- 1Fujian Provincial Key Laboratory of Haixia Applied Plant Systems, FAFU and UIUC-SIB Joint Center for Genomics and Biotechnology, College of Crop Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Resources and Environmental Sciences, Gansu Agricultural University, Lanzhou, China
- 3Fujian Provincial Key Laboratory of Plant Functional Biology, College of Life Science, Fujian and Agriculture and Forestry University, Fujian, China
- 4Department of Plant Biology, The University of Illinois at Champaign-Urbana, Champaign, IL, United States
Bio-based fuels have become popular being efficient, cost-effective, and eco-friendly alternatives to fossil fuels. Among plant sources exploited as feedstocks, C4 grasses, such as sugarcane, maize, sorghum, and miscanthus, are highly resourceful in converting solar energy into chemical energy. For a sustainable and reliable supply of feedstocks for biofuels, we expect dedicated bioenergy crops to produce high biomass using minimum input resources. In recent years, molecular and genetic advancements identified various factors regulating growth, biomass accumulation, and assimilate partitioning. Here, we reviewed important genes involved in cell cycle regulation, hormone dynamics, and cell wall biosynthesis. A number of important transcription factors and miRNAs aid in activation of important genes responsible for cell wall growth and re-construction. Also, environmental components interacting with genetic controls modulate plant biomass by modifying gene expression in multiple interacting pathways. Finally, we discussed recent progress using hybridization and genome editing techniques to improve biomass yield in C4 grasses. This review summarizes genes and environmental factors contributing biomass yield in C4 biofuel crops which can help to discover and design bioenergy crops adapting to changing climate conditions.
Introduction
The world population is expected to reach 9 billion by 2050, which was only ∼1.9 billion in the twentieth century (Mullet, 2017). Rapid population expansion and rise in energy demands have heightened research interests to build more sustainable, cost-effective, and eco-friendly energy sources. The gradual effects of using rapidly depleting finite fossil fuels turned out in global warming (greenhouse effect) and subsequent climate change that is ongoing. Owing to intensive industrialization and urbanization, the level of greenhouse gases (GHGs) has increased by 50 times in the atmosphere, which is among the primary drivers of climate change (Subramaniam et al., 2020). International Energy Agency predicted that from 2020 to 2025, the mean annual near-surface temperature would increase by 1°C with a range of 0.9–1.8°C in comparison with pre-industrial time-temperature level (time spanning from 1850 to 1900) along with the onset of frequent tropical cyclones (IEA, 2018). To minimize the disastrous effects of climate change and fulfill energy demands, International Energy Agency (IEA) has devised to exploit renewable energy sources. Major renewable energy resources, such as wind, solar, plant biomass, ocean, and hydropower being sustainable sources, can also aid in reducing the use of fossil fuels and consequently GHGs. In the year 2019, a 12.5% increase in biofuel production was observed from 142.6 to 160.9 million liters, whereas in 2020 due to the global pandemic situation, compromised prices of fossil crude oil have made transport biofuels less competitive. However, future predictions suggest that the average global output of bioethanol will further rise to 182 billion liters which was 160 million liters in 2019, whereas the United States and ASEAN region will contribute for biodiesel and Hydro-treated vegetable oil (HVO) (Lauf et al., 2021).
Biofuels are the fuels derived from biological substances (e.g., agricultural wastes, animal matter, algae, forest vegetation, and energy crops) (Koçar and Civaş, 2013). In this review, we will primarily focus on the high biomass yielding energy crops which are commercially used for energy generation. Presently, regarding biofuel crops, there is a concern of conflict about their use for food, feed, and energy generation. However, several plant species are efficient in accumulating high biomass with minimal inputs. Among them, ryegrass (Lolium perenne), bamboo (Bambusa vulgaris), poplar (Populus deltoides), and willow (Salix) are from C3 category of photosynthesis while giant miscanthus (Miscanthus giganteus), sweet sorghum (Sorghum bicolor), pearl millet (Pennisetum glaucum), napier grass (Pennisetum purpureum), maize (Zea mays), sugarcane (Saccharum), and switchgrass (Panicum virgatum) are ideal C4 energy crops (Byrt et al., 2011). C4 grasses take more advantage of biomass accumulation, owing to higher energy-conserving photosynthetic machinery, stress tolerance, water, and nitrogen use efficiency (Somerville et al., 2010). C4 photosynthesis system possesses specialized biochemistry and anatomical modifications that protect the oxygenation of RUBISCO. Whereas, the PEP enzyme, instead of RUBISCO serves as the first substrate of CO2 in mesophyll cells that reduces the energy losses caused by photorespiration (van der Weijde et al., 2013). This structural collaboration of mesophyll (M) and bundle sheath (BS) enables C4 plants to harvest more solar energy with improved water use efficiency (WUE) and nitrogen use efficiency (NUE).
In a couple of decades, the increasing trend of biofuel use in developed countries motivated many researchers to focus their interests on biofuel crops and their bio-products. Among C4 crops, Miscanthus and switchgrass have been extensively studied for this purpose in the United States and Europe. Sugarcane is contributing a major share of biofuels in Brazil. Sorghum is also a promising competitor as a bio-energy crop owing to drought tolerance and genetic diversity in sweet and grain sorghum (Byrt et al., 2011). Advancement in recent technologies of saccharification and lignocellulose digestion, cultivation of sugarcane is widely practiced as a biofuel crop (Carvalho-Netto et al., 2014). Scientists are more concerned with introducing specialized hybrids or transgene as energy crops to meet the objectives of sustainable energy by biofuels. Plant biomass depends on genetic, physiological, edaphic and environmental factors. Few publications encompassed basis of biomass accumulation at genetic level but a comprehensive study of genetic, physiological and environmental factors constituting to biomass, was lacking (Endo et al., 2009; Byrt et al., 2011; Lima et al., 2017). Therefore, besides biological and genetic basis we addressed environmental modulations-based plant biomass accumulation. Considering the general overview of the need for bioenergy and the importance of biomass crops with special concern for C4 crops, this review aims at identifying the genes involved in different growth-related processes and how growth patterns are modified in changed climatic conditions. Furthermore, possible tools and strategies are discussed that are effective in opting for increasing biomass in C4 crops.
Genetic Basis for Biomass
In plants, several genes belonging to different functional and structural categories are involved in vegetative development. Owning to advancement in molecular and genetic studies, many genes and transcription factors have been identified in contributing growth from juvenile to vegetative stage as previously covered in the reviews (Demura and Ye, 2010; Lima et al., 2017; Kandel et al., 2018). Following sections supplicated the existing literature about genetic aspects of plant growth regulation.
Growth and Developmental Regulation
Cell Cycle Machinery
Plant organ size control is a central component of biomass productivity. In many animals and plants, overall organ growth rate and size are associated with cell number that is controlled by strict action of cell division together with cell expansion (Vernoux et al., 2000). As in other eukaryotes, the cell cycle in plants consists of DNA-replication (S-phase), and mitosis (M-phase), which are separated by postmitotic (G1) and pre-mitotic interphase (G2) gap phases, respectively (Scofield et al., 2014). Cell cycle machinery is strongly modulated at different points to verify the fidelity of chromosome duplication and cell division. Highly conserved control mechanisms, the checkpoints G1/S and G2/M transitions confirm that either cell cycle process has been precisely accomplished at each phase before entering the next phase or not (Barnum and O’Connell, 2014). Different core cell cycle protein groups including CYCLINS (CYCs) complexed with CYCLIN-DEPENDENT KINASES (CDKs), the E2F/DIMERISATION PROTEIN (DP) transcriptional regulatory proteins, KIP-RELATED PROTEIN/INTERACTOR OF CDKs (KRP/ICK), RETINOBLASTOMA-RELATED (RBR), SIAMESE/SIAMESE-RELATED (SIM/SMR), proteins and the multi-subunit E3 ubiquitin ligase ANAPHASE-PROMOTING COMPLEX/CYCLOSOME (APC/C) control the progression of events involved different phases (Inz, 2006; Inagaki and Umeda, 2011). Genetic modulation of these proteins to enhance biomass has been reported in model plant species (Arabidopsis, tobacco, rice), with few C4 plants (sorghum and maize). Elevated growth rate in tobacco has been resulted from overexpressing the cyclin D-type (CycD2) gene from Arabidopsis. These plants were found to have normal cell and meristem size but taller stem overall, showing increased growth rate from seedling to maturity (Boucheron et al., 2005). Defective shoot and root formation, as well as a reduction in endoreduplication, were noticed in tobacco ectopically expressing CycA3; 2 (Yu et al., 2003). Transcriptome analysis identified elevated levels of cell cycle (i.e., cyclins) genes in bioenergy sorghum immature internodes which shows that their initial increase in size is due to cell-division coupled growth (Kebrom et al., 2017). Further, quadruple (ick1/krp1, ick2/krp2, ick6/krp3, ick7/krp4) and quintuple (ick1/krp1, ick2/krp2, ick6/krp3, ick7/krp4, ick5/krp7) mutants of CDK negative regulator ICK/KRP genes reported to have stimulated CDK activity and cell proliferation that resulted in increased fresh and dry weights; larger cotyledons; leaves; petals and seeds (Cheng et al., 2013). Overexpression of novel Arabidopsis ABAP1 protein decreased cell proliferation by limiting mitotic DNA replication in negative feedback loop during leaf development, by repressing transcription of pre-replication complex (pre-RC) genes (Masuda et al., 2008).
Another essential component of cell cycle machinery is the anaphase-promoting complex (APC), a multi-subunit E3 ligase that modulates cyclins (Cyc) and CDKs activity in checkpoints to ensure the maintenance of cell division rate (Bodrug et al., 2021). APC/C-subunits remain conserved throughout evolution, however, gene duplication of different subunits has been observed in some plants (de Lima et al., 2010). Overexpression of Arabidopsis CDC27a/APC3a in tobacco was associated with apical meristem restructuration, altered cell-cycle marker expression, and accelerated plant growth up to 30% at the flowering time leading to increased biomass production (Rojas et al., 2009). While APC10 overexpression in Arabidopsis causes CYCB1;1 protein degradation, thereby accelerating the transition through mitosis (Eloy et al., 2011). Transgenic tobacco plants overexpressing the APC10 gene are taller with larger leaves, produce more seed capsules, and have an augmented biomass accumulation. Furthermore, a cross between APC3a- and APC10- overexpressing tobacco T1 plants have enhanced growth phenotype compared to the overexpression of single APC/C subunits (de Freitas Lima et al., 2013). Down-regulation of rice OsCCS52A, an APC/C subunit resulted in reduced plant height and smaller seeds with an endosperm defect in endoreduplication (Su’udi et al., 2012). Semi-dwarfism and reduced leaf size are also observed in CCS52A ortholog rice tillering and dwarf 1 (tad1) mutant (Xu et al., 2012). SAMBA negatively modulates cell proliferation through APC/C interaction. In maize, samba-1 and samba-3 mutants showed developmental defects, involving short plant height, reduced leaf size due to an altered cell expansion and cell division rate (Gong et al., 2021). In addition, several DRP family members like DRP1A, DRP1E, DRP2A, DRP2B, and DRP5B are regarded as SAMBA interactors. All of these proteins, except DRP5B, are localized to the cell plate and mutations in DRP1E and DRP1A resulted in defective cell plate assembly and cytokinesis, as well as defects in cell expansion (Hong et al., 2003; Kang et al., 2003; Fujimoto et al., 2008).
Long Vegetative Duration
The plant life cycle is divided into vegetative, transition, and reproductive developmental phases. The vegetative phase is associated with meristems producing stems and leaves. The transition phase is related to an elevation of the apical meristem, while the reproductive phase centers on meristems capable of producing flowers or reproductive organs. The vegetative phase starts at germination and continues through tillering, the meristems actively produce a stem, buds, internodes, and leaves. The long vegetative phase establishes continuous leaf development needed to capture sunlight for photosynthesis that supplies nutrients for expansion of roots for anchoring, storage, and uptake of minerals for increased biomass production. Some high-yielding C4 crops are Miscanthus x giganteus, Sorghum bicolor, Pennisetum, and sugarcane (Saccharum spp.) genotypes, characterized by enriched canopies, taller stems, and longer growing seasons (Somerville et al., 2010; Mullet et al., 2014). The biomass yield of Miscanthus × giganteus in some mid-west United States locations reached 44–61 Mg/ha at peak biomass accumulation (Heaton et al., 2008). Pennisetum purpureum and Pennisetum typhoides reached their record yields of ∼88 and 80 Mg/ha, respectively during longer growing seasons (Somerville et al., 2010). Similarly, high-biomass first-generation sorghum hybrids accumulated ∼40–50 dry Mg/ha during ∼180 days growing season when grown in the south-central United States (Olson et al., 2012). Biomass varies among types of C4 crops for various reasons like stem sink strength, shoot/root partitioning, and season length. For example, Miscanthus × giganteus produced higher biomass than switchgrass and maize when these were grown in similar regions in the United States due to differences in shoot/root biomass partitioning (Anderson et al., 2011). High-biomass sorghum hybrids with long growing seasons produce twice shoot biomass (∼40–50 Mg/ha) when compared to grain sorghum hybrids in optimum growth conditions and ∼30% more biomass in rain-fed conditions when both hybrids are grown in similar regions in the south-central United States (Olson et al., 2012; Truong et al., 2017). The increased biomass yield of high-biomass sorghum hybrids was because of delayed flowering initiation resulting in prolonged vegetative growth duration that increased total light energy capture, improved radiation interception and use efficiency, and elevated biomass partitioning.
Further, delayed flowering in long days concomitant with an extensive period of vegetative growth resulted in increased biomass yield as observed in several C4 grasses. Photoperiod regulated flowering in sorghum is extensively studied by modulating flowering regulators florigen related genes (CN8, CN12, CN15), upstream activators (CONSTANS (CO) and EARLY HEADING DATE 1 (EHD1)), and repressors (PRR37 and GHD7) of these genes that are regulated by photoperiod and output from the circadian clock, once sorghum exits in juvenile phase (Murphy et al., 2011, 2014; Dong et al., 2012). Prolonged vegetative meristem activity with increased biomass yield was observed in several plants by overexpressing flowering-time genes (Demura and Ye, 2010). Indeed, the activation of the flowering promoting factor-like1, flowering locus T1, C-like MADS-box protein, early flowering 3 as well as embryo flowering 1-like protein in tomato IL2-6 cultivar, supported late-maturing performance (Caruso et al., 2016). Overexpression of gibberellin 20-oxidase-1 and ARGOS also resulted in an extended growing period by delaying the flowering time to give rise to larger organ size and taller plants (Hu et al., 2003; Voorend et al., 2016). Regulation switch from vegetative to reproductive phase can be controlled by manipulating genes from several developmental pathways, for example, gibberellin, circadian, and flowering-related genes.
