
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 23 March 2022
Sec. Plant Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.836267
This article is part of the Research Topic Plant RNA Structure View all 7 articles
 Junfei Ma
Junfei Ma Ying Wang*
Ying Wang*RNAs play essential roles in various biological processes. Mounting evidence has demonstrated that RNA subcellular localization and intercellular/systemic trafficking govern their functions in coordinating plant growth at the organismal level. While numerous types of RNAs (i.e., mRNAs, small RNAs, rRNAs, tRNAs, and long noncoding RNAs) have been found to traffic in a non-cell-autonomous fashion within plants, the underlying regulatory mechanism remains unclear. Viroids are single-stranded circular noncoding RNAs, which entirely rely on their RNA motifs to exploit cellular machinery for organelle entry and exit, cell-to-cell movement through plasmodesmata, and systemic trafficking. Viroids represent an excellent model to dissect the role of RNA three-dimensional (3D) structural motifs in regulating RNA movement. Nearly two decades of studies have found multiple RNA 3D motifs responsible for viroid nuclear import as well as trafficking across diverse cellular boundaries in plants. These RNA 3D motifs function as “keys” to unlock cellular and subcellular barriers and guide RNA movement within a cell or between cells. Here, we summarize the key findings along this line of research with implications for future studies on RNA trafficking in plants.
Multicellular organisms evolve diverse mechanisms to integrate individual cells during development and in response to environmental cues. Cellular boundaries function in this integration through balancing cell autonomy and communication among cells (Ding and Wang, 2009). In plants, neighboring cells are connected via plasmodesmata (PD), which are micro-channels crossing cell walls. The vascular system, including xylem and phloem, mediates the systemic transportation of molecules. The xylem system is mainly responsible for the transportation of water and minerals, while the phloem system transports photosynthates and macromolecules. Various proteins, RNAs, as well as viruses and viroids can be found in the translocation stream of phloem (Ding et al., 2003; Liu and Chen, 2018).
Most cellular RNAs are transcribed in the nucleus and then are either retained in the nucleus or transported to the cytoplasm for function. RNAs in the cytoplasm can participate in diverse biological activities and processes or are transported to the nucleus performing various functions, or even traffic to neighboring cells to act as non-cell-autonomous regulators (Ye et al., 2012; Axtell, 2013; Chantarachot and Bailey-Serres, 2018; Reagan et al., 2018; Long et al., 2021). Non-cell autonomous RNAs widely exist in plants, and there are many types of those trafficking RNAs, including various small RNAs, mRNAs, tRNAs, rRNAs, and infectious RNAs from viruses and viroids (Ding et al., 2003; Liu and Chen, 2018). Some mobile RNAs will move short distance across several cells, while others move through various tissues to traffic systemically in plants (Kim, 2005; Qin et al., 2012; Spiegelman et al., 2013; Thieme et al., 2015; Yang et al., 2015; Ham and Lucas, 2017; Reagan et al., 2018).
Non-cell autonomous RNAs serve as critical signals to regulate plant development and responses to biotic and abiotic challenges (Ding et al., 2003; Wang and Ding, 2010; Liu and Chen, 2018). Using grafting experiments, some mRNAs are found to traffic long distance across the grafting junctions in regulating plant development, such as tuber formation in potato (Banerjee et al., 2006) and leaf morphogenesis in tomato (Kim et al., 2001; Haywood et al., 2005). Related to plant physiology, a microRNA (miR399) has been found to move from shoot to root contributing to the maintenance of phosphate homeostasis in Arabidopsis (Lin et al., 2008; Pant et al., 2008). Numerous small RNAs, including miRNAs and short interfering RNAs (siRNAs), serve as long distance epigenetic signals coordinating gene expression and antiviral defense (Ding and Voinnet, 2007; Kalantidis et al., 2008; Chen and Rechavi, 2021).
A key question remains regarding how RNA is selected for trafficking (including intracellular, intercellular, and systemic trafficking). A recent report showed that m5C methylation is highly enriched in mobile mRNAs. Loss-of-methylation inhibits the non-cell-autonomous behavior of some mobile mRNAs (Yang et al., 2019). This finding provides mechanistic insights into the selection specificity of mobile transcripts. However, it is unclear whether m5C methylation ensures the accurate transportation of RNAs to their proper destiny within plants. RNAs by themselves also contain signals in regulating long distance RNA trafficking. For example, a cis element cloned from the 5′ untranslated region (UTR) of a potexviral RNA mediates cell-to-cell movement of the fused GFP reporter RNA (Lough et al., 2006). In addition, the UTRs of potato BEL5 mRNA possess the regulatory elements for long distance trafficking (Banerjee et al., 2006). The detailed molecular basis underlying these functional structures remains to be determined.
Viroids, single-stranded circular noncoding RNAs, harness cellular machinery to target specific organelles for replication, invade neighboring cells through PD, and spread systemically via phloem (Ding, 2009). Within phloem, viroids likely form an RNA-protein complex with phloem pectin PP2 for long distance translocation (Gómez and Pallás, 2001; Owens et al., 2001; Gomez and Pallas, 2004). During systemic infection, viroid RNAs move across various cellular boundaries (Ding and Wang, 2009; Wang and Ding, 2010). In a simplified view, viroids will traffic from epidermis, through palisade and spongy mesophyll and bundle sheath, to enter phloem. They also traffic in a reverse direction in systemic leaves (Takeda and Ding, 2009). Viroids accumulate to high levels in plant cells, and it is easy to engineer various mutants for functional analyses (Wang and Ding, 2010). There is no endogenous background signal interfering with analyses on viroid RNAs. Therefore, viroid infection provides a valuable experimental system to dissect the factors and regulatory mechanisms underlying RNA movement in plants.
Given that viroids do not encode any proteins, their RNA genomes must contain explicit information to guide cellular machinery for accurate localization and trafficking. Using potato spindle tuber viroid (PSTVd) as a model, specific RNA three-dimensional (3D) motifs responsible for crossing multiple cellular boundaries have been identified, providing solid genetic evidence that those RNA motifs guide specific trafficking in plants (Ding, 2009; Wang et al., 2018, 2021). It is noteworthy that the secondary structure of viroid RNAs is among the best-known structures thanks to the extensive chemical mapping analyses (Gast et al., 1996; Xu et al., 2012; Giguere et al., 2014; Giguere and Perreault, 2017; Lopez-Carrasco and Flores, 2017), which paves the way to further pinpoint the functional structures of local RNA 3D motifs.
