
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 13 May 2022
Sec. Plant Abiotic Stress
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.844201
This article is part of the Research Topic Advances in Plant Hormone Research in the Face of a Changing Environment View all 6 articles
Coordinated transcriptional regulation of stress-responsive genes orchestrated by a complex network of transcription factors (TFs) and the reprogramming of metabolism ensure a plant’s continued growth and survival under adverse environmental conditions (e.g., abiotic stress). DNA-binding with one finger (Dof) proteins, a group of plant-specific TF, were identified as one of several key components of the transcriptional regulatory network involved in abiotic stress responses. In many plant species, Dofs are often activated in response to a wide range of adverse environmental conditions. Dofs play central roles in stress tolerance by regulating the expression of stress-responsive genes via the DOFCORE element or by interacting with other regulatory proteins. Moreover, Dofs act as a key regulatory hub of several phytohormone pathways, integrating abscisic acid, jasmonate, SA and redox signaling in response to many abiotic stresses. Taken together, we highlight a unique role of Dofs in hormone and stress signaling that integrates plant response to adverse environmental conditions with different aspects of plant growth and development.
Transcription factors (TFs) are proteins that bind to specific DNA motifs in the promoter regions of target genes, thereby regulating their transcription. In plants, on average 5% of the protein-coding genes encode TFs (Jin J. et al., 2014). Plant TFs were classified into 58 families (and some into six superfamilies) based on their DNA-binding domains (Jin et al., 2014, 2016). Many TFs such as Dof, WRKY, ERF, NAC, GRAS, and MYB (Dubos et al., 2010; Nakashima et al., 2012; Muller and Munne-Bosch, 2015; Hakoshima, 2018; Ruta et al., 2020) play a crucial role in plant signaling and regulatory networks relevant to abiotic/biotic stress responses and many developmental/physiological processes (Lindemose et al., 2013; Gupta et al., 2015; Ramirez et al., 2021; Wang Z. et al., 2021).
In particular, one plant-specific TF contains a highly conserved DNA-binding domain and has been designated the DNA-binding with one finger (Dof) domain (Yanagisawa, 1995; Yanagisawa and Schmidt, 1999). The first Dof protein, ZmDof1 was identified to contain the CX2CX21CX2C protein motif in maize (Yanagisawa and Izui, 1993). The evolutionary analysis of Dofs among plant species has been studied extensively in model plants such as Arabidopsis and Oryza sativa (Lijavetzky et al., 2003; Khan et al., 2021) but also non-model systems like apple (Zhang Z. et al., 2018), physic nut and castor bean (Zou and Zhang, 2019), grape (Wang Z. et al., 2021) and between multiple plant species (Noguero et al., 2013; Ma et al., 2015). Generally, the Dof gene family contains a highly conserved Dof domain at the N-terminus of approximately 50 residues in length, an oligomerization site, a transcription regulation domain and a nuclear localization signal. Most Dofs have only one type of DNA binding region and oligomerization region, although some lack a transcription regulation domain or a specific DNA binding region (Krohn et al., 2002). Dof DNA-binding activities have been analyzed by different in vitro and in vivo approaches revealing that all the Dof TFs tested bind to a DOFCORE element (5′-(A/T)AAAG-3′) (Yanagisawa, 2016). In Arabidopsis, a systematic study of 529 TFs using a DNA affinity purification-sequencing (DAP-seq) technique revealed that all 39 Dof TFs evaluated had the aforementioned promoter binding preferences (O’Malley et al., 2016). Similar observations were also reported using independent in vitro assays in Arabidopsis (Imaizumi et al., 2005; Corrales et al., 2017) but also other plants such as potato (Kloosterman et al., 2013) and tomato (Corrales et al., 2014; Ewas et al., 2017).
It is well-known that the Dof domain also mediates protein-protein interactions (Krohn et al., 2002). In maize, Dof1 not only self-associates but interacts with another Dof protein, Dof2 (Yanagisawa and Izui, 1993; Cavalar et al., 2003; Yanagisawa, 2004). Interaction with other TF families such as Dof–bZIP (Vicente-Carbajosa et al., 1997) and Dof–MYB interaction (Diaz et al., 2002) were also observed. The C-terminal motifs of the Dof domain are also pivotal for various protein-protein interactions. For example, some cycling Dof factors (CDF) proteins typically contain specific domains in their C-terminal region such as the clock gene GIGANTEA (GI) and FLAVIN BINDING KELCH REPEAT F-BOX PROTEIN 1 (FKF1) -binding domains (Renau-Morata et al., 2020) that are known to participate in the control post-translational regulation by protein-protein interactions (Imaizumi et al., 2005; Kloosterman et al., 2013; Corrales et al., 2014). Nonetheless, it is noteworthy that factors such as the location of binding and the interaction with other factors such as chromosomal high-mobility group proteins (HMGBs) (Krohn et al., 2002; Cavalar et al., 2003) might determine the capacity of DNA binding and thus, transcriptional control at precise sites in the genome. Dofs are well known for their roles in growth and development processes such as seed germination (da Silva et al., 2016), flowering (Corrales et al., 2017; Liu X. et al., 2020), and leaf senescence (Shim et al., 2019; Zhuo et al., 2020). However, recent studies have also revealed pivotal roles of Dof TFs in various abiotic and biotic stresses but also hormone responses (Qin et al., 2019; Li et al., 2021; Ramirez et al., 2021; Wang P. et al., 2021). In this perspective article, we highlight 68 Dof proteins (as in Table 1 and Supplementary Figure 1) with potential roles in stress and hormone responses in a wide range of plants.
Adverse environmental conditions such as extreme temperatures (e.g., freezing and heat), water status (e.g., drought and flood) and salinity affect the survival, growth and reproduction of plants. Expression of Dofs have been reported to be induced by salt, drought, cold, heat stress, and changes in light availability in various species (Table 1). One common theme is that Dof genes are often multi-abiotic stress-responsive: salt, drought, cold, and heat stress treatment (4 studies); drought, cold, and heat stress treatment (6 studies); salt and drought (7 studies), among others. In various plant systems (Ravindran et al., 2020; Li et al., 2021; Ramirez et al., 2021; Zhu et al., 2021), overexpression, silencing, and mutational experiments of Dof genes have also established their role in various abiotic stress tolerance (Supplementary Table 1).
