
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 08 March 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.844149
This article is part of the Research Topic New Insights into Mechanisms of Epigenetic Modifiers in Plant Growth and Development, Volume II View all 6 articles
MicroRNAs (miRNAs), a class of endogenous, non-coding RNAs, which is 20–24 nucleotide long, regulate the expression of its target genes post-transcriptionally and play critical roles in plant normal growth, development, and biotic and abiotic stresses. In cells, miRNA biogenesis and stability control are important in regulating intracellular miRNA abundance. In addition, research on these two aspects has achieved fruitful results. In this review, we focus on the recent research progress in our understanding of miRNA biogenesis and their stability control in plants.
MicroRNAs (miRNAs), which are 20–24 nucleotide long, endogenous, non-coding RNAs, repress its target gene expression through sequence complementarity. miRNAs play an important role in all aspects of normal plant growth, development, and biotic and abiotic stress (Wu et al., 2009; Budak et al., 2015; Feng et al., 2016; Shriram et al., 2016; Li et al., 2017b; Brant and Budak, 2018; Ayubov et al., 2019; Song et al., 2019). Thus, the spatiotemporal expression of miRNAs is regulated at multiple levels to ensure the fine regulation of target genes and maintain normal life activities.
The first miRNA lin4 was discovered in Caenorhabditis elegans in the early 1990s (Lee et al., 1993). Researchers focus on the importance of miRNA in regulating gene expression. MIRNA (MIR) genes are transcribed by DNA-dependent RNA Polymerase II (Pol II), and the primary transcripts of MIRs (known as pri-miRNAs) are 5′ capped, 3′ polyadenylated, and/or spliced, similar to message RNA (mRNA) (Xie et al., 2005; Rogers and Chen, 2013; Hunt, 2014; Ramanathan et al., 2016; Deng and Cao, 2017).
Pri-miRNAs, a typical hairpin-like structure RNAs, can be recognized by the processing complex, subsequently undergoing two major nuclear processing steps mediated by Dicer-like protein DCLs and two important cofactors, Serrate (SE) and Hyponastic leaves 1 (HYL1). Dicer-like 1 (DCL1) is primarily responsible for miRNA production. Notably, the production of miR839 and miR822 is dependent on DCL4 but not DCL1 (Rajagopalan et al., 2006). After the two processes, the miRNA/miRNA* duplex, which contends 2 nt overhang at its 3′ end, is released either from Loop to Base or Base to Loop (Kurihara et al., 2006; Addo-Quaye et al., 2009; Bologna et al., 2009, 2013; Song et al., 2010; Figure 1).
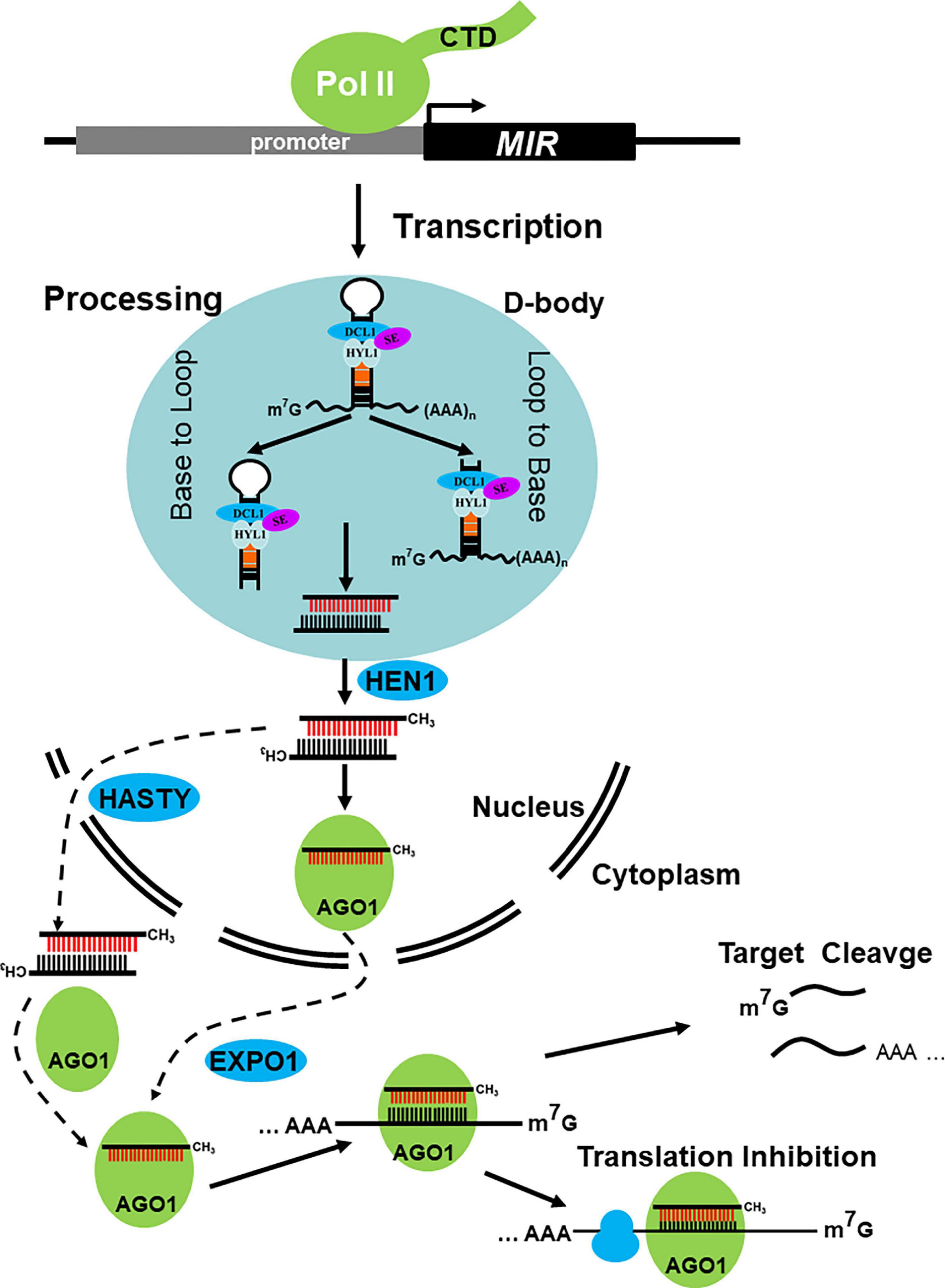
Figure 1. Major steps in miRNA biogenesis and action in plants. MIR genes are transcripted by DNA dependent RNA Polymerase II (Pol II), with that the primary transcripts (also knowed as pri-miRNAs) are processed to the miRNA/miRNA* duplex by dicing complex in D-bodies, neither from Base to Loop nor from Loop to Base. The resulting miRNA/miRNA* duplex is then methylated by terminal methyltransferase HEN1, which happens in nucleus. And then, the miRNA load into AGO1 protein and export via EXPO1 or the methylated duplex translocated from nucleus to cytoplasm via HASTY, here load into AGO. Both to form RISC to direct target cleavge and/or translational inhibition.
The miRNA/miRNA* duplex is methylated by the Hua Enhancer 1 (HEN1), a terminal methyltransferase, which methylates miRNA/miRNA* and siRNA/siRNA* duplexes on its 2′ OH of the 3′-terminal nucleotide particularly in plants (Yu et al., 2005; Yang Z. et al., 2006; Baranauskė et al., 2015). Then, the miRNA/miRNA* duplexes are thought to be translocated from the nucleus to the cytoplasm by HASTY (HST), a homologous gene of animal Exportin 5 (EXPO5) (Park et al., 2005). Afterward, the guide strand (miRNA) is loaded into Argonaute (AGO) to form RNA-induced silencing complex (RISC), whereas the passenger strand (miRNA*) is removed and degraded (Figure 1). This process is called Argonaute loading and sorting. After the formation of RISC, mature miRNAs can search its targets by base pairing. miRNA-target base pairing strengthens with nearly perfect match in plants but relatively loosens in animals. After the research period, the mode of miRNA’s action on its targets is clear, primarily in target cleavage and/or translation inhibition (Baumberger and Baulcombe, 2005; Figure 1). In addition, some miRNAs, such as miR390, miR173, and miR828 can trigger the production of secondary siRNAs, which are called phasiRNAs and/or tasiRNAs (Peragine et al., 2004; Vazquez et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005; Montgomery et al., 2008; Zhang et al., 2012; Fei et al., 2013; Deng et al., 2018). Maintenance of miRNA abundance is the prerequisite for its function, and the biogenesis and stability control are the two important coincides of miRNA abundance. After nearly 30 years of intensive research, dozens of genes involved in miRNA biogenesis and stability control have been identified, and the pathway has been gradually revealed. In this review, we focus on the recent research progress in our understanding of miRNA biogenesis and their stability control in plants and pay more attention to the regulation of genes that affect the abundance of miRNA.
