
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 17 February 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.825477
This article is part of the Research Topic Systemic RNA Signalling in Plants View all 6 articles
Small RNAs (sRNAs) encoded by plant genomes have received widespread attention because they can affect multiple biological processes. Different sRNAs that are synthesized in plant cells can move throughout the plants, transport to plant pathogens via extracellular vesicles (EVs), and transfer to mammals via food. Small RNAs function at the target sites through DNA methylation, RNA interference, and translational repression. In this article, we reviewed the systematic processes of sRNA biogenesis, trafficking, and the underlying mechanisms of its functions.
Excessive use of pesticides and chemical fertilizers has already caused dramatic damages to the ecological environment, yet some pests and plant diseases are still not completely controlled and prevented. Thus, extensive attentions have recently been paid on biological control approaches (Ma et al., 2021; Peng et al., 2021). Biological control approaches are highly effective to prevent plant diseases and increase the crop yields. In most eukaryotes, small RNAs (sRNAs) are generated by the ribonuclease III-like enzyme dicer or dicer-like (DCL) proteins and are incorporated into argonaute (AGO) proteins to induce gene silencing in a sequence-specific manner (Huang et al., 2019a). Small RNAs are widely present in plants and have been gradually utilized to control plant diseases and insect pests because they can regulate various biological processes, e.g., plant growth, development, and stress response (Si et al., 2020; Mekapogu et al., 2021).
To identify functional sRNAs, it is necessary to construct an sRNA library which is often completed by sRNA sequencing, DNA microarray, and shotgun cloning (Vogel et al., 2003; Boccara et al., 2017; Wu et al., 2020). The presence of sRNA in plants can be verified by RNA blotting, quantitative PCR and other techniques (Cai et al., 2018; Cui et al., 2020). Dual-luciferase reporter assay, gene transient expression analysis, and degradome sequencing are usually used to verify sRNA binding sites (Hu et al., 2020; Xie et al., 2021). The function of sRNAs can be analyzed by constructing transgenic plants using short tandem target mimic (STTM), CRISPR-Cas9, homologous recombination and other gene editing technologies (Cui et al., 2020; Qiao et al., 2020; Ji et al., 2021). To better understand the function of sRNA and promote the use of sRNA in agricultural production, we reviewed the process of sRNA biogenesis, trafficking and the underlying mechanisms of its functions.
Plant sRNAs are generally divided into two main categories, microRNAs (miRNAs) and small interfering RNAs (siRNAs). In plants, siRNAs can be generated through multiple biogenesis pathways (Borges and Martienssen, 2015). However, the pathway for miRNA biogenesis is unique. Based on the biogenesis and biosynthesis, siRNAs can be further divided into natural antisense transcript small interfering RNA (natsiRNA), heterochromatic small interfering RNA (hcsiRNA), virus-derived small interfering RNA (vsiRNA) and secondary siRNA (Song et al., 2019; Zhang et al., 2019; Middleton et al., 2021).
Transcription of miRNA genes (MIRs) in euchromatic regions of plant chromosomes is catalyzed by DNA-dependent RNA Polymerase II (Pol II; Figure 1A; Xie et al., 2005). The primary transcript of miRNA (pri-miRNAs) contains at least one characteristic hairpin-like structure. Subsequently, pri-miRNAs are loaded into nuclear dicing bodies (D-bodies) including DCL1, HYPONASTIC LEAVES 1 (HYL1), SERRATE (SE) and TOUGH (TGH; Fang and Spector, 2007). Then, DCL1 cuts the hairpin structure on the pri-miRNA through two consecutive cleavage steps, resulting in a miRNA duplex of approximately 21 nucleotides (nt; Kurihara and Watanabe, 2004). Following pri-miRNA processing, HUA ENHANCER 1 (HEN1) catalyzes 2’-O-methylation at the 3′-ends of miRNA duplex so that miRNAs are more stable (Huang et al., 2009). This mature miRNA duplexes are loaded into AGO1 protein and form an miRNA-induced silencing complex (miRISC) with the assistance of heat shock protein (HSP70/HSP90) and Constitutive Alterations in the Small RNAs Pathways9 (CARP9; Bologna et al., 2018; Tomassi et al., 2020). In RISC, only one strand from the miRNA duplex was usually loaded, and the other strand with higher thermodynamic stability at the 5′-end was degraded. For miRNA/miRNA* duplex, the miRNA* strand was usually degraded. However, miRNA* strand can also be accumulated and loaded into AGO protein (Eamens et al., 2009; Song et al., 2019). In addition, other models also propose that DCL3 can produce 24-nt miRNA. The trafficking and function of 24-nt miRNA are different from those of 21-nt miRNA (Cervantes-Pérez et al., 2021).
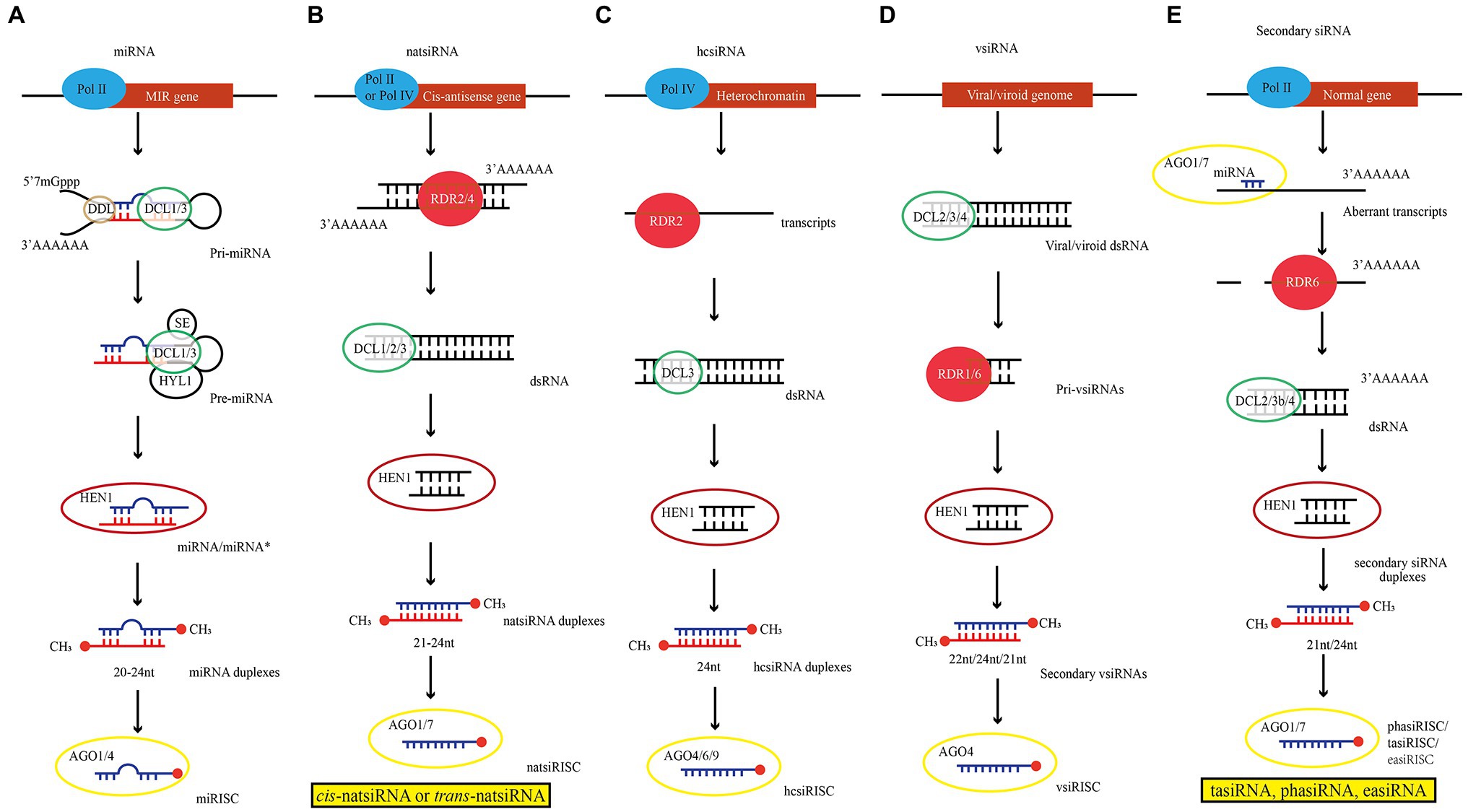
Figure 1. Illustrations of siRNA biogenesis in plants (Ref Borges et al., 2018; Singh et al., 2021). (A) miRNA biogenesis model derived from MIR gene. (B) natsiRNA biogenesis model derived from Cis-antisense gene. (C) hcsiRNA biogenesis model derived from Heterochromatin. (D) vsiRNA biogenesis model derived from Viral/viroid genome. (E) Secondary siRNA biogenesis model derived from Normal gene. Pol: RNA Polymerase; DDL: Dawdle; DCL: RNase III enzyme DICER-LIKE; SE: SERRATE; HYL1: HYPONASTIC LEAVES1; HEN1: HUA ENHANCER 1; RDR: RNA-DEPENDENT RNA POLYMERASE.
In general, MIR gene is not static and can evolve with the changing environment. Because MIR gene is evolving, the lineage-specific miRNAs between species are created and may guide the co-evolution of mRNA target sequences (Cui et al., 2017). Biogenesis of miRNAs can be regulated by both transcriptional and post-transcriptional factors (Voinnet, 2009; Li and Yu, 2021). For instance, 3′-phosphoadenosine 5′-phosphate (PAP) and Tocopherols (vitamin E) can protect pri-miRNAs from being degraded and promote the production of mature miRNA in Arabidopsis thaliana (Fang et al., 2019). RNA adenosine methylase (MTA) catalyzes the formation m6A on pri-miRNAs to modulate miRNA biogenesis (Bhat et al., 2020). The abundance of miR156 is positively regulated by AGL15 because AGL15 can inhibit the expression of DCL1 and SERRATE genes (Nowak et al., 2020). Nucleoplasmic exosome protein is an RNA processing complex containing 3′-5’exoribonuclease. HYL1 can promote pri-miRNA processing and prevent the attack from exosome (Gao et al., 2020)
Biosynthesis of miRNA is regulated not only by genetic factors under normal conditions but also by environmental factors, e.g., changes in the environmental stress. For example, strontium stress inhibits the biogenesis of miRNA by reducing the level of HYL1 protein in Arabidopsis (Pyo et al., 2020). Under environmental stress, MPK3 and SnRK2 can phosphorylate and inactivate cofactors, e.g., HYL1 and SE, leading to the decreases in the production of miRNA (Manavella et al., 2019). The activity of mitochondria is greatly inhibited by hypoxia condition, which triggers the biogenesis of miRNAs responsible for hypoxia tolerance (Betti et al., 2020). Under the environmental stress, plants can make corresponding adjustments via regulating the biosynthesis of miRNAs to maintain their own life activities.