GIGANTEA is central in diverse signaling pathways, including circadian clock regulation, sugar, and light signaling pathways, photoperiodic and stress responses (including drought, salinity, and low temperature) (Han et al., 2013; Kim et al., 2013; Fornara et al., 2015; Park et al., 2016; Cao et al., 2020). Emerging results indicate a GI-CDF module where CDFs regulate genes involved in abiotic stress responses through regulation of the C-REPEAT BINDING FACTOR (CBF) regulon (Corrales et al., 2017; Xu et al., 2017; Shi et al., 2018; Tominaga et al., 2021). In some cases, CDFs have been identified as part of the CBF/DREB transcriptional regulatory network involved in controlling abiotic stress responses (Corrales et al., 2017; Renau-Morata et al., 2017; Liu et al., 2019; Ramirez et al., 2021). For example, Arabidopsis AtCDF3 regulates several stress-response TFs (e.g., CBFs, DREB2A, and ZAT10) which involve both GI dependent and independent pathways (Corrales et al., 2017; Renau-Morata et al., 2017). Similarly, overexpression of Brassica napus BnCDF1, which is homologous to Arabidopsis Cycling DOF Factor 1, promotes freezing tolerance via the regulation of different abiotic-stress-responsive genes such as CBF1, CBF2, COR15A, and RESPONSIVE TO DESICCATION 29A (RD29A; Xu and Dai, 2016). Thus the data suggest that regulation of CBF genes by Dof/CDFs might be the initial step in the abiotic stress response signaling cascade (Figure 1A).
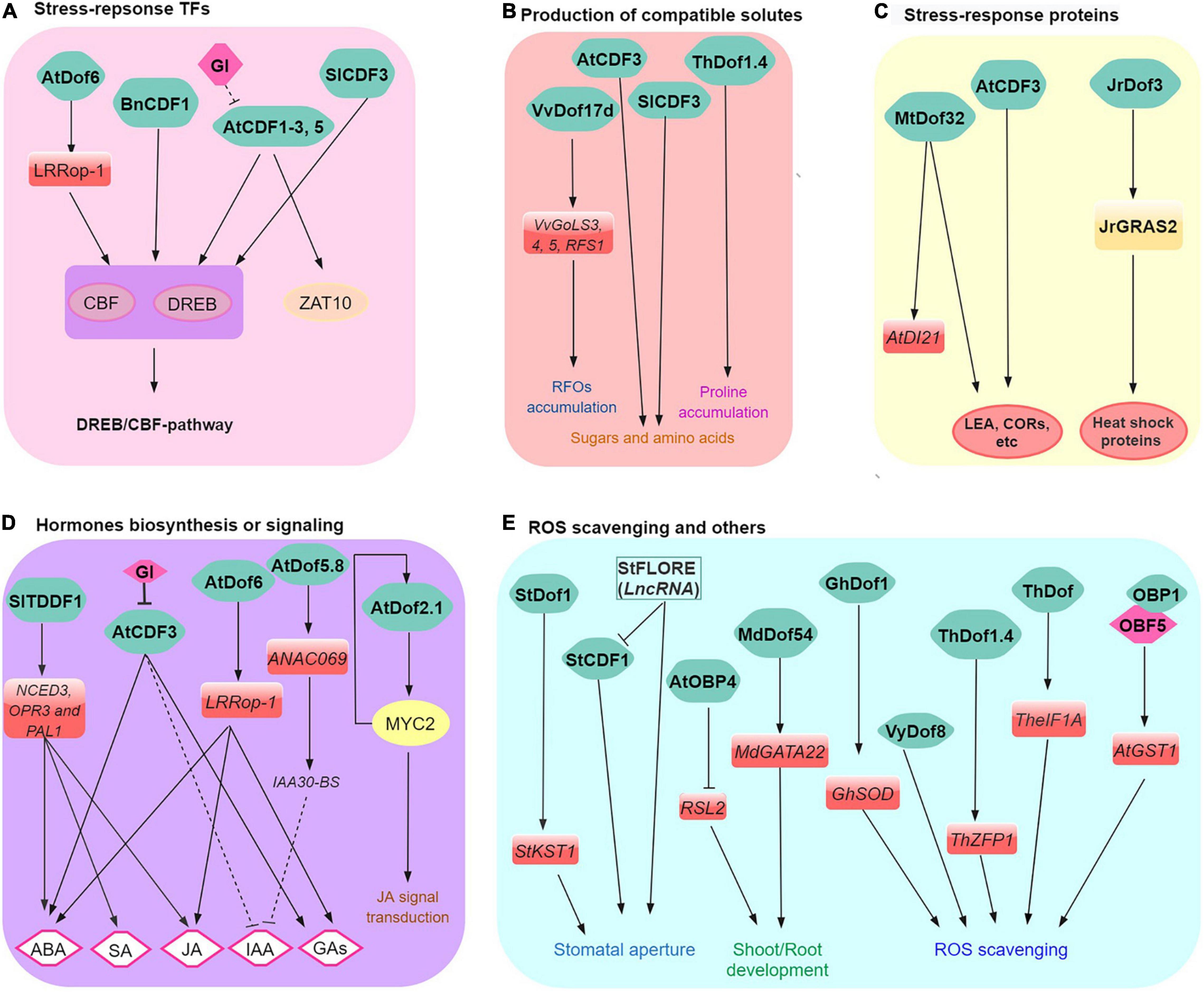
Figure 1. A simplified model of Dof functions in plant responses to abiotic stress. Dofs regulate (A) stress-related TFs (CBF, DREB, and ZAT10, etc., abiotic stress modules), (B) production of compatible solutes, (C) stress-related proteins (e.g., HSP and COR, etc.), (D) stress-related hormone biosynthesis or signaling, and (E) ROS scavenging and other biological processes. Positive and negative regulation are indicated by arrows and broken lines, respectively.
