ROS Damage in OsSAP Genome-Editing Lines
Superoxide dismutase and CAT are important enzymes that protect plant cells from oxidative damage caused by ROS: SOD acts as an antioxidant defense mechanism in almost all living cells by catalyzing the conversion of superoxide (O2−) to O2 and endogenous H2O2; CAT then catalyzes the conversion of H2O2 to H2O and O2−. To analyze the ROS scavenging capacity of OsSAP under drought stress, GeD, and OsSAP-T6 lines were subjected to drought stress until the soil moisture content reached 20%. Under control conditions, the H2O2 contents of GeD, and OsSAP-T6 lines were the same (Figure 4). However, under the drought stress conditions, the amount of H2O2 increased and the H2O2 increase rate differed in each line. In OsSAP-T6, H2O2 increased by 31.6 ± 0.9% due to drought stress. However, in the GeD, H2O2 increased by an average of 71.6 ± 4.5%. Under drought stress, the GeD lines generated more H2O2 than OsSAP-T6. All lines of GeD, and OsSAP-T6 had similar levels of malondialdehyde (MDA) content under control conditions. However, when drought stress was applied, MDA content increased in all analyzed lines relative to the control conditions. However, the OsSAP-T6 lines increased by only 6.5 ± 0.1% on average, whereas the GeD lines showed an average increase of 12.6 ± 3.2%. Proline production by drought treatment was also different in the GeD lines, compared to the OsSAP-T6 lines. Under control conditions, almost no proline was generated in all analyzed lines. However, when drought stress was applied, proline content increased overall in all analyzed lines: the average increase in the OsSAP-T6 line was 494.7 ± 9.1%, which represented an increase of 493.7% ± 9.1%. The average proline content of GeD lines was 283.9 ± 43.9%, which was an increase of 282.9 ± 43.9% when compared with the control. Thus, proline content increased more in the OsSAP-T6 lines following drought stress application. These findings suggest that the expression of OsSAP reduces cell damage in leaves when a rice plant in under drought stress conditions. Under normal conditions, the level of SOD activity was consistent in all lines. However, when drought stress was applied, SOD activity increased in all lines: the SOD activity was highest in the OsSAP-T6 lines and lowest in the GeD lines. POD activity was also the same in all analyzed lines under normal conditions; however, when drought stress was applied, POD activity was highest in the OsSAP-T6 lines and lowest in the GeD lines. CAT showed a similar trend to that of SOD: it was highest in OsSAP-T6 lines and lowest in GeD lines when drought stress was applied. These results suggest the possibility that OsSAP increases drought resistance by acting on the oxidative stress response.
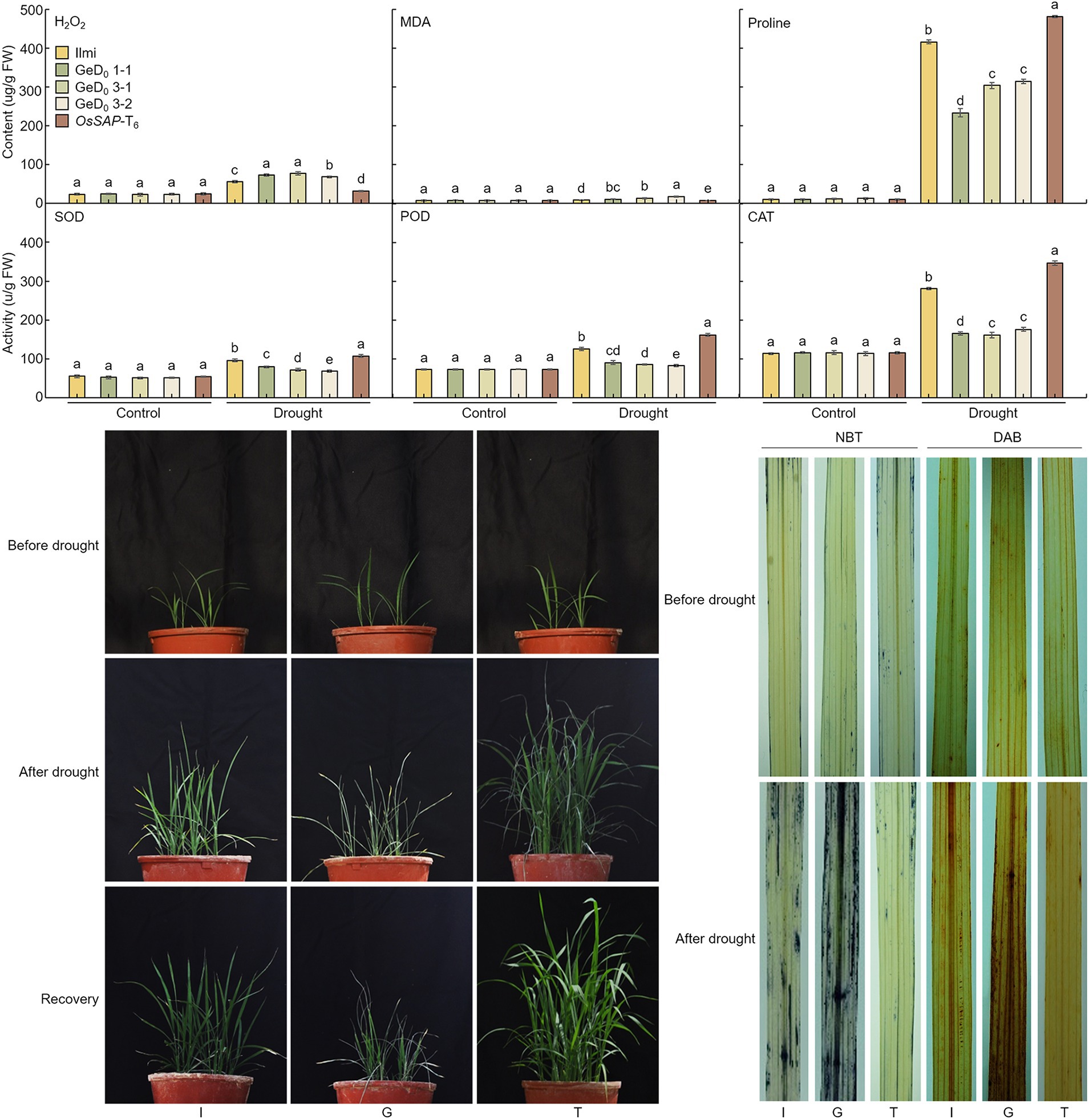
Figure 4. Effects of OsSAP on cellular antioxidants and reactive oxygen species (ROS) levels in Ilmi, GeD, and OsSAP-T6 lines under normal and drought conditions. Under drought conditions, the effects of OsSAP on the level of endogenous hydrogen peroxide (H2O2), the accumulation of malondialdehyde (MDA), the accumulation of proline (proline), the activity of superoxide dismutase (SOD), the activity of peroxidase (POD), and the activity of catalase (CAT). Data presented are expressed as mean ± SD from five independent biological experiments per line. Bars represent means ± SE. Means denoted by the same letter are not significantly different (p < 0.05) as evaluated by Duncan’s multiple range test. The GeD lines showed reduced drought tolerance, whereas the OsSAP-T6 lines showed increased drought tolerance before drought stress, and 20 days after germination. Drought stress treatment; phenotypic changes at 20% soil moisture content. Phenotypes after 7 days of recovery achieved by watering (recovery from 20% soil moisture content). Phenotypes of 3,3-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) staining when the soil moisture content was 20%. After drought stress treatment, H2O2 was visualized as brown spots by DAB, and O2− as blue spots by NBT. The picture presented is the most representative of five independent biological experiments per each line. I: Ilmi. G: GeD0 1–1. T: OsSAP-T6. Different letters on columns represent significant (p < 0.05) difference between rice lines based on Duncan’s test.