RNA molecules form various helices and loops in their secondary structures. Helices are composed of contiguous Watson-Crick (WC) base pairs [i.e., adenine (A)–uridine (U), guanine (G)–cytosine (C), and GU base pairs]. In contrast, loop regions are composed of diverse non-WC base pairs that are highly arranged (Wang et al., 2018). Many loop motifs can be found at nonhomologous positions of diverse RNA species but largely keep the base pair geometries and interaction details, so they are also termed recurring loop motifs (Khisamutdinov et al., 2021). RNA 3D loop motifs provide recognition sites for specific RNA-protein, RNA–RNA, and RNA-ligand interactions. This is feasible thanks to the non-WC-WC base pairs that widen the major groove of RNAs and expose distinct WC edges of four bases (Wang et al., 2018).
Each RNA base has three edges (i.e., the WC, Hoogsteen, and Sugar edges) that can participate in interaction with other base edges to form non-WC-WC base pairs in loop motifs (Figure 1). Adding to the complexity are the two possible orientations (cis as the same orientation and trans as the opposite orientation) of glycosidic bonds for each base pair. Therefore, there are 12 base-pairing geometries considering the combinations of interacting edges and glycosidic bond orientations (Stombaugh et al., 2009; Wang et al., 2018). Two base pairs are isosteric to each other if they are formed via the same base edge interactions, share the same glycosidic bond orientations, and possess highly similar distances between C1’ carbon atoms (Stombaugh et al., 2009; Wang et al., 2018). Isosteric base pairs have the potential to substitute for each other without disrupting functions. These geometries and isostericity in various loop motifs have been empirically observed in detail through atomic-resolution crystallography and NMR spectroscopy studies (Stombaugh et al., 2009; Wang et al., 2018). Efforts have been made to summarize those observed geometries, which are deposited in the RNA Basepair Catalog (Stombaugh et al., 2009). In addition, some prediction programs have been developed to facilitate the annotation of RNA loop motifs in terms of base geometries and interactions therein (Sarver et al., 2008; Zirbel et al., 2015). Therefore, it is possible to deduce the base interaction geometry within RNA loops and design structure-disruptive or structure-maintaining mutants for functional validations of predicted structural models of RNA loop motifs, providing genetic evidence in supporting some analyzed RNA 3D motifs.
Viroids of the family Pospiviroidae all enter the nucleus for replication. How RNA nuclear import is regulated remains as an interesting question. Most RNAs are made in the nucleus, and the prevailing view is that those RNAs either traffic to the cytoplasm or remain in the nucleus for function. In recent years, more and more RNAs, besides viroids, have been found to enter the nucleus (Rudt and Pieler, 1996; Chou et al., 1998; Gao et al., 2012; Ye et al., 2012; Kramer and Hopper, 2013; Chaturvedi et al., 2014; Long et al., 2021). Using PSTVd as a model, we found that viroids exploit cellular importin alpha-4 (IMPa-4)-based pathway and a viroid-binding protein (Virp1) to achieve nuclear import (Ma et al., 2021).
Previous studies have mapped a region in viroid RNA genomes responsible for Virp1 binding, termed RY motif (Gozmanova et al., 2003; Maniataki et al., 2003). However, the molecular basis of the RY motif for Virp1 recognition remains elusive. Recently, we carefully re-examine the RY motif in PSTVd and uncovered a C-loop structure within the region. C-loop is an asymmetric loop that has been found in many rRNAs, mammalian noncoding RNAs and one bacterial mRNA (Torres-Larios et al., 2002; Lescoute et al., 2005; Iacoangeli and Tiedge, 2013; Drsata et al., 2017). C-loop has the following features: (A) “C” is often the first base in the longer strand; (B) two bases in the longer strand form non-WC base pairs with bases in the opposite strand (cis-WC-sugar and trans-WC-Hoogsteen base-pairings); (C) two triads are formed through base-pairings from two strands; and (D) this motif is often found in hairpin stem-loop structure (Lescoute et al., 2005; Drsata et al., 2017). PSTVd C-loop is composed of C189-A173 cis-WC-sugar base-pairing and A171-U187 trans-WC-Hoogsteen base-pairing (Figure 2), which is supported by chemical mapping data and functional mutagenesis analyses (Ma et al., 2021). Our data showed that C-loop is pivotal for Virp1-binding, viroid nuclear accumulation, and infectivity. Interestingly, C-loop can be found in nearly all, except one, formal members of the family Pospiviroidae as well as a viral satellite RNA that relies on Virp1 for nuclear import (Ma et al., 2021). Therefore, C-loop is probably a widely used RNA motif for the nuclear import of subviral RNAs. Given that the nuclear import of RNAs is not limited to subviral agents, this finding will encourage new efforts to cast a wider net in search for more regulatory RNA motifs responsible for nuclear import.