In general, dsRNAs that are the precursors of siRNAs are produced through two different pathways. The first is from the abnormal transcripts of genes (including the hybridization of sense and antisense transcripts, the folding back of an inverted-repeat sequence, and the hybridization of unrelated RNA molecules with sequence complementarity) that are subsequently processed and loaded by RDRs and SGS3 (Marchais et al., 2019), and the second is from single-stranded RNA after being processed and loaded by RNA polymerase IV (Pol IV) and RDRs. The dsRNA formed by these two pathways is processed by DCL2/3/4 into 21 ~ 24 nt siRNA (Yao et al., 2020).
Natural antisense transcripts (NATs) are formed by annealing of two complementary and separately transcribed RNA strands. According to their genomic origin, they can be divided into cis-NAT and trans-NAT. Cis-NAT is transcribed from the same genomic locus, forming a completely complementary dsRNA between the two transcript sequences. In contrast, trans-NAT constitutes highly complementary dsRNAs encoded by two distant genomic sites. Amplification of these two dsRNAs requires the participation of RDR2/4(Yu et al., 2016). Subsequently, with the participation of cofactors, e.g., RDR6, SGS3, and DNA directed RNA polymerase IV subunit 1 (NRPD1), DCL1/2/3 cleave natsiRNA precursor, leading to the production of 21 ~ 24 nt cis-natsiRNA or trans-natsiRNA (Zhang et al., 2012; Figure 1B). The hcsiRNA is derived from repetitive sequences on chromatin and transposable elements (TE). Its biogenesis also requires RNA Pol IV-mediated transcription and RDR2-mediated formation of dsRNA (Parent et al., 2015). Finally, DCL3 processes the dsRNA into 24 nt siRNA duplexes, and HEN1 methylates the siRNA duplexes to form 24 nt hcsiRNA (Chen et al., 2021a; Figure 1C). Biosynthesis of vsiRNA also requires the participation of DCLs, AGOs, and RDR proteins. The difference is that vsiRNA originates from abnormal transgene (produced by viral DNA) or viral RNA in plants after virus infection. RNA-dependent RNA polymerase (RdRP) may recognize and use these abnormal RNAs as templates to synthesize antisense RNA and form dsRNA (Leonetti et al., 2020). These dsRNAs are processed by DCL2/3/4 to produce 22, 24, and 21 nt primary vsiRNAs, respectively, which are subsequently amplified by RDRs and loaded into AGOs to form vsiRNA (Garcia-Ruiz et al., 2010; Vivek et al., 2020; Figure 1D).
PolII catalyzes the transcription of plant genes (including PHAS loci, TAS gene, and active retrotransposons). After the transcripts are cleaved by sRNA, the 5′ end fragment of the transcript is degraded, while the 3′ end fragment is converted into dsRNA by RDR6 and becomes the precursor of secondary siRNA. Subsequently, dsRNA is processed by DCL2/4 to generate 21 ~ 24 nt siRNA (Figure 1E). They are subdivided into phased siRNA (phasiRNA), trans-acting siRNAs (tasiRNA) and epigenetically activated siRNAs (easiRNA). All the three subclasses of siRNAs are generated via different biogenetic pathways. For example, there are two mechanisms for the biogenesis of phasiRNA: “one-hit” and “two-hit” modes (Liu et al., 2020). In the “one-hit” mode, the 22 nt miRNA cleaves the mRNA from the 3′ end at the single target site to generate phasiRNAs with the participation of factors, e.g., RDR6, DCL4, and DCL3b (Tian et al., 2021). However, in the “two-hit” mode, although mRNA contains two miRNA target sites, only one site can be cleaved (usually at the 3’end site), and mRNA is cleaved successively by DCL4 to produce 21 nt phasiRNA (Axtell et al., 2006). As one of phasiRNAs, tasiRNA is produced by miRNA-guided cleavage of long and noncoding precursor transcripts. The cleaved fragments are then converted to dsRNAs by RDR6 and processed into 21 nt siRNAs by DCL4 (Yang et al., 2021b). Moreover, easiRNA is originated from active transposons in plants and is also produced by DCL2/4 processing (Creasey et al., 2014; Wu et al., 2020). SGS3 mediates the specific recognition of RDR6, which specifically recognizes the transposon RNA and synthesizes dsRNA (Kim et al., 2021).
The biosynthesis of siRNA and miRNA is also regulated by environmental factors. AGO1 could accumulate miRNA into the membrane-bound polysomes (MBPs) and cleave the targeted transcripts to produce phasiRNA. In other words, under specific condition, AGO1 affects the synthesis of phasiRNAs by regulating the membrane binding of miRNAs (Li et al., 2016; Komiya, 2017). NOT1, as a component of CCR4-NOT complex, regulates DNA methylation and transcriptional silencing by promoting the production of Pol-IV-dependent siRNA (Zhou et al., 2020). Under stress conditions, plants preferentially accumulate 22 nt siRNA from NIA1/2 gene to inhibit plant growth and enhance stress response (Wu et al., 2020). Thus, biogenesis of siRNA in plants is also regulated to ensure its rational synthesis.
miRNA and siRNA are the two common sRNAs in plants. However, to meet additional regulations, other sRNAs are also generated. For example, tRNA-derived RNA fragments (tRFs) are generated by excising from mature tRNA or produced as a by-product of pre-tRNA processing (Megel et al., 2019). According to the cleavage sites, tRF can be divided into tRF-5a and tRF-3a (Park and Kim, 2018). At present, the biogenesis of tRFs in plants is not clear. However, it is known that tRFs are also loaded onto AGO1/2/4, indicating that rRFs execute their functions of gene silencing similarly as miRNA and siRNA (Ren et al., 2019).
siRNAs independent of DCLs (sidRNAs) is considered as a new type of sRNA, which is mainly originated from the sidRNA loci on transposons, intergenic sequences or transgenes (Ye et al., 2016). The sidRNA loci are transcribed to form precursors under the action of PolIV and RDR2. Subsequently, 24 nt sidRNA is produced by 3′-5′ exonuclease, and gene Atrimmer may be a potential splicing site. SideRNAs recruits AGO4 to the target sites, implying similar mode with other sRNAs (Ye et al., 2016).
The sRNA produced by the plant factory is either transported to subcellular areas, or exposed to ZSWIM8 ubiquitin ligase in the cytoplasm, and then degraded after the 5′ end cap structure is removed by RDR6 (Baeg et al., 2017; Han et al., 2020).
One of the most fascinating aspects of sRNA is its mobility, in other words, its ability to spread from one cell to its neighboring cells (Liu and Chen, 2018). In early studies, transfer pathway of sRNA in plants was described as “particle bombardment with siRNA/transgenics” (Agrawal et al., 2003). The logistics network of sRNA has been recently elucidated with the development of various sRNA tracing technologies (Molnar et al., 2010; Huang et al., 2019b). In addition to the trafficking of sRNA within plants, external movements of sRNA have also been observed in plants (Cai et al., 2021).
After sRNA is synthesized in the cell nucleus, it is loaded into the AGO protein. Then, the nuclear localization signal and nuclear export signal (NES) directly guide the nucleocytoplasmic shuttle of RISC (Bologna et al., 2018). Subsequently, there are three main forms of sRNA involved in transfer: naked sRNAs, sRNAs bound to RNA-binding proteins (RBPs) and sRNAs inside vesicles (Wang and Dean, 2020). Short-range movement between cells occurs through plasmodesmata (PD; Garnelo Gómez et al., 2021). PD is a membrane channel that passes through the cell wall and connects adjacent cells through the plasma membrane (PM). In the channel, there is a specialized-cylindrical structure called desmotubule (DM). DM is derived from the smooth endoplasmic reticulum and can connect the endoplasmic reticulum of two cells. sRNA could move between cells through PD pore or along the desmosomes (Di Donato and Amari, 2014; Figure 2A, pathway 1, 2). The dominant sRNA involved in short distance movement is the 21-nt sRNA (Tamiru et al., 2018). SUC-SUL and SUC-PDS are artificial siRNA reporter systems. In both systems, long inverted-repeat dsRNAs are expressed in phloem companion cells. siRNA can be produced and diffused to 10–15 neighboring cells, reflecting its local cell-to-cell movement (Liu and Chen, 2018). It is worth noting that sRNA can bind RBPs to form sRNA ribonucleoprotein complexes (sRNPC), which are co-transported between cells. A recent study identified a conserved RBP (SRBP1) in the phloem of cucurbit, which mediates the trafficking of siRNA between cells (Yan et al., 2020). Therefore, detailed information on RBPs would facilitate the understanding on the regulation of sRNA movement. In addition to the mainstream PD transport pathway, both naked sRNAs and sRNAs inside vesicles can be secreted directly from PM and spread between plant cells (Weiberg et al., 2013; Cai et al., 2019; Figure 2A, pathway 3).
