- 1College of Life Science and Food Engineering, Huaiyin Institute of Technology, Huai’an, China
- 2Huai’an Key Laboratory for Facility Vegetables, Huaiyin Institute of Agricultural Sciences of Xuhuai Region in Jiangsu, Huai’an, China
CONSTANS-like (COL) genes play important regulatory roles in multiple growth and development processes of plants but have rarely been studied in Capsicum annuum. This study explored the evolutionary relationship and expression patterns of COL genes from C. annuum. A total of 10 COL genes were identified in the genome of the cultivated pepper Zunla-1 and were named CaCOL01–10. These genes were unequally distributed among five chromosomes and could be divided into three groups based on differences in gene structure characteristics. During evolutionary history, duplications and retentions were divergent among different groups of COL genes. Tandem duplication caused amplification of group I genes. Genetic distance among COL genes was the largest in group III, suggesting that group III genes undergo more relaxed selection pressure compared with the other groups. Expression patterns of CaCOLs in tissues were significantly different, with CaCOL08 exhibiting the highest expression in stem and leaf. Some COL orthologous genes showed markedly different expression patterns in pepper compared with tomato, such as COL_1 orthologs, which may be involved in fruit development in pepper. In addition, CaCOLs participated in the regulation of abiotic stresses to varying degrees. Five CaCOL genes were induced by cold, and CaCOL02 and CaCOL03 were specifically upregulated by cold and downregulated by heat. This study provides a theoretical basis for the in-depth understanding of the functions of COL genes in pepper and their molecular mechanisms involved in growth and development and responses to abiotic stresses.
Introduction
CONSTANS (CO), a crucial gene in the photoperiod flowering induction pathway of plants, is located in the circadian clock output pathway and promotes the flowering of plants by activating expression of the downstream floral gene FLOWERING LOCUS T (FT) (Putterill et al., 1995). CO was subsequently found to belong to a class of genes called the CONSTANS-like (COL) gene family (Griffiths et al., 2003). The COL gene family is widely distributed and the members vary greatly among different plant species. For example, 17 COL genes were identified in Arabidopsis thaliana (Griffiths et al., 2003), 9 in barley (Griffiths et al., 2003), 16 in rice (Griffiths et al., 2003), 26 in soybean (Wu et al., 2014), 42 in cotton (Cai et al., 2017), 19 in maize (Song et al., 2018), 20 in radish (Hu et al., 2018), 13 in tomato (Yang et al., 2020), and 22 in sunflower (Niu et al., 2022). COL genes are characterized based on two conservative domains. The first is the CONSTANS, CONSTANS-LIKE, TOC1 (CCT) domain at the C-terminus, which comprises 43 amino acids and conservatively plays a role in promoter elements binding, transcription regulation, and nuclear localization. The N-terminus of COL genes generally contains one to two B-box type zinc finger structures that can interact with ubiquitin ligase, receptor proteins, and transcriptional regulators (Robson et al., 2001; Valverde et al., 2004). According to the number and conservation of B-boxes, the A. thaliana COL gene family can be divided into three groups (Griffiths et al., 2003). Furthermore, in most other studied plants, the COL gene family clusters into three groups with strong bootstrap values. Both group I and group III genes stably contain two B-box domains in terrestrial plants before Amborella trichopoda. After the angiosperm whole-genome duplication (WGD) events, the first B-box domain in group I and the second B-box domain in group III were lost in some plant species (Song et al., 2015). Differentiation of the B-box domain has a crucial role in the phyletic evolution of COL genes (Song et al., 2015; Niu et al., 2022).
As transcription factors, CO was the first cloned in A. thaliana, regulating the flowering time in the photoperiod pathway (Putterill et al., 1995; Li and Xu, 2017). The function of CO may be conserved across the angiosperms. For example, CrCO (of Chlamydomonas reinhardtii) was overexpressed in A. thaliana to make flowering in advance (Tenorio et al., 2003), and PtCO was found to promote the early flowering of plants (Bohlenius et al., 2006). However, during evolutionary history, COL genes often exist in plants as multiple copies, displaying apparent functional differentiation in different plants. Moreover, not all the genes of this family are photoperiod related; several reports have demonstrated that COL genes are associated with plant development and even abiotic stress responses. CrCO may also participate in cell division and regulation of starch synthesis (Tenorio et al., 2003). In Populus, PtCO was involved in plant growth and bud differentiation (Bohlenius et al., 2006). AtCOL3 promoted the development of lateral roots and shoot branches (Datta et al., 2006). VvCOL1 was involved in regulating the maintenance and induction of bud dormancy in grape (Almada et al., 2009). MaCOL1, a transcriptional activator in banana, was reported to be associated with fruit ripening and abiotic stress tolerance (Chen et al., 2012). AtCOL4 was proven to positively regulate tolerance to abiotic stresses (acid, salt, and osmotic stress) in the plant (Min et al., 2015). In rice, Ghd2 participated in regulating leaf senescence and drought tolerance (Liu J. et al., 2016). OsCOL9 was reported to regulate disease resistance dependent on ethylene and salicylic acid (SA) signaling pathways (Liu H. et al., 2016). BnCOL2 in Brassica napus responded to drought stress (Liu et al., 2020), while HaCOL3, HaCOL6, and HaCOL19 were induced by cadmium (Cd), heat, and drought stresses in sunflower (Niu et al., 2022). In adapting to adverse environments, COL genes have developed essential new functions in response to abiotic stresses.
CONSTANS-like genes have been studied in many plants, but there are no systematic reports of COLs in Capsicum annuum, a vital vegetable crop. Publication of the whole-genome sequence and high-quality physical map of C. annuum has laid the foundation for phylogenetic evolution and functional research of COL genes (Kim et al., 2014; Qin et al., 2014). Systematically analyzing conserved domains, evolutionary relationships, and the functional expression of CaCOL genes would provide theoretical support for further investigation of the molecular mechanism of CaCOL genes and vegetable-breeding research.
Results
Members of the CaCOL Gene Family
Based on genome-wide identification and validation, all 10 COL gene family members were identified in the C. annuum cv. Zunla-1 genome and were named CaCOL01-CaCOL10 in order of chromosomal location (Table 1). Simultaneously, nine COLs were identified in C. annuum cv. CM334 (Supplementary Table 1). The lengths of the 10 COL genes in the “Zunla-1” genome varied, with a maximum of 1410 bp (CaCOL09) and a minimum of 1,032 bp (CaCOL01). The molecular weights of CaCOLs were predicted to be between 37.93∼51.67 kD, and the isoelectric points were concentrated in 4.85∼7.07 (Table 1). Comparative analysis indicated that CaCOL genes and AtCOL genes had high sequence similarity (E-value < 5E-44).
Phylogenetic and Structure Characterization of CaCOLs
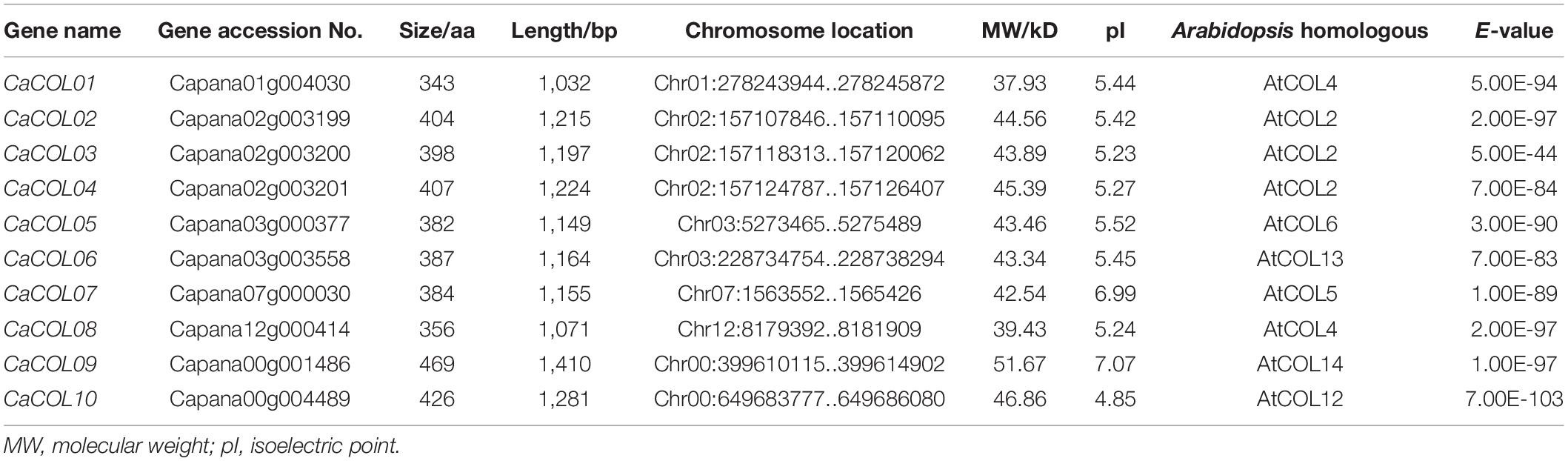
To understand the phylogenetic evolution of CaCOLs, the COL sequences of A. thaliana, tomato, and C. annuum cv. Zunla-1 and CM334 were selected for analysis. The COLs were divided into three groups (I, II, and III). In “Zunla-1,” group I contained six COLs (CaCOL01, CaCOL02, CaCOL03, CaCOL04, CaCOL07, and CaCOL08), which was similar to the number of COLs in group I of A. thaliana and tomato. Group II of “Zunla-1” contained only one gene (CaCOL05), while group III contained three COLs (CaCOL06, CaCOL09, and CaCOL10) (Figure 1A). In the “CM334” genome, there were five COLs in group I, with one missing compared to “Zunla-1,” groups II and III contained the same number of COLs as “Zunla-1.” There were eight pairs of homologous COLs in the two varieties of C. annuum (Figure 1A).
