- 1Department of Biological Sciences, Mississippi State University, Starkville, MS, United States
- 2Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology, Mississippi State University, Starkville, MS, United States
- 3Department of Entomology and Plant Pathology, Auburn University, Auburn, AL, United States
- 4Department of Computer and Information Sciences, Towson University, Towson, MD, United States
- 5USDA ARS NEA BARC Molecular Plant Pathology Laboratory, Beltsville, MD, United States
- 6Center for Computational Sciences High Performance Computing Collaboratory, Mississippi State University, Starkville, MS, United States
Glycine max root cells developing into syncytia through the parasitic activities of the pathogenic nematode Heterodera glycines underwent isolation by laser microdissection (LM). Microarray analyses have identified the expression of a G. max DOESN'T MAKE INFECTIONS3 (DMI3) homolog in syncytia undergoing parasitism but during a defense response. DMI3 encodes part of the common symbiosis pathway (CSP) involving DMI1, DMI2, and other CSP genes. The identified DMI gene expression, and symbiosis role, suggests the possible existence of commonalities between symbiosis and defense. G. max has 3 DMI1, 12 DMI2, and 2 DMI3 paralogs. LM-assisted gene expression experiments of isolated syncytia under further examination here show G. max DMI1-3, DMI2-7, and DMI3-2 expression occurring during the defense response in the H. glycines-resistant genotypes G.max[Peking/PI548402] and G.max[PI88788] indicating a broad and consistent level of expression of the genes. Transgenic overexpression (OE) of G. max DMI1-3, DMI2-7, and DMI3-2 impairs H. glycines parasitism. RNA interference (RNAi) of G. max DMI1-3, DMI2-7, and DMI3-2 increases H. glycines parasitism. The combined opposite outcomes reveal a defense function for these genes. Prior functional transgenic analyses of the 32-member G. max mitogen activated protein kinase (MAPK) gene family has determined that 9 of them act in the defense response to H. glycines parasitism, referred to as defense MAPKs. RNA-seq analyses of root RNA isolated from the 9 G. max defense MAPKs undergoing OE or RNAi reveal they alter the relative transcript abundances (RTAs) of specific DMI1, DMI2, and DMI3 paralogs. In contrast, transgenically-manipulated DMI1-3, DMI2-7, and DMI3-2 expression influences MAPK3-1 and MAPK3-2 RTAs under certain circumstances. The results show G. max homologs of the CSP, and defense pathway are linked, apparently involving co-regulated gene expression.
Introduction
Plants are sessile organisms that respond to biotic and abiotic conditions, accordingly, to the best of their capability. In some instances, plants undergo symbiotic interactions which facilitate the improvement of their existence and fitness due to the availability of needed materials (Stanley et al., 1993). Plants also undergo pathogenic interactions which impair their ability, and detrimentally affect their fitness (Traw et al., 2007). Therefore, the processes are contrasting in nature (Lewin, 1982). Furthermore, plants can undergo these contrasting processes simultaneously, leading to a disruption of normal cell biological and physiological activities (Barker et al., 1971, 1972; Lehman et al., 1971; Hussey and Barker, 1976; Huang and Barker, 1983; Ko et al., 1984). In these instances, plants may have to choose between symbiosis and defense reactions to varying degrees, considering both processes involve the invasion of plant tissue and interaction with specific cell types, leading to impaired symbiosis at the expense of the successful pathogen. Plants even make choices between different types of symbioses that are sustained in their tissue (Bethlenfalvay et al., 1985, 1987; Hohnjec et al., 2005). These results indicate there are contrasting ways in which the underlying genetic programs function during these different processes while also being shared as important actors. Such considerations are important not only from a metabolite assimilation standpoint, but even more broadly as it relates to climate change. For example, the production of commercial synthetic nitrogen-containing fertilizers consumes 1–2% of the earth's used energy each year while also being the leading source of CO2 generation, contributing between 1 and 2% of worldwide emissions (Boerner, 2019). Furthermore, to combat pathogens globally, approximately 3 billion kilograms (kg) of pesticides are applied annually, costing nearly $40 billion (Pimentel, 2005; Sharma et al., 2019). Knowledge of genetic pathways functioning in both processes, and with the capability of being fine-tuned to function more effectively, is of urgent need (Ried et al., 2014; Saha et al., 2014).
An important model used to understand symbioses is Glycine max (soybean), undergoing such relationships with arbuscular mycorrhizal (AM) fungi as well as rhizobia bacteria that lead to the production of nodules (Bethlenfalvay et al., 1985, 1987; van Kessel et al., 1985). The AM relationship, occurring within >80% of land plants, and nodulation occurring in the Fabaceae, represent the two most important symbiotic interactions that happen between the plant root and microbes (Zhu et al., 2006). AM associations are the more ancient form, first evolving around 460 million years ago (mya) in all land plant lineages and are believed to have allowed their ancestors to colonize land (Remy et al., 1994; Redecker et al., 2000; Heckman et al., 2001; Brundrett, 2002). Nodule-forming relationships are the less ancient form that evolved ~60–70 mya in angiosperms in the Fabaceae (legumes) (Doyle and Luckow, 2003). AM fungi enhance nutrient availability, mainly inorganic phosphate, to the plants whereas legume plants form an intricate symbiotic relationship with specific soil bacteria (i.e., rhizobia) forming specialized structures called nodules which fix atmospheric nitrogen for their host (Zhu et al., 2006; Ferguson et al., 2010). In AM and nodule-forming symbiosis the microbe colonizes the plant tissue. The plant permits their entry and establishment of the microorganism so it can gain from their activities. Experiments show the AM and nodulation processes that permit microorganism (symbiont) entry and maintenance are linked genetically, revealing the molecular components are equally ancient even though they provide different beneficial metabolites (Catoira et al., 2000; Ané et al., 2002, 2004; Zhu et al., 2006; Wang et al., 2010). These observations allow a generalization of the molecular nature of symbiosis as it relates to plant defense.
About half of the non-nodulating legume mutants isolated so far are also defective in the AM symbiosis, implying that the wild-type copies of those genes are required for both processes (Catoira et al., 2000; Ané et al., 2002, 2004). The signal transduction pathway mediated by those genes is denoted as the common symbiosis pathway (CSP) (Kouchi et al., 2010). The range of symbiosis-defective phenotypes of the CSP genes leads to their grouping into two categories; one is positioned upstream of divalent calcium (Ca2+) spiking (upstream genes) and the other is positioned downstream of Ca2+ spiking (downstream genes), which is a central physiological reaction in the CSP (Ehrhardt et al., 1996; Miwa et al., 2006). Following the perception of nodulation (nod) factors (NFs) through Lysin motif receptor-like kinases (LysM-RLKs), biphasic Ca2+ signaling is induced in root hair cells (i.e., a rapid influx of Ca2+ into the root hair cells) and then the occurrence of a periodical oscillation of cytosolic Ca2+ concentrations at the perinuclear region (i.e., Ca2+ spiking). Ca2+ spiking is also induced in response to AM infection, critical for AM symbiosis as well as nodule symbiosis (Kosuta et al., 2008).
Study of the CSP in model legumes has led to the identification of three genes designated as DOESN'T MAKE INFECTIONS1, 2, and 3 (DMI1-3) (Catoira et al., 2000; Ané et al., 2002, 2004). DMI genes control the NF signaling pathway leading to nodulation, and are required for formation of mycorrhiza, indicating that the symbiotic signaling pathway activated by both the rhizobial and fungal symbionts share common steps (Oláh et al., 2005).
DMI1 encodes a putative cation channel protein that is localized to the nuclear periphery (Riely et al., 2007). The Medicago truncatula (alfalfa) Mt-DMI1, and Pisum sativum (pea) Ps-DMI1/SYM8 are putative orthologs of L. japonicus LJ-POLLUX, and Lj-CASTOR (Zhu et al., 2006; Edwards et al., 2007). The DMI1 (M. truncatula) and DMI1/SYM8 (P. sativum) proteins have the capacity to compensate for the loss of both CASTOR and POLLUX ion channels in L. japonicus in both AM and nodule symbioses (Venkateshwaran et al., 2012). Lj-CASTOR and Lj-POLLUX are non-selective ion channels with a preference for K+ over anions (Charpentier et al., 2008). Along with these physiological changes is the involvement of Ca2+. There appears to be 2 different Ca2+-involved processes functioning early during the symbiotic relationship. One process occurs at the root hair tip and is the generation of a Ca2+ gradient that is important for infection thread development while the second process involves the generation of Ca2+ spikes of in the nuclear region (Cardenas et al., 1999; Shaw and Long, 2003). Interestingly, dmi1 mutants interfere with the generation of Ca2+ spikes (Ehrhardt et al., 1996; Wais et al., 2000; Shaw and Long, 2003; Lévy et al., 2004).
DMI2 exists in other plant systems as the symbiosis (SYM) nodulation receptor kinase (NORK), and Symbiosis receptor-like kinase (SYMRK) genes. DMI2 orthologs in P. sativum, M. truncatula, and L. japonicus, respectively, include PsSYM19/Ms-NORK/Lj-SYMRK that encode receptor-like kinases with leucine-rich-repeat (LRR) domains in the predicted extracellular region and possibly transmit the NF signal to the nuclear localized ion channel DMI1 (Ané et al., 2002; Endre et al., 2002; Stracke et al., 2002; Limpens et al., 2005; Riely et al., 2007; Smit et al., 2007). DMI2 is indispensable for AM- and plant-Frankia symbioses, and mutations in DMI2 lead to the abortion of rhizobia infection at a very early stage (Endre et al., 2002; Stracke et al., 2002; Gherbi et al., 2008). The DMI2 protein contains an intracellular kinase domain, a transmembrane domain, and the extracellular portion, including a region with LRRs and a malectin-like domain (MLD) (Pan et al., 2018). Overexpressing the full-length SYMRK/DMI2 or the intracellular kinase domain of SYMRK/DMI2 results in the spontaneous nodule formation even in the absence of rhizobia (Ried et al., 2014; Saha et al., 2014). Like dmi1, dmi2 mutants interfere with the generation of Ca2+ spikes (Ehrhardt et al., 1996; Wais et al., 2000; Shaw and Long, 2003; Lévy et al., 2004).
Mt-DMI3 and its ortholog Ps-SYM9 encode proteins with strong similarity to Ca (2+)/calmodulin-dependent protein kinase (CCaMK) (Lévy et al., 2004; Mitra et al., 2004). DMI3 plays a role downstream of the generation of Ca2+ spikes and is hypothesized to translate the information encoded in the Ca2+ spikes into one or more phosphorylation events (Ehrhardt et al., 1996; Wais et al., 2000; Lévy et al., 2004; Mitra et al., 2004). DMI3 appears to decode and transmit the information DMI3 encoded in the Ca2+ spikes but does not generate them since dmi3 mutants have no effect on Ca2+ spiking (Ehrhardt et al., 1996; Wais et al., 2000; Lévy et al., 2004). This observation contrasts with those made for dmi1 and dmi2 mutants which perturb Ca2+ spiking (Ehrhardt et al., 1996; Cardenas et al., 1999; Wais et al., 2000; Shaw and Long, 2003). Furthermore, dmi3 mutants exhibit increased sensitivity to NFs indicating signal transduction occurs at or downstream of DMI3 (Oldroyd et al., 2001; Shaw and Long, 2003). The functional analyses reveal the central regulatory position of CCaMK in connecting the infection and organogenetic pathways in L. japonicus. Additionally, components of the CSP upstream of Ca2+ spiking are only required for activation of CCaMK (Horváth et al., 2011). Mt-DMI1 and Mt-DMI2, acting upstream of Ca2+ spiking, suggests that Ca2+ is a component of the NF signal-transduction pathway (Oldroyd and Downie, 2004). In contrast, Mt-DMI3 lying downstream of Ca2+ spiking suggests for its possible role in perceiving the Ca2+ signal, decoding, and transducing the signal into an output response (Oldroyd and Downie, 2004). The absence of an Mt-DMI3 ortholog in Arabidopsis thaliana may explain why it cannot establish symbiosis with AM fungi (Zhu et al., 2006). In addition to promoting downstream gene expression, DMI3 negatively regulates upstream signaling events, as dmi3 mutants show an increased sensitivity for Ca2+ spiking in response to NFs and altered transcription (Oldroyd et al., 2001; Czaja et al., 2012).
AM and nodule symbioses are under competition by pathogenic organisms that can detrimentally affect their development (Winkler et al., 1994; Kennedy et al., 1999; Todd et al., 2001). One of the best examples of these detrimental relationships is with endoparasitic nematodes (EPNs) that in some cases produce a nurse cell through their interactions with the plant cell from which they feed. Like symbiosis, scientific descriptions have identified EPN-governed nurse cell formation occurring in all groups of land plants including bryophytes, ferns, gymnosperms, angiosperms, and even multicellular algae (Cobb, 1890, 1893, 1930; Barton, 1892; Dixon, 1908; Bird and DiGennaro, 2012). These observations indicate that a common and ancient circuitry is in place that regulates these processes, but unlike symbiosis, EPNs would co-opt them to facilitate parasitism. Several studies show nodulation and other symbiotic processes to be affected by EPN infection, indicating the organisms affect the same metabolic processes (Barker et al., 1971, 1972; Lehman et al., 1971; Hussey and Barker, 1976; Huang and Barker, 1983; Ko et al., 1984; Pawlowski and Hartman, 2020). This outcome is not surprising since AM and nodulation involve inner cortical root cells occurring in the vicinity of where syncytia (pericycle and surrounding cells) are produced by syncytium-forming EPNs like Heterodera glycines and vascular cells interacting with giant-cell producing EPNs such as Meloidogyne sp.
Very little functional information exists on the genetic program that underlies the compatibility of plants to EPNs even though many plants are susceptible to their infection. In this regard, G. max has become an important model for studying plant-EPN activities because it can undergo compatible and incompatible interactions with both giant cell and syncytium-forming EPNs while still being able to undergo interactions with symbiotic organisms (Rebois et al., 1970; Kirkpatrick and May, 1989; Opperman and Bird, 1998; Pueppke et al., 1998; Machado and Krishnan, 2003; Niblack et al., 2006; Matsye et al., 2011, 2012; Cook et al., 2012; Liu et al., 2012). EPN-induced nurse cell formation, leading to the production of a syncytium or giant cell-containing galls, involves reprogramming the metabolic processes of those specific root cells to provide the EPN with its nutritional needs (Balasubramanian and Rangaswami, 1962; Chitwood and Lusby, 1991). The plant cell can interfere with the deployment and engagement of these injected materials by apparently transducing signals in some manner as an effective defense response (Ross, 1958; Endo and Veech, 1970; Gipson et al., 1971; Riggs et al., 1973; Endo, 1991). Such expressed defense genes in G. max include alpha soluble NSF attachment protein (α-SNAP), present in the resistance to heterodera glycines 1 (rhg1) locus, and serine hydroxymethyltransferase (Rhg4) (Matsye et al., 2011, 2012; Cook et al., 2012; Liu et al., 2012; Matthews et al., 2013). Meanwhile, plants preserve their ability to engage in symbiosis if they are genetically capable of doing so (Kennedy et al., 1999).
Plant defense occurs through its perception of the pathogen. One defense signaling branch involves plant pathogen recognition receptors (PRRs) that recognize pathogen associated molecular patterns (PAMPs) to effect PAMP triggered immunity (PTI) (Jones and Dangl, 2006). Part of this response involves the release of Ca2+ into the cytoplasm (Ranf et al., 2014). A second branch, involving the perception of pathogen effectors, leads to effector triggered immunity (ETI) which if strong enough results in a hypersensitive response (HR) that leads to the sacrifice (death) of the affected cells/tissues (Jones and Dangl, 2006). PTI and ETI receptors signal through MAPKs leading to defense to H. glycines (Gopalan et al., 1996; Desikan et al., 1998; Asai et al., 2002; Day et al., 2006; Chinchilla et al., 2007; Aljaafri et al., 2017; McNeece et al., 2017, 2019; Klink et al., 2021a). MAPK expression leads to increased relative transcript abundances (RTAs) of genes within the H. glycines-induced syncytia undergoing a defense response that also have a demonstrated function in the defense response (Matsye et al., 2012; Pant et al., 2014; Sharma et al., 2016, 2020; Klink et al., 2017, 2021a; McNeece et al., 2017, 2019). Consequently, the identification of CSP gene expression occurring in syncytia undergoing the defense response indicates they may have a dual function in symbiosis and defense (Klink et al., 2009, 2010a,b, 2011, 2021a,b). Importantly, symbiosis and defense recruit calcium signaling with plant defense processes recruiting Ca2+ signaling in the cytoplasm while symbiosis employs nuclear Ca2+ signaling (Lévy et al., 2004; Mitra et al., 2004; Kwaaitaal et al., 2011; Ranf et al., 2011, 2014; Maintz et al., 2014; Keinath et al., 2015). Consequently, commonalities between the two processes likely involve Ca2+ at some level.
In the analysis presented here, G. max DMI1, DMI2, and DMI3 genes are shown to be expressed during its defense response to H. glycines in parasitized root cells undergoing a defense response. Several paralogs compose each of the G. max DMI1, DMI2, and DMI3 gene families. Through transgenic analyses, the DMI1-3, DMI2-7, and DMI3-2 paralogs expressed within the H. glycines-parasitized syncytia undergoing the defense response are shown to function in the defense process. RNA sequencing (RNA-seq) analyses of RNA isolated from roots overexpressing defense MAPKs also exhibit increased RTAs of some DMI paralogs (McNeece et al., 2019). Furthermore, transgenic roots overexpressing DMI1-3, DMI2-7, and DMI3-2 in some cases exhibit increased MAPK3 RTAs. In contrast, transgenic roots undergoing RNAi of DMI1-3, DMI2-7, and DMI3-2 in some cases exhibit decreased MAPK3 RTAs. The combined results indicate DMI gene expression correlates with their ability to function in the defense response to H. glycines parasitism. MAPKs which function in the defense response are shown to regulate the expression of some of these DMI genes. Lastly, DMI genes are shown to regulate the expression of MAPK3 which functions in the defense response possibly indicating the DMI3-2 and MAPK3-1 genes function in a co-regulated signal transduction loop.
