- 1Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
- 2Guangdong Key Laboratory of New Technology in Rice Breeding, Guangzhou, China
- 3Guangdong Rice Engineering Laboratory, Guangzhou, China
Pericarp colors (PC) in rice are determined by the types and content of flavonoids in the pericarp. The flavonoid compounds have strong antioxidant activities and are beneficial to human health. However, the genetic basis of PC in rice is still not well-understood. In this study, a genome-wide association study (GWAS) of PC was performed in a diverse rice collection consisting of 442 accessions using different phenotyping methods in two locations over 2 years. In the whole population consisting of white and colored pericarp rice, a total of 11 quantitative trait loci (QTLs) were identified using two phenotyping methods. Among these QTLs, nine were identified using the phenotypes represented by the presence and absence of pigmentation in pericarp, while 10 were identified using phenotypes of the degree of PC (DPC), in which eight are common QTLs identified using the two phenotyping methods. Using colored rice accessions and phenotypes based on DPC, four QTLs were identified, and they were totally different from the QTLs identified using the whole population, suggesting the masking effects of major genes on minor genes. Compared with the previous studies, 10 out of the 15 QTLs are first reported in this study. Based on the differential expression analysis of the predicted genes within the QTL region by both RNA-seq and real-time PCR (RT-PCR) and the gene functions in previous studies, LOC_Os01g49830, encoding a RAV transcription factor was considered as the candidate gene underlying qPC-1, a novel QTL with a large effect in this study. Our results provide a new insight into the genetic basis of PC in rice and contribute to developing the value-added rice with optimized flavonoid content through molecular breeding.
Introduction
Rice is one of the most important crops, serving as the staple food for more than half of the world’s population. For a long time, the majority of rice planted and consumed worldwide is white rice. The grains of most rice cultivars have white pericarps, but some have colored pericarps, such as brown, red, or black (purple). The colored pericarps accumulate abundant flavonoid compounds, which have strong antioxidant activities and are beneficial to human health (Sangma and Parameshwari, 2021). Therefore, colored rice is arousing more interest and attention.
Previous studies suggest that the types and content of flavonoids determine the pericarp color (PC) in rice grains (Joseph et al., 1998; Furukawa et al., 2006). Red and black rice are the most common types of colored rice, which accumulate proanthocyanidins and anthocyanins in their pericarps, respectively (Reddy et al., 1996; Oki et al., 2002). Proanthocyanidins and anthocyanidins are produced through a special branch of the flavonoid pathway, and they share most of the biosynthetic genes, which have been well-studied (Winkel-Shirley, 2001). Some structural and regulatory genes related to their biosynthesis have been isolated, and their functions have been confirmed. So far, several structural genes have been identified in rice, such as the genes encoding chalcone synthase (OsCHS1 and OsCHS2) (Reddy et al., 1996; Shih et al., 2008), chalcone isomerase (OsCHI) (Druka et al., 2002), flavanone 3-hydroxylase (OsF3H) (Kim et al., 2008), flavonoid 3-hydroxylase (OsF3′H) (Shih et al., 2008), dihydroflavonol 4-reductase (OsDFR/Rd) (Furukawa et al., 2006), anthocyanidin synthase (OsANS) (Shih et al., 2008), and UDP-dependent glucosyltransferase (UGT) (Ko et al., 2008). These structural genes are predominantly regulated by the MYB (v-myb avian myeloblastosis viral oncogene homolog), bHLH (basic helix-loop-helix) and WD40 (tryptophan-aspartate repeats) transcription factors (Koes et al., 2005; Morishita et al., 2009; Hichri et al., 2011; Kim et al., 2011; Czemmel et al., 2012; James et al., 2017; Lloyd et al., 2017).
Classical genetic analysis indicated that two genes, Pb on chromosome 4 and Pp on chromosome 1, are required for the pericarp pigmentation with anthocyanins in black rice (Causse et al., 1994; Yoshimura et al., 1997), while red pericarp is controlled by two major genes, Rd on chromosome 1 and Rc on chromosome 7 (Furukawa et al., 2006; Sweeney et al., 2006). Rd encoding a dihydroflavonol-4-reductase and Rc encoding a bHLH domain-containing transcription factor are involved in the synthesis of proanthocyanidins. The loss-of-function allele, rc, characterized by the 14-bp deletion in Rc that causes a frameshift mutation and a premature stop codon, results in the pericarp pigmentation from red to white. The Rcrd genotype produces brown pericarp, while RcRd produces red pericarp. Introduction of Rd into Rcrd rice changes the PC from brown to red, while rcRd or rcrd produces the white pericarp (Furukawa et al., 2006; Sweeney et al., 2006). So far, studies have identified several allelic variations in Rc that cause the changes in pericarp pigmentation in rice. Except for the 14-bp deletion resulting in a frameshift mutation, one-base transversion also causes premature termination of the Rc protein, leading to white rice (Sweeney et al., 2006; Gross et al., 2010), while a G base deletion in conjunction with the 14-bp deletion restores the reading frame of the gene and reverted the pericarp pigmentation to red (Brooks et al., 2008; Lee et al., 2009).
The development of molecular marker technology provides a powerful tool for dissecting the genetic basis of quantitatively inherited traits. A few QTLs associated with PC in rice have been reported using biparental QTL analysis (Sweeney et al., 2006; Wang and Shu, 2007; Dong et al., 2008; Maeda et al., 2014). With the rapid development of genome sequencing technology, a genome-wide association study (GWAS) based on linkage disequilibrium (LD) in a diverse natural population and using highly dense markers, such as single-nucleotide polymorphisms (SNPs), has been developed and proved to be a powerful approach for identifying genes that control complex traits, such as PC in rice, on a large scale (Wang et al., 2020). Using the GWAS approach, more QTLs for PC were identified in rice (Huang et al., 2010; Shao et al., 2011; Wang et al., 2016; Yang et al., 2018; Wang et al., 2020). Huang et al. (2010, 2011) identified three QTLs associated with PC and two genes, Os02g0650900 and Os08g0301500, encoding glutamate dehydrogenase and sucrose-phosphate synthase, were considered to be candidate genes underlying QTLs on chromosomes 2 and 8, respectively. Wang et al. (2016) identified a new QTL for PC, qPc10, which is unique to Aus, and the gene Os10g0536400, encoding flavanone 3-hydroxylase (F3H), was the candidate gene underlying qPc10. F3H catalyzes the reaction from flavanone to dihydroflavonol, the first committed step of the biosynthesis of proanthocyanidins and anthocyanins (Kim et al., 2008). In another study, 10 QTLs for PC were detected, and MYB, bHLH, and WD transcription factors were speculated as candidate genes for the corresponding QTLs (Yang et al., 2018). Although significant progress has been made in the discovery of QTLs or genes associated with PC in rice, only a few functional genes have been identified. In addition, the masking effect of major genes on QTLs with minor effects is common in complex traits in rice, such as disease resistance (Yu et al., 2003; Liu et al., 2004) and yield-related traits (Zhong et al., 2011). However, rare studies have considered this problem. The QTLs that truly contribute to the degree of PC (DPC) might not be identified because of the masking effect of major genes.