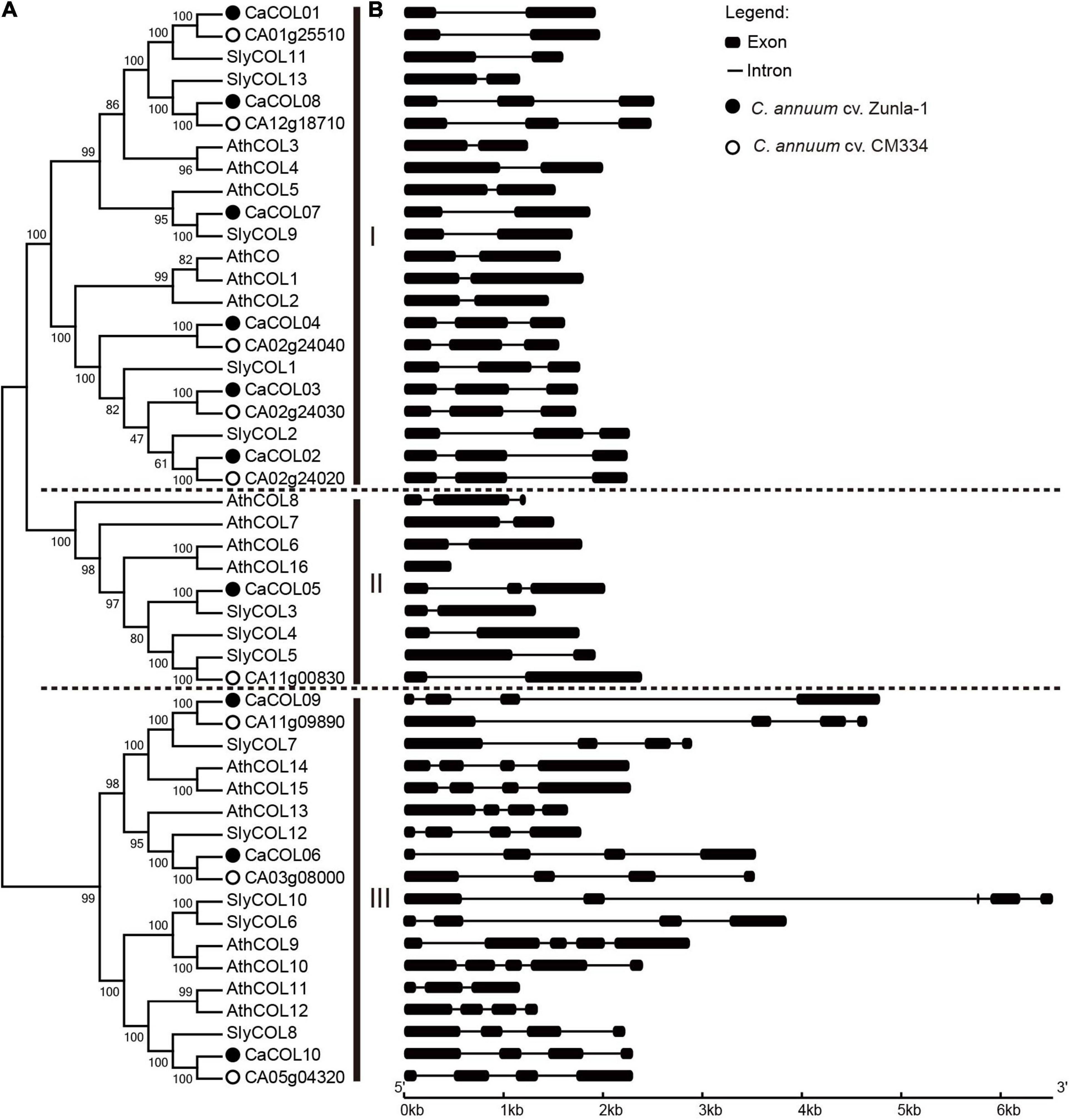
Figure 1. Phylogenetic analysis (A) and gene structures (B) of COL homologs in Capsicum annuum, Solanum lycopersicum, and Arabidopsis thaliana. The phylogenetic relationship of COLs categorized them into three groups (I, II, and III). COLs of C. annuum Zunla-1 and CM334 are indicated by black balls and hollow circles, respectively. Exons and introns are represented by black boxes and black lines, respectively.
There was apparent differentiation in gene structure characteristics among the three subgroups of COLs, but a relatively similar gene structure was present within the groups (Figure 1B). The number of exons in groups I and II was approximately two to three but reached approximately five in group III. In addition, the intron lengths of COLs in groups I and II were similar. CaCOL09 and SlyCOL10, contained longer introns than the other genes, exceeding 4 kb. Such phenomenon of intron-exon structural alienation may be due to differentiation of gene function during evolution.
Through analysis of the conserved domains of A. thaliana and C. annuum cv. Zunla-1 COL proteins (Figure 2), 10 conserved motifs were identified and annotated. Each COL protein contained Motif 1 and 2, indicating that the CCT domain and the B-box domain were the essential functional domains. However, in the three subgroups, there was significant differentiation of B-box domains. Groups I and III both contained two B-boxes, but only one B-box domain was conserved, with the first B-box in group I and the second B-box in group III exhibiting markedly different structures. There was only one B-box domain in group II (Figure 2). Among the three subgroups, group I had the most complex structure and contained seven types of motifs, group II had the simplest motif type, with only three types, and group III contained four types of motifs. COL homologous proteins of A. thaliana and C. annuum had similar conserved domains, indicating that they may have similar functions (Figure 2).
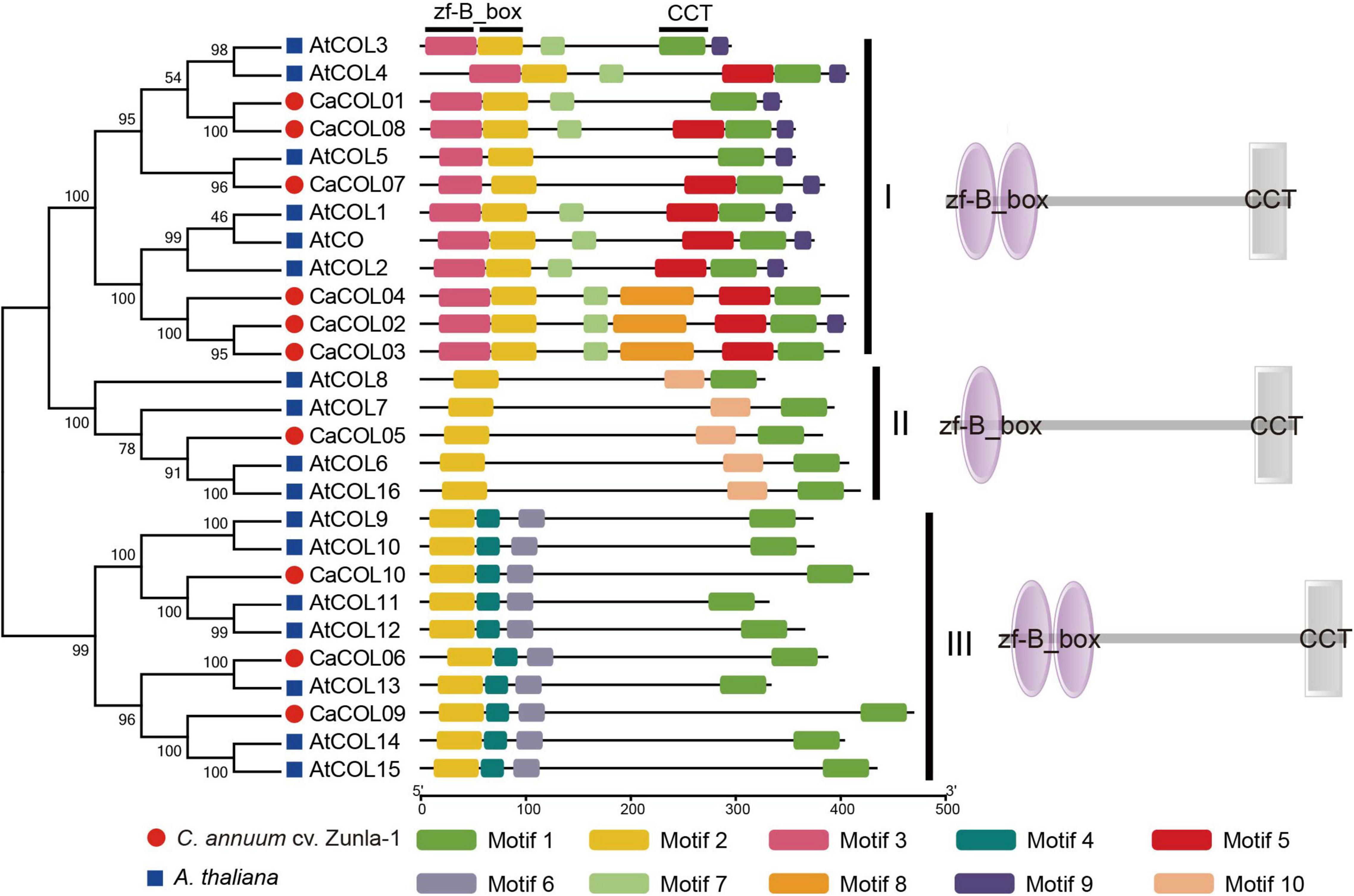
Figure 2. Analysis of conserved domains of COL proteins in Capsicum annuum and Arabidopsis thaliana. CaCOLs and AtCOLs are indicated by red balls and blue boxes, respectively. B-box and CCT domains were annotated by the SMART database.
Cis-Elements Analysis in the Promoters of CaCOL Genes
The cis-elements in the upstream 2-kb regions of the CaCOL gene sequences were analyzed to predict the functions of CaCOLs. The promoter regions of CaCOL genes contained different types and numbers of cis-elements (Figure 3). All 10 genes contained light response cis-elements, the stress response cis-element (MYB), and the cold resistance cis-element recognition site (MYC). In addition, two of the CaCOL genes contained the drought response cis-element (MBS), and nine of the genes contained the abscisic acid response cis-element (ABRE). The CaCOL genes also included a variety of hormone response cis-elements. These findings indicated that CaCOLs might participate in plant growth, development, and resistance to adversity stresses.
