
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 February 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.815859
This article is part of the Research TopicStructure and Function of Chloroplasts, Volume IIIView all 13 articles
Although numerous studies have been carried out on chloroplast development and biogenesis, the underlying regulatory mechanisms are still largely elusive. Here, we characterized a chloroplast stromal protein Chloroplast Development and Biogenesis1 (CDB1). The knockout cdb1 mutant exhibits a seedling-lethal and ivory leaf phenotype. Immunoblot and RNA blot analyses show that accumulation of chloroplast ribosomes is compromised in cdb1, resulting in an almost complete loss of plastid-encoded proteins including the core subunits of the plastid-encoded RNA polymerase (PEP) RpoB and RpoC2, and therefore in impaired PEP activity. Orthologs of CDB1 are found in green algae and land plants. Moreover, a protein shows high similarity with CDB1, designated as CDB1-Like (CDB1L), is present in angiosperms. Absence of CDB1L results in impaired embryo development. While CDB1 is specifically located in the chloroplast stroma, CDB1L is localized in both chloroplasts and mitochondria in Arabidopsis. Thus, our results demonstrate that CDB1 is indispensable for chloroplast development and biogenesis through its involvement in chloroplast ribosome assembly whereas CDB1L may fulfill a similar function in both mitochondria and chloroplasts.
Chloroplasts are the sites of photosynthesis in eukaryotic cells and arose from a cyanobacterium-like ancestor through endosymbiosis. In higher plants, light triggers chloroplast development from undifferentiated small organelles called proplastids in meristematic cells (Pogson and Albrecht, 2011). During differentiation and biogenesis, chloroplasts transcribe their own genome into mRNAs for protein synthesis. These proteins are essential for the development of functional chloroplasts (Sakamoto et al., 2008). Chloroplast genomes of green plants comprise ~120 genes encoding the components of the gene expression system (RNA polymerase core subunits, ribosomal proteins, tRNAs, and rRNAs), subunits of the photosynthetic machinery [Rubisco, photosystem I and II (PSI and PSII), cytochrome b6f complex (Cyt b6f), ATP synthase, and NAD(P)H dehydrogenase-like (NDH) complex] as well as some other proteins involved in various metabolic processes in chloroplasts (Dobrogojski et al., 2020). In addition to the plastid-encoded proteins, chloroplasts also contain ~3,000 nucleus-encoded proteins which are synthesized on cytosolic ribosomes and then imported into chloroplasts (Friso et al., 2004; Heazlewood et al., 2006). Thus, chloroplast development and biogenesis require tight coordination of plastid and nuclear gene expression.
Chloroplast transcription involves the interplay of two types of RNA polymerases: nuclear-encoded phage-type RNA polymerase (NEP) and plastid-encoded bacterial-type RNA polymerase (PEP; Shiina et al., 2005; Börner et al., 2015). In Arabidopsis, NEP is encoded by the nuclear genes RpoTp and RpoTmp, and is essential for transcription of the plastid PEP transcription machinery including rpoA and rpoB-C1-C2 as well as some plastid housekeeping genes (Steiner et al., 2011; Pfalz and Pfannschmidt, 2013). Plastid genes involved in photosynthesis are primarily transcribed by the PEP transcription machinery (Swiatecka-Hagenbruch et al., 2007; Liere et al., 2011). PEP is composed of four catalytic core subunits (α, β, β′, and β″), which are encoded by rpoA, rpoB, rpoC1, and rpoC2, respectively. Besides its four core subunits, PEP activity is also regulated by at least 12 PEP-associated proteins (PAPs; Pfalz and Pfannschmidt, 2013). Intriguingly, most mutants of these PEP complex subunits display an albino/ivory leaf phenotype with arrested plastid development and strongly decreased expression of PEP-dependent genes (Pfalz and Pfannschmidt, 2013).
Translation in chloroplasts is performed by prokaryotic-type 70S ribosomes composed of a large 50S and a small 30S subunit. Biochemical analysis showed that, in spinach, the 50S subunit contains 23S, 5S, and 4.5S rRNA as well as 33 ribosomal proteins whereas the 30S subunit consists of 16S rRNA and 25 ribosomal proteins (Manuell et al., 2007; Graf et al., 2016; Perez Boerema et al., 2018). Because of their common origin, chloroplast and bacterial ribosomes exhibit some conserved structural and functional features such as the mRNA decoding (30S small subunit) and the peptide bond synthesis (50S large subunit) functions (Manuell et al., 2007; Sharma et al., 2007). Besides the classical ribosomal proteins which have orthologs in Escherichia coli, five plastid-specific ribosomal proteins (PSRPs) with essential functional roles were also found in plant plastid ribosomes (Sharma et al., 2010; Tiller et al., 2012). Although the overall structural organization of chloroplast ribosomes has been characterized, little is known about the molecular mechanisms underlying their assembly.
Ribosome biogenesis is a complicated process that comprises the transcription of a large pre-rRNA precursor, its processing and folding and the assembly of the ribosomal proteins with the mature rRNAs (Shajani et al., 2011; Weis et al., 2015). So far, a dozen of factors appears to be required for the maturation of rRNAs in chloroplasts, such as RNase R homolog 1 (RNR1) which is involved in the maturation of 23S, 16S, and 5S rRNAs (Bollenbach et al., 2005). Loss of endonucleases RNase E in Arabidopsis causes defective rRNA processing and subsequent plastid ribosome deficiency (Schein et al., 2008; Walter et al., 2010). The DEAD-box RNA helicases RH22 and RH39 function in the assembly of the 50S ribosomal subunit and 23S rRNA processing, respectively (Nishimura et al., 2010; Chi et al., 2012). A conserved protein with an unknown functional DUF177 domain is specifically required for the accumulation of 23S rRNA (Yang et al., 2016). In addition to the factors required for chloroplast rRNA maturation, there are many factors involved in the biogenesis of chloroplast ribosomes. For example, ObgC is a GTPase that associates with chloroplast 50S ribosomal subunits through 23S rRNA (Bang et al., 2012). Pro-rich protein CGL20 is also required for the assembly of the 50S ribosomal subunit (Reiter et al., 2020).
Here, we have characterized an Arabidopsis mutant that displays an ivory leaf phenotype with arrested chloroplast development. We designated this mutant chloroplast development and biogenesis1 (cdb1). The CDB1 gene encodes a protein with unknown function that is targeted to the chloroplast stroma. Loss of CDB1 leads to defects in accumulation of plastid-encoded proteins, including chloroplast ribosomes. These data suggest that CDB1 is indispensable for chloroplast development through its involvement in chloroplast ribosome assembly. We also provide evidence that the paralog of CDB1, CDB1-Like (CDB1L), is located both in chloroplasts and mitochondria and essential for embryo development in Arabidopsis.
Mutants of cdb1 (SALK_080811C) and cdb1l (GK-844F05) were obtained from the Nottingham Arabidopsis Stock Center (NASC). The T-DNA insertions were confirmed by PCR analysis and subsequent sequencing with the primers CDB1-TF and CDB1-TR, CDB1L-TF, and CDB1L-TR (for all primer sequences, see Supplementary Table 3), respectively. Seeds were planted in Murashige and Skoog (MS) culture medium (pH 5.8) with 3% sucrose and 0.7% agar at 4°C in the dark for 48 h. Then the plants were cultured under long-day conditions (16 h-light/8 h-dark) at 23°C with an irradiance of 50 μmol photons m−2s−1 for 3–4 weeks. For complementation of the mutants, genomic sequences of CDB1 (AT4G37920) and CDB1L (AT1G36320) plus the upstream promoters were cloned into pCAMBIA1301 to produce transgenic lines. The genomic sequence of CDB1 was fused with the HA tag and cloned into the pCAMBIA1301 vector to generate CDB1-HA transgenic plants. All the above vectors were transferred into Agrobacterium tumefaciens strain GV3101 and then introduced into Arabidopsis by the floral-dip method (Clough and Bent, 1998).
Leaves from 2-week-old plants grown on MS medium with 3% sucrose were fixed with 2.5% glutaraldehyde in phosphate buffer (pH 7.2) for 24 h at 4°C. After washing three times with the same buffer, the fixed samples were dehydrated with a series of ethanol solutions (15, 30, 50, 70, 80, 90, and 100%). Then, the dehydrated samples were infiltrated with a series of epoxy resin in epoxy propane (25, 50, 75, and 100%), and embedded in Epon 812 resin. The samples were cut through an ultramicrotome, and images were taken with a transmission electron microscope (Phillips CM120).
To study the subcellular location of CDB1 and CDB1L, full cDNA sequences of CDB1 and CDB1L were cloned into the pBI221-GFP vector. The chloroplast localization control RbcS-GFP was constructed as described previously (Zhang et al., 2018). The constructs were transformed into Arabidopsis protoplasts by PEG-mediated protoplast transformation. The mitochondrial marker MitoTracker Red CMXRos at a final concentration of 100 nM was incubated with the protoplasts for 15 min in the dark and washed twice before imaging. GFP signals were captured by confocal laser scanning microscopy (LSM 780, Zeiss). The experiment was repeated twice independently with similar results.
Sequences encoding the mature CDB1 and CDB1L proteins were amplified and cloned into the pET-28a expression vector (Merck Millipore) to express the recombinant proteins in the E. coli strain BL21 (DE3) in the presence of 0.5 mM IPTG. Recombinant proteins were purified using Ni-NTA agarose (Qiagen) and used to produce rabbit polyclonal antiserum (PhytoAB). Antiserum was employed in dilutions of 1:1,000. For examination of the specificity of CDB1 and CDB1L antibodies, immunoblots were performed using the recombinant CDB1 and CDB1L proteins. No cross reaction between the two proteins was detected when as much as 8 ng of recombinant CDB1 and CDB1L proteins was loaded (Supplementary Figure 3), indicating the specificity of these two antibodies.
Antibodies against HA tag (PhytoAB, PHY5011), D1 (PhytoAB, PHY0057), D2 (PhytoAB, PHY0323), LHCII (made in our lab), PetA (PhytoAB, PHY0321), PetC (PhytoAB, PHY0163), PetD (PhytoAB, PHY0354), PsaA (PhytoAB, PHY0342), PsaD (PhytoAB, PHY0343), CF1γ (PhytoAB, PHY0161), CF1ε (PhytoAB, PHY0315), RbcL (PhytoAB, PHY0066), phosphoglycerate kinase (PGK1; PhytoAB, PHY0405), ribulose phosphate epimerase (RPE; PhytoAB, PHY0616), RpoB (PhytoAB, PHY1700), RpoC2 (PhytoAB, PHY0382), RPS2 (PhytoAB, PHY0427), RPS4 (PhytoAB, PHY0428), PSRP2 (PhytoAB, PHY0420), RPL1 (PhytoAB, PHY0421), RPL6 (PhytoAB, PHY0411), RPL10 (PhytoAB, PHY0423), RPL11 (PhytoAB, PHY0413), and RPL18 (PhytoAB, PHY0414) were purchased from a commercial supplier and used at a 1:1,000 dilution.
