
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 April 2022
Sec. Plant Metabolism and Chemodiversity
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.833811
This article is part of the Research TopicDiscovery, Biosynthesis, Regulation, Transport, Release and Engineering of Plant Natural ProductsView all 13 articles
 Beixuan He1†
Beixuan He1† Yanjie Zhang1†
Yanjie Zhang1† Lunuan Wang1†
Lunuan Wang1† Dandan Guo1
Dandan Guo1 Xinlei Jia1
Xinlei Jia1 Jianhui Wu1
Jianhui Wu1 Shuyi Qi1
Shuyi Qi1 Hong Wu2*
Hong Wu2* Yue Gao1*
Yue Gao1* Meili Guo1*
Meili Guo1*The unique flavonoids, quinochalcones, such as hydroxysafflor yellow A (HSYA) and carthamin, in the floret of safflower showed an excellent pharmacological effect in treating cardiocerebral vascular disease, yet the regulating mechanisms governing the flavonoid biosynthesis are largely unknown. In this study, CtACO3, the key enzyme genes required for the ethylene signaling pathway, were found positively related to the flavonoid biosynthesis at different floret development periods in safflower and has two CtACO3 transcripts, CtACO3-1 and CtACO3-2, and the latter was a splice variant of CtACO3 that lacked 5’ coding sequences. The functions and underlying probable mechanisms of the two transcripts have been explored. The quantitative PCR data showed that CtACO3-1 and CtACO3-2 were predominantly expressed in the floret and increased with floret development. Subcellular localization results indicated that CtACO3-1 was localized in the cytoplasm, whereas CtACO3-2 was localized in the cytoplasm and nucleus. Furthermore, the overexpression of CtACO3-1 or CtACO3-2 in transgenic safflower lines significantly increased the accumulation of quinochalcones and flavonols. The expression of the flavonoid pathway genes showed an upward trend, with CtCHS1, CtF3H1, CtFLS1, and CtDFR1 was considerably induced in the overexpression of CtACO3-1 or CtACO3-2 lines. An interesting phenomenon for CtACO3-2 protein suppressing the transcription of CtACO3-1 might be related to the nucleus location of CtACO3-2. Yeast two-hybrid (Y2H), glutathione S-transferase (GST) pull-down, and BiFC experiments revealed that CtACO3-2 interacted with CtCSN5a. In addition, the interactions between CtCSN5a and CtCOI1, CtCOI1 and CtJAZ1, CtJAZ1 and CtbHLH3 were observed by Y2H and GST pull-down methods, respectively. The above results suggested that the CtACO3-2 promoting flavonoid accumulation might be attributed to the transcriptional activation of flavonoid biosynthesis genes by CtbHLH3, whereas the CtbHLH3 might be regulated through CtCSN5-CtCOI1-CtJAZ1 signal molecules. Our study provided a novel insight of CtACO3 affected the flavonoid biosynthesis in safflower.
Flavonoids, as a group of secondary metabolites widely existing in plants, improve the adaptation ability in the volatile and complex environment of plants (Tahara, 2007; Petroni and Tonelli, 2011). Meanwhile, the flavonoid content in medicinal plants received much attention because of its beneficial health properties against a number of diseases (Rukh et al., 2019). Carthamus tinctorius L., commonly known as safflower, is an important medicinal plant and widely used in treating cardiocerebral vascular disease in China. The flavonoids in safflower are the main pharmacologically active compounds, especially the unique quinochalcones, such as hydroxysafflor yellow A (HSYA) and carthamin, which have high commercial and medicinal value (Tu et al., 2015). For the deep investigation and wide application of flavonoids in safflower, it is important to explore their biosynthesis mechanism and further improve the flavonoid content in plants.
The biosynthesis pathway of basic flavonoids skeleton and the enzymes related to it have been well characterized, including phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydrolase (C4H), 4-coumarate:CoA ligase (4CL), chalcone synthase (CHS), 2-hydroxyisoflavanone synthase (IFS), 2-hydroxyisoflavanone dehydratase (HID), chalcone isomerase (CHI), flavone synthase (FNS), flavanone 3-hydroxylase (F3H), flavonol synthase (FLS), flavonoid 3’-hydroxylase (F3’H), flavonoid 3’,5’-hydroxylase (F3’5’H), dihydroflavonol 4-reducatase (DFR), anthocyanidin synthase (ANS), leucoanthocyanidin 4-reductase (LAR), especially in model plant Arabidopsis thaliana, and a range of crop species, such as bean, tomato, maize, and rice (Tohge et al., 2017). The most of channel enzyme genes in flavonoids biosynthesis have been identified depending on transcriptome of safflower, and the function in the HSYA accumulation of CtCHS1, CtCHS4, CtCHI1, and CtF3H have been proved in vivo of safflower (Tu et al., 2016; Guo et al., 2017, 2019; He et al., 2018). There is broad consensus that the flavonoid pathways are regulated mostly through the coordinated transcription of structural genes by the interaction of MBW complex, such as R2R3 MYB transcription factors, basic helix–loop–helix (bHLH) transcription factors, and WD40 proteins (Payne et al., 2000; Ramsay and Glover, 2005). Some studies have reported that bHLH3, a bHLH transcription factor, played an important role in regulating the anthocyanins, flavones, and flavonols biosynthesis through the downstream channel enzyme genes (Rahim et al., 2014; Li et al., 2020; An et al., 2021).
In addition to the regulation of flavonoid metabolism channel enzyme genes and transcription factors in plants, flavonoid biosynthesis is also affected by plant hormones signaling pathway, such as methyl jasmonate (JA), auxin, and ethylene. Ethylene participates in many plants’ developmental processes and stress responses, such as plant growth, germination, flowering, fruit ripening, and senescence (Lin et al., 2009; Van de Poel et al., 2015; Wen, 2015). It is worth to mention that ethylene positivity regulates the accumulation of flavonoids implicated with various evidence. Treatment with ethylene and its precursor, 1-aminocyclopropane carboxylic acid (ACC), induced the flavonol and anthocyanin accumulation in Arabidopsis, apple, black carrot, and tea (Watkins et al., 2014; Barba-Espín et al., 2017; An et al., 2018; Ke et al., 2018), and isoflavone accumulation in soybean (Yuk et al., 2016). In A. thaliana, the anthocyanin levels were lower in the ein1-1, ein2-1, and the ein3/eil1 double mutant than in normal plants. In apple, EIN3-like1, MYB1, and ERF3 together modulate anthocyanin accumulation (An et al., 2018). Meanwhile, others reported that ethylene negatively affects anthocyanin biosynthesis. The inhibition of ethylene synthesis by aminoethoxyvinylglycine treatment increased the anthocyanin content in black rice at dark (Kumar et al., 2019). ACC acid treatment suppressed the sugar and light-inducible anthocyanin synthesis in Arabidopsis plants (Jeong et al., 2010). Ethylene treatment inhibited the light-induced anthocyanin and biosynthesis through the PpCTR1/PpETR1 system in the red pear fruits (Ni et al., 2020). Tobacco plant carrying the mutated melon CmETR1/H69A showed higher anthocyanin level than normal plant (Keita et al., 2005). The ethylene treatment influenced the accumulation of anthocyanin in blueberry, which might depend on the cultivar (Costa et al., 2018). Accordingly, ethylene regulated the biosynthesis of flavonols and anthocyanins, in which ethylene signaling pathway has been suggested to be a regulator of anthocyanin accumulation. However, these studies have not come to an accordant conclusion, and there is little evidence demonstrating the regulatory effect of ethylene on another flavonoid biosynthesis, especially chalcones, which is the principal component in safflower.
Increasing evidence showed that ACO (ACC oxidase) has a rate-limiting role in ethylene biosynthesis, which belongs to a multigene family (Zhang et al., 2012; Houben and Van de Poel, 2019). Most ACOs were the biosynthetic structure genes of ethylene, whereas others displayed some different functions. For instance, SlACO5 and CsACO2, respectively, played vital roles in low oxygen response in tomato (Sell and Hehl, 2005) and sex determination in cucumber flowers (Chen et al., 2016). In addition, the overexpression of PtACO1 in poplar caused cambial cell division (Jonathan et al., 2009). In our previous study, the possibility of ethylene synthesis pathway regulating the accumulation of flavonoids was concerned in safflower. The overexpression of CtACO1 reduced the accumulation of quinochalcone HSYA and carthamin, kaempferol, and its glycosylated derivatives, whereas it increased quercetin and its glycosylated derivatives (Tu et al., 2019). In the present study, it was found that the expression of CtACO3 was closely associated with flavonoid accumulation in the floret of safflower at different development periods. The overexpression of two CtACO3 splice variants, CtACO3-1 and CtACO3-2, significantly increased the accumulation of quinochalcone HSYA and carthamin, flavonol kaempferol glycosylated derivatives, and quercetin glycosylated derivatives. An interesting phenomenon for CtACO3-2 protein suppressing the transcription of CtACO3-1 was also found. Furthermore, a possible route of CtACO3-2 influenced flavonoid biosynthesis pathway was preliminarily explored. The following is our first report of the study.
The safflower plant ZHH0119 (C. tinctorius L.), which floret with orange-yellow color and major quinochalcones, was collected from the Chinese Safflower Germplasm Resources in the Academy of Agricultural Sciences of Xinjiang. The safflower was identified by Prof. Meili Guo. It was repeatedly purified in our laboratory. The plant was grown in the greenhouse at 23 ± 2°C under the light of circadian rhythm (16-h/8- light–dark cycle) in the Naval Medical University (Shanghai, China). The voucher specimen was SMMU171201 and deposited at the Naval Medical University.
The CDS of CtACO3-1 was cloned with primers (PMT39-CtACO3-1F and CtACO3-1-PMT39R; Supplementary Table 1); CtACO3-2 was cloned with primers (PMT39-CtACO3-2F and CtACO3-2-PMT39R; Supplementary Table 1), and empty vector PMT39 (pCAMBIA-1380-CaMV35S-MCS-EGFP-NOS) was digested by NcoI and made a green fluorescent protein (GFP) tag fused to the CDS of CtACO3-1 and CtACO3-2 and downstream of 35S promoter (Guo et al., 2017). The Agrobacterium strain (GV3101) including the above vector was introduced into safflower plants according to previous methods to generate overexpressing safflowers (Guo et al., 2017). Then, initial screening analyzed T1 transformants according to previous methods, the identification using primers (35SIDF, CtACO3-1IDR1, and CtACO3-2IDR1; Supplementary Table 1), the forward primer is on CaMV35S promoter, and reverse primer is on the CDS of target gene (Tu et al., 2019).