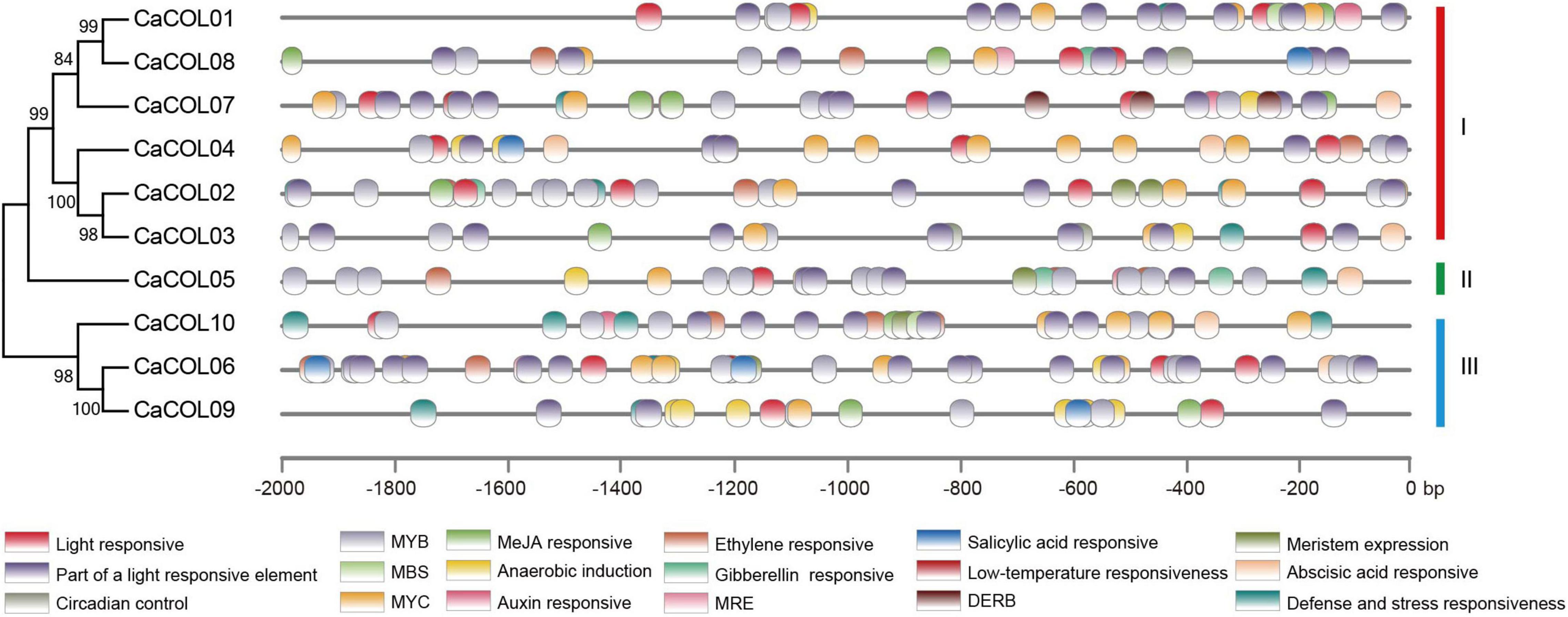
Figure 3. Predicted cis-elements in the promoter regions of CaCOL genes. Promoter sequences (-2000 bp) of CaCOL genes were analyzed. The elements are identified as different colored boxes.
Chromosomal Location and Homologous Genes of CaCOL Genes
Chromosome location analysis showed that eight of the ten CaCOL genes were anchored on five chromosomes of C. annuum—Chr01, Chr02, Chr03, Chr07, and Chr12 (Figure 4A). Three tandem duplicated genes were found on Chr02 (Figure 4A).
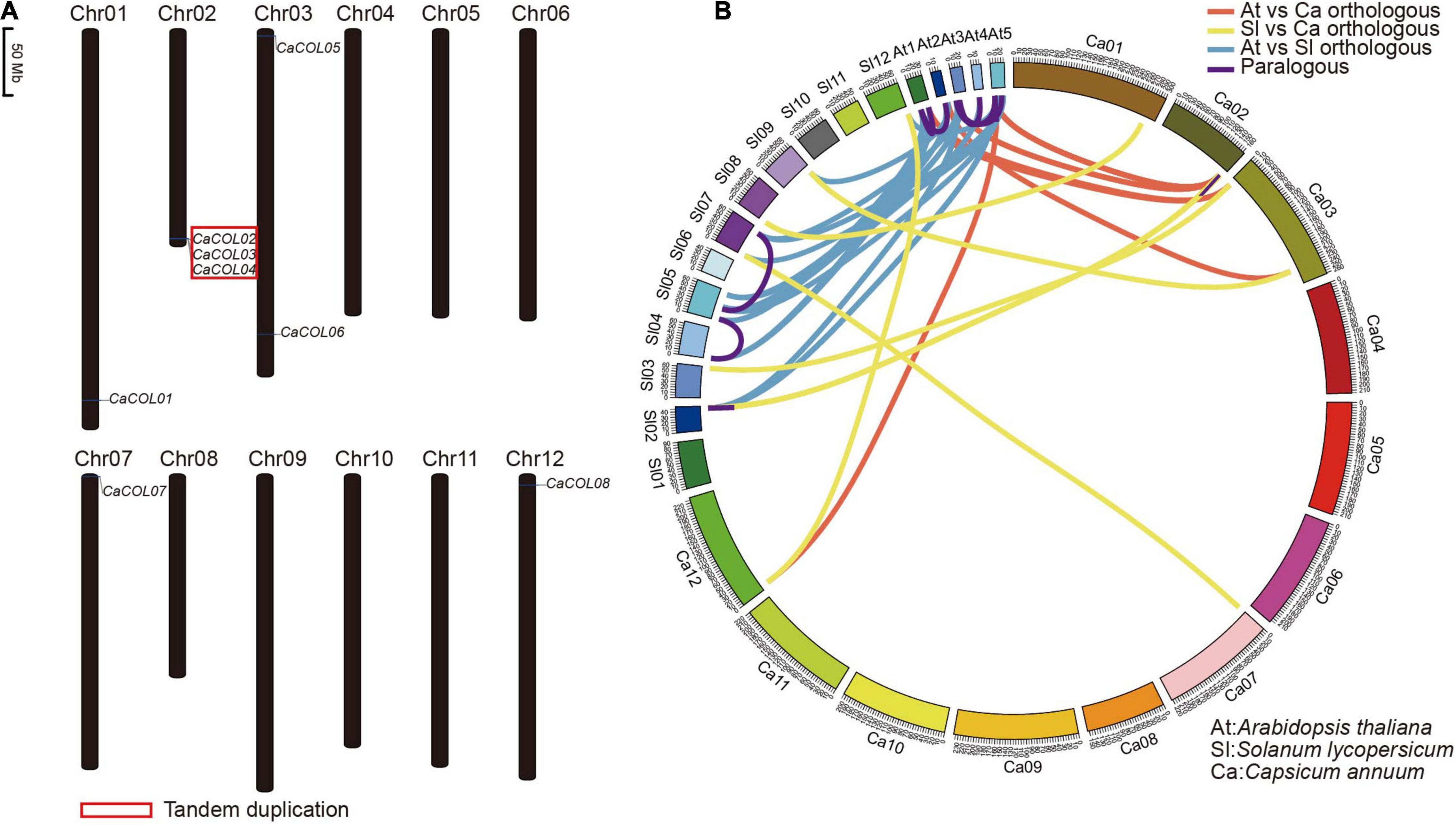
Figure 4. Chromosomal localization of CaCOL genes (A) and the relationship of COL homologous genes among Capsicum annuum, Solanum lycopersicum, and Arabidopsis thaliana (B). The red box indicates tandem duplicated genes. Lines showing orthologous gene pairs between A. thaliana and C. annuum are red; those between S. lycopersicum and C. annuum are yellow; and those between A. thaliana and S. lycopersicum are blue. Lines showing paralogous gene pairs are pink.
COL homologous analysis identified six orthologous gene pairs between A. thaliana and C. annuum, eight pairs between A. thaliana and tomato, and 13 pairs between C. annuum and tomato. In addition, only three paralogous gene pairs were found in C. annuum and tomato, while seven were identified in A. thaliana (Figure 4B and Supplementary Table 2).
Evolution Pattern Analysis of CONSTANS-Like Genes
CONSTANS-like gene family members of 12 plant species (Arabidopsis thaliana, Brassica rapa, Carica papaya, Populus trichocarpa, Fragaria vesca, Vitis vinifera, Solanum tuberosum, Solanum lycopersicum, C. annuum cv. Zunla-1, Zea mays, Oryza sativa, and Amborella trichopoda) were selected to reconstruct a phylogenetic tree (Supplementary Table 3) and further explore the evolutionary history of COLs. In the chosen angiosperms, A. trichopoda is one of the species known to exist for the longest time among existing angiosperms without experiencing ancient hexaploid (γ) events, while C. papaya, P. trichocarpa, and V. vinifera did not undergo α and β duplications, and Solanaceae crops have experienced unique whole-genome triplication (WGT) events. All COL genes were divided into three subgroups: groups I, II, and III (Figure 5A). Genetic distance was analyzed to further determine the relationship among the three subgroups (Figure 5B). The box plot revealed that genetic distances of the COL genes were the smallest in group I and the largest in group III, indicating that the degree of sequence divergence of group I was the lowest. The genetic distance of group II versus group III was higher than those of group I versus group II and group I versus group III, indicating that groups II and III may be differentiated from group I during the evolutionary process. The genetic distance was bigger among the three groups than within the three groups, confirming that the COL gene family was divided into three groups.
