
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 17 February 2022
Sec. Crop and Product Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.841228
This article is part of the Research Topic Molecular Interactions between Crops and Phytopathogens, Volume II: Rice View all 8 articles
Rice sheath blight (ShB) caused by Rhizoctonia solani is one of the most destructive diseases in rice. Fungicides are widely used to control ShB in agriculture. However, decades of excessive traditional fungicide use have led to environmental pollution and increased pathogen resistance. Generally, plant elicitors are regarded as environmentally friendly biological pesticides that enhance plant disease resistance by triggering plant immunity. Previously, we identified that the plant immune inducer ZhiNengCong (ZNC), a crude extract of the endophyte, has high activity and a strong ability to protect plants against pathogens. Here, we further found that guanine, which had a significant effect on inducing plant resistance to pathogens, might be an active component of ZNC. In our study, guanine activated bursts of reactive oxygen species, callose deposition and mitogen-activated protein kinase phosphorylation. Moreover, guanine-induced plant resistance to pathogens depends on ethylene and jasmonic acid but is independent of the salicylic acid signaling pathway. Most importantly, guanine functions as a new plant elicitor with broad-spectrum resistance to activate plant immunity, providing an efficient and environmentally friendly biological elicitor for bacterial and fungal disease biocontrol.
Rice is one of the most important food crops in the world, and the future demands for rice will be higher due to the trend of increasing populations worldwide. Plant pathogens not only reduce rice yield but also produce toxins, resulting in food contamination, which further threatens human health. Rice sheath blight (ShB) is caused by the necrotrophic fungus Rhizoctonia solani (R. solani); ShB is one of the most destructive diseases and a serious threat to the stability of rice production worldwide (Sathe et al., 2021). The cultivation of high-yielding varieties and heavy use of nitrogen fertilizers have caused a sharp rise in ShB prevalence (Savary et al., 1995). ShB can develop water-soaked, oval-shaped or irregularly elongated spots on leaf sheaths and leaf blades that are infected by hyphae or sclerotia (Lin et al., 2020; Molla et al., 2020). In China, the straw return policy returns almost all sclerotia produced back to rice fields. In addition, no effective ShB-resistant cultivars have been applied to rice agricultural production to date (Chen et al., 2012; Molla et al., 2020). At present, chemical fungicides are widely used to minimize the damage caused by ShB; however, pathogens that are resistant to fungicides have emerged (Casida and Durkin, 2017; Singh et al., 2019; Abbas et al., 2021), and these pathogens markedly reduce chemical fungicide efficacy. To make matters worse, chemical pesticides pollute the soil, water resources and crops, posing risks to human health and the ecological environment (Rodriguez-Salus et al., 2016; Casida and Durkin, 2017; Kim et al., 2017). Together, these challenges require pollution-free and efficient biological fungicides to control ShB.
In previous study, fungal biocontrol agents derived from Aspergillus, Clonostachys, Chaetomium, Trichoderma, and Streptoverticillium were used for seed germination pretreatment, root dipping and foliar application (Park et al., 2002, 2005; Naeimi et al., 2010; Shasmita et al., 2019). Unfortunately, no effective commercial biocontrol agent has been successfully used for ShB control thus far. To reduce the use of chemical fungicides, plant immune elicitors (PIEs) have become developing trend in elicitor-based biopesticide biopesticides (Schwessinger and Ronald, 2012). The most important feature of PIEs is their activation of plant immunity instead of killing pathogens directly, in contrast to traditional chemical fungicides. Generally, PIEs are classified as oligosaccharides, peptides, proteins, lipids and small-molecule metabolite elicitors (Schwessinger and Ronald, 2012; Yakhin et al., 2016). For instance, the lipid elicitor lipopolysaccharide, protein-like elicitor BcGs1 and carbohydrate elicitor β-glucan can protect plants against pathogen invasion by triggering innate plant immunity (Yamaguchi et al., 2000; Ma et al., 2015; Ranf et al., 2015).
Many studies have shown that PIEs enhance plant immunity by directly or indirectly activating the signal transduction pathway of plant endogenous hormones such as jasmonic acid (JA), salicylic acid (SA), and ethylene (ET). For instance, copper ions (Cu2+) can trigger plant immunity by upregulating AtACS8 gene expression through CuRE cis-acting elements, further promoting the early biosynthesis of ethylene and promoting the development of the ethylene pathway in Arabidopsis (Liu et al., 2015; Zhang et al., 2018). In addition, rutin enhances plant resistance to bacterial pathogens by activating the SA signaling pathway (Yang et al., 2016). A recent study has shown that silicon can enhance the resistance of potatoes to late blight, which depends on the JA/ET signaling pathway (Xue et al., 2021). Validamycin A induced broad-spectrum disease resistance in both dicots and monocots through the SA and JA/ET signaling pathways (Bian et al., 2020). ET and JA are widely involved in the interactions between plants and pathogens. JA activates plant resistance to necrotrophic pathogens and chewing insects (Howe and Jander, 2008; Brenya et al., 2020). In tomatoes, MYC2 positively regulates pathogen-responsive genes, thereby improving plant resistance to Botrytis cinerea (Du et al., 2017). The ERF1 and ERF6 ethylene-responsive factors can enhance the resistance of Arabidopsis to the soilborne fungus Fusarium oxysporum and the necrotrophic fungus powdery mildew, respectively (Berrocal-Lobo and Molina, 2004; Meng et al., 2013). TaCRK3 mediates wheat resistance to Rhizoctonia cerealis by enhancing the expression of genes involved in ET signaling pathway defense (Guo et al., 2021).
Nucleotide metabolism is a fundamental biological process in many organisms and plays a vital role in plant development and plant responses to biotic and abiotic stresses (Dahncke and Witte, 2013). A recent study has shown that nucleotides are also involved in plant immune responses. Both cyclic GMP (cGMP) and cyclic nucleotide monophosphates (cNMPs) are essential secondary messengers in plants and have been reported to be involved in plant immune responses (Gehring and Turek, 2017; Isner and Maathuis, 2018). In addition, nicotinamide adenine dinucleotide (NAD) is a common electronic carrier involved in the plant response to pathogen invasion, extracellular NAD-induced transcriptional changes and disease resistance to citrus canker (Noctor et al., 2006; Hashida et al., 2009; Alferez et al., 2018). Similarly, adenosine-5′-triphosphate (ATP) is the most direct energy source in most organisms, and extracellular ATP (eATP) can be recognized by the receptor-like kinase DORN1 located on the surface of the plant cell membrane, inducing immune responses such as calcium ion (Ca2+) bursts and mitogen-activated protein kinase (MAPK) phosphorylation in plants. Furthermore, eATP can improve plant resistance to Botrytis cinerea by activating JA signaling (Tripathi et al., 2018). A previous study has shown that guanosine phosphate nucleotides play a significant role in affecting plant–pathogen interactions and responses to plant hormones, such as SA, ET, jasmonic acid JA, and abscisic acid (ABA; Takahashi et al., 2004; Abdelkefi et al., 2018). Nevertheless, the role of nucleobases in regulating plant immunity has not been reported.
Guanine is one of the primary components of DNA. Previous research on guanine has been conducted mainly in the medical field. Guanine is the most easily oxidized base. Guanine oxidation produces 8-oxoguanine, which can pair with adenine and induce guanine-thymine mutations in DNA (Freudenthal et al., 2015). A guanine analog, favipiravir, was approved for influenza treatment and can effectively inhibit the RNA-dependent RNA polymerase of RNA viruses such as influenza, Ebola and COVID-19 (Furuta et al., 2017; Ghasemnejad-Berenji and Pashapour, 2021). In this study, we found that guanine enhanced plant resistance to pathogens and induced reactive oxygen species bursts, callose deposition and MAPK phosphorylation. Comprehensive transcriptome analysis and the transcription of signaling pathway genes with guanine-induced plant disease resistance were analyzed, and further examination found that guanine failed to activate rice resistance to ShB in the COI1b-RNAi plants and etr2, etr3 mutant plants. Moreover, we propose that guanine protected rice against ShB invasion depending on the ET receptor genes OsETR2 and OsETR3 and JA receptor gene OsCOI1b. In addition, guanine enhanced Arabidopsis resistance to Pst DC3000 and depended on AtETR1, AtEIN2, and AtJAR1. These results suggest that guanine induced plant immunity depending on JA and ET signaling. Our study preliminarily revealed the molecular mechanism by which guanine not only activates plant immunity but also provides an environmentally friendly, efficient prevention and control strategy for many diseases, including ShB.
In this study, the japonica rice (Oryza sativa) cultivar (Zhonghua 11, ZH11) was used as the wild type in rice experiments, the T-DNA insertion mutants etr2 and etr3 had a Dongjin (DJ) background, and the RNA interference lines COI1b-RNAi and SA-deficient NahG transgenic plants had a Nipponbare (NIP) background. Arabidopsis thaliana ecotype Columbia (Col-0) was used as a wild-type control in Arabidopsis experiments, and all Arabidopsis mutants used in this research had a Col-0 background. All rice plants were grown at 28°C with 75% relative humidity and a 16/8 h light/dark photoperiod. All Arabidopsis seedlings were grown at 22°C with 70% relative humidity and a 12/12 h light/dark photoperiod. In our study, 4-week-old rice or Arabidopsis plants were sprayed with guanine solution containing 0.05% silwet-L 77 or 0.05% Tween-20, respectively.
The R. solani strain YWK196 was incubated on potato dextrose agar (PDA) medium at 28°C for 2 days, transferred to potato dextrose medium containing matchsticks and grown at 28°C until the mycelium was wrapped around the matchstick. The matchsticks with hyphae were inserted into plant sheaths after guanine treatment for 2 h, and the lesion length was recorded at 3–5 days postinoculation in the field. Leaf inoculation with YWK196 involved the following steps. The clumps of R. solani were placed onto the middle of leaves, which were kept moist in a square petri dish (30 cm × 30 cm). The lesion areas were photographed and measured by ImageJ.1 Inoculation of Xanthomonas oryzae pv. oryzicola (Xoc) strain RS105, which causes bacterial leaf streak in rice, and Xanthomonas oryzae pv. oryzae (Xoo) strain PX099A (the causative agent of bacterial blight in rice) was performed according to previously reported methods (Ju et al., 2017; Li et al., 2018). Pseudomonas syringae pv. tomato (Pst) strain DC3000 was cultured on potato saccharose agar (PSA) medium at 28°C for 24 h. Before inoculation, the bacterial suspension was adjusted to an OD600 of 0.002. The bacterial suspension was infiltrated into plant leaves with a sterile needleless syringe after guanine treatment for 2 h. The lesion area was observed, and the bacterial growth assay was performed after 3 days.
For the quantification of bacteria, six diseased leaves from different plants were harvested and weighed. Then, the diseased leaves were surface sterilized with 75% alcohol for 1 min and washed with sterile distilled water three times. After that, the diseased leaves were ground with a mortar, and a gradient dilution (1:10) of the homogenate was performed. Finally, 10 μL of homogenate was plated on PSA medium and cultured at 28°C, and the number of colonies was calculated after 24 h.