Materials and Methods
Selection of Candidate Genes
Laser microdissection (LM) of H. glycines-induced feeding structures (syncytia) developing from parasitized root cells (pericycle) undergoing the process of defense is part of the experimental process used to identify the DMI genes under study (Klink et al., 2005, 2010a, 2021a). Two different H. glycines-resistant G. max genotypes are experimented on to identify consistently expressed candidate defense genes (Klink et al., 2011, 2021a; Matsye et al., 2011). To identify the DMI genes, H. glycines-resistant G.max[Peking/PI548402] and G.max[PI88788] are infected with H.glycines[NL1−Rhg/HG−type7/race3], generating a defense response (Klink et al., 2021a). Roots are processed for paraffin-embedding and histology, followed by LM (Klink et al., 2005, 2007, 2009, 2010a,b, 2011, 2017, 2021a,b). RNA is isolated from LM-collected control cells (pericycle) sampled at 0-days post infection (dpi), prior to infection, and syncytia undergoing the process of defense at 3- and 6 dpi. The 3-dpi time point occurs prior to the onset of visible (histological) signs of a defense response while by 6-dpi the defense response is clearly different than a susceptible reaction (Ross, 1958; Endo, 1965, 1991; Pant et al., 2014). RNA is isolated using the PicoPure RNA Isolation kit (Molecular Devices®) with a DNAse treatment added just before the second column wash using DNAfree® (Ambion®). RNA yield and quality are determined using the RNA 6000 Pico Assay® (Agilent Technologies®) using the Agilent 2100 Bioanalyzer® according to manufacturer protocol. The cDNA probe preparation and hybridization on the Affymetrix® Soybean GeneChip® are performed according to Affymetrix® guidelines (Affymetrix®), run in triplicate for each of the G.max[Peking/PI548402] and G.max[PI88788] genotypes (Klink et al., 2007, 2009, 2010a,b, 2011; Matsye et al., 2011). Genes are considered expressed at a particular time point in the detection call methodology (DCM) if probe signal is measurable above threshold on all three arrays for that time point for both G.max[Peking/PI548402] and G.max[PI88788] (6 total arrays), p < 0.05 using the Bioconductor implementation of the standard Affymetrix® DCM (Klink et al., 2010a). The standard Affymetrix® microarray DCM analysis done in Bioconductor consists of four steps including (1) saturated probe removal, (2) discrimination score calculation, (3) Wilcoxon's rank test p-value calculation, and (4) detection call assignment. The quantitative procedure determines if the gene's expression is provably different from zero (present [P]), has uncertain measurement (marginal [G]), or is not provably different from zero (absent [A]). In the analysis presented here, a DMI gene meets the measured [M] criteria when the probe signal is detectable above threshold (p < 0.05) on all 6 arrays for a given time point. In contrast, the expression of a DMI gene is considered not measured (NM) if probe signal is not detected at a statistically significant level (p ≥ 0.05) on any one of the 6 arrays using the Mann–Whitney–Wilcoxon (MWW) Rank-Sum Test (Mann and Whitney, 1947). The MWW Rank Sum Test is a non-parametric test of the null hypothesis not requiring the assumption of normal distributions (Mann and Whitney, 1947). Some genes have no probe set fabricated onto the microarray. Consequently, gene expression is not determined and is not applicable (n/a). Gene accession numbers are provided from 2 different G. max genome annotations. For the microarray analysis, the Affymetrix annotations are mapped to the original G. max genome release (Accession 1) Wm82.a1.v1.1 (2010). This annotation had to be used at that time of publication of the work of Matsye et al. (2011) because just that annotation had been available. These older annotations have undergone a comparison here to update the accessions to the more recent G. max Wm82.a2.v1 (2015) genome assembly and annotation (Accession 2).
G. max DMI Gene Identification
All of the G. max DMI1, DMI2, and DMI3 protein family sequences are identified at Phytozome (https://phytozome.jgi.doe.gov) using the 881 amino acid (aa) M. truncatula DMI1 (AAS49490), the 925 aa DMI2 (Q8L4H4), and the 523 aa DMI3 (Q6RET7) protein sequences (Schmutz et al., 2010; Goodstein et al., 2012). The processes involved in the G. max proteome query include the use of the Basic Local Alignment Search Tool program (BLAST) (Altschul et al., 1990). The parameters of the BLAST query are the default settings, including Target type: Proteome; Program: BLASTP-protein query to protein database; Expect (E) threshold:−1; Comparison matrix: BLOSUM62; Word (W) length: default = 3; number of alignments to show: 100 allowing for gaps and filter query, in order that they appear on the BLAST program.
PCR Primer Design
The G. max DMI1, DMI2, and DMI3 cDNA sequences are acquired from Phytozome (Schmutz et al., 2010; Goodstein et al., 2012). DNA primer sequences are designed for OE of the full length targeted DMI genes in the pRAP15-ccdB destination vector and RNAi in the pRAP17-ccdB RNAi destination vector (Supplementary Table 1) (Klink et al., 2009, 2021a; Matsye et al., 2012). The nucleotide sequence, CACC, is added to the 5' end of the forward PCR primer for directional cloning into the pENTR™ entry vector (Invitrogen). The control used for the RT-qPCR analyses is the G. max ribosomal protein gene RPS21 (Glyma.15G147700), proven to be transcribed into mRNA and translated into protein (Morita-Yamamuro et al., 2004; Klink et al., 2005; Matsye et al., 2012).
DMI Gene Cloning
The RNeasy Plus Mini Kit and protocol (Qiagen®) are used to isolate mRNA. SuperScript First Strand Synthesis System for RT-PCR (Invitrogen®) with oligo d(T) as the primer are used with their protocol to make cDNA template for PCR cloning using the appropriate primers (Supplementary Table 1). Using designed PCR primers, genes are PCR-amplified from the cDNA template (Niraula et al., 2020). PCR amplification of targeted DMI genes occurs with high fidelity Platinum® taq (Invitrogen®) according to their protocol. PCR conditions include DNA dissociation for 10 min at 96°C with subsequent PCR cycling and temperature set for denaturation for 30 s at 96°C, annealing for 60 s at 55°C and extension for 30 s at 72°C for 35 cycles, terminating at 4°C. PCR reactions, separated by gel electrophoresis, are run on a 1% TAE agarose gel. DMI gene products (amplicons) corresponding to correct size are excised from the gel with a fresh, unused, sterile razor blade. The DMI DNA amplicons are isolated from the agarose gel using the X-TRACTA gel extractor (USA Scientific) and purified using the QIAquick Gel Extraction Kit (Qiagen®) according to their protocol. The purified DMI DNA is used for Gateway® cloning (Karimi et al., 2002, 2007; Curtis and Grossniklaus, 2003). The purified, PCR-generated, DMI amplicons are directionally cloned into the pENTR™ entry vector following the pENTR™/D-TOPO® protocol (Invitrogen). Transformation of the entry vector containing the DMI amplicon into One Shot® chemically competent E. coli cells is immediately followed by selection on LB-kanamycin (LB-kan) plates, 50 μg/ml (Invitrogen). Subsequently, the selected colony is transferred to LB-kan broth, 50 μg/ml, followed by incubation for 12–14 h in a 37°C shaker at 225 rpm. Plasmid DNA is isolated from selected colonies using the Wizard Plus SV Minipreps DNA Purification System (Promega) according to the manufacturer's instructions. The DMI genes are engineered into the pRAP15-ccdB destination vector for OE of the gene and the pRAP17-ccdB destination vector for RNAi of the gene following the LR Clonase II kit and protocol (Invitrogen). The LR clonase reaction replaces the ccdB gene with the DMI amplicon. The pRAP15-ccdB and pRAP17-ccdB vectors have the figwort mosaic virus (FMV) sub-genomic transcript (Sgt) promoter to drive target (DMI) gene expression (Klink et al., 2009, 2021a; Matsye et al., 2012). The FMV-Sgt sequence is a 301-bp FMV-Sgt promoter fragment (sequence −270 to +31 from the transcription start site [TSS]) (Bhattacharyya et al., 2002). The pRAP15 and pRAP17 plasmids contain an enhanced green fluorescent protein (eGFP) gene driven by the rolD promoter and which is terminated by t35S translational terminator for effective visual reporting in plant tissue (White et al., 1985; Elmayan and Tepfer, 1995; Haseloff et al., 1997; Klink et al., 2021a). The reaction contents containing the pRAP15 and pRAP17 destination vectors undergoing the LR reaction to ligate the DMI gene amplicons are then transformed using One Shot® chemically competent E. coli cells and protocol (Invitrogen). Colony selection is performed on LB-tet plates, 5 μg/ml. Transformed One Shot® E. coli bacteria having the plasmid DNA are used to inoculate LB-tet broth, 5 μg/ml, incubated for 12–14 h in a 37°C shaker at 225 rpm. The plasmid DNA is isolated as described and confirmed for the presence of the DMI gene by PCR using the appropriate PCR primers. The DMI gene-containing destination vector DNA is used to transform Agrobacterium rhizogenes (K599) using the freeze thaw method (Hofgen and Willmitzer, 1988). Colonies are selected on LB-tet plates, 5 μg/ml. Positive colonies are tested for the presence of eGFP, root inducing (Ri) plasmid, and the DMI gene by PCR using the appropriate primers (Ryder et al., 1985; Haseloff et al., 1997; Hodges et al., 2004; Pant et al., 2016; McNeece et al., 2019) (Supplementary Table 1). Details of the pRAP15 and pRAP17 plasmids are available (Klink et al., 2021a).
G. max Genetic Transformations
Transgenic OE and RNAi for the respective DMI genes began when the G. max root of a 1-week-old plant is removed at the hypocotyl with a new, sterile razor blade (Pant et al., 2014). The H. glycines-susceptible, resistant to Heterodera glycines1 (rhg1)-lacking (-/-), (rhg1−/−), G.max[Williams82/PI518671] genetic background is used in the OE experiments of DMI1-3, DMI2-7, and DMI3-2 (Bernard and Cremeens, 1988; Atkinson and Harris, 1989; Schmutz et al., 2010). The H. glycines-resistant rhg1-containing (+/+), (rhg1+/+), G.max[Peking/PI548402] genetic background is used for RNAi studies of DMI1-3, DMI2-7, and DMI3-2 (Ross, 1958). To control for non-specific effects of target gene expression, controls for each experiment are constructed by transforming the respective genotype with pRAP15 or pRAP17 vector having the ccdB gene in place of the DMI gene sequence (pRAP15-ccdB or pRAP17-ccdB) (Pant et al., 2014). The pRAP15-ccdB OE control is produced in G.max[Williams82/PI518671]. The pRAP17-ccdB RNAi control is produced in G.max[Peking/PI548402]. The hypocotyl is immersed in the transformed K599 cell solution in Murashige and Skoog (MS) medium in suspension in a Petri dish with the root then being removed to permit the transformed K599 cells to gain access to the plant tissue (Murashige and Skoog, 1962). A group of 25 root-less plants is placed in a 140-ml glass beaker containing 25 ml of transformed K599 cells in MS medium in suspension. The plants are placed under vacuum using the VP60 Two Stage Vacuum Pump (CPS Products, Inc.) in a Bel-Art Space Saver polycarbonate vacuum desiccator with a clear polycarbonate bottom for 5 min and then left under vacuum for 10 min. The vacuum is then slowly released to allow the transformed K599 cells to further enter the plant tissue. After this cocultivation period, the cut ends of the root-less plants are individually placed 3–4 cm deep into fresh coarse grade A-3 vermiculite (Palmetto Vermiculite). The vermiculite is placed in and then pre-wetted with distilled water in 50-cell propagation trays (725602C) held in standard fats (710245C) with holes in the bottom (T.O. Plastics). The plant trays are placed in Sterlite® 25-qt./23-L modular latched boxes then covered with their lids. The covered modular latched boxes are placed 20 cm from standard fluorescent cool white 4,100-K/32-W bulbs emitting 2,800 lumens (Sylvania). The boxes remain under the lights for 5 days at ambient laboratory temperature (22°C). The plants are subsequently transferred to the greenhouse where the plants in the trays are removed from the modular latched boxes. The plants recover in the greenhouse for 1 week. Visual selection of transgenic G. max roots is carried out using the eGFP reporter, employing a Dark Reader Spot Lamp (SL10S) (Clare Chemical Research) (Klink et al., 2021a). Roots exhibiting eGFP reporter expression also possess the DMI gene expression cassette, each having their own promoter and terminator sequences (Klink et al., 2021a). Gene transfer happens because the K599 cells have the capability to facilitate the transport of the DNA cassettes present between the left and right borders of the pRAP15 and pRAP17 destination vectors into the somatic root cell chromosomal DNA. Even though the DNA cassette is not incorporated into the germline, the result is a stable transformation event occurring in the root somatic cell. Roots subsequently develop from the transgenic cell over a period of a few weeks. The resultant genetically mosaic plants have a non-transgenic shoot with a transgenic root system. Therefore, each individual transgenic root system is an independent transformant line. The transgenic plants are each planted in a Ray Leach Conetainer (SC10) (Stuewe and Sons, Inc.) having a cell diameter of 3.81 cm (1.5 in), a depth of 20.96 cm (8.25 in), and a volume of 164 ml in sandy (93.00% sand, 5.75% silt, and 1.25% clay) soil and allowed to recover for 2 weeks prior to the start of the experiment. The conetainers are secured in a Ray Leach Tray (RL98) (Stuewe and Sons, Inc.). The functionality of the genetic constructs (i.e., RTA increased in OE roots and RTA decreased in RNAi roots) in G. max is confirmed by real-time quantitative PCR [RT-qPCR].
Real-Time Quantitative PCR
Gene-specific RT-qPCR primers are designed for the DMI1-3, DMI2-7, and DMI3-2 gene family members, to carry out the RTA analyses (Supplementary Table 1). The cDNA prepared in the experiments is constructed from mRNA collected from the transgenic roots at 0-dpi, prior to infection but mock-inoculated, and used to confirm the expression of the respective targeted OE or RNAi of the DMI transgenes as described. The RTA of the candidate defense genes in the transgenic roots is confirmed using the already-described G. max RPS21. Analyses of the non-targeted DMI genes, and remaining genes in the G. max genome is beyond the scope of the confirmation process. The RT-qPCR experiments utilize the Taqman 6 carboxyfluorescein (6-FAM) probes with the Black Hole Quencher (BHQ1) (MWG Operon; Birmingham, AL). The qPCR reaction is accomplished by preincubation at 50°C for 2 min, followed by 95°C for 10 min. Proceeding from this step is alternating 95°C for 15 s then 60°C for 1 min for 40 cycles (Matsye et al., 2012). The statistical analysis using 2−ΔΔCT to calculate fold change is followed according to the derived formula presented in Livak and Schmittgen (2001). The results have been tested statistically using the Student's t-test (p < 0.05) (Yuan et al., 2006).
The Infection of G. max by H. glycines, Cyst Extraction, Female Index Calculation and Root Mass Determination
The H.glycines[NL1−Rhg/HG−type7/race3] is used for the infection of the transgenic roots. This choice is made because of its effectiveness in parasitizing G.max[Williams82/PI518671] and failure to successfully parasitize G.max[Peking/PI548402]. H. glycines females are isolated by sucrose flotation (Jenkins, 1964; Matsye et al., 2012). The standard H. glycines-susceptible G.max[Williams82/PI518671] is used for experiments requiring a susceptible genotype. The H. glycines-resistant G.max[Peking/PI548402] is used for experiments requiring a resistant genotype. The second stage juveniles (J2s) are hatched and concentrated to a final inoculum concentration of 2,000 J2s/ml (Matsye et al., 2012). The inoculum (1 ml) is dispensed into 7 mm diameter holes made near the base of the plant. This procedure directs the J2s to the root system. Once the inoculum is dispensed and absorbed into the soil, the holes are covered to prevent expulsion of the nematodes by subsequent watering. After 30 days, test roots are stained with acid fuschin to confirm infection (Byrd et al., 1983). The remaining roots from that replicate experiment are then processed for extraction of cysts from the soil to calculate the female index (FI) (Golden et al., 1970). H. glycines cyst extraction involves taking each individual plant and massaging the transgenic root to release the cysts from the soil/root system (Klink et al., 2009). The soil containing the cysts is repeatedly washed and the rinsed water filtered over a 20-mesh sieve nested within a 100-mesh sieve (Matsye et al., 2012). The outcome is the collection of all cysts (Matsye et al., 2012).
H. glycines cyst count and root mass are enumerated for each plant and used to calculate the female index (FI) as it relates to the whole root (wr) system and cysts per gram (pg) of root system (McNeece et al., 2019). The FI is the community standard, acknowledged evaluation for interpreting the effects of a condition on H. glycines (Golden et al., 1970). The approach is used in order to standardize the enumeration of cysts for the FI calculation. The wr analysis procedure is the historically performed method used to enumerate cysts which does not consider the effect the plant genotype or transgenic event has on H. glycines parasitism (McNeece et al., 2019). The pg analysis procedure is employed in order to consider the effect the plant genotype or transgenic event has on H. glycines parasitism since the calculation of the FI adjusts for root mass (McNeece et al., 2019).
The FI is calculated as FI = (Nx/Ns) X 100 (Golden et al., 1970). Nx is the average number of females on the test cultivar (Golden et al., 1970). Ns is the average number of females on the standard susceptible cultivar (Golden et al., 1970). Nx in the experiments presented here accounts for the pRAP15 containing the DMI where it is being overexpressed (DMI-OE) for its targeted increase in RTA or pRAP17 containing the DMI RNAi (DMI-RNAi) where the gene is targeted to decrease its RTA. Ns accounts for the engineered OE control containing the pRAP15-ccdB or RNAi control containing the pRAP17-ccdB non-engineered, empty vectors described previously. The pRAP15 and pRAP17 plasmids are not empty per se as they have the ccdB gene that functions as a control. The wr and pg FI calculations are tested statistically using the Mann–Whitney–Wilcoxon (MWW) Rank-Sum Test, p < 0.05 (Mann and Whitney, 1947; Matsye et al., 2012). The study incorporates three biological replicates, with at least 10 individual experimental replicates in each biological replicate. In the study presented here, the number of analyzed transgenic roots for DMI1-3-OE (n = 30), DMI2-7-OE (n = 30), and DMI3-2 -OE (n = 30), (10 roots per replicate) are compared to the pRAP15-ccdB control (n = 36) (at least 10 roots per replicate). The number of transgenic roots analyzed for DMI1-3-RNAi (n = 30), DMI2-7-RNAi (n = 30), and DMI3-2-RNAi (n = 30), (10 roots per replicate) are compared to the pRAP17-ccdB control (n = 34), having at least 10 roots per replicate.
RNA-Seq Gene Expression Analyses of Transgenic Root RNA
Prior functional transgenic analyses of the 32-member G. max MAPK gene family determine that 9 of them function in the defense response to H. glycines parasitism and are referred to as defense MAPKs (McNeece et al., 2019). The defense MAPKs undergoing overexpression (OE) or RNA interference (RNAi) include MAPK2 (Glyma.06G029700), MAPK3-1 (Glyma.U021800), MAPK 3-2 (Glyma.12G073000), MAPK 4-1 (Glyma.07G066800), MAPK 5-3 (Glyma.08G017400), MAPK6-2 (Glyma.07G206200), MAPK 13-1 (Glyma.12G073700), MAPK16-4 (Glyma.07G255400), and MAPK20-2 (Glyma.14G028100) with OE and RNAi root samples collected from each for RNA-seq. The pRAP15-ccdB and pRAP17-ccdB control root samples are collected for RNA-seq (Alshehri et al., 2019). RNA is isolated from the collected root samples as already described. The collected samples are validated, sequenced, and analyzed, producing Illumina® RNA-seq data for use in examining gene expression of the 55,022 genes in the G. max genome (Alshehri et al., 2019). The RNA-seq fold change (FC) data, representing the relative transcript abundance (RTA) is mined specifically for DMI gene paralog expression (Wang and Wang, 2021). The FC for the OE experiments is determined in comparisons of the transgenic MAPK-OE RNA-seq data as compared to the RNA-seq data obtained from the transgenic pRAP15-ccdB (overexpression) control (Wang and Wang, 2021). The FC for the RNAi experiments is determined in comparisons of the transgenic MAPK-RNAi RNA-seq data as compared to the RNA-seq data obtained from the transgenic pRAP17-ccdB (RNAi) control (Wang and Wang, 2021). When presented, confirmation of the RNA-seq RTAs, given as FC, is performed by RT-qPCR as described (Livak and Schmittgen, 2001; Klink et al., 2021a).