Chloroplast stromal proteins and thylakoid membranes were extracted from 4-week-old plants. Intact chloroplasts were isolated using isolation buffer (0.33 M sorbitol and 20 mM HEPES/KOH, pH 7.6) and then osmotically ruptured in 20 mM HEPES/KOH (pH 7.6; Zhang et al., 2018). To separate the thylakoid membranes and stromal proteins, the ruptured chloroplasts were centrifuged at 12,000 × g for 10 min at 4°C. Clear supernatant containing stromal proteins was quantified with a Protein Assay Kit (Bio-Rad Laboratories), and the pellet containing thylakoid membrane was solubilized in 20 mM HEPES/KOH (pH 7.6). Both stromal and thylakoid proteins were solubilized in 2 × sample buffer (50 mM Tris–HCl, pH 6.8, 5% SDS, 20% glycerol, 8 M urea, 5% 2-mercaptoethanol, and 1% bromophenol blue).
Mitochondria were isolated as previously described (Sweetlove et al., 2007). Four week-old WT plants were fully homogenized in isolation buffer (0.3 M sucrose, 5 mM tetrasodium pyrophosphate, 10 mM KH2PO4, pH 7.5, 2 mM EDTA, 1% PVP40, 1% BSA, 5 mM cysteine, and 20 mM ascorbic acid), filtered through Miracloth, and then centrifuged at 5,000 × g for 10 min at 4°C. The supernatant was collected and centrifuged at 20,000 × g for 10 min at 4°C. The pellet containing crud mitochondria was resuspended in buffer with 0.3 M sucrose, 1 mM EGTA, and 10 mM MOPS/KOH, pH 7.2 and further centrifuged through a Percoll density gradient consisting of 18, 25, and 50% Percoll solution at 40,000 × g for 55 min at 4°C. Intact mitochondria at the 25%–50% Percoll interface were collected. Protein concentration was determined using a DC Protein Assay kit (BioRad, 5000116). Mitochondrial proteins were solubilized in 2 × sample buffer.
Protein samples were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membrane was incubated with specific antibodies, and then the signals were detected by the LuminoGraph WSE-6100 (ATTO Technology).
Total RNA was extracted from WT and cdb1 leaves using Trizol reagent kit (Invitrogen, Carlsbad, CA, United States), and rRNAs were removed by Ribo-Zero™ Magnetic Kit (Epicentre, Madison, WI, United States) to retain mRNAs and other RNAs. Enriched RNAs were fragmented and reverse transcribed into cDNA with random primers. Second-strand cDNA was synthesized, end repaired, and ligated to Illumina sequencing adapters using NEB#7490 kit (NEB E7490L, New England Biolabs). The ligation products of 300–500 bp were selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeq2500 (Gene Denovo Biotechnology Co. Ltd.).
Raw Reads were filtered by fastp (version 0.18.0) to get high quality clean reads (Chen et al., 2018). Clean reads were mapped to the Arabidopsis reference genome (TAIR10) using HISAT2.2.4 (Kim et al., 2019) and then sorted according to chromosome and physical position of the reference genome by samtools (Li et al., 2009). Reads were counted by featureCounts in Rsubread (version: 2.2.6) R package and normalized to get Transcripts Per Kilobase of exon model per Million mapped reads (TPM) and trimmed mean of M value (TMM; Liao et al., 2019). Differentially expressed genes (DEGs) were analyzed using DESeq2 (version: 1.28.1) applying a |log2(FC)| > 1 and an adjusted p < .05 parameters (Love et al., 2014). Gene Ontology (GO) enrichment analysis for DEGs was performed using clusterProfiler package (Wu et al., 2021). A false discovery rate (FDR) of <0.05 was considered for threshold.
The RNA-seq data have been uploaded in the NCBI Sequence Read Archive under accession number PRJNA781386.
Total RNA of WT and cdb1 was extracted from 4-week-old Arabidopsis plants using Trizol reagent (Thermo Fisher Scientific). The rRNAs were detected by ethidium bromide staining. A total of 5 μg RNA of WT and cdb1 samples were separated by electrophoresis on 1.4% (w/v) agarose-formaldehyde gels and subsequently blotted onto a nylon membrane (GE Healthcare). RNA was fixed on the nylon membrane by UV irradiation (UVP Hybridizer Oven). The membrane was hybridized with the specific probes labeled with digoxigenin, and the signals were visualized by the LuminoGraph WSE-6100 (ATTO Technology). Primers used to generate probes for the chloroplast rrn operons are shown in Supplementary Table 3.
Chlorophyll fluorescence was measured using the MAXI version of the Imaging-PAM M-Series chlorophyll fluorescence system with default settings. Before measurement, the plants were kept in the dark for 20 min. Protein alignment and evolutionary tree were produced using MEGA6 (Tamura et al., 2013).
In recent years, many mutants with albino or ivory phenotype were reported. Analysis of these mutants by transmission electron microscopy revealed defects in plastid development and biogenesis as for pdm4 and several pTAC mutants (Pfalz et al., 2006; Wang et al., 2020). To further investigate the underlying mechanisms of chloroplast development and biogenesis, we characterized a T-DNA insertion mutant (SALK_080811C) with an ivory phenotype (Figures 1A,B). PCR product sequencing showed that the T-DNA was inserted in the sixth exon of AT4G37920 (Figure 1A). This mutant could grow on MS medium supplemented with 3% sucrose but could not survive photoautotrophically when transplanted in soil. The leaves of this mutant display an ivory phenotype (Figure 1B). By transmission electron microscopy observation, we found that, unlike the chloroplasts in WT with well-organized thylakoid membranes, the plastids of this mutant did not exhibit organized membrane structures (Figure 1C). These results indicate that chloroplast development and biogenesis is arrested. Accordingly, we named this mutant cdb1.
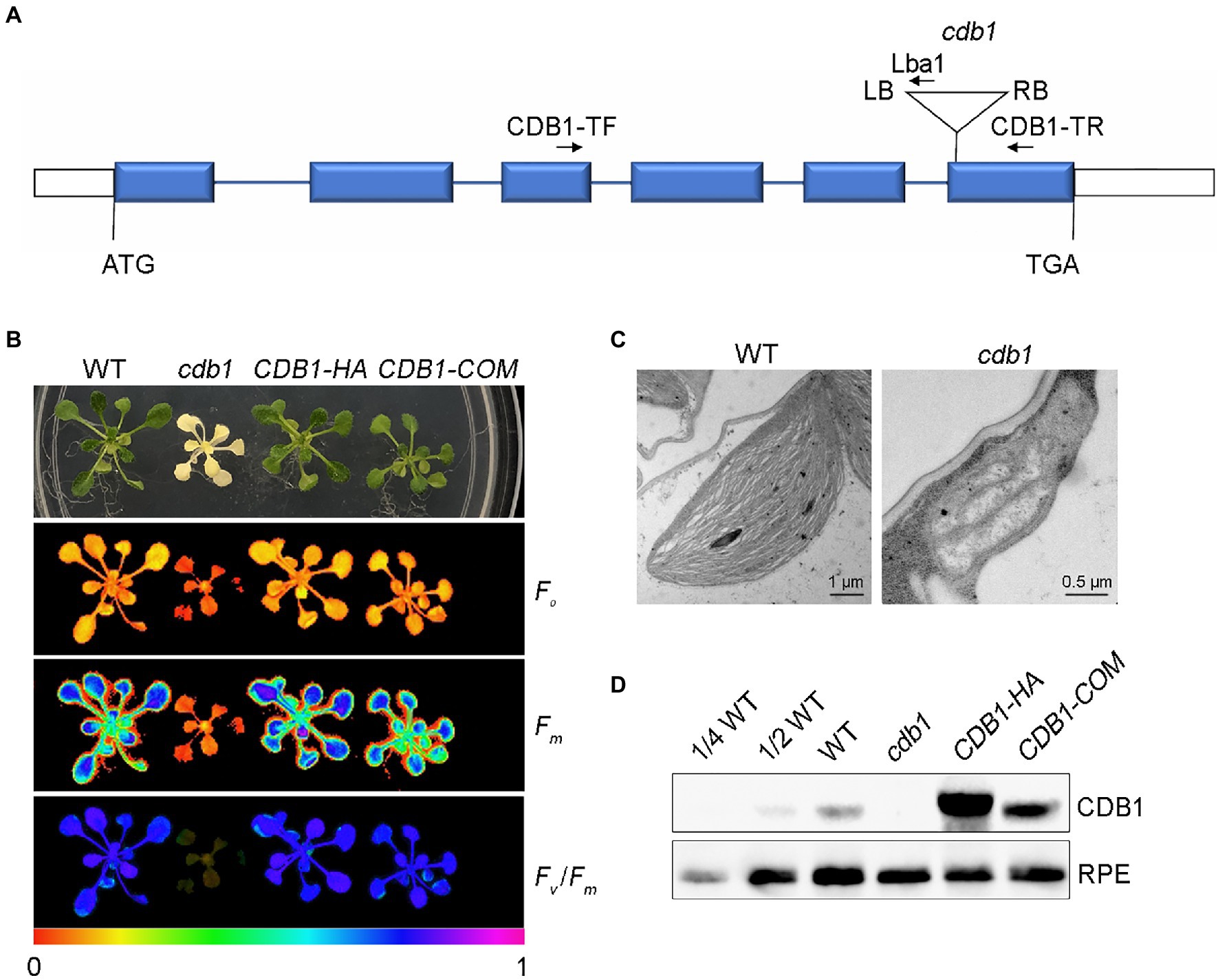
Figure 1. Characterization of the cdb1mutant. (A) Scheme of the structure of the Chloroplast Development and Biogenesis1 (CDB1) gene with the T-DNA insertion. The six blue boxes indicate exons. 5′ and 3′ UTRs are shown as white boxes and introns as black lines. The T-DNA insertion is indicated by a triangle. The forward (CDB1-TF) and reverse primers (CDB1-TR and Lba1) used for PCR analysis are indicated with arrows. (B) Chlorophyll fluorescence images of 4-week-old WT (wild-type), cdb1 and complemented plants CDB1-HA and CDB1-COM. The minimal fluorescence (Fo), the maximal fluorescence (Fm) and Fv/Fm of WT, cdb1 and two complemented plants were measured by Imaging-PAM. Fluorescence was visualized using a pseudocolor index from red (0) to purple (1) as indicated at the bottom. (C) Transmission electron micrographs of chloroplasts in leaves from 2-week-old WT and cdb1 mutant. Scale bars are indicated. (D) Protein accumulation of CDB1 in total protein extracts from 4-week-old leaves of WT, cdb1 and complemented plants CDB1-HA and CDB1-COM. Immunoblotting was performed with a CDB1-specific antibody. An antibody specific for ribulose phosphate epimerase (RPE) was used as loading control.
Chlorophyll fluorescence of WT and cdb1 plants grown on MS medium was analyzed. In WT, the minimal fluorescence (Fo) and the maximal fluorescence (Fm) were 0.17 ± 0.01 and 0.75 ± 0.01, respectively. The maximum quantum efficiency of PSII (Fv/Fm) was 0.77 ± 0.01. Consistent with the ivory phenotype of cdb1, almost no chlorophyll fluorescence was detected in the cdb1 mutant as Fo and Fm were 0.069 ± 0.020 and 0.086 ± 0.020, respectively (Figure 1B). The value of Fv/Fm is close to 0, indicating no PSII activity in cdb1.