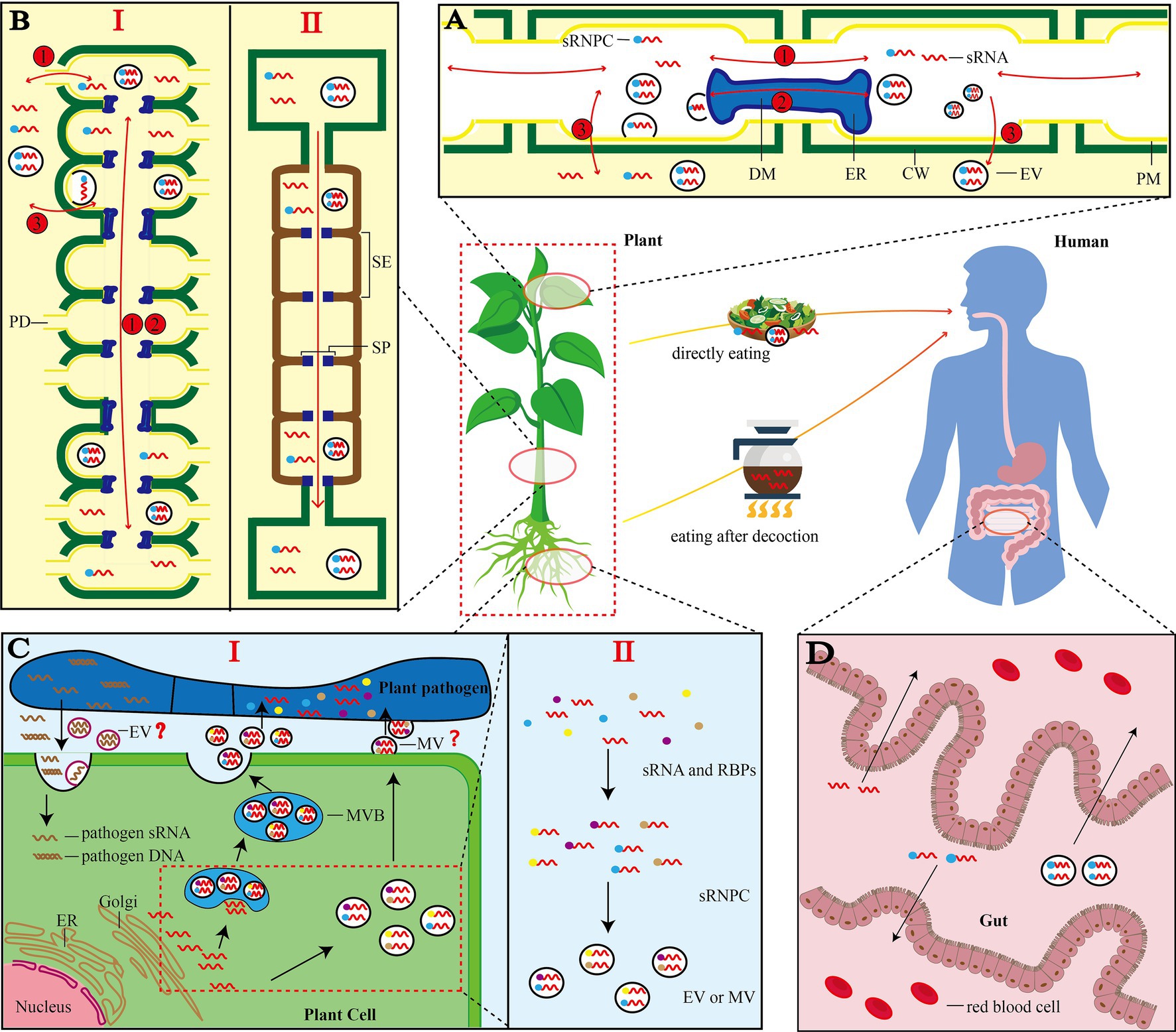
Figure 2. Illustrations of siRNA trafficking in plants. (A) Movement between plant cells (Wang and Dean, 2020): Pathway 1, naked small RNAs, small RNAs bound to RNA binding protein (RBP) and small RNAs enclosed in vesicles can pass through plasmodesmata (PD) moves between cells; pathway 2, desmotubules (DM) connects the endoplasmic reticulum (ER) of two adjacent cells, and small RNA can be transported through DM; pathway 3, small RNA can be directly secreted from PM and spread in plants. Note: It is unknown whether vesicles can be transported via DM. (B) Long-distance movement in plants (Tamiru et al., 2018): I: Long-distance movement occurs through the repetitive mechanism of pathways 1 and 2 in the plasmodesmata; II: sRNA enters the phloem sieve tube through the plasmodesmata (PD), and transports it quickly from top to bottom with the phloem sap. (C) Cross-domain transport of sRNA by EV (Huang et al., 2019a): I: Free sRNA in plant cytoplasm can be packaged by Golgi and transported to the outside of the cell to be absorbed by Plant pathogen. At the same time, Plant pathogen also produces sRNA and delivers its own DNA or sRNA to plant cells; II: When vesicles are formed, sRNA needs to be combined with RBPs before it can be selectively loaded into EVs for cross-domain transport. Note: Plant pathogen here is only a type of organism that absorbs vesicles. Many organisms can absorb vesicles provided by plants or produce sRNA and transfer them to plant cells. “?” indicates whether plants can produce MV and whether other organisms can produce EVs is not yet clear; (D) Transfer of sRNA to animal cells: Plants deliver sRNA to the human body through decoction or raw food directly. After different forms of sRNA enter the gut, it is absorbed into the blood through the villi of the small intestine and circulates throughout the body with the blood. Human here refers to mammals. sRNA: small RNA; MVB: multivesicular bodies; sRNPC: sRNA ribonucleoprotein complex; DM: desmotubule; ER: endoplasmic reticulum; CW: cell wall; EV: extracellular vesicles; PM: plasma membrane; PD: plasmodesmata; SE: sieve tube elements; SP: sieve tube plates; MV: microvesicles.
The most intuitive model for the long-distance movement of sRNA is the molecular signal transmission model between scions and rootstocks of grafted plants. The transfer of transgene-derived siRNA from rootstock to scion makes non-transgenic cherry scion resistant to the Prunus necrotic ringspot virus. 24-nt sRNA can also be transferred from cherry scion to the rootstock, which potentially affects the rootstock (Zhao and Song, 2014; Zhao et al., 2020). Long-distance root-to-shoot movement occurred intercellularly via plasmodesmata by a repeating mechanism (Tamiru et al., 2018). In this mechanism, the three forms of sRNA could still be transported via PD or directly across the PM (Figures 2A,B, I). Notably, the permeability of PD is strongly regulated by several factors, e.g., endogenous reactive oxygen species (ROS; Welchen and Gonzalez, 2021), light and circadian clock (Brunkard and Zambryski, 2019).
There is a special pathway in the long-distance transportation of sRNA from the top to the bottom of plants (Figure 2B, II). The phloem protein kinase PSRPK1 is phosphorylated by PSRP1 to form sRNA ribonucleoprotein complex (sRNPC) after sRNA is produced in the cell. sRNPC passes through the PD and enters the phloem sieve tube. During the long-distance movement, PSRP1-sRNPC is stable against the phloem phosphatase activity (Ham et al., 2014). Small RBP-bound RNAs pass through septum (SP) and are rapidly transported to target tissues by phloem sap. Subsequently, they are unloaded into the surrounding cells followed by decomposition of PSRP1-SrNPC complex (Ham et al., 2014). For the transport of sRNAs inside the vesicles, only vesicles are found in the phloem, while sRNA inside the vesicles needs to be further identified and characterized (Chukhchin et al., 2019). The long-distance transportation of sRNA from the bottom to the top of the plant also has a special route based on the xylem catheter, which is not well characterized. Cadmium treatment can alter miRNAs in leaves of maize and xylem sap, indicating that sRNA is involved in the stress response of plants, and xylem catheter can transport these sRNAs (Wang et al., 2019).
It is initially believed that the outer vesicles are just a way for cells to discharge metabolic waste, but further researches reveal that they are rich in protein and sRNA and could function in a cross-regional or cross-species manner (Thomma and Cook, 2018). The discovery of extracellular vesicles is a breakthrough in the field of secretion, as it provides a new mechanism for releasing components into the extracellular environment (Shao et al., 2018). The most classic study showed that host Arabidopsis cells can secrete extracellular vesicles to deliver sRNAs into fungal pathogen Botrytis cinerea (Cai et al., 2018). This mechanism has been discovered in plants such as sunflower, tomato and olive, indicating that the precise cross-kingdom targeting transport of plant sRNA is mediated by EV (Prado et al., 2014; Regente et al., 2017; De Palma et al., 2020; Figure 2C, I). In addition, fungi, bacterial and parasitic plants can also transport sRNA or genes into plants as one of the sources of vsiRNA (Shahid et al., 2018; Dunker et al., 2020; Ji et al., 2021; Figures 1D, 2C, I).
In the past, it was not clear if sRNAs in plant EVs are selectively loaded. In mammalian cells, the mechanism of sRNA loading has been revealed. In cancer cell EVs, members of the hnRNP family, as well as other molecules, e.g., YBX1, HUR, and AGO2 are used as RBPs (Fabbiano et al., 2020). In liver cells, SYNCRIP, involved in the exosomal sorting of miRNAs, interacts with specific miRNAs and binds extra-seed sequence (hEXO Motif), which regulates the localization of miRNAs (Santangelo et al., 2016). During autophagy, specific RBPs need to be loaded into extracellular vesicles through LC3 coupling mechanism (Leidal et al., 2020). A recent groundbreaking study demonstrated for the first time that RBPs affect EV loading of sRNA in plants. Several RBPs in the EV of Arabidopsis have been identified, including RBPs Ago1, RHs and ANNs. Studies on gene knockout experiments indicate that these RBPs may contribute to sRNA sorting and stabilization (He et al., 2021). These EV-coated Ago1, RH11 and RH37 may contribute to the selective sRNA sorting and stability in EVs. However, ANN1 and ANN2 only stabilize sRNA in EV, indicating that the vesicle transfer pathway of sRNA requires RBPs for selective loading and cross-domain transport (Figure 2C, II).
It has been controversial over whether plant-derived sRNA could pass through the mammalian gastrointestinal tract and enter the bloodstream because there are a series of obstacles in the mouth, stomach, large intestine, and small intestine (Dávalos et al., 2019; Figure 2D). Although it is theoretically difficult, it was found in 2012 that MIR168a carried by ginger-derived nanoparticles could travel to the liver after being absorbed from the gastrointestinal tract (Zhuang et al., 2015). Since then, extensive studies on the transfer of sRNA from plants to mammals have been carried out (Mar-Aguilar et al., 2020; Chen et al., 2021b). For example, some studies showed that sRNA from strawberries, blueberries and other plants have significant effect on human health (De Robertis et al., 2020; Alfieri et al., 2021; Perut et al., 2021). However, other studies argued that the cross-kingdom transfer of exogenous sRNAs was insignificant and biologically irrelevant, and the results lacked reproducibility (Mar-Aguilar et al., 2020). The cross-kingdom transfer of sRNA observed in these studies might be due to experimental artifacts and contaminations (Witwer, 2018).
Surprisingly, some high-temperature-resistant sRNAs in some plants especially Chinese medicinal materials can be preserved after decoction and can be absorbed by the intestines to achieve their potential functions. For example, MIR2911 is not significantly degraded after boiling, and can inhibit the expression of Enterovirus 71 (EV71) and VP1 protein in vitro and in vivo (Zhou et al., 2015; Li et al., 2018). Similarly, the unique miRNAs of Gastrodia elata including GAS-mir01 and gas-mir02 are stable during decoction and long-term preservation, and both could target the human A20 gene in vitro (Xia et al., 2020). High GC content in the miRNAs might be the reason of high stability after decoction in these studies. Overall, the evidence for the transfer of miRNAs from diet to blood remains inconclusive, and definitive evidence and reproducible findings are needed (Mar-Aguilar et al., 2020).
After sRNAs are biosynthesized and transported, they function at the target site. It functions only when it binds to the target sites based on AGO-guided watson-crick base pairing rules (Fei et al., 2021). miRNA and siRNA can not only mediate transcriptional gene silencing through RNA-directed DNA methylation (RdDM), but also perform post-transcriptional gene silencing through cleavage and translational inhibition without changing the DNA sequence (Borges and Martienssen, 2015).
sRNAs that can mediate DNA methylation are derived from the short transcripts of methylated templates (Matzke and Mosher, 2014). Among all kinds of sRNA, 24 nt sRNA is classified as sidRNA (Ye et al., 2016). sRNA can mediate transcriptional gene silencing by RdDM, which includes the initial recruitment of DNA methyltransferase and subsequent catalytic de novo DNA methylation of cytosine in all sequences after pairing of AGO loaded siRNA with Pol V transcribed scaffold RNA (Huang et al., 2021; Figure 3A).
