- 1Key Laboratory of Jiangsu Province for Agrobiology, Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences, Nanjing, China
- 2State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, Nanjing, China
Chloroplast development is crucial for photosynthesis and plant growth and many factors are involved in its regulation. The regulatory mechanism differs in different green tissues, and previous studies have focused on chloroplasts in leaves. In this study, a mutant with sepal-specific chlorophyll-deficiency was observed in Brassica napus and named as df74. Genetic analysis indicated that the phenotype was controlled by a single recessive nuclear gene. The gene was located on chromosome C08 by bulked-segregant analysis with whole-genome sequencing, which was designated as BnC08.cds. To fine-map the BnC08.cds, a F2 population was created from the cross of df74 and Zhongshuang11 (ZS11). Finally, the BnC08.cds was fine-mapped in the region between the single-nucleotide polymorphism (SNP) markers M5 and M6, corresponding to a 228.72 kb interval of the B. napus “ZS11” genome. Eighteen genes were predicted in the target region, and it was speculated that BnaC08G0442100ZS was the most likely candidate gene based on the results of transcriptome analyses and sequence variation analyses. These results provide a foundation to explore the regulation of chloroplast development in sepals.
Introduction
Green tissues are the photosynthetic organs in plants and their color is affected by the chlorophyll (Chl) content, which is the key factor for photomorphogenesis and photosynthesis. The biosynthesis and accumulation of Chl is a complex physiological and biochemical process. The mutation of any gene in this process may affect the content of Chl, so color mutants can readily form in artificial or spontaneous mutagenesis. The mutants are diverse, such as etiolation (Zhu et al., 2014), albino (Wang et al., 2016), white to green (Zhao et al., 2013; Ma et al., 2017), and stay-green (Jiang et al., 2007). These mutants are important for the study of Chl biosynthesis and degradation, chloroplast development, and chloroplast-nuclear signal transmission (Fu et al., 2019).
According to previous research, the factors involved in Chl content can be roughly divided into three categories: biosynthesis and degradation of Chl, metabolism of heme, and chloroplast development. In the first category, biosynthesis of Chl involves 16 enzymes encoded by 28 genes in Arabidopsis thaliana (Tanaka and Tanaka, 2007). The mutation of genes in this pathway may hinder the synthesis of Chl and cause chlorosis. On the contrary, mutations in genes related to the pathway of Chl degradation often trigger stay-green (Thomas and Ougham, 2014). Genes involved in metabolism of heme belong to the second category. The biosynthesis of Chl and heme are two branches of the tetrapyrrole biosynthetic pathway. They share precursors from 5-aminolevulinic acid to protoporphyrin IX, and they inhibit each other. If the content of heme increases, the excess heme will inhibit the biosynthesis of Chl via feedback, resulting in reduced Chl which causes the color mutation (Brzezowski et al., 2015). The third category is genes involved in chloroplast development. This is a complex network including chloroplast and nuclear gene transcription, RNA modification and processing, protein translation, folding and transport, and thylakoid formation (Beck, 2001). Mutations in any one of the genes may cause the blocking of chloroplast development and lead to color mutation.
The biogenesis and development of chloroplasts differ between organs and species (Pogson and Albrecht, 2011). It is useful to understand the molecular mechanisms underlying tissue-specific chloroplast development. Genetic studies have demonstrated that chloroplast development proceeds differently in cotyledons and true leaves. The var2 mutants produce green cotyledons but chlorotic true leaves (Liu et al., 2010). Conversely, the snowy cotyledon (sco) mutants have green true leaves but chlorotic or bleached cotyledons (Albrecht et al., 2006, 2008, 2010). Burley is another important tissue-specific chlorophyll-deficient mutant. It is a special type of tobacco (Nicotiana tabacum L.) cultivar that is characterized by white stem and leaf midvein. A two-step mutation process in the double White Stem 1 homologs has driven the evolution of burley tobacco (Wu et al., 2020). A similar phenotype was identified from a tomato mutant with albinic stems and this is caused by a mutation of SlARC6, which leads to the tissue-specific defects in chloroplast development (Chang et al., 2021).
Sepal is a kind of green tissue in many higher plants. It plays an important role in protecting the other floral organs. Sepals are thought to resemble leaves morphologically because of their green color and Chl content. In this study, we obtained a rapeseed mutant (named df74) with Chl-deficient sepals by ethyl methane sulfonate (EMS) mutagenizing. The mutant produces pale yellow sepals while the color of other green tissues stays normal. This kind of sepal-specific Chl-deficiency, as far as we know, has rarely been reported and it is a good candidate for studying the regulation of chloroplast development in sepals. Our genetic analysis has revealed that the phenotype of df74 is controlled by a single recessive nuclear gene. The gene was mapped on chromosome C08, in an interval of 228.72 kb physical distance. The comparative transcriptomic analysis showed that three differentially expressed genes (DEGs), which encode proteins participate in Chl breakdown were upregulated in df74, and one DEG participating in Chl biosynthesis was downregulated. The results will promote the map-based cloning of the candidate gene as well as the understanding of the regulation of chloroplast development in sepals of Brassica napus.
Materials and Methods
Plant Materials and Growth Conditions
The rapeseed mutant (df74) with Chl-deficient sepals was obtained from the conventional variety Ningyou 18 (NY18), whose seeds were treated with 1% EMS for 12 h. The mutant with a pale-yellow sepal phenotype was identified and was selfed to generate an inbred line. The df74 is smaller and shorter than NY18. The investigation of the F2 individuals derived from the cross of df74 and NY18 showed that the dwarfism and sepal-specific chlorophyll-deficiency are not linked to each other. So, we focused on the sepal-specific chlorophyll-deficiency phenomenon in this study. The cross between df74 and NY18 was carried out, and the F2 population was derived from the self-pollination of F1 plants. The BC1 was derived by the backcrosses of F1 to df74. The phenotype of the reciprocal hybrid F1 and the segregation ratio of F2 and BC1 population were used to detect the genetic pattern of the Chl-deficient sepal mutant. Another conventional variety Zhongshuang11 (ZS11) with normal green sepals was used to cross with df74 to construct a F2 population for fine mapping the candidate gene. All the plants were grown in fields located in Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu Province, China, under normal cultivation.
Measurement of Photosynthetic Pigments and Transmission Electron Microscopy Analysis
For pigment extraction, 200 mg fresh weight of sepals were harvested from df74 and NY18, respectively, and then extracted with 5 ml 95% ethanol. Specific Chl contents were measured at wavelengths of 665 and 649 nm with UV-2450 UV/Vis Spectrophotometers (Shimadzu Corporation, Kyoto, Japan) according to the method of Lichtenthaler (1987). Each measurement involved three biological replicates.
For transmission electron microscopy (TEM) analysis, sections were manufactured as described by Wang et al. (2016). Sepals at the same stage on df74 and NY18 were fixed in 2.5% glutaraldehyde and further fixed in 1% osmic acid, dehydrated in gradient acetone, embedded in Epon812 and sectioned, and then double stained with 2% uranyl acetate and lead citrate. The sections were observed and photographed under a Hitachi H-7650 TEM (Hitachi, Tokyo, Japan).