To confirm that the ivory leaf phenotype was due to the disruption of CDB1 in cdb1, we complemented the mutants with the genomic gene sequence including its native promoter (CDB1-COM) and the full-length coding region of CDB1 fused to the HA tag at its C terminus (CDB1-HA). The CDB1-COM and CDB1-HA complemented plants displayed a phenotype similar to WT and the ivory leaf and fluorescence emission defects were all rescued (Figure 1B). A specific antibody raised against CDB1 was used to detect the CDB1 protein by immunoblotting using a total protein extract from 4-week-old leaves of WT, CDB1-HA, and CDB1-COM plants (Figure 1D). The CDB1 gene encodes a 48.73 kDa protein with a predicted chloroplast transit peptide (cTP). A signal corresponding to a molecular mass of about 42 kDa was detected consistent with the predicted molecular mass of CDB1 without cTP (Figure 1D). CDB1-HA fusion protein was detected in the CDB1-HA complemented plants with a slightly larger molecular mass than the native CDB1 protein (Figure 1D). CDB1 was absent in the cdb1 mutant due to the T-DNA insertion in AT4G37920. These results indicate that cdb1 is a null mutation and CDB1 is responsible for chlorophyll accumulation and chloroplast development.
Since the cdb1 mutant showed an ivory phenotype, it was necessary to investigate chloroplast protein accumulation in vivo. Total protein from 3-week-old leaves of WT and cdb1 was isolated and tested by immunoblot analysis using antibodies against subunits of the major photosynthetic complexes. Accumulation of thylakoid membrane proteins, including D1, D2, and LHCII of the PSII complex, PetA (Cyt f) and PetD of Cyt b6f, and PsaA and PsaD of PSI were all undetectable in the cdb1 mutant and only trace amounts of CF1γ and CF1ε of ATP synthase and PetC of Cyt b6f were detected (Figure 2). Abundance of three enzymes involved in the photosynthetic carbon reduction Calvin cycle was also analyzed (Figure 2). While a trace amount of plastid-encoded RuBisCO large subunit (RbcL) was detected in the cdb1 mutant, the levels of the nuclear-encoded PGK1 and RPE were comparable in the mutant to those of WT (Figure 2).
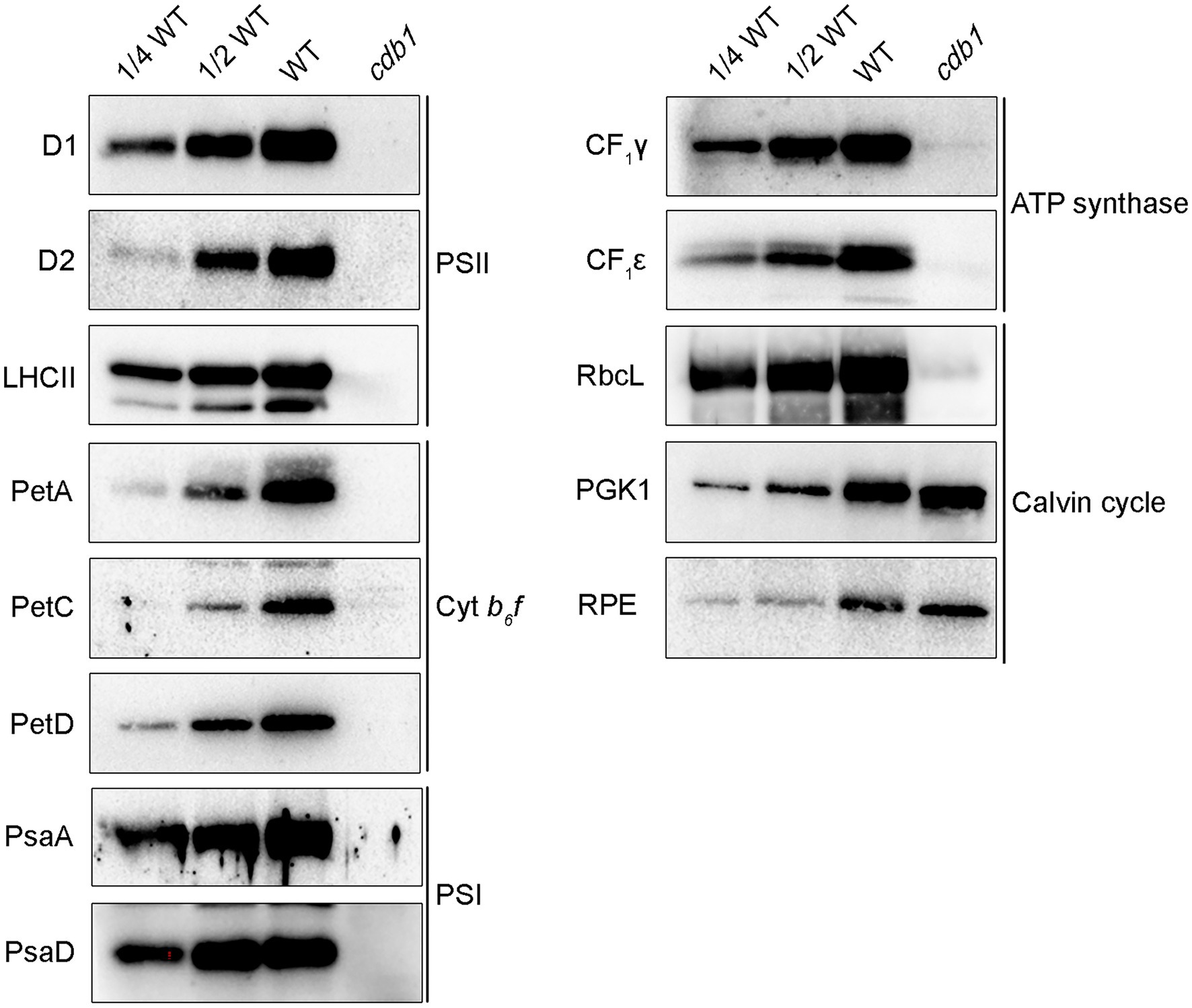
Figure 2. Comparative analysis of subunits of photosynthetic complexes from WT and cdb1. Total protein isolated from 3-week-old WT and cdb1 plants was analyzed by immunoblotting with antibodies against representative subunits of the multiprotein complexes of the thylakoid membrane: D1, D2, and LHCII from photosystem II (PSII), PetA, PetC, and PetD from Cyt b6f, PsaA and PsaD from PSI, CF1γ, and CF1ε from ATP synthase. Antibodies against RbcL, phosphoglycerate kinase (PGK1), and RPE were used as representative components of the Calvin cycle.
These results indicate that all plastid-encoded proteins analyzed do not accumulate or are only present in tiny amounts in cdb1 as shown for D1, D2, PetA, PsaA, CF1ε, and RbcL. Nucleus-encoded chloroplast proteins whose stable accumulation is independent of the presence of other plastid-encoded proteins were present in normal amounts in the cdb1 mutant as seen for PGK1 and RPE. These results agree with earlier studies showing that the stable accumulation of proteins belonging to a photosynthetic complex depends on the presence of all core subunits and especially on those that are plastid-encoded (Naver et al., 2001; Peng et al., 2006). Therefore, a possible explanation for the immunoblot results of cdb1 is that synthesis of all plastid-encoded proteins is blocked in cdb1 resulting not only in the absence of these proteins but also of their nucleus-encoded partner proteins of the same photosynthetic complex. In contrast, other nucleus-encoded chloroplast proteins such as PGK1 and RPE are unaffected in cdb1 (Figure 2).
To determine the function of CDB1 in chloroplast development, we carried out an RNA-seq analysis on 4-week-old leaves of cdb1 and WT. Transcriptional profiles of nucleus- and chloroplast-encoded genes were compared using RNA-seq. A total of 3,515 genes were differentially expressed more than 2-fold in cdb1 compared to WT (Figure 3A). They include 1,395 upregulated and 2,120 downregulated genes (Figure 3A). Gene Ontology (GO) analysis revealed that a large proportion of the DEGs are related to the nucleus and chloroplast (Figure 3B). Among 618 DEGs related to the chloroplast (GO:0009507), 272 nuclear and 30 chloroplast DEGs are upregulated whereas 296 nuclear and 18 chloroplast DEGs are downregulated (Supplementary Figure 1).