Galactinol synthases (GolS) and raffinose synthases (RFS) are closely related glycosyltransferases/hydrolases involved in the biosynthesis of raffinose family oligosaccharides (RFOs) that function as osmoprotectants that promote plant abiotic stress tolerance (Sprenger et al., 2018; Chai et al., 2019). In the Amur grape, transcriptomic analysis revealed that GolS and RFS are among the most cold-induced genes (Xin et al., 2013; Sun et al., 2018; Wang Z. et al., 2021). Overexpression and loss-of-function mutation (introduced by CRISPR/Cas9-mediated mutagenesis) of VaDof17d significantly induced and repressed expression of cold-induced GolS and RFS/RAF genes compared to wild-type plants, respectively. During cold stress, VaDof17d-ED loss-of-function mutants concordantly show poorer cold tolerance and decreased RFOs levels compared to normal plants (Wang Z. et al., 2021). These results highlight one alternative mechanism cold tolerance could be achieved in grapes (Figure 1B).
Furthermore, tomato SlCDF1–5 genes were differentially induced in response to salt, drought, and heat stress (Corrales et al., 2014). Tomato plants overexpressing the SlCDF3 showed enhanced biomass production and yield under salinity stress conditions (Renau-Morata et al., 2017). This has been attributed to higher sucrose, GABA, and asparagine in vegetative tissues of SlCDF3-overexpressing plants. These metabolites also act as cellular osmoprotectants in various abiotic stress conditions in plants (Mittler et al., 2011; Mittler, 2017). These reports point to a key role of Dof in the regulation of various compatible solutes under specific stress (e.g., cold and highly saline) conditions (Figure 1B).
Heat shock proteins (HSPs) expression underlies adaptive responses to many environmental stress in plants. Briefly, HSPs function as molecular chaperones that encompasses many functions such as ensuring proper folding of other proteins but also assist in the deterrence, reduction, and/or degradation of stress-damaged proteins (Chen et al., 2018). Like HSPs, LATE EMBRYOGENESIS ABUNDANT (LEA) proteins also participate in acclimation and/or in the adaptive response to various stress (Battaglia et al., 2008). LEAs are a type of highly hydrophilic glycine-rich proteins with antioxidant, metal ion binding, membrane and protein stabilization, hydration buffering, and DNA and RNA interaction properties. In walnut, the heat-stress responsive JrGRAS2 TF regulates a multitude of heat shock proteins and is pivotal for heat stress response. Interestingly, walnut JrDof3 activates JrGRAS2, thereby reinforcing the regulation of HSP and heat stress response (Yang et al., 2018). In addition, Medicago MtDof32 plays a positive role in regulating the expression of stress-related genes (i.e., upregulation of DROUGHT-INDUCED 21, AtDI21; SENESCENCE-ASSOCIATED GENE 21; AtCOR15A) leading to enhanced osmotic and salt tolerances in Arabidopsis (Guo et al., 2021). These examples point to the role of Dof in the regulation of stress-related genes and potentially enhanced abiotic stress tolerance especially heat (Figure 1C).
Coordinated regulation of multiple hormonal signaling pathways enables plants to respond and adapt to adverse environmental conditions by regulating the expression of TF genes in a finely tuned manner (Yang et al., 2019). Indeed, exogenous application of various phytohormones induces the expression of many Dof genes (Table 1) involved in hormone biosynthesis or signaling in many plants. Abscisic acid (ABA) is one key hormone that regulates plant responses to abiotic stresses (Tuteja, 2007). The grape Vitis yeshanensis VyDof8 showed multi-abiotic stress-responsive expression patterns. Overexpression of VyDof8 in tobacco (Nicotiana benthamiana) significantly enhanced ABA accumulation and drought tolerance during prolonged drought compared to control plants (Li et al., 2021).
In plants, jasmonate acid (JA) and its derivatives (e.g., MeJA and JA-Ile) also play diverse roles in response to abiotic stress (Chini et al., 2016; Wasternack and Feussner, 2018; Yang et al., 2019). With regards to the involvement of Dof proteins, Dof2.1 and MYC2 (MYC2, which encodes a central regulator of JA responses) form a feed-forward transcriptional loop (MYC2–Dof2.1–MYC2) that enhances jasmonate-induced leaf senescence in Arabidopsis (Zhuo et al., 2020). The integration of auxin and salt signals by ANAC069 in the regulation of Arabidopsis seed germination have been demonstrated (Park et al., 2011). Interestingly, subsequent studies revealed that Arabidopsis AtDof5.8 is the upstream regulator of ANAC069 that indirectly also mediates the regulation of salinity and osmotic stress tolerance (He et al., 2015, 2017). Salicylic acid has long been known to play a role in the induction of defense mechanisms in plants; however, its participation in abiotic stress signaling is slowly emerging (Siboza et al., 2014; Zhang L. et al., 2018; Yang et al., 2019). OBP1, OBP2, and OBP3 were initially identified as partners of OBF4 that interacts with salicylic acid-responsive ocs elements (Kang and Singh, 2000; Kang et al., 2003), thus potentially linking these Dofs in SA regulation.
In addition, Dof has been also reported to be involved in multiple hormonal pathways during abiotic stress (Figure 1D). For example, Dof6 induced LRRop-1 negatively regulates abiotic stress responses during Arabidopsis seed germination by altering endogenous hormone levels of ABA, GA, and JA (Ravindran et al., 2020). In tomato, TDDF1 positively regulates gene expression patterns for NCED3, OPR3, and PAL1, the key genes of ABA, JA, and SA biosynthesis pathway under drought and salt stress (Ewas et al., 2017). Together, these examples reveal that Dofs mediates multiple hormone responses linked to abiotic and biotic stress responses in a wide range of plants.
Antioxidant genes are also involved in Dof-mediated stress responses (e.g., salt, drought) in some plants (Figure 1E). The reactive oxygen species (ROS, such as H2O2 and O2–) are considered to be both beneficial (e.g., signaling) and potentially damaging molecules which result in cell membrane damage by stimulating membrane lipid peroxidation, and even cell death (Mittler et al., 2011; Devireddy et al., 2020). Overexpression of Tamarix hispida ThDof was found to enhance drought tolerance in transgenic tobacco plants by indirectly regulating the expression of stress-related genes (including ThSOD; peroxidase ThPOD and glutathione peroxidase ThGPX), resulting in improved reactive ROS scavenging and tolerance to salt and osmotic stress (Yang et al., 2017). Additionally, transient overexpression and silencing of another Dof, ThDof1.4 in T. hispida plants promoted and repressed the expression of ThZFP1, respectively. Together, ThDof1.4 and ThZFP1 form a transcriptional regulatory cascade and coordinately regulate the expression of T. hispida PODs, SODs, and proline biosynthesis-related genes leading to increased proline, enhanced ROS scavenging capacities. This finding point to a role of ThDof1.4 in salt and osmotic stress tolerance in T. hispida (Zang et al., 2017).