Similar to the formation of message RNA (mRNA), the formation of pri-miRNAs follows the same processes including transcription, capping, 3′ polyadenylation, and splicing. MIR genes are transcribed by DNA-dependent RNA polymerase II (Pol II). Thus, some factors associated with Pol II may also affect MIR transcription (Figure 2A). The mediator complex plays a critical role in recruiting Pol II to the promoters of MIR genes, such as MEDIATOR 20A (MED20A), MEDIATOR 17 (MED17), and MEDIATOR 18 (MED18). In these mutants, the transcription level of pri-miRNAs is suppressed (Chadick and Asturias, 2005; Kim et al., 2011). Tho2/Hpr2 Phenotype 1 (THP1) and Yeast Sac3 Homolog A (SAC3A), two core subunits of the Transcription Coupled Export 2 (TREX-2) complex, interact and colocalize with Pol II to promote MIR gene transcription (Zhang et al., 2020). The C-terminal domain (CTD) of Pol II can be phosphorylated by Cyclin-Dependent Kinase Ds (CDKDs) and Cyclin-Dependent Kinase (CDKF;1), and the phosphorylation of CTD facilitates MIR transcription, 5′-end capping, cotranscriptional RNA processing, and 3′-end polyadenylation (Hajheidari et al., 2012). Histone modification and chromatin remodeling are two important epigenetic modifications for gene activation and/or inhibition, and they are important for the transcription of MIR genes. For example, General Control Non-repressed protein 5 (GCN5), a histone acetyltransferase, targets a number of MIR genes, and it is responsible for the acetylation of H3K14 of these loci (Kim et al., 2009) and others, including SWINGER (SWN), CURLY LEAF (CLF), and PICKLE (PKL), which regulate MIR transcription as well. Different from the above, these three epigenetic factors affect only a few MIRs, for example, MIR156A/C, and accelerate the transition of juvenile to adult phase (Xu et al., 2016). Chromatin Remodeling factor 2 (CHR2), an ATPase subunit of the large Switch/Sucrose Non-Fermentable (SWI/SNF) chromatin-remodeling complex, positively regulates transcription of MIR loci by its chromatin remodeling activity (Wang Z. et al., 2018). Some transcription factors, such as Cell Division Cycle 5 (CDC5) and Cycling DOF Factor 2 (CDF2), also impair MIR transcription. CDC5, an MYB-related protein, positively regulates the occupancy of Pol II at MIR promoters and its activities (Zhang et al., 2013). CDF2, a member of DNA binding with One Finger (DOF) gene family, regulates MIR transcription positively and negatively by recruiting different transcriptional machinery to different MIR promoters (Sun et al., 2015). Apart from transcription factors, other regulators play an important role in MIR transcription, including Negative On TATA less2 (NOT2), ELONGATOR PROTEIN 2/5 (ELP2/5), Suppressor of npr1-1 Constitutive 1 (SNC1), Topless Related 1 (TPR1), SMALL1 (SMA1), Short Valve 1 (STV1), Increased Level of Polyploidy1-1D (ILP1), NTC-Related protein 1 (NTR1), and HASTY (HST). NOT2a/2b, a pair of NOT2_3_5 domain-containing proteins, promotes the transcription of MIR genes via interacting with Pol II (Wang et al., 2013). ELP2/5, which are two elongator proteins, and loss functions of ELP2/5 reduce Pol II occupancy at miRNA loci and pri-miRNA transcription (Fang et al., 2015a). SNC1, a disease-resistant gene, and its transcriptional corepressor TPR1 repress the transcription of MIR genes (Cai et al., 2018). Recent research suggests that STV1 impresses the transcription of MIR by affecting the binding of Pol II to the promoter of MIR genes and also SMA1 (Li et al., 2017c,2018). ILP1/NTR1, which are two conserved disassembly factors of the Intron-Lariat Spliceosome complex, positively regulate miRNA biogenesis by facilitating the transcriptional elongation of MIR genes (Wang et al., 2019a). HST, which is the ortholog of EXPORTIN5, is considered as an exportin of plant miRNAs from the nucleus to the cytoplasm for the last two decades. However, at present, Cambiagno et al. (2021) have suggested that HST plays a role in promoting MIR gene transcription.
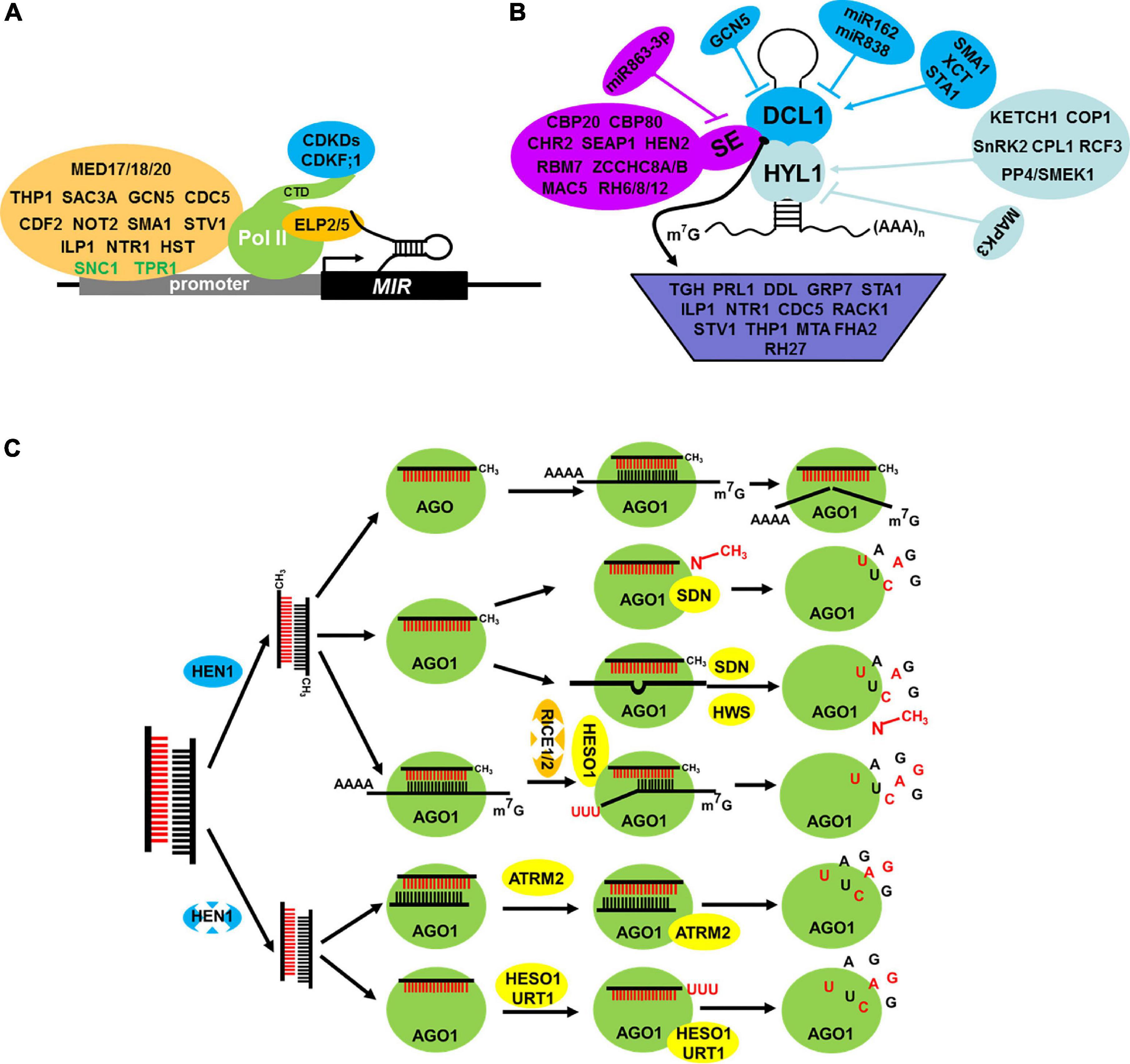
Figure 2. Factors involved in the regulation of MIR transcription (A), pri-miRNA processing (B) and miRNA stability control (C). (A) Factor in regulating MIR transcription. Several factors interact with MIR promoter and Pol II can active (orange with black text) or repress (orange with green text) the MIR transcription. Moreover, Pol II activity is subjected to phosphorylation at its C-terminal domain (CTD) via CDKDs and CDKF; 1 (blue). Elongator factor 2/5 can interact with pri-miRNAs and Pol II in transcription elongation stage to promote transcription. (B) Factors in regulating core processing machinery component DCL1 (blue), HYL1 (aqua), and SE (purple) and also factors that influence pri-miRNA structure, stability, splicing, loading to the processor and the processor activity (indigo blue). (C) Factors in regulating miRNA stability control. The AGO1 protein can protect miRNA degradation and also recruit some degradation factors to the RISC complex. HESO1 and URT1 catalyze the 3′ terminal uridylation of unmethylated miRNA and trigger their degradation, but ATRM2 degradate unmethylated miRNA/miRNA* duplex. Additionally, SDN1 degradate methylated nucleotide of 3′end of miRNA, but some factors also degradate uridilated cleavage products and also unoptimizable RISCs (RICE1/2 and HWS, respectively), all of these aspects affect the stability of miRNA and its abundance.