Following guanine treatment for 24 h, 4-week-old rice or Arabidopsis leaves were harvested and put into ethanol:phenol [3:1 (v/v)] solution, vacuumed for 30 min, and then transferred into a water bath at 60°C until the leaves became white and transparent. After being washed with water three times, the samples were stained in aniline blue solution (0.1% w/v aniline blue in 150 mM K2HPO4, pH = 9.5) in the dark overnight at room temperature, and then the samples were washed with deionized water three times to remove excess dye solution. Callose deposition was photographed under a fluorescence microscope.
Four-week-old Arabidopsis seedlings were selected for protein extraction after guanine treatment for 0, 10, 30, and 60 min. First, leaf samples were rapidly frozen in liquid nitrogen and ground to fine powder. Protein extraction was performed using plant protein extraction reagent (CWBIO, China) according to the manufacturer’s instructions. In addition, phosphorylated MAP kinases were detected with anti-phospho-p44/42 MAP kinase primary antibodies (Cell Signaling Technology, United States).
The production of hydrogen peroxide (H2O2) in leaves was detected using 3,3-diaminobenzidine (DAB) as an indicator. Four-week-old rice and Arabidopsis leaves were harvested after 2 h of guanine treatment, and the samples were immediately transferred into a DAB (0.5 mg/mL) staining solution and then vacuum-infiltrated for 30 min. The samples were washed three times with deionized water and then incubated for 8 h under light at room temperature. Subsequently, the samples were decolorized with boiled ethanol. Finally, the samples were imaged by using a stereomicroscope (Lu et al., 2019). To detect ROS production by using H2DCFDA under confocal microscopy, the plants were treated with guanine (100 ng/mL) for 0, 2, 4, 8, 12 and 24 h, 10 μM H2DCFDA solution was infiltrated into plant leaves, and the fluorescence signal was detected 20 min later using an EnSpire Plate Reader (PerkinElmer, United States) with 502 nm excitation and 530 nm emission.
Rice leaf total RNA was extracted using TRIzol reagent (T9424, Sigma-Aldrich, United States) following the kit instructions provided by the manufacturer. Reverse transcription was performed using HiScript III-RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, China). Genomic DNA was first removed from 1 μg of total RNA using 4 × gDNA wiper mix and then reverse transcribed with 5 × HiScriptIII® qRT SuperMix at 37°C for 15 min and 85°C for 5 s. Quantitative real-time PCR was carried out on the QuantStudio 6 Flex Real-Time PCR System (Life Technologies, United States) according to the method provided in the UltraSYBR Mixture (CWBIO, China) instructions. Expression of the rice endogenous actin gene Actin (Accession No: AY212324) was used for internal analysis of the results. The quantitative qRT-PCR primers are shown in Supplementary Table 1.
Four-week-old rice leaves were harvested at 0 h, 2 h, or 24 h after guanine treatment with three replicates of each treatment for a total of nine samples. RNA samples were constructed as previously reported (Feng et al., 2016; Wu et al., 2019), and sequencing was performed with a BGISEQ-500 platform by Wuhan Genomic Institution (2 BGI, Shenzhen, China). The expression levels of individual genes were normalized to FKPM (fragments per kilobase million) reads by RNA-seq through the expectation maximization algorithm. All differential gene expression was based on the following standard: the t significantly differential expression of genes was defined with the absolute value of log2 ratio≥1 and P ≤ 0.05. Heatmap analysis of data was performed via the BAR Heatmapper tool3.
A previous study showed that ZhiNengCong (ZNC), an extract of the endophyte Paecilomyces variotii, can protect plants against pathogenic infection (Lu et al., 2019; Cao et al., 2021). To investigate the effective immune fraction of ZNC, high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC–MS) were performed. Many nucleotides and their derivatives were identified as the active fractions of ZNC, including guanine (Supplementary Table 2). To determine whether guanine stimulates disease resistance in plants and at what concentration, 1 ng/mL, 10 ng/mL, and 100 ng/mL guanine were selected to perform the following experiments. The test was first performed on Arabidopsis. Our results showed that guanine treatment enhanced the resistance of Arabidopsis to Pst DC3000, and the increase in Pst DC3000 resistance was consistent with an increase in guanine treatment concentration (Supplementary Figures 1A,B). The lowest effective concentration, 100 ng/mL, was selected for further experiments. To investigate whether guanine can enhance rice resistance to ShB, inoculation experiments with the R. solani strain YWK196 were conducted in rice. The results indicated that rice leaves treated with guanine exhibited weaker disease symptoms, which showed shorter lesions and smaller areas of disease in the greenhouse (Figures 1A,B), suggesting that guanine significantly enhanced the resistance of rice to ShB. In the field, the average ShB lesion length on guanine-treated rice showed a 37% reduction compared with that of the mock rice (Figures 1C,D). In addition, 100 ng/mL guanine enhanced the resistance of rice to Xanthomonas oryzae pv. oryzicola (Xoc) and Xanthomonas oryzae pv. oryzae (Xoo), which caused rice bacterial leaf streak and bacterial blight, respectively (Figures 1E–H). To further determine whether guanine directly inhibits pathogen growth, pathogen growth experiments were conducted in solid medium containing 0, 1, 10, 100, or 200 ng/mL guanine. As shown in Supplementary Figure 2, guanine failed to inhibit the growth of RS105, PXO99A, YWK196, and Pst DC3000, which indicated that guanine conferred plant resistance to pathogens independently of antimicrobial activity. According to these results, we speculated that guanine may enhance disease resistance by stimulating immunity in plants.
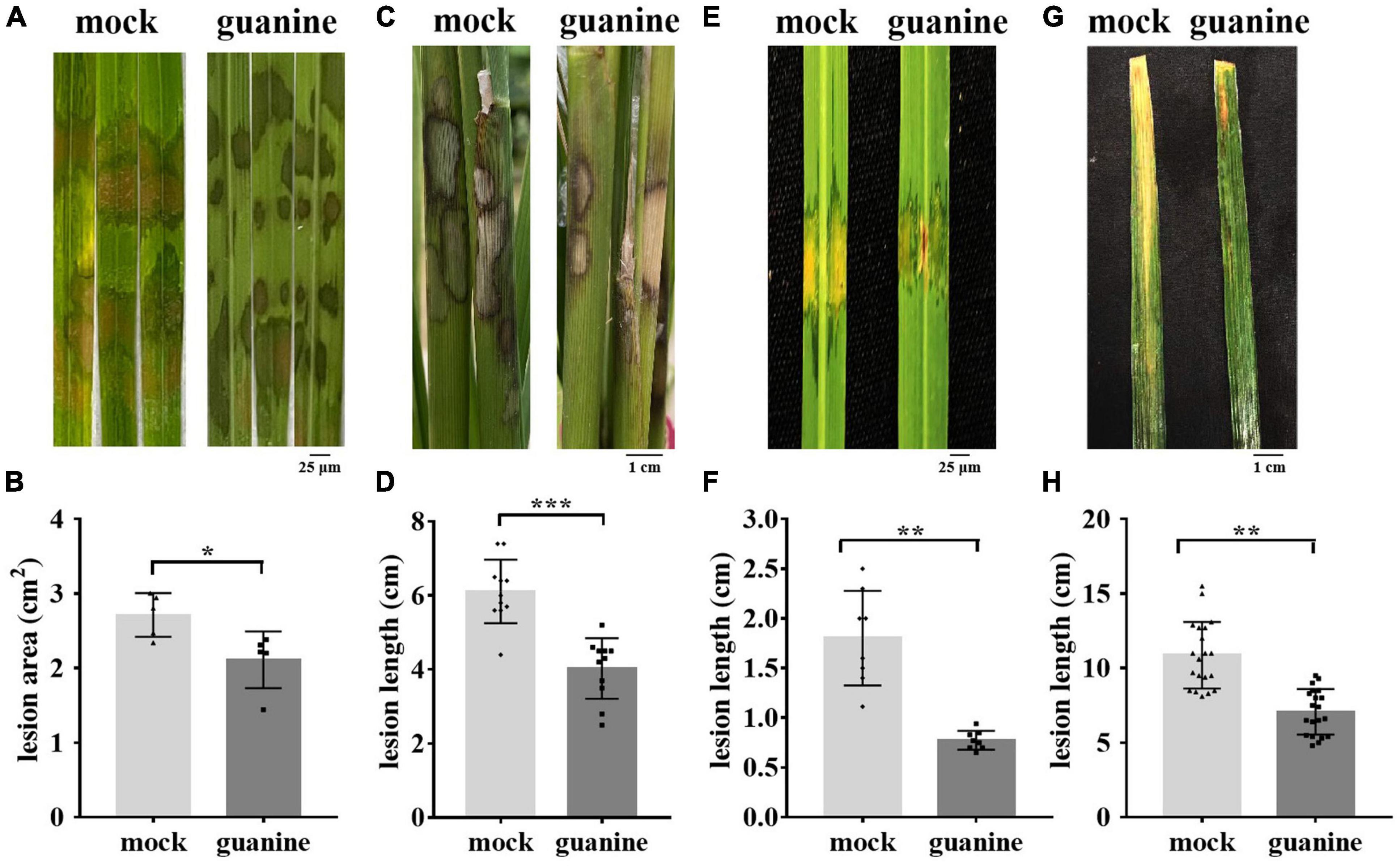
Figure 1. Guanine improved the resistance of rice to R. solani. (A) Rice sheath blight symptoms on the leaves after indoor guanine treatment. The images were photographed at 3 dpi. (B) Lesion area after 3 days of inoculation with R. solani after guanine treatment. Error bars indicate the SD (n = 5). Asterisks indicate P < 0.05 (*) in Student’s t-test analysis. (C) Phenotypes of the rice sheaths inoculated with R. solani after guanine treatment in the field. The images were photographed at 3 dpi (n = 11). (D) Rice sheath blight disease lesion length 3 days after R. solani inoculation after rice sheath guanine treatment. Error bars indicate the SD (n = 11). Asterisks indicate P < 0.001 (***) in Student’s t-test analysis. (E) Bacterial blight disease symptoms on rice leaves after guanine treatment. The images were photographed at 10 days postinoculation. (F) Treatment with guanine 2 h before inoculation with PXO99A and lesion length measurement at 10 days postinoculation (n = 8). Asterisks indicate P < 0.01 (**) in Student’s t-test analysis. (G) Rice leaves treated with 100 ng/mL guanine 2 h before Xoc strain RS105 inoculation. The images were photographed at 15 days postinoculation. (H) Lesion lengths after guanine treatment compared with that of the mock at 15 dpi with Xoc strain RS105 (n = 20). Asterisks indicate P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), and P < 0.001 (***) in Student’s t-test analysis. The experiment was repeated three times with similar results.