Hormones Dynamics and Primary Growth
Plant hormones are a diverse group of chemical substances controlling growth and development-related events in plants by regulating meristematic cell division and cell elongation. These chemical signals modulate microtubule and cell plate formation, cell wall constituent deposition, and remodeling which are key factors of growth and thus biomass accumulation. During the green revolution, scientists exploited the traditional plant breeding approaches for the selection of short stature, higher grain yield producing cultivars, which were low in levels of endogenous hormones like gibberellin (Sánchez-Rodríguez et al., 2010), auxin (Vanneste and Friml, 2009), and brassinolide (Müssig, 2005) that are important as growth regulators and performing growth regulatory functions from cellular to developmental levels (Table 1).
Gibberellin
In Gibberellin (GA) signaling pathways, manipulation of both positive and negative regulators employed positive effects on growth and biomass accumulation. One control point of biomass can be the substantial increase of GA rate-limiting enzyme GIBBERELLIN 20-OXIDASE (GA 20-OXIDASE) which is involved in the last steps of GA biosynthesis in the cytoplasm. One of the primitive functional evidence of GA20ox in Arabidopsis highlights the accelerated shoot growth, elongated hypocotyls, and onset of early flowering (Coles et al., 1999). In a potential biofuel crop, switchgrass, ectopic expression of ZmGA20ox resulted in increased growth and biomass-related traits (Do et al., 2016). Recently, nine genes of GA20-ox1 were identified in sweet sorghum (bioenergy sorghum) and showed differential spatiotemporal patterns of expression while SbGA20ox1 was predominantly related to increased stem biomass and assimilates partitioning (Wang et al., 2020). “Green revolution” gene GA20-oxidase is involved in the synthesis of principal biopolymer in the cell wall, i.e., cellulose in sorghum, whereas dwarf1-1 cellulose deficient and male gametophyte-dysfunctional mutant showed ablation of GA and altered expression of three CESA genes generating cellulose deficient and dwarf phenotypes (Petti et al., 2015). Similarly, plants overexpressing ZmGA20ox displayed longer internodes and leaves, more tillers, and twofold increase in maize biomass (Voorend et al., 2016). Secondly, GA-INSENSITIVE DWARF1 (GID1) is the first receptor of bioactive GA in the signaling pathway, which shows the highest affinity for GA4 (bioactive form). Overexpressing the GID1 gene shows a substantial increase in shoot elongation and growth in Arabidopsis, rice, and poplar (Sakamoto et al., 2004; Hirano et al., 2008; Mauriat and Moritz, 2009). The third main player in the gibberellin signaling pathway is the DELLA repressor gene which hinders the transcription of GA receptor, i.e., GID1. DELLA proteins act as a feedback regulatory control in the GA signaling pathway and are implicated in dwarf phenotype in maize with shifts in flowering time (Lawit et al., 2010). A recently conducted study on sugarcane affirmed the similar inhibitory roles of DELLA proteins. DELLA proteins interact with PIF4 and elements in the ethylene signaling pathway ScEIN3/ScEIL1, moreover, DELLA silenced lines showed changes in carbon allocation in storage and structural molecules and increased culm growth (Garcia Tavares et al., 2018).
Auxin
Auxin is a very important hormone involved in the growth process and cell wall architecture. Numerous mutants related to auxin synthesis, transport, and signaling showed overall dwarf phenotypes, defects in tropisms, and altered organ morphology (Vanneste and Friml, 2009). Auxin influx facilitator AUXIN1/LIKE-AUX1 (AUX/LAX) is involved in inflorescence development and root gravitropism. It is reported that mutations in homologs of AUX1 genes in maize (ZmAUX1) and Setaria viridis (SvAUX1) resulted in defective branch development of inflorescence, reduced plant height, increased panicle length, and sparse panicle phenotype (Huang et al., 2017). Aux/IAA homolog ROOTLESS WITH UNDETECTABLE MERISTEMS 1 (RUM1) in maize is involved in seminal and lateral roots formation. Transcriptome analysis of rum1 showed down-regulated expression of like-auxin1 (lax1), the plethora genes plethora 1 (plt1), baby boom 1 (bbm1), and heat shock complementing factor 1 (hscf1), and the auxin response factors arf8 and arf37 (Zhang et al., 2015). In maize, ARF5 (MONOPTEROS) is involved in vascular cells differentiation and rum1 showed defective xylem organization and more lignin deposition in root cells (Zhang et al., 2014).
Brassinosteroid
Brassinosteroids (BR) comprise an important group of steroidal hormones originally isolated from the pollen of brassica plants (Rehman et al., 2022a). Brassinolide (BL) is the biologically active BR that is synthesized from compound campesterol with the aid of a cytochrome P450-mediated pathway (Bishop, 2007). This was a relatively novel and less studied hormone in the past, but recently it has gained attention as an active growth-promoting hormone owing to its involvement in many physiological functions (Fridman and Savaldi-Goldstein, 2013; Rehman et al., 2022b). Several genes are involved in the signaling pathways of brassinolide from BL perception to activation of responsive genes for example receptor-like kinase BRASSINOSTEROID-INSENSITIVE 1 (BRI1), BRI1-ASSOCIATED RECEPTOR KINASE 1, SOMATIC EMBROGENESIS RECEPTOR KINASE 1, and a repressor gene GSK3-like kinase BIN2 (BRASSINOSTEROID-INSENSITIVE 2) (Sánchez-Rodríguez et al., 2010). After the discovery of BL, mutant analysis in Arabidopsis revealed that plants deficient in the genes related to BL biosynthesis or signaling pathways showed dwarf phenotype, compromised male fertility, delay in flowering time, altered patterns of vascular development, and impaired photomorphogenesis (Feldmann et al., 1989). In a very recent study on maize, an endoplasmic reticulum localized gene, i.e., ZmD11 related to the biosynthesis of BL rescued the panicle architecture and plant height in cpb1 mutant in maize and rice. ZmD11 increased seed length, seed weight, and both seed starch and protein contents in rice and maize crops (Sun et al., 2021). brassinosteroid-deficient dwarf1 (brd1) gene encoding brassinosteroid C-6 oxidase having a maize lilliputian1 allele (lil-1) caused alteration in gravitropic response of root, leaf cell density, and more wax deposition conferring the adaptive mechanism to stress (Castorina et al., 2018). BR receptor, i.e., BRASSINOSTEROID INSENSITIVE1 (BRI1) RNA interference (RNAi) knock-out mutants in maize showed overall dwarf stature, shortened internodes, folded dark green leaves, decreased auricle formation, and feminization of male flowers (Kir et al., 2015). Similarly, in sorghum, the nuclear localization of BRASSINOSTEROID INSENSITIVE 2 (BIN2) was inhibited by DW1 indicating its role in BR signaling. Sorghum lines harboring mutated Dw1 (dw1) showed impaired skotomorphogenesis, lamina joint bending, and insensitive to BR gene regulation and feedback (Hirano et al., 2017) (Figure 1A).
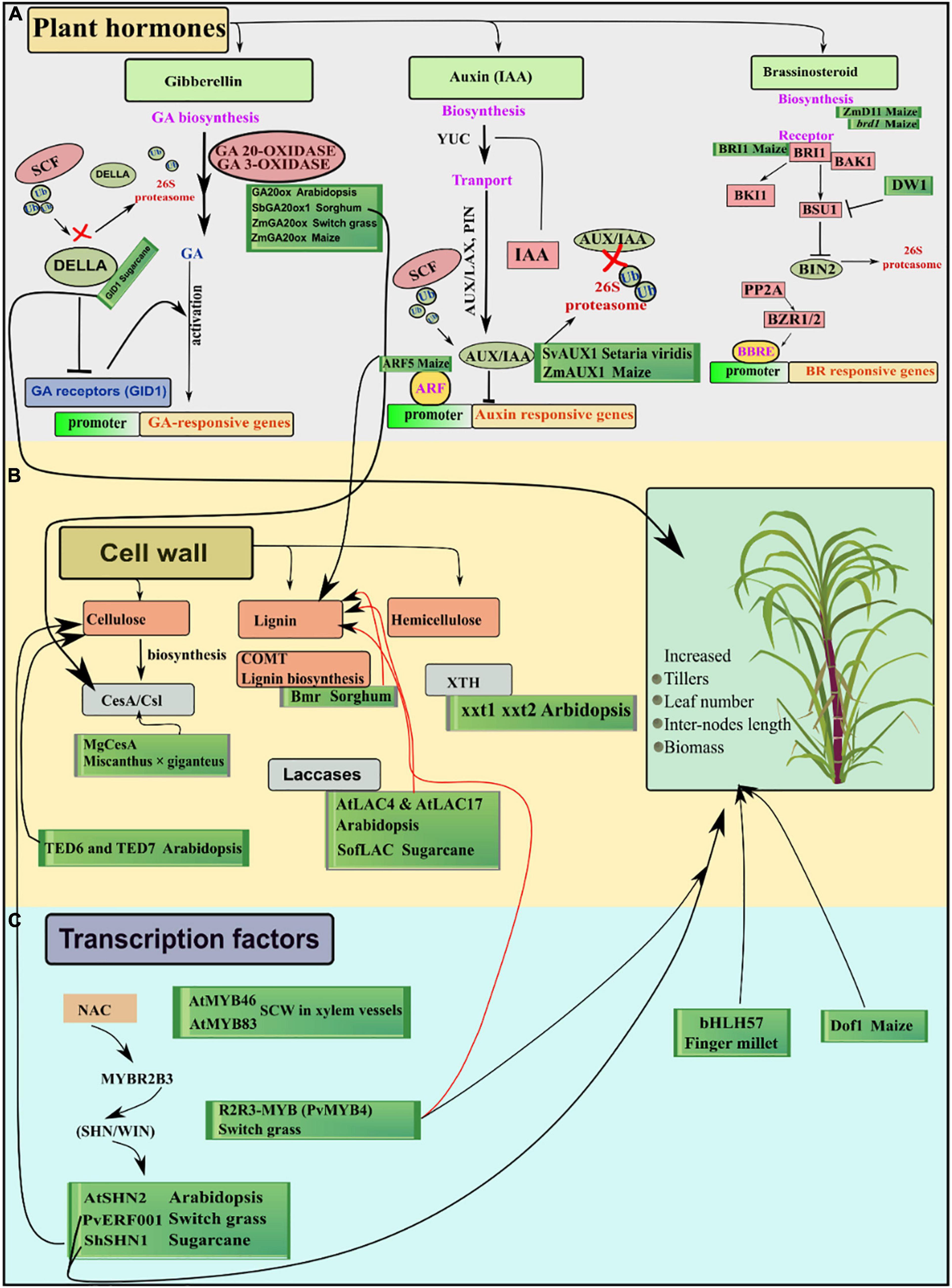
Figure 1. Overview of the genes involved in different pathways in important C4 biomass plants. (A) Highlights the hormone genes directly or indirectly related to growth and cell wall functioning. (B) Shows multiple genes involved in biosynthesis and remodeling of different cell wall-related components. (C) Transcription factors regulate many genes involved in different pathways of secondary cell wall synthesis leading to modified cell wall components improving saccharification efficiency. Black arrows show positive, red arrows show negative growth impacts and T lines show inhibitory influence on other genes. Whereas, green boxes enclose the functionally characterized genes in their respective pathways.
This cluster of genes involved in biosynthesis and signaling of the important growth-promoting hormones highlights the connections with the cell wall, carbohydrates, and photosynthesis-related pathways. Further studies need to elucidate growth patterns of double or triple mutants from multiple hormone pathways at transcriptional and biochemical levels for efficient biomass response.
Cell Wall-Related Genes
The cell wall is a non-living protective cell layer that comprises 70% of the world’s plant biomass (Poorter and Villar, 1997; Pauly and Keegstra, 2008). Second-generation cellulosic biofuel (bioethanol, biohydrogen, and biomethanol) produced from the plant biomass mostly comes from the cell wall. During plant growth and cell extensibility, several processes are involved among which cell wall loosening and rearrangement strongly contribute in plant biomass traits. In plant species, numerous gene families related to cell wall biogenesis, membrane trafficking, remodeling, secondary cell wall synthesis, and signaling comprise ∼10% of plant genomes (Lauter et al., 2005; Yong et al., 2005; McCann and Carpita, 2007).
Many studies involving plant biomass engineering techniques showed a strong effect on cell wall-related genes on growth and biomass accumulation processes in C4 biofuel crops e.g., miscanthus (van der Weijde et al., 2013), sorghum (Scully et al., 2016; Xia et al., 2018; Tetreault et al., 2020), and sugarcane (Jung et al., 2012; Bottcher et al., 2013). Genes responsible for cellulose synthesis mainly include members of cellulose synthase (CesA) and cellulose synthase-like (Csl) families. A recently published comparative study have identified 77, 35, and 109 CesA/Csl genes in Miscanthus floridulus, S. bicolor, and S. spontaneum, respectively. Among the 10 groups of CesA genes classified by phylogenetic approaches, a new group was identified in Miscanthus floridulus, i.e., CesAX which was not present in C3 rice. Higher expression of CesA genes and their duplicates mainly followed by WGD (Whole Genome Duplication) showed the additive effects in gene expression levels resulting in more cellulose accumulation (Zhang et al., 2021). Silencing or mutations of CesA genes in Arabidopsis and certain other monocots have resulted in certain functional deformities but there is no authentic evidence that over-expression of CesA will certainly increase the cellulose content of the cell wall. In Miscanthus × giganteus, cloning of six MgCesA genes showed the involvement of MgCesA2, 3, 4, 7, and 8 in primary cell wall biosynthesis and rest in (MgCesA10, 11, 12) secondary cell wall synthesis and formation of cellulose synthase complex (Zeng et al., 2020).
Two cellulose interacting genes TED6 and TED7 enhanced the cellulose synthesis in xylem vessel elements, and lack of function mutants resulted in the failed secondary cell wall formation in Arabidopsis (Endo et al., 2009). Similarly, expression profiling of interspecific sugarcane hybrids showed upregulation of CesA, laccases, and callose synthase-related genes in high biomass extreme F2 segregants (Wai et al., 2017). Lignin has been an undesirable component of the cell walls in terms of bioenergy generation, although it accounts for ∼30% terrestrial organic carbon fixation in the biosphere (Battle et al., 2000). In sorghum, brown midrib mutant (bmr) showed decreased levels of lignin and the formation of an altered subunit. The Bmr gene is a biosynthesis gene in monolignol units, which yield the hydroxycinnamic subunits of lignin. Another class of genes, laccases, are involved in the oxidation of monolignol units before the incorporation into cell wall polymers (Bonawitz and Chapple, 2010). In vitro oxidation of lignin precursors (Liang et al., 2006) and localization in lignin synthesizing tissues (Ranocha et al., 2002; Caparrós-Ruiz et al., 2006) have been experimentally proved by laccases in plants. In Arabidopsis, T-DNA insertion lines exhibited the reduced lignin content in single mutants of AtLAC4 and AtLAC17 whereas, double mutants displayed 40% reduced lignin but with irregular xylem tissues (Berthet et al., 2011). Sugarcane, a benchmark of biomass-derived biofuels showed the strong interactions of SofLAC genes with phenylpropanoid biosynthesis genes in a co-expression network. For the confirmation of monolignols oxidation, complementation of the SofLAC gene under the native promoter AtLAC17 was performed in Arabidopsis lac17 mutants having reduced lignin. SofLAC repaired the lignin content in Arabidopsis but lignin composition was altered in complemented lac17 mutant lines (Cesarino et al., 2013). Xyloglucans (XyG) comprise a major class in hemicellulose proportion of cell wall and are extensively found in primary cell walls of eudicots and non-gramineous monocots. In Arabidopsis, double mutant xxt1 xxt2 displayed a considerable reduction in detectable xyloglucan and altered mechanical properties (Cavalier et al., 2008). For an ideal bioenergy crop, higher lignin content is an undesirable trait due to its recalcitrance to degradation, whereas higher crystalline cellulose content is favored due to its digestibility. Conversely, hemicelluloses are crosslinking both lignin and cellulose causing a decrease in cellulose crystallinity, but a reduced level of hemicellulose branching ensures easy separation of cell wall components (Torres et al., 2015). However, molecular alteration of hemicellulose is handicapped due to its vague and complex biosynthesis and subsequent pathways. Research advances to this field are nevertheless confined at molecular levels in model species whereas, application of this knowledge in bioenergy-related species is the main goal (Figure 1B).