To address the above issues, 442 rice accessions from 61 countries, which were selected from the Rice Diversity Panel 2 (RDP2) and genotyped by 700K SNPs (McCouch et al., 2016), were used for genetic dissection of PC in two locations over 2 years in this study. To identify the QTLs that determine the presence and absence of PC, and the QTLs for the degree of PC (DPC), two different phenotyping methods were used, i.e., presence and absence of pigmentation in the pericarp (PA method) and DPC by comparing with a visual card (DPC method). To eliminate the masking effect of the major genes and identify the QTLs controlling DPC, the colored rice accessions and their DPC were further used for GWAS. Our results suggest that the environment does not affect the presence and absence of PC in rice but the DPC. In total, fifteen QTLs for PC were identified through GWAS in the whole population and the colored rice accessions. Among them, 10 QTLs were identified for the first time in this study, and the other five QTLs were co-localized with the previously identified QTLs or genes including Rc. Interestingly, four QTLs for DPC identified in the colored rice accessions were totally different from those identified in the whole population using phenotype of either DPC or presence and absence of pigmentation, suggesting that there exist masking effect of major genes on QTLs controlling DPC in rice. Based on gene differential expression analysis, gene annotation, and literature, LOC_Os01g49830, encoding a RAV transcription factor, was considered as the candidate gene underlying qPC-1, a novel QTL with a large effect in this study. This study provides new insight into the genetic basis of PC in rice and contributes to molecular breeding for colored rice.
Materials and Methods
Plant Materials
A subset of the RDP2 consisting of 442 rice accessions from 61 countries was used for GWAS in this study (Supplementary Table 1). These rice accessions were selected from the RDP2 consisting of 1,568 rice accessions based on their origins and diversity, including three groups of Indica (218 accessions), Japonica (148 accessions), and Aus (76 accessions) (McCouch et al., 2016).
Evaluation of Pericarp Colors
The 442 rice accessions were planted in the experimental bases of Guangzhou (2016GZ) and Yangjiang (2018YJ) in Guangdong Province, China, in the second cropping season in 2016 and 2018, respectively. The experiments were arranged in a randomized complete block design with two replicates. The field management, including irrigation, fertilization, and disease and pest control, followed the conventional practice in rice production. At complete maturity, rice grains for each accession were harvested and dried naturally. A total of 50 healthy grains for each accession were randomly selected and carefully hulled. In this study, two phenotyping methods were used to evaluate the PC. First, the PC of each accession was evaluated based on the presence (1) or absence (0) of pigmentation in the pericarp (PA method). To better exhibit the PC variations in the diverse rice panel, we developed a visual card with scores from 1 (white) to 7 (dark brown), according to the DPC from light to dark (DPC method, Figure 1A). Then the DPC was assessed by comparing with the visual card and given a score for each accession (Figure 1A). DPC of each accession was scored separately by three individuals, and the consistent scores were used for GWAS analysis.
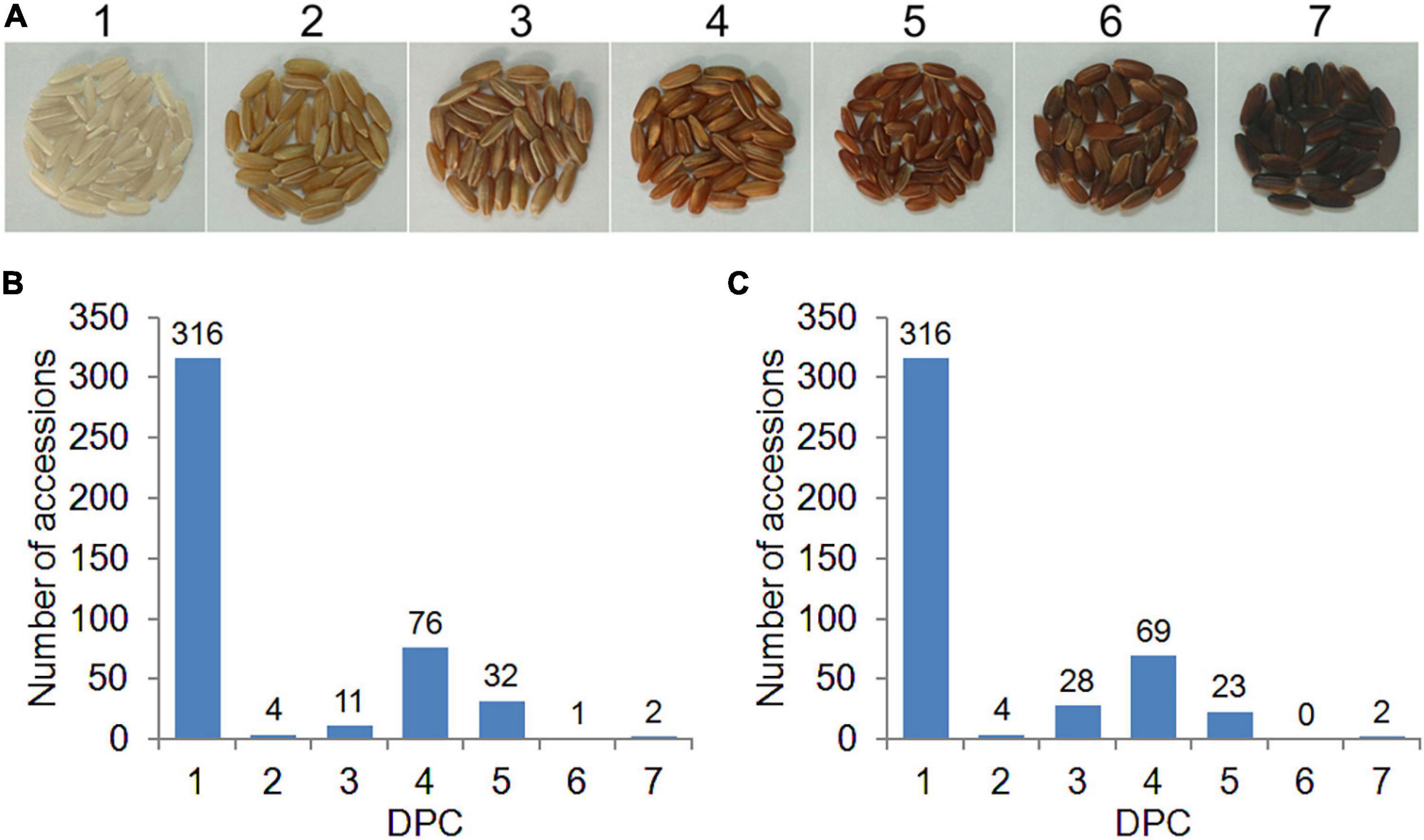
Figure 1. Distribution of pericarp color (PC) in 442 rice accessions. (A) The visual card with scores from 1 (white) to 7 (dark brown) is based on the degree of PC (DPC) from light to dark. (B,C) Distribution of DPC in 2016GZ and 2018YJ, respectively.
Genome-Wide Association Study Analysis and Quantitative Trait Loci Delimitation
Genome-wide association study analysis was performed as described in our previous study (Zhao et al., 2018) by using GAPIT version 2 software (Tang et al., 2016) and high-density rice array (HDRA) dataset consisting of 700K SNPs (McCouch et al., 2016). Briefly, SNPs were filtered using the criteria of having less than 30% of missing data and minor allele frequency (MAF) > 0.05 (McCouch et al., 2016). GWAS was conducted using the mixed linear model (MLM) with kinship matrix, and the principal component was set to 3 in GAPIT. R-package qqman (Turner, 2014) was used to produce Manhattan and QQ plots. A region having two or more than two significant SNPs (p < 0.0001) (Zhao et al., 2018; Yang et al., 2021) within 160 kb is considered as one QTL, according to 80 kb of the LD decay in the population used in this study (Supplementary Figure 1).