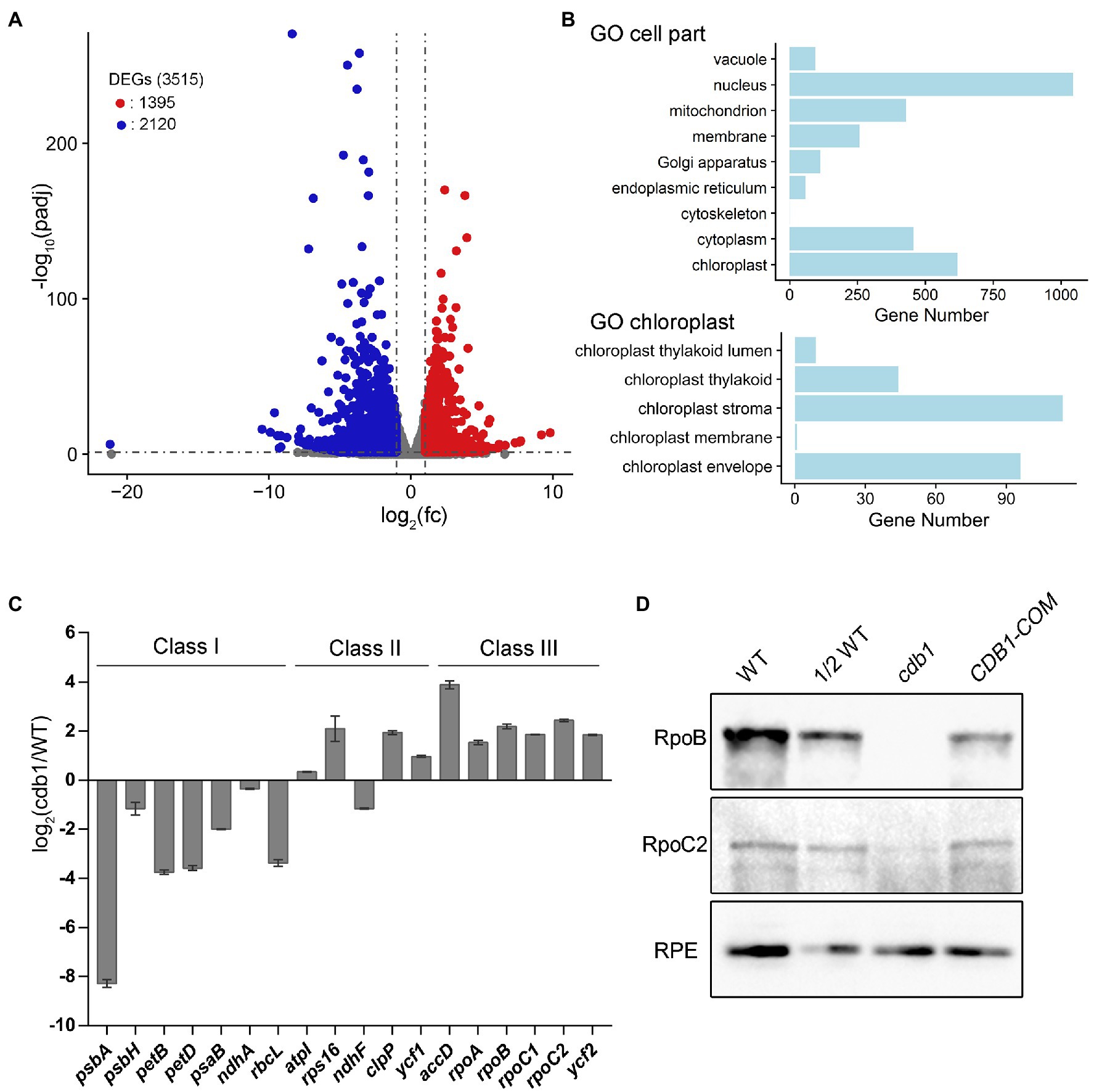
Figure 3. Transcriptome analysis of WT and cdb1 plants. (A) Volcano plot showing the differentially expressed genes (DEGs) in WT and cdb1 [p adj < 0.05, fold change (fc) > 2]. Total RNA was extracted from 4-week-old leaves of cdb1 and WT. The constructed cDNA library was sequenced using lllumina HiSeq 2,500. DEGs were identified by DESeq2 according to the parameter of fold change >2 and adjusted p value below 0.05. Volcano plot was generated by R package ggplot2. Down- and upregulated genes are indicated by blue and red dots, respectively. Three biological replicates of WT and cdb1 were used for RNA-seq experiment. (B) Diagram showing gene numbers relative to “cell part” and “chloroplast” in Gene Ontology (GO) enrichment of DEGs in cdb1 vs. WT. GO enrichment of DEGs was performed using clusterProfiler. (C) Plastid gene transcript levels. Differential expression of plastid genes in cdb1 vs. WT is represented according to the RNA-seq data. The representative plastid genes include Class I genes (psbA, psbH, petB, petD, psaB, ndhA, and rbcL), Class II genes (atpI, rps16, ndhF, clpP, and ycf1), and Class III genes (accD, rpoA, rpoB, rpoC1, rpoC2, and ycf2). Log2[fold change(cdb1/WT)] ± SD values are from three biological replicates. (D) Immunoblot analysis of RpoB and RpoC2. Total protein isolated from 3-week-old WT, cdb1 and CDB1-COM were hybridized with antibodies against RpoB and RpoC2. RPE was used as a loading control.
The plastid genes can be divided into three classes according to which plastid RNA polymerase transcribes them. Class I and III are transcribed by the PEP and NEP, respectively. Transcription of class II depends on both PEP and NEP. RNA-seq results showed that expression of class I genes (e.g., psbA, psbH, petB, petD, ndhA, and rbcL) was significantly reduced whereas expression of Class III genes (e.g., accD, rpoA, rpoB, rpoC1, rpoC2, and ycf2) and most of Class II genes (e.g., atpI, rps16, clpP, and ycf1) was greatly upregulated (Figure 3C; Supplementary Table 1). The upregulated NEP-dependent Class III genes and the differentially expressed nuclear genes of chloroplast proteins may possibly participate in a compensating response to the decreased expression of Class I genes. These results suggest that transcription of the PEP-dependent genes is impaired in the absence of CDB1. Immunoblot analysis showed that the core subunits of the PEP complex RpoB is undetectable and RpoC2 is only present in a tiny amount in cdb1 (Figure 3D), indicating that absence of the functional PEP complex is responsible for the impaired transcription of the PEP-dependent genes in cdb1.
Although drastic reductions of RpoB and RpoC2 were observed in cdb1, the levels of their mRNAs are higher in cdb1 than in WT (Figures 3C,D). This observation implies that translation of rpoB and rpoC2 by ribosomes or assembly of the PEP complex is impaired in cdb1. To address the possible role of CDB1 in chloroplast ribosome accumulation, we examined the levels of chloroplast ribosomal proteins and rRNA in cdb1 and WT. Immunoblot analysis showed that the amount of 30S ribosomal protein RPS2, RPS4, and PSRP2 and 50S ribosomal subunits RPL1, RPL6, RPL10, RPL11, and RPL18 are barely detectable or greatly reduced in cdb1 (Figure 4A). Chloroplast rRNAs are transcribed from a single transcription unit that includes the 16S, 23S, 4.5S, and 5S rrn genes (Figure 4B). The RNA precursors are then processed to produce mature 16S, 23S, 4.5S, and 5S rRNA (Figure 4B). RNA blotting revealed that the amounts of 1.5-kb 16S, 1.1 and 1.3-kb 23S, 0.1-kb 4.5S, and 0.12-kb 5S mature rRNAs are all drastically reduced (Figure 4C). These results indicate that accumulation of chloroplast ribosomes is impaired in cdb1, and explains why accumulation of the chloroplast-encoded proteins is reduced in cdb1 (Figure 2). Taken together, we propose that CDB1 is essential for the assembly of chloroplast ribosomes.
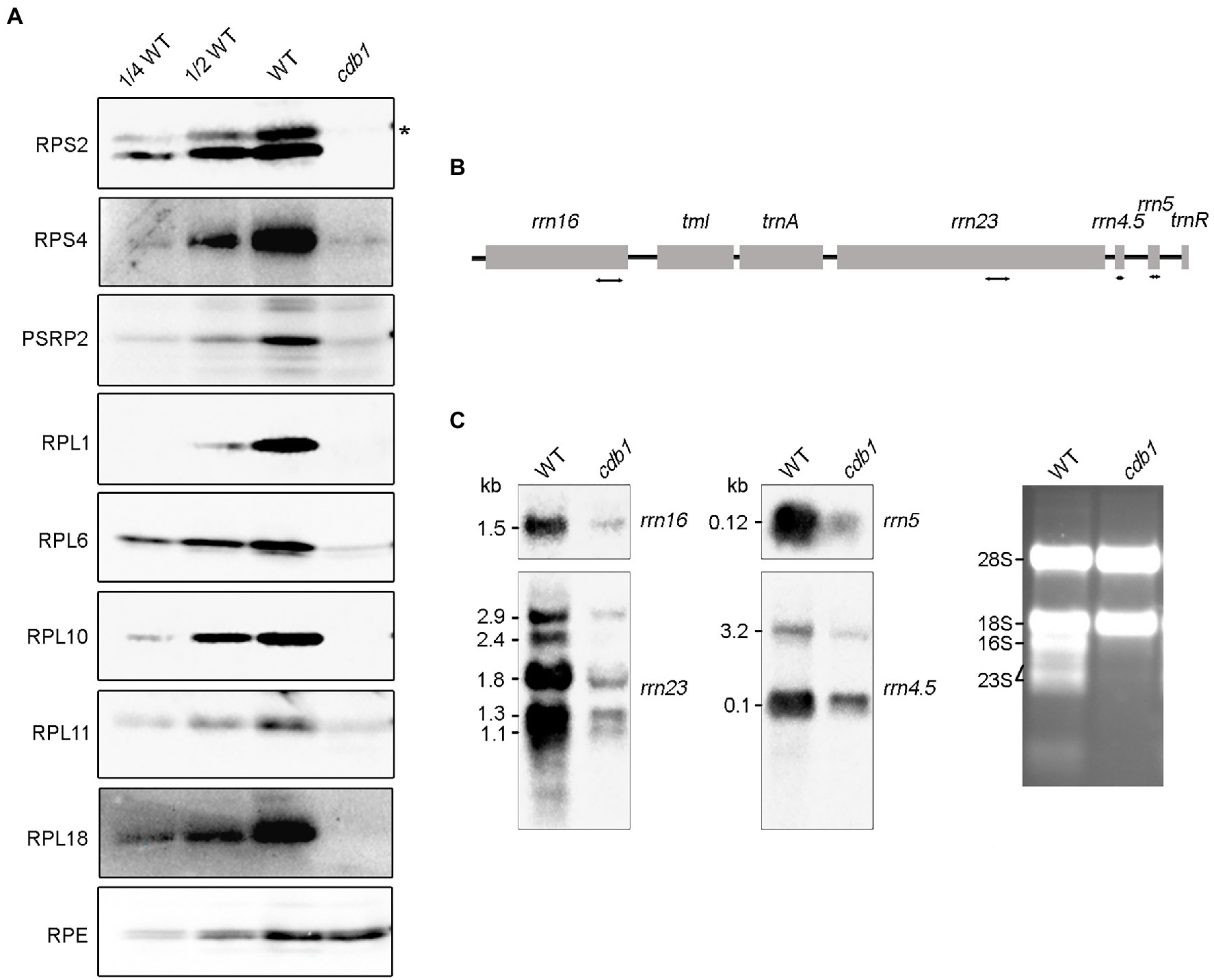
Figure 4. Accumulation of chloroplast ribosomes in WT and cdb1 plants. (A) Immunoblot analysis of representative 50S and 30S ribosomal subunit proteins. Total protein isolated from 3-week-old WT and cdb1 plants was analyzed by immunoblotting with antibodies against RPS2, RPS4, PSRP2, RPL1, RPL6, RPL10, RPL11, and RPL18. RPE was used as a loading control. Black asterisk indicates the authentic protein. (B) Scheme of the chloroplast rrn operon with the four probes used for the RNA blots. The four probes for rrn16, rrn23, rrn4.5, and rrn5 are marked by double-arrow lines under the chloroplast rrn operon. (C) Accumulation of rRNA in WT and cdb1. Total RNA from leaves of 3-week-old WT and cdb1 was subjected to RNA blot analysis with specific probes against 16S, 23S, 4.5S, and 5S. Sizes of the distinct forms of the rRNA species are indicated on the left; rRNAs of WT and cdb1 stained with ethidium bromide were used as loading control.
The CDB1 gene consists of six exons separated by five introns. It encodes a 427-amino acid protein with an unknown function. SMART search-based analysis1 and the annotation of The Plant Proteome Database (PPDB)2 suggest that CDB1 contains a putative cTP at its N-terminus (1–62 aa) and a predicated coiled-coil domain (135–168 aa; Figure 5A). No other assigned functional motif was found in CDB1.