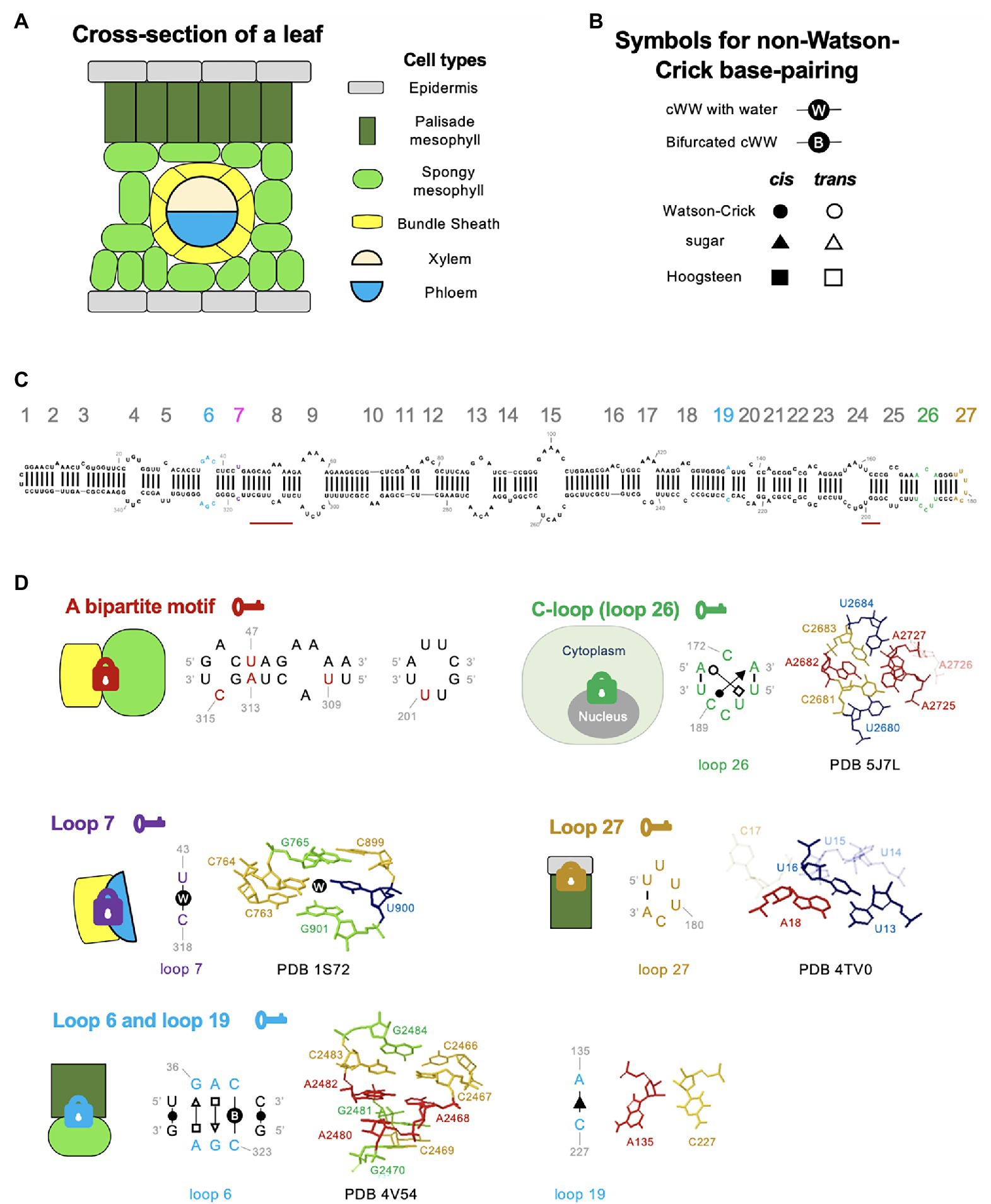
Figure 2. RNA 3D motif-mediated viroid RNA trafficking in plants. (A) A cartoon illustration of the cross-section of plant leaves. (B) Symbols for annotating RNA nucleotide edges. cWW, cis-Watson Crick-Watson Crick base-pairing. (C) The secondary structure of the PSTVdInt genome, drawing by using RNA2Drawer (Johnson et al., 2019). Loops and bulges are numbered from left to right as 1 to 27. Key structural motifs illustrated in (D) are highlighted in colors (nucleotides and loop numbers). The nucleotides of the bipartite motif (position indicated by two red bars) are not color-highlighted in the secondary structure because this motif was only found in the PSTVdNB strain, whose secondary structure has not been confirmed by chemical probing. (D) Loop structures critical for regulating trafficking across cellular boundaries. Locks with different colors indicate distinct barriers that restrain the trafficking of RNAs without necessary RNA motifs (“keys”). The annotated base-pairings were validated by chemical mapping and functional mutagenesis. The C-loop structure model is based on regions 2,680–2,684 (5′-UCACU-3′) and 2,725–2,727 (5′-AAA-3′) of bacterial 23S rRNA (PDB 5J7L; Cocozaki et al., 2016). The loop 7 structural model is based on regions 763–765 (5′-CCG-3′) and 899–901 (5′-CUG-3′) of Haloarcula marismortui 23S rRNA (PDB 1S72; Klein et al., 2004). The loop 27 model is based on the region 13–18 (5′-UUUUCA-3′) of a Drosophila histone mRNA (PDB 4TV0; Zhang et al., 2014). The loop 6 model is based on regions 2,466–2,470 (5′-CCACG-3′) and 2,480–2,484 (5′-AGACG-3′) of bacterial 23S rRNA (PDB 4V54; Borovinskaya et al., 2007). Note that C2483 and C2467 form a bifurcated cWW base-pairing. The illustration for loop 19 is extracted from the RNA Basepair Catalog. Nucleotides in transparent indicate that they are not involved in base interactions (e.g., A2726 in PDB 5J7L).
It is intuitive to reason that viroid RNAs contain the necessary information for cell-to-cell and even long distance trafficking. Early work using PSTVd sequence in chimeric RNAs supports this hypothesis (Ding et al., 1997). Analyses on PSTVdNT and PSTVdNB strains provided the first empirical evidence illustrating an RNA motif responsible for regulating trafficking across cellular boundaries (Qi et al., 2004). PSTVdNT harbors a spontaneous nucleotide substitution C259U in the tomato isolate PSTVdKF440-2 that enables infection in tobacco (Nicotiana tabacum; Wassenegger et al., 1996). PSTVdNB accumulated five more spontaneous mutations (A47U, G210U, A309U, U313A, and U315C) during vegetative propagation through cuttings from PSTVdNT-infected tobacco plants (Qi et al., 2004). PSTVdNT and PSTVdNB bear similar replication efficiency in protoplasts but display different accumulation levels in systemic leaves. In situ hybridization analyses showed that PSTVdNB, but not PSTVdNT, can exit the bundle sheath to invade more cells in systemic leaves (Qi et al., 2004).
Mutational analyses found that four out of the five spontaneous mutations in PSTVdNB are both required and sufficient to enable bundle sheath exit in tobacco. Interestingly, these substitutions are clustered in two discrete regions in the PSTVd genome forming a bipartite motif (Figure 2; Qi et al., 2004). The detailed structural basis of this bipartite motif remains elusive. It is noteworthy that this bipartite motif appears to be required for unidirectional bundle sheath exit only in tobacco but not in tomato or N. benthamiana, indicating that those RNA motifs can evolve very fast in nature and influence the host tropism of infectious RNAs.