Transcriptional Regulation and miRNA Role
Transcription factors are the genes encoding proteins (besides RNA polymerase) that are essentially required for transcription. Owing to the regulatory role in transcription activity, they are capable of controlling the expression of many downstream key genes related to growth and development. In plants, DNA transcription involves more than 1,500 TFs to regulate target genes by binding with cis-regulatory elements in the promoter region (Singh et al., 2002). Secondary cell wall formed between the primary cell wall and cell membrane strongly contributes to the development and is an important attribute for the biofuel industry. Biosynthesis and remodeling of cell wall components are achieved through an orchestrated action of TFs and downstream genes. Therefore, detailed knowledge of transcription factors controlling secondary cell wall initiation genes, polysaccharides synthesis, lignification process, and a parallel process of programmed cell death (PCD) of xylem cells (Ohashi-Ito et al., 2010) is important to dissect for biomass regulation (Table 1).
Transcription factors of NAC (NAM—No Apical Meristem, ATAF, CUC—CUP/SHAPED Cotyledon) family activates a nexus of downstream transcription factors for example MYBR2B3, and act as master switches by binding with cell wall biosynthetic genes (Martins et al., 2018). SHINE “SHINE/WAX INDUCER” (SHN/WIN) TF is a member of ETHYLENE RESPONSIVE FACTOR (ERF) that functions as a regulator of cell wall biosynthesis genes, resulting in increased cellulose and decreased lignin contents (Ambavaram et al., 2011). In switchgrass (Panicum virgatum) PvERF001 gene which is the homolog of AtSHN2 conferred activation of cell wall synthesis and accumulation of biomass (Wuddineh et al., 2015). Likewise, sugarcane transcription factor ShSHN1 overexpressed in rice induced changes in cell wall composition and increase in biomass by (91–340%), pectin (26–209%), cellulose content (10–22%), and saccharification efficiency (5–53%) in rice transgenic plants (Martins et al., 2018). McCarthy reported that AtMYB46 and its paralog AtMYB83 are found to function as activators of the secondary cell walls and are expressed in xylem fibers and vessels during secondary cell wall development (McCarthy et al., 2009). In Arabidopsis myb46/83 double mutant, maize orthologs of AtMYB46/83 successfully complemented the secondary cell wall synthesis and thickening after rescuing the defected walls (Zhong et al., 2011). Similarly, in switchgrass, overexpression transgenic lines of R2R3-MYB (PvMYB4) showed increased biomass up to ∼63% and reduced lignin content around 50% (Shen et al., 2012). Some MYB genes in grasses, e.g., maize and switchgrass function in lineage-specific fashion regarding lignin biosynthesis regulation (Agarwal et al., 2016). For example, co-immunoprecipitation and ChIP-seq assays (co-IP) assays showed that ZmMYB11, ZmMYB31, and ZmMYB42 induced reduction in expression of lignin biosynthesis-related genes COMT (caffeic acid-O-methyltransferase) and 4CL2 in maize (Vélez-Bermúdez et al., 2015). ZmMYB31 and ZmMYB42 in exogenous expression in Arabidopsis redirected phenylpropanoid and lignin-related genes in Arabidopsis contrary to maize. Moreover, ZmMYB31 and ZmMYB42 didn’t down regulate the ZmF5H (ferulate-5-hydroxylase) gene in maize as compared to Arabidopsis, leading to a decrease in S/G ratio (S, syringl units; G, guaiacyl units) (Sonbol et al., 2009; Fornalé et al., 2010; Vélez-Bermúdez et al., 2015). Ectopic expression of transcription factor SbMyb60 in sorghum showed involvement in monolignol biosynthesis pathways and increased lignin concentration and plant biomass. Constitutively overexpressing SbMyb60 displayed enhanced lignification in leaf midribs and soluble phenolic compounds in plant biomass (Scully et al., 2016). This suggests that in monocot grasses the route of MYB TFs and their regulatory pathways are more diverse and need to be investigated considering the models from grasses. Furthermore, the differences in C3 and C4 photosynthetic regulatory pathways should be studied in detail to increase the cellulose and hemicellulose contents and decreased contents of recalcitrant, i.e., lignin. (Figure 2B). Finger millet transgenic plants over-expressing bHLH57 (BASIC HELIX-LOOP-HELIX) depicted resistance to salinity stress with enhanced photosynthetic efficiency and increased biomass (Babitha et al., 2015). AHL (AT-HOOK MOTIF NUCLEAR LOCALIZED) family of transcription factors in Arabidopsis controls the petiole and rosette growth and architecture by antagonizing the role of growth-promoting PHYTOCHROMEINTERACTING FACTORS (PIFs) (Favero et al., 2020). Maize zinc finger protein Dof1 transcription factor increased the nitrogen use efficiency in transgenic sorghum and wheat. Tissue-specific expression of ZmDof 1 under rbcS1(maize) promoter increased growth by activation of carbon skeleton metabolism, i.e., PEPC activity (Peña et al., 2017) (Figure 1C).
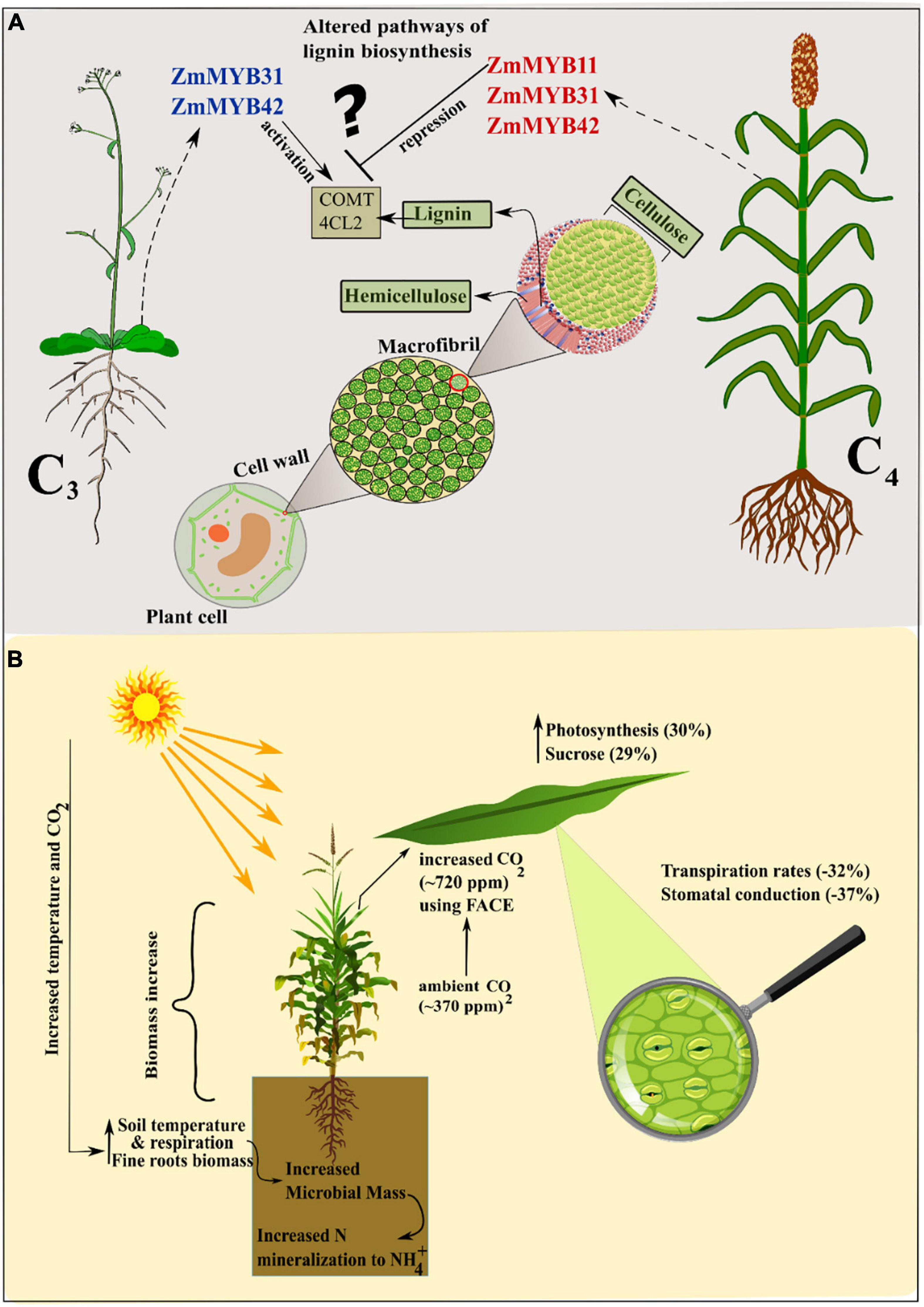
Figure 2. (A) Contrasting transcriptional regulation patterns of MYB TFs in C3 and C4 plants. In C3 (Arabidopsis) ectopic expression of maize TFs enhance lignin biosynthesis whereas in C4 (Maize) lignin content is reduced. This gives rise to lineage-specific transcription in C4 plants. (B) Increase in temperature and CO2 enhances photosynthesis and altered control of stomatal aperture enhancing WUE. Soil microbiota mass in the rhizosphere is also increasingly responsible for modifications in the nutrient pool.
SQUAMOSA Promoter-Binding Protein-Like (SPL) transcription factors are regulated by microRNAs (miRNA), i.e., miR156 and AtSPL9 which positively regulate another miRNA miR172 (Chen et al., 2010). AtSPL9 TF binds to the promoter region of the MIR172 gene and induces the transcription activity of downstream genes and repress adult-related characteristics in Arabidopsis. At the later plant stage, the miR156-AtSPL9-miR172 regulatory pathway progresses with the decrease in miR156 levels and increase in miR172 leading to repression of FLOWERING LOCUS T (FT) gene. This event allows entering the plant to the reproductive phase and the same regulatory pathway is conserved in maize where miR172 represses Glossy15, an AP2-like TF (Lauter et al., 2005; Chuck et al., 2007). This interactive regulatory role of transcription factor and microRNA is an effective molecular tool to extend the vegetative phase for enhanced sink capacity and biomass accumulation. Dumitrache et al. (2017) developed independently overexpressing (OE) and silenced (KD) transgenic switchgrass lines of specific genes and miRNA GAUT4-KD, miRNA156-OE, MYB4-OE, COMT-KD, and FPGS-KD (Dumitrache et al., 2017). Continuous monitoring of 2-year ratoon transgenes showed increased contents of carbohydrates by 12% and ethanol yields by 21% as compared to controlled conditions. In Arabidopsis, the use of zinc finger artificial transcription factor (ZF-ATF)-mediated interrogation lines helped in understanding growth and biomass-related characteristics. Introgression lines of two Arabidopsis genomes harboring 3F-EAR encoding T-DNA and 3F-VP16 encoding T-DNA constructs showed substantially large phenotypes. Whereas, 3F-EAR is Arabidopsis based ERF-associated Amphiphilic Repression (EAR) motif and acts as a dominant repressor evident from the previous studies and VP16 protein originated from the herpes simplex virus as a transcriptional activator (Ohta et al., 2001; Hiratsu et al., 2003; McCarthy et al., 2009). Further research is needed to elucidate the differences in transcriptional regulation of Arabidopsis and C4 grasses, involving the promoter analysis to identify important cis-elements and the spatio-temporal regulation of TFs, affecting development-related genes.
Hybridity and Polyploidy
Hybrids and polyploids are common in plants. Hybridization between and within species is a natural process and is estimated to occur in ∼25% of plant species (Mallet, 2005). Hybrid vigor is a common consequence of hybridization and refers to superior hybrid performance in yield, biomass, or other agronomic parameters. Polyploidy refers to a cell or organism having two or more sets of basic chromosomes. An autopolyploid is derived from genome duplication within the same species, such as alfalfa (Medicago sativa), sugarcane (Saccharum), and potato (Solanum tuberosum), while allopolyploids are formed by chromosome doubling following hybridization between species. Allopolyploid is a “doubled interspecific hybrid,” which leads to heterozygosity and hybrid vigor fixation. Many crops like cotton (Gossypium hirsutum), bread wheat (Triticum aestivum), and oilseed rape (Brassica napus) are cultivated as allopolyploids while rice (Oryza sativa), and maize (Zea mays) are mainly grown as hybrids. Both ploidy and hybridity affect growth vigor and cell size which are directly associated with plant biomass production (Chen, 2010).
In maize, increased ploidy had a detrimental effect on plant size which increases from haploid to triploid, but reduces in tetraploid (Riddle et al., 2006), and is consistent with smaller haploid Arabidopsis plants than diploids (Ravi and Chan, 2010). Induced polyploidy in hybrids may facilitate improving yield components, diminish hybrid vigor breaks in subsequent generations and restore inter-subspecific hybrids fertility (Miller et al., 2012). In sorghum, the colchicine-treated polyploidy induced plants showed high biomass with longer leaf length and stronger root system (Ardabili et al., 2015). A triploid Miscanthus × giganteus, C4 grass is considered an excellent bioenergy crop due to its capacity to capture greenhouse gases by sequestering carbon in underground rhizomes and high biomass production when compared to diploid Miscanthus species (Chae et al., 2013). The modern sugarcanes are polyploid interspecific hybrids combining disease resistance, hardiness, and ratooning of Saccharum spontaneum and high sugar content from Saccharum officinarum. Genome restructuring and gene expression modifications in these cultivars due to polyploidy provide a selective advantage for a wider geographical adaptation, increased vigor, sucrose, and fiber content (Ming et al., 2001; Hoang et al., 2015). There is increased global demand for alternative fuel sources, and sugarcane is gaining importance as a biofuel crop with its high biomass production potential, besides being a major sugar crop.