Proteome Mining
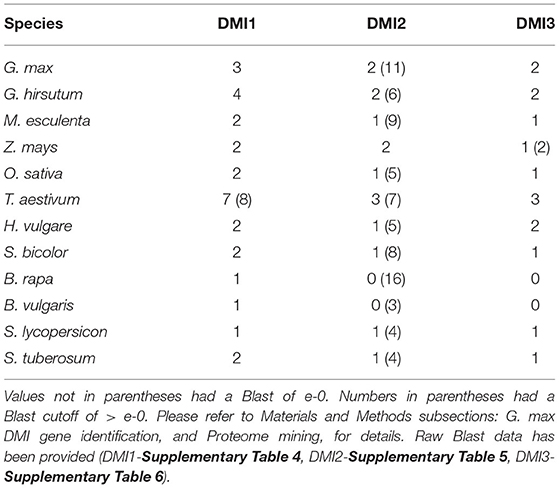
DMI homologs and splice variants from various agricultural crops of international importance and select importance in the U.S. are identified (Tilman et al., 2011; Ray et al., 2013, 2019; Burkhead and Klink, 2018). Analyses are performed by BLASTing selected conceptually translated genes to the described protein coding regions of genomes with the M. truncatula DMI1, DMI2, and DMI3 protein sequences. The proteomes, in addition to G. max (G.max Wm82.a2.v1), include Manihot esculenta (M.esculenta v8.1), Zea mays (Z.mays RefGen_V4), Oryza sativa (O.sativa v7.0), Triticum aestivum (T.aestivum v2.2), Hordeum vulgare (H.vulgare r1), Sorghum bicolor (S.bicolor v3.1.1), Brassica rapa (B.rapaFPsc v1.3), Solanum tuberosum (S.tuberosum v6.1), S. lycopersicum (S.lycopersicum ITAG4.0), Gossypium hirsutum (G.hirsutum v2.1), and B. vulgaris (B.vulgaris EL10_1.0) which are housed at Phytozome under default settings (Goodstein et al., 2012).
Results
The Identification of DMI Gene Families and Root Cell-Specific Expression
An analysis was performed that compared the gene expression which occurred in control (pericycle) cells (0 dpi), and within H. glycines-induced syncytia at 3, and 6 dpi in 2 different genotypes (G.max[Peking/PI548402] and G.max[PI88788]), each capable of mounting a defense response. The two different H. glycines-resistant G. max genotypes were experimented on to identify consistently expressed candidate defense genes (Klink et al., 2011, 2021a; Matsye et al., 2011). The results from those experiments led to the identification of an annotated G. max gene homologous to the M. truncatula DMI3. DMI3 did not exhibit expression at the 0 dpi time point but exhibited expression in syncytia that has undergone the defense response in G.max[Peking/PI548402] and G.max[PI88788] at 3, and 6 dpi (Table 1 and Supplementary Table 2). The CSP has 3 different DMI genes, DMI1, DMI2, and DMI3. BLAST analyses of the G. max proteome identified 3 DMI1, 12 DMI2, and an additional DMI3 paralog (Supplementary Tables 2, 3). Further examination of previously generated transcriptomic data identified the expression activity for the G. max DMI1, DMI2, and DMI3 paralogs that occurred while a defense response was mounted to H. glycines parasitism (Table 1 and Supplementary Tables 2, 3). The experiment demonstrated at least one paralog for each DMI gene family gene underwent expression during the defense response.
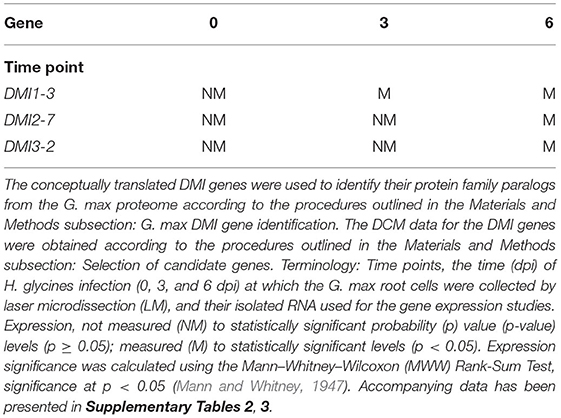
Table 1. G. max DMI genes identified as being expressed in syncytia that were undergoing a defense response to H. glycines parasitism.
BLAST queries of globally important crop proteomes, and some with more importance to U.S. production, identified DMI1, DMI2, and DMI3 homologs, and in some cases, additional paralogs (Table 2 and Supplementary Tables 4–6). Genomic data on alternative splice variants were also identified, provided here since alternate splice variants of other G. max genes function in the defense response that G. max has to H. glycines (Supplementary Tables 4–6). The DMI1-3 (Glyma.19G263500), DMI2-7 (Glyma.11G246200), and DMI3-2 (Glyma.15G222300) paralogs that exhibited expression during the defense response in syncytia were selected for functional transgenic experiments as the approach serves as an effective strategy in identifying defense genes.
DMI Relative Transcript Abundance Changes Occurred When Experimentally Targeted
The DMI1-3, DMI2-7, and DMI3-2 genes were targeted for experimentally altering their RTA through transgenic manipulation, presented as a fold change (FC) in expression as compared to the appropriate control. The DMI1-3, DMI2-7, and DMI3-2 genes were engineered for OE in the H. glycines-susceptible G.max[Williams82/PI518671], based on the hypothesis that their increase in RTA would make the H. glycines-susceptible G.max[Williams82/PI518671] resemble the observed H. glycines defense response that occurs in G.max[Peking/PI548402]. In contrast, DMI1-3, DMI2-7, and DMI3-2 genes were engineered for RNAi in the H. glycines-resistant G.max[Peking/PI548402], based on the hypothesis that their decrease in RTA would make the H. glycines-resistant G.max[Peking/PI548402] resemble the observed H. glycines susceptibility that occurs in G.max[Williams82/PI518671]. The combination of the hypothesized increase in DMI1-3, DMI2-7, and DMI3-2 RTA in G.max[Williams82/PI518671] that would lead to more H. glycines-resistant roots, and decrease in DMI1-3, DMI2-7, and DMI3-2 RTA in the G.max[Peking/PI548402] that would lead to more H. glycines-susceptible roots would be evidence that the targeted gene functioned in the defense response. Transgenic DMI1-3, DMI2-7, and DMI3-2 -OE roots, and their respective transgenic pRAP15-ccdB OE control roots, were made (Figure 1). Transgenic DMI1-3, DMI2-7, and DMI3-2-RNAi roots, and their respective transgenic pRAP17-ccdB RNAi control roots, had then also been made (Figure 1). The expected change in DMI1-3, DMI2-7, and DMI3-2 RTA, presented as a FC in gene expression for the -OE and -RNAi roots as compared to their respective transgenic controls using the G. max RPS21 were determined (Figure 1). The increase in RTA in the DMI1-3, DMI2-7, and DMI3-2 -OE roots ranged from 3 to 18-fold while the change in RTA in the DMI1-3, DMI2-7, and DMI3-2-RNAi roots ranged from−4 to−9-fold as compared to their controls (Figure 1). DMI1-3, DMI2-7, and DMI3-2 RTAs have also been determined at 6 dpi H. glycines infection in cDNA generated from their respective RNAs isolated from their OE and RNAi transgenic roots as compared to their controls (Supplementary Table 1). An examination of the effect that DMI1-3, DMI2-7, and DMI3-2 -OE or-RNAi had on the remaining DMI genes were not performed.
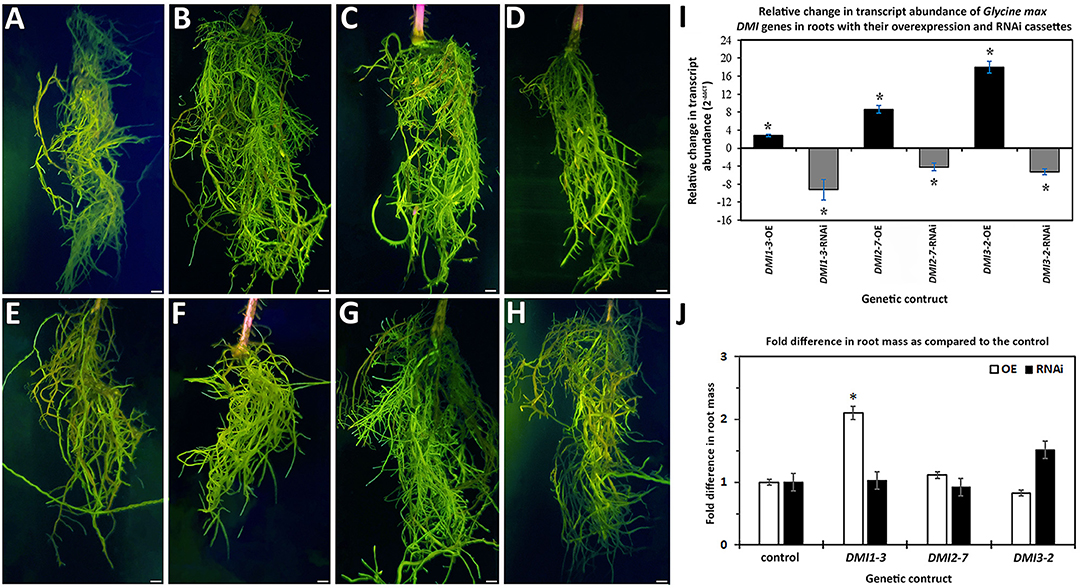
Figure 1. Transgenic roots obtained from chimeric plants having genetically engineered roots and un-engineered shoots that were generated through the K599-mediated genetic transformation procedure. Please refer to Materials and Methods subsections: DMI gene cloning, and G. max genetic transformations, for details. The presented values represent the numbers of plants used in the image generation, and those roots studied in the H. glycines infection analyses. Terminology: overexpression (OE); RNA interference (RNAi); relative transcript abundance (RTA); fold change (FC); probability (p) value (p-value;); female index (FI). (A) pRAP15-ccdB (OE control) (n = 36); (B) DMI1-3-OE (n = 30); (C) DMI2-7-OE (n = 30); (D) DMI3-2 -OE (n = 30); (E) pRAP17-ccdB (RNAi control) (n = 34); (F) DMI1-3-RNAi (n = 30); (G) DMI2-7-RNAi (n = 30); (H) DMI3-2-RNAi (n = 30). Bars = 1 cm. (I) The expected change in DMI1-3, DMI2-7, and DMI3-2 RTA, presented as FC, for the -OE and -RNAi roots, as compared to their pRAP15-ccdB and pRAP17-ccdB overexpression and RNAI controls, respectively, was calculated by 2−ΔΔCT to determine FC (Livak and Schmittgen, 2001; Klink et al., 2021a). The RTA of the candidate defense genes, presented as a FC, in the transgenic roots was compared using the G. max ribosomal protein gene RPS21 (Glyma.15G147700). The RT-qPCR analyses examined 3 experimental replicates (individual root systems) of DMI1-3, DMI2-7, and DMI3-2 -OE or -RNAi roots as compared to their pRAP15-ccdB, and pRAP17-ccdB controls, respectively, from each of the 3 biological replicates. Each experimental replicate was run in triplicate using the same RNA. (*), statistical significance of p < 0.05, Student's t-test (Yuan et al., 2006). Please refer to Materials and Methods subsection: Real-time quantitative PCR (RT-qPCR), for details. (J) The effect that the expression of the DMI1-3, DMI2-7, or DMI3-2 -OE or -RNAi cassettes had on root mass as compared to their respective pRAP15-ccdB or pRAP17-ccdB controls. The statistical significance (*) for the change in root mass was determined using the MWW Rank-Sum Test, p < 0.05 (Mann and Whitney, 1947). Please refer to Materials and Methods subsection: The infection of G. max by H. glycines, cyst extraction, FI calculation, and root mass, for details.
An examination of root mass from DMI1-3, DMI2-7, and DMI3-2 -OE and -RNAi roots as compared to their respective pRAP15-ccdB OE and pRAP17-ccdB RNAi controls presented in Figure 1 were performed. The analyses identified DMI1-3-OE transgenic roots exhibited affected growth to a statistically significant level as compared to the respective transgenic pRAP15-ccdB OE control roots. In this case, the statistically significant change in root mass observed for DMI1-3-OE roots was an increase of 2.10 fold as compared to its control (Figure 1). The remaining analyses of the other DMI-OE and -RNAi roots did not lead to the identification of a statistically significant effect on root mass as compared to their appropriate controls.
Altering G. max DMI RTA Changes the Capability of H. glycines to Parasitize Roots
The FI studies of H. glycines parasitism in relation to the altered expression of the DMI1-3 genes were performed, examining their pathogenic ability in relation to the enumerated cyst numbers in the wr and pg of root analyses. The analyses were done in this manner to gain insight as to whether the expression of the DMI-OE or DMI-RNAi gene cassettes affected root mass as compared to their controls and therefore skewed the FI analysis. The generation of DMI1-3, DMI2-7, DMI3-2-OE, and pRAP15-ccdB OE control roots occurred in the H. glycines susceptible G.max[Williams 82/PI518671]. In comparison to the generated pRAP15-ccdB OE control, the experimentally increased DMI1-3, DMI2-7, and DMI3-2 RTA that occurred through their OE led to a statistically significant 52-63% suppression of H. glycines parasitism as compared to the control (Figure 2). In contrast, the generation of DMI1-3, DMI2-7, DMI3-2-RNAi, and pRAP17-ccdB RNAi control roots were made in the H. glycines-resistant G.max[Peking/PI548402]. In comparison to the pRAP17-ccdB RNAi controls, experimentally decreased DMI1-3, DMI2-7, and DMI3-2 RTA that occurred by RNAi led to a statistically significant 2.1-to-4.6-fold increase in H. glycines parasitism as compared to te control (Figure 2). The combination of the two outcomes, opposite in nature, is taken as evidence that the gene functions during the defense response.
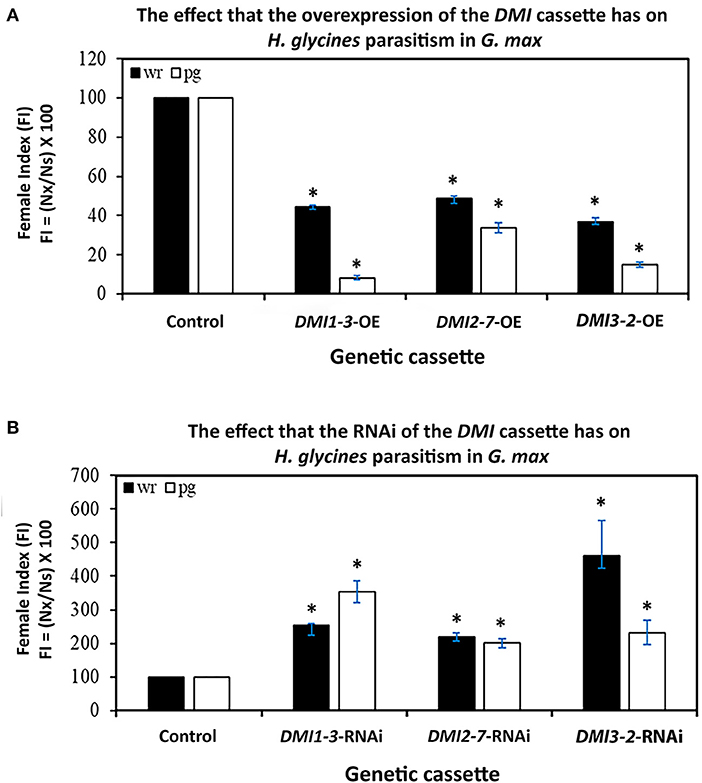
Figure 2. The FI for DMI1-3, DMI2-7, and DMI3-2 -OE and -RNAi roots as compared to their respective pRAP15-ccdB and pRAP17-ccdB controls. The statistical significance (*) was determined by the MWW Rank-Sum Test, p < 0.05 (Mann and Whitney, 1947). The analyzed transgenic roots for DMI1-3-OE (n = 30), DMI2-7-OE (n = 30), and DMI3-2-OE (n = 30) were compared to the pRAP15-ccdB control (n = 36). The analyzed transgenic roots for DMI1-3-RNAi (n = 30), DMI2-7-RNAi (n = 30), and DMI3-2-RNAi (n = 30) were compared to the pRAP17-ccdB control (n = 34). Please refer to Materials and Methods subsection: The infection of G. max by H. glycines, cyst extraction and female index calculation, for details.
MAPKs Influence DMI Expression
In G. max, MAPK gene expression had occurred in syncytia undergoing parasitism by H. glycines but while undergoing a defense response. These syncytium-expressed defense MAPK genes included MAPK2, MAPK3-1, MAPK4-1, MAPK6-2, MAPK13-1, MAPK16-4, and MAPK20-2. Two other MAPKs, MAPK3-2, and MAPK5-3, whose expression in syncytia that underwent the defense process could not be confirmed due to the original analysis procedures which lacked probe sets on the microarray, had also functioned in defense. RNA-seq data was available for G. max defense MAPK-OE and MAPK-RNAi roots, which included MAPK2, MAPK3-1, MAPK3-2, MAPK4-1, MAPK5-3, MAPK6-2, MAPK13-1, MAPK16-4, and MAPK20-2 as well as their OE (pRAP15-ccdB) and RNAi (pRAP17-ccdB) controls, respectively. The data was examined here for RTAs that occurred among the parologous members of the DMI1, DMI2, and DMI3 gene families (Supplementary Tables 7, 8). RT-qPCR confirmed the identified DMI1, DMI2, and DMI3 gene family member RTAs that exhibited differential expression in the MAPK-OE RNA-seq analyses as compared to the control (Figure 3). There were 153 MAPK-OE and MAPK-RNAi comparisons made between the 3 different DMI1 paralogs (n = 27), 12 different DMI2 paralogs (n = 108), and 2 different DMI3 paralogs (n = 18) (Supplementary Tables 7, 8). RNA-seq experiments of RNA that was isolated from the MAPK-OE roots had identified 20 instances of differentially expressed DMI genes that met the set criteria as compared to the control (Figure 3). Please refer to the figure legend and figure inset for the explanation of the examined RNA sample and tested DMI gene name designations found at the x-axis. The RTAs of those DMI genes as compared to their control were confirmed by RT-qPCR (Figure 3). RNA-seq data was also available for the 9 defense MAPKs undergoing RNAi. RNA-seq experiments of RNA isolated from the MAPK-RNAi roots identified 46 instances of differentially expressed DMI genes that met the set criteria as compared to their control (Figure 4). Please refer to the figure legend and figure inset for the explanation of the name designations found at the X-axis. Among the identified DMI genes that were found in the MAPK-OE analyses was one instance, DMI1-1, that had an altered RTA caused by MAPK3-1 OE that met the set criteria as compared to its control (Figure 3). There were 16 instances of a DMI2 gene, that involved DMI2-3, DMI2-4, DMI2-9, DMI2-10, and DMI2-11, that had an altered RTA caused by MAPK OE that met the set criteria as compared to its control (Figure 3). There were three instances of a DMI3 gene, that involved DMI3-1, and DMI3-2, that had an altered RTA caused by MAPK OE that met the set criteria as compared to its control (Figure 3). In contrast, there were 2 instances of a DMI1 gene, DMI1-1, and DMI1-2, that had an altered RTA that was caused by MAPK RNAi that met the set criteria as compared to its control (Figure 4). There were 35 instances of a DMI2 gene, that involved DMI2-3, DMI2-4, DMI2-6, DMI2-7, DMI2-8, DMI2-9, DMI2-10, DMI2-11, and DMI2-12, that had an altered RTA caused by MAPK RNAi that met the set criteria as compared to its control (Figure 4). There were 9 instances of a DMI3 gene, that involved DMI3-1 and DMI3-2, that had an altered RTA caused by MAPK RNAi that met the set criteria as compared to its control (Figure 4).
