- 1Department of Plant Pathology, College of Plant Protection, Nanjing Agricultural University, Nanjing, China
- 2Laboratory of Bio-interactions and Crop Health, Nanjing Agricultural University, Nanjing, China
- 3Institute of Industrial Crops, Shanxi Agricultural University, Taiyuan, China
- 4Department of Biotechnology, COMSATS University Islamabad Abbottabad Campus, Abbottabad, Pakistan
- 5College of Life Sciences, Zaozhuang University, Zaozhuang, China
Ascorbate peroxidases (APXs) maintain cellular reactive oxygen species (ROS) homeostasis through their peroxidase activity. Here, we report that OsAPX1 also promotes ROS production such that a delicate cellular ROS homeostasis is achieved temporally after Magnaporthe oryzae infection. OsAPX1 specifically induces ROS production through increasing respiratory burst oxidase homologs (OsRBOHs) expression and can be inhibited by DPI, a ROS inhibitor. The time-course experiment data show that the simultaneous induction of OsAPX1 and OsRBOHs leads to ROS accumulation at an early stage; whereas a more durable expression of OsAPX1 leads to ROS scavenging at a later stage. By the temporal switching between ROS inducer and eliminator, OsAPX1 triggers an instant ROS burst upon M. oryzae infection and then a timely elimination of ROS toxicity. We find that OsAPX1 is under the control of the miR172a-OsIDS1 regulatory module. OsAPX1 also affects salicylic acid (SA) synthesis and signaling, which contribute to blast resistance. In conclusion, we show that OsAPX1 is a key factor that connects the upstream gene silencing and transcription regulatory routes with the downstream phytohormone and redox pathway, which provides an insight into the sophisticated regulatory network of rice innate immunity.
Introduction
In plants, reactive oxygen species (ROS) production is considered to be an important defensive response to biotic and abiotic stresses (Jones and Dangl, 2006; Saxena et al., 2016). When plants are subject to biotic stress, elevated ROS production enhances plant resistance to pathogens. For example, after rice stripe virus (RSV) infection, rice L-ascorbate oxidase (AO) expression was induced, which boosts planta ROS accumulation and enhances resistance against RSV (Wu et al., 2017). The accumulation of ROS in rice also plays an important role in resistance against bacterial and fungal pathogens. For example, in rice over-expressing Triosephosphate Isomerase (OsTPI), ROS accumulated to a significantly higher level than the wild type, dramatically increasing rice resistance against Xanthomonas oryzae (Liu Y. et al., 2018). In rice over-expressing OsWRKY67 or mitogen-activated protein kinase 15 knockout mutants Osmpk15, resistance against M. oryzae and X. oryzae is enhanced, which is also associated with ROS accumulation (Vo et al., 2017; Hong et al., 2019).
ROS is produced in different plant subcellular compartments (e.g., chloroplasts, mitochondria, peroxisomes, and the apoplastic space) under different stressful conditions (Moller et al., 2007; Wrzaczek et al., 2013; Bose et al., 2014). Pathogen infection-triggered ROS burst happens in apoplast space by the membrane-localized NADPH-dependent oxidase system (also known as respiratory burst oxidase homologs (RBOHs)), which is in contrast to the abiotic stress-triggered ROS that is produced through organelles such as chloroplast, mitochondria, and peroxisome (Apel and Hirt, 2004). In Arabidopsis, rbohd, and rbohf mutants caused susceptibility to bacterial and oomycete infection due to low ROS levels in these mutants (Torres et al., 2002). OsRBOHb knockdown plants had lower content of ROS and were more susceptible to rice blast (Nagano et al., 2016).
Although ROS accumulation enhances plant resistance to pathogens, over-production of ROS can lead to detrimental effects such as membrane lipid peroxidation, protein denaturation, carbohydrate oxidation, pigment breakdown, and DNA damage (Moller et al., 2007). To maintain ROS homeostasis, plants have evolved multiple mechanisms to fine-tune ROS homeostasis so that normal biological processes and tolerance/resistance to stresses are delicately balanced. In green plants, ascorbate peroxidases (APXs) are the most significant components of the ROS detoxifying system (Shigeoka et al., 2002; Guan et al., 2015), which play an essential role in controlling intracellular hydrogen peroxide (H2O2; a ROS species) level (Munné-Bosch et al., 2013; Wang and Chu, 2020). APXs comprise a multigene family that encodes enzymes detoxifying H2O2. APXs utilize ascorbate as a specific electron donor to convert H2O2 to water, stabilizing cellular ROS to an ordinary level and maintaining regular metallic processes. For example, AtAPX1 and AtAPX2 (cytosolic) protect Arabidopsis against diverse abiotic stresses, such as high light, heat, wounding, and drought stress (Panchuk et al., 2005; Mullineaux et al., 2006; Rossel et al., 2006; Hu et al., 2011). In rice, OsAPX1 was found to be closely linked to chilling and high-temperature stresses (Saruyama and Tanida, 1995). It was reported that high temperature and subsequent chilling could affect OsAPX1 transcription and activity, indicating a role for OsAPX1 in the protection of rice seedlings against chilling injury (Sato et al., 2001). In agreement, OsAPX1-overexpressing rice also showed enhanced tolerance to chilling at the booting stage (Sato et al., 2011). Besides temperature, the OsAPX1 transcripts appeared to be up-regulated upon various abiotic stimuli such as wounding, salicylic acid (SA), ethylene, abscisic acid (ABA), H2O2, copper sulfate, and protein phosphatase (PP) inhibitors but not by jasmonic acid (JA), indicating its involvement in plant tolerance against a broad spectrum of abiotic stresses and stimuli (Agrawal et al., 2003).
Intriguingly, as a ROS scavenger that reduces excessive cellular H2O2, the induced expression of APXs in plants responding to biotic stresses has been widely documented. For example, sunflower APX1 was dramatically induced by sunflower chlorotic mottle virus (SuCMoV) infection (Rodriguez et al., 2012). In hot pepper, CaAPX1 was induced upon the Tobacco mosaic virus (TMV) and bacterial pathogen infection (Yoo et al., 2013). In rice, OsAPX1 and OsAPX2 transcripts were up-regulated by blast pathogen (M. oryzae) attack (Agrawal et al., 2003; Lin et al., 2018). OsAPX2 was induced in rice by brown planthopper (BPH) (Wei et al., 2009). OsAPX7 and OsAPX1 proteins were significantly accumulated in the resistant but not the susceptible rice line when infected by Rhizoctonia solani (Lee et al., 2006; Ma et al., 2019). Another proteomic study found that the OsAPX protein level was constantly induced by M. oryzae infection (Narula et al., 2019). Taken together, these observations imply that the induced expression of APXs upon a variety of biotic stresses is not a rare case, although the underlying mechanism is not yet clear.
In this study, we demonstrated that OsAPX1 plays a double-faced role in both promoting and eliminating ROS accumulation temporally against M. oryzae infection. At the early stage of infection, the expression of OsAPX1 and the membrane-localized ROS producer-OsRBOHs are induced, which collectively leads to ROS accumulation. At the later stage of M. oryzae infection, OsRBOHs expression decline but OsAPX1 expression remains active, which leads to ROS elimination. This delicate switch turns OsAPX1 from a ROS inducer at the early stage of M. oryzae infection into a ROS scavenger at the later stage, which ensures rice deploys a strong ROS burst to confine and eliminate M. oryzae right after infection but removes excessive ROS in time before damage to rice cells and tissue occurs. Our results reveal a new facet of OsAPX1, which advances our current understanding regarding the function and mechanism of APX. Our discovery is of great significance for understanding the crosstalk between plant responses against biotic and abiotic stresses.
Materials and Methods
Pathogen Inoculation
M. oryzae Guy11, Js153, and the eGFP-tagged Zhong1 (a gift from Dr. W-M Wang) strains were used for rice infection. M. oryzae strains were first inoculated on CM medium (complete medium) that grew at 28°C under a 12/12 (light/dark) condition. Spores were collected 2 weeks after inoculation, which were resuspended to 1 × 105 spores ml–1. Spores were spray-inoculated on three-leaf-stage rice seedlings. Disease symptoms were examined 5 days later. Genomic DNA was extracted from inoculated leaves for fungal accumulation examination (MoPot2; Supplementary Table 1). Disease resistance was also examined by using detached leaves from four-leaf-stage rice. 10 μl spores (1 × 105 spores ml–1) were added at two spots of each leaf, and kept in a culture dish with a wet filter. After incubation in a growth chamber at 28°C for 24 h in dark, the leaves were kept under a 12/12 h (light/dark) light rhythm for 6 days till disease symptoms were examined. For leaf sheath inoculation, leaf sheaths were prepared from four-leaf-stage rice. 10 μl spores (1 × 105 spores ml–1) were inoculated. Fungal hyphae development was observed under a microscope at 24 and 48 h post inoculation (hpi), respectively.
RNA Blotting
Total RNA extraction was performed as described previously (Zhang et al., 2018). In brief, the inoculated plants were used for RNA extraction at 0, 24, and 48 hpi. Total RNA was extracted using TRIzol reagent (Invitrogen, United States) following the manufacturer’s protocol. RNA was resolved on a 14% denaturing 8 M urea-PAGE gel and then transferred and UV cross-linked onto a Hybond N+ membrane (GE Healthcare Life Science, Beijing, China) using UV light. miRNA probes were end-labeled with [γ-32P] ATP by T4 polynucleotide kinase (New England Biolabs, Beijing, China). Expression levels were quantified using ImageJ as instructed.
Generating OsAPX1 Transgenic Plants
For generating OsAPX1 overexpression transgenic plants, the full-length CDS was cloned into a pCAM1300 vector driven by a CaMV35S promoter. For the OsAPX1 silencing mutant, highly specific target regions from the OsAPX1 CDS were cloned to the pYLCRISPR/Cas9Pubi vector. The construct was transferred to Agrobacterium strain EHA105, which was used for transgenic rice production. For verifying the OsAPX1 silencing mutant, genomic DNA was used to examine the OsAPX1 genomic DNA sequence. For verifying OsAPX1 over-expression plants, total protein samples from the transgenic plant were used for western blot; and total RNA was used to examine the transcript level of OsAPX1 by qRT-PCR.
qRT–PCR
1 μg total RNA was reverse transcribed into cDNA by using PrimeScript RT reagent Kit (Takara, Japan). The qRT–PCR was performed in 15 μl of reaction mixture consisting 1.5 μl 10 × SYBR Green (Invitrogen, United States), 1.5 μl PCR buffer, 0.3 μl 10 mM dNTPs (Takara, Japan), 0.3 μl Taq, 0.3 μl ROX DYE2 (Vazyme, China), 1.5 μl 2 mM each primer, and 2 μl appropriate diluted cDNA. The conditions for real-time RT-PCR were as follow: 94°C for 3 min, then 40 cycles at 94°C for 30 s and 58°C for 30 s followed by 72°C 35 s for PCR amplification. Transcript levels of each gene were measured by the Applied Biosystems 7500 (Applied Biosystems, United States) according to the manufacturer’s instructions. The data were normalized to the amplification of the rice 18sRNA gene. Real-time PCR primer sequences are available in Supplementary Table 1.
Measurement of H2O2 Accumulation
Leaf tissues and leaf sheaths were dipped into 50 ml solution containing 50 mg Diaminobenzidine (DAB), 25 μl Tween-20 and 2.5 ml 200 mM Na2HPO4 and vacuum infiltrated for 30 min followed by staining in dark at room temperature (25°C) overnight (10 h). The tissues were decolorized in 1:1:1 (v/v/v) acetic acid-ethanol-glycerol solution for 15–20 min at 90–95°C and visualized afterward (ThordalChristensen et al., 1997). Decolored leaves or leaf sheath tissues were examined for H2O2 accumulation around the inoculating loci by using a microscope.