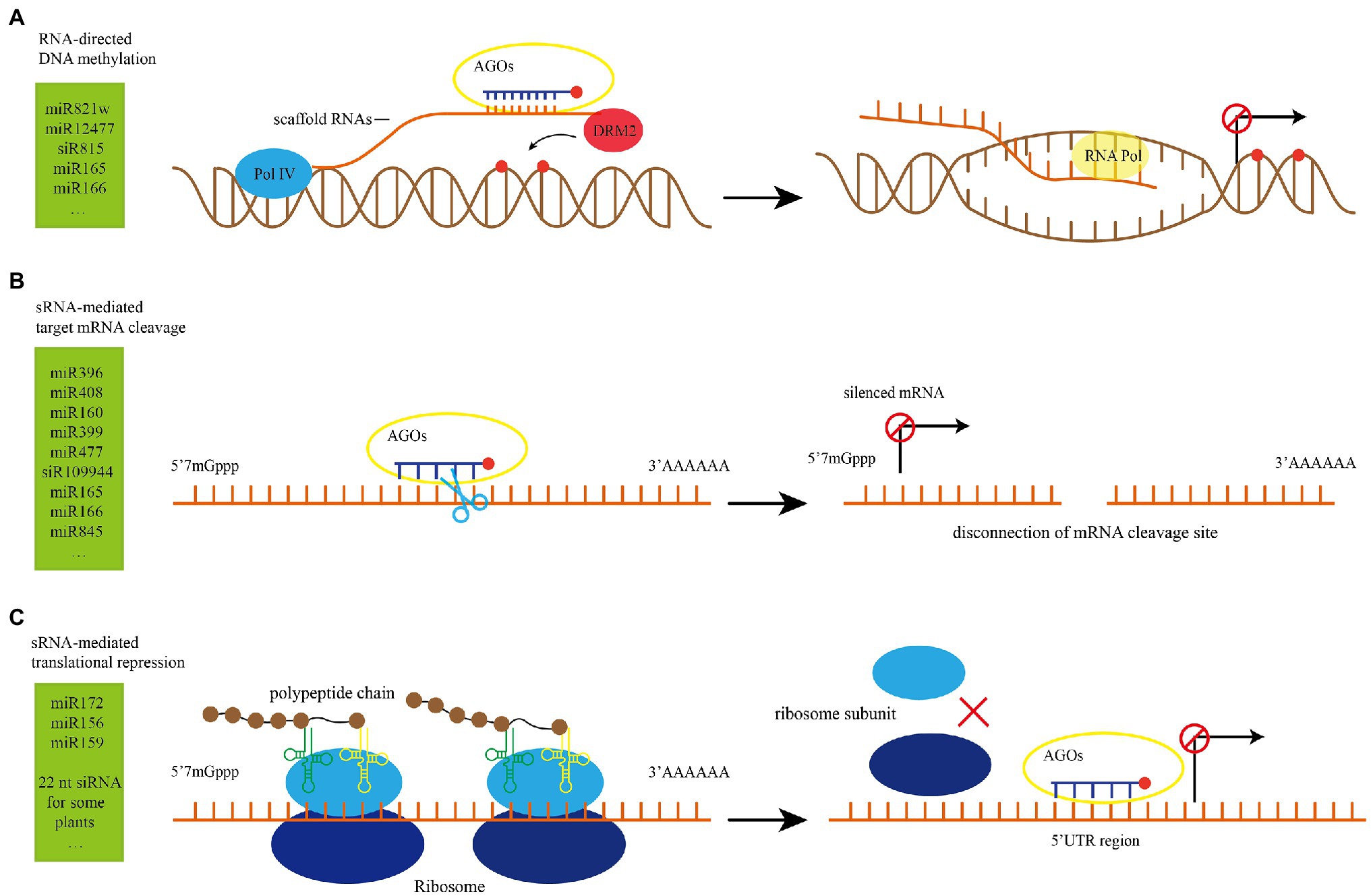
Figure 3. Illustrations of sRNA function. (A) RNA-directed DNA methylation (RdDM) model (Tang, 2020). The scaffold RNAs are produced by Pol V, recruit their complementary sRNA to the RdDM target loci and guides DRM2 to catalyze DNA methylation. (B) sRNA-mediated target mRNA cleavage model. RISC is paired with target mRNAs according to the principle of base complementary pairing. The PIWI domain of AGO proteins has slicer endonuclease activity, and the paired regions are cleaved with the participation of AGO protein. (C) sRNA-mediated translational repression model (Simone et al., 2021): RISC competes with ribosomes for binding to the UTR region of mRNA, which affects the translation process and inhibits the formation of polypeptide chains. RISC: sRNAs carried by RNA-induced silencing complex; UTR: untranslational region; DRM2: DOMAINS REARRANGED METHYLTRANSFERASE 2.
RdDM can not only maintain long-term genome stability by inhibiting transposable elements but also ensure plants’ life activities under stress conditions by regulating gene expression (Rymen et al., 2020; Guo et al., 2021). High temperature stress usually promotes RdDM (Singh et al., 2021), which inhibits the expression of ROS1 gene due to hypermethylation and affects seed germination (Malabarba et al., 2021). The slowdown of life activities may be beneficial to tolerate high temperature environments. Moreover, in the chilling environment, sRNA mediates hypermethylation of the dormancy-related gene DAM and initiates dormancy (Zhu et al., 2020a). Meanwhile, Osa-miR12477 regulates the expression of gene LAO to tolerate salt and reduces oxidative damage (Parmar et al., 2020). It has been reported that RdDM occurs in plants in response to abiotic stress environments, e.g., drought and salinization and biological stresses (Erdmann and Picard, 2020; Liu and He, 2020; Kumar and Mohapatra, 2021; Table 1). In general, when plants are subjected to environmental stress, sRNA can promote plant adaptability via DNA methylation and maintain life activities.
One characteristic feature of sRNA functioning through RdDM is that parental DNA methylation markers can be maintained to the next generation without alteration of DNA sequence (Quadrana et al., 2016). For example, sRNA of Trichoderma spp. can participate in epigenetic regulation of plants through RdDM and induce immune response to protect plants (Morán-Diez et al., 2021). Trichoderma atroviride induces resistance to root-knot nematodes (RKN) in tomato, and importantly, the first generation of the tomato (F1) inherited resistance to RKN (Medeiros et al., 2017). After Arabidopsis is exposed to infection by biotrophic or necrotrophic pathogens, its progeny inherited resistance to biotrophic or necrotrophic pathogens across generations (López Sánchez et al., 2021). Trans-generational epigenetic inheritance of RdDM makes it possible to carry out plant genetic modification without changing the genotype and provides fundamental bases for the development of next-generation plant engineering approaches (Srikant and Drost, 2021).
Because of its role in DNA methylation-mediated transgenerational inheritance, sRNA can not only be applied to develop retro-resistant crops, but also to prevent triploid arrest, that is, to restore seed activity after hybridization of plants with different chromosome numbers. According to RdDM theory, 24 nt sRNA maintains TEs methylation. However, 21–22 nt easiRNA exists in Arabidopsis pollen, which is produced by miR845 targeting the tRNAMet primer binding site (PBS) of the long terminal repeat (LTR) retrotransposon (Borges et al., 2018), this paternal easiRNA can prevent DNA methylation on TEs, leading to the overexpression of PEGs, failure of endosperm cellularization and seed abortion (Martinez et al., 2018). 22 nt easiRNA is generally increased in tetraploid pollen, so easiRNA is also considered to be a quantitative marker of paternal chromosome number (Martinez et al., 2018). Studies have shown that NRPD1a inhibits easiRNA formation and saves triploid seeds (Satyaki and Gehring, 2019). Nrpd1 inbreeding mutants have a continuously enhanced ability to inhibit triploid block due to the increased loss of DNA methylation at sites that are co-regulated by Chrome methylases 2 and 3 (CMT2/3), which further reflects the inter-generational inheritance of RdDM. Elucidation of the function of sRNA-mediated DNA methylation will benefit plant cultivation and agricultural production (Wang et al., 2021b).
sRNA-mediated target mRNA cleavage is also known as RNA interference (RNAi) in which sRNAs carried by RNA-induced silencing complex (RISC) are paired with target mRNAs according to the principle of base complementary pairing and the paired regions are cleaved with the participation of AGO protein (Figure 3B). This results in 5′ and 3′ end cleavages and post-transcriptional gene silencing for mRNA (Tan et al., 2020). sRNA-mediated target cleavage is widely used in the regulation of plant growth and development under normal conditions and plays an important role in abiotic and biotic stress response (Table 1). For example, MiR1885 in Brassica is naturally maintained at a low level, while it cleaves mRNA of R gene BraTNL1 and keeps the R protein BraTNL1 at a controlled level to maintain basic immunity and nutritional development. After infection with Turnip mosaic virus (TuMV), HC-Pro protein suppressor blocks the miR1885-dependent inhibition of R gene and promotes the induction of BraTNL1, leading to the increased aggregation of immune receptors. Meanwhile, TuMV infection promotes the biosynthesis of miR1885 and triggers the synthesis of phasiR130-4 through the secondary siRNA biogenesis pathway (Figure 1E). Subsequently, phasiR130-4 mediates the silencing of photosynthesis-related gene BraCP24, leading to the acceleration of the floral transition and developmental defects in responding to viral infection (Cui et al., 2020). These examples also illustrate that one sRNA can target different sites, which complicates the regulatory network of sRNA.
As shown in Figure 2C, cross-kingdom RNAi of plant sRNA relies on EVs, which has been reported in plant- fungal pathogens interactions (Middleton et al., 2021). Plant-derived sRNA is contained in EVs and is easily absorbed by fungal cells (Cai et al., 2018). EVs in tomato root inhibit the spore germination and mycelia development of the plant pathogens Fusarium oxysporum, Botrytis cinerea and Alternaria alternate. Although studies have only focused on the protein cargo in EVs, we speculate that sRNA may play an important role in the suppression of tomato pathogens by EVs (De Palma et al., 2020). After EVs of sunflower are ingested by the fungal pathogens Sclerotiania sclerotiorum, the spores exhibit growth inhibition, morphological changes, and cell death (Regente et al., 2017). Studies have shown that many low-abundant sRNAs found in plants are abundant in fungi, and these sRNAs usually target genes important for infection to reduce the virulence of fungi (Cai et al., 2018; Thomma and Cook, 2018). When Arabidopsis is infected with B. cinerea, plant sRNAs, e.g., TAS1c-siR483 and TAS2-siR45, are delivered by EVs to cleave the mRNA of Bc-Vps51, Bc-DCTN1 and Bc-SAC1 in B. cinerea, leading to the silence of these target genes and reduced pathogenicity of B. cinerea (Cai et al., 2018). The targeting of the sRNA loaded in EV makes it possible to determine the key virulence factors of fungal pathogens by analyzing EV cargoes. However, it is still unknown how the EVs is accurately localized and how the fungus absorbs the EVs.