After transcription, the pri-miRNAs with a typical stem-loop structure are produced in the nucleus. Then, pri-miRNAs follow two constitutive cuts to produce pre-miRNAs and to release the miRNA/miRNA* duplex. This process is called processing, which occurs in two distinct subnuclear bodies, namely, Dicing-bodies (D-bodies), or SmD3/SmB bodies (Fang and Spector, 2007; Fujioka et al., 2007). DCL1, HYL1, and SE are the three core components of a microprocessor. DCL1, an RNAse III endoribonuclease, is responsible for the cleavage of pri-/pre-miRNAs. The knockout of Arabidopsis DCL1 is embryo lethal (Errampalli et al., 1991). HYL1 and SE are the two cofactors of DCL1, which interact with each other in D-bodies to ensure the accuracy and efficiency of processing (Fang and Spector, 2007; Dong et al., 2008; Liu et al., 2012; Zhu et al., 2013). Increasing studies have been conducted on DCL1, HYL1, and SE because of its importance in processing (Figure 2B).
DCL1 was first identified in pri-miRNAs and pre-miRNAs processing (Kurihara and Watanabe, 2004). Several factors that could regulate the transcription of DCL1, including GCN5, Stabilized 1 (STA1), and XAP5 Circadian Timekeeper (XCT), were identified (Figure 2B); the first one is a negative factor and the last two are positive ones (Kim et al., 2009; Ben Chaabane et al., 2013; Fang et al., 2015b). Otherwise, the abundance of DCL1 is fine-tuned by two feedback regulating levels. On the one hand, the biogenesis of miR162 is dependent on DCL1, but the increase of miR162 suppresses the abundance of DCL1mRNA by target cleavage. On the other hand, the generation of miR838 depends on the processing of DCL1 pre-mRNA, which leads to a decrease in productive mRNA processing of DCL1 (Xie et al., 2003; Rajagopalan et al., 2006). Except for the above, a recent research suggested that SMA1 not only affects MIR transcription but also plays an important role in the correct splicing of DCL1 (Li et al., 2018).
HYL1, an important chaperone of DCL1, encodes a double-stranded RNA-binding protein and plays a critical role in processing accuracy and efficiency. In hyl1 mutant, more pri-miRNAs but less mature miRNAs accumulate (Vazquez et al., 2004). HYL1 protein comprises three representative domains, including two double-stranded RNA-binding domains (dsRBD1 and dsRBD2) in its N-terminal and one protein–protein interaction domain in its C-terminal. Yang et al. (2010, 2014) suggested that HYL1 binds to the double-stranded region of pri-miRNAs in dimer and the homodimer functions as a molecular anchor for correct DCL1 cleavage. In the past few years, several groups approached forward genetic screen of hyl1 mutant for its suppressor. Several dominant DCL1 alleles were identified, and all of them can rescue the phenotypes of hyl1 mutant defects in varying degrees (Tagami et al., 2009; Liu et al., 2012; Gao et al., 2020). Notably, apart from DCL1, Gao et al. (2020) identified new suppressors such as SOP1 (Suppressor of pas2 1), which is a novel factor of hyl1. Further study showed that HYL1 antagonizes nuclear exosome to protect pri-miRNAs from degradation. In recent years, considerable studies have been conducted on the function of HYL1, particularly in D-body formation, nuclear-cytoplasm translocation, and post translational modification (Figure 2B). MOS4-associated Complex 7 (MAC7), a component of the MAC complex, promotes the localization of HYL1 to D-bodies. In mac7 mutant, the number of D-bodies is decreasing, which may decrease the miRNA level (Jia et al., 2017). In addition, considerable research has shown that HYL1 can localize not only in the nucleus but also in the cytoplasm. Karyopherin Enabling the Transport of the Cytoplasmic HYL1 (KETCH 1), a member of the importin β-family, transports HYL1 from the cytoplasm to the nucleus to take part in miRNA biogenesis (Zhang et al., 2017a). In dark conditions, HYL1 suffers from degradation in the cytoplasm, whereas in light conditions, Constitutive Photomorphogenic 1 (COP1), which is a RING-finger E3 ligase, translocates into the cytoplasm from the nucleus and protects the degradation of HYL1 by suppressing the activity of an unknown protease (Cho et al., 2014). Phosphorylation plays an important role in regulating the activity and stability of HYL1 (Figure 2B). The activity and stability of the phosphorylation of HYL1 affect the accurate processing of miRNA. Phosphorylated HYL1 is non-functional. C-terminal domain Phosphatase-Like 1 (CPL1) shares a domain with yeast and human Fcp1 phosphatases (Koiwa et al., 2002) and dephosphorylates HYL1, thereby leading to accurate processing and strand selection (Manavella et al., 2012). Similar to CPL1, a PP4/Suppressor of MEK 1 (SMEK1) complex can also dephosphorylate and stabilize HYL1 (Su et al., 2017). Some kinases, such as Mitogen-Activated Protein Kinase (MAPK3) and SNF1-related Protein Kinase 2 (SnRK2), can phosphorylate HYL1 and waken the position of HYL1 during miRNA processing (Raghuram et al., 2015; Yan et al., 2017). The regulator of CBF gene expression 3 (RCF3), a KH-domain protein, interacting with phosphatases CPL1 and CPL2, mediates the dephosphorylation of HYL1 and then increases its activity in specific tissues (Karlsson et al., 2015).
SE, another cofactor of DCL1, encodes a C2H2 zinc finger protein. se mutant exhibits some developmental defects similar to hyl1 mutant. Further analysis suggested that SE plays a critical role in pri-miRNA processing (Yang L. et al., 2006). After years of studies, research on SE has made great progress (Figure 2B). The transcription level of SE is regulated by miR863-3P in a feedback loop manner (Niu et al., 2016). Apart from transcriptional regulation, post translational modification is important for SE. SnRK2 can phosphorylate HYL1 and SE in vitro, but the real function in vivo is largely unknown (Yan et al., 2017). Several SE-interacting proteins have been identified for the past few years. CHR2, a typical chromatin remodeling factor, which can interact with SE to remodel pri-miRNAs’ conformation and impair its processing (Wang Z. et al., 2018). MAC5, a component of MOS4-associated complex (MAC), interacts with SE and protects pri-miRNAs from nuclease degradation (Li et al., 2020). Recent study has found another SE-interacting protein, namely, Serrate-Associated Protein 1 (SEAP1), which positively promotes miRNA biogenesis by modulating pri-miRNA splicing, processing, and/or stability (Li M. et al., 2021). SE also interacts with the RNA helicase HEN2, RNA-binding protein RBM7, and one of the two ZCCHC8A/ZCCHC8B, which are members of the Nuclear Exosome Targeting (NEXT) complex. SE and NEXT complex promote the degradation of pre-miRNA, but unlike SE, the NEXT complex is not necessary for miRNA processing (Bajczyk et al., 2020). SE is not only involved in miRNA biogenesis, but also in regulating the expression of mRNA. First, SE promotes pre-mRNA splicing by interacting with Cap Binding Complex (CBC) (Laubinger et al., 2008). Second, SE plays an unexpected role in promoting the expression of intronless genes via promoting the association of Pol II with direct chromatin binding (Speth et al., 2018). Third, SE is also involved in the expression of TE by promoting H3K27me1 mediated by Trithorax Related Protein 5 and 6 (ATXR5/6) and by suppressing RNA silencing via RNA-dependent RNA polymerase 6 (RDR6) (Ma et al., 2018). Notably, SE also participates in phase separation. SE-mediated phase separation is necessary for the formation of D-bodies, in which accurate and efficient miRNA processing occurs (Xie et al., 2021). A recent study also reported that several RNA helicase RH6/8/12 interact with SE and promote the phase separation and the formation of D-bodies (Li Q. et al., 2021).