RNA-Sequencing
Four lines with consistent flowering time, including two lines with white pericarp (accessions 611 and 689) and two lines with red pericarp (accessions 414 and 752), were selected based on haplotype analysis to conduct RNA-sequencing. The spikes were sampled on the15th day after flowering, and total RNA was extracted from spikes using Trizol reagent (Invitrogen). RNA-seq was performed using Annoroad Gene Technology (Beijing, China), and data analysis was conducted using the HISAT2-Stringtie-Deseq2 pipeline (Pertea et al., 2016). Raw counts of each sample exported from Stringtie were imported and normalized by DEseq2. Genes with read count less than 30 were regarded as having no expression. Then, differentially expressed genes between two sets of contrasting samples were identified according to the criteria of p-value ≤ 0.05 and fold change ≥2 or ≤0.5 between white and red pericarp.
Real-Time PCR Analysis
cDNA synthesis was performed using the PrimeScript™ RT reagent kit (Takara, Japan). The PCR analysis was performed using the BioRad CFX 96 system. The primers were designed using the Primer designing tool on NCBI.1 The EF1α was used as the normalized genes for mRNA. The same RNA samples used in RNA-Seq assays were used to confirm the results of RNA-Seq, and all reactions were repeated three times. The gene-specific primers used in this study are listed in Supplementary Table 3.
Results
Phenotypic Variations of Pericarp Colors in 442 Rice Accessions
To better exhibit the PC variations of the diverse rice panel and identify the QTLs associated with DPC, we developed a visual card with scores of 1–7 based on the DPC from light to dark (Figure 1A). According to the scores of DPC, the whole population used in this study consisted of 316 white accessions (score 1) and 126 colored accessions (score 2–7), and the DPC in the 442 accessions displayed a continuous distribution. Large variations in DPC were observed in the colored rice accessions, ranging from score 2 to 7 (Figures 1B,C). Interestingly, the DPC of white rice accessions remained the same (score 1), while the DPC of about half of the colored rice accessions varied across environments (2016GZ and 2018YJ) (Supplementary Table 1 and Figures 1B,C). A highly and a moderately positive correlation was observed between the DPC in 2016GZ and 2018YJ in the whole population (r = 0.9627, p < 0.0001) and colored population (r = 0.5825, p < 0.0001), respectively, indicating that the grain pericarps with or without pigment in rice are highly hereditary, while the quantitative inheritance of DPC is affected by the environment.
Identification of Quantitative Trait Loci for Pericarp Colors by Genome-Wide Association Study
Based on the criteria of having less than 30% of missing data and MAF more than 5% in the whole population, 572,690 SNPs were selected for GWAS from the 700K SNP dataset in the Open Rice GWAS Platform (McCouch et al., 2016). GWAS of PC was conducted using the whole and the colored population, and phenotypes were generated using different phenotyping methods, respectively. According to the LD decay analysis, the LD distance of the population used in this study is about 80 kb (Supplementary Figure 1). Thus, a QTL was declared when there are two or more significant SNPs (Zhao et al., 2018; Yang et al., 2021) (p < 0.0001) within a 160-kb region centered on the most significant SNP.
In the whole population consisting of white and colored rice, a total of 11 QTLs were identified in two environments (2016GZ and 2018YJ) using two phenotyping methods: the presence and absence of pigmentation in the pericarp (PA method) and DPC method. Among these QTLs, nine QTLs (qPC-1, qPC-3, qPC-6, qPC-7, qPC-8a, qPC-8b, qPC-8c, qPC-9, and qPC-11) located on chromosomes 1, 3, 6, 7, 8, 9, and 11 were identified using the PA method, and all of them were identical in two locations over 2 years (2016GZ and 2018YJ) (Figure 2A and Table 1), while 10 QTLs were identified using the DPC method, in which qPC-2 and qPC-8d were only detected using the DPC method in 2016GZ and 2018YJ, respectively, and other eight QTLs (qPC-1, qPC-6, qPC-7, qPC-8a, qPC-8b, qPC-8c, qPC-9, and qPC-11) were detected in both environments (2016GZ and 2018YJ) (Figure 2B and Table 1). Notably, these eight QTLs were also identified using the PA method, and qPC-7, which overlapped with the major gene Rc, had the largest contribution to the phenotypic variations in the whole population.
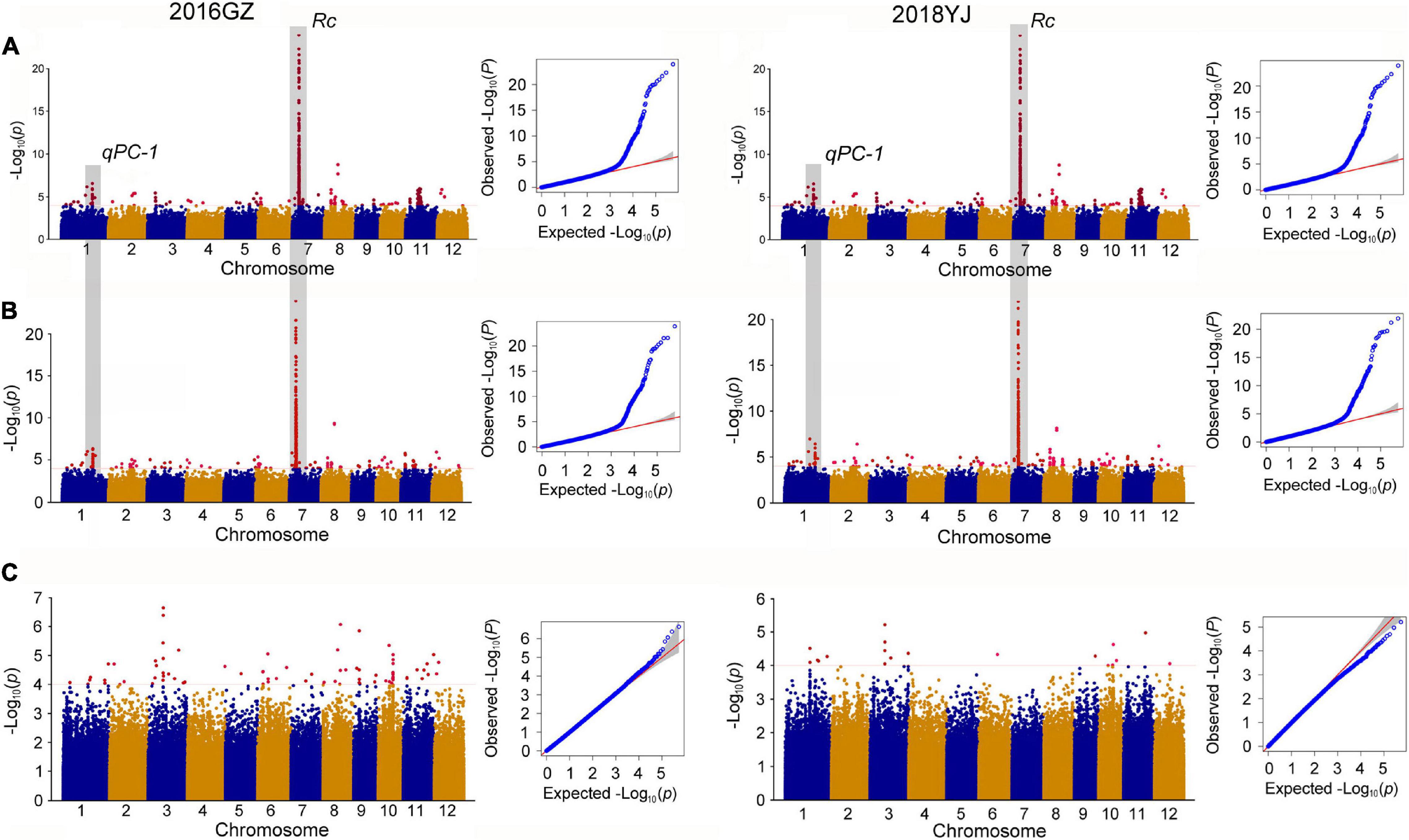
Figure 2. Genome-wide association study for PC. (A,B) Manhattan plots of GWAS in 12 chromosomes and QQ plots of p-values conducted in the whole population using the PA and DPC phenotyping method, respectively. PA method, pericarp color were evaluated based on presence or absence of pigmentation; DPC method, pericarp color were evaluated based on the degree of pericarp color (DPC) using the visual card with scores of 1–7. (C) Manhattan plots of GWAS in 12 chromosomes and QQ plot of p-value conducted in the colored population.