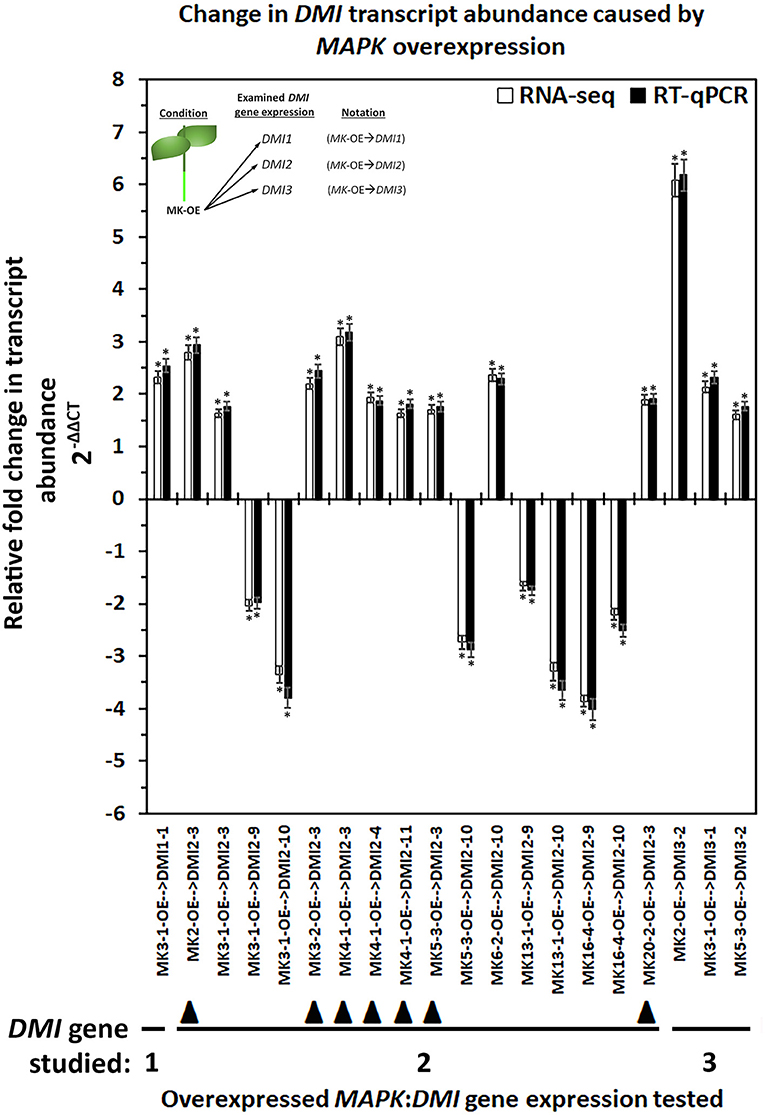
Figure 3. The RTA results, presented as FC, for the DMI1, DMI2, and DMI3 gene family members that had exhibited differential expression in the defense MAPK-OE roots measured by RNA-seq and confirmed by RT-qPCR (Livak and Schmittgen, 2001; Wang and Wang, 2021). The data presented in this figure analyzes the MAPK-OE root RNA by RNA-seq and RT-qPCR. The sample analysis type presented below the x-axis provides the analysis parameter as MAPK-OE with the presented gene expression for the specific DMI gene denoted after the arrow ( → ) giving the sample designation as MAPK(specific gene)-OE → DMI(specific gene). (*) The RNA-seq result was considered statistically significant at p < 0.001, Student's t-test; RT-qPCR expression was calculated by 2−ΔΔCT to enumerate the RTA, presented as FC in the y-axis (Livak and Schmittgen, 2001; Klink et al., 2021a). (*) The result is statistically significant at p < 0.05, Student's t-test (Yuan et al., 2006). The DMI genes that had contrasting expression (an FC that was increased [I] in the MAPK-OE RNA-seq and RT-qPCR analyses (Figure), and an FC that was decreased [D] in MAPK-RNAi RNA-seq, and RT-qPCR analyses (Figure 4) have been referred to as an MAPK-OE:RNAi couplet as designated in the Results subsection: Cross-comparative analyses involving MAPK-OE and MAPK-RNAi experiments) have been indicated with the designated black triangle (▴). This designation was made for MAPK2-OE:RNAi → DMI2-3 (I:D), MAPK3-2-OE:RNAi → DMI2-3 (I:D), MAPK4-1-OE:RNAi → DMI2-3 (I:D), MAPK4-1-OE:RNAi → DMI2-4 (I:D), MAPK4-1-OE:RNAi → DMI2-11 (I:D), MAPK5-3-OE:RNAi → DMI2-3 (I:D), and MAPK20-2-OE:RNAi → DMI2-3 (I:D). (Figure) presents data for 20 couplets. Details for these experiments can be found in Supplementary Table 6. Please refer to the Materials and Methods subsections: RNA-seq gene expression analyses of transgenic MAPK -OE and -RNAi root RNA, and Real-time quantitative PCR (RT-qPCR), for details. Inset, upper left corner of the histogram provides that flow chart that relates to the sample naming scheme found at the x-axis.
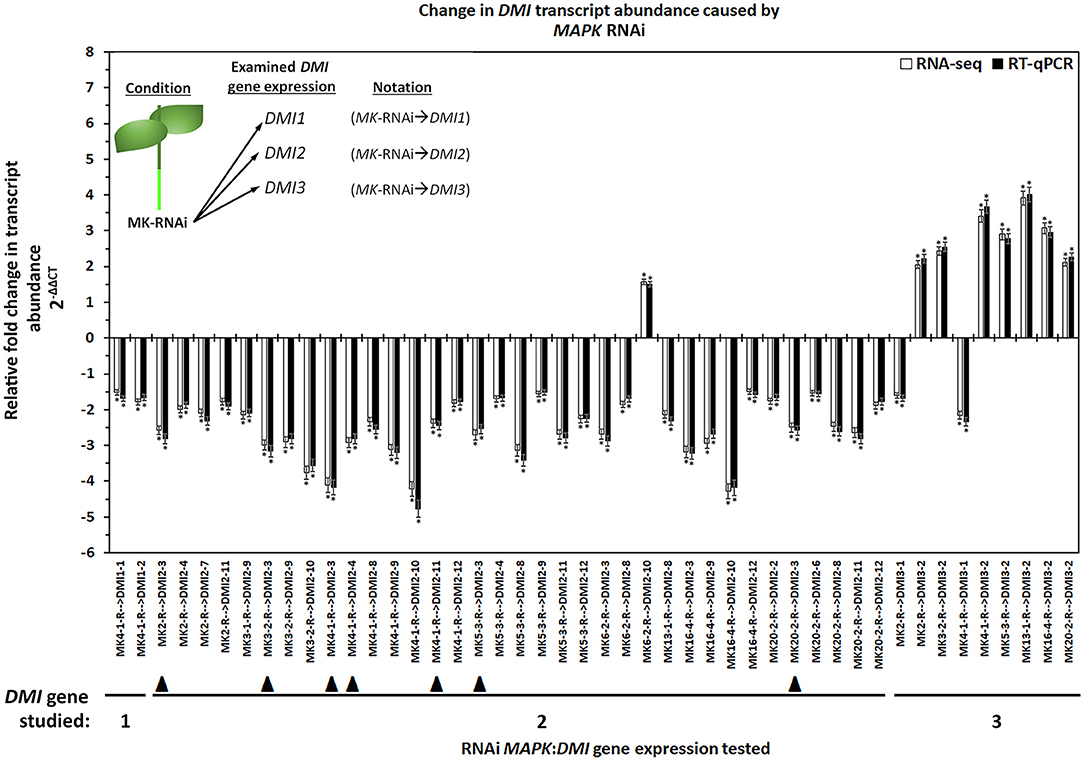
Figure 4. The results for the DMI1, DMI2, and DMI3 gene family members that had exhibited differential expression in the defense MAPK-RNAi roots measured by RNA-seq and confirmed by RT-qPCR (Livak and Schmittgen, 2001; Wang and Wang, 2021). The data presented in (Figure) analyzes the MAPK-RNAi root RNA by RNA-seq and RT-qPCR. The sample analysis type presented below the x-axis provides the analysis parameter as MAPK-RNAi with the presented gene expression for the specific DMI gene denoted by an arrow ( → ) giving MAPK(specific gene)-RNAi → DMI(specific gene). (*) The RNA-seq result is statistically significant at p < 0.001, Student's t-test. RT-qPCR expression was calculated by 2−ΔΔCT to enumerate the RTA as presented as the FC (Livak and Schmittgen, 2001; Klink et al., 2021a). (*) RT-qPCR result statistically significant at p < 0.05, Student's t-test (Yuan et al., 2006). The DMI genes that had contrasting expression (an FC that was increased [I] in the MAPK-OE RNA-seq and RT-qPCR analyses (Figure 3), and an FC that was decreased [D] in MAPK-RNAi RNA-seq, and RT-qPCR analyses (Figure 4), referred to as an MAPK-OE:RNAi couplet as designated in the Results subsection: Cross-comparative analyses involving MAPK-OE and MAPK-RNAi experiments) were indicated with a black triangle (▴). An arrow ( → ) denotes the DMI gene whose expression was examined in relation to the MAPK-OE:RNAi RNA-seq and RT-qPCR analyses. This designation was made for MAPK2-OE:RNAi → DMI2-3 (I:D), MAPK3-2-OE:RNAi → DMI2-3 (I:D), MAPK4-1-OE:RNAi → DMI2-3 (I:D), MAPK4-1-OE:RNAi → DMI2-4 (I:D), MAPK4-1-OE:RNAi → DMI2-11 (I:D), MAPK5-3-OE:RNAi → DMI2-3 (I:D) and MAPK20-2-OE:RNAi → DMI2-3 (I:D). This figure presents data for 46 couplets. Details have been provided in Supplementary Table 7. Please refer to Materials and Methods subsections: RNA-seq gene expression analyses of transgenic MAPK-OE and RNAi root RNA, and Real-time quantitative PCR (RT-qPCR), for details. Inset, upper left corner of the histogram provides that flow chart that relates to the sample naming scheme found at the x-axis.
Cross-Comparative Analyses Made Between the MAPK-OE and MAPK-RNAi Experiments
Cross-comparative analyses were done between the MAPK-OE, and MAPK-RNAi experiments. The analyses were done to determine whether the experimentally-altered MAPK expression (OE or RNAi) exerted contrasting effects on DMI RTAs as compared to their controls as might be expected (i.e., increased RTA in the OE sample and decreased RTA in the RNAi sample). Seven instances, referred to as couplets, were identified where the defense MAPK-OE increased the DMI RTA, while RNAi of that same MAPK led to a decreased DMI RTA as compared to their controls (Figures 3, 4). All of these instances involved the plasma membrane receptor DMI2. To describe these outcomes, the DMI genes that had contrasting expression (an FC that was increased [I] in the MAPK-OE RNA-seq and RT-qPCR analyses (Figure 3), and an FC that was decreased [D] in MAPK-RNAi RNA-seq and RT-qPCR analyses (Figure 4) had been referred to as an MAPK-OE:RNAi couplet, indicated with a black triangle [see figure legend]). A flow chart representing the analysis parameters that merge (cross-compare) the OE and RNAi analyses has been provided (Figure 5). An arrow ( → ) was used to denote the DMI gene whose expression was examined in relation to the MAPK-OE:RNAi RNA-seq and RT-qPCR analyses. This designation was made for MAPK2-OE:RNAi → F0E0DMI2-3 (I:D), MAPK3-2-OE:RNAi → F0E0DMI2-3 (I:D), MAPK4-1-OE:RNAi → F0E0DMI2-3 (I:D), MAPK4-1-OE:RNAi → F0E0DMI2-4 (I:D), MAPK4-1-OE:RNAi → F0E0DMI2-11 (I:D), MAPK5-3-OE:RNAi → F0E0DMI2-3 (I:D) and MAPK20-2-OE:RNAi → F0E0DMI2-3 (I:D) (Figures 3, 4). No instances existed under these conditions where defense MAPK OE decreased the DMI RTA, and MAPK RNAi increased the DMI RTA which was shown to happen in Type 1 expression found for some MAPKs (McNeece et al., 2019). In McNeece et al. (2019) Type 1 expression spanned the results of both the MAPK-OE and -RNAi experiments and was defined that Type 1-OE genes were induced (in their RTA [FC]) in the OE treatment, and were also suppressed in the RNAi treatment or suppressed in the OE treatment, and also induced in the RNAi treatment. Regarding the RNAi experiments, Type 1-RNAi expression was suppressed (in their RTA [FC]) in the RNAi treatment and induced in the OE treatment or induced in the RNAi treatment, and also suppressed in the OE treatment. There were 6 instances identified here whereby the DMI gene was differentially expressed in the same manner (increased, decreased) in the MAPK -OE, and -RNAi roots as compared to the control under the set criteria. Three of these instances involved increased DMI gene RTAs in the MAPK-OE, and -RNAi roots. Perhaps importantly, differences in the magnitude of these DMI RTAs were observed between the OE, and RNAi experiments and presented subsequently for clarity. The first occurrence was MAPK2-OE:RNAi → F0E0DMI3-2 that exhibited a lower RTA in the DMI3-2-RNAi samples that met the set criteria as compared to the controls (Figures 3, 4). The second occurrence was MAPK5-3-OE:RNAi → F0E0DMI3-2 that exhibited higher RTAs in the MAPK5-3-RNAi samples that met the set criteria as compared to the controls (Figures 3, 4). The third instance was MAPK6-2-OE:RNAi → F0E0DMI2-10, that exhibited lower RTAs in the MAPK6-2-RNAi samples which met the set criteria as compared to the controls (Figures 3, 4).
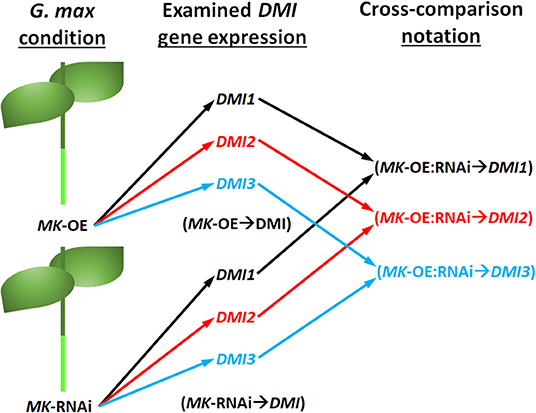
Figure 5. Flow chart of the cross comparison analyses made that used data presented in Figures 3, 4. The condition represents the MAPK (MK) -OE or -RNAi sample under study. The examined DMI gene expression represents the various examined DMI1, DMI2, or DMI3 paralogs. The black, red, and blue arrows and text were used to distinguish between the different DMI (DMI1, DMI2, and DMI3) genes. The notation of the cross comparisons took into consideration that data from both the MK-OE and MK-RNAi analyses were used in examining the DMI gene expression (i.e., MK-OE:RNAi → DMI).
Three instances involved decreased DMI RTAs as compared to the respective controls. The first instance was MAPK3-1-OE:RNAi → F0E0DMI2-9, that had about the same RTAs between the DMI2-9 -OE, and -RNAi samples and met the set criteria as compared to the controls (Figures 3, 4). The second instance was MAPK16-4-OE:RNAi → F0E0DMI29, that had lower RTAs in the MAPK16-4-RNAi samples, which met the set criteria as compared to the controls (Figures 3, 4). The third instance was MAPK16-4-OE:RNAi → F0E0DMI2-10, which had higher RTAs in the MAPK16-4-RNAi samples that met the set criteria as compared to the controls (Figures 3, 4).
The remaining DMI genes satisfied the set differential expression criteria in one of the two conditions (OE or RNAi) but not both, in the manner that Type 2 expression had been described in this pathosystem in McNeece et al. (2019) but for MAPKs (Figures 3, 4). In McNeece et al. (2019), Type 2 expression spanned the results of both OE and RNAi experiments. Type 2-OE was expression that was induced (in their RTA [FC]) in the OE treatment, and not differentially expressed in the RNAi treatment or was suppressed in the OE treatment, and not differentially expressed in RNAi treatment (McNeece et al., 2019). Within Type 2 expression, Type 2-RNAi had suppressed expression in the RNAi treatment while it was not differentially expressed in the OE treatment or was induced in the RNAi treatment and was not differentially expressed in the OE treatment (McNeece et al., 2019). An examination of MAPK-OE data was made that resulted in the identification that DMI1 had one case (DMI1-1) of an altered RTA but in a MAPK-OE (MAPK3-1-OE) sample, and which met the set criteria as compared to the controls (Figure 3). DMI2 had 9 cases involving DMI2-3, DMI2-4, DMI2-9, DMI2-10, and DMI2-11 of an altered RTA but in a MAPK-OE, and which met the set criteria as compared to the controls (Figure 3). DMI3 had 3 cases that involved DMI3-1, and DMI3-2 of an altered RTA but in a MAPK-OE, and which met the set criteria as compared to the controls (Figure 3). In contrast, an examination of MAPK-RNAi RNA-seq data was made. DMI1 had 2 cases (DMI1-1, and DMI1-2) of altered RTAs, that occurred in the MAPK4-1-RNAi sample, and which met the set criteria as compared to the controls (Figure 4). DMI2 had 28 cases that involved DMI2-2, DMI2-3, DMI2-4, DMI2-7, DMI2-8, DMI2-9, DMI2-10, DMI2-11, and DMI2-12 that had an altered RTA but occurred in an examined MAPK-RNAi sample, and which met the set criteria as compared to the controls (Figure 4). DMI3 had 9 cases that involved DMI3-1, and DMI3-2 that had an altered RTA but occurred in an examined MAPK-RNAi sample and met the set criteria as compared to the controls (Figure 4). These cases included the MAPK2-RNAi, MAPK3-2-RNAi, MAPK4-1-RNAi, MAPK5-3-RNAi, MAPK13-1-RNAi, MAPK16-4-RNAi, and MAPK20-2-RNAi samples (Figure 4). Other examples of differentially expressed DMI genes that were identified in the defense MAPK RNA-seq analyses existed, but they had not met the set FC criteria, and were not examined further (Supplementary Tables 7, 8). However, their expression could be important and potential targets of future analyses.
The Identification of MAPK-Altered DMI Expression to Below Experimental-Threshold Levels
The RTAs of DMI1-3, DMI2-7, and DMI3-2 used in the functional transgenic studies that examined H. glycines infection were identified in the MAPK-OE, and MAPK-RNAi RNA-seq experiments as compared to their respective transgenic pRAP15-ccdB and pRAP17-ccdB controls (Table 3 and Supplementary Tables 7, 8). A number of instances of statistically significant changes in DMI RTAs were noted but occurred below the set FC criteria. Therefore, while noteworthy were beyond the scope of further analysis here. Generally, the DMI RTAs that occurred in the RNA-seq analyses were lower than those found for the DMI1-3, DMI2-7, or DMI3-2 -OE or -RNAi roots in the RT-qPCR quality control analyses of the root RNA under comparison in the functional transgenic infection studies.