When the rice reached the four-leaf stage, the GeD and OsSAP-T6 lines were subjected to drought stress by stopping the water supply until the soil moisture content reached 20%. Before drought stress treatment, there was no difference in the phenotype of all lines and, the growth rate was similar (Figure 4). However, when water was supplied after drought stress, the recovery of the GeD lines was slow, whereas the recovery of the OsSAP-T6 lines was rapid. To determine whether ROS production was related to OsSAP-mediated resistance to drought stress (with moisture content at 20%, as described earlier), the H2O2 content of GeD, and OsSAP-T6 lines after drought stress treatment were analyzed using DAB staining. In the controls, all analyzed lines showed very little staining. Under drought stress, OsSAP-T6 lines were barely stained but the GeD lines produces many dark and brown spots were on their leaves; indeed, most of their leaves were covered with brown spots. In addition, NBT staining was analyzed to evaluate the relationship between OsSAP and O2− production under drought conditions. Under control conditions, almost no blue spots were observed in all lines; however, after drought stress, the OsSAP-T6 lines still showed a similar pattern to the control, whereas the incidence of blue spots increased in GeD lines compared to both. NBT and DAB staining indicated that drought stress generated H2O2 and O2− at increased levels in the GeD lines relative to the levels in the OsSAP-T6 lines, suggesting that OsSAP could control overproduction and regulation of H2O2 under drought stress. Thus, the expression of OsSAP could reduce peroxidation and ROS accumulation by enhancing the activity of major antioxidant enzymes under drought stress conditions.
OsSAP Is Involved in the Expression of Major Stress-Related Regulatory Genes During Drought Stress
To understand the molecular mechanism of OsSAP in drought stress, the gene expression level of transcription factors that respond to drought stresses was analyzed. Specifically, after drought stress on GeD, and OsSAP-T6 lines, relative expression levels of the stress-related transcription factors OsABI5, OsRAB16A, OsLEA3, OsLIP9, OsCATA, OsDREB2A, OsNAC5, and OsNAC6 were examined using qRT-PCR (Figure 5). All analyzed stress-related transcription factors were maintained at low levels or were significantly downregulated in all analyzed lines before drought stress treatment. However, after drought stress, all expression levels were upregulated. Expression levels of these stress-related transcription factors showed contrasting trends in OsSAP-T6 and GeD lines; significantly lower expression levels were observed in GeD compared to OsSAP-T6 lines. These results suggest that overexpression of OsSAP supports an increase in drought stress tolerance and enhances the expression of stress-related genes.
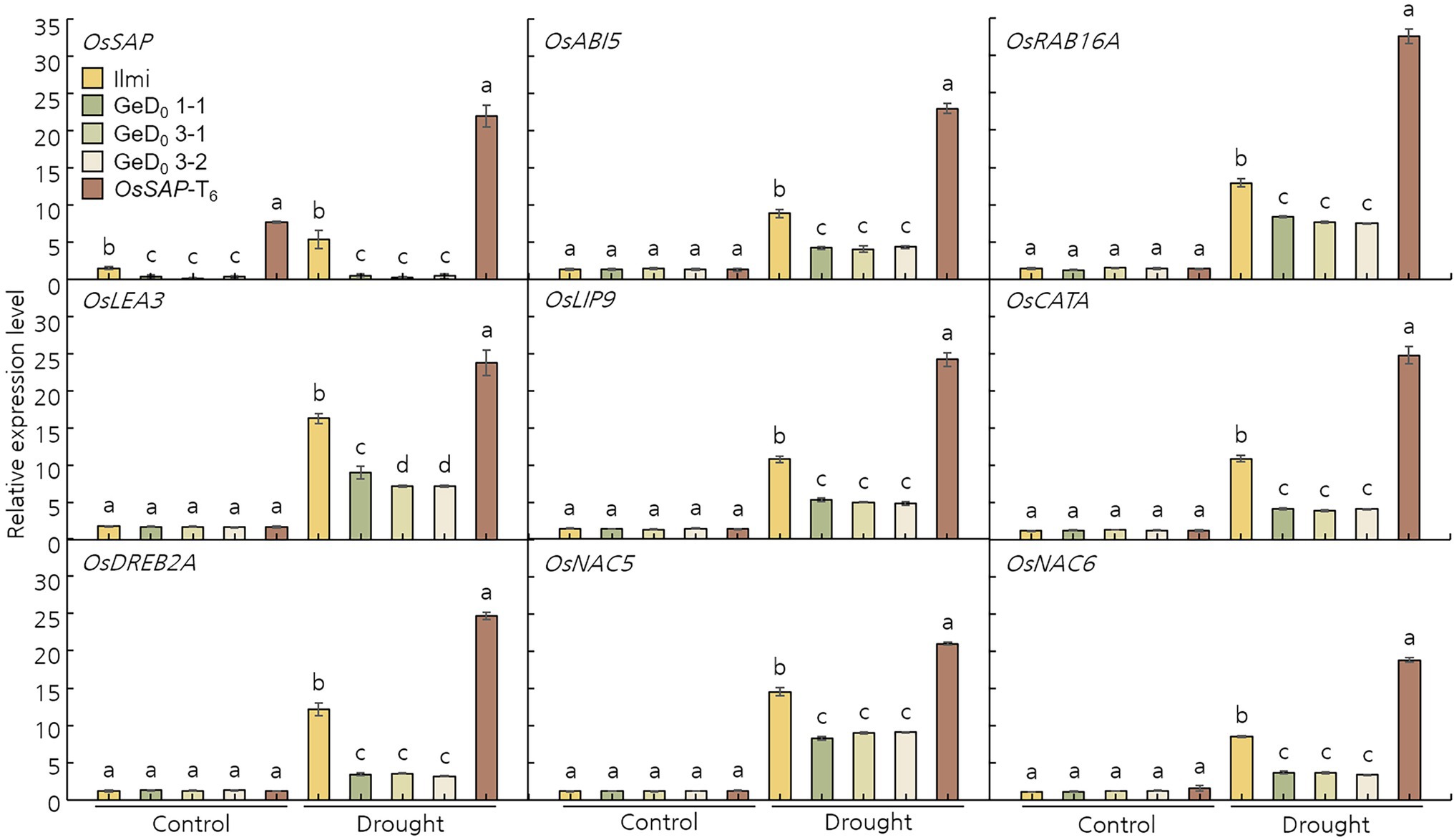
Figure 5. Oryza sativa Senescence-associated protein is involved in the expression levels of several critical stress-related transcription factors under drought conditions. Under normal conditions OsABI5, OsRAB16A, OsLEA3, OsLIP9, OsCATA, OsDREB2A, OsNAC5, and OsNAC6 expression levels were the same. However, overexpression of OsSAP under drought conditions improved the expression levels of the evaluated stress-related transcription factors. Bars represent mean ± SE. Means denoted by the same letter are not significantly different (p < 0.05) as evaluated by Duncan’s multiple range test. Different letters on columns represent significant (p < 0.05) difference between rice lines based on Duncan’s test.