Multiple sequence alignment was aligned in Geneious (v9.1) by the MUSCLE plugin. Then, the best-scoring maximum likelihood tree was built with 1,000 bootstrap replicates using Geneious (v9.1). Conserved protein domains were identified using SMART (Letunic et al., 2015).
The CDSs of CtACO3-1 and CtACO3-2 were cloned into the PMT-39 vector; the recombinant and control plasmids were transformed into the Agrobacterium strain GV3101. Positive Agrobacterium was cultured and cocultured with onion epidermal layers, and N. benthamiana leaves were injected according to previous methods (Guo et al., 2019; Tu et al., 2019). The GFP fluorescence of CtACO proteins was confirmed by a confocal microscope (Leica TCS SP5).
The florets of CtACO3-1, CtACO3-2 transgenic safflowers plants, and untreated safflower plants were collected at stage IV. Total RNA was extracted from safflower floret samples by TransZol reagents (the tubular flower without the ovary); first-strand cDNA was synthesized with the manufacturer’s instruction (TransGene Biotech, Beijing, China). Quantitative real-time PCR (qRT-PCR) was worked using TransStart Green qPCR Supermix (TransGene Biotech, Shanghai, China) with ABI7300 Real-Time PCR system (Applied Biosystems, Foster City, CA, United States). When designing Real-Time PCR primers for the transcription levels analysis of CtACO3-1 and CtACO3-2, there is one pair of primers in their same region and one in the CtACO3-1-specific region. The relative expression level of CtACO3-2 was confirmed by the difference between the expression level of CtACO3-1 and CtACO3-2 shared region and the expression level of the CtACO3-1-specific region. Primers used are listed in Supplementary Table 1. A quantitative reverse transcriptase–PCR thermal cycle was followed as per manufacturer’s instruction (Tm at 58°C). The results were calculated according to 2–ΔΔCt, whereas Ct60s gene (GenBank accession no. KJ634810) was used as a housekeeping gene.
The preparation of above plant samples, chemicals, and reagents was followed according to previous methods (Tu et al., 2019). Agilent 6538 Accurate Mass Quadrupole Time-of-Flight MS and Agilent 1290 Infinity LC System (Agilent, Santa Clara, CA, United States) was used for Ultra-high performance liquid chromatography with quadrupole time-of-flight mass spectrometry (UHPLC-QTOF-MS) analysis. XBridge TM BEH C18 column (2.5 μm, 2.1 mm × 100 mm; Waters, Milford, MA, United States) was used for chromatographic separations. Previous methods were followed for the methods, mass spectrometer, positive ion mode, and gradient elution used for the quantification (Guo et al., 2017). The eight standard compounds were confirmed, such as D-phenylalanine (m/z 165.079), kaempferol-3-O-glucoside (m/z 448.1006), quercetin-3-O-glucoside (m/z 464.0955), rutin (m/z 610.1534), and HSYA (m/z 612.1690), purchased from Yuanye Bio-Technology (Shanghai, China), and carthamin (m/z 910.2168) was extracted in our laboratory. Agilent MassHunter quantitative analysis software was used for metabolite data.
Yeast one-hybrid (Y1H) assays were performed according to the manufacturer’s instruction of Matchmaker One-Hybrid Library Construction and Screening Kit (Clontech). The safflower cDNA library cloned in the prey vector pGAD-T7 (AD) was made by the OE BioTech. In brief, the promoter fragment of CtACO3 (Figure 4A) was cloned into the pAbAi-bait vector, which was introduced into the yeast strain Y1H GOLD, and were cultured on SD/–Ura medium. Positive clones were sequence-verified by Matchmaker Insert Check PCR Mix 1 (Clontech), the yeast-based transcriptional activation test was followed. The screen was performed by using pAbAi-bait Y1H stain and the safflower cDNA-pGADT7-DEST library. These yeast strains were cultured on SD/–Leu medium containing 100 ng/ml AbA (Clontech). Positive clones were diluted and spotted on SD/–Leu medium containing 250 ng/ml AbA (Clontech), then sequence-verified by Matchmaker Insert Check PCR Mix 2 (Clontech).
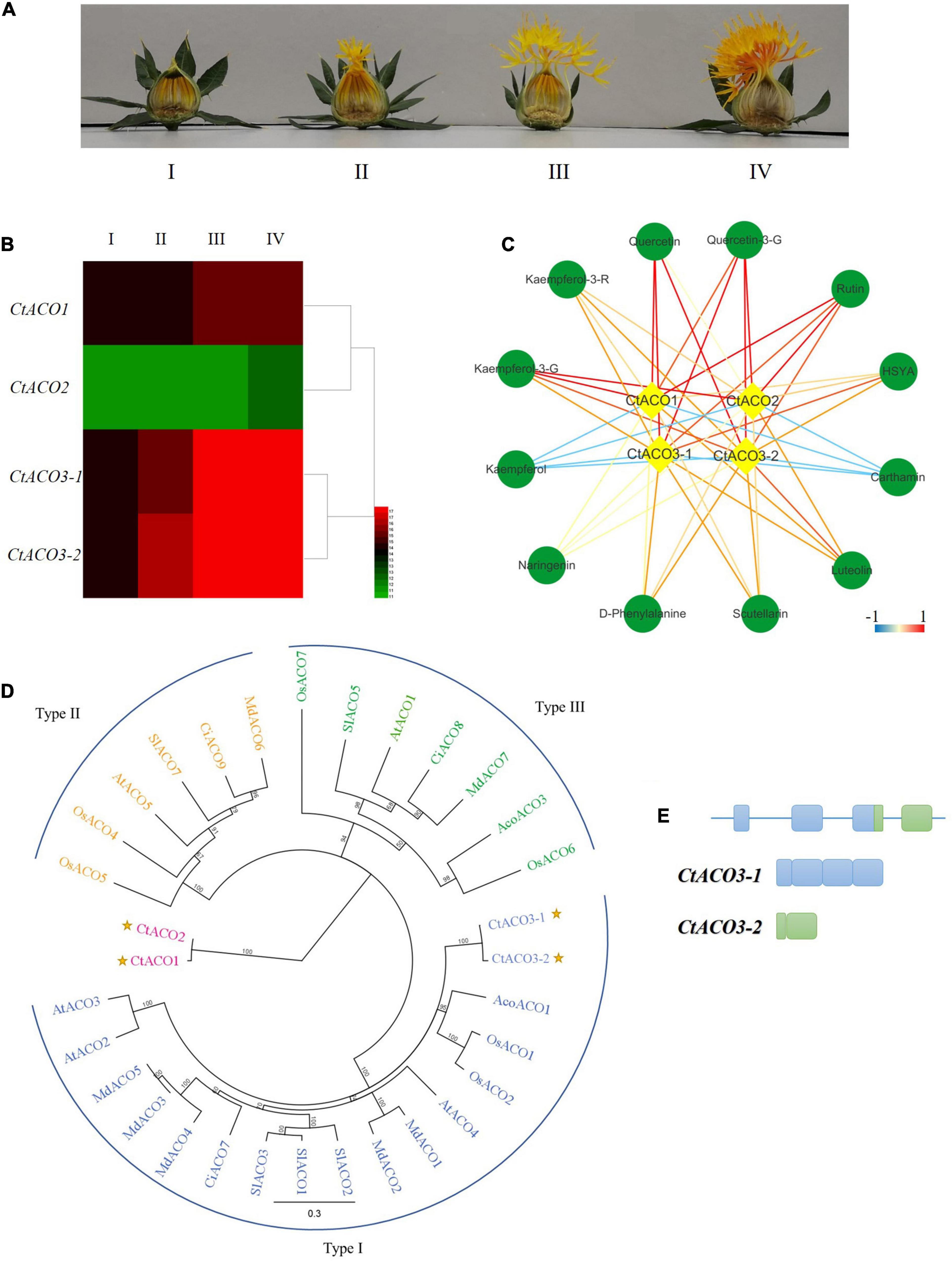
Figure 1. The expression pattern of ACO genes in safflower. (A) Different developmental stages of safflower lines. I: the day before flowering, II: the first day of flowering, III: the second to third day of flowering, IV: the full bloom. (B) Four CtACOs expression patterns at different developmental stages of safflower lines, signal values by gene chip. (C) The correlation coefficients between CtACOs and flavone glycoside compounds. The color key was set from –1 to +1. kaempferol-3-G: kaempferol-3-O-glucoside, kaempferol-3-R: kaempferol-3-rutinoside, quercetin-3-G: quercetin-3-O-glucoside. (D) Maximal likelihood phylogenetic tree for ACO protein sequences of Carthamus tinctorius (Ct), Arabidopsis thaliana (At), Solanum lycopersicum (Sl), Malus domestica (Md), Oryza sativa (Os), Ananas comosus (Aco), and Citrus sinensis (Ci). Type I ACO is shown in blue, Type II ACO is shown in yellow, and Type III ACO is shown in green, CtACOs are plotted. (E) Schematic diagram of CtACO3 splice variants indicating introns and exons.