Measurement of Salicylic Acid Concentrations
The free SA concentration in transgenic rice was measured as described (Liu et al., 2014). The rice tissues were homogenized in liquid nitrogen and then suspended in 90% (v/v) methanol. As an internal standard, 100 mg 3-hydroxy benzoic acid in 100% methanol was added to each sample. The SA solution was filtered and separated on a C18 analytical column using HPLC and detected using fluorescence (excitation at 305 nm, emission at 405 nm; Waters). The HPLC was programmed for isocratic conditions with a flow rate of 0.5 ml/min. The concentration of SA was quantified by area integration of the HPLC peaks.
Western Blot Analysis
Leaf tissues were snap-frozen in liquid nitrogen and ground into fine powder. The samples were added with 2 × SDS loading buffer, which was boiled at 100°C for 10 min. The supernatant, after 10,000 g centrifugation, was separated by 12% SDS-PAGE gels at 100 V for 1.5 h. The proteins were transferred to PVDF membrane (Bio-RAD, United States), blocked by using 5% dry milk for 30 min, which was followed by Flag antibody (Abmart, China) incubation for 2 h. The membranes were washed by using TBST buffer three times (5 min), followed by 2nd antibody incubation (Abmart, China) for 2 h. The protein signal was detected by chemoluminescence (Tanon, China).
Enzyme Activity
The APX enzyme activity was examined by using a kit (Beijing Solarbio Science and Technology Co, Beijing, China) measuring the oxidation rate of ascorbic acid within 2 min with a spectrophotometer at 290 nm.
Results
OsAPX1 Expression Is Responsive to Magnaporthe oryzae Infection
We challenged 4-week-old rice [the Japonica cultivar Nipponbare (NIP)] with the M. oryzae strain Guy11 (compatible strain) and Js153 (incompatible strain) (Supplementary Figure 1A). Compared with the mock, OsAPX1 expression was significantly induced at 24, 48, and 72 hpi after infection with both strains (Supplementary Figure 1B). At 72 hpi, OsAPX1 expression increased more than 3 folds. To examine the contribution of OsAPX1 in rice immunity against the blast disease, we first investigated whether an elevated OsAPX1 expression would lead to an altered rice blast resistance. Both OsAPX1 silencing (cas9-osapx1 #22 and #30; Supplementary Figure 2A) and over-expression (OsAPX1-OE #38 and #39; Supplementary Figures 2B,C) transgenic rice were constructed. Genotyping results indicated that cas9-osapx1 is a homozygous insertion mutant (Supplementary Figure 2A) with a T/C insertion to the 40th nucleotide of the second exon, leading to the complete silencing of OsAPX1 due to frameshifting. Cas9-osapx1 lines carried a 1-bp insertion causing protein truncation (Supplementary Figure 2A). Compared to NIP and cas9-osapx1 #22 rice, the OsAPX1-OE #38 rice were significantly higher in growth length, had longer roots (Supplementary Figure 2D), larger seed size (Supplementary Figure 2E), and greater seed weight (per 1,000 seeds) (Supplementary Figure 2F). In contrast, the cas9-osapx1 #22 rice was shorter and produced smaller seeds both in size and weight. However, neither measurable growth or developmental abnormality nor spontaneous lesions were observable on the leaf surface of the transgenic rice (Supplementary Figure 2D). There was no discernable difference in ROS accumulation between wild type and transgenic rice under normal growth conditions either (Supplementary Figure 2G), indicating that the rice innate immunity is not automatically activated without pathogen infection.
OsAPX1 Over-Expressing Plants Are More Resistant to Magnaporthe oryzae Infection
Detached leaves from transgenic plants were challenged by punching inoculation with Guy11 spores. The OsAPX1-OE #38 leaves developed less severe disease symptoms, manifested by significantly smaller necrosis size and lesser discoloration as observed on transgenic leaves when compared to leaves from control plants. Meanwhile, the lesion size on the cas9-osapx1 #22 leaves was significantly larger than that on the NIP leaves (Figure 1A). The propagation of M. oryzae on the infected leaves was quantified by qRT-PCR using primers specific to MoPot2, a M. oryzae housekeeping gene. In agreement with the resistant phenotype, less hyphae propagation was detected on leaves from the OsAPX1-OE #38 than the control, whereas the accumulation of hyphae increased on cas9-osapx1 #22 (Figure 1B), indicating that over-expression of OsAPX1 led to enhanced resistance to M. oryzae infection.
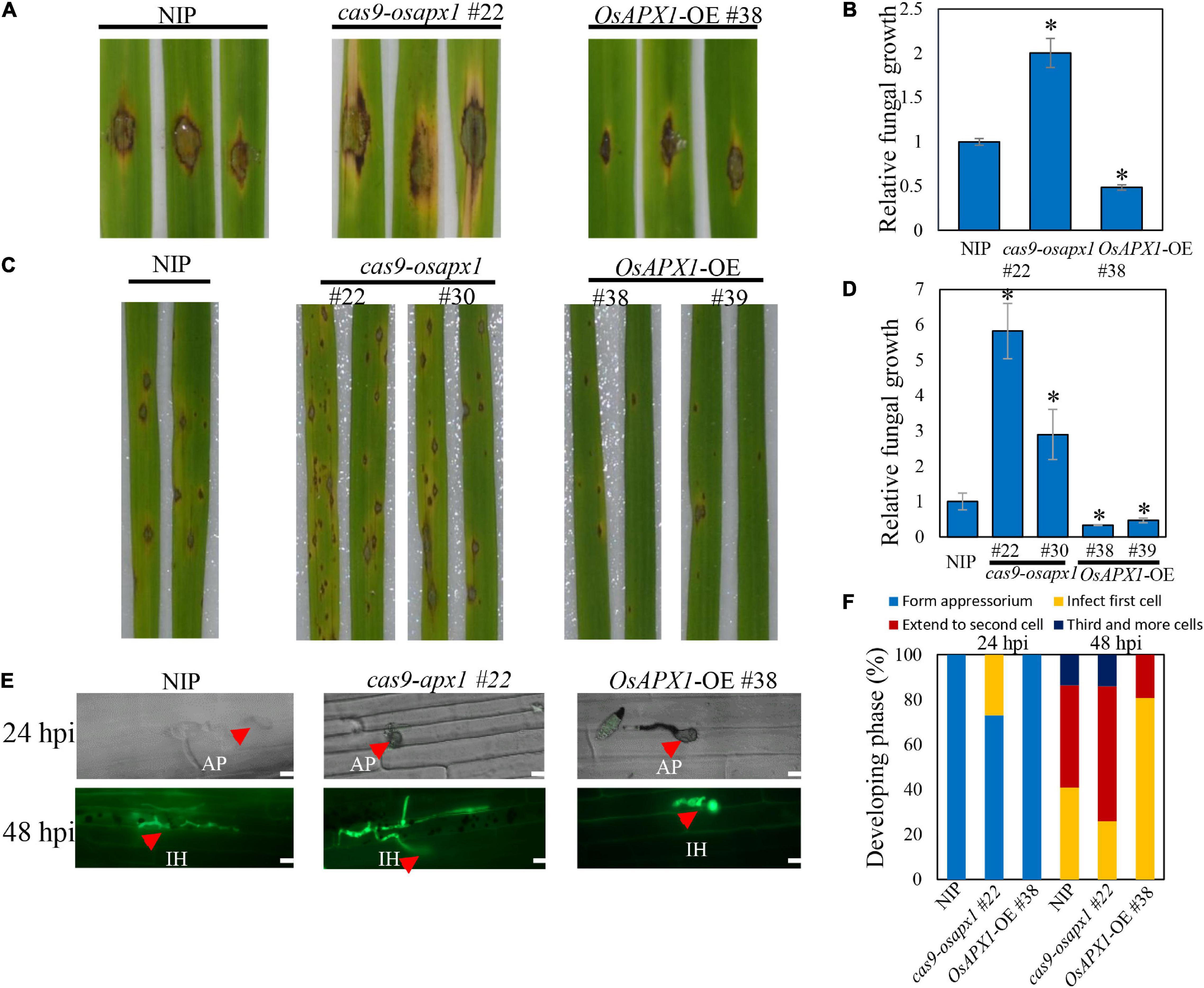
Figure 1. OsAPX1 overexpression enhances rice resistance to M. oryzae Guy11 infection. (A) Blast disease assay on the indicated lines. The phenotype of four-leaf stage leaves from wild type and the indicated transgenic lines cas9-osapx1 and OsAPX1-OE plants inoculated by the punch method with spores’ suspension of M. oryzae strain Guy11. (B) Relative fungal biomass is determined by examining the expression level of M. oryzae Pot2 gene against OsUbiquitin DNA level. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences according to the Student’s t-test (p < 0.05). (C) Rice pathogenicity assays. Leaf phenotypes were observed at 144 h post inoculation (hpi). (D) Relative fungal biomass is determined by examining the expression level of MoPot2 gene against OsUbiquitin DNA level at 144 hpi. Values are means of three replications. The error bars indicate ± SD. The asterisks indicate significant differences according to the Student’s t-test (p < 0.05). (E) Representative images of sheath cells from the indicated lines infected by eGFP-tagged blast isolate zhong1. Bar = 20 μm. (F) Quantification analysis on the progress of fungal infection at 24 and 48 hpi. All of the experiments were repeated three times with similar results.
The association between OsAPX1 expression level and disease resistance was further confirmed by Guy11 spore-spray inoculation on rice leaves. From 96 hpi on, both NIP and OsAPX1-OE or cas9-osapx1 rice developed typical blast disease symptoms such as scattered lesions on the leaf surface, cell death in the center of some lesions, and chlorosis on some leaves. Specifically, more lesions developed on the cas9-osapx1 leaves, accompanied by severer chlorosis, than NIP rice. In contrast, both lesion numbers and chlorosis were significantly milder on the OsAPX1-OE (#38, #39) leaves, when compared to both cas9-osapx1 (#22, #30) and NIP rice (Figure 1C). When hyphae growth was quantified, we detected more fungal hyphae on cas9-osapx1 (#22, #30) rice than both NIP and OsAPX1-OE (#38, #39) rice, whereas OsAPX1-OE (#38, #39) had the least hyphae development among them (Figure 1D). Our results indicate that OsAPX1 over-expression led to enhanced resistance against blast fungus infection. When rice sheath epidermal tissue was challenged with an eGFP-labeled M. oryzae strain (Zhong-1), we were able to observe and assess infection progress by quantifying the rate of appressorium formation, hyphae development, and invasive hyphae spreading (Figure 1E). At 24 hpi, 27% appressorium developed visible hyphae in cas9-osapx1 #22 sheath epidermal cells, and 0% hyphae development was recorded on either OsAPX1-OE #38 or NIP. At 48 hpi, with hyphae fully developed within the infected cells in all plants, about 60% hyphae were observed to spread to the adjacent cas9-osapx1 #22 sheath epidermal cells, and about 18% to the 3rd cells; on NIP rice, only about 45% of fungi spread to the adjacent cells and about 13% to the 3rd cells; on OsAPX1-OE #38 rice, only about 20% fungi spread to adjacent cells but none of them spread to a 3rd cell (Figure 1F). Our results indicate that OsAPX1 expression level is positively related to rice ability resisting fungal hyphae development in infected tissue.