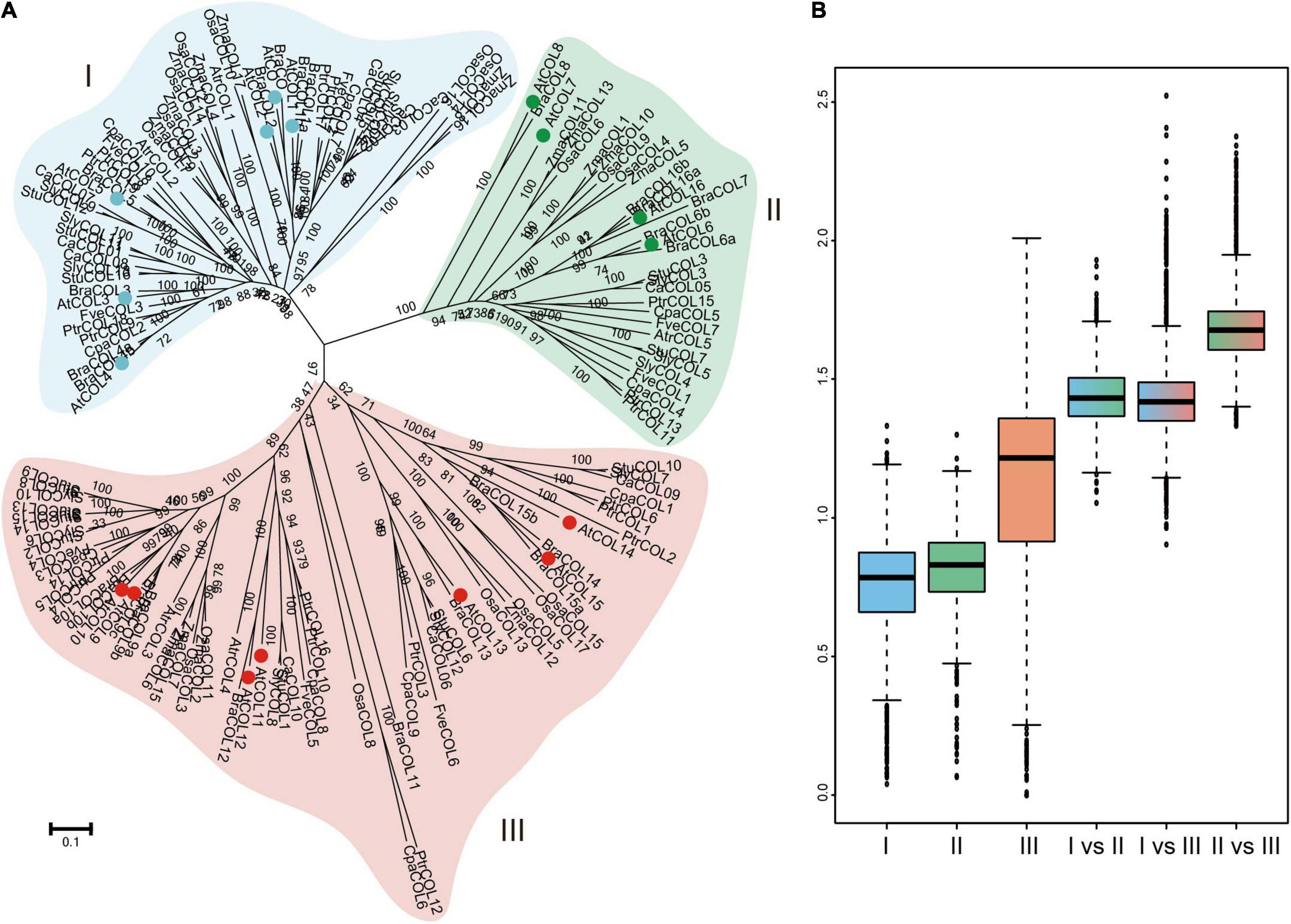
Figure 5. Phylogenetic relationship of COL gene family members in 12 selected plant species (A) and nucleotide distance of group I, II, and III COL genes (B). Group I, II and III of AtCOLs are indicated by blue, green and red balls, respectively.
Copy number variation of the COL family in the 12 selected plant species was subsequently determined (Figure 6). The COLs in A. trichopoda had already been divided into three subgroups (Figure 6). In A. thaliana and B. rapa, the cruciferous plants, groups I and III had the most apparent gene amplification (Figure 6). The number of COL genes in Z. mays and O. sativa was the same, but group III genes of O. sativa had been significantly amplified and differentiated, indicating that these genes may play a regulatory role in the growth of O. sativa (Figure 6). In the three Solanaceae crops, the copy number of COL genes was significantly different; S. tuberosum contained the most, while C. annuum had the least. Moreover, in C. annuum, only one group II gene was retained (Figure 6).
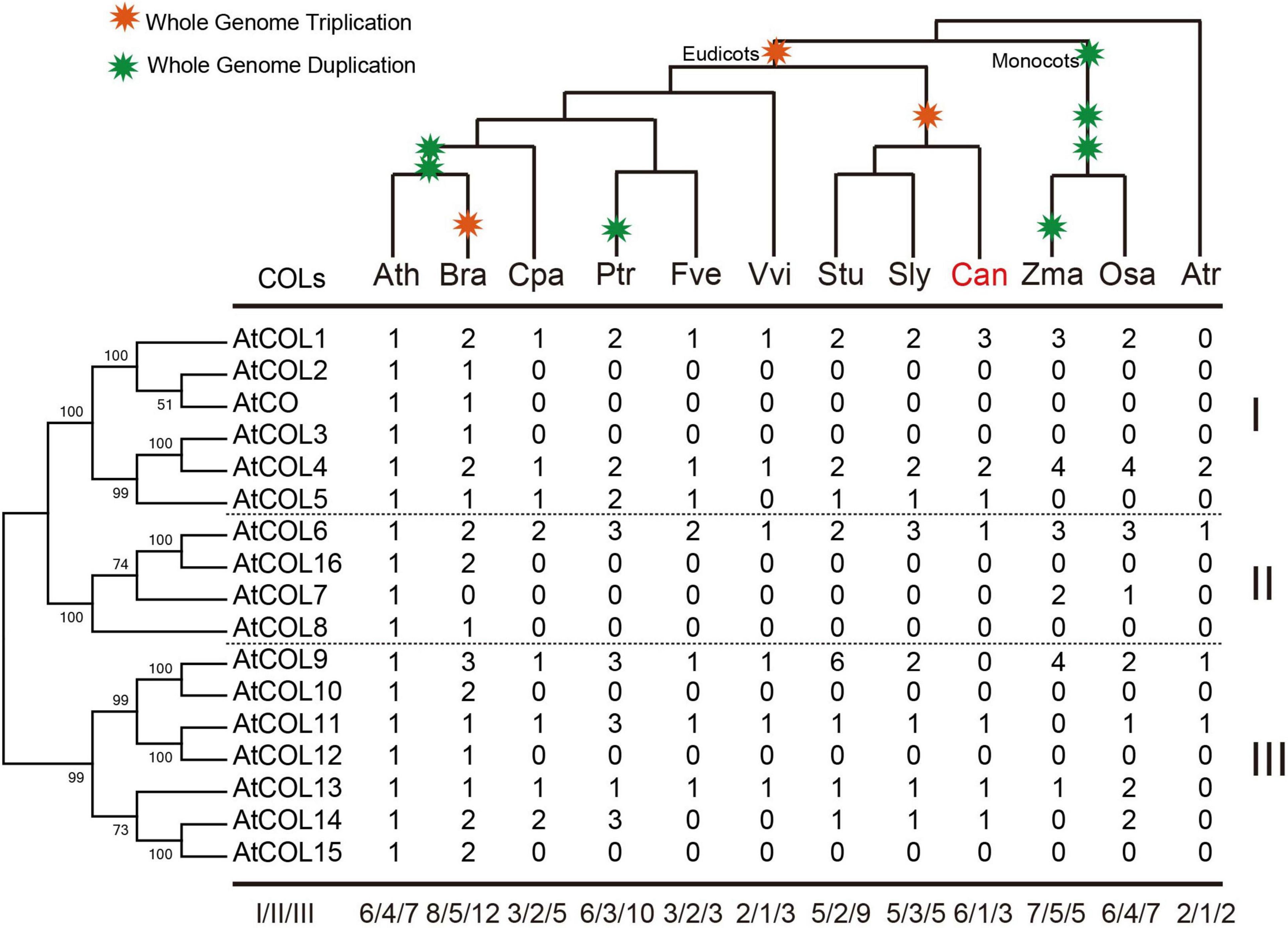
Figure 6. Copy number variation of the COL family in 12 selected plant species. The phylogenetic tree of AtCOLs is shown on the left, and the species tree is shown at the top. Whole-genome duplication and triplication events are indicated on the tree branches according to the Plant Genome Duplication Database. Numbers are copy numbers of each gene in Arabidopsis thaliana (Ath), Brassica rapa (Bra), Carica papaya (Cpa), Populus trichocarpa (Ptr), Fragaria vesca (Fve), Vitis vinifera (Vvi), Solanum tuberosum (Stu), Solanum lycopersicum (Sly), Capsicum annuum (Can), Zea mays (Zma), Oryza sativa (Osa), and Amborella trichopoda (Atr). Statistics of copy number variation of the COL genes are based on the phylogenetic relationships.
Expression Divergence of the CaCOL Genes in Different Tissues
To investigate the divergence of COL homologs, the expression patterns of CaCOL genes were analyzed based on the transcriptome data of different tissues and fruit development stages of C. annuum cv. Zunla-1. The expression patterns of CaCOL genes showed tissue specificity and a high degree of similarity between genes from the same clade of the phylogenetic tree (Figure 7A). In group I, the genes were clustered into two clades. The first clade included CaCOL01, CaCOL07, and CaCOL08, All three of these genes exhibited high expression in the primary tissues, with CaCOL08 showing the highest expression of all CaCOL genes in the stem and leaf, while CaCOL07 was expressed at a low level once the fruit had turned color. The second clade of group I comprised CaCOL02, CaCOL03, and CaCOL04, and these genes were either not expressed or were expressed at low levels in most of the investigated tissues, except for CaCOL02, which was only expressed at moderate levels in the stem. The sole gene of group II—CaCOL05—was expressed at high levels in the stem and leaf but exhibited low or no expression in other tissues. In group III, CaCOL06, CaCOL09, and CaCOL10 showed two distinct expression patterns. CaCOL10 was barely expressed in all tissues except in the stem, while COL06 and CaCOL09 were expressed at high or moderate levels in all tissues tested. These findings indicate that CaCOL genes play a crucial role in plant growth and undergo functional differentiation (Figure 7A).