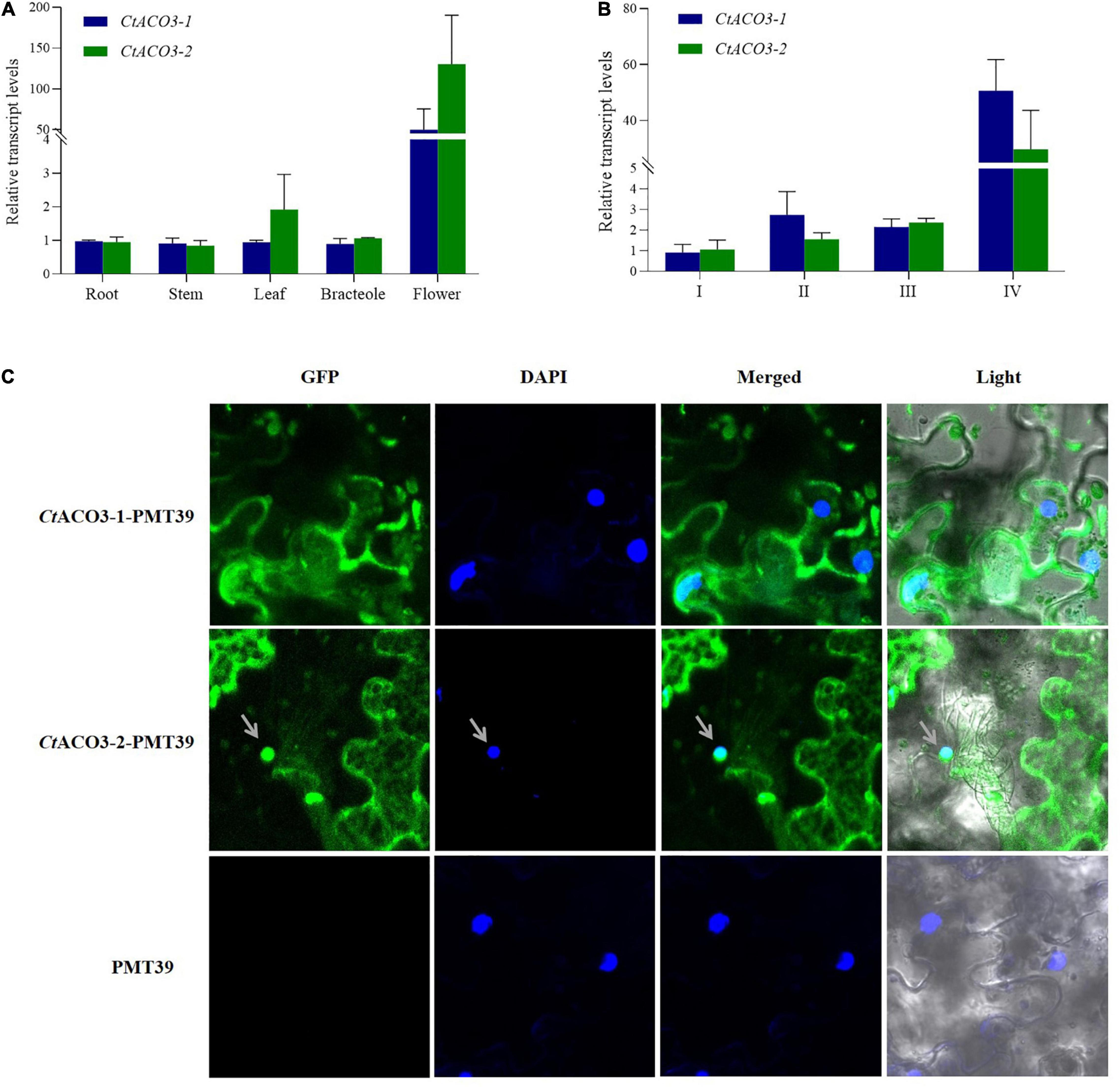
Figure 2. The molecular characterization of CtACO3 splice variants. (A) Relative abundance of CtACO3-1 and CtACO3-2 in different tissues. (B) Relative abundance of CtACO3-1 and CtACO3-2 in different floret developmental stages. (C) Subcellular localization of the CtACO3-1/-2-PMT39 fusion protein in Nicotiana benthamiana leaves. Data were expressed as means ± SD (n = 3).
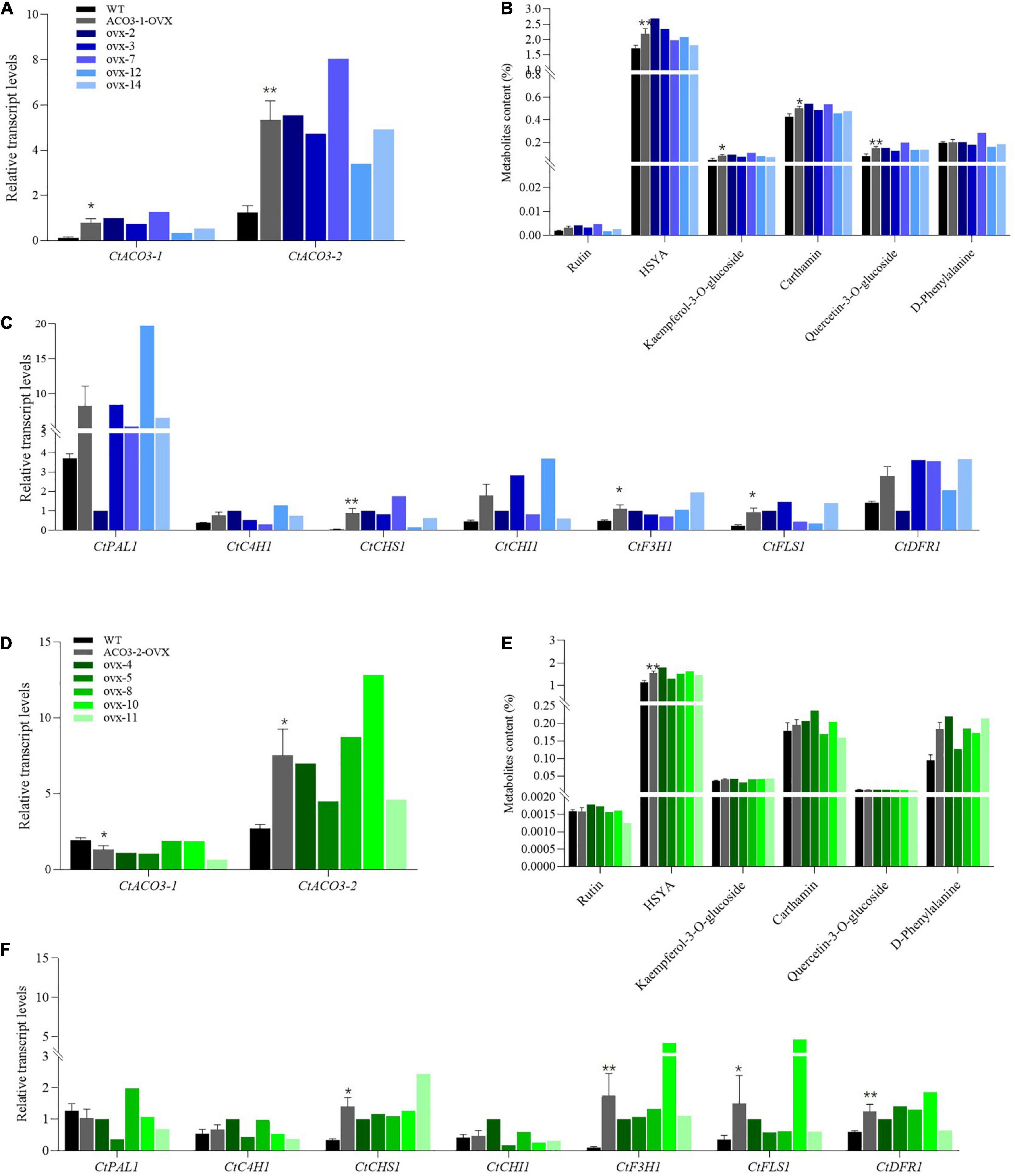
Figure 3. Stimulatory effect of CtACO3-1 overexpression and CtACO3-2 overexpression on main active compounds content in safflower. (A) Relative expression levels of CtACO3-1 and CtACO3-2 in WT and CtACO3-1 overexpression transgenic lines determined by quantitative PCR (qPCR). (B) Ultra-high performance liquid chromatography with quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS) analysis of main active compounds content in WT and CtACO3-1 overexpression transgenic lines. (C) The expression levels of flavonoid-related genes in WT and CtACO3-1 overexpression transgenic lines determined by qPCR. (D) The relative expression levels of CtACO3-1 and CtACO3-2 in WT and CtACO3-2 overexpression transgenic lines determined by qPCR. (E) UPLC-QTOF-MS analysis of main active compounds content in WT and CtACO3-2 overexpression transgenic lines. (F) The expression levels of flavonoid-related genes in WT and CtACO3-2 overexpression transgenic lines determined by qPCR. Data were expressed as mean ± SEM (n = 3), **p < 0.01, and *p < 0.05.
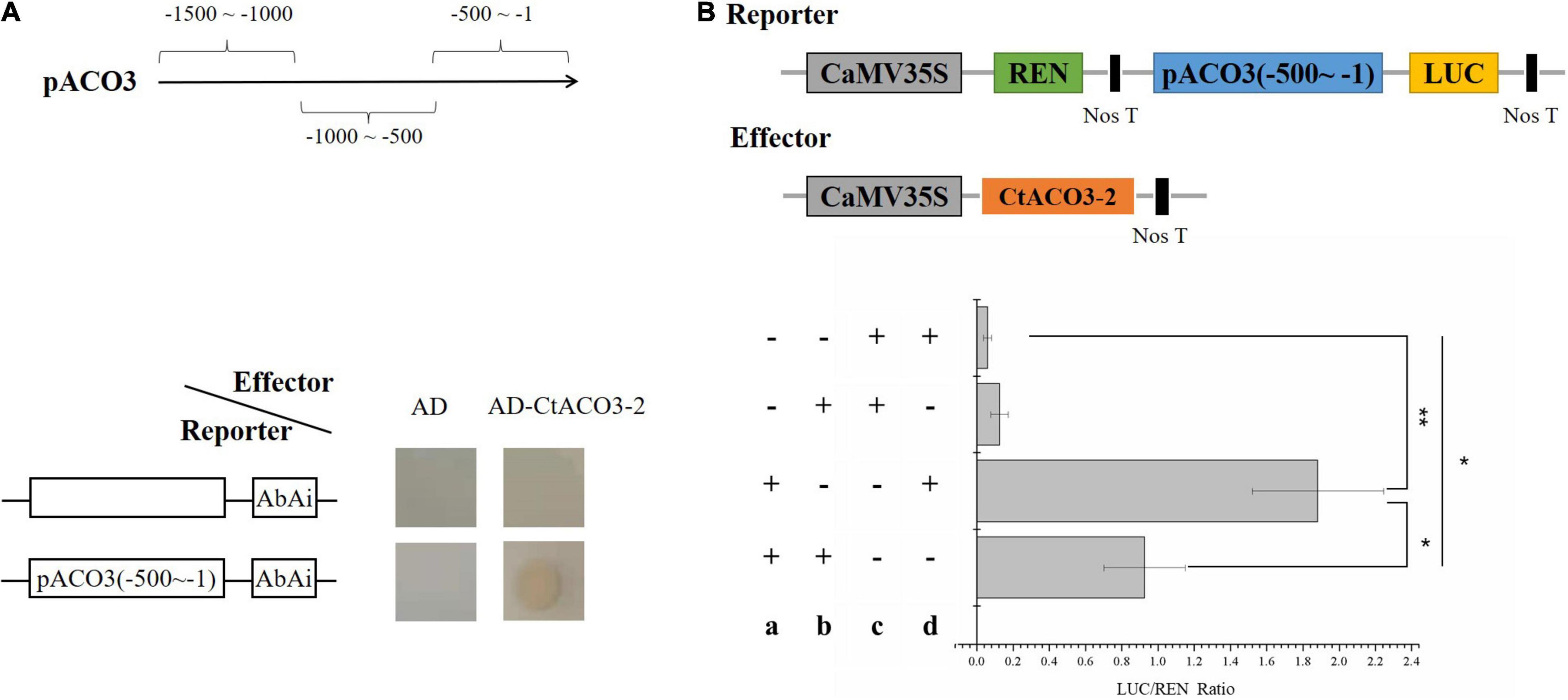
Figure 4. CtACO3-2 regulate the transcription of CtACO3-1. (A) Diagram of the promoter fragments of the CtACO3 promoter. CtACO3-2 binds to the -500∼-1 fragment of CtACO3 promoter using Y1H assay. Y1H assays were repeated for three times. (B) The effects of CtACO3-2 on CtACO3 promoter deactivation. CtACO3 promoter pCtACO3 (-500∼-1) was fused to LUC reporter and the promoter activity was determined by a transient Dual-LUC assay in N. benthamiana. The relative LUC activity was normalized to the reference Renilla (REN) luciferase. (a): Reporter, (b): effector, c: pGreenII0800-LUC, d: pGreenII 62-SK. Data were expressed as mean ± SD (n = 3), **p < 0.01, and *p < 0.05.