Bulked-Segregant Analysis With Whole-Genome Sequencing
Two contrasting DNA pools were constructed with equal amounts of DNA from 25 green-sepal plants (G-pool) and 25 yellow-sepal plants (Y-pool) from the F2 populations derived from df74 × NY18. The two bulks and two parents were sequenced and the data were generated using the Illumina HiSeq™ PE150 (Illumina, Inc.; San Diego, CA, United States) platform. The sequence depth of the parents were ×30 genome coverage and the two bulks × 45. Sequencing and data analysis were performed by Novogene Bioinformatics Technology Co. Ltd. (Beijing, China). The clean reads of each sample were aligned to the B. napus “Darmor-bzh” reference genome (Chalhoub et al., 2014) using BWA software (Li and Durbin, 2009), and multiple read pairs were removed by the SAMtools command “rmdup” (Li et al., 2009). Sequence variants including single-nucleotide polymorphisms (SNPs) and small insertions and deletions (INDELs) were called using the Unified Genotyper function in GATK software (McKenna et al., 2010). SNPs with read depth <8 or quality score in scale <20 were filtered out. Only the SNPs that were polymorphic between the parents and homozygous in either parent were selected for the further analysis. Using df74 as the reference parent, SNP-index of the two bulks were calculated and the corresponding SNP-index graphs were plotted to find the candidate regions responsible for phenotypic variation. The Δ(SNP-index) can be estimated by the formula: Δ(SNP-index) = SNP-index (G-pool) − SNP-index (Y-pool). The SNPs with significantly different Δ(SNP-index; p < 0.01) were considered closely linked to the causal gene (Yaobin et al., 2018).
Fine Mapping of the Candidate Gene
A F2 population with 3,911 individuals was derived from the cross between df74 and ZS11. The yellow-sepal individuals were selected for genotyping. According to the results of the sequencing of df74 and NY18, the SNPs between the two parents flanking the candidate interval were screened. The penta-primer amplification refractory mutation system (PARMS) was used to screen recombinant plants for fine-mapping. The primers were designed as described by Lu et al. (2020). The master mix for PARMS markers was purchased from Gentides Biotech Co. Ltd. (Wuhan, China). The parents, F1, 10 yellow-sepal individuals, and 10 green-sepal individuals were used to detect the reliability of PARMS markers. Then, two polymorphic markers (M1 and M2) were used to screen the yellow-sepal individuals of the F2 population. The recombinant individuals were selected to conduct the next round of genotyping. New PARMS SNP markers (M3–M10) were designed step by step based on the physical map of ZS11 (Song et al., 2020) to narrow the mapping interval gradually.
RNA Extraction and Sequencing
The sepals of NY18 and df74 were harvested and immediately frozen in liquid nitrogen and then stored at −80°C for RNA extraction and transcriptome analysis. Three biological replicates were performed. Total RNA was extracted using a MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China). RNA-sequencing (RNA-seq) was carried out by Novogene Bioinformatics Technology Co. Ltd. The RNA quality was determined by a Qubit RNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, United States), RNA Nano 6000 Assay Kit, and Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States). Then, six sequencing libraries were constructed and sequenced on Illumina Hiseq 2000 platform, and 100 bp paired-end reads were generated. These methods were described by Wang et al. (2020).
RNA-Seq Data Analysis
Raw RNA-seq reads were filtered by removing reads containing the adapter, and reads containing ploy-Ns and low-quality reads. Then, the clean reads were aligned to the B. napus “Darmor-bzh” reference genome using the hisat2 program (Wen, 2017). DESeq2 was used for filtering the DEGs with |logFC(log2fold change)| ≥ 1 and fdr < 0.05. The Gene Ontology (GO) annotation of DEGs was performed by the GOseq R package (Young et al., 2010), and GO terms with corrected p < 0.05 were considered as significantly enriched terms. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation was carried out with a BLAST search against the KEGG database.1 Pathway enrichment analysis was performed by KOBAS 2.0.2
Results
Phenotypic Characterization
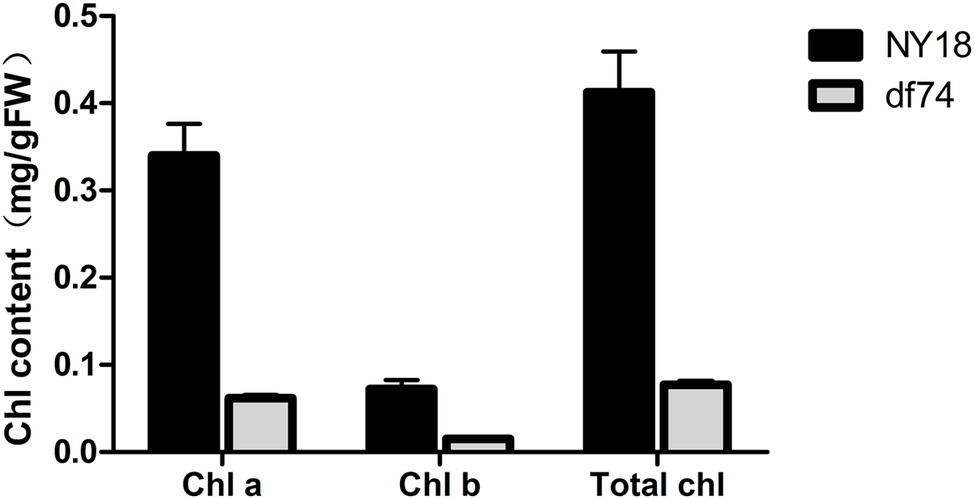
In the seedling stage, the leaves of df74 were as green as NY18, but smaller than NY18 (Figure 1A). The df74 entered the bolting stage and flowering stage 3–5 days later than NY18, and had smaller floral organs (Figures 1B–F). The sepals of df74 were pale yellow, but the other green tissues including leaves, stems, and carpels, had the same green color as NY18. The Chl content of sepals in df74 was significantly different from NY18. Sepals of df74 had 82, 71, and 80% reduction of Chl a, Chl b, and total Chl, respectively, compared with NY18 (Figure 2).

Figure 1. Phenotypic characterization of Ningyou 18 (NY18) and df74. (A) NY18 and df74 at seedling stage; (B) inflorescence of NY18 and df74; (C–F) sepal, petal, stamen, and carpel of NY18 and df74.
To investigate how the mutation affects chloroplast development, we observed the ultrastructure of the chloroplasts in the sepals of df74 and NY18 with TEM. In the cells of NY18, the chloroplasts were neatly arranged beside the cell walls and contained well-developed thylakoid membrane systems (Figures 3A,C). However, the cells of df74 showed plasmolysis, the number of chloroplasts was significantly less than that of NY18, and the chloroplasts were occupied by disordered thylakoids and 1–2 giant starch grains (Figures 3B,D).