Moreover, CDFs are key elements that integrate plant responses to adverse environmental conditions with different aspects of plant growth control and development like photoperiodism or root/shoot growth (Figure 1E). In Arabidopsis, OBP4/Dof5.4 also plays a central role in integrating developmental and environmental signals by mediating the transcriptional repression of RSL2 that ultimately contributes to the inhibition of root hair growth (Rymen et al., 2017). In cotton, overexpression of GhDof1 positively regulates GhP5CS, GhSOD and GhMYB, leading to improved salt tolerance and root growth over the wild-type plants (Su et al., 2017). Consistently, MdDof54 facilitated root development, stomatal conductance, transpiration, photosynthesis, and hydraulic conductivity, which lead to improved growth of apple trees under long-term drought stress (Chen et al., 2020).
In addition to the participation of Dofs in diverse abiotic stress responses and as a key regulatory hub of several phytohormone pathways (Figure 1 and Table 1), their roles in biotic stress response are slowly emerging. Phytocystatins (PhyCys) are a group of cystatin proteins in plants that specifically inhibits cysteine proteinases and peptidase that are often produced by pathogens to aid their colonization and proliferation in the host cells (Martinez et al., 2005). The role of Dof transcription factors in the regulation of PhyCys has been established in several plants for biotic stress tolerance. For example, rice OsDof1 (Park et al., 2014) show stress-responsive induction in response to plant wounding and pathogen challenge. In barley (Hordeum vulgare), a Dof transcription factor, SAD binds to the pyrimidine box in vitro and activates transcription of a protease promoter (of a cystatin gene) in bombarded aleurone layers (Isabel-LaMoneda et al., 2003). Additionally, another Dof TF, BPBF also interacts with oligonucleotides containing Dof binding sites derived from the cystatin gene promoter in vitro (Mena et al., 2002; Diaz et al., 2005). Furthermore, tomato Dof, TDDF1 may increase biotic resistance by regulating Pathogenesis Related Protein 1 (PR1) expression, a well-known resistance marker gene induced in response to pathogen and herbivores attack (Ewas et al., 2017). In watermelon, 22 and 34 (of 36) CsDof genes were upregulated or downregulated in response to downy mildew and watermelon mosaic virus inoculation, respectively (Wen et al., 2016). In pepper, RNA-seq analysis showed temporal and pathogen-specific variation of CaDofs during development and response to multiple biotic stresses such as two TMV strains, PepMoV and Phytophthora capsici infection (Kang et al., 2016). These results indicated that Dof may be involved biotic stress response in a wide range of plants.
In this perspective article, we present supporting evidence that the Dof family appears to show more multifaceted roles than previously expected. We present evidence that Dofs act as a key regulatory hub of several phytohormone pathways, integrating abscisic acid, jasmonate, SA and redox signaling in response to many abiotic stresses. However, many gaps in our knowledge on Dofs and hormonal interaction (such as Dof with BR, GA, etc.), and the answers to the remaining questions are likely important to increase our understanding of stress responses and adaptation. Beyond hormonal-related abiotic stress regulation, some of Dofs also seem to regulate ROS-responsive gene expression, but more evidence of synergistic Dofs- and ROS-responsive genes are needed in multiple plant systems.
Since TFs play an important role in increasing stress tolerance and developmental responses in plants, they can be targeted for the generation of improved varieties using transgenic technology (Wang et al., 2016). Yet, large numbers of Dofs remain uncharacterized in plants. Nonetheless, as exemplified with the success in small studies targeting individual Dofs (Ramirez et al., 2021; Wang Z. et al., 2021) but also large-scale attempts to generate knockout lines for all Dof members within a single plant species such as all 30 OsDofs in rice (Huang et al., 2019; Yu L. et al., 2021), CRISPR/Cas system is poised to be an effective and flexible way to study the function of Dofs through the generation of functional (e.g., loss- and/or gain-of-function) mutants. Long non-coding RNAs (lncRNAs) are a heterogeneous class of regulatory transcripts that represent a diverse class of regulatory loci with roles in development and stress responses in plants. To date, only one report has reported a link between Dofs and lncRNA (Figure 1E), therefore, it is of importance to determine whether functional roles between Dofs and lncRNA are more widespread than what is currently known. Therefore, additional studies are required to fully understand the molecular mechanisms by which Dofs orchestrate metabolic homeostasis, stress responses, crop improvement and plant growth and development.
ZW, XJ, and HS conceptualized the theme of the article and compiled the manuscript. ZW and DW prepared the first draft after collecting literature. ZC and WB added the work related to hormone and biotic stress. All authors read and approved the manuscript.