To confirm the interaction between CtACO3-2 and the promoter of CtACO3, the CDS of CtACO3-2 was inserted into pGreenII 62-SK, and the promoter pACO3 (-500 to -1) of CtACO3 was cloned into pGreen 0800-LUC.
The constructed effector pGreenII 62-SK-CtACO3-2, reporter plasmids pACO3-LUC and control vector pGreenII 62-SK, and pGreen 0800-LUC were introduced into Agrobacterium strain GV3101 (pSoup-19T), respectively. Mixed bacteria solution harbored the effector and reporter (1:1), which was injected into tobacco leaves. After 4 days, a dual-luciferase assay kit was used to measure LUC and REN luciferase activities (Promega) following Liu et al. (2013) Three biological repeats were assayed for each combination. The results were calculated using the ratio of LUC to REN.
Yeast two-hybrid (Y2H) screening and Y2H assays were performed following the manufacturer’s instructions (Clontech, Mountain View, CA, United States). The CDS of CtACO3-2, CtCSN5a, CtCOI1, CtJAZ1, and CtbHLH3; the C-terminal of CtCOI1 and CtbHLH3; and the N-terminal of CtCOI1 and CtbHLH3 were inserted into the pGBKT7 or pGADT7 vector to fuse with the DNA-BD and AD, respectively (primers are listed in Supplementary Table 1). Autoactivation and suppression of autoactivation of the bait constructs were tested by cultured in SD/-Trp medium 40 mg/ml X-α-Gal. Then, bait constructs without self-activation were transformed into Y2H GOLD strain with prey constructs using the lithium acetate method and cultured in DDO (SD/-Leu/-Trp) medium containing 125 ng/ml AbA and 40 mg/ml X-α-Gal for 5 days. Then, transformed positive colonies were plated onto QDO (SD/-Ade/-His/-Leu/-Trp) medium containing 125 ng/ml AbA and 40 mg/mL X-α-Gal, and positive clones were verified by using a Matchmaker Insert Check PCR Mix 2 (Clontech).
For the construction of GST-CtCSN5a, GST-CtCOI1, GST-CtbHLH3N, His-CtACO3-2, and His-CtJAZ1 expression vectors, the CDS of CtCSN5a, CtACO3-2, CtCOI1, CtJAZ1, and CtbHLH3N was cloned into pGEX-6P-1 or pET-32a, respectively. To test whether CtACO3-2 interacts with CtCSN5a protein, CtCSN5a interacts with CtCOI1, CtCOI1 interacts with CtJAZ1, and CtJAZ1 interacts with CtbHLH3N, according to the manufacturer’s instruction for the Pierce GST Protein Interaction Pull-Down Kit (Thermo Scientific). Briefly, His-bait fusion protein was incubated with GST-prey fusion proteins with slowly shaking for a night. Then, beads were washed five times and heated for 5 min in 100°C. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting were used to confirm the proteins by anti-GST (Beyotime, 1/1,000) and anti-His (Beyotime, 1/1,000) antibodies, respectively.
The CDSs of CtACO3-1 and CtACO3-2 were inserted in the pCAMBIA1300-35S-NY173 vector to create CtACO3-1-nYFP and CtACO3-2-nYFP constructs. Similarly, the CDSs of CtCSN5a were inserted in the pCAMBIA1300-35S-YC155 vector. The specific primers used for CtACO3-1-nYFP, CtACO3-2-nYFP, and CtCSN5a-cYFP construction are described in Supplementary Table 1. Then, the construction was introduced into Agrobacterium GV3101 strain subsequently. The mixed bacteria solution containing nYFP and cYFP pairs was injected into tobacco leaves with a syringe and grown for 4 days. The confocal microscope was used for YFP fluorescence detection (Leica TCS SP5).
The data are presented as mean ± standard deviation (SD) or mean ± standard error of mean (SEM) and analyzed using GraphPad Prism 8 software (GraphPad Software, La Jolla, CA, United States). A paired two-tailed Student’s t-test was used to compare group differences. The value of p < 0.05 was regarded as statistically significant.
In our previous study, a normalized cDNA library and gene chip data of safflower were analyzed systematically (Guo et al., 2017). Three genes were annotated as ACO enzymes in safflower line, there were two CtACO3 splice variants, CtACO3-1 and CtACO3-2. We analyzed the transcription levels of CtACOs and the contents of main flavonoids at different flowering times (Figures 1A,B). The coexpression analysis of “gene metabolites” is displayed in Figure 1C. Results indicate that CtACO family genes are positively related to most of flavonoid in safflower lines.
There were a few reports that classified three distinct phylogenetic groups in the ACOs (Jafari et al., 2012). CtACOs and some homologous proteins from other plants were analyzed by phylogenetic tree analysis, showing three clusters of ACOs (Houben and Van de Poel, 2019). CtACO3 within the type I ACO cluster which exhibits high sequence similarity with AcoACO1, OsACO1, and OsACO2, but CtACO1 and CtACO2 are not within any ACO clusters of these three types (Figure 1D). Furthermore, a detailed residue analysis of these ACO alignments is presented in Supplementary Figure 1. It has been reported that the ACO types can be classified by the intermediate residue presented in the conserved RXS motif, such as type I (R-M-S), type II (R-L/I-S), and type III (R-R-S) (Houben and Van de Poel, 2019). Interestingly, CtACO1 and CtACO2 consist of R-V-S, which was different from those three types. In parallel, all of CtACOs have conserved 2-His-1-carboxylate Fe (II) binding motif. Residues Q273, K284, K321, and F400 are conserved in CtACOs, which are important for ACO activity according to DR (Dilley et al., 2013). This could partially account for the different roles of CtACOs on flavonoid accumulation in safflower.
The CtACO3-1 (GenBank accession no. MH67444) is a full-length transcript corresponding to the coding sequence of CtACO3 and was predicted to encode a protein of 345 amino acids, with a molecular mass of 36.16 kD and a calculated pI of 6.57. The CtACO3-2 transcript (GenBank accession no. MW075467) encodes a truncated protein of 110 amino acid, with a molecular mass of 12.48 kD and a calculated isoelectric point (pI) of 7.40, in which the start codon is located at the 4th exon, lacking the 235 N-terminal residues (Figure 1E). Conserved domain analyses indicate that in the C-terminal regions, CtACOs contain a conserved C3HC4 RING finger domain (Supplementary Figure 1).
To detect the expression patterns of CtACO3-1 and CtACO3-2 at different flowering times (I, II, III, and IV) and specific tissues (flower, leaf, bracteole, and stem), the plant materials were collected. The transcript levels of CtACO3-1 and CtACO3-2 in flowers increased continuously with the floret flowering, shown in Figure 2A. Both CtACO3-1 and CtACO3-2 showed the highest transcript level in flower (Figure 2B), whereas CtACO1 had the highest level in leaf (Tu et al., 2019).
The CtACO3-1 and CtACO3-2 were coexpressed with GFP in onion epidermal cells and Nicotiana benthamiana leaves to identify the subcellular localization. The result indicates that CtACO3-1 localized to the cytoplasm, and CtACO3-2 localized to the cytoplasm and nucleus (Figure 2C and Supplementary Figure 2), which were different from the cytosol location of CtACO1.
To explore the function of CtACO3-1 and CtACO3-2 in vivo of safflower, transgenic safflower plants that overexpressed CtACO3-1 and CtACO3-2 under cauliflower mosaic virus (CaMV) 35S promoter were generated. In total, 10 independent positive CtACO3-1-overexpression transgenic lines and eight independent positive CtACO3-2-overexpression transgenic lines were screened out by genomic DNA PCR (Supplementary Figures 3, 4) and compared with untreated safflower lines (wild type); the relative transcription level of CtACO3-1 increased significantly in CtACO3-1-overexpression plants and had the highest expression level in ovx7 (∼10.5-fold), whereas a higher level was found in ovx2 (∼8.3-fold) and ovx3 (∼6.12-fold) lines (Figure 3A); the relative transcription level of CtACO3-2 showed the highest expression level in ovx10 (∼4.7-fold), whereas a higher level was found in ovx8 (∼3.2-fold) and ovx4 (∼2.5-fold) lines (Figure 3D). As shown in Supplementary Figure 5, there was almost no difference in plant appearance and growth status between the untreated and the transgenic plants. Five CtACO3-1-overexpression lines (nos. 2, 3, 4, 12, 14) and five CtACO3-2-overexpression lines (nos. 4, 5, 8, 10, and 11) were used to further analyze the profiling of flavonoids in safflower. The levels of flavonoid metabolites were measured by UPLC–electrospray ionization–QTOF-MS. It is shown that most of flavonoid accumulation enhanced in CtACO3-1-overexpression safflower lines and CtACO3-2-overexpression ones, especially the four mage compounds, quinochalcones (HSYA and carthamin), and flavonols (quercetin-3-O-glucoside and kaempferol-3-O-glucoside). In CtACO3-1-overexpression safflower lines, HSYA increased 56.79, 36.77, and 21.45% in ovx-2, ovx-3, and ovx-12 lines, respectively. Moreover, carthamin increased most robustly in the ovx-2 line (∼27.29%) and second most robustly in the ovx-7 line (∼26.21%). Quercetin-3-O-glucoside and kaempferol-3-O-glucoside were increased in each overexpression CtACO3 safflower plant (50–160%) (Figure 3B). Besides, in CtACO3-2-overexpression safflower lines, HSYA increased 59.34, 43.67, and 35.69% in ovx-4, ovx-10, and ovx-8 lines, respectively. Moreover, quercetin-3-O-glucoside increased most robustly in the ovx-11 line (∼600%) and second most robustly in the ovx-10 line (∼500%), whereas carthamin was slightly increased in each overexpression CtACO3-2 overexpression safflower plant (Figure 3E). In brief, the overexpression of CtACO3-1 and CtACO3-2 in safflower resulted in the most increase of flavonoids in flowers, and the metabolic flux of the flavonoid pathway was suggested to be directed into both the quinochacone and flavonol branch.