Application of Short Breeding Strategy With CRISPR/Cas9-Mediated Genome-Editing Rice
Expression levels of OsSAP and Cas9 were analyzed in GeD0 and GeD1 plants (Figure 6). In all GeD0 plants, Cas9 was expressed at a level similar to OsActin, but OsSAP was not expressed. In all GeD1 plants, OsSAP was not expressed; however, Cas9 was expressed in various concentrations throughout the GeD1 lines. Cas9 was very weakly expressed in GeD0 1–2, GeD0 1–3, and GeD0 1–4, and GeD0 1–1 and GeD0 1–5. OsSAP was not expressed in all OsSAP-G-2 lines; however, Cas9 expression of each GeD0 2 line was different. In GeD0 2–1, Cas9 was expressed very weakly compared to OsActin, and in GeD0 2–3, Cas9 was expressed at a level similar to that of OsActin. In addition, GeD0 2–2 showed weaker Cas9 expression than OsActin, but stronger Cas9 expression than GeD0 2–1. OsSAP was not expressed in GeD0 3 lines, and the Cas9 expression level of all GeD0 3 lines was weaker than that of OsActin. The expression levels of OsSAP and Cas9 in GeD2 seeds were also analyzed. OsSAP was not expressed in all lines of GeD2 seeds. In the GeD0 1-1-1, GeD0 1-2-1, and GeD0 1-4-1 lines, Cas9 expression was weaker than that of OsActin. Cas9 was strongly expressed in GeD0 1-4-1, and Cas9 expression of GeD0 1-3-1 and GeD0 1-5-1 is similar to that of OsActin. In all the GeD0 2 lines, the Cas9 expression level was lower than that of OsActin. In particular, Cas9 was not expressed in GeD0 2-3-1. In GeD0 3-1-1, the Cas9 expression level was lower than that of OsActin, but the Cas9 expression level of GeD0 3-3-1 and GeD0 3-6-1 was similar to that of OsActin. Agricultural traits of T0 plants, T1 plants, T1 seeds, and T2 seeds were investigated. The coefficient of variation (CV) of the heading date, plant height, and culm length of T0 plant were 2.5, 12.9, and 11.7%, respectively (Supplementary Figure S2). The CVs of the heading date, plant height, and culm length of T1 plant were 3.1, 2.2, and 2.6%, respectively. The CVs of grain length, grain width, and 1,000-grain weight of T1 seed were 4.5, 5.6, and 17.1%, respectively, and the CV of grain length, grain width, and 1,000-grain weight of T2 seed were 1.9, 3.4, and 4.1%, respectively.
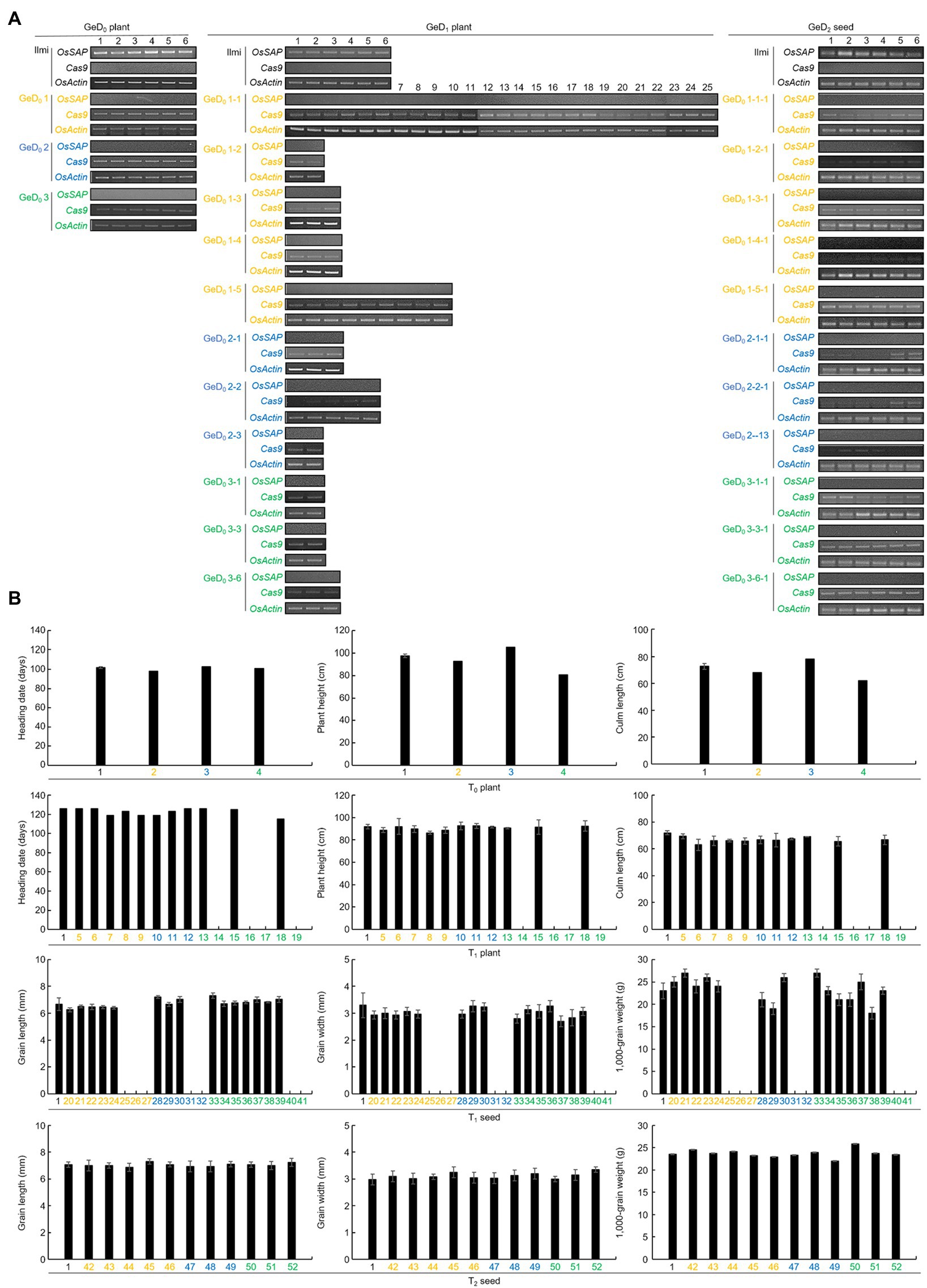
Figure 6. Analysis of the expression level of OsSAP and Cas9 in CRISPR/Cas9-mediated genome-editing lines in each generation through RT-PCR and analysis of agricultural traits of plants and seeds in each generation. (A) Expression levels of OsSAP and Cas9 were analyzed for each line in GeD0 plants, GeD1 plants, and GeD2 seeds, with OsActin used as a control. In the GeD0 plant, Cas9 expression was very strong in all lines with no expression of OsSAP. In GeD1 plants, OsSAP was not expressed. However, Cas9 was still strongly expressed, or the expression was weakened. In GeD2 seeds, OsSAP was not expressed, and Cas9 expression was weakened or maintained. (B) Agricultural traits of plants and seeds in each generation of genome-edited plants with CRISPR/Cas9. Since T0 plants are each different lines, there is no repeated experiment. T1 plants which did not germinate or died early in their growth stage were not investigated. T1 seed numbers were assigned to the tiller of the T0 plants. Effective tillering was investigated by harvesting the grains, but an accession number was only assigned to the non-productive tiller because there were no grains. For T2 seeds, grain length, grain width, and 1,000-grain weights were investigated. The 1 on the x-axis is Ilmi. Yellow color: Genome-editing lines using OsSAP-sgRNA1. Blue color: Genome-editing lines using OsSAP-sgRNA2. Green color: Genome-editing lines using OsSAP-sgRNA3.