In addition to the regulation of miRNA biogenesis core components, other regulatory factors may affect the pri-miRNA structure, stability, splicing, loading to the processor, and processor activity (Zhang et al., 2015; Wang Z. et al., 2018; Figure 2B). These processes may impress miRNA biogenesis. The lengths of plant pri-miRNAs hairpin structure range from 49 to 900 nt, which affect pri-miRNA processing pattern and efficiency (Bologna and Voinnet, 2014). To date, no regulatory factors influencing cleavage pattern determination have been identified. The stability of pri-miRNA also affects processing. For example, PRL1, a conserved WD-40 protein, promotes pri-miRNAs accumulation by stabilizing pri-miRNAs and enhancing the DCL1 activity (Zhang et al., 2014). Similar to PRL1, DAWDLE (DDL), a forkhead domain (FHA) containing protein, may stabilize pri-miRNAs but not affect MIR promoter activity (Yu et al., 2008). Plant MIRs also suffer from splicing and alternative splicing, which may impair pri-miRNA processing, for example, glycine-rich RNA-binding protein 7 (GRP7), STA1, and ILP1/NTR1. GRP7, a hnRNP-like glycine-rich RNA-binding protein, interacts with pri-miRNAs in vivo and promotes pri-miRNA splicing (Köster et al., 2014). ILP1/NTR1 also influences the alternative splicing of pri-miRNAs. However, in ILP1 or NTR1 mutant, pri-miRNAs with or without introns are globally downregulated (Wang et al., 2019a). Cap Binding Protein 20 (CBP20) and 80, which are two members of the CBC complex, are involved in mRNA and pri-miRNAs splicing, but they may have a direct role in pri-miRNA processing and splicing because spliced and unspliced pri-miRNAs are increased (Laubinger et al., 2008). Apart from the core component of dicing complex HYL1 and SE, many other proteins participate during processing. They regulate DCL1 activity and/or facilitate the loading of pri-miRNAs to the processing complex by interacting with these core factors. TOUGH (TGH), a G-patch domain-containing protein, is a known member in the DCL1 complex, which positively regulates miRNA and siRNA biogenesis (Ren et al., 2012a). CDC5 plays dual roles in miRNA biogenesis, positively regulates MIR transcription, and/or promotes pri-miRNAs processing by interacting with DCL1, thereby increasing its activity (Zhang et al., 2013). Receptor of Activated C Kinase 1 (RACK1), a partner protein of SE, promotes miRNA biogenesis by regulating processing (Speth et al., 2013). RH27, a DEAD-box RNA helicase, which associates with pri-miRNAs and interacts with miRNA-biogenesis components, including DDL, HYL1, and SE. In rh27-2, a large number of miRNAs and their pri-miRNAs are suppressed in shoot apices and root tips (Hou et al., 2021). Short Valve 1 (STV1), a conserved ribosomal protein, facilitates the recruitment of pri-miRNAs to HYL1 to promote miRNA biogenesis (Li et al., 2017c). miRNA processing occurs in D-bodies; thus, its formation directly influences the miRNA level. THP1, a subunit of the TREX-2 complex, interacts with SE and facilitates the formation of D-bodies, thereby promoting miRNA processing. Some factors in mRNA modification are important for miRNA biogenesis. mRNA adenosine methylase (MTA), a homolog of METTL3, introduces N6-methyladenosine (m6A) into pri-miRNAs and promotes miRNA biogenesis by interacting with Pol II and TOUGH, a known regulator in miRNA processing (Bhat et al., 2020). Recently, some researchers have proposed that miRNA biogenesis may be related to light signaling. Although the abundance of DCL1, HYL1, and SE is upregulated during de-etiolation, the levels of most miRNAs are not significantly increased owing to the reduction of miRNA processing activity through an unknown suppressor and the shortening of the half-life of some miRNAs via SMALL RNA DEGRADING NUCLEASE 1 (SDN1) (Choi et al., 2019). Secondly, FHA2, a forkhead-associated domain containing protein, which suppresses miRNA biogenesis in a light-dependent manner. FHA2 promotes HYL1 binding but inhibits the binding of DCL1 to pri-miRNAs (Park et al., 2021). Therefore, these regulatory proteins in miRNA biogenesis change pri-miRNA’s structure, stability, modification, splicing state, and binding to the processing machinery, thereby influencing miRNA biogenesis.
The miRNA/miRNA* duplex is released following transcription and processing. HEN1 methylates the miRNA/miRNA* duplexes at 2′ OH of the 3′-terminal nucleotide. Then, the duplexes export from the nucleus to the cytoplasm, and the guide strand (also known as miRNA) load into the effector protein AGO to form the RISC complex. AGO1 is the major effector protein for the vast majority of miRNAs. In recent years, relevant research in AGO1 transcriptional and translational regulation (Vaucheret et al., 2006; Bortolamiol et al., 2007; Chiu et al., 2010; Csorba et al., 2010; Earley et al., 2010; Derrien et al., 2012), AGO sorting, strand selection (Mi et al., 2008; Takeda et al., 2008), RISC assembly (Iki et al., 2010, 2012; Iwasaki et al., 2010; Earley and Poethig, 2011), RISC loading, and miRISCs export (Wang et al., 2011; Cui et al., 2016; Tomassi et al., 2020; Zhang et al., 2020) have made several achievements. Here, we do not expand, and we focus on the maintenance of miRNA stability.
Maintenance of intracellular miRNA homeostasis is important for plants in the adaption of environmental and developmental changes. Except for miRNA biogenesis, miRNA stability control also plays key roles in miRNA homeostasis regulation. Several aspects, including 3′-end modification (methylation and uridylation), endoribonuclease-mediated miRNA degradation, association with AGO protein, and miRNA-target interaction, can affect miRNA stability. The factors involved in these processes all affect the stability of miRNAs.
A number of factors, which regulate miRNA 3′ end modification such as methylation and uridylation, have been identified (Figure 2C). First, HEN1 catalyzed miRNA/miRNA* and siRNA/siRNA* duplex methylation; although the binding of its two dsRNA-binding domains to the RNA substrate is significant, the contribution is different (Baranauskė et al., 2015). After the action of HEN1, the methyl group is added at 2′ OH of 3′ terminal nucleotide of miRNA (Yu et al., 2005; Yang Z. et al., 2006). To date, HEN1-mediated methylation is considered as the core mechanism of miRNA stability regulation. In 2017, a group from Chinese Taipei suggested that light activated the expression of HEN1. Moreover, the activation depends on a number of photoreceptors and transcription factors HY5. Further analysis found that HY5 is also targeted by miR157d, whose abundance is regulated by HEN1 (Tsai et al., 2014). This project focuses on the transcriptional and post transcriptional regulation of HEN1. Apart from methylation and nuclease media degradation of methylated miRNAs (see blow, SDNs), miRNA also suffered from uridylation. In Arabidopsis, HEN1 Suppressor 1 (HESO1) is the first gene identified for miRNA uridylation (Li et al., 2005; Zhao et al., 2012). HESO1 catalyzes the addition of uracils to the 3′-end of miRNAs in the absence of HEN1. Therefore, HESO1 catalyzes unmethylated miRNAs. During the overexpression of HESO1 in hen1 mutant, the developmental defect was more serious than hen1 mutant. Also, the miRNAs level is less, indicating that HESO1 promotes miRNA degradation (Ren et al., 2012b). The second one is UTP: RNA uridylyltransferase 1 (URT1), a functional paralog of HESO1, which functionally reduant and cooperates with HESO1 (Tu et al., 2015; Wang et al., 2015). HESO1 and URT1 colocalize and interact with AGO1 and uridylate AGO1-bound miRNAs (Ren et al., 2014b; Tu et al., 2015; Wang et al., 2015). Although HESO1 and URT1 share many similarities in uridylation, there are also some differences. For miRNA, HESO1 has been proven to be the major enzyme responsible for uridylation of unmethylated miRNAs and siRNAs, while URT1 mainly uridylates miRNAs sequentially and cooperatively with HESO1 (Tu et al., 2015; Wang et al., 2015). For mRNA, URT1 becomes the main TUTase to cover 70%–80% of mRNAs uridylation; however, HESO1 targets mostly the mRNAs with short tails (Sement et al., 2013). HESO1 and URT1 also exhibit distinct substrate preferences, especially the 3′ end nucleotide of its substrate sRNAs, in detail, HESO1 has the strongest preference for U, while URT1 for A (Tu et al., 2015; Wang et al., 2015).
RNA degradation occurs either from its 5′ or 3′ terminal guide by exoribonucleases or cleavage by endoribonuclease theoretically. Different from mRNA, miRNA primarily includes 3′ to 5′ degradation, according to methylation mediated by HEN1. Small RNA-degrading nucleases (SDNs), a family of DEDDh 3′ to 5′ exonucleases, showed nuclease activity by removing 3′ end methylated nucleotides directly and promoted the degradation of some methylated miRNAs (Ramachandran and Chen, 2008; Yu et al., 2017; Figure 2C). SDN1 is responsible for the degradation of single-stranded small RNAs, which is 17–24 nt long. In addition, it is sensitive to methylated miRNAs but not to the miRNA/miRNA* duplex and long single-stranded RNAs (Ramachandran and Chen, 2008; Yu et al., 2017; Chen et al., 2018). Atrimmer 2 (ATRM2), a DEDDy type exoribonucleases, is involved in the degradation of unmethylated miRNA/miRNA* duplexes during RISC assembly. The loss function of ATRM2 in hen1 mutant partially rescues the developmental defects and the abundance of numerous miRNAs but decreases the expression of the corresponding miRNA targets (Wang X. et al., 2018; Figure 2C).