Fungal pathogens also deliver a series of sRNAs to plants to induce silencing of host immune genes. B. cinerea delivers Bc-siR3.1 to Arabidopsis and silences genes associated with oxidative stresses (Weiberg et al., 2013). The mechanism by which Arabidopsis-B.cinerea transmits sRNA to each other is designated as bidirectional cross-kingdom RNAi. This mechanism was also observed in cotton-Verticillium and wheat-Fusarium graminearum interactions (Zhang et al., 2016b; Jiao and Peng, 2018). This indicates that plant-fungal pathogens should be studied as an integral system. However, how to distinguish the origin of sRNA in this integral system has become a problem because both plants and fungal pathogens can produce sRNA. The genomes of most species have been sequenced, so the source of sRNA can usually be determined by homology search (Chen et al., 2021a). Using a sequential protoplast preparation method to purify fungal protoplasts from infected plant tissues, sRNAs that are transported from plant to fungal pathogen were identified (Cai et al., 2018). Fluorescent in situ hybridization can be used to study sRNA localization and expression (Huang et al., 2019b). The methods of fluorescein RNA label and fluorescent protein-sRNA vector construction are frequently used to study the absorption of sRNA (Wang et al., 2016a). The mechanisms by which EVs transport sRNA across the cell walls of plants and pathogens are still unknown. However, cell wall consists of interwoven fibrils and is incredibly elastic, which suggests that there is a potential way to control transport of sRNA by regulating the permeability of the cell wall (Coelho and Casadeval, 2019). Taken together, the bidirectional cross-kingdom RNAi mechanism requires to be further elucidated and discovered in more plant-fungal pathogens systems.
sRNA needs to form RISCs with AGO1 to exert its translational repression function. Specifically, RISCs target the 3′ or 5′ untranslational region (UTR) of mRNA or the open reading frame (ORF) and inhibit translation by affecting ribosome movement and translation process in the endoplasmic reticulum (ER; Song et al., 2019; Figure 3C). While miRNA-mediated translational repression has been extensively reported (Table 1), there are relatively few studies on siRNA-mediated translational repression. Recently, 22 nt siRNA-mediated translational repression has been reported. Normally, protein EIN5 and SKI2 inhibit siRNA to avoid endogenous gene silencing (Zhang et al., 2015). When nitrogen nutrition in the environment is limited, two genes that encode nitrate reductases NIA1 and NIA2 in Arabidopsis produce large amount of 22 nt siRNA. While 22 nt siRNA does not reduce the transcription level of NIA1 and NIA2 genes, but it significantly inhibits the translation of mRNA, indicating that 22 nt targets the translation rather than transcription of NIA1 and NIA2. This is a strategy for plants to adapt to the stress of nitrogen deficiency. Under the condition with limited nitrogen resources, the efficiency of protein translation and conversion is decreased, and the energy consumption is reduced in order to ensure the survival of the plants (Wu et al., 2020). In soybean, long inverted repeats (LIRs) located in the intron of a gene that is highly expressed in seed coat produces 22 nt siRNAs, which target the chalcone synthase (CHS) gene and trigger the biogenesis of secondary 21 nt siRNA. In the Gmdcl2a/2b mutant, 22 nt siRNAs and secondary 21 nt siRNAs cannot be produced, resulting in a significant increase in the accumulation of CHS mRNA in the seed coat, and changes of color of soybean seed coat from yellow to brown (Jia et al., 2020). sRNA can mediate translational inhibition, but the underlying mechanisms are still unclear, and need to be further explored (Ma et al., 2020). The sRNA-mediated translational inhibition has been widely used in various plant biological activities.
sRNA-mediated translational inhibition is also regulated by various proteins that are involved in the process of sRNA biogenesis. HYL1 is a member of D-body and mediates miRNA biosynthesis in the nucleus. However, recent studies have shown that HYL1 exists in the cytoplasm and ER, and HYL1 does not affect miRNA-mediated cleavage of target genes, but reduces the protein level of miRNA target genes by promoting translational repression (Yang et al., 2021a). Interestingly, it is generally believed that miRNA-mediated translational repression requires AMP1 gene, while siRNA-mediated translational repression does not require AMP1 gene (Wu et al., 2020; Yang et al., 2021a). However, recent studies have shown that AMP1 does not prevent translational repression of the SPL9 gene (target of miR156) or MYB33 gene (target of miR159), suggesting that AMP1 is not universally required for miRNA-mediated translational repression (Fouracre et al., 2020). In addition, sRNAs play an important role in the regulation of diverse plant phytohormones by controlling key factors involved in translational repression (Li et al., 2020). The plant hormone brassinosteroids (BRs) inhibit miRNA-mediated translational repression by negatively regulating the distribution of AGO1 in the ER of Arabidopsis. In BR-deficient mutants, the protein level of miRNA target genes is reduced, but can be recovered by BR treatment (Wang et al., 2021a). The controllability of sRNA-mediated translational inhibition suggests that it can be intervened manually, which provides novel strategies for the improvement of plant varieties.
Different types of sRNA are produced in the cell and reach the target site through different methods. Extensive studies on sRNA have formed a sRNA regulatory network. On one hand, based on this regulatory network, we can cultivate new traits of horticultural plants such as leaf development, flower development, fruit development and disease resistance by changing a certain process (Chen et al., 2018). On the other hand, we can actively promote population control and reduce the prevalence of plant diseases and insect pests through host-induced gene silencing (HIGS), nanoparticle-based exosome delivery of sRNA or spray-induced gene silencing (SIGS; Niu et al., 2021). However, there is still a long way to go before sRNA can be used in large-scale agriculture. New plant traits can be obtained by regulating sRNA’s ability to target mRNA, which requires more laboratory and field research in addition to the analysis of genomic results. The risks carried by genetically modified plants are also unpredictable. The study of plant EVs need to be rigorous and standardized (Pinedo et al., 2021), and the sRNA in a large number of plant EVs still need to be characterized. The application of SIGS needs to further optimize the stability of RNA in the environment and the delivery methods to improve the uptake efficiency by fungal pathogens (Qiao et al., 2021). It is highly expected that plant sRNA-based control strategies will be increasingly developed in the future to control plant diseases and insect pests and increase crop yields in an eco-friendly manner.
YT drafted the manuscript and the figures. XYa consulted the information on sRNA biogenesis and trafficking. CG consulted the information about the sRNA function. XYu conceived the idea and revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (81872951 and 82173920), Natural Science Foundation of Zhejiang Province (LGN21H280002), and Zhejiang Xinmiao Talents Program (2021R410063).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors appreciate the assistance from the Public Platform of the Medical Research Centre, Academy of Chinese Medical Science, and Zhejiang Chinese Medical University.
Agrawal, N., Dasaradhi, P. V., Mohmmed, A., Malhotra, P., Bhatnagar, R. K., and Mukherjee, S. K. (2003). RNA interference: biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. doi: 10.1128/MMBR.67.4.657-685.2003
Alfieri, M., Leone, A., and Ambrosone, A. (2021). Plant-derived nano and microvesicles for human health and therapeutic potential in nanomedicine. Pharmaceutics 13, 498–951. doi: 10.3390/pharmaceutics13040498
Axtell, M. J., Jan, C., Rajagopalan, R., and Bartel, D. P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. doi: 10.1016/j.cell.2006.09.032
Baeg, K., Iwakawa, H. O., and Tomari, Y. (2017). The poly(A) tail blocks RDR6 from converting self mRNAs into substrates for gene silencing. Nat. Plants 3, 17036–17040. doi: 10.1038/nplants.2017.36
Betti, F., Ladera-Carmona, M. J., Perata, P., and Loreti, E. (2020). RNAi mediated hypoxia stress tolerance in plants. Int. J. Mo.l Sci. 21, 9394–9410. doi: 10.3390/ijms21249394
Bhat, S. S., Bielewicz, D., Gulanicz, T., Bodi, Z., Yu, X., Anderson, S. J., et al. (2020). mRNA adenosine methylase (MTA) deposits m(6)A on pri-miRNAs to modulate miRNA biogenesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 117, 21785–21795. doi: 10.1073/pnas.2003733117
Boccara, M., Sarazin, A., Billoud, B., Bulski, A., Chapell, L., Baulcombe, D., et al. (2017). Analysis of small RNA populations using hybridization to DNA tiling arrays. Methods Mol. Biol. 1456, 127–139. doi: 10.1007/978-1-4899-7708-3_11
Bologna, N. G., Iselin, R., Abriata, L. A., Sarazin, A., Pumplin, N., Jay, F., et al. (2018). Nucleo-cytosolic shuttling of ARGONAUTE1 prompts a revised model of the plant microRNA pathway. Mol. Cell. 69, 709–719.e705. doi: 10.1016/j.molcel.2018.01.007
Borges, F., and Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Borges, F., Parent, J. S., van Ex, F., Wolff, P., Martínez, G., Köhler, C., et al. (2018). Transposon-derived small RNAs triggered by miR845 mediate genome dosage response in Arabidopsis. Nat. Genet. 50, 186–192. doi: 10.1038/s41588-017-0032-5
Brunkard, J. O., and Zambryski, P. (2019). Plant cell-cell transport via plasmodesmata is regulated by light and the circadian clock. Plant Physiol. 181, 1459–1467. doi: 10.1104/pp.19.00460
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Cai, Q., He, B., and Jin, H. (2019). A safe ride in extracellular vesicles - small RNA trafficking between plant hosts and pathogens. Curr. Opin. Plant Biol. 52, 140–148. doi: 10.1016/j.pbi.2019.09.001
Cai, Q., He, B., Wang, S., Fletcher, S., Niu, D., Mitter, N., et al. (2021). Message in a bubble: shuttling small RNAs and proteins between cells and interacting organisms using extracellular vesicles. Annu. Rev. Plant Biol. 72, 497–524. doi: 10.1146/annurev-arplant-081720-010616
Campo, S., Sánchez-Sanuy, F., Camargo-Ramírez, R., Gómez-Ariza, J., Baldrich, P., Campos-Soriano, L., et al. (2021). A novel transposable element-derived microRNA participates in plant immunity to rice blast disease. Plant Biotechnol. J. 19, 1798–1811. doi: 10.1111/pbi.13592
Cervantes-Pérez, S. A., Yong-Villalobos, L., Florez-Zapata, N. M. V., Oropeza-Aburto, A., Rico-Reséndiz, F., Amasende-Morales, I., et al. (2021). Atypical DNA methylation, sRNA-size distribution, and female gametogenesis in Utricularia gibba. Sci. Rep. 11:15725. doi: 10.1038/s41598-021-95054-y
Chen, C., Zeng, Z., Liu, Z., and Xia, R. (2018). Small RNAs, emerging regulators critical for the development of horticultural traits. Hortic. Res. 5, 63–77. doi: 10.1038/s41438-018-0072-8
Chen, C., Li, J., Feng, J., Liu, B., Feng, L., Yu, X., et al. (2021a). sRNAanno-a database repository of uniformly annotated small RNAs in plants. Hortic. Res. 8:45. doi: 10.1038/s41438-021-00480-8
Chen, X., Liu, L., Chu, Q., Sun, S., Wu, Y., Tong, Z., et al. (2021b). Large-scale identification of extracellular plant miRNAs in mammals implicates their dietary intake. PLoS One 16:e0257878. doi: 10.1371/journal.pone.0257878
Chukhchin, D. G., Bolotova, K., Sinelnikov, I., Churilov, D., and Novozhilov, E. (2019). Exosomes in the phloem and xylem of woody plants. Planta 251, 12–26. doi: 10.1007/s00425-019-03315-y
Coelho, C., and Casadeval, A. (2019). Answers to naysayers regarding microbial extracellular vesicles. Biochem. Soc. Trans. 47, 1005–1012. doi: 10.1042/BST20180252
Creasey, K. M., Zhai, J., Borges, F., Van Ex, F., Regulski, M., Meyers, B. C., et al. (2014). miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 508, 411–415. doi: 10.1038/nature13069
Cui, J., You, C., and Chen, X. (2017). The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 35, 61–67. doi: 10.1016/j.pbi.2016.11.006
Cui, C., Wang, J., Zhao, J., Fang, Y., He, X., Guo, H., et al. (2020). A brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol. Plant 13, 231–245. doi: 10.1016/j.molp.2019.11.010
Dai, X., Lu, Q., Wang, J., Wang, L., Xiang, F., and Liu, Z. (2021). MiR160 and its target genes ARF10, ARF16 and ARF17 modulate hypocotyl elongation in a light, BRZ, or PAC-dependent manner in Arabidopsis: miR160 promotes hypocotyl elongation. Plant Sci. 303:110686. doi: 10.1016/j.plantsci.2020.110686
Dávalos, A., Henriques, R., Latasa, M. J., Laparra, M., and Coca, M. (2019). Literature review of baseline information on non-coding RNA (ncRNA) to support the risk assessment of ncRNA-based genetically modified plants for food and feed. EFSA Supporting Publications 16, 1–220. doi: 10.2903/sp.efsa.2019.en-1688
De Palma, M., Ambrosone, A., Leone, A., Del Gaudio, P., Ruocco, M., Turiák, L., et al. (2020). Plant roots release small extracellular vesicles with antifungal activity. Plants 9, 1777–1791. doi: 10.3390/plants9121777
De Robertis, M., Sarra, A., D'Oria, V., Mura, F., Bordi, F., Postorino, P., et al. (2020). Blueberry-derived exosome-like nanoparticles counter the response to TNF-α-induced change on cene expression in EA.hy926 cells. Biomolecules 10, 742–759. doi: 10.3390/biom10050742
Di Donato, M., and Amari, K. (2014). “Analysis of the role of myosins in targeting proteins to plasmodesmata,” in Plasmodesmata: Methods and Protocols. ed. M. Heinlein (Strasbourg, France: IBMP-CNRS), 283–293.