To eliminate the masking effects of major genes on minor genes or minor effect QTLs and identify the QTLs controlling DPC, we removed the white rice accessions and used the colored rice and their DPC for GWAS. Indeed, the major gene Rc responsible for the production of pigment in rice (Furukawa et al., 2006; Sweeney et al., 2006) was not detected, but four QTLs for DPC were detected in this colored population (Figure 2C and Table 1). The four QTLs were mapped on chromosomes 3, 6, 9, and 10 and designated as qPC_C-3, qPC_C-6, qPC_C-9, and qPC_C-10. The qPC_C-3 could be detected in both environments, while the other three QTLs could only be detected in 2016GZ.
Candidate Gene Analysis of qPC-1
Among the stably expressed and newly identified QTLs using the whole population in this study, qPC-1 explained the largest phenotypic variation (Figures 2A,B and Table 1). The LD decay analysis in the QTL region indicated that an approximately 240 kb region (from 28.44 to 28.68 Mb on chromosome 1) was the putative region for qPC-1 (Figures 3A,B). Based on release 7 of the MSU Rice Genome Annotation Project2 (Kawahara et al., 2013), there are 31 annotated genes within the qPC-1 region (Supplementary Table 2). To reduce the number of candidate genes, we selected four lines with consistent flowering time, including two lines with white pericarp (accessions 611 and 689) and two lines with red pericarp (accessions 414 and 752) based on haplotype analysis of qPC-1, to conduct gene differential expression analysis. RNA-seq revealed that 17 genes were not expressed (Supplementary Table 2). Among the left 14 expressed genes, two genes, LOC_Os01g49740 and LOC_Os01g49830, were differentially expressed between the two sets of contrasting lines (Figures 3C,D and Supplementary Table 2). Furthermore, qRT-PCR assays also confirmed these results (Figures 3E,F). However, the two genes exhibited the opposite expression patterns: the expression levels of LOC_Os01g49740 in red pericarp lines were consistently and significantly lower than that in the white pericarp lines (p < 0.05), while LOC_Os01g49830 in the red pericarp lines were significantly higher than that in white pericarp lines (p < 0.05).
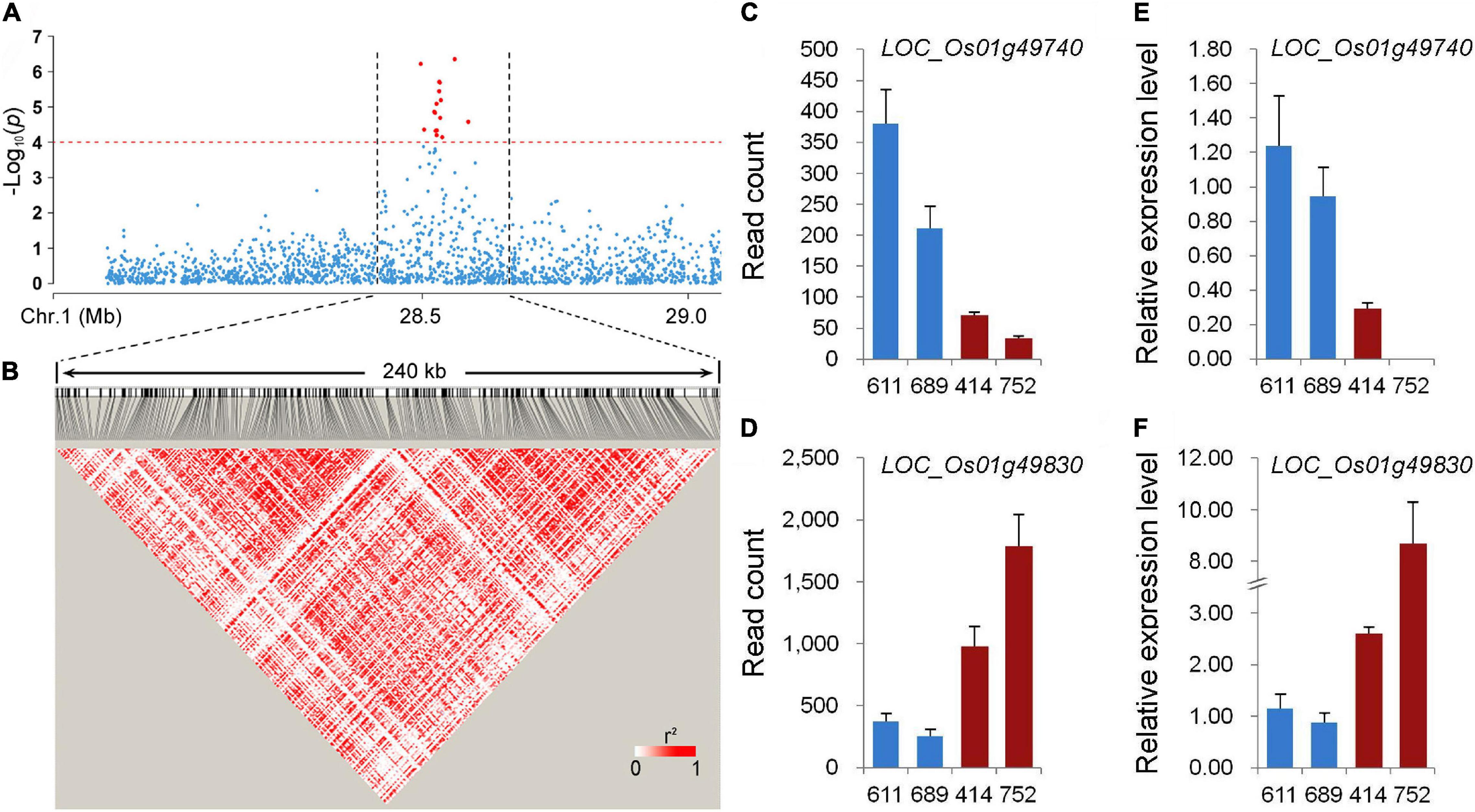
Figure 3. Candidate gene analysis of qPC-1. (A) Local Manhattan plot of GWAS for qPC-1. (B) Linkage disequilibrium (LD) heatmap around the peak. (C,E) The expression of LOC_Os01g49740 in lines with white pericarp (accessions 611 and 689) and red pericarp (accessions 414 and 752) measured using RNA-seq and qRT-PCR, respectively. (D,F) The expression of LOC_Os01g49830 in lines with white and red pericarp measured using RNA-seq and qRT-PCR, respectively.