OsAPX1 Is Induced by Magnaporthe oryzae Infection at Transcript Level via a miR172a-OsIDS1 Regulatory Module
It was reported that the OsAPX1 promoter is bound by transcription factor INDETERMINATE SPIKELET1 (OsIDS1), which inhibits the expression of OsAPX1 (Cheng et al., 2021). OsIDS1 is a target of miR172a, which silences OsIDS1 by reverse-complementary sequence match (Cheng et al., 2021). To check whether the induced expression of OsAPX1 is a result of variated expression of miR172a, we examined miR172a expression by a reverse complementary DNA probe through northern blot. We found that the expression of miR172a was induced significantly at 24 and 48 hpi (Figure 2A). The expression level of OsIDS1 decreased at both 24 and 48 hpi, while the expression of OsAPX1 increased at both 24 and 48 hpi, correspondingly (Figure 2B). If miR172a expression was inhibited (miR172a-KO), the expression of OsAPX1 did not significantly change along with M. oryzae infection further (Figure 2B).
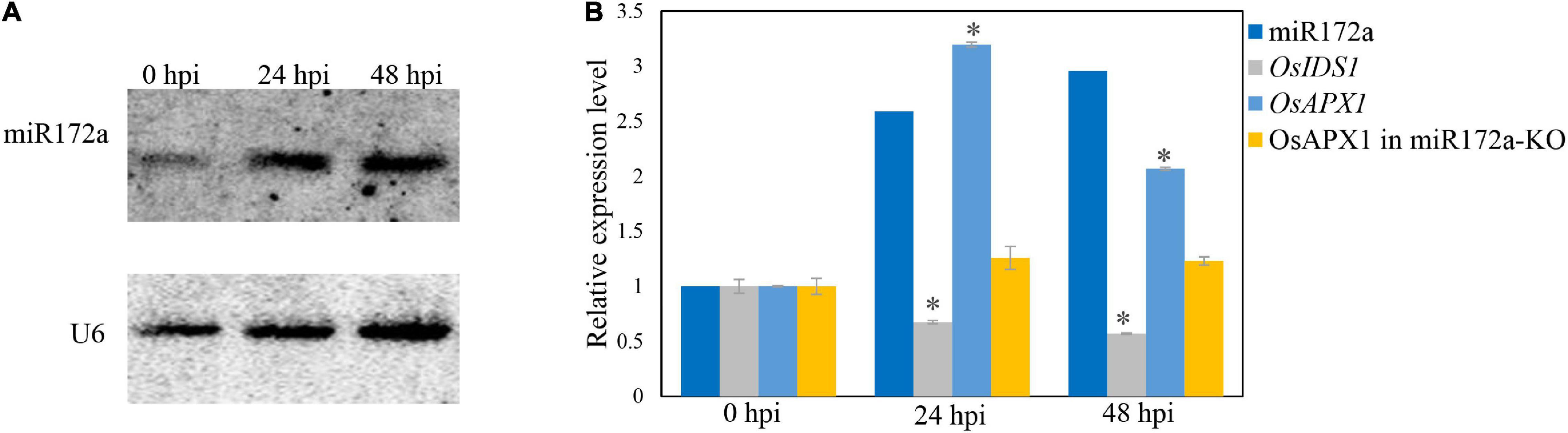
Figure 2. M. oryzae induces OsAPX1 transcription via a miR172a-OsIDS1 regulatory module. (A) RNA-blotting detection of mR172a at the indicated time points upon M. oryzae infection. U6 was used as a loading control. (B) miR172a relative abundance (from A) and qRT-PCR analysis of OsIDS1 and OsAPX1 expression (in both NIP and miR172a-KO rice) at the indicated time points upon M. oryzae infection. qRT-PCR values are means of three replications. Error bars indicate ± SD. Student’s t-test was used to determine the significance of differences between 0 hpi and the indicated time points. Asterisks indicate significant differences (p < 0.05). The qRT-PCR experiments were repeated three times with similar results.
We inoculated Guy11 spores on the detached leaves of both wild type and miR172a transgenic rice (a gift from Cheng) (Cheng et al., 2021). The lesions on the miR172a-OE rice leaves were significantly smaller than the WT rice. In contrast, miR172a-KO leaves developed significantly larger lesions than WT rice. Cell death and chlorosis were also very obvious on miR172a-KO leaves (Figure 3A). Guy11 developed more fungal mass on the miR172a-KO rice, while much lesser on the miR172a-OE leaves than the WT rice (Figure 3B). When the rice was spray-inoculated with Guy11 spores, there was almost no lesions development on miR172a-OE rice leaves, whereas miR172a-KO rice exhibited much more lesions than both the WT and miR172a-OE rice (Figure 3C). Quantification of hyphae in leaves also indicated that Guy11 was spread much more in tissues of miR172a-KO rice than the WT and miR172a-OE rice (Figure 3D). Compared with mock, the OsIDS1 and OsAPX1 gene expression levels were not significantly changed after M. oryzae infection in the miR172a-KO plant, but in the miR172a-OE plant, the expression of OsIDS1 reduced significantly and OsAPX1 was induced significantly (Figures 3E,F). In summary, our results indicate that OsAPX1 is subject to a miR172a-OsIDS1 regulatory module upon M. oryzae infection.
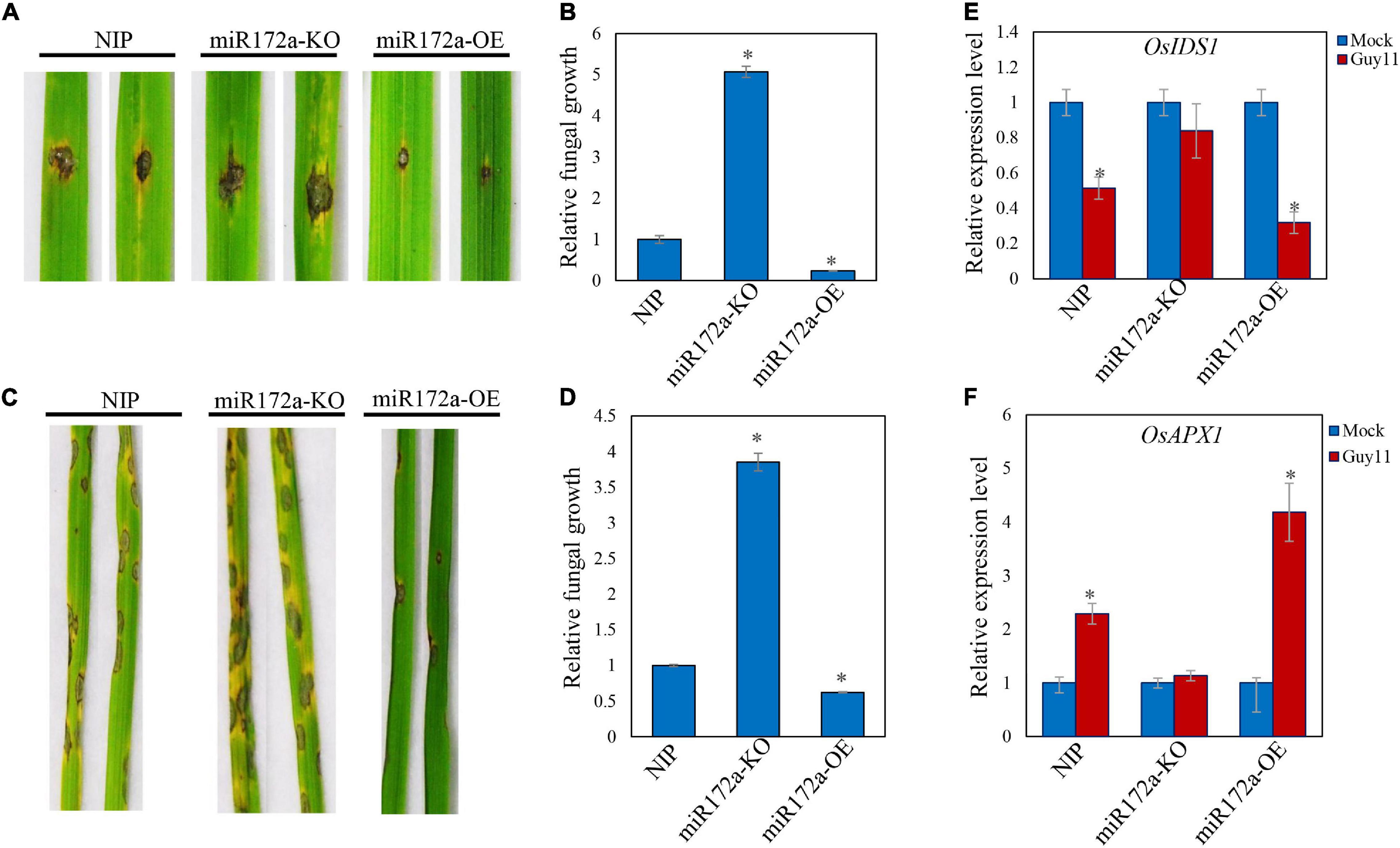
Figure 3. miR172a enhances rice resistance to M. oryzae. (A) Punch inoculation of M. oryzae strain Guy11 on four-leaf-stage leaves from wild type (NIP) and the indicated transgenic lines miR172a-KO and miR172a-OE. (B) Relative fungal biomass is determined by examining the expression level of MoPot2 gene against OsUbiquitin DNA level. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences according to the Student’s t-test (p < 0.05). (C) Rice pathogenicity assays. Leaf phenotypes were observed at 144 hpi. (D) Relative fungal biomass is determined by examining the expression level of MoPot2 gene against OsUbiquitin DNA level. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences according to the Student’s t-test (p < 0.05). All of the experiments were repeated three times with similar results. (E) OsIDS1 gene expression level in NIP, miR172a-KO, and miR172a-OE plants after M. oryzae infection. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences according to the Student’s t-test (p < 0.05). (F) OsAPX1 gene expression level in NIP, miR172a-KO, and miR172a-OE plants after M. oryzae infection. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences according to the Student’s t-test (p < 0.05).
OsAPX1 Temporally Fine-Tunes Reactive Oxygen Species
ROS production is a hallmark of plant early defense responses, which represents a successful pathogen recognition and the activation of plant defense response (Torres, 2010; Nie et al., 2017; Qi et al., 2017; Zhang et al., 2018). We checked the ROS accumulation in both transgenic and control plants. We found that ROS accumulation in OsAPX1-OE #38 and NIP plants could be observed around infection sites at 12 and 36 hpi, then reduced at 60 and 72 hpi. OsAPX1-OE #38 rice obviously accumulated more ROS at the penetrating sites than the NIP rice, whereas it was the least observed in cas9-osapx1 #22 rice (Figure 4B). The observation that Guy11 infection led to more ROS accumulation in OsAPX1-OE #38 rice than in WT rice challenges our current knowledge that OsAPX1 eliminates, instead of, induces ROS accumulation. However, numerous groups have reported that elevated APX expression is associated with ROS accumulation (details in discussion). We, therefore, hypothesized that there must be a mechanism that ROS accumulation and scavenging coalesce.