Discussion
Globally, unpredictable climate change has seriously threatened crop yields due to adverse biotic and abiotic conditions causing major harvest losses worldwide. The International Food Policy Research Institute predicts that rice demand will increase by 30% by 2050, and that drought stress will reduce rice yield by about 13%. CRISPR/Cas9 has developed rapidly in recent years and is now being applied to gene functional analysis and molecular breeding to combat future losses. Rice has adapted to various environments around the world, and as a result, it has become a very important staple food crop worldwide. Recently, however, the demand for new cultivar with improved resistance to abiotic/biotic stressors has increased and will continue to increase as climate change worsens over time. In this study, CRISPR/Cas9 was used to analyze gene function and to apply it to short breeding in the field. Research shows that ROS plays a key role in regulating the development and stress tolerance response of major crops in an era of unpredictable climate change (Moradi and Ismail, 2007; Steffens, 2014). Senescence-associated genes regulated by biotic stress play a key role in the production of ROS, and the sensitivity and function of stress responses vary according to plant types (Kajimura et al., 2010). Senescence-associated genes are reportedly involved not only in plant growth and development but also in plant resistance to various oxidative stresses (Li et al., 2021). Arabidopsis thaliana AtSOP1 and SAG29 (Sakuraba et al., 2017) act on salt-stress resistance, whereas OsDOF24 from rice is involved in the jasmonate-mediated pathway (Shim et al., 2019), and OsSRLK is involved in phytohormone-mediated chlorophyll degradation (Ying et al., 2020). These genes play an important role in conferring resistance to various biotic/abiotic stresses and establishing a complex signal transduction network in the aging and growth processes of plants.
We found that OsSAP genome editing make more susceptible against drought stress while decrease survival rate, shoot and root lengths, chlorophyll content, tiller number, and 1,000-grain weight. Previous studies have reported that introducing external genes and genome editing into rice and crops can negatively affect plant growth, development, and yield due to signal disturbance of internal genes (Baxter et al., 2015; Metje-Sprink et al., 2020). However, in our study, tiller number and 1,000-grain weight (which are major factors of quantity) were the same after genome editing of OsSAP under normal conditions as they were in Ilmi, that is, no effects were observed. However, under drought stress, OsSAP conferred drought tolerance by positively affecting these variables. Drought stress promoted the generation of excessive ROS in rice. H2O2 and O2−are both representative ROS caused by drought stress (Jira-Anunkul and Pattanagul, 2021; Mohammad et al., 2021). When ROS is generated in high amounts due to various stressors, antioxidant enzymes specifically act on each active oxygen to remove it (Dvořák et al., 2021). MDA, for example, inhibits cell function and induces cell membrane degradation while also acting as an important mediator in ROS scavenging (Sun et al., 2020). Often, the level of MDA in cells can be used to predict the occurrence of programmed cell death and analyze plant cytotoxicity (Wang et al., 2017). SOD specifically removes O2−, whereas CAT and POD are representative antioxidant enzymes that specifically remove H2O2 (Zhang et al., 2019). High levels of H2O2 were detected in the GeD lines, and numerous spots were formed as a result of DAB and NBT staining; this indicates that excessive H2O2 and O2− were present. However, in the OsSAP-T6 lines, H2O2 and O2− were hardly detected. The OsSAP-T6 lines also showed an increased activity of antioxidant enzymes, that is, CAT, SOD, and POD, whereas the GeD lines maintained antioxidant enzyme activity at a relatively low level. Therefore, OsSAP seems to improve drought tolerance by protecting against the oxidative damage caused by stress, which is achieved by improving the ability of plants to remove ROS when faced with drought stress (Qi et al., 2018). Plants have developed and evolved a complex antioxidant system to resist stress from various free radicals; consequently, they can maintain high expression levels of antioxidant enzymes, such as CAT, SOD, and POD, to confer stress resistance (Huang et al., 2019). We monitored the expression levels of various transcription factors related to drought stress, such as OsABI5 (Skubacz et al., 2016), OsRAB16A (Ganguly et al., 2012), OsLEA3 (Xiao et al., 2007), OsLIP9 (Chen et al., 2015), OsCATA (Yong et al., 2017), OsDREB2A (Agarwal et al., 2007), OsNAC5 (Song et al., 2011), and OsNAC6 (Lee et al., 2017), to understand molecular stress tolerance to OsSAP under drought conditions. The selected genes are known to be involved in resistance to abiotic stresses, such as drought stress, in various plants. These genes were downregulated in the GeD lines when compared with their expression in Ilmi under drought stress, which is consistent with the level of activity of ROS antioxidant enzymes observed. We found that overexpression of OsSAP improved the ability of plants to remove ROS and increased the transcript levels of genes required to resist drought stress. In addition, OsABI5, OsRAB16A, OsLEA3, OsLIP9, OsCATA, OsDREB2A, OsNAC5, and OsNAC6 are genes that improve the tolerance of various plants to abiotic stress. Senescence-associated genes belong to a large family with functions related to cell death under various abiotic/biotic stresses (Pan et al., 2016).