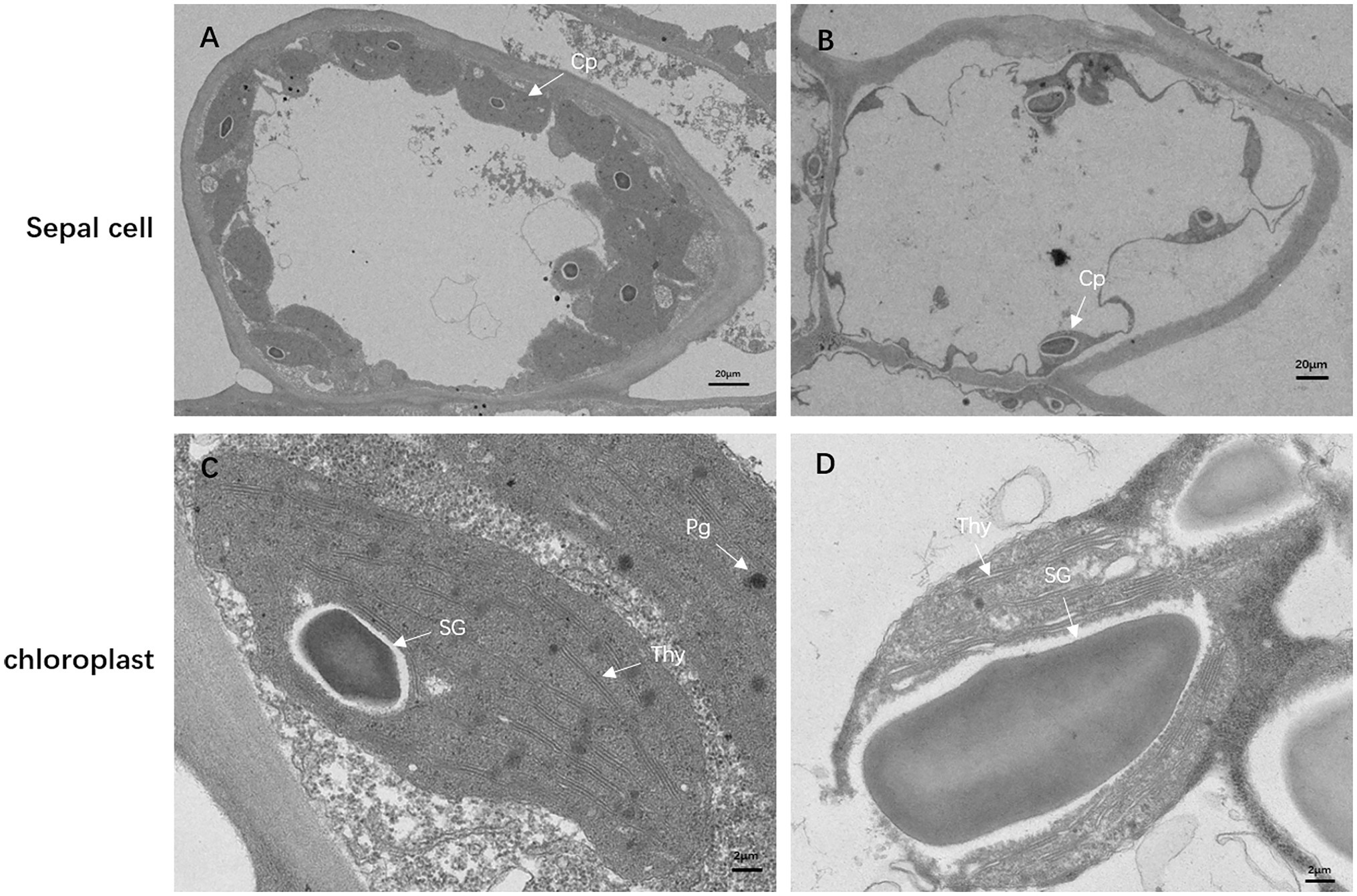
Figure 3. Transmission electron micrographs of chloroplasts from NY18 (A,C) and df74 (B,D). Examples of chloroplast (Cp), plastoglobule (Pg), starch granules (SG), and thylakoid (Thy). Bars: 20 μm (A,B), 2 μm (C,D).
Inheritance of the Chl-Deficient Sepal Trait
The phenotypes of the generations obtained by the crosses of df74 and NY18 were thoroughly investigated. The reciprocal F1 plants exhibited green sepals as wild type, which indicated that the Chl-deficient sepal trait was controlled by nuclear genes. An F2 population with 322 individuals contained 239 green-sepal and 83 yellow-sepal plants. A chi-squared test indicated that the segregation pattern agreed with the Mendelian segregation ratio of 3:1 (χ2 = 0.067, p > 0.05). In addition, the ratio of green-sepal plants to yellow-sepal plants in the BC1 progenies was approximately 1:1 (χ2 = 0.019, p > 0.05). These results indicated that the phenotype of df74 was controlled by a single recessive nuclear gene.
Bulked-Segregant Analysis With Whole-Genome Sequencing
Solexa sequencing of the two pools and two parental lines generated about 172 Gb clean data, with Q20 ≥ 97.97% and Q30 ≥ 93.08%. The data were aligned to the B. napus reference genome “Darmor-bzh.” The average read depth was 39.61-fold in NY18, 26.77-fold in df74, 35.94-fold in the G-pool, and 36.89-fold in the Y-pool. A total of 26,900 differential and homozygous SNP loci were detected between two parental lines. At a 99% significance level, a single peak in the 26.22–37.54 Mb region on chromosome C08 had an average Δ(SNP-index) of 0.55. The largest Δ(SNP-index) among this genomic region was 0.81 (Figure 4). These data confirmed the presence of a major gene located in this genomic region controlling Chl-deficient sepal phenotype, which was designated as BnC08.cds.