An early attempt to screen for PSTVd loss-of-function trafficking mutants identified two nucleotides (U43 and C318) that did not affect replication but abolished systemic trafficking (Zhong et al., 2007). Interestingly, U43 and C318 form a one base-pairing loop, loop 7 (Figure 2). Using in situ hybridization analyses found that loop “close” mutants (U43G or C318A) were present in epidermis, mesophyll, and bundle sheath cells but could not be loaded into phloem in inoculated leaves. It is unclear whether this regulation is unidirectional or bidirectional because transgenic expression of loop 7 mutants in companion cells all converted to wild-type sequences, rendering it difficult to assess the regulation of trafficking direction.
Using the FR3D program to search for similar loop structures obtained by highest-resolution X-ray crystallography in the Protein Data Bank (PDB) revealed that U43 and C318 form a cis-WC–WC base pair with a water molecule insertion (Figure 2; Zhong et al., 2007). All the mutants predicted to retain the structure for water insertion were able to traffic systemically in plants, whereas the mutants predicted to form canonical WC–WC base-pairing without water insertion failed to traffic out of inoculated leaves. The water molecule insertion widens the minor groove and increases the angle subtended by the glycosidic bonds, presumably favoring protein binding (Zhong et al., 2007). Indeed, similar structures in rRNAs and siRNA duplexes are involved in protein binding (Vargason et al., 2003; Klein et al., 2004).
The discovery of loop 7 and the bipartite motif inspired the idea that there exist elegant regulations at each cellular boundary regulating the exchange of cellular contents, including RNAs. Accordingly, multiple RNA motifs will work in concert to coordinate trafficking across those cellular boundaries. Therefore, genome-wide functional analyses of PSTVd loop motifs were performed to assess the role of each loop motif in regulating replication or systemic trafficking (Zhong et al., 2008). By replacing all possible noncanonical base-pairings in loop motifs with WC–WC base-pairings, a series of mutants were generated to “close” every loop motif except loop 15 and loop 7 that have been previously analyzed. These loop mutants were tested for replication ability in protoplasts and systemic trafficking in N. benthamiana plants. A total of 11 loop mutants were found to impair systemic trafficking (Zhong et al., 2008). Some of the loops, such as loop 6, loop 19, and loop 27, were subsequently found to regulate the trafficking across distinct cellular boundaries (see below for details). It is noteworthy that this analysis may overlook some more complex structures, such as the aforementioned bipartite motif. In addition, loop 26 mutant did not show trafficking because it regulates nuclear import as aforementioned (Ma et al., 2021). Nevertheless, this approach provides an overview of the genomic organization of viroids in controlling trafficking in N. benthamiana. Expanding this approach to other viroids, viruses, and diverse host-viroid combinations may achieve a much deeper understanding of structural motif-regulated RNA trafficking in plants.
Upon inoculation, viroid inoculum will initiate replication and then move through mesophyll layers to enter phloem. A recent study showed that the right terminal loop (loop 27) of PSTVd is critical for RNA moving from epidermis to palisade mesophyll (Wu et al., 2019). Mutagenesis analyses showed that most mutants disrupting this loop led to failure in replication, except for the U178G/U179G mutation. The U178G/U179G mutant could spread within epidermis of inoculated leaves but was restrained from entering the adjacent palisade mesophyll layer as observed via the in situ hybridization assay. Interestingly, needle punch delivery of this mutant into stems allowed mutant RNA to move across all cellular boundaries including from palisade mesophyll to epidermis in systemic leaves, indicating that this loop regulates unidirectional trafficking (Wu et al., 2019).
Using the JAR3D program (Zirbel et al., 2015), the terminal loop is predicted to be a UNCG-like motif.1 The exact homolog loop can be found in the 3′ UTR of a Drosophila histone mRNA, where the loop is involved in protein binding (Zanier et al., 2002). Within the loop region (nucleotides U177 to A182), U179 and C181 bulge outside of the motif. U180 stacks on the WC–WC pair (U177-A182) that closes the motif (Figure 2). It is believed that this loop does not contain stable noncanonical base-pairings when the protein partner is absent (Wu et al., 2019).
When a similar in situ hybridization analysis was performed using loop 6 mutants to determine the role of loop 6 in regulating viroid movements, those mutants, if replicable, were trapped in palisade mesophyll cells without entering spongy mesophyll (Takeda et al., 2011). A more recent work on loop 19 mutants also revealed a similar pattern that loop 19 mutants accumulated in palisade mesophyll but could not enter spongy mesophyll (Jiang et al., 2017). It represents the first example where two RNA motifs regulate trafficking across the same cellular boundary. However, it cannot be ruled out that these two motifs form a larger bipartite organization to coordinate functions.
PSTVd loop 6 contains three noncanonical base pairs: C323-C38 cis-WC–WC bifurcated pair, G324-A37 trans-Sugar-Hoogsteen (tSH) pair, and A325-G36 trans-Hoogsteen-Sugar (tHS) pair (Figure 2; Takeda et al., 2011). The C323-C38 cis-WC–WC bifurcated pair is so rigid that it cannot be replaced by any other substitution. The tSH and tHS pairs can be substituted by some but not all isosteric base-pairings, implying the existence of other selection pressures (Takeda et al., 2011). Loop 6 is conserved in viroids belonging to the genus Pospiviroid. Similar loop motifs can also be found in some 16S rRNAs, 23S rRNAs, and a group I intron, where this structural motif serves as a binding site for protein partners (Takeda et al., 2011). Loop 19 is a one base-pairing loop (Takeda et al., 2018). This motif can emerge through spontaneous base substitutions in plants inoculated with loop-close mutants. Mutational analyses found that loop 19 is likely composed of a cis-Sugar-Sugar base-pairing (Figure 2; Takeda et al., 2018).