Plant elicitors, such as flagellin 22 and Cu2+, generally trigger ROS bursts, callose deposition and MAPK phosphorylation in plant leaves (Liu et al., 2015). Thus, we determined whether guanine could induce plant immune responses. H2O2 is the main component of ROS, and its accumulation can be detected by diaminobenzidine (DAB) staining. In Arabidopsis, after guanine treatment, a deeper brown color was observed 2 h posttreatment (hpt) compared to that seen with the mock treatment (Figure 2A). Aniline blue staining was used to determine callose accumulation. As shown in Figures 2B,C, Arabidopsis leaves had significantly more callose deposits in response to treatment with guanine compared to the mock treatment (Figures 2B,C). In addition, elicitor-triggered plant immunity is often associated with MAPK phosphorylation activation (Asai et al., 2002), and guanine treatment significantly promoted the phosphorylation of MAPKs compared with the mock treatment at the indicated times (Figure 2D). Ca2+ is identified as an essential secondary messenger regulating multitudinous cellular processes (Bleecker and Kende, 2000), and Ca2+ influx is the first event in the activation of the defense response in plants. To determine whether Ca2+ influx is required for the guanine-induced defense response, a calcium channel blocker, lanthanum chloride (LaCl3), was used to treat Arabidopsis leaves in combination with guanine. The results showed that using LaCl3 significantly reduced guanine-induced disease resistance, implying that Ca2+ influx is necessary for the guanine-mediated defense response in Arabidopsis (Figure 2E). In rice, guanine also promoted the accumulation of ROS, as shown by DAB staining (Figure 2F). In addition, H2DCFDA fluorescence was used to detect ROS production. The results showed that guanine increased the accumulation of ROS within 24 h and peaked at 4 hpt (Figure 2G). The rice leaves showed a significant increase in callose deposition after guanine treatment (Figure 2H).
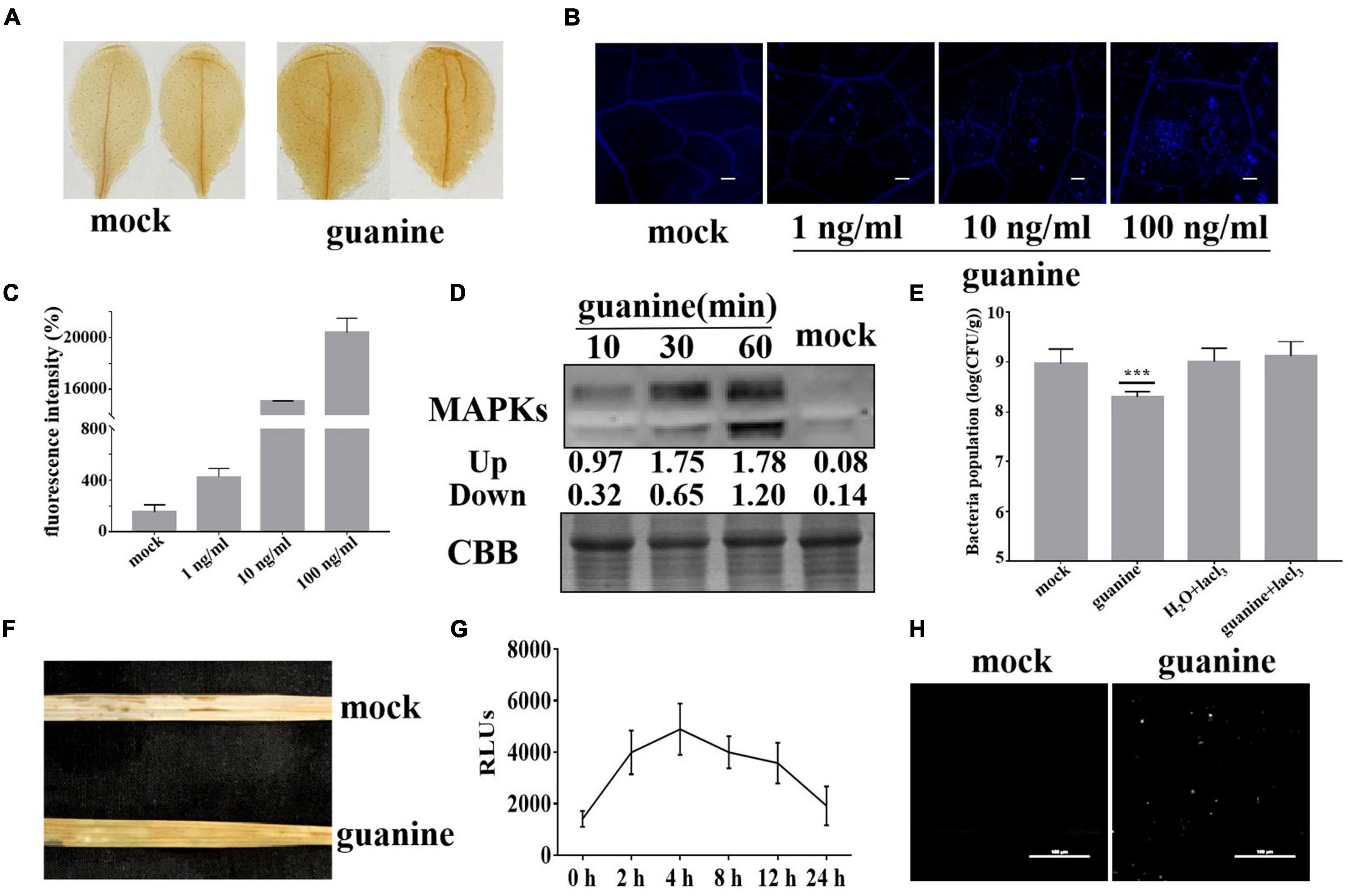
Figure 2. Guanine-induced plant immune responses. (A) DAB staining to detect H2O2 accumulation at 2 hpt after guanine treatment in Arabidopsis; brown deposits indicate H2O2 concentration. (B) Guanine-induced callose deposits in Arabidopsis. The leaves of 4-week-old Col-0 seedlings were treated with the mock and 1, 10, and 100 ng/mL guanine, and callose deposits were revealed by aniline blue staining. Scale bars = 25 μm. (C) Quantification of callose fluorescence intensity of guanine-treated Arabidopsis leaves was performed using ImageJ software. Error bars indicate the SD (n = 4). (D) Guanine-activated MAPK phosphorylation in Arabidopsis leaves. MAPK phosphorylation was detected by the phospho-p44/42 MPK antibody at the indicated times. Coomassie brilliant blue (CBB) staining of ribulose-1,5-bis-phosphate carboxylase/oxygenase was used to ensure equal protein loading in each lane. (E) Growth population of Pst DC3000 in the leaves of Arabidopsis plants treated with mock, guanine, H2O + lacl3 or guanine + lacl3. Error bars indicate the SD. Asterisks indicate P < 0.001 (***) in Student’s t-test analysis. (F) Accumulation of H2O2 in rice leaves after guanine treatment by DAB staining. (G) H2DCFDA was used to detect ROS accumulation in the rice leaves after guanine treatment at 0, 2, 4, 8, 12 and 24 h. Error bars indicate the SD (n = 5). (H) Callose deposits in guanine-treated rice leaves after aniline blue staining. The leaves of 4-week-old ZH-11 seedlings were treated with mock and 100 ng/mL guanine and then stained using aniline blue. Scale bars = 100 μm.
To further explore the role of guanine in regulating the expression of genes at the transcriptional level in rice, we performed an RNA sequencing transcriptome analysis of rice leaves treated with guanine (100 ng/mL) at 0, 2, and 24 hpt. The samples were divided into three groups with three biological replicates. Each sample produced at least 44 million high-quality clean reads with a clean date rate of 97% or higher for all samples, and an average of 83.36% reads were mapped to the rice genome, representing a total of 25045 genes expressed in all samples (Supplementary Tables 3, 4). Here, 3270 and 1606 genes were upregulated and 3251 and 794 genes were downregulated at 2 and 24 h after guanine treatment, respectively (Figures 3A–C). A set of differentially expressed genes (DEGs) that negatively regulate defense responses [namely, OsWRKY76 (Yokotani et al., 2013), OsWRKY45 (Tao et al., 2009), OsWRKY62 (Liang et al., 2017), OsMADS26 (Khong et al., 2015), OsTrxm (Hu et al., 2021), and OsHPL3 (Hu et al., 2021)] were found to be significantly downregulated with guanine treatment. Genes [namely, OsWRKY13 (Xiao et al., 2013), OsWRKY53 (Chujo et al., 2007), OsWRKY4 (Wang et al., 2015), OsWRKY19 (Du et al., 2021), OsWRKY30 (Peng et al., 2012), OsWRKY89 (Wang et al., 2007), OsWRKY71 (Liu et al., 2007), OsWRKY42 (Cheng et al., 2015), OsMYB30 (Li et al., 2020), OsMYB55 (Kishi-Kaboshi et al., 2018), OsMYB110 (Kishi-Kaboshi et al., 2018), OsJAMyb (Cao et al., 2015), OsRap2.6 (Wamaitha et al., 2012), OsNAC4 (Kaneda et al., 2009), and RAI1 (Kim et al., 2012)] that are involved in the positive regulation of plant disease resistance responses were markedly upregulated after guanine treatment (Figure 3D). These results showed that guanine could regulate the expression of disease resistance-related genes in rice.
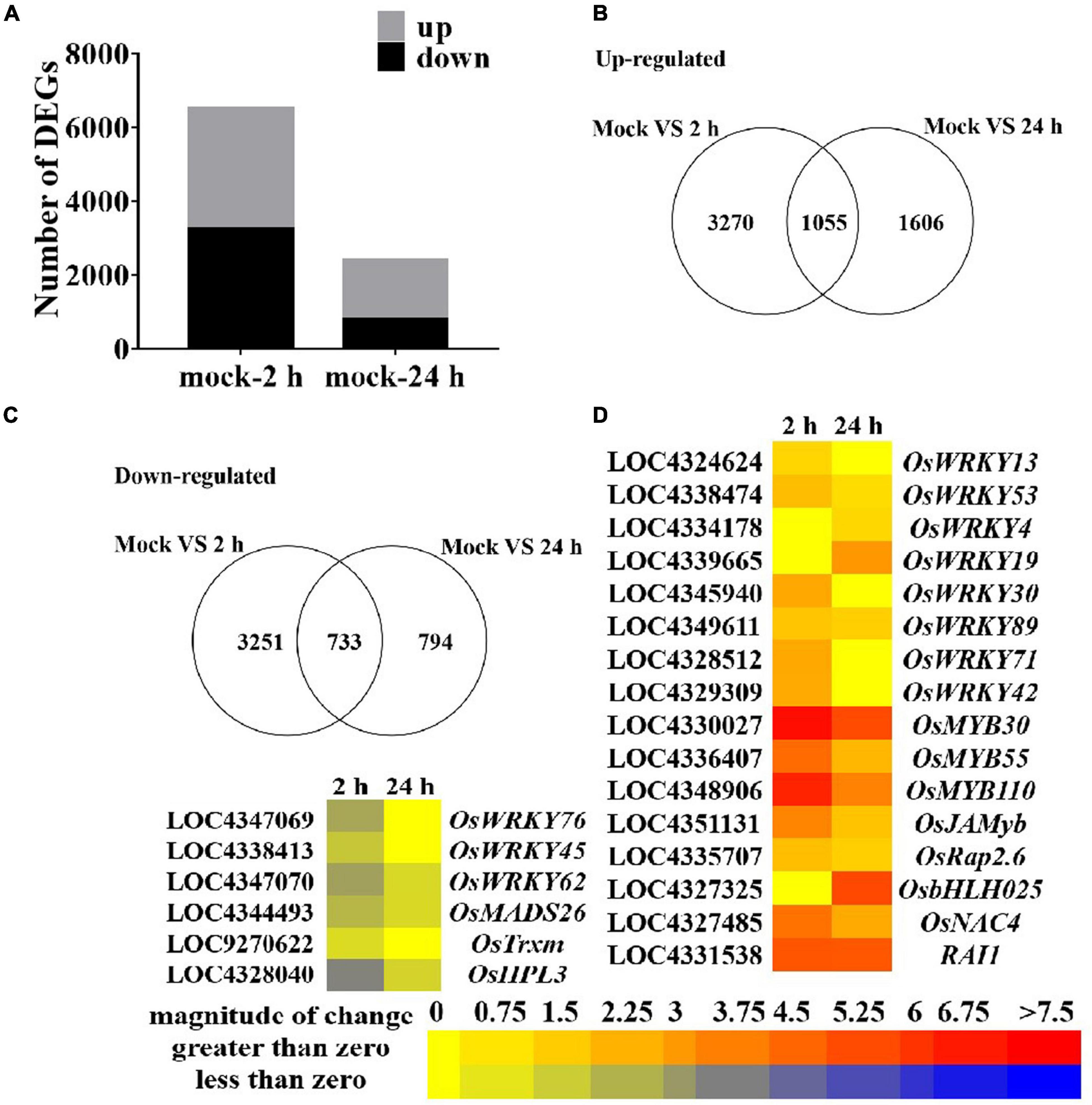
Figure 3. An overview of guanine-regulated gene expression in rice leaves. (A) Number of up- and downregulated genes after guanine treatment at 2 and 24 hpt. (B) Venn diagram of upregulated genes in the mock vs. guanine-2 h and mock vs. guanine-24 h groups. (C) Downregulated expression of genes after 2 and 24 hpt of guanine treatment. (D) Heatmap analysis of disease resistance-related gene expression after guanine treatment at 2 and 24 hpt.