Using these research results, we propose a response model for OsSAP under drought stress conditions (Supplementary Figure S3). Drought stress induces rapid overexpression of OsSAP, which in turn induces the expression of antioxidant enzymes, such as CAT, SOD, and POD, which increases the overall activity of these enzymes. Therefore, the scavenging ability of ROS caused by drought stress is improved, and drought tolerance emerges. OsSAP also increases drought tolerance by enhancing the expression of stress-related transcription factors. CRISPR/Cas9 can shorten the breeding period in rice (Figure 7). The CRISPR/Cas9 vector transformed into the callus by the Agrobacterium-mediated method; T0 plants were selected through T-DNA insertions and target gene editing sequences. Proper T0 plant selection and subsequent harvest of a T1 seed from a T0 plant takes 0.3 years. Here, we used the “ear-to-row method” to apply CRISPR/Cas9 to rice breeding. In this study, GeD0 1 had five spikes, GeD0 2 had three spikes, and GeD0 3 had seven spikes. In the field, T1 plants were transplanted in one row for each spike, and T2 seeds were harvested in bulk. Agricultural traits of the T0 plants, T1 plants, T1 seeds, and T2 seeds were investigated, and the CV of both T1 plants and T2 seeds were lower than 5%. Taken together, our results suggest that CRISPR/Cas9 can accurately genome edit homozygous target genes in T0 plants, and short breeding cycles are possible evidenced by the low CV in T1 plants and T2 seeds. Therefore, CRISPR/Cas9 can be used as a simple, rapid, and powerful tool in crop improvement, and will greatly advance molecular design in crop breeding. Our results suggest that a high efficiency of CRISPR/Cas9 genome editing and breeding in rice is possible, as a significant percentage of our T0 plants were homozygous for the target edited gene; however, not all target sequences were edited in T0 plants as evidenced by an abundance of heterozygotes and unedited genes.
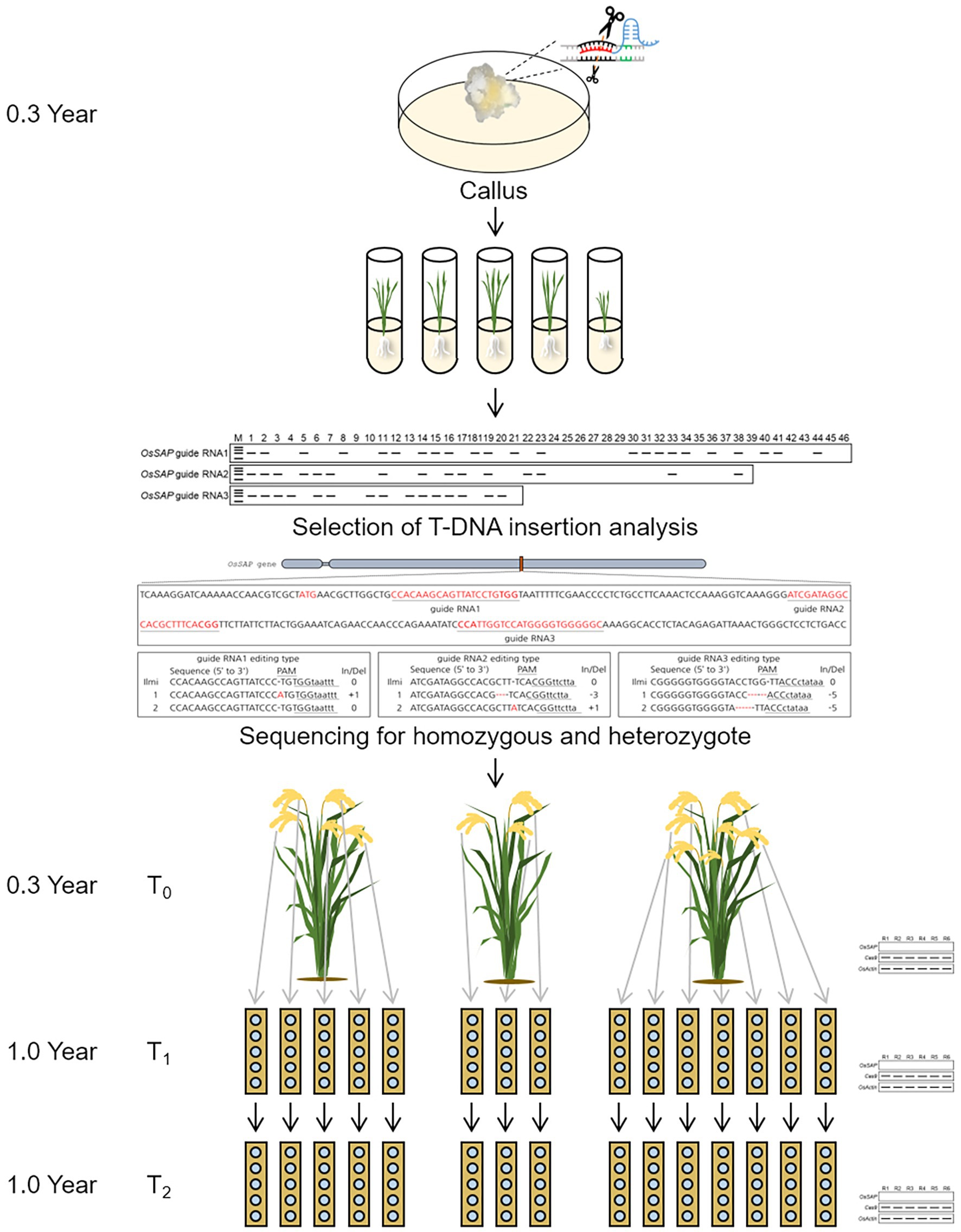
Figure 7. Scheme of short breeding cycle using CRISPR/Cas9 in rice. Agrobacterium-mediated transformation used for genome editing with CRISPR/Cas9. In regenerated plants, a gene was selected and edited by T-DNA insertion and sequencing. On the T0 plant, each spike is assigned a different accession number. T1 plants are transplanted to one row for each line. T2 seeds are harvested in bulk.
In this research, overexpression of OsSAP during drought stress increased drought resistance, whereas genome editing targeted at OsSAP increased drought susceptibility. Therefore, these findings provide evidence that OsSAP could be a potential candidate gene for the improvement of rice growth under drought stress. This would, in turn, improve the major agricultural traits of plants in stressed conditions, leaving them fairing as well as or better as plants in “normal” conditions (Sade et al., 2018; Piao et al., 2019). To resist severe abiotic/biotic stressors, cultivars are being developed with improved stress tolerance through the overexpression of stress-related genes and via genome editing. However, the continuous overexpression of such genes can induce the development of malformed plants and ultimately reduce their yield (Baxter et al., 2015). Thus, even if drought resistance is improved, the high proportion of malformed plants and lower yields would prevent their usage in farms. However, with overexpression of OsSAP, the characteristics of major agricultural traits, including Ilmi and yield amount, were the same as those found under normal conditions. Under drought conditions, when OsSAP was overexpressed, the major agricultural traits, including yield, were improved relative to those of Ilmi. In the era of unpredictable climate change, our research into OsSAP provides insights that could help resolve food shortages and food security, worldwide.