To further explore the flavonoid biosynthesis in CtACO3-1-overexpression safflower and CtACO3-2-overexpression safflower, the transgenic plants were used to investigate the transcript abundance of flavonoid biosynthesis–related genes, such as CtPAL1, CtC4H1, CtCHS1, CtCHI1, CtF3H1, CtFLS1, and CtDFR1, which displayed different expression pattern in CtACO3-1-overexpression safflower lines and CtACO3-2-overexpression safflower lines. As shown in Figure 3C, the transcript levels of upstream genes of the flavonoid pathway, such as CtPAL1, CtC4H1, and CtCHI1, shown an upward trend with CtCHS1 significantly increased in the CtACO3-1-overexpression lines. The expression of downstream genes CtF3H1 and CtFLS1 was considerably induced. Similarly, the transcript levels of CtC4H1 and CtCHI1 in CtACO3-2-overexpression safflower lines shown an upward trend as well, the expression of CtCHS1, CtF3H1, CtFLS1, and CtDFR1 was induced (Figure 3F). Overall, the transcript abundance of flavonoid biosynthesis–related genes performed similar trends after CtACO3-1 or CtACO3-2 overexpression. An additional interesting phenomenon was unraveled when we analyzed the transcript levels of other CtACOs in CtACO3-1 or CtACO3-2-overexpression safflower. Interestingly, CtACO3-1-overexpression safflowers had a higher transcript level of CtACO3-2, whereas the level of CtACO3-1 was lower in CtACO3-2-overexpression safflowers. These results indicate a feedback regulation between CtACO3-2 and CtACO3-1, and CtACO3-2 may be the reason that CtACO3-2 and CtACO3-1 overexpression resulted in different flavonoid accumulation.
The promoter of CtACO3-1 contained many cis-elements (Supplementary Figure 6), such as G-box (5′-CACGTG-3′). To determine whether the molecules related to flavonoid biosynthesis directly regulate the transcription of CtACO3-1, Y1H assays were conducted. The promoters of CtACO3 were divided into three fragments, namely, pCtACO3 (-1,500 to -1,000), pCtACO3 (-1,000 to -500), and pCtACO3 (-500 to -1), and fused to the pAbAi vector, respectively; only promoter pCtACO3 (-500 to -1) exhibited no transcriptional activation activity in the yeast-based transcriptional activation test. Moreover, the results showed that CtACO3-2 could specifically bind to the promoter pCtACO3 (-500 to -1) of CtACO3-1 (Figure 4A).
To further clarify the regulatory effect of CtACO3-2 on CtACO3-1 transcription, pCtACO3 (-500 to -1) was fused to the LUC to generate reporter constructs pCtACO3:LUC. Meanwhile, CtACO3-2 driven by CaMV 35S promoter was used as an effector construct. The pairs of effector and reporter were coexpressed in tobacco. When there was a presence of CtACO3-2 protein in the infiltration mixture, the luciferase (LUC)/Renilla (REN) values were significantly decreased by 49% for pCtACO3 (-500 to -1), compared with the control (Figure 4B). This resulted from the dual-luciferase assays that suggested CtACO3-2 downregulated CtACO3-1 at the transcriptional level.
To understand how CtACO3-1 and CtACO3-2 participate in affecting the flavonoid accumulation in safflower, we used the Y2H system to identify its potential interaction partners. The CDS of CtACO3-2 constructed a bait vector [CtACO3-2-binding domain (BD)]. The bait and a library of cDNAs containing inserts for prey proteins fused to GAL4–activation domain (AD) were cotransformed to Y2H GOLD. After screening, 12 independent clones were identified, and the information is shown in Supplementary Table 2 and Supplementary Figure 7. To confirm the interaction of the clones about flavonoid biosynthesis with CtACO3-2 in yeast, the CDS of CtCSN5a was fused to AD vector and cotransformed into Y2H GOLD with CtACO3-2-BD; the interactions were reconstructed (Figure 5A).
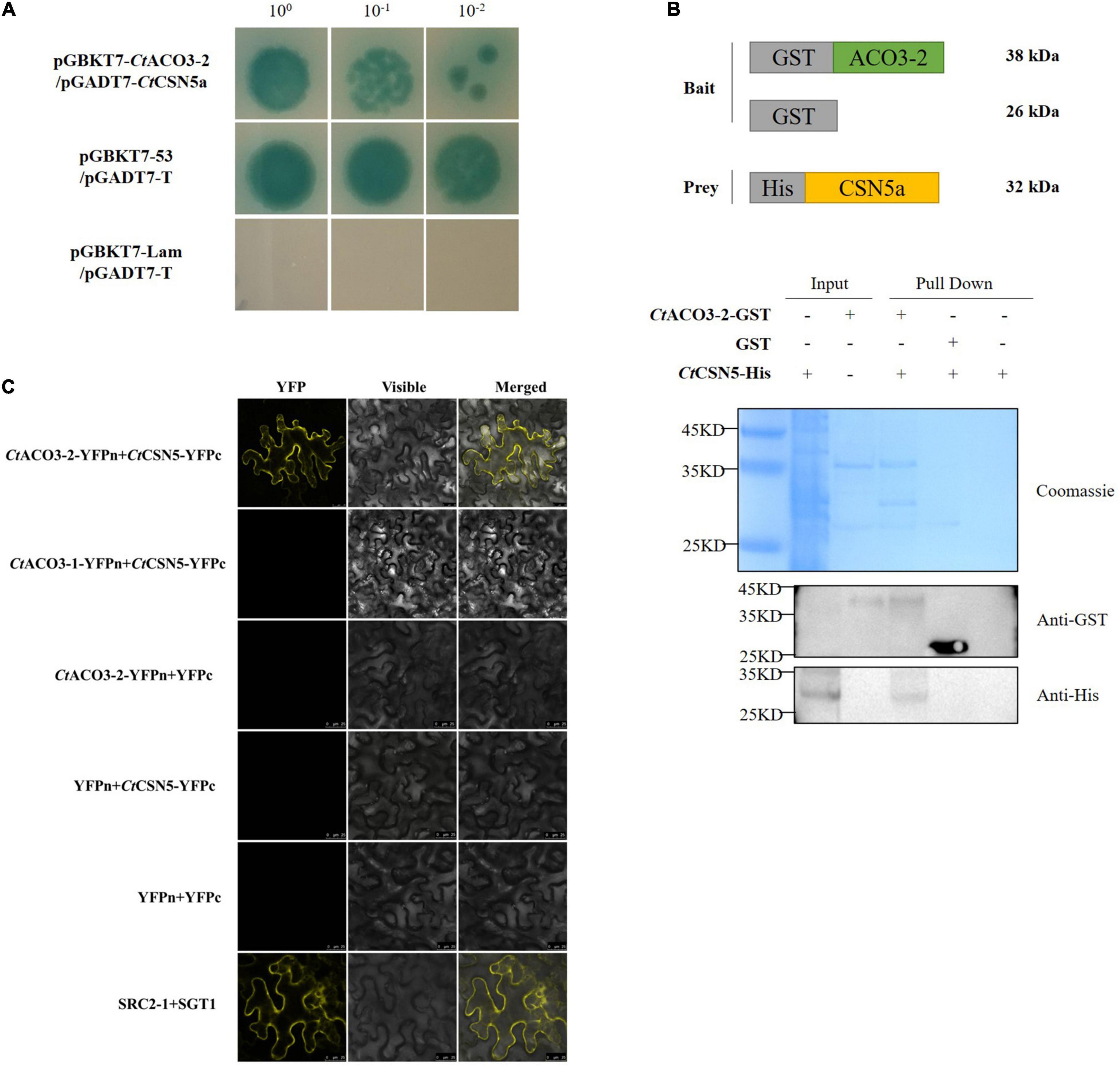
Figure 5. CtACO3-2 interacts with the COP9 signalosome (CSN) subunit 5. (A) Yeast two-hybrid (Y2H) assay showing that CtACO3-2 interact with CtCSN5a. (B) Glutathione S-transferase (GST)-pull down assay showing that CtCSN5 associates with CtACO3-2 in vitro. Purified GST-CtACO3-2 or GST proteins were bound to glutathione-sepharose beads, and then incubated with His-CtCSN5a. Here, we show the protein bands of resulting pull-down products after western blotting with anti-GST and anti-His antibodies. (C) Bifc assay showing that CtCSN5 interact with CtACO3-2 but not CtACO3-1 in vivo.
Our research then demonstrated that CtACO3-2 was associated with CtCSN5a (GenBank accession no. MW075465) using pull-down assay in vitro and BiFC assay in vivo (Figures 5B,C). Therefore, these results strongly indicate that CtACO3-2 is physically associated with CtCSN5a in vitro and in plant.
Wei et al. (2018) have reported that CSN subunit 5 enhanced MYB75 and suppressed GL2 and other genes associated with the TTG1/basic helix–loop–helix (bHLH)/MYB complexes to regulate anthocyanin accumulation. To understand how CtCSN5a participated in flavonoid accumulation regulation in safflower, Y2H screening assay was used to search the proteins that interact with CtCSN5a-BD. After screening and confirming, CtCOI1 (GenBank accession no. MW075466) interacted with CtCSN5a (Supplementary Table 3). COI1 as a subunit of SCF (COI1) E3 ubiquitin ligase encodes an F-box protein, which is required for JA responses. It has been reported that COI1 interacted directly with CSN (Feng et al., 2003). Therefore, we speculated that CtCSN5a regulated the accumulation of flavonoids in safflower through interaction with CtCOI1 (Figures 6A–C).