Emerging evidence supports the model that multiple structural motifs coordinate RNA subcellular localization and trafficking across different cellular boundaries within a plant. Those structural motifs act like “keys” to unlock restrictions at organellar gates as well as various cellular boundaries. A reasonable hypothesis is that those RNA 3D motifs are recognized by certain cellular proteins forming RNA-protein complexes, which will then be delivered to their destinations. In support of this model, the PSTVd C-loop serves a nuclear import signal recognized by cellular protein Virp1. The Virp1-PSTVd complex is then delivered into the nucleus via the IMPa-4 based nuclear import pathway (Ma et al., 2021). In addition, recent evidence supports that an Exportin 5 ortholog (HASTY) participates in miRNA cell-to-cell and vascular movement in plants (Brioudes et al., 2021). On the other hand, the molecular basis of those barriers at various cellular boundaries remains to be determined. It is intuitive to reason that PD may adopt different selectivity when connect various types of cells. Diverse groups of proteins contribute to cargo targeting to PD and/or PD gating, including specific β-1,3-Glucanases for callose deposition, and other PD-associated or mobile proteins (Haywood et al., 2002; Ding et al., 2003; Levy et al., 2007; Brunkard et al., 2015; Lee, 2015). Those components may have different homologs or activities in distinct tissues, which can explain the need of multiple RNA motifs for crossing various cellular boundaries.
The complexity in organizing the required RNA motifs for RNA trafficking is intriguing. Some RNA motifs act in a species-specific manner while others work in concert to cross one specific cellular boundary, reflecting the sophisticated design in maintaining the autonomy of various tissues in different plants. To gain a deeper understanding of the barriers of cellular boundaries in different plants, analyzing the requirement of PSTVd trafficking motifs in different host-viroid combinations will be a straightforward approach to provide informative insights. Given the importance of RNA 3D motifs in host-viroid interactions, they certainly play a role in constraining viroid evolution and adaptation to new hosts (Wang et al., 2018). On the other hand, emerging evidence (e.g., bipartite as well as Loops 6 and 19) supports that viroid RNAs may undergo significant changes in overall structure to carry out functions. Currently, viroid RNA structures are mostly probed using in vitro assays. It will be beneficial to gain more insights into viroid structures at distinct subcellular and cellular compartments using in vivo probing methods, particularly those that can achieve observation at the single molecular level to detect the transient structural changes (Stephenson et al., 2016; Yu and Lucks, 2019; Cawte et al., 2020; Schmidt et al., 2020).
It is desirable that the knowledge gained from the viroid model can facilitate the understanding of cellular RNA trafficking. Increasing evidence supports that regulatory RNA structures can control endogenous RNA trafficking in plants, especially those tRNA-like structures (TLS) that have been identified in many endogenous mobile transcripts (Zhang et al., 2016). However, the detailed 3D base-pairing geometries, which confers the regulatory function in RNA trafficking in plants, have not been annotated for those regulatory structures. This is likely due to the technical limitations that hinders the in-depth analyses on such functional motifs. First, recurring RNA motifs may not exert the same function in different RNAs. For example, both PSTVd and 5S rRNA contain the loop E motif (Branch et al., 1985). However, a cellular protein, TFIIIA-9ZF, only binds the loop E in 5S rRNA (Szymanski et al., 2003) but prefers a distinct region instead of the loop E in PSTVd (Wang et al., 2016). Therefore, it is difficult to predict the function of recurring RNA motifs in distinct RNAs at this stage. Second, most cellular RNAs do not have well-annotated secondary structure, except for rRNAs, tRNAs, etc. Well-annotated secondary structures are a prerequisite for analyzing local 3D motifs. With the rapid development of novel probing methods for analyzing RNA structures at the transcriptome level (Gosai et al., 2015; Tack et al., 2020; Liu et al., 2021), this limitation may soon be mitigated.
The discovery of m5C methylation as a regulatory mark for mRNA translocation crossing graft junctions is a significant advancement in understanding the trafficking of endogenous RNAs (Yang et al., 2019). However, the m5C mark is enriched mostly four nucleotides downstream of the start codon in plant mRNAs (Song et al., 2018) and promotes the efficiency of mRNA translation (Tang et al., 2020). How the two biological processes (i.e., translation and selection for trafficking) are balanced remains to be elucidated. Viroid RNAs do not possess m5C modification (Di Serio et al., 2019), demonstrating that more than one mechanism exists for selecting mobile RNAs. How these mechanisms are coordinated for accurate delivery of RNAs to destiny remains unexplored, which deserves future investigations.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Work from the author’s laboratory was supported by research grants from the US National Science Foundation (MCB-1906060 and MCB-2145967) and the US National Institutes of Health (R15GM135893). Funders had no role in the study design, in the collection, analysis, and interpretation of the data, in the writing of the report, and in the decision to submit the article for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
This review is dedicated to the late professors Biao Ding (Ohio State University) and Neocles B. Leontis (Bowling Green State University), who have significantly advanced the understanding of RNA motif-mediated RNA trafficking. The authors express gratitude to Shachinthaka D. Dissanayaka Mudiyanselage at Mississippi State University for critical reading of the manuscript. We apologize to colleagues whose work was not cited in this review due to the page limit.