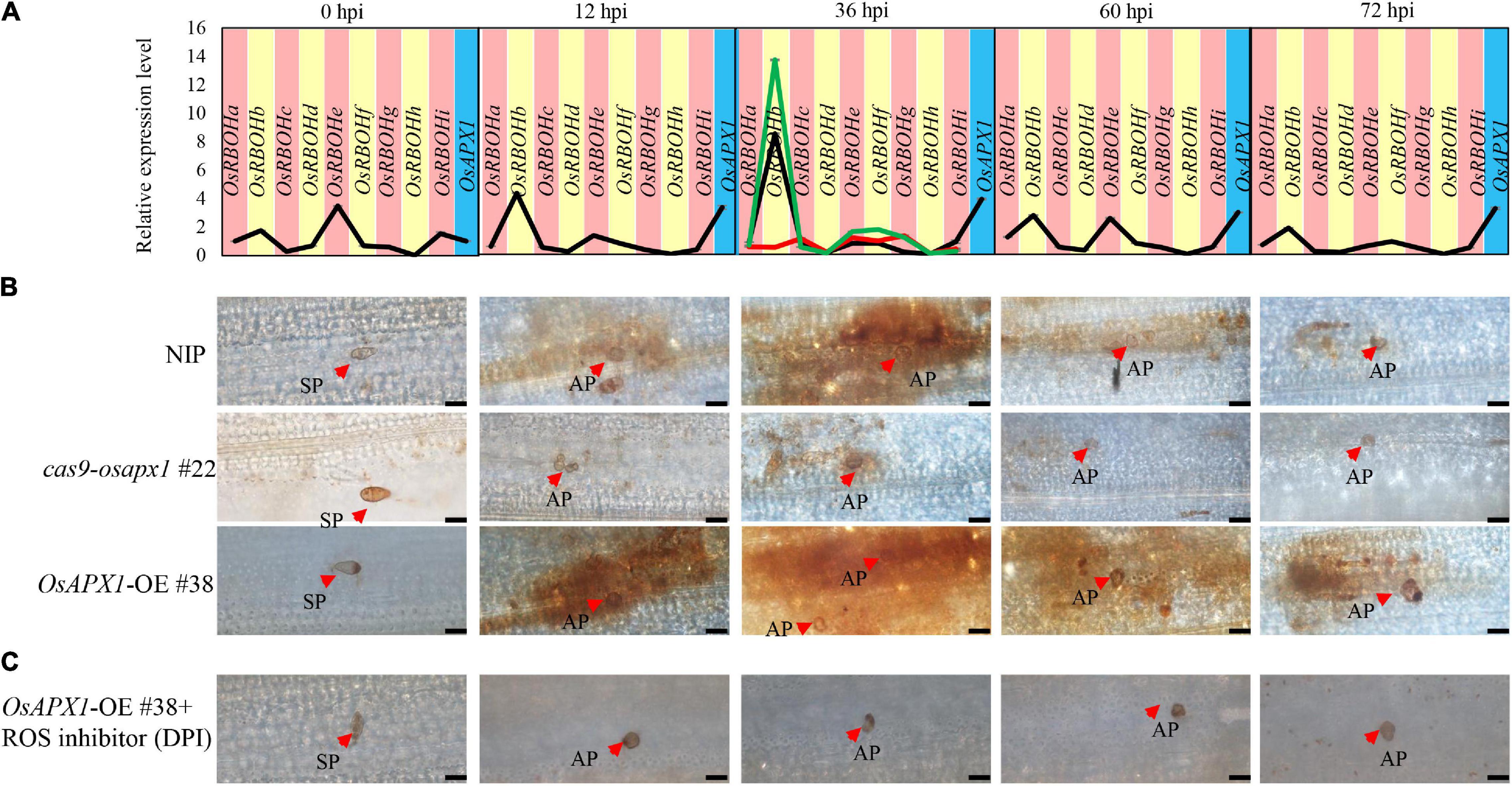
Figure 4. OsAPX1 temporally fine-tunes ROS. (A) OsRBOH genes expression level at the indicated time points by M. oryzae infected in NIP. Red-line in 36 hpi indicates OsRBOHs expression level in cas9-osapx1 rice at the time points infected by M. oryzae. Green-line in 36 hpi indicates OsRBOHs expression level in OsAPX1-OE rice at the time pointed infected by M. oryzae. Values are means of three replications. Error bars indicate ± SD. (B) ROS accumulation at the indicated time pointed by M. oryzae infected in the transgenic plant. Red arrows indicate infection loci. Bar = 20 μm. (C) The pictures show ROS accumulation at the infection sites of the OsAPX1-OE rice leaves at the indicated time pointed by DPI treatment. Red arrows indicate infection loci. Bar = 20 μm. All of the experiments were repeated three times with similar results.
Diphenyleneiodonium (DPI) is a flavoenzyme inhibitor that prevents the activation of NADPH oxidases necessary for ROS generation in plants (Cross and Jones, 1986; Li et al., 2020a). We found that ROS accumulation around the infection loci was dramatically reduced in OsAPX1-OE #38 rice after DPI treatment (Figure 4C), suggesting that these ROS may be produced by OsRBOHs.
OsRBOHs are crucial components in ROS accumulation upon rice M. oryzae infection (Dangol et al., 2019). OsRBOHs reduce cellular oxidation potential by catalyzing the transfer of electrons from NADPH to oxygen (O2), which generates superoxide radicals (O∙2–). A previous study reported that the lack of sAPX and tAPX drastically decreased the expression of H2O2 responsive genes in Arabidopsis under photooxidative stress (Maruta et al., 2010). Under flood stress, AtSUS1, AtPEPC, AtLDH gene expression increased in Arabidopsis plant that overexpression sponge gourd APX (LcAPX) (Chiang et al., 2017). Therefore, we hypothesized that OsAPX1 affects OsRBOHs gene expression under M. oryzae infection. To verify our hypothesis, we checked the OsRBOHs expression level in transgenic plants at 36 hpi, the time point when ROS accumulation peaked. Compared to NIP, OsRBOHs (especially OsRBOHb) expression was significantly induced in OsAPX1-OE #38 rice (green line) and significantly reduced in cas9-osapx1 #22 rice (red line) (Figure 4A). These results suggest that OsAPX1 may influence ROS production by affecting OsRBOH genes expression when rice is infected by M. oryzae.
The generation and removal of ROS are two parallel activities that maintain cellular ROS homeostasis (Li et al., 2016). We found that the expression of OsAPX1 (ROS removal) was constantly induced after M. oryzae infection, but ROS accumulation was temporal. Therefore, we speculated that ROS generation gene OsRBOHs expression is temporally induced. Indeed, the time-course experiment showed that OsRBOHs gene expression level was temporal during M. oryzae infection. At 0 hpi, OsRBOHb, OsRBOHe, and OsRBOHi expression were detectable, among which OsRBOHe is the major contributor of expressed OsRBOHs. Upon M. oryzae infection, OsRBOHb became a major contributor, which was induced at 12 hpi and peaked at 36 hpi. From 60 hpi on, expression of OsRBOH genes declined, especially OsRBOHb. It should be noticed that at the same infection period the expression of OsAPX1 was also dramatically induced (Figure 4A). The result indicates that OsAPX1 and OsRBOHb were simultaneously induced at the early stage after M. oryzae infection; at the later stage, the expression of OsAPX1 was constantly induced; however, OsRBOHb was declined. We also checked APX enzyme activity in NIP and transgenic plants. We found that APX enzyme activity was elevated both in the early and late-stage after M. oryzae infection (Supplementary Figure 3). In summary, these results showed that ROS generation activity masked ROS scavenging activity at the early stage after M. oryzae infection, which led to ROS accumulation; however, ROS scavenging activity prevailed at the later stage, which eventually led to ROS removal.
Salicylic Acid but Not Jasmonic Acid Signaling Pathway Activates in OsAPX1-OE Rice
ROS are crucial signal molecules that can activate phytohormone signaling pathways such as the salicylic acid and jasmonic acid pathway (Torres, 2010; Xu et al., 2015), which play important roles in plant innate immunity (Liu et al., 2016; Hong et al., 2019). To further investigate the mechanism underlying OsAPX1-mediated disease resistance against rice blast, we examined the expression of several key SA and JA signaling pathway genes. OsPR1a and OsPR1b are important SA signaling pathway reporter genes, which are induced by many pathogen infections. Our results revealed that in OsAPX1-OE rice the expression of OsPR1a was around 3-fold and 9-fold higher than that in NIP, whereas OsPR1b expression was around 2-fold and 4-fold higher than that in NIP (Figure 5A), indicating that SA signaling pathway was activated when OsAPX1 is over-expressed. Genes involved in SA synthesis and signal transduction were also checked. As shown in Figure 5A, genes associated or directly participated in SA synthesis such as OsPAD4, OsICS1, and OsPAL1 were significantly induced in the OsAPX1-OE rice, among which OsPAD4 showed an around the 3- and 1.5-fold increase. Expression of genes involved in SA signaling transduction, such as OsNPR1 and OsWRKY45, was also increased in OsAPX1-OE rice (Figure 5A). The expressions of these genes were reduced in the cas9-osapx1 rice (Figure 5A). Taken together, our results indicate that components involved in SA synthesis, signaling transduction are induced upon OsAPX1 over-expression, which may contribute to enhanced disease resistance.
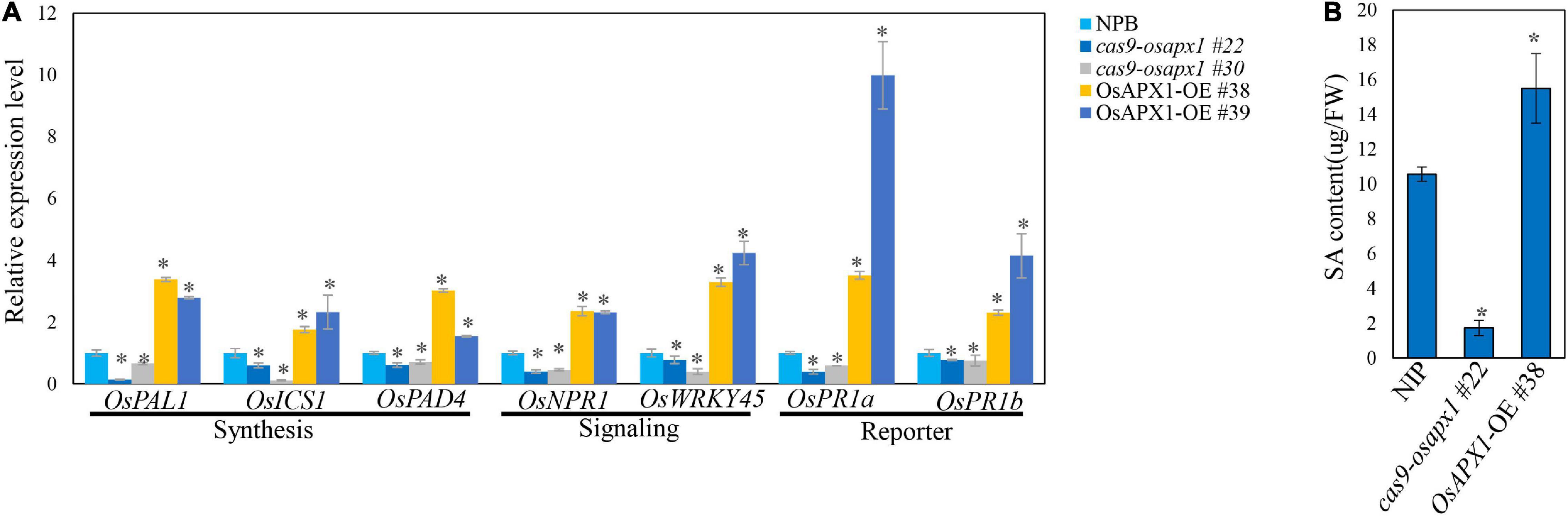
Figure 5. Overexpression of OsAPX1 enhanced SA signaling pathway gene expression and SA content. (A) The expression level of SA signaling pathway relative genes are compared between OsAPX1 transgenic rice and wild type rice by qRT-PCR. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences according to the Student’s t-test (p < 0.05). (B) The content of free SA in OsAPX1-OE, cas9-osapx1, and WT rice is measured by using HPLC. The OsAPX1-OE rice appears to accumulate a higher level of free SA than the WT and cas9-osapx1 rice. Measurement is repeated three times, error bars indicate ± SD, and asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). All of the experiments were repeated three times with similar results.