Dunker, F., Trutzenberg, A., Rothenpieler, J. S., Kuhn, S., Pröls, R., Schreiber, T., et al. (2020). Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. Elife 9:e56096. doi: 10.7554/eLife.56096
Eamens, A. L., Smith, N. A., Curtin, S. J., Wang, M., and Waterhouse, P. M. (2009). The Arabidopsis thaliana double-stranded RNA binding protein DRB1 directs guide strand selection from microRNA duplexes. RNA 15, 2219–2235. doi: 10.1261/rna.1646909
Erdmann, R. M., and Picard, C. L. (2020). RNA-directed DNA methylation. PLoS Genet. 16:e1009034. doi: 10.1371/journal.pgen.1009034
Fabbiano, F., Corsi, J., Gurrieri, E., Trevisan, C., Notarangelo, M., and D'Agostino, V. G. (2020). RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J. Extracell. Vesicles 10:e12043. doi: 10.1002/jev2.12043
Fang, X., Shi, Y., Lu, X., Chen, Z., and Qi, Y. (2015). CMA33/XCT regulates small RNA production through modulating the transcription of dicer-like genes in Arabidopsis. Mol. Plant 8, 1227–1236. doi: 10.1016/j.molp.2015.03.002
Fang, X., Zhao, G., Zhang, S., Li, Y., Gu, H., Li, Y., et al. (2019). Chloroplast-to-nucleus signaling regulates microRNA biogenesis in Arabidopsis. Dev. Cell 48, 371–382.e4. doi: 10.1016/j.devcel.2018.11.046
Fang, Y., and Spector, D. L. (2007). Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr. Biol. 17, 818–823. doi: 10.1016/j.cub.2007.04.005
Fei, Y., Nyikó, T., and Molnar, A. (2021). Non-perfectly matching small RNAs can induce stable and heritable epigenetic modifications and can be used as molecular markers to trace the origin and fate of silencing RNAs. Nucleic. Acids. Res. 49, 1900–1913. doi: 10.1093/nar/gkab023
Fouracre, J. P., Chen, V. J., and Poethig, R. S. (2020). ALTERED MERISTEM PROGRAM1 regulates leaf identity independently of miR156-mediated translational repression. Development 147:dev186874. doi: 10.1242/dev.186874
Gao, S., Wang, J., Jiang, N., Zhang, S., Wang, Y., Zhang, J., et al. (2020). Hyponastic leaves 1 protects pri-miRNAs from nuclear exosome attack. Proc. Natl. Acad. Sci. U. S. A. 117, 17429–17437. doi: 10.1073/pnas.2007203117
Garnelo Gómez, B., Rosas-Díaz, T., Shi, C., Fan, P., Zhang, D., Rufián, J. S., et al. (2021). The viral silencing suppressor P19 interacts with the receptor-like kinases BAM1 and BAM2 and suppresses the cell-to-cell movement of RNA silencing independently of its ability to bind sRNA. New Phytol. 229, 1840–1843. doi: 10.1111/nph.16981
Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J., et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during turnip mosaic virus infection. Plant Cell 22, 481–496. doi: 10.1105/tpc.109.073056
Grzybkowska, D., Nowak, K., and Gaj, M. D. (2020). Hypermethylation of auxin-responsive motifs in the promoters of the transcription factor genes accompanies the somatic embryogenesis induction in Arabidopsis. Int. J. Mol. Sci. 21, 6849–6871. doi: 10.3390/ijms21186849
Guo, W., Wang, D., and Lisch, D. (2021). RNA-directed DNA methylation prevents rapid and heritable reversal of transposon silencing under heat stress in Zea mays. PLoS Genet. 17:e1009326. doi: 10.1371/journal.pgen.1009326
Ham, B., Li, G., Jia, W., Leary, J., and Lucas, W. (2014). Systemic delivery of siRNA in pumpkin by a plant PHLOEM SMALL RNA-BINDING PROTEIN 1-ribonucleoprotein complex. Plant J. 80, 683–694. doi: 10.1111/tpj.12662
Han, J., LaVigne, C. A., Jones, B. T., Zhang, H., Gillett, F., and Mendell, J. T. (2020). A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming. Science 370:eabc9546. doi: 10.1126/science.abc9546
He, B., Cai, Q., Qiao, L., Huang, C., Wang, S., Miao, W., et al. (2021). RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat. Plants 7, 342–352. doi: 10.1038/s41477-021-00863-8
Hu, G., Hao, M., Wang, L., Liu, J., Zhang, Z., Tang, Y., et al. (2020). The cotton miR477-CBP60A module participates in plant defense against Verticillium dahlia. Mol.Plant Microbe. Interact. 33, 624–636. doi: 10.1094/MPMI-10-19-0302-R
Huang, C., Wang, H., Hu, P., Hamby, R., and Jin, H. (2019a). Small RNAs - big players in plant-microbe interactions. Cell Host. Microbe. 26, 173–182. doi: 10.1016/j.chom.2019.07.021
Huang, K., Baldrich, P., Meyers, B., and Caplan, J. L. (2019b). sRNA-FISH: versatile fluorescent in situ detection of small RNAs in plants. Plant J. 98, 359–369. doi: 10.1111/tpj.14210
Huang, P., Huang, H., Lin, X., Liu, P., Zhao, L., Nie, W., et al. (2021). MSI4/FVE is required for accumulation of 24-nt siRNAs and DNA methylation at a subset of target regions of RNA-directed DNA methylation. Plant J. 108, 347–357. doi: 10.1111/tpj.15441
Huang, Y., Ji, L., Huang, Q., Vassylyev, D. G., Chen, X., and Ma, J. (2009). Structural insights into mechanisms of the small RNA methyltransferase HEN1. Nature 461, 823–827. doi: 10.1038/nature08433
Ji, H., Mao, H., Li, S., Feng, T., Zhang, Z., Cheng, L., et al. (2021). Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol. 232, 705–718. doi: 10.1111/nph.17436
Jia, J., Ji, R., Li, Z., Yu, Y., Nakano, M., Long, Y., et al. (2020). Soybean DICER-LIKE2 regulates seed coat color via production of primary 22-nucleotide small interfering RNAs from long inverted repeats. Plant Cell 32, 3662–3673. doi: 10.1105/tpc.20.00562
Jiang, A., Guo, Z., Pan, J., Yang, Y., Zhuang, Y., Zuo, D., et al. (2021). The PIF1-miR408-PLANTACYANIN repression cascade regulates light-dependent seed germination. Plant Cell 33, 1506–1529. doi: 10.1093/plcell/koab060
Jiao, J., and Peng, D. (2018). Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J. Plant Interact. 13, 514–521. doi: 10.1080/17429145.2018.1528512
Kim, E. Y., Wang, L., Lei, Z., Li, H., Fan, W., and Cho, J. (2021). Ribosome stalling and SGS3 phase separation prime the epigenetic silencing of transposons. Nat. Plants 7, 303–309. doi: 10.1038/s41477-021-00867-4
Komiya, R. (2017). Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and dicer-processing. J. Plant Res. 130, 17–23. doi: 10.1007/s10265-016-0878-0
Kumar, S., and Mohapatra, T. (2021). Dynamics of DNA methylation and its functions in plant growth and development. Front. Plant Sci. 12:596236. doi: 10.3389/fpls.2021.596236
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U. S. A. 101, 12753–12761. doi: 10.1073/pnas.0403115101
Leidal, A. M., Huang, H., Marsh, T., Solvik, T., Zhang, D., Ye, J., et al. (2020). The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199. doi: 10.1038/s41556-019-0450-y
Leonetti, P., Miesen, P., van Rij, R. P., and Pantaleo, V. (2020). Viral and subviral derived small RNAs as pathogenic determinants in plants and insects. Adv. Virus Res. 107, 1–36. doi: 10.1016/bs.aivir.2020.04.001
Li, M., and Yu, B. (2021). Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 18, 2087–2096. doi: 10.1080/15476286.2021.1899491
Li, S., Le, B., Ma, X., Li, S., You, C., Yu, Y., et al. (2016). Biogenesis of phased siRNAs on membrane-bound polysomes in Arabidopsis. Elife 5:e22750. doi: 10.7554/eLife.22750
Li, X., Huang, Y., Sun, M., Ji, H., Dou, H., Hu, J., et al. (2018). Honeysuckle-encoded microRNA2911 inhibits Enterovirus 71 replication via targeting VP1 gene. Antiviral. Res. 152, 117–123. doi: 10.1016/j.antiviral.2018.02.015
Li, T., Gonzalez, N., Inzé, D., and Dubois, M. (2020). Emerging connections between small RNAs and phytohormones. Trends Plant Sci. 25, 912–929. doi: 10.1016/j.tplants.2020.04.004
Lin, Y., Zhu, Y., Cui, Y., Chen, R., Chen, Z., Li, G., et al. (2021). Derepression of specific miRNA-target genes in rice using CRISPR/Cas9. J. Exp. Bot. 72, 7067–7077. doi: 10.1093/jxb/erab336
Liu, L., and Chen, X. (2018). Intercellular and systemic trafficking of RNAs in plants. Nat. Plants 4, 869–878. doi: 10.1038/s41477-018-0288-5
Liu, J., and He, Z. (2020). Small DNA methylation, big player in plant abiotic stress responses and memory. Front. Plant Sci. 11:595603. doi: 10.3389/fpls.2020.595603
Liu, Y., Teng, C., Xia, R., and Meyers, B. C. (2020). PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell 32, 3059–3080. doi: 10.1105/tpc.20.00335
López Sánchez, A., Pascual-Pardo, D., Furci, L., Roberts, M. R., and Ton, J. (2021). Costs and benefits of transgenerational induced resistance in Arabidopsis. Front. Plant Sci. 12:644999. doi: 10.3389/fpls.2021.644999
Ma, X., Ibrahim, F., Kim, E. J., Shaver, S., Becker, J., Razvi, F., et al. (2020). An ortholog of the vasa intronic gene is required for small RNA-mediated translation repression in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U. S. A. 117, 761–770. doi: 10.1073/pnas.1908356117
Ma, S., Ji, Y., Dong, Y., Chen, S., Wang, Y., and Lü, S. (2021). An environmental-friendly pesticide-fertilizer combination fabricated by in-situ synthesis of ZIF-8. Sci. Total Environ. 789:147845. doi: 10.1016/j.scitotenv.2021.147845