To further investigate the molecular mechanism of guanine-induced disease resistance in rice, multiple disease signaling pathways were analyzed. The results showed that gene expression in the ET, JA, and SA signaling pathways was significantly upregulated. ET-JA and SA signaling pathways play an essential role in plant defense against biotrophic and necrotrophic pathogens (McDowell and Dangl, 2000; Berens et al., 2017; Zhao Z. X. et al., 2021). RNA sequencing analysis suggested that ethylene biosynthesis gene expression was upregulated by guanine at 2 and 24 hpt (Figure 4A), including OsSASM1, OsACS1, OsACS2, OsACS5, OsACO1, and OsACO7 (Li et al., 2011). Similarly, guanine increased the expression of ethylene-responsive transcriptional genes such as OsERF3 and OsERF62 (Hu et al., 2008; Lu et al., 2011; Figure 4A). Furthermore, as shown in Figure 4B, qRT-PCR experiments demonstrated that guanine treatment upregulated the expression of OsSASM1, OsACO7, and OsERF62, which was consistent with the RNA-seq data (Figure 4B). In rice, OsERS1, OsERS2, OsETR2, OsETR3, and OsETR4 have been identified as ET receptors (Wuriyanghan et al., 2009; Zhao H. et al., 2021). Here, the ET receptor T-DNA insertion mutants etr2 and etr3 were used to determine whether guanine induced rice resistance to R. solani that relies on ET signaling. As shown in Figures 4C,D, the lesion area was significantly smaller after guanine treatment compared to that in the mock in wild-type plants Dongjin (DJ); however, a similar lesion area between the mock and guanine treatment groups in etr2 and etr3 mutant plants was observed. In addition, the lesion areas of etr2 and etr3 were significantly greater than those of DJ, indicating that OsETR2 and OsETR3 function as positive regulators in ShB resistance and that guanine induces rice resistance to R. solani YWK196 in an OsETR2/OsETR3-dependent manner. Col-0, etr1 and ein2 mutants were injected with Pst DC3000 after 2 h of guanine and mock treatment. As a result, guanine failed to induce plant resistance to Pst DC3000 in the etr1 and ein2 mutants (Figure 4E), indicating that guanine activated plant immunity and was dependent on ethylene signaling in Arabidopsis.
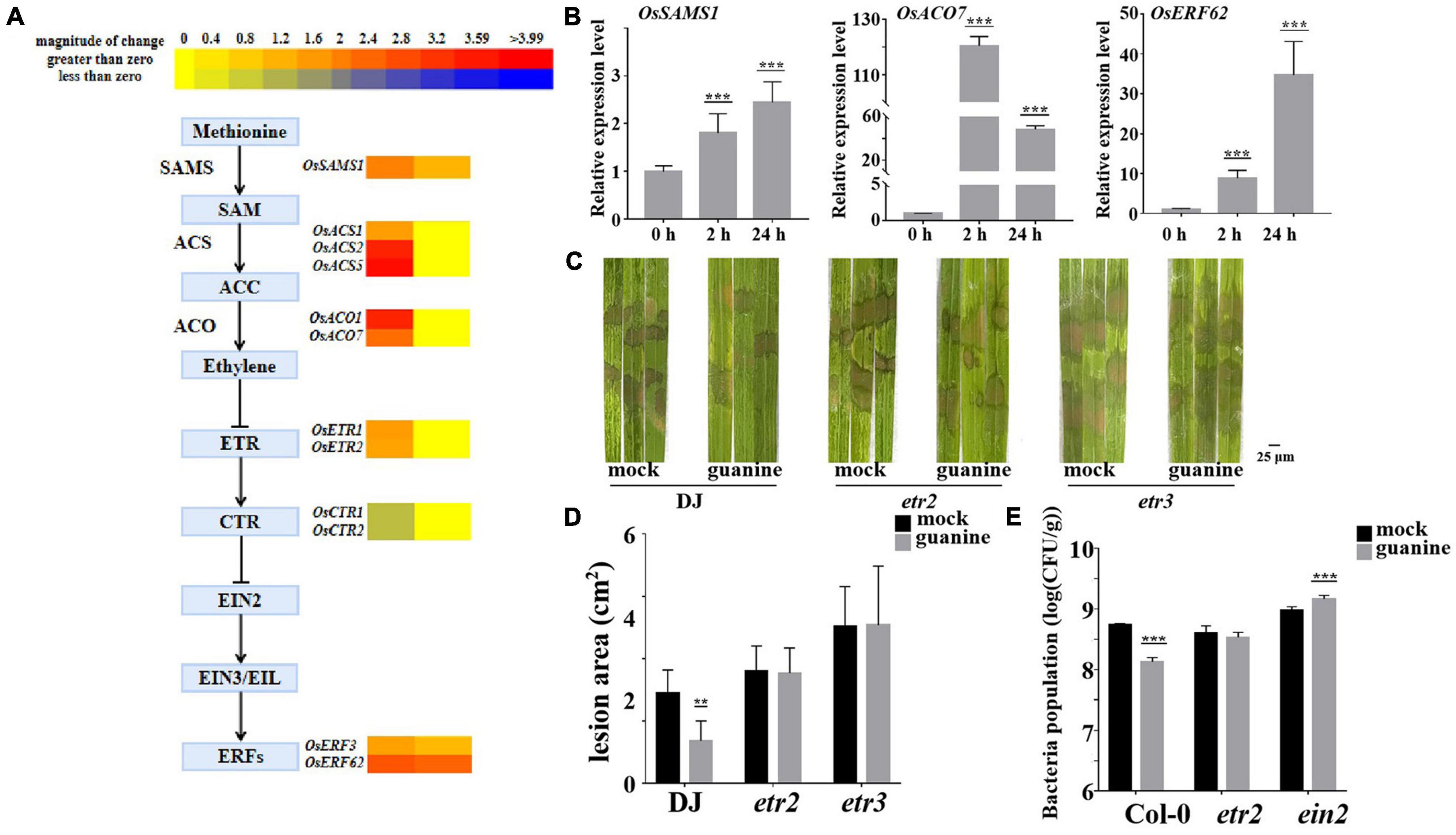
Figure 4. Guanine activated ET signaling pathway gene expression and enhanced plant disease resistance dependent on ethylene receptors. (A) Heatmap analysis of ET biosynthesis and response gene expression after guanine treatment at 2 and 24 hpt. (B) qRT-PCR for the detection of ethylene pathway genes OsSAMS1, OsACO7, and OsERF62. The horizontal coordinates indicate the time of guanine- and mock-treated rice, and the vertical coordinates indicate the relative gene expression levels of guanine compared to the mock at the same time point. (C) Susceptible phenotype of the etr2 and etr3 mutant plants. Leaves of the rice plants treated with the mock and 100 ng/mL guanine prior to R. solani strain YWK196 inoculation. The disease symptoms were photographed at 3 dpi. (D) Measurement of the area of (C) by ImageJ. Asterisks indicate P < 0.01 (**) in Student’s t-test analysis. (E) Bacterial population in Col-0, etr1 and ein2 plant leaves. Arabidopsis leaves treated with the mock and 100 ng/mL guanine prior to Pst DC3000 inoculation. The bacterial population was calculated at 3 dpi. Asterisks indicate P < 0.001 (***) in Student’s t-test analysis.
In addition, both RNA-seq and qRT-PCR showed that JA biosynthesis- and signal transduction-related gene expression were upregulated by guanine, as illustrated in Figures 5A,B. To explore whether JA signaling plays a role in the guanine-mediated resistance response, OsCOI1b-RNAi and NIP (wild type) were inoculated with R. solani that was treated with guanine and mock 2 h later. The results showed that guanine failed to reduce the lesion area in COI1b-RNAi plants (Figures 5C,D), suggesting that the JA receptor OsCOI1b was necessary for guanine-induced rice resistance to ShB. Furthermore, a Pst DC3000 inoculation experiment was conducted in a JA biosynthesis mutant in Arabidopsis. As shown in Figure 5E, guanine treatment did not reduce the bacterial population in jar1 mutant plants but significantly reduced the bacterial population in opr3. This difference indicated that guanine-induced resistance to Pst DC3000 is dependent on AtJAR1 but not AtOPR3. Taken together, these results suggested that JA signaling is essential for guanine-induced plant resistance.