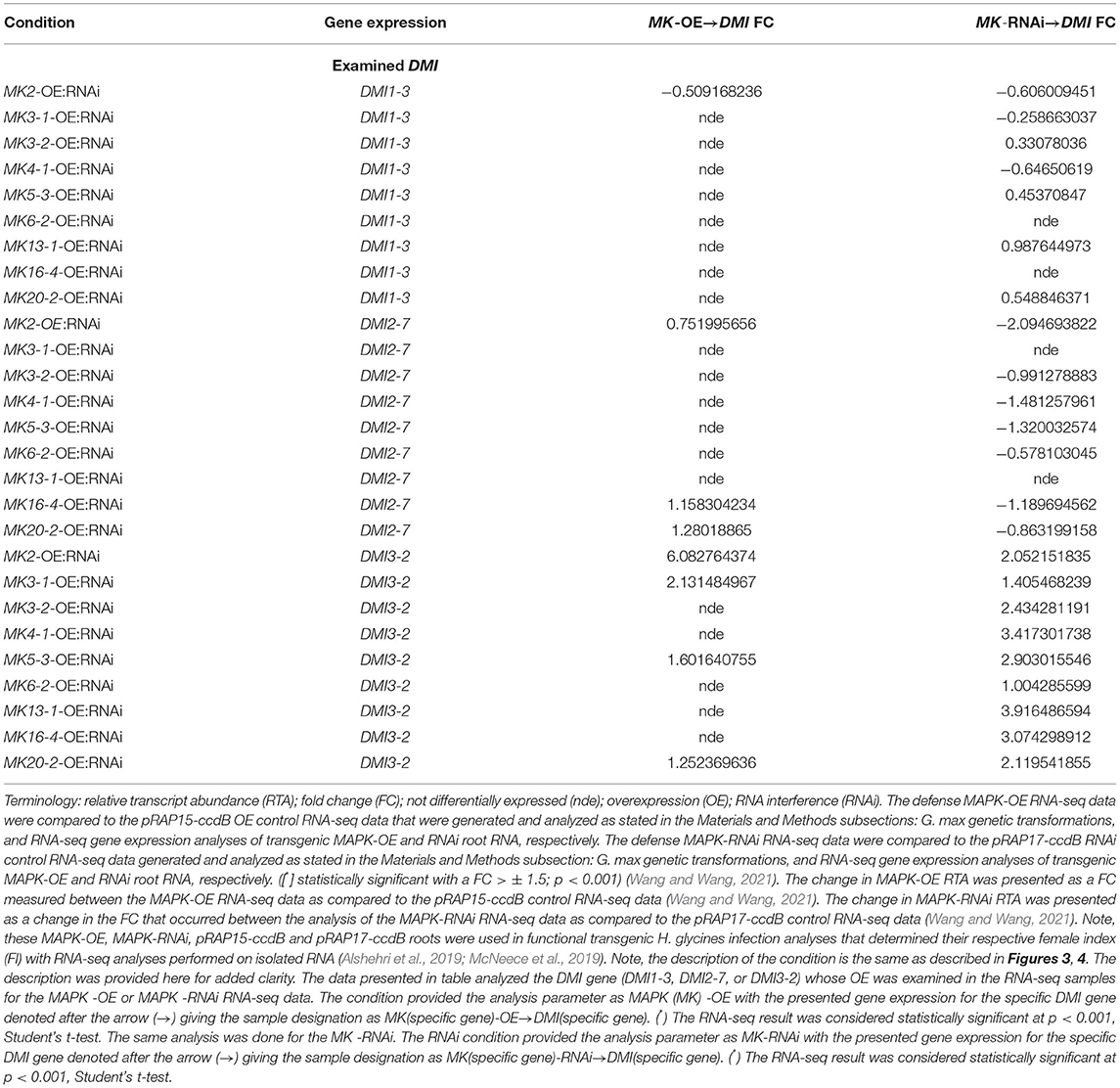
Table 3. RNA-seq differential expression measurements of G. max DMI-3, DMI2-7, and DMI3-2 gene family members in the defense MAPK-OE and MAPK-RNAi root RNA samples as compared to the pRAP15-ccdB (OE control) and pRAP17-ccdB (RNAi control) samples, p < 0.001 (Wang and Wang, 2021).
Altered MAPK3 RTAs Occur in the DMI1, DMI2, and DMI3 Transgenic Roots
The MAPK3 protein transduces signals from both PTI and ETI branches of defense signaling that leads to an output response that combats pathogen infection including parasitism by pathogenic nematodes. RT-qPCR experiments that targeted MAPK3-1 and MAPK3-2 were done that determined if genes functioning to transduce defense signaling through PTI and ETI were affected by OE, and/or RNAi of the syncytium-expressed DMI1-3, DMI2-7, and DMI3-2. Those results that employed RT-qPCR have been presented (Figure 6). The gene expression experiments showed DMI1-3-OE increased MAPK3-1 and MAPK3-2 RTA while, in contrast, DMI1-3-RNAi decreased MAPK3-1, and MAPK3-2 RTA as compared to the controls (Figure 6). DMI2-7-OE did not significantly affect MAPK3-1 or MAPK3-2 RTA, while DMI2-7-RNAi significantly decreased both MAPK3-1 and MAPK3-2 RTA as compared to the controls (Figure 6). DMI3-2 -OE increased MAPK3-1 RTA but had no effect on MAPK3-2 RTA as compared to the controls (Figure 6). DMI3-2-RNAi decreased MAPK3-1 RTA but had no effect on MAPK3-2 RTA as compared to the controls (Figure 6). The results indicate DMI1-3, DMI2-7, and DMI3-2 had functions in common with 2 major G. max defense genes (MAPK3-1, and MAPK3-2) that act in PTI and ETI, but that there are important differences possibly relating to the unique functions that the DMI genes have in relation to the respective MAPK3-1, and MAPK3-2 paralogs. The results provide clear evidence that DMI1-3, DMI2-7, and DMI3-2 influence the RTAs of MAPK genes that function downstream during PTI and ETI. The unique functions of the DMI paralogs as it relates to MAPK gene expression and gene expression in general are discussed subsequently.
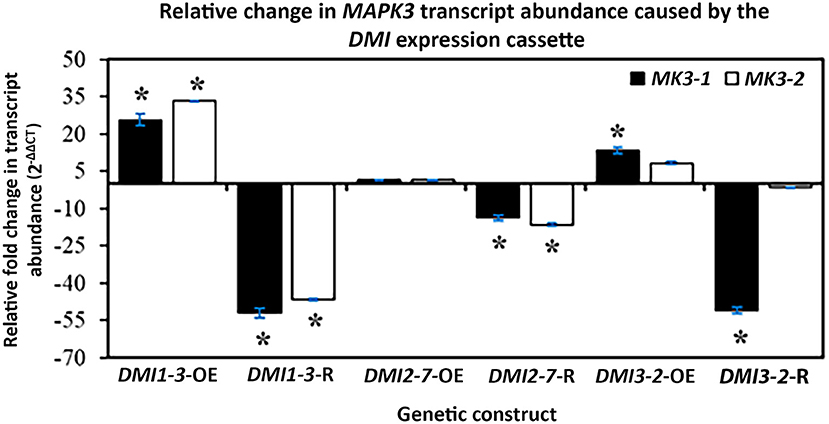
Figure 6. The effect that DMI1, DMI2, and DMI3 -OE or -RNAi had on the RTA, presented as FC, of MAPK3-1, and MAPK3-2, confirmed by RT-qPCR as calculated by 2−ΔΔCT (Livak and Schmittgen, 2001). (*) p < 0.05, Student's t-test (Yuan et al., 2006). Please refer to Materials and Methods subsection: Real-time quantitative PCR (RT-qPCR), for details.
Discussion
The capability of EPNs to impair symbioses indicates the molecular signaling machinery responses associated with symbiosis and pathogenicity are intertwined at some level. The knowledge of different DMI genes functioning at different points in the CSP and the expression of G. max homologs occurring within syncytia undergoing a defense response were a reasonable place to start an analysis. General aspects of plant signaling are accomplished through a number of basic pathways. Among these pathways is MAPK signaling, and several lines of evidence link symbiosis to MAPK signaling. Analyses in the G. max-H. glycines pathosystem demonstrate the importance of MAPK signaling to its defense response so it is examined further here in relation to the DMI genes.
Identification of G. max DMI Genes
The experimental analysis presented here explores a prior observation of DMI3 expression occurring in syncytia undergoing the defense process to H. glycines parasitism (Klink et al., 2010b). Analyses presented here reveal that the DMI3 identified in Klink et al. (2010b) is DMI3-2. The result indicates that genes acting in the CSP may also function in defense.. The experiments presented here focus in on a specific subset of CSP genes, DMI1, DMI2, and DMI3. G. max homologs of DMI1 (3 paralogs), DMI2 (12 paralogs), and DMI3 (2 paralogs), while splice variants are also identified. Analyses of other genomes of select agriculturally important crops identify their DMI1, DMI2, and DMI3 gene families, and splice variants. The examined genomes have between 1 and 7 (or more depending on stringency) DMI1 paralogs with the hexaploid T. aestivum having by far the most (7). The examined genomes have between 0 and 3 DMI2 and DMI3 paralogs with B. rapa (rape seed) and B. vulgaris (sugarbeet) having none. A. thaliana lacks DMI3, thought to explain one of the reasons why it cannot establish symbiosis with AM fungi (Zhu et al., 2006). This observation may also partially explain their absence in B. rapa and B. vulgaris. Splice variants of conserved genes are important in the plant defense process (Bazin et al., 2020). Alternative spliced transcripts are present within the syncytium and are an important feature that G. max employs in its defense response toward H. glycines infection (Lawaju et al., 2020; Klink et al., 2021b). The bioinformatics analyses presented here also identify numerous splice variants of the DMI genes. Some of the genomes, in particular T. aestivum, exhibit large numbers of predicted splice variants, their nature requiring functional experimentation to determine a biological role.
DMI Genes Are Expressed in Root Cells Undergoing a Defense Function
Since a goal here is to examine the function of the G. max DMI genes, comparisons to syncytium gene expression data leads to the confirmation of one paralog of each gene family being expressed within the syncytium undergoing the defense response. This observation allows for narrowing down the number of studied paralogs to one per gene family. However, the expression of a number of DMI genes are not determined because probe sets for those genes do not exist on those G. max microarrays and, thus, require further analysis beyond this works' scope. The functional examination of genes expressed in root cells undergoing defense to H. glycines is a sound approach to identify genes that function in impairing its parasitism (Matsye et al., 2011, 2012; McNeece et al., 2019; Klink et al., 2021a). Consequently, the DMI1-3, DMI2-7, and DMI3-2 genes have been cloned and engineered here for OE and RNAi into G. max in the targeted genotypes for functional transgenic studies as compared to their respective controls. Transgenic OE and RNAi of DMI1-3, DMI2-7, and DMI3-2 lead to the expected increase in their RTAs while their RNAi leads to their expected decreased RTAs. The effect that the DMI1-3, DMI2-7, and DMI3-2 OE and RNAi cassettes have on expression is further shown here to continue to occur at 6 dpi at a time point when the natural defense response has concluded. The effect that the DMI1-3, DMI2-7, and DMI3-2 OE and RNAi cassettes have on H. glycines parasitism as compared to their respective controls (discussed in the next section) are reflective of their expression within syncytia undergoing a defense response. Among the transgenic experiments, DMI1-3 OE leads to a 2.1-fold increase in root mass. This observation is not surprising since the stimulation of DMI1 and DMI2 signaling in M. truncatula results in an increase in lateral root formation (Oláh et al., 2005).
Functional Transgenic Analyses Reveal a Defense Role for G. max DMI Genes
The functional transgenic studies are the OE and RNAi of the DMI1-3, DMI2-7, and DMI3-2 genes as compared to their relevant transgenic controls. The analyses then follow infection by H. glycines during a 30 day time course, leading to the quantification of their effect on parasitism as measured through the FI. The expected experimentally-affected increase in DMI RTA in the OE roots is observed as compared to the controls, as demonstrated by RT-qPCR of RNA sample template isolated from in the H. glycines-susceptible G.max[Williams82/PI518671]. In contrast, a decrease in DMI RTA is observed as compared to the controls, as demonstrated by RT-qPCR using RNA template isolated from the roots of the DMI-RNAi root RNA of the H. glycines-resistant G.max[Peking/PI548402] (Livak and Schmittgen, 2001). These outcomes provide the set criteria required for the infection of transgenic DMI1-3, DMI2-7, and DMI3-2 -OE and -RNAi roots and respective controls by H. glycines. Hundreds of gene constructs, experimented on in this pathosystem once these criteria have been met, include genes functioning in the PTI and ETI defense branches which converge on the MAPK signaling platform (McNeece et al., 2019; Klink et al., 2021a). The experimentally-induced expression of G. max PTI components including BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) BAK1-1, and BOTRYTIS INDUCED KINASE1 (BIK1), BIK1-6, and ETI components including the bacterial effector harpin, NON-RACE SPECIFIC DISEASE RESISTANCE 1 (NDR1) NDR1-1, and the nitrate-induced (NOI) domain-containing, intrinsically disordered, molecular recognition feature (MoRF)-containing RPM1-INTERACTING PROTEIN 4 (RIN4) RIN4-4 all increase MAPK3-1 and MAPK3-2 RTA and function in the defense response that G. max has to H. glycines parasitism (Wei et al., 1992; Century et al., 1995, 1997; Gopalan et al., 1996; Li and Chory, 1997; Desikan et al., 2001; Li et al., 2002; Mackey et al., 2002, 2003; Day et al., 2006; Veronese et al., 2006; Pant et al., 2014; Sun et al., 2014; Aljaafri et al., 2017; McNeece et al., 2017, 2019; Klink et al., 2021a). The results of enumerating H. glycines from the transgenic roots demonstrates that the OE of DMI1-3, DMI2-7, and DMI3-2 genes leads to a significant decrease in H. glycines parasitism as compared to the pRAP15-ccdB OE control in the otherwise H. glycines-susceptible G.max[Williams82/PI518671] genetic background. In contrast, RNAi of DMI1-3, DMI2-7, and DMI3-2 genes, leads to an increase in H. glycines parasitism as compared to the transgenic pRAP17-ccdB RNAi control in the otherwise normally H. glycines-resistant G.max[Peking/PI548402] genetic background. The combination of decreased parasitism in the otherwise normally H. glycines-susceptible genetic background G.max[Williams82/PI518671] and increased parasitism in the otherwise normally H. glycines-resistant G.max[Peking/PI548402] genetic background are taken as evidence that the gene functions in the defense response because of the ability to obtain these opposite outcomes that are caused by target gene OE and RNAi (Pant et al., 2014; Sharma et al., 2016; McNeece et al., 2019; Klink et al., 2021a). DMI1, DMI2, and DMI3 function at different stages of the CSP (Catoira et al., 2000; Ané et al., 2002, 2004). This observation is similar to those made by McNeece et al. (2019) for the MAPK defense signaling pathway. Therefore, the CSP, containing the DMI genes, appears to function in complex ways during the defense process and whose understanding may benefit by examining their gene regulation in relation to MAPK signaling (Francia et al., 2011).
The determination that the G. max DMI genes function in complex ways began with the observation that a large difference exists in the DMI1-3-OE pg FI analysis as compared to the wr analysis. As stated, these observations are not surprising since stimulation of DMI1 and DMI2 signaling in M. truncatula results in an increase in lateral root formation which would be expected to lead to an increase in root mass (Oláh et al., 2005). The observation that the CCaMK DMI3 has a defense role is consistent with observations that a G. max calmodulin (Glyma.19G068300) also functions in the defense process (Matthews et al., 2013). With the demonstration that DMI1-3, DMI2-7, and DMI3-2 genes function in defense, further characterization of the genes began in order to understand the possible function(s) that they may have in signaling.
MAPKs and Symbiosis
Lupinus albus (white lupine) is capable of undergoing a salt-induced stress response that leads to the expression of MAPKs (Fernandez-Pascual et al., 2006). Some of these MAPKs are activated (phosphorylated) after host infection with the nodule-inducing Bradyrhizobium sp. (Fernandez-Pascual et al., 2006). The MAPK activation process happens in the root's infection zone and is impaired by MAPK inhibitors, indicating a positive role that MAPKs have in this nodulation process (Fernandez-Pascual et al., 2006). MAPKK inhibitors also alter the nodulation pattern (Fernandez-Pascual et al., 2006). The altered nodulation includes decreasing the number and mass of nodules in the upper root while increasing them in the lower register of the root zone, with MAPK inhibition blocking early infection events and delaying the nodule developmental process (Fernandez-Pascual et al., 2006).
In contrast, results demonstrate that MAPK signaling can also interfere with nodulation. The M. truncatula MtMKK5-MtMPK3/6 signaling module impairs the early symbiotic nodule formation process, and it is believed the MAPK system's role occurs upstream of ERF Required for Nodulation 1 (ERN1) and Nod factor Signaling Pathway 1 (NSP1) (Ryu et al., 2017). MtMKK5 OE in M. truncatula stimulates stress and defense signaling pathways, reducing nodule formation (Ryu et al., 2017). Furthermore, treatment with the MAPK U0126 inhibitor enhances nodule formation while increasing the RTA of the early M. truncatula Nodule Inception nodulation (MtNIN) (Ryu et al., 2017). MtMKK5 directly activates MtMPK3/6, promoting their physical interaction with the early nodulation ERN1 and NSP1 transcription factors (Ryu et al., 2017). The Sinorhizobium sp. strain NGR234 Nodulation outer protein L (NopL) is a Rhizobium-specific type 3 effector that acts in nodulation (Zhang et al., 2011). Treatment of Phaseolus vulgaris (common bean) with strain NGRΩnopL having a mutated nopL leads to the development of ineffective, senescing nodules, indicating that NopL antagonizes nodule senescence (Zhang et al., 2011). NopL interferes with MAPK signaling in Nicotiana tabacum (tobacco), suppressing apoptosis induced by the OE of the MAPK gene Salicylic acid-induced protein kinase (SIPK) OE or constitutive activity caused by a SIPK(DD) (mutation in the TXY motif) (Zhang et al., 2011). NopL acts by mimicking a MAPK substrate and by doing so, impairs MAPK signaling which results in suppressing premature nodule senescence (Zhang et al., 2011). A second type 3 secreted effector, the E3 ubiquitin ligase NopM, is shown through In planta NopM expression in N. tabacum to induce cell death, stimulating effector-triggered immunity responses and NopM expression in L. japonicus reduces nodule formation (Xu et al., 2018). Phosphorylation of NopM by the salicylic acid-induced protein kinase (NtSIPK) MAPK occurs in vitro (Xu et al., 2018). In L. japonicus, Symbiosis receptor-like kinase (SymRK) mediates the legume-Rhizobium symbiosis (Yin et al., 2019). SymRK is the ortholog of DMI2, interacting with NFR1 and NFR5 in promoting NF signaling. The MAPKK SIP2 is a SymRK-interacting protein that positively regulates nodule organogenesis (Yin et al., 2019). This result directly links the DMI2 membrane receptor to MAPK signaling. Furthermore, LjMPK6 is a phosphorylation target of SIP2 (Yin et al., 2019). LjMPK6 RNAi decreases nodule primordia (NP) and nodule number, while LjMPK6-OE increases nodule, infection threads (ITs), and NP numbers demonstrating a positive role for LjMPK6 in nodulation (Yin et al., 2019). Further exploration of the LjMPK6 phosphorylation in relation to nodulation identify the LjMPK6-interacting type 2C protein phosphatase, LjPP2C, which dephosphorylates LjMPK6 in vitro and in vivo in, functioning to fine-tune nodule development after rhizobia inoculation (Yan et al., 2020). Analysis of hairy root transformed plants demonstrate a non-phosphorylatable mutant LjMPK6 (T224A Y226F) mimics LjPP2C, illustrating LjPP2C phosphatase dephosphorylates LjMPK6 to fine tune nodule development in L. japonicus (Yan et al., 2020). The upstream SIMK activator SIMK kinase (SIMKK) infected with Sinorhizobium meliloti show in SIMKK RNAi lines exhibiting downregulation of both SIMKK and SIMK, having reduced root hair growth while also having a decrease in ability to form infection threads and nodules (Hrbáčková et al., 2021). Constitutive SIMK OE promotes root hair growth, infection thread, and nodule clustering (Hrbáčková et al., 2021). The results are consistent with the involvement of DMI genes in G. max in AM symbioses (Navazio et al., 2007). Furthermore, the results identify a link between DMI genes and MAPKs.