Analysis of the Allelic Variation of Rc Gene
A previous study demonstrated that the white pericarp is mainly caused by a 14-bp deletion in exon 6 of the Rc gene compared with the brown or red pericarp (Sweeney et al., 2006). To investigate allelic variations of the Rc gene, we used a pair of Indel primers designed based on the 14-bp deletion (HM1, Supplementary Table 3) (Yang et al., 2017) to genotype the 100 lines with white pericarp and the 126 lines with colored pericarp. The PCR assay showed that the three lines (accessions 536, 592, and 902 from Aus sub-group) with white pericarp exhibited the same genotype as those with red pericarp (Figures 4A,B), suggesting that the Rc gene in the three lines with white pericarp has no 14-bp deletion. We further sequenced the Rc gene using four pairs of primers (Os07g0211500-1∼4, Supplementary Table 3) (Yang et al., 2017) in the three lines and other 26 lines, including 12 lines with white pericarp and 14 lines with red pericarp. In fact, the sequencing results revealed that the 12 lines with white pericarp had a 14-bp deletion (CDS1408–CDS1421) in the Rc gene compared with 14 lines with red pericarp, while the three lines did not have the 14-bp deletion in the Rc gene (Figure 4C) but had a C-to-A transversion at CDS1353 in their Rc gene compared with the 26 lines, which results in a termination codon (TGA) and a premature protein (Figure 4C). This result is consistent with the previous studies (Sweeney et al., 2007).
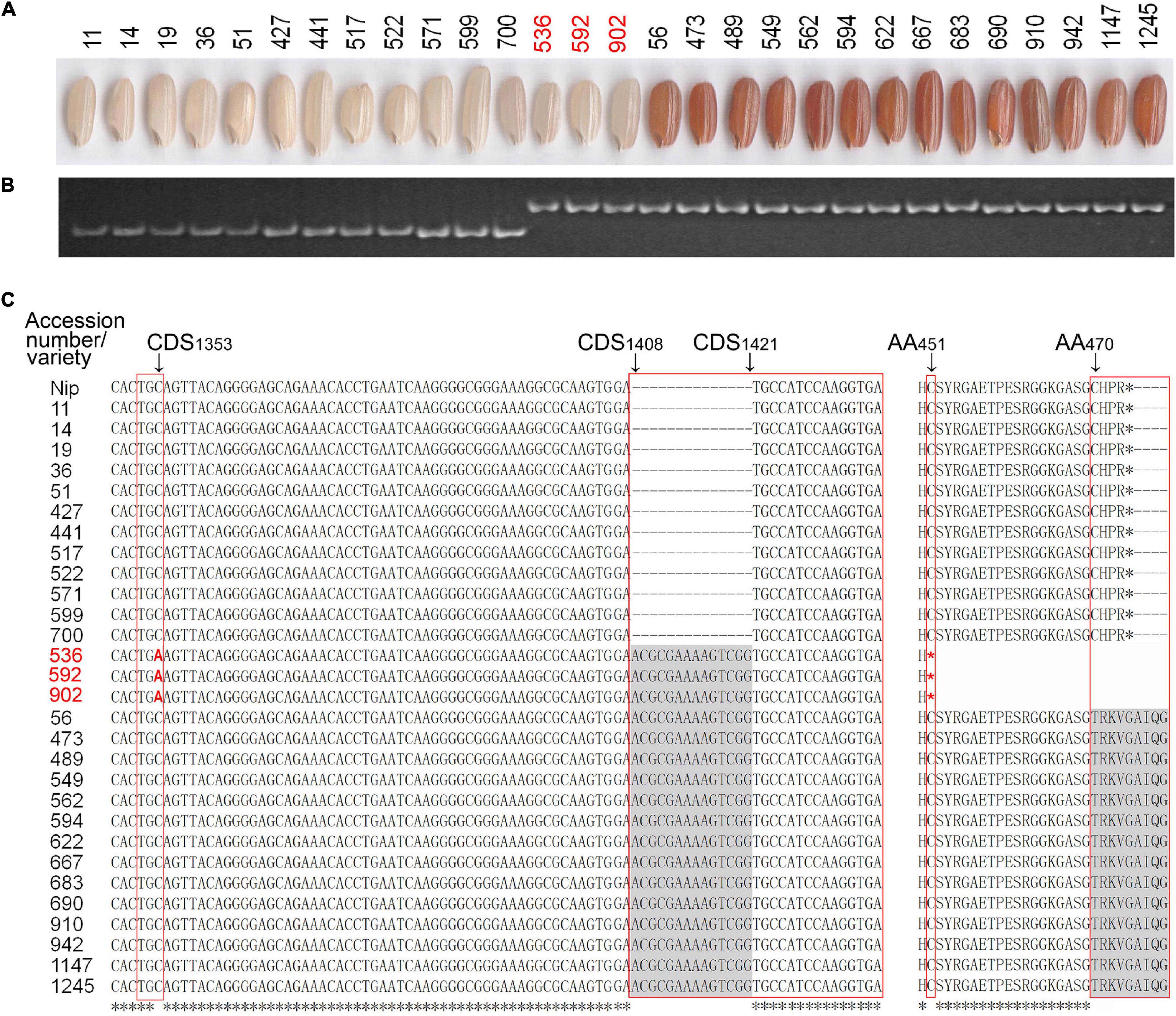
Figure 4. Analysis of allelic variation in Rc gene. (A) Exhibition of PC in the rice accessions. (B) Three rice accessions (accessions 536, 592, and 902) with white pericarp exhibited the same polymorphism as those with red pericarp in polyacrylamide gel electrophoresis. (C) Variation in the CDS region of the three and other rice accessions. The three rice accessions (accessions 536, 592, and 902) are highlighted in red fonts and the various regions are framed in red. Nip, Nippobare; CDS, coding sequence; AA, amino acid.