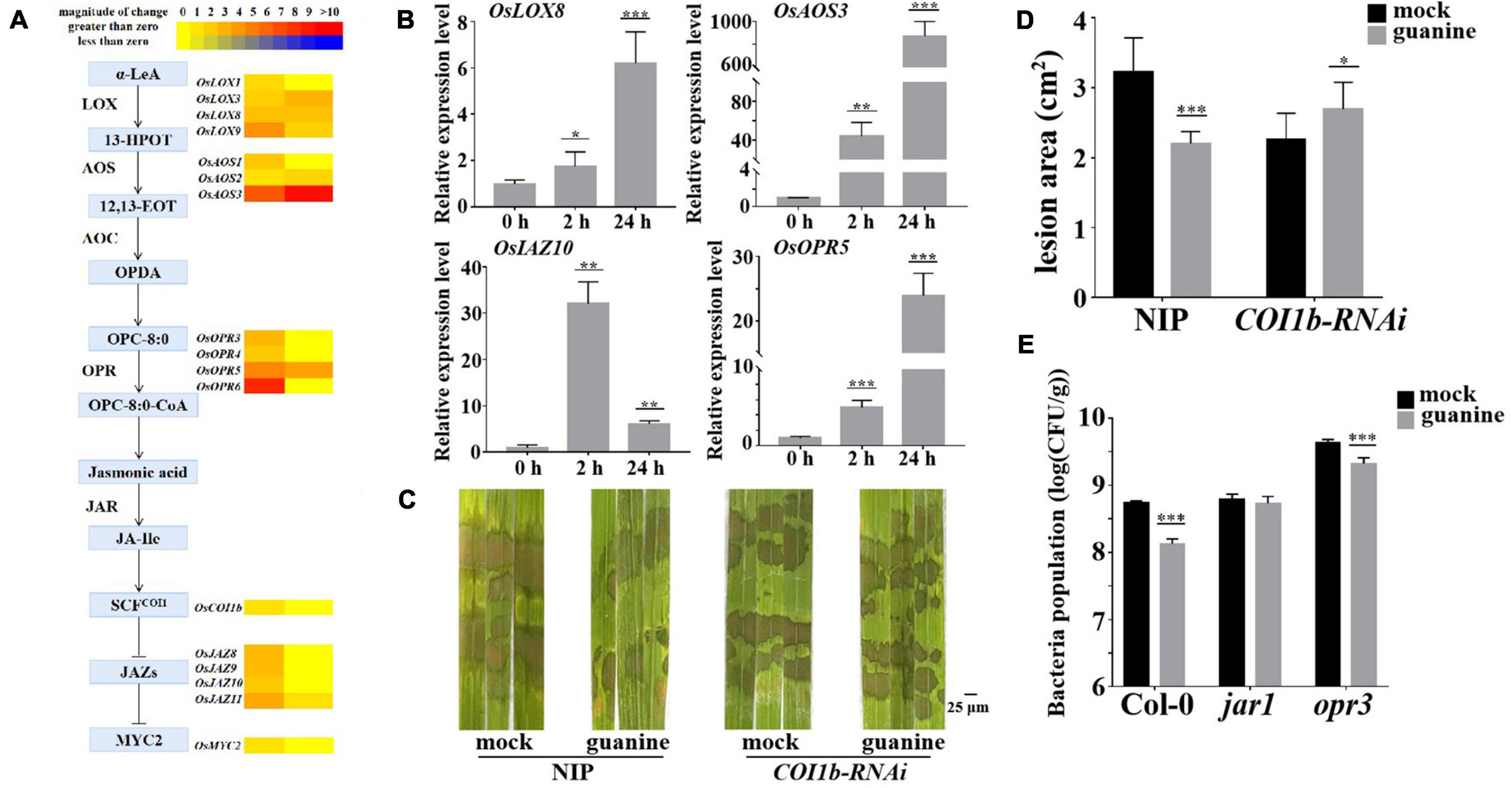
Figure 5. Guanine upregulated the expression of JA signaling pathway genes and enhanced plant disease resistance dependent on the JA signaling pathway. (A) Heatmap of JA-related gene expression after guanine treatment. (B) qRT-PCR for the detection of JA pathway genes OsLOX8, OsAOS3, OsOPR5, and OsJAZ10. The horizontal coordinates indicate the time of guanine- and mock-treated rice, and the vertical coordinates indicate the relative gene expression levels of guanine compared to the mock at the same time point. (C) Evaluation of the effect of guanine on COI1b-RNAi against R. solani. (D) Measurement of the lesion area by ImageJ. Lesion area after 3 days of inoculation with R. solani strain YWK196 after guanine treatment in NIP and COI1b-RNAi. Asterisks indicate P < 0.001 (***) and P < 0.05 (*) in Student’s t-test analysis. (E) Inoculation of Pst DC3000 after guanine treatment and bacterial growth in Col-0, jar1 and opr3 after 3 days. Asterisks indicate P < 0.001 (***) in Student’s t-test analysis.
A heatmap was used to determine the role of SA in guanine-mediated defense responses, and the results showed that OsPAL1, OsPAL3, and OsPAL7 (Tonnessen et al., 2015), which are involved in SA biosynthesis, were upregulated by guanine treatment. In addition, OsNPR1 and OsPR1a, marker genes of the SA pathway (Sohn et al., 2007; Yuan et al., 2007), were significantly upregulated by guanine treatment (Figure 6A). Furthermore, qRT-PCR analysis showed similar results as the transcriptome data (Figure 6B). Then, SA-deficient NahG transgenic plants were used for the following experiments. Guanine significantly reduced the lesion area in both NIP and NahG plants (Figures 6C,D). Similarly, guanine enhanced Arabidopsis resistance to Pst DC3000 in NahG and sid2 mutant plants (Figure 6E). These results suggested that SA signaling is not necessary for guanine-induced resistance to pathogens in plants.
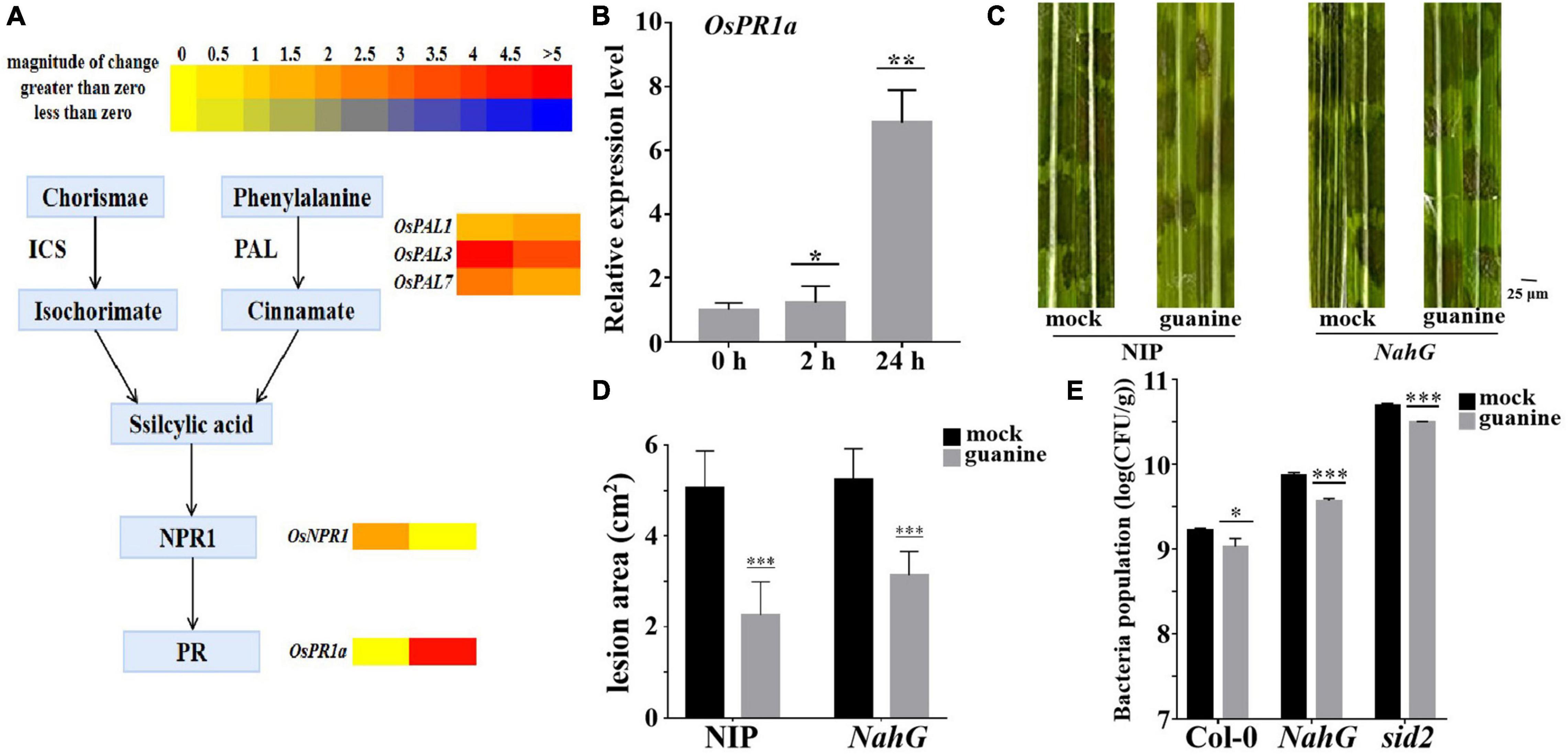
Figure 6. Guanine upregulated the expression of SA signaling pathway genes but still enhanced plant disease resistance in SA-deficient mutant plants. (A) Heatmap analysis of SA biosynthesis and responsive gene expression after guanine treatment at 2 and 24 hpt. (B) qRT-PCR for the detection of the SA pathway gene OsPR1a. The horizontal coordinates indicate the time of guanine- and mock-treated rice, and the vertical coordinates indicate the relative gene expression levels of guanine compared to the mock at the same time point. Asterisks indicate P < 0.05 (*), P < 0.01 (**) in Student’s t-test analysis. (C) Disease symptoms of the NahG transgenic plants. Leaves of the rice plants treated with the mock and 100 ng/mL guanine prior to R. solani strain YWK196 inoculation. The disease symptoms were photographed at 3 dpi. (D) Measurement of the lesion area of (C) by ImageJ. Asterisks indicate P < 0.001 (***) in Student’s t-test analysis. (E) Bacterial population in Col-0, NahG and sid2 plant leaves. Arabidopsis leaves treated with the mock and 100 ng/mL guanine prior to Pst DC3000 inoculation. Bacterial population was calculated at 3 dpi. Asterisks indicate P < 0.05 (*), P < 0.001 (***) in Student’s t-test analysis.
Here, we proposed a possible model to summarize the function of guanine in triggering plant immunity based on the above results (Figure 7). Guanine-enhanced rice resistance to ShB depends on the ET receptors OsETR2 and OsETR3 and the JA receptor OsCOI1b. In Arabidopsis, guanine-induced plant resistance to Pst DC3000 required AtJAR1, AtETR1, and AtEIN2. In summary, guanine activated jasmonic acid and ethylene signaling pathways to protect plants against pathogens. The mechanism of rice resistance to ShB is still largely unknown, and ShB control relies heavily on chemical fungicides (Zheng et al., 2013; Li et al., 2019; Wang et al., 2021). In this study, 100 ng/mL guanine significantly enhanced rice resistance to ShB, providing a novel, green and efficient strategy for ShB prevention and control.