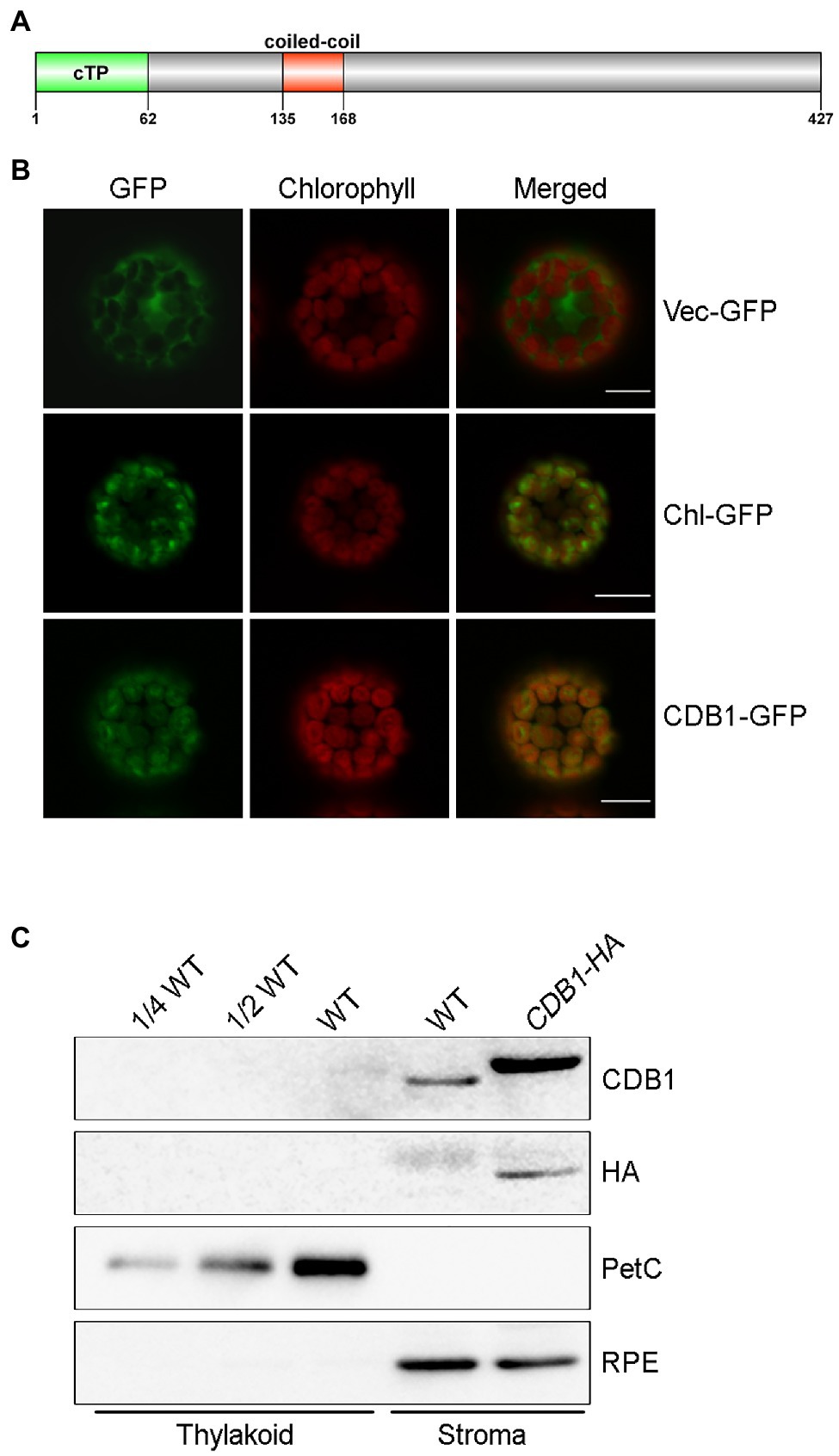
Figure 5. Subcellular localization of CDB1. (A) Protein structure of CDB1 protein. The CDB1 protein contains 427 amino acids. The light green part indicates the cTP region; the red box indicates the predicated coiled coil domain. (B) Subcellular localization of CDB1 fused with GFP. Images of GFP and chlorophyll fluorescence of protoplasts were captured by confocal laser scanning microscopy (LSM) and merged. The small subunit of ribulose-1, 5-bisphosphate carboxylase fused to GFP (Chl-GFP) was used as chloroplast-targeted control. Bars = 10 μm. (C) Immunoblot analysis of CDB1 in chloroplasts. Intact chloroplasts from WT and CDB1-HA complemented plants were separated into stroma and thylakoid membrane fractions and reacted with antibodies against CDB1 and HA. PetC and RPE were used as controls for the fractionation of thylakoids and stroma, respectively.
To determine the subcellular localization of CDB1, a construct containing 35S:CDB1-GFP was transformed into Arabidopsis protoplasts. Strong GFP signals were exclusively detected in chloroplasts with CDB1-GFP and they closely merged with chlorophyll autofluorescence (Figure 5B). Besides, the fluorescent signals of CDB1-GFP were similar to those from RbcS-GFP, the small subunit of ribulose-1, 5-bisphosphate carboxylase (RbcS) fused to GFP (Figure 5B). These results indicate that CDB1 is a chloroplast protein, consistent with the presence of cTP at its N-terminus (Figure 5A). To investigate its precise localization, immunoblots were performed using chloroplast stroma and thylakoids isolated from WT and CDB1-HA complemented plants. The results show that CDB1 and HA-tagged CDB1 are localized in the chloroplast stroma (Figure 5C).
To explore the phylogenic evolution of CDB1, its homologs were searched using BLAST-P against the NCBI database.3 Putative orthologs and paralogs were found in most photosynthetic eukaryotes (land plants and green algae) with a high conservation across the entire protein sequence except for the cTP region (Supplementary Figure 2). No homolog of CDB1 was found in cyanobacteria. A neighbor-joining phylogenetic tree for CDB1 homologs was constructed, containing 19 genes from 10 sequenced species representing green algae and land plants (Figure 6). Both green alga Chlamydomonas reinhardtii and the lycophyte Selaginella moellendorffii have one homolog of CDB1. A total of three homologous sequences were found in the moss Physcomitrella patens. No homologs were found in gymnosperms probably due to the incomplete genome information. Interestingly, besides CDB1, a second CDB1-related protein, designated as CDB1L, was found from both monocot and dicot plants. The mature Arabidopsis CDB1 and CDB1L proteins share 41.5% amino acid sequence identity (Figure 7). These imply that CDB1 proteins originated from green alga and were resolved into two clades in angiosperms during evolution.
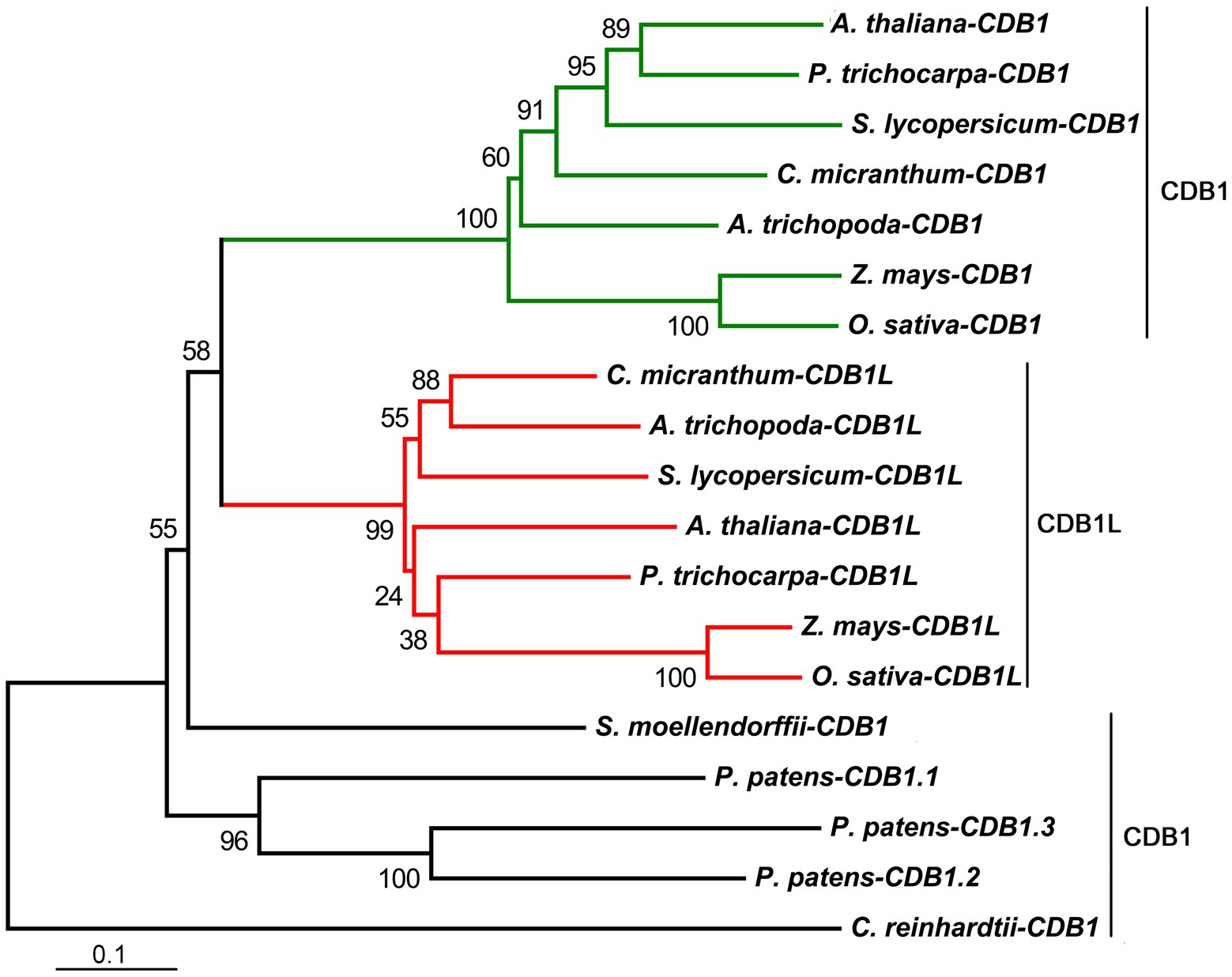
Figure 6. Phylogenetic tree of CDB1 homologs. Nineteen proteins of the CDB1 family were selected to infer the evolutionary history using the Neighbor-Joining method, with bootstrap values (%) from 1,000 replicates. Numbers at branches are percentage of replicate trees in the bootstrap test. The scale bar indicates the units of the number of amino acid substitutions per site. All the analyses were conducted in MEGA6. Alignments of all 19 sequences based on identity/similarity and structural properties are shown in Supplementary Figure 2.