Malabarba, J., Windels, D., Xu, W., and Verdier, J. (2021). Regulation of DNA (de)methylation positively impacts seed germination during seed development under heat stress. Genes 12, 457–478. doi: 10.3390/genes12030457
Manavella, P. A., Yang, S., and Palatnik, J. (2019). Keep calm and carry on: miRNA biogenesis under stress. Plant J. 99, 832–843. doi: 10.1111/tpj.14369
Mar-Aguilar, F., Arreola-Triana, A., Mata-Cardona, D., Gonzalez-Villasana, V., Rodríguez-Padilla, C., and Reséndez-Pérez, D. (2020). Evidence of transfer of miRNAs from the diet to the blood still inconclusive. PeerJ. 8:e9567. doi: 10.7717/peerj.9567
Marchais, A., Chevalier, C., and Voinnet, O. (2019). Extensive profiling in Arabidopsis reveals abundant polysome-associated 24-nt small RNAs including AGO5-dependent pseudogene-derived siRNAs. RNA 25, 1098–1117. doi: 10.1261/rna.069294.118
Martinez, G., Wolff, P., Wang, Z., Moreno-Romero, J., Santos-González, J., Conze, L., et al. (2018). Paternal easiRNAs regulate parental genome dosage in Arabidopsis. Nat. Genet. 50, 193–198. doi: 10.1038/s41588-017-0033-4
Matzke, M. A., and Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Medeiros, H. A., Araújo Filho, J. V., Freitas, L. G., Castillo, P., Rubio, M. B., Hermosa, R., et al. (2017). Tomato progeny inherit resistance to the nematode Meloidogyne javanica linked to plant growth induced by the biocontrol fungus Trichoderma atroviride. Sci. Rep. 7:40216. doi: 10.1038/srep40216
Megel, C., Hummel, G., Lalande, S., Ubrig, E., Cognat, V., Morelle, G., et al. (2019). Plant RNases T2, but not dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic. Acids. Res. 47, 941–952. doi: 10.1093/nar/gky1156
Mekapogu, M., Jung, J. A., Kwon, O. K., Ahn, M. S., Song, H. Y., and Jang, S. (2021). Recent progress in enhancing fungal disease resistance in ornamental plants. Int. J. Mol. Sci. 22, 7956–7976. doi: 10.3390/ijms22157956
Middleton, H., Yergeau, É., Monard, C., Combier, J. P., and El Amrani, A. (2021). Rhizospheric plant-microbe interactions: miRNAs as a key mediator. Trends. Plant Sci. 26, 132–141. doi: 10.1016/j.tplants.2020.09.005
Molnar, A., Melnyk, C. W., Bassett, A., Hardcastle, T. J., Dunn, R., and Baulcombe, D. C. (2010). Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328, 872–875. doi: 10.1126/science.1187959
Morán-Diez, M. E., Martínez de Alba, Á. E., Rubio, M. B., Hermosa, R., and Monte, E. (2021). Trichoderma and the plant heritable priming responses. J. Fungi. 7, 318–331. doi: 10.3390/jof7040318
Niu, D., Hamby, R., Sanchez, J. N., Cai, Q., Yan, Q., and Jin, H. (2021). RNAs—a new frontier in crop protection. Curr. Opin. Biotechnol. 70, 204–212. doi: 10.1016/j.copbio.2021.06.005
Nowak, K., Morończyk, J., Wójcik, A., and Gaj, M. D. (2020). AGL15 controls the embryogenic reprogramming of somatic cells in Arabidopsis through the histone acetylation-mediated repression of the miRNA biogenesis genes. Int. J. Mol. Sci. 21, 6733–6753. doi: 10.3390/ijms21186733
Ó’Maoiléidigh, D. S., van Driel, A. D., Singh, A., Sang, Q., Le Bec, N., Vincent, C., et al. (2021). Systematic analyses of the MIR172 family members of Arabidopsis define their distinct roles in regulation of APETALA2 during floral transition. PLoS Biol. 19:e3001043. doi: 10.1371/journal.pbio.3001043
Parent, J. S., Bouteiller, N., Elmayan, T., and Vaucheret, H. (2015). Respective contributions of Arabidopsis DCL2 and DCL4 to RNA silencing. Plant J. 81, 223–232. doi: 10.1111/tpj.12720
Park, E. J., and Kim, T. H. (2018). Fine-tuning of gene expression by tRNA-derived fragments during abiotic stress signal transduction. Int. J. Mol. Sci. 19, 518–529. doi: 10.3390/ijms19020518
Parmar, S., Gharat, S. A., Tagirasa, R., Chandra, T., Behera, L., Dash, S. K., et al. (2020). Identification and expression analysis of miRNAs and elucidation of their role in salt tolerance in rice varieties susceptible and tolerant to salinity. PLoS One 15:e0230958. doi: 10.1371/journal.pone.0230958
Peng, Y., Li, S., Yan, J., Tang, Y., Cheng, J., Gao, A., et al. (2021). Research progress on phytopathogenic fungi and their role as biocontrol agents. Front. Microbiol. 12:670135. doi: 10.3389/fmicb.2021.670135
Perut, F., Roncuzzi, L., Avnet, S., Massa, A., Zini, N., Sabbadini, S., et al. (2021). Strawberry-derived exosome-like nanoparticles prevent oxidative stress in human mesenchymal stromal cells. Biomolecules 11, 3390–3404. doi: 10.3390/biom11010087
Pinedo, M., de la Canal, L., and de Marcos Lousa, C. (2021). A call for rigor and standardization in plant extracellular vesicle research. J. Extracell. Vesicles 10:e12048. doi: 10.1002/jev2.12048
Prado, N., Alche Jde, D., Casado-Vela, J., Mas, S., Villalba, M., Rodriguez, R., et al. (2014). Nanovesicles are secreted during pollen germination and pollen tube growth: a possible role in fertilization. Mol. Plant 7, 573–577. doi: 10.1093/mp/sst153
Pyo, Y., Kim, G. M., Choi, S. W., Song, C. Y., Yang, S. W., and Jung, I. L. (2020). Strontium stress disrupts miRNA biogenesis by reducing HYL1 protein levels in Arabidopsis. Ecotoxicol. Environ. Saf. 204:111056. doi: 10.1016/j.ecoenv.2020.111056
Qiao, L., Zheng, L., Sheng, C., Zhao, H., Jin, H., and Niu, D. (2020). Rice siR109944 suppresses plant immunity to sheath blight and impacts multiple agronomic traits by affecting auxin homeostasis. Plant J. 102, 948–964. doi: 10.1111/tpj.14677
Qiao, L., Lan, C., Capriotti, L., Ah-Fong, A., Nino Sanchez, J., Hamby, R., et al. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 19, 1756–1768. doi: 10.1111/pbi.13589
Quadrana, L., Bortolini Silveira, A., Mayhew, G. F., LeBlanc, C., Martienssen, R. A., Jeddeloh, J. A., et al. (2016). The Arabidopsis thaliana mobilome and its impact at the species level. Elife 5:e15716. doi: 10.7554/eLife.15716
Regente, M., Pinedo, M., San Clemente, H., Balliau, T., Jamet, E., and de la Canal, L. (2017). Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 68, 5485–5495. doi: 10.1093/jxb/erx355
Ren, B., Wang, X., Duan, J., and Ma, J. (2019). Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365, 919–922. doi: 10.1126/science.aav8907
Rymen, B., Ferrafiat, L., and Blevins, T. (2020). Non-coding RNA polymerases that silence transposable elements and reprogram gene expression in plants. Transcription 11, 172–191. doi: 10.1080/21541264.2020.1825906
Santangelo, L., Giurato, G., Cicchini, C., Montaldo, C., Mancone, C., Tarallo, R., et al. (2016). The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 17, 799–808. doi: 10.1016/j.celrep.2016.09.031
Satyaki, P. R. V., and Gehring, M. (2019). Paternally acting canonical RNA-directed DNA methylation pathway genes sensitize Arabidopsis endosperm to paternal genome dosage. Plant Cell 31, 1563–1578. doi: 10.1105/tpc.19.00047
Shao, H., Im, H., Castro, C. M., Breakefield, X., Weissleder, R., and Lee, H. (2018). New technologies for analysis of extracellular vesicles. Chem. Rev. 118, 1917–1950. doi: 10.1021/acs.chemrev.7b00534
Shahid, S., Kim, G., Johnson, N. R., Wafula, E., Wang, F., Coruh, C., et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82–85. doi: 10.1038/nature25027
Si, F., Cao, X., Song, X., and Deng, X. (2020). Processing of coding and non-coding RNAs in plant development and environmental responses. Essays Biochem. 64, 931–945. doi: 10.1042/EBC20200029
Simone, R., Javad, F., Emmett, W., Wilkins, O. G., Almeida, F. L., Barahona-Torres, N., et al. (2021). MIR-NATs repress MAPT translation and aid proteostasis in neurodegeneration. Nature 594, 117–123. doi: 10.1038/s41586-021-03556-6
Singh, R. K., Prasad, A., Maurya, J., and Prasad, M. (2021). Regulation of small RNA-mediated high temperature stress responses in crop plants. Plant Cell Rep. doi: 10.1007/s00299-021-02745-x --> [Epub ahead of print]
Song, X., Li, Y., Cao, X., and Qi, Y. (2019). MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Srikant, T., and Drost, H. G. (2021). How stress facilitates phenotypic innovation through epigenetic diversity. Front. Plant Sci. 11:606800. doi: 10.3389/fpls.2020.606800
Tamiru, M., Hardcastle, T. J., and Lewsey, M. G. (2018). Regulation of genome-wide DNA methylation by mobile small RNAs. New Phytol. 217, 540–546. doi: 10.1111/nph.14874
Tan, H., Li, B., and Guo, H. (2020). The diversity of post-transcriptional gene silencing mediated by small silencing RNAs in plants. Essays Biochem. 64, 919–930. doi: 10.1042/EBC20200006
Tang, K. (2020). “Small RNA and DNA methylation in plants,” in Plant Small RNA: Biogenesis, Regulation and Application. eds. P. Guleria and V. Kumar (India: DAV University, Lovely Professional University), 353–376.