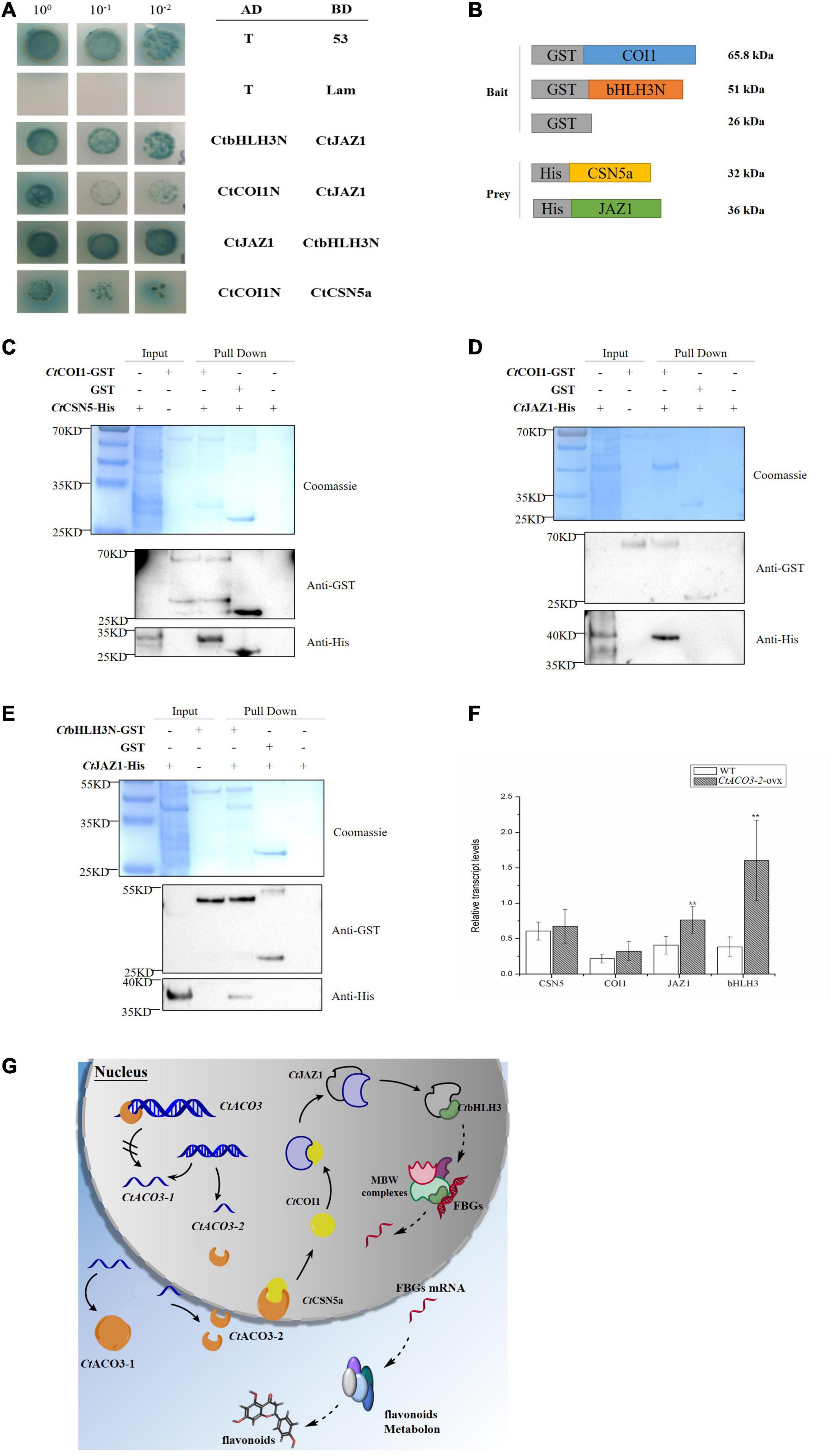
Figure 6. Preliminary molecular mechanism of CtACO3-2 regulates the biosynthesis pathway of flavonoids. (A) Yeast two-hybrid assay showing the protein–protein interaction of CtCSN5-CtCOI1-CtJAZ1-CtbHLH3. (B–E) Demonstration of CtCSN5-CtCOI1-CtJAZ1-CtbHLH3 interaction by GST pull-down assay. Purified GST-CtCOI1, GST-CtbHLH3N, or GST proteins were bound to glutathione-sepharose beads, and then incubated with His-CtCSN5a or His-CtJAZ1. Here, we show the protein bands of resulting pull-down products after western blotting with anti-GST and anti-His antibodies. (F) The relative transcript levels of CtCSN5a, CtCOI1, CtJAZ1, and CtbHLH3 in CtACO3-2 overexpression lines. Data were expressed as mean ± SD (n = 8), **p < 0.01. (G) Schematic diagram illustrating the preliminary molecular mechanism of CtACO3-2 regulate the biosynthesis pathway of flavonoids. The solid line represents the interactions have been proved in the study, the dotted line represents the interactions have not been proved. Arrowheads indicate activations and “//” on arrow indicate inhibitions. The left of figure is two transcripts of CtACO3, and CtACO3-2 inhibits the transcription of CtACO3-1; the right of figure is the network that CtACO3-2 influence the accumulation of flavonoids through CtCSN5-CtCOI1-CtJAZ1-CtbHLH3.
Jasmonate ZIM-domain (JAZ) protein family as a key regulator of JA signaling has been reported physically interacting with SCFCOI1 (Thines et al., 2007). There is broad consensus that the flavonoid pathways are regulated mostly through the coordinated transcription of structural genes by the interaction of MBW complex, such as R2R3 MYB transcription factors, bHLH transcription factors, and WD40 proteins (Payne et al., 2000; Ramsay and Glover, 2005). An et al. reported that JAZ protein interacts with MdbHLH3, which belongs to MBW complex, to regulate the accumulation of anthocyanins in apple (Ni et al., 2020). In the present study, the overexpression of CtACO3-2 resulted in the higher level of downstream structural genes. It is consistent with previous studies in which bHLH3 increased the transcription of downstream structural genes in mulberry fruits and apples (Li et al., 2020; An et al., 2021). The results therefore provided strong evidence for the hypothesis. To further test the hypothesis that JAZ1 interacts with SCFCOI1 and bHLH3 in safflower, we examined a possible physical interaction between CtCOI1 and CtJAZ1, CtbHLH3, and CtJAZ1 using the Y2H system, respectively (Figure 6A). To determine whether JAZ proteins interact with COI1 or bHLH3 in vitro, CtCOI1-GST and CtbHLH3N-GST were performed with CtJAZ1-HIS (Figures 6D,E). Taken together, it was demonstrated that CtJAZ1 (GenBank accession no. MW075468) could physically interact with CtCOI1 and CtbHLH3 (GenBank accession no. MW075469), respectively. It should be noted that these interactions have been examined only in vitro, and the preliminary model is novel and large (Figures 6F,G). The present study may provide novel ideas for flavonoids regulatory network in safflower, but further research is still required.
As a representative bulk Chinese medicine product, a growing number of research about safflower have been demonstrated from a molecular point of view. Results presented in this study revealed the diverse molecular characteristics of CtACO3-2, which influenced the flavonoid accumulation in safflower.
In safflower, the transcript levels of CtACOs in flower were all increased continuously with the floret flowering. However, the expression of different CtACOs showed tissue specificity; CtACO1 had the highest expression in leaf (Tu et al., 2019), whereas both CtACO3-1 and CtACO3-2 expression peaked in flower. These results were in line with the previous research, that ACO had multiple expression characteristics temporally and spatially (Barry et al., 1996; Nakatsuka et al., 1998; Brady et al., 2007; Park et al., 2018). CtACO3-1 is localized in cytosol, whereas CtACO3-2 is localized not only in cytosol but also in the nucleus. The different residues of CtACOs may be the reason for their different characteristics.
Only a few transcriptional factors have been identified for regulating ACO expression (Houben and Van de Poel, 2019), such as SlHB-1 in tomato (Lin et al., 2008), MaERF11 in banana (Han et al., 2016), CmEIN3-like in melon fruit (Huang et al., 2010), and CsWIP1in cucumber (Chen et al., 2016). In the present study, CtACO3-2 banded to the promoter of CtACO3 and repressed the transcription of CtACO3-1. CtACO3-2 is a splice variant of CtACO3 and lacked 5′ coding sequences, which might be the similar manner of TOC1, an autoregulatory response regulator, in Arabidopsis (Strayer et al., 2000). TOC1 encodes a nuclear protein and participates in a feedback loop to control its own expression, and CtACO3-2 encodes a nuclear protein to control CtACO3-1, the full-length transcript expression (Figure 6G). In present study, CtACO3-1-overexpression safflowers had a higher transcript level of CtACO3-2, whereas the level of CtACO3-1 was lower in CtACO3-2-overexpression safflowers. The above phenomenon may be because of the transcriptional regulation of CtACO3-2, while there is not sufficient evidence regarding the transcriptional factor function of CtACO3-2, but this finding deserves further exploration. In parallel, we do not discount the possibility of that the transcription level of CtACO3-1 and CtACO3-2 were coordinated by a transcriptional network as ACOs.
The overexpression of CtACO3-1 and CtACO3-2 promote the accumulation of quinochalcone and flavonol glycosylated derivatives, such as HSYA, carthamin, quercetin glycosylated derivatives, and kaempferol glycosylated derivatives in the present study, whereas CtACO1 suppressed the flavonoid accumulation (Tu et al., 2019). There have been numerous reports on the synthesis of flavonoids regulated by ethylene, CtACO as the key enzyme genes required for ethylene signaling pathway, which may further affect the flavonoid biosynthesis by regulating ethylene synthesis. Meanwhile, we preliminarily constructed a novel pathway, that CtACO3-2 regulated the biosynthesis of flavonoids by CtCSN5a. That may be the reason why CtACOs play various roles in flavonoid accumulation particularly in HSYA biosynthesis, which may be helpful in further work on studying the functions of ACOs, as well as regulating the metabolic flux of active compounds in safflower by appropriate genetic engineering strategies.
At the last step of the ethylene biosynthesis, ACO interacted with biomolecules mostly about that. For example, the flower senescence was affected by the interaction between ACO1 and GRL2 in petunia (Tan et al., 2014). In this study, we identified that CtEXLB (expansion-like) interacted with CtACO3-2 (Supplementary Table 2 and Supplementary Figure 7), as an effective factor of cell division participating in plant development and senescence. At the same time, CSN subunit 5 was found to interact with CtACO3-2 affecting flavonoid accumulation in safflower. Dohmann et al. (2005) reported that the CSN subunit 5 could enhance anthocyanin production in the loss-function Arabidopsis mutants (Dohmann et al., 2005), and CSN subunit 5 could also enhance MYB75 and suppress GL2 expressions associated with the MBW complexes through anthocyanin accumulation regulation (Wei et al., 2018). From the Y2H and GST pull-down results, it is indicated that CtCSN5a interacted with CtCOI1 protein, mediated the interaction between JAZ1 and bHLH3, directly bound to the promoter of flavonoid biosynthesis structural genes, and regulated their transcription. The protein interaction data in the current study provide CtCSN5a protein as the bridge of CtACO3-2 and CtCOI1 protein, whereas CtACO3-2 belongs to the ethylene biosynthesis pathway, and CtCOI1 belongs to the JA signaling pathway.