Environmental Cues Influencing Biomass Accumulation
Plants being sessile in nature are exposed to constantly changing environment enveloping around them. In the presence of judicious input resources and ideal genotypes, still plants are exposed to fluctuating environment in terms of CO2 concentration, irradiance, and temperature which are function of plant growth. Here we reviewed that how fluctuations in external environment alter gene expression of important pathways and eventually biomass accumulation patterns.
Ambient CO2 Fluctuations
With the gradual increase of GHGs in the atmosphere, the extent of CO2 is also increasing in the air as CO2 also comes under the category of greenhouse gases. This increase in GHGs tends to rise the global temperature which seriously changes the genetic and physiological attributes affecting the growing patterns of plants. This global warming accompanied by climate change has increased the variability of precipitation and a continuous increase in ambient CO2 concentration up to 490–1,260 ppm by the end of the twenty-first century (IPCC, 2007). Considering the global changes, fluctuations in CO2, light, and temperature are having both direct and indirect effects on the growth and biomass production of C3, C4, and CAM photosynthesis plants (Ainsworth and Long, 2005). Owing to anatomical and functional modifications in C4 species, it was assumed that C4 plants will be less affected by CO2 rise as ambient CO2 already meets the maximum saturation due to bundle sheath cells, as for C3 plants CO2 is a limiting factor for maximum photosynthesis. However, previous researches suggest different reasons for the increased response in terms of growth and productivity against elevated CO2 in C4 plants as (1) direct effect on Rubisco as CO2 saturation point increases (Ziska and Bunce, 1997), (2) leakiness of bundle sheath cells (Watling et al., 2000), (3) young leaves are supposed to undergo preliminary C3 photosynthesis system (Cousins and Bloom, 2003), (4) reduced stomatal aperture to enhance WUE or maintaining inner optimal temperature (Ghannoum et al., 2000).
Dieleman et al. (2012) published a meta-analysis of multifactorial experiments in which all treatments showed that increased CO2 and warming increased plant biomass and soil respiration. An increase in only CO2 treatment elicited more biomass of fine roots, soil respiration, and a decrease in foliar nitrogen (Dieleman et al., 2012). In another study De Souza et al. (2008) compared the effects of ambient (∼370 ppm) and increased (∼720 ppm) CO2 concentration on sugarcane growth and biomass. Elevated CO2 led to an increase of 17% in plant height, 30% in photosynthesis, 29% in sucrose contents, and 40% more accumulation of plant biomass (De Souza et al., 2008). This is thought to be achieved by physiological modifications regarding WUE as transpiration rates and stomatal conduction was reduced by –32 and –37%. An experiment involving free-air carbon enrichment (FACE) showed an elevated photosynthetic rate in young leaves and increased biomass and leaf number in sorghum and maize, respectively (Maroco et al., 1999; Cousins and Bloom, 2003). Recently, a study focused on the membrane properties and photosystem II activity of maize and pearl millet under elevated CO2 and temperature. The results showed that maize outperformed in biomass accumulation in the presence of high CO2 while pearl millet was more responsive in high temperature (Bordignon et al., 2019). Conversely, a comparative study conducted on two weedy species of C3 (Chenopodium album) and C4 (Setaria viridis) plants showed the decreased biomass at elevated temperature alone but a dramatic increase in biomass and seed yield by 33.9 and 114.4%, respectively, at increased temperature and CO2 concentrations in Chenopodium album. On the other hand, Setaria viridis showed 1.6- and 1.3-fold more biomass in an increased temperature and CO2 conditions as compared to control and only increased temperature (Lee, 2011). Experiments conducted on wheat and maize as the representatives of C3 and C4 plants showed that high CO2 in C3 (wheat) helps in ammonium NH4+ assimilation which was very less in ambient CO2 and showed declined rate of photosynthesis. Overall results showed that cellular and chloroplast CO2 enhanced electron flux in wheat as compared to maize. Several experiments showed that C3 plants are more responsive toward elevated CO2 as they reduce the extent of photorespiration and benefit more from enhanced CO2. However, recently reported from a 20-year continuous investigation using FACE experiment on 88 grassland plots, that for 12 years C3 plants showed increased biomass owing to higher eCO2 (Equivalent CO2) (Reich et al., 2018). Whereas, in subsequent 8 years a shift in this trend was seen from C3 to C4 plants, which resulted in enhanced biomass and soil nitrogen mineralization in C4 plants. These results are challenging to current concepts regarding the C3-C4 eCO2 paradigm (Figure 2A).
Circadian Rhythm Modulations by Environmental Factors
The instabilities of external cues of the environment, such as light, temperature, and nutrition, evoke a well-developed endogenous time-keeping mechanism in plants which is called the circadian cycle that allow in the modulation of energy and developmental metabolism. For example, fluctuations in the diurnal rhythm of light duration, temperature, and nutrition level modify intricate transcriptional and post-transcriptional loops in plants similar to animals (Harmer et al., 2001; Inoue et al., 2017). In Arabidopsis, the circadian clock starts by the mutualistic interaction of two transcription factors circadian clock associated 1 (CCA1) and late elongated hypocotyl (LHY) (Mizoguchi et al., 2002) during the morning hours. These two MYB TFs (CCA1, LHY) repress the transcription of TIMING OF CAB EXPRESSION 1 (TOC1) also known as PSEUDORESPONSE REGULATOR 1 (PRR1), which acts as a regulator of many downstream genes. CCA1 Hiking Expedition (CHE) (Pruneda-Paz et al., 2009), and GIGANTEA (GI) (Strayer et al., 2000) also acts as a positive regulator in circadian loops, whereas, CCA1 and LHY mRNA decrease during the mid-day by TOC1 homologs (PRR9 and PRR7) in feedback mechanism (Farré et al., 2005). During the evening, Lux Arrythmo (LUX) mediated transcriptional repression of PRR9 takes place by early flowering 4 (ELF4) and (ELF3) (McClung, 2014).
In grasses, growth activity is mediated by a rib zone called shoot apical meristem and is strongly influenced by light and shade conditions. In densely grown sorghum populations, leaves experience more shade conditions and inhibit shoot branching, more stem elongation, and early flowering. A combination of these responses is called Shade Avoidance Syndrome (SAS) (Casal, 2013), which is a survival strategy in plants for the quest for more sunlight and resources. Plant photoreceptors act to monitor the light environment and perception, and they work in synchrony with the circadian clock to regulate growth and development in plants (Devlin and Kay, 2001). Fluctuations in the light intensity change the expression of 24 circadian genes in bioenergy sorghum, among which CCA1 and LHY showed 12.7-fold lower and 5.9-fold higher expression in internodes of shade-treated plants in comparison to control. CCA1 and LHY regulate the expression of downstream genes for example in sorghum, the homolog of Arabidopsis Granule Bound Starch Synthase1 (GBSS1) gene which functions in starch biosynthesis was also down regulated by 6.6-fold. Other clock-related genes for example evening core clock genes were upregulated in shade-exposed plants. For example, the expression of BT2, and THIAMIN C SYNTHASE (THIC) function in important pathways related to hormones, sugars, and thianmin synthesis are regulated by circadian cycle genes (Mandadi et al., 2009). Similarly, 6 days of exposure to extended darkness resulted in malfunctioning in photosynthetic pigments, reduction in photoassimilates, and total soluble and insoluble carbohydrates in maize. However, CO2 exchange was not disturbed but transient carbon pools were largely consumed by elevated levels of nocturnal respiration rather than transport toward the sink. Underlying processes may involve signals by trehalose-6-phosphate and circadian rhythms which are controlling stress response in multi-dynamic pathways (Graf et al., 2010; Wingler et al., 2012). Light quality also mediates growth responses by genetic and physiological modifications. Transcriptome and metabolome profile of blue light exposed maize plants showed genes and metabolites related to stomatal, carotenoid, photosynthesis, and circadian cycle-related genes. CCA1 gene was upregulated in presence of blue light, this upregulation is consistent with the expression of many downstream genes related to photosynthesis, starch synthesis, and stomatal development processes (Liu and Zhang, 2021). In maize, the early stage of light exposure stimulates the circadian rhythm genes to increase light absorbance by regulation of photosynthetic genes (Khan et al., 2010). As C4 plants possess well-developed Kranz anatomy for the specialized storage of CO2 and water. In maize plant ZmPIP4c, a water transport aquaporin gene showed high expression profiles in bundle sheath cells during diurnal change and is potentially responsible for the transport of water in mesophyll cells. The synchronization in the expression of NAD-malic enzyme gene (NAD-ME), phosphoenolpyruvate carboxylase gene (PEPC), and carbonic anhydrase gene (CA) from base to tip is consistent with the circadian rhythm regulating cycle for efficient WUE in both light and dark conditions (Xiang et al., 2020). Moreover, as already discussed hybridization leads to heterosis, which is the outcome of enhanced photosynthesis and metabolism possibly influenced by CIRCADIAN CLOCK ASSOCIATED1 (CCA1). Two homologs of maize, ZmCCA1a, and ZmCCA1b are diurnally upregulated in Arabidopsis, cca1 Arabidopsis mutant was complemented by ZmCCA1. Whereas, ZmCCA1b showed disruption in circadian rhythms leading to reduced heterosis and plant height in the greenhouse and slightly compensated in field conditions upon light exposure. In hybrids, the temporal shift of ZmCCA1-binding targets suggest the activated photosynthesis and growth vigor genes in the morning phase relative to the inbred lines (Ko et al., 2016). The behavior of circadian genes is a good indicator to enhance our understanding of environment-influenced genetic modulations.
Tools and Strategies to Enhance Biomass
Hybridization and Molecular Techniques
Until now, some plant species have been given attention for the improvement of biomass which includes miscanthus, switchgrass, willow, poplar, and eucalyptus, with their improvement history dating back to the second half of the twentieth century (Allwright and Taylor, 2016; Clifton-Brown et al., 2019). Their improvement relied on hybridization or breeding methods (crossing of different strains, species, or lines) leading to heterosis or hybrid vigor of the F1 heterozygotes with higher fitness in the population. Heterotic fitness refers to superior growth, stature, fertility, and biomass in offsprings. Several factors, for example, transcriptional regulation and epigenetic changes drive improved characters in hybrids. We have already reviewed that breeding techniques are employed mostly in C4 grasses for achieving enhanced biomass. However, the approval and release of commercial cultivars take a long period of time, which factually delays the cultivation in agriculture systems and slows down the progress of conventional breeding (Clifton-Brown et al., 2019). Conventional breeding techniques advance our understanding toward marker-assisted selection for biomass-related traits, stress tolerance, and scarification in biofuel grasses and woody plants. In switchgrass, marker-assisted breeding enabled the understanding of substitution of cell wall hemicellulose polymers backbone and remodeling (De Souza et al., 2015) whereas identification of specific loci was identified from potential markers for high ethanol generation from switchgrass populations (Chen et al., 2016). Prairie cordgrass is a potential C4 bioenergy crop, and two clones of prairie cordgrass were crossed and developed SSR markers for marker-assisted selection of biomass traits (Gedye et al., 2012). Another study comprising 28 sugarcane genotypes identified simple sequence repeat (SSR) markers associated with stalk number and stalk volume (Bilal et al., 2015). In another experiment, 40 putative quantitative trait alleles (QTAs) were identified from a self-crossed (295) population of “R570,” with each allele contributing to phenotypic variation by 3–7% in sugarcane (Hoarau et al., 2002). Likewise, in sorghum, four QTLs were identified that control tiller number and formation (Hart et al., 2001). Recently research on the genotypes of M. sinensis indicated the genetic diversity of cell wall constituents and concluded that a higher ratio of para-coumaric acid to lignin contents and trans-ferulic acid (TFA) increased the saccharification efficacy (van der Weijde et al., 2017a). In sugarcane, stalk number is influenced by genes and their alleles with additive and non-additive effects or their interactions (Hongkai et al., 2009; Carvalho-Netto et al., 2014). Considering the use of genome editing techniques for bioenergy crops, the use of Transcription activator-like effector nucleases (TALENs) have been employed for targeted modification in the genome. High lignin content is an undesirable character for biofuel crops as in sugarcane, TALEN induced mutation in caffeic acid O-methyltransferase (COMT) sequences modified cell wall compositions. Pyrosequencing showed mutation frequencies up to 99% and revealed 29–32% reduced lignin and elevated hemicellulose contents (Jung and Altpeter, 2016). Similarly, RNAi-derived COMT silenced sugarcane callus-derived plants showed a 12% reduction in lignin and 32% improved scarification but compromised agronomic performance (Jung et al., 2013). CRISPR/Cas9 is a recent genome-editing technique expanding its applications to construct desirable genetic circuits (Khakhar et al., 2018). CRISPR/Cas9 system was employed in switchgrass for the reduction of lignin contents (Park et al., 2017). Knock-out mutant of the Pv4CL1 gene encoding 4-coumarate: coenzyme A ligase (4CL) displayed increased scarification as compared to wild. Tiller formation is one of the indices of biomass, two genes grassy tiller1 (gt1) and teosinte branched1 (tb1) control tiller formation in maize (Whipple et al., 2011). BRANCHED1 (BRC1) gene in sorghum is the homolog of tb1 gene, ectopic expression of tb1 gene in Arabidopsis promoted axillary buds formation Arabidopsis (Kebrom et al., 2006). Genome editing-based mutagenesis using CRISPR/Cas9 showed proliferated tillers in switchgrass as compared to wild plants (Clifton-Brown et al., 2019). In sorghum, by1 mutant obtained by knocking out BIOMASS YIELD 1 gene using CRISPR/Cas9 displayed reduced plant height narrow stem length, erect and narrow leaves, and abnormal floral organs. BIOMASS YIELD 1 gene translates into an enzymatic protein catalyzing the first step of the shikimate pathway (Chen et al., 2020). BY1 gene showed its role in primary metabolism and secondary metabolites for example flavonoids. In Arabidopsis hormone activated Cas9-based repressor (HACRs) showed significant results that can be utilized to achieve high grass biomass and economic yield (Khakhar et al., 2018). Similarly, in sugarcane, using transgenic and molecular techniques, an ortholog of the SLR1/D8/RHT1/GAI gene showed substantial stem growth and structural modifications in storage organs by regulating source-sink allocation changes (Garcia Tavares et al., 2018). Although several transgenic lines with enhanced features as bioenergy crops have been developed, there is a need for a suitable selection process and quality evaluation. However, due to cross-pollination in many grass species, transgenic lines pose a threat of seed contamination. Introgression and hybridization require labor-intensive and time-consuming efforts with uncertain outcomes. Therefore, recent genome editing techniques for example synthetic genetic circuits (SGC) or CRISPR offer sophisticated and foreseeable mutation induction in first-generation mutant lines (Scheben and Edwards, 2018). Moreover, Near-infrared spectroscopy (NIRS) and thermal aerial imaging technologies aid the phenotyping of constituents and high-throughput options to screen abiotic stress tolerance, respectively (van der Weijde et al., 2017b). Few examples in Table 2 highlight the use of integrated phenotyping and molecular technologies for biomass related research.