Conclusion
By applying CRISPR/Cas9, the functional loss of OsSAP interfered with the antioxidant defense by reducing the activation of SOD, POD, and CAT, and finally, the negative effects of various agricultural traits due to the accumulation of ROS and MDA resulted in leaf aging and reduction 1,000-grain weight. These results suggest that OsSAP plays an important role in inducing tolerance to drought stress, is related to leaf senescence, and can be used as a potential gene for high yield in drought stress when breeding new rice cultivars. Our research demonstrated promising applications of CRISPR/Cas9-mediated genomic engineering for basic and applied studies of rice and various plants in preparation for future unpredictable climate change. When a genome-edited rice plant is developed by CRISPR/Cas9 technology, homozygote type can be developed in T0 plants. In addition, the expression of the target gene was continuously removed from the T0 plant, and the expression of Cas9 remained stable or weakened as the course of generation advancement progressed. Although the expression of Cas9 was reduced, the expression of the target gene was still not analyzed. If CRISPR/Cas9 is applied to breeding, the desired trait can be improved by editing the target gene in early generation with homozygote type. Finally, CRISPR/Cas9 enables a short breeding cycle.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
J-RP and E-GK: conceptualization of experiments, methodology and research plan, and writing the original draft. Y-HJ: formal analysis. E-GK and Y-HJ: conducting experiments. RJ, MF, and MU: participating in phenotype evaluations. J-RP: reviewing and editing the writing. K-MK: project administration. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by a grant from the New Breeding Technologies Development Program (Project No. PJ016531012022), Rural Development Administration, Republic of Korea. This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2021M3E5E6022715).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.850441/full#supplementary-material
Footnotes
References
Agarwal, P., Agarwal, P. K., Nair, S., Sopory, S. K., and Reddy, M. K. (2007). Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA-binding activity. Mol. Gen. Genomics. 277, 189–198. doi: 10.1007/s00438-006-0183-z
Ariani, F., Mari, F., Pescucci, C., Longo, I., Bruttini, M., Meloni, I., et al. (2004). Real-time quantitative PCR as a routine method for screening large rearrangements in Rett syndrome: report of one case of MECP2 deletion and one case of MECP2 duplication. Hum. Mutat. 24, 172–177. doi: 10.1002/humu.20065
Bai, S., Yu, H., Wang, B., and Li, J. (2018). Retrospective and perspective of rice breeding in China. J. Genet. Genomics 45, 603–612. doi: 10.1016/j.jgg.2018.10.002
Barman, H. N., Sheng, Z., Fiaz, S., Zhong, M., Wu, Y., Cai, Y., et al. (2019). Generation of a new thermo-sensitive genic male sterile rice line by targeted mutagenesis of TMS5 gene through CRISPR/Cas9 system. BMC Plant Biol. 19, 109–111. doi: 10.1186/s12870-019-1715-0
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Baxter, H. L., Poovaiah, C. R., Yee, K. L., Mazarei, M., Rodriguez, M., Thompson, O. A., et al. (2015). Field evaluation of transgenic switchgrass plants overexpressing PvMYB4 for reduced biomass recalcitrance. Bioenergy Res. 8, 910–921. doi: 10.1007/s12155-014-9570-1
Beers, R. F., and Sizer, I. W. (1952). A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195, 133–140. doi: 10.1016/s0021-9258(19)50881-x
Cezary, W., Melanie, C., and Kangasjarvi, J. (2018). Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 145, 209–236. doi: 10.1242/dev.164376
Chen, H., Zhang, Q., Lu, Z., and Xu, F. (2018). Accumulation of ammonium and reactive oxygen mediated drought-induced rice growth inhibition by disturbed nitrogen metabolism and photosynthesis. Plant Soil 431, 107–117. doi: 10.1007/s11104-018-3752-0
Chen, M., Zhao, Y., Zhuo, C., Lu, S., and Guo, Z. (2015). Overexpression of a NF-YC transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 13, 482–491. doi: 10.1111/pbi.12270
Duan, J., Zhang, M., Zhang, H., Xiong, H., Liu, P., Ali, J., et al. (2012). OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci. 196, 143–151. doi: 10.1016/j.plantsci.2012.08.003
Dvořák, P., Krasylenko, Y., Zeiner, A., Šamaj, J., and Takáč, T. (2021). Signaling toward reactive oxygen species-scavenging enzymes in plants. Front. Plant Sci. 11:618835. doi: 10.3389/fpls.2020.618835
Fiaz, S., Ahmad, S., Noor, M. A., Wang, X., Younas, A., Riaz, A., et al. (2019). Applications of the CRISPR/Cas9 system for rice grain quality improvement: perspectives and opportunities. Int. J. Mol. Sci. 20, 1–18. doi: 10.3390/ijms20040888
Fiaz, S., Wang, X., Khan, S. A., Ahmar, S., Noor, M. A., Riaz, A., et al. (2021a). Novel plant breeding techniques to advance nitrogen use efficiency in rice: a review. GM Crops Food 25, 1–20. doi: 10.1080/21645698.2021.1921545
Fiaz, S., Wang, X., Younas, A., Alharthi, B., Riaz, A., and Ali, H. (2021b). Apomixis and strategies to induce apomixis to preserve hybrid vigor for multiple generations. GM Crops Food 12, 57–70. doi: 10.1080/21645698.2020.1808423
Fichman, Y., and Mittler, R. (2020). Rapid systemic signaling during abiotic and biotic stresses: is the ROS wave master of all trades? Plant J. 102, 887–896. doi: 10.1111/tpj.14685
Fitzgerald, H. A., Chern, M. S., Navarre, R., and Ronald, P. C. (2004). Overexpression of (At)NPR1 in rice leads to a BTH- and environment-induced lesion-mimic/cell death phenotype. Mol. Plant-Microbe Interact. 17, 140–151. doi: 10.1094/MPMI.2004.17.2.140
Ganguly, M., Datta, K., Roychoudhury, A., Gayen, D., Sengupta, D. N., and Datta, S. K. (2012). Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal. Behav. 7, 502–509. doi: 10.4161/psb.19646
Ganie, S. A., and Ahammed, G. J. (2021). Dynamics of cell wall structure and related genomic resources for drought tolerance in rice. Plant Cell Rep. 40, 437–459. doi: 10.1007/s00299-020-02649-2
Guo, H., Wang, R., Garfin, G. M., Zhang, A., Lin, D., Liang, Q., et al. (2021). Rice drought risk assessment under climate change: based on physical vulnerability a quantitative assessment method. Sci. Total Environ. 751:141481. doi: 10.1016/j.scitotenv.2020.141481