AGO protein not only serves as an effector, but also plays critical roles in maintaining the stability of miRNA (Figure 2C). As previously mentioned, several degradative factors, including HESO1, URT1, SDN1, and ATRM2, interact with AGO1 and play an important role in AGO1-bound miRNAs or unmethylated miRNA/miRNA* duplexes. Therefore, AGO1 protein can recruit these regulative factors for the degradation of AGO1-associated miRNAs or unmethylated miRNA/miRNA* duplexes (Ramachandran and Chen, 2008; Chen et al., 2018; Wang X. et al., 2018).
The Arabidopsis genome encodes 10 AGO proteins, and it can be grouped into three clades: AGO1/5/10, AGO2/3/7, and AGO4/6/8/9 (Vaucheret, 2008). Different AGO proteins had different functions on their bound miRNAs. AGO1 binds most miRNAs, and the 5′ end of these miRNAs tends to be uridine (Mi et al., 2008). In ago1 mutant, the abundance of miRNAs at a low level suggests that AGO1 can also stabilize miRNA (Vaucheret et al., 2004). AGO7 and AGO10 are primarily associated with miR390 and miR165/166, respectively. In ago7 mutant, the expression level of miR390 is high, but the cause remains unknown (Li et al., 2017a). Over-accumulation of miR165/166 in ago10 mutant is suppressed when AGO10 is overexpressed (Yu et al., 2017). In Oryza sativas, low abundant of miRNAs and phased secondary sRNAs in AGO18 loss function mutants suggested that AGO18 can also stabilize sRNAs including miRNAs (Das et al., 2020). In addition AGO18 can sequester miR168 and prevent it from repressing its target AGO1 essential for antiviral RNAi (Wu et al., 2015). Likely, AGO18 can sequesters miR528 apart from AGO1 to prevent the formation of an effective RISC, as a result, the function of miR528 targeted L-ascorbate oxidase (AO) is then released, following the initiation of ROS-mediated resistance against Rice stripe Tenuivirus (RSV) infection (Wu et al., 2017). In brief, AGO proteins exhibit different functions in miRNA stability control. On the one hand, it protects miRNAs from degradation. On the other hand, it can recruit some degradative factors facilitating the degradation of AGO-bound miRNAs.
miRNAs are associated with AGO proteins to direct post-transcriptional target gene repression by the manner of miRNA-target complementation. miRNA-target complementation can also trigger destabilization of miRNAs, for example, target mimicry, which is initially discovered in plants. Phosphate Starvation1 (IPS1), a non-coding RNA, whose sequence provides a complementary motif of miR399, due to its loosely complementary around cleavage site, as a result, miR399 is sequestered from its endogenous targets by IPS1 binding. Consequently, miR399 is sequestered from its endogenous targets (Franco-Zorrilla et al., 2007). Based on the abovementioned principles, some researchers designed target mimicry technologies artificially, such as MIM and Short Tandem Target Mimic (STTM), which can be used to reduce endogenous miRNA function (Todesco et al., 2010; Yan et al., 2012). In 2018, these two research groups found that the loss function of an F-box gene Hawaiian Skirt (HWS) leads to the recovery of MIM and STTM-induced developmental and molecular defects (Lang et al., 2018; Mei et al., 2019). In hws mutant, miRNA and its mimicry targets coexisted with AGO1 stably, showing that HWS may touch off unoptimizable RISCs degradation (Mei et al., 2019; Figure 2C). RISC interacts with Clearing 3′–5′ exoribonucleases 1/2 (RICE 1/2), which are two members of DnaQ-like exonucleases. Malfunction of RICE1 and RICE2 resulted in reduced miRNA levels. RICE1 and RICE2 interact with AGO1 and AGO10, thereby degrading uridylated 5′ products of miRNA cleavage products to maintain the normal function of RISC (Ren et al., 2014a; Zhang et al., 2017b; Zuber et al., 2018; Figure 2C). Therefore, the stability of RISC-associated miRNAs is impaired by the excessive accumulation of cleaved products.
The transcription, stability, processing of pri-miRNA, miRNA loading, and stability control require multiple proteins and their cooperation to maintain normal function. In recent years, a large number of factors involved in miRNA biogenesis and stability control have been identified, and the molecular mechanism has been revealed. However, many questions still persist. For example, pri-miRNA processing occurs in D-bodies; however, the formation of D-bodies and how the core members of the processing machinery located at D-bodies remain unknown. DCL1 plays a critical role in processing, but little is known about the post-translational modification of DCL1. Moreover, the regulation of DCL1 activity and HEN1 remains unknown. 3′-end modification, AGOs, and targets also affect the miRNA stability, but the detailed mechanisms remain unknown. Apart from terminal modification, other modifications (N6-methyladenosine, 5-methylcytidine, N1-methyladenosine, pseudouridine, and so on) for pri-miRNAs and/or miRNAs and their biological function remain to be further explored. Further studies should identify and characterize more factors to clarify the biological function and significance of these factors in all of these processes, particularly in miRNA stability control.
LZ and SG drafted the manuscript and revised it. YX, MS, and XJ retrieved and collected the references of miRNA biogenesis and drafted the first picture. SC and ZH retrieved and collected the other references and drafted the second picture. All authors have read and agreed to the publishment of this version.
This work was funded by the Natural Science Foundation of Zhejiang Province (LQY19C150001), the National Natural Science Foundation of China (32001477), and the Student Scientific Research Training Program of Zhejiang Agriculture and Forestry University (2021KX0193, 113-2013200128, and 2020KX0023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Addo-Quaye, C., Snyder, J. A., Park, Y. B., Li, Y.-F., Sunkar, R., and Axtell, M. J. (2009). Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA 15, 2112–2121. doi: 10.1261/rna.1774909
Allen, E., Xie, Z., Gustafson, A. M., and Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Ayubov, M. S., Mirzakhmedov, M. H., Sripathi, V. R., Buriev, Z. T., Ubaydullaeva, K. A., Usmonov, D. E., et al. (2019). Role of MicroRNAs and small RNAs in regulation of developmental processes and agronomic traits in Gossypium species. Genomics 111, 1018–1025. doi: 10.1016/j.ygeno.2018.07.012
Bajczyk, M., Lange, H., Bielewicz, D., Szewc, L., Bhat, S. S., Dolata, J., et al. (2020). SERRATE interacts with the nuclear exosome targeting (NEXT) complex to degrade primary miRNA precursors in Arabidopsis. Nucleic Acids Res. 48, 6839–6854. doi: 10.1093/nar/gkaa373
Baranauskė, S., Mickutė, M., Plotnikova, A., Finke, A., Venclovas, Č, Klimašauskas, S., et al. (2015). Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 43, 2802–2812. doi: 10.1093/nar/gkv102
Baumberger, N., and Baulcombe, D. C. (2005). Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. U. S. A. 102, 11928–11933. doi: 10.1073/pnas.0505461102
Ben Chaabane, S., Liu, R., Chinnusamy, V., Kwon, Y., Park, J.-H., Kim, S. Y., et al. (2013). STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 41, 1984–1997. doi: 10.1093/nar/gks1309
Bhat, S. S., Bielewicz, D., Gulanicz, T., Bodi, Z., Yu, X., Anderson, S. J., et al. (2020). mRNA adenosine methylase (MTA) deposits m(6)A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 117, 21785–21795. doi: 10.1073/pnas.2003733117
Bologna, N. G., Mateos, J. L., Bresso, E. G., and Palatnik, J. F. (2009). A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 28, 3646–3656. doi: 10.1038/emboj.2009.292
Bologna, N. G., Schapire, A. L., Zhai, J., Chorostecki, U., Boisbouvier, J., Meyers, B. C., et al. (2013). Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 23, 1675–1689. doi: 10.1101/gr.153387.112