Nitrogen Management
Plant biomass of crops including C4 plants is influenced by a variety of variables, i.e., plant genotype, photoperiod, solar radiation, soil temperature, soil humidity, and many more. Soil nutrient availability is one of the most significant variables determining the crop biomass in C4 crops. So, by regulating the optimal amounts of nutrient availability in soil, growers may optimize the biomass output for biofuels and of course economic gain (grain production). Soil degradation and low soil fertility status significantly minimize the nutrient availability in the soil to plants (Chatzistathis and Therios, 2013; Tanveer et al., 2019). Optimum fertilization appears to be the most common method chosen by farmers in instances of restricted nutrient availability in soils to improve the biomass of cultivated C4 crops. Macronutrients, i.e., nitrogen, phosphorous, magnesium, potassium, sulfur, and calcium, and micronutrients, i.e., copper, zinc, manganese, iron, chlorine, and molybdenum are classified as important nutrients for improving the biomass of C4 crops. Nutrient scarcity has a detrimental impact on biomass productivity (Anjum et al., 2019). Vegetation flushes of C4 crops are severely hampered by nitrogen shortage as nitrogen is an integral component of chlorophyll, pyrimidines, purines, amino acids, proteins, and nucleic acids in C4 crops. Borges et al. (2019) reported that the appropriate method of nitrogen application at the appropriate time (early season) significantly improved the biomass of sugarcane (Saccharum officinarum) by 30% as compared to traditional fertilizers practices. In addition, climate prediction models guided nitrogen management methods can be opted, as the climate is a significant determinant of crop growth, nitrogen demand, and nitrogen losses processes. Seasonal climatic projections might be used to establish nitrogen management plans for “dry” and “wet” years, directing application rate, timing, and frequency of nitrogen fertilizer application, as well as the advantages of employing different types of nitrogen fertilizer in C4 crops like sugarcane, maize (Zea mays), sorghum (Sorghum bicolor), pearl millet (Pennisetum glaucum), and Napier grass (Pennisetum purpureum) (Anjum et al., 2019). Moreover, Khan Khyber et al. (2010) reported that integrated application of 50% urea with 50% poultry manure significantly enhanced the grain yield of maize by 57.14%, respectively, as compared to plots having 0% nitrogen application. Although much effort has previously been done to maximize yields while maintaining high nutrient utilization efficiencies, more integrated approach results are still needed to minimize the nitrogen inputs while maintaining optimum usage efficiency in C4 crops (Noor, 2017). To attain the maximum production of biomass in C4 crops, maintaining the optimum levels of all the necessary soil nutrients should always be taken care of. However, determining the best nutrient prescription in terms of boosting productivity especially biomass production in C4 crops while also guaranteeing food security and environmental friendliness is a difficult task, and still needed further studies and detailed analysis.
Silicon Foliar Application
Silicon is a chemical element having atomic number 14 and is represented with the symbol Si. In plants application of silicon significantly enhances the crop biomass; improves the tolerance to biotic and abiotic stresses, and aid plant stability and protection (Zargar et al., 2019). In connection to the enhancement of cell wall elasticity and stiffness, silicon that is firmly linked to the cell walls is naturally present as a structural material. When the quantity of monosilicic acid in the xylem sap is high, it becomes a significant osmolyte, increasing the plant’s water and osmotic potential. Furthermore, in terms of structural material and osmolytes, Si consumes less energy than biomolecules like proline and lignin. As a result, for a cheap cost, silicon can enhance the homeostasis of C4 plants’ tolerance to a variety of biotic and abiotic stressors in terrestrial environments. C4 plant biomass recovery mediated by silicon is thought to have a bell-shaped response curve to abiotic stressors and an S-shaped response curve to biotic stresses. Silicon treatment to abiotic and biotic stressed crops can boost averaged plant biomass carbon and crop productivity by 35 and 24%, respectively. The efficacy of silicon-mediated restoration, on the other hand, varies substantially depending on the plant species and cultivars, the severity of abiotic and biotic stressors, and the amount of bio-available silicon. Ashraf et al. reported that the application of silicon significantly improved the biomass production in sugarcane by 77% under salinity stress (Ashraf et al., 2009). Similarly, the application of calcium silicate improved the crop biomass of sugarcane and enhanced the resistance in sugarcane against stem borer (Keeping and Meyer, 2002; Meyer and Keeping, 2005). A study conducted on maize reported that the application of silicon under water stress conditions significantly enhanced the crop biomass and nutrient uptake (Kaya et al., 2007). It was reported that the application of silicon significantly improved the plant biomass under agricultural soil contaminated with heavy metals like cadmium (Liang et al., 2005; Lukačová et al., 2013), and in arsenic (Ullah et al., 2016). Application of silicon is a significant option for improving the crop biomass of C4 crops, but still more research and detailed analysis should be done as most of the silicon application trials have so far been done in pots, field-scale to eco-system-scale investigation is needed. Moreover, several issues, such as the coupling relations between Si and plant essential elements, the efficiencies of Si-mediated plant biomass carbon restoration among plant species and stress intensities, and the relationship between the biogeochemical Si cycle and the resilience of terrestrial ecosystems, all require more research, particularly in fragmented landscapes.
Foliar Application of Plant Growth Regulators/Growth Hormones
Exogenous application of plant growth regulators/growth hormones (PGRs) has been found to improve plant stress tolerance and increase growth processes (Liu et al., 2019). Growth hormones are identified as playing a critical role in maintaining the plant morphology, flower blooming, development, photosynthetic activity, and stomatal closure in terms of physiological functions in C4 plants (Sharma et al., 2020). Exogenous growth hormones were also used to control seed germination, root elongation, cell development, and tiller formation in C4 plants cultivated under trace-metal contaminated soils (Maghsoudi et al., 2019). Similarly, in cereals like maize foliar application of plant growth hormones under abiotic stresses considerably improved the leaf area, plant growth, dry biomass, and stem diameter of C4 plants (Tran and Popova, 2013; Qandeel et al., 2020). It was reported that exogenous application of various types of brassinosteroids, i.e., 28-homobrassinolide, and 24-epibrassinolide significantly enhanced the biomass and productivity of maize, sugarcane, and sorghum grown under abiotic stresses, i.e., drought, salinity, and trace-metal contaminated soils (Tanveer et al., 2018, 2019). In another study, it was reported that application of 1-amino-cyclopropane-1-carboxylic-acid (ACC-deaminase), humic acid, and oxalic acid considerably enhanced bacterial community development in the rhizosphere, facilitating the remediation of organic pollutants, and improved the plant biomass, which had previously been hindered by the presence of organic contaminants in soil (Ping et al., 2006; Wen-Jie et al., 2011). Similarly, plant growth regulators like melatonin and indole acetic acid are also reported to significantly improve the plant biomass under various abiotic stresses (Rostami et al., 2021). Likewise, the application of abscisic acid, salicylic acid, auxins, cytokinin, methyl jasmonate, and ethylene are also documented to significantly improve the plant biomass under abiotic stresses (Hasan et al., 2019). Yet, to explain precise processes related to the impacts of growth hormones on plant biomass, integrative studies combining conventional and sequencing techniques are required.
Concluding Remarks and Prospects
Biofuels being an alternative to fossil fuels are considered an integral part of sustainable energy generation systems. To develop the biofuel industry on a sustainable basis, increasing plant biomass is a prime goal as feedstock in the biofuels industry. Biomass accumulation is a complex biological trait. However, advancements in genetics and biotechnology have deciphered that a plethora of genes are controlling growth and development starting from the cell cycle to the juvenile, vegetative, and reproductive maturity phase. Most of the pathways discussed highlight the important genes which have been exploited to tailor the bioenergy crops. Among them, genes involved in the cell cycle, cell wall, and hormone and related transcriptional factors considerably modify the carbohydrate allocation and improve photosynthetic efficiency. But the real challenge is the successful introduction of bioenergy specialized crops in fields on a sustainable basis. Moreover, we pointed out altered regulatory patterns of transcription activity of MYB TFs in C3 and C4 crops which indicate the lineage-specific carbohydrate storage biopolymer incorporation in both. Therefore, research focuses should be directed on C4 crops considering only C4 models in terms of biomass accumulation and later on energy generation.
Regarding environmental factors which are acting upon biomass accumulation, CO2, light, and temperature are among unavoidable stresses to threaten the growth process. For this, architecture for the maximum light interception, nutrient absorption traits, and certain anatomical changes can be engineered in wild plants to enable the cultivation of bioenergy and orphan lignocellulose crops on marginal lands (resource-deprived). Furthermore, optimization of locality-based bioenergy crops and cultural practices to enhance biomass is critical but not yet elaborated. The above-mentioned tools and practices including breeding, molecular methods (DNA-free genome editing method CRISPR/Cas9), high throughput sequencing, and cultural practices can be opted for engineering and validation of multiple genes from different pathways to generate climate-smart energy crops. The afore-mentioned strategies will only be realistic if they are part of an integrated approach to agriculture that is developed collaboratively with agronomists, engineers, and farmers to contribute to a bio-based economy.
Author Contributions
NA and RM conceptualized the review. NA wrote the review with the assistance of FH and MF. Habiba and NA designed the figures and tables. YZ improved and revised. All authors finally revised and approved the manuscript.
Funding
This work was supported by a startup fund from the Fujian Agriculture and Forestry University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ASEAN, Association of South East Asian Nations; eCO2, Equivalent CO2; GHGs, Greenhouse Gases; mg/h, megagrams per hectare; PEP, Phosphoenolpyruvate; RUBISCO, Ribulose Bisphosphate Carboxylase/Oxygenase; SQUAMOSA, SQUAMOSA Promoter-Binding Protein-Like (SPL); TFs, Transcription factors.
References
Agarwal, T., Grotewold, E., Doseff, A. I., and Gray, J. (2016). MYB31/MYB42 syntelogs exhibit divergent regulation of phenylpropanoid genes in maize, sorghum and rice. Sci. Rep. 6:28502. doi: 10.1038/srep28502
Ainsworth, E. A., and Long, S. P. (2005). What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 165, 351–372. doi: 10.1111/J.1469-8137.2004.01224.X
Allwright, M. R., and Taylor, G. (2016). Molecular breeding for improved second generation bioenergy crops. Trends Plant Sci. 21, 43–54. doi: 10.1016/j.tplants.2015.10.002
Ambavaram, M. M. R., Krishnan, A., Trijatmiko, K. R., and Pereira, A. (2011). Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol. 155, 916–931. doi: 10.1104/pp.110.168641
Anderson, E., Arundale, R., Maughan, M., Oladeinde, A., Wycislo, A., and Voigt, T. (2011). Growth and agronomy of Miscanthus × giganteus for biomass production. Biofuels 2, 167–183. doi: 10.4155/bfs.10.80
Anjum, K., Cheema, A., Farooq, M., Haider, F. U., Cheema, S. A., and Ur Rehman, H. (2019). Article citation: exploring the potential of selenium (Se) and Moringa (Moringa oleifera L.) leaf extract on the production and performance of Triticum aestivum L. Introduction. J. Res. Ecol. 7, 2390–2402.