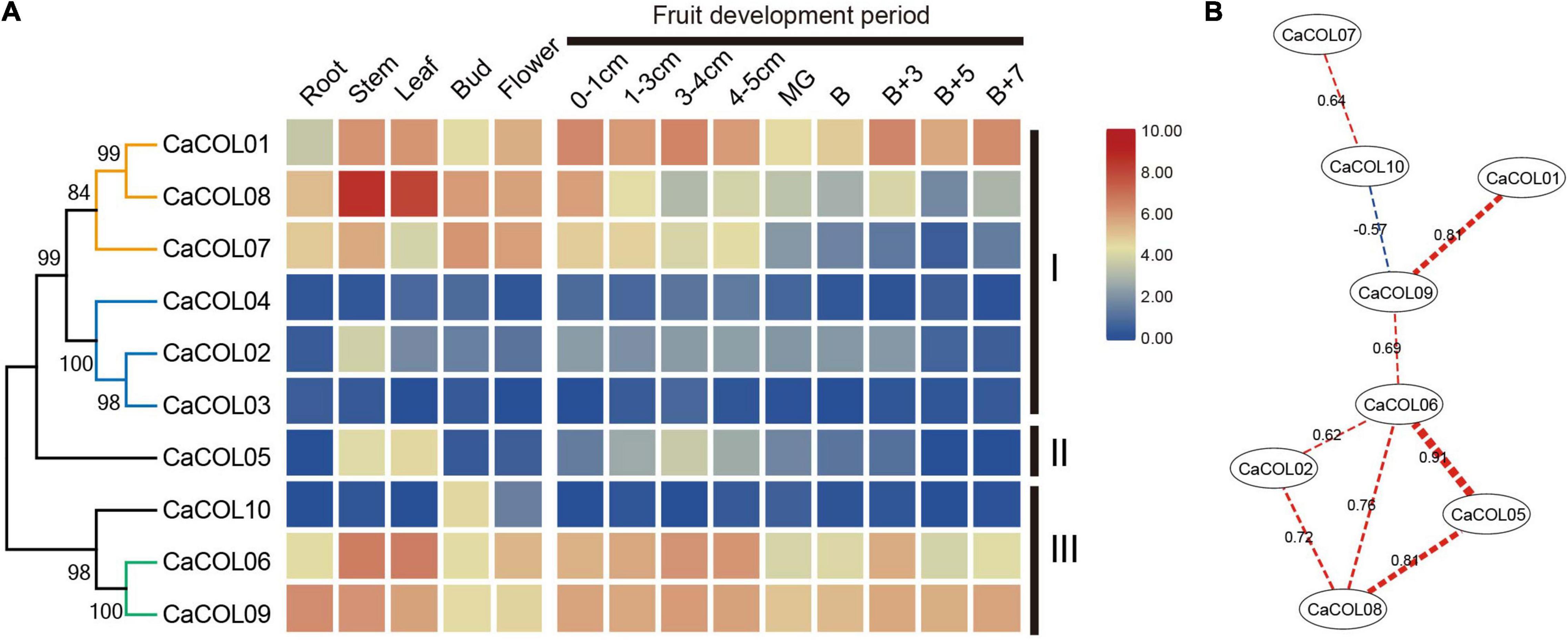
Figure 7. Expression patterns (A) and co-expression networks (B) of CaCOL genes in different tissues and at different stages of the fruit development process. Co-expression networks of CaCOL genes were established based on the PCCs of these gene pairs using transcriptome data, which involved eight nodes and nine regulatory edges.
To further understand the correlation between CaCOL genes and their homologous genes, correlation between the expression trends of CaCOL genes was explored by calculating Pearson Correlation Coefficient (PCC) values and then constructing a co-regulatory network by screening significantly co-expressed gene pairs (Figure 7B). CaCOL06 and CaCOL09 were at the central node of the co-expression network, and CaCOL06 had a strong co-expression relationship with four genes simultaneously, indicating that this gene may be at the central node position in the C. annuum growth and development regulatory network (Figure 7B).
Subsequently, transcriptome data of different tissues and developmental stages of C. annuum cv. CM334 and tomato were analyzed (Supplementary Figures 1, 2), and the expression patterns of seven groups of COL orthologous genes were compared (Supplementary Figure 3). The expression patterns of COL orthologous genes were both similar and opposite in pepper and tomato. Four of the seven groups of COL orthologous genes showed almost identical expression patterns, namely COL_2, COL_5, COL_6, and COL_7. Orthologous genes in the COL_2 group were highly expressed in leaf tissues of both tomato and pepper, and at low levels during fruit development, especially at breaker stage. In the COL_5 group, the orthologous genes were expressed highly in bud tissues. Both COL_6 and COL_7 orthologous genes were expressed in all tissues tested, but expression levels were higher in pepper compared with tomato (Supplementary Figure 3). The other three groups of orthologous genes (COL_1, COL_3, and COL_4) showed different expression patterns in the two plant species. COL_1 genes were highly expressed in pepper fruit but not in tomato fruit at the breaker stage, while COL_3 genes were the opposite, with very low expression or almost no expression in pepper fruit and higher expression in tomato fruit. COL_4 genes were barely expressed in all tissues of pepper but were expressed in tomato throughout fruit development (Supplementary Figure 3). The expression patterns of most genes were consistent in both pepper varieties, with only the COL_2 genes being expressed in the “Zunla-1” stem at higher levels than in “CM334” (Supplementary Figure 3).
Diurnal Rhythm Expression Patterns of CaCOL Genes
CONSTANS-like genes are reported to exhibit prominent circadian rhythm characteristics. Therefore, the expression patterns of 10 CaCOLs during 24 h were studied under long-day (LD) conditions. All genes, except CaCOL04 and CaCOL10, could be induced by light conditions (Figure 8). During the 24 h cycle, the expression of CaCOL01, CaCOL02, CaCOL03, CaCOL05, and CaCOL08 peaked and then decreased at 4 or 8 h after dawn and did not change significantly in the dark (Figure 8). CaCOL07 reached a peak at 12 h, while CaCOL06 peaked at 16 h, and CaCOL09 maintained a high expression level from 4 to 16 h (Figure 8).
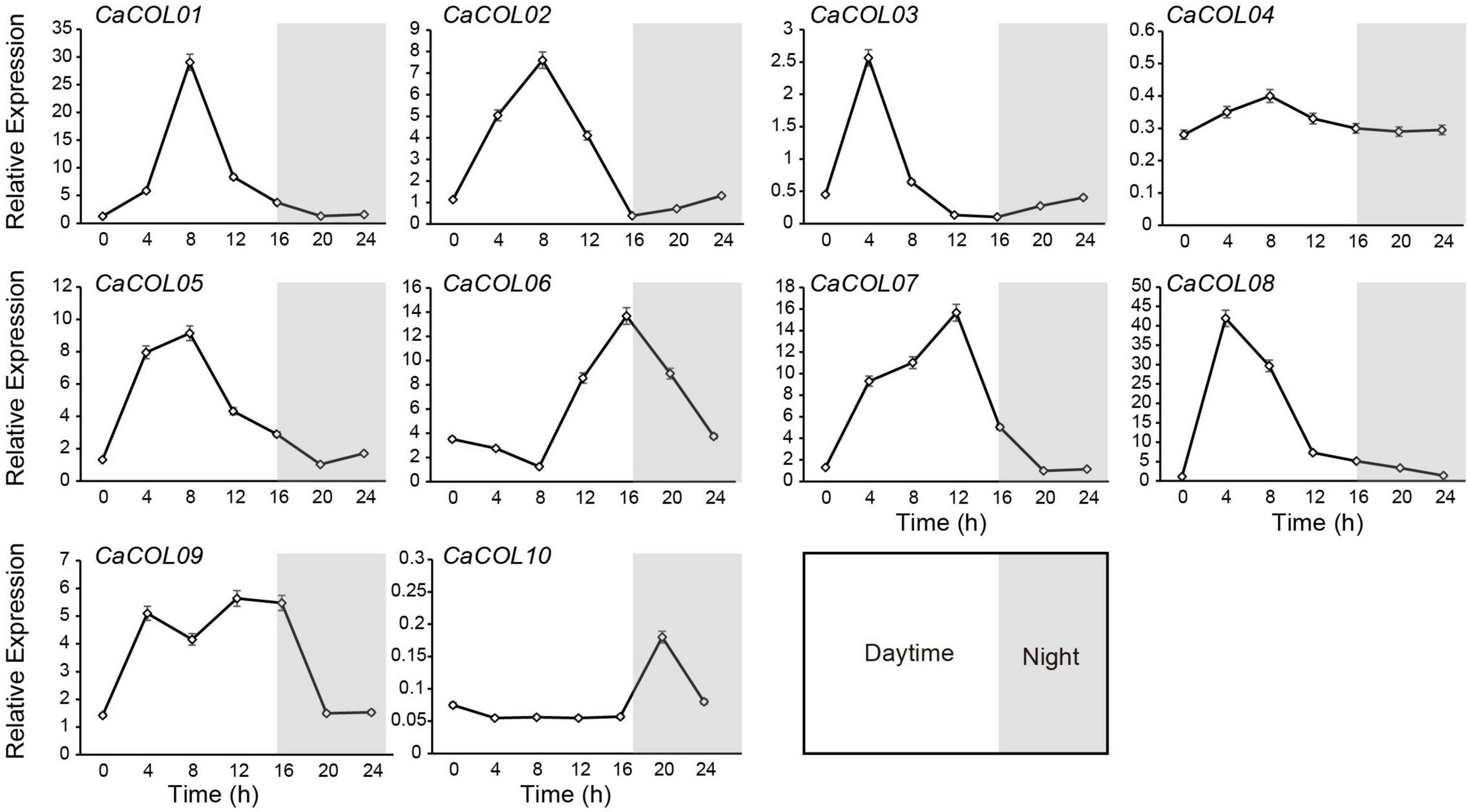
Figure 8. Effect of light on the expression patterns of COLs in pepper leaves. Line charts indicate transcript levels of COL genes. For each sample, transcript levels were normalized with those of actin (control). Data are the mean values ± standard deviation (SD) of three individual experiments (n = 3).
Expression Patterns of CaCOLs Under Abiotic Stresses
The expression patterns of the 10 CaCOL genes under four abiotic stresses (cold, heat, salt, and osmotic) were analyzed by qRT-PCR (Figure 9). Under cold treatment, five CaCOL genes were induced, among which the expression of CaCOL02, CaCOL03, CaCOL07, and CaCOL8 peaked at 12 h after treatment and then decreased, while CaCOL06 expression peaked at 24 h. Eight CaCOL genes participated in the resistance to high-temperature adversity to varying degrees—five genes (CaCOL01, CaCOL06, CaCOL07, CaCOL08, and CaCOL09) were upregulated, and three genes (CaCOL02, CaCOL03, and CaCOL05) were downregulated. Most genes did not respond strongly to NaCl treatment, with some genes presenting a downward trend in expression, including CaCOL02, CaCOL05, and CaCOL07. Under osmotic stress, the expression levels of CaCOL02, CaCOL03, CaCOL07, and CaCOL08 were upregulated after 2 h of treatment, while the other genes showed little or no change in expression (Figure 9).