Phosphoproteomic analyses made in conjunction with gene expression studies of NF-induced control and dmi3 mutants identify gene expression changes and phosphorylation events correlating with nodulation (Rose et al., 2012). Consequently, not only do the DMI genes function at specific stages of the CSP, but also govern gene expression. A dmi3-mediated feedback loop also exists and is present for interactions involving the response to symbiotic AM fungi (Rose et al., 2012). DMI3 plays a role in the epidermis and cortex during symbiosis (Rival et al., 2012). DMI functions in the activation of downstream genes that act in symbiosis (Czaja et al., 2012). Rhizobia and purified NFs induce the expression of M. truncatula DMI3 leading to the expression of AM symbiosis and nodulation ATP-binding cassette (ABC) B-type transporters including ABCB for mycorrhization and nodulation1-3 genes (AMN1, AMN2, and AMN3) (Roy et al., 2021). DMI is also regulated by plant hormones, including abscisic acid (ABA) (Ni et al., 2019). In O. sativa, ABA inhibits rice protein phosphatase PP45 which occurs through H2O2 leading to relieving DMI3 repression (Ni et al., 2019). The inactivation of DMI3 by PP45 occurs by dephosphorylating Thr-263 in DMI3. ABA-induced H2O2 production occurring through the NADPH oxidases inhibits the PP45 activity of PP45 by inhibiting PP45 expression and by oxidizing Cys-350 and Cys-428 residues, forming PP45 intermolecular dimers, blocking the interaction between PP45 and DMI3, preventing PP45-mediated DMI3 activity inhibition. DMI3 also plays roles in the regulation of genes whose expression is important to nodulation, including CCAAT-box-binding transcription factors (Laloum et al., 2014). Other downstream genes include M. truncatula auxin resistant 1/like aux1 (AUX1/LAX) influx carriers, LAX (MtLAX), M. truncatula pin-formed (PIN) efflux carriers (MtPIN), and M. truncatula MtABCB gene families (Shen et al., 2015). These genes, characterized in roots and shoots, indicate that three representative auxin transporters (MtLAX3, MtPIN7, and MtABCB1) have their plasma membrane localizations up-regulated in the roots by Sinorhizobium meliloti infection in the wild type (WT) but down-regulated in both the roots and shoots of dmi3 (Shen et al., 2015). The impairment of symbiosis by EPNs indicates the processes are antagonistic. Gene expression studies would provide some evidence supporting the hypothesis. The expression of the G. max DMI3-2 in syncytia experiencing defense is consistent with the observation that symbiosis genes can also have a dual function in defense (Mitra et al., 2004; Klink et al., 2010b). The experiments presented here place those observations into the context of gene defense function, gene expression, and gene co-regulation.
MAPKs Regulate DMI Gene Expression to Levels Below the Cutoff Threshold
An examination of the RNA-seq data identifies a lower threshold of MAPK-regulated DMI expression occurring at a statistically significant level for each DMI gene. This description focuses in on the syncytium-expressed DMI1-3, DMI2-7, and DMI3-2 genes. RNA-seq analyses show DMI1-3 RTAs are affected to statistically-significant levels by RNAi of MAPK3-1 (FC,−0.6060), MAPK3-2 (FC,−0.2587), MAPK4-1 (FC, 0.3308), MAPK5-3 (FC,−0.6466), MAPK13-1 (FC, 0.9876), and MAPK20-2 (FC, 0.5488) while none of the MAPK overexpressing roots affect DMI1-3 RTAs. Low levels of defense gene expression, including syncytium-expressed conserved oligomeric Golgi (COG) complex members, components of the exocyst, and various other genes are reported (Niraula et al., 2020; Sharma et al., 2020; Klink et al., 2021b).
For DMI2-7, MAPK2-RNAi leads to a decrease in RTA, confirmed by RT-qPCR and already discussed. However, RNA-seq analyses show that MAPK2-OE leads to a statistically significant increase in DMI2-7 RTA (FC, 0.7520). Therefore, MAPK2 OE leads to an increase in DMI2-7 RTA while its RNAi decreases its RTA. DMI2-7 is also affected by MAPK16-4-OE (FC, 1.1583) and MAPK16-4-RNAi (FC,−1.1897) MAPK20-2-OE (FC, 1.2802) and MAPK20-2-RNAi (FC,−0.8632). However, while MAPK3-1, MAPK3-2, MAPK4-1, and MAPK6-2 do not appear to affect DMI2-7 RTAs, expression of their RNAi cassettes appear to have a negative impact. For example, RNAi of MAPK3-2 (FC,−0.9913), MAPK4-1 (FC,−1.4813), MAPK5-3 (FC,−1.3200), and MAPK6-2 (FC,−0.5781) appear to decrease DMI2-7 RTAs.
For DMI3-2, the OE of MAPK2, and MAPK5-3 and -RNAi genetic cassettes have opposite effects on their RTA as described. Other effects on DMI3-2 RTA are caused by MAPK20-2-OE (FC, 1.2524), meaning its OE and RNAi have opposite effects as well. Furthermore, MAPK3-1 RNAi increases DMI3-2 RTA (FC, 1.4055), but not as much as MAPK3-1 OE. While MAPK6-2-OE has no effect on DMI3-2 RTA, its RNAi increases its RTA (FC, 1.0043).
MAPK3 As It Relates to DMI Expression
MAPK3 is one of the most studied MAPKs as it relates to plant defense (Desikan et al., 1998; Asai et al., 2002; McNeece et al., 2019). The expression of each of the G. max MAPK3 paralogs (MAPK3-1, and MAPK3-2) in each of the DMI1-3, DMI2-7, and DMI3-2 overexpressing and RNAi lines are examined here to determine whether a relationship exists between them. The results of those analyses show that DMI1-3 overexpressing roots have an increase in transcript abundance for MAPK3-1, and MAPK3-2. The results of those analyses also show that DMI1-3 RNAi roots have a decrease in RTA for MAPK3-1, and MAPK3-2. Consequently, it is evident that altered DMI1-3 expression leads to modulated MAPK3-1, and MAPK3-2 expression, linking the two pathways from a transcriptomic level. As stated earlier, DMI1 is a nuclear membrane protein that is necessary for the initiation of NF-induced Ca2+ spiking response in root hairs and functions upstream of DMI3, NSP1, and NSP2 (Wais et al., 2000).
An examination of the DMI2-7-OE RNA shows no effect on the RTA for MAPK3-1, and MAPK3-2. In contrast, DMI2-7 RNAi roots have a decrease in RTA for MAPK3-1, and MAPK3-2. Therefore, it is clear that decreased DMI2-7 RTAs leads to suppressed MAPK3-1, and MAPK3-2 expression in the RNAi roots, while no effect is observed in the DMI2-7-OE roots. These results also link the DMI and MAPK3 pathways from a transcriptomic standpoint. The observation that DMI2-7 OE does not affect MAPK3-1 or MAPK3-2 RTAs is noteworthy. It is possible that the maximum number of DMI2-7 plasma membrane receptors are already present, achieving maximum sensitivity. Consequently, a further increase through OE does not affect downstream MAPK3-1 or MAPK3-2 levels, indicating a possible negative feedback regulation in this case.
An examination of the DMI3-2 -OE RNA shows an increase of the RTA, but only for MAPK3-1 while MAPK3-2 is not affected. In contrast, an examination of the DMI3-2-RNAi RNA shows a decrease of the RTA, but only for MAPK3-1 while MAPK3-2 is not affected. This result demonstrates that there is a level of specificity in relation to the direction of influence that the DMI pathway has on defense. McNeece et al. (2019) demonstrates that the plasma membrane protein NDR1, also known as HARPIN INDUCED1 (HIN1), which can become increased in its RTA by the bacterial effector harpin, leads to co-regulated expression occurring between MAPK3-1, and NDR1. Furthermore, MAPK3-1 OE leads to increased RTA of the Rhg4 gene serine hydroxymethyltransferase, galactinol synthase (GS-3), reticuline oxidase (RO-40), and xyloglucan endotransglycosylase/hydrolase 43 (XTH43) (McNeece et al., 2019). Each of these genes function effectively in the defense response that G. max has toward H. glycines (Matthews et al., 2013; Pant et al., 2014; Klink et al., 2017). In the case of XTH43, it functions to shorten xyloglucan chains, create more of those shorter chains, and make more xyloglucan (Niraula et al., 2021). The result provides actual biochemical support to the histological and ultrastructural observations that the cell walls do not expand during the defense response which normally happens during a susceptible response (Ross, 1958; Endo and Veech, 1970; Gipson et al., 1971; Riggs et al., 1973; Endo, 1991; Niraula et al., 2021).
The results presented here demonstrate that crosstalk occurs between the DMI-containing CSP and MAPK pathways. These results are significant because they point to the possible interaction occurring between the CSP and defense signaling, two ancient pathways found in land plants, and it is already known that PTI and ETI function through MAPKs in the G. max-H. glycines pathosystem (Figure 7). The results provide a genetic basis to further examine the CSP. Functional experiments aimed at examining all of the DMI1, DMI2, and DMI3 paralogs would provide greater clarity on the roles of these genes in these processes. Determining whether DMI2 functions analogously as a co-receptor for NFR1 and NFR5 which bind NFs much like BAK1 does to various PRRs including FLAGELLIN-SENSITIVE 2 (FLS2), EF-Tu RECEPTOR (EFR) and the DAMP PEPTIDE 1 RECEPTOR (At-PEPR1) during defense processes is of interest (Li and Chory, 1997; Veronese et al., 2006; Zipfel et al., 2006; Chinchilla et al., 2007; Zhang et al., 2010; Liu et al., 2013). Like what has already been shown for H. glycines defense in G. max, an examination of other genes in the nodulation or CSP pathways would allow for greater resolution in understanding the defense process (Klink et al., 2021a). Lastly, transcriptomic analyses including RNA-seq of DMI1, DMI2, and DMI3 OE and RNAi roots may also provide greater resolution in understanding the intricacies of the roles these genes play in the defense process. The results are consistent with experiments that show symbiosis-inducing organisms (AM) are capable of propagating Ca2+ spikes in G. max while also inducing the expression of heterologously expressed M. truncatula DMI1, DMI2, and DMI3 (Navazio et al., 2007). Recent experiments show a reduction of 59-81% in cysts in AM-inoculated soybean, consistent with the results presented here where molecular context is provided (Pant et al., 2014; Aljaafri et al., 2017; McNeece et al., 2017, 2019; Pawlowski and Hartman, 2020; Klink et al., 2021a). An examination of other CSP genes and those that are not as well-conserved will provide additional, needed detail. Both defense and symbiosis processes involve the generation of Ca2+ signaling with defense involving cytoplasmic signals and symbiosis involving nuclear signals. The results presented here indicate the function and impact of the CSP regarding the interconnectedness of symbiosis and defense is far greater that previously appreciated. Part of its impact will be generating symbiotic interactions and defense processes where they currently do not exist or are not effective. Consequently, a functional transgenic examination of all DMI1-3 paralogs and additional CSP genes in relation to defense and symbiosis are of interest.
Figure 7. Model. (A) The plasma membrane (PM) receptor DMI2 which interacts with HMG-CoA reductase (3-hydroxy-3-methyl-glutaryl-coenzyme A reductase) (HMGR), and Nod factor receptors (NFR1 and NFR5), propagate signals that migrate either directly through the nuclear membrane or through nucleoporins (N), leading to the generation of calcium waves that involve the Type IIA calcium ATPase MCA8, and necessary to activate calmodulin and DMI3. The process concludes with an effective Rhizobium-legume symbiosis (RLS). The signaling process involves the engagement of the potassium channel DMI1. RNA-seq analyses, confirmed by RT-qPCR experiments, highlighted that some of the MAPKs undergoing transgenically-altered expression affects DMI RTAs as presented as their FC. Some of the involved MAPKs were determined to be expressed in syncytia undergoing the defense response (asterisk [*]) (McNeece et al., 2019). *expressed in syncytia undergoing a defense response. (adapted from Genre and Russo, 2016). (B) The G. max CSP, PTI, and ETI pathways converge on MAPK signaling, leading to downstream processes that function in defense to H. glycines parasitism. The hashed arrow between the CSP and PTI and ETI receptors is positioned because questions remain whether they directly influence their relative abundances (?).
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA664992.
Author Contributions
RK, NA, and VK designed experiments, performed experiments, ran analyses, and wrote manuscript. SP, KS, PN, BL, and KL performed experiments, ran analyses, and wrote manuscript. All authors contributed to the article and approved the submitted version.
Funding
VK: Cotton Incorporated (https://www.cottoninc.com/), grants 17-603, 19-603; Special Research Initiative, College of Arts and Sciences, Mississippi State University. KL: Hatch number: ALA015-2-14003. Mississippi Agricultural and Forestry Experiment Station (MAFES)-Special Research Initiative (SRI) grant.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
RK is thankful to his Ph.D. committee members Nancy Reichert and Dr. Ying Wang of the Department of Biological Sciences and Jiaxu Li and Shien Lu at the Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology at Mississippi State University who without their support the completion of this work would not be possible. Furthermore, RK and VK are thankful to Gary Lawrence (retired), Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology at Mississippi State University for all of his support over the years. Robert Nichols and Kater Hake are thanked for their support. Brant, McNeese and Yixiu (Jan) Pinnix are thanked for their support and as a lab member and technical staff, respectively. Jeff Dean, Chair, Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology at Mississippi State University is thanked for his generosity of providing greenhouse, head house, storage, and field space for the experiments and maintenance of plant stocks. The authors thank George Hopper, Wes Burger, Scott Willard, and Reuben Moore, Mississippi Agricultural and Forestry Experiment Station (MAFES), and Mississippi State University for support. VK has great gratitude to Giselle Thibaudeau, Mississippi State University for numerous conversations regarding research and approach. VK also has great gratitude to Greg Bohach, Mississippi State University for his time and willingness to go out of his way to help. VK thanks Brian Baldwin, Department of Biochemistry, Molecular Biology, Entomology and Plant Pathology at Mississippi State University for encouragement. VK thanks Mary Celeste Reese, Tom Holder and Donna Gordon, Department of Biological Sciences, Mississippi State University, whom without their discussions the work would not have been possible in its ultimate state. Undergraduate student help from the Department of Biological Sciences at Mississippi State University included Alexandra Rose, Courtney Borgognoni, and Holeh Heydari whose efforts were instrumental to the analysis. VK thanks James McCain, Melissa Davis, Cindy Palmer, Sara Burrell, Dragic Jeremic, Reynold Tan, Rob Allender, Russell Putnam, and Tom Clifton for their obvious contributions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.842597/full#supplementary-material
Supplementary Figure 1. Transgenic roots obtained from 6 dpi H. glycines-infected chimeric plants having genetically engineered roots and un-engineered shoots that were generated through the K599-mediated genetic transformation procedure. Please refer to Materials and Methods subsections: DMI gene cloning, and G. max genetic transformations, for details. FC was calculated by 2−ΔΔCT (Livak and Schmittgen, 2001; Klink et al., 2021a). The RTA of the candidate defense genes, presented as a FC, in the transgenic roots was compared using the G. max ribosomal protein gene RPS21 (Glyma.15G147700). The RT-qPCR analyses examined 3 experimental replicates (individual root systems) of DMI1-3, DMI2-7, and DMI3-2 -OE or -RNAi roots as compared to their pRAP15-ccdB, and pRAP17-ccdB controls, respectively, from each of the 3 biological replicates. Each experimental replicate was run in triplicate using the same RNA. (*), statistical significance of p < 0.05, Student's t-test (Yuan et al., 2006). Please refer to Materials and Methods subsection: Real-time quantitative PCR (RT-qPCR), for details.
Supplementary Table 1. PCR, and RT-qPCR primers used in the analysis.
Supplementary Table 2. Raw DMI DCM data. Each experiment was replicated independently 3 times. Consequently, each experiment included three independent biological replicates. The cell (pericycle control, or syncytium) samples, obtained at different times have been presented as their respective time point (dpi). The three independent biological replicates included RNA samples isolated from the collected cells for each uninfected control (0 dpi), and H. glycines-infected (3, and 6 dpi) sample for each G. max genotype (G.max[Peking/PI548402], and G.max[PI88788]). The replicated experiments, enacted at different times, were performed using three different microarrays (arrays) per H. glycines-resistant G. max genotype. These H. glycines-resistant G. max genotypes included G.max[Peking/PI548402], and G.max[PI88788]. Red, gene expression was measured (M); blue, gene expression was not measured (NM); gray, not applicable (n/a) because no probe set existed on the microarray to measure expression for that gene (Klink et al., 2010a). The DCM analysis that led to the results occurred using data derived from the three independent replicates with a statistical significance threshold of p < 0.05, Mann–Whitney–Wilcoxon (MWW) Rank Sum Test (Mann and Whitney, 1947). The genes highlighted in yellow were studied subsequently in the functional transgenic experiments (presented in Figures 1, 2). Accession 1: Wm82.a1.v1.1 (2010) annotation. Annotation 2: Wm82.a2.v1 (2015) annotation. Please refer to Materials and Methods subsection: The identification of DMI gene families and root cell-specific expression, for further details.
Supplementary Table 3. Summarized G. max DMI genes expressed in syncytia. Gene families and syncytium expression profiles were presented in Supplementary Table 2. The DMI protein family members were identified from the G. max genome through conceptual translation of their gene sequences according to the procedures outlined in the Materials and Methods subsection: Selection of candidate genes. The accessions had represented the gene identifier obtained from the G. max genome. The Accession 1 identifier was obtained from the later 2015 (Wm82.a2.v1) annotation. The Accession 2 identifier was obtained from the earlier 2010 (Wm82.a1.v1.1) annotation. Annotation 2 relates directly to the annotation used in the Klink et al. (2010a,b) Affymetrix® gene expression study employed to generate the gene expression data for the syncytium undergoing the defense process. Table header description: Affymetrix® p/s, probe set identifier relating to the probe set that was fabricated onto the microarray and used to generate the gene expression data. Annotation, the G. max genome annotation of the gene. Time points, the time point (dpi), of H. glycines infection at which time the G. max root cells was collected by LM with their isolated RNA used for the gene expression study. Homologs identified to be expressed in syncytia undergoing the defense response, and not in control cells, studied in the functional transgenic analyses was highlighted in yellow. Blue, expression not measured (NM) to statistically significant levels, p ≥ 0.05. Red, expression measured (M) to statistically significant level, p < 0.05. The statistical significance (*) was calculated using the Mann–Whitney–Wilcoxon (MWW) Rank-Sum Test, p < 0.05 (Mann and Whitney, 1947). Please refer to Materials and Methods subsection: The identification of DMI gene families and root cell-specific expression, for details.
Supplementary Table 4. DMI1 homologs that exist in select agriculturally important crops (Tilman et al., 2011; Ray et al., 2013, 2019). Values not in parentheses have a Blast of e-0. Numbers in parentheses have a Blast cutoff of > e-0. Please refer to Materials and Methods subsections: G. max DMI gene identification, and Proteome mining, for details.
Supplementary Table 5. DMI2 homologs that exist in select agriculturally important crops (Tilman et al., 2011; Ray et al., 2013, 2019). Values not in parentheses have a Blast of e-0. Numbers in parentheses have a Blast cutoff of > e-0. Please refer to Materials and Methods subsections: G. max DMI gene identification, and Proteome mining, for details.
Supplementary Table 6. DMI3 homologs that exist in agriculturally important crops (Tilman et al., 2011; Ray et al., 2013, 2019). Values not in parentheses have a Blast of e-0. Numbers in parentheses have a Blast cutoff of > e-0. Please refer to Materials and Methods subsections: G. max DMI gene identification, and Proteome mining, for details.
Supplementary Table 7. DMI gene expression in defense MAPK-overexpressing (MAPK-OE) root RNA as compared to the pRAP15-ccdB control root RNA. RNA-seq data whose differential expression is presented as an RTA that represents FC; obtained from Alshehri et al. (2019) (p < 0.001) (Wang and Wang, 2021). Please refer to Materials and Methods subsection: RNA-seq gene expression analyses of transgenic MAPK-OE and -RNAi root RNA, for details.
Supplementary Table 8. DMI gene expression in defense MAPK-RNA interference (MAPK-RNAi) root RNA as compared to the pRAP17-ccdB control root RNA. RNA-seq data whose differential expression was calculated as an RTA has been presented as FC; obtained from Alshehri et al. (2019) (p < 0.001) (Wang and Wang, 2021). Please refer to Materials and Methods subsection: RNA-seq gene expression analyses of transgenic MAPK-OE and -RNAi root RNA, for details.