The Scientific Research Start-up Fund by Gansu Agricultural University (GAU-KYQD-2020-26), Research Program Sponsored by State Key Laboratory of Aridland Crop Science, Gansu Agricultural University (No. GSCS-2020-07), Young Doctoral Fund by Education Department of Gansu Province (2021QB-031), and Youth Science and Technology Fund by Science and Technology Department of Gansu Province (21JR7RA844).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.844201/full#supplementary-material
Battaglia, M., Olvera-Carrillo, Y., Garciarrubio, A., Campos, F., and Covarrubias, A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. doi: 10.1104/pp.108.120725
Cai, M., Lin, J., Li, Z., Lin, Z., Ma, Y., Wang, Y., et al. (2020). Allele specific expression of Dof genes responding to hormones and abiotic stresses in sugarcane. PLoS One 15:e0227716. doi: 10.1371/journal.pone.0227716
Cao, B., Cui, Y., Lou, K., Luo, D., Liu, Z., and Zhou, Q. (2020). Genome-wide identification and expression analysis of the Dof gene family in Medicago sativa L. under various abiotic stresses. DNA Cell Biol. 39, 1976–1989. doi: 10.1089/dna.2020.5652
Carvallo, M. A., Pino, M. T., Jeknic, Z., Zou, C., Doherty, C. J., Shiu, S. H., et al. (2011). A comparison of the low temperature transcriptomes and CBF regulons of three plant species that differ in freezing tolerance: Solanum commersonii, Solanum tuberosum, and Arabidopsis thaliana. J. Exp. Bot. 62, 3807–3819. doi: 10.1093/jxb/err066
Cavalar, M., Möller, C., Offermann, S., Krohn, N. M., Grasser, K. D., and Peterhänsel, C. (2003). The interaction of DOF transcription factors with nucleosomes depends on the positioning of the binding site and is facilitated by maize HMGB5. Biochemistry 42, 2149–2157. doi: 10.1021/bi026761r
Chai, F., Liu, W., Xiang, Y., Meng, X., Sun, X., Cheng, C., et al. (2019). Comparative metabolic profiling of Vitis amurensis and Vitis vinifera during cold acclimation. Hortic. Res. 6:8. doi: 10.1038/s41438-018-0083-5
Chen, B., Feder, M. E., and Kang, L. (2018). Evolution of heat-shock protein expression underlying adaptive responses to environmental stress. Mol. Ecol. 27, 3040–3054. doi: 10.1111/mec.14769
Chen, P., Yan, M., Li, L., He, J., Zhou, S., Li, Z., et al. (2020). The apple DNA-binding one zinc-finger protein MdDof54 promotes drought resistance. Hortic. Res. 7:195. doi: 10.1038/s41438-020-00419-5
Chen, W., Chao, G., and Singh, K. B. (1996). The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 10, 955–966. doi: 10.1046/j.1365-313X.1996.10060955.x
Chen, Y., and Cao, J. (2014). Comparative analysis of Dof transcription factor family in maize. Plant Mol. Biol. Report. 33, 1245–1258. doi: 10.1007/s11105-014-0835-9
Cheng, Z., Hou, D., Liu, J., Li, X., and Jian, G. (2018). Characterization of moso bamboo (Phyllostachys edulis) Dof factors in floral development and abiotic stress responses. Genome 61, 151–156. doi: 10.1139/gen-2017-0189
Chini, A., Gimenez-Ibanez, S., Goossens, A., and Solano, R. (2016). Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 33, 147–156. doi: 10.1016/j.pbi.2016.07.005
Corrales, A. R., Carrillo, L., Lasierra, P., Nebauer, S. G., Dominguez-Figueroa, J., Renau-Morata, B., et al. (2017). Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 40, 748–764. doi: 10.1111/pce.12894
Corrales, A. R., Nebauer, S. G., Carrillo, L., Fernandez-Nohales, P., Marques, J., Renau-Morata, B., et al. (2014). Characterization of tomato cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 65, 995–1012. doi: 10.1093/jxb/ert451
da Silva, D. C., da Silveira Falavigna, V., Fasoli, M., Buffon, V., Porto, D. D., and Pappas, G. J. Jr., et al. (2016). Transcriptome analyses of the Dof-like gene family in grapevine reveal its involvement in berry, flower and seed development. Hortic. Res. 3:16042. doi: 10.1038/hortres.2016.42
Devireddy, A. R., Zandalinas, S. I., Fichman, Y., and Mittler, R. (2020). Integration of ROS and hormone signaling during abiotic stress. Plant J. 105, 459–476. doi: 10.1111/tpj.15010
Diaz, I., Martinez, M., Isabel-LaMoneda, I., Rubio-Somoza, I., and Carbonero, P. (2005). The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J. 42, 652–662. doi: 10.1111/j.1365-313X.2005.02402.x
Diaz, I., Vicente-Carbajosa, J., Abraham, Z., Martinez, M., Isabel-La Moneda, I., and Carbonero, P. (2002). The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 29, 453–464. doi: 10.1046/j.0960-7412.2001.01230.x
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Ewas, M., Khames, E., Ziaf, K., Shahzad, R., Nishawy, E., Ali, F., et al. (2017). The tomato DOF daily fluctuations 1, TDDF1 acts as flowering accelerator and protector against various stresses. Sci. Rep. 7:10299. doi: 10.1038/s41598-017-10399-7
Feng, B. H., Han, Y. C., Xiao, Y. Y., Kuang, J. F., Fan, Z. Q., Chen, J. Y., et al. (2016). The banana fruit Dof transcription factor MaDof23 acts as a repressor and interacts with MaERF9 in regulating ripening-related genes. J. Exp. Bot. 67, 2263–2275. doi: 10.1093/jxb/erw032
Fornara, F., de Montaigu, A., Sanchez-Villarreal, A., Takahashi, Y., Ver Loren van Themaat, E., Huettel, B., et al. (2015). The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J. 81, 695–706. doi: 10.1111/tpj.12759
Guo, T., Wang, S., Zhang, T., Xu, L., Li, Y., Chao, Y., et al. (2021). Expression of the Medicago truncatula MtDof32 transcription factor regulates plant growth and enhances abiotic stress tolerances in transgenic Arabidopsis. Environ. Exp. Bot. 183:104339. doi: 10.1016/j.envexpbot.2020.104339
Gupta, S., Malviya, N., Kushwaha, H., Nasim, J., Bisht, N. C., Singh, V. K., et al. (2015). Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 241, 549–562. doi: 10.1007/s00425-014-2239-3