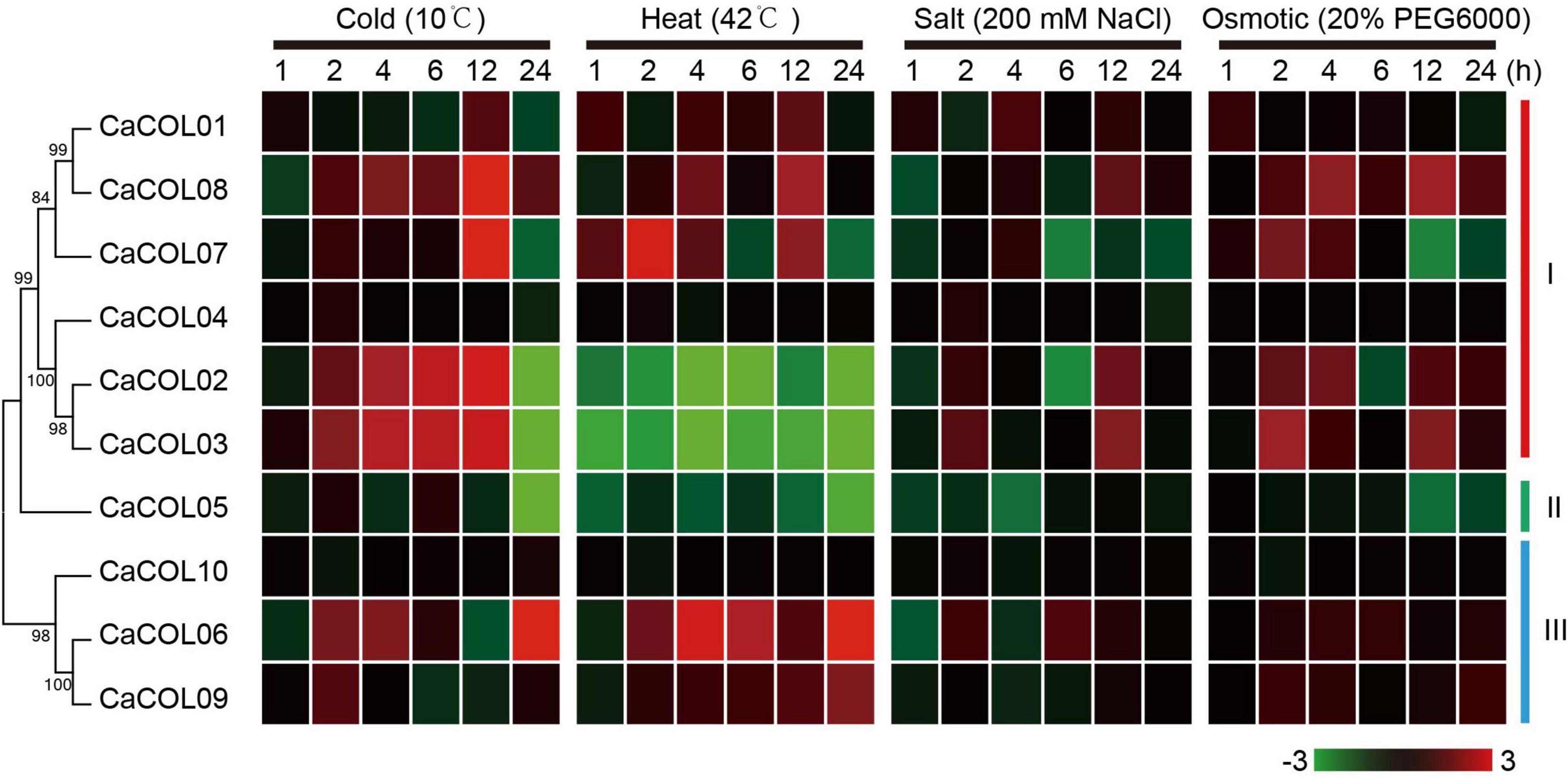
Figure 9. Expression of CaCOL genes under cold, heat, salt, and osmotic stress. The bar at the bottom of each heat map represents the relative expression values.
Discussion
To meet physiological needs and adapt to the external environment during the process of growth and development, plants usually control gene expression in vivo from multiple dimensions and levels, among which transcription level regulation is a crucial step (Jiao et al., 2007; Fornara et al., 2010). CO, located in the output pathway of the circadian clock, specifically activates the FT gene promoter to upregulate expression of the genes FT and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), thus CO is an essential gene that regulates the photoperiod pathway and plays a central regulatory role (Yoo et al., 2005; Jiao et al., 2007; Tiwari et al., 2010). Members of the COL gene family have been identified in detail in the model plant A. thaliana and many other species (Griffiths et al., 2003; Song et al., 2015). C. annuum is an important vegetable crop, but there are limited studies on COL genes in this species. In the current study, 10 and 9 COL genes were identified in the genomes of two varieties—“Zunla-1” and “CM334”—of C. annuum, respectively, and these genes could be divided into three subgroups. COL genes of “Zunla-1” were distributed on five chromosomes, while those of “CM334” were distributed on six chromosomes. This may be due to chromosomal splicing or gene differentiation between varieties of C. annuum.
CONSTANS-like family members possess three conserved domains, two B-box domains and a CCT domain (Griffiths et al., 2003). B-box domains were markedly differentiated among the CaCOL genes, while the CCT domain was relatively more conserved. Phylogenetic analysis indicated that the B-box domain plays a crucial role in the phyletic evolution of COL gene family, which is consistent with previous studies (Griffiths et al., 2003; Song et al., 2015). The evolution of plant regulatory genes is relatively fast and the evolutionary rate is significantly different among different regions of COL family genes (Lagercrantz and Axelsson, 2000). Although both group I and group III COLs contain B-box domains, the domains of the two groups differ significantly. B-box domains of groups I and III were missing in some dicots (Song et al., 2015), but such loss was not observed in C. annuum. Compared with A. thaliana, the gene structures of CaCOL genes were relatively conserved in groups I and II, while those of group III showed apparent differentiation. Investigation of genetic diversity was critical in linking selection pressure to specific genes (Fu et al., 2010; Fiil et al., 2011; Wei et al., 2011). The lower the nucleotide diversity, the greater the selection pressure (Whitt et al., 2002). In the current study, COLs of 12 representative plants (A. thaliana, B. rapa, C. Papaya, P. trichocarpa, F. vesca, V. vinifera, S. tuberosum, S. lycopersicum, C. annuum cv. Zunla-1, Z. mays, O. sativa, and A. trichopoda) were analyzed to further explore the evolutionary history of the COLs. Genetic distance between COL genes was the biggest in group III and the smallest in group I, suggesting that COL genes in group III undergo more relaxed selection pressure during evolution compared with genes of the other two groups. The more extensive selection pressure and structural differences in group III may account for the functional differentiation of group III COL genes. Among the three groups of COLs, genetic distance was the smallest between groups I and III, and the largest between groups II and III, suggesting that groups II and III may be differentiated from group I (Figure 5B).
During their long evolutionary history, angiosperms have undergone different WGD events that induced gene duplication and differentiation (Taylor and Raes, 2004; Lee et al., 2012; Rensing, 2014). Due to the different regions and external environments of different plants, homologous genes are partially lost and partially retained during evolution. Retained genes undergo functional differentiation, leading to gradually different regulatory mechanisms (Wang et al., 2015). Analysis of the replication and retention of COL genes in the 12 selected plants demonstrated that COL genes had also undergone duplication, loss, retention, and differentiation during the evolutionary history of the plants, especially during the genome duplication event. Starting from the relict plant A. trichopoda, angiosperm genomes have undergone different WGD or WGT events (Clark and Donoghue, 2018), with the COL genes of groups I, II, and III presenting unequal amplification among the 12 angiosperm species. Compared with other angiosperms, group I COL genes of C. annuum had the highest proportion of retained genes, which was mainly caused by tandem replication. However, the tandem duplication genes (CaCOL02, CaCOL03, and CaCOL04) in group I were not significantly expressed in different tissues but did respond to abiotic stresses, thus they may exist as backup genes in response to other environmental factors, as indicated in a previous study (Qian et al., 2010; Duan et al., 2016). Among the other species, group III had the most retained genes, especially in B. rapa, which had up to 25 genes. Segmental duplications may be one of the reasons for this phenomenon (Song et al., 2015). Therefore, amplification of group III is presumed to be the main reason for the increase in the number of genes in the COL family. COLs play an essential regulatory role in the photoperiod pathway, but in the 12 selected species, which included typical long- and short-daylight plants, no obvious retention rule was found for COL genes. This indicated that photoperiod regulation had not affected the number of retained COL genes. Specific gene structures or functional differentiation of individual genes may account for their functions.
Obvious functional differentiation was also found in COL orthologous genes of the two varieties of C. annuum. Among these orthologous genes, four pairs had an identity up to 100%, while the remaining four pairs exhibited an average identity of 97.79%. The eight pairs of orthologous genes displayed similar expression trends in leaves, but in root tissue, only one gene was highly expressed in “CM334” and three genes were highly expressed in “Zunla-1.” Most of the COL orthologous genes in C. annuum and tomato had similar expression patterns in different tissues and developmental stages, but with considerable differences in some expression levels. These findings and analyses demonstrated that COL genes display great conservation but have also experienced loss and differentiation. This may be because different selection pressures in different environments result in different degrees of retention of COL genes, and during the retention process, some genes have undergone new functionalization or sub-functionalization.