Ardabili, G. S., Zakaria, R. A., and Zare, N. (2015). In vitro induction of polyploidy in Sorghum bicolor L. Cytologia 80, 495–503. doi: 10.1508/CYTOLOGIA.80.495
Ashraf, M., Rahmatullah, Afzal, M., Ahmed, R., Mujeeb, F., Sarwar, A., et al. (2009). Alleviation of detrimental effects of NaCl by silicon nutrition in salt-sensitive and salt-tolerant genotypes of sugarcane (Saccharum officinarum L.). Plant Soil 326, 381–391. doi: 10.1007/S11104-009-0019-9
Babitha, K. C., Vemanna, R. S., Nataraja, K. N., and Udayakumar, M. (2015). Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS One 10:e0137098. doi: 10.1371/JOURNAL.PONE.0137098
Barnum, K. J., and O’Connell, M. J. (2014). Cell cycle regulation by checkpoints. Methods Mol. Biol. 1170, 29–40. doi: 10.1007/978-1-4939-0888-2_2
Battle, M., Bender, M. L., Tans, P. P., White, J. W. C., Ellis, J. T., Conway, T., et al. (2000). Global carbon sinks and their variability inferred from atmospheric O2 and δ13C. Science 287, 2467–2470. doi: 10.1126/SCIENCE.287.5462.2467
Berthet, S., Demont-Caulet, N., Pollet, B., Bidzinski, P., Cézard, L., Le Bris, P., et al. (2011). Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23, 1124–1137. doi: 10.1105/TPC.110.082792
Bilal, M., Saeed, M., Nasir, I. A., Tabassum, B., Zameer, M., Khan, A., et al. (2015). Association mapping of cane weight and tillers per plant in sugarcane. Biotechnol. Biotechnol. Equip. 29, 617–623. doi: 10.1080/13102818.2015.1008203
Bishop, G. J. (2007). Refining the plant steroid hormone biosynthesis pathway. Trends Plant Sci. 12, 377–380. doi: 10.1016/J.TPLANTS.2007.07.001
Bodrug, T., Welsh, K. A., Hinkle, M., Emanuele, M. J., and Brown, N. G. (2021). Intricate Regulatory mechanisms of the anaphase-promoting complex/cyclosome and its role in chromatin regulation. Front. Cell Dev. Biol. 9:687515. doi: 10.3389/FCELL.2021.687515
Bonawitz, N. D., and Chapple, C. (2010). The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363. doi: 10.1146/ANNUREV-GENET-102209-163508
Bordignon, L., Faria, A. P., França, M. G. C., and Fernandes, G. W. (2019). Osmotic stress at membrane level and photosystem II activity in two C4 plants after growth in elevated CO2 and temperature. Ann. Appl. Biol. 174, 113–122. doi: 10.1111/aab.12483
Borges, C. D., Carvalho, J. L. N., Kölln, O. T., Sanches, G. M., Silva, M. J., Castro, S. G. Q., et al. (2019). Can alternative N-fertilization methods influence GHG emissions and biomass production in sugarcane fields? Biomass Bioenergy 120, 21–27. doi: 10.1016/J.BIOMBIOE.2018.10.017
Bottcher, A., Cesarino, I., Brombini dos Santos, A., Vicentini, R., Mayer, J. L. S., Vanholme, R., et al. (2013). Lignification in sugarcane: biochemical characterization, gene discovery, and expression analysis in two genotypes contrasting for lignin content. Plant Physiol. 163, 1539–1557. doi: 10.1104/PP.113.225250
Boucheron, E., Healy, J. H. S., Bajon, C., Sauvanet, A., Rembur, J., Noin, M., et al. (2005). Ectopic expression of Arabidopsis CYCD2 and CYCD3 in tobacco has distinct effects on the structural organization of the shoot apical meristem. J. Exp. Bot. 56, 123–134. doi: 10.1093/JXB/ERI001
Byrt, C. S., Grof, C. P. L., and Furbank, R. T. (2011). C4 plants as biofuel feedstocks: optimising biomass production and feedstock quality from a lignocellulosic perspective. J. Integr. Plant Biol. 53, 120–135. doi: 10.1111/j.1744-7909.2010.01023.x
Caparrós-Ruiz, D., Fornalé, S., Civardi, L., Puigdomènech, P., and Rigau, J. (2006). Isolation and characterisation of a family of Laccases in maize. Plant Sci. 171, 217–225. doi: 10.1016/J.PLANTSCI.2006.03.007
Caruso, G., Gomez, L. D., Ferriello, F., Andolfi, A., Borgonuovo, C., Evidente, A., et al. (2016). Exploring tomato Solanum pennellii introgression lines for residual biomass and enzymatic digestibility traits. BMC Genet. 17:56. doi: 10.1186/s12863-016-0362-9
Carvalho-Netto, O. V., Bressiani, J. A., Soriano, H. L., Fiori, C. S., Santos, J. M., Barbosa, G. V., et al. (2014). The potential of the energy cane as the main biomass crop for the cellulosic industry. Chem. Biol. Technol. Agric. 1:20. doi: 10.1186/S40538-014-0020-2
Casal, J. J. (2013). Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 64, 403–427. doi: 10.1146/ANNUREV-ARPLANT-050312-120221
Castorina, G., Persico, M., Zilio, M., Sangiorgio, S., Carabelli, L., and Consonni, G. (2018). The maize lilliputian1 (lil1) gene, encoding a brassinosteroid cytochrome P450 C-6 oxidase, is involved in plant growth and drought response. Ann. Bot. 122, 227–238. doi: 10.1093/AOB/MCY047
Cavalier, D. M., Lerouxel, O., Neumetzler, L., Yamauchi, K., Reinecke, A., Freshour, G., et al. (2008). Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 20, 1519–1537. doi: 10.1105/TPC.108.059873
Cesarino, I., Araújo, P., Sampaio Mayer, J. L., Vicentini, R., Berthet, S., Demedts, B., et al. (2013). Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J. Exp. Bot. 64, 1769–1781. doi: 10.1093/jxb/ert045
Chae, W. B., Hong, S. J., Gifford, J. M., Lane Rayburn, A., Widholm, J. M., and Juvik, J. A. (2013). Synthetic polyploid production of Miscanthus sacchariflorus, Miscanthus sinensis, and Miscanthus x giganteus. GCB Bioenergy 5, 338–350. doi: 10.1111/j.1757-1707.2012.01206.x
Chatzistathis, T., and Therios, I. (2013). “How soil nutrient availability influences plant biomass and how biomass stimulation alleviates heavy metal toxicity in soils: the cases of nutrient use efficient genotypes and phytoremediators, respectively,” in Biomass Now – Cultivation and Utilization, ed. M. D. Matovic (London: IntechOpen), 428–448. doi: 10.5772/53594
Chen, J., Zhu, M., Liu, R., Zhang, M., Lv, Y., Liu, Y., et al. (2020). BIOMASS YIELD 1 regulates sorghum biomass and grain yield via the shikimate pathway. J. Exp. Bot. 71, 5506–5520. doi: 10.1093/JXB/ERAA275
Chen, S., Kaeppler, S. M., Vogel, K. P., and Casler, M. D. (2016). Selection signatures in four lignin genes from switchgrass populations divergently selected for in vitro dry matter digestibility. PLoS One 11:e0167005. doi: 10.1371/JOURNAL.PONE.0167005
Chen, X., Zhang, Z., Liu, D., Zhang, K., Li, A., and Mao, L. (2010). SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52, 946–951. doi: 10.1111/j.1744-7909.2010.00987.x
Chen, Z. J. (2010). Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 15, 57–71. doi: 10.1016/J.TPLANTS.2009.12.003
Cheng, Y., Cao, L., Wang, S., Li, Y., Shi, X., Liu, H., et al. (2013). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 75, 642–655. doi: 10.1111/TPJ.12228
Chuck, G., Meeley, R., Irish, E., Sakai, H., and Hake, S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517–1521. doi: 10.1038/ng.2007.20
Clifton-Brown, J., Harfouche, A., Casler, M. D., Dylan Jones, H., Macalpine, W. J., Murphy-Bokern, D., et al. (2019). Breeding progress and preparedness for mass-scale deployment of perennial lignocellulosic biomass crops switchgrass, miscanthus, willow and poplar. GCB Bioenergy 11, 118–151. doi: 10.1111/gcbb.12566
Coles, J. P., Phillips, A. L., Croker, S. J., García-Lepe, R., Lewis, M. J., and Hedden, P. (1999). Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 17, 547–556. doi: 10.1046/J.1365-313X.1999.00410.X
Cousins, A. B., and Bloom, A. J. (2003). Influence of elevated CO2 and nitrogen nutrition on photosynthesis and nitrate photo-assimilation in maize (Zea mays L.). Plant Cell Environ. 26, 1525–1530. doi: 10.1046/J.1365-3040.2003.01075.X
de Freitas Lima, M., Eloy, N. B., Bottino, M. C., Hemerly, A. S., and Ferreira, P. C. G. (2013). Overexpression of the anaphase-promoting complex (APC) genes in Nicotiana tabacum promotes increasing biomass accumulation. Mol. Biol. Rep. 40, 7093–7102. doi: 10.1007/s11033-013-2832-8
de Lima, M. F., Eloy, N. B., Pegoraro, C., Sagit, R., Rojas, C., Bretz, T., et al. (2010). Genomic evolution and complexity of the anaphase-promoting complex (APC) in land plants. BMC Plant Biol. 10:254. doi: 10.1186/1471-2229-10-254
De Souza, A. P., Gaspar, M., Da Silva, E. A., Ulian, E. C., Waclawovsky, A. J., Nishiyama, M. Y., et al. (2008). Elevated CO2 increases photosynthesis, biomass and productivity, and modifies gene expression in sugarcane. Plant Cell Environ. 31, 1116–1127. doi: 10.1111/j.1365-3040.2008.01822.x
De Souza, A. P., Kamei, C. L. A., Torres, A. F., Pattathil, S., Hahn, M. G., Trindade, L. M., et al. (2015). How cell wall complexity influences saccharification efficiency in Miscanthus sinensis. J. Exp. Bot. 66, 4351–4365. doi: 10.1093/JXB/ERV183
Demura, T., and Ye, Z. H. (2010). Regulation of plant biomass production. Curr. Opin. Plant Biol. 13, 298–303. doi: 10.1016/j.pbi.2010.03.002
Devlin, P. F., and Kay, S. A. (2001). Circadian photoperception. Annu. Rev. Physiol. 63, 677–694. doi: 10.1146/ANNUREV.PHYSIOL.63.1.677
Dieleman, W. I. J., Vicca, S., Dijkstra, F. A., Hagedorn, F., Hovenden, M. J., Larsen, K. S., et al. (2012). Simple additive effects are rare: a quantitative review of plant biomass and soil process responses to combined manipulations of CO2 and temperature. Glob. Chang. Biol. 18, 2681–2693. doi: 10.1111/j.1365-2486.2012.02745.x
Dimitroff, G., Little, A., Lahnstein, J., Schwerdt, J. G., Srivastava, V., Bulone, V., et al. (2016). (1,3;1,4)-β-Glucan biosynthesis by the CSLF6 enzyme: position and flexibility of catalytic residues influence product fine structure. Biochemistry 55, 2054–2061. doi: 10.1021/ACS.BIOCHEM.5B01384
Do, P. T., De Tar, J. R., Lee, H., Folta, M. K., and Zhang, Z. J. (2016). Expression of ZmGA20ox cDNA alters plant morphology and increases biomass production of switchgrass (Panicum virgatum L.). Plant Biotechnol. J. 14, 1532–1540. doi: 10.1111/pbi.12514
Dong, Z., Danilevskaya, O., Abadie, T., Messina, C., Coles, N., and Cooper, M. (2012). A gene regulatory network model for floral transition of the shoot apex in maize and its dynamic modeling. PLoS One 7:e43450. doi: 10.1371/JOURNAL.PONE.0043450
Dumitrache, A., Natzke, J., Rodriguez, M., Yee, K. L., Thompson, O. A., Poovaiah, C. R., et al. (2017). Transgenic switchgrass (Panicum virgatum L.) targeted for reduced recalcitrance to bioconversion: a 2-year comparative analysis of field-grown lines modified for target gene or genetic element expression. Plant Biotechnol. J. 15, 688–697. doi: 10.1111/PBI.12666
Eloy, N. B., Lima, M. D. F., Damme, V., Vanhaeren, H., and Gonzalez, N. (2011). The APC/C subunit 10 plays an essential role in cell proliferation during leaf development. Plant J. 68, 351–363. doi: 10.1111/j.1365-313X.2011.04691.x
Endo, S., Pesquet, E., Yamaguchi, M., Tashiro, G., Sato, M., Toyooka, K., et al. (2009). Identifying new components participating in the secondary cell wall formation of vessel elements in Zinnia and Arabidopsis. Plant Cell 21, 1155–1165. doi: 10.1105/TPC.108.059154
Farré, E. M., Harmer, S. L., Harmon, F. G., Yanovsky, M. J., and Kay, S. A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. doi: 10.1016/J.CUB.2004.12.067
Favero, D. S., Kawamura, A., Shibata, M., Takebayashi, A., Jung, J. H., Suzuki, T., et al. (2020). AT-hook transcription factors restrict petiole growth by antagonizing PIFs. Curr. Biol. 30, 1454–1466.e6. doi: 10.1016/J.CUB.2020.02.017
Feldmann, K. A., Marks, M. D., Christianson, M. L., and Quatrano, R. S. (1989). A dwarf mutant of Arabidopsis generated by T-DNA insertion mutagenesis. Science 243, 1351–1354. doi: 10.1126/SCIENCE.243.4896.1351
Fornalé, S., Shi, X., Chai, C., Encina, A., Irar, S., Capellades, M., et al. (2010). ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J. 64, 633–644. doi: 10.1111/J.1365-313X.2010.04363.X
Fridman, Y., and Savaldi-Goldstein, S. (2013). Brassinosteroids in growth control: how, when and where. Plant Sci. 209, 24–31. doi: 10.1016/j.plantsci.2013.04.002
Fujimoto, M., Arimura, S. I., Nakazono, M., and Tsutsumi, N. (2008). Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep. 27, 1581–1586. doi: 10.1007/S00299-008-0583-0/FIGURES/3
Garcia Tavares, R., Lakshmanan, P., Peiter, E., O’Connell, A., Caldana, C., Vicentini, R., et al. (2018). ScGAI is a key regulator of culm development in sugarcane. J. Exp. Bot. 69, 3823–3837. doi: 10.1093/JXB/ERY180
Gedye, K. R., Gonzalez-Hernandez, J. L., Owens, V., and Boe, A. (2012). Advances towards a marker-assisted selection breeding program in prairie cordgrass, a biomass crop. Int. J. Plant Genomics 2012:313545. doi: 10.1155/2012/313545
Ghannoum, O., von Caemmerer, S., Ziska, L. H., and Conroy, J. P. (2000). The growth response of C4 plants to rising atmospheric CO2 partial pressure: a reassessment. Plant Cell Environ. 23, 931–942. doi: 10.1046/J.1365-3040.2000.00609.X
Gong, P., Bontinck, M., Demuynck, K., de Block, J., Gevaert, K., Eeckhout, D., et al. (2021). SAMBA controls the rate of cell division in maize development through APC/C interaction. bioRxiv [Preprint]. doi: 10.1101/2021.04.22.440954
Graf, A., Schlereth, A., Stitt, M., and Smith, A. M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. U.S.A. 107, 9458–9463. doi: 10.1073/PNAS.0914299107
Guillaumie, S., Goffner, D., Barbier, O., Martinant, J.-P., Pichon, M., and Barrière, Y. (2008). Expression of cell wall related genes in basal and ear internodes of silking brown-midrib-3, caffeic acid O-methyltransferase (COMT) down-regulated, and normal maize plants. BMC Plant Biol. 8:71. doi: 10.1186/1471-2229-8-71
Harmer, S. L., Panda, S., and Kay, S. A. (2001). Molecular bases of circadian rhythms. Annu. Rev. Cell Dev. Biol. 17, 215–253. doi: 10.1146/ANNUREV.CELLBIO.17.1.215
Hart, G. E., Schertz, K. F., Peng, Y., and Syed, N. H. (2001). Genetic mapping of Sorghum bicolor (L.) Moench QTLs that control variation in tillering and other morphological characters. Theor. Appl. Genet. 103, 1232–1242. doi: 10.1007/S001220100582
Hasan, M. N., Hasan, M. R., Foysal, S. H., Hoque, H., Khan, M. F., Bhuiyan, M. F. H., et al. (2019). In-vitro regeneration of Citrus sinensis (L.) Osbeck from mature seed derived embryogenic callus on different solid basal media. Am. J. Plant Sci. 10, 285–297. doi: 10.4236/AJPS.2019.102022
Heaton, E. A., Dohleman, F. G., and Long, S. P. (2008). Meeting US biofuel goals with less land: the potential of Miscanthus. Glob. Chang. Biol. 14, 2000–2014. doi: 10.1111/J.1365-2486.2008.01662.X
Hirano, K., Kawamura, M., Araki-Nakamura, S., Fujimoto, H., Ohmae-Shinohara, K., Yamaguchi, M., et al. (2017). Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci. Rep. 7:126. doi: 10.1038/s41598-017-00096-w
Hirano, K., Ueguchi-Tanaka, M., and Matsuoka, M. (2008). GID1-mediated gibberellin signaling in plants. Trends Plant Sci. 13, 192–199. doi: 10.1016/J.TPLANTS.2008.02.005
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/J.1365-313X.2003.01759.X
Hoang, N. V., Furtado, A., Botha, F. C., Simmons, B. A., and Henry, R. J. (2015). Potential for genetic improvement of sugarcane as a source of biomass for biofuels. Front. Bioeng. Biotechnol. 3:182. doi: 10.3389/fbioe.2015.00182
Hoarau, J.-Y., Grivet, L., Offmann, B., Raboin, L.-M., Diorflar, J.-P., Payet, J., et al. (2002). Genetic dissection of a modern sugarcane cultivar (Saccharum spp.).II. Detection of QTLs for yield components. Theor. Appl. Genet. 105, 1027–1037. doi: 10.1007/S00122-002-1047-5
Hong, Z., Geisler-Lee, C. J., Zhang, Z., and Verma, D. P. S. (2003). Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol. Biol. 53, 297–312. doi: 10.1023/B:PLAN.0000006936.50532.3A
Hongkai, Z., Guifu, L., Jiannong, L., and Juemin, H. (2009). Genetic analysis of sugarcane biomass yield and its component traits using ADAA model. J. Trop. Agric. 47, 70–73.