To confirm the relationship between OsAPX1 and the enhanced expression of SA signaling pathway components, we further examined in vivo SA concentration in WT and transgenic plants by HPLC analysis. Free SA concentration was about 10 μg in each gram of fresh tissue (μg/FW) in NIP and was more than 15 μg/FW in OsAPX1-OE rice, which is about 1.5 times as much as in the NIP rice. In contrast, SA content in cas9-osapx1 rice was about 1.72 μg/FW, which is much lower than in the NIP rice (Figure 5B).
We also examined the expression of OsPDF1.2, the reporter gene of the JA signaling pathway. The expression of OsPDF1.2 was not changed measurably in OsAPX1-OE or cas9-osapx1 rice (Supplementary Figure 4), indicating that the JA signaling pathway was not affected by OsAPX1. In agreement, the expression of most key components participating in JA biosynthesis (e.g., OsLOX5, and OsAOS2) and signal transduction (e.g., OsJAZ8, OsCOL1b, OsMYC2) were not significantly changed between control and both OsAPX1-OE and cas9-osapx1 rice (Supplementary Figure 4), indicating that JA signaling pathway is not a major contributor to the OsAPX1-mediated defense against M. oryzae infection.
OsAPX1 May Play a Role in Broad-Spectrum Resistance
We also tested whether OsAPX1 responds to another rice disease. R. solani causes rice sheath blight, which is one of the devastating rice diseases. We challenged both transgenic and WT rice with R. solani. Cas9-osapx1 #22 rice exhibited significantly larger lesions and server chlorosis than the NIP rice, whereas OsAPX1-OE #38 rice had smaller lesions and weaker chlorosis (Figure 6A), indicating OsAPX1 may also play a role in resistance to sheath blight. Lesion size quantification clearly demonstrated a positive relationship between OsAPX1 expression level and disease symptoms (Figure 6B). When the rice was challenged by stem-inoculated R. solani, OsAPX1-OE #38 rice exhibited significantly smaller lesions than that from the NIP rice at 15 dpi, whereas cas9-osapx1 #22 leaf sheath had significantly larger lesions than the NIP rice (Figure 6C). Lesion size quantification confirmed our judgment that the expression level of OsAPX1 is positively related to resistance to sheath blight disease (Figure 6D).
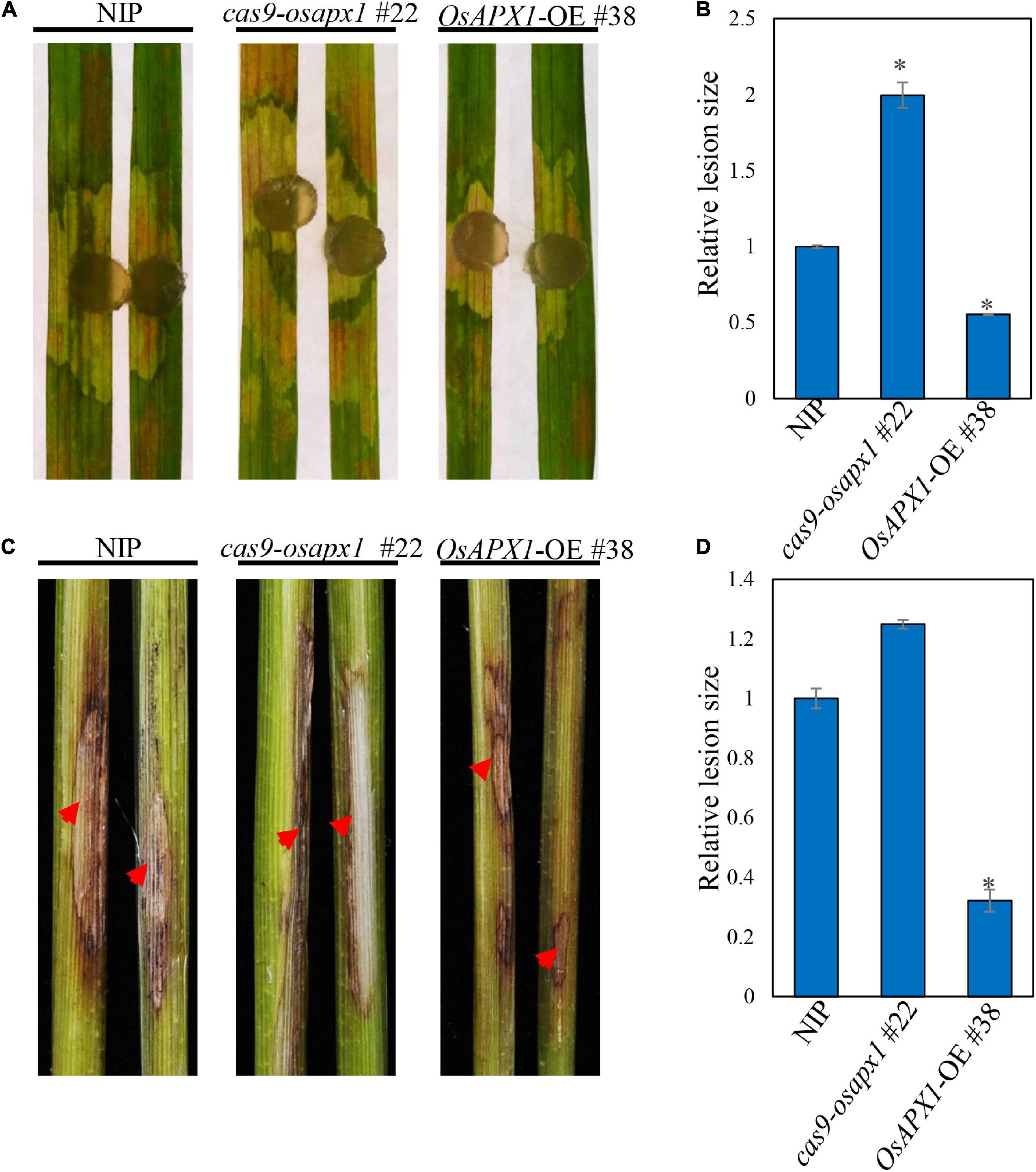
Figure 6. OsAPX1 positively regulates resistance to Rhizoctonia solani. (A) The disease phenotypes of the leaves of R. solani infected NIP, cas9-osapx1, and OsAPX1-OE at 48 hpi using a mycelium plug. (B) The relative lesion sizes were measured using ImageJ. Values are means of two replications. Error bars indicate ± SD. Asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). (C) The phenotypes of the stem of R. solani infected NIP, cas9-osapx1, and OsAPX1-OE at 15 days post inoculation (dpi). (D) The relative lesion sizes were measured using ImageJ. Values are means of two replications. Error bars indicate ± SD. Asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). Red arrows indicate infection loci. All of the experiments were repeated three times with similar results.
Discussion
It has been recognized that APXs can reduce excessive intracellular ROS elicited by different sorts of abiotic stresses, leading to an enhanced tolerance in different plant species (Ribeiro et al., 2012; Zhang et al., 2013; Guan et al., 2015; Yan et al., 2016). According to our previous knowledge, APXs’ function is associated with their peroxidase activity. In this study, we demonstrate that OsAPX1, together with OsRBOHb, maintains cellular ROS status by temporally balancing ROS generation and elimination, thereby enhancing rice resistance to rice blast. Our study reveals a delicate manipulation of cellular ROS homeostasis, which ensures rice battles off fungal pathogens at an early stage while protecting itself from excessive oxidative stress. Our discovery may lead to an in-depth understanding of APXs responding to both biotic and abiotic stresses.
OsAPX1 Regulates Reactive Oxygen Species Production by Affecting OsRBOHs Expression
Reactive oxygen species (ROS) burst is an important defense response upon pathogen infection, in which APX is supposed to play a ROS scavenger role. It appeared intriguing to us at the very beginning that OsAPX1-OE rice accumulates significantly more H2O2 than the WT plants while cas9-osapx1 showed the least H2O2 (Figure 4B). We later found that other groups also reported similar observations. For example, it was reported that SuCMoV-infected sunflower leaves demonstrated simultaneously increased expression of APX1 and elevated H2O2 accumulation (Rodriguez et al., 2012). In an NPR1-silencing tomato line that is highly resistant to Botrytis cinerea, both APX activity and H2O2 accumulation increased (Li et al., 2020b). Applying polyamine to apricot fruits not only enhanced resistance to black spot disease but also induced transcriptional expression of PaAPX and H2O2 accumulation (Li et al., 2019b). R. solani-infected beans exhibited both boosted APX activity and H2O2 accumulation during its infection (Keshavarz-Tohid et al., 2016). All these reports recorded simultaneous increases in both APX expression and ROS accumulation in multiple species.
When we carefully investigated the origin of ROS, we found that most of the ROS accumulation was demolished in the OsAPX1-OE plant by DPI treatment (Figure 4C). DPI is a flavoenzyme inhibitor that specifically prevents the activation of NADPH oxidases required for ROS generation (Cross and Jones, 1986). RBOHs are membrane-localized NADPH-dependent oxidases that catalyze the production of superoxide from oxygen and NADPH (Kaur et al., 2014; Li et al., 2019a). Increased OsRBOHs expression level very likely led to elevated ROS production. Taken together, we are confident to conclude that OsAPX1 contributes to cellular ROS homeostasis after M. oryzae infection. Our results may also explain the observation made on other APXs. For example, N. benthamiana leaves over-expressing sugarcane APX (ScAPX6) accumulated significantly more H2O2 and was more resistant to Fusarium solani var. coeruleum infection (Liu F. et al., 2018).
Our study observed that OsRBOHs genes expression levels increased in the OsAPX1-OE plant. The phenomenon is similar to other studies. For example, overexpressing ScAPX6 in N. benthamiana leaves, result in NtPR1a, NtPR3, and NtEFE26 gene expression levels were increased after Fusarium solani var. coeruleum infection (Liu F. et al., 2018). Transcriptome analysis showed that lignin biosynthesis relative genes induced in Rheum austral APX overexpression line under salt stress (Shafi et al., 2015). However, the mechanism of gene expression level increase needs further study.
OsAPX1 Temporally Regulates Reactive Oxygen Species Production
We found both OsAPX1 and OsRBOHs were involved in ROS homeostasis management. Both expressions were induced almost simultaneously, but the induced expression of OsRBOHs terminated shortly after M. oryzae infection while the expression of OsAPX1 was sustained. Previous studies reported that plant APX activity increases and more ROS accumulation were found under biotic stress (Rodriguez et al., 2012; Liu F. et al., 2018). It indicates that ROS generation and elimination co-exist at the same time, and ROS accumulation was temporal during the M. oryzae infection (Figure 4B). From 0 to 36 hpi, both OsRBOHs (especially OsRBOHb) and OsAPX1 are induced, during this period, the peroxidase activity is masked by the oxidase activity such that ROS homeostasis leans toward production rather than degradation. After 36 hpi, the expression of OsRBOHb gradually begins to decline but OsAPX1’s peroxidase activity remains strong, which favors ROS degradation over its production. The overlapping expression between OsRBOHs (especially OsRBOHb) and OsAPX1 corresponds very well with cellular ROS accumulation patterns (Figure 4A), manifesting a significant role played by OsRBOHb. This is further supported by the failed ROS accumulation in cas9-osapx1 rice. Although some OsRBOH members expressed normally, or even slightly increased, reduced OsRBOHb expression played a dominant role and led to failed overall ROS accumulation.