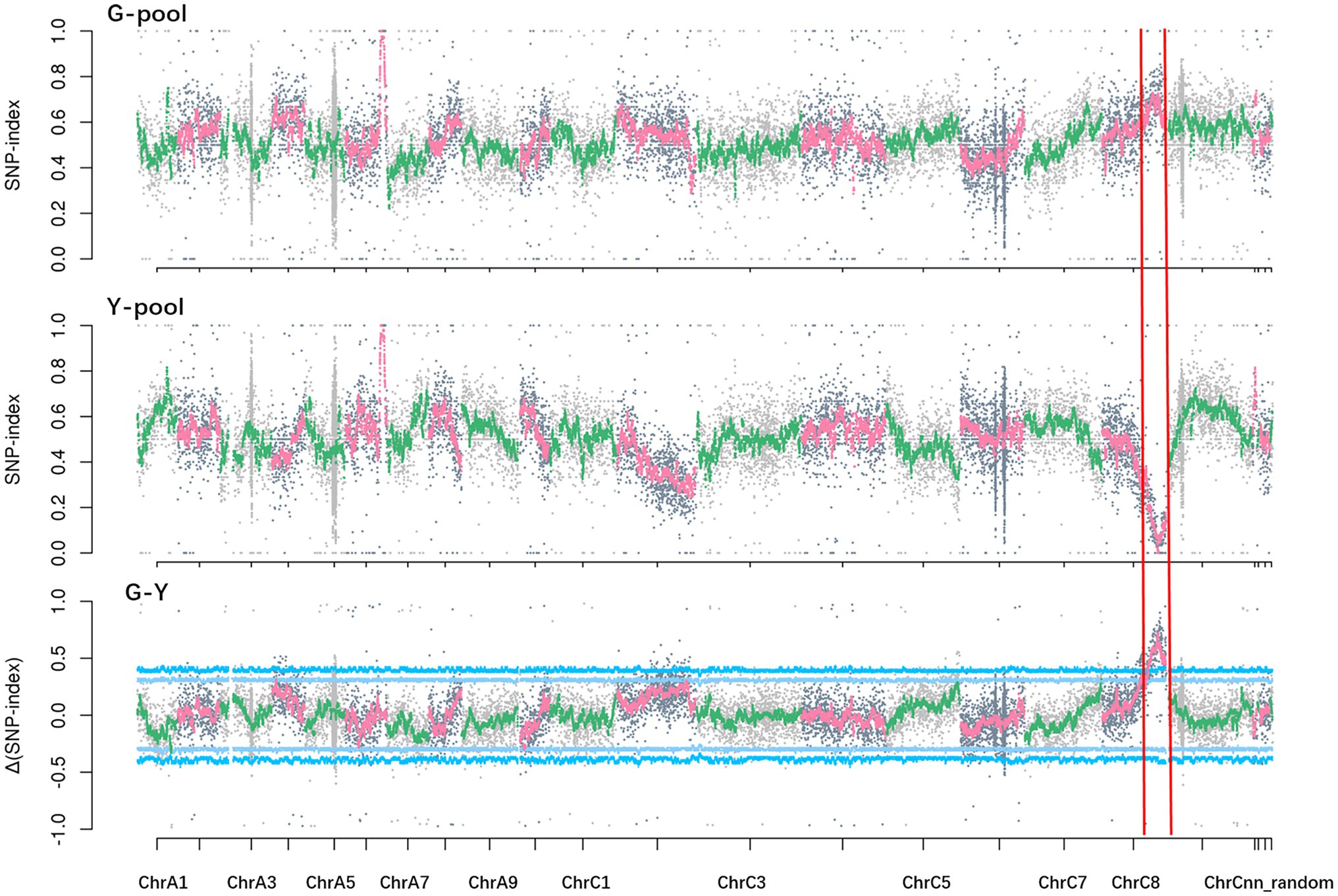
Figure 4. SNP-index (G-pool, Y-pool) and Δ(SNP-index; G-Y) graphs from the BSA-seq analysis. The x axes represent the Brassica napus chromosomes, and the y axes represent the SNP-index or Δ(SNP-index).
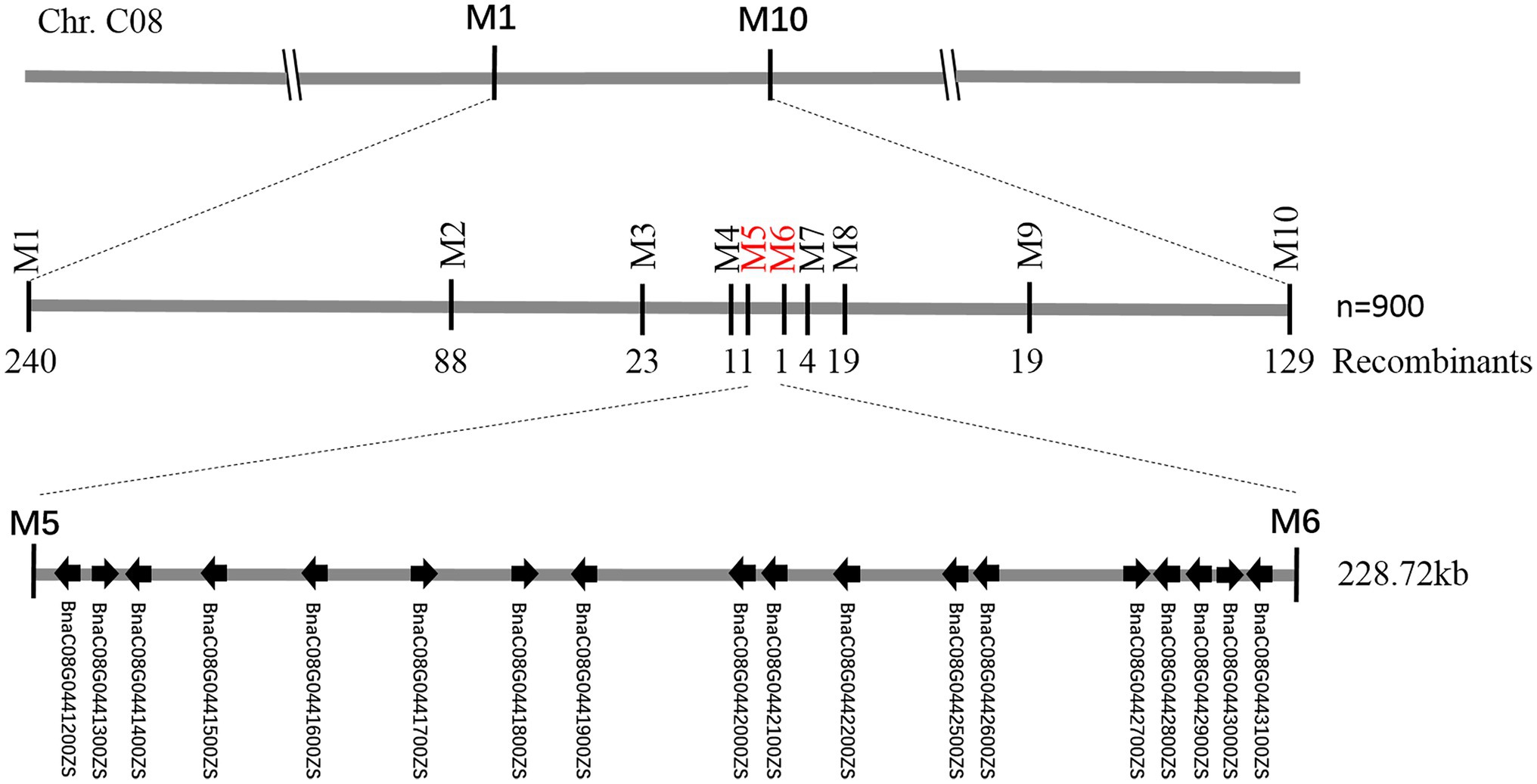
Figure 5. Fine-mapping of BnC08.cds. Using 900 recessive individuals of F2 population derived from the cross between df74 and ZS11, the BnC08.cds was mapped to a 228.72 kb interval on Chromosome C08, restricted by PARMS SNP markers M5 and M6.