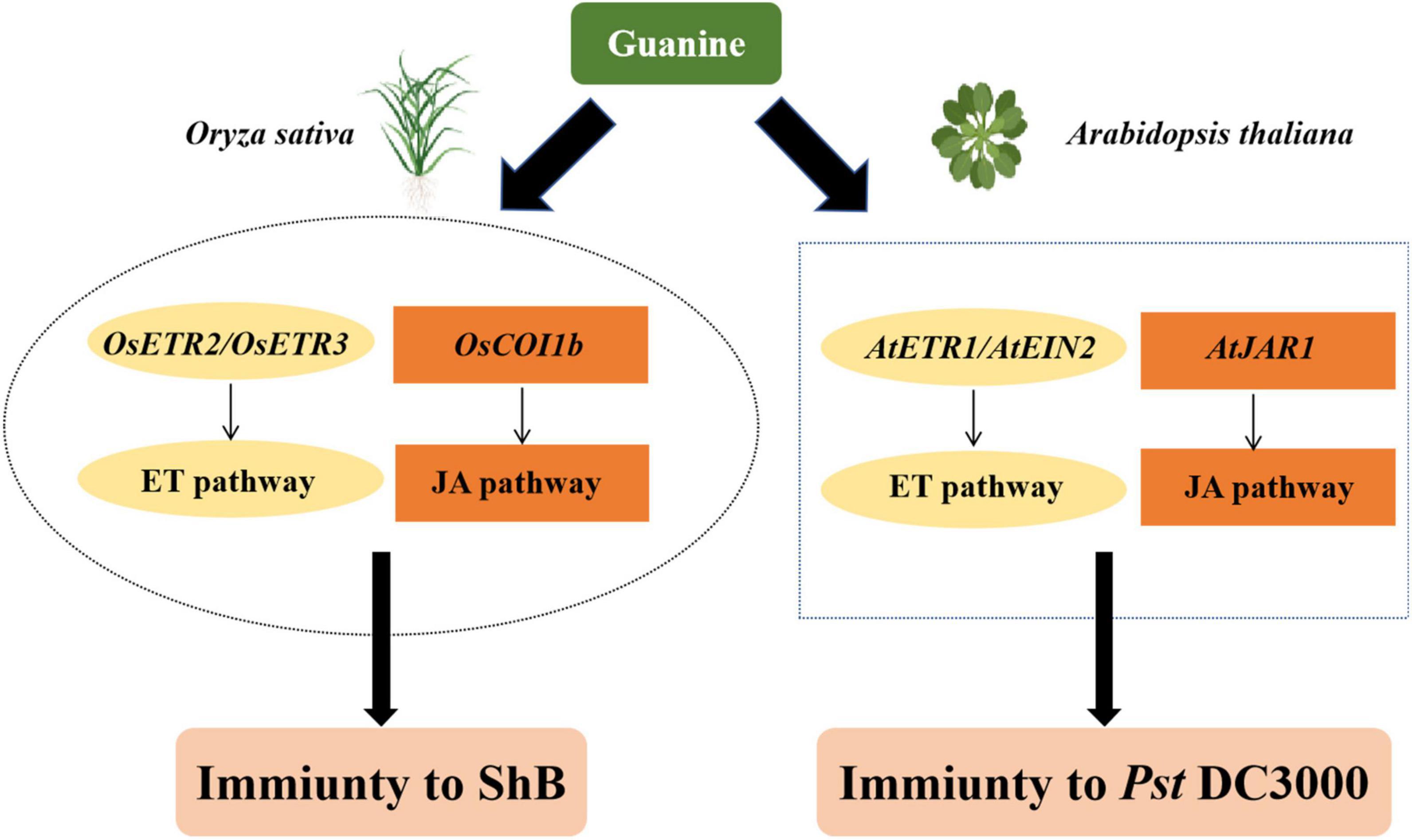
Figure 7. A proposed model of guanine-mediated plant immunity. In rice, OsETR2, OsETR3, and OsCOI1b are required for guanine-induced plant immunity to ShB. Similarly, guanine also needs AtETR1, AtEIN2, and AtJAR1 to enhance plant resistance to pst DC3000 in Arabidopsis.
At present, biological agents are usually used for the prevention and treatment of ShB in agricultural production. Rice ShB was effectively controlled by 0.1 mg/L Cu-based water-dispersible humic acid (Cu-WH) fungicide (Tang et al., 2019). In addition, previous studies have demonstrated that extracts of Ginkgo biloba outer seedcoats can inhibit the growth of R. solani Kuhn AG-1 and are considered most effective at a concentration of 250 μg/mL (Oh et al., 2015). Similarly, the rhizobacterium Bacillus amyloliquefaciens (SN13), as a biocontrol agent, can enhance immune responses against R. solani in rice by modulating various physiological, metabolic and molecular functions (Srivastava et al., 2016). In this study, 100 ng/mL guanine significantly enhanced the resistance of rice to ShB in both greenhouses and fields by triggering rice immunity (Figure 1). Furthermore, the concentration of guanine (100 ng/mL) also activated the ROS burst, MAPK phosphorylation, callose deposition and the expression of pathogen resistance-associated genes (Figures 2, 3), which was similar to the effects of Cu2+ and flg22. In the field, R. solani was inoculated on rice after 2 h of guanine treatment. The guanine-treated rice maintained better resistance to ShB after 3–5 days than the mock rice (Figures 1C,D). This finding indicates that the resistance of guanine-induced plants to ShB can be maintained for at least 3–5 days in the field. Thus, guanine is a highly efficient and environmentally friendly plant elicitor for ShB prevention and control in agricultural plant production.
In general, JA- and ET-mediated signaling pathways are considered to play a synergistic role in defense responses to necrotizing pathogens such as R. solani (Glazebrook, 2005). The JA/ET signaling pathway plays a crucial role in WRKY4-mediated defense responses to rice sheath blight (Wang et al., 2015). ET also plays an important role in plant growth and development (van Loon et al., 2006). ET is synthesized from the precursor intermediate 1-aminocyclopropane-1-carboxylate (ACC), which is catalyzed by ACC oxidase and derived from S-adenosylmethionine (SAM) via a reaction catalyzed by the ACC synthase ACS (Bleecker and Kende, 2000). A previous study showed that ET plays an essential role in rice immunity. For example, exogenous ET application induces pathogenesis-related (PR) gene expression in rice (Agrawal et al., 2001). In addition, overexpression of the ET biosynthetic gene OsACS2 can enhance rice resistance to ShB (Helliwell et al., 2013). These studies indicate that the ET pathway can positively regulate rice resistance to ShB. Similarly, our results confirmed that guanine regulates rice disease resistance through the ET pathway. Transcriptome analysis showed that guanine treatment increased the upregulated expression of the ET biosynthesis genes OsSAMS1 and OsACO7 and ET response transcription factors OsERF62 (Figures 4A,B). Interestingly, the ET receptors OsETR2 and OsETR3 were more sensitive to R. solani than the wild-type plants, and guanine treatment did not increase plant resistance to R. solani (Figures 4C,D). A possible explanation for this might be that guanine-induced resistance to ShB mainly depends on ET signal transduction. In Arabidopsis, guanine failed to induce plant resistance to Pst DC3000 in the etr1 and ein2 mutants (Figure 4E). These results suggested that guanine-mediated resistance to Pst DC3000 depends on ET signaling.
On the other hand, jasmonic acid plays a crucial role in plant resistance to insect and fungal pathogens. Exogenous JA can activate the expression of hundreds of defense-related genes, and the resulting feedback leads to the upregulation of the biosynthesis of resistance proteins and metabolites, including antifungal proteins and plant antitoxins (Ferrari et al., 2003; Srivastava et al., 2018; Ruan et al., 2019; Hu et al., 2021). In addition, WRKY30 overexpression in rice enhances resistance to the rice sheath blight fungus R. solani, which also depends on the JA pathway (Peng et al., 2012). This study found that guanine was involved in the induction of JA biosynthesis and responses in plants, which was largely dependent on the receptor OsCOI1b for the regulation of rice resistance to ShB. Furthermore, guanine-enhanced plant resistance to pathogens also required AtJAR1 but not the JA biosynthesis gene AtOPR3 in Arabidopsis (Figure 5). Hence, we proposed that guanine-enhanced resistance to pathogens mainly depends on the ethylene and jasmonic acid signaling pathways.
Our results demonstrated that guanine enhances the resistance of plants against bacteria and fungi. The ET and JA phytohormone signaling pathways are involved in guanine-mediated immunity. ET receptors OsETR2 and OsETR3 and JA receptor OsCOI1b are necessary for guanine-enhanced rice resistance to ShB. In Arabidopsis, guanine-induced plant resistance to Pst DC3000 depends on AtJAR1, AtETR1, and AtEIN2.
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA767282.
XD designed the research and wrote the manuscript. LW performed the experiments, analyzed the data and wrote the manuscript. HL performed experiments and wrote the manuscript. CL and QW performed experiments. ZY and YL wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Natural Science Foundation of China (32072500, 31801867, and 31872925), Shandong Provincial Key Research and Development Plan (2019JZZY020608, 2019GNC106152, and 2020CXGC010803), Natural Science Foundation of Shandong Province (ZR2020QC130 and ZR2020MC117), and Modern Agricultural Technology Industry System of Shandong province (SDAIT-17-06).
QW is employed by Shandong Pengbo Biotechnology Co., Ltd., Tai’an, Shandong, China.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Shimin Zuo at Yang Zhou University for kindly providing ET mutants etr2 and etr3. We also thank Jinlong Qiu at the Institute of Microbiology, Chinese Academy of Sciences, for providing NahG mutants.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.841228/full#supplementary-material
Supplementary Figure 1 | Guanine enhanced Arabidopsis resistance to Pst DC3000. (A) Disease phenotype in Col-0 plants after treatment with 0, 1, 10, and 100 ng/mL guanine at 3 days. (B) Bacterial growth in Col-0 plants after treatment with 0, 1, 10, and 100 ng/mL guanine at 3 days (n = 6).
Supplementary Figure 2 | Guanine failed to inhibit the growth of fungal and bacterial pathogens. (A) RS105 was grown in PSA medium containing 0, 10, and 100 ng/mL guanine. (B) PXO99A was grown in PSA medium containing 0, 10, and 100 ng/mL guanine. (C) YWK196 was grown in PDA medium containing 0, 1, 10, 100, and 200 ng/mL guanine. (D) Pst DC3000 was grown in PSA medium containing 0, 1, 10, 100, and 200 ng/mL guanine.
Supplementary Table 2 | Quantitative metabolomic analysis and overall metabolite identification based on HPLC-LC/MS with ZNC.