Axtell, M. J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. doi: 10.1146/annurev-arplant-050312-120043
Banerjee, A. K., Chatterjee, M., Yu, Y., Suh, S. G., Miller, W. A., and Hannapel, D. J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18, 3443–3457. doi: 10.1105/tpc.106.042473
Borovinskaya, M. A., Pai, R. D., Zhang, W., Schuwirth, B. S., Holton, J. M., Hirokawa, G., et al. (2007). Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat. Struct. Mol. Biol. 14, 727–732. doi: 10.1038/nsmb1271
Branch, A. D., Benenfeld, B. J., and Robertson, H. D. (1985). Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. U. S. A. 82, 6590–6594. doi: 10.1073/pnas.82.19.6590
Brioudes, F., Jay, F., Sarazin, A., Grentzinger, T., Devers, E. A., and Voinnet, O. (2021). HASTY, the Arabidopsis EXPORTIN5 ortholog, regulates cell-to-cell and vascular microRNA movement. EMBO J. 40:e107455. doi: 10.15252/embj.2020107455
Brunkard, J. O., Runkel, A. M., and Zambryski, P. C. (2015). The cytosol must flow: intercellular transport through plasmodesmata. Curr. Opin. Cell Biol. 35, 13–20. doi: 10.1016/j.ceb.2015.03.003
Cawte, A. D., Unrau, P. J., and Rueda, D. S. (2020). Live cell imaging of single RNA molecules with fluorogenic mango II arrays. Nat. Commun. 11:1283. doi: 10.1038/s41467-020-14932-7
Chantarachot, T., and Bailey-Serres, J. (2018). Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 176, 254–269. doi: 10.1104/pp.17.01468
Chaturvedi, S., Kalantidis, K., and Rao, A. L. (2014). A bromodomain-containing host protein mediates the nuclear importation of a satellite RNA of cucumber mosaic virus. J. Virol. 88, 1890–1896. doi: 10.1128/JVI.03082-13
Chen, X., and Rechavi, O. (2021). Plant and animal small RNA communications between cells and organisms. Nat. Rev. Mol. Cell Biol. 23, 185–203. doi: 10.1038/s41580-021-00425-y
Chou, H. C., Hsieh, T. Y., Sheu, G. T., and Lai, M. M. (1998). Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J. Virol. 72, 3684–3690. doi: 10.1128/JVI.72.5.3684-3690.1998
Cocozaki, A. I., Altman, R. B., Huang, J., Buurman, E. T., Kazmirski, S. L., Doig, P., et al. (2016). Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors. Proc. Natl. Acad. Sci. U. S. A. 113, 8188–8193. doi: 10.1073/pnas.1605127113
Di Serio, F., Torchetti, E. M., Daros, J. A., and Navarro, B. (2019). Reassessment of viroid RNA cytosine methylation status at the single nucleotide level. Viruses 11:11. doi: 10.3390/v11040357
Ding, B. (2009). The biology of viroid-host interactions. Annu. Rev. Phytopathol. 47, 105–131. doi: 10.1146/annurev-phyto-080508-081927
Ding, B., Itaya, A., and Qi, Y. (2003). Symplasmic protein and RNA traffic: regulatory points and regulatory factors. Curr. Opin. Plant Biol. 6, 596–602. doi: 10.1016/j.pbi.2003.09.010
Ding, B., Kwon, M. O., Hammond, R., and Owens, R. (1997). Cell-to-cell movement of potato spindle tuber viroid. Plant J. 12, 931–936. doi: 10.1046/j.1365-313X.1997.12040931.x
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Ding, B., and Wang, Y. (2009). Viroids: uniquely simple and tractable models to elucidate regulation of cell-to-cell trafficking of RNA. DNA Cell Biol. 28, 51–56. doi: 10.1089/dna.2008.0811
Drsata, T., Reblova, K., Besseova, I., Sponer, J., and Lankas, F. (2017). rRNA C-loops: mechanical properties of a recurrent structural motif. J. Chem. Theory Comput. 13, 3359–3371. doi: 10.1021/acs.jctc.7b00061
Gao, R., Liu, P., and Wong, S. M. (2012). Identification of a plant viral RNA genome in the nucleus. PLoS One 7:e48736. doi: 10.1371/journal.pone.0048736
Gast, F. U., Kempe, D., Spieker, R. L., and Sanger, H. L. (1996). Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J. Mol. Biol. 262, 652–670. doi: 10.1006/jmbi.1996.0543
Giguere, T., Adkar-Purushothama, C. R., and Perreault, J. P. (2014). Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae. PLoS One 9:e98655. doi: 10.1371/journal.pone.0098655
Giguere, T., and Perreault, J. P. (2017). Classification of the pospiviroidae based on their structural hallmarks. PLoS One 12:e0182536. doi: 10.1371/journal.pone.0182536
Gómez, G., and Pallás, V. (2001). Identification of an in vitro ribonucleoprotein complex between a viroid RNA and a phloem protein from cucumber plants. Mol. Plant-Microbe Interact. 14, 910–913. doi: 10.1094/MPMI.2001.14.7.910
Gomez, G., and Pallas, V. (2004). A Long-distance translocatable phloem protein from cucumber forms a ribonucleoprotein complex in vivo with hop stunt viroid RNA. J. Virol. 78, 10104–10110. doi: 10.1128/JVI.78.18.10104-10110.2004
Gosai, S. J., Foley, S. W., Wang, D., Silverman, I. M., Selamoglu, N., Nelson, A. D., et al. (2015). Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol. Cell 57, 376–388. doi: 10.1016/j.molcel.2014.12.004
Gozmanova, M., Denti, M. A., Minkov, I. N., Tsagris, M., and Tabler, M. (2003). Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 31, 5534–5543. doi: 10.1093/nar/gkg777
Ham, B. K., and Lucas, W. J. (2017). Phloem-mobile RNAs as systemic signaling agents. Annu. Rev. Plant Biol. 68, 173–195. doi: 10.1146/annurev-arplant-042916-041139
Haywood, V., Kragler, F., and Lucas, W. J. (2002). Plasmodesmata: pathways for protein and ribonucleoprotein signaling. Plant Cell 14 (Suppl. 1), S303–S325. doi: 10.1105/tpc.000778
Haywood, V., Yu, T. S., Huang, N. C., and Lucas, W. J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42, 49–68. doi: 10.1111/j.1365-313X.2005.02351.x
Iacoangeli, A., and Tiedge, H. (2013). Translational control at the synapse: role of RNA regulators. Trends Biochem. Sci. 38, 47–55. doi: 10.1016/j.tibs.2012.11.001
Jiang, D., Wang, M., and Li, S. (2017). Functional analysis of a viroid RNA motif mediating cell-to-cell movement in Nicotiana benthamiana. J. Gen. Virol. 98, 121–125. doi: 10.1099/jgv.0.000630