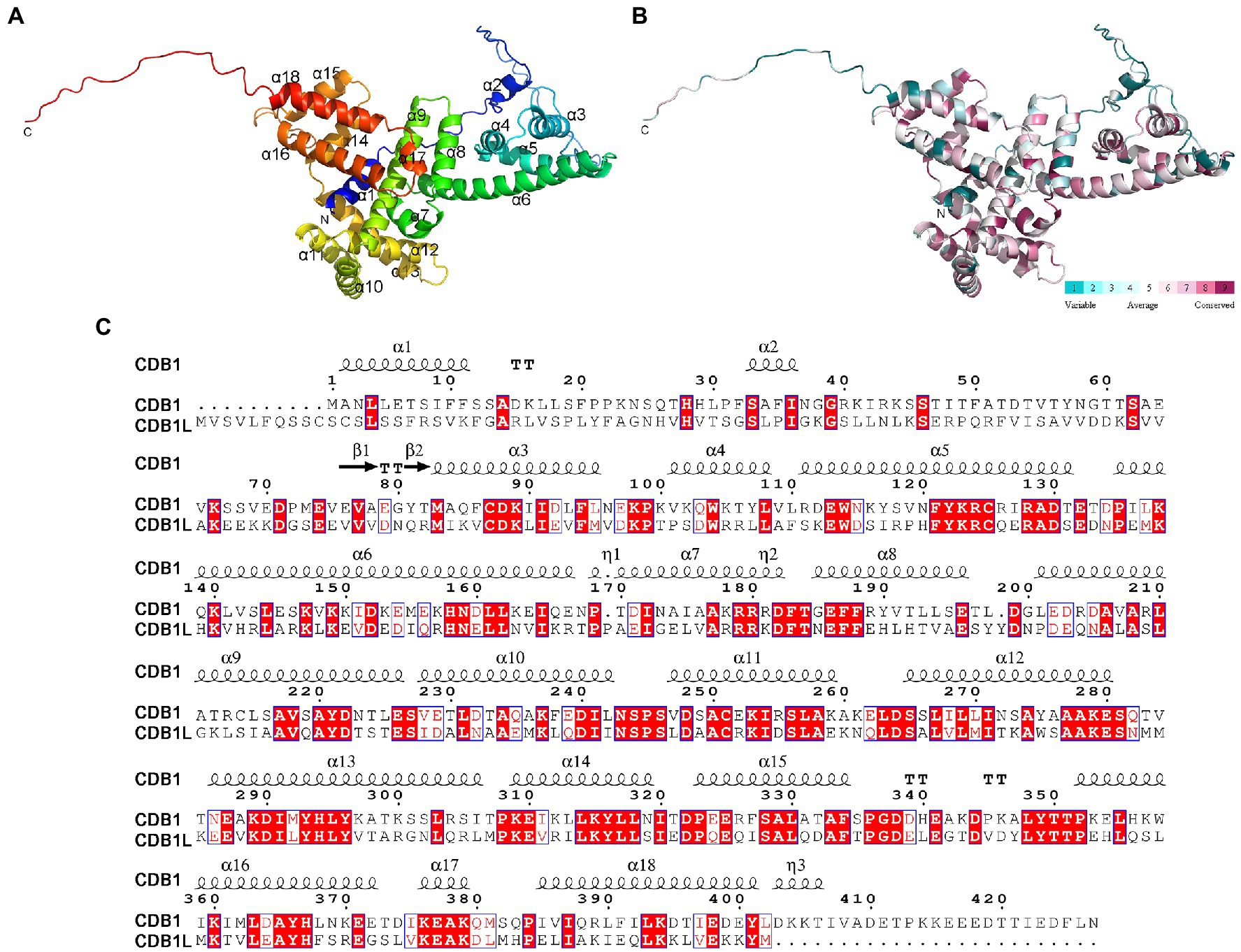
Figure 7. Overall structure of CDB1. (A) Crystal structure of CDB1 predicted by AlphaFold2. The α-helices and N, C-terminus are indicated. (B) Surface conservation analysis of CDB1 using ConSurf web server. The cartoon structure of CDB1 was colored according to the conservation score from the 19 CDB1 family homologs. (C) Sequence alignment of CDB1 and CDB1L from Arabidopsis. Secondary structure elements above the alignment were generated using ESPript (https://espript.ibcp.fr). Numbers indicate the original amino acid positions in CDB1. Highly conserved residues are represented by white letters with a red background.
The elucidation of the function of CDB1 remains challenging because no homolog or similar domain was characterized in previous reports. To gain insights into this question, the three-dimensional structure of Arabidopsis CDB1 was predicted by AlphaFold24 (Jumper et al., 2021; Figure 7A). The predicted structure of mature CDB1 contains two very short β-sheets (β1 and β2) and a total of 17 helical structures, including 14 α-helices (α3–17) and three 310-helices (Figures 7A,C). Based on the sequence identity to CDB1 homologs (Supplementary Figure 2), the evolutionary conservation scores were mapped onto the Arabidopsis CDB1 structure using the ConSurf server. The results show that the conserved residues in the CDB1 protein family are clustered in the secondary structure elements of mature Arabidopsis CDB1 (Figure 7B; Supplementary Figure 2). This suggests that the members of the CDB1 protein family might have similar biological functions.
To investigate the functions of the CDB1 paralog which was designated as CDB1L in Arabidopsis, the cdb1l (GK-844F05) mutant was obtained. Sequencing of PCR products showed that the T-DNA was inserted into the sixth exon of the CDB1L (Figure 8A). Because we were unable to obtain cdb1l homozygous mutants, we determined the segregation ratio of heterozygous to WT phenotype in the progeny of cdb1l heterozygous plants and found that it was close to 2:1 (Supplementary Table 2). Thus, it is likely that homozygous lethality occurs during embryo development of cdb1l homozygous seeds. To test this possibility, we examined the seeds in developing siliques from the cdb1l heterozygous plants. A deficiency in endosperm or embryo development of some seeds was detected in these plants (Figure 8B). Some of the affected seeds gave rise to intact seeds but they were pale and became withered at a later stage of development (Figure 8B). It is interesting that some seeds of the cdb1 heterozygous plants also exhibit a pale color (Figure 8B). These seeds are likely to be the cdb1 homozygous seeds defective in chloroplast biogenesis.
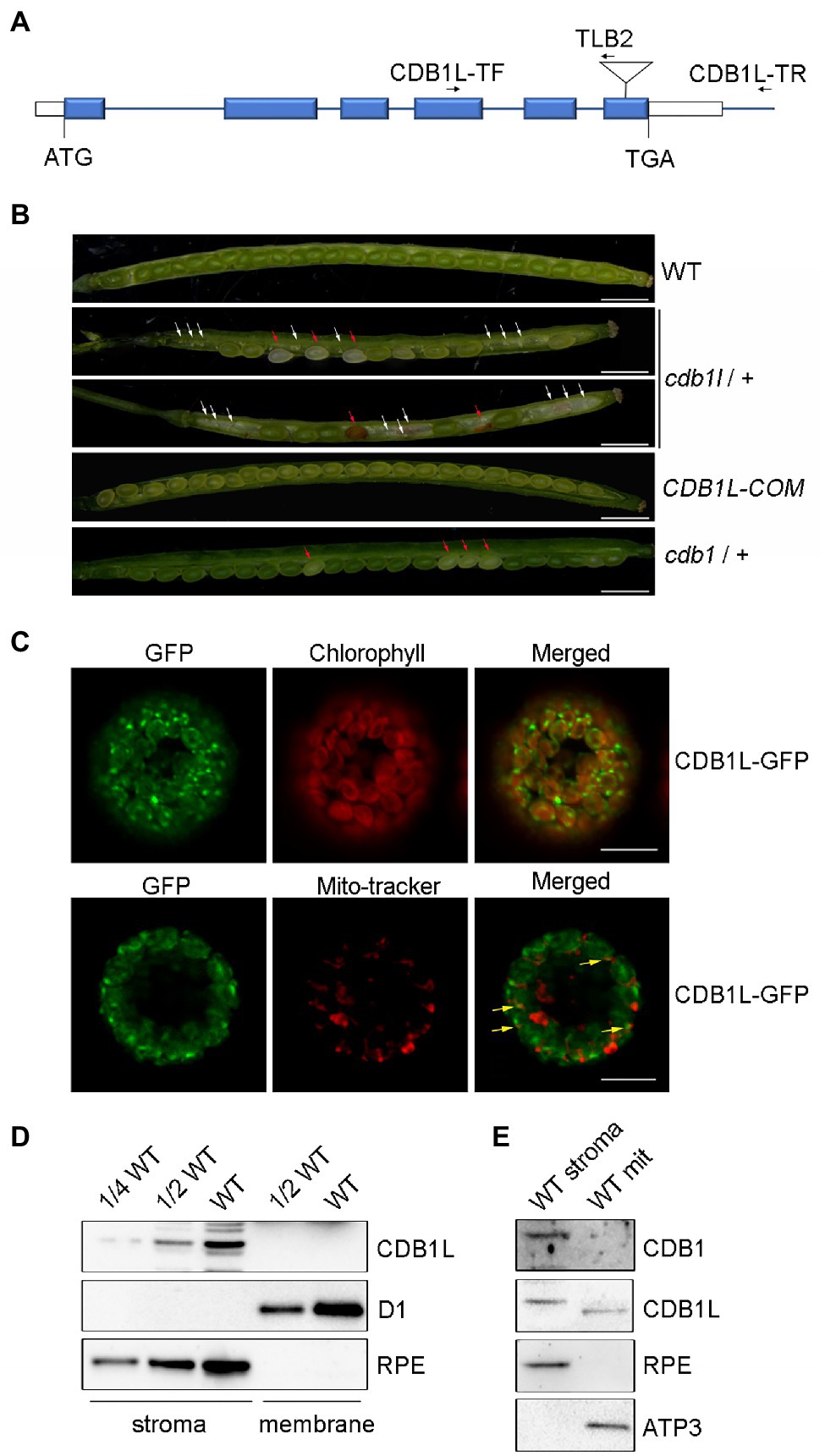
Figure 8. Characterization of the CDB1L in Arabidopsis. (A) Scheme of the structure of the CDB1L gene and its T-DNA insertion. The six blue boxes indicate exons. 5′ and 3′ UTRs are shown as white boxes. The T-DNA insertion is indicated by a triangle. Forward (CDB1L-TF) and reverse primers (CDB1L-TR and TLB2) used for the PCR analysis are indicated with arrows. (B) Siliques of WT, cdb1l heterozygotes, CDB1L-COM, and cdb1 heterozygotes after flowering. Seeds of early developing siliques of WT and CDB1L-COM were all green. Pale and green seeds were observed in early developing siliques of cdb1l heterozygous plants (top image of cdb1l/+) and of cdb1 heterozygous plants (cdb1/+). The pale seeds became withered at a later stage of seed development in cdb1l/+ (bottom image of cdb1l/+). White arrows highlight the aborted embryos in cdb1l/+. The pale and withered seeds in developing cdb1l/+ and cdb1/+ siliques are indicated with red arrows. Bars = 1 mm. (C) Subcellular localization of CDB1L fused with GFP. CDB1L-GFP, CDB1L-GFP fusion; Mito-tracker, Mito-tracker Red. The co-localization signals of CDB1L-GFP and Mito-tracker are indicated with yellow arrows. Bars = 10 μm. (D) Immunoblot analysis of CDB1L in chloroplasts. Intact chloroplasts from WT were separated into stroma and thylakoid membrane fractions and analyzed by immunoblotting with antibodies against CDB1L. D1 and RPE were used as controls for the fractionation of thylakoid and stroma, respectively. (E) Immunoblot analysis of CDB1L in mitochondria and chloroplast stroma. Stromal chloroplast and mitochondrial proteins from WT were separated and reacted with antibodies against CDB1 and CDB1L. RPE and ATP3 were used as controls for the fractionation of stroma and mitochondria, respectively.