Discussion
The PC in rice is a complex trait. Although it is well-known that proanthocyanidins and anthocyanins are responsible for PC and their biosynthetic pathway has been well-characterized, the genetic basis of PC in rice has not been well understood. In this study, we evaluated the PC of 442 rice accessions in two environments (2016GZ and 2018YJ) using different two phenotyping methods and GWAS of PC was conducted. Guangzhou belongs to the subtropical monsoon climate, while Yangjiang belongs to the subtropical maritime monsoon climate. The difference in light and temperature affects rice growth as manifested by the significant difference in growth periods of the rice accessions planted in Guangzhou (2016GZ) and Yangjiang (2018YJ) (p < 0.01, unpublished data). It is reported that light is an important factor affecting the expression of the flavonoid biosynthetic genes (Chen et al., 2013), suggesting that the difference in light and temperature may also lead to the variation in DPC of rice planted in the two places. Our results indicated that the white or colored pericarp of 442 rice accessions remain unchanged, while the DPC of the colored rice accessions varied across environments, suggesting that the presence and absence of PC in rice is a qualitative trait not affected by the environment, while the DPC of colored rice is a quantitative trait affected by the environment. In the whole population consisting of white and colored rice accessions, nine QTLs were identified using the phenotypes represented by presence and absence of pigmentation in pericarp, all of them were identical in both environments and determined the presence or absence of PC; while 10 QTLs were identified using DPC phenotypes. Interestingly, 8 out of 10 QTLs identified using DPC were the same as those identified using phenotypes of presence and absence of pigmentation, including qPC-7, which overlapped with the major gene Rc. After removing the white rice accessions, four QTLs for DPC were identified in the colored population. To our surprise, these four QTLs are totally different from the QTLs identified in the whole population using the phenotype of either DPC or the presence and absence of pigmentation. These results together suggest that there exist strong masking effects of major genes on QTLs with minor effects. This can explain why the QTLs for DPC identified in the colored rice population could not be detected in the whole population. Based on the coincidence of most of the QTLs identified in the whole population using either DPC phenotypes or the phenotypes represented by presence and absence of pigmentation and they were less affected by the environment, we believe that majority of QTLs identified in the whole population using DPC phenotypes could be still the QTLs determining the presence or absence of PC because of the masking effects of major genes. A previous study identified the same QTLs for DPC using a backcross-recombinant inbred line population consisting of white and colored rice lines in two different environments (China and Japan) (Dong et al., 2008), which might also be the evidence to support our inference. Therefore, it is critical to eliminate the masking effects of major genes before the QTLs controlling DPC in rice can be identified.
In total, 15 QTLs associated with PC in rice were identified in this study. Compared with the previous studies, 10 out of the 15 QTLs are first reported in this study, and the other five QTLs are co-localized with the previously identified QTLs or genes (Table 1), suggesting the reliability of the results and the diversity of the rice germplasm used in this study. The qPC-7 overlapped with the Rc gene, which encodes a bHLH transcription factor regulating proanthocyanidin synthesis (Sweeney et al., 2006). The qPC-8c was co-localized with the previously reported QTL, whose candidate gene was Os08g0301500, which encodes a sucrose-phosphate synthase (Huang et al., 2010, 2011). The qPC-8d was co-localized with the previously reported QTL associated with grain color and flavonoid content (Shao et al., 2011). The qPC_C-6 was co-localized with OsC1, an MYB transcription factor, which regulates anthocyanin biosynthesis in rice apiculus and sheath (Gao et al., 2011; Hu et al., 2020). Furthermore, qPC_C-9 overlaps with qDRC-9 for the degree of red coloration in rice pericarp (Dong et al., 2008).
Among the newly identified QTLs for PC using the whole population in this study, qPC-1 could be detected across environments using different phenotyping methods and explain the largest phenotypic variation (Table 1). To identify the gene underlying qPC-1, we delimited it to a 240-kb region containing 31 putative genes based on the LD decay analysis (Figures 3A,B and Supplementary Table 2). Only two genes, LOC_Os01g49740 and LOC_Os01g49830, were differentially expressed between the two sets of contrasting lines with white and red pericarps (Figures 3C–F) based on RNA-seq and qRT-PCR assays. LOC_Os01g49740 encodes a domain of the unknown function (DUF) domain-containing protein. It has been reported that DUF proteins are involved in plant growth, development, and abiotic stress (Li et al., 2012; Ganie et al., 2017). LOC_Os01g49830 encodes a Related to ABI3/VP1 (RAV) transcription factor, which belongs to the subfamily member of the AP2/ERF family. Himi et al. (2011) isolated three F3H genes, which were highly expressed in red grains and red coleoptiles but not in white tissues in wheat and found an RAV1 binding site in their promoters. Recently, two groups simultaneously reported that RAV transcription factors were involved in the regulation of flavonoid biosynthesis in fruit plants (Zhang et al., 2020; Zhao et al., 2020). Zhao et al. (2020) identified three AP2/ERF transcription factors (CitRAV1, CitERF32, and CitERF33), which positively regulate the expression of CHI (CitCHIL1) in citrus. CitERF32 and CitERF33 activated the transcription by directly binding to the promoter, while CitRAV1 formed a transcription complex with CitERF33 to strongly enhance the activation efficiency and flavonoid accumulation. The other study reported that FaRAV1 stimulated anthocyanin accumulation in strawberries by directly activating transcription of FaMYB10 and the structural genes of anthocyanins biosynthesis, CHS, F3H, DFR, and GT (Zhang et al., 2020). The CHS, CHI, F3H, and DFR are the important structural genes in pathways of flavonoid biosynthesis (Winkel-Shirley, 2001; Kim et al., 2008). Based on gene differential expression analysis in this study and the gene functions in the previous studies, LOC_Os01g49830 is more likely the candidate gene underlying qPC-1. However, further transgenic experiments are needed to confirm its function on PC in rice.
Among the four QTLs for DPC identified in the colored population, qPC_C-3 explained the largest phenotypic variation and was the only QTL detected across environments. Therefore, it has a great potential value in rice breeding for DPC. By analyzing the distribution of nucleotides at the most significant SNP position of qPC_C-3 in the rice accessions with different DPC, it was found that most of the rice accessions (78.6%) with dark PC (DPC 5-7) harbor the “T,” while only 13.8% of the rice accessions with light PC (DPC 2-4) harbor the “T.” The result provides valuable information for the development of the SNP marker for qPC_C-3 selection in rice breeding.
Rc is the most important regulatory gene involved in the proanthocyanidin synthesis in rice pericarp. Compared with red pericarp, the most white pericarp is caused by a loss-of-function allele, rc, characterized by a 14-bp deletion in exon 6 of Rc gene (Furukawa et al., 2006; Sweeney et al., 2007). In this study, an allele of the Rc gene in white pericarp without a 14-bp deletion in exon 6 was identified in the three Aus accessions. Further sequence analysis showed that a C-to-A transversion at CDS1353 exists in the allele of the Rc gene, which causes early termination of translation (Figure 4). Previous studies also identified this allelic variation in Rc, namely Rc-s, which is mainly found in the Aus subpopulation and produces white pericarps but light-red pericarps in some genetic backgrounds (Sweeney et al., 2006, 2007). Recently, Wang et al. (2016) identified an Aus-specific QTL, qPc10, and indicated that the presence or absence of qPc10 resulted in light-red or white pericarps, suggesting that interaction between Rc alleles and other loci may also affect the PC in different lineages. Except for rc and Rc-s, the rc-gl characterized by an A-to-T substitution at CDS1207 in Rc also results in a premature stop-codon and a white pericarp in the domesticated African rice (Gross et al., 2010). Furthermore, Rcr and Rc-g, having a G base deletion at the 20 or 44 bp upstream of the 14-bp deletion, cause reverse mutation to red pericarp (Brooks et al., 2008; Lee et al., 2009). This inspires us that these mutant regions are potentially important as cis-regulatory regions. It remains to be studied whether mutations in other genes also cause a difference in PC.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
WY, BL, and SZ conceived and designed the research. WY, LC, and JZ conducted the experiments, performed data analysis, and wrote the manuscript. JW, WL, TY, JD, YM, LZ, JC, and WW participated in material development, sample preparation, and data analysis. WY and BL drafted proposals and corrected the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Scientific and Technological Plan of Guangzhou (201804020078 and 202102021005), the Innovation Team Project of Guangdong Modern Agricultural Industrial System (2021KJ106), the Guangdong Basic and Applied Basic Research Foundation (2020A1515010906), the Special Fund for Scientific Innovation Strategy-Construction of High-Level Academy of Agriculture Science (R2021PY-QF002, 202027, and R2018QD-010), and the Guangdong Key Laboratory of New Technology in Rice Breeding (2020B1212060047).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.841191/full#supplementary-material
Supplementary Figure 1 | The linkage disequilibrium (LD) decay in the 442 rice accessions.