Thomma, B., and Cook, D. E. (2018). Targeting microbial pathogens. Science 360, 1070–1071. doi: 10.1126/science.aat9343
Tian, P., Zhang, X., Xia, R., Liu, Y., Wang, M., Li, B., et al. (2021). Evolution and diversification of reproductive phased small interfering RNAs in Oryza species. New Phytol. 229, 2970–2983. doi: 10.1111/nph.17035
Tomassi, A. H., Re, D. A., Romani, F., Cambiagno, D. A., Gonzalo, L., Moreno, J. E., et al. (2020). The intrinsically disordered protein CARP9 bridges HYL1 to AGO1 in the nucleus to promote microRNA activity. Plant Physiol. 184, 316–329. doi: 10.1104/pp.20.00258
Vivek, A. T., Zahra, S., and Kumar, S. (2020). From current knowledge to best practice: a primer on viral diagnostics using deep sequencing of virus-derived small interfering RNAs (vsiRNAs) in infected plants. Methods 183, 30–37. doi: 10.1016/j.ymeth.2019.10.009
Vogel, J., Bartels, V., Tang, T., Churakov, G., Slagter-Jäger, J. G., Hüttenhofer, A., et al. (2003). RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003, 6435–6443. doi: 10.1093/nar/gkg867
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wang, M., and Dean, R. A. (2020). Movement of small RNAs in and between plants and fungi. Mol. Plant Pathol. 21, 589–601. doi: 10.1111/mpp.12911
Wang, M., Weiberg, A., Lin, F., Thomma, B. P., Huang, H., and Jin, H. (2016a). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2:16151. doi: 10.1038/nplants.2016.151
Wang, Y., Wang, Y., Song, Z., and Zhang, H. (2016b). Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. doi: 10.1016/j.molp.2016.07.003
Wang, B., Cheng, D., Chen, Z., Zhang, M., Zhang, G., Jiang, M., et al. (2019). Bioinformatic exploration of the targets of xylem sap miRNAs in maize under cadmium stress. Int. J. Mol. Sci. 20, 1474–1490. doi: 10.3390/ijms20061474
Wang, T., Zheng, Y., Tang, Q., Zhong, S., Su, W., and Zheng, B. (2021a). Brassinosteroids inhibit miRNA-mediated translational repression by decreasing AGO1 on the endoplasmic reticulum. J. Integr. Plant Biol. 63, 1475–1490. doi: 10.1111/jipb.13139
Wang, Z., Butel, N., Santos-González, J., Simon, L., Wärdig, C., and Köhler, C. (2021b). Transgenerational effect of mutants in the RNA-directed DNA methylation pathway on the triploid block in Arabidopsis. Genome. Biol. 22, 141–158. doi: 10.1186/s13059-021-02359-2
Weiberg, A., Wang, M., Lin, F., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Welchen, E., and Gonzalez, D. H. (2021). Breaking boundaries: exploring short- and long-distance mitochondrial signalling in plants. New Phytol. 232, 494–501. doi: 10.1111/nph.17614
Witwer, K. W. (2018). Alternative miRNAs? Human sequences misidentified as plant miRNAs in plant studies and in human plasma. F1000Res 7, 244–255. doi: 10.12688/f1000research.14060.1
Wu, H., Li, B., Iwakawa, H. O., Pan, Y., Tang, X., Ling-Hu, Q., et al. (2020). Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature 581, 89–93. doi: 10.1038/s41586-020-2231-y
Xia, C., Zhou, H., Xu, X., Jiang, T., Li, S., Wang, D., et al. (2020). Identification and investigation of miRNAs from Gastrodia elata blume and their potential function. Front. Pharmacol. 11:542405. doi: 10.3389/fphar.2020.542405
Xie, Y., Liu, G., Zang, X., Hu, Q., Zhou, C., Li, Y., et al. (2021). Differential expression pattern of goat uterine fluids extracellular vesicles miRNAs during peri-implantation. Cells 10, 2308–2315. doi: 10.3390/cells10092308
Xie, Z., Allen, E., Fahlgren, N., Calamar, A., Givan, S. A., and Carrington, J. C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138, 2145–2154. doi: 10.1104/pp.105.062943
Xin, Y., Ma, B., Zeng, Q., He, W., Qin, M., and He, N. (2021). Dynamic changes in transposable element and gene methylation in mulberry (Morus notabilis) in response to Botrytis cinerea. Hortic. Res. 8, 154–168. doi: 10.1038/s41438-021-00588-x
Yan, Y., Ham, B. K., Chong, Y., Yeh, S. D., and Lucas, W. J. (2020). A plant SMALL RNA-BINDING PROTEIN 1 family mediates cell-to-cell trafficking of RNAi signals. Mol. Plant 13, 321–335. doi: 10.1016/j.molp.2019.12.001
Yang, X., Dong, W., Ren, W., Zhao, Q., Wu, F., and He, Y. (2021a). Cytoplasmic HYL1 modulates miRNA-mediated translational repression. Plant Cell 33, 1980–1996. doi: 10.1093/plcell/koab090
Yang, X., You, C., Wang, X., Gao, L., Mo, B., Liu, L., et al. (2021b). Widespread occurrence of microRNA-mediated target cleavage on membrane-bound polysomes. Genome. Biol. 22, 15–46. doi: 10.1186/s13059-020-02242-6
Yao, W., Li, Y., Xie, W., and Wang, L. (2020). Features of sRNA biogenesis in rice revealed by genetic dissection of sRNA expression level. Comput. Struct. Biotechnol. J. 18, 3207–3216. doi: 10.1016/j.csbj.2020.10.012
Ye, R., Chen, Z., Lian, B., Rowley, M. J., Xia, N., Chai, J., et al. (2016). A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol. Cell 61, 222–235. doi: 10.1016/j.molcel.2015.11.015
Yu, D., Meng, Y., Zuo, Z., Xue, J., and Wang, H. (2016). NATpipe: an integrative pipeline for systematical discovery of natural antisense transcripts (NATs) and phase-distributed nat-siRNAs from de novo assembled transcriptomes. Sci. Rep. 6:21666. doi: 10.1038/srep21666
Zhang, X., Xia, J., Lii, Y. E., Barrera-Figueroa, B. E., Zhou, X., Gao, S., et al. (2012). Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome. Biol. 13:R20. doi: 10.1186/gb-2012-13-3-r20
Zhang, X., Zhu, Y., Liu, X., Hong, X., Xu, Y., Zhu, P., et al. (2015). Plant biology. Suppression of endogenous gene silencing by bidirectional cytoplasmic RNA decay in Arabidopsis. Science 348, 120–123. doi: 10.1126/science.aaa2618
Zhang, H., Tao, Z., Hong, H., Chen, Z., Wu, C., Li, X., et al. (2016a). Transposon-derived small RNA is responsible for modified function of WRKY45 locus. Nat. Plants 2:16016. doi: 10.1038/nplants.2016.16
Zhang, T., Zhao, Y., Zhao, J., Wang, S., Jin, Y., Chen, Z., et al. (2016b). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2:16153. doi: 10.1038/nplants.2016.153
Zhang, C., Fan, L., Le, B., Ye, P., Mo, B., and Chen, X. (2020). Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev. Cell 55, 603–616.e605. doi: 10.1016/j.devcel.2020.10.019
Zhang, Z., Teotia, S., Tang, J., and Tang, G. (2019). Perspectives on microRNAs and phased amall interfering RNAs in maize (Zea mays L.): functions and big impact on agronomic traits enhancement. Plants 8, 170–187. doi: 10.3390/plants8060170
Zhao, D., and Song, G. (2014). Rootstock-to-scion transfer of transgene-derived small interfering RNAs and their effect on virus resistance in nontransgenic sweet cherry. Plant Biotechnol J 12, 1319–1328. doi: 10.1111/pbi.12243
Zhao, D., Zhong, G. Y., and Song, G. Q. (2020). Transfer of endogenous small RNAs between branches of scions and rootstocks in grafted sweet cherry trees. PLoS One 15:e0236376. doi: 10.1371/journal.pone.0236376
Zhou, Z., Li, X., Liu, J., Dong, L., Chen, Q., Liu, J., et al. (2015). Honeysuckle-encoded atypical microRNA2911 directly targets influenza a viruses. Cell Res. 25, 39–49. doi: 10.1038/cr.2014.130
Zhou, H., Lin, R., Huang, H., Li, L., Cai, T., Zhu, J., et al. (2020). The CCR4-NOT complex component NOT1 regulates RNA-directed DNA methylation and transcriptional silencing by facilitating pol IV-dependent siRNA production. Plant J. 103, 1503–1515. doi: 10.1111/tpj.14818
Zhu, H., Chen, Y., Zhong, S., Dardick, C., Callahan, A., An, Y. Q., et al. (2020a). Thermal-responsive genetic and epigenetic regulation of DAM cluster controlling dormancy and chilling requirement in peach floral buds. Hortic. Res. 7, 114–128. doi: 10.1038/s41438-020-0336-y
Zhu, J., Park, J., Lee, S., Lee, J. H., Hwang, D., Kwak, J. M., et al. (2020b). Regulation of stomatal development by stomatal lineage miRNAs. Proc. Natl. Acad. Sci. U. S. A. 117, 6237–6245. doi: 10.1073/pnas.1919722117
Keywords: small RNA, biogenesis, trafficking, functions, DNA methylation, RNA interference, translational repression
Citation: Tang Y, Yan X, Gu C and Yuan X (2022) Biogenesis, Trafficking, and Function of Small RNAs in Plants. Front. Plant Sci. 13:825477. doi: 10.3389/fpls.2022.825477
Received: 30 November 2021; Accepted: 19 January 2022;
Published: 17 February 2022.
Edited by:
Jie Qiu, Shanghai Normal University, ChinaReviewed by:
Collin Hudzik, The Pennsylvania State University (PSU), United StatesCopyright © 2022 Tang, Yan, Gu and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofeng Yuan, eXVhbnhpYW9mZW5nQHpjbXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.