Abbas, A., Fu, Y. P., Qu, Z., Zhao, H. Z., Sun, Y. J., Lin, Y., et al. (2021). Isolation and evaluation of the biocontrol potential of Talaromyces spp. against rice sheath blight guided by soil microbiome. Environ. Microbiol. 23, 5946–5961. doi: 10.1111/1462-2920.15596
Abdelkefi, H., Sugliani, M., Ke, H., Harchouni, S., Soubigou-Taconnat, L., Citerne, S., et al. (2018). Guanosine tetraphosphate modulates salicylic acid signalling and the resistance of Arabidopsis thaliana to Turnip mosaic virus. Mol. Plant Pathol. 19, 634–646. doi: 10.1111/mpp.12548
Agrawal, G. K., Rawkwal, R., and Jwa, N. S. (2001). Differential induction of three pathogenesis-related genes, PR10, PR1b, and PR5 by the ethylene generator ethephon under light and dark in rice (Oryza sativa L.) seedlings. J. Plant Physiol. 158, 133–137. doi: 10.1078/0176-1617-00186
Alferez, F. M., Gerberich, K. M., Li, J. L., Zhang, Y., Graham, J. H., and Mou, Z. (2018). Exogenous nicotinamide adenine dinucleotide induces resistance to citrus canker in citrus. Front. Plant Sci. 9:1472. doi: 10.3389/fpls.2018.01472
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Berens, M. L., Berry, H. M., Mine, A., Argueso, C. T., and Tsuda, K. (2017). Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401–425. doi: 10.1146/annurev-phyto-080516-035544
Berrocal-Lobo, M., and Molina, A. (2004). Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 17, 763–770. doi: 10.1094/MPMI.2004.17.7.763
Bian, C. H., Duan, Y. B., Wang, J. Y., Xiu, Q., Wang, J. X., Hou, Y. P., et al. (2020). Validamycin A induces broad-spectrum resistance involving salicylic acid and jasmonic acid/ethylene signaling pathways. Mol. Plant Microbe Interact. 33, 1424–1437. doi: 10.1094/MPMI-08-20-0211-R
Bleecker, A. B., and Kende, H. (2000). Ethylene: a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Brenya, E., Chen, Z. H., Tissue, D., Papanicolaou, A., and Cazzonelli, C. I. (2020). Prior exposure of Arabidopsis seedlings to mechanical stress heightens jasmonic acid-mediated defense against necrotrophic pathogens. BMC Plant Biol. 20:548. doi: 10.1186/s12870-020-02759-9
Cao, J., Liu, B. Y., Xu, X. N., Zhang, X. Y., Zhu, C. X., Li, Y., et al. (2021). Plant endophytic fungus extract ZNC improved potato immunity, yield, and quality. Front. Plant Sci. 12:707256. doi: 10.3389/fpls.2021.707256
Cao, W. L., Chu, R. Z., Zhang, Y., Luo, J., Su, Y. Y., Xie, L. J., et al. (2015). OsJAMyb, a R2R3-type MYB transcription factor, enhanced blast resistance in transgenic rice. Physiol. Mol. Plant Pathol. 92, 154–160. doi: 10.1016/j.pmpp.2015.04.008
Casida, J. E., and Durkin, K. A. (2017). Pesticide chemical research in toxicology: lessons from nature. Chem. Res. Toxicol. 30, 94–104. doi: 10.1021/acs.chemrestox.6b00303
Chen, Y., Zhang, A. F., Wang, W. X., Zhang, Y., and Gao, T. C. (2012). Baseline sensitivity and efficacy of thifluzamide in Rhizoctonia solani. Ann. Appl. Biol. 161, 247–254. doi: 10.1111/j.1744-7348.2012.00569.x
Cheng, H. T., Liu, H. B., Deng, Y., Xiao, J. H., Li, X. H., and Wang, S. P. (2015). The WRKY45-2 WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. doi: 10.1104/pp.114.256016
Chujo, T., Takai, R., Akimoto-Tomiyama, C., Ando, S., Minami, E., Nagamura, Y., et al. (2007). Involvement of the elicitor-induced gene OsWRKY53 in the expression of defense-related genes in rice. Biochim. Biophys. Acta 1769, 497–505. doi: 10.1016/j.bbaexp.2007.04.006
Dahncke, K., and Witte, C. P. (2013). Plant purine nucleoside catabolism employs a guanosine deaminase required for the generation of xanthosine in Arabidopsis. Plant Cell 25, 4101–4109. doi: 10.1105/tpc.113.117184
Du, D., Zhang, C. W., Xing, Y. D., Lu, X., Cai, L. J., Yun, H., et al. (2021). The CC-NB-LRR OsRLR1 mediates rice disease resistance through interaction with OsWRKY19. Plant Biotechnol. J. 19, 1052–1064. doi: 10.1111/pbi.13530
Du, M. M., Zhao, J. H., Tzeng, D. T. W., Liu, Y. Y., Deng, L., Yang, T. X., et al. (2017). MYC2 orchestrates a hierarchical transcriptional cascade that regulates jasmonate-mediated plant immunity in tomato. Plant Cell 29, 1883–1906. doi: 10.1105/tpc.16.00953
Feng, C. S., Zhang, X., Wu, T., Yuan, B., Ding, X. H., Yao, F. Y., et al. (2016). The polygalacturonase-inhibiting protein 4 (OsPGIP4), a potential component of the qBlsr5a locus, confers resistance to bacterial leaf streak in rice. Planta 243, 1297–1308. doi: 10.1007/s00425-016-2480-z
Ferrari, S., Plotnikova, J. M., De Lorenzo, G., and Ausubel, F. M. (2003). Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J. 35, 193–205. doi: 10.1046/j.1365-313x.2003.01794.x
Freudenthal, B. D., Beard, W. A., Perera, L., Shock, D. D., Kim, T., Schlick, T., et al. (2015). Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature 517, 635–639. doi: 10.1038/nature13886
Furuta, Y., Komeno, T., and Nakamura, T. (2017). Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 93, 449–463. doi: 10.2183/pjab.93.027
Gehring, C., and Turek, I. S. (2017). Cyclic nucleotide monophosphates and their cyclases in plant signaling. Front. Plant Sci. 8:1704. doi: 10.3389/fpls.2017.01704
Ghasemnejad-Berenji, M., and Pashapour, S. (2021). Favipiravir and COVID-19: a simplified summary. Drug Res. (Stuttg.) 71, 166–170. doi: 10.1055/a-1296-7935
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Guo, F. L., Wu, T. C., Shen, F. D., Xu, G. B., Qi, H., and Zhang, Z. Y. (2021). The cysteine-rich receptor-like kinase TaCRK3 contributes to defense against Rhizoctonia cerealis in wheat. J. Exp. Bot. 72, 6904–6919. doi: 10.1093/jxb/erab328
Hashida, S. N., Takahashi, H., and Uchimiya, H. (2009). The role of NAD biosynthesis in plant development and stress responses. Ann. Bot. 103, 819–824. doi: 10.1093/aob/mcp019
Helliwell, E. E., Wang, Q., and Yang, Y. (2013). Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnol. J. 11, 33–42. doi: 10.1111/pbi.12004
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Hu, B., Zhou, Y., Zhou, Z. H., Sun, B., Zhou, F., Yin, C. X., et al. (2021). Repressed OsMESL expression triggers reactive oxygen species-mediated broad-spectrum disease resistance in rice. Plant Biotechnol. J. 19, 1511–1522. doi: 10.1111/pbi.13566
Hu, Y. B., Chong, K., and Wang, T. (2008). OsRAF is an ethylene responsive and root abundant factor gene of rice. Plant Growth Regul. 54, 55–61. doi: 10.1007/s10725-007-9228-5
Isner, J. C., and Maathuis, F. J. M. (2018). cGMP signalling in plants: from enigma to main stream. Funct. Plant Biol. 45, 93–101. doi: 10.1071/FP16337
Ju, Y. H., Tian, H. J., Zhang, R. H., Zuo, L. P., Jin, G. X., Xu, Q., et al. (2017). Overexpression of OsHSP18.0-CI enhances resistance to bacterial leaf streak in rice. Rice 10:12. doi: 10.1186/s12284-017-0153-6
Kaneda, T., Taga, Y., Takai, R., Iwano, M., Matsui, H., Takayama, S., et al. (2009). The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 28, 926–936. doi: 10.1038/emboj.2009.39
Khong, G. N., Pati, P. K., Richaud, F., Parizot, B., Bidzinski, P., Mai, C. D., et al. (2015). OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol. 169, 2935–2949. doi: 10.1104/pp.15.01192
Kim, K. H., Kabir, E., and Jahan, S. A. (2017). Exposure to pesticides and the associated human health effects. Sci. Total Environ. 575, 525–535. doi: 10.1016/j.scitotenv.2016.09.009
Kim, S. H., Oikawa, T., Kyozuka, J., Wong, H. L., Umemura, K., Kishi-Kaboshi, M., et al. (2012). The bHLH Rac Immunity1 (RAI1) is activated by OsRac1 via OsMAPK3 and OsMAPK6 in rice immunity. Plant Cell Physiol. 53, 740–754. doi: 10.1093/pcp/pcs033
Kishi-Kaboshi, M., Seo, S., Takahashi, A., and Hirochika, H. (2018). The MAMP-responsive MYB transcription factors MYB30, MYB55 and MYB110 activate the HCAA synthesis pathway and enhance immunity in rice. Plant Cell Physiol. 59, 903–915. doi: 10.1093/pcp/pcy062
Li, N., Lin, B., Wang, H., Li, X. M., Yang, F. F., Ding, X. H., et al. (2019). Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 51, 1540–1548. doi: 10.1038/s41588-019-0503-y
Li, N., Wei, S. T., Chen, J., Yang, F. F., Kong, L. G., Chen, C. X., et al. (2018). OsASR2 regulates the expression of a defence-related gene, Os2H16, by targeting the GT-1 cis-element. Plant Biotechnol. J. 16, 771–783. doi: 10.1111/pbi.12827
Li, W. T., Wang, K., Chern, M., Liu, Y. C., Zhu, Z. W., Liu, J., et al. (2020). Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol. 226, 1850–1863. doi: 10.1111/nph.16505
Li, W. X., Han, Y. Y., Tao, F., and Chong, K. (2011). Knockdown of SAMS genes encoding S-adenosyl-l-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J. Plant Physiol. 168, 1837–1843. doi: 10.1016/j.jplph.2011.05.020
Liang, X. X., Chen, X. J., Li, C., Fan, J., and Guo, Z. J. (2017). Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci. Rep. 7:2474. doi: 10.1038/s41598-017-02643-x
Lin, F. F., Guo, S., Tan, C. W., Zhou, X. G., and Zhang, D. Y. (2020). Identification of rice sheath blight through spectral responses using hyperspectral images. Sensors 20:6243. doi: 10.3390/s20216243
Liu, H. F., Zhang, B. G., Wu, T., Ding, Y. L., Ding, X. H., and Chu, Z. H. (2015). Copper ion elicits defense response in Arabidopsis thaliana by activating salicylate- and ethylene-dependent signaling pathways. Mol. Plant 8, 1550–1553. doi: 10.1016/j.molp.2015.07.008
Liu, X. Q., Bai, X. Q., Wang, X. J., and Chu, C. C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Lu, C. C., Liu, H. F., Jiang, D. P., Wang, L. L., Jiang, Y. K., Tang, S. Y., et al. (2019). Paecilomyces variotii extracts (ZNC) enhance plant immunity and promote plant growth. Plant Soil 441, 383–397.