Fine-Mapping of the BnC08.cds Gene
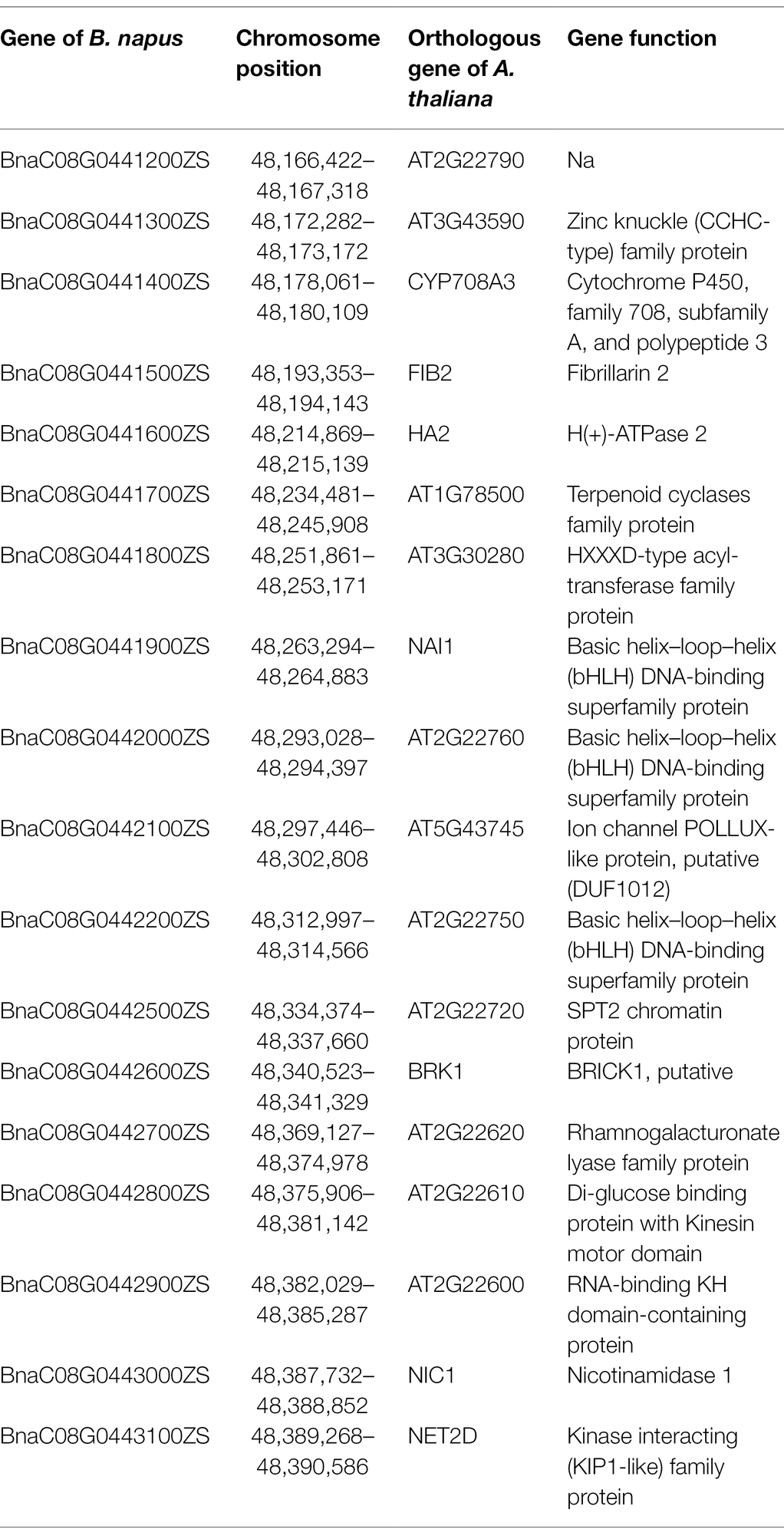
To fine-map the BnC08.cds gene, the candidate region derived from BSA-seq was aligned to the reference genome sequence of B. napus “ZS11.” According to the result, two flanking PARMS SNP markers, M1 (physical position of the B. napus “ZS11” genome: 44420751) and M10 (50875750) were first designed to investigate the recombinants among the 900 yellow-sepal individuals in the (ZS11 × df74) F2 population. According to the result, 369 individuals containing recombinants between the two markers were selected. Furthermore, eight polymorphic PARMS SNP markers (M2–M9) were designed step by step according to the genome sequencing data (Table 1), and the recombinant individuals were analyzed. The BnC08.cds was finally mapped in an interval between M5 and M6 (Figure 5), both resulting in one recombinant. The region had a 228.72 kb physical distance and 18 candidate genes (Table 2) corresponding to the B. napus “ZS11” genome (Song et al., 2020).
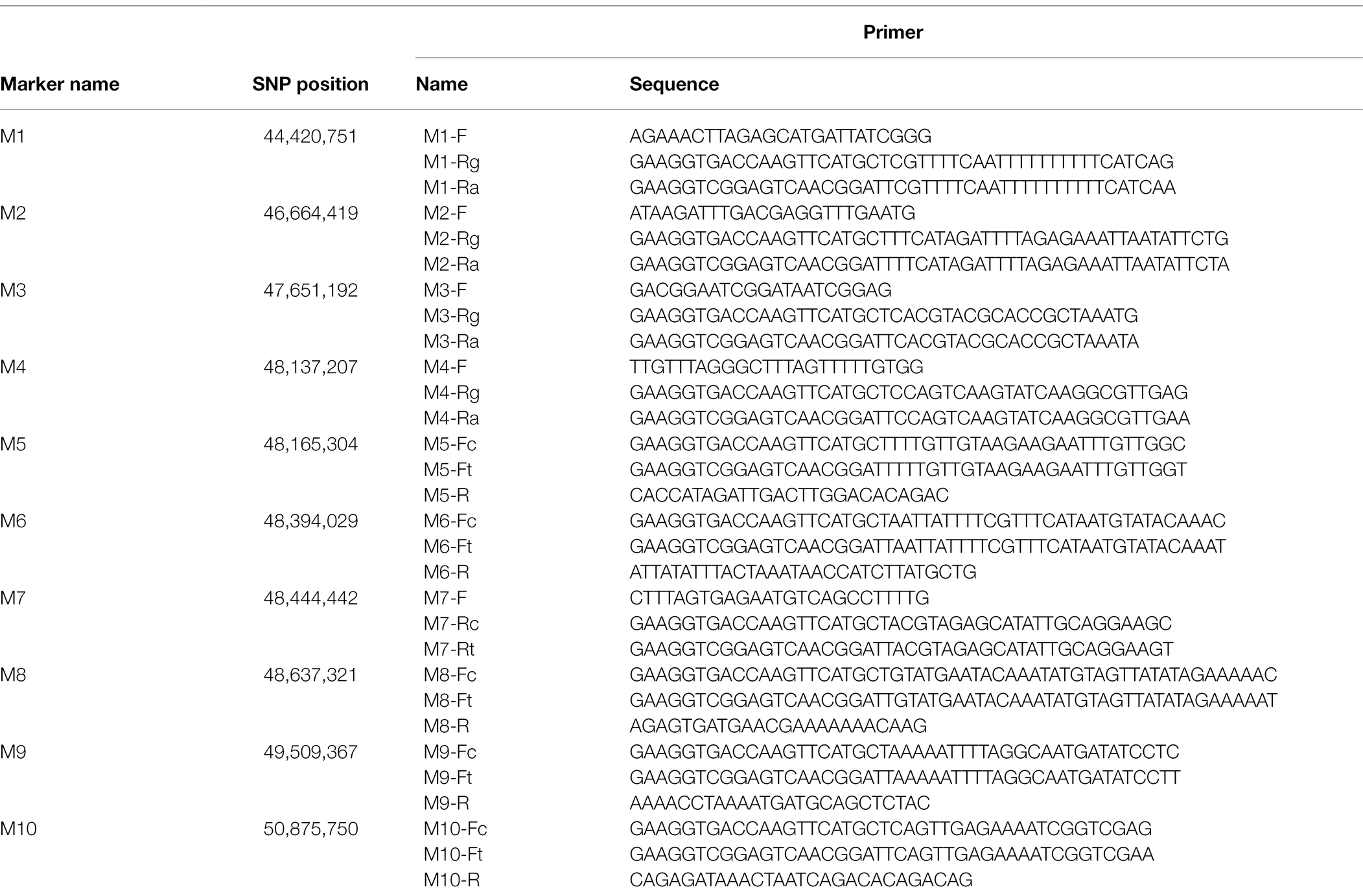
Table 1. Penta-primer amplification refractory mutation system (PARMS) single-nucleotide polymorphism (SNP) markers used for fine-mapping of the BnC08.cds gene.