COLs are instrumental in photoperiod regulation. Previous studies showed that transcription levels of COLs in tomato and Arabidopsis were regulated by circadian rhythms (Suarez-Lopez et al., 2001; Steinbach, 2019; Yang et al., 2020). In the current study, the expression patterns of CaCOL genes also exhibited a clear circadian rhythm under light induction. Under LD conditions, the expression trends of most homologous genes in pepper and tomato were similar, but they differed significantly from those in Arabidopsis (Suarez-Lopez et al., 2001; Yang et al., 2020). Expression patterns analysis showed that expression of CaCOL01, CaCOL02, CaCOL03, and CaCOL08 peaked at 4 or 8 h after dawn, and their homologs in tomato had similar expression trends (Yang et al., 2020). However, CaCOL01 and CaCOL08, as homologous genes of AtCOL4, were expressed at different levels in LD conditions. Additionally, the CaCOL genes showed different circadian rhythms (Figure 8), suggesting functional divergence of these genes in multiple aspects of plant development in response to the photoperiod. This may be because different species have different photoperiod requirements and that structurally similar genes have diverged functionally within or between species. In addition, COLs are also involved in the regulation of the development of different tissues, such as GmCOL9 in soybeans and MaCOL1 in bananas (Huang et al., 2011; Chen et al., 2012). In C. annuum, CaCOL01, CaCOL06, CaCOL08, and CaCOL09 were highly expressed in stem and leaf. In addition, CaCOL01, CaCOL08, and CaCOL07 were homologous to CO have higher expression in flower buds and flower tissues, and CaCOL01, CaCOL06, and CaCOL09 were highly expressed during fruit development. These findings indicated that CaCOL genes play an important regulatory role in C. annuum growth and development.
Analysis of the promoter regions of the CaCOL genes revealed that the regions contained not only light response elements but also a variety of adversity stress response elements, suggesting CaCOL genes may be associated with resistance to abiotic stress. COL homologous genes, such as MaCOL1 (Chen et al., 2012), AtCOL4 (Min et al., 2015), OsCOL9 (Liu H. et al., 2016), Ghd2 (Liu J. et al., 2016), and BnCOL2 (Liu et al., 2020), were reported to be involved in adversity expression in other plant species. In pepper, five CaCOL genes (CaCOL02, CaCOL03, CaCOL06, CaCOL07, and CaCOL08) were expressed under the four adversity treatments (cold, heat, salt, and osmotic stress), with all five genes significantly expressed in the temperature treatments. CaCOL02 and CaCOL03 were upregulated in low temperatures and downregulated in high temperatures. A previous study showed that the CO-HOS1 module was crucial for fine-tuning of photoperiodic flowering under short-term temperature fluctuations (Jung et al., 2012). This may be one of the reasons why COL genes respond to temperature regulation. The CaCOL genes may help plants better adapt to the survival environment by changing the flowering time in response to adversity. Results from the current study provide a specific direction for studying stress-tolerance mechanisms in plants.
Conclusion
This study investigated the evolutionary characteristics and expression function of C. annuum COL genes and included structural characteristics, replication types, evolutionary patterns, and expression analysis. Findings from this study lay the foundation for the in-depth understanding of the function of the COL gene family members in C. annuum and will facilitate research on the molecular response mechanism of these genes during growth and development.
Materials and Methods
Identification of CONSTANS-Like Gene Family in Pepper
Pepper (C. annuum cv. Zunla-1 and CM334) genome data were downloaded from Solanaceae Genomics Network1 (Kim et al., 2014; Qin et al., 2014). To identify all COL family genes in the pepper genome, a batch Basic Local Alignment Search Tool (BLAST) search was performed against the pepper genome data using the full-length amino acid sequences of COs and COLs of Arabidopsis (Griffiths et al., 2003) as queries with an E-value cut-off of 1 × 10––10. All retrieved protein sequences were submitted to Pfam2 databases to annotate the protein domains. Only proteins containing the N-terminal B-box domain (PF00643.19) and C-terminal CCT domain (PF06203.9) were considered to be COL homologs in pepper. Candidate genes were verified in Simple Modular Architecture Research Tool (SMART) and National Center for Biotechnology Information (NCBI) databases to derive the final sequences.
Physical and chemical properties of the CaCOL proteins, including molecular weight (MV) and isoelectric point (pI), were determined and analyzed using EMBOSS programs3.
Phylogenetic Tree, Gene Structure, and Conserved Domain Analysis of CONSTANS-Like Gene Family Members
Multiple sequence alignment analyses were performed using the CLUSTALW program (Thompson et al., 2002). Phylogenetic trees were then reconstructed by the Neighbor-Joining (NJ) method with a bootstrap value of 1000, using MEGA 6.0 software. The genetic distance of COL genes was also calculated using MEGA 6.0 software (Tamura et al., 2013).
Gene structure was plotted using the online tool Gene Structure Display Server (GSDS)4. Based on the Arabidopsis thaliana General Feature Format (GFF) file and C. annuum genome annotation information, the local Perl program was used to extract the location information of exons and introns and convert them into a GSDS readable bed. Online software MEME 4.9.0 was used to predict and analyze structural domains of COL protein sequences of A. thaliana and C. annuum; the motif search value was set to 10, the structural domain width was set to a minimum of 10 and a maximum of 100, and the other parameters used default settings. MEME structures were plotted using Tbtool software (Chen et al., 2020). The COL domain model was analyzed through the SMART database5.
Chromosome Location and Orthologous and Paralogous Genes of the CaCOL Gene Family
Chromosome position information of CaCOL genes was obtained through the GFF file of annotation information of the C. annuum genome. The local Perl program was used to extract location information and construct the C. annuum chromosome map. The duplication type of CaCOL genes was analyzed by duplicate_gene_classifier, the downstream program of MCScanX (Wang et al., 2012).
OrthoMCL6 software was used to identify orthologous and paralogous COL genes of C. annuum, S. lycopersicum, and A. thaliana (Li et al., 2003). Orthologous and paralogous relationship diagrams of COL genes in the three genomes were plotted using Circos software7 (Krzywinski et al., 2009).
Prediction of Cis-Elements in Promoters of CaCOL Genes
Based on the genome sequence of C. annuum cv. Zunla-1 (Qin et al., 2014), the upstream 2-kb sequences of CaCOL genes were extracted using the local Perl program and submitted to the PlantCARE website8 for prediction of cis-acting elements. Finally, a cis-element distribution map was plotted by Tbtool software (Chen et al., 2020).
Analysis of Expression of CaCOL Genes in Different Tissues
Transcriptome data from different tissues and fruit development stages of C. annuum cv. Zunla-1, C. annuum cv. CM334, and tomato were explored in this study (Tomato Genome Consortium, 2012; Kim et al., 2014; Qin et al., 2014). An expression heatmap was plotted using R software (R Core Team, 2013). Pearson correlation coefficient (PCC) values of the expression relationships among CaCOL genes in different tissues were calculated by the local Perl program, and the co-expression network was constructed with the Cytoscape 3.7 program (Shannon et al., 2003).
Planting and Treating of Pepper
The pepper cultivar “Xianhong No. 1” was used for expression pattern analysis. Seeds were disinfected (2% sodium hypochlorite) and sown in the growth room of Huaiyin Institute of Technology, Jiangsu Province, China at 25/18°C day/night temperature, and with a 16/8-h light/dark cycle, relative humidity of 60–70%, and light intensity of 6000 Lux. Various treatments were applied to 45-day old seedlings and samples for RNA extraction were collected from leaves. Consistently robust 45-day-old plants were selected for 24 h sampling, collecting leaf tissues from the same leaf position at an interval of 4 h and performing three biological replicates. For abiotic stress treatments, the seedlings were placed over a plastic box filled with Japanese garden test nutrient solution, pH 6.0. Salt stress was applied by adding NaCl to a final concentration of 200 mmol/L in the nutrient solution; osmotic stress was imitated by adding polyethylene glycol 6000 (PEG6000) to a final concentration of 20%; cold or heat stress treatments were applied by transferring the seedlings into a growth chamber at 10 or 42°C, respectively. The illumination, photoperiod, and relative humidity of the treated plants were kept the same as the plants without abiotic stress treatments. A parallel control (normal growth) was set for each treatment. The second true leaves were sampled at 0, 1, 2, 4, 6, 12, and 24 h after treatment, and three biological replicates were performed at each time point. All samples were wrapped in tin foil, quickly placed in liquid nitrogen, and then stored in a refrigerator at –80°C.
RNA Extraction and qRT-PCR Expression Analysis
A Simple Total RNA Kit (TIANGEN) was used to extract high-quality RNA from pepper leaves, and cDNA was derived by reverse transcription using a PrimeScript RT reagent kit (TaKaRa kit). Actin (GenBank accession number GQ339766.1) was used as an internal reference gene. qRT-PCR was performed (three technical repeats) according to the instructions of the SYBR premix Ex Taq kit (TaKaRa). The 2–ΔΔCT method was used to calculate the relative expression of CaCOL genes (Heid et al., 1996). Primers used for the expression analysis are listed in Supplementary Table 4.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
WD and ZH conceived the study and collected the public dataset of the researched species. XB and ZH contributed to data analysis, bioinformatics analysis, and manuscript preparation. GC, BX, and RC participated in the qRT-PCR experiment. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31902021, 31701931, and 31801829).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Our deepest gratitude goes to the reviewers for their careful work and thoughtful suggestions that have helped to improve the manuscript substantially.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.828209/full#supplementary-material
Footnotes
- ^ https://solgenomics.net/organism/Capsicum_annuum/genome
- ^ http://pfam.xfam.org/
- ^ https://www.ebi.ac.uk/Tools/emboss/
- ^ http://gsds.cbi.pku.edu.cn
- ^ http://smart.embl-heidelberg.de
- ^ http://orthomcl.org/orthomcl/
- ^ http://circos.ca/
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
References
Almada, R., Cabrera, N., Casaretto, J. A., Ruiz-Lara, S., and Gonzalez Villanueva, E. (2009). VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 28, 1193–1203. doi: 10.1007/s00299-009-0720-4
Bohlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Cai, D., Liu, H., Sang, N., and Huang, X. (2017). Identification and characterization of CONSTANS-like (COL) gene family in upland cotton (Gossypium hirsutum L.). PLoS One 12:e0179038. doi: 10.1371/journal.pone.0179038
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Chen, J. Y., Wang, J. N., Kuang, J. F., Shan, W., and Lu, W. J. (2012). Molecular characterization and expression profiles of MaCOL1, a CONSTANS-like gene in banana fruit. Gene 496, 110–117. doi: 10.1016/j.gene.2012.01.008
Clark, J. W., and Donoghue, P. C. J. (2018). Whole-genome duplication and plant macroevolution. Trends Plant Sci. 23, 933–945. doi: 10.1016/j.tplants.2018.07.006
Datta, S., Hettiarachchi, G. H., Deng, X. W., and Holm, M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18, 70–84. doi: 10.1105/tpc.105.038182
Duan, W., Ren, J., Li, Y., Liu, T., Song, X., Chen, Z., et al. (2016). Conservation and expression patterns divergence of ascorbic acid d-mannose/l-galactose pathway genes in brassica rapa. Front. Plant Sci. 7:778. doi: 10.3389/fpls.2016.00778
Fiil, A., Lenk, I., Petersen, K., Jensen, C. S., Nielsen, K. K., Schejbel, B., et al. (2011). Nucleotide diversity and linkage disequilibrium of nine genes with putative effects on flowering time in perennial ryegrass (Lolium perenne L.). Plant Science 180, 228–237. doi: 10.1016/j.plantsci.2010.08.015
Fornara, F., De Montaigu, A., and Coupland, G. (2010). SnapShot: control of flowering in arabidopsis. Cell 141, e551–e552. doi: 10.1016/j.cell.2010.04.024
Fu, Z., Yan, J., Zheng, Y., Warburton, M. L., Crouch, J. H., and Li, J.-S. (2010). Nucleotide diversity and molecular evolution of the PSY1 gene in zea mays compared to some other grass species. Theor. Appl. Genet. 120, 709–720. doi: 10.1007/s00122-009-1188-x
Griffiths, S., Dunford, R. P., Coupland, G., and Laurie, D. A. (2003). The evolution of CONSTANS-like gene families in barley, rice, and arabidopsis. Plant Physiol. 131, 1855–1867. doi: 10.1104/pp.102.016188
Heid, C. A., Stevens, J., Livak, K. J., and Williams, P. M. (1996). Real time quantitative PCR. Genome Res. 6, 986–994.