Johnson, P. Z., Kasprzak, W. K., Shapiro, B. A., and Simon, A. E. (2019). RNA2Drawer: geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences. RNA Biol. 16, 1667–1671. doi: 10.1080/15476286.2019.1659081
Kalantidis, K., Schumacher, H. T., Alexiadis, T., and Helm, J. M. (2008). RNA silencing movement in plants. Biol. Cell. 100, 13–26. doi: 10.1042/BC20070079
Khisamutdinov, E. F., Sweeney, B. A., and Leontis, N. B. (2021). Context-sensitivity of isosteric substitutions of non-Watson-Crick basepairs in recurrent RNA 3D motifs. Nucleic Acids Res. 49, 9574–9593. doi: 10.1093/nar/gkab703
Kim, J. Y. (2005). Regulation of short-distance transport of RNA and protein. Curr. Opin. Plant Biol. 8, 45–52. doi: 10.1016/j.pbi.2004.11.005
Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. doi: 10.1126/science.1059805
Klein, D. J., Moore, P. B., and Steitz, T. A. (2004). The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177. doi: 10.1016/j.jmb.2004.03.076
Kramer, E. B., and Hopper, A. K. (2013). Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 110, 21042–21047. doi: 10.1073/pnas.1316579110
Lee, J. Y. (2015). Plasmodesmata: a signaling hub at the cellular boundary. Curr. Opin. Plant Biol. 27, 133–140. doi: 10.1016/j.pbi.2015.06.019
Lescoute, A., Leontis, N. B., Massire, C., and Westhof, E. (2005). Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 33, 2395–2409. doi: 10.1093/nar/gki535
Levy, A., Erlanger, M., Rosenthal, M., and Epel, B. L. (2007). A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. 49, 669–682. doi: 10.1111/j.1365-313X.2006.02986.x
Lin, S. I., Chiang, S. F., Lin, W. Y., Chen, J. W., Tseng, C. Y., Wu, P. C., et al. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746. doi: 10.1104/pp.108.116269
Liu, L., and Chen, X. (2018). Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4, 869–878. doi: 10.1038/s41477-018-0288-5
Liu, Z., Liu, Q., Yang, X., Zhang, Y., Norris, M., Chen, X., et al. (2021). In vivo nuclear RNA structurome reveals RNA-structure regulation of mRNA processing in plants. Genome Biol. 22:11. doi: 10.1186/s13059-020-02236-4
Long, J., Walker, J., She, W., Aldridge, B., Gao, H., Deans, S., et al. (2021). Nurse cell--derived small RNAs define paternal epigenetic inheritance in Arabidopsis. Science 373:eabh0556. doi: 10.1126/science.abh0556
Lopez-Carrasco, A., and Flores, R. (2017). Dissecting the secondary structure of the circular RNA of a nuclear viroid in vivo: A “naked” rod-like conformation similar but not identical to that observed in vitro. RNA Biol. 14, 1046–1054. doi: 10.1080/15476286.2016.1223005
Lough, T. J., Lee, R. H., Emerson, S. J., Forster, R. L., and Lucas, W. J. (2006). Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351, 455–465. doi: 10.1016/j.virol.2006.03.043
Ma, J., Mudiyanselage, S. D. D., Park, W. J., Wang, M., Takeda, R., Liu, B., et al. (2021). A nuclear import pathway exploited by viroid RNAs. bioRxiv [Preprint].
Maniataki, E., Martinez De Alba, A. E., Sagesser, R., Tabler, M., and Tsagris, M. (2003). Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9, 346–354. doi: 10.1261/rna.2162203
Owens, R. A., Blackburn, M., and Ding, B. (2001). Possible involvement of the phloem lectin in long-distance viroid movement. Mol. Plant-Microbe Interact. 14, 905–909. doi: 10.1094/MPMI.2001.14.7.905
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Qi, Y., Pélissier, T., Itaya, A., Hunt, E., Wassenegger, M., and Ding, B. (2004). Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16, 1741–1752. doi: 10.1105/tpc.021980
Qin, C., Shi, N., Gu, M., Zhang, H., Li, B., Shen, J., et al. (2012). Involvement of RDR6 in short-range intercellular RNA silencing in Nicotiana benthamiana. Sci. Rep. 2:467. doi: 10.1038/srep00467
Reagan, B. C., Ganusova, E. E., Fernandez, J. C., Mccray, T. N., and Burch-Smith, T. M. (2018). RNA on the move: the plasmodesmata perspective. Plant Sci. 275, 1–10. doi: 10.1016/j.plantsci.2018.07.001
Rudt, F., and Pieler, T. (1996). Cytoplasmic retention and nuclear import of 5S ribosomal RNA containing RNPs. EMBO J. 15, 1383–1391. doi: 10.1002/j.1460-2075.1996.tb00480.x
Sarver, M., Zirbel, C. L., Stombaugh, J., Mokdad, A., and Leontis, N. B. (2008). FR3D: finding local and composite recurrent structural motifs in RNA 3D structures. J. Math. Biol. 56, 215–252. doi: 10.1007/s00285-007-0110-x
Schmidt, A., Gao, G., Little, S. R., Jalihal, A. P., and Walter, N. G. (2020). Following the messenger: recent innovations in live cell single molecule fluorescence imaging. Wiley Interdiscip. Rev. RNA 11:e1587. doi: 10.1002/wrna.1587
Song, J., Zhai, J., Bian, E., Song, Y., Yu, J., and Ma, C. (2018). Transcriptome-wide annotation of m(5)C RNA modifications using machine learning. Front. Plant Sci. 9:519. doi: 10.3389/fpls.2018.00519
Spiegelman, Z., Golan, G., and Wolf, S. (2013). Don't kill the messenger: Long-distance trafficking of mRNA molecules. Plant Sci. 213, 1–8. doi: 10.1016/j.plantsci.2013.08.011
Stephenson, J. D., Kenyon, J. C., Symmons, M. F., and Lever, A. M. (2016). Characterizing 3D RNA structure by single molecule FRET. Methods 103, 57–67. doi: 10.1016/j.ymeth.2016.02.004
Stombaugh, J., Zirbel, C. L., Westhof, E., and Leontis, N. B. (2009). Frequency and isostericity of RNA base pairs. Nucleic Acids Res. 37, 2294–2312. doi: 10.1093/nar/gkp011
Szymanski, M., Barciszewska, M. Z., Erdmann, V. A., and Barciszewski, J. (2003). 5 S rRNA: structure and interactions. Biochem. J. 371, 641–651. doi: 10.1042/bj20020872
Tack, D. C., Su, Z., Yu, Y., Bevilacqua, P. C., and Assmann, S. M. (2020). Tissue-specific changes in the RNA structurome mediate salinity response in Arabidopsis. RNA 26, 492–511. doi: 10.1261/rna.072850.119