Genome-Wide Transcriptomic Analysis of NY18 and df74
To further understand the molecular mechanisms of the difference between NY18 and df74, high-throughput RNA-seq was performed using Illumina technology. After the raw data were trimmed, 19.90–23.54 million clean reads for each sample were aligned to the B. napus “Darmor-bzh” genome. A total of 4,108 DEGs between NY18 and df74 were identified, including 1,550 genes upregulated and 2,558 downregulated in the sepals of df74. To investigate the function of the DEGs, GO enrichment analysis was performed. DEGs were mainly categorized and highly enriched as follows, “cellular process,” “metabolic process,” and “single-organism process” in the “Biological Process” category, and “binding” and “catalytic activity” in the “Molecular Function” category. The most abundant terms in the “Cellular Component” category were “cell,” “cell part,” “membrane,” “organelle,” and “membrane part” (Supplementary Additional File 1). Functional annotation of the DEGs was performed against the KEGG database to further investigate the functions of DEGs. The 1,522 up- and 2,527 downregulated DEGs were classified into 116 and 117 pathways, respectively. The result showed the most significantly enriched KEGG pathways were pentose and glucuronate interconversions, starch and sucrose metabolism, and metabolic pathways (Supplementary Additional File 2).
As color change is related to Chl content, we focused on the porphyrin and Chl metabolism pathways. Three DEGs that encode proteins participating in Chl breakdown were upregulated in df74, and one DEG participating in Chl biosynthesis, was downregulated. The three upregulated genes included BnaA08g30360D, BnaC01g20010D, and BnaC01g01800D. The first two genes were both homologous genes of Methylesterase family member 16 (MES16) in Arabidopsis, which catalyzes the demethylation of fluorescent Chl catabolite (Li and Pu, 2016). The third, BnaC01g01800D, was homologous to accelerated cell death gene ACD2 in Arabidopsis, which encodes a Chl breakdown enzyme to protect cells from programmed cell death caused by porphyrin-related molecules (Pattanayak et al., 2012). The downregulated gene was BnaC09g47630D, which is the homologous gene of HEMC that encodes the porphobilinogen deaminase function in Chl biosynthesis (Lim et al., 1994). These pathway annotations of the DEGs revealed the functions of BnC08.cds.
Identification of the Candidate Genes in the Mapped Interval
According to the fine-mapping results, the BnC08.cds was located in an interval with a 228.72 kb physical distance and 18 candidate genes (Table 2). The RNA-seq analysis indicated that none of the 18 genes had significantly different expression levels between the sepals of df74 and NY18. Six genes (BnaC08G0441400ZS, BnaC08G0441600ZS, BnaC08G0441700ZS, BnaC08G0441800ZS, BnaC08G0442000ZS, and BnaC08G0442200ZS) were not expressed in either df74 or NY18. These genes were excluded from the candidate genes.
In this interval, there was an enrichment region of basic helix–loop–helix (bHLH) DNA-binding superfamily genes, including BnaC08G0441900ZS, BnaC08G0442000ZS, and BnaC08G0442200ZS. The bHLH superfamily is one of the largest transcription factor gene families in Arabidopsis (Heim, 2003). Members in the phytochrome-interacting factor family that belongs to the bHLH superfamily VII can negatively regulate Chl biosynthesis (Huq et al., 2004; Shin et al., 2009). In view of this previous research, we cloned and aligned the sequence of the three genes with bHLH domain in df74 and NY18. There was no difference in the three genes between df74 and NY18; therefore, these genes were excluded as candidate genes for BnC08.cds.
BnaC08G0441400ZS is homologous to CYP708A3 in Arabidopsis, which belongs to the cytochrome P450 gene family and functions in heme binding. We sequenced the BnaC08G0441400ZS in df74 and NY18. There is only one base mutation in the intron region, so it was also excluded as a candidate gene for BnC08.cds.
According to the result of resequencing, there was only one missense mutation in the 18 genes. In BnaC08G0442100ZS, there was a single nucleotide substitution (G–A) in the seventh exon, which changed serine into asparagine at the 232nd amino acid site. The mutation was confirmed by sequencing the gene in df74 and NY18. The orthologous gene of BnaC08G0442100ZS in A. thaliana is AT5G43745, which encodes an ion channel POLLUX-like protein and is located in the chloroplast and chloroplast envelope. Therefore, we speculated that BnaC08G0442100ZS may be the most likely candidate gene of BnC08.cds.
Discussion
According to the pigment measurements, the Chl content of sepals in df74 was significantly lower than NY18. However, the fine-mapping revealed there was no missense mutant gene that directly participated in the biosynthesis and degradation of Chl or heme in the candidate region on chromosome C08. Therefore, the BnC08.cds might belong to the third category and function in chloroplast development. The factors involved in chloroplast development are numerous and varied. For example, the YSA gene in rice, which encodes a pentapeptide repetitive (PPR) protein, is required for chloroplast development in early seedling leaves (Su et al., 2012). The ysa mutant displays a stage-specific albino. Leaves of the mutant appear albino before the three-leaf stage, but gradually turn green from the four-leaf stage and return to normal green after the six-leaf stage. The PPR family is one of the largest families in plants. It functions in the regulation of gene expression in plastids, including transcription, splicing, RNA cleavage, RNA editing, translation, and RNA stabilization (Schmitz-Linneweber and Small, 2008). These kinds of PPRs involved in chloroplast development have been found in other species, including DG1 (Chi et al., 2008) and YS1 (Zhou et al., 2010) in Arabidopsis, and PPR4 (Schmitz-Linneweber et al., 2006) and PPR10 (Prikryl et al., 2011) in maize. As a tissue-specific albino, a tomato mutant suffulta (su) with albinic stems and visually normal leaves were collected (Chang et al., 2021). Map-based cloning showed that Su encodes a DnaJ heat shock protein, and the homolog gene AtARC6 (accumulation and replication of chloroplasts) in Arabidopsis is involved in chloroplast division. Previous observations in Arabidopsis arc6 mutants suggest that chloroplasts can divide during cell division (Marrison et al., 1999). By contrast, stem cells only undergo cell expansion, while leaf growth involves both cell division and cell expansion. These differences may lead to the tissue specificity of chloroplast development. In this study, the Chl-deficiency is sepal-specific. More research is needed to reveal the difference in chloroplast development between sepal and other green tissues.