Hu, T., Wei, Q., Wang, W., Hu, H., Mao, W., Zhu, Q., et al. (2018). Genome-wide identification and characterization of CONSTANS-like gene family in radish (Raphanus sativus). PLoS One 13:e0204137. doi: 10.1371/journal.pone.0204137
Huang, G., Ma, J., Han, Y., Chen, X., and Fu, Y.-F. (2011). Cloning and expression analysis of the soybean CO-like gene GmCOL9. Plant Mol. Biol. Rep. 29, 352–359. doi: 10.1007/s11105-010-0240-y
Jiao, Y., Lau, O. S., and Deng, X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet 8, 217–230. doi: 10.1038/nrg2049
Jung, J.-H., Seo, P. J., and Park, C.-M. (2012). The E3 ubiquitin ligase HOS1 regulates arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287, 43277–43287. doi: 10.1074/jbc.M112.394338
Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M., Lee, H. A., et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in capsicum species. Nat. Genet. 46, 270–278. doi: 10.1038/ng.2877
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Lagercrantz, U., and Axelsson, T. (2000). Rapid evolution of the family of CONSTANS LIKE genes in plants. Mol. Biol. Evol. 17, 1499–1507. doi: 10.1093/oxfordjournals.molbev.a026249
Lee, T.-H., Tang, H., Wang, X., and Paterson, A. H. (2012). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158. doi: 10.1093/nar/gks1104
Li, L., Stoeckert, C. J. Jr., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
Li, Y., and Xu, M. (2017). CCT family genes in cereal crops: a current overview. Crop J. 5, 449–458. doi: 10.1016/j.cj.2017.07.001
Liu, H., Dong, S., Sun, D., Liu, W., Gu, F., Liu, Y., et al. (2016). CONSTANS-Like 9 (OsCOL9) interacts with receptor for activated C-kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS One 11:e0166249. doi: 10.1371/journal.pone.0166249
Liu, J., Shen, J., Xu, Y., Li, X., Xiao, J., and Xiong, L. (2016). Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 67, 5785–5798. doi: 10.1093/jxb/erw344
Liu, L., Ding, Q., Liu, J., Yang, C., Chen, H., Zhang, S., et al. (2020). Brassica napus COL transcription factor BnCOL2 negatively affects the tolerance of transgenic arabidopsis to drought stress. Environ. Exp. Bot. 178:104171. doi: 10.1016/j.envexpbot.2020.104171
Min, J. H., Chung, J. S., Lee, K. H., and Kim, C. S. (2015). The CONSTANS-like 4 transcription factor, AtCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in arabidopsis. J. Int. Plant Biol. 57, 313–324. doi: 10.1111/jipb.12246
Niu, T., Wang, X., Abbas, M., Shen, J., Liu, R., Wang, Z., et al. (2022). Expansion of CONSTANS-like genes in sunflower confers putative neofunctionalization in the adaptation to abiotic stresses. Industr. Crops Prod. 176:114400. doi: 10.1016/j.indcrop.2021.114400
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Qian, W., Liao, B. Y., Chang, A. Y., and Zhang, J. (2010). Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 26, 425–430. doi: 10.1016/j.tig.2010.07.002
Qin, C., Yu, C., Shen, Y., Fang, X., Chen, L., Min, J., et al. (2014). Whole-genome sequencing of cultivated and wild peppers provides insights into capsicum domestication and specialization. Proc. Natl. Acad. Sci. U.S.A. 111, 5135–5140. doi: 10.1073/pnas.1400975111
Rensing, S. A. (2014). Gene duplication as a driver of plant morphogenetic evolution. Curr. Opin. Plant Biol. 17, 43–48. doi: 10.1016/j.pbi.2013.11.002
Robson, F., Costa, M. M., Hepworth, S. R., Vizir, I., Pineiro, M., Reeves, P. H., et al. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28, 619–631. doi: 10.1046/j.1365-313x.2001.01163.x
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Song, N., Xu, Z., Wang, J., Qin, Q., Jiang, H., Si, W., et al. (2018). Genome-wide analysis of maize CONSTANS-LIKE gene family and expression profiling under light/dark and abscisic acid treatment. Gene 673, 1–11. doi: 10.1016/j.gene.2018.06.032
Song, X., Duan, W., Huang, Z., Liu, G., Wu, P., Liu, T., et al. (2015). Comprehensive analysis of the flowering genes in Chinese cabbage and examination of evolutionary pattern of CO-like genes in plant kingdom. Sci. Rep. 5:14631. doi: 10.1038/srep14631
Steinbach, Y. (2019). The Arabidopsis thalianaconstans-LIKE 4 (COL4)–a modulator of flowering time. Front. Plant Sci. 10:651.
Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. doi: 10.1038/35074138
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Taylor, J. S., and Raes, J. (2004). Duplication and divergence: the evolution of new genes and old ideas. Ann. Rev. Genet. 38, 615–643. doi: 10.1146/annurev.genet.38.072902.092831
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Tenorio, G., Orea, A., Romero, J. M., and Merida, A. (2003). Oscillation of mRNA level and activity of granule-bound starch synthase I in arabidopsis leaves during the day/night cycle. Plant Mol. Biol. 51, 949–958. doi: 10.1023/a:1023053420632
Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2002). Multiple sequence alignment using clustalw and clustalx. Curr. Protoc. Bioinform. Chapter 2:Unit2.3. doi: 10.1002/0471250953.bi0203s00
Tiwari, S. B., Shen, Y., Chang, H. C., Hou, Y., Harris, A., Ma, S. F., et al. (2010). The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66. doi: 10.1111/j.1469-8137.2010.03251.x
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485:635. doi: 10.1038/nature11119
Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. doi: 10.1126/science.1091761
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, Z., Zhou, Z., Liu, Y., Liu, T., Li, Q., Ji, Y., et al. (2015). Functional evolution of phosphatidylethanolamine binding proteins in soybean and arabidopsis. Plant Cell 27, 323–336. doi: 10.1105/tpc.114.135103
Wei, S., Peng, Z., Zhou, Y., Yang, Z., Wu, K., and Ouyang, Z. (2011). Nucleotide diversity and molecular evolution of the WAG-2 gene in common wheat (Triticum aestivum L) and its relatives. Genet. Mol. Biol. 34, 606–615. doi: 10.1590/S1415-47572011000400013
Whitt, S. R., Wilson, L. M., Tenaillon, M. I., Gaut, B. S., and Buckler, E. S. (2002). Genetic diversity and selection in the maize starch pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 12959–12962. doi: 10.1073/pnas.202476999
Wu, F., Price, B. W., Haider, W., Seufferheld, G., Nelson, R., and Hanzawa, Y. (2014). Functional and evolutionary characterization of the CONSTANS gene family in short-day photoperiodic flowering in soybean. PLoS One 9:e85754. doi: 10.1371/journal.pone.0085754
Yang, T., He, Y., Niu, S., Yan, S., and Zhang, Y. (2020). Identification and characterization of the CONSTANS (CO)/CONSTANS-like (COL) genes related to photoperiodic signaling and flowering in tomato. Plant Sci. 301:110653. doi: 10.1016/j.plantsci.2020.110653
Keywords: COL, phylogenetic relationship, expression pattern, abiotic stress, Capsicum annuum
Citation: Huang Z, Bai X, Duan W, Chen B, Chen G, Xu B, Cheng R and Wang J (2022) Genome-Wide Identification and Expression Profiling of CONSTANS-Like Genes in Pepper (Capsicum annuum): Gaining an Insight to Their Phylogenetic Evolution and Stress-Specific Roles. Front. Plant Sci. 13:828209. doi: 10.3389/fpls.2022.828209
Received: 03 December 2021; Accepted: 14 January 2022;
Published: 17 February 2022.
Edited by:
Pei Xu, China Jiliang University, ChinaReviewed by:
Arun K. Pandey, China Jiliang University, ChinaYuan Cheng, Zhejiang Academy of Agricultural Sciences, China
Copyright © 2022 Huang, Bai, Duan, Chen, Chen, Xu, Cheng and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weike Duan, d2Vpa2VkdWFuQDEyNi5jb20=
†These authors have contributed equally to this work
 Zhinan Huang1†
Zhinan Huang1† Weike Duan
Weike Duan Guodong Chen
Guodong Chen Rui Cheng
Rui Cheng