References
Aljaafri, W. A. R., McNeece, B. T., Lawaju, B. R., Sharma, K., Niruala, P. M., Pant, S. R., et al. (2017). A harpin elicitor induces the expression of a coiled-coil nucleotide binding leucine rich repeat (CC-NB-LRR) defense signaling gene and others functioning during defense to parasitic nematodes. Plant Physiol. Biochem. 121, 161–175. doi: 10.1016/j.plaphy.2017.10.004
Alshehri, H. A., Alkharouf, N. W., Darwish, O., McNeece, B. T., and Klink, V. P. (2019). MAPKDB: A MAP kinase database for signal transduction element identification. Bioinformation 15, 338–341. doi: 10.6026/97320630015338
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ané, J. M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E. D., Ayax, C., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367. doi: 10.1126/science.1092986
Ané, J. M., Lévy, J., Thoquet, P., Kulikova, O., De Billy, F., Penmetsa, V., et al. (2002). Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol. Plant-Microbe Interact. 15, 1108–1118. doi: 10.1094/MPMI.2002.15.11.1108
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Atkinson, H. J., and Harris, P. D. (1989). Changes in nematode antigens recognized by monoclonal antibodies during early infections of soya beans with the cyst nematode Heterodera glycines. Parasitology 98, 479–487. doi: 10.1017/S0031182000061576
Balasubramanian, M., and Rangaswami, G. (1962). Presence of indole compound in nematode galls. Nature 194, 774–775. doi: 10.1038/194774a0
Barker, K. R., Huisingh, D., and Johnston, S. A. (1972). Antagonistic interaction between Heterodera glycines and Rhizobium japonicum on soybean. Phytopathology 62, 1–1205. doi: 10.1094/Phyto-62-1201
Barker, K. R., Lehman, P. S., and Huisingh, D. (1971). Influence of nitrogen and Rhizobium iaponicum on the activity of Heteroidera glycines. Nematologica 17, 377–385. doi: 10.1163/187529271X00602
Barton, E. S.. (1892). On malformations of Ascophyllum and Desmarestia. Phycol. Mem. London 1892, 21–24.
Bazin, J., Mariappan, K., Jiang, Y., Blein, T., Voelz, R., Crespi, M., et al. (2020). Role of MPK4 in pathogen-associated molecular pattern-triggered alternative splicing in Arabidopsis. PLoS Pathog. 16, e1008401. doi: 10.1371/journal.ppat.1008401
Bernard, R. L., and Cremeens, C. R. (1988). Registration of ‘Williams 82' Soybean. Crop Sci. 28, 1027–1028. doi: 10.2135/cropsci1988.0011183X002800060049x
Bethlenfalvay, G. J., Brown, M. S., Mihara, K. L., and Stafford, A. E. (1987). Glycine-glomus-rhizobium symbiosis: V. Effects of mycorrhiza on nodule activity and transpiration in soybeans under drought stress. Plant Physiol. 85, 115–119. doi: 10.1104/pp.85.1.115
Bethlenfalvay, G. J., Brown, M. S., and Stafford, A. E. (1985). Glycine-Glomus-Rhizobium Symbiosis: II. Antagonistic effects between mycorrhizal colonization and nodulation. Plant Physiol. 79, 1054–1058. doi: 10.1104/pp.79.4.1054
Bhattacharyya, S., Dey, N., and Maiti, I. B. (2002). Analysis of cis-sequence of subgenomic transcript promoter from the Figwort mosaic virus and comparison of promoter activity with the cauliflower mosaic virus promoters in monocot and dicot cells. Virus Res. 90, 47–62. doi: 10.1016/S0166-0934(02)00146-5
Bird, D. M., and DiGennaro, P. M. (2012). “Nematode Communication with Plants is Surprisingly Complex and Multidimensional,” in Biocommunication of Plants, eds G. Witzany and F. Baluška (Berlin: Springer Berlin Heidelberg) 213–230. doi: 10.1007/978-3-642-23524-5_12
Boerner, L. K.. (2019). Industrial ammonia production emits more CO2 than any other chemical-making reaction. Chem. Eng. News 97, 18–24.
Brundrett, M. C.. (2002). Coevolution of roots and mycorrhizas of land plants. New Phytol. 154, 275–304. doi: 10.1046/j.1469-8137.2002.00397.x
Burkhead, T. R., and Klink, V. P. (2018). American agricultural commodities in a changing climate. AIMS Agri. Food 3, 406–425. doi: 10.3934/agrfood.2018.4.406
Byrd, D. W., Kirkpatrick, T., and Barker, K. R. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142–143.
Cardenas, L., Feijo, J. A., Kunkel, J. G., Sanchez, F., Holdaway-Clarke, T., Hepler, P. K., et al. (1999). Rhizobium nod factors induce increases in intracellular free calcium and extracellular calcium influxes in bean root hairs. Plant J. 19, 347–352. doi: 10.1046/j.1365-313X.1999.00517.x
Catoira, R., Galera, C., De Billy, F., Penmetsa, R. V., Journet, E. P., Maillet, F., et al. (2000). Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12, 1647–1665. doi: 10.1105/tpc.12.9.1647
Century, K. S., Holub, E. B., and Staskawicz, B. J. (1995). NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc. Natl. Acad. Sci. U.S.A. 92, 6597–6601. doi: 10.1073/pnas.92.14.6597
Century, K. S., Shapiro, A. D., Repetti, P. P., Dahlbeck, D., Holub, E., and Staskawicz, B. J. (1997). NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278, 1963–1965. doi: 10.1126/science.278.5345.1963
Charpentier, M., Bredemeier, R., Wanner, G., Takeda, N., Schleiff, E., and Parniske, M. (2008). Lotus japonicus Castor and Pollux are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20, 3467–3479. doi: 10.1105/tpc.108.063255
Chinchilla, D., Zipfel, C., Robatzek, S., Kemmerling, B., Nürnberger, T., and Jones, J. D. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448, 497–500. doi: 10.1038/nature05999
Chitwood, D. J., and Lusby, W. R. (1991). Metabolism of plant sterols by nematodes. Lipids 26, 619–627. doi: 10.1007/BF02536426
Cobb, N. A.. (1890). Tylenchus and Root-gall. Agricultural gazette of New South Wales. doi: 10.5962/bhl.title.66060
Cobb, N. A.. (1893). Nematodes : mostly Australian and Fijian. Sydney: F. Cunninghame & Co., printers. doi: 10.5962/bhl.title.56231
Cook, D. E., Lee, T. G., Guo, X., Melito, S., Wang, K., Bayless, A. M., et al. (2012). Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. doi: 10.1126/science.1228746
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Czaja, L. F., Hogekamp, C., Lamm, P., Maillet, F., Martinez, E. A., Samain, E., et al. (2012). Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol. 159, 1671–1685. doi: 10.1104/pp.112.195990
Day, B., Dahlbeck, D., and Staskawicz, B. J. (2006). NDR1 interaction with RIN4 mediates the differential activation of multiple disease resistance pathways in Arabidopsis. Plant Cell 18, 2782–2791. doi: 10.1105/tpc.106.044693
Desikan, R., Hancock, J. T., Ichimura, K., Shinozaki, K., and Neill, S. J. (2001). Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol. 126, 1579–1587. doi: 10.1104/pp.126.4.1579
Desikan, R., Reynolds, A., Hancock, T. J., and Neill, J. S. (1998). Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 330, 115–120. doi: 10.1042/bj3300115
Dixon, H. N.. (1908). Nematode galls on mosses. Bryologist 11, 31–31. doi: 10.1639/0007-2745(1908)1131:NGOM2.0.CO
Doyle, J. J., and Luckow, M. A. (2003). The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 131, 900–910. doi: 10.1104/pp.102.018150
Edwards, A., Heckmann, A. B., Yousafzai, F., Duc, G., and Downie, J. A. (2007). Structural implications of mutations in the pea SYM8 symbiosis gene, the DMI1 ortholog, encoding a predicted ion channel. Mol. Plant-Microbe Interact. 20, 1183–1191. doi: 10.1094/MPMI-20-10-1183
Ehrhardt, D. W., Wais, R. J., and Long, S. R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85, 673–681. doi: 10.1016/S0092-8674(00)81234-9
Elmayan, T., and Tepfer, M. (1995). Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res. 4, 388–396. doi: 10.1007/BF01973757
Endo, B. Y.. (1965). Histological responses of resistant and susceptible soybean varieties and backcross progeny to entry and development of Heterodera glycines. Phytopathology 55:375.
Endo, B. Y.. (1991). Ultrastructure of initial responses of susceptible and resistant soybean roots to infection by Heterodera glycines. Rev. Nématol. 14, 73–94.
Endo, B. Y., and Veech, J. A. (1970). Morphology and histochemistry of soybean roots infected with Heterodera glycines. Phytopathology 60, 1493–1498. doi: 10.1094/Phyto-60-1493
Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kal,ó, P., and Kiss, G. B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966. doi: 10.1038/nature00842
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M. H., Lin, Y. H., Reid, D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. doi: 10.1111/j.1744-7909.2010.00899.x
Fernandez-Pascual, M., Lucas, M. M., de Felipe, M. R., Boscá, L., Hirt, H., and Golvano, M. P. (2006). Involvement of mitogen-activated protein kinases in the symbiosis Bradyrhizobium-Lupinus. J. Exp. Bot. 57, 2735–2742. doi: 10.1093/jxb/erl038
Francia, D., Chiltz, A., Lo Schiavo, F., Pugin, A., Bonfante, P., and Cardinale, F. (2011). AM fungal exudates activate MAP kinases in plant cells in dependence from cytosolic Ca(2+) increase. Plant Physiol. Biochem. 49, 963–969. doi: 10.1016/j.plaphy.2011.04.008
Genre, A., and Russo, G. (2016). Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 7, 96. doi: 10.3389/fpls.2016.00096
Gherbi, H., Markmann, K., Svistoonoff, S., Estevan, J., Autran, D., Giczey, G., et al. (2008). SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankia bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 4928–4932. doi: 10.1073/pnas.0710618105
Gipson, I., Kim, K. S., and Riggs, R. D. (1971). An ultrastructural study of syncytium development in soybean roots infected with Heterodera glycines. Phytopathology 61, 7–353. doi: 10.1094/Phyto-61-347
Golden, A. M., Epps, J. M., Riggs, R. D., Duclos, L. A., Fox, J. A., and Bernard, R. L. (1970). Terminology and identity of infraspecific forms of the soybean cyst nematode (Heterodera glyecines'). Plant Dis. Rep. 54, 544–546.
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 40, gkr944. doi: 10.1093/nar/gkr944
Gopalan, S., Wei, W., and He, S. Y. (1996). hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J. 10, 591–600. doi: 10.1046/j.1365-313X.1996.10040591.x
Haseloff, J., Siemering, K. R., Prasher, D. C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. U.S.A. 94, 2122–2127. doi: 10.1073/pnas.94.6.2122
Heckman, D. S., Geiser, D. M., Eidell, B. R., Stauffer, R. L., Kardos, N. L., and Hedges, S. B. (2001). Molecular evidence for the early colonization of land by fungi and plants. Science 293, 1129–1133. doi: 10.1126/science.1061457
Hodges, L. D., Cuperus, J., and Ream, W. (2004). Agrobacterium rhizogenes GALLS protein substitutes for Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J. Bacteriol. 186, 3065–3077. doi: 10.1128/JB.186.10.3065-3077.2004
Hofgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16, 69877. doi: 10.1093/nar/16.20.9877
Hohnjec, N., Vieweg, M. F., Pühler, A., Becker, A., and Küster, H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 137, 1283–1301. doi: 10.1104/pp.104.056572
Horváth, B., Yeun, L. H., Domonkos, Á., Halász, G., Gobbato, E., Ayaydin, F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant-Microbe Interact. 24, 1345–1358. doi: 10.1094/MPMI-01-11-0015
Hrbáčková, M., Luptovčiak, I., Hlaváčková, K., Dvorák, P., Tichá, M., Šamajová, O., et al. (2021). Overexpression of alfalfa SIMK promotes root hair growth, nodule clustering and shoot biomass production. Plant Biotechnol. J. 19, 767–784. doi: 10.1111/pbi.13503
Huang, J. S., and Barker, K. R. (1983). Influence of Heterodera glycines on leghemoglobins of soybean nodules. Phytopathology 73, 1002–1004. doi: 10.1094/Phyto-73-1002
Hussey, R. S., and Barker, K. R. (1976). Influence of nematodes and light sources on growth and nodulation of soybean. J. Nematol. 8, 48–52.
Jenkins, W. R.. (1964). A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Rep. 48, 692.
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Karimi, M., Depicker, A., and Hilson, P. (2007). Recombinational cloning with plant gateway vectors. Plant Physiol. 145, 1144–1154. doi: 10.1104/pp.107.106989
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Keinath, N. F., Waadt, R., Brugman, R., Schroeder, J. I., Grossmann, G., Schumacher, K., et al. (2015). Live cell imaging with R-GECO1 sheds light on flg22- and chitin-induced transient [Ca(2+)]cyt patterns in arabidopsis. Mol. Plant. 8, 1188–1200. doi: 10.1016/j.molp.2015.05.006
Kennedy, M. J., Niblack, T. L., and Krishnan, H. B. (1999). Infection by Heterodera glycines elevates isoflavonoid production and influences soybean nodulation. J. Nematol. 31, 341–347.
Kirkpatrick, T. L., and May, M. L. (1989). Host suitability of soybean cultivars for Meloidogyne incognita and M. arenaria. J. Nematol. 21, 666–670.
Klink, V. P., Alkharouf, N. W., MacDonald, M. H., and Matthews, B. F. (2005). Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode). Plant Mol. Biol. 59, 965–979. doi: 10.1007/s11103-005-2416-7
Klink, V. P., Darwish, O., Alkharouf, N. W., Lawaju, B. R., Khatri, R., and Lawrence, K. S. (2021b). Conserved oligomeric Golgi (COG) complex genes functioning in defense are expressed in root cells undergoing a defense response to a pathogenic infection and exhibit regulation my MAPKs. PLoS ONE 16, e0256472. doi: 10.1371/journal.pone.0256472
Klink, V. P., Darwish, O., Alkharouf, N. W., and Lawrence, K. S. (2021a). The impact of pRAP vectors on plant genetic transformation and pathogenesis studies including an analysis of BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1)-mediated resistance. J. Plant Interact. 16, 270–283. doi: 10.1080/17429145.2021.1940328
Klink, V. P., Hosseini, P., Matsye, P., Alkharouf, N. W., and Matthews, B. F. (2009). A gene expression analysis of syncytia laser microdissected from the roots of the Glycine max (soybean) genotype PI 548402 (Peking) undergoing a resistant reaction after infection by Heterodera glycines (soybean cyst nematode). Plant Mol. Biol. 71, 525–567. doi: 10.1007/s11103-009-9539-1
Klink, V. P., Hosseini, P., Matsye, P. D., Alkharouf, N. W., and Matthews, B. F. (2010a). Syncytium gene expression in Glycine max[PI 88788] roots undergoing a resistant reaction to the parasitic nematode Heterodera glycines. Plant Physiol. Biochem. 48, 176–193. doi: 10.1016/j.plaphy.2009.12.003
Klink, V. P., Hosseini, P., Matsye, P. D., Alkharouf, N. W., and Matthews, B. F. (2011). Differences in gene expression amplitude overlie a conserved transcriptomic program occurring between the rapid and potent localized resistant reaction at the syncytium of the Glycine max genotype Peking (PI 548402) as compared to the prolonged and potent PI 88788. Plant Mol. Biol. 75, 141–165. doi: 10.1007/s11103-010-9715-3
Klink, V. P., Overall, C. C., Alkharouf, N. W., MacDonald, M. H., and Matthews, B. F. (2007). Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226, 1389–1409. doi: 10.1007/s00425-007-0578-z
Klink, V. P., Overall, C. C., Alkharouf, N. W., MacDonald, M. H., and Matthews, B. F. (2010b). Microarray detection call methodology as a means to identify and compare transcripts expressed within syncytial cells from soybean (Glycine max) roots undergoing resistant and susceptible reactions to the soybean cyst nematode (Heterodera glycines). J. Biomed. Biotechnol. 2010, e491217. doi: 10.1155/2010/491217
Klink, V. P., Sharma, K., Pant, S. R., McNeece, B., Niraula, P., and Lawrence, G. W. (2017). Components of the SNARE-containing regulon are co-regulated in root cells undergoing defense. Plant Signal. Behav. 12, e1274481. doi: 10.1080/15592324.2016.1274481
Ko, M. P., Barker, K. R., and Huang, J. S. (1984). Nodulation of soybeans as affected by half-root infection with heterodera glycines. J. Nematol. 16, 97–105.