Footnotes
References
Brooks, S. A., Yan, W., Jackson, A. K., and Deren, C. W. (2008). A natural mutation in rc reverts white-rice-pericarp to red and results in a new. dominant, wild-type allele: Rc-g. Theor. Appl. Genet. 117, 575–580. doi: 10.1007/s00122-008-0801-8
Causse, M. A., Fulton, T. M., Cho, Y. G., Ahn, S. N., Chunwongse, J., Wu, K. S., et al. (1994). Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138, 1251–1274. doi: 10.1093/genetics/138.4.1251
Chen, X. Q., Itani, T., Wu, X. J., Chikawa, Y., and Irifune, K. (2013). Physiological factors affecting transcription of genes involved in the flavonoid biosynthetic pathway in different rice varieties. Plant Signal. Behav. 8:e27555. doi: 10.4161/psb.27555
Czemmel, S., Heppel, S. C., and Bogs, J. (2012). R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma 249, S109–S118. doi: 10.1007/s00709-012-0380-z
Dong, Y. J., Xu, J. L., Xiao, K., Zhang, Y. J., Zhang, J. Z., Luo, L. J., et al. (2008). Genomic regions associated with the degree of red coloration in pericarp of rice (Oryza sativa L.). J. Cereal Sci. 48, 556–560. doi: 10.1016/j.jcs.2007.11.011
Druka, A., Kudrna, D., Rostoks, N., Brueggeman, R., Von-wettstein, D., and Kleinhofs, A. (2002). Chalcone isomerase gene from rice (Oryza sativa) and barley (Hordeum vulgare): physical, genetic and mutation mapping. Gene 302, 171–178. doi: 10.1016/s0378-1119(02)01105-8
Furukawa, T., Maekawa, M., Oki, T., Suda, I., Iida, S., Shimada, H., et al. (2006). The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 49, 91–102. doi: 10.1111/j.1365-313X.2006.02958.x
Ganie, S. A., Pani, D. R., and Monda, T. K. (2017). Genome-wide analysis of DUF221 domain-containing gene family in Oryza species and identification of its salinity stress-responsive members in rice. PLoS One 12:e0182469. doi: 10.1371/journal.pone.0182469
Gao, D. Y., He, B., Zhou, Y. H., and Sun, L. H. (2011). Genetic and molecular analysis of a purple sheath somaclonal mutant in japonica rice. Plant Cell Rep. 30, 901–911. doi: 10.1007/s00299-011-1004-3
Gross, B. L., Steffen, F. T., and Olsen, K. M. (2010). The molecular basis of white pericarps in African domesticated rice: novel mutations at the Rc gene. J. Evol. Biol. 23, 2747–2753. doi: 10.1111/j.1420-9101.2010.02125.x
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 2465–2483. doi: 10.1093/jxb/erq442
Himi, E., Maekawa, M., and Noda, K. (2011). Differential expression of three flavanone 3-hydroxylase genes in grains and coleoptiles of wheat. Int. J. Genomics 2011:369460. doi: 10.1155/2011/369460
Hu, W., Zhou, T. H., Han, Z. H., Tan, C., and Xing, Y. Z. (2020). Dominant complementary interaction between OsC1 and two tightly linked genes, Rb1 and Rb2, controls the purple leaf sheath in rice. Theor. Appl. Genet. 133, 2555–2566. doi: 10.1007/s00122-020-03617-w
Huang, X. H., Wei, X. H., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. doi: 10.1038/ng.695
Huang, X. H., Zhao, Y., Wei, X. H., Li, C. Y., Wang, A., Zhao, Q., et al. (2011). Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 44, 32–39. doi: 10.1038/ng.1018
James, A. M., Ma, D., Mellway, R., Gesell, A., Yoshida, K., Walker, V., et al. (2017). Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol. 174, 154–171. doi: 10.1104/pp.16.01962
Joseph, M., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. doi: 10.1007/978-94-011-4661-6_134
Kawahara, Y., Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Kim, C. K., Cho, M. A., Choi, Y. H., Kim, J. A., Kim, Y. H., Kim, Y. K., et al. (2011). Identification and characterization of seed-specific transcription factors regulating anthocyanin biosynthesis in black rice. J. Appl. Genet. 52, 161–169. doi: 10.1007/s13353-011-0027-3
Kim, J. H., Lee, Y. J., Kim, B. G., Lim, Y., and Ahn, J. H. (2008). Flavanone 3β-hydroxylases from rice: Key enzymes for favonol and anthocyanin biosynthesis. Mol. Cells 25, 312–316. doi: 10.1016/j.febslet.2008.03.057
Ko, J. H., Kim, B. G., Kim, J. H., Kima, H., Lima, C. E., Lima, J., et al. (2008). Four glucosyltransferases from rice: cDNA cloning, expression, and characterization. J. Plant Physiol. 165, 435–444. doi: 10.1016/j.jplph.2007.01.006
Koes, R., Verweij, W., and Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242. doi: 10.1016/j.tplants.2005.03.002
Lee, D., Lupotto, E., and Powell, W. (2009). G-string slippage turns white rice red. Genome 52, 490–493. doi: 10.1139/g09-025
Li, X. J., Sun, L. J., Tan, L. B., Liu, F. X., Zhu, Z. F., Fu, Y. C., et al. (2012). TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol. Biol. 78, 351–359. doi: 10.1007/s11103-011-9868-8
Liu, B., Zhang, S. H., Zhu, X. Y., Yang, Q. Y., Wu, S. Z., Mei, M. T., et al. (2004). Candidate defense genes as predictors of quantitative blast resistance in rice. Mol. Plant Microbe Interact. 17, 1146–1152. doi: 10.1094/MPMI.2004.17.10.1146
Lloyd, A., Brockman, A., Aguirre, L., Campbell, A., Bean, A., Cantero, A., et al. (2017). Advances in the MYB–bHLH–WD repeat MBW pigment regulatory model: addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 58, 1431–1441. doi: 10.1093/pcp/pcx075
Maeda, H., Yamaguchi, T., Omoteno, M., Takarada, T., Fujita, K., Murata, K., et al. (2014). Genetic dissection of black grain rice by the development of a near isogenic line. Breed. Sci. 64, 134–141. doi: 10.1270/jsbbs.64.134
McCouch, S. R., Wright, M. H., Tung, C. W., Maron, L. G., McNally, K. L., Fitzgerald, M., et al. (2016). Open access resources for genome-wide association mapping in rice. Nat. Commun. 7:10532. doi: 10.1038/ncomms10532
Morishita, T., Kojima, Y., Maruta, T., Nishizawa-Yokoi, A., Yabuta, Y., and Shigeoka, S. (2009). Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 50, 2210–2222. doi: 10.1093/pcp/pcp159