Therefore, we concluded that it is the unique expression patterns of OsRBOH and OsAPX1 that is governing the ROS rhythmic generation and elimination upon M. oryzae infection. At the early stage of M. oryzae infection, OsAPX1 and OsRBOHb co-expressed. OsAPX1’s peroxidase activity was masked by OsRBOH activity, which leads to ROS accumulation; at the later stage, OsRBOHb expression declined while OsAPX1 expression remained constantly activated, which led to ROS elimination. This hypothesis was further supported by the DPI treatment that specifically inhibits ROS. DPI treatment functionally mimics the earlier termination of RBOH activity. In OsAPX1-OE rice, DPI treatment destroys RBOH activity such that OsAPX1 scavenger activity was unmasked, demonstrated by the absence of ROS accumulation around the infection loci (Figure 4C). In contrast, when the OsRBOH activity was not offset by the DPI treatment, ROS accumulation peaked at 36 hpi before it dropped.
Our study showed that OsAPX1 can induce OsRBOHs expression after M. oryzae infection. The dynamic OsRBOHb expression was also reported, in which OsRBOHb was induced at 24 hpi but then declined at 48 hpi upon M. oryzae infection (Yang et al., 2017). It was reported that OsRBOHs expression could be regulated by other factors. For example, OsEIL1 binds OsRBOHb promoter and regulates its expression (Yang et al., 2017). OsHXK1 can regulate OsRBOHs gene expression through an unknown mechanism (Zheng et al., 2019); auxin can induce OsRBOHs expression (Zhang et al., 2019). Therefore, we speculate that the OsRBOHs expression pattern in responding to M. oryzae infection is intricately regulated, in which OsAPX1 plays an unstated role.
OsAPX1 Expression Is Sophisticatedly Regulated
We previously showed that OsAPX1 proteins were induced by M. oryzae infection as early as 24 hpi (Lin et al., 2018). In this study, we further confirmed that OsAPX1 was induced at the early infecting stage at transcription level as well (Figure 2B and Supplementary Figure 1B), suggesting that OsAPX1 participates in blast resistance from a very early stage. The induced expression of OsAPX1 and other rice APXs has been reported by multiple groups. For example, Agrawal and colleagues revealed that OsAPX1/2 transcripts were up-regulated by M. oryzae infection (Agrawal et al., 2003). OsAPX1/2 were differentially expressed by some defense-related phytohormone treatments (Durner and Klessig, 1995; Chandrashekar and Umesha, 2014). OsAPX1 protein is induced by R. solani in resistant cultivars but not in susceptible cultivars (Ma et al., 2019). OsAPX7 protein was significantly induced by a necrotrophic pathogen, R. solani (Lee et al., 2006). Similarly, OsAPX8 transcription was dramatically induced when rice was infected by Xanthomonas (Jiang et al., 2016). However, the mechanism underlying these inductions is not clear to date.
Our study showed that OsAPX1 expression upon M. oryzae infection is regulated by the miR172a/OsIDS1 module. miR172a has been reported to play a role in immunity, such as in tomato (Solanum lycopersicum), in which over-expressing miR172a and miR172b enhance resistance to Phytophthora infestans by inhibiting the expression of AP2/ERF (Luan et al., 2018). Immunity-related genes expressed significantly higher in miR172b-OE Arabidopsis than in wild type after flg22 treatment (Zou et al., 2020). The above evidence supports the notion that miR172 is a positive immune regulator in multiple plants. In this study, we found that miR172a was significantly induced after M. oryzae infection (Figure 2A), accompanied by reduced expression of OsIDS1, its target gene. Most interestingly, the fluctuated expression of miR172a and OsIDS1 correspond very well with OsAPX1, their downstream target gene (Figure 2B). Our results are consistent with an observation made by Cheng and her colleagues that miR172 was induced by salt stress and contributes to salt stress through miR172a/OsIDS1 module (Cheng et al., 2021). Whether rice resistance/tolerance converges at miR172a/IDS1/OsAPX1 module is worth further exploration.
OsAPX1 Affects Defense Responses Through Salicylic Acid Signaling Pathway
Other than the direct elimination of pathogens, ROS also serves as a signal molecule that is involved in multiple innate immunity signaling pathways, including SA and JA. SA and JA signaling pathways are an important way to respond to biotic stress (Van Camp et al., 1998; Quan et al., 2008; Torres, 2010; Xu et al., 2015; Tian et al., 2016). In the current study, we were able to show that the JA signaling pathway was not changed measurably (Supplementary Figure 4). Instead, several key components involved in both SA synthesis and signaling were upregulated in OsAPX1-OE plants but were downregulated in cas9-osapx1 plants. OsPAD4, OsPAL1, and OsICS1 are genes involved in SA synthesis, among which OsPAD4 is involved in SA regulation whereas OsPAL1 and OsICS1 are the two key components directly involved in SA synthesis. It was proposed that OsICS1 is an important factor that contributes to most of the induced SA production upon biotic challenge (Wildermuth et al., 2001; Loake and Grant, 2007). Consistently, in OsAPX1-OE plants we observed a remarkable stronger induction of OsICS1 than OsPAL1, indicating OsAPX1 contributes to the risen SA synthesis primarily through OsICS1 induction (Figure 5A). Key components involved in SA signaling such as OsNPR1 and OsWRKY45 were also differentially expressed in OsAPX1-OE plants. In OsAPX1-OE plants, both OsNPR1 and OsWRKY45 branches were up-regulated. In cas9-osapx1 plants, SA signaling pathway components behaved to an opposite profile as in the OsAPX1-OE plants, indicating a reliable connection between OsAPX1 and SA signaling pathway. The induction of OsWRKY45 was slightly stronger than the expression of OsNPR1 (Figure 5A), suggesting that OsWRKY45 is favored by OsAPX1.
We observed that OsAPX1-OE leaves had much higher SA content, whereas cas9-osapx1 leaves had much lower SA contents, compared to the WT plants (Figure 5B). Rice accumulates high basal levels of SA (8–37 μg g–1 fresh weight) that do not change significantly upon pathogen attack (Silverman et al., 1995), which leads to the misconception that the SA signaling pathway is unrelated to rice defense against blast infection. High basal endogenous SA content does not mean that rice is insensitive to SA signaling. For example, exogenously administrated SA triggers resistance to M. oryzae in adult plants (Iwai et al., 2007). In rice mutant with constantly activated SA signaling pathway (i.e., osmpk15), rice resistance against blast disease is enhanced (Hong et al., 2019). Moreover, synthetic SA analogs such as probenazole, benzothiadiazole (BTH), and tiadinil can induce rice defense response to a wide range of pathogens, ranging from (hemi) biotrophic M. oryzae and bacterial leaf blight pathogen X. oryzae pv. oryzae (Xoo), to necrotrophic root pathogens such as Hirschmanniella oryzae (Shimono et al., 2007; De Vleesschauwer et al., 2008, 2012; Nahar et al., 2012; Xu et al., 2013).
Based on the results, we propose that rice can sense M. oryzae infection and induce the expression of a set of immune regulating factors. miR172a was induced upon M. oryzae infection. The induced expression of miR172a leads to the suppressed expression of OsIDS1, which encodes a transcription factor and enhances OsAPX1 transcription as well. Meanwhile, OsRBOHb was induced at the early stage, but it was reduced at the later stage. At the early stage, OsRBOH activity masked OsAPX activity that results in ROS generation; at the later stage, OsAPX activity was unmasked by OsRBOH activity that leads to ROS elimination. By a delicately regulated sequential expression, OsAPX1 and OsRBOHs manipulate ROS homeostasis temporally. ROS accumulates shortly after M. oryzae infection, which is at the earliest time and the most imminent frontier. After the initial ROS burst, OsAPX1 removes excessive ROS to prevent the rice from ROS toxicity (Figure 7). It would be interesting to investigate whether OsAPX1 plays a role in tolerance against abiotic stresses employing a similar mechanism.
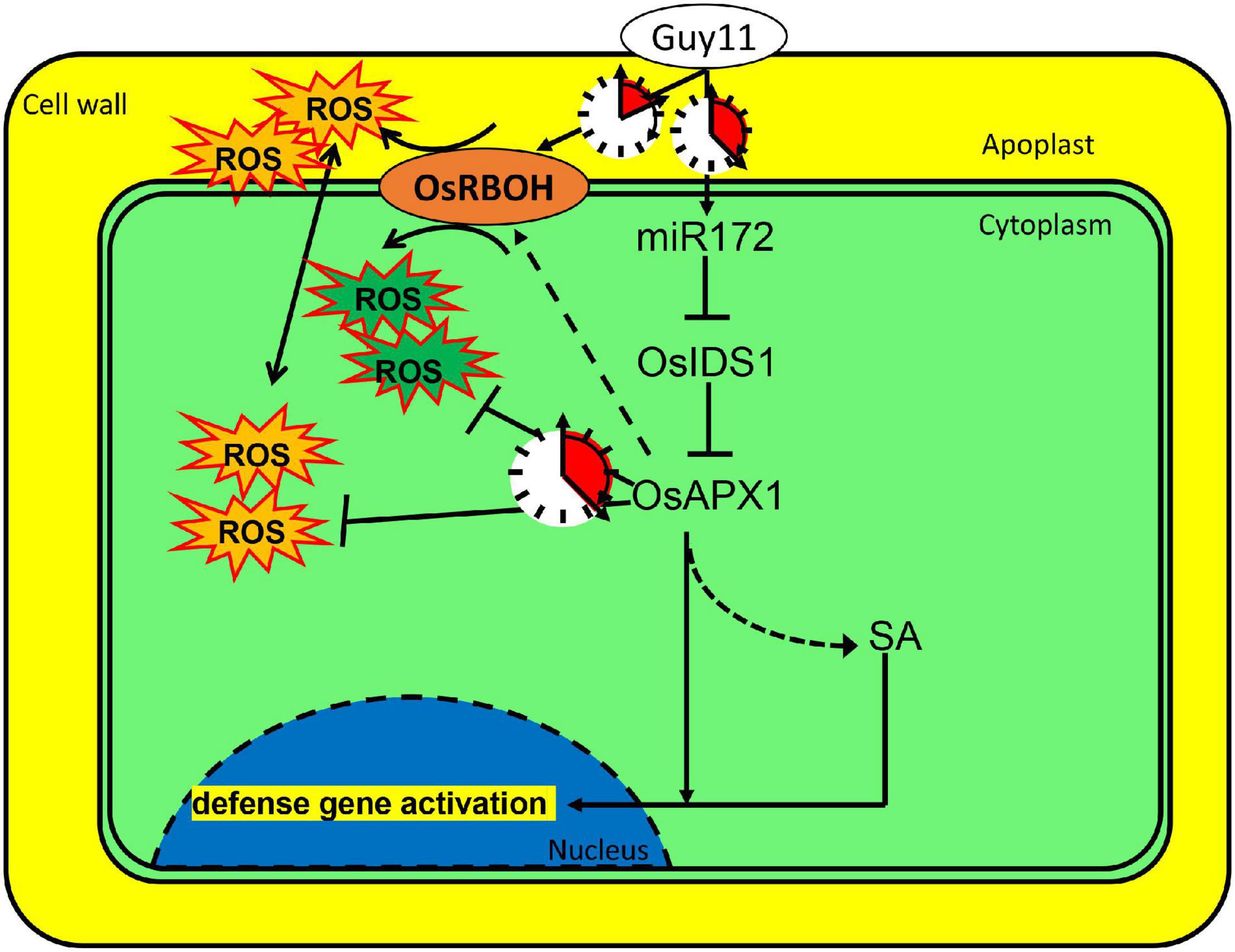
Figure 7. OsAPX1 manipulates cellular ROS homeostasis upon M. oryzae infection. A model demonstrating how OsAPX1 might be involved in rice resistance against the blast disease. Guy11 infection induces miR172a expression, which releases OsAPX1 expression from the restriction of a transcription factor, OsIDS1. OsAPX1 overexpression increase OsRBOHs expression in the early stage that promotes ROS production. ROS is eliminated by OsAPX1 at a later stage that protects rice from its toxicity. OsAPX1 also activates both SA synthesis and signaling, which leads to downstream defense gene activation.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
CS and HZ designed the experiments and wrote the manuscript. CS, DY, XL, HY, YZ, XZ, MS, AB, HM, and PN performed the experimental work. CS, AB, and HZ performed the data analysis.