Heath, R. L., and Packer, L. (1968). Photoperoxidation in isolated chloroplasts. Arch. Biochem. Biophys. 125, 189–198. doi: 10.1016/0003-9861(68)90654-1
Hu, X., Meng, X., Liu, Q., Li, J., and Wang, K. (2018). Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice. Plant Biotechnol. J. 16, 292–297. doi: 10.1111/pbi.12771
Huang, H., Ullah, F., Zhou, D. X., Yi, M., and Zhao, Y. (2019). Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 10:800. doi: 10.3389/fpls.2019.00800
Ilyas, M., Nisar, M., Khan, N., Hazrat, A., Khan, A. H., Hayat, K., et al. (2021). Drought tolerance strategies in plants: a mechanistic approach. J. Plant Growth Regul. 40, 926–944. doi: 10.1007/s00344-020-10174-5
Jiang, Y., Ling, L., Zhang, L., Wang, K., Li, X., Cai, M., et al. (2018). Comparison of transgenic Bt rice and their non-Bt counterpart in yield and physiological response to drought stress. Field Crop Res. 217, 45–52. doi: 10.1016/j.fcr.2017.12.007
Jira-Anunkul, W., and Pattanagul, W. (2021). Effects of hydrogen peroxide application on agronomic traits of rice (Oryza sativa L.) under drought stress. Plant Soil Environ. 67, 221–229. doi: 10.17221/628/2020-PSE
Kajimura, T., Mizuno, N., and Takumi, S. (2010). Utility of leaf senescence-associated gene homologs as developmental markers in common wheat. Plant Physiol. Biochem. 48, 851–859. doi: 10.1016/j.plaphy.2010.08.014
Lee, D. K., Chung, P. J., Jeong, J. S., Jang, G., Bang, S. W., Jung, H., et al. (2017). The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 15, 754–764. doi: 10.1111/pbi.12673
Li, L., He, Y., Zhang, Z., Shi, Y., Zhang, X., Xu, X., et al. (2021). OsNAC109 regulates senescence, growth and development by altering the expression of senescence- and phytohormone-associated genes in rice. Plant Mol. Biol. 105, 637–654. doi: 10.1007/s11103-021-01118-y
Li, N., Kong, L., Zhou, W., Zhang, X., Wei, S., Ding, X., et al. (2013). Overexpression of Os2H16 enhances resistance to phytopathogens and tolerance to drought stress in rice. Plant Cell Tissue Organ Cult. 115, 429–441. doi: 10.1007/s11240-013-0374-3
Li, C., Li, W., Zhou, Z., Chen, H., Xie, C., and Lin, Y. (2020). A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 18, 313–315. doi: 10.1111/pbi.13217
Li, J., Sun, Y., Du, J., Zhao, Y., and Xia, L. (2017). Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 10, 526–529. doi: 10.1016/j.molp.2016.12.001
Liu, J., Hasanuzzaman, M., Wen, H., Zhang, J., Peng, T., Sun, H., et al. (2019). High temperature and drought stress cause abscisic acid and reactive oxygen species accumulation and suppress seed germination growth in rice. Protoplasma 256, 1217–1227. doi: 10.1007/s00709-019-01354-6
Liu, H., Ma, Y., Chen, N., Guo, S., Liu, H., Guo, X., et al. (2014). Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 37, 1144–1158. doi: 10.1111/pce.12223
Liu, Y., Wang, W., Yang, B., Currey, C., and Fei, S. Z. (2020). Functional analysis of the teosinte branched 1 gene in the tetraploid switchgrass (Panicum virgatum L.) by CRISPR/Cas9-directed mutagenesis. Front. Plant Sci. 11:572193. doi: 10.3389/fpls.2020.572193
Metje-Sprink, J., Sprink, T., and Hartung, F. (2020). Genome-edited plants in the field. Curr. Opin. Biotechnol. 61, 1–6. doi: 10.1016/j.copbio.2019.08.007
Mikami, M., Toki, S., and Endo, M. (2015). Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 88, 561–572. doi: 10.1007/s11103-015-0342-x
Mohammad, A. H., Lutful, H., Khandakar, M. I., Arvind, K., and Robert, H. (eds.) (2021). “Molecular breeding for rice abiotic stress tolerance and nutritional quality,” in Molecular Breeding for Improving Drought Tolerance in Rice: Recent Progress and Future Perspective. (West Sussex, UK: John Wiley & Sons Ltd), 53–74.
Moradi, F., and Ismail, A. M. (2007). Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice. Ann. Bot. 99, 1161–1173. doi: 10.1093/aob/mcm052
Munro, D., and Treberg, J. R. (2017). A radical shift in perspective: mitochondria as regulators of reactive oxygen species. J. Exp. Biol. 220, 1170–1180. doi: 10.1242/jeb.132142
Nahar, S., Vemireddy, L. R., Sahoo, L., and Tanti, B. (2018). Antioxidant protection mechanisms reveal significant response in drought-induced oxidative stress in some traditional rice of Assam, India. Rice Sci. 25, 185–196. doi: 10.1016/j.rsci.2018.06.002
Pan, Y. J., Liu, L., Lin, Y. C., Zu, Y. G., Li, L. P., and Tang, Z. H. (2016). Ethylene antagonizes salt-induced growth retardation and cell death process via transcriptional controlling of ethylene-, BAG- and senescence-associated genes in Arabidopsis. Front. Plant Sci. 7:696. doi: 10.3389/fpls.2016.00696
Park, J., Bae, S., and Kim, J. S. (2015). Cas-designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016. doi: 10.1093/bioinformatics/btv537
Passioura, J. (2007). The drought environment: physical, biological and agricultural perspectives. J. Exp. Bot. 58, 113–117. doi: 10.1093/jxb/erl212
Piao, W., Sakuraba, Y., and Paek, N. C. (2019). Transgenic expression of rice MYB102 (OsMYB102) delays leaf senescence and decreases abiotic stress tolerance in Arabidopsis. BMB Rep. 52, 653–658. doi: 10.5483/BMBRep.2019.52.11.071
Prabnakorn, S., Maskey, S., Suryadi, F. X., and de Fraiture, C. (2018). Rice yield in response to climate trends and drought index in the Mun river basin, Thailand. Sci. Total Environ. 621, 108–119. doi: 10.1016/j.scitotenv.2017.11.136
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Qi, J., Wang, J., Gong, Z., and Zhou, J. M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Rao, M. V., Lee, H. I., Creelman, R. A., Mullet, J. E., and Davis, K. R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12, 1633–1646. doi: 10.1105/tpc.12.9.1633
Rehman, F., Gong, H., Bao, Y., Zeng, S., Huang, H., and Wang, Y. (2022). CRISPR gene editing of major domestication traits accelerating breeding for Solanaceae crops improvement. Plant Mol. Biol. 108, 157–173. doi: 10.1007/s11103-021-01229-6
Sade, N., Del Mar Rubio-Wilhelmi, M., Umnajkitikorn, K., and Blumwald, E. (2018). Stress-induced senescence and plant tolerance to abiotic stress. J. Exp. Bot. 69, 845–853. doi: 10.1093/jxb/erx235