Hakoshima, T. (2018). Structural basis of the specific interactions of GRAS family proteins. FEBS Lett. 592, 489–501. doi: 10.1002/1873-3468.12987
Han, Y., Zhang, X., Wang, W., Wang, Y., and Ming, F. (2013). The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS One 8:e73541. doi: 10.1371/journal.pone.0073541
He, L., Shi, X., Wang, Y., Guo, Y., Yang, K., and Wang, Y. (2017). Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol. Biol. 93, 369–387. doi: 10.1007/s11103-016-0567-3
He, L., Su, C., Wang, Y., and Wei, Z. (2015). ATDOF5.8 protein is the upstream regulator of ANAC069 and is responsive to abiotic stress. Biochimie 110, 17–24. doi: 10.1016/j.biochi.2014.12.017
Huang, W., Huang, Y., Li, M. Y., Wang, F., Xu, Z. S., and Xiong, A. S. (2016). Dof transcription factors in carrot: genome-wide analysis and their response to abiotic stress. Biotechnol. Lett. 38, 145–155. doi: 10.1007/s10529-015-1966-2
Huang, Y., Han, Z., Cheng, N., Luo, M., Bai, X., and Xing, Y. (2019). Minor effects of 11 Dof family genes contribute to the missing heritability of heading date in rice (Oryza sativa L.). Front. Plant Sci. 10:1739. doi: 10.3389/fpls.2019.01739
Imaizumi, T., Schultz, T. F., Harmon, F. G., Ho, L. A., and Kay, S. A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. doi: 10.1126/science.1110586
Isabel-LaMoneda, I., Diaz, I., Martinez, M., Mena, M., and Carbonero, P. (2003). SAD: a new DOF protein from barley that activates transcription of a cathepsin B-like thiol protease gene in the aleurone of germinating seeds. Plant J. 33, 329–340. doi: 10.1046/j.1365-313X.2003.01628.x
Jin, J., Tian, F., Yang, D.-C., Meng, Y.-Q., Kong, L., Luo, J., et al. (2016). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Jin, J., Zhang, H., Kong, L., Gao, G., and Luo, J. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, D1182–D1187. doi: 10.1093/nar/gkt1016
Jin, Z., Chandrasekaran, U., and Liu, A. (2014). Genome-wide analysis of the Dof transcription factors in castor bean (Ricinus communis L.). Genes Genomics 36, 527–537. doi: 10.1007/s13258-014-0189-6
Kang, H. G., Foley, R. C., Onate-Sanchez, L., Lin, C., and Singh, K. B. (2003). Target genes for OBP3, a Dof transcription factor, include novel basic helix-loop-helix domain proteins inducible by salicylic acid. Plant J. 35, 362–372. doi: 10.1046/j.1365-313x.2003.01812.x
Kang, H. G., and Singh, K. B. (2000). Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J. 21, 329–339. doi: 10.1046/j.1365-313x.2000.00678.x
Kang, W. H., Kim, S., Lee, H. A., Choi, D., and Yeom, S. I. (2016). Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep. 6:33332. doi: 10.1038/srep33332
Khan, I., Khan, S., Zhang, Y., and Zhou, J. (2021). Genome-wide analysis and functional characterization of the Dof transcription factor family in rice (Oryza sativa L.). Planta 253:101. doi: 10.1007/s00425-021-03627-y
Kim, W. Y., Ali, Z., Park, H. J., Park, S. J., Cha, J. Y., Perez-Hormaeche, J., et al. (2013). Release of SOS2 kinase from sequestration with GIGANTEA determines salt tolerance in Arabidopsis. Nat. Commun. 4:1352. doi: 10.1038/ncomms2357
Kloosterman, B., Abelenda, J. A., Gomez Mdel, M., Oortwijn, M., de Boer, J. M., Kowitwanich, K., et al. (2013). Naturally occurring allele diversity allows potato cultivation in northern latitudes. Nature 495, 246–250. doi: 10.1038/nature11912
Krohn, N. M., Yanagisawa, S., and Grasser, K. D. (2002). Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. J. Biol. Chem. 277, 32438–32444. doi: 10.1074/jbc.M203814200
Li, G., Xu, W., Jing, P., Hou, X., and Fan, X. (2021). Overexpression of VyDOF8, a Chinese wild grapevine transcription factor gene, enhances drought tolerance in transgenic tobacco. Environ. Exp. Bot. 190:104592. doi: 10.1016/j.envexpbot.2021.104592
Lijavetzky, D., Carbonero, P., and Vicente-Carbajosa, J. (2003). Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol. Biol. 3:17. doi: 10.1186/1471-2148-3-17
Lindemose, S., O’Shea, C., Jensen, M. K., and Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Liu, J., Cheng, Z., Xie, L., Li, X., and Gao, J. (2019). Multifaceted role of PheDof12-1 in the regulation of flowering time and abiotic stress responses in moso bamboo (Phyllostachys edulis). Int. J. Mol. Sci. 20:424. doi: 10.3390/ijms20020424
Liu, X., Liu, Z., Hao, Z., Chen, G., Qi, K., Zhang, H., et al. (2020). Characterization of Dof family in Pyrus bretschneideri and role of PbDof9.2 in flowering time regulation. Genomics 112, 712–720. doi: 10.1016/j.ygeno.2019.05.005
Liu, Y., Liu, N., Deng, X., Liu, D., Li, M., Cui, D., et al. (2020). Genome-wide analysis of wheat DNA-binding with one finger (Dof) transcription factor genes: evolutionary characteristics and diverse abiotic stress responses. BMC Genomics 21:276. doi: 10.1186/s12864-020-6691-0
Ma, J., Li, M. Y., Wang, F., Tang, J., and Xiong, A. S. (2015). Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16:33. doi: 10.1186/s12864-015-1242-9
Martinez, M., Rubio-Somoza, I., Fuentes, R., Lara, P., Carbonero, P., and Diaz, I. (2005). The barley cystatin gene (Icy) is regulated by DOF transcription factors in aleurone cells upon germination. J. Exp. Bot. 56, 547–556. doi: 10.1093/jxb/eri033
Mena, M., Cejudo, F. J., Isabel-Lamoneda, I., and Carbonero, P. (2002). A role for the DOF transcription factor BPBF in the regulation of gibberellin-responsive genes in barley aleurone. Plant Physiol. 130, 111–119. doi: 10.1104/pp.005561
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Molina-Hidalgo, F. J., Medina-Puche, L., Canete-Gomez, C., Franco-Zorrilla, J. M., Lopez-Vidriero, I., Solano, R., et al. (2017). The fruit-specific transcription factor FaDOF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 68, 4529–4543. doi: 10.1093/jxb/erx257