Bologna, N. G., and Voinnet, O. (2014). The Diversity, Biogenesis, and Activities of Endogenous Silencing Small RNAs in Arabidopsis. Annu. Rev. Plant Biol. 65, 473–503. doi: 10.1146/annurev-arplant-050213-035728
Bortolamiol, D., Pazhouhandeh, M., Marrocco, K., Genschik, P., and Ziegler-Graff, V. (2007). The Polerovirus F Box Protein P0 Targets ARGONAUTE1 to Suppress RNA Silencing. Curr. Biol. 17, 1615–1621. doi: 10.1016/j.cub.2007.07.061
Brant, E. J., and Budak, H. (2018). Plant Small Non-coding RNAs and Their Roles in Biotic Stresses. Front. Plant Sci. 9, 1038–1038. doi: 10.3389/fpls.2018.01038
Budak, H., Kantar, M., Bulut, R., and Akpinar, B. A. (2015). Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 235, 1–13. doi: 10.1016/j.plantsci.2015.02.008
Cai, Q., Liang, C., Wang, S., Hou, Y., Gao, L., Liu, L., et al. (2018). The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat. Commun. 9, 5080–5080. doi: 10.1038/s41467-018-07516-z
Cambiagno, D. A., Giudicatti, A. J., Arce, A. L., Gagliardi, D., Li, L., Yuan, W., et al. (2021). HASTY modulates miRNA biogenesis by linking pri-miRNA transcription and processing. Mol. Plant 14, 426–439. doi: 10.1016/j.molp.2020.12.019
Chadick, J. Z., and Asturias, F. J. (2005). Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30, 264–271. doi: 10.1016/j.tibs.2005.03.001
Chen, J., Liu, L., You, C., Gu, J., Ruan, W., Zhang, L., et al. (2018). Structural and biochemical insights into small RNA 3′ end trimming by Arabidopsis SDN1. Nat. Commun. 9, 3585–3585. doi: 10.1038/s41467-018-05942-7
Chiu, M.-H., Chen, I. H., Baulcombe, D. C., and Tsai, C.-H. (2010). The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 11, 641–649. doi: 10.1111/j.1364-3703.2010.00634.x
Cho, S. K., Chaabane, S. B., Shah, P., Poulsen, C. P., and Yang, S. W. (2014). COP1 E3 ligase protects HYL1 to retain microRNA biogenesis. Nat. Commun. 5:5867. doi: 10.1038/ncomms6867
Choi, S. W., Ryu, M. Y., Viczián, A., Jung, H. J., Kim, G. M., Arce, A. L., et al. (2019). Light triggers the miRNA-biogenetic inconsistency for de-etiolated seedling survivability in Arabidopsis thaliana. Mol. Plant 13, 431–445. doi: 10.1016/j.molp.2019.10.011
Csorba, T., Lózsa, R., Hutvágner, G., and Burgyán, J. (2010). Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 62, 463–472. doi: 10.1111/j.1365-313X.2010.04163.x
Cui, Y., Fang, X., and Qi, Y. (2016). TRANSPORTIN1 Promotes the Association of MicroRNA with ARGONAUTE1 in Arabidopsis. Plant Cell 28, 2576–2585. doi: 10.1105/tpc.16.00384
Das, S., Swetha, C., Pachamuthu, K., Nair, A., and Shivaprasad, P. V. (2020). Loss of function of Oryza sativa Argonaute 18 induces male sterility and reduction in phased small RNAs. Plant Reprod. 33, 59–73. doi: 10.1007/s00497-020-00386-w
Deng, P., Muhammad, S., Cao, M., and Wu, L. (2018). Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol. J. 16, 965–975. doi: 10.1111/pbi.12882
Deng, X., and Cao, X. (2017). Roles of pre-mRNA splicing and polyadenylation in plant development. Curr. Opin. Plant Biol. 35, 45–53. doi: 10.1016/j.pbi.2016.11.003
Derrien, B., Baumberger, N., Schepetilnikov, M., Viotti, C., De Cillia, J., Ziegler-Graff, V., et al. (2012). Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. U. S. A. 109, 15942–15946. doi: 10.1073/pnas.1209487109
Dong, Z., Han, M. H., and Fedoroff, N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U. S. A. 105, 9970–9975. doi: 10.1073/pnas.0803356105
Earley, K., Smith, M., Weber, R., Gregory, B., and Poethig, R. (2010). An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1:15. doi: 10.1186/1758-907x-1-15
Earley, K. W., and Poethig, R. S. (2011). Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J. Biol. Chem. 286, 38184–38189. doi: 10.1074/jbc.M111.290130
Errampalli, D., Patton, D., Castle, L., Mickelson, L., Hansen, K., Schnall, J., et al. (1991). Embryonic Lethals and T-DNA Insertional Mutagenesis in Arabidopsis. Plant Cell 3, 149–157. doi: 10.1105/tpc.3.2.149
Fang, X., Cui, Y., Li, Y., and Qi, Y. (2015a). Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nat. Plants 1:15075. doi: 10.1038/nplants.2015.75
Fang, X., Shi, Y., Lu, X., Chen, Z., and Qi, Y. (2015b). CMA33/XCT Regulates Small RNA Production through Modulating the Transcription of Dicer-Like Genes in Arabidopsis. Mol. Plant 8, 1227–1236. doi: 10.1016/j.molp.2015.03.002
Fang, Y., and Spector, D. L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823. doi: 10.1016/j.cub.2007.04.005
Fei, Q., Xia, R., and Meyers, B. C. (2013). Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25, 2400–2415. doi: 10.1105/tpc.113.114652
Feng, S., Xu, Y., Guo, C., Zheng, J., Zhou, B., Zhang, Y., et al. (2016). Modulation of miR156 to identify traits associated with vegetative phase change in tobacco (Nicotiana tabacum). J. Exp. Bot. 67, 1493–1504. doi: 10.1093/jxb/erv551
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I, Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet 39, 1033–1037. doi: 10.1038/ng2079
Fujioka, Y., Utsumi, M., Ohba, Y., and Watanabe, Y. (2007). Location of a Possible miRNA Processing Site in SmD3/SmB Nuclear Bodies in Arabidopsis. Plant Cell Physiol. 48, 1243–1253. doi: 10.1093/pcp/pcm099
Gao, S., Wang, J., Jiang, N., Zhang, S., Wang, Y., Zhang, J., et al. (2020). Hyponastic Leaves 1 protects pri-miRNAs from nuclear exosome attack. Proc. Natl. Acad. Sci. U. S. A. 117, 17429–17437. doi: 10.1073/pnas.2007203117
Hajheidari, M., Farrona, S., Huettel, B., Koncz, Z., and Koncz, C. (2012). CDKF;1 and CDKD protein kinases regulate phosphorylation of serine residues in the C-terminal domain of Arabidopsis RNA polymerase II. Plant Cell 24, 1626–1642. doi: 10.1105/tpc.112.096834
Hou, X., Chen, W., Hou, Y., Gong, H., Sun, J., Wang, Z., et al. (2021). DEAD-BOX RNA HELICASE 27 regulates microRNA biogenesis, zygote division, and stem cell homeostasis. Plant Cell 33, 66–84. doi: 10.1093/plcell/koaa001
Hunt, A. G. (2014). The Arabidopsis polyadenylation factor subunit CPSF30 as conceptual link between mRNA polyadenylation and cellular signaling. Curr. Opin. Plant Biol. 21, 128–132. doi: 10.1016/j.pbi.2014.07.002
Iki, T., Yoshikawa, M., Meshi, T., and Ishikawa, M. (2012). Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 31, 267–278. doi: 10.1038/emboj.2011.395
Iki, T., Yoshikawa, M., Nishikiori, M., Jaudal, M. C., Matsumoto-Yokoyama, E., Mitsuhara, I., et al. (2010). In Vitro Assembly of Plant RNA-Induced Silencing Complexes Facilitated by Molecular Chaperone HSP90. Mol. Cell 39, 282–291. doi: 10.1016/j.molcel.2010.05.014
Iwasaki, S., Kobayashi, M., Yoda, M., Sakaguchi, Y., Katsuma, S., Suzuki, T., et al. (2010). Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 39, 292–299. doi: 10.1016/j.molcel.2010.05.015
Jia, T., Zhang, B., You, C., Zhang, Y., Zeng, L., Li, S., et al. (2017). The Arabidopsis MOS4-Associated Complex Promotes MicroRNA Biogenesis and Precursor Messenger RNA Splicing. Plant Cell 29, 2626–2643. doi: 10.1105/tpc.17.00370
Karlsson, P., Christie, M. D., Seymour, D. K., Wang, H., Wang, X., Hagmann, J., et al. (2015). KH domain protein RCF3 is a tissue-biased regulator of the plant miRNA biogenesis cofactor HYL1. Proc. Natl Acad. Sci. U. S. A. 112, 14096–14101. doi: 10.1073/pnas.1512865112
Kim, W., Benhamed, M., Servet, C., Latrasse, D., Zhang, W., Delarue, M., et al. (2009). Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Res. 19, 899–909. doi: 10.1038/cr.2009.59
Kim, Y. J., Zheng, B., Yu, Y., Won, S. Y., Mo, B., and Chen, X. (2011). The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30, 814–822. doi: 10.1038/emboj.2011.3
Koiwa, H., Barb, A. W., Xiong, L., Li, F., Mccully, M. G., Lee, B. H., et al. (2002). C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc. Natl. Acad. Sci. U. S. A. 99, 10893–10898. doi: 10.1073/pnas.112276199
Köster, T., Meyer, K., Weinholdt, C., Smith, L. M., Lummer, M., Speth, C., et al. (2014). Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res. 42, 9925–9936. doi: 10.1093/nar/gku716
Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212. doi: 10.1261/rna.2146906