Sakuraba, Y., Bülbül, S., Piao, W., Choi, G., and Paek, N. C. (2017). Arabidopsis EARLY FLOWERING3 increases salt tolerance by suppressing salt stress response pathways. Plant J. 92, 1106–1120. doi: 10.1111/tpj.13747
Shamloo-Dashtpagerdi, R., Lindlöf, A., Aliakbari, M., and Pirasteh-Anosheh, H. (2020). Plausible association between drought stress tolerance of barley (Hordeum vulgare L.) and programmed cell death via MC1 and TSN1 genes. Physiol. Plant. 170, 46–59. doi: 10.1111/ppl.13102
Shi, H., Wang, Y., Cheng, Z., Ye, T., and Chan, Z. (2012). Analysis of natural variation in bermudagrass (Cynodon dactylon) reveals physiological responses underlying drought tolerance. PLoS One 7:e53422. doi: 10.1371/journal.pone.0053422
Shim, Y., Kang, K., An, G., and Paek, N. C. (2019). Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 60, 2065–2076. doi: 10.1093/pcp/pcz105
Siebert, S., Webber, H., and Rezaei, E. E. (2017). Weather impacts on crop yields—searching for simple answers to a complex problem. Environ. Res. Lett. 12:081001. doi: 10.1088/1748-9326/aa7f15
Singh, A., Kumar, A., Yadav, S., and Singh, I. K. (2019). Reactive oxygen species-mediated signaling during abiotic stress. Plant Gene 18:100173. doi: 10.1016/j.plgene.2019.100173
Skubacz, A., Daszkowska-Golec, A., and Szarejko, I. (2016). The role and regulation of ABI5 (ABA-insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7:1884. doi: 10.3389/fpls.2016.01884
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., and Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234, 331–345. doi: 10.1007/s00425-011-1403-2
Steffens, B. (2014). The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 5:685. doi: 10.3389/fpls.2014.00685
Sun, Y., Wang, C., Chen, H. Y. H., and Ruan, H. (2020). Response of plants to water stress: a meta-analysis. Front. Plant Sci. 11:978. doi: 10.3389/fpls.2020.00978
Tsai, S. Q., and Joung, J. K. (2016). Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat. Rev. Genet. 17, 300–312. doi: 10.1038/nrg.2016.28
Ubaidillah, M., Kim, K. A., Kim, Y. H., Lee, I. J., Yun, B. W., Kim, D. H., et al. (2013). Identification of a drought-induced rice gene, OsSAP, that suppresses Bax-induced cell death in yeast. Mol. Biol. Rep. 40, 6113–6121. doi: 10.1007/s11033-013-2723-z
Uji, Y., Akimitsu, K., and Gomi, K. (2017). Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 245, 1241–1246. doi: 10.1007/s00425-017-2697-5
Verma, G., Srivastava, D., Tiwari, P., and Chakrabarty, D. (2019). “ROS modulation in crop plants under drought stress,” in Reactive Oxygen, Nitrogen and Sulfur Species in Plants: Production, Metabolism, Signaling and Defense Mechanisms. eds. K. Nahar, M. Fujita, M. Hasanuzzaman, and V. Fotopoulos (West Sussex, UK: John Wiley & Sons Ltd), 311–336.
Wang, S. S., Zhang, Y. X., Yang, F., Huang, Z. Q., Tang, J., Hu, K.Di, et al. (2017). Sulfur dioxide alleviates programmed cell death in barley aleurone by acting as an antioxidant. PLoS One 12:e0188289. doi: 10.1371/journal.pone.0188289
Weydert, C. J., and Cullen, J. J. (2010). Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 5, 51–66. doi: 10.1038/nprot.2009.197
Xiao, B., Huang, Y., Tang, N., and Xiong, L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46. doi: 10.1007/s00122-007-0538-9
Yang, X., Wang, B., Chen, L., Li, P., and Cao, C. (2019). The different influences of drought stress at the flowering stage on rice physiological traits, grain yield, and quality. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-40161-0
Ying, Y., Liu, F. F., Li, G. P., Zheng, Q., Li, B., Li, Z. S., et al. (2020). Silencing of the receptor-like cytoplasmic kinase gene tarkl1 reduces photosynthetic capacity in wheat. Photosynthetica 58, 1188–1199. doi: 10.32615/ps.2020.069
Yong, B., Wang, X., Xu, P., Zheng, H., Fei, X., Hong, Z., et al. (2017). Isolation and abiotic stress resistance analyses of a catalase gene from Ipomoea batatas (L.) Lam. Biomed. Res. Int. 2017:6847532. doi: 10.1155/2017/6847532
Yukoh, H., and Toshihiko, K. (2008). Arobacterium-mediated transformation: rice transformation; agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat. Protoc. 3, 824–834. doi: 10.1038/nprot.2008.46
Zhang, H., Li, Y., and Zhu, J. K. (2018). Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 4, 989–996. doi: 10.1038/s41477-018-0309-4
Zhang, W., Liu, S., Li, C., Zhang, P., and Zhang, P. (2019). Transcriptome sequencing of Antarctic moss under salt stress emphasizes the important roles of the ROS-scavenging system. Gene 696, 122–134. doi: 10.1016/j.gene.2019.02.037
Zhou, J., Deng, K., Cheng, Y., Zhong, Z., Tian, L., Tang, X., et al. (2017). CRISPR-cas9 based genome editing reveals new insights into microRNA function and regulation in rice. Front. Plant Sci. 8:1598. doi: 10.3389/fpls.2017.01598
Keywords: abiotic stress, agricultural trait, generation, OsSAP, reactive oxygen species
Citation: Park J-R, Kim E-G, Jang Y-H, Jan R, Farooq M, Ubaidillah M and Kim K-M (2022) Applications of CRISPR/Cas9 as New Strategies for Short Breeding to Drought Gene in Rice. Front. Plant Sci. 13:850441. doi: 10.3389/fpls.2022.850441
Edited by:
Qian-Hao Zhu, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaReviewed by:
Sajid Fiaz, University of Haripur, PakistanMuntazir Mushtaq, National Bureau of Plant Genetic Resources (ICAR), India
Copyright © 2022 Park, Kim, Jang, Jan, Farooq, Ubaidillah and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyung-Min Kim, a2ttQGtudS5hYy5rcg==
†ORCID: Jae-Ryoung Park, orcid.org/0000-0003-2207-5313
Eun-Gyeong Kim, orcid.org/0000-0002-8532-608X
Yoon-Hee Jang, orcid.org/0000-0002-6282-9557
Rahmatullah Jan, orcid.org/0000-0001-5788-3017
Muhammad Farooq, orcid.org/0000-0002-4917-4402
Mohammad Ubaidillah, orcid.org/0000-0003-4745-3878
Kyung-Min Kim, orcid.org/0000-0003-4812-6297
‡These authors have contributed equally to this work
 Jae-Ryoung Park
Jae-Ryoung Park Eun-Gyeong Kim
Eun-Gyeong Kim Yoon-Hee Jang
Yoon-Hee Jang Rahmatullah Jan
Rahmatullah Jan Muhammad Farooq
Muhammad Farooq Mohammad Ubaidillah
Mohammad Ubaidillah Kyung-Min Kim
Kyung-Min Kim