Funding
This work was supported by a grant from the Natural Science Foundation of China (31872040), the Fundamental Research Funds for the Central Universities (KYXK202009 and JCQY201904) to HZ and the Natural Science Foundation of Shandong Province of China (ZR2019PC049).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We appreciate the great gifts from Xiliu Cheng (miR172 transgenic lines) and W.-M. Wang [M. oryzae strain (Zhong-1)]. We are grateful for technical assistance from Chen Dai, Jiao Li, and Tao Li.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.843271/full#supplementary-material
Supplementary Figure 1 | OsAPX1 transcript induces by M. oryzae infection. (A) Rice (Oryzae sativa cv. Nipponbare) infected with strain Guy11 and Js153. The disease symptom is recorded at 72 hpi. (B) Relative transcriptional level of OsAPX1 in rice infected by mock, Guy11, and Js153 at 24, 48, and 72 hpi (OsAPX1/18srRNA). Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). All of the experiments were repeated three times with similar results.
Supplementary Figure 2 | Validation of OsAPX1 transgenic rice. (A) Sequence confirmation of the homogenous cas9-osapx1 mutant lines. (B) Protein confirmation of OsAPX1-OE lines. (C) OsAPX1 transcript level in OsAPX1-OE and NIP rice. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). (D) The phenotype of the NIP, cas9-osapx1, and OsAPX1-OE rice at 45 days post-sowing. (E) Comparison of grain length in OsAPX1-OE, cas9-osapx1, and NIP rice. (F) Weight of 1,000 seeds of OsAPX1-OE, cas9-osapx1, and NIP rice. Values are means of three replications. Error bars indicate ± SD. Asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). (G) DAB staining shows H2O2 accumulation of transgenic and wild-type rice upon normal conditions. All of the experiments were repeated three times with similar results.
Supplementary Figure 3 | APX activity was affected by M. oryzae infected. The APX enzyme activity in indicated lines upon Guy11 treatment. Values are means of three replications. The error bars indicate ± SD. The asterisks indicate significant differences between samples according to the Student’s t-test (p < 0.05). The experiment was repeated three times with similar results.
Supplementary Figure 4 | OsAPX1 do not affect JA signaling pathway. The expression levels of JA signaling pathway relative genes are compared between OsAPX1 transgenic rice and wild type rice by qRT-PCR. Values are means of three replications. Error bars indicate + SD. ns indicates no significant difference between samples according to the Student’s t-test (p < 0.05). The experiment was repeated three times with similar results.
References
Agrawal, G. K., Jwa, N. S., Iwahashi, H., and Rakwal, R. (2003). Importance of ascorbate peroxidases OsAPX1 and OsAPX2 in the rice pathogen response pathways and growth and reproduction revealed by their transcriptional profiling. Gene 322, 93–103. doi: 10.1016/j.gene.2003.08.017
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bose, J., Rodrigo-Moreno, A., and Shabala, S. (2014). ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 65, 1241–1257. doi: 10.1093/jxb/ert430
Chandrashekar, S., and Umesha, S. (2014). 2,6-Dichloroisonicotinic acid enhances the expression of defense genes in tomato seedlings against Xanthomonas perforans. Physiol. Mol. Plant Pathol. 86, 49–56. doi: 10.1016/j.pmpp.2014.03.003
Cheng, X., He, Q., Tang, S., Wang, H., Zhang, X., Lv, M., et al. (2021). The miR172/IDS1 signaling module confers salt tolerance through maintaining ROS homeostasis in cereal crops. New phytol. 230, 1017–1033. doi: 10.1111/nph.17211
Chiang, C.M., Chen, C.C., Chen, S.P., Lin, K.H., Chen, L.R., Su, Y.H., et al. (2017). Overexpression of the ascorbate peroxidase gene from eggplant and sponge gourd enhances flood tolerance in transgenic Arabidopsis. J. Plant Res. 130, 373–386. doi: 10.1007/s10265-016-0902-4
Cross, A. R., and Jones, O. T. (1986). The effect of the inhibitor diphenylene iodonium on the superoxide-generating system of neutrophils. Specific labelling of a component polypeptide of the oxidase. Biochem. J. 237, 111–116. doi: 10.1042/bj2370111
Dangol, S., Chen, Y., Hwang, B.K., and Jwa, N.S. (2019). Iron- and reactive oxygen species-dependent ferroptotic cell death in rice-Magnaporthe oryzae interactions. Plant Cell 31, 189–209. doi: 10.1105/tpc.18.00535
De Vleesschauwer, D., Djavaheri, M., Bakker, P. A. H. M., and Hoefte, M. (2008). Pseudomonas fluorescens WCS374r-induced systemic resistance in rice against Magnaporthe oryzae is based on Pseudobactin-mediated priming for a salicylic acid-repressible multifaceted defense response. Plant Physiol. 148, 1996–2012. doi: 10.1104/pp.108.127878
De Vleesschauwer, D., Van Buyten, E., Satoh, K., Balidion, J., Mauleon, R., Choi, I.-R., et al. (2012). Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 158, 1833–1846. doi: 10.1104/pp.112.193672
Durner, J., and Klessig, D. F. (1995). Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 92, 11312–11316. doi: 10.1073/pnas.92.24.11312
Guan, Q.J., Wang, Z.J., Wang, X.H., Takano, T., and Liu, S.K. (2015). A peroxisomal APX from Puccinellia tenuiflora improves the abiotic stress tolerance of transgenic Arabidopsis thaliana through decreasing of H2O2 accumulation. J. Plant Physiol. 175, 183–191. doi: 10.1016/j.jplph.2014.10.020
Hong, Y., Liu, Q., Cao, Y., Zhang, Y., Chen, D., Lou, X., et al. (2019). The OsMPK15 negatively regulates Magnaporthe oryza and Xoo disease resistance via SA and JA signaling pathway in rice. Front. Plant Sci. 10:752. doi: 10.3389/fpls.2019.00752
Hu, L., Liang, W., Yin, C., Cui, X., Zong, J., Wang, X., et al. (2011). Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23, 515–533. doi: 10.1105/tpc.110.074369
Iwai, T., Seo, S., Mitsuhara, I., and Ohashi, Y. (2007). Probenazole-induced accumulation of salicylic acid confers resistance to Magnaporthe grisea in adult rice plants. Plant Cell Physiol. 48, 915–924. doi: 10.1093/pcp/pcm062
Jiang, G.H., Yin, D.D., Zhao, J.Y., Chen, H.L., Guo, L.Q., Zhu, L.H., et al. (2016). The rice thylakoid membrane-bound ascorbate peroxidase OsAPX8 functions in tolerance to bacterial blight. Sci. Rep. 6:26104. doi: 10.1038/srep26104
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaur, G., Sharma, A., Guruprasad, K., and Pati, P.K. (2014). Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnol. Adv. 32, 551–563. doi: 10.1016/j.biotechadv.2014.02.002
Keshavarz-Tohid, V., Taheri, P., Taghavi, S. M., and Tarighi, S. (2016). The role of nitric oxide in basal and induced resistance in relation with hydrogen peroxide and antioxidant enzymes. J. Plant Physiol. 199, 29–38. doi: 10.1016/j.jplph.2016.05.005
Lee, J., Bricker, T.M., Lefevre, M., Pinson, S.R.M., and Oard, J.H. (2006). Proteomic and genetic approaches to identifying defence-related proteins in rice challenged with the fungal pathogen Rhizoctonia solani. Mol. Plant Pathol. 7, 405–416. doi: 10.1111/j.1364-3703.2006.00350.x
Li, P., Cai, Q., Wang, H., Li, S., Cheng, J., Li, H., et al. (2020a). Hydrogen peroxide homeostasis provides beneficial micro-environment for SHR-mediated periclinal division in Arabidopsis root. New Phytol. 228, 1926–1938. doi: 10.1111/nph.16824
Li, R., Wang, L., Li, Y., Zhao, R., Zhang, Y., Sheng, J., et al. (2020b). Knockout of SlNPR1 enhances tomato plants resistance against Botrytis cinerea by modulating ROS homeostasis and JA/ET signaling pathways. Physiol. Plant. 170, 569–579. doi: 10.1111/ppl.13194
Li, Y. B., Han, L. B., Wang, H. Y., Zhang, J., Sun, S. T., Feng, D. Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in Cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Li, Y., Ma, Y., Zhang, T., Bi, Y., Wang, Y., and Prusky, D. (2019b). Exogenous polyamines enhance resistance to Alternaria alternata by modulating redox homeostasis in apricot fruit. Food Chem. 301:125303. doi: 10.1016/j.foodchem.2019.125303
Li, Y., Cao, X. L., Zhu, Y., Yang, X. M., Zhang, K. N., Xiao, Z. Y., et al. (2019a). Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 222, 1507–1522. doi: 10.1111/nph.15678
Lin, S.Y., Nie, P.P., Ding, S.C., Zheng, L.Y., Chen, C., Feng, R.Y., et al. (2018). Quantitative proteomic analysis provides insights into rice defense mechanisms against Magnaporthe oryzae. Int. J. Mol. Sci. 19:1950. doi: 10.3390/ijms19071950
Liu, F., Huang, N., Wang, L., Ling, H., Sun, T. T., Ahmad, W., et al. (2018). A novel L-ascorbate peroxidase 6 Gene, ScAPX6, plays an important role in the regulation of response to biotic and abiotic stresses in Sugarcane. Front. Plant Sci. 8:2262. doi: 10.3389/fpls.2017.02262
Liu, L.J., Sonbol, F.M., Huot, B., Gu, Y.N., Withers, J., Mwimba, M., et al. (2016). Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 7:13099. doi: 10.1038/ncomms13099
Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. doi: 10.1038/ncomms5686
Liu, Y., Cao, Y., Zhang, Q., Li, X., and Wang, S. (2018). A cytosolic triosephosphate isomerase is a key component in XA3/XA26-mediated resistance. Plant Physiol. 178, 923–935. doi: 10.1104/pp.18.00348
Loake, G., and Grant, M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi: 10.1016/j.pbi.2007.08.008
Luan, Y., Cui, J., Li, J., Jiang, N., Liu, P., and Meng, J. (2018). Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta 247, 127–138. doi: 10.1007/s00425-017-2773-x