Flavonoid biosynthesis is regulated by diverse plant hormones, such as ethylene and JA (Flores and Ruiz del Castillo, 2014; An et al., 2018). Rudell and Mattheis reported that JA and ethylene could induce anthocyanin accumulation in apple fruits, synergistically (Rudell and Mattheis, 2008). Ni et al. (2020) found that ethylene could mediate the branching of the JA-induced flavonoid biosynthesis pathway in the red Chinese pear fruit. In safflower, exogenous application of methyl JA increases the accumulation of mostly flavonoids in safflower shown in our previous study (He et al., 2018). This study offers new insight for the common effects of ethylene and JA on flavonoid accumulation in safflower (Figure 7). The accumulation of active ingredients in botanicals is regulated by extensive networks, such as salvianolic acid in Salvia miltiorrhiza, Artemisinin in Artemisia annua (Lv et al., 2017; Deng et al., 2020a,b; Fu et al., 2020; Hao et al., 2020). Therefore, there is a long way to study the acting factors and regulatory networks of flavonoids biosynthesis in safflower.
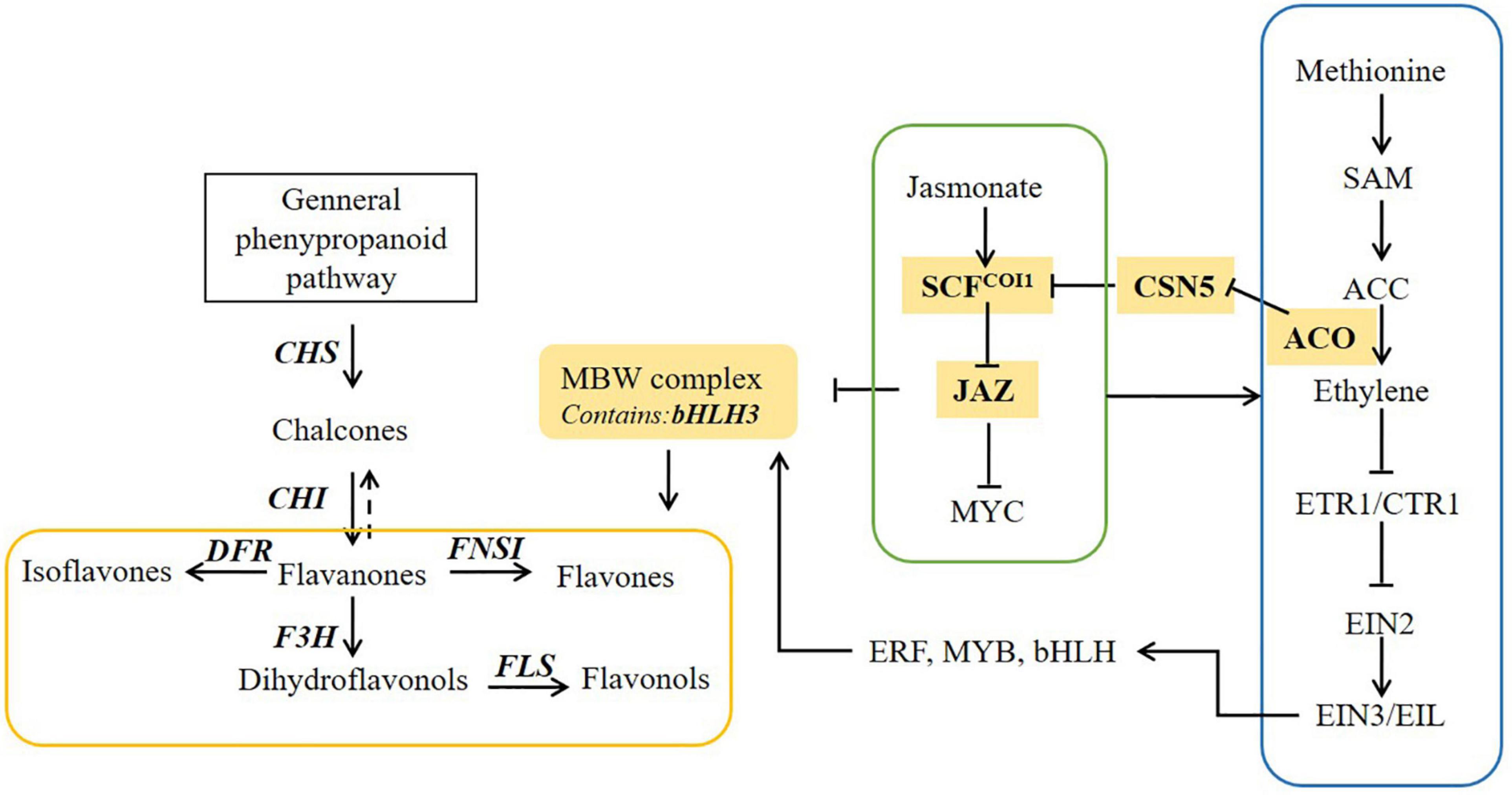
Figure 7. The proposed pathway underlying ethylene and jasmonate (JA) pathway induced flavonoid biosynthesis in safflower.
This study is limited by that the genetic background remains unclear, the medicinal ingredients accumulate in flower and the tissue culture system still difficult. So, in the present study, we were unable to achieve the knockout or knockdown to further verify the function of gene from the opposite side. Besides, the principal limitations of the present study were that the validation of the regulatory network that CtACO3-2 regulates flavonoid synthesis via CtCSN5a was performed only in vitro experiments, further research in vivo is required.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
BH, YZ, and LW contributed to most of the experiments, data analysis, and writing of the manuscript. BH, DG, XJ, JW, and SQ contributed with reagents, materials, and assisted in doing the experiment. MG, YG, and HW designed the experiments, suggested the manuscript outline and guided the writing of the manuscript, and data analysis. All authors read and approved the final manuscript.
This work was sponsored by the National Key R&D Program of China (2019YFC1711100) and the National Natural Science Foundation of China (Grant Nos. 81973421 and 81473300).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.833811/full#supplementary-material
Supplementary Figure 1 | Sequence alignment for ACO protein sequences of Carthamus tinctorius (Ct), Arabidopsis thaliana (At), Solanum lycopersicum (Sl), Malus domestica (Md), Oryza sativa (Os), Ananas comosus (Aco), and Citrus sinensis (Ci). Type I ACO is shown in blue, Type II ACO is shown in yellow, and Type III ACO is shown in green, CtACOs are boxed.
Supplementary Figure 2 | Subcellular localization of the CtACO3-PMT39 fusion protein in onion epidermal cells.
Supplementary Figure 3 | Representative PCR analyses for the specific genes of CtACO3-1-overexpressing plants.
Supplementary Figure 4 | Representative PCR analyses for the specific genes of CtACO3-2-overexpressing plants.
Supplementary Figure 5 | Phenotypic difference between WT and CtACO-overexpressing plants.
Supplementary Figure 6 | Cis-elements in the promoter region of CtACO3-1.
Supplementary Figure 7 | Proteins interact with CtACO3-2 by Y2H.
Supplementary Table 1 | PCR primers used in the text.
Supplementary Table 2 | Proteins interact with CtACO3-2 by Y2H.
Supplementary Table 3 | Proteins interact with CtCSN5a by Y2H.
An, J. P., Wang, X. F., Li, Y. Y., Song, L. Q., Zhao, L. L., You, C. X., et al. (2018). EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 178, 808–823. doi: 10.1104/pp.18.00068
An, J. P., Zhang, X. W., Liu, Y. J., Wang, X. F., You, C. X., and Hao, Y. J. (2021). ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 72, 1460–1472. doi: 10.1093/jxb/eraa525
Barba-Espín, G., Glied, S., Crocoll, C., Dzhanfezova, T., Joernsgaard, B., Okkels, F., et al. (2017). Foliar-applied ethephon enhances the content of anthocyanin of black carrot roots (Daucus carota ssp. sativus var. atrorubens Alef.). BMC Plant Biol. 17:70. doi: 10.1186/s12870-017-1021-7
Barry, C. S., Blume, B., Bouzayen, M., Cooper, W., Hamilton, A. J., and Grierson, D. (1996). Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 9, 525–535. doi: 10.1046/j.1365-313x.1996.09040525.x
Brady, S. M., Orlando, D. A., Lee, J. Y., Wang, J. Y., Koch, J., Dinneny, J. R., et al. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. doi: 10.1126/science.1146265
Chen, H., Sun, J., Li, S., Cui, Q., Zhang, H., Xin, F., et al. (2016). An ACC oxidase gene essential for cucumber carpel development. Mol. Plant. 9, 1315–1327. doi: 10.1016/j.molp.2016.06.018
Costa, D. V., Almeida, D. P., and Pintado, M. (2018). Effect of postharvest application of ethylene on the profile of phenolic acids and anthocyanins in three blueberry cultivars (Vaccinium corymbosum). J. Sci. Food Agric. 98, 5052–5061. doi: 10.1002/jsfa.9042
Deng, C., Shi, M., Fu, R., Zhang, Y., Wang, Q., Zhou, Y., et al. (2020a). ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 71, 5948–5962. doi: 10.1093/jxb/eraa295
Deng, C., Wang, Y., Huang, F., Lu, S., Zhao, L., Ma, X., et al. (2020b). SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 62, 1688–1702. doi: 10.1111/jipb.12943
Dilley, D. R., Wang, Z., Kadirjan-Kalbach, D. K., Ververidis, F., Beaudry, R., and Padmanabhan, K. (2013). 1-Aminocyclopropane-1-carboxylic acid oxidase reaction mechanism and putative post-translational activities of the ACCO protein. AoB Plants 5:plt031. doi: 10.1093/aobpla/plt031
Dohmann, E. M., Kuhnle, C., and Schwechheimer, C. (2005). Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17, 1967–1978. doi: 10.1105/tpc.105.032870
Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S. P., Wei, N., et al. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15, 1083–1094. doi: 10.1105/tpc.010207
Flores, G., and Ruiz del Castillo, M. L. R. (2014). Influence of preharvest and postharvest methyl jasmonate treatments on flavonoid content and metabolomic enzymes in red raspberry. Postharvest Biol. Technol. 97, 77–82. doi: 10.1016/j.postharvbio.2014.06.009
Fu, R., Shi, M., Deng, C., Zhang, Y., Zhang, X., Wang, Y., et al. (2020). Improved phenolic acid content and bioactivities of Salvia miltiorrhiza hairy roots by genetic manipulation of RAS and CYP98A14. Food Chem. 331:127365. doi: 10.1016/j.foodchem.2020.127365
Guo, D., Gao, Y., Liu, F., He, B., Jia, X., Meng, F., et al. (2019). Integrating molecular characterization and metabolites profile revealed CtCHI1’s significant role in Carthamus tinctorius L. BMC Plant Biol. 19:376. doi: 10.1186/s12870-019-1962-0
Guo, D., Xue, Y., Li, D., He, B., Jia, X., Dong, X., et al. (2017). Overexpression of CtCHS1 increases accumulation of quinochalcone in safflower. Front. Plant Sci. 8:1409. doi: 10.3389/fpls.2017.01409
Han, Y. C., Kuang, J., and Chen, J. (2016). Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol. 171, 1070–1084. doi: 10.1104/pp.16.00301
Hao, X., Pu, Z., Cao, G., You, D., Zhou, Y., Deng, C., et al. (2020). Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J. Adv. Res. 23, 1–12. doi: 10.1016/j.jare.2020.01.012
He, B., Xue, Y., Tu, Y., Gao, Y., and Guo, M. (2018). CtCHS4 induces the accumulation of safflower quinone chalcones in response to methyl jasmonate induction. Acta Pharm. Sin. 53, 636–645.