Kosuta, S., Hazledine, S., Sun, J., Miwa, H., Morris, R. J., Downie, J. A., et al. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. U.S.A. 105, 9823–9828. doi: 10.1073/pnas.0803499105
Kouchi, H., Imaizumi-Anraku, H., Hayashi, M., Hakoyama, T., Nakagawa, T., Umehara, Y., et al. (2010). How many peas in a pod? Legume genes responsible for mutualistic symbioses underground. Plant Cell Physiol. 51, 1381–1397. doi: 10.1093/pcp/pcq107
Kwaaitaal, M., Huisman, R., Maintz, J., Reinstädler, A., and Panstruga, R. (2011). Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem. J. 440, 355–365. doi: 10.1042/BJ20111112
Laloum, T., Baudin, M., Frances, L., Lepage, A., Billault-Penneteau, B., Cerri, M. R., et al. (2014). Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 79, 757–768. doi: 10.1111/tpj.12587
Lawaju, B. R., Niraula, P., Lawrence, G. W., Lawrence, K. S., and Klink, V. P. (2020). The Glycine max conserved oligomeric golgi (COG) complex functions during a defense response to Heterodera glycines. Front. Plant Sci. 11, 564495. doi: 10.3389/fpls.2020.564495
Lehman, P. S., Huisingh, D., and Barker, K. R. (1971). Influence of races of Heterodera glycines on nodulation and nitrogen-fixing capacity of soybean. Phytopathology 61, 1239–1244. doi: 10.1094/Phyto-61-1239
Lévy, J., Bres, C., Geurts, R., Chalhoub, B., Kulikova, O., Duc, G., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364. doi: 10.1126/science.1093038
Lewin, R. A.. (1982). Symbiosis and parasitism: Definitions and evaluations. Bioscience 32, 254–260. doi: 10.2307/1308530
Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, J., Wen, J., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/S0092-8674(02)00812-7
Limpens, E., Mirabella, R., Fedorova, E., Franken, C., Franssen, H., Bisseling, T., et al. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. U.S.A. 102, 10375–10380. doi: 10.1073/pnas.0504284102
Liu, S., Kandoth, P. K., Warren, S. D., Yeckel, G., Heinz, R., Alden, J., et al. (2012). A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 492, 256–260. doi: 10.1038/nature11651
Liu, Z., Wu, Y., Yang, F., Zhang, Y., Chen, S., Xie, Q., et al. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. U.S.A. 110, 6205–6210. doi: 10.1073/pnas.1215543110
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Machado, D., and Krishnan, H. B. (2003). nodD alleles of Sinorhizobium fredii USDA191 differentially influence soybean nodulation, nodC expression, and production of exopolysaccharides. Curr. Microbiol. 47, 134–137. doi: 10.1007/s00284-002-3972-6
Mackey, D., Belkhadir, Y., Alonso, J. M., Ecker, J. R., and Dangl, J. L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389. doi: 10.1016/S0092-8674(03)00040-0
Mackey, D., Holt, B. F. 3rd., Wiig, A., and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
Maintz, J., Cavdar, M., Tamborski, J., Kwaaitaal, M., Huisman, R., Meesters, C., et al. (2014). Comparative analysis of MAMP-induced calcium influx in Arabidopsis seedlings and protoplasts. Plant Cell Physiol. 55, 1813–1825. doi: 10.1093/pcp/pcu112
Mann, H. B., and Whitney, D. R. (1947). On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 18, 50–60. doi: 10.1214/aoms/1177730491
Matsye, P. D., Kumar, R., Hosseini, P., Jones, C. M., Tremblay, A., Alkharouf, N. W., et al. (2011). Mapping cell fate decisions that occur during soybean defense responses. Plant Mol. Biol. 77, 513–528. doi: 10.1007/s11103-011-9828-3
Matsye, P. D., Lawrence, G. W., Youssef, R. M., Kim, K. H., Lawrence, K. S., Matthews, B. F., et al. (2012). The expression of a naturally occurring, truncated allele of an α-SNAP gene suppresses plant parasitic nematode infection. Plant Mol. Biol. 80, 131–155. doi: 10.1007/s11103-012-9932-z
Matthews, B. F., Beard, H., MacDonald, M. H., Kabir, S., Youssef, R. M., Hosseini, P., et al. (2013). Engineered resistance and hypersusceptibility through functional metabolic studies of 100 genes in soybean to its major pathogen, the soybean cyst nematode. Planta 237, 1337–1357. doi: 10.1007/s00425-013-1840-1
McNeece, B. T., Pant, S. R., Sharma, K., Niruala, P., Lawrence, G. W., and Klink, V. P. (2017). A Glycine max homolog of NON-RACE SPECIFIC DISEASE RESISTANCE 1 (NDR1) alters defense gene expression while functioning during a resistance response to different root pathogens in different genetic backgrounds. Plant Physiol. Biochem. 114, 60–71. doi: 10.1016/j.plaphy.2017.02.022
McNeece, B. T., Sharma, K., Lawrence, G. W., Lawrence, K. S., and Klink, V. P. (2019). The mitogen activated protein kinase (MAPK) gene family functions as a cohort during the Glycine max defense response to Heterodera glycines. Plant Physiol. Biochem. 137, 25–41. doi: 10.1016/j.plaphy.2019.01.018
Mitra, R. M., Gleason, C. A., Edwards, A., Hadfield, J., Downie, J. A., Oldroyd, G. E. D., et al. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. U.S.A. 101, 4701–4705. doi: 10.1073/pnas.0400595101
Miwa, H., Sun, J., Oldroyd, G. E. D., and Downie, A. J. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48, 883–894. doi: 10.1111/j.1365-313X.2006.02926.x
Morita-Yamamuro, C., Tsutsui, T., Tanaka, A., and Yamaguchi, J. (2004). Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant Cell Physiol. 45, 781–788. doi: 10.1093/pcp/pch093
Murashige, T., and Skoog, F. A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Navazio, L., Moscatiello, R., Genre, A., Novero, M., Baldan, B., Bonfante, P., et al. (2007). A diffusible signal from arbuscular mycorrhizal fungi elicits a transient cytosolic calcium elevation in host plant cells. Plant Physiol. 144, 673–681. doi: 10.1104/pp.106.086959
Ni, L., Fu, X., Zhang, H., Li, X., Cai, X., Zhang, P., et al. (2019). Abscisic acid inhibits rice protein phosphatase PP45 via H(2)O(2) and relieves repression of the Ca(2+)/CaM-Dependent protein kinase DMI3. Plant Cell 31, 128–152. doi: 10.1105/tpc.18.00506
Niblack, T. L., Lambert, K. N., and Tylka, G. L. (2006). A Model Plant pathogen from the kingdom animalia: Heterodera glycines, the soybean cyst nematode. Annu. Rev. Phytopathol. 44, 283–303. doi: 10.1146/annurev.phyto.43.040204.140218
Niraula, P. M., Sharma, K., McNeece, B. T., Troell, H. A., Darwish, O., Alkharouf, N. W., et al. (2020). Mitogen activated protein kinase (MAPK)-regulated genes with predicted signal peptides function in the Glycine max defense response to the root pathogenic nematode Heterodera glycines. PLoS ONE 15, e0241678. doi: 10.1371/journal.pone.0241678
Niraula, P. M., Zhang, X., Jeremic, D., Lawrence, K. S., and Klink, V. P. (2021). Xyloglucan endotransglycosylase/hydrolase increases tightly-bound xyloglucan and chain number but decreases chain length contributing to the defense response that Glycine max has to Heterodera glycines. PLoS ONE 16, e0244305. doi: 10.1371/journal.pone.0244305
Oláh, B., Brière, C., Bécard, G., Dénarié, J., and Gough, C. (2005). Nod factors and a diffusible factor from arbuscular mycorrhizal fungi stimulate lateral root formation in Medicago truncatula via the DMI1/DMI2 signalling pathway. Plant J. 44, 195–207. doi: 10.1111/j.1365-313X.2005.02522.x
Oldroyd, G. E. D., and Downie, J. A. (2004). Calcium, kinases and nodulation signalling in legumes. Nat. Rev. Mol. Cell Biol. 5, 566–576. doi: 10.1038/nrm1424
Oldroyd, G. E. D., Mitra, R. M., Wais, R. J., and Long, S. R. (2001). Evidence for structurally specific negative feedback in the nod factor signal transduction pathway. Plant J. 28, 191–199. doi: 10.1046/j.1365-313X.2001.01149.x
Opperman, C. H., and Bird, D. M. (1998). The soybean cyst nematode, Heterodera glycines: a genetic model system for the study of plant-parasitic nematodes. Curr. Opin. Plant Biol. 1, 342–346. doi: 10.1016/1369-5266(88)80057-8
Pan, H., Stonoha-Arther, C., and Wang, D. (2018). Medicago plants control nodulation by regulating proteolysis of the receptor-like kinase DMI2. Plant Physiol. 177, 792–802. doi: 10.1104/pp.17.01542
Pant, S. R., Matsye, P. D., McNeece, B. T., Sharma, K., Krishnavajhala, A., Lawrence, G. W., et al. (2014). Syntaxin 31 functions in Glycine max resistance to the plant parasitic nematode Heterodera glycines. Plant Mol. Biol. 85, 107–121. doi: 10.1007/s11103-014-0172-2
Pant, S. R., McNeece, B. T., Sharma, K., Niruala, P., Burson, H. E., Lawrence, G. W., et al. (2016). The heterologous expression of a Glycinemax homolog of NONEXPRESSOR OF PR1 (NPR1) and α-hydroxynitrile glucosidase suppresses parasitism by the root pathogen Meloidogyne incognita in Gossypium hirsutum. J. Plant Interact. 11, 41–52. doi: 10.1080/17429145.2016.1163423
Pawlowski, M. L., and Hartman, G. L. (2020). Impact of arbuscular mycorrhizal species on Heterodera glycines. Plant Dis. 104, 2406–2410. doi: 10.1094/PDIS-01-20-0102-RE
Pimentel, D.. (2005). Environmental and economic costs of the application of pesticides primarily in the United States? Environ. Dev. Sustain. 7, 229–252. doi: 10.1007/s10668-005-7314-2
Pueppke, S. G., Bolanos-Vasquez, M. C., Werner, D., Bec-Ferte, M. P., Prome, J. C., and Krishnan, H. B. (1998). Release of flavonoids by the soybean cultivars McCall and peking and their perception as signals by the nitrogen-fixing symbiont sinorhizobium fredii. Plant Physiol. 117, 599–606. doi: 10.1104/pp.117.2.599
Ranf, S., Eschen-Lippold, L., Fröhlich, K., Westphal, L., Scheel, D., and Lee, J. (2014). Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14, 374. doi: 10.1186/s12870-014-0374-4
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., and Scheel, D. (2011). Interplay between calcium signalling and early signalling elements during defence responses to microbe- or damage-associated molecular patterns. Plant J. 68, 100–113. doi: 10.1111/j.1365-313X.2011.04671.x
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS ONE 8, e0066428. doi: 10.1371/journal.pone.0066428
Ray, D. K., West, P. C., Clark, M., Gerber, J. S., Prishchepov, A. V., and Chatterjee, S. (2019). Climate change has likely already affected global food production. PLoS ONE 14, e0217148. doi: 10.1371/journal.pone.0217148
Rebois, R. V., Epps, J. M., and Hartwig, E. E. (1970). Correlation of resistance in soybeans to Heterodera glycines and Rotylenchulus reniformis. Phytopathology 60, 695–700. doi: 10.1094/Phyto-60-695
Redecker, D., Kodner, R., and Graham, L. E. (2000). Glomalean fungi from the ordovician. Science 289, 1920–1921. doi: 10.1126/science.289.5486.1920
Remy, W., Taylor, T. N., Hass, H., and Kerp, H. (1994). Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U.S.A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Ried, M. K., Antolín-Llovera, M., and Parniske, M. (2014). Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. Elife 3, 1–17. doi: 10.7554/eLife.03891
Riely, B. K., Lougnon, G., Ané, J. M., and Cook, D. R. (2007). The symbiotic ion channel homolog DMI1 is localized in the nuclear membrane of Medicago truncatula roots. Plant J. 49, 208–216. doi: 10.1111/j.1365-313X.2006.02957.x
Riggs, R. D., Kim, K. S., and Gipson, I. (1973). Ultrastructural changes in Peking soybeans infected with Heterodera glycines. Phytopathology 63, 76–84. doi: 10.1094/Phyto-63-76
Rival, P., de Billy, F., Bono, J.-J., Gough, C., Rosenberg, C., and Bensmihen, S. (2012). Epidermal and cortical roles of NFP and DMI3 in coordinating early steps of nodulation in Medicago truncatula. Development 139, 3383–3391. doi: 10.1242/dev.081620
Rose, C. M., Venkateshwaran, M., Volkening, J. D., Grimsrud, P. A., Maeda, J., Bailey, D. J., et al. (2012). Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11, 724–744. doi: 10.1074/mcp.M112.019208
Ross, J. P.. (1958). Host-parasite relationship of the soybean cyst nematode in resistant soybean roots. Phytopathology 48, 578–579.
Roy, S., Breakspear, A., Cousins, D., Torres-Jerez, I., Jackson, K., Kumar, A., et al. (2021). Three common symbiotic ABC subfamily B transporters in Medicago truncatula are regulated by a NIN-independent branch of the symbiosis signaling pathway. Mol. Plant-Microbe Interact. 34, 939–951. doi: 10.1094/MPMI-02-21-0036-R
Ryder, M. H., Tate, M. E., and Kerr, A. (1985). Virulence properties of strains of agrobacterium on the apical and basal surfaces of carrot root discs. Plant Physiol. 77, 215–221. doi: 10.1104/pp.77.1.215
Ryu, H., Laffont, C., Frugier, F., and Hwang, I. (2017). MAP kinase-mediated negative regulation of symbiotic nodule formation in Medicago truncatula. Mol. Cells 40, 17–23. doi: 10.14348/molcells.2017.2211
Saha, S., Dutta, A., Bhattacharya, A., and DasGupta, M. (2014). Intracellular catalytic domain of symbiosis receptor kinase hyperactivates spontaneous nodulation in absence of rhizobia. Plant Physiol. 166, 1699–1708. doi: 10.1104/pp.114.250084
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Sharma, A., Kumar, V., Shahzad, B., Tanveer, M., Preet, G., Sidhu, S., et al. (2019). Worldwide pesticide usage and its impacts on ecosystem. SN Appl. Sci. 1, 1446. doi: 10.1007/s42452-019-1485-1
Sharma, K., Niraula, P. M., Troell, H. A., Adhikari, M., Alshehri, H. A., Alkharouf, N. W., et al. (2020). Exocyst components promote an incompatible interaction between Glycine max (soybean) and Heterodera glycines (the soybean cyst nematode). Sci. Rep. 10, 15003. doi: 10.1038/s41598-020-72126-z
Sharma, K., Pant, S. R., McNeece, B. T., Lawrence, G. W., and Klink, V. P. (2016). Co-regulation of the Glycine max soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor (SNARE)-containing regulon occurs during defense to a root pathogen. J. Plant Interact. 11, 74–93. doi: 10.1080/17429145.2016.1195891
Shaw, S. L., and Long, S. R. (2003). Nod factor elicits two separable calcium responses in Medicago truncatula root hair cells. Plant Physiol. 131, 976–984. doi: 10.1104/pp.005546
Shen, C., Yue, R., Bai, Y., Feng, R., Sun, T., Wang, X., et al. (2015). Identification and analysis of Medicago truncatula auxin transporter gene families uncover their roles in responses to Sinorhizobium meliloti infection. Plant Cell Physiol. 56, 1930–1943. doi: 10.1093/pcp/pcv113
Smit, P., Limpens, E., Geurts, R., Fedorova, E., Dolgikh, E., Gough, C., et al. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145, 183–191. doi: 10.1104/pp.107.100495
Stanley, M. R., Koide, R. T., and Shumway, D. L. (1993). Mycorrhizal symbiosis increases growth, reproduction and recruitment of Abutilon theophrasti Medic. in the field. Oecologia 94, 30–35. doi: 10.1007/BF00317297
Stracke, S., Kistner, C., Yoshida, S., Mulder, L., Sato, S., Kaneko, T., et al. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417, 959–962. doi: 10.1038/nature00841
Sun, X., Greenwood, D. R., Templeton, M. D., Libich, D. S., McGhie, T. K., Xue, B., et al. (2014). The intrinsically disordered structural platform of the plant defence hub protein RPM1-interacting protein 4 provides insights into its mode of action in the host-pathogen interface and evolution of the nitrate-induced domain protein family. FEBS Lett. 281, 3955–3979. doi: 10.1111/febs.12937
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Todd, T. C., Winkler, H. E., and Wilson, G. W. (2001). Interaction of Heterodera glycines and Glomus mosseae on soybean. J. Nematol. 33, 306–310.
Traw, M. B., Kniskern, J. M., and Bergelson, J. (2007). SAR increases fitness of Arabidopsis thaliana in the presence of natural bacterial pathogens. Evolution 61, 2444–2449. doi: 10.1111/j.1558-5646.2007.00211.x
van Kessel, C., Singleton, P. W., and Hoben, H. J. (1985). Enhanced N-transfer from a soybean to maize by vesicular arbuscular mycorrhizal (VAM) fungi. Plant Physiol. 79, 562–563. doi: 10.1104/pp.79.2.562
Venkateshwaran, M., Cosme, A., Han, L., Banba, M., Satyshur, K. A., Schleiff, E., et al. (2012). The recent evolution of a symbiotic ion channel in the legume family altered ion conductance and improved functionality in calcium signaling. Plant Cell 24, 2528–2545. doi: 10.1105/tpc.112.098475
Veronese, P., Nakagami, H., Bluhm, B., AbuQamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 18, 257–273. doi: 10.1105/tpc.105.035576
Wais, R. J., Galera, C., Oldroyd, G. E. D., Catoira, R., Penmetsa, R. V., Cook, D. R., et al. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 97, 13407–13412. doi: 10.1073/pnas.230439797
Wang, B., Yeun, L. H., Xue, J. Y., Liu, Y., An,é, J. M., and Qiu, Y. L. (2010). Presence of three mycorrhizal genes in the common ancestor of land plants suggests a key role of mycorrhizas in the colonization of land by plants. New Phytol. 186, 514–525. doi: 10.1111/j.1469-8137.2009.03137.x
Wang, L., and Wang, X. (2021). DEGseq: Identify Differentially Expressed Genes from RNA-seq data. R package version 1.48.40.
Wei, Z. M., Laby, R. J., Zumoff, C. H., Bauer, D. W., He, S. Y., Collmer, A., et al. (1992). Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257, 85–88. doi: 10.1126/science.1621099
White, F. F., Taylor, B. H., Huffman, G. A., Gordon, M. P., and Nester, E. W. (1985). Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 164, 33–44. doi: 10.1128/jb.164.1.33-44.1985
Winkler, H. E., Hetrick, B. A., and Todd, T. C. (1994). Interactinos of Heterodera glycines, Macrophomina phaseolina, and mycorrhizal fungi on soybean in Kansas. J. Nematol. 26, 675–682.
Xu, C.-C., Zhang, D., Hann, D. R., Xie, Z.-P., and Staehelin, C. (2018). Biochemical properties and in planta effects of NopM, a rhizobial E3 ubiquitin ligase. J. Biol. Chem. 293, 15304–15315. doi: 10.1074/jbc.RA118.004444
Yan, Z., Cao, J., Fan, Q., Chao, H., Guan, X., Zhang, Z., et al. (2020). Dephosphorylation of LjMPK6 by phosphatase LjPP2C is involved in regulating nodule organogenesis in Lotus japonicus. Int. J. Mol. Sci. 21, 5565. doi: 10.3390/ijms21155565
Yin, J., Guan, X., Zhang, H., Wang, L., Li, H., Zhang, Q., et al. (2019). An MAP kinase interacts with LHK1 and regulates nodule organogenesis in Lotus japonicus. Sci. China Life Sci. 62, 1203–1217. doi: 10.1007/s11427-018-9444-9
Yuan, J. S., Reed, A., Chen, F., and Stewart, C. N. (2006). Statistical analysis of real-time PCR data. BMC Bioinform. 7, 85. doi: 10.1186/1471-2105-7-85
Zhang, J., Li, W., Xiang, T., Liu, Z., Laluk, K., Ding, X., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 7, 290–301. doi: 10.1016/j.chom.2010.03.007
Zhang, L., Chen, X.-J., Lu, H.-B., Xie, Z.-P., and Staehelin, C. (2011). Functional analysis of the type 3 effector nodulation outer protein L (NopL) from Rhizobium sp. NGR234: symbiotic effects, phosphorylation, and interference with mitogen-activated protein kinase signaling. J. Biol. Chem. 286, 32178–32187. doi: 10.1074/jbc.M111.265942
Zhu, H., Riely, B. K., Burns, N. J., and Ané, J. M. (2006). Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172, 2491–2499. doi: 10.1534/genetics.105.051185
Keywords: plant parasitic nematode, pathogen recognition receptor (PRR), effector triggered immunity (ETI), pathogen associated molecular pattern (PAMP), PAMP triggered immunity (PTI), Glycine max, common symbiosis pathway (CSP), DOESN'T MAKE INFECTIONS (DMI)
Citation: Khatri R, Pant SR, Sharma K, Niraula PM, Lawaju BR, Lawrence KS, Alkharouf NW and Klink VP (2022) Glycine max Homologs of DOESN'T MAKE INFECTIONS 1, 2, and 3 Function to Impair Heterodera glycines Parasitism While Also Regulating Mitogen Activated Protein Kinase Expression. Front. Plant Sci. 13:842597. doi: 10.3389/fpls.2022.842597
Received: 23 December 2021; Accepted: 21 March 2022;
Published: 04 May 2022.
Edited by:
Marc Libault, University of Nebraska-Lincoln, United StatesReviewed by:
Senjuti Sinharoy, National Institute of Plant Genome Research (NIPGR), IndiaPingchuan Deng, Northwest A&F University, China
Copyright © 2022 Khatri, Pant, Sharma, Niraula, Lawaju, Lawrence, Alkharouf and Klink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vincent P. Klink, dmluY2VudC5rbGlua0B1c2RhLmdvdg==
†ORCID: Rishi Khatri orcid.org/0000-0003-4985-0607
Shankar R. Pant orcid.org/0000-0003-3755-7075
Keshav Sharma orcid.org/0000-0002-1137-5663
Prakash M. Niraula orcid.org/0000-0002-3145-6364
Bisho R. Lawaju orcid.org/0000-0001-8563-1036
‡Present address: Shankar R. Pant, Pebble Labs, Los Alamos, NM, United States
Keshav Sharma, Cereal Disease Laboratory, Saint Paul, MN, United States
Prakash M. Niraula, Department of Biological Sciences, Delaware State University, Dover, DE, United States
Bisho R. Lawaju, Department of Plant Pathology, North Dakota State University, Fargo, ND, United States
 Rishi Khatri1†
Rishi Khatri1† Shankar R. Pant
Shankar R. Pant Keshav Sharma
Keshav Sharma Prakash M. Niraula
Prakash M. Niraula Bisho R. Lawaju
Bisho R. Lawaju Vincent P. Klink
Vincent P. Klink