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U. S. A. 101, 12753–12758. doi: 10.1073/pnas.0403115101
Lang, P. L. M., Christie, M. D., Dogan, E. S., Schwab, R., Hagmann, J., Van De Weyer, A.-L., et al. (2018). A Role for the F-Box Protein HAWAIIAN SKIRT in Plant microRNA Function. Plant physiol. 176, 730–741. doi: 10.1104/pp.17.01313
Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J. U., Rätsch, G., et al. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 105, 8795–8800. doi: 10.1073/pnas.0802493105
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-y
Li, S., Castillo-González, C., Yu, B., and Zhang, X. (2017b). The functions of plant small RNAs in development and in stress responses. Plant J. 90, 654–670. doi: 10.1111/tpj.13444
Li, S., Liu, K., Zhang, S., Wang, X., Rogers, K., Ren, G., et al. (2017c). STV1, a ribosomal protein, binds primary microRNA transcripts to promote their interaction with the processing complex in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 1424–1429. doi: 10.1073/pnas.1613069114
Li, J., Wang, Z., Hu, Y., Cao, Y., and Ma, L. (2017a). Polycomb Group Proteins RING1A and RING1B Regulate the Vegetative Phase Transition in Arabidopsis. Front. Plant Sci. 8:867. doi: 10.3389/fpls.2017.00867
Li, J., Yang, Z., Yu, B., Liu, J., and Chen, X. (2005). Methylation Protects miRNAs and siRNAs from a 3′-End Uridylation Activity in Arabidopsis. Curr. Biol. 15, 1501–1507. doi: 10.1016/j.cub.2005.07.029
Li, M., Yu, H., Liu, K., Yang, W., Zhou, B., Gan, L., et al. (2021). Serrate-Associated Protein 1, a splicing-related protein, promotes miRNA biogenesis in Arabidopsis. New Phytol. 232, 1959–1973. doi: 10.1111/nph.17691
Li, Q., Liu, N., Liu, Q., Zheng, X., Lu, L., Gao, W., et al. (2021). DEAD-box helicases modulate dicing body formation in Arabidopsis. Sci. Adv. 7:eabc6266. doi: 10.1126/sciadv.abc6266
Li, S., Li, M., Liu, K., Zhang, H., Zhang, S., Zhang, C., et al. (2020). MAC5, an RNA-binding protein, protects pri-miRNAs from SERRATE-dependent exoribonuclease activities. Proc. Natl. Acad. Sci. U. S. A. 117, 23982–23990. doi: 10.1073/pnas.2008283117
Li, S., Xu, R., Li, A., Liu, K., Gu, L., Li, M., et al. (2018). SMA1, a homolog of the splicing factor Prp28, has a multifaceted role in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 46, 9148–9159. doi: 10.1093/nar/gky591
Liu, C., Axtell, M. J., and Fedoroff, N. V. (2012). The helicase and RNaseIIIa domains of Arabidopsis Dicer-Like1 modulate catalytic parameters during microRNA biogenesis. Plant Physiol. 159, 748–758. doi: 10.1104/pp.112.193508
Ma, Z., Castillo-González, C., Wang, Z., Sun, D., Hu, X., Shen, X., et al. (2018). Arabidopsis Serrate Coordinates Histone Methyltransferases ATXR5/6 and RNA Processing Factor RDR6 to Regulate Transposon Expression. Dev. Cell 45, 769–784.e6. doi: 10.1016/j.devcel.2018.05.023
Manavella, P. A., Hagmann, J., Ott, F., Laubinger, S., Franz, M., Macek, B., et al. (2012). Fast-Forward Genetics Identifies Plant CPL Phosphatases as Regulators of miRNA Processing Factor HYL1. Cell 151, 859–870. doi: 10.1016/j.cell.2012.09.039
Mei, J., Jiang, N., and Ren, G. (2019). The F-box protein HAWAIIAN SKIRT is required for mimicry target-induced microRNA degradation in Arabidopsis. J. Integr. Plant Biol. 61, 1121–1127. doi: 10.1111/jipb.12761
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Montgomery, T. A., Howell, M. D., Cuperus, J. T., Li, D., Hansen, J. E., Alexander, A. L., et al. (2008). Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141. doi: 10.1016/j.cell.2008.02.033
Niu, D., Lii, Y. E., Chellappan, P., Lei, L., Peralta, K., Jiang, C., et al. (2016). miRNA863-3p sequentially targets negative immune regulator ARLPKs and positive regulator SERRATE upon bacterial infection. Nat. Commun. 7:11324. doi: 10.1038/ncomms11324
Park, M. Y., Wu, G., Gonzalez-Sulser, A., Vaucheret, H., and Poethig, R. S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 102, 3691–3696. doi: 10.1073/pnas.0405570102
Park, S. J., Choi, S. W., Kim, G. M., Møller, C., Pai, H.-S., and Yang, S. W. (2021). Light-stabilized FHA2 suppresses miRNA biogenesis through interactions with DCL1 and HYL1. Mol. Plant 14, 647–663. doi: 10.1016/j.molp.2021.01.020
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L., and Poethig, R. S. (2004). SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Gene Dev. 18, 2368–2379. doi: 10.1101/gad.1231804
Raghuram, B., Sheikh, A. H., Rustagi, Y., and Sinha, A. K. (2015). MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 282, 521–536. doi: 10.1111/febs.13159
Rajagopalan, R., Vaucheret, H., Trejo, J., and Bartel, D. P. (2006). A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Gene Dev. 20, 3407–3425. doi: 10.1101/gad.1476406
Ramachandran, V., and Chen, X. (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321, 1490–1492. doi: 10.1126/science.1163728
Ramanathan, A., Robb, G. B., and Chan, S. H. (2016). mRNA capping: biological functions and applications. Nucleic Acids Res. 44, 7511–7526. doi: 10.1093/nar/gkw551
Ren, G., Chen, X., and Yu, B. (2012a). Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr.Biol. 22, 695–700. doi: 10.1016/j.cub.2012.02.052
Ren, G., Xie, M., Dou, Y., Zhang, S., Zhang, C., and Yu, B. (2012b). Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 109, 12817–12821. doi: 10.1073/pnas.1204915109
Ren, G., Chen, X., and Yu, B. (2014a). Small RNAs meet their targets: when methylation defends miRNAs from uridylation. RNA Biol. 11, 1099–1104. doi: 10.4161/rna.36243
Ren, G., Xie, M., Zhang, S., Vinovskis, C., Chen, X., and Yu, B. (2014b). Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. U. S. A. 111, 6365–6370. doi: 10.1073/pnas.1405083111
Rogers, K., and Chen, X. (2013). Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell 25, 2383–2399. doi: 10.1105/tpc.113.113159
Sement, F., Ferrier, E., Zuber, H., Merret, R., Alioua, M., Deragon, J., et al. (2013). Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res. 41, 7115–7127. doi: 10.1093/nar/gkt465
Shriram, V., Kumar, V., Devarumath, R. M., Khare, T. S., and Wani, S. H. (2016). MicroRNAs As Potential Targets for Abiotic Stress Tolerance in Plants. Front. Plant Sci. 7:817. doi: 10.3389/fpls.2016.00817
Song, L., Axtell, M. J., and Fedoroff, N. V. (2010). RNA Secondary Structural Determinants of miRNA Precursor Processing in Arabidopsis. Curr. Biol. 20, 37–41. doi: 10.1016/j.cub.2009.10.076
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and Their Regulatory Roles in Plant–Environment Interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Speth, C., Szabo, E. X., Martinho, C., Collani, S., Zur Oven-Krockhaus, S., Richter, S., et al. (2018). Arabidopsis RNA processing factor SERRATE regulates the transcription of intronless genes. Elife 7:e37078. doi: 10.7554/eLife.37078
Speth, C., Willing, E.-M., Rausch, S., Schneeberger, K., and Laubinger, S. (2013). RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 76, 433–445. doi: 10.1111/tpj.12308
Su, C., Li, Z., Cheng, J., Li, L., Zhong, S., Liu, L., et al. (2017). The Protein Phosphatase 4 and SMEK1 Complex Dephosphorylates HYL1 to Promote miRNA Biogenesis by Antagonizing the MAPK Cascade in Arabidopsis. Dev. Cell 41, 527–539.e5. doi: 10.1016/j.devcel.2017.05.008
Sun, Z., Guo, T., Liu, Y., Liu, Q., and Fang, Y. (2015). The Roles of Arabidopsis CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs. PLoS Genet. 11:e1005598. doi: 10.1371/journal.pgen.1005598
Tagami, Y., Motose, H., and Watanabe, Y. (2009). A dominant mutation in DCL1 suppresses the hyl1 mutant phenotype by promoting the processing of miRNA. RNA 15, 450–458. doi: 10.1261/rna.1297109
Takeda, A., Iwasaki, S., Watanabe, T., Utsumi, M., and Watanabe, Y. (2008). The mechanism selecting the guide strand from small RNA duplexes is different among argonaute proteins. Plant Cell Physiol. 49, 493–500. doi: 10.1093/pcp/pcn043