Takeda, R., and Ding, B. (2009). Viroid intercellular trafficking: RNA motifs, cellular factors and broad impacts. Viruses 1, 210–221. doi: 10.3390/v1020210
Takeda, R., Petrov, A. I., Leontis, N. B., and Ding, B. (2011). A three-dimensional RNA motif in potato spindle tuber viroid mediates trafficking from palisade mesophyll to spongy mesophyll in Nicotiana benthamiana. Plant Cell 23, 258–272. doi: 10.1105/tpc.110.081414
Takeda, R., Zirbel, C. L., Leontis, N. B., Wang, Y., and Ding, B. (2018). Allelic RNA motifs in regulating systemic trafficking of potato spindle tuber viroid. Viruses 10. doi: 10.3390/v10040160
Tang, Y., Gao, C. C., Gao, Y., Yang, Y., Shi, B., Yu, J. L., et al. (2020). OsNSUN2-mediated 5-methylcytosine mRNA modification enhances rice adaptation to high temperature. Dev. Cell 53, 272–286.e7. doi: 10.1016/j.devcel.2020.03.009
Thieme, C. J., Rojas-Triana, M., Stecyk, E., Schudoma, C., Zhang, W., Yang, L., et al. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1:15025. doi: 10.1038/nplants.2015.25
Torres-Larios, A., Dock-Bregeon, A. C., Romby, P., Rees, B., Sankaranarayanan, R., Caillet, J., et al. (2002). Structural basis of translational control by Escherichia coli threonyl tRNA synthetase. Nat. Struct. Biol. 9, 343–347. doi: 10.1038/nsb789
Vargason, J. M., Szittya, G., Burgyan, J., and Hall, T. M. (2003). Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115, 799–811. doi: 10.1016/S0092-8674(03)00984-X
Wang, Y., and Ding, B. (2010). Viroids: small probes for exploring the vast universe of RNA trafficking in plants. J. Integr. Plant Biol. 52, 28–39. doi: 10.1111/j.1744-7909.2010.00900.x
Wang, T., Li, X., Zhang, X., Wang, Q., Liu, W., Lu, X., et al. (2021). RNA motifs and modification involve in RNA long-distance transport in plants. Front. Cell Dev. Biol. 9:651278. doi: 10.3389/fcell.2021.651278
Wang, Y., Qu, J., Ji, S., Wallace, A. J., Wu, J., Li, Y., et al. (2016). A land plant-specific transcription factor directly enhances transcription of a pathogenic noncoding RNA template by DNA-dependent RNA polymerase II. Plant Cell 28, 1094–1107. doi: 10.1105/tpc.16.00100
Wang, Y., Zirbel, C. L., Leontis, N. B., and Ding, B. (2018). RNA 3-dimensional structural motifs as a critical constraint of viroid RNA evolution. PLoS Pathog. 14:e1006801. doi: 10.1371/journal.ppat.1006801
Wassenegger, M., Spieker, R. L., Thalmeir, S., Gast, F. U., Riedel, L., and Sanger, H. L. (1996). A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226, 191–197. doi: 10.1006/viro.1996.0646
Wu, J., Leontis, N. B., Zirbel, C. L., Bisaro, D. M., and Ding, B. (2019). A three-dimensional RNA motif mediates directional trafficking of potato spindle tuber viroid from epidermal to palisade mesophyll cells in Nicotiana benthamiana. PLoS Pathog. 15:e1008147. doi: 10.1371/journal.ppat.1008147
Xu, W., Bolduc, F., Hong, N., and Perreault, J. P. (2012). The use of a combination of computer-assisted structure prediction and SHAPE probing to elucidate the secondary structures of five viroids. Mol. Plant Pathol. 13, 666–676. doi: 10.1111/j.1364-3703.2011.00776.x
Yang, Y., Mao, L., Jittayasothorn, Y., Kang, Y., Jiao, C., Fei, Z., et al. (2015). Messenger RNA exchange between scions and rootstocks in grafted grapevines. BMC Plant Biol. 15:251. doi: 10.1186/s12870-015-0626-y
Yang, L., Perrera, V., Saplaoura, E., Apelt, F., Bahin, M., Kramdi, A., et al. (2019). m(5)C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 29, 2465–2476.e5. doi: 10.1016/j.cub.2019.06.042
Ye, R., Wang, W., Iki, T., Liu, C., Wu, Y., Ishikawa, M., et al. (2012). Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell 46, 859–870. doi: 10.1016/j.molcel.2012.04.013
Yu, A. M., and Lucks, J. B. (2019). Tracking RNA structures as RNAs transit through the cell. Nat. Struct. Mol. Biol. 26, 256–257. doi: 10.1038/s41594-019-0213-2
Zanier, K., Luyten, I., Crombie, C., Muller, B., Schumperli, D., Linge, J. P., et al. (2002). Structure of the histone mRNA hairpin required for cell cycle regulation of histone gene expression. RNA 8, 29–46. doi: 10.1017/S1355838202014061
Zhang, J., Tan, D., Derose, E. F., Perera, L., Dominski, Z., Marzluff, W. F., et al. (2014). Molecular mechanisms for the regulation of histone mRNA stem-loop-binding protein by phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 111, E2937–E2946. doi: 10.1073/pnas.1406381111
Zhang, W., Thieme, C. J., Kollwig, G., Apelt, F., Yang, L., Winter, N., et al. (2016). tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28, 1237–1249. doi: 10.1105/tpc.15.01056
Zhong, X., Archual, A. J., Amin, A. A., and Ding, B. (2008). A genomic map of viroid RNA motifs critical for replication and systemic trafficking. Plant Cell 20, 35–47. doi: 10.1105/tpc.107.056606
Zhong, X., Tao, X., Stombaugh, J., Leontis, N., and Ding, B. (2007). Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 26, 3836–3846. doi: 10.1038/sj.emboj.7601812
Keywords: viroid, RNA motif, RNA nuclear import, RNA cell-to-cell trafficking, RNA systemic trafficking
Citation: Ma J and Wang Y (2022) Studies on Viroid Shed Light on the Role of RNA Three-Dimensional Structural Motifs in RNA Trafficking in Plants. Front. Plant Sci. 13:836267. doi: 10.3389/fpls.2022.836267
Received: 15 December 2021; Accepted: 23 February 2022;
Published: 23 March 2022.
Edited by:
Yiliang Ding, John Innes Centre, United KingdomReviewed by:
Yueying Zhang, John Innes Centre, United KingdomCopyright © 2022 Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Wang, d2FuZ0BiaW9sb2d5Lm1zc3RhdGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.