To confirm that the seed abortion phenotype was due to the disruption of CDB1L, we complemented the cdb1l heterozygous mutant with the full-length genomic CDB1L sequence driven by its authentic promoter. As shown in Figure 8B, the complemented plants (CDB1L-COM) produced well-developed seeds similar to WT, confirming that the aborted seeds in the cdb1l heterozygous mutants resulted from the disruption of CDB1L.
To determine the subcellular localization of CDB1L, a construct containing its coding region fused with GFP at the C-terminus was transformed into Arabidopsis. The CDB1L-GFP signal co-localized with chlorophyll (Figure 8C), suggesting that CDB1L is localized in chloroplasts. However, some of the CDB1L-GFP signals were also found outside chloroplasts and co-localized with mito-tracker red, indicating that CDB1L is also localized in mitochondria (Figure 8C). To further confirm the location of CDB1L, a specific antibody against CDB1L was raised to detect CDB1L accumulation in chloroplasts and mitochondria (Figures 8D,E; Supplementary Figure 3). The stroma and thylakoid fractions of chloroplasts isolated from WT leaves were used for immunoblot analysis. The results showed that CDB1L is localized in the chloroplast stroma (Figure 8D). These results are consistent with the localization of CDB1L-GFP (Figure 8C) and the detection of CDB1L in chloroplasts by mass spectrometry (The Plant Proteome Database).5
Analysis of proteins from purified mitochondria showed that CDB1L is also present in mitochondria. However, the molecular mass of mitochondrial-localized CDB1L is slightly less than that of chloroplast-localized CDB1L (Figure 8E). This might be due to the different length of the transit peptide in CDB1L, which is removed after entering into chloroplasts and mitochondria. No CDB1 signal was found in the mitochondrial samples (Figure 8E), indicating that CDB1 is not a mitochondrial protein but specifically localized in chloroplasts (Figure 5). In summary, these results indicate that the CDB1L protein has a dual localization in chloroplasts and mitochondria. Loss of CDB1L leads to a deficiency of seed development in Arabidopsis.
Chloroplast development is a programmed and complicated process regulated by numerous nuclear and chloroplast genes (Pogson and Albrecht, 2011). Previous studies have identified many nuclear factors that are involved in the regulatory mechanisms of chloroplast development. Here, we report the existence of a novel chloroplast stromal protein CDB1 that is essential for chloroplast development and biogenesis. Absence of CDB1 leads to an ivory seedling phenotype of Arabidopsis and the seedlings cannot survive autotrophically in the soil (Figure 1). Consistent with its ivory and chlorophyll fluorescence phenotype, the cdb1 mutant is defective in the accumulation of thylakoid protein complexes and RuBisCO complex in the chloroplast stroma (Figure 2). These results suggest that CDB1 is essential for chloroplast development and plant growth.
RNA-seq analysis revealed that plastid gene expression was dramatically altered in cdb1 leaves (Supplementary Figure 1). Transcript levels of PEP-dependent genes were all decreased (Class I genes, including psbA, psbH, petB, petD, psaB, and rbcL); on the contrary, those of NEP-dependent genes were unchanged or even increased (Class III genes, including rpoA, rpoB, rpoC1, rpoC2, clpP, ycf1, and ycf2; Figure 3C). These results indicate that cdb1 mutant is severely impaired in PEP activity. A similar molecular phenotype has been observed in mutants lacking PEP components or regulatory factors, such as ptac2/ptac6/ptac12/ptac14 (Pfalz et al., 2006; Gao et al., 2012), sig6 (Loschelder et al., 2006; Chi et al., 2010); PPR genes, such as pdm2, pdm3, and pdm4 (Du et al., 2017; Zhang et al., 2017; Wang et al., 2020); and some other factors, such as ys1 (Zhou et al., 2009), clb19 (Chateigner-Boutin et al., 2008), and dg1 (Chi et al., 2008). In contrast to these mutants, however, the cdb1 mutant accumulates no RpoB and only a trace amount of RpoC2 (Figure 3D), indicating the impaired formation of the PEP complex. Because the expression levels of the genes encoding the core PEP subunits (rpoA, rpoB, rpoC1, and rpoC2) are highly increased in cdb1 (Figure 3C; Supplementary Figure 1), it is likely that the translation of the PEP subunits or assembly of the PEP complex is impaired in the chloroplasts of cdb1.
Further immunoblot and RNA analysis showed that the accumulation of proteins of the 30S and 50S ribosomal subunits as well as ribosomal rRNAs (16S, 23S, 4.5S, and 5S) is dramatically decreased in cdb1 (Figure 4). These results imply that chloroplast ribosomes cannot assemble in this mutant. This conclusion could also be confirmed by the analysis of the levels of chloroplast proteins. Plastid-encoded proteins (such as D1, D2, PetA, PsaA, CF1ε, and RbcL) and nucleus-encoded chloroplast proteins (such as PetC, PsaD, and CF1γ) which together with their plastid-encode partner proteins form stable complexes are absent or barely detectable in cdb1 (Figure 2). In contrast, nucleus-encoded chloroplast proteins which do not assemble in complexes with other plastid-encoded proteins accumulate normally in the mutant (Figure 2). We therefore conclude that the absence of CDB1 compromises chloroplast ribosome assembly, which in turn affects the translation of plastid-encoded mRNAs, notably those of the PEP complex and ultimately impairs chloroplast development during the early stages of seedling growth.
RNA-seq data revealed that almost all chloroplast ribosomal protein genes were upregulated in the cdb1 mutant except rps14, a plastid gene encoding a 30S ribosomal subunit (Supplementary Figure 1). The level of rps14 transcript in cdb1 was reduced to ~1/4 of wild type (Supplementary Figure 1). However, this moderate reduction in rps14 mRNA is unlikely to be the direct reason for the severe deficiency in accumulation of chloroplast ribosomes. Previous studies demonstrated that RPS14 could also be transcribed to some extent in PEP-deficient mutants (Legen et al., 2002), and reduction of rps14 transcript can be explained by the absence of the PEP complex. Hence, we propose that CDB1 participates in chloroplast ribosome biogenesis. It may function as a molecular chaperone to assist chloroplast ribosome assembly or maintain the structural stability of ribosomes during their biogenesis. It is also possible that CDB1 is involved in the maturation of the chloroplast rRNAs.
Phylogenetic analysis revealed that the CDB1 paralog CDB1L is present in angiosperms (Figure 6). Knockout of CDB1L results in embryo abortion (Figure 8B). Subcellular localization indicated that CDB1L is dually localized in chloroplasts and mitochondria (Figures 8C–E). The sequence and structural similarity of CDB1L and CDB1 indicates that mitochondria-localized CDB1L may perform a similar function during mitochondrial ribosome biogenesis. Previous reports have confirmed that loss of mitochondrial function usually results in arrested embryo development as observed in mutants deficient in Atp11 and Atp12 that are essential for mitochondrial ATP synthase assembly (Duan et al., 2020) and in AARS proteins that are required for translation in mitochondria (Berg et al., 2005). In cdb1l heterozygous plants, some of the seeds can mature but they have a pale color (Figure 8B). Similar seeds were also found in the cdb1 heterozygous plants (Figure 8B), as well as in the pmd1, pmd2, pmd3, and pmd4 mutants, in which chloroplast PEP activity is impaired and chloroplast development and biogenesis is arrested (Pyo et al., 2013; Du et al., 2017; Zhang et al., 2017; Wang et al., 2020). These observations suggest that chloroplast development is affected in the pale seeds of cdb1 and cdb1l heterozygous plants. It is possible that chloroplast-localized CDB1L plays an essential role during chloroplast biogenesis and this function cannot be complemented by its CDB1 paralog.
Phylogenetic tree analysis showed that CDB1 homologs can be found in most photosynthetic eukaryotes from green algae to land plants (Figure 6). The conserved residues in the CDB1 family proteins could be clustered to the secondary structure elements of the CDB1 protein structure predicted by AlphaFold2 (Supplementary Figure 2), suggesting that the CDB1 family proteins might have similar biological functions. Interestingly, homologs in angiosperms were further resolved into two clades, containing CDB1 and CDB1L, respectively. The phylogenetic tree showed that CDB1 proteins from lower photosynthetic Viridiplantae are closer to CDB1L than to CDB1 in angiosperms (Figure 6). This suggests that CDB1L proteins originated from green alga and that the CDB1 proteins in monocot and dicot plants probably evolved through gene duplication during the emergence of angiosperms, where they have assumed new functions in chloroplast ribosome biogenesis. The dual-localized CDB1L proteins in angiosperms may have a similar role as CDB1 from lower photosynthetic Viridiplantae, and operate both in chloroplasts and mitochondria.
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA781386.
QX conceived the study and designed experiments and produced the figures. WC, JH, and SC performed experiments. WC, JH, SC, LZ, J-DR, LP, and QX analyzed the data. QX, J-DR, and LP wrote the manuscript. QX and LP supervised the whole study. All authors contributed to the article and approved the submitted version.
This work was supported by the fund of Shanghai Engineering Research Center of Plant Germplasm Resources (17DZ2252700).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the Nottingham Arabidopsis Stock Center (NASC) for providing the mutant seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.815859/full#supplementary-material
Supplementary Figure 1 | Transcript levels of all sequenced plastid genes. Differential expression of plastid-encoded genes in cdb1 vs. WT is represented according to the RNA-seq data. The log2[fold change(cdb1/WT)] ± SD values are from the three biological replicates.
Supplementary Figure 2 | Sequences of CDB1 family proteins from Arabidopsis thaliana (CDB1: AT4G37920; CDB1L: AT1G36320), Populus trichocarpa (CDB1: XP_024461654.1; CDB1L: XP_024450195.1), Solanum lycopersicum (CDB1: XP_004231977.1; CDB1L: XP_004237860.1), Cinnamomum micranthum f. kanehirae (CDB1: RWR82510.1; CDB1L: RWR94545.1), Amborella trichopoda (CDB1: XP_006856624.2; CDB1L: XP_020521452.1), Zea mays (CDB1: NP_001144269.1; CDB1L: NP_001358639.1), Oryza sativa Japonica (CDB1: XP_015623683.1; CDB1L: XP_015622979.1), Physcomitrella patens (CDB1.1: XP_024388931.1; CDB1.2: XP_024392048.1; and CDB1.3: XP_024372527.1), Selaginella moellendorffii (CDB1: XP_002962701.2), and Chlamydomonas reinhardtii (CDB1: PNW69832.1). Secondary structure elements above the alignment were generated by ESPript (https://espript.ibcp.fr). Numbers indicate the original amino acid positions in CDB1. Highly conserved residues are represented in white letters with red background.
Supplementary Figure 3 | Test of CDB1 and CDB1L antibodies. Mature CDB1 (corresponding to amino acids 63–427 of CDB1) and CDB1L (corresponding to amino acids 13–414 of CDB1L) were used to raise antibodies in rabbits. Recombinant protein of CDB1 and CDB1L (8 ng) was separated by SDS–PAGE and detected with antibodies against CDB1 (left) and CDB1L (right), respectively. Recombinant proteins with predicted molecular mass are indicated with arrows.