Todesco, M., Rubio-Somoza, I., Paz-Ares, J., and Weigel, D. (2010). A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 6:e1001031. doi: 10.1371/journal.pgen.1001031
Tomassi, A. H., Re, D. A., Romani, F., Cambiagno, D. A., Gonzalo, L., Moreno, J. E., et al. (2020). The Intrinsically Disordered Protein CARP9 Bridges HYL1 to AGO1 in the Nucleus to Promote MicroRNA Activity. Plant Physiol. 184, 316–329. doi: 10.1104/pp.20.00258
Tsai, H.-L., Li, Y.-H., Hsieh, W.-P., Lin, M.-C., Ahn, J. H., and Wu, S.-H. (2014). HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell 26, 2858–2872. doi: 10.1105/tpc.114.126722
Tu, B., Liu, L., Xu, C., Zhai, J., Li, S., Lopez, M. A., et al. (2015). Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 11:e1005119. doi: 10.1371/journal.pgen.1005119
Vaucheret, H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13, 350–358. doi: 10.1016/j.tplants.2008.04.007
Vaucheret, H., Mallory, A. C., and Bartel, D. P. (2006). AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22, 129–136. doi: 10.1016/j.molcel.2006.03.011
Vaucheret, H., Vazquez, F., Crété, P., and Bartel, D. P. (2004). The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Gene Dev. 18, 1187–1197. doi: 10.1101/gad.1201404
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Wang, J., Chen, S., Jiang, N., Li, N., Wang, X., Li, Z., et al. (2019a). Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana. Nucleic Acids Res. 47, 7886–7900. doi: 10.1093/nar/gkz526
Wang, J., Mei, J., and Ren, G. (2019b). Plant microRNAs: biogenesis, Homeostasis, and Degradation. Front. Plant Sci. 10:360. doi: 10.3389/fpls.2019.00360
Wang, L., Song, X., Gu, L., Li, X., Cao, S., Chu, C., et al. (2013). NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell 25, 715–727. doi: 10.1105/tpc.112.105882
Wang, W., Ye, R., Xin, Y., Fang, X., Li, C., Shi, H., et al. (2011). An importin β protein negatively regulates MicroRNA activity in Arabidopsis. Plant Cell 23, 3565–3576. doi: 10.1105/tpc.111.091058
Wang, X., Wang, Y., Dou, Y., Chen, L., Wang, J., Jiang, N., et al. (2018). Degradation of unmethylated miRNA/miRNA*s by a DEDDy-type 3′ to 5′ exoribonuclease Atrimmer 2 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 115, E6659–E6667. doi: 10.1073/pnas.1721917115
Wang, Z., Ma, Z., Castillo-González, C., Sun, D., Li, Y., Yu, B., et al. (2018). SWI2/SNF2 ATPase CHR2 remodels pri-miRNAs via Serrate to impede miRNA production. Nature 557, 516–521. doi: 10.1038/s41586-018-0135-x
Wang, X., Zhang, S., Dou, Y., Zhang, C., Chen, X., Yu, B., et al. (2015). Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet. 11:e1005091. doi: 10.1371/journal.pgen.1005091
Wu, G., Park, M. Y., Conway, S. R., Wang, J. W., Weigel, D., and Poethig, R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. doi: 10.1016/j.cell.2009.06.031
Wu, J., Yang, R., Yang, Z., Yao, S., Zhao, S., Wang, Y., et al. (2017). ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 3:16203. doi: 10.1038/nplants.2016.203
Wu, J., Yang, Z., Wang, Y., Zheng, L., Ye, R., Ji, Y., et al. (2015). Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. Elife 4:e05733. doi: 10.7554/eLife.05733
Xie, D., Chen, M., Niu, J., Wang, L., Li, Y., Fang, X., et al. (2021). Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 23, 32–39. doi: 10.1038/s41556-020-00606-5
Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S. A., and Carrington, J. C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138, 2145–2154. doi: 10.1104/pp.105.062943
Xie, Z., Kasschau, K. D., and Carrington, J. C. (2003). Negative Feedback Regulation of Dicer-Like1 in Arabidopsis by microRNA-Guided mRNA Degradation. Curr. Biol. 13, 784–789. doi: 10.1016/S0960-9822(03)00281-1
Xu, M., Hu, T., Smith, M. R., and Poethig, R. S. (2016). Epigenetic Regulation of Vegetative Phase Change in Arabidopsis. Plant Cell 28, 28–41. doi: 10.1105/tpc.15.00854
Yan, J., Gu, Y., Jia, X., Kang, W., Pan, S., Tang, X., et al. (2012). Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24, 415–427. doi: 10.1105/tpc.111.094144
Yan, J., Wang, P., Wang, B., Hsu, C.-C., Tang, K., Zhang, H., et al. (2017). The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 13:e1006753. doi: 10.1371/journal.pgen.1006753
Yang, L., Liu, Z., Lu, F., Dong, A., and Huang, H. (2006). SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 47, 841–850. doi: 10.1111/j.1365-313X.2006.02835.x
Yang, Z., Ebright, Y. W., Yu, B., and Chen, X. (2006). HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675. doi: 10.1093/nar/gkj474
Yang, S. W., Chen, H.-Y., Yang, J., Machida, S., Chua, N.-H., and Yuan, Y. A. (2010). Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18, 594–605. doi: 10.1016/j.str.2010.02.006
Yang, X., Ren, W., Zhao, Q., Zhang, P., Wu, F., and He, Y. (2014). Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res. 42, 12224–12236. doi: 10.1093/nar/gku907
Yoshikawa, M., Peragine, A., Park, M. Y., and Poethig, R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. doi: 10.1101/gad.1352605
Yu, B., Bi, L., Zheng, B., Ji, L., Chevalier, D., Agarwal, M., et al. (2008). The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 105, 10073–10078. doi: 10.1073/pnas.0804218105
Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. doi: 10.1126/science.1107130
Yu, Y., Ji, L. J., Le, B. H., Zhai, J. X., Chen, J. Y., Luscher, E., et al. (2017). ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 15:e2001272. doi: 10.1371/journal.pbio.2001272
Zhang, B., You, C., Zhang, Y., Zeng, L., Hu, J., Zhao, M., et al. (2020). Linking key steps of microRNA biogenesis by TREX-2 and the nuclear pore complex in Arabidopsis. Nat. Plants 6, 957–969. doi: 10.1038/s41477-020-0726-z
Zhang, C., Ng, D. W.-K., Lu, J., and Chen, Z. J. (2012). Roles of target site location and sequence complementarity in trans-acting siRNA formation in Arabidopsis. Plant J. 69, 217–226. doi: 10.1111/j.1365-313X.2011.04783.x
Zhang, S., Liu, Y., and Yu, B. (2014). PRL1, an RNA-binding protein, positively regulates the accumulation of miRNAs and siRNAs in Arabidopsis. PLoS Genet. 10:e1004841. doi: 10.1371/journal.pgen.1004841
Zhang, S., Liu, Y., and Yu, B. (2015). New insights into pri-miRNA processing and accumulation in plants. Wiley Interdiscip. Rev. RNA 6, 533–545. doi: 10.1002/wrna.1292
Zhang, S., Xie, M., Ren, G., and Yu, B. (2013). CDC5, a DNA binding protein, positively regulates posttranscriptional processing and/or transcription of primary microRNA transcripts. Proc. Natl. Acad. Sci. U. S. A. 110, 17588–17593. doi: 10.1073/pnas.1310644110
Zhang, Z., Guo, X., Ge, C., Ma, Z., Jiang, M., Li, T., et al. (2017a). KETCH1 imports HYL1 to nucleus for miRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 4011–4016. doi: 10.1073/pnas.1619755114
Zhang, Z., Hu, F., Sung, M. W., Shu, C., Castillo-González, C., Koiwa, H., et al. (2017b). RISC-interacting clearing 3′- 5′ exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana. Elife 6:e24466. doi: 10.7554/eLife.24466
Zhao, Y., Yu, Y., Zhai, J., Ramachandran, V., Dinh, T. T., Meyers, B. C., et al. (2012). The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 22, 689–694. doi: 10.1016/j.cub.2012.02.051
Zhu, H., Zhou, Y., Castillo-González, C., Lu, A., Ge, C., Zhao, Y.-T., et al. (2013). Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat. Struct. Mol. Biol. 20, 1106–1115. doi: 10.1038/nsmb.2646
Keywords: miRNA biogenesis, processing, transcription, stability control, Argonaute
Citation: Zhang L, Xiang Y, Chen S, Shi M, Jiang X, He Z and Gao S (2022) Mechanisms of MicroRNA Biogenesis and Stability Control in Plants. Front. Plant Sci. 13:844149. doi: 10.3389/fpls.2022.844149
Received: 27 December 2021; Accepted: 27 January 2022;
Published: 08 March 2022.
Edited by:
Ming Luo, South China Botanical Garden (CAS), ChinaReviewed by:
Zhong-Hui Zhang, South China Normal University, ChinaCopyright © 2022 Zhang, Xiang, Chen, Shi, Jiang, He and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuai Gao, MjAyMTAwMzNAemFmdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.