Floral organs are believed to be modified versions of a ground-state organ similar to the leaf. The modifications are led by different combinations of MADS-domain transcription factors encoded by floral organ identity genes (Sablowski, 2015). The model system of flower development is well-established, but how the floral organ identity genes perform their functions and create the diverse morphology of floral organs is little known. Among the floral organs of most angiosperms, the sepals and stigmas have chloroplasts, while the petals and stamens rarely have. In petals and stamens, photosynthetic capacity is lost during modification. Researchers have speculated that the B-function genes suppress the differentiation of photosynthetic tissues. Two genes, GNC and GNL, which were implicated in the regulation of Chl biosynthesis, were both directly and negatively regulated by AP3/PI in petals and stamens (Mara and Irish, 2008). Another direct target of organ identity genes related to light responses is BHLH161/BANQUO3 (BNQ3; Mara et al., 2012). The bnq3 mutants have pale-green sepals and carpels and decreased Chl levels. The BNQ3 is widely expressed in shoots but is directly repressed by AP3/PI in developing petals. Thus, one way in which B-function genes suppress the photosynthetic capacity in petals and stamens is by repressing regulatory genes involved in light response and chloroplast development. However, the photosynthetic capacity in sepals of most angiosperms persists during modification. This may have led to the assumption that the floral organ identity genes may not participate in the regulation of chloroplast development in sepals. But, in our study, the chlorosis in the mutant is sepal-specific. We speculate that the A-function genes may be involved in chloroplast development in sepals, and the BnC08.cds gene may be a target gene that regulated by floral organ identity genes, directly or indirectly.
The model system of flower development is well-established from the original ABC model (Coen and Meyerowitz, 1991) to the ABCE model (Theissen, 2001), new (A)BC model (Causier et al., 2010), and floral quartet model (Theißen et al., 2016). But how the floral organ identity genes perform their functions is little known. In recent years, it has become increasingly clear that the downstream genes of these master regulators execute many of the functions originally attributed to the floral organ identity factors (Thomson et al., 2017). Studies on the gene-regulatory networks and molecular mechanisms of reproductive development in plants have made tremendous progress in recent years (Wils and Kaufmann, 2017). Research on the downstream genes of the floral organ identity factors has become a focus of interest. The df74 with Chl-deficient sepals is valuable because this kind of sepal-specific chlorosis has rarely been reported. The BnC08.cds which controls the sepal-specific Chl-deficiency in df74 will be a good subject to study the regulation network of chloroplast development in sepals.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA807748.
Author Contributions
JZ and XW designed the research. WZ led and coordinated the overall research. FC, HZ, RS, CS, SC, and MH performed the research. WZ and XZ co-wrote the manuscript. XW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The work was supported by National Natural Science Foundation of China (32001580), China Agriculture Research System of MOF and MARA (CARS-12), and Key Laboratory of Jiangsu Province for Agrobiology (JKLA2021-ZD02). All sources of funding received for the research were submitted.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.850330/full#supplementary-material
Supplementary Additional File 1 | Go classification of DEGs between sepals of NY18 and df74.
Supplementary Additional File 2 | Pathway enrichment of DEGs between sepals of NY18 and df74.
Footnotes
References
Albrecht, V., Ingenfeld, A., and Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: the impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60, 507–518. doi: 10.1007/s11103-005-4921-0
Albrecht, V., Ingenfeld, A., and Apel, K. (2008). Snowy cotyledon 2: the identification of a zinc finger domain protein essential for chloroplast development in cotyledons but not in true leaves. Plant Mol. Biol. 66, 599–608. doi: 10.1007/s11103-008-9291-y
Albrecht, V., Šimková, K., Carrie, C., Delannoy, E., Giraud, E., Whelan, J., et al. (2010). The cytoskeleton and the peroxisomal-targeted snowy cotyledon 3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22, 3423–3438. doi: 10.1105/tpc.110.074781
Beck, C. F. (2001). Signaling pathways in chloroplast-to-nucleus communication. Protist 152, 175–182. doi: 10.1078/1434-4610-00056
Brzezowski, P., Richter, A. S., and Grimm, B. (2015). Regulation and function of tetrapyrrole biosynthesis in plants and algae. BBA-Bioenergetics 1847, 968–985. doi: 10.1016/j.bbabio.2015.05.007
Causier, B., Schwarz-Sommer, Z., and Davies, B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21, 73–79. doi: 10.1016/j.semcdb.2009.10.005
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chang, J., Zhang, F., Qin, H., Liu, P., Wang, J., and Wu, S. (2021). Mutation of SlARC6 leads to tissue-specific defects in chloroplast development in tomato. Hortic. Res. 8:127. doi: 10.1038/s41438-021-00567-2
Chi, W., Ma, J., Zhang, D., Guo, J., Chen, F., Lu, C., et al. (2008). The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584. doi: 10.1104/pp.108.116194
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Fu, W., Ye, X., Ren, J., Li, Q., Du, J., Hou, A., et al. (2019). Fine mapping of lcm1, a gene conferring chlorophyll-deficient golden leaf in chinese cabbage (Brassica rapa ssp. Pekinensis). Mol. Breed. 39, 1–12. doi: 10.1007/s11032-019-0945-z
Heim, M. A. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P. H. (2004). PHYTOCHROME-INTERCATING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305, 1937–1941. doi: 10.1126/science.1099728
Jiang, H., Li, M., Liang, N., Yan, H., Wei, Y., Xu, X., et al. (2007). Molecular cloning and function analysis of the stay green gene in rice. Plant J. 52, 197–209. doi: 10.1111/j.1365-313X.2007.03221.x
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and samtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, H., and Pu, H. (2016). Crystal structure of methylesterase family member 16 (MES16) from Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 474, 226–231. doi: 10.1016/j.bbrc.2016.04.115