Lu, J., Ju, H. P., Zhou, G. X., Zhu, C. S., Erb, M., Wang, X. P., et al. (2011). An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 68, 583–596. doi: 10.1111/j.1365-313X.2011.04709.x
Ma, Z. C., Song, T. Q., Zhu, L., Ye, W. W., Wang, Y., Shao, Y. Y., et al. (2015). A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell 27, 2057–2072. doi: 10.1105/tpc.15.00390
McDowell, J. M., and Dangl, J. L. (2000). Signal transduction in the plant immune response. Trends Biochem. Sci. 25, 79–82. doi: 10.1016/s0968-0004(99)01532-7
Meng, X. Z., Xu, J., He, Y. X., Yang, K. Y., Mordorski, B., Liu, Y. D., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Molla, K. A., Karmakar, S., Molla, J., Bajaj, P., Varshney, R. K., Datta, S. K., et al. (2020). Understanding sheath blight resistance in rice: the road behind and the road ahead. Plant Biotechnol. J. 18, 895–915. doi: 10.1111/pbi.13312
Naeimi, S., Okhovvat, S. M., Javannikkhah, M., Vagvolgyi, C., Khosravi, V., and Kredics, L. (2010). Biological control of Rhizoctonia solani AG1-1A, the causal agent of rice sheath blight with Trichoderma strains. Phytopathol. Mediterr. 49, 287–300. doi: 10.14601/Phytopathol_Mediterr-3437
Noctor, G., Queval, G., and Gakiere, B. (2006). NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 57, 1603–1620. doi: 10.1093/jxb/erj202
Oh, T. S., Koo, H. M., Yoon, H. R., Jeong, N. S., Kim, Y. J., and Kim, C. H. (2015). Antifungal action of Ginkgo biloba outer seedcoat on rice sheath blight. Plant Pathol. J. 31, 61–66. doi: 10.5423/PPJ.NT.03.2014.0021
Park, J. H., Choi, G. J., Jang, K. S., Lim, H. K., Kim, H. T., Cho, K. Y., et al. (2005). Antifungal activity against plant pathogenic fungi of chaetoviridins isolated from Chaetomium globosum. FEMS Microbiol. Lett. 252, 309–313. doi: 10.1016/j.femsle.2005.09.013
Park, J. O., El-Tarabily, K. A., Ghisalberti, E. L., and Sivasithamparam, K. (2002). Pathogenesis of Streptoverticillium albireticuli on Caenorhabditis elegans and its antagonism to soil-borne fungal pathogens. Lett. Appl. Microbiol. 35, 361–365. doi: 10.1046/j.1472-765x.2002.01194.x
Peng, X. X., Hu, Y. J., Tang, X. K., Zhou, P. L., Deng, X. B., Wang, H. H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498. doi: 10.1007/s00425-012-1698-7
Ranf, S., Gisch, N., Schaffer, M., Illig, T., Westphal, L., Knirel, Y. A., et al. (2015). A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 16, 426–433. doi: 10.1038/ni.3124
Rodriguez-Salus, M., Bektas, Y., Schroeder, M., Knoth, C., Vu, T., Roberts, P., et al. (2016). The synthetic elicitor 2-(5-bromo-2-hydroxy-phenyl)-thiazolidine-4-carboxylic acid links plant immunity to hormesis. Plant Physiol. 170, 444–458. doi: 10.1104/pp.15.01058
Ruan, J. J., Zhou, Y. X., Zhou, M. L., Yan, J., Khurshid, M., Weng, W. F., et al. (2019). Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 20:2479. doi: 10.3390/ijms20102479
Sathe, A. P., Kumar, A., Mandlik, R., Raturi, G., Yadav, H., Kumar, N., et al. (2021). Role of silicon in elevating resistance against sheath blight and blast diseases in rice (Oryza sativa L.). Plant Physiol. Biochem. 166, 128–139. doi: 10.1016/j.plaphy.2021.05.045
Savary, S., Castilla, N., Elazegui, F., McLaren, C., Ynalvez, M., and Teng, P. (1995). Direct and indirect effects of nitrogen supply and disease source structure on rice sheath blight spread. Phytopathology 85, 959–965.
Schwessinger, B., and Ronald, P. C. (2012). Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. doi: 10.1146/annurev-arplant-042811-105518
Shasmita, H. S., Soumendra, K. N., and Arup, K. M. (2019). Comparative analysis of different biotic and abiotic agents for growth promotion in rice (Oryza sativa L.) and their effect on induction of resistance against Rhizoctonia solani: a soil borne pathogen. Biol. Control 133, 123–133. doi: 10.1016/j.biocontrol.2019.02.013
Singh, P., Mazumdar, P., Harikrishna, J. A., and Babu, S. (2019). Sheath blight of rice: a review and identification of priorities for future research. Planta 250, 1387–1407. doi: 10.1007/s00425-019-03246-8
Sohn, S. I., Kim, Y. H., Kim, B. R., Lee, S. Y., Lim, C. K., Hur, J. H., et al. (2007). Transgenic tobacco expressing the hrpN(EP) gene from Erwinia pyrifoliae triggers defense responses against botrytis cinerea. Mol. Cells 24, 232–239.
Srivastava, A. K., Orosa, B., Singh, P., Cummins, I., Walsh, C., Zhang, C., et al. (2018). SUMO suppresses the activity of the jasmonic acid receptor CORONATINE INSENSITIVE1. Plant Cell 30, 2099–2115. doi: 10.1105/tpc.18.00036
Srivastava, S., Bist, V., Srivastava, S., Singh, P. C., Trivedi, P. K., Asif, M. H., et al. (2016). Unraveling aspects of Bacillus amyloliquefaciens mediated enhanced production of rice under biotic stress of Rhizoctonia solani. Front. Plant Sci. 7:587. doi: 10.3389/fpls.2016.00587
Takahashi, K., Kasai, K., and Ochi, K. (2004). Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. U. S. A. 101, 4320–4324. doi: 10.1073/pnas.0308555101
Tang, Y. F., Hou, S. M., Yang, Y. C., Cheng, D. D., Gao, B., Wan, Y. W., et al. (2019). Cu(II)-based water-dispersible humic acid: synthesis, characterizations, and antifungal and growth-promoting performances. J. Agric. Food Chem. 67, 12987–13000. doi: 10.1021/acs.jafc.9b05145
Tao, Z., Liu, H. B., Qiu, D. Y., Zhou, Y., Li, X. H., Xu, C. G., et al. (2009). A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol. 151, 936–948. doi: 10.1104/pp.109.145623
Tonnessen, B. W., Manosalva, P., Lang, J. M., Baraoidan, M., Bordeos, A., Mauleon, R., et al. (2015). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9
Tripathi, D., Zhang, T., Koo, A. J., Stacey, G., and Tanaka, K. (2018). Extracellular ATP acts on jasmonate signaling to reinforce plant defense. Plant Physiol. 176, 511–523. doi: 10.1104/pp.17.01477
van Loon, L. C., Geraats, B. P., and Linthorst, H. J. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. doi: 10.1016/j.tplants.2006.02.005
Wamaitha, M. J., Yamamoto, R., Wong, H. L., Kawasaki, T., Kawano, Y., and Shimamoto, K. (2012). OsRap2.6 transcription factor contributes to rice innate immunity through its interaction with receptor for Activated Kinase-C 1 (RACK1). Rice 5:35. doi: 10.1186/1939-8433-5-35
Wang, A. J., Shu, X. Y., Jing, X., Jiao, C. Z., Chen, L., Zhang, J. F., et al. (2021). Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol. J. 19, 1553–1566. doi: 10.1111/pbi.13569
Wang, H. H., Hao, J. J., Chen, X. J., Hao, Z. N., Wang, X., Lou, Y. G., et al. (2007). Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol. Biol. 65, 799–815. doi: 10.1007/s11103-007-9244-x
Wang, H. H., Meng, J., Peng, X. X., Tang, X. K., Zhou, P. L., Xiang, J. H., et al. (2015). Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. doi: 10.1007/s11103-015-0360-8
Wu, T., Peng, C. E., Li, B. B., Wu, W., Kong, L. G., Liu, F., et al. (2019). OsPGIP1-mediated resistance to bacterial leaf streak in rice is beyond responsive to the polygalacturonase of Xanthomonas oryzae pv. oryzicola. Rice 12:90. doi: 10.1186/s12284-019-0352-4
Wuriyanghan, H., Zhang, B., Cao, W. H., Ma, B., Lei, G., Liu, Y. F., et al. (2009). The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21, 1473–1494. doi: 10.1105/tpc.108.065391
Xiao, J., Cheng, H. T., Li, X. H., Xiao, J. H., Xu, C. G., and Wang, S. P. (2013). Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 163, 1868–1882. doi: 10.1104/pp.113.226019
Xue, X. J., Geng, T. T., Liu, H. F., Yang, W., Zhong, W. R., Zhang, Z. L., et al. (2021). Foliar application of silicon enhances resistance against Phytophthora infestans through the ET/JA- and NPR1- dependent signaling pathways in potato. Front. Plant Sci. 12:609870. doi: 10.3389/fpls.2021.609870
Yakhin, O. I., Lubyanov, A. A., Yakhin, I. A., and Brown, P. H. (2016). Biostimulants in plant science: a global perspective. Front. Plant Sci. 7:2049. doi: 10.3389/fpls.2016.02049
Yamaguchi, T., Yamada, A., Hong, N., Ogawa, T., Ishii, T., and Shibuya, N. (2000). Differences in the recognition of glucan elicitor signals between rice and soybean: beta-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultured rice cells. Plant Cell 12, 817–826. doi: 10.1105/tpc.12.5.817
Yang, W., Xu, X. N., Li, Y., Wang, Y. Z., Li, M., Wang, Y., et al. (2016). Rutin-mediated priming of plant resistance to three bacterial pathogens initiating the early SA signal pathway. PLoS One 11:e0146910. doi: 10.1371/journal.pone.0146910
Yokotani, N., Sato, Y., Tanabe, S., Chujo, T., Shimizu, T., Okada, K., et al. (2013). WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J. Exp. Bot. 64, 5085–5097. doi: 10.1093/jxb/ert298
Yuan, Y. X., Zhong, S. H., Li, Q., Zhu, Z. R., Lou, Y. G., Wang, L. Y., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zhang, B. G., Liu, H. F., Ding, X. H., Qiu, J. J., Zhang, M., and Chu, Z. H. (2018). Arabidopsis thaliana ACS8 plays a crucial role in the early biosynthesis of ethylene elicited by Cu2+ ions. J. Cell Sci. 131:jcs202424. doi: 10.1242/jcs.202424
Zhao, H., Yin, C. C., Ma, B., Chen, S. Y., and Zhang, J. S. (2021). Ethylene signaling in rice and Arabidopsis: new regulators and mechanisms. J. Integr. Plant Biol. 63, 102–125. doi: 10.1111/jipb.13028
Zhao, Z. X., Feng, Q., Liu, P. Q., He, X. R., Zhao, J. H., Xu, Y. J., et al. (2021). RPW8.1 enhances the ethylene-signaling pathway to feedback-attenuate its mediated cell death and disease resistance in Arabidopsis. New Phytol. 229, 516–531. doi: 10.1111/nph.16857
Keywords: elicitor, ethylene (ET) signaling, guanine, jasmonic acid (JA) signaling, nucleobases, rice sheath blight (ShB)
Citation: Wang L, Liu H, Yin Z, Li Y, Lu C, Wang Q and Ding X (2022) A Novel Guanine Elicitor Stimulates Immunity in Arabidopsis and Rice by Ethylene and Jasmonic Acid Signaling Pathways. Front. Plant Sci. 13:841228. doi: 10.3389/fpls.2022.841228
Received: 22 December 2021; Accepted: 10 January 2022;
Published: 17 February 2022.
Edited by:
Xiaodong Wang, Agricultural University of Hebei, ChinaReviewed by:
Bing Wang, Hunan Agricultural University, ChinaCopyright © 2022 Wang, Liu, Yin, Li, Lu, Wang and Ding. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhua Ding, eGhkaW5nQHNkYXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.