Hu, Y., Xie, Q., and Chua, N. H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961. doi: 10.1105/tpc.013557
Huang, P., Jiang, H., Zhu, C., Barry, K., Jenkins, J., Sandor, L., et al. (2017). Sparse panicle1 is required for inflorescence development in Setaria viridis and maize. Nat. Plants 3:17054. doi: 10.1038/nplants.2017.54
Inagaki, S., and Umeda, M. (2011). Cell-cycle control and plant development. Int. Rev. Cell Mol. Biol. 291, 227–261. doi: 10.1016/B978-0-12-386035-4.00007-0
Inoue, K., Araki, T., and Endo, M. (2017). Integration of input signals into the gene network in the plant circadian clock. Plant Cell Physiol. 58, 977–982. doi: 10.1093/PCP/PCX066
Inz, D. (2006). Cell cycle regulation in plant development. Annu. Rev. Genet. 40, 77–105. doi: 10.1146/annurev.genet.40.110405.090431
IPCC (2007). Climate Change 2007: Synthesis Report. Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Core Writing Team, eds R. K. Pachauri and A. Reisinger (Geneva: IPCC), 104.
Jung, J. H., and Altpeter, F. (2016). TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 92, 131–142. doi: 10.1007/S11103-016-0499-Y
Jung, J. H., Fouad, W. M., Vermerris, W., Gallo, M., and Altpeter, F. (2012). RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 10, 1067–1076. doi: 10.1111/J.1467-7652.2012.00734.X
Jung, J. H., Vermerris, W., Gallo, M., Fedenko, J. R., Erickson, J. E., and Altpeter, F. (2013). RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol. J. 11, 709–716. doi: 10.1111/PBI.12061
Kandel, R., Yang, X., Song, J., and Wang, J. (2018). Potentials, challenges, and genetic and genomic resources for sugarcane biomass improvement. Front. Plant Sci. 9, 1–14. doi: 10.3389/fpls.2018.00151
Kang, B.-H., Busse, J. S., and Bednarek, S. Y. (2003). Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15, 899–913. doi: 10.1105/TPC.009670
Kaya, C., Tuna, L., and Higgs, D. (2007). Effect of silicon on plant growth and mineral nutrition of maize grown under water-stress conditions. J. Plant Nutr. 29, 1469–1480. doi: 10.1080/01904160600837238
Kebrom, T. H., Burson, B. L., and Finlayson, S. A. (2006). Phytochrome B represses teosinte branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 140, 1109–1117. doi: 10.1104/PP.105.074856
Kebrom, T. H., McKinley, B., and Mullet, J. E. (2017). Dynamics of gene expression during development and expansion of vegetative stem internodes of bioenergy sorghum. Biotechnol. Biofuels 10:159. doi: 10.1186/S13068-017-0848-3
Keeping, M. G., and Meyer, J. H. (2002). Calcium silicate enhances resistance of sugarcane to the African stalk borer Eldana saccharina walker (Lepidoptera: Pyralidae). Agric. For. Entomol. 4, 265–274. doi: 10.1046/J.1461-9563.2002.00150.X
Khakhar, A., Leydon, A. R., Lemmex, A. C., Klavins, E., and Nemhauser, J. L. (2018). Synthetic hormone-responsive transcription factors can monitor and reprogram plant development. Elife 7:e34702. doi: 10.7554/ELIFE.34702
Khan, S., Rowe, S. C., and Harmon, F. G. (2010). Coordination of the maize transcriptome by a conserved circadian clock. BMC Plant Biol. 10:126. doi: 10.1186/1471-2229-10-126
Khan Khyber, H., Wahid, M. A., Rasul, F., and Mohkum Hammad, H. (2010). Nitrogen management strategies for sustainable maize production assessing the impact of climate change on wheat & cotton grown under different types of soils in various agro-environmental conditions of southern Punjab-Pakistan using crop simulation models. View project effect of nitrogen on seed production view project. Crop Environ. 1, 49–52.
Kir, G., Ye, H., Nelissen, H., Neelakandan, A. K., Kusnandar, A. S., Luo, A., et al. (2015). RNA interference knockdown of BRASSINOSTEROID INSENSITIVE1 in maize reveals novel functions for brassinosteroid signaling in controlling plant architecture. Plant Physiol. 169, 826–839. doi: 10.1104/PP.15.00367
Ko, D. K., Rohozinski, D., Song, Q., Taylor, S. H., Juenger, T. E., Harmon, F. G., et al. (2016). Temporal shift of circadian-mediated gene expression and carbon fixation contributes to biomass heterosis in maize hybrids. PLoS Genet. 12:e1006197. doi: 10.1371/journal.pgen.1006197
Koçar, G., and Civaş, N. (2013). An overview of biofuels from energy crops: current status and future prospects. Renew. Sustain. Energy Rev. 28, 900–916. doi: 10.1016/J.RSER.2013.08.022
Lauf, T., Memmler, M., and Schneider, S. (2021). Emissions Balance of Renewable Energy Sources Determination of Avoided Emissions in 2020 (Emissionsbilanz Erneuerbarer Energieträger Bestimmung Der Vermiedenen Emissionen Im Jahr 2020). Available online at: http://inis.iaea.org/search/search.aspx?orig_q=RN:53029699
Lauter, N., Kampani, A., Carlson, S., Goebel, M., and Moose, S. P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. U.S.A. 102, 9412–9417. doi: 10.1073/PNAS.0503927102
Lawit, S. J., Wych, H. M., Xu, D., Kundu, S., and Tomes, D. T. (2010). Maize DELLA proteins dwarf plant8 and dwarf plant9 as modulators of plant development. Plant Cell Physiol. 51, 1854–1868. doi: 10.1093/PCP/PCQ153
Lee, J. S. (2011). Combined effect of elevated CO2 and temperature on the growth and phenology of two annual C3 and C4 weedy species. Agric. Ecosyst. Environ. 140, 484–491. doi: 10.1016/j.agee.2011.01.013
Liang, M., Davis, E., Gardner, D., Cai, X., and Wu, Y. (2006). Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224, 1185–1196. doi: 10.1007/S00425-006-0300-6
Liang, Y., Wong, J. W. C., Wei, L., Liang, Y., Wong, J. W. C., and Wei, L. (2005). Silicon-mediated enhancement of cadmium tolerance in maize (Zea mays L.) grown in cadmium contaminated soil. Chemosphere 58, 475–483. doi: 10.1016/J.CHEMOSPHERE.2004.09.034
Lima, M. d. F., Eloy, N. B., Siqueira, J. A. B. d., Inzé, D., Hemerly, A. S., and Ferreira, P. C. G. (2017). Molecular mechanisms of biomass increase in plants. Biotechnol. Res. Innov. 1, 14–25. doi: 10.1016/j.biori.2017.08.001
Liu, T., and Zhang, X. (2021). Transcriptome and metabolomic analyses reveal regulatory networks controlling maize stomatal development in response to blue light. Int. J. Mol. Sci. 22:5393. doi: 10.3390/ijms22105393
Liu, Y., Chen, X., Wang, X., Fang, Y., Zhang, Y., Huang, M., et al. (2019). The influence of different plant hormones on biomass and starch accumulation of duckweed: a renewable feedstock for bioethanol production. Renew. Energy 138, 659–665. doi: 10.1016/J.RENENE.2019.01.128
Lukačová, Z., Švubová, R., Kohanová, J., and Lux, A. (2013). Silicon mitigates the Cd toxicity in maize in relation to cadmium translocation, cell distribution, antioxidant enzymes stimulation and enhanced endodermal apoplasmic barrier development. Plant Growth Regul. 70, 89–103. doi: 10.1007/S10725-012-9781-4
Maghsoudi, K., Arvin, M. J., and Ashraf, M. (2019). Mitigation of arsenic toxicity in wheat by the exogenously applied salicylic acid, 24-Epi-brassinolide and silicon. J. Soil Sci. Plant Nutr. 20, 577–588. doi: 10.1007/S42729-019-00147-3
Mallet, J. (2005). Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. doi: 10.1016/j.tree.2005.02.010
Mandadi, K. K., Misra, A., Ren, S., and McKnight, T. D. (2009). BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 150, 1930–1939. doi: 10.1104/PP.109.139220
Maroco, J. P., Edwards, G. E., and Ku, M. S. B. (1999). Photosynthetic acclimation of maize to growth under elevated levels of carbon dioxide. Planta 210, 115–125. doi: 10.1007/S004250050660
Martins, A. P. B., Brito, M. S., Mayer, J. L. S., Llerena, J. P. P., Oliveira, J. F., Takahashi, N. G., et al. (2018). Ectopic expression of sugarcane SHINE changes cell wall and improves biomass in rice. Biomass Bioenergy 119, 322–334. doi: 10.1016/j.biombioe.2018.09.036
Masuda, H. P., Cabral, L. M., de Veylder, L., Tanurdzic, M., Engler, J. A., Geelen, D., et al. (2008). ABAP1 is a novel plant armadillo BTB protein involved in DNA replication and transcription. EMBO J. 27, 2746–2756. doi: 10.1038/EMBOJ.2008.191
Mauriat, M., and Moritz, T. (2009). Analyses of GA20ox- and GID1-over-expressing aspen suggest that gibberellins play two distinct roles in wood formation. Plant J. 58, 989–1003. doi: 10.1111/J.1365-313X.2009.03836.X
McCann, M., and Carpita, N. (2007). Looking for invisible phenotypes in cell wall mutants of Arabidopsis thaliana. Plant Biosyst. 139, 80–83. doi: 10.1080/11263500500059801
McCarthy, R. L., Zhong, R., and Ye, Z.-H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/PCP/PCP139
McClung, C. R. (2014). Wheels within wheels: new transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 6:2. doi: 10.12703/P6-2
Meyer, J. H., and Keeping, M. G. (2005). Impact of silicon in alleviating biotic stress in sugarcane in South Africa. Proc. S. Afr. Sugar Technol. Assoc. 23, 14–18.
Miller, M., Zhang, C., and Chen, Z. J. (2012). Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 Genes Genomes Genet. 2, 505–513. doi: 10.1534/G3.112.002162
Ming, R., Liu, S. C., Moore, P. H., Irvine, J. E., and Paterson, A. H. (2001). QTL analysis in a complex autopolyploid: genetic control of sugar content in sugarcane. Genome Res. 11, 2075–2084. doi: 10.1101/gr.198801
Mizoguchi, T., Wheatley, K., Hanzawa, Y., Wright, L., Mizoguchi, M., Song, H.-R., et al. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2, 629–641. doi: 10.1016/s1534-5807(02)00170-3
Mullet, J., Morishige, D., McCormick, R., Truong, S., Hilley, J., McKinley, B., et al. (2014). Energy sorghum—a genetic model for the design of C4 grass bioenergy crops. J. Exp. Bot. 65, 3479–3489. doi: 10.1093/JXB/ERU229
Mullet, J. E. (2017). High-biomass C4 grasses—filling the yield gap. Plant Sci. 261, 10–17. doi: 10.1016/j.plantsci.2017.05.003
Murphy, R. L., Klein, R. R., Morishige, D. T., Brady, J. A., Rooney, W. L., Miller, F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U.S.A. 108, 16469–16474. doi: 10.1073/PNAS.1106212108
Murphy, R. L., Morishige, D. T., Brady, J. A., Rooney, W. L., Yang, S., Klein, P. E., et al. (2014). Ghd7 (Ma6) represses sorghum flowering in long days: Ghd7 alleles enhance biomass accumulation and grain production. Plant Genome 7:plantgenome2013.11.0040. doi: 10.3835/PLANTGENOME2013.11.0040
Müssig, C. (2005). Brassinosteroid-promoted growth. Plant Biol. 7, 110–117. doi: 10.1055/S-2005-837493
Noor, M. A. (2017). Nitrogen management and regulation for optimum NUE in maize – a mini review. Cogent Food Agric. 3:1348214. doi: 10.1080/23311932.2017.1348214
Ohashi-Ito, K., Oda, Y., and Fukuda, H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. doi: 10.1105/TPC.110.075036
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/TPC.010127
Olson, S. N., Ritter, K., Rooney, W., Kemanian, A., McCarl, B. A., Zhang, Y., et al. (2012). High biomass yield energy sorghum: developing a genetic model for C4 grass bioenergy crops. Biofuels Bioprod. Biorefin. 6, 640–655. doi: 10.1002/BBB.1357
Park, J.-J., Yoo, C. G., Flanagan, A., Pu, Y., Debnath, S., Ge, Y., et al. (2017). Defined tetra-allelic gene disruption of the 4-coumarate:coenzyme A ligase 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol. Biofuels 10:284. doi: 10.1186/S13068-017-0972-0
Pauly, M., and Keegstra, K. (2008). Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 54, 559–568. doi: 10.1111/J.1365-313X.2008.03463.X
Peña, P. A., Quach, T., Sato, S., Ge, Z., Nersesian, N., Changa, T., et al. (2017). Expression of the maize dof1 transcription factor in wheat and sorghum. Front. Plant Sci. 8:434. doi: 10.3389/fpls.2017.00434
Petti, C., Hirano, K., Stork, J., and DeBolt, S. (2015). Mapping of a cellulose-deficient mutant named dwarf1-1 in Sorghum bicolor to the green revolution gene gibberellin20-oxidase reveals a positive regulatory association between gibberellin and cellulose biosynthesis. Plant Physiol. 169, 705–716. doi: 10.1104/pp.15.00928
Ping, L., Luo, Y., Wu, L., Qian, W., Song, J., and Christie, P. (2006). Phenanthrene adsorption by soils treated with humic substances under different pH and temperature conditions. Environ. Geochem. Health 28, 189–195. doi: 10.1007/s10653-005-9030-0
Poorter, H., and Villar, R. (1997). The Fate of Acquired Carbon in Plants: Chemical Composition and Construction Costs. Available online at: https://agris.fao.org/agris-search/search.do?recordID=US1997062597 (accessed September 6, 2021).