1. ^http://smart.embl-heidelberg.de/
2. ^http://ppdb.tc.cornell.edu/
3. ^http://blast.ncbi.nlm.nih.gov
Bang, W. Y., Chen, J., Jeong, I. S., Kim, S. W., Kim, C. W., Jung, H. S., et al. (2012). Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J. 71, 122–134. doi: 10.1111/j.1365-313X.2012.04976.x
Berg, M., Rogers, R., Muralla, R., and Meinke, D. (2005). Requirement of aminoacyl-tRNA synthetases for gametogenesis and embryo development in Arabidopsis. Plant J. 44, 866–878. doi: 10.1111/j.1365-313X.2005.02580.x
Bollenbach, T. J., Lange, H., Gutierrez, R., Erhardt, M., Stern, D. B., and Gagliardi, D. (2005). RNR1, a 3′-5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana. Nucleic Acids Res. 33, 2751–2763. doi: 10.1093/nar/gki576
Börner, T., Aleynikova, A. Y., Zubo, Y. O., and Kusnetsov, V. V. (2015). Chloroplast RNA polymerases: role in chloroplast biogenesis. BBA-Bioenergetics 1847, 761–769. doi: 10.1016/j.bbabio.2015.02.004
Chateigner-Boutin, A.-L., Ramos-Vega, M., Guevara-García, A., Andrés, C., De La Luz Gutiérrez-Nava, M., Cantero, A., et al. (2008). CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J. 56, 590–602. doi: 10.1111/j.1365-313X.2008.03634.x
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chi, W., He, B., Mao, J., Li, Q., Ma, J., Ji, D., et al. (2012). The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol. 158, 693–707. doi: 10.1104/pp.111.186775
Chi, W., Ma, J., Zhang, D., Guo, J., Chen, F., Lu, C., et al. (2008). The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584. doi: 10.1104/pp.108.116194
Chi, W., Mao, J., Li, Q., Ji, D., Zou, M., Lu, C., et al. (2010). Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 64, 14–25. doi: 10.1111/j.1365-313X.2010.04304.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dobrogojski, J., Adamiec, M., and Luciński, R. (2020). The chloroplast genome: a review. Acta Physiol. Plant 42:98. doi: 10.1007/s11738-020-03089-x
Du, L., Zhang, J., Qu, S., Zhao, Y., Su, B., Lv, X., et al. (2017). The pentratricopeptide repeat protein pigment-defective Mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 58, 747–759. doi: 10.1093/pcp/pcx0004
Duan, Z., Li, K., Zhang, L., Che, L., Lu, L., Rochaix, J.-D., et al. (2020). F-type ATP synthase assembly factors Atp11 and Atp12 in Arabidopsis. Front. Plant Sci. 11:522753. doi: 10.3389/fpls.2020.522753
Friso, G., Giacomelli, L., Ytterberg, A. J., Peltier, J.-B., Rudella, A., Sun, Q., et al. (2004). In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell 16, 478–499. doi: 10.1105/tpc.017814
Gao, Z. P., Chen, G. X., and Yang, Z. N. (2012). Regulatory role of Arabidopsis pTAC14 in chloroplast development and plastid gene expression. Plant Signal. Behav. 7, 1354–1356. doi: 10.4161/psb.21618
Graf, M., Arenz, S., Huter, P., Dönhöfer, A., Nováček, J., and Wilson, D. N. (2016). Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions. Nucleic Acids Res. 45, gkw1272–gkw2896. doi: 10.1093/nar/gkw1272
Heazlewood, J. L., Verboom, R. E., Tonti-Filippini, J., Small, I., and Millar, A. H. (2006). SUBA: the Arabidopsis subcellular database. Nucleic Acids Res. 35, D213–D218. doi: 10.1093/nar/gkl863
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Legen, J., Kemp, S., Krause, K., Profanter, B., Herrmann, R. G., and Maier, R. M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31, 171–188. doi: 10.1046/j.1365-313X.2002.01349.x
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liao, Y., Smyth, G. K., and Shi, W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47:e47. doi: 10.1093/nar/gkz114
Liere, K., Weihe, A., and Börner, T. (2011). The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J. Plant Physiol. 168, 1345–1360. doi: 10.1016/j.jplph.2011.01.005
Loschelder, H., Schweer, J., Link, B., and Link, G. (2006). Dual temporal role of plastid sigma factor 6 in Arabidopsis development. Plant Physiol. 142, 642–650. doi: 10.1104/pp.106.085878
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Manuell, A. L., Quispe, J., and Mayfield, S. P. (2007). Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol. 5:e209. doi: 10.1371/journal.pbio.0050209
Naver, H., Boudreau, E., and Rochaix, J.-D. (2001). Functional studies of Ycf3: its role in assembly of photosystem I and interactions with ssome of its subunits. Plant Cell 13, 2731–2745. doi: 10.1105/tpc.010253
Nishimura, K., Ashida, H., Ogawa, T., and Yokota, A. (2010). A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J. 63, 766–777. doi: 10.1111/j.1365-313X.2010.04276.x
Peng, L., Ma, J., Chi, W., Guo, J., Zhu, S., Lu, Q., et al. (2006). LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18, 955–969. doi: 10.1105/tpc.105.037689
Perez Boerema, A., Aibara, S., Paul, B., Tobiasson, V., Kimanius, D., Forsberg, B. O., et al. (2018). Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat. Plants 4, 212–217. doi: 10.1038/s41477-018-0129-6
Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K.-J., and Oelmüller, R. (2006). pTAC2, −6, and −12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197. doi: 10.1105/tpc.105.036392
Pfalz, J., and Pfannschmidt, T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18, 186–194. doi: 10.1016/j.tplants.2012.11.003
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Pyo, Y. J., Kwon, K.-C., Kim, A., and Cho, M. H. (2013). Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 163, 1844–1858. doi: 10.1104/pp.113.227199
Reiter, B., Vamvaka, E., Marino, G., Kleine, T., Jahns, P., Bolle, C., et al. (2020). The Arabidopsis protein CGL20 is required for plastid 50S ribosome biogenesis. Plant Physiol. 182, 1222–1238. doi: 10.1104/pp.19.01502
Sakamoto, W., Miyagishima, S.-Y., and Jarvis, P. (2008). Chloroplast biogenesis: control of plastid development, protein import, division and inheritance. Arabidopsis Book 6:e0110. doi: 10.1199/tab.0110
Schein, A., Sheffy-Levin, S., Glaser, F., and Schuster, G. (2008). The RNase E/G-type endoribonuclease of higher plants is located in the chloroplast and cleaves RNA similarly to the E. coli enzyme. RNA 14, 1057–1068. doi: 10.1261/rna.907608
Shajani, Z., Sykes, M. T., and Williamson, J. R. (2011). Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80, 501–526. doi: 10.1146/annurev-biochem-062608-160432
Sharma, M. R., Dönhöfer, A., Barat, C., Marquez, V., Datta, P. P., Fucini, P., et al. (2010). PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G) 2. J. Biol. Chem. 285, 4006–4014. doi: 10.1074/jbc.M109.062299
Sharma, M. R., Wilson, D. N., Datta, P. P., Barat, C., Schluenzen, F., Fucini, P., et al. (2007). Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc. Natl. Acad. Sci. U. S. A. 104, 19315–19320. doi: 10.1073/pnas.0709856104
Shiina, T., Tsunoyama, Y., Nakahira, Y., and Khan, M. S. (2005). Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int. Rev. Cytol. 244, 1–68. doi: 10.1016/S0074-7696(05)44001-2
Steiner, S., Schröter, Y., Pfalz, J., and Pfannschmidt, T. (2011). Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157, 1043–1055. doi: 10.1104/pp.111.184515
Sweetlove, L. J., Taylor, N. L., and Leaver, C. J. (2007). Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol. Biol. 372, 125–136. doi: 10.1007/978-1-59745-365-3_9
Swiatecka-Hagenbruch, M., Liere, K., and Börner, T. (2007). High diversity of plastidial promoters in Arabidopsis thaliana. Mol. Gen. Genomics 277, 725–734. doi: 10.1007/s00438-007-0222-4
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tiller, N., Weingartner, M., Thiele, W., Maximova, E., Schöttler, M. A., and Bock, R. (2012). The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 69, 302–316. doi: 10.1111/j.1365-313X.2011.04791.x
Walter, M., Piepenburg, K., Schöttler, M. A., Petersen, K., Kahlau, S., Tiller, N., et al. (2010). Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 64, 851–863. doi: 10.1111/j.1365-313X.2010.04377.x
Wang, X., Zhao, L., Man, Y., Li, X., Wang, L., and Xiao, J. (2020). PDM4, a pentatricopeptide repeat protein, affects chloroplast gene expression and chloroplast development in Arabidopsis thaliana. Front. Plant Sci. 11:1198. doi: 10.3389/fpls.2020.01198
Weis, B. L., Kovacevic, J., Missbach, S., and Schleiff, E. (2015). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20, 729–740. doi: 10.1016/j.tplants.2015.07.003
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovations 2:100141. doi: 10.1016/j.xinn.2021.100141
Yang, J., Suzuki, M., and McCarty, D. R. (2016). Essential role of conserved DUF177A protein in plastid 23S rRNA accumulation and plant embryogenesis. J. Exp. Bot. 67, 5447–5460. doi: 10.1093/jxb/erw311
Zhang, L., Pu, H., Duan, Z., Li, Y., Liu, B., Zhang, Q., et al. (2018). Nucleus-encoded protein BFA1 promotes efficient assembly of the chloroplast ATP synthase coupling factor 1. Plant Cell 30, 1770–1788. doi: 10.1105/tpc.18.00075
Zhang, J., Xiao, J., Li, Y., Su, B., Xu, H., Shan, X., et al. (2017). PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 68, 5615–5627. doi: 10.1093/jxb/erx360
Zhou, W., Cheng, Y., Yap, A., Chateigner-Boutin, A.-L., Delannoy, E., Hammani, K., et al. (2009). The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. Plant J. 58, 82–96. doi: 10.1111/j.1365-313X.2008.03766.x
Keywords: chloroplast, ribosome, mitochondria, CDB1, CDB1L
Citation: Chen W, Huang J, Chen S, Zhang L, Rochaix J-D, Peng L and Xin Q (2022) Stromal Protein Chloroplast Development and Biogenesis1 Is Essential for Chloroplast Development and Biogenesis in Arabidopsis thaliana. Front. Plant Sci. 13:815859. doi: 10.3389/fpls.2022.815859
Received: 15 November 2021; Accepted: 17 January 2022;
Published: 10 February 2022.
Edited by:
Hongbo Gao, Beijing Forestry University, ChinaReviewed by:
Peng Wang, Humboldt University of Berlin, GermanyCopyright © 2022 Chen, Huang, Chen, Zhang, Rochaix, Peng and Xin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Xin, eGlucWlhbmcwMzI3QHNobnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.