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lim, S. H., Witty, M., Wallace-Cook, A. D. M., Ilag, L. I., and Smith, A. G. (1994). Porphobilinogen deaminase is encoded by a single gene in Arabidopsis thaliana and is targeted to the chloroplasts. Plant Mol. Biol. 26, 863–872. doi: 10.1007/BF00028854
Liu, X., Yu, F., and Rodermel, S. (2010). Arabidopsis chloroplast FtsH, var2 and suppressors of var2 leaf variegation: a review. J. Integr. Plant Biol. 52, 750–761. doi: 10.1111/j.1744-7909.2010.00980.x
Lu, J., Hou, J., Ouyang, Y., Luo, H., Zhao, J., Mao, C., et al. (2020). A direct PCR-based SNP marker-assisted selection system (D-MAS) for different crops. Mol. Breed. 40, 1–10. doi: 10.1007/s11032-019-1091-3
Ma, F., Hu, Y., Ju, Y., Jiang, Q., Cheng, Z., Zhang, Q., et al. (2017). A novel tetratricopeptide repeat protein, white to green1, is required for early chloroplast development and affects RNA editing in chloroplasts. J. Exp. Bot. 68, 5829–5843. doi: 10.1093/jxb/erx383
Mara, C. D., Huang, T., and Irish, V. F. (2012). The Arabidopsis floral homeotic proteins APETALA3 and PISTILLATA negatively regulate the BANQUO genes implicated in light signaling. Plant Cell 22, 690–702. doi: 10.1105/tpc.109.065946
Mara, C. D., and Irish, V. F. (2008). Two Gata transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis. Plant Physiol. 147, 707–718. doi: 10.1104/pp.107.115634
Marrison, J. L., Rutherford, S. M., Robertson, E. J., Lister, C., Dean, C., and Leech, R. M. (1999). The distinctive roles of five different ARC genes in the chloroplast division process in Arabidopsis. Plant J. 18, 651–662. doi: 10.1046/j.1365-313x.1999.00500.x
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Pattanayak, G. K., Venkataramani, S., Hortensteiner, S., Kunz, L., Christ, B., Moulin, M., et al. (2012). ACCELERATED CELL DEATH 2 suppresses mitochondrial oxidative bursts and modulates cell death in Arabidopsis. Plant J. 69, 589–600. doi: 10.1111/j.1365-313X.2011.04814.x
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Prikryl, J., Rojas, M., Schuster, G., and Barkan, A. (2011). Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U. S. A. 108, 415–420. doi: 10.1073/pnas.1012076108
Sablowski, R. (2015). Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 66, 1065–1073. doi: 10.1093/jxb/eru514
Schmitz-Linneweber, C., and Small, I. (2008). Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. doi: 10.1016/j.tplants.2008.10.001
Schmitz-Linneweber, C., Williams-Carrier, R. E., Williams-Voelker, P. M., Kroeger, T. S., Vichas, A., and Barkan, A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18, 2650–2663. doi: 10.1105/tpc.106.046110
Shin, J., Kim, K., Kang, H., Zulfugarov, I. S., Bae, G., Lee, C. H., et al. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. U.S.A. 106, 7660–7665. doi: 10.1073/pnas.0812219106
Song, J. M., Guan, Z., Hu, J., Guo, C., and Guo, L. (2020). Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34–45. doi: 10.1038/s41477-019-0577-7
Su, N., Hu, M., Wu, D., Wu, F., Fei, G., Lan, Y., et al. (2012). Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol. 159, 227–238. doi: 10.1104/pp.112.195081
Tanaka, R., and Tanaka, A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. doi: 10.1146/annurev.arplant.57.032905.105448
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/s1369-5266(00)00139-4
Theißen, G., Melzer, R., and Rümpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Thomas, H., and Ougham, H. (2014). The stay-green trait. J. Exp. Bot. 65, 3889–3900. doi: 10.1093/jxb/eru037
Thomson, B., Zheng, B., and Wellmer, F. (2017). Floral organogenesis: when knowing your ABCs is not enough. Plant Physiol. 173, 56–64. doi: 10.1104/pp.16.01288
Wang, Y., Wang, C., Zheng, M., Lyu, J., Xu, Y., Li, X., et al. (2016). WHITE PANICLE1, a val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiol. 170, 2110–2123. doi: 10.1104/pp.15.01949
Wang, X., Zheng, M., Liu, H., Zhang, L., Chen, F., Zhang, W., et al. (2020). Fine-mapping and transcriptome analysis of a candidate gene controlling plant height in Brassica napus L. Biotechnol. Biofuels 13:42. doi: 10.1186/s13068-020-01687-y
Wen, G. (2017). “A Simple Process of RNA-Sequence Analyses by Hisat2, Htseq and DESeq2.” in The 2017 International Conference. September 14, 2017.
Wils, C. R., and Kaufmann, K. (2017). Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. BBA-Gene Regul. Mech. 1860, 95–105. doi: 10.1016/j.bbagrm.2016.07.014
Wu, X., Gong, D., Xia, F., Dai, C., Zhang, X., Gao, X., et al. (2020). A two-step mutation process in the double WS1 homologs drives the evolution of burley tobacco, a special chlorophyll-deficient mutant with abnormal chloroplast development. Planta 251:10. doi: 10.1007/s00425-019-03312-1
Yaobin, Q., Peng, C., Yi, C., Yue, F., De, H., Tingxu, H., et al. (2018). QTL-seq identified a major QTL for grain length and weight in rice using near isogenic F2 population. Rice Sci. 25, 121–131. doi: 10.1016/j.rsci.2018.04.001
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhao, H., Yu, L., Huai, Z., Wang, X., Ding, G., Chen, S., et al. (2013). Mapping and candidate gene identification defining BnChd1-1, a locus involved in chlorophyll biosynthesis in Brassica napus. Acta Physiol. Plant. 36, 859–870. doi: 10.1007/s11738-013-1464-x
Zhou, W., Cheng, Y., Yap, A., Chateigner-Boutin, A. L., and Huang, J. (2010). The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96. doi: 10.1111/j.1365-313X.2008.03766.x
Keywords: Brassica napus L, chlorophyll-deficient mutant, fine-mapping, RNA-sequencing, yellow-sepal, candidate gene
Citation: Zhang W, Zhou X, Chen F, Zhu H, Shi R, Sun C, Chen S, Hu M, Zhang J and Wang X (2022) Fine Mapping and Candidate Gene Analysis of BnC08.cds, a Recessive Gene Responsible for Sepal-Specific Chlorophyll-Deficiency in Brassica napus L. Front. Plant Sci. 13:850330. doi: 10.3389/fpls.2022.850330
Edited by:
Byoung-Cheorl Kang, Seoul National University, South KoreaReviewed by:
Chuchuan Fan, Huazhong Agricultural University, ChinaDengfeng Hong, Huazhong Agricultural University, China
Copyright © 2022 Zhang, Zhou, Chen, Zhu, Shi, Sun, Chen, Hu, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Wang, eGR3YW5nMTIwQDE2My5jb20=; Jiefu Zhang, amllZnVfekAxNjMuY29t
†These authors have contributed equally to this work
 Wei Zhang
Wei Zhang Xiaoying Zhou1†
Xiaoying Zhou1† Chengming Sun
Chengming Sun Jiefu Zhang
Jiefu Zhang Xiaodong Wang
Xiaodong Wang