Houben, M., and Van de Poel, B. (2019). 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 10:695. doi: 10.3389/fpls.2019.00695
Huang, S., Sawaki, T., Takahashi, A., Mizuno, S., Takezawa, K., Matsumura, A., et al. (2010). Melon EIN3-like transcription factors (CmEIL1 and CmEIL2) are positive regulators of an ethylene- and ripening-induced 1-aminocyclopropane-1-carboxylic acid oxidase gene (CM-ACO1). Plant Sci. 178, 251–257. doi: 10.1016/j.plantsci.2010.01.005
Jafari, Z., Haddad, R., Hosseini, R., and Garoosi, G. (2012). Cloning, identification and expression analysis of ACC oxidase gene involved in ethylene production pathway. Mol. Biol. Rep. 40, 1341–1350. doi: 10.1007/s11033-012-2178-7
Jeong, S. W., Das, P. K., Jeoung, S. C., Song, J. Y., Lee, H. K., Kim, Y. K., et al. (2010). Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 154, 1514–1531. doi: 10.1104/pp.110.161869
Jonathan, L., Simon, B., Jorma, V., Magnus, H., Jaakko, K., and Björn, S. (2009). Ethylene is an endogenous stimulator of cell division in the cambial meristem of Populus. Proc. Natl. Acad. Sci. U.S.A. 106, 5984–5989. doi: 10.1073/pnas.0811660106
Ke, S., Chen, G., Chen, C., Tzen, J., and Yang, C. (2018). Ethylene signaling modulates contents of catechin and ability of antioxidant in Camellia sinensis. Bot. Stud. 59:11. doi: 10.1186/s40529-018-0226-x
Keita, T., Kentaro, I., Kiwamu, M., Hiroshi, K., and Hiroshi, E. (2005). Expression of a mutated melon ethylene receptor gene Cm-ETR1/H69A affects stamen development in Nicotiana tabacum. Plant Sci. 169, 935–942. doi: 10.1016/j.plantsci.2005.06.012
Kumar, D., Sarmah, B. K., and Das, P. K. (2019). Ethylene mediates repression of anthocyanin accumulation in black rice pericarps in the absence of light. J. Plant Physiol. 236, 34–38. doi: 10.1016/j.jplph.2019.02.011
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, H., Yang, Z., Zeng, Q., Wang, S., Luo, Y., Huang, Y., et al. (2020). Abnormal expression of bHLH3 disrupts a flavonoid homeostasis network, causing differences in pigment composition among mulberry fruits. Hortic. Res. 7:83. doi: 10.1038/s41438-020-0302-8
Lin, Z., Hong, Y., Yin, M., Li, C., Zhang, K., and Grierson, D. (2008). A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 55, 301–310. doi: 10.1111/j.1365-313X.2008.03505.x
Lin, Z., Zhong, S., and Grierson, D. (2009). Recent advances in ethylene research. J. Exp. Bot. 60, 3311–3336. doi: 10.1093/jxb/erp204
Liu, H., Wang, Q., Liu, Y., Zhao, X., Imaizumi, T., Somers, D. E., et al. (2013). Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 110, 17582–17587. doi: 10.1073/pnas.1308987110
Lv, Z., Zhang, L., and Tang, K. (2017). New insights into artemisinin regulation. Plant Signal. Behav. 12:e1366398. doi: 10.1080/15592324.2017.1366398
Nakatsuka, A., Murachi, S., Okunishi, H., Shiomi, S., Nakano, R., Kubo, Y., et al. (1998). Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant Physiol. 118, 1295–1305. doi: 10.1104/pp.118.4.1295
Ni, J., Zhao, Y., Tao, R., Yin, L., Gao, L., and Strid, Å (2020). Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol. J. 18, 1223–1240. doi: 10.1111/pbi.13287
Park, C. H., Roh, J., Youn, J. H., Son, S. H., Park, J. H., Kim, S. Y., et al. (2018). Arabidopsis ACC oxidase 1 coordinated by multiple signals mediates ethylene biosynthesis and is involved in root development. Mol. Cells 41, 923–932.
Payne, C. T., Zhang, F., and Lloyd, A. M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. doi: 10.1093/genetics/156.3.1349
Petroni, K., and Tonelli, C. (2011). Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 181, 219–229. doi: 10.1016/j.plantsci.2011.05.009
Rahim, M. A., Busatto, N., and Trainotti, L. (2014). Regulation of anthocyanin biosynthesis in peach fruits. Planta 240, 913–929. doi: 10.1007/s00425-014-2078-2
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Rudell, D. R., and Mattheis, J. P. (2008). Synergism exists between ethylene and methyl jasmonate in artificial light-induced pigment enhancement of ‘Fuji’ apple fruit peel. Postharvest Biol. Technol. 47, 136–140. doi: 10.1016/j.postharvbio.2007.05.021
Rukh, G., Ahmad, N., Rab, A., Ahmad, N., Fazal, H., Akbar, F., et al. (2019). Photo-dependent somatic embryogenesis from non-embryogenic calli and its polyphenolics content in high-valued medicinal plant of Ajuga bracteosa. J. Photochem. Photobiol. B 190, 59–65. doi: 10.1016/j.jphotobiol.2018.11.012
Sell, S., and Hehl, R. (2005). A fifth member of the tomato 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase gene family harbours a leucine zipper and is anaerobically induced. DNA Seq. 16, 80–82. doi: 10.1080/10425170500050817
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Más, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Tahara, S. (2007). A journey of twenty-five years through the ecological biochemistry of flavonoids. Biosci. Biotechnol. Biochem. 71, 1387–1404. doi: 10.1271/bbb.70028
Tan, Y., Liu, J., Huang, F., Guan, J., Zhong, S., Tang, N., et al. (2014). PhGRL2 protein, interacting with PhACO1, is involved in flower senescence in the petunia. Mol. Plant 7, 1384–1387. doi: 10.1093/mp/ssu024
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Tohge, T., de Souza, L. P., and Fernie, A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68, 4013–4028. doi: 10.1093/jxb/erx177
Tu, Y., He, B., Gao, S., Guo, D., Jia, X., Dong, X., et al. (2019). CtACO1 overexpression resulted in the alteration of the flavonoids profile of safflower. Molecules 24:1128. doi: 10.3390/molecules24061128
Tu, Y., Liu, F., Guo, D., Fan, L., Zhu, Z., Xue, Y., et al. (2016). Molecular characterization of flavanone 3-hydroxylase gene and flavonoid accumulation in two chemotyped safflower lines in response to methyl jasmonate stimulation. BMC Plant Biol. 16:132. doi: 10.1186/s12870-016-0813-5
Tu, Y., Xue, Y., Guo, D., Sun, L., and Guo, M. (2015). Carthami flos: a review of its ethnopharmacology, pharmacology and clinical applications. Rev. Bras. Farmacogn. 25, 553–566. doi: 10.1016/j.bjp.2015.06.001
Van de Poel, B., Smet, D., and Van Der Straeten, D. (2015). Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72. doi: 10.1104/pp.15.00724
Watkins, J., Hechler, P., and Muday, G. (2014). Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiol. J. 164, 1707–1717. doi: 10.1104/pp.113.233528
Wei, S., Li, X., Gruber, M. Y., Feyissa, B. A., Amyot, L., and Hannoufa, A. (2018). COP9 signalosome subunit 5A affects phenylpropanoid metabolism, trichome formation and transcription of key genes of a regulatory tri-protein complex in Arabidopsis. BMC Plant Biol. 18:134. doi: 10.1186/s12870-018-1347-9
Yuk, H., Song, Y., Curtis-Long, M., Kim, D., Woo, S., Lee, Y., et al. (2016). Ethylene induced a high accumulation of dietary isoflavones and expression of isoflavonoid biosynthetic genes in soybean (Glycine max) leaves. J. Agric. Food Chem. 64, 7315–7324. doi: 10.1021/acs.jafc.6b02543
Keywords: safflower (Carthamus tinctorius L.), flavonoids biosynthesis, HSYA, regulating mechanism, CtACO3
Citation: He B, Zhang Y, Wang L, Guo D, Jia X, Wu J, Qi S, Wu H, Gao Y and Guo M (2022) Both Two CtACO3 Transcripts Promoting the Accumulation of the Flavonoid Profiles in Overexpressed Transgenic Safflower. Front. Plant Sci. 13:833811. doi: 10.3389/fpls.2022.833811
Received: 12 December 2021; Accepted: 23 February 2022;
Published: 06 April 2022.
Edited by:
Wei Zhou, Zhejiang Chinese Medical University, ChinaReviewed by:
Jiang Chen, Chengdu University of Traditional Chinese Medicine, ChinaCopyright © 2022 He, Zhang, Wang, Guo, Jia, Wu, Qi, Wu, Gao and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Wu, ZG9jdG9yd2g2NjZAMTI2LmNvbQ==; Yue Gao, Z2FveXVlMjAwMEBob3RtYWlsLmNvbQ==; Meili Guo, bWxndW9AMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.