Pruneda-Paz, J. L., Breton, G., Para, A., and Kay, S. A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. doi: 10.1126/SCIENCE.1167206
Qandeel, M., Jabbar, A., Haider, F. U., Virk, A. L., and Ain, N. U. (2020). Effects of plant growth regulators and dates planting on spring maize production under agro-climatic conditions of Faisalabad, Pakistan. Pak. J. Agric. Agric. Eng. Vet. Sci. 36, 120–128. doi: 10.47432/2020.36.2.5
Ranocha, P., Chabannes, M., Chamayou, S., Danoun, S., Jauneau, A., Boudet, A.-M., et al. (2002). Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 129, 145–155. doi: 10.1104/PP.010988
Ravi, M., and Chan, S. W. L. (2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464, 615–618. doi: 10.1038/nature08842
Rehman, A., Shahzad, B., Haider, F. U., Ibraheem Ahmed, H. A., Lee, D.-J., Im, S. Y., et al. (2022a). “Chapter 1 - An introduction to brassinosteroids: history, biosynthesis, and chemical diversity,” in Brassinosteroids in Plant Developmental Biology and Stress Tolerance, eds J. Q. Yu, G. J. Ahammed, and P. Krishna (Amsterdam: Elsevier), 1–14. doi: 10.1016/B978-0-12-813227-2.00006-0
Rehman, A., Shahzad, B., Haider, F. U., Moeen-ud-din, M., Ullah, A., and Khan, I. (2022b). “Chapter 8 - Brassinosteroids in plant response to high temperature stress,” in Brassinosteroids in Plant Developmental Biology and Stress Tolerance, ed. G. J. A. P. K. Jing Quan Yu (Amsterdam: Elsevier), 173–187. doi: 10.1016/B978-0-12-813227-2.00014-X
Reich, P. B., Hobbie, S. E., Lee, T. D., and Pastore, M. A. (2018). Response to comment on “Unexpected reversal of C3 versus C4 grass response to elevated CO2 during a 20-year field experiment”. Science 361, 317–320. doi: 10.1126/science.aau8982
Riddle, N. C., Kato, A., and Birchler, J. A. (2006). Genetic variation for the response to ploidy change in Zea mays L. Theor. Appl. Genet. 114, 101–111. doi: 10.1007/s00122-006-0414-z
Rojas, C. A., Eloy, N. B., De Freitas Lima, M., Rodrigues, R. L., Franco, L. O., Himanen, K., et al. (2009). Overexpression of the Arabidopsis anaphase promoting complex subunit CDC27a increases growth rate and organ size. Plant Mol. Biol. 71, 307–318. doi: 10.1007/S11103-009-9525-7
Rostami, S., Azhdarpoor, A., Ali, M., Dehghani, M., Reza, M., Jaskulak, M., et al. (2021). The effects of exogenous application of melatonin on the degradation of polycyclic aromatic hydrocarbons in the rhizosphere of Festuca *. Environ. Pollut. 274:116559. doi: 10.1016/j.envpol.2021.116559
Sakamoto, T., Miura, K., Itoh, H., Tatsumi, T., Ueguchi-Tanaka, M., Ishiyama, K., et al. (2004). An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134, 1642–1653. doi: 10.1104/PP.103.033696
Sánchez-Rodríguez, C., Rubio-Somoza, I., Sibout, R., and Persson, S. (2010). Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 15, 291–301. doi: 10.1016/j.tplants.2010.03.002
Sattler, S. E., Saballos, A., Xin, Z., Funnell-Harris, D. L., Vermerris, W., and Pedersen, J. F. (2014). Characterization of novel sorghum brown midrib mutants from an EMS-mutagenized population. G3 Genes Genomes Genet. 4, 2115–2124. doi: 10.1534/G3.114.014001
Scheben, A., and Edwards, D. (2018). Towards a more predictable plant breeding pipeline with CRISPR/Cas-induced allelic series to optimize quantitative and qualitative traits. Curr. Opin. Plant Biol. 45, 218–225. doi: 10.1016/J.PBI.2018.04.013
Scofield, S., Jones, A., and Murray, J. A. H. (2014). The plant cell cycle in context. J. Exp. Bot. 65, 2557–2562. doi: 10.1093/JXB/ERU188
Scully, E. D., Gries, T., Sarath, G., Palmer, N. A., Baird, L., Serapiglia, M. J., et al. (2016). Overexpression of SbMyb60 impacts phenylpropanoid biosynthesis and alters secondary cell wall composition in Sorghum bicolor. Plant J. 85, 378–395. doi: 10.1111/tpj.13112
Sharma, A., Sidhu, G. P. S., Araniti, F., Bali, A. S., Shahzad, B., Tripathi, D. K., et al. (2020). The role of salicylic acid in plants exposed to heavy metals. Molecules 25:540. doi: 10.3390/molecules25030540
Shen, H., He, X., Poovaiah, C. R., Wuddineh, W. A., Ma, J., Mann, D. G. J., et al. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193, 121–136. doi: 10.1111/J.1469-8137.2011.03922.X
Singh, K. B., Foley, R. C., and Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/S1369-5266(02)00289-3
Somerville, C., Youngs, H., Taylor, C., Davis, S. C., and Long, S. P. (2010). Feedstocks for lignocellulosic biofuels. Science 329, 790–792. doi: 10.1126/SCIENCE.1189268
Sonbol, F.-M., Fornalé, S., Capellades, M., Encina, A., Touriño, S., Torres, J.-L., et al. (2009). The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 70, 283–296. doi: 10.1007/S11103-009-9473-2
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Mas, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/SCIENCE.289.5480.768
Subramaniam, Y., Masron, T. A., and Azman, N. H. N. (2020). Biofuels, environmental sustainability, and food security: a review of 51 countries. Energy Res. Soc. Sci. 68:101549. doi: 10.1016/j.erss.2020.101549
Sun, H., Xu, H., Li, B., Shang, Y., Wei, M., Zhang, S., et al. (2021). The brassinosteroid biosynthesis gene, ZmD11, increases seed size and quality in rice and maize. Plant Physiol. Biochem. 160, 281–293. doi: 10.1016/J.PLAPHY.2021.01.031
Su’udi, M., Cha, J. Y., Jung, M. H., Ermawati, N., Han, C. D., Kim, M. G., et al. (2012). Potential role of the rice OsCCS52A gene in endoreduplication. Planta 235, 387–397. doi: 10.1007/S00425-011-1515-8
Tanveer, M., Shahzad, B., Sharma, A., Biju, S., and Bhardwaj, R. (2018). 24-Epibrassinolide; an active brassinolide and its role in salt stress tolerance in plants: a review. Plant Physiol. Biochem. 130, 69–79. doi: 10.1016/j.plaphy.2018.06.035
Tanveer, M., Shahzad, B., Sharma, A., and Khan, E. A. (2019). 24-Epibrassinolide application in plants: an implication for improving drought stress tolerance in plants. Plant Physiol. Biochem. 135, 295–303. doi: 10.1016/J.PLAPHY.2018.12.013
Tetreault, H. M., O’Neill, P., Toy, J., Gries, T., Funnell-Harris, D. L., and Sattler, S. E. (2020). Field evaluation of sorghum (Sorghum bicolor) lines that overexpress two monolignol-related genes that alter cell wall composition. Bioenergy Res. 14, 1–12. doi: 10.1007/s12155-020-10218-4
Torres, A. F., Visser, R. G. F., and Trindade, L. M. (2015). Bioethanol from maize cell walls: genes, molecular tools, and breeding prospects. GCB Bioenergy 7, 591–607. doi: 10.1111/gcbb.12164
Tran, T. A., and Popova, L. P. (2013). Functions and toxicity of cadmium in plants: recent advances and future prospects. Turk. J. Bot. 37, 1–13.
Truong, S. K., McCormick, R. F., and Mullet, J. E. (2017). Bioenergy sorghum crop model predicts VPD-limited transpiration traits enhance biomass yield in water-limited environments. Front. Plant Sci. 8:335. doi: 10.3389/FPLS.2017.00335
Ullah, A., Rolf Richter, P., Ahmed, H., Pratap Singh, V., Mohan Prasad, S., Kumar Chauhan, D., et al. (2016). Silicon nanoparticles more efficiently alleviate arsenate toxicity than silicon in maize cultiver and hybrid differing in arsenate tolerance. Front. Environ. Sci. 4:46. doi: 10.3389/fenvs.2016.00046
van der Weijde, T., Alvim Kamei, C. L., Torres, A. F., Vermerris, W., Dolstra, O., Visser, R. G. F., et al. (2013). The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 4:107. doi: 10.3389/fpls.2013.00107
van der Weijde, T., Kamei, C. L. A., Severing, E. I., Torres, A. F., Gomez, L. D., Dolstra, O., et al. (2017a). Genetic complexity of miscanthus cell wall composition and biomass quality for biofuels. BMC Genomics 18:406. doi: 10.1186/s12864-017-3802-7
van der Weijde, T., Kiesel, A., Iqbal, Y., Muylle, H., Dolstra, O., Visser, R. G. F., et al. (2017b). Evaluation of Miscanthus sinensis biomass quality as feedstock for conversion into different bioenergy products. GCB Bioenergy 9, 176–190. doi: 10.1111/GCBB.12355
Vanneste, S., and Friml, J. (2009). Auxin: a trigger for change in plant development. Cell 136, 1005–1016. doi: 10.1016/J.CELL.2009.03.001
Vélez-Bermúdez, I.-C., Salazar-Henao, J. E., Fornalé, S., López-Vidriero, I., Franco-Zorrilla, J.-M., Grotewold, E., et al. (2015). A MYB/ZML complex regulates wound-induced lignin genes in maize. Plant Cell 27, 3245–3259. doi: 10.1105/TPC.15.00545
Vernoux, T., Autran, D., and Traas, J. (2000). Developmental control of cell division patterns in the shoot apex. Plant Mol. Biol. 43, 569–581. doi: 10.1023/A:1006464430936
Voorend, W., Nelissen, H., Vanholme, R., De Vliegher, A., Van Breusegem, F., Boerjan, W., et al. (2016). Overexpression of GA20-OXIDASE1 impacts plant height, biomass allocation and saccharification efficiency in maize. Plant Biotechnol. J. 14, 997–1007. doi: 10.1111/pbi.12458
Wai, C. M., Zhang, J., Jones, T. C., Nagai, C., and Ming, R. (2017). Cell wall metabolism and hexose allocation contribute to biomass accumulation in high yielding extreme segregants of a Saccharum interspecific F2 population. BMC Genomics 18:773. doi: 10.1186/s12864-017-4158-8
Wang, Y., Sun, J., Ali, S. S., Gao, L., Ni, X., Li, X., et al. (2020). Identification and expression analysis of Sorghum bicolor gibberellin oxidase genes with varied gibberellin levels involved in regulation of stem biomass. Ind. Crops Prod. 145:111951. doi: 10.1016/j.indcrop.2019.111951
Watling, J. R., Press, M. C., and Quick, W. P. (2000). Elevated CO2 induces biochemical and ultrastructural changes in leaves of the C4 cereal sorghum. Plant Physiol. 123, 1143–1152. doi: 10.1104/PP.123.3.1143
Wen-Jie, W., Ling, Q., Yuan-Gang, Z., Dong-Xue, S., Jing, A., Hong-Yan, W., et al. (2011). Changes in soil organic carbon, nitrogen, pH and bulk density with the development of larch (Larix gmelinii) plantations in China. Glob. Chang. Biol. 17, 2657–2676. doi: 10.1111/j.1365-2486.2011.02447.x
Whipple, C. J., Kebrom, T. H., Weber, A. L., Yang, F., Hall, D., Meeley, R., et al. (2011). grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. U.S.A. 108, E506–E512. doi: 10.1073/PNAS.1102819108
Wingler, A., Delatte, T. L., O’Hara, L. E., Primavesi, L. F., Jhurreea, D., Paul, M. J., et al. (2012). Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol. 158, 1241–1251. doi: 10.1104/PP.111.191908
Wuddineh, W. A., Mazarei, M., Turner, G. B., Sykes, R. W., Decker, S. R., Davis, M. F., et al. (2015). Identification and molecular characterization of the switchgrass AP2/ERF transcription factor superfamily, and overexpression of PvERF001 for improvement of biomass characteristics for biofuel. Front. Bioeng. Biotechnol. 3:101. doi: 10.3389/FBIOE.2015.00101
Xia, J., Zhao, Y., Burks, P., Pauly, M., and Brown, P. J. (2018). A sorghum NAC gene is associated with variation in biomass properties and yield potential. Plant Direct 2:e00070. doi: 10.1002/pld3.70
Xiang, L., Yuling, L., Zhiying, M., and Zheng, L. (2020). Identification of maize aquaporin gene ZmPIP4c as a signature of C4 traits. Mol. Plant Breed. 11, 2819–2825. doi: 10.5376/mgg.2020.11.0002
Xu, C., Wang, Y., Yu, Y., Duan, J., Liao, Z., Xiong, G., et al. (2012). Degradation of MONOCULM 1 by APC/CTAD1 regulates rice tillering. Nat. Commun. 3:750. doi: 10.1038/NCOMMS1743
Yong, W., Link, B., O’Malley, R., Tewari, J., Hunter, C. T., Lu, C.-A., et al. (2005). Genomics of plant cell wall biogenesis. Planta 221, 747–751. doi: 10.1007/S00425-005-1563-Z
Yu, Y., Steinmetz, A., Meyer, D., Brown, S., and Shen, W.-H. (2003). The tobacco A-type cyclin, Nicta;CYCA3;2, at the nexus of cell division and differentiation. Plant Cell 15, 2763–2777. doi: 10.1105/TPC.015990
Zargar, S. M., Mahajan, R., Bhat, J. A., Nazir, M., and Deshmukh, R. (2019). Role of silicon in plant stress tolerance: opportunities to achieve a sustainable cropping system. 3 Biotech 9:73. doi: 10.1007/S13205-019-1613-Z
Zeng, X., Sheng, J., Zhu, F., Zhao, L., Hu, X., Zheng, X., et al. (2020). Differential expression patterns reveal the roles of cellulose synthase genes (CesAs) in primary and secondary cell wall biosynthesis in Miscanthus × giganteus. Ind. Crops Prod. 145:112129. doi: 10.1016/J.INDCROP.2020.112129
Zhang, G., Ge, C., Xu, P., Wang, S., Cheng, S., Han, Y., et al. (2021). The reference genome of Miscanthus floridulus illuminates the evolution of Saccharinae. Nat. Plants 7, 608–618. doi: 10.1038/s41477-021-00908-y
Zhang, Y., Paschold, A., Marcon, C., Liu, S., Tai, H., Nestler, J., et al. (2014). The Aux/IAA gene rum1 involved in seminal and lateral root formation controls vascular patterning in maize (Zea mays L.) primary roots. J. Exp. Bot. 65, 4919–4930. doi: 10.1093/jxb/eru249
Zhang, Y., von Behrens, I., Zimmermann, R., Ludwig, Y., Hey, S., and Hochholdinger, F. (2015). LATERAL ROOT PRIMORDIA 1 of maize acts as a transcriptional activator in auxin signalling downstream of the Aux/IAA gene rootless with undetectable meristem 1. J. Exp. Bot. 66, 3855–3863. doi: 10.1093/JXB/ERV187
Zhong, R., Lee, C., McCarthy, R. L., Reeves, C. K., Jones, E. G., and Ye, Z.-H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. doi: 10.1093/PCP/PCR123
Keywords: biomass accumulation, C4 crops, hormone dynamics, cell wall growth, circadian rhythms, fossil fuels
Citation: Ain N-u, Haider FU, Fatima M, Habiba, Zhou Y and Ming R (2022) Genetic Determinants of Biomass in C4 Crops: Molecular and Agronomic Approaches to Increase Biomass for Biofuels. Front. Plant Sci. 13:839588. doi: 10.3389/fpls.2022.839588
Received: 20 December 2021; Accepted: 17 January 2022;
Published: 23 June 2022.
Edited by:
Xingtan Zhang, Agricultural Genomics Institute at Shenzhen (CAAS), ChinaReviewed by:
Sajid Fiaz, The University of Haripur, PakistanXiaomin Feng, Guangdong Academy of Sciences, China
Copyright © 2022 Ain, Haider, Fatima, Habiba, Zhou and Ming. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ray Ming, cmF5bWluZ0BpbGxpbm9pcy5lZHU=
 Noor-ul- Ain1
Noor-ul- Ain1 Fasih Ullah Haider
Fasih Ullah Haider Habiba
Habiba Yongmei Zhou
Yongmei Zhou Ray Ming
Ray Ming