Oki, T., Masuda, M., Kobayashi, M., Nishiba, Y., Furuta, S., Suda, I., et al. (2002). Polymeric procyanidins as radical-scavenging components in red hulled rice. J. Agr. Food Chem. 50, 7524–7529. doi: 10.1021/jf025841z
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11, 1650–1659. doi: 10.1038/nprot.2016.095
Reddy, A. R., Scheffler, B., Madhuri, G., Srivastava, M. N., Kumar, A., Sathyanarayanan, P. V., et al. (1996). Chalcone synthase in rice (Oryza sativa L.): detection of the CHS protein in seedlings and molecular mapping of the chs locus. Plant Mol. Biol. 32, 735–743. doi: 10.1007/BF00020214
Sangma, H. C. R., and Parameshwari, S. (2021). Health benefits of black rice (Zizania aqatica) – a review. Mater. Today doi: 10.1016/j.matpr.2021.07.257
Shao, Y. F., Jin, L., Zhang, G., Lu, Y., Shen, Y., and Bao, J. S. (2011). Association mapping of grain color, phenolic content, flavonoid content and antioxidant capacity in dehulled rice. Theor. Appl. Genet. 122, 1005–1016. doi: 10.1007/s00122-010-1505-4
Shih, C. H., Chu, H., Tang, L. K., Sakamoto, W., Maekawa, M., Chu, I. K., et al. (2008). Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228, 1043–1054. doi: 10.1007/s00425-008-0806-1
Sweeney, M. T., Thomson, M. J., Cho, Y. J., Park, Y. J., Williamson, S. H., Bustamante, C. D., et al. (2007). Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 3:e133. doi: 10.1371/journal.pgen.0030133
Sweeney, M. T., Thomson, M. J., Pfeil, B. E., and McCouch, S. (2006). Caught red-handed: rc encodes a basic helixloop-helix protein conditioning red pericarp in rice. Plant Cell 18, 283–294. doi: 10.1105/tpc.105.038430
Tang, Y., Liu, X. L., Wang, J. B., Li, M., Wang, Q. S., Tian, F., et al. (2016). GAPIT version 2: an enhanced integrated tool for genomic association and prediction. Plant Genome 9:2. doi: 10.3835/plantgenome2015.11.0120
Turner, S. D. (2014). Qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. BioRxiv [Preprint]. doi: 10.1101/005165
Wang, C. X., and Shu, Q. Y. (2007). Fine mapping and candidate gene analysis of purple pericarp gene Pb in rice (Oryza sativa L.). Chinese Sci. Bull. 52, 3097–3104. doi: 10.1007/s11434-007-0472-x
Wang, H., Xu, X., Vieira, F. G., Xiao, Y., Li, Z., Wang, J., et al. (2016). The power of inbreeding: NGS-based GWAS of rice reveals convergent evolution during rice domestication. Mol. Plant 9, 975–985. doi: 10.1016/j.molp.2016.04.018
Wang, W. J., Zhao, M. H., Zhang, G. C., Liu, Z. M., Hua, Y. C., Jia, X. T., et al. (2020). Weedy rice as a novel gene resource: a genome-wide association study of anthocyanin biosynthesis and an evaluation of nutritional quality. Front. Plant Sci. 11:878. doi: 10.3389/fpls.2020.00878
Winkel-Shirley, B. (2001). Flavonoid biosynthesis, a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. doi: 10.1104/pp.126.2.485
Yang, W., Zhao, J. L., Zhang, S. H., Chen, L., Yang, T. F., Dong, J. F., et al. (2021). Genome-wide association mapping and gene expression analysis reveal the negative role of OsMYB21 in regulating bacterial blight resistance in rice. Rice 14:58. doi: 10.1186/s12284-021-00501-z
Yang, X. H., Nong, B. X., Xia, X. Z., Zhang, Z. Q., Zeng, Y., Liu, K. Q., et al. (2017). Validation of the red pericarp gene from 419 rice landrace core collection in Guangxi using genome-wide association study and discovery of two novel Rc Alleles. Mol. Plant Breed. 15, 1–6.
Yang, X. H., Xia, X. Z., Zeng, Y., Nong, B. X., Zhang, Z. Q., Wu, Y. Y., et al. (2018). Identification of candidate genes for gelatinization temperature, gel consistency and pericarp color by GWAS in rice based on SLAF-sequencing. PLoS One 13:e0196690. doi: 10.1371/journal.pone.0196690
Yoshimura, A., Ideta, O., and Iwata, N. (1997). Linkage map of phenotype and RFLP markers in rice. Plant Mol. Biol. 35, 49–60. doi: 10.1023/A:1005764026871
Yu, Y. C., Teng, S., Zeng, D. L., Dong, G. J., Qian, Q., Huang, D. N., et al. (2003). Analysis of QTLs for resistance to rice bacterial blight. Chin. J. Rice Sci. 17, 315–318.
Zhang, Z. Y., Shi, Y. N., Ma, Y. C., Yang, X. F., Yin, X. R., Zhang, Y. Y., et al. (2020). The strawberry transcription factor FaRAV1 positively regulates anthocyanin accumulation by activation of FaMYB10 and anthocyanin pathway genes. Plant Biotechnol. J. 18, 2267–2279. doi: 10.1111/pbi.13382
Zhao, C. N., Liu, X. J., Gong, Q., Cao, J. P., Shen, W. X., Yin, X. R., et al. (2020). Three AP2/ERF family members modulate flavonoid synthesis by regulating type IV chalcone isomerase in citrus. Plant Biotechnol. J. 19, 671–688. doi: 10.1111/pbi.13494
Zhao, J. L., Yang, W., Zhang, S. H., Yang, T. F., Liu, Q., Dong, J. F., et al. (2018). Genome-wide association study and candidate gene analysis of rice cadmium accumulation in grain in a diverse rice collection. Rice 11:61. doi: 10.1186/s12284-018-0254-x
Keywords: rice, pericarp color, quantitative trait locus, candidate gene, genome-wide association study
Citation: Yang W, Chen L, Zhao J, Wang J, Li W, Yang T, Dong J, Ma Y, Zhou L, Chen J, Wu W, Zhang S and Liu B (2022) Genome-Wide Association Study of Pericarp Color in Rice Using Different Germplasm and Phenotyping Methods Reveals Different Genetic Architectures. Front. Plant Sci. 13:841191. doi: 10.3389/fpls.2022.841191
Received: 22 December 2021; Accepted: 02 February 2022;
Published: 09 March 2022.
Edited by:
Guoyou Ye, International Rice Research Institute (IRRI), PhilippinesReviewed by:
Mingyong Zhang, South China Botanical Garden (CAS), ChinaJingguang Chen, Sun Yat-sen University, China
Copyright © 2022 Yang, Chen, Zhao, Wang, Li, Yang, Dong, Ma, Zhou, Chen, Wu, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaohong Zhang, c3poYW5nZ3pAdG9tLmNvbQ==; Bin Liu, bGJnejEwMDlAMTYzLmNvbQ==
 Wu Yang
Wu Yang Luo Chen1,2,3
Luo Chen1,2,3 Junliang Zhao
Junliang Zhao Tifeng Yang
Tifeng Yang Lian Zhou
Lian Zhou Shaohong Zhang
Shaohong Zhang Bin Liu
Bin Liu