Ma, H. Y., Sheng, C., Qiao, L. L., Zhao, H. W., and Niu, D. D. (2019). A comparative proteomic approach to identify defence-related proteins between resistant and susceptible rice cultivars challenged with the fungal pathogen Rhizoctonia solani. Plant Growth Regul. 90, 73–88. doi: 10.1007/s10725-019-00551-w
Maruta, T., Tanouchi, A., Tamoi, M., Yabuta, Y., Yoshimura, K., Ishikawa, T., et al. (2010). Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant Cell Physiol. 51, 190–200. doi: 10.1093/pcp/pcp177
Moller, I. M., Jensen, P. E., and Hansson, A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481. doi: 10.1146/annurev.arplant.58.032806.103946
Mullineaux, P.M., Karpinski, S., and Baker, N.R. (2006). Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant Physiol. 141, 346–350. doi: 10.1104/pp.106.078162
Munné-Bosch, S., Queval, G., and Foyer, C. H. (2013). The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol. 161, 5–19. doi: 10.1104/pp.112.205690
Nagano, M., Ishikawa, T., Fujiwara, M., Fukao, Y., Kawano, Y., Kawai-Yamada, M., et al. (2016). Plasma membrane microdomains are essential for Rac1-RbohB/H-mediated immunity in rice. Plant Cell 28, 1966–1983. doi: 10.1105/tpc.16.00201
Nahar, K., Kyndt, T., Nzogela, Y. B., and Gheysen, G. (2012). Abscisic acid interacts antagonistically with classical defense pathways in rice-migratory nematode interaction. New Phytol. 196, 901–913. doi: 10.1111/j.1469-8137.2012.04310.x
Narula, K., Choudhary, P., Ghosh, S., Elagamey, E., Chakraborty, N., and Chakraborty, S. (2019). Comparative nuclear proteomics analysis provides insight into the mechanism of signaling and immune response to blast disease caused by Magnaporthe oryzae in rice. Proteomics 19:e1800188. doi: 10.1002/pmic.201800188
Nie, P. P., Li, X., Wang, S. N., Guo, J. H., Zhao, H. W., and Niu, D. D. (2017). Induced systemic resistance against Botrytis cinerea by Bacillus cereus AR156 through a JA/ET- and NPR1-dependent signaling pathway and activates PAMP-triggered immunity in Arabidopsis. Front. Plant Sci. 8:238. doi: 10.3389/fpls.2017.00238
Panchuk, II, Zentgraf, U., and Volkov, R.A. (2005). Expression of the Apx gene family during leaf senescence of Arabidopsis thaliana. Planta 222(5), 926–932. doi: 10.1007/s00425-005-0028-8
Qi, J., Wang, J., Gong, Z., and Zhou, J. M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Quan, L.J., Zhang, B., Shi, W.W., and Li, H.Y. (2008). Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. doi: 10.1111/j.1744-7909.2007.00599.x
Ribeiro, C. W., Carvalho, F. E. L., Rosa, S. B., Alves-Ferreira, M., Andrade, C. M. B., Ribeiro-Alves, M., et al. (2012). Modulation of genes related to specific metabolic pathways in response to cytosolic ascorbate peroxidase knockdown in rice plants. Plant Biol. 14, 944–955. doi: 10.1111/j.1438-8677.2012.00587.x
Rodriguez, M., Munoz, N., Lenardon, S., and Lascano, R. (2012). The chlorotic symptom induced by Sunflower chlorotic mottle virus is associated with changes in redox-related gene expression and metabolites. Plant Sci. 196, 107–116. doi: 10.1016/j.plantsci.2012.08.008
Rossel, J. B., Walter, P. B., Hendrickson, L., Chow, W. S., Poole, A., Mullineaux, P. M., et al. (2006). A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 29, 269–281. doi: 10.1111/j.1365-3040.2005.01419.x
Saruyama, H., and Tanida, M. (1995). Effect of chilling on activated oxygen-scavenging enzymes in low temperature-sensitive and -tolerant cultivars of rice (Oryza sativa L.). Plant Sci. 109, 105–113. doi: 10.1016/0168-9452(95)04156-o
Sato, Y., Masuta, Y., Saito, K., Murayama, S., and Ozawa, K. (2011). Enhanced chilling tolerance at the booting stage in rice by transgenic overexpression of the ascorbate peroxidase gene, OsAPXa. Plant Cell Rep. 30, 399–406. doi: 10.1007/s00299-010-0985-7
Sato, Y., Murakami, T., Funatsuki, H., Matsuba, S., Saruyama, H., and Tanida, M. (2001). Heat shock-mediated APX gene expression and protection against chilling injury in rice seedlings. J. Exp. Bot. 52, 145–151. doi: 10.1093/jexbot/52.354.145
Saxena, I., Srikanth, S., and Chen, Z. (2016). Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 7:570. doi: 10.3389/fpls.2016.00570
Shafi, A., Chauhan, R., Gill, T., Swarnkar, M.K., Sreenivasulu, Y., Kumar, S., et al. (2015). Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 87, 615–631. doi: 10.1007/s11103-015-0301-6
Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. doi: 10.1093/jexbot/53.372.1305
Shimono, M., Sugano, S., Nakayama, A., Jiang, C.-J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell 19, 2064–2076. doi: 10.1105/tpc.106.046250
Silverman, P., Seskar, M., Kanter, D., Schweizer, P., Metraux, J. P., and Raskin, I. (1995). Salicylic acid in rice (biosynthesis, conjugation, and possible role). Plant Physiol. 108, 633–639. doi: 10.1104/pp.108.2.633
ThordalChristensen, H., Zhang, Z.G., Wei, Y.D., and Collinge, D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11, 1187–1194. doi: 10.1046/j.1365-313X.1997.11061187.x
Tian, S., Wang, X., Li, P., Wang, H., Ji, H., Xie, J., et al. (2016). Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 171, 1635–1650. doi: 10.1104/pp.15.01237
Torres, M. A., Dangl, J. L., and Jones, J. D. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522. doi: 10.1073/pnas.012452499
Torres, M.A. (2010). ROS in biotic interactions. Physiol. Plant. 138, 414–429. doi: 10.1111/j.1399-3054.2009.01326.x
Van Camp, W., Van Montagu, M., and Inze, D. (1998). H2O2 and NO: redox signals in disease resistance. Trends Plant Sci. 3, 330–334. doi: 10.1016/s1360-1385(98)01297-7
Vo, K. T. X., Kim, C. Y., Hoang, T. V., Lee, S. K., Shirsekar, G., Seo, Y. S., et al. (2017). OsWRKY67 plays a positive role in basal and XA21-mediated resistance in rice. Front. Plant Sci. 8:2220. doi: 10.3389/fpls.2017.02220
Wang, Y., and Chu, C. (2020). S-Nitrosylation control of ROS and RNS homeostasis in plants: the switching function of catalase. Mol. Plant 13, 946–948. doi: 10.1016/j.molp.2020.05.013
Wei, Z., Hu, W., Lin, Q. S., Cheng, X. Y., Tong, M. J., Zhu, L. L., et al. (2009). Understanding rice plant resistance to the Brown Planthopper (Nilaparvata lugens): a proteomic approach. Proteomics 9, 2798–2808. doi: 10.1002/pmic.200800840
Wildermuth, M.C., Dewdney, J., Wu, G., and Ausubel, F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Wrzaczek, M., Brosche, M., and Kangasjarvi, J. (2013). ROS signaling loops - production, perception, regulation. Curr. Opin. Plant Biol. 16, 575–582. doi: 10.1016/j.pbi.2013.07.002
Wu, J., Yang, R., Yang, Z., Yao, S., Zhao, S., Wang, Y., et al. (2017). ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 3:16203. doi: 10.1038/nplants.2016.203
Xu, E., Vaahtera, L., and Brosche, M. (2015). Roles of defense hormones in the regulation of ozone-induced changes in gene expression and cell death. Mol. Plant 8, 1776–1794. doi: 10.1016/j.molp.2015.08.008
Xu, J., Audenaert, K., Hofte, M., and De Vleesschauwer, D. (2013). Abscisic acid promotes susceptibility to the rice leaf blight pathogen Xanthomonas oryzae pv oryzae by suppressing salicylic acid-mediated defenses. PLos One 8:e67413. doi: 10.1371/journal.pone.0067413
Yan, H., Li, Q., Park, S. C., Wang, X., Liu, Y. J., Zhang, Y. G., et al. (2016). Overexpression of CuZnSOD and APX enhance salt stress tolerance in sweet potato. Plant Physiol. Biochem. 109, 20–27. doi: 10.1016/j.plaphy.2016.09.003
Yang, C., Li, W., Cao, J., Meng, F., Yu, Y., Huang, J., et al. (2017). Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. 89, 338–353. doi: 10.1111/tpj.13388
Yoo, T. H., Park, C. J., Lee, G. J., Shin, R., Yun, J. H., Kim, K. J., et al. (2013). A hot pepper cDNA encoding ascorbate peroxidase is induced during the incompatible interaction with virus and bacteria (Retraction of vol 14, pg 75, 2002). Mol. Cells 35:175. doi: 10.1007/s10059-013-3069-z
Zhang, H., Tan, X., Li, L., He, Y., Hong, G., Li, J., et al. (2019). Suppression of auxin signalling promotes rice susceptibility to rice black streaked dwarf virus infection. Mol. Plant Pathol. 20, 1093–1104. doi: 10.1111/mpp.12814
Zhang, X., Bao, Y., Shan, D., Wang, Z., Song, X., Wang, Z., et al. (2018). Magnaporthe oryzae induces the expression of a MicroRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. doi: 10.1104/pp.17.01665
Zhang, Z.G., Zhang, Q., Wu, J.X., Zheng, X., Zheng, S., Sun, X.H., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLos One 8:e57472. doi: 10.1371/journal.pone.0057472
Zheng, S., Li, J., Ma, L., Wang, H., Zhou, H., Ni, E., et al. (2019). OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc. Natl. Acad. Sci. U.S.A. 116, 7549–7558. doi: 10.1073/pnas.1817675116
Keywords: rice blast resistance, ascorbate peroxidases, OsAPX1, miR172a, ROS homeostasis, salicylic acid
Citation: Sheng C, Yu D, Li X, Yu H, Zhang Y, Saqib Bilal M, Ma H, Zhang X, Baig A, Nie P and Zhao H (2022) OsAPX1 Positively Contributes to Rice Blast Resistance. Front. Plant Sci. 13:843271. doi: 10.3389/fpls.2022.843271
Received: 25 December 2021; Accepted: 28 January 2022;
Published: 21 March 2022.
Edited by:
Wen-Ming Wang, Sichuan Agricultural University, ChinaReviewed by:
Yanjun Kou, China National Rice Research Institute (CAAS), ChinaZhixue Zhao, Sichuan Agricultural University, China
Xuli Wang, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Sheng, Yu, Li, Yu, Zhang, Saqib Bilal, Ma, Zhang, Baig, Nie and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongwei Zhao, aHpoYW9AbmphdS5lZHUuY24=
 Cong Sheng
Cong Sheng Dongli Yu
Dongli Yu Xuan Li1,2
Xuan Li1,2 Hongyu Ma
Hongyu Ma Xin Zhang
Xin Zhang Hongwei Zhao
Hongwei Zhao