Muller, M., and Munne-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nan, H., Ludlow, R. A., Lu, M., and An, H. (2021). Genome-wide analysis of Dof genes and their response to abiotic stress in rose (Rosa chinensis). Front. Genet. 12:538733. doi: 10.3389/fgene.2021.538733
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Park, H. J., Kim, W. Y., Pardo, J. M., and Yun, D. J. (2016). Molecular interactions between flowering time and abiotic stress pathways. Int. Rev. Cell Mol. Biol. 327, 371–412. doi: 10.1016/bs.ircmb.2016.07.001
Park, J., Kim, Y. S., Kim, S. G., Jung, J. H., Woo, J. C., and Park, C. M. (2011). Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol. 156, 537–549. doi: 10.1104/pp.111.177071
Park, S. H., Lim, H., Hyun, S. J., Yun, D.-W., Yoon, U.-H., Ji, H., et al. (2014). Wound-inducible expression of the OsDof1 gene promoter in a Ds insertion mutant and transgenic plants. Plant Biotechnol. Rep. 8, 305–313. doi: 10.1007/s11816-014-0316-5
Qin, H., Wang, J., Chen, X., Wang, F., Peng, P., Zhou, Y., et al. (2019). Rice OsDOF15 contributes to ethylene-inhibited primary root elongation under salt stress. New Phytol. 223, 798–813. doi: 10.1111/nph.15824
Ramirez, G. L., Shi, L., Bergonzi, S. B., Oortwijn, M., Franco-Zorrilla, J. M., Solano-Tavira, R., et al. (2021). Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 105, 855–869. doi: 10.1111/tpj.15093
Ravindran, P., Yong, S. Y., Mohanty, B., and Kumar, P. P. (2020). An LRR-only protein regulates abscisic acid-mediated abiotic stress responses during Arabidopsis seed germination. Plant Cell Rep. 39, 909–920. doi: 10.1007/s00299-020-02538-8
Renau-Morata, B., Carrillo, L., Dominguez-Figueroa, J., Vicente-Carbajosa, J., Molina, R. V., Nebauer, S. G., et al. (2020). CDF transcription factors: plant regulators to deal with extreme environmental conditions. J. Exp. Bot. 71, 3803–3815. doi: 10.1093/jxb/eraa088
Renau-Morata, B., Molina, R. V., Carrillo, L., Cebolla-Cornejo, J., Sanchez-Perales, M., Pollmann, S., et al. (2017). Ectopic expression of CDF3 genes in tomato enhances biomass production and yield under salinity stress conditions. Front. Plant Sci. 8:660. doi: 10.3389/fpls.2017.00660
Ruta, V., Longo, C., Lepri, A., De Angelis, V., Occhigrossi, S., Costantino, P., et al. (2020). The DOF transcription factors in seed and seedling development. Plants 9:218. doi: 10.3390/plants9020218
Rymen, B., Kawamura, A., Schafer, S., Breuer, C., Iwase, A., Shibata, M., et al. (2017). ABA suppresses root hair growth via the OBP4 transcriptional regulator. Plant Physiol. 173, 1750–1762. doi: 10.1104/pp.16.01945
Shaw, L. M., McIntyre, C. L., Gresshoff, P. M., and Xue, G. P. (2009). Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct. Integr. Genomics 9, 485–498. doi: 10.1007/s10142-009-0130-2
Shi, Y., Ding, Y., and Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant. Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Shim, Y., Kang, K., An, G., and Paek, N. C. (2019). Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 60, 2065–2076. doi: 10.1093/pcp/pcz105
Siboza, X. I., Bertling, I., and Odindo, A. O. (2014). Salicylic acid and methyl jasmonate improve chilling tolerance in cold-stored lemon fruit (Citrus limon). J. Plant Physiol. 171, 1722–1731. doi: 10.1016/j.jplph.2014.05.012
Skirycz, A., Reichelt, M., Burow, M., Birkemeyer, C., Rolcik, J., Kopka, J., et al. (2006). DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 47, 10–24. doi: 10.1111/j.1365-313X.2006.02767.x
Song, A., Gao, T., Li, P., Chen, S., Guan, Z., Wu, D., et al. (2016). Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front. Plant Sci. 7:199. doi: 10.3389/fpls.2016.00199
Sprenger, H., Erban, A., Seddig, S., Rudack, K., Thalhammer, A., Le, M. Q., et al. (2018). Metabolite and transcript markers for the prediction of potato drought tolerance. Plant Biotechnol. J. 16, 939–950. doi: 10.1111/pbi.12840
Su, Y., Liang, W., Liu, Z., Wang, Y., Zhao, Y., Ijaz, B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017
Sun, X., Matus, J. T., Wong, D. C. J., Wang, Z., Chai, F., Zhang, L., et al. (2018). The GARP/MYB-related grape transcription factor AQUILO improves cold tolerance and promotes the accumulation of raffinose family oligosaccharides. J. Exp. Bot. 69, 1749–1764. doi: 10.1093/jxb/ery020
Tominaga, Y., Suzuki, K., Uemura, M., and Kawamura, Y. (2021). In planta monitoring of cold-responsive promoter activity reveals a distinctive photoperiodic response in cold acclimation. Plant Cell Physiol. 62, 43–52. doi: 10.1093/pcp/pcaa138
Tuteja, N. (2007). Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2, 135–138. doi: 10.4161/psb.2.3.4156
Vicente-Carbajosa, J., Moose, S. P., Parsons, R. L., and Schmidt, R. J. (1997). A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. U.S.A. 94, 7685–7690. doi: 10.1073/pnas.94.14.7685
Wang, H., Wang, H., Shao, H., and Tang, X. (2016). Recent advances in utilizing transcription factors to improve plant abiotic stress tolerance by transgenic technology. Front. Plant Sci. 7:67. doi: 10.3389/fpls.2016.00067
Wang, P., Yan, Z., Zong, X., Yan, Q., and Zhang, J. (2021). Genome-wide analysis and expression profiles of the dof family in Cleistogenes songorica under temperature, salt and ABA treatment. Plants 10:850. doi: 10.3390/plants10050850
Wang, Z., Wang, Y., Tong, Q., Xu, G., Xu, M., Li, H., et al. (2021). Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 253:55. doi: 10.1007/s00425-021-03574-8
Washio, K. (2003). Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol. 133, 850–863. doi: 10.1104/pp.103.027334
Wasternack, C., and Feussner, I. (2018). The oxylipin pathways: biochemistry and function. Annu. Rev. Plant Biol. 69, 363–386. doi: 10.1146/annurev-arplant-042817-040440
Wen, C. L., Cheng, Q., Zhao, L., Mao, A., Yang, J., Yu, S., et al. (2016). Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 6:23072. doi: 10.1038/srep23072
Wu, Q., Li, D., Li, D., Liu, X., Zhao, X., Li, X., et al. (2015). Overexpression of OsDof12 affects plant architecture in rice (Oryza sativa L.). Front. Plant Sci. 6:833. doi: 10.3389/fpls.2015.00833
Wu, Z., Cheng, J., Cui, J., Xu, X., Liang, G., Luo, X., et al. (2016). Genome-wide identification and expression profile of dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant Sci. 7:574. doi: 10.3389/fpls.2016.00574
Xin, H., Zhu, W., Wang, L., Xiang, Y., Fang, L., Li, J., et al. (2013). Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress. PLoS One 8:e58740. doi: 10.1371/journal.pone.0058740
Xu, J., and Dai, H. (2016). Brassica napus cycling Dof factor1 (BnCDF1) is involved in flowering time and freezing tolerance. Plant Growth Regul. 80, 315–322. doi: 10.1007/s10725-016-0168-9
Xu, X. X., Hu, Q., Yang, W. N., and Jin, Y. (2017). The roles of call wall invertase inhibitor in regulating chilling tolerance in tomato. BMC Plant Biol. 17:195. doi: 10.1186/s12870-017-1145-9
Yanagisawa, S. (1995). A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 23, 3403–3410. doi: 10.1093/nar/23.17.3403
Yanagisawa, S. (2004). Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiol. 45, 386–391. doi: 10.1093/pcp/pch055
Yanagisawa, S. (2016). “Structure, function, and evolution of the Dof transcription factor family,” in Plant Transcription Factors, ed. D. H. Gonzalez (Cambridge, MA: Academic Press), 183–197. doi: 10.1016/b978-0-12-800854-6.00012-9
Yanagisawa, S., and Izui, K. (1993). Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J. Biol. Chem. 268, 16028–16036. doi: 10.1016/s0021-9258(18)82353-5
Yanagisawa, S., and Schmidt, R. J. (1999). Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 17, 209–214. doi: 10.1046/j.1365-313x.1999.00363.x
Yang, G., Gao, X., Ma, K., Li, D., Jia, C., Zhai, M., et al. (2018). The walnut transcription factor JrGRAS2 contributes to high temperature stress tolerance involving in Dof transcriptional regulation and HSP protein expression. BMC Plant Biol. 18:367. doi: 10.1186/s12870-018-1568-y
Yang, G., Yu, L., Wang, Y., Wang, C., and Gao, C. (2017). The translation initiation factor 1A (TheIF1A) from Tamarix hispida is regulated by a Dof transcription factor and increased abiotic stress tolerance. Front. Plant Sci. 8:513. doi: 10.3389/fpls.2017.00513
Yang, J., Duan, G., Li, C., Liu, L., Han, G., Zhang, Y., et al. (2019). The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10:1349. doi: 10.3389/fpls.2019.01349
Yang, J., Yang, M. F., Wang, D., Chen, F., and Shen, S. H. (2010). JcDof1, a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol. Plant. 139, 324–334. doi: 10.1111/j.1399-3054.2010.01363.x
Yu, H., Ma, Y., Lu, Y., Yue, J., and Ming, R. (2021). Expression profiling of the Dof gene family under abiotic stresses in spinach. Sci. Rep. 11:14429. doi: 10.1038/s41598-021-93383-6
Yu, L., Ma, S., Zhang, X., Tian, D., Yang, S., Jia, X., et al. (2021). Ancient rapid functional differentiation and fixation of the duplicated members in rice Dof genes after whole genome duplication. Plant J. 108, 1365–1381. doi: 10.1111/tpj.15516
Yu, Y. H., Bian, L., Wan, Y. T., Jiao, Z. L., and Guo, D. L. (2019). Grape (Vitis vinifera) VvDOF3 functions as a transcription activator and enhances powdery mildew resistance. Plant Physiol. Biochem. 143, 183–189. doi: 10.1016/j.plaphy.2019.09.010
Zang, D., Wang, L., Zhang, Y., Zhao, H., and Wang, Y. (2017). ThDof1.4 and ThZFP1 constitute a transcriptional regulatory cascade involved in salt or osmotic stress in Tamarix hispida. Plant Mol. Biol. 94, 495–507. doi: 10.1007/s11103-017-0620-x
Zhang, L., Cheng, J., Sun, X., Zhao, T., Li, M., Wang, Q., et al. (2018). Overexpression of VaWRKY14 increases drought tolerance in Arabidopsis by modulating the expression of stress-related genes. Plant Cell Rep. 37, 1159–1172. doi: 10.1007/s00299-018-2302-9
Zhang, Z., Yuan, L., Liu, X., Chen, X., and Wang, X. (2018). Evolution analysis of Dof transcription factor family and their expression in response to multiple abiotic stresses in Malus domestica. Gene 639, 137–148. doi: 10.1016/j.gene.2017.09.039
Zhou, Y., Cheng, Y., Wan, C., Li, J., Yang, Y., and Chen, J. (2020). Genome-wide characterization and expression analysis of the Dof gene family related to abiotic stress in watermelon. PeerJ 8:e8358. doi: 10.7717/peerj.8358
Zhu, X., Hong, X., Liu, X., Li, S., Yang, J., Wang, F., et al. (2021). Calcium-dependent protein kinase 32 gene maintains photosynthesis and tolerance of potato in response to salt stress. Sci. Hortic. 285:110179. doi: 10.1016/j.scienta.2021.110179
Zhuo, M., Sakuraba, Y., and Yanagisawa, S. (2020). A jasmonate-activated MYC2-Dof2.1-MYC2 transcriptional loop promotes leaf senescence in Arabidopsis. Plant Cell 32, 242–262. doi: 10.1105/tpc.19.00297
Keywords: transcription factor, Dof, hormonal signaling, transcriptional regulation, abiotic stresses
Citation: Wang Z, Wong DCJ, Chen Z, Bai W, Si H and Jin X (2022) Emerging Roles of Plant DNA-Binding With One Finger Transcription Factors in Various Hormone and Stress Signaling Pathways. Front. Plant Sci. 13:844201. doi: 10.3389/fpls.2022.844201
Received: 27 December 2021; Accepted: 25 March 2022;
Published: 13 May 2022.
Edited by:
Luisa M. Sandalio, Department of Biochemistry, Cell and Molecular Biology of Plants, Experimental Station of Zaidín (CSIC), SpainReviewed by:
Dierk Wanke, University of Tübingen, GermanyCopyright © 2022 Wang, Wong, Chen, Bai, Si and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zemin Wang, d3ptNDA2NDMzMjU0QDEyNi5jb20=; Huaijun Si, aGpzaUBnc2F1LmVkdS5jbg==; Xin Jin, amlueEBnc2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.